- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Institute of Agro-product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2College of Agriculture and Animal Husbandry, Qinghai University, Xining, China
- 3State Key Laboratory of Microbial Metabolism, School of Agriculture and Biology, MOST-USDA Joint Research Center for Food Safety, Shanghai Jiao Tong University, Shanghai, China
- 4College of Veterinary Medicine, Northwest A&F University, Yangling, China
Antimicrobial resistance (AMR) may lead to worldwide epidemics through human activities and natural transmission, posing a global public safety threat. Colistin resistance mediated by the mcr-1 gene is the most prevalent among animal-derived Escherichia coli, and mcr-1-carrying E. coli have been frequently detected in central-eastern China. However, animal-derived E. coli with AMR and the prevalence of mcr-1 in the Qinghai Plateau have been rarely investigated. Herein, 375 stool samples were collected from 13 poultry farms in Qinghai Province and 346 E. coli strains were isolated, of which eight carried mcr-1. The AMR rates of the E. coli strains to ampicillin, amoxicillin/clavulanic acid, and tetracycline were all above 90%, and the resistance rates to ciprofloxacin, cefotaxime, ceftiofur, and florfenicol were above 70%. Multidrug-resistant strains accounted for 95.66% of the total isolates. Twelve E. coli strains showed colistin resistance, from which a total of 46 AMR genes and 36 virulence factors were identified through whole-genome sequencing. The mcr-1 gene resided on the IncHI2, IncI2-type and IncY-type plasmids, and mcr-1 was located in the nikA-nikB-mcr-1-pap2 gene cassette (three strains) or the pap2-mcr-1-ISApl1 structure (one strain). Completed IncI2-type plasmid pMCR4D31–3 sequence (62,259 bp) revealed that it may cause the horizontal transmission of mcr-1 and may increase the risk of its spread through the food chain. Taken together, the AMR of chicken-derived E. coli in the plateau is of concern, suggesting that it is very necessary for us to strengthen the surveillance in various regions under the background of one health.
Introduction
With rising antimicrobial resistance (AMR) to less toxic antibiotics, colistin is used as a backup drug to treat infections caused by carbapenem-resistant Enterobacteriaceae pathogens (Tonny and Anti, 2015; Sarica and Yurttutan, 2018). Due to the unregulated use of colistin in livestock before the antibiotics ban, colistin-resistant Enterobacteriaceae have already become prevalent. The gene mcr-1(1,626 bp), which mediates colistin resistance, was first reported on the IncI2-type plasmid of Escherichia coli (SHP45; Liu et al., 2016). Subsequently, the mcr gene series (mcr-1 to mcr-10) that mediates colistin resistance in plasmids of enterobacteria has been successively reported in more than 50 countries (Ling et al., 2020; Yang et al., 2020; Pan et al., 2021; Xu et al., 2021). The mcr-1 gene is present in various types of E. coli plasmids, including IncI, IncX, IncHI, IncP, and IncF (Wang et al., 2017). Of these, IncI2, IncX4, and IncHI2 are common and were the earliest reported plasmid types carrying the mcr-1 gene. The IncHI2-type plasmids from different origins are similar to each other and have become the most common effective carrier for the spread of mcr-1 in humans, animals, and food (Chang et al., 2020).
Escherichia coli is a common pathogenic bacteria in human infections and has caused various medical and community public safety issues. Manges et al. retrospectively analyzed relevant studies from 1995 to 2018 and found that the epidemiology and population dynamics of E. coli are complex and dynamic (Manges et al., 2019). It is worth noting that E. coli, as an indicator of the prevalence of Gram-negative bacteria, acts as a rich antibiotic resistance gene pool and a mobile center of resistance gene exchange (Poirel et al., 2018). The high prevalence of mcr-1, a colistin resistance gene in E. coli, poses a clinical threat, and its diverse genetic mechanisms have jointly promoted its spread and long-term presence (Wang et al., 2018; Zhong et al., 2018). China and Vietnam are the two countries with the most mcr-1-positive isolates verified through genome sequencing (Wang et al., 2018). However, studies on mcr-1 in China have been mostly concentrated in the central, eastern, and southern regions, while data in the northwestern region, especially in Qinghai Province, are still limited. This disparity may be related to the high altitude, low population density, and low avicultural intensity of Qinghai Province, allowing for poultry farming to be kept away from other human activities in most areas in this region. In the past, studies related to animal-derived mcr-1-carrying E. coli in Qinghai included four E. coli strains isolated from sheep diarrhea samples (Tang et al., 2019b), 14 E. coli strains isolated from pig feces specimens, and six extended-spectrum beta-lactamases (ESBLs)-carrying E. coli strains found in pigs and yaks in Xining and Haidong, respectively (Tong et al., 2018; Wang et al., 2020). Of the above-described resistance gene-carrying E. coli strains, only the sheep-derived E. coli strain has been fully genome sequenced. Moreover, animal-derived drug-resistant E. coli strains in this region have rarely been reported, and the scattered published results are insufficient for comprehensively understanding animal-derived AMR in Qinghai Province and its risks.
Surveillance of AMR in China is mainly in developed provinces and rarely involved in remote areas. Qinghai Province represents a unique geographical unit and is located in the hinterland of the Qinghai-Tibet Plateau, which is known as the third pole of the earth and the roof of the world. As one of the areas in China least disturbed by human activities, Qinghai is the origin of three rivers, i.e., the Yangtze River, the Yellow River, and the Lancang River, and has rich biodiversity (Wu et al., 2016). From 2012 to 2013, an E. coli strain obtained from wild Marmota himalayana in Yushu, Qinghai, was found to be sensitive to 23 antibiotics including penicillins, cephalosporins, and carbapenems (Bai et al., 2016; Lu et al., 2016). However, all E. coli strains obtained from wild plateau pika (Ochotona curzoniae) in Qinghai Province in 2016 were penicillin-resistant and some strains were cephalosporin- and streptomycin-resistant (Bai et al., 2016). Therefore, the evaluation of the AMR of zoonotic E. coli in Qinghai Province can help us understand the status of antibiotic pollution in animal husbandry and apicultural environments and provide references for formulating animal husbandry and natural environmental protection policies in this unique area.
In this study, we collected fresh stool samples from chickens of different varieties and different ages under different farming environments in Qinghai Province (at altitudes of ~2,500–4,000 m) from June 2020 to July 2021 and analyzed drug-resistant E. coli strains in the samples to address the inadequacy of previous studies on drug-resistant bacteria of animal origin in Qinghai Province. We performed whole-genome sequencing on the colistin-resistant E. coli isolates to reveal the prevalence of the animal-derived AMR gene mcr-1 and its transmission patterns in the environment.
Materials and methods
Isolate information
From June 2020 to July 2021, 375 cloaca stool samples were collected from 13 chicken farms in Qinghai Province (Figure 1A; Supplementary Table 1). Then, they were cultured in 5 ml of sterile buffered peptone water (BPW) at 37°C for enrichment, and then sequentially cultured in MacConkey agar (MacConkey, MAC) and eosin methylene blue (EMB) agar medium for identification. Putative E. coli colonies were selected and cultured on Luria broth (LB) plates and purified through three rounds of streaking, from which single E. coli colonies were determined through Bruker time-of-flight mass spectrometry (MALDI-TOF MS, Germany; Kostrzewa, 2018; Tang et al., 2021). The identified E. coli strains were stored at −80°C in a freezer in a 25% glycerol solution.
