- 1Department of Entomology, South China Agricultural University, Guangzhou, China
- 2State Key Laboratory of Rice Biology, Institute of Insect Sciences, Zhejiang University, Hangzhou, China
- 3Department of Agriculture and Forestry, Kohsar University Murree, Punjab, Pakistan
- 4Department of Life Sciences, Khwaja Fareed University of Engineering and Information Technology, Rahim Yar Khan, Pakistan
- 5Department of Entomology, Pir Mehr Ali Shah Arid Agriculture University, Rawalpindi, Pakistan
- 6Key Laboratory of Integrated Pest Management of Crop in South China, Ministry of Agriculture and Rural Affairs, Guangzhou, China
- 7Key Laboratory of Natural Pesticide and Chemical Biology, Ministry of Education, South China Agricultural University, Guangzhou, China
- 8Department of Entomology, University of Faisalabad, Faisalabad, Pakistan
Insect pests cause significant agricultural and economic losses to crops worldwide due to their destructive activities. Pesticides are designed to be poisonous and are intentionally released into the environment to combat the menace caused by these noxious pests. To survive, these insects can resist toxic substances introduced by humans in the form of pesticides. According to recent findings, microbes that live in insect as symbionts have recently been found to protect their hosts against toxins. Symbioses that have been formed are between the pests and various microbes, a defensive mechanism against pathogens and pesticides. Insects’ guts provide unique conditions for microbial colonization, and resident bacteria can deliver numerous benefits to their hosts. Insects vary significantly in their reliance on gut microbes for basic functions. Insect digestive tracts are very different in shape and chemical properties, which have a big impact on the structure and composition of the microbial community. Insect gut microbiota has been found to contribute to feeding, parasite and pathogen protection, immune response modulation, and pesticide breakdown. The current review will examine the roles of gut microbiota in pesticide detoxification and the mechanisms behind the development of resistance in insects to various pesticides. To better understand the detoxifying microbiota in agriculturally significant pest insects, we provided comprehensive information regarding the role of gut microbiota in the detoxification of pesticides.
Introduction
Insects are the world’s most diverse and abundant animals in terms of species diversity and body mass in all ecological habitats (Nagarajan et al., 2022). Their numerous interactions with beneficial microbes are essential for survival and diversity. Microbes that are living in the guts of insects play a vital role in the biology and behavior of their hosts, including assisting in the digestion of recalcitrant food components, upgrading nutrient-poor diets, modulating the immune response, and protecting from predators, parasites, pathogens, and disease vectors. Other functions include facilitating plant specialization, governing mating preference and reproductive systems, and contributing to inter- and intraspecific communication (Sharon et al., 2010; Engel et al., 2012; Tokuda et al., 2018; Xia et al., 2018).
Many studies describing symbiotic connections between microbes and insects have been published (Funaro et al., 2011; Dang et al., 2017; Nicoletti and Becchimanzi, 2022). Most insects are thought to be in symbiotic partnerships with microbes, with estimates ranging from 15 to 20% of the total (Zhou et al., 2021). The role of microorganisms, particularly gut microbes, in insect function is important from various viewpoints, including agriculture, ecology, and medicine. Few insects are good laboratory models for studying microbe populations and their associations with hosts, especially immunology and metabolic associations (Hamilton and Perlman, 2013). Entomological studies of parasitic and mutualistic connections have focused on social insects like ants, which have evolved diverse interactions with other species at various levels, including individual and community interactions. These interactions can occur between bacteria and different insects and plants (Moreau, 2020).
Symbiotic bacteria can affect the efficacy of disease vectors or their developmental time, making them possible targets for disease control (Chouaia et al., 2012; Ricci et al., 2012). Microorganisms allied with pollinators and herbivores, and insects that feed on them are likely to impact the agricultural crops’ health substantially. Insects and their gut microbial populations play vital roles in the nitrogen cycle and the decomposition of plant material in natural and human-impacted ecosystems (Fox-Dobbs et al., 2010; Engel and Moran, 2013). A symbiotic relationship with very adaptable bacteria may have opened new ecological niches and unbalanced food sources like plant sap or blood (Sudakaran et al., 2017). Mutualism between insects and microbes is unquestionably one of the primary drivers of insect evolution. It is one of the most important factors contributing to the remarkable success of this gigantic group of animals. Mutualism is described as an interaction between various species mutually advantageous to both parties (Armitage et al., 2022). Several fitness traits of insects are heavily influenced by associated microbiota (de Almeida et al., 2017). The association of insects with microbiota is very important for the evolution of ecological features and feeding habits in which insects exchange nutrients or specific functions, such as protection from adversaries or transit between parties (Kikuchi et al., 2012; Suárez-Moo et al., 2020). Symbiotic-associated bacteria allow insects to feed on hard-to-digest and nutritionally poor diets (Salem and Kaltenpoth, 2022). However, insects may be associated with various microbes that also play an important role in degrading pesticides.
Pesticides may have unintended harmful impacts on humans, non-target creatures, and the environment (surface, soils, and groundwater), as the products are designed to be poisonous and are intentionally discharged into the environment (Kamal et al., 2020). Pesticide hazard is a function of the pesticide’s (eco) toxicological qualities and the pesticide’s ability to harm humans, flora, and animals (Müller et al., 2014). In modern farming systems, pesticides have become an important part of the process. As a result of persistent pests’ resurgence, the overreliance on pesticides for pest control may not end soon. Consequently, various biological and ecological factors mediate several available reports on insect pests’ resistance against different pesticides (Table 1). As a result of the overdependence on synthetic pesticides, numerous concerns have been raised in lieu of their side effects, such as the development of resistance in the target insects, the pollution of the environment, and the effects on human health (Du et al., 2020). It has also been suggested that pesticide resistance may be influenced by gut microbiota, which adds another degree of complexity to the processes of resistance (Gressel, 2018). Bacteria have been demonstrated to directly break down organic pesticides such as chlorpyrifos, dimethoate, and ethoprophos (Nayak et al., 2018; Chen et al., 2020; Gunstone et al., 2021). Furthermore, agricultural pests regularly acquire these bacteria after ingesting them from various sources, including food and the environment (Kikuchi et al., 2012). The gut microbiome may also potentially aid in detoxification by modulating the immune system of the host (Xia et al., 2018). Gut bacteria that produce nutrients and other beneficial chemicals may help the host develop better and increased tolerance to food poisons, although direct experimental data remains sparse (Kohl and Dearing, 2016; Mason et al., 2019).
Increasing apprehensions about the dramatic upsurge in pesticide resistance in pests have prompted researchers to better understand the mechanisms through which insect gut microbiome may confer resistance. Insect gut microbial populations have been studied for their potential role in pesticide resistance—for example, in Riptortus pedestris, Burkholderia symbionts have been demonstrated to promote pesticide resistance, and fenitrothion-degrading Burkholderia strains can also be shifted horizontally to other insects (Kikuchi and Yumoto, 2013). Similarly, Cheng et al. (2017) found that trichlorfon-degrading Citrobacter sp. (CF-BD) isolated from the gut of Bactrocera dorsalis increased pesticide resistance in the cockroach gut. In addition, many non-septate fungi and bacteria, assumed to be mutualistic, were found in the small intestines of workers of cephalotinid ants. These bacteria live as a moderately dense flora that contains a diverse range of bacterial species, including gram-positive and gram-negative coccobacilli and anaerobes similar to Bacterioides and Clostridia species (Donelli et al., 2012).
We already know that the environment in the insect gut regulates or even determines the shape of the community microbiota diversity and its metabolic activities, which might cause physical consequences for insects (Tang et al., 2012; Xia et al., 2018). Variations in environmental situations have been shown to affect the microbiota interrelationships among insects and their microbiota and related gene expression (Possemiers et al., 2011; Stencel and Wloch-Salamon, 2018). Recently, emerging research have suggested associations between insect gut microbiome and pesticide resistance. Several studies ranging from community diversity surveys to molecular analyses have focused on the gut bacteria’s interactions with the host immune systems (Kikuchi et al., 2012; Engel and Moran, 2013; Xia et al., 2013; Chmiel et al., 2019).
However, despite compelling reasons to further understand the roles played by insect gut microorganisms and a recent increase in research on microbes that live in insect guts, there has been little progress in expanding the available knowledge on the role of insect gut microbiota in the degradation of pesticides. Currently, pest resistance issues need to be addressed, so the current review will explore the functions and mechanism of pesticide resistance aided by gut microbiota and elaborate their role in pesticide degradation.
Insect Gut Structure and Functions
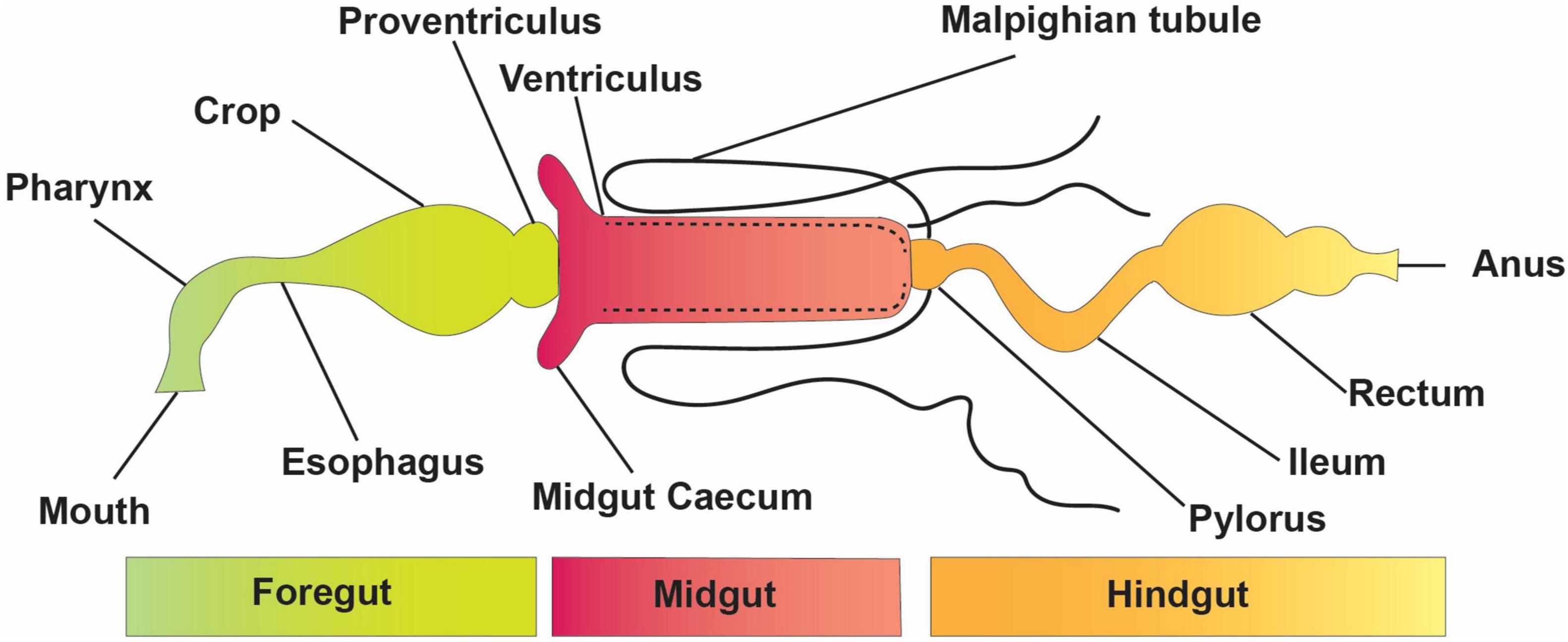
The elementary structure of the intestinal system is alike among insects, even though they have a variety of alterations connected with adaptation to diverse feeding styles and environmental conditions (Figure 1). The digestive tract is divided into three basic regions: the foregut, the midgut, and the hindgut (Simpson, 2013). The foregut and hindgut originate from the embryonic epithelium and are protected from pathogens by an exoskeleton of chitin and integument glycoproteins. This exoskeleton is shed at each ecdysis, separating the gastrointestinal lumen from the epithelia. When divided into functionally different subgroups, the foregut is frequently distinguished by another diverticula or crop for impermanent food storage (Linser and Dinglasan, 2014). The hindgut includes distinct portions like fermentation compartments and a distinct rectum for retaining feces during earlier evacuation, among other things. In many insects, the midgut is the main location of absorption and digestion. It lacks an exoskeletal lining and develops from endodermal cells rather than the rest of the body (Engel and Moran, 2013). A protective envelope known as the peritrophic matrix (or peritrophic membrane) is released by the midgut epithelial cells of many insects. This envelope, constantly being renewed as lost, is essential for the insect’s survival. The midgut has two parts: the endo- and ectoperitrophic space. Microorganisms are generally kept in the endo-peritrophic area, which prevents them from coming into direct contact with the epithelium. Peritrophic matrixes are classified into two discrete categories, namely, type I and type II. Type I refers to the whole midgut and is occasionally active when particular foods are consumed, whereas type II is in the remote location of the anterior mid-gut (Engel and Moran, 2013).
The peritrophic matrix shields the epithelium against mechanical injury by food elements, toxins in food, invasive microbes, and absorbed food and digestive enzymes (Kuraishi et al., 2013; Dastranj et al., 2016). In other circumstances, the peritrophic medium wraps around the undigested food mass as it passes along the digestive tract. Tiny pores in the peritrophic matrix prevent most microbes from passing through while allowing enzymes and small molecules from digested food to get through (Terra and Ferreira, 2012; Engel and Moran, 2013). Several insect species, including most sap-feeding species (Hemiptera), various other species of family Formicidae, and order Coleoptera (Nardi and Bee, 2012), that rely solely on cell sap or honeydew do not form a peritrophic matrix (Engel and Moran, 2013).
The Malpighian tubules of insects are excretory structures that extend from the anterior hindgut into the body void and ingest wastes, such as uric acid supplied to the hindgut (Figure 1). As a result, the hindgut of insects comprises a distinct nutritional environment which is well documented for water resorption (Simpson, 2013); the hindgut might function as a location of nutrient assimilation, as verified for different insect pests, including termites (Ayitso and Onyango, 2016), crickets (Smith et al., 2017), cockroaches (Tinker and Ottesen, 2016), and heteropteran (Gutiérrez-Cabrera et al., 2016)—for instance, intercellular passages in the hindgut membrane of several cockroaches permit nutrients, such as amino acids and fatty acids made via the biota in the hindgut, to flow from the hindgut lumen to the insect hemolymph (O’Donnell and Donini, 2017). The basic form of an insect gut has undergone numerous alterations due to adaptations to specialized niches and eating patterns.
Insect Gut Microbiome Composition
A wide range of parameters can influence gut microbiota composition, including insect growth, biochemical changes in different intestinal areas, and the insect’s ability to obtain available resources (Bruno et al., 2019b). The hindgut of insects, which serves as an extension of the body cavity, is one of these structures that collect dietary waste. Therefore, it provides a great food environment to the gut microbiota, encouraging their proliferation and diversification (Engel and Moran, 2013; Bruno et al., 2019a).
The insect gut microbiome includes protozoa, fungus, archaea, and bacteria. Protists occupy almost 90% of the hindgut of subterranean termites—for example, lower and higher termites’ guts include bacteria and archaea (Hongoh, 2010). Scientists revealed that the digestive regions of adult workers of honeybee (Apis mellifera) are dominated by a diverse group of nine bacterial species (five of which are Snodgrassella alvi and Gilliamella apicola, two species of Lactobacillus, and a species of Bifidobacterium) (Douglas, 2018). Additionally, the gut microbiota is rarely directly touched with intestinal epithelial cells due to their unique placement. Most of the time, bacteria that live in the gut are found in the lumen of the endoperitrophic space, a chitinous barrier that lines the middle of the gut (Erlandson et al., 2019). Yun et al. (2014) have comprehensively categorized and thoroughly defined the insect-associated gut bacteria of 305 samples belonging to 218 species in 21 taxonomic orders. The results indicated that Proteobacteria and Firmicutes were found to make up 62.1 and 20.7% of the total reads in the insect gut microbiota, respectively. Moreover, Wolbachia made up 14.1% of the total reads.
