- 1Department of Environmental Health Science, Konkuk University, Seoul, South Korea
- 2School of Biological Sciences and Technology, Chonnam National University, Gwangju, South Korea
- 3Division of Environmental and Forest Science, Gyeongsang National University, Jinju, South Korea
Bacterial cell-based biosensors have been widely developed for detecting environmental toxic materials. The znt-operon in Escherichia coli is a Zn(II)-responsive genetic system and is employed in Zn(II), Cd(II), and Hg(II)-sensing biosensors. In this study, point mutations were introduced in the regulatory protein ZntR to modulate its target selectivity, and metal ion-exporting genes, such as copA and zntA, in host cells were deleted to increase cellular metal ion levels and enhance specificity. Thus, the overall responses of the E. coli cell-based biosensors toward metal(loid) ions were increased, and their selectivity, which was originally for Cd(II) and Hg(II), was shifted to Pb(II). The gene encoding ZntA, known as the Zn(II)-translocating P-type ATPase, showed an impact on the ability of E. coli to export Pb(II), whereas copA deletion showed no significant impact. Noteworthily, the newly generated biosensors employing ZntR Cys115Ile showed the capacity to detect under 5 nM Pb(II) in solution, without response to other tested metal ions within 0–100 nM. To understand the marked effect of single point mutations on ZntR, computational modeling was employed. Although it did not provide clear answers, changes in the sequences of the metal-binding loops of ZntR modulated its transcriptional strength and target selectivity. In summary, the approaches proposed in this study can be valuable to generate new target-sensing biosensors with superior selectivity and specificity, which can in turn broaden the applicability of cell-based biosensors to monitor Pb(II) in environmental systems.
Highlights
- The regulatory protein ZntR was mutated to modulate the target selectivity.
- Metal ion-exporting genes were deleted in host cells to improve specificity.
- The combination of these two elements led to enhanced detection potential toward Pb(II).
Introduction
Lead (Pb) is a heavy metal widely used in diverse industries, including printing, construction, automobile manufacturing, rechargeable batteries, and others. Along with its widespread use, Pb has been released and is accumulated in environmental systems and is considered a risk for human health (Hernberg, 2000; Gould, 2009). It had been also reported that lead induces toxic stress in plants, resulting in the inhibition of seed germination and growth (Shu et al., 2012; Hadi and Aziz, 2015). Although leaded petrol and coal combustion have been the main contamination sources, it is also important to consider the effects of other potential contamination sources. Recently, shifts toward sustainable and green energy sources have occurred globally to protect the environment. In this regard, many companies are currently pursuing carbon neutrality, actively investing efforts into developing rechargeable batteries, which can be widely used in portable electronics and to replace internal combustion engines, especially, in vehicles. However, the massive production and usages of these batteries may also be potential sources of environmental pollutants such as diverse heavy metal(loid)s, including lithium, lead, and cobalt, among others (Nnorom and Osibanjo, 2009; Palacin, 2009; Yang et al., 2010; Placke et al., 2017). Thus, it is necessary to have tools for monitoring heavy metals from the wastes of rechargeable batteries and to recycle them.
Heavy metals in environmental systems, such as water, soil, and air, are typically analyzed by analytical instruments, including inductively coupled plasma-atomic spectroscopy and mass spectroscopy. Instrumental analysis shows high sensitivity and accuracy, but it can be time-consuming and expensive. Therefore, diverse metal sensing tools have been reported based on different techniques as potential alternatives to instrumental analysis approaches. Among these new techniques, bacterial cell-based biosensors have been actively investigated to quantify heavy metals (Magrisso et al., 2008; Kim et al., 2020a; Kannappan and Ramisetty, 2021). Importantly, bacterial cell-based biosensors are inexpensive and simple tools for fast detection of heavy metals, thereby overcoming the limitations of instrument-based analysis methods. Therefore, several target-detecting biosensors have been developed to monitor hazardous materials, including heavy metals, antibiotics, and chemicals, in diverse environmental systems (Elcin et al., 2021; Kannappan and Ramisetty, 2021; Shpigel et al., 2021; Zeng et al., 2021).
Recently, the targets of bacterial cell-based biosensors have been diversified upon findings of new genetic systems that respond to toxic materials, including formaldehyde, explosives, petroleum hydrocarbon, and nitrification inhibitors (Hynninen and Virta, 2009; Elcin et al., 2021; Henshke et al., 2021; Zappi et al., 2021). Owing to their simplicity and applicability, bacterial cell-based biosensors are considered alternative tools to monitor and assess risks for environmental pollutants. However, the number of available genetic systems is relatively limited compared with the number of pollutants resulting from industrial developments. Moreover, most genetic systems show a broad target specificity owing to the nature of living organisms, which may be disadvantageous. Hence, many researchers have made efforts to minimize the interference caused by nontarget materials. Noteworthily, enhancement of target specificity and selectivity was achieved by genetic engineering of regulatory proteins and host bacterial cells, rearrangement of genetic circuits, or by improving the detection techniques used (Nivens et al., 2004; Hynninen et al., 2010; Van Der Meer and Belkin, 2010; Changye et al., 2021).
In this study, we presented a strategy to generate target-specific biosensors using existing genetic systems. Briefly, by engineering the regulatory protein ZntR and deleting the metal ion-translocating P-type ATPases in host cells, a Pb(II)-specific biosensor was derived from Cd(II) sensing biosensors employing znt-operon as a genetic system as an alternative to the pbr-operon, which had been the only genetic system reported for Pb(II) detection (Rensing et al., 1998; Taghavi et al., 2009; Wei et al., 2014). Furthermore, we also demonstrated that by mutating ZntR, it is possible to achieve superior selectivity and specificity toward Pb(II) from the structural point of view. Hence, these novel strategies to generate biosensors may expand the targets for biosensors and thereby contribute to environmental monitoring and risk assessment of diverse pollutants.
Materials and Methods
Materials and Escherichia coli Strains
All heavy metal(loid)s used (e.g., AsCl3, CdCl2, CrSO4, HgCl2, PbSO4, MnCl2, AuCl, and ZnCl2) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Novagen pCDF-Duet and pET-21(a) vectors (Sigma-Aldrich) were used to carry the sensing domain, zntR and mutants, and the reporter domain, zntA promoter fused with egfp, respectively (Yoon et al., 2016). PfuTurbo DNA polymerase (Agilent, Santa Clara, CA, USA) was used for site-directed mutagenesis of zntR, and all restriction enzymes and T4 DNA ligase were purchased from Takara Korea Biomedical (Seoul, Korea). E. coli DH5α and BL21(DE3) strains were used for plasmid constructions and as host cells of the biosensors, respectively. E. coli strains, BL21-copA/zntR and BL21-zntA/zntR used in previous studies (Kang et al., 2018a; Kim et al., 2020b) and BL21-copA/zntA/zntR, newly generated in this study, were used as host cells for the biosensors. Endogenous gene deletion was performed using a Quick and Easy E. coli Gene Deletion Kit (Gene Bridges, Heidelberg, Germany). All strains used in the study are listed in Online Resource 1 with a description of their genetic properties.
Endogenous Gene Deletion in E. coli BL21
Triple-gene-deficient E. coli was generated by deleting copA from BL21-zntA/zntR using a Quick & Easy E. coli Gene Deletion Kit. Briefly, the FRT-flanked PGK-gb2-neo cassette and copA sequences were amplified by a polymerase chain reaction and electroporated into E. coli BL21-zntA/zntR cells harboring the pRedET plasmid, which has the recombinase-encoding gene. Electroporation was performed with an Electroporator 2510 (Eppendorf, Hamburg, Germany) at 1,350 V, 10 μF, and 600 Ω (Kang et al., 2018b). The cells were then induced by adding 10% of arabinose to replace copA with the kanamycin resistance gene (kan). Gene deletion was further verified by polymerase chain reaction.