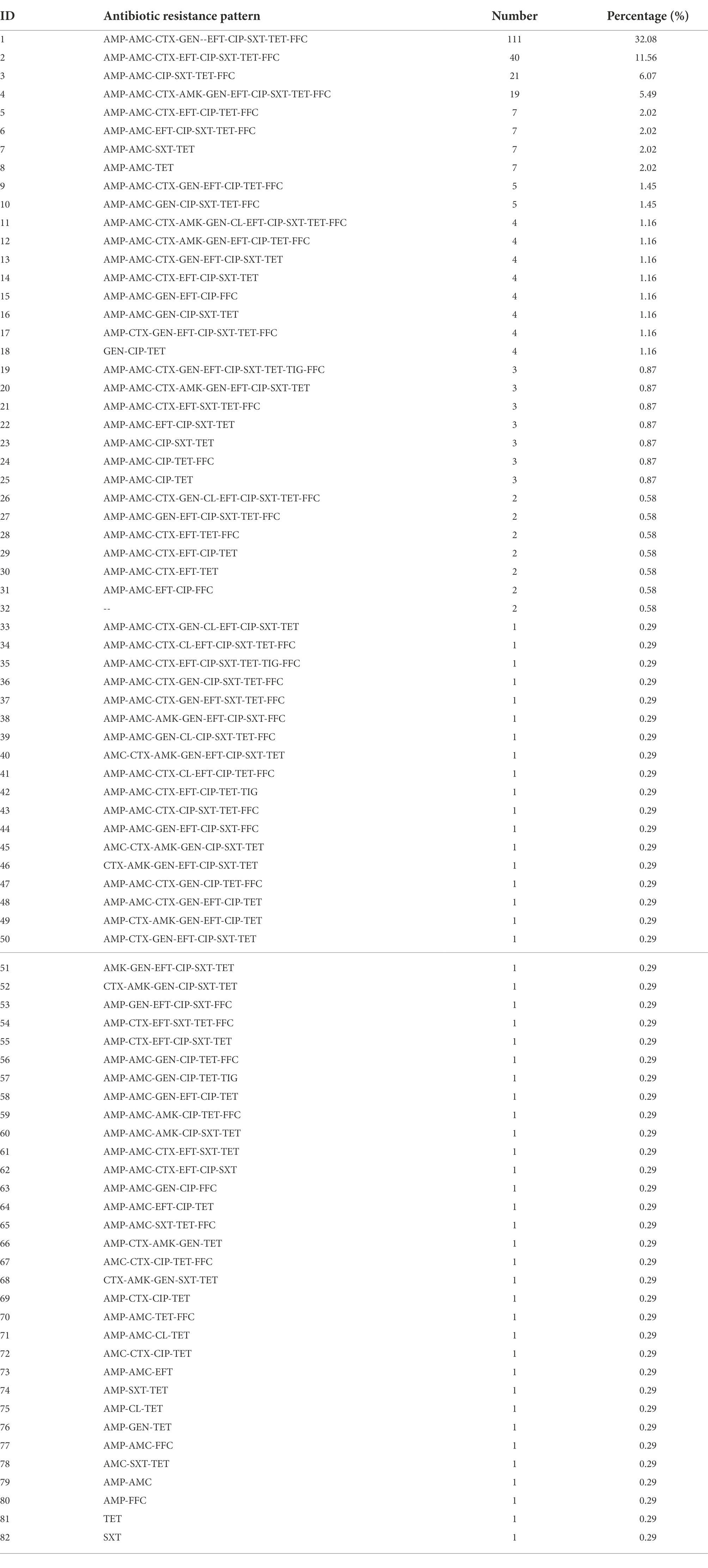
Figure 1. The sampling locations and susceptibility test results of 346 strains of Escherichia coli to 13 antibiotics. (A) Sampling areas in Qinghai Province, China. (B) AMR rates of E. coli. (C) The distribution of multidrug-resistant strains. (D) MIC distributions of 13 antibiotics in isolated E. coli strains.
Antimicrobial susceptibility testing
All isolated strains of E. coli were tested for antibiotic resistance using the standard broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The minimal inhibitory concentration (MIC) of 13 antibiotics was detected using a drug susceptibility panel for Gram-negative aerobic bacteria. The panel of antimicrobial compounds tested included ampicillin (AMP; 2–128 μg/ml), amoxicillin/clavulanate potassium (AMC; 4/2–128/64 μg/ml), cefotaxime (CTX; 0.06–8 μg/ml), meropenem (MEM; 0.5–16 μg/ml), amikacin (AMK; 2–64 μg/ml), gentamicin (GEN; 0.25–32 μg/ml), colistin (CL; 0.125–8 μg/ml), ceftiofur (EFT; 0.25–32 μg/ml), ciprofloxacin (CIP; 0.06–8 μg/ml), sulfamethoxazole (SXT; 9.5–304 μg/ml), tetracycline (TET; 0.25–64 μg/ml), tigecycline (TIG; 0.25–32 μg/ml), and florfenicol (FFC; 2–128 μg/ml; Tang et al., 2020).
To determine the bacterial viability of E. coli, serially diluted mid-log cultures were spotted on LB plates supplemented with different concentrations of colistin (0, 0.25, 0.5, 1, 2, 4, 8, and 16 μg/ml) and incubated at 37°C for 12 h. The E. coli strain ATCC 25922 was used as a negative control (Tang et al., 2019a).
A MIC test strip (Liofilchem®, Italy) was used to detect the MIC value of colistin-resistant E. coli and strain ATCC 25922 in this experiment. A sterile cotton swab was immersed in 0.5 McFarland standard suspension, which was then spread evenly on the MH (OXOID, England) agar medium (Tang et al., 2019a; Chang et al., 2020).
Whole-genome sequencing, identification of antimicrobial resistance genes, multilocus sequence typing analysis, virulence factors, and plasmid replicon types
DNA was extracted from strains of E. coli isolates showing colistin resistance using a commercially available bacterial genomic DNA isolation kit (Generay, China), according to the manufacturer’s instructions. The concentration of DNA in the extracted samples was detected with a NanoDrop 2000 spectrophotometer (Thermo Fisher, United States). The presence of the mcr-1 gene in E. coli isolates was detected by amplification using mcr-1-specific primers (Chang et al., 2020).
Indexed Illumina sequencing libraries were prepared using a TruSeq DNA PCR-free sample preparation kit (Illumina Inc., San Diego, CA) following the standard protocol, and samples were sequenced on the Illumina NovaSeq 6,000 platform according to the manufacturer’s protocols, thus producing 150-bp paired-end reads. The sequence was assembled with a CLC Genomics Workbench 12. Libraries of genomic DNA of representative strain were prepared by SQK-LSK109 kit (Oxford Nanopore Technologies, United Kingdom) and sequenced using R9.4 flow cell technology on a GridION sequencer. The reads were assembled using Unicycler 0.4.8. The NCBI Prokaryotic Genome Annotation Pipeline was used for gene prediction and annotation of the genome.
Antimicrobial resistance genes were predicted using ResFinder 4.1 software (Zankari et al., 2012).1 Virulence factors were predicted using Virulence Finder 2.0 software2 and the virulence factor database.3 Plasmid replicon types were predicted using PlasmidFinder 2.1 software.4 Multilocus sequence typing (MLST) was carried out as previously described using MLST 2.0 software.5 The BLAST Ring Image Generator (BRIG) and Easyfig software were used in the comparative analysis of plasmids (Alikhan et al., 2011; Sullivan et al., 2011).
S1-pulsed-field gel electrophoresis, Southern blotting, and conjugation transfer
The Salmonella enterica serovar Braenderup H9812 (as size marker) and E. coli cells harboring the mcr-1 gene were fixed by SeaKem Gold Agarose (Lonza Group AG, United States) and subsequently lysed. The embedded DNAs were digested using Xba I or S1 nuclease (Takara Bio, China) in a 37°C water bath for 3 h. The restricted DNA fragments were separated in 0.5 × Tris-Borate-EDTA buffer (Sangon Biotech, Shanghai, China) at 14°C for 18 h using a pulsed-field electrophoresis system (CHEF Mapper, Bio-Rad, United States) with pulse times of 2.2 to 63.8 s. The gel block was stained and observed with a gel imager (Tang et al., 2019a; Liang et al., 2021b). The mcr-1-specific probe was labeled using a DIG High Prime DNA Labeling and Detection Starter Kit (Roche, Sant Cugat del Vallés, Spain) following the manufacturer’s instructions. E. coli J53 was used as the recipient strain, and E. coli strains harboring mcr-1 were used as the donor strain in the conjugation transfer assay. Donor and recipients bacteria were grown together overnight and then inoculated onto LB plates with colistin and sodium azide.
Statistical analysis
The error values of conjugation transfer efficiency were calculated by GraphPad Prism software. The heatmap of clustering of strains and AMR phenotypes were performed by TBtools. In this analysis, the presence of the above resistance phenotype received a score of 2, intermediate received a score of 1, and the susceptibility received a score of 0.
Results
Isolation of Escherichia coli and antimicrobial susceptibility
A total of 346 strains of E. coli were isolated and identified from 375 stool samples (Figure 1A) collected in Qinghai Province, with an isolation rate of 92.27%. The sensitivity test results of 346 of the E. coli strains to 13 antibiotics are shown in Figures 1B,C. In terms of the MIC distribution (Figure 1D), the MIC values of the antibiotics AMP, AMC, CIP, CTX, EFT, TET, and FFC were significantly higher, indicating that the isolated E. coli strains are highly resistant to these antibiotics. Among these, AMP, AMC, and TET showed the highest resistance rates of over 90% and were followed by that of CIP, at 88.15%, while CTX, EFT, and FFC exhibited resistance rates greater than 70%. Among the isolates, strains with multidrug resistance (MDR) (resistance to more than three kinds of antibiotics) accounted for 95.66% of the total isolates. The tested strains showed a high resistance rate to GEN (58.09%) and a low resistance rate to AMK (11.85%). In addition, their resistance rates to colistin and TIG were 3.47% and 1.73%, respectively. All of the tested E. coli strains were sensitive to MEM. These results indicate that the AMR of chicken-derived E. coli in Qinghai Province is serious.
