
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 July 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.878912
This article is part of the Research Topic Drug Resistance, Global Epidemiology and Virulence of Acinetobacter View all 14 articles
In Acinetobacter baumannii (A. baumannii), a wide repertoire of resistance genes is often carried within genomic resistance islands (RIs), particularly in high-risk global clones (GCs). As the first in Egypt, the current study aimed at exploring the diversity and genetic configuration of RIs in the clinical isolates of A. baumannii. For this purpose, draft genomes of 18 isolates were generated by Illumina sequencing. Disk diffusion susceptibility profiling revealed multidrug resistance (MDR) and extensive drug resistance (XDR) phenotypes in 27.7 and 72.2%, respectively. The highest susceptibility was noted for tigecycline (100.0%) followed by colistin (94.4%), for which an MIC50 of 0.25 μg/ml was recorded by the broth microdilution assay. Sequence typing (ST) showed that the majority of the isolates belonged to high-risk global clones (GC1, GC2, and GC9). A novel Oxford sequence type (ST2329) that also formed a novel clonal complex was submitted to the PubMLST database. A novel blaADC variant (blaADC−258) was also identified in strain M18 (ST85Pas/1089Oxf). In addition to a wide array of resistance determinants, whole-genome sequencing (WGS) disclosed at least nine configurations of genomic RIs distributed over 16/18 isolates. GC2 isolates accumulated the largest number of RIs (three RIs/isolate) followed by those that belong to GC1 (two RIs/isolate). In addition to Tn6022 (44.4%), the comM gene was interrupted by AbaR4 (5.5%) and three variants of A. baumannii genomic resistance island 1(AbGRI)-type RIs (44.4%), including AbaR4b (16.6%) and two novel configurations of AbGRI1-like RIs (22.2%). Three of which (AbaR4, AbaR4b, and AbGRI1-like-2) carried blaOXA−23 within Tn2006. With less abundance (38.8%), IS26-bound RIs were detected exclusively in GC2 isolates. These included a short version of AbGRI2 (AbGRI2-15) carrying the genes blaTEM−1 and aphA1 and two variants of AbGRI3 RIs carrying up to seven resistance genes [mphE-msrE-armA-sul1-aadA1-catB8-aacA4]. Confined to GC1 (22.2%), sulfonamide resistance was acquired by an ISAba1 bracketed GIsul2 RI. An additional RI (RI-PER-7) was also identified on a plasmid carried by strain M03. Among others, RI-PER-7 carried the resistance genes armA and blaPER−7. Here, we provided a closer view of the diversity and genetic organization of RIs carried by a previously unexplored population of A. baumannii.
In the last decades, Acinetobacter baumannii infections have moved to the forefront of challenges encountered by clinicians worldwide. It is mainly recognized for causing a wide range of difficult-to-treat hospital-acquired infections, particularly in critically ill patients (Morris et al., 2019). Working in concert, the remarkable capacity for upregulating intrinsic resistance mechanisms and acquisition of foreign resistance genes contributed to an ever-expanding spectrum of antimicrobial resistance in A. baumannii. Leaving behind limited or no antimicrobial treatment options, extensively drug-resistant (XDR) and pandrug-resistant strains have been increasingly reported from different parts of the world (Hsueh et al., 2002; Leite et al., 2016; Hamidian and Nigro, 2019). Most of them are members of the high-risk global clones (GCs) 1 and 2 (also known as international clones; ICs) (Karah et al., 2012). Genome sequencing of the earliest strains of the high-risk GCs uncovered a wide repertoire of resistance genes being associated with genomic resistance islands (RIs) (Hamidian and Hall, 2017b). These are genomic regions encompassing variable assortments of transposons and integrons loaded with specific resistance genes (Fournier et al., 2006). They are one of the hallmarks of the horizontal transfer of resistance genes (Carraro et al., 2017).
The first known genomic RI, designated AbaR1 (A. baumannii Resistance 1), was identified in A. baumannii strain AYE from France carrying antimicrobial and heavy metal resistance genes within transposon fragments (Fournier et al., 2006). With a wide variability in size, genetic structure, and insertion sites (Bi et al., 2020), at least seven families of genomic RIs are currently known. These include AbaR-type islands (Post et al., 2010), AbaR4 (Hamidian and Hall, 2011), and A. baumannii genomic resistance islands (AbGRIs) types 1 to 5 (Nigro and Hall, 2012b; Nigro et al., 2013; Wright et al., 2014; Blackwell et al., 2017; Chan et al., 2020; Hua et al., 2021). Any or more than one RIs may be carried by MDR A. baumannii strains (Chan et al., 2015; Hamidian and Hall, 2017b). In addition to the Tn6019 backbone, AbaR contains multiple antibiotic resistance regions (MARRs) enclosed by two copies of Tn6018. AbaR-type RIs are commonly inserted within the ATPase-coding gene comM (Hamidian and Hall, 2018) often in GC1 strains. In the same location, two other RIs were identified. These include AbaR4, in which Tn2006 is inserted in a Tn6022 backbone (Hamidian and Hall, 2011), and AbGRI1, identified in GC2 strains (Nigro and Hall, 2012a,b; Hamidian and Hall, 2017b). AbGRI1 consists of Tn6022 (or its deletion derivatives) and Tn6172 joined by a plasmid-derived linker (Hamidian and Hall, 2017b). AbGRI1 variants often carry the resistance genes sul2, tet(B), strA, strB, and sometimes blaOXA−23 (Bi et al., 2020). The three RIs, AbaR, AbaR4, and AbGRI1, are complex class III transposons carrying the transposition genes tniCABDE that target the comM gene for insertion (Hamidian and Hall, 2017b). The other four types of AbGRIs are IS26-bound transposons harboring variable combinations of resistance genes that are characteristic for each type. They are commonly identified in the chromosomes of A. baumannii strains that belong to GC2. They include AbGRI2 (Nigro et al., 2013), AbGRI3 (Blackwell et al., 2017), AbGRI4 (Chan et al., 2020), and AbGRI5 (Hua et al., 2021). AbGRI2 characteristically carries all or some of the resistance genes blaTEM, aphA1, catA1, and a class I integron carrying sul1, aacC1, and aadA1 (Nigro et al., 2013). AbGRI3 commonly inserts within a putative GNAT family N-acetyltransferase gene. In addition to armA conferring resistance to all clinically useful aminoglycosides, AbGRI3 also carries msrE and mphE, with or without class I integron carrying the resistance genes aacA4, catB8, aadA, and sul1. In some cases, IS26-bracketed aphA1 also integrates into AbGRI3 (Blackwell et al., 2017). Recently, AbGRI4 was identified in GC2 and non-GC2 strains carrying the resistance genes aadB, aadA2, and sul1 in a class I integron. AbGRI4 uniquely targets an α/β-hydrolase gene (Chan et al., 2020). AbGRI5 is the latest RI to be identified in A. baumanni that resembles AbGRI3 in harboring armA, msrE-mphE, sul1, and class I integron that carries a different array of resistance genes [blaPER−1-blaCARB−2-aadA2-cmlA1-aadA1] compared to AbGRI3. In addition, AbGRI5 distinctively carries the macrolide resistance gene ere(B) (Hua et al., 2021).
Even though reports about the structure of RIs carried by strains of this extremely problematic pathogen were published from several parts of the world (Lee et al., 2016; Blackwell et al., 2017; Kim et al., 2017; Chan et al., 2020; Leal et al., 2020; Hua et al., 2021), little is known about those circulating in Egyptian hospitals. Here, we used whole-genome sequencing (WGS) to analyze the diversity and configuration of RIs carried by 18 strains of A. baumannii isolated from patients admitted to one of the largest tertiary university hospitals in Cairo, Egypt, in 2020.
The current study included 20 non-duplicate clinical isolates of carbapenem-resistant A. baumannii from patients admitted to Kasr Al-Ainy University Hospital, Cairo, Egypt. The isolates were recovered from clinical specimens received by the clinical pathology laboratory for bacteriological analysis in the period from July to October 2020. They were identified to species level using the VITEK®2 automated identification system (bioMérieux, Marcy l'Etoile, France) before polymerase chain reaction (PCR) amplification of the blaOXA−51-like genes, as described before (Turton et al., 2006). Identification was further confirmed by WGS using the Speciesfinder tool hosted by the Center of Genomic Epidemiology (http://www.genomicepidemiology.org/).
Broth microdilution assay was used for the determination of the minimum inhibitory concentrations (MICs) of colistin (Sigma-Aldrich, St Louis, MO, USA) in a concentration range of 128-0.125 μg/ml. Susceptibility to other antimicrobial agents was inferred by Kirby–Bauer disc diffusion test. These included amikacin (30 μg), amoxicillin/clavulanic acid (20/10 μg), ampicillin (10 μg), cefepime (30 μg), cefotaxime (30 μg), cefoxitin (30 μg), ceftriaxone (30 μg), imipenem (10 μg), levofloxacin (5 μg), meropenem (10 μg), piperacillin/tazobactam (10/100 μg), tetracycline (30 μg), tigecycline (15 μg), and trimethoprim/sulfamethoxazole (1.25/23.75 μg). Both susceptibility tests were performed and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020) for all antimicrobial agents except tigecycline for which susceptibility breakpoints recommended by EUCAST v11.0 for Enterobacterales were used (EUCAST, 2021). For quality control purposes, Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used.
