
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 June 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.871855
Escherichia coli is a problematic pathogen that causes life-threatening diseases, being a frequent causative agent of several nosocomial infections such as urinary tract and bloodstream infections. Proper and rapid bacterial identification is critical for allowing prompt and targeted antimicrobial therapy. (Bacterio)phage receptor-binding proteins (RBPs) display high specificity for bacterial surface epitopes and, therefore, are particularly attractive as biorecognition elements, potentially conferring high sensitivity and specificity in bacterial detection. In this study, we elucidated, for the first time, the potential of a recombinant RBP (Gp17) to recognize E. coli at different viability states, such as viable but not culturable cells, which are not detected by conventional techniques. Moreover, by using a diagnostic method in which we combined magnetic and spectrofluorimetric approaches, we demonstrated the ability of Gp17 to specifically detect E. coli in various human specimens (e.g., whole blood, feces, urine, and saliva) in about 1.5 h, without requiring complex sample processing.
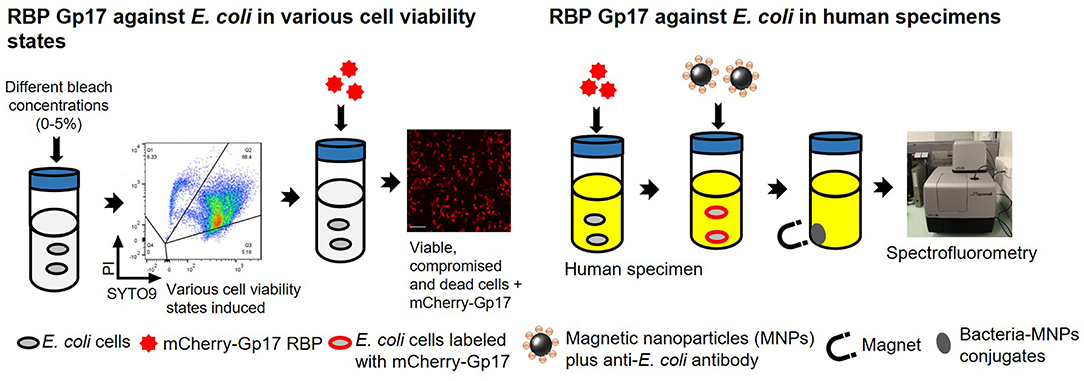
Graphical Abstract. Overview of the experimental studies. The RBP mCherry-Gp17 was tested against E. coli at different viability states and used as a recognition molecule for the detection of E. coli in human specimens.
Bacterial infections are among the primary causes of death worldwide, and bacterial agents are mainly responsible for healthcare-associated infections (HCAI) that are acquired in hospital environments or other healthcare facilities (World Health Organization, 2011; Haque et al., 2018). These infections arise more than 48 h after hospital admission or within 30 days after receiving medical treatment and have been a worrisome issue (World Health Organization, 2011; Cassini et al., 2016). Escherichia coli is an important pathogen responsible for several life-threatening diseases such as pneumonia, meningitis, sepsis, enteric/diarrheal diseases, and urinary tract infections (UTIs) (Kaper et al., 2004). It is the most frequent Gram-negative bacterium isolated from bloodstream infections (BSIs) and the principal causative agent of UTIs (ECDC, 2019).
For effectively controlling these diseases and avoiding the subsequent serious complications, the early diagnosis is of great importance, and it also contributes to the prompt targeted antimicrobial therapy, helps decrease the medical and financial burden, and prevents the spread of antimicrobial resistance (Bereket et al., 2012). The gold standard methods for detecting bacteria are mainly based on culture and colony counting and biochemical tests for the identification of microorganisms (Laupland and Valiquette, 2013; Abayasekara et al., 2017). However, these methodologies are laborious, can be affected by non-microbial material, and require lengthy enrichment steps as a result of the minimal concentration of pathogens in samples and their fastidious growth (Váradi et al., 2017; Rajapaksha et al., 2019). Moreover, bacteria are reported to enter a state, known as viable but non-culturable (VBNC), where they remain metabolically active, keeping their virulence, but cannot grow in standard solid culture media. This compromised state has been described for several pathogenic bacteria and arises due to the influence of different physiological stress conditions (Oliver, 2005, 2010). The appearance of VBNC bacteria may represent a serious threat to public health (Oliver, 2005; Castellani et al., 2013; Pienaar et al., 2016; Zhao et al., 2017) since these cells are present in samples from clinical environments, water distribution systems, and the food industry, but they cannot be detected by conventional culturing detection methods. Moreover, since antibiotics can act as inducers for the VBNC state (Mason et al., 1995; Rivers and Steck, 2001; Ayrapetyan et al., 2015), these cells can remain present and resistant to these compounds, but when the treatment is stopped, they can resuscitate and regain their virulence and thus lead to chronic recurring infections in patients who were considered cured (Rivers and Steck, 2001; Pasquaroli et al., 2013; Ayrapetyan et al., 2018). Additionally, Wilks et al. (2021) reported the appearance of VBNC E. coli in urinary catheters resulting from the biofilm development.
In an effort to overcome the disadvantages of the gold standard methods, other technologies like the enzyme-linked immunosorbent assay (ELISA) (Verma et al., 2013), nucleic acid hybridization (McLoughlin, 2011; Frickmann et al., 2017), matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) (Singhal et al., 2015), and polymerase chain reaction (PCR) (Mothershed and Whitney, 2006) have emerged. Nonetheless, these methods tend to be expensive, involve several steps, suffer from amplification inhibition or infidelity issues, and require prior knowledge of the target DNA sequence or mass spectrometry profile (Ferreira et al., 2011; Marder et al., 2017; Rajapaksha et al., 2019). Another important concern is the fact that these techniques exhibit some issues when applied to complex matrices (Stevens and Jaykus, 2004). Human biological samples, in particular, fecal specimens or whole blood, are among the most complex samples to analyze and detect bacteria due to the presence of numerous inhibitory compounds that interfere in the analysis, especially in the case of amplification- or immunology-based methods (Holland et al., 2000; Stevens and Jaykus, 2004; Opota et al., 2015), making these approaches not attractive for a real monitoring application.
Magnetic trapping (MT) has been widely used for several years as a sample preparation method due to its attractive features (Cudjoe et al., 1995; Kretzer et al., 2007; Qiu et al., 2009; Yang et al., 2013; Lopes et al., 2016). The basic principle is the use of magnetic nanoparticles (MNPs) functionalized with affinity molecules for the target bacterial cells, such as antibodies, to provide a specific capture and isolation of intact cells directly from a complex sample without the need for centrifugation or chromatography methodologies, which are laborious or require specific equipment (Cudjoe et al., 1995; Stevens and Jaykus, 2004). MT presents advantages over other methods, including the effective separation of target bacteria from competitive microflora, the removal of matrix components and potential inhibitors, and the reduction of the sample volume (Stevens and Jaykus, 2004).
The choice of the biorecognition molecule in a diagnostic assay is of great importance to the specificity and robustness of the method (Stevens and Jaykus, 2004; Singh et al., 2012). Several recognition elements for bacteria have been described, such as antibodies, enzymes, aptamers, proteins, or (bacterio)phages (Singh et al., 2013). Receptor-binding proteins (RBPs) from phages, which are responsible for the binding of the phage to the bacterial surface receptors in the initial step of the phage infection (Singh et al., 2010; Schmelcher and Loessner, 2014), have been appointed as great candidates to replace the traditional recognition elements. RBPs are involved in phage adsorption by specifically binding to receptors on the bacterial surfaces, such as proteins, polysaccharides, lipopolysaccharides (LPS), and carbohydrate moieties, dictating the phage infection spectrum (Leiman et al., 2003; Rakhuba et al., 2010). While common receptors for phages infecting Gram-negative bacteria are LPS, capsular polysaccharides (CPS), or bacterial surface proteins (porins and transport proteins), the most frequent in Gram-positive bacteria are the peptidoglycan, teichoic acids, or exposed polysaccharides (Rakhuba et al., 2010; Latka et al., 2017; Dowah and Clokie, 2018). RBPs offer several benefits over other recognition elements like antibodies, such as comparable or higher specificity, high sensitivity, ease of genetic modification, enabling its fusion with fluorescent reporter tags, low production costs, small size, and high chemical and physical resistance (Barbirz et al., 2009; Singh et al., 2010, 2012; Simpson et al., 2015). Therefore, in the past few years, several successful applications of the RBPs for bacterial detection have been reported in the literature, particularly when combined with biosensors (Tay et al., 2012; Shin and Lim, 2018), spectrophotometry (Kunstmann et al., 2018; Santos et al., 2020), ELISA (Denyes et al., 2017; Górska et al., 2018), or lab-on-chip systems (Cunha et al., 2021).
In this study, we unveiled the potential of a recombinant RBP (Gp17) derived from the T7-like polyvalent phage 285p (Xu et al., 2014) as a recognition probe for E. coli cells at different viability states (viable, compromised, dead) by using flow cytometry and fluorescence microscopy. Some studies reported the detection of all these cell states by whole phages but are contradictory (Krueger, 1931; Watanabe, 1976; Oda et al., 2004; Awais et al., 2006; Hu et al., 2012; Tlili et al., 2013; Fernandes et al., 2014). Therefore, it was important to assess the binding capacity of a recombinant RBP to these cells, which, to the best of our knowledge, has never been completely elucidated. In this study, we also leveraged the capacity of Gp17 to specifically recognize and detect E. coli in complex human biological samples. For this, we combined the benefits of the MT and spectrofluorometry techniques, enabling the separation and detection of E. coli in 1.5 h directly from saliva, urine, feces, and whole blood, without requiring complex sample processing.