Interaction of Insect and Their Related Microbiota
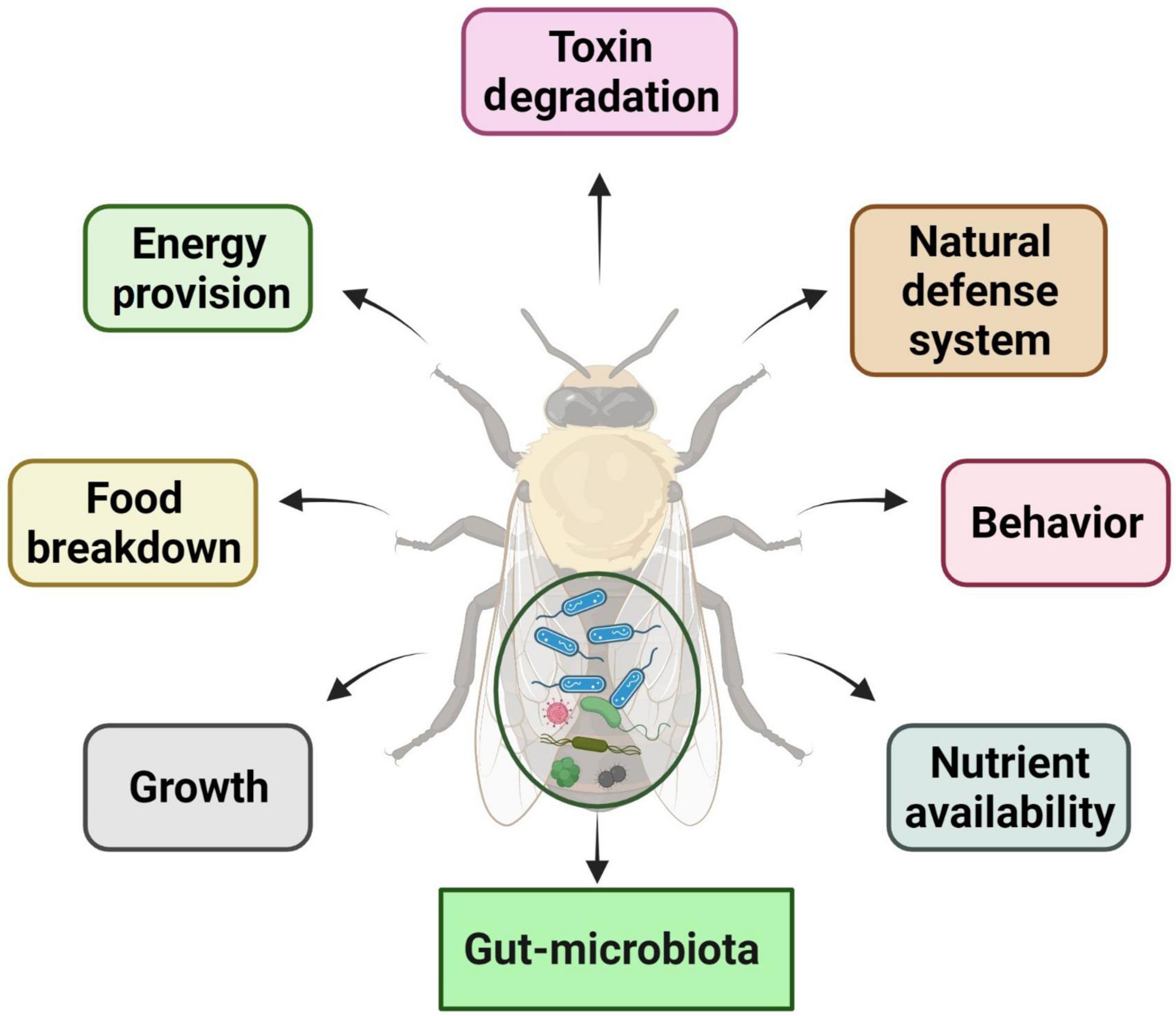
Insect–microbiota interactions are quite diverse. Insects rely on symbiotic bacteria for a variety of essential activities. Symbiotic bacteria can be critical for host survival and growth (Consortium, 2012; Douglas, 2015; Berasategui et al., 2016). They can help break down food, provide energy, make vitamins, and even help shape the body’s natural defenses (Cheng et al., 2019; Figure 2). Microbial symbionts have been proven to have many consequences on insect health and behavior (Sampson and Mazmanian, 2015). Certain insects have specialized organs that can only house a few symbiont species, while others have a far more diverse and variable flora in their guts and other internal organs. Numerous associations are developed with a sole or a few species of microbiota. They might require establishing specialized insect organs and cells (i.e., subsequent midgut crypts, mycangia, and microbiome) to house definite obligate symbionts (Zaidman-Rémy et al., 2018; Kuechler et al., 2019; Maire et al., 2019; Trappeniers et al., 2019). In these partnerships, the genetic integral of biochemical processes essential for the persistence of both interrelating groups is frequently observed (Hansen and Moran, 2011).
Some insect species are more involved in symbiotic associations with bacteria than others. Among the insects, three taxonomic groups are regularly involved. These groups include Blattaria, Coleoptera, Homoptera, and Hymenoptera. Additionally, certain bacteria seem to be particularly adept at symbiotic interactions. Numerous arthropods carry representatives of the Wolbachia genus (Aikawa et al., 2022), which is closely linked to pathogenic Rickettsia (Shan et al., 2021) and is categorized in Proteobacteria’s subgroup. The subgroup contains symbiotic organisms closely related to significant human diseases, such as Francisella tulariensis, Coxiella burnetii, and several Enterobacteriaceae (Dang et al., 2017). Symbionts of mealybugs and the protist family Trypanosomatidae are members of the Proteobacteria β-subgroup (Boursaux-Eude and Gross, 2000). Cockroach mycetocyte symbionts (Blattaria) belong to the Flavobacterium–Bacteroides group (Guzman and Vilcinskas, 2020).
The maize weevil Sitophilus zeamais, for example, needs nutrients made by its endosymbiont Sodalis pierantonius to stay healthy. The symbionts’ innate immune system is generally activated by the weevils’ secretion of an antimicrobial peptide (AMP) in the microbiome, which prevents the weevils from generating a systemic antibacterial response against them (Wang et al., 2017; Maire et al., 2019; Trappeniers et al., 2019). When it comes to digesting plant tissues that are resistant to digestion, termites require more composite mutualism with lots of digestive-zone bacterial or protist species (Tokuda et al., 2018; Liu et al., 2019), and many of these microorganisms are termite-specific symbionts with a high degree of niche specialization (Bourguignon et al., 2018; Hervé et al., 2020). The microbiota of other various insects may be more varied and adaptable, as they do not rely on explicit critical symbionts (Coon et al., 2016; Scolari et al., 2019). The gut biota is critical for most insects’ digestion, fertility, fecundity, and immunity (Heys et al., 2018; Salcedo-Porras et al., 2020), as growing axenic insects can be deadly (Flury et al., 2019). Insects need to get several symbionts that successfully make good and functional microbiota.
Primary symbionts are more common in insects having particularly nutrient-deficient foods (obligate hematophagy or phytophagy). In contrast, secondary symbionts are more common in polyphagous and omnivorous insects, which obtain a diverse microbiota from their surroundings (Salcedo-Porras et al., 2020). While most primary symbionts are internal (endosymbionts), secondary symbionts are external. There may be an association between the symbiont acquisition or transmission and the nature of the interactions between insects and symbionts. It is usual for female germline transmission to occur vertically through the female germline as with primary mutualists, for example, those present in aphids and weevils (Caspi-Fluger et al., 2012; Douglas, 2015; Hassan et al., 2020). It is common for environmental microorganisms to be transmitted across internal organs, some of which can form secondary symbioses without specialized organs.
Additionally, insect growth affects the time during which microbiota are acquired horizontally. Except for vertically transmitted microorganisms, most insects hatch practically germ-free and obtain their microbiome by cannibalism, trophallaxis, coprophagy, or ingesting their contaminated eggshells (Taylor et al., 2014; Salcedo-Porras et al., 2020). Holometabolous insects pupate in a nearly axenic state, and adults re-acquire some of their gut microbiota from the environment (Powell et al., 2014; Hammer and Moran, 2019; Rolff et al., 2019) after emerging from the pupal stage. The microbiota of adults in some species may differ greatly from the microbiome of the immature stages or may acquire a similar gut microbiota from the conspecifics or environment (Johnston et al., 2019; Majumder et al., 2020; Suárez-Moo et al., 2020). On the contrary, microorganisms attained after egg hatching can be preserved in hemimetabolous insects for an extended period (Rodríguez-Ruano et al., 2018; Hammer and Moran, 2019). Finally, social insects, whether hemimetabolous or holometabolous, can get microbiota from each other repeatedly, choosing and keeping a specific microbiota (Onchuru et al., 2018; Tokuda et al., 2018; Liu et al., 2019).
Acquisition Resistance Characteristics of Native Gut Bacteria
The increased predominance of naturally existing inhibitory gut bacteria could be a viable alternative to para-transgenic techniques for reducing pathogen burden in natural populations of insects’ vector. The configuration of the gut microbiome regulates vector capability by modulation of immunological reactions, competition for positions, or production of inhibitory compounds (Cirimotich et al., 2011; Boissière et al., 2012). The practical investigation of the gut microbiome to understand its contact with the parasite and host might lead to the development of innovative and more effective techniques to regulate vector-borne infections. As a result, future plant pest control efforts should consider this. Numerous microbial plant inflammations are conveyed via insect vectors, and the identification of these insects’ intestinal bacteria has been conducted to create techniques to prevent pathogen spread (Raddadi et al., 2011; Engel and Moran, 2013). An excellent example is a disease (Pierce) of grapes produced via pathogenic Xylella fastidiosa. Alcaligenes xylosoxidans was isolated as a bacterial symbiont since the sharpshooter (Cicadellidae) spreads X. fastidiosa.
These bacteria live in insect’s foregut, where they share space with X. fastidiosa, a bacterium that can be harmful to people. Because A. xylosoxidans are elated into the plants’ xylem by insects feeding on sap, it is more likely to spread to other insects. These properties make A. xylosoxidans a promising option for use as a bio-control mediator against X. fastidiosa establishment through modest position elimination or as a para-transgenic conveyer for providing anti-Xylella drugs among other applications (Miller, 2011).
Impacts of Gut Microbiota on the Activity of Pesticides
The insect-associated microbial community is dynamic and responsive to various stressors (Zhang et al., 2022). The related microbiota, like the insect, is subject to natural selection pressure, and its composition can be influenced by variables such as dietary changes, food scarcity, and exposure to toxic substances (Adair and Douglas, 2017; Akami et al., 2022). The microbiota of hosts exposed to pesticides as a source of selection pressure may also assist the host in metabolizing these substances. It may act as a source of variation, resulting in the host’s reduced susceptibility to pesticides (Akami et al., 2019a,b). Pesticide-degrading bacteria are prevalent throughout nature and have been identified in a variety of insect orders, including Lepidoptera (Ramya et al., 2016b; de Almeida et al., 2017), Hemiptera (Kikuchi et al., 2012), Diptera (Cheng et al., 2017), and Coleoptera (Akami et al., 2019b). There has been evidence that resistant strains of bacteria from the gut of Plutella xylostella Linnaeus (Xia et al., 2018) and Spodoptera frugiperda (de Almeida et al., 2017) have the capacity to breakdown many pesticides (Gomes et al., 2020). The selection of S. frugiperda strains based on pesticide-guided selection led to selecting pesticide-degrading bacteria absent in the microbiota of vulnerable, unselected larvae (de Almeida et al., 2017).
The microbial population of an insect’s digestive tract comprises bacteria belonging to the phyla Firmicutes, Proteobacteria, Actinobacteria, and Bacterioidetes, all of which can impact the biology of hosts (Paniagua Voirol et al., 2018; Gomes et al., 2020). Research on Spodoptera littoralis (Boisduval) found that the microbial community was mostly made up of Firmicutes, especially Enterococcus (Chen et al., 2016; Higuita Palacio et al., 2021). Firmicutes are found in the digestive tracts of many lepidopteran larvae, even though the digestive tracts of larvae are suggested to be not very suitable for bacteria to live. This includes Spodoptera litura Fabricius (Thakur et al., 2016), Manduca sexta Linnaeus (Holt, 2013), Helicoverpa armigera Hubner (Yuan et al., 2021), and many other lepidopteran species (Mereghetti et al., 2017; Gomes et al., 2020). Bacteria belonging to the genus Enterococcus are known to create a variety of bacteriocins, which are potent antibacterial chemicals that can influence the composition of the gut microbial communities (Van Arnam et al., 2018). The highest relative amount of Enterococcus was reported in S. frugiperda populations from the laboratory and from natural fields (Gomes et al., 2020).
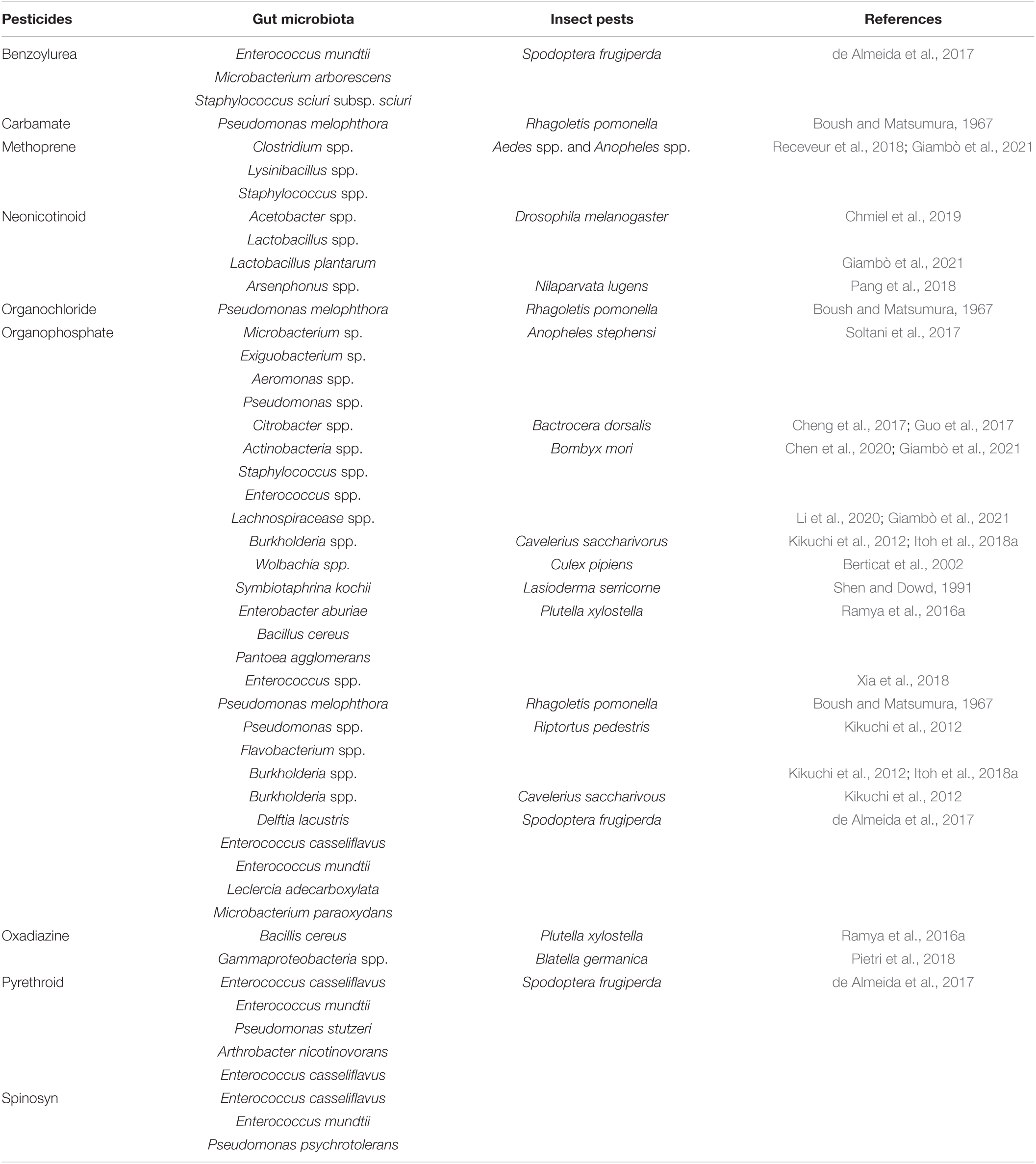
According to various studies, the intestinal bacteria of insects have been shown to break down multiple pesticides and interfere with the effectiveness of pesticides used to control them (Ramya et al., 2016a; Cheng et al., 2017; de Almeida et al., 2017). The Proteobacteria families (Enterobacteria, Pseudomonada, and Burkholderia) could break down acephate, chlorpyrifos, trichlorfon, lambda-cyhalothrin, and Spinosad, respectively (Kikuchi et al., 2012; de Almeida et al., 2017; Itoh et al., 2018b; Gomes et al., 2020). Similarly, Actinobacteria and Firmicutes bacteria have also been shown to have a role in the process of removing toxins from the environment (de Almeida, 2013; Ramya et al., 2016b). The resistant strain of S. frugiperda harbor gut bacteria Enterococcus (Firmicutes) that were able to break down the pesticides (chlorpyrifos, lambda-cyhalothrin, deltamethrin, spinosad, and lufenuron) (Gomes et al., 2020). According to previous studies, there are several gut symbionts of different insects (orders Coleoptera, Diptera, Hemiptera, and Lepidoptera) that detoxify the pesticides (classes Benzoylurea, Carbamate, Methoprene, Neonicotinoid, Organochloride, and Organophosphate) by the different species of genera Acetobacter, Actinobacteria, Aeromonas, Arsenphonus, Burkholderia, Citrobacter, Clostridium, Enterococcus, Exiguobacterium, Lachnospiracease, Lactobacillus, Lysinibacillus, Microbacterium, Pseudomonas, Staphylococcus, Symbiotaphrina, and Wolbachia (Table 2).
Microorganisms’ ability to utilize pesticides as a carbon source is contingent upon the coding of the biochemical systems required to cope with these substrates (Lourthuraj et al., 2022). Temperature and pH, nutrition availability, chemical concentration, and the size of the bacterial population all influence pesticide metabolization (Russell et al., 2011; Gomes et al., 2020). The pesticides’ chemical composition and complexity play a role in how quickly and effectively bacteria use them as a food source (Hubbard et al., 2014). Microorganisms use a wide range of metabolic pathways to break down and change xenobiotics when they grow rapidly (Itoh et al., 2018b; Bhatt et al., 2019, 2021; Gangola et al., 2022)—for example, Pseudomonas spp. and Ensifer adhaerens metabolized the thiamethoxam pesticide. The principal metabolic pathway involves the transition of its N-nitroimino group (= N-NO2) to N-nitrosimine/nitrosoguanidine (= N-NO, THX-II) and urea (= O; THX-III) metabolites (Hussain et al., 2016), which is shown in Figure 3. Another example is the symbionts species of genera Arsenophonus (Pang et al., 2018) and Pseudomonas (Pang et al., 2020b); Ensifer spp., Stenotrophomonas spp., Variovorax spp. (Hussain et al., 2016) have been reported to degrade imidacloprid. The mechanisms and associated metabolic pathways are shown in Figure 4, which indicates that nitro-reduction and oxidation are two of the main ways that bacteria break down imidacloprid (Lu et al., 2016; Fusetto et al., 2017). The gut microbiota produces enzymes that detoxify pesticides like pyrethroids, carbamates, diamides, and organochlorines, which have been identified (Russell et al., 2011; Khalid et al., 2016; Gomes et al., 2020; Lin et al., 2022).