Engineering of ZntR
Engineered ZntRs were generated by site-directed mutagenesis of the metal-binding loop (MBL) region, consisting of 12 amino acids. The nucleotide sequences for the amino acid residues surrounding the MBL were mutated to encode specific amino acids using designed primers, which was confirmed by DNA sequencing. The mutation targets were selected upon analysis of the three-dimensional structure of the ZntR. The engineered ZntRs used (Table 1) were tested as sensing domains for metal ion detection in E. coli BL21-zntR, BL21-copA/zntR, BL21-zntA/zntR, and BL21-copA/zntA/zntR. The primers for gene cloning and point mutations are listed in Online Resource 2.
Biosensor Assay
Escherichia coli cell-based biosensors were generated by introducing a pair of plasmids carrying zntAp::egfp and engineered zntR in genetically modified E. coli BL21 strains, including E. coli-zntR, E. coli-zntR/copA, E. coli-zntR/zntA, and E. coli-zntR/copA/zntA. The biosensor assay was conducted as previously described with some modifications (Kang et al., 2018b; Yoon et al., 2018). Briefly, the biosensor cells were grown at 37°C in a shaking incubator overnight, and 1 ml of the cultures was then inoculated in 50 ml of fresh Luria broth until it reached an OD600 value of 0.10. Next, the cells were placed in test tubes and directly exposed to metal ions. After 2 h of incubation, the E. coli cells were harvested, and the cell pellet was resuspended in Tris buffer (50 mM Tris-HCl, 160 mM KCl, pH 7.4) before measuring fluorescence intensity.
Fluorescence Intensity Analysis
The intensity of the fluorescence signals generated by the biosensors after heavy metal exposure was measured using fluorescence spectroscopy (FC-2; Scinco, Seoul, Korea). The expression of the enhanced green fluorescent protein (eGFP) was measured at 480 and 510 nm for excitation and emission wavelength, respectively. The fluorescence signals induced by heavy metal exposure were represented as the induction coefficient values defined as (fluorescence intensity of metal-exposed biosensors)/(fluorescence intensity of non-exposed biosensors). The detection limit (DL) for biosensor assay was calculated by the following: DL = 3 s/m (s: standard deviation obtained from multiplicated measurements; m: slope of the standard curve).
Structural Analysis
The crystal X-ray structure of ZntR was obtained from the Protein Data Bank (PDB ID:1Q08) and modified amino acids by PyMol (Schrödinger, New York, NY, USA). Sybyl 7.3 (Tripos, St. Louis, MO, USA) was used for energy minimization processing. In the protein structures, water molecules and phosphate ions were removed, and hydrogens were added. After modification, they were minimized using the Tripos force field. The energy minimization process was terminated at the convergence criteria of the total energy (0.05 kcal/molÅ). Chimera 1.11 software (https://www.cgl.ucsf.edu/chimera/download.html) was used for protein visualization and analysis.
Data Analysis
R v4.1.0 (https://cran.r-project.org) and DescTools package were used for the statistical analysis of all data when the minimum difference between the control and treatment groups was statistically significant (Team, 2014; Signorell et al., 2019).
Results
Effects of Metal-Translocating ATPases on Metal Selectivity of Biosensors
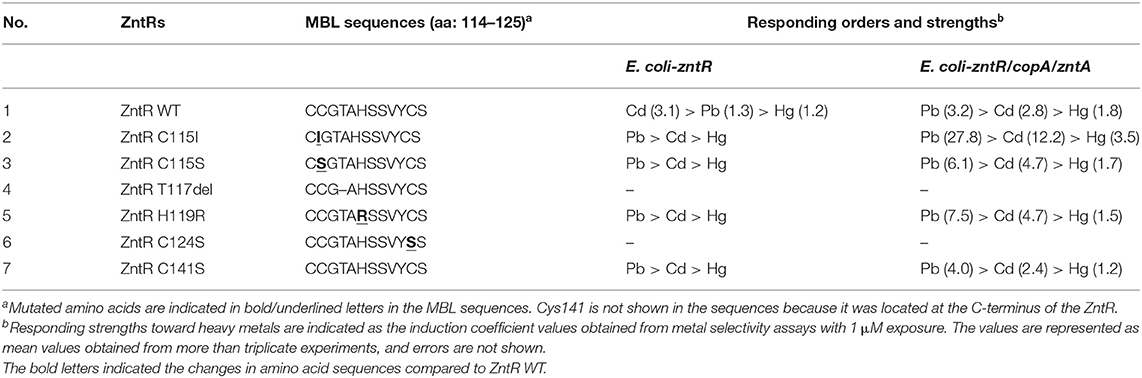
Since biosensors based on zntA-deficient strains showed strong fluorescence signals even without metal exposure, the experimental conditions used in this study for heavy metal selectivity were modified from those previously reported (Yoon et al., 2016; Kang et al., 2018a). The metal selectivity of the biosensors using E. coli-zntR, E. coli-zntR/zntA, and E. coli-zntR/copA/zntA with wild-type (WT) ZntR was tested under optimized experimental conditions. Since further incubation was required before exposition to metal ions, the responses indicated as induction coefficient values and background fluorescent signals were decreased. Various metal ion concentrations (0.5, 1.0, 5.0, and 10 μM) were tested; the induction coefficient values induced by 1 μM metal ions are shown in Figure 1. As reported previously, E. coli WT-based biosensors showed Cd(II)-specific response although the induction coefficient values were decreased (Figure 1A; Yoon et al., 2016). Hence, the optimized experimental conditions were reliable to test the detection potential of the biosensors. Interestingly, Pb(II) responses were enhanced by copA or zntA deletions, whereas Cd(II) responses were slightly decreased (Figures 1B,C). Moreover, the order of strength was inversed as Pb(II) > Cd(II). In turn, the Pb(II) responses were clearly enhanced, and the responding order to heavy metals was changed to Pb(II) > Cd(II) > Hg(II) when both copA and zntA were deleted (Figure 1D). In addition, it was necessary to mention that the responses toward Zn(II) were not significant. Although the znt-operon had been identified as a Zn(II)-responsive genetic system, the regulatory protein, ZntR, prefers Cd(II) and Hg(II) rather than Zn(II). It was consistent with our previous data (Yoon et al., 2018). Thus, deleting zntA in E. coli led to increased Pb(II) responses, although the induction coefficient values varied. The effects of copA deletion on metal responses were less significant than those obtained upon zntA deletion, which suggests that zntA plays more relevant roles in divalent metal homeostasis in E. coli cells. In fact, it was unclear how metal-translocating channels affect the metal selectivity of E. coli-based biosensors. Nevertheless, metal selectivity and sensitivity can be modulated by disrupting metal homeostasis in the host cells.
