- 1Forest Department, Forestry College, Hebei Agricultural University, Baoding, China
- 2State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
- 3Hebei Key Laboratory for Tree Genetic Resources and Forest Protection, Baoding, China
- 4Key Laboratory of Tree Breeding and Cultivation of State Forestry Administration, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
Wetwood disease of poplar limits the processing and manufacturing of poplar, and the pathogenic bacteria of wet heartwood are poorly known. We used high-throughput sequencing methods to analyze the bacterial community of the heartwood, sapwood, root tissue, and rhizosphere soil of Populus × euramericana cv. “74/76” (poplar 107) in wetwood trees and healthy trees to explore the cause of poplar wetwood disease. Bacterial diversity and community structure were analyzed, and the correlation between wood properties and bacterial relative abundance was analyzed to explore their relationship. Two alpha-diversity indices of endophytic bacteria in the heartwood of wetwood trees were significantly (p < 0.05) lower than that in the heartwood of healthy trees, and the community structure between the two types of trees was significantly different. No significant differences in the alpha-diversity indices nor community structure were observed in the sapwood, root tissue, or rhizosphere bacterial community of diseased and healthy trees. The distribution of dominant bacteria genus in the heartwood of diseased and healthy trees differed. Proteiniphilum, Actinotalea, and Methanobacterium were the dominant genera in diseased trees’ heartwood. Proteiniphilum, Dysgonomonas, and Bacteroides were the dominant genera in healthy trees’ heartwood. The relative abundance of Proteiniphilum, Actinotalea, and Methanobacterium was significantly higher in the heartwood of wetwood trees than those of healthy trees. A db-RDA analysis found that these three bacterial genera were positively correlated with the rate of wet heartwood. These three bacterial genera may be the main pathogens causing poplar wetwood disease.
Introduction
Wet heartwood, also called wetwood disease, is an abnormal tissue formed in living standing trees’ heartwood. Its typical characteristics are higher water content than healthy heartwood, a darker tissue color, and a stench of fatty acids. Wetwood is a common tree disease that occurs in coniferous and broad-leaved trees (Alizadeh et al., 2017a,b; Basavand et al., 2021; Martin et al., 2021). The wetwood disease has been studied since the 20th century in species such as white fir (Wilcox and Pong, 1971; Wilcox and Oldham, 1972), Abies lasiocarpa [Hook] Nutt (Zhang et al., 2006), Pinus sylveatris L. (Kovaleva et al., 2015), Abies balsamea (L.) Mill. (Jeremic et al., 2004), Abies alba Mill. (Streichan, 1986; Martin et al., 2021), and other coniferous species, as well as P. deltoides cv. Lux ex. I-69/55 (Yu et al., 2019), Betula pendula roth (Goychuk et al., 2020), Ulmus spp. L. (Alizadeh et al., 2017a,b), Morus (Basavand et al., 2021), Casuarina equisetifolia (Ayin et al., 2019), Gmelina arboreaI (Moya et al., 2009), and other broad-leaved tree species. Many things have been investigated to identify the causes for development of wetwood, such as genetic variability, bacterial and fungal communities, wood physicochemical properties, and wood processing and utilization (Schroeder and Kozlik, 1972; Schink et al., 1981a; Xu et al., 2001; Jeremic et al., 2004; Zhang et al., 2005). Wetwood is mainly caused by a bacterial infection, related to physical, mechanical, and biological damage (Moya et al., 2009; Alizadeh et al., 2017a; Martin et al., 2021). Some researchers have reported that pathogens responsible for the development of wetwood are endophytic bacteria of the tree. And they thought that bacteria were part of the natural microbes of the normal wood, generally in a dormant state, and only became active when the tree species changed in a way that would provide nutrients for its inhabitants, leading to the development of wetwood (Zu, 2000). Wetwood has been researched extensively but its cause and underlying process of formation remain unclear (Wang et al., 2008). Wetwood disease is an important problem for the forest and lumber products industry because it often takes longer to dry and may crack, yielding a lumber with diminished quality (Jeremic et al., 2004; Roque et al., 2009). Furthermore, wet heartwood may deteriorate glue bond strength, lowering the quality of plywood and reconstituted wood products (Xu et al., 2010).
One study compared the anatomical structure of wet and healthy heartwood in A. balsamea (L.) Mill., and reported no significant differences in their characteristics except for the higher frequency of bacteria (Jeremic et al., 2004). A variety of pathogenic bacteria have been isolated from wet heartwood tissues. Strains of Enterobacteriaceae are most often isolated from diseased trees, particularly Erwinia and Klebsiella, as well as Enterobacter, Pantoea, Citrobacter, Edwardsiella, Lactobacillus, and Kosakonia (Carter, 1945; Hartley et al., 1961; Knutson, 1973; Schink et al., 1981b; Murdoch, 1983; Diamandis and Koukos, 1992; Zheng et al., 2006; Ayin et al., 2019; Goychuk et al., 2020). Other bacteria are also often separated from the wet heartwood of conifers and broadleaf trees, such as Bacillus, Clostridium, Bacteroides, Ralstonia, Xanthomonas, and Pseudomonas (Carter, 1945; Knutson, 1973; Schink et al., 1981b; Murdoch, 1983; Warshaw et al., 1985; Diamandis and Koukos, 1992; Kovaleva et al., 2015; Ayin et al., 2019; Goychuk et al., 2020).
The symptoms of wet heartwood occur in many tree species, including Leuce, Aigeiros, Tacamahaca, and Populus (Song et al., 2017). Poplar is one of the most important species for short-cycle industrial raw materials and for constructing ecologically protected forests in China (Song et al., 2017). Wet heartwood is prone to cracking, warping, shrinkage, and other defects during the drying process, which reduces the value of the wood (Jeremic et al., 2004; Roque et al., 2009). Traditional bacterial culture methods do not fully reflect the composition and diversity of the bacterial communities of wetwood trees, which limits the study of the disease. In this study, Populus × euramericana cv. “74/76” (poplar 107) from 25 plantations were used as research materials. Wood properties of diseased and healthy trees were analyzed and compared to explore the effect of wetwood disease on wood properties. Additionally, the composition and diversity of the bacterial community in the heartwood, sapwood, root and rhizosphere soil of the wetwood and healthy trees were evaluated by high-throughput sequencing. The bacteria possibly involved in wetwood formation were analyzed by comparing bacterial communities between different locations of diseased and healthy trees. The results have important implications for further understanding the development of wetwood disease and also provide new insights into the role of bacterial communities in plant health.
Materials and Methods
Sample Collection
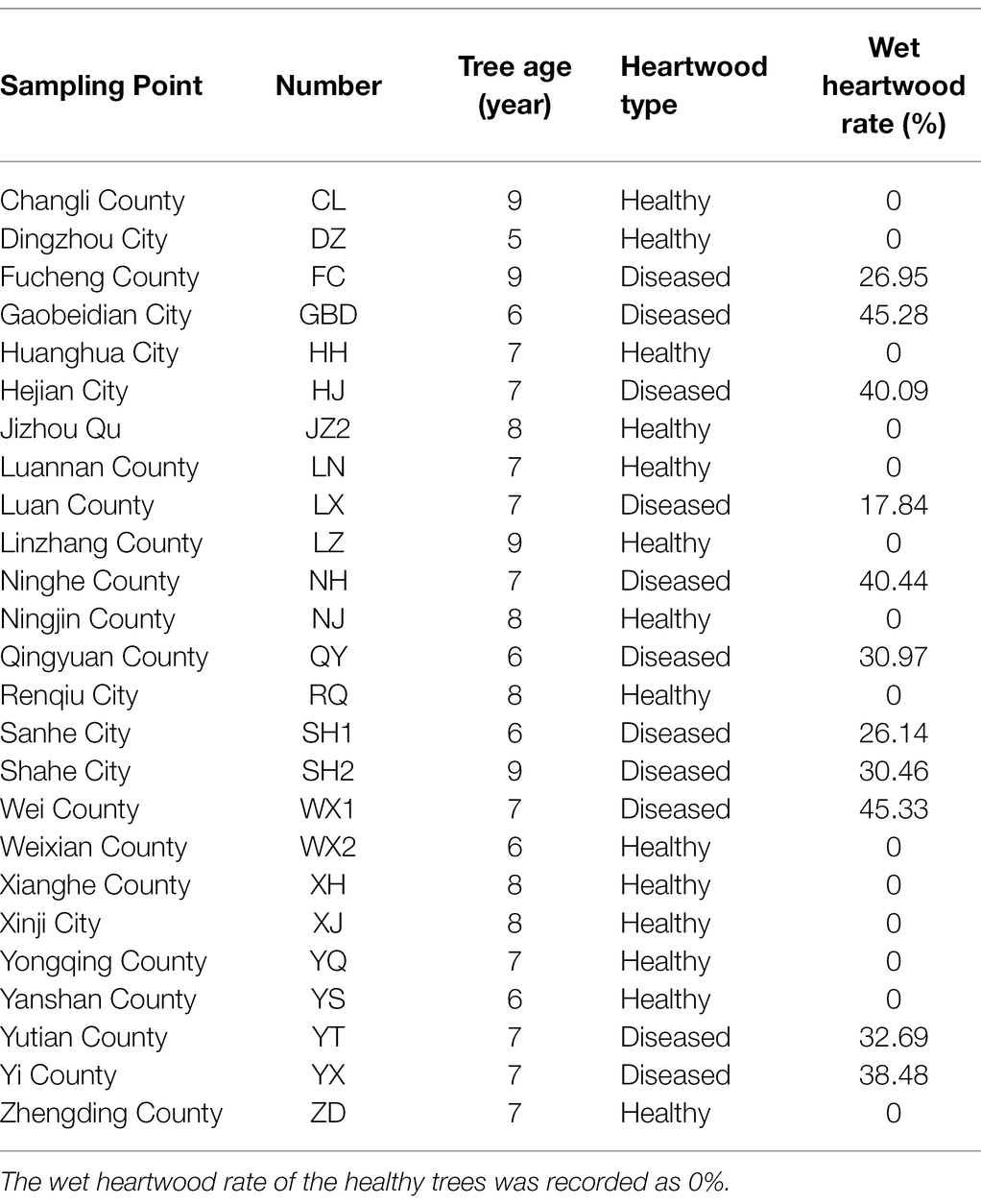
In July 2017, a random sampling method was used to investigate 25 poplar plantation in Hebei Plain, China. One standard tree was selected from each of the 25 plantations wood property testing. The plantation trees were 5–9 years of age and were not maintained. The forest density was 802 trees/h·m−2. The location of each sample is shown in Figure 1, and the basic information of the sample wood is shown in Table 1. After the standard wood was felled, the wet heartwood area was immediately visible. About 11 of the 25 trees were diseased trees, and the other 14 were healthy trees (Figure 2). A 5 cm thick disk was sawed at the base of the trunk (0 m above ground) to determine the color difference and the wet heartwood ratio of the wood. Two 5 cm thick disks were sawed at breast height (1.3 m above ground). One disk was used to determine the chemical properties of the wood, and the other disk was stored at −80°C to determine the bacterial community composition. A 1 m long peeling block was sawed at 2–3 and at 3–4 m from the base of each tree to determine physical and mechanical properties of the wood. Rhizosphere soil samples were collected by the shaking soil method (Hua et al., 2012). The samples were taken 40 cm from the base of the target plant at each cardinal point (east, west, south, and north) and at a soil depth of 20 cm. The nonrhizosphere soil without the roots was shaken off, and sterile tweezers were used to gently peel off the soil attached to the surface of the root system. After the rhizosphere soil was obtained, filled a 10 ml centrifuge tube with the soil by the quarter method (Bao, 2000), and stored at −80°C prior to DNA extraction. Root and soil samples were taken at the same sampling point. The tender roots were taken from the root tips, placed in a 10 ml centrifuge tube, and stored at −80°C prior to DNA extraction.
