
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 August 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.895533
This article is part of the Research Topic Environmental Influences on the Host-associated Microbiomes View all 13 articles
The availability of limiting nutrients plays a crucial role in shaping communities of endophytes. Moreover, whether fungal endophytes are host-specific remains controversial. We hypothesized that in a harsh and nitrogen (N)-deficient area, diversity and community composition of foliar endophytic fungi (FEFs) varied substantially among plots with experimentally elevated levels of macronutrients, and thus, N availability, instead of host species identity, would have a greater influence in structuring fungal communities at different scales. We also expected an important subset of taxa shared among numerous host species and N gradients to form a community-wide core microbiome. We measured the leaf functional traits and community structures of FEFs of three commonly seen species in an alpine meadow nested with a long-term N fertilization experiment. We found that host plant identity was a powerful factor driving the endophytic fungal community in leaves, even in habitats where productivity was strongly limited by nitrogen (p < 0.001). We also found that within the same host, nitrogen was an important driving force for the composition of the endophytic fungi community (p < 0.05). In addition, the leaf carbon content was the most important functional trait that limited the diversity of endophytic fungi (p < 0.001). Finally, we documented a distinct core microbiome shared among our three focal species and N gradients.
Ecosystems worldwide have been undergoing severe nitrogen (N) deposition over the last few decades (Cardinale et al., 2011; IPCC, 2014). In terrestrial ecosystems, N availability is supposed to be a primary limitation to plant growth (Vitousek and Howarth, 1991). Previous research has shown that N deposition, simulated by N fertilization experiments, increases plant growth and crop yields. However, N losses from industry and agriculture alter the plant community structure and reduce terrestrial biodiversity (Stevens et al., 2004; Bobbink et al., 2010). N deposition also results in an increase in air pollution (EPA, 2013) and soil acidification (Sullivan et al., 2013) and is hence considered a major threat to ecosystem functioning and services. While N deposition has declined in some parts of the world due to legislation, it is estimated that soil communities, ecosystem productivity, and nutrient cycling continue to be affected by historical soil nitrogen loads (Gilliam et al., 2019; Crawford et al., 2020).
Although the effect of atmospheric N deposition on plant ecology has been widely studied (Ellison and Gotelli, 2002; Scarpitta et al., 2017; Vallicrosa et al., 2022), our understanding of how N shapes the microbial community, particularly the fungal community, inside plants remains elusive. Plants do not exist as axenic organisms but are closely associated with large quantities of microorganisms. Leaves comprise one of the world's largest terrestrial habitats and provide a key location for interactions between plants and their associated microbiomes, yet we know relatively little about this association (Vacher et al., 2016). Foliar endophytic fungi (FEFs) spend all or part of their lifetime colonizing plant leaves and may increase, decrease, or show no apparent impact on host performance, as reviewed by Rodriguez et al. (2009) and Hardoim et al. (2015). Such mutualistic parasitic symbiosis depends on the host–endophyte genotype–genotype interaction, environmental conditions, and the state of the host's health (Stevens et al., 2004). However, the mechanism of this complex interactive network remains unclear. Moreover, although existing research indicates that FEFs mediate plant N uptake (Buckley et al., 2019; Christian et al., 2019), how N input affects the community structure of the FEF remains relatively unexplored. FEFs represent a ubiquitous and highly diversified fungal guild (Rodriguez et al., 2009) and a substantial and mostly under-explored microbiota critical for ecosystem functioning (Hardoim et al., 2015). Despite the ecological importance, the factors that shape FEF communities, either biotic or abiotic, are largely unknown.
Plant species shape biotic and abiotic environments in which the fungi live, and hence, different FEF species might be a function of host plant phylogeny (Apigo and Oono, 2018). However, the degree that FEFs are host-specific is controversial. Some endophytes were unique to divergent host clades in boreal and arctic sites (Higgins et al., 2007; Zhang and Yao, 2015), and they also showed a degree of host specificity in rainforest trees (Vincent et al., 2016), tropical trees (Arnold and Lutzoni, 2007), and grasses (Higgins et al., 2011, 2014). If high specificity is prevalent, this may have profound implications for plant–fungus coevolution, as plants can vary dramatically in susceptibility to their parasitic fungal companions, and plants prefer harboring mutualistic fungal companions (Clay and Schardl, 2002; Saikkonen et al., 2004; Chen et al., 2019; Razali et al., 2019). Conversely, it was also found that distantly related pine species hosted the same FEF species, indicating host species did not influence the microbial genetic structure (Oono et al., 2014). This might be explained by the fact that FEFs are often horizontally transmitted (Arnold et al., 2009; Rodriguez et al., 2009); thus, some populations may be panmictic, reflecting generalist associations with multiple host species (Dunham et al., 2013). Altogether, much evidence needs to be accumulated to understand the host specificity of FEF communities. Understanding the host specificity of FEFs will also provide critical insights into plant performance, competitive abilities among species, and the FEF dispersal strategy.
N input increases plant biomass and enhances plant growth. Changes in plant growth may further lead to variations in the plant-associated fungal community structure (Seghers et al., 2004; Larkin et al., 2012; Suryanarayanan and Shaanker, 2021). Furthermore, over different growth stages of the host plant, the plant endophytic fungal community is likely subject to dynamic changes over time (Singha et al., 2021; Sosso et al., 2021; Zheng et al., 2021). Several studies have provided some insights into how FEFs respond to high N supply. An inoculation experiment showed that endophyte (Neotyphodium lolii) concentration was reduced by 40% under high N supply in perennial ryegrass (Lolium perenne) cultivars (Rasmussen et al., 2007). Data from several sources also identified that FEFs enhanced plant N uptake and growth based on inoculation experiments (Newman et al., 2003; Knoth et al., 2013; Christian et al., 2019). However, all these conclusions were drawn based on greenhouse-based experiments with inoculation of specific species of endophytic fungi. How macronutrients, particularly N, regulate endophytic fungal communities in natural systems remains controversial. For example, Larkin et al. (2012) reported that nitrate in leaves was correlated with differences in the endophyte community, while in another study, Huang et al. (2016) found no correlation between foliar N and endophytic abundance or diversity in one host species, but a negative correlation with endophytic abundance in another species was found.
Independent of whether FEFs are highly host-specific, there still may be a subset of taxa that are shared among co-occurring host species. For example, Zhang and Yao (2015) found that 7.6% of total FEF operational taxonomic units (OTUs) were shared by all four plant species in the study of vascular plants in the high arctic zone. Thus, a very small group of fungi occurred repeatedly among plant species. Some subsets of fungi that comprise the shared OTUs (or, “core microbiome”, see the explanation from Vandenkoornhuyse et al., 2015 and Shade and Handelsman, 2012) may have traits that enhance their ability to colonize the interior of a phylogenetically diverse group of plant species, indicating their ecological competitiveness and importance. However, such investigations are very few, therefore, we cannot draw general conclusions on commonly shared FEF OTUs among plant species in different habitats.
The Qinghai–Tibet Plateau (QTP) is one of the hot spots for research, yet little is known about the endophytic fungal community and its response to global change in this region. Learning endophytic fungal host specificity and simulating the effects of N deposition on endophytic fungal communities will help us better identify the ecological and evolutionary constraints that limit symbiont distribution. To fill this knowledge gap, we established the following three hypotheses: (1) In alpine meadows on the QTP, FEFs vary substantially among different host plant species, (2) a subset of microbial taxa will be shared among all host plant species, and (3) FEF communities vary substantially among plots with experimentally elevated levels of N under N-deficient conditions, and nutrient availability would have a greater influence in structuring fungal communities than host identity. We assume that if phylogenetically conserved traits serve as biological filters for endophyte colonization and establishment, the endophytic fungal community may vary considerably in species with different morphology, structure, and tissue texture, as suggested by Zhang and Yao (2015). A study in southern Chile, on the other hand, found that the homogeneity of endophytic fungal communities might be due to the filtering effect of physical and chemical traits of leaves independent of the evolutionary history of the host (Gonzalez-Teuber et al., 2020). Therefore, we hypothesize that in alpine meadows on the QTP, where productivity is strongly limited by N availability (Xu et al., 2015), N will transcend host identity in shaping the community composition of FEFs. To address these hypotheses, we used high-throughput sequencing to characterize the FEF community composition of three grass species in 4-year N addition treatments in an alpine meadow ecosystem on the QTP. To our knowledge, this is the first study to explore the responses of FEF communities to N fertilization in an alpine meadow ecosystem. This research provides better understanding of the response of plant-associated fungi to changing global scenarios.
