
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 11 April 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.869441
This article is part of the Research TopicInnovative Vaccine Strategies for Enhanced Flavivirus ImmunizationView all 3 articles
MicroRNAs (miRNAs) are small non-coding RNA that affect mRNA abundance or translation efficiency by binding to the 3′UTR of the mRNA of the target gene, thereby participating in multiple biological processes, including viral infection. Flavivirus genus consists of small, positive-stranded, single-stranded RNA viruses transmitted by arthropods, especially mosquitoes and ticks. The genus contains several globally significant human/animal pathogens, such as Dengue virus, Japanese encephalitis virus, West Nile virus, Zika virus, Yellow fever virus, Tick-borne encephalitis virus, and Tembusu virus. After flavivirus invades, the expression of host miRNA changes, exerting the immune escape mechanism to create an environment conducive to its survival, and the altered miRNA in turn affects the life cycle of the virus. Accumulated evidence suggests that host miRNAs influence flavivirus replication and host–virus interactions through direct binding of viral genomes or through virus-mediated host transcriptome changes. Furthermore, miRNA can also interweave with other non-coding RNAs, such as long non-coding RNA and circular RNA, to form an interaction network to regulate viral replication. A variety of non-coding RNAs produced by the virus itself exert similar function by interacting with cellular RNA and viral RNA. Understanding the interaction sites between non-coding RNA, especially miRNA, and virus/host genes will help us to find targets for antiviral drugs and viral therapy.
MicroRNAs (miRNA) is a class of small non-coding RNA, about 20–24 nucleotides in length. miRNA is first transcribed from the genome to form primary miRNA (pri-miRNA), and then cleaved by Drosha enzyme and Dicer enzyme successively to form mature miRNA. One strand of mature miRNA typically degrades mRNA or inhibits translation by binding to the 3′untranslated region (UTR) of the target gene (Bartel, 2004). Not only in diverse biological pathways, cellular miRNAs also play vital roles in virus-host interactions, which are post-transcriptional regulators in directing regulation of virus proliferation (Wong et al., 2020).
Flavivirus genus contains more than 50 arthropod-borne viruses (arboviruses), of which important family members include Dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus (WNV), Zika virus (ZIKV), Yellow fever virus (YFV), Tick- borne encephalitis virus (TBEV) and Tambusu virus (TMUV). Flavivirus is a positive-sense RNA virus with a full length of about 11 kb. The structure of viral genome contains a single open reading frame (ORF) that encodes multiple proteins, cleaved by host and viral proteases into three structural proteins (C, prM, E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), flanked by a 5′UTR and a 3′UTR, which is conserved throughout the Flavivirus genus (Heiss et al., 2011; Figure 1).
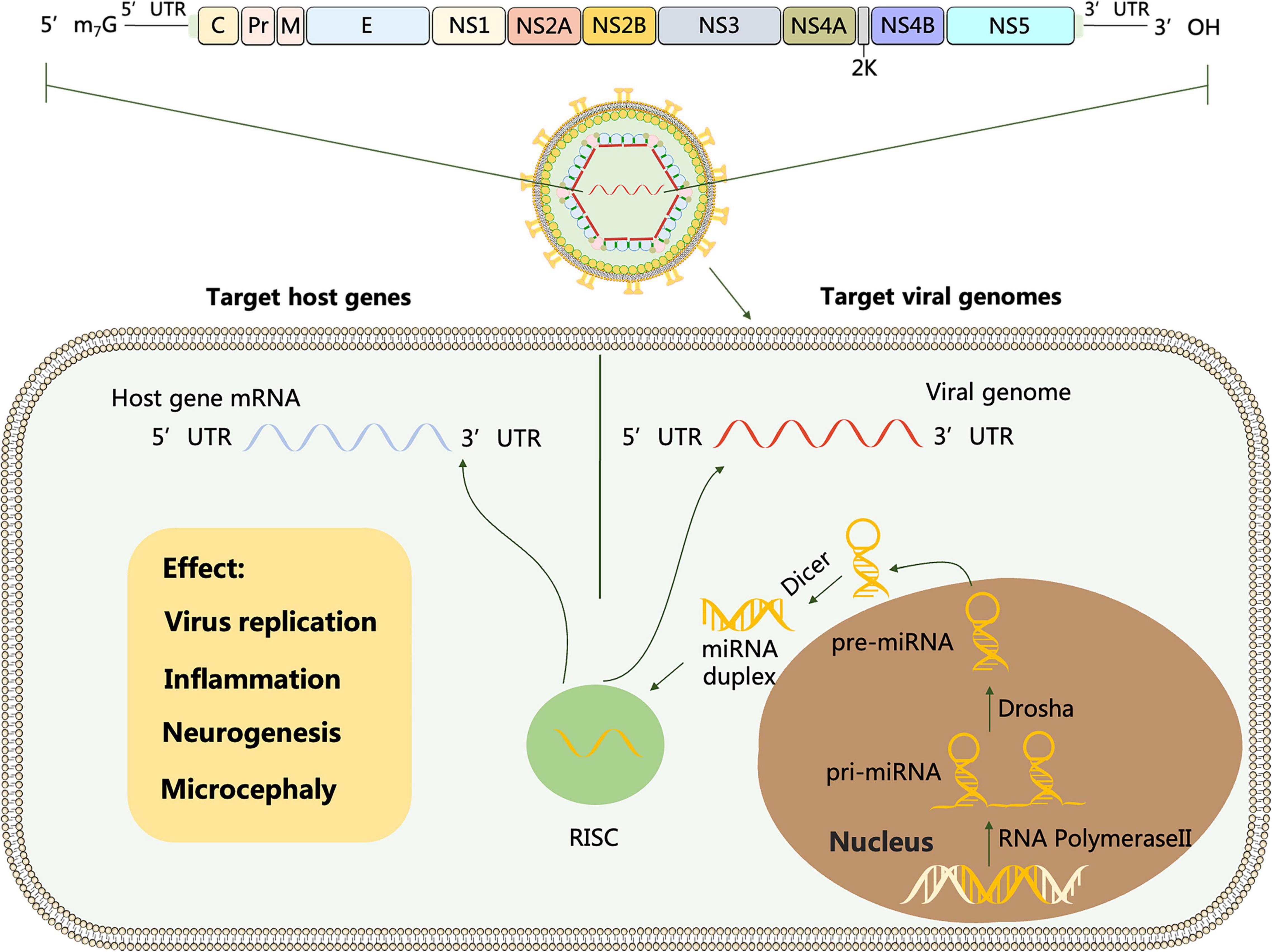
Figure 1. Schematic representation of flavivirus genome. The polyprotein encoded by the genome is cleaved by host and viral proteases to form three structural proteins: capsid (C), membrane (M) and envelope (E) and 7 non-structural proteins (NS-1, 2A, 2B, 3, 4A, 4B and 5). miRNA is transcribed from host mRNA to primary miRNA (pri-miRNA) in the nucleus by RNA polymerase II (Pol II). Pre-miRNA is transported from the nucleus to the cytoplasm and cleaved by the Dicer enzyme into miRNA double strands, one of which forms an RNA-induced silencing complex (RISC). Target viral genomes: After flavivirus infection, the RNA viral genome releases into the cytoplasm, and RISC carrying miRNA complements the non-coding and coding sequences of the virus through miRNA seed sequences to regulate viral replication. Target host genes: After flavivirus invasion, miRNAs can activate or repress specific cellular signaling pathways and ultimately mediate viral replication by regulating the target genes they interact with.
The expression of host miRNA can be altered by flavivirus infection (Campbell et al., 2014; Kumari et al., 2016; McLean et al., 2017; Cui et al., 2018; Bagasra et al., 2021). Further, differentially expressed miRNAs can directly regulate virus replication by complementing viral genome or indirectly by binding to crucial host genes, which has been reported in various viruses (Smith et al., 2012). For instance, Kumar et al. (2018) confirmed that miR-324-5p suppresses H5N1 highly pathogenic influenza A virus (HPAIV) replication through directly binding within the viral genome, thereby inhibiting viral gene expression. miR-140 inhibits classical swine fever virus (CSFV) genome amplification via interacting with an oncogene Rab25 in swine umbilical vein endothelial cells (Xu et al., 2020). Moreover, it is worth mentioning that miRNAs are usually negatively correlated with target genes. Nevertheless, miR-122 is a special case, as it specifically expresses and is highly abundant in human livers, while promoting the Hepatitis C virus (HCV) life cycle by targeting its 5′UTR (Jopling et al., 2005; Kunden et al., 2020).
