
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 29 April 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.895537
The human oral cavity provides a habitat for oral microbial communities. The complexity of its anatomical structure, its connectivity to the outside, and its moist environment contribute to the complexity and ecological site specificity of the microbiome colonized therein. Complex endogenous and exogenous factors affect the occurrence and development of the oral microbiota, and maintain it in a dynamic balance. The dysbiotic state, in which the microbial composition is altered and the microecological balance between host and microorganisms is disturbed, can lead to oral and even systemic diseases. In this review, we discuss the current research on the composition of the oral microbiota, the factors influencing it, and its relationships with common oral diseases. We focus on the specificity of the microbiota at different niches in the oral cavity, the communities of the oral microbiome, the mycobiome, and the virome within oral biofilms, and interventions targeting oral pathogens associated with disease. With these data, we aim to extend our understanding of oral microorganisms and provide new ideas for the clinical management of infectious oral diseases.
- The communities of bacteria, fungi, and viruses in the oral cavity are discussed.
- The regulatory mechanisms linking oral microorganisms with host health and disease states are described.
- The specificity of the oral microbiota at different niches is summarized.
- Interventions targeting oral diseases caused by oral pathogens are discussed.
Oral microorganisms, including bacteria, archaea, fungi, viruses, and protozoa, are closely associated with oral disease processes. It has been demonstrated that the diversity of the oral microbiome in children increases after they acquire their initial colonizing microorganisms (Gomez and Nelson, 2017). A homeostatic balance is maintained between the host and the oral microbial community through a variety of bidirectional communication and regulatory mechanisms during a person’s life. However, oral infectious diseases, such as caries, periodontal disease, and oral candidiasis, can be induced by the dysbiosis of the oral microbiota (Lamont et al., 2018). The development of cancers [e.g., head and neck squamous cell cancer (Hayes et al., 2018), pancreatic cancer (Fan et al., 2018), and colorectal cancer (Flemer et al., 2018)] and various systemic diseases, including rheumatoid arthritis (Rodríguez-Lozano et al., 2019), hypertension (Czesnikiewicz-Guzik et al., 2019), Alzheimer’s disease (Liu et al., 2019), and systemic lupus erythematosus (SLE) (Pessoa et al., 2019), have been shown to be related to the oral microbiota. In the past, the limitations of traditional pure culture techniques have meant that studies of the oral microbiome focused mainly on bacteria. However, in recent years, with the development of sequencing technologies and molecular investigative methods, oral fungi, the candidate phyla radiation (CPR) group (a unique class of bacteria that co-parasitize the host), and viruses are becoming hot topics in the ecology of the human oral microbiota (Baker et al., 2017). However, until now, determining the communication and regulatory mechanisms of oral microorganisms and oral diseases has remained a challenge.
In this review, the composition of the oral microbiota and the correlations between it and oral and systemic diseases are examined. The specificity of the microbiota at different niches in the oral cavity and the interaction mechanisms between oral microorganisms and their host are also highlighted. These data should provide support for the prevention and treatment of oral diseases.
There are about 1,000 species of bacteria in the oral cavity, mainly including the phyla Actinobacteria, Bacteroidetes, Chlamydia, Euryarchaeota, Fusobacteria, Firmicutes, Proteobacteria, Spirochaetes, and Tenericutes (Dewhirst et al., 2010). There are also a few lesser-known phyla and candidate divisions in the oral cavity, including Chloroflexi, Chlorobi, GN02, Synergistetes, SR1, TM7, and WPS-2 (Camanocha and Dewhirst, 2014). Among these, GN02, SR1, and TM7 belong to the CPR. Oral CPR members are thought to influence the oral microbial ecology by modulating the structural hierarchy and functions of the oral microbiome (Bor et al., 2019), which have been shown to correlate with oral diseases, such as periodontitis (Griffen et al., 2012) and halitosis (Takeshita et al., 2012). However, the pure culture of the CPR is difficult, and only TM7 had been cultured from the human oral cavity until November 2021 (Bor et al., 2020).
Oral archaea are far less numerous and diverse than oral bacteria. Although oral archaea were originally thought to be exclusively methanogens (Wade, 2013), many studies have detected DNA sequences of non-methanogenic bacteria in the oral cavity (Li et al., 2009; Dame-Teixeira et al., 2020). Surprisingly, archaea are currently recognized as non-pathogenic, although they have been detected in inflamed pulp tissue (Efenberger et al., 2015), in subgingival biofilms from patients with peri-implantitis and periodontitis (Aleksandrowicz et al., 2020), and in caries biofilm samples (Dame-Teixeira et al., 2020). These findings suggest that archaea are involved in the pathogenic processes of oral diseases. However, their potential pathogenicity requires further investigation.
There are approximately 100 species of fungi in the oral cavity. The common genera are Aspergillus, Aureobasidium, Candida, Cladosporium, Cryptococcus, Fusarium, Gibberella, Penicillium, Rhodotorula, Saccharomycetales, and Schizophyllum (Ghannoum et al., 2010; Peters et al., 2017). It has been confirmed that fungi account for 0.004% of the overall oral microorganisms and have only been detected in specimens from the hard palate, supragingival plaque, and oral rinses, possibly due to the nature of the participants and the low prevalence of fungi at other sites (Caselli et al., 2020). However, recent investigation detected a broad range of fungi in saliva samples and identified two well-demarcated genus-level community types (Malassezia and Candida) (Hong et al., 2020). It is noteworthy that oral fungal species show high interindividual variability in oral rinse samples from healthy individuals (Ghannoum et al., 2010).
The oral virome consists of eukaryotic viruses and phages (Caselli et al., 2020). These eukaryotic viruses mainly include Anelloviridae, Herpesviridae, and Papillomaviridae (Bottalico et al., 2011; Wylie et al., 2014), whereas the phages are more diverse and have been mainly studied because their capacity for bacterial lysis allow them to be used to treat bacterial infectious diseases (Shen et al., 2018; Chen J. et al., 2020). In one study, many oral viruses were not transient members of the oral ecosystem but persisted throughout the study period (Abeles et al., 2014). It has also been suggested that the viruses in saliva act as hosts for pathogenic genes in the oral environment (Pride et al., 2012). At present, the consensus on oral viruses is that they are personal, persistent, and gender-specific. Viral communities are also highly similar in the same environments (e.g., belong to the same family) (Robles-Sikisaka et al., 2013; Abeles et al., 2014).
The Human Microbiome Project (HMP) originally raised the question of whether a core human microbiome exists. A “core microbiome” is defined as the set of shared genes found in a given habitat in all humans (Turnbaugh et al., 2007). The term has evolved into five different types: the common core, temporal core, ecological core, functional core, and host-adapted core. These are defined according to their spatial distribution, temporal stability, ecological impact, and effects on host functions and health (Risely, 2020). Therefore, the oral core microbiome is the collection of microorganisms colonizing the oral cavity and their genomes, which are organized according to the five definitions mentioned above. Caselli et al. showed that there were no statistically significant differences in the microbiomes of the subjects enrolled in their study, supporting the idea that the oral core microbiome can be used as a reference for a definition of eubiosis (Caselli et al., 2020). A 16S rDNA analysis showed that Actinobacteria (genera Corynebacterium, Rothia, and Actinomyces), Bacteroidetes (genera Prevotella, Capnocytophaga, and Porphyromonas), Firmicutes (genera Streptococcus and Granulicatella), Fusobacteria (genus Fusobacterium), and Proteobacteria (genera Neisseria and Haemophilus) are predominant in the microbiomes of healthy people (Zaura et al., 2009). The oral microbiota also differs according to the oral niches and changes dynamically across life-history stages. For example, an analysis of saliva, supragingival, and mucosal plaque samples from healthy volunteers at different ages and stages of dentition revealed that the oral cavity is a highly heterogeneous ecological system containing distinct niches with significantly different microbial communities (Xu et al., 2015). Physiological hormones cause changes in Campylobacter, Haemophilus, Oribacterium, and Prevotella, during the female menstrual cycle (Bostanci et al., 2021). The oral abundances of Neisseria, Porphyromonas, and Treponema are also higher in pregnant women than in non-pregnant women. However, the abundances of Streptococcus and Veillonella are richer in non-pregnant women than in pregnant women (Saadaoui et al., 2021). These data suggest that the core oral microbiota should be defined based on the life-history stage of the host and its specific ecological locus in the oral cavity. The association between the core oral microbiota and host microecological homeostasis, health, and disease status can help us better understand the interactions among various oral microorganisms and between oral microorganisms and their hosts. Many studies are investigating the precise composition of the core oral microbiota to confirm the functional combinations of microorganisms involved in complex microecological and disease processes (Jiang et al., 2015; Oliveira et al., 2021). However, any individually defined core microbiota will not include all key microorganisms throughout the host’s entire life (Risely, 2020).
In recent years, two major oral microbiome databases have been established, the Human Microbiome Project (HMP) and the Human Oral Microbiome Database (HOMD). HMP aims to characterize the ecology of human microbial communities and to analyze the roles of microorganisms in human health and diseases. It maps the metagenomics of microorganisms in five major parts of the human body: the oral cavity, nasal cavity, vagina, intestine, and skin (Integrative HMP (iHMP) Research Network Consortium, 2019). The oral microbiome is one of the research priorities of HMP. Nine specimens are collected from the oral cavity and oropharynx, including the saliva, buccal mucosa, keratinized gingiva, palate, tonsil, throat, tongue soft tissues, supragingival dental plaque, and subgingival dental plaque (The Human Microbiome Project Consortium, 2012). HOMD is a database of oral microorganisms launched in 2008 by the National Institute of Dental and Craniofacial Research (Bethesda Maryland, United States). Since then, the expanded Human Oral Microbiome Database (eHOMD) has provided comprehensive information on the bacterial species present in the human aerodigestive tract. The eHOMD already contains 775 microbial species, 687 of which are from HOMD version 14.51.
In summary, the development of DNA sequencing technologies and data analysis methods has greatly extended our understanding of the oral microbiome, mycobiome, and virome. Many studies have grossly underestimated the complexity of the human oral microbiome (Keijser et al., 2008; Caselli et al., 2020). Oral microorganisms often colonize different sites in the oral cavity and form distinct microbial communities on living or non-living surfaces. A comparative analysis of the microbiomes at different niches in the oral cavity has demonstrated the significant spatial specificity of the oral microbiota (Xu et al., 2015).
To explore the information related to the composition of oral microbiota at different oral niches and the influence of the niches on the composition and structure of the oral microbiota, the oral cavity is usually divided into several parts for sampling, mainly the saliva, hard tissue surface of teeth, soft tissue surfaces of the tongue, buccal mucosa, palate, and gingiva (Zhang et al., 2018).
The microbial composition of saliva includes the microorganisms shed from various oral niches. Therefore, saliva is considered to be a reservoir of the oral microbiota and also a fingerprint of the entire oral microbiota. However, recent studies have shown that the salivary microbiome does not represent the entire oral microbiome (Simón-Soro et al., 2013; Ren et al., 2017; Shi et al., 2018), but is significantly less representative than the microbiome of the supragingival plaque (Shi et al., 2018). Firmicutes are more abundant in the saliva, whereas Actinobacteria and Fusobacteria are more abundant in dental plaque (Shi et al., 2018). Further analysis has confirmed that the microbiota in oral rinse fluid samples is more representative of the entire site-specific microbiota than that in the saliva (Caselli et al., 2020). A study of the salivary microbiome in 997 members of the Qatari population found that the predominant member of the salivary microbiota was Bacteroidetes, and that Gemella, Haemophilus, Neisseria, Porphyromonas, Prevotella, Streptococcus, and Veillonella were the most prevalent genera (Murugesan et al., 2020). However, the microbial composition of the saliva from Japanese (Takeshita et al., 2016) and Chinese populations (Xun et al., 2018) differed from these results. These differences may be influenced by genetic, dietary, and environmental factors (Shaw et al., 2017). Nevertheless, the changes in the composition of the salivary microbiome can be used as a biomarker to monitor the prevalence and prognosis of diseases such as caries, oral cancer, and periodontal disease, and to reflect the oral and general health status (Lee et al., 2017; Yamashita and Takeshita, 2017).
Saliva is thought to greatly influence the colonization and clearance of microorganisms and to play important roles in the physical, chemical, and immunological defenses of the oral cavity (Fábián et al., 2012). It is involved in the formation of a protective liquid layer, the acquired pellicle, which participates in the microbial adhesion and colonization processes (Lynge Pedersen and Belstrøm, 2019). Saliva also contains a variety of defense proteins that are involved in the immunity of the oral mucosa, such as salivary immunoglobulins, antimicrobial peptides, lysozyme, α-amylase, mucin, peroxidases, and statherin (Fábián et al., 2012). For example, anxiety and depressive symptoms can affect the abundances of Actinomyces, Fusobacterium, Leptotrichia, Spirochaetaceae, and Treponema by regulating cortisol and C-reactive protein in the saliva (Simpson et al., 2020). Endogenous antimicrobial lipids also have antibacterial, antifungal, antiviral, and antiparasitic activities (Brasser et al., 2011; Fischer, 2020). The host-derived tRNA-derived small RNAs (tsRNAs) in saliva, which share sequence similarities with bacterial tRNAs, act as cross-domain mediators to regulate host-associated bacterial growth (He et al., 2018).
The tooth surface is the only non-shedding surface in the oral cavity and therefore provides an ideal environment for bacterial growth and the formation of dental plaque. It has been shown that dental plaque has higher α-diversity, microbial richness, and evenness than samples of saliva and tongue (Ren et al., 2017). It is noteworthy that the plaque on the supragingival enamel surface is formed by the acquired pellicle (Lynge Pedersen and Belstrøm, 2019), whereas at subgingival sites, the gingival crepuscular fluid acts as a special source of nutrition, and its serum proteins combine with salivary proteins to form a unique proteinaceous film. The physical barrier effect of the gingiva also significantly reduces the oxygen tension and shear forces at subgingival sites, causing the compositions and structures of the supragingival and subgingival plaque microbiotas to differ (Costalonga and Herzberg, 2014). For example, anaerobic bacteria, including Actinomyces, Fusobacterium, and Veillonella, are predominantly found in the subgingival plaque (Caselli et al., 2020). The oral microbial composition on tooth surface is also influenced by the anatomy and physiology of the tooth surfaces and the perigingival area. The crown surface of the same tooth can be divided into five different areas: the occlusal or chewing surface, the proximal surface or contact point between teeth, the supragingival surface, the buccal or cheek-contacting surface, and the lingual surface proximate to the tongue (Costalonga and Herzberg, 2014). There are significant differences in the bacterial compositions at these different locations (Simón-Soro et al., 2013; Costalonga and Herzberg, 2014). For example, Streptococcus spp. are found on the labial surface of incisors and cuspids in 40–70% of cases, but hardly ever on the lingual surface (Fábián et al., 2012; Simón-Soro et al., 2013).
