
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 June 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.881043
This article is part of the Research Topic Biofilms from a Food Microbiology Perspective: Structures, Functions and Control Strategies, Volume II View all 7 articles
The extracellular matrix is essential for the biofilm formation of food spoilers. Pseudomonas fluorescens PF07 is a previous isolate from spoiled marine fish; however, the genes involved in the extracellular matrix formation of PF07 biofilms remain poorly defined. In this study, PF07 formed a wrinkled macrocolony biofilm through the high production of extracellular matrix. The genes involved in biofilm matrix formation and regulation were screened and identified by RNA-seq-dependent transcriptomic analysis and gene knock-out analysis. The macrocolony biofilms of PF07 grown for 5 days (PF07_5d) were compared with those grown for 1 day (PF07_1d). A total of 1,403 genes were significantly differentially expressed during biofilm formation. These mainly include the genes related to biofilm matrix proteins, polysaccharides, rhamnolipids, secretion system, biofilm regulation, and metabolism. Among them, functional amyloid genes fapABCDE were highly upregulated in the mature biofilm, and the operon fapA-E had a –24/–12 promoter dependent on the sigma factor RpoN. Moreover, the RNA-seq analyses of the rpoN mutant, compared with PF07, revealed 159 genes were differentially expressed in the macrocolony biofilms, and fapA-E genes were positively regulated by RpoN. In addition, the deletion mutants of fapC, rpoN, and brfA (a novel gene coding for an RpoN-dependent transcriptional regulator) were defective in forming mature macrocolony biofilms, solid surface-associated (SSA) biofilms, and pellicles, and they showed significantly reduced biofilm matrices. The fap genes were significantly downregulated in ΔbrfA, as in ΔrpoN. These findings suggest that the functional amyloid Fap is the main component of PF07 biofilm matrices, and RpoN may directly regulate the transcription of fap genes, in conjunction with BrfA. These genes may serve as potential molecular targets for screening new anti-biofilm agents or for biofilm detection in food environments.
Biofilms are complex communities embedded in a self-produced extracellular matrix composed of proteins, exopolysaccharides, and extracellular DNAs (Flemming and Wingender, 2010). Bacteria form different types of biofilms depending on the environment. They include macrocolony biofilms that form on semi-solid agar plates for extended time, pellicles that form at the air–liquid interface of a standing culture, and solid surface-associated (SSA) submerged biofilms that formed on abiotic solid surfaces (Serra and Hengge, 2014). During food production, processing, and distribution, bacteria can form macrocolony biofilms or pellicles on the surfaces of various food materials and can form SSA biofilms on the surfaces of food industry equipment (Galié et al., 2018; Alvarez-Ordóñez et al., 2019). Biofilm formation provides physical resistance, mechanical resistance, or chemical protection to the bacterial cells, creating a serious challenge for the food industry in removing these food spoilers and pathogens (Bridier et al., 2015).
The extracellular matrix is essential for the biofilm formation of bacteria (Flemming and Wingender, 2010). The composition of the extracellular matrix formed by different strains varies greatly due to the genetic background and environment conditions. In the model bacterium Escherichia coli K12, amyloid curli fibers and cellulose are main extracellular matrix components (Mika and Hengge, 2014). The opportunistic pathogen Pseudomonas aeruginosa strains PAO1 and PA14 use polysaccharides Psl or Pel as the main components of the extracellular matrices, respectively (Ma et al., 2009; Jennings et al., 2015). The rhizobacteria P. fluorescens strains Pf0-1 and SBW25 produce adhesion protein LapA and cellulose Wss as the key components of the extracellular matrices, respectively (Spiers et al., 2003; Fazli et al., 2014). In P. fluorescens UK4 isolated from a reservoir, the functional amyloid Fap was identified from SSA biofilms (Dueholm et al., 2010). Recombinant overexpression of fap genes resulted in highly aggregative phenotypes with enhanced biofilm forming capacity in Pseudomonas strains; however, the Δfap mutants formed SSA biofilms similar to the wild types, indicating that Fap is not an absolute requirement for biofilm formation of these wild-type strains under the experimental conditions (Dueholm et al., 2013; Zeng et al., 2015). Several other extracellular matrix components of Pseudomonas have been described using in silico methods, such as Flp/Tad pilus and polysaccharide poly-N-acetyl-glucosamine (PNAG) (Blanco-Romero et al., 2020).
Biofilm matrix production is controlled by complex regulatory networks that integrate multiple environmental signals by transcription factors, the second messenger cyclic diguanylate monophosphate (c-di-GMP), and sRNA (Mika and Hengge, 2014). Transcription factors are key regulators for the transcription of biofilm matrix genes. In E. coli K12, the transcription factor CsgD directly regulates the transcription of csgBAC coding for Curli (Mika and Hengge, 2014). In P. aeruginosa PA14, the transcription factor FleQ directly regulates the transcription of the pel genes (Hickman and Harwood, 2008). In P. fluorescens Pf0-1 and SBW25, the direct transcription factors for lapA and wss are currently unknown. In P. fluorescens UK4 cultured in liquid fish juice at 4°C, the transcription of fap genes were positively regulated by the alternative sigma factor RpoN, but the understanding of the transcription regulation of the fap gene in biofilm remains limited (Liu et al., 2021). In addition, c-di-GMP has effects on the production of biofilm matrices. C-di-GMP is produced and degraded by multiple diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively. The production of curli amyloids in E. coli biofilms, Pel polysaccharides in P. aeruginosa biofilms, and LapA, and Wss in P. fluorescens biofilms are all regulated by c-di-GMP (Serra et al., 2013; Okegbe et al., 2017; Collins et al., 2020). Moreover, sRNA is also involved in the regulation of the biofilm matrix production (Fazli et al., 2014; Mika and Hengge, 2014).
Pseudomonas spp. contribute significantly to the spoilage of food. They are the predominant spoilers of proteinaceous raw foods stored under aerobic refrigerated conditions (Remenant et al., 2015). P. fluorescens is currently recognized as one of the most abundant spoilers of this genus, and it has been extensively isolated from dairy, fish, and meat (Nogarol et al., 2013; Wang et al., 2018b,2020). P. fluorescens is a strong biofilm producer, and the extracellular matrix of the biofilm has a structural role, enhancing the persistence of these biofilms in the food industry (Galié et al., 2018; Zhu et al., 2019). Particularly in the fish industries, P. fluorescens adheres to the surface of processing equipment in the form of a biofilm, even after cleaning and disinfecting the equipment (Bagge-Ravn et al., 2003; Langsrud et al., 2016). Moreover, the extracellular matrix of its biofilm can provide shelter for foodborne pathogen Listeria monocytogenes (Puga et al., 2018).
The P. fluorescens PF07 strain was isolated from spoiled large yellow croaker in our previous work, and it exhibited high spoilage potential (Wang et al., 2021). However, the genetic basis for the formation of extracellular matrices in PF07 biofilms remains poorly defined. In this study, PF07 was identified as a strong biofilm producer, as it can form dense macrocolony biofilms, SSA biofilms, and pellicles. The genes involved in biofilm matrix formation and regulation were screened and identified. First, RNA-seq-dependent transcriptomic analysis of macrocolony biofilms (PF07_5d vs. PF07_1d, ΔrpoN vs. PF07) was performed to identify genes related to the biofilm matrix formation. Moreover, by promoter, gene knock-out, and biofilm phenotype analyses, functional amyloid Fap is the main component of PF07 biofilm matrices, and RpoN may directly regulate the transcription of fap genes, in conjunction with a novel RpoN-dependent transcriptional regulator D7M10_RS14495, which we have named BrfA for “biofilm regulator of fap.”
All bacterial strains and plasmids used in this study are presented in Table 1. P. fluorescens PF07 and its mutants were grown in Luria-Bertani (LB) or tryptone broth (10 g/L tryptone) medium at 28°C.
