- 1Biotechnology Division, CSIR-Institute of Himalayan Bioresource Technology, Palampur, India
- 2Academy of Scientific and Innovative Research (AcSIR), CSIR–Human Resource Development Centre (CSIR-HRDC), Ghaziabad, India
Bacterial adaption to the multiple stressed environments of high-altitude niches in the Himalayas is intriguing and is of considerable interest to biotechnologists. Previously, we studied the culturable and unculturable metagenome microbial diversity from glacial and kettle lakes in the Western Himalayas. In this study, we explored the adaptive strategies of a unique Himalayan eurypsychrophile Iodobacter sp. PCH194, which can synthesize polyhydroxybutyrate (PHB) and violacein pigment. Whole-genome sequencing and analysis of Iodobacter sp. PCH194 (4.58 Mb chromosome and three plasmids) revealed genetic traits associated with adaptive strategies for cold/freeze, nutritional fluctuation, defense against UV, acidic pH, and the kettle lake's competitive environment. Differential proteome analysis suggested the adaptive role of chaperones, ribonucleases, secretion systems, and antifreeze proteins under cold stress. Antifreeze activity inhibiting the ice recrystallization at −9°C demonstrated the bacterium's survival at subzero temperature. The bacterium stores carbon in the form of PHB under stress conditions responding to nutritional fluctuations. However, violacein pigment protects the cells from UV radiation. Concisely, genomic, proteomic, and physiological studies revealed the multiple adaptive strategies of Himalayan Iodobacter to survive the high-altitude stresses.
Introduction
The Himalayas host a range of environmental niches such as permafrost, glacial streams, lakes, sediments, meadows, and deserts that contain distinct microbial communities (Stres et al., 2013; Kumar et al., 2018, 2019, 2022; Thakur et al., 2018; Dhakar and Pandey, 2020). High-altitude Himalayan environments are characterized by multiple stress factors such as high UV irradiation, oxidative stress, frequent freeze-thaw cycles, desiccation, oxygen limitation, nutritional fluctuation, and perennially cold temperatures (Singh and Singh, 2004; Stres et al., 2013). Microbes thriving in permanently cold environments have evolved many adaptive strategies at the molecular and physiological levels (Mykytczuk et al., 2013; De Maayer et al., 2014; Goordial et al., 2016; Kumar et al., 2020a). Thus, the microbes thriving in high-altitude Himalayan niches provide an interesting model system to study microbial adaptation strategies in response to the fluctuating environment.
Global warming has led to the rapid meltdown and receding of Himalayan glaciers (Bolch et al., 2012; Maurer et al., 2019), resulting in new environmental niches that provide opportunities to study their microbial ecology (Hotaling et al., 2017). Glacial meltdown forms many enclosed depressions in a geographic area called kettle holes or kettle lakes (Corti et al., 2012). In fact, kettle lakes are exposed to frequent changes in physicochemical factors such as thermal, hydrodynamics, and nutrients (Reverey et al., 2018). Hence, such niches are a goldmine for studying biogeochemical cycles (Reverey et al., 2016, 2018), distinct microbial communities (Kumar et al., 2022), and commercially important bacteria (Kumar et al., 2021).
A recent glacial retreat has created a kettle lake at 4,200 m above sea level in the Sach Pass area of the Western Himalayas (Himachal Pradesh, India). The kettle lake is exposed to daily and seasonal fluctuations in thermal, hydrological, nutritional, and other physicochemical factors. The eurypsychrophilic bacterial strain Iodobacter sp. PCH194, which coproduces PHB and violacein pigment, was isolated from a sediment sample of this lake (Kumar et al., 2021). The genus Iodobacter is a minor member of microbial communities inhabiting rivers, streams, and low-temperature niches such as Arctic glacial lake sediment and the Antarctic Peninsula (Logan, 1989; Leblanc et al., 2009; Su et al., 2013; Atalah et al., 2020). Iodobacter and other violacein-producing bacteria are distinct in cold-water bodies and glacial environments. There are reports on violacein-producing bacteria from cold niches, studying the adaptive and ecological function of violacein pigment (Matz et al., 2004; Pantanella et al., 2007; Deines et al., 2009; Abboud and Arment, 2013; Mojib et al., 2013; Atalah et al., 2020). However, the unique features displayed by Himalayan Iodobacter, such as coproducing PHB and violacein and surviving in the freezing environment of high-altitude kettle lake, intrigue to explore its survival strategies.
This study investigated the adaptive strategies of Himalayan Iodobacter sp. PCH194 using genomic, proteomic, and physiological approaches. The bacterium possesses multiple genomic traits to cope with cold and freezing conditions, oxygen depletion, oxidative stress, starvation, acidic pH, and high UV irradiance in its high-altitude niche. Differential proteome and antifreeze activity analysis provides insight into a bacterial response to cold and freezing. In addition, physiological experiments demonstrated polyhydroxyalkanoate synthesis and violacein pigment-containing microbial mat formation as a key survival strategy. Our findings identify the multiple adaptive strategies of Iodobacter sp. PCH194 enable it to survive in the kettle lake environment of the high-altitude Himalaya.
Materials and Methods
Phenotypic and Biochemical Characterization of Iodobacter sp. PCH194
Iodobacter sp. PCH194, isolated previously from high-altitude kettle lake sediment (Kumar et al., 2021), was used for the study. The pure culture was submitted in patent deposit to the Microbial Type Culture Collection (MTCC number 25171) at CSIR-IMTECH, Chandigarh, India. The colony morphology of the bacterium was observed after 72 h of agar plate incubation at 20°C on the Antarctic bacterial medium (AMB) (0.5% peptone, 0.2% yeast extract, and 2.0% agar). The carbohydrate and sugar utilization tests and catalase, oxidase, and nitrite reduction activity assays were performed using HiMedia kits (HiMedia, Mumbai, India).
Whole-Genome Sequencing and Genomic Insights of Iodobacter sp. PCH194
Genomic DNA isolation and library construction for Iodobacter sp. PCH194 were performed as described previously (Kumar et al., 2020a). The genomic library was evaluated using Qubit 2.0 (Invitrogen, Carlsbad, California, USA) and Bioanalyser (Agilent Technologies, Santa Clara, California, USA). Whole-genome sequencing (WGS) was carried out using the PacBio RSII system (PacBio, Menlo Park, California, USA), and genome assembly was performed using the RS Hierarchical Genome Assembly Process (HGAP) protocol version 3.0 (Chin et al., 2013). The complete genome sequence was submitted to the NCBI database with genome assembly accession number GCA_004194535.1, BioSample ID SAMN08324479, and BioProject ID PRJNA428922. Genome annotation was performed using Rapid Annotation and Subsystem Technology (RAST) (Aziz et al., 2008) and Prokaryotic Genome Annotation Pipeline (PGAP) version 3.1 (Tatusova et al., 2016). DNA-DNA hybridization (DDH) and average nucleotide identity (ANI) analyses were performed using online tools, viz., Genome-to-Genome Distance Calculator (GGDC) (http://ggdc.dsmz.de/) (Meier-Kolthoff et al., 2013) and ANI calculator of Kostas lab (http://enve-omics.ce.gatech.edu/ani/) (Goris et al., 2007), respectively. Phylogenetic trees for the members of the genus Iodobacter were constructed based upon the available whole-genome sequences in the NCBI genome database. A phylogenetic tree was constructed using Type (Strain) Genome Server (TYGS) (https://tygs.dsmz.de/) by manually restricting the job to the input sequences (Meier-Kolthoff and Göker, 2019). Genes associated with adaptive traits such as carbon storage, respiration, chemotaxis, cold stress and oxidative stress response, acidic pH adaptation, defense against UV, and interspecies competition were mined by analyzing subsystem categories in RAST and NCBI PGAP files (Aziz et al., 2008; Tatusova et al., 2016).
Growth of Iodobacter sp. PCH194 at low Temperatures
Growth of Iodobacter sp. PCH194 was monitored in nutrient broth medium (g/l; meat extract 1.0, peptone 5.0, yeast extract 2.0, NaCl 5.0) at a temperature range of −5 to 37°C by observing the OD600 and OD460. The specific growth rate (μ) of the bacterium was calculated in a static condition at temperatures of 0, 4, and 20°C and in a shaking condition at 20°C.
Proteomic Analysis of Iodobacter sp. PCH194 Growing at Different Temperatures
The bacterium culture was inoculated in nutrient broth and incubated at 20°C to prepare a seed culture of 1.0 OD460. Seed culture (1.0%, v/v) was transferred to the same medium and incubated at temperatures of 20, 10, and 4°C. After 3 days of cold acclimation at 4°C (OD460 0.567), the culture was transferred to 0°C and the growth was monitored. To further evaluate the adaptive changes, a differential proteome study was performed.
Protein Extraction and In-Gel Digestion and MALDI-TOF-TOF Identification
Iodobacter sp. PCH194 was grown at 0, 4, and 20°C, and the culture was centrifuged at 9,600 g for 10 min to obtain the pellet. The bacterial pellet was resuspended in 10 mM Tris-HCl, pH 8.0, and total protein was extracted by sonicating the bacterial pellet. The protein was quantified using the Bradford assay with a standard curve of BSA at various concentrations. An equal amount of protein was loaded onto the 10% SDS PAGE to visualize differentially expressed proteins. The differentially expressed polypeptides from SDS-PAGE were excised, washed in deionized water, and destained in 100 mM NH4HCO3/50% acetonitrile for 15 min at room temperature (RT). Polypeptides were reduced with 10 mM dithiothreitol and alkylated with 55 mM iodoacetamide. The resulting alkylated polypeptides were dehydrated using acetonitrile followed by overnight digestion with a freshly prepared trypsin solution (25 mM NH4HCO3 with 5 ng/μl of trypsin, Sigma, USA) at 37°C. Trypsin was inactivated by incubating peptide samples in 0.1% formic acid (1.0 μl) for 40 min at 37°C. The digested peptides were extracted using extraction buffer (50% trifluoroacetic acid/50% acetonitrile) in an ultrasonic water bath at RT. Extracted peptides were mixed with MALDI matrix (0.5 μl) α-cyano-4-hydroxycinnamic acid (20 mg/ml) in 0.1% trifluoroacetic/30% (v/v) acetonitrile (1:2) and dried at RT followed by mass spectrometric analysis by MALDI-TOF/TOF-MS/MS using an UltrafleXtremeTM mass spectrometer (Bruker Daltonics Inc., Billerica, Massachusetts, USA). The instrument was set in positive ionization mode and was previously calibrated with a mass standard starter kit and standard tryptic BSA digest (Bruker). Proteins were identified using Mascot Software version 3.5 (https://matrixxscience.com) (Matrix Science, London, UK) at 1.2 Da MS/MS mass tolerance, in which SwissProt database was searched among Proteobacteria and one missed cleavage was allowed. Oxidation of methionine and carbamidomethylation of cysteine were considered variable and fixed modifications, respectively. Proteins identified from Mascot were functionally annotated by the UniProt database (https://www.uniprot.org).