The structural characteristics of the oral mucosal epithelial layer mean that it is shed continuously. Although it is still continuously colonized by oral microorganisms, these microorganisms are relatively restricted (Costalonga and Herzberg, 2014). Streptococcus is the most abundant genus in mucosal tissue (Caselli et al., 2020). Interestingly, the tongue has a higher density and greater diversity of microorganisms than other mucosal surfaces (Zhang et al., 2018). Facultative and obligate anaerobes, including Actinomyces, Porphyromonas, Prevotella, Streptococcus, and Veillonella, are the main microorganisms in the tongue coating, probably because an anaerobic environment is provided by the tongue papillae (Ren et al., 2017). Haemophilus, Leptotrichia, and Neisseria are also abundant in tongue samples. Notably, the tongue biofilm is closely associated with halitosis (Zhang et al., 2021b).
It is worth noting that the oral mucosa, as the main anatomical barrier of the oral cavity, has both microbial immune barrier and physical barrier functions. The immunity of the oral mucosa is part of the body’s immune system, which also influences the composition of oral microorganisms (Fábián et al., 2012). Pattern recognition receptors are mainly expressed on immune cells, and their recognition of pathogen-associated molecules activates a variety of intracellular signaling pathways, which trigger proinflammatory and antimicrobial responses (Şenel, 2021). For instance, oral immune cells recognize the cell wall components of Candida albicans, and then exert antifungal effects (Zhou Y. et al., 2021). In a healthy host, C. albicans exists as a non-toxic yeast that induces little immune response (Garcia-Rubio et al., 2019). However, when it transforms into the mycelial state, it activates the interleukin 17 (IL17)/Th17 immune pathway (Gaffen and Moutsopoulos, 2020). A deficiency in the Th17 pathway is associated with fungal overgrowth, which may be one reason that immunocompromised people are more susceptible to oral candidiasis (Smeekens et al., 2014).
In response to salivary flow, mastication, chewing, and other factors, these locations in the oral cavity have different physicochemical properties, such as nutrients, pH, oxygen tension, and shear forces, which select suitable microorganisms for each oral niche. As a result, these oral niches display different compositions of oral microbiota. Furthermore, oral niches, such as the tongue and teeth, also provide physical support for the formation of oral microbial biofilms.
Oral microbial biofilms are complex ecological environments with plentiful and diverse oral microorganisms, which are associated with various oral diseases (Rath et al., 2021). Based on the acquired pellicle formed by salivary proteins, the initial colonizing bacteria, such as Streptococcus gordonii, Streptococcus mitis, Streptococcus oralis, and Streptococcus sanguinis, bind specifically to their complementary salivary receptors through their surface adhesins (Kolenbrander et al., 2010). Extracellular polysaccharides, structural proteins, cell fragments, and nucleic acids then contribute to extracellular polymeric substances (EPS) (Heller et al., 2017). Ultimately, a three-dimensional ecosystem is formed, composed of a variety of microorganisms, EPS, proteins, and lipids from food and saliva, as well as voiding and conduit systems (Bowen et al., 2018; Rath et al., 2021).
Because of its strong and extensive adhesive ability, Fusobacterium nucleatum establishes interdependent relationships with other members of the oral microbiota, playing an bridging role among the early and later bacterial colonizers (Kriebel et al., 2018). The adhesion ability of F. nucleatum mainly depends on its surface outer membrane proteins, which can be divided into the lactose inhibitory type (such as Fap2) and the amino acid inhibitory type (such as RadD and FomA) (Brennan and Garrett, 2019). RadD generally mediates colonization by Gram-positive bacteria, such as Staphylococcus aureus, Streptococcus gordonii, and S. mutans, and even C. albicans (Mutha et al., 2018; Lima et al., 2019). The coadhesion ability of a radD-gene-deleted F. nucleatum strain with S. gordonii was reduced. The lack of fad-I, a gene expressed immediately upstream from radD, increases the expression of radD, and then increases the coaggregation of F. nucleatum with S. gordonii and subsequent biofilm formation. These findings suggest that the lipoprotein Fad-I plays an important role in the regulation of RadD adhesion (Shokeen et al., 2020). Another small fusobacterial lipoprotein, Aid1, also participates in the interspecies interactions of F. nucleatum, especially with oral streptococci and related Gram-positive species (Kaplan et al., 2014). Moreover, the protein FomA (Puth et al., 2019), the autonomic transporter Fap2 (Silbergleit et al., 2020), and the newly discovered arginine inhibitory adhesive designated “coaggregation mediating protein A” (Lima et al., 2017) mediate the interaction between F. nucleatum and different oral microorganisms and also affect the formation of oral biofilms. And the biofilm thickness, stability, and architecture in five subspecies of F. nucleatum are varied, suggesting their different adhesion abilities and roles in biofilm formation (Muchová et al., 2022). Corynebacterium and Veillonella also have been confirmed to play a bridging role in oral biofilm development (Kriebel et al., 2018; Zhou P. et al., 2021).
During the maturation of oral microorganism biofilms, the main bacteria of the biofilms change from Streptococcus in the early stage to obligate anaerobes later, such as Capnocytophaga, Fusobacterium, Porphyromonas, and Prevotella, and especially Actinomyces (Takeshita et al., 2015). They gradually form complex multispecies structures, such as corncobs, palisades, and test tubes, which provide the best biochemical conditions for these members of the microbiota (Zijnge et al., 2010; Keijser et al., 2018). It has discovered a distinctive, multigenus consortium in the dental plaque, which shows the significance of microbial biogeography at the micron scale. The highly organized hedgehog structure mainly consist nine taxa, representing a radially organized spatial framework, in which Corynebacterium filaments radiate outward from near the center, with other species and their fibers as well as corncob structure around the distal tips of it (Mark Welch et al., 2016). In accordance with their biochemical characteristics, obligately anaerobic microorganisms tend to occur internally, whereas facultatively or obligately aerobic microorganisms tend to occur peripherally, and the microorganisms that consume or produce the same metabolite tend to occur in close proximity to each other (Mark Welch et al., 2016; Kriebel et al., 2018). In terms of bacterial morphologies and fluorescence intensities, subgingival plaque can be divided into four different layers: eukaryotic cells, top layer, intermediate layer, and basal layer (Zijnge et al., 2010). The micron-scale structure elaborates the roles of individual oral microorganisms and provides a comprehensive understanding of their community ecology and potential pathogenicity (Mark Welch et al., 2020). In addition, using multiscale imaging, sequencing analysis, and other techniques, recent investigation found that an alternative biofilm development process in multiple species, even containing human epithelial cells, shape polymicrobial communities at various spatial and taxonomic scales (Simon-Soro et al., 2022).
Coaggregation occurs when two paired individual bacteria are suspended in the planktonic phase. However, “coadhesion” usually refers to a situation in which one microorganism is fixed to a surface and the coadhering microorganism is suspended (Ruhl et al., 2014). Binding is extremely important in the formation of multispecies biofilms, and depends on the adhesins expressed on the surface of one bacterium and the complementary polysaccharide-containing receptors expressed by the other species (Chen et al., 2019). S. gordonii, which occurs widely in oral microbial biofilms, can coaggregate with Actinomyces (Mohammed et al., 2018), Fusobacterium (Mutha et al., 2018), Streptococcus (Choo et al., 2021), and Veillonella (Zhou P. et al., 2021), and plays an important role in the formation of biofilms. A molecular analysis showed that the antigen I/II (AgI/II) family proteins, with highly conserved structures and basic sequences, are detectable on the surfaces of all Streptococcus in the human oral cavity (Choo et al., 2021). SspA protein, a member of the AgI/II family of S. gordonii, recognizes a polysaccharide of Actinomyces oris T14V containing glucose, mannose, and galactose, which facilitates the coaggregation of the two bacteria. Significantly, this combination has a certain specificity. For instance, this polysaccharide only binds to SspB expressed by Lactococcus lactis, but does not bind to other L. lactis AgI/II family proteins (Back et al., 2015). A recent study showed that the novel intrageneric coadhesion between S. agalactiae and S. mutans is promoted by glucoside transferase B (GtfB) and GtfC (Liu et al., 2020). Another investigation showed that none of the six Lactobacillus strains tested, including clinical isolates from children with caries, could form biofilms in vitro. However, when grown with S. mutans, all Lactobacillus showed a significant increase in biofilm formation. This phenomenon was related the transfer of GtfB from S. mutans to the Lactobacillus species (Wen et al., 2017).
Communication between the same or different species is mediated by chemical signals synthesized and secreted by bacteria. These signals can be divided into two groups. One group is related to cell density and quorum sensing (QS), and the other group involves signals produced by bacteria at various stages of growth (Jayaraman and Wood, 2008). QS is a microbial cell-to-cell communication process principally dependent upon the population density. As the population density increases, signals accumulate locally, and once a threshold concentration is reached, the autoinducer interacts with the receptor protein. When different target genes are activated, the physiological behavior of the bacteria, such as their virulence, competition, pathogenicity, and drug resistance, is altered (Abisado et al., 2018). Recent studies have mainly focused on the roles of acyl-homoserine lactones produced by Gram-negative bacteria, autoinducing peptides produced by Gram-positive bacteria, the LuxS/AI-2 QS system, and other signaling molecules (Wu et al., 2020). Autoinducer-2 (AI-2), detected in multiple oral pathogens and symbiotic bacteria, is regarded as a universal signaling molecule in interactions between bacterial species (Muras et al., 2022). A scanning electron microscopic analysis showed that the LuxS/AI-2 QS system had no effect on the morphology of S. mutans, but affected its growth (Yuan et al., 2021). Another investigation found that the LuxS/AI-2 QS system of S. gordonii influenced the structure and composition of the dual-species biofilm, formed with S. mutans, and its sensitivity to chlorhexidine. AI-2 synthesized in vitro mediated S. mutans biofilm formation and the expression of its virulence genes (Wang et al., 2017).
Candida albicans, the most abundant fungus in the oral cavity, has been studied extensively (Ghannoum et al., 2010; Ellepola et al., 2019). The formation of mycelia and its transformation from yeast into mycelia are considered to be its key virulence factors (Priya and Pandian, 2020). In the oral cavity of children with severe early childhood caries, the presence of C. albicans significantly increases the abundance of highly acidogenic and acid-tolerant bacteria, such as S. mutans, Lactobacillus, and Scardovia species, and the activity of Gtfs in the dental plaque (Xiao et al., 2018a). The production of EPS is correspondingly increased, which could accommodate more caries-active S. mutans (Falsetta et al., 2014; Xiao et al., 2018a). An analysis of the interaction between C. albicans and S. mutans revealed that when the two strains are mixed and cultured, the carbohydrate-metabolism-related genes and proteins of C. albicans are significantly enhanced. The concentrations of other substances, such as mannan and glucan, also increased. These data indicate that mixed incubation can enhance fungal activity (Ellepola et al., 2019). A further investigation also showed that the GtfB secreted by S. mutans mainly binds to the C. albicans mannan layer, and that this mechanism greatly promotes the formation of the extracellular matrix and the development of mixed-species biofilms (Hwang et al., 2017).
The interactions between mixed bacteria are not unidirectional, but are interdependent and mutually beneficial. Symbiotic cooperative relationships depend to some extent on AgI/II. When S. mutans was cultured with C. albicans, the total biofilm biomass increased significantly. However, the loss of spaP, a gene encoding AgI/II in S. mutans, apparently reduced the abundance of C. albicans both in vitro and in vivo. Interestingly, the number of spaP mutant cells did not change in vitro, but decreased significantly in vivo when coinfected with C. albicans. These results suggest that AgI/II is required for the promotion of the two species biofilm (Yang C. et al., 2018). Another study found that the induction of SigX, an alternative sigma factor of S. mutans induced by QS signals, was observed in dual-species biofilms, but not in single-species biofilms (Sztajer et al., 2014). It has been demonstrated that the recognition of C. albicans by S. gordonii involves Als3 and SspB interaction, providing a new mechanism for the communication between fungi and bacteria (Silverman et al., 2010). As well as its interaction with S. mutans, C. albicans also interacts with Actinomyces naeslundii (Arzmi et al., 2015). Compared with single-species biofilms, the cross-kingdom dual-species biofilm formed by A. viscosus and C. albicans showed significantly enhanced cariogenic virulence (Deng et al., 2019).
In recent years, research into oral microbial biofilms has shifted from the detection and investigation of the microbes involved to the analysis of the EPS, extracellular proteins, and other matrix components. The matrix helps cells survive in their respective niches, plays a key role in meshing microbial cells, provides three-dimensional scaffolds to limit the cells’ diffusion, and ensures microenvironmental heterogeneity (Castillo Pedraza et al., 2017). In particular, the highly structured matrix in dental plaque produced by pathogenic S. mutans provides binding sites for other microorganisms (Bowen et al., 2018; Cugini et al., 2019). With a better understanding of the functions and effects of various matrix components, the development of matrix inhibitors has recently become a hot topic.
After the colonization of the oral cavity, oral microorganisms undergo ecological succession and gradually form stable microbial communities. However, the ecological balance is also influenced by a variety of endogenous and exogenous factors. These factors, including drug use, host lifestyle, environment, and host status, contribute to differences in disease susceptibility by influencing the composition, structure, and metabolic functions of oral microorganisms.
Antibiotics not only affect the composition and functions of the oral microbiome, but also induce specific metabolic changes during antibiotic interventions (Moraes et al., 2020). Like previous studies (Monroy-Pérez et al., 2020), a prospective cohort study found that Shannon’s diversity index was reduced by treatment with amoxicillin relative to that in the untreated group, and decreased continuously for 6 months (Menon et al., 2019). It is noteworthy that the relative abundance of Actinobacteria decreased by 60% in the antibiotic-treated group after 1 week and remained 50% lower than baseline after 6 months. The abundances of Bacteriodetes and Fusobacteria also decreased significantly. Surprisingly, the abundance of Proteobacteria increased at 1 month and Aggregatibacter paraphrophilus was detected at 1 month and persisted to the 3- and 6-month time points. However, no operational taxonomic units persisted up to 6 months in the control group (Menon et al., 2019). The use of amoxicillin has also been shown to reduce the abundances of Neisseria, Streptococcus, and Veillonella in the oral microbiota (Larsson Wexell et al., 2016; Moraes et al., 2020). In another study, a group of neonates was enriched in lipopolysaccharide biosynthesis and amino-acid-related metabolic functions after maternal antibiotic exposure, whereas the group of unexposed neonates was enriched in carbohydrate metabolic pathways (Li H. et al., 2019).
In addition to the commonly used amoxicillin, antibiotics with different pharmacological mechanisms of action also affect the oral microbiome and increase the abundance of genes associated with antibiotic resistance (Zaura et al., 2015; Moraes et al., 2020). In recent years, it has been reported that some common oral fungi are resistant to antifungal azoles (Gao et al., 2018). Another study showed that genes associated with resistance to ketoconazole and fluconazole were highly expressed in C. albicans isolated from patients with periodontal disease. These data suggest that these drug-resistant Candida strains play an important role in acute and chronic periodontal infections (Monroy-Pérez et al., 2020). However, antibiotics have a much smaller impact on the oral microbiome than on the gut microbiome (Zaura et al., 2015; Menon et al., 2019).
