
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 May 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.876272
This article is part of the Research TopicAlgal PhotosynthesisView all 13 articles
Cyanobacteria evolved an inorganic carbon-concentrating mechanism (CCM) to perform effective oxygenic photosynthesis and prevent photorespiratory carbon losses. This process facilitates the acclimation of cyanobacteria to various habitats, particularly in CO2-limited environments. To date, there is limited information on the CCM of thermophilic cyanobacteria whose habitats limit the solubility of inorganic carbon. Here, genome-based approaches were used to identify the molecular components of CCM in 17 well-described thermophilic cyanobacteria. These cyanobacteria were from the genus Leptodesmis, Leptolyngbya, Leptothermofonsia, Thermoleptolyngbya, Thermostichus, and Thermosynechococcus. All the strains belong to β-cyanobacteria based on their β-carboxysome shell proteins with 1B form of Rubisco. The diversity in the Ci uptake systems and carboxysome composition of these thermophiles were analyzed based on their genomic information. For Ci uptake systems, two CO2 uptake systems (NDH-13 and NDH-14) and BicA for HCO3– transport were present in all the thermophilic cyanobacteria, while most strains did not have the Na+/HCO3– Sbt symporter and HCO3– transporter BCT1 were absent in four strains. As for carboxysome, the β-carboxysomal shell protein, ccmK2, was absent only in Thermoleptolyngbya strains, whereas ccmK3/K4 were absent in all Thermostichus and Thermosynechococcus strains. Besides, all Thermostichus and Thermosynechococcus strains lacked carboxysomal β-CA, ccaA, the carbonic anhydrase activity of which may be replaced by ccmM proteins as indicated by comparative domain analysis. The genomic distribution of CCM-related genes was different among the thermophiles, suggesting probably distinct expression regulation. Overall, the comparative genomic analysis revealed distinct molecular components and organization of CCM in thermophilic cyanobacteria. These findings provided insights into the CCM components of thermophilic cyanobacteria and fundamental knowledge for further research regarding photosynthetic improvement and biomass yield of thermophilic cyanobacteria with biotechnological potentials.
Thermophilic cyanobacteria are photoautotrophic prokaryotes with a cosmopolitan distribution in diverse thermal environments (Tang et al., 2018a,b; Alcorta et al., 2020). With the increase of studies on these unique microorganisms and their niches, their importance as primary photosynthetic producers of the geothermal ecosystems becomes apparent. The thermophilic cyanobacteria account for a large part of the thermal ecosystem biomass and productivity. From an applicative perspective, thermophilic cyanobacteria can be explored for high value-added products relevant to agricultural, pharmaceutical, nutraceutical, and industrial applications (Patel et al., 2019). Particularly, the potential contribution of thermophilic cyanobacteria to mitigating carbon dioxide becomes attractive to researchers in the context of global warming and greenhouse gas emissions. Thermophilic cyanobacteria may facilitate the collection of CO2 right from locations where it was generated, e.g., fossil fuels power plants, thanks to their higher thermostability of photosynthetic apparatus and resistance to photosynthetic inhibitors such as NOx and SOx found in high levels in the flue gases (Liang et al., 2019; Chen and Wang, 2020).
The CO2-concentrating mechanism (CCM) is a vital biological process for CO2 utilization by cyanobacteria under conditions of CO2 limitation. Cyanobacterial CCM enables adequate photosynthetic CO2 fixation by elevating the CO2 level near the active site of Rubisco, thus providing adaptation to various CO2-limited niches (Clement et al., 2016; Raven et al., 2017). For most thermophilic cyanobacteria, the aquatic environments where they live suffer from low availability of inorganic carbon (Ci), primarily due to external factors, e.g., temperature, pH and gas exchange (Durall and Lindblad, 2015).
The dissolution of gaseous CO2 in water starts from CO2 (g) transferring to CO2 (aq), i.e., CO2 mass transfer from gas to liquid phase (Figure 1A). Further, the small fraction of dissolved CO2 reacts in the liquid phase via two parallel paths depending on pH and subsequent availability of OH–. One leads to H+ and HCO3– formation with the unstable intermediate product – carbonic acid H2CO3*, while the second with hydroxide ions also forms bicarbonate ions which further can dissociate into H+ and CO3–. When the pH of the aqueous solution is lower than 8, CO2 hydration follows the first path, and due to the very low value of the dissociation constant of the bicarbonate, the formation of carbonate ions can be neglected (Kierzkowska-Pawlak et al., 2022). The direct data on CO2 dissociation and different species of inorganic carbon present in hot springs have not been much discussed in the literature yet. However, the known equations of dissociation constants of CO2 in water and saline solutions are sufficient approximations to understand what inorganic carbon species are available across different pH ranges of those hot springs. The most commonly used equations for dissociation constants of CO2 in water (both fresh and saline) are applicable up to 40–50°C (Mook, 2000; Woosley, 2021). Based on those works, Bjerrum plot (Figure 1) of the carbonate system can be further extrapolated to 72°C and adjusted for salinity (Mook, 2000; Solomon, 2001; Tremaine et al., 2004). The solubility of CO2 in water is a function of temperature and do lower extend salinity (Figure 1), the higher the temperature, the lower the carbon dioxide solubility, elevating the temperature from 30 to 60°C results in 40% lower availability of CO2. Further decrease of that solubility occurs due to increased salinity or presence of other dissolved elements ubiquitous in hot springs. Meanwhile the distribution of carbon species across different pH is also affected by the temperature and to lesser extend salinity (Figure 1). Whilst bicarbonate (HCO3–) remains a dominant dissolved inorganic specie between pH 6 and 10; its ratio with other carbon species is temperature sensitive. The equilibrium between carbonic acid, bicarbonate and carbonate species is acid-shifted with an increase of temperature. Therefore, to survive in their unique aquatic habitat, the thermophilic cyanobacteria utilize CCM to ensure that the Rubisco with low affinity for CO2 is surrounded by high CO2 levels and regularly function regardless of thermal stress.
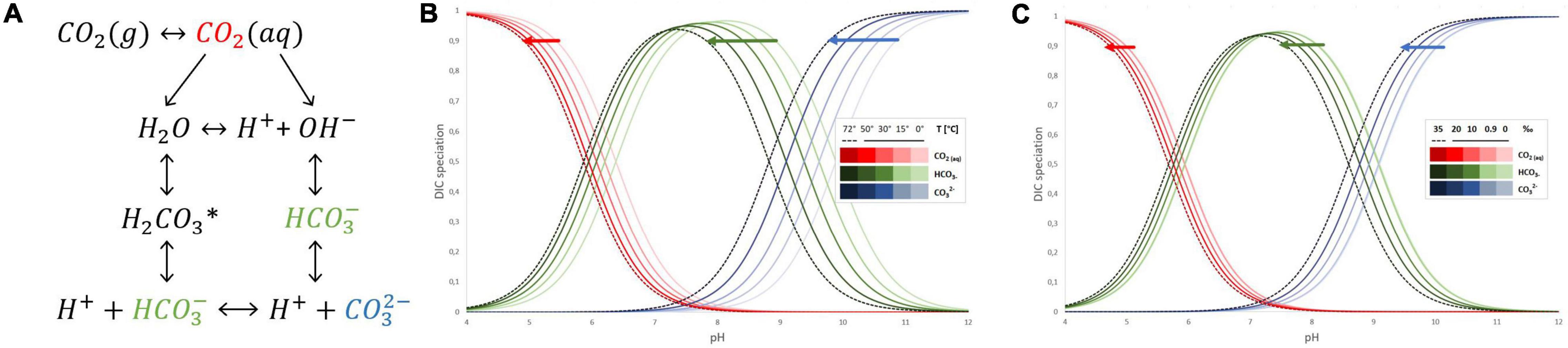
Figure 1. Interconversions of inorganic carbon species in aqueous solution (A). The effect of temperature on inorganic carbon species distribution across different pH in aqueous solution (B). The effect of salinity in ‰ on inorganic carbon species distribution across different pH in aqueous solution (C).
Prior studies on the effect of temperature on Rubisco, including that of thermophilic origin revealed that the maximal of rates of carboxylation reaction are typically higher than optimal growth temperatures of an organism (Galmés et al., 2015, 2016). This discrepancy in is partially due to the decreased affinity of Rubisco and lower specificity of carboxylation versus oxidation reaction at higher temperatures (Sc/o) (Gubernator et al., 2008; Galmés et al., 2015, 2016). All in all thermophilic cyanobacteria must operate carbon concentration mechanisms to mitigate both challenges associated with lower carbon availability resulting from its poor solubility, and lower specificity of Rubisco toward CO2 at elevated temperatures.
