
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 June 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.896916
This article is part of the Research TopicInsights in Antimicrobials, Resistance & Chemotherapy: 2021View all 14 articles
 Weiwei Li1,2
Weiwei Li1,2 Jiamin Hu1
Jiamin Hu1 Ling Li1
Ling Li1 Mengge Zhang1
Mengge Zhang1 Qingyu Cui1
Qingyu Cui1 Yanan Ma1
Yanan Ma1 Hainan Su1
Hainan Su1 Xuhua Zhang3*
Xuhua Zhang3* Hai Xu1*
Hai Xu1* Mingyu Wang1*
Mingyu Wang1*Daptomycin (DAP), a last-resort antibiotic for treating Gram-positive bacterial infection, has been widely used in the treatment of vancomycin-resistant enterococci (VRE). Resistance to both daptomycin and vancomycin leads to difficulties in controlling infections of enterococci. A clinical multidrug-resistant Enterococcus faecium EF332 strain that shows resistance to both daptomycin and vancomycin was identified, for which resistance mechanisms were investigated in this work. Whole-genome sequencing and comparative genomic analysis were performed by third-generation PacBio sequencing, showing that E. faecium EF332 contains four plasmids, including a new multidrug-resistant pEF332-2 plasmid. Two vancomycin resistance-conferring gene clusters vanA and vanM were found on this plasmid, making it the second reported vancomycin-resistant plasmid containing both clusters. New mutations in chromosomal genes cls and gdpD that, respectively, encode cardiolipin synthase and glycerophosphoryl diester phosphodiesterase were identified. Their potential roles in leading to daptomycin resistance were further investigated. Through molecular cloning and phenotypic screening, two-dimensional thin-layer chromatography, fluorescence surface charge test, and analysis of cardiolipin distribution patterns, we found that mutations in cls decrease surface negative charges of the cell membrane (CM) and led to redistribution of lipids of CM. Both events contribute to the DAP resistance of E. faecium EF332. Mutation in gdpD leads to changes in CM phospholipid compositions, but cannot confer DAP resistance. Neither mutation could result in changes in cellular septa. Therefore, we conclude that the daptomycin resistance of E. faecium EF332 is conferred by new cls mutations. This work reports the genetic basis for vancomycin and daptomycin resistance of a multidrug-resistant E. faecium strain, with the finding of new mutations of cls that leads to daptomycin resistance.
Enterococcus faecium, a Gram-positive coccus, is an important nosocomial pathogen that can cause fatal bacteremia and myocarditis (Arias and Murray, 2012). The emergence of multidrug-resistant (MDR) E. faecium, particularly vancomycin-resistant enterococci (VRE), led to high morbidity and mortality in hospitalized patients (Tran et al., 2013a, 2015; Herc et al., 2017; Lebreton et al., 2018). To make matters worse, the horizontal transfer of antibiotic resistance genes (ARGs) directly led to the rapid increase in VRE. In recent decades, VRE was gradually recognized as a major cause of MDR hospital infection in many countries (Uttley et al., 1988; Cattoir and Giard, 2014). A rapid increase of VRE undoubtedly brings more difficulties for the treatment of E. faecium infections (Depardieu et al., 2007; Lebreton et al., 2018). Initially, linezolid and quinupristin–dalfopristin were used to treat VRE infections, but their use was hampered by their strong adverse drug effects (Carver et al., 2003; Hogan et al., 2010; Matsumoto et al., 2010). By contrast, daptomycin (DAP) was generally considered to be a safer antimicrobial agent and gradually became the front-line drug for the patients who have VRE infections (Chow et al., 2016; Gonzalez-Ruiz et al., 2016; Herc et al., 2017; Suleyman et al., 2017).
Daptomycin is a cyclic lipopeptide antibiotic produced by Streptomyces roseosporus (Heidary et al., 2018), which has a special mechanism that binds to Ca2+ and inserts into the cell membrane (CM) causing leakage of intracellular ions and ATP, thus killing bacteria (LaPlante and Rybak, 2004; Heidary et al., 2018). Unfortunately, the widespread use of daptomycin resulted in the emergence of daptomycin non-susceptible VRE (DNVRE), which undoubtedly further reduced the option of treatment for VRE infections (Munoz-Price et al., 2005; Lellek et al., 2015; Chow et al., 2016; Hussain et al., 2016; Herc et al., 2017). Multiple studies suggested that the mechanism of daptomycin resistance is mainly related to mutations in chromosomal genes, including mprF, gdpD, yycG, rpoB, rpoC, pgsA, cls, liaFSR, etc. (Fischer et al., 2011; Tran et al., 2013a,b; Heidary et al., 2018). With the increase of reports on DNVRE, a series of investigations were carried out to try to find the resistance mechanisms of daptomycin and effective applications of DAP (Munoz-Price et al., 2005; Lellek et al., 2015; Chow et al., 2016; Hussain et al., 2016; Suleyman et al., 2017). However, the mechanism of DAP resistance has not been fully elucidated: One view is that DAP is diverted from effective targets at the septum by redistribution of anionic phospholipids; another view is that resistant bacteria achieve electrostatic repulsion to DAP by reducing the negative surface charge of CM (Heidary et al., 2018). In summary, the reports on DAP resistance mainly focus on the interaction between CM and DAP–Ca2+ complex.
In view of this, in the present study, a clinical multidrug-resistant E. faecium from a hospital showing daptomycin resistance and high-level vancomycin resistance was identified. Whole-genome sequencing and subsequent mechanistic investigations were performed, hoping to find the answers to the following questions: (1) the genetic basis of the multidrug resistance; (2) the risk of multiple resistance transmission; and (3) the mechanism of daptomycin resistance.
Enterococcus faecium EF332 strain used in this study was isolated in the Laboratory Medicine Center of the Second Hospital of Shandong University. The laboratory maintains an opportunistic pathogen library isolated from patient samples and routinely screens them for possible new findings. The strain was identified by the analysis of 16S rDNA sequence.
Antibiotic susceptibility tests were conducted according to CLSI/EUCAST guidelines (EUCAST, 2017; CLSI, 2018). The tested antibiotics include: penicillin (PEN), ampicillin (AMP), ciprofloxacin (CIP), gatifloxacin (GAT), chloramphenicol (CHL), tetracycline (TET), tigecycline (TGC), fosfomycin (FOF), erythromycin (ERY), linezolid (LZD), rifampicin (RIF), vancomycin (VAN), and daptomycin (DAP). K-B Disk diffusion assays were performed as previously documented (Vading et al., 2011). Agar dilution method (for FOF) and broth microdilution method (for the rest antibiotics) were adopted to determine MICs of antibiotics. Staphylococcus aureus ATCC 25923 and Enterococcus faecalis ATCC 29212 were used as control strains for disk diffusion and dilution method, respectively, according to CLSI and EUCAST standards (EUCAST, 2017; CLSI, 2018).
The genomic DNA of E. faecium EF332 and constructed strains (29212-pDL278, EFDO-cls, EF332-cls) was extracted following previous reported protocol (Qian and Zhao, 2014), and the purity and integrity of the DNA were confirmed by agarose gel electrophoresis. DNA samples of E. faecium EF332 were constructed into a 10-kb SMRTbell DNA library, and PacBio single-molecule sequencing was used to obtain at least 50× of sequencing data. In addition, a 350-bp small fragment library was constructed, and Illumina NovaSeq PE150 platform was used for paired-end sequencing to obtain at least 100× clean data for auxiliary assembly. DNA samples of constructed strains were used as templates for quantitative polymerase chain reaction (qPCR) to detect the relative copy number of plasmids.
To verify the transferability of plasmid pEF332-2, we conducted conjugation and transformation experiments. A chloramphenicol-resistant and vancomycin-sensitive E. faecalis 3–147 strain was used as the recipient for conjugations, and vancomycin-sensitive E. faecalis ATCC 29212 was used as recipient for transformation. Transconjugants were identified on the plates containing 32 μg/ml of vancomycin and 50 μg/ml of chloramphenicol, and transformants were identified on the plates containing 32 μg/ml of vancomycin.
Genomic DNA of E. faecium EF332 strain was used as template to amplify the presumed DAP resistance genes cls and gdpD by PCR using Phanta super-fidelity DNA polymerase (Vazyme Biotech Co. Ltd. Nanjing, China). All the PCR products were analyzed by 1% agarose gel electrophoresis and purified by DNA clean-up kit (TIANGEN, Beijing, China).
All primer sequences are shown in Supplementary Table 1, in which the cleavage sites of BamHI and SalI were added to the 5′ ends for the next step of cloning. The shuttle plasmid pDL278 with spectinomycin resistance was used for molecular cloning (Genbank accession number AF216802.1). The cls, gdpD fragments and pDL278 vector, were digested by BamHI and SalI endonuclease, respectively, and then, the gene fragments were ligated to pDL278 vector by T4 DNA ligase to generate recombinant shuttle plasmids (pDL278-cls, pDL278-gdpD). These recombinant plasmids were transferred into Escherichia coli DH5α chemically competent cells, respectively, according to the manufacturer’s protocols, and Luria-Bertani (LB) agar plates with 500 mg/L spectinomycin were used to select for positive transformants. The plasmids of the positive clones were extracted and transformed into the electroporation competent cells of E. faecalis ATCC 29212, and tryptic soy broth (TSB) agar plates with 1,000 mg/L spectinomycin were used to select for positive transformants. The successfully transferred gene fragments were validated by Sanger sequencing using primer M13 (as shown in Supplementary Table 1). Finally, we obtained the EF332-cls and EF332-gdpD strains for downstream analysis. A summary of constructed strains is shown in Table 1 for easier comparison.
In order to further confirm the role of mutations in cls and gdpD genes, site-directed mutagenesis was performed using overlap extension PCR (SOE-PCR; Ho et al., 1989). The cls gene was divided into four fragments at three mutated sites, and gdpD gene was divided into two fragments at one mutated site. The primers used are shown in Supplementary Table 1.
The SOE-PCR process consisted of two consecutive reactions. First-round PCR: Genomic DNA of E. faecium EF332 was used as a template to obtain the corresponding gene fragments. PCR products obtained were purified using the DNA purification kit (TIANGEN, Beijing, China). Second-round PCR: The purified PCR products obtained from the previous step were used as templates, and corresponding primers were added to the reaction system for amplification.