To develop a diet conducive to oral health, many scholars have investigated the relationship between diet and the oral microbiota. An investigation of the oral microbiota in infants showed that breast-fed infants had a higher proportion of Streptococcus, whereas formula-fed infants had higher proportions of Actinomyces and Prevotella. Moreover, breast- and mixed-fed infants developed oral candidiasis at a lower frequency than infants who were fed solid food (Oba et al., 2020). A further investigation confirmed that the significant differences in the oral microorganisms of breast- and formula-fed infants were associated with the production of the antibacterial compound hydrogen peroxide (Sweeney et al., 2018). Even in adults, dietary changes can modulate the oral microbiota and prevent the development of related diseases (Anderson et al., 2020). Fiber, medium-chain fatty acids, piscine monounsaturated fatty acids, and polyunsaturated fatty acids are associated with the diversity and community structure of the oral microbiota (Hansen et al., 2018), and the intake of sugar and refined carbohydrates is associated with the abundance of oral bacteria. For instance, carbonated beverage consumption is positively related to the abundance of Bacteroidetes, Gammaproteobacteria, Fusobacterium, and Veillonella (Chumponsuk et al., 2021). It is generally accepted that the consumption of high-sugar foods provides a favorable environment for caries-associated bacteria, thereby increasing the prevalence of the disease. Sugar alcohols, such as erythritol, xylitol, and sorbitol, have been widely used as sugar substitutes for the prevention of caries. The daily consumption of these sugar alcohols was found to have a specific effect on the composition of the salivary microbiome (Štšepetova et al., 2019). Like sugar, excessive alcohol consumption can also cause an imbalance of the oral microbiome (Yussof et al., 2020). However, on the contrary, some believe that red wine and enological extracts exert some antibacterial effects (Sanchez et al., 2019), and that regular and moderate red wine consumption does not alter the overall diversity and stability of representative bacterial groups in human saliva (Barroso et al., 2015). Further research is required to confirm whether alcohol consumption is beneficial to oral health.
Smoking is widely believed to affect the composition of the oral microbiota. For instance, the Proteobacteria of the oral microbiome are significantly reduced, whereas Firmicutes and Actinobacteria are increased in current smokers (Wu et al., 2016). Interestingly, a study found no significant difference in terms of microbial diversity or richness between smokers and non-smokers (Al Bataineh et al., 2020), in contrast to the results of Jia et al. (2021) and Yu et al. (2017). This discrepancy may be attributable to multiple influential factors, including diet, ethnicity, and environment. However, it is clear that the effect of smoking on the oral microbiota is stable and persists for several years after quitting. Investigations have confirmed that after a certain period of smoking cessation, the overall composition of smoker’s oral microbiota is similar to that of never smokers, suggesting that quitting smoking is beneficial in restoring a healthy phenotype (Wu et al., 2016; Yang et al., 2019; Jia et al., 2021). However, the overall composition of the microbiota of former smokers who quit more than a year earlier tended to be more similar to that of current smokers than to that of never smokers (Jia et al., 2021). Recent investigations have demonstrated that the potential effects of smoking on the oral microbiome include the creation of an anaerobic environment, damage to the host’s immunity, alteration of the pH of the oral saliva and the adhesion of oral bacteria, and the antibacterial effects of the toxic substances in cigarette smoke (Wu et al., 2016; Jia et al., 2021).
The diseases associated with oral microorganisms involve multiple systems, including the cardiovascular system, digestive system, endocrine system, and other systems. For example, the salivary microbiome of obese patients differs from that of normal-weight hosts, and also changes during the period of weight loss (Džunková et al., 2020). However, it remains unclear whether the dietary changes or obesity itself affects the microbiome. Primary sclerosing cholangitis is a chronic, progressive cholestatic liver disease, characterized by progressive inflammation and fibrosis of bile ducts and multi-focal bile duct strictures, in which the salivary microbiota is significantly altered and may invade the gut (Lapidot et al., 2021). Interestingly, the relationships between systemic diseases and the oral microbiome differ. Diabetes, rheumatoid arthritis (RA), and SLE are all associated with inflammatory responses and the development of periodontal disease. One study found that the levels of Capnocytophaga, Porphyromonas, and Pseudomonas are elevated in diabetic patients, whereas the levels of Prevotella and Selenomonas are increased in SLE and the levels of Leptotrichia and Prevotella are increased in RA (Graves et al., 2019). It has also been reported that the development of tumors is related to changes in oral microbes (Fan et al., 2018; Flemer et al., 2018; Hayes et al., 2018). However, the mechanisms underlying the correlation between tumors and oral microorganisms are still unclear (Irfan et al., 2020). Oncological treatments can lead to changes in oral microorganisms. For instance, Enterococcus, Lactobacillus, Staphylococcus, S. mutans, and C. albicans increase after radiotherapy (Sami et al., 2020). Chemotherapy can also alter the composition of the oral microbiota. And the changes in bacteria are more obvious than the changes in fungi (Hong et al., 2019).
In the oral cavity, a variety of endogenous and exogenous factors can alter the abundance and composition of the oral microbial communities, leading to microbial dysbiosis, the occurrence of oral caries, periodontitis, and other diseases, and even causing systemic diseases (Sampaio-Maia et al., 2016; Valm, 2019).
Traditionally, oral pathogens have been considered key factors in the occurrence and development of oral diseases. In recent years, based on 16S rRNA gene sequencing, it has been shown that oral diseases are not caused by a single pathogen, but by microbiotal dysbiosis (Sanz-Martin et al., 2017; Hurley et al., 2019; Qian et al., 2019). One study confirmed that the composition and structure of supragingival plaque is mainly related to caries, whereas subgingival plaque is mainly related to periodontal disease (Costalonga and Herzberg, 2014). Furthermore, when people are in a pathological state, microbial biofilms in the gingival sulcus and periodontal pocket are more likely to cause periodontal tissue inflammation (Vartoukian et al., 2009). This disordered subgingival plaque damages the immune response within the gum tissue. In turn, this compromised immune response promotes a further imbalance in host–bacterial interactions and can lead to severe microbiotal dysbiosis (Johnston et al., 2021).
Streptococcus mutans has long been regarded as the pathogen of caries, given its acidogenicity, acidurance, and adhesion (Baker et al., 2021). However, a study demonstrated that S. mutans is present in all healthy oral samples, but is not detected in all caries-affected dental samples (Bansal et al., 2020). In recent years, molecular biological detection techniques have shown that the occurrence and development of caries are also closely associated with Actinomyces, Lactobacillus, Neisseria, Prevotella, Propionibacterium, Scardovia, and even C. albicans and Epstein-Barr virus (EBV) (Hurley et al., 2019; Baker et al., 2021; Zheng et al., 2021). Furthermore, the dominant bacteria gradually change from high levels of S. mutans in the early stage to high levels of Actinomycetes and Prevotella in deep caries (Johansson et al., 2016). These data strongly support the notion that the interactions among multiple microorganisms and the dysbiosis of the entire microbial community contribute to the progression of caries. The host’s sugar consumption also determines the fate of caries by modifying the oral microbiota (Zhan, 2018). When sugar is consumed frequently, more-aciduric strains increase and the acidogenicity and acidurance of other bacteria are adaptively enhanced. The demineralization–remineralization equilibrium is disrupted by these changes (Takahashi and Nyvad, 2011). An in vivo investigation, based on 16S rRNA gene sequencing, also confirmed the association between sugar intake and the oral microbiotal community (Esberg et al., 2020).
Dental plaque is a key factor initiating periodontal disease (Seneviratne et al., 2011; Valm, 2019). As well as red complex bacteria (including Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola), Aggregatibacter actinomycetemcomitans, Eubacterium nodatum, Filifactor alocis, Selenomonas sputigena, TM7, Treponema socranskii, and several Bacteroides, Campylobacter, Desulfobulbus, and Prevotella species are also regarded as biomarkers of periodontitis (Abusleme et al., 2013; Galimanas et al., 2014). Recently, Candidatus Bacteroides periocalifornicus (Torres et al., 2019), Catonella (Kumar et al., 2005), TG5 (Schaumann et al., 2014), and the viral family Redondoviridae (Abbas et al., 2019) have also been shown to be associated with dental plaque in periodontal disease. The proportions of Bacteroidetes, Fusobacteria, Spirochaetes, and TM7 increase and those of Actinobacteria and Proteobacteria decrease as the depth of the probing pocket increases (Cai Z. et al., 2021). Interestingly, increased microbial diversity is a feature that markedly distinguishes periodontal disease from caries and other infectious diseases (Curtis et al., 2020). The hypothesis that the dysbiosis of dental plaque results from changes in the dominant species, rather than colonization by new strains, is thus further validated (Abusleme et al., 2013).
In addition to caries and periodontal diseases, other oral diseases, including oral cancer (Zhang et al., 2019), periapical periodontitis (Zargar et al., 2020), and recurrent oral ulcers (Zhu et al., 2021), are also related to changes in the oral microbiome. To identify possible representative target microorganisms, the changes in microbial diversity that occur in oral diseases are summarized in Table 1. Many investigations have confirmed that herpes simplex virus (HSV) and especially EBV play important roles in the occurrence and development of periodontitis and oral cancer (Blankson et al., 2019; Fakhry et al., 2019; Imai and Ogata, 2020). The presence of HSV can increase the virulence of F. periodonticum, P. gingivalis, S. aureus, and T. forsythia, leading to the development of generalized invasive periodontitis (Passariello et al., 2017). A recent study found that latent membrane protein 1, encoded by EBV, induces IL8 production, which promotes the progression of periodontitis in human gingival cells (Watanabe et al., 2019). However, the role of EBV in the etiology of periodontitis has not been clarified (Imai and Ogata, 2020; Tonoyan et al., 2020). In summary, the roles of viruses in oral diseases are extremely important. However, the relevant research into fungi and viruses is very limited compared with the many studies of the roles of bacteria in oral diseases.
Oral antibiotics specifically directed against oral caries can disrupt and destroy the signaling systems of pathogenic bacteria (Cui et al., 2019). For example, sulfated vizantin inhibits the extracellular release of cell-free glucosyltransferase, enhances the accumulation of cell-associated glucosyltransferase, and inhibits the maturation of S. mutans biofilms (Oda et al., 2020). Walkmycin C inhibits the biofilm formation and acid resistance of S. mutans (Eguchi et al., 2011). Moreover, the combination of amoxicillin and metronidazole exerted greater antimicrobial effects on subgingival biofilms of bacterial species in vitro than either drug alone (Soares et al., 2015). Further clinical trials have also shown that the administration of amoxicillin and metronidazole combined with scaling and root planning (SRP) was significantly superior to SRP alone in reducing the probing depth and clinical attachment loss, as well as clinical improvement in both the subgingival region and saliva (Hagenfeld et al., 2018; Lu et al., 2021), especially in the treatment of periodontitis in type 2 diabetic patients (Cruz et al., 2021) or aggressive periodontitis (Ardila et al., 2020).
In recent years, the development of antibiotic resistance has been accelerated by the abuse of antibiotics, especially in dentistry (Loffler and Bohmer, 2017). The effectiveness of antibiotics is rapidly being eroded by drug resistance, and the discovery of other unique antibiotics that are specific to pathogens is urgently required (Singh et al., 2017).
“Periodontal intervention therapy” mainly refers to supragingival and subgingival SRP in patients with periodontal disease, which remove periodontal pathogens and calculus and create a relatively healthy environment. This therapy not only affects the composition and structure of the oral microbiome, but also influences the interactions between microorganisms (Zhang et al., 2021a). Periodontal treatment with oral mechanical cleaning and adjuvant systemic antibiotics significantly reduces the abundances of A. actinomycetemcomitans and F. alocis, whereas the abundances of health-related microorganisms, such as Streptococcus spp., Rothia spp., and Prevotella spp. increase (Velsko et al., 2020). After a periodontal intervention, the microbial abundance and biodiversity in the dental plaque decreased, whereas those in the saliva did not change (Yamanaka et al., 2012). Although the antimicrobial effect of periodontal intervention is often non-specific, its combination with adjuvant antibiotic therapy can achieve better dental plaque control than the antibiotic alone. Therefore, periodontal intervention therapy remains an essential technique for the treatment of oral diseases.
Antimicrobial photodynamic therapy (PDT) is a technique in which the excitation of light at an appropriate wavelength is used to irradiate a photosensitizer and release energy (Lu et al., 2021). The oxygen in tissues produces unstable singlet oxygen (1O2), cytotoxic reactive oxygen, and other substances that non-specifically attack the microbiota, resulting in the rapid oxidation of bacterial lipids and ultimately bacterial death (Dobson and Wilson, 1992; Qi et al., 2019). The bacteria in dental plaque are so sensitive to PDT, making it an effective bactericidal technique. In a randomized controlled clinical trial of periodontal disease, gingival index, probing depth, clinical attachment level, and the A. actinomycetemcomitans and P. gingivalis counts were significantly lower in the test group (SRP followed by PDT) than in the control group (SRP alone), after follow-up for 1, 3, 6, and 9 months (Gandhi et al., 2019). As in that clinical trial, the combination of PDT and photobiomodulation therapy accelerated the healing process after oral mucositis, reducing the remission time from 15 to 11 days (Pires Marques et al., 2020). Clinical trials of pulpitis (da Mota et al., 2015), periapical periodontitis (Moreira et al., 2015), oral leukoplakia (Li Y. et al., 2019), peri-implantitis (Al Habashneh et al., 2015), and adverse biofilm changes caused by orthodontic brackets (Rosa et al., 2020) showed that PTD was more effective than traditional treatments. However, further clinical trials are required to establish whether PTD can be used as an alternative mechanical debridement process in the prevention of oral infectious diseases (Javali et al., 2019).
Probiotics are non-pathogenic living microorganisms that have both preventive and therapeutic effects on oral infectious diseases (Zaura and Twetman, 2019). The bacteriocins released by probiotics effectively antagonize acidic dental plaque, and produce glucanase and urease after their colonization of the oral mucosa, which can counteract plaque formation and saliva acidity, respectively (Di Pierro et al., 2015). Probiotics can induce the expression of the virulence genes of S. mutans, including acid tolerance genes (atpD and aguD), EPS production genes (gtfB and sacB), and QS genes (vicKR and comCD), and inhibit the immunomodulatory effects of interferon γ (IFN-γ) and IL10 (Wasfi et al., 2018). Probiotics have also been shown to play an important role in the treatment of chronic periodontal disease. For example, patients with generalized chronic periodontitis treated with SRP and probiotic lozenges had significantly reduced levels of periodontal pathogenic red and orange complexes, tumor necrosis factor α (TNF-α), and IL1β (Invernici et al., 2018). Probiotics can also play a therapeutic role in patients with oral candidiasis, reducing the number of C. albicans (Mundula et al., 2019). Both L. casei and L. rhamnosus exert strong antifungal activity against the growth and biofilm formation of C. albicans but do not affect the surface roughness of denture base resin. Therefore, they are ideal probiotics for the prevention and treatment of denture-related stomatitis (Song and Lee, 2017). Bacillus subtilis and S. thermophilus also inhibit the growth and biofilm formation of C. albicans in a dose-dependent manner, and can be used for the treatment of oral candidiasis (Zhao et al., 2016; Azad et al., 2021). Clinical trials have shown that the oral intake of probiotics reduces the prevalence of oral candidiasis in the frail elderly (Kraft-Bodi et al., 2015). A recent study indicated that S. parasanguinis, a symbiotic bacterium, directly inhibits the activity of GTF, which can prevent C. albicans binding to glucan (Huffines and Scoffield, 2020). Although many studies have suggested that probiotics are effective in the treatment of caries, several problems must still be considered. For example, the common probiotic Lactobacillus also promotes the development of caries (Hurley et al., 2019; Zheng et al., 2021), and a report indicated that the oral health status was not significantly improved after the administration of probiotics (Sivamaruthi et al., 2020).
