- Tissue Culture Laboratory, School of Biotechnology, Shri Mata Vaishno Devi University, Katra, India
Endophytic bacteria isolated from medicinal plants are crucial for the production of antimicrobial agents since they are capable of possessing bioactive compounds with diverse structures and activities. Cordia dichotoma, a plant of medicinal importance native to the Jammu region of India, was selected for the isolation and characterization of culturable endophytic bacteria and evaluation of their antimicrobial activities. Standardized surface sterilization methods were employed to isolate thirty-three phenotypically distinguishable endophytic bacteria from the root, stem, and leaf parts of the plant. Shannon Wiener diversity index clearly divulged diverse endophytes in roots (0.85), stem (0.61), and leaf (0.54) tissues. Physio-biochemical features of the isolates differentiated the distinct variations in their carbohydrate utilization profile and NaCl tolerance. The endophytes produced an array of enzymes, namely, catalase, oxidase, amylase, cellulase, nitrate reductase, and lipase. The bacterial isolates belong to the genera Bacillus, Pseudomonas, Paenibacillus, Acidomonas, Streptococcus, Ralstonia, Micrococcus, Staphylococcus, and Alcalignes predominantly. However, the antibiotic susceptibility pattern indicated that the isolates were mostly sensitive to erythromycin and streptomycin, while they were resistant to rifampicin, amoxicillin, and bacitracin. Interestingly, majority of the bacterial endophytes of C. dichotoma showed antimicrobial activity against Bacillus subtilis followed by Klebsiella pneumoniae. The 16S rRNA sequence of Bacillus thuringiensis has been deposited in the NCBI GenBank database under accession number OM320575. The major compounds of the crude extract derived from endophytic B. thuringiensis OM320575, according to the metabolic profile examination by GC-MS, are dibutyl phthalate, eicosane, tetrapentacontane, heneicosane, and hexadecane, which possessed antibacterial activities. In conclusion, results indicated the potential of C. dichotoma to host a plethora of bacterial endophytes that produce therapeutic bioactive metabolites.
Introduction
Global health problems due to the evolution of defiance to accessible antibiotics by pathogenic fungi and bacteria, the inefficacy of present antifungal and antibacterial agents to various fungal and bacterial infections, and the emergence of life-threatening viruses require a critical need to search for novel and effective antimicrobial agents (Monowar et al., 2018). Numerous factors are responsible for antibiotic resistance, such as poor hygienic conditions, inappropriate use of antibiotics, late diagnosis of infections, and immunocompromised patients (Subramani et al., 2017). Medicinal plants are a source of diverse compounds that can be used for the treatment of human illnesses (Daniel et al., 2018), and due to their biological friendly nature and bioactive compounds, they are used as pharmaceutical agents (Waheed et al., 2013). Recently, natural compounds obtained from plants, fungi, and bacteria have been sourced to treat multidrug-resistant contagious pathogens singly or in combination with antibiotics (Mai et al., 2019). The use of natural antimicrobial agents is also preferred because they form protein-protein bonds during the interaction, and thus, microbes infrequently develop resistance against them (Nisa et al., 2020). Recently, search for novel curative agents has been focused toward endophytes from plant hosts because of their numerous applications of novel and interesting bioactive compounds.
Endophytes are microbes (bacteria and fungi) that colonize inner healthy plant tissues without causing them any pathogenicity (Wilson, 1995). Nearly all plants are thought to associate with endophytic microbes, yet some plant species have never been entirely studied for endophytes (Mengistu, 2020). Endophytic microbes are considered natural reservoirs due to their ability to produce myriad bioactive compounds (Gouda et al., 2016). Valuable bioactive metabolites, such as alkaloids, steroids, terpenoids, lactones, quinines, lignans, and phenols, have been isolated from endophytic fungi and bacteria (Deshmukh et al., 2015). Endophytic bacteria from medicinal plants have also been considered for their antimicrobial activities (Wang et al., 2019; Xu et al., 2020). Bacterial endophytes could also produce metabolites alike or with additional prominent activity than that of their respective hosts (Venieraki et al., 2017).
Bacterial endophytes are thought to have a symbiotic relationship with plants. In this mutualistic relationship, plants give shelter and nutrients to the endophytes (Liarzi et al., 2016), while host plants are protected from herbivores and pathogens (Qin et al., 2017). Additionally, by producing phytohormones, endophytic bacteria also promote plant growth, thus enhancing their resistance to various abiotic stresses, i.e., heavy metal toxicity and salinity (Khan et al., 2014), and they can be used in agriculture, industry, and medicine (Ryan et al., 2008). Studies on the isolation of bioactive products from bacterial endophytes can help in the discovery of several new compounds that can also be developed as antimicrobial agents to manage antibiotic resistance.
Cordia dichotoma, commonly known as Lasura, is the main plant with ethnobotanical importance belonging to the family Boraginaceae and is widespread in different parts of India. Its bark paste is useful to treat stomach disorders and to relieve chest pain. Traditionally, all parts of the plant are used to treat various illnesses, such as wound healing, antiulcer, antihelmintic, urinary infections, analgesic, antitumor, antifertility, antimicrobial, dysentery, dyspepsia, cough, and jaundice (Ragasa et al., 2015; Kumari et al., 2016). Due to its diverse therapeutic uses, this plant was selected to study its related bacterial endophytes and to screen the strains for their antimicrobial potential against a panel of clinically serious human pathogens. This plant has not been estimated for the isolation of bacterial endophytes and their activities. In this study, we estimated the diversity and antibacterial potential of bacterial endophytes colonizing C. dichotoma.
Materials and Methods
Collection of Plant Sample
Healthy plant parts, i.e., root, stem, and leaf, were collected from three individual plants of C. dichotoma growing in the Herbal Garden of Shri Mata Vaishno Devi University, Katra, Jammu and Kashmir (32.9418°N and 74.9541°E, elevation 754 m), India, in sterile polythene bags and brought immediately to the laboratory and processed to reduce the risk of contamination. The specimen was deposited at the Department of Botany, University of Jammu, with accession number 16613.
Isolation of Endophytic Bacteria
Surface Sterilization
The collected plant samples (i.e., root, stem, and leaf) were washed under running tap water to remove the dust and debris present on the surface, followed by washing in distilled water. They were carefully excised and exposed to four different reported surface sterilization methods. In Method I, samples were surface sterilized with 70% ethanol for 8 min (Tian et al., 2007). In Method II, samples were immersed in 70% ethanol for 2 min, then washed with 1% sodium hypochlorite for 3 min, and finally rinsed with 70% ethanol for 1 min (Khanam and Chandra, 2017). In Method III, samples were surface sterilized with 0.1% mercuric chloride for 2 min, 70% ethanol for 1 min, and 2% sodium hypochlorite for 5 min (Baldan et al., 2014). In Method IV, samples were immersed in Tween 20 (0.1% in 100 ml sterile distilled water), then washed with 4% sodium hypochlorite for 10 min, and finally rinsed with 70% ethanol for 3 min (Ramalashmi et al., 2018). After each step, all plant samples were washed with sterile distilled water, and each method was carried out in triplicates.
Sterility Test
Plant samples were washed in sterile distilled water at every step of the surface sterilization process. To assess the effectiveness of the surface sterilization method, a sterility check was performed for every sample. For this surface sterilization, plant samples were placed on nutrient agar, incubated at 37°C for 48 h, and checked for feasible microbial growth. Plant samples were used for further experimentation only if no growth was observed (Hallmann et al., 2006).
Isolation, Purification, and Preservation
The surface-sterilized plant segments were cut into approximately 6 mm diameter disks under aseptic conditions and placed on nutrient agar plates, and they were incubated for 48 h at 37°C in an inverted position. After 48 h, the plates were observed for bacterial growth surrounding the root, stem, and leaf sections. Endophytic bacteria emerging from the surface of plant segments were collected using an inoculation needle and further subcultured to obtain pure colonies. Single colonies acquired were streaked on fresh nutrient agar plates for further purification. These pure colonies were preserved in 50% glycerol stock and used for further experimental procedures.
Characterization of Bacterial Isolates
All selected bacterial isolates were identified through morphological characteristics, such as colony color, margin, consistency, and texture, and microscopic characterization, such as gram staining, endospore staining, and motility. The biochemical and physiological tests like indole utilization, methyl red, and sugar utilization were performed according to standard procedures (Pielou, 1975; Smibert and Krieg, 1994).
Enzyme Activity
The endophytic bacterial strains were screened for the presence of several enzyme activities, such as catalase, oxidase, amylase, cellulase, and lipase. The monitoring was done by streaking the selected endophytic bacteria on culture agar media with the substrate (Table 1 in Supplementary Material). For lipase, cellulase, and amylase activities, standard substrates like tributyrin, carboxymethyl cellulose, and starch were used, respectively. Enzyme activity was observed by flooding the plates with Congo red for cellulase and iodine reagent for amylase, and the presence of clear zones was confirmed for lipase. Catalase and oxidase activities of the bacterial isolates were determined by adding hydrogen peroxide (H2O2) and oxidase reagent (1% tetramethyl-p-phenylenediamine dihydrochloride), respectively, on the smear of freshly grown endophytic bacterial cultures in nutrient agar plates. The presence of oxygen bubbles and purple color, respectively, was considered as positive.
Salt Tolerance
To examine the effect of sodium chloride on the growth of endophytic bacterial isolates, a nutrient agar medium was prepared by adding different concentrations of NaCl (0–12%). Endophytic bacterial strains were inoculated into the plates and incubated at 37°C for 6 days, and observations were recorded every 24 h.