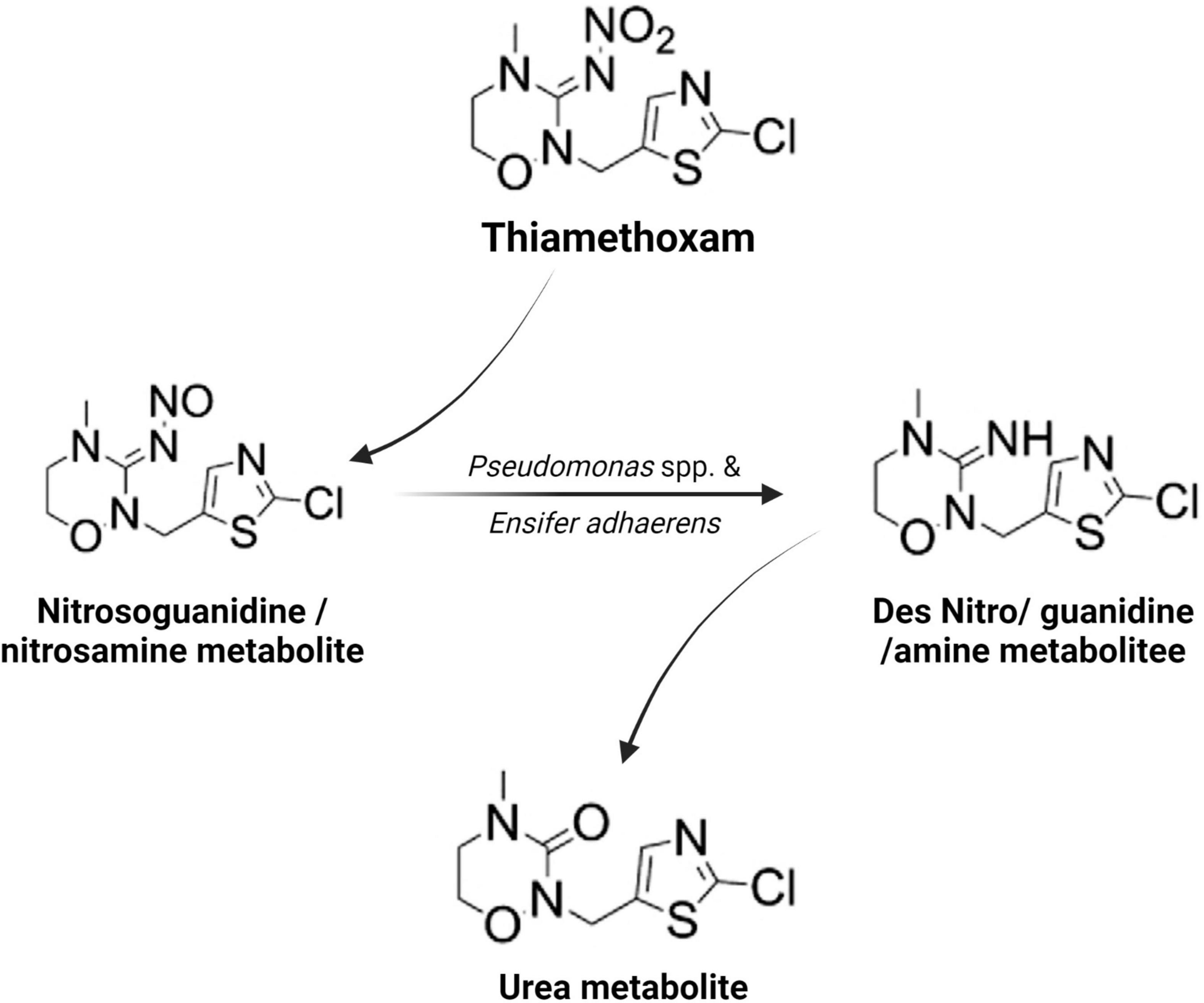
Figure 3. Metabolic routes for bacterial degradation of the insecticide thiamethoxam (Hussain et al., 2016).
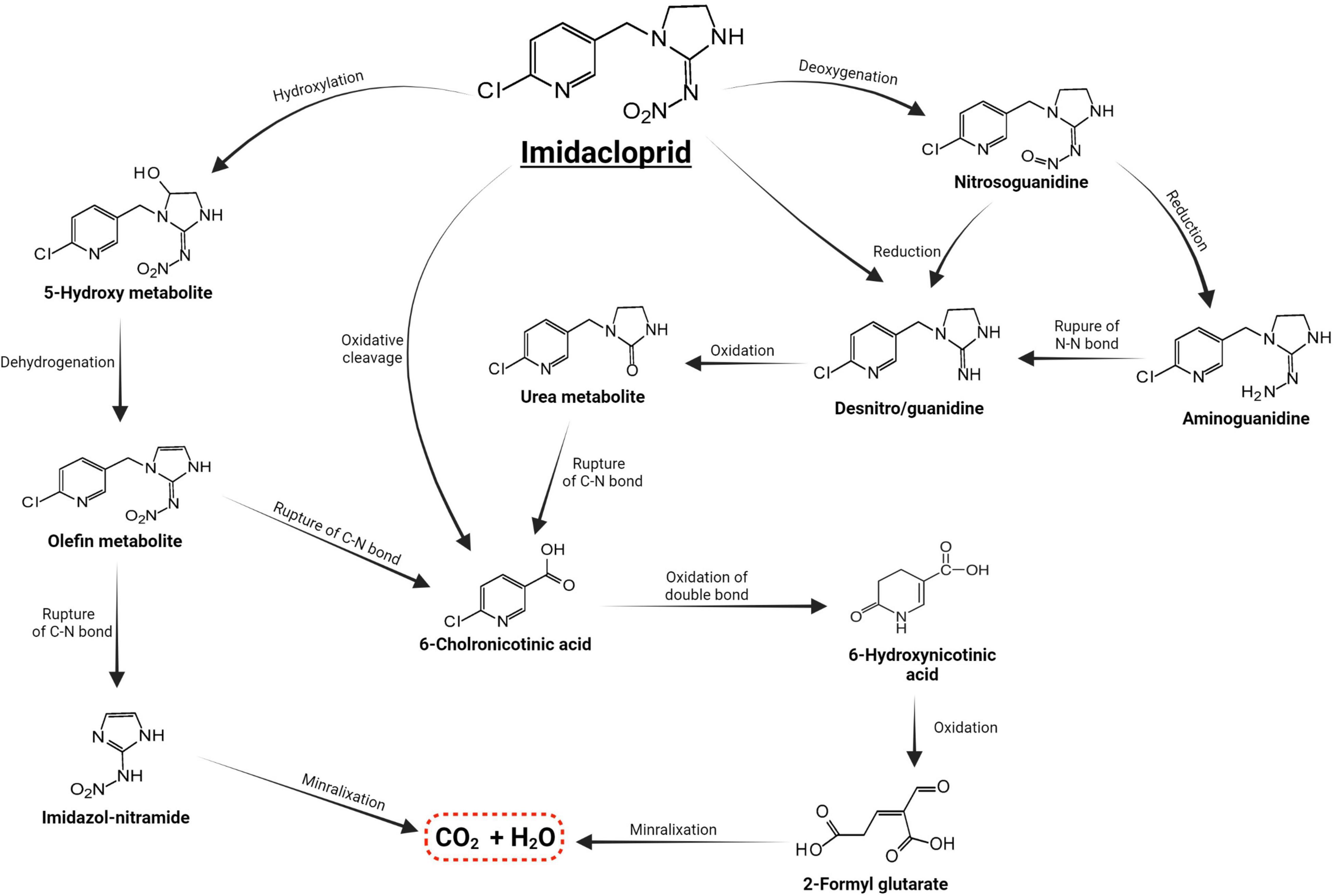
Figure 4. Metabolic routes for bacterial degradation of the insecticide imidacloprid (Pang et al., 2020a).
Moreover, Plant secondary components, such as terpenes, alkaloids, glycosides, and phenolic compounds, are degraded by Proteobacteria in the presence of insects (Mereghetti et al., 2017; Gomes et al., 2020). Proteobacteria have the most diverse morphology and adaptability of all bacterial phylum, which offers them an advantage in various ecological niches (Shin et al., 2015). Proteobacteria may thus act as a source of available variety and a tool for host adaptation in nature when they interact with other organisms (Bradley and Pollard, 2017; Degli Esposti and Martinez Romero, 2017; Hauffe and Barelli, 2019).
The gut microbiota has been linked to promoting the insecticidal action of Bacillus thuringiensis, the frequently used biological pesticide for herbivore pest management in agriculture (Mason et al., 2011; Eski et al., 2018). According to a study, when the gut microbial population was removed from gypsy moth larvae, the B. thuringiensis pesticide no longer worked as intended, whereas when some microbiota of the gut microbiome was added back in, the B. thuringiensis-facilitated mortality was reestablished (Polenogova et al., 2021). Several insect species have similar mechanisms for degrading imidacloprid (Thurman et al., 2013). Additionally, the Cyp6g1 gene discovered in Drosophila is critical for imidacloprid breakdown in animals, regulating and promoting the generation of metabolites in the oxidation pathway (Fusetto et al., 2017). These findings indicate the importance of considering the gut microbiome of insects in the development of novel pest control methods.
Symbiont-Mediated Pesticide Resistance
The rapid emergence of pesticide resistance in a wide range of organisms is a cause for concern, and it merits additional studies. Several mechanisms of pesticide resistance are ascribed to the physiology at the host level (Tabashnik and Carrière, 2010; van den Bosch and Welte, 2017), but few researchers have argued in recent years that some pesticides’ resistance might be ascribed by symbiont detoxification. Detoxifying enzymes targeting harmful allelochemicals and pesticides have been found in fungal symbionts isolated from insects (Tabashnik and Carrière, 2010; Naik et al., 2018). Evidence showing that changes in mutualist-level physiology can cause pesticide resistance is very limited—for example, the Burkholderia mutualist in midgut crypts acquired via the environment in each generation instead of the “traditional” mode of vertical maternal spread, as is the case with humans (Garcia, 2015). The pesticide fenitrothion, a common organophosphorus chemical in agriculture, can be degraded by the symbiotic Burkholderia (Kikuchi et al., 2012).
Furthermore, the R. pedestris pest bug easily forms symbiotic relationships through fenitrothion, degrading Burkholderia mutualists, and have significantly more persistence rates on fenitrothion-treated plants than insects by non-demeaning Burkholderia mutualists (Kikuchi et al., 2012). Spraying of fenitrothion to the field enabled more bacteria to degrade fenitrothion in soil, which is thought to impact the dynamics of symbiotic-degrading Burkholderia spread via soil to stinkbugs (Tago et al., 2015). These discoveries imply that pesticide resistance may mature in the absence of pest insects in a field and then spread rapidly within sole insect pest generation (Kikuchi et al., 2012). In addition, Burkholderia symbionts of the established R. pedestris model give an excellent chance for research on microbial symbiotic aspects at the molecular level since they are cultivable and genetically manipulable (Kim and Lee, 2015). These studies help develop an ecological pesticide that uses gut symbionts to control insects. Such studies could be useful to learn about pesticide resistance mechanisms that have not been found yet.
Molecular Mechanisms by Which Enzymes Mediate Pesticide Detoxification
Detoxification enzymes occur naturally in various biological processes, functioning on the target sites to neutralize various toxins prevalent in the insect body (Lin et al., 2015; Bhandari et al., 2021; Siddiqui et al., 2022). According to some previous studies, the biochemical characterization of insect resistance to pesticides is connected to pesticide sensitivity at the target site and pesticide detoxification by metabolic enzymes (acetylcholinesterase, carboxylesterase, glutathione S-transferase, and cytochrome P450) (Wu et al., 2014; Ismail, 2020; Yang et al., 2021; Siddiqui et al., 2022). These enzymes are crucial in detoxifying xenobiotics (Hu et al., 2014), where their hosts can utilize these enzymes as biological indicators during pesticide detoxification (Khan et al., 2021)—for instance, Zhang et al. (2016) reported that detoxification enzymes (cytochrome P450 genes) were detected during detoxification of fipronil in the red imported fire ants (Solenopsis invicta Buren), where up to 36.4-fold rise in resistance was recorded following exposure of the ants to fipronil. Another related study has also linked cytochrome P450 enzymes with fluralaner detoxification in S. invicta (Xiong et al., 2020).
Additionally, these enzymes may raise the responder gene’s copy number, mRNA levels, and coding sequence diversity by introducing point mutations (Pang et al., 2020a). These enzymes are involved in various processes, including biosynthesis and the metabolism of invading species, among others—for instance, P450 CYP6ER1 in Nilaparvata lugens and CYP6CM1 in Bemisia tabaci were used to characterize and assess imidacloprid metabolism. These findings revealed that amino acid changes in the binding site enhanced imidacloprid metabolism (Bao et al., 2016; Pang et al., 2016; Hamada et al., 2019). Puinean et al. (2010) discovered that the CYP6Y3 gene in Myzus persicae could confer resistance to neonicotinoids. Moreover, CYP353D1v2 was found to be overexpressed in several imidacloprid-resistant Laodelphax striatellus strains, and silencing this gene greatly reduced resistance (Elzaki et al., 2017). The effective suppression of CYP6CY14 transcription by RNAi in the overexpressed P450 gene of the CYP3 clade greatly improved the vulnerability of pesticide-resistant cotton aphids to thiamethoxam (Wu Y. et al., 2018).
In order to detoxify xenobiotics in the gut lumen, insects can employ various techniques. They can do so by creating an acidic environment and supplying a complex of enzymes (monooxygenases and esterases) that can cleave or alter the xenobiotic in preparation for excretion (de Almeida et al., 2017). It has been confirmed that microbial enzymatic activity in the gut lumen contributes to the breakdown of pesticides consumed by the host. The hydrolysis of these compounds provides resources for the microbiota to thrive (Mohammadi et al., 2021). The diversity and differences in prokaryote- and eukaryote-produced enzymes suggest that microbial enzymes could play a significant role in pesticide metabolization in contaminated insects (Russell et al., 2011; de Almeida et al., 2017).
Pesticide Degradation by Symbionts in Invasive Species
Multiple resistance mechanisms have been functionally recognized as conveying pesticide resistance in several invasive insects, including penetration resistance via cuticle thickening or remodeling, metabolic resistance via the amplified activity of detoxification enzymes (e.g., esterases and cytochrome P450 monooxygenases), and knockdown resistance via kdr transmutations (Khan et al., 2021; Rigby et al., 2021). There are also possible behavioral and physiological resistance mechanisms. These include point mutations that make esterases more active, GST, target place insensitivity, reformed AChE, GABA receptor insensitivity, and transformed nAChRs (Dang et al., 2017). The diamondback moth, Plutella xylostella, is an example of invasive species that act as a significant universal pest of various crops (Nakaishi et al., 2018). P. xylostella also generates an enzyme that avoids the generation of dietetic isothiocyanates that act as plant defense compounds emitted by the host plant and regulate feeding behavior of diamondback moths female (Hussain et al., 2019, 2020). Furthermore, P. xylostella has been discovered to be resistant to a wide range of chemical pesticides. Only three other pest species have established resistance to Bacillus thuringiensis-based pest control technologies, which is one of them (Furlong et al., 2013). The quick evolution of extremely resistant phenotypes in P. xylostella is partly ascribed to insect pests, including altered carbamate and organophosphate target locations, parathion metabolism by GST, and pyrethroid detoxification by P-450 monooxygenases (Ramya et al., 2016a). The indoxacarb-degrading microbiota (B. cereus bacteria) identified in the digestive tract of P. xylostella was found to degrade the pesticide by converting it into food (van den Bosch and Welte, 2017). Another pesticide, acephate, was quickly degraded by gut bacteria obtained from diamondback moth intestines. Together with earlier research on the gut microbiota of stinkbugs showing pesticide resistance (Kikuchi et al., 2012), these findings suggest that the gut biota might have a greater part in pesticide resistance than formerly assumed.
Role of Gut Microbiota in Tolerance and Resistance
Insect’s digestive systems are equipped with a multilevel defensive system, likely a primary driver in structuring gut microbiome communities. Different aspects of such a defensive system provide the host’s ability to tolerate and reject harmful bacteria in the gut through various processes. While tolerance reduces the detrimental effects of a bacterial burden on the host’s health, resistance reduces the bacterial burden so as not to harm the host (Moreno-García et al., 2014). Most immunological research have concentrated on resistance mechanisms, and there is little knowledge about the processes that mediate tolerance. However, host–microorganism associations in the insect gut are frequently commensalism or mutualism.
