
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 28 April 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.889835
Autophagy is an evolutionarily conserved lysosomal degradation system which can recycle multiple cytoplasmic components under both physiological and stressful conditions. Autophagy could be highly selective to deliver different cargoes or substrates, including protein aggregates, pathogenic proteins or superfluous organelles to lysosome using a series of cargo receptor proteins. During viral invasion, cargo receptors selectively target pathogenic components to autolysosome to defense against infection. However, viruses not only evolve different strategies to counteract and escape selective autophagy, but also utilize selective autophagy to restrict antiviral responses to expedite viral replication. Furthermore, several viruses could activate certain forms of selective autophagy, including mitophagy, lipophagy, aggrephagy, and ferritinophagy, for more effective infection and replication. The complicated relationship between selective autophagy and viral infection indicates that selective autophagy may provide potential therapeutic targets for human infectious diseases. In this review, we will summarize the recent progress on the interplay between selective autophagy and host antiviral defense, aiming to arouse the importance of modulating selective autophagy as future therapies toward viral infectious diseases.
Macroautophagy, hereafter simply referred to as autophagy, is an ancient and highly conserved catabolic degradative process for the elimination of cytoplasmic components through lysosome (Mizushima and Levine, 2010; Mizushima et al., 2011; Nakatogawa, 2020). Cytoplasmic materials destined for autophagic disposal are engulfed by a specialized double-membrane vesicle called autophagosome (Dikic and Elazar, 2018). Autophagosomes then fuse with lysosomes and form autolysosomes through membrane remodeling (Zhao et al., 2021). Autophagy is initially regarded as a starvation-associated bulk degradation process, while it becomes increasingly appreciated that autophagy can target particular substrates through a series of specific cargo receptors. SQSTM1/p62, CALCOCO2/NDP52, OPTN, and NBR1 are well-known cargo receptors that possess both ubiquitin-binding domains (UBDs) and LC3-interacting regions (LIRs) to ensure their capacity to deliver dedicated substrates to autophagosomes (Rogov et al., 2014; Kirkin and Rogov, 2019). Selective autophagy could be classified into multiple subsets according to their degradative substrates, including xenophagy/virophagy for invading intracellular pathogens, mitophagy for mitochondria, aggrephagy for protein aggregates, ER-phagy for endoplasmic reticulum, lipophagy for lipid droplets, and ferritinophagy for ferritin. Based on the subsets of selective autophagy, diverse groups of specific cargo receptors associate with degradative cargoes and deliver their cargoes to autophagy machinery (Table 1).
As a critical intracellular degradative process, autophagy also participates in cell-intrinsic defense by eliminating invading cytosolic microbes (Deretic et al., 2013; Huang and Brumell, 2014; Choi et al., 2018). Given the powerful cleavage capability, autophagy is generally activated to degrade and dispose of invading viruses during virus infection (Choi et al., 2018). Autophagy also promotes antigen processing to adaptive immune responses against invading viruses in the late stage of infection (Paludan et al., 2005). As counteractions, viruses have also evolved strategies to resist, escape, subvert or even hijack autophagy for their replication (Choi et al., 2018; Viret et al., 2021). For example, viruses could escape from selective autophagic degradation via cleaving cargo receptors, restricting autophagy activation and disturbing autophagy pathway. In addition, viruses could promote selective autophagy of key molecules in interferon (IFN) pathway to evade host antiviral innate responses. Viruses could also take advantages of specific types of selective autophagy to enhance viral replication (Choi et al., 2018; Viret et al., 2021; Vo and Choi, 2021).
In this review, we will focus on the involvement of selective autophagy as antiviral defense mechanism and discussed the strategies against selective autophagy by viruses. Additionally, we will discuss whether targeting selective autophagy possesses the potential to develop host-directed therapies as treatment options against viral infectious diseases.
In addition to remove cytoplasmic material such as protein aggregates and damaged or superfluous organelles, selective autophagy has also been reported to mediate autophagic elimination of viral constituents or virions via several specific cargo receptors. Selective autophagy contributes to host antiviral responses through directly targeting viral components for degradation as well as facilitating host innate and adaptive immunity.
As a part of host immune responses, the fundamental properties of selective autophagy are to sense viral particles or viral components and deliver them to autolysosome for degradation (Liang et al., 2021). Ref(2)P, the p62 ortholog in Drosophila melanogaster, was first identified to restrict sigma virus replication through binding its capsid protein (Dru et al., 1993; Wyers et al., 1993), and it could also restrict the replication of vesicular stomatitis virus (VSV) and flavivirus zika virus (ZIKV) (Shelly et al., 2009; Liu et al., 2018). In mammalian, autophagy was first observed to defense virus infection against Sindbis virus (SINV), as defective autophagic pathways enhanced fatal SINV encephalitis in mice (Liang et al., 1998; Mizushima et al., 2001). Subsequently, p62 also exhibited an antiviral role during SINV infection in the central nervous system as it could deliver SINV capsid protein to autophagosome for viral protein clearance (Orvedahl et al., 2010). In addition, p62 was also involved in selective autophagy targeting against double-stranded DNA herpes simplex virus type 1 (HSV-1), adenovirus (AdV) (Orvedahl et al., 2011; Sparrer et al., 2017), positive-sense RNA virus Chikungunya alphavirus (CHIKV), foot-and-mouth disease virus (FMDV) (Berryman et al., 2012; Judith et al., 2013), negative-sense RNA virus measles virus (MeV) (Richetta et al., 2013) and double-stranded RNA virus infectious bursal disease virus (IBDV) (Li Y. et al., 2020). In most case, p62 prefers to target viral capsid or nucleocapsid protein with ubiquitin tags, however, the mechanisms on these preferences still remain unclear (Viret et al., 2021). Besides sensing ubiquitinated capsid proteins, p62 could sense VP1 and VP3 from Seneca Valley virus (SVV) in UBA domain independent manner to inhibit SVV replication (Wen et al., 2021). In addition to ubiquitin tag, Galectin decoration serves as a novel cargo receptor recognition signal (Montespan et al., 2017; Miyakawa et al., 2022). p62 is reported to target hepatitis B virus (HBV) after Galectin-9 decoration (Miyakawa et al., 2022). Reminiscent of the role in targeting viral components, p62 also binds to host factors hijacked by viruses for their replication (Sagnier et al., 2015). In CD4+ T lymphocytes, p62 could target human immunodeficiency virus (HIV) transactivator Tat for autophagic degradation, thereby inhibiting HIV replication (Sagnier et al., 2015).
Along with prototypic roles of p62, several other cargo receptors also participate in autophagic degradation of viral components. OPTN targets HSV-1 tegument protein, VP16, and the fusion glycoprotein, gB, for degradation by selective autophagy in the central nervous system (Ames et al., 2021). NBR can target pararetrovirus cauliflower mosaic virus (CaMV) for autophagic degradation (Hafren et al., 2017). Unlike the other antiviral autophagic receptors, NDP52 seems to promote CHIKV replication through interaction with CHIKV NSP2 in a ubiquitination-dependent manner (Judith et al., 2013). Besides the well-known cargo receptors, several host molecules also act as autophagy receptor-like factors. Fanconi anemia group C protein (FANCC), a factor controlling mitophagy, directly links SINV and HSV-1 capsid with autophagosomes for degradation (Minton, 2016; Sumpter et al., 2016). SMAD ubiquitin regulatory factor 1 (SMURF1) could directly interact with SINV and HSV-1 capsid proteins and mediate their autophagic degradation (Orvedahl et al., 2011; Sumpter et al., 2016). Endoplasmic reticulum (ER) protein SCOTIN restricts hepatitis C virus (HCV) replication by delivering its NS5A to autophagosomes for degradation (Kim N. et al., 2016). Host antiviral factor TRIM5α is also shown to behave as an autophagic receptor by bridging ATG8 factors and HIV capsid factor p24 in cooperation with p62 (Mandell et al., 2014). Hence, host cargo receptors and receptor-like factors direct viral components to autophagosomes for selective degradation, thus contributing to cell-intrinsic defense against viruses (Figure 1).
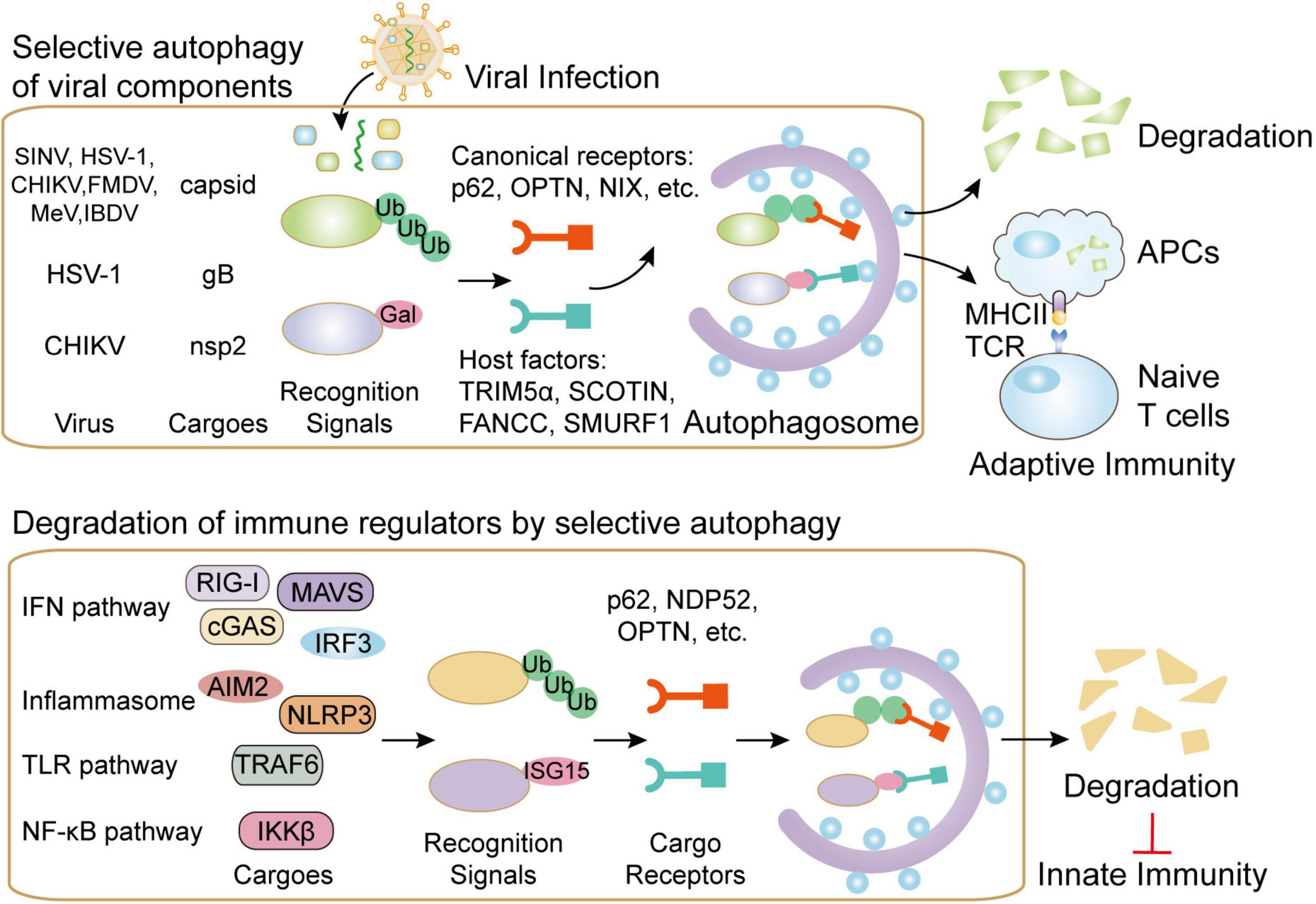
Figure 1. The functions of selective autophagy during viral infection. Selective autophagy targets invading viruses by directly delivering them to autophagosomes for degradation. Several canonical autophagy cargo receptors such as p62 could bind to ubiquitin-coated viral particles or components. Moreover, several host factors, including TRIM5α, SCOTIN, FANCC, and SMURF1, are also reported to assist the transportation of viral components to autophagosomes. In addition to directly targeting viral particles for degradation, selective autophagy also contributes to viral presentation on MHC-II and MHC-I in APCs, thus initiating antiviral adaptive immunity. On the contrary, cargo receptors in selective autophagy pathway could also control autophagic degradation of key molecules in IFN, inflammasome, TLR and NF-κB pathway in order to reduce excessive innate immunity during viral infection.