Recently, interaction networks composed of non-coding RNAs, including miRNA, long non-coding RNA (lncRNA) and circular RNA (circRNA), have attracted increasing attention for their important roles in various biological processes such as viral infection. lncRNA/circRNA, as competing endogenous RNA (ceRNA), competitively bind to miRNA through their miRNA recognition element (MRE), thereby regulating the expression level of miRNA target mRNA (Salmena et al., 2011). Hence, lncRNA/circRNA–miRNA–mRNA interaction may be an important mechanism of flavivirus pathogenesis. In addition, the virus itself produces a variety of non-coding RNAs, such as subgenomic flaviviral RNAs (sfRNAs) and viral small RNAs (vsRNAs), the latter also include viral small interfering RNAs (siRNAs) and virus-encoded miRNAs. All of them can interact with cellular and viral RNA to regulate flavivirus replication (Schuessler et al., 2012; Hussain and Asgari, 2014; Zeng et al., 2021).
Based on the character that flavivirus completes its life cycle and pathogenicity is highly dependent on the metabolism of living host cells, clarifying the effect of host miRNA as well as other non-coding RNA on host–virus interactions will help us better understand the pathogenesis of flavivirus and novel candidates for the development of therapeutic interventions (Smith et al., 2017). In this review, we elaborate how miRNAs could relate to flavivirus infections and summarize the potential value of miRNAs in combating flavivirus.
Dengue viruses are four closely related but antigenically distinct viral serotypes (DENV1, DENV2, DENV3, and DENV4) that cause very similar disease spectrum in humans. It is the tenth leading cause of death and morbidity in developing countries and the leading cause of death among children under 15 years of age in some South-East Asian countries (Lim, 2019). At present, DENV is also the most widely studied virus in the Flavivirus genus and has been reported to regulate its own replication by using host miRNA (Zhou et al., 2014; Castillo et al., 2016; Diosa-Toro et al., 2017; Kanokudom et al., 2017; Mishra et al., 2020).
Several independent studies have suggested that miRNAs can complementarily bind to non-coding regions or even coding regions of DENV genome and consequently influencing viral replication, while other research results imply that DENV infection regulates cellular miRNAs expression for their own interests (Miranda et al., 2006; Pasquinelli, 2012; Yan et al., 2014; Zhou et al., 2014; Wen et al., 2015; Castillo et al., 2016; Castrillón-Betancur and Urcuqui-Inchima, 2017; Kanokudom et al., 2017; Table 1).
Non-coding regions of flavivirus, the 5′UTR and 3′UTR, have significant impact in translation, replication and assembly of virus. miR-484, miR-744, and miR-133a are downregulated at early stage during DENV infection in Vero cells (Castillo et al., 2016; Castrillón-Betancur and Urcuqui-Inchima, 2017). Moreover, these three miRNAs facilitate DENV replication through targeting and increasing the 3′UTR of viral genome. Furthermore, miR-281 and miR-548g-3p have been shown to complement the 5′UTR of DENV, but when both two miRNAs are up-regulated, their effects on viral proliferation are opposite (Pasquinelli, 2012; Zhou et al., 2014; Wen et al., 2015). miR-281, which is abundantly expressed in the female mosquitoes’ midgut, promotes the replication of DENV by increasing its 5′UTR, while the up-regulated miR-548g-3p inhibits viral genome amplification by impairing DENV 5′UTR. It is worth mentioning that the mechanism of miR-281 is contrast to the classical pathways in which miRNAs interact with target genes to degrade mRNA or inhibit translation. miRNAs can also bind complementary to coding regions of viral genomes, although this is less common. Down-regulation of miR-252 by a synthetic inhibitor enhances DENV-2 transmembrane envelop protein (E protein), a protein involved in virus invasion, as well as virus titers (Yan et al., 2014). miR-21 shows a trend of up-regulation after DENV infection, and promotes DENV-2 proliferation in HepG2 cells. Unfortunately, the mechanism remains unclear, but may be related to the potential interaction of miR-21 with viral NS1 sequence (Miranda et al., 2006; Kanokudom et al., 2017). NS1 exerts multiple roles in the viral life cycle, especially in virus propagation and immune evasion of the complement pathway (Glasner et al., 2018).
More recently, it has been achieved to control the virus tissue tropism by incorporating a specific miRNA recognition element (MRE) from the viral genome into engineered viruses. This has been implemented as a neoteric strategy to exploit secure vaccines against pathogenic viruses (Wong et al., 2020). For instance, inserting the MRE for the hepatic-specific miR-122 in the 3′UTR of DENV replicon could block DENV infection of vital tissues/organs (Lee et al., 2010). Similarly, the insertion of the MRE for the hematopoietic specific miR-142 into the DENV-2 genome restricts replication of the virus in dendritic cells and macrophages, but not in non-hematopoietic cell types (Pham et al., 2012).
After DENV infection, miRNAs can regulate series of key genes related to innate immunity. For example, miR-146a-targeted TNFR-associated factor 6 (TRAF6) and miR-155-targeted BTB domain and CNC homolog 1 (BACH1) can promote viral replication by dampening IFN-β (Wu et al., 2013; Arboleda et al., 2019; Su et al., 2020). TRAF6, which mediates multiple immune signaling pathways, is essential for the production of type I IFN, and the characteristic of miR-146a is consistent with reports in JEV and ZIKV (Wu et al., 2013; Sharma et al., 2015; Shukla et al., 2021; Xie et al., 2021). Furthermore, miR-146a is also associated with anti-inflammatory effects in JEV and ZIKV infection, but has not been reported in DENV. Pro-DENV activity of downregulated miR-155 is due to augment BACH1, and thus contributing to the production of the heme oxygenase-1 (HO-1) mediated promotion of DENV NS2B/NS3 protease activity and inhibit IFN responses (Pulkkinen et al., 2011; Su et al., 2020). Nevertheless, let-7c exerts a role in the restriction of viral replication by down-regulating BACH1 (Escalera-Cueto et al., 2015). The depletion of miR-223 or overexpression of Stathmin 1 (STMN1) enhances DENV2 replication in EAhy926 cells (a human endothelial-like cell line) (Wu et al., 2014). Function as a microtubule (MT)-destabilizing protein, up-regulated STMN1 is likely to influence actin function by changing MT dynamics, which provides advantageous conditions for virus invasion. Abundant extracellular vesicles (EVs) load with miR-148a in DENV-infected monocytes and DENV-NS1-transfected cells. Whereafter, miR-148a represses the expression levels of Ubiquitin-specific peptidase (USP33), which in turn weakens the stability of ATF3 protein, an inhibitor molecule of pro-inflammatory pathways, by deubiquitylation, ultimately preparing the uninfected cells for more favorable DENV infection and triggering a massive production of proinflammatory genes (Mishra et al., 2020). miR-375, miR-927, and miR-4728-5p are detected and their expression levels increased in the blood fed mosquitoes infected DENV (Hussain et al., 2013; Su et al., 2017; Avila-Bonilla et al., 2020). miR-375 controls two genes involved in immunity, CACTUS versus REL1 via targeting their respective 5′UTR but not 3′UTR and further suppresses phosphorylation of the NF-kB, it is possible to enhance DENV infection. Similarly, the virus utilizes miR-927 and miR-4728-5p to boost its own proliferation, but neither miR-927 nor miR-4728-5p are known about the specific mechanism of DENV-host interaction.
Previous studies have clarified overexpressed miR-30* and miR-424 knockdown IκBα and Siah E3 ubiquitin protein ligase 1 (SIAH1) genes related to inherent immunity. Subsequently, miR-30* mediated NF-κB/IκBα negative feedback loop is disrupted with an excessive activation of NF-κB and increases IFN-β and the downstream ISGs, eventually leading to inhibition of DENV replication (Jiang et al., 2012; Zhu et al., 2014). As an E3 ubiquitin ligase, SIAH1 generally induces DENV infection by activating the unfolded protein response (UPR). While SIAH1 expression is suppressed, proliferation of DENV is also restricted (Murphy Schafer et al., 2020). DENV replication is reduced by miR-3614-5p overexpression, a miRNA that binds to and degrades the DENV pro-viral protein Adenosine Deaminase Acting on RNA 1 (ADAR1) in macrophages (Diosa-Toro et al., 2017), while down-regulated miR-378 achieves the same effect in DENV-infected NK cells by promoting granzyme B (GrzB), a serine protease released by NK cells that inhibits viruses (Liu et al., 2016). miR-34 family, as valid anti-flaviviral miRNAs, has been confirmed that miR-34a mimic transfection effectively impaired infection by DENV, JEV and WNV(Smith et al., 2017). Another member of the miR-34 family, miR-34c, also plays a role in neurogenesis and pro-inflammatory response, which will be elucidated in JEV, WNV and ZIKV in detail.