After DNA extraction using the QIAGEN DNA purification kit (Qiagen, Valencia, CA) and library preparation using the Nextera DNA Sample Preparation kit (Nextera, USA), WGS was performed on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). Pre-assembly processing of the generated reads was carried out by FastQC (Andrews, 2010) for quality assessment and Trimmomatic v0.32 (Bolger et al., 2014) for the trimming of low-quality reads. De novo assembly of trimmed reads was carried out using SPAdes 3.14.1 (Bankevich et al., 2012). Post-assembly metrics were generated by QUAST v5.0.2 (Gurevich et al., 2013). Draft genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016). Plasmids were assembled from Illumina reads using PlasmidSPAdes (Antipov et al., 2016), a software for reading coverage-assisted plasmid identification. Assembly graphs (Fastg files) generated by PlasmidSPAdes were visualized on a bandage (Wick et al., 2015). Plasmid sequences were extracted from circular contigs or groups of contigs forming circular paths containing plasmid replication and/or mobilization genes. Contigs forming circular but overlapping paths were BLASTed for closest plasmids that were subsequently used for reference mapping using the short reads mapping tool BWA-MEM (Li and Durbin, 2010).
Two sequence-based typing methods were used for the epidemiology analysis of the isolates. These included multilocus sequence typing (MLST) and single-nucleotide polymorphism (SNP)-based phylogeny analysis.
The draft genomes were uploaded to the PubMLST server (https://pubmlst.org/abaumannii/) for assigning STs for the isolates according to both Pasteur (Diancourt et al., 2010) and Oxford schemes (Bartual et al., 2005). Allocation of the isolates into clonal complexes (CCs) was done by goeBURST analysis. For this purpose, all allelic profiles defined by both schemes were retrieved from the PubMLST database (accessed on 30 April 2021). Together with the allelic profiles of the isolates studied here, they were used as inputs for Phyloviz software for the generation of minimum spanning trees using the goeBURST algorithm (Ribeiro-Goncalves et al., 2016).
Using the default setting parameters, the CSI phylogeny 1.4 online tool (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) was employed in inferring the phylogeny of the isolates based on the concatenated alignment of the high-quality SNPs. A. baumannii ATCC17978 was used as a reference for the analysis. The analysis initially included 44 complete and draft genomes of A. baumannii strains that belong to STs identified here obtained from the NCBI and PubMLST databases. For easier visualization, the final phylogenetic tree was constructed using a smaller number of genomes of A. baumannii strains that were clustered with our isolates. Interactive tree of life (iTOL) v3 software (https://itol.embl.de/) (Letunic and Bork, 2016) was used for visualization and editing of the phylogenetic tree.
Genes with a minimum of 80% coverage and 95% identity to known resistance genes were identified using the Comprehensive Antibiotic Resistance Database (CARD) server (https://card.mcmaster.ca/analyze/rgi) (Alcock et al., 2020). Point mutations of the genes relevant to fluoroquinolones (gyrA and parC) and colistin (lpxACD and pmrABC) resistance were extracted from the assemblies for pairwise comparison to the corresponding genes of the reference strain A. baumannii ATCC 19606 (GenBank accession: CP045110.1).
The context of resistance genes was examined by visualizing the annotated contigs using SnapGene software v5.1.3.1 by Insightful Science (http://www.snapgene.com). Insertion sequences (ISs) were identified by BLAST analysis against the nucleotide database of the NCBI and novel ISs were named by the ISFinder database team (http://www-is.biotoul.fr). Resistance islands were predicted using the webserver IslandViewer4 webtool (http://www.pathogenomics.sfu.ca/islandviewer/) (Bertelli et al., 2017), through which the draft genomes were mapped against different reference genomes. For RIs fragmented into multiple contigs, assembly gaps were filled by mapping raw reads against the closest RI using BWA (Li and Durbin, 2010).
The Whole Genome Shotgun project including Fastq files generated by the Illumina sequencer and the assembled draft genomes were submitted to GenBank database under the BioProject number PRJNA690827. The nucleotide sequence of the novel blaADC−258 variant was submitted to the NCBI GenBank database under the accession number (MZ224612.1).
Twenty carbapenem-resistant A. baumannii isolates were received by the clinical pathology laboratory of Kasr Al-Ainy University Hospital, Cairo, Egypt, during the study period. All were preserved with the purpose of a WGS-based analysis of RIs. Having successfully passed the post-assembly quality control criteria, only 18 strains were selected for further analysis. Post-assembly and annotation metrics of the generated draft genomes are shown in Supplementary Table 1. Half of the strains selected for the study were isolated from patients in critical care units and at least 22.2% were from pediatric patients. Clinical data of all isolates are shown in Table 1.
The MLST analysis revealed that the isolates belonged to six Pasteur and nine Oxford STs (Figure 1). GoeBURST analysis (Supplementary Figure 1) showed that the majority of the isolates belonged to the high-risk global clones 1, 2, and 9. Predominantly, seven isolates (38.8%) belonged to GC2 distributed over two CCs (CC208 and CC546) according to the Oxford scheme. GC9 (CC464Pas/1078Oxf) and GC1 (CC1Pas/231Oxf) were represented by three and two isolates, respectively. M14 had a novel Oxford ST (2329) that also formed a novel clonal complex to which 11 STs including that of M03 (ST2246Oxf) belonged. The SNP-based phylogenetic analysis generated a seven-cluster phylogenetic tree (Figure 1). Notably, isolates that shared an Oxford ST were clustered together. The isolates M06 and M09 with undetermined STs were found to be phylogenetically related to GC1 isolates. The tree also showed that our isolates were clustered with other strains isolated in different parts of the world.
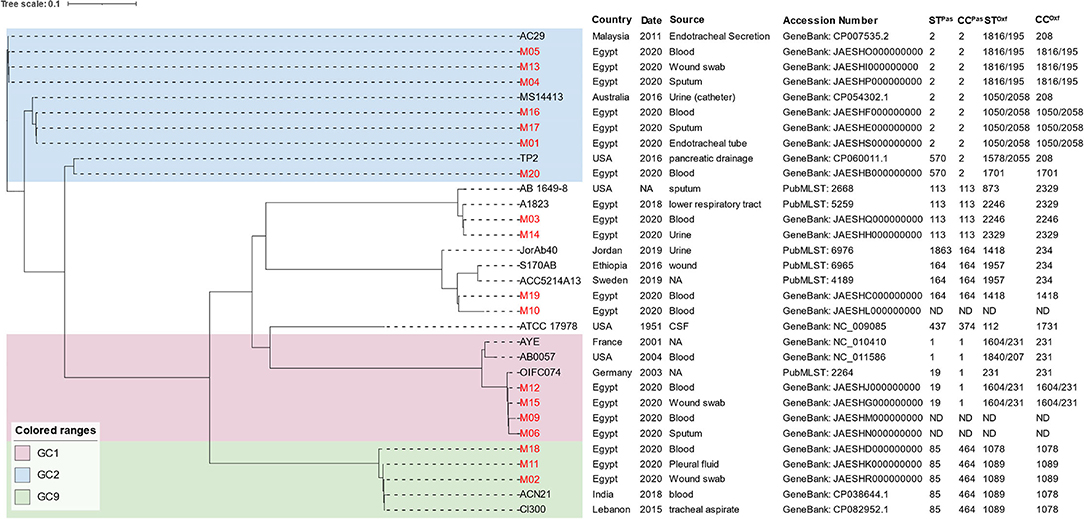
Figure 1. SNP-based phylogenetic tree of A. baumannii draft genomes sequenced in the current study compared to other strains obtained from the NCBI and PubMLST databases. Labels of the A. baumannii strains sequenced in the current study are written in red. STPas, sequence type according to Pasteur scheme; STOxf, sequence type according to Oxford scheme; GC, global clone.
Based on the definitions proposed by Magiorakos et al. (2012) for MDR and XDR, the majority of the isolates were XDR (13/18, 72.2%), and only five isolates (27.7%) showed an MDR phenotype. All GC2 and GC9 isolates were XDR, while MDR isolates belonged to GC1 as well as STs that do not belonging to high-risk clones. Susceptibility to tigecycline was retained by all isolates. Except for one isolate (5.5%), all were susceptible to colistin with an MIC50 of 0.25 μg/ml. Only five isolates (27.7%) were susceptible to amikacin, and one isolate (5.5%) was susceptible to trimethoprim/sulfamethoxazole. All isolates were nonsusceptible to all other tested antimicrobials. A wide repertoire of resistance genes was identified in our isolates, most of which were associated with mobile elements. As many as 38 resistance determinants were identified in combinations of up to 28 determinants per isolate. These included genes coding for antibiotic inactivation, target protection, target alteration, target replacement, and antibiotic efflux. The largest number of co-existing resistance genes was found in GC2 isolates (25-28 determinants/isolate), followed by those that belonged to GC9 (21–23 determinants/isolate). Antimicrobial susceptibility profiles and resistance determinants of all isolates are shown in Figure 2.