This study was conducted in 2014 at the Research Station of Alpine Meadow and Wetland Ecosystems of Lanzhou University, Maqu County, China. The study site is located in the eastern QTP (33°40′N, 101°51′E, altitude 3,500 m a.s.l, Supplementary Figure S1), with an average precipitation of 620 mm (mostly occurs in summer). The mean annual temperature is about 1.2°C (ranging from −10°C in winter to 11.7°C in summer), and the frost-free season lasts about 95 days (Wu et al., 2010). The mean aboveground primary productivity is 280–400 g m−2 (dry weight), and the species richness is, on average, 20–35 per 0.25 m2 (Yang et al., 2011). The vegetation of the meadow is mainly dominated by Kobresia capillifolia (Cyperaceae), Elymus nutans Griseb (Poaceae), and Anemone rivularis Buch.-Ham. (Ranunculaceae). The soil type in the study area is Mattic Cryic Cambisols (Gong and Li, 2001). Total N deposition in this region is estimated to range from 14.26 to 18.65 kg ha−1 yr−1 (Lu and Tian, 2007).
The experimental site was used for grazing in the past and was fenced in 2011 to prevent grazing by large animals, such as yak and sheep. To understand how plants and related microorganisms respond to N addition in the field, NH4NO3 fertilizer was applied annually at the beginning of the growing season (usually in May) since then. The field experiment was a complete randomized block design, with 24 plots (20 × 10 m each, 1-m buffer strips) arranged in a regular 6 × 4 matrix. N gradients were generated with 0, 5, 10, and 15 g N (NH4NO3) m−2 yr−1 referring to N0 (control), N5, N10, and N15 treatments, and each treatment had six replicates (Supplementary Figure S1). After 4 years of N addition, available soil N concentration was increased significantly (F = 11.1, p < 0.001, Supplementary Table S1).
Samples were collected on 25 July 2014. We targeted three commonly observed plant species, namely, A. rivularis, E. nutans, and Thermopsis lanceolata. In each plot, six individuals of each species were randomly selected. For individuals of A. rivularis and T. lanceolata, five random mature leaves devoid of visible pathogen damage were collected from each individual. For individuals of E. nutans, the second leaf from the bottom of each individual was collected. The leaves collected from the same species in a plot were mixed as a sample. In total, we collected 71 samples (3 species × 24 plots, T. lanceolata was not found in one of the N15 plots). The samples were placed in zip-lock plastic bags and stored in coolers equipped with ice packs, transferred to the laboratory in 18 h, and stored at 4°C.
The leaves were thoroughly washed in tap water, patted dry, and divided into two subsamples: One subsample was sterilized for DNA extraction within 36 h of collection, and the other was stored at −20°C until the determination of the physical and chemical properties of the leaves. Before DNA extraction, the leaves were immersed in 75% ethanol for 1 min, in 1% sodium hypochlorite for 2 min, and in 75% ethanol for 30 s to eliminate microorganisms on leaf surfaces. The surface-sterilized tissues were then rinsed with sterile water for 30 s three times and then patted dry with a sterile filter paper. Before sample grinding, ~1 ml of the final rinse water of each sample was plated on potato dextrose agar (PDA) and cultured in the dark to validate the effect of surface sterilization. The leaves were ground with liquid nitrogen using mortars and pestles in a sterile room. The mortars and pestles were sterilized before use and re-sterilized between samples to avoid cross-contamination.
Total genomic DNA was extracted using 100 mg leaf tissue powder from each sample using a Plant DNA Extraction Kit following the manufacturer's instructions (Tiangen Biotech, Beijing, China). The extracted DNA samples were frozen and shipped to Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China. Fungal diversity was determined by sequencing the internal transcribed spacer (ITS) region with primers ITS1F (CTTGGTCATTTAGAGGAAGTAA)/ITS2 (GCTGCGTTCTTCATCGATGC) (Gardes and Bruns, 1993) on an Illumina MiSeq platform (2 × 250 PE).
Sequences were pre-processed using the Trimmomatic tool (v 0.36, Bolger et al., 2014). 3′ or 5′ ends with a Phred quality score lower than 20 were trimmed, and sequences of <200 bp in length and with an average quality score of <20 on a window of 50 bases were discarded. The remaining sequences were imported into QIIME2 version 2019.10 for bioinformatics analyses (Bolyen et al., 2019). The qiime2-dada2 plugin was used for denoising, dereplication, merging paired-end reads, and removing chimeras (Callahan et al., 2016). Molecular singletons were removed from the downstream analysis to minimize the possibility of sequencing artifacts (Unterseher et al., 2011). Taxonomic assignments were determined using the qiime2-feature-classifier (Bokulich et al., 2018) classify-sklearn against species hypotheses (SH) of UNITE's database version 8.2 (Koljalg et al., 2020). Click or tap here to enter text. Trained with the Naive Bayes classifier with a confidence threshold of 97%. All representative sequencing data were submitted to the National Center for Biotechnology Information (NCBI) under accession numbers OM744714-OM745695.
The leaf traits were measured using the second subsample stored at −20°C. This work was completed within 10 days of sample collection. We focused on nitrogen and carbon fractions. Total leaf carbon and nitrogen per tree were determined using a C/N elemental analyzer, Vario MAXCN (Elementar, Hessia, Germany). Low-molecular weight (LMW) carbohydrates (mostly glucose, fructose, and sucrose) and high-molecular weight (HMW) carbohydrates (mainly fructans) were extracted and quantified from aboveground tissues, as previously described (Hunt et al., 2005; Rasmussen et al., 2007). Total free amino acids were analyzed from the ethanol fraction used for carbohydrate extraction and were determined colorimetrically with ninhydrin, as described previously (Yemm et al., 1955). Soluble proteins were extracted using 0.1% mercaptoethanol in 100 mM potassium phosphate buffer. Extracts were centrifuged, and the resulting supernatant was analyzed according to Bradford (1976) with absorbance measured at 595 nm using bovine serum albumin (BSA) as a standard.
The averages and standard deviations of the tree traits and leaf chemicals in each plot were calculated. Diversity assessment used Fisher's alpha (a richness index) and Shannon-index (considering both richness and abundance). Analyses were combined with ANOVA of the multivariate generalized linear models and randomized species accumulation curves. The abundance-based Bray–Curtis similarity coefficient was used to examine the dissimilarity of community composition and turnover. The distinctiveness of leaf endophytic fungi in different hosts and under different N conditions, and independence of the interactions of hosts and N were tested using a permutational multivariate analysis of variance (PERMANOVA) using the distance metrics mentioned earlier. The beta diversity of FEFs was analyzed using non-metric multidimensional scaling (NMDS) by R 4.1.1 statistical software (R Core Team, 2019). In addition, the environmental variables were screened using bioenv, mantel, and vif.cca functions, and the selected environmental variables were fitted onto the NMDS ordination plot using the envfit function in the vegan package v2.5 (Oksanen et al., 2019). A Mantel test was used in the vegan package to identify the plant traits that significantly correlated with the FEF community composition. Pearson's correlation analyses were used to identify the plant traits that significantly correlated with the FEF diversity indices in R. To reflect the OTU composition similarity among different samples or host identities, we performed network analysis. Sample-level OTUs by occurrence frequency were first filtered. A correlation between the two samples was considered statistically robust if Spearman's correlation coefficient (ρ) was >0.4 and the P-value was <0.05. All the robust correlations identified from pairwise comparison of OTU abundance form a correlation network, where each node represents one sample and each edge stands for a strong and significant correlation between nodes. Network analyses used the R packages vegan, igraph (Csardi and Nepusz, 2006), and Hmisc (Harrell and Dupont, 2018). Network visualization was then conducted using the interactive platform Gephi (Bastian et al., 2009). All variables were transformed, where necessary. Prior to all analyses, data were rarefied to 1,944 sequence counts (minimum sequencing depth) using rrarefy function in the vegan package.
In total, 71 genomic DNA samples were sequenced, generating a total of 1,284,298 sequences after quality filtering. A total of 1,457 OTUs were clustered at the 97% sequence identity level. Of these, 475 (32.6%) OTUs were non-fungal taxa and were removed from the pool, the remaining 966,780 sequences; varied between 1,944 and 49,914 per sample; and clustered into 982 OTUs (Supplementary Figure S2a). Specifically, 337,614 sequences were observed in A. rivularis, 509,880 sequences were found in E. nutans, and 119,286 sequences were recovered in T. lanceolata, clustered into 505, 507, and 280 OTUs, respectively (Supplementary Table S2). The rarefaction curves of the observed OTU richness failed to approach the asymptote (Supplementary Figure S2b). This implies that a few rare fungal taxa were not observed, due to either insufficient sampling or sequencing depths. Nevertheless, valid comparisons can still be drawn at the species or at the plot level.
Endophytic fungal communities were strongly dominated by ascomycetes and basidiomycetes, together accounting for 99.4% of the sequences (Figure 1A; Supplementary Figure S3a; Supplementary Table S3). Members of those classified as “Basidiomycota” were present primarily in E. nutans and T. lanceolata, but ascomycetes are more prevalent in A. rivularis (Figure 1A). Early diverging lineages, including Chytridiomycota, Mortierellomycota, and Zoopagomycota (862 sequences, comprising 0.09% of the total sequences), were also observed (Supplementary Table S3). Unidentified OTUs (i.e., those classified as fungi but failed to be assigned to a phylum) comprised 0.51% of the total sequences (Supplementary Table S3). The most dominant class across our dataset, Dothideomycetes, comprised ~35% of all assigned sequence reads, followed by Tremellomycetes (29.2%), Sordariomycetes (16.0%), and Eurotiomycetes [(6.1%), Figure 1B; Supplementary Figure S3b; Supplementary Table S4].
