
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 May 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.871152
This article is part of the Research Topic Microbial interactions of Clostridioides difficile View all 7 articles
 Andrew E. Clark1
Andrew E. Clark1 Chelsea C. Adamson1
Chelsea C. Adamson1 Katelyn E. Carothers1
Katelyn E. Carothers1 Bryan Angelo P. Roxas1
Bryan Angelo P. Roxas1 V. K. Viswanathan1,2,3†
V. K. Viswanathan1,2,3† Gayatri Vedantam1,2,3,4*†
Gayatri Vedantam1,2,3,4*†The alternative sigma factor SigL (Sigma-54) facilitates bacterial adaptation to the extracellular environment by modulating the expression of defined gene subsets. A homolog of the gene encoding SigL is conserved in the diarrheagenic pathogen Clostridioides difficile. To explore the contribution of SigL to C. difficile biology, we generated sigL-disruption mutants (sigL::erm) in strains belonging to two phylogenetically distinct lineages—the human-relevant Ribotype 027 (strain BI-1) and the veterinary-relevant Ribotype 078 (strain CDC1). Comparative proteomics analyses of mutants and isogenic parental strains revealed lineage-specific SigL regulons. Concomitantly, loss of SigL resulted in pleiotropic and distinct phenotypic alterations in the two strains. Sporulation kinetics, biofilm formation, and cell surface-associated phenotypes were altered in CDC1 sigL::erm relative to the isogenic parent strain but remained unchanged in BI-1 sigL::erm. In contrast, secreted toxin levels were significantly elevated only in the BI-1 sigL::erm mutant relative to its isogenic parent. We also engineered SigL overexpressing strains and observed enhanced biofilm formation in the CDC1 background, and reduced spore titers as well as dampened sporulation kinetics in both strains. Thus, we contend that SigL is a key, pleiotropic regulator that dynamically influences C. difficile's virulence factor landscape, and thereby, its interactions with host tissues and co-resident microbes.
Clostridioides difficile, an anaerobic, spore-forming, Gram-positive bacillus, is a significant cause of healthcare-associated infections (Stewart et al., 2020). C. difficile infection manifests as a diverse spectrum of diarrheal diseases with possible progression toward more serious sequelae, including pseudomembranous colitis or fatal toxic megacolon (George et al., 1978; Dudukgian et al., 2009). Ingested spores germinate into vegetative cells upon exposure to specific bile salt and amino acid triggers in the mammalian host. Disruption or suppression of the endogenous microbiota by antibiotics, immunosuppressive drugs, or host factors, facilitate vegetative cell colonization of the intestine. Vegetative cells from toxigenic C. difficile strains produce two major enterotoxins (TcdA and TcdB) that glucosylate host Rho GTPases, leading to actin cytoskeleton reorganization, solute transport disruption, and consequent symptomatic disease (Vedantam et al., 2012). C. difficile toxin synthesis increases as bacteria enter the stationary phase of growth; thus pathogen metabolism and virulence are linked (Voth and Ballard, 2005; Dineen et al., 2010; Bouillaut et al., 2015).
Sigma factors are dissociable components of the RNA polymerase holoenzyme complex that recognize and bind defined promoter sequences, thereby dictating cellular transcriptional programs (Merrick, 1993; Feklístov et al., 2014). Canonical sigma-70 family members engage promoters at −35 and −10 regions relative to the transcriptional start site, induce promoter open-complex formation, and initiate transcription (Zhang and Buck, 2015). In contrast, the structurally unrelated sigma-54 family members recognize unique −24 and −12 promoter regions; however, the formation of a repressive fork junction structure near −12 prevents the recruited holoenzyme from forming an open complex unassisted (Rappas et al., 2007; Zhang and Buck, 2015). Conversion to an open complex requires energy transfer from unique bacterial enhancer-binding proteins (EBPs, or sigma-54 activators) that bind conserved enhancer sequences upstream of the promoter. The formation of a hairpin-like structure, assisted by DNA-bending proteins, such as integration host factor (IHF), allows enhancer-bound EBPs to interact with the promoter-bound sigma-54 closed complex (Zhang and Buck, 2015). Activator-mediated ATP hydrolysis results in loss of the fork junction structure and isomerization to an open complex, leading to transcription initiation.
Nie et al. used comparative genomics of 57 species from the Clostridiales to reconstruct sigma-54 regulons and identify EBPs and their regulatory modules (Nie et al., 2019). Of the 24 predicted EBPs detected in C. difficile, the roles of two, CdsR and PrdR, involved in cysteine and proline catabolism, respectively, were experimentally validated (Bouillaut et al., 2013; Gu et al., 2017). To comprehensively characterize the SigL regulon, Soutourina et al. used in silico analysis, transcriptome analyses, and transcription start site (TSS) mapping of the laboratory-adapted C. difficile strain 630Δerm (Soutourina et al., 2020). These studies revealed that the SigL regulon includes genes involved in the oxidation and reduction of amino acids to the corresponding organic acids (Stickland metabolism). Stickland reactions generate ATP and NAD+ and regulate C. difficile toxin production and virulence (Stickland, 1934; Neumann-Schaal et al., 2019).
To expand these observations to clinically-contemporary isolates, and further characterize the role of SigL in C. difficile biology, we generated isogenic insertion mutants (sigL::erm) in two genetically-divergent strains of human and veterinary origin: BI-1 (flagellated) and CDC1 (non-flagellated), belonging to the lineages (Ribotypes) RT027 and RT078, respectively. The epidemic-associated RT027 strains are often responsible for the most severe human C. difficile infections (CDI). RT078 strains, in contrast, while sharing several attributes with RT027 strains, are frequently associated with livestock infections (Elliott et al., 2017). We performed phenotypic assays and comparative proteomics studies on parental and sigL::erm derivatives. SigL modulated the C. difficile proteome in a temporal manner, and loss of sigL had profound but contrasting and pleiotropic effects on the two strains. Our studies support a key role for SigL in C. difficile gene regulation, including strain-specific roles in the modulation of virulence factor expression, central metabolic functions, and sporulation.
All C. difficile strains utilized in this study (Supplementary Table S4) were grown at 37°C using a Type B Coy Laboratory Anaerobic Chamber under anaerobic conditions in an 85% N2, 10% H2, 5% CO2 gas mixture. Bacteria were recovered from the freezer storage in glycerol and routinely maintained on BHIS agar plates (Brain-Heart Infusion base media supplemented with 5 g/L Yeast Extract and 0.1% L-Cysteine). All assays requiring liquid culture were performed using Bacto™ BHI unless otherwise indicated.
Antibiotics were used as necessary for C. difficile: (15 μg/mL thiamphenicol, 20 μg/mL lincomycin, 0.8 μg/mL cefoxitin, 25 μg/mL cycloserine) added to either BHI or BHIS. E. coli Top10 (Invitrogen) used for cloning and plasmid construction, and CA434 used for conjugation were routinely grown at 37°C on LB media using antibiotics as necessary: (20 μg/mL chloramphenicol, 100 μg/mL spectinomycin, 150 μg/ml ampicillin). The C. difficile clinical isolates used in this study were from the Hines VA Hospital culture collection of Dr. Dale Gerding or from the collection of Dr. Glenn Songer.
C. difficile genome sequences were maintained and annotated using Rapid Annotation using Subsystem Technology (RAST) bioinformatics web tools (Overbeek et al., 2014). sigL promoter prediction reported here was undertaken using Microbes Online web tools (Dehal et al., 2010). Functional domains of C. difficile SigL reported here were analyzed using the Similar Modular Architecture Research Tool (SMART) (Letunic et al., 2015).
C. difficile BI-1 and CDC1 strains were grown to mid-exponential (OD600 ~ 0.6), late-exponential (OD600 ~ 0.8), and stationary phase (OD600 ~ 1.0). At each phase of growth, cultures were diluted 1:2 in RNAlater RNA stabilization reagent (Sigma Aldrich) and incubated anaerobically for 1 h, before pelleting and freezing bacteria at −80°C. RNA was extracted from cell pellets using the RNeasy Plus Mini Kit (Qiagen) according to manufacturer instructions, with modifications. Briefly, beat-beading tubes were filled 1/3 with beads and soaked overnight in 500 μl buffer RLT (Qiagen). Tubes were spun at full speed and excess buffer was removed. Bacterial pellets were lysed by bead beating for 2 min in 600 μl buffer RLT and 2-Mercaptoethanol (10 μl/mL) per sample. Tubes were centrifuged at max speed for 3 min, and the supernatant was removed for RNA extraction according to manufacturer instructions. RNA samples were treated with DNase I to remove genomic DNA contamination, and RNA yields were quantified on a Nanodrop instrument. cDNA synthesis from the RNA samples was performed using the SuperScript III First-Strand Synthesis System (Invitrogen) according to manufacturer instructions and yields were quantified using Nanodrop. Standard PCR was performed to confirm the expression of sigL and 16S (housekeeping gene), with Taq 2× Master Mix, using AC106 sigL-F/AC106 sigL-R and 16SRT_F/16SRT_R primer pairs respectively (Supplementary Table S5). Quantitative real-time PCR was performed using iTaq Universal SYBR Green Supermix (BioRad) on the StepOnePlus instrument (Applied Biosystems) according to manufacturer instructions. Expression fold changes were determined using ΔΔCT calculations (Livak and Schmittgen, 2001).
Isogenic sigL mutants were created in C. difficile BI-1 and CDC1 using the ClosTron system (Kuehne and Minton, 2012). Briefly, retargeted ClosTron introns were designed using the Perutka algorithm at Clostron.com and synthesized by DNA2.0 (DNA2.0, Melo Park, CA). The resulting constructs were electrotransformed into E. coli CA434 donor strain which was recovered on LB supplemented with spectinomycin and chloramphenicol. Isolated colonies were inoculated into BHIS broth supplemented with chloramphenicol and grown overnight at 37°C with shaking. One milliliter of culture was pelleted at 1,500 × g for 1 min, washed in BHIS, centrifuged again for 1 min at 1,500 × g, and transferred to an anaerobic chamber. The pellet was subsequently resuspended with 200 μl of C. difficile overnight recipient culture and incubated overnight on BHIS agar. Growth was recovered in one milliliter of PBS, and 200 μl aliquots were inoculated to BHIS Tetrazolium Chloride (TCC) and incubated overnight at 37°C. Thiamphenicol-resistant colonies were isolated on BHIS supplemented with lincomycin to isolate potential integrant clones. Integration of the intron was confirmed by PCR using primers that detect both the integrated form of the intron, as well as primers AC55 sigL-F and AC56 sigL-R which amplify the intron/sigL junction (Supplementary Table S5, Supplementary Figure S2). Loss of SigL expression was verified by RT-PCR using primers AC106 sigL-F and AC107 sigL-R (Supplementary Figure S2), and sigL disruption mutants were cured of the Clostron plasmid by serial passage on BHIS without selection.
