
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 11 May 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.883513
This article is part of the Research Topic Advances and Challenges in the Detection and Treatment of Pathogenic Microorganisms in Infectious Disease Control View all 25 articles
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) is the leading lethal infectious disease with 1.3 million deaths in 2020. Despite significant advances have been made in detection techniques and therapeutic approaches for tuberculosis, no suitable diagnostic tools are available for early and precise screening. Many studies have reported that Long non-coding RNAs (lncRNAs) play a regulatory role in gene expression in the host immune response against Mtb. Dysregulation of lncRNAs expression patterns associated with immunoregulatory pathways arose in mycobacterial infection. Meanwhile, host-induced lncRNAs regulate antibacterial processes such as apoptosis and autophagy to limit bacterial proliferation. In this review, we try to summarize the latest reports on how dysregulated expressed lncRNAs influence host immune response in tuberculosis infection. We also discuss their potential clinical prospects for tuberculosis diagnosis and development as molecular biomarkers.
LncRNAs, frequently defined as non-protein-coding transcripts greater than 200 nucleotides in length (McFadden and Hargrove, 2016), were initially thought to be “transcriptional noise” (Ulitsky and Bartel, 2013). Recent studies have unraveled the biogenesis of lncRNAs, which are distinct from that of mRNAs. Those lncRNAs, generally display lower sequence conservation (Pang et al., 2006) and contain fewer exons (Washietl et al., 2014; Guo et al., 2020), and their functions are typically linked to their specific subcellular localizations (Statello et al., 2021). In addition, lncRNAs have been reported to include small open reading frames (SMORFs) encoding short functional peptides (Kim et al., 2014; Huang et al., 2017; Fathizadeh et al., 2020; Zhu et al., 2020). Accumulated evidence demonstrates that they are emerging as essential regulators in gene regulation, such as chromatin modification, mRNA stability, transcriptional, translation and post-translation activation/inhibition (Quinn and Chang, 2016; Yao et al., 2019). Meanwhile, lncRNAs have various regulatory functions in biological processes, including apoptosis (Heydarnezhad Asl et al., 2022).
Tuberculosis, one of the leading lethal infectious diseases worldwide caused by Mtb, has been considered a public health emergency that concerns the world. According to the WHO global TB report, there were 5.8 million newly sick and 1.5 million TB-related deaths in 2021 (WHO, 2021, no date). Approximately one-third of the world’s population has a latent TB infection (LTBI), and about 10% can develop active tuberculosis (ATB) with impairment in the immune system (Pai et al., 2016). In the host-pathogen interactions, intracellular survival and replication of Mtb after macrophage phagocytosis leads to an immune response that converges on granuloma formation and/or disease (McCaffrey et al., 2022; Medley et al., 2022). The host cells have adopted a series of clearance mechanisms to facilitate intracellular bacterial killing, such as apoptosis, autophagy, inflammation, and macrophage polarization. Mtb has also evolved with a set of almost perfect immune escape mechanisms to evade the host immune system (Stanley and Cox, 2013). Growing evidence has delineated that the expression of many lncRNAs is involved in TB with a definitive role in orchestrating the biological processes from immune response to host-pathogen interactions. In addition, tissue-specific and condition-specific expression patterns suggest that lncRNAs provide potential biomarkers (Statello et al., 2021). Therefore, in-depth elucidation of the effect and mechanism of lncRNAs may develop clinical prospects for precise TB diagnosis and treatments (Lyu et al., 2021; Wei et al., 2021). In this paper, we review the progress of lncRNA roles in Mtb infection from host immunity to biomarkers and discuss its potential in clinical diagnosis.
Depending on specific interactions with DNA, RNA, and proteins, lncRNAs can modulate diverse biological processes through complex and diverse mechanisms, such as execute-as signals, decoys, guides, scaffolds to regulate target genes (Wang and Chang, 2011; Kazemzadeh et al., 2015; Statello et al., 2021).
A feature of lncRNAs is that they can generate a hybrid structure with DNA to influence gene expression. They mediate DNA methylation and transcriptional inhibition by complementary pairing with unstranded DNA bases. In addition, lncRNAs can inhibit the recruitment of Pol II or alter the binding of transcription factors at the promoter sequences whose function is blocked, modulating downstream target genes transcription (Figure 1A; Hainer and Martens, 2011; Schmitz et al., 2016). Recent findings suggest that the DNA-mediated innate immune response underpins antibacterial defense. In this regard, an investigation identified a multi-subunit complex built around HEXIM1 and lncRNA NEAT1, containing DNA-PK subunits and paraspeckle proteins, as a key nuclear regulator of DNA-mediated activation of innate immune response (Morchikh et al., 2017). Also, RNA–DNA complex formation has been proposed as an example of lncRNA–DNA interplay in mediating gene silencing or activation (Mondal et al., 2015; O’Leary et al., 2015). For instance, lncRNA PARTICLE, forming a DNA-lncRNA triplex, repress the tumor suppressor methionine adenosyltransferase (MAT2A) via methylation in response to irradiation, implicating it as a recruitment platform for gene-silencing machinery (O’Leary et al., 2015).