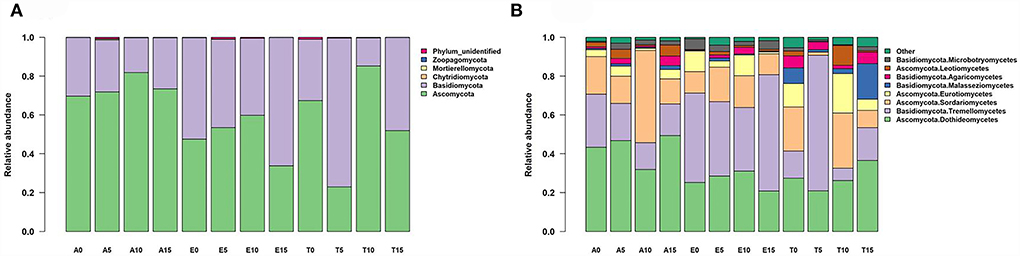
Figure 1. Taxonomic composition, (A) phylum level, and (B) class level of the fungal communities recovered from leaf interiors of three species along the nitrogen enrichment gradients in the study site. Bars show relative abundance of different taxonomic groups. The fungal endophyte classes with relative abundances of <0.1% were assigned to “Other”. A, Anemone rivularis; E, Elymus nutans; T, Thermopsis lanceolata. 0 (control), 5, 10, and 15 referring to 0, 5, 10, and 15 g N (NH4NO3) m−2 yr−1 treatments, respectively.
Diversity measures displayed a significant difference among the three hosts. A. rivularis samples showed higher phylotype richness and Shannon diversity than E. nutans and T. lanceolata (Figure 2A). Statistical tests confirmed a significant effect of host types (GLM, p < 0.01, Table 1). N input showed a relatively weak effect on fungal diversity (Figure 2B; Table 1). However, at a smaller scale (i.e., within-host scale), N input decreased the fungal diversity of A. rivularis but increased that of T. lanceolata (Figure 3; Supplementary Table S5). Thus, fungal diversity can be maintained across the plot scale, except at the smaller, within-host scale under the background of N enrichment.
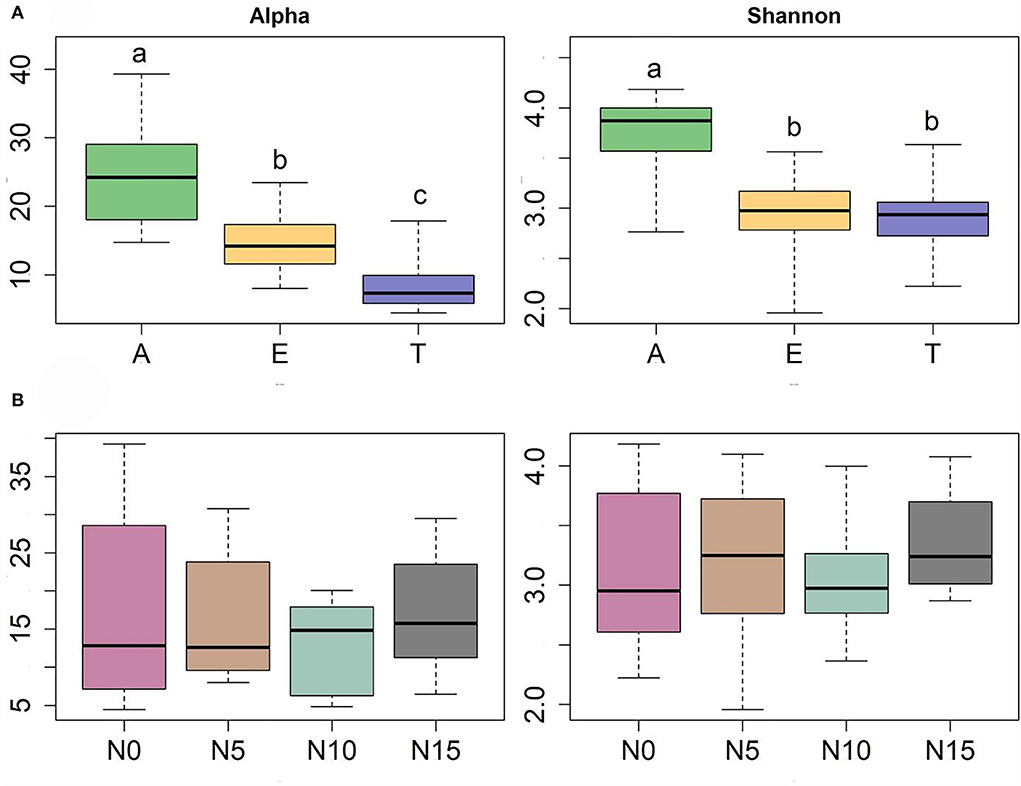
Figure 2. Diversity patterns of the entire fungal community of three species against host (A) and N gradients (B) based on linear models. Alpha is the number of observed species, and Shannon is the Shannon diversity index. The sequences were rarified to 1,944, n = 71. Significant differences of each variable are indicated by dissimilar letters above boxes. A, Anemone rivularis; E, Elymus nutans; T, Thermopsis lanceolata. N0, N5, N10, and N15 referring to 0, 5, 10, and 15 g N (NH4NO3) m−2 yr−1 treatments, respectively.
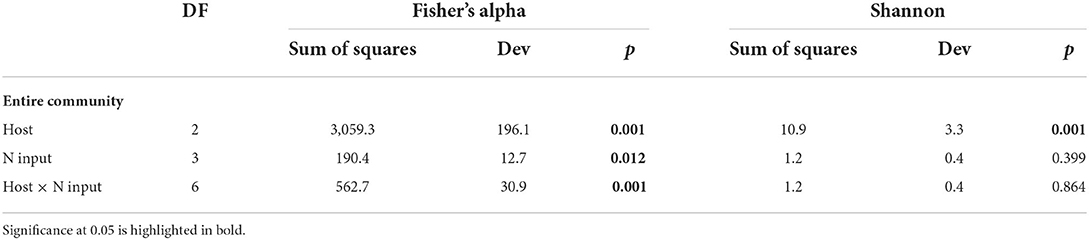
Table 1. Diversity statistics for the fungal endophytes in three hosts based on a multispecies generalized linear model.
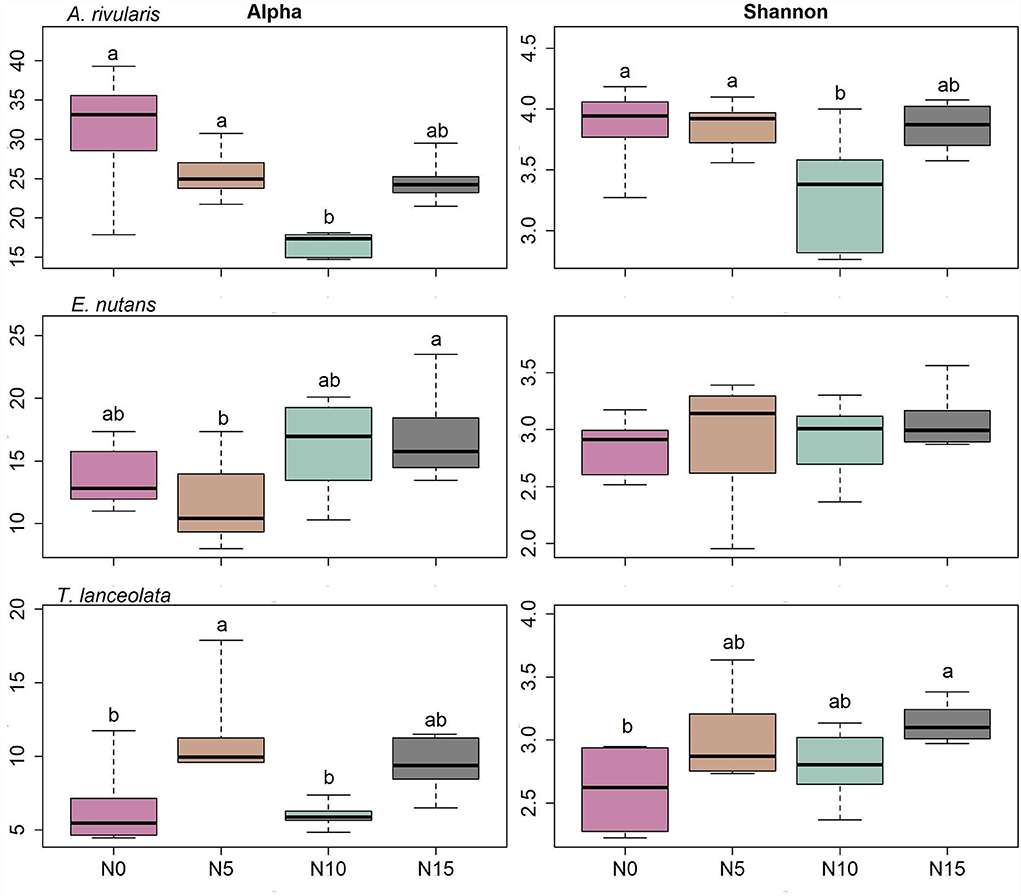
Figure 3. Diversity patterns of the fungal community within each host against N gradients. Alpha is the number of observed species, and Shannon is the Shannon diversity index. The sequences were rarified to 1,944, n = 71. Significant differences of each variable are indicated by dissimilar letters above boxes. N0, N5, N10, and N15 referring to 0, 5, 10, and 15 g N (NH4NO3) m−2 yr−1 treatments, respectively.