The pMTL8000-series of shuttle vectors was used for the construction of sigL complementation and overexpression plasmids. Primers AC55 sigL-F and AC56 sigL-R were designed to amplify the coding sequence ~300 base pairs upstream of the sigL gene. The resulting PCR products were cleaned and digested with restriction enzymes allowing them to be cloned into either pMTL82151 and expressed under the control of their endogenous promoter (complementation) or pMTL82153 and expressed under the control of a constitutive promoter (Pfdx, overexpression). pMTL82151 and pMTL82153 contain the same backbone, including the Gram(+)Rep (pBP1), marker (catP), and Gram(–)Rep (ColE1+tra). The resulting plasmids were transformed into CA434 and conjugated into C. difficile sigL mutants. Successfully complemented strains were selected on BHIS containing thiamphenicol.
Twelve putative promoter sequences for genes or operons with previously established downregulation in sigL mutants (Soutourina et al., 2020) were used to generate a positional weight matrix (PWM). Genomes of 630 (NCBI Assembly GCA_000009205.2), BI-1 (GCA_000211235.1), and an RT078 strain, QCD-23m63 (GCA_000155065.1), were searched for promoter sites based on the C. difficile sigL PWM using FIMO (Find Individual Motif Occurrences) (Grant et al., 2011). Identified sigL sites in 630, BI-1, and RT078 with FIMO q-values <0.05 located <200 bp upstream of the gene and/or operon were compiled, tabulated, and cross-referenced to previous C. difficile 630 sigL promoters, enhancer-binding protein (EBP) upstream activator sequences (UAS), and sigL::erm microarray results (Nie et al., 2019; Soutourina et al., 2020). The sequence and position of EBP UAS identified by Nie et. al. in BI-1 and RT078 genomes were determined using BLASTn.
C. difficile was grown to an OD600 of either 0.6 (mid-exponential), 0.8 (late-exponential) or 1 (stationary) in BHI. Cell pellets were resuspended in 400 μl of 0.5 M triethylammonium bicarbonate (TEAB) buffer and sonicated (five pulses of 15 s duration each) at an amplitude setting of 4 for lysis. Lysates were centrifuged at 20,000 × g for 30 min at 4°C to clear debris. Total recovered proteins were quantitated using a BCA Protein Assay (Pierce, Rockford, IL).
A total of 100 μg of protein in 20 μl of 0.5 M TEAB from each sample was denatured (1 μl of 2% SDS), reduced (1 μl of 100 mM tris-(2-carboxyethyl) phosphine), and alkylated (1 μl of 84 mM iodoacetamide). Trypsin was added at a ratio of 1:10 and the digestion reaction was incubated for 18 h at 48°C. iTRAQ reagent labeling was performed according to the 8-plex iTRAQ kit instructions (AB SCIEX, Framingham, MA).
Strong cationic exchange (SCX) fractionation was performed on a passivated Waters 600E HPLC system, using a 4.6 × 250 mm polysulfoethyl aspartamide column (PolyLC, Maryland, USA) at a flow rate of 1 ml/min. Fifteen SCX fractions were collected and dried down completely, then resuspended in 9 μl of 2% (v/v) acetonitrile (ACN) and 0.1% (v/v) trifluoroacetic acid (TFA).
For the second-dimension fractionation by reverse phase C18 nanoflow-LC, each SCX fraction was autoinjected onto a Chromolith CapRod column (150 × 0.1 mm, Merck) using a 5 μl injector loop on a Tempo LC MALDI Spotting system (ABI-MDS/Sciex). Separation was over a 50 min solvent gradient from 2% ACN and.1% TFA (v/v) to 80% ACN and 0.1% TFA (v/v) with a flow rate of 2.5 μl/min. An equal flow of MALDI matrix solution was added post-column (7 mg/ml recrystallized CHCA (α-cyano-hydroxycinnamic acid), 2 mg/ml ammonium phosphate, 0.1% TFA, 80% ACN). The combined eluent was automatically spotted onto a stainless steel MALDI target plate every 6 s (0.6 μl per spot), for a total of 370 spots per original SCX fraction.
MALDI target plates were analyzed in a data-dependent manner on an ABSciexTripleTOF 5,600+ mass spectrometer (AB SCIEX, Framingham, MA). MS spectra were acquired from each sample spot using 500 laser shots per spot, laser intensity of 3,200. The highest peak of each observed m/z value was selected for subsequent MS/MS analysis.
Up to 2,500 laser shots at laser power 4,200 were accumulated for each MS/MS spectrum. Protein identification and quantitation were performed using the Paragon algorithm implemented in Protein Pilot 3.0 software by searching the acquired MS and MS/MS spectra from all 15 plates against the C. difficile strain BI-1 protein database, or the 078 representative strain QCD-23m63 database, plus common contaminants. The Protein Pilot Unused score cutoff of >0.82 (1% global false discovery rate) was calculated from the slope of the accumulated Decoy database hits by the Proteomics System Performance Evaluation Pipeline (PSPEP) program [72]. Proteins with at least one peptide >95% confidence (score based on the number of matches between the data and the theoretical fragment ions) and a Protein Pilot Unused score of >0.82 were considered as valid identifications (IDs).
A pooled sample consisting of equal amounts of each protein sample was used as a technical replicate. iTRAQ tags 113 and 121 were used to tag equal amounts of the same pooled sample. A receiver operating characteristic (ROC) curve was plotted using the technical replicate ratios, 119:121 and 121:119, as true negatives and all studied ratios as true positives. A 2-fold change cut-off was set to define proteins altered in abundance between wt and respective sigL::erm mutants.
PSII extraction and quantitation were performed by previously published methods (Chu et al., 2016) with minor modifications. C. difficile vegetative cells were grown to an OD600 = 1.2 in BHI at 37°C. Cells were pelleted by centrifugation for 10 min at 4,000 rpm, and the supernatant was discarded. Pellets were resuspended in 0.1 M EDTA-triethanolamine buffer pH 7.0 for 20 min, and subsequently centrifuged for 10 min at 4,000 rpm. The supernatant was collected, serially diluted, and dot-blotted onto activated PVDF membranes, and probed with PSII-specific antiserum. Antibodies were detected using anti-Rabbit HRP secondary and FemtoWest Maximum sensitivity substrate (Pierce).
Autolysis of wild-type and sigL mutant strains was measured using the method described previously (El Meouche et al., 2013). Fifty milliliters C. difficile cultures were grown in BHI to an OD600 = 1.2 and were centrifuged for 10 min at 3,200 × g to pellet. The harvested cells were washed twice in PBS and resuspended in 50 ml of 50 mM phosphate buffer containing 0.01% of Triton X-100. Absorbance values at OD600 of resuspended cells were recorded, and cultures were placed back into an anaerobic environment at incubated at 37°C. Time points were harvested every 15 min, and autolysis was monitored by measuring absorbance values as a percentage of the initial OD600. Each assay was performed in biological triplicate and photographed at the termination of the experiment.
Autoaggregation was measured by the method of Faulds-Pain et al. (2014) with modifications. Overnight cultures of C. difficile wild type and sigL mutants were diluted at 1:50 in BHI, and 5 ml of culture was separated into pre-reduced glass culture tubes and incubated upright at 37°C. Each hour, six culture tubes were removed from incubation and the OD600 was determined. The OD600 was directly read from the top 1 mL of culture in three “un-mixed” fractions per time point, and the remaining three tubes were vortexed and the 0D600 was measured constituting “mixed” fractions. Aggregated cells settle from suspension and are not recorded in the OD600 from “unmixed” fractions but are detected in the “mixed” fractions. The percent agglutination of aggregative C. difficile vegetative cells was calculated as the difference between the OD600 of the whole (mixed) solution and the OD600 of the top 1 ml (unmixed) as a percentage of the whole (mixed) solution OD600.
The influence of SigL expression on C. difficile biofilm formation was evaluated using a biofilm assay as previously reported (Pantaléon et al., 2015) with modifications. Overnight C. difficile cultures were diluted 1:50 in BHI broth and distributed into 12-well plates, 2 ml per well. Plates were wrapped to prevent evaporation and incubated anaerobically at 37°C for 72 h. Following incubation, the culture supernatant was removed and the biofilms were washed twice with PBS and then incubated at 37°C with 0.2% crystal violet to fix and stain. The crystal violet was subsequently removed, and the biofilms were washed an additional two times with PBS and photographed. To measure biofilm formation, crystal violet retained by the biomass was released with acetone/ethanol and quantitated by taking absorbance measurements at 570 nm in a technical quadruplicate for three individual wells.
C. difficile attachment to human colonic epithelial cells was performed using the modified methods of Merrigan et al. (2013). C2BBe1 cells were obtained from the American Type Culture Collection (ATCC) and were grown to confluent monolayers in 5% CO2 in DMEM +10% fetal bovine serum, 20 μM HEPES, and 100 IU/mL Penicillin and Streptomycin in 6 well tissue culture plates. Twenty-four hours prior to the attachment assay, cells were washed and serum-free DMEM was applied. All additional assay solutions used were pre-reduced in the anaerobic chamber for at least 14 h. C. difficile strains to be analyzed were subcultured from overnight growth in BHI 1:50 into BHI and grown to an OD600 = 0.4 under anaerobic conditions. C2BBe1 cells were enumerated and multiplicity of infection (MOI) was calculated for each individual strain tested with a target of MOI = 50. C. difficile was harvested at OD600 = 0.4 and centrifuged for 5 min at 3,200 × g to remove BHI. C. difficile pellets and confluent monolayers were brought into the anaerobic chamber. C. difficile pellets were resuspended in pre-reduced serum-free DMEM+25 mM CaCl2 and placed on the monolayers to adhere for a total of 40 min at 37°C anaerobically. The inoculum was serially diluted and titered on BHIS with antibiotics as appropriate to calculate MOI. Following attachment, the supernatant was removed, and the monolayers were washed twice with pre-reduced PBS to remove non-adherent bacteria. Monolayers were resuspended in 1 ml PBS and scraped up to quantify attachment. Recovered C. difficile were serially diluted in PBS and plated on BHIS with antibiotics as appropriate to enumerate adherent levels of bacteria. The percent adherence was calculated as the ratio of recovered C. difficile to input C. difficile, multiplied by 100.
To determine the kinetics of C. difficile sporulation, overnight cultures of C. difficile in BHIS were subcultured 1:50 into BHI and grown to an OD600 = 0.4. These cultures were used to inoculate 50 ml of BHI per time point analyzed. At each time point, cultures were harvested by centrifugation for 10 min at 3,200 × g, and the supernatant was removed. Cell pellets were washed once in 1 ml PBS, centrifuged, and resuspended in 1 ml PBS prior to heat-shock for 20 min at 60°C. Following heat shock to inactivate remaining vegetative cells, the remaining spores were serially diluted in PBS and plated on BHIS-Taurocholate plates. Plates were incubated anaerobically for 24 h at 37°C and enumerated. Sporulation efficiency was calculated as the percentage of spores obtained per total terminal vegetative cell count (Merrigan et al., 2013).