In this study, E. coli isolates showed a broad spectrum of resistance to most antibiotics, as shown in Table 1; Figure 2, among which the AMP-AMC-CTX-GEN-EFT-CIP-SXT-TET-FFC-resistant phenotype was the highest proportion (32.08%) out of a total of 111 strains. Among the 346 E. coli isolates, only two (0.58%) were sensitive to all of the antibiotics assayed, and the rest exhibited 81 types of resistance across a spectrum. Six of these strains (1.73%) showed resistance to eight of the tested antibiotic categories, with two types of resistance, AMP-AMC-CTX-AMK-GEN-CL-EFT-CIP-SXT-TET-FFC-resistant (four strains) and AMP-AMC-CTX-GEN-CL-EFT-CIP-SXT-TET-FFC-resistant (two strains). One hundred forty-three strains (41.33%) showed resistance to seven of the tested antibiotic categories, with seven types of resistance. Seventy-nine strains (22.83%) showed resistance to six of the tested antibiotic categories, with 18 types of resistance. In the multidrug resistance assay, 95.66% of the isolates showed resistance to the 13 tested antibiotics, indicating that in Qinghai Province, multidrug-resistant chicken-derived E. coli strains are common and have diverse and wide AMR spectra.

Figure 2. Antimicrobial resistance profiles of 346 Escherichia coli strains. Vertical and horizontal trees represent clustering relationships. The abscissa is the antibiotic, and the ordinate is the E. coli isolate.
Through the agar dilution method (Figure 3A), we found that the control strain ATCC 25922, could grow normally on plates without colistin but could not grow on plates with colistin (≥0.25 μg·ml−1). With 8 μg·ml−1 of colistin, the growth of QH20-4D3-1, QH20-5 T-2, QH21-3-15, QH21-5-14, QH21-2-13, QH21-3-13, and QH21-7-8 was completely inhibited.
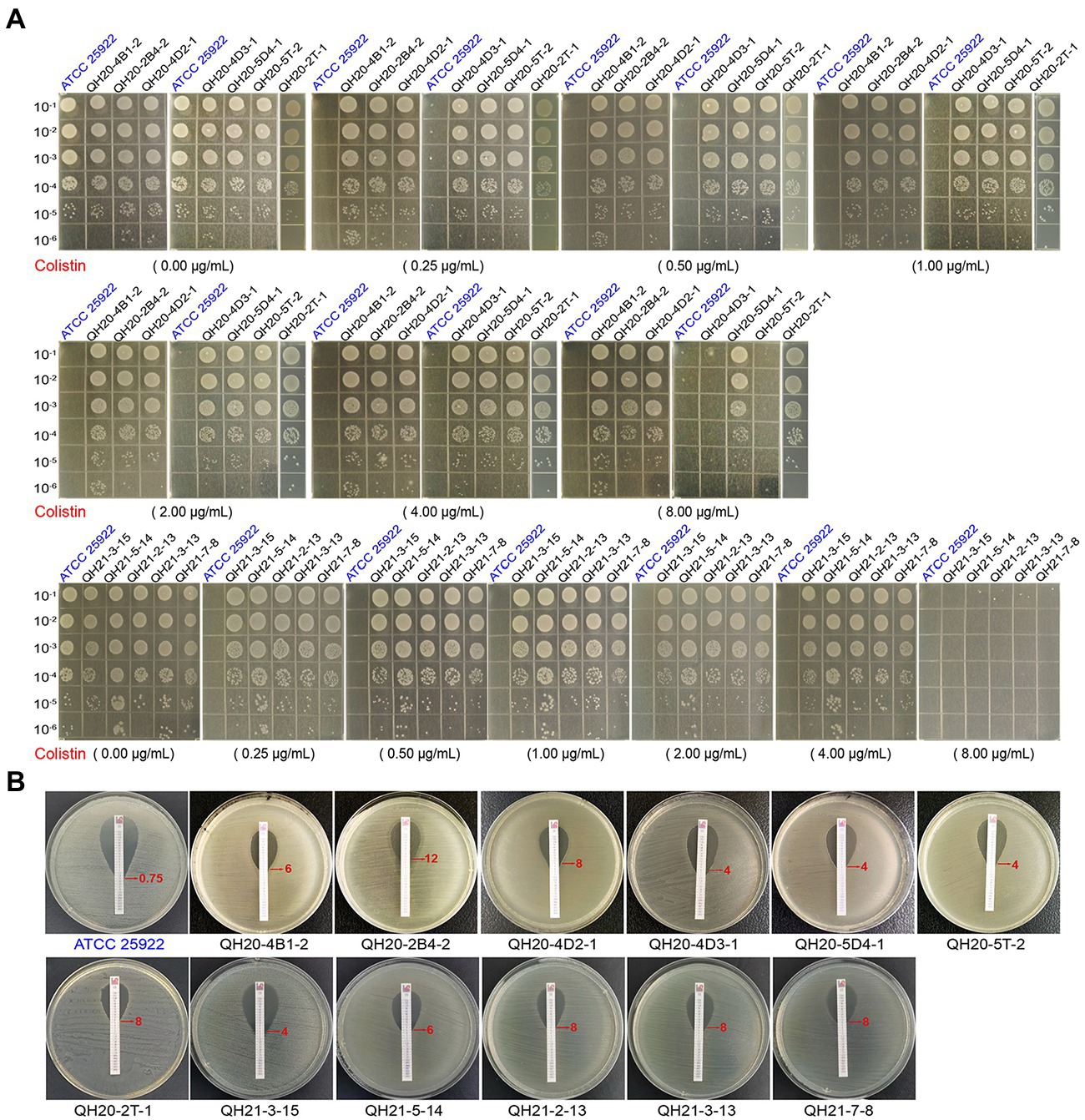
Figure 3. The insusceptibility of mcr-1-harboring Escherichia coli strains to colistin. (A) Bacterial viability of E. coli strains on LB plates containing different concentrations of colistin. The diluted suspension (10 μl) was inoculated onto the prepared LB plates. Escherichia coli ATCC 25922 was used as a negative control. (B) E-test to compare the MIC of colistin in E. coli strains. Escherichia coli ATCC 25922 was used as a negative control.
The colistin resistance of eight mcr-1-carrying E. coli strains was verified with the E-test strip: (0.016–256 μg·mL−1 of colistin; Figure 3B). The MIC of the negative control strain ATCC 25922 was 0.75 μg·ml−1, while those of the mcr-1-carrying E. coli strains were all ≥4 μg·ml−1, demonstrating colistin resistance.
Distribution of antimicrobial resistance genes, STs, virulence factors, and plasmid replicon types of Escherichia coli isolates
We sequenced the genomes of all 12 colistin-resistant E. coli isolates, from which 46 AMR genes were predicted, as shown in Figure 4A. The complete genome sequence of strain QH20-4D3-1 was performed by third-generation sequencing. Among them, 15 aminoglycoside resistance genes [aac(3)-IIa, aac(3)-IV, aadA1, aadA2, aadA5, aadA16, aadA22, aadA24, aadA2b, aph(3′)-Ia, aph(3′)-IIa, aph(3″)-Ib, aph(4)-IA, aph(6)-Id, and rmtB], eight beta-lactam resistance genes (blaCTX-M-55, blaCTX-M-64, blaCTX-M-65, blaOXA-10, blaTEM-141, blaTEM-206, blaTEM-214, blaTEM-1B, and blaTEM-1C), seven folate antagonist pathway resistance genes (dfrA12, dfrA14, dfrA17, dfrA27, sul1, sul2, and sul3), four fluoroquinolone resistance genes (oqxA, oqxB, qnrS1, and qnrS2), one lincosamide resistance gene [lnu(F)], two macrolide resistance genes [mdf(A) and mph(A)], one peptide resistance gene (mcr-1), two phenol resistance genes (cmlA1 and floR), one fosfomycin resistance gene (fosA3), two rifampicin resistance genes (arr-2 and arr-3), and two TET resistance genes [tet(A) and tet(M)] were detected. These strains were predicted to have nine different ST types (ST29735, ST43, ST10, ST93, ST162, ST2329, ST189, ST4689, and ST259; Supplementary Table 2), indicating that the E. coli isolates in this study are highly diverse. 8 of these 12 strains harbored the mcr-1 gene.