Compared to insects with sparsely populated digestive tracts, those with vast bacterial communities are more likely to be tolerant and less likely to be resistant to bacteria in their guts. As a result, the gut immunity mechanisms of diverse insects may be tailored to the definite desires of their hosts. As mentioned previously, the midguts of most insects produce a peritrophic medium composed of a network of chitin microfibrils implanted in a protein–carbohydrate medium (Muthukrishnan et al., 2012). The peritrophic medium is semi-permeable, allowing nutrients, digestive enzymes, and defense chemicals to flow while protecting the epithelial cell layer from a direct microbe or toxin exposure. The cuticle layer bordering the epithelial cell layer in the foregut and hindgut may have comparable protective roles.
These physiological barriers among the lumen and epithelium are decent instances of tolerance mechanisms since they minimize the influence of bacteria on the host rather than reduce the bacterial load in the gut. Certain parts of the insect gut can have a low or high pH or contain enzymes that target bacterial cell wall components, such as peptidoglycan or lysozymes (PGN) hydrolases (Liu et al., 2014; Moreno-García et al., 2014). Such systems can cause deliberate resistance by reducing the number of bacterial communities in specific parts of the gut, but they may be useful in bacterial cell breakdown to enhance nutrition.
Bacterial endosymbionts have been extensively examined in the context of biological invasions to detect or quantify their impact in increasing the invasion process of imported species (Klock et al., 2015; Taerum et al., 2016; Cheng et al., 2019). There is still a lack of understanding of the mechanisms that drive the responses of native species to invasive species’ selective pressures. A deeper knowledge of the structure and function of bacterial mutualists, on the other hand, may disclose possible mechanisms for inhabitant hosts to adapt to exotic species, as variations in bacterial mutualists have been revealed to correlate with variations in food sources in both invertebrates and vertebrates (Rokhsefat et al., 2016; Shapira, 2016). Furthermore, identifying and describing the bacterial mutualists of inhabitant species may supply vital hints about handling exotic species in the future.
Plant defenses and pesticides can potentially interact with and supplement host immune systems (Mason, 2020). Secondary metabolites play a critical role in protecting plants from arthropod herbivores. Secondary chemicals play an important role in insect resistance and vulnerability (Erb and Kliebenstein, 2020). Plant secondary metabolites with antinutritive, deterring, antibacterial, and poisonous properties frequently affect the growth and productivity of phytophagous insects feeding on various host plants (Puri et al., 2022). In nature, the level of plant-defensive compounds varies by species and is determined by the plant’s genotype, growing circumstances, and phenology. Plant allelochemicals impose a very strong selection pressure on herbivorous insects and the microbiota in their guts, which is particularly important for their survival (Douglas, 2015; Chen et al., 2022), for example, a study discovered that symbionts such as Phenylobacterium, Ochrobactrum, Erwinia, Amycolatopsis, and Sediminibacterium spp. may play critical roles in the metabolism of tea saponins, according to the findings. Two of them, Acinetobacter calcoaceticus and Acinetobacter oleivorans, were very important in the degradation of tea saponins (Zhang et al., 2020b).
The gut microorganisms’ digesting abilities can also assist in the removal or inactivation of toxic compounds in food (Schmidt and Engel, 2021). Detoxification symbioses have been observed in a wide range of hosts, even though certain insects have these functions encoded in their genomes (Itoh et al., 2018b). They are particularly important for herbivorous insects since plants produce a diverse spectrum of phytotoxins that are toxic to them (Itoh et al., 2018b). Adaptation to the highly toxic terpenoids present in the bark of pine trees has been achieved by cooperation between the mountain pine beetle (Dendroctonus ponerosae) and the pine weevil (Hylbius abietis) and their gut microbiota. Gammaproteobacteria, in particular, play an important role in the degradation of diterpenes (Berasategui et al., 2017; Schmidt and Engel, 2021).
An example of a social insect belonging to the genera Apis and Bombus harbors gut microbiota that plays important roles in their health, with a possible impact on pathogen protection and nutrient acquisition (Engel et al., 2016; Zheng et al., 2016). Glycolysis pathways and phosphotransferase systems have been found in the genomes of Gilliamella apicola, indicating that this bacterium functions as a saccharolytic fermenter that aids in the digestion of the host’s carbohydrate-rich meal (Tilottama et al., 2021). The pollen grain of G. apicola was subjected to a metagenomic investigation, and the results revealed the presence of genes encoding pectin-degrading enzymes. These enzymes play a vital role in breaking down the stiff polysaccharide walls of pollen grains and release constituent monosaccharides (Zheng et al., 2016).
The coffee borer beetle (Hypothenemus hampei) engages in a detoxifying symbiosis to facilitate nutritional adaption to coffee beans, which contain high quantities of the poisonous alkaloid caffeine (Mejía-Alvarado et al., 2021). It was discovered that the beetle’s gut microbiota is dominated by Pseudomonas species, which are seen in beetles from several coffee-producing countries (Ceja-Navarro et al., 2015). Beetle pseudomonad spores were able to develop on caffeine alone, and they were able to restore the breakdown of caffeine in beetles that had been previously treated with antibiotics (Schmidt and Engel, 2021). In addition to promoting nutritional adaptability, it has been observed that several pest species carry gut symbionts that are capable of degrading pesticides (de Almeida et al., 2017; Itoh et al., 2018b). The Burkholderia gut symbiont, R. pedestris, may degrade the pesticide fenitrothion and increase the survival of R. pedestris in soil infected with the pesticide (Itoh et al., 2018a). The wasp Nasonia vitripennis, for example, was found to have a greater survival rate in the presence of its gut microbiota in the exposure of atrazine (Wang et al., 2020). This research highlights the gut microbiota’s ability to boost the adaptive capabilities of its insect host, which has crucial implications for pest and pollinator insect control.
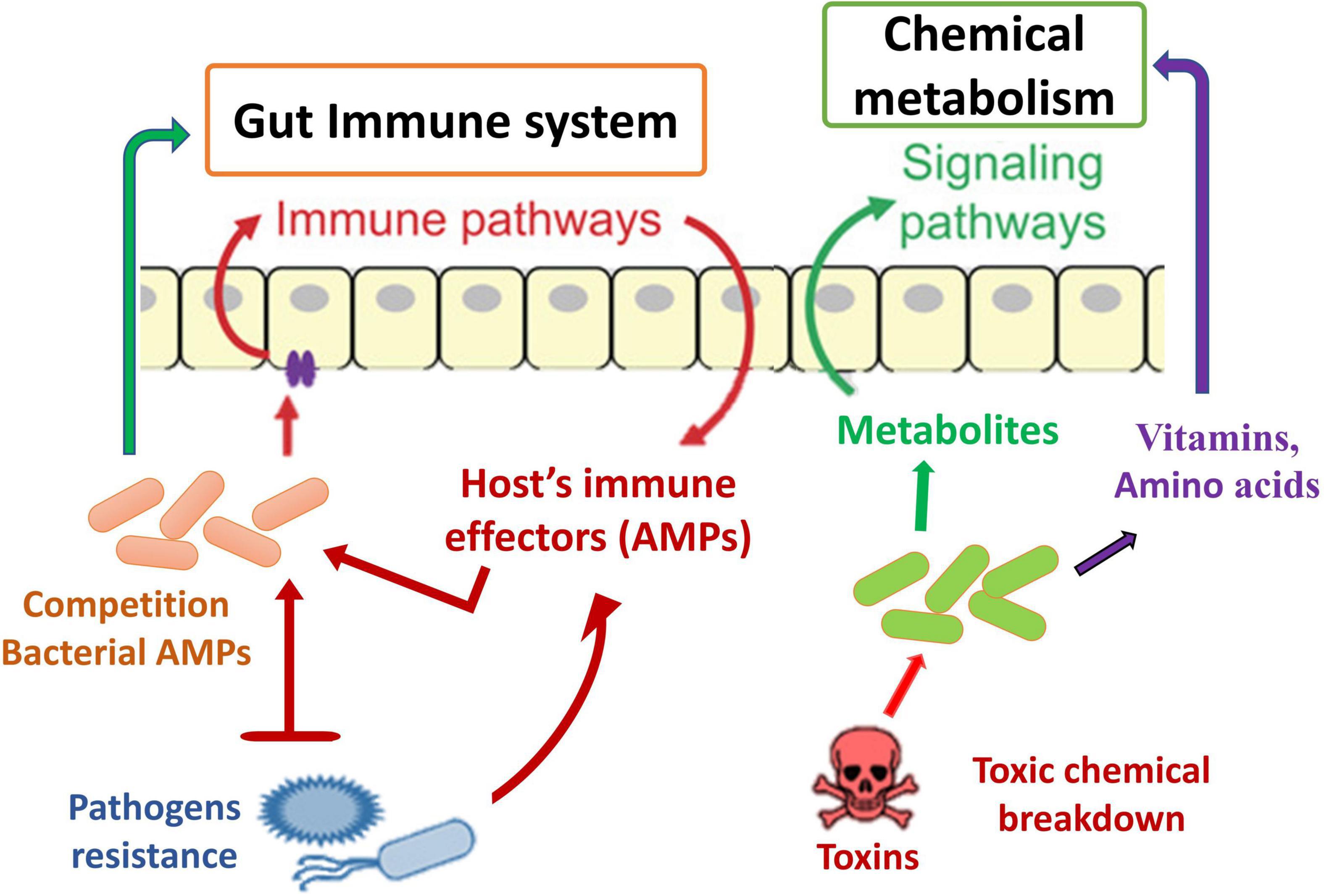
Another defense mechanism is the inherent immune system of insect species, which comprises numerous immunological responses (Chambers et al., 2012; Engel and Moran, 2013) and summarizes the general principles of innate immunity in insects. A key inducible response permitting resident immunity at the gut epithelial cell layer has been identified, mostly through experiments with D. melanogaster. These are the creation of amino acids (AMPs) (Figure 5) and the combination of reactive oxygen species (ROS). The generated reactions may altogether be considered traditional resistance mechanisms; nevertheless, they contain undesirable response circles and modulatory mechanisms, conveying host tolerance toward the commensal gut microbiota. The Toll and IMD signaling channels are two of the most important signaling mechanisms causing AMP synthesis in D. melanogaster’s systemic immune response (Hanson and Lemaitre, 2020). The reaction in the gut is distinct in that only the IMD pathway is activated, resulting in the induction of resident AMP reactions in response to pathogen stimulation (Nehme et al., 2007; Lee et al., 2013). Initiation happens when different types of bacterial PGN bind to receptors on the outside or inside of the body’s epithelium that belong to the peptidoglycan recognition protein (PGRP) family (Engel and Moran, 2013). Signaling downstream via the IMD pathway activates the transcriptome factor Relish, which stimulates the production of multiple AMPs and other immunity-associated genes (Figure 5). Introduction to pathogens also results in ROS formation in the gut of D. melanogaster through the membrane-related dual oxidase (DUOX) system (Alaraby et al., 2018). The PGN-independent and PGN-dependent signaling pathways are involved in this process (Charroux and Royet, 2012). In addition to the bacteria, the host’s epithelial cells are also subjected to oxidative stress when ROS are produced.
Immune catalases are activated in D. melanogaster to remove excess ROS (Buchon et al., 2014). Tolerance is improved due to this catalase synthesis, which reduces the immune response-induced self-harm (Simpson et al., 2015). How these enzymes protect the host cells without affecting the pathogens’ ability to produce ROS remains a mystery. One possible reason is that the catalase activity is constrained to a specific region of the epithelial surface—for instance, the vicinity of the epithelial surface. Immunological reactions to the related gut microbiome community have been confirmed in D. melanogaster by employing the DUOX system activity and IMD pathway at varying degrees. In the IMD pathway, the homeobox transcript factor caudal binds to the promoter regions of AMP genes in the gut and stops them from being made. The gut flora alters, and the epithelial cell layer breaks down in caudal-defective flies because of a constant generation of AMP (Ryu et al., 2008; Broderick and Lemaitre, 2012). As a result, it appears that caudal avoids over-encouragement of the immunity system by mutualistic gut biota. Additional immunological regulatory operations in D. melanogaster are regulated by amidases produced by the midgut cells of the epithelium and cleave pro-inflammatory PGN into passive systems (Bischoff et al., 2006; Zaidman-Rémy et al., 2006; Engel and Moran, 2013).
There are many ways in which obligatory insect-associated bacteria contribute to their host insect’s overall health and well-being; however, the primary contribution of these bacteria is connected only to their ability to provide nutrients. Secondary bacterial symbionts boost the host’s immunological response to entomophagy (Vorburger et al., 2010) and entomopathogens (Jaenike et al., 2010), impact host plant selection (Frago et al., 2012), defend against heat stress (Pons et al., 2022), and aid in the detoxification of compounds produced for herbivore defense (Hammer and Bowers, 2015). Microbes also plays a role in detoxifying xenobiotics by catabolizing organic compounds used in applied pest management, as demonstrated by degradation (Pietri et al., 2018).
Conclusion and Future Perspectives
Microbes are known to degrade a wide variety of allelochemicals and pesticides, providing numerous opportunities for insects to develop detoxifying symbiotic relationships. The gut microbiota plays various roles in the host’s physiology, including immunological modulation and toxin degradations. Arguably, the microbiota evolves more rapidly than their host insects, resulting in rapid pest adaptation to pesticides through the employment of mutualistic microbes. Additionally, insects can swiftly obtain novel metabolic activities and colonize new ecological niches through symbiotic interactions with microbiota that previously have fully developed well-tuned metabolic pathways. As results of the ever-dynamic climatic conditions and human populations, it is imperative that additional/novel insect pest management strategies are implemented to synergize the existing ones. Exploring symbiotic microorganisms as a means of managing their associated hosts could be one way to meet this need. Currently, sterile insect technology, introduction of natural enemies such as parasitoids or predators, application of entomopathogenic fungi or bacteria, etc., are some of the most commonly used integrated pest management techniques. Additional research into (detoxifying) symbiosis may result in environmentally acceptable and long-term ways of controlling large pest insect populations. Insect pest status, for example, may be heavily influenced by microbiota genotype, allowing for the identification and selection of genotypes most suited for addressing specific pest management priorities, ideally through low-tech means. In the same vein, detoxifying microbiota that can be isolated could be used in bioremediation or to treat pesticide poisoning. To better understand detoxifying microbiota in agriculturally significant pest insects, we provided comprehensive information regarding the role of gut microbiota in the detoxification of pesticides. Further investigation may be helpful to produce an effective integrated pest management program.
Author Contributions
JS, MK, BB, and MH wrote the initial draft. YX financially supported and supervised the manuscript. JS and YX conceptualized and developed the document. MQ and MTR provided critical feedback and reviewed the manuscript. MAR, SA, and MS revised the manuscript. All authors have read and agreed to the final version of the manuscript.
Funding
This study was supported by the National Key Research and Development Project (2021YFC2600404).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adair, K. L., and Douglas, A. E. (2017). Making a microbiome: the many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 35, 23–29. doi: 10.1016/j.mib.2016.11.002
Ahmad, M., Sayyed, A. H., Saleem, M. A., and Ahmad, M. (2008). Evidence for field evolved resistance to newer insecticides in Spodoptera litura (Lepidoptera: Noctuidae) from Pakistan. Crop Prot. 27, 1367–1372. doi: 10.1016/j.cropro.2008.05.003
Aikawa, T., Maehara, N., Ichihara, Y., Masuya, H., Nakamura, K., and Anbutsu, H. (2022). Cytoplasmic incompatibility in the semivoltine longicorn beetle Acalolepta fraudatrix (Coleoptera: Cerambycidae) double infected with Wolbachia. PLoS One 17:e0261928. doi: 10.1371/journal.pone.0261928
Akami, M., Andongma, A. A., Zhengzhong, C., Nan, J., Khaeso, K., Jurkevitch, E., et al. (2019a). Intestinal bacteria modulate the foraging behavior of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). PLoS One 14:e0210109. doi: 10.1371/journal.pone.0210109
Akami, M., Njintang, N. Y., Gbaye, O. A., Andongma, A. A., Rashid, M. A., Niu, C. Y., et al. (2019b). Gut bacteria of the cowpea beetle mediate its resistance to dichlorvos and susceptibility to Lippia adoensis essential oil. Sci. Rep. 9:6435. doi: 10.1038/s41598-019-42843-1
Akami, M., Ren, X., Wang, Y., Mansour, A., Cao, S., Qi, X., et al. (2022). Host fruits shape the changes in the gut microbiota and development of Bactrocera dorsalis (Diptera: Tephritidae) larvae. Int. J. Trop. Insect. Sci. 1–15. doi: 10.1007/s42690-022-00733-6
Alaraby, M., Hernández, A., and Marcos, R. (2018). Systematic in vivo study of NiO nanowires and nanospheres: biodegradation, uptake and biological impacts. Nanotoxicology 12, 1027–1044. doi: 10.1080/17435390.2018.1513091
de Almeida, L. G., de Moraes, L. A. B., Trigo, J. R. J. R., Omoto, C., et al. (2017). The gut microbiota of insecticide-resistant insects houses insecticide-degrading bacteria: A potential source for biotechnological exploitation. PLoS One 12:e0174754. doi: 10.1371/journal.pone.0174754
Armitage, S. A. O., Genersch, E., McMahon, D. P., Rafaluk-Mohr, C., and Rolff, J. (2022). Tripartite interactions: How immunity, microbiota and pathogens interact and affect pathogen virulence evolution. Curr. Opin. Insect. Sci. 50:100871. doi: 10.1016/j.cois.2021.12.011
Ayitso, A. S., and Onyango, D. M. (2016). Isolation and identification by morphological and biochemical methods of antibiotic producing microorganisms from the gut of Macrotermes michaelseni in maseno. Kenya. J. App. Bio. Biotech. 4, 27–33.
