- 1Key Laboratory of Medical Microbiology and Parasitology and Key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, School of Basic Medical Sciences, Guizhou Medical University, Guiyang, China
- 2Center for Yunnan Plateau Biological Resources Protection and Utilization, College of Biological Resource and Food Engineering, Qujing Normal University, Qujing, China
- 3Section of Genetics, Institute for Research and Development in Health and Social Care, Battaramulla, Sri Lanka
- 4Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 5Faculty of Science, Environmental Science Research Center, Chiang Mai University, Chiang Mai, Thailand
- 6Fungal Physiology, Westerdijk Fungal Biodiversity Institute and Fungal Molecular Physiology, Utrecht University, Utrecht, Netherlands
- 7Guizhou Provincial Academician Workstation of Microbiology and Health, Guizhou Academy of Tobacco Science, Guiyang, China
Traditional Chinese medicine is one of the ancient medicines which is popular in Asian countries, among which the residue produced by the use of anti-biodegradables is endless, and causes significant adverse impacts on the environment. However, the high acidity of anti-biodegradable residues and some special biological activities make it difficult for microorganisms to survive, resulting in a very low degradation rate of lignocellulose in naturally stacked residues, which directly impedes the degradation of residues. We aimed to identify the fungal strains that efficiently biodegrade anti-biodegradable residue and see the possibility to improve the biodegradation of it and other agricultural wastes by co-cultivating these fungi. We isolated 302 fungal strains from anti-biodegradable residue to test hydrolysis ability. Finally, we found Coniochaeta sp., Fomitopsis sp., Nemania sp., Talaromyces sp., Phaeophlebiopsis sp. which inhabit the anti-biodegradable residues are capable of producing higher concentrations of extracellular enzymes. Synergistic fungal combinations (viz., Fomitopsis sp. + Phaeophlebiopsis sp.; Talaromyces sp. + Coniochaeta sp. + Fomitopsis sp.; Talaromyces sp. + Fomitopsis sp. + Piloderma sp. and Talaromyces sp. + Nemania sp. + Piloderma sp.) have better overall degradation effect on lignocellulose. Therefore, these fungi and their combinations have strong potential to be further developed for bioremediation and biological enzyme industrial production.
Introduction
Biomass is the fourth largest energy source in parallel with wind energy, solar energy, nuclear energy, and other new energy sources. This is considered the main alternative energy source today (Sherwood, 2020). It includes lignocellulose in the waste generated by plants, animals, and agricultural production (Zeng et al., 2013). Lignocelluloses are composed of three macromolecules: cellulose, hemicellulose, and lignin (Ni and Tokuda, 2013), which constitute an anti-hydrolysis barrier and make it difficult to be hydrolyzed (Ojha et al., 2015; Barros et al., 2016). Among them, cellulose and hemicellulose can be converted into glucose after hydrolysis. Xylose and other fermentable sugars (monosaccharides) can be further used to produce biofuels (Tan et al., 2014; Taha et al., 2016).
It has been widely studied that the application of traditional industrial and agricultural biomass, and recognized by filamentous fungi fermentation method to produce extracellular enzymes for degradation of lignocellulose has a significant effect (Lange, 2017; Kwak et al., 2019). In addition, enzymes produced by filamentous fungi have been found in nature that can be used for bioremediation, such as the ligninolytic enzyme system, which can purify heavy metal dyes in sewage, and depolymerize chlorophenols, polycyclic aromatic hydrocarbons (PAHs), organophosphorus, and phenols (Falade et al., 2017). Currently, the enzyme-producing fungi include Trichoderma reesii, wood/white-rotting fungi, and Aspergillus (Namnuch et al., 2020; Silva et al., 2020; Zhuo and Fan, 2021), which are mostly isolated from agricultural wastes such as corn straw, bagasse, wheat straw, rice husk, animal feces and some terrestrial plants (Falade et al., 2017).
However, there are also quite a number of specialized biomasses in the world, which are difficult to degraded via ordinary exogenous microorganisms due to their various characteristics, such as some extreme conditions of medical waste biomass (Hutchins et al., 2019). Isolation and screening of filamentous fungi with strong survival ability from their own accumulation environment has been increasing, and many successful cases have been reported recently. For example, Ming et al. (Ming et al., 2019) screened out several enzyme-producing filamentous fungi with strong acid tolerance from liquor grains with high acidity.
Traditional Chinese Medicine (TCM) is one of the ancient medicines which influences China and other Southeast Asian countries. The States Environment Protection Agency (SEPA; States Environment Protection Agency [SEPA], 1997) has mentioned that 11,146 species of different herbs and plants are used in TCM of which 492 species are cultivated and the remaining 10,654 species are wild plants. However, with the population growth, the demand for TCM has increased, and TCM and its residues are causing different adverse impacts on the environment (e.g., over-exploitation of plant materials, soil degradation and erosion, chemical pollution in agricultural lands). From a conservative estimate, the Chinese pharmaceutical industry of TCM can produce 60–70 million tons of residues every year, among which the TCMR (Traditional Chinese Medicine Residues) with strong anti-microbial action residues constitutes a significant proportion (Lu and Li, 2021). At the same time, such pharmaceutical residues are often accompanied by extremely low pH values, which makes it difficult for ordinary microorganisms to survive, leading to a low degradation rate of natural stacking of TCMR and irreversible damage to the environment. However, it is also a kind of agricultural biomass, containing high lignocellulose (Čater et al., 2015; Zhang and Sun, 2018; Zhu and Sun, 2018). If used correctly, it has the potential to not only reduce the pollution created by the particular environment but also to find a way for the development of biomass energy.
Meanwhile, the structure of lignocellulose is complicated and different fungi have limitations in enzyme production. Common single fungi cannot achieve complete degradation of lignocellulose (Karuppiah et al., 2021). Research shows that different microorganisms in the microbial community operate synergistically through the secretion of a variety of biocatalysts, in order to achieve comprehensive enzyme production to degrade lignocellulose (Silva et al., 2021). Therefore, we can use the method of artificial screening and mixing, fungi producing different enzymes in the same medium for co-culture to improve the degradation rate of lignocellulose.
In order to solve these problems, we report the isolation of fungi from potent anti-biodegradable TCMR, explore new lignocellulose-degrading enzymes production strains, and by using the selected strain of hydrolysis ability to build synergistic fungal combinations (co-cultivation), realize the degradation of lignocellulose. The results here would be helpful to bioremediation and develop better enzyme producers for lignocellulosic biorefinery.
Materials and Methods
Materials
Fungal strains were directly isolated by the spread plate method from anti-biodegradable residue which was piled up for 2–5 years at Guizhou Shuangsheng Company (Guiyang, China). Glycine, 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid; ABTS), Avicel, Briffon-Robinson 1buffer (pH 4.5), Birchwood, pNP-β-D-glucopyranose (pNP-BGL), pNP-β-D-xylopyranose (pNP-BXL) and other chemicals were purchased from Sigma-Aldrich to determine enzymatic activity; Carboxymethyl cellulose and xylan for solid media were purchased from Hongdaer Biotechnology Company (Guiyang, China) for preliminary screening of fungi.
Analysis of Carbohydrate Composition and Lignin Content in Anti-biodegradable Residue
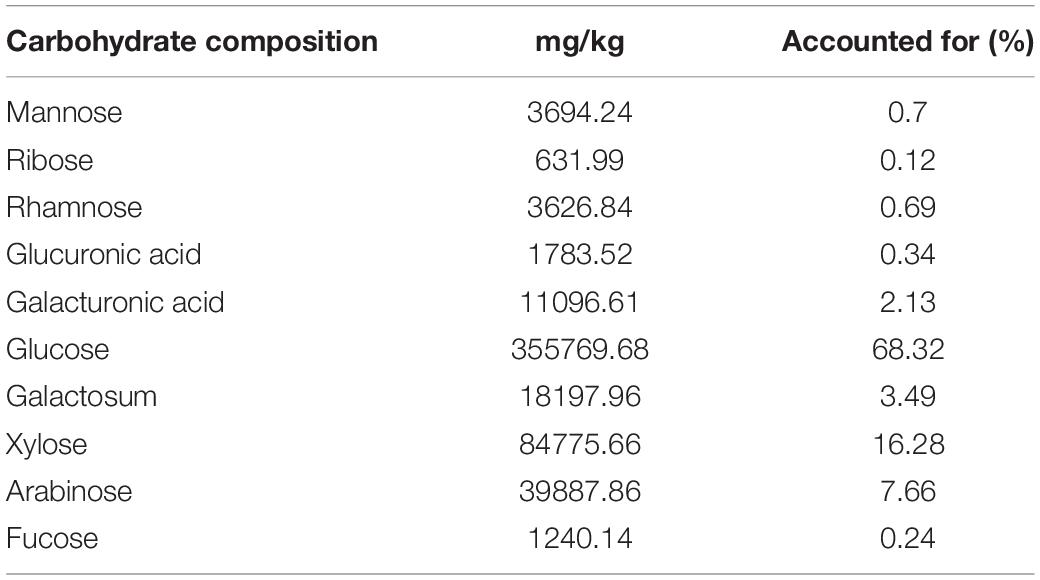
Fresh anti-biodegradable residue (younger than 2 months) was obtained from Guizhou Shuangsheng Company and ground in a blender (Conair Waring Pulverizer, Fisher Scientific), thereafter, 100 g was sieved through 50 mesh and was placed in a 40°C drying oven for 24 h. After grinding, carbohydrate composition was determined using high-performance liquid chromatography by Shandong Kechuang Quality Testing Company, and the lignin content was determined by a lignin assay kit (Solarbio, China) according to the instructions provided by the manufacturer.