Composition of leaf mycobiome differed significantly among hosts (Figure 4A). However, the effects of fertilization led to overlapped fungal communities across the plot scale (Figure 4B). The dissimilarities of communities of FEFs among hosts, N gradients, and the interaction of the two factors were also confirmed by a multispecies generalized linear model calculation and a permutational multivariate analysis of variance (PERMANOVA) (Table 2). Community turnover in each host showed stronger clustering with N enrichment (Figure 4C; Supplementary Table S6).
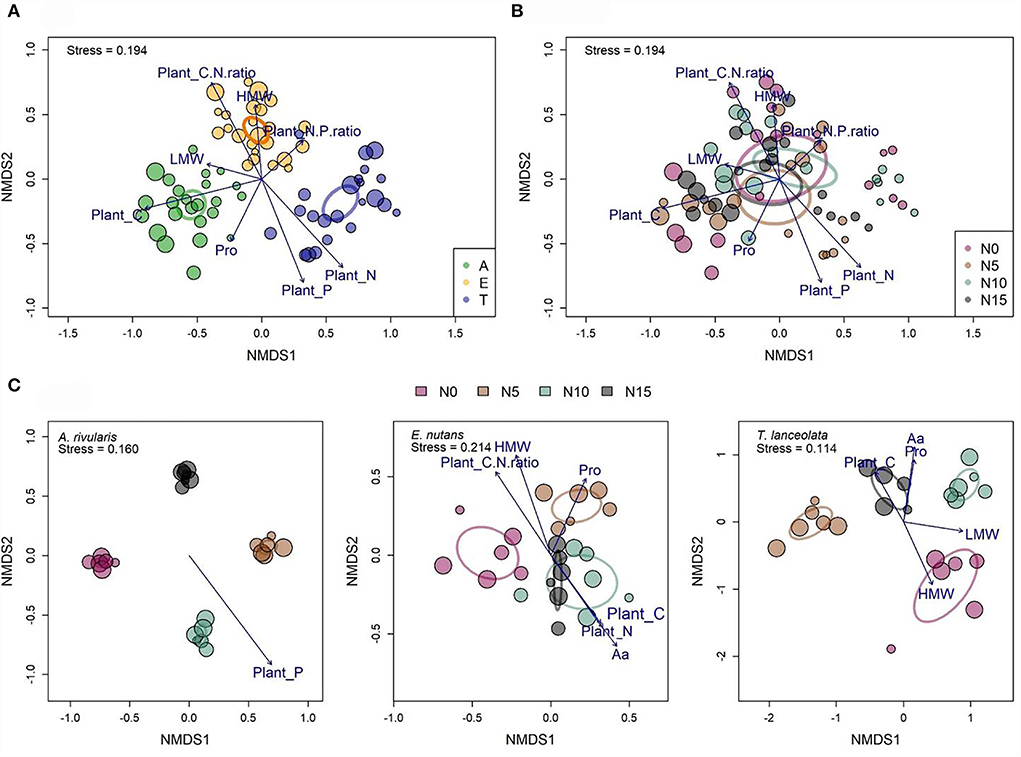
Figure 4. Community structure of fungal endophytes among different hosts (A), N gradients (B), and within each host along the N gradients (C) were assessed with non-metric multidimensional scaling (NMDS) (Bray–Curtis distance). Dot size is proportional to the operational taxonomic unit (OTU) richness of individual samples (n = 71). Ellipses with different colors indicate 95% confidence intervals. Significant plant variables that are correlated with each community ordination are shown. Pro: soluble proteins. LMW, low-molecular weight carbohydrates; HMW, high-molecular weight carbohydrates; Aa, total free amino acids; A, Anemone rivularis; E, Elymus nutans; T, Thermopsis lanceolata. N0, N5, N10, and N15 referring to 0, 5, 10, and 15 g N (NH4NO3) m−2 yr−1 treatments, respectively.
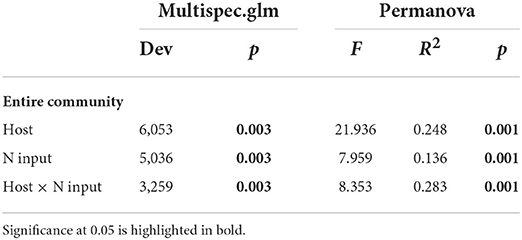
Table 2. Statistical testing of fungal compositional dissimilarity based on a multispecies generalized linear model calculation (multispec.glm) and a permutational multivariate analysis of variance (PERMANOVA) using Bray–Curtis distance metrics.
The effects of fertilization on plant measures varied among hosts. Only total free amino acids (Aa) and total soluble proteins (Pro) significantly changed in all three species with N fertilization (Supplementary Table S7). In E. nutans, both low-molecular weight (LMW) carbohydrates and high-molecular weight (HMW) carbohydrates decreased under high N treatment (F = 3.783, p = 0.027 and F = 3.253, p = 0.043, respectively). Although these water-soluble carbohydrates also decreased in A. rivularis and T. lanceolata under high N conditions, only HMW sugars in T. lanceolata showed a significant response (F = 7.547, p < 0.001).
Unlike observations in soil (Supplementary Table S1), only tissue N in E. nutans was found increased under the high N condition (F = 14.298, p < 0.001), leading to a significant decrease in the tissue C/N ratio (F = 18.287, p < 0.001) and an increase in the tissue N/P ratio (F = 4.129, p = 0.02). An increased tissue N/P ratio was observed in A. rivularis (F = 5.054, p = 0.009) due to a decreased tissue P concentration (F = 4.837, p = 0.011). At the plot scale, elevated nutrient supply can cause a significant change in Pro (F = 3.242, p = 0.04), Aa (F = 51.364, p < 0.001), and tissue N/P ratio (F = 5.854, p = 0.005).
Clearly, environmental variables were significantly correlated with both endophytic fungal diversity and community composition (Table 3). Pearson's correlation analyses showed that Pro, LMW sugars, tissue C, N, C/N ratio, and N/P ratio could be significantly related to the fungal alpha diversity indices to varying degrees (Table 3). There was a strong positive correlation between leaf carbon and endophytic fungal diversity indices, and LMW carbohydrates and Pro also showed a similar effect. Tissue N had a significant negative influence on endophytic fungal richness, but not on Shannon diversity. In addition, the leaf N/P ratio is negatively related to fungal diversity.
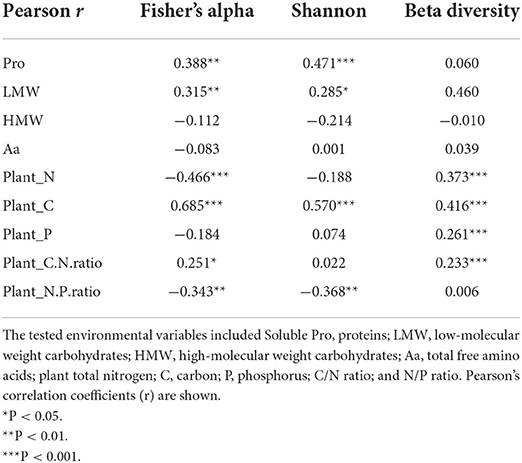
Table 3. Pearson's product–moment correlations between environmental variables and alpha (Fisher and Shannon indices) and beta diversity (Bray–Curtis distances) of fungal endophytes in three hosts.
The Mantel test revealed a significant correlation between the fungal community composition and the following environmental variables (listed from highest to lowest Pearson's correlation coefficients): tissue C > N > P > C/N ratio (Table 3). With the envfit function in the vegan package (Oksanen et al., 2019) of R, Pro, LMW sugars, tissue C, N, P, C/N ratio, and N/P ratio were chosen to fit the NMDS plot (Figures 4A,B). When controlling for species, OTU assemblage was significantly correlated with tissue P in A. rivularis. Pro, Aa, HMW, tissue C, N, and C/N ratio were correlated with the fungal community in E. nutans, and ordination of T. lanceolata displayed the significance of five parameters, namely, Aa, Pro, LMW, and HMW sugars, and tissue C (Figure 4C). In summary, among all the measured indices, leaf carbon was considered the most significant factor constraining alpha and beta diversity.
