
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 June 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.901793
This article is part of the Research Topic Microbial Co-cultures: A New Era of Synthetic Biology and Metabolic Engineering View all 16 articles
 Mritunjoy Barman1†
Mritunjoy Barman1† Snigdha Samanta1†
Snigdha Samanta1† Gouranga Upadhyaya2*
Gouranga Upadhyaya2* Himanshu Thakur3
Himanshu Thakur3 Swati Chakraborty4
Swati Chakraborty4 Arunava Samanta1
Arunava Samanta1 Jayanta Tarafdar4*
Jayanta Tarafdar4*Bemisia tabaci (whitefly) is one of the most detrimental agricultural insect pests and vectors of many plant viruses distributed worldwide. Knowledge of the distribution patterns and insecticide resistance of this cryptic species is crucial for its management. In this study, genetic variation of mitochondrial cytochrome oxidase subunit 1 (MtCoI) gene of B. tabaci was analyzed followed by a study of the infection profile of various endosymbionts in 26 whitefly populations collected from West Bengal, India. Phylogenetic analysis revealed Asia I as the major cryptic species (65.38%), followed by Asia II 5, China 3, and Asia II 7, which were diversified into 20 different haplotypes. In addition to the primary endosymbiont (C. poriera), each of the four whitefly species showed a variable population of three secondary endosymbionts, majorly Arsenophonus with the highest infection rate (73.07%), followed by Wolbachia and Rickettsia. Further phylogenetic analyses revealed the presence of two subgroups of Arsenophonus, viz., A1 and A2, and one each in Wolbachia (W1) and Rickettsia (R3). Resistance to thiamethoxam, imidacloprid, and acetamiprid insecticides was analyzed for a clear picture of pesticide resistance status. The highest susceptibility was noted toward thiamethoxam (LC50 = 5.36 mg/L), followed by imidacloprid and acetamiprid. The whitefly population from Purulia and Hooghly districts bearing Asia II 7 and Asia II 5 cryptic species, respectively, shows maximum resistance. The differences in mean relative titer of four symbiotic bacteria among field populations varied considerably; however, a significant positive linear correlation was observed between the resistance level and relative titer of Arsenophonus and Wolbachia in the case of imidacloprid and thiamethoxam, while only Wolbachia was found in case of acetamiprid. Expression analysis demonstrated differential upregulation of insecticide resistance genes with Purulia and Hooghly populations showing maximally upregulated P450 genes. Moreover, thiamethoxam and imidacloprid resistance ratio (RR) showed a significant correlation with CYP6CM1, CYP6DZ7, and CYP4C64 genes, while acetamiprid RR correlated with CYP6CX1, CYP6DW2, CYP6DZ7, and CYP4C64 genes. Taken together, these findings suggested that P450 mono-oxygenase and symbiotic bacteria together affected whitefly resistance to neonicotinoids. Hence, a symbiont-oriented management programme could be a better alternative to control or delay resistance development in whitefly and can be used for pesticide clean-up in an agricultural field.
The whitefly, Bemisia tabaci, is an economically important agricultural pest causing huge damage to crops worldwide. They inflict damage to plants directly and as a vector of several hundred viruses, with a majority (>320 species) of them belonging to the genus Begomovirus and other economically important viruses belonging to the genera Ipomovirus, Carlavirus, Crinivirus, Torradovirus, and Polerovirus (Jones, 2003; Mugerwa et al., 2021). Whitefly is composed of a complex group of genetically distinct species and/or biotypes that differ substantively by host plant preference, virus transmitting ability, and insecticide resistance (De Barro et al., 2011; Barman et al., 2022). It is listed as one of the top 100 dreadful alien invasive species due to its virus transmission ability and wide host adaptability (Lowe et al., 2000). Dinsdale et al. (2010) proposed a 3.5% pair-wise genetic divergence of mitochondrial cytochrome oxidase I (MtCoI), which led to the identification of 44 distinct species of B. tabaci. Additionally, two new species, Asia II 13 and Spain 1, have also been recently reported (Kanakala and Ghanim, 2019). The Indian geographical regions unveil a huge diversity of B. tabaci with the presence of 10 cryptic species out of 46 recorded species so far (Rehman et al., 2021). Precise knowledge of the distribution pattern of this cryptic species is crucial for the efficient management of this notorious pest worldwide.
Nonetheless, increasing reports of resistance development in B. tabaci to various organophosphates, synthetic pyrethroids, and other neonicotinoid compounds have been surfacing (Peshin and Zhang, 2014; Naveen et al., 2017; Wang et al., 2020). Several countries, namely, China (Wang et al., 2010), India (Kranthi et al., 2002), Iran (Basij et al., 2017), Israel (Alon et al., 2008), Malaysia (Shadmany et al., 2015), Pakistan (Ahmad et al., 2010), and the USA (Prabhaker et al., 2014), have reported the development of insecticide resistance in whitefly to even compounds of novel chemistry. Intensive and frequent use of insecticides, selection pressure, and choice of poor insecticides may be some of the other factors rendering resistance in whitefly both at field and laboratory levels (Sethi and Dilawari, 2008; Pietri and Liang, 2018). The neonicotinoid group of compounds, represented by imidacloprid, thiamethoxam, and others, are majorly used insecticides in controlling the whitefly population worldwide, including in India (Sethi and Dilawari, 2008; Yang et al., 2013; Pang et al., 2020). However, several countries reported neonicotinoid resistance in whitefly due to the overuse of this chemical compound (Nauen et al., 2002; Ma et al., 2007; Luo et al., 2010; Naveen et al., 2017).
Researchers have observed that increased metabolic detoxification may be due to gene amplification, overexpression, and modification of gene coding proteins of major detoxifying enzymes, such as cytochrome P450 and glutathione S-transferases (GSTs) (Karunker et al., 2008; Feyereisen, 2011; Elzaki et al., 2017). The involvement of P450 genes in conferring resistance to several insecticides has been listed in insects, such as Drosophila melanogaster to DDT and imidacloprid, brown planthopper (BPH) to imidacloprid, and so on (Daborn et al., 2001; Garrood et al., 2016). Another most notable aspect of this complex is the bacterial symbionts that infect its members. Considerable evidence exists regarding microbe-mediated effects in whitefly, ranging from host survival, development, and nutritional fitness to even insecticide resistance (Su et al., 2014; Li et al., 2018). The B. tabaci species complex carries a primary endosymbiont Candidatus Portiera aleyrodidarum that occurs in all individuals and is located in specialized cells termed as bacteriocytes (Sloan and Moran, 2012). The secondary symbionts, namely, Rickettsia (Gottlieb et al., 2006), Wolbachia (Nirgianaki et al., 2003), Hamiltonella, Arsenophonus (Thao and Baumann, 2004; Sloan and Moran, 2012), Cardinium (Weeks et al., 2003), Fritschea (Everett et al., 2005), and Hemipteriphilus (Bing et al., 2013) have been reported to be present in B. tabaci populations around the world. Unlike primary endosymbionts which have a direct mutualistic relationship with the host, the secondary symbionts form a less stable partnership with their host. The infection pattern of these secondary symbionts is highly complex and varies according to the geographical regions (Zchori-Fein et al., 2014) and the different genetic groups (Chiel et al., 2007; Ghosh et al., 2015; Lestari et al., 2021).
The development of resistance is generally manifested by a failure to control the pests at the field level. A very few studies from India were involved in determining the field level resistance in B. tabaci; however, there was no in-depth study explaining the exact mechanism behind such resistance development. Hence, a precise understanding of the mechanism of insecticide resistance is of great significance for the management or delay in the development of resistance. Within this framework, this work hypothesized that there are differences in the resistance levels of whitefly populations in different regions toward insecticides like neonicotinoids, and this variation might be affected by the expression of genes associated with cytochrome P450 mono-oxygenase or variation of symbiont titres, either separately or together. To test this hypothesis, the following objectives were considered: (1) characterization and resistance level of the whitefly field population toward imidacloprid, thiamethoxam, and acetamiprid, (2) infection pattern and the relative titer of the endosymbiotic bacteria (Portiera, Arsenophonus, Wolbachia, and Rickettsia) associated with the whitefly field population, and (3) gene expression associated with cytochrome P450 mono-oxygenase (CYP6CM1, CYP6CX1, CYP6CX5, CYP6CX3, CYP6DZ4, CYP4C64, CYP6DZ7, and CYP6DW2) and their relationship with resistance level. Our finding, for the first time, shall shed light on field evolved resistance in different whitefly cryptic species toward popularly used neonicotinoids, further attempting to predict the mechanism behind such resistance development, in particular, the involvement of endosymbionts and P450 genes.
The samples of B. tabaci used in the current study were collected from different locations across Bengal province, India (as detailed in Supplementary Table S1). Adult whiteflies (mixed sex) were collected from various crop plants (chili, eggplant, cucumber, tomato, lady's finger, and pointed gourd) and weed plants (cida and wild brinjal). The flies were pooled together inside ventilated insect-proof cages on the basis of location and further reared in the Molecular Biology laboratory, Directorate of Research, BCKV.
The genetic group identity of B. tabaci field population was examined by random sampling of 10 adult flies for each population using PCR amplification of the MtCoI gene and sequencing as described by Dinsdale et al. (2010). Genomic DNA was extracted from each specimen as per the manufacturer's instructions (Invitrogen, USA). PCR amplification using 20 ng of insect DNA was carried out in Veriti 96-Well Thermal Cycler (Applied Biosystems, USA). The endosymbionts (Portiera, Wolbachia, Arsenophonus, and Rickettsia) in the collected whitefly populations were also determined using specific primers (as listed in Supplementary Table S2). The amplified PCR products were excised from the gel and purified using the XcelGen DNA Gel/ PCR Purification mini kit (Xcelris genomics, India) following the manufacturer's instructions. The DNA sequence was obtained through Sanger dideoxy sequencing method from Chromus Biotech Laboratory, India.
Different neutrality statistics were performed to examine the demographic history of whitefly and their endosymbionts. Tajima's D (Tajima, 1989) and Fu's Fs (Fu, 1997) statistics were subsequently calculated to check whether the COI conformed to the expectations of neutrality by DnaSP (Librado and Rozas, 2009). The population pair-wise F-statistics (FST) and analysis of molecular variance (AMOVA) were calculated with the help of Arlequin v.3.5 (Excofer and Lischer, 2010). The haplotype network of the sequences was examined using a minimum spanning network relationship with the help of popART software. Multiple alignments of all the nucleotide sequences of whiteflies and endosymbionts were conducted using ClustalW, and analysis was done in Mega-X (Kimura, 1980; Kumar et al., 2018). The tree was verified using the maximum likelihood (ML) model, and a total 1,000 of bootstrap replicates were performed. The tree was processed through iTol (version 6.4.3) for graphical representation. All the nucleotide sequences of the whitefly population and endosymbionts were submitted to the NCBI GenBank database.
