
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 May 2022
Sec. Systems Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.892605
This article is part of the Research Topic Rumen Microbiome Dynamics and Their Implications in Health and Environment View all 16 articles
 Rajaraman Bharanidharan1
Rajaraman Bharanidharan1 Krishnaraj Thirugnanasambantham2,3,4
Krishnaraj Thirugnanasambantham2,3,4 Ridha Ibidhi2
Ridha Ibidhi2 Myunggi Baik1
Myunggi Baik1 Tae Hoon Kim5
Tae Hoon Kim5 Yookyung Lee6
Yookyung Lee6 Kyoung Hoon Kim2,5*
Kyoung Hoon Kim2,5*
We identified metabolites in the seeds of Pharbitis nil (PA) and evaluated their effects on rumen methanogenesis, fiber digestibility, and the rumen microbiome in vitro and in sacco. Four rumen-cannulated Holstein steers (mean body weight 507 ± 32 kg) were used as inoculum donor for in vitro trial and live continuous culture system for in sacco trial. PA was tested in vitro at doses ranging from 4.5 to 45.2% dry matter (DM) substrate. The in sacco trial was divided into three phases: a control phase of 10 days without nylon bags containing PA in the rumen, a treatment phase of 11 days in which nylon bags containing PA (180 g) were placed in the rumen, and a recovery phase of 10 days after removing the PA-containing bags from the rumen. Rumen headspace gas and rumen fluid samples were collected directly from the rumen. PA is enriched in polyunsaturated fatty acids dominated by linoleic acid (C18:2) and flavonoids such as chlorogenate, quercetin, quercetin-3-O-glucoside, and quinic acid derivatives. PA decreased (p < 0.001) methane (CH4) production linearly in vitro with a reduction of 24% at doses as low as 4.5% DM substrate. A quadratic increase (p = 0.078) in neutral detergent fiber digestibility was also noted, demonstrating that doses < 9% DM were optimal for simultaneously enhancing digestibility and CH4 reduction. In sacco, a 50% decrease (p = 0.087) in CH4 coupled with an increase in propionate suggested increased biohydrogenation in the treatment phase. A decrease (p < 0.005) in ruminal ammonia nitrogen (NH3-N) was also noted with PA in the rumen. Analysis of the rumen microbiome revealed a decrease (p < 0.001) in the Bacteroidetes-to-Firmicutes ratio, suggesting PA to have antiprotozoal potential. At the genus level, a 78% decrease in Prevotella spp. and a moderate increase in fibrolytic Ruminococcus spp. were noted in the treatment phase. In silico binding of PA metabolites to cyclic GMP-dependent protein kinase of Entodinium caudatum supported the antiprotozoal effect of PA. Overall, based on its high nutrient value and antiprotozoal activity, PA could probably replace the ionophores used for CH4 abatement in the livestock industry.
Ruminal CH4 production, in addition to its adverse effect on the environment, represents the loss of 3–10% of the gross energy intake of the animal (Appuhamy et al., 2016). Over the past 50 years, ruminant nutritionists have developed numerous strategies for enteric CH4 abatement (Beauchemin et al., 2020). A meta-analysis showed that dietary supplementation of ionophore antibiotics (e.g., monensin) decreases CH4 production by up to 15% and increases feed efficiency in beef cattle (Appuhamy et al., 2013). However, the use of ionophores has been banned in many countries, including South Korea, due to concern over their residues in livestock products (Maron et al., 2013; Lee et al., 2018). Similarly, consumer acceptance of 3-nitrooxypropanol, although a promising CH4 inhibitor, will influence the acceptance of inorganic synthetic compounds (Beauchemin et al., 2020). Therefore, studies have focused on plant-based organic strategies such as supplementing lipids and oil seeds rich in polyunsaturated fatty acids (PUFAs) or additives rich in flavonoids, tannins, and saponins, which reportedly reduce CH4 production (Hassan et al., 2020; Ku-Vera et al., 2020). Nevertheless, the adverse effects of PUFAs and other phyto-additives on nutrient digestibility, rumen fermentation, and animal performance have raised concerns over the feasibility of such strategies (Beauchemin et al., 2020). Natural seaweeds and seaweed bioactives can reduce CH4 production with fewer adverse effects on animal productivity (Abbott et al., 2020). However, concern over the safety of feeding bromoform-containing seaweeds to livestock, their associated toxicity to the environment (i.e., ozone depletion), and their net carbon footprint necessitates the discovery of new potential natural feed additives for CH4 abatement (Glasson et al., 2022).
Our previous in vitro screening study involving 152 plant extracts reported that ethanolic extracts from the seeds of Pharbitis nil (Pharbitis semen; PA) can decrease CH4 by up to 37% (Bharanidharan et al., 2021a). Pharbitis nil (Convolvulaceae) is an annual climbing herb widely distributed throughout Korea, Japan, and China, and PA is used as a purgative agent and in treating digestive disorders (Kim et al., 2020). Previous phytochemical investigations have reported the isolation of bioactive chemical constituents such as resin glycosides (Ono, 2017), monoterpenoids (Lee et al., 2017), diterpenoids (Ki et al., 2009; Woo et al., 2017), triterpenoid saponins (Jung et al., 2008), flavonoids, chlorogenic acid derivatives (Saito et al., 1994), and phenolic compounds (Kim et al., 2011) from different parts of P. nil. Similarly, we reported the enrichment of PUFAs in PA using gas chromatography-mass spectroscopy (GC-MS) without derivatization (Bharanidharan et al., 2021a). However, a complete secondary metabolite and fatty acid (FA) profile of PA is needed to understand their effects on methanogenesis and fermentation. Similarly, studies on the dose response effect of PA on CH4 production and fermentation characteristics are needed to optimize the additive dosage such that minimal adverse effects on rumen fermentation and nutrient digestibility are achieved. Furthermore, nutritional interventions with high dietary PUFAs should be evaluated, including their in vivo effect on the rumen microbiota based on their toxicity toward major cellulolytic bacteria (Beauchemin et al., 2020).
Artificial rumen replacements such as in vitro continuous or batch culture systems do not adequately simulate in vivo rumen gas and volatile fatty acid (VFA) production due to various factors such as lack of VFA absorption and feed passage in vitro (Krishnamoorthy et al., 2005; Amanzougarene and Fondevila, 2020). In sacco experimental approaches involving live continuous culture systems (LCCSs) have been used to assess the digestibility of feedstuffs (Krizsan et al., 2013). However, to the best our knowledge, only our previous work (Kim et al., 2016) has used this approach for the initial assessment of feed additives in terms of their effects on CH4 production and microbial population. Moreover, LCSS-based approaches have yet to be validated.
Here, we profiled PA metabolites using ultra-performance liquid chromatography high-resolution mass spectroscopy (UPLC-HRMS/MS) and GC-MS; evaluated the in vitro effect of ground PA on CH4 production, fermentation, and digestibility; and evaluated in sacco the effect of PA on CH4 production, fermentation, and rumen microbial dynamics using an LCCS.
Four cannulated Holstein steers (mean body weight 507 ± 32 kg), cared for in accordance with guidelines of the Animal Ethical Committee, Seoul National University, Republic of Korea (approval number SNU-210615-1), were used for the in vitro and in sacco trials. A diet comprising tall fescue hay and commercial concentrate pellets (33:67, w/w) was used as the basal diet of the donor steers and as substrate for in vitro incubations. The animals were fed 5 kg dry matter (DM) of the basal diet twice daily at 09:00 and 17:00 and had ad libitum access to fresh water. PA was purchased from a local market in Dongdaemun-gu, Seoul, Republic of Korea. The diet material and PA were dried in a forced-air oven at 65°C for 72 h to estimate the DM content, ground to pass through a 1-mm screen (Thomas Scientific Model 4, Swedesboro, NJ, United States), and stored at −20°C until in vitro incubations and chemical analyses. The ingredient and nutrient compositions of the basal diet/substrate are presented in Table 1, and its FA composition can be found in Supplementary Table 1.
The feed and PA samples were dried in a forced-air oven at 65°C for 72 h to estimate DM content and then ground to pass through a 1-mm screen (Thomas Scientific Model 4). The organic matter content was determined after ashing at 600°C for 3 h using a Nabertherm LE 14/11/R7 Compact Muffle Furnace (Lilienthal, Germany) (Undersander et al., 1993). The ether extract (EE) content was determined using an ANKOMXT15 Extractor (Ankom Technology Corp., Fairport, NY, United States) following a filter bag procedure (AM-5-04; 2001) with petroleum ether as the solvent. Neutral detergent fiber (NDF) and acid detergent fiber contents were measured using a filter bag technique with an Ankom A2000 Fiber Analyzer (ANKOM Technology Corp.). The neutral detergent fiber content was analyzed using heat-stable amylase and expressed exclusive of residual ash (Van Soest et al., 1991). The analytical method for acid detergent fiber was based on Van Soest (1973), and the results are presented exclusive of residual ash. The nitrogen (N) content was determined using the Kjeldahl method (Kjeltec Auto Sampler System, 8400 Analyzer; Foss, Sweden) as described by (AOAC International, 2016). Crude protein (CP) was calculated as 6.25 × N. The gross energy content was determined using an automatic isoperibol calorimeter (6400EF, Parr Instrument Company, Moline, IL, United States). Concentrations of FAs in the feed and PA were analyzed using the direct methylation method of O’Fallon et al. (2007) and an Agilent 7890B GC system (Agilent Technologies, Santa Clara, CA, United States) with a flame ionization detector as described previously (Bharanidharan et al., 2021b). The FA content was expressed as mg/100 g DM.
Pharbitis nil samples (10 g) were ground into a homogenous fine powder using a mortar and pestle in liquid nitrogen, followed by extraction with 2 L of methanol-water (80:20, v/v) with continuous stirring for 24 h at room temperature. The resultant extract was filtered using Whatman No. 2 filter paper, and the residue was re-extracted using 1 L of methanol-water (80:20, v/v) with continuous stirring for 12 h at room temperature. Both extracts were pooled and concentrated using a rotary vacuum evaporator (Heidolph Instruments, Schawabatch, Germany) followed by freeze drying (FDFC-12012, Operon, Seoul, South Korea) for 48 h. The dry extract (yield 3.2% w/w) was stored at −80°C until analysis. For UPLC-HRMS analysis, 20 mg of dry extract was dissolved in 2 mL of methanol-water (80:20, v/v) via sonication for 10 min (VCX130, SONICS Vibra-Cell™, Newtown, CT, United States), and the solution was passed through a 0.22-μm polyvinylidene fluoride syringe filter (Millex-GV, Millipore®, Darmstadt, Germany). The filtered samples were dried completely using a nitrogen evaporator (HyperVap HV-300, Labogene, Seoul, South Korea), and the dried extracts were reconstituted (5 mg/mL) in 80% HPLC-grade methanol (Sigma-Aldrich, St. Louis, MO, United States) in an autosample vial prior to analysis.
UPLC separation was performed using an UltiMate™ 3000 (Thermo Scientific, Waltham, MA, United States) UPLC system with a CORTECS C18 (2.1 mm × 150 mm, 1.6 μm; Waters, Milford, MA, United States) UPLC column. The flow rate was 0.3 mL/min, and the solvent system consisted of 0.1% aqueous formic acid (A) and acetonitrile with 0.1% formic acid (B). Linear gradient elution was applied as follows: 1% B at 0–1.0 min, 30% B at 1.0–25.0 min, 30-100% B at 25–50 min, and 100-1% B at 50–60 min. The column temperature was held at 45°C, and the injection volume was 5 μL.
Electrospray ionization (ESI)-MS was carried out using a hybrid triple quadrupole time-of-flight (TripleTOF ® 5600 + ; AB Sciex, Framingham MA, United States) mass spectrometer with MS1 and MS2 data recorded in both positive and negative ionization modes. The ion spray voltage in positive and negative mode was 5.5 and −4.5 kV, respectively. The desolvation gas (N2) temperature was set to 500°C. For MS acquisition, the nebulizer gas (N2), heating gas (N2), and curtain gas (N2) flow rates were 50, 50, and 25 psi, respectively. A declustering potential of ±60 V, collision energy of ±35 V, and collision energy spread of ±10 V were applied, in both positive and negative ionization modes. The MS analysis alternated between MS full scanning and information-dependent acquisition scanning.
Elements version 2.1.1 software (Proteome Software Inc., Portland, OR, United States) was used to process raw ion chromatograms. Raw data files were converted to mz5 format using ProteoWizard version pwiz_Reader_ABI: 3.0.9987. Raw profile data and converted data were imported into Elements software for peak identification, alignment, feature extraction, and area normalization, with separate analyses used for positive and negative ionization mode. Feature finding was conducted over a mass range of 30–1500 m/z as described previously (Lim, 2020).
Approximately 300 mL of ruminal fluid was collected from each donor steer before the morning feeding and strained through four layers of muslin before getting pooled into a prewarmed flask flushed with O2-free CO2. The fluid was then diluted with O2-free buffer (McDougall, 1948; adjusted to pH 7.0) at a ratio of 1:2 (v/v) and placed in a water bath pre-heated to 39°C with continuous CO2 flushing. The in vitro trial was performed as described in Bharanidharan et al. (2021a) to test the effect of ground PA. Briefly, incubation was carried out with three replicates with each comprising 30 mL of rumen fluid mixture in 60-mL serum bottles containing 210 mg DM of substrate. The experimental setup comprised a blank (i.e., only rumen fluid mixture without substrate and ground PA), a control (i.e., rumen fluid mixture with substrate but without ground PA), a positive control (i.e., rumen fluid mixture with substrate and 30 ppm monensin; CAS No. 22373-78-0, Sigma-Aldrich), and dietary treatments (i.e., rumen fluid mixture with substrate and 4.5, 9.0, 13.6, 18.1, 22.6, or 45.2% DM ground PA). After 24 h of incubation, the total gas production was measured using water displacement apparatus (Fedorah and Hrudey, 1983). CH4 concentration in the headspace gas and VFAs in the medium were determined using the Agilent 7890B GC system (Agilent Technologies, Santa Clara, CA, USA) with a flame ionization detector using methods described by Bharanidharan et al. (2021a). The pH was measured using a pH meter (model AG 8603; Seven Easy pH, Mettler-Toledo, Schwerzenbach, Switzerland) and the NH3-N concentration was determined using a modified colorimetric method (Chaney and Marbach, 1962).