The deletion mutants ΔrpoN, ΔfapC, and ΔbrfA of P. fluorescens PF07 were constructed by double-crossover allelic exchange, according to the method by Liu et al. (2018). All primers used in the mutant construction are listed in Supplementary Table 1. Briefly, two fragments flanking rpoN were amplified by PCR with 07rpoN-MF1/MR1 and 07rpoN-MF2/MR2 primers, respectively. The sequences of the two primers 07rpoN-MR1 and 07rpoN-MF2 were reverse and complementary. The two fragments were purified and fused in subsequent PCR reactions using 07rpoN-MF1/MR2 primers. In the same way, two fragments flanking the fapC or brfA were amplified and fused using the corresponding primers of each gene. The fused segment of the rpoN gene with a deletion of 1,258 bp in its 1,494-bp open reading frame (ORF), the fused segment of the fapC gene with a deletion of 676 bp in its 756-bp ORF, and the fused segment of the brfA gene with a deletion of 1,134 bp in its 1,326-bp ORF, were cloned into the suicide vector pLP12Cm using a ClonExpress II One Step Cloning Kit (Vazyme, China), respectively. The recombinant plasmids pLP12Cm-rpoN, pLP12Cm-fapC, and pLP12Cm-brfA were transferred from E. coli β2163 (Demarre et al., 2005) into P. fluorescens PF07 by conjugation. The single crossover transconjugants were selected on LB agar medium containing 20 μg/mL chloramphenicol and 0.3% D-glucose at 28°C. L-arabinose can induce the expression of the virulence gene vmi480 on the suicide vector; thus, the second crossover mutants were screened on LB agar with 0.4% L-arabinose. The PF07 ΔrpoN, ΔfapC, and ΔbrfA mutants were confirmed via PCR, using external primer pairs rpoN-TF/TR, fapC-TF/TR, and brfA-TF/TR, respectively. The deletion mutants were further confirmed by sequencing.
The overnight cultures of PF07 and the rpoN mutant (5 μL each) were spotted on tryptone plates (1% tryptone, 1% agar). After 1 day of incubation at 28°C, 24 colonies of PF07 were gently scraped from six tryptone plates and mixed in 10 mL of 0.9% NaCl. The sample was harvested by centrifugation at 7,000 × g for 10 min and frozen in liquid nitrogen prior to RNA isolation. After 5 days of incubation, macrocolonies of PF07 and the rpoN mutant were collected using the same method, respectively. Two biological replicates were prepared for each treatment. The obtained samples were sent to Novogene Co., Ltd. (Beijing, China) for RNA-seq analysis. Total RNA was isolated from the frozen samples using an RNAprep Pure Cell/Bacteria Kit and a miRcute miRNA Isolation Kit (Tiangen, China). The concentration and quality of total RNA were validated using a NanoDrop spectrophotometer (Thermo, United States) and Agilent 2100 Bioanalyzer (Agilent, United States), and the qualified RNA samples were used as input materials. mRNA was purified from total RNA using probes to remove rRNA. Then, strand-specific RNA sequencing libraries were constructed using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, United States) according to the manufacturer’s introductions. Following validation with an Agilent Bioanalyzer 2100 system (Agilent, United States), the libraries were sequenced on an Illumina Hiseq platform and paired-end reads were generated. The raw reads were filtered to remove adapter-containing reads, high N rate reads (N rates > 10%), and low-quality reads (50% bases with Q-score ≤ 5) using in-house Perl scripts. The genome sequence of P. fluorescens PF08, an isolate from refrigerated turbot, was used as the reference genome,1 and the obtained clean reads were mapped to the reference genome using Bowtie2 v2.2.3 (Langmead and Salzberg, 2012). The novel gene and sRNA were identified by Rockhopper (McClure et al., 2013). The gene expression levels were quantified, and differential expression analysis was performed using DESeq2 (Love et al., 2014). Genes with an adjusted P-value < 0.05 and | log2 fold change| ≥ 1 were considered to be differentially expressed (Wang et al., 2018a; Liu et al., 2019).
qRT-PCR analyses were performed with RNA samples under the same culture conditions used for the RNA-seq experiments. Total RNA was isolated with a TRIzol Plus RNA Purification Kit (Invitrogen, United States) and treated with RNase-Free DNase Set (Qiagen, Germany) to remove the contaminating DNA. cDNA was synthesized using the SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, United States), and qRT-PCR analyses were conducted using a CFX384 Touch Real-Time PCR Detection System (Bio-Rad, United States) with the Power SYBR1 Green PCR Master Mix (Applied Biosystems, United States). Primers for qRT-PCR are listed in Supplementary Table 2. The 16S rRNA gene was used as a reference control for sample normalization. The relative expression of the target genes was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). Three biological replicates were prepared, and samples were run in triplicate.
To identify the 5′-end of a gene, rapid amplification of 5′-cDNA ends (5′-RACE) was performed according to a method described previously (Liu et al., 2018). Briefly, the Vaccinia Capping System (NEB, United States) and GeneRacer Kit (Invitrogen, United States) were used according to the manufacturers’ instructions. The 5′-ends of the transcripts were capped with the Vaccinia Capping System, and the GeneRacer Kit ensured the amplification of only full-length transcripts via the elimination of truncated messages from the amplification process. The fapA specific primers used for 5′-RACE are listed in Supplementary Table 1. The 5′ RACE products were directly sequenced to identify the 5′-end of fapA.
Macrocolony biofilms were observed by Congo red assay and transmission electron microscopy (TEM), using a previously reported method (Liu et al., 2019). Congo red plates (1% tryptone, 1% agar, 40 μg/mL Congo red, and 10 μg/mL Coomassie brilliant blue G250) were used to assess macrocolony biofilm color and morphology. Cells of PF07 and the mutants were grown overnight in liquid LB medium with shaking at 28°C. A total of 5 μL of the overnight cultures was spotted on Congo red plates. The plates were incubated at 28°C for 1, 3, 5, and 7 days to observe the macrocolony morphology. For TEM analysis, the macrocolonies on day 5 were gently scraped from a tryptone plate (1% tryptone, 1% agar) and fixed in 2.5% glutaraldehyde for more than 4 h at room temperature. Then, the macrocolony biofilms were postfixed, dehydrated, embedded in TAAB resin (TAAB Laboratories, United Kingdom), ultrathin-sectioned, stained, and observed via TEM (Hitachi H-600, Japan).
Overnight cultures of PF07 and the mutants (6 μL each) were added into 6 mL of fresh sterile tryptone broth. The dilution culture was statically grown in a glass tube at 28°C for 3 days. Pellicles were assayed by visual inspection of the air–liquid interface of the standing culture.
Quantification of biofilms grown in microplates was performed as described by Liu et al. (2021). Tryptone broth was used as the growth medium, and cultures were incubated at 28°C for 6, 12, 24, and 48 h with shaking (150 rpm). After crystal violet staining, the absorbance at 595 nm was measured using a microplate reader (Infinite 200, Tecan, Switzerland). Eight individual samples were assessed for each strain at each time point, and the experiment was repeated at least twice.
Overnight cultures were inoculated into 12-well dishes containing tryptone broth at a 1:1,000 ratio. Glass coverslips (20 mm × 20 mm) were partially submerged in the medium and grown at 28°C with shaking (150 rpm). After incubation for 24 h, the glass slips were washed two times with 0.9% NaCl. The biofilm on the glass slips was successively stained with 150 ng/mL 4′,6-diamidino-2-phenylindole (DAPI) for 20 min and 2 mg/mL Congo red for 20 min in the dark at room temperature (Larsen et al., 2007). The coverslips were washed and dried, and then a drop of liquid Antifade mounting medium (Beyotime, China) was placed on the coverslips to prevent fluorescence quenching. All the coverslips with stained biofilms were placed on slides and imaged with a Nikon Eclipse Ti2 confocal laser scanning microscopy (CLSM) (Nikon, Japan) and a 63 × oil objective. Fluorescence signals were recorded in the blue channel (excitation 405 nm, emission 425–475 nm) for DAPI and in the red channel (excitation 561 nm, emission 570–620 nm) for Congo red, where at least 10 fields of view were captured. NIS-Elements AR confocal software was used to analyze the biofilm images, allowing for collection of z-stacks.