A quantitative assessment of C. difficile motility was performed as previously described (Baban et al., 2013). Briefly, overnight cultures of C. difficile grown in BHIS were diluted 1:50 in fresh BHI and grown to mid-exponential growth phase (OD600nm = 0.6) at 37°C in anaerobic conditions. Five μL of culture was then stab inoculated into the center of one well of a 6-well tissue culture plate filled with 20 ml of C. difficile motility agar (BHI+0.3% agar). Culture plates were wrapped in plastic wrap, placed into a container with damp paper towels to prevent evaporation, and grown anaerobically for 72 h at 37°C. Following incubation, the diameter of the swim radius was measured and averaged. Results represent the average of three independent experiments.
C. difficile toxin in the culture supernatant was detected by immunoblotting using monoclonal antibodies against both TcdA and TcdB. C. difficile cultures were grown in 35 ml quantities four times as indicated, and vegetative cells were removed by centrifugation at 4 °C for 10 min at 3,200 × g. Thirty ml of culture supernatant was harvested and concentrated 1:50 on a 100 kDa molecular weight cut-off column (EMD Millipore, Billerica, MA). Concentrated proteins were quantitated using a standard BCA assay (ThermoFisher, Waltham, MA), and 50 μg of protein was loaded onto 7.5% TGX acrylamide gels (BioRad, Hercules, CA) for electrophoresis. Proteins were transferred to nitrocellulose membranes (BioRad) and TcdA and TcdB were immunoblotted using specific monoclonal antisera (Abcam) at 1:5,000 dilution. Samples were detected using a secondary horseradish-peroxidase conjugated rabbit anti-mouse IgG antibody (BioRad) at 1:10,000 concentration. Antibodies were detected using FemtoWest Max Sensitivity substrate (ThermoFisher).
The Alere Wampole A/B Toxin ELISA kit (Alere, Atlanta GA) was utilized to determine relative levels of combined TcdA and TcdB in the supernatant of C. difficile culture. Strains were grown for 72 h in BHI broth, and the supernatant was harvested by centrifugation. Supernatants were diluted 1:10 and 1:100 in fresh BHI and processed for ELISA as per the manufacturer's instructions to determine the appropriate working concentration. Prior to analysis, the amount of total protein was normalized using the BCA assay (Pierce) to allow direct comparison of toxin levels among different C. difficile strains. Following assay processing as per the manufacturer's instructions, quantification of C. difficile toxin was performed by reading absorbance at 450 nm using an automated microtiter plate reader (Bio-Tek, Winooski VT).
The Excel-Stat application was used for statistical analysis. Student's t-tests were used to compute differences between two groups, and error bars were calculated from standard deviation(s). ANOVA was used to compare >2 groups, followed by Tukey's honest significant difference (HSD) test for post-hoc analysis.
The C. difficile BI-1 (RT027) sigL ortholog is located immediately downstream from a putative purine biosynthesis locus (Figure 1A). The encoded 454 amino acid protein includes conserved sigma-54 DNA-, RNA polymerase-, and transcriptional activator-binding domains. sigL is conserved among all sequenced C. difficile genomes, and the predicted amino acid sequence is identical between BI-1 and CDC1 strains.
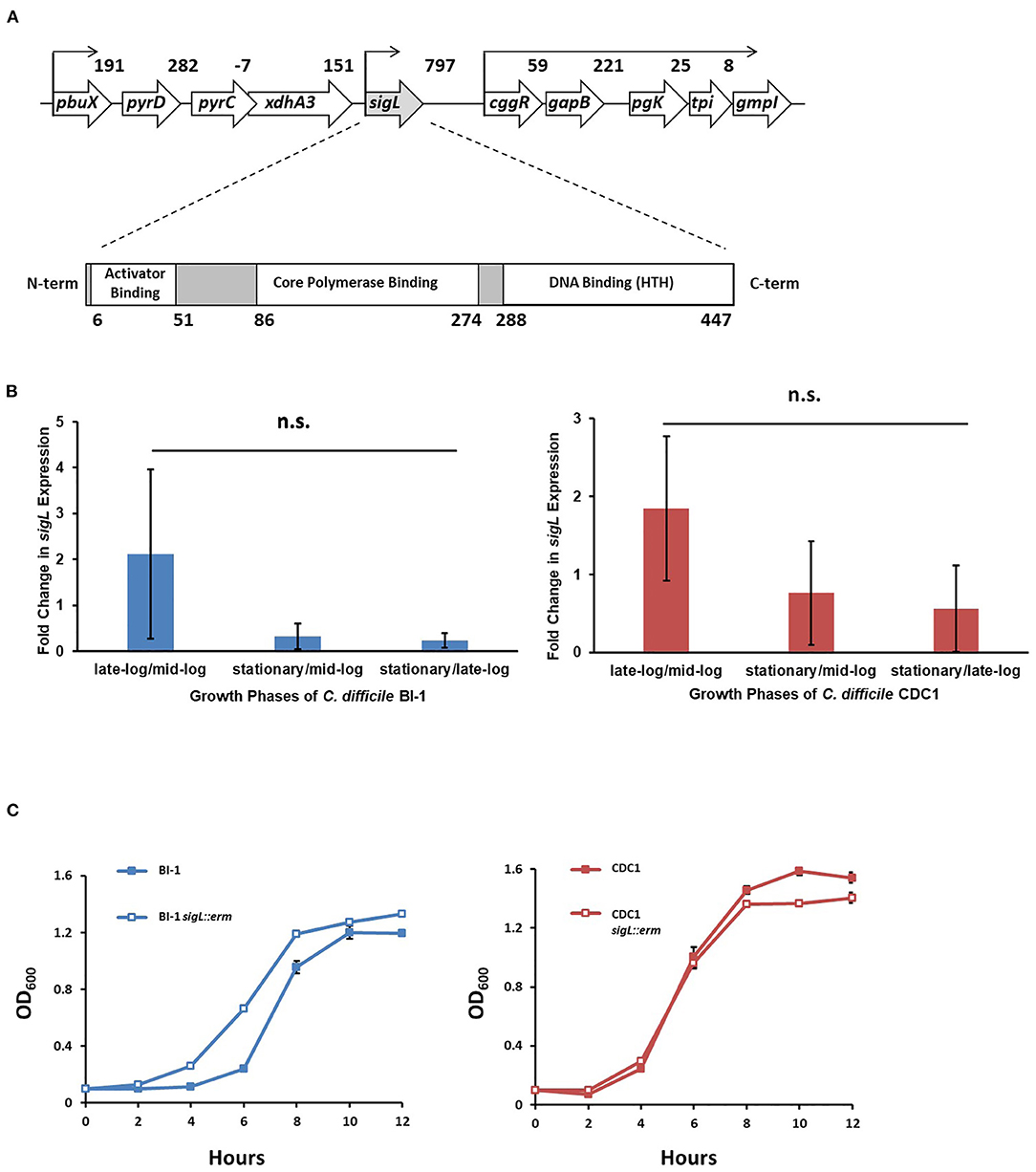
Figure 1. sigL loci, expression, and growth kinetics in C. difficile BI-1 and CDC1. (A) C. difficile sigL locus, showing predicted promoters, based on strain BI-1 genome sequence, and SigL conserved binding domains. Numbers in the upper schematic represent intergenic distances, and numbers below the lower schematic indicate amino acids. (B) Quantitative RT-PCR of sigL in BI-1 and CDC-1 across multiple growth phases. 16s was used as a housekeeping control for qRT-PCR. Fold changes in expression represent the average of three biological replicates with standard deviation. n.s. = not significant (C) Growth of BI-1 and CDC1 strains compared to respective sigL::erm mutants. Cultures were grown in triplicate.
During C. difficile growth, sigL expression was highly variable. While not significantly different between growth phases, expression trends showed the highest expression in the late-exponential phase, and lowest in early growth- and stationary phases. Importantly, the expression of sigL was not restricted to a specific phase of growth. These expression trends were similar in BI-1 and CDC1 strains (Figure 1B, Supplementary Figure S1). To interrogate the contribution of sigL to C. difficile biology, a lactococcal retrotransposition approach was used to generate isogenic mutants and corresponding complements in strains BI-1 and CDC1 (Kuehne and Minton, 2012) (Supplementary Figure S2). While the sigL::erm strains did not exhibit viability defects, they displayed modest, distinct, and reproducible differences in growth kinetics relative to the isogenic parent strains (Figure 1C).
Soutourina and colleagues identified thirty SigL-dependent promoter sequences controlling 95 genes in the C. difficile strain in the laboratory-adapted 630Δerm strain, using strain 630 as a reference genome. Twenty-four of the identified genes or operons were adjacent to enhancer-binding protein (EBP) genes involved in sigma-54-dependent activation (Soutourina et al., 2020). They also noted that transcription levels of genes or operons in 12 of the 30 putative sigL promoter sequences were downregulated in microarray analysis of the C. difficile 630Δerm sigL::erm mutant (GEO accession GPL10556). We used the 12 putative sigL promoter sequences whose genes or operons were downregulated to generate a sigL positional weight matrix (PWM); this was used for promoter-proximal SigL binding site determination by employing the Find Individual Motif Occurrences (FIMO) tool (Grant et al., 2011).
Consistent with Soutourina et al. (2020), FIMO identified all 30 putative sigL promoter sites in both the 630 and BI-1 genomes, and notable differences from these two strains in the RT078 isolate, CDC1 (Table 1, Supplementary Table S1). First, only 28 of the 30 sites were conserved in CDC1; CD0166-CD0165 (peptidase, amino acid transporter) and CD2699-CD2697 (membrane protein, peptidase) operons and their respective sigL promoter sequences were either missing or disrupted relative to 630 and BI-1 (Table 1). Second, the CD0284-CD0289 operon (PTS Mannose/fructose/sorbose family) exhibited a substantially lower FIMO score compared to 630 and BI-1, with an atypical G residue at position 18 of the PWM. Similarly, CD3094 (sigma-54-dependent transcriptional regulator) has an atypical G residue at position 17 in the CDC1 promoter site (Table 1). Third, all three strains exhibited unique insertions/deletions in the sigL-dependent prd operon, linked to proline metabolism (Supplementary Figure S3). CD3246, a putative surface protein, is inserted between prdC and prdR in C. difficile 630, and is absent or truncated in BI-1 and CDC1. prdB, encoding a proline reductase, is present in 630 and BI-1 but truncated and missing a functional domain in CDC1 (Supplementary Figure S3). Of the previously identified EBPs for sigL-dependent promoters, all 24 identified in C. difficile 630 (Nie et al., 2019; Soutourina et al., 2020) were conserved in BI-1, while two EBPs (CD0167 and CD2700) were missing in CDC1 (data not shown).