Label-Free Quantification
Label-free relative quantification was performed according to the method described by Sharma et al. (2014) to identify and analyze the differentially expressed proteins at a low temperature. The total protein of Iodobacter sp. grown at different temperatures was quantified by the Bradford method. Total protein (200 μg) from each sample was lyophilized and resuspended in 100 μl of ammonium bicarbonate (100 mM). Protein suspension was reduced with 10 mM dithiothreitol and alkylated with iodoacetamide (55 mM), followed by trypsinization (Promega, Madison, Wisconsin, USA) for 12–16 h at 37°C. Trypsin was inactivated by incubating the sample with 0.1% formic acid (1 μl) for 40 min at 37°C. Digested peptides thus obtained were lyophilized and dissolved in 0.1% formic acid. The resuspended sample (10 μl) was loaded on an Advance Bio Peptide column (2.1, 100 mm, 2.7 μm) (Agilent Technologies, Santa Clara, California, USA) equilibrated with 0.1% (v/v) formic acid (Solvent A) for 3 min with a flow rate of 5 μl/min. For peptide separation, a linear gradient starting from 5 to 45% of solvent B (0.1%, v/v) formic acid (in acetonitrile) for 75 min was employed. Three replicates per sample were used for all data acquisition using 6550 UHPLC-Q-TOF-IMS (Agilent Technologies, Santa Clara, California, USA). The following parameters were set in the acquisition mode; MS minimum range (m/z): 50, MS maximum range (m/z): 1,700, and MS and MS/MS scan rate (spectra/sec): 3. The following parameters were fixed in precursor selection; maximum precursor selection per cycle: 3, threshold (absolute): 5,000, threshold (relative %): 0.010, and target (counts/spectrum): 25,000. The instrument was set in positive ionization mode. For protein identification by Spectrum Mill software (Agilent Technologies, Santa Clara, USA), the following search parameters were used; NCBI database (version 2010_09), having 16,365 protein sequences; fixed modification was carbamidomethylation; N-terminal acetylation, N and Q deamidation, and M oxidation, as variable modifications; missed cleavage: 1 and fragment error tolerance: 0.5 Da. Accession numbers retrieved from Spectrum Mill software were annotated using NCBI. The proteins identified using label-free quantification and MALDI-TOF/TOF-MS/MS were analyzed with BLAST2GO analysis and the UniProt database, and the matched protein GIs were functionally categorized.
Ice Recrystallization Inhibition Activity
The IRI activity of lysate was analyzed using a splat assay following Knight and Duman (1986), with slight modifications as mentioned by Kashyap et al. (2020). Total protein (1.0 mg/ml) in 30% sucrose was sandwiched in two coverslips and was flash-frozen at −20°C. It was allowed to recrystallize at −9°C for 40 min using a nanolitre osmometer (Otago Instruments, Dunedin, New Zealand). The growth of ice crystals was monitored under an upright Eclipse Ci Microscope (Nikon, Tokyo, Japan) at 10X resolution. BSA (2.0 mg/ml) was used as a negative control. Heat denaturation of crude lysate at 100°C for 10 min was performed to confirm that the activity was because of the protein content. The area of crystals was quantified using ImageJ software.
PHA Production Under Different Physiological Conditions
In the PHA production medium, PHA production by Iodobacter sp. PCH194 was assessed (g/l; K2HPO4 6.0, KH2PO4 3.0, NH4Cl 0.5, NaCl 1.0, and 1.0 mM MgSO4, 0.1 mM CaCl2) under different environmental conditions, viz., temperature, aeration, and pH. The bacterium was also tested to synthesize PHA at varying nutritional conditions of carbon to nitrogen ratio from 0.2 to 60 (where NH4Cl concentration was kept constant at 0.05% and glucose concentration varied accordingly). Biomass and PHA production were quantified as described by us in earlier studies (Kumar et al., 2018, 2020a).
Morphology and Anatomy of Iodobacter sp. PCH194
The cell morphology of bacterial cells from the late log phase was examined using TEM analysis using JEOL JEM-1010 (JEOL, Akishima, Japan). The PHA granules formation by bacterial isolate was analyzed as per the methods described by Sandoval et al. (2007) with slight modification. The bacterial cells were grown to stationary phase in 2.0% glucose and harvested at 2,400 g, washed three times with phosphate buffer saline, and fixed by glutaraldehyde solution (2.0%, v/v) at 4°C. Samples were washed three times with PBS buffer and fixed with 1.0% osmium tetroxide for 60 min in the dark. The washing step was repeated, stained in blocks with uranyl acetate (30%), and embedded slowly in Embed 812 epoxy resin (Electron Microscopy Sciences, Hatfield, Pennsylvania, USA). Polymerization of blocks was carried out in an oven at 50°C for 72 h. Ultrathin sections of the samples were cut using an ultramicrotome, and further grids were prepared. Structural studies were performed with an FEI Technai G2200kV transmission electron microscope (FEI, Hillsboro, Oregon, USA).
Synthesis of Violacein Pigment by Iodobacter sp. PCH194 and Its UV Protective Role
Violacein was produced by Iodobacter sp. PCH194 growing in glucose and tryptone medium, and extracted as described by Kumar et al. (2021). For analyzing the protective role of violacein pigment against UV, the cells of Iodobacter sp. PCH194 were drawn at a different time interval at the initial log phase (when cells are non-pigmented), late log phase, and stationary phase (when cells produce pigments), washed in normal saline and diluted to 1.0 OD600. Different dilutions were spread on a nutrient agar plate and treated with 302 nm UV radiation (UV B302) from a distance of 1.4 feet by using UV torch (Analytik Jena, California, USA) for 1, 2, and 3 min. The dose of UV-B was 0.32 Js−1cm−2 (320 mWcm−2) as measured by a UV light meter (Model UV340A, Lutron Electronic, Pennsylvania, USA). UV-untreated plates from each growth phase served as controls. Non-pigmented cells of Iodobacter sp. PCH194 were harvested after 16 h of growth, amended with methanol-extracted violacein pigment (100 μg/ml), and compared to non-pigmented cells without violacein in a UV exposure experiment as described. All plates were further incubated at 20°C for 2–3 days, and the number of colonies was counted and recorded as log cfu/ml. The significance of the experiment was determined by the t-test, and the level of significance was expressed as P-value at *≤ 0.05, **≤ 0.01, and ***≤0.001.
Results
Sampling Site and Bacterium Description
The Bhoot ground lake at Sach Pass (Pangi Valley, Himachal Pradesh, India) is a shallow kettle lake formed due to the glacier retreat and remains frozen for almost half of the year (Figure 1A). The lake water temperature, pH, and UV radiation were 4°C, 4.5, and 50 mWcm−2, respectively; measured at the site during the sampling (18 November 2016). Available N, P, K (kg/Ha), electrical conductivity (μS/cm), and pH of dried mud vary from 65.86 to 194.9, 11.68 to 25.76, 68.26 to 162.13, 88 to 95, and 4.1 to 4.26, respectively (Supplementary Table 1). The isolated bacterium Iodobacter sp. PCH194 forms violet-pigmented colonies (Figure 1B). The cells were found to be in straight round-ended rods, Gram-negative, facultative anaerobes, oxidase and catalase-positive, and glucose-fermenting. The bacterium can grow at −5 to 25°C, with optimum growth at 20°C (Supplementary Table 2).
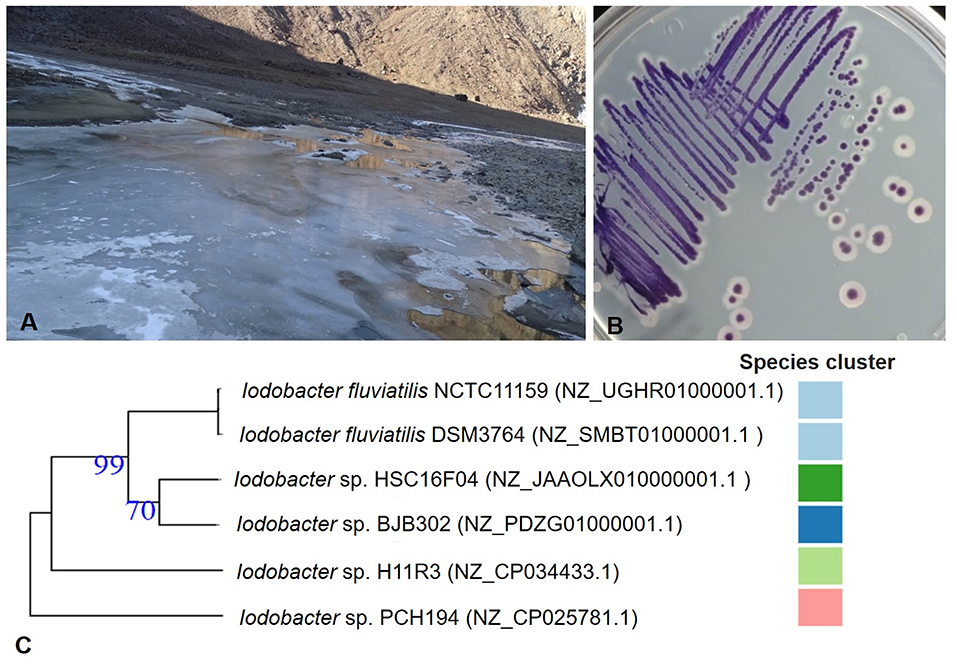
Figure 1. Sampling site and Himalayan bacterium Iodobacter sp. PCH194. (A) A view of partially frozen kettle lake (Picture was taken on 18 November 2016) at an altitude of 4,160 masl and geographical coordinates of 33.006831°N, 76.248209°E at Sach Pass area in Chamba district of Himachal Pradesh, India, (B) isolated single colonies of Iodobacter sp. PCH194, and (C) a phylogenetic tree for genus Iodobacter based on the available whole-genome sequences. The number in parenthesis is the NCBI reference number for the respective genome assembly, and the color code represents the species cluster, where different colors represent the different species. The tree was constructed using an online TYGS server.