To determine the current susceptibility status and resistant gene expression of whitefly, the field populations from the five most important regions (one population each from Kalimpong, Midnapore, Hooghly, Malda, and Purulia) were selected (as listed in Supplementary Table S3). For the bioassay, a total of approximately 800 adult flies were collected per location, and around 650 of them were used immediately in the bioassay study. The whitefly population collected from the university farm (B.C.K.V., Kalyani, India) was chosen to be reared for 10 additional generations without any exposure to insecticides in the laboratory and was considered as the susceptible population (SP). Three insecticides, namely, imidacloprid 17.8 SL (Confidor) obtained from Bayer Co., Ltd., thiamethoxam 25 WG (Actara) from Syngenta Chemical Ltd., and acetamiprid 20SP (Pyramid) from Hifield-AG Chem. Pvt. Ltd., were used for bioassay studies against the adult whitefly population. The toxicity of imidacloprid, thiamethoxam, and acetamiprid was evaluated following a modified leaf dip bioassay method by the Insecticide Resistance Action Committee (Naveen et al., 2017). To evaluate the toxicity, final doses were decided based on the prior experiments conducted on lab populations, and five different concentrations of each insecticide were used. In each insecticide treatment, fresh brinjal leaves (5 × 5 cm2) were dipped in the respective insecticide dilutions for 30 ± 2 s in a corning glass Petri plate (dia. = 15 cm) and air-dried, whereas the control leaves were dipped in water alone. After drying the solution on the leaf surface, the treated leaves were transferred to the corning glass Petri plate (dia. = 9 cm) containing a thin 2% agar layer at the bottom. Each treatment was replicated five times, and six adult whiteflies were released per replication (thirty in each treatment) and plates were covered with ventilated lids. The experiment was conducted under laboratory conditions (26 ± 1°C, 70–80% RH, 16L: 8D photoperiod), and the observations regarding the mortality were taken at 72 h after feeding (HAF).
Total genomic DNA (for quantification of symbiont titer) and RNA (for gene expression) were extracted from field-collected whitefly (adult) population using Insect DNA and RNA Isolation Kit (Thermo Fisher Scientific, USA) following the manufacturer's protocol. Quantity was assessed with a Qubit Flex Fluorometer, and cDNA was prepared using 1 μl of total RNA according to the manufacturer's instructions (GeneSure H-Minus First Strand cDNA Synthesis Kit, Genetix Biotech Asia Pvt. Ltd.).
The relative abundance of targeted symbionts (Portiera, Arsenophonus, Wolbachia, and Rickettsia) and expression pattern of cytochrome P450 genes, namely, CYP6CM1, CYP6CX1, CYP6CX5, CYP6CX3, CYP6DZ4, CYP4C64, CYP6DZ7, and CYP6DW2, were examined using qRT-PCR (Wang et al., 2020). qRT-PCR was performed in Agilent Technologies Stratagene (Model-Mx3000P) using SYBR Green Master Mix (Applied Biosystems, USA). Primer details and annealing temperatures are mentioned in Supplementary Table S2. The relative expression of each target gene was calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001). The Actin gene was used as an internal control.
To determine the lethal concentration (LC) values, the observations recorded on mortality data were corrected using Abbott's formula (Abbott, 1925). The data were subjected to probit analysis (Finney, 1971) using SPSS. The resistance ratio (RR) for each population was calculated by the ratio: LC50 value of the population/LC50 value of the susceptible population. The RRs were then classified as follows: RR < 5 indicated low resistance; RR = 5–10 indicated moderate resistance, and RR > 10 indicated high resistance (Mazzarri and Georghiou, 1995; World Health Organization, 2016). The qRT-PCR experiments were conducted at least thrice with a minimum of three individual biological replicates. Data have been represented as mean ± SD (n = 3) values. Statistical significance was analyzed using the GraphPad Prism 8.0 software by one-way analysis of variance (ANOVA), followed by Dunnett's multiple comparison test. Significant differences with the susceptible population have been represented as *p < 0.01, **p < 0.001, and ***p < 0.0001. Linear Model II regression analysis was used to determine the functional relationship between the resistance level of the population and the mean normalized expression value of each gene and endosymbiont in different populations using SPSS (SPSS for Windows, Rel. 17.0.0 2009. Chicago: SPSS Inc.).
In totality, 26 whitefly populations collected from different host crops across different regions of Bengal province (as shown in Figure 1A) were identified using the primer pair (C1-J-2195 F/L2-N-3014 R) of the universal Mt-co1 gene. The phylogenetic analysis of the determined COI sequences divided B. tabaci into four different cryptic species, with Asia I being the most predominant species followed by Asia II 5, China 3, and Asia II 7 at rates of 65.38, 19.23, 11.53, and 3.84%, respectively (Figure 1A inset). To show the distribution of collected samples, a maximum likelihood phylogenetic tree of the COI sequences of Bemisia tabaci collected from different locations has been drawn with previously reported GenBank sequences, wherein the bold text COI sequences represent collected samples used in this study (Figure 1B). Subsequently, diagnostic PCR confirmed the presence of primary endosymbiont Portiera and secondary endosymbionts Wolbachia, Arsenophonus, and Rickettsia in the selected whitefly population.
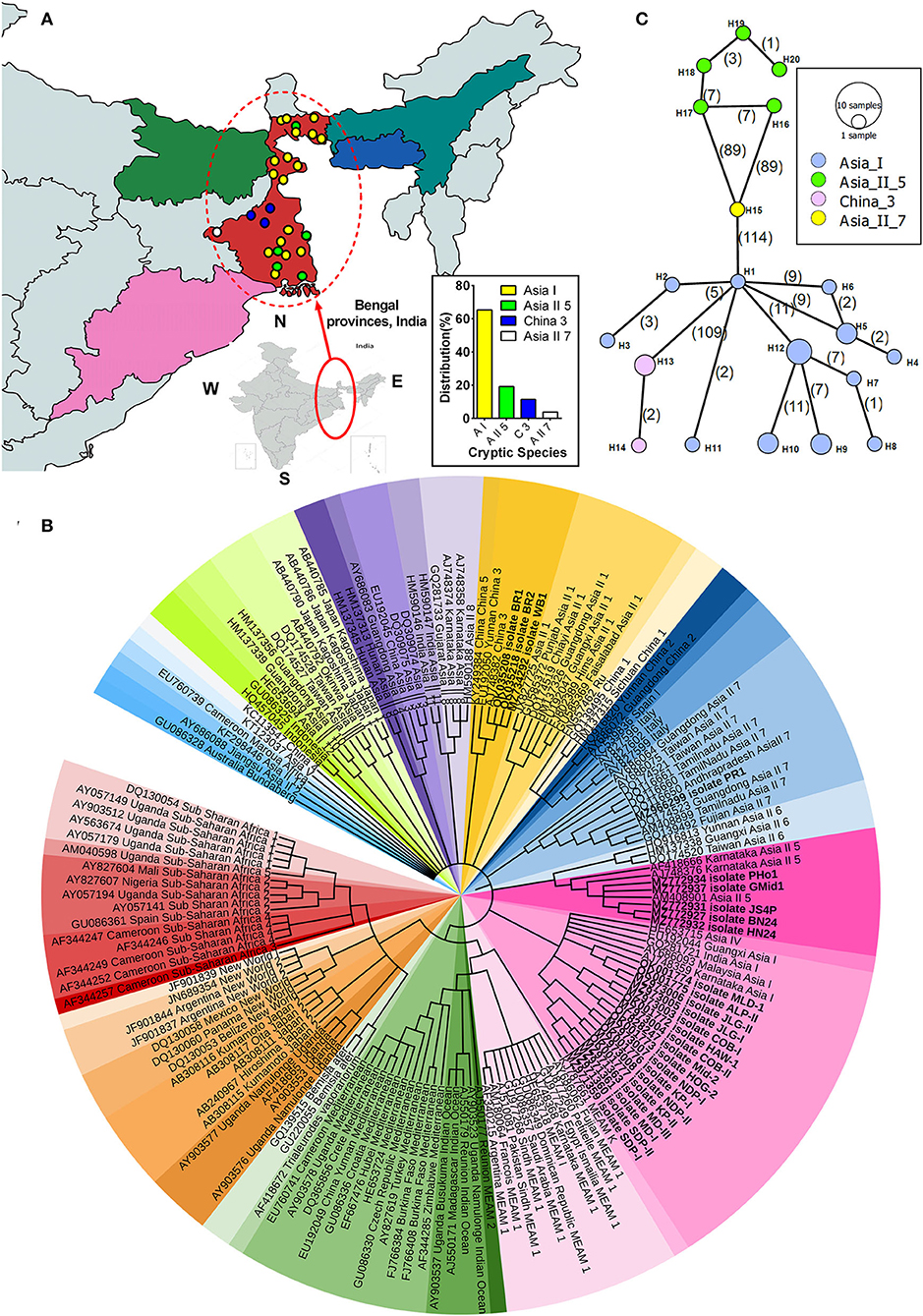
Figure 1. Phylogenetic analysis of whitefly (Bemisia tabaci) based on COI gene. (A) Map indicating different locations of B. tabaci sampling and distribution across different genetic groups in Bengal provinces, India. Inset shows the percentage distribution of various cryptic species of B. tabaci from collected specimens. (B) The maximum likelihood phylogenetic tree of the COI sequences of Bemisia tabaci collected from different locations. The bold text COI sequences represent collected samples used in this study, and the rest represent reference sequences obtained from the GenBank database. A total of 159 nucleotide sequences were selected to construct the tree, wherein Bemisia afer and Trialeurodes vaporariorom were taken as an out-group. Different color sheds indicate the different cryptic species of whitefly distributed worldwide. Hasegawa-Kishnio-Yano HKY850 model and gamma distribution rate of variation among sites were implemented to construct the phylogenetic tree in MEGA X. (C) Minimum spanning network of different B. tabaci cryptic species haplotypes from the studied samples. The four different colors indicate the haplotype of B. tabaci cryptic species. Each circle represents a unique haplotype, the frequency of which is proportional to the diameter of the circle. The number of mutations is represented by numbers in parenthesis.