In parallel procedures, another fermentation run was conducted to determine the effects of PA on in vitro DM and NDF digestibility. Incubation was carried out with three replicates with each comprising 60 mL of rumen fluid mixture in 120-mL serum bottles containing 420 mg DM of substrate. The same experimental setup with the same dietary treatments as above was used for the trial. After 24 h of incubation, the incubation medium was transferred to 50-mL centrifuge tubes. Any particles attached to the walls of the serum bottles were washed off with distilled H2O and transferred to centrifuge tubes. The tubes were centrifuged at 3000 × g (ScanSpeed 1580R, Labogene, Seoul, South Korea) for 20 min, and the supernatants were discarded. The tubes containing the pellets were dried in a forced-air oven at 65°C for 48 h to determine the residual DM weights and NDF digestibility. The digestibility of DM and NDF was calculated based on the proportion of the initial weight incubated lost. The DM content of the blank was used for normalization. The incubation procedure was repeated three times in separate weeks to assess the repeatability.
The same four cannulated Holstein steers used as rumen fluid donors were allocated to individual feeders equipped with steel stanchions, and the experiment was carried out as described previously (Kim et al., 2016). A total of twelve polyethylene/nylon bags (10 × 20 cm, pore size 300 ± 20 μm, FILTRA-BAG®, Mfr. No: EFT-1250A, Thomas Scientific) were filled with 60 g of unground PA each (three bags per steer). Each bag was sealed using a heat sealer and placed in a large retaining sac (20 × 30 cm, pore size 3 × 5 mm). A 3-m-long nylon cable was attached to one end of the retaining sac, and a cannula stopper was attached to the other end. The steers were fed the basal diet during a 14-day adaptation period before beginning the trial. The experiment was divided into three phases: a control phase of 10 days without sacs containing PA in the rumen, a treatment phase of 11 days in which three sacs containing PA were placed in three positions in the rumen (total 180 g per steer), and a recovery phase of 10 days after the sacs containing PA were removed from the rumen. The steers were locked in position by metal stanchions and allowed to stand until sampling was complete to minimize changes in ruminal volume and shape. Sampling for ruminal headspace gas was conducted on 11 occasions beginning 3 days before the initiation of the treatment phase (day 0). Gas samples from four steers were collected on days −3, −2, −1, 3, 5, 7, 9, 11, 14, 17, and 21 of the feeding period. On day 12, the steers began the recovery phase. Samples of rumen head space gas were taken at 1, 2, and 3 h after morning feeding by passing a 20-mL graduated syringe connected to a two-way stopcock (KOVAX, Seoul, South Korea) into the rumen through the cannula stopper. Six samples (three each during rumen contraction and relaxation) per sampling period were collected and immediately transferred to a vacuum tube (ref 364979, BD Vacutainer, Becton Dickinson, NJ, United States) for CH4 analysis as described previously (Bharanidharan et al., 2021a). Ruminal fluid samples were collected on days −1, 3, 11, and 21 at 3.5 h after morning feeding. After immediately measuring the pH using a pH meter (model AG 8603; Seven Easy pH, Mettler-Toledo, Schwerzenbach, Switzerland), ruminal fluid was transferred to a 50-mL centrifuge tube and centrifuged at 12,000 × g for 10 min (Supra-22K, Hanil Science Industrial, Gimpo, South Korea). The supernatant was transferred to a 15-mL centrifuge tube and stored at −20°C until analysis of NH3-N and VFA concentrations as described previously (Bharanidharan et al., 2018).
For microbial analysis, ruminal fluid samples collected on days −1, 11, and 21 were snap-frozen in liquid nitrogen and stored at −80°C until DNA extraction and next-generation sequencing using previously described methods and primers (Bharanidharan et al., 2018). Primers targeting the variable region V4 (515F/806R) were used because they cover both bacterial and archaeal populations, producing an amplicon size amenable for Illumina MiSeq sequencing (Caporaso et al., 2011; Kozich et al., 2013). The raw Illumina MiSeq reads generated were demultiplexed according to their barcodes, and the sequences were quality-filtered (≥Q20) based on the quality control process in Quantitative Insights into Microbial Ecology (QIIME) version 1.9.0 software with a filtered length of ≥ 200 bp as described previously (Bharanidharan et al., 2021c). The processed paired reads were concatenated into a single read, and each single read was screened for operational taxonomic unit (OTU) picking using the de novo clustering method CD-HIT-OTU embedded in QIIME 1.9.0 with reference to the National Center for Biotechnology Information (NCBI) database (NCBI_16S_20190602, 97% nucleotide identity).
Cyclic GMP (cGMP)-dependent protein kinase (cGK), the central regulator of cGMP signaling in malaria parasites and a potential target for new antimalarial drugs (Baker et al., 2020), was used to evaluate the potential antiprotozoal effects of major PA metabolites. Because the rumen protozoa-specific protein sequence of cGK was not available in non-redundant databases, we used a manual method to determine the protein sequence. The partial mRNA sequence of cGK 9-1(XM_001017123) from Tetrahymena thermophila SB210, a widely used protozoal model, was retrieved from the NCBI database (accessed on 02 September 2021). The non-annotated genome assembly of E. caudatum MZG-1 (NBJL00000000.3) was downloaded from the NCBI database (accessed on 02 September 2021), and a local nucleotide database was created in BioEdit version 7.2.5 (Scripps Research, La Jolla, CA, United States) (Hall, 1999). A local nucleotide BLAST (BLASTn) search of the T. thermophila SB210 cGK nucleotide sequence was performed against the E. caudatum strain MZG-1 genome database. The result revealed nucleotide homology with E. caudatum strain MZG-1 (NBJL03004678.1). Furthermore, a protein homolog of the E. caudatum strain MZG-1 protein was searched for using the BLASTx algorithm1. The results revealed that the sequence of E. caudatum strain MZG-1 was 67% homologous (e-value = 0) to Bacillus cyclic nucleotide-binding domain-containing protein (MBP3841481) and its corresponding nucleotide (JAGAWU010000032) sequences. The corresponding aligned nucleotide sequence of E. caudatum strain MZG-1 was extracted and translated to an amino-acid sequence using the BLASTx algorithm. The amino-acid sequence was confirmed to encode cGK and was later used as the query for structure prediction.
The primary structural parameters of the E. caudatum strain MZG-1 specific cGK (query) protein were determined using ProtParam2. The physicochemical characters such as molecular weight, theoretical isoelectric point, amino-acid composition, extinct coefficient, estimated half-life, instability index, aliphatic index, and grand average of hydropathicity were computed. Secondary structure conformational parameters related to the presence of α-helices, β-turns, extended strands, and random coils were computed using the self-optimized prediction method with alignment (SOPMA) tool3. The similarity of the query protein with publicly available homologs was assessed by searching against non-redundant databases, including NCBI and Protein Data Bank (PDB) databases. The online protein structure prediction tool PS24 was used to predict the three-dimensional structure of the protein. Superimposition of predicted models of the query onto the template was performed using SALIGN5 and visualized using the UCSF Chimera package (Pettersen et al., 2004). The models were analyzed based on the DOPE score, and their stereochemical quality was validated by generating a Ramachandran plot using PROCHECK6 on the SAVES server7.
For molecular docking studies, the chemical structures of the major metabolites (ligands) of PA (dicaffeoyl quinic acid, quercetin-3-O-glucoside, chlorogenate, palmitic acid, linoleic acid, and oleic acid) were retrieved from the PubChem compound database8. The retrieved ligand structures in .sdf format were converted to .pdb format using PyMOL version 1.7.4.5 software (DeLano Scientific LLC, San Carlos, CA, United States). Docking analysis was carried out using AutoDockTools version 1.5.4 and AutoDock version 4.2 software (Scripps Research Institute Molecular Graphics Laboratory, La Jolla, CA, United States) as described previously (Arokiyaraj et al., 2015). The docked complexes were visualized using Discovery Studio Visualizer version 4.5 (BIOVIA, San Diego, CA, United States).
In vitro data were analyzed using the PROC MIXED procedure of SAS version 9.4 (SAS Institute, Cary, NC, United States) with a Tukey-Kramer adjustment. The dose was considered a fixed effect and each incubation run a random effect. Linear, quadratic, and cubic components of the response to increasing doses of PA were evaluated using orthogonal polynomial contrasts. The CONTRAST option of the MIXED procedure uses the coefficient matrix generated in PROC IML for unequally spaced treatments. Pearson’s correlation coefficients were calculated to identify correlations between substrate FA concentration, CH4 production, fermentation characteristics, and digestibility using the PROC CORR function in SAS.
In sacco data were also analyzed using the PROC MIXED procedure of SAS. The statistical model for rumen microbial composition data included treatment type and period as fixed effects and each animal as a random effect. The headspace CH4 concentration, ruminal fermentation characteristics (pH, VFAs, and NH3), and microbial abundance were subjected to repeated-measures analysis of variance. The statistical model for fermentation parameters used the same model with the inclusion of day as a fixed effect. Within the period, each day was taken as a repeated measurement, and the treatment × day interaction was not evaluated. For the CH4 data, both day and time were included as fixed effects in the model and were considered repeated measurements. The treatment, day, and time effects as well as treatment × day and treatment × time interactions were evaluated. The Akaike information criterion was used to identify the covariance structure with the best fit, and the autoregressive and compound symmetry covariance structures were used to analyze the fermentation characteristics and CH4 data, respectively. Means were calculated using the LSMEANS function and were compared using the PDIFF option in SAS. Significant treatment effects were detected by pairwise comparisons employing Tukey’s test. Regarding differences according to treatment, p < 0.05 was taken to indicate significance and 0.05 < p < 0.1 was considered to indicate a trend toward significance. To identify bacterial lineages that drive the clustering of microbial communities in different phases, we performed principal component analysis (PCA) using the fviz_pca_biplot function in the FactoMineR package (Husson et al., 2020) in R version 4.0.3 software (The R Foundation for Statistical Computing, Vienna, Austria).
The proximate and FA composition of PA are shown in Table 2. The CP, NDF, and EE contents of PA were 235.0, 421.0, and 129.4 g/kg DM, respectively. The analysis of the FA composition of PA revealed enrichment of saturated FAs (SFAs; 3.5 g/kg DM), monounsaturated fatty acids (MUFAs; 2.3 g/kg DM), and PUFAs (5.1 g/kg DM) dominated by linoleic acid (C18:2; 4.5 g/kg DM). The omega-6:omega-3 FA ratio was 10.2 with enrichment of omega-6 FAs (4.6 g/kg DM).
Altogether, 147 significant analytes were identified in the PA methanolic extract, 59 of which were identified in ESI positive ion mode (Figure 1A and Table 3), 54 in ESI negative ion mode (Figure 1B and Table 4), and 34 in both ESI positive and negative ion mode (Table 5). The identified analytes can be classified into saccharides and their derivatives, phenolic compounds and their derivatives, alkaloids, flavonoids, triterpenoids, coumarins, quinic acids, FAs, organosulfonic acids, and others. FAs and their derivatives, especially linoleic acid derivatives, were the main constituents in PA. Analytes such as caffeic acid, chlorogenate, quercetin, quercetin-3-O-glucoside, betaine, wilforlide A, 1,28-dicaffeoyloctacosanediol, hederagenin base + O-dHex-Hex-Hex, soyasaponin V, osmanthuside H, ginsenoside Ro, cinncassiol D2 glucoside, sarasinoside A1, and dioctyl sulfosuccinate were present in high proportions. Precursor organic acids including nicotinic acid, coumaric acids, dicaffeoylquinic acid, echinocystic acid, koetjapic acid, linoelaidic acid, and bovinic acid were identified. Finally, the phytosterols fucosterol, campesterol, and sitosterol were detected.