Bao, H., Gao, H., Zhang, Y., Fan, D., Fang, J., and Liu, Z. (2016). The roles of CYP6AY1 and CYP6ER1 in imidacloprid resistance in the brown planthopper: Expression levels and detoxification efficiency. Pestic. Biochem. Physiol. 129, 70–74. doi: 10.1016/j.pestbp.2015.10.020
Barber, M. D., Moores, G. D., Tatchell, G. M., Vice, W. E., and Denholm, I. (1999). Insecticide resistance in the currant–lettuce aphid, Nasonovia ribisnigri (Hemiptera: Aphididae) in the UK. Bull. Entomol. Res. 89, 17–23. doi: 10.1017/s0007485399000036
Basit, M., Saeed, S., Saleem, M. A., Denholm, I., and Shah, M. (2013). Detection of resistance, cross-resistance, and stability of resistance to new chemistry insecticides in Bemisia tabaci (Homoptera: Aleyrodidae). J. Econ. Entomol. 106, 1414–1422. doi: 10.1603/ec12414
Bass, C., Denholm, I., Williamson, M. S., and Nauen, R. (2015). The global status of insect resistance to neonicotinoid insecticides. Pestic. Biochem. Physiol. 121, 78–87. doi: 10.1016/j.pestbp.2015.04.004
Berasategui, A., Salem, H., Paetz, C., Santoro, M., Gershenzon, J., Kaltenpoth, M., et al. (2017). Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 26, 4099–4110. doi: 10.1111/mec.14186
Berasategui, A., Shukla, S., Salem, H., and Kaltenpoth, M. (2016). Potential applications of insect symbionts in biotechnology. Appl. Microbiol. Biotechnol. 100, 1567–1577. doi: 10.1007/s00253-015-7186-9
Berticat, C., Rousset, F., Raymond, M., Berthomieu, A., and Weill, M. (2002). High Wolbachia density in insecticide–resistant mosquitoes. Proc. R. Soc. London. Ser. B Biol. Sci. 269, 1413–1416. doi: 10.1098/rspb.2002.2022
Bhandari, G., Sharma, M., Negi, S., Gangola, S., Bhatt, P., and Chen, S. (2021). System biology analysis of endosulfan biodegradation in bacteria and its effect in other living systems: modeling and simulation studies. J. Biomol. Struct. Dyn. 8, 1–13. doi: 10.1080/07391102.2021.1982773
Bhatt, P., Pal, K., Bhandari, G., and Barh, A. (2019). Modelling of the methyl halide biodegradation in bacteria and its effect on environmental systems. Pestic. Biochem. Physiol. 158, 88–100. doi: 10.1016/j.pestbp.2019.04.015
Bhatt, P., Rene, E. R., Huang, Y., Lin, Z., Pang, S., Zhang, W., et al. (2021). Systems biology analysis of pyrethroid biodegradation in bacteria and its effect on the cellular environment of pests and humans. J. Environ. Chem. Eng. 9:106582. doi: 10.1016/j.jece.2021.106582
Bischoff, V., Vignal, C., Duvic, B., Boneca, I. G., Hoffmann, J. A., and Royet, J. (2006). Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2:e14. doi: 10.1371/journal.ppat.0020014
Boissière, A., Tchioffo, M. T., Bachar, D., Abate, L., Marie, A., Nsango, S. E., et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 8:e1002742. doi: 10.1371/journal.ppat.1002742
Bourguignon, T., Lo, N., Dietrich, C., Šobotník, J., Sidek, S., Roisin, Y., et al. (2018). Rampant host switching shaped the termite gut microbiome. Curr. Biol. 28, 649–654. doi: 10.1016/j.cub.2018.01.035
Boursaux-Eude, C., and Gross, R. (2000). New insights into symbiotic associations between ants and bacteria. Res. Microbiol. 151, 513–519. doi: 10.1016/s0923-2508(00)00221-7
Boush, M. G., and Matsumura, F. (1967). Insecticidal degradation by Pseudomonas melophthora, the bacterial symbiote of the apple maggot. J. Econ. Entomol. 60, 918–920. doi: 10.1093/jee/60.4.918
Bradley, P. H., and Pollard, K. S. (2017). Proteobacteria explain significant functional variability in the human gut microbiome. Microbiome 5, 1–23. doi: 10.1186/s40168-017-0244-z
Broderick, N. A., and Lemaitre, B. (2012). Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307–321. doi: 10.4161/gmic.19896
Bruno, D., Bonelli, M., Cadamuro, A. G., Reguzzoni, M., Grimaldi, A., Casartelli, M., et al. (2019a). The digestive system of the adult Hermetia illucens (Diptera: Stratiomyidae): morphological features and functional properties. Cell Tissue Res. 378, 221–238. doi: 10.1007/s00441-019-03025-7
Bruno, D., Bonelli, M., De Filippis, F., Di Lelio, I., Tettamanti, G., Casartelli, M., et al. (2019b). The Intestinal Microbiota of Hermetia i llucens Larvae Is Affected by Diet and Shows a Diverse Composition in the Different Midgut Regions. Appl. Environ. Microbiol. 85, e1864–e1818. doi: 10.1128/AEM.01864-18
Buchon, N., Silverman, N., and Cherry, S. (2014). Immunity in Drosophila melanogaster—from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810. doi: 10.1038/nri3763
Byrne, F. J., Toscano, N. C., Urena, A. A., and Morse, J. G. (2005). Quantification of imidacloprid toxicity to avocado thrips, Scirtothrips perseae Nakahara (Thysanoptera: Thripidae), using a combined bioassay and ELISA approach. Pest Manag. Sci. Former. Pestic. Sci. 61, 754–758. doi: 10.1002/ps.1052
Caspi-Fluger, A., Inbar, M., Mozes-Daube, N., Katzir, N., Portnoy, V., Belausov, E., et al. (2012). Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc. R. Soc. B Biol. Sci. 279, 1791–1796. doi: 10.1098/rspb.2011.2095
Ceja-Navarro, J. A., Vega, F. E., Karaoz, U., Hao, Z., Jenkins, S., Lim, H. C., et al. (2015). Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 6:7618. doi: 10.1038/ncomms8618
Chambers, M. C., Lightfield, K. L., and Schneider, D. S. (2012). How the fly balances its ability to combat different pathogens. PLoS Pathog. 8:e1002970. doi: 10.1371/journal.ppat.1002970
Charroux, B., and Royet, J. (2012). “Gut-microbiota interactions in non-mammals: what can we learn from Drosophila? Semi. Immunol. 24, 17–24. doi: 10.1016/j.smim.2011.11.003
Chaudhari, V. K., Desai, H. R., and Patel, N. M. (2015). Assessment of the insecticide resistance builds up on cotton leaf hopper, Amarasca biguttula (Ishida). Int. J. Adv. Multidiscip. Res. 2, 4–8.
Chen, B., Teh, B.-S., Sun, C., Hu, S., Lu, X., Boland, W., et al. (2016). Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6:29505. doi: 10.1038/srep29505
Chen, B., Zhang, N., Xie, S., Zhang, X., He, J., Muhammad, A., et al. (2020). Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ. Int. 143:105886. doi: 10.1016/j.envint.2020.105886
Chen, C., Shan, T., Liu, Y., Wang, C., Shi, X., and Gao, X. (2019). Identification and functional analysis of a cytochrome P450 gene involved in imidacloprid resistance in Bradysia odoriphaga Yang et Zhang. Pestic. Biochem. Physiol. 153, 129–135. doi: 10.1016/j.pestbp.2018.11.009
Chen, H., Singh, H., Bhardwaj, N., Bhardwaj, S. K., Khatri, M., Kim, K.-H., et al. (2022). An exploration on the toxicity mechanisms of phytotoxins and their potential utilities. Crit. Rev. Environ. Sci. Technol. 52, 395–435. doi: 10.1080/10643389.2020.1823172
Chen, X. D., Neupane, S., Gossett, H., Pelz-Stelinski, K. S., and Stelinski, L. L. (2021). Insecticide rotation scheme restores insecticide susceptibility in thiamethoxam-resistant field populations of Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Liviidae), in Florida. Pest Manag. Sci. 77, 464–473. doi: 10.1002/ps.6039
Cheng, D., Chen, S., Huang, Y., Pierce, N. E., Riegler, M., Yang, F., et al. (2019). Symbiotic microbiota may reflect host adaptation by resident to invasive ant species. PLoS Pathog. 15:e1007942. doi: 10.1371/journal.ppat.1007942
Cheng, D., Guo, Z., Riegler, M., Xi, Z., Liang, G., and Xu, Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5, 1–12. doi: 10.1186/s40168-017-0236-z
Chmiel, J. A., Daisley, B. A., Burton, J. P., and Reid, G. (2019). Deleterious effects of neonicotinoid pesticides on Drosophila melanogaster immune pathways. mbio 10, e1395–e1319. doi: 10.1128/mBio.01395-19
Chouaia, B., Rossi, P., Epis, S., Mosca, M., Ricci, I., Damiani, C., et al. (2012). Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 12:S2. doi: 10.1186/1471-2180-12-S1-S2
Cirimotich, C. M., Ramirez, J. L., and Dimopoulos, G. (2011). Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 10, 307–310. doi: 10.1016/j.chom.2011.09.006
Consortium, H. M. P. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486:207. doi: 10.1038/nature11234
Coon, K. L., Brown, M. R., and Strand, M. R. (2016). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 25, 5806–5826. doi: 10.1111/mec.13877
Dang, K., Doggett, S. L., Singham, G. V., and Lee, C.-Y. (2017). Insecticide resistance and resistance mechanisms in bed bugs. Cimex spp. (Hemiptera: Cimicidae). Parasit. Vectors 10:318.
Dângelo, R. A. C., Michereff-Filho, M., Campos, M. R., Da Silva, P. S., and Guedes, R. N. C. (2018). Insecticide resistance and control failure likelihood of the whitefly Bemisia tabaci (MEAM1; B biotype): a Neotropical scenario. Ann. Appl. Biol. 172, 88–99. doi: 10.1111/aab.12404
Dastranj, M., Gharechahi, J., and Salekdeh, G. H. (2016). “Insect pest proteomics and its potential application in pest control management,” in Agricultural Proteomics Volume 2, G. H. Salekdeh ed (Berlin: Springer), 267–287. doi: 10.1007/978-3-319-43278-6_12
de Almeida, L. G. (2013). Simbiontes de Spodoptera Frugiperda (Je Smith, 1797) (Lepidoptera: Noctuidae): Potencial Biotecnológico Para Biorremediação e Implicações na Metabolização de Inseticidas Pelo Hospedeiro. São Paulo: Diss. Universidade de São Paulo.
Degli Esposti, M., and Martinez Romero, E. (2017). The functional microbiome of arthropods. PLoS One 12:e0176573. doi: 10.1371/journal.pone.0176573
Donelli, G., Vuotto, C., Cardines, R., and Mastrantonio, P. (2012). Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol. Med. Microbiol. 65, 318–325. doi: 10.1111/j.1574-695X.2012.00962.x
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Douglas, A. E. (2018). Omics and the metabolic function of insect–microbial symbioses. Curr. Opin. insect Sci. 29, 1–6. doi: 10.1016/j.cois.2018.05.012
Du, Y., Grodowitz, M. J., and Chen, J. (2020). Insecticidal and enzyme inhibitory activities of isothiocyanates against red imported fire ants, Solenopsis invicta. Biomolecules 1:716. doi: 10.3390/biom10050716
Elzaki, M. E. A., Miah, M. A., Wu, M., Zhang, H., Pu, J., Jiang, L., et al. (2017). Imidacloprid is degraded by CYP353D1v2, a cytochrome P450 overexpressed in a resistant strain of Laodelphax striatellus. Pest Manag. Sci. 73, 1358–1363. doi: 10.1002/ps.4570
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Engel, P., Kwong, W. K., McFrederick, Q., Anderson, K. E., Barribeau, S. M., Chandler, J. A., et al. (2016). The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mbio 7, e2164–e2115. doi: 10.1128/mBio.02164-15
Engel, P., Martinson, V. G., and Moran, N. A. (2012). Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. 109, 11002–11007. doi: 10.1073/pnas.1202970109
Erb, M., and Kliebenstein, D. J. (2020). Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol. 184, 39–52. doi: 10.1104/pp.20.00433
Erlandson, M. A., Toprak, U., and Hegedus, D. D. (2019). Role of the peritrophic matrix in insect-pathogen interactions. J. Insect Physiol. 117:103894. doi: 10.1016/j.jinsphys.2019.103894
Eski, A., Demir, I., Güllü, M., and Demirbağ, Z. (2018). Biodiversity and pathogenicity of bacteria associated with the gut microbiota of beet armyworm, Spodoptera exigua Hübner (Lepidoptera: Noctuidae). Microb. Pathog. 121, 350–358. doi: 10.1016/j.micpath.2018.05.012
Ferguson, J. S. (2004). Development and stability of insecticide resistance in the leafminer Liriomyza trifolii (Diptera: Agromyzidae) to cyromazine, abamectin, and spinosad. J. Econ. Entomol. 97, 112–119. doi: 10.1093/jee/97.1.112
Flury, P., Vesga, P., Dominguez-Ferreras, A., Tinguely, C., Ullrich, C. I., Kleespies, R. G., et al. (2019). Persistence of root-colonizing Pseudomonas protegens in herbivorous insects throughout different developmental stages and dispersal to new host plants. ISME J. 13, 860–872. doi: 10.1038/s41396-018-0317-4
Foster, S. P., Paul, V. L., Slater, R., Warren, A., Denholm, I., Field, L. M., et al. (2014). A mutation (L1014F) in the voltage-gated sodium channel of the grain aphid, Sitobion avenae, is associated with resistance to pyrethroid insecticides. Pest Manag. Sci. 70, 1249–1253. doi: 10.1002/ps.3683
Fox-Dobbs, K., Doak, D. F., Brody, A. K., and Palmer, T. M. (2010). Termites create spatial structure and govern ecosystem function by affecting N2 fixation in an East African savanna. Ecology 91, 1296–1307. doi: 10.1890/09-0653.1
Frago, E., Dicke, M., and Godfray, H. C. J. (2012). Insect symbionts as hidden players in insect–plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Funaro, C. F., Kronauer, D. J. C., Moreau, C. S., Goldman-Huertas, B., Pierce, N. E., and Russell, J. A. (2011). Army ants harbor a host-specific clade of Entomoplasmatales bacteria. Appl. Environ. Microbiol. 77, 346–350. doi: 10.1128/AEM.01896-10
Furlong, M. J., Wright, D. J., and Dosdall, L. M. (2013). Diamondback moth ecology and management: problems, progress, and prospects. Annu. Rev. Entomol. 58, 517–541. doi: 10.1146/annurev-ento-120811-153605
Fusetto, R., Denecke, S., Perry, T., Richard, A. J., Batterham, P., O’Hair, R. A. J., et al. (2017). Partitioning the roles of CYP6G1 and gut microbes in the metabolism of the insecticide imidacloprid in Drosophila melanogaster. Sci. Rep. 7:11339. doi: 10.1038/s41598-017-09800-2
Gangola, S., Bhatt, P., Kumar, A. J., Bhandari, G., Joshi, S., Punetha, A., et al. (2022). Biotechnological tools to elucidate the mechanism of pesticide degradation in the environment. Chemosphere 8:133916. doi: 10.1016/j.chemosphere.2022.133916
Garcia, J. (2015). Acquisition and Maintenance of Specific Bacterial Symbionts in Vertical and Horizontal Symbioses. [Ph.D thesis] (Atlanta: Diss. Emory University).
