
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 13 May 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.899563
This article is part of the Research TopicSynergistic Interaction of Plants and Microbes for Removal of Toxic Elements/Chemicals: Multidisciplinary approaches for a Sustainable EnvironmentView all 9 articles
The growth and stress responses developed by the plant in virtue of the action of PGPR are dictated by the changes in hormone levels and related signaling pathways. Each plant possesses its specific type of microbiota that is shaped by the composition of root exudates and the signal molecules produced by the plant and microbes. Plants convey signals through diverse and complex signaling pathways. The signaling pathways are also controlled by phytohormones wherein they regulate and coordinate various defense responses and developmental stages. On account of improved growth and stress tolerance provided by the PGPR to plants, there exist crosstalk of signaling events between phytohormones and other signaling molecules secreted by the plants and the PGPR. This review discusses some of the important aspects related to the ambiguities of signaling events occurring in plants, allowing the interaction of PGPR with plants and providing stress tolerance to the plant.
Plant growth-promoting rhizobacteria (PGPR) are known to positively stimulate the growth of plants by providing important nutrients and stress tolerance (Chen et al., 2006; Vansuyt et al., 2007; Tang et al., 2020). As biotic stress management, PGPR are known to suppress the growth of plant pathogenic microbes and are known to activate the plant's defense responses against the pathogens (Niu et al., 2011; Prasannakumar et al., 2015). Apart from biotic stresses, PGPR also have a positive role in conferring tolerance to the abiotic stressors like salinity, drought, and heavy metal stress (Gupta et al., 2002; Lim and Kim, 2013; Kim et al., 2014). The growth and stress response developed by the plant in virtue of the action of PGPR is dictated by the changes in hormone levels and related signaling pathways.
Each plant possesses its specific type of microbiota, and the structure of microbiota in the rhizosphere of the plant is shaped by the composition of root exudates secreted by the plant roots and the signal molecules produced by the plant and microbes (Chaparro et al., 2014; Nelson and Sadowsky, 2015; Zhang et al., 2017; Jalmi, 2020). With the help of these signaling molecules, different types of associations are made between plants' roots and microbes.
The plant consists of diverse signaling networks and pathways transmitting various extracellular and intracellular signals. Based on the stimuli or the signaling components, there are various signaling pathways like mitogen-activated protein kinase (MAPK) pathways and calcium-dependent protein kinase (CDPK) pathways working inside the plant (Hamel et al., 2006; Dodd et al., 2010a; Sinha et al., 2011; Bredow and Monaghan, 2019). These signaling pathways are very well-explored for their role in abiotic or biotic stress per se. They are also controlled by phytohormones wherein they regulate and coordinate various defense responses and developmental stages. Of these several signaling components, MAPKs are highly conserved signaling molecules consisting of three-tier components, MAPKKK, MAPKK, and MAPK, transmitting signals by phosphorelay mechanisms (Hamel et al., 2006). MAPKs are activated by specific cues; however, at times crosstalks occur in these pathways but often end up with specific responses (Sözen et al., 2020). MAPKs also are known to interact with other signaling molecules to transmit the stimuli, giving a specific response (Jalmi and Sinha, 2015; Jalmi et al., 2018). MAPKs are involved in the transmission of diverse stresses, both abiotic and biotic, as well as developmental cues (Sinha et al., 2011; Sethi et al., 2014; Jalmi and Sinha, 2016; Singh and Sinha, 2016; Verma et al., 2020). MAPKs play an important role in pathogen defense, wherein studies have been carried out in understanding upstream receptors/sensors and downstream targets of MAPKs (Meng and Zhang, 2013). In this regard, MAPKs such as MPK3, MPK4, MPK6, and MPK7 have been reported in imparting resistance to pathogenic microbes (Sheikh et al., 2013; Jalmi and Sinha, 2016). Under any stress, cells can quickly respond to the environment, which is due to the fluctuation of cytosolic calcium and the presence of Ca2+ sensing proteins like calmodulins and calcium-dependent protein kinases (CDPKs). In addition to MAPKs, CDPKs have also shown an important role in plant growth and development under abiotic stresses by modulating abscisic acid (ABA) signaling and regulating reactive oxygen species (ROS) accumulation (Asano et al., 2012). Calcium signaling also plays a crucial role in defense against plant pathogens wherein the CDPKs transmit the signals related to pathogens regulating the immune response of plants (Sardar et al., 2017; Bredow and Monaghan, 2019). However, the involvement of MAPKs and CDPKs in transmitting signals related to beneficial microbes has not been detailed completely.
Pathogen defense in the plant has been very well-evolved, with a powerful immune system consisting of two tiers of defense: PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is triggered by conserved microbial signatures called pathogen/microbe-associated molecular patterns (PAMP/MAMP), which are perceived by pattern recognition receptor (PRR) at the cell surface, leading to a basal level of the immune response. The second level of immunity, ETI, is developed against effector molecules injected by pathogens inside the plant to counteract the PTI. ETI is also called the R gene-dependent resistance or gene for gene resistance (Jones and Dangl, 2006). Defense responses in the plant are also controlled by plant hormones like salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). These defense responses are systemic acquired resistance (SAR) and induced systemic resistance (ISR). SAR is induced by long-distance signaling by salicylic acid, resulting in the upregulation of defense response genes like pathogenesis-related genes (PR), whereas ISR is induced by jasmonic acid (JA) and ethylene in plants (Glazebrook, 2005; Jones and Dangl, 2006; Fu and Dong, 2013). Many PGPRs are studied to induce SAR by producing SA at the root interface, and some of the rhizobacteria induce SA independent ISR. Pseudomonas spp. and Bacillus spp. induce ISR and SAR in Arabidopsis and many other plants, thus providing resistance to a broad range of plant pathogens (Haas and Défago, 2005). Signaling events and the signaling components involved in plants during interaction with beneficial microbiota lack extensive information. Signaling components participating in symbiotic association with plants have been explored, but signaling phenomena occurring in non-symbiotic association need detailed understanding. Questions also arise about whether the same receptors and signaling components transmit the signals of beneficial microbes to plants as they do for pathogenic microbes. Also, if this is the fact, then how are the responses different for pathogenic and beneficial microbes? This review discusses some of the important aspects related to the crosstalk of signaling events occurring in plants, allowing the interaction of PGPR with plants, thus providing stress tolerance to the plant.
The most important molecular signals regulating plant growth and development are phytohormones. They have shown a profound role in various environmental stress and developmental signaling (Bedini et al., 2018). PGPR are known to produce a wide range of hormones acting as signal molecules in the rhizospheric region, thus allowing the interaction of PGPR with the plant roots and plant development. It has been studied that PGPR including Bacillus amyloliquefacians, Pseudomonas fluorescence, and Bradyrhizobium japonicum significantly produce plant growth hormones such as Indole-3-acetic acid (IAA), Gibberellic acid (GA), zeatin, ET, and ABA (Boiero et al., 2007) (Figure 1).

Figure 1. Signaling compounds produced by PGPR and plants for setting up the beneficial rhizospheric association. The compounds produced by PGPR include hormones (IAA, GA, Zeatin, ABA), ACC deaminase, VOCs (Alkanes, Ketones, Terpenoids, Alcohols, 2-heptanol, 2-endecanone, Pentadecane), cyclopeptides (CDPs), acyl homoserine lactones (AHLs) like 3-oxo-C6HL, 3-oxo-C8HL, which triggers plant signaling, helping in plant growth promotion and stress tolerance. Similarly, plants produce signaling molecules like plant growth hormones (SA, JA, CK, IAA) in response to PGPR, helping in their signaling and stress response. The associated PGPR improves plant growth by providing essential minerals through nitrogen fixation, ion uptake (Fe, Zn, micronutrients), and phosphate solubilization.
Indole-3-acetic acid produced by rhizobacteria is an essential hormone for root nodulation, vascular bundle formation, and cell division and differentiation. The biosynthesis of IAA by rhizobacteria is influenced by environmental factors and genetic factors (Spaepen et al., 2007). The majority of rhizobacteria studied are capable of producing IAA via indole-3-pyruvic acid and indole-3-acetic aldehyde pathway, some of which include: Azorhizobium caulinodans, B. japonicum, Rhizobium japonicum, Rhizobium leguminosarum, Rhizobium meliloti, Rhizobium phaseoli, Rhizobium trifolii, and Sinorhizobium meliloti (Yanni et al., 2001; Naidu et al., 2004; Boiero et al., 2007; Senthilkumar et al., 2009; Chi et al., 2010) (Table 1).
Cytokinins (CKs) are important hormones regulating seed germination, apical dominance, senescence, and interaction of the plant with microbes (Akhtar et al., 2020). Strains of rhizobacteria are also reported to efficiently produce cytokinin hormone, wherein they enhance the growth of shoot, lateral roots, and increase secretion of root exudates, further increasing the beneficial bacteria-plant interaction (García de Salamone et al., 2001; Senthilkumar et al., 2009; Liu et al., 2013; Asari et al., 2017) (Table 1).