Adaptive immune response is an indispensable step for effectively eliminating invaded viruses. Accumulating evidence reveals that autophagy has been extensively involved in antigen presentation (Ireland and Unanue, 2011; Puleston et al., 2014). During viral infection, autophagy is also responsible for MHC-II-meditated antigen presentation (Liang et al., 2021). Epstein–Barr virus (EBV) is presented by MHC-II molecules and then recognized by CD4+ T cells, which could be inhibited by blocking of autophagy (Paludan et al., 2005). HIV is also targeted to autophagosome, in which HIV Gag-derived proteins are processed and presented on MHC-II molecules (Kyei et al., 2009). Autophagy also participates in antigen’s internalization and the formation of MHC-I molecules. The latency-associated protein, pUL138 from human cytomegalovirus (HCMV) could be presented by MHC-I through non-conventional transporter associated with antigen processing-independent pathways, which is mediated by autophagy (Tey and Khanna, 2012; Figure 1). Autophagy could process HSV-1 antigens loading on MHC class I molecules, thereby presenting to CD8+ cells (English et al., 2009). Despite the incontrovertible roles of autophagy in antigen presentation during viral infection, the participations of cargo receptors in this process have been seldom mentioned. Diverse viral components are selectively presented by MHC-I and MHC-II molecules and then recognized by CD8+ and CD4+ T cells, however, the contribution of selective autophagy in antigen presentation during infection remains to be illustrated.
Although selective autophagy restricts viral replication via degrading viral particles and enhancing host immune responses, viruses have evolved multiple strategies to escape or subvert such restrictions. The strategies could be generally summed up as (1) directly cleavage of autophagic cargo receptors; (2) inhibition of autophagy activation through ATG proteins; and (3) restricting autophagy pathway through host inhibitory factors (Yordy and Iwasaki, 2011; Liang et al., 2021). To optimize replication capacity, several viruses also utilize selective autophagy to minimize host antiviral responses (Viret et al., 2021; Figure 2 and Table 2).
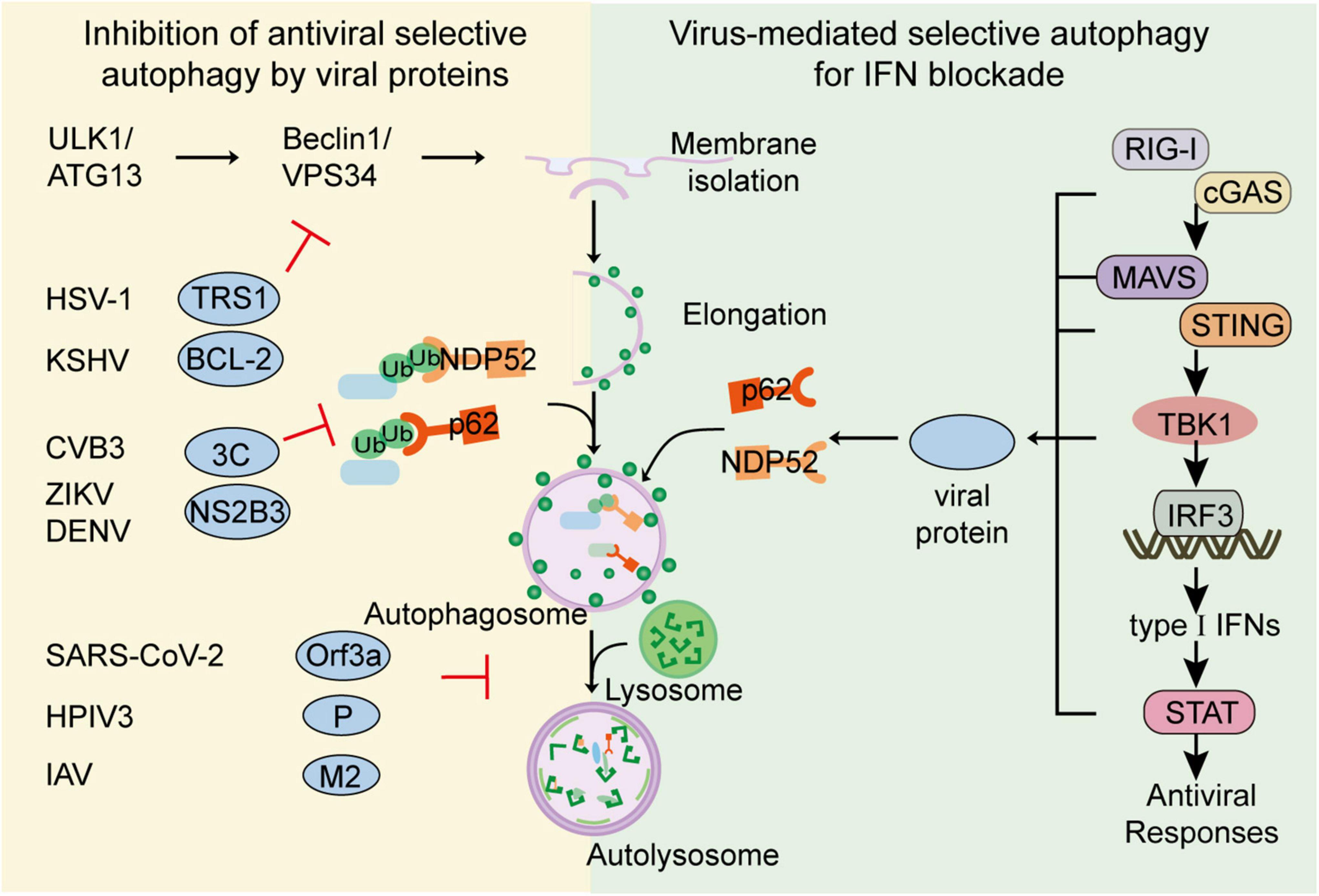
Figure 2. Viral evasion against selective autophagy. To escape the restriction of selective autophagy, viruses have evolved multiple strategies to subvert selective autophagy. Viruses such as CVB3, DENV could directly cleave cargo receptors. Viruses such as HSV-1 or KSHV could inhibit autophagy activation through ATG proteins. Viruses such as SARS-CoV-2 and IAV could restrict the fusion of autophagosome and lysosome for their replication. Additionally, to optimize replication capacity, viruses including H7N9, SARS-CoV-2 and BTV, could also utilize selective autophagy to minimize host antiviral responses. Viruses such as HSV-1, H7N9, and SARS-CoV-2, could utilize cargo receptors to target IFN signal molecules for autophagic degradation.
As the co-evolution between virus and host has taken place over a long period, several viruses have developed various strategies to counteract the restriction initiated by autophagy. The direct strategy employed by viruses to reduce selective autophagy is inducing cargo receptors cleavage. The picornavirus, coxsackievirus B3 (CVB3) utilizes its protease 2A and 2C to cleave cargo receptors p62, NBR and NDP52, which can target CVB3 VP1 capsid protein for degradation (Shi et al., 2013, 2014; Mohamud et al., 2019). SVV 3Cpro could target p62 for cleavage, to abolish the capacity of selective autophagy (Wen et al., 2021). Aichi virus 3C protease reduces p62-mediated antiviral response by targeting p62 and LC3 (Kung et al., 2020). Dengue virus (DENV) is shown to promote proteasomal degradation of p62 to restrict autophagic flux (Metz et al., 2015). NS2B3 from DENV and ZIKV can cleave ER-phagy receptor FAM134B to facilitate viral replication (Lennemann and Coyne, 2017).
In addition to cleave cargo receptors, viruses also decrease autophagic flux by inhibiting the activation of ATG proteins. HSV-1 could use its ICP34.5, called TRS1, to restrict autophagy by targeting Beclin-1 (Orvedahl et al., 2007). Kaposi’s sarcoma-associated herpesvirus (KSHV) bears viral homologues of BCL-2, which can compete with cellular BCL-2 for Beclin-1 binding, thereby antagonizing autophagy (Cuconati and White, 2002; Pattingre et al., 2005; Ku et al., 2008). HIV Nef is reported to reduce autophagic maturation by targeting Beclin-1, thus restricting autophagic processing of HIV (Kyei et al., 2009). 2AB protein of Seneca virus A (SVA) is reported to antagonize selective autophagy process by degrading LC3 and inhibit autophagic degradation of viral 3C protein (Sun et al., 2021).
Viruses could also impair selective autophagic degradation by recruiting different host factors. For example, an accessory protein Orf3a of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) recruits and sequestrates VPS39, the component of host HOPS complex and inhibits the association between HOPS and STX17 or RAB7, thereby preventing the autophagosome-lysosome fusion (Hayn et al., 2021; Koepke et al., 2021; Miao et al., 2021). Similarly, viral phosphoprotein (P) of human parainfluenza virus type 3 (HPIV3) could interact with SNAP29, which restricts the association between SNAP29 and STX17 and inhibits the fusion between autophagosome and lysosome (Ding et al., 2014). Matrix protein 2 of influenza A virus (IAV) is also shown to promote accumulation of autophagosomes by blocking the fusion of autolysosome, thus inhibiting autophagosome degradation and compromising the survival of IAV-infected cells (Gannage et al., 2009; Figure 2).