Isolation and Identification of Fungi
A suitable anti-biodegradable residue tissue block of 10 g was selected and washed with sterile normal saline three times, fungi were isolated by plant tissue separation (Makhuvele et al., 2017; Thi Minh Le et al., 2019), the sample was repeated three times. Genomic DNA of the purified fungi was extracted using Ezup Column Fungal genomic DNA Extraction Kit (Qingke, Chongqing) and used as a template for internal transcribed spacer (ITS) region amplification (Bellemain et al., 2010). Prime pair ITS5 and ITS4 (Mishra et al., 2017) was used to amplify the ITS regions (Thermal Cycler PCR instrument BIO-RAD T100, Japan). The PCR products were sequenced and used for the identification of fungi by nucleotide BLAST (Altschul et al., 1990) against the NCBI databases. The living strains were deposited at Guizhou Medical University. Phylogenetic tree (Supplementary Figure 1) was constructed by using ITS regional sequences of 302 residue isolates and by MEGA10 using the neighbor-joining (NJ) method after multiple alignments of sequences data (Saitou and Nei, 1987). The corrected evolutionary distance was evaluated according to the p-distance model (Nei and Kumar, 2000). In order to estimate the consensus of the branching, the bootstrap resampling analysis of the phylogenetic tree was employed with 1000 replicates of the data set (Felsenstein, 1985). According to the results of phylogenetic analysis, the pie chart was constructed with Adobe Illustrator CS5 software and Office.
Preliminary Screening of Fungi on Special Media
The fungi were pre-grown on a PDA agar medium at 25°C for 3–7 days, then mycelium was transferred to three different sole carbon source media. (1) carboxymethyl cellulose Congo red agar medium (CMC-red): 1% sodium CMC, 0.05% KH2PO4, 0.05% MgSO4⋅7H2O, 1.5% agar, 0.02% Congo red, 0.2% gelatin, pH 6.8–7.2; Gupta et al., 2012; Fen et al., 2014; Wu et al., 2016); (2) xylan Congo red agar medium (Xyl-red): 0.5% xylan, 0.01% Congo red, 0.1% MgSO4⋅7H2O, 0.5% yeast extract, 0.1% KH2PO4, 1.5% agar, 0.1% (NH4)2SO4, 0.5% NaCl, pH 6.5–7; Liu et al., 2010), and (3) potato dextrose agar containing 0.01% aniline blue (PDA-blue), pH 6–7 (Wang H. et al., 2009). The ratio of the hydrolytic circle (D) to fungal colony diameter (d) was measured after 5 days of constant temperature incubation. Analysis of hydrolytic efficacy was performed by division of the diameter of mycelium growth by that of the clearance zone.
Re-screening of Fungi in Liquid Culture Medium of Anti-biodegradable Residue
Fungi with high activity were cultured on a PDA plate to produce mycelium and spores. The culture medium with fungus was then placed in a sterile beaker, and spores and mycelia were separated by Tween80. Adding 2 × 106 spores (Li et al., 2016) of each fungus into minimal medium (MM; composed of 4 g/L glucose and 10 g/L peptone), after shaking for 3–7 days at 28°C and a rotational speed of 30× g, mycelia were filtered through a 200-mesh (10 cm diameter) sieve and collected, and fully washed with sterile water. Mycelia weighing 2 g (wet weight) was transferred into a 50 ml culture medium with anti-biodegradable residue (the residue was filtered through a 50-mesh sieve) as the only carbon source (anti-biodegradable residue 3 g, Peptone 0.5 g, Yeast powder 0.25 g, NaCl 0.25 g, KH2PO4 0.05 g, MgSO4⋅7H2O 0.025 g, (NH4)2SO4 0.25 g, CuSO4 0.0003 g; Wang J. F. et al., 2009; Cheng et al., 2012; Zhang et al., 2015) at 28°C, 30× g, 2 mL supernatant was taken at the same time on d 3, 5, 7, 9, 11, centrifuged at 4°C, 21,100× g for 10 min, and the clarified supernatant was taken and stored in −80°C for use.
Construction of Synergistic Fungal Combinations (Co-cultivation)
The single filamentous fungi, which can degrade cellulose hemicellulose lignin effectively, were paired by the permutation group (Ming et al., 2019) method, in order to construct fungal combinations that can degrade lignocellulose comprehensively. The specific method is to use a sterile knife to cut the thriving fungal culture medium into 5 mm square pieces, and transfer them to a new PDA culture medium. Each piece of fungus is separated by about 1 cm. Then, the medium is sealed in a 28°C medium for 3–7 days to observe the ability of fungi to grow in co-cultivation conditions.
Enzyme Activity Assay
β-glucosidase and β-xylosidase activities were measured with the substrates para-nitrophenyl β-glucoside (pNP-BGL) and para-nitrophenyl β-xyloside (pNP-BXL), respectively. The reaction contained 40 μl culture supernatant, 10 μl 0.1% pNP-BGL or pNP-BXL, and 50 μl 100 mM sodium acetate, pH 5.0. The reactions were performed overnight (∼16 h) at 25°C followed by the addition of 100 μl 250 mM Na2CO3 prior to measurement of the absorbance at 405 nm using a microtiter plate reader (biotek epoch2, US). Para-nitrophenol (0–100 μM) was used as the standard (Patyshakuliyeva et al., 2016). One unit was defined as the amount of enzyme releasing 1 μmol pNP from pNP-BGL or pNP-BXL under the assay condition.
Cellulase and xylanase activities were measured toward avicel cellulose and birchwood xylan, respectively. The reaction contained 20 μl culture supernatant and 180 μl 1% avicel or birch xylan in 50 mM sodium acetate, pH 5.0, and was incubated overnight (∼16 h) at 25°C. Mixing 100 μl of the reaction mixture with 150 μl 3,5-dinitrosalicylic acid solution (1% 3,5-dinitrosalicylic acid, 0.2% phenol, 0.05% Na2SO3, and 1% NaOH), incubated at 95°C, 30 min and cooled on ice prior to measurement of the absorbance at 560 nm using a microtiter plate reader (Miller, 1959). 2–20 mM D-glucose or D-xylose was used as standard (Peciulyte et al., 2017).
Peroxidase and laccase activities were measured toward 2,2′-azinobis (3-ethylbenzthiazoline-6-sulfonic acid; ABTS) with or without supplement of hydrogen peroxide, respectively. The peroxidase reaction contained 20 μl culture supernatant, 20 μl 140 μM ABTS, 10 μl 3% H2O2, 25 μl 400 mM Britton–Robinson buffer, pH 4.5, and 125 μl ddH2O. Detection of initial rates of oxidation was performed at 440 nm with a 2 min interval up to 60 min, at 25°C using a microtiter plate reader. The laccase reaction contained 20 μl culture supernatant, 20 μl 140 μM ABTS, 20 μl 500 mM glycine-HCl, pH 3.0, and 140 μl ddH2O. The initial rates of hydrolysis were determined by measuring absorbance at 440 nm in a 2 min interval for 30 min, at 25°C. Laccase and peroxidase activities were calculated based on Lambert-Beer law, where the extinction coefficient of 3.6× 104 M–1 cm–1 was used as mentioned in Childs and Bardsley (1975) and Srinivasan et al. (1995).
Hydrolytic Weight Loss Ability Test
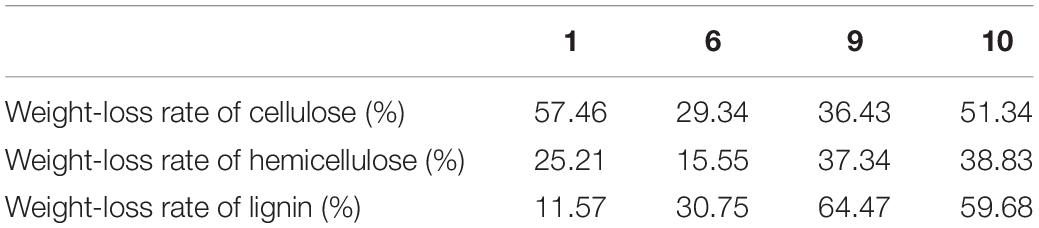
The selected co-cultivation was incubated at 28°C for 15 days, and the actual hydrolysis capacity of the residue was tested. The residue was taken from the liquid medium and thoroughly dried before weighing to determine the total weight loss rate of the residue as well as the degradation rates of cellulose, hemicellulose, and lignin. Each strain was repeated three times, and the residue liquid medium without inoculation was used as a negative control. The negative control M0 was used to calculate the hydrolysis rate of residue and cellulose, instead of residue weight before inoculation of fungi M1, in order to eliminate the natural loss of residue powder placed in the solution during the experiment and ensure the strictest of the experiment. The formula for calculating the total weight loss rate of drug residue hydrolysis is (M0-M2)/M0, where M0 represents the dry weight of drug residue after 15 days of culture in a negative control group under the same conditions, and M2 represents the weight of drug residue after hydrolysis (Ming et al., 2017). Hemicellulose and lignin content in the hydrolyzed residue was detected by the content detection Kit (Solarbio BC4445, Solarbio BC4205) and cellulose content detection kit (ZCIBIO ZC-S0876). The specific operation method is described in the instruction, three biological replicates were performed.