With the increase in antibiotic-resistant bacteria, and because the formation and development of oral biofilms are closely associated with bacterial QS, quorum quenching (QQ), which interferes with microbial communication, is another alternative treatment for oral infections (Paluch et al., 2020). Without eradicating any oral bacteria, QQ maintains the balance of the oral microflora and simply inhibits the formation of biofilms, which prevents overgrowth of C. albicans and S. epidermidis (Weiland-Brauer et al., 2019). For example, D-galactose interrupts the AI-2 QS system by interfering with the biochemical synthesis of AI-2 proteins, thereby reducing biofilm formation (Ryu et al., 2020). Furanone compounds and D-ribose are QS inhibitors that reduced bacterial infection and the destruction of periodontal tissue caused by coinfection with F. nucleatum and P. gingivalis (Ben Amara et al., 2018). The inhibition of the peptidase domain of the ATP-binding box transporter ComA effectively inhibited streptococcal QS pathway, leading to phenotypic and/or behavioral changes (Long et al., 2020). In summary, these data confirm that disrupting dental plaque formation with QS inhibitors (or QQ) is a promising method for controlling oral-biofilm-related diseases.
Given their highly specific effects on bacterial species and their coevolution with their host bacteria, phage can kill multidrug-resistant bacteria and even effectively destroy biofilms (Baker et al., 2017; Shlezinger et al., 2017; Torres-Barcelo, 2018). Phage therapy makes use of strictly lytic phages and is regarded as a novel method for the prevention, control, and treatment of oral infectious diseases (Shlezinger et al., 2017; Harada et al., 2018). The construction of a bacteriophage T4 RNA ligase 1 (T4 Rnl1) expressing system in S. mutans showed that T4 Rnl1 exerts membrane-directed antimicrobial activity against S. mutans and affects the spatial structure of its extracellular polysaccharide (Chen J. et al., 2020; Wolfoviz-Zilberman et al., 2021). Therefore, phages can be used as a new preventive treatment for caries. In the presence of phage ϕAPCM01, the metabolic activity of the S. mutans biofilm was reduced after 24 h of contact and the live cells in the biofilm decreased by at least 5log cfu/ml (Dalmasso et al., 2015). Recently, novel bacteriophages in the family Siphoviridae were shown to cause a 70% reduction in the biomass of F. nucleatum biofilms and to damage their membrane structure (Kabwe et al., 2019). A selection of phages suitable for the treatment of specific pathogens can be used as an alternative therapy or in combination with broad-spectrum antibacterial agents (Shlezinger et al., 2017). At present, phages associated with Actinobacteria (Hatfull, 2020), Firmicutes (Wolfoviz-Zilberman et al., 2021), and Fusobacteria (Machuca et al., 2010) have been successfully used for the treatment of oral infectious diseases. Therefore, the construction of phage libraries against oral pathogens will offer excellent opportunities for more-personalized dentistry and oral microbiome engineering (Shlezinger et al., 2017). However, there have been insufficient clinical trials to confirm the feasibility of these phages.
Fluoride nanophase materials and oral hygiene products based on natural extracts have been widely used to prevent caries. Fluoride, which exerts an anti-caries effect, not only promotes the remineralization of the enamel, but also reduces carbohydrate-decomposing microorganisms and inhibits the carbohydrate fermentation pathways of the oral microbiota (Staun Larsen et al., 2018; Lopez-Lopez and Mira, 2020). It also regulates oral biofilm formation by changing the hydrophobicity and aggregation of bacteria (Lopez-Lopez and Mira, 2020). Nanoparticles (NPs) can attach to and destroy bacterial cell walls, successfully disrupt the biofilm matrix, release related ions that damage cell functions, improve the water solubility of drugs, and carry, retain, and release drugs at fixed points and scheduled times (Gupta et al., 2016; Wang et al., 2016). Moreover, binding traditional drugs to NPs protects them from pH changes and degradation by the enzymes specific to different ecological sites (Lu et al., 2016). These advantages confer potential utility on NPs in the prevention and treatment of oral diseases. To inhibit the occurrence of secondary caries, silver ions and some antibacterial agents are conventionally added to the resin and adhesive (Dias et al., 2019). In recent years, the discovery of quaternary ammonium salts has offered a new method for the prevention and treatment of secondary caries (Beyth et al., 2006). Dimethylaminododecyl methacrylate, a quaternary ammonium salt, has pH-dependent antifungal effects on C. albicans (Chen H. et al., 2020). As natural antibacterial agents, extracts from plants exert important effects on the oral microbiota. For instance, the essential oil of Thymus capitatus strongly affects the growth of S. mutans and C. albicans, and an extract of Citrus limon var. pompia exerts bactericidal effects on S. mutans, although it only weakly affects C. albicans (Pinna et al., 2019). Catechol inhibits the development of oral pathogens such as A. actinomycetemcomitans, P. gingivalis, and Prevotella intermedia, and also inhibits the growth, adhesion, and acid production of acid-producing oral Streptococcus (Fournier-Larente et al., 2016; Higuchi et al., 2019). These data provide promising methods for the clinical treatment of dental caries, periodontitis, pulpitis, oral mucosal diseases, and halitosis.
In this review, we have summarized recent studies of the communities of oral microbiota and the endogenous and exogenous factors that influence its composition (Figure 1). The development of metagenomics, metaproteomics, and metabonomics has provided a more comprehensive understanding of oral microbiotal communities. Increasing evidence supports the notion that various internal and external factors cause the dysbiosis of the oral microbiota, which contributes greatly to oral and systematic diseases. Therefore, more attention must be paid to the mechanisms underlying the interactions among oral microorganisms within these communities and their interactions with their host, rather than merely identifying the composition of the oral microbiome. Current studies still focus on bacteria and C. albicans, and identifying the communications and regulatory mechanisms within and between the mycobiome, virome, and CPR remains a challenge. We have also highlighted recent promising therapeutic strategies for oral diseases. Few investigations into the effects of these strategies on oral dysbiosis have produced consistent results, given the limitations of sampling procedures and the difficulties of enrolling subjects with heterogeneous clinical traits. To adequately mimic both the host and microbial behavior during the therapeutic process, an effective approach or model is required in which to analyze the shifts in compositions of the microbial communities.
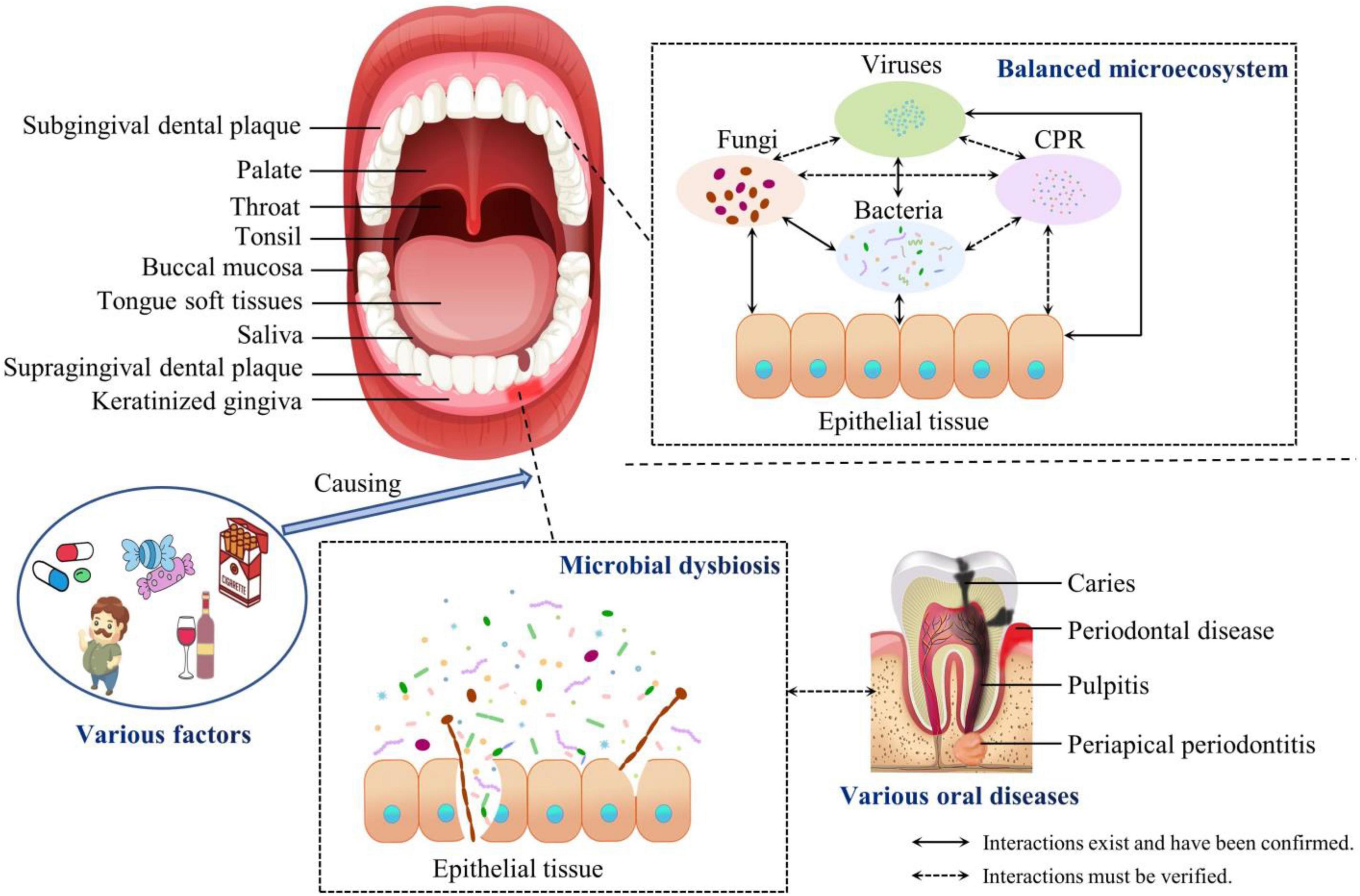
Figure 1. Compositions of the balanced oral microbiota and during dysbiosis. The oral cavity is divided into nine niches. The composition of the oral microbiota and the structure of the oral biofilm adapt specifically to these different microecological environments. The communities of oral microorganisms and their interactions with the host maintain the oral microecosystem in a dynamic balance. However, various factors cause the dysbiosis of the oral microbiota, which contributes to oral and even systemic diseases. CPR, candidate phyla radiation.
ZS conceived and designed the manuscript. XL and YL wrote the manuscript. CL, XY, and ZS critically revised and supervised it. All authors contributed to the article and approved the submitted version.
