- College of Food Science, Fujian Agriculture and Forestry University, Fuzhou, China
Cronobacter sakazakii is a common foodborne pathogen, and the mortality rate of its infection is as high as 40–80%. SdiA acts as a quorum sensing regulator in many foodborne pathogens, but its role in C. sakazakii remains unclear. Here, we further determined the effect of the sdiA gene in C. sakazakii pathogenicity. The SdiA gene in C. sakazakii was knocked out by gene editing technology, and the biological characteristics of the ΔsdiA mutant of C. sakazakii were studied, followed by transcriptome analysis to elucidate its effects. The results suggested that SdiA gene enhanced the drug resistance of C. sakazakii but diminished its motility, adhesion and biofilm formation ability and had no effect on its growth. Transcriptome analysis showed that the ΔsdiA upregulated the expression levels of D-galactose operon genes (including dgoR, dgoK, dgoA, dgoD and dgoT) and flagella-related genes (FliA and FliC) in C. sakazakii and downregulated the expression levels of related genes in the type VI secretion system (VasK gene was downregulated by 1.53-fold) and ABC transport system (downregulated by 1.5-fold), indicating that SdiA gene was related to the physiological metabolism of C. sakazakii. The results were useful for clarifying the pathogenic mechanism of C. sakazakii and provide a theoretical basis for controlling bacterial infection.
Introduction
Cronobacter sakazakii, a spiral-shaped flagellated Gram-negative rod-shaped bacilli bacterium, is an emerging food-borne opportunistic pathogen that is primarily parasitic in human and animal intestines (Farmer et al., 1980; Guo et al., 2019). Cronobacter sakazakii comes from a wide range of sources, and Powdered Infant Formula (PIF) is considered the primary source of infection and medium for transmission (Chap et al., 2009; Ogrodzki and Forsythe, 2017; Odeyemi and Sani, 2019). In addition, C. sakakazaki was also isolated from cheese, grains, fruits, vegetables, herbs and meats (Jung and Park, 2006; Jaradat et al., 2009). Neonates and young infants are susceptible to the bacterium that could cause severe clinical presentations of septicemia, meningitis and necrotizing enterocolitis and produce possible life-threatening chronic neurologic sequelae (Hariri et al., 2013; Hunter and Bean, 2013). Unfortunately, the death rate is as high as 40–80% (Wang et al., 2019). In 2022, the U. S. FDA reported four infant illnesses from three states, and found the four cases related to PIF produced in Abbott nutrition’s facility in Sturgis, Michigan. Four hospitalizations resulted from PIF produced environmental contamination by C. sakakazaki, and C. sakakazaki may have contributed to an infant death in one case (U.S. FDA, 2022).
Quorum sensing (QS) is a collaborative regulation system of bacterial density dependent on multi-gene expression, an environmental signal sensing system for bacteria to monitor their own population density, an important regulatory mechanism involved in adhesion, biofilm formation, virulence and so on (Williams et al., 2000; Anetzberger et al., 2009; Almeida et al., 2016; Stefany et al., 2017). In most Gram-negative bacteria, two major QS systems are autoinducer-1 (AI-1) and autoinducer-2 (AI-2). The AI-1-type QS system is mainly responsible for communication between Gram-negative bacteria. LuxI and LuxR are two key proteins: LuxI synthase produces N-acyl homoserine lactones (AHLs) as its own inducers, and the LuxR transcription factor is their homologous receptor (Lars et al., 2001; Assia et al., 2017; Vadakkan et al., 2018; Wang et al., 2018; Pacheco et al., 2021). The AI-2-type QS system is present in both Gram-negative and Gram-positive bacteria, and LuxS is a key protein (Ahmer, 2010; Papenfort and Bassler, 2016).
Interestingly, some Gram-negative bacteria encode LuxR receptors but do not produce AHLs because they lack LuxI synthase. For instance, Klebsiella pneumoniae, Salmonella and Enterobacterium do not have the LuxI synthase gene, and they do not produce AHL signaling molecules (Almeida et al., 2016; Pacheco et al., 2021). However, there is a chromosomal LuxR homolog called SdiA of these bacteria; these bacteria can use the SdiA sensor to sense the signaling molecules produced by other bacteria in the environment and then regulate the expression of their own related genes (Sharma and Bearson, 2013).
SdiA was identified as “a quorum-sensing regulator” that regulates the transcription of ftsQAZ operons involved in cell division of klebsiella pneumoniae. It has been noted that in enterohemorrhagic Escherichia coli (EHEC) and Enterobacteriaceae, SdiA is involved in the regulation of many virulence factors, such as pili production, biofilm formation, adhesion and motility of cells (Sharma and Bearson, 2013; Culler et al., 2018; Ma et al., 2020). In addition, Salmonella utilizes SdiA proteins to detect AHLs synthesized by other species and enhances srgE and RCK operon expression (Ahmer, 2010; Almeida et al., 2016). Recent studies have reported that SdiA has an effect on the survival of C. sakazakii under different environmental stresses, with improvement of C. sakazakii tolerance to heat, desiccation, osmotic and acid stress conditions (Cao et al., 2021). Nevertheless, the function of the QS receptor SdiA in C. sakazakii and its underlying mechanisms remain largely unknown. More studies are required to elucidate the exact functions of SdiA on the physiology of these microorganisms since a great level of complexity is observed as revealed by these previous works.
In this study, gene editing technology was used for the construction of an SdiA gene deletion mutant to investigate the role of SdiA in C. sakazakii pathogenicity by assessing the growth, biofilm formation, motility, adhesion and multidrug resistance of the QS receptor SdiA in C. sakazakii. Meanwhile, transcriptome analysis was conducted to elucidate the expression of SdiA-related genes.
Materials and Methods
Bacterial Strains and Culture Conditions
Cronobacter sakazakii CICC 21550 was obtained from the China Center of Industrial Culture Collection (CICC). Cronobacter sakazakii CICC 21550 wild-type (WT) and its mutant (ΔsdiA) strains were grown in trypticase soy broth (TSB) or on trypticase soy agar (TSA) at 37°C. Stock cultures of C. sakazakii CICC 21550 (WT) and mutant (ΔsdiA) strains were maintained in brain heart infusion (BHI) broth with 20% glycerol at −80°C. Activation of the strains was achieved by streaking the stock culture onto violet red bile glucose agar (VRBGA) and incubating for 24 h at 37°C. The WT and ΔsdiA strains were stored on VRBGA at 4°C to maintain cell viability.
Equations Sequence Alignment, Construction and Identification of the SdiA Deletion Mutant
The nucleotide sequences of the SdiA gene and its upstream and downstream genes were obtained based on the whole genome sequencing results of C. sakazakii CICC 21550. Primers were designed using Primer 5.0 software (as shown in Table 1). All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Cronobacter sakazakii CICC 21550 genomic DNA was isolated using an Ezup Column Bacteria Genomic DNA Purification kit (Sangon Biotech Co., Ltd, Shanghai, China) according to the manufacturer’s recommended protocol and stored at −20°C.
The SdiA in-frame deletion mutant was constructed using the suicide plasmid pCVD442, plasmid pKD3 of the chloramphenicol resistance gene and E. coli strain β2155. These plasmids and E. coli strain β2155 were purchased from BioVector (Biovector NTCC Inc., China). The gene deletion construct was generated according to homologous recombination as previously described (Sharma and Bearson, 2013; Zhang et al., 2018; Cao et al., 2021). Briefly, SdiA gene upstream and downstream homologous recombination arms were amplified by PCR from C. sakazakii CICC 21550 genomic DNA using primers SdiA-5F/SdiA-5R and SdiA-3F/SdiA-3R (Table 1). The Cm resistance cassette from plasmid pKD3 was PCR amplified with primers CmSeqF2 and CmSeqR2 (Table 1).
The fusion PCR approach was used to construct the SdiA gene-targeting vector ΔSdiA:Cm (2650 bp, Figure 1A), and the fusion PCR product was transformed into the suicide plasmid pCVD442. The plasmids were introduced by electroporation into E. coli strain β2155 and plated on TSA containing ampicillin (100 μg/ml) and 0.5 mM 2,6-diaminoheptanedioic acid (DAP) for selection at 37°C. After ligation, the pCVD442-ΔSdiA:Cm plasmid was transformed from β2155 into C. sakazakii CICC 21550 through conjugation (Zhang et al., 2018). Finally, 100 μL was plated on TSA containing chloramphenicol (17 μg/ml) agar plates and cultured in an incubator at 30°C until colonies formed.