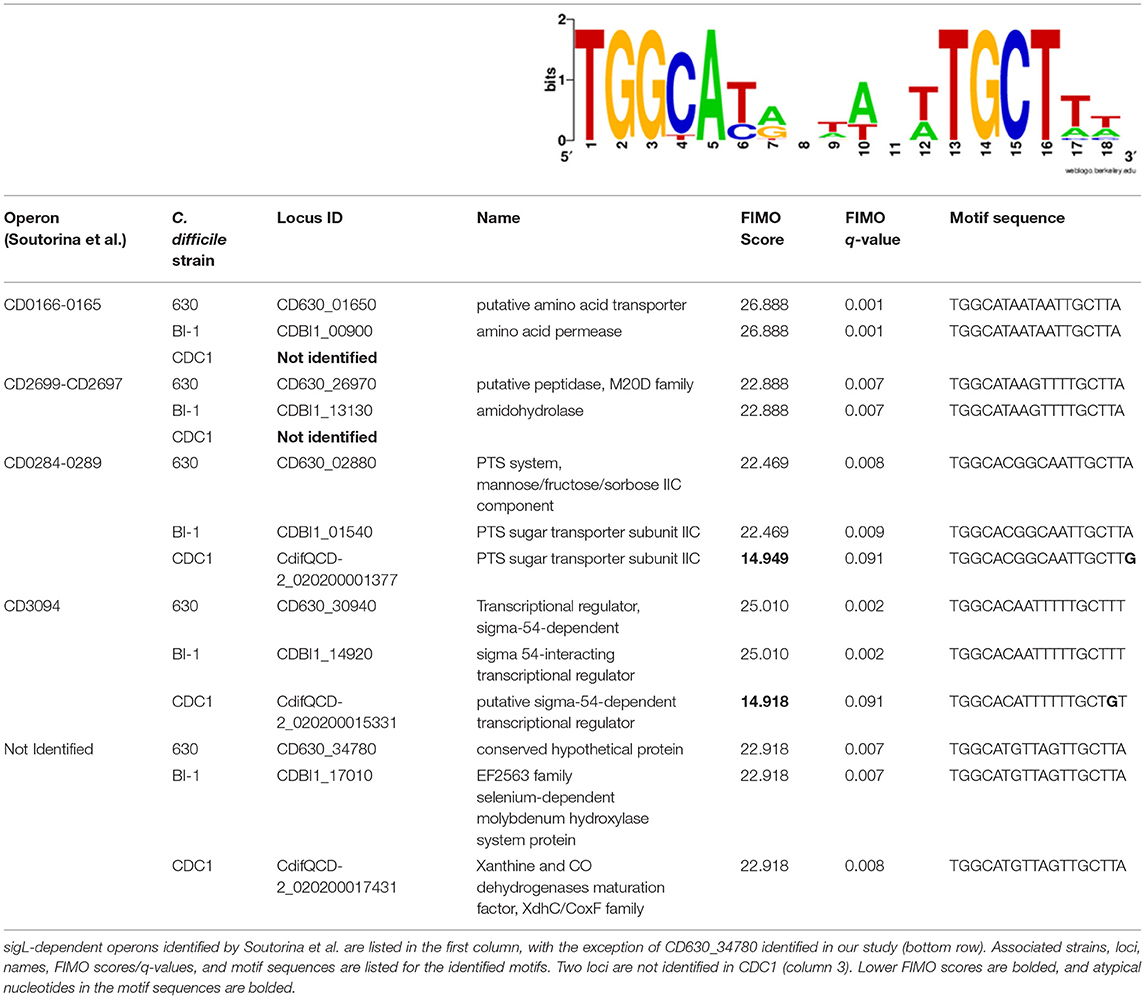
Table 1. sigL positional weight matrix and sigL motif sequences with FIMO scores in C. difficile strains 630, BI-1, and CDC1.
To gain a comprehensive appreciation of SigL-dependent regulation in ribotype RT027 and RT078 C. difficile strains, total protein abundances of BI-1, CDC1, and their respective sigL::erm derivatives were quantitated at mid-exponential (OD600.6 0.6), late-exponential (OD600.8 0.8), and stationary (OD600 1) growth phases using quantitative mass spectrometry-based proteomics, and multiple stringent statistical tests. Overall, relative to the isogenic parent strains, more proteins were differentially abundant in CDC1 sigL::erm than in BI-1 sigL::erm across all three growth phases, but especially during late-exponential growth (Figure 2, Supplementary Table S2). For BI-1, 335 of 1426 identified proteins were altered in abundance in sigL::erm in at least one growth phase relative to WT. This corresponds to 24.9% of all expressed proteins for this strain. For CDC1, 516 proteins were altered in abundance in sigL::erm, corresponding to 73.5% of all expressed proteins for this strain. There was also a marked decrease in the abundance of proteins implicated in isoleucine fermentation and short-chain fatty acid metabolism in sigL::erm for both strains, and across all growth phases (Supplementary Table S2). Based on the markedly different protein profiles between the two strains, we further explored strain-specific SigL-dependent regulation of C. difficile physiology, including cell surface-related phenotypes, sporulation kinetics, biofilm formation, and toxin production.
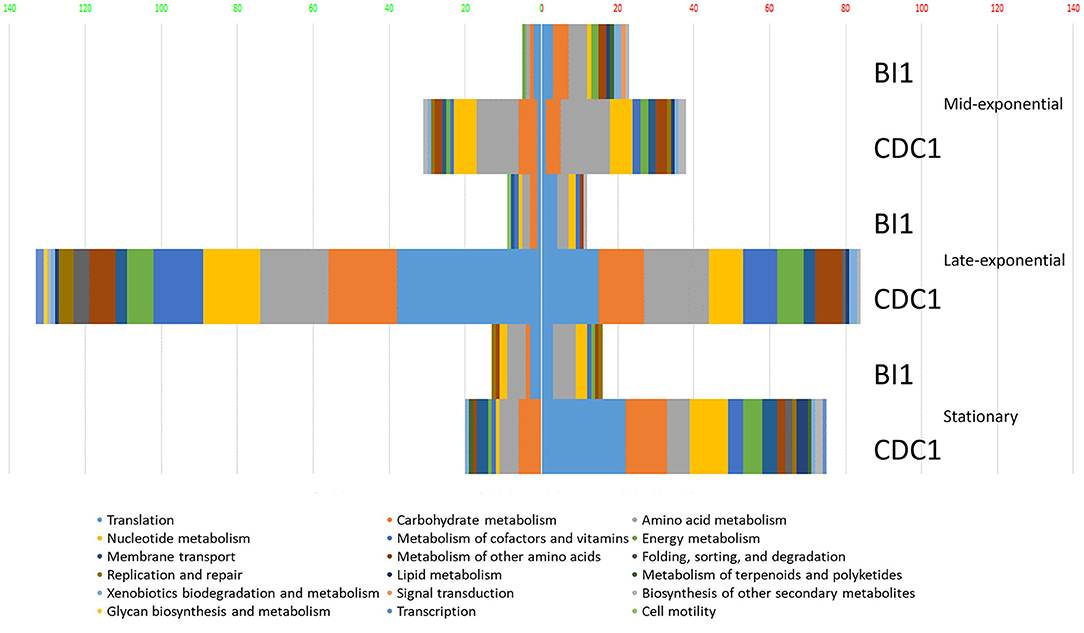
Figure 2. Functional classification of dysregulated proteins in C. difficile strains BI-1 and CDC1 resulting from sigL disruption. Functional classification analysis was performed on proteins identified as altered in sigL disruption mutants using an arbitrary fold change cut-off of 2. The x-axis is the number of dysregulated proteins. Numbers on the left in (green) are decreased in abundance, on the right (red) are increased in abundance. The colors of the columns represent different functional categories based on RAST classification system.
Relative to the BI-1 WT strain, sigL::erm had 47 surface-associated proteins with altered abundance (21 upregulated, 28 downregulated), and for CDC1, sigL::erm had 61 surface-associated proteins with altered abundance (44 upregulated, 34 downregulated) (Supplementary Table S3). Proteomics findings also revealed that dysregulated molecules included those predicted to be involved in S-layer display, host-cell adhesion, antimicrobial peptide resistance, and transport (Supplementary Table S3). Therefore, we probed the surface properties of the sigL::erm mutants relative to the respective isogenic parent strains.
PSII, an antigenically-important surface polysaccharide unique to C. difficile strains, is hypothesized to assemble on the inner face of the cell membrane and translocated to the cell surface via the action of a flippase, MviN (Willing et al., 2015; Chu et al., 2016). Perturbations in PSII biogenesis and/or cell surface display result in diminished extractability of this molecule (Chu et al., 2016). Our proteomics findings revealed alterations in abundance of cell wall proteins Cwp2 and SlpA, to which PSII binds (Willing et al., 2015). Immunoblotting revealed diminished surface-extractable PSII in BI-1 sigL::erm compared to the isogenic parent strain. In contrast, however, PSII was more readily released from CDC1 sigL::erm strain relative to the isogenic parent (Figure 3). Thus, SigL influences expression, export and/or assembly of key C. difficile surface molecules in a strain-specific manner.
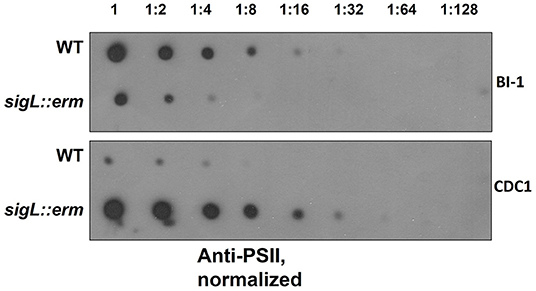
Figure 3. PSII abundance in sigL::erm differs with respect to wild-type strain BI-1 and CDC1. Serially-diluted cell extracts of WT and sigL::erm derivatives (normalized to OD600 = 1.2 in BHI) were spotted on PVDF membranes and immunoblotted using PSII-specific antiserum.
Changes in bacterial surface architecture impact autolysis susceptibility; this has implications for toxin release in C. difficile (Tsuchido et al., 1990; Wydau-Dematteis et al., 2018; Mayer et al., 2019). Surface composition dictates the ease with which detergents, such as Triton X-100, interact with lipoteichoic acid from Gram-positive bacterial cell walls, which in turn influences the activation of autolytic enzymes (Komatsuzawa et al., 1994; Fujimoto and Bayles, 1998). To establish the influence of SigL on C. difficile cell surface properties, we quantitatively measured rates of autolysis between wild type and sigL::erm mutants of both BI-1 and CDC1. While autolysis rates were similar between the BI-1 WT and sigL::erm strains at early time points, the mutant demonstrated decreased autolysis at later time points. In contrast, the CDC1 sigL::erm strain exhibited a more rapid rate of autolysis than the WT strain, and underwent complete autolysis in 90 min (Figures 4A,B).