Antibacterial Evaluation
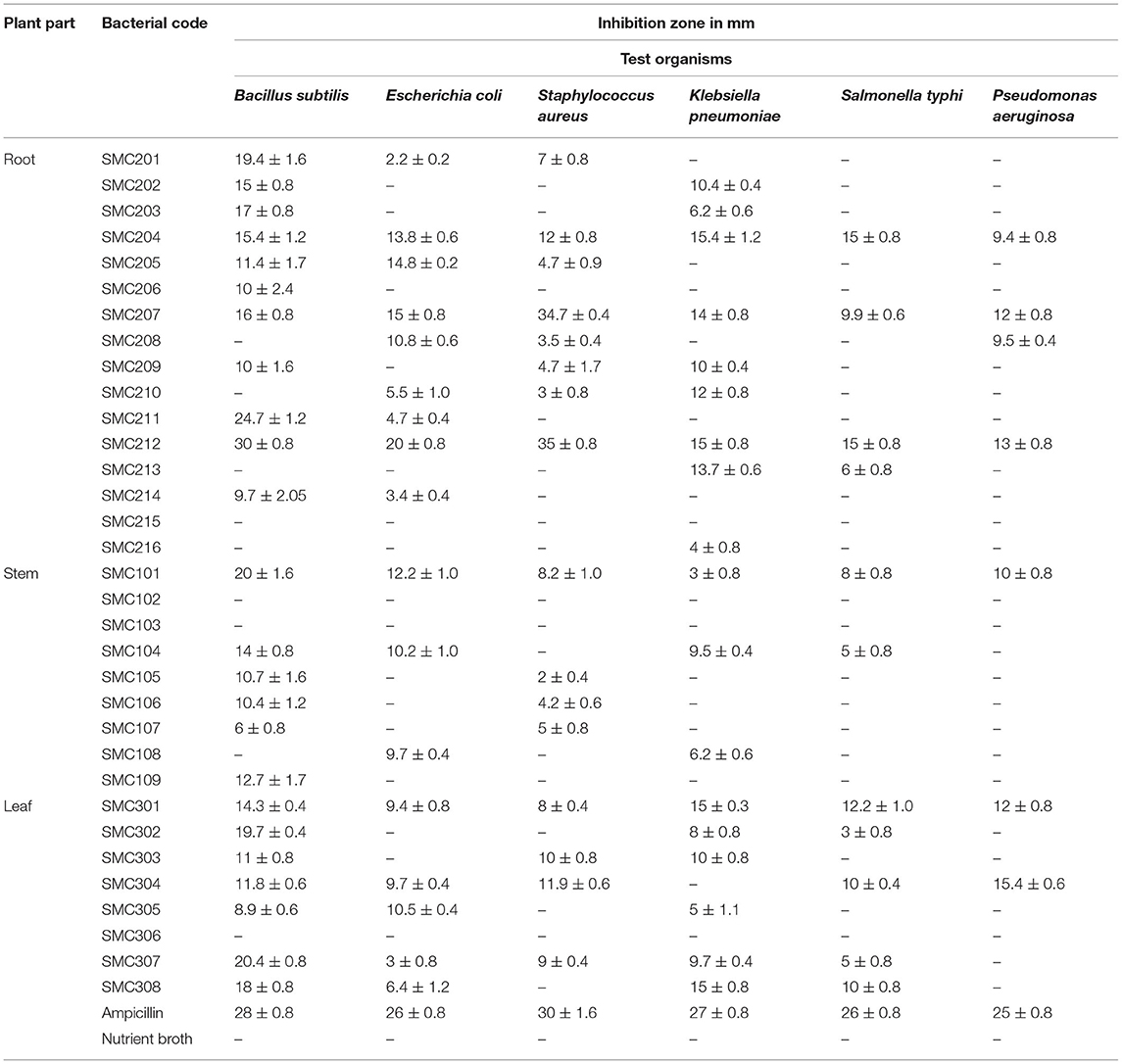
All endophytic bacterial strains were grown in nutrient broth medium for 5 days at 37°C, and antimicrobial activity was determined by agar diffusion method against six pathogenic bacteria, namely, Staphylococcus aureus MTCC 737, Escherichia coli MTCC 1687, Bacillus subtilis MTCC 1789, Pseudomonas aeruginosa MTCC 1688, Klebsiella pneumonia MTCC 432, and Salmonella typhi MTCC 733. Each endophytic bacterial culture (10 ml) was centrifuged at 1,000 rpm for 10 min and the supernatant was used to determine antibacterial activity. Overnight grown cultures of the test organisms were spread by sterile cotton swabs onto the surface of the Muller Hinton agar plates. Wells (6 mm) were made and 100 μl of endophytic bacterial strain was added into it; an equal volume of sterile nutrient broth instead of bacterial endophyte was used as a negative control, and ampicillin was used as a positive control. All plates were wrapped with parafilm and incubated for 24 h at 37°C and observed for the zone of inhibition of pathogenic bacteria. Antibacterial activity was evaluated by measuring the diameter of the clear zone of inhibition (Mohamad et al., 2018).
Antibiotic Susceptibility Assay
Antibiotic susceptibility test of endophytic bacterial strains was performed by adopting the Kirby-Bauer disk diffusion method (Bauer, 1966). Overnight nutrient broth culture of endophytic bacteria was prepared, and all the endophytic bacterial isolates were inoculated into Muller Hinton agar plates using a sterile cotton swab. Standard antibiotic disks, such as ampicillin (10 μg/disk), streptomycin (30 μg/disk), neomycin (30 μg/disk), chloramphenicol (30 μg/disk), bacitracin (10 U/disk), rifampicin (5 μg/disk), erythromycin (15 μg/disk), kanamycin (30 μg/disk), amoxycillin (30 μg/disk), and tetracycline (30 μg/disk), were placed on Muller Hinton agar plates and incubated 37°C for 24 h. After incubation, the antibiotic susceptibility pattern was determined by measuring the inhibition zone. Based on the diameter of the zone of inhibition recorded to the nearest mm, the organisms were designated as sensitive, intermediate, and resistant following DIFCO Manual 10th edition (1984).
Molecular Identification of Endophytic Bacterial Isolate
Bacterial isolates with high antibacterial activity were chosen for molecular identification using 16S rDNA sequence analysis. A loopful of freshly grown bacterial cells was dissolved in tubes with 500 μl of Tris-EDTA for DNA extraction. Then, 500 μl of phenol: chloroform: isoamyl alcohol (25:24:1) was added. This mixture was vortexed and centrifuged at 14,000 g for 5 min. The upper aqueous phase was transferred to a fresh tube, and 800 μl of chilled isopropanol was added. The samples were centrifuged again for 5 min at 14,000 g, and the supernatant was discarded. The pellets were dried at room temperature overnight before being eluted in 70 μl of TE buffer. A NanoDrop spectrophotometer was used to measure the absorbance at 260 nm to determine the concentration of genomic DNA. The primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used to amplify the 16S rRNA gene. Total DNA (50–500 ng) was added to the PCR mix (30 μl), which contained 24 μl of DreamTaq PCR master mixture, 1 μl of 10 nm each primer, 2 μl of DNA, and 7.5 μl of 10X buffer in a total volume of 30 μl. This reaction was performed under the following conditions: one cycle of 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, and a final extension of 10 min at 72°C. The amplified DNA products were separated and visualized under UV light using agarose gel electrophoresis. The amplicons were purified using the Genei PureTM quick PCR purification kit and quantified using a spectrophotometer at 260 nm. Biologia Research India sequenced the purified partial 16S rDNA amplicons. The sequences were assembled, edited, and aligned in MEGA11 before comparison with those in the GenBank database with the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine the sequence homology with closely related organisms. In this study, the microorganisms with the highest level of identity (100%) were chosen as the closest match, and isolated bacteria were classified to the species level based on the information of the closest microbes.
Estimation of Chemical Components Using GC-MS Analysis
To analyze the various volatile bioactive constituents, an ethyl acetate extract of SMC212 was subjected to gas chromatography-mass spectrometry (GC-MS). The Indian Institute of Technology, Jammu, conducted the GC-MS analysis. SHIMADZU, QP2010 PLUS was used for GC-MS analysis. The injecting temperature was set to 250°C, the column temperature was set to 50°C, the pressure was set to 29.7 kPa, and the column flow rate was set to 0.72 ml/min. The sample's total running time was 40 min. The phytochemical compounds in ethyl acetate extract were identified based on retention time by matching MS with available standards using the Willey and NIST libraries. The constituents' names, molecular formulas, and molecular weights were determined.
Statistical Analysis
Colonization rate was determined as the total number of plant segments colonized by bacteria divided by the total number of incubated samples. Isolation rate was calculated as the number of bacterial strains obtained from plant segments divided by the total number of incubated segments. The Shannon Weiner diversity index (H /) was calculated as:
where Pi is the relative abundance of species and i contributes to the total number of species (Di Bitetti, 2000). All of the experiments were carried out in triplicate, and the data reported are the averages of the results. The SPSS-22 statistical software (SPSS, Inc., Chicago, IL, USA) was used to calculate the means and standard deviation.
Results
Efficiency of Surface Sterilization Methods
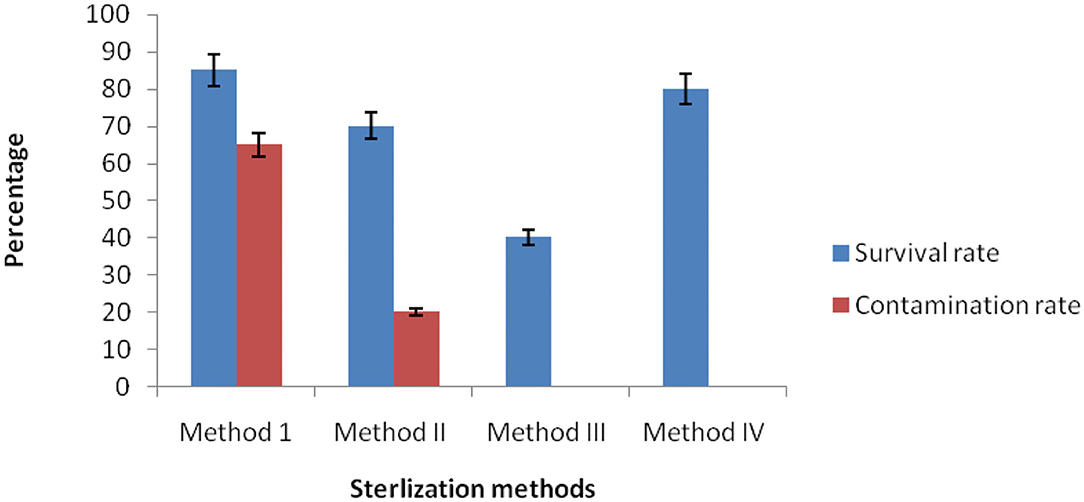
To get pure endophytes from the inner plant tissues of C. dichotoma, epiphytic microorganisms and other contamination must be removed or killed through the surface sterilization method. For this, plant samples (i.e., root, stem, and leaf) were treated separately by a different amalgam of chemical disinfectants. Method I (70% ethanol) was not found productive individually as a high percentage of contamination was noticed along with the growth of endophytes. While in Methods II, III, and IV, plant samples were treated with different mixtures and duration of sodium hypochlorite, ethanol, and mercuric chloride to attain a sufficient result. In Method III, mercuric chloride was successful in removing the contamination, yet the survival percentage of endophytic bacteria declined. Our result shows that only Method IV (0.1% Tween 20, 4% sodium hypochlorite for 10 min, and 70% ethanol for 3 min) was found effective for surface sterilization of C. dichotoma plant tissues, with no contamination and a high percentage of survival rate. Reports are indicative of surface sterilization methods used in dye-yielding plants for the isolation of endophytic bacteria with 70% ethanol for 6–8 min and 0.1% mercuric chloride for 5–10 min showing promising results (Khanam and Chandra, 2017). It was found that surface sterilization by 2% sodium hypochlorite and 0.1% Tween 20 for 3 min followed by ethanol 70% for 1 min was well-suited for the isolation of endophytic microorganisms from Acalypha indica (Ramalashmi et al., 2018). The results for optimization of surface sterilization are shown in Figure 1.
