
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 25 April 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.867342
This article is part of the Research Topic Insights in Microbial Physiology and Metabolism: 2021 View all 13 articles
Methyl-coenzyme M reductase (MCR) is an archaeal enzyme that catalyzes the final step of methanogenesis and the first step in the anaerobic oxidation of methane, the energy metabolisms of methanogens and anaerobic methanotrophs (ANME), respectively. Variants of MCR, known as alkyl-coenzyme M reductases, are involved in the anaerobic oxidation of short-chain alkanes including ethane, propane, and butane as well as the catabolism of long-chain alkanes from oil reservoirs. MCR is a dimer of heterotrimers (encoded by mcrABG) and requires the nickel-containing tetrapyrrole prosthetic group known as coenzyme F430. MCR houses a series of unusual post-translational modifications within its active site whose identities vary depending on the organism and whose functions remain unclear. Methanogenic MCRs are encoded in a highly conserved mcrBDCGA gene cluster, which encodes two accessory proteins, McrD and McrC, that are believed to be involved in the assembly and activation of MCR, respectively. The requirement of a unique and complex coenzyme, various unusual post-translational modifications, and many remaining questions surrounding assembly and activation of MCR largely limit in vitro experiments to native enzymes with recombinant methods only recently appearing. Production of MCRs in a heterologous host is an important step toward developing optimized biocatalytic systems for methane production as well as for bioconversion of methane and other alkanes into value-added compounds. This review will first summarize MCR catalysis and structure, followed by a discussion of advances and challenges related to the production of diverse MCRs in a heterologous host.
Methyl-coenzyme M reductase (MCR) catalyzes the final methane-forming step of methanogenesis in methanogens, and the initial methane activation step in the anaerobic oxidation of methane (AOM) in anaerobic methanotrophic archaea (ANME). MCR is generally highly conserved in sequence and structure among methanogens and ANME, where it consists of three different subunits, α (McrA), β (McrB), and γ (McrG), arranged in a α2β2γ2 configuration harboring two active sites (Ermler et al., 1997; Shima et al., 2012; Wagner et al., 2017). Each active site contains F430, the nickel hydrocorphin prosthetic group (Figure 1). Methanogenic MCR has been extensively studied in the methane formation direction, where it catalyzes the conversion of methyl-coenzyme M (CH3-S-CoM) and coenzyme B (HS-CoB) to methane and a CoM-S-S-CoB heterodisulfide (Bobik et al., 1987; Ellermann et al., 1987, 1988; Figure 1). This reaction is proposed to occur in reverse in ANME that anaerobically oxidize methane to CO2 via reverse methanogenesis (Hallam et al., 2004; Shima and Thauer, 2005; Scheller et al., 2010; Timmers et al., 2017). In addition to methane formation and oxidation, variants of MCR are also involved in the anaerobic oxidation of short- and long-chain alkanes (Borrel et al., 2019; Wang et al., 2021; Zhou et al., 2022).
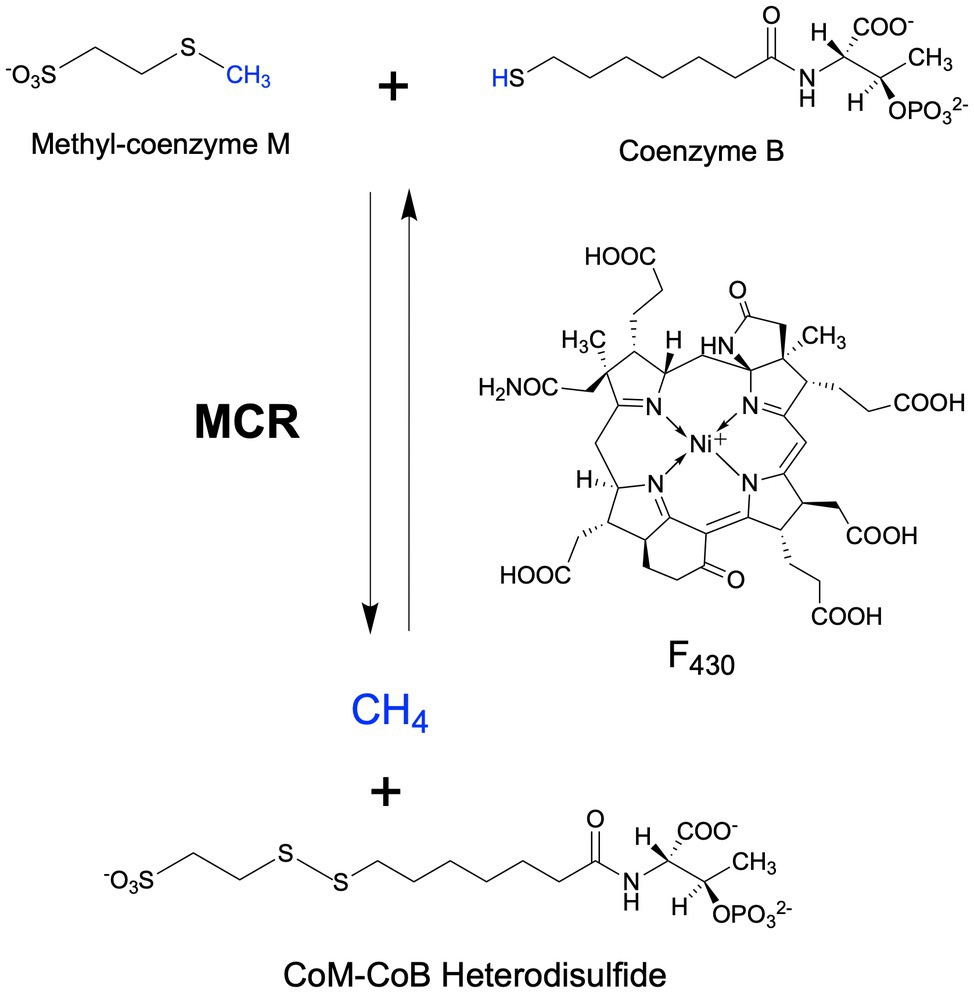
Figure 1. MCR-catalyzed reactions in the final step of methanogenesis in methanogens and the first step of the anaerobic oxidation of methane in anaerobic methanotrophs.
Given the remarkable chemistry catalyzed by MCR as well as its central importance in the global carbon cycle and potential for bioenergy applications, this enzyme has been of interest to enzymologists since its initial discovery in the 1970s (McBride and Wolfe, 1971; Gunsalus and Wolfe, 1976). Much of what is known about MCR catalysis comes from work by several groups on the natively purified MCR from the methanogen, Methanothermobacter marburgensis. This organism grows to high cell densities (3 g dry mass per L) with a doubling time of less than 2 h (Kaster et al., 2011) and, most importantly, effective procedures have been developed for the isolation of an active MCR from this organism.
A major challenge in the MCR field is the heterologous production of recombinant MCRs. This is due to many reasons including, but not limited to, the heterooligomeric structure of MCR that may require chaperones for proper assembly, the requirement of the unique and complex coenzyme F430, the presence of several unusual post-translational modifications that are organism-specific, and the lack of knowledge surrounding proteins required for activation and incorporation of F430. Although undoubtedly a difficult task, successful development of heterologous expression systems for MCRs would transform the field, allowing further investigation into the catalytic properties and mechanistic aspects of different MCRs, as well as facilitate the development of optimized biocatalytic systems for methane production or methane conversion applications.
In this review, we will provide an overview of the most relevant aspects of MCR structure and catalysis, and then will focus on considerations and perspectives related to the production of MCR in a heterologous host. For more detailed reviews on MCR biochemistry, the reader is referred to recent excellent reviews by Thauer (2019) and Ragsdale et al. (2017).