Japanese encephalitis virus is a neurotropic virus that infects the Central Nervous System (CNS) and causes fatal encephalitis. Accumulating evidence points to microglia activation and bystander damage as the main pathogenesis of Japanese encephalitis and its complications (Chang et al., 2021). After viral infection, miRNAs expression alters mostly occur in human/mouse microglia and subsequently respond to viral invasion by influencing viral replication and inflammation in the body.
miR-301a (Hazra et al., 2017), miR-22 (Zhou et al., 2020), miR-432 (Sharma et al., 2016), miR-370-5p (Li W. et al., 2016), miR-374b-5p (Rastogi and Singh, 2019) are shown to exert the proviral effect by directly targeting immunity-related genes or interferon-induced genes, such as IRF1, SOCS5, MAVS, STAT1, and PTEN, which further mediated the expression of type I IFN (Table 2). It is quite interesting that down-regulated miR-370-5p and up-regulated miR-374b-5p increase the level of type I interferon in JEV infected cells, while other miRNAs above mentioned promote JEV replication by impairing the level of it. Moreover, miR-33a-5p degradation contributes to viral replication via increasing the intracellular EEF1A1, an interaction partner of the JEV proteins NS3 and NS5 in replicase complex (Chen et al., 2016). Jiang et al. (2020) also found JEV NS3 can suppress the expression level of miR-466d-3p and enhance JEV amplification by targeting IL-1β, but the mechanism remains to be explored.
Persistently JEV infection increases miR-125b-5p expression and limits viral replication by dampening STAT3, MAP2K7, TRIAP1 production, which genes are associated with apoptosis and cell growth (Huang et al., 2021). Overexpression of miR-124 in PK-15 cells is found to decrease the expression level of DNM2, a GTPase responsible for vesicle scission, further impairing JEV proliferation (Yang et al., 2016). Work by Pareek et al. (2014) delineated that increased miR-155 levels versus decreased IRF8 levels can dramatically inhibit NF-κB signaling pathway and JEV multiplication, while the effect of miR-146a on virus inhibition is not significant. On the contrary, reports by Sharma et al. (2015) have shown that miR-146a participates in the reduction of NF-κB activity by inhibiting specific genes (STAT1, IRAK1, IRAK2, TRAF6) in the TLR mediated NF-κB pathway. Subsequently this miRNA exerts an anti-inflammatory role and promote viral replication, similar cases appear in ZIKV infection as well (Shukla et al., 2021).
During JEV infection, host miRNAs can not only regulate JEV replication, but also increase microglial activation and accelerate the production of inflammatory cytokines by influencing important genes in the pro-inflammatory pathway (Lawrence, 2009; Thounaojam et al., 2014a,b; Ashraf et al., 2016; Hazra et al., 2019; Mukherjee et al., 2019). Highly expressed miR-15b (Zhu et al., 2015) and miR-155 (Thounaojam et al., 2014b) are considered to exert the pro-inflammatory effect by directly targeting the 3′UTR of RNF125 and SHIP1 respectively, leading to the degradation of both two genes. This results in the subsequent phosphorylation of NF-κB (Lawrence, 2009). Similarly, significantly increased expression level of miR-19b-3p (Ashraf et al., 2016), miR-29b (Thounaojam et al., 2014a), and miR-301a (Hazra et al., 2019) stimulate microglia secrete inflammatory mediators in JEV-infected BV2 cells, including TNF-α, IL-6, and IL-1β, via separately decreasing RNF11, TNFAIP3 and NKRF, which are all negative regulators of NF-κB signaling pathway. Mukherjee et al. (2019) substantiated that JEV infection results in a time-dependent increase in the let-7a/b expression, both two miRNAs act as a TLR7 ligand and activate NOTCH, a pathway involved inflammatory cytokine production. Simultaneously, they also found down-regulation of miR-34b-5p increases NOTCH1 expression challenged with JEV, which is contrary to the upregulation of this miRNA expression in WNV infection (Kumari et al., 2016).
West Nile virus belongs to the JEV serotype, which maintains in the environment by an avian–mosquito life cycle (Barrows et al., 2018). The neuro-invasive disease of WNV can be fatal, or recovery from the infection may require specialized care, with symptoms lasting more than a year (Petersen et al., 2013). Such neurological symptoms are also reflected in the concentration of targets of differentially expressed miRNAs.
Slonchak et al. (2014, 2015) manifested that up-regulated miR-532-5p and down-regulated miR-2940 suppress WNV proliferation in cells by inhibiting their target genes SESTD1, TAB3 and MetP, all of which are required for efficient WNV replication (Table 3). Hs_154, is significantly produced by WNV in various cell lines and occurred in vivo. Upregulation of Hs_154 and subsequent suppression of CTCF and ECOP, two genes related to anti-apoptosis, cause a significant reduction in viral replication through facilitating WNV-dependent induction of apoptosis (Smith et al., 2012).
Kumar and Nerurkar (2014) conducted an integrated analysis of miRNAs and their disease related targets in mice brain with WNV infection. The network of miRNA interactions with presumptive target genes is demonstrated that miR-196a, miR-202-3p, miR-449c, and miR-125a-3p are downregulated, while miR-155 and miR-32 are adverse. All these miRNAs are positive correlated with protein levels of neuroinflammatory genes. In addition, WT and miR-155–/– mice was selected to further and affirm the role of miR-155 in suppressing virus replication and promoting anti-viral immune response, which is similar to JEV, but the target gene for its action has yet to be identified.
Zika virus was first isolated from the blood of a non-human primate in 1947 and from Aedes africanus mosquitoes 1 year later, which is mainly transmitted by Aedes albopictus and Aedes aegypti (Musso and Gubler, 2016; Lin et al., 2018). As shown in a variety of experimental models and human samples, ZIKV infection induces neuronal apoptosis and disrupts cellular homeostasis, resulting in microcephaly, anencephaly, ventricular enlargement and calcification (Polonio and Peron, 2021).
miR-142-5p has been confirmed to be a factor that accelerates the expression of IL-6 signal transducer (L6ST) and Integrin subunit alpha V (ITGAV), then inhibits ZIKV replication, possibly through activation of innate immune antiviral response and phagosome pathway (Seong et al., 2020; Table 4).
Moreover, due to the close relationship between ZIKV and mosquitoes, multiple studies reported changes in miRNA expression in mosquitoes after ZIKV infection. Saldaña et al. (2017) and Yen et al. (2019) predicted the putative binding sites of three arboviruses (CHIKV, DENV, ZIKV) to mosquito-derived miRNAs, and found eight miRNA-binding sites have low minimum free energy (MFE), suggesting they are more likely to constitute miRNA-viral RNA complexes and subsequently participate in viral replication theoretically.
Based on ZIKV’s special preference for neural stem cells (NSCs), this cell is the best choice to study the effects of ZIKV infection on neurogenesis in vitro. Bhagat et al. revealed miR-1273g-3p as well as miR-204-3p are upregulated in response to ZIKV envelope E protein, a viral component responsible for attachment and entry into host cell (Bhagat et al., 2018). Further, these two miRNAs directly influenced developmental genes PAX3 and NOTCH2 respectively, which contributes to developmental defects in fNSCs (Bhagat et al., 2018). Interestingly, miR-204-5p, also a member of the miR-204 family, constitutes the miR-204-5p/WNT2 axis after ZIKV E protein invasion, and is involved in ZIKV induced impairment in the proliferation and immature differentiation of NSCs. miR-34c was identified as a ZIKV-regulated miRNA with a potential role in restricting NSC growth (Iannolo et al., 2019). This may be dependent on the inhibition of in anti-apoptosis gene Bcl2 and genes involved in stemness maintenance and nervous system development, such as NOTCH and NUMB.