Venn diagrams illustrated 927 OTUs (rarefied data) partitioning among the plant species and N gradients (Supplementary Figure S4). A total of 94 OTUs were shared among three species (10.14%, Supplementary Figure S4a), and 95 OTUs appeared in all treatment plots (10.25%, Supplementary Figure S4b). At the within-host scale, 47 (9.46%), 58 (11.79%), and 18 (6.43%) OTUs in A. rivularis, E. nutans, and T. lanceolata were shared among all N gradients, respectively (Supplementary Figure S5). We found evidence for a core microbiome, whereby nine OTUs were present among 90% or more of samples in the N0 (control) plot. These nine OTUs comprised ~30% of the total reads (Supplementary Table S8), and taxonomic assignments were made using the National Center for Biotechnology Information (NCBI) nucleotide database. Most of the putative dominant fungi were previously found in close association with plants, which corroborated their potential symbiotic capability. In addition, among the 10 OTUs, seven were found among 90% or more of all individuals in all plots, indicating their prevalence in varied hosts along the environmental gradient.
For the network analysis of FEFs, 378 pairs of significant correlations were identified from 71 samples (Figure 5). Nodes of A. rivularis and T. lanceolata were highly interconnected (clustered) at each N level. Unique fungal taxon co-occurrence patterns were also found in E. nutans but were not affected by N input. Inter-host co-occurrence patterns were also observed; however, the correlations were relatively loose (18 edges), with Spearman's correlation coefficients ranging from 0.40 to 0.44. Overall, co-occurring FEFs tend to be tightly related to host identity, and the effect of N varied among species.
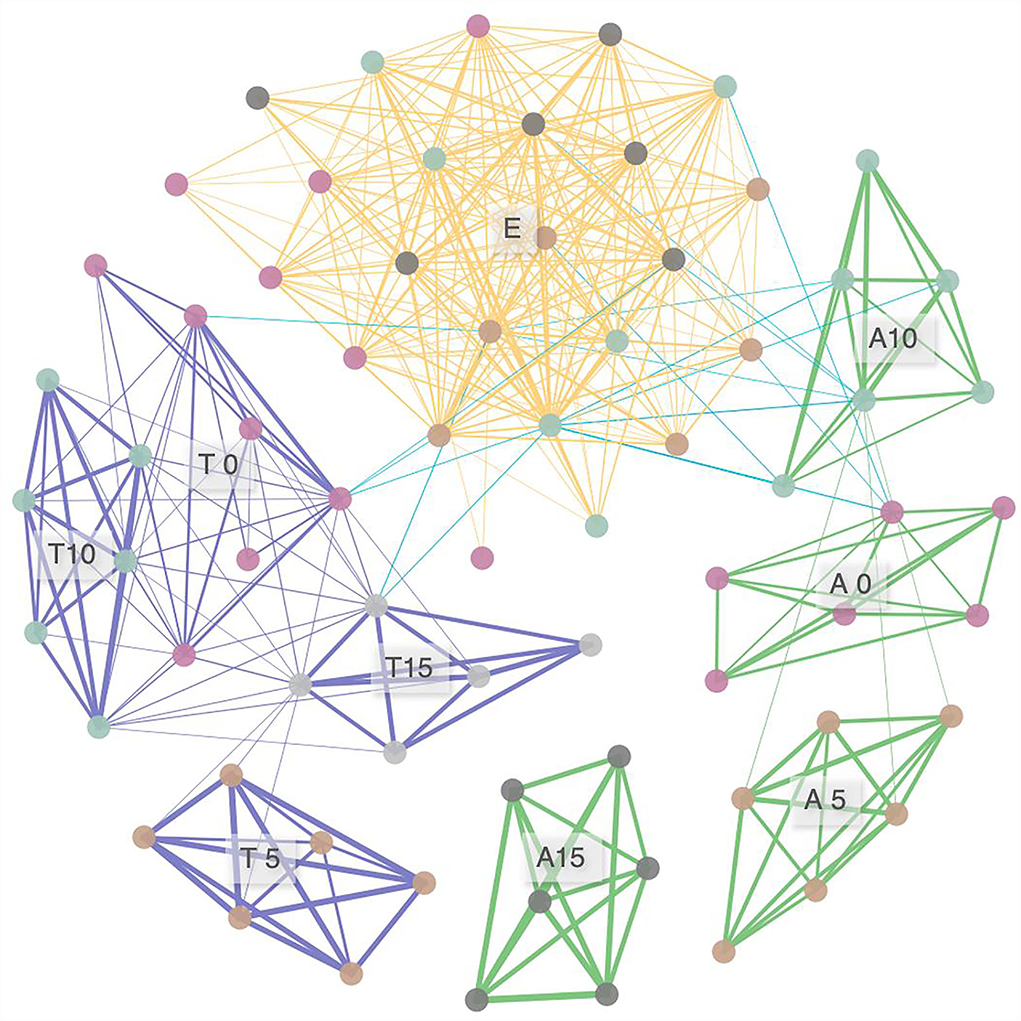
Figure 5. Networks of taxa similarity in three species along the nitrogen enrichment gradients, based on correlation analysis. A connection stands for a correlation with Spearman's ρ > 0.4 and p < 0.05. The thickness of each connection between two nodes (i.e., edge) is proportional to the value of Spearman's correlation coefficients ranging from 0.40 to 0.93. A, Anemone rivularis; E, Elymus nutans; T, Thermopsis lanceolata. 0 (control), 5, 10, and 15 referring to 0, 5, 10, and 15 g N (NH4NO3) m−2 yr−1 treatments, respectively.
E. nutans is dominant perennial grass in the QTP due to its high adaptability (Miao J. et al., 2011; Miao Q. et al., 2011), and A. rivularis and T. lanceolata are widespread forbs in this area. A recent survey on 596 publications on endophyte biodiversity revealed that forbs and graminoid-inhabiting fungal endophytes are largely understudied, especially in multiple hosts (Harrison and Griffin, 2020). In this study, diverse FEFs were found in three herbaceous hosts, suggesting a complex fungal network within QTP plant tissues, despite the geographic isolation and extreme environmental conditions. In a recent study, 210 endophytic fungal OTUs were identified in E. nutans collected from a nearby field site using similar study methods (Guo et al., 2021). Here, we identified 576 OTUs (492 OTUs after data rarefaction) in leaves of E. nutans, challenging the argument put forward by Harrison and Griffin (2020) that roots were the tissue that harbors the richest endophytes in graminoids. In terrestrial plants, environmental differences were proven to contribute to differences between organs in endophyte flora (Fisher et al., 1992). Higher fungal richness in leaves of E. nutans might be caused by the more dynamic aboveground environment, since environmental variability could promote diversity (Hutchinson, 1961; Chesson, 2000). During the short growing season in the QTP, leaves are more biochemically active than roots and may provide more available nutrients to attract microorganisms. Alternatively, low temperature can limit microbial activity, reducing the transmission of endophytes in QTP soil. Given the limited amount of published data on endophytes of alpine plants, it is difficult identify which patterns of distribution are normal.
To our knowledge, this is the first study to apply high-throughput sequencing to explore the endophyte communities in A. rivularis and T. lanceolata. Here, a taxonomically diverse assemblage of FEFs was recovered from A. rivularis, whereas by cultivation-dependent methods, only 23 fungal taxa were isolated from the leaves of Anemone tomentosa in a study in west China. We screened relatively less fungal taxa in T. lanceolata (280 OTUs), possibly due to the reduced abundance of T. lanceolata after N enrichment in the study site (Jiang et al., 2018). Abundant hosts have been proven to support a greater number and diversity of symbiotic fungi (Gilbert et al., 2007). Thus, we suggest that host abundance plays an important role in structuring endophytic microbiomes.
Ascomycota was described as the predominant endophytic fungal phyla in biomes ranging from tropical rainforests to harsh Antarctica in previous works (Arnold and Lutzoni, 2007; Zhang and Yao, 2015; Teasdale et al., 2018; Whitaker et al., 2020). Using 454 pyrosequencing, only 0.4% endophytic Basidiomycota was recovered from leaves of Metrosideros polymorpha in the tropics (Zimmerman and Vitousek, 2012). Conversely, 8.6% endophytic Basidiomycota was found in subalpine timberline ecotone (Yang et al., 2016), and 19.4% of FEFs in four species in a high arctic zone are basidiomycetous fungi (Zhang and Yao, 2015). Research on fungal endophytes has expanded dramatically in recent years, but the ecological roles of endophytic basidiomycetes are still unknown. Here, we found as high as 36.75% endophytic Basidiomycota, adding evidence to the importance of Basidiomycota in high-altitude or high-latitude areas.