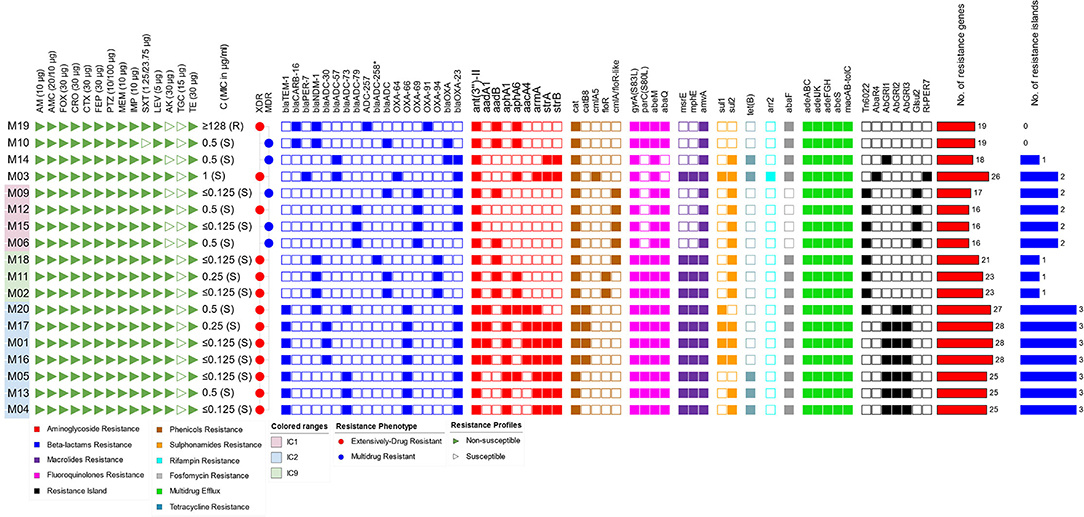
Figure 2. Overview of the antimicrobial susceptibility profiles and resistance determinants. The isolates were ordered according to their SNP-based phylogenetic relationship. Labels of GC1 isolates (together with their phylogenetically related isolates) are highlighted in pink and those of GC2 and GC9 are highlighted in blue and green, respectively. The map is divided into panels corresponding to susceptibility to antimicrobial agents (triangular icons), resistance phenotypes (circular icons), resistance determinants (square icons), and the number of resistance determinants and islands (bars). AM, ampicillin; AMC, amoxicillin/clavulanic acid; FOX, cefoxitin; CRO, ceftriaxone; CTX, cefotaxime; FEP, cefepime; PTZ, piperacillin/tazobactam; MEM, meropenem; IMP, imipenem; SXT, sulfamethoxazole/trimethoprim; LEV, levofloxacin; AK, amikacin; TGC, tigecycline; TE, tetracycline; C, colistin; XDR, extensive drug resistance; MDR, multidrug resistance.
In addition to the intrinsic resistance genes (blaADC and blaOXA−51−like) to β-lactams, five acquired β-lactamase-coding genes, encompassing blaTEM−1, blaCARB−16, blaNDM−1, blaPER−7, and blaOXA−23, were identified. Up to five β-lactamase coding genes co-existed in the tested isolates. At least six blaADC variants were identified, including a novel variant (blaADC−258) carried by M18. blaADC−258 showed 99.74% similarity to blaADC−176 with the amino acid alterations Q2R and D24G. Meanwhile, blaADC variants carried by four isolates could not be identified due to insertion sequence (IS) interruption (M02 and M11) or assembly gaps (M09 and M10). The N terminus of the blaADC−73 carried by M20 was interrupted by an unknown sequence, as described before (Zafer et al., 2021). Interestingly, isolates of the same Oxford ST carried the same blaADC variants. An upstream ISAba1 was confirmed for only eight isolates (M01, M04, M06, M12, M13, M15, M16, and M17), all belonging to GCs 1 and 2. On their chromosomes, the isolates also carried five alleles of the intrinsic β-lactamase-coding gene blaOXA−51−like. Isolates of the same clonal complex (Pasteur or Oxford) shared the same blaOXA−51−like variant. Of all acquired β-lactamase-coding genes, blaOXA−23 (class D β-lactamase-coding gene) was the most prevalent (12/18, 66.6%) either within RIs (5/18, 27.7%) or more frequently bracketed by ISAba1 in Tn2006 (7/18, 38.8%). The blaOXA−23-positive isolates belonged to GC2 and GC1, and two isolates (M03 and M14) belonged to the novel CC113Pas/2329 The gene blaOXA−23 was carried within Tn2006 in GC1 isolates and GC2 isolates that belonged to the Oxford STs ST1050/2058 and ST1701. Meanwhile, in GC2 isolates of the ST1816/195Oxf, blaOXA−23 was hosted by AbaR4b. M03 and M14 carried blaOXA−23 within AbaR4 and an AbGRI1-like-2 RI, respectively. Harbored by an AbGRI2-15 and exclusively in GC2, the class A β-lactamase-coding gene blaTEM−1 was found in seven isolates (38.8%). Among our isolates were six (33.3%) NDM-1 producers. These included all GC9 isolates, one GC2 isolate (M20), as well as M19 (ST164Pas/1418Oxf) and its phylogenetically related isolate M10. The genetic environment of blaNDM−1 was described in our previous study (Zafer et al., 2021). We reported, for the first time, a novel transposon in which both blaNDM−1 and aphA6 were enclosed by two direct copies of ISAba14. The transposition potential of the transposon was later demonstrated using bioinformatic tools (unpublished data). This environment was described only for GC9 isolates that belonged to ST1089Oxf as well as M10. While the right arm of the ISAba14-bracketed transposon carrying blaNDM−1 was found in other NDM producers, the full sequence of the transposon could not be spotted.
The isolates M19 and M10 also carried blaCARB−16 in contigs showing 100% similarity to a 63,650 kb plasmid carried by A. baumannii strain DT01139C (GenBank accession: CP053220.1) isolated in Tanzania in 2017. However, blaCARB−16-positive plasmids could not be identified either by PlasmidSPAdes de novo assembly or by mapping the raw reads against the DT01139C plasmid. Finally, with the lowest prevalence, blaPER−7 was identified only in M03. Together with six more resistance genes, blaPER−7 was carried within RI-PER-7.
Resistance genes to aminoglycosides were found in abundance in our collection. Of them, armA, conferring resistance to all clinically relevant aminoglycosides, was the most prevalent and was carried by 8/18 (44.4%) isolates. These included all GC2 isolates in which it was carried on AbGRI3 and M03 in which armA was hosted by RI-PER-7. The amikacin resistance gene aphA6 was carried by 7/18 (38.8%) isolates that belonged to different STs. Most commonly, aphA6 was bracketed by ISAba14 and ISAba125 in the blaNDM−1-positive isolates either within the composite ISAba14 bracketed transposon described above (M02, M10, and M11) or not (M19 and M20). In M05, aphA6 was carried on a 70,101-bp RepAci6 plasmid closely similar to pACICU2 (GenBank accession: CP031382.1). The context of aphA6 in M03 could not be defined. On pRAY plasmid derivatives, aadB was carried by 7/18 (38.8%) isolates that belonged to different sequence types except for those of GC2. Within AbGRI3, GC2 isolates of the STs ST1050/2058Oxf and ST1701Oxf carried aacA4 and aadA1 that were undetectable in other GC2 strains carrying a shorter version of AbGRI3. Other detected aminoglycoside resistance genes included the intrinsic gene ant(3”)-II (Zhang et al., 2017) and other acquired resistance genes, such as aphA1(38.8%%), strA (44.4%), strB (44.4%), and aadA1 (22.2%). In all aphA1-positive isolates that all belonged to GC2, the gene was carried on AbGRI2-15 together with blaTEM−1. The genes strA and strB co-existed in all GC2 isolates that belonged to CC208Oxf in which they were carried on variants of AbGRI1. They also co-existed in CC113Pas/2329Oxf isolates (M03 and M14) but within different environments. In M14, strA and strB were carried on AbGRI1-type RI. In M03, they were harbored by a transposon (~ 24 Kb) also carrying sul2 and tet(B) and bracketed by ISAba1 and IS26 that were shared with an adjacent RI-PER-7. The whole genetic structure showed 99.9% similarity to a 226,394-bp plasmid pPM194229_1 (GenBank accession: CP050433.1). Using the plasmid sequence as a reference for mapping produced a sequence with 99.99% identity to the reference plasmid with a coverage of 90.0%. Although the full sequence of the plasmid could not be identified, the de novo assembled fragment revealed genetic rearrangement compared to the closely matched plasmid. This included insertion of [tet(B)-tetR(B)] downstream to sul2 through homologous recombination that was also associated with ISVsa3-mediated deletion, as shown in Figure 3. The context of β-lactams and aminoglycoside resistance determinants in all isolates are summarized in Table 2.
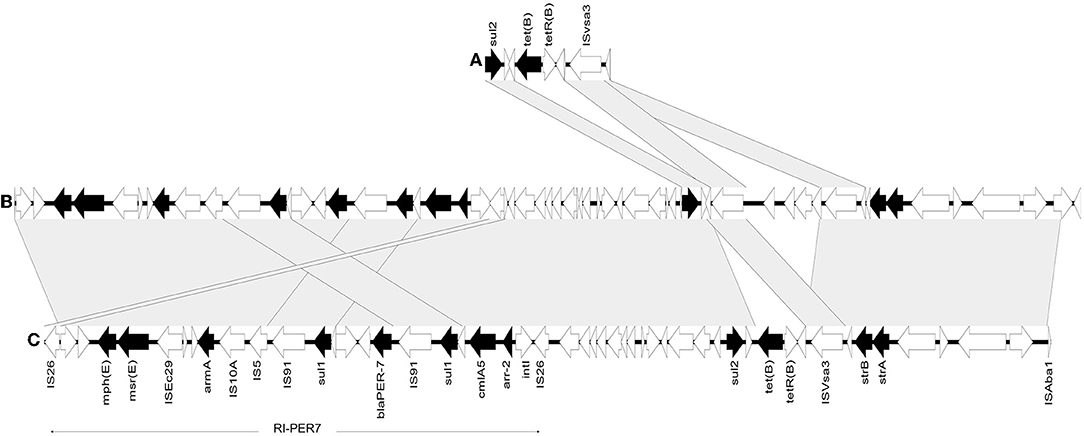
Figure 3. Comparative genomic analysis of the partial sequence of M03 plasmid and the closely similar plasmid pPM194229_1(CP050433.1). (A) pPM194229_1 plasmid sequence region [218446-224334 bp]. (B) pPM194229_1 plasmid sequence region [1-50187 bp]. (C) M03 plasmid partial sequence. Arrows correspond to open reading frames, and black ones denote resistance genes. Gray shadings highlight regions of 99% or more nucleotide identity.