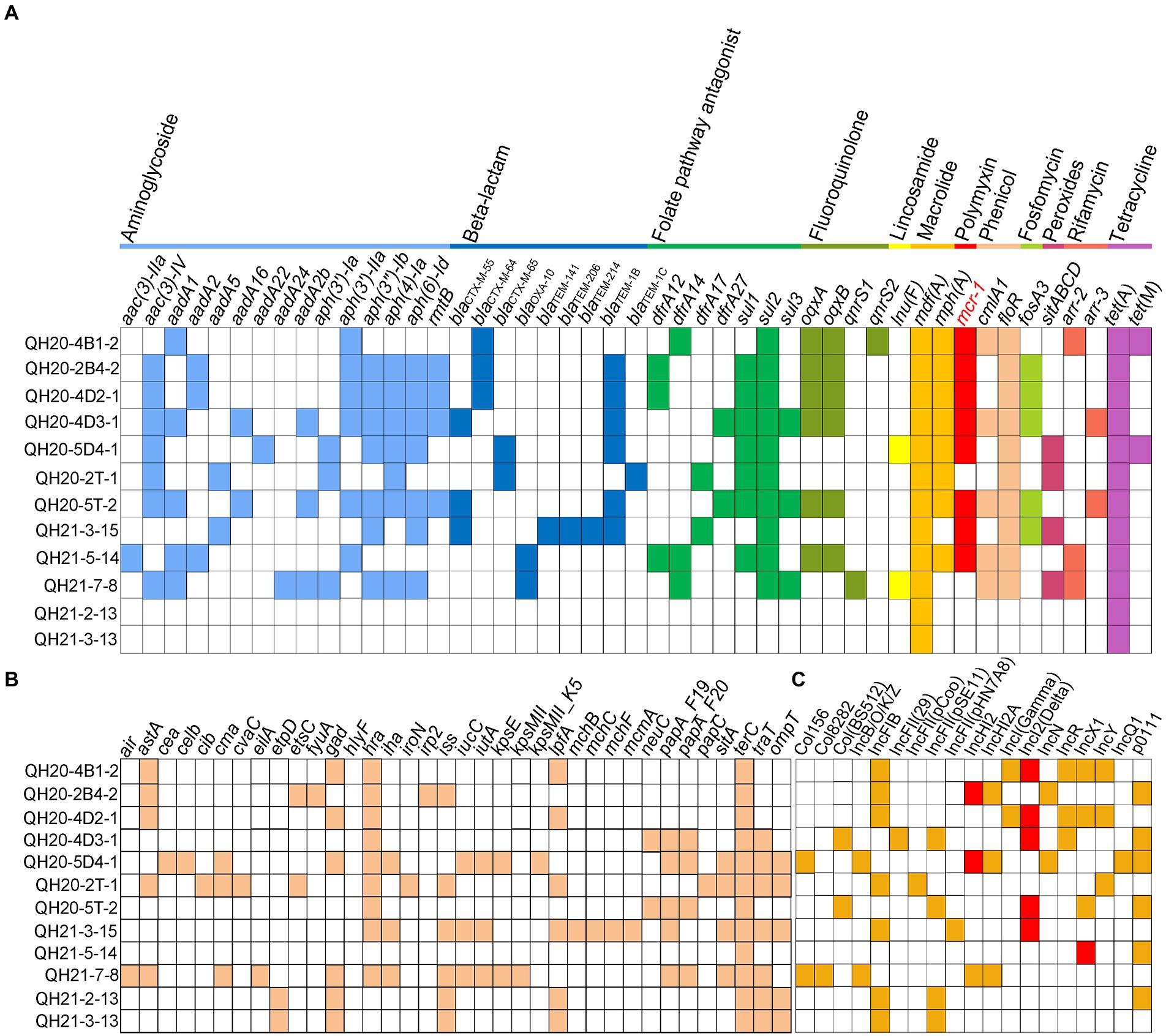
Figure 4. Acquired resistance genes, virulence genes, and plasmid replicon types of Escherichia coli strains with whole-genome sequences. (A) Antimicrobial resistance (AMR) gene profiles. Various types of AMR genes are labeled with different colors. White squares indicate no AMR genes. (B) Virulence gene profiles. Colored squares represent virulence genes, and white squares indicate no virulence genes. (C) Plasmid replicon profiles. Colored squares represent the presence of plasmids, and white squares indicate no plasmids.
A total of 36 virulence factors (Figure 4B) and 19 plasmid replicons (Figure 4C) were predicted from the 12 colistin-resistant E. coli isolates. Among them, the tellurium ion resistance virulence gene terC was found in all 12 strains, and the virulence gene astA, which is related to the EAST-1 heat-stable toxin and is a typical marker for entero-aggregating E. coli (EAEC), was found in QH20-4B1-2, QH20-2B4-2, QH20-4D2-1, QH20-2 T-1, and QH21-7-8. The plasmid replicators were as follows: Col156, Col8282, Col (BS512), IncB/O/K/Z, IncFIB (AP001918), IncFII (29), IncFII (pCoo), IncFII (pSE11), IncFII (pHN7A8), IncHI2, IncHI2A, INCI (Gamma), IncI2-type (Delta), IncN, INCR, IncX1, IncY, IncQ1, and po111. In the eight mcr-1-containing colistin-resistant isolates, the mcr-1 gene was located on the IncHI2-, IncI2-, or IncY-type plasmid (Figure 4C). Plasmids carrying the mcr-1 gene were verified by S1-PFGE and Southern blot (Supplementary Figure 1). The conjugation transfer frequency was ~10−2–10−6 for four strains QH20-4D2-1, QH21-5-14, QH20-4D3-1, and QH20-5 T-2, with QH20-4D2-1 exhibiting the highest frequency (2.97 ± 0.15) × 10−2. Whereas, the mcr-1-harboring plasmid in strains QH21-3-15, QH20-4B1-2, QH20-2B4-2, and QH20-5D4-1 were not found to be conjugatively transferred (Supplementary Figure 2).
In QH20-2 T-1, the etsC, hlyF, and ompT virulence genes all resided on the replicon of the IncFIB-type plasmid. The virulence gene etpD was also predicted to be on the IncFIB-type plasmid replicon of QH21-2-13 and QH21-3-13, while the virulence gene traT was predicted to be on the IncFII replicon of Plasmid 4 of QH21-3-13 and QH20-4D3-1. The virulence genes traT and ompT are related to the activity of biofilm outer membrane proteins, etsC confers resistance to the serum complement, and the ets operon and ompT and hlyF are related to the conserved virulence region of the FIB replicon (Moran and Hall, 2018). Therefore, different types of plasmid replicons facilitate the horizontal spread of virulence genes.
Sequence alignment analysis of homologous IncI2-type and IncHI2-type plasmids carrying the mcr-1 gene
In different isolates, the contig on which mcr-1 resides ranged from 11,011–123,144 bp in size. In QH21-5-14, the contig that contains mcr-1 was too short, making it impossible to analyze the sequence. In the other seven E. coli isolates, the mcr-1 gene was located on the IncI2-type (4 strains) or IncHI2-type (1 strain) plasmid. Here, we selected the sequences of the mcr-1-carrying IncI2-type plasmid pHNSHP45 (Acc. No. KP347127) and the IncHI2-type plasmid pHNSHP45-2 (Acc. No. KU341381) that were the earliest to be reported for sequence alignment analysis.
