
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 April 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.890807
This article is part of the Research TopicViruses in Agricultural Systems: Interactions with Plants, Insect Pollinators and FungiView all 10 articles
 Aarthi Nekkanti1,2
Aarthi Nekkanti1,2 Prosenjit Chakraborty1
Prosenjit Chakraborty1 Amalendu Ghosh1*
Amalendu Ghosh1* Mir Asif Iquebal3
Mir Asif Iquebal3 Sarika Jaiswal3
Sarika Jaiswal3 Virendra Kumar Baranwal1
Virendra Kumar Baranwal1
Bemisia tabaci (Hemiptera: Aleyrodidae) is a highly efficient vector in the spread of chilli leaf curl virus (ChiLCV, Begomovirus) which is a major constraint in the production of chilli in South Asia. Transcriptome analysis of B. tabaci post-6 h acquisition of ChiLCV showed differential expression of 80 (29 upregulated and 51 downregulated) genes. The maximum number of DEGs are categorized under the biological processes category followed by cellular components and molecular functions. KEGG analysis of DEGs showed that the genes are involved in the functions like metabolism, signaling pathways, cellular processes, and organismal systems. The expression of highly expressed 20 genes post-ChiLCV acquisition was validated in RT-qPCR. DEGs such as cytosolic carboxypeptidase 3, dual-specificity protein phosphatase 10, 15, dynein axonemal heavy chain 17, fasciclin 2, inhibin beta chain, replication factor A protein 1, and Tob1 were found enriched and favored the virus infection and circulation in B. tabaci. The present study provides an improved understanding of the networks of molecular interactions between B. tabaci and ChiLCV. The candidate genes of B. tabaci involved in ChiLCV transmission would be novel targets for the management of the B. tabaci-begomovirus complex.
Transmission of a plant virus within or between the fields is often dependent upon a mobile vector. Insects are the most efficient vectors of plant viruses because of their abundance and feeding behavior (Milenovic et al., 2019). About 70% of reported insect vectors are hemipterans (Fereres and Raccah, 2015). Silverleaf whitefly, Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae) is an invasive insect pest and vector of several plant viruses (Legarrea et al., 2015). Transmission of several begomoviruses, carlaviruses, criniviruses, cytorhabdoviruses, ipomoviruses, poleroviruses, and torradoviruses by B. tabaci causes economic losses exceeding billions of US$ and threatens food security (Dasgupta et al., 2003; Jones, 2003; Navas-Castillo et al., 2011; Brown et al., 2015; Orfanidou et al., 2016; Saeed and Samad, 2017; Zanardo and Carvalho, 2017; Ghosh et al., 2019; Costa et al., 2020; Pinheiro-Lima et al., 2020; Cornejo-Franco et al., 2022).
The spectrum of diseases caused by begomoviruses is a continuing challenge to crop production worldwide (family: Geminiviridae) (Navas-Castillo et al., 2011). These diseases cause an estimated yield loss of 50–90% in tomatoes, chilli, and other crops including beans, cassava, cotton, cucurbits, eggplant, papaya, and potatoes (Briddon et al., 2010; Saeed and Samad, 2017). To date, 445 begomovirus species have been reported1. One of the begomoviruses, i.e., chilli leaf curl virus (ChiLCV) is a major threat to chilli production in tropical and sub-tropical countries (Senanayake et al., 2007, 2012; Menike and De Costa, 2017; Oraonand and Tarafdar, 2018; Thakur et al., 2018). ChiLCV has caused several epidemics in India and Sri Lanka (Senanayake et al., 2007, 2012). The disease is typically manifested in the infected plants as upward curling, puckering, and bunching of leaves. The leaves become smaller and severely affected plants produce fewer and deformed fruits. Yield loss of 20–50% has been recorded in chilli by ChiLCV (Thakur et al., 2018) which may rise to 100% with the infestation of thrips and mites (Menike and De Costa, 2017). Control options for B. tabaci-ChiLCV are very limited as insecticides continue to lose their efficacy due to the emergence of resistant B. tabaci populations (Barman et al., 2021). Additionally, insecticides adversely affect the environment and human health. Understanding the molecular interactions between B. tabaci and ChiLCV and interrupting the interrelationship is a promising approach to manage the virus-vector complex. The present understanding of B. tabaci-begomovirus interaction is largely based on the study of B. tabaci cryptic species MEAM1 and MED and tomato yellow leaf curl virus (TYLCV). Once ingested by B. tabaci, virus particles pass through the midgut, where they move across the midgut membrane into the hemolymph, possibly via receptor-mediated endocytosis, and circulate to the primary salivary glands, from where they are egested along with saliva during feeding (Czosnek and Ghanim, 2012). During the entire process, the viral proteins need to interact with several proteins in the midgut, hemolymph, and primary salivary glands (Wei et al., 2014). B. tabaci MEAM1 heat shock proteins (Hsp), cyclophilins, and peptidoglycan recognition protein interact with TYLCV coat protein (CP) for successful internalization (Götz et al., 2012; Kanakala and Ghanim, 2016; Wang et al., 2016). Silencing of hsp70 in B. tabaci Asia II 1 inhibits transmission of ChiLCV (Chakraborty and Ghosh, 2022). A GroEL homolog produced by C-type endosymbionts in B. tabaci MEAM1 is known to transport TYLCV particles through the hemolymph of B. tabaci in coated vesicles (Bragard et al., 2013).
Over the past 5 years, transcriptomic analysis of B. tabaci has enabled us to study the differential expression of genes that are involved in virus transmission (Luan et al., 2011; Kaur et al., 2017; Xia et al., 2017; Hasegawa et al., 2018; Ding et al., 2019). However, these studies were limited to gene regulations in MEAM1 and MED cryptic species of B. tabaci in response to infection of TYLCV, tomato yellow leaf curl China virus (TYLCCV), and a crinivirus-tomato chlorosis virus (ToCV). To date, 46 morphologically indistinguishable cryptic species of B. tabaci are known (Rehman et al., 2021) that differ in genetic structure, chemical resistance, adaptability, and virus transmission (Brown, 2000; Gorman et al., 2010; Wang et al., 2010; Qin et al., 2016). The genes of B. tabaci involved in the transmission of begomoviruses are not conserved across all the B. tabaci cryptic species–begomovirus combinations. Also, little is known about the role of putative genes of B. tabaci Asia II 1 in response to ChiCLV transmission. The present study reports the candidate genes of B. tabaci Asia II 1 that are regulated at an early stage of ChiLCV infection. Identification of candidate genes involved in key physiological processes and ChiLCV infection would be novel targets for the management of the B. tabaci- ChiLCV complex.
A homogeneous population of B. tabaci Asia II 1 maintained at Advanced Centre for Plant Virology, Indian Agricultural Research Institute (IARI), New Delhi since 2015 was used in the present study. The homogeneous population was developed from a single adult female on healthy eggplants. The cryptic identity of the B. tabaci was confirmed by PCR amplification of the mitochondrial cytochrome oxidase subunit I (mtCOI) using C1-J-2195 and L2-N-3014 primers (Simon et al., 1994; Supplementary Table 1) and sequencing. DNA was isolated from randomly collected adult flies from the homogeneous population using CTAB extraction buffer as described by Rehman et al. (2021). PCR was performed in a 25 μl reaction mixture comprised of 1x DreamTaq buffer, 0.1 mM dNTPs (Thermo Fisher Scientific, Waltham, MA, United States), 10 picomoles forward and reverse primers, 1.25 U DreamTaq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, United States), and 50 ng of template DNA. Thermal cycling was followed as initial denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s and polymerization at 72°C for 1 min, followed by final extension step at 72°C for 10 min. PCR products were visualized on 1% agarose gel stained with GoodView™ (BR Biochem, New Delhi, India). The purified PCR products were sequenced bidirectionally. The sequences were processed by BioEdit and species homology was checked in BLASTn. A consensus sequence was submitted to GenBank. The genotype or cryptic species of the B. tabaci population was confirmed based on Bayesian Inference phylogeny considering a genetic divergence cutoff of 4% as described by Rehman et al. (2021). The population was maintained under controlled environmental conditions at 28 ± 2°C, 60 ± 10% relative humidity, and 16 h light- 8 h dark photoperiod.