Results
Identification of Strains
The ITS sequences were used to identify the strains from mega blast results in GenBank. In this preliminary study, we only intended to check whether there are fungal taxa that are inhabiting TCM residues. In Supplementary Table 1, we provide results that were generated in the mega blast. It provides the closest hits which are similar to our selected strains.
Determination of the Composition of Anti-biodegradable Residue
The determination of glycosylated components in the residue showed that glucose, accounting for 68.32%, glucose is the reducing sugar of cellulose, indicating that the sample contains a lot of cellulose. Plus, there were xylose, arabinose, mannose, galactose, and other five-carbon sugars, accounting for about 30% (Table 1), and the analysis of lignin content in the residue showed that there was 13.66% lignin. The lignin content test suggests the anti-biodegradable residue contains a moderate amount of lignin in liquid samples. In addition, 50 ml of 6% residue is pH 4.9.
Biodiversity and Phylogenetic Analysis of Isolated Filamentous Fungi
A total of 302 fungi were isolated and labeled as ZYJHYZ01-ZYJHYZ302 (Supplementary Table 2). Phylogenetic analysis showed that they belong to 30 genera in 3 phyla. Among the isolates, 72.19% of strains (218 isolates of 16 genera) were identified as Ascomycota, while 19.87% of strains (60 isolates of 12 genera) were identified as Basidiomycota and 7.95% strains (24 isolates of two genera) were identified as Mucoromycota (Figure 1A). At the genus level, 40.07% of isolates (n = 121) belong to Aspergillus, 7.62% to Coniochaeta (n = 23), 6.29% to Filobasidium (n = 19), 5.96% to Penicillium (n = 18), and 5.30% to Mucor (n = 16; Figure 1B).
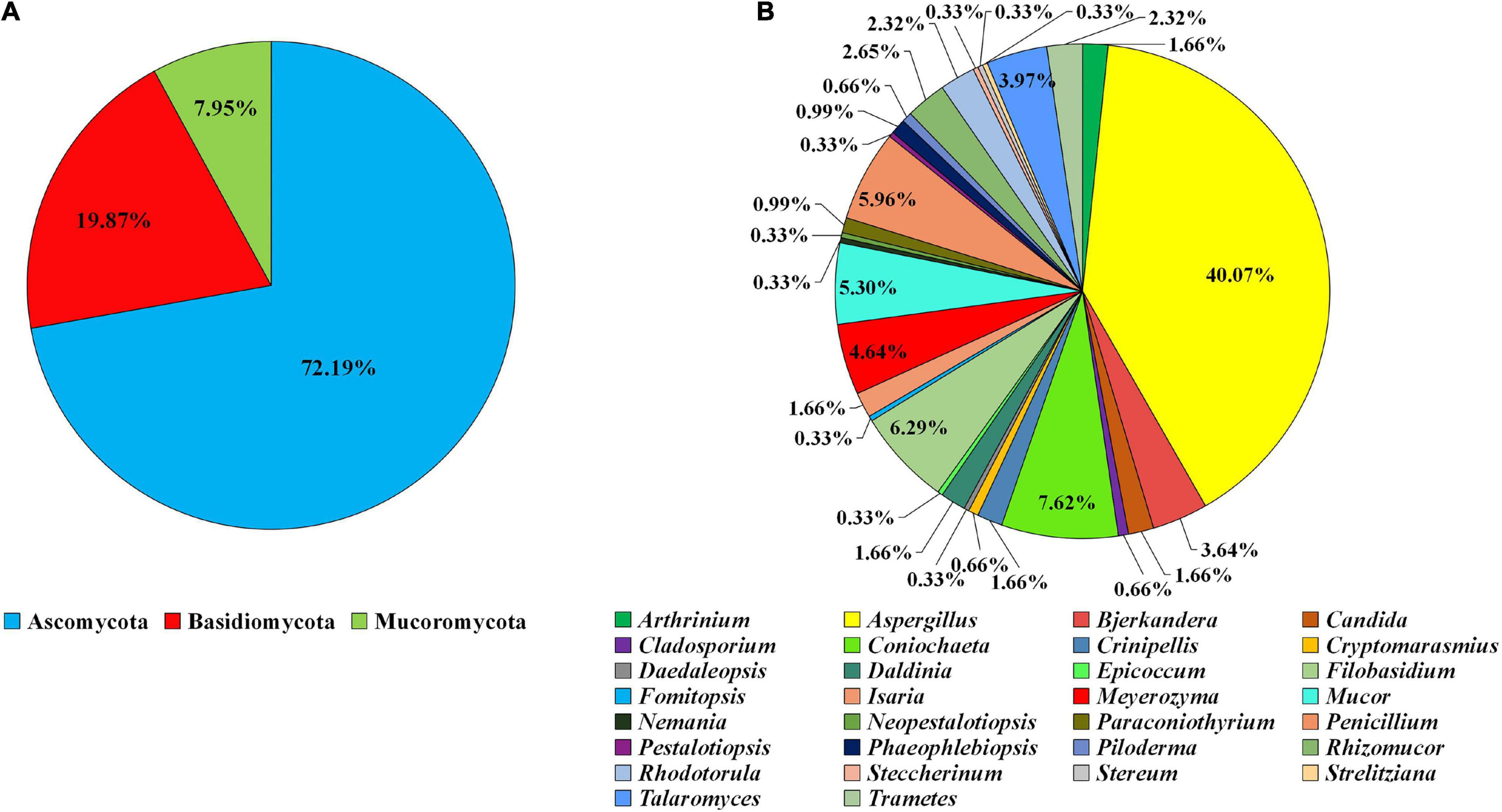
Figure 1. Composition of the filamentous fungal strains isolated from anti-biodegradable residue. (A) The classification was shown at the phyla level. (B) Generic level.
A Single Polymer Was Used as the Sole Carbon Source in Solid Medium With Indicator
Three-hundred and two (302) isolates were screened by inoculating onto a solid medium with a single polymer (CMC, xylan, or glucose) as the sole carbon source and indicator added: CMC-red, PDA-blue, Xyl-red, 25°C, the ratio of the hydrolytic circle (D) to fungal colony diameter (d) was measured after 5 days of constant temperature incubation. From this analysis, we were able to find that 35 strains produced transparent circles while 24 strains showed a ratio greater than or equal to 2 (Supplementary Table 3).
Among them, the D/d ratio of ZYJHYZ163, ZYJHYZ254, ZYJHYZ257, ZYJHYZ265, and ZYJHYZ270 were greater than other strains. According to the mega blast results, Aspergillus fumigatus (ZYJHYZ279) and Aspergillus flavus (ZYJHYZ260) were indicated as harmful to human and animal health, thus, further experimental studies were abandoned. Therefore, fungi with a total score of 2 were preliminarily screened through three kinds of solid media for the next experiment, and a total of 24 strains were identified: ZYJHYZ257, ZYJHYZ259, ZYJHYZ261, ZYJHYZ263, ZYJHYZ265, ZYJHYZ267, ZYJHYZ268, ZYJHYZ269, ZYJHYZ270, ZYJHYZ271, ZYJHYZ272, ZYJHYZ273, ZYJHYZ274, ZYJHYZ53, ZYJHYZ163, ZYJHYZ244, ZYJHYZ242, ZYJHYZ28, ZYJHYZ246, ZYJHYZ254, ZYJHYZ255, ZYJHYZ240, ZYJHYZ247, and ZYJHYZ256 (Figure 2).