General Description of the Genome and Phylogenomic Analysis
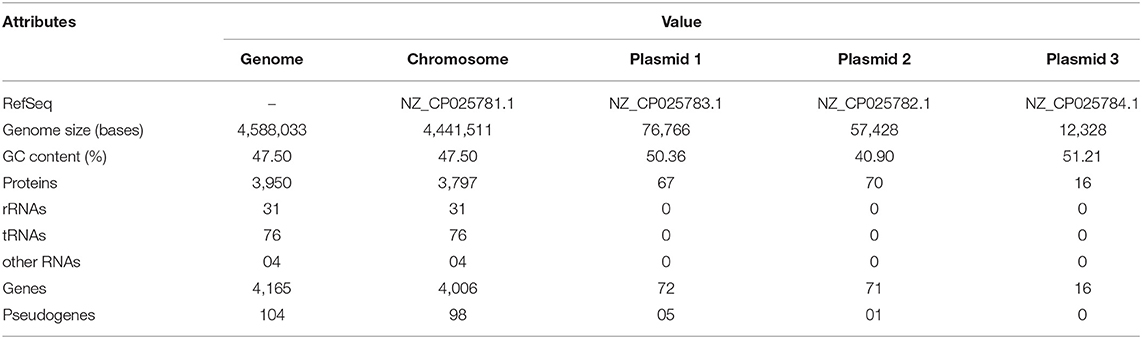
Whole-genome sequencing of Iodobacter sp. PCH194 has resulted in a single high-quality contig of a 4.44 Mb chromosome and three sizes of 76.7, 57.4, and 12.3 kb plasmids. The NCBI predicted 4,165 genes, 3,950 proteins, 31 rRNA, 76 tRNA, 04 ncRNAs, and 104 pseudogenes (Table 1). The phylogenomic analysis of Iodobacter sp. PCH194 with reference strains I. fluviatilis DSM 3764 and I. fluviatilis NCTC11159 showed a low value of ANI (80.81%) and DDH (25.90% and 26.40%). Similarly, other strains of Iodobacter, i.e., BJB302, H11R3, and HSC-16F04, also showed a low percentage in ANI and DDH analysis (Supplementary Table 3). The phylogenetic tree was constructed using the TYGS server based on the available whole-genome sequences in the NCBI database. Iodobacter sp. PCH194 showed a separate and distinct clade compared to the rest of the Iodobacter spp. (Figure 1C). The low ANI (79.99–80.81), DDH (25.70–26.40) score, and phylogenetic analysis of strain PCH194 clearly describe it as a putative novel species of the genus Iodobacter.
Genomic Insights for Adaptive Strategies of Iodobacter sp. PCH194
Genomic analysis of Iodobacter sp. PCH194 revealed multiple genes/proteins for stress adaptation (Table 2). The adaptive genomic traits are described in detail in the following subheads and are tabled in Supplementary Data Set 1.
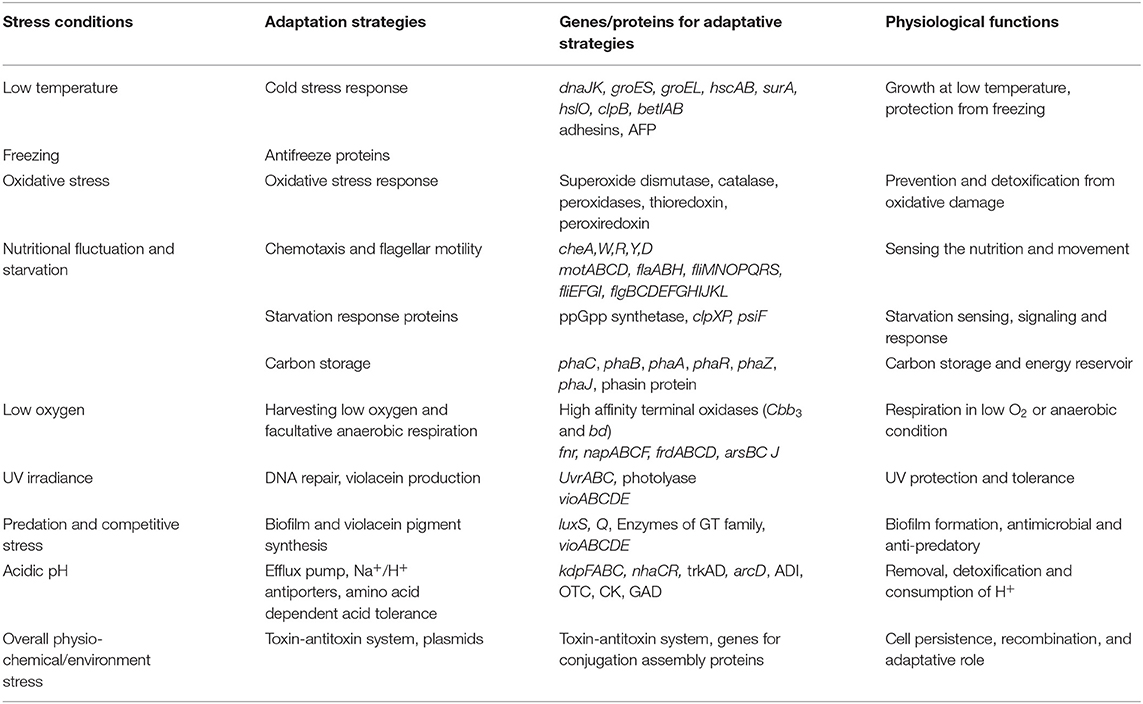
Table 2. Adaptive genomic traits of Iodobacter sp. PCH194. Adaptive strategies to stress conditions and related genes/proteins are tabulated along with their putative physiological and ecological functions.
Starvation and Storage of Carbon
The genome of Iodobacter sp. PCH194 showed the presence of key genes involved in nutritional starvation, viz., carbon sensing and downstream response mechanisms such as carbon starvation protein A (cstA), sigma factor (rpoS), stringent starvation proteins A and B (sspAB), bifunctional ppGpp synthetase, Clp proteases (clpA,B,S,X,P), and phosphate starvation-inducible protein (psiF). The genes for polyhydroxyalkanoate (PHA) synthesis, namely, polyhydroxyalkanoate synthase (phaC) and acetyl-3-hydroxybutyryl-CoA dehydrogenase (phaB), were found in two copies each, while acetyl-CoA-acyltransferase or thiolase (phaA) was found in a single copy. Additionally, one copy of PHA depolymerase (phaZ) and PHA regulatory gene (phaR), two genes encoding for phasin family protein, and three copies of enoyl CoA (fadB or phaJ) were also present.
Respiration
The genomic insights revealed the presence of genes for various high-affinity terminal oxidases, viz., cytochrome cbb3 and cytochrome bd, and an operon of NADH-quinone oxidoreductase (nuoABCEFGHIJKLMN). The genome analysis also showed various genes encoding for fumarate/nitrate reduction transcriptional regulator Fnr (fnr), two-component system response regulator NarL (narL), and various reductases such as periplasmic nitrate reductase (napABCF), fumarate reductase (frdABCD), and arsenate reductase (arsBCJ).
Genomic Features for Cold, UV, Oxidative, and Osmotic Stress
NCBI annotation revealed genes for cold stress, i.e., cold-shock proteins, molecular chaperones, co-chaperone, chaperonin, fatty acid desaturase, and stress proteins. The genes encoding for the proteins involved in oxidative stress include superoxide dismutase, catalase, peroxiredoxin, thioredoxin, and various peroxidases, and the excinuclease repair system (uvrABCD) were also present in the genome. Additionally, genes encoding for choline ABC transporters and permease, betaine aldehyde dehydrogenase, and choline dehydrogenase involved in osmolyte glycine-betaine synthesis were present.
Violacein Biosynthesis and Biofilm Formation
A complete operon containing five genes, vioABCDE encoding enzymes involved in violacein pigment biosynthesis and quorum sensing regulatory genes (luxS and luxQ), was present in the genome. The bacterium possesses genes (epsDEF GI) for the biosynthesis of polysaccharides and export. Furthermore, the dbCAN meta server found genes encoding for 13 different glycosyltransferase (GT) family proteins in the genome.
Additional Adaptive Features
The genome of Iodobacter sp. PCH194 possesses key genes for chemotaxis (cheA, D, W, R, Y) and flagellar assembly (motABCD, fliMNOPQRS, fliEFGI, flaABH, flgBCDEFGHIJKL). The genome showed the presence of genes encoding for the cation-efflux pump, cation transporters and Na+/H+ (sodium:proton) antiporters, potassium channel protein, potassium transporter (Kup), and an operon kdpFABC encoding for the ATP-dependent potassium transporter system. The bacterium also possesses the genes encoding for the enzymes involved in amino acid-dependent acid tolerance, i.e., arginine-ornithine antiporter (ArcD), arginine deiminase (ADI), ornithine carbamoyltransferase (OTC) and carbamate kinase (CK), glutaminase, glutamate decarboxylase, lysine decarboxylase, and histidine decarboxylase. Apart from other mentioned strategies, the genome of Iodobacter sp. PCH194 possesses genes encoding for the toxin-antitoxin (TA) system on chromosomes and plasmid. Additionally, three plasmids possess various genes encoding for replication proteins, transposase, recombinase, replicase, mobilization protein, conjugal transfer protein TraA, and transcriptional regulators.
Proteomic Response of Iodobacter sp. PCH194 to Cold and Freezing Conditions
Growth Kinetics and General Proteome Response to Cold and Freeze
Iodobacter sp. PCH194 grew at a temperature ranging from −5 to 25°C, with optimum growth at 20°C. The growth rate (μ) of the bacterium calculated in NB medium (pH: 7.0) at 0, 4, and 20°C in static, and at 20°C with 150 rpm was 0.008, 0.038, 0.049, and 0.096 h−1, respectively (Figure 2A). The bacterium under cold stress (at 4 and 0°C conditions) underwent proteome changes as visualized using SDS-PAGE (Supplementary Figure 1). A total of 19 differentially expressed polypeptides were observed and subjected to mass spectrometric identification using MALDI-TOF/TOF. After Mascot analysis, 68 upregulated and 17 downregulated protein targets were identified (Supplementary Data Set 2).