The clinical strains used to test the binding spectrum (Supplementary Table S1) including the E. coli HB104 (used as the target bacteria) and Staphylococcus aureus HB22 (used as the negative control) were provided by the Hospital of Braga (Portugal). E. coli CECT 515 was acquired from the Spanish Type Culture Collection, University of Valencia. The strains were routinely grown overnight in Tryptic Soy Broth (TSB) (Liofilchem) and Luria Bertani (LB) (Liofilchem) at 37°C under agitation (120 rpm) or in solid plates, obtained by adding 12 g/L of agar (Liofilchem).
The gp17 was identified as a gene encoding for a tail fiber protein (Gp17) derived from the 285p T7-like polyvalent bacteriophage belonging to the PYO97_8 phage cocktail (Georgian Eliava Institute of Bacteriophage, Microbiology and Virology) (Xu et al., 2014; Villarroel et al., 2017). The protein sequence was analyzed using the Basic Local Alignment Search Tool Protein (BLASTp) non-redundant protein sequence database and expected functional domains were found through Pfam (Finn et al., 2014) and InterProScan (Jones et al., 2014) using the default parameters of the programs. To determine the molecular weight and isoelectric point, the Compute pI/Mw program ExPASy (Artimo et al., 2012) was employed.
The gene gp17 encoding a potential RBP was synthesized, fused with the mCherry gene derived from Discosoma sp. at the N-terminus, and cloned into the expression vector pHTP1 (NZYTech) containing a poly-histidine (6xHis) sequence tag at N-terminus. The vector containing the red fluorescent protein mCherry-Gp17 was chemically transformed into E. coli BL21 (DE3) cells (Invitrogen). Expression of the fusion protein was carried out as previously described (Costa et al., 2020). In brief, E. coli BL21 cells carrying the recombinant plasmid were cultivated at 37°C in an LB medium, adding 50 μg/μl of kanamycin until an optical density (OD) at 600 nm (OD600 nm) of 0.6 was reached. The induction of protein overexpression was achieved by adding 1 mM isopropyl-β-D-thiogalactopyranoside (Sigma-Aldrich) and subsequent overnight incubation at 16°C, 120 rpm. Afterward, to lyse the cells, centrifugation was performed (9,000 × g, 10 min, 4°C), phosphate lysis buffer (20 mM sodium dihydrogen phosphate, 500 mM sodium chloride, pH 7.4) was used for cell resuspension, and three freeze-thaw cycles and sonication at 12% amplitude (Disintegrator Ultrasonic Mod. 450, Branson) for 15 min (10 s ON and 10 s OFF) were conducted. Then centrifugation (9,000 × g, 15 min, 4°C) was carried out to collect the supernatant enriched with soluble proteins, and the protein was purified using a nickel-nitrilotriacetic acid (Ni-NTA) column (Thermo Fisher Scientific). After the washing steps, the elution of proteins was completed with 300 mM imidazole, and fractions were analyzed with SDS-PAGE (12% (w/v) acrylamide) after Blue Safe staining (NZYTech). The protein dialysis and concentration were performed with 0.1 M phosphate buffer pH 7.2 (PB) by using centrifugal filters (Amicon Ultra, 0.5 ml MWCO 10 KDa, Merck Millipore) and proteins were stored at 4°C. The BCA Protein Assay Kit (Thermo Fisher Scientific) assisted in the protein concentration assessment.
To assess the ability of the mCherry-Gp17 to bind E. coli cells, fluorescence microscopy assays were conducted against E. coli HB104, Klebsiella pneumoniae HB11, and S. aureus HB22 as the negative controls, following the procedure previously described with minor modifications (Costa et al., 2020). In brief, overnight bacterial cells grown in TSB at 37°C were centrifuged (6,000 × g, 10 min) and the OD600 nm adjusted to 0.5. Then 400 μl of cultures were centrifuged at 9,000 × g for 5 min and resuspended in 40 μl of PB. Then, 20 μl of mCherry-Gp17 at a final concentration of 20 μM was added and incubated at room temperature (RT) for 30 min. Cells were centrifuged at equal conditions, and two washes were done with PB to remove unbound protein and resuspended in 10 μl of PB. Samples were observed at the confocal microscope LSM780 (Zeiss) equipped with a 5 mW 488–645 nm light source in brightfield or under a laser source at 561 nm (DPSS 561-10) for mCherry excitation and setting a bandpass filter (604–735 nm) in the Zeiss ZEN 2010 software for visualization of the protein emission. Control samples of bacterial cells without the addition of the recombinant protein were prepared simultaneously. mCherry alone was used as a negative control.
Suspensions of clinical strains, which are listed in Supplementary Table S1, were prepared as described in the previous section. The spectrofluorimetric analysis was accomplished according to the protocol described elsewhere (Santos et al., 2020). Suspensions of bacteria (400 μl) were centrifuged at 9,000 × g for 5 min and resuspended in 40 μl of PB. Then, 20 μl of mCherry-Gp17 at a final concentration of 5 μM was added and incubated at RT for 30 min. Cells were washed twice by centrifugation (9,000 × g for 5 min) using PB to eliminate protein debris and resuspended in 100 μl of PB. The samples were transferred to a black 96-well microplate and examined at a BioTek™ Synergy H1 Hybrid Multi-Mode Microplate Reader with the BioTek Gen5 software. Excitation/emission wavelengths were defined as 570/610 nm (gain 100), and the fluorescence intensity was displayed in arbitrary units (a.u.).
Bacterial suspensions of E. coli HB104 (used as the target) and of the negative control bacteria were prepared as described in the functional analysis section. Bacterial cells were subjected to different concentrations of sodium hypochlorite (commercial bleach with a stock concentration of 5%) to induce the cells to enter a compromised state (Liu et al., 2009; Fernandes et al., 2014; Ye et al., 2020). Dead cells were prepared by incubation with 5% of bleach (Aranke et al., 2021).
Bacterial suspensions (1 ml) were centrifuged at 7,000 × g for 10 min and resuspended in 1 ml of each of the several concentrations of commercial bleach used, which were prepared in PB: 0.000, 0.006, 0.007, 0.020, 0.030, 0.050, and 5.000% (v/v). Samples were incubated for 10 min, then centrifuged at 7,000 × g for 10 min at 4°C and washed twice with PB. The culturable cells were quantified after each treatment through colony forming units (CFU) counting.
The viability of the cells was estimated after subjecting bacteria to the aforementioned bleach concentrations and staining the cells with the LIVE/DEAD BacLight Bacterial Viability and Counting Kit (Molecular Probes) according to the manufacturer's instructions (Berney et al., 2007; Singh et al., 2012). Later, cells were examined either by confocal microscopy (Zeiss LSM780) or by flow cytometry (Bio-Rad S3e Cell Sorter). In the flow cytometry analysis, the gating strategy was defined according to the LIVE/DEAD BacLight kit (Berney et al., 2007). Microspheres of 6 μm diameter were used as the standard for absolute cell quantification. The results obtained in flow cytometry were analyzed using the FlowJo software (Tree Star, Ashland, OR).
The labeling of cells in different viability states with mCherry-Gp17 was carried out as described above for the spectrofluorimetric analysis. The samples were analyzed by flow cytometry using a Bio-Rad S3e Cell Sorter equipped with a dual laser (488/561 nm) and a 615/25 nm filter, allowing the mCherry fluorescence to be detected on the FL3 channel. According to our previous work (Costa et al., 2020), a total of 45,000 events were acquired with a sample flow rate of 10 μl/min. Data analysis was performed using FLOWJO-Single Cell Analysis Software v10 (BD, New Jersey, USA).
The spectrofluorimetric magnetic sandwich assay was set up starting with the incubation of cells with the mCherry-Gp17 for 30 min and then magnetically labeled with antibody-functionalized MNPs.
To prepare the MNPs, 50 μl of commercial 250 nm streptavidin-coated MNPs (Nanomag-D, Micromod, 4.9 × 1011 particles/ml) were rinsed twice with 500 μl of 0.1 M PB Tween 20 (0.05%, v/v) by removing the supernatant with a magnetic concentrator (Dynal-Biotech). Then, 500 μl of the biotinylated E. coli polyclonal antibody [LSBio (LifeSpan) Cat# LS-C56164-1, RRID:AB_1509874] at a final concentration of 50 μg/ml were mixed with the MNPs and left on an orbital shaker at 500 rpm, 20°C for 2 h of incubation. Following functionalization, the same process was employed to withdraw the supernatant containing the unbound antibody. The MNPs blocking was carried out by adding 500 μl of bovine serum albumin (BSA) at 5% (w/v) in PB and incubating for 1 h under equal conditions. After supernatant removal, the antibody-conjugated MNPs were washed with PB Tween, resuspended in 50 μl, and kept at 4°C as stock.