Zika virus infection was found to increase the incidence of microcephaly in fetuses born to mothers infected with ZIKV in 2015. Then the association of ZIKV infection and microcephaly was confirmed in a ZIKV-infected mouse model the following year (Cugola et al., 2016; Li C. et al., 2016; Li H. et al., 2016; Mlakar et al., 2016; Shao et al., 2016). Current studies have shown that miRNA-mRNA network is dysregulated during ZIKV infection, resulting in microcephaly. By evaluating neurosphere growth dynamics, it is speculated that miR-124-3p mediates microcephaly by inhibiting its target gene related to cell-cycle regulation TFRC (Dang et al., 2019). Zhang et al. (2019) demonstrated miR-9 is overexpressed after ZIKV infection and identified down-regulated Glial cell-derived neurotrophic factor (GDNF) is a target of miR-9, GDNF can protect neural progenitors from miR-9-induced apoptosis, which perhaps related to the microcephaly phenotype. Two upregulated miRNAs, miR-145 and miR-148a, are screened involving in cellular migration in brain samples from stillborn with congenital Zika syndrome and 13 common targets genes related to microcephaly, lissencephaly and other neuronal malformation for both miRNAs are predicted by bioinformatics software (Castro et al., 2019). Another bioinformatics study predicted potential genome-wide binding sites between miRNA and ZIKV, and found significant homology between 6 miRNAs and ZIKV genome and 12 autosomal recessive primary microcephaly (MCPH) genes, which may contribute to MCPH production during human fetal brain development (McLean et al., 2017).
In addition to the above widely influential zoonoses, other members of the Flavivirus genus whose infection is also closely related to cellular miRNA. TMUV is an emerging and re-emerging zoonotic pathogen that adversely affects poultry industry in recent years, especially the duck breeding industry (Yang et al., 2021). Cui et al. (2018, 2020) and Huang et al. (2020) first employed deep sequencing to investigate miRNA profiles in DEF cells after DTMUV infection and revealed a novel host evasion mechanism adopted by DTMUV via miR-221-3p inhibiting the negative regulator of host immune response, SOCS5. Subsequently, this miRNA hinders JAK-STAT signaling mediated IFN response and promote virus replication, while miR-148a-5p has a similar effect on the virus replication by modulating SOCS1. TBEV causes neurological symptoms that can lead to permanent disability or death. Although there have been no studies on miRNA expression after TBEV infection, several studies have shown that host-specific miRNAs target sequences integrated into viral genomes (such as tick-specific miR-275 and neuro-specific miR-124a) can effectively control the escape of TBEV chimeric viruses or LGTV (a naturally attenuated member of the TBEV serum complex) and reversal of neurotoxic phenotype (Heiss et al., 2011, 2012; Tsetsarkin et al., 2016). Hermance et al. (2019) confirmed that miRNA changes can regulate Powassan virus (POWV) replication in the host through high-throughput sequencing and transfection of specific miRNA inhibitors in vitro.
Since lncRNA/circRNA can competitively bind with miRNA, the expression level of target mRNA can be indirectly regulated through miRNA. ERG-associated lncRNA (lncRNA-ERGAL) was proved to be involved in promoting stability and integrity of vascular endothelial barrier during DENV infection by sponging miR-183-5p (Zheng et al., 2020). Li et al. (2020) established circRNA–miRNA–mRNA interaction network following JEV infection and unraveled cirC_0000220 binds to miR-326-3p to restrict BCL3, MK2 and TRIM25 mediated inflammatory response.
sfRNA is a product of incomplete degradation of viral genomic RNA by host cellular 5′-3′ exonuclease XRN-1 (Slonchak and Khromykh, 2018). However, the production of sfRNA is limited to Flavivirus genus and not found in the other two genera (Schnettler et al., 2012). This non-coding RNA generally promotes flavivirus pathogenesis by impairing the IFN response, as well documented in DENV, WNV (Schuessler et al., 2012; Pompon et al., 2017). But the specific affinity between ZIKV sfRNA and mosquito DEAD-Box helicase ME31B inhibits viral RNA replication and virion production (Göertz et al., 2019). Only when loaded into Argonaute (AGO), siRNAs and miRNAs perform their functions. Hence, Zeng et al. (2021) constructed AGO-associated viral siRNA profiles and completed a network between ZIKV siRNAs and their probable viral RNA (vRNA) targets via AGO-associated RNA sequencing. Twenty-nine siRNAs and 114 potential targets were obtained from the ZIKV genome, and further functional characterization of ZIKV siRNAs need to be verified. Hussain and Asgari (2014) obtained six miRNA-like vsRNAs mapped to the viral 5′ and 3′UTR regions, termed vsRNA1-6, in Ae. aegypti cells after DENV-2 infection. As vsRNA5 has the most significant effect on virus replication, it was selected for further study. The results showed that vsRNA5 with characteristics similar to miRNAs with AGO-2 targets the viral genome NS1 and inhibits viral replication. The same research group also utilized bioinformatics analysis to identify and coin a WNV-encoded small regulatory viral RNA, KUN-miR-1, from viral 3′UTR (Hussain et al., 2012). The transcription factor GATA4 is identified as a target of KUN-miR-1. GATA mRNA expression also increases after KUN-miR-1 RNA mimic is transfected into Ae. aegypti C6/36 cells, which seems to be necessary for efficient WNV replication.
To understand host–virus interactions is critical to antiviral response. Based on miRNAs regulate a variety of intracellular biological pathways, including virus infection, which has been demonstrated in many viruses (Patil and Karpe, 2020; Song et al., 2020; Zhao et al., 2021). Hence, we emphasized the interaction between flavivirus and host miRNA. Two primary regulatory approaches are adopted by miRNAs in response to viral infection. Due to the high mutation rate of RNA viruses, antiviral effects resulting from direct binding of miRNAs to viral genomes may be rare, unless the viral genome is extremely capable of binding to miRNA (Trobaugh and Klimstra, 2017). This may well explain the lack of coverage, only in DENV (Zhou et al., 2014; Wen et al., 2015; Castillo et al., 2016; Castrillón-Betancur and Urcuqui-Inchima, 2017). Moreover, it has been widely reported that viruses can manipulate the expression levels of specific miRNAs to establish an environment conducive to virus survival and transmission (Hussain et al., 2013; Wu et al., 2014; Chen et al., 2016; Sharma et al., 2016). Meanwhile, the host can also interrupt the virus life cycle by altering miRNA expression levels. In addition to host miRNA, host/virus also produces some other non-coding RNAs during viral infection, which all play different roles in response to viral invasion (Hussain et al., 2012; Ding et al., 2018; Göertz et al., 2019; Zheng et al., 2020; Zeng et al., 2021).
In this view, we collected the differentially expressed miRNAs infected by Flavivirus genus members, mainly DENV, JEV, WNV, and ZIKV. Through the classification of regulatory effects, it is found that these miRNAs principally participate in the process of viral replication, neuroinflammation, neurogenesis and so on (Escalera-Cueto et al., 2015; Liu et al., 2016; Castrillón-Betancur and Urcuqui-Inchima, 2017; Dang et al., 2019; Hazra et al., 2019; Mukherjee et al., 2019; Bhagat et al., 2021). Furthermore, several miRNAs show identical or opposite expression by different flavivirus infection and thus play similar or opposite roles. miR-146a, upregulated after DENV, JEV and ZIKV infections, promotes viral replication by targeting TRAF6 in different cell lines (Wu et al., 2013; Sharma et al., 2015; Shukla et al., 2021). As a member of the antiviral miR-34 family, miR-34c is down-regulated after JEV infection, while up-regulated after WNV and ZIKV infection. But they all work by targeting the NOTCH gene, therefore, it can be speculated that one of the reasons for this differential expression is the difference in cell lines. Although the anti-viral effect of miR-155 has been extensively studied, its expression is reduced during DENV infection to promote viral survival, but with distinct outcomes in JEV and WNV (Pareek et al., 2014; Arboleda et al., 2019; Natekar et al., 2019; Su et al., 2020). Overall, miR-146a, miR-155, and miR-34 family are expected to be potential antiviral targets, the mechanism by which they control the virus should be further investigated in the future. Beyond that, expressing tissue-specific miRNA recognition element has been used as an effective mechanism to exploit attenuated vaccines, helping to reduce the transmission of flavivirus (Lee et al., 2010; Pham et al., 2012; Yen et al., 2018; Buchman et al., 2019). Based on the close association between flavivirus and vector mosquitoes, a large number of mosquito-derived miRNAs have been reported (Skalsky et al., 2010; Hussain et al., 2013; Avila-Bonilla et al., 2020). These host-specific miRNAs also rely on binding to host genes to regulate virus infection. It is worth mentioning that Zhu et al. (2021) discovered a human blood-derived miRNA (miR-150-5p), which interferes with mosquito antiviral system to promote flavivirus infection through cross-species RNAi mechanism, determining the transmission ability of flavivirus in nature.