These point mutations led to the amino acid substitutions T269I, V203I, T298S in Cls, and S201P in GdpD. Plasmids carrying these mutated gene fragments were transformed to obtain EFDO-cls and EFDO-gdpD strains by the method described above.
Poly-L-lysine conjugated to fluorescein isothiocyanate (PLL:FITC) was used to assay cell surface charges according to previous studies (Prater et al., 2021). In brief, the strains grew until OD600 reached 0.5 in TSB. Cells were then washed three times with sterilized HEPES buffer (20 mM, pH 7.0) and diluted to OD600 of 0.1. Cells were incubated with 50 μg/ml PLL:FITC (final concentration) at room temperature with shaking for 10 min and were then washed once using HEPES buffer to remove unbound PLL:FITC. Treated cells were spread on poly-L-lysine-treated glass slides, followed by the addition of 20 μl fluorescent anti-quenching agent. Fluorescence images were observed under NIKON Ti-E inverted fluorescence microscope (Nikon Instruments Co., LTD, Japan) and quantified by ImageJ, with duplicates per strain.
The fluorescent probe 10-N-nonyl acridine orange (NAO) specifically binds to cardiolipin, and the change of cardiolipin distribution caused by cls mutation can be determined by observing the NAO fluorescence distribution on the membrane (Mileykovskaya et al., 2001). NAO fluorescence assay was performed according to previous studies (Tran et al., 2013b). In brief, the bacteria were activated in TSB overnight and transferred to new TSB to grow to exponential phase (OD600 of ~0.5), after which NAO was added to the medium at a final concentration of 2.5 μM. The sample was oscillated at 100 rpm and 37°C for 4 h under dark conditions. The cells were washed three times (5,000 rpm, 3 min) with sterilized 0.9% saline and immediately mixed with 2× volume of fluorescent anti-quenching agent and fixed on the poly-L-lysine-treated glass slides. Fluorescence observation was performed using a laser scanning confocal microscope with 100× objective and airyscan detector (LSM880, ZEISIS, Germany).
Membrane lipids were extracted using previous methods (Komagata and Suzuki, 1988). The lipid components of tested strains were separated by two-dimensional thin-layer chromatography (2D-TLC, Silica 60 F254 TLC plates; Merck) and stained with phosphomolybdic acid (blue for all lipids) and ninhydrin (red for lipids with amino groups), respectively, with three replicates for each strain. The first dimension was developed with chloroform/methanol/water (65:25:4, by volume), and the second dimension was developed with chloroform/acetic acid/methanol/water (80:15:12:4, by volume). The dried TLC plates were heated for 15 min in a 110°C oven to find phospholipid composition.
Bacteria were inoculated in 30-ml TSB and grew to exponential phase by shaking at 160 rpm and 37°C. The cultures were centrifuged at 5,000 rpm for 3 min to pellet cells. Cells were fixed at 4°C for 1 h with 500 μl 2.5% glutaraldehyde, washed with PBS (pH 7.2, 0.01 M) for five times (20 min each), fixed with 1% osmium acid at 4°C for 1 h, and washed with PBS (pH 7.2, 0.01 M) for three times, 15 min each. The samples were sequentially dehydrated with 30, 50, 70, 90, and 100% ethanol (twice) and replaced twice by acetone, 15 min each time. Different proportions of embedding agent (EPON812: acetone =1:3,1:1,3:1) were used to penetrate for 1 h successively, and then, pure embedding agent (100% EPON812) was used for penetration overnight. The samples were embedded in EPON812 and then subjected to temperature-programmed curing for 48 h before ultra-thin section. The sections were stained with 2% uranium acetate solution for 20 min (in dark), washed with ddH2O three times for 5 min each, stained with 1% lead citrate solution for 10 min, and then washed with ddH2O three times for 5 min each. The prepared samples were observed by Focused Ion Beam-Scanning Transmission Electron Microscope (Crossbeam 550, ZEISIS, Germany).
qPCRs were used to assay the copy numbers of pDL278 vector and variants in E. faecium strains (29212-pDL278, EFDO-cls, EF332-cls) to verify the stability of this vector. Plasmid levels were calculated using the 2−ΔΔCt method. 16S rDNA was used as the housekeeping gene. The primers used are listed in Supplementary Table 1. The qPCR system (20 μl) contains the following: 10 μl of 2× SYBR green premix pro taq HS premix (Accurate Biotechnology Co., Ltd., China), 7.8 μl dd H2O, 0.4 μl of forward and reverse primers each (10 μM), 0.4 μl of ROX reference dye (20 μM, Accurate Biotechnology Co., Ltd., China), and 1 μl template DNA (100 ng/μl). The qPCRs were performed with the following programs: denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 30 s. Three biological replicates were performed for each sample, and each qPCR was conducted in triplicate using the ABI StepOnePlus system (Applied Biosystems Inc., Waltham, MA, United States).
Survival analysis was performed according to the previously published method to compare the survival of different strains under daptomycin stress (Grein et al., 2020). After the bacteria grew to exponential phase (OD600 = 0.5) in 5 ml Müller Hinton broth containing 0.05 g/L Ca2+ (CAMHB), 100 μl of the cultures were diluted 105-fold. One hundred microliters of the diluted bacterial solution was inoculated on TSB agar plates (three replications) for CFU calculation. The remaining cultures were incubated with 32 μg/ml (4 × MIC) daptomycin (final concentration) at 37°C with shaking at 180 rpm for 1 h. During incubation, 100 μl cultures were taken every 15 min, diluted, and counted as described above. In order to avoid the influence of the change of bacterial solution volume during incubation, an equal volume of CAMHB was added after 100 μl bacterial solution was taken each time. Colony counts at different time points were recorded after overnight incubation at 37°C, and survival rate was calculated as the percentage of colony counts to those without treatment of daptomycin.
Whole-genome sequencing reads were assembled into a preliminary genome using SMRT Link v5.1.0 software (Ardui et al., 2018), and arrow software was used to optimize the assembly results. The optimized assembly results were analyzed and compared with original data to discriminate chromosomes and plasmid sequences. Final circular genomes were obtained, followed by gene prediction using GeneMarkS v4.17 software (Besemer et al., 2001). Genomic Islands (GIs) were predicted using IslandPath-DIOMB v0.12 software (Hsiao et al., 2003). Prophage prediction was completed by online prediction server PHASTER (Zhou et al., 2011; Arndt et al., 2016). Functional annotation of coding genes was performed using GO (Ashburner et al., 2000), COG (Galperin et al., 2014), NR (Li et al., 2002), Pfam (Punta et al., 2011), TCDB (Saier et al., 2013), and SWISS-PROT (Bairoch and Apweiler, 2000) databases. ARG annotation used Resistance Gene Identifier (RGI) v5.1.0 software provided by CARD database (Jia et al., 2016). Multilocus sequence typing (MLST) and plasmid classification of E. faecium EF332 were performed using MLST1 and PlasmidFinder databases,2 respectively, provided by Center for Genomic Epidemiology. AlphaFold 2 software was used to construct Cls structure model (Jumper et al., 2021). Pfam database was used to predict Cls protein domain structure (Mistry et al., 2021), and DOG 2.0 software was used to draw the protein domain structure map (Ren et al., 2009). The relative intensity of fluorescence was quantified by ImageJ 1.52a, resulting in the mean of two biological duplicates, and the significance level was analyzed by Student’s t-test. The ggplot2 package in R Studio 1.3.929 software (based on R 3.6.3) was used to draw the box charts of fluorescence intensity, and the gggenes and ggplot2 packages were used to generate the gene structure diagrams.
Enterococcus faecium EF332 strain used in this study was isolated in the Laboratory Medicine Center of the Second Hospital of Shandong University during routine screening of pathogens. Antimicrobial susceptibility tests were performed for this strain using 13 antibiotics in 10 classes by both Kirby–Bauer disk diffusion method and determination of minimum inhibitory concentration (MIC) levels. It was shown that E. faecium EF332 was resistant to most antibiotics (Table 2), suggesting that it is multidrug-resistant. A striking observation is that E. faecium EF332 is resistant to both last-line antibiotics against enterococcal infections: vancomycin and daptomycin (Bender et al., 2018). This unusual phenomenon drove us to further investigate the mechanisms underlying multidrug resistance of this strain, particularly for the two last-resort antibiotics vancomycin and daptomycin.
To fully understand the genetic basis of multidrug resistance in E. faecium EF332, whole-genome sequences were obtained with third-generation PacBio sequencing accompanied by second-generation Illumina sequencing (The sequencing data were deposited in GenBank with accession numbers CP058891-CP058895). Full sequences for a circular chromosome and four plasmids were obtained. The size of the chromosome is 2,755,510 bp, similarly to previously sequenced E. faecium strains. The four plasmids, pEF332-1, pEF332-2, pEF332-3, and pEF332-4, are, respectively, 12,341, 87,675p, 28,308, and 203,652 bp long (Supplementary Figure 1; Supplementary Table 2). Multilocus sequence typing (MLST) result showed that E. faecium EF332 belongs to ST192, one type of the CC17 clone complex (clade A1), which is believed to have evolved in infections of hospital environment (Freitas et al., 2010).
With accurate gene prediction algorithms, a total of 3,121 genes were predicted to be encoded by E. faecium EF332, of which 2,735 are chromosome-borne and 386 are plasmid-borne (Supplementary Table 2). Annotation with Comprehensive Antibiotic Resistance Database (CARD) showed that 9, 0, 12, 0, and 2 ARGs were encoded on the chromosome, pEF332-1, pEF332-2, pEF332-3, and pEF332-4, respectively (Table 3; Supplementary Table 2). Determinants for all antibiotics were found, most of which are chromosome-borne. Only pEF332-2 and pEF332-4 are antibiotic-resistant plasmids. While pEF332-4 only carries ARGs for aminoglycosides and was previously found in E. faecium VRE1 (Sun et al., 2020), pEF332-2 is a new plasmid that carries both vanA and vanM gene clusters. To the best of our knowledge, plasmids carrying both vanA and vanM genes are very rare, and only one such plasmid (pELF1) has been found so far in Japan, which is associated with high vancomycin resistance and high risk of transmission (Hashimoto et al., 2019). pEF332-2 is therefore the second plasmid that bears both vanA and vanM clusters identified so far. This is also in consistency with the high level of vancomycin resistance (128 μg/ml) observed with E. faecium EF332. In addition, vanA gene clusters are encoded on a genomic island GIs011 that is also part of a multidrug-resistant prophage (Figure 1; Supplementary Table 3). This is a strong implication that these ARGs are acquired by horizontal gene transfer. Therefore, this new pEF332-2 plasmid is a strong vancomycin-resistant plasmid that also has a strong implication for ARG mobility. Further phylogenetic analysis of E. faecium EF332 plasmids and 55 other plasmids, along with subtyping of pEF332-2 with PlasmidFinder, suggests pEF332-2 belongs to type rep2 (Figure 2). Previous report suggested type rep2 plasmids originate from a variety of sources, including hospitals, poultry, and foods (Schwarz et al., 2001; Sletvold et al., 2007; Sun et al., 2020), suggesting that these plasmids may spread between multiple environments. Nevertheless, attempts to transfer pEF332-2 from the host E. faecium EF332 strain to another bacterium via conjugation or transformation were unsuccessful, showing the limited efficiency of horizontal gene transfer.
