- 1Innovative Team of Antimicrobial Peptides and Alternatives to Antibiotics, Gene Engineering Laboratory, Feed Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Key Laboratory of Feed Biotechnology, Ministry of Agriculture and Rural Affairs, Beijing, China
- 3Machine Biology Group, Departments of Psychiatry and Microbiology, Institute for Biomedical Informatics, Institute for Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
- 4Departments of Bioengineering and Chemical and Biomolecular Engineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, PA, United States
- 5Penn Institute for Computational Science, University of Pennsylvania, Philadelphia, PA, United States
- 6S-Inova Biotech, Universidade Católica Dom Bosco, Campo Grande, Brazil
- 7Centro de Análises Proteômicas e Bioquímicas Programa de Pós-Graduação em Ciências Genômicas e Biotecnologia, Universidade Católica de Brasília, Brasília, Brazil
Editorial on the Research Topic
Antimicrobial Peptides: Molecular Design, Structure-Function Relationship, and Biosynthesis Optimization
The long-term use of antibiotics has accelerated the emergence of antibiotic-resistant bacteria (ARB). Annual global death due to antibiotic resistance is over 700,000 in 2014, it is predicted that a continued rise in resistance would lead to annual death rate of 10 million by 2050 (O'Neill, 2014; WHO, 2014). Antimicrobial peptides (AMPs) are a class of small molecules produced by numerous living organisms as part of their host innate immune response to infection (Loose et al., 2006; Torres and de la Fuente-Nunez, 2019; Cesaro et al., 2022). AMPs evolution in insects and other species have demonstrated the ability of these molecules to eliminate invading pathogens and thus have generated a great excitement at the prospect of developing AMPs as alternatives to antibiotics (ATAs) for years (Czaplewski et al., 2016; Magana et al., 2020).
The field of AMP research started in the 1980s owing to the discoveries of insect cecropins by Hans Boman, human α-defensins by Robert Lehrer, and magainins by Michael Zasloff (Wang et al., 2016). Over 3,300 kinds of AMPs have now been found in a wide range of biological sources, ranging from microbes, plants, to animals (Torres et al., 2022). These peptides may possess optimal properties for further drug development, including their ability to permeabilize and disrupt the bacterial membrane, capability of regulating the immune system and also broad-spectrum antibiofilm activity, and reduced propensity to select for bacterial resistance (de la Fuente-Núñez et al., 2012, 2016; Yang et al., 2019; Wang et al., 2022). These advantages of AMPs over conventional antibiotics have attracted attention despite that their use have primarily been limited to topical infections due to their relative narrow druggability and lack of special unique protocols to assess pharmacodynamics (Liu et al., 2021a; Ma et al., 2021; Dos Santos-Silva et al.). The highlights of 21 papers in this topic will be briefly presented and reviewed as follows.
Discovering New Natural AMPs
Natural product and their special functional structures or domains have traditionally played a significant role in drug discovery and development (Newman and Cragg, 2012). Plant AMPs are rich in diversity and have been reported to have antimicrobial activity against infections caused by pathogens (Dos Santos-Silva et al.). Lee et al. isolated a novel peptide PN5 from pine needles of Pinus densiflora. Sieb. et Zucc, exhibiting a strong antimicrobial activity against foodborne bacteria, and no detectable cytotoxicity. In fact, animals, plants, and microbiota harbor a large number of bacteria that they compete for nutrients and space and exchange biomolecules (Tobias et al., 2017). In vivo competition and interspecies exchange involve the synthesis and continuous evolution of many previously unknown AMPs. Ngashangva et al. isolated a new AMP from a bacterial endophyte derived from the medicinal plant Millettia achycarpa Benth and analyzed its biosynthetic gene cluster by collective analysis of both genomic and proteomic data. In addition, Brevibacillin 2V, a novel lipo-tridecapeptide with a strong antimicrobial activity against antibiotic-resistant Staphylococcus aureus ATCC15975 (MRSA) was reported to show much lower hemolytic activity and cytotoxicity toward eukaryotic cells than previously reported non-ribosomally produced peptides from the lipo-tridecapeptide family, making it a promising candidate for new peptide antibiotic development (Zhao et al.). However, AMPs' toxicity, instability, low yield obtained by recombinant expression, and high cost of chemical synthesis have hindered their application and translation into the clinic. Recent advances in data mining, artificial intelligence (Porto et al., 2018b; de la Fuente-Nunez, 2019; Torres et al., 2022), synthetic biology, chemical biology, and interdisciplinary tools are greatly pushing the pace of new discoveries and solutions to overcome these drawbacks.