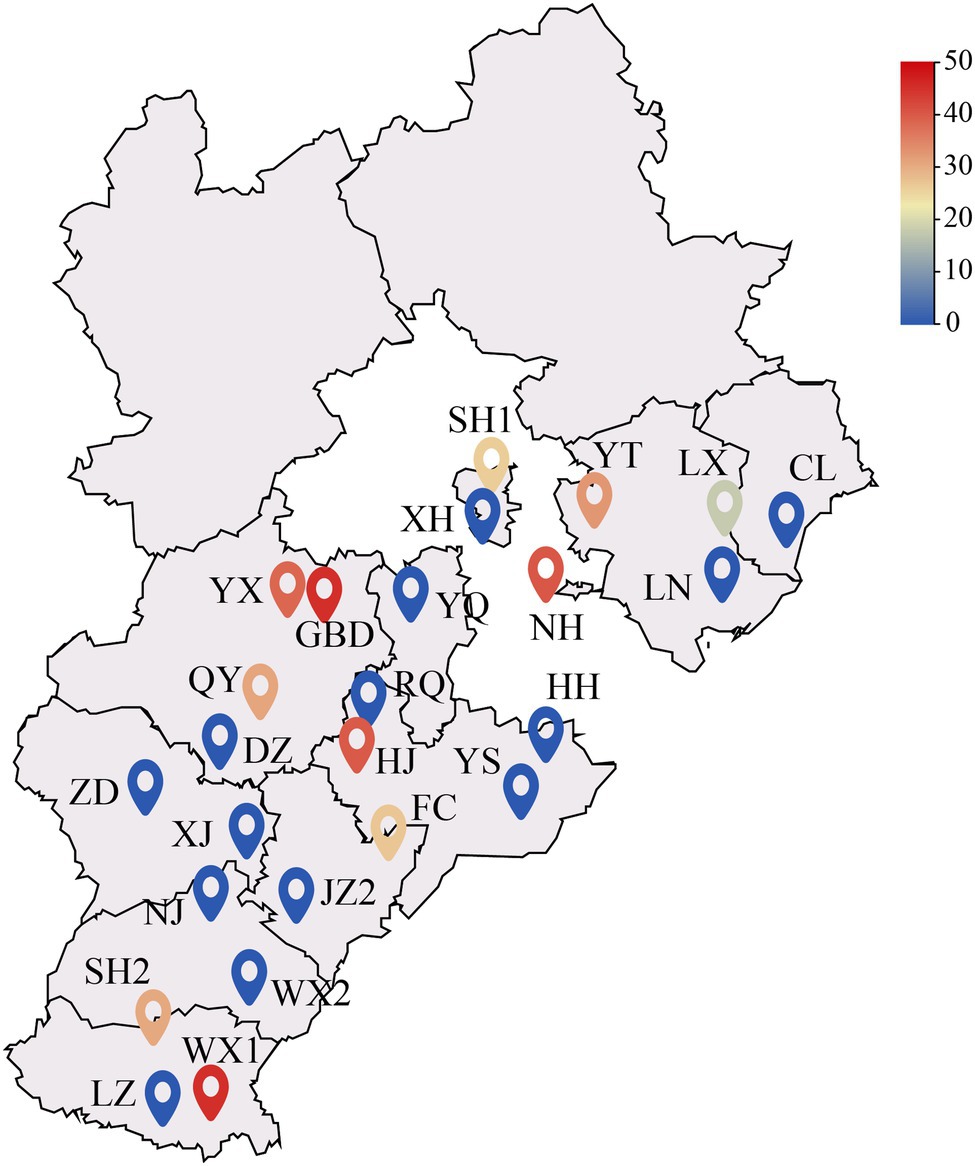
Figure 1. Distribution of the sampling sites. The color of the sampling point icon indicates the size of the wet heartwood rate at that site. The smaller the wet heartwood percentage, the bluer the color of the icon, the higher the wet heartwood percentage, and the redder the color of the icon.

Figure 2. The disease condition of wet heartwood disease in different degrees. (A) Healthy heartwood; (B) Wet heartwood with mild symptoms; and (C) Wet heartwood with more severe symptoms.
Determining the Color Difference and Wet Heartwood Rate of the Wood
The surface of the wood disk was scraped after natural air drying. According to the standard colorimetric theory of the International Commission on Illumination (CIE) Lab (Li et al., 2018), the chroma values L*, a*, and b* of the heartwood were measured with an automatic colorimeter, and each disk was measured three times.
A ruler was used to measure the diameter of the disk (with the bark removed) and the diameter of the wet heartwood part of the disk. The diameter of the wet heartwood of the healthy trees was recorded as 0 mm. Each disk was measured twice, and the average value was taken to obtain the total radius R1 and the wet heartwood radius R2 of the disk. The wet heartwood rate was calculated according to the following formula:
Determining the Chemical, Physical, and Mechanical Properties of the Wood
A disk was cut at breast height and the heartwood was separated from the sapwood. The test material was air-dried, cut into small slices, crushed with a pulveriser, and passed through a 60-mesh sieve prior to analyses. The contents of cellulose, hemicellulose, and lignin were measured in the heartwood and sapwood using the method described by Yin et al. (2017). The contents of cold/hot water and 1% NaOH extractive were measured following the guidelines GB/T 2677.4-1993 [Standardization Administration of China (SAC), 1993a] and GB/T 2677.5-1993 [Standardization Administration of China (SAC), 1993b], respectively. Each sample was measured three times, and the average value was taken.
The wood density and shrinkage rate of the heartwood and sapwood were measured in accordance with GB/T 1933-2009 [Standardization Administration of China (SAC), 2009a] and GB/T 1932-2009 [Standardization Administration of China (SAC), 2009b], respectively. Bending strength, the bending elastic modulus, and compressive strength parallel to the grain of the heartwood and sapwood were measured in accordance with GB/T 1936-1-2009 [Standardization Administration of China (SAC), 2009c], GB/T 1936-2-2009 [Standardization Administration of China (SAC), 2009d], and GB/T 1935-2009 [Standardization Administration of China (SAC), 2009e], respectively.
DNA Extraction and High-Throughput Sequencing
DNA was extracted from the heartwood (H), sapwood (S), root (R), and rhizosphere soil (SP) samples using a modified CTAB method (Porebski et al., 1997). The 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNG GGTATCTAAT-3′) primers were used for PCR amplification of 16S rRNA. After successful amplification, the PCR products were purified by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions and quantified using ABI StepOnePlus Real-Time PCR System (Life Technologies, Foster City, United States). Purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 250 bp) on an Illumina MiSeq platform according to the standard protocols. The Illumina data generated in this study were deposited in the NCBI Sequence Read Archive and are available under the project number PRJNA832960.