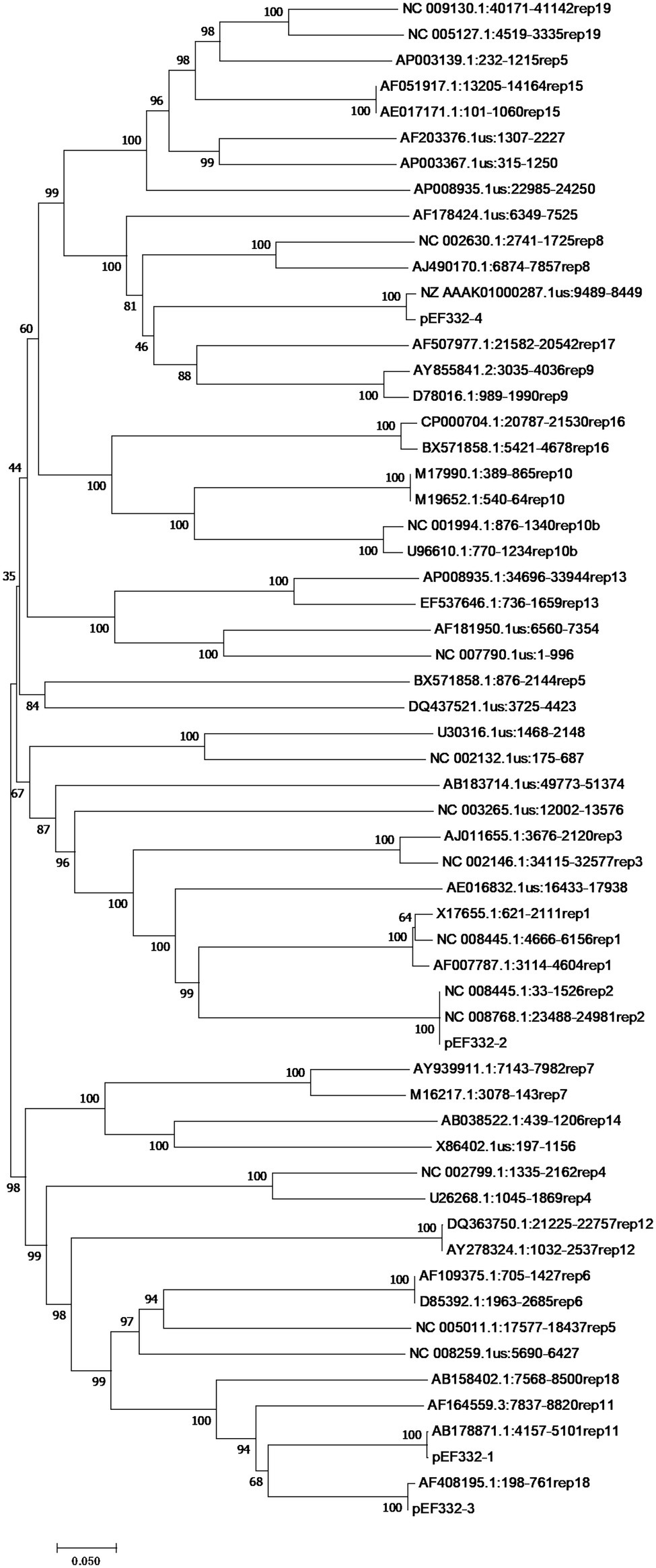
Figure 2. Phylogenetic analysis of rep genes of Enterococcus faecium EF332 plasmids. The bootstrap values are displayed on the branches with 1,000 times of replications. Bar, evolutionary distance.
While comparing the genomic sequences of E. faecium EF332 and daptomycin-sensitive E. faecium DO strain, mutations were found on cls and gdpD that, respectively, encode cardiolipin synthase and glycerophosphoryl diester phosphodiesterase (Table 4). It was previously reported that mutations in these genes can lead to daptomycin resistance (Humphries et al., 2012; Davlieva et al., 2013; Kelesidis et al., 2013; Lellek et al., 2015; Prater et al., 2019). However, known mutations that confer DAP resistance on these genes were not present in E. faecium EF332, leading to the hypothesis that new mutations of the two genes could lead to daptomycin resistance.
In order to find out which mutations on cls and/or gdpD lead to daptomycin resistance in E. faecium EF332 strains, a series of clones were constructed using daptomycin-sensitive E. faecalis ATCC 29212 as the parent strain: the EF332-cls strain that harbors cls from E. faecium EF332; the EFDO-cls strain that harbors cls from E. faecium EF332 with identified mutations (V203, T269, T298) reverted to I203, I269, and S298; the EF332-gdpD strain that harbors gdpD from E. faecium EF332; and the EFDO-gdpD strain that harbors gdpD from E. faecium EF332 with identified mutation S201 reverted to P201; the 29212-pDL278 strain that harbors pDL278 empty vector as control for constructed strains (Table 1). It is shown that the DAP MIC of E. faecalis ATCC 29212 increased from 1 to 8 with the introduction of pDL278 vector (Supplementary Table 4). A twofold increase in MIC values for daptomycin was found for EF332-cls strain (16 μg/ml) over the EFDO-cls strain (8 μg/ml), whereas no change of MIC values for daptomycin was found for EF332-gdpD strain (8 μg/ml) over the EFDO-gdpD strain (8 μg/ml; Supplementary Table 4). These results suggest that mutations in cls can lead to daptomycin resistance, while mutations in gdpD cannot.
Further susceptibility tests including K-B disk diffusion assays and survival assays support this finding. The inhibition zones of daptomycin with the EF332-cls strain are significantly smaller than the EFDO-cls strain (p = 2.09 × 10−5, Figure 3), while no significant difference was found between EFDO-cls strain and 29212-pDL278. Similarly, in survival assays, a significantly higher daptomycin resistance strength was found for EF332-cls in comparison with EFDO-cls, while EFDO-cls and 29212-pDL278 respond to daptomycin stress similarly (Figure 4). Additional analysis of plasmid copy numbers confirmed that introducing variants of cls did not lead to plasmid copy number fluctuation (Figure 5). It is further interesting to find that in both liquid-media-based analyses, aka the MIC determination and survival analysis, pDL278 introduction increased daptomycin resistance (Figure 4), while in the plate-based K-B disk diffusion assay, pDL278 introduction led to little difference in daptomycin resistance (Figure 3). This prompted us to hypothesize that maybe pDL278 can result in growth status-specific response to daptomycin, but without further evidence we cannot suggest the specific mechanism behind this phenomenon. Despite this difference, in all scenarios and in all assays, introducing cls from E. faecium EF332 leads to significantly stronger daptomycin resistance than introducing cls from E. faecium DO, confirming the above suggestion that mutations in cls found in E. faecium EF332 lead to daptomycin resistance.
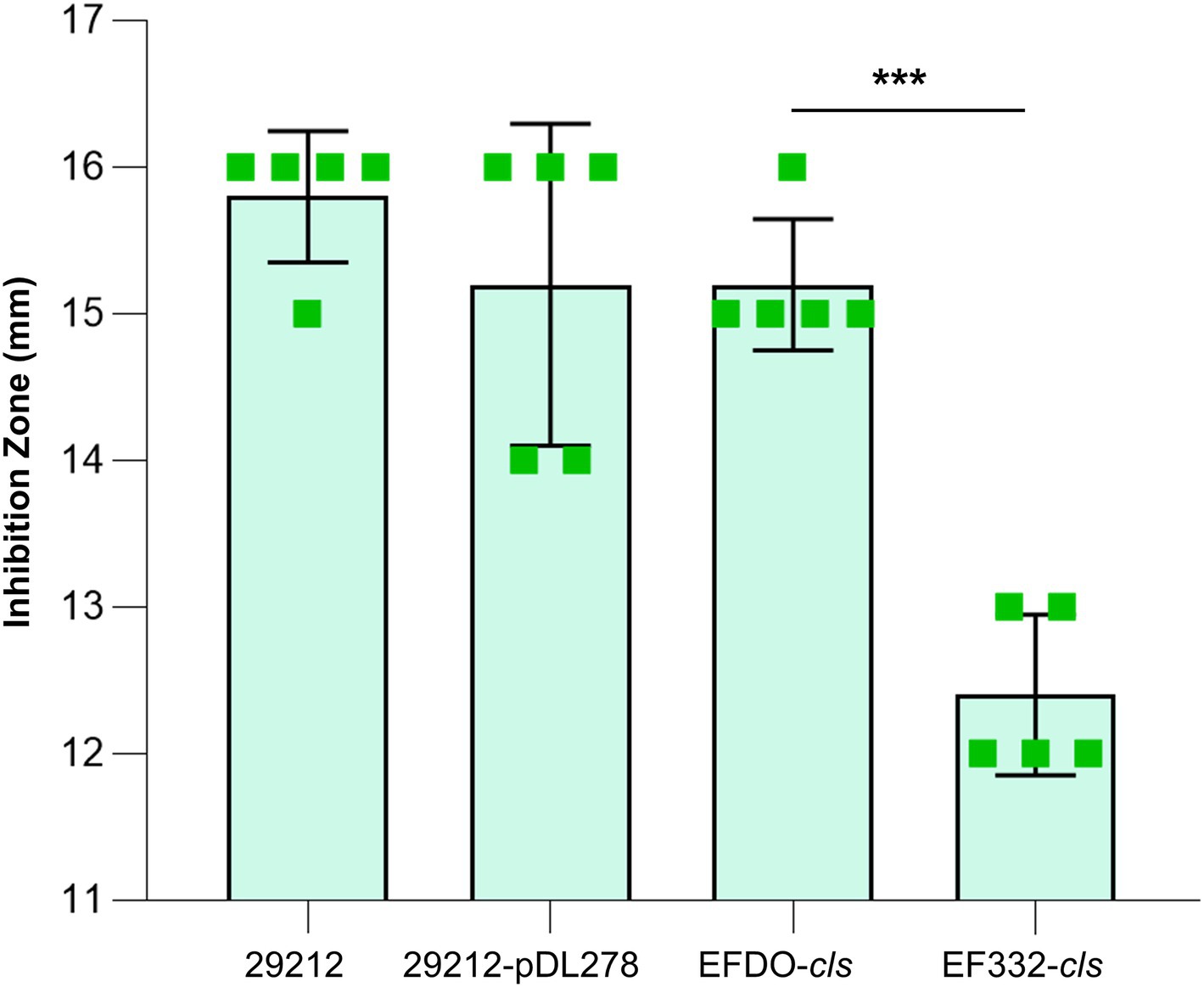
Figure 3. Susceptibility tests of strains based on K-B disk diffusion method. 29212, Enterococcus faecalis ATCC 29212; ***p<0.001; bar, standard deviation.