Other host-derived RNAs, such as lncRNA and circRNA, often regulate target mRNAs by forming a functional grid together with miRNA. During flavivirus infection, lncRNA-ERGAL and cirC_0000220 can regulate the biological processes of infected cells by associating with miR-183-5p and miR-326-3p, respectively (Li et al., 2020; Zheng et al., 2020). But this type of reporting is still limited and a lot of work needs to be done. Increasing evidence points to that viruses themselves can produce non-coding RNAs in addition to their hosts, such as sfRNAs, siRNAs and viral miRNAs mentioned in this review. The interaction between these non-coding RNAs and host/virus has expanded the novel strategy to manage flavivirus (Hussain et al., 2012; Hussain and Asgari, 2014; Slonchak and Khromykh, 2018; Göertz et al., 2019).
In brief, we discussed the regulatory role of host miRNAs after infection with Flavivirus genus members, mainly through miRNA directly binds to the viral genome or vital host genes, although the latter is more widely reported. Understanding host–virus interactions will help us discover promising targets of antiviral drugs and viral therapeutic, which will provide new thinking directions for preventing viral infection.
WC and YP contributed ideas for the review, wrote the manuscript, and produced the figures. AC, MW, ZY, and RJ edited and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32172833), and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2021-18), and by China Agriculture Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We apologize to the authors of articles reporting relevant research that were not cited in this manuscript due to limited space.
5 ′ UTR, 5 ′ untranslated region; 3 ′ UTR, 3 ′ untranslated region; DENV, Dengue virus; WNV, West Nile virus; ZIKV, Zika virus; JEV, Japanese encephalitis virus; YFV, Yellow Fever virus; TBEV, Tick-borne encephalitis virus; LGTV, Langat virus; TMUV, Tembusu; AGO, Argonaute; IFN, interferon; ISG, IFN-stimulating genes; TLRs, toll-like receptors; RIG-I, retinoic acid-inducible gene I; NF- κ B, nuclear factor- κ B; CHME3, human microglial cells; C6/36, Ae. albopictus cell line; C7/10, Ae. Albopictus cell line; THP-1, human monocytic cell line; hNSCs, human neural stem cells; fNSCs, fetal neural stem cells; GSCs, glioblastoma stem cells; N2A, neuro-2A; PK-15, porcine kidney-15 cells; MEFs, mouse embryonic fibroblasts; DEF, Duck embryo fibroblasts; PBMCs, peripheral blood mononuclear cells; HO-1, heme oxygenase-1.
Alpuche-Lazcano, S. P., Saliba, J., Costa, V. V., Campolina-Silva, G. H., Marim, F. M., Ribeiro, L. S., et al. (2021). Profound downregulation of neural transcription factor Npas4 and Nr4a family in fetal mice neurons infected with Zika virus. PLoS Negl. Trop. Dis. 15:e0009425. doi: 10.1371/journal.pntd.0009425
Arboleda, J. F., Fernandez, G. J., and Urcuqui-Inchima, S. (2019). Vitamin D-mediated attenuation of miR-155 in human macrophages infected with dengue virus: implications for the cytokine response. Infect. Genet. Evol. 69, 12–21. doi: 10.1016/j.meegid.2018.12.033
Ashraf, U., Zhu, B., Ye, J., Wan, S., Nie, Y., Chen, Z., et al. (2016). MicroRNA-19b-3p Modulates Japanese Encephalitis Virus-Mediated Inflammation via Targeting RNF11. J. Virol. 90, 4780–4795. doi: 10.1128/JVI.02586-15
Avila-Bonilla, R. G., Yocupicio-Monroy, M., Marchat, L. A., Pérez-Ishiwara, D. G., and Cerecedo-Mercado, D. A. (2020). Del Ángel RM, et al. miR-927 has pro-viral effects during acute and persistent infection with dengue virus type 2 in C6/36 mosquito cells. J. Gen. Virol. 101, 825–839. doi: 10.1099/jgv.0.001441
Bagasra, O., Shamabadi, N. S., Pandey, P., Desoky, A., and McLean, E. (2021). Differential expression of miRNAs in a human developing neuronal cell line chronically infected with Zika virus. Libyan J. Med. 16:1909902. doi: 10.1080/19932820.2021.1909902
Barrows, N. J., Campos, R. K., Liao, K.-C., Prasanth, K. R., Soto-Acosta, R., Yeh, S.-C., et al. (2018). Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 118, 4448–4482. doi: 10.1021/acs.chemrev.7b00719
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/s0092-8674(04)00045-5
Bhagat, R., Prajapati, B., Narwal, S., Agnihotri, N., Adlakha, Y. K., Sen, J., et al. (2018). Zika virus E protein alters the properties of human fetal neural stem cells by modulating microRNA circuitry. Cell Death Differ. 25, 1837–1854. doi: 10.1038/s41418-018-0163-y
Bhagat, R., Rajpara, P., Kaur, G., Gupta, K., and Seth, P. (2021). Zika virus E protein dysregulate mir-204/WNT2 signalling in human fetal neural stem cells. Brain Res. Bull. 176, 93–102. doi: 10.1016/j.brainresbull.2021.08.009
Bhardwaj, U., and Singh, S. K. (2021). Zika Virus NS1 Suppresses VE-Cadherin and Claudin-5 via hsa-miR-101-3p in Human Brain Microvascular Endothelial Cells. Mol. Neurobiol. 58, 6290–6303. doi: 10.1007/s12035-021-02548-x
Buchman, A., Gamez, S., Li, M., Antoshechkin, I., Li, H.-H., Wang, H.-W., et al. (2019). Engineered resistance to Zika virus in transgenic expressing a polycistronic cluster of synthetic small RNAs. Proc. Natl. Acad. Sci. U.S.A. 116, 3656–3661. doi: 10.1073/pnas.1810771116
Campbell, C. L., Harrison, T., Hess, A. M., and Ebel, G. D. (2014). MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 23, 132–139. doi: 10.1111/imb.12070
Castillo, J. A., Castrillón, J. C., Diosa-Toro, M., Betancur, J. G., St Laurent, G., Smit, J. M., et al. (2016). Complex interaction between dengue virus replication and expression of miRNA-133a. BMC Infect. Dis. 16:29. doi: 10.1186/s12879-016-1364-y
Castrillón-Betancur, J. C., and Urcuqui-Inchima, S. (2017). Overexpression of miR-484 and miR-744 in Vero cells alters Dengue virus replication. Mem. Inst. Oswaldo. Cruz. 112, 281–291. doi: 10.1590/0074-02760160404
Castro, F. L., Geddes, V. E. V., Monteiro, F. L. L., Gonçalves, R. M. D. T., Campanati, L., Pezzuto, P., et al. (2019). MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection. ASN Neuro. 11:1759091419850983. doi: 10.1177/1759091419850983
Chang, C.-Y., Wu, C.-C., Wang, J.-D., Li, J.-R., Wang, Y.-Y., Lin, S.-Y., et al. (2021). DHA attenuated Japanese Encephalitis virus infection-induced neuroinflammation and neuronal cell death in cultured rat Neuron/glia. Brain Behav. Immun. 93, 194–205. doi: 10.1016/j.bbi.2021.01.012
Chen, R.-F., Yang, K. D., Lee, I.-K., Liu, J.-W., Huang, C.-H., Lin, C.-Y., et al. (2014). Augmented miR-150 expression associated with depressed SOCS1 expression involved in dengue haemorrhagic fever. J. Infect. 69, 366–374. doi: 10.1016/j.jinf.2014.05.013