Bioinformatics and Statistical Analysis
After the raw reads were obtained by sequencing, the low-quality reads were filtered using FASTP software, and the paired-end reads were spliced into raw tags using FLASH (v1.2.11) software. The raw tags were filtered using Qiime (v1.9.1) to obtain clean tags, and chimeras were removed based on the UCHIME algorithm in USEARCH software. The Uparse algorithm in Usearch software was used to cluster the effective tag sequences, and cluster them as OTUs with a similarity of 97% by default. Alpha-diversity indices were calculated using an iterative subsampling procedure based on the samples with the lowest reads counts (23,000 sequences) as implemented in QIIME in order to account for differences in read counts across samples. The differences in alpha-diversity indices between healthy trees and wetwood trees were analyzed using the Wilcoxon rank-sum test after checking normal distributions of data using the Shapiro–Wilk test and analyzing homoscedasticity of variances of data using the Bartlett’s test in the Vegan package of R language. The differences in bacterial relative abundance between healthy trees and wetwood trees were analyzed using the Wilcoxon rank-sum test in the Vegan package of R language. Non-metric multidimensional scaling (NMDS) analysis was performed using the Bray–Curtis distance. SPSS (v26.0) software was used to calculate Spearman correlation coefficient between microorganisms and the wood chroma values, wet heartwood rate, chemical properties, and physical and mechanical properties of the wood. The variables with a significant correlation were selected and the Cytoscape software was used to draw correlation network diagrams. A db-RDA analysis were executed in R project Vegan package (version 2.5.3) to clarify the influence of endophytic bacteria of heartwood and sapwood on wood variables of heartwood and sapwood. Parametric assumptions test and independent samples t-test were performed with SPSS (v26.0) software to analyze the differences in the chemical properties and physical and mechanical properties of the wood between the groups. Cluster analysis of the chroma value and wet heartwood rate of wood was performed using Past (v4.0) software.
Results and Analysis
Chroma Value and Wet Heartwood Rate of the Wood
The heartwood chroma values L*, a*, and b* of 25 samples were measured using an automatic colorimeter. The L* value (70.14) of the wetwood trees (WW) was lower than that of healthy trees (NW; 71.29), indicating that the heartwood of WW was less bright and darker in color. The a* value (7.16) and b* value (14.79) of the WW were greater than those of NW (6.91 and 14.64, respectively), indicating that the color of the heartwood of WW was reddish-brown (Supplementary Figure S1). The wet heartwood rate of the WW ranged from 17.84 to 45.34%, and it was significantly larger than NW (p < 0.05; Table 1). Cluster analysis of 25 samples based on the chroma value and the wet heartwood rate showed that the WW samples were clustered into one category except for FC, and the NW samples were clustered into another category (Supplementary Figure S2). Therefore, our subsequent analyses excluded the FC sample.
Chemical, Physical, and Mechanical Properties of the Wood
The hemicellulose content of wet heartwood was significantly (p < 0.05) higher than healthy heartwood (Figure 3A) and the bending elastic modulus of the heartwood was significantly (p < 0.05) lower than normal heartwood (Figure 3D). However, the other indicators of heartwood chemical composition (Figure 3A), heartwood dry shrinkage rate (Figure 3B), heartwood density (Figure 3C), and heartwood bending elastic modulus (Figure 3D) were not significantly different between healthy and diseased trees.

Figure 3. Chemical properties and physical and mechanical indicators of the heartwood. (A) Chemical composition of the heartwood; (B) Dry shrinkage rate of the heartwood; (C) Density of the heartwood; and (D) Mechanical indicators of the heartwood. The letter represents a significant difference at the p < 0.05 level. WW and NW represent diseased and healthy trees, respectively.
The compressive strength parallel to the grain of the sapwood in diseased trees was significantly (p < 0.05) lower than in healthy trees (Figure 4D). However, the other indicators of sapwood chemical composition (Figure 4A), sapwood dry shrinkage rate (Figure 4B), sapwood density (Figure 4C), and sapwood bending elastic modulus (Figure 4D) were not significantly different between healthy and diseased trees.
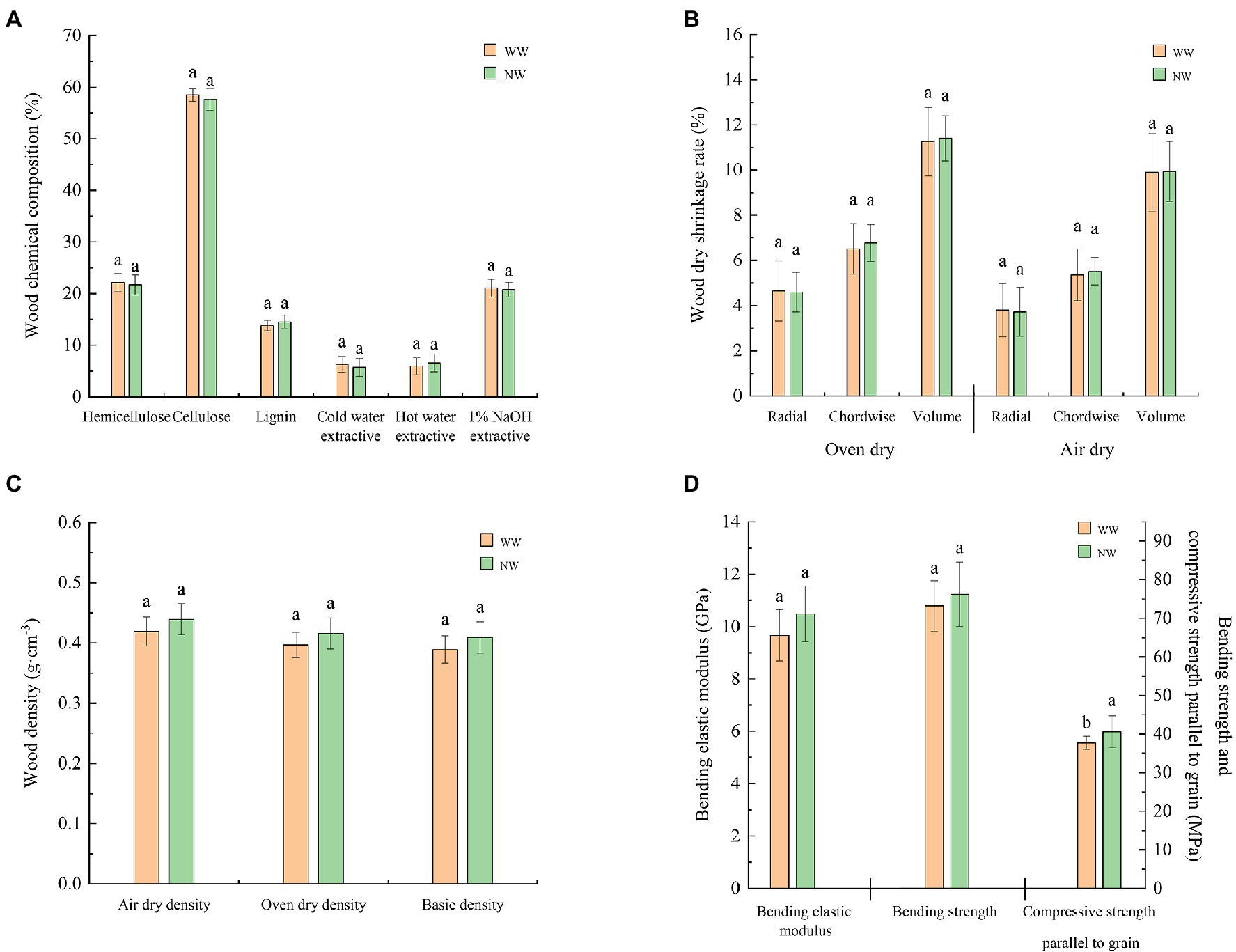
Figure 4. Chemical properties and physical and mechanical indicators of the sapwood. (A) Chemical composition of the sapwood; (B) Dry shrinkage rate of the sapwood; (C) Density of the sapwood; and (D) Mechanical indicators of the sapwood. The letter represents a significant difference at the p < 0.05 level. WW and NW represent diseased and healthy trees, respectively.
Microbial Community Diversity
The number of effective tags ranged from 103,280 to 191,493 after strictly filtering the original data. The number of OTUs ranged from 1,034 to 4,987, and the good coverage rate ranged from 0.985 to 0.997. The number of WW OTUs at the different sampling locations was less than those of NW. Regardless of WW or NW, the number of OTUs in the rhizosphere soil was the highest, followed by the heartwood. The number of OTUs in the sapwood was the smallest (Supplementary Table S1). The Wilcoxon rank-sum test was used to analyze the differences between the alpha-diversity indices of WW and NW, which showed that the Pielou’s evenness index, Shannon index, and Simpson index of WW-H were significantly lower than those of NW-H (Figures 5B–D). No significant differences in the richness, evenness, or diversity indices were detected in sapwood, root, and rhizosphere samples between WW and NW (Figure 5).