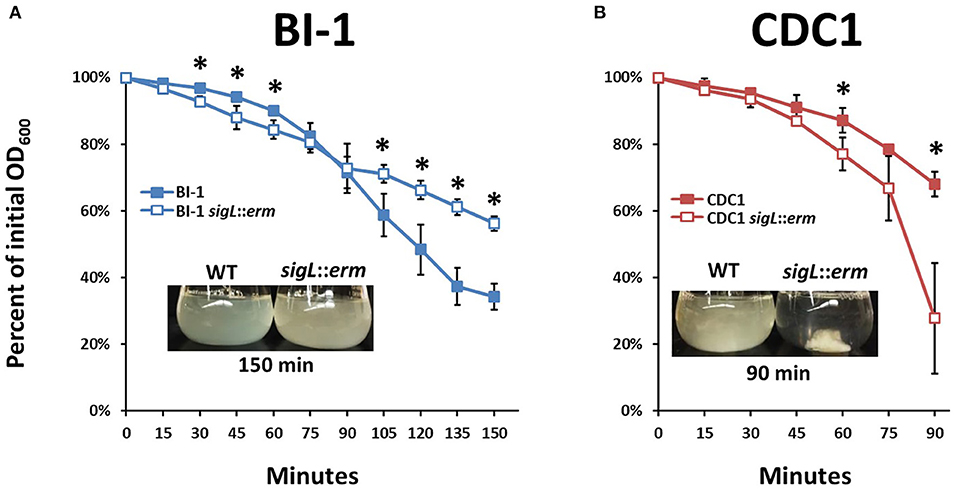
Figure 4. Autolysis rates of C. difficile strains and sigL mutants differ between BI-1 and CDC1. Cultures were grown to OD600 level of approximately 1.2, and OD600 values were measured as percent of initial OD over time following the addition of Triton X-100. Visualization of autolysis in (A). BI-1 and (B). CDC1 compared to respective sigL::erm mutants are show as insets. Each assay was performed in biological triplicate. *p < 0.05.
Changes in cell surface protein composition, including variation in charge and hydrophobicity, can dictate bacterial aggregation which, in turn, can influence virulence (Misawa and Blaser, 2000). Therefore, we monitored aggregation rates of both BI-1 and CDC1 sigL::erm mutants during growth in Brain Heart Infusion (BHI). While the aggregation rate of C. difficile BI-1 and the isogenic sigL::erm remained unchanged throughout growth, CDC1 sigL::erm was more aggregative than the parent WT strain during late-exponential and stationary growth phases (Figures 5A,B). These phenotypes were independently verified via autoagglutination assays using plate-grown cultures. Again, CDC1 sigL::erm agglutinated significantly more than the cognate wildtype strain at multiple time points (Figure 5C). Autoagglutination rates of the BI-1 sigL::erm were not different from wild type with the exception of two time points at 8 and 10 h (Figure 5C).
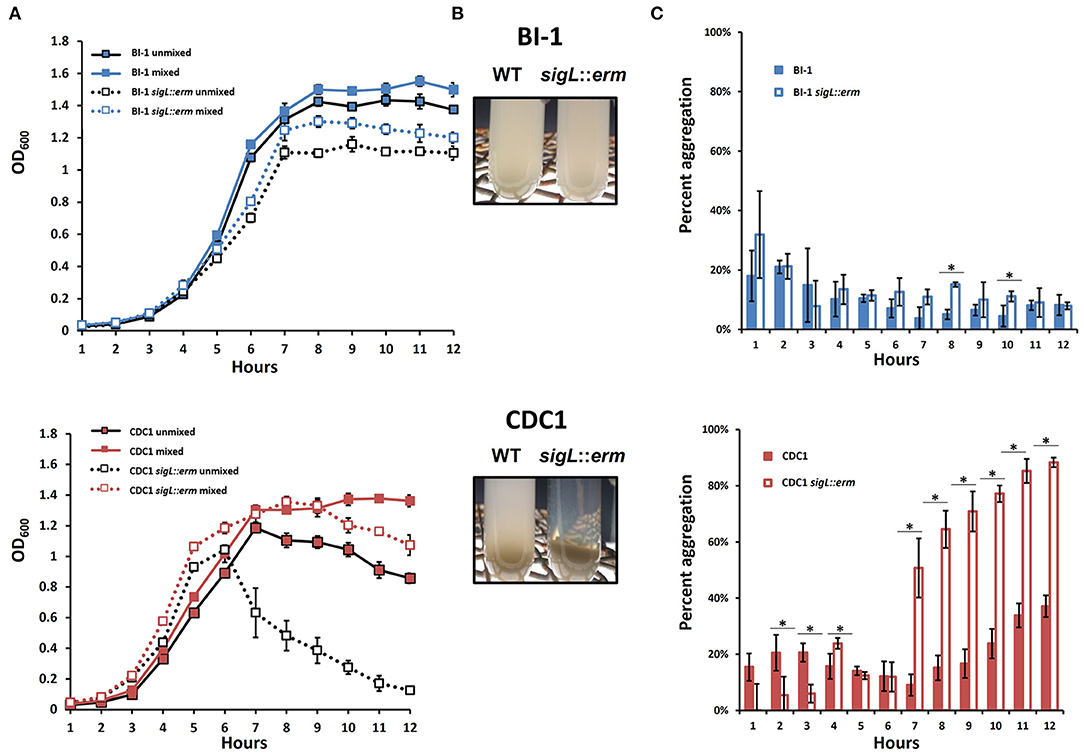
Figure 5. Aggregation of C. difficile strains and respective sigL::erm mutants differs between BI-1 and CDC1. (A) OD600 values of the top 1 ml of culture (unmixed) and a replicate vortexed culture sample (mixed) were read hourly to determine sedimentation curves. (B) Visualization of aggregation in BI-1 (top) and CDC1 (bottom) WT and sigL::erm strains.(C) Percent aggregation of C. difficile strains and respective sigL::erm mutants, calculated as the difference in OD600 values between the whole (mixed) culture and top 1 ml (unmixed) culture, as a percentage of the mixed culture OD600 value. Cultures were grown in biological triplicate. WT and sigL::erm pairs were compared at each time point using Student's t-test. *p < 0.05.
Altered surface properties can influence the ability to form biofilms; in C. difficile, this can impact pathogen persistence in the gut (Pantaléon et al., 2015; Fernández Ramírez et al., 2018; Chamarande et al., 2021; JaneŽ et al., 2021; Kumari and Kaur, 2021). Disruption of SigL had no discernable impact on the ability of BI-1 strains to form biofilms grown in liquid culture on polystyrene plates (Figure 6A). In contrast, CDC1 sigL::erm had decreased biofilm density relative to the isogenic parent strain (Figure 6B). Complementation restored the CDC1 phenotype (Supplementary Figure S4A), and overexpression of SigL enhanced the ability of the CDC1 strain, but not BI-1, to form a biofilm (Supplementary Figures S4B,C).
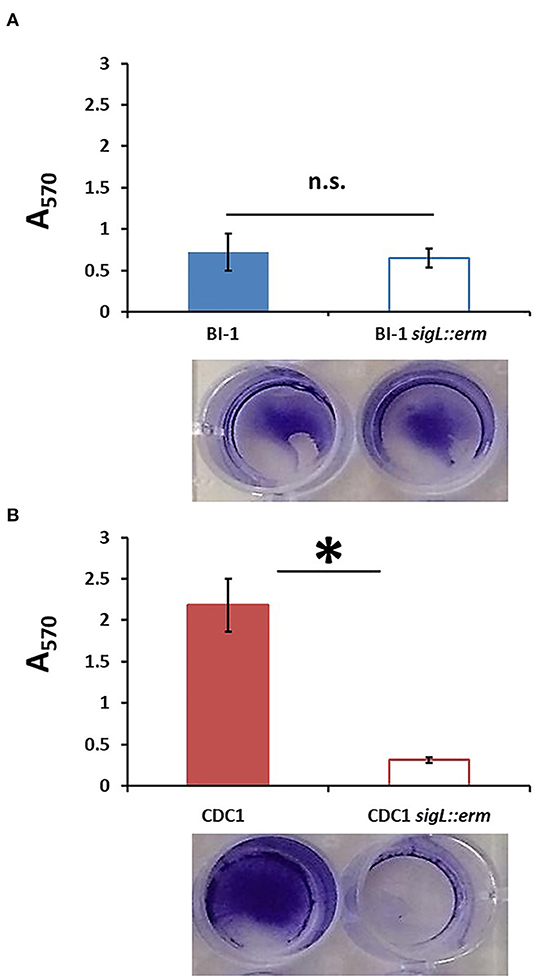
Figure 6. Biofilm formation is reduced in CDC1 sigL::erm, but not in BI-1 sigL::erm. Biofilm was quantified by the release of crystal violet retained by the C. difficile biomass after 72 h. Absorbance was measured at 570 nm. (A) Biofilm formation in BI-1 and BI-1 sigL::erm. (B) Biofilm formation in CDC1 and CDC1 sigL::erm. Samples were grown in biological triplicate and compared using Student's t-test. *p < 0.05.
Variations in cell surface composition leading to alterations in phenotypic properties, including aggregation, can affect bacterial attachment to host cells (Burdman et al., 2000; Merrigan et al., 2013; Foster, 2019), and this has profound implications for C. difficile host establishment (Vedantam et al., 2018). We, therefore, compared the adherence of parental and sigL mutant strains to Caco-2BBe intestinal epithelial cells. Two different growth phases were chosen for analysis representing differential protein expression in sigL::erm strains (Figure 7): OD600 = 0.4, mid-exponential, and OD600 = 1.2, stationary. For mid-exponential phase bacteria, compared to the respective parent strains, BI-1 sigL::erm exhibited diminished levels of attachment, whereas CDC1 sigL::erm attachment was more robust (Figure 7A). For bacteria in the stationary phase, there was no apparent difference in attachment between the WT and the mutant in the BI-1 background, but CDC1 sigL::erm was less adherent to host cells relative to the isogenic WT strain (Figure 7B). Thus, the contribution of SigL toward the ability of C. difficile to attach to biotic surfaces is both strain- and growth phase-dependent.