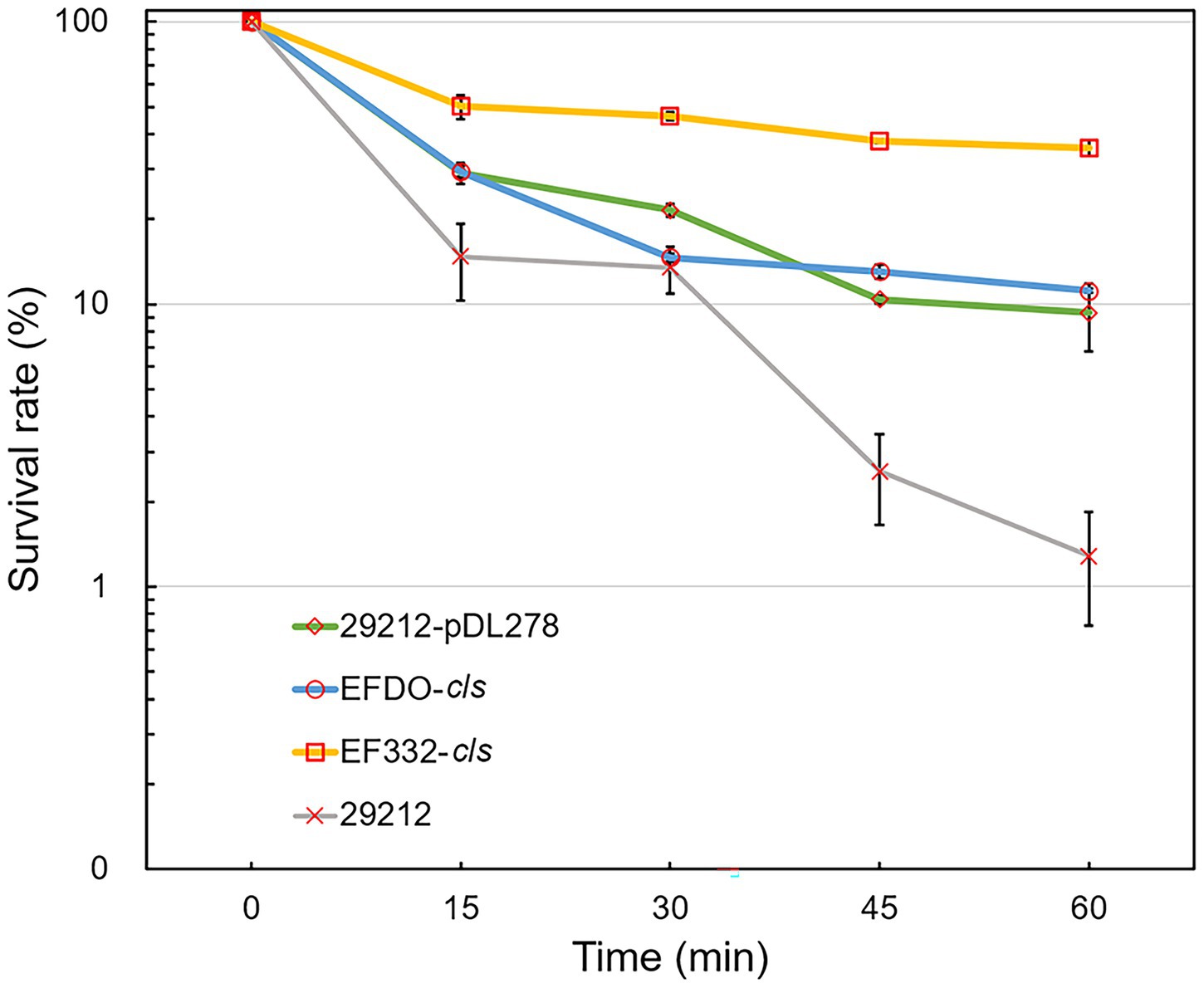
Figure 4. Daptomycin survival assays of Enterococcus faecalis strains. 29212, E. faecalis ATCC 29212; bar, standard deviation.
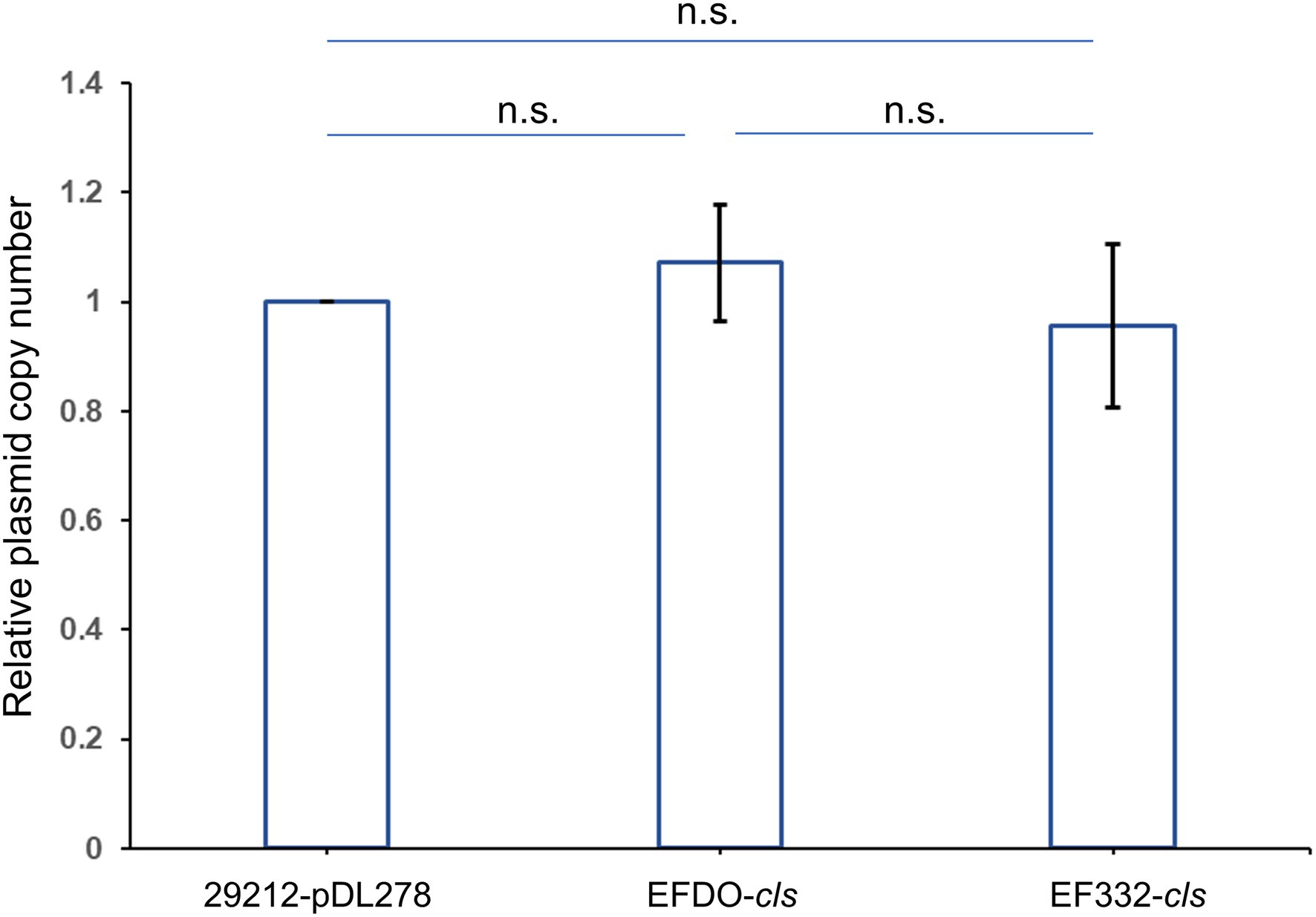
Figure 5. Relative plasmid copy numbers. Relative plasmid copy numbers are defined as relative normalized plasmid copy numbers to 29212-pDL278 strain. Bar, standard deviation; n.s., not significant.
Both cls and gdpD genes are related to phospholipid metabolism in CM: The cls gene encodes cardiolipin synthase, and gdpD gene encodes glycerophosphoryl diester phosphodiesterase (Arias et al., 2011). We therefore hypothesized that mutations in these two genes alter membrane properties, which in turn leads to alteration in membrane–daptomycin interactions. To confirm this, two-dimensional thin-layer chromatography (2D-TLC) was performed to analyze the changes of membrane lipids components caused by mutations. As shown in Figure 6, mutation in gdpD altered the aminolipid composition of CM, while mutations in cls did not significantly alter the lipid composition of CM. This finding suggests a putative role of gdpD in the biosynthesis of relatively poorly characterized aminolipid. Furthermore, membrane surface charges were quantified using positively charged fluorescence dye poly-L-lysine conjugated to fluorescein isothiocyanate (PLL: FITC). A significant reduction of binding to PLL: FITC was found between EF332-cls and E. faecalis ATCC 29212 (p = 6.27 × 10−65), as well as between EF332-cls and EFDO-cls (p = 2.20 × 10−101; Figure 7). This significant change suggests that a reduction of membrane negative charges followed cls mutations, which can lead to weakened binding of positively charged DAP-Ca2+ to the membrane, the target for the bactericidal effect of daptomycin. This finding suggests that cls mutations lead to daptomycin resistance by reduction of membrane negative charges. Only minor changes of membrane negative potentials were found for gdpD mutation strain (Figure 7), in consistent with the finding that gdpD mutation did not lead to increase of daptomycin resistance.