Garrood, W. T., Zimmer, C. T., Gorman, K. J., Nauen, R., Bass, C., and Davies, T. G. E. (2016). Field-evolved resistance to imidacloprid and ethiprole in populations of brown planthopper Nilaparvata lugens collected from across South and East Asia. Pest Manag. Sci. 72, 140–149. doi: 10.1002/ps.3980
Garrood, W. T., Zimmer, C. T., Gutbrod, O., Lüke, B., Williamson, M. S., Bass, C., et al. (2017). Influence of the RDL A301S mutation in the brown planthopper Nilaparvata lugens on the activity of phenylpyrazole insecticides. Pestic. Biochem. Physiol. 142, 1–8. doi: 10.1016/j.pestbp.2017.01.007
Giambò, F., Teodoro, M., Costa, C., and Fenga, C. (2021). Toxicology and Microbiota: How Do Pesticides Influence Gut Microbiota? A Review. Int. J. Environ. Res. Public Health 18:5510. doi: 10.3390/ijerph18115510
Gomes, A. F. F., Omoto, C., and Cônsoli, F. L. (2020). Gut bacteria of field-collected larvae of Spodoptera frugiperda undergo selection and are more diverse and active in metabolizing multiple insecticides than laboratory-selected resistant strains. J. Pest Sci. 93, 833–851. doi: 10.1007/s10340-020-01202-0
Gressel, J. (2018). Microbiome facilitated pest resistance: potential problems and uses. Pest Manag. Sci. 74, 511–515. doi: 10.1002/ps.4777
Gunstone, T., Cornelisse, T., Klein, K., Dubey, A., and Donley, N. (2021). Pesticides and soil invertebrates: A hazard assessment. Front. Environ. Sci. 9:122. doi: 10.3389/fenvs.2021.643847
Guo, Z., Lu, Y., Yang, F., Zeng, L., Liang, G., and Xu, Y. (2017). Transmission modes of a pesticide-degrading symbiont of the oriental fruit fly Bactrocera dorsalis (Hendel). Appl. Microbiol. Biotechnol. 101, 8543–8556. doi: 10.1007/s00253-017-8551-7
Gutiérrez-Cabrera, A. E., Córdoba-Aguilar, A., Zenteno, E., Lowenberger, C., and Espinoza, B. (2016). Origin, evolution and function of the Hemipteran perimicrovillar membrane with emphasis on Reduviidae that transmit Chagas disease. Bull. Entomol. Res. 106, 279–291. doi: 10.1017/S0007485315000929
Guzman, J., and Vilcinskas, A. (2020). Bacteria associated with cockroaches: health risk or biotechnological opportunity? Appl. Microbiol. Biotechnol. 104, 10369-10387. doi: 10.1007/s00253-020-10973-6
Hafeez, M., Li, X., Zhang, Z., Huang, J., Wang, L., Zhang, J., et al. (2021). De novo transcriptomic analyses revealed some detoxification genes and related pathways responsive to poposion yihaogong® 5% ec (Lambda-cyhalothrin 5%) exposure in Spodoptera frugiperda third-instar larvae. Insects 12:132. doi: 10.3390/insects12020132
Hafeez, M., Liu, S., Jan, S., Ali, B., Shahid, M., Fernández-Grandon, G. M., et al. (2019). Gossypol-induced fitness gain and increased resistance to deltamethrin in beet armyworm, Spodoptera exigua (Hübner). Pest Manag. Sci. 75, 683–693. doi: 10.1002/ps.5165
Hafeez, M., Liu, S., Yousaf, H. K., Jan, S., Wang, R. L., Fernández-Grandon, G. M., et al. (2020). RNA interference-mediated knockdown of a cytochrome P450 gene enhanced the toxicity of α-cypermethrin in xanthotoxin-fed larvae of Spodoptera exigua (Hübner). Pestic. Biochem. Physiol. 162, 6–14. doi: 10.1016/j.pestbp.2019.07.003
Hamada, A., Stam, L., Nakao, T., Kawashima, M., and Banba, S. (2020). Differential metabolism of neonicotinoids by brown planthopper, Nilaparvata lugens, CYP6ER1 variants. Pestic. Biochem. Physiol. 165:104538. doi: 10.1016/j.pestbp.2020.02.004
Hamada, A., Wahl, G. D., Nesterov, A., Nakao, T., Kawashima, M., and Banba, S. (2019). Differential metabolism of imidacloprid and dinotefuran by Bemisia tabaci CYP6CM1 variants. Pestic. Biochem. Physiol. 159, 27–33. doi: 10.1016/j.pestbp.2019.05.011
Hamilton, P. T., and Perlman, S. J. (2013). Host defense via symbiosis in Drosophila. PLoS Pathog. 9:e1003808. doi: 10.1371/journal.ppat.1003808
Hammer, T. J., and Bowers, M. D. (2015). Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia 179, 1–14.
Hammer, T. J., and Moran, N. A. (2019). Links between metamorphosis and symbiosis in holometabolous insects. Philos. Trans. R. Soc. B 374:20190068. doi: 10.1098/rstb.2019.0068
Hansen, A. K., and Moran, N. A. (2011). Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci.U.S.A. 108, 2849–2854. doi: 10.1073/pnas.1013465108
Hanson, M. A., and Lemaitre, B. (2020). New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 62, 22–30. doi: 10.1016/j.coi.2019.11.008
Hassan, B., Siddiqui, J. A., and Xu, Y. (2020). Vertically Transmitted Gut Bacteria and Nutrition Influence the Immunity and Fitness of Bactrocera dorsalis Larvae. Front. Microbiol. 11:596352. doi: 10.3389/fmicb.2020.596352
Hauffe, H. C., and Barelli, C. (2019). Conserve the germs: the gut microbiota and adaptive potential. Conserv. Genet. 20, 19–27. doi: 10.1007/s10592-019-01150-y
He, M., Ai, Z., Jiang, Z., Long, Y., Zhang, Y., and He, P. (2015). Biochemistry and molecular characterisation of chlorpyrifos resistance in field strains of the white-backed planthopper Sogatella furcifera (Hemiptera: Delphacidae). Austral Entomol. 54, 376–384. doi: 10.1111/aen.12140
Hegeto, L. A., Ronqui, L., Lapenta, A. S., and Albuquerque, F. A. (2015). Identification and functional characterization of esterases in Euschistus heros (Hemiptera, Pentatomidae) and their relationship with thiamethoxam and lambda-cyhalothrin. Genet Mol Res 14, 11079–11088. doi: 10.4238/2015.September.22.1
Hervé, V., Liu, P., Dietrich, C., Sillam-Dussès, D., Stiblik, P., Šobotník, J., et al. (2020). Phylogenomic analysis of 589 metagenome-assembled genomes encompassing all major prokaryotic lineages from the gut of higher termites. PeerJ 8:e8614. doi: 10.7717/peerj.8614
Heys, C., Lizé, A., Blow, F., White, L., Darby, A., and Lewis, Z. J. (2018). The effect of gut microbiota elimination in Drosophila melanogaster: A how-to guide for host–microbiota studies. Ecol. Evol. 8, 4150–4161. doi: 10.1002/ece3.3991
Higuita Palacio, M. F., Montoya, O. I., Saldamando, C. I., García-Bonilla, E., Junca, H., Cadavid-Restrepo, G. E., et al. (2021). Dry and rainy seasons significantly alter the gut microbiome composition and reveal a key Enterococcus sp. (Lactobacillales: Enterococcaceae) core component in Spodoptera frugiperda (Lepidoptera: Noctuidae) corn strain from Northwestern Colombia. J. Insect Sci. 21:10. doi: 10.1093/jisesa/ieab076
Højland, D. H., Nauen, R., Foster, S. P., Williamson, M. S., and Kristensen, M. (2015). Incidence, spread and mechanisms of pyrethroid resistance in European populations of the cabbage stem flea beetle, Psylliodes chrysocephala L. (Coleoptera: Chrysomelidae). PLoS One 10:e0146045. doi: 10.1371/journal.pone.0146045
Holt, J. F. (2013). Ecology of the Manduca Sexta gut Microbiota. Madison, WI: The University of Wisconsin-Madison*.
Hongoh, Y. (2010). Diversity and genomes of uncultured microbial symbionts in the termite gut. Biosci. Biotechnol. Biochem. 74, 1145–1151. doi: 10.1271/bbb.100094
Hu, Z., Xia, F., Lin, Q., Chen, H., Li, Z., Fei, Y. I. N., et al. (2014). Biochemical mechanism of chlorantraniliprole resistance in the diamondback moth, Plutella xylostella Linnaeus. J. Integr. Agric. 13, 2452–2459. doi: 10.1016/s2095-3119(14)60748-6
Hubbard, M., Hynes, R. K., Erlandson, M., and Bailey, K. L. (2014). The biochemistry behind biopesticide efficacy. Sustain. Chem. Process. 2:18.
Hussain, M., Debnath, B., Qasim, M., Bamisile, B. S., Islam, W., Hameed, M. S., et al. (2019). Role of saponins in plant defense against specialist herbivores. Molecules 24:2067. doi: 10.3390/molecules24112067
Hussain, M., Gao, J., Bano, S., Wang, L., Lin, Y., Arthurs, S., et al. (2020). Diamondback moth larvae trigger host plant volatiles that lure its adult females for oviposition. Insects 11:725. doi: 10.3390/insects11110725
Hussain, S., Hartley, C. J., Shettigar, M., and Pandey, G. (2016). Bacterial biodegradation of neonicotinoid pesticides in soil and water systems. FEMS Microbiol. Lett. 363:fnw252. doi: 10.1093/femsle/fnw252
İşci, M., and Ay, R. (2017). Determination of resistance and resistance mechanisms to thiacloprid in Cydia pomonella L. (Lepidoptera: Tortricidae) populations collected from apple orchards in Isparta Province, Turkey. Crop Prot. 91, 82–88. doi: 10.1016/j.cropro.2016.09.015
Ishtiaq, M., Saleem, M. A., and Razaq, M. (2012). Monitoring of resistance in Spodoptera exigua (Lepidoptera: Noctuidae) from four districts of the Southern Punjab, Pakistan to four conventional and six new chemistry insecticides. Crop Prot. 33, 13–20. doi: 10.1016/j.cropro.2011.11.014
Ismail, S. M. (2020). Effect of sublethal doses of some insecticides and their role on detoxication enzymes and protein-content of Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae). Bull. Natl. Res. Cent 44:35.
Itoh, H., Hori, T., Sato, Y., Nagayama, A., Tago, K., Hayatsu, M., et al. (2018a). Infection dynamics of insecticide-degrading symbionts from soil to insects in response to insecticide spraying. ISME J. 12, 909–920. doi: 10.1038/s41396-017-0021-9
Itoh, H., Tago, K., Hayatsu, M., and Kikuchi, Y. (2018b). Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 35, 434–454. doi: 10.1039/c7np00051k
Jaenike, J., Unckless, R., Cockburn, S. N., Boelio, L. M., and Perlman, S. J. (2010). Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. doi: 10.1126/science.1188235
Jin, J.-X., Jin, D.-C., Li, W.-H., Cheng, Y., Li, F.-L., and Ye, Z.-C. (2017). Monitoring trends in insecticide resistance of field populations of Sogatella furcifera (Hemiptera: Delphacidae) in Guizhou province, China, 2012–2015. J. Econ. Entomol. 110, 641–650. doi: 10.1093/jee/tox027
Johnston, P. R., Paris, V., and Rolff, J. (2019). Immune gene regulation in the gut during metamorphosis in a holo-versus a hemimetabolous insect. Philos. Trans. R. Soc. B 374, 20190073. doi: 10.1098/rstb.2019.0073
Kalsi, M., and Palli, S. R. (2017). Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 83, 1–12. doi: 10.1016/j.ibmb.2017.02.002
Kamal, A., Ahmad, F., and Shafeeque, M. A. (2020). Toxicity of pesticides to plants and non-target organism: A comprehensive review. Iran. J. Plant Physiol. 10, 3299–3313.
Khalid, M., Rasul, S., Hussain, J., Ahmad, R., Zia, A., Bilal, M., et al. (2016). Biodegradation of Organophosphorus Insecticides, Chlorpyrifos, by Pseudomonas putida CP-1. Pak. J. Zool. 48, 1453–1458.
Khan, H. A. A., Akram, W., Khan, T., Haider, M. S., Iqbal, N., and Zubair, M. (2016). Risk assessment, cross-resistance potential, and biochemical mechanism of resistance to emamectin benzoate in a field strain of house fly (Musca domestica Linnaeus). Chemosphere 151, 133–137. doi: 10.1016/j.chemosphere.2016.02.077
Khan, M. M., Kaleem-ullah, R. M., Siddiqui, J. A., and Ali, S. (2020). Insecticide Resistance and Detoxification Enzymes Activity in Nilaparvata lugens Stål Against Neonicotinoids. J. Agric. Sci. 12, 24–36. doi: 10.5539/jas.v12n5p24
Khan, M. M., Khan, A. H., Ali, M. W., Hafeez, M., Ali, S., Du, C., et al. (2021). Emamectin benzoate induced enzymatic and transcriptional alternation in detoxification mechanism of predatory beetle Paederus fuscipes (Coleoptera: Staphylinidae) at the sublethal concentration. Ecotoxicology 30, 1227–1241. doi: 10.1007/s10646-021-02426-1
Kikuchi, Y., and Yumoto, I. (2013). Efficient colonization of the bean bug Riptortus pedestris by an environmentally transmitted Burkholderia symbiont. Appl. Environ. Microbiol. 79, 2088–2091. doi: 10.1128/AEM.03299-12
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci.U.S.A. 109, 8618–8622. doi: 10.1073/pnas.1200231109
Kim, J. K., and Lee, B. L. (2015). Symbiotic factors in Burkholderia essential for establishing an association with the bean bug, Riptortus pedestris. Arch. Insect Biochem. Physiol. 88, 4–17. doi: 10.1002/arch.21218
Kim, J., Il, Kwon, M., Kim, G.-H., Kim, S. Y., and Lee, S. H. (2015). Two mutations in nAChR beta subunit is associated with imidacloprid resistance in the Aphis gossypii. J. Asia. Pac. Entomol. 18, 291–296. doi: 10.1016/j.aspen.2015.01.010
Klock, M. M., Barrett, L. G., Thrall, P. H., and Harms, K. E. (2015). Host promiscuity in symbiont associations can influence exotic legume establishment and colonization of novel ranges. Divers. Distrib. 21, 1193–1203. doi: 10.1111/ddi.12363
Kohl, K. D., and Dearing, M. D. (2016). The woodrat gut microbiota as an experimental system for understanding microbial metabolism of dietary toxins. Front. Microbiol. 7:1165. doi: 10.3389/fmicb.2016.01165
Koo, H.-N., An, J.-J., Park, S.-E., Kim, J.-I., and Kim, G.-H. (2014). Regional susceptibilities to 12 insecticides of melon and cotton aphid, Aphis gossypii (Hemiptera: Aphididae) and a point mutation associated with imidacloprid resistance. Crop Prot. 55, 91–97. doi: 10.1016/j.cropro.2013.09.010
Kuechler, S. M., Fukatsu, T., and Matsuura, Y. (2019). Repeated evolution of bacteriocytes in lygaeoid stinkbugs. Environ. Microbiol. 21, 4378–4394. doi: 10.1111/1462-2920.14804
Kuraishi, T., Hori, A., and Kurata, S. (2013). Host-microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 4:375. doi: 10.3389/fphys.2013.00375
Lai, T., and Su, J. (2011). Assessment of resistance risk in Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae) to chlorantraniliprole. Pest Manag. Sci. 67, 1468–1472. doi: 10.1002/ps.2201
Lee, K.-A., Kim, S.-H., Kim, E.-K., Ha, E.-M., You, H., Kim, B., et al. (2013). Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811. doi: 10.1016/j.cell.2013.04.009
Li, F., Li, M., Mao, T., Wang, H., Chen, J., Lu, Z., et al. (2020). Effects of phoxim exposure on gut microbial composition in the silkworm, Bombyx mori. Ecotoxicol. Environ. Saf. 189:110011. doi: 10.1016/j.ecoenv.2019.110011
Lin, T., Cai, Z., and Wu, H. (2015). Transcriptome analysis of the Japanese pine sawyer beetle, Monochamus alternatus (Coleoptera: Cerambycidae) by high-throughput Illumina sequencing. J. Asia. Pac. Entomol. 18, 439–445. doi: 10.1016/j.aspen.2015.04.011
Lin, Z., Pang, S., Zhou, Z., Wu, X., Li, J., Huang, Y., et al. (2022). Novel pathway of acephate degradation by the microbial consortium ZQ01 and its potential for environmental bioremediation. J. Hazard. Mater. 426:127841. doi: 10.1016/j.jhazmat.2021.127841
Linser, P. J., and Dinglasan, R. R. (2014). Insect gut structure, function, development and target of biological toxins. Adv. In Insect Phys. 47, 1–37. doi: 10.1016/b978-0-12-800197-4.00001-4
Liu, N., Li, H., Chevrette, M. G., Zhang, L., Cao, L., Zhou, H., et al. (2019). Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J. 13, 104–117. doi: 10.1038/s41396-018-0255-1
Liu, X., Grabherr, H. M., Willmann, R., Kolb, D., Brunner, F., Bertsche, U., et al. (2014). Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. elife 3:e01990. doi: 10.7554/eLife.01990
Longhurst, C., Babcock, J. M., Denholm, I., Gorman, K., Thomas, J. D., and Sparks, T. C. (2013). Cross-resistance relationships of the sulfoximine insecticide sulfoxaflor with neonicotinoids and other insecticides in the whiteflies Bemisia tabaci and Trialeurodes vaporariorum. Pest Manag. Sci. 69, 809–813. doi: 10.1002/ps.3439
Lourthuraj, A. A., Hatshan, M. R., and Hussein, D. S. (2022). Biocatalytic degradation of organophosphate pesticide from the wastewater and hydrolytic enzyme properties of consortium isolated from the pesticide contaminated water. Environ. Res. 205:112553. doi: 10.1016/j.envres.2021.112553
Lu, T.-Q., Mao, S.-Y., Sun, S.-L., Yang, W.-L., Ge, F., and Dai, Y.-J. (2016). Regulation of hydroxylation and nitroreduction pathways during metabolism of the neonicotinoid insecticide imidacloprid by Pseudomonas putida. J. Agric. Food Chem. 64, 4866–4875. doi: 10.1021/acs.jafc.6b01376
Maire, J., Vincent-Monégat, C., Balmand, S., Vallier, A., Hervé, M., Masson, F., et al. (2019). Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc. Natl. Acad. Sci.U.S.A. 116, 5623–5632. doi: 10.1073/pnas.1821806116
Majumder, R., Sutcliffe, B., Taylor, P. W., and Chapman, T. A. (2020). Microbiome of the Queensland fruit fly through metamorphosis. Microorganisms 8:795. doi: 10.3390/microorganisms8060795
Malekmohammadi, M., and Galehdari, H. (2016). Target site insensitivity mutations in the AChE enzyme confer resistance to organophosphorous insecticides in Leptinotarsa decemlineata (Say). Pestic. Biochem. Physiol. 126, 85–91. doi: 10.1016/j.pestbp.2015.08.002
Mason, C. J. (2020). Complex relationships at the intersection of insect gut microbiomes and plant defenses. J. Chem. Ecol. 46, 793–807. doi: 10.1007/s10886-020-01187-1
Mason, C. J., Jones, A. G., and Felton, G. W. (2019). Co-option of microbial associates by insects and their impact on plant–folivore interactions. Plant. Cell Environ. 42, 1078–1086. doi: 10.1111/pce.13430
Mason, K. L., Stepien, T. A., Blum, J. E., Holt, J. F., Labbe, N. H., Rush, J. S., et al. (2011). From commensal to pathogen: translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. MBio 2, e65–e11. doi: 10.1128/mBio.00065-11
Mejía-Alvarado, F. S., Ghneim-Herrera, T., Góngora, C. E., Benavides, P., and Navarro-Escalante, L. (2021). Structure and Dynamics of the Gut Bacterial Community Across the Developmental Stages of the Coffee Berry Borer, Hypothenemus hampei. Front. Microbiol 12:639868. doi: 10.3389/fmicb.2021.639868
Mereghetti, V., Chouaia, B., and Montagna, M. (2017). New insights into the microbiota of moth pests. Int. J. Mol. Sci. 18:2450. doi: 10.3390/ijms18112450
Miller, T. A. (2011). Paratransgenesis as a potential tool for pest control: review of applied arthropod symbiosis. J. Appl. Entomol. 135, 474–478. doi: 10.1111/j.1439-0418.2010.01600.x
Mohammadi, M., Shadnoush, M., Sohrabvandi, S., Yousefi, M., Khorshidian, N., and Mortazavian, A. M. (2021). Probiotics as potential detoxification tools for mitigation of pesticides: a mini review. Int. J. Food Sci. Technol. 56, 2078–2087. doi: 10.1111/ijfs.14880
Moreau, C. S. (2020). Symbioses among ants and microbes. Curr. Opin. Insect Sci. 39, 1–5. doi: 10.1016/j.cois.2020.01.002
Moreno-García, M., Condé, R., Bello-Bedoy, R., and Lanz-Mendoza, H. (2014). The damage threshold hypothesis and the immune strategies of insects. Infect. Genet. Evol. 24, 25–33. doi: 10.1016/j.meegid.2014.02.010
Müller, K., Tiktak, A., Dijkman, T. J., Green, S., and Clothier, B. (2014). “Advances in pesticide risk reduction”. Enceclopedia of Agriculture and Food Systems. N. V Alfen (Berlin: Elsevier) 17–34. doi: 10.1016/b978-0-444-52512-3.00242-4
Musa Khan, M., Hafeez, M., Ali Siddiqui, J., Ullah, F., Shah, S., Iftikhar, A., et al. (2021). Residual toxicity and low lethal effects of fenvalerate on the development and physiology of Spodoptera exigua reared on different hosts. J. King Saud Univ.Sci. 33:101593. doi: 10.1016/j.jksus.2021.101593
Muthukrishnan, S., Merzendorfer, H., Arakane, Y., and Kramer, K. J. (2012). “Chitin metabolism in insects,” in Insect Molecular Biology and Biochemistry. L I. Gilbert ed (Berlin: Elsevier), 193–235. doi: 10.1016/b978-0-12-384747-8.10007-8
Naeem, A., Freed, S., Jin, F. L., Akmal, M., and Mehmood, M. (2016). Monitoring of insecticide resistance in Diaphorina citri Kuwayama (Hemiptera: Psyllidae) from citrus groves of Punjab, Pakistan. Crop Prot. 86, 62–68. doi: 10.1016/j.cropro.2016.04.010
Nagarajan, V. M., Yuvan, M., Srinivasan, R., Satagopan, N. R., Asokan, A., and Anooja, A. (2022). Status of important coastal habitats of North Tamil Nadu: Diversity, current threats and approaches for conservation. Reg. Stud. Mar. Sci. 49:102106. doi: 10.1016/j.rsma.2021.102106
Naik, V. C. B., Kumbhare, S., Kranthi, S., Satija, U., and Kranthi, K. R. (2018). Field-evolved resistance of pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae), to transgenic Bacillus thuringiensis (Bt) cotton expressing crystal 1Ac (Cry1Ac) and Cry2Ab in India. Pest Manag. Sci. 74, 2544–2554. doi: 10.1002/ps.5038
Nakaishi, Y., Sato, M., Bando, H., and Asano, S. (2018). A mutation in Plutella xylostella ABCC2 causes resistance to Bacillus thuringiensis Cry1Ac by interfering with its receptor function. J. Insect Biotechnol. Sericology 87, 45–42.