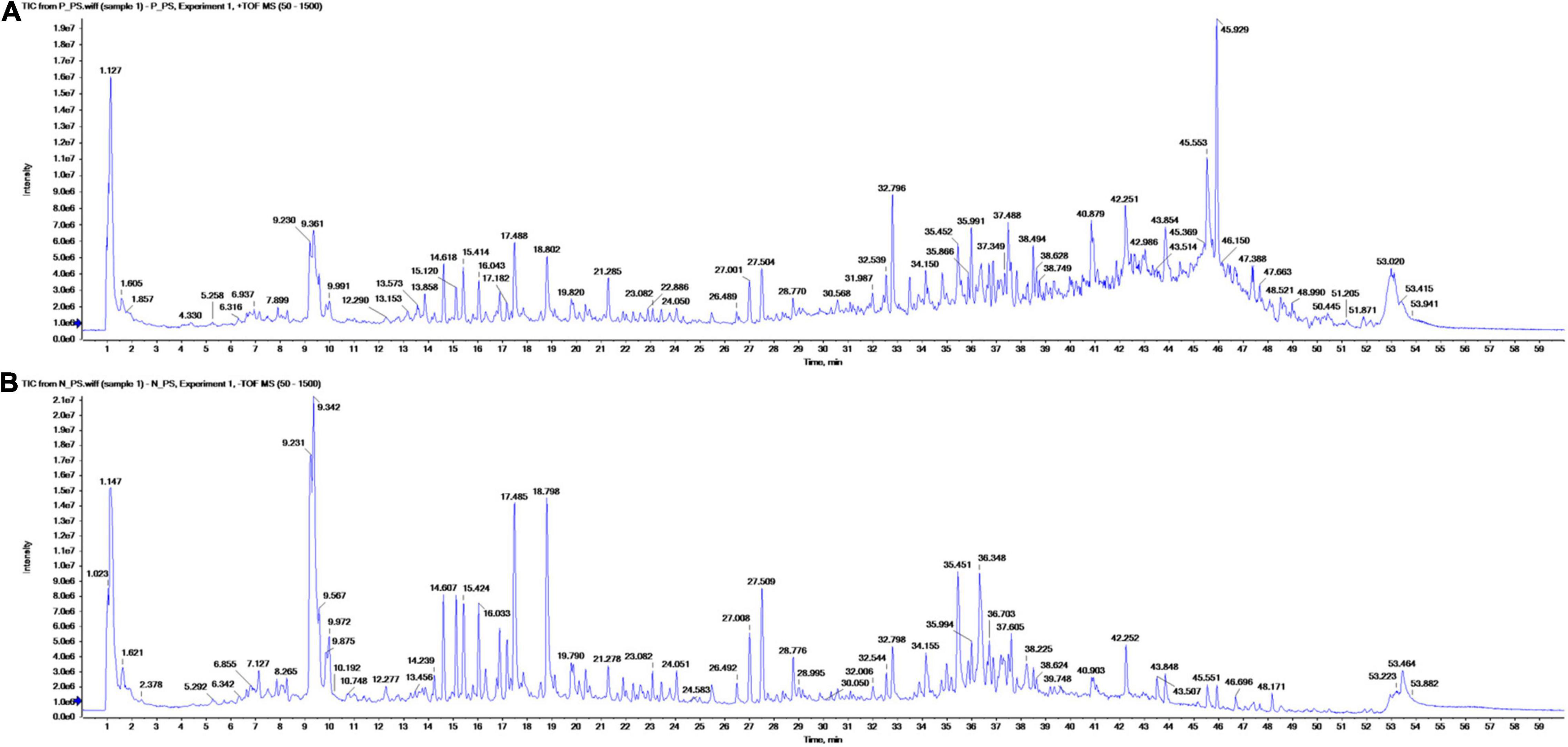
Figure 1. Total ion chromatograms of metabolites in the methanol extract of Pharbitis nil seeds in positive (A) and negative ionization (B) mode from ultra-performance liquid chromatography high-resolution mass spectroscopy analysis.
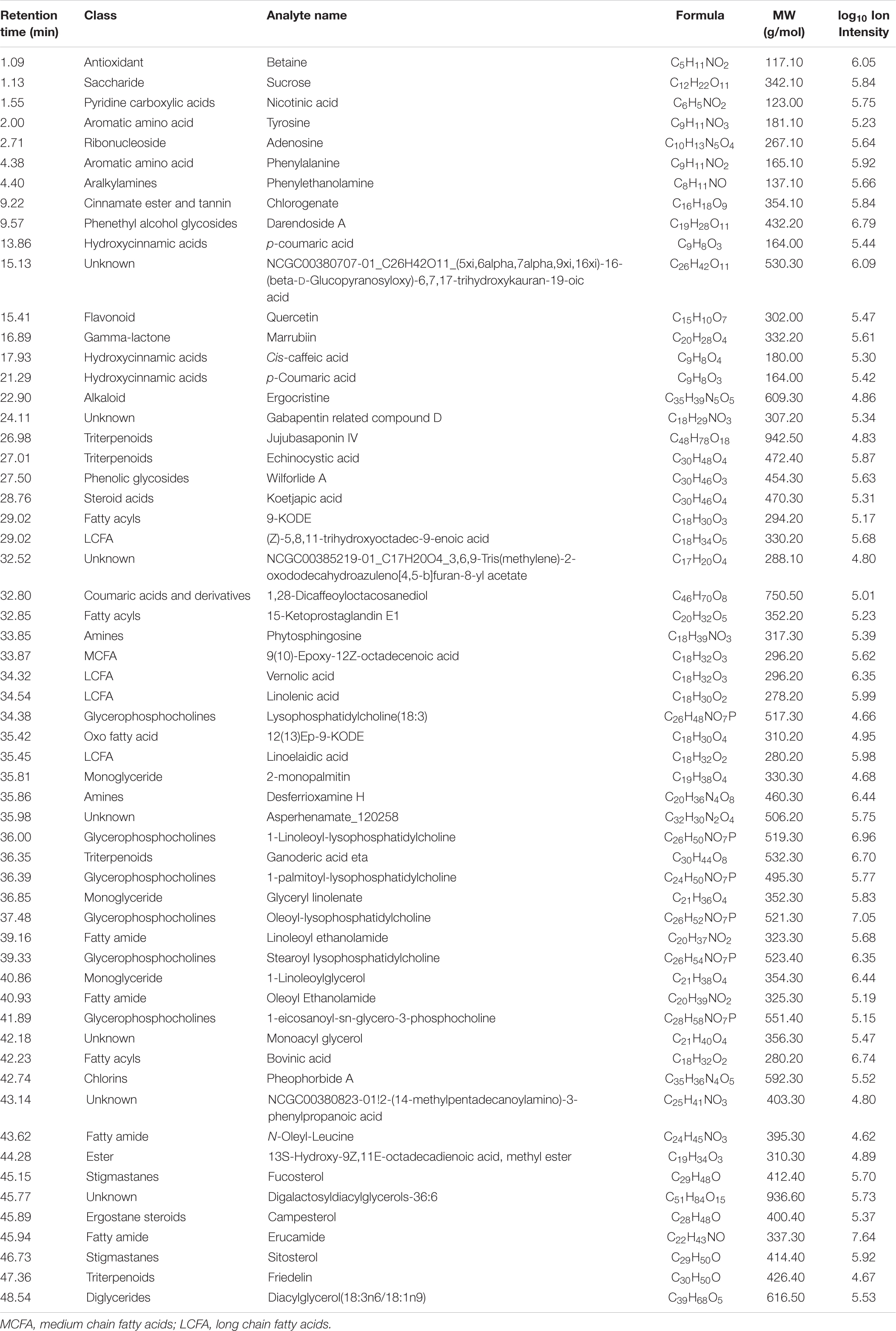
Table 3. Metabolites in the total methanolic extract of P. nil seeds identified using ultra-performance liquid chromatography high-resolution mass spectroscopy (UPLC-HRMS/MS) in positive ion mode.
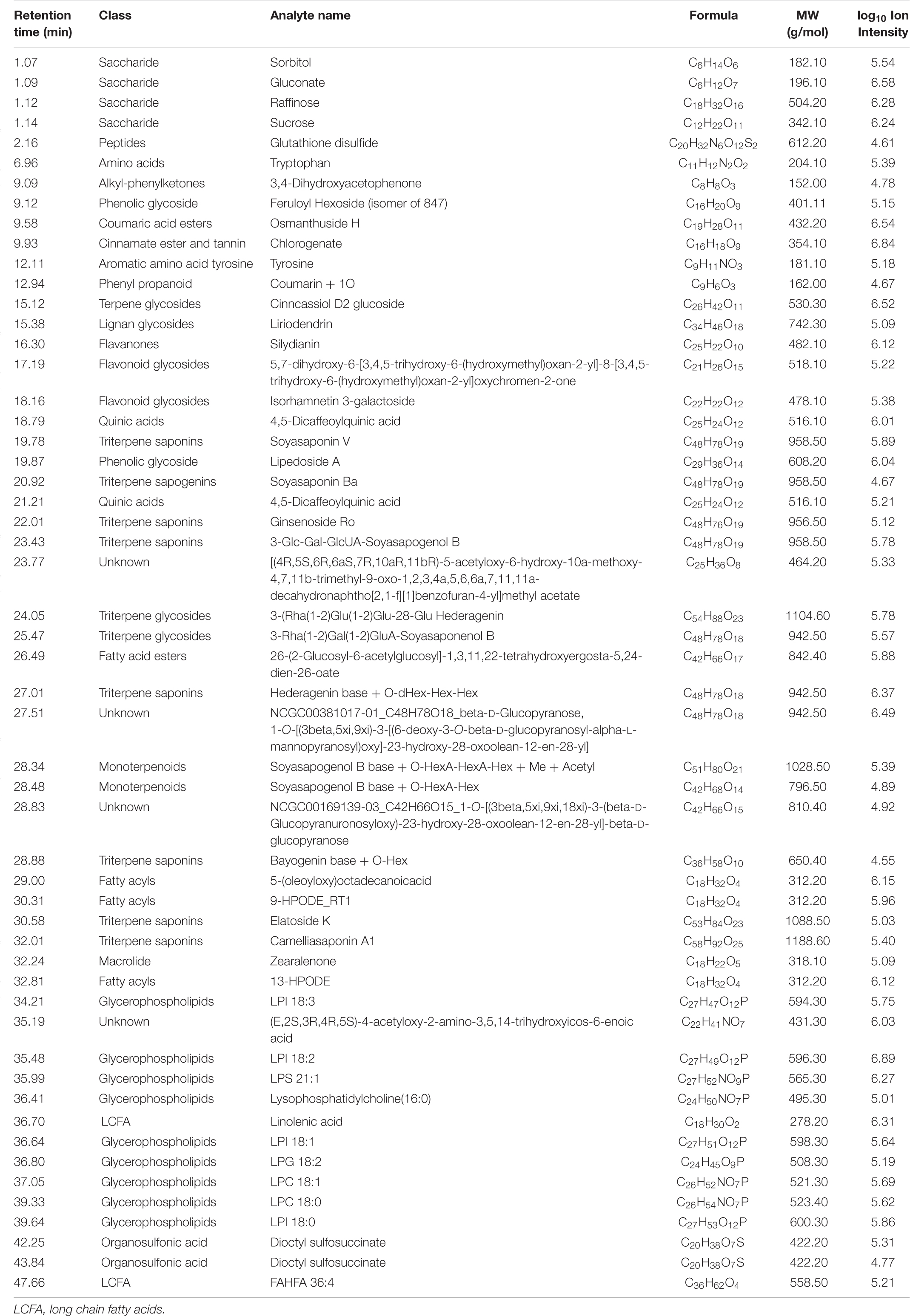
Table 4. Metabolites in the total methanolic extract of P. nil seeds identified using UPLC-HRMS/MS in negative ion mode.
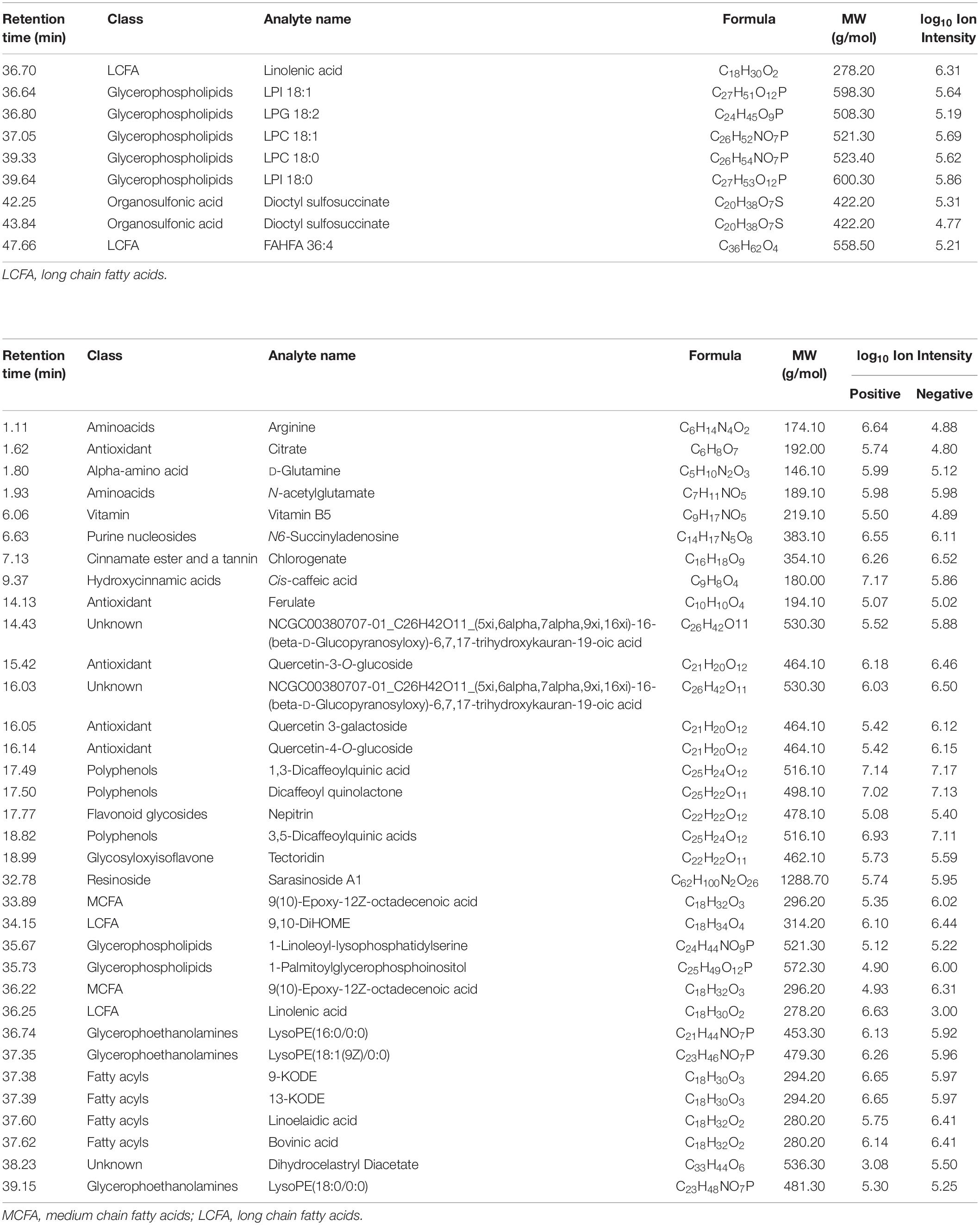
Table 5. Metabolites in the total methanolic extract of P. nil seeds identified using UPLC-HRMS/MS in both positive and negative ion modes.