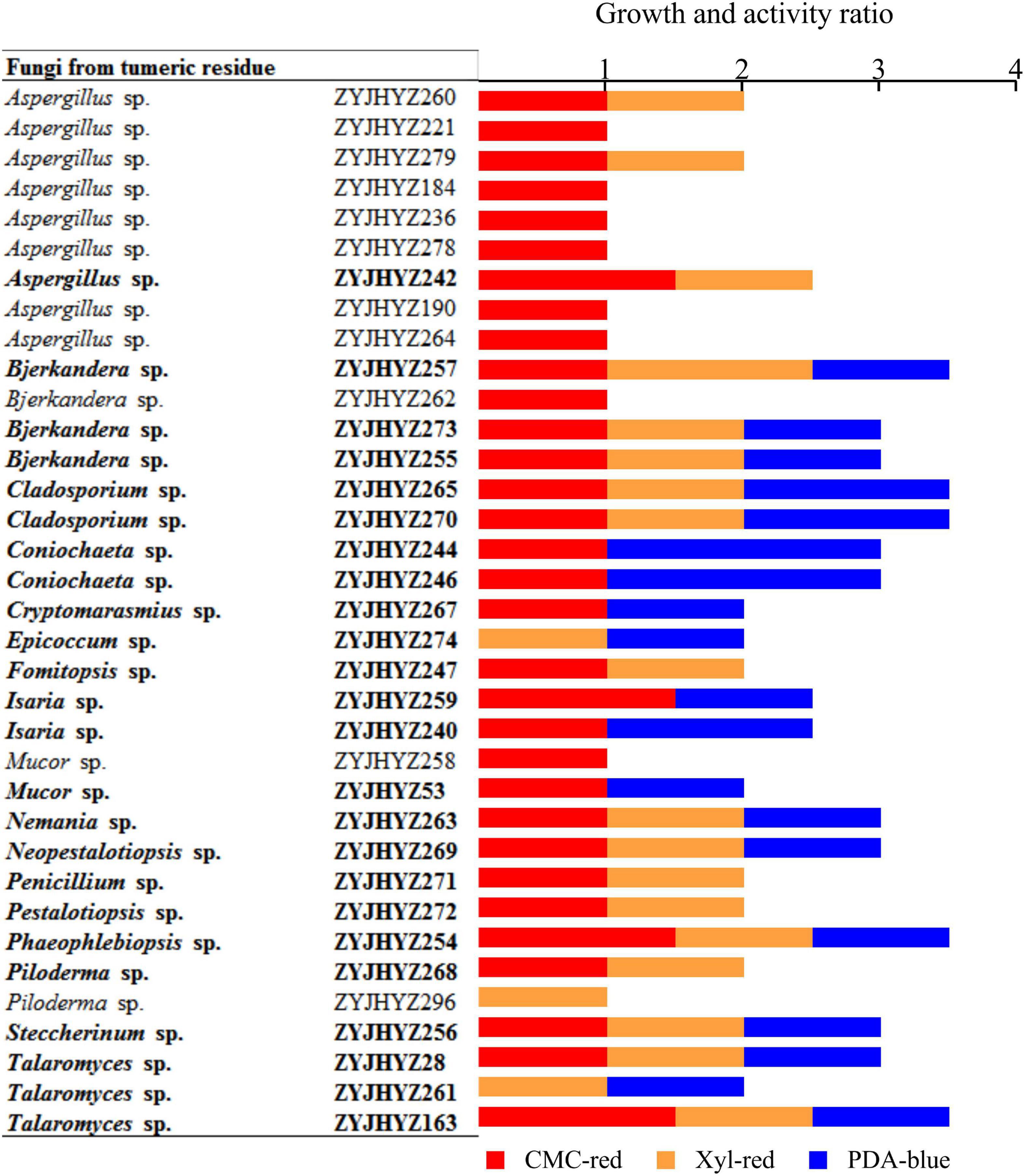
Figure 2. Growth and activity ratio of the selected fungi grown on solid media. The media contains 1% cellulose (CMC-red), 0.5% xylan (Xyl-red) or 0.01% aniline blue (PDA-blue). Fungi in bold showed > 2.0 growth ratio and were chosen for cultivation in liquid media. The growth and activity ratio was estimated by hydrolytic diameter/colony diameter after 5 days.
Comparison of Lignocellulose Degrading Enzyme Capacity of Single-Cultured Fungi
All fungi produce different levels of enzymes with different activities. Here, we screened for six different enzymes, viz., β-glucosidase, cellulase, β-xylosidase, xylanase, laccase, and peroxidase. β-glucosidase – In general, most fungi can produce higher β-glucosidase activity on days 7–9 (Figure 3A), among which Fomitopsis sp. (ZYJHYZ247) produces the highest β-glucosidase level on day 7 (151.68 U/L) followed by Nemania sp. (ZYJHYZ263) reached 134.22 U/L on day 9. Cellulase – However, the peak time of production was different among different fungal strains, among which Aspergillus niger (ZYJHYZ242) reached 66.55 U/L on day 7, and Fomitopsis sp. (ZYJHYZ247) reached 64.63 U/L on day 9 (Figure 3B). β-xylosidase – Figure 3C, showed that multiple fungi had the same enzyme production, among which the best enzyme-producing fungus was Coniochaeta sp. (ZYJHYZ246) and Phaeophlebiopsis sp. (ZYJHYZ254). Coniochaeta sp. (ZYJHYZ246) had the highest enzyme production at 80.97 U/L on the eighth day while Phaeophlebiopsis sp. (ZYJHYZ254) had the highest enzyme production at 79.92 U/L on the nineth day. Xylanase – In Figure 3D, Phaeophlebiopsis sp. (ZYJHYZ254) had the highest enzyme production, with the highest enzyme production time reaching 81.59 U/L on the 5th day, Bjerkandera sp. (ZYJHYZ257) and Isaria sp. (ZYJHYZ259) performed higher xylanase activity at day 9 and 7 with 76.14 and 72.98 U/L, respectively, in addition, Talaromyces sp. (ZYJHYZ163) also showed excellent and stable β-xylosidase and xylanase activity. Laccase – From Figure 3E, strain Piloderma sp. (ZYJHYZ268) reached the highest enzyme production (99.39 U/L) on the fifth day, followed by Coniochaeta sp. (ZYJHYZ246) and Cladosporium sp. (ZYJHYZ265) with 99.31 and 99.24 U/L, respectively. Peroxidase – From Figure 3F, including Phaeophlebiopsis sp. (ZYJHYZ254), Piloderma sp. (ZYJHYZ268), Coniochaeta sp. (ZYJHYZ246), Coniochaeta sp. (ZYJHYZ244), Aspergillus sp. (ZYJHYZ242) showed significantly higher enzyme activity than other fungi, with Phaeophlebiopsis sp. (ZYJHYZ254) having the highest enzyme activity, reaching 602.96 U/L on day 7, followed by Coniochaeta sp. (ZYJHYZ246) and Aspergillus sp. (ZYJHYZ242) with 580.42 and 561.06 U/L, respectively.
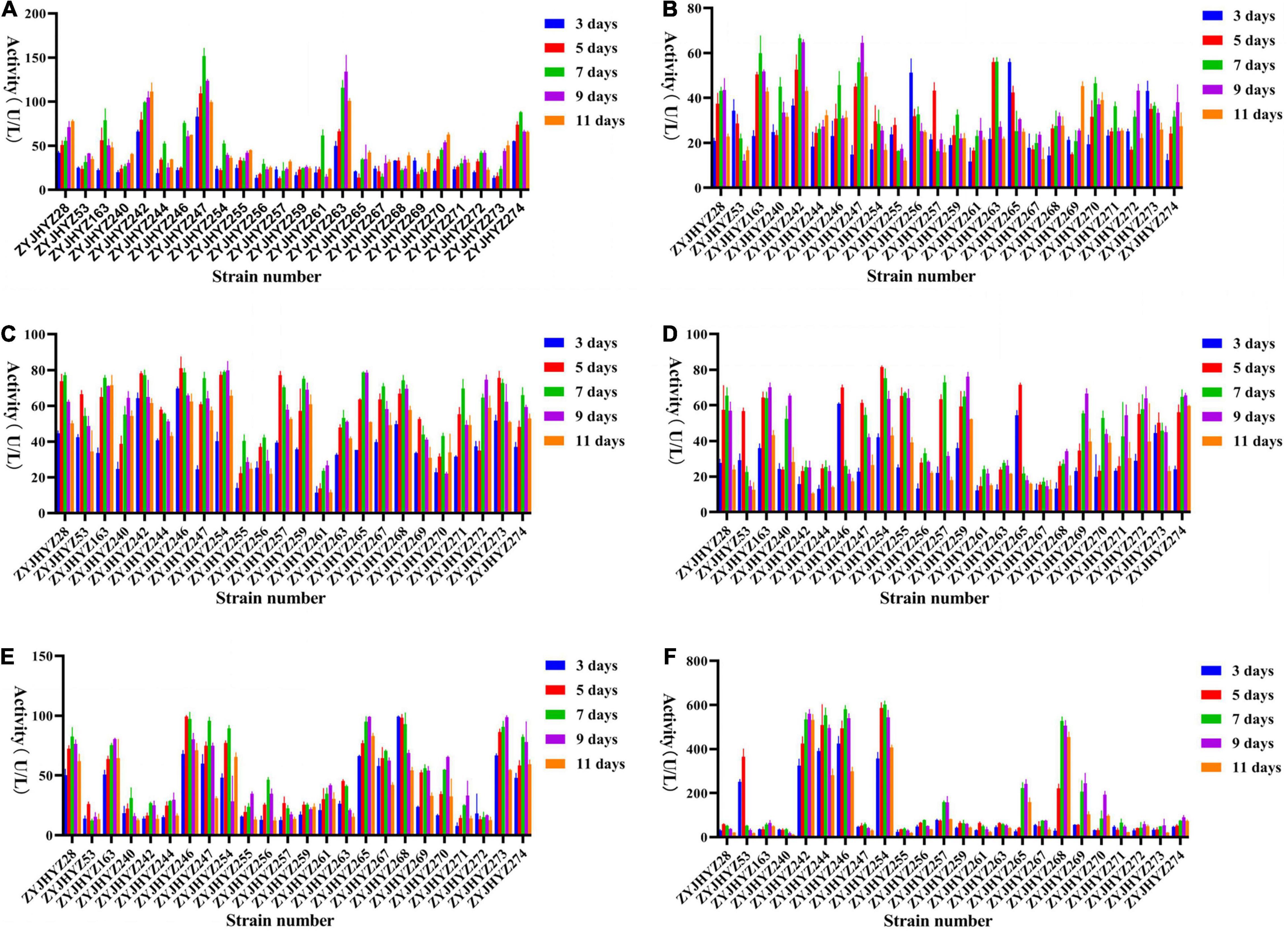
Figure 3. Extracellular enzyme activities of selected fungi for pre-screening. (A) β-glucosidase, (B) Cellulase, (C) β-xylosidase, (D) Xylanase, (E) Laccase, and (F) Peroxidase activity of culture supernatant of fungi grew in 6% anti-biodegradable residue (in water) as the sole carbon source. The culture supernatants were collected on 3, 5, 7, 9, and 11 days after the inoculation. Three biological replicates were performed for each of the above data.