The pair-wise divergence in the MtCoI nucleotide sequence among the four identified genetic groups varied both at intraspecific and interspecific levels (Supplementary Table S4). Intraspecific variation was recorded to be the highest in Asia I (0.24–2.66%), and the lowest was observed in China 3 (0.11–0.23%). While considering the interspecific variation, Asia I and Asia II 5 (17.47–20.80%) were higher than that of Asia I and China 3 (14.63–17.09%). However, the lowest variation was observed between China 3 and Asia II 7 (16.95–17.30%).
Among the 20 haplotypes (based on MtCoI sequence), the highest number was recorded in Asia I (12), followed by Asia II 5 (5) and China 3 (2) (Supplementary Table S5). Asia II 7 was excluded from the genetic diversity study, as only one such sample was found in our survey. Haplotype diversity was higher in Asia II 5 (1) when compared to Asia I (0.956), whereas nucleotide diversity was higher in Asia I (0.014) in comparison to Asia II 5 (0.008). Results obtained from the neutrality test (Fu and Li's D*, Fu and Li's F*, and Tajima's D) recorded a significant negative value for Asia II 5 and China 3, indicating that these two cryptic species across the regions might be undergoing expansion.
Minimum spanning network analysis indicated four cryptic species to be diversified into 20 haplotypes, which are quite distant from one another (Figure 1C). Among the 12 haplotypes of Asia I, H1 occupied the central position of the network and was the linking haplotype between China 3 (H13) and Asia II 7 (H15) with the mutational differences of 109 and 114, respectively. Furthermore, hierarchical AMOVA revealed that most of the evident genetic diversity is collectively accredited to highly significant genetic differences between the four whitefly cryptic species (Fst: 91.83%; P < 0.001), and the haplotypes within these cryptic species contributed to 8.16% of the variation (Supplementary Table S6). Moreover, the genetic differentiation among the population (pair-wise Fst value) was >0.90 for all the four cryptic species (Supplementary Table S7).
Primary endosymbiont (C. poriera) was present in all the test samples; however, the infection profiles of three secondary endosymbionts (Arsenophonus, Wolbachia, and Rickettsia) showed a substantial variation in each of the four identified cryptic whitefly species. Overall, Arsenophonus showed the highest infection rate (73.07%), followed by Rickettsia (65.38%) and Wolbachia (53.84%). Arsenophonus infection was found in Asia I, Asia II 5, and Asia II 7 population but not in China 3 population, with Asia I bearing the highest infection rate (Supplementary Table S8).
Furthermore, based on the criterion of Kanakala and Ghanim (2019), the secondary endosymbionts were classified at the subgroup level. The phylogenetic tree analysis revealed the presence of only one subgroup of Wolbachia (W1) and one subgroup of Rickettsia (R3) that infected all the four cryptic whitefly species (Figures 2A,B, respectively). The phylogenetic tree also revealed the presence of two subgroups of Arsenophonus: A1 and A2 (Figure 2C). The infection rate of A1 was always higher than A2 in Asia I population, which was in contrast to Asia II 5 population, where A2 was higher. The infection rate of Wolbachia was 40–100%, with the highest rate occurring in Asia II 7 and China 3 populations. The infection rate of Rickettsia was the highest in China 3 and Asia II 7 populations, followed by Asia I and Asia II 5 populations, respectively. Thus, as observed, all the whitefly individuals were infected with the variable combinations of the three secondary endosymbionts (Supplementary Table S9).
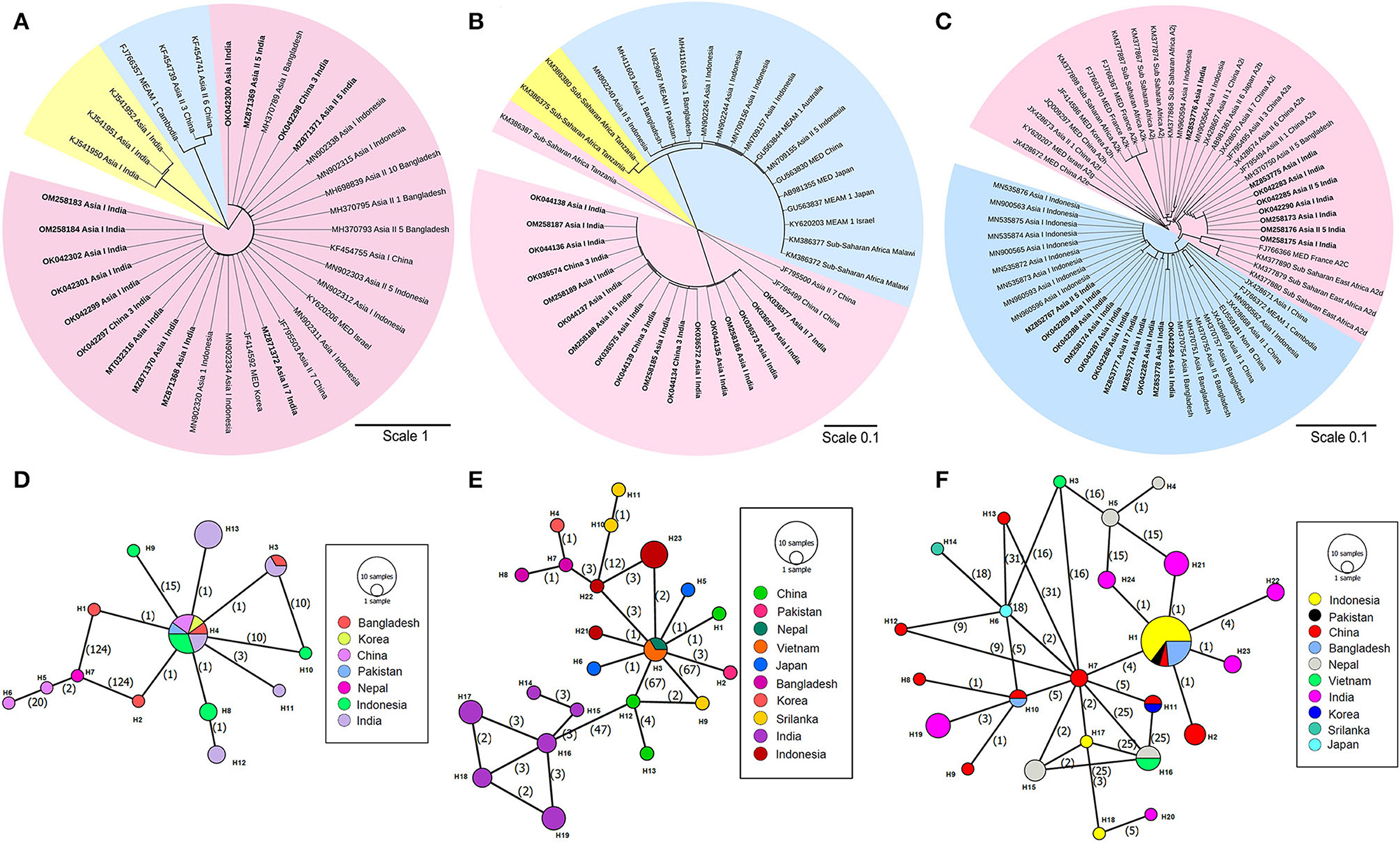
Figure 2. Phylogenetic tree and network analysis of secondary endosymbiont based on 16S rDNA (Wolbachia and Rickettsia) and 23S rDNA (Arsenophonus) gene segments. (A) The maximum likelihood phylogenetic tree of the 16S rDNA sequences of Wolbachia sp. infecting different whitefly populations. Different color shades indicate the subgroup of Wolbachia species (W1 subgroup-pink shade, W2 subgroup-blue shade, W3 subgroup-yellow shade) distributed worldwide. (B) The maximum likelihood phylogenetic tree of the 16S rDNA sequences of Rickettsia sp. infecting different whitefly populations. Different color shades indicate the subgroup of Rickettsia species (R1 subgroup-blue shade, R2 subgroup-yellow shade, and R3 subgroup-pink shade) distributed worldwide. (C) The maximum likelihood phylogenetic tree of the 23S rDNA sequences of Arsenophonus sp. infecting different whitefly populations. Different color shades indicate the subgroup of Arsenophonus species (A1 subgroup-blue shade and A2 subgroup-pink shade) distributed worldwide. Hasegawa-Kishnio-Yano HKY850 model and gamma distribution rate of variation among sites were implemented to construct all the phylogenetic trees in MEGA X. Minimum spanning network of different Wolbachia species (D), Rickettsia species (E), and Arsenophonus species (F) present in collected whitefly samples from India and other Asian countries. The different colors indicate the haplotype of each species. Each circle represents a unique haplotype, the frequency of which is proportional to the diameter of the circle. The number of mutations is represented by numbers in parenthesis.
In the current study, an intriguing facet was to comprehend the genetic variation of secondary symbionts with that of the symbionts in other whitefly species complex. This is anticipated to further assist in the discovery of the population demography of whitefly and the relationship between different sub-populations. The haplotype diversity and nucleotide diversity of Wolbachia in our collected samples were 0.803 and 0.006, respectively (Supplementary Table S10), which further diversified into five different haplotypes (Figure 2D). On the other hand, the neutrality test revealed the haplotype and nucleotide diversity of Rickettsia to be 0.879 and 0.003, respectively, with the presence of six haplotypes (Figure 2E). Subsequently, the presence of six haplotypes was observed among the Arsenophonus sequences: the A2 subgroup of Arsenophonus had only two haplotypes, and the A1 subgroup consisted of four haplotypes (Figure 2F). Interestingly, the network analysis also revealed a considerable overlap among the symbiont haplotypes from different Asian countries, sometimes with a little variation.
The demographic history when analyzed using Fu and Li's D*, Fu and Li's F*, and Tajima's D tests, each returned a positive value for the sequences of Arsenophonus and Rickettsia (interpreted as a sudden population contraction) used in the current study, but since the value was not significant, the null hypothesis regarding the neutrality of the population is accepted. However, Tajima's D test statistic showed a negative but non-significant value for Wolbachia specimens. Overall, the Tajima's D values were negative but not significant in all the symbionts outside India. As a test for a recent population expansion, the mismatch distribution analysis indicated a non-significant multimodal distribution for the genes of three endosymbiotic bacteria. However, the distribution observed was higher than the expected one.