Our results negate a dose–response relationship between PA and in vitro CH4 production, fermentation parameters, and digestibility (Table 6). Dietary SFA, MUFA, and PUFA concentrations increased with an increasing PA dose, whereas total gas production decreased linearly (p < 0.001). The production of CH4 (mmol/g DM) decreased by 23.7–69.1% compared with the control at doses of 4.5–45.2% DM. At doses ≤ 9% DM, enhancement of DM digestibility (DMD; p = 0.052) and NDF digestibility (NDFD; p = 0.073) by 9.6–15.2% was noted compared to the control, in which 31.1–50.5% decreases (p < 0.001) in CH4 production expressed as mmol/g digestible DM (CH4/dDM) or NDF (CH4/dNDF) were observed. At doses ≥ 13.6% DM, DMD and NDFD decreased but remained higher than in the control. Although DM and NDF degradation was unaffected at low doses of PA, the production of total VFAs and NH3-N decreased (p < 0.001). Conversely, the molar proportions of the VFAs propionate and butyrate increased linearly (p < 0.001) with an increasing dose. By contrast, the proportions of acetate, iso-butyrate, valerate, and iso-valerate decreased linearly (p < 0.001).
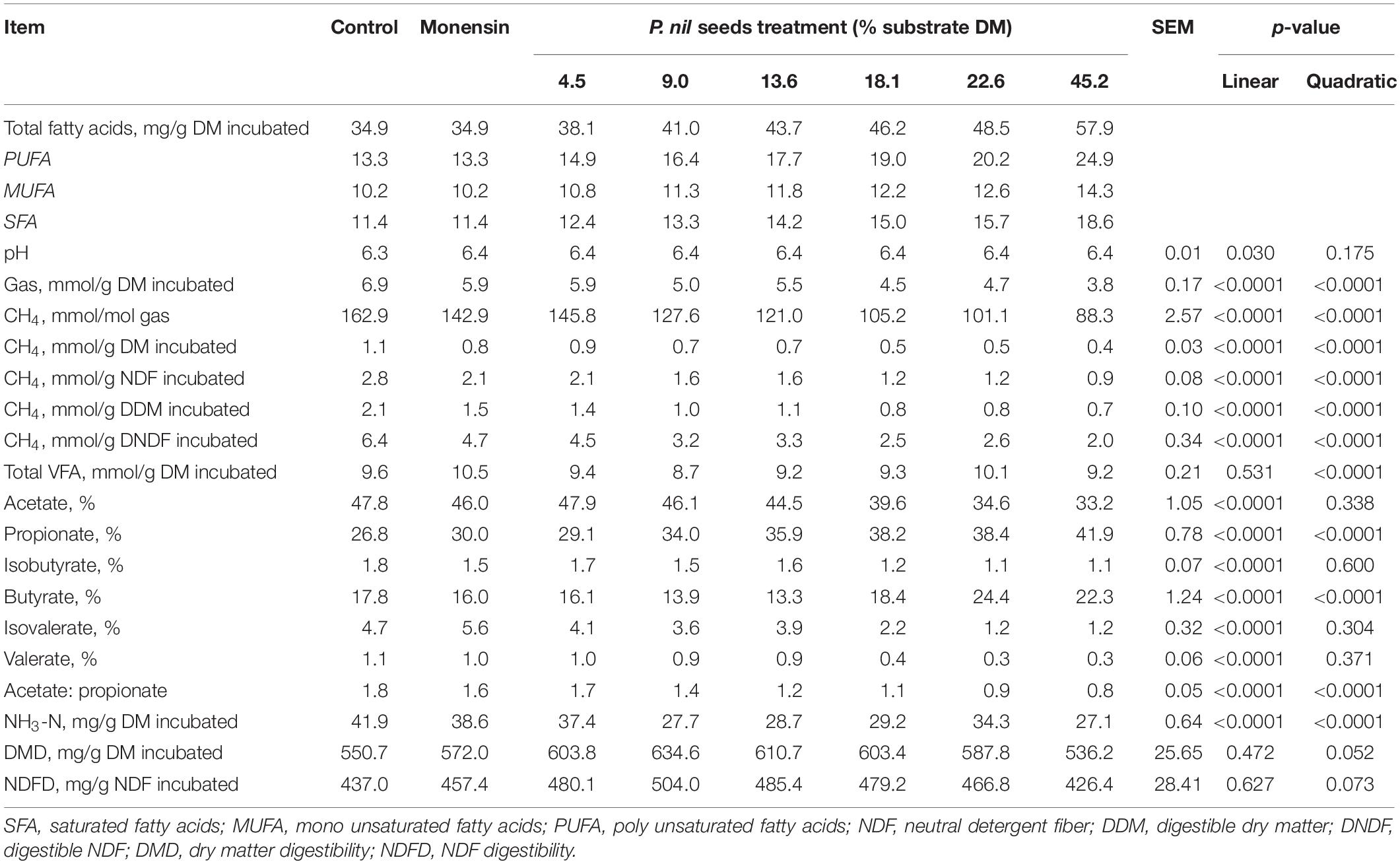
Table 6. Dose–response effect of P. nil seeds on in vitro methane (CH4) production, fermentation parameters, and digestibility (nreplicate = 4).
Pearson’s correlation coefficients for the tested parameters are listed in Table 7. Strong negative associations of dietary FA concentrations with total gas production (r = −0.85; p < 0.005), total CH4 production (r = − 0.85; p < 0.005), and CH4 production per g dNDF (r = −0.76; p < 0.005) were noted in vitro. The dietary FA concentration was strongly associated with propionate production (r = 0.89; p < 0.005) and the NH3-N concentration (r = −0.71; p < 0.005). However, no association (p > 0.05) was noted between the dietary FA concentration and DMD or NDFD. The proportion of propionate was strongly negatively associated with total CH4 production (r = − 0.95; p < 0.005) and CH4 production per g dNDF (r = − 0.89; p < 0.005).
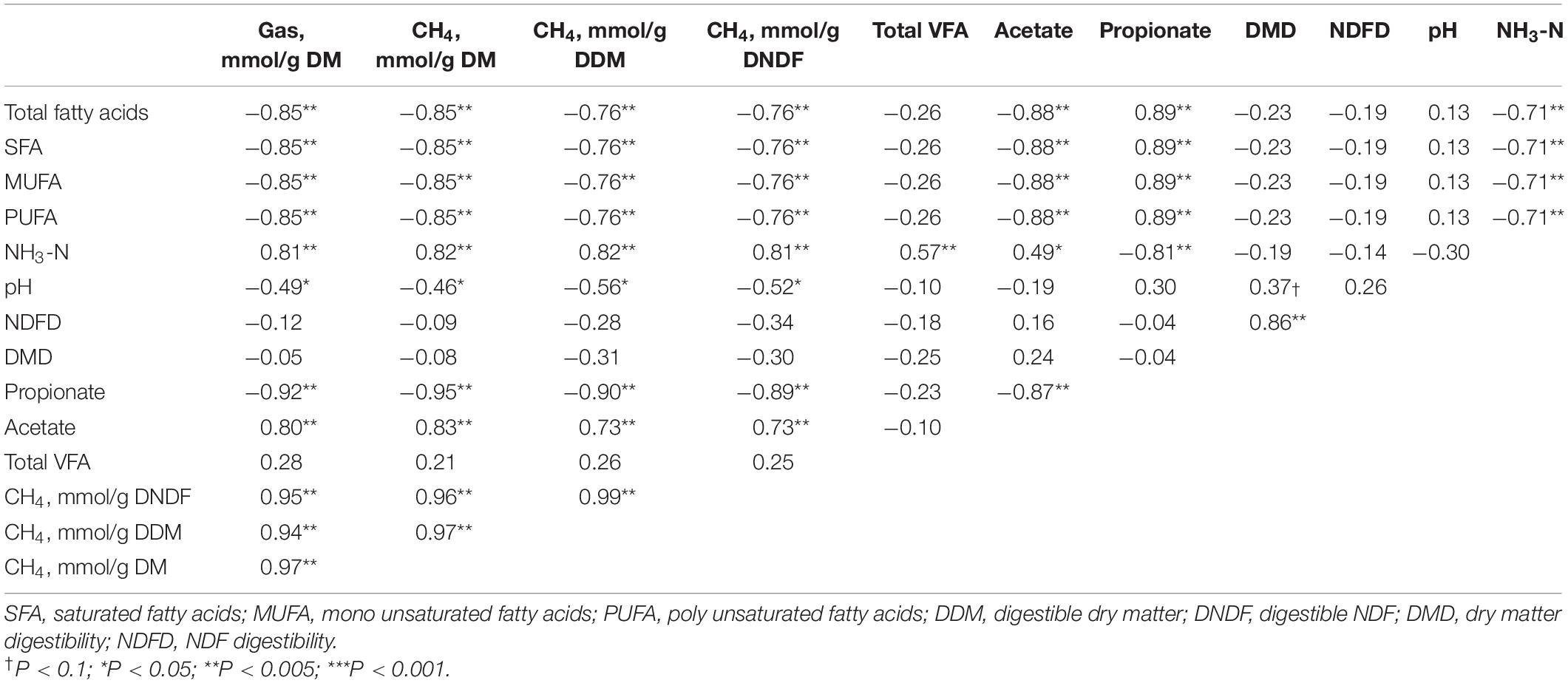
Table 7. Pearson correlation coefficients for associations between dietary fatty acid content, CH4 production, fermentation parameters, and digestibility.
In sacco, the average CH4 concentration in the rumen headspace gas sample decreased (p = 0.087) from 11.3 to 5.5% in the treatment phase and then steadily increased to 8.9% in the recovery phase (Figure 2). An interaction between treatment and sampling day (p < 0.05) was noted for the CH4 concentration. No interaction (p > 0.05) between treatment and sampling time was noted. Ruminal fluid pH and NH3-N concentration decreased (p < 0.005) and remained relatively low at the end of the treatment phase, then increased in the recovery phase (Table 8). The total VFA concentration (p < 0.05) and the proportions of propionate (p = 0.089) and butyrate (p = 0.078) increased during the treatment phase and recovered toward the control level during the recovery phase. The acetate proportion exhibited the opposite pattern (p < 0.05).
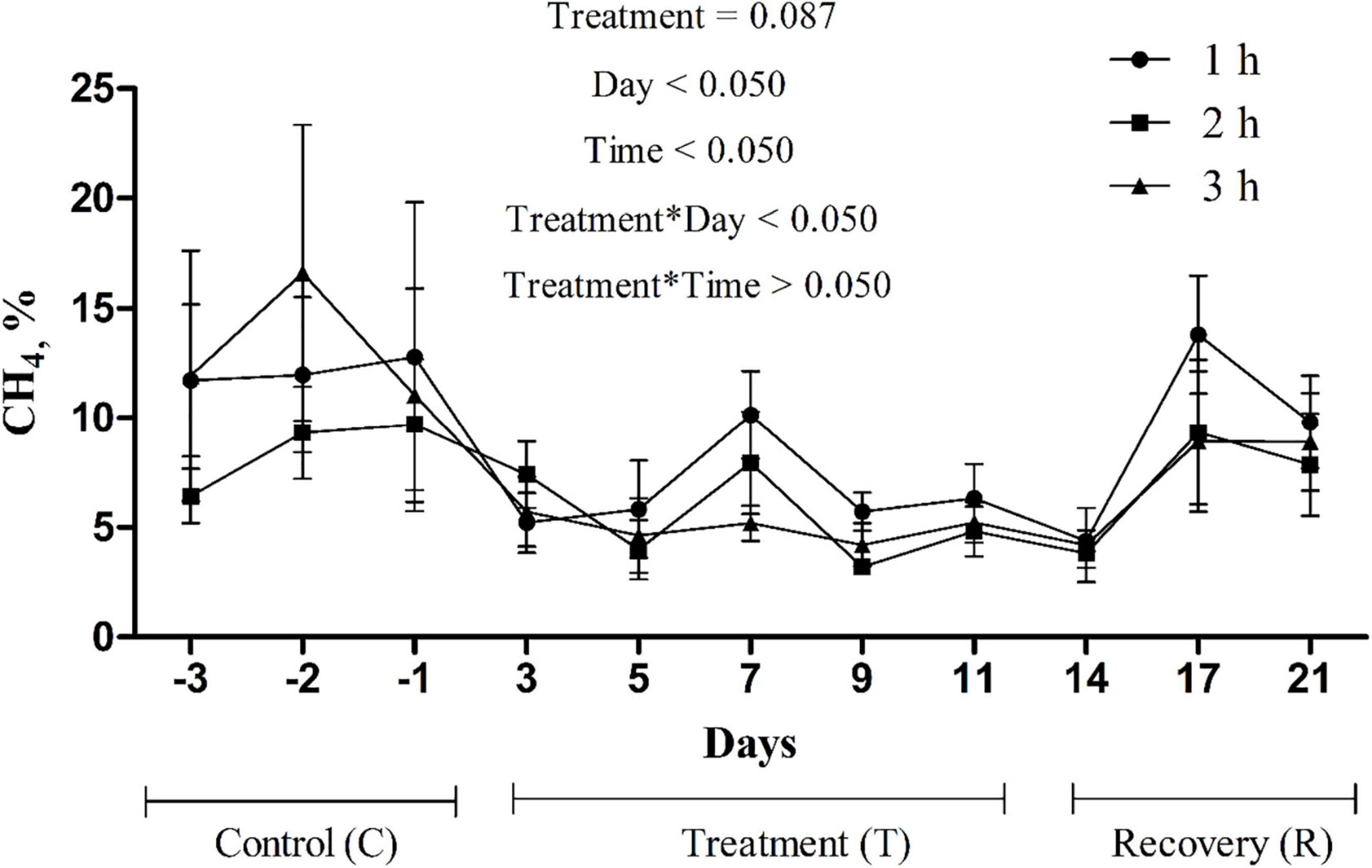
Figure 2. Changes in methane (CH4) concentration in rumen headspace gas due to the addition of P. nil seeds (n = 4 heads).