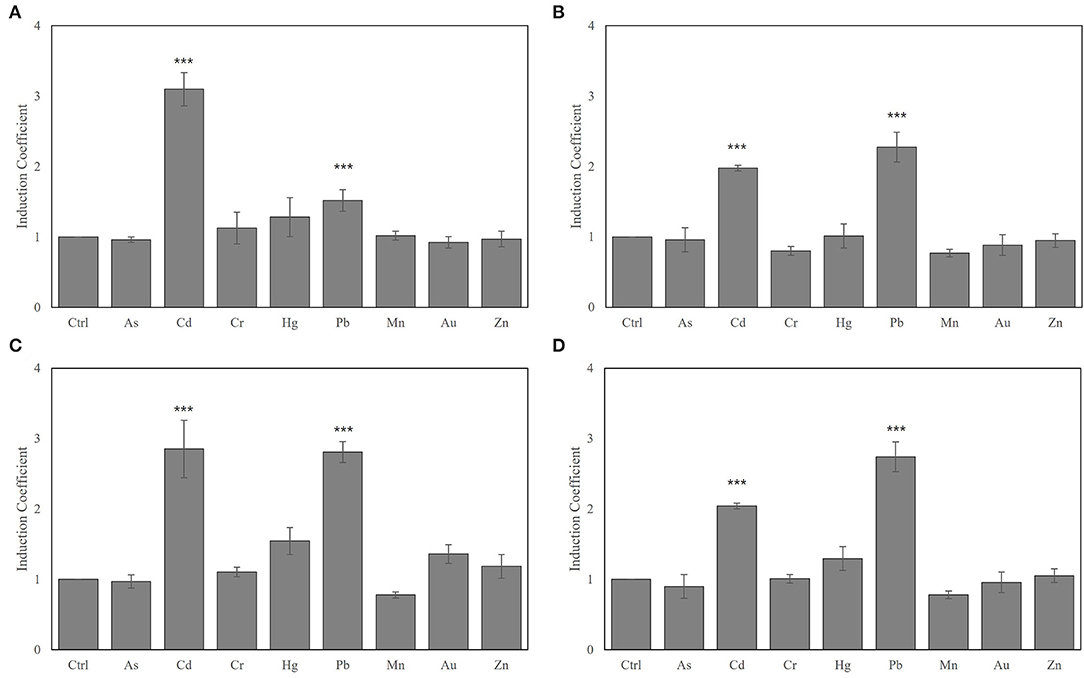
Figure 1. Metal selectivity of biosensors using (A) E. coli-zntR, (B) E. coli-zntR/copA, (C) E. coli-zntR/zntA, and (D) E. coli-zntR/copA/zntA. The biosensors were exposed to 1 μM of tested metal ions for 2 h, and the eGFP expression levels were represented as induction coefficient values. The values were obtained from more than three times of replicated experiments. The asterisk means the data were significantly higher than the control (Dunnett's test, ***p < 0.001).
Effects of Genetic Engineering on ZntRs
Since the E. coli biosensors employed the znt operon, the target selectivity and sensitivity were determined by using ZntR, a known regulatory protein of the znt operon (Brocklehurst et al., 1999). Thus, the amino acids in the MBL regions of ZntR were selected as targets for genetic engineering to modulate its metal selectivity, and the engineered ZntRs were introduced into E. coli-zntR/copA/zntA strains with zntAp::egfp to generate the enhanced biosensors (Table 1).
The biosensor cells were exposed to 1 μM of heavy metal ions, including As(III), Cd(II), Cr(II), Hg(II), Pb(II), Mn(II), Au(I), and Zn(II), and the fluorescence signals were measured after 2 h of exposure (Figure 2). Using ZntR WT as regulatory protein produced similar responses to Cd(II) and Pb(II), reaching ~3-fold induction coefficient values, and the same strength to Hg(II) and Au(I) as observed in the E. coli-zntR/copA/zntA strains (Figure 2A). Moreover, the orders of response strength to heavy metals were changed to Pb(II) > Cd(II) > Hg(II) in the systems engineered with mutated ZntR (Cys115Ile, Cys115Ser, His119Arg, and Cys141Ser) (Figure 2). Although it was unclear why the target selectivity of the biosensors shifted due to the point mutations, it was hypothesized that mutations on the metal-binding sites altered the ability of the cell to interact with the metal ions and induce conformational changes to selectively prefer Pb(II) over Cd(II). Since Cys115 and His119 directly mediate the conformational interaction with the metal ions, the selectivity of the engineered ZntRs was modulated by these amino acids. In addition, the Cys141 mutation, which is located in the C-terminal region of ZntR, also shifted the target selectivity from Cd(II) to Pb(II). Although this outcome was unexpected, the Cys141Ser mutation may induce conformational changes that affect metal selectivity. However, the biosensors employing ZntR Thr117 deletion and Cys124Ser lost their capacity to respond to the presence of the metal ions (Figures 2D,F).
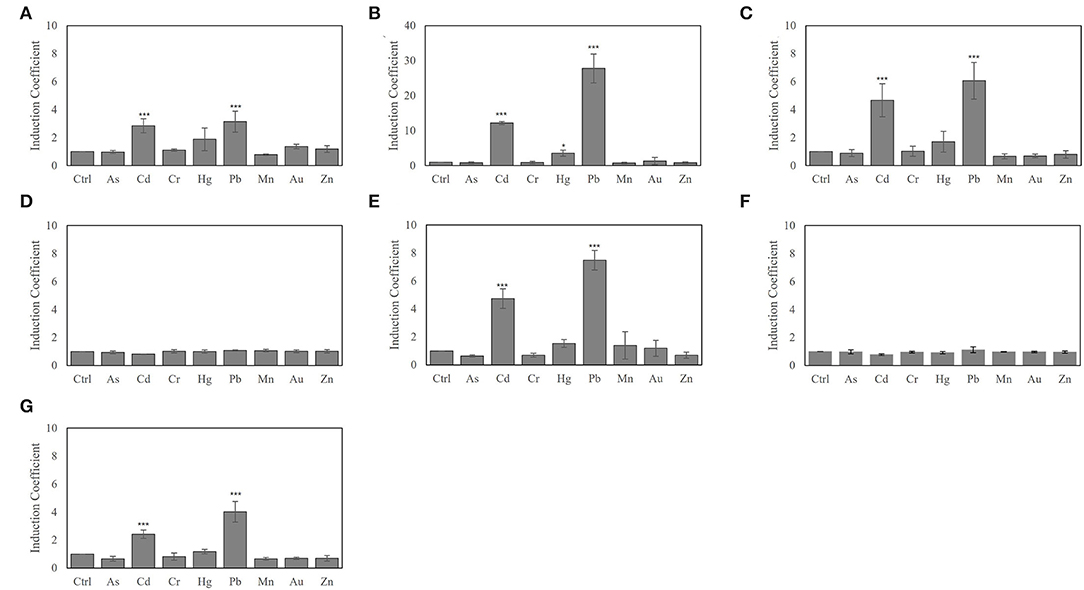
Figure 2. Modulating metal selectivity of biosensors based on E. coli-zntR/copA/zntA harboring pZnt-eGFP and ZntR WT (A), C115I (B), C115S (C), T117del (D), H119R (E), C124S (F), and C141S (G). The biosensors were exposed to 1 μM of tested metal ions for 2 h, and the eGFP expression levels were represented as induction coefficient values. The asterisk means the data were significantly higher than the control (Dunnett's test, ***p < 0.001, *p < 0.005).
Most interestingly, it was noticed that biosensors having ZntR Cys115Ile as a regulatory protein showed extremely strong responses to Pb(II) at induction coefficient values of over 35 (Figure 2B). Although it showed strong responses to Cd(II) and Hg(II) compared with ZntR WT, the response to Pb(II) was significant. In this regard, the E. coli-zntR/copA/zntA biosensors having ZntR C115I were investigated to further elucidate the potential as Pb(II)-specific biosensors and to understand the mechanisms of superior responses to Pb(II).