Overnight cultures were diluted 1:100 into 150 mL fresh tryptone broth medium, and cultivated at 28°C with constant shaking (220 rpm) for 24 h. The optical absorbance at 600 nm was measured at 4-h intervals. Three independent experiments were performed, and the mean optical absorbance values at each time point were used to determine the growth rate.
P. fluorescens PF07 is a potent biofilm producer, and it formed a red, wrinkled macrocolony on a Congo red plate after at least 3 days of growth. The red and wrinkled macrocolony of bacteria often results from the production of large amounts of extracellular matrices (Friedman and Kolter, 2004). In the present work, RNA-seq-dependent transcriptomic analysis was utilized to identify genes involved in biofilm matrix formation and regulation. The macrocolony biofilms of PF07 grown on tryptone plates for 5 days (PF07_5d) were compared with those grown for 1 day (PF07_1d). RpoN is an important sigma factor involved in biofilm regulation of bacteria (Xu et al., 2019; Liu et al., 2021); therefore, the rpoN mutant of PF07 was constructed to determine the regulatory role of RpoN in biofilm matrix formation (Supplementary Figure 1). The raw sequencing data of RNA-seq were deposited in the Sequence Read Archive,2 with an accession number PRJNA800221. After the raw reads were filtered, the clean reads were mapped to the reference genome sequence of P. fluorescens PF08, with a total mapped rate of at least 96.31% for all samples. The high mapped rates suggest that PF07 strain is very closely related to the reference strain PF08, an isolate from refrigerated turbot (Xu et al., 2021). The obtained read data and mapping data are shown in Supplementary Tables 3, 4. According to the screening criteria for differentially expressed genes (DEGs) (| log2 fold change| ≥ 1, padj ≤ 0.05), a total of 1,403 DEGs were identified, including 641 significantly upregulated genes and 762 significantly downregulated genes in PF07_5d, compared with PF07_1d (Figure 1A). In addition, 159 DEGs were identified by comparing the macrocolony biofilms of rpoN mutant and PF07 on day 5, including 29 significantly upregulated genes and 130 significantly downregulated genes (Figure 1B). The detailed information on the two groups of DEGs is summarized in Supplementary Tables 5, 6. The relationship between the two groups of DEGs was further analyzed, and a total of 80 DEGs were obtained in the overlap of the two groups (Figure 1C).
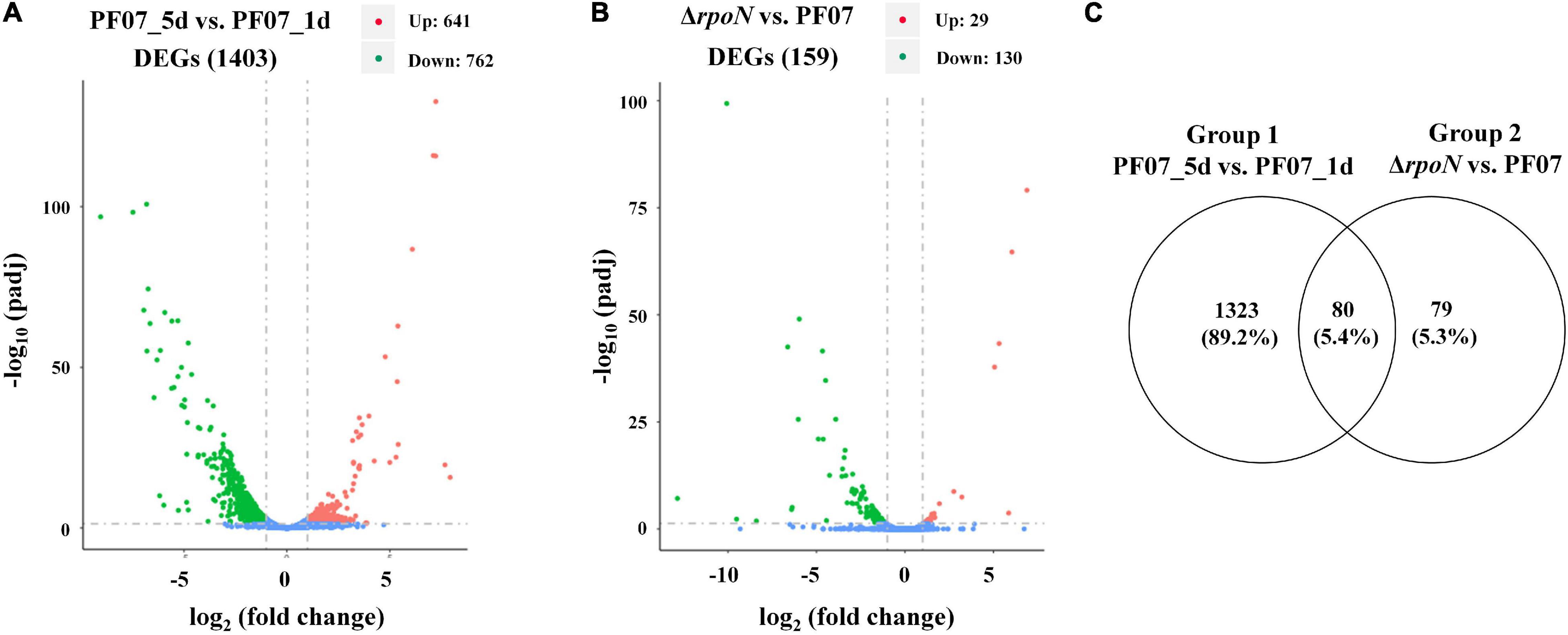
Figure 1. DEGs from RNA-seq analyses. (A) DEGs between macrocolony biofilms of PF07_5d and PF07_1d (Group 1). (B) DEGs between macrocolony biofilms of ΔrpoN and PF07 after 5 days of incubation (Group 2). The horizontal axis represents fold changes of gene expression, and the vertical axis represents the level of statistical significance. The red dots represent significantly upregulated genes, and the green dots represent significantly downregulated genes. (C) Venn diagram of Group 1 and Group 2. This figure was drawn using Venny 2.0. (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Based on the functional annotation, chromosomal location, and expression pattern, the representative DEG clusters or DEGs are listed in Table 2, and the results were validated by qRT-PCR. The qRT-PCR results agreed with the expression patterns from the RNA-seq analyses, except for pgaABCD, a gene cluster related to the biosynthesis of the extracellular polysaccharide PNAG. The main components of the biofilm matrix are proteins or extracellular polysaccharides, and the genes coding for these components are usually present in the form of gene clusters (Blanco-Romero et al., 2020). The proteinaceous components of the biofilm matrix mainly include secreted extracellular proteins, cell surface adhesins and protein subunits of cell appendages, such as flagella and pili (Fong and Yildiz, 2015). As shown in Table 2, the main DEGs related to matrix proteins are fapABCDE, flp/tad, and flagellar genes. Interestingly, the gene cluster fapA-E, encoding functional amyloids, was significantly upregulated in PF07_5d compared with PF07_1d, and significantly downregulated in ΔrpoN compared with PF07 (Table 2). This gene cluster is likely to be involved in the formation of biofilm matrix and is positively regulated by RpoN. However, the flp/tad gene cluster, encoding a special type IVb pilus, was significantly downregulated in PF07_5d compared with PF07_1d. The expression of genes associated with flagella was also significantly downregulated in PF07_5d, compared with PF07_1d, as well as in ΔrpoN compared with PF07 (Table 2). In addition to the matrix protein genes, the representative DEGs related to polysaccharide metabolism are pgaABCD and glycogen metabolism genes. According to the results of RNA-seq, the pga gene cluster was upregulated in PF07_5d compared with PF07_1d, but the qRT-PCR results showed downregulation. The discrepancy suggests that the expression of pga genes in biofilms may lack repeatability as the qRT-PCR tests were performed using independently extracted RNAs. The expression of the genes associated with glycogen metabolism was also downregulated in PF07_5d compared with PF07_1d. Moreover, the gene cluster rhlABRI was significantly downregulated in PF07_5d compared with PF07_1d, but upregulated in the rpoN mutant compared with PF07_1d. The gene cluster encodes enzymes (RhlAB) for rhamnolipid synthesis and the quorum sensing system (RhlRI). Finally, the bacterial secretion system is also likely to be associated with biofilm formation. As shown in Table 2, the type II secretion system was upregulated, while another possible secretion system was downregulated at the transcription level in PF07_5d, compared with PF07_1d.