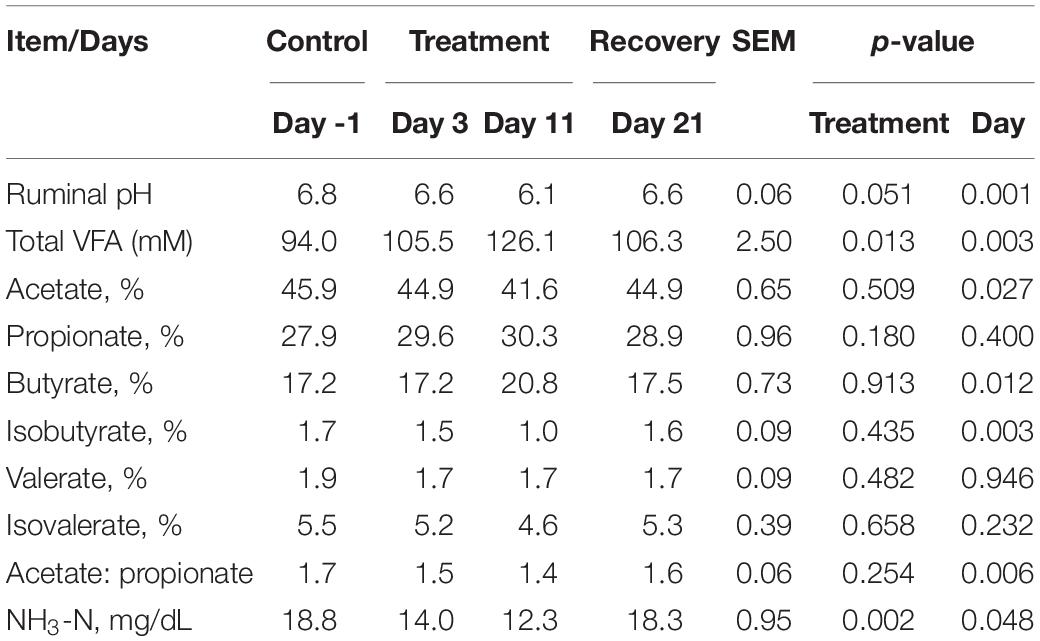
Table 8. Effects of P. nil seeds on fermentation parameters in sacco in a live continuous culture system (n = 4 heads).
Analysis of the rumen microbial reads at a depth of 46,860 quality reads yielded an average of 886 OTUs (range 711–985 OTUs; Supplementary Figure 1). Diversity analysis of the rumen microbiota revealed that the total microbial species richness (alpha diversity) was unaffected by PA (Supplementary Figure 1). Analysis of the rumen microbial composition revealed marked shifts in relative abundance at the phylum and species levels in the post-treatment period (Figures 2, 3 and Supplementary Table 2). Decreased abundances of sequences assigned to the phyla Bacteroidetes (p < 0.05), Spirochaetes (p < 0.05), and Proteobacteria (p = 0.062) and an increase (p < 0.05) in the phylum Firmicutes were observed post-treatment (Figures 3, 4 and Supplementary Table 2). Consequently, a decrease (p < 0.005) in the Bacteroidetes:Firmicutes ratio was noted (Figure 5). Regarding beta diversity, PCA revealed that all rumen samples from steers at different phases clustered separately, with 43.9% of the variation explained (Figure 6). PA led to a 78% decrease (p < 0.05) in the abundance of OTUs assigned to the most abundant bacterial species, Prevotella ruminicola (18.1%), compared to the control phase (Supplementary Table 2). Other species in the families Prevotellaceae and Paraprevotellaceae exhibited decreases of similar magnitude. In terms of fibrolytic microorganisms, OTUs assigned to Butyrivibrio fibrisolvens, Ruminococcus albus, Ruminococcus bromii, and Ruminococcus lactaris increased (p < 0.05) several fold post-treatment (Supplementary Table 2). The abundance of Streptococcus sp. increased several-fold. Although the abundance of Phylum Euryarchaeota was unchanged, OTUs assigned to the genus Methanobrevibacter and Methanosphaera increased (p < 0.05) post-treatment (Supplementary Table 2).
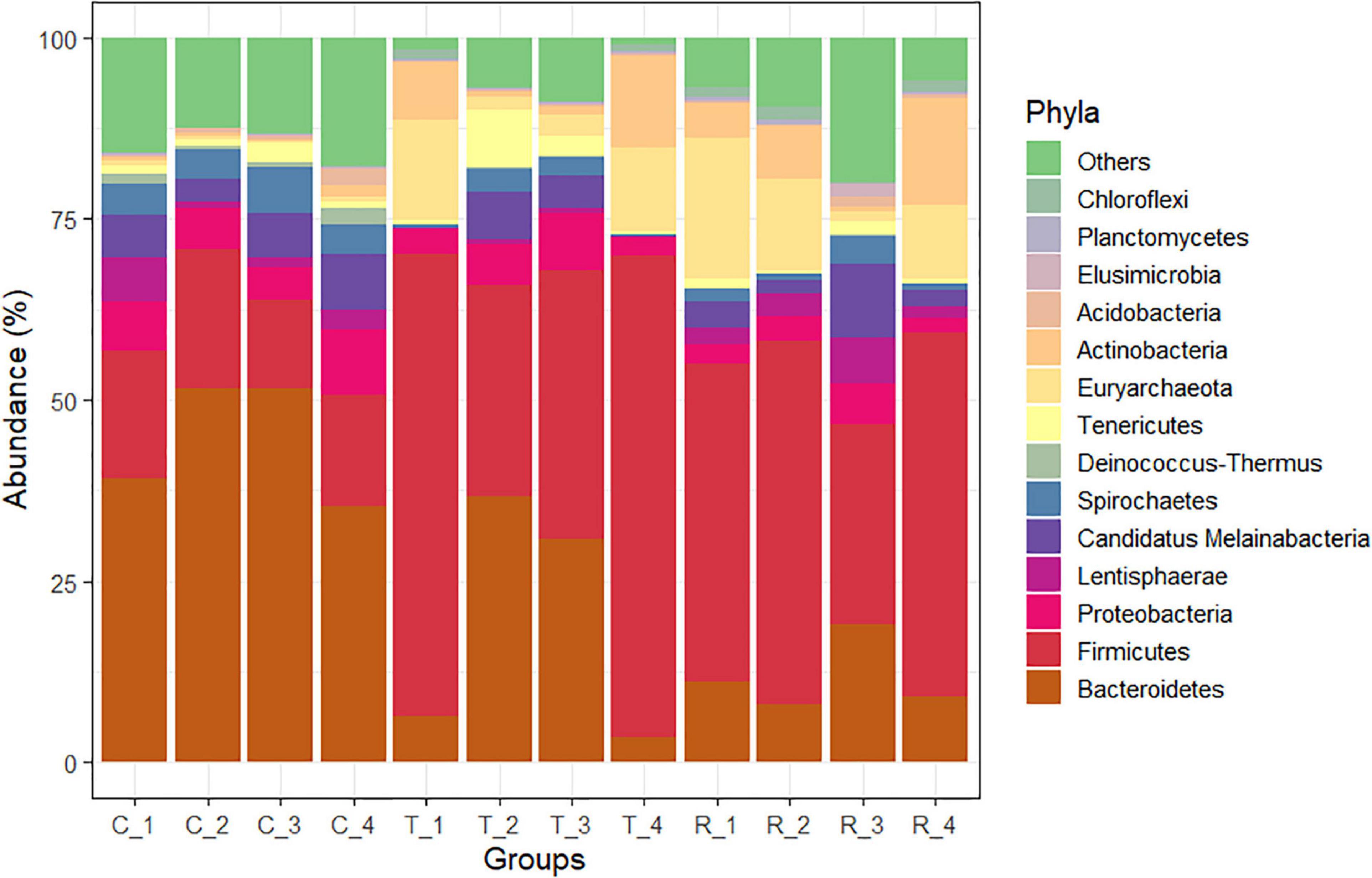
Figure 3. Phylum-level changes in the rumen microbiome of cannulated Holstein steers due to P. nil seeds. C, control; T, treatment; R, recovery.
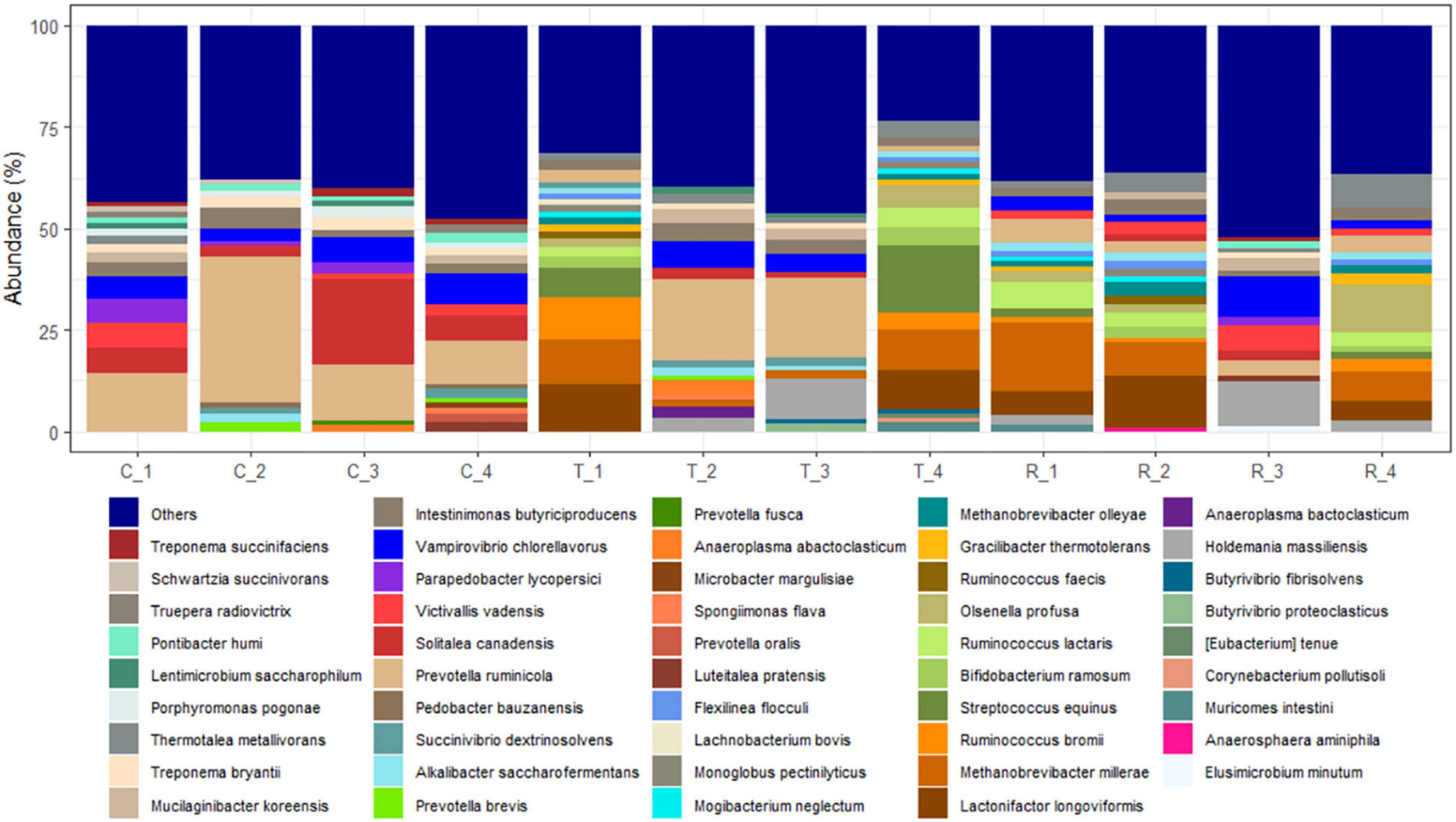
Figure 4. Species-level changes in the rumen microbiome of cannulated Holstein steers due to P. nil seeds. C, control; T, treatment; R, recovery.