Gibberellic acid is another phytohormone reported to be produced by PGPR which is responsible for leaf expansion and stem elongation. PGPR including Azospirillum spp., Azotobacter spp., Bacillus spp., and rhizobia produce GA (Dodd et al., 2010b). GA3 production by rhizobacteria B. japonicum was first reported by Katznelson and Cole (1965) and later reported by Rhizobium species and S. meliloti (Arshad and Frankenberger, 1997; Boiero et al., 2007). GA producing bacteria exhibit beneficial effects on plants promoting root and shoot growth and also improving seedling vigor (Yanni et al., 2001). Apart from GA3, Rhizobium and B. japonicum also produce a significant amount of ABA, which is an important hormone in providing tolerance to plants under drought stress (Boiero et al., 2007). Another study reports an increase in the level of ABA in plants when inoculated with Azospirillum spp. (Cohen et al., 2008) (Table 1).
The negative effects of ET hormone, i.e., inhibition of root elongation, transport of auxins, and leaf senescence were nullified by PGPR by producing enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase controlling excessive ethylene production (Figure 1). Bacteria producing ACC deaminase are known to take up ACC produced by plants from the rhizosphere and convert it into ketobutyrate and NH3. The strains studied to be producers of ACC deaminase are R. leguminosarum, R japonicum, Rhizobium gallicum, B. japonicum, Bradyrhizobium elkanii, S. meliloti, and Variovorax sp. (Duan et al., 2009; Onofre-Lemus et al., 2009; Gupta and Pandey, 2019; Bessadok et al., 2020) (Table 1). Bacteria producing ACC deaminase enhanced root and shoot elongation, increased nodulation in legumes, and increased mineral uptake. This explains the fact that PGPR protects the plant from environmental stresses by lowering the levels of phytohormone ethylene. Other than the phytohormones, PGPR also produces volatile organic compounds (VOCs) which are used for the intra-bacterial communication. These VOCs include alkanes, ketones, terpenoids, alcohols, sulfur compounds, and so on, of which 2-heptanol, 2-endecanone, and pentadecane are studied to act as signals for communication with plants (Zhang et al., 2007; Farag et al., 2013; Pérez-Flores et al., 2017; Ayaz et al., 2021) (Figure 1). These aromatic lipophilic compounds act as signals to cognate receptors for cell-cell communication and are also used as signals for communication with plants (Table 1). Upon sensing these VOCs by the plant root system, it modulates the root architecture showing an ecological significance for strengthening of mutual interaction between plant roots and PGPR. A study in Arabidopsis reported that VOCs, such as aldehyde, 1-butanol, and ketones, secreted by Bacillus spp. caused modulation of root architecture and increased root biomass (Pérez-Flores et al., 2017). Apart from this, VOCs such as acetoin and 2,3-butanediol produced by Bacillus spp. had a stimulatory effect on plant growth. This stimulatory effect by VOCs could be due to the activation of hormonal signaling (Farag et al., 2013; Pérez-Flores et al., 2017). Yet another VOC, Butyrolactone, acts as an inducer of quorum sensing in bacteria, thereby stimulating more bacterial communications and plant root interactions (Polkade et al., 2016).
Besides signaling molecules produced by PGPR, the plant secretes low molecular weight, high molecular weight, volatile, and non-volatile compounds in the root exudates, which influences the interaction of various microbes in the rhizosphere. Studies have shown that signaling molecules such as coumarins, triterpenes, flavonoids, and benzoxazinoids allow the growth and inhibition of specific microbes in rhizospheric regions of plants (Hu et al., 2018; Cotton et al., 2019).
The roots of the majority of land plants produce a group of carotenoid-derived novel metabolites playing a role as phytohormones called Strigolactones (SLs). The SLs are involved in many of the important aspects of plant development earlier defined as ex-planta signaling molecules secreted by plants' roots inducing germination of parasitic plants. Later, they were found to help in establishing a symbiotic relationship between plants and arbuscular mycorrhizal fungi (Mori et al., 2016). The reports also suggest the role of SLs in regulating the positive interaction of S. meliloti with Medicago sativa (Soto et al., 2010).
It is now clear that the multitude of chemical compounds secreted by the PGPR and plant act as signaling molecules in influencing the rhizospheric associations and growth of the plant. With the help of metabolites produced, plants can influence microbiota and in turn microbiota influence the metabolites produced by the plants (Pang et al., 2021). Many studies have shown the positive effect of a secreted chemical compound on plant growth. Some studies have identified the PGPR organism and the related transcriptional activity without identifying secreted chemical compounds. The improvement in plant growth under environmental stress conditions imparted by PGPR is done by modulating intricate signaling and transcriptional activities, resulting in differential expression of genes. Studies show that PGPR ameliorate the stress conditions by modulating the expression of genes involved in hormonal biosynthesis, such as ACO and ACS genes (ethylene biosynthesis), MYC2 (Jasmonate), PR1 (SA), several genes encoding antioxidant enzymes (SOD, CAT, APX, GST), transcription factor NAC1, and so on (Tiwari et al., 2017). These studies need a correlation in order to understand the signaling networks and the mechanisms involved. Studies are required to identify the developmental signaling pathway in plants induced by signaling molecules secreted by the PGPR. This possibility is shown by a study of versatile signaling molecule cyclodipeptides (CDPs) involved in quorum sensing and PGPR interaction. One of the PGPR, P. aeruginosa, secreted CDPs that affected lateral root development in plants by acting as auxin-like signal and activating the auxin signaling pathway (Ortiz-Castro and López-Bucio, 2019) (Table 1).
Lipo-chitooligosaccharide (LCO) also called Nod factors are signaling molecules, originally studied to be produced by Rhizobia, arbuscular, and ecto mycorrhizal fungi, helping in the nodulation process and establishing a symbiotic relationship with plants (Oldroyd and Downie, 2008; Maillet et al., 2011). Many other fungi belonging to Ascomycetes and Basidiomycetes are studied to be the producers of LCOs, wherein they act as signals regulating fungal growth and development (Rush et al., 2020). Apart from their important role in plant symbiotic association, they also have a direct impact on plant growth and development. They have been shown to increase the growth of the plant in stressed conditions and stimulate the lateral root formation (Zipfel and Oldroyd, 2017; Buendia et al., 2018). Studies show that the lateral root formation by the LCO is through the regulation of auxin homeostasis (Buendia et al., 2018) (Table 1). This opens an interesting avenue of the effects of microbial signaling molecules on hormone signaling in plants.
Lipo-chitooligosaccharides produced by beneficial microbes are recognized by plant receptors, lysin motif receptor-like kinase (LysM-RLKs), containing oligosaccharide-binding LysM domain. It is presumed that binding of LCOs with LysM receptors activates the signaling pathway, leading to the oscillation of nuclear calcium concentration, leading to the formation of nodules, and promoting the symbiotic association. The symbiosis established by Rhizobia with leguminous plants is with the help of recognition of LCOs/Nod factors (NFs) by Nod factor receptors (NFRs) of leguminous plants. Two NFR receptors, NFR1 and NFR5, bind Nod factors directly (Oldroyd, 2013). A study reports that LYK10, LysM receptor-like kinases, show a high affinity for LCOs, and their interaction is essential for root nodule symbiosis (Girardin et al., 2019). Several LysM-RLKs have been identified in many plants like OsCERK1 in rice, SlLYK10 and SlLYK12 in tomato, and MtLYK9 in Medicago truncatula. OsCERK1 is known to perceive various other ligands like such as chitins, peptidoglycans, and chitiologosaccarides (COs), which shows the dual functions, i.e., in symbiosis as well as in plant defense (Ao et al., 2014). Here, the questions on how these interactions lead to two different responses, i.e., establishing a symbiotic association and initiating plant defense against pathogens, or of does pathogen utilize the same signaling pathways in establishing their own colonization arise. This fact has been explained very well in a study by Feng et al. (2019), wherein they suggest that AM fungi promote symbiotic signaling through a combination of LCOs and COs. Different CO molecules show different responses, and short-chain COs like CO4 and CO5 activate symbiotic signaling, whereas long-chain COs like CO8 induce immune signaling (Cao et al., 2017). However, Feng et al. (2019) suggested that there exists no difference in the perception and signaling of COs from CO4 to CO8. LCOs together with COs play a role in symbiosis and can suppress immunity signaling, thus promoting symbiotic outcomes. While the perception of only COs is required for immune responses, an important receptor kinase DMI2 plays an important role in the activation of symbiotic signaling in response to LCOs and COs but shows no role in plant immune signaling (Feng et al., 2019). LCO and CO perceived by DMI2 receptor kinase promote a symbiotic association possibly by creating nuclear-associated calcium oscillations. Perception of COs by CERK1 and LYK receptors activates plant defense by activating calcium influx across the plasma membrane, producing ROS, further activating MAPK signaling (Cao et al., 2017) (Figure 2). LCOs are also known to block the formation of ROS in response to pathogens without affecting plants' immunity (Rey et al., 2019). Accounting for the role of LCOs produced by both Rhizobia and AM fungi in symbiosis, the activation of common symbiotic genes and corresponding pathways by LCOs of both these microbes has been suggested by a previous study. However, it raises the question on specificity in activating these two pathways from different microbial symbionts. A study performed in the identification of specific components reported specific function of two important genes RAM1 (reduced arbuscular mycorrhization) and RAM2 in AM fungal symbiosis. RAM1 encodes the GRAS-transcription factor which regulates the expression of RAM2 encoding Glycerol-3-phosphate Acyl transferase (GPAT). Both RAM1 and RAM2 are specifically required for AM symbiosis and not in rhizobial symbiosis, of which RAM2 is very critical for induction of hyphopodia in AM fungi and also appressoria formation in Phytophtora infection (Gobbato et al., 2012, 2013) (Figure 2).