Some genes that may be involved in the regulation of biofilm matrix synthesis were also identified (Table 2). A novel RpoN-dependent transcription factor D7M10_RS14495, which we named BrfA, was significantly upregulated in PF07_5d compared with PF07_1d. Some genes encoding diguanylate cyclase and phosphodiesterase (D7M10_RS04965, D7M10_RS06580, and D7M10_RS22095) were significantly altered during the formation of the biofilm. The second messenger c-di-GMP regulates the expression of biofilm matrix components in most bacteria, and the level of intracellular c-di-GMP concentration is controlled by diguanylate cyclase and phosphodiesterase (Fazli et al., 2014). In addition, the transcript levels of several novel sRNAs were also altered during the formation of biofilms (Supplementary Table 5). However, these novel sRNAs require further confirmation.
With regard to the usage of carbon and nitrogen sources, we found that genes related to galactose catabolism (D7M10_RS00150-65) were significantly upregulated in the mature biofilm, but genes related to amino acid utilization (D7M10_RS13750-85, arcDABC) were significantly downregulated during the biofilm formation. In addition, a gene cluster with unknown function (D7M10_RS18830-15) was highly upregulated in PF07_5d compared with PF07_1d, and significantly downregulated in ΔrpoN compared with PF07 (Table 2).
The RNA-seq results indicated the gene cluster fapA-E was positively regulated by RpoN. RpoN recognizes promoter sequences with conserved GG and GC elements located –24 and –12 nucleotides upstream from the transcriptional start site (Barrios et al., 1999). According to chromosomal location and expression pattern, fapA-E are arrayed in an operon, and fapA is the first gene of the operon. The 5′-RACE method was performed to identify the transcriptional start site of fapA. As shown in Figure 2, a conserved –24/–12 element was present upstream from the transcriptional start site. This result suggests that fapA-E genes are directly regulated by RpoN.
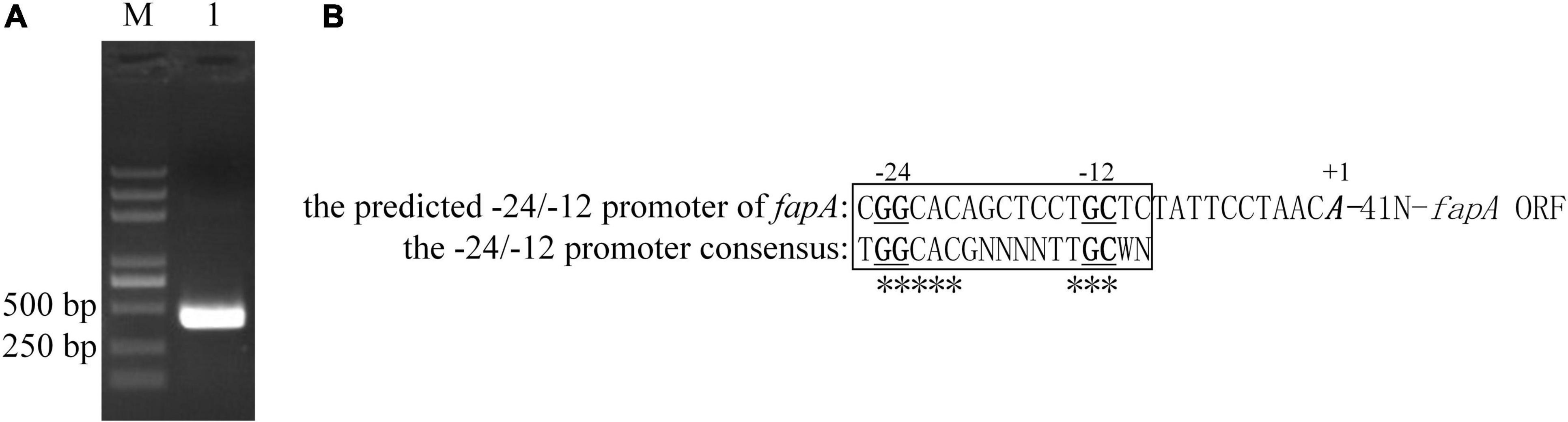
Figure 2. Transcription initiation site and RpoN-dependent promoter of fapA. (A) The 5′-RACE-PCR products of fapA (Lane 1). Lane M, DL2000 DNA marker (Takara, China). (B) The translation start site is indicated by bold and italic text. Nucleotides that were 100% conserved in all examined RpoN-dependent promoters by Barrios et al. (1999) are labeled with asterisks (*). The GG and GC elements are indicated by bold and underlined text.
The fap cluster contains six ORFs fapABCDEF. According to the results of the RNA-seq, the transcription of fapA-E was significantly upregulated during biofilm formation, but the expression of fapF was not upregulated (Table 2). The time-course analysis of fap gene expression during biofilm formation by qRT-PCR further supported the RNA-seq results (Figure 3A). These results suggest differences in the expression patterns of fapF and fapA-E. FapC is the main component of the Fap fibers (Bleem et al., 2018); thus, the fapC mutant was constructed to observe the role of the Fap amyloids during biofilm formation (Supplementary Figure 1). Biofilm morphology assays on Congo red plates were performed (Figure 3B). After 1 day of growth, the colonies of the fapC mutant were the same as the wild-type colonies and characterized as pale and smooth. After 3 days, the colonies of the wild type started to become red and wrinkled, but those of the fapC mutant remained pale and smooth. On days 5 and 7, the wild-type strain formed mature macrocolony biofilms, which were redder and more wrinkled than the fapC mutant. Additionally, we used TEM to visualize the structure of the macrocolony biofilms. After 5 days, the wild-type cells were encased in a basket-like matrix, whereas the mutant cells generated much less extracellular matrix (Figure 3B). These results suggest that PF07 can develop wrinkled macrocolony biofilms through the high production of the extracellular matrix, and that Fap amyloid is the main component of PF07 biofilm matrices.