Chen, Z., Ye, J., Ashraf, U., Li, Y., Wei, S., Wan, S., et al. (2016). MicroRNA-33a-5p Modulates Japanese Encephalitis Virus Replication by Targeting Eukaryotic Translation Elongation Factor 1A1. J. Virol. 90, 3722–3734. doi: 10.1128/JVI.03242-15
Cugola, F. R., Fernandes, I. R., Russo, F. B., Freitas, B. C., Dias, J. L. M., Guimarães, K. P., et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271. doi: 10.1038/nature18296
Cui, M., Chen, S., Zhang, S., Cheng, A., Pan, Y., Huang, J., et al. (2020). Duck Tembusu Virus Utilizes miR-221-3p Expression to Facilitate Viral Replication Targeting of Suppressor of Cytokine Signaling 5. Front. Microbiol. 11:596. doi: 10.3389/fmicb.2020.00596
Cui, M., Jia, R., Huang, J., Wu, X., Hu, Z., Zhang, X., et al. (2018). Analysis of the microRNA expression profiles in DEF cells infected with duck Tembusu virus. Infect. Genet. Evol. 63, 126–134. doi: 10.1016/j.meegid.2018.05.020
Dang, J. W., Tiwari, S. K., Qin, Y., and Rana, T. M. (2019). Genome-wide Integrative Analysis of Zika-Virus-Infected Neuronal Stem Cells Reveals Roles for MicroRNAs in Cell Cycle and Stemness. Cell Rep. 27, 3618–3628.e5. doi: 10.1016/j.celrep.2019.05.059
Ding, S.-W., Han, Q., Wang, J., and Li, W.-X. (2018). Antiviral RNA interference in mammals. Curr. Opin. Immunol. 54, 109–114. doi: 10.1016/j.coi.2018.06.010
Diosa-Toro, M., Echavarría-Consuegra, L., Flipse, J., Fernández, G. J., Kluiver, J., van den Berg, A., et al. (2017). MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression. PLoS Negl. Trop Dis. 11:e0005981. doi: 10.1371/journal.pntd.0005981
Escalera-Cueto, M., Medina-Martínez, I., del Angel, R. M., Berumen-Campos, J., Gutiérrez-Escolano, A. L., and Yocupicio-Monroy, M. (2015). Let-7c overexpression inhibits dengue virus replication in human hepatoma Huh-7 cells. Virus Res. 196, 105–112. doi: 10.1016/j.virusres.2014.11.010
Glasner, D. R., Puerta-Guardo, H., Beatty, P. R., and Harris, E. (2018). The Good, the Bad, and the Shocking: The Multiple Roles of Dengue Virus Nonstructural Protein 1 in Protection and Pathogenesis. Annu. Rev. Virol. 5, 227–253. doi: 10.1146/annurev-virology-101416-041848
Göertz, G. P., van Bree, J. W. M., Hiralal, A., Fernhout, B. M., Steffens, C., Boeren, S., et al. (2019). Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by. Proc. Natl. Acad. Sci. U.S.A. 116, 19136–19144. doi: 10.1073/pnas.1905617116
Hazra, B., Chakraborty, S., Bhaskar, M., Mukherjee, S., Mahadevan, A., and Basu, A. (2019). miR-301a Regulates Inflammatory Response to Japanese Encephalitis Virus Infection via Suppression of NKRF Activity. J. Immunol. 203, 2222–2238. doi: 10.4049/jimmunol.1900003
Hazra, B., Kumawat, K. L., and Basu, A. (2017). The host microRNA miR-301a blocks the IRF1-mediated neuronal innate immune response to Japanese encephalitis virus infection. Sci. Signal. 10:eaaf5185. doi: 10.1126/scisignal.aaf5185
Heiss, B. L., Maximova, O. A., and Pletnev, A. G. (2011). Insertion of microRNA targets into the flavivirus genome alters its highly neurovirulent phenotype. J. Virol. 85, 1464–1472. doi: 10.1128/JVI.02091-10
Heiss, B. L., Maximova, O. A., Thach, D. C., Speicher, J. M., and Pletnev, A. G. (2012). MicroRNA targeting of neurotropic flavivirus: effective control of virus escape and reversion to neurovirulent phenotype. J. Virol. 86, 5647–5659. doi: 10.1128/JVI.07125-11
Hermance, M. E., Widen, S. G., Wood, T. G., and Thangamani, S. (2019). Ixodes scapularis salivary gland microRNAs are differentially expressed during Powassan virus transmission. Sci. Rep. 9:13110. doi: 10.1038/s41598-019-49572-5
Huang, C.-W., Tsai, K.-N., Chen, Y.-S., and Chang, R.-Y. (2021). Differential miRNA Expression Profiling Reveals Correlation of miR125b-5p with Persistent Infection of Japanese Encephalitis Virus. Int. J. Mol. Sci. 22:4218. doi: 10.3390/ijms22084218
Huang, S., Cheng, A., Cui, M., Pan, Y., Wang, M., Huang, J., et al. (2020). Duck Tembusu virus promotes the expression of suppressor of cytokine signaling 1 by downregulating miR-148a-5p to facilitate virus replication. Infect. Genet. Evol. 85:104392. doi: 10.1016/j.meegid.2020.104392
Hussain, M., and Asgari, S. (2014). MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc. Natl. Acad. Sci. U.S.A. 111, 2746–2751. doi: 10.1073/pnas.1320123111
Hussain, M., Torres, S., Schnettler, E., Funk, A., Grundhoff, A., Pijlman, G. P., et al. (2012). West Nile virus encodes a microRNA-like small RNA in the 3’ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 40, 2210–2223. doi: 10.1093/nar/gkr848
Hussain, M., Walker, T., O’Neill, S. L., and Asgari, S. (2013). Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector. Aedes aegypti. Insect Biochem. Mol. Biol. 43, 146–152. doi: 10.1016/j.ibmb.2012.11.005
Iannolo, G., Sciuto, M. R., Cuscino, N., Pallini, R., Douradinha, B., Ricci Vitiani, L., et al. (2019). Zika virus infection induces MiR34c expression in glioblastoma stem cells: new perspectives for brain tumor treatments. Cell Death Dis. 10:263. doi: 10.1038/s41419-019-1499-z
Jiang, H., Bai, L., Ji, L., Bai, Z., Su, J., Qin, T., et al. (2020). Degradation of MicroRNA miR-466d-3p by Japanese Encephalitis Virus NS3 Facilitates Viral Replication and Interleukin-1β Expression. J. Virol. 94, e294–e220. doi: 10.1128/JVI.00294-20
Jiang, L., Lin, C., Song, L., Wu, J., Chen, B., Ying, Z., et al. (2012). MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-κB/IκBα negative feedback loop. J. Clin. Invest. 122, 33–47. doi: 10.1172/jci58849
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M., and Sarnow, P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581. doi: 10.1126/science.1113329
Kanokudom, S., Vilaivan, T., Wikan, N., Thepparit, C., Smith, D. R., and Assavalapsakul, W. (2017). miR-21 promotes dengue virus serotype 2 replication in HepG2 cells. Antiviral Res. 142, 169–177. doi: 10.1016/j.antiviral.2017.03.020
Kumar, A., Kumar, A., Ingle, H., Kumar, S., Mishra, R., Verma, M. K., et al. (2018). MicroRNA hsa-miR-324-5p Suppresses H5N1 Virus Replication by Targeting the Viral PB1 and Host CUEDC2. J. Virol. 92, e1057–e1018. doi: 10.1128/JVI.01057-18
Kumar, M., and Nerurkar, V. R. (2014). Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology 45, 143–151. doi: 10.1016/j.virol.2014.01.004
Kumari, B., Jain, P., Das, S., Ghosal, S., Hazra, B., Trivedi, A. C., et al. (2016). Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese Encephalitis Virus in microglial cells. Sci. Rep. 6:20263. doi: 10.1038/srep20263
Kunden, R. D., Khan, J. Q., Ghezelbash, S., and Wilson, J. A. (2020). The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int. J. Mol. Sci. 21:5677. doi: 10.3390/ijms21165677
Lawrence, T. (2009). The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 1:a001651.