Heavy Metal Sensitivity of Biosensors With Engineered ZntRs
The biosensors generated with the engineered ZntRs and E. coli-zntR/copA/zntA showed modulated metal selectivity and responses to heavy metals (1 μM) upon 2 h of exposure. To elucidate the sensitivity to each heavy metal tested, the responses of the biosensors exposed to 0–10 μM of Pb(II), Cd(II), and Hg(II) were determined under the same experimental conditions (Figure 3). The biosensors employing ZntR WT, Cys115Ser, His119Arg, and Cys141Ser showed responses to all three tested metal ions in the order of induction coefficient values of Pb(II) ≈ Cd(II) > Hg(II). Noteworthy, the biosensor employing ZntR Cys115Ile showed superior signals toward Pb(II), Cd(II), and Hg(II); however, it had superior specificity toward Pb(II) over the other heavy metals at 0.1 μM and below (Figure 3). The induction coefficient value for Pb(II) was ~8, whereas it was less than 2 for Cd(II) and Hg(II). Thus, E. coli-zntR/copA/zntA biosensors with ZntR Cys115Ile as the regulatory protein may be Pb(II)-specific biosensors within certain concentration ranges.
Figure 3. The metal specificity of biosensors based on E. coli-zntR/copA/zntA harboring pZnt-eGFP and ZntRs toward lead (A) cadmium (B) and mercury (C). The biosensors were exposed to 0 to 10 μM ranges of metal ions and determined the induction coefficient values after 2 h exposures.
Characterization of Pb(II)-Specific Biosensor
Since the newly generated biosensor employing ZntR Cys115Ile showed responses toward Pb(II) only at 0.1 μM, the properties of the biosensors were further characterized. The responses of the biosensor to Pb(II) were determined at concentrations below 100 nM (Figure 4). Overall, the biosensor responded in a concentration-dependent manner, with its detection limit being calculated at ~5 nM Pb(II) and statistical significance being observed at 1 nM Pb(II) (Figure 4, inbox). Moreover, the relationship between the induction coefficient values and Pb(II) concentration showed a linear correlation, thereby allowing for quantitative analysis.
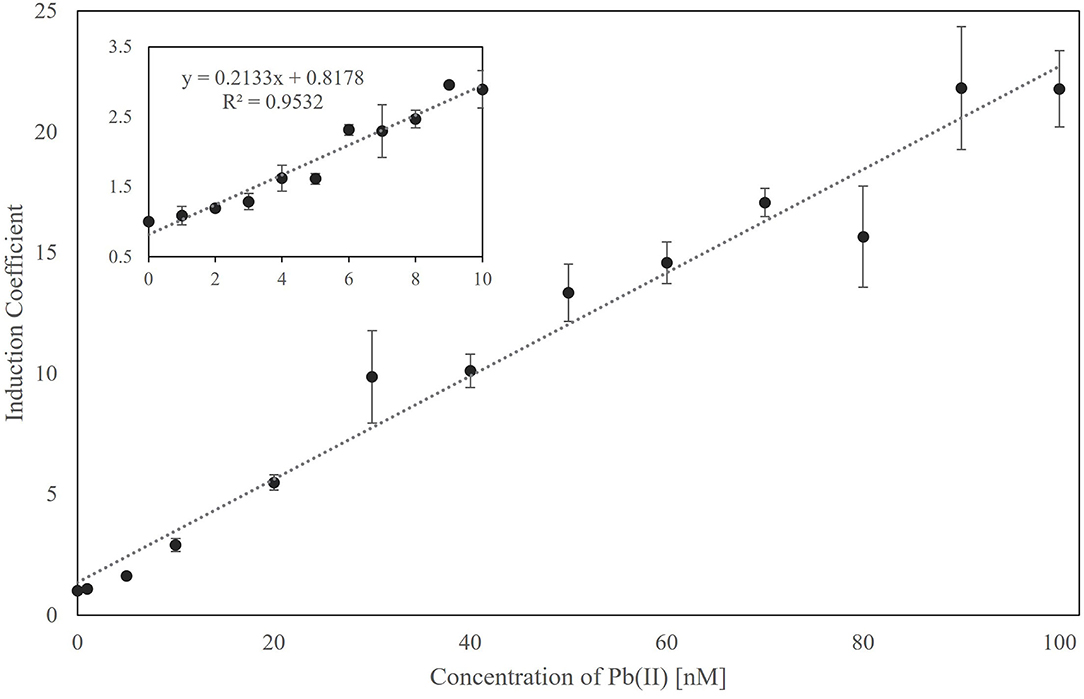
Figure 4. The relationship between the responses of biosensors employing ZntR-C115I as a regulatory protein and Pb(II) concentration ranged from 0 to 100 nM. The responses from 0 to 10 nM of Pb(II) were shown in the inbox, and the relationship was analyzed by a linear regression fit with 0.953 of R2 value.
Interference of the other heavy metals, such as Cd(II) and Hg(II), on Pb(II) detection was also investigated to elucidate the robustness of the biosensor to serve as a Pb(II) detector. The biosensors were exposed to 10 nM of Pb(II), Cd(II), and Hg(II) alone or in combination (Figure 5). Biosensors exposed to Cd(II) and Hg(II) showed 0.12- and 0.25-fold increased induction coefficient values, respectively, whereas Pb(II) increased the fluorescence signal by 2.2-fold. When exposed to a combination of heavy metal ions, such as Cd(II)/Pb(II) or Hg(II)/Pb(II), the biosensor showed 2.4-fold induction coefficient values, which were comparable to those obtained with Pb(II) alone. Although the signals were slightly increased by Cd(II) and Hg(II) presence, the new biosensor was shown to have superior Pb(II) selectivity and specificity at certain concentration ranges.
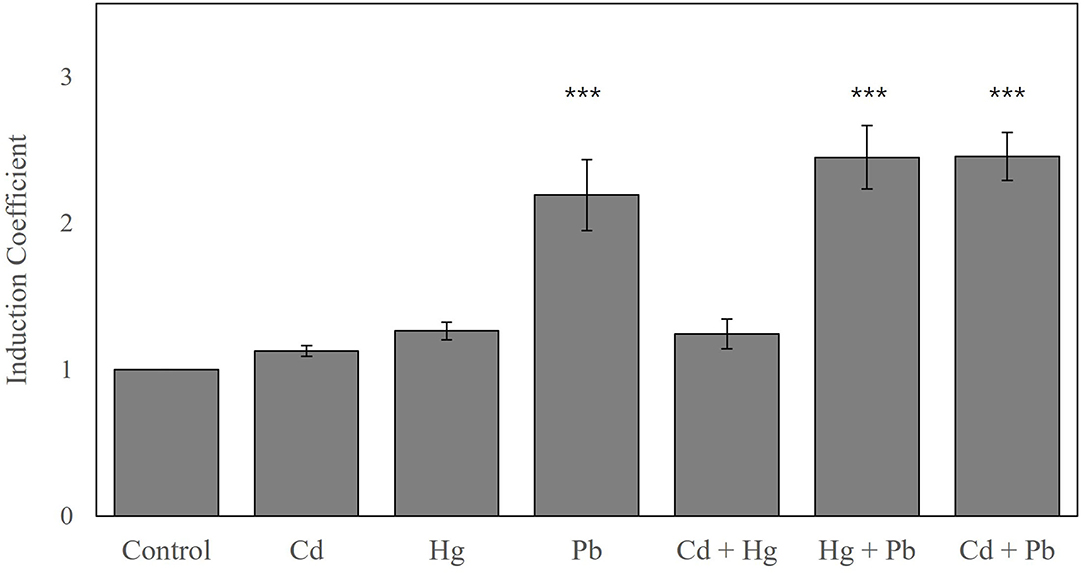
Figure 5. The comparative and combined exposures of biosensors E. coli-zntR/copA/zntA harboring pZnt-eGFP and ZntR C115I to Pb(II), Cd(II), and Hg(II). The biosensors were exposed to 10 nM of metal ions comparatively and combinedly, and the responses were represented as induction coefficient values. The asterisk means the data were significantly higher than the control (Dunnett's test, ***p < 0.001).