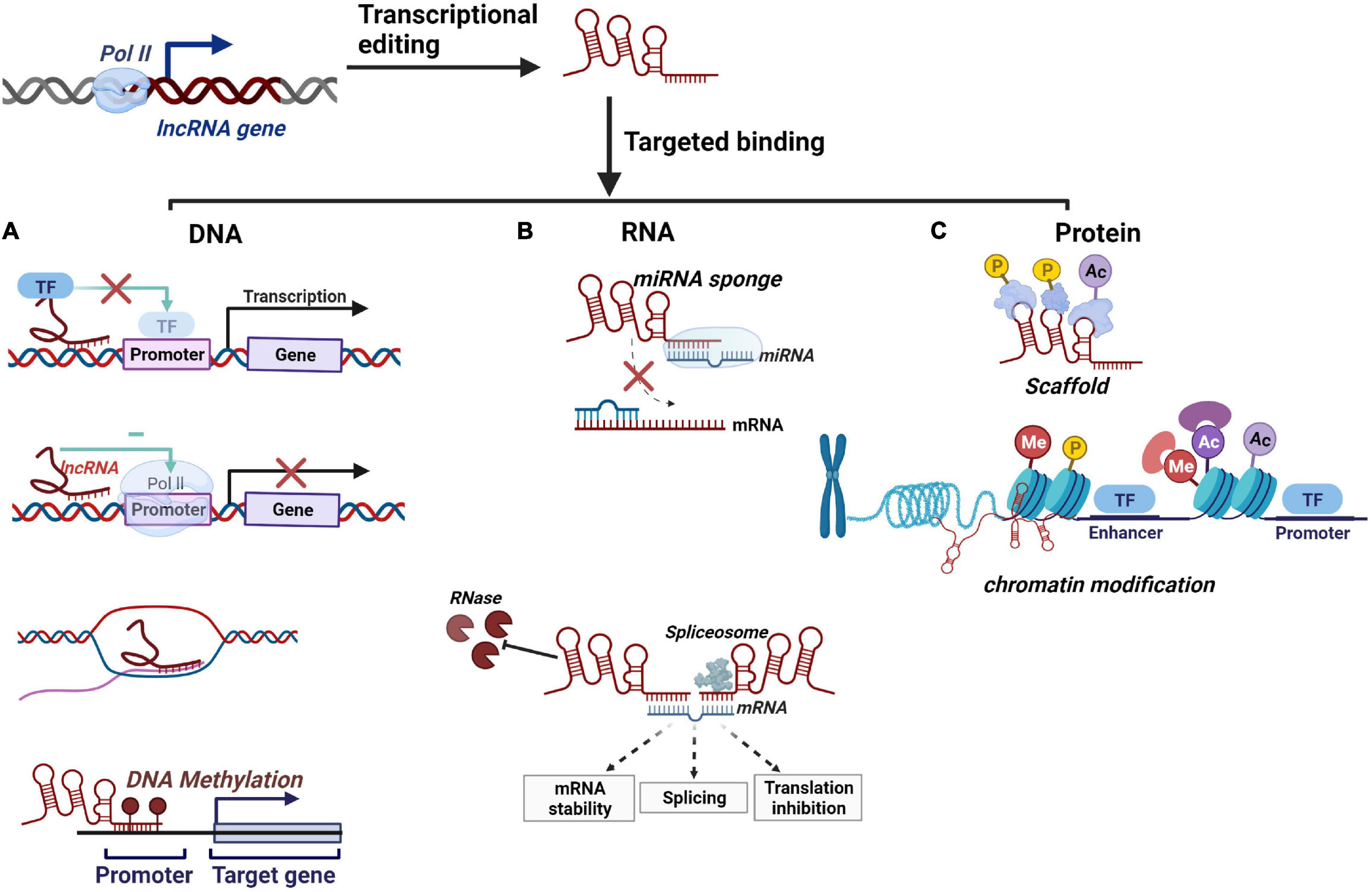
Figure 1. Schematic overview of lncRNAs’ molecular mechanisms. (A) lncRNAs regulate downstream gene transcription by binding to transcription factors, repression of RNA Pol II, complementary pairing with unstranded DNA bases, and DNA methylation. (B) lncRNAs serve as miRNA sponges to competitively bind with miRNA. lncRNAs regulate mRNA transcription by modulating mRNA stability and pre-mRNA splicing. lncRNAs can inhibit RNase degradation and promote mRNA stability. (C) lncRNAs can serve as a scaffold for protein phosphorylation and ubiquitination and regulate chromatin modification by employing histone-modifying complex.
At the RNA level, lncRNAs often function as lncRNA-miRNA-mRNA competing endogenous RNAs (ceRNAs) that serve as miRNA sponges and inhibit their regulatory effect on the target gene (Figure 1B; Salmena et al., 2011; Wang M. et al., 2019; Wang et al., 2020). For example, a ceRNA network constructed from pulmonary tuberculosis (PTB) patients suggested that lncRNAs regulate mRNAs expression may mediate by acting as sponged miRNAs (Zhang et al., 2020). Furthermore, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), pandemic H1N1, and H7N9 infection induced upregulation of lncRNA-34087.27, which could serve as ceRNA, competitively binding with miR-302b-3p to stabilize IRF1 mRNA (Yang et al., 2022). Additionally, mRNA splicing is one of the biological functions that can be affected by lncRNAs through forming complementary double strands with transcripts (Figure 1B; De Troyer et al., 2020, p. 3). For instance, lncRNA MALAT1 can modulate the levels of serine/arginine (SR) proteins, thereby regulating the alternative splicing (AS) of pre-mRNA (Tripathi et al., 2010). Moreover, with lncRNA-mRNA dimer formation, lncRNAs can also inhibit RNase degradation and alter the stability and translation of cytoplasmic mRNAs (Figure 1B; Statello et al., 2021).
Numerous lncRNAs localize on chromatin, interacting with proteins to facilitate or inhibit their binding and activity at targeted DNA regions. On the one hand, it can participate in protein phosphorylation by protein kinase or ubiquitin modification of a protein by ubiquitin-modified enzymes (Statello et al., 2021). For instance, HOTAIR is known to act at the posttranslational level by serving as an assembly scaffold for protein ubiquitination (Figure 1C). In addition, lncRNAs could regulate histone modification and chromatin accessibility, thereby regulating transcription (Figure 1C). A well-described example is the X chromosome dosage compensation process. The X-inactive-specific transcript (Xist) recruits polycomb repressive complex 2 (PRC2) and triggers large numbers of histones methylated as well as a cascade of events that entails chromosome remodeling to achieve stable silencing (Creamer and Lawrence, 2017; Jégu et al., 2017). Similarly, HOTAIR interacts with PRC2 favors epigenetic silencing through Enhance of Zeste2 (EZH2) catalyzed deposition of H3K27me3, facilitating the survival of virulent Mtb (Table 1; Subuddhi et al., 2020). Moreover, lncRNAs can be used as a protein structural component to affect protein spatial conformation.
Different expression patterns of lncRNAs are emerging as critical regulators to control the function of innate and adaptive immune cell types and initiate effective defense mechanisms in TB (Figure 2 and Table 1; Chen Y. G. et al., 2017; Wei et al., 2021).