Isolation of Endophytic Bacteria
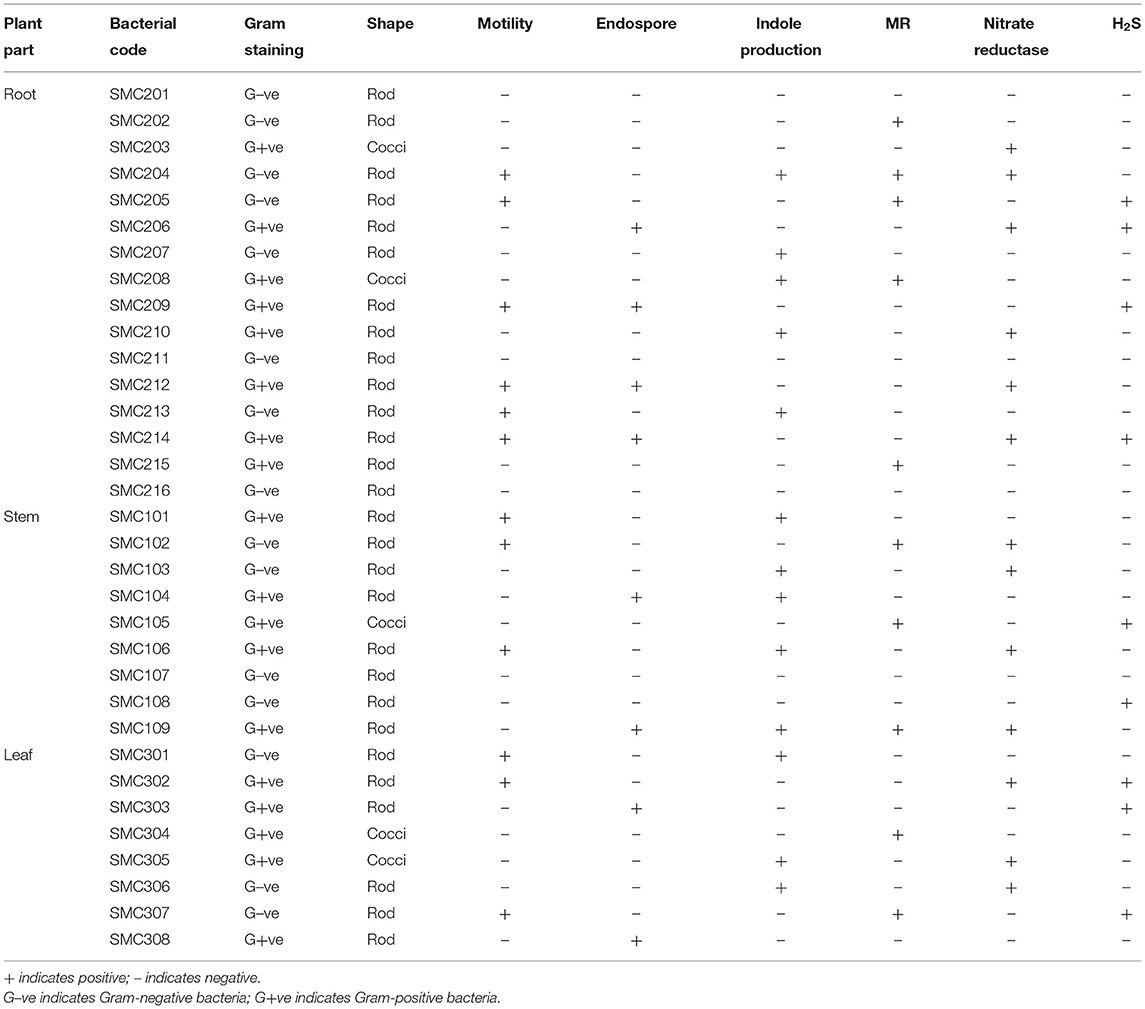
Segments of surface-sterilized root, stem, and leaf of C. dichotoma incubated on nutrient agar plates showed growth of morphologically distinct bacterial colonies surrounding the plant segments after 48–96 h. To avoid the collection of repetitive strains, a total of 33 endophytic bacteria were isolated in pure form from 170 segments (i.e., 62 root, 54 stem, and 54 leaf segments) of C. dichotoma of which 16, 9, and 8 isolates were obtained from root, stem, and leaf segments, respectively (Table 1; Figure 2 in Supplementary Material). Colonization frequency calculated in percentage was highest in leaf samples (37.03) but gradually decreased in stem (35.18) and root (32.25) samples, while the isolation rate was lower in leaf (0.14) as compared to stem (0.16) and root (0.25) samples. The Shannon Wiener diversity index showed that roots (0.85) of C. dichotoma harbor diverse types of endophytic bacteria as compared with stem (0.61) and leaf (0.54) which are in line with the available reports for Chinese medicinal plants Berberis poiretii, Eucommia ulmoides, and Rhus potanini showing a high colonization frequency (47–63%) and isolation rates (0.7–0.9) of endophytic fungi (Sun et al., 2008).
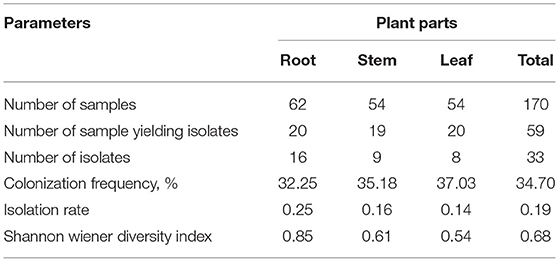
Table 1. Diversity of endophytic bacterial isolates in root, stem, and leaf tissues of C. dichotoma.
Characterization of Endophytic Bacteria
The bacterial endophytes of C. dichotoma were characterized based on morphological (Table 3 in Supplementary Material), microscopical, and biochemical features (Table 2); evaluation of enzymatic activity; and carbohydrate utilization (Table 3). Cultural characteristics of all the isolated endophytic bacteria showed that out of 33 isolates, 18 pigmented and 15 non-pigmented organisms were identified. Microscopic characteristics of all endophytic bacteria showed that out of 33 isolates, 18 were Gram-positive (5 cocci and 13 rods) and 15 were Gram-negative rods. Filamentous forms were not observed in either root, stem or leaf samples. Twelve isolates out of 33 were motile, and 8 isolates showed endospore formation, and they were suspected as Bacillus species. Enzyme profile of endophytic bacterial isolates showed that all endophytic bacteria produced oxidase, and 87% of them produced catalase. The isolates also showed lipase (69%), amylase (78%), cellulase (87%), nitrate reductase (39%), methyl red (30%), and indole (39%) production. The isolated endophytes were also screened for their ability to utilize carbohydrates, i.e., dextrose, fructose, sucrose, maltose, and lactose, in phenol red agar medium supplemented with 1% sugar. Dextrose was found to be the best-utilized carbon source by most of the bacterial isolates; bacterial isolates were moderate in fermenting fructose and sucrose, while very few isolates were fermenting maltose (6) and lactose (7).
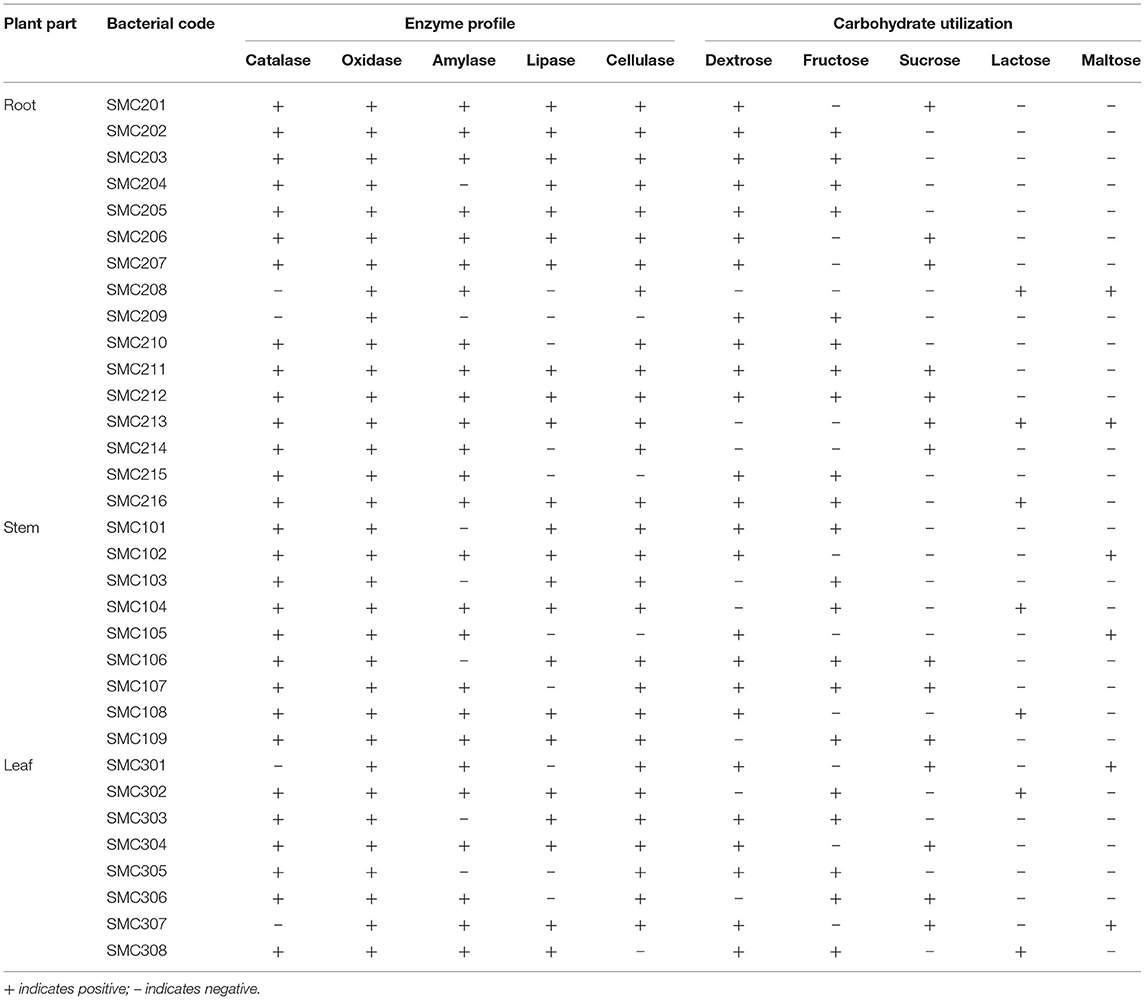
Table 3. Evaluation of enzymatic activity and carbohydrate utilization of endophytic bacteria isolated from root, stem, and leaf tissues of C. dichotoma.