Methanogenic archaea (“methanogens”) are ancient and diverse microorganisms within the archaeal domain of life (Battistuzzi et al., 2004; Adam et al., 2017). They are found in a wide range of anaerobic environments including marine and freshwater habitats, anoxic soils, and as important components of animal microbiomes (Moissl-Eichinger et al., 2018; Lyu et al., 2018a; Borrel et al., 2020). As their sole source of energy, methanogens carry out a form of anaerobic respiration known as methanogenesis, which reduces simple oxidized carbon compounds to generate methane as an end product. There are three main types of methanogenic metabolism depending on the substrate used for methanogenesis (Liu and Whitman, 2008; Costa and Leigh, 2014; Yan and Ferry, 2018). Hydrogenotrophic methanogenesis involves the reduction of CO2 to CH4, usually with H2 as the electron donor. Some hydrogenotrophic methanogens are also able to use other electron donors, such as formate, CO, alcohols, and iron (Dolfing et al., 2008; Ferry, 2010; Kurth et al., 2020). Methylotrophic methanogenesis involves the activation of methylated compounds, such as methanol and trimethylamine via substrate-specific corrinoid proteins, which then transfer the methyl group into methanogenesis via CH3-S-CoM. The more recently discovered methoxydotrophic pathway involves methanogenesis from methoxylated aromatic compounds (Mayumi et al., 2016), where the methyl group is transferred to tetrahydromethanopterin instead of coenzyme M (HS-CoM; Kurth et al., 2021a). Finally, acetoclastic methanogenesis utilizes acetate as a methanogenesis substrate, where the carboxyl group is oxidized to CO2 and the methyl group is reduced to CH4. Although acetoclastic methanogenesis is the least bioenergetically favorable, 2/3 of biologically derived methane comes from acetate (Lyu et al., 2018a). While there are notable distinctions across the three methanogenic pathways, the key methane-generating step is always catalyzed by MCR (Figure 1).
Methanogenesis produces nearly a billion tons of methane each year, which accounts for at least 70% of global methane emissions (Conrad, 2009; Kirschke et al., 2013; Jackson et al., 2020). About half of this methane is consumed by methanotrophic microorganisms, while the remainder unfortunately escapes to our atmosphere (Kirschke et al., 2013). Although much less abundant compared to CO2, methane is a more potent greenhouse gas since it has at least a 25-fold higher global warming potential than CO2 over a 100-year period (Montzka et al., 2011). The rising methane concentration is believed to account for ~20% of the current global warming trend (Lyu et al., 2018a). Thus, the development of strategies to curb methane emissions is essential to mitigate climate change. Indeed, MCR is a highly pursued target for developing inhibitors toward biological methane production (Duin et al., 2016; Yu et al., 2021).
Anaerobic methanotrophic archaea (ANME) are related to methanogens and are capable of oxidizing methane in the absence of O2, consuming substantial amounts of methane in anaerobic environments and thus playing a critical role in the global methane budget (Knittel and Boetius, 2009). Metagenome and gene/protein expression data have revealed that ANME contain and express previously characterized methanogenic genes, indicating that they utilize a reverse methanogenesis pathway to oxidize methane to CO2 (Hallam et al., 2004; Meyerdierks et al., 2010; Stokke et al., 2012; Timmers et al., 2017). Most commonly, ANME exist with syntrophic sulfate-reducing bacteria that allow AOM to be coupled with sulfate reduction (Nauhaus et al., 2002; Knittel and Boetius, 2009; Holler et al., 2011; Wegener et al., 2016). In these consortia, ANME carry out the oxidation reactions, with MCR presumably catalyzing the initial methane activation step (Scheller et al., 2010; Figure 1). The reducing equivalents generated throughout methane oxidation to CO2 are transferred to the bacteria, likely mediated by multi-heme c-type cytochromes, for use in sulfate reduction (McGlynn et al., 2015; Wegener et al., 2015, 2016). Additionally, single archaeal populations have been identified that have the genes necessary for performing AOM as well as sulfite reduction (McKay et al., 2019) or nitrate/nitrite reduction (Haroon et al., 2013), or may transfer electrons directly to metals (He et al., 2018), indicating that there may be exceptions to the paradigm of interspecies redox coupling. On the basis of metagenomic data, ANME are separated into four main clades: ANME-1 (Hinrichs et al., 1999), ANME-2 (Orphan et al., 2001, 2002), ANME-2d (Raghoebarsing et al., 2006; more recently referred to as Ca. Methanoperedenaceae; Haroon et al., 2013), and ANME-3 (Knittel et al., 2005; Niemann et al., 2006).