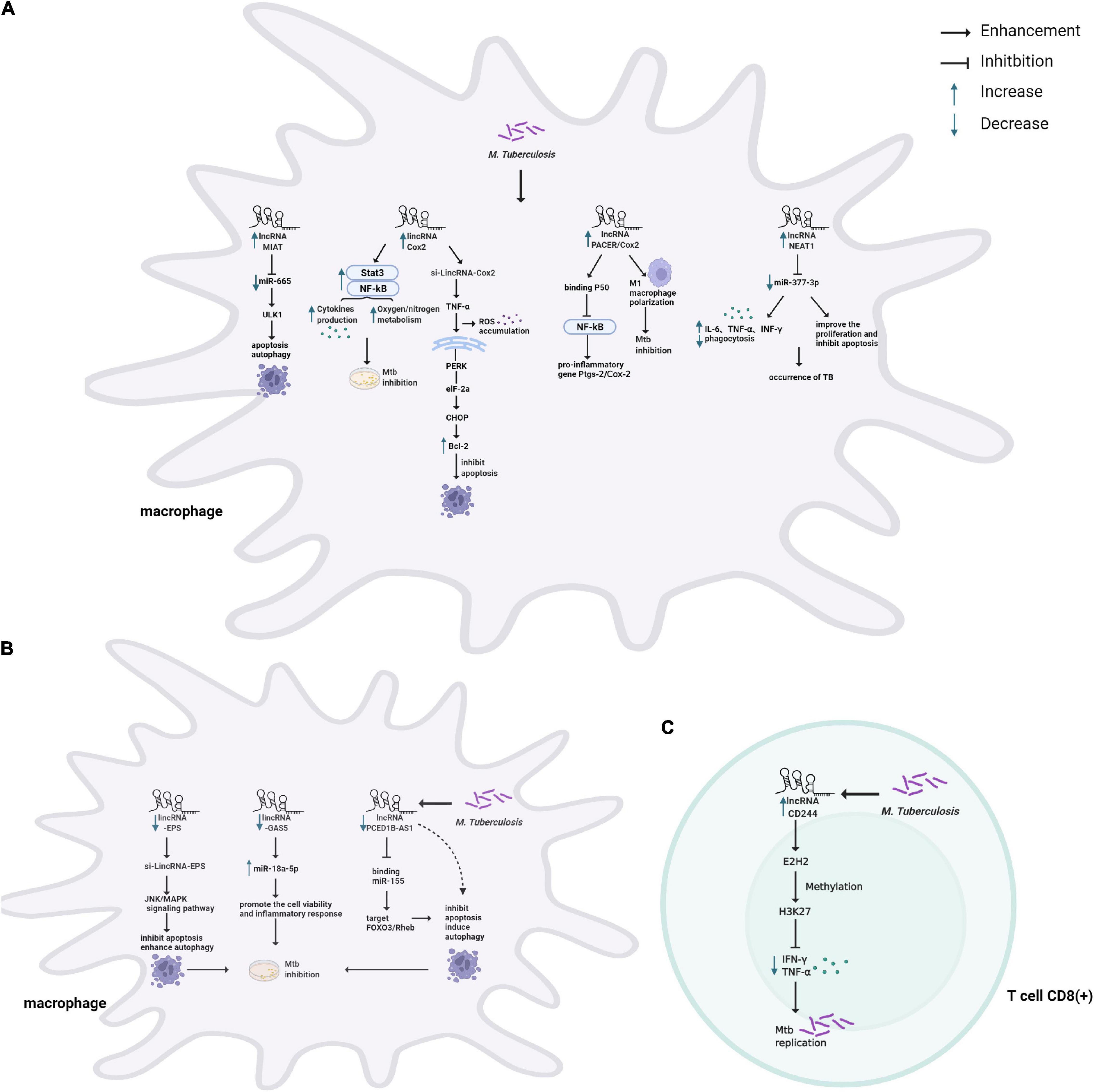
Figure 2. Schematic representation of the role of lncRNAs in host immune system during Mtb infection. (A) TB infection causes upregulation of lncRNAs modulating the immune system of macrophages. lncRNA MIAT modulates apoptosis and autophagy through the miR-665/ULK1 crosstalk. COX-2 modulates inflammatory response by activating NF-κb/STAT3, associating with p50 (a repressive subunit of NF-κb), and M1 macrophage polarization, and induces apoptosis by activating the PERK-eIF-2α-CHOP signaling pathway. NEAT1 modulates inflammatory response and apoptosis through targeting miR-377-3p. (B) TB infection causes downregulation of lncRNAs modulating the immune system of macrophages. LincRNA-EPS inhibited apoptosis and enhanced autophagy by activating the JNK/MAPK signaling pathway. GAS5 promotes the cell vitality and inflammatory response by sponging miR-18a-5p. PCED1B-AS1 can directly bind to miR-155 to inhibit apoptosis and induce autophagy. (C) TB infection causes upregulation of lncRNA-CD244 to interact with EZH2 and mediates H3K27 trimethylation at ifng/tnfa loci, thereby suppressing IFN-γ/TNF-α expression and increasing bacterial proliferation in T cells.
Accumulating lines of evidence have revealed that lncRNAs play an important role in regulating the innate immune response of host macrophages, which are the sentinels to phagocytose and eliminate Mtb (Figure 2A,B). For one thing, lncRNAs participate in macrophage-mediated immune-inflammatory responses that an inducible program of inflammatory gene expression is central to anti-microbial defenses. For example, lincRNA-Cox2 mediates the activation/repression of immune genes that may activate NF-κB and STAT3 to regulate inflammatory responses for resistance of Mtb infection (Figure 2A; Carpenter et al., 2013; Li D. et al., 2020). In addition, lncRNA-PACER (also known as lncRNA-Cox2) was induced in Mtb infected macrophages, acting as a positive regulator of its proximal pro-inflammatory gene Ptgs-2 (also known as Cox2). Via mechanisms involving the sequestration of repressive NF-κB subunit p50 away from Ptgs-2 promoter (Krawczyk and Emerson, 2014; Tamgue et al., 2021). LncRNA-Cox2 can promote the activation of macrophages toward the pro-inflammatory M1 phenotype known to be efficient in killing Mtb (Figure 2A; Ye et al., 2018; Tamgue et al., 2021). Recently, some studies have found that in Mtb infected macrophages, the down-regulated lncRNA GAS5 might facilitate the cell vitality and the inflammatory response by sponging miR-18a-5p (Figure 2A; Li et al., 2021), and NEAT1 participates in inflammatory response through targeted regulation of miR-377-3p (Figure 2A; Sun et al., 2021).
For another, previous studies indicated that massive lncRNAs could regulate Mtb-induced apoptosis and autophagy of macrophages, playing a vital role in the pathogenesis of TB (Behar et al., 2011; Kim et al., 2019). For example, the Bacillus Calmette-Guerin (BCG)-infected macrophages induced apoptosis with upregulated lincRNA-Cox2 expression. Knockdown of Cox2 aggravated reactive oxygen species (ROS) accumulation and initiated apoptosis by activating the PERK-eIF-2α-CHOP signaling pathway (Figure 2B; Xu et al., 2021). A similar mechanism has been described for lncRNA-EPS, modulating apoptosis and autophagy by activating the JNK/MAPK signaling pathway in macrophages (Figure 2B; Ke et al., 2020). Additionally, down-regulation of lncRNA-MEG3 in infected macrophages induced autophagy and enhanced eradication of intracellular Mycobacterium Bovis BCG (Pawar et al., 2016). Recent studies have revealed that several lncRNAs act as ceRNAs to regulate apoptosis and autophagy. For example, lncRNA PCED1B-AS1 decreased in ATB patients compared to healthy individuals. The suppression of PCED1B-AS1 significantly attenuated apoptosis and enhanced autophagy in macrophages by sponging the miR-155 and inhibiting its targets FOXO3/Rheb (Figure 2B; Li et al., 2019). Moreover, lncRNA MIAT modulated macrophage apoptosis and autophagy upon BCG infection through the miR-665/ULK1 crosstalk (Figure 2A; Jiang et al., 2021).
Once the innate immune system is breached, the “human guardians,” including T and B cells, immediately enter a fighting state and initiate adaptive immunity. LncRNAs also regulate T cell-mediated immune regulation in TB (Figure 2C). CD4 + T cells play a dominant role in the host immune response of TB (Jasenosky et al., 2015). The study analyzed the lncRNA profile in CD4 + T cells and revealed that compared with healthy controls, lncRNAs showed abnormal expression in ATB and LTBI (Yi et al., 2014). In addition, significantly enriched signaling pathways based on deregulated mRNAs were cytokine-cytokine receptor interaction, mitogen-activated protein kinase (MAPK), and TLR signaling pathway (Yi et al., 2014).