Structural Aspects of Engineered ZntRs
As described above, the engineered ZntR granted improved target selectivity and specificity to the biosensors while using E. coli-zntR/copA/zntA as host cells. To answer why and how the specificity and selectivity were modulated, it was necessary to analyze the fluorescence intensity, since the responses were determined as induction coefficient values. Hence, the fluorescence signals from Pb(II)-exposed and Pb(II)-non-exposed biosensors were screened (Figure 6). In the case of Thr117 deletion and Cys124Ser variant, the signals were similar regardless of the presence or absence of the metal ion, resulting in no increase of the induction coefficient value. Nonetheless, the other biosensors showed similar selectivity toward Pb(II), Cd(II), and Hg(II) although the induction coefficient values varied (Figure 2). As shown in Figure 6, the ZntR WT in E. coli-zntR/copA/zntA showed a strong signal even without Pb(II) exposure, which may be explained by the increased level of endogenous metal ions due to the deletion of the metal exporters that consequently turned on the egfp transcription. However, this assumption would not be accepted because the background signals of ZntR Cys15Ile were much lower even if they responded to metal ions better. In addition, the mutations on the MBL, such as His119Arg and Cys115Ser, showed less background and induced signals compared with ZntR WT in E. coli-zntR/copA/zntA. Furthermore, Cys141Ser located at the C-terminal region of ZntR showed no contribution for weaker background signals. Therefore, it was assumed that conformational changes on ZntRs were responsible for the variable strength observed to initiate the transcription of the reporter gene.
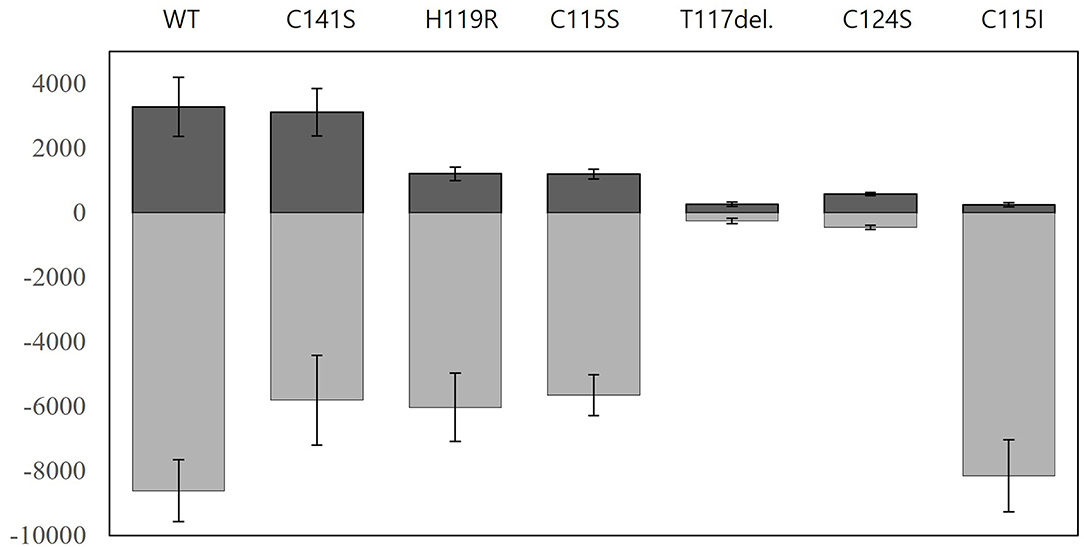
Figure 6. Nascent fluorescence signals of biosensors employing ZntR WT and mutants. The fluorescence intensities of biosensors based on E. coli-zntR/copA/zntA harboring pZntA-eGFP and ZntRs were measured after 2 h exposures to 0 μM and 1 μM of Pb(II). Positive (dark gray) and negative (light gray) values indicate the arbitrary units of eGFPs induced by non-exposure and exposure, respectively. The experiments were replicated more than three times and the standard deviations were indicated as error bars.
To elucidate this assumption, structural analysis was performed by computational three-dimensional modeling and simulation. Overall, the MBL of ZntR C115I was relatively looser compared with that of the WT because of the hydrophobic nature of isoleucine (Figures 7A,B). It was clear when only MBLs were shown as stick representation that three cysteines and one histidine were placed closer to two zinc ions in the WT compared with those amino acids of the mutant C115I (Figures 7C,D). Additionally, the distances between the zinc ions and residues around the MBL, including three cysteines and one histidine, were measured (Table 2). Taken together, the enlargement of the metal-binding site may be the reason why ZntR Cys115Ile showed enhanced Pb(II) specificity. However, it was still unclear why the different mutations led to variable transcription levels. In fact, we expected that the mutations on the metal-binding site could lead to conformational changes of the ZntR DNA-binding domains, but no significant differences were observed in the model structures.
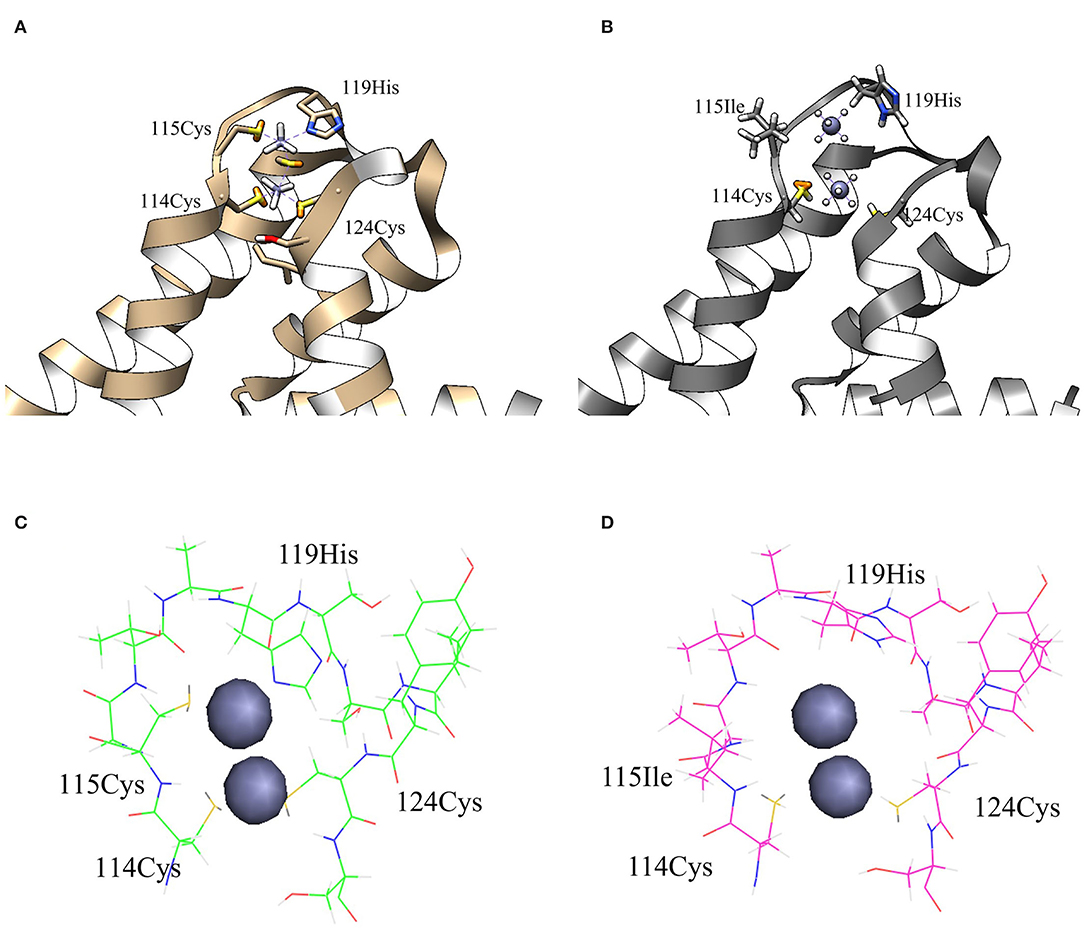
Figure 7. The structures of metal-binding loop regions for ZntR WT and C115I. The structures of MBLs for ZntR WT (A) and C115I (B) with stick representation of 4 residues interacting with zinc ions. The stick representation of amino acids consisted of MBLs for ZntR WT (C) and ZntR C115I (D). The zinc-binding sites of wild type (green) and mutant of C115I (magenta) are shown as the stick representation.