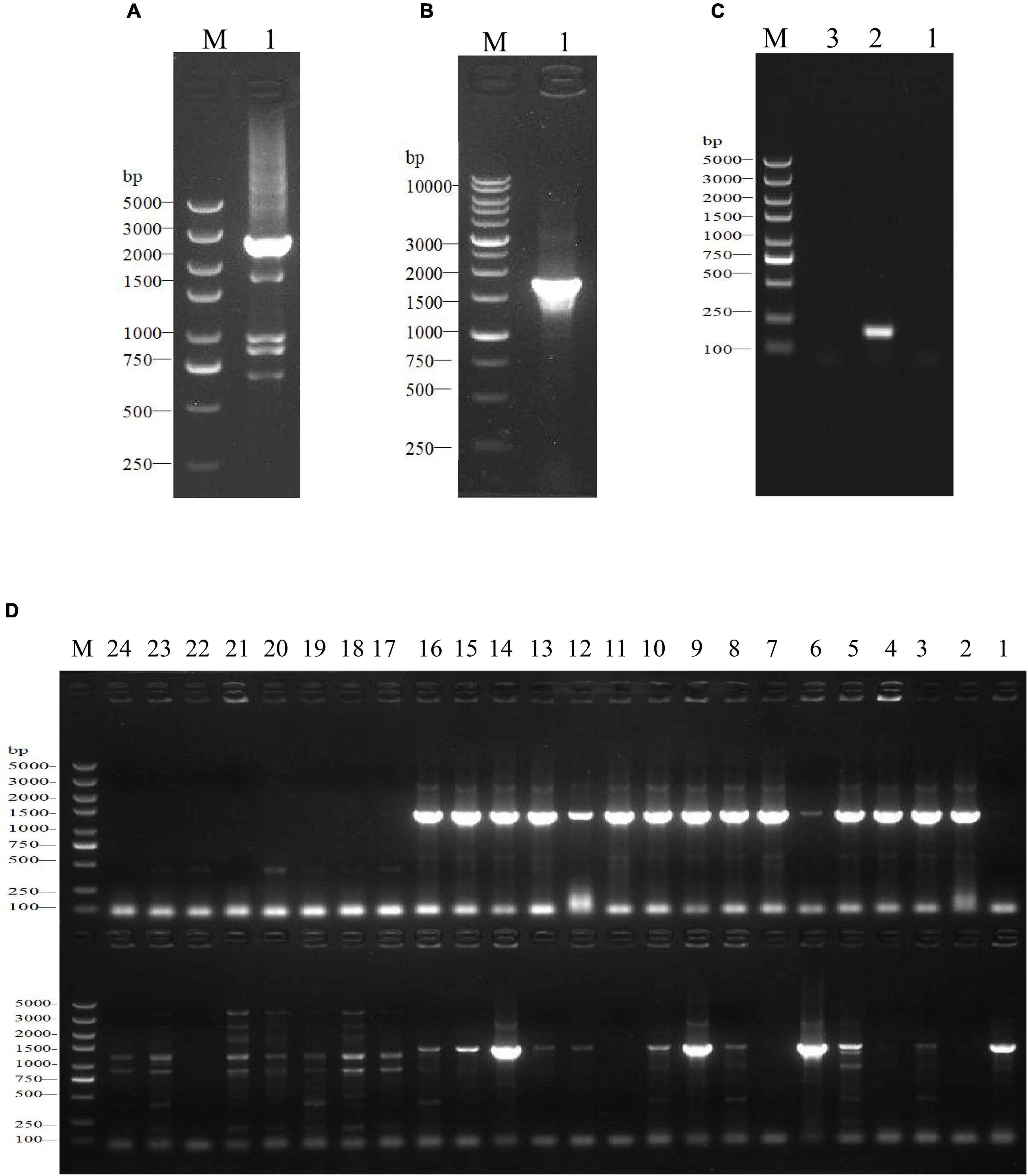
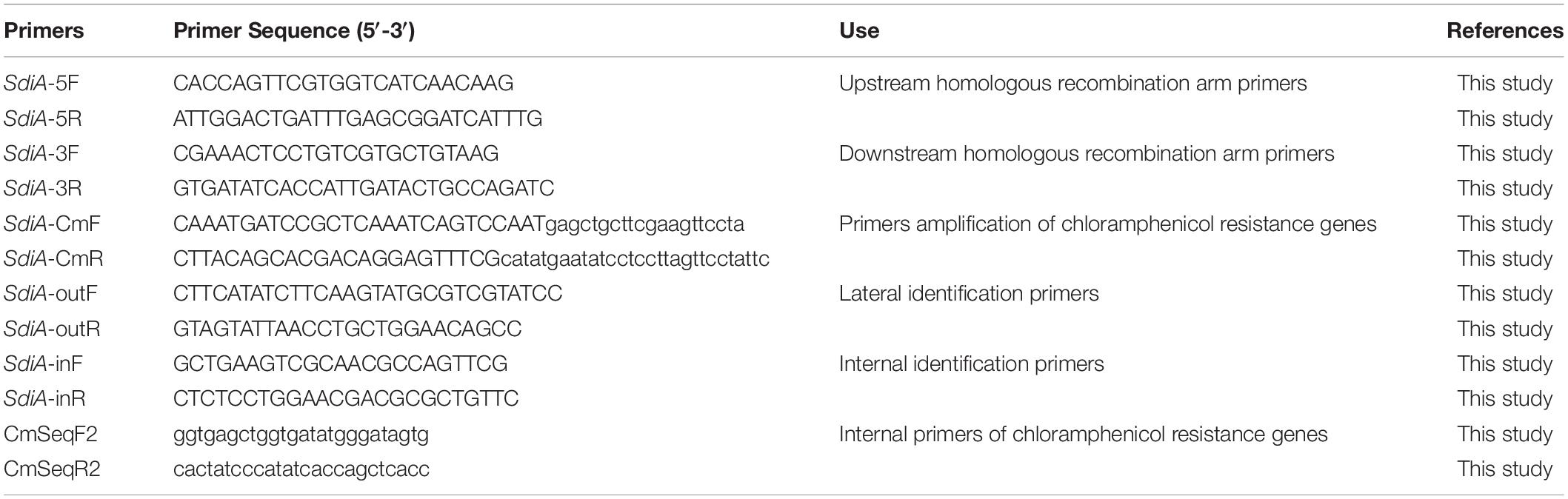
Figure 1. Construction and identification of the SdiA deletion mutant of C. sakazakii by gel electrophoresis. (A) Construction of SdiA gene target shooting fragment. M: DL5000 DNA marker; 1: SdiA gene target shooting fragment ΔsdiA:Cm (2650 bp). (B,C) Chloramphenicol resistance gene removal. (B) M: DL10000 DNA Marker; 1: Results of identification of lateral primers for colonies growing on non-resistant a TSA plates. (C) M: DL5000 DNA marker; 1: Identification results of lateral primers for colonies growing on non-resistant a LB plates; 2: Internal primer amplification results of the original strain (193 bp); 3: No template negative control amplification results. (D) Screening of SdiA gene knockout strain. M: DL5000 DNA marker; 1–24: Amplification results of primer pairs on both sides of 24 single colonies (Amplification results on both sides of 5′ endpoints above; Amplification results on both sides of the 3′ endpoint are shown below).
The mutant strain was confirmed by PCR analysis using the Table 1 primer pair. Resistant colonies were streaked onto TSA plates without NaCl but supplemented with 10% sucrose (wt/vol) and chloramphenicol (17 μg/ml) to select for cells in which recombination and loss of the pCVD442 vector occurred. The chloramphenicol resistance gene was removed from the selected mutant using the pCP20 plasmid (Biovector NTCC Inc., China). Transformants were selected at 30°C on TSA plates containing 100 μg/ml ampicillin, and integration was confirmed by PCR with primers SdiA-outF/SdiA-outR (Table 1).
Growth Study
The powdered infant formula (PIF, 0–6 months, Yili Jinlingguanzhenhu) was sterilized by Co60 irradiation (8 kGy, Fujian Compton Irradiation Technology Co., Fuzhou, China) and then sealed and preserved. A single colony of each strain from VRBGA was grown in 10 ml of TSB on a shaker at 130 rpm for 18–20 h at 37°C. Bacterial cells were harvested by centrifugation at 5000 rpm for 15 min at 4°C. Ten milliliters of 0.1% (wt/vol) peptone water (PW)-resuspended bacterial cells were washed off their surface toxins, and the supernatant was discarded. Then, the samples were resuspended in 10 ml of PW, and the bacterial washing step was repeated once (Xie et al., 2020). Subsequently, the solution was resuspended in 5 ml of PW and then diluted with PW to obtain an inoculum containing approximately 104.0–4.5 CFU/ml or 4.0–4.5 log CFU/ml of C. sakazakii. According to the instructions of PIF, sterilized deionized water was used at high temperature and high pressure (121°C, 20 min) to prepare recovered milk powder. Then, 10 mL of milk was added to sterilizated empty test tubes with 0.1 mL aliquots of WT and ΔsdiA mutant cultures.
According to the growth parameters of C. sakazakii (Fang et al., 2012), the growth experiment with WT and ΔsdiA mutant strains was conducted at fluctuating temperatures. The inoculated samples were incubated at fluctuating temperatures of 8.4°C–48°C, 10°C–47.8°C, and 11°C–35°C. Samples were taken at predesignated time intervals and then serially diluted and plated on TSA plates overnight and enumerated.
Scanning Electron Microscopy
Cells were grown and harvested by the above methods, resuspended in 2.5% glutaraldehyde fixative and fixed at 4°C for 10 h. Subsequently, the cells were washed in 0.1 M phosphate buffered saline (PBS) and dehydrated through graded ethanol solutions. Before EM-30AX scanning electron microscopy (SEM, COXEM, Daejeon, South Korea) observation, the samples were freeze-dried for 8 h and coated with gold.
Swimming and Swarming Motility Assays
The effect of WT and ΔsdiA mutant strains on swimming motility was assessed by examining swimming motility on TSA containing 0.3% (wt/vol) agar. Cronobacter sakazakii was cultivated in TSB overnight at 37°C in a shaking incubator at 130 rpm. Bacterial cells were diluted to 106 CFU/ml after stab-inoculation into 0.3% (wt/vol) TSA agar plates and incubated for 6 h–24 h at 30°C. Motility was assessed quantitatively by measuring the circular swimming motion of the growing motile C. sakazakii every 3 h. The effect of WT and ΔsdiA mutant strains on swarming motility was assessed by examining swarming motility on TSA containing 0.5% (wt/vol) agar. The same dilution (as mentioned above) was used to inoculate 0.5% (wt/vol) TSA agar plates and incubated for 6 h–36 h at 30°C. Motility was assessed quantitatively by measuring the circular swarming motion of the growing motile C. sakazakii every 6 h. The motility assays were repeated in three separate experiments.
Crystal Violet Staining Biofilm Assay
The biofilm forming capacities of the WT and ΔsdiA mutant strains were assessed by a crystal violet staining assay (Liu et al., 2018) with slight modifications. Bacterial cells were cultivated in TSB overnight at 37°C until an optical density at 595 nm (OD595) of 0.5 or 109 CFU/ml was reached. Thirty microliters of bacterial overnight culture was inoculated into 150 μL of TSB liquid broth in sterile 96-well plates and incubated for 24 h at 37°C. Then, 200 μL of 99% methanol (vol/vol) was added to each well for 15 min to fix the biofilms. After washing three times with 200 μL of 0.1 M PBS and dried in a hot air oven at 55°C for 1 h. The plates were stained with 200 μL of 1% crystal violet for 30 min, washed with 0.1 M PBS two times and then air dried at 37°C. Finally, the remaining crystal violet was resuspended in 200 μL of 95% ethanol and measured at 570 nm (OD570) using a microplate reader (SpectraMax®i3x, Molecular Devices, United States). In addition, 200 μL of TSB was used as a blank control, and each sample was set up with six replicates. The biofilm assay was repeated in four separate experiments.