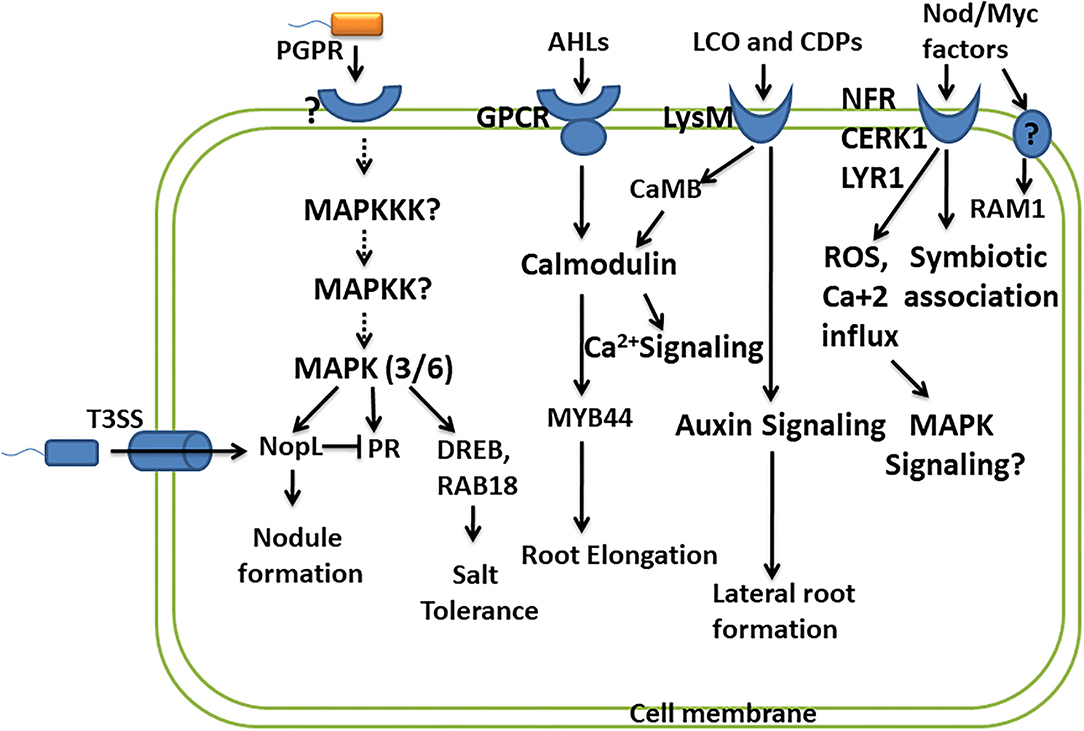
Figure 2. Convergence of PGPR and plant signaling pathways. This depicts the studies carried out in understanding the signaling molecules/pathways involved in perceiving PGPR and their response in promoting plant growth and development. The important signaling pathways involved are MAPK signaling, calcium signaling, and hormone signaling. However, many other components of these pathways and their role in response to PGPR remain to be studied.
Like pathogenic bacteria which use type 3 secretion system (T3SS) to deliver virulence factors (T3 effector proteins), certain rhizobial strains also produce effector protein called nodulation outer protein L (NopL), affecting the nodule formation in symbiosis. NopL bypass the NFR for the formation of the nodule in the legume. NopL affects the process of nodule formation by interacting with plant signaling pathways. A study was performed to show that phosphorylation of NopL was inhibited by Ser/Thr kinase inhibitor and MAPKK inhibitor, suggesting NopL to be a substrate of MAPK (Bartsev et al., 2003; Ge et al., 2016) (Figure 2). NopL suppresses premature nodule senescence by interfering with MAPK signaling (Zhang et al., 2011). NopL also shows indirect interaction with the MAPK pathway by blocking the transcription of PR proteins like glucanase and chitinase, both of which are regulated by the MAPK pathway (Bartsev et al., 2004; Ge et al., 2016). These studies show the importance of the MAPK pathway as a convergent point involved in transmitting the signals for establishing symbiosis (Figure 2). Another study revealed that Bel 2-5 effectors produced by Rhizobia hijack the legume nodulation signaling by Nod factors and NFRs. Bel 2-5 resembles the XopD effector produced by plant pathogen Xanthomonas campestris. The induction of nodulation by Bel 2-5 effector is through the expression of cytokinin-related genes involved in nodule organogenesis and repressing ethylene and defense-related genes (Ratu et al., 2021).
As described earlier, LCOs influence the plant growth promotional activities by altering the plant hormone homeostasis, leading to improved photosynthesis and enhanced resistance to the environmental stresses. For LCOs to exert their effect, plants perceive these signaling molecules through the plant receptors. These receptors are LysM and belong to the lysine motif containing receptor-like kinase family (Liang et al., 2014). They are present in all microbial domains except Archea and are studied to interact with and respond to microbial-associated molecular pattern (MAMPs) (Gust et al., 2012). Intense studies determining the signal transduction by the perception of LCOs by LysM receptors are missing. However, a transcriptome analysis in response to LCO in plants suggested a strong induction of genes encoding calmodulin-binding protein CaMB (Zeng et al., 2015) (Fig. 2). Calmodulin binding proteins are major calcium sensors and regulate broad spectrum of target proteins. Induction of CaMB by LCO shows its possible involvement in calcium signaling in plants. Signaling of AHLs like 3-oxo-C6-HL and 3-oxo-C8-HL is mediated through G-protein coupled receptors in plants, wherein they exert unknown effect on the activity of calmodulin and transcription factor MYB44 involved in root elongation response (Zhao et al., 2015, 2016) (Figure 2). Another R2R3 MYB-like transcription factor MYB72 exhibits its role in early signaling of rhizobacteria-mediated jasmonate/ethylene-dependent ISR (Van der Ent et al., 2008) (Figure 2).
A few studies have reported the interlinking of signaling components in stress tolerance mediated by PGPR. A very important CBL-interacting protein kinase CIPK24 involved in calcium signaling is required for the proper functioning of K+ transporter by phosphorylating SOS1 (Na+/H+ antiporter) (Halfter et al., 2000). Also, another CBL-interacting protein kinase, CIPK12, imparts tolerance to salt stress in Arabidopsis. This suggests the involvement of calcium signaling components in the regulation of ion transporters and salt tolerance. As PGPR provides salt tolerance by various mechanisms by controlling ion uptake and homeostasis, this could be due to the effect of PGPR on such signaling pathways which are known to be involved in providing salt tolerance.
The stress tolerance is ameliorated by PGPR by modulating several genes related to hormonal biosynthesis and defense response, which propose an interlink between PGPR and signaling pathways. Enterobacter species ameliorated the salt stress by inducing salt stress-responsive genes such as DRE-binding protein (DREB), late embryogenesis abundant (RAB18), and also the important players of MAPK signaling cascade MPK3 and MPK6, thus increasing the ROS scavenging activity (Ilangumaran and Smith, 2017) (Figure 2). These reports suggest connecting bridges between rhizobacteria perception by signaling pathways and its subsequent transmission for improving the health of the plants in environmental stresses. However, information is still required to fill the necessary gaps.
Plants being sessile are exposed to a wide range of environmental stresses generally grouped into abiotic and biotic stresses. Work is being carried out to study different mechanisms of stress tolerance. Plant breeders use the long, capital-intensive breeding method to obtain tolerant plant varieties. However, beneficial PGPR plays an important role in inducing stress tolerance, thus giving importance to stress management. PGPR are known to produce an array of compounds that act as signaling molecules activating and altering several cellular processes, thus conferring stress tolerance to plants. Here, with the help of some reports, we will discuss how stress tolerance is obtained in PGPR-plant interaction.
One of the most important PGPR, Pseudomonas putida strain KT 2440, has agronomical importance and has been studied to impart a wide range of tolerance to abiotic as well as toward certain pathogens. Being salt-tolerant bacteria, it improves plant growth and seed germination under saline conditions. The salinity tolerance exhibited by P. putida KT2440 is due to the biosynthesis of lipopolysaccharides (LPS) by the enzyme phosphoethanolamine-lipid-A transferase (EptA) (Costa-Gutierrez et al., 2020). PGPR also provide salt stress tolerance by producing exopolysaccharide (EPS) that covers the root surface and prevents the influx of Na+ ions into the root cells. A study reported two PGPR species, Aeromonas hydrophilla and Bacillus sp., that colonies the wheat roots and produces EPS, thus trapping Na+ and making it unavailable for the plant (Ashraf et al., 2004) (Table 2). However, the biosynthesis of EPS is regulated by the regulation of the gene encoding enzyme EptA. The eptA gene is induced by two components signaling involving PmrA and PmrB, with them being the major regulators. In this regulatory signaling component, PmrA is a cytoplasmic response regulator and PmrB is a membrane-bound sensor kinase (Chen and Groisman, 2013). PmrB kinase is studied to be activated by various signals like high metal ions, low pH, and vanadate, which phosphorylates PmrA and causes the activation of the eptA gene, further causing EPS production (Gunn, 2008). In response to various environmental stresses, the bacteria synthesizes EPS, which gives protection toward stresses. The major regulatory components, PmrA/PmrB and EptA enzymes, could be under the regulation of various other signaling molecules that could be explored in depth.