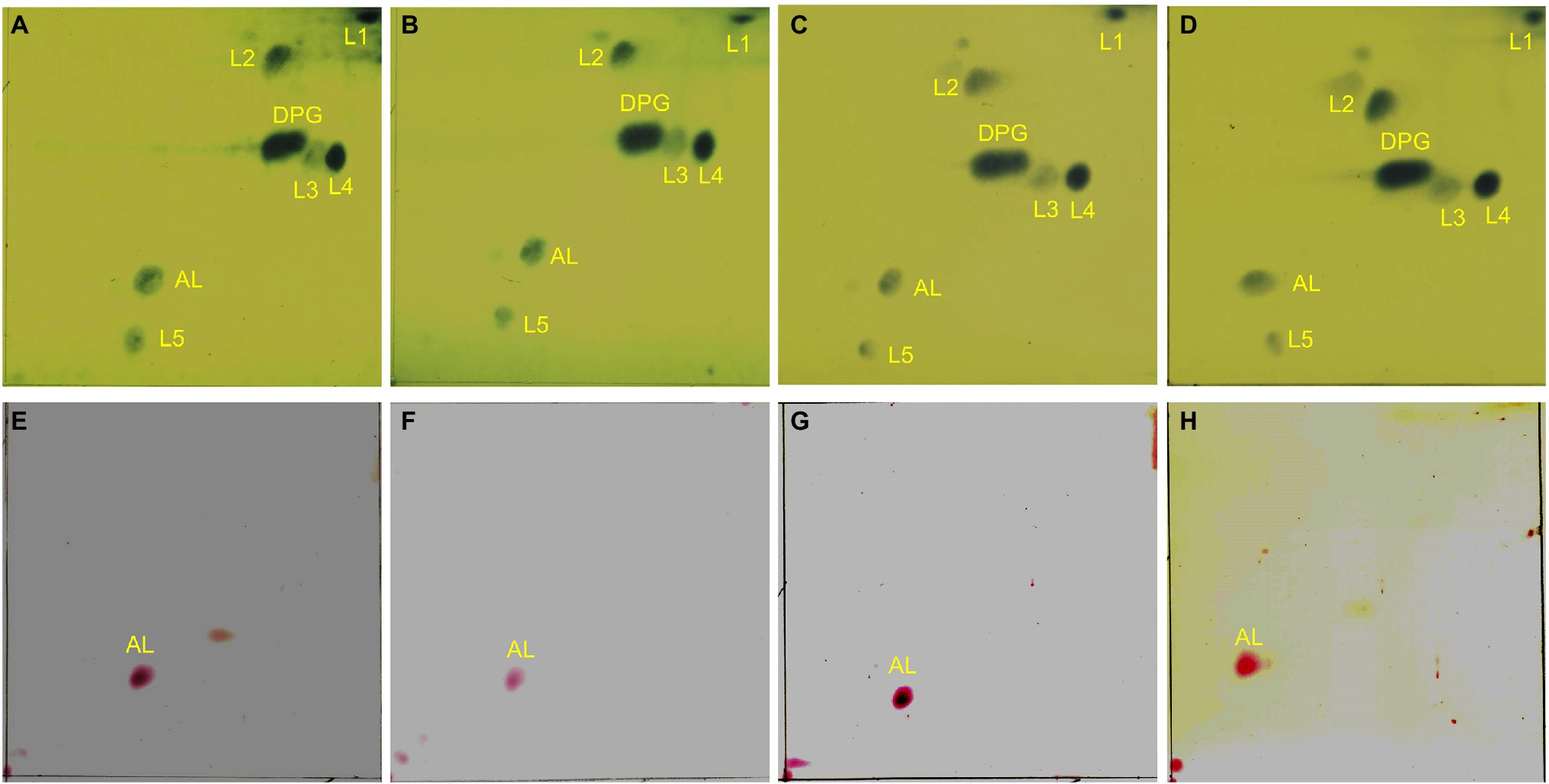
Figure 6. Two-dimensional thin-layer chromatography of phospholipids in Enterococcus faecium EF332 strains. Panels (A–D) are results of phosphomolybdic acid staining, and all lipids are dyed blue; Panels (E–H) are results of ninhydrin staining, and aminolipids are dyed red. Panels (A,E) EFDO-cls; Panels (B,F) EF332-cls; Panels (C,G) EFDO-gdpD; Panels (D,H) EF332-gdpD. DPG: diphosphatidyl glycerol (cardiolipin); AL, aminolipids; L1–L5, lipids.
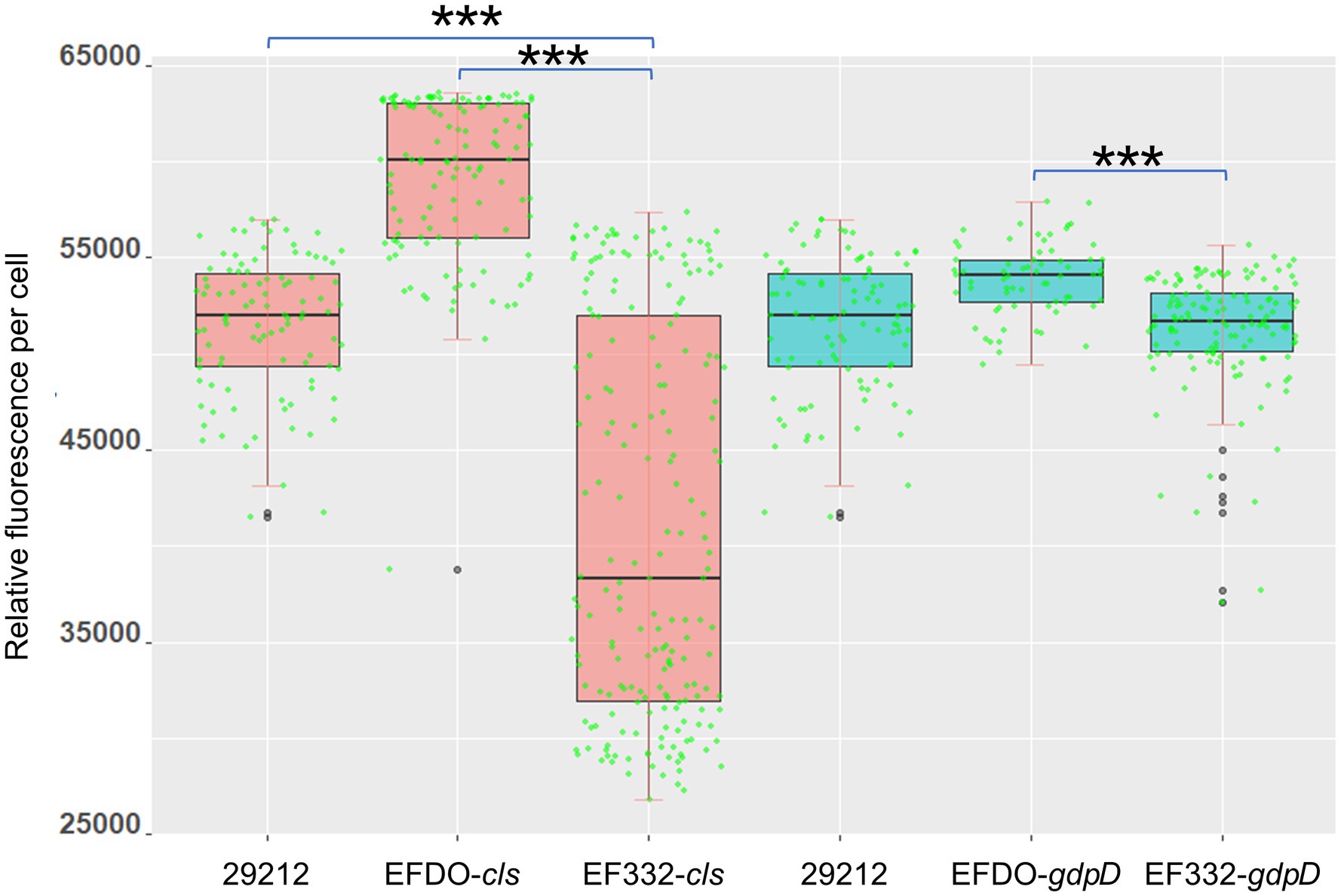
Figure 7. Cell surface charge measured with PLL: FITC fluorescence. 29212, Enterococcus faecalis ATCC 29212, ***p <0.001.
We further sought to investigate whether cls mutations also alter the distribution of cardiolipin produced by cls-encoded cardiolipin synthase. A specific cardiolipin binding fluorescent probe, 10-N-nonyl acridine orange (NAO), was used to observe the distribution of cardiolipins in CM. A redistribution of cardiolipin in CM was found (Figure 8). In E. faecalis ATCC 29212 (Figures 8A,D) and EFDO-cls (Figures 8B,E), the cardiolipin-rich regions were mainly located at the cell septum or at both poles, while in the cls-mutant strain EF332-cls, the cardiolipin-rich regions were scattered all over the cell, leading to a more even distribution and reduced abundance at septa (Figures 8C,F). This is in agreement with previous suggestion that daptomycin resistance may be resulted from redistribution of phospholipids and diverting DAP from effective targets at the septum (Heidary et al., 2018).