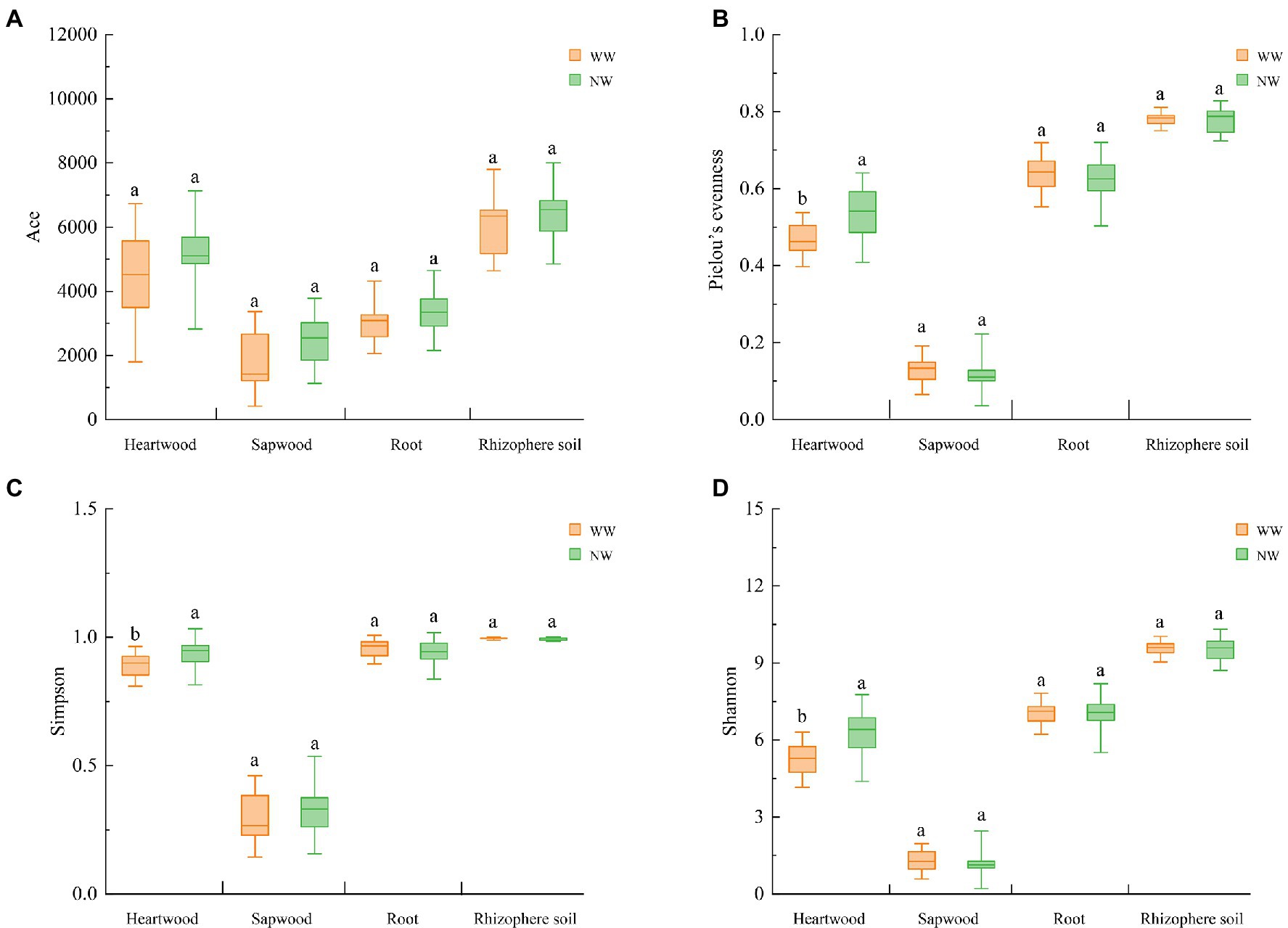
Figure 5. Alpha indices of the different samples. (A) Ace index; (B) Pielou’s evenness index; (C) Simpson index; and (D) Shannon index. The letter represents a significant difference at the p < 0.05 level. WW and NW represent diseased and healthy trees, respectively.
To further compare the bacterial community structure between the WW and NW samples, a NMDS analysis combined with ANOSIM was performed (Figure 6). The results showed that bacterial community structure differed between healthy and diseased trees in the heartwood only, while sapwood, root, and rhizosphere soil bacterial communities were similar.
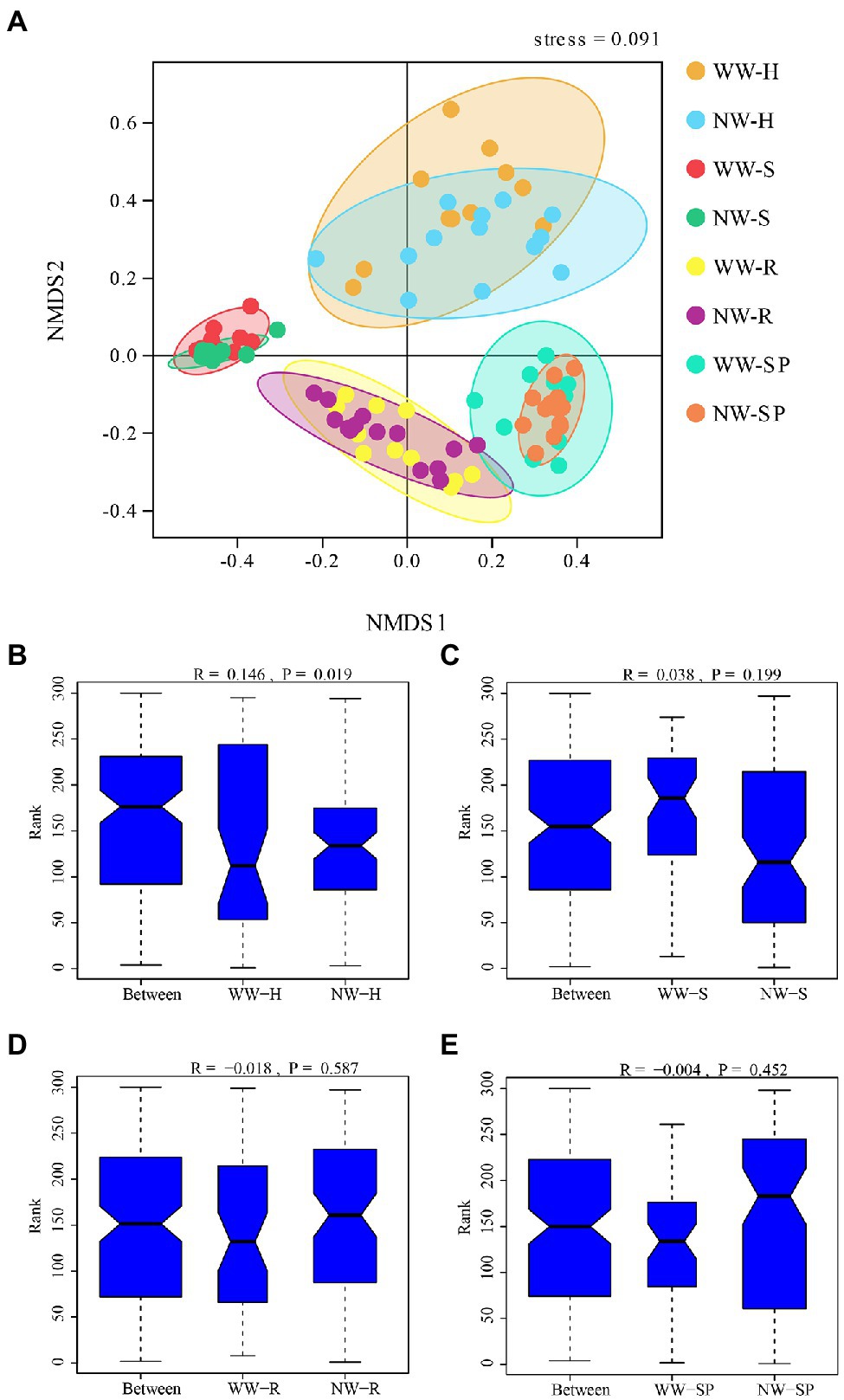
Figure 6. Beta diversity of the bacterial communities in each sample. (A) Non-Metric Multidimensional Scaling (NMDS) analysis of the bacterial community structure based on Bray–Curtis dissimilarity at the OTU level; (B) ANOSIM analysis of WW-H and NW-H; (C) ANOSIM analysis of the sapwood from wetwood trees (WW-S) and the sapwood from healthy trees (NW-S); (D) ANOSIM analysis of root tissue from wetwood trees (WW-R) and root tissue from healthy trees (NW-R); and (E) ANOSIM analysis of rhizosphere soil from wetwood trees (WW-SP) and rhizosphere soil from healthy trees (NW-SP).
Taxonomic Composition of the Bacterial Communities
A total of 41 phyla were identified from all samples. Bacteroidetes, Firmicutes, Euryarchaeota, Proteobacteria, and Cyanobacteria were the dominant bacterial phyla in the heartwood samples. The same was true for sapwood (excluding Euryarchaeota) and root tissue (excluding Euryarchaeota but including Actinobacteria). The main phyla were Proteobacteria, Actinobacteria, Acidobacteria, and Bacteroidetes in rhizosphere soil samples (Figure 7A).