Cell Adhesion Assay
The cell adhesion capacities were measured according to the method of Fabíola (Cacciatore et al., 2020) with slight modifications. The bacterial cells were diluted with PW to 108 CFU/mL. The 2 cm × 2 cm samples (made of glass, plastic, silicone hydrogel and stainless steel) were soaked in 10 ml of bacterial solution, incubated for 15 min at 30°C, and then washed three times with 10 ml of 0.01 M PBS to remove unbound cells. Then, 10 ml of 0.01 M sterile PBS was added for full oscillation to release the adhered cells into the PBS. The adhesion value was determined by counting viable cells on TSA plates. The cell adhesion assay was repeated in three separate experiments.
Tolerance and Resistance Assays
The susceptibility of the WT and ΔsdiA mutant strains to different antibiotics was evaluated by the disc diffusion method. The bacterial cells were diluted with PW to 108CFU/mL, and 0.1 mL of bacterial culture was plated onto a TSA plate. The antibiotic resistance was checked against 6 different antibiotics (200 μg/mL and 500 μg/mL): kanamycin, chloramphenicol, rifampicin, erythromycin, tetracycline hydrochloride and penicillin (Macklin Inc., Shanghai, China). Sterilized deionized water was used as a blank control. The sterilized circular filter paper (Φ = 8 mm) was soaked in antibiotic solutions and deionized water for 10 min. Then, the filter papers were removed with sterile tweezers, affixed to a dried TSA plate and incubated for 24 h at 37°C, and the diameter of the antibacterial coil was measured. The tolerance and resistance assay was repeated in three separate experiments. Results are presented as the mean ± SD.
RNA-Seq Analysis
Cronobacter sakazakii WT (W1) and ΔsdiA mutant strains (S1) were selected for RNA-Seq analysis. Total RNA was extracted by a UNIQ-10 column TRIzol total RNA extraction kit (Sangon Biotech Co., Ltd, China) using cultures (130 rpm at 37°C) from the logarithmic growth stage. A Ribo-off rRNA Depletion Kit (Bacteria) (Vazyme Biotech Co., Ltd, Nanjing, China) was used to remove rRNA from total RNA samples. The cDNA libraries of RNA samples were constructed using the VAHTS™ Stranded mRNA-seq V2 Library Prep Kit for Illumina® (Vazyme Biotech Co., Ltd, Nanjing, China). The amplified cDNA library was visualized by 8% polyacrylamide gel electrophoresis (PAGE) (Shanghai Furi Technology Co., Ltd, China).
High-throughput sequencing was performed using the Illumina HISeq™ 2500 Platform.
Illumina HISeq™ raw image data files were analyzed by CASAVA Base calling and transformed into sequenced reads. The resulting sequences were then referred to as Raw Data or Raw Reads, and the results were stored in FASTQ file format. Trimmomatic software was used for data processing, the original data quality value and other information were counted, and FastQC software was used for visual evaluation of the sequencing data quality of the samples. Reference to the whole genome sequencing information of the C. sakazakii CICC21550 strain for gene alignment analysis, gene expression level analysis and differentially expression analysis. ClusterProfiler software was used for functional enrichment analysis and the main biological functions of differential expressed genes were obtained by comparing the GO (Gene Ontology) database. The related biological system information was obtained by comparative analysis with the KEGG (Kyoto Encyclopedia of Genes and Genomes) database. All experiments were performed in triplicate.
Statistical Analysis
All experimental results are presented as the mean ± standard deviation, and statistical analysis was performed with SPSS 18.0. The significance of the WT and ΔsdiA mutant strains was established through a t-test using P values < 0.05.
Results
Construction of a Cronobacter sakazakii CICC 21550 Mutant Strain
In this study, we constructed a C. sakazakii CICC 21550 strain in which the SdiA gene was deleted by gene replacement. Twenty-four single colonies were randomly selected from TSA containing chloramphenicol (17 μg/ml) agar plates for PCR analysis to screen the SdiA gene knockout strain. At the 5′ endpoint of the target fragment, a 1368 bp fragment was generated by pairing SdiA-outF and CmSeqF2 primers. At the 3′ endpoint, CmSeqR2 and SdiA-outR primer pairs produced a 1391 bp fragment. When both fragments were present, the chloramphenicol resistance gene fragment replaced the SdiA gene fragment. The results indicated that both sides of clones 9 and 14 were amplified with strong specific PCR products in accordance with the expected length (Figure 1D). After the chloramphenicol resistance gene was deleted, the length of the amplified product of the lateral primer was shortened to 1802 bp (Figures 1B,C). This assay confirmed that we successfully constructed the SdiA deletion strain of C. sakazakii CICC 21550.
Growth of Cronobacter sakazakii Wild-Type and ΔsdiA Mutant Strains in Powdered Infant Formula
The growth model of the WT strain and ΔsdiA mutant strains at fluctuating temperatures was constructed using a one-step data analysis method (Jia et al., 2020). Figure 2D shows the growth rate distribution of the ΔsdiA mutant and WT strains at 4–50°C, and their growth parameters were very similar. There were no significant differences, in that the absence of the SdiA gene had no effect on the growth of C. sakakazakii. Therefore, we combined the data of the WT and ΔsdiA mutant strains to analyze their growth (a total of 260 data points) in the recovered PIF at fluctuating temperatures of 8.4–48°C based on the no lag-Cardinal model and Baranyi-Cardinal models of the one-step data analysis method (Fang and Huang, 2014; Huang, 2017). The statistical results and estimated values of each parameter are shown in Tables 2, 3. The observed values and model prediction curves of the WT and ΔsdiA mutant strains at each temperature are shown in Figures 2A–C. Figure 2E shows the effect of temperature on the growth rate of the WT and ΔsdiA mutant strains. The results showed that no lag-Cardinal model and Baranyi-Cardinal model had the same fitting effect on the growth of the C. sakazakii WT and ΔsdiA mutant strains.
Figure 2. One-step analysis for C. sakazakii wild-type and ΔsdiA mutant strains. (A–C) One-step fitting analysis of the growth curve of C. sakazakii WT and ΔsdiA mutant strains in powdered infant formula recovery. (D) Growth rate distribution of ΔsdiA mutant and WT strains. (E) One-step analysis: Effect of temperature on growth rate of WT and ΔsdiA mutant strains.
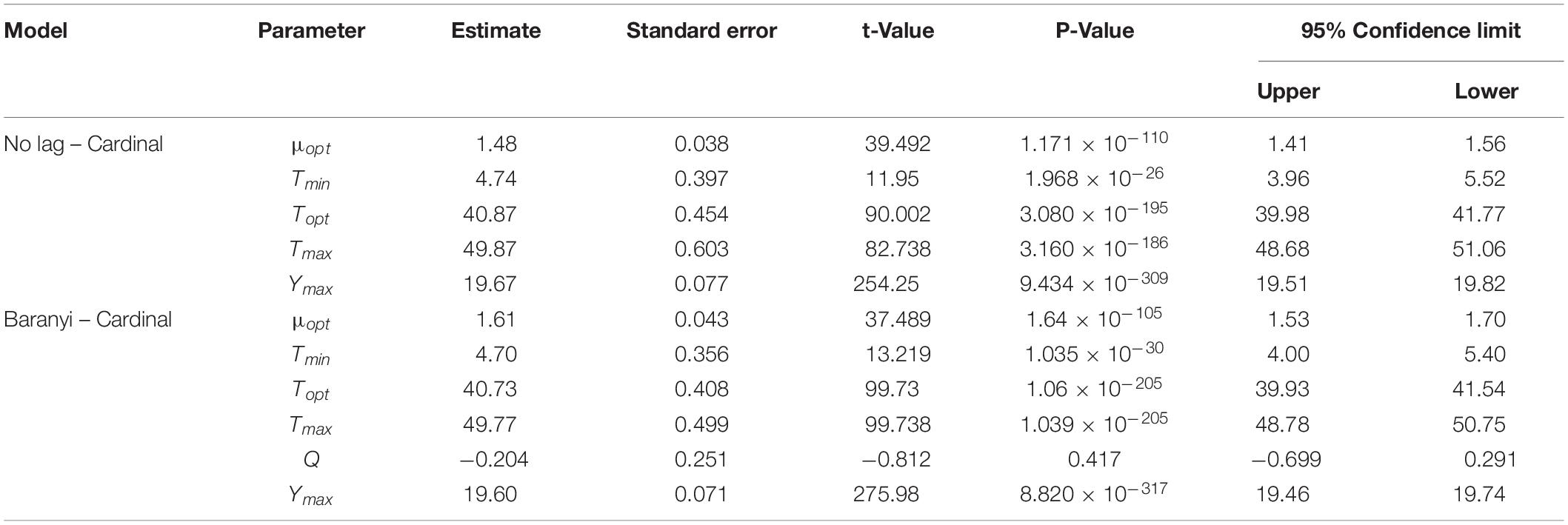
Table 3. Parameter estimation of No lag–Cardinal and Baranyi–Cardinal models of wild-type and ΔsdiA mutant strains.
Hence, we further calculated the influence of SdiA gene deletion on the growth parameters of C. sakazakii and the growth changes of C. sakazakii in PIF. A mathematical model for the growth of C. sakazakii WT and ΔsdiA mutant strains in the recovered PIF was established by a one-step method. In this study, C. sakazakii WT and ΔsdiA mutant strain parameters were combined to be estimated by the no lag-Cardinal model and Baranyi-Cardinal model; the minimum growth temperatures estimated by the two models were 4.74°C and 4.70°C, respectively, and the highest growth temperatures were 49.87°C and 49.77°C, respectively. The optimum growth temperatures were 40.87°C and 40.73°C, respectively, and the optimum growth rates were 1.48 h–1 and 1.62 h–1, respectively. In addition, the maximum growth concentrations estimated by the no lag-Cardinal model and Baranyi-Cardinal model were 19.67 Ln CFU/ml (8.54 log CFU/ml) and 19.60 Ln CFU/ml (8.51 log CFU/ml), respectively.