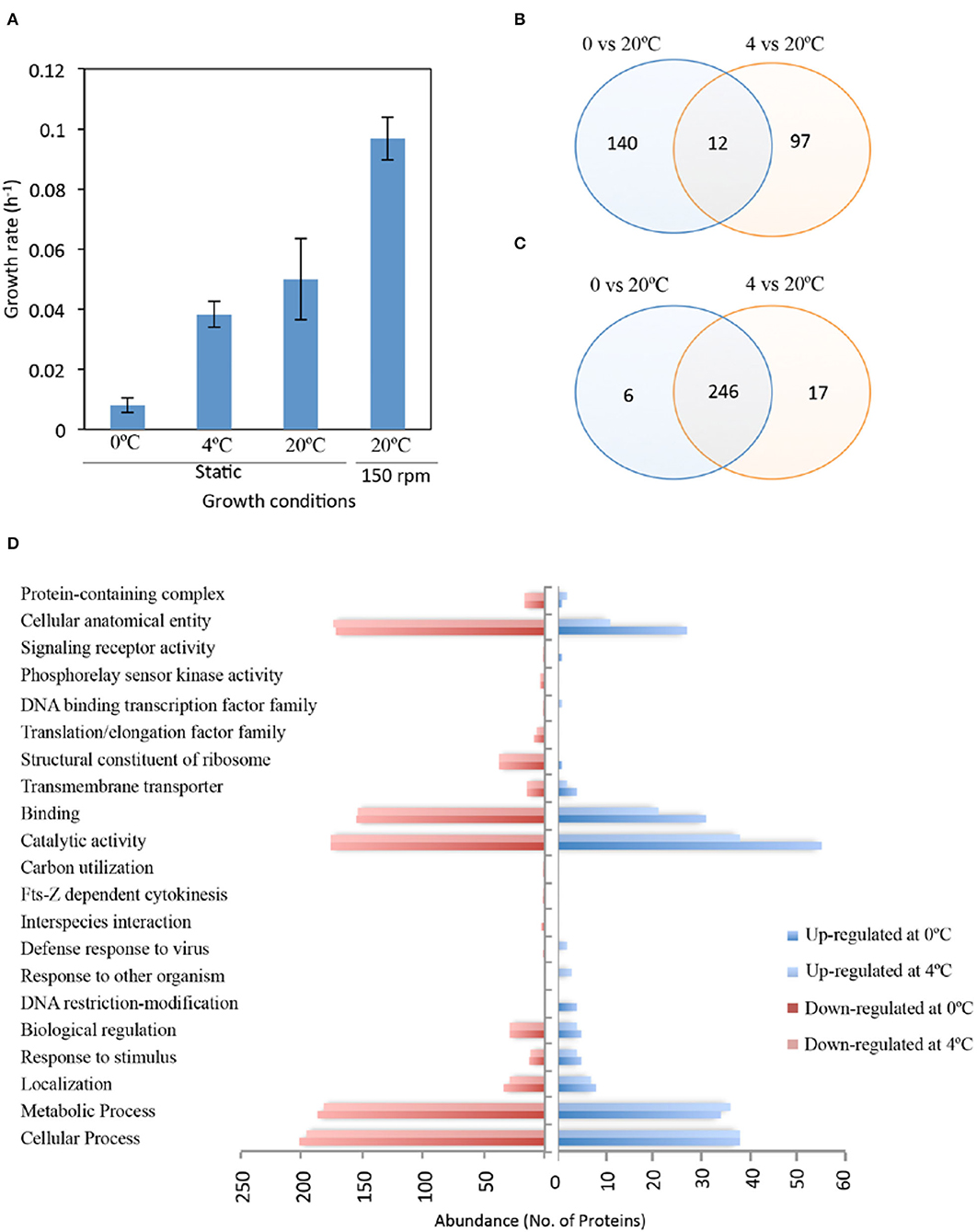
Figure 2. Growth and proteomic response of Iodobacter sp. PCH194 in cold/freeze conditions. (A) Growth rate (OD460) of Iodobacter sp. PCH194 at different temperatures in nutrient broth medium, (B) Venn diagram showing the total and shared number of upregulated, and (C) downregulated protein targets in response to cold (4°C) and freeze (0°C) compared to 20°C identified by gel-based and gel-free proteomics approaches, and (D) the abundance of upregulated and downregulated proteins with functional categorization by UniProt.
In contrast, the label-free proteomics data led to the quantitative identification of 558 proteins (p < 0.05), showing a ≥1.5-fold increase and ≤0.75 decrease in abundance upon cold/freeze stress (Supplementary Data Set 3). Based on the gel and gel-free approaches, 152 and 109 proteins were upregulated at 0 and 4°C, respectively, compared to 20°C (Figure 2B). Among all proteins that were upregulated at cold temperatures, 125 and 64 proteins were identified exclusively at 0 and 4°C, respectively. The bacterium also revealed downregulation of 252 and 263 proteins at 0 and 4°C, respectively (Figure 2C). These downregulated proteins include the majority of proteins expressed at 20°C. The gene ontology (GO) study of differently expressed proteins under cold (4°C) and freezing (0°C) conditions revealed upregulated and downregulated targets in the cellular component and biological process category (Supplementary Figure 2). Cellular components that were upregulated at 0°C as compared to 4°C included a higher number of membrane proteins and intrinsic components of membrane proteins. Other proteins belonging to these categories were found to be downregulated in both cold and freezing conditions. These results indicate an important role for membrane dynamics during freeze stress. The upregulated and downregulated protein targets were further functionally categorized using the UniProt database (Figure 2D, Supplementary Data Set 4). Upon cold stress, the bacterium showed differential regulation in cellular and metabolic processes such as nucleotide biosynthesis, transcription, translation, DNA recombination and repair, peptidoglycan, and fatty acid metabolism. Additionally, proteins involved in maintaining the structural and functional integrity of cells (cellular anatomical entity) and transmembrane transporters also exhibit different protein regulations during low temperatures. Since these categories show differential proteome responses, their functions in Iodobacter sp. PCH194 were further analyzed to reveal cold-adaptive strategies.
Differential Proteome Response for Important Functional Categories to Cold and Freeze
The proteome changes in Iodobacter sp. PCH194 in response to cold/freezing stress were functionally categorized (Supplementary Table 4). Cold and freeze stress cause misfolding of proteins. Therefore, chaperones and proteases play a crucial role during cold/freeze stress conditions. Iodobacter sp. PCH194 showed an abundance of molecular chaperones DnaJ and HscA at freezing temperatures. However, chaperone GroEL and co-chaperone GrpE exhibited a decreased abundance, while chaperone DnaK was slightly reduced during cold stress. Additionally, ATP-dependent Clp proteases were also found to be upregulated in both gel-based and gel-free studies during low-temperature stress. Cold/freeze stress-regulated not only chaperones and proteases but also RNA metabolism, mediated by cold shock proteins (CSPs) and ribonucleases (RNase). RNase E and RNase Z were found abundant at 0°C in Iodobacter sp. PCH194, whereas RNase II and RNase BN were not detected at low-temperature conditions.
Interestingly, the proteins of secretion system VI and IV families were found to be upregulated during cold/freeze stress. In addition, T4SS and T6SS, phage tail proteins closely resembling T6SS, were also upregulated at freezing temperature (0°C). Besides, the glycosyltransferase (family 1) protein that participated in biofilm formation was also upregulated during low temperatures. One of the strategies to survive under subzero temperatures is the production of antifreeze proteins such as adhesins. In Iodobacter sp. PCH194, adhesin was detected in all three temperatures, but its expression decreased during 4°C stress and then increased at 0°C. Furthermore, MafB adhesion was detected only at 0°C, indicating that it might also have a role as an antifreeze protein during freezing conditions.
Ice Recrystallization Inhibition Activity in Iodobacter sp. PCH194
Antifreeze activity in crude lysate proteins (1.0 mg/ml) of Iodobacter sp. PCH194 was validated by inhibition of ice recrystallization at −9°C, when Iodobacter sp. PCH194 lysate decreased the average area of ice crystals by a factor of 4 compared to BSA (1.0 mg/ml) as a negative control. Heat denaturation of the protein lysate resulted in the loss of the IRI activity and produced a 5-fold increase in the average area of ice crystals (Figure 3). The IRI activity in crude lysate indicated that antifreeze proteins are constitutively present in Iodobacter sp. PCH194.
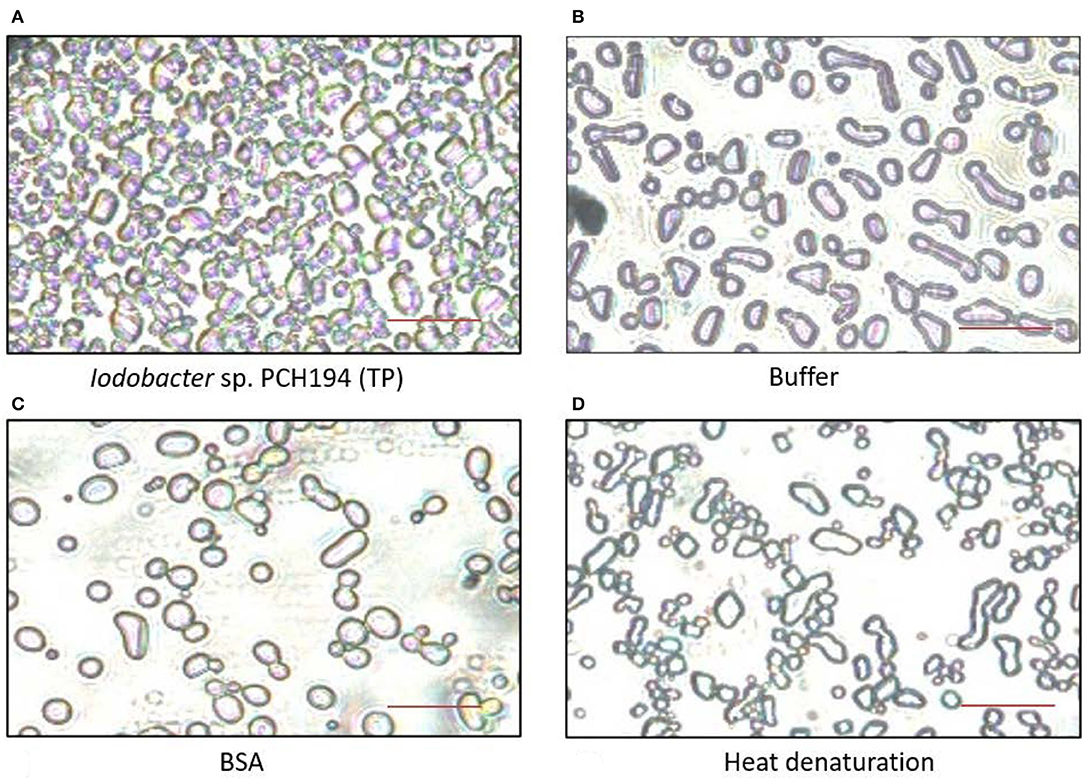
Figure 3. Antifreeze activity exhibited by Iodobacter sp. PCH194. The ice recrystallization inhibition (IRI) was observed at −9°C after 40 min. (A) Iodobacter sp. PCH194 [total cell lysate protein (TP), 1.0 mg/ml] showed smaller ice crystals upon recrystallization in comparison to (B) buffer (20 mM Tris-HCl, pH 8.0), and (C) BSA (1.0 mg/ml) as a negative control, and (D) heat denaturation (at 100°C for 10 min) of Iodobacter sp. PCH194 (TP) resulted in the loss of activity. Scale bar = 200 μm.