Lee, T.-C., Lin, Y.-L., Liao, J.-T., Su, C.-M., Lin, C.-C., Lin, W.-P., et al. (2010). Utilizing liver-specific microRNA-122 to modulate replication of dengue virus replicon. Biochem. Biophys. Res. Commun. 396, 596–601. doi: 10.1016/j.bbrc.2010.04.080
Li, C., Xu, D., Ye, Q., Hong, S., Jiang, Y., Liu, X., et al. (2016). Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 19, 120–126. doi: 10.1016/j.stem.2016.04.017
Li, H., Saucedo-Cuevas, L., Regla-Nava, J. A., Chai, G., Sheets, N., Tang, W., et al. (2016). Zika Virus Infects Neural Progenitors in the Adult Mouse Brain and Alters Proliferation. Cell Stem Cell 19, 593–598. doi: 10.1016/j.stem.2016.08.005
Li, W., Cheng, P., Nie, S., and Cui, W. (2016). miR-370 mimic inhibits replication of Japanese encephalitis virus in glioblastoma cells. Neuropsychiatr. Dis. Treat. 12, 2411–2417. doi: 10.2147/NDT.S113236
Li, Y., Ashraf, U., Chen, Z., Zhou, D., Imran, M., Ye, J., et al. (2020). Genome-wide profiling of host-encoded circular RNAs highlights their potential role during the Japanese encephalitis virus-induced neuroinflammatory response. BMC Genom. 21:409. doi: 10.1186/s12864-020-06822-5
Lim, S. P. (2019). Dengue drug discovery: progress, challenges and outlook. Antiviral Res. 163, 156–178. doi: 10.1016/j.antiviral.2018.12.016
Lin, H.-H., Yip, B.-S., Huang, L.-M., and Wu, S.-C. (2018). Zika virus structural biology and progress in vaccine development. Biotechnol. Adv. 36, 47–53. doi: 10.1016/j.biotechadv.2017.09.004
Liu, S., Chen, L., Zeng, Y., Si, L., Guo, X., Zhou, J., et al. (2016). Suppressed expression of miR-378 targeting gzmb in NK cells is required to control dengue virus infection. Cell Mol. Immunol. 13, 700–708. doi: 10.1038/cmi.2015.52
McLean, E., Bhattarai, R., Hughes, B. W., Mahalingam, K., and Bagasra, O. (2017). Computational identification of mutually homologous Zika virus miRNAs that target microcephaly genes. Libyan. J. Med. 12:1304505. doi: 10.1080/19932820.2017.1304505
Miranda, K. C., Huynh, T., Tay, Y., Ang, Y.-S., Tam, W.-L., Thomson, A. M., et al. (2006). A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217. doi: 10.1016/j.cell.2006.07.031
Mishra, R., Lahon, A., and Banerjea, A. C. (2020). Dengue Virus Degrades USP33-ATF3 Axis via Extracellular Vesicles to Activate Human Microglial Cells. J. Immunol. 205, 1787–1798. doi: 10.4049/jimmunol.2000411
Mlakar, J., Korva, M., Tul, N., Popović, M., Poljšak-Prijatelj, M., Mraz, J., et al. (2016). Zika Virus Associated with Microcephaly. N. Engl. J. Med. 374, 951–958. doi: 10.1056/NEJMoa1600651
Mukherjee, S., Akbar, I., Kumari, B., Vrati, S., Basu, A., and Banerjee, A. (2019). Japanese Encephalitis Virus-induced let-7a/b interacted with the NOTCH-TLR7 pathway in microglia and facilitated neuronal death via caspase activation. J. Neurochem. 149, 518–534. doi: 10.1111/jnc.14645
Murphy Schafer, A. R., Smith, J. L., Pryke, K. M., DeFilippis, V. R., and Hirsch, A. J. (2020). The E3 Ubiquitin Ligase SIAH1 Targets MyD88 for Proteasomal Degradation During Dengue Virus Infection. Front. Microbiol. 11:24. doi: 10.3389/fmicb.2020.00024
Natekar, J. P., Rothan, H. A., Arora, K., Strate, P. G., and Kumar, M. (2019). Cellular microRNA-155 Regulates Virus-Induced Inflammatory Response and Protects against Lethal West Nile Virus Infection. Viruses 12:9. doi: 10.3390/v12010009
Pareek, S., Roy, S., Kumari, B., Jain, P., Banerjee, A., and Vrati, S. (2014). MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J. Neuroinflam. 11:97. doi: 10.1186/1742-2094-11-97
Pasquinelli, A. E. (2012). MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282. doi: 10.1038/nrg3162
Patil, R. N., and Karpe, Y. A. (2020). Uncovering the Roles of miR-214 in Hepatitis E Virus Replication. J. Mol. Biol. 432, 5322–5342. doi: 10.1016/j.jmb.2020.07.015
Petersen, L. R., Brault, A. C., and Nasci, R. S. (2013). West Nile virus: review of the literature. JAMA 310, 308–315. doi: 10.1001/jama.2013.8042
Pham, A. M., Langlois, R. A., and TenOever, B. R. (2012). Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 8:e1002465. doi: 10.1371/journal.ppat.1002465
Polonio, C. M., and Peron, J. P. S. (2021). ZIKV Infection and miRNA Network in Pathogenesis and Immune Response. Viruses 13:1992. doi: 10.3390/v13101992
Pompon, J., Manuel, M., Ng, G. K., Wong, B., Shan, C., Manokaran, G., et al. (2017). Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog. 13:e1006535. doi: 10.1371/journal.ppat.1006535
Pulkkinen, K. H., Ylä-Herttuala, S., and Levonen, A.-L. (2011). Heme oxygenase 1 is induced by miR-155 via reduced BACH1 translation in endothelial cells. Free Radic Biol. Med. 51, 2124–2131. doi: 10.1016/j.freeradbiomed.2011.09.014
Rastogi, M., and Singh, S. K. (2019). Modulation of Type-I Interferon Response by hsa-miR-374b-5p During Japanese Encephalitis Virus Infection in Human Microglial Cells. Front. Cell Infect. Microbiol. 9:291. doi: 10.3389/fcimb.2019.00291
Saldaña, M. A., Etebari, K., Hart, C. E., Widen, S. G., Wood, T. G., Thangamani, S., et al. (2017). Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Negl. Trop. Dis. 11:e0005760. doi: 10.1371/journal.pntd.0005760
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. A. (2011). ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Schnettler, E., Sterken, M. G., Leung, J. Y., Metz, S. W., Geertsema, C., Goldbach, R. W., et al. (2012). Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J. Virol. 86, 13486–13500. doi: 10.1128/JVI.01104-12
Schuessler, A., Funk, A., Lazear, H. M., Cooper, D. A., Torres, S., Daffis, S., et al. (2012). West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86, 5708–5718. doi: 10.1128/JVI.00207-12
Seong, R.-K., Lee, J. K., Cho, G. J., Kumar, M., and Shin, O. S. (2020). mRNA and miRNA profiling of Zika virus-infected human umbilical cord mesenchymal stem cells identifies miR-142-5p as an antiviral factor. Emerg. Microb. Infect. 9, 2061–2075. doi: 10.1080/22221751.2020.1821581
Shao, Q., Herrlinger, S., Yang, S.-L., Lai, F., Moore, J. M., Brindley, M. A., et al. (2016). Zika virus infection disrupts neurovascular development and results in postnatal microcephaly with brain damage. Development 143, 4127–4136. doi: 10.1242/dev.143768
Sharma, N., Kumawat, K. L., Rastogi, M., Basu, A., and Singh, S. K. (2016). Japanese Encephalitis Virus exploits the microRNA-432 to regulate the expression of Suppressor of Cytokine Signaling (SOCS) 5. Sci. Rep. 6:27685. doi: 10.1038/srep27685
Sharma, N., Verma, R., Kumawat, K. L., Basu, A., and Singh, S. K. (2015). miR-146a suppresses cellular immune response during Japanese encephalitis virus JaOArS982 strain infection in human microglial cells. J. Neuroinflam. 12:30. doi: 10.1186/s12974-015-0249-0
Shukla, A., Rastogi, M., and Singh, S. K. (2021). Zika virus NS1 suppresses the innate immune responses via miR-146a in human microglial cells. Int. J. Biol. Macromol. 193, 2290–2296. doi: 10.1016/j.ijbiomac.2021.11.061
Skalsky, R. L., Vanlandingham, D. L., Scholle, F., Higgs, S., and Cullen, B. R. (2010). Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genom. 11:119. doi: 10.1186/1471-2164-11-119
Slonchak, A., Hussain, M., Torres, S., Asgari, S., and Khromykh, A. A. (2014). Expression of mosquito microRNA Aae-miR-2940-5p is downregulated in response to West Nile virus infection to restrict viral replication. J. Virol. 88, 8457–8467. doi: 10.1128/JVI.00317-14