The impact of human development on endophyte biodiversity in wilderness areas remains understudied (Harrison and Griffin, 2020). We hypothesized that N could lead to significant community turnover at both plot and within-host scale. We found that fungal diversity for all samples (or all individuals) pooled was not dominantly shaped by N supply at the plot scale after 4 years of fertilization treatment (although some indices showed a significant effect of N, clearly lower deviances, F, and R2 statistics were observed, cf. Tables 1, 2; Figures 2, 4). The findings agree with culture-based experimental evidence from Sweden recently (Witzell et al., 2022) that N input did not alter the total richness and Shannon diversity of FEFs in 12 aspen genotypes. We suggest that in the alpine meadow ecosystems, total fungal communities harbored in diverse groups of plants tend to persist under increasing N deposition. However, it is still not clear if plant microbiomes could be affected by anthropogenic environmental changes at larger spatial scales across the QTP landscapes. Further studies incorporating additional sites (or testing for spatial autocorrelation) are required in the QTP in order to make a strong inference or generalizations on these fungal communities.
In the present study, the effects of elevated N are more pronounced at the within-host scale (Supplementary Tables S5, S6; Figures 3–5). Fungal assemblages of A. rivularis exhibited decreased species diversity (Shannon diversity and richness) across the N enrichment gradients. However, an increased fungal diversity pattern was observed in T. lanceolata as compared to the control group. By contrast, FEF communities of E. nutans were not significantly affected by N. A study of root-associated arbuscular mycorrhizal (AM) fungi in E. nutans within the same experimental plots sampled at the same time found that N addition reduced AM fungal abundance (Jiang et al., 2018). These findings indicate that the effects of N input on plant microbiomes are multifarious and may depend upon host identity and tissue types. The multispecies generalized linear model, PERMANOVA model together with NMDS, and network analyses demonstrated that endophyte community composition varies significantly among host species, supporting findings of previous studies (e.g., Arnold and Lutzoni, 2007; Higgins et al., 2007; Dastogeer et al., 2018; Liu et al., 2019; Chen et al., 2020). Perennial hosts were suggested to cultivate and nurture their microbial surroundings (Baltrus, 2017). Thus, it was possible that in the stressful environment in the QTP, the fungal communities may have adapted to different types of leaves over evolutionary timescales. We reinforce the view that host plant identity is the significant driver of endophyte assemblage patterns, and we suggest host identity supersedes N supply in shaping the community composition of FEFs.
Consistent with previous studies (Rasmussen et al., 2007; Ryan et al., 2014), leaf carbohydrates (LMW and HMW sugars) decreased with N input. Leaves provide shelter and nutrition for endophytes, and foliar carbon was known to correlate with plant function (Wright et al., 2004) and life history strategy (Wright et al., 2010), yet how leaf chemicals, especially leaf carbon, affect communities of fungal endophytes is not well studied. We found a strong relation between tissue carbon and communities of FEFs in the study site (Table 3). The NMDS plot with fitted environmental variables also showed that the carbon-associated factors, including tissue C, C/N ratio, and LMW and HMW sugar, significantly correlated with FEF community assemblages along the N gradient (Figures 4A,B). In addition, these carbon-associated factors correlated well with the dissimilarities among fungal assemblages in E. nutans and T. lanceolata. Similar trends were also found in the leaves of Betula (Yang et al., 2016) and Ficus (Liu et al., 2019), emphasizing the universal relationship between carbon source availability and this heterogeneous ecological guild in plants.
We demonstrated the existence of a fairly dominant core microbiome: three focal species shared one-third of all endophyte sequences, seven OTUs were prevalent in all sampled hosts along the N gradient, and two prevalent OTUs assigned to uncultured fungus in this study were also observed in the phyllosphere of trees in different ecosystems (Supplementary Table S8), indicating their potential wide niche range. Some subsets of fungi that comprise the core microbiome may have functional roles that enhance their ability to colonize the interior of a phylogenetically diverse group of plant species (Vandenkoornhuyse et al., 2015). These traits likely include high habitat adaptation (Pereira et al., 2019) and metabolite (enzymes or hormone) production that induces stomatal opening for fungi entry (Hardoim et al., 2015). The same microbial taxa may reach varied plant species in varied environmental conditions, and hence, searching for endophytes from the core microbiome of wild plants adapted to unhospitable habitats will provide solutions for the study of stress tolerance of plants.
This study provides insights into the changes in plant fungal microbiome in response to host identity and environmental conditions. We report two key findings: first, in the harsh habitat of the Qinghai–Tibetan Plateau, host identity, rather than N supply, is a stronger factor for driving communities of foliar fungal endophytes, even in an alpine meadow ecosystem where productivity is strongly limited by N availability; and second, N addition has varying effects on both α and β diversity of within-host scale fungal endophytes. We demonstrated leaf carbon and carbohydrates significantly affected fungal communities in host leaves, indicating FEFs may play a pivotal role in the carbon cycling of alpine meadow ecosystems. In addition, we found unequivocal evidence for a core microbiome present among 90% or more of all control samples, representing one-third of the total reads. The results suggest that even though soil resource availability often mediates plant performance, environmental modifications on fungal microbiomes likely depend on the host species. We suggest that future studies quantify the nutritional requirements of endophytes, at both small and large scales. The results implement theoretical frameworks of community ecology and gain new sights into the dynamics of endophyte communities in grasses and forbs driven by environment and host functional traits.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OM744714:OM745695.
Conceptualization: HF, QZ, and YL. Methodology: GD, HF, and YL. Software, formal analysis, and visualization: YM. Investigation: GS, YL, QZ, and YM. Resources: GD. Data curation: QZ and YM. Original draft preparation: HF, QZ, and YM. Project administration: QZ and HF. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was financially supported by the Second Tibetan Plateau Scientific Expedition and Research Program (2019QZKK0301) and the National Natural Science Foundation of China (31971445, 31870494, 31860146, 32171579, and U21A20186).
We thank the following members for assistance with fieldwork: Shengjing Jiang, Mingsen Qin, Yuxing Chai, and Jiajia Luo. YM is grateful to Sanni Färkkilä, Slendy Rodriguez, Iris Reinula, and Kai-Yun Li for their suggestions on writing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.895533/full#supplementary-material
Apigo, A., and Oono, R. (2018). Dimensions of host specificity in foliar fungal endophytes. Endop. Forest Trees Biol. Appl. 86, 15–42. doi: 10.1007/978-3-319-89833-9_2
Arnold, A. E., and Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Arnold, A. E., Miadlikowska, J., Higgins, K. L., Sarvate, S. D., Gugger, P., Way, A., et al. (2009). A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? System. Biol. 58, 283–297. doi: 10.1093/sysbio/syp001
Baltrus, D. A. (2017). Adaptation, specialization, and coevolution within phytobiomes. Curr. Opin. Plant Biol. 38, 109–116. doi: 10.1016/j.pbi.2017.04.023
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the International AAAI Conference on Web and Social Media, Vol. 3, 361–362.
Bobbink, R., Hicks, K., Galloway, J., Spranger, T., Alkemade, R., Ashmore, M., et al. (2010). Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis. Ecol. Appl. 20, 30–59. doi: 10.1890/08-1140.1
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6, 90. doi: 10.1186/s40168-018-0470-z
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 (vol 37, pg 852, 2019). Nat. Biotechnol. 37, 1091–1091. doi: 10.1038/s41587-019-0252-6
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Buckley, H., Young, C. A., Charlton, N. D., Hendricks, W. Q., Haley, B., Nagabhyru, P., et al. (2019). Leaf endophytes mediate fertilizer effects on plant yield and traits in northern oat grass (Trisetum spicatum). Plant Soil 434, 425–440. doi: 10.1007/s11104-018-3848-6
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581. doi: 10.1038/nmeth.3869
Cardinale, B. J., Matulich, K. L., Hooper, D. U., Byrnes, J. E., Duffy, E., Gamfeldt, L., et al. (2011). The functional role of producer diversity in ecosystems. Am. J. Bot. 98, 572–592. doi: 10.3732/ajb.1000364
Chen, J. H., Akutse, K. S., Saqib, H. S. A., Wu, X. L., Yang, F. Y., Xia, X. F., et al. (2020). Fungal endophyte communities of crucifer crops are seasonally dynamic and structured by plant identity, plant tissue and environmental factors. Front. Microbiol. 11, 01519. doi: 10.3389/fmicb.2020.01519
Chen, X. X., Luo, X. L., Fan, M. M., Zeng, W. L., Yang, C. R., Wu, J. R., et al. (2019). Endophytic fungi from the branches of Camellia taliensis (W. W. Smith) Melchior, a widely distributed wild tea plant. World J. Microbiol. Biotechnol. 35, 113. doi: 10.1007/s11274-019-2686-x
Chesson, P. (2000). Mechanisms of maintenance of species diversity. Ann. Rev. Ecol. System. 343–366. doi: 10.1146/annurev.ecolsys.31.1.343
Christian, N., Herre, E. A., and Clay, K. (2019). Foliar endophytic fungi alter patterns of nitrogen uptake and distribution in Theobroma cacao. New Phytol. 222, 1573–1583. doi: 10.1111/nph.15693
Clay, K., and Schardl, C. (2002). Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Natur. 160, S99–S127. doi: 10.1086/342161
Crawford, J. T., Hinckley, E. L. S., and Neff, J. C. (2020). Long-term trends in acid precipitation and watershed elemental export from an alpine catchment of the colorado rocky mountains, USA. J. Geophys. Res. Biogeosci. 125, e2020JG005683. doi: 10.1029/2020JG005683
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Inter J. Complex Syst. 1695, 1–9.