Based on the micromorphological and physio-biochemical analysis, the isolated endophytic bacteria were tentatively identified as species of Bacillus, Pseudomonas, Paenibacillus, Acidomonas, Streptococcus, Ralstonia, Micrococcus, Staphylococcus, and Alcalignes. Similar endophytic bacteria have been reported from medicinal plants like Azadirachta indica, Gynura procumbens, Tephrosia apolline, Phyllanthus emblica, and Vitis vinifera (Baldan et al., 2014; Khan et al., 2014). However, several authors have reported the presence of endophytic bacteria inside plants belonging to the genera Pantoea, Bacillus, Microbacterium, Paenibacillus, and Sphingomonas (Rijavec et al., 2007).
Effect of Sodium Chloride
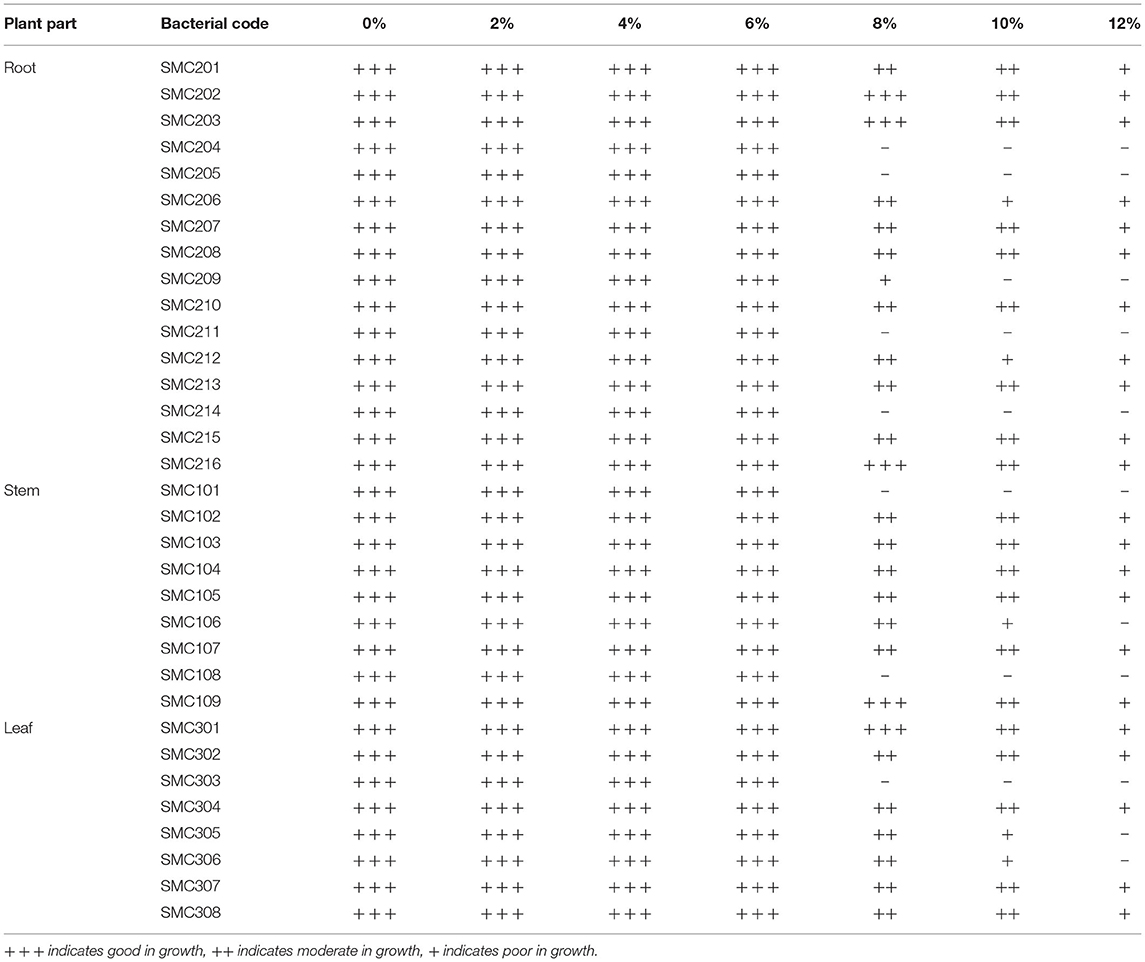
The growth of isolated endophytic bacteria at different sodium chloride concentrations (Table 4) indicates that all the 33 isolates showed good growth at 0–6% NaCl concentration. Only 22 isolates showed sustained growth at 12% NaCl concentration. Thus, it can be inferred that an increase in NaCl concentration causes a proportional decrease in growth rate.
Antibiotic Susceptibility Pattern
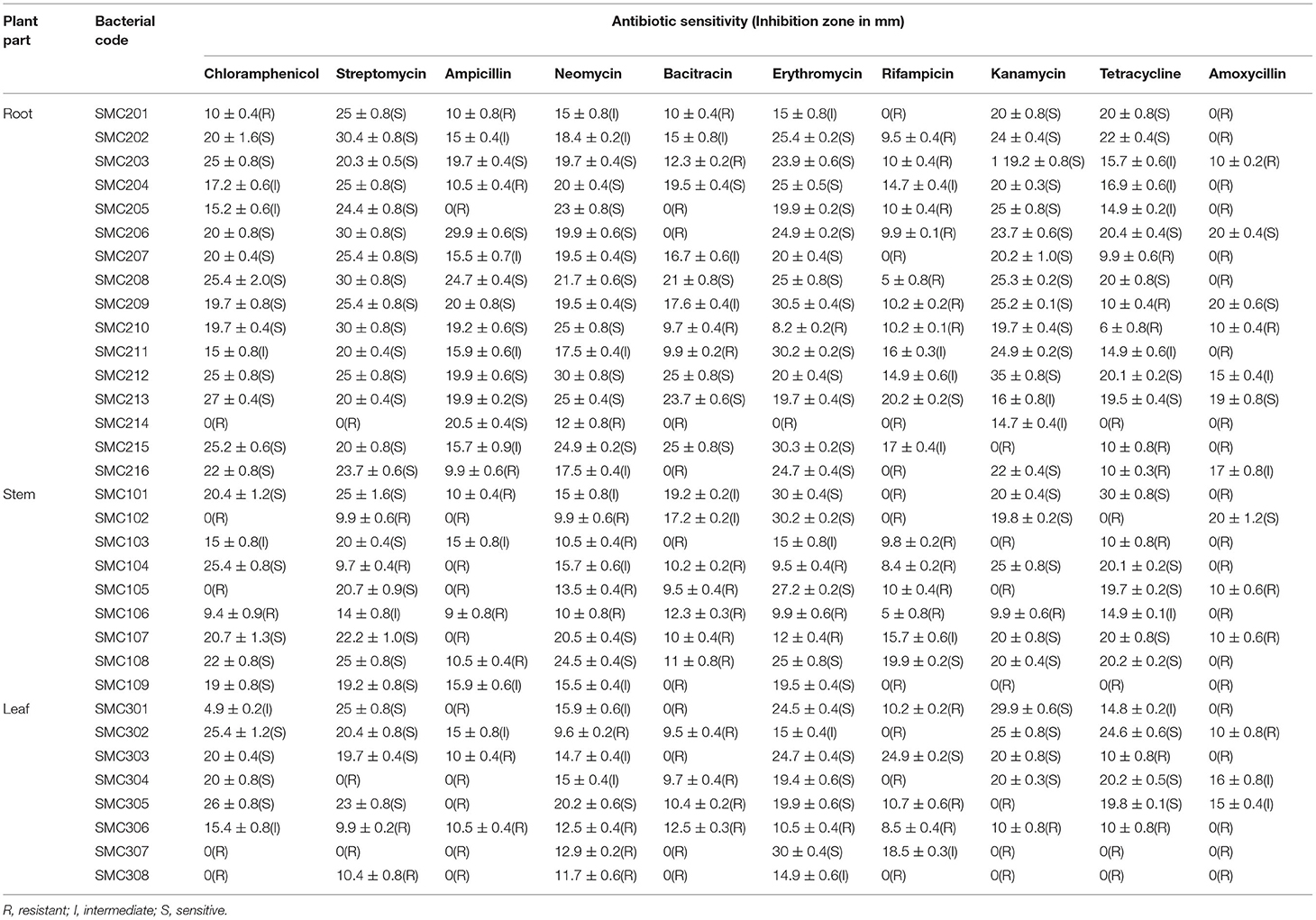
The antibiotic susceptibility pattern of the selected endophytic bacteria was determined against ten different antibiotics by the disk diffusion method, which shows that endophytic bacteria from root, stem, and leaf of C. dichotoma were mostly resistant to rifampicin, amoxicillin, and bacitracin, while they were susceptible to erythromycin and streptomycin. Out of 33 isolates, 10 and 7 bacteria showed an intermediate response to neomycin and ampicillin, respectively. Out of 33 isolates, 3 isolates were sensitive to rifampicin, 5 isolates were sensitive to amoxicillin and bacitracin, 8 isolates were sensitive to ampicillin, 14 isolates were sensitive to tetracycline and neomycin, 20 isolates were sensitive to chloramphenicol, 22 isolates were sensitive to kanamycin, 23 isolates were sensitive to erythromycin, and 25 isolates were sensitive to streptomycin. Antibiotic sensitivity patterns of isolated endophytic bacteria with zone size in mm are shown in Table 5.
Antibacterial Activity
Antibacterial properties of all isolated endophytic bacteria were assessed against clinical strains of both Gram-negative and Gram-positive bacteria. The endophytic bacteria which inhibited the growth of any of the test organisms were considered to have antibacterial activity, and the zone of inhibition length was measured in mm (Table 6). Out of 33 endophytes screened, the majority showed antibacterial activity against B. subtilis followed by K. pneumonia. Bacterial strains, i.e., SMC204, SMC212, SMC101, and SMC301, exhibited antibacterial activity among all the six test organisms. The best activity was expressed by SMC212 against S. aureus, B. subtilis, E. coli, S. typhi, K. pneumonia, and Pseudomonas aeruginosa with a zone of inhibition having a diameter of 35, 30, 20, 15, 15, and 13 mm, respectively. Although several reports showed the antimicrobial potential of endophytic fungi from medicinal plants (Kuo et al., 2021), the antimicrobial evaluation of endophytic bacteria is rare (Cardoso et al., 2020). Li et al. (2008) investigated endophytic Actinomycetes associated with medicinal plants in the rainforest of Yunnan, China, and recognized that endophytic Streptomyces exhibit antimicrobial activities against S. aureus, E. coli, and Candida albicans.