Molecular Design of AMPs
Advances in methodological design are necessary to discover and advance new AMPs. De novo design involving the latest theoretical knowledge is being applied to determine and screen quickly an economically feasible strategy for short candidate AMPs sequences with basic antibacterial ability and high selectivity (Wang et al.). In recent years, with the development of big data and artificial intelligence, database screening and mining technology have become an exciting tool for drug discovery. Based on the AMP data pool APD, new target peptides with specific potent properties were designed and candidates with the most likely target activity based on key parameters were efficiently identified (Mishra and Wang, 2012; Magana et al., 2020). Using this method, Bobde et al. screened eight novel AMPs, termed PHNX peptides, against Gram-negative bacteria. Advances in the development of computational tools have greatly facilitated the discovery of novel peptide-based drugs. For example, a generative model (Porto et al., 2018a) was used to yield synthetic peptides with anti-infective efficacy in mouse models (Porto et al., 2018b; Torres et al., 2021). Recently, an algorithmic approach was used to explore the human body as a source of peptide antibiotics (Torres et al., 2022). In this issue, Dean et al. built an improved automated semi-supervised approach termed PepVAE for generating promising new sequences using a variational autoencoder.
Molecular Modification of AMPs
Recently, the design and modification of AMPs have been extensively used for drug development purposes. Molecular modification is no longer limited to site substitution of select residues (Han et al.; Zhao et al.). An increasing number of additional tools have been developed such as chemical modification, cyclization, insertion of large hydrophobic side-chains (Liu et al., 2021a), chimera (Yu et al.) and polymerization, as well as their combination or cross (Hao et al., 2017; Li et al., 2018a,b; Wang et al., 2020). The conformational and physicochemical properties of AMPs play important roles in determining antibacterial activity, toxicity and bioavailability. The distribution of positive charge and hydrophobic amino acid within the helical wheel seems to be the key factors determining the antibacterial activity of AMPs, and their selectivity can be effectively improved by adjusting amphiphilicity (Luo et al.). Steigenberger et al. designed a membrane-permeabilizing lipopeptide with different chain-length and found its antimicrobial specificity depending on membrane difference of the target bacteria. These results reveal the importance and weight of taking into account bacterial membrane composition when designing AMPs.
Nanotechnology Applied to AMPs
Most AMPs, if administered systemically into the body, may cause side effects or be degraded through multiple proteolytic cleavage. A gradual decrease, distribution, and slow accumulation of AMPs level released in vivo may lead to the induction of bacterial resistance at sub-inhibitory concentrations (Tan et al., 2021). To overcome these potential issues, nanotechnology has entered the realm of AMP application, and nanocarriers can be used as delivery systems in order to enhance therapeutic effects and minimize above undesirable side-effects (Magana et al., 2020). Nanocarriers can help improve the pharmacokinetic/pharmacodynamic profiles of AMPs, extending shelf life and half-life, stability and bioavailability. With the development of nanotechnology, a variety of peptide-based antibacterial nanomaterials have been produced as metal nanoparticles or carbon nanotubes conjugated AMPs, polymeric material nanosystem containing AMPs, and self-assembled AMPs (Yang et al.). More and better exciting new results in this field are expected in the near future.
Recombinant Expression of AMPs
Considering the high cost of new drug development and feasible competition with existing antibiotics, mass production of AMPs at low cost is necessary and essential for deployment of these agents in the population. AMPs are produced at very low levels in living organisms, and their extraction is difficult, inefficient, expensive, and time consuming. Recombinant expression of peptides in DNA holds promise to enable large-scale production of AMPs at an affordable cost. Wang' team (Zhang et al., 2011, 2014; Cao et al., 2015; Li et al., 2017, 2020; Yang et al., 2019; Liu et al., 2020, 2021b; Shen et al.) and others (Cao et al., 2018) have been working in this area for decades, obtaining exciting peptide yields higher than 1 g (target peptide)/L(ferment supernatant) secreted from yeast cells (Zasloff, 2016; Sampaio de Oliveira et al., 2020).