Isolates carrying single mutations (gyrA; S83L) belonged to ST113Oxf (M03 and M14) and ST231Oxf (M12 and M15) and their phylogenetically related strains (M06 and M09). Genes coding the quinolone efflux pumps abeM and abaQ were found in 100.0% and 88.8% of the isolates, respectively.
The chloramphenicol acetyltransferase-coding gene cat was carried in the chromosomes of all isolates not associated with any mobile elements. Meanwhile, catB8 was identified in GC2 isolates within AbGRI3 except those carrying the short version of the island (M04, M05, and M13). Genes coding the chloramphenicol efflux pumps FloR and CmlA5 were also identified. FloR efflux pump-coding gene was carried by M02 and M11 (2/18, 11.1%) in association with sul2 gene and ISs, as described before (Zafer et al., 2021). A novel cmlA/floR-like gene variant was identified in five isolates (27.7%). These included GC1 isolates M06, M09, M12, and M15 and the GC9 isolate M18. The gene was associated with a novel downstream 1,206-bp long IS designated ISAba61. Together with the passenger cmlA/floR-like gene, ISAba61 was distinctively inserted within a molybdopterin-dependent oxidoreductase-coding gene, as shown in Supplementary Figure 2. As a transposition signature, the insertion of ISAba61 generated an 8-bp TGAAAATA duplication in the target site. Only one isolate (M03) carried cmlA5 within RI-PER-7.
In addition to the efflux pump gene amvA, macrolide resistance was coded by msrE and mphE that co-existed in 61.1% of the isolates. They were associated with the resistance islands AbGRI3 and RI-PER-7 in GC2 isolates and M03, respectively. In GC9 isolates, msrE and mphE were located outside the RIs enclosed by the insertion sequences ISNCY and ISAba1 in the upstream and downstream regions, respectively.
As many as 15 isolates (83.3%) carried at least one sulfonamide resistance gene. More frequently, the isolates carried sul2, which was identified in 14 isolates with no clonal bias. The gene was hosted by two types of RIs, namely, GIsul2 in GC1 (M12 and M15) and phylogenetically related isolates (M06 and M09) and AbGRI1-type RIs in M01, M14, M16, and M17. Only five isolates carried sul1 that was associated with AbGRI3 in GC2 isolates (Oxford STs ST1050/2058 and ST1701) and RI-PER-7 in M03.
Resistance to levofloxacin was associated with gyrA mutations encoding S83L amino acid alterations in all isolates. Meanwhile, missense mutations (S80L) in the topoisomerase-coding gene (parC) were identified in only 12 (66.6%) isolates.
The gene tet(B) was the only tetracycline resistance gene identified in the isolates (5/18, 27.7%). In association with the resistance genes strA, strB, and blaOXA−23, it was located within AbGRI1 in all GC1 isolates and M14. A different environment was found for tet(B) in M03 in which it was carried on a plasmid whose sequence was partially identified, as described above. Except for GC1 isolates, genes coding the fosfomycin major facilitator superfamily (MFS) transporter AbaF were carried on the chromosomes of all isolates not associated with any mobile elements. Within RI-PER-7, the rifampin resistance gene arr-2 was only carried by M03. Colistin resistance in M19 was found to be associated with mutations in pmrB (H89L), pmrC (I42V, I212V, R323K, A354S, and V470I), IpxA (Y131H and Y231H), IpxC (C120R, N287D, and K130T), and lpxD (V631 and E117K), as described before (Zafer et al., 2021).
Whole-genome sequencing of the isolates disclosed at least nine configurations of genomic RIs. An additional RI, RI-PER-7, was found to be carried on a plasmid whose structure was only partially identified. GC2 isolates accumulated the largest number of RIs (three RIs/isolate) followed by GC1 isolates in which two genomic RIs co-existed.
For the detection of AbaR-, AbaR4-, and AbGRI-type RIs, comM gene integrity was checked in all isolates. The gene was found to be interrupted in 16/18 (88.8%) isolates (all except M10 and M19) most frequently by Tn6022 (the backbone of AbaR4). Lacking any resistance genes, Tn6022 was carried by eight isolates (44.4%), including all GC1 and GC9 isolates as well as M20 that belonged to GC2. Less often, comM was interrupted by AbGRI1-type RIs carrying resistance genes exclusively within GC2 isolates (except M20), while AbaR4 was only found in M03 that belongs to the ST113Pas/2246Oxf. AbGRI1 was found in three variant structures shown in Figure 4. The first variant was AbaR4b, also called AbGRI1-1/Tn2006 and Tn6166/Tn2006, that lacked the typical AbGRI1 structure [Tn6022-plasmid linker-Tn6172]. Instead, AbaR4b was formed of Tn6166 in which Tn6022Δ1 was interrupted by Tn2006. AbaR4b was identified in the chromosomes of GC2 isolates of the Oxford ST 1816/195. Another AbGRI1 variant, named here AbGRI1-like-1, was similar to AbGRI1-0, but Tn6022 was replaced by Tn6022Δ1, and the integrase-coding gene (int) of the plasmid linker was uniquely interrupted by and ISAba125 element. The island carried three resistance genes (sul2, strA, and strB). Of all abGRI1-type RIs, AbGRI1-like-2 carried the largest number of resistance genes (blaOXA−23, tet(B), sul2, strA, and strB). Compared to AbGRI1-0, this version distinctively carried Tn6022 in which sup gene was interrupted by Tn2006. In addition, [tet(B)-tetR(B)] element was also inserted within Tn6172.
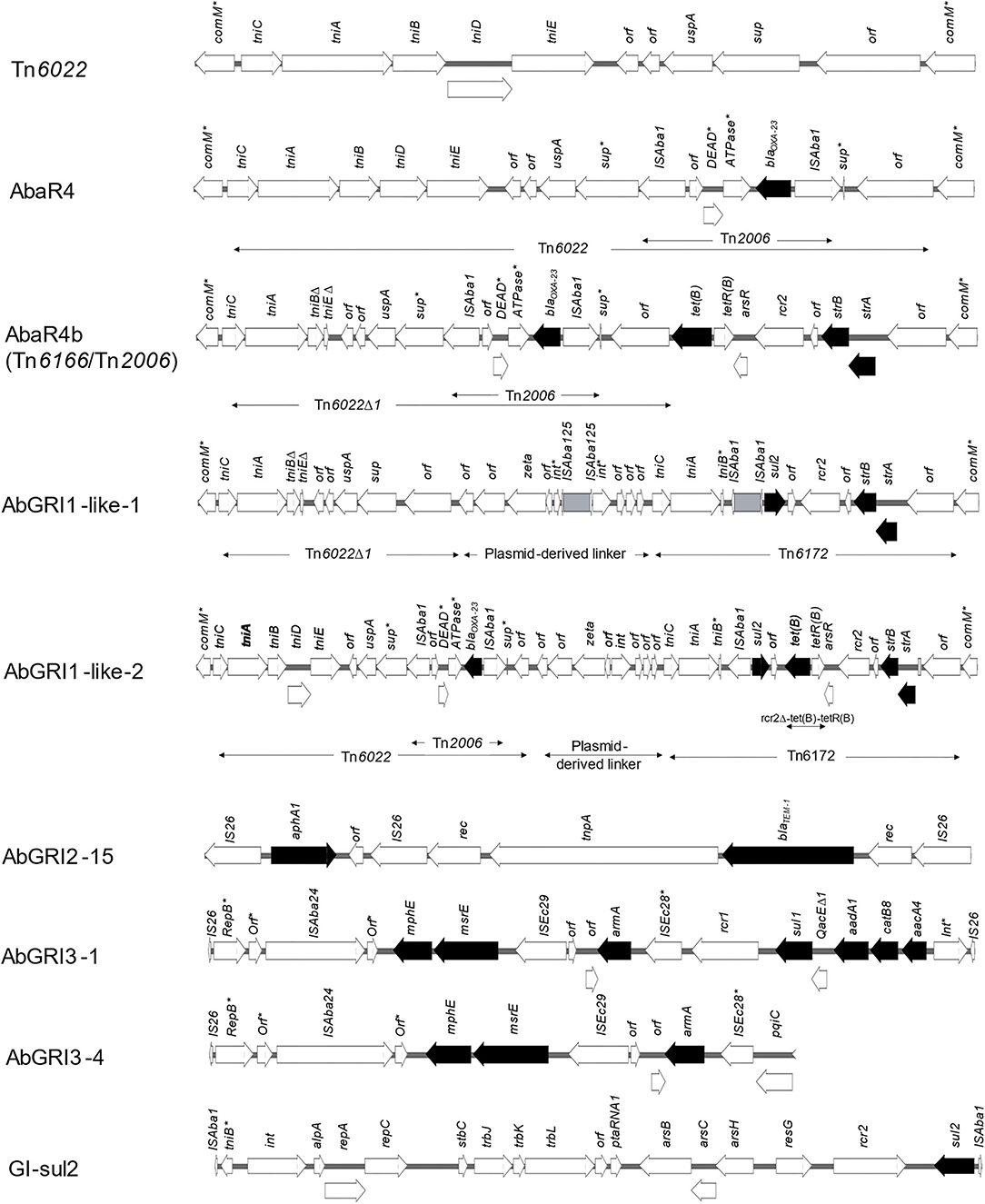
Figure 4. Structures of resistance islands identified in the current study. Nine types of genomic RIs were identified in the isolates. These included Tn6022, AbaR4, three AbGRI1 variants, one AbGRI2, two variants of AbGRI3, and GIsul2. Arrows denote open reading frames. Antimicrobial resistance genes are represented by black arrows. Asterisks indicate interrupted genes.