In conclusion, strains belonging to Coniochaeta, Fomitopsis, Phanerochaete, and Piloderma are capable of producing a higher amount of lignocellulose degrading enzymes compared to other strains.
Construction of Fungal Strains and Their Co-cultivation Potential to Improve Enzymatic Degradation
Through the screening for six fungal extracellular enzymes, the first three fungal strains with the highest activity of each enzyme were selected for further co-cultivation; Aspergillus sp. (ZYJHYZ242), Fomitopsis sp. (ZYJHYZ247), and Nemania sp. (ZYJHYZ263) for cellulose degradation. Talaromyces sp. (ZYJHYZ163), Phaeophlebiopsis sp. (ZYJHYZ254), and Bjerkandera sp. (ZYJHYZ257) for hemicellulose degradation. High lignin-degrading strains numbered as Coniochaeta sp. (ZYJHYZ246), Phaeophlebiopsis sp. (ZYJHYZ254), and Piloderma sp. (ZYJHYZ268), the BLAST results from NCBI as Supplementary Table 3. A total of 27 fungal co-cultivation was constructed by permutation and combination of the above fungi, in order to screen out fungi that can comprehensively degrade lignocellulose. Twelve co-cultivation were growing together through the symbiosis test (Figure 4) and their enzyme activities showed in Figure 5. The other strain combinations were excluded from liquid cultivation.
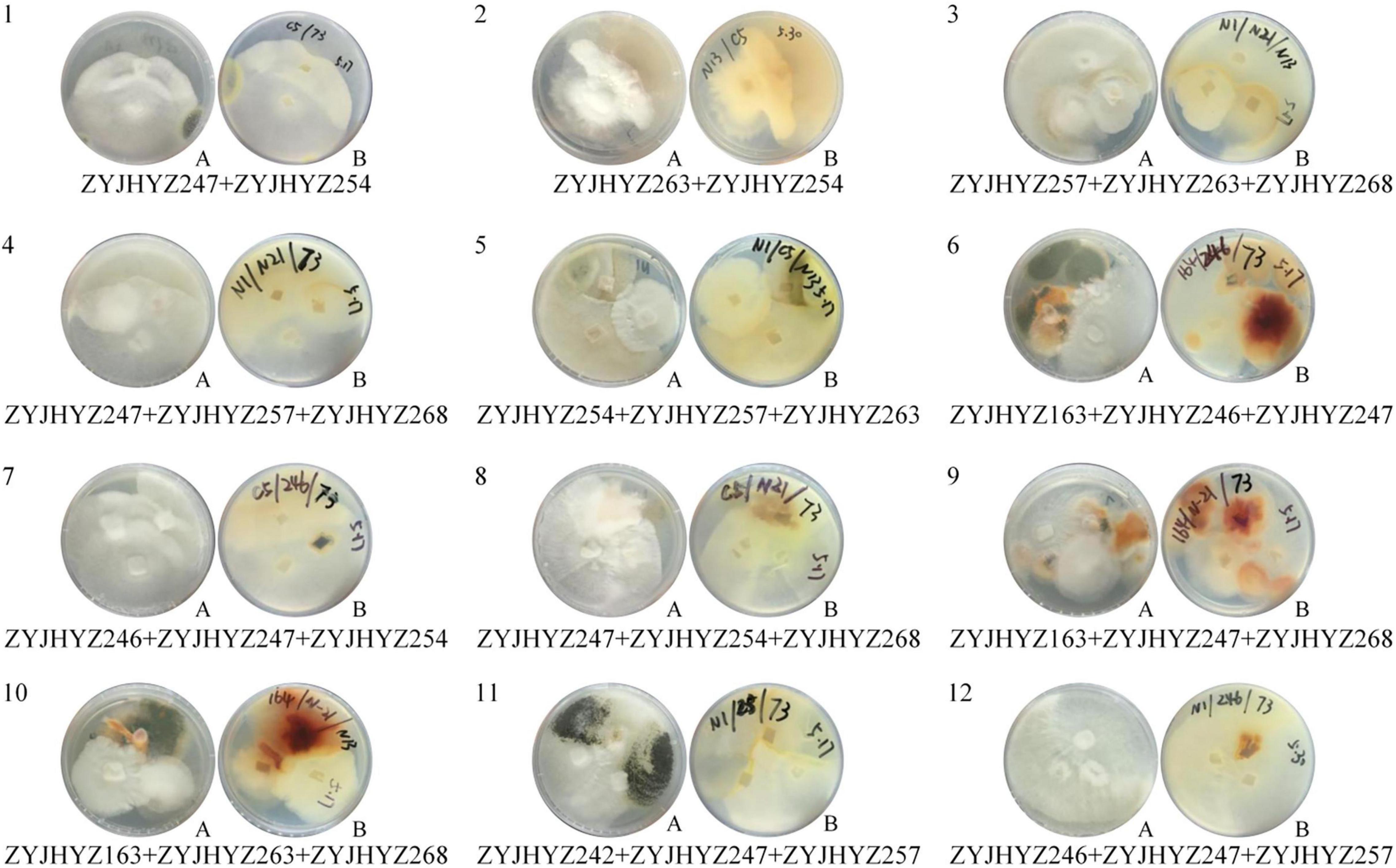
Figure 4. Growth of fungi in synergistic combinations (co-cultivation). 1, 2, 3, 4… is co-cultivation code; (A). from above, (B). from below.
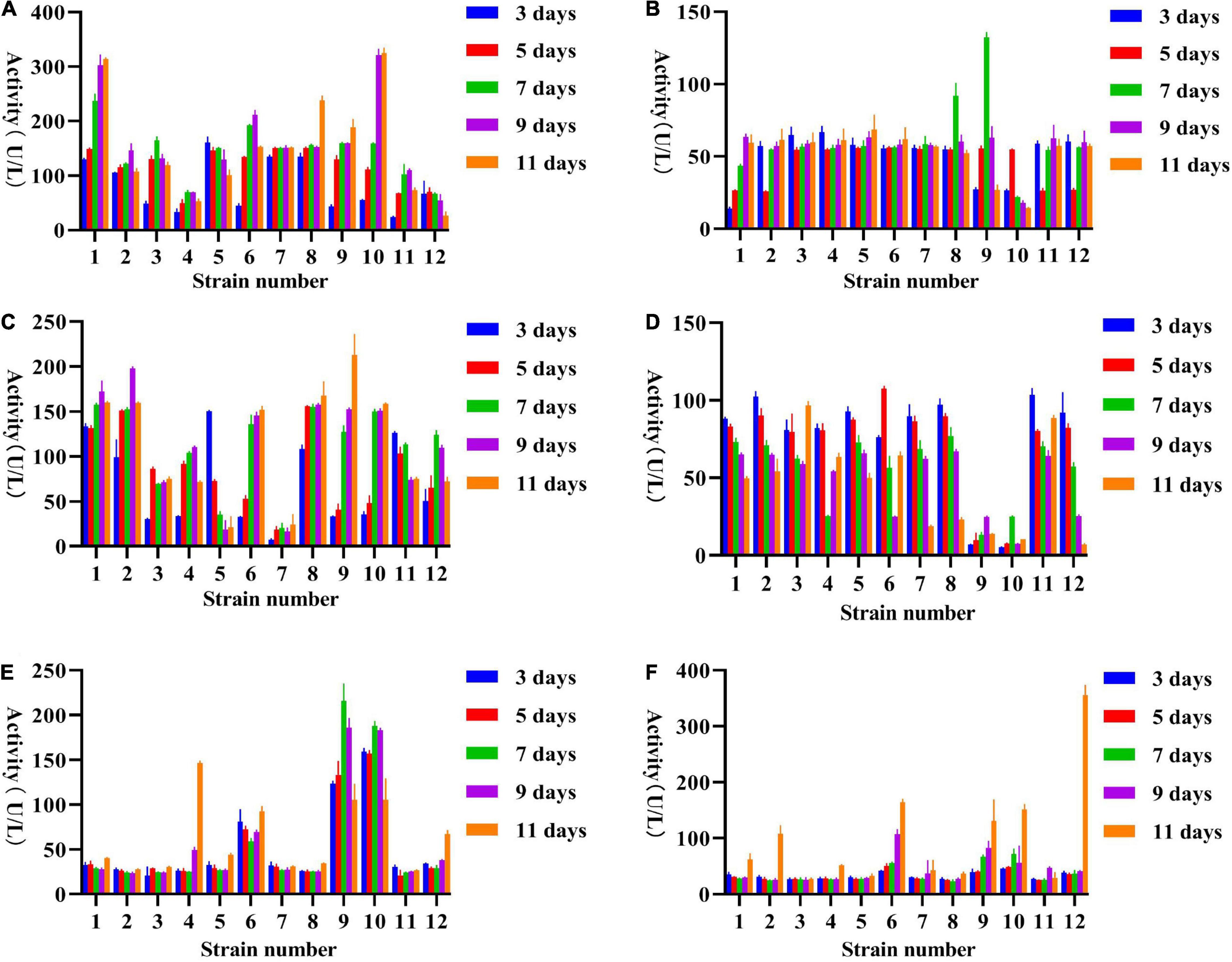
Figure 5. Extracellular enzyme activities of sixteen fungal co-cultivation supernatant at days 3, 5, 7, 9, and 11 after inoculation in water with 6% anti-biodegradable residue; (A) β-glucosidase, (B) Cellulase, (C) β-xylosidase, (D) Xylanase, (E) Laccase, and (F). Peroxidase. Three biological replicates were performed for each of the above data.