A pure culture of ChiLCV maintained on chilli plant (var. Preeti, Nunhems, Haelen, Netherlands) by B. tabaci inoculation under insect-proof conditions was used in the study. ChiLCV was confirmed by amplifying DNA-A using Begomo F and Begomo R primers (Akhter et al., 2009) (Supplementary Table 1) and nucleotide sequencing. To establish the ChiLCV-exposed and non-exposed B. tabaci populations, freshly emerged female adults were collected and released onto ChiLCV-infected and virus-free chilli plants at 4–6 leaf stage for 6 h in three biological replicates. A 6 h acquisition was found adequate for the successful transmission of ChiLCV by B. tabaci Asia II 1 (Senanayake et al., 2012; Roy et al., 2021). After 6 h of acquisition, ChiLCV-exposed (BtTrI1, BtTrI2, and BtTrI3) and non-exposed (BtTrH1, BtTrH2, and BtTrH3) B. tabaci adults were collected. ChiLCV infection in B. tabaci populations was confirmed by randomly collecting 10 flies from each population and testing in PCR using ChiLCV-specific primers (AG149F-AG150R) (Roy et al., 2021, Supplementary Table 1). Each population was divided into two parts. One part was utilized for RNA-Seq and another part was preserved in −80°C for gene expression analysis in a reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Total RNA was isolated from each ChiLCV-exposed (BtTrI1, BtTrI2, and BtTrI3) and non-exposed (BtTrH1, BtTrH2, and BtTrH3) B. tabaci populations using Trizol reagent (Invitrogen, Waltham, MA, United States) following manufacturer’s protocol. In brief, 50 B. tabaci were crushed in 1 mL Trizol and kept at room temperature for 10 min. Two hundred microliter of chloroform was added to the mixture, vortexed for <10 s, and incubated at room temperature for 10 min. The mixture was then centrifuged at 16,000 xg for 10 min at 4°C. The upper aqueous phase was transferred to a fresh tube and 0.8 volume of ice-chilled isopropanol was added, mixed properly, and incubated at 4°C for 10 min. The mixture was again centrifuged at 16,000 xg for 10 min at 4°C and the supernatant was discarded. The pellet was finally washed with 70% ethanol, air-dried, and resuspended in 30 μl nuclease-free water. RNaseZAP (Thermo Fisher Scientific, Waltham, MA, United States) was used for decontamination, RNase-free tips, microfuge tubes, and water were used throughout the experiment. Total RNA quality was measured using RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, United States) on 2100 Bioanalyzer (Agilent Technologies) with a minimum RNA Integrity Number (RIN) value of 7. RNA concentrations were determined using a NanoDrop ND-8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
RNA-Seq libraries for all samples were prepared using NEBNext UltraII RNA library preparation kit for Illumina (New England Biolabs, Ipswich, MA, United States) following manufacturer’s protocol and sequencing was done in a single HiSEQ 4000 (Illumina Inc., San Diego, CA, United States) lane using 150 bp paired-end chemistry. The library preparation and sequencing were done by commercial service providers (NxGenBio Life Sciences, Delhi, India). Briefly, mRNA was purified using oligo-dT beads. Magnetic beads were used for the second round of purification. During the second elution of the poly-A RNA, the RNA was also fragmented into short stretches of 200–500 bp at 94°C for 5 min using an ultrasonicator in presence of divalent cations. The cleaved RNA fragments were copied into first-strand cDNA using SuperScript-II reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, United States) with random primers. After second-strand cDNA synthesis, fragments were end-repaired, A-tailed, and ligated to indexed adapters. The products were purified and enriched with PCR to create the final cDNA library. The tagged cDNA libraries were pooled in equal ratios and used for 2 × 150 bp paired-end sequencing on a single lane of the Illumina HiSEQ 4000. Illumina clusters were generated and were loaded onto Illumina Flow Cell and sequenced. After sequencing, the samples were demultiplexed and the indexed adapter sequences were trimmed using the CASAVA v1.8.2 software (Illumina Inc.).
The ambiguous “N” nucleotides with a ratio of “N” > 5%, reads with adaptor sequences, and low-quality reads with a quality score < 20% were removed by the Trim Galore v0.4.1 to get the high-quality reads. Reference genome index was established using BWAv0.7.5 and the clean reads were mapped to the reference genome of B. tabaci MEAM1 (Chen et al., 2016). Read numbers, mapped to every gene were counted using Samtools v0.1.19. Differential expression between the ChiLCV-exposed (BtTrI1, BtTrI2, and BtTrI3) and non-exposed (BtTrH1, BtTrH2, and BtTrH3) B. tabaci populations was analyzed using the DESeq R package2. Significant differential gene expression that was consistent among the biological replicates, was counted with ≥ log2 2-fold change and p < 0.05.
Gene annotations and functional enrichment analysis including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathways were used to identify the differentially expressed genes (DEGs) that were significantly enriched in GO terms or biological pathways post-6 h of ChiLCV acquisition. Gene annotations against the Uniprot GO database3 were performed by aligning DEGs to the NR database using the blast v 2.6.0+ programme. KEGG pathway enrichment analysis of DEGs was performed using the KEGG database resource4 by KAAS (Moriya et al., 2007) to identify the pathways that were differentially regulated between ChiLCV-exposed and non-exposed B. tabaci with p-value < 0.05.
To validate the RNA-Seq data, 20 putative genes of B. tabaci Asia II 1 with ≥log2 2-fold change values were selected to assess the gene expression in RT-qPCR. Two sets of primers were designed for each of the target and endogenous control (β-actin) genes. The primer pairs were initially optimized in a gradient PCR. One set of primers for each target gene was selected based on the efficiency of PCR amplification at the same PCR conditions for the endogenous control primers. A part of the same ChiLCV-exposed and non-exposed B. tabaci populations used for RNA-Seq was preserved for RT-qPCR analysis. Total RNA was isolated from ChiLCV-exposed (BtTrI1, BtTrI2, and BtTrI3) and non-exposed (BtTrH1, BtTrH2, and BtTrH3) B. tabaci populations as described earlier. cDNA was synthesized using the FIREScript RT cDNA synthesis kit (Solis BioDyne, Estonia). The reaction mixture contained 1X RT reaction buffer, 1.0 μg template RNA, 5.0 μM oligo dT primer, 500 μM dNTP mix, 10-unit FIREScript RT, and 1-unit RiboGrip RNase inhibitor. The reverse transcription was carried out in a thermocycler (T100, Bio-Rad, Hercules, CA, United States) at 42°C for 60 min followed by enzyme inactivation at 85°C for 5 min.
The relative RT-qPCR was carried out in an InstaQ 48 real-time PCR (Himedia, Mumbai, India) with 20 μl reaction volume containing 10 μl of 2X Maxima SYBR green master mix, 10 μM ROX passive reference dye, 10 picomoles each forward and reverse primer (Supplementary Table 1), and 2 μl template cDNA. Thermal cycling was performed as initial denaturation at 94°C for 5 min, 30 cycles of 94°C for 30 sec, 56°C for 30 sec, and 72°C for 30 sec. The dissociation or melting stage was carried out after every reaction to determine the specificity of the amplicons in RT-qPCR using a computer interface programme for InstaQ 48M2 (Himedia). The RT-qPCR was performed with three biological and two technical replicates. The expression change of the target gene was normalized by excluding the changes in cycle threshold (CT) value of endogenous control, β-actin. Log2-fold change value was calculated and relative expression of mRNA was determined by normalizing the log2 values of the ChiLCV-exposed populations with non-exposed using the 2–ΔΔ CT method (Livak and Schmittgen, 2001) in Mircosoft Excel 2016.
An isofemale line of B. tabaci Asia II 1 (Accession No. MT920041), maintained on healthy eggplant at Advanced Centre for Plant Virology, IARI was used to generate ChiLCV-exposed and non-exposed B. tabaci populations. The identity of the B. tabaci cryptic species was confirmed by sequencing the mtCOI gene. PCR amplification of B. tabaci mtCOI with C1-J-2195 and L2-N-3014 primers showed an expected amplicon of ∼860 bp on an agarose gel. The nucleotide (nt) sequence showed 99.99% homology in BLASTn analysis with other B. tabaci sequences in NCBI. The sequence can be retrieved using the GenBank Accession No. MT920041. Bayesian Inference phylogeny considering genetic divergence cutoff of 4% revealed that the population belonged to the cryptic species B. tabaci Asia II 1 (data not shown).
PCR amplification of the DNA-A using Begomo F and Begomo R primer produced a 2.7 kb product as visualized on 1% agarose gel. Bidirectional sequencing of the cloned products produced an 1896 nt sequence, comprising complete AV1 and AV2 genes, and partial AC1, AC2, AC3, and AC4 genes, that was 100% similar to ChiLCV isolates upon BLASTn analysis. The sequence can be retrieved by GenBank Accession No. MW399222.
ChiLCV-exposed and non-exposed B. tabaci populations were developed by allowing the freshly emerged adult flies to feed on infected and healthy chilli plants for 6 h. A few randomly collected B. tabaci adults were tested in PCR with ChiLCV-specific primers, AG149F and AG150R (Supplementary Table 1). A product of 290 bp was visualized on agarose gel that confirmed the virus infection in ChiLCV-exposed B. tabaci adults. B. tabaci populations (BtTrH1, BtTrH2, and BtTrH3) that were exposed to healthy chilli plants did not produce any ChiLCV-specific amplification in PCR.
Total RNA was extracted from three ChiLCV-exposed (BtTrI1, BtTrI2, and BtTrI3) and non-exposed (BtTrH1, BtTrH2, and BtTrH3) B. tabaci Asia II 1 populations. For RNA-Seq analysis of ChiLCV-exposed and non-exposed B. tabaci, a total of six cDNA libraries were constructed and used for 2 × 150 bp pair-end sequencing on a single HiSEQ4000 lane. About 26–33 million raw reads were generated from each library (Table 1). Of which, 99.58–99.70% reads were clean reads. The cleaned reads of all the six libraries were mapped with the reference genome of B. tabaci MEAM1 (Chen et al., 2016). The mapping percent for all six libraries ranged from 87.21 to 93.16% of clean reads.