Synergism and Combination of AMPs Administration
AMPs have been shown to potentiate the activity of conventional antibiotics to target pathogens (Reffuveille et al., 2014; de la Fuente-Núñez et al., 2015). Synergism can enhance the antimicrobial activity of peptides (Zhang et al., 2014; Zhao et al., 2019; Ma et al., 2021) while reducing the amount of drug dosage needed to kill bacteria both by the peptide and antibiotic, markedly reducing the risk of ARB development. The specific molecular mechanisms underlying the synergistic effects between AMPs and other antibiotics are still unclear, although it has been hypothesized that the ability of peptides to penetrate into bacterial cells is a key contributor (Reffuveille et al., 2014). Wu et al. attempted to uncover more details into this by examining the effects of bulky non-natural amino acid end tagging strategy of short AMPs on their combination with conventional antibiotics against resistant bacteria.
Conclusion
AMPs constitute the first line of innate defense against infection. These molecules are attractive drug candidates due to their multifactorial mechanism of action, low propensity to select for bacterial resistance, among other things (Hao et al., 2017; Wang et al., 2018, 2022; Liu et al., 2021a; Zheng et al., 2021). A reasonable equilibrium between antimicrobials and infectious pathogens is critical in order to control high resistance and high variation among pathogens, and can potentially be achieved by the appropriate use of AMPs, antibiotic and vaccines, as the iron triangle theory summarized in Figure 1. Future work should focus on determining the sequence requirements underlying for special unique PK/PD profiles of peptides, along with formulation, and ADME-Toxicity studies (Andes et al., 2009; Xiong et al., 2011; Magana et al., 2020) in order to translate AMPs into the clinic as soon as possible.
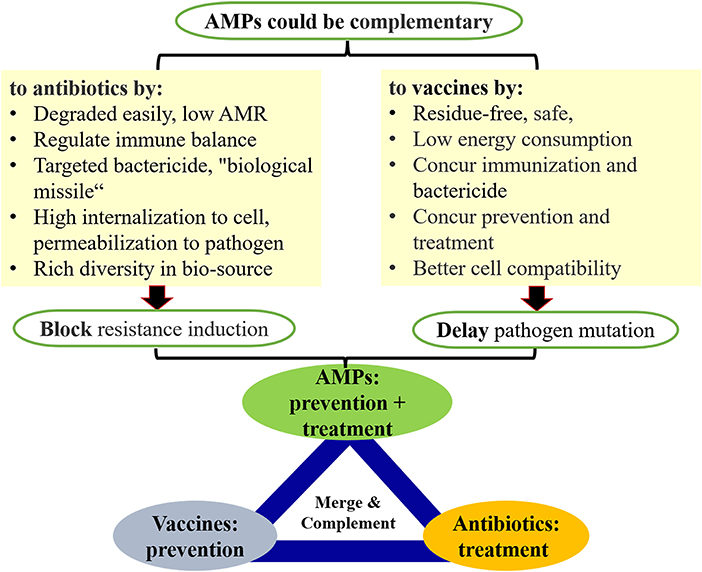
Figure 1. An iron triangle of health protection from AMPs, antibiotics and vaccines for maintaining a reasonable equilibrium among pathogens, antimicrobials and drug resistances is important and essential for one health management at safety level (Hao et al., 2017; Wang et al., 2018, 2022; Liu et al., 2021a; Zheng et al., 2021).