The susceptibility of six whitefly populations has been evaluated against three different insecticides, namely, thiamethoxam, imidacloprid, and acetamiprid (Figure 3). The results (detailed in Supplementary Table S11) indicate that the toxicity (LC50) level varies between 5.36 and 52.14 mg/L for thiamethoxam, 12.87 and 112.09 mg/L for imidacloprid, and 13.49 and 60.04 mg/L for acetamiprid, signifying the varying susceptibility of these populations toward the neonicotinoid group of insecticides. Based on the LC50 value of the susceptible laboratory population, thiamethoxam (LC50 = 5.36 mg/L) was found to be the most toxic, followed by imidacloprid (12.87 mg/L) and acetamiprid (13.49 mg/L). The populations belonging to Midnapore and Kalimpong exhibited a similar toxicity trend against these insecticides (detailed in Supplementary Table S11). On the contrary, populations from Hooghly, Malda, and Purulia followed a slightly different trend, wherein the highest toxicity was observed in the case of thiamethoxam, followed by acetamiprid (LC50 = 60.04, 24.37, and 52.23 mg/L) and imidacloprid (LC50 = 77.79, 44.53, and 112.09 mg/L, respectively) (detailed in Supplementary Table S11).
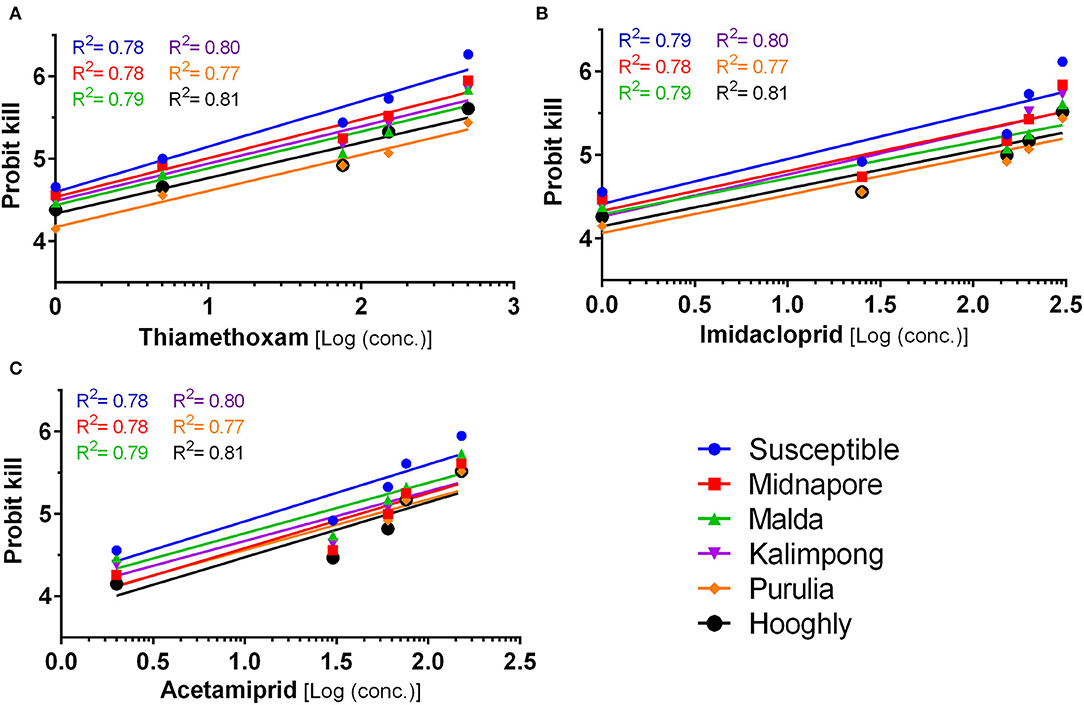
Figure 3. Mortality response of whitefly (Bemisia tabaci) population to insecticides. Increasing concentrations of three insecticides, thiamethoxam (A), imidacloprid (B), and acetamiprid (C) was tested against different populations of whitefly labeled with separately colored lines, as marked in the legend to analyse the mortality response. The X-axis represents the logarithmic dose of every insecticide concentration tested, while the Y-axis represents the probit kill values. The straight line represents the best fitting probit regression line between insecticide log dose and mortality probits (working probits) for adult whitefly. The R2 of each individual line in the graphs represents the regression coefficient. Slopes of the lines are given in Supplementary Table S11.
On comparing the susceptibility of different populations against an insecticide, the data indicated that the resistance ratio (RR value) is in the range of 1.00–9.73, 1.00–8.71, and 1.00–4.45 for thiamethoxam, imidacloprid, and acetamiprid, respectively (Supplementary Table S11). The maximum RR value for thiamethoxam was recorded in the population of Purulia (9.73), where the lowest (1 ppm) and highest (300 ppm) tested concentration resulted in the working probit kill of 4.15 and 5.44, respectively. On the other hand, susceptible (Nadia population) working probit kill was 4.66 and 6.27, respectively, for the same concentrations (Figure 3A). Toxicity comparison with the laboratory population for imidacloprid indicated the highest RR value (8.71) for the Purulia population (Supplementary Table S11). The population of Purulia showed working probit kill values of 4.15 and 5.44 for the lowest (2 ppm) and highest (450 ppm) test concentrations of imidacloprid, respectively, whereas the susceptible population (Nadia) showed working probit kill values of 4.56 and 6.12, respectively (Figure 3B). In the case of acetamiprid, the maximum RR value (4.45) was observed in the Hooghly population (Supplementary Table S11), wherein the lowest (4 ppm) and highest (400 ppm) tested concentrations resulted in a working probit kill of 4.15 and 5.52 (Figure 3C), respectively. Based on the guidelines by World Health Organization (2016) and studies by Mazzarri and Georghiou, 1995 and Yi et al., 2020, the resistance to thiamethoxam and imidacloprid can be considered to exist at a moderate level (i.e., 8.7–9.7), while resistance to acetamiprid was comparatively lower (i.e., 4.45).
Therefore, the bioassay study indicates that the whitefly population from Purulia and Hooghly, consisting of Asia II 7 and Asia II 5 cryptic species populations, respectively, have developed maximum resistance toward the selected neonicotinoids (Supplementary Table S3).
A comparative study involving symbiont titres in different field populations of whitefly revealed varying patterns in their quantitative analysis. The relative titer of P-symbiont (Portiera) differed significantly among the experimental whitefly population. Significantly higher members of Portiera were observed in the population of Hooghly, followed by the population of Malda and Purulia when compared to the susceptible reference population (Nadia) (Figure 4A). The results also revealed varying patterns in titer level of S-endosymbionts, that is, Arsenophonus, Wolbachia, and Rickettsia. A significantly higher abundance of Arsenophonus and Wolbachia was observed in the population of Purulia, followed by the population of Hooghly and Malda when compared to the reference population (Figures 4B,C). Contrary to the other symbionts, the maximum titer of Rickettsia was observed in the population of Midnapore and Kalimpong (Figure 4D).
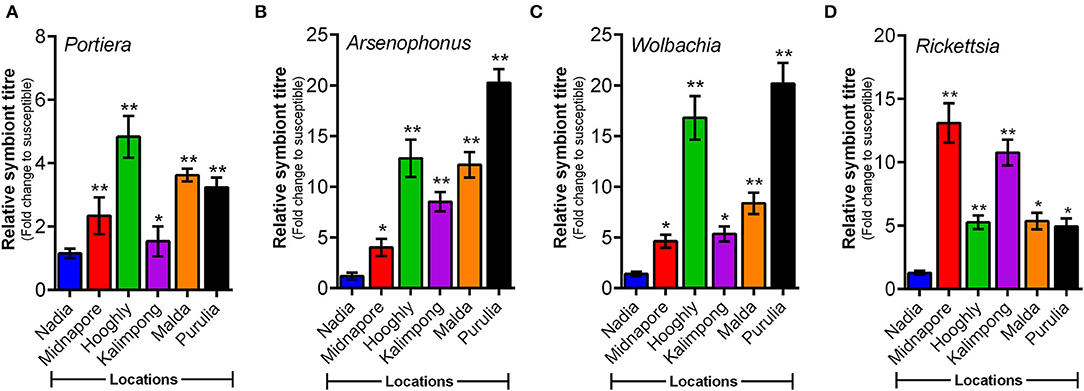
Figure 4. Relative titer of endosymbiotic bacteria (normalized to the whitefly's nuclear β-actin gene) in different whitefly populations as quantified by qPCR. Titer of four endosymbiotic bacteria (Portiera, Arsenophonus, Wolbachia, and Rickettsia) in whitefly populations (as listed along the X-axes). (A) Portiera, (B) Arsenophonus, (C) Wolbachia, and (D) Rickettsia. Values represent mean ± SE values for three independent replicates. Statistical significance of the experimental sets as compared to the susceptible population has been marked with ***p ≤ 0.0001, **p ≤ 0.001, and *p ≤ 0.01.
The correlation coefficient between resistance ratio (RR) values of whitefly field population and relative abundance of symbiont titer is presented in Figures 5A–D. The titer level of Arsenophonus was significantly correlated with RR of imidacloprid and thiamethoxam, with correlation coefficients of 0.88 (p = 0.005) and 0.88 (p = 0.006), respectively (Figure 5B). Moreover, the resistant ratio of all the three neonicotinoids exhibited a positive correlation with the titer level of Wolbachia. The correlation coefficients were 0.97 (p = < 0.001) for imidacloprid, 0.96 (p = <0.001) for thiamethoxam, and 0.65 (p = 0.050) for acetamiprid, respectively (Figure 5C). The titer level of Portiera shows positive but no significant relation with RR among all the treated insecticides.