In general, the cyanobacterial CCM comprises two primary components: Ci uptake systems and carboxysomes. Cyanobacteria have been reported to have up to five different systems to actively acquire and transport Ci into the cells, namely two for uptake of CO2 and three for transport of HCO3– (Pronina et al., 2017). The two CO2 uptake systems that convert cytosolic CO2 into HCO3– rely on plastoquinone oxidoreductase NADPH dehydrogenase respiratory complexes (Price, 2011). The complexes include NDH-13, a low-CO2 inducible high-affinity CO2 uptake system encoded by ndhD3, ndhF3 and cupA (chpY) genes, and NDH-14, a constitutive low-affinity CO2 uptake system encoded by ndhD4, ndhF4, and cupB (chpX) genes (Kaplan, 2017). The transport of HCO3– takes place at the plasma membrane and is performed by three transporters, including bicA (a sulP-type sodium-dependent HCO3– transporter), sbtA (a sodium-dependent HCO3– symporter), and BCT1 (an ATP-binding cassette ABC-type HCO3– transporter) (Raven et al., 2017). Inside the cyanobacterial cell, the HCO3– is further transported into the carboxysomes that comprise protein shells and two encapsulated enzymes, Rubisco and carbonic anhydrase (CA) (Kerfeld and Melnicki, 2016). CA catalyzes the conversion of HCO3– into CO2, which is a substrate for Rubisco. Rubisco consisting in cyanobacteria of eight small (rbcS) and eight large (rbcL) subunits, catalyzes CO2 fixation reaction to generate 3-phosphoglycerate for subsequent rearrangement through the Calvin-Benson-Bassham cycle (Hill et al., 2020). There are two types of shell proteins encoded by cso operon and ccmKLMNO operon, termed α-carboxysomes and β-carboxysomes, respectively. Based on this criterion, the cyanobacterial species carrying form 1A of Rubisco within α-carboxysomes are classified as α-cyanobacteria, while the species containing form 1B of Rubisco within β-carboxysomes as β-cyanobacteria (Melnicki et al., 2021). The ccmP was another β-carboxysome component, forming a bilayered shell protein (Cai et al., 2013). In addition, various carboxysomal CAs have been reported, including β-CA (ccaA) and γ-CA (ccmM) in β-cyanobacteria and β-CA (csoSCA) in α-cyanobacteria (MacCready et al., 2020). Two types of non-carboxysomal CAs were also reported in β-cyanobacteria, namely ecaB (β-CA) and ecaA (α-CA) (Sun et al., 2019).
Although CCM of freshwater, marine and alkaliphilic cyanobacteria have been reported (Burnap et al., 2015; Kupriyanova and Samylina, 2015; Klanchui et al., 2017), no comprehensive investigation on CCM of thermophilic cyanobacteria have been performed. Moreover, previous studies mainly focused on cyanobacterial CCM related to environments with distinct Ci content, salinity and especially pH that strongly influenced the CCM due to the equilibrium of Ci species (Mangan et al., 2016). Therefore, it is indispensable to survey the CCM of thermophilic strains in relation to their habitats that limit the solubility of inorganic carbon (Kamennaya et al., 2012). In recent years, next-generation sequencing (NGS) has been extensively employed to investigate the cyanobacterial genomes in thermal environments (Walter et al., 2017; Alcorta et al., 2020; Chen et al., 2021). This offers an opportunity to explore the thermophilic cyanobacterial CCM and the relationship with their niches at the genomic level. Furthermore, the studies on the molecular components of cyanobacterial CCM in relation to their specific habitats may improve photosynthetic CO2 fixation and biomass production in cyanobacteria (Noreña-Caro and Benton, 2018) and crop plants (Hennacy and Jonikas, 2020) through genetic engineering.
In the present study, genome sequences of 17 thermophilic cyanobacterial strains were used to investigate the molecular basis of CCM by computational identification. The relationship between CCM components and adaptations of these thermophilic cyanobacteria were further discussed. The insights into the CCM components lay a solid foundation for future research regarding photosynthetic improvement and biomass yield of thermophilic cyanobacteria with biotechnological potentials.
According to the genomic resources of the NCBI at the time of this study (2021/05/20), cyanobacteria with available genomes and closely related to thermophilic or hot-spring strains were first retrieved as a preliminary dataset. To ensure the accurate collection of thermophilic cyanobacteria, these strains were further filtered by conducting literature searches, contacting culture collections, leading authors of the manuscripts and submissions to retain thermophilic cyanobacteria with definite information about their thermophilic characteristics. Then, quality control for all genomes was performed based on the following criteria to reduce data redundancy and biased genome representation of cyanobacteria. First, the whole-genome average nucleotide identity (ANI) for each pair of genomes was calculated using the ANI calculator with default settings1. Genomes with an ANI value greater than 99.9% were considered as redundant genomes, and then only one of these genomes was randomly kept for the analysis. Second, the quality of the genomes was evaluated using CheckM (Parks et al., 2015) to ensure a more reliable genome dataset with near completeness (≥95%) and low contamination (<5%). These analyses finally generated a dataset comprising 16 thermophilic cyanobacteria. The genome, protein sequences and genomic annotations of the thermophilic cyanobacteria studied were downloaded from the database of NCBI. The genomes with no or incomplete annotations were annotated using the RAST annotation system (Aziz et al., 2008), provided in Supplementary Table 1.
Information regarding the 16 thermophilic strains was summarized in Table 1. Briefly, the 16 thermophiles were affiliated to four families of order Pseudanabaenales and Synechococcales, including Leptolyngbyaceae: Leptodesmis sichuanensis A121 (Tang et al., 2022a), Leptolyngbya sp. JSC-1 (Brown et al., 2010), Leptothermofonsia sichuanensis E412 (Tang et al., 2022b); Oculatellaceae: Thermoleptolyngbya sp. O-77 (Yoon et al., 2017), T. sichuanensis A183 (Tang et al., 2021); Synechococcaceae: Synechococcus sp. 60AY4M2, 63AY4M2, 65AY6A5, and 65AY6Li (Olsen et al., 2015), Thermostichus sp. JA-2-3B and JA-3-3Ab (Bhaya et al., 2007); Thermosynechococcaceae: Thermosynechococcus sp. CL-1 (Cheng et al., 2020) and TA-1 (Leu et al., 2013), T. vestitus BP-1 (Nakamura et al., 2002) and E542 (Liang et al., 2019), T. vulcanus NIES-2134 (Liang et al., 2018).
Additional to the 16 downloaded genomes, we sequenced the genome of another thermophilic cyanobacterium, purchased from Pasteur Culture Collection of Cyanobacteria as Synechococcus lividus PCC 6715. Whole-genome sequencing was carried out using integrated sequencing strategies, including PacBio and Illumina HiSeq Technology. The PacBio SMRT sequencing of PCC 6715 yielded 96,587 adapter-trimmed reads (subreads) with an average read length of approximately 11 kbp, corresponding to 400-fold coverage. De novo assembly was performed using the hierarchical genome assembly process (HGAP) method implemented in SMRT analysis v2.3.0 (Chin et al., 2013), generating two contigs. The Illumina sequencing of PCC 6715 generated 5,566,668 filtered paired-end reads (clean data), providing approximately 520-fold coverage of the genome. The clean data of short reads generated by Illumina were assembled into contigs using SOAPdenovo v2.04 (Luo et al., 2012) with default parameters. Based on the contigs from SOAPdenovo assembler, the contigs derived from the HGAP method were comparatively examined to determine their continuity and concatenated into one closed circular chromosome. The genome obtained was mapped with Illumina reads using BWA v0.7.17 (Li and Durbin, 2009) and then Pilon v1.23 (Walker et al., 2014) to correct any assembly and sequence errors. The genome of PCC 6715 was annotated as described above and deposited in the NCBI under the accession number CP018092.
Amino acid sequences of 28 proteins involved in CCM of Synechocystis sp. PCC 6803 were downloaded from the CyanoBase2 as a reference protein set. Based on the bidirectional best hit (BBH) criterion (Brilli et al., 2010), orthologous proteins involved in CCM of the studied species were identified using BLASTP with the following thresholds: E-value cut-off of 1E-6, ≥30% identity and 70% coverage. The identified proteins were homologous to these proteins in Synechocystis PCC 6803, namely bicA1 (sll0834), bicA2 (slr0096), ccaA (slr1347), ccmK1 (sll1029), ccmK2 (sll1028), ccmK3 (slr1838), ccmK4 (slr1839), ccmL (sll1030), ccmM (sll1031), ccmN (sll1032), ccmO (slr0436), ccmP (slr0169), cmpA (slr0040), cmpB (slr0041), cmpC (slr0043), cmpD (slr0044), cupA (sll1734), cupB (slr1302), ecaB (slr0051), ndhD3 (sll1733), ndhD4 (sll0027), ndhF3 (sll1732), ndhF4 (sll0026), rbcL (slr0009), rbcS (slr0012), rbcX (slr0011), sbtA (slr1512), and SbtB (slr1513). The homologs of ecaA protein were searched using the experimentally confirmed protein ecaA (all2929) of Anabaena PCC 7120 (Soltes-Rak et al., 1997) as a query. The italic number in bracket mentioned above refers to the gene ID in CyanoBase (see text footnote 2). The accession numbers of orthologous proteins identified in each genome of the thermophilic cyanobacteria studied was summarized in Supplementary Table 2.
Amino acid sequences of Rubisco large subunit (rbcL) were collected for the studied species and reference cyanobacteria. The phylogram of rbcL was used to infer the protein function and classification among the strains. Protein sequences of genes ccmK, -L, -M, -N, -O, and -P encoding carboxysome shell proteins, cmpA, -B, -C, and -D encoding the HCO3– transporter BCT1 and nrtA, -B, -C, and -D of the nitrite/nitrate assimilation were also collected for phylogenetic reconstruction. All the protein sequences used were retrieved from the public databases, CyanoBase (see text footnote 2) or the NCBI3. All the phylogenetic analyses were performed using the following pipeline: multiple sequence alignments by Muscle complemented in Mega7 (Kumar et al., 2016) and Maximum-Likelihood (ML) inference of phylogenies by PhyML v3.3 (Guindon et al., 2010). Parameter settings in PhyML and bootstrap analysis of phylogenies were followed as described (Tang et al., 2019).