A total of 15,514 genes in adult B. tabaci Asia II 1 were found to be differentially expressed post 6 h of ChiLCV exposure. Out of which, 7,193 genes were upregulated and 8,321 genes were downregulated. However, only a total of 80 (0.52%) genes showed significant regulations with ≥log2 2-fold change in expression level at a significant p-value of ≤0.05. Among the significant differentially expressed genes (DEGs), 29 genes were upregulated and 51 genes were downregulated in combined BtTrI as compared to BtTrH (Figure 1). The top upregulated and downregulated genes of B. tabaci in response to ChiLCV-infection are listed in Table 2. The DEGs of B. tabaci were involved in receptor binding, antigen binding, epithelial cell differentiation, extracellular matrix organization, cell-to-cell, and cell-surface receptor signaling. Besides, a large number of orphan genes were significantly regulated in B. tabaci post 6 h of ChiLCV acquisition.
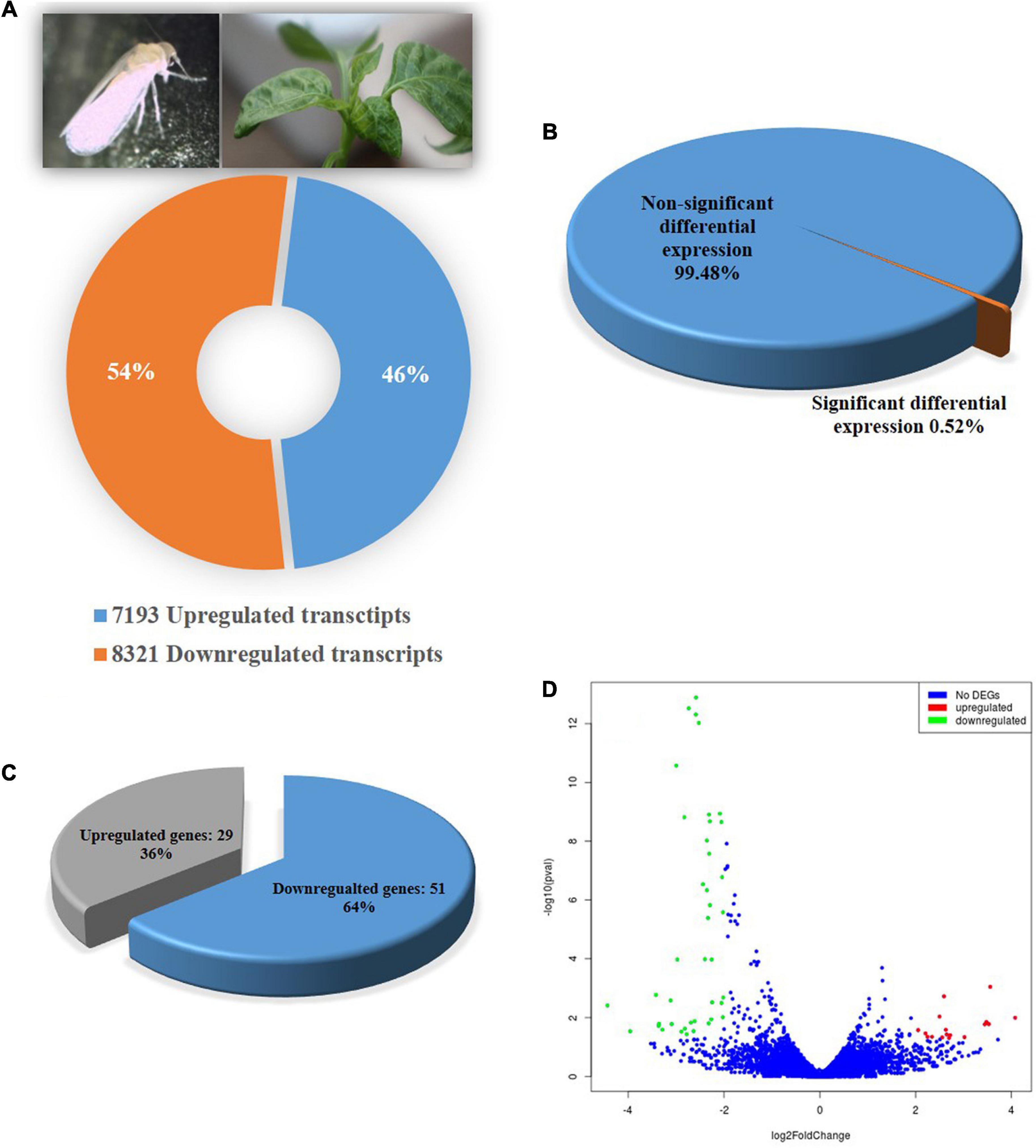
Figure 1. Differential gene expression of B. tabaci Asia II1 in response to ChiLCV infection. (A) Percentage of upregulated and down-regulated transcripts; (B) Percentage differentially expressed genes of B. tabaci at the expression level of ≥log2 2-fold and p-value ≤ 0.05; (C) Proportion of differentially expressed genes (DEGs) in RNA-Seq analysis. A total of 29 DEGs were found upregulated while 51 were downregulated; (D) Volcano plot of DEGs. The x-axis shows the fold change in gene expression between different samples, and the y-axis shows the statistical significance of the differences. Significantly up- and downregulated genes with log2 FC ≥ 2 are highlighted in red and green, respectively. The blue dots represent non-significant DEGs.

Table 2. Differentially expressed upregulated and downregulated genes of B. tabaci Asia II 1 in response to ChiLCV infection.
A total of 16 known genes were found to be upregulated in adult B. tabaci at an early stage of ChiLCV infection. The key upregulated genes included dual-specificity protein phosphatase 10 (DUSP10)-like, dual-specificity protein phosphatase 15 (DUSP15)-like, estrogen sulfotransferase (Ste), cytosolic carboxypeptidase 3 (AGBL3)-like, fasciclin 2 (Fas2), Tob1, GMP reductase 1 (GMPR), dentin sialophosphoprotein (DSPP), toll receptor 3 (TLR3), dynein axonemal heavy chain 17 (DNAH17), ATP-dependent DNA helicase (DDX11), WAS/WASL-interacting protein family member 1 (WIPF1), proline-rich extensin protein EPR1 (EPR1), CG13607, and glutamyl-tRNA(Gln) amidotransferase subunit A (QRSL1).
Twenty-three genes were significantly downregulated in B. tabaci adults post 6 h of ChiLCV acquisition. Putative genes of B. tabaci such as nose resistant to fluoxetine protein 6 (nrf-6), protein masquerade (mas), protein argonaute 2 (AGO2), endochitinase A (CHIA), pupal cuticle protein 36 (PCP36), keratinocyte proline-rich protein (KPRP), adenylate cyclase, juvenile hormone acid O-methyltransferase (jhamt), T-box transcription factor TBX20 (TBX20), 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC), AGAP004475-PA, protein phosphatase 1L (PPM1L), neurobeachin-like protein 1 (NBEAL1), major royal jelly protein (MRJP), prolyl 4-hydroxylase alpha-1 subunit (P4HA1), AT-rich interactive domain-containing protein (ARID), Klingon (klg), inhibin beta chain (Actbeta), adenosine deaminase (ADA), chloroquine resistance marker protein, leptin receptor (LEPR), and homeobox protein Hox-A2 (HOXA2) could be seen significantly downregulated in response to ChiLCV infection.
Among the 80 differentially expressed transcripts, 50 (16 upregulated and 34 downregulated) transcripts could not be annotated based on nucleotide similarity against B. tabaci genome database5. When searched for nucleotide similarity at NCBI RefSeq non-redundant protein database, 43 (16 upregulated and 27 downregulated) transcripts did not show any similarity to known proteins. Out of the 16 upregulated genes, 9 genes showed similarity with unknown protein-coding genes. Whereas, 8 genes out of 27 downregulated genes showed similarity with unknown protein-coding genes. In common, 13 upregulated and 22 downregulated transcripts in adult B. tabaci could not be assigned any annotation. These genes were depicted as orphan genes that were differentially expressed in response to ChiLCV infection.
Based on the GO study, the DEGs were categorized into cellular components, biological processes, and molecular functions (Figure 2). A total of 38 genes were categorized under biological processes (47.5%) followed by cellular components (24 genes, 30%) and molecular functions (18 genes, 22.5%). In the biological processes, genes involved in the metabolic process (24%), cellular process (24%), biological regulation (16%), regulation of biological process (16%), signaling (8%), response to stimulus (8%), growth (2%), and cell proliferation (2%) were highly enriched. Similarly, genes associated with catalytic activity (28%), protein-containing complex (12%), organelle (12%), cell part (12%), cell (12%), membrane (6%), membrane part (6%), extracellular regions (6%), organelle part (3%), and extracellular region part (3%) were differentially enriched under cellular components category. In the molecular functions category, binding (67%), transcription regulator activity (22%), and molecular function regulator (11%) were enriched significantly.