As shown in Figure 5A, the alignment of the IncI2-type plasmid pHNSHP45 sequence with the sequences of plasmids pMCR4D31–3 in QH20-4D3-1, QH21-3-15, QH20-5 T-2, QH20-4D2-1, and QH20-4B1-2 (123.144 kp, GC content: 42.6%, CP085520), QH21-3-15 contig_75 (≈59.073 kp, GC content: 42.2%, JAJDKL010000075), QH20-5 T-2 contig_2 (≈56.045 kp, GC content: 42.2%, JAJDKK010000002), QH20-4D2-1 contig_81 (32.841 kp, GC content: 42.6%, JAJDKI010000081), and QH20-4B1-2 (≈11.011 kp, GC content: 42.3%, JAJDKH010000139) indicates that all sequences were highly homologous, contained the backbone of the typical IncI2-type plasmid, and included the replication-related gene (repA), conjugative transfer-related pilus gene cluster genes (tra, pil), and the plasmid stability-associated gene (parA). The backbone region of the IncI2-type plasmid is conserved, containing a shufflon region led by the site-specific recombinase gene (rci; Sekizuka et al., 2017). Moreover, upstream of mcr-1, all sequences lacked the insertion element ISApl1. The pap2 gene was always present downstream of mcr-1 except for QH20-4D2-1 contig_81 (JAJDKI010000081), which contained blaCTX-M-64, an independent resistance region of the ESBL resistance gene. This region was located in ISEcp1-blaCTX-M-64-Δorf477, a typical transposon unit, of which the ISEcp1 insertion sequence can mediate the transposition of many types of the blaCTX-M gene (Zhao et al., 2020). Except in the case of QH20-4B1-2 contig_139 (JAJDKH010000139), the resistance gene mcr-1 was integrated at a location downstream to the nikB gene, which is identical to that of the majority of mcr-1-positive IncI2-type plasmids reported previously. The other five sequences all lacked the IS683 insertion sequence present in pHNSHP45, while two IS2 insertion elements were present in the transposase genes insD and insC downstream of mcr-1 in pMCR4D31–3. In pMCR4D31–3, QH21-3-15 contig_75 (JAJDKL010000075) and QH20-5 T-2 contig_2 (JAJDKK010000002), a virB/virD4 secretion system similar to that of pHNSHP45 was present.
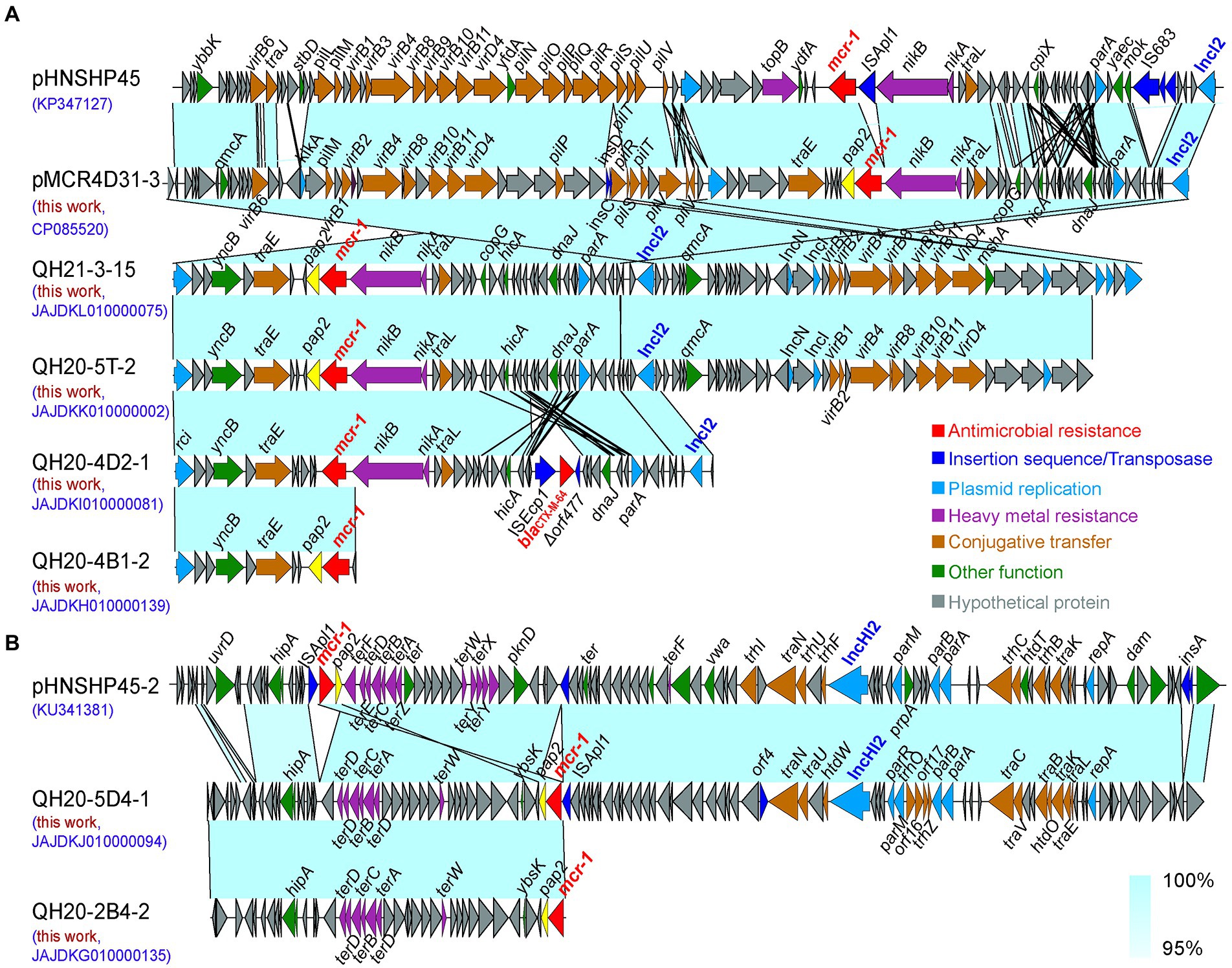
Figure 5. Sequence alignment analysis of IncI2 (A) and IncHI2-type (B) plasmids carrying the mcr-1 gene. The arrows indicate the direction of gene transcription. Functional genes are shown in different colors. Regions of >95% nucleotide homology are indicated by colored shading.
As shown in Figure 5B, the sequence alignment of the IncHI2-type plasmid pHNSHP45-2 and QH20-5D4-1 contig_94 (≈107.905 kp, GC content: 44.0%, JAJDKJ010000094) and QH20-2B4-2 contig_135 (≈38.480 kp, GC content: 49.1%, JAJDKG010000135) indicates that the three sequences were highly homologous. In the two IncHI2-type mcr-1-containing plasmids obtained in this study, the pHNSHP45-2 toxin-antitoxin system (higB and hipB-A) and the region mediating resistance to heavy metal tellurium (terDDCBAD) were present. QH20-5D4-1 contig_94 (JAJDKJ010000094) carried four partition-associated genes (parR, parM, parB, and parA) and 16 plasmid-independent conjugation genes (traN, traU, traC, traV, traB, traK, traE, traL, htdO, htdW, orf4, orf17, orf16, trhP, trhO, and trhZ). Moreover, in all cases, the pap2 gene was present downstream; however, in QH20-2B4-2 contig_135 (JAJDKG010000135), the insertion element ISApl1 present in QH20-5D4-1 contig_94 was absent. In the QH20-2B4-2 contig_135 sequence, the 3-terminus of mcr-1 was incomplete, and we were therefore unable to obtain information about the genetic elements of plasmid replicon. However, in the QH20-5D4-1 contig_94 sequence, the pap2-mcr-1-ISApl1 structure that mediates the horizontal transfer of mcr-1 and is present in pHNSHP45-2 was observed.
Analysis of IncI2-type mcr-1-carrying plasmids from different sources
For sequence alignment analysis, we selected nine plasmids from the NCBI database with the highest homology to the sequence of pMCR4D31–3 carried in strain QH20-4D3-1 (Figure 6) and found that the homology between pMCR4D31–3 and all the selected plasmids was over 99% (Supplementary Table 3). Among the selected plasmids, pEC13-1 was from a water sample in a Malaysian pond (Yu et al., 2017), p2018-10-2CC was from a fecal sample of healthy residents in a rural community in Vietnam (Yamaguchi et al., 2020), p1106-IncI2-type and pAH62-1 were from poultry meat samples of Anhui Province, China (Liu et al., 2019; Yin et al., 2021), pHLJ109-11 was from the cecal content of chickens that had died of illness in Heilongjiang Province, China (Li et al., 2020), and the other four plasmids, pColR644SK1, pL889-MCR1, p5CRE51-MCR-1, and p778, were from patients from Switzerland, Zhejiang, China, Taiwan, China, and Ecuador, respectively (Zurfluh et al., 2017; Lin et al., 2019; Loayza-Villa et al., 2020; Liang et al., 2021a). Unfortunately, the patient from which p5CRE51-MCR-1 was isolated died of illness despite treatment. Seven plasmids, including pMCR4D31–3, all contained the nikA-nikB-mcr-1-pap2 structure. Among the other three plasmids, pColR644SK1 and pEC13-1 (from Switzerland and Malaysia, respectively) lacked the nikA gene in the mcr-1 cassette; p2018-10-2CC only contained the pap2 gene in sequences flanking the mcr-1 gene. Overall, the mcr-1 gene in pMCR4D31–3 showed a genetic neighborhood similar to that of the IncI2-type plasmids from various origins, emphasizing the feasibility of horizontal transmission.