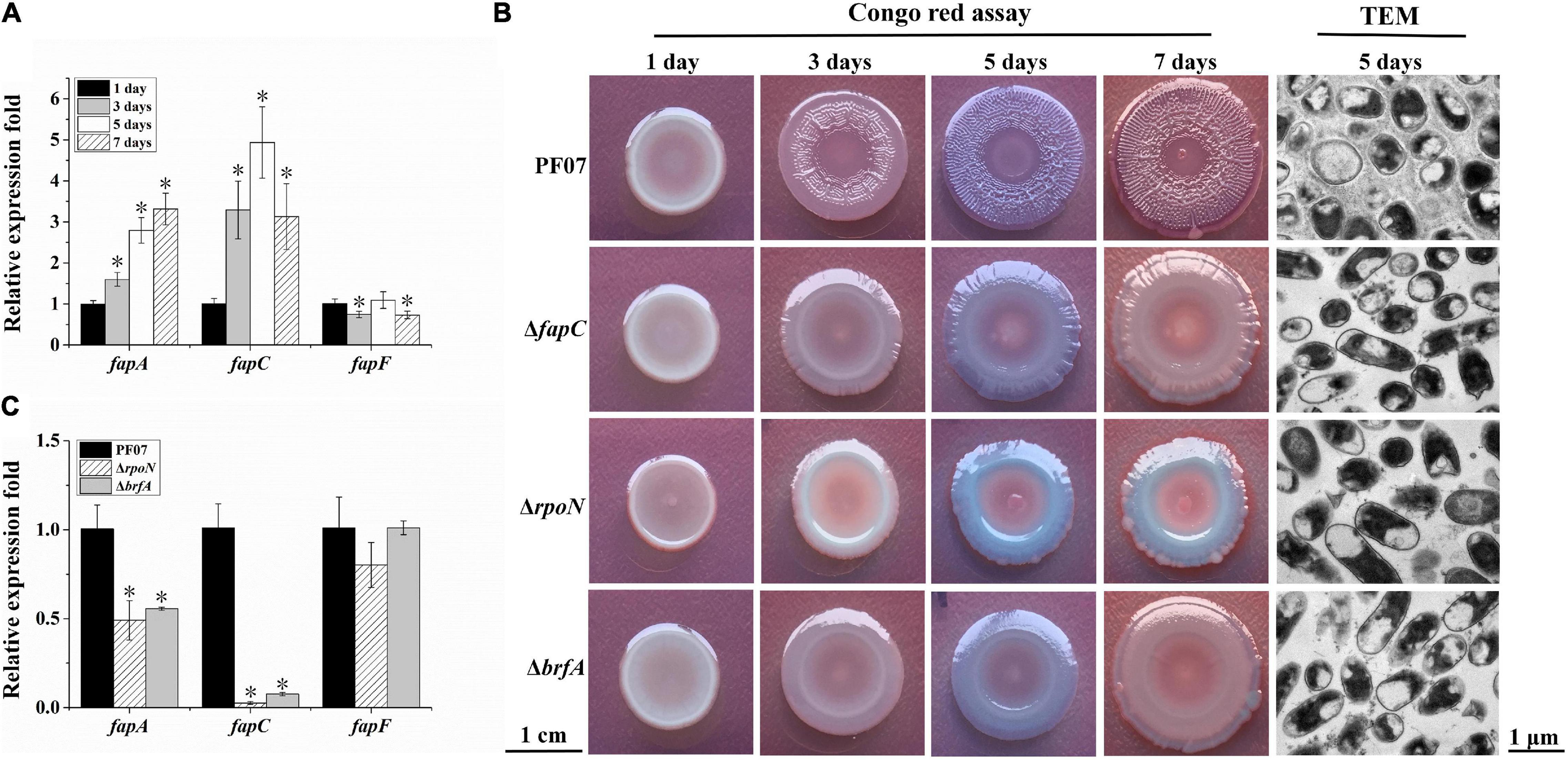
Figure 3. Expression analyses of fap genes and macrocolony biofilm observation. (A) Expression changes of fap genes over time for PF07 on tryptone plates. (B) Macrocolony biofilms observed by Congo red assay (scale bar = 1 cm) and TEM (scale bar = 1 μm). (C) Expression changes of fap genes in 5-day-old macrocolony biofilms of △rpoN and △brfA, compared to PF07. The expression levels of genes were assayed by qRT-PCR. Data are presented as the mean ± SD (n = 9). Significant differences compared to 1 day (A) or compared to PF07 (C) were analyzed using one-way ANOVA (analysis of variance), *P < 0.01.
Similar to the fapC mutant, the rpoN mutant also failed to form red and wrinkle macrocolonies, and it failed to generate an extracellular matrix in the biofilm (Figure 3B). The RNA-seq results show the cells in the macrocolony biofilm of the rpoN mutant significantly downregulated the transcription of fapA-E genes (Table 2). These results indicate that RpoN regulates the production of the biofilm matrix via regulation of the fap gene cluster.
Downstream of the fapA-F gene sequence of PF07, there is an adjacent reverse gene, brfA (D7M10_RS14495), which encodes an RpoN-dependent Fis family transcriptional regulator and is upregulated in mature biofilms (Table 2). The deletion mutant of brfA was constructed (Supplementary Figure 1), and ΔbrfA showed macrocolony biofilms similar to ΔfapC and ΔrpoN (Figure 3B). In addition, the transcription of fapA and fapC was also significantly down-regulated in ΔbrfA as in ΔrpoN (Figure 3C). These results suggest BrfA is likely to act in conjunction with RpoN to regulate the transcription of fap genes.
P. fluorescens PF07 formed robust pellicles when grown in standing culture (1% tryptone) after 3 days of growth. However, the mutants ΔfapC, ΔrpoN, and ΔbrfA grew as turbid cultures and failed to form robust pellicles, especially the rpoN mutant (Figure 4A). Thus, the three genes are required for pellicle formation by P. fluorescens PF07.
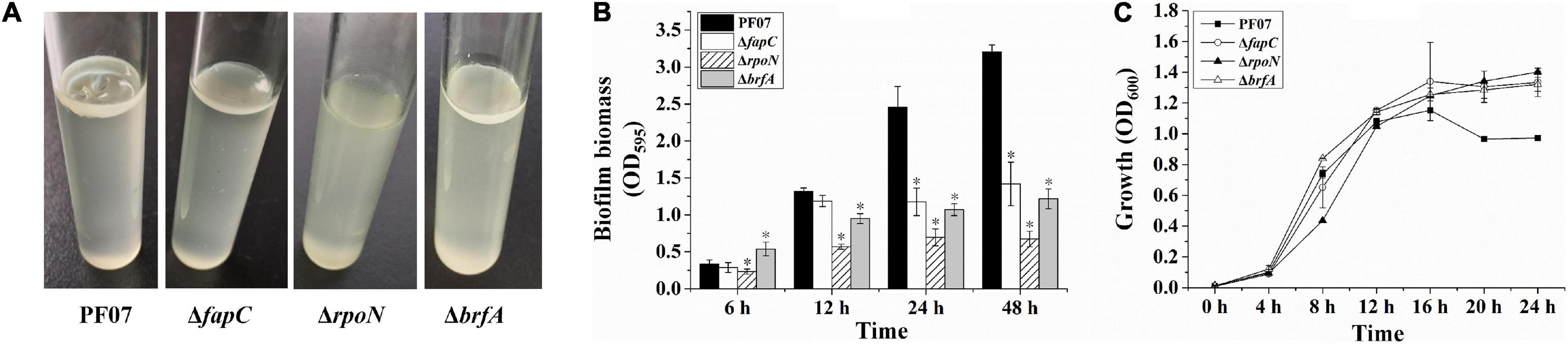
Figure 4. Pellicle and SSA biofilm formation of PF07, ΔfapC, ΔrpoN, and ΔbrfA. (A) Pellicle formed at the air–liquid interface of the standing culture. (B) Crystal violet assay for SSA biofilms in microplates. The biofilm biomass was quantified by crystal violet staining. Data are expressed as mean ± SD of eight replicates. Significant differences between PF07 and the mutants were analyzed by one-way ANOVA, *P < 0.01. (C) The growth curves of PF07 and the mutants in tryptone broth medium with shaking. Data are expressed as mean ± SD of three replicates.
The ability of the bacteria to form SSA biofilms in plastic microplates was analyzed by crystal violet staining. As shown in Figure 4B, after 6 h of growth, ΔrpoN showed a slight defect in the initial adhesion. After 12 h, the biofilm biomass of ΔrpoN and ΔbrfA was significantly reduced compared with the wild type. After 24 and 48 h, the three mutants produced significantly less biofilm than the wild type, by at least 50%. To determine whether the three mutants had growth defects, their growth curves were plotted. As shown in Figure 4C, all the strains started to enter the stationary phase after 12 h of growth. During the logarithmic growth phase (4–12 h), the growth of ΔrpoN was slower than the other strains. After 16 h, the optical density of the wild type was lower than the mutants. These results suggest that fapC, rpoN, and brfA genes are required for SSA biofilm maturation in P. fluorescens PF07. However, FapC and BrfA may not be involved in the initial adhesion process of biofilm formation.
The biofilms formed on glass coverslips were stained with both Congo red and DAPI to show the structure of the biofilm by CLSM. Congo red is frequently used for amyloid and polysaccharide staining in biofilm matrices, and the DNA stain DAPI (blue) is used to visualize the bacterial cells (Larsen et al., 2007). As shown in Figure 5, PF07 formed thick and rough biofilms covering most of the surface after 24 h of incubation. The cells with blue fluorescence were wrapped and covered by extracellular matrices with red fluorescence. Compared to PF07, ΔfapC, ΔrpoN, and ΔbrfA formed notably thinner biofilms that sparsely covered the surface, and the red fluorescent signal was significantly reduced. The CLSM results are consistent with the results of the crystal violet assay in plastic microplates. These results further confirm that Fap amyloid is the main component of the SSA biofilm matrices, and both RpoN and BrfA regulate the production of biofilm matrices by controlling the transcription of the fap genes.