Author Contributions
The first draft text of this editorial was written by Ph.D. student YH as assistant of JW with his guide and direction. All authors listed have made substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
JW was supported by the National Natural Science Foundation of China (Grant no. 31872393), the Agricultural Science and Technology Innovation Program (ASTIP) in CAAS (CAAS-ASTIP-2017-FRI-02), and its key projects (CAAS-ZDXT2018008 and CAAS-ZDRW202111). OF was also supported by CAPES, CNPq, FAPDF, and FUNDECT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to sincerely thank a total of 141 authors of 21 papers and over 70 peer reviewers for their valuable professional contributions into the first issue of this Research Topic Antimicrobial Peptides: Molecular Design, Structure-Function Relationship, and Biosynthesis Optimization, along with Prof. Rustam Aminov as SCE and the staff of Frontiers in Microbiology, and also team supports of three editors JW, CF-N, and OF, including Dr. Ruoyu Mao's contribution into the first draft text of about this Research Topic of this topic as assistant of JW with his guide and direction.
References
Andes, D., Craig, W., Nielsen, L. A., and Kristensen, H. H. (2009). In vivo pharmacodynamic characterization of a novel plectasin antibiotic, NZ2114, in a murine infection model. Antimicrob. Agents Chemother. 53, 3003–3009. doi: 10.1128/AAC.01584-08
Cao, J., de la Fuente-Nunez, C., Ou, R. W., Torres, M. T., Pande, S. G., Sinskey, A. J., et al. (2018). Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth. Biol. 7, 896–902. doi: 10.1021/acssynbio.7b00396
Cao, X., Zhang, Y., Mao, R., Teng, D., Wang, X., and Wang, J. (2015). In vitro and in vivo characterization of a new recombinant antimicrobial peptide, MP1102, against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 99, 2649–2662. doi: 10.1007/s00253-014-6077-9
Cesaro, A., Torres, M. T., and de la Fuente-Nunez, C. (2022). Methods for the design and characterization of peptide antibiotics. Methods Enzymol. 663, 303–326. doi: 10.1016/bs.mie.2021.11.003
Czaplewski, L., Bax, R., Clokie, M., Dawson, M., Fairhead, H., Fischetti, V. A., et al. (2016). Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251. doi: 10.1016/S1473-3099(15)00466-1
de la Fuente-Nunez, C. (2019). Toward autonomous antibiotic discovery. mSystems 4, e00151–e00119. doi: 10.1128/mSystems.00151-19
de la Fuente-Núñez, C., Cardoso, M. H., de Souza Cândido, E., Franco, O. L., and Hancock, R. E. (2016). Synthetic antibiofilm peptides. Biochim. Biophys. Acta 1858, 1061–1069. doi: 10.1016/j.bbamem.2015.12.015
de la Fuente-Núñez, C., Korolik, V., Bains, M., Nguyen, U., Breidenstein, E. B., Horsman, S., et al. (2012). Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56, 2696–2704. doi: 10.1128/AAC.00064-12
de la Fuente-Núñez, C., Reffuveille, F., Mansour, S. C., Reckseidler-Zenteno, S. L., Hernández, D., Brackman, G., et al. (2015). D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205. doi: 10.1016/j.chembiol.2015.01.002
Hao, Y., Yang, N., Wang, X., Teng, D., Mao, R., Wang, X., et al. (2017). Killing of Staphylococcus aureus and Salmonella enteritidis and neutralization of lipopolysaccharide by 17-residue bovine lactoferricins: improved activity of Trp/Ala-containing molecules. Sci. Rep. 7, 44278. doi: 10.1038/srep44278
Li, B., Yang, N., Wang, X., Hao, Y., Mao, R., Li, Z., et al. (2020). An enhanced variant designed from DLP4 cationic peptide against Staphylococcus aureus CVCC 546. Front. Microbiol. 11, 1057. doi: 10.3389/fmicb.2020.01057
Li, Z, Mao, R, Teng, D, Hao, Y, Chen, H, Wang, X, et al. (2017). Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 7, 12124. doi: 10.1038/s41598-017-10839-4
Li, Z., Teng, D., Mao, R., Wang, X, Hao, Y., Wang, X., and Wang, J. (2018b). Improved antibacterial activity of the marine peptide N6 against intracellular Salmonella Typhimurium by conjugating with the cell-penetrating peptide Tat11 via a cleavable linker. J. Med. Chem. 61, 7991–8000. doi: 10.1021/acs.jmedchem.8b01079
Li, Z., Wang, X., Teng, D., Mao, R., Hao, Y., Yang, N., et al. (2018a). Improved antibacterial activity of a marine peptide-N2 against intracellular Salmonella typhimurium by conjugating with cell-penetrating peptides- bLFcin6/Tat11. Eur. J. Med. Chem. 145, 263–272. doi: 10.1016/j.ejmech.2017.12.066
Liu, H., Yang, N., Mao, R., Teng, D., Hao, Y., Wang, X., et al. (2020). A new high- yielding antimicrobial peptide NZX and its antibacterial activity against Staphylococcus hyicus in vitro/vivo. Appl. Microbiol. Biotechnol. 104, 1555–1568. doi: 10.1007/s00253-019-10313-3
Liu, H., Yang, N., Teng, D., Mao, R., Hao, Y., Ma, X, et al. (2021a), Fatty acid modified-antimicrobial peptide analogues with potent antimicrobial activity topical therapeutic efficacy against Staphylococcus hyicus. Appl. Microbiol. Biotechnol. 105, 5845–5859. doi: 10.1007/s00253-021-11454-0.