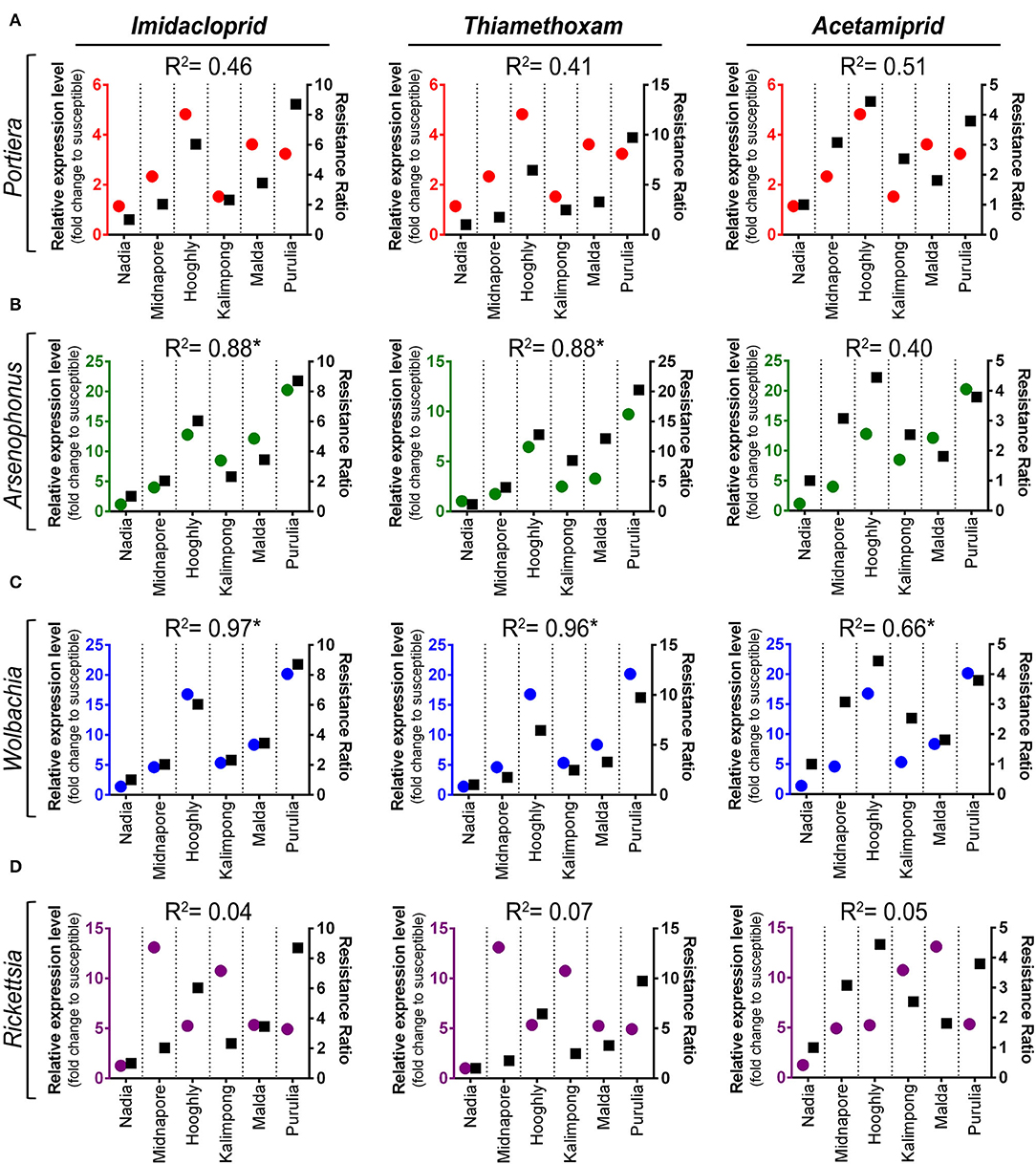
Figure 5. Linear regression analysis between insecticides resistance level and relative symbiont titer of different whitefly populations. Linear regression analysis was used to test for a functional relationship between the resistance level of each insecticide (imidacloprid, thiamethoxam, and acetamiprid) and the mean normalized expression value of each symbiotic bacteria in different whitefly populations: (A) Portiera, (B) Arsenophonus, (C) Wolbachia, and (D) Rickettsia. Circles in each graph represent the relative titer of each symbiont (marked on the left Y-axis), while squares depict the resistance ratio of each insecticide (labeled on the right Y-axis).
The expression of eight cytochrome P450 genes was compared among five whitefly populations against the susceptible reference population (Nadia), wherein upregulation of all the eight genes was noted at varying degrees. In the case of CYP6CX5, Kalimpong, Purulia, and Hooghly populations showed maximum upregulation (4–5-fold), followed by Malda and Midnapore populations that depicted a 3-fold upregulation (Figure 6A). Subsequently, for the CYP6CM1 transcript level, the Purulia population exhibited the highest upregulation (41-fold), followed by Hooghly (23-fold) and Malda (15-fold) (Figure 6B). The relative expression of CYP6DZ4 was low in all the populations, with maximum expression observed in the Malda population (3-fold) (Figure 6C). Next, for the CYP6CX3 transcript level, the Purulia population exhibited the highest upregulation (21-fold), followed by Kalimpong (15-fold) and Midnapore (5-fold) (Figure 6D). Both CYP6CX1 and CYP4C64 transcript levels were maximum for the Hooghly population, with an increase of 60-fold and 17-fold, respectively (Figures 6E,F). Lastly, the relative expression of CYP6DZ7 and CYP6DW2 was moderately high in the population of Purulia and Hooghly (20-fold and 18-fold; 10-fold and 13-fold, respectively) (Figures 6G,H). Overall, the results show that Purulia and Hooghly populations harbored maximally upregulated insecticide resistance P450 genes.
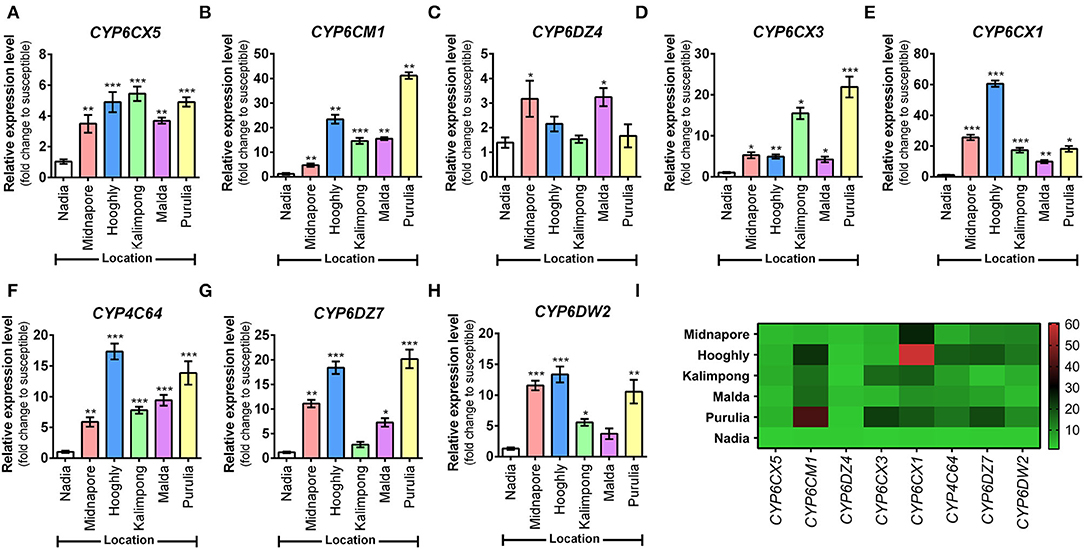
Figure 6. Expression analysis of different cytochrome P450 genes in whitefly (Bemisia tabaci) population. Expression profiles of eight P450 genes governing insecticide resistance in whitefly populations (as listed along the X-axes). (A) CYP6CX5, (B) CYP6CM1, (C) CYP6DZ4, (D) CYP6CX3, (E) CYP6CX1, (F) CYP4C64, (G) CYP6DZ7, and (H) CYP6DW2. Relative gene expression was measured by qRT-PCR analysis. Nuclear β-actin has been used as the internal control. The Ct values for tested genes were normalized to the Ct value of actin and calculated relative to a calibrator using the formula 2−ΔΔCt. Values represent mean ± SE for three independent replicates. Statistical significance of the experimental sets as compared to the susceptible population has been marked with ***p ≤ 0.0001, **p ≤ 0.001, and *p < 0.01. (I) Heatmap represents an overview of the expression level of each gene in a variable whitefly population, with the color gradient provided alongside. Gene expression profiles are represented as a relative expression when compared to the susceptible population, obtained from the qRT-PCR experiment.
The correlation coefficient between RR values of the whitefly field population and the relative expression of detoxifying genes (P450s) is presented in Figures 7A–H. The expression level of CYP6CM1 significantly correlated with the RR of imidacloprid and thiamethoxam. The correlation coefficients were 0.94 (p = 0.012) and 0.95 (p = < 0.001), respectively (Figure 7B). The expression level of both CYP6CX1 and CYP6DW2 genes was positively correlated with the RR of acetamiprid (R2 = 0.72, p = 0.030; R2 = 0.91, p = 0.003) (Figures 7E,H).
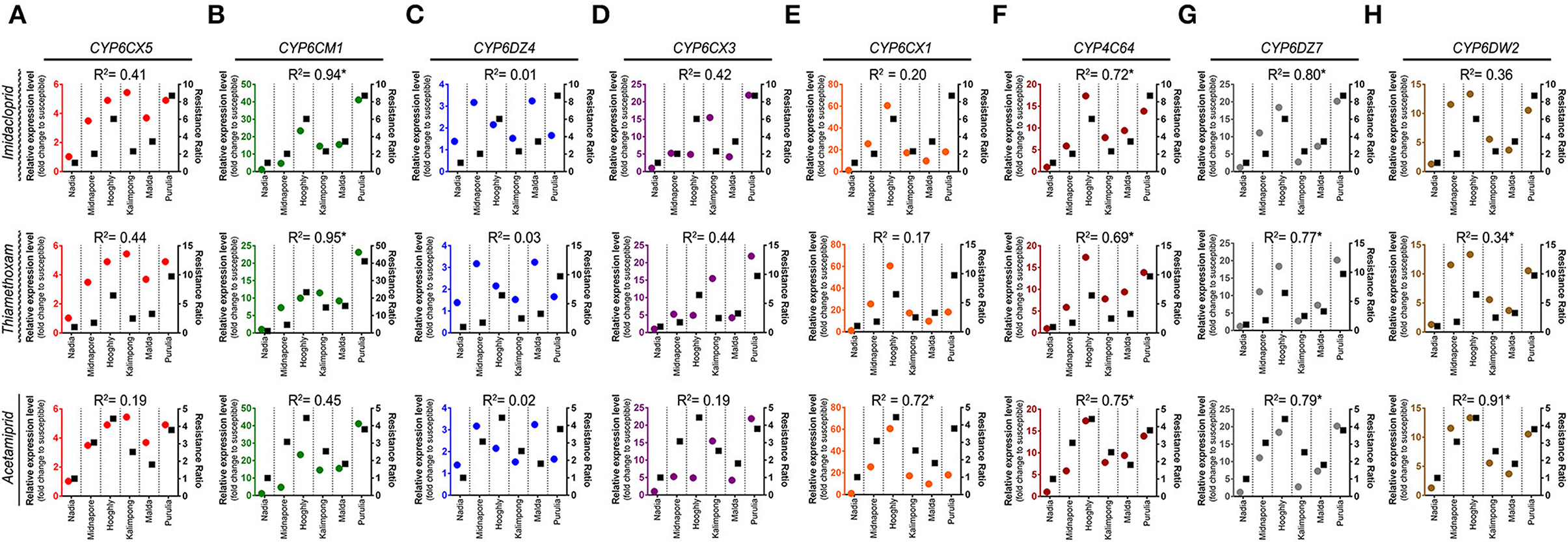
Figure 7. Linear regression analysis between insecticide resistance level and relative expression of insecticide resistance genes (P450s). Linear regression analysis was used to test for a functional relationship between the resistance level of each insecticide (imidacloprid, thiamethoxam, and acetamiprid) and the mean normalized expression value of each P450s gene in different whitefly populations: (A) CYP6CX5, (B) CYP6CM1, (C) CYP6DZ4, (D) CYP6CX3, (E) CYP6CX1, (F) CYP4C64, (G) CYP6DZ7, and (H) CYP6DW2. Circles in each graph represent the gene expression values (marked on the left Y-axis), while squares depict the resistance ratio of each insecticide (labeled on the right Y-axis).