Physiological Insights for Adaptive Strategies of Iodobacter sp. PCH194
Carbon Storage Under Different Physiological Conditions
Iodobacter sp. PCH194 showed the ability to synthesize PHB in different conditions of pH (4–10), temperature (4–25°C), varied carbon to nitrogen ratio, and in static as well as shaking conditions (Figure 4). The bacterium synthesizes 40–60% PHB to its dry cell mass in a wide range of carbon to nitrogen ratios of 0.2–60 (glucose and NH4Cl as sole carbon and nitrogen sources). Though the growth is slow in the medium with a C/N ratio of 0.2, the bacterium can synthesize 40% of PHB to its dry cell mass. The TEM analysis of an ultrathin section of the stationary phase showed the formation of intercellular PHB granules.
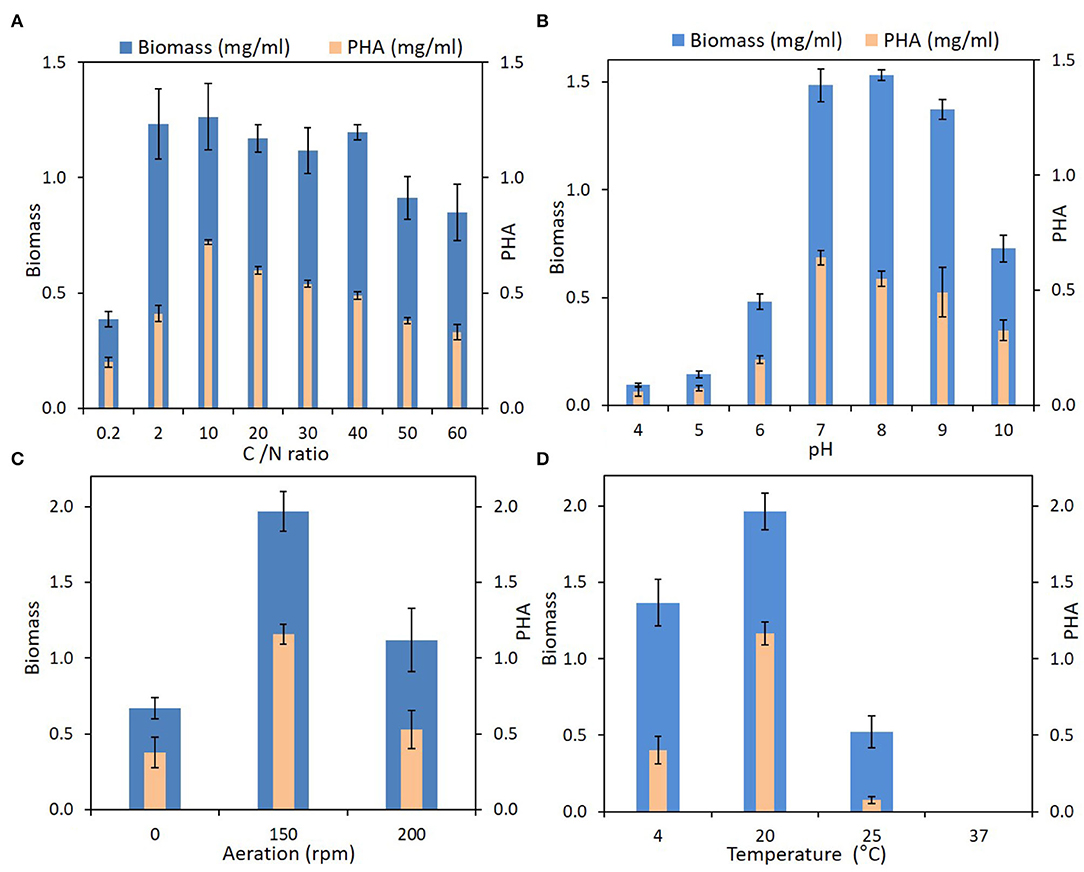
Figure 4. PHA production by Iodobacter sp. PCH194 at different physicochemical and environmental conditions. (A) Carbon to nitrogen (g/g) ratio, (B) pH, (C) aeration, and (D) temperature.
Violacein Pigment Production and Formation of Biofilm
Physiological experiments showed that Iodobacter sp. PCH194 can grow and produce violacein pigment under static and shaking conditions in the NB medium. The cells adhered to the media's surface under static conditions and formed a violet-colored mat in 7–8 days of incubation at 4°C (Figure 5A). The bacterium also formed a violet-colored biofilm around cells under shaking conditions as well. The TEM analysis of stationary phase bacteria grown in NB at 20°C and 150 rpm showed intercellular PHB granules and a clear sheath-like structure around the cells (Figures 5B,C).
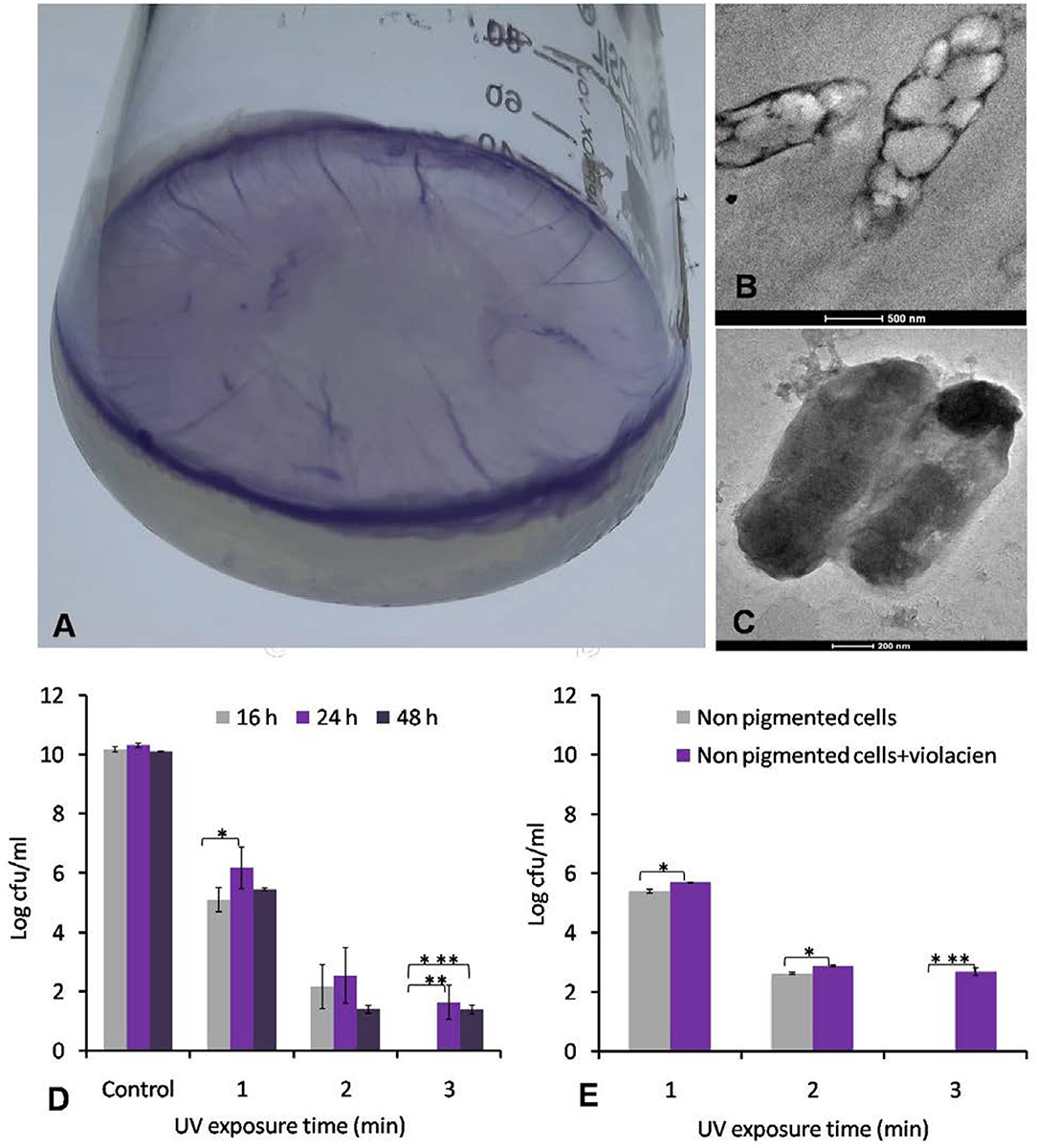
Figure 5. Formation of violate biofilm by Iodobacter sp. PCH194 and UV protective role of violacein pigment. (A) Formation of violet-colored mat on the surface of medium when growing at 4°C under static conditions, (B) TEM image of ultrathin sections of cells showing the presence of PHA granules, (C) morphology of bacterial cells showing the presence of a sheath around the cell, (D) effect of UV irradiance to cells of Iodobacter sp. PCH194 harvested at different growth phases, viz., non-pigmented cells of the initial log phase and pigmented cells of exponential log phase and stationary phase (expressing violacein pigment 43–50 and 41–45 μg/ml/OD460), and (E) effect of violacein (100 μg/ml) addition to UV tolerance of non-pigmented cells. The level of significance was expressed as a P-value as *≤0.05, **≤0.01, and ***≤0.001.
UV Protective Role of Violacein Pigment
The colonies of Iodobacter sp. PCH194 are violet pigmented, and they also produce violet pigment in a liquid medium. During undisturbed growth at 4°C, the bacterium produces a violet-colored mat at the medium's surface (Figure 5A). Violacein is likely to protect bacterial cells against UV irradiation at high altitudes. Therefore, to validate the role of violacein pigment against UV stress, the non-pigmented and pigmented cells of Iodobacter sp. PCH194 were subjected to a UVB irradiance of 320 mWcm−2. The effect of UV irradiance on the bacterial cells from different growth stages showed that the pigmented cells have higher tolerance and survival under UV. Pigmented cells from the late log phase showed 21% higher survival at 1 and 2 min and 10% higher survival at 3 min of UV exposure. Similarly, pigmented cells from the stationary phase showed 13%, 7%, and 6% higher survival than the non-pigmented cells in 1, 2, and 3 min of UV exposure (Figure 5D). The violacein pigment treated (100 μg/ml) non-pigmented cells of Iodobacter sp. PCH194 showed better UV tolerance than untreated cells (Figure 5E).