Besides producing EPS and protecting plant roots against adverse effects of stresses, PGPR also use other mechanisms for providing tolerance. In plants, high-affinity K+ transporter (HKT) controls the Na+ import and overexpression of this transporter fails to impart salt tolerance, making plants more susceptible (Zhang et al., 2008). It is studied that PGPR Bacillus subtilis GB03 inoculation downregulates the HKT expression, thereby reducing the Na+ uptake and providing salt tolerance to plants (Zhang et al., 2008). More specifically, it is reported that Bacillus subtilis GB03 downregulates HKT2 but upregulates HKT1 and SOS1, thereby imparting tolerance (Niu et al., 2016). Also, further restricting the uptake of Na+ by the roots causes induction of HKT, causing transport of Na+ from the leaves to the roots, thereby reducing the salt stress effects (Qin and Huang, 2020). Apart from restricting the uptake of Na+, some PGPR-like Azotobacter strains enhance the uptake of K+ that leads to the accumulation of proline, polyphenols, and so on (Rojas-Tapias et al., 2012) (Table 2). To understand this mechanism of salt tolerance in the presence of PGPR, it will be important to understand the signaling components regulating the HKT and other important transporters. A MAPK signaling cascade studied to provide salt tolerance by regulating proline levels includes MKK3-MPK6-MYC2, which points out one possible signaling pathway that could regulate this process (Verma et al., 2020).
Yet another mechanism for salt tolerance is the production of ACC deaminase. Rhizospheric bacteria, Variovorax paradoxus 5C-2, has been studied to produce enzyme 1-aminocyclopropane-1-carboxylate deaminase, which promotes the growth of plants by lowering the ABA levels in plants, limiting Na+ accumulation under salt stress. This effect was studied in the Pisum sativum plant and was studied to have increased photosynthesis and biomass under salt stress when treated with V. paradoxus (Wang et al., 2016). It has been studied that the ACC deaminase produced by PGPR lowers the level of ABA, whereas other growth hormones produced by PGPR promote plant growth and impart salt tolerance by regulating the production of secondary metabolites (Kang et al., 2019). Another ACC deaminase producing rhizobacteria of Enterobacter species enhances salt tolerance by increasing the activity of antioxidant enzymes, such as SOD, APX, CAT, and upregulation of ROS pathway genes (Habib et al., 2016) (Table 2). ROS are one of the important messengers activating many cellular signaling pathways such as MAPK signaling pathways, involved in combating environmental stresses (Jalmi and Sinha, 2015). This could be yet another point of convergence of bacterial and plant signaling which needs further exploration.
Hormones play a very important role in combating stress. Several hormones are already reported to have a major role in stress tolerance (Ryu and Cho, 2015). PGPR are very well-discussed in hormone production. Production of IAA, cytokinin, gibberellins, and ABA by PGPR was found to impart salt tolerance in many plant species (Dodd et al., 2010b). According to the reports, around 80% of rhizobacterial isolates possess the ability to produce IAA as one of the secondary metabolites. The IAA secreted by bacteria acts as a signaling molecule in plants and alters many of the plant developmental processes, including the endogenous plant IAA levels (Spaepen et al., 2007). Cellular auxin homeostasis becomes necessary to have proper morphogenesis and stress responses because of overproduction or uptake of auxin developmental defects and impaired stress responses in plants. In this process, many of the plant signalings like MAPK and calcium signaling are involved in maintaining the auxin levels. In turn, auxin mediates the induction of these signaling pathways, and they further alter many cellular processes during the responses to a wide range of environmental stresses (Mockaitis and Howell, 2000; Vanneste and Friml, 2013). A recent study suggests the role of a halotolerant bacteria, Bacillus spizizenii FMH45, in the alleviation of salt stress in tomato plants. This bacterium is studied to be a potential producer of IAA, siderophores, and hydrolytic enzymes and is capable of phosphate solubilization, thus improving chlorophyll content, membrane integrity, and phenol peroxidase levels in imparting salt tolerance to plant (Masmoudi et al., 2021).
Apart from IAA production, rhizobacteria are also known to be producers of other stress hormones. The strain of P. putida (H-2-3) was reported to improve salt and drought tolerance by reprogramming the biosynthesis of stress hormones, such as salicylic acid and abscisic acid, chlorophyll content, and expression of antioxidants (Kang et al., 2014). These ABA and SA signaling pathways are identified as central regulators of abiotic and biotic stresses, wherein they induce various other plant signaling pathways like MAPK and calcium signaling and alter the gene expression of many stress response genes (Danquah et al., 2014; Edel and Kudla, 2016).
A growing body of studies of different strains of P. putida reveals its role in imparting tolerance to abiotic stresses other than salinity. A strain MTCC5279 mitigated drought stress in Cicer arietinum by modulating osmolyte accumulation, ROS scavenging ability, and membrane integrity. Also, modulation of gene expression involved in hormone biosynthesis and antioxidant enzyme suggested the involvement of stress hormone and antioxidant enzyme in giving stress tolerance (Tiwari et al., 2016) (Table 2). Apart from salinity and drought tolerance, PGPR also provide tolerance to submergence stress. A study demonstrated that Pseudomonas fluorescence REN1, when inoculated on rice seedlings, improved the growth in flooded conditions by increasing root elongation, possibly by producing ACC deaminase (Etesami et al., 2014). PGPR are also equally known to provide tolerance to heat and cold stress. Gibberellin-producing PGPR Serratia nematodiphila helped in better growth of the plant under low temperatures. This effect was due to the production of hormones GA and ABA and the lowering of the levels of SA and JA (Asaf et al., 2017). Similarly, Burkholderia phytofirmans PsJN and Pseudomonas species provided cold stress tolerance by modulating carbohydrate metabolism and increasing the expression of cold acclimation genes and antioxidant activity (Fernandez et al., 2012; Subramanian et al., 2015) (Table 2).
One of the mechanisms shown by PGPR in promoting plant growth is through suppressing plant diseases; this can occur through microbial antagonism or by inducing resistance in plants against pathogenic microbes. Resistance conferred is due to the priming of effective resistant mechanisms possibly by ISR and SAR, both of which represent basal resistance depending on signaling hormones jasmonic acid and salicylic acid (Van Loon et al., 1998). The phenomenon of induction of ISR by PGPR was first reported by Van Peer et al. (1991) and Wei et al. (1991), wherein some of the rhizobacterial strains provided resistance against pathogens Fusarium oxysporum and Colletotrichum orbiculare. The mechanisms of resistance shown by the PGPR against plant pathogens are the production of hydrolytic enzymes, siderophores, antibiotics, and regulation of plant ethylene levels (Glick and Bashan, 1997; Neeraja et al., 2010). The most important PGPR studied to induce ISR belong to Pseudomonas, Serratia, and Bacillusspp (Van Wees et al., 2008).
It has been difficult to point out and differentiate the ISR induced by beneficial microbes and pathogenic microbes since ISR can be induced by several beneficial and pathogenic microorganisms and also other stresses activating the same type of response. Over past decades, several bacterial components have been identified to trigger ISR, which include lipopolysaccharides in cell envelop, flagella, AHLs, exopolysaccarides, secreted metabolites, and quorum sensing molecules (Ryu et al., 2004; Ortmann et al., 2006). Defense mechanisms shown by the plant in response to non-pathogenic microbes are callose deposition, production of phytoalexin, and pathogenic-related proteins (PR proteins), similar to responses shown by pathogenic infection (Hammond-Kosack and Jones, 1996; Raj et al., 2012). Cell wall modification through lignin deposition is one of the mechanisms of defense response shown by PGPR. This response was observed in Bacillus pumilus, B. subtilis, Pseudomonas fluorescence, and P. putida (Anderson and Guerra, 1985; García-Gutiérrez et al., 2013). Bacillus amyloliquefaciens (SN13) has been reported to act as a biocontrol agent apart from its beneficial role in imparting salt tolerance. This bacterium exerts its effect by modulating phytohormone signaling, producing secondary metabolites, osmoprotectants, and scavenging ROS (Chen et al., 2016; Srivastava et al., 2016; Tiwari et al., 2017) (Table 2). Some isolates of Bacillus spp. improved plant growth and provided resistance by inducing ISR against PepGMV, wherein there was improved photosynthetic performance, photochemical parameters, and gas exchange (Samaniego-Gámez et al., 2021) (Table 2).