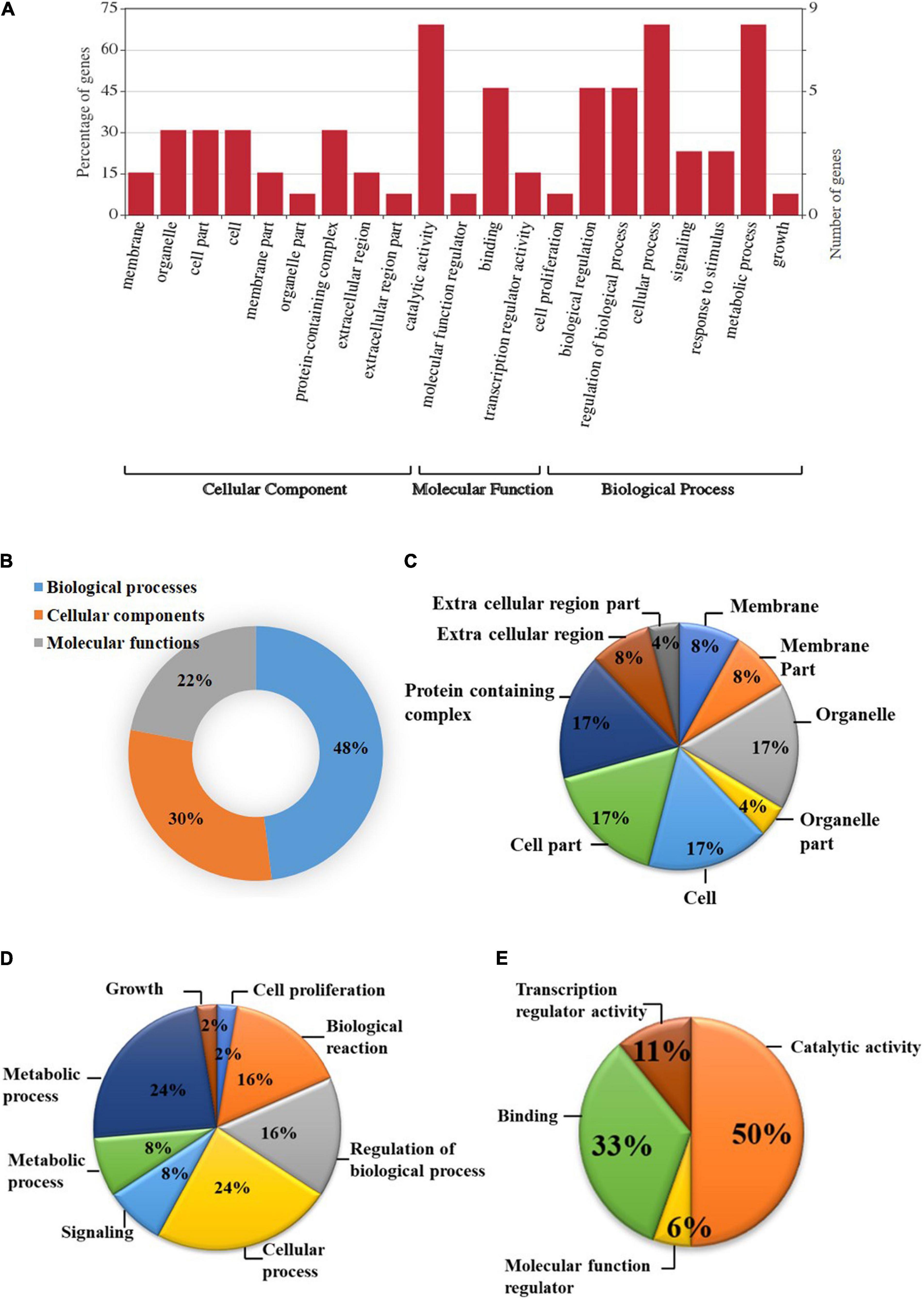
Figure 2. Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs). (A) DEGs were characterized under the cellular components, molecular functions, and biological processes based on GO analysis; (B) Pie chart describing the Gene Ontology (GO) analysis of differentially expressed genes (DEGs) and their distribution under different GO terms. DEGs were categorized majorly under biological processes (48%) followed by cellular components (30%), and molecular functions (22%); (C) A major category of DEGs fall under cellular processes in which most of the DEGs belongs to cell, cell part, organelle, and protein-containing complex (17%) followed by membrane, membrane part, and extracellular region (8%); (D) In biological processes, cellular and metabolic processes (24%), regulation of biological process, and biological reaction (16%), etc. were enriched; (E) DEGS categorized under molecular functions were mainly distributed under catalytic activity (50%), binding (33%), transcription regulator activity (11%), and molecular function regulator (6%).
KEGG pathway analysis of differentially expressed genes showed that a total of 17 DEGs in B. tabaci were involved in different functions like metabolism, signaling pathways, and cellular processes. The metabolic pathways that were majorly affected by the DEGs were metabolic pathways, TGF-beta signaling pathway, signaling pathways regulating pluripotency of stem cells, RNA degradation, regulation of actin cytoskeleton, purine metabolism, phospholipase D signaling pathway, glycerophospholipid metabolism, glycerolipid metabolism, fat digestion and absorption, endocytosis, cytokine-cytokine receptor interaction, cell adhesion molecules, arginine and proline metabolism, and adherens junction (Figure 3). The pathways identified in GO enrichment analysis were consistent with the findings of the KEGG pathway study.
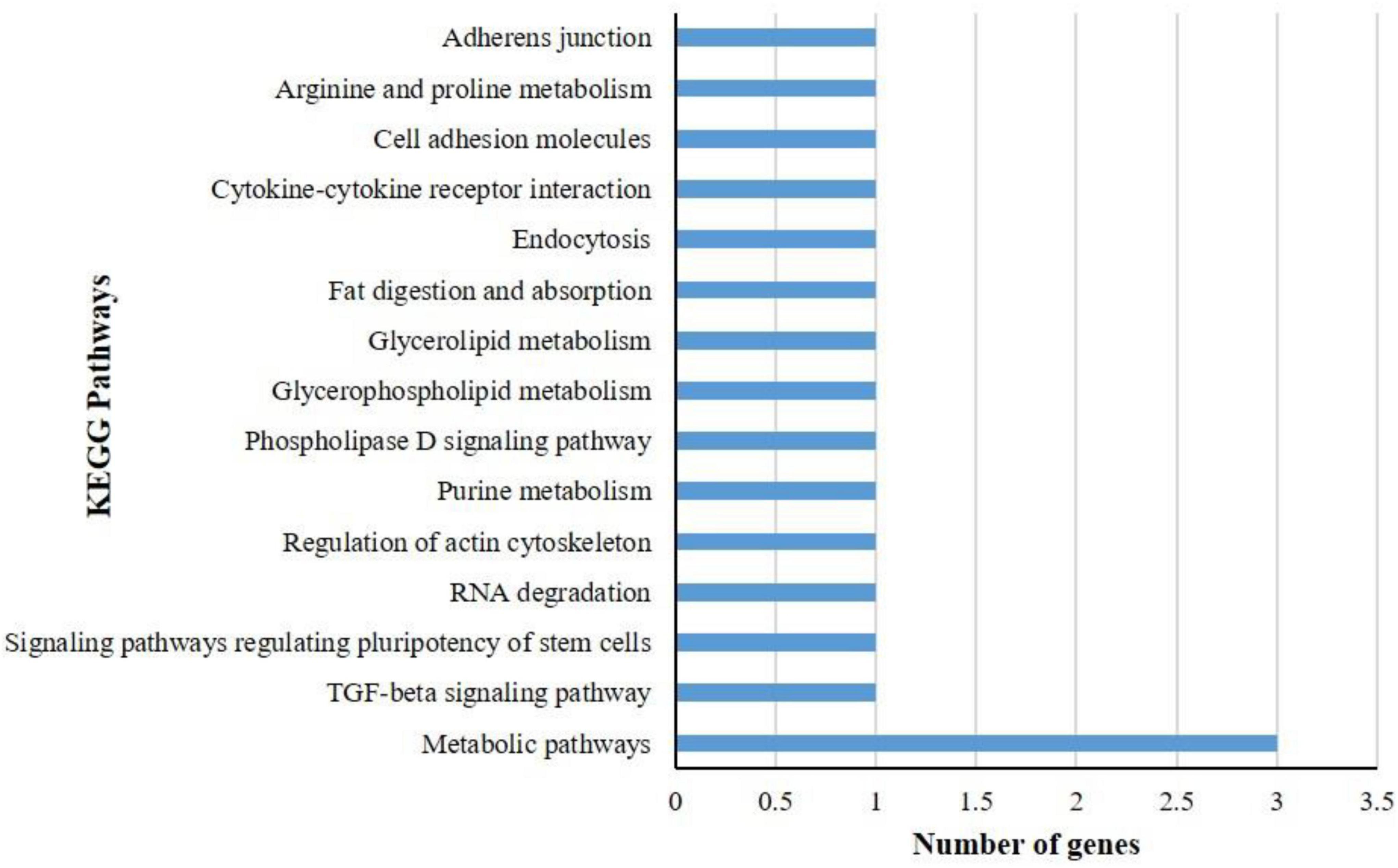
Figure 3. KEGG pathway analysis of differentially expressed genes (DEGs) of B. tabaci Asia II 1 at an early stage of ChiLCV infection. KEGG pathway analysis of DEGs showed that the genes were involved in the functions like metabolic pathways, biosynthesis of secondary metabolites, signaling pathways, and actin cytoskeleton regulation in which mostly DEGs were involved in metabolic pathways.