Comparison of Lignocellulose Degrading Enzyme Capacity of Co-cultivation Fungi
According to Figure 5, the activities of 5 enzymes (except for lignin peroxidase) have been greatly improved over those strains in single culture conditions. β-glucosidase – contained more than 100 U/L activities in most fungal combinations while only three single-cultured fungi were capable of production at this level. The highest β-glucosidase was produced on day 11 of incubation by fungal co-cultivation no. 1 (313.63 U/L), followed by no. 10 (325.03 U/L; Figure 5A). Cellulase – showed that multiple fungi had the same enzyme production, the cellulase activity performed highest on the seventh after inoculation by fungal combination nos. 8 and 9 with 91.86 and 132.43 U/L, respectively (Figure 5B), the highest enzyme activity of no. 9 reached 132.43 U/L, was about twice that of a single fungus. β-xylosidase – the peak of enzyme production in combinations nos. 5 and 11 was on the third day, and then decreased with time, while the other fungal combinations had an opposite trend, mostly reaching the peak around the ninth day, and β-xylosidase activity performed highest at day 11 and 9 after inoculation by fungal combination nos. 9 and 2 with 213.07 and 197.86 U/L, respectively (Figure 5C). Xylanase – most fungal combinations showed the highest xylanase activity from day 3 to day 5, among which the highest was no. 6, which reached 107.47 U/L on day 5, followed by no. 11 (103.55 U/L at day 3; Figure 5D). Laccase – from Figure 5E, the enzyme activity of no. 4, 9, and 10 were significantly higher than that of other combinations. The enzyme activity of no. 9 reached 215.46 U/L on the seventh day of inoculation, more than twice that of a single fungus, followed by no. 10 (187.69 U/L). Peroxidase – it can be seen from Figure 5F that the peak of enzyme production of almost all fungal combinations was on day 11, and the enzyme activity of fungal combination no. 12 on day 11 was at least twice that of other fungal combinations, reaching 355.65 U/L. In addition, the enzyme activity of fungal combinations no. 6, 9, and 10 are also relatively advantageous, but not as high as that of single fungi (Figure 5F).
Thus, we suggest using co-cultivation viz., no. 1 Fomitopsis sp. (ZYJHYZ247) + Phaeophlebiopsis sp. (ZYJHYZ254); no. 6 Talaromyces sp. (ZYJHYZ163) + Coniochaeta sp. (ZYJHYZ246) + Fomitopsis sp. (ZYJHYZ247); no. 9 Talaromyces sp. (ZYJHYZ163) + Fomitopsis sp. (ZYJHYZ247) + Piloderma sp. (ZYJHYZ268); no. 10 Talaromyces sp. (ZYJHYZ163) + Nemania sp. (ZYJHYZ263) + Piloderma sp. (ZYJHYZ268) to degrade the anti-biodegradable residue rather introducing them as a monoculture.
Hydrolytic Weight Loss Ability Test of Co-cultivation
In the co-cultivation, fermentation broth with anti-biodegradable residue as the sole carbon source was added at a constant temperature of 30× g at 28°C for 15 days (Figure 6). Compared with the negative control, about half of the residue was left in nos. 1 and 6, and about one-third of residue was left in nos. 9 and 10. The overall weight loss rate of TCMR is shown in Table 2. According to Table 2, the weight loss rate of no. 10 hydrolysis for 15 days is the highest, reaching 60.34%. The overall weight loss data is also consistent (Figure 6). Results of lignocellulosic content are shown in Table 3. It can be seen that the weight loss rate of cellulose in no. 1 after 15 days of hydrolysis is the highest, reaching 57.46%. Nos. 9 and 10 had the strongest hydrolysis ability for hemicellulose, and the weight loss rate of no. 10 reached 38.83%. No. 9 had the strongest hydrolysis capacity of lignin, reaching 64.47%.

Figure 6. Residue after 15 days of fermentation in liquid medium. (A) and (D) Negative control, (B) no. 1, (C) no. 6, (E) no. 9, and (F) no. 10.
Discussion
Herbal medicine plays a vital role in Asian countries and has a long history. Kloos mentioned that TCM has an average of 15% of annual growth between 2011 and 2016 (Kloos, 2017). Besides, Dang et al. (2016) reported that the value of different products of TCM is over 120 billion USD by the end of 2014 which represents 31% of the total pharmaceutical industry market in China. Hence, TCM is still in considerable demand. This also directly leads to the discharge of a large number of TCMR, long-term accumulation is currently the most adopted method, for environmental protection and economic development are great challenges. In recent years, they are often used for fermentation and composting research, due to the special properties of the TCMR (such as high acidity and strong bacteriostasis). In this study, the characteristics of the residue itself are taken as the starting point to screen out the fungi that can resist such an unusual environment, so as to solve the environmental pressure and lay a foundation for the development of bioenergy.
Three hundred and two (302) strains that belong to 30 genera have been isolated. A total of 35 strains with lignocellulosic degradation ability were obtained by single polymer solid medium screening. According to the diameter ratio, 24 strains were further screened out for 6 plant-degrading enzyme activities: β-glucosidase, cellulase, β-xylosidase, xylanase, laccase, and peroxidase. Among these six enzymes, we found that most fungi in 7–9 days can produce higher β-glucosidase and Cellulase. Aspergillus sp. (ZYJHYZ242) and Fomitopsis sp. (ZYJHYZ247) had the highest enzyme activity and the best cellulose degradation potential. Aspergillus is an important industrial fermentation strain that can produce cellulose (Van Munster et al., 2020; Zhao et al., 2020). Fomitopsis has also been repeatedly reported as a model brown rot fungus that causes destructive wood decay based on cellulase systems, producing cellulolytic enzymes and lignin decomposing enzymes (Hong et al., 2017). In the detection process of enzymes related to lignin degradation, it was found that the peak of enzyme production of most strains was 5–7 days, among which Phaeophlebiopsis sp. (ZYJHYZ254) had the highest peroxidase production, which reached 602.96 U/L on the seventh day, which was two to three times of the fungi with high total lignin peroxidase production reported in the current literature (Saito et al., 2018; Su et al., 2018). In addition, Phaeophlebiopsis sp. (ZYJHYZ254), Piloderma sp. (ZYJHYZ268), and Coniochaeta sp. (ZYJHYZ246) were also the three strains that produced the highest lignin-degrading enzymes. Coniochaeta was also reported to produce β-glucosidase, cellulase, hemicellulase and laccase (Mondo et al., 2019). Piloderma is a common ectomycorrhizal fungus (EMF) in forest soil. It is well known that EMF can produce a large number of peroxidase to degrade the complex structure containing lignin (Heinonsalo et al., 2015). Recently, Rineau et al. also found more than one laccase gene was encoded in the genome of Piloderma (Rineau et al., 2012). In the hemicellulase activity results, Phaeophlebiopsis sp. (ZYJHYZ254) had the highest xylanase production, and the highest enzyme production time was on the fifth day, and β-xylosidase production was similar among the fungi. The best enzyme-producing fungi was Coniochaeta sp. (ZYJHYZ246) and Phaeophlebiopsis sp. (ZYJHYZ254).