Induced systemic resistance mediated by PGPR resembles the SAR induced by the pathogen, in that, in both responses, plants get more resistant to pathogenic microbes and one strain induces broad-spectrum resistance toward several pathogens in the same plant. In SAR, the SA produced activates specific sets of defense-related PR proteins that enhance the defensive capacity of the plant, which is normally not observed in ISR induced by the pathogen (Van Loon, 2007). Non-pathogenic microbes are generally studied to induce ISR consisting of jasmonate, and ethylene response and pathogenic microbes are known to induce SAR response (Van Loon et al., 1998). However, a study by Maurhofer et al. (1994) suggested the accumulation of SA-induced PR protein upon inoculation with PGPR P. fluorescence CHA0 (Table 2). The ISR by non-pathogenic microbes is dependent on NPR1 protein like that of pathogen-induced SAR and unlike that of ISR (Ryu et al., 2004). NPR1 along with the transcription factor TGA is a master regulator protein required for the signal transmission of SA in SAR (Shah et al., 1997). The NPR1 and TGA regulators are known to be regulated by members of the MAPK signaling pathway (Ekengren et al., 2003). The role of MAPK signaling and calcium signaling in plants is very well-explored in activating the SA- and JA-mediated SAR and ISR resistance (Tena et al., 2011). Many of the MAPK members have been studied to be activators of PR genes leading to the development of induced disease resistance (Meng and Zhang, 2013). More and more signaling pathways working in PGPR mediated ISR needs to be explored further. Also, the specificity in receptors and signaling molecules that differentiates pathogenic microbes and PGPR responses remains an unrevealed area.
These reports suggest that signaling pathways that are activated by the plant in response to pathogenic microbes and PGPR overlap with each other and indicate that they are regulated and balanced. The signaling pathways are controlled by hormones, and they do show interactions with each other. Over past years, several regulatory molecules have been identified in the interplay between pathways leading to SAR and ISR; however, their exact role remains to be elucidated.
Surveying the eco-friendly plant growth-promoting activities of PGPR, these bacteria can be exploited as efficient biotechnological tools in improving plant growth and crop yield in a stressful environment Qin et al., 2016. These bacteria are acting as promising fertilizers in agriculture and even as potential cleaners of the toxic environment by the process of remediation. The use of biocontrol PGPR replacing chemical pesticides is considered to be a better option for sustainable crop production (Bhardwaj et al., 2014). Some of the techniques used for sustainable agriculture are the use of these microbes or genetically engineered microbes as biofertilizers to enhance plant growth. PGPR-based biofertilizers are much safer for human health and the environment and are more readily degraded in soil. Biofertilizers with a single strain of PGPR or a consortium of multiple strains have been successfully used on various crop plants against various plant pathogens in multiple modes of application. For the commercialization of PGPR, it has to be passed from different stages, starting from its production in the laboratory to the farmer. Factors to take into consideration for the selection of strain or consortia and its commercialization are the type of crop, ecological zones, climatic, and soil conditions. All these factors affect the performance and effectiveness of the PGPR (Compant et al., 2010). Due to the variability and inconsistency observed in the performance of PGPR in the laboratory and in the field, efforts have been made to overcome this drawback by use of biotechnological techniques such as micro-encapsulation and nano-encapsulation. The stages involved in commercializing the PGPR strain as a biofertilizer includesL: field survey, isolation and characterization of PGPR based on soil properties and type of crop, method of inoculation and the carrier material to be used along, and extensive pot trials and field trials (Tabassum et al., 2017). The promising strains of PGPR used commercially as biofertilizers are the strains of Pseudomonas, Paenibacillus, Bacillus, Brevibacillus, Achromobacter, Trichoderma, and Enterobacter for various crop species like rice, soybean, pearl millet, peas, tomato, peanut, cotton, potato, and so on (Tabassum et al., 2017).
Plant growth-promoting rhizobacteria are known to show various direct and indirect mechanisms of enhancing plant growth and development. Their interaction with plants and the type of microbes present in the rhizospheric region of plant roots are specified by the chemical compounds acting as signaling molecules, secreted by both microbes and plant roots. PGPR produce diverse sets of signaling molecules at the plant root interface, which is essential to set up beneficial associations. These signaling molecules are perceived, and signals are transmitted by plant signaling pathways, to stimulate and alter genetic and molecular mechanisms leading to beneficial responses. The responses, such as enhanced plant growth and stress tolerance, also suggest the regulation of hormonal pathways. Plant signaling has been very well-studied in different environmental stresses and is known to play an important role in combating plant stress. The signaling pathways working in the perception and transmission of signals from pathogens are well-known; however, information regarding the perception and transmission of signals for beneficial microbes is limited. Extensive studies revealing the mechanisms by which plant determines the interaction with specific microbes and the factors involved in their coexistence is needed. Further, studies determining exudates dependent changes in plant microbiota and the screening of interaction of plant signaling molecules with microbial components will help provide further insights. This review is an attempt to focus on plant signaling interplay in PGPR association so that we can further explore the mechanisms and interlinks of signaling networks. Understanding the action of PGPR signaling molecules will allow more effective application of PGPR in the field.
SJ conceptualized the idea, wrote the manuscript, and prepared the illustrations. AS wrote and edited the manuscript. Both authors contributed to the article and approved the submitted version.
The SJ Lab was supported by start-up grant (UGC-BSR) from University Grant Commission, Govt. of India, and DST-SERB start-up grant (File no. SRG/2021/001830-G) Govt. of India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
SJ is thankful to Goa University for financial support, while AS thanks the National Institute of Plant Genome Research, New Delhi and the Department of Biotechnology, Government of India for the support.
Akhtar, S. S., Mekureyaw, M. F., Pandey, C., and Roitsch, T. (2020). Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sci. 10, 1777. doi: 10.3389/fpls.2019.01777
Anderson, A. J., and Guerra, D. (1985). Responses of bean to root colonization with Pseudomonas putida in a hydroponic system. Phytopathology 75, 992–995. doi: 10.1094/Phyto-75-992
Ao, Y., Li, Z., Feng, D., Xiong, F., Liu, J., Li, J. F., et al. (2014). Os CERK 1 and Os RLCK 176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084. doi: 10.1111/tpj.12710
Arshad, M., and Frankenberger, W. T. Jr. (1997). Plant growth-regulating substances in the rhizosphere: microbial production and functions. Adv. Agron. 62, 45–151. doi: 10.1016/S0065-2113(08)60567-2
Asaf, S., Khan, M. A., Khan, A. L., Waqas, M., Shahzad, R., Kim, A. Y., et al. (2017). Bacterial endophytes from arid land plants regulate endogenous hormone content and promote growth in crop plants: an example of Sphingomonas sp. and Serratia marcescens. J. Plant Interact. 12, 31–38. doi: 10.1080/17429145.2016.1274060
Asano, T., Hayashi, N., Kikuchi, S., and Ohsugi, R. (2012). CDPK-mediated abiotic stress signaling. Plant Signal. Behav. 7, 817–821. doi: 10.4161/psb.20351
Asari, S., Tarkowsk,á, D., Rolčík, J., Novák, O., Palmero, D. V., Bejai, S., et al. (2017). Analysis of plant growth-promoting properties of Bacillusamyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 245, 15–30. doi: 10.1007/s00425-016-2580-9
Ashraf, M., Hasnain, S., Berge, O., and Mahmood, T. (2004). Inoculating wheat seedlings with exopolysaccharide-producing bacteria restricts sodium uptake and stimulates plant growth under salt stress. Biol. Fertil. Soils 40, 157–162. doi: 10.1007/s00374-004-0766-y