The 17 thermophilic cyanobacteria collected in this study all originated from hot springs (Table 1). Although the niche temperatures showed significant variation, these thermophilic cyanobacteria were defined based on their ability to grow above 50°C. Two types of morphology were characterized for these thermophilic cyanobacteria, comprising unicellular Synechococcus, Thermostichus and Thermosynechococcus, and filamentous Leptodesmis, Leptolyngbya, Leptothermofonsia, and Thermoleptolyngbya. In addition, strains JA-2-3Ba and JA-3-3Ab, previously known as Synechococcus strains, have been reclassified into a newly delineated taxon, genus Thermostichus (Komárek et al., 2020). According to the suggested values for species (ANI > 96%, AAI ≥ 95%) and genus (ANI < 83%, AAI ≤ 70%) delimitation (Jain and Rodriguez, 2018), the result of genome-wide ANI and AAI indicated that strain JA-2-3Ba and JA-3-3Ab were different species (ANI = 85.6%, AAI = 87.8%) within Thermostichus, while strain JA-3-3Ab, plus the other four strains from Yellowstone National Park (Table 1), belong to the same species (ANI/AAI > 98%) also within Thermostichus. However, the case of S. lividus PCC 6715 was contradictory. Between S. lividus PCC 6715 and the five Thermosynechococcus strains, the values of ANI and ANI were ∼ 77 and ∼ 81% (Supplementary Table 3), respectively. To resolve the dilemma, a third approach was employed to determine the percentages of conserved proteins (POCP) between genomes. The POCP values (Supplementary Table 3) between S. lividus PCC 6715 and the Thermosynechococcus strains ranged from 83.4 to 84.0%, all far beyond the threshold (50%) for the definition of a prokaryotic genus (Qin et al., 2014). Taken together, strain PCC 6715 might be a member of Thermosynechococcus, and was proposed to be Thermosynechococcus lividus. However, this conclusion requires validation with comprehensive investigation beyond the genomic data.
The carboxysome type of the 17 thermophilic cyanobacteria was investigated based on the phylogenetic analysis of a frequently used molecular marker, rbcL (Klanchui et al., 2017). The ML phylogram of rbcL categorized the 35 cyanobacteria into two categories (Figure 2), α-cyanobacteria and β-cyanobacteria, according to their Rubisco forms. There was only one exception that Synechococcus PCC 7942 was solely placed in a separate branch. The 17 thermophilic cyanobacteria surveyed were all grouped into the β-cyanobacteria category, indicating the presence of Rubisco 1B form in these thermophiles. However, considerable genetic diversity of rbcL amino acid sequences was also shown among these thermophilic cyanobacteria, distributing the strains into two clades (Figure 2). Clade A included only Thermostichus strains, also known as early-branching or early-divergent Synechococcus (Blank and Sánchez-Baracaldo, 2010), whereas clade B contained thermophiles from a different genus. In addition, the clustering of strains did not completely comply with morphology, e.g., filamentous strains clustered with unicellular strains (clade B). Intriguingly, the 17 thermophilic strains did not group with any non-thermophilic strains, suggesting rbcL specificity of thermophiles. Although phylogenetic analysis of rbcL sequences can be used for the classification of cyanobacterial types, the evolutionary relationship based on habitats or morphology cannot be elucidated from the present rbcL phylogram. This is consistent with the conclusions in previous studies that phylogenetic inference of molecular markers may be improper for the elaboration of evolutionary relationship with cyanobacterial habitat and environments (Komárek, 2016; Klanchui et al., 2017).
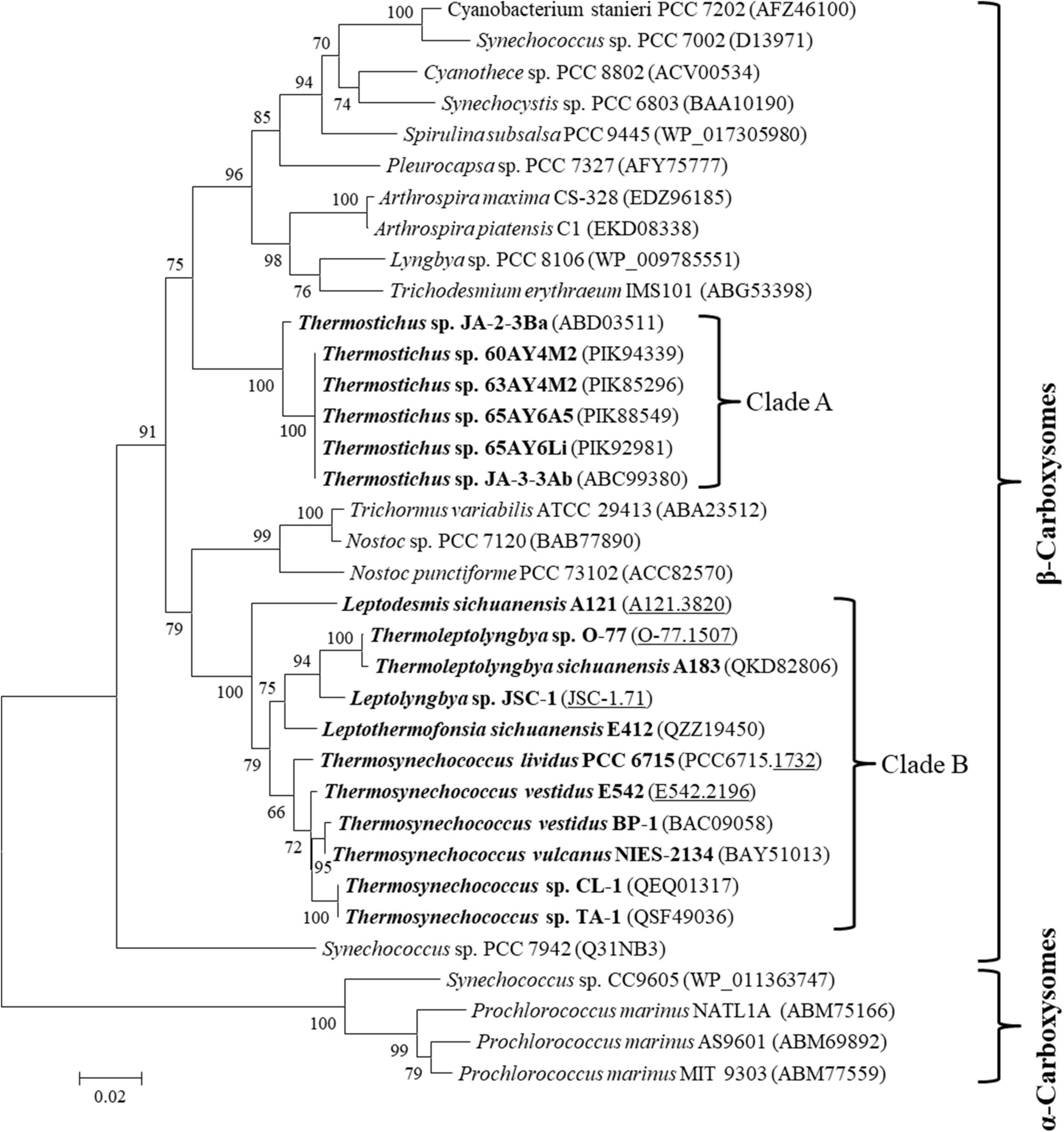
Figure 2. Phylogenetic inference of Rubisco large subunit protein sequences. The thermophilic cyanobacteria investigated in this study are indicated in bold. The accession numbers underscored refers to the gene IDs in Supplementary Table 1. Only bootstrap values > 50% are indicated at nodes. Scale bar = 1% substitutions per site.
Five different transport systems (Table 2) have been identified in the 17 thermophilic cyanobacteria. The two CO2 uptake systems, NDH-13 and NDH-14 complex, were present in all the surveyed thermophilic cyanobacteria. Both CO2 uptake systems in these thermophilic cyanobacteria were consistent with the previous reports that both CO2 transporters were present in β-cyanobacteria living in freshwater, brackish or eutrophic lakes (Sandrini et al., 2014; Durall and Lindblad, 2015). In contrast, oceanic α-cyanobacteria (e.g., Prochlorococcus species) and marine β-cyanobacteria (e.g., Trichodesmium species) contained only one or even lacked them entirely (Price et al., 2007; Pronina et al., 2017). These results strongly indicated that the presence of NDH-13 and NDH-14 might be relevant to the environments where these cyanobacteria live. Thermophilic cyanobacteria, similar to freshwater and estuarine strains, contained essential components of CO2 uptake and bicarbonate transport genes. Both constitutive systems NDH-14 complex and bicA were ubiquitous in all known thermophilic strains. The inducible NDH-13 complex is also universal suggesting that these strains are prepared for occasional carbon dioxide shortages. Unfortunately, it was not verified based on the current data that the niche temperature directly affected the existence of the two CO2 uptake systems in light of their near-ubiquity in non-thermophilic cyanobacteria. The protein sequences of five genes encoding the two systems showed different identities with the sequences of non-thermophilic reference cyanobacteria (Synechocystis PCC 6803), ranging from 50.4 to 64.7% (Table 2), but a high degree of homology (≥70% identity) was noticed only among cupA genes. In addition, intragenus and intergenic variations of protein sequences were revealed by the different identities (Table 2). Moreover, the distinct property of the two complexes, a low-CO2 inducible high-affinity CO2 uptake system (NDH-13 complex) and a constitutive low-affinity CO2 uptake system (NDH-14 complex), may confer the thermophilic cyanobacteria with more alternative strategies to survive in environments with significant CO2 fluctuation, particularly in hot springs.