Liu, H., Yang, N., Teng, D., Mao, R., Hao, Y., Ma, X., et al. (2021b). Design and pharmacodynamics of recombinant fungus defensin NZL with improved activity against Staphylococcus hyicus in vitro and in vivo. Int. J. Mol. Sci. 22, 5435. doi: 10.3390/ijms22115435
Loose, C., Jensen, K., Rigoutsos, I., and Stephanopoulos, G. (2006). A linguistic model for the rational design of antimicrobial peptides. Nature 443, 867–869. doi: 10.1038/nature05233
Ma, X., Yang, N., Mao, R., Hao, Y., Yan, X., Teng, D., et al. (2021). The pharmacodynamics study of insect defensin DLP4 against toxigenic Staphylococcus hyicus ACCC 61734 in vitro and vivo. Front. Cell Infect. Microbiol. 11, 638598. doi: 10.3389/fcimb.2021.638598
Magana, M., Pushpanathan, M., Santos, A. L., Leanse, L., Fernandez, M., Ioannidis, A., et al. (2020). The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 20, e216–e230. doi: 10.1016/S1473-3099(20)30327-3
Mishra, B., and Wang, G. (2012). Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 134, 12426–12429. doi: 10.1021/ja305644e
Newman, D. J., and Cragg, G. M. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335. doi: 10.1021/np200906s
O'Neill, J. (2014). Antimicrobial resistance: tackling a crisis for the health and wealth of nations. UK review on antimicrobial resistance, December 2014. Available online at: http://amr-review.org/Publications
Porto, W. F., Fensterseifer, I. C. M., Ribeiro, S. M., and Franco, O. L. (2018a). Joker: an algorithm to insert patterns into sequences for designing antimicrobial peptides. Biochim. Biophys. Acta Gen. Subj. 1862, 2043–2052. doi: 10.1016/j.bbagen.2018.06.011
Porto, W. F., Irazazabal, L., Alves, E. S. F., Ribeiro, S. M., Matos, C. O., Pires, Á. S., et al. (2018b). In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 9, 1490. doi: 10.1038/s41467-018-03746-3
Reffuveille, F., de la Fuente-Núñez, C., Mansour, S., and Hancock, R. E. (2014). A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371. doi: 10.1128/AAC.03163-14
Sampaio de Oliveira, K. B., Leite, M. L., Rodrigues, G. R., Duque, H. M., da Costa, R. A., Cunha, V. A., et al. (2020). Strategies for recombinant production of antimicrobial peptides with pharmacological potential. Expert. Rev. Clin. Pharmacol. 13, 367–390. doi: 10.1080/17512433.2020.1764347
Tan, P., Fu, H., and Ma, X. (2021). Design, optimization, and nanotechnology of antimicrobial peptides: from exploration to applications. Nano Today 39, 101229. doi: 10.1016/j.nantod.2021.101229
Tobias, N. J., Wolff, H., Djahanschiri, B., Grundmann, F., Kronenwerth, M., Shi, Y. M., et al. (2017). Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2, 1676–1685. doi: 10.1038/s41564-017-0039-9
Torres, M. D. T., Cao, J., Franco, O. L., Lu, T. K., and de la Fuente-Nunez, C. (2021). Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano 15, 2143–64. doi: 10.1021/acsnano.0c09509
Torres, M. D. T., and de la Fuente-Nunez, C. (2019). Reprogramming biological peptides to combat infectious diseases. Chem. Commun. 55, 15020–15032. doi: 10.1039/C9CC07898C
Torres, M. D. T., Melo, M. C. R., Crescenzi, O., Notomista, E., and de la Fuente-Nunez, C. (2022). Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng. 6, 67–75. doi: 10.1038/s41551-021-00801-1