Lack of SdiA Increases the Swarming and Swimming Motility of Cronobacter sakazakii
According to Figure 3A, after deletion of the SdiA gene in C. sakazakii, the swimming motility of the mutant strain (ΔsdiA) was markedly enhanced (p < 0.01). Nonetheless, the WT strain barely spread after 21 h of culture on TSA plates with 0.3% agar. The ΔsdiA mutant strain showed a trend of spreading after 6 h of culture. After incubation for 21 h, the ΔsdiA mutant strain spread to the entire plate, and the swimming motility of the ΔsdiA mutant strain showed a linear increase (Figure 3C). Furthermore, the ΔsdiA mutant strain showed significantly higher swarming motility than the WT strain and rapidly spread after 18 h of culture (p < 0.05), showing an exponential growth trend. Although the WT strain indicated a tiny trend of outward diffusion, their movement was still not visible after 36 h of culture (Figures 3A,D). In the transcriptomics study of C. sakazakii WT and ΔsdiA mutant strains, we found that the expression levels of FliA and FliC-related genes regulating the flagellate of C. sakazakii were significantly increased (Supplementary Tables 1, 3, 4), which resulted in enhanced flagellate activity and significantly enhanced swarming and swimming motility of C. sakazakii.
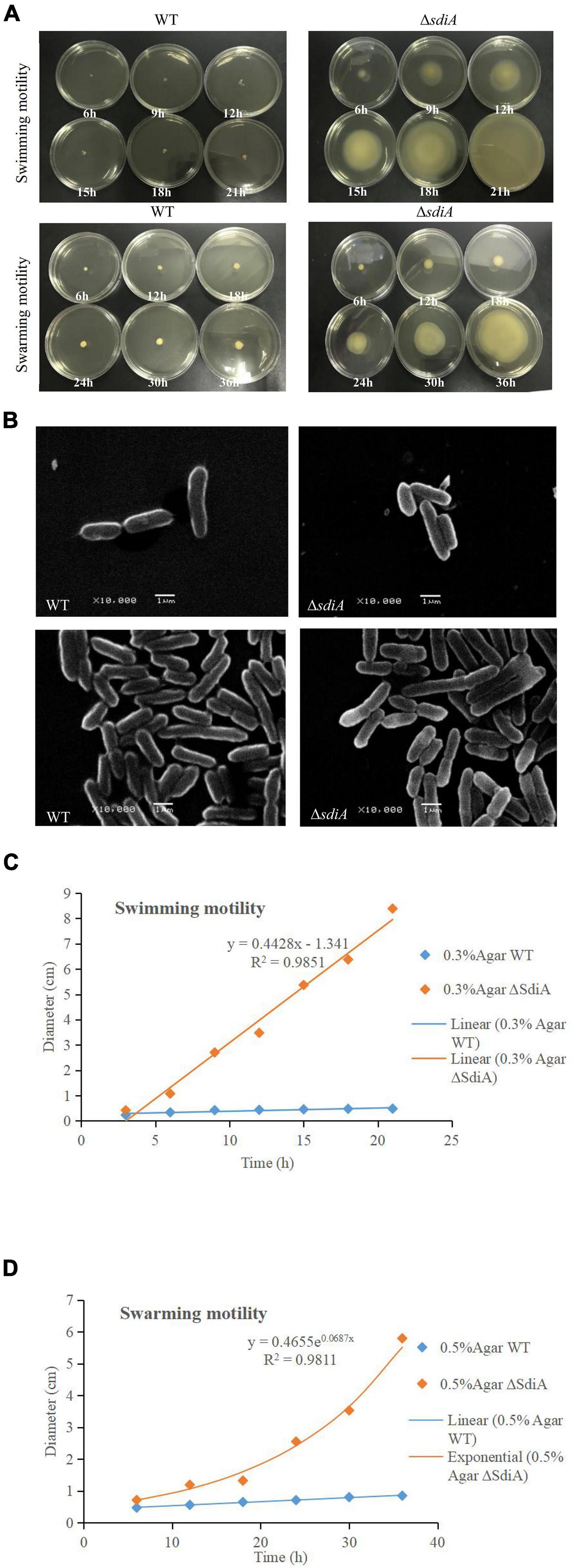
Figure 3. C. sakazakii WT and ΔsdiA mutant strains cell viability and morphology differences. (A) WT and ΔsdiA mutant strains motility assay. (B) WT and ΔsdiA mutant strains scanning electron microscopy (SEM) images. (C,D) WT and ΔsdiA mutant strains motility assay curve.
Lack of SdiA Increases the Biofilm Formation and Surface Adhesion Capabilities of Cronobacter sakazakii
To evaluate whether SdiA was also involved in the biofilm formation of C. sakazakii, we compared the biofilm formation ability of WT and ΔsdiA mutant strains. Compared with the WT strain of C. sakazakii, the amount of biofilm formation of the SdiA gene deletion strain increased by 77.34%, and the ΔsdiA mutant strain showed more biofilm formation (p < 0.01) (Figure 4A).
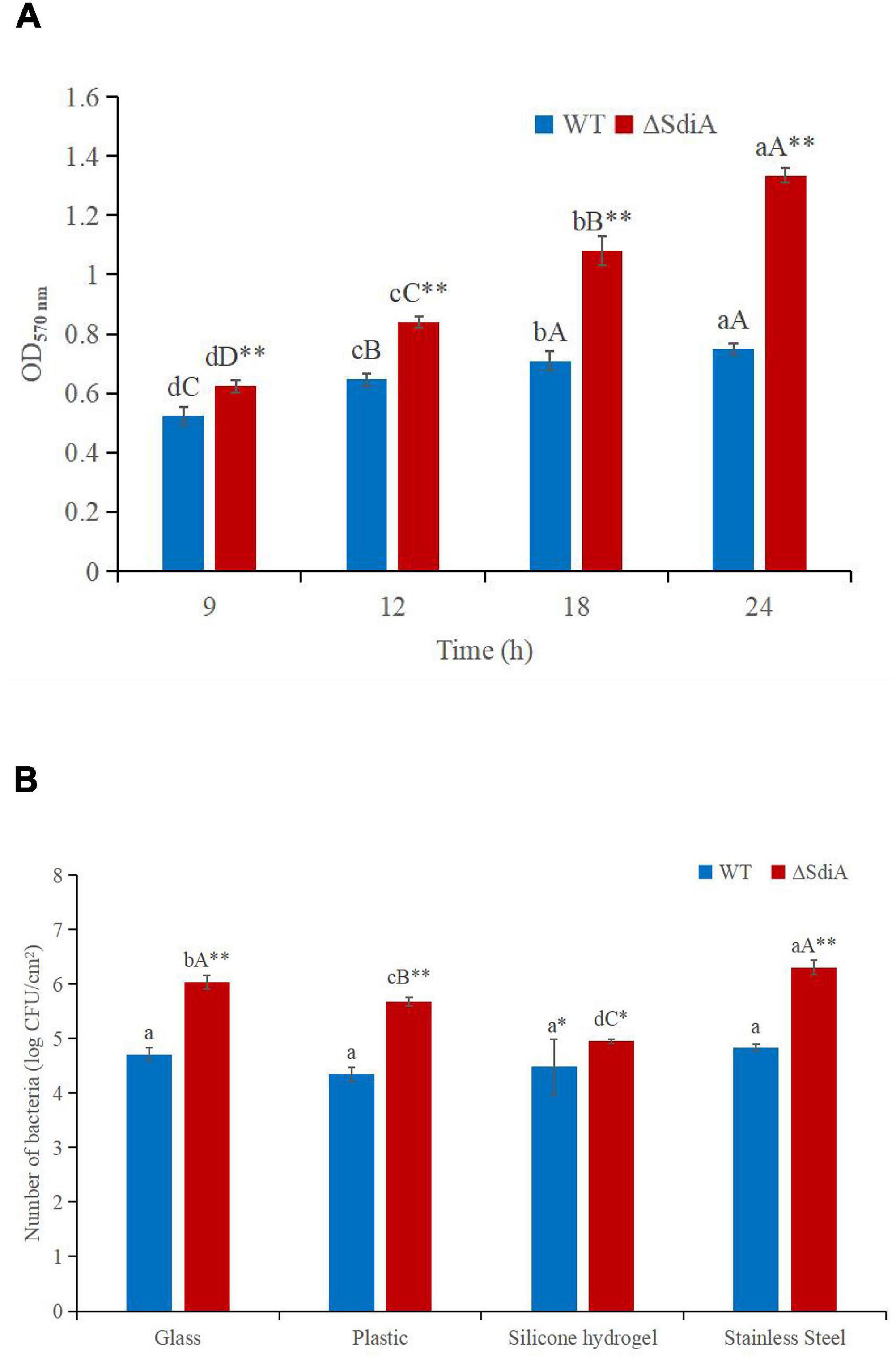
Figure 4. C. sakazakii WT and ΔsdiA mutant strains biofilm formation and surface adhesion capabilities assay. (A) Optical density values of CV solutions obtained from biofilms of C. sakazakii WT and ΔsdiA mutant strains grown in the TSB at 37°C. a– dMean values differ significantly in different time of each strain (p < 0.05), A– Dmeans (p < 0.01); ** Mean values differ significantly between two strains in the same time (p < 0.01). (B) Number of adhered C. sakazakii WT and ΔsdiA mutant strains to different substratum. aMean values differ significantly in different substratum of each strain (p < 0.05), A– C means (p < 0.01); *Mean values differ significantly between two strains in the same substratum (p < 0.05), **means (p < 0.01). Error bars represent standard deviation. All experimental data’s were plotted as mean ± standard from triplicate determinations and statistical analysis was performed with SPSS 18.0 Significance of WT and ΔsdiA mutant strains were established through t-test using P values < 0.05.