To validate the differential expression of B. tabaci genes in response to ChiLCV infection, 20 highly regulated genes of B. tabaci were selected and mRNA expression levels were quantified in RT-qPCR. The primer pairs that produced a single sharp amplicon at the same PCR conditions for endogenous control (β-actin) primers were selected for RT-qPCR assay. Primer pair, AG177F and AG178R (Supplementary Table 1) specific to β-actin produced sharp bands at annealing temperatures of 54–59°C. The annealing temperature of target genes was standardized within the same temperature range. One primer pair for each of the target genes was optimized for RT-qPCR assay (Supplementary Table 1). The relative expression of a target gene was estimated using the 2–ΔΔ CT method (Roy et al., 2021). The CT value of β-actin was used to normalize variation among biological replicates. Among the highly regulated genes that were selected for the RT-qPCR assay, the genes like Fas2-like (Bta00469) showed the highest regulation at an early stage of ChiLCV infection. The mRNA expression level of Fas2-like was upregulated by 4.518-fold in the RT-qPCR assay. TLR3 (Bta12916) was another upregulated gene with log2 3.517-fold change in mRNA expression in response to ChiLCV infection. The mRNA expression of replication factor A protein 1 (RFA1) was upregulated by log2 0.403-fold in ChiLCV-exposed B. tabaci adults in comparison to non-exposed adults. As a result of exposure to ChiLCV, the expression of GMPR of B. tabaci (Bta00655) was elevated by a factor of log2 2.246-fold. Expression of protein Tob1 (Bta07280) was also augmented by log2 1.34-fold at an early stage of ChiLCV infection. Likewise, the mRNA expression levels of a few other genes like AGBL3-like (Bta06925), DUSP10-like (Bta09526), QRSL1 (Bta13949) were elevated by log2 0.168, 2.593, and 0.081-fold, respectively, in the RT-qPCR assay. An uncharacterized protein, CG13607 (Bta13910) was also upregulated by log2 1.74-fold in the ChiLCV-exposed B. tabaci population in comparison to non-exposed populations.
Among the highly downregulated genes, expression of B. tabaci Actbeta (Bta15272) decreased by log2 5.982-fold in the RT-qPCR assay at an early stage of ChiLCV infection. The next highly downregulated genes in RT-qPCR were TBX20 (Bta05443) and HOXA2 (Bta14472). Expression of TBX20 and HOXA2 declined by log2 3.517, 2.378-fold, respectively in response to ChiLCV infection. The mRNA expression level of NBEAL1 (Bta07414) was also downregulated by log2 1.146-fold in ChiLCV-exposed B. tabaci in comparison to non-exposed populations. A log2 0.793-fold downregulation for MRJP (Bta05465) was also recorded in response to ChiLCV infection. Few other genes of B. tabaci like ARID (Bta03804), anther specific protein (sf2)-like (Bta01185), plsC (Bta15094) showed significant downregulations by log2 0.241, 0.036, and 1.687-fold, respectively, in RT-qPCR assay post-ChiLCV exposure.
All the primer pairs for the target and endogenous control genes produced single specific peaks without any secondary amplifications in the RT-qPCR melting curve analysis that indicated the specificity of the reactions. The melting temperatures of RT-qPCR amplicons were listed in Supplementary Table 1. The melt curve of each reaction has been provided as Supplementary Figure 1. The relative expression of selected B. tabaci genes in response to ChiLCV infection substantiated the RNA-Seq analysis (Figure 4).
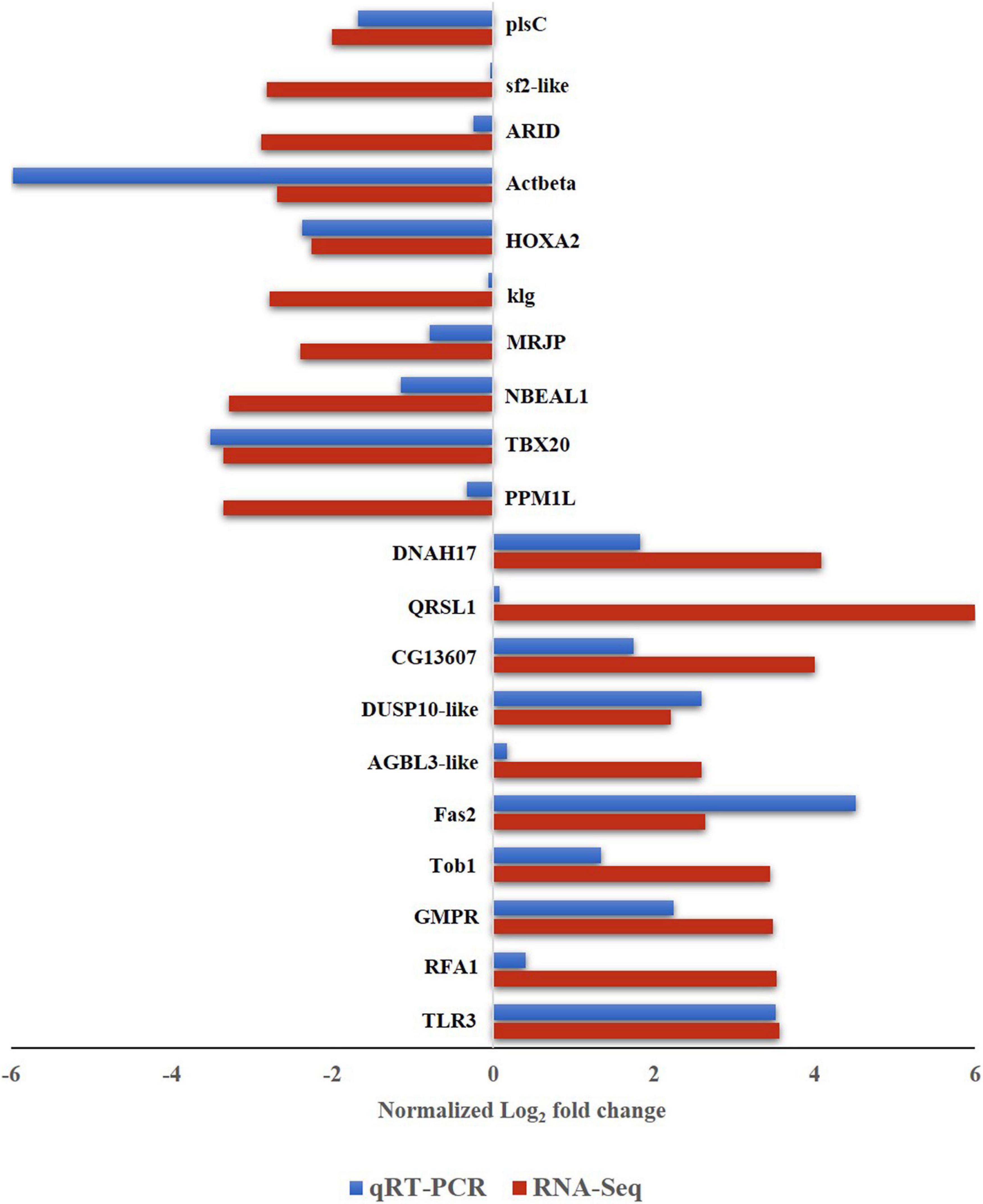
Figure 4. Expression of B. tabaci Asia II 1 putative genes in response to ChiLCV infection in RNA-Seq and RT-qPCR. The values of log2-fold changes calculated in RNA-Seq analysis were in accordance with the RT-qPCR fold change values.
The molecular interactions between B. tabaci cryptic species and begomoviruses are not universal (Czosnek et al., 2017) and not much is known about the interaction of ChiLCV with B. tabaci. For successful transmission, the begomovirus particles need to reach the salivary glands of B. tabaci. It takes 4–7 h after the onset of feeding to be circulated through hemolymph in coated vesicles and translocated into the primary salivary glands (Czosnek et al., 2002). In our recent study, ChiLCV copies were estimated in individual B. tabaci at different exposure of active acquisition feeding (Roy et al., 2021). A 6 h sacquisition was found adequate for the successful transmission of ChiLCV by B. tabaci Asia II 1 (Senanayake et al., 2012; Roy et al., 2021). About 7.86 × 1013 copies of ChiLCV can be acquired by an individual B. tabaci adult female during 6 h of feeding on an infected chilli leaf (Roy et al., 2021). Comparison between ChiLCV-exposed and non-exposed B. tabaci transcripts post-6 h of acquisition revealed differential expression of 80 DEGs involved in replication factor, cell adhesion receptor, and intracellular trafficking. GO analysis indicated the majority of the DEGs were involved in biological processes followed by cellular components and molecular functions. KEGG pathway analysis showed that the majority of the DEGs were involved in the metabolic pathways of B. tabaci.
The key upregulated genes included TLR3, Fas2, GMPR, RFA1, Tob 1, DUSP, DNAH17, AGBL3, and QRSL1. Several genes of B. tabaci that were related to immune response pathways such as TLR3, Fas2, RFA1, and GMPR were induced in response to ChiLCV infection. Toll receptors are important parts of the insect’s innate immune system by triggering a cascade of signaling pathways. In mammals, it induces interferons to confer antiviral resistance (Ozato et al., 2002). Toll receptors might have a function in limiting the virus infection in vector and upregulation of TLR3 might be activated upon ChiLCV infection to preclude the adverse effect of the virus on the vector. Another component of B. tabaci immunoglobulin (Ig)-related superfamily, Fas2 was upregulated post-exposure to the ChiLCV in the present study. Fas2 belongs to the superfamily of cell adhesion receptors with structural similarity to the vertebrate receptor NCAM (Grenningloh et al., 1990; Brummendorf and Rathjen, 1993). NCAM molecules also have a role in viral attachment to promote virus penetration into the host cells but inhibit the replication of rabies virus via induction of interferon β (Hotta et al., 2007), which is mainly involved in innate immunity against viral infection. Silencing Fas 2 using dsRNA results in an increase of ChiLCV copies in B. tabaci Asia II 1 (Chakraborty and Ghosh, 2022). This suggests that upregulation of Fas2 transcripts in B. tabaci post-ChiLCV infection is due to the innate immune response against the virus infection. GMPR plays an important role in the purine salvage pathway and regulates intracellular purine nucleotides. Besides, it has a role in host innate immunity (Imamura et al., 2020). Upregulation of this gene after ChiLCV exposure indicated induction of defense in B. tabaci upon ChiLCV exposure. RFA1 is an ssDNA binding protein (Wold, 1997). Although replication factors play an important role in DNA replication, recombination, and repair (Vanoli et al., 2010; Yamamoto et al., 2019), ChiLCV-DNA does not replicate within the vector (Rosen et al., 2015; Ghosh et al., 2021). This rejected the possibility of involvement of RFA1 in virus multiplication in B. tabaci. Although not confirmed, upregulation of RFA1 the ChiLCV-exposure might be due to the employment of innate immune response by B. tabaci to protect itself from virus nuclease attack.