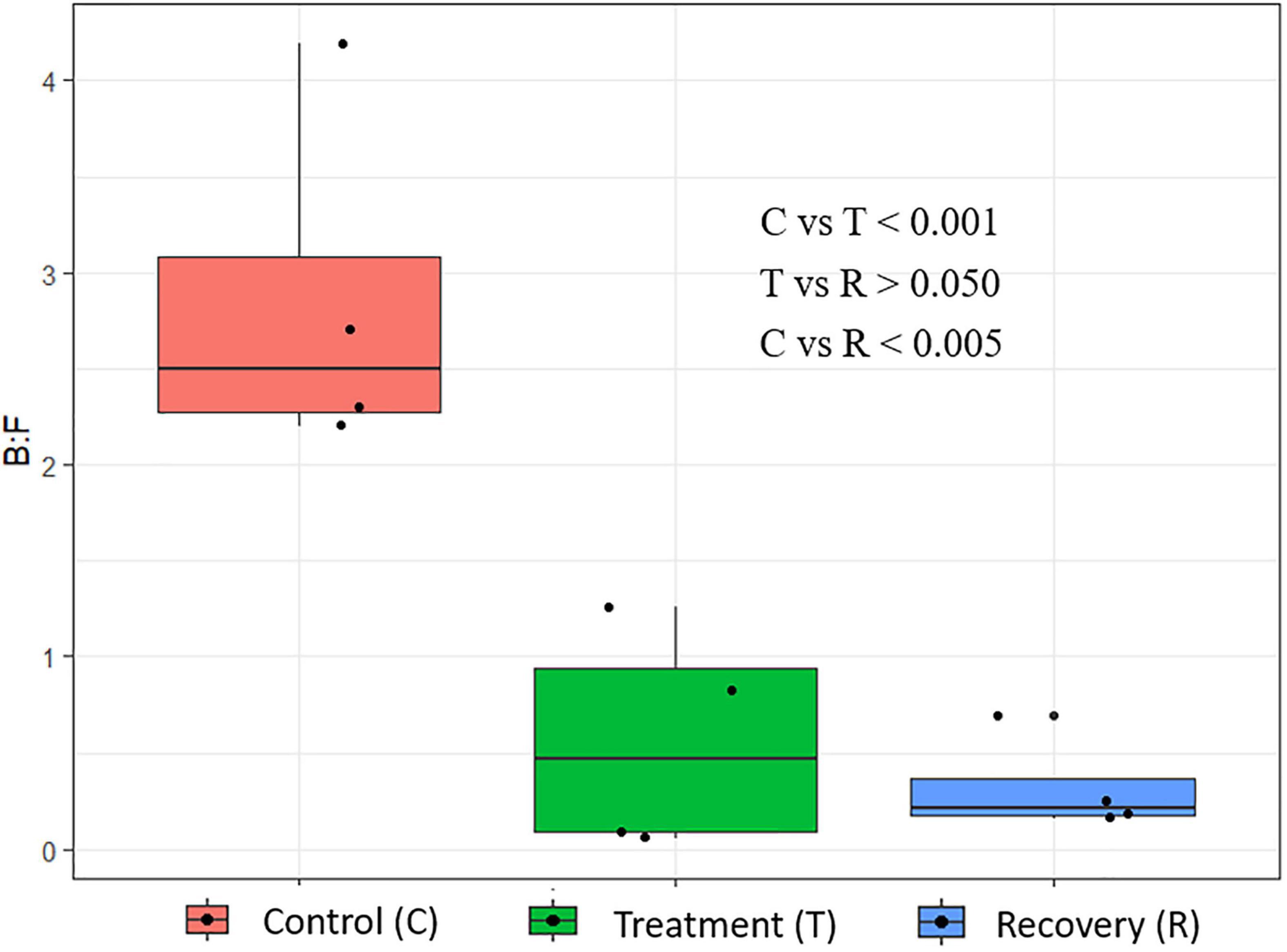
Figure 5. Changes in the Bacteroidetes:Firmicutes ratio in the rumen of cannulated Holstein steers due to P. nil seeds.

Figure 6. Principal component analysis results reflecting correlations between rumen bacterial/archaeal communities, CH4 yield, and fermentation parameters in sacco.
The results of a primary structural evaluation of the deduced cGK protein using ProtParam are shown in Supplementary Tables 3, 4. Evaluation of secondary structure conformational parameters using the SOPMA tool indicated that cGK is composed of 29.06% loops, 46.39% helices, 16.83% extended strands, and 7.72% β-turns (Supplementary Figure 2). Homology modeling of cGK performed based on the crystal structure of the A chain of Plasmodium vivax Sal-1 cGK (PDB ID: 4RZ7) as the template yielded sequence identity and e-values of 41.8 and 2e-94, respectively (Supplementary Figure 3A). The predicted model of cGK was superimposed on the cGK template (PDB ID: 4RZ7; Supplementary Figure 3B). PROCHECK analysis to assess the quality of the predicted model using a Psi/Phi Ramachandran plot indicated that 85.2% of the residues were in the most-favored regions, 10.1% were within additionally allowed regions, and 2.3% were in disallowed regions (Supplementary Table 5 and Supplementary Figure 3C). The G-factor for the modeled cGK was −0.27, revealing the predicted model to be of high quality (Supplementary Table 6). Therefore, model validation suggested that the model adequately represented the native protein.
Analysis of docked protein-ligand complexes based on binding affinity revealed that the predicted protein model of E. caudatum cGK has the highest binding energies of −6.83, −6.75, and −5.92 kcal/mol with dicaffeoyl quinic acid, quercetin-3-O-glucoside, and chlorogenate, respectively (Supplementary Table 7). Long-chain FAs such as linoleic acid, oleic acid, and palmitic acid also exhibited fair binding to cGK with moderate energy values of −4.75, −4.52, and −4.14 kcal/mol, respectively (Supplementary Table 7). Hydrogen bond interactions between the ligands and the active residues of cGK were noted (Figures 7A–F and Supplementary Table 7).
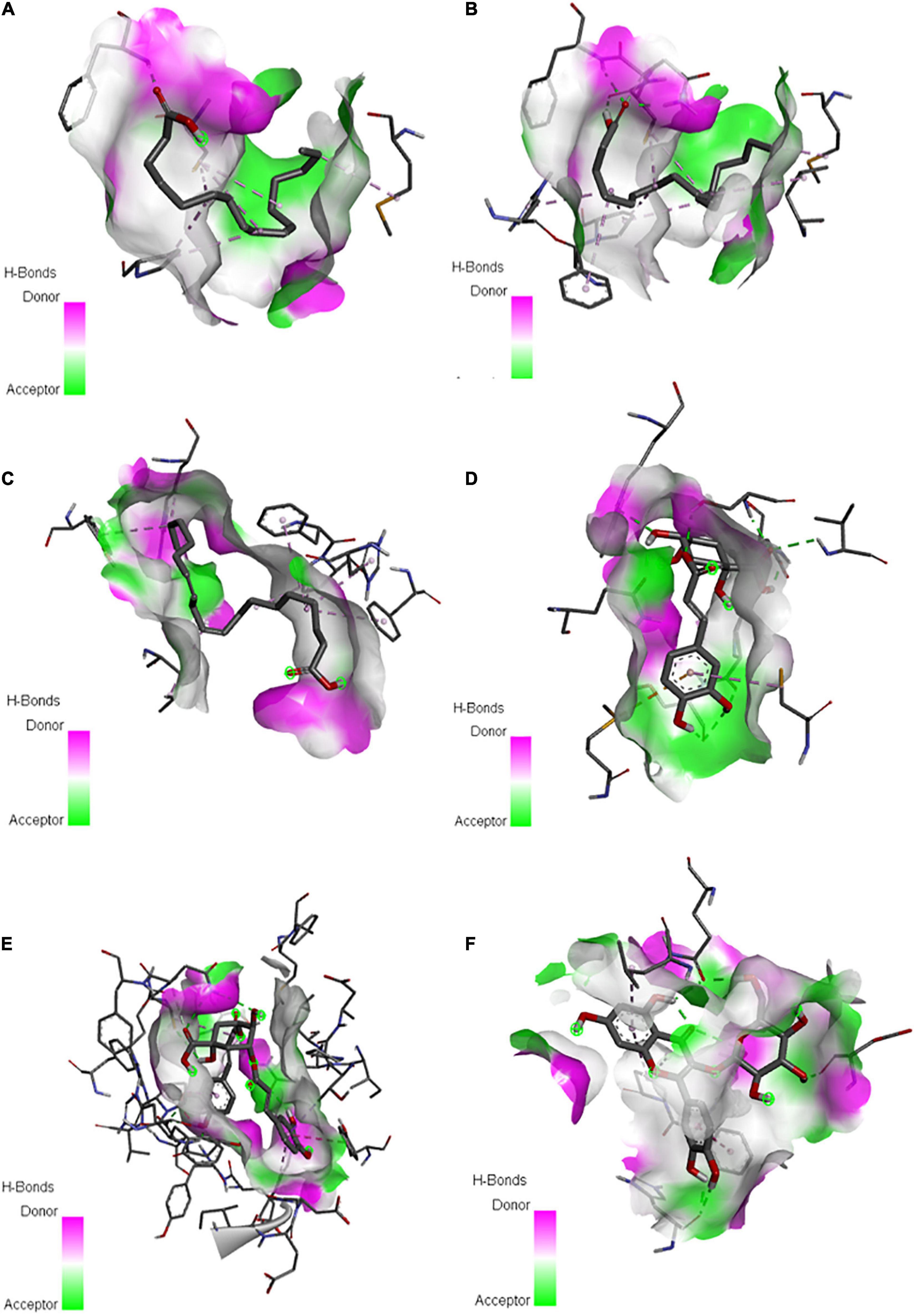
Figure 7. Putative binding sites of palmitic acid (A), oleic acid (B), linoleic acid (C), chlorogenate (D), dicaffeoyl quinic acid (E), and quercetin-3-O-glucoside (F) from P. nil seeds with cyclic GMP-dependent protein kinase of the rumen protozoan Entodinium caudatum.
Research on plant-based natural feed additives has been promoted due to restrictions on the non-therapeutic use of antibiotics in livestock production. Research into natural resources with high nutritional value and bioactive compounds can enhance productivity and mitigate the environmental footprint of livestock systems. Following our previous work (Bharanidharan et al., 2021a), we investigated for the first time the nutritive value and secondary metabolites of PA. We found high contents of CP, NDF, fat, and FAs in PA, suggesting that it has high nutritive value. The greater contents of FAs identified using GC-MS and FA derivatives identified using UPLC-HRMS/MS corroborate our prior finding (Bharanidharan et al., 2021a) that 60% of the compounds identified using GC-MS were derivatives of long-chain FAs. The fat content (129 g/kg DM) of PA is similar to that of grapeseed (Kapcsándi et al., 2021), whereas the FA profile (C16:0 [21%], C18:0 [21%], C18:2 [41%]) is comparable with that of sunflower seeds (Akkaya, 2018). Moreover, PA, as a source of omega-3 and -6 FAs (predominantly linoleic acid), can promote animal and human health (Haug et al., 2007). Plant secondary metabolites (PSMs) such as polyphenols reportedly benefit the health and productivity of ruminants (Olagaray and Bradford, 2019). The identification of flavonoids such as osmanthuside H, p-coumaric acid, quercetin, quercetin-3-O-glucoside, quercetin-4-O-glucoside, caffeic acid, ferulic acid, chlorogenic acid, and their derivatives in PA is consistent with previous studies (Saito et al., 1994; Kim et al., 2011). However, several metabolites (including betaine, wilforlide A, 1,28-dicaffeoyloctacosanediol, ginsenoside Ro, and other saturated and unsaturated FAs) are reported for the first time in this study. Notably, dioctyl sulfosuccinate, an anionic surfactant that is used as a laxative and stool softener, was identified in PA for the first time. PA is used in traditional medicine for its gastroprokinetic effect (Kim et al., 2020). In addition, the identified metabolites, including the FAs, have been shown to exhibit antimicrobial activity against bacteria (McGaw et al., 2002; Vasta et al., 2019) and protozoa (Matsumoto et al., 1991; Ayemele et al., 2020). Therefore, these compounds could be used to manipulate the rumen microbiome to mitigate CH4 emissions and optimize feed efficiency.
It is valuable to identify and assess the effects of metabolites present in plant materials on rumen fermentation characteristics and microbes than testing the raw plant material directly.
It is because the environmental factors such as soil conditions could greatly affect the concentration of the active metabolite in that plant. This in turn would influence the optimal dose determination and practical application of the plant material. However, it is also important to consider the synergic effects from other components which could be achieved when plant raw materials are used directly. Because the effects of crude PA extract was investigated at one dose in our previous study (Bharanidharan et al., 2021a), we used raw plant material at range of doses here. The decrease in CH4 production in response to PA in vitro confirms our previous observation (Bharanidharan et al., 2021a). At doses as low as 4.5% DM, PA decreased the production of CH4 (mmol/g DM incubated) by 24% in vitro, similar to 30 ppm monensin. The same effect was evident in sacco; the CH4 concentration in the rumen headspace gas decreased by 50% after 10 days. However, this could not be considered as a decrease in absolute CH4 production, since the total gas production was not quantified. This observed effect of PA on methanogenesis might have been related to the higher concentrations of FAs (3.8–5.8% DM) in vitro. The maximum decrease in CH4 production was 69% in vitro, and there were strong negative associations between the FA concentration and gas parameters, as reported previously (Patra, 2013). Patra (2013) suggested that for each percentage point increase in fat in the diet, CH4 emission decreased by 3.77% in vivo with a maximum reduction of 15.1% at 6% dietary DM, which is lower than our finding. It is possible that other PSMs in PA might have contributed to the effect (Ku-Vera et al., 2020). In addition, in vitro culture systems can overestimate such effects compared to in vivo systems (Machmüller, 2006).