Ayaz, M., Ali, Q., Farzand, A., Khan, A. R., Ling, H., and Gao, X. (2021). Nematicidal volatiles from Bacillus atrophaeus GBSC56 promote growth and stimulate induced systemic resistance in tomato against meloidogyne incognita. Int. J. Mol. Sci. 22, 5049. doi: 10.3390/ijms22095049
Bartsev, A. V., Boukli, N. M., Deakin, W. J., Staehelin, C., and Broughton, W. J. (2003). Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 554, 271–274. doi: 10.1016/S0014-5793(03)01145-1
Bartsev, A. V., Deakin, W. J., Boukli, N. M., McAlvin, C. B., Stacey, G., Malnoë, P., et al. (2004). NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134, 871–879. doi: 10.1104/pp.103.031740
Bedini, A., Mercy, L., Schneider, C., Franken, P., and Lucic-Mercy, E. (2018). Unraveling the initial plant hormone signaling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front. Plant Sci. 9, 1800. doi: 10.3389/fpls.2018.01800
Bessadok, K., Navarro-Torre, S., Pajuelo, E., Mateos-Naranjo, E., Redondo-Gómez, S., Caviedes, M. Á., et al. (2020). The ACC-deaminase producing bacterium Variovorax sp. CT7.15 as a tool for improving Calicotomevillosa nodulation and growth in arid regions of Tunisia. Microorganisms 8, 541. doi: 10.3390/microorganisms8040541
Bhardwaj, D., Ansari, M. W., Sahoo, R. K., and Tuteja, N. (2014). Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 13, 1–10. doi: 10.1186/1475-2859-13-66
Boiero, L., Perrig, D., Masciarelli, O., Penna, C., Cassán, F., and Luna, V. (2007). Phytohormone production by three strains of Bradyrhizobium japonicum and possible physiological and technological implications. Appl. Microbiol. Biotechnol. 74, 874–880. doi: 10.1007/s00253-006-0731-9
Bredow, M., and Monaghan, J. (2019). Regulation of plant immune signaling by calcium-dependent protein kinases. Mol. Plant-Microbe Interact. 32, 6–19. doi: 10.1094/MPMI-09-18-0267-FI
Buendia, L., Girardin, A., Wang, T., Cottret, L., and Lefebvre, B. (2018). LysM receptor-like kinase and LysM receptor-like protein families: an update on phylogeny and functional characterization. Front. Plant Sci. 9, 1531. doi: 10.3389/fpls.2018.01531
Cao, Y., Halane, M. K., Gassmann, W., and Stacey, G. (2017). The role of plant innate immunity in the legume-rhizobium symbiosis. Annu. Rev. Plant Biol. 68, 535–561. doi: 10.1146/annurev-arplant-042916-041030
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chen, H. D., and Groisman, E. A. (2013). The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67, 83–112. doi: 10.1146/annurev-micro-092412-155751
Chen, L., Liu, Y., Wu, G., VeronicanNjeri, K., Shen, Q., Zhang, N., et al. (2016). Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 158, 34–44. doi: 10.1111/ppl.12441
Chen, Y. P., Rekha, P. D., Arun, A. B., Shen, F. T., Lai, W. A., and Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 34, 33–41. doi: 10.1016/j.apsoil.2005.12.002
Chi, F., Yang, P., Han, F., Jing, Y., and Shen, S. (2010). Proteomic analysis of rice seedlings infected by Sinorhizobiummeliloti 1021. Proteomics 10, 1861–1874. doi: 10.1002/pmic.200900694
Cohen, A. C., Bottini, R., and Piccoli, P. N. (2008). AzospirillumbrasilenseSp 245 produces ABA in chemically-defined culture medium and increases ABA content in arabidopsis plants. Plant Growth Regul. 54, 97–103. doi: 10.1007/s10725-007-9232-9
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Costa-Gutierrez, S. B., Lami, M. J., Caram-Di Santo, M. C., Zenoff, A. M., Vincent, P. A., Molina-Henares, M. A., et al. (2020). Plant growth promotion by Pseudomonas putida KT2440 under saline stress: role of eptA. Appl. Microbiol. Biotechnol. 104, 4577–4592. doi: 10.1007/s00253-020-10516-z
Cotton, T. E., Pétriacq, P., Cameron, D. D., Meselmani, M. A., Schwarzenbacher, R., Rolfe, S. A., et al. (2019). Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 13, 1647–1658. doi: 10.1038/s41396-019-0375-2
Danquah, A., de Zelicourt, A., Colcombet, J., and Hirt, H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52. doi: 10.1016/j.biotechadv.2013.09.006
Dodd, A. N., Kudla, J., and Sanders, D. (2010a). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Dodd, I. C., Zinovkina, N. Y., Safronova, V. I., and Belimov, A. A. (2010b). Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. doi: 10.1111/j.1744-7348.2010.00439.x
Duan, J., Müller, K. M., Charles, T. C., Vesely, S., and Glick, B. R. (2009). 1-aminocyclopropane-1-carboxylate (ACC) deaminase genes in rhizobia from southern Saskatchewan. Microb. Ecol. 57, 423–436. doi: 10.1007/s00248-008-9407-6
Edel, K. H., and Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Ekengren, S. K., Liu, Y., Schiff, M., Dinesh-Kumar, S. P., and Martin, G. B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905–917. doi: 10.1046/j.1365-313X.2003.01944.x
Etesami, H., Mirseyed Hosseini, H., and Alikhani, H. A. (2014). Bacterial biosynthesis of 1-aminocyclopropane-1-caboxylate (ACC) deaminase, a useful trait to elongation and endophytic colonization of the roots of rice under constant flooded conditions. Physiol. Mol. Biol. Plants 20, 425–434. doi: 10.1007/s12298-014-0251-5
Farag, M. A., Zhang, H., and Ryu, C. M. (2013). Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J. Chem. Ecol. 39, 1007–1018. doi: 10.1007/s10886-013-0317-9
Feng, F., Sun, J., Radhakrishnan, G. V., Lee, T., Bozsóki, Z., Fort, S., et al. (2019). A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat. Commun. 10, 5047. doi: 10.1038/s41467-019-12999-5
Fernandez, O., Theocharis, A., Bordiec, S., Feil, R., Jacquens, L., Clément, C., et al. (2012). BurkholderiaphytofirmansPsJN acclimates grapevine to cold by modulating carbohydrate metabolism. Mol. Plant-Microbe Interact. 25, 496–504. doi: 10.1094/MPMI-09-11-0245
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fukuda, H., Takahashi, M., Fujii, T., and Ogawa, T. (1989). Ethylene production from L-methionine by Cryptococcus albidus. J. Ferment. Bioeng. 67, 173–175. doi: 10.1016/0922-338X(89)90117-7
García de Salamone, I. E., Hynes, R. K., and Nelson, L. M. (2001). Cytokinin production by plant growth promoting rhizobacteria and selected mutants. Can. J. Microbiol. 47, 404–411. doi: 10.1139/w01-029
García-Gutiérrez, L., Zeriouh, H., Romero, D., Cubero, J., de Vicente, A., and Pérez-García, A. (2013). The antagonistic strain Bacillus subtilis UMAF 6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate-and salicylic acid-dependent defence responses. Microb. Biotechnol. 6, 264–274. doi: 10.1111/1751-7915.12028
Ge, Y. Y., Xiang, Q. W., Wagner, C., Zhang, D., Xie, Z. P., and Staehelin, C. (2016). The type 3 effector NopL of Sinorhizobium sp. strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Botany 67, 2483–2494. doi: 10.1093/jxb/erw065
Girardin, A., Wang, T., Ding, Y., Keller, J., Buendia, L., Gaston, M., et al. (2019). LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Curr. Biol. 29, 4249–4259. doi: 10.1016/j.cub.2019.11.038
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Glick, B. R., and Bashan, Y. (1997). Genetic manipulation of plant growth-promoting bacteria to enhance biocontrol of phytopathogens. Biotechnol. Adv. 15, 353–378. doi: 10.1016/S0734-9750(97)00004-9
Gobbato, E., Marsh, J. F., Vernié, T., Wang, E., Maillet, F., Kim, J., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22, 2236–2241. doi: 10.1016/j.cub.2012.09.044
Gobbato, E., Wang, E., Higgins, G., Bano, S. A., Henry, C., Schultze, M., et al. (2013). RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal. Behav. 8, e26049. doi: 10.4161/psb.26049
Govindasamy, V., Senthilkumar, M., Magheshwaran, V., Kumar, U., Bose, P., Sharma, V., et al. (2010). “Bacillus and Paenibacillus spp.: potential PGPR for sustainable agriculture,” in Plant Growth and Health Promoting Bacteria, ed D. K. Maheshwari (Berlin, Heidelberg: Springer), 333–364. doi: 10.1007/978-3-642-13612-2_15
Gunn, J. S. (2008). The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16, 284–290. doi: 10.1016/j.tim.2008.03.007
Gupta, A., Meyer, J. M., and Goel, R. (2002). Development of heavy metal-resistant mutants of phosphate solubilizing Pseudomonas sp. NBRI 4014 and their characterization. Curr. Microbiol. 45, 323–327. doi: 10.1007/s00284-002-3762-1
Gupta, S., and Pandey, S. (2019). ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front. Microbiol. 10, 1506. doi: 10.3389/fmicb.2019.01506
Gust, A. A., Willmann, R., Desaki, Y., Grabherr, H. M., and Nürnberger, T. (2012). Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502. doi: 10.1016/j.tplants.2012.04.003
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Habib, S. H., Kausar, H., and Saud, H. M. (2016). Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed Res. Int. 2016, 6284547. doi: 10.1155/2016/6284547