We found that the SdiA gene regulates the adhesion of C. sakazakii to stainless steel, glass and plastics. The adhesion of the WT strain to glass, plastic, silicone hydrogel and stainless steel surfaces was 4.71, 4.35, 4.49 and 4.83 log CFU/cm2, respectively, at 30°C for 15 min. The adhesion of the ΔsdiA mutant strain to glass, plastic, silicone hydrogel and stainless steel was 6.04, 5.68, 4.95 and 6.31 log CFU/cm2, respectively (Figure 4B). The SdiA deletion resulted in increased adhesion of C. sakazakii on glass, plastic and stainless steel (p < 0.01), but there was no significant difference in adhesion between the WT and ΔsdiA mutant strains on the silica hydrogel surface. The adhesion of C. sakazakii was affected by the dielectric surface, and the adhesion of WT on the plastic surface was lower than that of the other three dielectric materials. However, the adhesion of the ΔsdiA mutant strain on the silica hydrogel surface was lower than that of the other three media materials. Through our transcriptomic studies, we also found that the expression of the ATP-active membrane-associated protein VasK in the VI secretion system, which is important to the regulation of C. sakazakii adhesion, was downregulated by 1.53-fold in SdiA mutant strains (Supplementary Tables 2–4). PIF packaging, brewing and drinking bottles were mainly made of stainless steel, glass, plastic and silicone hydrogel materials, and C. sakazakii was the main food-borne pathogen in PIF. By studying the adhesion of the SdiA mutant and WT strains to these materials, we can make it clear that the key to controlling the contamination of C. sakazakii during the processing and brewing of PIF was the targeted regulation of the SdiA gene.
Lack of SdiA Decreases the Multidrug Resistance of Cronobacter sakazakii
As shown in Figure 5, when the SdiA gene was deleted, antibiotic resistance of C. sakazakii showed a general decrease. With the increase in antibiotic concentration, the antibacterial zone of antibiotics showing antibacterial effect was larger, among which kanamycin showed the most obvious antibacterial effect and showed relatively good antibacterial effect on WT and ΔsdiA mutant strains. Second, penicillin has a certain bacteriostatic effect, but compared with kanamycin, it does not show a strong bacteriostatic effect. Chloramphenicol, rifampicin and tetracycline hydrochloride exhibited approximately the same antibacterial effect, with a slight antibacterial effect. Among the 6 antibiotics, erythromycin showed poor antibacterial activity against C. sakazakii, and neither WT nor ΔsdiA mutant strains showed an antibacterial zone, showing no visible antibacterial effect. By studying the changes in the transcript level of the C. sakazakii SdiA mutant and the wild-type strains, we found that the expression level of related genes in the ABC transport system regulating drug resistance of C. sakazakii was downregulated by 1.5-fold (Supplementary Tables 2–4) (Wichelecki et al., 2015; Christoph and Robert, 2018), which revealed the mechanism by which the SdiA gene enhances drug resistance in C. sakazakii.
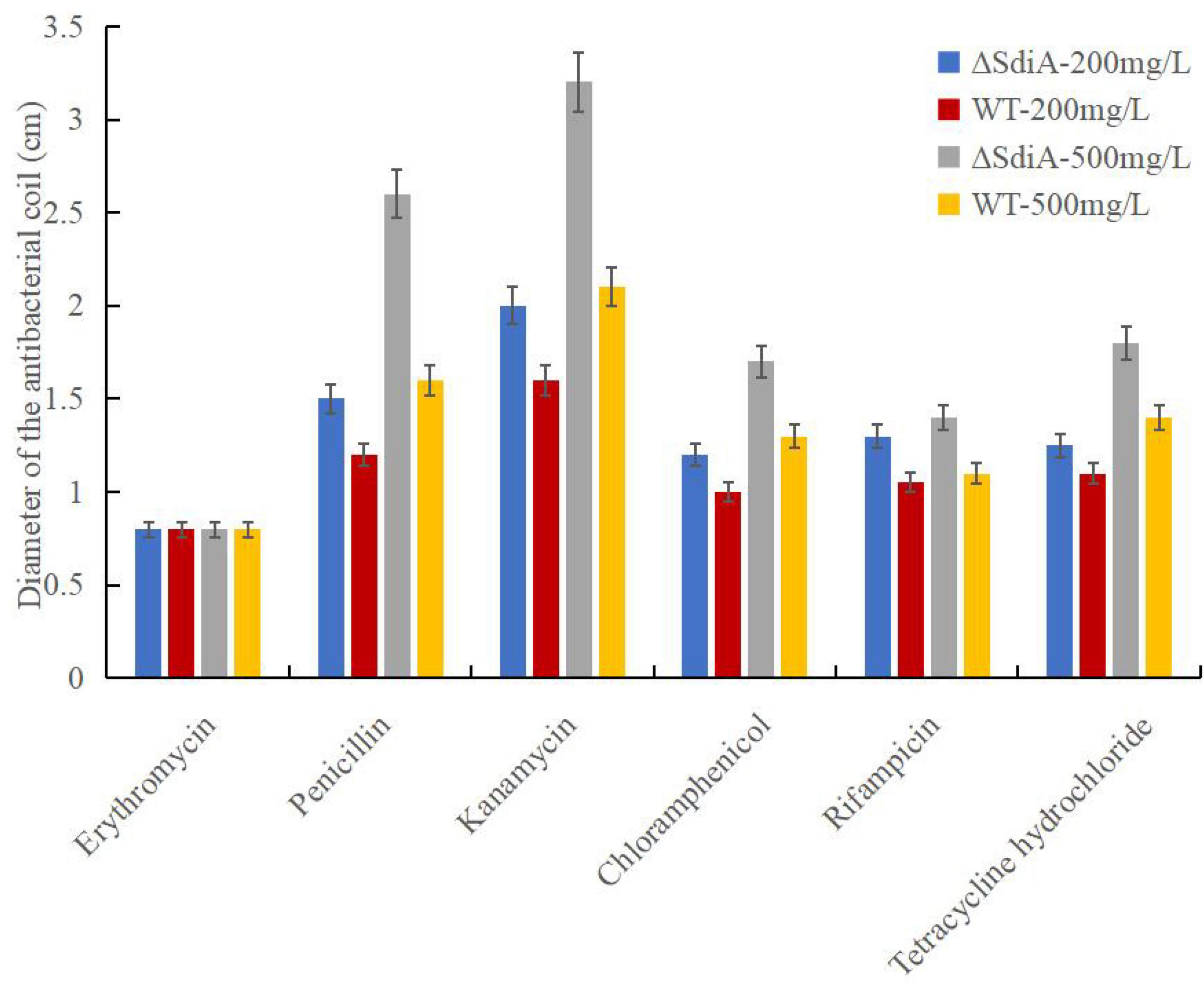
Figure 5. Cronobacter sakazakii WT and ΔsdiA mutant strains antibiotic resistance assay. Error bars represent standard deviation. All experimental data’s were plotted as mean ± standard from triplicate determinations and statistical analysis was performed with SPSS 18.0 Significance of WT and ΔsdiA mutant strains were established through t-test using P values < 0.05.
Lack of SdiA Alters the Expression of Genes Involved in Motility, the Virulence Factor and Antimicrobial Resistance of Cronobacter sakazakii
By sequencing us through the transcriptome compared with the WT strain, the ΔsdiA mutant strain in normal culture showed significant differences in the expression of 244 genes, including 50 genes upregulated and 194 genes downregulated (Figure 6). The ΔsdiA mutant strain showed significant expression differences in small molecule catabolic (14 genes), polysaccharide metabolic (15 genes), polysaccharide biosynthetic (15 genes), lipid metabolic (19 genes), cellular carbohydrate metabolic (23 genes) and carbohydrate metabolic (32 genes), as well as response to virus and phage shock protein (3 genes), as shown in Figures 7A,C (p < 0.01). As shown in Figures 7B,D, the ΔsdiA mutant showed significant differences in lipid and sugar metabolic pathways, including sphingolipid metabolism (2 genes), fructose and mannose metabolism (5 genes), amino sugar and nucleotide sugar metabolism (5 genes) (p < 0.01), drug metabolism - cytochrome P450 (2 genes), ether lipid metabolism (1 gene), retinol metabolism (2 genes) (p < 0.05) (Supplementary Tables 3, 4). These results indicate that the QS transcription regulator SdiA plays an important role in the cellular activities of C. sakazakii, and the mechanism of SdiA in each metabolic pathway of C. sakazakii needs to be revealed in the future.
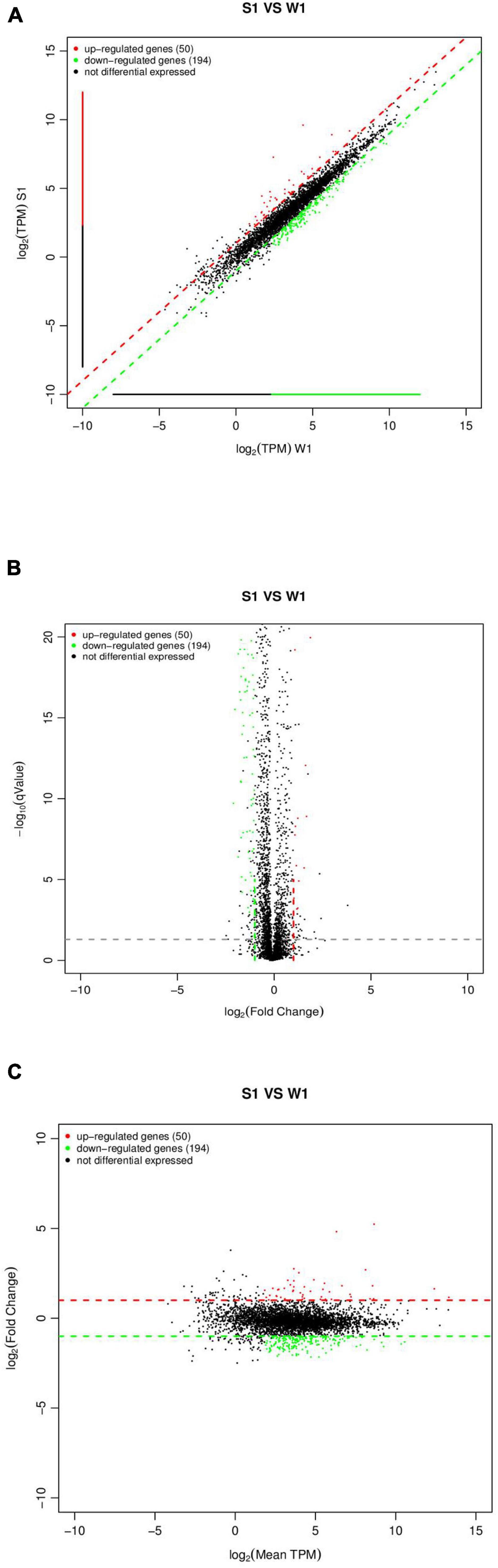
Figure 6. Expression difference analysis for transcriptional profiles [C. sakazakiiΔsdiA mutant (S1) compared to WT (W1) strains]. (A) Comparison group expression difference scatter plot. MeanTPM: Expression amount of a group. The horizontal and vertical axes are, respectively, the log2 (TPM) values of two groups of samples. Each point in the figure represents a gene, and the closer the point expression is to the origin the lower. The vertical sample is relative to the horizontal sample. (B) Comparison group expression difference volcano diagram. The horizontal axis represents the fold-change [log2 (B/A)] value of gene expression difference between different groups of samples. The vertical axis represents gene expression pValue, the smaller the pValue, the greater the −log10 (pValue), and the more significant the difference. (C) MA diagram of expression difference between groups was compared. The horizontal axis is the mean value of log2 (TPM) of the two groups of samples {[log2 (A) + log2 (B)]/2}. The vertical axis is log2 (Fold Change) [that is, log2 (B/A)] values.