More variations among the 17 thermophilic cyanobacteria were noticed in HCO3– transport systems than in CO2 uptake systems (Table 2 and Figure 3). Association of the abundance of these mechanisms with their respective environmental niches is inconclusive. Similar repertoires of CO2 and bicarbonate uptake genes were observed in L. sichuanensis A121 with the lowest habitat temperature and the most thermophilic strains of genus Thermostichus (Table 2 and Figure 3). When it comes to the pH of the environmental niches, no evident niches preference of genes was observed. Genetic repertoire of strains isolated from acidic and alkaline niches appeared to be unrelated to the niche pH. For bicA transporter, only Leptothermofonsia E412 and Thermoleptolyngbya A183 possessed both bicA1 and bicA2, while the other strains contained bicA1 or bicA2 (Table 2). The amino acid sequence identities of bicA genes were around 60% between the thermophilic cyanobacteria and Synechocystis PCC 6803. The presence of bicA1 and/or bicA2 was consistent within each genus except for Thermoleptolyngbya (strain A183 and O-77). Sequence analysis suggested that a putative protein-coding gene of O-77 (O-77.5066, Supplementary Table 1) showed very low amino acid coverage (46.9%) to bicA2 of A183, but the aligned region was almost identical to each other (Supplementary Figure 1), indicating this short CDS might be a partial region of bicA2. Such discrepancy in CDS length of bicA2 within Thermoleptolyngbya requires future studies to elucidate the possible causes, e.g., mutations, gene lost, or sequencing errors. For sbt regulator, both sbtA and sbtB were found only in six thermophilic cyanobacteria (Table 2). Furthermore, considerable variations existed within the genus Thermosynechococcus. Strain E542, PCC 6715 and CL-1 possessed both sbtA and sbtB. TA-1 had only sbtB, while the other two Thermosynechococcus strains contained none. This result implied that the sbt genes of these Thermosynechococcus strains might be acquired by horizontal gene transfer or vertically inherited but lost during the evolutionary process. In addition, it appeared that the thermophilic cyanobacteria typically have bicA rather than sbt as revealed by the dominant presence (Table 2). This could be ascribed to the distinct traits of the two transporters and the habitat of these cyanobacteria. First, bicA shows low affinity and high flux rate to HCO3– and the genes encoding bicA were found to be primarily constitutively expressed (Price et al., 2004). Second, sbt was highly inducible under carbon-limited conditions and had a relatively high affinity to HCO3– (Price, 2011). The sbt may therefore become accessory when cyanobacteria live in alkaline ecosystems typically rich in HCO3–, which was in accordance with most cases in this study (Table 1), e.g., Thermostichus strains. Meanwhile, it was evident that thermophilic cyanobacteria with both transporters might be advantageous to acclimatize to the highly dynamic shift of exogenous HCO3– by selective utilization of the two HCO3– transport systems.
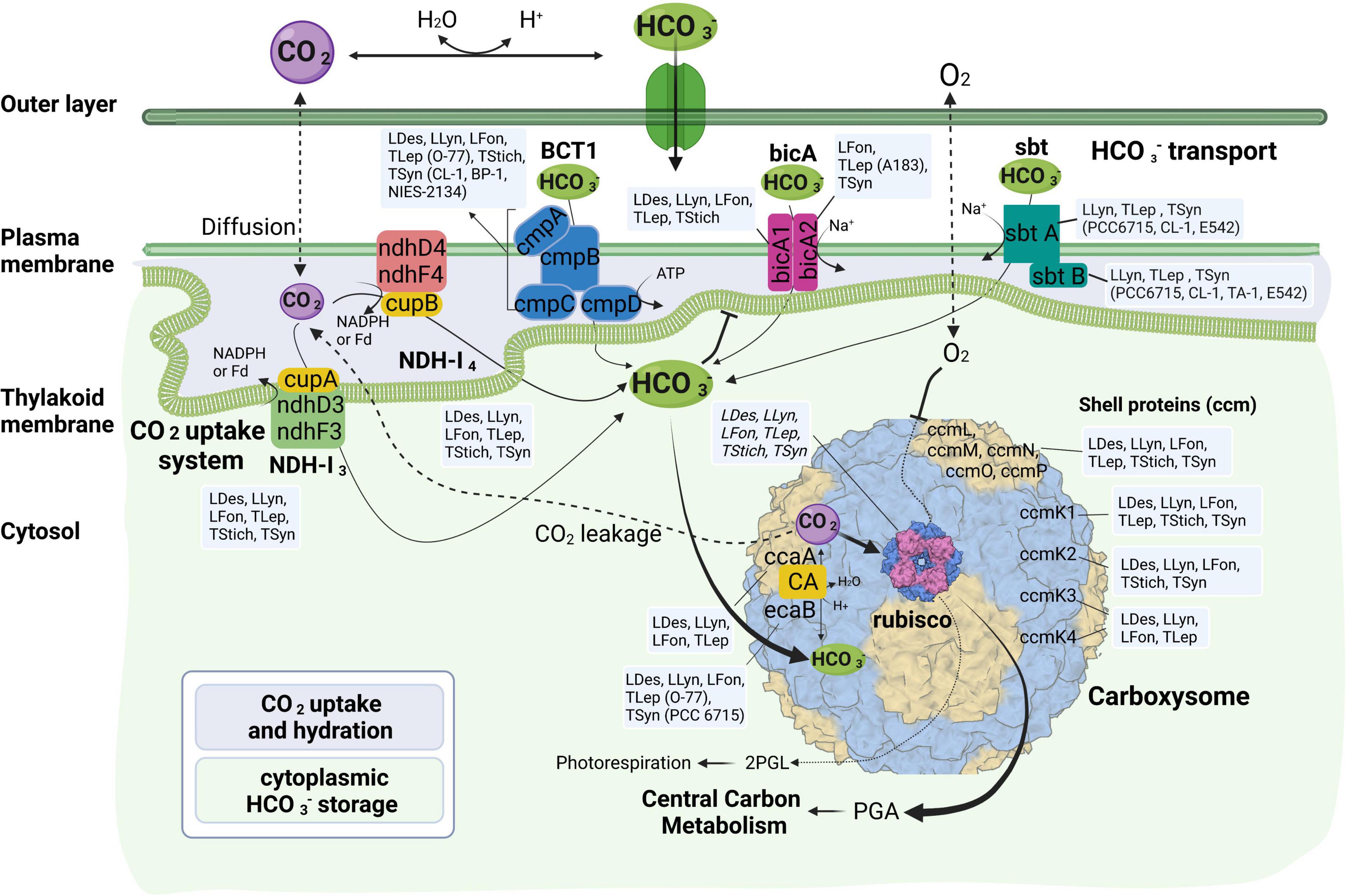
Figure 3. Molecular components of CCM characteristic for various strains of thermophilic cyanobacteria divided according to their function. LDes – Leptodesmis, LLyn – Leptolyngbya, LFon – Leptothermofonsia, TLep – Thermoleptolyngbya, TStich – Thermostichus, TSyn – Thermosynechococcus; 2PGL – 2-phosphoglycolate; PGA – phosphoglyceric acid. Figure created with BioRender.com.
As for the third HCO3– transporter, BCT1 encoded by cmpA, -B, -C, and -D and with high affinity for HCO3– was induced under low levels of Ci and enhanced by high light conditions (Omata et al., 2002). The cmp genes were detected in all thermophilic cyanobacteria except for Thermoleptolyngbya A183 and Thermosynechococcus E542, PCC 6715 and TA-1 (Table 2). Intriguingly, strains without cmp genes all possessed sbt, which might to some extent compensate the function of cmp genes for adaptation to thermal environments limited in inorganic carbon (Kamennaya et al., 2012). The identified cmp genes showed amino acid sequence identity of 59.5–75.8% with the reference proteins (Table 2). In addition, it was reported that nrtA, -B, -C, and -D (nitrate/nitrite transport system) shared high sequence similarity with cmpA, -B, -C, and -D (Omata, 1995). Therefore, we also identified the nrt genes in the 17 thermophilic cyanobacteria, and phylogenetic analysis based on amino acid sequences of cmp and nrt genes was performed to verify the annotations with the help of experimentally confirmed cmp proteins of Synechococcus PCC 7942 (Omata et al., 1999). As shown in Figure 4, the cmp proteins of thermophilic cyanobacteria clustered with that of Synechococcus PCC 7942 and other reference strains, while the nrt proteins grouped into clades or clusters separated from cmp proteins. Furthermore, sequence conservation was low, as suggested by long branches among the clusters formed in the phylogram (Figure 4). Surprisingly, the genome of Thermoleptolyngbya O-77 was the only genome without the complete set of nrtA, -B, -C, and -D, nrtC of which was not found by BLASTP. However, sequence analysis (Supplementary Figure 2) showed that two putative genes both shared high similarity of protein sequence with nrtC of Thermoleptolyngbya A183, suggesting unknown interruption within CDS of nrtC in O-77 genome. This might be ascribed to mutations, partial gene lost, or sequencing errors. Intriguingly, the discrepancy of the presence of cmp genes within genus Thermoleptolyngbya and Thermosynechococcus again suggested that these genes might be acquired by horizontal gene transfer or vertically inherited but lost during the evolutionary process.