The magnetic sandwich assay was first performed in buffer and in defibrinated horse blood (Probiológica), with cultures of E. coli HB104 (used as the target) and S. aureus HB22 (used as the negative control) that were prepared as aforementioned. Control samples without any bacteria added were prepared at the same time and submitted to the same process. The bacterial suspensions (400 μl) in PB or horse blood were centrifuged at 9,000 × g for 5 min and resuspended in 40 μl of PB. Then, 20 μl of mCherry-Gp17 at a final concentration of 5 μM was added and incubated at RT for 30 min. Then, 4 μl of the antibody-conjugated MNPs stock was added to the samples, completing the volume to 400 μl with PB. The MNPs and bacteria were incubated for 1 h in an orbital shaker at 500 rpm, 20°C. After, the supernatants were gently removed using the magnetic concentrator, and the MNPs were washed twice with 400 μl of PB Tween. Both supernatants and washes were stored for CFUs assessment. Finally, the MNPs were resuspended in 100 μl of PB and immediately analyzed on the spectrofluorometer according to the settings described above. The bacterial capture efficiency was calculated using the following equation:
Here, CFUs I represent the CFUs initially added, and the CFUs S and CFUs W represent the CFUs from supernatants and washes, respectively.
All human biological samples used in this study were collected from healthy adult volunteers upon written informed consent. The same type of biological samples were mixed and thus processed de-identified in this study, being the data and samples fully anonymized. Samples of saliva, feces, and urine were collected using a plastic tube. Whole blood was collected using EDTA blood collection tubes (BD Vacutainer). After collection, fecal samples were filtered. Bacterial suspensions (E. coli HB104 as target and S. aureus HB22 as negative control) were prepared as described before, and 1 ml of each suspension was centrifuged (9,000 × g, 5 min) and resuspended in 1 ml of each human specimen. Then, the magnetic sandwich assay was performed as described in the previous section. Bacteria in buffer and control samples without any bacteria added were prepared at the same time and submitted to the same process.
All data are represented as mean ± SD (standard deviation). For Figures 4, 5, multiple comparisons of means were performed using two-way ANOVA followed by Sidak's multiple comparison test (p-value < 0.0001) and Dunnett's multiple comparison test (p-value < 0.0001), respectively.
Phage RBPs are regularly related to tail fiber proteins, tail spikes, or spike proteins (Simpson et al., 2016). The gene gp17 was identified as encoding a tail fiber protein in the E. coli phage 285p from a pyophage (PYO) cocktail (Xu et al., 2014; Villarroel et al., 2017). To further confirm, the sequence of the encoded protein (Gp17) was compared in terms of homology with other phage tail proteins deposited at the National Center for Biotechnology Information (NCBI) database. The results indicate that Gp17 has homology with tail fiber proteins from other E. coli phages such as phage BA14 (72% homology), phage PhiV-1 (71%), or phage P483 (60%). Also, the Gp17 has homology with tails from phages infecting Enterobacter (Enterobacter phage PZJ0206 tail, 95%), Salmonella (Salmonella phage BSP161 tail, 88%), Yersinia (Yersinia phage PYPS50 tail, 87%), and Erwinia (Erwinia phage FE44 tail, 73%). The search for domains and families found hits with the phage_T7_tail_fiber family at N-terminus.
After the identification of the Gp17 as a potential RBP, the protein was synthesized, expressed in E. coli, and purified using Ni-NTA columns (Supplementary Figure S1). The functional analysis of this protein was performed to confirm its recognition binding ability to the target bacteria and other bacterial species. This was assessed by fluorescence microscopy through the observation of cells emitting red fluorescence since Gp17 was fused with the reporter protein, mCherry (mCherry-Gp17). As shown in Figure 1, this protein can effectively bind to E. coli HB104 cells (Figure 1A) and not to the non-target bacteria K. pneumoniae HB11 (Figure 1B) or S. aureus HB22 (Figure 1C).
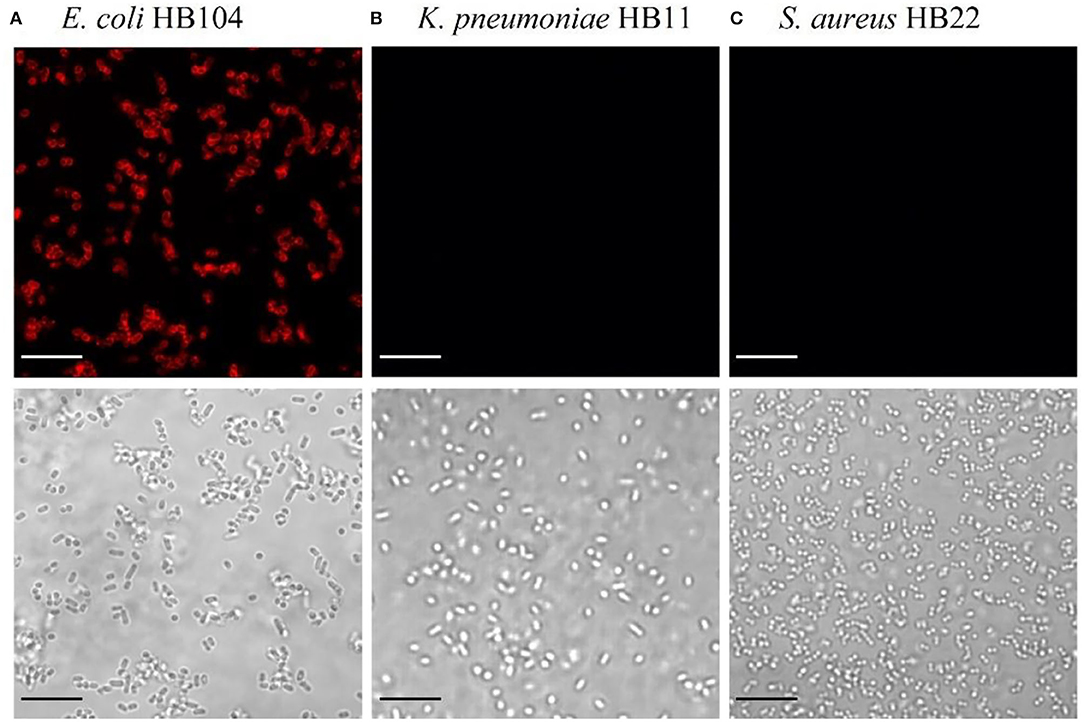
Figure 1. Functional analysis of the RBP mCherry-Gp17. Fluorescence microscopy images after incubation of mCherry-Gp17 with bacterial cells of the target E. coli HB104 (A) and the non-target K. pneumoniae HB11 (B) and S. aureus HB22 (C). The scale bar represents 10 μm.
In order to evaluate the binding spectrum of mCherry-Gp17 against a panel of strains of E. coli and from other genera, spectrofluorimetric analysis was carried out after incubation of cells with mCherry-Gp17. The results are represented in Supplementary Table S1 and indicate that mCherry-Gp17 was able to bind to 59% of the E. coli strains tested without showing unspecific binding to strains belonging to other genera (such as S. aureus, Enterobacter aerogenes, Pseudomonas aeruginosa, or K. pneumoniae). Most of these strains presented a fluorescent signal ratio of 0.4–0.6, thus the clinical isolate E. coli HB104 was selected as the representative strain for further assays.
To assess if the protein mCherry-Gp17 could recognize different cell viability states, an assay was first developed and optimized to induce the cells to enter these states (e.g., live, compromised, dead) by using different concentrations of sodium hypochlorite (commercial bleach) (see Supplementary Figure S2). The cell viability was determined through flow cytometry and fluorescence microscopy after incubation of cells with the viability dyes SYTO9 and propidium iodide (PI), together with the CFU validation. These assays permitted to conclude that the treatment with 0.03% and 5% of bleach was the best to induce most of the cells into the compromised and dead states, respectively (Supplementary Figure S2).
Figures 2A–C depicts the representative dot plots showing the percentage of cells in each quadrant, defined according to the gating strategy (Supplementary Material) and for the different treatments that were submitted: 0% of bleach (Figure 2A), 0.03% (Figure 2B), and 5% (Figure 2C). These cells were observed by fluorescence microscopy, and the results corroborate the flow cytometry assays. The cells that were not subjected to any treatment appeared green (Figure 2D), and most of the cells (≈92%, Figure 2A) were considered viable. Also, after bleach treatment, some cells appeared double-labeled (≈90%, Figure 2B), showing intermediate colors from yellow to orange by microscopy (Figure 2E), indicating the presence of compromised cells which on CFU counting revealed to be unable to grow on standard solid media (Supplementary Figure S2), implying that these cells are in the VBNC state (Stiefel et al., 2015; Truchado et al., 2020). Moreover, cells submitted to 5% of bleach appeared as dead bacteria cells by flow cytometry (≈99%, Figure 2C), emitting red fluorescence (Figure 2F).