Halfter, U., Ishitani, M., and Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Nat. Acad. Sci. U.S.A. 97, 3735–3740. doi: 10.1073/pnas.97.7.3735
Hamel, L. P., Nicole, M. C., Sritubtim, S., Morency, M. J., Ellis, M., Ehlting, J., et al. (2006). Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. doi: 10.1016/j.tplants.2006.02.007
Hammond-Kosack, K. E., and Jones, J. D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773. doi: 10.2307/3870229
Hu, L., Robert, C. A., Cadot, S., Zhang, X. I., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738. doi: 10.1038/s41467-018-05122-7
Ilangumaran, G., and Smith, D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 8, 1768. doi: 10.3389/fpls.2017.01768
Jalmi, S. K. (2020). Rhizosphere signaling nurturing phyto-microbiome niche. Tropic. Plant Res. 7, 522–528. doi: 10.22271/tpr.2020.v7.i2.064
Jalmi, S. K., Bhagat, P. K., Verma, D., Noryang, S., Tayyeba, S., Singh, K., et al. (2018). Traversing the links between heavy metal stress and plant signaling. Front. Plant Sci. 9, 12. doi: 10.3389/fpls.2018.00012
Jalmi, S. K., and Sinha, A. K. (2015). ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front. Plant Sci. 6, 769. doi: 10.3389/fpls.2015.00769
Jalmi, S. K., and Sinha, A. K. (2016). Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonasoryzae in rice. Sci. Rep. 6, 37974. doi: 10.1038/srep37974
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, S. M., Radhakrishnan, R., Khan, A. L., Kim, M. J., Park, J. M., Kim, B. R., et al. (2014). Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 84, 115–124. doi: 10.1016/j.plaphy.2014.09.001
Kang, S. M., Shahzad, R., Bilal, S., Khan, A. L., Park, Y. G., Lee, K. E., et al. (2019). Indole-3-acetic-acid and ACC deaminase producing Leclerciaadecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 19, 1–14. doi: 10.1186/s12866-019-1450-6
Katznelson, H., and Cole, S. E. (1965). Production of gibberellin-like substances by bacteria and actinomycetes. Canad. J. Microbiol. 11, 733–741. doi: 10.1139/m65-097
Kim, K., Jang, Y. J., Lee, S. M., Oh, B. T., Chae, J. C., and Lee, K. J. (2014). Alleviation of salt stress by Enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol. Cells 37, 109. doi: 10.14348/molcells.2014.2239
Lian, B., Souleimanov, A., Zhou, X., and Smith, D. L. (2002). In vitro induction of lipo-chitooligosaccharide production in Bradyrhizobium japonicum cultures by root extracts from non-leguminous plants. Microbiol. Res. 157, 157–160. doi: 10.1078/0944-5013-00145
Liang, Y., Tóth, K., Cao, Y., Tanaka, K., Espinoza, C., and Stacey, G. (2014). Lipochitooligosaccharide recognition: an ancient story. New Phytol. 204, 289–296. doi: 10.1111/nph.12898
Lim, J. H., and Kim, S. D. (2013). Induction of drought stress resistance by multi-functional PGPR Bacillus licheniformis K11 in pepper. Plant Pathol. J. 29, 201. doi: 10.5423/PPJ.SI.02.2013.0021
Liu, F., Xing, S., Ma, H., Du, Z., and Ma, B. (2013). Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladusorientalis container seedlings. Appl. Microbiol. Biotechnol. 97, 9155–9164. doi: 10.1007/s00253-013-5193-2
Maillet, F., Poinsot, V., André, O., Puech-Pagès, V., Haouy, A., Gueunier, M., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. doi: 10.1038/nature09622
Masmoudi, F., Tounsi, S., Dunlap, C. A., and Trigui, M. (2021). Halotolerant Bacillus spizizenii FMH45 promoting growth, physiological, and antioxidant parameters of tomato plants exposed to salt stress. Plant Cell Rep. 40, 1199–1213. doi: 10.1007/s00299-021-02702-8
Maurhofer, M., Hase, C., Meuwly, P., Metraux, J. P., and Defago, G. (1994). Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: influence of the gacA gene and of pyoverdine production. Phytopathology 84, 139–146. doi: 10.1094/Phyto-84-139
Meng, X., and Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Mockaitis, K., and Howell, S. H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796. doi: 10.1046/j.1365-313x.2000.00921.x
Mori, N., Nishiuma, K., Sugiyama, T., Hayashi, H., and Akiyama, K. (2016). Carlactone-type strigolactones and their synthetic analogues as inducers of hyphal branching in arbuscular mycorrhizal fungi. Phytochemistry 130, 90–98. doi: 10.1016/j.phytochem.2016.05.012
Naidu, V. S. G. R., Panwar, J. D. S., and Annapurna, K. (2004). Effect of synthetic auxins and Azorhizobiumcaulinodans on growth and yield of rice. Ind. J. Microbiol. 44, 211–213.
Neeraja, C., Anil, K., Purushotham, P., Suma, K., Sarma, P. V. S. R. N., Moerschbacher, B. M., et al. (2010). Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit. Rev. Biotechnol. 30, 231–241. doi: 10.3109/07388551.2010.487258
Nelson, M. S., and Sadowsky, M. J. (2015). Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 6, 491. doi: 10.3389/fpls.2015.00491
Niu, D. D., Liu, H. X., Jiang, C. H., Wang, Y. P., Wang, Q. Y., Jin, H. L., et al. (2011). The plant growth–promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate-and jasmonate/ethylene-dependent signaling pathways. Mol. Plant-Microbe Interact. 24, 533–542. doi: 10.1094/MPMI-09-10-0213
Niu, S. Q., Li, H. R., Par,é, P. W., Aziz, M., Wang, S. M., Shi, H., et al. (2016). Induced growth promotion and higher salt tolerance in the halophyte grass Puccinelliatenuiflora by beneficial rhizobacteria. Plant Soil 407, 217–230. doi: 10.1007/s11104-015-2767-z
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oldroyd, G. E., and Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
Onofre-Lemus, J., Hernández-Lucas, I., Girard, L., and Caballero-Mellado, J. (2009). ACC (1-aminocyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Appl. Environ. Microbiol. 75, 6581. doi: 10.1128/AEM.01240-09
Ortiz-Castro, R., Díaz-Pérez, C., Martínez-Trujillo, M., Rosa, E., Campos-García, J., and López-Bucio, J. (2011). Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. Proc. Nat. Acad. Sci. 108, 107253–107258. doi: 10.1073/pnas.1006740108
Ortiz-Castro, R., and López-Bucio, J. (2019). Phytostimulation and root architectural responses to quorum-sensing signals and related molecules from rhizobacteria. Plant Sci. 284, 135–142. doi: 10.1016/j.plantsci.2019.04.010
Ortmann, I., Conrath, U., and Moerschbacher, B. M. (2006). Exopolysaccharides of Pantoea agglomerans have different priming and eliciting activities in suspension-cultured cells of monocots and dicots. Febs. Lett. 580, 4491–4494. doi: 10.1016/j.febslet.2006.07.025
Pang, Z., Chen, J., Wang, T., Gao, C., Li, Z., Guo, L., et al. (2021). Linking plant secondary metabolites and plant microbiomes: a review. Front. Plant Sci. 12, 300. doi: 10.3389/fpls.2021.621276
Pérez-Flores, P., Valencia-Cantero, E., Altamirano-Hernández, J., Pelagio-Flores, R., López-Bucio, J., García-Juárez, P., et al. (2017). Bacillus methylotrophicus M4-96 isolated from maize (Zea mays) rhizoplane increases growth and auxin content in Arabidopsis thaliana via emission of volatiles. Protoplasma 254, 2201–2213. doi: 10.1007/s00709-017-1109-9
Polkade, A. V., Mantri, S. S., Patwekar, U. J., and Jangid, K. (2016). Quorum sensing: an under-explored phenomenon in the phylum Actinobacteria. Front. Microbiol. 7, 131. doi: 10.3389/fmicb.2016.00131
Prasannakumar, S. P., Gowtham, H. G., Hariprasad, P., Shivaprasad, K., and Niranjana, S. R. (2015). Delftiatsuruhatensis WGR–UOM–BT1, a novel rhizobacterium with PGPR properties from Rauwolfia serpentina (L.) Benth. ex Kurz also suppresses fungal phytopathogens by producing a new antibiotic—AMTM. Lett. Appl. Microbiol. 61, 460–468. doi: 10.1111/lam.12479
Qin, H., and Huang, R. (2020). The phytohormonal regulation of Na+/K+ and reactive oxygen species homeostasis in rice salt response. Mol. Breed. 40, 1–13. doi: 10.1007/s11032-020-1100-6
Qin, Y., Druzhinina, I. S., Pan, X., and Yuan, Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34, 1245–1259. doi: 10.1016/j.biotechadv.2016.08.005
Raj, S. N., Lavanya, S. N., Amruthesh, K. N., Niranjana, S. R., Reddy, M. S., and Shetty, H. S. (2012). Histo-chemical changes induced by PGPR during induction of resistance in pearl millet against downy mildew disease. Biol. Control 60, 90–102. doi: 10.1016/j.biocontrol.2011.10.011
Ratu, S. T. N., Teulet, A., Miwa, H., Masuda, S., Nguyen, H. P., Yasuda, M., et al. (2021). Rhizobia use a pathogenic-like effector to hijack leguminous nodulation signalling. Sci. Rep. 11, 2034. doi: 10.1038/s41598-021-81598-6
Rey, T., André, O., Nars, A., Dumas, B., Gough, C., Bottin, A., et al. (2019). Lipo-chitooligosaccharidesignalling blocks a rapid pathogen-induced ROS burst without impeding immunity. New Phytol. 221, 743–749. doi: 10.1111/nph.15574