Nevertheless, studies have shown that a single strain cannot degrade lignocellulose completely (Karuppiah et al., 2021). Hence, it is recommended to construct co-cultivation to achieve complete degradation of lignocellulose. The main aim of the co-cultivation is to improve the overall lignocellulolytic activity, as lignocellulose is the main component in agricultural waste (Čater et al., 2015; Ming et al., 2019). However, it is not feasible to predict which strains will be able to grow together. Hence, we first grew different combinations on solid complex media (Hu et al., 2011). Strain combinations showing a separation zone or barrage line indicated incompatibility among the strains. These strain combinations were excluded from liquid cultivation. The enzyme activity of co-cultivation (12 groups) that can grow together was detected. It was found that except for lignin peroxidase, the enzyme activity of the other 5 enzymes had a significant increase, with the same amount of mycelium inoculation as that of a single strain, the effect of enzyme production of the co-cultivation was two to three times higher than that of a single strain, indicating that the co-cultivation was successfully constructed. However, it should be noted that the enzyme activity of the co-cultivation of Figure 5F was generally lower than that of the single fungi, which may be due to the interaction of various chemical substances produced by fungi, resulting in low overall enzyme activity.
To sum up, we believe that co-cultivation viz., no. 1 Fomitopsis sp. (ZYJHYZ247) + Phaeophlebiopsis sp. (ZYJHYZ254); no. 6 Talaromyces sp. (ZYJHYZ163) + Coniochaeta sp. (ZYJHYZ246) + Fomitopsis sp. (ZYJHYZ247); no. 9 Talaromyces sp. (ZYJHYZ163) + Fomitopsis sp. (ZYJHYZ247) + Piloderma sp. (ZYJHYZ268); no. 10 Talaromyces sp. (ZYJHYZ163) + Nemania sp. (ZYJHYZ263) + Piloderma sp. (ZYJHYZ268) has a better overall degradation potential of lignocellulose. Based on these results, we conclude that wood white rots fungi and other inhabiting fungi are capable of producing higher concentrations of extracellular enzymes, such as Coniochaeta sp. (ZYJHYZ246), Fomitopsis sp. (ZYJHYZ247), and Phaeophlebiopsis sp. (ZYJHYZ254).
Nevertheless, as this is not a taxonomy paper, we did only preliminary identification for strains. It is essential to carry out multi-gene analyses to identify cryptic species in Aspergillus and other prominent species which are inhabiting species complexes. Current results showed that the all isolated strains are acid-resistant, and with the extension of residue stacking time, their pH value will be lower, the diversity of fungi will gradually decline, and the nutritional type of strains will tend to be single. Therefore, it can be predicted that the longer the residue stacking time, the more likely it is to isolate the lignocellulose-degrading enzymes strain. It is generally believed that the unique high acidity and bacteriostatic environment of TCMR tend to give it special characteristics of its own microorganisms, so the special functions of isolated strains can be further discussed.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
MC, RdV, YL, and XG performed the material preparation, data collection, and analysis. MC, NW, and IP wrote the first draft of the manuscript. FW, YL, GL, MW, QL, and YK advanced the manuscript revision and examination. All authors contributed to the conception and design of the study, commented on previous versions of the manuscript, and read and approved the final manuscript.
Funding
This work was supported by grants from Guizhou Scientific Plan Project [(2019) 2873], the 111 Project (D20009), the National Natural Science Foundation of China (32060034), the Guizhou Scientifc Plan Project ([2020]4Y220), the International Science and Technology Cooperation Base of Guizhou Province [(2020) 4101], and Guizhou Provincial Academician Workstation of Microbiology and Health [(2020) 4004].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
NW thanks the High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program and “High-End Foreign Experts” Program) and the Institute of Research and Development in Sri Lanka. IP, is grateful to Chiang Mai University for its partial support of this research work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.877884/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Barros, V. G., Duda, R. M., and Oliveira, R. A. (2016). Biomethane production from vinasse in upflow anaerobic sludge blanket reactors inoculated with granular sludge. Braz. J. Microbiol. 47, 628–639. doi: 10.1016/j.bjm.2016.04.021
Bellemain, E., Carlsen, T., Brochmann, C., Coissac, E., Taberlet, P., and Kauserud, H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10:189. doi: 10.1186/1471-2180-10-189
Čater, M., Fanedl, L., Malovrh, Š, and Marinšek Logar, R. (2015). Biogas production from brewery spent grain enhanced by bioaugmentation with hydrolytic anaerobic bacteria. Bioresour. Technol. 186, 261–269. doi: 10.1016/j.biortech.2015.03.029
Cheng, C., Wu, H. C., Liu, X. J., Yang, X. Y., Zhou, X., and Fan, H. B. (2012). Screening of cellulose degrading bacteria from distiller’s grains. J. Anhui. Agri. Sci 40, 7892–7893.
Childs, R. E., and Bardsley, W. G. (1975). The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem. J. 145, 93–103. doi: 10.1042/bj1450093
Dang, H., Wang, Q., Wang, H., Yan, M., and Liu, X. (2016). The integration of Chinese material medica into the Chinese health care delivery system, an update. Phytother. Res 30, 292–297. doi: 10.1002/ptr.5530
Falade, A. O., Nwodo, U. U., Iweriebor, B. C., Green, E., Mabinya, L. V., and Okoh, A. I. (2017). Lignin peroxidase functionalities and prospective applications. Microbiol. 6:e00394. doi: 10.1002/mbo3.394
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fen, L., Xuwei, Z., Nanyi, L., Puyu, Z., Shuang, Z., Xue, Z., et al. (2014). Screening of lignocellulose degrading superior mushroom strains and determination of their CMCase and laccase activity. Sci. World. J. 2014:763108. doi: 10.1155/2014/763108
Gupta, P., Samant, K., and Sahu, A. (2012). Isolation of cellulose degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012:578925. doi: 10.1155/2012/578925
Heinonsalo, J., Sun, H., Santalahti, M., Bäcklund, K., Hari, P., and Pumpanen, J. (2015). Evidences on the ability of mycorrhizal genus Piloderma to use organic nitrogen and deliver it to scots pine. PloS One 10:e0131561. doi: 10.1371/journal.pone.0131561
Hong, C. Y., Lee, S. Y., Ryu, S. H., and Kim, M. (2017). Whole genome de novo sequencing of wood rot fungus Fomitopsis palustris (ATCC62978) with both a cellulolytic and ligninolytic enzyme system. J. biotechnol. 251, 156–159. doi: 10.1016/j.jbiotec.2017.04.009
Hu, H. L., van den Brink, J., Gruben, B. S., Wösten, H. A. B., Gu, J. D., and de Vries, R. P. (2011). Improved enzyme production by co-cultivation of Aspergillus niger and Aspergillus oryzae and with other fungi. Int. Biodeterior. Biodegrad. 65, 248–252. doi: 10.1016/j.ibiod.2010.11.008
Hutchins, D. A., Jansson, J. K., Remais, J. V., Rich, V. I., Singh, B. K., and Trivedi, P. (2019). Climate change microbiology problems and perspectives. Nat. Rev. Microbiol. 17, 391–396. doi: 10.1038/s41579-019-0178-5
Karuppiah, V., Zhixiang, L., Liu, H., Murugappan, V., Kumaran, S., Perianaika Anahas, et al. (2021). Co-cultivation of T. asperellum GDFS1009 and B. amyloliquefaciens 1841: strategy to regulate the production of ligno-cellulolytic enzymes for the lignocellulose biomass degradation. J. Environ. Manage. 27:113833. doi: 10.1016/j.jenvman.2021.113833
Kloos, S. (2017). The pharmaceutical assemblage. Rethinking sowa rigpa and the herbal pharmaceutical industry in Asia. Curr. Anthropol. 58, 693–717. doi: 10.1086/693896
Kwak, S., Jo, J. H., Yun, E. J., Jin, Y. S., and Seo, J. H. (2019). Production of biofuels and chemicals from xylose using native and engineered yeast strains. Biotechnol. Adv. 37, 271–283. doi: 10.1016/j.biotechadv.2018.12.003
Lange, L. (2017). Fungal enzymes and yeasts for conversion of plant biomass to bioenergy and high value products. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.FUNK-0007-2016
Li, Y. H., Zhang, X. Y., Zhao, X. Q., and Bai, F. W. (2016). Optimization of fermentation medium for β-glucosidase production by penicillus and preparation of soluble inducers for cellulase. Chin. J. Appl. Environ. Biol. 22, 0377–0381. doi: 10.3724/SP.J.1145.2015.09015
Liu, D. T., Yang, Q., Wang, Y. J., and Zhang, X. D. (2010). Identification and enzymatic analysis of an alcohol stable hemicellulose degradation bacterium. Acta. Energiae. Solaris. Sin 31, 107–112.