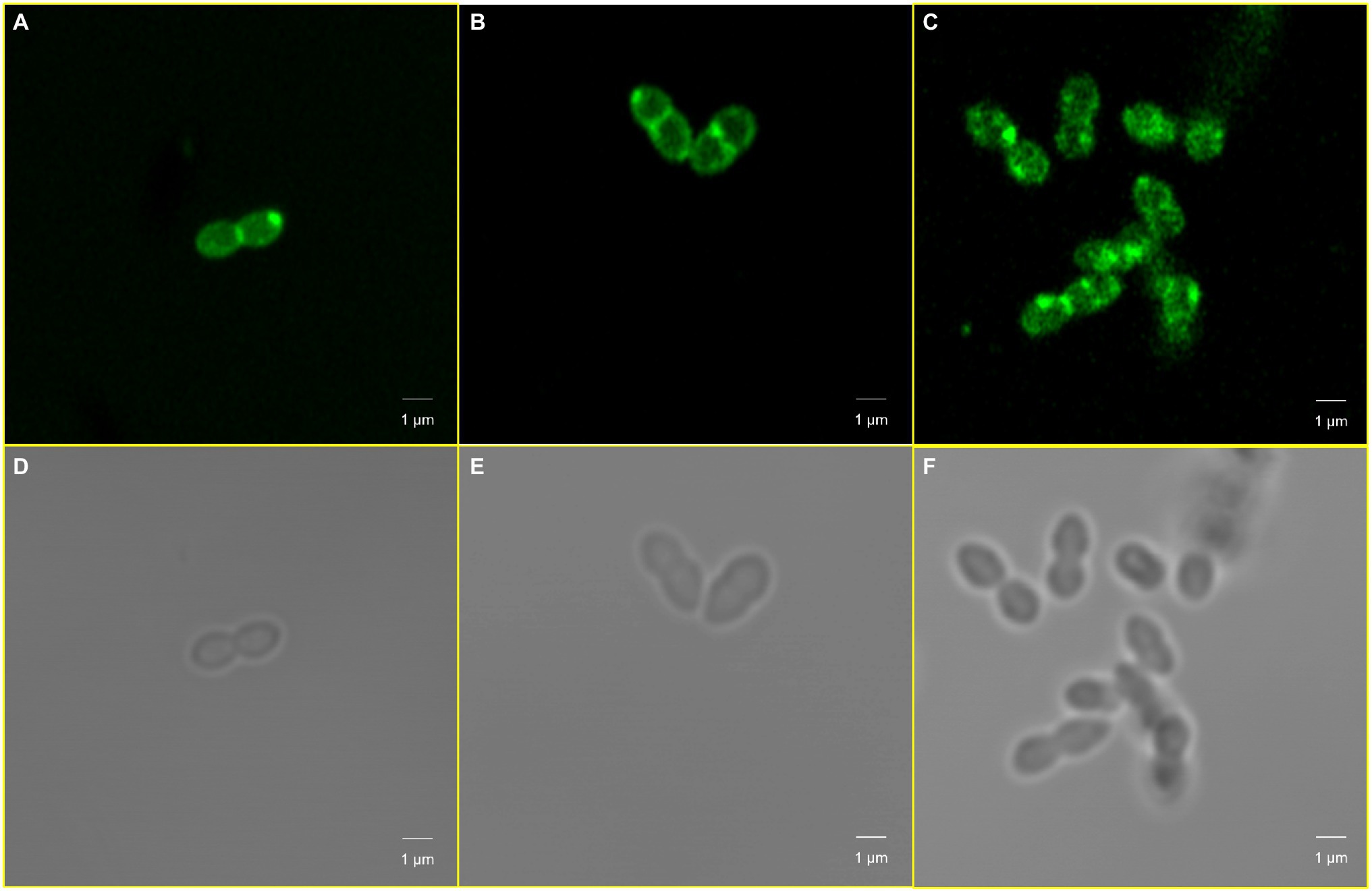
Figure 8. Cardiolipin redistribution caused by cls mutations. Panels (A,D) Enterococcus faecalis ATCC 29212; Panels (B,E) EFDO-cls; Panels (C,F) EF332-cls.
Pfam was used to predict Cls domain structures, leading to the finding that the identified I269T mutation in this work is located in one of phospholipase D-like domains (Figure 9). As a three-dimensional structure of Cls is not available, we predicted its three-dimensional structure by AlphaFold 2 (Figure 10). Interestingly, T269 of the Cls mutant is located at the bottom of a pocket that could potentially bind substrate for catalysis (Figure 10B). This is in agreement with domain structure binding, which leads to the suggestion that mutations of Cls may affect the phospholipase activity of this cardiolipin synthase.
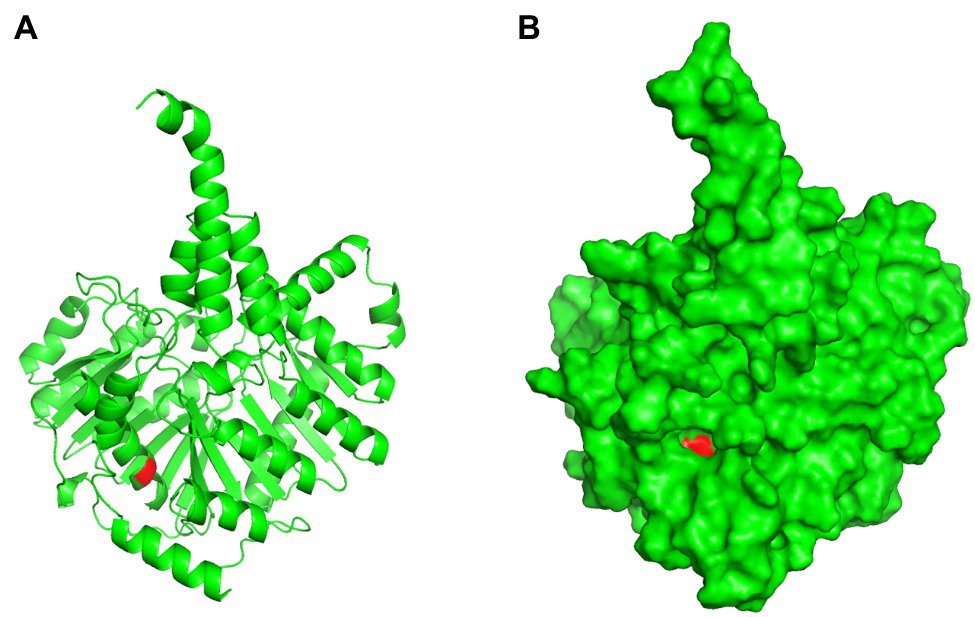
Figure 10. Three-dimensional structure prediction of Cls I269T mutant. Panel (A), cartoon presentation; Panel (B), surface presentation. Red color indicates T269 residue.
Using transmission electron microscopy, we were able to compare whether cls mutations caused changes in cell envelope and septa between cls mutant and parent strains. As shown in Figure 11, for parent strain E. faecalis ATCC 29212 and mutant strain EF332-cls, the cell surfaces and septum of both strains were smooth, and both single-septum (red arrows) and multiple-septum morphology (blue arrows) were observed and their morphology were similar. This result suggests that cls mutations does not change the structure of cell envelope.
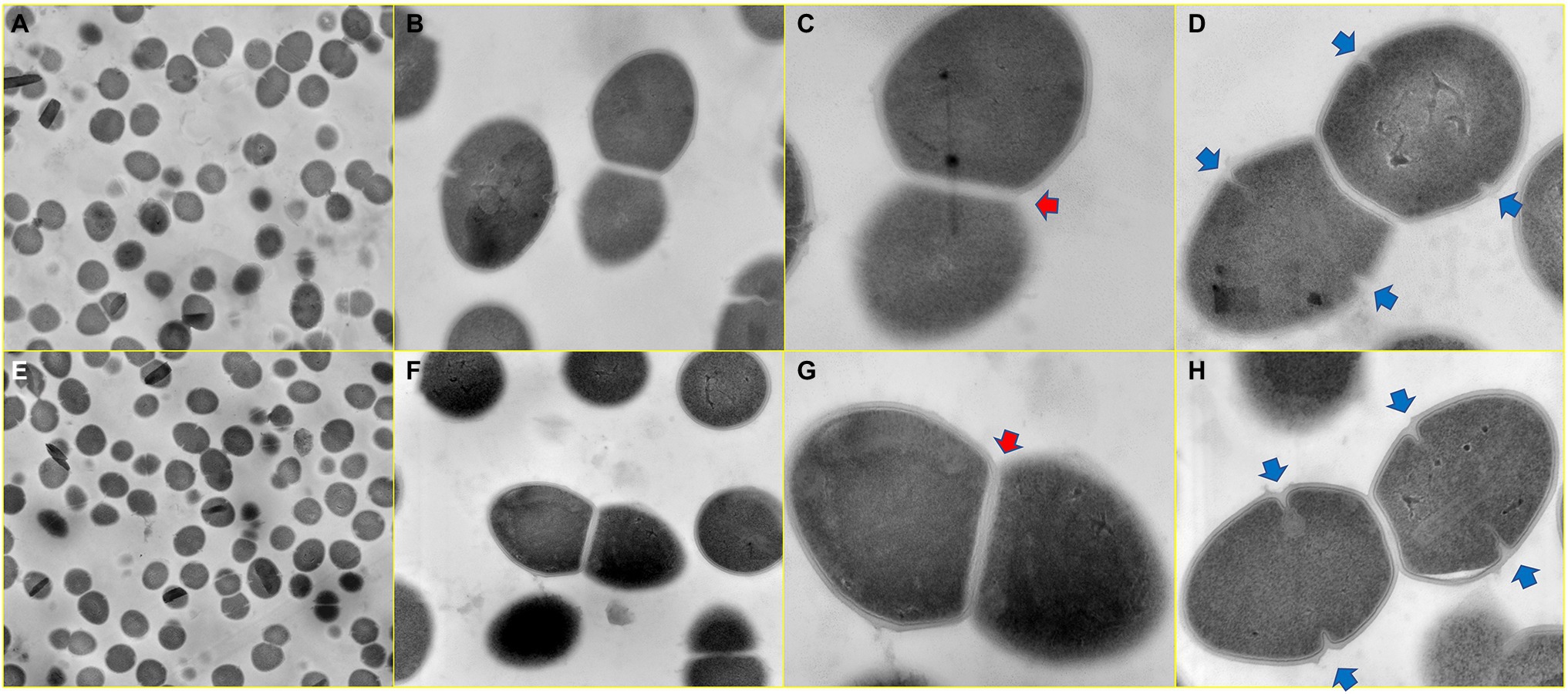
Figure 11. Ultrastructure of cls parent and mutant strains. Panels (A–D) Enterococcus faecalis ATCC 29212; Panels (E–H) EF332-cls. Image magnifications for Panels (A,E), Panels (B,F), Panels (C,G), and Panels (D,H) are 5,000, 20,000, 50,000, and 50,000, respectively. The red arrow marks a single-septum structure, and the blue arrow marks multiple-septum structures.