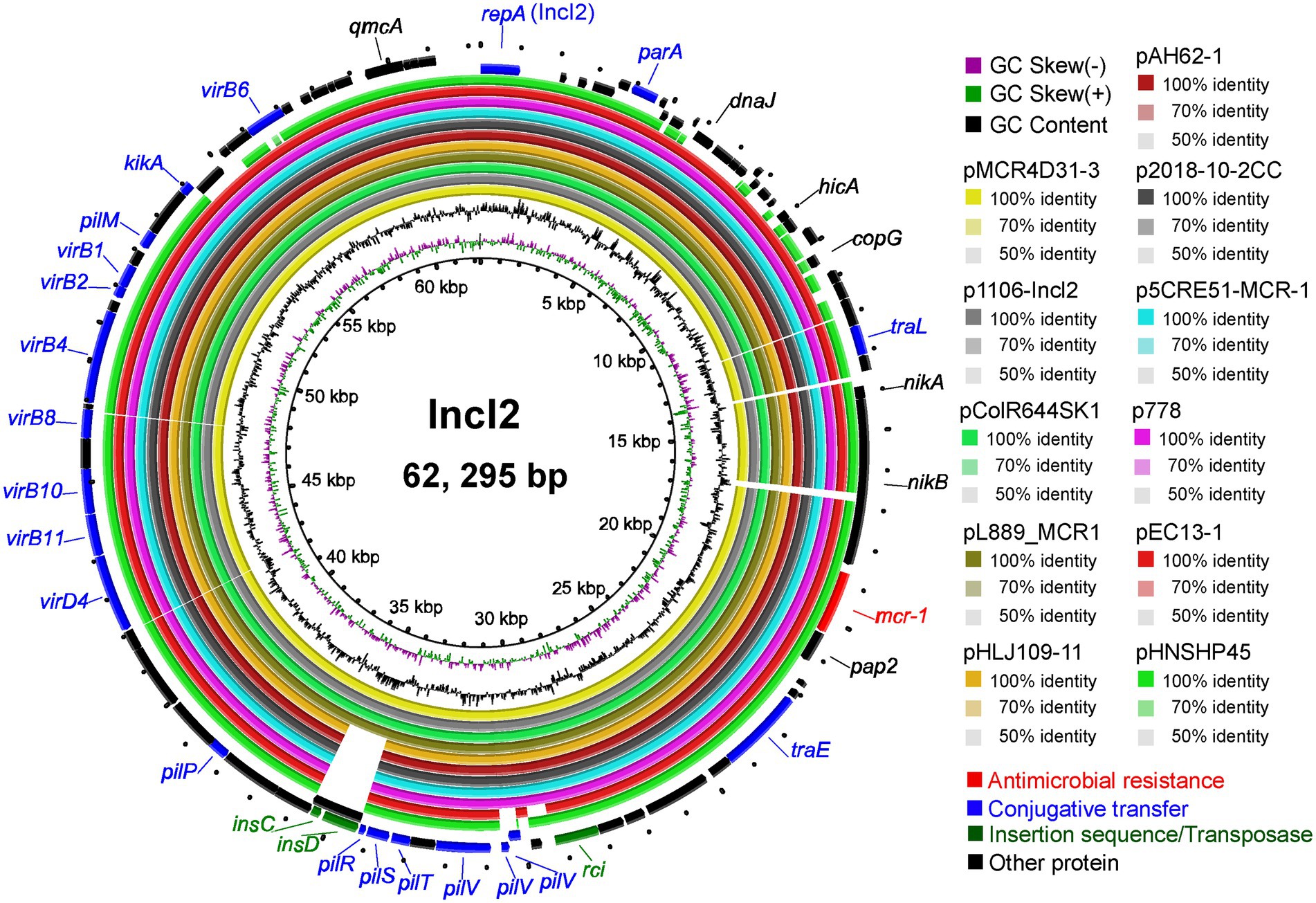
Figure 6. Circular diagram of the IncI2 plasmids. Circles, from inside to outside, indicate the GC skew, GC content, and ORFs in the positive and negative DNA strands. Genes are shown in different colors.
Discussion
In the 21st century, antimicrobial resistance is considered to be one of the most serious global threats to human health. The AMR of E. coli is closely associated with the abuse of antibiotics, with a high rate of resistance to antibiotics that have been used for a long time in poultry farming but a very low rate of resistance to antibiotics that are prohibited or restricted in poultry farming (Rhouma et al., 2016; Shen et al., 2016). Poultry farming varies profoundly in different regions of China in terms of scale and method, and the use of antibiotics also varies greatly among regions due to differences in the geographic environment and farming methods. In this study, we found that the antibiotic resistance rate of E. coli in poultry farms of Qinghai Province was high, with a resistance rate of 3.47% to colistin and a resistance rate of 1.73% to TIG, and all the E. coli isolates in this study were sensitive to MEM. Although poultry farming in Qinghai Province is smaller in scale, shorter in history, and has a more independent poultry farming environment, the drug-resistant E. coli in Qinghai is as serious as that in inland areas, suggesting antibiotic abuses in poultry farming in this region. Given that Qinghai Province has a small population and a small number of poultry farms in the western region, and the samples of this study were mostly collected from the eastern areas of the province, the data were limited, and more follow-up monitoring is needed on the drug-resistant E. coli strains from chickens in China’s plateau region.
Colistin resistance derived from the horizontal transfer of the mcr-1 gene has led to an epidemic risk of colistin-resistant bacteria (Wang et al., 2017). In 8022 stool samples, Zhong et al. found a mcr-1 positivity rate of up to 6.2%. It is the diverse genetic mechanisms of the mcr-1-carrying IncI and IncHI2/HI2A plasmids that cause the high horizontal transfer rate of mcr-1, e.g., the mcr-1-related insertion element ISApl1 (1,070 bp; Zhong et al., 2018), which was originally discovered in Actinobacillus pleuropneumoniae (Tegetmeyer et al., 2008), is the most important mobile element of the IS30 family mediating the spread of mcr-1. The Tn6330 transposon (ISApl1-pap2-mcr-1-ISApl1) can mediate the horizontal transfer of mcr-1, in which ISApl1 forms a ring-shaped intermediate in a “cut-and-paste” manner to mediate the spread of mcr-1 among different types of plasmids (Girardello et al., 2021). In the QH20-5D4-1 isolate, the transposon element ISApl1 was present downstream of mcr-1, and the pap2 gene present upstream of mcr-1 encodes the phospholipase closely related to the horizontal transmission of mcr-1 (Ma et al., 2021), both playing a very important role in mcr-1 mediated bacterial colistin resistance (Choi et al., 2020). Although we did not observe the Tn6330 transposon unit that mediates the rapid spread of the mcr-1 gene in the poultry farms of Qinghai Province, the insertion element ISApl1 and the pap2 gene neighboring mcr-1 similarly mobilized the plasmid-mediated horizontal spread of the mcr-1 gene.
With the exception of QH20-4D2-1 contig_81 (JAJDKI010000081), all the sequences of the mcr-1-carrying E. coli isolates contained the pap2 gene nearby; this gene is associated with colistin resistance and the horizontal transmission of mcr-1. However, only in QH20-5D4-1 contig_94 (JAJDKJ010000094), the pap2-mcr-1-ISApl1 structure was present near mcr-1, and Wang et al. noted that the missing ISApl1 insertion sequence is a remanent of the frequent shuffling of the pap2-mcr-1-ISApl1 structure (Wang et al., 2017). It is worth noting that in the QH20-4D2-1 contig_81 sequence downstream to mcr-1, the ESBL gene blaCTX-M-64, which is capable of degrading the third and fourth generations of cephalosporins, was observed. This gene also mediates resistance to amoxicillin, AMP, aztreonam, cefepime, CTX, ceftazidime, ceftriaxone, piperacillin, ticarcillin, and other antibiotics. This gene is located in ISEcp1-blaCTX-M-64-Δorf477, a typical transposon unit, in which ISEcp1 and ISApl1 are two important insertion sequences for the horizontal transfer of mcr-1 (Wang et al., 2017). In 2016, the co-occurrence of blaCTX-M and mcr-1 in the same E. coli strain was first reported (Zhang et al., 2016), and the IncI2-type E. coli plasmids simultaneously carrying the two genes have since been continuously reported (Gao et al., 2016; Sun et al., 2016), possibly because the IncI2-type plasmids are also important carriers and transmission vectors of the blaCTX-M resistance gene (Wu et al., 2018). The co-presence of mcr-1 and blaCTX-M-64 on the same plasmid may accelerate the spread of the two genes through co-selection (Sun et al., 2016), which provides a way for the bacterium to acquire multidrug resistance more quickly. Consequently, the risk and threat of the epidemic spread of multidrug-resistant bacteria is increased, further suggesting that E. coli with the mcr-1 gene may have pan-drug resistance (PDR) and cause infections that are difficult to treat clinically. To our knowledge, this study represents the first report on the co-existence of the animal-derived ESBL resistance gene and mcr-1 resistance gene in the same E. coli plasmid in Qinghai Province.