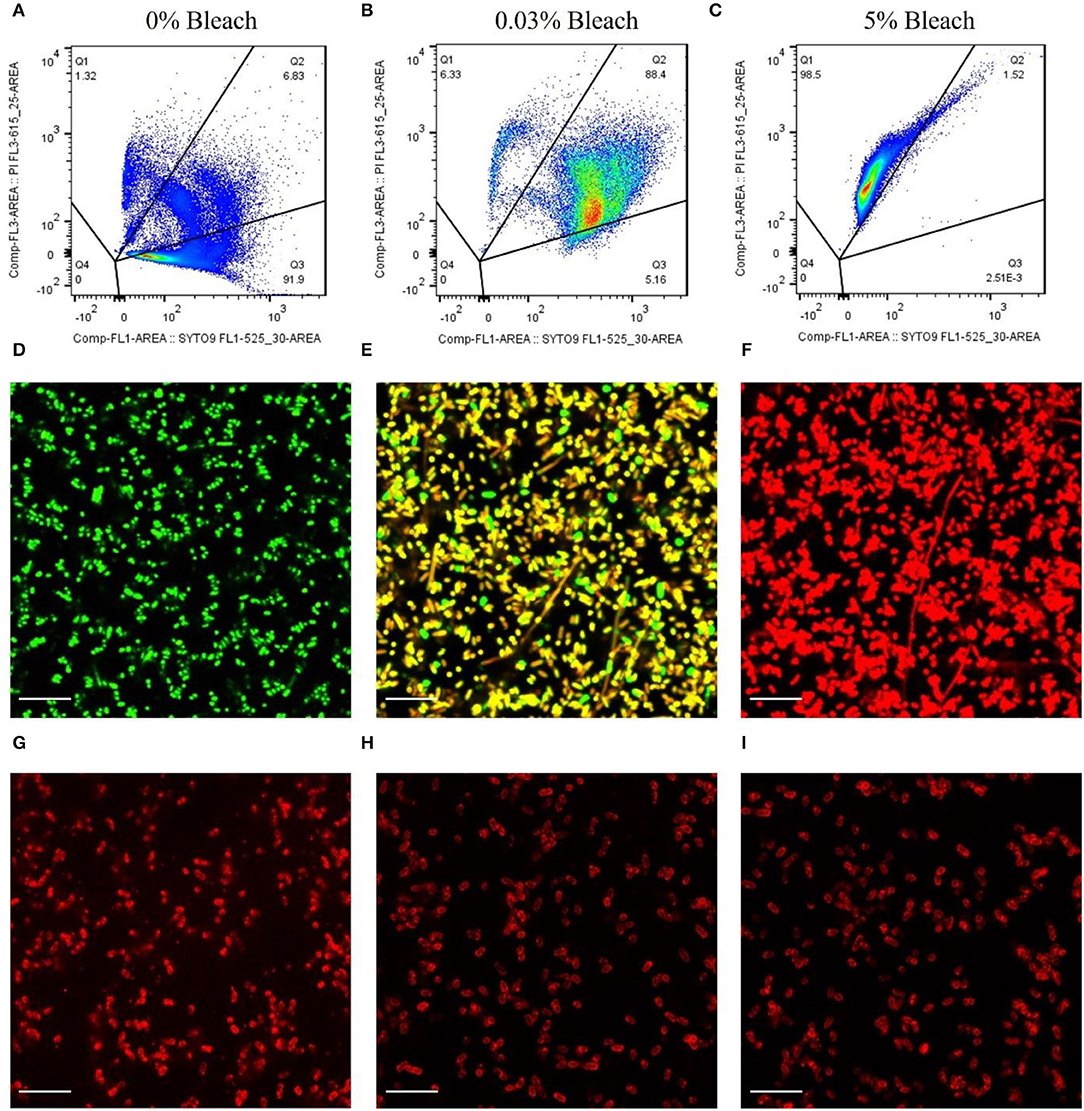
Figure 2. Assessment of the binding ability of mCherry-Gp17 to E. coli cells in different viability states. (A–C) Representative flow cytometry plots showing the percentage of the dead (Q1), compromised (Q2), and viable (Q3) cells present after treatment with bleach at different concentrations. Cells were labeled with SYTO 9 (green) and PI (red). (D–F) Fluorescence microscopy images of E. coli cells treated with 0% bleach (viable), 0.03% bleach (compromised), and 5% bleach (dead) stained with SYTO 9 (green) and PI (red). (G–I) Fluorescence microscopy images of cells treated with 0% bleach (viable), 0.03% bleach (compromised), and 5% bleach (dead) after incubation with mCherry-GP17.
After optimizing the procedure for converting the cells into compromised (such as VBNC) and dead cells, the ability of the RBP mCherry-Gp17 to recognize and bind to cells at different viability states was studied (Figures 2G–I, 3). The results revealed that the protein was able to recognize viable (Figures 2G, 3A), compromised (Figures 2H, 3B), and dead cells (Figures 2I, 3C). The percentage of positive events was similar between the different types of samples (≈90%) (Figures 3A–C), suggesting that the RBPs recognized the cells at different viability states in the same manner. In the case of the negative control, which corresponds to S. aureus HB22 cells labeled with mCherry-Gp17 (non-target bacteria, Figure 3D), just a neglected number of cells were recorded as positive (0.37%).
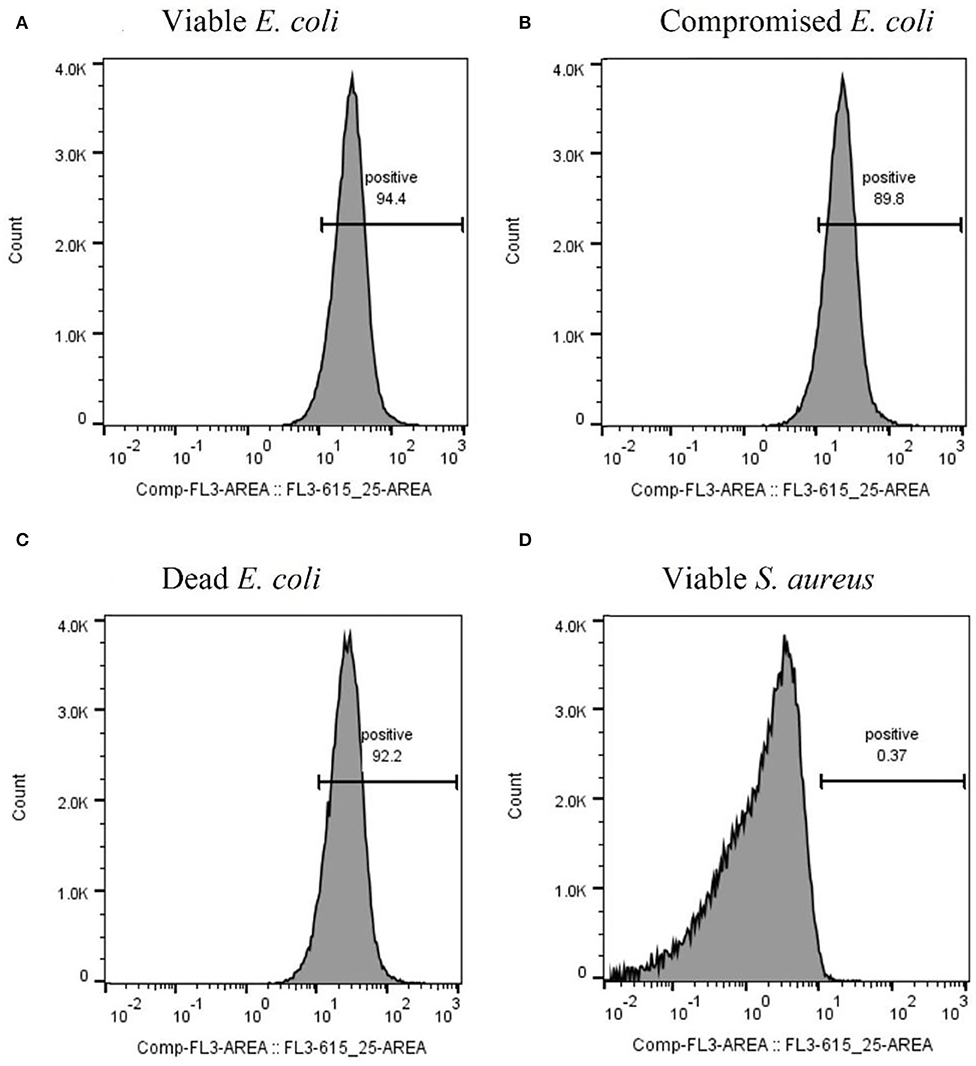
Figure 3. Assessment of the binding ability of mCherry-Gp17 to cells in different viability states by flow cytometry. Representative histograms were obtained from the analysis of viable (A), compromised (B), and dead (C) E. coli HB104 cells and viable S. aureus HB22 (D) (negative control) after incubation with mCherry-Gp17.
Since HCAIs can be presented in different sites of the body (Haque et al., 2018), it is important to assess the ability of the RBP to detect pathogens in different types of human biological specimens. After obtaining the promising results from the preliminary experiments performed in buffer and horse blood (Supplementary Figure S3), samples of urine, blood, feces, and saliva provided by healthy volunteers were spiked with E. coli HB104 cells or with S. aureus HB22 as the negative control.
The samples were first incubated with mCherry-Gp17 and then cells were magnetically separated using MNPs functionalized with an anti-E. coli antibody, and the resulting fluorescent signals were measured on the spectrofluorometer.
After magnetic enrichment, supernatants and washes were analyzed by CFU counting, and the bacterial capture efficiencies are displayed in Figure 4. The results demonstrated high bacterial capture efficiency (more than 87%) for E. coli HB104 present in different human biological samples, proving the efficacy of this methodology to recover bacterial cells from complex sample matrices. Moreover, although some unspecific capture occurred for the negative control with S. aureus, especially in whole blood (30% of capture efficiency), these results were significantly different (p-value < 0.0001) from the values obtained for E. coli.