Lu, Q., and Li, C. (2021). Comprehensive utilization of Chinese medicine residues for industry and environment protection: turning waste into treasure. J. Clea. Prod. 279:123856. doi: 10.1016/j.jclepro.2020.123856
Makhuvele, R., Ncube, I., Jansen, van Rensburg, E. L., and La Grange, D. C. (2017). Isolation of fungi from dung of wild herbivores for application in bioethanol production. Braz. J. Microbiol. 48, 648–655. doi: 10.1016/j.bjm.2016.11.013
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Ming, C. Y., Adiphol, D., Zou, C. G., Liao, W. Q., Zhao, L., Wang, M. Z., et al. (2019). The quest for fungal strains and their co-cultivation potential to improve enzymatic degradation of Chinese distiller’s grain and other agricultural wastes. Int. Biodeter. Biodegr. 144:104765. doi: 10.1016/j.ibiod.2019.104765
Ming, C. Y., Wang, Y. Y., Cao, Y., Qiu, T. T., Xia, M. L., Niu, X. K., et al. (2017). The study on hydrolysis ability of several fungi to cellulose in waste distiller’s grains. J. Guizhou. Med. Univers 42, 1274–1278. doi: 10.19367/j.cnki.1000-2707.2017.11.008
Mishra, V., Jana, A. K., Jana, M. M., and Gupta, A. (2017). Fungal pretreatment of sweet sorghum bagasse with supplements: improvement in lignin degradation, selectivity and enzymatic saccharification. 3 Biotech. 7:110. doi: 10.1007/s13205-017-0719-4
Mondo, S. J., Jiménez, D. J., Hector, R. E., Lipzen, A., Yan, M., LaButti, K., et al. (2019). Genome expansion by allopolyploidization in the fungal strain Coniochaeta 2T2.1 and its exceptional lignocellulolytic machinery.. Biotechnol. Biofuels 12:229. doi: 10.1186/s13068-019-1569-6
Namnuch, N., Thammasittirong, A., and Thammasittirong, S. N. (2020). Lignocellulose hydrolytic enzymes production by Aspergillus flavus KUB2 using submerged fermentation of sugarcane bagasse waste. Mycology 12, 119–127. doi: 10.1080/21501203.2020.1806938
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. Oxford: Oxford University Press.
Ni, J., and Tokuda, G. (2013). Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv 31, 838–850. doi: 10.1016/j.biotechadv.2013.04.005
Ojha, S. K., Mishra, S., Kumar, S., Mohanty, S. S., Sarkar, B., Singh, M., et al. (2015). Performance evaluation of vinasse treatment plant integrated with physico chemical methods.. J. Environ. Biol. 36, 1269–1275.
Patyshakuliyeva, A., Arentshorst, M., Allijn, I. E., Ram, A. F., de Vries, R. P., and Gelber, I. B. (2016). Improving cellulase production by Aspergillus niger using adaptive evolution. Biotechnol. Lett. 38, 969–974. doi: 10.1007/s10529-016-2060-0
Peciulyte, A., Pisano, M., de Vries, R. P., and Olsson, L. (2017). Hydrolytic potential of five fungal supernatants to enhance a commercial enzyme cocktail. Biotechnol. Lett. 39, 1403–1411. doi: 10.1007/s10529-017-2371-9
Rineau, F., Roth, D., Shah, F., Smits, M., Johansson, T., Canbäck, B., et al. (2012). The ectomycorrhizal fungus Paxillus involutus converts organic matter in plant litter using a trimmed brown rot mechanism involving Fenton chemistry. Environ. Microbiol. 14, 1477–1487. doi: 10.1111/j.1462-2920.2012.02736.x
Saito, Y., Tsuchida, H., Matsumoto, T., Makita, Y., Kawashima, M., Kikuchi, J., et al. (2018). Screening of fungi for decomposition of lignin derived products from Japanese cedar. J. Biosci. Bioeng. 126, 573–579. doi: 10.1016/j.jbiosc.2018.05.001
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
SEPA. (1997). National Status Report on Biodiversity of China, States Environment Protection Agency (SEPA). Beijing: China Environment Press.
Sherwood, J. (2020). The significance of biomass in a circular economy. Bioresour. Technol. 300:122755. doi: 10.1016/j.biortech.2020.122755
Silva, J., Salgado, J., Vici, A. C., Ward, R. J., Polizeli, M., Guimarães, L., et al. (2020). A novel Trichoderma reesei mutant RP698 with enhanced cellulase production. Braz. J. Microbiol. 51, 537–545. doi: 10.1007/s42770-019-00167-2
Silva, J. P., Ticona, A., Hamann, P., Quirino, B. F., and Noronha, E. F. (2021). Deconstruction of lignin: from enzymes to microorganisms. Molecules 26:2299. doi: 10.3390/molecules26082299
Srinivasan, C., Dsouza, T. M., Boominathan, K., and Reddy, C. A. (1995). Demonstration of laccase in the white rot Basidiomycete Phanerochaete chrysosporium BKM-F1767. Appl. Environ. Microb 61, 4274–4277. doi: 10.1128/aem.61.12.4274-4277.1995
Su, Y., Yu, X., Sun, Y., Wang, G., Chen, H., and Chen, G. (2018). Evaluation of screened lignin-degrading fungi for the biological pretreatment of corn stover. Sci. Rep. 8:5385. doi: 10.1038/s41598-018-23626-6
Taha, M., Foda, M., Shahsavari, E., Aburto-Medina, A., Adetutu, E., and Ball, A. (2016). Commercial feasibility of lignocellulose biodegradation: possibilities and challenges. Curr. Opin. Biotech. 38, 190–197. doi: 10.1016/j.copbio.2016.02.012
Tan, L., Sun, Z., Zhang, W., Tang, Y., Morimura, S., and Kida, K. (2014). Production of biofuel ethanol from distilled grain waste eluted from Chinese spirit making process. Bioproc. Biosyst. Eng. 37, 2031–2038. doi: 10.1007/s00449-014-1178-5
Thi Minh Le, T., Thi Hong, Hoang, A., Thi Bich, Le, T., et al. (2019). Isolation of endophytic fungi and screening of Huperzine A-producing fungus from Huperzia serrata in Vietnam. Sci. Rep. 9:16152. doi: 10.1038/s41598-019-52481-2
Van Munster, J. M., Daly, P., Blythe, M. J., Ibbett, R., Kokolski, M., Gaddipati, S., et al. (2020). Succession of physiological stages hallmarks the transcriptomic response of the fungus Aspergillus niger to lignocellulose. Biotechnol. Biofuels 13:69. doi: 10.1186/s13068-020-01702-2
Wang, H., Liu, X., Luo, H., and Yang, G. (2009). Screening of lignin degrading fungi and study of lignin degradation of vineyard pruning. Acta. Agricul. Boreali-Occidentalis. Sin. 18, 268–302.
Wang, J. F., Zhang, W., Jiang, L., and Jun, R. (2009). Study on laccase production by liquid fermentation of porus infundiformis. Mycosystema 28, 440–444. doi: 10.13346/j.mycosystema.2009.03.009
Wu, X., Gan, B., Huang, Z., and Tan, W. (2016). Characteristics and identification of cellulase decomposing strain DBJ. Southwest Chin 29, 81–84. doi: 10.16213/j.cnki.scjas.2016.01.016
Zeng, M., Li, C., and Zhou, L. (2013). Progress and prospective on the police system of renewable energy in China. Renew. Sust. Rev. 20, 36–44. doi: 10.1016/j.rser.2012.11.048
Zhang, L., and Sun, X. (2018). Effects of waste lime and Chinese medicinal herbal residue amendments on physical, chemical, and microbial properties during green waste composting. Environ. Sci. pollut. R. 25, 31381–31395. doi: 10.1007/s11356-018-3085-9
Zhang, Q. F., Ma, J. L., Kong, X. Q., Jia, X. N., Chen, J. X., and Zhao, X. (2015). Screening, identification and production conditions of hemicellulase high yielding strains. China. Biogas 33:5.
Zhao, J., Shi, D., Yang, S., Lin, H., and Chen, H. (2020). Identification of an intracellular β-glucosidase in Aspergillus niger with transglycosylation activity. Appl. Microbiol. Biotech. 104, 8367–8380. doi: 10.1007/s00253-020-10840-4
Zhu, M. Q., and Sun, R. C. (2018). Eucommia ulmoides oliver: a potential feedstock for bioactive products. J. Agr. Food. Chem. 66, 5433–5438. doi: 10.1021/acs.jafc.8b01312
Keywords: CAZymes, filamentous fungi, fungal diversity, hydrolysis, ITS, synergistic fungal combinations
Citation: Cheng M, Wijayawardene NN, Promputtha I, de Vries RP, Lan Y, Luo G, Wang M, Li Q, Guo X, Wang F, Liu Y and Kang Y (2022) Potential Fungi Isolated From Anti-biodegradable Chinese Medicine Residue to Degrade Lignocellulose. Front. Microbiol. 13:877884. doi: 10.3389/fmicb.2022.877884
Received: 17 February 2022; Accepted: 30 March 2022;
Published: 10 May 2022.
Edited by:
Dimitris G. Hatzinikolaou, National and Kapodistrian University of Athens, GreeceReviewed by:
Danushka Sandaruwan Tennakoon, Chiang Mai University, ThailandThushara Chathuranga Bamunuarachchige, Rajarata University of Sri Lanka, Sri Lanka
Chang-lin Zhao, Southwest Forestry University, China
Copyright © 2022 Cheng, Wijayawardene, Promputtha, de Vries, Lan, Luo, Wang, Li, Guo, Wang, Liu and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingqian Kang, a2FuZ3lpbmdxaWFuQGdtYy5lZHUuY24=
 Min Cheng
Min Cheng Nalin N. Wijayawardene
Nalin N. Wijayawardene Itthayakorn Promputtha
Itthayakorn Promputtha Ronald P. de Vries
Ronald P. de Vries Yongzhe Lan1
Yongzhe Lan1 Gang Luo
Gang Luo Xinyao Guo
Xinyao Guo