Nardi, J. B., and Bee, C. M. (2012). Regenerative cells and the architecture of beetle midgut epithelia. J. Morphol. 273, 1010–1020. doi: 10.1002/jmor.20038
Nayak, S. K., Dash, B., and Baliyarsingh, B. (2018). Icrobial Remediation Of Persistent Agro-Chemicals By Soil Bacteria: An Overview. In Microbial Biotechnology. Patra J., Das G., Shin H. S. eds (Singapore: Springer). 275–301. doi: 10.1007/978-981-10-7140-9_13
Nazemi, A., Khajehali, J., and Van Leeuwen, T. (2016). Incidence and characterization of resistance to pyrethroid and organophosphorus insecticides in Thrips tabaci (Thysanoptera: Thripidae) in onion fields in Isfahan. Iran. Pestic. Biochem. Physiol. 129, 28–35. doi: 10.1016/j.pestbp.2015.10.013
Nehme, N. T., Liégeois, S., Kele, B., Giammarinaro, P., Pradel, E., Hoffmann, J. A., et al. (2007). A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3:e173. doi: 10.1371/journal.ppat.0030173
Nicoletti, R., and Becchimanzi, A. (2022). Ecological and Molecular Interactions between Insects and Fungi. Microorganisms 10:96. doi: 10.3390/microorganisms10010096
O’Donnell, M. J., and Donini, A. (2017). “Nitrogen excretion and metabolism in insects,” in Acid-Base Balance and Nitrogen Excretion in Invertebrates, (Berlin: Springer), 109–126. doi: 10.1007/978-3-319-39617-0_4
Onchuru, T. O., Martinez, A. J., Ingham, C. S., and Kaltenpoth, M. (2018). Transmission of mutualistic bacteria in social and gregarious insects. Curr. Opin. Insect Sci. 28, 50–58. doi: 10.1016/j.cois.2018.05.002
Pang, R., Chen, M., Liang, Z., Yue, X., Ge, H., and Zhang, W. (2016). Functional analysis of CYP6ER1, a P450 gene associated with imidacloprid resistance in Nilaparvata lugens. Sci. Rep. 6:34992. doi: 10.1038/srep34992
Pang, R., Chen, M., Yue, L., Xing, K., Li, T., Kang, K., et al. (2018). A distinct strain of Arsenophonus symbiont decreases insecticide resistance in its insect host. PLoS Genet. 14:e1007725. doi: 10.1371/journal.pgen.1007725
Pang, S., Lin, Z., Zhang, W., Mishra, S., Bhatt, P., and Chen, S. (2020a). Insights into the microbial degradation and biochemical mechanisms of neonicotinoids. Front. Microbiol. 11:868. doi: 10.3389/fmicb.2020.00868
Pang, S., Lin, Z., Zhang, Y., Zhang, W., Alansary, N., Mishra, S., et al. (2020b). Insights into the toxicity and degradation mechanisms of imidacloprid via physicochemical and microbial approaches. Toxics 8:65. doi: 10.3390/toxics8030065
Paniagua Voirol, L. R., Frago, E., Kaltenpoth, M., Hilker, M., and Fatouros, N. E. (2018). Bacterial symbionts in Lepidoptera: their diversity, transmission, and impact on the host. Front. Microbiol. 9:556. doi: 10.3389/fmicb.2018.00556
Panini, M., Dradi, D., Marani, G., Butturini, A., and Mazzoni, E. (2014). Detecting the presence of target-site resistance to neonicotinoids and pyrethroids in Italian populations of Myzus persicae. Pest Manag. Sci. 70, 931–938. doi: 10.1002/ps.3630
Patil, S. S., Ugale, T. B., Lande, G. K., and Barkhade, U. P. (2011). Resistance to emamamectin benzoate in Plutella xylostella collected from different geographic locations. Int. J. Plant Prot. 4, 168–171.
Pietri, J. E., Tiffany, C., and Liang, D. (2018). Disruption of the microbiota affects physiological and evolutionary aspects of insecticide resistance in the German cockroach, an important urban pest. PLoS One 13:e0207985. doi: 10.1371/journal.pone.0207985
Polenogova, O. V., Noskov, Y. A., Yaroslavtseva, O. N., Kryukova, N. A., Alikina, T., Klementeva, T. N., et al. (2021). Influence of Bacillus thuringiensis and avermectins on gut physiology and microbiota in Colorado potato beetle: Impact of enterobacteria on susceptibility to insecticides. PLoS One 16:e0248704. doi: 10.1371/journal.pone.0248704
Pons, I., Scieur, N., Dhondt, L., Renard, M.-E., Renoz, F., and Hance, T. (2022). Pervasiveness of the symbiont Serratia symbiotica in the aphid natural environment: distribution, diversity and evolution at a multitrophic level. FEMS Microbiol. Ecol. 98:fiac012. doi: 10.1093/femsec/fiac012
Possemiers, S., Bolca, S., Verstraete, W., and Heyerick, A. (2011). The intestinal microbiome: a separate organ inside the body with the metabolic potential to influence the bioactivity of botanicals. Fitoterapia 82, 53–66. doi: 10.1016/j.fitote.2010.07.012
Powell, J. E., Martinson, V. G., Urban-Mead, K., and Moran, N. A. (2014). Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387. doi: 10.1128/AEM.01861-14
Puinean, A. M., Foster, S. P., Oliphant, L., Denholm, I., Field, L. M., Millar, N. S., et al. (2010). Amplification of a cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. PLoS Genet. 6:e1000999. doi: 10.1371/journal.pgen.1000999
Puri, S., Singh, S., and Sohal, S. K. (2022). Oviposition behaviour and biochemical response of an insect pest, Zeugodacus cucurbitae (Coquillett) (Diptera: Tephritidae) to plant phenolic compound phloroglucinol. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 255:109291. doi: 10.1016/j.cbpc.2022.109291
Raddadi, N., Gonella, E., Camerota, C., Pizzinat, A., Tedeschi, R., Crotti, E., et al. (2011). ‘Candidatus Liberibacter europaeus’ sp. nov. that is associated with and transmitted by the psyllid Cacopsylla pyri apparently behaves as an endophyte rather than a pathogen. Environ. Microbiol. 13, 414–426. doi: 10.1111/j.1462-2920.2010.02347.x
Ramya, S. L., Venkatesan, T., Murthy, K. S., Jalali, S. K., and Varghese, A. (2016a). Degradation of acephate by Enterobacter asburiae, Bacillus cereus and Pantoea agglomerans isolated from diamondback moth Plutella xylostella (L), a pest of cruciferous crops. J. Environ. Biol. 37:611.
Ramya, S. L., Venkatesan, T., Srinivasa Murthy, K. S., Jalali, S. K., and Verghese, A. (2016b). Detection of carboxylesterase and esterase activity in culturable gut bacterial flora isolated from diamondback moth, Plutella xylostella (Linnaeus), from India and its possible role in indoxacarb degradation. Brazilian J. Microbiol. 47, 327–336. doi: 10.1016/j.bjm.2016.01.012
Receveur, J. P., Pechal, J. L., Benbow, M. E., Donato, G., Rainey, T., and Wallace, J. R. (2018). Changes in larval mosquito microbiota reveal non-target effects of insecticide treatments in hurricane-created habitats. Microb. Ecol. 76, 719–728. doi: 10.1007/s00248-018-1175-3
Ricci, I., Damiani, C., Capone, A., DeFreece, C., Rossi, P., and Favia, G. (2012). Mosquito/microbiota interactions: from complex relationships to biotechnological perspectives. Curr. Opin. Microbiol. 15, 278–284. doi: 10.1016/j.mib.2012.03.004
Rigby, L. M., Johnson, B. J., Rašić, G., Peatey, C. L., Hugo, L. E., Beebe, N. W., et al. (2021). The presence of knockdown resistance mutations reduces male mating competitiveness in the major arbovirus vector, Aedes aegypti. PLoS Negl. Trop. Dis. 15:e0009121. doi: 10.1371/journal.pntd.0009121
Roditakis, E., Steinbach, D., Moritz, G., Vasakis, E., Stavrakaki, M., Ilias, A., et al. (2017). Ryanodine receptor point mutations confer diamide insecticide resistance in tomato leafminer, Tuta absoluta (Lepidoptera: Gelechiidae). Insect Biochem. Mol. Biol. 80, 11–20. doi: 10.1016/j.ibmb.2016.11.003
Roditakis, E., Vasakis, E., Garcia-Vidal, L., del Rosario, Martinez-Aguirre, M., Rison, J. L., et al. (2018). A four-year survey on insecticide resistance and likelihood of chemical control failure for tomato leaf miner Tuta absoluta in the European/Asian region. J. Pest Sci. 91, 421–435. doi: 10.1007/s10340-017-0900-x
Rodríguez-Ruano, S. M., Škochová, V., Rego, R. O. M., Schmidt, J. O., Roachell, W., Hypša, V., et al. (2018). Microbiomes of North American Triatominae: the grounds for Chagas disease epidemiology. Front. Microbiol. 9:1167. doi: 10.3389/fmicb.2018.01167
Rokhsefat, S., Lin, A., and Comelli, E. M. (2016). Mucin–microbiota interaction during postnatal maturation of the intestinal ecosystem: clinical implications. Dig. Dis. Sci. 61, 1473–1486. doi: 10.1007/s10620-016-4032-6
Rolff, J., Johnston, P. R., and Reynolds, S. (2019). Complete metamorphosis of insects. Philos. Trans. R. Soc. B 374:20190063. doi: 10.1098/rstb.2019.0063
Russell, R. J., Scott, C., Jackson, C. J., Pandey, R., Pandey, G., Taylor, M. C., et al. (2011). The evolution of new enzyme function: Lessons from xenobiotic metabolizing bacteria versus insecticide-resistant insects. Evol. Appl. 4, 225–248. doi: 10.1111/j.1752-4571.2010.00175.x
Ryu, J.-H., Kim, S.-H., Lee, H.-Y., Bai, J. Y., Nam, Y.-D., Bae, J.-W., et al. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. doi: 10.1126/science.1149357
Salcedo-Porras, N., Umaña-Diaz, C., Bitencourt, R., de, O. B., and Lowenberger, C. (2020). The role of bacterial symbionts in triatomines: An evolutionary perspective. Microorganisms 8:1438. doi: 10.3390/microorganisms8091438
Saleem, M. A., Ahmad, M., Ahmad, M., Aslam, M., and Sayyed, A. H. (2008). Resistance to selected organochlorin, organophosphate, carbamate and pyrethroid, in Spodoptera litura (Lepidoptera: Noctuidae) from Pakistan. J. Econ. Entomol. 101, 1667–1675. doi: 10.1603/0022-0493(2008)101[1667:rtsooc]2.0.co;2
Salem, H., and Kaltenpoth, M. (2022). Beetle–bacterial symbioses: endless forms most functional. Annu. Rev. Entomol 67, 201–219 doi: 10.1146/annurev-ento-061421-063433
Sampson, T. R., and Mazmanian, S. K. (2015). Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 17, 565–576. doi: 10.1016/j.chom.2015.04.011
Schmidt, K., and Engel, P. (2021). Mechanisms underlying gut microbiota-host interactions in insects. J. Exp. Biol. 224:jeb207696 doi: 10.1242/jeb.207696
Scolari, F., Casiraghi, M., and Bonizzoni, M. (2019). Aedes spp. and their microbiota: a review. Front. Microbiol. 10:2036. doi: 10.3389/fmicb.2019.02036
Shah, R. M., Shad, S. A., and Abbas, N. (2017). Methoxyfenozide resistance of the housefly, Musca domestica L. (Diptera: Muscidae): cross-resistance patterns, stability and associated fitness costs. Pest Manag. Sci. 73, 254–261. doi: 10.1002/ps.4296
Shan, H., Wu, W., Sun, Z., Chen, J., and Li, H. (2021). The gut microbiota of the insect infraorder Pentatomomorpha (Hemiptera: Heteroptera) for the light of ecology and evolution. Microorganisms 9:464. doi: 10.3390/microorganisms9020464
Shapira, M. (2016). Gut microbiotas and host evolution: scaling up symbiosis. Trends Ecol. Evol. 31, 539–549. doi: 10.1016/j.tree.2016.03.006
Sharon, G., Segal, D., Ringo, J. M., Hefetz, A., Zilber-Rosenberg, I., and Rosenberg, E. (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci.U.S.A. 107, 20051–20056. doi: 10.1073/pnas.1009906107
Shen, S. K., and Dowd, P. F. (1991). Detoxification spectrum of the cigarette beetle symbiont Symbiotaphrina kochii in culture. Entomol. Exp. Appl. 60, 51–59. doi: 10.1111/j.1570-7458.1991.tb01522.x
Shin, N.-R., Whon, T. W., and Bae, J.-W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Siddiqui, J. A., Zhang, Y., Luo, Y., Bamisile, B. S., Rehman, N. U., Islam, W., et al. (2022). Comprehensive Detoxification Mechanism Assessment of Red Imported Fire Ant (Solenopsis invicta) against Indoxacarb. Molecules 27:870. doi: 10.3390/molecules27030870
Silva, W. M., Berger, M., Bass, C., Williamson, M., Moura, D. M. N., Ribeiro, L. M. S., et al. (2016). Mutation (G275E) of the nicotinic acetylcholine receptor α6 subunit is associated with high levels of resistance to spinosyns in Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae). Pestic. Biochem. Physiol. 131, 1–8. doi: 10.1016/j.pestbp.2016.02.006
Simpson, S. J. (2013). “Mouthparts and feeding,” in ‘the Insects. Structure and Function.’, 5th Edn, eds S. J. Simpson and A. E. Douglas (Cambridge: Cambridge University Press)15–45. doi: 10.1017/cbo9781139035460.005
Simpson, S. J., Clissold, F. J., Lihoreau, M., Ponton, F., Wilder, S. M., and Raubenheimer, D. (2015). Recent advances in the integrative nutrition of arthropods. Annu. Rev. Entomol. 60, 293–311. doi: 10.1146/annurev-ento-010814-020917
Smith, C. C., Srygley, R. B., Healy, F., Swaminath, K., and Mueller, U. G. (2017). Spatial structure of the mormon cricket gut microbiome and its predicted contribution to nutrition and immune function. Front. Microbiol. 8:801. doi: 10.3389/fmicb.2017.00801
Soltani, A., Vatandoost, H., Oshaghi, M. A., Enayati, A. A., and Chavshin, A. R. (2017). The role of midgut symbiotic bacteria in resistance of Anopheles stephensi (Diptera: Culicidae) to organophosphate insecticides. Pathog. Glob. Health 111, 289–296. doi: 10.1080/20477724.2017.1356052
Sonoda, S., Shi, X., Song, D., Zhang, Y., Li, J., Wu, G., et al. (2012). Frequencies of the M918I mutation in the sodium channel of the diamondback moth in China, Thailand and Japan and its association with pyrethroid resistance. Pestic. Biochem. Physiol. 102, 142–145. doi: 10.1016/j.pestbp.2011.12.005
Sosa-Gómez, D. R., and da Silva, J. J. (2010). Neotropical brown stink bug (Euschistus heros) resistance to methamidophos in Paraná, Brazil. Pesqui. Agropecuária Bras. 45, 767–769. doi: 10.1590/s0100-204x2010000700019