The inhibitory effects of fats, FAs, and other PSMs on rumen methanogenesis are related to altered H2 thermodynamics in the rumen caused by a decrease in the populations of ciliate protozoa, their associated methanogens and other H2 producing bacteria (Newbold et al., 2015; Toprak, 2015). Although the protozoal population was not quantified in the current study, the decrease in methanogenesis in the current study could be attributed to the 90% decrease in total ciliate protozoan population as observed upon addition of ethanol extract of PA in our previous in vitro study (Bharanidharan et al., 2021a). Generally, rumen protozoa are associated with and engulf rumen bacteria and archaea (Park and Yu, 2018). Rumen protozoa degrade the proteins of engulfed microbial cells into oligopeptides and free amino acids, which are deaminated to produce free NH3-N, leading to inefficient dietary N utilization (Newbold et al., 2015). A recent study also reported that the presence or absence of different protozoal genera differentially modulate rumen prokaryotic community ecological structure (Solomon et al., 2021). Therefore, in this study, the shift in Bacteroidetes to Firmicutes ratio in the treatment phase could also be partially related to the defaunation effect of PA. However, other reasons including the specific binding of FAs in PA to the lipid bilayer membrane of Gram-negative bacteria that leads to cell death (McGaw et al., 2002) could also be attributed to the microbial shift. Huws et al. (2015) reported a 65% decrease in the Prevotella population upon adding C18:3-rich flax seed oil to the diet, supporting our hypothesis. Consistent with our results, Park et al. (2019) indicated a decrease in the Bacteroidetes:Firmicutes ratio as well as decreased Prevotella and increased Streptococcus abundances upon inhibition of E. caudatum. However, it could not be completely related to the results observed in the current study as different Entodinium spp. that exhibit different predatory behavior are represented in different proportions in the rumen (Kišidayová et al., 2021). Furthermore, the decrease in ruminal NH3-N in this study indicates reduced microbial protein turnover due to the inhibition of protozoa (Dai and Faciola, 2019). However, Prevotella species reportedly degrade dietary protein to produce NH3-N (Griswold et al., 1999); thus, the decrease in their relative abundance might have contributed to the reduced NH3-N. Although rumen protozoa population is associated with butyrate production, the increase in butyrate proportion upon adding PA in vitro may be partially related with the increase in relative abundance of butyrate producers such as Butyrivibrio fibrisolvens and Pseudobutyrivibrio ruminis in treatment phase of in sacco trial. The strong binding of PUFAs such as oleic acid, palmitic acid, and linoleic acid to cGK of E. caudatum in silico further provided insight into the antiprotozoal effect of PA, as these FAs are reportedly toxic to rumen protozoa (Dohme et al., 2008). This is the first study to predict the structure of E. caudatum cGK, which is involved in cell replication and other cell-cycle processes. Furthermore, in an evaluation of Calotropis gigantea extract rich in quercetin, quercetin-3-O-glucoside, and other hydrocinnamic acid derivatives for its effect on protozoa, Ayemele et al. (2021) reported a 50% decrease in the abundance of E. caudatum coupled with a decrease in NH3-N. This is consistent with the strong binding of quercetin-3-O-glucoside to E. caudatum cGK. Jin et al. (2021) reported the antimethanogenic potential of caffeic acid, in agreement with the large proportion of that flavonoid in PA in this study. Therefore, the major metabolites of PA may be responsible, at least in part, for inhibiting E. caudatum, thereby decreasing CH4 and NH3-N production.
Despite the symbiotic relationship between protozoa and methanogens (Levy and Jami, 2018), protozoal inhibition increased methanogen abundance, consistent with our previous study (Bharanidharan et al., 2021a). Similar responses to defaunation (Mosoni et al., 2011) and the addition of oils (Nur Atikah et al., 2018) have been reported. The proportion of methanogens that associates with protozoa is not large (Belanche et al., 2014), and other planktonic methanogens in the rumen might not be affected by PA. The decrease in CH4 caused by feed with increased lipid content is related to the increased production of propionic acid via biohydrogenation of unsaturated FAs (Hook et al., 2010). This is consistent with the strong associations between dietary PUFA content, propionate proportion, and CH4 production. This suggests that propionate synthesis was the major hydrogen sink despite the greater abundance of methanogens, which use H2 for CH4 production. Similarly, a monensin-mediated decrease in CH4 via increased biohydrogenation was caused by the inhibition of protozoa, whereas the effect on methanogens was minimal (Hook et al., 2010). In addition, the abundance of B. fibrisolvens, which is involved in the initial step of rumen biohydrogenation of PUFA to vaccenic acid (McKain et al., 2010), increased in the treatment phase. Moreover, decreased CH4 production due to linseed and coconut oil rich in PUFAs is reportedly not clearly linked to methanogenic gene abundance or changes in the archaeal community (Patra and Yu, 2013; Martin et al., 2016). It is also possible that the PA metabolites hindered the gene activity of methanogens for reducing H2 to CH4 in the rumen. However, after withdrawing PA (recovery phase) in sacco, the CH4 concentration in the headspace gas and other fermentation parameters started reverting to their initial values. This might have been due to the recovery of rumen microbes to their initial composition as a result of probable increase in rumen protozoal population in absence of PA. In vitro batch or continuous culture systems cannot be used to evaluate short- and long-term responses together with re-adaptation of microbes, and typically do not yield consistent results, unlike in sacco methods involving LCCSs. However, the outward and inward flow of gases through the cannula needs to be considered if this method is to be used to evaluate the effects of plant materials on methanogenesis and other fermentation parameters (Wang et al., 2019). In addition, lack of models for quantifying the total gas production has limited the LCCS approach to determine only the rumen headspace CH4 concentration rather than the absolute CH4 production.
Methane mitigation strategies involving plant additives are useful only if nutrient digestibility and other fermentation parameters are not concomitantly compromised. Dietary supplementation of lipids and the elimination of rumen protozoa are associated with decreased nutrient digestibility (Patra, 2013; Dai and Faciola, 2019). However, DMD and NDFD were increased by PA, likely because of the increased populations of fibrolytic bacteria such as B. fibrisolvens, R. albus, R. bromii, and R. lactaris. This is in agreement with the increase in R. albus and R. flavefaciens observed after defaunation (Mosoni et al., 2011; Park et al., 2019) and the addition of oil to a sheep diet (Adeyemi et al., 2016). Doreau et al. (2009) noted that dietary addition of oils did not negatively affect organic matter fermentation in the rumen, consistent with this work and a prior defaunation study (Park et al., 2019). It is also possibly a result of the protozoal groups inhibited; Epidinium, Polyplastron, and Eudiplodinium are cellulolytic, whereas Entodinium is weakly hemicellulolytic and so its contribution to fiber digestion is minimal (Takenaka et al., 2004). However, in this study, the magnitude of increase in digestibility declined at doses > 9% DM (>4.1% FAs), thereby suggesting that a dose of < 9% DM is optimal for decreasing CH4 production in vitro while enhancing digestibility. Moreover, meta-analyses (Hess et al., 2008; Knapp et al., 2014) have shown that dietary addition of up to 6% fats does not affect feed digestibility. Although an increase in digestibility was noted, the decrease in total VFA concentration and increase in pH suggest inhibition of rumen microbial fermentation in vitro, consistent with our previous study (Bharanidharan et al., 2021a). However, an inverse effect was observed in sacco—a decrease in ruminal pH and an increase in total VFAs. This variation in fermentation characteristics between culture systems further suggests the importance of using LCCSs, as an increase in the partial pressure of headspace gas inhibits fermentation in in vitro batch culture systems (Yang, 2017).
Our results highlight the advantages of LCCSs for in sacco investigations over in vitro batch culture systems for studying fermentation characteristics. We determined the optimal dose of PA for decreasing CH4 production in vitro and the doses that modulate fermentation characteristics and digestibility. At doses < 9% DM, PA may be a potential additive for mitigating livestock CH4 emissions as it also increased the DMD and NDFD in vitro. PA contains bioactive compounds such as PUFAs and flavonoids that may reduce the rumen ciliate protozoan population, potentially decreasing intra-ruminal protein recycling and increasing biohydrogenation. However, re-adaptation of rumen microbes after the withdrawal of supplementation hampers commercial application of this strategy. Thus, the treatment duration and effects on individual protozoan genera should be addressed in future studies. Considering its high nutritive value, PA can be used in total mixed rations or as a substitute for grains in concentrate feed, making it a new source of functional feed for ruminants. However, in vivo studies are needed to evaluate other important nutritional traits, such as palatability, growth performance, nutrient digestibility, energy partitioning, and N utilization.
The datasets generated for this study can be found in online repositories. The name of the repository and accession number can be found below: NCBI, PRJNA789417 (https://www.ncbi.nlm.nih.gov/sra/PRJNA789417).
The animal study was reviewed and approved by Institutional Animal Care and Use of Seoul National University (SNU-210615-1).
RB and KK designed and conceptualized the experiment. RB performed the management of steers, in vitro and in sacco trial, and sample collection. RB, KT, RI, and TK performed laboratory analyses. KT performed the in silico docking analysis. MB and YL supervised the experiment. RB organized the data, performed the microbial data processing, bioinformatics, statistical analyses and visualization. RB wrote the first draft of the manuscript including tables and figures, which was revised and edited by RB and KK. All authors read and approved the final manuscript.
The present study was supported by the National Institute of Animal Science, Ministry of Rural Development Administration, South Korea (research project no. PJ0149402022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.892605/full#supplementary-material
Abbott, D. W., Aasen, I. M., Beauchemin, K. A., Grondahl, F., Gruninger, R., Hayes, M., et al. (2020). Seaweed and seaweed bioactives for mitigation of enteric methane: challenges and opportunities. Animals 10, 1–28. doi: 10.3390/ani10122432
Adeyemi, K. D., Ahmed, M. A., Jotham, S., Roslan, N. A., Jahromi, M. F., Samsudin, A. A., et al. (2016). Rumen microbial community and nitrogen metabolism in goats fed blend of palm oil and canola oil. Ital. J. Anim. Sci. 15, 666–672. doi: 10.1080/1828051X.2016.1222245
Akkaya, M. R. (2018). Prediction of fatty acid composition of sunflower seeds by near-infrared reflectance spectroscopy. J. Food Sci. Technol. 55, 2318–2325. doi: 10.1007/s13197-018-3150-x
Amanzougarene, Z., and Fondevila, M. (2020). Fitting of the in vitro gas production technique to the study of high concentrate diets. Animals 10, 1–13. doi: 10.3390/ani10101935
Appuhamy, J. A. D. R. N., France, J., and Kebreab, E. (2016). Models for predicting enteric methane emissions from dairy cows in North America, Europe, and Australia and New Zealand. Glob. Chang. Biol. 22, 3039–3056. doi: 10.1111/gcb.13339
Appuhamy, J. A. D. R. N., Strathe, A. B., Jayasundara, S., Wagner-Riddle, C., Dijkstra, J., and France, J. (2013). Anti-methanogenic effects of monensin in dairy and beef cattle: a meta-analysis. J. Dairy Sci. 96, 5161–5173. doi: 10.3168/jds.2012-5923
Arokiyaraj, S., Choi, S. H., Lee, Y., Bharanidharan, R., Hairul-Islam, V. I., Vijayakumar, B., et al. (2015). Characterization of ambrette seed oil and its mode of action in bacteria. Molecules 20, 384–395. doi: 10.3390/molecules20010384
Ayemele, A. G., Ma, L., Li, X., Yang, P., Xu, J., Yu, Z., et al. (2021). Identification of bioactive phytochemicals from six plants: mechanistic insights into the inhibition of rumen protozoa, ammoniagenesis, and α-glucosidase. Biology 10:1055. doi: 10.3390/biology10101055
Ayemele, A. G., Ma, L., Park, T., Xu, J., Yu, Z., and Bu, D. (2020). Giant milkweed (Calotropis gigantea): a new plant resource to inhibit protozoa and decrease ammoniagenesis of rumen microbiota in vitro without impairing fermentation. Sci. Total Environ. 743:140665. doi: 10.1016/j.scitotenv.2020.140665
Baker, D. A., Matralis, A. N., Osborne, S. A., Large, J. M., and Penzo, M. (2020). Targeting the Malaria Parasite cGMP-Dependent Protein Kinase to Develop New Drugs. Front. Microbiol. 11:602803. doi: 10.3389/fmicb.2020.602803
Beauchemin, K. A., Ungerfeld, E. M., Eckard, R. J., and Wang, M. (2020). Review: fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal 14, S2–S16. doi: 10.1017/S1751731119003100
Belanche, A., de la Fuente, G., and Newbold, C. J. (2014). Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol. Ecol. 90, 663–677. doi: 10.1111/1574-6941.12423
Bharanidharan, R., Arokiyaraj, S., Baik, M., Ibidhi, R., Lee, S. J., Lee, Y., et al. (2021a). In vitro screening of east asian plant extracts for potential use in reducing ruminal methane production. Animals 11:1020. doi: 10.3390/ani11041020
Bharanidharan, R., Thirugnanasambantham, K., Ibidhi, R., Bang, G., Jang, S. S., Baek, Y. C., et al. (2021b). Effects of Dietary Protein Concentration on Lipid Metabolism Gene Expression and Fatty Acid Composition in 18-23-Month-Old Hanwoo Steers. Animals 11:3378. doi: 10.3390/ANI11123378
Bharanidharan, R., Lee, C. H., Thirugnanasambantham, K., Ibidhi, R., Woo, Y. W., Lee, H. G., et al. (2021c). Feeding Systems and Host Breeds Influence Ruminal Fermentation. Front. Microbiol. 12:701081. doi: 10.3389/fmicb.2021.701081
Bharanidharan, R., Arokiyaraj, S., Kim, E. B., Lee, C. H., Woo, Y. W., Na, Y., et al. (2018). Ruminal methane emissions, metabolic, and microbial profile of Holstein steers fed forage and concentrate, separately or as a total mixed ration. PLoS One 13:e0202446. doi: 10.1371/journal.pone.0202446
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/PNAS.1000080107
Chaney, A. L., and Marbach, E. P. (1962). Modified reagents for determination of urea and ammonia. Clin. Chem. 8, 130–132. doi: 10.1093/clinchem/8.2.130
Dai, X., and Faciola, A. P. (2019). Evaluating Strategies to Reduce Ruminal Protozoa and Their Impacts on Nutrient Utilization and Animal Performance in Ruminants – A Meta-Analysis. Front. Microbiol. 10:2648. doi: 10.3389/fmicb.2019.02648
Dohme, F., Machmüller, A., Wasserfallen, A., and Kreuzer, M. (2008). Ruminal methanogenesis as influenced by individual fatty acids supplemented to complete ruminant diets. Lett. Appl. Microbiol. 32, 47–51. doi: 10.1111/j.1472-765x.2001.00863.x
Doreau, M., Aurousseau, E., and Martin, C. (2009). Effects of linseed lipids fed as rolled seeds, extruded seeds or oil on organic matter and crude protein digestion in cows. Anim. Feed Sci. Technol. 150, 187–196. doi: 10.1016/j.anifeedsci.2008.09.004
Fedorah, P. M., and Hrudey, S. E. (1983). A simple apparatus for measuring gas production by methanogenic cultures in serum bottles. Environ. Technol. Lett. 4, 425–432. doi: 10.1080/09593338309384228
Glasson, C. R. K., Kinley, R. D., de Nys, R., King, N., Adams, S. L., Packer, M. A., et al. (2022). Benefits and risks of including the bromoform containing seaweed Asparagopsis in feed for the reduction of methane production from ruminants. Algal Res. 64:102673. doi: 10.1016/j.algal.2022.102673
Griswold, K. E., White, B. A., and Mackie, R. I. (1999). Diversity of extracellular proteolytic activities among Prevotella species from the rumen. Curr. Microbiol. 39, 187–194. doi: 10.1007/s002849900443
Hall, T. A. (1999). BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hassan, F. U., Arshad, M. A., Ebeid, H. M., ur Rehman, M. S., Khan, M. S., Shahid, S., et al. (2020). Phytogenic Additives Can Modulate Rumen Microbiome to Mediate Fermentation Kinetics and Methanogenesis Through Exploiting Diet–Microbe Interaction. Front. Vet. Sci. 7:575801. doi: 10.3389/fvets.2020.575801
Haug, A., Høstmark, A. T., and Harstad, O. M. (2007). Bovine milk in human nutrition - A review. Lipids Health Dis. 6, 1–16. doi: 10.1186/1476-511X-6-25
Hess, B. W., Moss, G. E., and Rule, D. C. (2008). A decade of developments in the area of fat supplementation research with beef cattle and sheep. J. Anim. Sci. 86, E188–E204. doi: 10.2527/jas.2007-0546
Hook, S. E., Wright, A. D. G., and McBride, B. W. (2010). Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010:945785. doi: 10.1155/2010/945785
Husson, F., Josse, J., Le, S., and Maintainer, J. M. (2020). Multivariate exploratory data analysis and data mining. Cran 1, 1–130.