Dastogeer, K. M. G., Li, H., Sivasithamparam, K., Jones, M. G. K., and Wylie, S. J. (2018). Host specificity of endophytic mycobiota of wild nicotiana plants from arid regions of Northern Australia. Microbial. Ecol. 75, 74–87. doi: 10.1007/s00248-017-1020-0
Dunham, S. M., Mujic, A. B., Spatafora, J. W., and Kretzer, A. M. (2013). Within-population genetic structure differs between two sympatric sister-species of ectomycorrhizal fungi, Rhizopogon vinicolor and R. vesiculosus. Mycologia 105, 814–826. doi: 10.3852/12-265
Ellison, A. M., and Gotelli, N. J. (2002). Nitrogen availability alters the expression of carnivory in the northern pitcher plant, Sarracenia purpurea. Proc. Natl. Acad. Sci. U. S. A. 99, 4409–4412. doi: 10.1073/pnas.022057199
EPA (2013). Reactive Nitrogen in the United States: An Analysis of Inputs, Flows, Consequences, and Management Options: A Report of the EPA Science Advisory Board. Washington, DC: EPA.
Fisher, P. J., Petrini, O., and Scott, H. M. L. (1992). The distribution of some fungal and bacterial endophytes in maize (Zea-Mays L). New Phytol. 122, 299–305. doi: 10.1111/j.1469-8137.1992.tb04234.x
Gardes, M., and Bruns, T. D. (1993). Its primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gilbert, G. S., Reynolds, D. R., and Bethancourt, A. (2007). The patchiness of epifoliar fungi in tropical forests: host range, host abundance, and environment. Ecology 88, 575–581. doi: 10.1890/05-1170
Gilliam, F. S., Burns, D. A., Driscoll, C. T., Frey, S. D., Lovett, G. M., and Watmough, S. A. (2019). Decreased atmospheric nitrogen deposition in eastern North America: Predicted responses of forest ecosystems. Environ. Pollut. 244, 560–574. doi: 10.1016/j.envpol.2018.09.135
Gonzalez-Teuber, M., Vilo, C., Guevara-Araya, M. J., Salgado-Luarte, C., and Gianoli, E. (2020). Leaf resistance traits influence endophytic fungi colonization and community composition in a South American temperate rainforest. J. Ecol. 108, 1019–1029. doi: 10.1111/1365-2745.13314
Guo, J., Bowatte, S., and Hou, F. (2021). Diversity of endophytic bacteria and fungi in seeds of Elymus nutans growing in four locations of Qinghai Tibet Plateau, China. Plant Soil 459, 49–63. doi: 10.1007/s11104-020-04608-y
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttila, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Harrell, F. E., and Dupont, C. (2018). Hmisc: Harrell miscellaneous. R package version 4.1-1. R Found. Stat. Comput. Avaible online at: https://CRAN.R-project.org/package=Hmisc (accessed February 16, 2018).
Harrison, J. G., and Griffin, E. A. (2020). The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: how far have we come and where do we go from here? Environ. Microbiol. 22, 2107–2123. doi: 10.1111/1462-2920.14968
Higgins, K. L., Arnold, A. E., Coley, P. D., and Kursar, T. A. (2014). Communities of fungal endophytes in tropical forest grasses: highly diverse host- and habitat generalists characterized by strong spatial structure. Fungal Ecol. 8, 1–11. doi: 10.1016/j.funeco.2013.12.005
Higgins, K. L., Arnold, A. E., Miadlikowska, J., Sarvate, S. D., and Lutzoni, F. (2007). Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol. Phylogen. Evol. 42, 543–555. doi: 10.1016/j.ympev.2006.07.012
Higgins, K. L., Coley, P. D., Kursar, T. A., and Arnold, A. E. (2011). Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycologia 103, 247–260. doi: 10.3852/09-158
Huang, Y. L., Devan, M. M. N., U'Ren, J. M., Furr, S. H., and Arnold, A. E. (2016). Pervasive effects of wildfire on foliar endophyte communities in montane forest trees. Microbial. Ecol. 71, 452–468. doi: 10.1007/s00248-015-0664-x
Hunt, M. G., Rasmussen, S., Newton, P. C. D., Parsons, A. J., and Newman, J. A. (2005). Near-term impacts of elevated CO2, nitrogen and fungal endophyte-infection on Lolium perenne L. growth, chemical composition and alkaloid production. Plant Cell Environ. 28, 1345–1354. doi: 10.1111/j.1365-3040.2005.01367.x
IPCC (2014). Climate Change 2014 – Impacts, Adaptation and Vulnerability: Part A: Global and Sectoral Aspects: Working Group II Contribution to the IPCC Fifth Assessment Report: Volume 1: Global and Sectoral Aspects (Vol. 1). Cambridge; New York, NY: Cambridge University Press.
Jiang, S. J., Liu, Y. J., Luo, J. J., Qin, M. S., Johnson, N. C., Opik, M., et al. (2018). Dynamics of arbuscular mycorrhizal fungal community structure and functioning along a nitrogen enrichment gradient in an alpine meadow ecosystem. New Phytol. 220, 1222–1235. doi: 10.1111/nph.15112
Knoth, J. L., Kim, S. H., Ettl, G. J., and Doty, S. L. (2013). Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. Global Change Biol. Bioenergy 5, 408–418. doi: 10.1111/gcbb.12006
Koljalg, U., Nilsson, H. R., Schigel, D., Tedersoo, L., Larsson, K. H., May, T. W., et al. (2020). The taxon hypothesis paradigm-on the unambiguous detection and communication of Taxa. Microorganisms 8, 910. doi: 10.3390/microorganisms8121910
Larkin, B. G., Hunt, L. S., and Ramsey, P. W. (2012). Foliar nutrients shape fungal endophyte communities in Western white pine (Pinus monticola) with implications for white-tailed deer herbivory. Fungal Ecol. 5, 252–260. doi: 10.1016/j.funeco.2011.11.002
Liu, J. W., Zhao, J., Wang, G., and Chen, J. (2019). Host identity and phylogeny shape the foliar endophytic fungal assemblages of Ficus. Ecol. Evol. 9, 10472–10482. doi: 10.1002/ece3.5568
Lu, C. Q., and Tian, H. Q. (2007). Spatial and temporal patterns of nitrogen deposition in China: Synthesis of observational data. J. Geophys. Res. Atmosp. 112, D22. doi: 10.1029/2006JD007990
Miao, J., Zhang, X., Chen, S., Ma, X., Chen, Z., Zhong, J., et al. (2011). Gliadin analysis of Elymus nutans Griseb. from the Qinghai–Tibetan Plateau and Xinjiang, China. Grassland Sci. 57, 127–134. doi: 10.1111/j.1744-697X.2011.00219.x
Miao, Q., Qin, S., Bian, G.-K., Yuan, B., Xing, K., Zhang, Y.-J., et al. (2011). Amycolatopsis endophytica sp. nov., a novel endophytic actinomycete isolated from oil-seed plant Jatropha curcas L. Antonie van Leeuwenhoek 100, 333–339. doi: 10.1007/s10482-011-9588-8
Newman, J. A., Abner, M. L., Dado, R. G., Gibson, D. J., Brookings, A., and Parsons, A. J. (2003). Effects of elevated CO2, nitrogen and fungal endophyte-infection on tall fescue: growth, photosynthesis, chemical composition and digestibility. Global Change Biol. 9, 425–437. doi: 10.1046/j.1365-2486.2003.00601.x
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). Vegan: Community Ecology Package (Version 2.5-6). The Comprehensive R Archive Network. Available online at: https://CRAN.R-project.org/package=vegan (accessed July 1, 2019).
Oono, R., Lutzoni, F., Arnold, A. E., Kaye, L., U'Ren, J. M., May, G., et al. (2014). Genetic variation in horizontally transmitted fungal endophytes of pine needles reveals population structure in cryptic species. Am. J. Bot. 101, 1362–1374. doi: 10.3732/ajb.1400141
Pereira, E., de Aldana, B. R. V., San Emeterio, L., and Zabalgogeazcoa, I. (2019). A survey of culturable fungal endophytes from festuca rubra subsp. pruinosa, a grass from marine cliffs, reveals a core microbiome. Front. Microbiol. 9, e03321. doi: 10.3389/fmicb.2018.03321
R Core Team (2019). R: A Language and Environment for Statistical Computing. In (Version 3.6.2) R Foundation for Statistical Computing. Vienna: R Core Team.