Discussion
This study unveiled multiple adaptive strategies of a unique Himalayan bacterium, Iodobacter sp. PCH194, ensuring its survival in high-altitude stresses (Figure 6). It also provides a complete genome sequence of Himalayan Iodobacter and highlights the adaptive traits in its genome. Furthermore, differential proteome response to cold and physiological experiments of antifreeze activity, PHB, and violacein synthesis supports its survival in the stress environment of high-altitude Himalayas. Additionally, the phylogenomic analysis suggested that Iodobacter sp. PCH194 is a putative novel bacterium in the genus Iodobacter with biotechnological importance. Herein, we discuss multiple adaptive strategies of Iodobacter sp. PCH194, supported by its genomic, proteomic, and physiological data.
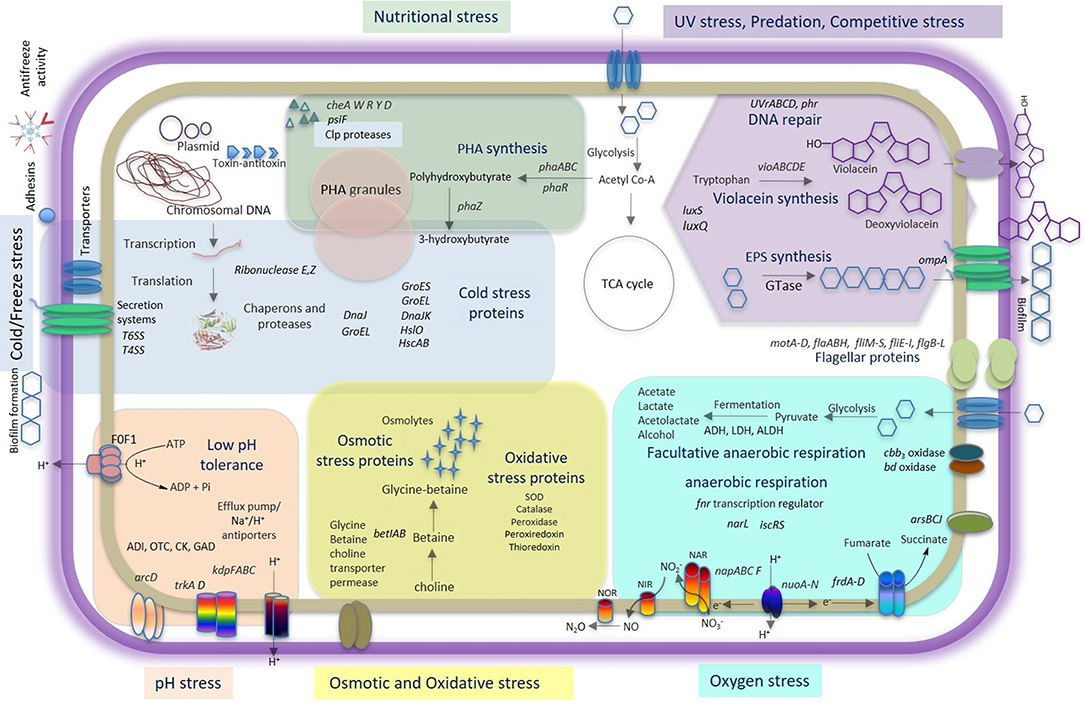
Figure 6. Comprehensive model of Iodobacter sp. PCH194 adaptive strategies. The model is based on genomic, proteomic, and physiological data showing multiple strategies of Iodobacter sp. PCH194 to adapt to the predominantly frozen kettle lake environment in the high-altitude Himalayas.
Cold stress is one of the major environmental stresses in the high-altitude Himalayan lakes. Iodobacter sp. PCH194 showed adaptation to changes in temperature and can grow from subzero to 25°C. Wide temperature adaptability was supported by many genes encoding for cold/freeze and other stress adaptation functions. Previous studies also reported a high copy number of genes for general stress, cold shock, osmotic, and oxidative stress in bacteria from the Antarctic and Arctic cryo-environments (Mykytczuk et al., 2013; Goordial et al., 2016). In response to the cold, Iodobacter sp. PCH194 revealed a differential proteome expression. During cold stress, bacteria reduce cell growth, and there is a downshift in the efficiency of transcription and translational machinery with an increase in the Csps proteins (Polissi et al., 2003). The proteins that show the most significant changes upon cold stress have been known to be involved in nucleotide biosynthesis, translation processes, DNA recombination and repair, peptidoglycan, and fatty acid metabolism (De Maayer et al., 2014; Baraúna et al., 2017). Proteins regulating all these functions were differentially regulated in Iodobacter sp. PCH194 upon low-temperature stress. The roles of the proteins in these categories are well-established in many cold-loving bacteria, such as Planococcus halocryophilus (Mykytczuk et al., 2013), Sphingopyxis alaskensis (Ting et al., 2010), and Psychrobacter sp. (Koh et al., 2017).
Chaperones and proteases play an important role in the cold adaptation of bacteria. Iodobacter sp. PCH194 chaperone system responded to low-temperature stress by increasing the expression of DnaJ, while the expression of DnaK did not show much change. DnaJ is known to interact with unfolded proteins and prevent their aggregation (Han and Christen, 2004). Besides, it also acts as Hsp40 and interacts with Hsp70 heat shock proteins (Hennessy et al., 2005). An interesting observation during the cold stress response of Iodobacter sp. PCH194 was the abundance of RNase E and RNase Z at 0°C. Low-temperature stress triggers the RNase to maintain the optimum transcript levels. DEAD-box helicases are associated with RNase E-based degradosomes under cold shock conditions to degrade structured RNAs (Prud'homme-Genereux et al., 2004; Barria et al., 2013). Furthermore, the stress response regulator rpoS is known to be degraded by RNase E and RNase III (Basineni et al., 2009). The RNA polymerase sigma factor RpoD responsible for transcription of constitutive genes was detected in Himalayan Iodobacter while growing at 4°C, but RpoS was not found at cold or freeze stress. This might be because of rpoS degradation by upregulated expression of RNase E. Therefore, RNases seem to play an important role in the cold adaptation of Iodobacter sp. PCH194. In addition, upregulated T4SS and phage tail proteins closely resembling T6SS (Pukatzki et al., 2007) play a role in communication with their population, competing with others, and interacting with the environment (Russell et al., 2014).
Various organisms produce antifreeze proteins to cope up with the freezing conditions. During freezing, smaller crystals are formed in huge numbers. Their formation does not damage cells. However, large crystals harm cells during a process called recrystallization. During recrystallization, the smaller ice crystals grow by absorbing water and merging smaller ice crystals into larger ones. Antifreeze proteins (AFP) bind to ice crystals and prevent their growth during recrystallization, which results in an increased number of smaller ice crystals. Hence, AFP mitigate the damage caused to the cells. AFP are known to act extracellularly, for example, in plant apoplast space (Gupta and Deswal, 2012), while they operate in the periplasm and as adhesins in bacteria (Gilbert et al., 2005). The upregulation of adhesins in Iodobacter at 0°C indicated that they might play a role in freeze protection similar to MpAFP from the Antarctic bacterium Marinomonas primoryensis (Gilbert et al., 2005; Guo et al., 2017). The presence of IRI activity in Iodobacter sp. PCH194 validated the antifreeze proteins as one of the survival strategies to cope with the freezing climate of the Himalayan kettle lake. Bacterial AFP protect the cells from frost damage by inhibiting ice crystal growth during recrystallization. This study reports antifreeze activity in a Himalayan bacterium for the first time. Furthermore, antifreeze activity without cold acclimation indicates that the activity is constitutively inherited in PCH194 to cope with a frequent drop in daily temperature in the kettle lake.
Kettle lakes have frequent changes in physicochemical environments (Reverey et al., 2018); thus, nutritional fluctuation is inevitable. The presence of genes for chemotaxis and flagellar assembly in the bacterium suggested its ability to sense chemical and nutritional change and swim using flagella (Bren and Eisenbach, 2000). I. fluviatilis, I. limnosediminis, and Iodobacter sp. 7MAnt showed polar and lateral flagella (Logan, 1989; Su et al., 2013; Atalah et al., 2020). PHA synthesis to store carbon is a central strategy of many microbes in response to environmental and nutritional stress (Obruca et al., 2018; Kumar et al., 2020b). Iodobacter sp. PCH194 can synthesize and utilize PHA as a carbon reserve, evident from the genome analysis showing a complete set of genes for PHA synthesis (phaABC) and PHA depolymerization (phaZ). Accordingly, physiological experiments with various carbon/nitrogen ratios and environmental conditions revealed that the bacterium could synthesize PHB in the kettle lake environment. This has suggested the central role of PHB for Iodobacter sp. survival in the kettle lake. Apart from its role as a carbon reserve, PHA provides resistance to bacterial cells against multiple environmental stresses, including cold, freezing, oxidative, osmotic, and UV (Slaninova et al., 2018; Sedlacek et al., 2019; Obruca et al., 2020). Thus, PHA metabolism is one of the key metabolic strategies of Iodobacter sp. PCH194 to counter the multiple stresses in the Himalayan kettle lake.
Since the high-altitude Himalaya receives high amounts of UV irradiation (Singh and Singh, 2004), the inhabitant microflora must have adaptive strategies to deal with it. Consistently, Iodobacter sp. PCH194 possesses genes for the excinuclease system and deoxyribodipyrimidine photolyase to repair UV-induced DNA damage, as explained in the high-altitude bacterium Acinetobacter sp. Ver3 (Kurth et al., 2015). Additionally, PCH194 synthesizes violacein pigment as a UV protectant. The physiological experiment showed that violet-pigmented bacterium cells are more tolerant to UV (Figure 5D) than non-pigmented cells, demonstrating better survival upon adding methanol-extracted violacein pigment (Figure 5E). A literature survey supports the photoprotective role of violacein for Janthinobacterium spp. under UVB and UVC exposure (Abboud and Arment, 2013; Mojib et al., 2013). Iodobacter sp. PCH194 forms a violet microbial mat containing violacein at the surface of the medium. The phenomenon can be explained because violacein biosynthesis requires O2, which would be higher on the surface than in sediments. Apart from UV tolerance, violacein production and biofilm formation may be responsible for protecting the bacterium from other environmental stresses, similar to Janthinobacterium lividum DSM1522 (Pantanella et al., 2007). Violacein is also known as a defense against bacterivorous nanoflagellates and other planktonic community members (Matz et al., 2004; Deines et al., 2009; Batista et al., 2017) and has a strong antibacterial effect against gram-positive bacteria (Kumar et al., 2021). Violacein synthesis also served as a mechanism of interspecies interaction in Chromobacterium violaceum (Lozano et al., 2020). Thus, violacein production and biofilm formation constitute essential survival strategies of Himalayan Iodobacter spp. against abiotic and biotic stresses in its high-altitude lake habitat.