MCR is a dimer of heterotrimers with a α2β2γ2 configuration (Figure 2A), harboring two active sites that are only accessible through a 50 Å channel (Ermler et al., 1997). Each active site contains the nickel hydrocorphin prosthetic group, coenzyme F430 (Ellefson et al., 1982, Pfaltz et al., 1982, Livingston et al., 1984, Farber et al., 1991; Figures 1 and 2). The active form of MCR contains F430 in the Ni(I) oxidation state (Goubeaud et al., 1997). In the methane-forming direction, MCR catalyzes the conversion of CH3-S-CoM and HS-CoB to methane and the CoM-S-S-CoB heterodisulfide (Figure 1). Only one site is activated at any given time (“half-of-the-sites reactivity”), and thus the two active sites are proposed to function similar to a two-stroke engine where binding of the substrates in one active site induces a conformational change that provides energy for the heterodisulfide product to be expelled in the other active site (Goenrich et al., 2005; Scheller et al., 2013). Although any ANME MCR has yet to be enzymatically investigated in vitro, MCR from M. marburgensis can catalyze the reverse methane oxidation reaction at rates comparable to those measured in AOM consortia in vivo (Scheller et al., 2010), supporting the proposal that ANME utilize MCR to oxidize methane.
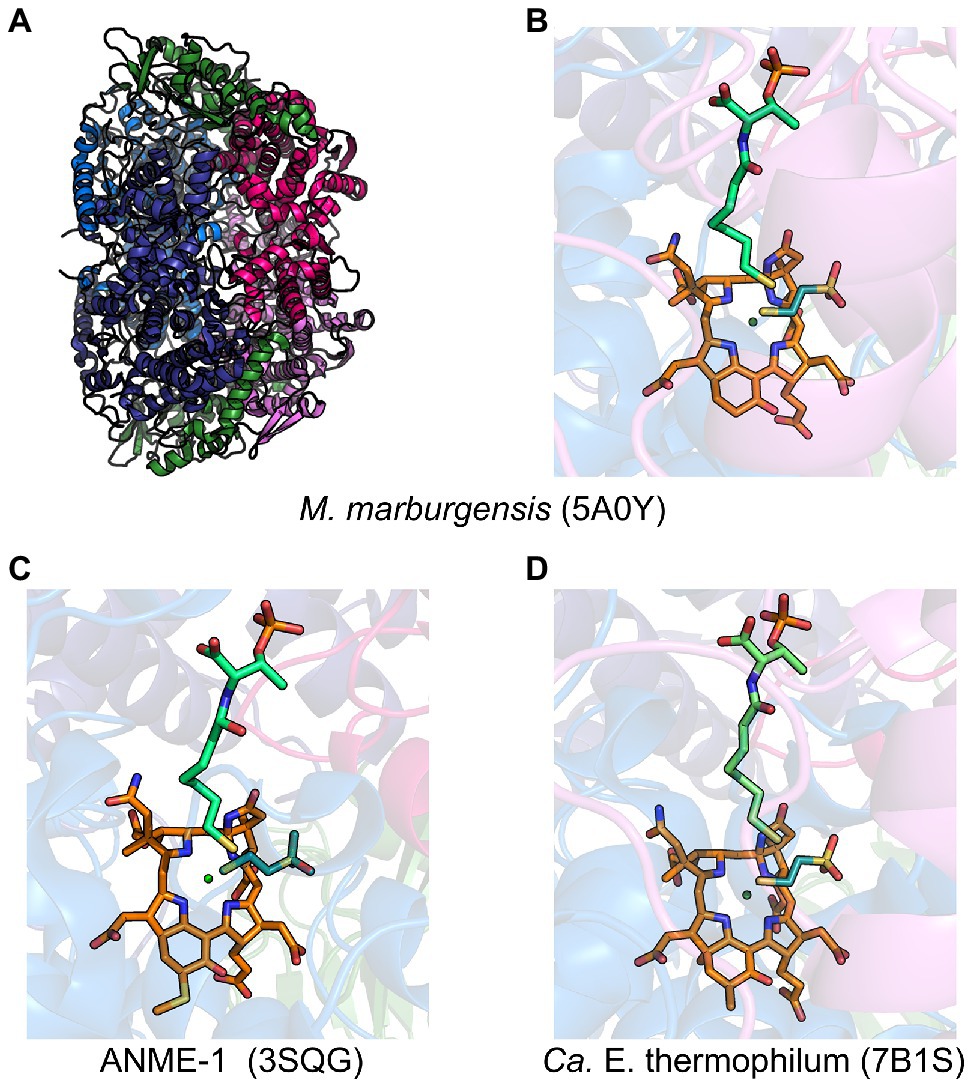
Figure 2. Representative crystal structures of MCRs and ECR. (A) MCR from M. marburgensis and (B) the associated active site with F430, HS-CoB, and HS-CoM. (C) Black Sea mat ANME-1 MCR active site with 172-methylthio-F430, HS-CoB, and HS-CoM. (D) Ca. E. thermophilum ECR active site with dimethyl-F430, HS-CoB, and HS-CoM. α subunits are shown in marine and deep blue, β subunits in hot pink and violet, γ subunits in forest green, F430 in orange, coenzyme M in deep teal, and coenzyme B in lime green.