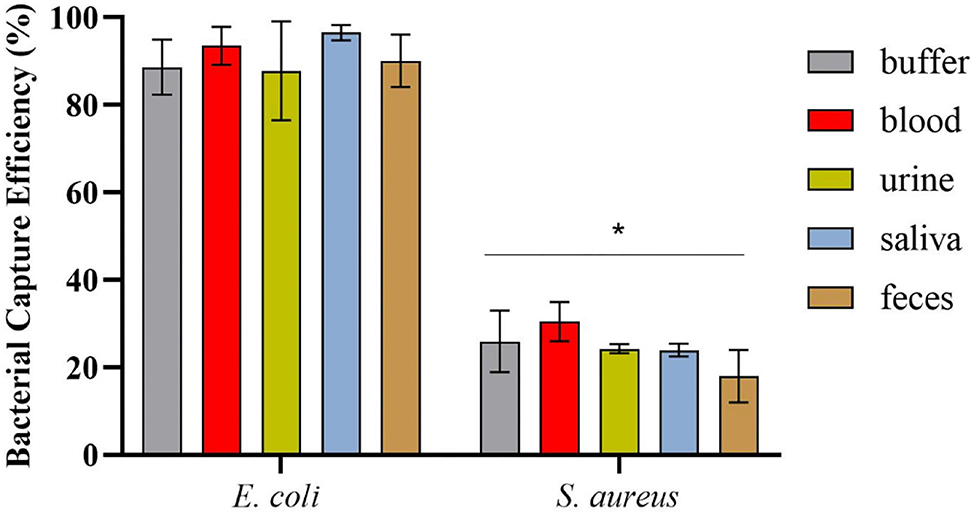
Figure 4. Bacterial capture efficiencies (in percentage) obtained in the magnetic sandwich assay performed in the different types of human specimens for E. coli HB104 (target bacteria) and S. aureus HB22 (non-target bacteria), assessed by CFU counting. Errors bars represent the standard deviation of the average of three independent experiments (n = 3). *Statistical analysis was performed comparing the percentage of bacterial capture obtained for the target bacteria E. coli and the negative control S. aureus in each of the human specimens. Multiple comparisons were done using the two-way ANOVA with Sidak's multiple comparison test (p-value < 0.0001).
The spectrofluorimetric results, which correspond to the fluorescence signals derived from the conjugated mCherry-Gp17-bacteria-MNPs that remained after the washing steps, are shown in Figure 5. It was possible to observe a statistically significant difference (p-value < 0.0001) between the fluorescence signals acquired for E. coli HB104 in the different human biological samples (fluorescent signals over 900 a.u.) and the negative controls, namely the non-target S. aureus HB22 and the samples without bacteria that were subjected to the same process (means of fluorescence less than 300 a.u.) (Figure 5). Also, in the fluorescence microscopy analysis of conjugated mCherry-Gp17-bacteria-MNPs obtained in an assay performed in the blood (Supplementary Figure S4), it was not possible to see any fluorescence signal for the negative control bacteria S. aureus HB22, which is visible in the case of E. coli HB104 cells.
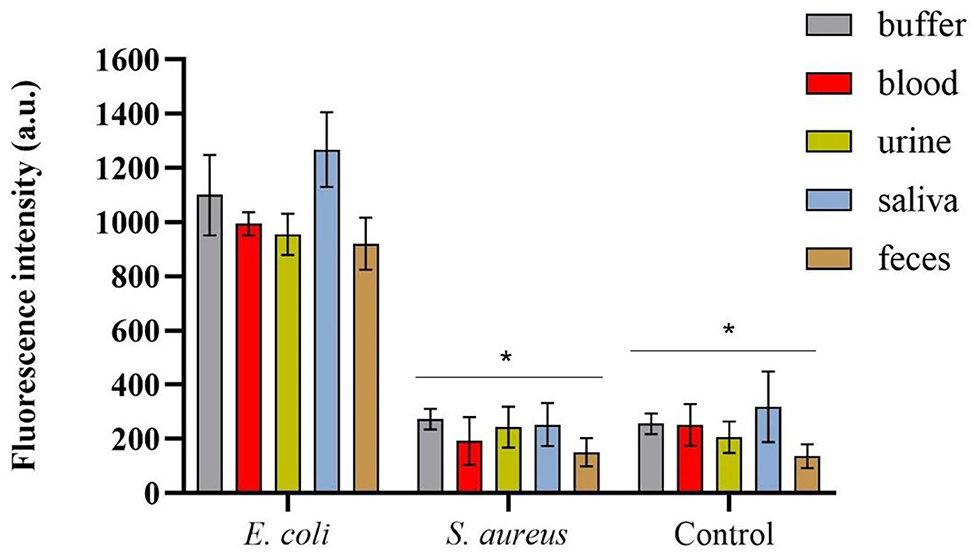
Figure 5. Spectrofluorimetric results of the magnetic sandwich assay performed in the different types of human specimens for E. coli HB104 (used as target bacteria), S. aureus HB22 (used as non-target bacteria), and for the control without bacteria added. Errors bars represent the standard deviation of the average of three measurements in the three independent assays (n = 3). *Statistical analysis was done comparing the signals in each of the human specimens obtained for S. aureus and control (without bacteria) with the signal of E. coli (target bacteria), defined as “positive” control, in the same samples. Multiple comparisons were done using the two-way ANOVA test (p-value < 0.0001) followed by Dunnett's multiple comparison test (p-value < 0.0001). a.u. stands for arbitrary units.
Considering that E. coli is among the most common microorganisms isolated from HCAIs, particularly being an important pathogen responsible for BSIs and UTIs (ECDC, 2019), we have assessed the potential of a phage RBP as a recognition probe for E. coli detection in different human biological samples. Accordingly, after identifying Gp17 as a potential RBP, this protein was synthesized and fused with mCherry as a reporter protein. The binding ability of this protein was tested first by fluorescence microscopy and then by spectrofluorometry against a larger panel of clinically isolated strains, derived from blood, urine, and skin exudates, among others. The results revealed that Gp17 successfully recognize 60% of E. coli strains and did not show affinity against other bacterial species that are also prevalent causative agents of HCAIs (Bassetti et al., 2017; Lee et al., 2018; ECDC, 2019).
Besides the affinity for viable cells, which has been extensively studied for RBPs (Fujinami et al., 2007; Javed et al., 2013; Denyes et al., 2017; He et al., 2018; Santos et al., 2020), we have tested the binding capacity of the recombinant Gp17 against other cell viability states, namely dead and compromised cells, which to the best of our knowledge have never been completely addressed. The results demonstrated that Gp17 can recognize cells in all these cell states, which may indicate that the receptors involved in the RBP's cell recognition and binding remained available independently of the cell viability. T-even phage adsorption is mediated by long-tail fibers, which target and bind to host physiological receptors such as LPS and outer membrane proteins (Drexler et al., 1989; Yoichi et al., 2005; González-García et al., 2015a,b; Lupo et al., 2015), commonly known as the receptors of RBPs (Rakhuba et al., 2010). According to some studies that have focused on the use of whole phages against VBNC and dead cells, this thematic seems to be controversial once some authors have shown that phages attach to dead cells (Krueger, 1931; Watanabe, 1976; Oda et al., 2004; Awais et al., 2006; Hu et al., 2012), in contrast with others (Tlili et al., 2013; Fernandes et al., 2014). The only consensus was the ability of the phages to adsorb to VBNC cells (Oda et al., 2004; Awais et al., 2006; Fernandes et al., 2014). Akusobi et al. (2018) reported that E. coli phage PP01 was able to effectively adsorb to dead cells, and this effect was enhanced through natural point mutations occurring on its long tail fiber (gp38). This protein was previously described as responsible for the reversible binding of the phage PP01 to the outer membrane protein C (OmpC) on E. coli O157:H7's cell surface (Yoichi et al., 2005).
The ability of Gp17 to detect viable and compromised cells can be considered an advantage for real-time monitoring of bacterial infections. Particularly, the recognition of compromised cells like VBNC is of extreme importance since these cells, which were not detected by the standard culture methods, can “resuscitate” when in favorable conditions and cause diseases (Du et al., 2007; Li et al., 2014). Nonetheless, the use of Gp17 as a recognition molecule should be thoroughly considered once a bacterial pre-enrichment step may be required to overcome the possibility of detection of dead cells, potentially causing false positives (Denyes et al., 2017).
The application of phage RBPs as recognition elements for bacterial detection in human specimens is limited, with only a few studies reporting their use in urine (He et al., 2018; Shi et al., 2021). Therefore, it was important to prove the applicability of Gp17 for the detection of E. coli in biological samples and thus evaluate whether these elements can bind bacterial cells even in the presence of many other irrelevant components, known as having interferent or inhibitory activity in other methodologies, such as in PCR-based and immunology techniques (Holland et al., 2000; Stevens and Jaykus, 2004; Opota et al., 2015). To achieve this goal, we used the Gp17 as a probe and combined it with the advantages of MT and spectrofluorometry techniques for bacterial capture and detection, respectively. The use of MT as a sample preparation approach assisted in the efficient isolation of the target bacterial cells (more than 87%) from the human specimens, allowing the cleaning of the interfering components in a fast and simple way, without the need for laborious centrifugation steps. MT has been shown as a promising sample preparation approach in diverse systems (Favrin et al., 2001; Schmelcher et al., 2010; Wang and Alocilja, 2015; Ngamsom et al., 2016; Chen et al., 2020), including some using RBPs to functionalize the MNPs (Kretzer et al., 2007; Denyes et al., 2017; Cunha et al., 2021). The results from the MT also revealed some capture for the non-target bacteria S. aureus. This cross-reactivity was expected since MNPs were functionalized with a polyclonal antibody, which is commonly associated with cross-reactivity issues (Frank, 2002). This broad capture was not a problem in the further spectrofluorimetric assays, because the Gp17 only recognized E. coli, guaranteeing the specificity of the detection assay. Indeed, the results obtained from these measurements revealed distinguishable signals derived from the detection of E. coli (around 1,000 a.u.) in several human biological specimens compared to the signals obtained in the controls (about 300 a.u.). This background signal can be the result of some residual binding occurring between the sample components and the Gp17, or due to remaining components that were not fully washed and that can have auto-fluorescence, such as the red blood cells (Azevedo et al., 2011; Skvarc et al., 2013). Nonetheless, the fluorescence microscopy analysis of the conjugated mCherry-Gp17-bacteria-MNPs revealed no fluorescence for the negative control bacteria S. aureus HB22, which is visible in the case of E. coli HB104 cells. This suggests that the residual signal obtained in the spectrofluorimetric measurements was not due to the non-specificity of the Gp17 but rather to the sensitivity of the methodology.