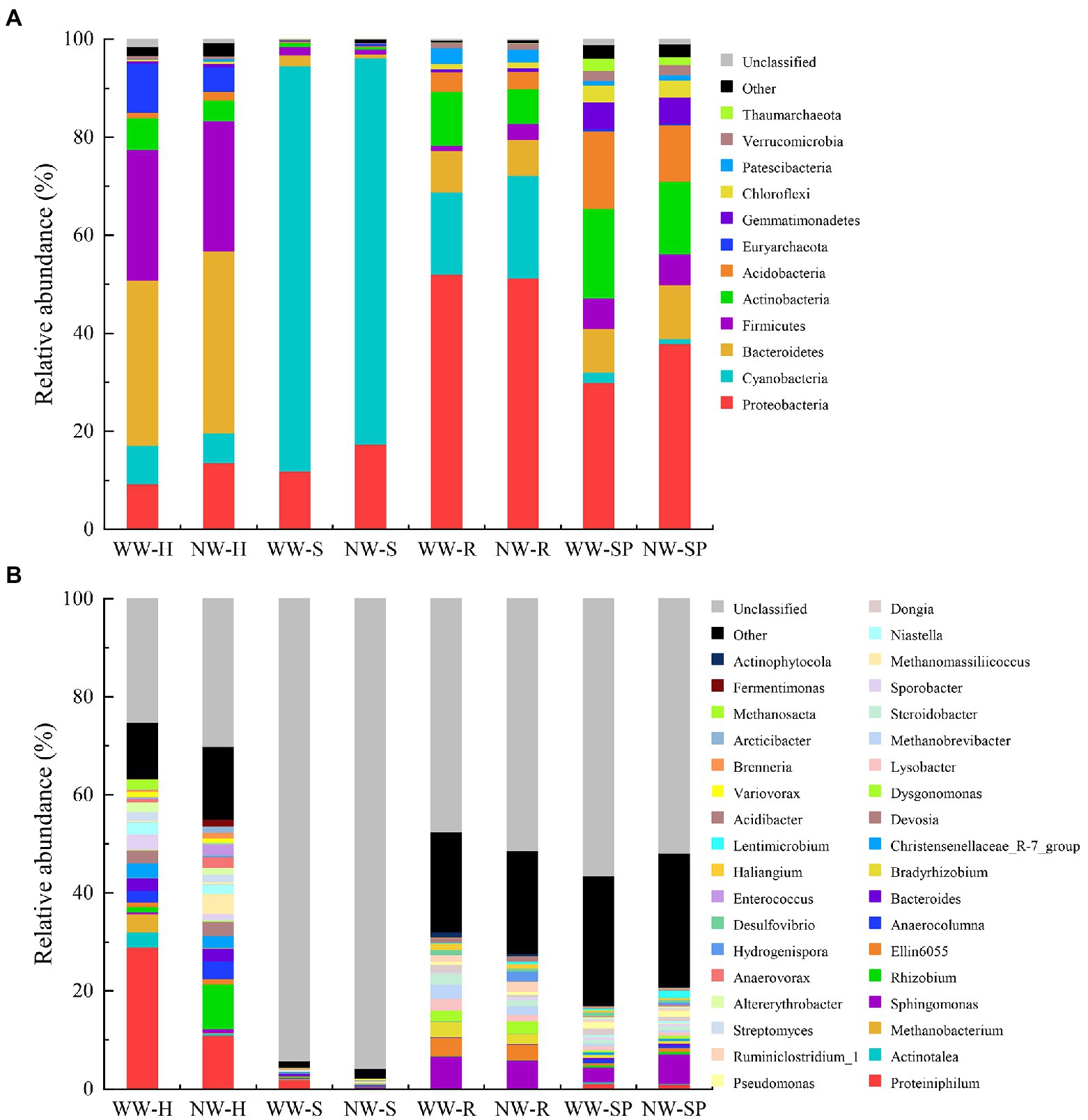
Figure 7. Relative abundance of species in the samples. (A) Relative abundance of bacterial taxa at the phylum level; (B) Relative abundance of bacterial taxa at the genus level. The figure shows species with relative abundance >1% of each group at each taxonomic level. WW-H, NW-H, WW-S, NW-S, WW-R, NW-R, WW-SP, and NW-SP represent heartwood of diseased trees, heartwood of healthy trees, sapwood of diseased trees, sapwood of healthy trees, root of diseased trees, root of healthy trees, rhizosphere soil of diseased trees, and rhizosphere soil of healthy trees, respectively.
Bacterial abundance at the genus level for all samples was shown in Figure 7B for breakdown. Proteiniphilum (29.01%), Actinotalea (3.70%), and Methanobacterium (3.08%) were the dominant genus in WW-H. Proteiniphilum (10.91%), Dysgonomonas (9.10%), and Bacteroides (4.05%) were the dominant genus in NW-H. Proteiniphilum (1.97%), Anaerocolumna (0.47%), and Sphingomonas (0.21%) were the dominant genus in WW-S. Methanobrevibacter (0.47%), Christensenellaceae_R-7_group (0.47%), and Sphingomonas (0.47%) were the dominant genus in NW-S. Sphingomonas was the dominant genre in root and rhizosphere soil of diseased and healthy trees.
Significant differences in the relative abundance of bacterial phyla and genera between the samples were revealed according to a Wilcoxon rank-sum test. There were significant differences in the abundances of a few number of phyla between diseased and healthy trees (p < 0.05, Supplementary Table S2). There were 73 genus had significant differences between WW-H and NW-H (p < 0.05, Supplementary Table S3). For example, the relative abundances of Proteiniphilum, Actinotalea, and Methanobacterium were significantly higher in WW-H than NW-H. For samples from sapwood, root and rhizosphere soil, we observed significant differences of 41, 25, and 29 genus between diseased and healthy trees, respectively.
Correlation Between Endophytic Bacteria and the Chroma Value, Wet Heartwood Rate, and the Wood Properties
We performed Spearman correlation analysis to study the relationship between endophytic bacteria and the chroma value, wet heartwood rate, and wood properties. The results showed that most of the heartwood endophytic bacteria were negatively correlated with the chroma value, wet heartwood rate, and heartwood wood properties. Proteiniphilum was highly significantly positively correlated with wet heartwood rate, significantly negatively correlated with compressive strength parallel to the grain of the heartwood, and highly significantly negatively correlated with the bending elastic modulus of the heartwood. Methanobacterium was significantly positively correlated with the wet heartwood rate, significantly negatively correlated with a* and oven-dried radial shrinkage of the heartwood, and highly significantly negatively correlated with oven-dried volume shrinkage and air-dried radial shrinkage of the heartwood (Figures 8A–C; Supplementary Table S4).
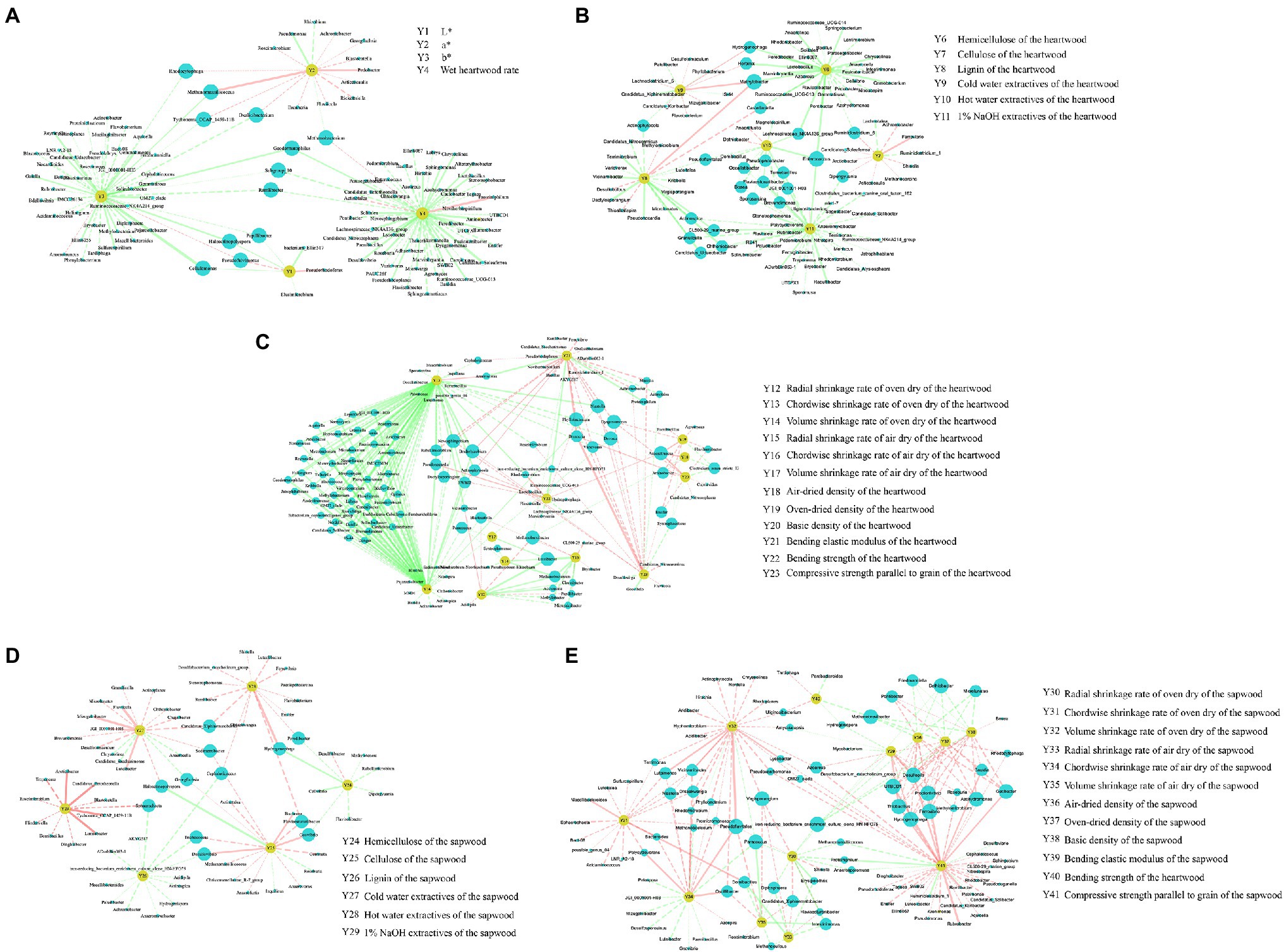
Figure 8. Spearman correlation network diagram of the endophytic bacteria and the chroma value, wet heartwood rate, and the wood properties. (A) Spearman correlation between the heartwood endophytic bacteria and the chroma value and wet heartwood rate; (B) Spearman correlation between heartwood endophytic bacteria and the chemical properties of the heartwood; (C) Spearman correlation between the heartwood endophytic bacteria and the physical and mechanical properties of the heartwood; (D) Spearman correlation between the sapwood endophytic bacteria and the chemical properties of sapwood; (E) Spearman correlation between sapwood endophytic bacteria and the physical and mechanical properties of sapwood. The blue nodes represent the bacterial genus, and the yellow nodes represent the wood property indicators. The size of the blue node represents the number of connections to that node. The solid line represents highly significant, and the dashed line represents significant. The red line represents a positive correlation, and the green line represents a negative correlation. The thickness of the connecting line represents the strength of the correlation.