In general, CD8 + T cells were previously thought to be less critical than CD4 + T cells in the immune response to TB. It has been newly stated its non-redundant role with a specific CD8 + T cell response (Lin and Flynn, 2015). A recent study confirmed that lncRNA-CD244 epigenetically repressed the IFN-γ and TNF-α expression in CD8 + T cells (Figure 2C; Wang et al., 2015). Adoptive transfer of CD244–depressed CD8 + T cells to Mtb-infected mice reduced infection and pathology compared to mice transplanted with wild-type CD8 + cells (Wang et al., 2015). Moreover, Heme Oxygenase 1 (HMOX1) was increased after Mtb infection and can distinguish LTBI from ATB in the previous research (Costa et al., 2016). Also, 328 differentially expressed lncRNAs were found in CD8 + T cells’ response to ATB. Among them, lincRNA XLOC_014219 was upregulated, while its nearby protein-coding gene HMOX1 was significantly decreased. It is essential to uncover why HMOX1 is downregulated and whether lincRNA XLOC_014219 relates to it, which is ultimately involved in the dysfunction of CD8 + T cells (Fu et al., 2017a).
It has come to light that lncRNAs can influence antibodies produced by B cells and impact B cell biology by regulating survival signals during activation (Zeni and Mraz, 2021). For instance, a study reported that 844 lncRNAs differentially expressed in B cell samples. Additionally, SOCS3 is an essential negative regulator of cytokine response to Mtb infection, and its upstream lncRNA XLOC_012582 highly increased in B cells of ATB. Whether upregulation of XLOC_012582 leads to overexpression of SOCS3 and ultimately participates in the progression of TB needs in-depth investigation (Fu et al., 2017b). Findings provided new insight into the pathogenesis of TB. However, discoveries related to B cells are like a tip of an iceberg.
In summary, the immune protection mechanism of TB is complex and comprehensive. Although there have been preliminary studies on the role of lncRNAs in different host cells to Mtb, their specific functions in immunity are mainly unexplored.
So far, most clinical diagnosis methods have inherent limitations (Pai et al., 2016). Methods like smear microscopy and mycobacterium culture have insufficient sensitivity and timeliness and have a poor detection rate of smear-negative PTB (Walzl et al., 2011; Fang et al., 2021; Mirzaei et al., 2021), while interferon-gamma release assays (IGRA) unable to discriminate between LTBI and ATB (Walzl et al., 2018). The recent recommendations of the WHO include non-pathogen-based detection to improve the identification of clinically diagnosed TB with rapid and universal methods (Martinez and Andrews, 2019). Therefore, biomarkers of the host immune responses might provide critical insights to solve this problem. Some ncRNAs are considered to be biomarkers (Beermann et al., 2016), such as miR-889 targets that can be manipulated for antimycobacterial therapeutic purposes and candidate biomarkers for LTBI (Chen et al., 2020). Relative to investigated in high detail miRNAs, the roles of lncRNAs remain largely elusive (Lee, 2012). Nevertheless, a rising number of dysregulation processes indicate that lncRNAs are highly promising as biomarkers of TB (Table 2).
Accumulating lines of studies have revealed the abnormally expressed lncRNAs in peripheral blood mononuclear cells (PBMCs) of TB patients (Table 2). A study reported that two significantly aberrantly expressed lncRNA (MIR3945HG V1 and MIR3945HG V2) in PBMCs samples from active PTB patients have the potential to be novel diagnostic biomarkers (Yang et al., 2016). Furthermore, the expression of NEAT1 (both NEAT1_1 and NEAT1_2) in TB patients was higher than healthy control, declined gradually with treatment, and was restored to the normal level. This dynamic change could reflect the efficacy of anti-TB therapy. Therefore, NEAT1 may serve as a potential indicator for patient prognosis of TB (Huang et al., 2018). Additionally, the study suggested that downregulated PCED1B-AS1 in PBMCs and THP-1 show promise as a new early diagnostic biomarker for ATB (Li et al., 2019). Nevertheless, little information has been done on the underlying mechanisms of lncRNAs above. To enhance the PTB identification, lncRNA n344917 was confirmed down-regulated in PBMC of PTB. Hence, a web-based prediction model combining the molecular biomarker n344917, laboratory, and EHR variables was constructed and could serve as a user-friendly, accurate platform to improve the clinical diagnosis of PTB (Meng et al., 2021).
Differentially expressed lncRNAs in the plasma or serum of TB patients have also been explored as potential diagnostic biomarkers (Table 2). For instance, expression levels of ENST00000354432 and ENST00000427151 were suggested to act as biomarkers for the early detection of TB (He et al., 2017). LOC152742 in plasma had higher specificity in ATB and gradually downregulated in the treatment. Hence it could serve as a novel biomarker for the diagnosis and therapy of ATB (Wang L. et al., 2019). Also, plasma lncRNAs might act as potential biomarkers to evaluate TB cure in an efficient and precise manner. LncRNAs uc.48 + and NR_105053 may serve as biomarkers to distinguish between untreated TB patients and cured TB subjects (Li Z.-B. et al., 2020). Recently, studies reported some lncRNA sets with high diagnostic sensitivity and specificity. For example, ceRNA analysis of four differentially expressed lncRNAs (NR_038221, NR_003142, ENST00000570366, and ENST00000422183) was demonstrated to discriminate PTB from healthy individuals. The results showed that NR_038221 was the most significantly associated with TB (Chen Z.-L. et al., 2017). Their previous study had verified hsa-miR-378a-3p as a potential biomarker for PTB, which was associated with NR_038221, indicating that NR_038221 and hsa-miR-378a-3p might play a similar function during the biological process of PTB (Zhang et al., 2013; Chen Z.-L. et al., 2017). At present, the development of high-throughput experimental technologies has led to a rapid expansion of lncRNA research. For example, an integrated analysis of the GEO dataset and the NON-CODE database identified four significantly downregulated lncRNAs (NON-HSAT101518.2, NON-HSAT067134.2, NON-HSAT148822.1, and NON-HSAT078957.2) in serum exosomes of ATB patients. ROC curve analysis suggests that these four lncRNAs can discriminate ATB from healthy individuals with high specificity and sensitivity (Fang et al., 2021).