Two types of IS26-bound genomic RIs were also identified. These included AbGRI2- and AbGRI3-type RIs that coexisted in all GC2 isolates. Only one version of AbGRI2 (AbGRI2-15) (Liepa et al., 2021) was identified. It harbored the two resistance genes blaTEM−1 and aphA1. While two versions of AbGRI3 were identified, an expanded form (designated before AbGRI3-1) was the dominant one (Blackwell et al., 2017). AbGRI3-1 carried seven resistance genes [mphE-msrE-armA-sul1-aadA1-catB8-aacA4]. A shorter version, designated before as AbGRI3-4 (Blackwell et al., 2017), was confined to GC2 isolates of the ST1816/195Oxf with only three resistance genes onboard [mphE, msrE, and armA]. Bracketed by two inversely oriented copies of ISAba1, GIsul2 RI carrying sul2 as a sole resistance gene co-existed with Tn6022 in all GC1 isolates and their phylogenetically related isolates M06 and M09.
As described above, RI-PER-7 was identified on a plasmid carried by M03 whose sequence was partially identified. The island hosted the largest combination of resistance genes compared to other RIs identified in the current study. These included arr-2, cmlA5, blaPER−7, armA, msrE, mphE, and two copies of sul1. RIs correlated to different STs are shown in Table 3.
Updates on the genetic background of MDR and XDR A. baumannii are continuously being published from different parts of the world (Hamidian and Nigro, 2019; Gheorghe et al., 2021; Wareth et al., 2021) and from Egypt as well (Hassan et al., 2021; Jalal et al., 2021; Wasfi et al., 2021). However, reports about the association of resistance genes with RIs are relatively scarce. This is in part due to the need for multistep PCR mapping or whole-genome sequencing. To the best of our knowledge, this is the first report about the diversity and the genetic configuration of genomic RIs carried by A. baumannii isolates from Egypt. For this purpose, draft genomes of 18 non-duplicate MDR and XDR isolates were generated. Mostly from ICUs, the isolates were recovered from patients with bloodstream, respiratory tract, and wound infections. Very few treatment options were available. Draft genomes were employed for MLST analysis using both Pasteur and Oxford schemes. Oxford scheme-based analysis revealed the co-existence of two different alleles of the gdhB locus in 7/18 isolates generating two STs per isolate, a previously reported drawback for the scheme (Tomaschek et al., 2016). Nevertheless, MLST profiles generated by the Oxford scheme showed superior discrimination and concordance with the SNP-based phylogeny results. This was in line with other reports as well (Tomaschek et al., 2016; Gaiarsa et al., 2019). As reported before (Karah et al., 2012), most of the MDR and XDR A. baumannii strains belong to the Pasteur CCs 1 and two widely known as GC1 and GC2. In agreement with other studies (Al-Hassan et al., 2019; Fam et al., 2020; Wasfi et al., 2021), MLST analysis revealed the predominance of GC2 isolates in our collection (38.8%). Nevertheless, GC1 strains outweighed those that belonged to GC2 studied by others in our region (Ghaith et al., 2017; Jalal et al., 2021). Less representation (16.6%) was noted for the recently described GC9 (Müller et al., 2019) known to be endemic in Middle East countries (Al-Hassan et al., 2013, 2021; Bonnin et al., 2013; Ghaith et al., 2017; Jaidane et al., 2018; Salloum et al., 2018). Notably, a low prevalence of high-risk GCs was reported by older studies in Egypt (Al-Hassan et al., 2013; El Bannah et al., 2018), reflecting the progressive expansion of high-risk global clones in Egyptian hospitals over years. The emergence of successful STs not assigned to any high-risk GCs was also evident in the current study. These included the novel clonal complex CC2329Oxf (CC113Pas) represented by M03 (ST1132246Oxf) and M14 (ST113 ST2329Oxf). Isolates that belong to this CC were reported from Egypt (Jalal et al., 2021), Saudi Arabia (Lopes et al., 2015), and Brazil (Leal et al., 2020). Another ST identified here, ST164Pas/1418 was reported in the isolates from Myanmar (Tada et al., 2020), Vietnam (Wareth et al., 2021), Brazil (Coelho-Souza et al., 2013), Kenya (Musila et al., 2021), and as a major clone in Thailand (Khuntayaporn et al., 2021). SNP-based phylogenetic analysis showed that our isolates were clustered with others from different parts of the world. Transmission of genetically related strains over continents might be facilitated by travel-associated fecal colonization (Ostholm-Balkhed et al., 2013) and medical tourism (Benenson et al., 2018). Together with international strains with similar STs, our isolates were distributed over seven clusters in a pattern that fully matched the Oxford scheme-based STs.
The detailed profiling of the resistance determinants and correlation to STs and resistance phenotypes was presented in the current study with special emphasis on resistance to β-lactams and aminoglycosides. The epidemiological linkage between specific blaOXA−51−like variants and certain GCs was reported before (Zander et al., 2012; Karah et al., 2016; Jaidane et al., 2018) and was also evident in our isolates. This was noted for blaOXA−64,blaOXA−66,and blaOXA−94 that were linked to GC1, GC2, and GC9, respectively. In agreement with others (Jalal et al., 2021), blaADC−79 was linked to GC1. Except for those that belonged to ST1050/2058Oxf which carried blaADC−30, GC2 isolates carried blaADC−73, a single-nucleotide variant of blaADC−30 (Karah et al., 2016). ISAba1-amplified ADC-type β-lactamases, previously coupled to high-level cephalosporin resistance (Corvec et al., 2003), were confined to GC1 and GC2 isolates. Carbapenem resistance in our collection was mediated by two carbapenem-hydrolyzing enzymes, namely, OXA-23 and NDM-1. OXA-23 was the most common of all acquired β-lactamases produced by our isolates. It is also the most frequently described carbapenemase globally (Hamidian and Nigro, 2019). The gene blaOXA−23 existed in Tn2006 that, in turn, was sometimes embedded in RIs. Despite the variety of the genetic platforms known to harbor blaOXA−23, Tn2006 is the dominant vehicle for the acquisition of the gene worldwide (Nigro and Hall, 2016; Hamidian and Nigro, 2019). Less frequently, carbapenem resistance was mediated by NDM-1 (class B β-lactamases). In addition to one GC2 isolate (M20) and the phylogenetically related isolates M10 and M19, blaNDM−1 was carried by all GC9 (ST85Pas) isolates. GC9 was claimed to act as a reservoir for blaNDM−1 in our region (Bonnin et al., 2013; Jaidane et al., 2018; Salloum et al., 2018; Al-Hassan et al., 2021). As described in our previous study (Zafer et al., 2021), blaNDM−1 laid within an ISAba14-bracketed transposon also carrying aphA6 in M02, M11, and M10. However, the transposon could not be identified in other NDM-producer strains in our collection. Large-scale screening of this novel transposon is, therefore, recommended. Although reported in a higher prevalence in Egypt (El-Sayed-Ahmed et al., 2015; Abouelfetouh et al., 2019; Wasfi et al., 2021), a combination of the carbapenemase-coding genes blaOXA−23 and blaNDM−1 was found in only one isolate (M20). Class A β-lactamases, including TEM-1, CARB-16, and PER-7, had a considerable share in our collection too. While the exact environment of blaCARB−16 could not be identified, both blaTEM−1 and blaPER−7 were exclusively carried within RIs, including AbGRI2 and RI-PER-7, respectively. The latter was first described as Tn1548-like-2 transposon by Karah et al. (2016) and later designated RI-PER-7 by Adams et al. (2020). Both reported the island in isolates that had Pasteur ST25, a double-locus variant of ST113Pas in which RI-PER-7 was identified here. Shorter versions of the island were also reported to be carried by members of the family Enterobacteriaceae (Adams et al., 2020). WGSs of four isolates of the ST ST2246Oxf were generated in a recent study in Egypt (Jalal et al., 2021). While the authors did not report the RI-PER-7 in their isolates, the signature genes of the island [mphE, msrE, armA, sul1, blaPER−7, cmlA5, and arr2] were identified in 3/4 (75%) isolates. This newly emerging ST might thus serve as a reservoir for RI-PER-7. While the RI-PER-7-positive contig of M03 showed the highest similarity to a plasmid sequence and was thus anticipated to be plasmid-mediated, the exact location of the island in ST2246Oxf is yet to be confirmed.
Of all the aminoglycoside resistance genes identified here, those conferring resistance to the clinically relevant aminoglycosides, amikacin, gentamicin, and tobramycin, were of much concern. Most importantly, the broad-spectrum armA gene conferring high-level resistance to all aminoglycosides (Galimand et al., 2003) was carried by all GC2 isolates. A high prevalence of armA has been previously reported in Egypt (El-Sayed-Ahmed et al., 2015). In addition, co-existing blaOXA−23, blaNDM−1, and armA were found in one GC2 isolate (M20). This combination was identified on plasmids of 8/25 (32%) isolates in an older study in Egypt (El-Sayed-Ahmed et al., 2015). Amikacin resistance in our isolates was also coded by aphA6 in 38.8% of the isolates. The gene is commonly identified in A. baumannii within the composite transposon TnaphA6 in which it is bracketed by two directly oriented ISAba125 elements (Nigro et al., 2011). Nevertheless, here it was most commonly identified within a bracket of an upstream ISAba14 element and a downstream ISAba125 element that was sometimes part of a composite transposon also carrying blaNDM−1, as described above. Given that the ISAba125 elements are thought to drive the overexpression of aphA6 imparting high-level amikacin resistance, the effect of the upstream ISAba14 insertion is questionable. Notably, this unusual environment was associated with unrelated STs. While frequently found on plasmids (Nigro and Hall, 2012b; Hamidian and Hall, 2014; Hamidian et al., 2014), plasmid-associated aphA6 was found in one isolate (M05) in our collection. Encoding resistance to both gentamicin and tobramycin, aadB was spotted in 7/18 isolates on pRAY plasmid derivatives. In A. baumannii, aadB was identified within a class 1 integron or more commonly on the globally distributed pRAY plasmid derivatives (Hamidian et al., 2012). While the GC1 isolates M12 and M15 did not carry any pRAY plasmids or aadB, such plasmids were carried by their phylogenetically related isolates M06 and M09. Other pRAY-positive STs included ST85Pas and ST164Pas. In addition to armA, the long version of AbGRI3-1 RIs also carried aac4 that encodes the aminoglycoside modifying enzyme AAC(6')-Ib', conferring resistance to gentamicin rather than amikacin modified by AAC(6')-Ib (Ramirez and Tolmasky, 2010). In agreement with a recent study from Egypt, aac4 was merely carried by a subset of GC2 strains, perhaps carrying the same version of AbGRI3 (Jalal et al., 2021). In addition, a wide array of resistance genes to other antimicrobial classes including multidrug efflux pumps was abundantly identified in our collection.