Propionivibrio, Candidatus_Xiphinematobacter, Desulfopila, and Thiobacillus. Proteiniphilum was significantly negatively correlated with oven-dried radial shrinkage of the sapwood, and highly significantly negatively correlated with compressive strength parallel to the grain of the sapwood. Methanobacterium was significantly negatively correlated with oven-dried volume shrinkage and air-dried chordwise shrinkage of the sapwood. Methanoculleus was significantly negatively correlated with air-dried radial shrinkage and air-dried volume shrinkage of the sapwood (Figures 8D,E; Supplementary Table S5).
The db-RDA analysis was performed to identify the most important taxa liked to the wood variables (Figure 9). The main endophytic bacteria affecting the wood variables of the heartwood were Desulfovibrio, Actinotalea, Proteiniphilum and Methanomassiliicoccus. Additionally, Proteiniphilum, Actinotalea, Methanosaeta, and Methanobacterium were positively correlated with wet heartwood rate, but they were negatively correlated with cellulose of the heartwood (Figure 9A). In comparison, endophytic bacteria of the sapwood had little effect on wood variables of sapwood (Figure 9B).
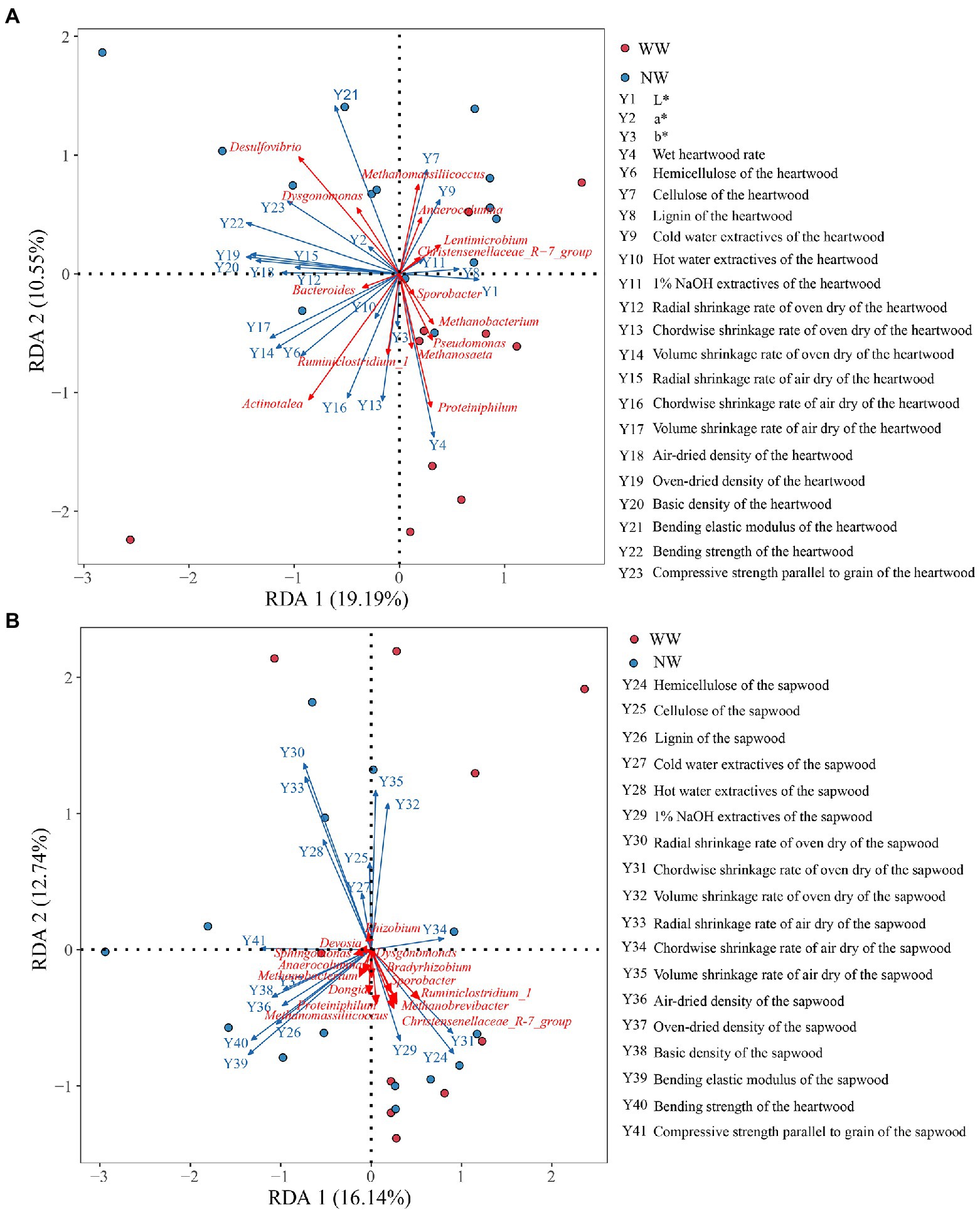
Figure 9. db-RDA analysis of the endophytic bacteria and the chroma value, wet heartwood rate, and the wood properties. (A) The relationship between endophytic bacteria of the heartwood and wood variables of the heartwood; (B) The relationship between endophytic bacteria of the sapwood and wood variables of the sapwood. WW and NW represent diseased and healthy trees, respectively.
Discussion
In this study, we showed that Populus × euramericana cv. “74/76” presents typical characteristics of the wet heartwood disease, such as dark heartwood color and a foul odor of fatty acids. The chromophore and auxochrome in the wood structure are connected with an aromatic ring structure to form a conjugated system, which absorbs and reflects visible light to make the wood reflect different colors. These groups are mainly present in lignin and the extractives (Yao et al., 2012). Most heartwood extractives include compounds, such as polyphenols, resins, organic acid salts, and fats, which also cause a darkening of the wet heartwood (Zhu Ge et al., 1997; Burtin et al., 2000).
Most studies have used traditional culture methods to isolate and purify the dominant bacterial strains from the heartwood tissue of wetwood (Alizadeh et al., 2017a; Song et al., 2018; Basavand et al., 2021), or to study the microorganisms by comparing the microbial communities of the wetwood and normal sapwood tissues of diseased trees (Streichan, 1986; Kovaleva et al., 2015). In this study, the high-throughput sequencing method was used to identify and compare the bacterial microbial diversity of heartwood, sapwood, root tissue, and rhizosphere soil of diseased and healthy trees. The bacterial evenness and diversity indices in the heartwood of diseased trees were significantly lower than that of healthy trees. However, the alpha-diversity indices of sapwood, root, and rhizosphere soil between diseased and healthy trees were no significant differences. These results suggest that wetwood affected the diversity and evenness of the endophytic bacterial community in the heartwood, but had no effect on the richness, evenness, and diversity of endophytic bacteria in the sapwood, endophytic bacteria of the root tissue, or rhizosphere soil bacteria. Endophytic bacteria, make-up part of plants microbiome, these microorganisms are associated to every single plant organ, where they are protected from environmental changes and therefore mostly might be affected by changes occurring within plant tissues (Ryan et al., 2007). The higher diversity of endophytic bacteria in heartwood of healthy trees could be an indication of pathogen suppression which delayed or prevented the diseased expression. Suhaimi et al. (2017) have also reported that the higher diversity of endophytic bacteria in the non-symptomatic bacterial wilt-diseased banana plants compared to the symptomatic plant.
A NMDS analysis revealed a significant difference in the bacterial community structure of heartwood between the diseased and healthy trees, indicating that the functions of the two microenvironments differ. And the bacterial community structure of sapwood, root tissue, and rhizosphere soil samples were not significantly different between diseased and healthy trees, indicating that wet heartwood disease could only cause changes in the endophytic bacterial community in the heartwood, which may be one of the characteristics of trees suffering from wet heartwood disease.