Accurate diagnosis of complete inactivation of TB lesions remains a challenge concerning sputum-negative TB, one of the significant factors for the development and spread of ATB (Horsburgh, 2004). Clinically diagnosed PTB patients without microbiological evidence of Mtb often lead to misdiagnosis or delayed diagnosis. Therefore, A study validated the lncRNAs and corresponding predictive models to effectively diagnose these patients, finding differentially expressed lncRNAs (ENST00000497872, n333737, n335265) between PTB patients and the healthy group. These lncRNAs might be used as putative biomarkers to facilitate the early identification of PTB cases among suspected patients with negative microbiological evidence (Table 2; Hu et al., 2020). In addition, lncRNAs (ENST00000429730.1 and MSTRG.93125.4) were upregulated in lung tissues with high metabolic activity demonstrated by fluorine-18-fluorodeoxyglucose positron emission tomography/computed tomography from sputum-negative TB patients compared with low metabolic activity. Two lncRNAs might be considered potential metabolic activity indicators in TB lesions for sputum-negative TB (Table 2; Wang et al., 2021).
Abundant evidence from the investigations suggests that host genetic factors contribute to determine susceptibility to TB disease (Möller and Hoal, 2010). Recent advances in lncRNA research have preliminarily explored the relationship between lncRNA single nucleotide polymorphisms (SNPs), TB susceptibility, and clinical manifestations in TB patients of the western Chinese Han population (Table 2). For example, a study uncovered that the lncRNA AC079767.4 might be involved in the progression of TB infection. In addition, the C allele of SNP rs12477677 in AC079767.4 was associated with reduced susceptibility to PTB (Zhao et al., 2017). Moreover, the SNPs (rs12477677 and rs1055229) may influence clinical TB disease. Conceivably, AC079767.4 polymorphisms may serve as novel biomarkers for TB diagnostic and even as therapeutic targets (Zhao et al., 2017). Similarly, the potential TB-associated promoting effects were identified for the decreased expression levels of lnc-AC145676.2.1-6 and lnc-TGS-1 (Bai et al., 2019). Recently, the SNPs of rs39509 G allele in PIPK2 Near gene-3 region, lncRNA - HNF1B - 3:1, lncRNA - RP11-37 b2. 1, lncRNA - CASC8 has also been proved to be associated with TB susceptibility (Song et al., 2019a,b; Liu et al., 2020; Wu et al., 2020). These lncRNA polymorphisms are promising molecular biomarkers for clinical TB infection and/or efficacy evaluation. However, there are some questions need to be answered, such as are there sex and ethnic differences in differentially expressed lncRNAs?
Overall, the advancement of sequencing technology has facilitated the discovery of lncRNAs with unknown functions. These lncRNAs can thwart disease or lead to disease progression. Importantly, lncRNAs have presented a diverse perspective on the regulation of TB, especially complicated immune regulation for multiple biomolecular interactions. By high-throughput sequencing and deepening validation, lncRNAs have been found as potential biomarkers for diagnosis, monitoring progression, and clinical efficacy of TB. However, the complexity of the lncRNAs themselves and the lack of accurate databases for the lncRNAs discovered are restricting research to the single analysis of differences in gene expression. We are still far from completely understanding how lncRNAs influence complex physiopathological processes of TB, and several aspects of lncRNA remain enigmatic. For instance, whether lncRNA regulates Mtb infection as an infection phenomenon or a specific bacterial infection phenomenon under certain conditions. In addition, there is a lack of using lncRNAs as biomarkers to indicate progression from latent infections to clinical disease. Despite the questions, lncRNAs are highly ideal biomarkers. Further validation studies on different ethnic populations and function experiments in a large-scale cohort help to confirm the roles of the lncRNAs, and may also help identify biomarkers for latent infections. Most of the research on ncRNAs mainly focuses on miRNA, but understanding the lncRNA function can only be achieved from more studies on a case-by-case basis.
Several new biotechnologies such as Xpert MTB/RIF Ultra have been developed in this context. It has increased sensitivity for diagnosis but is sophisticated. More accurate, rapid, and cost-effective tests are needed to improve TB detection. Various platforms have explored other possible biomarkers, such as host marker signatures, including host gene expression, protein, and metabolites. Some have combined omics techniques which have made some progress in distinguishing LTBI from ATB to explore progression to TB. However, such measurement of expression of markers requires substantial laboratory infrastructure and is time-consuming. Additionally, the most work done so far has been small case-control studies, and the diagnostic potential of these markers still has to be confirmed. The continued study of TB-associated lncRNAs will reveal more unanticipated biomarkers for predicting progression, response to treatment, and relapse. Moreover, the personalized lncRNA-targeted drugs in host-directed therapy (HDTS) are under development.
XZ collected the related manuscript and drafted the manuscript. XZ, CC, and YX discussed and developed its conceptual framework. CC and YX proofread the manuscript. YX and XZ revised the review. All authors contributed to the article and approved the submitted version.
This study was supported by grants from the National Natural Science Foundation of China (No. 82000017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the reviewers for their valuable suggestions. Figures were created with BioRender.com.
Bai, H., Wu, Q., Hu, X., Wu, T., Song, J., Liu, T., et al. (2019). ‘Clinical significance of lnc-AC145676.2.1-6 and lnc-TGS1-1 and their variants in western Chinese tuberculosis patients’. Int. J. Infect. Dis. 84, 8–14. doi: 10.1016/j.ijid.2019.04.018
Beermann, J., Piccoli, M.-T., Viereck, J., and Thum, T. (2016). ‘Non-coding RNAs in development and disease: background. mechanisms, and therapeutic approaches’, Physiol. Rev. 96, 1297–1325. doi: 10.1152/physrev.00041.2015
Behar, S. M., Martin, C. J., Booty, M. G., Nishimura, T., Zhao, X., Gan, H.-X., et al. (2011). ‘Apoptosis is an innate defense function of macrophages against mycobacterium tuberculosis’. Mucosal Immunol. 4, 279–287. doi: 10.1038/mi.2011.3