Type I IFN responses provide the first line of defense against invading viruses. By recognizing cytosolic viral nucleic acid, host RNA sensors including retinoic acid-inducible gene I (RIG-I) -I-like receptors (RLRs) and DNA sensors including cyclic GMP-AMP Synthase (cGAS), initiate signaling cascade through their adaptor proteins, mitochondrial antiviral signaling srotein (MAVS) and stimulator of interferon genes (STING), respectively. Once activated, these adaptors recruit downstream kinase TANK binding kinase 1 (TBK1) to the signalosomes, which subsequently phosphorylate transcription factor interferon regulatory factor 3 (IRF3) to activate type I IFN transcriptions. After binding to a series of cognate receptors, secreted IFN productions induce the expression of various IFN-stimulated genes and initiate antiviral responses (Akira et al., 2006; Loo and Gale, 2011; Schneider et al., 2014). Although signal transmission of type I IFN is critical for antiviral responses, excessive IFN responses would trigger severe immunopathology. Thus, type I IFN signaling requires stringent regulation to ensure effective but not self-injurious antiviral responses. Accumulating evidence reveals that selective autophagy is extensively involved in the regulation of host immunity (Wu and Cui, 2019; Viret et al., 2021). To avoid hyperactivation of IFN responses, several cargo receptors are reported to mediate the selective autophagy of the key signal components at almost each level of IFN signaling pathway, including RIG-I, cGAS, MAVS, STING, TBK1, and IRF3 (Chen et al., 2016; Jin et al., 2017; Du et al., 2018; Wu and Cui, 2019; Wu et al., 2021; Xie et al., 2022). However, recent studies revealed several viruses are capable to subvert selective autophagy to counteract IFN signaling cascade by promoting autophagic degradation of key components in IFN pathway. To attenuate the activation of IFN signaling, porcine circoviruses type 2 (PCV2) could induce phosphorylation of cGAS at S278 via activation of PI3K/Akt signaling which abolishes cGAS’s catalytic activity, and subsequently promote the K48-linked ubiquitination of cGAS, which is targeted by HDAC6 and delivered to autolysosome for degradation (Wang Z. et al., 2021). In addition to degrade viral receptors, several viruses promote the autophagic degradation of adaptor proteins in IFN pathway to block signal amplification. PB1 protein of influenza A virus subtype H7N9 (H7N9) is shown to recruit host E3 ligase RNF5 to induce K27-linked ubiquitination of MAVS at its Lys362 and Lys461, which will subsequently be recognized by cargo receptor NBR1 and lead to its autophagic degradation. Consequently, H7N9 PB1 protein-mediated degradation of MAVS leads to lessened RIG-I-MAVS IFN signaling and enhanced H7N9 infection (Zeng et al., 2021). SARS-CoV-2 helicase NSP13 can promote the autophagic degradation of TBK1, the central kinase in IFN pathway in cooperation with cargo receptor p62, thus in turn inhibiting IFN productions (Sui et al., 2022). Besides restricting the generation of type I IFN, viruses may also enforce selective autophagy to further soften IFN signal transduction. Bluetongue virus (BTV) NS3 is responsible to recruit host E3 ligase to ubiquitinate transcription factor STAT2 and induce its autophagic degradation for IFN blockade (Avia et al., 2019). Apart from cutting off IFN antiviral responses, viruses can compel cargo receptors to degrade host factors directly to enhance replication. HIV gp120 interacts with stathmin and trigger its autophagic degradation to enhance mucosal permeability for viral infection (Xie et al., 2020), and stress-induced corticosterone can induce autophagic degradation of PML, a famous antiviral ISGs, to increase susceptibility to HSV-1 in brain (Maarifi et al., 2014; Li W. et al., 2020). Collectively, virus-utilized selective autophagy may be involved in every step of IFN antiviral process from IFN production to antiviral factors, thus benefiting viral infection (Figure 2).
According to the category of degradative substrates, autophagy can be divided into several specific subsets, including mitophagy for mitochondria, aggrephagy for aggregated proteins, nucleophagy for nucleus, ferritinophagy for ferritin, and lipophagy for lipid droplets (Stolz et al., 2014; Kirkin, 2020). In order to ensure efficient replication, viruses employ and even activate certain forms of organelles-specific autophagy (Vo and Choi, 2021, Table 2).
Mitophagy, the most-studied organelle-specific autophagy, is the selective clearance of damaged or surplus mitochondria process, which acts as an important role in mitochondrial quality and quantity control. Hypoxia, mitochondrial depolarization and viral infection can induce mitophagy. In mammalian cells, mitophagy can be processed in E3 ligase parkin RBR E3 ubiquitin protein ligase (PRKN)-dependent and independent way (Kim et al., 2007; Youle and Narendra, 2011). In PRKN-dependent mitophagy, PINK1 accumulates on the outer membrane of mitochondria and recruits PRKN, which can link more ubiquitin chains to mitochondria to promote the engulfment of mitochondria by phagophores to form mitophagosomes (Kondapalli et al., 2012). In PRKN-independent mitophagy, several mitophagy receptors located on the outer membrane of mitochondria, including FUNDC1, NIX, BCL2L13, and BNIP3, can interact with LC3 through LIR motif and promote the engulfment of mitochondria (Schweers et al., 2007; Zhang and Ney, 2009; Liu et al., 2012; Murakawa et al., 2019). After the engulfment of mitochondria, the phagophores form mature mitophagosomes, then fuse with lysosomes and degrade the mitochondria (Youle and Narendra, 2011).
During viral infection, portions of viral proteins can influence the process of mitophagy to shape an environment for viral replication (Zhang L. et al., 2018). The activation of RIG-I-mediated IFN antiviral responses depends on the oligomerization of MAVS, which located on mitochondrial outer membrane, therefore mitophagy suppresses the activation of IFN responses to a certain extent (Jacobs and Coyne, 2013). Accordingly, RNA viruses evolved mechanisms to achieve immune evasion by inhibiting the activation of IFN pathway through promoting mitophagy. For example, HPIV3 matrix protein and IAV F2 protein can interact with mitochondrial elongation factor TUFM and LC3 to induce mitophagy, which inhibits the activation of IFN responses (Ding et al., 2017; Wang R. et al., 2021). NS1 of influenza virus could disrupt activity of mitochondria and enhance mitophagy via host ULK1 and BNIP3 (Lee et al., 2021). Moreover, DNA virus human herpes virus 8 (HHV-8) can also induce mitophagy through the interaction between viral protein vIRF-1 and mitophagy receptor NIX, which can promote cell survival via reducing mitochondria-induced cytotoxicity (Vo et al., 2019). HHV-8 vIRF-1 is also noted to promote mitophagy and suppress caspase-8-mediated apoptosis through interacting with TUFM, thus promoting the survival of lytic infected cells (Choi et al., 2022).
Apoptosis is a classic and well-studied programmed cell death modality. It has been widely reported that the apoptosis of viral infected cells is one of the crucial immune responses against viral infection (Kvansakul, 2017). The initiation of apoptosis is also dependent on mitochondria, which could be degraded and inhibited by mitophagy to promote viral replication (Galluzzi et al., 2008). Indeed, several viruses have developed strategies to prevent apoptosis via inducing mitophagy. Classic swine fever virus (CSFV) NS3 can interact with lactate dehydrogenase B (LDHB) and decrease the expression of LDHB, which leads to the activation of mitophagy and inhibition of apoptosis, thus enhancing the replication of CSFV (Fan et al., 2021). As the reinforcement of mitophagy in promoting viral propagation, targeting mitophagy may become a potential therapeutic treatment against certain viral infectious diseases.
Protein aggregation is generated and accumulated as a result of improper folding or misfolding due to mutation, incomplete translation, abnormal post translational modification and oxidative stress (Dobson, 2003). There are two major protein quality control mechanisms including molecular chaperones and ubiquitin-proteasome system (UPS) (Kaufman et al., 2002). Chaperones are proteins that assist in the assembly and folding of proteins, which can recognize and repair misfolded proteins (Bukau and Horwich, 1998). UPS is another major protein degradation system, which can recognize those ubiquitinated soluble misfolded proteins (Kaufman et al., 2002). Protein aggregation can also be recognized and eliminated by selective autophagy, which called aggrephagy (Lamark and Johansen, 2012). The initiation of aggrephagy is mediated by several autophagy receptors including Toll interacting protein (TOLLIP), p62/SQSTM1, NBR1, and OPTN (Kirkin et al., 2009; Clausen et al., 2010; Korac et al., 2013; Muscolino et al., 2020). It has been reported that viruses can utilize aggrephagy to escape from immune surveillance. HCMV M45 protein can induce the aggregation of host proteins receptor-interacting serine/threonine-protein kinase 1 (RIPK1) and NF-kappaB essential modulator (NEMO) (Muscolino et al., 2020), which functions in the activation of NF-κB signaling pathway and apoptosis/necroptosis pathway through its IPAM motif (Kondylis et al., 2017). The aggregation of RIPK1 and NEMO would abolish their functions and induces the selective degradation, thereby resulting in the inhibition of premature cell death and viral replication (Muscolino et al., 2020). In addition, HSV-1 ICP6 protein is demonstrated to induce RIPK1 aggregation and degradation in a similar pattern as HCMV M45 does, suggesting the pro-viral role of aggrephagy in virus infection (Muscolino et al., 2020).
Lipid droplets (LDs), organelles with a monolayer phospholipid membrane, are the main storage site of intracellular neutral lipids (Olzmann and Carvalho, 2019). Multiple lipids including cholesterol esters and triglycerides are stored within LDs. The reservation of lipids is indispensable for the energy metabolism and lipid homeostasis, however, excessive accumulation of lipid can lead to lipotoxicity and cell damage, therefore, the biogenesis and degradation of LDs are required to be an issue of dynamic equilibrium (Jones, 2016). Lipophagy is a subset of selective autophagy for degradation of LDs. Cells can regulate lipid metabolism through selectively degrading triglycerides stored in lipid droplets via lipophagy to free fatty acids by the lysosomal acid lipase (LAL) and generating ATP (Singh and Cuervo, 2012; Ward et al., 2016). Several cargo receptors are involved in lipophagy (Shin, 2020). Adipose triglyceride lipase (ATGL) can bind to LDs and interact with LC3 via LIR motif, straightly inducing lipophagy (Sathyanarayan et al., 2017). Small G protein Rab family and lipase located on LDs surface such as patatin-like phospholipase domain-containing enzyme 5 (PNPLA5) and patatin-like phospholipase domain-containing enzyme 8 (PNPLA8) also contribute to the recognition and engulfment process of lipophagy (Shin, 2020). Considering the momentous role of lipophagy in lipid metabolism regulation, and given the close relationship between lipid and the regulation of immune system (Yan and Horng, 2020), it is necessary to comprehensively explore the association between lipophagy and viral infection.