A db-RDA found that the wet heartwood rate was positively correlated with the relative abundance of Proteiniphilum, Actinotalea, and Methanobacterium. The cellulose content of diseased trees’ heartwood was lower than that of healthy trees’ heartwood, and cellulose content was negatively correlated with the relative abundances of Proteiniphilum, Actinotalea, and Methanobacterium. This may be due to the small sample size or the uneven distribution of these three bacteria in samples. The relative abundances of Proteiniphilum, Actinotalea, and Methanobacterium in diseased trees’ heartwood were significantly higher than in healthy trees’ heartwood, particularly Proteiniphilum, which was significantly enriched in diseased trees’ heartwood. According to reports, the main gases in wet heartwood are CH4, N2, and CO2, with a small amount of H2 but no O2 (Schink et al., 1981a; Schink and Ward, 1984). Proteiniphilum is a facultative anaerobe and potent cellulolytic bacterium that directly degrades lignocellulose to CO2, formate, and acetate via aerobic respiration and anaerobic fermentation (Hahnke et al., 2016; Wu et al., 2020). Actinotalea is an anaerobic bacterium from Cellulomonadaceae family. Cellulomonadaceae family was known as the functional bacteria to degrade cellulose, and the intermediate metabolites could be used as carbon source for denitrification (Rusznyak et al., 2011; Feng et al., 2017). Wetwood is an environment in which anaerobic cellulolytic bacteria might be expected to thrive. And Warshaw et al. (1985) isolated obligately anaerobic, mesophilic, cellulolytic bacteria from the wetwood of elm and maple trees. Methanobacterium is a typical hydrogen-oxygen methanogen that produces CH4 from formate and H2/CO2 and is often isolated from wetwood (Zeikus and Ward, 1974; Yu et al., 2019; Zhang et al., 2019). In addition to Methanobacterium, the relative abundances of Methanobrevibacter, Methanosaeta, and Methanoculleus of diseased trees’ heartwood were also greater than those of healthy trees’ heartwood. These bacteria belong to Euryarchaeota, which converts acetate, H2 and CO2 to CH4 by acetic acid and hydrogenotrophic methanogenesis, respectively (Wagner et al., 2013; Oszust et al., 2018; Lee and Hwang, 2019). Based on the function of these bacteria, this study speculated that anaerobic bacteria with specific functions such as Proteiniphilum and Actinotalea hydrolyzed cellulose and easily degradable carbohydrates in wood into sugars, amino acids, and long-chain fatty acids. These products provided methanogens with the substrates they need to grow and convert methane. However, these speculations need further verification.
In this study, wetwood disease caused a shift in the heartwood bacterial community of Populus × euramericana cv. “74/76,” but the disease had little effect on the bacterial communities of sapwood, root tissue, or rhizosphere soil. Song et al. studied the infection route of the pathogenic bacteria causing poplar wetwood disease and found that the stem wound infection was the main pathway for pathogenic bacteria to infect poplars, and neither leaf cuttings nor root irrigation infected the poplars (Song et al., 2017). Therefore, carrying out poplar infection test can further explore the pathogenic mechanisms of poplar wetwood disease. In addition, the use of the antagonism between microorganisms to screen the bacteria antagonistic to the pathogenic bacteria of wet heartwood would be of great significance for the biological control of poplar wetwood disease.
Data Availability Statement
The datasets presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, accession number PRJNA832960.
Author Contributions
JF conceived the study, analyzed the data, and edited the manuscript. SW and CD collected and analyzed the data. CM and XC analyzed the data. JW, MY, and XS designed the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Key Research and Development Program of China (grant number 2021YFD2201200) and Province Key Research and Development Program of HeBei (grant number 21326301D).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb. 2022.868078/full#supplementary-material
References
Alizadeh, M., Khakvar, R., and Ahari, A. B. (2017a). Isolation and characterization of bacterial agents associated of wetwood disease on elm trees in Iran. Acta Phytopathol. Entomol. Hung. 52, 1–11. doi: 10.1556/038.52.2017.028
Alizadeh, M., Moharrami, M., and Rasouli, A. A. (2017b). Geographic information system (Gis) as a tool in the epidemiological assessment of wetwood disease on elm trees in Tabriz City, Iran. Agron. Res. Mold. 50, 91–100. doi: 10.1515/cerce-2017-0018
Ayin, C. M., Alvarez, A. M., Awana, C., Schleinzer, F. M., Marx, B. D., and Schlub, R. L. (2019). Ralstonia solanacearum, Ganoderma australe, and bacterial wetwood as predictors of ironwood tree (Casuarina equisetifolia) decline in Guam. Australas. Plant Pathol. 48, 625–636. doi: 10.1007/s13313-019-00666-8
Basavand, E., Khodaygan, P., Rahimian, H., Ganjeh, A., Molaei, S., and Firouzianbandpey, S. (2021). Identification and characterization of Klebsiella oxytoca strains associated with wetwood disease of Morus trees. Indian Phytopathol. 74, 1123–1127. doi: 10.1007/s42360-021-00360-0
Burtin, P., Jay-Allemand, C., Charpentier, J. P., and Janin, G. (2000). Modification of hybrid walnut (Juglans nigra 23 × Juglans regia) wood colour and phenolic composition under various steaming condition. Holzforschung 54, 33–38. doi: 10.1515/HF.2000.006
Diamandis, S., and Koukos, P. (1992). Effect of bacteria on the mechanical and chemical properties of wood in poplars damaged by frost cracks. Eur. J. Plant Pathol. 22, 362–370. doi: 10.1111/j.1439-0329.1992.tb00308.x
Feng, L. J., Chen, K., Han, D. D., Zhao, J., Lu, Y., Yang, G. F., et al. (2017). Comparison of nitrogen removal and microbial properties in solid-phase denitrification systems for water purification with various pretreated lignocellulosic carriers. Bioresour. Technol. 224, 236–245. doi: 10.1016/j.biortech.2016.11.002
Goychuk, A. F., Drozda, V. F., Shvets, M. V., and Kulbanska, I. (2020). Bacterial wetwood of silver birch (Betula pendula Roth): symptomology, etiology and pathogenesis. Folia For. Pol. A 62, 145–159. doi: 10.2478/ffp-2020-0015
Hahnke, S., Langer, T., Koeck, D. E., and Klocke, M. (2016). Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. Nov., sp. nov. isolated from mesophilic laboratory-scale biogas reactors and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 66, 1466–1475. doi: 10.1099/ijsem.0.000902
Hartley, C., Davidson, R., and Crandall, B. S. (1961). Wet wood, bacteria, and increased pH in trees. Usda for serv for product lab rep.
Hua, J. L., Liu, G. R., and Huang, J. S. (2012). Effect of continuous cropping of sesame on rhizospheric microbial communities. Acta Ecol. Sin. 32, 2936–2942. doi: 10.5846/stxb201104010422
Jeremic, D., Cooper, P., and Srinivasan, U. (2004). Comparative analysis of balsam fir wetwood, heartwood, and sapwood properties. Can. J. For. Res. 34, 1241–1250. doi: 10.1139/x03-287
Knutson, D. M. (1973). The bacteria in sapwood, wetwood, and heartwood of trembling aspen (Populus tremuloides). Can. J. Bot. 51, 498–500. doi: 10.1139/b73-060
Kovaleva, V., Shalovylo, Y., Gorovik, Y. N., Lagonenko, A., Evtushenkov, A., and Gout, R. T. (2015). Bacillus pumilus—a new phytopathogen of scots pine—short communication. J. For. Sci. 61, 131–137. doi: 10.17221/16/2014-JFS
Lee, J., and Hwang, S. (2019). Single and combined inhibition of Methanosaeta concilii by ammonia, sodium ion and hydrogen sulfide. Bioresour. Technol. 281, 401–411. doi: 10.1016/j.biortech.2019.02.106
Li, R. R., Xu, W., Wang, X. D., and Wang, C. G. (2018). Modeling and predicting of the color changes of wood surface during CO2 laser modification. J. Clean. Prod. 183, 818–823. doi: 10.1016/j.jclepro.2018.02.194
Martin, L., Cochard, H., Mayr, S., and Badel, E. (2021). Using electrical resistivity tomography to detect wetwood and estimate moisture content in silver fir (Abies alba mill.). Ann. For. Sci. 78:65. doi: 10.1007/s13595-021-01078-9
Moya, R., Muñoz, F., Jeremic, D., and Berrocal, A. (2009). Visual identification, physical properties, ash composition, and water diffusion of wetwood in Gmelina arborea. Can. J. For. Res. 39, 537–545. doi: 10.1139/X08-193
Murdoch, C. W. (1983). Bacterial species associated with wetwood of elm. Phytopathology 73:1270. doi: 10.1094/Phyto-73-1270
Oszust, K., Gryta, A., Ziemiński, K., Bilińska-Wielgus, N., Gałązka, R., and Frac, M. (2018). Characterization of microbial functional and genetic diversity as a novel strategy of biowaste ecotoxicological evaluation. Int. J. Environ. Sci. Technol. 16, 4261–4274. doi: 10.1007/s13762-018-2066-3
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 15, 8–15. doi: 10.1007/BF02772108
Roque, R. M., Muńoz-Acosta, F., Jeremic, D., and Berrocal, A. (2009). Visual identification, physical properties, ash composition, and water diffusion of wetwood in Gmelina arborea. Can. J. For. Res. 39, 537–545. doi: 10.1139/X08-193
Rusznyak, A., Toth, E. M., Schumann, P., Spröer, C., Makk, J., Szabo, G., et al. (2011). Cellulomonas phragmiteti sp. nov., a cellulolytic bacterium isolated from reed (Phragmites australis) periphyton in a shallow soda pond. Int. J. Syst. Evol. Microbiol. 61, 1662–1666. doi: 10.1099/ijs.0.022608-0
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2007). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Schink, B., and Ward, J. C. (1984). Microaerobic and anaerobic bacterial activities involved in formation of wetwood and discoloured wood. IAWA J. 5, 105–109. doi: 10.1163/22941932-90000872
Schink, B., Ward, J. C., and Zeikus, J. G. (1981a). Microbiology of wetwood: importance of pectin degradation and clostridium species in living trees. Appl. Environ. Microbiol. 42, 526–532. doi: 10.1007/BF00443249
Schink, B., Ward, J. C., and Zeikus, J. G. (1981b). Microbiology of wetwood: role of anaerobic bacterial populations in living trees. J. Gen. Microbiol. 123, 313–322. doi: 10.1099/00221287-123-2-313
Schroeder, H. A., and Kozlik, C. J. (1972). The characterization of wetwood in western hemlock. Wood Sci. Technol. 6, 85–94. doi: 10.1007/BF00350823
Song, C. C., Mei, L., Cai, L., Zhang, S., and Fan, X. P. (2018). Identification of pathogen bacteria and antagonistic bacteria of wet-heart wood disease in Populus. J Southwest For. Univ. 38, 134–144.