Carpenter, S., Aiello, D., Atianand, M. K., Ricci, E. P., Gandhi, P., Hall, L. L., et al. (2013). ‘A long noncoding RNA mediates both activation and repression of immune response genes’. Science 341, 789–792. doi: 10.1126/science.1240925
Chen, D.-Y., Chen, Y.-M., Lin, C.-F., Lo, C.-M., Liu, H.-J., and Liao, T.-L. (2020). ‘MicroRNA-889 inhibits autophagy to maintain mycobacterial survival in patients with latent tuberculosis infection by targeting TWEAK’. mBio 11, e3045–e3019. doi: 10.1128/mBio.03045-19
Chen, Y. G., Satpathy, A. T., and Chang, H. Y. (2017). ‘Gene regulation in the immune system by long noncoding RNAs’. Nat. Immunol. 18, 962–972. doi: 10.1038/ni.3771
Chen, Z.-L., Wei, L.-L., Shi, L.-Y., Li, M., Jiang, T.-T., Chen, J., et al. (2017). ‘Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis’. Sci. Rep. 7:16751. doi: 10.1038/s41598-017-17146-y
Costa, D. L., Namasivayam, S., Amaral, E. P., Arora, K., Chao, A., Mittereder, L. R., et al. (2016). ‘Pharmacological inhibition of host heme oxygenase-1 suppresses mycobacterium tuberculosis infection in vivo by a mechanism dependent on T lymphocytes’. mBio 7, e1675–e1616. doi: 10.1128/mBio.01675-16
Creamer, K. M., and Lawrence, J. B. (2017). ‘XIST RNA: a window into the broader role of RNA in nuclear chromosome architecture’. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372:20160360. doi: 10.1098/rstb.2016.0360
De Troyer, L., Zhao, P., Pastor, T., Baietti, M. F., Barra, J., Vendramin, R., et al. (2020). ‘Stress-induced lncRNA LASTR fosters cancer cell fitness by regulating the activity of the U4/U6 recycling factor SART3’. Nucleic Acids Res. 48, 2502–2517. doi: 10.1093/nar/gkz1237
Fang, Y., Zhao, J., Wang, X., Wang, X., Wang, L., Liu, L., et al. (2021). Identification of differentially expressed lncRNAs as potential plasma biomarkers for active tuberculosis. Tuberculosis 128:102065. doi: 10.1016/j.tube.2021.102065
Fathizadeh, H., Hayat, S. M. G., Dao, S., Ganbarov, K., Tanomand, A., Asgharzadeh, M., et al. (2020). ‘Long non-coding RNA molecules in tuberculosis’. Int. J. Biol. Macromol. 156, 340–346. doi: 10.1016/j.ijbiomac.2020.04.030
Fu, Y., Gao, K., Tao, E., Li, R., and Yi, Z. (2017a). ‘Aberrantly expressed long non-coding RNAs in CD8+ T cells response to active tuberculosis’. J. Cell. Mol. Biochem. 118, 4275–4284. doi: 10.1002/jcb.26078
Fu, Y., Xu, X., Xue, J., Duan, W., and Yi, Z. (2017b). ‘Deregulated lncRNAs in B cells from patients with active tuberculosis’. PLoS One 12:e0170712. doi: 10.1371/journal.pone.0170712
Guo, C.-J., Ma, X.-K., Xing, Y.-H., Zheng, C.-C., Xu, Y.-F., Shan, L., et al. (2020). Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell 181, 621.e–636.e. doi: 10.1016/j.cell.2020.03.006
Hainer, S. J., and Martens, J. A. (2011). ‘Transcription of ncDNA: many roads lead to local gene regulation’. Transcription 2, 120–123. doi: 10.4161/trns.2.3.15684
He, J., Ou, Q., Liu, C., Shi, L., Zhao, C., Xu, Y., et al. (2017). ‘Differential expression of long non-coding RNAs in patients with tuberculosis infection’. Tuberculosis 107, 73–79. doi: 10.1016/j.tube.2017.08.007
Heydarnezhad Asl, M., Pasban Khelejani, F., Bahojb Mahdavi, S. Z., Emrahi, L., Jebelli, A., and Mokhtarzadeh, A. (2022). The various regulatory functions of long noncoding RNAs in apoptosis, cell cycle, and cellular senescence. J. Cell. Biochem. [Preprint] doi: 10.1002/jcb.30221
Horsburgh, C. R. (2004). ‘Priorities for the treatment of latent tuberculosis infection in the United States’. N. Engl. J. Med. 350, 2060–2067. doi: 10.1056/NEJMsa031667
Hu, X., Liao, S., Bai, H., Gupta, S., Zhou, Y., Zhou, J., et al. (2020). ‘Long noncoding RNA and predictive model to improve diagnosis of clinically diagnosed pulmonary tuberculosis’. J. Clin. Microbiol. 58, e1973–e1919. doi: 10.1128/JCM.01973-19
Huang, J.-Z., Chen, M., Chen, D., Gao, X.-C., Zhu, S., Huang, H., et al. (2017). A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell 68, 171.e–184.e. doi: 10.1016/j.molcel.2017.09.015
Huang, S., Huang, Z., Luo, Q., and Qing, C. (2018). ‘The Expression of lncRNA NEAT1 in human tuberculosis and its antituberculosis effect’. Biomed. Res. Int. 2018:9529072. doi: 10.1155/2018/9529072
Jasenosky, L. D., Scriba, T. J., Hanekom, W. A., and Goldfeld, A. E. (2015). ‘T cells and adaptive immunity to mycobacterium tuberculosis in humans’. Immunol. Rev. 264, 74–87. doi: 10.1111/imr.12274
Jégu, T., Aeby, E., and Lee, J. T. (2017). ‘The X chromosome in space’. Nat. Rev. Genet. 18, 377–389. doi: 10.1038/nrg.2017.17
Jiang, F., Lou, J., Zheng, X.-M., and Yang, X.-Y. (2021). ‘LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by mycobacterium tuberculosis through the miR-665/ULK1 signaling axis’. Mol. Immunol. 139, 42–49. doi: 10.1016/j.molimm.2021.07.023
Kazemzadeh, M., Safaralizadeh, R., and Orang, A. V. (2015). ‘LncRNAs: emerging players in gene regulation and disease pathogenesis’. J. Genet. 94, 771–784. doi: 10.1007/s12041-015-0561-6
Ke, Z., Lu, J., Zhu, J., Yang, Z., Jin, Z., and Yuan, L. (2020). ‘Down-regulation of lincRNA-EPS regulates apoptosis and autophagy in BCG-infected RAW264.7 macrophages via JNK/MAPK signaling pathway’. Infect. Genet. Evol. 77:104077. doi: 10.1016/j.meegid.2019.104077
Kim, M.-S., Pinto, S. M., Getnet, D., Nirujogi, R. S., Manda, S. S., Chaerkady, R., et al. (2014). ‘A draft map of the human proteome’. Nature 509, 575–581. doi: 10.1038/nature13302
Kim, Y. S., Silwal, P., Kim, S. Y., Yoshimori, T., and Jo, E.-K. (2019). ‘Autophagy-activating strategies to promote innate defense against mycobacteria’. Exp. Mol. Med. 51, 1–10. doi: 10.1038/s12276-019-0290-7
Krawczyk, M., and Emerson, B. M. (2014). ‘p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes’. Elife 3:e01776. doi: 10.7554/eLife.01776
Lee, J. T. (2012). ‘Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439. doi: 10.1126/science.1231776
Li, D., Gao, C., Zhao, L., and Zhang, Y. (2020). ‘Inflammatory response is modulated by lincRNACox2 via the NF−κB pathway in macrophages infected by mycobacterium tuberculosis’. Mol. Med. Rep. 21, 2513–2521. doi: 10.3892/mmr.2020.11053
Li, M., Cui, J., Niu, W., Huang, J., Feng, T., Sun, B., et al. (2019). ‘Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis’. Biochem. Biophys. Res. Commun. 509, 803–809. doi: 10.1016/j.bbrc.2019.01.005
Li, Y., Sun, L., Liu, J., Xu, G., Hu, Y., and Qin, A. (2021). ‘Down-regulation of GAS5 has diagnostic value for tuberculosis and regulates the inflammatory response in mycobacterium tuberculosis infected THP-1 cells’. Tuberculosis 132:102141. doi: 10.1016/j.tube.2021.102141