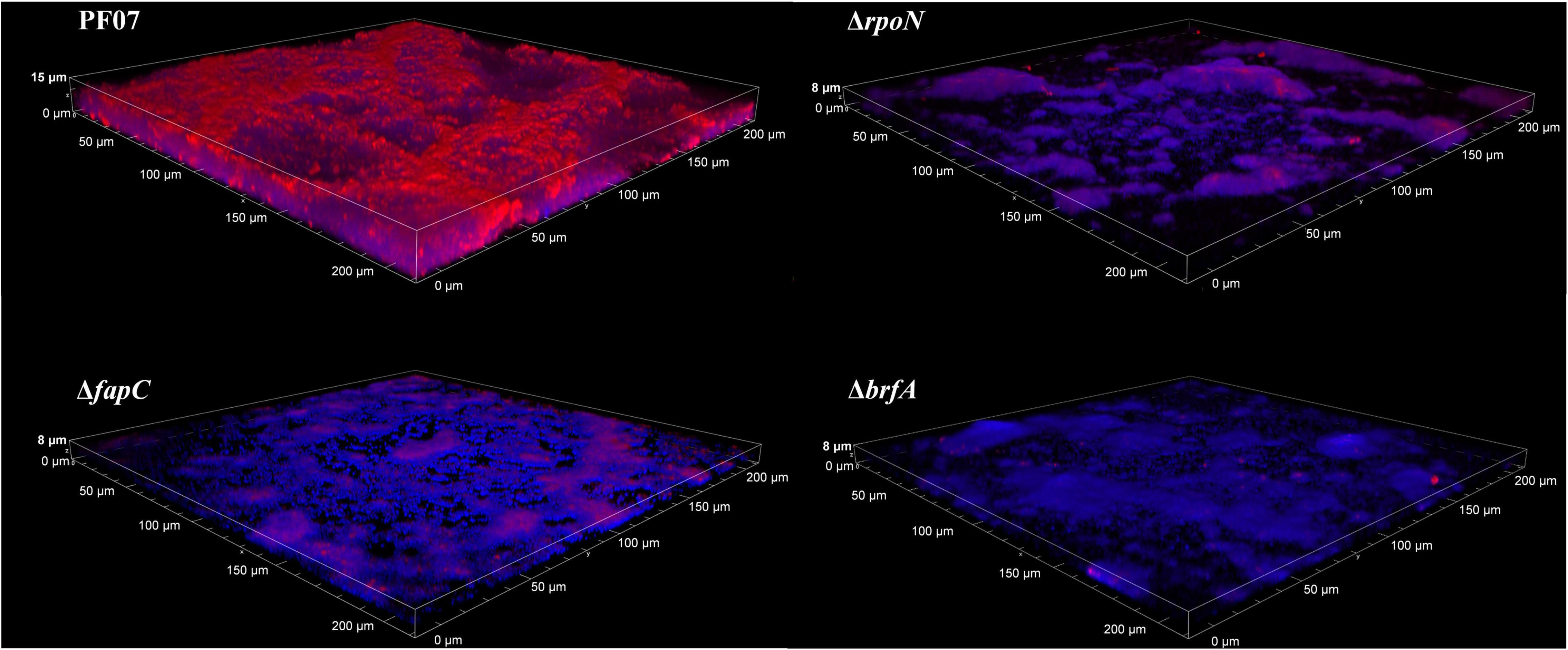
Figure 5. CLSM observation of biofilm on glass slips of PF07, ΔfapC, ΔrpoN, and ΔbrfA. The biofilms were grown along the air-liquid interface of 20-mm glass coverslips. At 24 h the coverslips were removed, washed in 0.9% NaCl, and stained with DAPI for cells (blue) and Congo red for extracellular matrix (Red). Shown are representative images of each strain.
The biofilm matrix encases the constituent cells, providing a scaffold for the biofilm architecture (Flemming and Wingender, 2010). In the present work, P. fluorescens PF07 developed wrinkled macrocolony biofilms on tryptone plates for an extended time through the high production of extracellular matrices. The genes differentially expressed during macrocolony biofilm formation may be related to extracellular matrix production. In addition, the rpoN mutant was deficient in forming wrinkled macrocolony biofilms and in producing extracellular matrices (Figure 3B). Thus, we explored the genes involved in biofilm matrix formation by RNA-seq-dependent transcriptomic analyses of macrocolony biofilm of P. fluorescens PF07 (PF07_5d vs. PF07_1d, ΔrpoN vs. PF07), and we identified several gene clusters or genes that may be associated with the formation and regulation of the biofilm matrix (Table 2).
The fap gene cluster is the most intriguing. Dueholm et al. (2010) first showed that the Fap amyloid in Pseudomonas is expressed from the gene cluster fapABCDEF. Detailed biophysical investigations of purified fibrils confirmed that FapC represents the major subunit of the mature Fap fibril, and FapB is a minor constituent. However, little is known about the expression regulation of fap genes. In the present work, the transcription of fapA-E genes was significantly induced during macrocolony biofilm formation of PF07, an isolate from spoiled fish (Table 2 and Figure 3A). In addition, we showed that the operon fapABCDE has an RpoN-dependent promoter, and RpoN may directly regulate the transcription of fap genes in PF07 (Figure 2). RpoN recognizes a characteristic –24/–12 promoter and requires an associated activator to initiate the transcription of specific genes (Bush and Dixon, 2012). The brfA gene is an adjacent reverse gene downstream of the fapA-F gene sequence of PF07, which encodes a novel RpoN-dependent Fis family transcription regulator and is upregulated in mature biofilms (Table 2). Moreover, we found the mutants ΔfapC, ΔrpoN, and ΔbrfA were defective in producing the biofilm matrixes, and the fap genes were positively regulated by BrfA (Figures 3–5). Thus, BrfA is likely to act in conjunction with RpoN to regulate the transcription of fapA-E. The regulation mode of BrfA is different from that of the master regulators CsgD in E. coli and FleQ in P. aeruginosa, which regulate the expression of biofilm matrices without RpoN (Baraquet et al., 2012; Mika and Hengge, 2014). These results suggest that the Fap amyloids are the main components of the biofilm matrices, and that RpoN and BrfA are essential regulators for the transcription of fap genes in PF07.
In this work, several DGC or PDE genes were significantly upregulated or downregulated in the PF07_5d biofilm compared to the PF07_1d biofilm, such as D7M10_RS04965, D7M10_RS06580, and D7M10_RS22095 (Table 2). The DGCs or PDEs coded by these genes may regulate the production of biofilm matrices by changing the internal levels of c-di-GMP. In P. aeruginosa PA14, there were 40 proteins predicted to produce or degrade c-di-GMP, and eight of them take part in regulating the expression of pel genes (Okegbe et al., 2017). As the regulation of the DGC and PDE genes to the biofilm matrix formation is often complicated, we intend to study these genes in depth in the future.