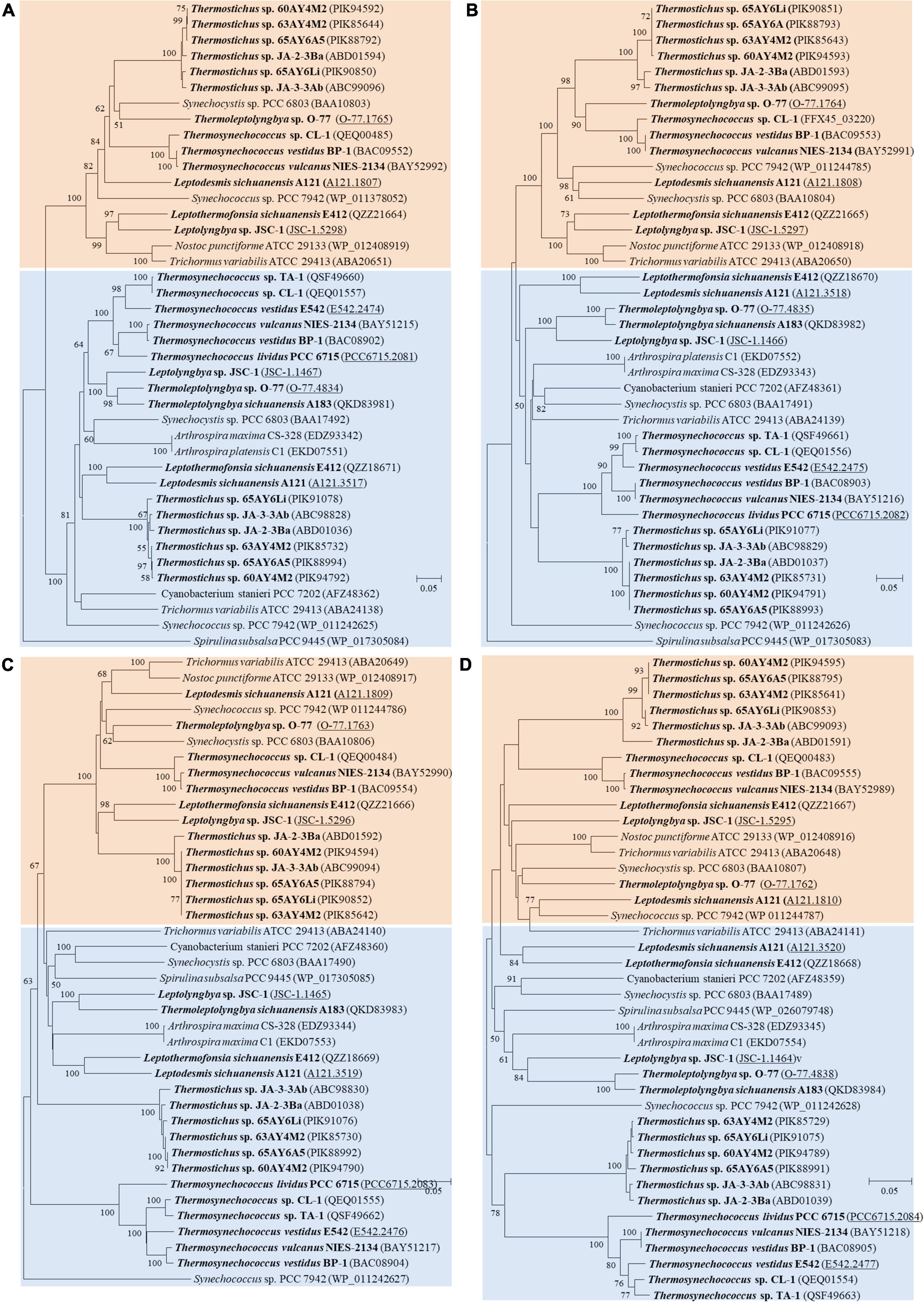
Figure 4. Phylogenetic inference of protein sequences of cmp and nrt genes. (A) cmpA and nrtA; (B) cmpB and nrtB; (C) cmpC and nrtC; (D) cmpD and nrtD. The thermophilic cyanobacteria investigated in this study are indicated in bold. The accession numbers underscored refers to the gene IDs in Supplementary Table 1. Only bootstrap values > 50% are indicated at nodes.
In summary, the Ci uptake systems could assign the 17 thermophilic cyanobacteria into three genotypes: (I) strains comprising NDH-13, NDH-14, bicA, sbt, and BCT1; (II) NDH-13, NDH-14, bicA, and sbt; and (III) NDH-13, NDH-14, bicA, and BCT1. This result suggested that the same CO2 uptake systems and different HCO3– transport systems were shared by these thermophilic cyanobacteria. Different genotypes were primarily attributed to the presence/absence of sbt and BCT1 (Table 2). Only three strains, namely Leptolyngbya JSC-1, Thermoleptolyngbya O-77, and Thermosynechococcus CL-1, contained both types of high-affinity HCO3– transporters, indicating that these strains may have a higher capacity for HCO3– uptake under various growth environments. Definitely, owning one of sbt and BCT1 appeared to be sufficient for thermophilic strains to cope with extremely low inorganic carbon and ions in thermal aquatic environments. Future experimental studies should be carried out to elucidate the function of these in silico identified proteins in thermophilic cyanobacteria.
Carboxysomes in cyanobacteria are bacterial microcompartments (BMCs) that provide an environment for enhancing the catalytic capabilities of the CO2 fixing enzyme, Rubisco (Cai et al., 2009). A typical β-cyanobacterial genome usually contains ccmK1-4 (MacCready et al., 2020). However, only three out of 17 thermophilic cyanobacteria studied showed such diversity of the shell proteins, while Thermoleptolyngbya strains lacked ccmk2, and Thermostichus and Thermosynechococcus strains contained none of ccmk3 and ccmK4 (Table 3). Previous studies showed that ccmK1 and ccmK2 were the main structural proteins of the carboxysome shell (Kerfeld et al., 2005; Rae et al., 2013). Phylogenetic analysis and identity calculation suggested particularly high sequence conservation between the two homologs of ccmK1/2, as revealed by low amino acid substitution rates and short branches (Figure 5A) and high identity (Table 3). In contrast, phylogenetic branches of the ccmK3 and ccmK4 proteins were much longer, and residues were less conserved (Figures 5B,C and Table 3). Although deletion of the ccmK2 gene in Synechococcus PCC 7942 that lacked ccmK1 caused a carboxysome-less phenotype (Rae et al., 2012), this experiment did not verify whether normal phenotype can be achieved with the presence of sole ccmK1 homolog. Besides, the main distinguishing feature between ccmK1 and ccmK2 was the ∼10 amino acid long C-terminal extension of ccmK1 (Kerfeld et al., 2005), which has been proven to show limited structural relevance for shell assembly (Cai et al., 2016). Instead, it might be just the genomic proximity of the two ccmK genes, as indicated by its high degree of conservation (15/17 genomes, Table 3). Future studies are required to carefully investigate the specificity regarding the structure and function of ccmk1 in the Thermoleptolyngbya strains. Regarding ccmK3 and ccmK4, the widespread absence of the two genes was not surprising. The previous studies showed they are similar molecular components (Sommer et al., 2017) and suggested functional redundancy that play only a minor structural role (Rae et al., 2012). On the other hand, a recent study proposed a functional scenario in which minor ccmK3/K4 incorporation in shells would introduce sufficient local disorder to allow shell remodeling (Garcia-Alles et al., 2019). Another study indicated that ccmK3-ccmK4 heterohexamers potentially expand the range of permeability properties of metabolite channels in carboxysome shells (Sommer et al., 2019). Taken together, these findings suggested that ccmK3/K4 present in Leptodesmis A121, Leptolyngbya JSC-1, Leptothermofonsia E412, and Thermoleptolyngbya A183 and O-77 may be utilized to adjust the properties of carboxysome for rapid adaptation to environmental changes that happen in thermal regions. In addition to ccmK, other genes encoding carboxysome shell proteins, ccmL, -M, -N, -O, and -P were also found to be present in all 17 thermophilic cyanobacteria (Table 3). Only ccmN exhibited weak homologs (<40%). All the surveyed thermophilic cyanobacteria showed a high degree of homology (67.6–76.1%) with the ccmP of Synechocystis PCC 6803, and similarly identity range of 65.8–75.5% to the ccmP of Synechococcus PCC 7942, an experimentally confirmed protein (Cai et al., 2013). Further phylogenetic analysis suggested extensive genetic diversity in these genes as indicated by the assignments of these thermophilic cyanobacteria into different clusters or clades (Supplementary Figure 3).

Figure 5. Phylogenetic inference of protein sequences of ccmK1/2 (A), ccmK3 (B) and ccmK4 (C). The thermophilic cyanobacteria investigated in this study are indicated in bold. The accession numbers underscored refers to the gene IDs in Supplementary Table 1. Only bootstrap values > 50% are indicated at nodes.
The subunits of Rubisco, rbcL and rbcS, and Rubisco assembly chaperone, rbcX, were present in all the 17 thermophilic cyanobacteria (Table 3). The protein sequences of rbcL in thermophilic cyanobacteria were highly conserved to the reference protein (>83% identity), while that of rbcS was moderately conserved (∼ 65% identity). The protein sequences of rbcX were more variable, showing ∼ 45% identity with the reference protein. In regards to β-CA, carboxysomal ccaA and/or non-carboxysomal ecaB proteins with low sequence identity (38.1–49.7%) were detected only in six thermophilic strains (Table 3), respectively. Apart from the functions as shell protein, an additional function of ccmM (γ-CA) as CA activity was reported in cyanobacteria lacking ccaA (Peña et al., 2010; de Araujo et al., 2014). Thus, we compared γ-CA-like domain in N-terminal of ccmM in thermophilic cyanobacteria to the functional γ-CA in Thermosynechococcus BP-1 (Peña et al., 2010) and Nostoc PCC 7120 (de Araujo et al., 2014) as well as non-functional γ-CA in Synechocystis PCC 6803 (Cot et al., 2008) and Synechococcus PCC 7942 (So and Espie, 2005). As shown in Figure 6, strains from Thermostichus and Thermosynechococcus exhibited similar primary structure of amino acid residues in γ-CA-like domain of ccmM to that with CA activity in Thermosynechococcus BP-1 and Nostoc PCC 7120. The result suggested that these ccmM proteins may play a role in CA activity in light of the absence of a carboxysomal CA (ccaA) (Table 3) in these thermophilic cyanobacteria. Although strains from Leptodesmis, Leptolyngbya, Leptothermofonsia, and Thermoleptolyngbya had both ccmM and ccaA, it is likely that only ccaA function as CA activity as indicated by the primary structure analysis, which was consistent with the result found in haloalkaliphilic cyanobacteria Microcoleus sp. IPPAS B-353 (Kupriyanova et al., 2016). In addition, strains living in high temperatures above 50°C (Table 1) appeared to tend to lose ccaA. Meanwhile, high pH conditions were speculated to be an important factor in ccaA absence (Klanchui et al., 2017). However, the reasons were still unclear. Certainly, the co-evolution of ccmM and ccaA is crucial for carboxysome to regularly function in extreme environments regardless of high temperature or alkalinity. As for α-CA, non-carboxysomal ecaA was not found in all the 17 thermophilic cyanobacteria.