Slonchak, A., and Khromykh, A. A. (2018). Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antiviral Res. 159, 13–25. doi: 10.1016/j.antiviral.2018.09.006
Slonchak, A., Shannon, R. P., Pali, G., and Khromykh, A. A. (2015). Human MicroRNA miR-532-5p Exhibits Antiviral Activity against West Nile Virus via Suppression of Host Genes SESTD1 and TAB3 Required for Virus Replication. J. Virol. 90, 2388–2402. doi: 10.1128/JVI.02608-15
Smith, J. L., Grey, F. E., Uhrlaub, J. L., Nikolich-Zugich, J., and Hirsch, A. J. (2012). Induction of the cellular microRNA, Hs_154, by West Nile virus contributes to virus-mediated apoptosis through repression of antiapoptotic factors. J. Virol. 86, 5278–5287. doi: 10.1128/JVI.06883-11
Smith, J. L., Jeng, S., McWeeney, S. K., and Hirsch, A. J. (2017). A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 91, e2388–e2316. doi: 10.1128/JVI.02388-16
Song, J., Sun, H., Sun, H., Jiang, Z., Zhu, J., Wang, C., et al. (2020). Swine MicroRNAs and Restrict the Cross-Species Infection of Avian Influenza Virus. J. Virol. 94, e1700–e1720. doi: 10.1128/JVI.01700-20
Su, J., Li, C., Zhang, Y., Yan, T., Zhu, X., Zhao, M., et al. (2017). Identification of microRNAs expressed in the midgut of Aedes albopictus during dengue infection. Parasit. Vectors 10:63. doi: 10.1186/s13071-017-1966-2
Su, Y.-C., Huang, Y.-F., Wu, Y.-W., Chen, H.-F., Wu, Y.-H., Hsu, C.-C., et al. (2020). MicroRNA-155 inhibits dengue virus replication by inducing heme oxygenase-1-mediated antiviral interferon responses. FASEB J. 34, 7283–7294. doi: 10.1096/fj.201902878R
Thounaojam, M. C., Kaushik, D. K., Kundu, K., and Basu, A. (2014a). MicroRNA-29b modulates Japanese encephalitis virus-induced microglia activation by targeting tumor necrosis factor alpha-induced protein 3. J. Neurochem. 129, 143–154. doi: 10.1111/jnc.12609
Thounaojam, M. C., Kundu, K., Kaushik, D. K., Swaroop, S., Mahadevan, A., Shankar, S. K., et al. (2014b). MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting Src homology 2-containing inositol phosphatase 1. J. Virol. 88, 4798–4810. doi: 10.1128/JVI.02979-13
Trobaugh, D. W., and Klimstra, W. B. (2017). MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 23, 80–93. doi: 10.1016/j.molmed.2016.11.003
Tsetsarkin, K. A., Liu, G., Kenney, H., Hermance, M., Thangamani, S., and Pletnev, A. G. (2016). Concurrent micro-RNA mediated silencing of tick-borne flavivirus replication in tick vector and in the brain of vertebrate host. Sci. Rep. 6:33088. doi: 10.1038/srep33088
Wen, W., He, Z., Jing, Q., Hu, Y., Lin, C., Zhou, R., et al. (2015). Cellular microRNA-miR-548g-3p modulates the replication of dengue virus. J. Infect. 70, 631–640. doi: 10.1016/j.jinf.2014.12.001
Wong, R. R., Abd-Aziz, N., Affendi, S., and Poh, C. L. (2020). Role of microRNAs in antiviral responses to dengue infection. J. Biomed Sci. 27:4. doi: 10.1186/s12929-019-0614-x
Wu, N., Gao, N., Fan, D., Wei, J., Zhang, J., and An, J. (2014). miR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926 cells. Microb. Infect. 16, 911–922. doi: 10.1016/j.micinf.2014.08.011
Wu, S., He, L., Li, Y., Wang, T., Feng, L., Jiang, L., et al. (2013). miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J. Infect. 67, 329–341. doi: 10.1016/j.jinf.2013.05.003
Xie, L.-M., Yin, X., Bi, J., Luo, H.-M., Cao, X.-J., Ma, Y.-W., et al. (2021). Identification of potential biomarkers in dengue via integrated bioinformatic analysis. PLoS Negl. Trop. Dis. 15:e0009633. doi: 10.1371/journal.pntd.0009633
Xu, P., Jia, S., Wang, K., Fan, Z., Zheng, H., Lv, J., et al. (2020). MiR-140 inhibits classical swine fever virus replication by targeting Rab25 in swine umbilical vein endothelial cells. Virulence 11, 260–269. doi: 10.1080/21505594.2020.1735051
Yan, H., Zhou, Y., Liu, Y., Deng, Y., and Chen, X. (2014). miR-252 of the Asian tiger mosquito Aedes albopictus regulates dengue virus replication by suppressing the expression of the dengue virus envelope protein. J. Med. Virol. 86, 1428–1436. doi: 10.1002/jmv.23815
Yang, S., Huang, Y., Shi, Y., Bai, X., Yang, P., and Chen, Q. (2021). Tembusu Virus entering the central nervous system caused nonsuppurative encephalitis without disrupting the blood-brain barrier. J. Virol. 95, e2191–e2120. doi: 10.1128/JVI.02191-20
Yang, S., Pei, Y., Li, X., Zhao, S., Zhu, M., and Zhao, A. (2016). miR-124 attenuates Japanese encephalitis virus replication by targeting DNM2. Virol. J. 13, 105. doi: 10.1186/s12985-016-0562-y
Yen, P.-S., Chen, C.-H., Sreenu, V., Kohl, A., and Failloux, A.-B. (2019). Assessing the Potential Interactions between Cellular miRNA and Arboviral Genomic RNA in the Yellow Fever Mosquito. Viruses 11:540. doi: 10.3390/v11060540
Yen, P.-S., James, A., Li, J.-C., Chen, C.-H., and Failloux, A.-B. (2018). Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito. Commun. Biol. 1:11. doi: 10.1038/s42003-017-0011-5
Zeng, J., Luo, Z., Dong, S., Xie, X., Liang, X., Yan, Y., et al. (2021). Functional Mapping of AGO-Associated Zika Virus-Derived Small Interfering RNAs in Neural Stem Cells. Front. Cell Infect. Microbiol. 11:628887. doi: 10.3389/fcimb.2021.628887
Zhang, H., Chang, Y., Zhang, L., Kim, S.-N., Otaegi, G., Zhang, Z., et al. (2019). Upregulation of MicroRNA miR-9 Is Associated with Microcephaly and Zika Virus Infection in Mice. Mol. Neurobiol. 56, 4072–4085. doi: 10.1007/s12035-018-1358-4
Zhao, W., Hou, P., Ma, W., Jiang, C., Wang, H., and He, H. (2021). Bta-miR-101 suppresses BEFV replication via targeting NKRF. Vet. Microbiol. 259:109127. doi: 10.1016/j.vetmic.2021.109127
Zheng, B., Wang, H., Cui, G., Guo, Q., Si, L., Yan, H., et al. (2020). ERG-Associated lncRNA (ERGAL) Promotes the Stability and Integrity of Vascular Endothelial Barrier During Dengue Viral Infection via Interaction With miR-183-5p. Front. Cell Infect. Microbiol. 10:477. doi: 10.3389/fcimb.2020.00477
Zhou, D., Li, Q., Jia, F., Zhang, L., Wan, S., Li, Y., et al. (2020). The Japanese Encephalitis Virus NS1’ Protein Inhibits Type I IFN Production by Targeting MAVS. J. Immunol. 204, 1287–1298. doi: 10.4049/jimmunol.1900946
Zhou, Y., Liu, Y., Yan, H., Li, Y., Zhang, H., Xu, J., et al. (2014). miR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication. Parasit. Vectors 7:488. doi: 10.1186/s13071-014-0488-4
Zhu, B., Ye, J., Nie, Y., Ashraf, U., Zohaib, A., Duan, X., et al. (2015). MicroRNA-15b Modulates Japanese Encephalitis Virus-Mediated Inflammation via Targeting RNF125. J. Immunol. 195, 2251–2262. doi: 10.4049/jimmunol.1500370
Zhu, X., He, Z., Hu, Y., Wen, W., Lin, C., Yu, J., et al. (2014). MicroRNA-30e* suppresses dengue virus replication by promoting NF-κB-dependent IFN production. PLoS Negl. Trop. Dis. 8:e3088. doi: 10.1371/journal.pntd.0003088
Keywords: flaviviruses, host miRNAs, host–virus interaction, virus replication, inflammatory
Citation: Cai W, Pan Y, Cheng A, Wang M, Yin Z and Jia R (2022) Regulatory Role of Host MicroRNAs in Flaviviruses Infection. Front. Microbiol. 13:869441. doi: 10.3389/fmicb.2022.869441
Received: 04 February 2022; Accepted: 16 March 2022;
Published: 11 April 2022.
Edited by:
Michael Thomas Wolfinger, University of Vienna, AustriaReviewed by:
Joanna Sztuba-Solinska, Auburn University, United StatesCopyright © 2022 Cai, Pan, Cheng, Wang, Yin and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=; Renyong Jia, amlhcnlAc2ljYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.