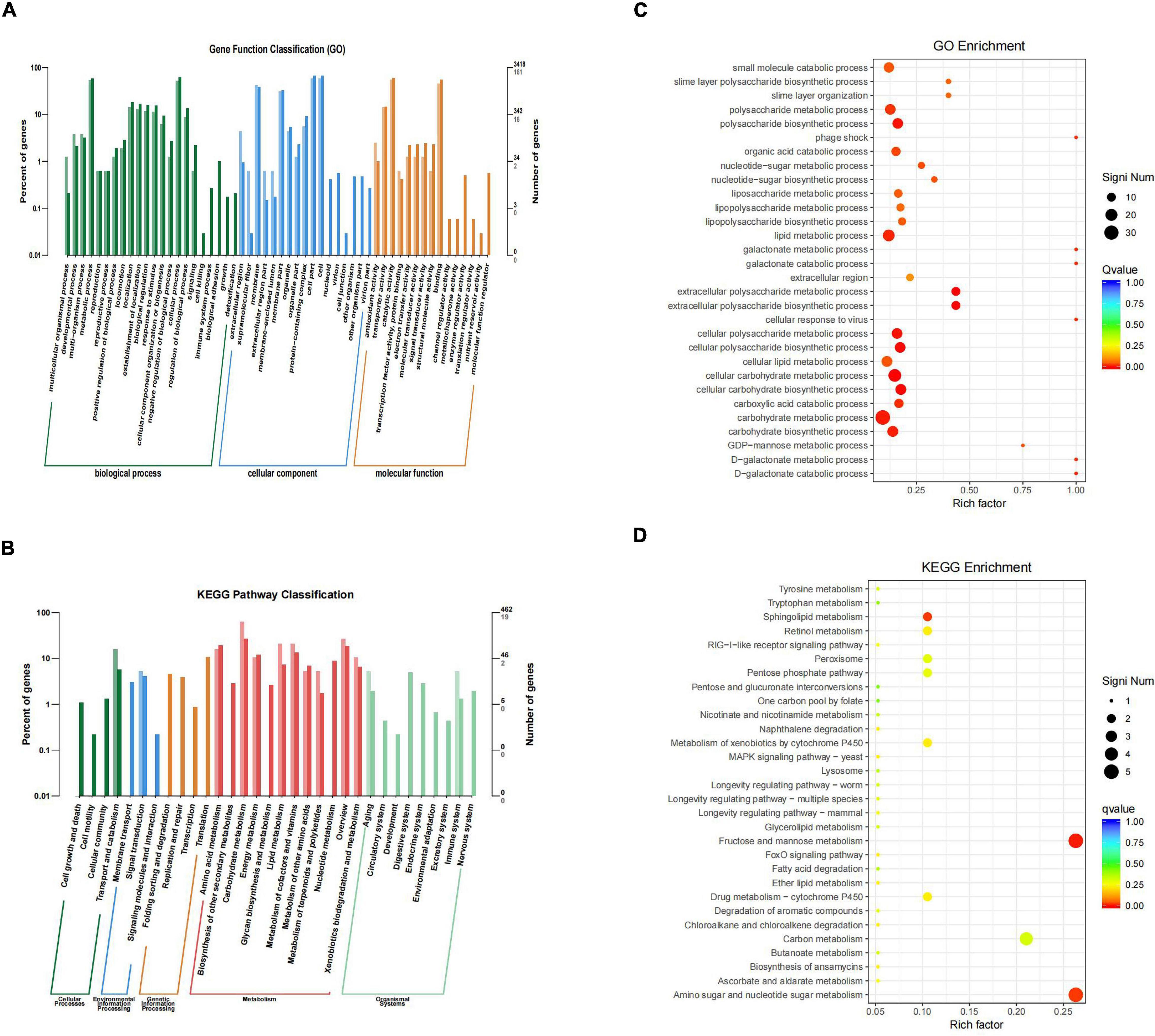
Figure 7. Differential genes classification and enrichment analysis in C. sakazakii ΔsdiA mutant compared to WT strains. (A) Differential genes GO functional classification. (B) Differential genes KEGG pathway classification. For A and B, the horizontal axis is a functional classification, and the vertical axis is the number of genes in the classification (right) and their percentage in the total number of genes annotated (left). Different colors represent different categories. Light colors represent different genes and dark colors represent all genes. (C) GO function enrichment analysis of C. sakazakiiΔsdiA mutant compared to WT strains. (D) KEGG pathway enrichment of C. sakazakiiΔsdiA mutant compared to WT strains. For C, the vertical axis represents the annotation of the function; for D, the vertical axis represents the annotation of the metabolic pathway. Rich factor represents the ratio of the number of GO enriched or KEGG enriched in the pathway to the number of annotated genes. The larger the Rich factor, the greater the degree of enrichment. The size of the q value is shown by the color of the dot as shown. The number of differential genes contained under each function or metabolic pathway is shown by the size of dots. The closer q value is to 0, the more significant the enrichment (Only the top 30 GO and KEGG with the highest enrichment degree were selected to draw).
Compared to the WT strain, the expression of D-galactose operon-related genes in the ΔsdiA deletion strain was markedly upregulated, including the dgoR, dgoK, dgoA, dgoD and dgoT genes. In addition, the expression levels of the flagellate-related genes FliA and FliC were significantly increased, and the activity of flagella was enhanced. Consequently, the ΔsdiA mutant strain showed significantly enhanced motor activity. Among the upregulated genes, the 6-phospho-glucosidase gene glvA was the most significantly upregulated by 5.24-fold (Supplementary Tables 1, 3, 4). 6-phosphoglucosidase has oxidoreductase activity, acting on CH-OH donors, NAD or NADP as receptors, and can bind to metal ions to express corresponding functions (Chen et al., 2018).
The differential gene expression of the SdiA gene deletion strain was markedly downregulated, which involved the VI secretion system and ABC transport system (Papenfort and Bassler, 2016). The type VI secretion system is a multifunctional protein secretion system that can directly deliver toxins to eukaryotic cells and other bacteria, and its function is related to virulence host immune resistance and bacterial interactions (Wang et al., 2018). In the downregulated gene expression, the key function of the VI secretory system was the ATP-active membrane-associated protein VasK, whose gene expression was downregulated by 1.53-fold. The type VI secretion system plays an important role in the cell adhesion, virulence, invasion and proliferation of C. sakazakii; this system also produces protein toxin, which is a potential toxic factor (Chen et al., 2018). Moreover, the expression level of related genes in the ABC transport system was downregulated by 1.5-fold, which may affect the pathogenesis and virulence of C. sakazakii and reduce its ability to infect host cells and bacteria in drug resistance (Supplementary Tables 2–4) (Wichelecki et al., 2015; Christoph and Robert, 2018).
Discussion
Homologous recombination is a conventional method for constructing gene deletion strains. Based on the principle of DNA homologous recombination, gene knockout technology can be roughly classified as homologous recombination, site-specific recombination and transposable recombination (Zhang et al., 2018). QS is a process of communication between microbial cells, which is closely related to the secretion, colonization, invasion and infection of virulence factors of pathogenic bacteria and the formation of biofilms (Papenfort and Bassler, 2016; Stefany et al., 2017). Nonetheless, the regulatory mechanism of QS on C. sakazakii remains largely unknown (Cao et al., 2021). To determine the role of QS in C. sakazakii-induced foodborne diseases, it is necessary to construct mutant strains of QS regulatory factors. Cao et al. successfully constructed SdiA frame deletion mutants using the suicide T vector pLP12 plasmid and Escherichia coli strain β2163 by sequence alignment based on the homologous recombination principle. Additionally, the QS receptor (SdiA) of C. sakazakii was screened out and the survival impact of SdiA on C. sakazakii under different environmental stresses was verified (Sabag et al., 2015; Cao et al., 2021). In this research, a mutant strain of the C. sakazakii SdiA gene was successfully established by homologous recombination and electrical transformation. Moreover, the mechanism of SdiA action on the growth, drug resistance, motility, biofilm formation and adhesion of C. sakazakii was verified, which laid a foundation for SdiA targeted inhibition of the pathogenicity of C. sakazakii in PIF food.