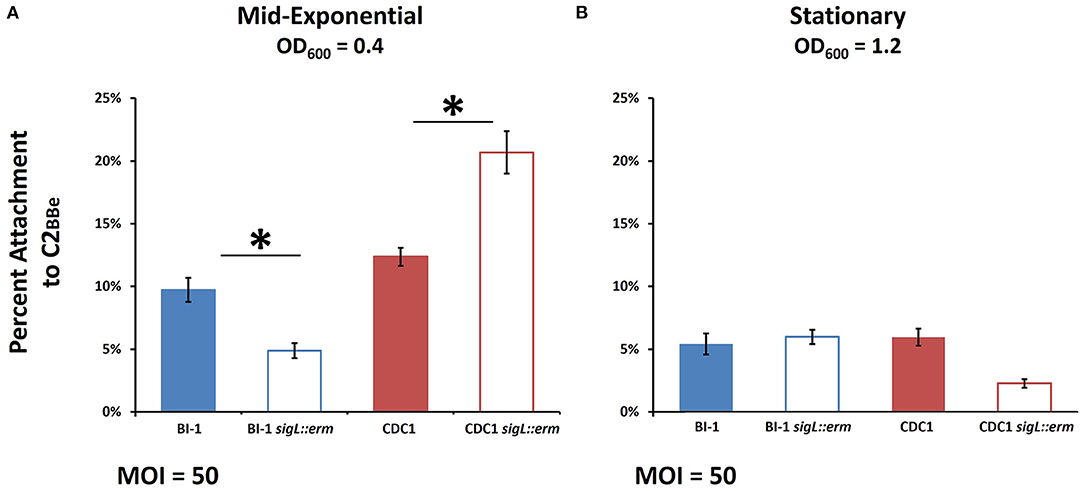
Figure 7. Host cell attachment in sigL::erm is phase- and ribotype-dependent. C. difficile cultures were grown to mid-exponential or stationary phase and added to Caco-2BBe intestinal epithelial cell monolayers. Percent attachment was calculated by adhered CFUs as a percentage of the initial inoculum CFUs. (A) Attachment to Caco-2BBe intestinal epithelial cells in mid-exponential phase. (B) Attachment to Caco-2BBe intestinal epithelial cells in stationary phase. The assays were conducted in biological triplicate and samples were compared using ANOVA and Tukey's HSD post-hoc test. *p < 0.05.
C. difficile sporulates in response to a combination of unfavorable extracellular environment and nutritional/metabolic deprivation, and sporulation efficiency can impact bacterial gut persistence (Paredes-Sabja et al., 2014; Shen et al., 2019). We next assessed if loss of SigL influences C. difficile sporulation kinetics, by measuring the outgrowth of preformed spores following the heat shock of vegetative cells BI-1 and BI-1-sigL::erm had comparable spore titer levels over a period of 5 days, indicating that SigL does not influence sporulation processes in this strain (Figure 8A). In contrast, CDC1 sigL::erm had higher spore titers than the WT strain at 24 and 48 h, but the difference resolved at later time points (72–120 h) (Figure 8B). Sporulation efficiency was also calculated in our C. difficile strains and respective sigL::erm derivatives, with spore titers as a percent of terminal vegetative cell titers. These data show higher sporulation efficiencies at earlier time points in the CDC1 sigL::erm strain (Supplementary Figure S5), commensurate with the higher spore titers observed in Figure 8B; these also demonstrate a SigL-dependent effect on sporulation efficiency in CDC1 that is absent in BI-1 (Supplementary Figure S5). Plasmid-driven SigL overexpression, using pMTL82153, which expresses sigL under a constitutive promoter, reduced sporulation of both the CDC1- and BI-1-sigL::erm mutants, as well as the corresponding WT strains (Figures 8C,D).
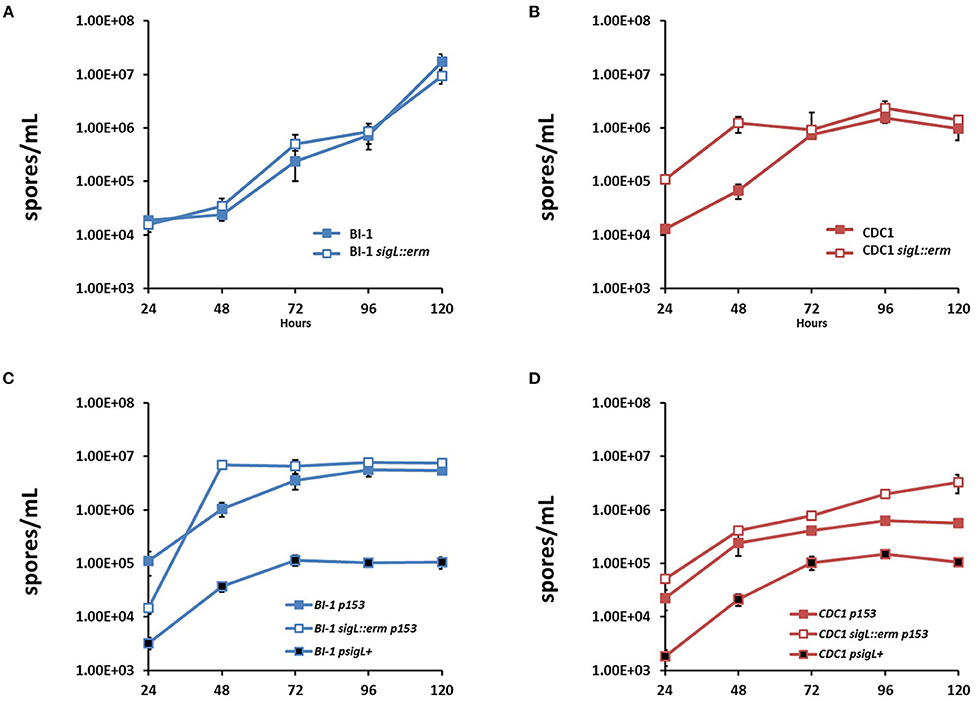
Figure 8. Sporulation kinetics are altered in sigL::erm in a ribotype-dependent manner. Spores were enumerated by heat shocking and plating cultures on BHIS-Taurocholate at each time point. For overexpression, WT C. difficile strains were conjugated with empty vector pMTL82153 (p153). sigL::erm in BI-1 and CDC1 backgrounds were conjugated with empty vector or p153 expressing sigL under a constitutive promoter (psigL+). (A) Spore enumeration over 120 h in BI-1 and BI-1 sigL::erm strains. (B) Spore enumeration over 120 h in CDC1 and CDC1 sigL::erm strains. Cultures were grown in biological triplicate. (C) Spore enumeration over 120 h in BI-1 sigL overexpression strains. (D) Spore enumeration over 120 h in CDC1 sigL overexpression strains. Cultures were grown in biological triplicate.
We next assessed if the increased sporulation phenotype of CDC1 sigL::erm could be reversed by plasmid-complementation, using pMTL82151, which expresses sigL under a native promoter. Plasmid (empty vector)-harboring WT- and mutant strains recapitulated the sporulation phenotypes for CDC1 though differences in overall kinetics were observed (likely due to the presence of the antibiotic used for plasmid maintenance). This was reversed by a sigL-encoding plasmid (Supplementary Figure S6). Spore production was also reduced in BI-1 with plasmid complementation.
Sigma-54 is widely recognized as an important regulator of flagellar biosynthesis in many Gram-negative species, but its role is non-uniform among different families of organisms (Tsang and Hoover, 2014). To evaluate the role of SigL in the regulation of flagellar synthesis in C. difficile, motility assays were performed using the motile strain BI-1. CDC1 is aflagellate due to the loss of a substantial part of the flagellar locus encoding early structural flagellar genes (Stabler et al., 2009), a hallmark of the RT078 ribotype, and was used as a negative control. In contrast to other bacterial systems whereby motility is influenced by sigma-54 expression, the motility and flagellar expression of C. difficile BI-1 were not significantly affected by sigL inactivation (Supplementary Figure S7).
C. difficile toxin expression is influenced by both metabolic factors and genetic determinants (Bouillaut et al., 2015; Martin-Verstraete et al., 2016; Ransom et al., 2018). Given the wide-ranging impact of SigL presence on C. difficile metabolic enzymes, we used immunoblot analysis to monitor TcdA and TcdB in WT and mutant culture supernatants from 24 to 72 h. Loss of SigL dramatically increased supernatant TcdA and TcdB levels in BI-1 but decreased the levels of the two toxins in the CDC1 background (Figure 9). This was confirmed by the toxin ELISA (Supplementary Figure S8). Correspondingly, complementation decreased supernatant toxin levels by BI-1 sigL::erm, but increased TcdA and TcdB levels in the CDC1 background (Supplementary Figure S8).

Figure 9. Supernatant toxin level is differentially altered in sigL::erm in BI-1 and CDC1 backgrounds. BI-1 and CDC1 C. difficile strains and respective sigL::erm mutants were grown for 24, 48, and 72 h. Vegetative cells were removed by centrifugation and the supernatant was assayed for toxins by immunoblotting using monoclonal antibodies against TcdA and TcdB. Blots are representative of three experiments.
Although rarely an essential protein, no unified functional theme has been identified concerning the sigma-54 dependent metabolic repertoire. This is in contrast with the sigma-70 counterpart, which often serves dedicated functions. Sigma-54-dependent proteins have pleiotropic functions in bacterial systems, with roles ranging from carbohydrate and nitrogen metabolism to modification of cell surface structures, including flagella and pili (Alm et al., 1993; Francke et al., 2011; Hayrapetyan et al., 2015). In Gram-positive organisms, the Sigma-54, SigL, regulates adaptation to environmental conditions, including cold shock (Wiegeshoff et al., 2006), osmotic stress (Okada et al., 2006), and oxidative stress (Stevens et al., 2010), and is involved in amino acid catabolism (Debarbouille et al., 1999), acetoin cycle regulation (Ali et al., 2001; Francke et al., 2011), and flagellar synthesis (Hayrapetyan et al., 2015).
A previous report revealed that C. difficile sigL transcript levels were highest during the stationary phase of growth (Karlsson et al., 2008). In this study, we observed a trend of higher SigL expression during the late-exponential phase, though the expression was present and highly variable during all phases of growth, indicative of the biological relevance of SigL throughout the C. difficile life cycle. Due to its requirement for activation prior to transcription, it is likely C. difficile SigL function is regulated through the coordinated expression of EBPs, driving SigL transcription of target operons when physiologically necessary. The number of EBPs roughly correlates with genome size, with C. difficile strains 630 and BI-1 harboring 23 such genes, two of which are absent in CDC1 (Nie et al., 2019).