The most studied viruses associated with lipophagy are flaviviruses. DENV NS4A and NS4B can interact with LDs surface protein AUP1 and inhibit the acyltransferase activity of AUP1, thus facilitating lipophagy and promoting the generation of ATP for virus replication (Heaton and Randall, 2010; Zhang J. et al., 2018). It is noteworthy that DENV infection activates lipophagy in an AMP-activated protein kinase (AMPK)-dependent manner, which is unnecessary in autophagy activation (Jordan and Randall, 2017). Meanwhile, NS5 of HCV and capsid protein of DENV can accumulate on the surface of LDs, which can promote the assembly of virions (Samsa et al., 2009; Filipe and McLauchlan, 2015). Thus, the association between virus-induced lipophagy and LDs accumulation requires further investigations. In addition to flaviviruses, porcine reproductive and respiratory syndrome virus (PRRSV) infection can decrease the expression of N-Myc downstream-regulated gene 1 (NDRG1) to promote lipophagy and therefore enhance virus replication (Wang et al., 2019). Future studies should focus on how to inhibit the viral-advantageous lipophagy, while attenuating the lipotoxicity caused by the LDs accumulation is a potential therapeutic goal.
Ferritinophagy is another subset of selective autophagy, refer to the selective autophagic degradation of ferritin, an iron-sequestering protein complex (Kaur and Debnath, 2015). Iron is an important nutrient that can affect the activity of iron-dependent enzymes and then influence a variety of different biochemical reaction processes in cells (Cassat and Skaar, 2013). Excessive free iron will induce the accumulation of lipid peroxidation product and cause the oxidative stress and cytotoxicity (Pantopoulos et al., 2012). The balance of intracellular iron metabolism, an essential cellular process for normal cell physiology, which is controlled by ferritinophagy through selective degradation of ferritin (Kaur and Debnath, 2015). NCOA4, the specific ferritinophagy receptor, can interact with FTH1, a subunit of ferritin complex and autophagy protein LC3, hence promotes the engulfment of ferritin complex by phagophores. Upon fusion with lysosomes, ferritin complexes are degraded, thereby resulting in the release of free iron (Mancias et al., 2014). Ferritinophagy mediates the degradation of ferritin and the release of free iron, which contributes ferroptosis and ROS generation via fenton reaction (Ajoolabady et al., 2021). Although it has been reported that viruses can utilize ferroptosis to spread infection (Ashida et al., 2011), ferroptosis can play a positive role in antiviral immune response (Jorgensen et al., 2017). In order to ensure replication, viruses have developed several ways to inhibit ferroptosis caused by ferritinophagy. HCMV pUL38 can interact with host deubiquitinase USP24 which can stabilize ferritinophagy receptor NCOA4. By interacting with USP24, pUL38 can restrict the functions of USP24 and inhibit ferroptosis, thus promoting the proliferation of HCMV (Sun et al., 2018). Moreover, the V protein of human parainfluenza virus 2 (hPIV2) can bind to NOCA4 and ferritin subunit FTH1, thus competitively suppressing ferritinophagy and ferroptosis, resulting in efficient amplification of hPIV2 (Ohta et al., 2021).
As discussed above, selective autophagy plays a vital role during viral infection, which leads us to explore the antiviral interventions by targeting selective autophagy. However, as the double-edged sword effects of autophagy during viral infection, it remains to be challenged to distinguish whether autophagy needs to be upregulated or restricted. Additionally, accumulating evidence reveals that non-specific induction of autophagy as antiviral therapies leads to unexpected side effects with limiting effects on viral evasion. Thus, it is essential for therapy development against infectious diseases by targeting autophagy in a more selective and controlled way with specific degradative cargoes (Table 3).
Macroautophagy is the best-characterized form of autophagy, involving a specialized double-membrane vesicle known as the autophagosome which delivers virus-derived antigens for presentation to T lymphocytes. The outbreak of the novel coronavirus SARS-CoV-2 has infected thousands of people since the end of 2019 and caused worldwide social and economic disruption. Sirolimus (also known as rapamycin) has been shown to be a possible respiratory syndrome inhibitor against virus such as SARS-CoV-2, MERS as well as influenza A virus subtype H1N1 (H1N1) (Keating et al., 2013; Kindrachuk et al., 2015; Husain and Byrareddy, 2020). Beyond that, metformin which has been well-known for its pleiotropic effects also has a possible role in combating SARS-CoV-2 as well as HCV, HBV or HIV through increasing insulin sensitivity and autophagy (Chen et al., 2018; Sharma et al., 2020). As an autophagy inducer, ivermectin is likely to have a significant impact on SARS-CoV-2 by inhibiting the AKT phosphorylation, which has been demonstrated in numerous clinical trials (Bryant et al., 2021). It has been reported that pharmacological inhibition of autophagy using wortmannin—an inhibitor of autophagy by blocking PI3K—is associated to the reduction of ZIKV replication in human umbilical vein endothelial cells (Peng et al., 2018).
Apart from traditional mTOR inhibitors, further investigations had brought the use of autophagy activators to the forefront. A representative compound, CSC27 (manassantin B), abrogates EBV lytic replication by specifically blocking mTORC2-mediated phosphorylation and suppresses the expression of EBV immediate-early gene BZLF1, providing a candidate compound for further development of a new drug with minimal cytotoxicity to treat EBV-associated diseases (Wang Q. et al., 2020). Reminiscent of the role in targeting immune system, corticosteroids is known to inhibit autophagy by blocking LC3 recruitment (Kyrmizi et al., 2013), and its clinical roles in viral infection have been investigated in H1N1, SARS, MERS and SARS-CoV-2 (Auyeung et al., 2005; Arabi et al., 2018; Lipworth et al., 2020).
Chaperon-mediated autophagy (CMA) directly delivers substrate proteins into the lysosome lumen using the cytosolic chaperone HSC70 and the lysosomal receptor LAMP-2A for degradation. Recent studies have shown that HSC70 can be a promising target for antivirals. A monoclonal antibody against HSC70 could block the infectivity of Rotavirus (Pérez-Vargas et al., 2006). Similarly, a selective HSC70 inhibitor, oxymatrine, suppresses de novo synthesis of both the wild-type HBV strain and strains resistant to 3TC, ADV, and ETV, presumably through destabilization of Hsc70 mRNA (Gao et al., 2011). As an ATP analog, adenosine derivative compound VER-155008 acts as a competitive inhibitor to bind the HSP70 family and induces the conformational changes of HSC70, making it impossible for HSC70 to interact with IBDV VP2 protein and thus inhibiting virus replication (Chen et al., 2020).
Chloroquine (CQ) and hydroxychloroquine (HCQ) have been used as antimalarial agents for many years. Based on its effects on the annulation of endosomal/lysosomal acidification, CQ showed its antiviral effects against ZIKV, CHIKV, and HIV (Al-Bari, 2017; Peng et al., 2018). A randomized, triple-blind, placebo-controlled pilot trial reports that treatment with chloroquine reduces the HCV viral RNA levels in non-responders to other viral treatments (Peymani et al., 2016). In 2020, CQ and HCQ were approved for the U.S. Food and Drug Administration (FDA) for the treatment of infected patients with the coronavirus SARS-CoV-2 (Martinez et al., 2020), and its effect had been proved in abundant clinical trials and researches in vitro (Wang M. et al., 2020; Yao et al., 2020). However, the potential adverse effect of CQ and HCQ cannot be ignored since they may make acute kidney injury (AKI) and other organ failures worse (Edelstein et al., 2020). Recent years, more efficient small molecules are under development for instead of CQ and HCQ. ROC-325, a dimeric small molecule containing core motifs of HCQ and lucanthone, has approximately 10-fold greater anticancer activity and autophagic inhibition than HCQ (Jones et al., 2019), and may go into a phase I clinical trial in the near future.
Transcription factor EB (TFEB) is a key transcription molecule that regulates autophagy at the transcriptional level. Given the modulation of TFEB is known to regulate lysosome biogenesis and autophagy, inhibiting TFEB to block the lysosome production may be a promising strategy for host against viruses. Moringa A, a new compound from Moringa oleifera seeds, can inhibit the expression and nuclear transfer of TFEB and weaken the autophagy in infected cells, which could be an important antiviral mechanism (Xiong et al., 2021). Trehalose can activate PIKfyve leading to TFEB nuclear translocation in MCOLN1-dependent manner to induce autophagy. Remarkably, trehalose significantly reduced HIV-p24 levels in ex-vivo-infected PBMCs or PBMCs from therapy naïve HIV patients (Sharma et al., 2021).
Two years of scientific exploration has attested that autophagy plays an essential role in SARS-CoV-2-mediated COVID-19, and modulating autophagy by using various drugs that act on potential targets of the virus seems to be an undoubtedly promising therapeutic solution for SARS-COV2 infection and replication. Preclinical studies have suggested repurposing a few FDA-approved drugs to treat COVID-19, and almost half of these well-recognized medications are thought to be inhibitors of the pathways involved in autophagy (Mijaljica and Klionsky, 2020). As mentioned above, treatments toward macroautophagy have a splendid contribution to anti-coronaviral therapy. However, clinical trials showed that the potential side effects of autophagy inhibitors destroyed more than virus. CQ cannot be regarded as an ideal option as it may cause simultaneous tissue damage when the therapeutic goal was to decelerate replication (Edelstein et al., 2020), while corticosteroid-treated patients were also more likely to develop myocardial injury (15.6 vs. 10.4%, P = 0.041), acute liver injury (18.3 vs. 9.9%, P = 0.001), and shock (22.0 vs. 12.6%, P < 0.001) compared with controls during hospitalization (Liu et al., 2020). Thus, it desires an urgently needed solution for antiviral therapy to look for more specific drug targets with limiting influences on normal physiological processes, in which selective autophagy may play a considerable role. Enhancing p62-mediated selective autophagy, as we mentioned above, may accelerate virus protein clearance of SINV, HSV-1, and AdV (Viret et al., 2021). Stabilization of novel types of autophagosomes involving in the presentation of various phagocytosed antigens by MHC class I and II molecules may help us fighting with virus infection through coordinating adaptive immunity. When viruses apply selective autophagy for IFN blockade, autophagy inhibitors specifically targeting cargo receptors which are involved in the autophagic degradation of key components in IFN pathway could reverse IFN level and enhance antiviral response. However, it is necessary to foster and extend robust research involving all the pros and cons of selective autophagy therapy before it eventually becomes a priority.
Targeting organelle-specific selective autophagy might also provide a potential therapeutic strategy against infectious diseases. During infection, viruses altered mitochondrial dynamics in order to modulate mitochondria-mediated antiviral immune responses via the alteration of mitochondrial events such as mitophagy to facilitate their proliferation (Singh et al., 2021). Valinomycin stimulates mitophagy by loss of MMP, which is shown to inhibit the replication of coronaviruses (Wu et al., 2004; Klein et al., 2011; Zhang et al., 2020). Agonists of the sigma-1 receptor could provide protection of the mitochondria by activating mitophagy to remove damaged and leaking mitochondria. Agonists of the sigma-1 receptor are also shown to prevent cell death in response to HCV and SARS-CoV-2 infection by inducing autophagy (Friesland et al., 2013; Brimson et al., 2021).