The advances in the next-generation sequencing technology made possible the genome-wide resistome analysis of different bacterial species and the investigation of the association of resistance genes with mobile genetic elements. Despite the difficulties imparted by the fragmented nature of contigs assembled from the short-read sequencer output, we used a combined approach of de novo assembly and reference mapping to uncover the configurations of RIs carried by our isolates. Examining the integrity of the comM gene, the hotspot for insertion of AbaR, AbaR4, and AbGRI1-type islands was the first step in RI analysis. The gene was found to be interrupted in the majority of the isolates (16/18) equally by the resistance genes-free transposon Tn6022 and either AbaR4 or AbGRI1-like RIs (44.4%). A similar prevalence (46.7%) was reported for Tn6022 in A. baumannii isolates from Korea (Kim et al., 2017). Tn6022, the backbone of AbaR4, comprises five transposition genes (tniCABDE), a universal stress protein-encoding gene (uspA), and a sulfate permease-coding gene (sup) (Hamidian and Hall, 2011). It has been widely identified in lineage 2 GC1 isolates (Hamidian and Hall, 2011). Here, the transposon was carried by GC1 isolates and their phylogenetically related ones (M06 and M09). In addition, the transposon interrupted the comM gene in all GC9 isolates and only one GC2 isolate (M20). In Tn6022, the gene sup is a hotspot for insertion of Tn2006 with an embedded blaOXA−23 generating AbaR4. While widely described in GC2 isolates (Kim et al., 2012, 2013), AbaR4 was identified here within the chromosome of M03 only. Interestingly, co-existing chromosomal and plasmid copies of AbaR4 were recently reported in Proteus mirabilis (Octavia et al., 2020). While not self-transmissible (Bi et al., 2019), AbaR4 mobility to the chromosome of P. mirabilis was proposed to be mediated by plasmids (Octavia et al., 2020). This demonstrates the impending threat of interspecies plasmid-mediated transfer of AbaR4.
Twenty-two AbGRI1 configurations with different backbones were characterized by Bi et al. (2020). Here, we identified three variant structures of AbGRI1. Of them, two variants had the typical AbGRI1 backbone [Tn6022 (or the deletion derivative, Tn6022Δ)-linker-Tn6172]. Despite their unique configurations, they were named here as AbGRI1-like-1 and AbGRI1-like-2 due to the lack of a universal nomenclature system for A. baumannii genomic RIs (Hamidian and Hall, 2018). AbGRI1-like-1 was identified in ST1050/2058Oxf GC2 isolates. It was made up of Tn6022Δ, a plasmid-derived linker with int gene uniquely interrupted by an ISAba125 element and Tn6172. The ISAba125 element interrupting the int gene might act as a hotspot for the insertion of resistance elements flanked by two direct copies of ISAba125 (such as TnaphA6) by homologous recombination. The second variant AbGRI1-like-2 was formed of AbaR4, plasmid-derived linker, and Tn6172 to which a ΔISCR2-ΔTn10 fragment carrying tet(B) gene was inserted (Hamidian and Hall, 2017b). This configuration resembled that of an unnamed RI identified by Bi et al. (2020) that also carried a second copy of AbaR4 inserted within tet(B) gene. AbGRI1-like-2 was solely carried by M14. The third variant of AbGRI1 was a Tn2006-interrupted AbGRI1-1. It had an atypical AbGRI1 structure lacking the plasmid-derived linker. Even though AbaR4 was reported not to have any variants (Hamidian and Hall, 2018), the Tn2006-interrupted AbGRI1-1 was named by Seputiene et al. (2012) as AbaR4b. For convenience, this name was used in the context of this study. In addition to conferring resistance to carbapenems, AbaR4b confers resistance to tetracycline and streptomycin/spectinomycin and was first identified in A. baumannii strains from Lithuania (Seputiene et al., 2012). In Korea, AbaR4b was found in 22.4% of the tested isolates, and all belonged to GC2 (Kim et al., 2017).
The IS26-bracketed RIs AbGRI2 and AbGRI3 co-existed in all GC2 isolates. While AbGRI2 occurred in only one configuration (AbGRI2-15), two variants of AbGRI3 with different resistance spectra were found. AbGRI2 has been reported most commonly in GC2 isolates carrying all or some of the resistance genes blaTEM, aphA1, catA1, and [sul1, aadA1, and aacC1] within a class I integron (Nigro et al., 2013). AbGRI2-15 was first identified by Liepa et al. (2021) in A. baumannii strain collected from a patient in Lebanon. The island is a short version of AbGRI2 in which only aphA1 and blaTEM−1 remained after IS26-mediated deletion and recombination events, as reported by the author. Each of the GC2 isolates carried either of the two variants AbGRI3-1 or AbGRI3-4. AbGRI3-1, also known as Tn6180, was first identified in A. baumannii strains MDR-TJ (GenBank accession: CP003500.1) and TYTH-1 (GenBank accession: CP003856.1) conferring resistance to four classes of antimicrobial agents (Blackwell et al., 2017). It was the least frequently identified among AbGRI3-type islands identified in a collection of A. baumannii isolates from Singapore in a study by Blackwell et al. (2017). In the same study, AbGRI3-4 was the dominant one identified in 46.6% of isolates. AbGRI3-4 is a short version of AbGRI3, most importantly, retaining armA. Blackwell et al. (2017) also highlighted the successful dispersion of AbGRI3-4 in isolates from India and Sweden.
Genomic island sul2 (GIsul2) is an integrating element first described by Nigro and Hall (2011) in the chromosome of A. baumannii strain ATCC 17978 isolated in France in 1951. Within GIsul2 is located a dihydropteroate synthase type 2-coding gene (sul2) preceded by an ISAba1. While thought to provide a promotor for sul2 (Nigro and Hall, 2011), high-level sulfonamide resistance in ATCC 19606 conferred by sul2 was not accompanied by an upstream ISAba1 (Hamidian and Hall, 2017a). GIsul2 was reported as the main vehicle for the mobilization of the sul2 gene (Hamidian and Hall, 2017a). It is also the donor of [CR2-sul2] elements in AbGRI1-type RIs integrated through homologous recombination. Here, GIsul2 was solely identified in GC1 isolates and those found to be phylogenetically related.
Beyond merely defining the resistance genes standing behind MDR and XDR in A. baumannii, WGS was used in the current study for defining the genomic RIs carried by clinical isolates of A. baumannii from Egypt. Epidemiology analysis of the isolates showed a significant representation of the high-risk GCs, particularly GC2. Novel ST (ST2329Oxf), resistance gene (blaADC−258), and IS (ISAba61) were identified. RIs were carried by the majority of the isolates most frequently co-existing in GC2. They were loaded with several genes conferring resistance to various antimicrobial classes and hotspots for the acquisition of more resistance genes. AbGRI1-type RIs showed up with the widest diversity, including two novel configurations.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The study was performed in accordance with relevant guidelines and regulations, and no experiments were performed on humans and/or human tissue samples. The study was approved by the local Ethical Committee of the Clinical and Chemical Pathology Department, Kasr Al-Ainy Hospital, Cairo University. Bacterial isolates were only collected as part of the routine patient care, and informed consent was not required.
MZ, AH, MA-A, HR, and SH contributed to the study design, performance of experiments, and data analysis. SH performed the genome assembly, bioinformatics analysis, and wrote the first draft of the manuscript. All authors read and approved the final version of the manuscript.
The authors thank the Deanship of Scientific Research at King Saud University for funding this work through Project No. RGP-038.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.878912/full#supplementary-material
Abouelfetouh, A., Torky, A. S., and Aboulmagd, E. (2019). Phenotypic and genotypic characterization of carbapenem-resistant Acinetobacter baumannii isolates from Egypt. Antimicrob. Resist. Infect. Control 8, 185. doi: 10.1186/s13756-019-0611-6
Adams, M. D., Pasteran, F., Traglia, G. M., Martinez, J., Huang, F., Liu, C., et al. (2020). Distinct mechanisms of dissemination of NDM-1 metallo-beta-lactamase in acinetobacter species in argentina. Antimicrob. Agents Chemother. 64. doi: 10.1128/AAC.00324-20
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Al-Hassan, L., El Mehallawy, H., and Amyes, S. G. (2013). Diversity in Acinetobacter baumannii isolates from paediatric cancer patients in Egypt. Clin. Microbiol. Infect. 19, 1082–1088. doi: 10.1111/1469-0691.12143
Al-Hassan, L., Elbadawi, H., Osman, E., Ali, S., Elhag, K., Cantillon, D., et al. (2021). Molecular epidemiology of carbapenem-resistant acinetobacter baumannii from Khartoum State, Sudan. Front. Microbiol. 12, 628736. doi: 10.3389/fmicb.2021.628736
Al-Hassan, L., Zafer, M. M., and El-Mahallawy, H. (2019). Multiple sequence types responsible for healthcare-associated Acinetobacter baumannii dissemination in a single centre in Egypt. BMC Infect. Dis. 19, 829. doi: 10.1186/s12879-019-4433-1
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. (accessed December 27, 2020).