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., and Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Rush, T. A., Puech-Pagès, V., Bascaules, A., Jargeat, P., Maillet, F., Haouy, A., et al. (2020). Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 11, 3897. doi: 10.1038/s41467-020-17615-5
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., and Paré, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Ryu, H., and Cho, Y. G. (2015). Plant hormones in salt stress tolerance. J. Plant Biol. 58, 147–155. doi: 10.1007/s12374-015-0103-z
Samaniego-Gámez, B. Y., Garruña, R., Tun-Suárez, J. M., Moreno-Valenzuela, O. A., Reyes-Ramírez, A., Valle-Gough, R. E., et al. (2021). Healthy photosynthetic mechanism suggests ISR elicited by Bacillus spp. in Capsicum chinense plants infected with PepGMV. Pathogens 10, 455. doi: 10.3390/pathogens10040455
Sardar, A., Nandi, A. K., and Chattopadhyay, D. (2017). CBL-interacting protein kinase 6 negatively regulates immune response to Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 68, 3573–3584. doi: 10.1093/jxb/erx170
Senthilkumar, M., Madhaiyan, M., Sundaram, S. P., and Kannaiyan, S. (2009). Intercellular colonization and growth promoting effects of Methylobacterium sp. with plant-growth regulators on rice (Oryza sativa L. Cv CO-43). Microb. Res. 164, 92–104. doi: 10.1016/j.micres.2006.10.007
Sethi, V., Raghuram, B., Sinha, A. K., and Chattopadhyay, S. (2014). A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26, 3343–3357. doi: 10.1105/tpc.114.128702
Shah, J., Tsui, F., and Klessig, D. F. (1997). Characterization of a s alicylic a cid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. doi: 10.1094/MPMI.1997.10.1.69
Sheikh, A. H., Raghuram, B., Jalmi, S. K., Wankhede, D. P., Singh, P., and Sinha, A. K. (2013). Interaction between two rice mitogen activated protein kinases and its possible role in plant defense. BMC Plant Biol. 13, 121. doi: 10.1186/1471-2229-13-121
Singh, P., and Sinha, A. K. (2016). A positive feedback loop governed by SUB1A1 interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 imparts submergence tolerance in rice. Plant Cell 28, 1127–1143. doi: 10.1105/tpc.15.01001
Sinha, A. K., Jaggi, M., Raghuram, B., and Tuteja, N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6, 196–203. doi: 10.4161/psb.6.2.14701
Soto, M. J., Fernández-Aparicio, M., Castellanos-Morales, V., García-Garrido, J. M., Ocampo, J. A., Delgado, M. J., et al. (2010). First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biol. Biochem. 42, 383–385. doi: 10.1016/j.soilbio.2009.11.007
Sözen, C., Schenk, S. T., Boudsocq, M., Chardin, C., Almeida-Trapp, M., Krapp, A., et al. (2020). Wounding and insect feeding trigger two independent MAPK pathways with distinct regulation and kinetics. Plant Cell 32, 1988–2003. doi: 10.1105/tpc.19.00917
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Srivastava, S., Bist, V., Srivastava, S., Singh, P. C., Trivedi, P. K., Asif, M. H., et al. (2016). Unraveling aspects of Bacillus amyloliquefaciens mediated enhanced production of rice under biotic stress of Rhizoctoniasolani. Front. Plant Sci. 7, 587. doi: 10.3389/fpls.2016.00587
Subramanian, P., Mageswari, A., Kim, K., Lee, Y., and Sa, T. (2015). Psychrotolerant endophytic Pseudomonas sp. strains OB155 and OS261 induced chilling resistance in tomato plants (Solanum lycopersicum Mill.) by activation of their antioxidant capacity. Mol. Plant-Microbe Interact. 28, 1073–1081. doi: 10.1094/MPMI-01-15-0021-R
Tabassum, B., Khan, A., Tariq, M., Ramzan, M., Khan, M. S. I., Shahid, N., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 121, 102–117. doi: 10.1016/j.apsoil.2017.09.030
Tang, A., Haruna, A. O., Majid, N. M. A., and Jalloh, M. B. (2020). Potential PGPR properties of cellulolytic, nitrogen-fixing, phosphate-solubilizing bacteria in rehabilitated tropical forest soil. Microorganisms 8, 442. doi: 10.3390/microorganisms8030442
Tena, G., Boudsocq, M., and Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529. doi: 10.1016/j.pbi.2011.05.006
Timmusk, S., Nicander, B., Granhall, U., and Tillberg, E. (1999). Cytokinin production by Paenibacilluspolymyxa. Soil Biol. Biochem. 31, 1847–1852. doi: 10.1016/S0038-0717(99)00113-3
Tiwari, S., Lata, C., Chauhan, P. S., and Nautiyal, C. S. (2016). Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 99, 108–117. doi: 10.1016/j.plaphy.2015.11.001
Tiwari, S., Prasad, V., Chauhan, P. S., and Lata, C. (2017). Bacillus amyloliquefaciens confers tolerance to various abiotic stresses and modulates plant response to phytohormones through osmoprotection and gene expression regulation in rice. Front. Plant Sci. 8, 1510. doi: 10.3389/fpls.2017.01510
Van der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. doi: 10.1104/pp.107.113829
Van Loon, L. C. (2007). “Plant responses to plant growth-promoting rhizobacteria,” in New Perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research eds P. A. H. M. Bakker, J. M. Raaijmakers, G. Bloemberg, M. Hofte, P. Lemanceau, and B. M. Cooke (Dordrecht: Springer), 243–254. doi: 10.1007/978-1-4020-6776-1_2
Van Loon, L. C., Bakker, P. A. H. M., and Pieterse, C. M. J. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. doi: 10.1146/annurev.phyto.36.1.453
Van Peer, R., Niemann, G. J., and Schippers, B. (1991). Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS 417 r. Phytopathology 81, 728–734. doi: 10.1094/Phyto-81-728
Van Wees, S. C., Van der, S., and Pieterse, C. M. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. doi: 10.1016/j.pbi.2008.05.005
Vanneste, S., and Friml, J. (2013). Calcium: the missing link in auxin action. Plants 2, 650–675. doi: 10.3390/plants2040650
Vansuyt, G., Robin, A., Briat, J. F., Curie, C., and Lemanceau, P. (2007). Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol. Plant-Microbe Interact. 20, 441–447. doi: 10.1094/MPMI-20-4-0441
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., and Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 287, 2560–2576. doi: 10.1111/febs.15157
Wang, Q., Dodd, I. C., Belimov, A. A., and Jiang, F. (2016). Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct. Plant Biol. 43, 161–172. doi: 10.1071/FP15200
Wei, G., Kloepper, J. W., and Tuzun, S. (1991). Induction of systemic resistance of cucumber to Colletotrichumorbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology 81, 1508–1512. doi: 10.1094/Phyto-81-1508
Yanni, Y. G., Rizk, R. Y., Abd El-Fattah, F. K., Squartini, A., Corich, V., Giacomini, A., et al. (2001). The beneficial plant growth-promoting association of Rhizobium leguminosarumbv. trifolii with rice roots. Funct. Plant Biol. 28, 845–870. doi: 10.1071/PP01069
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6, 600. doi: 10.3389/fpls.2015.00600
Zhang, H., Kim, M. S., Krishnamachari, V., Payton, P., Sun, Y., Grimson, M., et al. (2007). Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226, 839–851. doi: 10.1007/s00425-007-0530-2
Zhang, H., Kim, M. S., Sun, Y., Dowd, S. E., Shi, H., and Paré, P. W. (2008). Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant-Microbe Interact. 21, 737–744. doi: 10.1094/MPMI-21-6-0737
Zhang, L., Chen, X. J., Lu, H. B., Xie, Z. P., and Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Zhang, X., Zhang, R., Gao, J., Wang, X., Fan, F., Ma, X., et al. (2017). Thirty-one years of rice-rice-green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol. Biochem. 104, 208–217. doi: 10.1016/j.soilbio.2016.10.023
Zhao, Q., Li, M., Jia, Z., Liu, F., Ma, H., Huang, Y., et al. (2016). AtMYB44 positively regulates the enhanced elongation of primary roots induced by N-3-oxo-hexanoyl-homoserine lactone in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 29, 774–785. doi: 10.1094/MPMI-03-16-0063-R
Zhao, Q., Zhang, C., Jia, Z., Huang, Y., Li, H., and Song, S. (2015). Involvement of calmodulin in regulation of primary root elongation by N-3-oxo-hexanoyl homoserine lactone in Arabidopsis thaliana. Front. Plant Sci. 5, 807. doi: 10.3389/fpls.2014.00807
Keywords: microbial signaling, PGPR, plant signaling, hormones, stress response, plant development
Citation: Jalmi SK and Sinha AK (2022) Ambiguities of PGPR-Induced Plant Signaling and Stress Management. Front. Microbiol. 13:899563. doi: 10.3389/fmicb.2022.899563
Received: 18 March 2022; Accepted: 08 April 2022;
Published: 13 May 2022.
Edited by:
Surendra Pratap Singh, Chhatrapati Shahu Ji Maharaj University, IndiaReviewed by:
Krishan M. Rai, CoverCress Inc., United StatesCopyright © 2022 Jalmi and Sinha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siddhi Kashinath Jalmi, c2lkZGhpQHVuaWdvYS5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.