Sparks, T. C., Dripps, J. E., Watson, G. B., and Paroonagian, D. (2012). Resistance and cross-resistance to the spinosyns–a review and analysis. Pestic. Biochem. Physiol. 102, 1–10. doi: 10.1016/j.pestbp.2021.104924
Steinbach, D., Gutbrod, O., Lümmen, P., Matthiesen, S., Schorn, C., and Nauen, R. (2015). Geographic spread, genetics and functional characteristics of ryanodine receptor based target-site resistance to diamide insecticides in diamondback moth. Plutella xylostella. Insect Biochem. Mol. Biol. 63, 14–22. doi: 10.1016/j.ibmb.2015.05.001
Stencel, A., and Wloch-Salamon, D. M. (2018). Some theoretical insights into the hologenome theory of evolution and the role of microbes in speciation. Theor. Biosci. 137, 197–206. doi: 10.1007/s12064-018-0268-3
Suárez-Moo, P., Cruz-Rosales, M., Ibarra-Laclette, E., Desgarennes, D., Huerta, C., and Lamelas, A. (2020). Diversity and composition of the gut microbiota in the developmental stages of the dung beetle Copris incertus Say (Coleoptera. Scarabaeidae). Front. Microbiol. 11:1698. doi: 10.3389/fmicb.2020.01698
Sudakaran, S., Kost, C., and Kaltenpoth, M. (2017). Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 25, 375–390. doi: 10.1016/j.tim.2017.02.014
Sun, H., Yang, B., Zhang, Y., and Liu, Z. (2017). Metabolic resistance in Nilaparvata lugens to etofenprox, a non-ester pyrethroid insecticide. Pestic. Biochem. Physiol. 136, 23–28. doi: 10.1016/j.pestbp.2016.08.009
Tabashnik, B. E., and Carrière, Y. (2010). Field-evolved resistance to Bt cotton: bollworm in the US and pink bollworm in India. Southwest. Entomol. 35, 417–424. doi: 10.3958/059.035.0326
Taerum, S. J., Konečný, A., De Beer, Z. W., Cibrián-Tovar, D., and Wingfield, M. J. (2016). Population genetics and symbiont assemblages support opposing invasion scenarios for the red turpentine beetle (Dendroctonus valens). Biol. J. Linn. Soc. 118, 486–502. doi: 10.1111/bij.12781
Tago, K., Kikuchi, Y., Nakaoka, S., Katsuyama, C., and Hayatsu, M. (2015). Insecticide applications to soil contribute to the development of Burkholderia mediating insecticide resistance in stinkbugs. Mol. Ecol. 24, 3766–3778. doi: 10.1111/mec.13265
Tang, J., Li, J., Shao, Y., Yang, B., and Liu, Z. (2010). Fipronil resistance in the white backed planthopper (Sogatella furcifera): possible resistance mechanisms and cross-resistance. Pest Manag. Sci. Former. Pestic. Sci. 66, 121–125. doi: 10.1002/ps.1836
Tang, X., Freitak, D., Vogel, H., Ping, L., Shao, Y., Cordero, E. A., et al. (2012). Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. PLoS One 7:e36978. doi: 10.1371/journal.pone.0036978
Taylor, C. M., Coffey, P. L., DeLay, B. D., and Dively, G. P. (2014). The importance of gut symbionts in the development of the brown marmorated stink bug, Halyomorpha halys (Stål). PLoS One 9:e90312. doi: 10.1371/journal.pone.0090312
Terra, W. R., and Ferreira, C. (2012). “Biochemistry and molecular biology of digestion,” in Insect Molecular Biology and Biochemistry. L. I. Gilbert ed (Berlin: Elsevier), 365–418. doi: 10.1016/b978-0-12-384747-8.10011-x
Thakur, A., Dhammi, P., Saini, H. S., and Kaur, S. (2016). Effect of antibiotic on survival and development of Spodoptera litura (Lepidoptera: Noctuidae) and its gut microbial diversity. Bull. Entomol. Res. 106, 387–394. doi: 10.1017/S0007485316000031
Thurman, E. M., Ferrer, I., Zavitsanos, P., and Zweigenbaum, J. A. (2013). Identification of imidacloprid metabolites in onion (Allium cepa L.) using high-resolution mass spectrometry and accurate mass tools. Rapid Commun. Mass Spectrom. 27, 1891–1903. doi: 10.1002/rcm.6637
Tilottama, M., Teh, B. S., Aishwarya, M., Schmidt-Heck, W., Schlenker, Y., Vogel, H., et al. (2021). Transcriptomics Reveal the Survival Strategies of Enterococcus mundtii in the Gut of Spodoptera littoralis. J. Chem. Ecol. 47, 227–241. doi: 10.1007/s10886-021-01246-1
Tinker, K. A., and Ottesen, E. A. (2016). The core gut microbiome of the American cockroach, Periplaneta americana, is stable and resilient to dietary shifts. Appl. Environ. Microbiol. 82, 6603–6610. doi: 10.1128/AEM.01837-16
Tiwari, S., Mann, R. S., Rogers, M. E., and Stelinski, L. L. (2011). Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 67, 1258–1268. doi: 10.1002/ps.2181
Toda, S., and Morishita, M. (2009). Identification of three point mutations on the sodium channel gene in pyrethroid-resistant Thrips tabaci (Thysanoptera: Thripidae). J. Econ. Entomol. 102, 2296–300. doi: 10.1603/029.102.0635
Tokuda, G., Mikaelyan, A., Fukui, C., Matsuura, Y., Watanabe, H., Fujishima, M., et al. (2018). Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci.U.S.A. 115, E11996–E12004. doi: 10.1073/pnas.1810550115
Trappeniers, K., Matetovici, I., Van Den Abbeele, J., and De Vooght, L. (2019). The tsetse fly displays an attenuated immune response to its secondary symbiont, Sodalis glossinidius. Front. Microbiol. 10:1650. doi: 10.3389/fmicb.2019.01650
Van Arnam, E. B., Currie, C. R., and Clardy, J. (2018). Defense contracts: molecular protection in insect-microbe symbioses. Chem. Soc. Rev. 47, 1638–1651. doi: 10.1039/c7cs00340d
van den Bosch, T. J. M., and Welte, C. U. (2017). Detoxifying symbionts in agriculturally important pest insects. Microb. Biotechnol. 10, 531–540. doi: 10.1111/1751-7915.12483
Vorburger, C., Gehrer, L., and Rodriguez, P. (2010). A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol. Lett. 6, 109–111. doi: 10.1098/rsbl.2009.0642
Wan, Y., Yuan, G., He, B., Xu, B., Xie, W., Wang, S., et al. (2018). Foccα6, a truncated nAChR subunit, positively correlates with spinosad resistance in the western flower thrips, Frankliniella occidentalis (Pergande). Insect Biochem. Mol. Biol. 99, 1–10. doi: 10.1016/j.ibmb.2018.05.002
Wang, G.-H., Berdy, B. M., Velasquez, O., Jovanovic, N., Alkhalifa, S., Minbiole, K. P. C., et al. (2020). Changes in microbiome confer multigenerational host resistance after sub-toxic pesticide exposure. Cell Host Microbe. 27, 213–224. doi: 10.1016/j.chom.2020.01.009
Wang, S., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Wang, X., Wu, S., Gao, W., and Wu, Y. (2016a). Dominant inheritance of field-evolved resistance to fipronil in Plutella xylostella (Lepidoptera: Plutellidae). J. Econ. Entomol. 109, 334–338. doi: 10.1093/jee/tov317
Wang, Z., Gong, Y., Jin, G., Li, B., Chen, J., Kang, Z., et al. (2016b). Field-evolved resistance to insecticides in the invasive western flower thrips Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae) in China. Pest Manag. Sci. 72, 1440–1444. doi: 10.1002/ps.4200
Wei, Y., Appel, A. G., Moar, W. J., and Liu, N. (2001). Pyrethroid resistance and cross-resistance in the German cockroach. Blattella germanica (L). Pest Manag. Sci. Former. Pestic. Sci. 57, 1055–1059. doi: 10.1002/ps.383
Wu, H., Gao, C., Guo, Y., Zhang, Y., Zhang, J., and Ma, E. (2014). Acute toxicity and sublethal effects of fipronil on detoxification enzymes in juvenile zebrafish (Danio rerio). Pestic. Biochem. Physiol. 115, 9–14. doi: 10.1016/j.pestbp.2014.07.010
Wu, S. F., Zeng, B., Zheng, C., Mu, X. C., Zhang, Y., Hu, J., et al. (2018). The evolution of insecticide resistance in the brown planthopper (Nilaparvata lugens Stål) of China in the period 2012–2016. Sci. Rep. 8:4586.
Wu, Y., Xu, H., Pan, Y., Gao, X., Xi, J., Zhang, J., et al. (2018). Expression profile changes of cytochrome P450 genes between thiamethoxam susceptible and resistant strains of Aphis gossypii Glover. Pestic. Biochem. Physiol. 149, 1–7. doi: 10.1016/j.pestbp.2018.05.007
Xia, X., Sun, B., Gurr, G. M., Vasseur, L., Xue, M., and You, M. (2018). Gut microbiota mediate insecticide resistance in the diamondback moth. Plutella xylostella (L.). Front. Microbiol. 9, 25. doi: 10.3389/fmicb.2018.00025
Xia, X., Zheng, D., Zhong, H., Qin, B., Gurr, G. M., Vasseur, L., et al. (2013). DNA Sequencing Reveals the Midgut Microbiota of Diamondback Moth. Plutella xylostella (L.) and a Possible Relationship with Insecticide Resistance. PLoS One 8:e68852. doi: 10.1371/journal.pone.0068852
Xiong, T., Ling, S., and Liu, J. (2020). Insecticidal and P450 mediate mechanism of uralaner against red imported fire Ant, Solenopsis invicta (Hymenoptera: Formicidae). Sci. Rep. 25.
Yang, Y. X., Lin, R. H., Li, Z., Wang, A. Y., Xue, C., Duan, A. L., et al. (2021). Function Analysis of P450 and GST Genes to Imidacloprid in Aphis craccivora (Koch). Front. Physiol. 11:1822. doi: 10.3389/fphys.2020.624287
Yuan, C., Xing, L., Wang, M., Hu, Z., and Zou, Z. (2021). Microbiota modulates gut immunity and promotes baculovirus infection in Helicoverpa armigera. Insect Sci. 28, 1766–1779. doi: 10.1111/1744-7917.12894
Yun, J. H., Roh, S. W., Whon, T. W., Jung, M. J., Kim, M. S., Park, D. S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264. doi: 10.1128/AEM.01226-14
Zaidman-Rémy, A., Hervé, M., Poidevin, M., Pili-Floury, S., Kim, M.-S., Blanot, D., et al. (2006). The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473. doi: 10.1016/j.immuni.2006.02.012
Zaidman-Rémy, A., Vigneron, A., Weiss, B. L., and Heddi, A. (2018). What can a weevil teach a fly, and reciprocally? Interaction of host immune systems with endosymbionts in Glossina and Sitophilus. BMC Microbiol. 18:150. doi: 10.1186/s12866-018-1278-5
Zhang, B., Zhang, L., Cui, R., Zeng, X., and Gao, X. (2016). Cloning and expression of multiple cytochrome P450 genes: Induction by Fipronil in Workers of the Red Imported Fire Ant (Solenopsis invicta Buren). PLoS One 11:e0150915. doi: 10.1371/journal.pone.0150915
Zhang, B.-Z., Su, X., Xie, L.-F., Zhen, C.-A., Hu, G.-L., Jiang, K., et al. (2020a). Multiple detoxification genes confer imidacloprid resistance to Sitobion avenae Fabricius. Crop Prot. 128:105014. doi: 10.1016/j.cropro.2019.105014
Zhang, S., Shu, J., Xue, H., Zhang, W., Zhang, Y., Liu, Y., et al. (2020b). The gut microbiota in Camellia Weevils are influenced by plant secondary metabolites and contribute to saponin degradation. Msystems 5, e692–e619. doi: 10.1128/mSystems.00692-19
Zhang, K., Zhang, W., Zhang, S., Wu, S.-F., Ban, L.-F., Su, J.-Y., et al. (2014). Susceptibility of Sogatella furcifera and Laodelphax striatellus (Hemiptera: Delphacidae) to six insecticides in China. J. Econ. Entomol. 107, 1916–1922. doi: 10.1603/EC14156
Zhang, J., Ai, Z., Liu, H., Tang, D. W. S., Yang, X., Wang, G., et al. (2022). Short-term N addition in a Pinus tabuliformis plantation: Microbial community composition and interactions show different linkages with ecological stoichiometry. Appl. Soil Ecol. 174:104422. doi: 10.1016/j.apsoil.2022.104422
Zhang, X., Liao, X., Mao, K., Yang, P., Li, D., Ali, E., et al. (2017). Neonicotinoid insecticide resistance in the field populations of Sogatella furcifera (Horváth) in Central China from 2011 to 2015. J. Asia. Pac. Entomol. 20, 955–958. doi: 10.1016/j.aspen.2017.07.004
Zheng, H., Nishida, A., Kwong, W. K., Koch, H., Engel, P., Steele, M. I., et al. (2016). Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. MBio 7, e1326–e1316. doi: 10.1128/mBio.01326-16
Zhou, L.-F., Wu, J., Li, S., Li, Q., Jin, L.-P., Yin, C.-P., et al. (2021). Antibacterial Potential of Termite-Associated Streptomyces spp. ACS omega 6, 4329–4334. doi: 10.1021/acsomega.0c05580
Zhu, F., Lavine, L., O’Neal, S., Lavine, M., Foss, C., and Walsh, D. (2016). Insecticide resistance and management strategies in urban ecosystems. Insects 7:2. doi: 10.3390/insects7010002
Zimmer, C. T., and Nauen, R. (2011). Pyrethroid resistance and thiacloprid baseline susceptibility of European populations of Meligethes aeneus (Coleoptera: Nitidulidae) collected in winter oilseed rape. Pest Manag. Sci. 67, 599–608. doi: 10.1002/ps.2137
Keywords: toxicology, microbial detoxification, insecticide degradation, resistant species, symbiotic bacteria
Citation: Siddiqui JA, Khan MM, Bamisile BS, Hafeez M, Qasim M, Rasheed MT, Rasheed MA, Ahmad S, Shahid MI and Xu Y (2022) Role of Insect Gut Microbiota in Pesticide Degradation: A Review. Front. Microbiol. 13:870462. doi: 10.3389/fmicb.2022.870462
Received: 06 February 2022; Accepted: 25 February 2022;
Published: 03 May 2022.
Edited by:
Cormac Murphy, University College Dublin, IrelandReviewed by:
Mubasher Hussain, Guangdong Academy of Agricultural Sciences (GDAAS), ChinaWaqar Islam, Fujian Agriculture and Forestry University, China
Abrar Muhammad, Zhejiang University, China
Geeta Bhandari, Swami Rama Himalayan University, India
Kalpana Bhatt, Gurukul Kangri Vishwavidyalaya, India
Copyright © 2022 Siddiqui, Khan, Bamisile, Hafeez, Qasim, Rasheed, Rasheed, Ahmad, Shahid and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junaid Ali Siddiqui, anVuYWlkYWxpMjA2QGdtYWlsLmNvbQ==; Yijuan Xu, eHV5aWp1YW5AeWFob28uY29t
 Junaid Ali Siddiqui
Junaid Ali Siddiqui Muhammad Musa Khan
Muhammad Musa Khan Bamisope Steve Bamisile
Bamisope Steve Bamisile Muhammad Hafeez
Muhammad Hafeez Muhammad Qasim3
Muhammad Qasim3 Muhammad Tariq Rasheed
Muhammad Tariq Rasheed Muhammad Atif Rasheed
Muhammad Atif Rasheed Sajjad Ahmad
Sajjad Ahmad Yijuan Xu
Yijuan Xu