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., and Pevzner, P. A. (2016). plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 32, 3380–3387. doi: 10.1093/bioinformatics/btw493
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A., Wisplinghoff, H., and Rodriguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005
Benenson, S., Nir-Paz, R., Golomb, M., Schwartz, C., Amit, S., Moses, A. E., et al. (2018). Carriage of multi-drug resistant bacteria among foreigners seeking medical care. Sci. Rep. 8, 9471. doi: 10.1038/s41598-018-27908-x
Bertelli, C., Laird, M. R., Williams, K. P., Simon Fraser University Research Computing G., Lau, B. Y., and Hoad, G. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Bi, D., Xie, R., Zheng, J., Yang, H., Zhu, X., Ou, H. Y., et al. (2019). Large-scale identification of AbaR-Type genomic islands in acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob. Agents Chemother. 63. doi: 10.1128/AAC.02526-18
Bi, D., Zheng, J., Xie, R., Zhu, Y., Wei, R., Ou, H. Y., et al. (2020). Comparative analysis of AbaR-type genomic islands reveals distinct patterns of genetic features in elements with different backbones. mSphere. 5. doi: 10.1128/mSphere.00349-20
Blackwell, G. A., Holt, K. E., Bentley, S. D., Hsu, L. Y., and Hall, R. M. (2017). Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 72, 1031–1039. doi: 10.1093/jac/dkw542
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonnin, R. A., Cuzon, G., Poirel, L., and Nordmann, P. (2013). Multidrug-resistant Acinetobacter baumannii clone, France. Emerging Infect. Dis. 19, 822–823. doi: 10.3201/eid1905.121618
Carraro, N., Rivard, N., Burrus, V., and Ceccarelli, D. (2017). Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob. Genet. Elements 7, 1–6. doi: 10.1080/2159256X.2017.1304193
Chan, A. P., Choi, Y., Clarke, T. H., Brinkac, L. M., White, R. C., Jacobs, M. R., et al. (2020). AbGRI4, a novel antibiotic resistance island in multiply antibiotic-resistant Acinetobacter baumannii clinical isolates. J. Antimicrob. Chemother. 75, 2760–2768. doi: 10.1093/jac/dkaa266
Chan, A. P., Sutton, G., Depew, J., Krishnakumar, R., Choi, Y., Huang, X. Z., et al. (2015). A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 16, 143. doi: 10.1186/s13059-015-0701-6
CLSI (2020). “Performance standards for antimicrobial susceptibility testing; 30th edition,” in CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Coelho-Souza, T., Reis, J. N., Martins, N., Martins, I. S., Menezes, A. O., Reis, M. G., et al. (2013). Longitudinal surveillance for meningitis by Acinetobacter in a large urban setting in Brazil. Clin. Microbiol. Infect. 19, E241–244. doi: 10.1111/1469-0691.12145
Corvec, S., Caroff, N., Espaze, E., Giraudeau, C., Drugeon, H., and Reynaud, A. (2003). AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52, 629–635. doi: 10.1093/jac/dkg407
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE. 5, e10034. doi: 10.1371/journal.pone.0010034
El Bannah, A. M. S., Nawar, N. N., Hassan, R. M. M., and Salem, S. T. B. (2018). Molecular epidemiology of carbapenem-resistant acinetobacter baumannii in a tertiary care hospital in Egypt: clonal spread of blaOXA-23. Microb. Drug Resist. 24, 269–277. doi: 10.1089/mdr.2017.0057
El-Sayed-Ahmed, M. A., Amin, M. A., Tawakol, W. M., Loucif, L., Bakour, S., and Rolain, J. M. (2015). High prevalence of bla(NDM-1) carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical Isolates in Egypt. Antimicrob. Agents Chemother. 59, 3602–3605. doi: 10.1128/AAC.04412-14
EUCAST (2021). Breakpoint tables for interpretation of MICs and zone diameters, version 11.0. Available online at: http://www.eucast.org (accessed May 1, 2021).
Fam, N. S., Gamal, D., Mohamed, S. H., Wasfy, R. M., Soliman, M. S., El-Kholy, A. A., et al. (2020). Molecular characterization of carbapenem/colistin-resistant acinetobacter baumannii clinical isolates from Egypt by whole-genome sequencing. Infect. Drug Resist. 13, 4487–4493. doi: 10.2147/IDR.S288865
Fournier, P. E., Vallenet, D., Barbe, V., Audic, S., Ogata, H., Poirel, L., et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2, e7. doi: 10.1371/journal.pgen.0020007
Gaiarsa, S., Batisti Biffignandi, G., Esposito, E. P., Castelli, M., Jolley, K. A., Brisse, S., et al. (2019). Comparative analysis of the two acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front. Microbiol. 10, 930. doi: 10.3389/fmicb.2019.00930
Galimand, M., Courvalin, P., and Lambert, T. (2003). Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47, 2565–2571. doi: 10.1128/AAC.47.8.2565-2571.2003
Ghaith, D. M., Zafer, M. M., Al-Agamy, M. H., Alyamani, E. J., Booq, R. Y., and Almoazzamy, O. (2017). The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann. Clin. Microbiol. Antimicrob. 16, 34. doi: 10.1186/s12941-017-0208-y
Gheorghe, I., Barbu, I. C., Surleac, M., Sarbu, I., Popa, L. I., Paraschiv, S., et al. (2021). Subtypes, resistance and virulence platforms in extended-drug resistant Acinetobacter baumannii Romanian isolates. Sci. Rep. 11, 13288. doi: 10.1038/s41598-021-92590-5
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hamidian, M., and Hall, R. M. (2011). AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 66, 2484–2491. doi: 10.1093/jac/dkr356
Hamidian, M., and Hall, R. M. (2014). pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J. Antimicrob. Chemother. 69, 1146–1148. doi: 10.1093/jac/dkt488
Hamidian, M., and Hall, R. M. (2017a). Acinetobacter baumannii ATCC 19606 carries GIsul2 in a genomic island located in the Chromosome. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.01991-16
Hamidian, M., and Hall, R. M. (2017b). Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J. Antimicrob. Chemother. 72, 2944–2947. doi: 10.1093/jac/dkx206
Hamidian, M., and Hall, R. M. (2018). The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 - structure, origin and evolution. Drug Resist. Updat. 41, 26–39. doi: 10.1016/j.drup.2018.10.003
Hamidian, M., Holt, K. E., Pickard, D., Dougan, G., and Hall, R. M. (2014). A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J. Antimicrob. Chemother. 69, 955–958. doi: 10.1093/jac/dkt454
Hamidian, M., and Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 5. doi: 10.1099/mgen.0.000306
Hamidian, M., Nigro, S. J., and Hall, R. M. (2012). Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J. Antimicrob. Chemother. 67, 2833–2836. doi: 10.1093/jac/dks318
Hassan, R. M., Salem, S. T., Hassan, S. I. M., Hegab, A. S., and Elkholy, Y. S. (2021). Molecular characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates from Egyptian patients. PLoS ONE. 16, e0251508. doi: 10.1371/journal.pone.0251508
Hsueh, P. R., Teng, L. J., Chen, C. Y., Chen, W. H., Yu, C. J., Ho, S. W., et al. (2002). Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerging Infect. Dis. 8, 827–832. doi: 10.3201/eid0805.020014
Hua, X., Moran, R. A., Xu, Q., He, J., Fang, Y., Zhang, L., et al. (2021). Acquisition of a genomic resistance island (AbGRI5) from global clone 2 through homologous recombination in a clinical Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 76, 65–69. doi: 10.1093/jac/dkaa389
Jaidane, N., Naas, T., Oueslati, S., Bernabeu, S., Boujaafar, N., Bouallegue, O., et al. (2018). Whole-genome sequencing of NDM-1-producing ST85 Acinetobacter baumannii isolates from Tunisia. Int. J. Antimicrob. Agents 52, 916–921. doi: 10.1016/j.ijantimicag.2018.05.017
Jalal, D., Elzayat, M. G., Diab, A. A., El-Shqanqery, H. E., Samir, O., Bakry, U., et al. (2021). Deciphering multidrug-resistant acinetobacter baumannii from a pediatric cancer hospital in Egypt. mSphere 6, e0072521. doi: 10.1128/mSphere.00725-21
Karah, N., Dwibedi, C. K., Sjostrom, K., Edquist, P., Johansson, A., Wai, S. N., et al. (2016). Novel aminoglycoside resistance transposons and transposon-derived circular forms detected in carbapenem-resistant acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 60, 1801–1818. doi: 10.1128/AAC.02143-15
Karah, N., Sundsfjord, A., Towner, K., and Samuelsen, O. (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001
Khuntayaporn, P., Kanathum, P., Houngsaitong, J., Montakantikul, P., Thirapanmethee, K., and Chomnawang, M. T. (2021). Predominance of international clone 2 multidrug-resistant Acinetobacter baumannii clinical isolates in Thailand: a nationwide study. Ann. Clin. Microbiol. Antimicrob. 20, 19. doi: 10.1186/s12941-021-00424-z
Kim, D. H., Choi, J. Y., Kim, H. W., Kim, S. H., Chung, D. R., Peck, K. R., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 57, 5239–5246. doi: 10.1128/AAC.00633-13