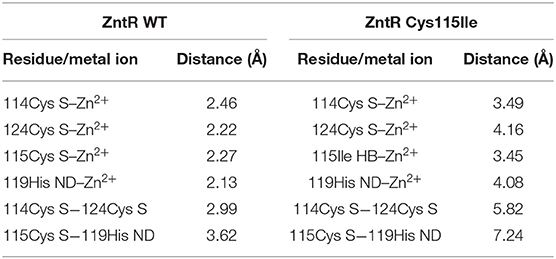
Table 2. Distances between the metal ions and residues located at the metal-binding loops of ZntR wild-type (WT) and Cys115Ile mutant measured from the model structures.
Discussion
In this study, we presented the development of novel Pb(II)-specific biosensors from existing genetic systems by engineering regulatory proteins and host cells. Although diverse bacterial cell-based biosensors have been reported for target metals and with corresponding genetic systems (Leveau and Lindow, 2002; Harms et al., 2006; Hynninen and Virta, 2009), heavy metals without corresponding genetic systems were excluded as targets of bacterial cell-based biosensors. In the case of Pb, the lead-resistant operon, pbr of Cupriavidus metallidurans CH34, was used for bacterial cell-based biosensors (Taghavi et al., 2009; Wei et al., 2014). Except for this previously described approach, there was no genetic system reported to date for obtaining a specific Pb(II) response. Therefore, the approaches described in this study to generate Pb(II)-specific biosensors are of great relevance, as they can represent an alternative bacterial cell-based biosensor for Pb(II) detection.
In fact, commercially available strains such as DH5α and BL21(DE3) could be used as host cells for biosensors. However, the BL21(DE3) was selected as a host cell because it possesses the IPTG-mediated overexpression systems, although the overexpression of regulatory genes was not necessary in this study. E. coli cell-based biosensors employing znt-operon showed strong responses toward Cd(II) and Hg(II), as reported previously (Yoon et al., 2016; Kang et al., 2018a). In addition, the recombinant ZntR was shown to effectively restore the metal-sensing properties of biosensors using zntR-deficient E. coli (E. coli-zntR) as host cells, with similar responses to those of the znt-operon-based system (data not shown). To evaluate the effects of metal ion channels (metal ion-translocating P-type ATPases), gene-deleted E. coli strains (E. coli-zntR/copA, E. coli-zntR/zntA, and E. coli-zntR/copA/zntA) were generated and used as host cells for the biosensors. Recombinant zntR WT was introduced as zntAp::egfp to the gene-deficient E. coli strains, and their metal selectivity was tested. However, this engineering approach was not suitable for evaluating the effects of the channels, since zntA deletion in the host cells induced eGFP overexpression even without metal exposure, thereby resulting in low induction coefficient values upon metal exposure. To overcome this problem, the initial cell density and duration for metal exposure were optimized. Briefly, the 0.05–0.30 of OD600 values were tested as initial cell density before metal ion exposure, and the fluorescence intensity was measured after 0.5–4 h. Since cell density of OD600 of 0.10 with 2 h exposure showed desirable results, it was selected as the optimal condition. Then, the metal selectivity of E. coli cell-based biosensors employing ZntR and zntAp::egfp as sensing and reporter domains, respectively, was changed by deleting the metal-exporting proteins (Figure 1). Deletion of copA and zntA, which encode CopA and ZntA, would impair the activity of Cu(I)- and Zn(II)-translocating P-type ATPases and consequently disrupt the metal homeostasis, resulting in the accumulation of metal ions in host cells (Sharma et al., 2000; Mitra and Sharma, 2001; Liu et al., 2006). The effects of CopA deletion weakened the responses of the biosensors toward Cd(II), whereas ZntA deletion enhanced their responses to Pb(II). In concordance with these results, weakened responses to Cd(II) and enhanced responses to Pb(II) were observed in both CopA- and ZntR-deleted strains (Figure 1D).
Next, the effects of genetic engineering of ZntR on target selectivity were investigated. Since the metal selectivity of the biosensors was determined by the target affinity of the regulatory proteins, they were selected as targets for genetic engineering to modulate metal selectivity (Ibáñez et al., 2015; Kang et al., 2018a; Lee et al., 2020). The E. coli cell-based biosensors investigated in this study employed znt-operon as a genetic system; thus, ZntR was the transcriptional regulator controlling the expression of the reporter gene (egfp). ZntR is one of the MerR family proteins known to unwind DNA and break two central base pairs upon target binding, thereby triggering transcriptional initiation (Heldwein and Brennan, 2001; Chen and He, 2004). Therefore, it was necessary to consider that mutations on its metal-binding site could not affect the function of the transcriptional factors. The MBL of the ZntR is composed of 12 amino acids, with three cysteines and one histidine, and two Zn(II) ions were placed on each loop region of ZntR (Changela et al., 2003; Pennella and Giedroc, 2005). As listed in Table 1, the numbers and positions of the cysteines were changed, and point mutations were introduced. Among the tested mutants, engineered ZntR Cys115Ser, H119R, and C141S showed stronger responses to Pb(II) rather than to Cd(II) (Figure 2). However, ZntR Cys115Ile showed the most marked changes concerning metal response compared with the other E. coli-zntR/copA/zntA systems. Although it showed enhanced response to Cd(II) compared with the WT, the response to Pb(II) was extremely increased to about 30-fold of the induction coefficient value. Nonetheless, this biosensor showed a broad specificity toward Pb(II), Cd(II), and Hg(II) at the tested concentration. To verify its specificity toward Pb(II), the biosensors were exposed to lower concentration ranges of the heavy metals (alone or in combination). Overall, the new biosensor responded only to Pb(II) at 0.1 μM and showed concentration-dependent responses from 0 to 100 nM of Pb(II) (Figures 3, 4). In addition, the other metal ions showed only marginal interference on signal acquisition at 10 nM (Figure 5), but no interference was observed at 5 nM of Cd(II) and Hg(II).
Finally, structural analysis was performed to better understand how the newly engineered biosensor achieved the observed superior selectivity and specificity toward Pb(II). The preference toward Pb(II) over Cd(II) was explained by the structural changes introduced around the MBLs of the ZntR (Figure 6). In particular, the 115Cys mutation disrupted the structural coordination with the zinc ions, thus resulting in decreased interaction with Cd(II) as compared with its originally stronger interaction. It was also noticed that the distances between the zinc ions and amino acids related to zinc binding, such as 114Cys, 115Cys, 124Cys, and 119His, were increased (Table 2), and the functions of the ZntRs as repressors were enhanced by the mutations (Figure 6). Although it was expected that conformational changes in the DNA-binding regions of ZntR could be responsible for the variations detected in the ZntR repressing activity, no significant structural differences around the DNA binding regions were observed. Thus, further investigations concerning the protein structures of the ZntRs are still warranted.