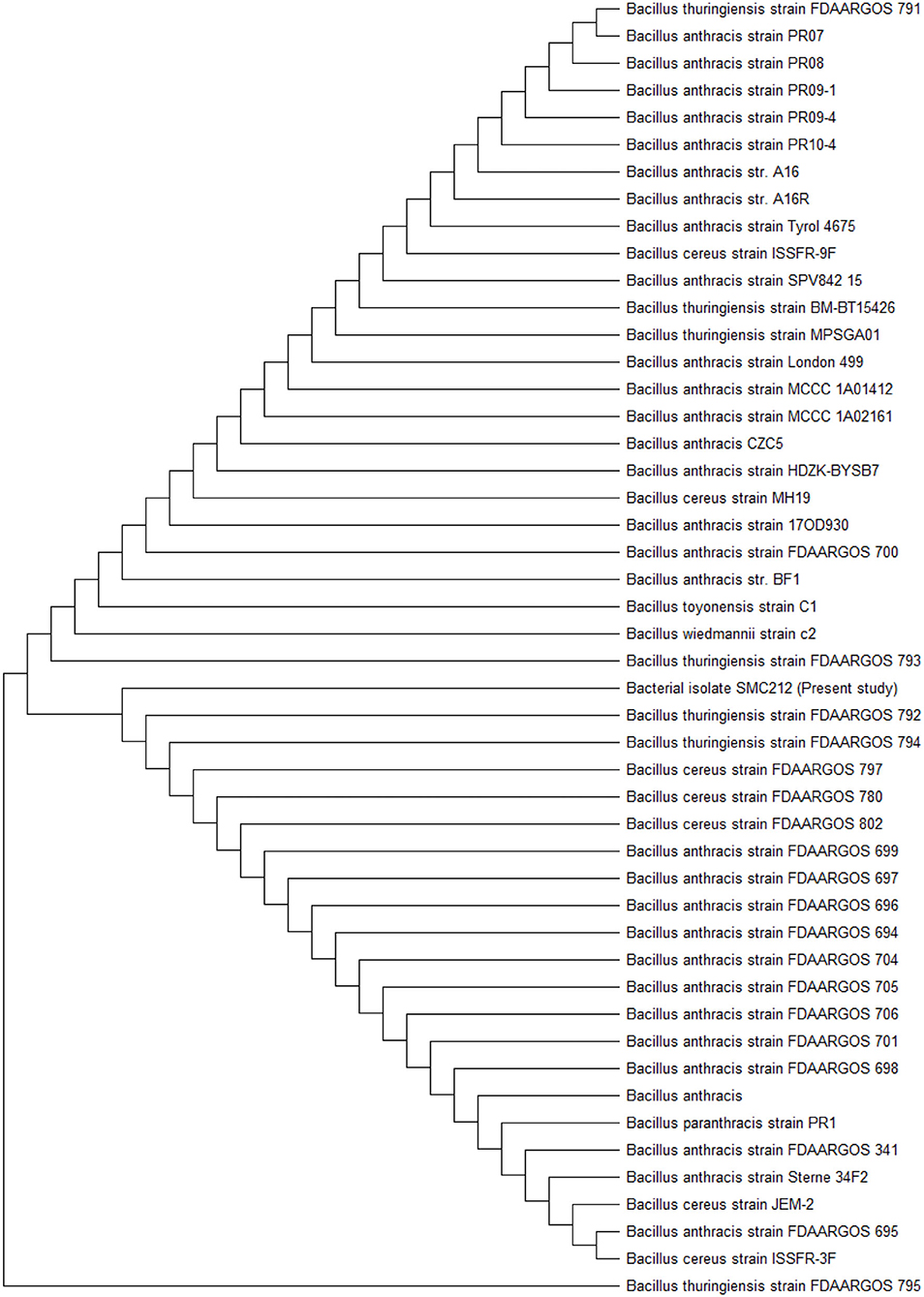
Molecular Identification of Endophytic Bacteria
The 16S rRNA gene sequencing was used to characterize the endophytic bacterium SMC212, and a phylogenetic tree was constructed using MEGA 11 software. Using MEGA 11, the maximum likelihood tree of SMC 212, constructed based on 16s rDNA gene sequences analysis, was based on the Tamura 3-parameter model with the lowest BIC and highest AIC values. All spots with gaps and missing data were removed. Pairwise deletion was used to close gaps, and the estimated transition/transversion bias (R) was 2.2. The evolutionary history was inferred by using the maximum likelihood method and the bootstrap consensus tree inferred from 1,000 to 3,000 iterations. The evolutionary relationship is represented as a dendrogram (Figure 2), which clearly shows that SMC212 is related to Bacillus thuringiensis. To the best of our knowledge, this is the first article to report the isolation of an endophytic bacterium from C. dichotoma that has been identified and showed similarity to B. thuringiensis. The sequences obtained in this study have been deposited in GenBank under accession number OM320575. Numerous previous studies investigated the diversity of bacterial endophytes in medicinal plants (Hamayun et al., 2021). Pseudomonas sp., Paenibacillus sp., and Bacillus megaterium were previously identified as Korean ginseng root endophytes in the bacterial population isolated from Plectranthus tenuiflorus (Cho et al., 2007). Paenibacillus has been discovered as an endophyte in various woody plants such as coffee, pine, and poplar (Bent and Chanway, 2002).
Chemical Constituents Using GC-MS Analysis
The results pertaining to GC-MS analysis (Figure 3) of the ethyl acetate crude extract of B. thuringiensis OM320575 were analyzed using GC-MS which led to the identification of 38 different compounds. Table 7 shows the retention time (RT), molecular formula, and molecular weight. The NIST program was used to deconvolute the GC-MS data, and the obtained mass spectra were matched to entries in the compound library. The chemical structures of these compounds are also depicted in Figure 4 in the Supplementary Material. The compounds that occupied major percentage in the extract are dibutyl phthalate (32.53%) and eicosane (13.59%) at various time intervals; tetrapentacontane (6.88%) at various time intervals; heneicosane (6.58%) at various time intervals; hexadecane (5.57%); tetradecane (5.08%); silane, trichlorooctadecyl (2.6%); and 2,4-di-tert-butylphenol (2.27%). The bacterium B. thuringiensis OM320575 produced bioactive chemicals with antimicrobial, antioxidant, and anticancer properties, according to GC-MS analyses.
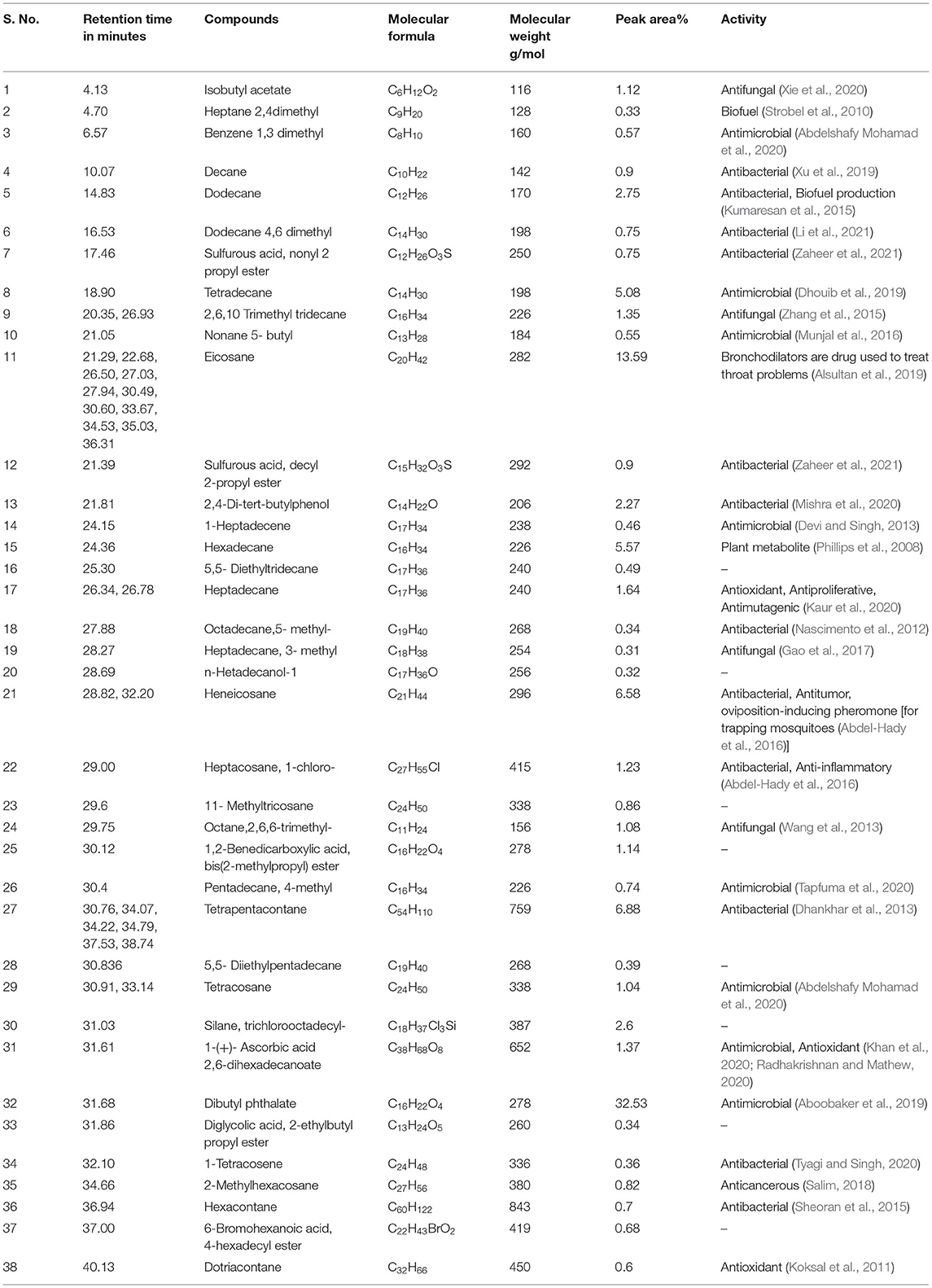
Table 7. Chemical composition of ethyl acetate extract of Bacillus thuringiensis SMC212 from GCMS analysis.
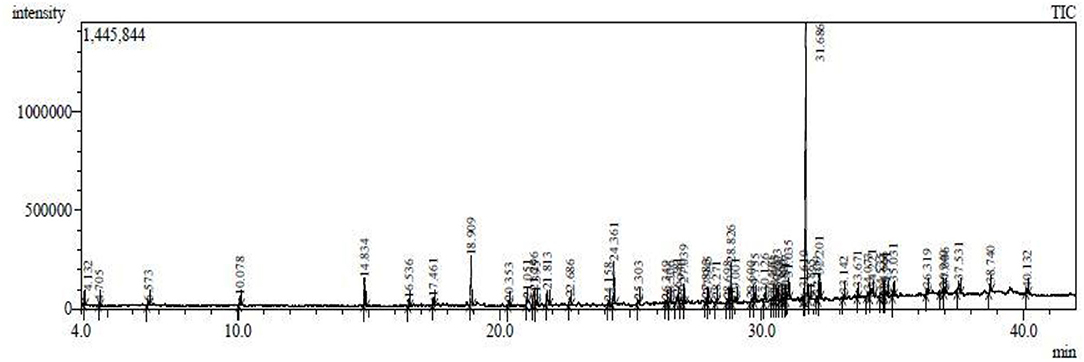
Figure 3. Abundance of the chemical compounds present in ethyl acetate extract of Bacillus thuringiensis SMC212.