Kim, D. H., Jung, S. I., Kwon, K. T., and Ko, K. S. (2017). Occurrence of diverse AbGRI1-type genomic islands in acinetobacter baumannii global clone 2 isolates from South Korea. Antimicrob. Agents Chemother. 61. doi: 10.1128/AAC.01972-16
Kim, D. H., Park, Y. K., and Ko, K. S. (2012). Variations of AbaR4-type resistance islands in Acinetobacter baumannii isolates from South Korea. Antimicrob. Agents Chemother. 56, 4544–4547. doi: 10.1128/AAC.00880-12
Leal, N. C., Campos, T. L., Rezende, A. M., Docena, C., Mendes-Marques, C. L., De Sa Cavalcanti, F. L., et al. (2020). Comparative genomics of acinetobacter baumannii clinical strains from brazil reveals polyclonal dissemination and selective exchange of mobile genetic elements associated with resistance genes. Front. Microbiol. 11, 1176. doi: 10.3389/fmicb.2020.01176
Lee, Y., D'souza, R., Yong, D., and Lee, K. (2016). Prediction of putative resistance islands in a carbapenem-resistant acinetobacter baumannii global clone 2 clinical isolate. Ann. Lab. Med. 36, 320–324. doi: 10.3343/alm.2016.36.4.320
Leite, G. C., Oliveira, M. S., Perdigao-Neto, L. V., Rocha, C. K., Guimaraes, T., Rizek, C., et al. (2016). Antimicrobial combinations against pan-resistant acinetobacter baumannii isolates with different resistance mechanisms. PLoS ONE. 11, e0151270. doi: 10.1371/journal.pone.0151270
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–245. doi: 10.1093/nar/gkw290
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Liepa, R., Mann, R., Osman, M., Hamze, M., Gunawan, C., and Hamidian, M. (2021). Cl415, a carbapenem-resistant Acinetobacter baumannii isolate containing four AbaR4 and a new variant of AbGRI2, represents a novel global clone 2 strain. J. Antimicrob. Chemother. 77, 345–350. doi: 10.1093/jac/dkab399
Lopes, B. S., Al-Agamy, M. H., Ismail, M. A., Shibl, A. M., Al-Qahtani, A. A., Al-Ahdal, M. N., et al. (2015). The transferability of blaOXA-23 gene in multidrug-resistant Acinetobacter baumannii isolates from Saudi Arabia and Egypt. Int. J. Med. Microbiol. 305, 581–588. doi: 10.1016/j.ijmm.2015.07.007
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Morris, F. C., Dexter, C., Kostoulias, X., Uddin, M. I., and Peleg, A. Y. (2019). The Mechanisms of disease caused by Acinetobacter baumannii. Front. Microbiol. 10, 1601. doi: 10.3389/fmicb.2019.01601
Müller, C., Stefanik, D., Wille, J., Hackel, M., Higgins, P. G., and Siefert, H. (2019). “Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii clinical isolates and identification of the novel international clone IC9: results from a worldwide surveillance study (2012–2016)”, in Paper presented at the ECCMID 2019: Proceeding of the 29th European Congress of Clinical Microbiology & Infectious Diseases. Amsterdam, Netherlands.
Musila, L., Kyany'a, C., Maybank, R., Stam, J., Oundo, V., and Sang, W. (2021). Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target gram-negative bacteria in Kenya. PLoS ONE. 16, e0246937. doi: 10.1371/journal.pone.0246937
Nigro, S. J., Farrugia, D. N., Paulsen, I. T., and Hall, R. M. (2013). A novel family of genomic resistance islands, AbGRI2, contributing to aminoglycoside resistance in Acinetobacter baumannii isolates belonging to global clone 2. J. Antimicrob. Chemother. 68, 554–557. doi: 10.1093/jac/dks459
Nigro, S. J., and Hall, R. M. (2011). GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J. Antimicrob. Chemother. 66, 2175–2176. doi: 10.1093/jac/dkr230
Nigro, S. J., and Hall, R. M. (2012a). Antibiotic resistance islands in A320 (RUH134), the reference strain for Acinetobacter baumannii global clone 2. J. Antimicrob. Chemother. 67, 335–338. doi: 10.1093/jac/dkr447
Nigro, S. J., and Hall, R. M. (2012b). Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J. Antimicrob. Chemother. 67, 1342–1346. doi: 10.1093/jac/dks037
Nigro, S. J., and Hall, R. M. (2016). Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 71, 1135–1147. doi: 10.1093/jac/dkv440
Nigro, S. J., Post, V., and Hall, R. M. (2011). Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66, 1504–1509. doi: 10.1093/jac/dkr163
Octavia, S., Xu, W., Ng, O. T., Marimuthu, K., Venkatachalam, I., Cheng, B., et al. (2020). Identification of AbaR4 Acinetobacter baumannii resistance island in clinical isolates of blaOXA-23-positive Proteus mirabilis. J. Antimicrob. Chemother. 75, 521–525. doi: 10.1093/jac/dkz472
Ostholm-Balkhed, A., Tarnberg, M., Nilsson, M., Nilsson, L. E., Hanberger, H., Hallgren, A., et al. (2013). Travel-associated faecal colonization with ESBL-producing Enterobacteriaceae: incidence and risk factors. J. Antimicrob. Chemother. 68, 2144–2153. doi: 10.1093/jac/dkt167
Post, V., White, P. A., and Hall, R. M. (2010). Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 1162–1170. doi: 10.1093/jac/dkq095
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ribeiro-Goncalves, B., Francisco, A. P., Vaz, C., Ramirez, M., and Carrico, J. A. (2016). PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 44, W246–251. doi: 10.1093/nar/gkw359
Salloum, T., Tannous, E., Alousi, S., Arabaghian, H., Rafei, R., Hamze, M., et al. (2018). Genomic mapping of ST85 blaNDM-1 and blaOXA-94 producing Acinetobacter baumannii isolates from Syrian Civil War Victims. Int. J. Infect. Dis. 74, 100–108. doi: 10.1016/j.ijid.2018.07.017
Seputiene, V., Povilonis, J., and Suziedeliene, E. (2012). Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob. Agents Chemother. 56, 1969–1973. doi: 10.1128/AAC.05678-11
Tada, T., Uchida, H., Hishinuma, T., Watanabe, S., Tohya, M., Kuwahara-Arai, K., et al. (2020). Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates from hospitals in Myanmar. J Glob Antimicrob Resist 22, 122–125. doi: 10.1016/j.jgar.2020.02.011
Tatusova, T., Dicuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tomaschek, F., Higgins, P. G., Stefanik, D., Wisplinghoff, H., and Seifert, H. (2016). Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of acinetobacter baumannii outbreak and sporadic isolates. PLoS ONE. 11, e0153014. doi: 10.1371/journal.pone.0153014
Turton, J. F., Woodford, N., Glover, J., Yarde, S., Kaufmann, M. E., and Pitt, T. L. (2006). Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44, 2974–2976. doi: 10.1128/JCM.01021-06
Wareth, G., Linde, J., Nguyen, N. H., Nguyen, T. N. M., Sprague, L. D., Pletz, M. W., et al. (2021). WGS-based analysis of carbapenem-resistant Acinetobacter baumannii in Vietnam and molecular characterization of antimicrobial determinants and MLST in Southeast Asia. Antibiotics (Basel) 10. doi: 10.3390/antibiotics10050563
Wasfi, R., Rasslan, F., Hassan, S. S., Ashour, H. M., and Abd El-Rahman, O. A. (2021). Co-Existence of Carbapenemase-Encoding GeJ. Clin. Microbiol.nes in Acinetobacter baumannii from Cancer Patients. Infect Dis Ther. 10, 291–305. doi: 10.1007/s40121-020-00369-4
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wright, M. S., Haft, D. H., Harkins, D. M., Perez, F., Hujer, K. M., Bajaksouzian, S., et al. (2014). New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio. 5, e00963–e00913. doi: 10.1128/mBio.00963-13
Zafer, M. M., Hussein, A. F. A., Al-Agamy, M. H., Radwan, H. H., and Hamed, S. M. (2021). Genomic characterization of extensively drug-resistant NDM-producing acinetobacter baumannii clinical isolates with the emergence of novel bla ADC-257. Front. Microbiol. 12, 736982. doi: 10.3389/fmicb.2021.736982
Zander, E., Nemec, A., Seifert, H., and Higgins, P. G. (2012). Association between beta-lactamase-encoding bla(OXA-51) variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J. Clin. Microbiol. 50, 1900–1904. doi: 10.1128/JCM.06462-11
Zhang, G., Leclercq, S. O., Tian, J., Wang, C., Yahara, K., Ai, G., et al. (2017). A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3”)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 13, e1006602. doi: 10.1371/journal.pgen.1006602
Keywords: Acinetobacter baumannii, whole genome sequencing, resistance islands, AbaR4, AbGRI1, AbGRI2, AbGRI3, RI-PER-7
Citation: Hamed SM, Hussein AFA, Al-Agamy MH, Radwan HH and Zafer MM (2022) Genetic Configuration of Genomic Resistance Islands in Acinetobacter baumannii Clinical Isolates From Egypt. Front. Microbiol. 13:878912. doi: 10.3389/fmicb.2022.878912
Received: 18 February 2022; Accepted: 22 June 2022;
Published: 22 July 2022.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Sara Domingues, University of Coimbra, PortugalCopyright © 2022 Hamed, Hussein, Al-Agamy, Radwan and Zafer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mai M. Zafer, bWFpemFmZXJAYWN1LmVkdS5lZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.