Promoters recognized by members of the sigma-54 family have highly conserved −24 and −12 regions upstream of the transcriptional start site, including a GC dinucleotide at position −12 and GG at position −24 (Barrios et al., 1999; Zhang and Buck, 2015). Barrios et al. identified 85 SigL promoter sequences in multiple bacterial species, with the consensus sequence “TTGGCATNNNNNTTGCT” used to inform more recent studies to search for promoter candidates in Clostridiales genomes (Barrios et al., 1999; Nie et al., 2019). While C. difficile SigL interaction with specific promoters was described previously (Nie et al., 2019; Soutourina et al., 2020), the relative strength of the promoters, or the essentiality of specific residues in the promoter for interaction, has not been defined.
We identified SigL-dependent promoter sequences in BI-1 and CDC1 strains in agreement with previous studies in C. difficile 630Δerm, using strain 630 as a reference strain, with SigL-dependent regulation of genes involved in amino acid metabolism and sugar transport, in addition to novel promoter sequences not identified in previous 630 studies (Soutourina et al., 2020). In BI-1, a promoter sequence for an EF2563 family selenium-dependent molybdenum hydroxylase (CD630_3478/CDBI1_17010) was uniquely identified. This same EF2563 protein was identified in CDC-1 (CdifQCD-2_020200017431) (Table 1, Supplementary Table S1). Other strain-specific differences noted in the repertoire of SigL regulated genes included the absence of two SigL-dependent promoters in CDC1 compared to BI-1 and 630, corresponding to an amino acid transporter and a membrane protein/peptidase, respectively. Additionally, a G residue replaces the typical A or T at position 18 in the positional weight matrix (PWN) for the CD0284-CD0289 operon (PTS Mannose/fructose/sorbose family) and position 17 for the CD3094 operon, resulting in a lower FIMO score suggestive of decreased SigL interaction. These unique promoter features noted for BI-1 and CDC-1 extended to other published sequences of strains belonging to the RT027 and RT078 lineages, respectively.
In our proteomics analysis of BI-1 and CDC1, we observed profoundly pleiotropic, strain-specific, and growth-phase-specific SigL-dependent regulation. In the late-exponential phase, 224 proteins were downregulated in the CDC1::erm strain, compared to only 76 downregulated proteins in the BI-1 background for the same growth phase (Supplementary Table S2). Our proteomics data showed that several hydroxyisocaproyl-CoA dehydratase (Had) proteins, associated with L-leucine fermentation, were downregulated in sigL::erm in both strain backgrounds and across all growth phases, consistent with established roles for sigma-54 in amino acid fermentation (Gardan et al., 1995, 1997; Debarbouille et al., 1999). Our data also show that SigL-dependent protein regulation is not only highly pleiotropic within a strain, but widely differential between C. difficile 027 and 078 strains.
The SigL regulon is remarkably distinct between BI-1 (RT027) and CDC1 (RT078), and this is manifested via distinct phenotypes. Such differences have been shown for C. difficile strains R20291 (RT027) and 630Δerm (RT012) specifically in the context of flagellar protein mutants (Baban et al., 2013), but comparative studies for sigma-54 mutants are lacking. A recent large-scale study identified evolutionary relationships between sigma-54 genes and the synthesis and transport of elements of the bacterial cell surface and exterior, including exopolysaccharides, lipids, lipoproteins, and peptidoglycan (Francke et al., 2011). In agreement, several dysregulated protein targets observed between the C. difficile sigL mutants were either components of, or associated with, the cell surface indicating dynamic compositional regulation by SigL. Phenotypic analysis of the sigL mutants of the two ribotypes confirms strain-specific surface alterations. Differential cell surface phenotypes with respect to aggregation, attachment to biotic surfaces, exopolysaccharide levels, and autolysis were observed.
Metabolic events and environmental conditions encountered during the stationary phase determine whether C. difficile vegetative cells will form biofilms, produce toxins and/or initiate sporulation. In this work, a strong correlation was observed between the expression of SigL, sporulation, biofilm, and toxin phenotypes. In CDC1, there is an inverse association between sporulation and toxin production: in CDC1 sigL::erm, spore titers and sporulation efficiency are increased, and biofilm and toxin accumulation in the supernatant are decreased relative to the parent strain. In BI-1, the relationship appears more complex. Sporulation and biofilm formation are not influenced by the loss of sigL expression in the BI-1 background; therefore, the reason for the accompanying increase in supernatant toxin in the BI-1 sigL mutant remains unknown, but our observed strain-specific differences in SigL-dependent operons may yield some insight. The prd locus is associated with proline metabolism, which along with other amino acids can be metabolized by C. difficile via Stickland reactions. This amino acid metabolism is associated with reduced toxin production (Bouillaut et al., 2013). We identified a SigL-dependent promoter for the prd locus; therefore, a contributing factor to the increased toxin in BI-1 sigL::erm may be differences in SigL-dependent proline metabolism. Protein levels in members of the prd locus, including the proline reductase PrdA and proline racemase PrdF, were shown to be similarly decreased in sigL::erm in both the BI-1 and CDC1 backgrounds (Supplementary Table S2), but interestingly, we observed a difference in the architecture of the prd locus in CDC1 compared to BI-1, with a truncation in prdB (Supplementary Figure S3). How this truncation may impact proline metabolism and toxin production in CDC1 requires further investigation. There are also potential epistatic effects of other regulators, such as SigD, that may affect toxin production.
A recent study in C. difficile 630Δerm showed major proteomic differences in two models of biofilm-like growth: aggregate biofilms and colony biofilms, and demonstrated that SigL was significantly induced in aggregate biofilms (Brauer et al., 2021). The proteomic and phenotypic differences in our respective 027 and 078 sigL::erm mutants may be partially explained by strain-specific differences in the ability to aggregate and form biofilms, resulting in differential SigL induction. Indeed, WT BI-1 shows reduced baseline biofilm formation compared to WT CDC1 (Figure 6).
It is well-appreciated that strain-level variations may influence C. difficile virulence and epidemiology in fundamental ways, but less is known about the regulatory networks underlying these differences. Our studies suggest that the conserved regulator SigL has profoundly differential impacts on gene expression, including toxin production, in the predominantly human- and veterinary-associated C. difficile lineages RT027 and RT078, respectively. Future studies, including animal experiments, will be instructive in determining the consequences of these regulatory differences to disease establishment, progression, asymptomatic carriage, recurrence, and/or response to treatment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CA, AC, and KC performed the experiments. AC and KC analyzed the data and drafted the manuscript. BR performed most of the informatic analyses. VV and GV designed the study, obtained research funding, and supervised all aspects of the study. GV and VV contributed equally as senior authors to this study. All authors approved the submitted version of the manuscript.
This work was supported by funding from the National Institutes of Health [R33AI121590531 (GV)] and the US Department of Veterans Affairs [Research Career Scientist Award IK6BX003789 (GV) and Merit Award I01BX001183 (GV)].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge Aaron Brussels for expert technical assistance, the University of Arizona Mass Spectrometry Consortium for mass spectrometry studies and data compilation, and members of the Vedantam and Viswanathan laboratories for helpful discussions and critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.871152/full#supplementary-material
Ali, N. O., Bignon, J., Rapoport, G., and Debarbouille, M. (2001). Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J. Bacteriol. 183, 2497–2504. doi: 10.1128/JB.183.8.2497-2504.2001
Alm, R. A., Guerry, P., and Trust, T. J. (1993). The Campylobacter sigma 54 flaB flagellin promoter is subject to environmental regulation. J. Bacteriol. 175, 4448–4455. doi: 10.1128/jb.175.14.4448-4455.1993
Baban, S. T., Kuehne, S. A., Barketi-Klai, A., Cartman, S. T., Kelly, M. L., Hardie, K. R., et al. (2013). The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS ONE 8, e73026. doi: 10.1371/journal.pone.0073026
Barrios, H., Valderrama, B., and Morett, E. (1999). Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27, 4305–4313. doi: 10.1093/nar/27.22.4305
Bouillaut, L., Dubois, T., Sonenshein, A. L., and Dupuy, B. (2015). Integration of metabolism and virulence in Clostridium difficile. Res. Microbiol. 166, 375–383. doi: 10.1016/j.resmic.2014.10.002
Bouillaut, L., Self, W. T., and Sonenshein, A. L. (2013). Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 195, 844–854. doi: 10.1128/JB.01492-12
Brauer, M., Lassek, C., Hinze, C., Hoyer, J., Becher, D., Jahn, D., et al. (2021). What's a biofilm?-how the choice of the biofilm model impacts the protein inventory of Clostridioides difficile. Front. Microbiol. 12, 682111. doi: 10.3389/fmicb.2021.682111
Burdman, S., Okon, Y., and Jurkevitch, E. (2000). Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Crit. Rev. Microbiol. 26, 91–110. doi: 10.1080/10408410091154200
Chamarande, J., Cunat, L., Caillet, C., Mathieu, L., Duval, J. F. L., Lozniewski, A., et al. (2021). Surface properties of Parabacteroides distasonis and impacts of stress-induced molecules on its surface adhesion and biofilm formation capacities. Microorganisms 9, 1602. doi: 10.3390/microorganisms9081602
Chu, M., Mallozzi, M. J., Roxas, B. P., Bertolo, L., Monteiro, M. A., Agellon, A., et al. (2016). A Clostridium difficile cell wall glycopolymer locus influences bacterial shape, polysaccharide production and virulence. PLoS Pathog. 12, e1005946. doi: 10.1371/journal.ppat.1005946
Debarbouille, M., Gardan, R., Arnaud, M., and Rapoport, G. (1999). Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181, 2059–2066. doi: 10.1128/JB.181.7.2059-2066.1999