Huws, S. A., Kim, E. J., Cameron, S. J. S., Girdwood, S. E., Davies, L., Tweed, J., et al. (2015). Characterization of the rumen lipidome and microbiome of steers fed a diet supplemented with flax and echium oil. Microb. Biotechnol. 8, 331–341. doi: 10.1111/1751-7915.12164
Jin, Q., You, W., Tan, X., Liu, G., Zhang, X., Liu, X., et al. (2021). Caffeic acid modulates methane production and rumen fermentation in an opposite way with high-forage or high-concentrate substrate in vitro. J. Sci. Food Agric. 101, 3013–3020. doi: 10.1002/jsfa.10935
Jung, D. Y., Ha, H., Lee, H. Y., Kim, C., Lee, J. H., Bae, K. H., et al. (2008). Triterpenoid saponins from the seeds of Pharbitis nil. Chem. Pharm. Bull. 56, 203–206. doi: 10.1248/cpb.56.203
Kapcsándi, V., Hanczné Lakatos, E., Sik, B., and Linka, L. Á, and Székelyhidi, R. (2021). Characterization of fatty acid, antioxidant, and polyphenol content of grape seed oil from different Vitis vinifera L. varieties. OCL Oilseeds Fats Crop Lipids 28:30. doi: 10.1051/ocl/2021017
Ki, H. K., Sang, U. C., and Kang, R. L. (2009). Diterpene glycosides from the seeds of Pharbitis nil. J. Nat. Prod. 72, 1121–1127. doi: 10.1021/np900101t
Kim, K. H., Arokiyaraj, S., Lee, J., Oh, Y. K., Chung, H. Y., Jin, G. D., et al. (2016). Effect of rhubarb (Rheum spp.) root on in vitro and in vivo ruminal methane production and a bacterial community analysis based on 16S rRNA sequence. Anim. Prod. Sci. 56, 402–408. doi: 10.1071/AN15585
Kim, K. H., Ha, S. K., Choi, S. U., Kim, S. Y., and Lee, K. R. (2011). Bioactive phenolic constituents from the seeds of Pharbitis nil. Chem. Pharm. Bull. 59, 1425–1429. doi: 10.1248/cpb.59.1425
Kim, Y. S., Kim, J. W., Ha, N. Y., Kim, J., and Ryu, H. S. (2020). Herbal Therapies in Functional Gastrointestinal Disorders: a Narrative Review and Clinical Implication. Front. Psychiatry 11:601. doi: 10.3389/fpsyt.2020.00601
Kišidayová, S., Durkaj, D., Mihaliková, K., Váradyová, Z., Puchalska, J., Szumacher-Strabel, M., et al. (2021). Rumen Ciliated Protozoa of the Free-Living European Bison (Bison bonasus, Linnaeus). Front. Microbiol. 12:658448. doi: 10.3389/fmicb.2021.658448
Knapp, J. R., Laur, G. L., Vadas, P. A., Weiss, W. P., and Tricarico, J. M. (2014). Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J. Dairy Sci. 97, 3231–3261. doi: 10.3168/JDS.2013-7234
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Krishnamoorthy, U., Rymer, C., and Robinson, P. H. (2005). The in vitro gas production technique: limitations and opportunities. Anim. Feed Sci. Technol. 123, 1–7. doi: 10.1016/j.anifeedsci.2005.04.015
Krizsan, S. J., Janèík, F., Ramin, M., and Huhtanen, P. (2013). Comparison of some aspects of the in situ and in vitro methods in evaluation of neutral detergent fiber digestion. J. Anim. Sci. 91, 838–847. doi: 10.2527/jas.2012-5343
Ku-Vera, J. C., Jiménez-Ocampo, R., Valencia-Salazar, S. S., Montoya-Flores, M. D., Molina-Botero, I. C., Arango, J., et al. (2020). Role of Secondary Plant Metabolites on Enteric Methane Mitigation in Ruminants. Front. Vet. Sci. 7:584. doi: 10.3389/fvets.2020.00584
Lee, H. J., Cho, S. H., Shin, D., and Kang, H. S. (2018). Prevalence of antibiotic residues and antibiotic resistance in isolates of chicken meat in Korea. Korean J. Food Sci. Anim. Resour. 38, 1055–1063. doi: 10.5851/kosfa.2018.e39
Lee, S. R., Moon, E., and Kim, K. H. (2017). Neolignan and monoterpene glycoside from the seeds of Pharbitis nil. Phytochem. Lett. 20, 98–101. doi: 10.1016/j.phytol.2017.04.019
Levy, B., and Jami, E. (2018). Exploring the Prokaryotic Community Associated With the Rumen Ciliate Protozoa Population. Front. Microbiol. 9:2526. doi: 10.3389/fmicb.2018.02526
Lim, J. H. (2020). Relationship between Glossiness and Cooking Liquid Components of Temperate japonica Rice. Available Online at: https://s-space.snu.ac.kr/handle/10371/166743 (accessed on 25 Feb, 2022).
Machmüller, A. (2006). Medium-chain fatty acids and their potential to reduce methanogenesis in domestic ruminants. Agric. Ecosyst. Environ. 112, 107–114. doi: 10.1016/j.agee.2005.08.010
Maron, D. F., Smith, T. J. S., and Nachman, K. E. (2013). Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Glob. Health 9:48. doi: 10.1186/1744-8603-9-48
Martin, C., Ferlay, A., Mosoni, P., Rochette, Y., Chilliard, Y., and Doreau, M. (2016). Increasing linseed supply in dairy cow diets based on hay or corn silage: effect on enteric methane emission, rumen microbial fermentation, and digestion. J. Dairy Sci. 99, 3445–3456. doi: 10.3168/jds.2015-10110
Matsumoto, M., Kobayashi, T., Takenaka, A., and Itabashi, H. (1991). Defaunation effects of medium-chain fatty acids and their derivatives on goat rumen protozoa. J. Gen. Appl. Microbiol. 37, 439–445. doi: 10.2323/jgam.37.439
McDougall, E. I. (1948). The composition and output of sheep’s saliva. Biochem. J. 43, 99–109. doi: 10.1042/bj0430099
McGaw, L. J., Jäger, A. K., and Van Staden, J. (2002). Antibacterial effects of fatty acids and related compounds from plants. S. Afr. J. Bot. 68, 417–423. doi: 10.1016/S0254-6299(15)30367-7
McKain, N., Shingfield, K. J., and Wallace, R. J. (2010). Metabolism of conjugated linoleic acids and 18: 1 fatty acids by ruminal bacteria: products and mechanisms. Microbiology 156, 579–588. doi: 10.1099/mic.0.036442-0
Mosoni, P., Martin, C., Forano, E., and Morgavi, D. P. (2011). Long-term defaunation increases the abundance of cellulolytic ruminococci and methanogens but does not affect the bacterial and methanogen diversity in the rumen of sheep. J. Anim. Sci. 89, 783–791. doi: 10.2527/jas.2010-2947
Newbold, C. J., De la Fuente, G., Belanche, A., Ramos-Morales, E., and McEwan, N. R. (2015). The role of ciliate protozoa in the rumen. Front. Microbiol. 6:1313. doi: 10.3389/fmicb.2015.01313
Nur Atikah, I., Alimon, A. R., Yaakub, H., Abdullah, N., Jahromi, M. F., Ivan, M., et al. (2018). Profiling of rumen fermentation, microbial population and digestibility in goats fed with dietary oils containing different fatty acids. BMC Vet. Res. 14:334. doi: 10.1186/S12917-018-1672-0/TABLES/5
O’Fallon, J. V., Busboom, J. R., Nelson, M. L., and Gaskins, C. T. (2007). A direct method for fatty acid methyl ester synthesis: application to wet meat tissues, oils, and feedstuffs. J. Anim. Sci. 85, 1511–1521. doi: 10.2527/jas.2006-491
Olagaray, K. E., and Bradford, B. J. (2019). Plant flavonoids to improve productivity of ruminants – A review. Anim. Feed Sci. Technol. 251, 21–36. doi: 10.1016/j.anifeedsci.2019.02.004
Ono, M. (2017). Resin glycosides from Convolvulaceae plants. J. Nat. Med. 71, 591–604. doi: 10.1007/s11418-017-1114-5
Park, T., and Yu, Z. (2018). Do Ruminal Ciliates Select Their Preys and Prokaryotic Symbionts? Front. Microbiol. 9:1710. doi: 10.3389/fmicb.2018.01710
Park, T., Yang, C., and Yu, Z. (2019). Specific inhibitors of lysozyme and peptidases inhibit the growth of the rumen protozoan Entodinium caudatum without decreasing feed digestion or fermentation in vitro. J. Appl. Microbiol. 127, 670–682. doi: 10.1111/jam.14341
Patra, A. K. (2013). The effect of dietary fats on methane emissions, and its other effects on digestibility, rumen fermentation and lactation performance in cattle: a meta-analysis. Livest. Sci. 155, 244–254. doi: 10.1016/j.livsci.2013.05.023
Patra, A. K., and Yu, Z. (2013). Effects of coconut and fish oils on ruminal methanogenesis, Fermentation, And abundance and diversity of microbial populations in vitro. J. Dairy Sci. 96, 1782–1792. doi: 10.3168/jds.2012-6159
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Saito, N., Cheng, J., Ichimura, M., Yokoi, M., Abe, Y., and Honda, T. (1994). Flavonoids in the acyanic flowers of Pharbitis nil. Phytochemistry 35, 687–691. doi: 10.1016/S0031-9422(00)90588-0
Solomon, R., Wein, T., Levy, B., Eshed, S., Dror, R., Reiss, V., et al. (2021). Protozoa populations are ecosystem engineers that shape prokaryotic community structure and function of the rumen microbial ecosystem. ISME J. 16, 1187–1197. doi: 10.1038/s41396-021-01170-y
Takenaka, A., Tajima, K., Mitsumori, M., and Kajikawa, H. (2004). Fiber Digestion by Rumen Ciliate Protozoa. Microbes Environ. 19, 203–210. doi: 10.1264/jsme2.19.203
Toprak, N. N. (2015). Do fats reduce methane emission by ruminants? - A review. Anim. Sci. Pap. Rep. 33, 305–321.
Undersander, D., Mertens, D. R., and Thiex, N. (1993). Forage analysis procedures. Omaha, NE: National Forage Testing Association.
Van Soest, P. J. (1973). Collaborative Study of Acid-Detergent Fiber and Lignin. J. AOAC Int. 56, 781–784. doi: 10.1093/jaoac/56.4.781
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Vasta, V., Daghio, M., Cappucci, A., Buccioni, A., Serra, A., Viti, C., et al. (2019). Invited review: plant polyphenols and rumen microbiota responsible for fatty acid biohydrogenation, fiber digestion, and methane emission: experimental evidence and methodological approaches. J. Dairy Sci. 102, 3781–3804. doi: 10.3168/jds.2018-14985
Wang, R., Wang, M., Zhang, X. M., Wen, J. N., Ma, Z. Y., Long, D. L., et al. (2019). Effects of rumen cannulation on dissolved gases and methanogen community in dairy cows. J. Dairy Sci. 102, 2275–2282. doi: 10.3168/jds.2018-15187
Woo, K. W., Park, K. J., Choi, S. Z., Son, M. W., Kim, K. H., and Lee, K. R. (2017). A New ent-kaurane Diterpene Glycoside from Seeds of Pharbitis nil. Chem. Nat. Compd. 53, 468–471. doi: 10.1007/s10600-017-2024-1
Keywords: methane, rumen, in silico, culture systems, PUFA, quercetin, Entodinium caudatum
Citation: Bharanidharan R, Thirugnanasambantham K, Ibidhi R, Baik M, Kim TH, Lee Y and Kim KH (2022) Metabolite Profile, Ruminal Methane Reduction, and Microbiome Modulating Potential of Seeds of Pharbitis nil. Front. Microbiol. 13:892605. doi: 10.3389/fmicb.2022.892605
Received: 09 March 2022; Accepted: 04 April 2022;
Published: 09 May 2022.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Gabriel De La Fuente Oliver, Universitat de Lleida, SpainCopyright © 2022 Bharanidharan, Thirugnanasambantham, Ibidhi, Baik, Kim, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyoung Hoon Kim, a2hoa2ltQHNudS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.