Prediction of microbiology is a research field that applies mathematical models to predict the growth, inactivation and survival of microorganisms in food (Huang and Hwang, 2017). The two-step method is a classical method to predict microbial modeling, but due to the two-step process, large errors will accumulate in the calculation (Baranyi and Roberts, 1995). A one-step method is to construct the primary model and the secondary model simultaneously according to the observed values. Due to the one-step computation, the accumulated error is small, the fitting is excellent, and the growth parameters of microorganisms can be obtained more precisely (Huang, 2017). In recent years, the one-step predictive microbial model has been reported in the relevant literature, and a good fitting effect has been obtained. For example, the one-step kinetic analysis method was used to simultaneously analyze the growth curve of Clostridium botulinum in cooked beef (Huang, 2017) and the growth of Salmonella in liquid eggs (Huang, 2015). Fang et al. studied the growth kinetics of thermally damaged C. sakazakii in restored infant formula (RPIF) and established a mathematical model to forecast its growth (Fang et al., 2012). In RPIF, a logistic model was used to calculate the minimum and maximum growth temperatures of untreated cells as 6.5°C and 51.4°C, respectively. The Huang model was used to calculate the minimum and maximum growth temperatures of C. sakazakii with heat treatment as 6.9°C and 50.1°C, respectively. Although relevant literature has demonstrated the growth model of C. sakazakii in recovery PIF, the verification of the model by fluctuating temperature is deficient (Wei et al., 2017). The actual growth of microbes in food is based on practically a dynamic model. Therefore, we studied the actual growth of C. sakazakii WT and ΔsdiA mutant strains in PIF based on one-step dynamic analysis. Pacheco et al. also found that no significant changes were observed in the growth curves of Klebsiella pneumoniae SdiA mutant and wild-type strains. However, cell division was impaired, and cell morphology was abnormal (Pacheco et al., 2021). This is similar to the results found in this study (Figure 3B).
The increased motility of the SdiA mutant strain may be due to the influence of the SdiA gene on the expression of flagellate-related gene in C. sakazakii, which enhanced the motility of the mutant strain. Culler et al. studied the atypical enteropathogenic Escherichia coli SdiA gene and discovered that SdiA gene deletion demonstrated stronger motility (Culler et al., 2018). Kanamaru et al. showed that the flagellate-related gene FliC of Escherichia coli O157:H7 was negatively regulated by the SdiA gene (Kanamaru et al., 2010), which was similar to the conclusion reached in this study. In addition, the cell morphology of the WT and ΔsdiA mutant strains was observed by scanning electron microscopy (Figure 3B). When the SdiA gene of C. sakazakii was removed, the overall morphology of the cells was not altered much. However, the ΔsdiA mutant strain was more elongated than the WT strain. This may help to enhance the swarming and swimming motility of C. sakazakii. In addition to quorum sensing, which can regulate cell movement, virulence factor production and biofilm formation, some similar substances also play such roles. Papenfort and Bassler showed that cyclic dimeric guanosine monophosphate (c-di-GMP) and cyclic adenosine monophosphate (cAMP), as intracellular second-messenger signaling molecules, regulate bacterial virulence and biofilms (Papenfort and Bassler, 2016).
Culler et al. (2018) studied biofilm formation of atypical enteropathogenic Escherichia coli wild-type, SdiA mutant and complementary strains. The results showed that the SdiA mutant was able to form thicker biofilm structures and exhibit stronger motility. In addition, the transcription of CsgA, CsgD and FliC was increased in the mutant strain, and the addition of AHLs reduced biofilm formation and the transcription of CsgD, CsgA and FimA in the wild-type strain. Sabag et al. (2015) showed that AHL response genes and SdiA-dependent genes were identified in the Enterobacter cloacae mouse strain, and the presence of the SdiA gene inhibited the formation of biofilms. The conclusions from these studies are similar to those from this experiment, indicating that the SdiA gene may reduce the amount of bacterial biofilm formation. In this study, the expression levels of the FliA and FliC genes regulating the flagella of the C. sakazakii SdiA deletion strain increased at the transcriptional level, and the flagella activity was enhanced, which strengthened the adhesion process of the SdiA deletion strain in biofilm formation and was conducive to the formation of microcolonies. In a plant-related Escherichia coli, the SdiA mutant terminated the csgBAC operon, resulting in excessive pili production and increased adherence and biofilm formation of the SdiA mutant strain (Tavío et al., 2010). Sharma and Bearson studied the effect of the QS transcriptional regulator SdiA on the retention of Escherichia coli O157:H7 in weaned calves and found that SdiA could sense AHLs in the rumen of cattle and enhance the acid resistance of Escherichia coli O157:H7 to allow Escherichia coli O157:H7 to persist and colonize the bovine intestine for a period of time (Sharma and Bearson, 2013).
Stefany et al. (2017) showed that bacteria sense environmental changes by secreting their own inducers or receiving external environmental signals through the QS system to cope with the influence of bacterial population density, external pH and antibiotics, such as the antibiotic stress response. Tavío et al. (2010) studied the role of SdiA in multidrugresistant strains of E. coli and found that the expression of the SdiA gene promoted the expression of the multidrug resistance-related gene acrAB, leading to the strengthening of drug resistance in E. coli. In addition, amplification of SdiA can enhance resistance to mitomycin C by increasing the ability of chromosome replication and repair.
Carneiro et al. (2020) examined the expression of SdiA and genes associated with the nucleotide salvage pathway in Salmonella by qRT-PCR and found that SdiA affects glucose consumption, metabolism and gene expression in Salmonella and is closely related to metabolic pathways associated with glycine, purine, amino acid and aminoacyl tRNA biosynthesis. It plays a role in metabolic optimization under a high population density of Salmonella. Cao et al. (2022) found that SdiA may inhibit the formation of C. sakazakii biofilms by regulating biosynthesis of flagella and extracellular polymers. Pacheco et al. (2021) that Klebsiella pneumoniae SdiA binds to the promoter regions imA, LuxS, LSR-LSRA and ftsQAZ, and this binding is independent of AHLs. In addition, a lack of SdiA can increase the formation of Klebsiella pneumoniae biofilms and agglutination of yeast cells and lead to downregulation of type 3 and upregulation of type 1 pili expression. More importantly, SdiA inhibits bacterial adhesion and biofilm aggregation (Papenfort and Bassler, 2016; Stefany et al., 2017; Pacheco et al., 2021).
Summary
In summary, we constructed a mutant strain with the SdiA gene deletion of C. sakazakii CICC21550 using the principle of homologous recombination. The function of the QS-related gene SdiA in C. sakazakii was analyzed to obtain the relationship between the SdiA gene and pathogenicity. The results demonstrated that SdiA enhanced the drug resistance of C. sakazakii but diminished its motility, adhesion and biofilm formation ability and had no effect on its growth. It can regulate the expression of D-galactose operon genes and flagellum-related gene upregulation and VI secretory system and ABC transport system-related gene expression downregulation. These results are helpful to further explore the function of the SdiA gene, revealing the pathogenic mechanism of C. sakazakii. Our data provide a new target for therapeutic interventions targeting the pathogenicity of C. sakazakii and developing quorum-sensing inhibitors.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject – PRJNA820965.
Author Contributions
CC conceptualized the experiments, draft preparation, and analyzed the data. XY performed the experiments and collected the experimental data. BL supervised the experimentation and analyzed the data. TJ and ZZ collected and interpretated the data. FG and QZ assisted with critical revision of the manuscript. CL designed the experiment, analyzed data, and revised the manuscript. TF designed the experiment, secured funding, and revised the manuscript. All authors reviewed, revised, and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (NSFC 31601393, 31401597), the National and Science Foundation of Fujian Province (2018J01696), Fujian Agricultural and Forestry University (KXb16012A) and Fujian Education Department (JAT160147), 13th Five-year Plan on Fuzhou Marine Economic Innovation and Development Demonstration City Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like thank Yanhong Liu and Lihan Huang at the U.S. Department of Agriculture for critical review of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.901912/full#supplementary-material
References
Ahmer, B. (2010). Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52, 933–945. doi: 10.1111/j.1365-2958.2004.04054.x
Almeida, F., Pinto, U. M., and Vanetti, M. (2016). Novel insights from molecular docking of SdiA from Salmonella enteritidis and Escherichia coli with quorum sensing and quorum quenching molecules. Microb. Pathog. 99, 178–190. doi: 10.1016/j.micpath.2016.08.024
Anetzberger, C., Pirch, T., and Jung, K. (2009). Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol. Microbiol. 73, 267–277. doi: 10.1111/j.1365-2958.2009.06768.x
Assia, G., Laure, P., Janek, B., Pauline, J., Benjamin, R., Mikael, E., et al. (2017). Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front. Microbiol. 8:227. doi: 10.3389/fmicb.2017.00227
Baranyi, J., and Roberts, T. A. (1995). Mathematics of predictive food microbiology. Int. J. Food Microbiol. 26, 199–218. doi: 10.1016/0168-1605(94)00121-L
Cacciatore, F. A., Dalmás, M., Maders, C., Isaía, H. A., and Malheiros, P. D. S. (2020). Carvacrol encapsulation into nanostructures: characterization and antimicrobial activity against foodborne pathogens adhered to stainless steel. Food Res. Int. 133:109143. doi: 10.1016/j.foodres.2020.109143
Cao, Y., Li, L., Zhang, Y., Liu, F., Xiao, X., Li, X., et al. (2022). Evaluation of Cronobacter sakazakii biofilm formation after sdiA knockout in different osmotic pressure conditions. Food Res. Int. 151:110886. doi: 10.1016/j.foodres.2021.110886
Cao, Y., Li, L., Zhang, Y., Liu, F., and Yu, Y. (2021). SdiA plays a crucial role in stress tolerance of C. sakazakii CICC 21544. LWT Food Sci. Technol. 143:111189. doi: 10.1016/j.lwt.2021.111189
Carneiro, D. G., Almeida, F. A. D., Aguilar, A. P., Nívea, M. V., and Vanetti, M. C. D. (2020). Salmonella enterica optimizes metabolism after addition of acyl-homoserine lactone under anaerobic conditions. Front. Microbiol. 11:1459. doi: 10.3389/fmicb.2020.01459
Chap, J., Jackson, P., Siqueira, R., Gaspar, N., Quintas, C., Park, J., et al. (2009). International survey of Cronobacter sakazakii and other Cronobacter spp. in follow up formulas and infant foods. Int. J. Food Microbiol. 136, 185–188. doi: 10.1016/j.ijfoodmicro.2009.08.005
Chen, Q., Zhu, Y., Qin, Z., Qiu, Y., and Zhao, L. (2018). Cronobacter spp., foodborne pathogens threatening neonates and infants. Front. Agric. Sci. Eng. 5:330–339. doi: 10.15302/J-FASE-2018208
Christoph, T., and Robert, T. (2018). Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr. Opin. Struct. Biol. 51, 116–128. doi: 10.1016/j.sbi.2018.03.016
Culler, H. F., Couto, S. C. F., Higa, J. S., Ruiz, R. M., and Sircili, M. P. (2018). Role of sdiA on biofilm formation by atypical enteropathogenic Escherichia coli. Genes 9:253. doi: 10.3390/genes9050253
Fang, T., Gurtler, J. B., and Huang, L. (2012). Growth kinetics and model comparison of Cronobacter sakazakii in reconstituted powdered infant formula. J. Food Sci. 77, 1–9. doi: 10.1111/j.1750-3841.2012.02873.x
Fang, T., and Huang, L. (2014). Growth and survival kinetics of Listeria monocytogenes in cooked egg whites. Food Control 36, 191–198. doi: 10.1016/j.foodcont.2013.08.034
Farmer, J. J., Asbury, M. A., Hickman, F. W., and Brenner, D. J. (1980). Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Bacteriol. 30, 569–584. doi: 10.1099/00207713-30-3-569
Guo, D., Wang, S., Li, J., Bai, F., and Shi, C. (2019). The antimicrobial activity of coenzyme Q0 against planktonic and biofilm forms of Cronobacter sakazakii. Food Microbiol. 86:e103337. doi: 10.1016/j.fm.2019.103337
Hariri, S., Joseph, S., and Forsythe, S. J. (2013). Cronobacter sakazakii ST4 strains and neonatal meningitis United States. Emerg. Infect. Dis. 19, 175–177. doi: 10.3201/eid1901.120649
Huang, L. (2015). Direct construction of predictive models for describing growth of Salmonella enteritidis in liquid eggs – a one-step approach. Food Control 57, 76–81. doi: 10.1016/j.foodcont.2015.03.051
Huang, L. (2017). Dynamic identification of growth and survival kinetic parameters of microorganisms in foods. Curr. Opin. Food Sci. 14, 85–92. doi: 10.1016/j.cofs.2017.01.013
Huang, L. (2018). Growth of non-toxigenic Clostridium botulinum mutant LNT01 in cooked beef: one-step kinetic analysis and comparison with C. sporogenes and C. perfringens. Food Res. Int. 107, 248–256. doi: 10.1016/j.foodres.2018.02.028
Huang, L., and Hwang, A. (2017). Practical Methods in Predictive Food Microbiology. Boca Raton, FL: CRC Press.