Li, Z.-B., Han, Y.-S., Wei, L.-L., Shi, L.-Y., Yi, W.-J., Chen, J., et al. (2020). Screening and identification of plasma lncRNAs uc.48+ and NR_105053 as potential novel biomarkers for cured pulmonary tuberculosis. Int. J. Infect. Dis. 92, 141–150. doi: 10.1016/j.ijid.2020.01.005
Lin, P. L., and Flynn, J. L. (2015). ‘CD8 T cells and mycobacterium tuberculosis infection’. Semin. Immunopathol. 37, 239–249. doi: 10.1007/s00281-015-0490-8
Liu, G., Xia, R., Wang, Q., Wang, Z., Ying, B., and Yan, H. (2020). ‘Significance of LncRNA CASC8 genetic polymorphisms on the tuberculosis susceptibility in Chinese population’. J. Clin. Lab. Anal. 34:e23234. doi: 10.1002/jcla.23234
Lyu, M., Cheng, Y., Zhou, J., Chong, W., Wang, Y., Xu, W., et al. (2021). ‘Systematic evaluation, verification and comparison of tuberculosis-related non-coding RNA diagnostic panels’. J. Cell. Mol. Med. 25, 184–202. doi: 10.1111/jcmm.15903
Martinez, L., and Andrews, J. R. (2019). ‘Improving tuberculosis case finding in persons living with advanced HIV through new diagnostic algorithms’. Am. J. Respir. Crit. Care Med. 199, 559–560. doi: 10.1164/rccm.201809-1702ED
McCaffrey, E. F., Donato, M., Keren, L., Chen, Z., Delmastro, A., Fitzpatrick, M. B., et al. (2022). ‘The immunoregulatory landscape of human tuberculosis granulomas’. Nat. Immunol. 23, 318–329. doi: 10.1038/s41590-021-01121-x
McFadden, E. J., and Hargrove, A. E. (2016). ‘Biochemical methods to investigate lncRNA and the influence of lncRNA:Protein complexes on chromatin’. Biochemistry 55, 1615–1630. doi: 10.1021/acs.biochem.5b01141
Medley, J., Goff, A., Bettencourt, P. J. G., Dare, M., Cole, L., Cantillon, D., et al. (2022). ‘Dissecting the mycobacterium bovis BCG response to macrophage infection to help prioritize targets for anti-tuberculosis drug and vaccine discovery’. Vaccines 10:113. doi: 10.3390/vaccines10010113
Meng, Z., Wang, M., Guo, S., Zhou, Y., Lyu, M., Hu, X., et al. (2021). ‘Novel long non-coding RNA and LASSO prediction model to better identify pulmonary tuberculosis: a case-control study in China’. Front. Mol. Biosci. 8:632185. doi: 10.3389/fmolb.2021.632185
Mirzaei, R., Babakhani, S., Ajorloo, P., Ahmadi, R. H., Hosseini-Fard, S. R., Keyvani, H., et al. (2021). ‘The emerging role of exosomal miRNAs as a diagnostic and therapeutic biomarker in mycobacterium tuberculosis infection’. Mol. Med. 27:34. doi: 10.1186/s10020-021-00296-1
Möller, M., and Hoal, E. G. (2010). ‘Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis’. Tuberculosis 90, 71–83. doi: 10.1016/j.tube.2010.02.002
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Reinius, B., et al. (2015). ‘MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures’. Nat. Commun. 6:7743. doi: 10.1038/ncomms8743
Morchikh, M., Cribier, A., Raffel, R., Amraoui, S., Cau, J., Severac, D., et al. (2017). HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 67, 387.e–399.e. doi: 10.1016/j.molcel.2017.06.020
O’Leary, V. B., Ovsepian, S. V., Carrascosa, L. G., Buske, F. A., Radulovic, V., Niyazi, M., et al. (2015). ‘PARTICLE, a triplex-forming long ncRNA, Regulates locus-specific methylation in response to low-dose irradiation’. Cell Rep. 11, 474–485. doi: 10.1016/j.celrep.2015.03.043
Pai, M., Behr, M. A., Dowdy, D., Dheda, K., Divangahi, M., Boehme, C. C., et al. (2016). ‘Tuberculosis’. Nat. Rev. Dis. Primers 2:16076. doi: 10.1038/nrdp.2016.76
Pang, K. C., Frith, M. C., and Mattick, J. S. (2006). ‘Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function’. Trends Genet. 22, 1–5. doi: 10.1016/j.tig.2005.10.003
Pawar, K., Hanisch, C., Palma Vera, S. E., Einspanier, R., and Sharbati, S. (2016). ‘Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy’. Sci. Rep. 6:19416. doi: 10.1038/srep19416
Quinn, J. J., and Chang, H. Y. (2016). ‘Unique features of long non-coding RNA biogenesis and function’. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). ‘A ceRNA hypothesis: the rosetta stone of a hidden RNA language?’. Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). ‘Mechanisms of long noncoding RNA function in development and disease’. Cell. Mol. Life Sci. 73, 2491–2509. doi: 10.1007/s00018-016-2174-5
Song, J., Liu, T., Jiao, L., Zhao, Z., Hu, X., Wu, Q., et al. (2019a). ‘RIPK2 polymorphisms and susceptibility to tuberculosis in a Western Chinese Han population’. Infect. Genet. Evol. 75:103950. doi: 10.1016/j.meegid.2019.103950
Song, J., Liu, T., Zhao, Z., Hu, X., Wu, Q., Peng, W., et al. (2019b). ‘Genetic polymorphisms of long noncoding RNA RP11-37B2.1 associate with susceptibility of tuberculosis and adverse events of antituberculosis drugs in west China’. J. Clin. Lab. Anal. 33:e22880. doi: 10.1002/jcla.22880
Stanley, S. A., and Cox, J. S. (2013). ‘Host-pathogen interactions during mycobacterium tuberculosis infections’. Curr. Top. Microbiol. Immunol. 374, 211–241. doi: 10.1007/82_2013_332
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). ‘Gene regulation by long non-coding RNAs and its biological functions’. Nat. Rev. Mol. Cell. Biol. 22, 96–118. doi: 10.1038/s41580-020-00315-9
Subuddhi, A., Kumar, M., Majumder, D., Sarkar, A., Ghosh, Z., Vasudevan, M., et al. (2020). ‘Unraveling the role of H3K4 trimethylation and lncRNA HOTAIR in SATB1 and DUSP4-dependent survival of virulent mycobacterium tuberculosis in macrophages’. Tuberculosis 120:101897. doi: 10.1016/j.tube.2019.101897
Sun, Q., Shen, X., Ma, J., Lou, H., and Sha, W. (2021). LncRNA NEAT1 participates in inflammatory response in macrophages infected by mycobacterium tuberculosis through targeted regulation of miR-377-3p. Microb. Pathog. 150:104674. doi: 10.1016/j.micpath.2020.104674
Tamgue, O., Chia, J. E., and Brombacher, F. (2021). ‘Triptolide modulates the expression of inflammation-associated lncRNA-PACER and lincRNA-p21 in mycobacterium tuberculosis-infected monocyte-derived macrophages’. Front. Pharmacol. 12:618462. doi: 10.3389/fphar.2021.618462
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). ‘The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation’. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Ulitsky, I., and Bartel, D. P. (2013). ‘lincRNAs: genomics, evolution, and mechanisms’. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
WHO (2021). Global Tuberculosis Report 2021 Available Online at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2021 (accessed October 30, 2021).