Figure 6. Partial alignments of ccmM amino acid sequences representing 17 thermophilic cyanobacteria, Nostoc PCC 7120, and Synechocystis PCC 6803 and Synechococcus PCC 7942 as outgroup. As suggested by Peña et al. (2010), red boxes refer to conserved regions of the N-terminal domain of ccmM necessary for CA activity, while shaded cysteine amino acids indicate essential residues participating in the disulfide bond in the C-termini of active ccmM protein.
The genomic location of CCM-related genes in thermophilic cyanobacteria may provide insights into the function and evolution of these genes. In the present study, the genomic organization of CCM-related genes was illustrated in Figure 7. Genes encoding each NDH-1 complex were all clustered together, except for the ndhD4 of Thermosynechococcus TA-1. This gene was remote from the other genes of NDH-14 complex (Figure 7). In light of the coordination of NDH and cup proteins in CO2 uptake (Han et al., 2017), whether and how such organization in Thermosynechococcus TA-1 affects the co-regulation requires further experimental elucidation. In addition, the genes encoding bicA1 and bicA2 were positionally scattered in the genomes of Leptothermofonsia E412 and Thermoleptolyngbya A183, whereas genes encoding cmpABCD and sbtA/B were located together in genomes (Figure 7), respectively.

Figure 7. Genomic organization of CCM-related genes in the 17 thermophilic cyanobacteria studied. Solid arrow boxes refer to genes and the direction of transcription.
Focusing on the gene organization of the carboxysome, there appeared to be the main carboxysome locus (MCL) in the genomes of all the studied thermophilic cyanobacteria, comprising one to two ccmK1/2 genes, always followed by ccmL, -M, -N and in the majority of genomes (11 out of 17), ccmO (Figure 7). The sequential arrangement of ccmK2 and ccmK1 in the MCL might facilitate protein complex assembly or help to balance the shell protein stoichiometry during translation of MCL genes (Cai et al., 2016). Moreover, previous studies suggested that ccmK, -L, -M, and -N were co-regulated in an MCL as an operon (Rae et al., 2013; Billis et al., 2014). In addition to the MCL, satellite loci were found in several genomes. Satellite locus encoding the ccmP protein was found in all the 17 thermophilic cyanobacteria. The satellite loci formed by ccmK3 and ccmK4 was present only in five genomes (Figure 7). The organization of K1/K2 and K3/K4 paralogs in separated operons might relate to structural segregation of the two groups (Sommer et al., 2019). Although ccmK3 and ccmK4 clustered in operon, it was reported that the stoichiometry of incorporation of ccmK3 in associations with ccmK4 was low in Synechocystis PCC6803 (Garcia-Alles et al., 2019). However, the expression of ccmK3 and ccmK4 from a satellite locus may increase the flexibility of carboxysome shell assembly and permeability (Sommer et al., 2019), and may provide differing metabolite selectivities (Sommer et al., 2017). Another satellite locus, ccmO, was positionally variable (Figure 7). In 11 genomes, ccmO was the terminal gene in the MCL. However, ccmO in six Thermosynechococcus genomes was located as a satellite locus, remote from other carboxysome genes, and this ccmO formed a distinct clade as revealed by the phylogenetic analysis (Supplementary Figure 3D). Previous transcriptome analysis demonstrated that the ccmO gene was co-regulated with MCL genes only when it was associated with the MCL (Sommer et al., 2017). Therefore, the dispersal of ccmO as well as other carboxysome genes across the genome may increase the plasticity of individual regulation of their expression, perhaps in response to changes in environmental conditions, and vice versa, diverse environmental conditions appear to promote relocation of these genes driven by evolutionary force (Cai et al., 2012; Shih et al., 2013).
Genes encoding rbcL, rbcS, and rbcX for Rubisco were localized together in all of the 17 thermophilic cyanobacteria, which was in line with the previous consensus that the three genes were clustered in an operon in β-cyanobacteria (Badger et al., 2002). The Rubisco gene cluster was all apart from the MCL in each genome, indicating that it, as satellite loci to MCL, might regulate its expression independently. This was supported by the results reported by Billis et al. (2014) that ccmO, rbcL, and rbcS were less strictly co-regulated with the MCL genes. As for the chaperone rbcX, sometimes it was non-essential for the assembly of an active Rubisco enzyme, e.g., in Synechococcus PCC 7942 (Emlyn-Jones et al., 2006). The role of rbcX in these thermophilic cyanobacteria requires future investigations. In regards to β-CAs, scattered distribution of ccaA and ecaB was present in the genomes.
The overall composition of the CCM system presented in Figure 3 contains all typical components found in other cyanobacteria, including the complete set of genes for carboxysome assembly. A comparison of these components among β-cyanobacteria was performed between thermophilic cyanobacteria and non-thermophilic cyanobacteria. For a precise comparison, we divided the non-thermophilic cyanobacteria into three groups: freshwater, marine, and alkaliphilic cyanobacteria. The overall compositions of CCM components in thermophilic cyanobacteria were more similar to the freshwater and alkaliphilic than the marine groups (Klanchui et al., 2017). The thermophilic, freshwater and alkaliphilic cyanobacteria possessed both CO2 uptake systems, NDH-13 and NDH-14, whereas most marine cyanobacteria lacked the NDH-13. In light of the HCO3– uptake, marine and some thermophilic/alkaliphilic cyanobacteria lacked the BCT1 transporter. Although freshwater cyanobacteria consistently possessed BCT1 transporter, some thermophilic cyanobacteria with freshwater origins lacked that. The relatively high diversity of bicarbonate transporters is a distinguishing factor for certain thermophilic strains probably as indicator of their specific location and not their thermophilic character. However, there is no apparent correlation between the pH of the isolation sources of these strains and the presence or absence of specific bicarbonate uptake systems (Figure 3 and Table 1). More thorough studies are required to characterize the functions of these transporters in the future. In addition, the freshwater cyanobacteria possessed the highest abundance of CAs, β-CA (ccaA and ecaB), α-CA (ecaA), and γ-CA (ccmM), while the thermophilic cyanobacteria with freshwater origins were likely to possess up to two CAs (Table 3 and Figure 3). Notably, 11 out of the 17 investigated thermophiles appeared to have only γ-CA (ccmM). On the other hand, the diversity of carboxysome shell proteins appears to be lower in thermophilic strains than many of their mesophilic counterparts, with multiple strains lacking at least one of the four ccmK genes.
In the present study, we investigated the molecular components and distribution of CCM in 17 thermophilic cyanobacteria. The diversity in the Ci uptake systems and carboxysome of these thermophiles were observed. Particularly, the presence of HCO3– transporters, ccmK2/3/4 and CAs tremendously varied among these thermophiles. Several strains have more Ci uptake-related components, indicating the capabilities of these strains to acclimate and establish competitive growth by using different Ci uptake strategies in response to environmental fluctuation. The distinct genomic distribution of CCM-related genes among the thermophiles probably suggested various regulations of expression. Overall, the comparative genomic analysis revealed distinct molecular components and organization of CCM in thermophilic cyanobacteria. These findings provided insights into the CCM components of thermophilic cyanobacteria and fundamental knowledge for further research regarding photosynthetic improvement and biomass yield of thermophilic cyanobacteria with biotechnological potentials.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, CP018092.