The involvements of selective autophagy have been largely established during viral infection. A set of cargo receptors are involved in autophagic clearance viral factors, thus restricting viral replication. The presentation of viral antigens to active host adaptive immunity also requires the participation of autophagy, and the contributions of autophagic receptors still need to be evaluated. On the contrary, viruses have also evolved strategies to counteract selective autophagy, through degrading cargo receptors or inhibiting receptor’s activities through autophagic flux or other host factors. Autophagic receptors-mediated infection-related calibration of IFN signaling is also disturbed by viruses, which could lead to the reduction of IFN antiviral responses. Moreover, several viruses can induce and utilize certain forms of selective autophagy, including mitophagy, aggrephagy, and lipophagy, to benefit viral replication.
Considering close relationship between selective autophagy and viral replication, antiviral interventions targeting selective autophagy are still at a very early stage. How to effectively modulate autophagy, even selective autophagy, remains to be a long-standing challenge in the implementation of autophagy-based therapies. Given the devastating consequences of the current COVID-19 pandemic and its long-lasting impact on all of us, viral infection and specific treatments toward it are with no doubt of high concern. Researchers have attested that cellular degradation mechanism known as autophagy might be a beneficial approach to fight numerous viruses by using inhibitors or enhancers. As autophagy acts as a double-edged sword during viral invasion, potent regulators based on overall induction of autophagy may have the potential to cause unexpected side effects such as shock, superinfection or even life-threaten multiple organ failure. Instead, more selective approaches that allow only particular autophagic pathways or steps to be targeted has come to the center stage of antiviral therapy. Further robust and interdisciplinary investigations are in great need to looking for more appropriate therapies based on specific autophagy modulation, so that they can be implemented to treat highly contagious virus in the future.
JC and YW initiated and designed the project. JW and XG provided suggestions. JH and SJ provided technical assistance. JH draw the schematic diagram. YW, TZ, and JH collect the references. JC, XG, YW, YL, and TZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by Guangdong Clinical Research Center for Critical Care Medicine (2020B1111170005). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
UBD, Ubiquitin-binding domain; LIR, LC3-interacting region; IFN, Interferon; VSV, Vesicular stomatitis virus; ZIKV, Zika virus; SINV, Sindbis virus; HSV-1, Herpes simplex virus type 1; AdV, Adenovirus; CHIKV, Chikungunya alphavirus; FMDV, Foot-and-mouth disease virus; MeV, Measles virus; IBDV, Infectious bursal disease virus; SVV, Seneca valley virus; UBA, Ubiquitin associated domain; HBV, Hepatitis B virus; HIV, Human immunodeficiency virus; CaMV, Cauliflower mosaic virus; FANCC, Fanconi anemia group C protein; SMURF1, SMAD ubiquitin regulatory factor 1; ER, Endoplasmic reticulum; HCV, Hepatitis C virus; MHC, Major Histocompatibility complex; EBV, Epstein-Barr virus; HCMV, Human cytomegalovirus; TAP, Transporters associated with antigen processing; CVB3, Coxsackievirus; AIV, Aichi virus; DENV, Dengue virus; ATGs, Autophagy-related genes; KSHV, Kaposi’s sarcoma-associated herpesvirus; SVA, Seneca virus A; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus-2; HOPS, Homotypic fusion and protein sorting; HPIV3, Human Parainfluenza virus 3; IAV, Influenza A virus; RLRs, Retinoic acid-inducible gene I (RIG-I) -I-like receptors; cGAS, cyclic GMP-AMP Synthase; MAVS, Mitochondrial Antiviral Signaling Protein; STING, Stimulator of Interferon Genes; TBK1, TANK Binding Kinase 1; IRF3, Interferon Regulatory Factor 3; PCV2, Porcine circoviruses type 2; H7N9, influenza A virus subtype H7N9; BTV, Bluetongue virus; ISGs, Interferon-stimulated genes; PRKN, Parkin RBR E3 Ubiquitin Protein Ligase; HHV-8, Human herpes virus 8; CSFV, Classic swine fever virus; UPS, Ubiquitin-proteasome system; TOLLIP, Toll Interacting Protein; RIPK1, Receptor-interacting serine/threonine-protein kinase 1; IPAM, Induced protein aggregation motif; LDs, Lipid droplets; LAL, Lysosomal acid lipase; ATGL, Adipose triglyceride lipase; PRRSV, Porcine reproductive and respiratory syndrome virus; ROS, Reactive oxygen species; HPIV2, Human parainfluenza virus 2; MERS, Middle east respiratory syndrome coronavirus; H1N1, influenza A virus subtype H1N1; CMA, Chaperon-mediated autophagy; 3TC, Lamivudine; ADV, Adefovir; ETV, Entecavir; CQ, Chloroquine; HCQ, Hydroxychloroquine; FDA, U.S food and drug administration; AKI, Acute kidney injury; TFEB, Transcription factor EB; PBMCs, Peripheral blood mononuclear cells; MMP, Mitochondrial membrane potential.
Ajoolabady, A., Aslkhodapasandhokmabad, H., Libby, P., Tuomilehto, J., Lip, G. Y. H., Penninger, J. M., et al. (2021). Ferritinophagy and ferroptosis in the management of metabolic diseases. Trends Endocrinol. Metab. 32, 444–462. doi: 10.1016/j.tem.2021.04.010
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Al-Bari, M. A. A. (2017). Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 5:e00293. doi: 10.1002/prp2.293
Ames, J., Yadavalli, T., Suryawanshi, R., Hopkins, J., Agelidis, A., Patil, C., et al. (2021). OPTN is a host intrinsic restriction factor against neuroinvasive HSV-1 infection. Nat. Commun. 12:5401. doi: 10.1038/s41467-021-25642-z
Arabi, Y. M., Mandourah, Y., Al-Hameed, F., Sindi, A. A., Almekhlafi, G. A., Hussein, M. A., et al. (2018). Corticosteroid therapy for critically Ill patients with middle east respiratory syndrome. Am. J. Respir. Crit. Med. 197, 757–767. doi: 10.1164/rccm.201706-1172OC
Ashida, H., Mimuro, H., Ogawa, M., Kobayashi, T., Sanada, T., Kim, M., et al. (2011). Cell death and infection: a double-edged sword for host and pathogen survival. J. Cell Biol. 195, 931–942. doi: 10.1083/jcb.201108081
Auyeung, T. W., Lee, J. S. W., Lai, W. K., Choi, C. H., Lee, H. K., Lee, J. S., et al. (2005). The use of corticosteroid as treatment in SARS was associated with adverse outcomes: a retrospective cohort study. J. Infect. 51, 98–102.
Avia, M., Rojas, J. M., Miorin, L., Pascual, E., Van Rijn, P. A., Martin, V., et al. (2019). Virus-induced autophagic degradation of STAT2 as a mechanism for interferon signaling blockade. EMBO Rep. 20:e48766. doi: 10.15252/embr.201948766
Berryman, S., Brooks, E., Burman, A., Hawes, P., Roberts, R., Netherton, C., et al. (2012). Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J. Virol. 86, 12940–12953. doi: 10.1128/JVI.00846-12
Bhujabal, Z., Birgisdottir, A. B., Sjottem, E., Brenne, H. B., Overvatn, A., Habisov, S., et al. (2017). FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 18, 947–961. doi: 10.15252/embr.201643147
Brimson, J. M., Prasanth, M. I., Malar, D. S., Brimson, S., Thitilertdecha, P., and Tencomnao, T. (2021). Drugs that offer the potential to reduce hospitalization and mortality from SARS-CoV-2 infection: the possible role of the sigma-1 receptor and autophagy. Expert Opin. Therap. Target. 25, 435–449. doi: 10.1080/14728222.2021.1952987
Bryant, A., Lawrie, T. A., Dowswell, T., Fordham, E. J., Mitchell, S., Hill, S. R., et al. (2021). Ivermectin for prevention and treatment of COVID-19 infection: a systematic review, meta-analysis, and trial sequential analysis to inform clinical guidelines. Am. J. Therap. 28, e434–e460. doi: 10.1097/MJT.0000000000001402
Bukau, B., and Horwich, A. L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366. doi: 10.1016/s0092-8674(00)80928-9
Cassat, J. E., and Skaar, E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. doi: 10.1016/j.chom.2013.04.010
Chen, C., Qin, Y., Qian, K., Shao, H., Ye, J., and Qin, A. (2020). HSC70 is required for infectious bursal disease virus (IBDV) infection in DF-1 cells. Virol. J. 17:65. doi: 10.1186/s12985-020-01333-x
Chen, M., Meng, Q., Qin, Y., Liang, P., Tan, P., He, L., et al. (2016). TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell 64, 105–119. doi: 10.1016/j.molcel.2016.08.025
Chen, Q., Xiao, Y., Chai, P., Zheng, P., Teng, J., and Chen, J. (2019). ATL3 Is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 29, 846–855 e846. doi: 10.1016/j.cub.2019.01.041
Chen, Y., Gu, F., and Guan, J.-L. (2018). Metformin might inhibit virus through increasing insulin sensitivity. Chin. Med. J. 131, 376–377. doi: 10.4103/0366-6999.223856
Chino, H., Hatta, T., Natsume, T., and Mizushima, N. (2019). Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell 74:906–921.e6. doi: 10.1016/j.molcel.2019.03.033
Choi, C. Y., Vo, M. T., Nicholas, J., and Choi, Y. B. (2022). Autophagy-competent mitochondrial translation elongation factor TUFM inhibits caspase-8-mediated apoptosis. Cell Death Differ. 29, 451–464. doi: 10.1038/s41418-021-00868-y
Choi, Y., Bowman, J. W., and Jung, J. U. (2018). Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 16, 341–354. doi: 10.1038/s41579-018-0003-6
Clausen, T. H., Lamark, T., Isakson, P., Finley, K., Larsen, K. B., Brech, A., et al. (2010). p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344. doi: 10.4161/auto.6.3.11226
Cuconati, A., and White, E. (2002). Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 16, 2465–2478. doi: 10.1101/gad.1012702
Deretic, V., Saitoh, T., and Akira, S. (2013). Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737. doi: 10.1038/nri3532
Dikic, I., and Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364. doi: 10.1038/s41580-018-0003-4