Some genes such as Tob1 and DUSP10 having a functional role in the virus lifecycle were also upregulated to favor virus transcription. Gene encoding Tob1 protein was found upregulated in B. tabaci post-ChiLCV exposure. Overexpression of Tob1 suppresses cell growth (Shan et al., 2009). The upregulation of this gene in the current study might lead to cellular dysfunction in B. tabaci to favor ChiLCV infection. DUSP10, also called MPK5 is involved in the regulation of mitogen-activated protein kinases. These proteins are the major modulators of critical signaling pathways that are dysregulated in various biotic stresses (Patterson et al., 2009). DUSP facilitates vaccinia virus transcription (Liu et al., 1995). Upregulation of this gene indicated the activation of signaling cascades in response to ChiLCV infection and functional role in the virus lifecycle in B. tabaci.
Expression of B. tabaci putative genes like DNAH17, AGBL3, and QRSL1 was manipulated by ChiLCV to facilitate its circulation within the vector. Several viruses access dynein to mediate the viral replication processes. Silencing of dynein reduced the infection of Murine leukemia virus (Valle-Tenney et al., 2016). Carboxypeptidases play an important role in the stimulation of slow migrating forms of cowpea mosaic virus (CPMV) to fast migrating CPMV leading to increased infectivity (Niblett and Semancik, 1969). Upregulation of DNAH17 and AGBL3 post-ChiLCV exposure in the present study might be due to the manipulation of B. tabaci genes by ChiLCV to facilitate the infection and circulation of the virus within the vector. QRSL1 is a highly conserved protein throughout eukaryotes and prokaryotes. This gene is essential for the proper translation of the proteins (Morris et al., 2008). Upregulation of this gene post-ChiLCV exposure might be due to the utilization of host translation apparatus by the virus.
Other than the above-mentioned upregulated genes, a few immune-associated genes like AGO2, PPM1L, NBEAL1, TBX20, and Actbeta were downregulated in B. tabaci upon ChiLCV exposure. AGO2 is a major component of the RNA-induced silencing complex (RISC). In Drosophila, loss of function of AGO2 leads to susceptibility against Drosophila C virus and Cricket paralysis virus (Van Rij et al., 2006). PPM1L is known to be associated with the replication of viruses like HIV-1, HIV-2, Ebola virus, papovavirus, adenovirus, and rift valley fever virus (Brown et al., 1994; Modrof et al., 2002; Nekhai et al., 2007; Zeng et al., 2009; Baer et al., 2016). Neurobeachin is a peripheral membrane protein of the BEACH domain protein family that is involved in the subcellular targeting of membrane proteins (De Lozanne, 2003). Increased expression of neurobeachin was recorded in stimulated immune cells (Wang et al., 2001, 2004). As an essential factor of autoimmunity, TBX20 produces IgG2a via activation of B cells during viral infection (Rubtsova et al., 2013). Inhibin plays an important function in immunological cell development (Licona-Limon and Soldevila, 2007). Lack of plsC activity increased the virulence and infectivity of coxsackievirus (Karlsson et al., 2009). Downregulation of these immune-associated genes in B. tabaci post-ChiLCV acquisition indicated a possibility of suppression of vector immune response by ChiLCV to become circulative in its vector.
Transcription regulatory genes like HOXA2, klg, and ARID were also downregulated to support ChiLCV entry and movement within B. tabaci. Homeobox proteins are transcriptional factors that regulate several genes in insects (Cillo et al., 2001). In tomatoes, homeobox protein was found to be involved in limiting programmed cell death that limits pathogens (Mayda et al., 1999). In ChiLCV-exposed B. tabaci, downregulation of HOXA2 might be due to the suppression activity of ChiLCV that triggered the repression in B. tabaci. Klg is a CAM orthologue in insects that is required for photoreceptors and the prevention of excessive synapses (Shimozono et al., 2019). Upon infection of human cytomegalovirus (HCMV), klg is downregulated and causes increased adhesion of the virus to the tumor cells and transendothelial penetration (Blaheta et al., 2004). Downregulation of klg might help the binding and cellular entry of ChiLCV. ARID contains a DNA binding domain (Patsialou et al., 2005) and is an important factor for development, tissue-specific gene expression, and cell growth regulation (Kortschak et al., 2000; Wilsker et al., 2002). This gene is also involved in transcriptional activation and repression of genes through chromatin remodeling (Zhang et al., 2016). Repression of ARID in B. tabaci might favor ChiLCV entry and movement within the vector.
Putative genes that code for anther specific protein, major royal jelly protein, and CG13607 were also differentially expressed in B. tabaci at an early stage of ChiLCV infection. Although these genes have been characterized in other insects, their functions in B. tabaci and begomovirus transmission are yet to be explored. The expression of several orphan genes in B. tabaci Asia II 1 was also modulated upon ChiLCV exposure. Functional validation of these orphan genes in B. tabaci Asia II 1 and their role in virus transmission need further in-depth study.
In conclusion, we have assembled a whole-body transcriptome of B. tabaci Asia II 1. Several genes of B. tabaci associated with innate immune response, cell adhesion, and intracellular trafficking were regulated in response to ChiLCV infection to facilitate its survival and circulation within the vector, B. tabaci. The present study helps in understanding the network of molecular interactions between B. tabaci Asia II 1 and ChiLCV. Data generated in this study will enrich genomic information of whitefly and will enable future functional studies. The candidate genes of B. tabaci that are involved in key physiological processes and ChiLCV transmission would be novel targets for sustainable management of the whitefly-begomovirus complex.
The datasets generated during the current study are available in the NCBI with BioProject ID PRJNA759071.
AG conceived and designed the research and wrote and edited the final manuscript. AN and PC conducted the experiments, recorded the experimental data, and wrote the draft manuscript. AN, PC, MI, and SJ performed the analysis. AG and VB reviewed the data. All authors read and approved the manuscript.