Many genomic traits of Iodobacter sp. PCH194 (Table 2) that were not experimentally validated might play important roles in survival under different environmental stresses. For instance, Iodobacter sp. PCH194 possesses genes encoding high-affinity terminal oxidases, suggesting its ability to harvest O2 at low concentrations for respiration (Morris and Schmidt, 2013). Genes encoding for periplasmic nitrate reductases, fumarate reductases, and transcriptional regulator (FNR) suggested that Himalayan Iodobacter can use nitrate or fumarate as a terminal electron acceptor under complete or near anaerobic conditions (Stewart et al., 2002). FNR is reported to act as an oxygen sensor and a molecular switch between aerobic and anaerobic respiration (Reinhart et al., 2008). Thus, Iodobacter has the genetic machinery for living under low oxygen concentrations and adapting to oxic-anoxic fluctuations at the sediment–water interface of the kettle lake. The various efflux pumps, porter and antiporters, and enzymes for amino acid-dependent acid tolerance might provide a survival advantage under acidic conditions (Guan and Liu, 2020). Furthermore, the toxin-antitoxin (TA) system encoded in this bacterium's chromosomal and plasmid DNA might play a crucial role in cell persistence, biofilm formation, tolerance to antibiotics, and other environmental stresses, as observed in many microbes (Page and Peti, 2016). The presence of plasmids and genes for conjugal transfer indicates the exchange of genomic information. Studies have demonstrated plasmids' adaptive, defensive, and metabolic roles in psychrophilic and psychrotolerant bacteria (Dziewit and Bartosik, 2014; Ciok et al., 2018).
Conclusion
The high-altitude Himalaya imposes numerous stresses on its bacterial communities. Therefore, it is vital to understand bacterial adaptation to the extreme environments of the Himalayas. The bacterium Iodobacter sp. PCH194 possesses multiple genomic and metabolic traits, which might enable its survival under cold and freezing temperatures, nutrient fluctuations, and high UV irradiation (Figure 6). Survival at subzero temperatures was further supported by antifreeze activity of the cell extract and a differential proteome response to cold and freezing conditions. PHB synthesis under different physicochemical conditions supports adaptation to fluctuating substrate and nutrient availability. Violacein pigment protects against UV radiation. The genomic and metabolic traits found in Iodobacter sp. PCH194 explain its survival under challenging high-altitude conditions and provide opportunities for exploring biotechnological applications and microbial community interactions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DS conceived the study. VK, VT, and DS did the field trip and sampling. VK, PK, and DS designed the study and analyzed the data and wrote and finalized the manuscript. VK performed microbial, genomic, and physiological experiments. PK performed proteomic and AFP experiments. VT and SuK assisted in genomic and physiological experiments, respectively. SaK did scientific discussion and research support. All authors reviewed and approved the final manuscript.
Funding
The study was funded by Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India under the scheme of National Post-Doctoral Fellowship by Grant Nos. PDF/2016/000508 and PDF/2016/003805, Scheme for Young Scientist and Technologist (SYST) Grant No. SP/YO/2019/1261. The study was generously funded to DS by the Council of Scientific and Industrial Research (CSIR), New Delhi, India with a Grant No. MLP0143 under the Niche Creating Project scheme.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors duly acknowledge the technical assistance provided by Mr. Mohit Swarnkar for whole-genome sequencing, Dr. Vishal Acharya for genome assembly, Dr. Robin Joshi for MALDI-TOF-TOF, and UHPLC-Q-TOF-IMS analysis, Dr. Rimpy Dhiman for ultramicrotomy, and Dr. Avnesh Kumari for TEM analysis. In addition, the authors duly acknowledge handling editor Andreas Teske and reviewers for their critical comments and suggestions on the manuscript. The manuscript represents the CSIR-IHBT communication number 4840.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.881873/full#supplementary-material
References
Abboud, A. N., and Arment, A. (2013). The protective effects of the violacein pigment against UV-C irradiation in Chromobacterium violaceum. Ohio J. Sci. 111, 28–32. Available online at: http://hdl.handle.net/1811/53721
Atalah, J., Blamey, L., Munoz-Ibacache, S., Gutierrez, F., Urzua, M., Encinas, M. N., et al. (2020). Isolation and characterization of violacein from an Antarctic Iodobacter: a non-pathogenic psychrotolerant microorganism. Extremophiles 24, 43–52. doi: 10.1007/s00792-019-01111-w
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edward, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Baraúna, R. A., Freitas, D. Y., Pinheiro, J. C., Folador, A. R., and Silva, A. (2017). A proteomic perspective on the bacterial adaptation to cold: integrating OMICs data of the psychrotrophic bacterium Exiguobacterium antarcticum B7. Proteomes 5, 9. doi: 10.3390/proteomes5010009
Barria, C., Malecki, M., and Arraiano, C. M. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. doi: 10.1099/mic.0.052209-0
Basineni, S. R., Madhugiri, R., Kolmsee, T., Hengge, R., and Klug, G. (2009). The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol. 6, 584–594. doi: 10.4161/rna.6.5.10082
Batista, A. H. M., Moreira, A. C. D., de Carvalho, R. M., Sales, G. W. P., Nogueira, P. C. N., Grangeiro, T. B., et al. (2017). Antimicrobial effects of violacein against planktonic cells and biofilms of Staphylococcus aureus. Molecules 22, 1534. doi: 10.3390/molecules22101534
Bolch, T., Kulkarni, A., Kääb, A., Huggel, C., Paul, F., Cogley, J. G., et al. (2012). The state and fate of Himalayan glaciers. Science 336, 310–314. doi: 10.1126/science.1215828
Bren, A., and Eisenbach, M. (2000). How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182, 6865–6873. doi: 10.1128/JB.182.24.6865-6873.2000
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Ciok, A., Budzik, K., Zdanowski, M. K., Gawor, J., Grzesiak, J., Decewicz, P., et al. (2018). Plasmids of psychrotolerant Polaromonas spp. isolated from Arctic and Antarctic glaciers–diversity and role in adaptation to polar environments. Front. Microbiol. 9, 1285. doi: 10.3389/fmicb.2018.01285
Corti, G., Cocco, S., Basili, M., Cioci, C., Warburton, J., and Agnelli, A. (2012). Soil formation in kettle holes from high altitudes in central Apennines, Italy. Geoderma 170, 280–294. doi: 10.1016/j.geoderma.2011.10.016
De Maayer, P., Anderson, D., Cary, C., and Cowan, D. A. (2014). Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 15, 508–517. doi: 10.1002/embr.201338170
Deines, P., Matz, C., and Jürgens, K. (2009). Toxicity of violacein-producing bacteria fed to bacterivorous freshwater plankton. Limnol. Oceanogr. 54, 1343–1352. doi: 10.4319/lo.2009.54.4.1343
Dhakar, K., and Pandey, A. (2020). Microbial ecology from the Himalayan cryosphere perspective. Microorganisms 8, 257. doi: 10.3390/microorganisms8020257
Dziewit, L., and Bartosik, D. (2014). Plasmids of psychrophilic and psychrotolerant bacteria and their role in adaptation to cold environments. Front. Microbiol. 5, 596. doi: 10.3389/fmicb.2014.00596
Gilbert, J. A., Davies, P. L., and Laybourn-Parry, J. (2005). A hyperactive. Ca2+ dependent antifreeze protein in an Antarctic bacterium. FEMS Microbiol. Lett. 245, 67–72. doi: 10.1016/j.femsle.2005.02.022
Goordial, J., Raymond-Bouchard, I., Zolotarov, Y., de Bethencourt, L., Ronholm, J., Shapiro, N., et al. (2016). Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol. Ecol. 92, 154. doi: 10.1093/femsec/fiv154
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Guan, N., and Liu, L. (2020). Microbial response to acid stress: mechanisms and applications. Appl. Microbiol. Biotechnol. 104, 51–65. doi: 10.1007/s00253-019-10226-1
Guo, S., Stevens, C. A., Vance, T. D. R., Olijve, L. L. C., Graham, L. A., Campbell, R. L., et al. (2017). Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci. Adv. 3, e1701440. doi: 10.1126/sciadv.1701440
Gupta, R., and Deswal, R. (2012). Low temperature stress modulated secretome analysis and purification of antifreeze protein from Hippophae rhamnoides, a Himalayan wonder plant. Proteome Res. 11, 2684–2696. doi: 10.1021/pr200944z
Han, W., and Christen, P. (2004). cis-Effect of DnaJ on DnaK in ternary complexes with chimeric DnaK/DnaJ-binding peptides. FEBS Lett. 563, 146–150. doi: 10.1016/S0014-5793(04)00290-X
Hennessy, F., Nicoll, W. S., Zimmermann, R., Cheetham, M. E., and Blatch, G. L. (2005). Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 14, 1697–1709. doi: 10.1110/ps.051406805
Hotaling, S., Hood, E., and Hamilton, T. L. (2017). Microbial ecology of mountain glacier ecosystems: biodiversity, ecological connections and implications of a warming climate. Environ. Microbiol. 19, 2935–2948. doi: 10.1111/1462-2920.13766
Kashyap, P., Kumar, S., and Singh, D. (2020). Performance of antifreeze protein HrCHI4 from Hippophae rhamnoides in improving the structure and freshness of green beans upon cryopreservation. Food Chem. 320, 126599. doi: 10.1016/j.foodchem.2020.126599
Knight, C. A., and Duman, J. G. (1986). Inhibition of the recrystallization of ice by insect thermal hysteresis proteins: a possible cryoprotective role. Cryobiology 23, 256–262. doi: 10.1016/0011-2240(86)90051-9