JT: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing – original draft, writing, review, editing, visualization, supervision, project administration, and funding acquisition. HZ: formal analysis, investigation, data curation, and visualization. DYa: formal analysis, investigation, and data curation. SR: formal analysis, data curation, and visualization. DYo: formal analysis, visualization, writing, review, and editing. AK-S: formal analysis, investigation, and funding acquisition. MD: conceptualization, methodology, resources, data curation, writing – original draft, writing, review, editing, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (31970092 and 32071480), Shenzhen Fundamental Research Program (GXWD20201231165807007-20200806170221001), National Science Centre (Poland) through project no. 2018/31/B/ST8/00822, and Tenure-Track Fund to MD. Funding bodies had no influence over the design and execution of this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Dr. Igor Brown and the culture collections of PCC and NIES for the assistance in verifying the actual thermophilicity of certain strains.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.876272/full#supplementary-material
Alcorta, J., Alarcón-Schumacher, T., Salgado, O., and Díez, B. (2020). Taxonomic novelty and distinctive genomic features of hot spring cyanobacteria. Front. Genet. 11:568223. doi: 10.3389/fgene.2020.568223
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Badger, M. R., Hanson, D., and Price, G. D. (2002). Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 29, 161–173. doi: 10.1071/PP01213
Bhaya, D., Grossman, A. R., Steunou, A.-S., Khuri, N., Cohan, F. M., Hamamura, N., et al. (2007). Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 1, 703–713. doi: 10.1038/ismej.2007.46
Billis, K., Billini, M., Tripp, H. J., Kyrpides, N. C., and Mavromatis, K. (2014). Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation. PLoS One 9:e109738. doi: 10.1371/journal.pone.0109738
Blank, C. E., and Sánchez-Baracaldo, P. (2010). Timing of morphological and ecological innovations in the cyanobacteria–a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23. doi: 10.1111/j.1472-4669.2009.00220.x
Brilli, M., Fondi, M., Fani, R., Mengoni, A., Ferri, L., Bazzicalupo, M., et al. (2010). The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst. Biol. 4:52. doi: 10.1186/1752-0509-4-52
Brown, I. I., Bryant, D. A., Casamatta, D., Thomas-Keprta, K. L., Sarkisova, S. A., Shen, G., et al. (2010). Polyphasic characterization of a thermotolerant siderophilic filamentous cyanobacterium that produces intracellular iron deposits. Appl. Environ. Microbiol. 76, 6664–6672. doi: 10.1128/aem.00662-10
Burnap, R. L., Hagemann, M., and Kaplan, A. (2015). Regulation of CO2 concentrating mechanism in cyanobacteria. Life 5, 348–371. doi: 10.3390/life5010348
Cai, F., Bernstein, S. L., and Wilson, S. C. (2016). Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol. 170, 1868–1877. doi: 10.1104/pp.15.01822
Cai, F., Kerfeld, C. A., and Sandh, G. (2012). “Bioinformatic identification and structural characterization of a new carboxysome shell protein,” in Functional Genomics and Evolution of Photosynthetic Systems, eds R. Burnap and W. Vermaas (Dordrecht: Springer Netherlands), 345–356. doi: 10.1016/j.jmb.2009.03.056
Cai, F., Menon, B. B., Cannon, G. C., Curry, K. J., Shively, J. M., and Heinhorst, S. (2009). The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521. doi: 10.1371/journal.pone.0007521
Cai, F., Sutter, M., Cameron, J. C., Stanley, D. N., Kinney, J. N., and Kerfeld, C. A. (2013). The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-Carboxysome, forms a subcompartment within a microcompartment. J. Biol. Chem. 288, 16055–16063. doi: 10.1074/jbc.M113.456897
Chen, H., and Wang, Q. (2020). Microalgae-based nitrogen bioremediation. Algal Res. 46:101775. doi: 10.1016/j.algal.2019.101775
Chen, M.-Y., Teng, W.-K., Zhao, L., Hu, C.-X., Zhou, Y.-K., Han, B.-P., et al. (2021). Comparative genomics reveals insights into cyanobacterial evolution and habitat adaptation. ISME J. 15, 211–227. doi: 10.1038/s41396-020-00775-z
Cheng, Y.-I., Chou, L., Chiu, Y.-F., Hsueh, H.-T., Kuo, C.-H., and Chu, H.-A. (2020). Comparative genomic analysis of a novel strain of Taiwan hot-spring cyanobacterium Thermosynechococcus sp. CL-1. Front. Microbiol. 11:82. doi: 10.3389/fmicb.2020.00082
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Meth. 10, 563–569. doi: 10.1038/nmeth.2474
Clement, R., Dimnet, L., Maberly, S. C., and Gontero, B. (2016). The nature of the CO2-concentrating mechanisms in a marine diatom, Thalassiosira pseudonana. New Phytol. 209, 1417–1427. doi: 10.1111/nph.13728
Cot, S. S. W., So, A. K. C., and Espie, G. S. (2008). A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J. Bacteriol. 190, 936–945. doi: 10.1128/jb.01283-07
de Araujo, C., Arefeen, D., Tadesse, Y., Long, B. M., Price, G. D., Rowlett, R. S., et al. (2014). Identification and characterization of a carboxysomal γ-carbonic anhydrase from the cyanobacterium Nostoc sp. PCC 7120. Photosynth. Res. 121, 135–150. doi: 10.1007/s11120-014-0018-4
Durall, C., and Lindblad, P. (2015). Mechanisms of carbon fixation and engineering for increased carbon fixation in cyanobacteria. Algal Res. 11, 263–270. doi: 10.1016/j.algal.2015.07.002
Emlyn-Jones, D., Woodger, F. J., and Whitney, P. S. M. (2006). RbcX can function as a rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol. 47, 1630–1640. doi: 10.1093/pcp/pcl028
Galmés, J., Hermida Carrera, C., Laanisto, L., and Niinemets, Ü (2016). A compendium of temperature responses of Rubisco kinetic traits: variability among and within photosynthetic groups and impacts on photosynthesis modeling. J. Exp. Bot. 67:erw267. doi: 10.1093/jxb/erw267
Galmés, J., Kapralov, M. V., Copolovici, L. O., Hermida-Carrera, C., and Niinemets, Ü (2015). Temperature responses of the Rubisco maximum carboxylase activity across domains of life: phylogenetic signals, trade-offs, and importance for carbon gain. Photosynth. Res. 123, 183–201. doi: 10.1007/s11120-014-0067-8
Garcia-Alles, L., Root, K., Maveyraud, L., Aubry, N., Lesniewska, E., Mourey, L., et al. (2019). Occurrence and stability of hetero-hexamer associations formed by β-carboxysome CcmK shell components. PLoS One 14:e0223877. doi: 10.1101/730861
Gubernator, B., Bartoszewski, R., Kroliczewski, J., Wildner, G., and Szczepaniak, A. (2008). Ribulose-1,5-bisphosphate carboxylase/oxygenase from thermophilic cyanobacterium Thermosynechococcus elongatus. Photosynth. Res. 95, 101–109. doi: 10.1007/s11120-007-9240-7
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Han, X., Sun, N., Xu, M., and Mi, H. (2017). Co-ordination of NDH and Cup proteins in CO2 uptake in cyanobacterium Synechocystis sp. PCC 6803. J. Exp. Bot. 68, 3869–3877. doi: 10.1093/jxb/erx129
Hennacy, J. H., and Jonikas, M. C. (2020). Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yields. Annu. Rev. Plant Biol. 71, 461–485. doi: 10.1146/annurev-arplant-081519-040100
Hill, N. C., Tay, J. W., Altus, S., Bortz, D. M., and Cameron, J. C. (2020). Life cycle of a cyanobacterial carboxysome. Sci. Adv. 6:eaba1269. doi: 10.1126/sciadv.aba1269
Jain, C., and Rodriguez, R. L. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1101/225342
Kamennaya, N. A., Ajo-Franklin, C. M., Northen, T., and Jansson, C. (2012). Cyanobacteria as biocatalysts for carbonate mineralization. Minerals 2, 338–364. doi: 10.3390/min2040338
Kaplan, A. (2017). On the cradle of CCM research: discovery, development, and challenges ahead. J. Exp. Bot. 68, 3785–3796. doi: 10.1093/jxb/erx122
Kerfeld, C. A., and Melnicki, M. R. (2016). Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75. doi: 10.1016/j.pbi.2016.03.009
Kerfeld, C. A., Sawaya, M. R., Tanaka, S., Nguyen, C. V., Phillips, M., Beeby, M., et al. (2005). Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938. doi: 10.1126/science.1113397
Kierzkowska-Pawlak, H., Kruszczak, E., and Tyczkowski, J. (2022). Catalytic activity of plasma-deposited Co3O4-based thin films for CO2 hydration – A new approach to carbon capture applications. Appl. Catal. B Environ. 304:120961. doi: 10.1016/j.apcatb.2021.120961
Klanchui, A., Cheevadhanarak, S., Prommeenate, P., and Meechai, A. (2017). Exploring components of the CO2-concentrating mechanism in alkaliphilic cyanobacteria through genome-based analysis. Comput. Struct. Biotechnol. J. 15, 340–350. doi: 10.1016/j.csbj.2017.05.001
Komárek, J. (2016). A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. Eur. J. Phycol. 51, 346–353. doi: 10.1080/09670262.2016.1163738
Komárek, J., Johansen, J., Smarda, J., and Strunecký, O. (2020). Phylogeny and taxonomy of Synechococcus-like cyanobacteria. Fottea 20, 171–191. doi: 10.5507/fot.2020.006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kupriyanova, E. V., Cho, S. M., Park, Y.-I., Pronina, N. A., and Los, D. A. (2016). The complete genome of a cyanobacterium from a soda lake reveals the presence of the components of CO2-concentrating mechanism. Photosynth. Res. 130, 151–165. doi: 10.1007/s11120-016-0235-0
Kupriyanova, E. V., and Samylina, O. S. (2015). CO2-concentrating mechanism and its traits in haloalkaliphilic cyanobacteria. Microbiology 84, 112–124. doi: 10.1134/S0026261715010075
Leu, J.-Y., Lin, T.-H., Selvamani, M. J. P., Chen, H.-C., Liang, J.-Z., and Pan, K.-M. (2013). Characterization of a novel thermophilic cyanobacterial strain from Taian hot springs in Taiwan for high CO2 mitigation and C-phycocyanin extraction. Process Biochem. 48, 41–48. doi: 10.1016/j.procbio.2012.09.019
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liang, Y., Kaczmarek, M. B., Kasprzak, A. K., Tang, J., Shah, M. M. R., Jin, P., et al. (2018). Thermosynechococcaceae as a source of thermostable C-phycocyanins: properties and molecular insights. Algal Res. 35, 223–235. doi: 10.1016/j.algal.2018.08.037
Liang, Y., Tang, J., Luo, Y., Kaczmarek, M. B., Li, X., and Daroch, M. (2019). Thermosynechococcus as a thermophilic photosynthetic microbial cell factory for CO2 utilisation. Bioresour. Technol. 278, 255–265. doi: 10.1016/j.biortech.2019.01.089
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1:18. doi: 10.1186/2047-217x-1-18
MacCready, J. S., Basalla, J. L., and Vecchiarelli, A. G. (2020). Origin and evolution of carboxysome positioning systems in cyanobacteria. Mol. Biol. Evol. 37, 1434–1451. doi: 10.1093/molbev/msz308
Mangan, N. M., Flamholz, A., Hood, R. D., Milo, R., and Savage, D. F. (2016). pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, 5354–5362. doi: 10.1073/pnas.1525145113
Melnicki, M. R., Sutter, M., and Kerfeld, C. A. (2021). Evolutionary relationships among shell proteins of carboxysomes and metabolosomes. Curr. Opin. Microbiol. 63, 1–9. doi: 10.1016/j.mib.2021.05.011
Mook, W. G. (2000). Environmental Isotopes in the Hydrological Cycle - Principles and Applications. Paris: UNESCO-IHP.