Conclusion
In this study, we described the generation of Pb(II)-specific biosensors, derived from existing genetic systems that can also be used as Cd(II) and Hg(II) biosensors. Briefly, the target selectivity of the regulatory protein, ZntR, was modulated by introducing point mutations on the metal-binding site, and the metal-exporting proteins were deleted from the host cells (Figure 8). The biosenor-based native znt-operon system shows the best response toward Cd(II), but its selectivity and specificity were shifted toward Pb(II) via genetic engineering. With the engineered ZntR C115I and copA/zntA-deficient E. coli, the novel biosensors possess superior selectivity and specificity to Pb(II), with a detection limit <5 nM. The Pb(II)-sensing capability of biosenors was sufficient to monitor the tolerance limit of lead in foods, which is set as 0.1–0.5 ppm (corresponding to ~1.2–6.0 μM) as defined by the Ministry of Food and Drug Safety in Korea. Thus, the Pb(II)-specific biosensor can be used as an alternative tool to monitor lead in foods and environmental samples. Although these bacterial cell-based biosensors were not actively applied to real samples, the engineering strategy used here may be of value to further enhance the capability and broaden the applications of cell-derived biosensors.
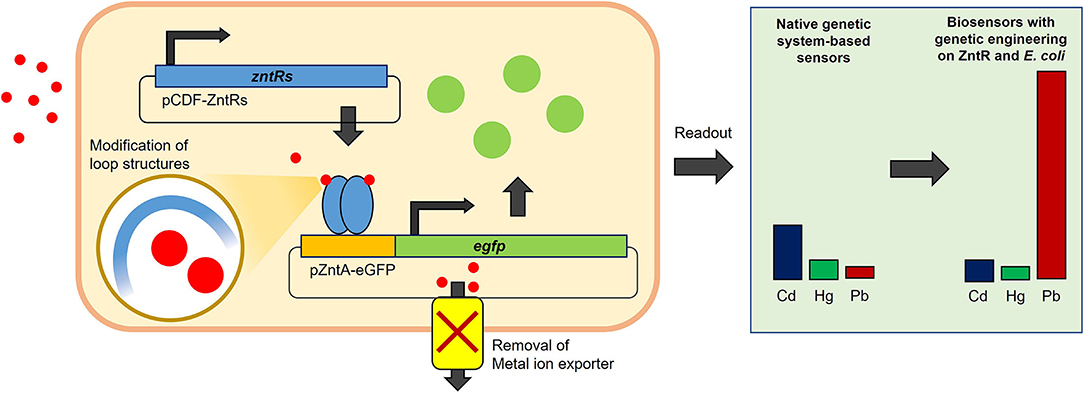
Figure 8. A schematic figure for the working mechanism of biosensor system. The regulatory protein, ZntR, repressed the expression of eGFP by binding on zntA promoter region without metal ions, while ZntR induced the expression of eGFP upon conformational change by metal ion binding. The zntR deleted E. coli BL21 harboring pCDF-ZntR WT and pZntA-eGFP showed responses in the order of Cd, Hg, and Pb, while the responding order and strength were modulated with modification of ZntR loop structure and with deleting metal exporting channel in host cells.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YY: conceptualization. YL and YJ: methodology, investigation, and data analysis. YY and B-GK: writing-original draft preparation. YY and GJ: writing-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Konkuk University in 2019 (to YY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.881050/full#supplementary-material
References
Brocklehurst, K. R., Hobman, J. L., Lawley, B., Blank, L., Marshall, S. J., Brown, N. L., et al. (1999). ZntR is a Zn (II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31, 893–902. doi: 10.1046/j.1365-2958.1999.01229.x
Changela, A., Chen, K., Xue, Y., Holschen, J., Outten, C. E., O'halloran, T. V., et al. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387. doi: 10.1126/science.1085950
Changye, H., Yan, G., Jian, W., Lisa, L., Xueqin, Y., Xiang, G., et al. (2021). Detection of bioavailable cadmium by double-color fluorescence based on a dual-sensing bioreporter system. Front. Microbiol. 12:1915. doi: 10.3389/fmicb.2021.696195
Chen, P., and He, C. (2004). A general strategy to convert the MerR family proteins into highly sensitive and selective fluorescent biosensors for metal ions. J. Am. Chem. Soc. 126, 728–729. doi: 10.1021/ja0383975
Elcin, E., Ayaydin, F., and Øktem, H.A. (2021). Formaldehyde biosensing in air using fluorescent bacterial bioreporter cells. Int. J. Environ. Analyt. Chem, 2021, 1–19. doi: 10.1080/03067319.2021.1998474
Gould, E. (2009). Childhood lead poisoning: conservative estimates of the social and economic benefits of lead hazard control. Environ. Health Perspectiv. 117, 1162–1167. doi: 10.1289/ehp.0800408
Hadi, F., and Aziz, T. (2015). A mini review on lead (Pb) toxicity in plants. J. Biol. Life Sci. 6, 91–101. doi: 10.5296/jbls.v6i2.7152
Harms, H., Wells, M. C., and Van Der Meer, J. R. (2006). Whole-cell living biosensors—are they ready for environmental application? Appl. Microbiol. Biotechnol. 70, 273–280. doi: 10.1007/s00253-006-0319-4
Heldwein, E. E. Z., and Brennan, R. G. (2001). Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409, 378–382. doi: 10.1038/35053138
Henshke, Y., Shemer, B., and Belkin, S. (2021). The Escherichia coli azoR gene promoter: a new sensing element for microbial biodetection of trace explosives. Curr. Res. Biotechnol. 3, 21–28. doi: 10.1016/j.crbiot.2021.01.003
Hernberg, S. (2000). Lead poisoning in a historical perspective. Am. J. Indus. Med. 38, 244–254. doi: 10.1002/1097-0274(200009)38:3<244::AID-AJIM3>3.0.CO;2-F
Hynninen, A., Tõnismann, K., and Virta, M. (2010). Improving the sensitivity of bacterial bioreporters for heavy metals. Bioeng. Bugs 1, 132–138. doi: 10.4161/bbug.1.2.10902
Hynninen, A., and Virta, M. (2009). Whole-cell bioreporters for the detection of bioavailable metals. Whole Cell Sens. Syst. II 31–63. doi: 10.1007/10_2009_9
Ibáñez, M. M., Checa, S. K., and Soncini, F. C. (2015). A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J. Bacteriol. 197, 1606–1613. doi: 10.1128/JB.02565-14
Kang, Y., Lee, W., Jang, G., Kim, B.-G., and Yoon, Y. (2018a). Modulating the sensing properties of Escherichia coli-based bioreporters for cadmium and mercury. Appl. Microbiol. Biotechnol. 102, 4863–4872. doi: 10.1007/s00253-018-8960-2
Kang, Y., Lee, W., Kim, S., Jang, G., Kim, B.-G., and Yoon, Y. (2018b). Enhancing the copper-sensing capability of Escherichia coli-based whole-cell bioreporters by genetic engineering. Appl. Microbiol. Biotechnol. 102, 1513–1521. doi: 10.1007/s00253-017-8677-7
Kannappan, S., and Ramisetty, B. C. M. (2021). Engineered whole-cell-based biosensors: sensing environmental heavy metal pollutants in water—a review. Appl. Biochem. Biotechnol. 2, 1–27. doi: 10.1007/s12010-021-03734-2
Kim, H., Jang, G., and Yoon, Y. (2020a). Specific heavy metal/metalloid sensors: current state and perspectives. Appl. Microbiol. Biotechnol. 104, 907–914. doi: 10.1007/s00253-019-10261-y
Kim, H., Jeon, Y., Lee, W., Jang, G., and Yoon, Y. (2020b). Shifting the specificity of E. coli biosensor from inorganic arsenic to phenylarsine oxide through genetic engineering. Sensors 20:3093. doi: 10.3390/s20113093