pMCR4D31–3 shows high homology to the IncI2-type mcr-1-carrying plasmids that are geographically distant and different in species. Loayza-Villa et al. discovered the first IncI2-type mcr-1-carrying E. coli plasmid (p778) in Ecuador, which was isolated from a 14-year-old boy. Meanwhile, three non-identical E. coli clones isolated from the chickens and dogs of the boy’s family contained similar IncI2-type plasmids, which supports the notion that mcr-1 can be horizontally transmitted between animals and humans living in the same environment (Loayza-Villa et al., 2020). When pHNSHP45, a plasmid carrying the mcr-1 gene, was first reported, Liu et al. proposed that given that the proportion of mcr-1-positive samples in animals is far greater than that of mcr-1-positive samples in humans, the mcr-1-mediated colistin resistance most likely originated in animals and was then transmitted to humans (Liu et al., 2016). Zurfluh et al. analyzed the mcr-1-carrying IncI2-type plasmids isolated from the patient’s urethra, diarrhea patients, and those with travel history to Asian countries (pColR644SK1) as well as from retail poultry meat and turkey meat, and found that the horizontal transfer of the mcr-1 gene via plasmids is the primary method of spread along the food chain, indicating that the food chain may be an important transmission route of mcr-1-carrying plasmids (Zurfluh et al., 2017). It is worth noting that p778 and pColR644SK1 have high homology to pMCR4D31–3 isolated in this study, with similar mcr-1 gene cassettes; thus, it is necessary to prevent and control the pMCR4D31-3-mediated horizontal transmission of the mcr-1 gene between animals and humans in the same environment and the risk of its spread along the food chain.
Conclusion
In this study, we provide data on the AMR of chicken-derived E. coli in Qinghai Province and the genetic environment of the mcr-1 gene and for the first time, generated a complete plasmid profile of the IncI2-type mcr-1-carrying plasmid from Qinghai, which helps us understand the prevalence of mcr-1 in animal-derived E. coli in this area. In summary, our study indicates that it is necessary to continuously monitor the AMR of E. coli and the prevalence of mcr-1 in the plateau region, which is of great significance to understanding and controlling the spread of bacterial antibiotic resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Zhejiang Academy of Agricultural Sciences. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
BT: conceptualization. HY: funding acquisition. BT, JC, XZ, JM, JuW, XJ, and BD: investigation. JiW, XZ, and BD: methodology. BT and BD: supervision. JM, JiW, and BT: visualization. BT and JiW: writing—original draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Research and Development Program of Zhejiang Province (2020C02031), the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2010DS700124-ZZ2008, ZZ2102), and the Collaborative Extension Plan of Major Agricultural Technologies in Zhejiang Province (2021XTTGXM03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.885132/full#supplementary-material
Footnotes
1. ^https://cge.cbs.dtu.dk/services/ResFinder/
2. ^https://cge.cbs.dtu.dk/services/VirulenceFinder/
3. ^http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Bai, X., Zhang, W., Tang, X., Xin, Y., Xu, Y., Sun, H., et al. (2016). Shiga toxin-producing E. coli in plateau Pika (O. curzoniae) on the Qinghai-Tibetan plateau, China. Front. Microbiol. 7:375. doi: 10.3389/fmicb.2016.00375
Chang, J., Tang, B., Chen, Y., Xia, X., Qian, M., and Yang, H. (2020). Two IncHI2 plasmid-mediated colistin-resistant E. coli strains from the broiler chicken supply chain in Zhejiang Province, China. J. Food Prot. 83, 1402–1410. doi: 10.4315/JFP-20-041
Choi, Y., Lee, J. Y., Lim, S. K., and Ko, K. S. (2020). Intact pap2 downstream of mcr-1 appears to be required for colistin resistance. Diagn. Microbiol. Infect. Dis. 97:114997. doi: 10.1016/j.diagmicrobio.2020.114997
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and mechanism for the mcr-1 Colistin resistance. PLoS Pathog. 12:e1005957. doi: 10.1371/journal.ppat.1005957
Girardello, R., Piroupo, C. M., Martins, J. Jr., Maffucci, M. H., Cury, A. P., Franco, M. R. G., et al. (2021). Genomic characterization of mcr-1.1-Producing E. coli recovered from human infections in São Paulo, Brazil. Front. Microbiol. 12:663414. doi: 10.3389/fmicb.2021.663414
Kostrzewa, M. (2018). Application of the MALDI Biotyper to clinical microbiology: progress and potential. Expert Rev. Proteomics 15, 193–202. doi: 10.1080/14789450.2018.1438193
Li, X., Sun, R., Song, J., Fang, L., Zhang, R., Lian, X., et al. (2020). Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int. J. Antimicrob. Agents 55:105806. doi: 10.1016/j.ijantimicag.2019.09.010
Liang, Z., Pang, J., Hu, X., Nie, T., Lu, X., Li, X., et al. (2021b). Low prevalence of mcr-1 Among clinical Enterobacteriaceae isolates and co-transfer of mcr-1 and blaNDM-1 from separate donors. Microb. Drug Resist. 27, 476–484. doi: 10.1089/mdr.2020.0212
Liang, G., Rao, Y., Wang, S., Chi, X., Xu, H., and Shen, Y. (2021a). Co-occurrence of NDM-9 and MCR-1 in a human gut Colonized E. coli ST1011. Infect. Drug Resist. 14, 3011–3017. doi: 10.2147/IDR.S321732
Lin, Y. C., Kuroda, M., Suzuki, S., and Mu, J. J. (2019). Emergence of an E. coli strain co-harbouring mcr-1 and blaNDM-9 from a urinary tract infection in Taiwan. J. Glob. Antimicrob. Resist. 16, 286–290. doi: 10.1016/j.jgar.2018.10.003
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, X., Geng, S., Chan, E. W.-C., and Chen, S. (2019). Increased prevalence of E. coli strains from food carrying blaNDM and mcr-1-bearing plasmids that structurally resemble those of clinical strains, China, 2015 to 2017. Euro Surveill. 24:1800113. doi: 10.2807/1560-7917.ES.2019.24.13.1800113
Liu, Y., Wang, Y., Walsh, T. R., Yi, L., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Loayza-Villa, F., Salinas, L., Tijet, N., Villavicencio, F., Tamayo, R., Salas, S., et al. (2020). Diverse E. coli lineages from domestic animals carrying colistin resistance gene mcr-1 in an Ecuadorian household. J. Glob. Antimicrob. Resist. 22, 63–67. doi: 10.1016/j.jgar.2019.12.002
Lu, S., Jin, D., Wu, S., Yang, J., Lan, R., Bai, X., et al. (2016). Insights into the evolution of pathogenicity of E. coli from genomic analysis of intestinal E. coli of M. himalayana in Qinghai-Tibet plateau of China. Emerg. Microbes Infect. 5, 1–9. doi: 10.1038/emi.2016.122
Ma, Z., Liu, J., Chen, L., Liu, X., Xiong, W., Liu, J., et al. (2021). Rapid increase in the IS26-mediated cfr gene in E. coli isolates with IncP and IncX4 plasmids and co-existing cfr and mcr-1 genes in a swine farm. Pathogens 10:33. doi: 10.3390/pathogens10010033
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. D. (2019). Global Extraintestinal Pathogenic E. coli (ExPEC) lineages. Clin. Microbiol. Rev. 32, e00135–e00118. doi: 10.1128/cmr.00135-18
Moran, R. A., and Hall, R. M. (2018). Evolution of regions containing antibiotic resistance genes in FII-2-FIB-1 ColV-Colla virulence plasmids. Microb. Drug Resist. 24, 411–421. doi: 10.1089/mdr.2017.0177