Nakamura, Y., Kaneko, T., Sato, S., Ikeuchi, M., Katoh, H., Sasamoto, S., et al. (2002). Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9, 123–130. doi: 10.1093/dnares/9.4.123
Noreña-Caro, D., and Benton, M. G. (2018). Cyanobacteria as photoautotrophic biofactories of high-value chemicals. J. CO2 Util. 28, 335–366. doi: 10.1016/j.jcou.2018.10.008
Olsen, M. T., Nowack, S., Wood, J. M., Becraft, E. D., LaButti, K., Lipzen, A., et al. (2015). The molecular dimension of microbial species: 3. Comparative genomics of Synechococcus strains with different light responses and in situ diel transcription patterns of associated putative ecotypes in the Mushroom Spring microbial mat. Front. Microbiol. 6:604. doi: 10.3389/fmicb.2015.00604
Omata, T. (1995). Structure, function and regulation of the nitrate transport system of the cyanobacterium Synechococcus sp. PCC7942. Plant Cell Physiol. 36, 207–213. doi: 10.1093/oxfordjournals.pcp.a078751
Omata, T., Price, G. D., Badger, M. R., Okamura, M., Gohta, S., and Ogawa, T. (1999). Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc. Natl. Acad. Sci. U.S.A. 96, 13571–13576. doi: 10.1073/pnas.96.23.13571
Omata, T., Takahashi, Y., Yamaguchi, O., and Nishimura, T. (2002). Structure, function and regulation of the cyanobacterial high-affinity bicarbonate transporter, BCT1. Funct. Plant Biol. 29, 151–159. doi: 10.1071/pp01215
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genom. Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Patel, A., Matsakas, L., Rova, U., and Christakopoulos, P. (2019). A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour. Technol. 278, 424–434. doi: 10.1016/j.biortech.2019.01.063
Peña, K. L., Castel, S. E., de Araujo, C., Espie, G. S., and Kimber, M. S. (2010). Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. U.S.A. 107, 2455–2460. doi: 10.1073/pnas.0910866107
Price, G. D. (2011). Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 109, 47–57. doi: 10.1007/s11120-010-9608-y
Price, G. D., Badger, M. R., Woodger, F. J., and Long, B. M. (2007). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59, 1441–1461. doi: 10.1093/jxb/erm112
Price, G. D., Woodger, F. J., Badger, M. R., Howitt, S. M., and Tucker, L. (2004). Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 101, 18228–18233. doi: 10.1073/pnas.0405211101
Pronina, N. A., Kupriyanova, E. V., and Igamberdiev, A. U. (2017). “Photosynthetic carbon metabolism and CO2-concentrating mechanism of cyanobacteria,” in Modern Topics in the Phototrophic Prokaryotes: Metabolism, Bioenergetics, and Omics, ed. P. C. Hallenbeck (Cham: Springer International Publishing), 271–303. doi: 10.1007/978-3-319-51365-2_8
Qin, Q., Xie, B., Zhang, X., Chen, X., Zhou, B., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/jb.01688-14
Rae, B., Long, B. M., Badger, M. R., Dean, P. G., and Lei, B. (2012). Structural determinants of the outer shell of β-Carboxysomes in Synechococcus elongatus PCC 7942: Roles for ccmK2, K3-K4, ccmO, and ccmL. PLoS One 7:e43871. doi: 10.1371/journal.pone.0043871
Rae, B. D., Long, B. M., Badger, M. R., and Price, G. D. (2013). Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379. doi: 10.1128/MMBR.00061-12
Raven, J. A., John, B., and Patricia, S. B. (2017). The possible evolution and future of CO2-concentrating mechanisms. J. Exp. Bot. 68, 3701–3716. doi: 10.1093/jxb/erx110
Sandrini, G., Matthijs, H. C. P., Verspagen, J. M. H., Muyzer, G., and Huisman, J. (2014). Genetic diversity of inorganic carbon uptake systems causes variation in CO2 response of the cyanobacterium Microcystis. ISME J. 8, 589–600. doi: 10.1038/ismej.2013.179
Shih, P. M., Dongying, W., Latifi, A., Axen, S. D., Fewer, D. P., Talla, E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058. doi: 10.1073/pnas.1217107110
So, A. K.-C., and Espie, G. S. (2005). Cyanobacterial carbonic anhydrases. Can. J. Bot. 83, 721–734. doi: 10.1139/b05-057
Solomon, T. (2001). The Definition and Unit of Ionic Strength. J.Chem. Educ. 78, 1691. doi: 10.1021/ed078p1691
Soltes-Rak, E., Mulligan, M. E., and Coleman, J. R. (1997). Identification and characterization of a gene encoding a vertebrate-type carbonic anhydrase in cyanobacteria. J. Bacteriol. 179, 769–774. doi: 10.1128/jb.179.3.769-774.1997
Sommer, M., Cai, F., Melnicki, M., and Kerfeld, C. A. (2017). β-Carboxysome bioinformatics: identification and evolution of new bacterial microcompartment protein gene classes and core locus constraints. J. Exp. Bot. 68, 3841–3855. doi: 10.1093/jxb/erx115
Sommer, M., Sutter, M., Gupta, S., Kirst, H., Turmo, A., Lechno-Yossef, S., et al. (2019). Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of Beta carboxysome shells. Plant Physiol. 179, 156–167. doi: 10.1104/pp.18.01190
Sun, N., Han, X., Xu, M., Kaplan, A., Espie, G. S., and Mi, H. (2019). A thylakoid-located carbonic anhydrase regulates CO2 uptake in the cyanobacterium Synechocystis sp. PCC 6803. New Phytol. 222, 206–217. doi: 10.1111/nph.15575
Tang, J., Du, L., Li, M., Yao, D., Waleron, M., Waleron, K. F., et al. (2022a). Characterization of a novel hot-spring cyanobacterium Leptodesmis sichuanensis sp. nov. and genomic insights of molecular adaptations into its habitat. Front. Microbiol. 12:739625. doi: 10.3389/fmicb.2021.739625
Tang, J., Shah, M. R., Yao, D., Du, L., Zhao, K., Li, L., et al. (2022b). Polyphasic identification and genomic insights of Leptothermofonsia sichuanensis gen. sp. nov., a novel thermophilic cyanobacteria within Leptolyngbyaceae. Front. Microbiol. 13:765105. doi: 10.3389/fmicb.2022.765105
Tang, J., Du, L., Liang, Y., and Daroch, M. (2019). Complete genome sequence and comparative analysis of Synechococcus sp. CS-601 (SynAce01), a cold-adapted cyanobacterium from an oligotrophic Antarctic habitat. Int. J. Mol. Sci. 20:152. doi: 10.3390/ijms20010152
Tang, J., Jiang, D., Luo, Y., Liang, Y., Li, L., Shah, M., et al. (2018a). Potential new genera of cyanobacterial strains isolated from thermal springs of western Sichuan, China. Algal Res. 31, 14–20. doi: 10.1016/j.algal.2018.01.008
Tang, J., Liang, Y., Jiang, D., Li, L., Luo, Y., Shah, M. M. R., et al. (2018b). Temperature-controlled thermophilic bacterial communities in hot springs of western Sichuan, China. BMC Microbiol. 18:134. doi: 10.1186/s12866-018-1271-z
Tang, J., Li, L., Li, M., Du, L., Shah, M. R., Waleron, M., et al. (2021). Description, taxonomy, and comparative genomics of a novel species, Thermoleptolyngbya sichuanensis sp. nov., isolated from Hot Springs of Ganzi, Sichuan, China. Front. Microbiol. 12:696102. doi: 10.3389/fmicb.2021.696102
Tremaine, P., Zhang, K., Bénézeth, P., and Xiao, C. (2004). “Ionization equilibria of acids and bases under hydrothermal conditions,” in Aqueous Systems at Elevated Temperatures and Pressures, eds D. A. Palmer, R. Fernández-Prini, and A. H. Harvey (London: Academic Press), 441–492. doi: 10.1016/b978-012544461-3/50014-4
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Walter, J. M., Coutinho, F. H., Dutilh, B. E., Swings, J., Thompson, F. L., and Thompson, C. C. (2017). Ecogenomics and taxonomy of cyanobacteria phylum. Front. Microbiol. 8:2132. doi: 10.3389/fmicb.2017.02132
Woosley, R. J. (2021). Evaluation of the temperature dependence of dissociation constants for the marine carbon system using pH and certified reference materials. Mar. Chem. 229:103914. doi: 10.1016/j.marchem.2020.103914
Keywords: thermophilic cyanobacteria, inorganic carbon uptake, CO2-concentrating mechanisms (CCMs), carboxysomes, photosynthesis, Rubisco
Citation: Tang J, Zhou H, Yao D, Riaz S, You D, Klepacz-Smółka A and Daroch M (2022) Comparative Genomic Analysis Revealed Distinct Molecular Components and Organization of CO2-Concentrating Mechanism in Thermophilic Cyanobacteria. Front. Microbiol. 13:876272. doi: 10.3389/fmicb.2022.876272
Received: 15 February 2022; Accepted: 04 April 2022;
Published: 06 May 2022.
Edited by:
Qiang Wang, Henan University, ChinaReviewed by:
John Albert Raven, University of Dundee, United KingdomCopyright © 2022 Tang, Zhou, Yao, Riaz, You, Klepacz-Smółka and Daroch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurycy Daroch, bS5kYXJvY2hAcGt1c3ouZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.