The fellowship of AN was supported by Indian Council of Agricultural Research, New Delhi.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful to Subhash Chander (NRCIPM) and Vinay Kalia (IARI) for their advisories. The support received from IARI, New Delhi is thankfully acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.890807/full#supplementary-material
Akhter, A., Qazi, J., Saeed, M., and Mansoor, S. (2009). A severe leaf curl disease on chilies in Pakistan is associated with multiple begomovirus components. Plant Dis. 93, 962–962. doi: 10.1094/PDIS-93-9-0962B
Baer, A., Shafagati, N., Benedict, A., Ammosova, T., Ivanov, A., Hakami, R. M., et al. (2016). Protein phosphatase-1 regulates Rift Valley fever virus replication. Antiv. Res. 127, 79–89. doi: 10.1016/j.antiviral.2016.01.007
Barman, M., Samanta, S., Thakur, H., Chakraborty, S., Samanta, A., Ghosh, A., et al. (2021). Effect of neonicotinoids on bacterial symbionts and insecticide-resistant gene in whitefly, Bemisia tabaci. Insects 12:742. doi: 10.3390/insects12080742
Blaheta, R. A., Beecken, W. D., Engl, T., Jonas, D., Oppermann, E., Hundemer, M., et al. (2004). Human cytomegalovirus infection of tumor cells downregulates NCAM (CD56): a novel mechanism for virus-induced tumor invasiveness. Neoplasia 6, 323–331. doi: 10.1593/neo.03418
Bragard, C., Caciagli, P., Lemaire, O., Lopez-Moya, J. J., MacFarlane, S., Peters, D., et al. (2013). Status and prospects of plant virus control through interference with vector transmission. Annu. Rev. Phytopathol. 51, 177–201. doi: 10.1146/annurev-phyto-082712-102346
Briddon, R. W., Patil, B. L., Bagewadi, B., Nawaz-ul-Rehman, M. S., and Fauquet, C. M. (2010). Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 10:97. doi: 10.1186/1471-2148-10-97
Brown, J. K. (2000). Molecular markers for the identification and global tracking of whitefly vector–Begomovirus complexes. Virus Res. 71, 233–260. doi: 10.1016/s0168-1702(00)00221-5
Brown, J. K., Zerbini, F. M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J. C., et al. (2015). Revision of begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 160, 1593–1619. doi: 10.1007/s00705-015-2398-y
Brown, S. M., Harland, J., MacLean, A. R., Podlech, J., and Clements, J. B. (1994). Cell type and cell state determine differential in vitro growth of non-neurovirulent ICP34. 5-negative herpes simplex virus types 1 and 2. J. Gen. Virol. 75, 2367–2377. doi: 10.1099/0022-1317-75-9-2367
Brummendorf, T., and Rathjen, F. G. (1993). Axonal glycoproteins with immunoglobulin−and fibronectin type III−related domains in vertebrates: structural features, binding activities, and signal transduction. J. Neurochem. 61, 1207–1219. doi: 10.1111/j.1471-4159.1993.tb13611.x
Chakraborty, P., and Ghosh, A. (2022). Topical spray of dsRNA induces mortality and inhibits chilli leaf curl virus transmission by Bemisia tabaci Asia II 1. Cells 11:833. doi: 10.3390/cells11050833
Chen, W., Hasegawa, D. K., Kaur, N., Kliot, A., Pinheiro, P. V., Luan, J., et al. (2016). The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 14:110. doi: 10.1186/s12915-016-0321-y
Cillo, C., Cantile, M., Faiella, A., and Boncinelli, E. (2001). Homeobox genes in normal and malignant cells. J. Cell. Physiol. 188, 161–169. doi: 10.1002/jcp.1115
Cornejo-Franco, J. F., Reyes-Proaño, E. G., Mollov, D., Mowery, J., and Quito-Avila, D. F. (2022). Transmission and pathogenicity of papaya virus E: insights from an experimental papaya orchard. Plant Dis. 106:685–690. doi: 10.1094/PDIS-08-21-1785-RE
Costa, T. M., Inoue-Nagata, A. K., Vida, A. H., Ribeiro, S. G., and Nagata, T. (2020). The recombinant isolate of cucurbit aphid-borne yellows virus from Brazil is a polerovirus transmitted by whiteflies. Plant Pathol. 69, 1042–1050. doi: 10.1111/ppa.13186
Czosnek, H., and Ghanim, M. (2012). Back to basics: are begomoviruses whitefly pathogens. J. Integr. Agric. 11, 225–234. doi: 10.1016/S2095-3119(12)60007-0
Czosnek, H., Ghanim, M., and Ghanim, M. (2002). The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci-insights from studies with tomato yellow leaf curl virus. Ann. Appl. Biol. 140, 215–231. doi: 10.1111/j.1744-7348.2002.tb00175.x
Czosnek, H., Hariton-Shalev, A., Sobol, I., Gorovits, R., and Ghanim, M. (2017). The incredible journey of begomoviruses in their whitefly vector. Viruses 9:273. doi: 10.3390/v9100273
Dasgupta, I., Malathi, V. G., and Mukherjee, S. K. (2003). Genetic engineering for virus resistance. Curr. Sci. 84, 341–354.
De Lozanne, A. (2003). The role of BEACH proteins in Dictyostelium. Traffic 4, 6–12. doi: 10.1034/j.1600-0854.2003.40102.x
Ding, T. B., Li, J., Chen, E. H., Niu, J. Z., and Chu, D. (2019). Transcriptome profiling of the whitefly Bemisia tabaci MED in response to single infection of tomato yellow leaf curl virus, tomato chlorosis virus, and their co-infection. Front. Physiol. 10:302. doi: 10.3389/fphys.2019.00302
Fereres, A., and Raccah, B. (2015). Plant Virus Transmission by Insects, eLS. Chichester: John Wiley & Sons, doi: 10.1002/9780470015902.a0000760.pub3
Ghosh, A., Roy, B., Nekkanti, A., Dhar, S., and Mukherjee, S. (2021). Transovarial transmission of dolichos yellow mosaic virus by its vector, Bemisia tabaci Asia II 1. Front. Microbiol. 12:755155. doi: 10.3389/fmicb.2021.755155
Ghosh, S., Kanakala, S., Lebedev, G., Kontsedalov, S., Silverman, D., Alon, T., et al. (2019). Transmission of a new polerovirus infecting pepper by the whitefly Bemisia tabaci. J. Virol. 93:e00488-19. doi: 10.1128/JVI.00488-19
Gorman, K., Slater, R., Blande, J. D., Clarke, A., Wren, J., McCaffery, A., et al. (2010). Cross-resistance relationships between neonicotinoids and pymetrozine in Bemisia tabaci (Hemiptera: Aleyrodidae). Pest Manage. Sci. 66, 1186–1190. doi: 10.1002/ps.1989
Götz, M., Popovski, S., Kollenberg, M., Gorovits, R., Brown, J. K., Cicero, J. M., et al. (2012). Implication of Bemisia tabaci heat shock protein 70 in begomovirus-whitefly interactions. J. Virol. 86, 13241–13252. doi: 10.1128/JVI.00880-12
Grenningloh, G., Bieber, A. J., Rehm, E. J., Snow, P. M., Traquina, Z. R., Hortsch, M., et al. (1990). Molecular genetics of neuronal recognition in Drosophila: evolution and function of immunoglobulin superfamily cell adhesion molecules. Cold Spring Harb. Symp. Quant. Biol. 55, 327–340. doi: 10.1101/sqb.1990.055.01.034
Hasegawa, D. K., Chen, W., Zheng, Y., Kaur, N., Wintermantel, W. M., Simmons, A. M., et al. (2018). Comparative transcriptome analysis reveals networks of genes activated in the whitefly, Bemisia tabaci when fed on tomato plants infected with tomato yellow leaf curl virus. Virology 513, 52–64. doi: 10.1016/j.virol.2017.10.008
Hotta, K., Motoi, Y., Okutani, A., Kaku, Y., Noguchi, A., Inoue, S., et al. (2007). Role of GPI-anchored NCAM-120 in rabies virus infection. Microbes Infect. 9, 167–174. doi: 10.1016/j.micinf.2006.11.003
Imamura, A., Okada, T., Mase, H., Otani, T., Kobayashi, T., Tamura, M., et al. (2020). Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-β-synthase domain. Nat. Commun. 11:1837. doi: 10.1038/s41467-020-15611-3
Kanakala, S., and Ghanim, M. (2016). Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of tomato yellow leaf curl virus. Front. Plant Sci. 7:1702. doi: 10.3389/fpls.2016.01702
Karlsson, E. A., Wang, S., Shi, Q., Coleman, R. A., and Beck, M. A. (2009). Glycerol-3-phosphate acyltransferase 1 is essential for the immune response to infection with coxsackievirus B3 in mice. J. Nutr. 139, 779–783. doi: 10.3945/jn.108.101683
Kaur, N., Chen, W., Zheng, Y., Hasegawa, D. K., Ling, K. S., Fei, Z., et al. (2017). Transcriptome analysis of the whitefly, Bemisia tabaci MEAM1 during feeding on tomato infected with the crinivirus, Tomato chlorosis virus, identifies a temporal shift in gene expression and differential regulation of novel orphan genes. BMC Genomics 18:370. doi: 10.1186/s12864-017-3751-1
Kortschak, R. D., Tucker, P. W., and Saint, R. (2000). ARID proteins come in from the desert. Trends Biochem. Sci. 25, 294–299. doi: 10.1016/s0968-0004(00)01597-8
Legarrea, S., Barman, A., Marchant, W., Diffie, S., and Srinivasan, R. (2015). Temporal effects of a Begomovirus infection and host plant resistance on the preference and development of an insect vector, Bemisia tabaci, and implications for epidemics. PLoS One 10:e0142114. doi: 10.1371/journal.pone.0142114
Licona-Limon, P., and Soldevila, G. (2007). The role of TGF-β superfamily during T cell development: new insights. Immun. Lett. 109, 1–12. doi: 10.1016/j.imlet.2006.12.010
Liu, K., Lemon, B., and Traktman, P. (1995). The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 69, 7823–7834. doi: 10.1128/JVI.69.12.7823-7834.1995
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, J. B., Li, J. M., Varela, N., Wang, Y. L., Li, F. F., Bao, Y. Y., et al. (2011). Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl china virus reveals the relationship of coevolved adaptations. J. Virol. 85, 3330–3340. doi: 10.1128/JVI.02507-10