Pan, Y., Fang, Y., Song, X., Lyu, N., Chen, L., Feng, Y., et al. (2021). Co-occurrence of mcr-9, extended spectrum beta-lactamase (ESBL) and AmpC genes in a conjugative IncHI2A plasmid from a multidrug-resistant clinical isolate of Salmonella diarizonae. J. Infect. 82, 84–123. doi: 10.1016/j.jinf.2020.11.008
Poirel, L., Madec, J. Y., Lupo, A., Schink, A. K., Kieffer, N., Nordmann, P., et al. (2018). Antimicrobial Resistance in E. coli. Microbiol. Spectr. 6: ARBA-0026-2017. doi: 10.1128/microbiolspec.ARBA-0026-2017
Rhouma, M., Beaudry, F., Theriault, W., and Letellier, A. (2016). Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front. Microbiol. 7:1789. doi: 10.3389/fmicb.2016.01789
Sarica, S., and Yurttutan, S. (2018). An evaluation of hearing in infants administered with colistin in the premature neonatal intensive care unit. J. Matern. Fetal Neonatal Med. 31, 2918–2922. doi: 10.1080/14767058.2018.1479388
Sekizuka, T., Kawanishi, M., Ohnishi, M., Shima, A., Kato, K., Yamashita, A., et al. (2017). Elucidation of quantitative structural diversity of remarkable rearrangement regions, shufflons, in IncI2 plasmids. Sci. Rep. 7:928. doi: 10.1038/s41598-017-01082-y
Shen, Z., Wang, Y., Shen, Y., Shen, J., and Wu, C. (2016). Early emergence of mcr-1 in E. coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/S1473-3099(16)00061-X
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Li, X., Yang, R., Fang, L., Huo, W., Li, S., et al. (2016). Complete nucleotide sequence of an IncI2 plasmid Coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 60, 5014–5017. doi: 10.1128/aac.00774-16
Tang, B., Chang, J., Cao, L., Luo, Q., Xu, H., Lyu, W., et al. (2019a). Characterization of an NDM-5 carbapenemase-producing E. coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol. 19:82. doi: 10.1186/s12866-019-1454-2
Tang, B., Chang, J., Zhang, L., Liu, L., Xia, X., Hassan, B. H., et al. (2020). Carriage of distinct mcr-1-harboring plasmids by unusual serotypes of Salmonella. Adv. Biosyst. 4:e1900219. doi: 10.1002/adbi.201900219
Tang, B., Ma, Y., He, X., Zhou, Q., Chang, J., Qian, M., et al. (2021). Similar antimicrobial resistance of E. coli strains isolated from retail chickens and poultry farms. Foodborne Pathog. Dis. 18, 489–496. doi: 10.1089/fpd.2021.0019
Tang, F., Wang, J., Li, D., Gao, S., Ren, J., Ma, L., et al. (2019b). Comparative genomic analysis of 127 E. coli strains isolated from domestic animals with diarrhea in China. BMC Genomics 20:212. doi: 10.1186/s12864-019-5588-2
Tegetmeyer, H. E., Jones, S. C., Langford, P. R., and Baltes, N. (2008). ISApl1, a novel insertion element of A. pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 128, 342–353. doi: 10.1016/j.vetmic.2007.10.025
Tong, H., Liu, J., Yao, X., Jia, H., Wei, J., Shao, D., et al. (2018). High carriage rate of mcr-1 and antimicrobial resistance profiles of mcr-1-positive E. coli isolates in swine faecal samples collected from eighteen provinces in China. Vet. Microbiol. 225, 53–57. doi: 10.1016/j.vetmic.2018.09.018
Tonny, L., and Anti, D. (2015). Colistin: an antibiotic and its role in multiresistant gram-negative infections. Acta Med. Indones. 47, 157–168.
Wang, Q., Sun, J., Li, J., Ding, Y., Li, X. P., Lin, J., et al. (2017). Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 5:70. doi: 10.1186/s40168-017-0288-0
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wang, Y., Zhou, J., Li, X., Ma, L., Cao, X., Hu, W., et al. (2020). Genetic diversity, antimicrobial resistance and extended-spectrum beta-lactamase type of E. coli isolates from chicken, dog, pig and yak in Gansu and Qinghai provinces, China. J. Glob. Antimicrob. Resist. 22, 726–732. doi: 10.1016/j.jgar.2020.06.028
Wu, J., Duan, D., Lu, J., Luo, Y., Wen, X., Guo, X., et al. (2016). Inorganic pollution around the Qinghai-Tibet plateau: An overview of the current observations. Sci. Total Environ. 550, 628–636. doi: 10.1016/j.scitotenv.2016.01.136
Wu, C., Wang, Y., Shi, X., Wang, S., Ren, H., Shen, Z., et al. (2018). Rapid rise of the ESBL and mcr-1 genes in E. coli of chicken origin in China, 2008-2014. Emerg. Microbes Infect. 7:30. doi: 10.1038/s41426-018-0033-1
Xu, T., Zhang, C., Ji, Y., Song, J., Liu, Y., Guo, Y., et al. (2021). Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ. Pollut. 268:115706. doi: 10.1016/j.envpol.2020.115706
Yamaguchi, T., Kawahara, R., Hamamoto, K., Hirai, I., Khong, D. T., Nguyen, T. N., et al. (2020). High prevalence of Colistin-Resistant E. coli with chromosomally carried mcr-1 in healthy residents in Vietnam. mSphere 5, e00117–e00120. doi: 10.1128/mSphere.00117-20
Yang, X., Peng, K., Zhang, Y., Liu, L., and Li, R. (2020). Characterization of a novel mcr-8.2-bearing plasmid in ST395 Klebsiella pneumoniae of chicken origin. Infect. Drug Resist. 13, 1781–1784. doi: 10.2147/IDR.S256544
Yin, D., Cheng, B., Yang, K., Xue, M., Lin, Y., Li, Z., et al. (2021). Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in avian pathogenic E. coli isolates from diseased chickens in Anhui Province in China. mSphere 6, e01135–e01120. doi: 10.1128/mSphere.01135-20
Yu, C. Y., Ang, G. Y., Chong, T. M., Chin, P. S., Ngeow, Y. F., Yin, W. F., et al. (2017). Complete genome sequencing revealed novel genetic contexts of the mcr-1. J. Antimicrob. Chemother. 72, 1253–1255. doi: 10.1093/jac/dkw541
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, H., Seward, C. H., Wu, Z., Ye, H., and Feng, Y. (2016). Genomic insights into the ESBL and MCR-1-producing ST648 Escherichiacoli with multi-drug resistance. Sci. Bull. 61, 875–878. doi: 10.1007/s11434-016-1086-y
Zhao, Q., Chen, P., Yang, L., Cai, R., Zhu, J., Fang, L., et al. (2020). Transmission of plasmid-borne and chromosomal blaCTX-M-64 among E. coli and Salmonella isolates from food-producing animals via ISEcp1-mediated transposition. J. Antimicrob. Chemother. 75, 1424–1427. doi: 10.1093/jac/dkaa044
Zhong, L., Phan, H. T. T., Shen, C., Vihta, K. D., Sheppard, A. E., Huang, X., et al. (2018). High rates of human fecal carriage of mcr-1-positive multidrug-resistant Enterobacteriaceae emerge in China in association With successful plasmid families. Clin. Infect. Dis. 66, 676–685. doi: 10.1093/cid/cix885
Keywords: Escherichia coli, antimicrobial resistance, Qinghai Plateau, genome sequences, mcr-1
Citation: Tang B, Wang J, Zheng X, Chang J, Ma J, Wang J, Ji X, Yang H and Ding B (2022) Antimicrobial resistance surveillance of Escherichia coli from chickens in the Qinghai Plateau of China. Front. Microbiol. 13:885132. doi: 10.3389/fmicb.2022.885132
Edited by:
Ziad Daoud, Central Michigan University, United StatesReviewed by:
Ximin Zeng, The University of Tennessee, Knoxville, United StatesEvelyn Madoroba, University of Zululand, South Africa
Copyright © 2022 Tang, Wang, Zheng, Chang, Ma, Wang, Ji, Yang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Biao Tang, dGFuZ2JpYW9AemFhcy5hYy5jbg==; Baoan Ding, ZGluZ2Jhb2FuMTk2N0AxNjMuY29t
†These authors have contributed equally to this work
 Biao Tang
Biao Tang Jingge Wang1,2†
Jingge Wang1,2† Jiang Chang
Jiang Chang Jiangang Ma
Jiangang Ma Juan Wang
Juan Wang Hua Yang
Hua Yang