As VRE has spread extensively and treatment options have become increasingly limited, DAP is considered to be one of last resort antibiotics for VRE. Assessing DAP resistance, particularly in a vancomycin-resistant strain, is therefore of particular interest. In this work, a DAP-resistant VRE (E. faecium EF332) was identified and investigated to understand its mechanism for DAP resistance. Based on whole-genome sequencing and genome comparison, new mutations of two genes associated with phospholipid metabolism, cls and gdpD, were identified and suspected to be involved in DAP resistance. Mutations in the two genes were shown to be necessary for DAP resistance in previous studies (Arias et al., 2011). Therefore, investigations of the identified new mutations were carried out to verify their relationship with DAP resistance.
Results of this work suggest that mutations in cls alone can confer DAP resistance. The cls gene encodes a cardiolipin synthase, which catalyzes the production of cardiolipin from two molecules of phosphatidylglycerol. Previous studies have suggested that an increase in the concentration of cardiolipin in CM can divert more DAP from its target septum to other sites, thereby improving DAP resistance (Zhang et al., 2014; Tran et al., 2015). Some bacteria can change the membrane anionic microdomain to resist DAP by reducing the negatively charged membrane phospholipid composition (Mishra et al., 2017). While analysis of phospholipid composition by 2D-TLC showed that the cls mutations did not change the phospholipid composition of CM (including cardiolipin levels; Figure 6), the cls mutations led to decrease in negative charges on the cell surface (Figure 7) and may lead to a reduced adhesion to DAP. It has been reported that membrane lipids are asymmetrically distributed in the inner and outer leaflet of CM (Tannert et al., 2003). We therefore postulated that cls mutations do not change the level of cardiolipin synthesized but change the distribution of cardiolipin in CM, leading to reduction of cardiolipin in the outer leaflet of CM, thereby reducing membrane surface negative charges and the adhesion of DAP (Mukhopadhyay et al., 2007). It was previously suggested that DAP resistance can also be caused by redistribution of anionic phospholipids and diverting from effective targets at the septum (Tran et al., 2013b; Heidary et al., 2018). In this work, mutations of cls led to a more evenly distributed cardiolipins and reduction in septa (Figure 8C), in agreement with this theory. Therefore, we suspect the resistance to DAP found in this work is also attributable to the redistribution of cardiolipin away from the cell septa. Both events, reduction of cellular negative charge and redistribution of phospholipids, may work together or even synergistically to complete the development of DAP resistance in E. faecium. We also found that cls mutations did not lead to changes in cell ultrastructure, which is inconsistent with the previously reported results observed in S613 (DAP susceptible) and R712 (DAP-resistant, deletion of Lys at position 61) strains (Arias et al., 2011). This may be due to different effects of different mutated sites on the cell envelope and septum.
Out of the three mutations in cls, both I203V and S298T are mutations to similar amino acids, while I269T is a mutation from hydrophobic residue to polar residue. We therefore speculate that I269T is the effective mutation conferring DAP resistance. This is further supported by predictions of domain structure (Figure 9) and three-dimensional structure (Figure 10), which suggests that I269T mutation may alter the catalytic activity of cardiolipin synthase. Another DAP-resistant strain of E. faecium found by our research team also showed the same I269T mutation (data not shown), which also supports our prediction.
Although the MIC of DAP on E. faecium EF332 is not high (8 μg/ml), it has a high potential transmission risk. MLST analysis revealed that the sequence type of E. faecium EF332 is ST192, belonging to epidemic hospital strains (clade A1), which included polyclonal complex 17 (CC17; Lebreton et al., 2013, 2018). Enterococcus faecium of CC17 is high-risk clonal lineages, which is often associated with nosocomial VRE outbreak and leads to severe morbidity and mortality (Lee et al., 2019). Worryingly, plasmid pEF332-2 in this study carried two high-risk gene clusters vanA and vanM. VanA gene clusters were also found on genomic islands GIs011 and prophage (Figure 1). This means that vancomycin resistance of this strain could easily disseminate, leading to higher levels of prevalence. One more thing to note is vanM gene has only been previously reported in Shanghai, Chengdu, Hangzhou, and Beijing in China (Chen et al., 2014, 2015; Sun et al., 2019a,b). Enterococcus faecium EF332 carrying vanM genes in this study was found in Jinan of China, suggesting that vanM gene may have already spread nationwide.
In this study, a clinical DAP-resistant VRE was identified. Whole-genome sequencing revealed the genetic background for multidrug resistance. A new plasmid pEF332-2 was found to carry two vancomycin resistance clusters, a rare phenomenon reported only twice so far. The genetic basis for daptomycin was investigated, suggesting new mutations in cls gene confer DAP resistance. The mechanisms of DAP resistance were further investigated, showing that cls mutations lead to significant decrease of membrane surface negative charges and the redistribution of cardiolipin in CM, both of which contribute to DAP resistance. This work reports the genetic basis of multidrug resistance of a daptomycin- and vancomycin-resistant E. faecium, and new mutations of cls that leads to resistance of daptomycin, a key last-resort antibiotic.
To fully understand the genetic basis of multidrug resistance in E. faecium EF332, whole-genome sequences were obtained with third-generation PacBio sequencing accompanied by second-generation Illumina sequencing (The sequencing data were deposited in GenBank with accession numbers CP058891-CP058895).
HX and MW designed this study and revised the manuscript. XZ was responsible for sample collection and strain identification and participated in experimental design. HS helped to analyze membrane lipid compositions. JH, LL, MZ, QC, and YM participated in the experiments and assisted in collecting experimental data. WL performed experiments, collected and analyzed the data, and wrote the draft of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (grant number 2021YFE0199800), Key R&D Program of Shandong Province (grant number 2020CXGC011305); the National Natural Science Foundation of China (Grant numbers 31770042 and 31770043); and Shandong Provincial Natural Science Foundation (grant number ZR2020MH308).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Sen Wang, Haiyan Yu, and Yuyu Guo of the Core Facilities for Life and Environmental Sciences, State Key Laboratory of Microbial Technology of Shandong University for focused ion beam-scanning transmission electron microscope, inverted fluorescence microscope, and laser scanning confocal microscope analysis. We also thank Jingyi Zhu from State Key Laboratory of Microbial Technology of Shandong University for protein structure prediction.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.896916/full#supplementary-material
Ardui, S., Ameur, A., Vermeesch, J. R., and Hestand, M. S. (2018). Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi: 10.1093/nar/gky066
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138.Genetic
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bairoch, A., and Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48. doi: 10.1128/AAC.02435-16
Bender, J. K., Cattoir, V., Hegstad, K., Sadowy, E., Coque, T. M., Westh, H., et al. (2018). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist. Updat. 40, 25–39. doi: 10.1016/j.drup.2018.10.002
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Carver, P. L., Whang, E., VandenBussche, H. L., Kauffman, C. A., and Malani, P. N. (2003). Risk factors for arthralgias or myalgias associated with quinupristin-dalfopristin therapy. Pharmacotherapy 23, 159–164. doi: 10.1592/phco.23.2.159.32078
Cattoir, V., and Giard, J. C. (2014). Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev. Anti-Infect. Ther. 12, 239–248. doi: 10.1586/14787210.2014.870886
Chen, C., Sun, J., Guo, Y., Lin, D., Guo, Q., Hu, F., et al. (2015). High prevalence of vanM in vancomycin-resistant Enterococcus faecium isolates from Shanghai, China. Antimicrob. Agents Chemother. 59, 7795–7798. doi: 10.1128/AAC.01732-15
Chen, C., Xu, X., Qu, T., Yu, Y., Ying, C., Liu, Q., et al. (2014). Prevalence of the fosfomycin-resistance determinant, fosB3, in Enterococcus faecium clinical isolates from China. J. Med. Microbiol. 63, 1484–1489. doi: 10.1099/jmm.0.077701-0
Chow, A., Win, N. N., Ng, P. Y., Lee, W., and Win, M. K. (2016). Vancomycin-resistant enterococci with reduced daptomycin susceptibility in Singapore: prevalence and associated factors. Epidemiol. Infect. 144, 2540–2545. doi: 10.1017/S0950268816000923
CLSI (2018). Performance standards for antimicrobial susceptibility testing. Clin. Lab. Stand. Inst. 1–296.
Davlieva, M., Zhang, W., Arias, C. A., and Shamoo, Y. (2013). Biochemical characterization of cardiolipin synthase mutations associated with daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 57, 289–296. doi: 10.1128/AAC.01743-12
Depardieu, F., Podglajen, I., Leclercq, R., Collatz, E., and Courvalin, P. (2007). Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20, 79–114. doi: 10.1128/CMR.00015-06
EUCAST (2017). The European committee on antimicrobial susceptibility testing. Eur. Comm. Antimicrob. Susceptibility Test 1–146.