Overall, the RBP-based methodology enabled the specific detection of E. coli in about 1.5 h directly in human specimens, without the need for complex sample processing steps that are laborious and time-consuming. Although the inherent detection limit of the equipment restricted the application of the methodology in lower bacterial contents (less than ≈ 107 CFU), this can be overcome by using more sensitive analytical methods, like optical sensors (Jin et al., 2017).
The developed method can be adapted for the detection of other bacterial species. The broad capture achieved by the functionalized MNPs may be advantageous once there is no need for another antibody targeting other bacterial species, possibly decreasing the costs associated with the detection assay. Thus, it can be a benefit for multiplex bacterial detection, once other phage proteins, providing the high specificity, can be used to target other bacterial species that may be present at the same time in these human biological samples. Also, since this methodology detects and identifies the specific causative agents without compromising their viability, it enables their use for subsequent antibiotic susceptibility tests. This is an important feature of the designed assay since these tests are not possible to be conducted with many other techniques, for instance, nucleic acid-based, that can cause cell degradation or death. This is essential for choosing the most appropriate antimicrobial treatment and to support an efficient therapy, thereby reducing the overuse and misuse of antibiotics and associated adverse outcomes (Afshari et al., 2012).
Further evaluation of our assay in bacteria in biofilm growth mode (Wilks et al., 2021) and mixed bacterial populations, preferentially in combination with different probes specific to other pathogens, would greatly contribute to a more precise assessment of the feasibility of this method. Moreover, it would be interesting to test the RBP against bacteria submitted to other conditions that affect their viability (Liu et al., 2008; Zhao et al., 2013; Wei and Zhao, 2018) or cells under different physiological states, namely, induced by prolonged substrate limitation (Hadas et al., 1997; Nabergoj et al., 2018).
In this study, we provided further insights into the ability of an RBP to recognize with high specificity E. coli strains at different viability states. Also, when this protein was combined with the outstanding benefits of spectrophotometry and magnetic approaches, it enabled the detection of E. coli in different human biological matrices, proving the feasibility of the use of RBPs as exceptional probing elements in biosensing technologies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethical review and approval was not required for this study in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study and the data of the participants were made anonymous in such a way that the data subject can no longer be identified.
SC and AC performed the laboratory work. SC, PF, and CC were responsible for the conceptualization of the study. SC analyzed the data and wrote the original draft of the manuscript. SC and CC performed the review and editing of the manuscript. All authors read and approved the manuscript.
This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the project Phages-on-chip PTDC/BTM-SAL/32442/2017 (POCI-01-0145-FEDER-032442) and the strategic funding of the research units CEB (UIDB/04469/2020) and INESC MN (UID/05367/2020) through the pluriannual BASE and PROGRAMATICO financing and BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2020, Programa Operacional Regional do Norte. SC was supported by the FCT grant SFRH/BD/130098/2017.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to acknowledge Centro Clinico Académico (2CA-Braga) and Dr. Alberta Faustino from the Hospital of Braga for gently providing the clinical bacterial isolates used in this study. We thank the INL Nanophotonics & Bioimaging Facility for their help in microscopy image acquisition and flow cytometry assays.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.871855/full#supplementary-material
Abayasekara, L. M., Perera, J., Chandrasekharan, V., Gnanam, V. S., Udunuwara, N. A., Liyanage, D. S., et al. (2017). Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: a comparative study. BMC Infect. Dis. 17, 1–11. doi: 10.1186/s12879-017-2727-8
Afshari, A., Schrenzel, J., Ieven, M., and Harbarth, S. (2012). Bench-to-bedside review: Rapid molecular diagnostics for bloodstream infection - a new frontier? Crit. Care. 16, 222. doi: 10.1186/cc11202
Akusobi, C., Chan, B. K., Williams, E. S. C. P., Wertz, J. E., and Turner, P. E. (2018). Parallel evolution of host-attachment proteins in phage PP01 populations adapting to escherichia coli O157:H7. Pharmaceutical. 11, 1–14. doi: 10.3390/ph11020060
Aranke, M., Moheimani, R., Phuphanich, M., Kaye, A. D., Ngo, A. L., Viswanath, O., et al. (2021). Disinfectants In Interventional Practices. Curr. Pain Headache Rep. 25, 21. doi: 10.1007/s11916-021-00938-3
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Awais, R., Fukudomi, H., Miyanaga, K., Unno, H., and Tanji, Y. (2006). A recombinant bacteriophage-based assay for the discriminative detection of culturable and viable but nonculturable Escherichia coli O157:H7. Biotechnol. Prog. 22, 853–859. doi: 10.1021/bp060020q
Ayrapetyan, M., Williams, T., and Oliver, J. D. (2018). Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol. 200, e00249–18. doi: 10.1128/JB.00249-18
Ayrapetyan, M., Williams, T. C., and Oliver, J. D. (2015). Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 23, 7–13. doi: 10.1016/j.tim.2014.09.004
Azevedo, N. F., Jardim, T., Almeida, C., Cerqueira, L., Almeida, A. J., Rodrigues, F., et al. (2011). Application of flow cytometry for the identification of Staphylococcus epidermidis by peptide nucleic acid fluorescence in situ hybridization (PNA FISH) in blood samples. Int. J. Gen. Mol. Microbiol. 100, 463–470. doi: 10.1007/s10482-011-9595-9
Barbirz, S., Becker, M., Freiberg, A., and Seckler, R. (2009). Phage tailspike proteins with beta-solenoid fold as thermostable carbohydrate binding materials. Macromol. Biosci. 9, 169–173. doi: 10.1002/mabi.200800278
Bassetti, M., Carnelutti, A., and Righi, E. (2017). The role of methicillin-resistant Staphylococcus aureus in skin and soft tissue infections. Curr. Opin. Infect. Dis. 30, 150–157. doi: 10.1097/QCO.0000000000000353
Bereket, W., Hemalatha, K., Getenet, B., Wondwossen, T., Solomon, A., Zeynudin, A., et al. (2012). Update on bacterial nosocomial infections. Eur. Rev. Med. Pharmacol. Sci. 16, 1039–1044.
Berney, M., Hammes, F., Bosshard, F., Weilenmann, H.-U., and Egli, T. (2007). Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73, 3283–3290. doi: 10.1128/AEM.02750-06
Cassini, A., Plachouras, D., Eckmanns, T., Sin, M. A., Blank, H. -P., Ducomble, T., et al. (2016). Burden of six healthcare-associated infections on european population health: Estimating incidence-based disability-adjusted life years through a population prevalence-based modelling study. PLoS Med. 13, e1002150. doi: 10.1371/JOURNAL.PMED.1002150
Castellani, F., Ghidini, V., Tafi, M. C., Boaretti, M., and Lleo, M. M. (2013). Fate of pathogenic bacteria in microcosms mimicking human body sites. Microb. Ecol. 66, 224–231. doi: 10.1007/s00248-013-0239-7
Chen, X., Miller, A., Cao, S., Gan, Y., Zhang, J., He, Q., et al. (2020). Rapid escherichia coli trapping and retrieval from bodily fluids via a three-dimensional bead-stacked nanodevice. ACS Appl. Mater. Interfaces 12, 7888–7896. doi: 10.1021/acsami.9b19311
Costa, S. P., Dias, N. M., Melo, L. D. R., Azeredo, J., Santos, S. B., and Carvalho, C. M. (2020). A novel flow cytometry assay based on bacteriophage-derived proteins for Staphylococcus detection in blood. Sci. Rep. 10, 6260. doi: 10.1038/s41598-020-62533-7
Cudjoe, K. S., Hagtvedt, T., and Dainty, R. (1995). Immunomagnetic separation of Salmonella from foods and their detection using immunomagnetic particle (IMP)-ELISA. Int. J. Food Microbiol. 27, 11–25. doi: 10.1016/0168-1605(94)00134-R
Cunha, A. P., Henriques, R., Cardoso, S., Freitas, P. P., and Carvalho, C. M. (2021). Rapid and multiplex detection of nosocomial pathogens on a phage-based magnetoresistive lab-on-chip platform. Biotechnol. Bioeng. 118, 3164–3174. doi: 10.1002/bit.27841
Denyes, J. M., Dunne, M., Steiner, S., Mittelviefhaus, M., Weiss, A., Schmidt, H., et al. (2017). Modified bacteriophage S16 long tail fiber proteins for rapid and specific immobilization and detection of Salmonella cells. Appl. Environ. Microbiol. 83, 1–15. doi: 10.1128/AEM.00277-17
Dowah, A. S. A., and Clokie, M. R. J. (2018). Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 10, 535–542. doi: 10.1007/s12551-017-0382-3
Drexler, K., Riede, I., Montag, D., Eschbach, M. L., and Henning, U. (1989). Receptor specificity of the Escherichia coli T-even type phage Ox2. Mutational alterations in host range mutants. J. Mol. Biol. 207, 797–803. doi: 10.1016/0022-2836(89)90245-3
Du, M., Chen, J., Zhang, X., Li, A., Li, Y., and Wang, Y. (2007). Retention of Virulence in a Viable but Nonculturable Edwardsiella tarda Isolate. Appl. Environ. Microbiol. 73, 1349. doi: 10.1128/AEM.02243-06
ECDC (2019). “Healthcare-associated infections acquired in intensive care units”, in Annual Epidemiological Report for 2017. Stockholm: ECDC.