Ding, B., Zhang, G., Yang, X., Zhang, S., Chen, L., Yan, Q., et al. (2014). Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 15, 564–577. doi: 10.1016/j.chom.2014.04.004
Ding, B., Zhang, L., Li, Z., Zhong, Y., Tang, Q., Qin, Y., et al. (2017). The matrix protein of human Parainfluenza Virus type 3 induces mitophagy that suppresses interferon responses. Cell Host Microbe 21, 538–547.64. doi: 10.1016/j.chom.2017.03.004
Dru, P., Bras, F., Dezelee, S., Gay, P., Petitjean, A. M., Pierre-Deneubourg, A., et al. (1993). Unusual variability of the Drosophila melanogaster ref(2)P protein which controls the multiplication of sigma rhabdovirus. Genetics 133, 943–954. doi: 10.1093/genetics/133.4.943
Du, Y., Duan, T., Feng, Y., Liu, Q., Lin, M., Cui, J., et al. (2018). LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 37, 351–366. doi: 10.15252/embj.201796781
Dupont, N., Chauhan, S., Arko-Mensah, J., Castillo, E. F., Masedunskas, A., Weigert, R., et al. (2014). Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 24, 609–620. doi: 10.1016/j.cub.2014.02.008
Edelstein, C. L., Venkatachalam, M. A., and Dong, Z. (2020). Autophagy inhibition by chloroquine and hydroxychloroquine could adversely affect acute kidney injury and other organ injury in critically ill patients with COVID-19. Kidney Int. 98, 234–235. doi: 10.1016/j.kint.2020.05.001
English, L., Chemali, M., Duron, J., Rondeau, C., Laplante, A., Gingras, D., et al. (2009). Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10, 480–487. doi: 10.1038/ni.1720
Fan, S., Wu, K., Zhao, M., Yuan, J., Ma, S., Zhu, E., et al. (2021). LDHB inhibition induces mitophagy and facilitates the progression of CSFV infection. Autophagy 17, 2305–2324. doi: 10.1080/15548627.2020.1823123
Filipe, A., and McLauchlan, J. (2015). Hepatitis C virus and lipid droplets: finding a niche. Trends Mol. Med. 21, 34–42. doi: 10.1016/j.molmed.2014.11.003
Friesland, M., Mingorance, L., Chung, J., Chisari, F. V., and Gastaminza, P. (2013). Sigma-1 receptor regulates early steps of viral RNA replication at the onset of hepatitis C virus infection. J. Virol. 87, 6377–6390. doi: 10.1128/JVI.03557-12
Galluzzi, L., Brenner, C., Morselli, E., Touat, Z., and Kroemer, G. (2008). Viral control of mitochondrial apoptosis. PLoS Pathog. 4:e1000018. doi: 10.1371/journal.ppat.1000018
Gannage, M., Dormann, D., Albrecht, R., Dengjel, J., Torossi, T., Ramer, P. C., et al. (2009). Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6, 367–380. doi: 10.1016/j.chom.2009.09.005
Gao, L.-M., Han, Y.-X., Wang, Y.-P., Li, Y.-H., Shan, Y.-Q., Li, X., et al. (2011). Design and synthesis of oxymatrine analogues overcoming drug resistance in hepatitis B virus through targeting host heat stress cognate 70. J. Med. Chem. 54, 869–876. doi: 10.1021/jm101325h
Grumati, P., Morozzi, G., Holper, S., Mari, M., Harwardt, M. I., Yan, R., et al. (2017). Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6:e25555. doi: 10.7554/eLife.25555
Hafren, A., Macia, J. L., Love, A. J., Milner, J. J., Drucker, M., and Hofius, D. (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. U.S.A. 114, E2026–E2035. doi: 10.1073/pnas.1610687114
Hayn, M., Hirschenberger, M., Koepke, L., Nchioua, R., Straub, J. H., Klute, S., et al. (2021). Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 35:109126. doi: 10.1016/j.celrep.2021.109126
Heaton, N. S., and Randall, G. (2010). Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8, 422–432. doi: 10.1016/j.chom.2010.10.006
Huang, J., and Brumell, J. H. (2014). Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microbiol. 12, 101–114. doi: 10.1038/nrmicro3160
Husain, A., and Byrareddy, S. N. (2020). Rapamycin as a potential repurpose drug candidate for the treatment of COVID-19. Chem. Biol. Interact. 331:109282. doi: 10.1016/j.cbi.2020.109282
Ireland, J. M., and Unanue, E. R. (2011). Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J. Exp. Med. 208, 2625–2632. doi: 10.1084/jem.20110640
Jacobs, J. L., and Coyne, C. B. (2013). Mechanisms of MAVS regulation at the mitochondrial membrane. J. Mol. Biol. 425, 5009–5019. doi: 10.1016/j.jmb.2013.10.007
Jin, S., Tian, S., Luo, M., Xie, W., Liu, T., Duan, T., et al. (2017). Tetherin suppresses Type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol. Cell 68, 308–322.e304. doi: 10.1016/j.molcel.2017.09.005
Jones, J. G. (2016). Hepatic glucose and lipid metabolism. Diabetologia 59, 1098–1103. doi: 10.1007/s00125-016-3940-5
Jones, T. M., Espitia, C., Wang, W., Nawrocki, S. T., and Carew, J. S. (2019). Moving beyond hydroxychloroquine: the novel lysosomal autophagy inhibitor ROC-325 shows significant potential in preclinical studies. Cancer Commun. (London, England) 39:72. doi: 10.1186/s40880-019-0418-0
Jordan, T. X., and Randall, G. (2017). Dengue virus activates the AMP Kinase-mTOR axis to stimulate a proviral lipophagy. J. Virol. 91:e02020-16. doi: 10.1128/JVI.02020-16
Jorgensen, I., Rayamajhi, M., and Miao, E. A. (2017). Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164. doi: 10.1038/nri.2016.147
Judith, D., Mostowy, S., Bourai, M., Gangneux, N., Lelek, M., Lucas-Hourani, M., et al. (2013). Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep. 14, 534–544. doi: 10.1038/embor.2013.51
Kaufman, R. J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C. Y., et al. (2002). The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell Biol. 3, 411–421. doi: 10.1038/nrm829
Kaur, J., and Debnath, J. (2015). Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 16, 461–472. doi: 10.1038/nrm4024
Keating, R., Hertz, T., Wehenkel, M., Harris, T. L., Edwards, B. A., McClaren, J. L., et al. (2013). The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat. Immunol. 14, 1266–1276. doi: 10.1038/ni.2741
Khaminets, A., Heinrich, T., Mari, M., Grumati, P., Huebner, A. K., Akutsu, M., et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. doi: 10.1038/nature14498
Kim, I., Rodriguez-Enriquez, S., and Lemasters, J. J. (2007). Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253. doi: 10.1016/j.abb.2007.03.034
Kim, K. Y., Jang, H. J., Yang, Y. R., Park, K. I., Seo, J., Shin, I. W., et al. (2016). SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci. Rep. 6:35732. doi: 10.1038/srep35732
Kim, N., Kim, M. J., Sung, P. S., Bae, Y. C., Shin, E. C., and Yoo, J. Y. (2016). Interferon-inducible protein SCOTIN interferes with HCV replication through the autolysosomal degradation of NS5A. Nat. Commun. 7:10631. doi: 10.1038/ncomms10631
Kindrachuk, J., Ork, B., Hart, B. J., Mazur, S., Holbrook, M. R., Frieman, M. B., et al. (2015). Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Chemother. 59, 1088–1099. doi: 10.1128/AAC.03659-14
Kirkin, V. (2020). History of the selective autophagy research: how did it begin and where does it stand today? J. Mol. Biol. 432, 3–27. doi: 10.1016/j.jmb.2019.05.010
Kirkin, V., Lamark, T., Sou, Y. S., Bjorkoy, G., Nunn, J. L., Bruun, J. A., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516. doi: 10.1016/j.molcel.2009.01.020
Kirkin, V., and Rogov, V. V. (2019). A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell 76, 268–285. doi: 10.1016/j.molcel.2019.09.005
Klein, B., Wörndl, K., Lütz-Meindl, U., and Kerschbaum, H. H. (2011). Perturbation of intracellular K(+) homeostasis with valinomycin promotes cell death by mitochondrial swelling and autophagic processes. Apoptosis 16, 1101–1117. doi: 10.1007/s10495-011-0642-9
Koepke, L., Hirschenberger, M., Hayn, M., Kirchhoff, F., and Sparrer, K. M. (2021). Manipulation of autophagy by SARS-CoV-2 proteins. Autophagy 17, 2659–2661. doi: 10.1080/15548627.2021.1953847
Kondapalli, C., Kazlauskaite, A., Zhang, N., Woodroof, H. I., Campbell, D. G., Gourlay, R., et al. (2012). PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2:120080. doi: 10.1098/rsob.120080
Kondylis, V., Kumari, S., Vlantis, K., and Pasparakis, M. (2017). The interplay of IKK. NF-kappaB and RIPK1 signaling in the regulation of cell death, tissue homeostasis and inflammation. Immunol. Rev. 277, 113–127. doi: 10.1111/imr.12550
Korac, J., Schaeffer, V., Kovacevic, I., Clement, A. M., Jungblut, B., Behl, C., et al. (2013). Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 126(Pt 2), 580–592. doi: 10.1242/jcs.114926
Ku, B., Woo, J. S., Liang, C., Lee, K. H., Hong, H. S., and E X, et al. (2008). Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 4:e0040025. doi: 10.1371/journal.ppat.0040025
Kung, M. H., Lin, Y. S., and Chang, T. H. (2020). Aichi virus 3C protease modulates LC3- and SQSTM1/p62-involved antiviral response. Theranostics 10, 9200–9213. doi: 10.7150/thno.47077
Kyei, G. B., Dinkins, C., Davis, A. S., Roberts, E., Singh, S. B., Dong, C., et al. (2009). Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186, 255–268. doi: 10.1083/jcb.200903070
Kyrmizi, I., Gresnigt, M. S., Akoumianaki, T., Samonis, G., Sidiropoulos, P., Boumpas, D., et al. (2013). Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J. Immunol. (Baltimore, MD: 1950) 191, 1287–1299. doi: 10.4049/jimmunol.1300132
Lamark, T., and Johansen, T. (2012). Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012:736905. doi: 10.1155/2012/736905
Lazarou, M., Sliter, D. A., Kane, L. A., Sarraf, S. A., Wang, C., Burman, J. L., et al. (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. doi: 10.1038/nature14893
Lee, J. H., Oh, S. J., Yun, J., and Shin, O. S. (2021). Nonstructural protein NS1 of Influenza virus disrupts mitochondrial dynamics and enhances mitophagy via ULK1 and BNIP3. Viruses 13:1845. doi: 10.3390/v13091845
Lennemann, N. J., and Coyne, C. B. (2017). Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 13, 322–332. doi: 10.1080/15548627.2016.1265192