In a study of other biofilm matrix proteins, the presence Flp/Tad pili at the surface of bacterial cells was associated with the formation of extremely tenacious biofilms on a variety of solid surfaces in Actinobacillus actinomycetemcomitans (Bhattacharjee et al., 2001). Flagella participate in initial attachment to surfaces, migration along the surfaces, and matrix formation of biofilms in some microorganisms (Mika and Hengge, 2014; Fong and Yildiz, 2015). In addition, polysaccharides constitute a predominant fraction of the biofilm matrix, including Psl or Pel polysaccharides in P. aeruginosa (Fazli et al., 2014), as well as cellulose and PNAG polysaccharides in E. coli (Mika and Hengge, 2014). However, in this work, the genes related to Flp/Tad pili, flagella, and polysaccharides were downregulated, rather than upregulated, in the mature macrocolony biofilm of PF07 (Table 2). Therefore, these genes may not be essential for matrix formation of macrocolony biofilms. However, our results show ΔrpoN had more severe defects in forming pellicles and SSA biofilms than ΔfapC and ΔbrfA (Figure 4A). This is likely because RpoN also regulates the synthesis of flagella (Table 2), and flagella may be important to the formation of pellicles and SSA biofilms (Klausen et al., 2003; Yamamoto et al., 2012). Moreover, rhamnolipids are synthesized by rhamnosyltransferases RhlA and RhlB in P. aeruginosa (Davey et al., 2003). The synthesis of rhamnolipids is regulated by quorum sensing system RhlRI, based on acylated homoserine lactones (Pearson et al., 1997). Low concentration of rhamnolipids increases the affinity for initial adherence of cells to a surface, but the overproduction of rhamnolipids inhibits the formation of mature biofilms (Nickzad and Déziel, 2014). In this work, the expression of gene cluster rhlABRI was significantly downregulated in the mature biofilm, and it was upregulated in the rpoN mutant. This agrees with the fact that high levels of rhamnolipids are detrimental to the formation of mature biofilms.
The type II secretion (T2S) system directs the transport of a variety of proteins from Gram-negative bacteria, including virulence factors, extracellular enzymes and biofilm matrix proteins (Johnson et al., 2014; Cianciotto and White, 2017). It was reported that the T2S system delivered biofilm matrix proteins RbmA, RbmC, and Bap1 for biofilm formation in Vibrio cholera (Johnson et al., 2014). In the present work, the genes for the T2S system were strongly induced in the mature biofilm. The role of the T2S system in PF07 may be similar to that in Vibrio cholera. However, the substrates for the T2S system in PF07 need to be further defined.
In addition to biofilm matrix formation, bacterial metabolism was remodeled in adaptation to biofilm growth. According to our results, the genes related to galactonate degradation were significantly upregulated, whereas the genes related to biosynthesis and secretion of extracellular protease and the genes related to arginine degradation were significantly downregulated during the biofilm formation (Table 2). The metabolic alterations may be associated with limited availability of specific nutrients in the biofilm. However, different bacteria show different biofilm metabolism adaptation. For example, in P. aeruginosa, the genes related to anthranilate degradation were overexpressed in biofilms (Vital-Lopez et al., 2015). In B. subtilis, the activity of the tricarboxylic acid cycle increased during early pellicle formation, and a shift from fatty acid biosynthesis to fatty acid degradation was detected (Pisithkul et al., 2019).
In conclusion, the expression of a large set of genes changed significantly during macrocolony biofilm formation, mainly including the genes related to matrix proteins, polysaccharides, rhamnolipids, secretion systems, biofilm regulation, and metabolism. These genes may be involved in biofilm matrix formation directly or indirectly. In addition, we first identified the functional amyloid Fap as the main component of PF07 biofilm matrixes, and the transcription of fapA-E genes may be directly regulated by RpoN in conjunction with BrfA. These genes may serve as potential molecular targets for screening new anti-biofilm agents or for biofilm detection in food environments. For example, by qRT-PCR detection of the target genes, the agents that inhibit the transcription of these genes may be screened out, and the formation of the biofilms may be detected quickly.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
MG and ST performed the experiments. JZ, AS, and XL analyzed the data. PD revised the manuscript. XL conceived, designed the experiments, analyzed the data, and wrote the manuscript. All the authors read and approved the final version of the manuscript.
This research was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LY20C200002) and the Basic Scientific Research Funds of the Department of Education of Zhejiang Province (No. KYYB202104).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Accdon (www.accdon.com) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.881043/full#supplementary-material
ANOVA, analysis of variance; c-di-GMP, cyclic diguanylate monophosphate; CLSM, confocal laser scanning microscopy; DAPI, 4′,6-diamidino-2-phenylindole; DEGs, differentially expressed genes; DGC, diguanylate cyclase; 5′-RACE, rapid amplification of 5′-cDNA ends; LB, Luria-Bertani; PDE, phosphodiesterases; PNAG, polysaccharide poly-N-acetyl-glucosamine; qRT-PCR, quantitative reverse transcription-PCR; SSA, solid surface-associated; TEM, transmission electron microscopy; T2S, type II secretion.
Alvarez-Ordóñez, A., Coughlan, L. M., Briandet, R., and Cotter, P. D. (2019). Biofilms in food processing environments: challenges and opportunities. Annu. Rev. Food Sci. Technol. 10, 173–195. doi: 10.1146/annurev-food-032818-121805
Bagge-Ravn, D., Ng, Y., Hjelm, M., Christiansen, J. N., Johansen, C., and Gram, L. (2003). The microbial ecology of processing equipment in different fish industries-analysis of the microflora during processing and following cleaning and disinfection. Int. J. Food Microbiol. 87, 239–250. doi: 10.1016/s0168-1605(03)00067-9
Baraquet, C., Murakami, K., Parsek, M. R., and Harwood, C. S. (2012). The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218. doi: 10.1093/nar/gks384
Barrios, H., Valderrama, B., and Morett, E. (1999). Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27, 4305–4313. doi: 10.1093/nar/27.22.4305
Bhattacharjee, M. K., Kachlany, S. C., Fine, D. H., and Figurski, D. H. (2001). Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183, 5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001
Blanco-Romero, E., Garrido-Sanz, D., Rivilla, R., Redondo-Nieto, M., and Martín, M. (2020). In silico characterization and phylogenetic distribution of extracellular matrix components in the model rhizobacteria Pseudomonas fluorescens F113 and other Pseudomonads. Microorganisms 8, 1740–1764. doi: 10.3390/microorganisms8111740
Bleem, A., Christiansen, G., Madsen, D. J., Maric, H., Strømgaard, K., Bryers, J. D., et al. (2018). Protein engineering reveals mechanisms of functional amyloid formation in Pseudomonas aeruginosa biofilms. J. Mol. Biol. 430, 3751–3763. doi: 10.1016/j.jmb.2018.06.043
Bridier, A., Sanchez-Vizuete, P., Guilbaud, M., Piard, J. C., Naïtali, M., and Briandet, R. (2015). Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 45, 167–178. doi: 10.1016/j.fm.2014.04.015
Bush, M., and Dixon, R. (2012). The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529. doi: 10.1128/MMBR.00006-12
Cianciotto, N. P., and White, R. C. (2017). Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 85:e00014-17. doi: 10.1128/IAI.00014-17
Collins, A. J., Smith, T. J., Sondermann, H., and O’Toole, G. A. (2020). From input to output: the Lap/c-di-GMP biofilm regulatory circuit. Annu. Rev. Microbiol. 74, 607–631. doi: 10.1146/annurev-micro-011520-094214
Davey, M. E., Caiazza, N. C., and O’Toole, G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003
Demarre, G., Guérout, A. M., Matsumoto-Mashimo, C., Rowe-Magnus, D. A., Marlière, P., and Mazel, D. (2005). A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPa) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 156, 245–255. doi: 10.1016/j.resmic.2004.09.007
Dueholm, M. S., Petersen, S. V., Sønderkaer, M., Larsen, P., Christiansen, G., Hein, K. L., et al. (2010). Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020.
Dueholm, M. S., Søndergaard, M. T., Nilsson, M., Christiansen, G., Stensballe, A., Overgaard, M. T., et al. (2013). Expression of Fap amyloids in Pseudomonas aeruginosa. P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2, 365–382. doi: 10.1002/mbo3.81
Fazli, M., Almblad, H., Rybtke, M. L., Givskov, M., Eberl, L., and Tolker-Nielsen, T. (2014). Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16, 1961–1981.
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.3390/nano10081527
Fong, J. N. C., and Yildiz, F. H. (2015). Biofilm matrix proteins. Microbiol. Spectr. 3:10.1128/microbiolspec.MB-0004-2014.
Friedman, L., and Kolter, R. (2004). Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. doi: 10.1046/j.1365-2958.2003.03877.x
Galié, S., García-Gutiérrez, C., Miguélez, E. M., Villar, C. J., and Lombó, F. (2018). Biofilms in the food industry: health aspects and control methods. Front. Microbiol. 9:898. doi: 10.3389/fmicb.2018.00898
Hickman, J. W., and Harwood, C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389.