Favrin, S. J., Jassim, S. A., and Griffiths, M. W. (2001). Development and optimization of a novel immunomagnetic separation- bacteriophage assay for detection of Salmonella enterica serovar enteritidis in broth. Appl. Environ. Microbiol. 67, 217–224. doi: 10.1128/AEM.67.1.217-224.2001
Fernandes, E., Martins, V. C., Nóbrega, C., Carvalho, C. M., Cardoso, F. A., et al. (2014). A bacteriophage detection tool for viability assessment of Salmonella cells. Biosens. Bioelectron. 52, 239–246. doi: 10.1016/j.bios.2013.08.053
Ferreira, L., Sánchez-Juanes, F., Porras-Guerra, I., García-García, M. I., García-Sánchez, J. E., González-Buitrago, J. M., et al. (2011). Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17, 546–551. doi: 10.1111/j.1469-0691.2010.03257.x
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Frank, S. A. (2002). “Specificity and Cross-Reactivity,” in Immunology and Evolution of Infectious Disease (Princeton, NJ: Princeton University Press).
Frickmann, H., Zautner, A., Moter, A., Kikhney, J., Hagen, R., Stender, H., et al. (2017). Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review. Crit. Rev. Microbiol. 43, 263–293. doi: 10.3109/1040841X.2016.1169990
Fujinami, Y., Hirai, Y., Sakai, I., Yoshino, M., and Yasuda, J. (2007). Sensitive detection of Bacillus anthracis using a binding protein originating from γ-phage. Microbiol. Immunol. 51, 163–169. doi: 10.1111/j.1348-0421.2007.tb03894.x
González-García, V. A., Bocanegra, R., Pulido-Cid, M., Martín-Benito, J., Cuervo, A., and Carrascosa, J. L. (2015a). Characterization of the initial steps in the T7 DNA ejection process. Bacteriophage 5, e1056904. doi: 10.1080/21597081.2015.1056904
González-García, V. A., Pulido-Cid, M., Garcia-Doval, C., Bocanegra, R., Van Raaij, M. J., Martín-Benito, J., et al. (2015b). Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J. Biol. Chem. 290, 10038–10044. doi: 10.1074/jbc.M114.614222
Górska, S., Rydosz, A., Brzozowska, E., Drab, M., Wincza, K., Gamian, A., et al. (2018). Effectiveness of sensors contact metallization (Ti, Au, and Ru) and biofunctionalization for Escherichia coli detection. Sensors. 18, 2912. doi: 10.3390/s18092912
Hadas, H., Einav, M., Fishov, I., and Zaritsky, A. (1997). Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 143, 179–185. doi: 10.1099/00221287-143-1-179
Haque, M., Sartelli, M., McKimm, J., and Bakar, M. A. (2018). Health care-associated infections – an overview. Infect. Drug Resist. 11, 2321. doi: 10.2147/IDR.S177247
He, Y., Shi, Y., Liu, M., Wang, Y., Wang, L., Lu, S., et al. (2018). Nonlytic Recombinant Phage Tail Fiber Protein for Specific Recognition of Pseudomonas aeruginosa. Anal. Chem. 90, 14462–14468. doi: 10.1021/acs.analchem.8b04160
Holland, J. L., Louie, L., Simor, A. E., and Louie, M. (2000). PCR detection of Escherichia coli O157:H7 directly from stools: Evaluation of commercial extraction methods for purifying fecal DNA. J. Clin. Microbiol. 38, 4108–4113. doi: 10.1128/JCM.38.11.4108-4113.2000
Hu, J., Miyanaga, K., and Tanji, Y. (2012). Diffusion of bacteriophages through artificial biofilm models. Biotechnol. Prog. 28, 319–326. doi: 10.1002/btpr.742
Javed, M. A., Poshtiban, S., Arutyunov, D., Evoy, S., and Szymanski, C. M. (2013). Bacteriophage receptor binding protein based assays for the simultaneous detection of campylobacter jejuni and campylobacter coli. PLoS ONE. 8, e69770. doi: 10.1371/journal.pone.0069770
Jin, B., Wang, S., Min Lin, Y. J., Zhang, S., Cui, X., Gong, Y., et al. (2017). Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection. Biosens. Bioelectron. Apr 15, 525–533. doi: 10.1016/j.bios.2016.10.029
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kaper, J. B., Nataro, J. P., and Mobley, H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Kretzer, J. W., Lehmann, R., Schmelcher, M., Banz, M., Kim, K.-P. P., Korn, C., et al. (2007). Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 73, 1992–2000. doi: 10.1128/AEM.02402-06
Krueger, A. P. (1931). The sorption of bacteriophage by living and dead susceptible bacteria: I. Equilibrium conditions. J. Gen. Physiol. 14, 493–516. doi: 10.1085/jgp.14.4.493
Kunstmann, S., Scheidt, T., Buchwald, S., Helm, A., Mulard, L. A., Fruth, A., et al. (2018). Bacteriophage Sf6 tailspike protein for detection of shigella flexneri pathogens. Viruses. 10, 431. doi: 10.3390/v10080431
Latka, A., Maciejewska, B., Majkowska-Skrobek, G., Briers, Y., and Drulis-Kawa, Z. (2017). Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 101, 3103. doi: 10.1007/s00253-017-8224-6
Laupland, K. B., and Valiquette, L. (2013). The changing culture of the microbiology laboratory. Can. J. Infect. Dis. Med. Microbiol. 24, 125. doi: 10.1155/2013/101630
Lee, A. S., De Lencastre, H., Garau, J., Kluytmans, J., Malhotra-Kumar, S., Peschel, A., et al. (2018). Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 4, 1–23. doi: 10.1038/nrdp.2018.33
Leiman, P. G., Kanamaru, S., Mesyanzhinov, V. V., Arisaka, F., and Rossmann, M. G. (2003). Structure and morphogenesis of bacteriophage T4. Cell. Mol. Life Sci. 60, 2356–2370. doi: 10.1007/s00018-003-3072-1
Li, L., Mendis, N., Trigui, H., Oliver, J. D., and Faucher, S. P. (2014). The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5, 1–1. doi: 10.3389/fmicb.2014.00258
Liu, Y., Gilchrist, A., Zhang, J., and Li, X. F. (2008). Detection of viable but nonculturable escherichia coli O157:H7 bacteria in drinking water and river water. Appl. Environ. Microbiol. 74, 1502–1507. doi: 10.1128/AEM.02125-07
Liu, Y., Wang, C., Tyrrell, G., Hrudey, S. E., and Li, X. F. (2009). Induction of Escherichia coli O157:H7 into the viable but non-culturable state by chloraminated water and river water, and subsequent resuscitation. Environ. Microbiol. Rep. 1, 155–161. doi: 10.1111/j.1758-2229.2009.00024.x
Lopes, A. L. K., Cardoso, J., dos Santos, F. R. C. C., Silva, A. C. G., Stets, M. I., Zanchin, N. I. T., et al. (2016). Development of a magnetic separation method to capture sepsis associated bacteria in blood. J. Microbiol. Methods 128, 96–101. doi: 10.1016/j.mimet.2016.07.012
Lupo, D., Leptihn, S., Nagler, G., Haase, M.J, Molineux, I., et al. (2015). The T7 ejection nanomachine components gp15–gp16 form a spiral ring complex that binds DNA and a lipid membrane. Virology 486, 263–271. doi: 10.1016/j.virol.2015.09.022
Marder, E. P., Cieslak, P. R., Cronquist, A. B., Dunn, J., Lathrop, S., Rabatsky-Ehr, T., et al. (2017). Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance — foodborne diseases active surveillance network, 10 U.S. Sites, 2013–2016. MMWR. Morb. Mortal. Wkly. Rep. 66, 397–403. doi: 10.15585/mmwr.mm6615a1
Mason, D. J., Power, E. G. M., Talsania, H., Phillips, I., and Gant, V. A. (1995). Antibacterial action of ciprofloxacin. Antimicrob. Agents Chemother. 39, 2752–2758. doi: 10.1128/AAC.39.12.2752
McLoughlin, K. S. (2011). Microarrays for pathogen detection and analysis. Brief. Funct. Genomics 10, 342. doi: 10.1093/bfgp/elr027
Mothershed, E. A., and Whitney, A. M. (2006). Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clin. Chim. Acta 363, 206–220. doi: 10.1016/j.cccn.2005.05.050
Nabergoj, D., Modic, P., and Podgornik, A. (2018). Effect of bacterial growth rate on bacteriophage population growth rate. Microbiologyopen 7, e00558. doi: 10.1002/mbo3.558
Ngamsom, B., Esfahani, M. M. N., Phurimsak, C., Lopez-Martinez, M. J., Raymond, J. C., Broyer, P., et al. (2016). Multiplex sorting of foodborne pathogens by on-chip free-flow magnetophoresis. Anal. Chim. Acta 918, 69–76. doi: 10.1016/j.aca.2016.03.014
Oda, M., Morita, M., Unno, H., and Tanji, Y. (2004). Rapid detection of Escherichia coli O157:H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl. Environ. Microbiol. 70, 527–534. doi: 10.1128/AEM.70.1.527-534.2004
Oliver, J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34, 415–425. doi: 10.1111/j.1574-6976.2009.00200.x
Opota, O., Jaton, K., and Greub, G. (2015). Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood. Clin. Microbiol. Infect. 21, 323–331. doi: 10.1016/j.cmi.2015.02.005