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093. doi: 10.1093/nar/gkv1278
Wang, X., Wang, X., Teng, D., Mao, R., Hao, Y., Yang, N., et al. (2018). Increased intracellular activity of MP1102 and NZ2114 against Staphylococcus aureus in vitro and in vivo. Sci. Rep. 8, 4204. doi: 10.1038/s41598-018-22245-5
Wang, Z., Liu, X., Teng, D., Mao, R., Hao, Y., Yang, N., et al. (2020). Development of chimeric peptides to facilitate the neutralisation of lipopolysaccharides during bactericidal targeting of multidrug-resistant Escherichia coli. Commun. Biol. 3, 41. doi: 10.1038/s42003-020-0761-3
Wang, Z., Yang, N., Teng, D., Hao, Y., Li, T., Han, H., et al. (2022). Resistance response to arenicin derivatives in Escherichia coli. Appl. Microbiol. Biotechnol. 106, 211–226. doi: 10.1007/s00253-021-11708-x
Xiong, Y. Q., Hady, W. A., Deslandes, A., Rey, A., Fraisse, L., Kristensen, H. H., et al. (2011). Efficacy of NZ2114, a novel plectasin-derived cationic antimicrobial peptide antibiotic, in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 5325–5330. doi: 10.1128/AAC.00453-11
Yang, N., Teng, D., Mao, R., Hao, Y., Wang, X., Wang, Z., et al. (2019). A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biotechnol. 103, 5193–5213. doi: 10.1007/s00253-019-09785-0
Zasloff, M. (2016). “Antimicrobial peptides: do they have a future as therapeutics?” in Antimicrobial Peptides, editors J. Harder and J. M. Schröder (Basel: Springer Press), 147–154.
Zhang, J., Yang, Y., Teng, D., Tian, Z., Wang, S., and Wang, J. (2011). Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr. Purif. 78, 189–196. doi: 10.1016/j.pep.2011.04.014
Zhang, Y., Teng, D., Mao, R., Wang, X., Xi, D., Hu, X., et al. (2014). High expression of a plectasin-derived peptide NZ2114 in Pichia pastoris and its pharmacodynamics, postantibiotic and synergy against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 98:681–694. doi: 10.1007/s00253-013-4881-2
Zhao, F., Yang, N., Wang, X., Mao, R., Hao, Y., Li, Z., et al. (2019). In vitro/vivo mechanism of action of MP1102 with low/nonresistance against Streptococcus suis Type 2 strain CVCC 3928. Front. Cell Infect. Microbiol. 9, 48. doi: 10.3389/fcimb.2019.00048
Keywords: antimicrobial peptides (AMPs), molecular design and modification, structure-function relationship, biosynthesis optimization, drug combination
Citation: Hao Y, Wang J, de la Fuente-Nunez C and Franco OL (2022) Editorial: Antimicrobial Peptides: Molecular Design, Structure-Function Relationship, and Biosynthesis Optimization. Front. Microbiol. 13:888540. doi: 10.3389/fmicb.2022.888540
Received: 03 March 2022; Accepted: 03 March 2022;
Published: 11 April 2022.
Edited and reviewed by: William James Hickey, University of Wisconsin-Madison, United States
Copyright © 2022 Hao, Wang, de la Fuente-Nunez and Franco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Wang, d2FuZ2ppYW5odWFAY2Fhcy5jbg==; d2FuZ2ppYW5odWEucGVraW5nQHFxLmNvbQ==
 Ya Hao1,2
Ya Hao1,2 Jianhua Wang
Jianhua Wang Cesar de la Fuente-Nunez
Cesar de la Fuente-Nunez Octavio Luiz Franco
Octavio Luiz Franco