Discussion
In this study, a total of 33 bacterial strains were recovered from plant segments collected from selected C. dichotoma plant parts such as root, stem, and leaf collected from SMVDU, Katra, Jammu, for the isolation of endophytic bacteria. In India, a countable number of reports showed the diversity of endophytic bacteria and fungi in medicinal plants (Praptiwi et al., 2020), but to our best of knowledge, this is the first-ever report on endophytic bacteria from C. dichotoma, particularly in Jammu region. In this study, the surface sterilization method was optimized to get maximum endophytic bacteria from the medicinal plant C. dichotoma. The population density of epiphytes or rhizospheric bacteria is more as compared with endophytic bacteria. To avoid contamination for the isolation of endophytic bacteria, plant samples must be properly surface sterilized before inoculating them into the nutrient agar medium. The simple and powerful method of surface sterilization was applied for the isolation of endophytic bacteria from the root, stem, and leaf tissues. The study found that surface sterilization of C. dichotoma medicinal plant for the isolation of endophytic bacteria using 70% ethanol was not effective to remove contamination on the plant surface. Hence, it was evaluated that 70% ethanol was not efficient in removing epiphytic bacteria. Although mercuric chloride was found to be a good decontaminating agent, surface sterilization of C. dichotoma using mercuric chloride was not found effective because the survival percentage of endophyte decreased. In our study, a high concentration of (4%) sodium hypochlorite was found to be more effective than the low concentration of (1%) in removing plant surface microorganisms. Hence, sodium hypochlorite showed very low contamination because it is very effective as a disinfectant agent against many contaminated bacteria as previously reported (Khanam and Chandra, 2017). Hypochlorite acts as a strong oxidant that can denature by aggregating necessary proteins of bacteria as reported earlier (Winter et al., 2008). Our results are in acceptance with previous studies on attempts using several sterilization methods (Daud et al., 2012). In brief, a combination of sodium hypochlorite, ethanol, and Tween 20 was found to be very effective in removing epiphytic microorganisms. The earlier study used 70% alcohol for 2 min, 2% sodium hypochlorite for 3 min, and sterile distilled water for 2 min for surface sterilization (Cardoso et al., 2020).
Another part of the study was a preliminary characterization of isolated endophytic bacteria. Endophytic bacteria exhibited a broad range of morphological and biochemical characteristics which designated that they are different bacterial species. The endophytic bacteria multiply and inhabit in the plants where the environment carries high ionic strength. Earlier, many studies reported that the endophytic bacteria tolerate the high salt concentration (Kumar et al., 2015). In this study, the bacterial endophytes were able to grow differentially at different salt concentrations. In this study, 22 of 33 bacterial isolates exhibited sustained growth at a NaCl concentration of 12%. The earlier study found that Bacillus sp. tolerated up to 2% NaCl, while Pseudomonas sp. 4% NaCl (Rashid et al., 2012). The endophytic bacterial isolates of Momordica charentia exhibited tolerance to NaCl concentration of 4–10% (Singh et al., 2013).
In plants, endophytes are chemical synthesizers. They are capable of producing bioactive compounds that can be used by plants for defense against pathogens, and some of these products have been proved for useful drug discovery (Bungtongdee et al., 2019). To date, most of the natural compounds from endophytes are used as anticancerous, antibacterial, antifungal, antiviral, antidiabetic, and other bioactive products because of their different functional roles (Guo et al., 2008). In this study, out of 33 isolates, 4 showed antibacterial activity against all the six test organisms, i.e., B. subtilis, E. coli, K. pneumonia, S. aureus, S. typhi, and P. aeruginosa, which is similar to an earlier report (Sun et al., 2013). Endophytic bacterial variety was found to be abundant in C. dichotoma. Bacillus sp., a Gram-positive bacterium, was found as endophytic with considerable antibacterial activity. Bacillus sp. was shown to be the closest homolog to B. thuringiensis, an endophytic Bacillus. In many instances, isolates belonging to this genus have been found to produce antimicrobial and other bioactive compounds (Hateet, 2020). According to Beiranvand et al. (2017), endophytic B. thuringiensis isolated from Iranian medicinal plants produced a broad range of antimicrobial compounds. Similarly, Islam et al. (2019) discovered that B. thuringiensis isolated from several gymnosperms and angiosperms has antibacterial activity. Endophytic bacteria found in plant tissues could be a new source of bioactive compounds, according to these studies.
The appearance of antibiotic resistance among pathogenic microorganisms restricts treatment alternatives (Mengoni et al., 2014). In addition to clinical pathogens, antibiotic-resistant genes are also present in environmental strains that are horizontally transferred to other microorganisms (Christina et al., 2013). In this study, susceptibility pattern of isolated endophytic bacteria was studied using 10 different antibiotics. Most of the isolates were resistant to rifampicin, amoxicillin, and bacitracin, while they were susceptible to erythromycin and streptomycin. Kumar et al. (2015) found that the bacterial strain Pseudomonas sp. from Cassia tora was resistant to chloramphenicol and amoxicillin.
The substances found in B. thuringiensis OM320575 crude extracts are predominantly alcohols, terpenes, alkaloids, hydrocarbons, and their derivatives. These chemicals are renowned for their therapeutic effects and have been found in endophytes isolated from medicinal plants (Tapfuma et al., 2020). Some of these chemicals are separated and utilized as antimicrobials individually in extracts. The principal chemical components found in the B. thuringiensis OM320575 ethyl acetate extract include dibutyl phthalate, eicosane, tetrapentacontane, heneicosane, hexadecane, tetradecane, silane, trichlorooctadecyl, and 2,4-di-tert-butylphenol. Dibutyl phthalate is one of the most significant chemicals found in the strain SMC 212, and it may play a role in microbial inhibition. Similarly, Wilkins et al. (2000) reported that Trichoderma viride produced pathogen-inhibiting volatile metabolites such as 2-propanol, 3-methylfuran, methyl-1-propanol, 1-pentanol, and 2-hexanone. Additionally, pentanones, octanones, non-anones, and undecanones have been described in T. atroviride culture (Nemčovič et al., 2008) and heptanone by T. viride (Siddiquee et al., 2012). Sulfurous acid, heptadecane, and octane identified in this investigation have fumigant, insecticidal, and fungicidal activities. Of these, octadecane, heptadecane, and di-tert-butylphenol are known to be emitted by plants under stress. The antifungal activity of eicosane (C20H42) and dibutyl phthalate (C16H22O4) was discovered in larger percentages in the ethyl acetate fraction of Streptomyces strain (Ahsan et al., 2017).
Conclusion
This may be the first-ever report on endophytic bacteria isolated from C. dichotoma in Jammu region, and our findings indicate the high diversity of endophytic bacterial strains associated with the root, stem, and leaf of the medicinal plant that differed appreciably in their morphological, physiological, and biochemical features. This study revealed that C. dichotoma is a potential but underexploited resource for bioactive bacterial endophytes since the exploited bacteria isolated from C. dichotoma showed promising results for antimicrobial and enzymatic activities, utilization of various carbon sources, and tolerance for high salt concentration (12% NaCl). Antimicrobial estimation revealed that bacterial endophytes showed significant antibacterial activity against S. typhi, E. coli, P. aeruginosa, B. subtilis, S. aureus, and K pneumonia. The therapeutic properties of C. dichotoma may be a consequence of its endophytic microorganisms producing biologically active products. The bacterial strains were sensitive to antibiotic erythromycin and streptomycin, whereas most of them were resistant to rifampicin, amoxicillin, and bacitracin. Endophytes have been found to be abundant sources of novel natural chemicals with a wide range of biological functions and a high level of structural diversity. By synthesizing dibutyl phthalate; eicosane; tetrapentacontane; heneicosane; hexadecane; tetradecane; silane, trichlorooctadecyl; and 2,4-di-tert-butylphenol as bioactive chemicals, one potential endophyte isolated from C. dichotoma and identified as B. thuringiensis OM320575 by 16S rRNA demonstrated considerable antibacterial activity against pathogenic bacteria. Further exploration would provide us an insight into the potential use of isolated bacterial endophytes, and it will lead to the discovery of various high-value metabolites.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
MS conducted the experiments, recorded the observations, analyzed the results, and prepared the draft manuscript. SM contributed to designing the experiments, providing the facilities, analyzing the results, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflictof interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.879386/full#supplementary-material
References
Abdel-Hady, H., Abdel-Wareth, M. T. A., El-Wakil, E. A., and Helmy, E. A. (2016). Identification and evaluation of antimicrobial and cytotoxic activities of Penicillium islandicum and Aspergillus tamarii ethyle acetate extracts. Pharmaceuticals. 6, 2021–2039. doi: 10.20959/wjpps20169-7674
Abdelshafy Mohamad, O. A., Ma, J.-B., Liu, Y.-H., Zhang, D., Hua, S., Bhute, S., et al. (2020). Beneficial endophytic bacterial populations associated with medicinal plant Thymus vulgaris alleviate salt stress and confer resistance to Fusarium oxysporum. Front. Plant Sci. 11, 47. doi: 10.3389/fpls.2020.00047
Aboobaker, Z., Viljoen, A., Chen, W., Crous, P., Maharaj, V., and Van Vuuren, S. (2019). Endophytic fungi isolated from Pelargonium sidoides DC: antimicrobial interaction and isolation of a bioactive compound. South Afr. J. Bot. 122, 535–542. doi: 10.1016/j.sajb.2019.01.011
Ahsan, T., Chen, J., Zhao, X., Irfan, M., and Wu, Y. (2017). Extraction and identification of bioactive compounds (eicosane and dibutyl phthalate) produced by Streptomyces strain KX852460 for the biological control of Rhizoctonia solani AG-3 strain KX852461 to control target spot disease in tobacco leaf. AMB Express. 7, 1–9. doi: 10.1186/s13568-017-0351-z
Alsultan, W., Vadamalai, G., Khairulmazmi, A., Saud, H. M., Al-Sadi, A. M., Rashed, O., et al. (2019). Isolation, identification and characterization of endophytic bacteria antagonistic to Phytophthora palmivora causing black pod of cocoa in Malaysia. Eur. J. Plant Pathol. 155, 1077–1091. doi: 10.1007/s10658-019-01834-8
Baldan, E., Nigris, S., Populin, F., Zottini, M., Squartini, A., and Baldan, B. (2014). Identification of culturable bacterial endophyte community isolated from tissues of Vitis vinifera “Glera”. Plant Biosyst. 148, 508–516. doi: 10.1080/11263504.2014.916364
Bauer, A. T.. (1966). Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 45, 149–158. doi: 10.1093/ajcp/45.4_ts.493
Beiranvand, M., Amin, M., Hashemi-Shahraki, A., Romani, B., Yaghoubi, S., and Sadeghi, P. (2017). Antimicrobial activity of endophytic bacterial populations isolated from medical plants of Iran. IJM 9, 11–18.