Pasquaroli, S., Zandri, G., Vignaroli, C., Vuotto, C., Donelli, G., and Biavasco, F. (2013). Antibiotic pressure can induce the viable but non-culturable state in Staphylococcus aureus growing in biofilms. J. Antimicrob. Chemother. 68, 1812–1817. doi: 10.1093/jac/dkt086
Pienaar, J. A., Singh, A., and Barnard, T. G. (2016). The viable but non-culturable state in pathogenic Escherichia coli: a general review. Afr. J. Lab. Med. 5, 2225–2002. doi: 10.4102/ajlm.v5i1.368
Qiu, J., Zhou, Y., Chen, H., and Lin, J. M. (2009). Immunomagnetic separation and rapid detection of bacteria using bioluminescence and microfluidics. Talanta 79, 787–795. doi: 10.1016/j.talanta.2009.05.003
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., Chapman, J., et al. (2019). A review of methods for the detection of pathogenic microorganisms. Analyst 144, 396–411. doi: 10.1039/C8AN01488D
Rakhuba, D. V., Kolomiets, E. I., Dey, E. S., and Novik, G. I. (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Polish J. Microbiol. 59, 145–155. doi: 10.33073/pjm-2010-023
Rivers, B., and Steck, T. R. (2001). Viable but nonculturable uropathogenic bacteria are present in the mouse urinary tract following urinary tract infection and antibiotic therapy. Urol. Res. 29, 60–66. doi: 10.1007/s002400000151
Santos, S. B., Cunha, A. P., Macedo, M., Nogueira, C. L., Brandão, A., Costa, S. P., et al. (2020). Bacteriophage-receptor binding proteins for multiplex detection of Staphylococcus and Enterococcus in blood. Biotechnol. Bioeng. 117, 3286–3298. doi: 10.1002/bit.27489
Schmelcher, M., and Loessner, M. J. (2014). Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 4, e28137. doi: 10.4161/bact.28137
Schmelcher, M., Shabarova, T., Eugster, M. R., Eichenseher, F., Tchang, V. S., Banz, M., et al. (2010). Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl. Environ. Microbiol. 76, 5745–5756. doi: 10.1128/AEM.00801-10
Shi, Y., He, Y., Zhang, L., Wang, L., and Fu, Z. (2021). Dual-site recognition of Pseudomonas aeruginosa using polymyxin B and bacteriophage tail fiber protein. Anal. Chim. Acta 1180, 338855. doi: 10.1016/j.aca.2021.338855
Shin, H. J., and Lim, W. K. (2018). Rapid label-free detection of E. coli using a novel SPR biosensor containing a fragment of tail protein from phage lambda. Prep. Biochem. Biotechnol. 48, 498–505. doi: 10.1080/10826068.2018.1466154
Simpson, D. J., Sacher, J. C., and Szymanski, C. M. (2015). Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 34, 69–77. doi: 10.1016/j.sbi.2015.07.006
Simpson, D. J., Sacher, J. C., and Szymanski, C. M. (2016). Development of an assay for the identification of receptor binding proteins from bacteriophages. Viruses 8. doi: 10.3390/v8010017
Singh, A., Arutyunov, D., Szymanski, C. M., and Evoy, S. (2012). Bacteriophage based probes for pathogen detection. Analyst 137, 3405. doi: 10.1039/c2an35371g
Singh, A., Arya, S. K., Glass, N., Hanifi-Moghaddam, P., Naidoo, R., Szymanski, C. M., et al. (2010). Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 26, 131–138. doi: 10.1016/j.bios.2010.05.024
Singh, A., Poshtiban, S., and Evoy, S. (2013). Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 13, 1763–1786. doi: 10.3390/s130201763
Singhal, N., Kumar, M., Kanaujia, P., and Virdi, J. (2015). MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 6, 791. doi: 10.3389/fmicb.2015.00791
Skvarc, M., Stubljar, D., Rogina, P., and Kaasch, A. J. (2013). Non-culture-based methods to diagnose bloodstream infection: does it work? Eur. J. Microbiol. Immunol. 3, 97–104. doi: 10.1556/EuJMI.3.2013.2.2
Stevens, K. A., and Jaykus, L. A. (2004). Bacterial separation and concentration from complex sample matrices: A review. Crit. Rev. Microbiol. 30, 7–24. doi: 10.1080/10408410490266410
Stiefel, P., Schmidt-Emrich, S., Maniura-Weber, K., and Ren, Q. (2015). Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 15, 1–9. doi: 10.1186/s12866-015-0376-x
Tay, L. L., Huang, P. J., Tanha, J., Ryan, S., Wu, X., Hulse, J., et al. (2012). Silica encapsulated SERS nanoprobe conjugated to the bacteriophage tailspike protein for targeted detection of Salmonella. Chem. Commun. 48, 1024–1026. doi: 10.1039/C1CC16325F
Tlili, C., Sokullu, E., Safavieh, M., Tolba, M., Ahmed, M. U., and Zourob, M. (2013). Bacteria screening, viability, and confirmation assays using bacteriophage-impedimetric/loop-mediated isothermal amplification dual-response biosensors. Anal. Chem. 85, 4893–4901. doi: 10.1021/ac302699x
Truchado, P., Gil, M. I., Larrosa, M., and Allende, A. (2020). Detection and Quantification Methods for Viable but Non-culturable (VBNC) Cells in Process Wash Water of Fresh-Cut Produce: Industrial Validation. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00673
Váradi, L., Luo, J. L., Hibbs, D. E., Perry, J. D., Anderson, R. J., Orenga, S., et al. (2017). Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 46, 4818–4832. doi: 10.1039/C6CS00693K
Verma, J., Saxena, S., and Babu, S. G. (2013). “ELISA-Based Identification and Detection of Microbes,” in Analyzing Microbes (Springer Protocols) 169–186. doi: 10.1007/978-3-642-34410-7_13
Villarroel, J., Larsen, M. V., Kilstrup, M., and Nielsen, M. (2017). Metagenomic analysis of therapeutic PYO phage cocktails from 1997 to 2014. Viruses. 9, 328. doi: 10.3390/v9110328
Wang, Y., and Alocilja, E. C. (2015). Gold nanoparticle-labeled biosensor for rapid and sensitive detection of bacterial pathogens. J. Biol. Eng. 9, 1–7. doi: 10.1186/S13036-015-0014-Z
Watanabe, T. (1976). Role of lipopolysaccharide in adsorption of coliphage T4D to Escherichia coli B. Can. J. Microbiol. 22, 745–751. doi: 10.1139/m76-108
Wei, C., and Zhao, X. (2018). Induction of Viable but Nonculturable Escherichia coli O157:H7 by Low Temperature and Its Resuscitation. Front. Microbiol. 9, 2728. doi: 10.3389/fmicb.2018.02728
Wilks, S. A., Koerfer, V. V., Prieto, J. A., Fader, M., and Keevil, C. W. (2021). Biofilm development on urinary catheters promotes the appearance of viable but nonculturable bacteria. MBio 12, e03584–e03520. doi: 10.1128/mBio.03584-20
World Health Organization (2011). Report on the Burden of Endemic Health Care-Associated Infection Worldwide. Geneva: WHO Document Production Services.
Xu, B., Ma, X., Xiong, H., and Li, Y. (2014). Complete genome sequence of 285P, a novel T7-like polyvalent E. coli bacteriophage. Virus Genes. 48, 528–533. doi: 10.1007/s11262-014-1059-7
Yang, Y., Xu, F., Xu, H., Aguilar, Z. P., Niu, R., Yuan, Y., et al. (2013). Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella Typhimurium, Escherichia coli O157: H7 and Listeria monocytogenes in food products. Food Microbiol. 34, 418–424. doi: 10.1016/j.fm.2013.01.004
Ye, C., Lin, H., Zhang, M., Chen, S., and Yu, X. (2020). Characterization and potential mechanisms of highly antibiotic tolerant VBNC Escherichia coli induced by low level chlorination. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-58106-3
Yoichi, M., Abe, M., Miyanaga, K., Unno, H., and Tanji, Y. (2005). Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J. Biotechnol. 115, 101–107. doi: 10.1016/j.jbiotec.2004.08.003
Zhao, F., Bi, X., Hao, Y., and Liao, X. (2013). Induction of viable but nonculturable escherichia coli O157:H7 by high pressure CO2 and its characteristics. PLoS ONE 8, e62388. doi: 10.1371/journal.pone.0062388
Keywords: healthcare-associated infections, cell viability states, diagnostic method, human biological samples, receptor-binding proteins, Escherichia coli
Citation: Costa SP, Cunha AP, Freitas PP and Carvalho CM (2022) A Phage Receptor-Binding Protein as a Promising Tool for the Detection of Escherichia coli in Human Specimens. Front. Microbiol. 13:871855. doi: 10.3389/fmicb.2022.871855
Received: 08 February 2022; Accepted: 26 April 2022;
Published: 01 June 2022.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Ewa Brzozowska, Polish Academy of Sciences (PAN), PolandCopyright © 2022 Costa, Cunha, Freitas and Carvalho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carla M. Carvalho, Y2FybGEuY2FydmFsaG9AaW5sLmludA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.