Fischer, A., Yang, S. J., Bayer, A. S., Vaezzadeh, A. R., Herzig, S., Stenz, L., et al. (2011). Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J. Antimicrob. Chemother. 66, 1696–1711. doi: 10.1093/jac/dkr195
Freitas, A. R., Tedim, A. P., Novais, C., Ruiz-Garbajosa, P., Werner, G., Laverde-Gomez, J. A., et al. (2010). Global spread of the hylEfm colonization-virulence gene in megaplasmids of the Enterococcus faecium CC17 polyclonal subcluster. Antimicrob. Agents Chemother. 54, 2660–2665. doi: 10.1128/AAC.00134-10
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2014). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Gonzalez-Ruiz, A., Seaton, R. A., and Hamed, K. (2016). Daptomycin: an evidence-based review of its role in the treatment of gram-positive infections. Infect. Drug Resist. 9, 47–58. doi: 10.2147/IDR.S99046
Grein, F., Müller, A., Scherer, K. M., Liu, X., Ludwig, K. C., Klöckner, A., et al. (2020). Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 11:1455. doi: 10.1038/s41467-020-15257-1
Hashimoto, Y., Taniguchi, M., Uesaka, K., Nomura, T., Hirakawa, H., Tanimoto, K., et al. (2019). Novel multidrug-resistant enterococcal mobile linear plasmid pELF1 encoding vanA and vanM gene clusters from a Japanese vancomycin-resistant enterococci isolate. Front. Microbiol. 10:2568. doi: 10.3389/fmicb.2019.02568
Heidary, M., Khosravi, A. D., Khoshnood, S., Nasiri, M. J., Soleimani, S., and Goudarzi, M. (2018). Daptomycin. J. Antimicrob. Chemother. 73, 1–11. doi: 10.1093/jac/dkx349
Herc, E. S., Kauffman, C. A., Marini, B. L., Perissinotti, A. J., and Miceli, M. H. (2017). Daptomycin nonsusceptible vancomycin resistant Enterococcus bloodstream infections in patients with hematological malignancies: risk factors and outcomes. Leuk. Lymphoma 58, 2852–2858. doi: 10.1080/10428194.2017.1312665
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Hogan, H. L., Hachem, R. Y., Neuhauser, M., Raad, I. I., and Coyle, E. (2010). Clinical experience of linezolid in bone marrow transplantation patients. J. Pharm. Pract. 23, 352–357. doi: 10.1177/0897190009358773
Hsiao, W., Wan, I., Jones, S. J., and Brinkman, F. S. L. (2003). IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19, 418–420. doi: 10.1093/bioinformatics/btg004
Humphries, R. M., Kelesidis, T., Tewhey, R., Rose, W. E., Schork, N., Nizet, V., et al. (2012). Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 56, 6051–6053. doi: 10.1128/AAC.01318-12
Hussain, K., Ullah, S., Tahir, H., Alkilani, W. Z., Naeem, M., Vinod, N. R., et al. (2016). Daptomycin- vancomycin-resistant Enterococcus faecium native valve endocarditis: successfully treated with off-label quinupristin-dalfopristin. J. Investig. Med. High Impact Case Rep. 4, 3–5. doi: 10.1177/2324709616665408
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2016). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kelesidis, T., Tewhey, R., and Humphries, R. M. (2013). Evolution of high-level daptomycin resistance in Enterococcus faecium during daptomycin therapy is associated with limited mutations in the bacterial genome. J. Antimicrob. Chemother. 68, 1926–1928. doi: 10.1093/jac/dkt117
Komagata, K., and Suzuki, K. I. (1988). 4 lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207. doi: 10.1016/S0580-9517(08)70410-0
LaPlante, K. L., and Rybak, M. J. (2004). Daptomycin – a novel antibiotic against gram-positive pathogens. Expert. Opin. Pharmacother. 5, 2321–2331. doi: 10.1517/14656566.5.11.2321
Lebreton, F., Valentino, M. D., Schaufler, K., Earl, A. M., Cattoir, V., and Gilmore, M. S. (2018). Transferable vancomycin resistance in clade B commensal-type Enterococcus faecium. J. Antimicrob. Chemother. 73, 1479–1486. doi: 10.1093/jac/dky039
Lebreton, F., van Schaik, W., Manson McGuire, A., Godfrey, P., Griggs, A., Mazumdar, V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4:00534-13. doi: 10.1128/mBio.00534-13
Lee, T., Pang, S., Abraham, S., and Coombs, G. W. (2019). Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present and the future. J. Glob. Antimicrob. Resist. 16, 36–47. doi: 10.1016/j.jgar.2018.08.016
Lellek, H., Franke, G. C., Ruckert, C., Wolters, M., Wolschke, C., Christner, M., et al. (2015). Emergence of daptomycin non-susceptibility in colonizing vancomycin-resistant Enterococcus faecium isolates during daptomycin therapy. Int. J. Med. Microbiol. 305, 902–909. doi: 10.1016/j.ijmm.2015.09.005
Li, W., Jaroszewski, L., and Godzik, A. (2002). Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18, 77–82. doi: 10.1093/bioinformatics/18.1.77
Matsumoto, K., Takeshita, A., Ikawa, K., Shigemi, A., Yaji, K., Shimodozono, Y., et al. (2010). Higher linezolid exposure and higher frequency of thrombocytopenia in patients with renal dysfunction. Int. J. Antimicrob. Agents 36, 179–181. doi: 10.1016/j.ijantimicag.2010.02.019
Mileykovskaya, E., Dowhan, W., Birke, R. L., Zheng, D., Lutterodt, L., and Haines, T. H. (2001). Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 507, 187–190. doi: 10.1016/S0014-5793(01)02948-9
Mishra, N. N., Tran, T. T., Seepersaud, R., Garcia-De-La-Maria, C., Faull, K., Yoon, A., et al. (2017). Perturbations of phosphatidate cytidylyltransferase (CdsA) mediate daptomycin resistance in Streptococcus mitis/oralis by a novel mechanism. Antimicrob. Agents Chemother. 61, 1–13. doi: 10.1128/AAC.02435-16
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mukhopadhyay, K., Whitmire, W., Xiong, Y. O., Molden, J., Jones, T., Peschel, A., et al. (2007). In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrene phospholipid composition and asymmetry. Microbiology 153, 1187–1197. doi: 10.1099/mic.0.2006/003111-0
Munoz-Price, L. S., Lolans, K., and Quinn, J. P. (2005). Emergence of resistance to daptomycin during treatment of vancomycin-resistant Enterococcus faecalis infection. Clin. Infect. Dis. 41, 565–566. doi: 10.1086/432121
Prater, A. G., Mehta, H. H., Beabout, K., Supandy, A., Miller, W. R., Tran, T. T., et al. (2021). Daptomycin resistance in Enterococcus faecium can be delayed by disruption of the LiaFSR stress response pathway. Antimicrob. Agents Chemother. 65, 1–6. doi: 10.1128/AAC.01317-20
Prater, A. G., Mehta, H. H., Kosgei, A. J., Miller, W. R., Tran, T. T., Arias, C. A., et al. (2019). Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob. Agents Chemother. 63, 3–5. doi: 10.1128/AAC.00790-19
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2011). The Pfam protein families database. Nucleic Acids Res. 32, 138D–1141D. doi: 10.1093/nar/gkh121
Qian, Y., and Zhao, X. (2014). A comparison of methods for the extraction of bacterial genomic DNA in rat feces. Sci. Technol. Food Ind. 35, 166–169. doi: 10.13386/j.issn1002-0306.2014.04.005
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. doi: 10.1038/cr.2009.6
Saier, M. H. Jr., Reddy, V. S., Tamang, D. G., and Västermark, Å. (2013). The transporter classification database. Nucleic Acids Res. 42, D251–D258. doi: 10.1093/nar/gkt1097
Schwarz, F. V., Perreten, V., and Teuber, M. (2001). Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46, 170–187. doi: 10.1006/plas.2001.1544
Sletvold, H., Johnsen, P. J., Simonsen, G. S., Aasnæs, B., Sundsfjord, A., and Nielsen, K. M. (2007). Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob. Agents Chemother. 51, 736–739. doi: 10.1128/AAC.00557-06
Suleyman, G., Mahan, M., and Zervos, M. J. (2017). Comparison of daptomycin and linezolid in the treatment of vancomycin-resistant Enterococcus faecium in the absence of endocarditis. Infect. Dis. Clin. Pract. 25, 151–154. doi: 10.1097/IPC.0000000000000482
Sun, H. L., Liu, C., Zhang, J. J., Zhou, Y. M., and Xu, Y. C. (2019a). Molecular characterization of vancomycin-resistant enterococci isolated from a hospital in Beijing, China. J. Microbiol. Immunol. Infect. 52, 433–442. doi: 10.1016/j.jmii.2018.12.008
Sun, L., Qu, T., Wang, D., Chen, Y., Fu, Y., Yang, Q., et al. (2019b). Characterization of vanM carrying clinical Enterococcus isolates and diversity of the suppressed vanM gene cluster. Infect. Genet. Evol. 68, 145–152. doi: 10.1016/j.meegid.2018.12.015
Sun, L., Xu, J., Wang, W., and He, F. (2020). Emergence of vanA-type vancomycin-resistant Enterococcus faecium ST 78 strain with a rep2-type plasmid carrying a Tn1546-like element isolated from a urinary tract infection in China. Infect. Drug Resist. 13, 949–955. doi: 10.2147/IDR.S247569
Tannert, A., Pohl, A., Pomorski, T., and Herrmann, A. (2003). Protein-mediated transbilayer movement of lipids in eukaryotes and prokaryotes: the relevance of ABC transporters. Int. J. Antimicrob. Agents 22, 177–187. doi: 10.1016/S0924-8579(03)00217-6
Tran, T. T., Munita, J. M., and Arias, C. A. (2015). Mechanisms of drug resistance: daptomycin resistance. Ann. N. Y. Acad. Sci. 1354, 32–53. doi: 10.1111/nyas.12948
Tran, T. T., Panesso, D., Gao, H., Roh, J. H., Munita, J. M., Reyes, J., et al. (2013a). Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57, 261–268. doi: 10.1128/AAC.01454-12
Tran, T. T., Panesso, D., Mishra, N. N., Mileykovskaya, E., Guan, Z., Munita, J. M., et al. (2013b). Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio 4:e00281-13. doi: 10.1128/mBio.00281-13
Uttley, A. C., Collins, C. H., Naidoo, J., and George, R. C. (1988). Vancomycin-resistant enterococci. Lancet 331, 57–58. doi: 10.1016/S0140-6736(88)91037-9
Vading, M., Samuelsen, H. B., Sundsfjord, A. S., and Giske, C. G. (2011). Comparison of disk diffusion, Etest and VITEK2 for detection of carbapenemase-producing Klebsiella pneumoniae with the EUCAST and CLSI breakpoint systems. Clin. Microbiol. Infect. 17, 668–674. doi: 10.1111/j.1469-0691.2010.03299.x
Zhang, T. H., Muraih, J. K., Tishbi, N., Herskowitz, J., Victor, R. L., Silverman, J., et al. (2014). Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J. Biol. Chem. 289, 11584–11591. doi: 10.1074/jbc.M114.554444
Keywords: vancomycin-resistant enterococci, Enterococcus faecium, daptomycin resistance, vancomycin resistance, membrane surface charge, gdpD, cls
Citation: Li W, Hu J, Li L, Zhang M, Cui Q, Ma Y, Su H, Zhang X, Xu H and Wang M (2022) New Mutations in cls Lead to Daptomycin Resistance in a Clinical Vancomycin- and Daptomycin-Resistant Enterococcus faecium Strain. Front. Microbiol. 13:896916. doi: 10.3389/fmicb.2022.896916
Received: 15 March 2022; Accepted: 19 May 2022;
Published: 21 June 2022.
Edited by:
Hidetada Hirakawa, Gunma University, JapanCopyright © 2022 Li, Hu, Li, Zhang, Cui, Ma, Su, Zhang, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuhua Zhang, Y2hoODEwNUAxMjYuY29t; Hai Xu, aGFpeHVAc2R1LmVkdS5jbg==; Mingyu Wang, d2FuZ21pbmd5dUBzZHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.