This research was supported financially by Sichuan Province Administration of Traditional Chinese Medicine (2020JC0129), Science and Technology Project of Luzhou (2021-JYJ-73), Foundation of Southwest Medical University (2021ZKMS008), and College Students’ Innovation and Entrepreneurship Training Program (S202110632135).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbas, A. A., Taylor, L. J., Dothard, M. I., Leiby, J. S., Fitzgerald, A. S., Khatib, L. A., et al. (2019). Redondoviridae, a family of small, circular DNA viruses of the human oro-respiratory tract associated with periodontitis and critical illness. Cell Host Microbe 25, 719–729.e4. doi: 10.1016/j.chom.2019.04.001
Abeles, S. R., Robles-Sikisaka, R., Ly, M., Lum, A. G., Salzman, J., Boehm, T. K., et al. (2014). Human oral viruses are personal, persistent and gender-consistent. ISME J. 8, 1753–1767. doi: 10.1038/ismej.2014.31
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9:e02331-17. doi: 10.1128/mBio.02331-17
Abusleme, L., Dupuy, A. K., Dutzan, N., Silva, N., Burleson, J. A., Strausbaugh, L. D., et al. (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025. doi: 10.1038/ismej.2012.174
Al Bataineh, M. T., Dash, N. R., Elkhazendar, M., Alnusairat, D. M. H., Darwish, I. M. I., Al-Hajjaj, M. S., et al. (2020). Revealing oral microbiota composition and functionality associated with heavy cigarette smoking. J. Transl. Med. 18:421. doi: 10.1186/s12967-020-02579-3
Al Habashneh, R., Asa’ad, F. A., and Khader, Y. (2015). Photodynamic therapy in periodontal and peri-implant diseases. Quintessence Int. 46, 677–690. doi: 10.3290/j.qi.a34078
Aleksandrowicz, P., Brzezińska-Błaszczyk, E., Dudko, A., and Agier, J. (2020). Archaea occurrence in the subgingival bioflm in patients with peri-implantitis and periodontitis. Int. J. Periodontics Restorative Dent. 40, 677–683. doi: 10.11607/prd.4670
Anderson, A. C., Rothballer, M., Altenburger, M. J., Woelber, J. P., Karygianni, L., Vach, K., et al. (2020). Long-term fluctuation of oral biofilm microbiota following different dietary phases. Appl. Environ. Microbiol. 86:e0142120. doi: 10.1128/aem.01421-20
Ardila, C. M., Florez-Florez, J., Castaneda-Parra, L. D., Guzman, I. C., and Bedoya-Garcia, J. A. (2020). Moxifloxacin versus amoxicillin plus metronidazole as adjunctive therapy for generalized aggressive periodontitis: a pilot randomized controlled clinical trial. Quintessence Int. 51, 612–621. doi: 10.3290/j.qi.a44715
Arzmi, M. H., Dashper, S., Catmull, D., Cirillo, N., Reynolds, E. C., and McCullough, M. (2015). Coaggregation of Candida albicans, Actinomyces naeslundii and Streptococcus mutans is Candida albicans strain dependent. FEMS Yeast Res. 15:fov038. doi: 10.1093/femsyr/fov038
Azad, A., Ranjbaran, A., Zareshahrabadi, Z., Mehrabani, D., Zahed Zahedani, M., Talebanpour, A., et al. (2021). Protective effects of the probiotic bacterium Streptococcus thermophilus on Candida albicans morphogenesis and a murine model of oral candidiasis. Iran. J. Med. Sci. 46, 207–217. doi: 10.30476/ijms.2020.82080.0
Back, C. R., Douglas, S. K., Emerson, J. E., Nobbs, A. H., and Jenkinson, H. F. (2015). Streptococcus gordonii DL1 adhesin SspB V-region mediates coaggregation via receptor polysaccharide of Actinomyces oris T14V. Mol. Oral Microbiol. 30, 411–424. doi: 10.1111/omi.12106
Baker, J. L., Bor, B., Agnello, M., Shi, W., and He, X. (2017). Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 25, 362–374. doi: 10.1016/j.tim.2016.12.012
Baker, J. L., Morton, J. T., Dinis, M., Alvarez, R., Tran, N. C., Knight, R., et al. (2021). Deep metagenomics examines the oral microbiome during dental caries, revealing novel taxa and co-occurrences with host molecules. Genome Res. 31, 64–74. doi: 10.1101/gr.265645.120
Bansal, K., Chaudhary, R., Mathur, V. P., and Tewari, N. (2020). Comparison of oral micro-flora in caries active and caries free Indian children using culture techniques and PCR analysis. Indian J. Dent. Res. 31, 420–425. doi: 10.4103/ijdr.IJDR_39_19
Barroso, E., Martín, V., Martínez-Cuesta, M. C., Peláez, C., and Requena, T. (2015). Stability of saliva microbiota during moderate consumption of red wine. Arch. Oral Biol. 60, 1763–1768. doi: 10.1016/j.archoralbio.2015.09.015
Ben Amara, H., Song, H. Y., Ryu, E., Park, J. S., Schwarz, F., Kim, B. M., et al. (2018). Effects of quorum-sensing inhibition on experimental periodontitis induced by mixed infection in mice. Eur. J. Oral Sci. 126, 449–457. doi: 10.1111/eos.12570
Beyth, N., Yudovin-Farber, I., Bahir, R., Domb, A. J., and Weiss, E. I. (2006). Antibacterial activity of dental composites containing quaternary ammonium polyethylenimine nanoparticles against Streptococcus mutans. Biomaterials 27, 3995–4002. doi: 10.1016/j.biomaterials.2006.03.003
Blankson, P. K., Blankson, H. N. A., Obeng-Nkrumah, N., Turkson, A. A., Tormeti, D., Adamafio, M., et al. (2019). Detection of herpes viruses in Ghanaian patients with periodontitis. J. Investig. Clin. Dent. 10:e12386. doi: 10.1111/jicd.12386
Bor, B., Bedree, J. K., Shi, W., McLean, J. S., and He, X. (2019). Saccharibacteria (TM7) in the human oral microbiome. J. Dent. Res. 98, 500–509. doi: 10.1177/0022034519831671
Bor, B., Collins, A. J., Murugkar, P. P., Balasubramanian, S., To, T. T., Hendrickson, E. L., et al. (2020). Insights obtained by culturing Saccharibacteria with their bacterial hosts. J. Dent. Res. 99, 685–694. doi: 10.1177/0022034520905792
Bostanci, N., Krog, M. C., Hugerth, L. W., Bashir, Z., Fransson, E., Boulund, F., et al. (2021). Dysbiosis of the human oral microbiome during the menstrual cycle and vulnerability to the external exposures of smoking and dietary sugar. Front. Cell. Infect. Microbiol. 11:625229. doi: 10.3389/fcimb.2021.625229
Bottalico, D., Chen, Z., Dunne, A., Ostoloza, J., McKinney, S., Sun, C., et al. (2011). The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J. Infect. Dis. 204, 787–792. doi: 10.1093/infdis/jir383
Bowen, W. H., Burne, R. A., Wu, H., and Koo, H. (2018). Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242. doi: 10.1016/j.tim.2017.09.008
Brasser, A. J., Barwacz, C. A., Dawson, D. V., Brogden, K. A., Drake, D. R., and Wertz, P. W. (2011). Presence of wax esters and squalene in human saliva. Arch. Oral Biol. 56, 588–591. doi: 10.1016/j.archoralbio.2010.12.002
Brennan, C. A., and Garrett, W. S. (2019). Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Cai, W. Y., Zhou, S., and Lin, L. Y. (2021). Effect of triple antibiotic paste on microorganisms in root canals of deciduous teeth. Shanghai Kou Qiang Yi Xue 30, 374–378. doi: 10.19439/j.sjos
Cai, Z., Lin, S., Hu, S., and Zhao, L. (2021). Structure and function of oral microbial community in periodontitis based on integrated data. Front. Cell. Infect. Microbiol. 11:663756. doi: 10.3389/fcimb.2021.663756
Camanocha, A., and Dewhirst, F. E. (2014). Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J. Oral Microbiol. 6:25468. doi: 10.3402/jom.v6.25468
Caselli, E., Fabbri, C., D’Accolti, M., Soffritti, I., Bassi, C., Mazzacane, S., et al. (2020). Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 20:120. doi: 10.1186/s12866-020-01801-y
Castillo Pedraza, M. C., Novais, T. F., Faustoferri, R. C., Quivey, R. G. Jr., Terekhov, A., Hamaker, B. R., et al. (2017). Extracellular DNA and lipoteichoic acids interact with exopolysaccharides in the extracellular matrix of Streptococcus mutans biofilms. Biofouling 33, 722–740. doi: 10.1080/08927014.2017.1361412
Chen, H., Zhou, Y., Zhou, X., Liao, B., Xu, H. H. K., Chu, C. H., et al. (2020). Dimethylaminododecyl methacrylate inhibits Candida albicans and oropharyngeal candidiasis in a pH-dependent manner. Appl. Microbiol. Biotechnol. 104, 3585–3595. doi: 10.1007/s00253-020-10496-0
Chen, J., Chen, Z., Yuan, K., Huang, Z., and Mao, M. (2020). Recombinant bacteriophage T4 Rnl1 impacts Streptococcus mutans biofilm formation. J. Oral Microbiol. 13:1860398. doi: 10.1080/20002297.2020.1860398
Chen, W. P., Chang, S. H., Tang, C. Y., Liou, M. L., Tsai, S. J., and Lin, Y. L. (2018). Composition analysis and feature selection of the oral microbiota associated with periodontal disease. Biomed Res. Int. 2018:3130607. doi: 10.1155/2018/3130607
Chen, Y., Li, Y., and Zou, J. (2019). Intrageneric and intergeneric interactions developed by oral streptococci: pivotal role in the pathogenesis of oral diseases. Curr. Issues Mol. Biol. 32, 377–434. doi: 10.21775/cimb.032.377
Choo, S. W., Mohammed, W. K., Mutha, N. V. R., Rostami, N., Ahmed, H., Krasnogor, N., et al. (2021). Transcriptomic responses to coaggregation between Streptococcus gordonii and Streptococcus oralis. Appl. Environ. Microbiol. 87:e0155821. doi: 10.1128/AEM.01558-21
Chumponsuk, T., Gruneck, L., Gentekaki, E., Jitprasertwong, P., Kullawong, N., Nakayama, J., et al. (2021). The salivary microbiota of Thai adults with metabolic disorders and association with diet. Arch. Oral Biol. 122:105036. doi: 10.1016/j.archoralbio.2020.105036
Costalonga, M., and Herzberg, M. C. (2014). The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162, 22–38. doi: 10.1016/j.imlet.2014.08.017
Cruz, D. F. D., Duarte, P. M., Figueiredo, L. C., da Silva, H. D. P., Retamal-Valdes, B., Feres, M., et al. (2021). Metronidazole and amoxicillin for patients with periodontitis and diabetes mellitus: 5-year secondary analysis of a randomized controlled trial. J. Periodontol. 92, 479–487. doi: 10.1002/JPER.20-0196
Cugini, C., Shanmugam, M., Landge, N., and Ramasubbu, N. (2019). The role of exopolysaccharides in oral biofilms. J. Dent. Res. 98, 739–745. doi: 10.1177/0022034519845001
Cui, T., Luo, W., Xu, L., Yang, B., Zhao, W., and Cang, H. (2019). Progress of antimicrobial discovery against the major cariogenic pathogen Streptococcus mutans. Curr. Issues Mol. Biol. 32, 601–644. doi: 10.21775/cimb.032.601
Curtis, M. A., Diaz, P. I., and Van Dyke, T. E. (2020). The role of the microbiota in periodontal disease. Periodontol 2000 83, 14–25. doi: 10.1111/prd.12296
Czesnikiewicz-Guzik, M., Osmenda, G., Siedlinski, M., Nosalski, R., Pelka, P., Nowakowski, D., et al. (2019). Causal association between periodontitis and hypertension: evidence from Mendelian randomization and a randomized controlled trial of non-surgical periodontal therapy. Eur. Heart J. 40, 3459–3470. doi: 10.1093/eurheartj/ehz646
da Mota, A. C., Goncalves, M. L., Bortoletto, C., Olivan, S. R., Salgueiro, M., Godoy, C., et al. (2015). Evaluation of the effectiveness of photodynamic therapy for the endodontic treatment of primary teeth: study protocol for a randomized controlled clinical trial. Trials 16:551. doi: 10.1186/s13063-015-1086-2
Dalmasso, M., de Haas, E., Neve, H., Strain, R., Cousin, F. J., Stockdale, S. R., et al. (2015). Isolation of a novel phage with activity against Streptococcus mutans biofilms. PLoS One 10:e0138651. doi: 10.1371/journal.pone.0138651
Dame-Teixeira, N., de Cena, J. A., Côrtes, D. A., Belmok, A., Dos Anjos Borges, L. G., Marconatto, L., et al. (2020). Presence of archaea in dental caries biofilms. Arch. Oral Biol. 110:104606. doi: 10.1016/j.archoralbio.2019.104606
Deng, L., Li, W., He, Y., Wu, J., Ren, B., and Zou, L. (2019). Cross-kingdom interaction of Candida albicans and Actinomyces viscosus elevated cariogenic virulence. Arch. Oral Biol. 100, 106–112. doi: 10.1016/j.archoralbio.2019.02.008
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol. 192, 5002–5017. doi: 10.1128/jb.00542-10
Di Pierro, F., Zanvit, A., Nobili, P., Risso, P., and Fornaini, C. (2015). Cariogram outcome after 90 days of oral treatment with Streptococcus salivarius M18 in children at high risk for dental caries: results of a randomized, controlled study. Clin. Cosmet. Investig. Dent. 7, 107–113. doi: 10.2147/CCIDE.S93066
Dias, H. B., Bernardi, M. I. B., Marangoni, V. S., de Abreu Bernardi, A. C., de Souza Rastelli, A. N., and Hernandes, A. C. (2019). Synthesis, characterization and application of Ag doped ZnO nanoparticles in a composite resin. Mater. Sci. Eng. C Mater. Biol. Appl. 96, 391–401. doi: 10.1016/j.msec.2018.10.063
Dingsdag, S., Nelson, S., and Coleman, N. V. (2016). Bacterial communities associated with apical periodontitis and dental implant failure. Microb. Ecol. Health Dis. 27:31307. doi: 10.3402/mehd.v27.31307
Dobson, J., and Wilson, M. (1992). Sensitization of oral bacteria in biofilms to killing by light from a low-power laser. Arch. Oral Biol. 37, 883–887. doi: 10.1016/0003-9969(92)90058-g
Džunková, M., Lipták, R., Vlková, B., Gardlík, R., Čierny, M., Moya, A., et al. (2020). Salivary microbiome composition changes after bariatric surgery. Sci. Rep. 10:20086. doi: 10.1038/s41598-020-76991-6