Li, W., Luo, Z., Yan, C. Y., Wang, X. H., He, Z. J., Ouyang, S. H., et al. (2020). Autophagic degradation of PML promotes susceptibility to HSV-1 by stress-induced corticosterone. Theranostics 10, 9032–9049. doi: 10.7150/thno.46921
Li, Y., Hu, B., Ji, G., Zhang, Y., Xu, C., Lei, J., et al. (2020). Cytoplasmic cargo receptor p62 inhibits avibirnavirus replication by mediating autophagic degradation of viral protein VP2. J. Virol. 94:e01255-20. doi: 10.1128/JVI.01255-20
Liang, S., Wu, Y. S., Li, D. Y., Tang, J. X., and Liu, H. F. (2021). Autophagy in viral infection and pathogenesis. Front. Cell Dev. Biol. 9:766142. doi: 10.3389/fcell.2021.766142
Liang, X. H., Kleeman, L. K., Jiang, H. H., Gordon, G., Goldman, J. E., Berry, G., et al. (1998). Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72, 8586–8596. doi: 10.1128/JVI.72.11.8586-8596.1998
Lipworth, B., Kuo, C. R., Lipworth, S., and Chan, R. (2020). Inhaled Corticosteroids and COVID-19. Am. J. Respir. Crit. Care Med. 202, 899–900. doi: 10.1164/rccm.202005-2000LE
Liu, J., Zhang, S., Dong, X., Li, Z., Xu, Q., Feng, H., et al. (2020). Corticosteroid treatment in severe COVID-19 patients with acute respiratory distress syndrome. J. Clin. Invest. 130, 6417–6428. doi: 10.1172/JCI140617
Liu, L., Feng, D., Chen, G., Chen, M., Zheng, Q., Song, P., et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–185. doi: 10.1038/ncb2422
Liu, Y., Gordesky-Gold, B., Leney-Greene, M., Weinbren, N. L., Tudor, M., and Cherry, S. (2018). Inflammation-induced, STING-dependent autophagy restricts Zika Virus infection in the Drosophila brain. Cell Host Microbe 24, 57–68 e53. doi: 10.1016/j.chom.2018.05.022
Loi, M., Raimondi, A., Morone, D., and Molinari, M. (2019). ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat. Commun. 10:5058. doi: 10.1038/s41467-019-12991-z
Loo, Y. M., and Gale, M. Jr. (2011). Immune signaling by RIG-I-like receptors. Immunity 34, 680–692. doi: 10.1016/j.immuni.2011.05.003
Lu, K., Psakhye, I., and Jentsch, S. (2014). Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158, 549–563. doi: 10.1016/j.cell.2014.05.048
Maarifi, G., Chelbi-Alix, M. K., and Nisole, S. (2014). PML control of cytokine signaling. Cytokine Growth Factor Rev. 25, 551–561. doi: 10.1016/j.cytogfr.2014.04.008
Mancias, J. D., Wang, X., Gygi, S. P., Harper, J. W., and Kimmelman, A. C. (2014). Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109. doi: 10.1038/nature13148
Mandell, M. A., Jain, A., Arko-Mensah, J., Chauhan, S., Kimura, T., Dinkins, C., et al. (2014). TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 30, 394–409. doi: 10.1016/j.devcel.2014.06.013
Martinez, G. P., Zabaleta, M. E., Di Giulio, C., Charris, J. E., and Mijares, M. R. (2020). The role of chloroquine and hydroxychloroquine in immune regulation and diseases. Curr. Pharm. Des. 26, 4467–4485. doi: 10.2174/1381612826666200707132920
Metz, P., Chiramel, A., Chatel-Chaix, L., Alvisi, G., Bankhead, P., Mora-Rodriguez, R., et al. (2015). Dengue virus inhibition of autophagic flux and dependency of viral replication on proteasomal degradation of the autophagy receptor p62. J. Virol. 89, 8026–8041. doi: 10.1128/JVI.00787-15
Miao, G., Zhao, H., Li, Y., Ji, M., Chen, Y., Shi, Y., et al. (2021). ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell 56, 427–442.e5. doi: 10.1016/j.devcel.2020.12.010
Mijaljica, D., and Klionsky, D. J. (2020). Autophagy/virophagy: a “disposal strategy” to combat COVID-19. Autophagy 16, 2271–2272. doi: 10.1080/15548627.2020.1782022
Minton, K. (2016). Autophagy: inflammatory pathology of Fanconi anaemia. Nat. Rev. Mol. Cell Biol. 17, 330–331. doi: 10.1038/nrm.2016.64
Miyakawa, K., Nishi, M., Ogawa, M., Matsunaga, S., Sugiyama, M., Nishitsuji, H., et al. (2022). Galectin-9 restricts hepatitis B virus replication via p62/SQSTM1-mediated selective autophagy of viral core proteins. Nat. Commun. 13:531. doi: 10.1038/s41467-022-28171-5
Mizushima, N., and Levine, B. (2010). Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823–830. doi: 10.1038/ncb0910-823
Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., et al. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668. doi: 10.1083/jcb.152.4.657
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Mohamud, Y., Qu, J., Xue, Y. C., Liu, H., Deng, H., and Luo, H. (2019). CALCOCO2/NDP52 and SQSTM1/p62 differentially regulate coxsackievirus B3 propagation. Cell Death Differ. 26, 1062–1076. doi: 10.1038/s41418-018-0185-5
Montespan, C., Marvin, S. A., Austin, S., Burrage, A. M., Roger, B., Rayne, F., et al. (2017). Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog. 13:e1006217. doi: 10.1371/journal.ppat.1006217
Murakawa, T., Okamoto, K., Omiya, S., Taneike, M., Yamaguchi, O., and Otsu, K. (2019). A mammalian mitophagy receptor, Bcl2-L-13, recruits the ULK1 complex to induce mitophagy. Cell Rep. 26, 338–345 e336. doi: 10.1016/j.celrep.2018.12.050
Muscolino, E., Schmitz, R., Loroch, S., Caragliano, E., Schneider, C., Rizzato, M., et al. (2020). Herpesviruses induce aggregation and selective autophagy of host signalling proteins NEMO and RIPK1 as an immune-evasion mechanism. Nat. Microbiol. 5, 331–342. doi: 10.1038/s41564-019-0624-1
Nakatogawa, H. (2020). Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458. doi: 10.1038/s41580-020-0241-0
Negoita, F., Blomdahl, J., Wasserstrom, S., Winberg, M. E., Osmark, P., Larsson, S., et al. (2019). PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J. Cell Biochem. 120, 343–356. doi: 10.1002/jcb.27378
Ohta, K., Saka, N., and Nishio, M. (2021). Human Parainfluenza virus type 2 V protein modulates iron homeostasis. J. Virol. 95:e01861-20. doi: 10.1128/JVI.01861-20
Olzmann, J. A., and Carvalho, P. (2019). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155. doi: 10.1038/s41580-018-0085-z
Orvedahl, A., Alexander, D., Talloczy, Z., Sun, Q., Wei, Y., Zhang, W., et al. (2007). HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35. doi: 10.1016/j.chom.2006.12.001
Orvedahl, A., MacPherson, S., Sumpter, R. Jr., Talloczy, Z., Zou, Z., and Levine, B. (2010). Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7, 115–127. doi: 10.1016/j.chom.2010.01.007
Orvedahl, A., Sumpter, R. Jr., Xiao, G., Ng, A., Zou, Z., Tang, Y., et al. (2011). Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117. doi: 10.1038/nature10546
Paludan, C., Schmid, D., Landthaler, M., Vockerodt, M., Kube, D., Tuschl, T., et al. (2005). Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307, 593–596. doi: 10.1126/science.1104904
Pantopoulos, K., Porwal, S. K., Tartakoff, A., and Devireddy, L. (2012). Mechanisms of mammalian iron homeostasis. Biochemistry 51, 5705–5724. doi: 10.1021/bi300752r
Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., et al. (2005). Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939. doi: 10.1016/j.cell.2005.07.002
Peng, H., Liu, B., Yves, T. D., He, Y., Wang, S., Tang, H., et al. (2018). Zika virus induces autophagy in human umbilical vein endothelial cells. Viruses 10:259. doi: 10.3390/v10050259
Pérez-Vargas, J., Romero, P., López, S., and Arias, C. F. (2006). The peptide-binding and ATPase domains of recombinant hsc70 are required to interact with rotavirus and reduce its infectivity. J. Virol. 80, 3322–3331.
Peymani, P., Yeganeh, B., Sabour, S., Geramizadeh, B., Fattahi, M. R., Keyvani, H., et al. (2016). New use of an old drug: chloroquine reduces viral and ALT levels in HCV non-responders (a randomized, triple-blind, placebo-controlled pilot trial). Can. J. Physiol. Pharmacol. 94, 613–619. doi: 10.1139/cjpp-2015-0507
Puleston, D. J., Zhang, H., Powell, T. J., Lipina, E., Sims, S., Panse, I., et al. (2014). Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife 3, e03706. doi: 10.7554/eLife.03706
Richetta, C., Gregoire, I. P., Verlhac, P., Azocar, O., Baguet, J., Flacher, M., et al. (2013). Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 9:e1003599. doi: 10.1371/journal.ppat.1003599
Rogov, V., Dotsch, V., Johansen, T., and Kirkin, V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178. doi: 10.1016/j.molcel.2013.12.014
Sagnier, S., Daussy, C. F., Borel, S., Robert-Hebmann, V., Faure, M., Blanchet, F. P., et al. (2015). Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J. Virol. 89, 615–625. doi: 10.1128/JVI.02174-14
Samsa, M. M., Mondotte, J. A., Iglesias, N. G., Assuncao-Miranda, I., Barbosa-Lima, G., Da Poian, A. T., et al. (2009). Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. doi: 10.1371/journal.ppat.1000632
Sathyanarayan, A., Mashek, M. T., and Mashek, D. G. (2017). ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep. 19, 1–9. doi: 10.1016/j.celrep.2017.03.026
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Schweers, R. L., Zhang, J., Randall, M. S., Loyd, M. R., Li, W., Dorsey, F. C., et al. (2007). NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 104, 19500–19505. doi: 10.1073/pnas.0708818104
Sharma, S., Ray, A., and Sadasivam, B. (2020). Metformin in COVID-19: a possible role beyond diabetes. Diabetes Res. Clin. Pract. 164:108183. doi: 10.1016/j.diabres.2020.108183
Sharma, V., Makhdoomi, M., Singh, L., Kumar, P., Khan, N., Singh, S., et al. (2021). Trehalose limits opportunistic mycobacterial survival during HIV co-infection by reversing HIV-mediated autophagy block. Autophagy 17, 476–495. doi: 10.1080/15548627.2020.1725374
Shelly, S., Lukinova, N., Bambina, S., Berman, A., and Cherry, S. (2009). Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30, 588–598. doi: 10.1016/j.immuni.2009.02.009
Shi, J., Fung, G., Piesik, P., Zhang, J., and Luo, H. (2014). Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ. 21, 1432–1441. doi: 10.1038/cdd.2014.58