Bent, E., and Chanway, C. P. (2002). Potential for misidentification of a spore-forming Paenibacillus polymyxa isolate as an endophyte by using culture-based methods. Appl. Environ. Microbiol. 68, 4650–4652. doi: 10.1128/AEM.68.9.4650-4652.2002
Bungtongdee, N., Sopalun, K., Laosripaiboon, W., and Iamtham, S. (2019). The chemical composition, antifungal, antioxidant and antimutagenicity properties of bioactive compounds from fungal endophytes associated with Thai orchids. J Phytopathol. 167, 56–64. doi: 10.1111/jph.12773
Cardoso, V., Campos, F., Santos, A., Ottoni, M., Rosa, C., Almeida, V., et al. (2020). Biotechnological applications of the medicinal plant Pseudobrickellia brasiliensis and its isolated endophytic bacteria. J. Appl. Microbiol. 129, 926–934. doi: 10.1111/jam.14666
Cho, K. M., Hong, S. Y., Lee, S. M., Kim, Y. H., Kahng, G. G., Lim, Y. P., et al. (2007). Endophytic bacterial communities in ginseng and their antifungal activity against pathogens. Microb. Ecol. 54, 341–351. doi: 10.1007/s00248-007-9208-3
Christina, A., Christapher, V., and Bhore, S. J. (2013). Endophytic bacteria as a source of novel antibiotics: an overview. Pharmacogn. Rev. 7, 11. doi: 10.4103/0973-7847.112833
Daniel, J. J. Jr., Zabot, G. L., Tres, M. V., Harakava, R., Kuhn, R. C., and Mazutti, M. A. (2018). Fusarium fujikuroi: a novel source of metabolites with herbicidal activity. Biocatal. Agric. Biotechnol. 14, 314–320. doi: 10.1016/j.bcab.2018.04.001
Daud, N. H., Jayaraman, S., and Mohamed, R. (2012). Methods Paper: an improved surface sterilization technique for introducing leaf, nodal and seed explants of Aquilaria malaccensis from field sources into tissue culture. Aspac J. Mol. Biol. Biotechnol. 20, 55–58.
Deshmukh, S. K., Verekar, S. A., and Bhave, S. V. (2015). Endophytic fungi: a reservoir of antibacterials. Front. Microbiol. 5, 715. doi: 10.3389/fmicb.2014.00715
Devi, N. N., and Singh, M. S. (2013). GC-MS analysis of metabolites from endophytic fungus Colletotrichum gloeosporioides isolated from Phlogacanthus thyrsiflorus nees. Int J Pharm Sci 23, 392–395.
Dhankhar, S., Dhankhar, S. P., and Yadav, J. (2013). Investigating antimicrobial properties of endophytic fungi associated with Salvadora oleoides decne. Anti Infect. Agents 11, 48–58. doi: 10.2174/22113626130106
Dhouib, H., Zouari, I., Abdallah, D. B., Belbahri, L., Taktak, W., Triki, M. A., et al. (2019). Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biol. Control. 139, 104092. doi: 10.1016/j.biocontrol.2019.104092
Di Bitetti, M. S.. (2000). The distribution of grooming among female primates: testing hypotheses with the Shannon-Wiener diversity index. Behaviour 137, 1517–1540. doi: 10.1163/156853900502709
Gao, Z., Zhang, B., Liu, H., Han, J., and Zhang, Y. (2017). Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control. 105, 27–39. doi: 10.1016/j.biocontrol.2016.11.007
Gouda, S., Das, G., Sen, S. K., Shin, H.-S., and Patra, J. K. (2016). Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 7, 1538. doi: 10.3389/fmicb.2016.01538
Guo, B., Wang, Y., Sun, X., and Tang, K. (2008). Bioactive natural products from endophytes: a review. Appl. Biochem. Microbiol. 44, 136–142. doi: 10.1134/S0003683808020026
Hallmann, J., Berg, G., and Schulz, B. (2006). “Isolation procedures for endophytic microorganisms,” in Microbial Root Endophytes. eds B. Schulz, C. Boyle, and T. N. Sieber (Berlin; Heidelberg: Springer-Verlag), 299–319. doi: 10.1007/3-540-33526-9_17
Hamayun, M., Khan, N., Khan, M. N., Qadir, M., Hussain, A., Iqbal, A., et al. (2021). Antimicrobial and plant growth-promoting activities of bacterial endophytes isolated from Calotropis procera (Ait.) WT Aiton. Biocell 45, 363. doi: 10.32604/biocell.2021.013907
Hateet, R. R.. (2020). GC-MS analysis of extract for endophytic fungus acremonium coenophialum and its antimicrobial and antidiabetic. Res. J. Pharm. Technol. 13, 119–123. doi: 10.5958/0974-360X.2020.00024.4
Islam, M. N., Ali, M. S., Choi, S.-J., Hyun, J.-W., and Baek, K.-H. (2019). Biocontrol of citrus canker disease caused by Xanthomonas citri subsp. citri using an endophytic Bacillus thuringiensis. Plant Pathol. 35, 486–497. doi: 10.5423/PPJ.OA.03.2019.0060
Kaur, N., Arora, D. S., Kalia, N., and Kaur, M. (2020). Antibiofilm, antiproliferative, antioxidant and antimutagenic activities of an endophytic fungus Aspergillus fumigatus from Moringa oleifera. Mol. Biol. Rep. 47, 2901–2911. doi: 10.1007/s11033-020-05394-7
Khan, A. L., Waqas, M., Kang, S.-M., Al-Harrasi, A., Hussain, J., Al-Rawahi, A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52, 689–695. doi: 10.1007/s12275-014-4002-7
Khan, I. H., Javaid, A., Ahmed, D., and Khan, U. (2020). Identification of volatile constituents of ethyl acetate fraction of Chenopodium quinoa roots extract by GC-MS. Int. J. Biol. Biotechnol 17, 17–21.
Khanam, B., and Chandra, R. (2017). “Optimization of surface sterilization process of selected dye-yielding plants for isolation of bacterial endophytes,” in Applications of Biotechnology for Sustainable Development, ed K. Mukhopadhyay (Singapore: Springer), 45–50. doi: 10.1007/978-981-10-5538-6_7
Koksal, E., Bursal, E., Dikici, E., Tozoglu, F., and Gulcin, I. (2011). Antioxidant activity of Melissa officinalis leaves. J. Med. Plant Res. 5, 217–222. doi: 10.5897/JMPR.9000199
Kumar, V., Kumar, A., Pandey, K. D., and Roy, B. K. (2015). Isolation and characterization of bacterial endophytes from the roots of Cassia tora L. Ann. Microbiol. 65, 1391–1399. doi: 10.1007/s13213-014-0977-x
Kumaresan, S., Senthilkumar, V., Stephen, A., and Balakumar, B. (2015). GC-MS analysis and pass-assisted prediction of biological activity spectra of extract of Phomopsis sp. isolated from Andrographis paniculata. World J. Pharm. Res 4, 1035–1053.
Kumari, R. M., Thapa, N., Gupta, N., Kumar, A., and Nimesh, S. (2016). Antibacterial and photocatalytic degradation efficacy of silver nanoparticles biosynthesized using Cordia dichotoma leaf extract. Adv. Nat. Sci. Nanosci. Nanotechnol. 7, 045009. doi: 10.1088/2043-6262/7/4/045009
Kuo, J., Chang, C.-F., and Chi, W.-C. (2021). Isolation of endophytic fungi with antimicrobial activity from medicinal plant Zanthoxylum simulans Hance. Folia Microbiol. 66, 385–397. doi: 10.1007/s12223-021-00854-4
Li, J., Zhao, G. Z., Chen, H. H., Wang, H. B., Qin, S., Zhu, W. Y., et al. (2008). Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Lett. Appl. Microbiol. 47, 574–580. doi: 10.1111/j.1472-765X.2008.02470.x
Li, Y., Kumar, P. S., Tan, Q., Tan, X., Yuan, M., Luo, J., et al. (2021). Diversity and chemical fingerprinting of endo-metabolomes from endophytes associated with Ampelopsis grossedentata (Hand.-Mazz.) WT Wang possessing antibacterial activity against multidrug resistant bacterial pathogens. J. Infect. Public Health. 14, 1917–1926. doi: 10.1016/j.jiph.2021.10.019
Liarzi, O., Bucki, P., Braun Miyara, S., and Ezra, D. (2016). Bioactive volatiles from an endophytic Daldinia cf. concentrica isolate affect the viability of the plant parasitic nematode Meloidogyne javanica. PLoS ONE 11, e0168437. doi: 10.1371/journal.pone.0168437
Mai, P. Y., Levasseur, M., Buisson, D., Touboul, D., and Eparvier, V. (2019). Identification of antimicrobial compounds from Sandwithia guyanensis-associated endophyte using molecular network approach. Plants 9, 47. doi: 10.3390/plants9010047
Mengistu, A. A.. (2020). Endophytes: colonization, behaviour, and their role in defense mechanism. Int. J. Microbiol. 2020, 6927219. doi: 10.1155/2020/6927219
Mengoni, A., Maida, I., Chiellini, C., Emiliani, G., Mocali, S., Fabiani, A., et al. (2014). Antibiotic resistance differentiates Echinacea purpurea endophytic bacterial communities with respect to plant organs. Res. Microbiol. 165, 686–694. doi: 10.1016/j.resmic.2014.09.008
Mishra, R., Kushveer, J. S., Khan, M., Imran, K., Pagal, S., Meena, C. K., et al. (2020). 2, 4-Di-tert-butylphenol isolated from an endophytic fungus, Daldinia eschscholtzii, reduces virulence and quorum sensing in Pseudomonas aeruginosa. Front. Microbiol. 11, 1668. doi: 10.3389/fmicb.2020.01668
Mohamad, O. A., Li, L., Ma, J.-B., Hatab, S., Xu, L., Guo, J.-W., et al. (2018). Evaluation of the antimicrobial activity of endophytic bacterial populations from Chinese traditional medicinal plant licorice and characterization of the bioactive secondary metabolites produced by Bacillus atrophaeus against Verticillium dahliae. Front. Microbiol. 9, 924. doi: 10.3389/fmicb.2018.00924
Monowar, T., Rahman, M., Bhore, S. J., Raju, G., and Sathasivam, K. V. (2018). Silver nanoparticles synthesized by using the endophytic bacterium Pantoea ananatis are promising antimicrobial agents against multidrug resistant bacteria. Molecules 23, 3220. doi: 10.3390/molecules23123220
Munjal, V., Nadakkakath, A. V., Sheoran, N., Kundu, A., Venugopal, V., Subaharan, K., et al. (2016). Genotyping and identification of broad spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium BP17. Biol. Control. 92, 66–76. doi: 10.1016/j.biocontrol.2015.09.005
Nascimento, A. M., Conti, R., Turatti, I. C., Cavalcanti, B. C., Costa-Lotufo, L. V., Pessoa, C., et al. (2012). Bioactive extracts and chemical constituents of two endophytic strains of Fusarium oxysporum. Rev. Bras. Farmacogn. 22, 1276–1281. doi: 10.1590/S0102-695X2012005000106
Nemčovič, M., Jakubíková, L., Víden, I., and Farkaš, V. (2008). Induction of conidiation by endogenous volatile compounds in Trichoderma spp. FEMS Microbiol. Lett. 284, 231–236. doi: 10.1111/j.1574-6968.2008.01202.x
Nisa, S., Khan, N., Shah, W., Sabir, M., Khan, W., Bibi, Y., et al. (2020). Identification and bioactivities of two endophytic fungi Fusarium fujikuroi and Aspergillus tubingensis from foliar parts of Debregeasia salicifolia. Arab. J. Sci. Eng. 45, 1–11. doi: 10.1007/s13369-020-04454-1
Phillips, L. A., Germida, J. J., Farrell, R. E., and Greer, C. W. (2008). Hydrocarbon degradation potential and activity of endophytic bacteria associated with prairie plants. Soil Biol. Biochem. 40, 3054–3064. doi: 10.1016/j.soilbio.2008.09.006
Praptiwi, P., Fathoni, A., and Ilyas, M. (2020). Diversity of endophytic fungi from Vernonia amygdalina, their phenolic and flavonoid contents and bioactivities. Biodiversitas 21, 436–441. doi: 10.13057/biodiv/d210202
Qin, S., Feng, W.-W., Wang, T.-T., Ding, P., Xing, K., and Jiang, J.-H. (2017). Plant growth-promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant Soil 416, 117–132. doi: 10.1007/s11104-017-3192-2
Radhakrishnan, E. K. A. A., and Mathew, J. (2020). Characterization of biosurfactant produced by the endophyte Burkholderia sp. WYAT7 and evaluation of its antibacterial and antibiofilm potentials. J. Biotechnol. 313, 1–10. doi: 10.1016/j.jbiotec.2020.03.005
Ragasa, C. Y., Ebajo, V. Jr., Mariquit, M., Mandia, E. H., Tan, M. C. S., Brkljača, R., et al. (2015). Chemical constituents of Cordia dichotoma G. For. J. Appl. Pharm. Sci. 5, 16–21. doi: 10.7324/JAPS.2015.58.S3
Ramalashmi, K., Prasanna, V., Magesh, K., Sanjana, R., Siril, J., and Ravibalan, K. (2018). A potential surface sterilization technique and culture media for the isolation of endophytic bacteria from Acalypha indica and its antibacterial activity. J. Med. Plant Stud. 6, 181–184.