Song, C. C., Wang, K., Mei, L., Fan, X. P., Zhang, S., and Zhu, Y. J. (2017). Isolation, identification and infection route of pathogenic bacteria of poplar wet heartwood disease. J. Huazhong Agric. Univ. 36, 15–21. doi: 10.13300/j.cnki.hnlkxb.20170213.018
Standardization Administration of China (SAC) (1993a). Fibrous raw material-Determination of water solubility. GB/T 2677.4–1993. Beijing: Standards Press of China. p207-209.
Standardization Administration of China (SAC) (1993b). Fibrous raw material-Determination of one percent sodium hydroxide solubility. GB/T 2677.5–1993. Beijing: Standards Press of China. p204-205.
Standardization Administration of China (SAC) (2009a). Method for determination of the density of wood. GB/T 1933–2009. Beijing: Standards Press of China. p1-5.
Standardization Administration of China (SAC) (2009b). Method for determination of the shrinkage of wood. GB/T 1932–2009. Beijing: Standards Press of China. p1-3.
Standardization Administration of China (SAC) (2009c). Method of testing in bending strength of wood. GB/T 1936–1-2009. Beijing: Standards Press of China. p1-3.
Standardization Administration of China (SAC) (2009d). Method for determination of the modulus of elasticity in static bending of wood. GB/T 1936–2-2009. Beijing: Standards Press of China. p1-3.
Standardization Administration of China (SAC) (2009e). Method of testing in compressivs strength parallel to grain of wood. GB/T 1935–2009. Beijing: Standards Press of China. p1-2.
Streichan, M. (1986). Microbial populations in wetwood of european white fir (Abies alba mill.). FEMS Microbiol. Lett. 38, 141–150. doi: 10.1016/0378-1097(86)90045-5
Suhaimi, N. S. M., Goh, S. Y., Ajam, N., Othman, R. Y., Chan, K. G., and Thong, K. L. (2017). Diversity of microbiota associated with symptomatic and non-symptomatic bacterial wilt-diseased banana plants determined using 16S rRNA metagenome sequencing. World J. Microbiol. Biotechnol. 33:168. doi: 10.1007/s11274-017-2336-0
Wagner, A. O., Lins, P., Malin, C., Reitschuler, C., and Illmer, P. (2013). Impact of protein-, lipid- and cellulose-containing complex substrates on biogas production and microbial communities in batch experiments. Sci. Total Environ. 458-460, 256–266. doi: 10.1016/j.scitotenv.2013.04.034
Wang, X. Q., Jiang, Z. H., and Ren, H. Q. (2008). Distribution of wet heartwood in stems of Populus xiaohei from a spacing trial. Scand. J. For. Res. 23, 38–45. doi: 10.1080/02827580701763706
Warshaw, J. E., Leschine, S. B., and Canale-Parola, E. (1985). Anaerobic cellulolytic bacteria from wetwood of living trees. Appl. Environ. Microbiol. 50, 807–811. doi: 10.1128/aem.50.4.807-811.1985
Wilcox, W. W., and Oldham, N. D. (1972). Bacterium associated with wetwood in white fir. Phytopathology 62, 384–385. doi: 10.1094/Phyto-62-384
Wilcox, W. W., and Pong, W. Y. (1971). The effects of height, radial position, and wetwood on white fir wood properties. Wood Fiber Sci. 3, 47–55.
Wu, Z. Y., Nguyen, D., Lam, T. Y. C., Zhuang, H. C., Shrestha, S., Raskin, L., et al. (2020). Synergistic association between cytochrome bd-encoded Proteiniphilum and reactive oxygen species (Ros)-scavenging methanogens in microaerobic-anaerobic digestion of lignocellulosic biomass. Water Res. 190:116721. doi: 10.1016/j.watres.2020.116721
Xu, H. M., Hu, X. Y., Zheng, L. Y., and Fan, X. P. (2010). Reaearch progress of wetwood. Hubei For. Sci. Technol. 4, 39–42. doi: 10.3969/j.issn.1004-3020.2010.04.011
Xu, Z. C., Leininger, T. D., Lee, A. W. C., and Tainter, F. H. (2001). Chemical properties associated with bacterial wetwood in red oaks. Wood Fiber Sci. 33, 76–83.
Yao, C., Fan, Y., Gao, J., and Li, H. (2012). Coloring characteristics of in situ lignin during heat treatment. Wood Sci. Technol. 46, 33–40. doi: 10.1007/s00226-010-0388-5
Yin, S. P., Xiao, Z. H., Zhao, G. H., Zhao, X., Sun, X. Y., Zhang, Y., et al. (2017). Variation analyses of growth and wood properties of Larix olgensis clones in China. J. For. Res. 28, 687–697. doi: 10.1007/s11676-016-0359-2
Yu, X., Hu, X., Peng, Y., Wu, Z., Zhang, Q., Li, Z., et al. (2019). Amplicon sequencing reveals different microbial communities in living poplar wetwood and sapwood. Trees 33, 851–865. doi: 10.1007/s00468-019-01822-9
Zeikus, J. G., and Ward, J. C. (1974). Methane formation in living trees: a microbial origin. Science 184, 1181–1183. doi: 10.1126/science.184.4142.1181
Zhang, D. M., Bao, F. C., Zhang, Z. Y., and Huang, R. F. (2005). Genetic analysis of wetwood proportion on clone test stand of Populus tomentosa. Sci. Silvae Sin. 41, 140–144. doi: 10.11707/j.1001-7488.20050424
Zhang, Y., Cai, L., and Xu, Y. (2006). Aspirated pits in wetwood and micromorphology of microbial degradation in subalpine fir. Front. For. Chin. 4, 449–452. doi: 10.1007/s11461-006-0052-2
Zhang, W., Zhang, F., Li, Y. X., Jiang, Y., and Zeng, R. J. (2019). No difference in inhibition among free acids of acetate, propionate and butyrate on hydrogenotrophic methanogen of Methanobacterium formicicum. Bioresour. Technol. 294:122237. doi: 10.1016/j.biortech.2019.122237
Zheng, H. Y., Dai, Y. S., Xie, C. X., Guo, T. B., and Wang, Z. Y. (2006). Study on the cause of formation of poplar red cor wood. J. Nanjing For. Univ. 30, 82–84. doi: 10.3969/j.jssn.1000-2006.2006.02.020
Zhu Ge, Q., Huang, M. R., Pan, H. X., Li, H. G., and Feng, W. Z. (1997). Chemical properties and formation mechanism of poplar wet heartwood. Sci. Silvae Sin. 33, 259–266.
Keywords: Populus × euramericana cv. “74/76”, wetwood, heartwood, high-throughput sequencing, bacterial diversity, bacterial community structure
Citation: Fan J, Wang S, Ding C, Ma C, Chen X, Wang J, Yang M and Su X (2022) Correlation Analysis of the Bacterial Community and Wood Properties of Populus × euramericana cv. “74/76” Wet Heartwood. Front. Microbiol. 13:868078. doi: 10.3389/fmicb.2022.868078
Edited by:
Bernardo González, Adolfo Ibáñez University, ChileReviewed by:
Murali Gopal, Central Plantation Crops Research Institute (ICAR), IndiaKarelle Rheault, University of Copenhagen, Denmark
Copyright © 2022 Fan, Wang, Ding, Ma, Chen, Wang, Yang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinmao Wang, bHh3am1AaGViYXUuZWR1LmNu; Minsheng Yang, eWFuZ21zMTAwQDEyNi5jb20=; Xiaohua Su, c3V4aEBjYWYuYWMuY24=
†These authors have contributed equally to this work and share co-first authorship
 Jianmin Fan
Jianmin Fan Shijie Wang
Shijie Wang Changjun Ding
Changjun Ding Changming Ma
Changming Ma Xinghao Chen1,3
Xinghao Chen1,3 Jinmao Wang
Jinmao Wang Minsheng Yang
Minsheng Yang Xiaohua Su
Xiaohua Su