Efenberger, M., Agier, J., Pawłowska, E., and Brzezińska-Błaszczyk, E. (2015). Archaea prevalence in inflamed pulp tissues. Cent. Eur. J. Immunol. 40, 194–200. doi: 10.5114/ceji.2015.51358
Eguchi, Y., Kubo, N., Matsunaga, H., Igarashi, M., and Utsumi, R. (2011). Development of an antivirulence drug against Streptococcus mutans: repression of biofilm formation, acid tolerance, and competence by a histidine kinase inhibitor, Walkmycin C. Antimicrob. Agents Chemother. 55, 1475–1484. doi: 10.1128/AAC.01646-10
Ellepola, K., Truong, T., Liu, Y., Lin, Q., Lim, T. K., Lee, Y. M., et al. (2019). Multi-omics analyses reveal synergistic carbohydrate metabolism in Streptococcus mutans-Candida albicans mixed-species biofilms. Infect. Immun. 87:e0033919. doi: 10.1128/iai.00339-19
Esberg, A., Haworth, S., Hasslöf, P., Lif Holgerson, P., and Johansson, I. (2020). Oral microbiota profile associates with sugar intake and taste preference genes. Nutrients 12:681. doi: 10.3390/nu12030681
Fábián, T. K., Hermann, P., Beck, A., Fejérdy, P., and Fábián, G. (2012). Salivary defense proteins: their network and role in innate and acquired oral immunity. Int. J. Mol. Sci. 13, 4295–4320. doi: 10.3390/ijms13044295
Fakhry, C., Blackford, A. L., Neuner, G., Xiao, W., Jiang, B., Agrawal, A., et al. (2019). Association of oral human papillomavirus DNA persistence with cancer progression after primary treatment for oral cavity and oropharyngeal squamous cell carcinoma. JAMA Oncol. 5, 985–992. doi: 10.1001/jamaoncol.2019.0439
Falsetta, M. L., Klein, M. I., Colonne, P. M., Scott-Anne, K., Gregoire, S., Pai, C. H., et al. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82, 1968–1981. doi: 10.1128/IAI.00087-14
Fan, X., Alekseyenko, A. V., Wu, J., Peters, B. A., Jacobs, E. J., Gapstur, S. M., et al. (2018). Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67, 120–127. doi: 10.1136/gutjnl-2016-312580
Fischer, C. L. (2020). Antimicrobial activity of host-derived lipids. Antibiotics 9:75. doi: 10.3390/antibiotics9020075
Flemer, B., Warren, R. D., Barrett, M. P., Cisek, K., Das, A., Jeffery, I. B., et al. (2018). The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454–1463. doi: 10.1136/gutjnl-2017-314814
Fournier-Larente, J., Morin, M. P., and Grenier, D. (2016). Green tea catechins potentiate the effect of antibiotics and modulate adherence and gene expression in Porphyromonas gingivalis. Arch. Oral Biol. 65, 35–43. doi: 10.1016/j.archoralbio.2016.01.014
Gaffen, S. L., and Moutsopoulos, N. M. (2020). Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci. Immunol. 5:eaau4594. doi: 10.1126/sciimmunol.aau4594
Galimanas, V., Hall, M. W., Singh, N., Lynch, M. D., Goldberg, M., Tenenbaum, H., et al. (2014). Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome 2:32. doi: 10.1186/2049-2618-2-32
Gandhi, K. K., Pavaskar, R., Cappetta, E. G., and Drew, H. J. (2019). Effectiveness of adjunctive use of low-level laser therapy and photodynamic therapy after scaling and root planning in patients with chronic periodontitis. Int. J. Periodontics Restorative Dent. 39, 837–843. doi: 10.11607/prd.4252
Gao, J., Wang, H., Li, Z., Wong, A. H., Wang, Y. Z., Guo, Y., et al. (2018). Candida albicans gains azole resistance by altering sphingolipid composition. Nat. Commun. 9:4495. doi: 10.1038/s41467-018-06944-1
Garcia-Rubio, R., de Oliveira, H. C., Rivera, J., and Trevijano-Contador, N. (2019). The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 10:2993. doi: 10.3389/fmicb.2019.02993
Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., et al. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. doi: 10.1371/journal.ppat.1000713
Ghensi, P., Manghi, P., Zolfo, M., Armanini, F., Pasolli, E., Bolzan, M., et al. (2020). Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics. NPJ Biofilms Microbiomes 6:47. doi: 10.1038/s41522-020-00155-7
Gomez, A., and Nelson, K. E. (2017). The oral microbiome of children: development, disease, and implications beyond oral health. Microb. Ecol. 73, 492–503. doi: 10.1007/s00248-016-0854-1
Graves, D. T., Corrêa, J. D., and Silva, T. A. (2019). The oral microbiota is modified by systemic diseases. J. Dent. Res. 98, 148–156. doi: 10.1177/0022034518805739
Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185. doi: 10.1038/ismej.2011.191
Gupta, A., Landis, R. F., and Rotello, V. M. (2016). Nanoparticle-based antimicrobials: surface functionality is critical. F1000Res 5:F1000 Faculty Rev-364. doi: 10.12688/f1000research.7595.1
Hagenfeld, D., Koch, R., Jünemann, S., Prior, K., Harks, I., Eickholz, P., et al. (2018). Do we treat our patients or rather periodontal microbes with adjunctive antibiotics in periodontal therapy? A 16S rDNA microbial community analysis. PLoS One 13:e0195534. doi: 10.1371/journal.pone.0195534
Hansen, T. H., Kern, T., Bak, E. G., Kashani, A., Allin, K. H., Nielsen, T., et al. (2018). Impact of a vegan diet on the human salivary microbiota. Sci. Rep. 8:5847. doi: 10.1038/s41598-018-24207-3
Harada, L. K., Silva, E. C., Campos, W. F., Del Fiol, F. S., Vila, M., Dabrowska, K., et al. (2018). Biotechnological applications of bacteriophages: state of the art. Microbiol. Res. 212–213, 38–58. doi: 10.1016/j.micres.2018.04.007
Hatfull, G. F. (2020). Actinobacteriophages: genomics, dynamics, and applications. Annu. Rev. Virol. 7, 37–61. doi: 10.1146/annurev-virology-122019-070009
Hayes, R. B., Ahn, J., Fan, X., Peters, B. A., Ma, Y., Yang, L., et al. (2018). Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 4, 358–365. doi: 10.1001/jamaoncol.2017.4777
He, X., Li, F., Bor, B., Koyano, K., Cen, L., Xiao, X., et al. (2018). Human tRNA-derived small RNAs modulate host-oral microbial interactions. J. Dent. Res. 97, 1236–1243. doi: 10.1177/0022034518770605
Heller, D., Helmerhorst, E. J., and Oppenheim, F. G. (2017). Saliva and serum protein exchange at the tooth enamel surface. J. Dent. Res. 96, 437–443. doi: 10.1177/0022034516680771
Higuchi, T., Suzuki, N., Nakaya, S., Omagari, S., Yoneda, M., Hanioka, T., et al. (2019). Effects of Lactobacillus salivarius WB21 combined with green tea catechins on dental caries, periodontitis, and oral malodor. Arch. Oral Biol. 98, 243–247. doi: 10.1016/j.archoralbio.2018.11.027
Hong, B. Y., Hoare, A., Cardenas, A., Dupuy, A. K., Choquette, L., Salner, A. L., et al. (2020). The salivary mycobiome contains 2 ecologically distinct mycotypes. J. Dent. Res. 99, 730–738. doi: 10.1177/0022034520915879
Hong, B. Y., Sobue, T., Choquette, L., Dupuy, A. K., Thompson, A., Burleson, J. A., et al. (2019). Chemotherapy-induced oral mucositis is associated with detrimental bacterial dysbiosis. Microbiome 7:66. doi: 10.1186/s40168-019-0679-5
Huffines, J. T., and Scoffield, J. A. (2020). Disruption of Streptococcus mutans and Candida albicans synergy by a commensal streptococcus. Sci. Rep. 10:19661. doi: 10.1038/s41598-020-76744-5
Hurley, E., Barrett, M. P. J., Kinirons, M., Whelton, H., Ryan, C. A., Stanton, C., et al. (2019). Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health 19:13. doi: 10.1186/s12903-018-0693-1
Hwang, G., Liu, Y., Kim, D., Li, Y., Krysan, D. J., and Koo, H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13:e1006407. doi: 10.1371/journal.ppat.1006407
Ikeda, E., Shiba, T., Ikeda, Y., Suda, W., Nakasato, A., Takeuchi, Y., et al. (2020). Japanese subgingival microbiota in health vs disease and their roles in predicted functions associated with periodontitis. Odontology 108, 280–291. doi: 10.1007/s10266-019-00452-4
Imai, K., and Ogata, Y. (2020). How does Epstein-Barr Virus contribute to chronic periodontitis? Int. J. Mol. Sci. 21:1940. doi: 10.3390/ijms21061940
Integrative HMP (iHMP) Research Network Consortium (2019). The integrative human microbiome project. Nature 569, 641–648. doi: 10.1038/s41586-019-1238-8
Invernici, M. M., Salvador, S. L., Silva, P. H. F., Soares, M. S. M., Casarin, R., Palioto, D. B., et al. (2018). Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: a randomized clinical trial. J. Clin. Periodontol. 45, 1198–1210. doi: 10.1111/jcpe.12995
Irfan, M., Delgado, R. Z. R., and Frias-Lopez, J. (2020). The oral microbiome and cancer. Front. Immunol. 11:591088. doi: 10.3389/fimmu.2020.591088
Javali, M. A., AlQahtani, N. A., Ahmad, I., and Ahmad, I. (2019). Antimicrobial photodynamic therapy (light source; methylene blue; titanium dioxide): bactericidal effects analysis on oral plaque bacteria: an in vitro study. Niger. J. Clin. Pract. 22, 1654–1661. doi: 10.4103/njcp.njcp_189_19
Jayaraman, A., and Wood, T. K. (2008). Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu. Rev. Biomed. Eng. 10, 145–167. doi: 10.1146/annurev.bioeng.10.061807.160536
Jia, Y. J., Liao, Y., He, Y. Q., Zheng, M. Q., Tong, X. T., Xue, W. Q., et al. (2021). Association between oral microbiota and cigarette smoking in the Chinese population. Front. Cell. Infect. Microbiol. 11:658203. doi: 10.3389/fcimb.2021.658203
Jiang, W. X., Hu, Y. J., Gao, L., He, Z. Y., Zhu, C. L., Ma, R., et al. (2015). The impact of various time intervals on the supragingival plaque dynamic core microbiome. PLoS One 10:e0124631. doi: 10.1371/journal.pone.0124631
Johansson, I., Witkowska, E., Kaveh, B., Lif Holgerson, P., and Tanner, A. C. (2016). The microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 95, 80–86. doi: 10.1177/0022034515609554
Johnston, W., Rosier, B. T., Artacho, A., Paterson, M., Piela, K., Delaney, C., et al. (2021). Mechanical biofilm disruption causes microbial and immunological shifts in periodontitis patients. Sci. Rep. 11:9796. doi: 10.1038/s41598-021-89002-z
Kabwe, M., Brown, T. L., Dashper, S., Speirs, L., Ku, H., Petrovski, S., et al. (2019). Genomic, morphological and functional characterisation of novel bacteriophage FNU1 capable of disrupting Fusobacterium nucleatum biofilms. Sci. Rep. 9:9107. doi: 10.1038/s41598-019-45549-6
Kaplan, A., Kaplan, C. W., He, X., McHardy, I., Shi, W., and Lux, R. (2014). Characterization of aid1, a novel gene involved in Fusobacterium nucleatum interspecies interactions. Microb. Ecol. 68, 379–387. doi: 10.1007/s00248-014-0400-y
Keijser, B. J., Zaura, E., Huse, S. M., van der Vossen, J. M., Schuren, F. H., Montijn, R. C., et al. (2008). Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87, 1016–1020. doi: 10.1177/154405910808701104
Keijser, B. J. F., van den Broek, T. J., Slot, D. E., van Twillert, L., Kool, J., Thabuis, C., et al. (2018). The Impact of maltitol-sweetened chewing gum on the dental plaque biofilm microbiota composition. Front. Microbiol. 9:381. doi: 10.3389/fmicb.2018.00381
Kim, Y. J., Choi, Y. S., Baek, K. J., Yoon, S. H., Park, H. K., and Choi, Y. (2016). Mucosal and salivary microbiota associated with recurrent aphthous stomatitis. BMC Microbiol. 16(Suppl. 1):57. doi: 10.1186/s12866-016-0673-z
Kolenbrander, P. E., Palmer, R. J. Jr., Periasamy, S., and Jakubovics, N. S. (2010). Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8, 471–480. doi: 10.1038/nrmicro2381
Kraft-Bodi, E., Jorgensen, M. R., Keller, M. K., Kragelund, C., and Twetman, S. (2015). Effect of probiotic bacteria on oral Candida in frail elderly. J. Dent. Res. 94, 181S–186S. doi: 10.1177/0022034515595950
Kriebel, K., Hieke, C., Muller-Hilke, B., Nakata, M., and Kreikemeyer, B. (2018). Oral biofilms from symbiotic to pathogenic interactions and associated disease -connection of periodontitis and rheumatic arthritis by peptidylarginine deiminase. Front. Microbiol. 9:53. doi: 10.3389/fmicb.2018.00053
Kumar, P. S., Griffen, A. L., Moeschberger, M. L., and Leys, E. J. (2005). Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43, 3944–3955. doi: 10.1128/jcm.43.8.3944-3955.2005
Lamont, R. J., Koo, H., and Hajishengallis, G. (2018). The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759. doi: 10.1038/s41579-018-0089-x
Lapidot, Y., Amir, A., Ben-Simon, S., Veitsman, E., Cohen-Ezra, O., Davidov, Y., et al. (2021). Alterations of the salivary and fecal microbiome in patients with primary sclerosing cholangitis. Hepatol. Int. 15, 191–201. doi: 10.1007/s12072-020-10089-z
Larsson Wexell, C., Ryberg, H., Sjöberg Andersson, W. A., Blomqvist, S., Colin, P., Van Bocxlaer, J., et al. (2016). Antimicrobial effect of a single dose of amoxicillin on the oral microbiota. Clin. Implant. Dent. Relat. Res. 18, 699–706. doi: 10.1111/cid.12357
Lee, W. H., Chen, H. M., Yang, S. F., Liang, C., Peng, C. Y., Lin, F. M., et al. (2017). Bacterial alterations in salivary microbiota and their association in oral cancer. Sci. Rep. 7:16540. doi: 10.1038/s41598-017-16418-x
Li, C. L., Liu, D. L., Jiang, Y. T., Zhou, Y. B., Zhang, M. Z., Jiang, W., et al. (2009). Prevalence and molecular diversity of archaea in subgingival pockets of periodontitis patients. Oral Microbiol. Immunol. 24, 343–346. doi: 10.1111/j.1399-302X.2009.00514.x