Koh, H. Y., Park, H., Lee, J. H., Han, S. J., Sohn, Y. C., and Lee, S. G. (2017). Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ. Microbiol. 19, 628–644. doi: 10.1111/1462-2920.13578
Kumar, V., Darnal, S., Kumar, S., Kumar, S., and Singh, D. (2021). Bioprocess for co-production of polyhydroxybutyrate and violacein using Himalayan bacterium Iodobacter sp. PCH194. Bioresour. Technol. 319, 124235. doi: 10.1016/j.biortech.2020.124235
Kumar, V., Kumar, S., Darnal, S., Patial, V., Singh, A., Thakur, V., et al. (2019). Optimized chromogenic dyes-based identification and quantitative evaluation of bacterial l-asparaginase with low/no glutaminase activity bioprospected from pristine niches in Indian trans-Himalaya. 3 Biotech 9, 275. doi: 10.1007/s13205-019-1810-9
Kumar, V., Kumar, S., and Singh, D. (2020b). Microbial polyhydroxyalkanoates from extreme niches: bioprospection status, opportunities and challenges. Int. J. Biol. Macromol. 147, 1255–1267. doi: 10.1016/j.ijbiomac.2019.09.253
Kumar, V., Kumar, S., and Singh, D. (2022). Metagenomic insights into Himalayan glacial and kettle lake sediments revealed microbial community structure, function, and stress adaptation strategies. Extremophiles 26, 3. doi: 10.1007/s00792-021-01252-x
Kumar, V., Thakur, V., Ambika, K. S., and Singh, D. (2018). Bioplastic reservoir of diverse bacterial communities revealed along altitude gradient of Pangi-Chamba trans-Himalayan region. FEMS Microbiol. Lett. 365, fny144. doi: 10.1093/femsle/fny144
Kumar, V., Thakur, V., Ambika, K. V., Kumar, R., and Singh, S. (2020a). Genomic insights revealed physiological diversity and industrial potential for Glaciimonas sp. PCH181 isolated from Satrundi glacier in Pangi-Chamba Himalaya. Genomics 112, 637–646. doi: 10.1016/j.ygeno.2019.04.016
Kurth, D., Belfiore, C., Gorriti, M. F., Cortez, N., Farias, M. E., and Albarracín, V. H. (2015). Genomic and proteomic evidences unravel the UV-resistome of the poly-extremophile Acinetobacter sp. Ver3. Front. Microbiol. 6, 328. doi: 10.3389/fmicb.2015.00328
Leblanc, C., Sarcos, A. C., Comeau, A. M., and Krisch, H. M. (2009). Isolation and genomic characterization of the first phage infecting Iodobacteria: φPLPE, a myovirus having a novel set of features. Environ. Microbiol. Rep. 1, 499–509. doi: 10.1111/j.1758-2229.2009.00055.x
Logan, N. A. (1989). Numerical taxonomy of violet-pigmented, Gram-negative bacteria and description of Iodobacter fluviatile gen. nov., comb. nov. Int. J. Syst. Bacteriol. 39, 450–456. doi: 10.1099/00207713-39-4-450
Lozano, G. L., Guan, C., Cao, Y., Borlee, B. R., Broderick, N. A., Stabb, E. V., et al. (2020). A chemical counterpunch: Chromobacterium violaceum ATCC31532 produces violacein in response to translation-inhibiting antibiotics. mBio 11, e00948–e00920. doi: 10.1128/mBio.00948-20
Matz, C., Deines, P., Boenigk, J., Arndt, H., Eberl, L., Kjelleberg, S., et al. (2004). Impact of violacein-producing bacteria on survival and feeding of bacterivorous nanoflagellates. Appl. Environ. Microbiol. 70, 1593–1599. doi: 10.1128/AEM.70.3.1593-1599.2004
Maurer, M., Schaefer, J. M., Rupper, S., and Corley, A. (2019). Acceleration of ice loss across the Himalayas over the past 40 years. Sci. Adv. 5, 7266. doi: 10.1126/sciadv.aav7266
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14, 60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182. doi: 10.1038/s41467-019-10210-3
Mojib, N., Farhoomand, A., Andersen, D. T., and Bej, A. K. (2013). UV and cold tolerance of a pigment-producing Antarctic Janthinobacterium sp. Ant5-2. Extremophiles 17, 367–378. doi: 10.1007/s00792-013-0525-9
Morris, R., and Schmidt, T. (2013). Shallow breathing: bacterial life at low O2. Nat. Rev. Microbiol. 11, 205–212. doi: 10.1038/nrmicro2970
Mykytczuk, N., Foote, S., Omelon, C., Southam, G., Greer, C. W., and Whyte, L. G. (2013). Bacterial growth at−15 °C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 7, 1211–1226. doi: 10.1038/ismej.2013.8
Obruca, S., Sedlacek, P., Koller, M., Kucera, D., and Pernicova, I. (2018). Involvement of polyhydroxyalkanoates in stress resistance of microbial cells: biotechnological consequences and applications. Biotechnol. Adv. 36, 856–870. doi: 10.1016/j.biotechadv.2017.12.006
Obruca, S., Sedlacek, P., Slaninova, E., Fritz, I., Daffert, C., Meixner, K., et al. (2020). Novel unexpected functions of PHA granules. Appl. Microbiol. Biotechnol. 104, 4795–4810. doi: 10.1007/s00253-020-10568-1
Page, R., and Peti, W. (2016). Toxin–antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Pantanella, F., Berlutti, F., Passariello, C., Sarli, S., Morea, C., and Schippa, S. (2007). Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 102, 992–999. doi: 10.1111/j.1365-2672.2006.03155.x
Polissi, A., De Laurentis, W., Zangrossi, S., Briani, F., Longhi, V., Pesole, G., et al. (2003). Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res. Microbiol. 154, 573–580. doi: 10.1016/S0923-2508(03)00167-0
Prud'homme-Genereux, A., Beran, R. K., Iost, I., Ramey, C. S., Mackie, G. A., and Simons, R. W. (2004). Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome'. Mol. Microbiol. 54, 1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. PNAS 104, 15508–15513. doi: 10.1073/pnas.0706532104
Reinhart, F., Achebach, S., Koch, T., and Unden, G. (2008). Reduced apo-fumarate nitrate reductase regulator (ApoFNR) as the major form of FNR in aerobically growing Escherichia coli. J. Bacteriol. 190, 879–886. doi: 10.1128/JB.01374-07
Reverey, F., Ganzert, L., Lischeid, G., Ulrich, A., Premke, K., and Grossart, H. P. (2018). Dry-wet cycles of kettle hole sediments leave a microbial and biogeochemical legacy. Sci. Total Environ. 627, 985–996. doi: 10.1016/j.scitotenv.2018.01.220
Reverey, F., Grossart, H. P., Premke, K., and Lischeid, G. (2016). Carbon and nutrient cycling in kettle hole sediments depending on hydrological dynamics: a review. Hydrobiologia 775, 1–20. doi: 10.1007/s10750-016-2715-9
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Sandoval, A. E., Arias-Barrau, M., Arcos, G., Naharro, G., Olivera, E. R., and Luengo, J. M. (2007). Genetic and ultrastructural analysis of different mutants of Pseudomonas putida affected in the poly-3-hydroxy-n-alkanoate gene cluster. Environ. Microbiol. 9, 737–751. doi: 10.1111/j.1462-2920.2006.01196.x
Sedlacek, P., Slaninova, E., Koller, M., Nebesarova, J., Marova, I., Krzyzanek, V., et al. (2019). PHA granules help bacterial cells to preserve cell integrity when exposed to sudden osmotic imbalances. New Biotechnol. 49, 129–136. doi: 10.1016/j.nbt.2018.10.005
Sharma, S., Ray, S., Moiyadi, A., Sridhar, E., and Srivastava, S. (2014). Quantitative proteomic analysis of meningiomas for the identification of surrogate protein markers. Sci. Rep. 4, 7140. doi: 10.1038/srep07140
Singh, S., and Singh, R. (2004). High-altitude clear-sky direct solar ultraviolet irradiance at Leh and Henle in the western Himalayas: observations and model calculations. J. Geophys. Res. 109, D19201. doi: 10.1029/2004JD004854
Slaninova, E., Sedlacek, P., Mravec, F., Mullerova, L., Samek, O., Koller, M., et al. (2018). Light scattering on PHA granules protects bacterial cells against the harmful effects of UV radiation. Appl. Microbiol. Biotechnol. 102, 1923–1931. doi: 10.1007/s00253-018-8760-8
Stewart, V., Lu, Y., and Darwin, A. J. (2002). Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184, 1314–1323. doi: 10.1128/JB.184.5.1314-1323.2002
Stres, B., Sul, W. J., Murovec, B., and Tiedje, J. M. (2013). Recently deglaciated high-altitude soils of the Himalaya: Diverse environments, heterogenous bacterial communities and long-range dust inputs from the upper troposphere. PLoS ONE 8, e76440. doi: 10.1371/journal.pone.0076440
Su, W., Zhou, Z., Jiang, F., Chang, X. L., Liu, Y., Wang, S., et al. (2013). Iodobacter limnosediminis sp. nov., isolated from Arctic lake sediment. Int. J. Syst. Evol. Microbiol. 63, 1464–1470. doi: 10.1099/ijs.0.039982-0
Tatusova, T. M., DiCuccio, A., Badretdin, V., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acid Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thakur, V., Kumar, V., Kumar, S., and Singh, D. (2018). Diverse culturable bacterial communities with cellulolytic potential revealed from pristine habitat in Indian trans-Himalaya. Can. J. Microbiol. 28, 1–11. doi: 10.1139/cjm-2017-0754
Keywords: high-altitude Himalaya, bacterial adaptation, genomic traits, proteomic response, antifreeze proteins, polyhydroxybutyrate
Citation: Kumar V, Kashyap P, Kumar S, Thakur V, Kumar S and Singh D (2022) Multiple Adaptive Strategies of Himalayan Iodobacter sp. PCH194 to High-Altitude Stresses. Front. Microbiol. 13:881873. doi: 10.3389/fmicb.2022.881873
Received: 23 February 2022; Accepted: 01 June 2022;
Published: 06 July 2022.
Edited by:
Andreas Teske, University of North Carolina at Chapel Hill, United StatesReviewed by:
Grégoire Michoud, Swiss Federal Institute of Technology Lausanne, SwitzerlandSalma Mukhtar, Connecticut Agricultural Experiment Station, United States
Copyright © 2022 Kumar, Kashyap, Kumar, Thakur, Kumar and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dharam Singh, ZGhhcmFtc2luZ2hAaWhidC5yZXMuaW4=; ZGhhcmFtczE0QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Vijay Kumar
Vijay Kumar Prakriti Kashyap
Prakriti Kashyap Subhash Kumar
Subhash Kumar Vikas Thakur
Vikas Thakur Sanjay Kumar
Sanjay Kumar Dharam Singh
Dharam Singh