Rasmussen, S., Parsons, A. J., Bassett, S., Christensen, M. J., Hume, D. E., Johnson, L. J., et al. (2007). High nitrogen supply and carbohydrate content reduce fungal endophyte and alkaloid concentration in Lolium perenne. New Phytol. 173, 787–797. doi: 10.1111/j.1469-8137.2006.01960.x
Razali, N. M., Cheah, B. H., and Nadarajah, K. (2019). Transposable elements adaptive role in genome plasticity, pathogenicity and evolution in fungal phytopathogens. Int. J. Mol. Sci. 20, 597. doi: 10.3390/ijms20143597
Rodriguez, R. J., White, J. F., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Ryan, G. D., Rasmussen, S., Xue, H., Parsons, A. J., and Newman, J. A. (2014). Metabolite analysis of the effects of elevated CO2 and nitrogen fertilization on the association between tall fescue (Schedonorus arundinaceus) and its fungal symbiont Neotyphodium coenophialum. Plant Cell Environ. 37, 204–212. doi: 10.1111/pce.12146
Saikkonen, K., Wali, P., Helander, M., and Faeth, S. H. (2004). Evolution of endophyte-plant symbioses. Trends Plant Sci. 9, 275–280. doi: 10.1016/j.tplants.2004.04.005
Scarpitta, A. B., Bardat, J., Lalanne, A., and Vellend, M. (2017). Long-term community change: bryophytes are more responsive than vascular plants to nitrogen deposition and warming. J. Veget. Sci. 28, 1220–1229. doi: 10.1111/jvs.12579
Seghers, D., Wittebolle, L., Top, E. M., Verstraete, W., and Siciliano, S. D. (2004). Impact of agricultural practices on the Zea mays L. endophytic community. Appl. Environ. Microbiol. 70, 1475–1482. doi: 10.1128/AEM.70.3.1475-1482.2004
Shade, A., and Handelsman, J. (2012). Beyond the Venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Singha, K. M., Singh, B., and Pandey, P. (2021). Host specific endophytic microbiome diversity and associated functions in three varieties of scented black rice are dependent on growth stage. Sci. Rep. 11, 12259. doi: 10.1038/s41598-021-91452-4
Sosso, J., Zakeel, M. C. M., and Akinsanmi, O. A. (2021). Culturable fungal endophytes in Australian macadamia nursery plants. Austral. Plant Pathol. 50, 739–746. doi: 10.1007/s13313-021-00824-x
Stevens, C. J., Dise, N. B., Mountford, J. O., and Gowing, D. J. (2004). Impact of nitrogen deposition on the species richness of grasslands. Science 303, 1876–1879. doi: 10.1126/science.1094678
Sullivan, T. J., Lawrence, G. B., Bailey, S. W., McDonnell, T. C., Beier, C. M., Weathers, K. C., et al. (2013). Effects of acidic deposition and soil acidification on sugar maple trees in the Adirondack Mountains, New York. Environ. Sci. Technol. 47, 12687–12694. doi: 10.1021/es401864w
Suryanarayanan, T. S., and Shaanker, R. U. (2021). Can fungal endophytes fast-track plant adaptations to climate change? Fungal Ecol. 50, 101039. doi: 10.1016/j.funeco.2021.101039
Teasdale, S. E., Caradus, J. R., and Johnson, L. J. (2018). Fungal endophyte diversity from tropical forage grass Brachiaria. Plant Ecol. Diversity 11, 611–624. doi: 10.1080/17550874.2019.1610913
Unterseher, M., Jumpponen, A., Opik, M., Tedersoo, L., Moora, M., Dormann, C. F., et al. (2011). Species abundance distributions and richness estimations in fungal metagenomics - lessons learned from community ecology. Mol. Ecol. 20, 275–285. doi: 10.1111/j.1365-294X.2010.04948.x
Vacher, C., Hampe, A., Porte, A. J., Sauer, U., Compant, S., and Morris, C. E. (2016). The phyllosphere: microbial jungle at the plant-climate interface. Ann. Rev. Ecol. Evol. Syst. 47, 1–24. doi: 10.1146/annurev-ecolsys-121415-032238
Vallicrosa, H., Sardans, J., Maspons, J., and Penuelas, J. (2022). Global distribution and drivers of forest biome foliar nitrogen to phosphorus ratios (N:P). Global Ecol. Biogeogr. 31, 861–871. doi: 10.1111/geb.13457
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vincent, J. B., Weiblen, G. D., and May, G. (2016). Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol. Ecol. 25, 825–841. doi: 10.1111/mec.13510
Vitousek, P. M., and Howarth, R. W. (1991). Nitrogen limitation on land and in the sea - how can it occur. Biogeochemistry 13, 87–115. doi: 10.1007/BF00002772
Whitaker, B. K., Christian, N., Chai, Q., and Clay, K. (2020). Foliar fungal endophyte community structure is independent of phylogenetic relatedness in an Asteraceae common garden. Ecol. Evol. 10, 13895–13912. doi: 10.1002/ece3.6983
Witzell, J., Decker, V. H. G., Agostinelli, M., Romeralo, C., Cleary, M., and Albrectsen, B. R. (2022). Aspen leaves as a “chemical landscape” for fungal endophyte diversity-effects of nitrogen addition. Front. Microbiol. 13, 846208. doi: 10.3389/fmicb.2022.846208
Wright, I. J., Reich, P. B., Westoby, M., Ackerly, D. D., Baruch, Z., Bongers, F., et al. (2004). The worldwide leaf economics spectrum. Nature 428, 821–827. doi: 10.1038/nature02403
Wright, S. J., Kitajima, K., Kraft, N. J. B., Reich, P. B., Wright, I. J., Bunker, D. E., et al. (2010). Functional traits and the growth-mortality trade-off in tropical trees. Ecology 91, 3664–3674. doi: 10.1890/09-2335.1
Wu, G. L., Liu, Z. H., Zhang, L., Chen, J. M., and Hu, T. M. (2010). Long-term fencing improved soil properties and soil organic carbon storage in an alpine swamp meadow of western China. Plant Soil 332, 331–337. doi: 10.1007/s11104-010-0299-0
Xu, D. H., Fang, X. W., Zhang, R. Y., Gao, T. P., Bu, H. Y., and Du, G. Z. (2015). Influences of nitrogen, phosphorus and silicon addition on plant productivity and species richness in an alpine meadow. Aob Plants 7, plv125. doi: 10.1093/aobpla/plv125
Yang, T., Weisenhorn, P., Gilbert, J. A., Ni, Y., Sun, R., Shi, Y., et al. (2016). Carbon constrains fungal endophyte assemblages along the timberline. Environ. Microbiol. 18, 2455–2469. doi: 10.1111/1462-2920.13153
Yang, Z., van Ruijven, J., and Du, G. (2011). The effects of long-term fertilization on the temporal stability of alpine meadow communities. Plant Soil 345, 315–324. doi: 10.1007/s11104-011-0784-0
Yemm, E. W., Cocking, E. C., and Ricketts, R. E. (1955). The determination of amino-acids with ninhydrin. Analyst 80, 209–214. doi: 10.1039/an9558000209
Zhang, T., and Yao, Y. F. (2015). Endophytic fungal communities associated with vascular plants in the high arctic zone are highly diverse and host-plant specific. PLoS ONE 10, e0130051. doi: 10.1371/journal.pone.0130051
Zheng, Y., Li, X., Cao, H., Lei, L., Zhang, X., Han, D., et al. (2021). The assembly of wheat-associated fungal community differs across growth stages. Appl. Microbiol. Biotechnol. 105, 7427–7438. doi: 10.1007/s00253-021-11550-1
Keywords: foliar fungal endophytes, nitrogen deposition, Qinghai–Tibet Plateau, microbial community, host-specificity
Citation: Meng Y, Zhang Q, Shi G, Liu Y, Du G and Feng H (2022) Can nitrogen supersede host identity in shaping the community composition of foliar endophytic fungi in an alpine meadow ecosystem? Front. Microbiol. 13:895533. doi: 10.3389/fmicb.2022.895533
Received: 14 March 2022; Accepted: 29 July 2022;
Published: 22 August 2022.
Edited by:
Juan Sun, Qingdao Agricultural University, ChinaReviewed by:
Chao Yang, Qingdao Agricultural University, ChinaCopyright © 2022 Meng, Zhang, Shi, Liu, Du and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Zhang, emhhbmdxaUBsenUuZWR1LmNu; Huyuan Feng, ZmVuZ2h5QGx6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.