Li, H., Xiao, B., Zhang, Y., Xiao, S., Luo, J., and Huang, W. (2019). Impact of maternal intrapartum antibiotics on the initial oral microbiome of neonates. Pediatr. Neonatol. 60, 654–661. doi: 10.1016/j.pedneo.2019.03.011
Li, Y., Wang, B., Zheng, S., and He, Y. (2019). Photodynamic therapy in the treatment of oral leukoplakia: a systematic review. Photodiagnosis Photodyn. Ther. 25, 17–22. doi: 10.1016/j.pdpdt.2018.10.023
Lima, B. P., Hu, L. I., Vreeman, G. W., Weibel, D. B., and Lux, R. (2019). The oral bacterium Fusobacterium nucleatum binds Staphylococcus aureus and alters expression of the staphylococcal accessory regulator sarA. Microb. Ecol. 78, 336–347. doi: 10.1007/s00248-018-1291-0
Lima, B. P., Shi, W., and Lux, R. (2017). Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. Microbiologyopen 6:e00444. doi: 10.1002/mbo3.444
Liu, T., Liu, J., Liu, J., Yang, R., Lu, X., He, X., et al. (2020). Interspecies interactions between Streptococcus mutans and Streptococcus agalactiae in vitro. Front. Cell. Infect. Microbiol. 10:344. doi: 10.3389/fcimb.2020.00344
Liu, X. X., Jiao, B., Liao, X. X., Guo, L. N., Yuan, Z. H., Wang, X., et al. (2019). Analysis of salivary microbiome in patients with Alzheimer’s disease. J. Alzheimers Dis. 72, 633–640. doi: 10.3233/jad-190587
Loffler, C., and Bohmer, F. (2017). The effect of interventions aiming to optimise the prescription of antibiotics in dental care-a systematic review. PLoS One 12:e0188061. doi: 10.1371/journal.pone.0188061
Long, X., Zheng, R., Liu, M., Wu, C., and Bao, J. (2020). Identification potential inhibitors against the Streptococcus quorum-sensing signal pathway. J. Biomol. Struct. Dyn. 38, 2965–2975. doi: 10.1080/07391102.2019.1651674
Lopez-Lopez, A., and Mira, A. (2020). Shifts in composition and activity of oral biofilms after fluoride exposure. Microb. Ecol. 80, 729–738. doi: 10.1007/s00248-020-01531-8
Lu, H., He, L., Jin, D., Zhu, Y., and Meng, H. (2021). Effect of adjunctive systemic antibiotics on microbial populations compared with scaling and root planing alone for the treatment of periodontitis: a pilot randomized clinical trial. J. Periodontol. 93, 570–583. doi: 10.1002/JPER.20-0764
Lu, Y., Aimetti, A. A., Langer, R., and Gu, Z. (2016). Bioresponsive materials. Nat. Rev. Mater. 2:16075. doi: 10.1038/natrevmats.2016.75
Lynge Pedersen, A. M., and Belstrøm, D. (2019). The role of natural salivary defences in maintaining a healthy oral microbiota. J. Dent. 80(Suppl. 1), S3–S12. doi: 10.1016/j.jdent.2018.08.010
Machuca, P., Daille, L., Vines, E., Berrocal, L., and Bittner, M. (2010). Isolation of a novel bacteriophage specific for the periodontal pathogen Fusobacterium nucleatum. Appl. Environ. Microbiol. 76, 7243–7250. doi: 10.1128/AEM.01135-10
Mark Welch, J. L., Ramírez-Puebla, S. T., and Borisy, G. G. (2020). Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe 12, 160–168. doi: 10.1016/j.chom.2020.07.009
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., and Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800. doi: 10.1073/pnas.1522149113
Menon, R. K., Gomez, A., Brandt, B. W., Leung, Y. Y., Gopinath, D., Watt, R. M., et al. (2019). Long-term impact of oral surgery with or without amoxicillin on the oral microbiome-a prospective cohort study. Sci. Rep. 9:18761. doi: 10.1038/s41598-019-55056-3
Mohammed, W. K., Krasnogor, N., and Jakubovics, N. S. (2018). Streptococcus gordonii Challisin protease is required for sensing cell–cell contact with Actinomyces oris. FEMS Microbiol. Ecol. 94:fiy043. doi: 10.1093/femsec/fiy043
Monroy-Pérez, E., Rodríguez-Bedolla, R. M., Garzón, J., Vaca-Paniagua, F., Arturo-Rojas Jiménez, E., and Paniagua-Contreras, G. L. (2020). Marked virulence and azole resistance in Candida albicans isolated from patients with periodontal disease. Microb. Pathog. 148:104436. doi: 10.1016/j.micpath.2020.104436
Moraes, L. C., Lang, P. M., Arcanjo, R. A., Rampelotto, P. H., Fatturi-Parolo, C. C., Ferreira, M. B. C., et al. (2020). Microbial ecology and predicted metabolic pathways in various oral environments from patients with acute endodontic infections. Int. Endod. J. 53, 1603–1617. doi: 10.1111/iej.13389
Moreira, M. S., de FreitasArchilla, J. R., Lascala, C. A., Ramalho, K. M., Gutknecht, N., and Marques, M. M. (2015). Post-treatment apical periodontitis successfully treated with antimicrobial photodynamic therapy via sinus tract and laser phototherapy: report of two cases. Photomed. Laser Surg. 33, 524–528. doi: 10.1089/pho.2015.3936
Muchová, M., Balacco, D., Grant, M., Chapple, I., Kuehne, S., and Hirschfeld, J. (2022). Fusobacterium nucleatum subspecies differ in biofilm forming ability in vitro. Front. Oral Health 3:853618. doi: 10.3389/froh.2022.853618
Mundula, T., Ricci, F., Barbetta, B., Baccini, M., and Amedei, A. (2019). Effect of probiotics on oral candidiasis: a systematic review and meta-analysis. Nutrients 11:2449. doi: 10.3390/nu11102449
Muras, A., Mallo, N., Otero-Casal, P., Pose-Rodríguez, J. M., and Otero, A. (2022). Quorum sensing systems as a new target to prevent biofilm-related oral diseases. Oral Dis. 28, 307–313. doi: 10.1111/odi.13689
Murugesan, S., Al Ahmad, S. F., Singh, P., Saadaoui, M., Kumar, M., and Al Khodor, S. (2020). Profiling the salivary microbiome of the Qatari population. J. Transl. Med. 18:127. doi: 10.1186/s12967-020-02291-2
Mutha, N. V. R., Mohammed, W. K., Krasnogor, N., Tan, G. Y. A., Choo, S. W., and Jakubovics, N. S. (2018). Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation. Mol. Oral Microbiol. 33, 450–464. doi: 10.1111/omi.12248
Oba, P. M., Holscher, H. D., Mathai, R. A., Kim, J., and Swanson, K. S. (2020). Diet Influences the oral microbiota of infants during the first six months of life. Nutrients 12:3400. doi: 10.3390/nu12113400
Oda, M., Kurosawa, M., Yamamoto, H., Domon, H., Takenaka, S., Ohsumi, T., et al. (2020). Sulfated vizantin inhibits biofilm maturation by Streptococcus mutans. Microbiol. Immunol. 64, 493–501. doi: 10.1111/1348-0421.12797
Oliveira, S. G., Nishiyama, R. R., Trigo, C. A. C., Mattos-Guaraldi, A. L., Dávila, A. M. R., Jardim, R., et al. (2021). Core of the saliva microbiome: an analysis of the MG-RAST data. BMC Oral Health 21:351. doi: 10.1186/s12903-021-01719-5
Paluch, E., Rewak-Soroczynska, J., Jedrusik, I., Mazurkiewicz, E., and Jermakow, K. (2020). Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 104, 1871–1881. doi: 10.1007/s00253-020-10349-w
Passariello, C., Gigola, P., Testarelli, L., Puttini, M., Schippa, S., and Petti, S. (2017). Evaluation of microbiota associated with herpesviruses in active sites of generalized aggressive periodontitis. Ann. Stomatol. 8, 59–70. doi: 10.11138/ads/2017.8.2.071
Pérez-Chaparro, P. J., McCulloch, J. A., Mamizuka, E. M., Moraes, A., Faveri, M., Figueiredo, L. C., et al. (2018). Do different probing depths exhibit striking differences in microbial profiles? J. Clin. Periodontol. 45, 26–37. doi: 10.1111/jcpe.12811
Pessoa, L., Aleti, G., Choudhury, S., Nguyen, D., Yaskell, T., Zhang, Y., et al. (2019). Host-microbial Interactions in systemic lupus erythematosus and periodontitis. Front. Immunol. 10:2602. doi: 10.3389/fimmu.2019.02602
Peters, B. A., Wu, J., Hayes, R. B., and Ahn, J. (2017). The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 17:157. doi: 10.1186/s12866-017-1064-9
Pinna, R., Filigheddu, E., Juliano, C., Palmieri, A., Manconi, M., D’Hallewin, G., et al. (2019). Antimicrobial effect of Thymus capitatus and Citrus limon var. pompia as raw extracts and nanovesicles. Pharmaceutics 11:234. doi: 10.3390/pharmaceutics11050234
Pires Marques, E. C., Piccolo Lopes, F., Nascimento, I. C., Morelli, J., Pereira, M. V., Machado Meiken, V. M., et al. (2020). Photobiomodulation and photodynamic therapy for the treatment of oral mucositis in patients with cancer. Photodiagnosis Photodyn. Ther. 29:101621. doi: 10.1016/j.pdpdt.2019.101621
Pride, D. T., Salzman, J., Haynes, M., Rohwer, F., Davis-Long, C., and White, R. A. III, et al. (2012). Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6, 915–926. doi: 10.1038/ismej.2011.169
Priya, A., and Pandian, S. K. (2020). Piperine impedes biofilm formation and hyphal morphogenesis of Candida albicans. Front. Microbiol. 11:756. doi: 10.3389/fmicb.2020.00756
Puth, S., Hong, S. H., Na, H. S., Lee, H. H., Lee, Y. S., Kim, S. Y., et al. (2019). A built-in adjuvant-engineered mucosal vaccine against dysbiotic periodontal diseases. Mucosal Immunol. 12, 565–579. doi: 10.1038/s41385-018-0104-6
Qi, M., Chi, M., Sun, X., Xie, X., Weir, M. D., Oates, T. W., et al. (2019). Novel nanomaterial-based antibacterial photodynamic therapies to combat oral bacterial biofilms and infectious diseases. Int. J. Nanomed. 14, 6937–6956. doi: 10.2147/IJN.S212807
Qian, W., Ma, T., Ye, M., Li, Z., Liu, Y., and Hao, P. (2019). Microbiota in the apical root canal system of tooth with apical periodontitis. BMC Genomics 20:189. doi: 10.1186/s12864-019-5474-y
Rath, S., Bal, S. C. B., and Dubey, D. (2021). Oral biofilm: development mechanism, multidrug resistance, and their effective management with novel techniques. Rambam Maimonides Med. J. 12:e0004. doi: 10.5041/rmmj.10428
Ren, W., Zhang, Q., Liu, X., Zheng, S., Ma, L., Chen, F., et al. (2017). Exploring the oral microflora of preschool children. J. Microbiol. 55, 531–537. doi: 10.1007/s12275-017-6474-8
Risely, A. (2020). Applying the core microbiome to understand host-microbe systems. J. Anim. Ecol. 89, 1549–1558. doi: 10.1111/1365-2656.13229
Robles-Sikisaka, R., Ly, M., Boehm, T., Naidu, M., Salzman, J., and Pride, D. T. (2013). Association between living environment and human oral viral ecology. ISME J. 7, 1710–1724. doi: 10.1038/ismej.2013.63
Rodríguez-Lozano, B., González-Febles, J., Garnier-Rodríguez, J. L., Dadlani, S., Bustabad-Reyes, S., Sanz, M., et al. (2019). Association between severity of periodontitis and clinical activity in rheumatoid arthritis patients: a case-control study. Arthritis Res. Ther. 21:27. doi: 10.1186/s13075-019-1808-z
Rosa, E. P., Murakami-Malaquias-Silva, F., Schalch, T. O., Teixeira, D. B., Horliana, R. F., Tortamano, A., et al. (2020). Efficacy of photodynamic therapy and periodontal treatment in patients with gingivitis and fixed orthodontic appliances: protocol of randomized, controlled, double-blind study. Medicine 99:e19429. doi: 10.1097/MD.0000000000019429
Ruhl, S., Eidt, A., Melzl, H., Reischl, U., and Cisar, J. O. (2014). Probing of microbial biofilm communities for coadhesion partners. Appl. Environ. Microbiol. 80, 6583–6590. doi: 10.1128/AEM.01826-14
Ryu, E. J., An, S. J., Sim, J., Sim, J., Lee, J., and Choi, B. K. (2020). Use of d-galactose to regulate biofilm growth of oral streptococci. Arch. Oral Biol. 111:104666. doi: 10.1016/j.archoralbio.2020.104666
Saadaoui, M., Singh, P., and Al Khodor, S. (2021). Oral microbiome and pregnancy: a bidirectional relationship. J. Reprod. Immunol. 145:103293. doi: 10.1016/j.jri.2021.103293
Sami, A., Elimairi, I., Stanton, C., Ross, R. P., and Ryan, C. A. (2020). The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome-treatment axis. Int. J. Mol. Sci. 21:8061. doi: 10.3390/ijms21218061
Sampaio-Maia, B., Caldas, I. M., Pereira, M. L., Pérez-Mongiovi, D., and Araujo, R. (2016). The oral microbiome in health and its implication in oral and systemic diseases. Adv. Appl. Microbiol. 97, 171–210. doi: 10.1016/bs.aambs.2016.08.002
Sanchez, M. C., Ribeiro-Vidal, H., Esteban-Fernandez, A., Bartolome, B., Figuero, E., Moreno-Arribas, M. V., et al. (2019). Antimicrobial activity of red wine and oenological extracts against periodontal pathogens in a validated oral biofilm model. BMC Complement. Altern. Med. 19:145. doi: 10.1186/s12906-019-2533-5
Sanz-Martin, I., Doolittle-Hall, J., Teles, R. P., Patel, M., Belibasakis, G. N., Hämmerle, C. H. F., et al. (2017). Exploring the microbiome of healthy and diseased peri-implant sites using Illumina sequencing. J. Clin. Periodontol. 44, 1274–1284. doi: 10.1111/jcpe.12788
Schaumann, S., Staufenbiel, I., Scherer, R., Schilhabel, M., Winkel, A., Stumpp, S. N., et al. (2014). Pyrosequencing of supra- and subgingival biofilms from inflamed peri-implant and periodontal sites. BMC Oral Health 14:157. doi: 10.1186/1472-6831-14-157
Şenel, S. (2021). An overview of physical, microbiological and immune barriers of oral mucosa. Int. J. Mol. Sci. 22:7821. doi: 10.3390/ijms22157821
Seneviratne, C. J., Zhang, C. F., and Samaranayake, L. P. (2011). Dental plaque biofilm in oral health and disease. Chin. J. Dent. Res. 14, 87–94.