Taking together, the RR of all the tested neonicotinoids exhibited a positive correlation with the transcript levels of CYP4C64 and CYP6DZ7 genes (Figures 7F,G). The correlation coefficients were 0.72 (p = 0.030) and 0.80 (p = 0.015) for imidacloprid, 0.69 (p =0.030) and 0.77 (p = 0.020) for thiamethoxam, and 0.75 (p = 0.020) and 0.79 (p = 0.010) for acetamiprid, respectively.
“Species concept” has been one of the most controversial concepts in biology consistently. This statement holds true for B. tabaci, as it is recognized as a complex of cryptic species that are morphologically indistinguishable from one another but genetically diverse (Firdaus et al., 2013; Hadjistylli et al., 2016). The Indian region exhibits an enormous diversity of B. tabaci with the presence of 10 cryptic species among the 46 species recorded so far (Rehman et al., 2021). The vast array of genetic groups not only specialize in host plant preference, but also exhibit a diverse dispersal pattern and differential response to pesticides (Pashley et al., 1992; Adamczyk et al., 1997; Barman et al., 2022). In this regard, precise assessment of the genetic identity and insecticide resistance properties is imperative for the development of control strategies.
An extensive survey of whitefly across different regions of Bengal province in India revealed that the pair-wise genetic variation reached up to 20.80%, and the species complex was further diversified into four cryptic species, namely, Asia I, Asia II 5, Asia II 7, and China 3. Of all the reported four genetic groups, Asia I was found to be the major group in the collected whitefly samples. This group was previously reported to be prevalent across Asian countries, including Pakistan, Bangladesh, Malaysia, Singapore, Indonesia, and Cambodia (Dinsdale et al., 2010; Götz and Winter, 2016; Kanakala and Ghanim, 2019). The Asia II cryptic whitefly species is a genetically diverse group consisting of 13 cryptic species, namely, Asia II (1–13) (Kanakala and Ghanim, 2019). Among them, the genetic groups of only Asia II 5 and Asia II 7 were detected in the collected whitefly samples spanning across the Bengal province. Previous reports suggest that Asia II 5 was majorly prevalent in the South Asian countries like Bangladesh, Pakistan, and Nepal (Islam et al., 2018; Khatun et al., 2018; Acharya et al., 2020). In the survey conducted, Asia II 5 was found to be majorly distributed along the southern part of Bengal and associated with a single host crop, that is, eggplant. From a geographical point of view, the close proximity of Bangladesh and Bengal provinces of India might have paved a way for the rapid dispersion of this particular genetic group in India. On the other hand, Asia II 7 was specifically identified in one region, Bagmundi, Purulia, indicating that this cryptic species did not undergo rampant dispersion. The distribution of China 3 was also restricted to only the Birbhum region of Bengal, aligning with the reports of Ellango et al. (2015). Despite the extensive survey, this genetic group did not extend its distribution until now. Keeping in mind the rampant distribution of different cryptic species along with the prevalence of suitable hosts, B. tabaci has a high chance of developing adaptive advantage in different regions of the country.
Management of whiteflies is a grave task for farmers and scientists all around the globe, with chemical pesticides being the primary strategy in diverse agricultural systems. After gradual replacement with different groups of insecticides, the neonicotinoids, particularly imidacloprid, are extensively used against B. tabaci (Nauen and Denholm, 2005; Barman et al., 2021). Existing field problems, such as poor selection of insecticides and sub-standard application techniques, result in control failures of insecticides against this pest (Peshin and Zhang, 2014; Wang et al., 2020). Several global studies have highlighted the development of resistance in different whitefly populations to different groups of insecticides (Zhao et al., 2020). However, the occurrence of different cryptic forms makes the task further complex due to differential responses to insecticides. Furthermore, the increasing level of resistance observed in the whitefly population and the rapid evolution of insecticide resistance genes are also a matter of concern. Nonetheless, literature focusing on the susceptibility/resistance status of the whitefly population to insecticides in India is also limited. In this regard, field populations of whitefly surveyed from different geographical locations were tested for their susceptibilities to three commonly used neonicotinoids in these regions. The results revealed that among the three studied insecticides (thiamethoxam, imidacloprid, and acetamiprid), the high susceptibility of the population was recorded toward thiamethoxam. Variation in susceptibility status across different regions can be attributed to the prevalence of different genetic groups. The study data show that the whitefly population from Purulia and Hooghly, comprising Asia II 7 and Asia II 5 cryptic species, recorded higher resistance toward these novel compounds. Several global studies have documented resistance in B. tabaci MED and MEAM 1 genetic groups to different groups of insecticides across the continent (Whalon et al., 2013). However, there is limited literature available on the insecticide status of indigenous B. tabaci genetic groups in Asia. This study clearly provides the insecticide resistance/susceptibility status of Asian genetic groups like Asia I, Asia II 5, and Asia II 7 against the selected neonicotinoid compounds. As insecticide resistance is regarded by some workers as a major driving force for the selection and establishment of specific B. tabaci genetic groups in a region (Horowitz et al., 2005; Barman et al., 2021), there is a need for regular monitoring of insecticide resistance status in diverse B. tabaci genetic groups across India. Results from the present survey indicate the dominance of Asia I groups in most of the regions, with Asia II 5 ranking second on the basis of abundance. What is evident in the coming times is that if Asia II 5 or Asia II 7 becomes dominant in these regions, management using neonicotinoids would eventually become difficult. Therefore, further studies were carried out for unraveling the basis of insecticide resistance in different B. tabaci field populations, which is necessary for the future management of this pest.
Furthermore, endosymbionts also have an important role to play in protecting their hosts from different environmental stressors, including insecticides (Bhatt et al., 2021). This symbiont-mediated insecticide resistance or susceptibility is highly variable and depends on the species of symbiont and the chemical compound (Liu and Guo, 2019). Hence, the work further focussed on bacterial endosymbiont communities which infect different B. tabaci populations across a wide geographical range to gain a detailed understanding of the endosymbiont diversity and their association with insecticide resistance. The profiles of endosymbionts within the different whitefly genetic groups were highly variable. A significant positive association of Wolbachia titer was observed with the resistance level of the three test insecticides (imidacloprid, thiamethoxam, and acetamiprid) in different whitefly field populations. Earlier reports opined that the presence of Wolbachia increased the resistance of whiteflies to neonicotinoids, such as acetamiprid (Ghanim and Kontsedalov, 2009). On the other hand, Arsenophonus also correlated well with the resistance levels of imidacloprid and thiamethoxam. Although the co-existence of Arsenophonus with Rickettsia symbiont was reported to confer resistance to acetamiprid (Kontsedalov et al., 2008), no such significant correlation between Rickettsia titer and insecticide resistance level in whitefly was noted. Such a difference in symbiont performance can be attributed to symbiont species, host genotype, and co-infection with other symbionts (Ghanim and Kontsedalov, 2009; Hoffmann et al., 2015). The network analysis used to compare the genetic variation of secondary symbionts (study samples) with other Asian countries revealed considerable overlap between the haplotypes (Indonesia, Korea, China, Bangladesh, and Pakistan), sometimes with a little variation. These results are clearly suggestive of the fact that the symbionts of whitefly are effectively isolated from other locations and might be in the process of genetic divergence. In contrast, there appear to be significant interactions between the whitefly populations and other distant countries despite the stretches of kilometers between them. The mixing of whitefly populations seems unavoidable due to the migration of nymphs that can occur up to 7 km (Byrne et al., 1996). Hence, this is a source of concern, as it raises the possibility of resistance build up in whiteflies of other countries as well, provided the symbiont dynamics remain the same.
Overexpression and modification of gene encoding members of cytochrome P450, carboxylesterases, and glutathione S-transferases (GSTs) are associated with resistance to different groups of insecticides (Li et al., 2007). In the current study, expression of both CYP6DZ7 and CYP4C64 showed a good correlation with the resistance level of imidacloprid, thiamethoxam, and acetamiprid in different whitefly field populations. According to previous reports, expression of CYP4C64 positively correlated with imidacloprid resistance in whitefly; however, no such correlation was recorded in CYP6DZ7 (Yang et al., 2013). Possibly, the expression of CYP6DZ7 imparts resistance to imidacloprid in some whitefly field populations but not in others. As reported by Zhuang et al. (2011), CYP6CX1 is involved in imidacloprid resistance in both laboratory and field populations of whitefly. Present results revealed a direct correlation of CYP6CX1 with acetamiprid resistance. Based on the current and previous studies, it can be presumed that CYP6CX1 is an important gene in conferring resistance to whitefly against several neonicotinoid compounds. Moreover, similar to earlier reports, CYP6CM1 expression also correlated absolutely well with the resistance level of imidacloprid and thiamethoxam. Therefore, multiple symbiotic interactions with varying gene expression patterns might govern the differential susceptibility of the whitefly population against different test insecticides.