Walzl, G., McNerney, R., du Plessis, N., Bates, M., McHugh, T. D., Chegou, N. N., et al. (2018). ‘Tuberculosis: advances and challenges in development of new diagnostics and biomarkers’. Lancet Infect. Dis. 18, e199–e210. doi: 10.1016/S1473-3099(18)30111-7
Walzl, G., Ronacher, K., Hanekom, W., Scriba, T. J., and Zumla, A. (2011). ‘Immunological biomarkers of tuberculosis’. Nat. Rev. Immunol. 11, 343–354. doi: 10.1038/nri2960
Wang, K. C., and Chang, H. Y. (2011). ‘Molecular mechanisms of long noncoding RNAs’. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, L., Wen, Z., Ma, H., Wu, L., Chen, H., Zhu, Y., et al. (2021). ‘Long non-coding RNAs ENST00000429730.1 and MSTRG.93125.4 are associated with metabolic activity in tuberculosis lesions of sputum-negative tuberculosis patients’. Aging 13, 8228–8247. doi: 10.18632/aging.202634
Wang, L., Xie, B., Zhang, P., Ge, Y., Wang, Y., and Zhang, D. (2019). ‘LOC152742 as a biomarker in the diagnosis of pulmonary tuberculosis infection’. J. Cell. Biochem. 120, 8949–8955. doi: 10.1002/jcb.27452
Wang, M., Mao, C., Ouyang, L., Liu, Y., Lai, W., Liu, N., et al. (2019). ‘Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA’. Cell Death Differ. 26, 2329–2343. doi: 10.1038/s41418-019-0304-y
Wang, W., Hu, W., Wang, Y., An, Y., Song, L., Shang, P., et al. (2020). ‘Long non-coding RNA UCA1 promotes malignant phenotypes of renal cancer cells by modulating the miR-182-5p/DLL4 axis as a ceRNA’. Mol. Cancer 19:18. doi: 10.1186/s12943-020-1132-x
Wang, Y., Zhong, H., Xie, X., Chen, C. Y., Huang, D., Shen, L., et al. (2015). ‘Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection’. Proc. Natl. Acad. Sci. U.S.A 112, E3883–E3892. doi: 10.1073/pnas.1501662112
Washietl, S., Kellis, M., and Garber, M. (2014). ‘Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals’. Genome Res. 24, 616–628. doi: 10.1101/gr.165035.113
Wei, L., Liu, K., Jia, Q., Zhang, H., Bie, Q., and Zhang, B. (2021). ‘The roles of host noncoding RNAs in mycobacterium tuberculosis infection’. Front. Immunol. 12:664787. doi: 10.3389/fimmu.2021.664787
Wu, Q., Zhong, H., Bai, H., Liu, T., Song, J., Wen, Y., et al. (2020). ‘Clinical relevance of the lnc-HNF1B-3:1 genetic polymorphisms in Western Chinese tuberculosis patients’. J. Clin. Lab. Anal. 34:e23076. doi: 10.1002/jcla.23076
Xu, Y., Yu, J., Ma, C., Gong, Z., Wu, X., and Deng, G. (2021). ‘Impact of knockdown LincRNA-Cox2 on apoptosis of macrophage infected with Bacillus calmette-guérin’. Mol. Immunol. 130, 85–95. doi: 10.1016/j.molimm.2020.11.008
Yang, M., Li, J., Deng, S., Fan, H., Peng, Y., Ye, G., et al. (2022). Competitive endogenous RNA network activates host immune response in SARS-CoV- 2-, panH1N1 (A/California/07/2009)-, and H7N9 (A/Shanghai/1/2013)-infected cells. Cells 11:487. doi: 10.3390/cells11030487
Yang, X., Yang, J., Wang, J., Wen, Q., Wang, H., He, J., et al. (2016). ‘Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with mycobacterium tuberculosis’. Sci. Rep. 6:38963. doi: 10.1038/srep38963
Yao, R.-W., Wang, Y., and Chen, L.-L. (2019). ‘Cellular functions of long noncoding RNAs’. Nat. Cell Biol. 21, 542–551. doi: 10.1038/s41556-019-0311-8
Ye, Y., Xu, Y., Lai, Y., He, W., Li, Y., Wang, R., et al. (2018). ‘Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization’. J. Cell. Biochem. 119, 2951–2963. doi: 10.1002/jcb.26509
Yi, Z., Li, J., Gao, K., and Fu, Y. (2014). ‘Identifcation of differentially expressed long non-coding RNAs in CD4+ T cells response to latent tuberculosis infection’. J. Infect. 69, 558–568. doi: 10.1016/j.jinf.2014.06.016
Zeni, P. F., and Mraz, M. (2021). ‘LncRNAs in adaptive immunity: role in physiological and pathological conditions’. RNA Biol. 18, 619–632. doi: 10.1080/15476286.2020.1838783
Zhang, X., Guo, J., Fan, S., Li, Y., Wei, L., Yang, X., et al. (2013). ‘Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberc(walzl et al., 2018)ulosis diagnosis’. PLoS One 8:e81076. doi: 10.1371/journal.pone.0081076
Zhang, X., Liang, Z., Zhang, Y., Dai, K., Zhu, M., Wang, J., et al. (2020). ‘Comprehensive analysis of long non-coding RNAs expression pattern in the pathogenesis of pulmonary tuberculosis’. Genomics 112, 1970–1977. doi: 10.1016/j.ygeno.2019.11.009
Zhao, Z., Zhang, M., Ying, J., Hu, X., Zhang, J., Zhou, Y., et al. (2017). ‘Significance of genetic polymorphisms in long non-coding RNA AC079767.4 in tuberculosis susceptibility and clinical phenotype in Western Chinese Han population’. Sci. Rep. 7:965. doi: 10.1038/s41598-017-01163-y
Keywords: tuberculosis, lncRNA, immune response, diagnosis, biomarker
Citation: Zhang X, Chen C and Xu Y (2022) Long Non-coding RNAs in Tuberculosis: From Immunity to Biomarkers. Front. Microbiol. 13:883513. doi: 10.3389/fmicb.2022.883513
Received: 25 February 2022; Accepted: 24 March 2022;
Published: 11 May 2022.
Edited by:
Shuang-yong Xu, New England Biolabs, United StatesReviewed by:
Weiwei Yang, New England Biolabs, United StatesCopyright © 2022 Zhang, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuzhong Xu, MjU0NTAxNjUxQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.