Hunter, C. J., and Bean, J. F. (2013). Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J. Perinatol. 33, 581–585. doi: 10.1038/jp.2013.26
Jaradat, Z. W., Ababneh, Q. O., Saadoun, I. M., Samara, N. A., and Rashdan, A. M. (2009). Isolation of Cronobacter spp. (formerly Enterobacter sakazakii) from infant food, herbs and environmental samples and the subsequent identification and confirmation of the isolates using biochemical, chromogenic assays, PCR and 16S rRNA sequencing. BMC Microbiol. 9:225. doi: 10.1186/1471-2180-9-225
Jia, Z., Bai, W., Li, X., Tfa, B., and Cla, B. (2020). Assessing the growth of Listeria monocytogenes in salmon with or without the competition of background microflora-A one-step kinetic analysis. Food Control 114:107139. doi: 10.1016/j.foodcont.2020.107139
Jung, M. K., and Park, J. H. (2006). Prevalence and thermal stability of Enterobacter sakazakii from unprocessed ready-to-eat agricultural products and powdered infant formulas. Food Sci. Biotechnol. 15, 152–157. doi: 10.1016/j.foodpol.2005.08.003
Kanamaru, K., Tatsuno, I., Tobe, T., and Sasakawa, C. (2010). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38, 805–816. doi: 10.1046/j.1365-2958.2000.02171.x
Lars, R. A., Allan, B. C. B., Sren, M. B., Michael, G. B., and Lone, G. A. (2001). Methods for detecting acylated homoserine lactones produced by gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44, 239–251. doi: 10.1016/S0167-7012(01)00217-2
Liu, L., Yan, Y., Feng, L., and Zhu, J. (2018). Quorum sensing asaI mutants affect spoilage phenotypes, motility, and biofilm formation in a marine fish isolate of Aeromonas salmonicida. Food Microbiol. 76, 40–51. doi: 10.1016/j.fm.2018.04.009
Ma, X., Zhang, S., Xu, Z., Li, H., and Lu, Y. (2020). SdiA improves the acid tolerance of E. coli by regulating GadW and GadY expression. Front. Microbiol. 11:1078. doi: 10.3389/fmicb.2020.01078
Odeyemi, O. A., and Sani, N. A. (2019). Antibiotic resistance, putative virulence factors and curli fimbrination among Cronobacter species. Microb. Pathog. 136:103665. doi: 10.1016/j.micpath.2019.103665
Ogrodzki, P., and Forsythe, S. J. (2017). DNA-sequence based typing of the Cronobacter genus using MLST, CRISPR-cas array and capsular profiling. Front. Microbiol. 8:1875. doi: 10.3389/fmicb.2017.01875
Pacheco, T., Ana, R. I. G., Nathália, M. G. S., Assoni, L., Lúcio, F., and Caldas, F. (2021). SdiA, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front. Microbiol. 12:597735. doi: 10.3389/fmicb.2021.597735
Papenfort, K., and Bassler, B. (2016). Quorum sensing signal–response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Sabag, D. A., Dyszel, J. L., Gonzalez, J. F., Ali, M. M., and Ahmer, B. (2015). Identification of sdiA-regulated genes in a mouse commensal strain of Enterobacter cloacae. Front. Cell Infect. Microbiol. 5:47. doi: 10.3389/fcimb.2015.00047
Sharma, V. K., and Bearson, S. M. D. (2013). Evaluation of the impact of quorum sensing transcriptional regulator sdiA on long-term persistence and fecal shedding of Escherichia coli O157: H7 in weaned calves. Microb. Pathog. 57, 21–26. doi: 10.1016/j.micpath.2013.02.002
Stefany, M. G., Sorg, R. A., Domenech, A., Kjo, M., Weissing, F. J., Doorn, G. S. V., et al. (2017). Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun. 8, 1–12. doi: 10.1038/s41467-017-00903-y
Tavío, M. M., Aquili, V. D., Poveda, J. B., Antunes, N. T., and Sánchez, C. J. (2010). Quorum-sensing regulator sdiA and marA overexpression is involved in in vitro-selected multidrug resistance of Escherichia coli. J. Antimicrob. Chemother. 65, 1178–1186. doi: 10.1093/jac/dkq112
U. S. FDA (2022). FDA Warns Consumers not to Use Certain Powdered Infant Formula Produced in Abbott Nutrition’s Facility in Sturgis, Michigan. Available online at: https://www.fda.gov/news-events/press-announcements/fda-warns-consumers-not-use-certain-powdered-infant-formula-produced-abbott-nutritions-facility (accessed February 20, 2022).
Vadakkan, K., Choudhury, A. A., Gunasekaran, R., Janarthanam, H. B., and Selvaraj, V. A. (2018). Quorum sensing intervened bacterial signaling: pursuit of its cognizance and repression. J. Genet. Eng. Biotechnol. 16, 239–252. doi: 10.1016/j.jgeb.2018.07.001
Wang, M., Wang, L., Wu, P., Chen, T. T., Zhu, Y. M., Zhang, Y., et al. (2019). Genomics and experimental analysis reveal a novel factor contributing to the virulence of Cronobacter sakazakii strains associated with neonate infection. J. Infect. Dis. 220, 306–315. doi: 10.1093/infdis/jiz098
Wang, Y., Wang, Y., Sun, L., Daniel, G., and Li, Y. (2018). The LuxS/AI-2 system of Streptococcus suis. Appl. Microbiol. Biotechnol. 102, 7231–7238. doi: 10.1007/s00253-018-9170-7
Wei, Q., Gong, X. L., Lin, J. Y., Zhang, X. Y., Jiang, Y. J., and Fang, T. (2017). The biological properties of Cronobacter sakazakii and its predictive model in infant formula. J. Chin. Food Sci. Technol. 17, 58–64. doi: 10.16429/j.1009-7848.2017.04.008
Wichelecki, D. J., Vetting, M. W., Chou, L., Al-Obaidi, N., Bouvier, J. T., Almo, S. C., et al. (2015). TP-binding cassette (ABC) transport system solute-binding protein-guided identification of novel d-Altritol and galactitol catabolic pathways in Agrobacterium tumefaciens C58. J. Biol. Chem. 290, 28963–28976. doi: 10.1074/jbc.M115.686857
Williams, P., Camara, M., Hardman, A., Swift, S., Hope, V. J., Winzer, K., et al. (2000). Quorum sensing and population-dependent control of virulence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 667–680. doi: 10.1098/rstb.2000.0607
Xie, Z. P., Peng, Y. B., Li, C. C., Luo, X. J., Fang, T., and Huang, L. (2020). Growth kinetics of Staphylococcus aureus and background microorganisms in camel milk. J. Dairy Sci. 103, 9958–9968. doi: 10.3168/jds.2020-18616
Keywords: Cronobacter sakazakii, quorum sensing receptor, SdiA, transcriptomics, multidrug resistance, biofilm
Citation: Cheng C, Yan X, Liu B, Jiang T, Zhou Z, Guo F, Zhang Q, Li C and Fang T (2022) SdiA Enhanced the Drug Resistance of Cronobacter sakazakii and Suppressed Its Motility, Adhesion and Biofilm Formation. Front. Microbiol. 13:901912. doi: 10.3389/fmicb.2022.901912
Received: 22 March 2022; Accepted: 19 April 2022;
Published: 06 May 2022.
Edited by:
Ramona Iseppi, University of Modena and Reggio Emilia, ItalyReviewed by:
Tom Defoirdt, Ghent University, BelgiumQingli Dong, University of Shanghai for Science and Technology, China
Copyright © 2022 Cheng, Yan, Liu, Jiang, Zhou, Guo, Zhang, Li and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changcheng Li, Y2hhbmdjaGVuZ19saUBmYWZ1LmVkdS5jbg==; Ting Fang, ZmFuZ3Rpbmc5MzBAMTYzLmNvbQ==
 Chuansong Cheng
Chuansong Cheng Xiaotong Yan
Xiaotong Yan Fengting Guo
Fengting Guo Ting Fang
Ting Fang