Jennings, L. K., Storek, K. M., Ledvina, H. E., Coulon, C., Marmont, L. S., Sadovskaya, I., et al. (2015). Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 112, 11353–11358. doi: 10.1073/pnas.1503058112
Johnson, T. L., Fong, J. C., Rule, C., Rogers, A., Yildiz, F. H., and Sandkvist, M. (2014). The Type II secretion system delivers matrix proteins for biofilm formation by Vibrio cholerae. J. Bacteriol. 196, 4245–4252. doi: 10.1128/JB.01944-14
Klausen, M., Heydorn, A., Ragas, P., Lambertsen, L., Aaes-Jørgensen, A., Molin, S., et al. (2003). Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48, 1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Langsrud, S., Moen, B., Møretrø, T., Løype, M., and Heir, E. (2016). Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon-processing plants. J. Appl. Microbiol. 120, 366–378. doi: 10.1111/jam.13013
Larsen, P., Nielsen, J. L., Dueholm, M. S., Wetzel, R., Otzen, D., and Nielsen, P. H. (2007). Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x
Liu, X., Ji, L., Wang, X., Li, J., Zhu, J., and Sun, A. (2018). Role of RpoS in stress resistance, quorum sensing and spoilage potential of Pseudomonas fluorescens. Int. J. Food Microbiol. 270, 31–38. doi: 10.1016/j.ijfoodmicro.2018.02.011
Liu, X., Xu, J., Zhu, J., Du, P., and Sun, A. (2019). Combined transcriptome and proteome analysis of RpoS regulon reveals its role in spoilage potential of Pseudomonas fluorescens. Front. Microbiol. 10:94. doi: 10.3389/fmicb.2019.00094
Liu, X., Ye, Y., Zhu, Y., Wang, L., Yuan, L., Zhu, J., et al. (2021). Involvement of RpoN in regulating motility, biofilm, resistance, and spoilage potential of Pseudomonas fluorescens. Front. Microbiol. 12:641844. doi: 10.3389/fmicb.2021.641844
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, L., Conover, M., Lu, H., Parsek, M. R., Bayles, K., and Wozniak, D. J. (2009). Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354. doi: 10.1371/journal.ppat.1000354
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41, e140. doi: 10.1093/nar/gkt444
Mika, F., and Hengge, R. (2014). Small RNAs in the control of RpoS. CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 11, 494–507. doi: 10.4161/rna.28867
Nickzad, A., and Déziel, E. (2014). The involvement of rhamnolipids in microbial cell adhesion and biofilm development - an approach for control? Lett. Appl. Microbiol. 58, 447–453. doi: 10.1111/lam.12211
Nogarol, C., Acutis, P. L., Bianchi, D. M., Maurella, C., Peletto, S., Gallina, S., et al. (2013). Molecular characterization of Pseudomonas fluorescens isolates involved in the Italian “blue mozzarella” event. J. Food Prot. 76, 500–504. doi: 10.4315/0362-028X.JFP-12-312
Okegbe, C., Fields, B. L., Cole, S. J., Beierschmitt, C., Morgan, C. J., Price-Whelan, A., et al. (2017). Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc. Natl. Acad. Sci. U.S.A. 114, E5236–E5245. doi: 10.1073/pnas.1700264114
Pearson, J. P., Pesci, E. C., and Iglewski, B. H. (1997). Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179, 5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997
Pisithkul, T., Schroeder, J. W., Trujillo, E. A., Yeesin, P., Stevenson, D. M., Chaiamarit, T., et al. (2019). Metabolic remodeling during biofilm development of Bacillus subtilis. mBio 10:e00623-19.
Puga, C. H., Dahdouh, E., SanJose, C., and Orgaz, B. (2018). Listeria monocytogenes colonizes Pseudomonas fluorescens biofilms and induces matrix over-production. Front. Microbiol. 9:1706. doi: 10.3389/fmicb.2018.01706
Remenant, B., Jaffrès, E., Dousset, X., Pilet, M. F., and Zagorec, M. (2015). Bacterial spoilers of food: behavior, fitness and functional properties. Food Microbiol. 45, 45–53. doi: 10.1016/j.fm.2014.03.009
Serra, D. O., and Hengge, R. (2014). Stress responses go three dimensional - the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 16, 1455–1471. doi: 10.1111/1462-2920.12483
Serra, D. O., Richter, A. M., Klauck, G., Mika, F., and Hengge, R. (2013). Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13
Spiers, A. J., Bohannon, J., Gehrig, S. M., and Rainey, P. B. (2003). Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50, 15–27. doi: 10.1046/j.1365-2958.2003.03670.x
Vital-Lopez, F. G., Reifman, J., and Wallqvist, A. (2015). Biofilm formation mechanisms of Pseudomonas aeruginosa predicted via genome-scale kinetic models of bacterial metabolism. PLoS Comput. Biol. 11:e1004452. doi: 10.1371/journal.pcbi.1004452
Wang, H., Cai, L., Li, Y., Xu, X., and Zhou, G. (2018b). Biofilm formation by meat-borne Pseudomonas fluorescens on stainless steel and its resistance to disinfectants. Food Control 91, 397–403.
Wang, G., Ma, F., Chen, X., Han, Y., Wang, H., Xu, X., et al. (2018a). Transcriptome analysis of the global response of Pseudomonas fragi NMC25 to modified atmosphere packaging stress. Front. Microbiol. 9:1277. doi: 10.3389/fmicb.2018.01277
Wang, Y., Feng, L., Lu, H., Zhu, J., Kumar, V., and Liu, X. (2021). Transcriptomic analysis of the food spoilers Pseudomonas fluorescens reveals the antibiofilm of carvacrol by interference with intracellular signaling processes. Food Control 127:108115.
Wang, Y., Hong, X., Liu, J., Zhu, J., and Chen, J. (2020). Interactions between fish isolates Pseudomonas fluorescens and Staphylococcus aureus in dual-species biofilms and sensitivity to carvacrol. Food Microbiol. 91:103506. doi: 10.1016/j.fm.2020.103506
Xu, J., Li, Q., Zhang, J., Li, X., and Sun, T. (2021). In silico structural and functional analysis of cold shock proteins in Pseudomonas fluorescens PF08 from marine fish. J. Food Prot. 84, 1446–1454. doi: 10.4315/JFP-21-044
Xu, T., Yu, M., Liu, J., Lin, H., Liang, J., and Zhang, X. H. (2019). Role of RpoN from Labrenzia aggregata LZB033 (Rhodobacteraceae) in formation of flagella and biofilms, motility, and environmental adaptation. Appl. Environ. Microbiol. 85:e02844-18.
Yamamoto, K., Arai, H., Ishii, M., and Igarashi, Y. (2012). Involvement of flagella-driven motility and pili in Pseudomonas aeruginosa colonization at the air-liquid interface. Microbes Environ. 27, 320–323. doi: 10.1264/jsme2.me11322
Zeng, G., Vad, B. S., Dueholm, M. S., Christiansen, G., Nilsson, M., Tolker-Nielsen, T., et al. (2015). Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 6:1099. doi: 10.3389/fmicb.2015.01099
Keywords: Pseudomonas fluorescens, biofilm, extracellular matrix, food spoiler, functional amyloid, RpoN, BrfA
Citation: Guo M, Tan S, Zhu J, Sun A, Du P and Liu X (2022) Genes Involved in Biofilm Matrix Formation of the Food Spoiler Pseudomonas fluorescens PF07. Front. Microbiol. 13:881043. doi: 10.3389/fmicb.2022.881043
Received: 22 February 2022; Accepted: 17 May 2022;
Published: 06 June 2022.
Edited by:
Avelino Alvarez-Ordóñez, Universidad de León, SpainReviewed by:
Alexa Elena Alexandra, Teagasc Food Research Centre, Teagasc, IrelandCopyright © 2022 Guo, Tan, Zhu, Sun, Du and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxiang Liu, bGl1eGlhb3hpYW5nNDEzQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.