Shi, J., Wong, J., Piesik, P., Fung, G., Zhang, J., Jagdeo, J., et al. (2013). Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy 9, 1591–1603. doi: 10.4161/auto.26059
Shin, D. W. (2020). Lipophagy: molecular mechanisms and implications in metabolic disorders. Mol. Cells 43, 686–693. doi: 10.14348/molcells.2020.0046
Singh, R., and Cuervo, A. M. (2012). Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012:282041. doi: 10.1155/2012/282041
Singh, S. P., Amar, S., Gehlot, P., Patra, S. K., Kanwar, N., and Kanwal, A. (2021). Mitochondrial modulations, autophagy pathways shifts in viral infections: consequences of COVID-19. Int. J. Mol. Sci. 22:8180. doi: 10.3390/ijms22158180
Smith, M. D., Harley, M. E., Kemp, A. J., Wills, J., Lee, M., Arends, M., et al. (2018). CCPG1 Is a non-canonical autophagy cargo receptor essential for ER-Phagy and pancreatic ER proteostasis. Dev. Cell 44, 217–232 e211. doi: 10.1016/j.devcel.2017.11.024
Sparrer, K. M. J., Gableske, S., Zurenski, M. A., Parker, Z. M., Full, F., Baumgart, G. J., et al. (2017). TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol. 2, 1543–1557. doi: 10.1038/s41564-017-0017-2
Stolz, A., Ernst, A., and Dikic, I. (2014). Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16, 495–501. doi: 10.1038/ncb2979
Strappazzon, F., Di Rita, A., Peschiaroli, A., Leoncini, P. P., Locatelli, F., Melino, G., et al. (2020). HUWE1 controls MCL1 stability to unleash AMBRA1-induced mitophagy. Cell Death Differ. 27, 1155–1168. doi: 10.1038/s41418-019-0404-8
Sui, C., Xiao, T., Zhang, S., Zeng, H., Zheng, Y., Liu, B., et al. (2022). SARS-CoV-2 NSP13 inhibits Type I IFN production by degradation of TBK1 via p62-dependent selective autophagy. J. Immunol. 208, 753–761. doi: 10.4049/jimmunol.2100684
Sumpter, R. Jr., Sirasanagandla, S., Fernandez, A. F., Wei, Y., Dong, X., Franco, L., et al. (2016). Fanconi anemia proteins function in mitophagy and immunity. Cell 165, 867–881. doi: 10.1016/j.cell.2016.04.006
Sun, D., Kong, N., Dong, S., Chen, X., Qin, W., Wang, H., et al. (2021). 2AB protein of Senecavirus A antagonizes selective autophagy and type I interferon production by degrading LC3 and MARCHF8. Autophagy 18, 1–13. doi: 10.1080/15548627.2021.2015740
Sun, Y., Bao, Q., Xuan, B., Xu, W., Pan, D., Li, Q., et al. (2018). Human cytomegalovirus protein pUL38 prevents premature cell death by binding to ubiquitin-specific protease 24 and regulating iron metabolism. J. Virol. 92:e00191-18. doi: 10.1128/JVI.00191-18
Tey, S. K., and Khanna, R. (2012). Autophagy mediates transporter associated with antigen processing-independent presentation of viral epitopes through MHC class I pathway. Blood 120, 994–1004. doi: 10.1182/blood-2012-01-402404
Tumbarello, D. A., Manna, P. T., Allen, M., Bycroft, M., Arden, S. D., Kendrick-Jones, J., et al. (2015). The autophagy receptor TAX1BP1 and the molecular motor myosin VI Are required for clearance of Salmonella typhimurium by autophagy. PLoS Pathog. 11:e1005174. doi: 10.1371/journal.ppat.1005174
Verlhac, P., Viret, C., and Faure, M. (2015). Dual function of CALCOCO2/NDP52 during xenophagy. Autophagy 11, 965–966. doi: 10.1080/15548627.2015.1046672
Viret, C., Duclaux-Loras, R., Nancey, S., Rozieres, A., and Faure, M. (2021). Selective autophagy receptors in antiviral defense. Trends Microbiol. 29, 798–810. doi: 10.1016/j.tim.2021.02.006
Vo, M. T., and Choi, Y. B. (2021). Herpesvirus regulation of selective autophagy. Viruses 13:820. doi: 10.3390/v13050820
Vo, M. T., Smith, B. J., Nicholas, J., and Choi, Y. B. (2019). Activation of NIX-mediated mitophagy by an interferon regulatory factor homologue of human herpesvirus. Nat. Commun. 10:3203. doi: 10.1038/s41467-019-11164-2
Wang, J., Liu, J. Y., Shao, K. Y., Han, Y. Q., Li, G. L., Ming, S. L., et al. (2019). Porcine reproductive and respiratory syndrome virus activates lipophagy to facilitate viral replication through downregulation of NDRG1 expression. J. Virol. 93:e00526-19. doi: 10.1128/JVI.00526-19
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. doi: 10.1038/s41422-020-0282-0
Wang, Q., Zhu, N., Hu, J., Wang, Y., Xu, J., Gu, Q., et al. (2020). The mTOR inhibitor manassantin B reveals a crucial role of mTORC2 signaling in Epstein-Barr virus reactivation. J. Biol. Chem. 295, 7431–7441. doi: 10.1074/jbc.RA120.012645
Wang, R., Zhu, Y., Ren, C., Yang, S., Tian, S., Chen, H., et al. (2021). Influenza A virus protein PB1-F2 impairs innate immunity by inducing mitophagy. Autophagy 17, 496–511. doi: 10.1080/15548627.2020.1725375
Wang, Z., Chen, J., Wu, X., Ma, D., Zhang, X., Li, R., et al. (2021). PCV2 targets cGAS to inhibit type I interferon induction to promote other DNA virus infection. PLoS Pathog. 17:e1009940. doi: 10.1371/journal.ppat.1009940
Ward, C., Martinez-Lopez, N., Otten, E. G., Carroll, B., Maetzel, D., Singh, R., et al. (2016). Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta 1861, 269–284. doi: 10.1016/j.bbalip.2016.01.006
Wen, W., Li, X., Yin, M., Wang, H., Qin, L., Li, H., et al. (2021). Selective autophagy receptor SQSTM1/p62 inhibits Seneca Valley virus replication by targeting viral VP1 and VP3. Autophagy 17, 3763–3775. doi: 10.1080/15548627.2021.1897223
Wild, P., Farhan, H., McEwan, D. G., Wagner, S., Rogov, V. V., Brady, N. R., et al. (2011). Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233. doi: 10.1126/science.1205405
Wong, Y. C., and Holzbaur, E. L. (2014). Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. U.S.A. 111, E4439–E4448. doi: 10.1073/pnas.1405752111
Wu, C.-Y., Jan, J.-T., Ma, S.-H., Kuo, C.-J., Juan, H.-F., Cheng, Y.-S. E., et al. (2004). Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U.S.A. 101, 10012–10017.
Wu, Y., and Cui, J. (2019). Selective autophagy regulates innate immunity through cargo receptor network. Adv. Exp. Med. Biol. 1209, 145–166. doi: 10.1007/978-981-15-0606-2_9
Wu, Y., Jin, S., Liu, Q., Zhang, Y., Ma, L., Zhao, Z., et al. (2021). Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy 17, 1379–1392. doi: 10.1080/15548627.2020.1761653
Wyers, F., Dru, P., Simonet, B., and Contamine, D. (1993). Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J. Virol. 67, 3208–3216. doi: 10.1128/JVI.67.6.3208-3216.1993
Xie, W., Jin, S., Zhang, C., Yang, S., Wu, Y., Zhao, Y., et al. (2022). Selective autophagy controls the stability of TBK1 via NEDD4 to balance host defense. Cell Death Differ. 29, 40–53. doi: 10.1038/s41418-021-00833-9
Xie, W., Li, D., Dong, D., Li, Y., Zhang, Y., Duan, L., et al. (2020). HIV-1 exposure triggers autophagic degradation of stathmin and hyperstabilization of microtubules to disrupt epithelial cell junctions. Signal. Transduct. Target. Ther. 5:79. doi: 10.1038/s41392-020-0175-1
Xiong, Y., Rajoka, M. S. R., Mehwish, H. M., Zhang, M., Liang, N., Li, C., et al. (2021). Virucidal activity of Moringa A from Moringa oleifera seeds against Influenza A Viruses by regulating TFEB. Int. Immunopharmacol. 95:107561. doi: 10.1016/j.intimp.2021.107561
Yan, C., Gong, L., Chen, L., Xu, M., Abou-Hamdan, H., Tang, M., et al. (2020). PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 16, 419–434. doi: 10.1080/15548627.2019.1628520
Yan, J., and Horng, T. (2020). Lipid metabolism in regulation of macrophage functions. Trends Cell Biol. 30, 979–989. doi: 10.1016/j.tcb.2020.09.006
Yang, H., Ni, H. M., Guo, F., Ding, Y., Shi, Y. H., Lahiri, P., et al. (2016). Sequestosome 1/p62 protein is associated with autophagic removal of excess hepatic endoplasmic reticulum in mice. J. Biol. Chem. 291, 18663–18674. doi: 10.1074/jbc.M116.739821
Yao, X., Ye, F., Zhang, M., Cui, C., Huang, B., Niu, P., et al. (2020). In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 71, 732–739. doi: 10.1093/cid/ciaa237
Yordy, B., and Iwasaki, A. (2011). Autophagy in the control and pathogenesis of viral infection. Curr. Opin. Virol. 1, 196–203. doi: 10.1016/j.coviro.2011.05.016
Youle, R. J., and Narendra, D. P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14. doi: 10.1038/nrm3028
Zeng, Y., Xu, S., Wei, Y., Zhang, X., Wang, Q., Jia, Y., et al. (2021). The PB1 protein of influenza A virus inhibits the innate immune response by targeting MAVS for NBR1-mediated selective autophagic degradation. PLoS Pathog. 17:e1009300. doi: 10.1371/journal.ppat.1009300
Zhang, D., Ma, Z., Chen, H., Lu, Y., and Chen, X. (2020). Valinomycin as a potential antiviral agent against coronaviruses: a review. Biomed. J. 43, 414–423. doi: 10.1016/j.bj.2020.08.006
Zhang, J., Lan, Y., Li, M. Y., Lamers, M. M., Fusade-Boyer, M., Klemm, E., et al. (2018). Flaviviruses exploit the lipid droplet protein AUP1 to trigger lipophagy and drive virus production. Cell Host Microbe 23, 819–831 e815. doi: 10.1016/j.chom.2018.05.005
Zhang, J., and Ney, P. A. (2009). Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 16, 939–946. doi: 10.1038/cdd.2009.16
Zhang, L., Qin, Y., and Chen, M. (2018). Viral strategies for triggering and manipulating mitophagy. Autophagy 14, 1665–1673. doi: 10.1080/15548627.2018.1466014
Keywords: selective autophagy, virophagy, antiviral responses, macroautophagy (autophagy), viral infectious diseases
Citation: Liu Y, Zhou T, Hu J, Jin S, Wu J, Guan X, Wu Y and Cui J (2022) Targeting Selective Autophagy as a Therapeutic Strategy for Viral Infectious Diseases. Front. Microbiol. 13:889835. doi: 10.3389/fmicb.2022.889835
Received: 04 March 2022; Accepted: 29 March 2022;
Published: 28 April 2022.
Edited by:
Young Bong Choi, Johns Hopkins Medicine, United StatesReviewed by:
Ping Qian, Huazhong Agricultural University, ChinaCopyright © 2022 Liu, Zhou, Hu, Jin, Wu, Guan, Wu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangdong Guan, Z3VhbnhkQG1haWwuc3lzdS5lZHUuY24=; Yaoxing Wu, d3V5YW94NUBtYWlsLnN5c3UuZWR1LmNu; Jun Cui, Y3VpajVAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.