Dehal, P. S., Joachimiak, M. P., Price, M. N., Bates, J. T., Baumohl, J. K., Chivian, D., et al. (2010). MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38, D396–D400. doi: 10.1093/nar/gkp919
Dineen, S. S., Mcbride, S. M., and Sonenshein, A. L. (2010). Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192, 5350–5362. doi: 10.1128/JB.00341-10
Dudukgian, H., Sie, E., Gonzalez-Ruiz, C., Etzioni, D. A., and Kaiser, A. M. (2009). C. difficile colitis—predictors of fatal outcome. J. Gastrointest. Surg. 14, 315–322. doi: 10.1007/s11605-009-1093-2
El Meouche, I., Peltier, J., Monot, M., Soutourina, O., Pestel-Caron, M., Dupuy, B., et al. (2013). Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS ONE 8, e83748. doi: 10.1371/journal.pone.0083748
Elliott, B., Androga, G. O., Knight, D. R., and Riley, T. V. (2017). Clostridium difficile infection: evolution, phylogeny and molecular epidemiology. Infect. Genet. Evol. 49, 1–11. doi: 10.1016/j.meegid.2016.12.018
Faulds-Pain, A., Twine, S. M., Vinogradov, E., Strong, P. C., Dell, A., Buckley, A. M., et al. (2014). The post-translational modification of the Clostridium difficile flagellin affects motility, cell surface properties and virulence. Mol. Microbiol. 94, 272–289. doi: 10.1111/mmi.12755
Feklístov, A., Sharon, B. D., Darst, S. A., and Gross, C. A. (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376. doi: 10.1146/annurev-micro-092412-155737
Fernández Ramírez, M. D., Nierop Groot, M. N., Smid, E. J., Hols, P., Kleerebezem, M., and Abee, T. (2018). Role of cell surface composition and lysis in static biofilm formation by Lactobacillus plantarum WCFS1. Int. J. Food Microbiol. 271, 15–23. doi: 10.1016/j.ijfoodmicro.2018.02.013
Foster, T. J. (2019). Surface proteins of Staphylococcus aureus. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0046-2018
Francke, C., Groot Kormelink, T., Hagemeijer, Y., Overmars, L., Sluijter, V., Moezelaar, R., et al. (2011). Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12, 385. doi: 10.1186/1471-2164-12-385
Fujimoto, D. F., and Bayles, K. W. (1998). Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J. Bacteriol. 180, 3724–3726. doi: 10.1128/JB.180.14.3724-3726.1998
Gardan, R., Rapoport, G., and Débarbouillé, M. (1995). Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 249, 843–856. doi: 10.1006/jmbi.1995.0342
Gardan, R., Rapoport, G., and Débarbouillé, M. (1997). Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 24, 825–837. doi: 10.1046/j.1365-2958.1997.3881754.x
George, R., Symonds, J. M., Dimock, F., Brown, J., Arabi, Y., Shinagawa, N., et al. (1978). Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br. Med. J. 1, 695. doi: 10.1136/bmj.1.6114.695
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Gu, H., Yang, Y., Wang, M., Chen, S., Wang, H., Li, S., et al. (2017). Novel cysteine desulfidase cdsb involved in releasing cysteine repression of toxin synthesis in Clostridium difficile. Front Cell Infect. Microbiol. 7, 531. doi: 10.3389/fcimb.2017.00531
Hayrapetyan, H., Tempelaars, M., Nierop Groot, M., and Abee, T. (2015). Bacillus cereus ATCC 14579 RpoN (sigma 54) is a pleiotropic regulator of growth, carbohydrate metabolism, motility, biofilm formation and toxin production. PLoS ONE 10, e0134872. doi: 10.1371/journal.pone.0134872
JaneŽ, N., Škrlj, B., Sterniša, M., Klančnik, A., and Sabotič, J. (2021). The role of the Listeria monocytogenes surfactome in biofilm formation. Microb. Biotechnol. 14, 1269–1281. doi: 10.1111/1751-7915.13847
Karlsson, S., Burman, L. G., and Åkerlund, T. (2008). Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 154, 3430–3436. doi: 10.1099/mic.0.2008/019778-0
Komatsuzawa, H., Suzuki, J., Sugai, M., Miyake, Y., and Suginaka, H. (1994). The effect of Triton X-100 on the in-vitro susceptibility of methicillin-resistant Staphylococcus aureus to oxacillin. J. Antimicrob. Chemother. 34, 885–897. doi: 10.1093/jac/34.6.885
Kuehne, S. A., and Minton, N. P. (2012). ClosTron-mediated engineering of Clostridium. Bioengineered 3, 247–254. doi: 10.4161/bioe.21004
Kumari, B., and Kaur, J. (2021). Correlation of over-expression of rv1900c with enhanced survival of M. smegmatis under stress conditions: modulation of cell surface properties. Gene 791, 145720. doi: 10.1016/j.gene.2021.145720
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Research 43, D257–D260. doi: 10.1093/nar/gku949
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martin-Verstraete, I., Peltier, J., and Dupuy, B. (2016). The regulatory networks that control Clostridium difficile toxin synthesis. Toxins 8, 153. doi: 10.3390/toxins8050153
Mayer, C., Kluj, R. M., Mühleck, M., Walter, A., Unsleber, S., Hottmann, I., et al. (2019). Bacteria's different ways to recycle their own cell wall. Int. J. Med. Microbiol. 309, 151326. doi: 10.1016/j.ijmm.2019.06.006
Merrick, M. J. (1993). In a class of its own — the RNA polymerase sigma factor σ;54 (σN). Mol. Microbiol. 10, 903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x
Merrigan, M. M., Venugopal, A., Roxas, J. L., Anwar, F., Mallozzi, M. J., Roxas, B. A., et al. (2013). Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One 8, e78404. doi: 10.1371/journal.pone.0078404
Misawa, N., and Blaser, M. J. (2000). Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immunity 68, 6168–6175. doi: 10.1128/IAI.68.11.6168-6175.2000
Neumann-Schaal, M., Jahn, D., and Schmidt-Hohagen, K. (2019). Metabolism the difficile way: the key to the success of the pathogen Clostridioides difficile. Front. Microbiol. 10, 219. doi: 10.3389/fmicb.2019.00219
Nie, X., Dong, W., and Yang, C. (2019). Genomic reconstruction of σ(54) regulons in Clostridiales. BMC Genomics 20, 565. doi: 10.1186/s12864-019-5918-4
Okada, Y., Okada, N., Makino, S.-I., Asakura, H., Yamamoto, S., and Igimi, S. (2006). The sigma factor RpoN (σ54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol. Lett. 263, 54–60. doi: 10.1111/j.1574-6968.2006.00405.x
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pantaléon, V., Soavelomandroso, A. P., Bouttier, S., Briandet, R., Roxas, B., Chu, M., et al. (2015). The Clostridium difficile protease Cwp84 modulates both biofilm formation and cell-surface properties. PLoS ONE 10, e0124971. doi: 10.1371/journal.pone.0124971
Paredes-Sabja, D., Shen, A., and Sorg, J. A. (2014). Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 22, 406–416. doi: 10.1016/j.tim.2014.04.003
Ransom, E. M., Kaus, G. M., Tran, P. M., Ellermeier, C. D., and Weiss, D. S. (2018). Multiple factors contribute to bimodal toxin gene expression in Clostridioides (Clostridium) difficile. Mol. Microbiol. 110, 533–549. doi: 10.1111/mmi.14107
Rappas, M., Bose, D., and Zhang, X. (2007). Bacterial enhancer-binding proteins: unlocking σ54-dependent gene transcription. Curr. Opin. Struct. Biol. 17, 110–116. doi: 10.1016/j.sbi.2006.11.002
Shen, A., Edwards, A. N., Sarker, M. R., and Paredes-Sabja, D. (2019). Sporulation and germination in Clostridial pathogens. Microbiol. Spectr. 7, 10. doi: 10.1128/microbiolspec.GPP3-0017-2018
Soutourina, O., Dubois, T., Monot, M., Shelyakin, P. V., Saujet, L., Boudry, P., et al. (2020). Genome-Wide transcription start site mapping and promoter assignments to a sigma factor in the human enteropathogen Clostridioides difficile. Front. Microbiol. 11, 1939. doi: 10.3389/fmicb.2020.01939
Stabler, R. A., He, M., Dawson, L., Martin, M., Valiente, E., Corton, C., et al. (2009). Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10, R102. doi: 10.1186/gb-2009-10-9-r102
Stevens, M. J. A., Molenaar, D., De Jong, A., De Vos, W. M., and Kleerebezem, M. (2010). Involvement of the mannose phosphotransferase system of Lactobacillus plantarum WCFS1 in peroxide stress tolerance. Appl. Environ. Microbiol. 76, 3748–3752. doi: 10.1128/AEM.00073-10
Stewart, D., Anwar, F., and Vedantam, G. (2020). Anti-virulence strategies for Clostridioides difficile infection: advances and roadblocks. Gut Microbes 12, 1802865. doi: 10.1080/19490976.2020.1802865
Stickland, L. H. (1934). Studies in the metabolism of the strict anaerobes (genus Clostridium): THE chemical reactions by which Cl. sporogenes obtains its energy. Biochem. J. 28, 1746–1759. doi: 10.1042/bj0281746
Tsang, J., and Hoover, T. R. (2014). Themes and variations: regulation of RpoN-dependent flagellar genes across diverse bacterial species. Scientifica 2014, 681754. doi: 10.1155/2014/681754
Tsuchido, T., Svarachorn, A., Soga, H., and Takano, M. (1990). Lysis and aberrant morphology of Bacillus subtilis cells caused by surfactants and their relation to autolysin activity. Antimicrob. Agents Chemother. 34, 781–785. doi: 10.1128/AAC.34.5.781
Vedantam, G., Clark, A., Chu, M., Mcquade, R., Mallozzi, M., and Viswanathan, V. K. (2012). Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 3, 121–134. doi: 10.4161/gmic.19399
Vedantam, G., Kochanowsky, J., Lindsey, J., Mallozzi, M., Roxas, J. L., Adamson, C., et al. (2018). An engineered synthetic biologic protects against Clostridium difficile infection. Front. Microbiol. 9, 2080. doi: 10.3389/fmicb.2018.02080
Voth, D. E., and Ballard, J. D. (2005). Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18, 247–263. doi: 10.1128/CMR.18.2.247-263.2005
Wiegeshoff, F., Beckering, C. L., Debarbouille, M., and Marahiel, M. A. (2006). Sigma L is important for cold shock adaptation of Bacillus subtilis. J. Bacteriol. 188, 3130–3133. doi: 10.1128/JB.188.8.3130-3133.2006
Willing, S. E., Candela, T., Shaw, H. A., Seager, Z., Mesnage, S., Fagan, R. P., et al. (2015). Clostridium difficile surface proteins are anchored to the cell wall using CWB2 motifs that recognise the anionic polymer PSII. Mol. Microbiol. 96, 596–608. doi: 10.1111/mmi.12958
Wydau-Dematteis, S., El Meouche, I., Courtin, P., Hamiot, A., Lai-Kuen, R., Saubaméa, B., et al. (2018). Cwp19 is a novel lytic transglycosylase involved in stationary-phase autolysis resulting in toxin release in Clostridium difficile. Mbio 9, e00648–e00618. doi: 10.1128/mBio.00648-18
Keywords: Clostridium difficile, SigL, sigma 54, RpoN, sporulation, Clostridioides difficile
Citation: Clark AE, Adamson CC, Carothers KE, Roxas BAP, Viswanathan VK and Vedantam G (2022) The Alternative Sigma Factor SigL Influences Clostridioides difficile Toxin Production, Sporulation, and Cell Surface Properties. Front. Microbiol. 13:871152. doi: 10.3389/fmicb.2022.871152
Received: 07 February 2022; Accepted: 06 April 2022;
Published: 11 May 2022.
Edited by:
Shan Goh, University of Hertfordshire, United KingdomReviewed by:
Meera Unnikrishnan, University of Warwick, United KingdomCopyright © 2022 Clark, Adamson, Carothers, Roxas, Viswanathan and Vedantam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gayatri Vedantam, Z2F5YXRyaUBlbWFpbC5hcml6b25hLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.