Shaw, L., Ribeiro, A. L. R., Levine, A. P., Pontikos, N., Balloux, F., Segal, A. W., et al. (2017). The human salivary microbiome is shaped by shared environment rather than genetics: evidence from a large family of closely related individuals. mBio 8:e01237-17. doi: 10.1128/mBio.01237-17
Shen, M., Yang, Y., Shen, W., Cen, L., McLean, J. S., Shi, W., et al. (2018). A linear plasmid-like prophage of Actinomyces odontolyticus promotes biofilm assembly. Appl. Environ. Microbiol. 84:e0126318. doi: 10.1128/aem.01263-18
Shi, W., Tian, J., Xu, H., Zhou, Q., and Qin, M. (2018). Distinctions and associations between the microbiota of saliva and supragingival plaque of permanent and deciduous teeth. PLoS One 13:e0200337. doi: 10.1371/journal.pone.0200337
Shlezinger, M., Khalifa, L., Houri-Haddad, Y., Coppenhagen-Glazer, S., Resch, G., Que, Y. A., et al. (2017). Phage therapy: a new horizon in the antibacterial treatment of oral pathogens. Curr. Top. Med. Chem. 17, 1199–1211. doi: 10.2174/1568026616666160930145649
Shokeen, B., Park, J., Duong, E., Rambhia, S., Paul, M., Weinberg, A., et al. (2020). Role of FAD-I in fusobacterial interspecies interaction and biofilm formation. Microorganisms 8:70. doi: 10.3390/microorganisms8010070
Silbergleit, M., Vasquez, A. A., Miller, C. J., Sun, J., and Kato, I. (2020). Oral and intestinal bacterial exotoxins: potential linked to carcinogenesis. Prog. Mol. Biol. Transl. Sci. 171, 131–193. doi: 10.1016/bs.pmbts.2020.02.004
Silverman, R. J., Nobbs, A. H., Vickerman, M. M., Barbour, M. E., and Jenkinson, H. F. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78, 4644–4652. doi: 10.1128/IAI.00685-10
Simon-Soro, A., Ren, Z., Krom, B. P., Hoogenkamp, M. A., Cabello-Yeves, P. J., and Daniel, S. G. (2022). Polymicrobial aggregates in human saliva build the oral biofilm. mBio 13:e0013122. doi: 10.1128/mbio.00131-22
Simón-Soro, A., Tomás, I., Cabrera-Rubio, R., Catalan, M. D., Nyvad, B., and Mira, A. (2013). Microbial geography of the oral cavity. J. Dent. Res. 92, 616–621. doi: 10.1177/0022034513488119
Simpson, C. A., Adler, C., du Plessis, M. R., Landau, E. R., Dashper, S. G., Reynolds, E. C., et al. (2020). Oral microbiome composition, but not diversity, is associated with adolescent anxiety and depression symptoms. Physiol. Behav. 226:113126. doi: 10.1016/j.physbeh.2020.113126
Singh, S. B., Young, K., and Silver, L. L. (2017). What is an “ideal” antibiotic? Discovery challenges and path forward. Biochem. Pharmacol. 133, 63–73. doi: 10.1016/j.bcp.2017.01.003
Sivamaruthi, B. S., Kesika, P., and Chaiyasut, C. (2020). A review of the role of probiotic supplementation in dental caries. Probiotics Antimicrob. Proteins 12, 1300–1309. doi: 10.1007/s12602-020-09652-9
Smeekens, S. P., Huttenhower, C., Riza, A., van de Veerdonk, F. L., Zeeuwen, P. L., Schalkwijk, J., et al. (2014). Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J. Innate Immun. 6, 253–262. doi: 10.1159/000351912
Soares, G. M., Teles, F., Starr, J. R., Feres, M., Patel, M., Martin, L., et al. (2015). Effects of azithromycin, metronidazole, amoxicillin, and metronidazole plus amoxicillin on an in vitro polymicrobial subgingival biofilm model. Antimicrob. Agents Chemother. 59, 2791–2798. doi: 10.1128/AAC.04974-14
Song, Y. G., and Lee, S. H. (2017). Inhibitory effects of Lactobacillus rhamnosus and Lactobacillus casei on Candida biofilm of denture surface. Arch. Oral Biol. 76, 1–6. doi: 10.1016/j.archoralbio.2016.12.014
Staun Larsen, L., Baelum, V., Tenuta, L. M. A., Richards, A., and Nyvad, B. (2018). Fluoride in saliva and dental biofilm after 1500 and 5000 ppm fluoride exposure. Clin. Oral Investig. 22, 1123–1129. doi: 10.1007/s00784-017-2195-y
Stehlikova, Z., Tlaskal, V., Galanova, N., Roubalova, R., Kreisinger, J., Dvorak, J., et al. (2019). Oral microbiota composition and antimicrobial antibody response in patients with recurrent aphthous stomatitis. Microorganisms 7:636. doi: 10.3390/microorganisms7120636
Štšepetova, J., Truu, J., Runnel, R., Nõmmela, R., Saag, M., Olak, J., et al. (2019). Impact of polyols on oral microbiome of Estonian schoolchildren. BMC Oral Health 19:60. doi: 10.1186/s12903-019-0747-z
Sweeney, E. L., Al-Shehri, S. S., Cowley, D. M., Liley, H. G., Bansal, N., Charles, B. G., et al. (2018). The effect of breastmilk and saliva combinations on the in vitro growth of oral pathogenic and commensal microorganisms. Sci. Rep. 8:15112. doi: 10.1038/s41598-018-33519-3
Sztajer, H., Szafranski, S. P., Tomasch, J., Reck, M., Nimtz, M., Rohde, M., et al. (2014). Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271. doi: 10.1038/ismej.2014.73
Takahashi, N., and Nyvad, B. (2011). The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303. doi: 10.1177/0022034510379602
Takeshita, T., Kageyama, S., Furuta, M., Tsuboi, H., Takeuchi, K., Shibata, Y., et al. (2016). Bacterial diversity in saliva and oral health-related conditions: the Hisayama study. Sci. Rep. 6:22164. doi: 10.1038/srep22164
Takeshita, T., Suzuki, N., Nakano, Y., Yasui, M., Yoneda, M., Shimazaki, Y., et al. (2012). Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci. Rep. 2:215. doi: 10.1038/srep00215
Takeshita, T., Yasui, M., Shibata, Y., Furuta, M., Saeki, Y., Eshima, N., et al. (2015). Dental plaque development on a hydroxyapatite disk in young adults observed by using a barcoded pyrosequencing approach. Sci. Rep. 5:8136. doi: 10.1038/srep08136
The Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Tonoyan, L., Chevalier, M., Vincent-Bugnas, S., Marsault, R., and Doglio, A. (2020). Detection of Epstein-Barr virus in periodontitis: a review of methodological approaches. Microorganisms 9:72. doi: 10.3390/microorganisms9010072
Torres, P. J., Thompson, J., McLean, J. S., Kelley, S. T., and Edlund, A. (2019). Discovery of a novel periodontal disease-associated bacterium. Microb. Ecol. 77, 267–276. doi: 10.1007/s00248-018-1200-6
Torres-Barcelo, C. (2018). The disparate effects of bacteriophages on antibiotic-resistant bacteria. Emerg. Microbes Infect. 7:168. doi: 10.1038/s41426-018-0169-z
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Valm, A. M. (2019). The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J. Mol. Biol. 431, 2957–2969. doi: 10.1016/j.jmb.2019.05.016
Vartoukian, S. R., Palmer, R. M., and Wade, W. G. (2009). Diversity and morphology of members of the phylum “synergistetes” in periodontal health and disease. Appl. Environ. Microbiol. 75, 3777–3786. doi: 10.1128/aem.02763-08
Velsko, I. M., Harrison, P., Chalmers, N., Barb, J., Huang, H., Aukhil, I., et al. (2020). Grade C molar-incisor pattern periodontitis subgingival microbial profile before and after treatment. J. Oral Microbiol. 12:1814674. doi: 10.1080/20002297.2020.1814674
Wade, W. G. (2013). The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143. doi: 10.1016/j.phrs.2012.11.006
Wang, L. S., Gupta, A., and Rotello, V. M. (2016). Nanomaterials for the treatment of bacterial biofilms. ACS Infect. Dis. 2, 3–4. doi: 10.1021/acsinfecdis.5b00116
Wang, X., Li, X., and Ling, J. (2017). Streptococcus gordonii LuxS/autoinducer-2 quorum-sensing system modulates the dual-species biofilm formation with Streptococcus mutans. J. Basic Microbiol. 57, 605–616. doi: 10.1002/jobm.201700010
Wasfi, R., Abd El-Rahman, O. A., Zafer, M. M., and Ashour, H. M. (2018). Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell. Mol. Med. 22, 1972–1983. doi: 10.1111/jcmm.13496
Watanabe, N., Nodomi, K., Koike, R., Kato, A., Takeichi, O., Kotani, A. I., et al. (2019). EBV LMP1 in gingival epithelium potentially contributes to human chronic periodontitis via inducible IL8 production. In Vivo 33, 1793–1800. doi: 10.21873/invivo.11670
Weiland-Brauer, N., Malek, I., and Schmitz, R. A. (2019). Metagenomic quorum quenching enzymes affect biofilm formation of Candida albicans and Staphylococcus epidermidis. PLoS One 14:e0211366. doi: 10.1371/journal.pone.0211366
Wen, Z. T., Liao, S., Bitoun, J. P., De, A., Jorgensen, A., Feng, S., et al. (2017). Streptococcus mutans displays altered stress responses while enhancing biofilm formation by Lactobacillus casei in mixed-species consortium. Front. Cell. Infect. Microbiol. 7:524. doi: 10.3389/fcimb.2017.00524
Wolfoviz-Zilberman, A., Kraitman, R., Hazan, R., Friedman, M., Houri-Haddad, Y., and Beyth, N. (2021). Phage targeting Streptococcus mutans in vitro and in vivo as a caries-preventive modality. Antibiotics 10:1015. doi: 10.3390/antibiotics10081015
Wu, J., Peters, B. A., Dominianni, C., Zhang, Y., Pei, Z., Yang, L., et al. (2016). Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 10, 2435–2446. doi: 10.1038/ismej.2016.37
Wu, S., Liu, J., Liu, C., Yang, A., and Qiao, J. (2020). Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343. doi: 10.1007/s00018-019-03326-8
Wylie, K. M., Mihindukulasuriya, K. A., Zhou, Y., Sodergren, E., Storch, G. A., and Weinstock, G. M. (2014). Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 12:71. doi: 10.1186/s12915-014-0071-7
Xiao, J., Grier, A., Faustoferri, R. C., Alzoubi, S., Gill, A. L., Feng, C., et al. (2018a). Association between oral Candida and bacteriome in children with severe ECC. J. Dent. Res. 97, 1468–1476. doi: 10.1177/0022034518790941
Xiao, J., Huang, X., Alkhers, N., Alzamil, H., Alzoubi, S., Wu, T. T., et al. (2018b). Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52, 102–112. doi: 10.1159/000481833
Xu, X., He, J., Xue, J., Wang, Y., Li, K., Zhang, K., et al. (2015). Oral cavity contains distinct niches with dynamic microbial communities. Environ. Microbiol. 17, 699–710. doi: 10.1111/1462-2920.12502
Xun, Z., Zhang, Q., Xu, T., Chen, N., and Chen, F. (2018). Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front. Microbiol. 9:1136. doi: 10.3389/fmicb.2018.01136
Yamanaka, W., Takeshita, T., Shibata, Y., Matsuo, K., Eshima, N., Yokoyama, T., et al. (2012). Compositional stability of a salivary bacterial population against supragingival microbiota shift following periodontal therapy. PLoS One 7:e42806. doi: 10.1371/journal.pone.0042806
Yamashita, Y., and Takeshita, T. (2017). The oral microbiome and human health. J. Oral Sci. 59, 201–206. doi: 10.2334/josnusd.16-0856
Yang, C., Scoffield, J., Wu, R., Deivanayagam, C., Zou, J., and Wu, H. (2018). Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol. Oral Microbiol. 33, 283–291. doi: 10.1111/omi.12223
Yang, C. Y., Yeh, Y. M., Yu, H. Y., Chin, C. Y., Hsu, C. W., Liu, H., et al. (2018). Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front. Microbiol. 9:862. doi: 10.3389/fmicb.2018.00862
Yang, Y., Zheng, W., Cai, Q. Y., Shrubsole, M. J., Pei, Z., Brucker, R., et al. (2019). Cigarette smoking and oral microbiota in low-income and African-American populations. J. Epidemiol. Community Health 73, 1108–1115. doi: 10.1136/jech-2019-212474
Yang, Z., Cui, Q., An, R., Wang, J., Song, X., Shen, Y., et al. (2020). Comparison of microbiomes in ulcerative and normal mucosa of recurrent aphthous stomatitis (RAS)-affected patients. BMC Oral Health 20:128. doi: 10.1186/s12903-020-01115-5
Yu, G., Phillips, S., Gail, M. H., Goedert, J. J., Humphrys, M. S., Ravel, J., et al. (2017). The effect of cigarette smoking on the oral and nasal microbiota. Microbiome 5:3. doi: 10.1186/s40168-016-0226-6
Yuan, K., Hou, L., Jin, Q., Niu, C., Mao, M., Wang, R., et al. (2021). Comparative transcriptomics analysis of Streptococcus mutans with disruption of LuxS/AI-2 quorum sensing and recovery of methyl cycle. Arch. Oral Biol. 127:105137. doi: 10.1016/j.archoralbio.2021.105137
Yussof, A., Yoon, P., Krkljes, C., Schweinberg, S., Cottrell, J., Chu, T., et al. (2020). A meta-analysis of the effect of binge drinking on the oral microbiome and its relation to Alzheimer’s disease. Sci. Rep. 10:19872. doi: 10.1038/s41598-020-76784-x
Zargar, N., Ashraf, H., Marashi, S. M. A., Sabeti, M., and Aziz, A. (2020). Identification of microorganisms in irreversible pulpitis and primary endodontic infections with respect to clinical and radiographic findings. Clin. Oral Investig. 24, 2099–2108. doi: 10.1007/s00784-019-03075-9
Zaura, E., Brandt, B. W., Teixeira de Mattos, M. J., Buijs, M. J., Caspers, M. P., Rashid, M. U., et al. (2015). Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio 6:e01693-15. doi: 10.1128/mBio.01693-15
Zaura, E., Keijser, B. J., Huse, S. M., and Crielaard, W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. doi: 10.1186/1471-2180-9-259
Zaura, E., and Twetman, S. (2019). Critical appraisal of oral pre- and probiotics for caries prevention and care. Caries Res. 53, 514–526. doi: 10.1159/000499037
Zhan, L. (2018). Rebalancing the caries microbiome dysbiosis: targeted treatment and sugar alcohols. Adv. Dent. Res. 29, 110–116. doi: 10.1177/0022034517736498
Zhang, L., Liu, Y., Zheng, H. J., and Zhang, C. P. (2019). The oral microbiota may have Influence on oral cancer. Front. Cell. Infect. Microbiol. 9:476. doi: 10.3389/fcimb.2019.00476
Zhang, Y., Zhu, C., Feng, X., and Chen, X. (2021b). Microbiome variations in preschool children with halitosis. Oral Dis. 27, 1059–1068. doi: 10.1111/odi.13603
Zhang, Y., Qi, Y., Lo, E. C. M., McGrath, C., Mei, M. L., and Dai, R. (2021a). Using next-generation sequencing to detect oral microbiome change following periodontal interventions: a systematic review. Oral Dis. 27, 1073–1089. doi: 10.1111/odi.13405
Zhang, Y., Wang, X., Li, H., Ni, C., Du, Z., and Yan, F. (2018). Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 99, 883–893. doi: 10.1016/j.biopha.2018.01.146
Zhao, C., Lv, X., Fu, J., He, C., Hua, H., and Yan, Z. (2016). In vitro inhibitory activity of probiotic products against oral Candida species. J. Appl. Microbiol. 121, 254–262. doi: 10.1111/jam.13138
Zhao, H., Chu, M., Huang, Z., Yang, X., Ran, S., Hu, B., et al. (2017). Variations in oral microbiota associated with oral cancer. Sci. Rep. 7:11773. doi: 10.1038/s41598-017-11779-9
Zheng, H., Xie, T., Li, S., Qiao, X., Lu, Y., and Feng, Y. (2021). Analysis of oral microbial dysbiosis associated with early childhood caries. BMC Oral Health 21:181. doi: 10.1186/s12903-021-01543-x
Zheng, H., Xu, L., Wang, Z., Li, L., Zhang, J., Zhang, Q., et al. (2015). Subgingival microbiome in patients with healthy and ailing dental implants. Sci. Rep. 5:10948. doi: 10.1038/srep10948
Zheng, J., Wu, Z., Niu, K., Xie, Y., Hu, X., Fu, J., et al. (2019). Microbiome of deep dentinal caries from reversible pulpitis to irreversible pulpitis. J. Endod. 45, 302–309.e1. doi: 10.1016/j.joen.2018.11.017
Zhou, P., Manoil, D., Belibasakis, G. N., and Kotsakis, G. A. (2021). Veillonellae: beyond bridging species in oral biofilm ecology. Front. Oral Health 2:774115. doi: 10.3389/froh.2021.774115
Zhou, Y., Cheng, L., Lei, Y. L., Ren, B., and Zhou, X. (2021). The interactions between Candida albicans and mucosal immunity. Front. Microbiol. 12:652725. doi: 10.3389/fmicb.2021.652725
Zhu, Z., He, Z., Xie, G., Fan, Y., and Shao, T. (2021). Altered oral microbiota composition associated with recurrent aphthous stomatitis in young females. Medicine 100:e24742. doi: 10.1097/MD.0000000000024742
Keywords: oral microbiome, composition, biofilm, community, oral disease, microbial dysbiosis
Citation: Li X, Liu Y, Yang X, Li C and Song Z (2022) The Oral Microbiota: Community Composition, Influencing Factors, Pathogenesis, and Interventions. Front. Microbiol. 13:895537. doi: 10.3389/fmicb.2022.895537
Received: 14 March 2022; Accepted: 06 April 2022;
Published: 29 April 2022.
Edited by:
Fabian Cieplik, University Medical Center Regensburg, GermanyReviewed by:
Jonathan Vernon, University of Leeds, United KingdomCopyright © 2022 Li, Liu, Yang, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengwen Li, bGN3NzExMUBzd211LmVkdS5jb20=; Zhangyong Song, c3p5ODM1MjlAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.