Mayda, E., Tornero, P., Conejero, V., and Vera, P. (1999). A tomato homeobox gene (HD−Zip) is involved in limiting the spread of programmed cell death. Plant J. 20, 591–600. doi: 10.1046/j.1365-313x.1999.00633.x
Menike, G. D. N., and De Costa, D. M. (2017). Variation of field symptoms and molecular diversity of the virus isolates associated with chilli leaf curl complex in different agroecological regions of Sri Lanka. Trop. Agric. Res. 28, 144–161. doi: 10.4038/tar.v28i2.8192
Milenovic, M., Wosula, E. N., Rapisarda, C., and Legg, J. P. (2019). Impact of host plant species and whitefly species on feeding behavior of Bemisia tabaci. Front. Plant Sci. 10:1. doi: 10.3389/fpls.2019.00001
Modrof, J., Mu, E., Klenk, H. D., and Becker, S. (2002). Phosphorylation of VP30 impairs Ebola virus transcription. J. Biol. Chem. 277, 33099–33104. doi: 10.1074/jbc.M203775200
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Morris, J. Z., Bergman, L., Kruyer, A., Gertsberg, M., Guigova, A., Arias, R., et al. (2008). Mutations in the Drosophila mitochondrial tRNA amidotransferase, bene/gatA, cause growth defects in mitotic and endoreplicating tissues. Genetics 178, 979–987. doi: 10.1534/genetics.107.084376
Navas-Castillo, J., Fiallo-Olivé, E., and Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nekhai, S., Jerebtsova, M., Jackson, A., and Southerland, W. (2007). Regulation of HIV-1 transcription by protein phosphatase 1. Curr. HIV Res. 5, 3–9. doi: 10.2174/157016207779316279
Niblett, C. L., and Semancik, J. S. (1969). Conversion of the electrophoretic forms of cowpea mosaic virus in vivo and in vitro. Virology 38, 685–693. doi: 10.1016/0042-6822(69)90187-1
Oraonand, U. B., and Tarafdar, J. (2018). Occurrence and distribution of chilli leaf curl complex disease in West Bengal. Biomed. J. Sci. Tech. Res. 3, 3515–3519. doi: 10.26717/BJSTR.2018.3.000948
Orfanidou, C. G., Pappi, P. G., Efthimiou, K. E., Katis, N. I., and Maliogka, V. I. (2016). Transmission of Tomato Chlorosis Virus (ToCV) by Bemisia tabaci biotype Q and evaluation of four weed species as viral sources. Plant Dis. 100, 2043–2049. doi: 10.1094/PDIS-01-16-0054-RE
Ozato, K., Tsujimura, H., and Tamura, T. (2002). Toll-like receptor signaling and regulation of cytokine gene expression in the immune system. Biotechniques 33, S66–S75. doi: 10.2144/Oct0208
Patsialou, A., Wilsker, D., and Moran, E. (2005). DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33, 66–80. doi: 10.1093/nar/gki145
Patterson, K. I., Brummer, T., O’brien, P. M., and Daly, R. J. (2009). Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem. J. 418, 475–489. doi: 10.1042/bj20082234
Pinheiro-Lima, B., Pereira-Carvalho, R. C., Alves-Freitas, D. M. T., Kitajima, E. W., Vidal, A. H., Lacorte, C., et al. (2020). Transmission of the bean-associated cytorhabdovirus by the whitefly Bemisia tabaci MEAM1. Viruses 12:1028. doi: 10.3390/v12091028
Qin, L., Pan, L. L., and Liu, S. S. (2016). Further insight into reproductive incompatibility between putative cryptic species of the Bemisia tabaci whitefly complex. Insect Sci. 23, 215–224. doi: 10.1111/1744-7917.12296
Rehman, M., Chakraborty, P., Tanti, B., Mandal, B., and Ghosh, A. (2021). Occurrence of a new cryptic species of Bemisia tabaci (Hemiptera: Aleyrodidae): an updated record of cryptic diversity in India. Phytoparasitica 49, 869–882. doi: 10.1007/s12600-021-00909-9
Rosen, R., Kanakala, S., Kliot, A., Pakkianathan, B. C., Farich, B. A., Santana-Magal, N., et al. (2015). Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 15, 1–8. doi: 10.1016/j.coviro.2015.06.008
Roy, B., Chakraborty, P., and Ghosh, A. (2021). How many begomovirus copies are acquired and inoculated by its vector, whitefly (Bemisia tabaci) during feeding? PLoS One 16:e0258933. doi: 10.1371/journal.pone.0258933
Rubtsova, K., Rubtsov, A. V., Van Dyk, L. F., Kappler, J. W., and Marrack, P. (2013). T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl. Acad. Sci. 110, E3216–E3224. doi: 10.1073/pnas.1312348110
Saeed, S. T., and Samad, A. (2017). Emerging threats of begomoviruses to the cultivation of medicinal and aromatic crops and their management strategies. Virusdisease 28, 1–17. doi: 10.1007/s13337-016-0358-0
Senanayake, D. M. J. B., Mandal, B., Lodha, S., and Varma, A. (2007). First report of Chilli leaf curl virus affecting chilli in India. Plant Pathol. 56:343. doi: 10.1111/j.1365-3059.2007.01513.x
Senanayake, D. M. J. B., Varma, A., and Mandal, B. (2012). Virus–vector relationships, host range, detection and sequence comparison of chilli leaf curl virus associated with an epidemic of leaf curl disease of chilli in Jodhpur, India. J. Phytopathol. 160, 146–155. doi: 10.1111/j.1439-0434.2011.01876.x
Shan, J., Zhao, W., and Gu, W. (2009). Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell 36, 469–476. doi: 10.1016/j.molcel.2009.10.018
Shimozono, M., Osaka, J., Kato, Y., Araki, T., Kawamura, H., Takechi, H., et al. (2019). Cell surface molecule, Klingon, mediates the refinement of synaptic specificity in the Drosophila visual system. Genes Cells 24, 496–510. doi: 10.1111/gtc.12703
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651–701. doi: 10.1093/aesa/87.6.651
Thakur, H., Jindal, S. K., Sharma, A., and Dhaliwal, M. S. (2018). Chilli leaf curl virus disease: a serious threat for chilli cultivation. J. Plant Dis. Protect. 125, 239–249. doi: 10.1007/s41348-018-0146-8
Valle-Tenney, R., Opazo, T., Cancino, J., Goff, S. P., and Arriagada, G. (2016). Dynein regulators are important for ecotropic murine leukemia virus infection. J. Virol. 90, 6896–6905. doi: 10.1128/JVI.00863-16
Van Rij, R. P., Saleh, M. C., Berry, B., Foo, C., Houk, A., Antoniewski, C., et al. (2006). he RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995. doi: 10.1101/gad.1482006
Vanoli, F., Fumasoni, M., Szakal, B., Maloisel, L., and Branzei, D. (2010). Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genetics 6:e1001205. doi: 10.1371/journal.pgen.1001205
Wang, J. W., Gamsby, J. J., Highfill, S. L., Mora, L. B., Bloom, G. C., Yeatman, T. J., et al. (2004). Deregulated expression of LRBA facilitates cancer cell growth. Oncogene 23, 4089–4097. doi: 10.1038/sj.onc.1207567
Wang, J. W., Howson, J., Haller, E., and Kerr, W. G. (2001). Identification of a novel lipopolysaccharide-inducible gene with key features of both A kinase anchor proteins and chs1/beige proteins. J. Immunol. 166, 4586–4595. doi: 10.4049/jimmunol.166.7.4586
Wang, P., Ruan, Y. M., and Liu, S. S. (2010). Crossing experiments and behavioural observations reveal reproductive incompatibility among three putative species of the whitefly Bemisia tabaci. Insect Sci. 17, 508–516. doi: 10.1111/j.1744-7917.2010.01353.x
Wang, Z. Z., Shi, M., Huang, Y. C., Wang, X. W., Stanley, D., and Chen, X. X. (2016). A peptidoglycan recognition protein acts in whitefly (Bemisia tabaci) immunity and involves in Begomovirus acquisition. Sci. Rep. 6:37806. doi: 10.1038/srep37806
Wei, J., Zhao, J. J., Zhang, T., Li, F. F., Ghanim, M., Zhou, X. P., et al. (2014). Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of Begomoviruses. J. Virol. 88, 13460–13468. doi: 10.1128/JVI.02179-14
Wilsker, D., Patsialou, A., Dallas, P. B., and Moran, E. (2002). ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 13, 95–106.
Wold, M. S. (1997). Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Ann. Rev. Biochem. 66, 61–92. doi: 10.1146/annurev.biochem.66.1.61
Xia, W. Q., Wang, X. R., Liang, Y., Liu, S. S., and Wang, X. W. (2017). Transcriptome analyses suggest a novel hypothesis for whitefly adaptation to tobacco. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-12387-3
Yamamoto, K., Haque, M. R., and Saruta, F. (2019). Identification and expression analysis of the replication factor C protein in the silkworm, Bombyx mori. J. Insect Biotechnol. Sericol. 88, 1017–1020. doi: 10.11416/jibs.88.1_017
Zanardo, L. G., and Carvalho, C. M. (2017). Cowpea mild mottle virus (Carlavirus, Betaflexiviridae): a review. Trop. Plant Pathol. 42, 417–430. doi: 10.1007/s40858-017-0168-y
Zeng, Q., Huang, Y., Zeng, L., Lan, X., Huang, Y., He, S., et al. (2009). Effect of IPP5, a novel inhibitor of PP1, on apoptosis and the underlying mechanisms involved. Biotechnol. Appl. Biochem. 54, 231–238. doi: 10.1042/BA20090168
Keywords: ChiLCV, silverleaf whitefly, RNA-Seq, transcriptome, RT-qPCR, virus-vector relationship
Citation: Nekkanti A, Chakraborty P, Ghosh A, Iquebal MA, Jaiswal S and Baranwal VK (2022) Transcriptomic Changes of Bemisia tabaci Asia II 1 Induced by Chilli Leaf Curl Virus Trigger Infection and Circulation in Its Vector. Front. Microbiol. 13:890807. doi: 10.3389/fmicb.2022.890807
Received: 06 March 2022; Accepted: 25 March 2022;
Published: 28 April 2022.
Edited by:
Sean Michael Prager, University of Saskatchewan, CanadaReviewed by:
Susheel Kumar, National Botanical Research Institute (CSIR), IndiaCopyright © 2022 Nekkanti, Chakraborty, Ghosh, Iquebal, Jaiswal and Baranwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amalendu Ghosh, YW1hbDRlbnRvQGdtYWlsLmNvbQ==; QW1hbGVuZHUuR2hvc2hAaWNhci5nb3YuaW4=; orcid.org/0000-0001-6634-5771
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.