Lee, W., Kim, H., Jang, G., Kim, B.-G., and Yoon, Y. (2020). Antimony sensing whole-cell bioreporters derived from ArsR genetic engineering. Appl. Microbiol. Biotechnol. 104, 2691–2699. doi: 10.1007/s00253-020-10413-5
Leveau, J. H., and Lindow, S. E. (2002). Bioreporters in microbial ecology. Curr. Opin. Microbiol. 5, 259–265. doi: 10.1016/S1369-5274(02)00321-1
Liu, J., Dutta, S. J., Stemmler, A. J., and Mitra, B. (2006). Metal-binding affinity of the transmembrane site in ZntA: implications for metal selectivity. Biochemistry 45, 763–772. doi: 10.1021/bi051836n
Magrisso, S., Erel, Y., and Belkin, S. (2008). Microbial reporters of metal bioavailability. Microbial. Biotechnol. 1, 320–330. doi: 10.1111/j.1751-7915.2008.00022.x
Mitra, B., and Sharma, R. (2001). The cysteine-rich amino-terminal domain of ZntA, a Pb (II)/Zn (II)/Cd (II)-translocating ATPase from Escherichia coli, is not essential for its function. Biochemistry 40, 7694–7699. doi: 10.1021/bi010576g
Nivens, D. E., Mcknight, T., Moser, S., Osbourn, S., Simpson, M., and Sayler, G. (2004). Bioluminescent bioreporter integrated circuits: potentially small, rugged and inexpensive whole-cell biosensors for remote environmental monitoring. J. Appl. Microbiol. 96, 33–46. doi: 10.1046/j.1365-2672.2003.02114.x
Nnorom, I., and Osibanjo, O. (2009). Heavy metal characterization of waste portable rechargeable batteries used in mobile phones. Int. J. Environ. Sci. Technol. 6, 641–650. doi: 10.1007/BF03326105
Palacin, M. R. (2009). Recent advances in rechargeable battery materials: a chemist's perspective. Chem. Soc. Rev. 38, 2565–2575. doi: 10.1039/b820555h
Pennella, M. A., and Giedroc, D. P. (2005). Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals 18, 413–428. doi: 10.1007/s10534-005-3716-8
Placke, T., Kloepsch, R., Dühnen, S., and Winter, M. (2017). Lithium ion, lithium metal, and alternative rechargeable battery technologies: the odyssey for high energy density. J. Solid State Electrochem. 21, 1939–1964. doi: 10.1007/s10008-017-3610-7
Rensing, C., Sun, Y., Mitra, B., and Rosen, B. P. (1998). Pb (II)-translocating P-type ATPases. J. Biol. Chem. 273, 32614–32617. doi: 10.1074/jbc.273.49.32614
Sharma, R., Rensing, C., Rosen, B. P., and Mitra, B. (2000). The ATP hydrolytic activity of purified ZntA, a Pb (II)/Cd (II)/Zn (II)-translocating ATPase from Escherichia coli. J. Biol. Chem. 275, 3873–3878. doi: 10.1074/jbc.275.6.3873
Shpigel, E., Shemer, B., Elad, T., Glozman, A., and Belkin, S. (2021). Bacterial bioreporters for the detection of trace explosives: performance enhancement by DNA shuffling and random mutagenesis. Appl. Microbiol. Biotechnol. 105, 4329–4337. doi: 10.1007/s00253-021-11290-2
Shu, X., Yin, L., Zhang, Q., and Wang, W. (2012). Effect of Pb toxicity on leaf growth, antioxidant enzyme activities, and photosynthesis in cuttings and seedlings of Jatropha curcas L. Environ. Sci. Pollut. Res. 19, 893–902. doi: 10.1007/s11356-011-0625-y
Signorell, A., Aho, K., Alfons, A., Anderegg, N., Aragon, T., and Arppe, A. (2019). DescTools: Tools for Descriptive Statistics. R Package Version 0.99 28,17.
Taghavi, S., Lesaulnier, C., Monchy, S., Wattiez, R., Mergeay, M., and Van Der Lelie, D. (2009). Lead (II) resistance in Cupriavidus metallidurans CH34: interplay between plasmid and chromosomally-located functions. Antonie Van Leeuwenhoek 96, 171–182. doi: 10.1007/s10482-008-9289-0
Team, R. C. (2014). R: A Language and Environment for Statistical Computing. Vienna: Foundation for Statistical Computing. Available online at http://www.R-project.org/ (accessed July 28, 2015).
Van Der Meer, J. R., and Belkin, S. (2010). Where microbiology meets microengineering: design and applications of reporter bacteria. Nat. Rev. Microbiol. 8, 511–522. doi: 10.1038/nrmicro2392
Wei, W., Liu, X., Sun, P., Wang, X., Zhu, H., Hong, M., et al. (2014). Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ. Sci. Technol. 48, 3363–3371. doi: 10.1021/es4046567
Yang, S., Cui, G., Pang, S., Cao, Q., Kolb, U., Feng, X., et al. (2010). Fabrication of cobalt and cobalt Oxide/Graphene composites: towards High-Performance anode materials for lithium ion batteries. ChemSusChem 3, 236–239. doi: 10.1002/cssc.200900106
Yoon, Y., Kang, Y., Lee, W., Oh, K.-C., Jang, G., and Kim, B.-G. (2018). Modulating the properties of metal-sensing whole-cell bioreporters by interfering with Escherichia coli metal homeostasis. J. Microbiol. Biotechnol. 28, 323–329. doi: 10.4014/jmb.1710.10012
Yoon, Y., Kim, S., Chae, Y., Kang, Y., Lee, Y., Jeong, S.-W., et al. (2016). Use of tunable whole-cell bioreporters to assess bioavailable cadmium and remediation performance in soils. PLoS ONE 11:e0154506. doi: 10.1371/journal.pone.0154506
Zappi, D., Coronado, E., Basile, G., Varani, G., Turemis, M., and Giardi, M. T. (2021). A microbial sensor platform based on bacterial bioluminescence (luxAB) and green fluorescent protein (gfp) reporters for in situ monitoring of toxicity of wastewater nitrification process dynamics. Talanta 221:121438. doi: 10.1016/j.talanta.2020.121438
Keywords: bacterial cell-based biosensors, heavy metals, Zn(II)-translocating P-type ATPase, protein engineering, Pb(II)
Citation: Jeon Y, Lee Y, Jang G, Kim B-G and Yoon Y (2022) Design of Pb(II)-Specific E. coli-Based Biosensors by Engineering Regulatory Proteins and Host Cells. Front. Microbiol. 13:881050. doi: 10.3389/fmicb.2022.881050
Received: 22 February 2022; Accepted: 04 April 2022;
Published: 20 May 2022.
Edited by:
Yang-Chun Yong, Jiangsu University, ChinaReviewed by:
Shimshon Belkin, Hebrew University of Jerusalem, IsraelMarie J. Durand, Université de Nantes, France
Copyright © 2022 Jeon, Lee, Jang, Kim and Yoon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youngdae Yoon, eXlvb24yMUBnbWFpbC5jb20=
 Yangwon Jeon
Yangwon Jeon Yejin Lee
Yejin Lee Geupil Jang2
Geupil Jang2 Youngdae Yoon
Youngdae Yoon