Rashid, S., Charles, T. C., and Glick, B. R. (2012). Isolation and characterization of new plant growth-promoting bacterial endophytes. Agric. Ecosyst. Environ. Appl. Soil Ecol. 61, 217–224. doi: 10.1016/j.apsoil.2011.09.011
Rijavec, T., Lapanje, A., Dermastia, M., and Rupnik, M. (2007). Isolation of bacterial endophytes from germinated maize kernels. Can. J. Microbiol. 53, 802–808. doi: 10.1139/W07-048
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Salim, S. A.. (2018). Identification of active pharmaceutical ingredients in Thevetia neriifolia juss leaf callus using analysis of GC-MS. Indian J Public Health Res. Dev. 9, 1019–1023. doi: 10.5958/0976-5506.2018.01982.4
Sheoran, N., Nadakkakath, A. V., Munjal, V., Kundu, A., Subaharan, K., Venugopal, V., et al. (2015). Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds. Microbiol. Res. 173, 66–78. doi: 10.1016/j.micres.2015.02.001
Siddiquee, S., Cheong, B. E., Taslima, K., Kausar, H., and Hasan, M. M. (2012). Separation and identification of volatile compounds from liquid cultures of Trichoderma harzianum by GC-MS using three different capillary columns. J. Chromatogr. Sci. 50, 358–367. doi: 10.1093/chromsci/bms012
Singh, R., Kumar, A., Singh, M., and Pandey, K. (2013). “Effect of salt stress on endophytic bacteria isolated from root of Momordica charantia,” The Indian Society of Vegetable Science, National Symposium on Abiotic and Biotic Stress Management in Vegetable Crops (Bangalore).
Smibert, R., and Krieg, N. R. (1994). “Phenotypic characterization,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Kreig (American Society for Microbiology; Washington, DC), 607–654.
Strobel, G. A., Knighton, W. B., Kluck, K., Ren, Y., Livinghouse, T., Griffin, M., et al. (2010). The production of myco-diesel hydrocarbons and their derivatives by the endophytic fungus Gliocladium roseum (NRRL 50072). Microbiology 156, 3830–3833. doi: 10.1099/mic.0.30824-0
Subramani, R., Narayanasamy, M., and Feussner, K.-D. (2017). Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech 7, 1–15. doi: 10.1007/s13205-017-0848-9
Sun, H., He, Y., Xiao, Q., Ye, R., and Tian, Y. (2013). Isolation, characterization, and antimicrobial activity of endophytic bacteria from Polygonum cuspidatum. Afr. J. Microbiol. Res. 7, 1496–1504. doi: 10.5897/AJMR12.899
Sun, J., Guo, L., Zang, W., Ping, W., and Chi, D. (2008). Diversity and ecological distribution of endophytic fungi associated with medicinal plants. Sci. China Life Sci. 51, 751–759. doi: 10.1007/s11427-008-0091-z
Tapfuma, K. I., Nchabeleng, E. K., Adebo, O. A., Hussan, R., Williams, R. D., Ravuluvulu, A. B., et al. (2020). Antibacterial activity and gas chromatography mass spectrometry (GC–MS)-based metabolite profiles of Celtis africana and its endophytic extracts. Ind. Crops Prod. 157, 112933. doi: 10.1016/j.indcrop.2020.112933
Tian, X., Cao, L., Tan, H., Han, W., Chen, M., Liu, Y., et al. (2007). Diversity of cultivated and uncultivated actinobacterial endophytes in the stems and roots of rice. Microb. Ecol. 53, 700–707. doi: 10.1007/s00248-006-9163-4
Tyagi, S., and Singh, R. K. (2020). Chemical profile of the antibacterial component from Leptolyngbya sp. HNBGU 002 isolated from a hot spring of Garhwal Himalaya. Int. J. Pharm. Sci. Res. 11, 5225–5238. doi: 10.13040/IJPSR.0975-8232.11(10).5225-38
Venieraki, A., Dimou, M., and Katinakis, P. (2017). Endophytic fungi residing in medicinal plants have the ability to produce the same or similar pharmacologically active secondary metabolites as their hosts. HPPJ 10, 51–66. doi: 10.1515/hppj-2017-0006
Waheed, A., Bibi, Y., Nisa, S., Chaudhary, F. M., Sahreen, S., and Zia, M. (2013). Inhibition of human breast and colorectal cancer cells by Viburnum foetens L. extracts in vitro. Asian Pac. J. Trop. Dis. 3, 32–36. doi: 10.1016/S2222-1808(13)60007-9
Wang, C., Wang, Z., Qiao, X., Li, Z., Li, F., Chen, M., et al. (2013). Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol. Lett. 341, 45–51. doi: 10.1111/1574-6968.12088
Wang, S.-S., Liu, J.-M., Sun, J., Sun, Y.-F., Liu, J.-N., Jia, N., et al. (2019). Diversity of culture-independent bacteria and antimicrobial activity of culturable endophytic bacteria isolated from different dendrobium stems. Sci. Rep. 9, 1887. doi: 10.1038/s41598-019-46863-9
Wilkins, K., Larsen, K., and Simkus, M. (2000). Volatile metabolites from mold growth on building materials and synthetic media. Chemosphere 41, 437–446. doi: 10.1016/S0045-6535(99)00273-8
Wilson, D.. (1995). Endophyte: the evolution of a term, and clarification of its use and definition. Oikos 73, 274–276. doi: 10.2307/3545919
Winter, J., Ilbert, M., Graf, P., Özcelik, D., and Jakob, U. (2008). Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701. doi: 10.1016/j.cell.2008.09.024
Xie, S., Liu, J., Gu, S., Chen, X., Jiang, H., and Ding, T. (2020). Antifungal activity of volatile compounds produced by endophytic Bacillus subtilis DZSY21 against Curvularia lunata. Ann. Microbiol. 70, 1–10. doi: 10.1186/s13213-020-01553-0
Xu, J.-X., Li, Z.-Y., Lv, X., Yan, H., Zhou, G.-Y., Cao, L.-X., et al. (2020). Isolation and characterization of Bacillus subtilis strain 1-L-29, an endophytic bacteria from Camellia oleifera with antimicrobial activity and efficient plant-root colonization. PLoS ONE 15, e0232096. doi: 10.1371/journal.pone.0232096
Xu, L.-L., Chen, H.-L., Hai, P., Gao, Y., Xie, C.-D., Yang, X.-L., et al. (2019). (+)-and (–)-Preuisolactone A: a pair of caged norsesquiterpenoidal enantiomers with a tricyclo [4.4. 01, 6.02, 8] decane carbon skeleton from the endophytic fungus Preussia isomera. Org. Lett. 21, 1078–1081. doi: 10.1021/acs.orglett.8b04123
Zaheer, J., Najam-Us-Saqib, Q., Anwar, T., Khan, F. S., Akram, M., Munir, N., et al. (2021). Phytochemical profile of rock jasmine by using HPLC and GC–MS analyses. Arab. J. Sci. Eng. 46, 5385–5392. doi: 10.1007/s13369-020-05241-8
Keywords: bacterial endophytes, Lasura, antimicrobial activity, antibiotic susceptibility, surface sterilization
Citation: Sharma M and Mallubhotla S (2022) Diversity, Antimicrobial Activity, and Antibiotic Susceptibility Pattern of Endophytic Bacteria Sourced From Cordia dichotoma L. Front. Microbiol. 13:879386. doi: 10.3389/fmicb.2022.879386
Received: 22 February 2022; Accepted: 11 April 2022;
Published: 13 May 2022.
Edited by:
Ranjit Gurav, Konkuk University, South KoreaReviewed by:
Mahloro Hope Serepa-Dlamini, University of Johannesburg, South AfricaVineet Kumar Mishra, Indian Institute of Chemical Biology (CSIR), India
Copyright © 2022 Sharma and Mallubhotla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharada Mallubhotla, c2hhcmRhLnBAc212ZHUuYWMuaW4=
 Mahima Sharma
Mahima Sharma Sharada Mallubhotla
Sharada Mallubhotla