Assembling together all these pieces of information, this research article provides an in-depth notion of the current scenario of different whitefly cryptic species and their susceptibility status toward the selected neonicotinoids. The study further dwells on the infection patterns and role of endosymbionts in imparting insecticide resistance. Studies of symbiont-mediated resistance could provide an interesting prospect to determine biological phenomenona with implications that may extend beyond entomological research and further contribute to a greater understanding of bioremediation, human–microbiome interactions, pharmacology, etc. The use of neonicotinoid group of insecticides is employed as a major strategy for controlling insects like whiteflies; however, the deep-rooted negative environmental effects of neonicotinoids call for serious attention to some alternative management techniques for this pest. With further development, bioaugmentation of detoxifying bacteria could possibly serve to reduce pesticide residuals in the environment, henceforth improving the health of the ecosystem while eliminating the deleterious effects of these pesticides.
Furthermore, an understanding of different P450 genes and their positive correlation with insecticide resistance in different whitefly populations have also been depicted. Nonetheless, the heterogeneity observed in the collected whitefly population indicates toward the differential response it might show toward the management approaches, which is indeed a matter of concern for scientists and farming communities. A few vital questions that arise on our way stem from the basic evolution of symbiont-mediated insecticide resistance and the precise molecular interactions occurring between symbiont and host genes. Possibly through proper clarification of these vital issues, symbiont-targeted pest management will prove to be an effective control component for the future agricultural community. However, detailed basic work needs to be carried out before the large-scale application of symbiont-targeted pest management.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Conceptualization: MB and SS. Methodology: MB and HT. Original draft preparation and writing—reviewing and editing the manuscript: SS and GU. Validation: MB and GU. Resources: AS and JT. Investigation: JT and SC. All authors have read and agreed to the published version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thankfully acknowledge Bidhan Chandra Krishi Viswavidyalaya (ICAR-accredited State Agricultural University) for providing the University Research Scholarship to carry out the research work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.901793/full#supplementary-material
Abbott, W. S. (1925). A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 18, 265–267. doi: 10.1093/jee/18.2.265a
Acharya, R., Shrestha, Y. K., Sharma, S. R., and Lee, K. Y. (2020). Genetic diversity and geographic distribution of Bemisia tabaci species complex in Nepal. J. Asia Pac. Entomol. 23, 509–515. doi: 10.1016/j.aspen.2020.03.014
Adamczyk, J. J., Holloway, J. W., Leonard, B. R., and Graves, J. B. (1997). Susceptibility of fall armyworm collected from different plant hosts to selected insecticides and transgenic Bt cotton. J. Cotton. Sci. 1, 21–28.
Ahmad, M., Arif, M. I., and Naveed, M. (2010). Dynamics of resistance to organophosphate and carbamate insecticides in the cotton whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) from Pakistan. J. Pest. Sci. 83, 409–420. doi: 10.1007/s10340-010-0311-8
Alon, M., Alon, F., Nauen, R., and Morin, S. (2008). Organophosphates' resistance in the B-biotype of Bemisia tabaci (Hemiptera: Aleyrodidae) is associated with a point mutation in an ace1-type acetylcholinesterase and overexpression of carboxylesterase. Insect Biochem. Mol. Biol. 38, 940–949. doi: 10.1016/j.ibmb.2008.07.007
Barman, M., Samanta, S., Chakraborty, S., Samanta, A., and Tarafdar, J. (2022). Copy number variation of two begomovirus acquired and inoculated by different cryptic species of whitefly, Bemisia tabaci in Okra. PLoS ONE 17:e0265991. doi: 10.1371/journal.pone.0265991
Barman, M., Samanta, S., Thakur, H., Chakraborty, S., Samanta, A., Ghosh, A., et al. (2021). Effect of neonicotinoids on bacterial symbionts and insecticide-resistant gene in whitefly, Bemisia tabaci. Insects 12, 742. doi: 10.3390/insects12080742
Basij, M., Talebi, K., Ghadamyari, M., Hosseininaveh, V., and Salami, S. A. (2017). Status of resistance of Bemisia tabaci (Hemiptera: Aleyrodidae) to neonicotinoids in Iran and detoxification by cytochrome P450-dependent monooxygenases. Neotrop. Entomol. 46, 115–124. doi: 10.1007/s13744-016-0437-3
Bhatt, P., Rene, E. R., Huang, Y., Lin, Z., Pang, S., Zhang, W., et al. (2021). Systems biology analysis of pyrethroid biodegradation in bacteria and its effect on the cellular environment of pests and humans. J. Environ. Chem. Eng. 9, 106582. doi: 10.1016/j.jece.2021.106582
Bing, X. L., Yang, J., Zchori-Fen, E., Wang, X. W., and Liu, S. S. (2013). Characterization of a newly discovered symbiont of the whitefly Bemisia tabaci (Hemiptera Aleyrodidae). Appl. Environ. Microbiol. 72, 569–575. doi: 10.1128/AEM.03030-12
Byrne, D. N., Rathman, R. J., Orum, T. V., and Palumbo, J. C. (1996). Localized migration and dispersal by the sweet potato whitefly, Bemisia tabaci. Oecologia 105, 320–328. doi: 10.1007/BF00328734
Chiel, E., Gottlieb, Y., Zchori-Fein, E., Mozes-Daube, N., Katzir, N., Inbar, M., et al. (2007). Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull. Entomol. Res. 97, 407–413. doi: 10.1017/S0007485307005159
Daborn, P., Boundy, S., Yen, J., Pittendrigh, B., and ffrench-Constant, R. (2001). DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genom. 266, 556–563. doi: 10.1007/s004380100531
De Barro, P. J., Liu, S. S., Boykin, L. M., and Dinsdale, A. B. (2011). Bemisia tabaci: a statement of species status. Annu. Rev. Entomol. 56, 1–19. doi: 10.1146/annurev-ento-112408-085504
Dinsdale, A., Cook, L., Riginos, C., Buckley, Y. M., and De Barro, P. (2010). Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 103, 196–208. doi: 10.1603/AN09061
Ellango, R., Singh, S. T., Rana, V. S., Gayatri, P. N., Raina, H., Chaubey, R., et al. (2015). Distribution of Bemisia tabaci genetic groups in India. Environ. Entomol. 44, 1258–1264. doi: 10.1093/ee/nvv062
Elzaki, M. E. A., Miah, M. A., Wu, M., Zhang, H., Pu, J., Jiang, L., et al. (2017). Imidacloprid is degraded by CYP353D1v2, a cytochrome P450 overexpressed in a resistant strain of laodelphax striatellus. Pest. Manag. Sci. 73, 1358–1363. doi: 10.1002/ps.4570
Everett, K. D. E., Thao, M. L., Horn, M., Dyszynski, G. E., and Baumann, P. (2005). Novel chlamydiae in whiteflies and scale insects: endosymbionts ‘Candidatus Fritschea bemisiae' strain Falk and ‘Candidatus Fritschea eriococci' strain Elm. Int. J. Syst. Evol. Microbiol. 55, 1581–1587. doi: 10.1099/ijs.0.63454-0
Excofer, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Feyereisen, R. (2011). “Insect CYP genes and P450 enzymes,” in Insect Molecular Biology and Biochemistry, 1st Edn. ed L. Gilbert (Amsterdam: Elsevier), 115–128. doi: 10.1016/B978-0-12-384747-8.10008-X
Firdaus, S., Vosman, B., Hidayati, N., Jaya, S. E. D. G. F., Visser, R., et al. (2013). The Bemisia tabaci species complex: additions from different parts of the world. Insect Sci. 20, 723–733. doi: 10.1111/1744-7917.12001
Fu, Y. X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Garrood, W. T., Zimmer, C. T., Gorman, K. J., Nauen, R., Bass, C., and Davies, T. G. (2016). Field–evolved resistance to imidacloprid and ethiprole in populations of brown planthopper Nilaparvata lugens collected from across south and east Asia. Pest Manag. Sci. 72, 140–149. doi: 10.1002/ps.3980
Ghanim, M., and Kontsedalov, S. (2009). Susceptibility to insecticides in the Q biotype of Bemisia tabaci is correlated with bacterial symbiont densities. Pest Manag. Sci. 65, 939–942. doi: 10.1002/ps.1795
Ghosh, S., Bouvaine, S., and Maruthi, M. N. (2015). Prevalence and genetic diversity of endosymbiotic bacteria infecting cassava whiteflies in Africa. BMC Microbiol. 15:93. doi: 10.1186/s12866-015-0425-5
Gottlieb, Y., Ghanim, M., Chiel, E., Gerling, D., Portnoy, V., Steinberg, S., et al. (2006). Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72, 3646–3652. doi: 10.1128/AEM.72.5.3646-3652.2006
Götz, M., and Winter, S. (2016). Diversity of Bemisia tabaci in Thailand and Vietnam and indications of species replacement. J. Asia Pac. Entomol. 19, 537–543. doi: 10.1016/j.aspen.2016.04.017
Hadjistylli, M., Roderick, G. K., and Brown, J. K. (2016). Global population structure of a worldwide pest and virus vector: genetic diversity and population history of the Bemisia tabaci sibling species group. PLoS ONE 11:e0165105. doi: 10.1371/journal.pone.0165105
Hoffmann, A. A., Ross, P. A., and Rasic, G. (2015). Wolbachia strains for diseases control: ecological and evolutionary considerations. Evol. Appl. 8, 751–768. doi: 10.1111/eva.12286
Horowitz, A. R., Kontsedalov, S., Khasdan, V., and Ishaaya, I. (2005). Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Arch. Insect Biochem. Physiol. 58, 216–225. doi: 10.1002/arch.20044
Islam, W., Lin, W., Qasim, M., Islam, S. U., Ali, H., Adnan, M., et al. (2018). A nation-wide genetic survey revealed a complex population structure of Bemisia tabaci in Pakistan. Acta Trop. 183, 119–125. doi: 10.1016/j.actatropica.2018.04.015
Jones, D. R. (2003). Plant viruses transmitted by whiteflies. Eur. J. Plant Pathol. 109, 195–219. doi: 10.1023/A:1022846630513
Kanakala, S., and Ghanim, M. (2019). Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE 14:e0213946. doi: 10.1371/journal.pone.0213946
Karunker, I., Benting, J., and Lueke, B. (2008). Over-expression of cytochrome P450 CYP6CM1 is associated with high resistance to imidacloprid in the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Biochem. Mol. Biol. 38, 634–644 doi: 10.1016/j.ibmb.2008.03.008
Khatun, M. F., Jahan, S. H., Lee, S., and Lee, K. Y. (2018). Genetic diversity and geographic distribution of the Bemisia tabaci species complex in Bangladesh. Acta Trop. 187, 28–36. doi: 10.1016/j.actatropica.2018.07.021
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kontsedalov, S., Zchori-Fein, E., Chiel, E., Gottlieb, Y., Inbar, M., and Ghanim, M. (2008). The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag. Sci. 64, 789–792. doi: 10.1002/ps.1595
Kranthi, K. R., Jadhav, D. R., Kranthi, S., Wanjari, R. R., Ali, S. S., and Russell, D. A. (2002). Insecticide resistance in five major insect pests of cotton in India. Crop Prot. 21, 449–460. doi: 10.1016/S0261-2194(01)00131-4
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 6, 1547–1549. doi: 10.1093/molbev/msy096
Lestari, S. M., Hidayat, P., Hidayat, S. H., Shim, J. K., and Lee, K. Y. (2021). Bemisia tabaci in Java, Indonesia: genetic diversity and the relationship with secondary endosymbiotic bacteria. Symbiosis 83, 317–333. doi: 10.1007/s13199-021-00752-w
Li, X. C., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Li, Y., Liu, X., and Guo, H. (2018). Variations in endosymbiont infection between buprofezin-resistant and susceptible strains of Laodelphax striatellus (Fallén). Curr. Microbiol. 75, 709–715. doi: 10.1007/s00284-018-1436-x
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma. Appl. Note. 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, X. D., and Guo, H. F. (2019). Importance of endosymbionts Wolbachia and Rickettsia in insect resistance development. Curr. Opin. Insect. Sci. 33, 84–90. doi: 10.1016/j.cois.2019.05.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lowe, S., Browne, M., Boudjelas, S., and de Pooter, M. (2000). 100 of the World's Worst Invasive Alien Species a Selection from the Global Invasive Species Database. Auckland: The Invasive Species Specialist Group (ISSG).
Luo, C., Jones, C. M., and Devine, G. (2010). Insecticide resistance in Bemisia tabaci biotype Q (Hemiptera: Aleyrodidae) from China. Crop Prot. 29, 429–434. doi: 10.1016/j.cropro.2009.10.001
Ma, D. Y., Gorman, K., and Greg, D. (2007). The biotype and insecticide-resistance status of whiteflies, Bemisia tabaci (Hemiptera: Aleyrodidae), invading cropping systems in Xinjiang uygur autonomous region, northwestern China. Crop Prot. 26, 612–617. doi: 10.1016/j.cropro.2006.04.027
Mazzarri, M. B., and Georghiou, G. P. (1995). Characterization of resistance to organophosphate, carbamate, and pyrethroid insecticides in field populations of Aedes aegypti from Venezuela. J. Am. Mosq. Control Assoc.-Mosquito News 11, 315–322.
Mugerwa, H., Colvin, J., Alicai, T., Omongo, C. A., Kabaalu, R., Visendi, P., et al. (2021). Genetic diversity of whitefly (Bemisia spp.) on crop and uncultivated plants in Uganda: implications for the control of this devastating pest species complex in Africa. J. Pest Sci. 94, 1307–1330. doi: 10.1007/s10340-021-01355-6
Nauen, R., and Denholm, I. (2005). Resistance of insect pests to neonicotinoid insecticides: current status and future prospects. Arch. Insect Biochem. Physiol. 58, 200–215. doi: 10.1002/arch.20043
Nauen, R., Stumpf, N., and Elbert, A. (2002). Toxicological and mechanistic studies on neonicotinoid cross resistance in Q-type Bemisia tabaci (Hemiptera: Aleyrodidae). Pest Manag. Sci. 58, 868–875. doi: 10.1002/ps.557
Naveen, N. C., Chaubey, R., Kumar, D., Rebijith, K. B., Rajagopal, R., Subrahmanyam, B., et al. (2017). Insecticide resistance status in the whitefly, Bemisia tabaci genetic groups Asia-I, Asia-II-1 and Asia-II-7 on the Indian subcontinent. Sci. Rep. 7, 1–15. doi: 10.1038/srep40634
Nirgianaki, A., Banks, G. K., Frohlich, D. R., Veneti, Z., Braig, H. R., Miller, T. A., et al. (2003). Wolbachia infections of the whitefly Bemisia tabaci. Curr. Microbiol. 47, 93–101. doi: 10.1007/s00284-002-3969-1
Pang, S., Lin, Z., Zhang, W., Mishra, S., Bhatt, P., and Chen, S. (2020). Insights into the microbial degradation and biochemical mechanisms of neonicotinoids. Front. Microbiol. 11:868. doi: 10.3389/fmicb.2020.00868
Pashley, D. P., Hammond, A. M., and Hardy, T. N. (1992). Reproductive isolating mechanisms in fall armyworm host strains (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 85, 400–405. doi: 10.1093/aesa/85.4.400
Peshin, R., and Zhang, W. (2014). “Integrated pest management and pesticide use,” in Integrated Pest Management, eds D. Pimentel, and R. Peshin (Dordrecht: Springer), 1–46. doi: 10.1007/978-94-007-7796-5_1
Pietri, J. E., and Liang, D. (2018). The links between insect symbionts and insecticide resistance: causal relationships and physiological tradeoffs. Ann. Entomol. Soc. Am. 111, 92–97. doi: 10.1093/aesa/say009
Prabhaker, N., Castle, S., and Perring, T. M. (2014). Baseline susceptibility of Bemisia tabaci B biotype (Hemiptera: Aleyrodidae) populations from California and Arizona to spirotetramat. J. Econ. Entomol. 107, 773–780. doi: 10.1603/EC13429
Rehman, M., Chakraborty, P., Tanti, B., Mandal, B., and Ghosh, A. (2021). Occurrence of a new cryptic species of Bemisia tabaci (Hemiptera: Aleyrodidae): an updated record of cryptic diversity in India. Phytoparasitica 49, 869–882. doi: 10.1007/s12600-021-00909-9
Sethi, A., and Dilawari, V. K. (2008). Spectrum of insecticide resistance in whitefly from upland cotton in Indian subcontinent. J. Entomol. 5, 138–147. doi: 10.3923/je.2008.138.147
Shadmany, M., Omar, D., and Muhamad, R. (2015). Biotype and insecticide resistance status of Bemisia tabaci populations from Peninsular Malaysia. J. Appl. Entomol. 139, 67–75. doi: 10.1111/jen.12131
Sloan, D. B., and Moran, N. A. (2012). Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 8, 986–989. doi: 10.1098/rsbl.2012.0664
Su, Q., Xie, W., Wang, S. L., Wu, Q. J., Ghanim, M., and Zhang, Y. J. (2014). Location of symbionts in the whitefly Bemisia tabaci affects 498 their densities during host development and environmental stress. PLoS ONE 9:e91802. doi: 10.1371/journal.pone.0091802
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Thao, M. L., and Baumann, P. (2004). Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 70, 3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004
Wang, Q., Wang, M. N., Jia, Z. Z., Ahmat, T., Xie, L. J., and Jiang, W. H. (2020). Resistance to neonicotinoid insecticides and expression changes of eighteen cytochrome P450 genes in field populations of Bemisia tabaci from Xinjiang, China. Entomol. Res. 50, 205–211. doi: 10.1111/1748-5967.12427
Wang, Z. Y., Yan, H. F., and Yang, Y. H. (2010). Biotype and insecticide resistance status of the whitefly Bemisia tabaci from China. Pest Manag. Sci. 66, 1360–1366. doi: 10.1002/ps.2023
Weeks, A. R., Velten, R., and Stouthamer, R. (2003). Incidence of a new sex-ratiodistorting endosymbiotic bacterium among arthropods. Proc. Royal Soc. 270, 1857–1865. doi: 10.1098/rspb.2003.2425
Whalon, M. E., Mota-Sanchez, D., and Hollingworth, R. M. (2013). Gutierrez R. Michigan State University, Arthropod Pesticide Resistance Database. Available online at: http://www.pesticideresistance.com/ (accessed January 5, 2016).
World Health Organization (2016). Monitoring and Managing Insecticide Resistance in Aedes Mosquito Populations: Interim Guidance for Entomologists. Available online at: https://apps.who.int/iris/handle/10665/204588 (accessed January 2020).
Yang, X., Xie, W., Wang, S. L., Wu, Q. J., Pan, H. P., Li, R. M., et al. (2013). Two cytochrome P450 genes are involved in imidacloprid resistance in field populations of the whitefly, Bemisia tabaci, in China, Pestic. Biochem. Physiol. 107, 343–350. doi: 10.1016/j.pestbp.2013.10.002
Yi, T., Lei, L., He, L., Yi, J., Li, L., Dai, L., et al. (2020). Symbiotic fungus affected the asian citrus psyllid (ACP) resistance to imidacloprid and thiamethoxam. Front. Microbiol. 11:522164. doi: 10.3389/fmicb.2020.522164
Zchori-Fein, E., Lahav, T., and Freilich, S. (2014). Variations in the identity and complexity of endosymbiont combinations in whitefly hosts. Front. Microbiol. 5:310. doi: 10.3389/fmicb.2014.00310
Zhao, Y. X., Huang, J. M., Ni, H., Guo, D., Yang, F. X., Wang, X., et al. (2020). Susceptibility of fall armyworm, Spodoptera frugiperda (JE Smmith), to eight insecticides in China, with special reference to lambda-cyhalothrin. Pestic. Biochem. Physiol. 168, e104623. doi: 10.1016/j.pestbp.2020.104623
Keywords: whitefly, cryptic species, endosymbionts, genetic group, P450 monooxygenase, insecticide resistance, neonicotinoid, mtCOI
Citation: Barman M, Samanta S, Upadhyaya G, Thakur H, Chakraborty S, Samanta A and Tarafdar J (2022) Unraveling the Basis of Neonicotinoid Resistance in Whitefly Species Complex: Role of Endosymbiotic Bacteria and Insecticide Resistance Genes. Front. Microbiol. 13:901793. doi: 10.3389/fmicb.2022.901793
Received: 22 March 2022; Accepted: 25 May 2022;
Published: 23 June 2022.
Edited by:
Durgesh K. Jaiswal, Savitribai Phule Pune University, IndiaReviewed by:
Jie Wang, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Barman, Samanta, Upadhyaya, Thakur, Chakraborty, Samanta and Tarafdar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gouranga Upadhyaya, Z291ci5jdWJvdEBnbWFpbC5jb20=; Jayanta Tarafdar, amF5YW50YTk0YmNrdkBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.