Recent mechanistic studies have provided evidence that the reaction occurs by “mechanism II,” involving a methyl radical intermediate (Wongnate et al., 2016) that was originally proposed on the basis of quantum mechanical modeling studies (Pelmenschikov et al., 2002; Pelmenschikov and Siegbahn, 2003; Chen et al., 2012, 2014). The major alternative mechanism (“mechanism I”) involves nucleophilic chemistry with a Ni(III)-methyl intermediate (Yang et al., 2007; Dey et al., 2010). In the proposed radical mechanism, Ni(I) induces homolytic cleavage of the methyl-sulfur bond of CH3-S-CoM to generate a methyl radical and Ni(II). The methyl radical then reacts with HS-CoB to produce methane and a •S-CoB radical, which reacts with the Ni-bound CoM thiolate to generate a disulfide anion radical. One-electron transfer to Ni(II) then releases the heterodisulfide and regenerates the Ni(I) (Wongnate et al., 2016).
The structures of MCR for which crystal structures have been obtained from various methanogens and alkane-oxidizing organisms are all remarkably similar. Based on phylogenetic and structural comparison, MCRs from Methanobacteriales and Methanococcales were classified into MCR types I, II, and III (Wagner et al., 2017). The different MCR types have representative crystal structures and mainly differ in their electrostatic surface potentials, loop architectures, and the C-terminal end of their γ-subunits that interact with α and β subunits. Based on phylogenetic comparisons, MCRs from other organisms, such as Methanomicrobiales, Methanosarcinales, Methanocellales, and ANME-1, are distinct from the defined types I-III present in Methanobacteriales and Methanococcales (Wagner et al., 2017).
MCR crystal structures with HS-CoB and the substrate analog HS-CoM show the sulfhydryl group of HS-CoM serving as an axial ligand to the Ni(II) of F430 (Figure 2B). However, all MCR crystal structures are of the enzyme in its inactive Ni(II) state or a chemically modified methyl-Ni(III) state (Cedervall et al., 2011). Thus, the true coordination state of Ni(I) and the binding conformation of CH3-S-CoM remains unclear since a crystal structure with CH3-S-CoM has never been obtained. However, recent studies (Patwardhan et al., 2021) have provided new evidence for the possible orientation of CH3-S-CoM binding to the nickel center of F430. Interestingly, results indicate that there is no nickel-sulfur interaction and thus suggest that the thioether portion of the substrate does not bind to the Ni(I) (Patwardhan et al., 2021). Instead, CH3-S-CoM appears to bind to Ni(I) through the sulfonate group. This proposed alternate binding scenario puts the reactive portions of the two substrates in close proximity so that the subsequently generated proposed methyl radical is in position to abstract a hydrogen atom from HS-CoB (Patwardhan et al., 2021).
When the first crystal structure of MCR from M. marburgensis (Figures 2A,B) was solved, five unusual post-translational modifications (PTMs) were revealed in the α subunit near the active site (Ermler et al., 1997). Since then, subsequent work has discovered additional MCR PTMs, where the presence of specific PTMs varies depending on the organism (Table 1; Grabarse et al., 2000; Kahnt et al., 2007; Wagner et al., 2016; Wagner et al., 2017; Kurth et al., 2021b). PTMs found in methanogens include three strictly conserved modifications—N1-methylhistidine, 5-(S)-methylarginine, and thioglycine—as well as a handful of more variable modifications including S-methylcysteine, 2-(S)-methylglutamine, didehydroaspartate, and 6-hydroxytryptophan (Table 1 and Figure 3). The impacts of these PTMs remain a major area of research in the field as their precise role in catalysis and/or active site structure remains unclear. MCR PTMs were recently summarized in a mini-review (Chen et al., 2020), but we will outline key aspects here.
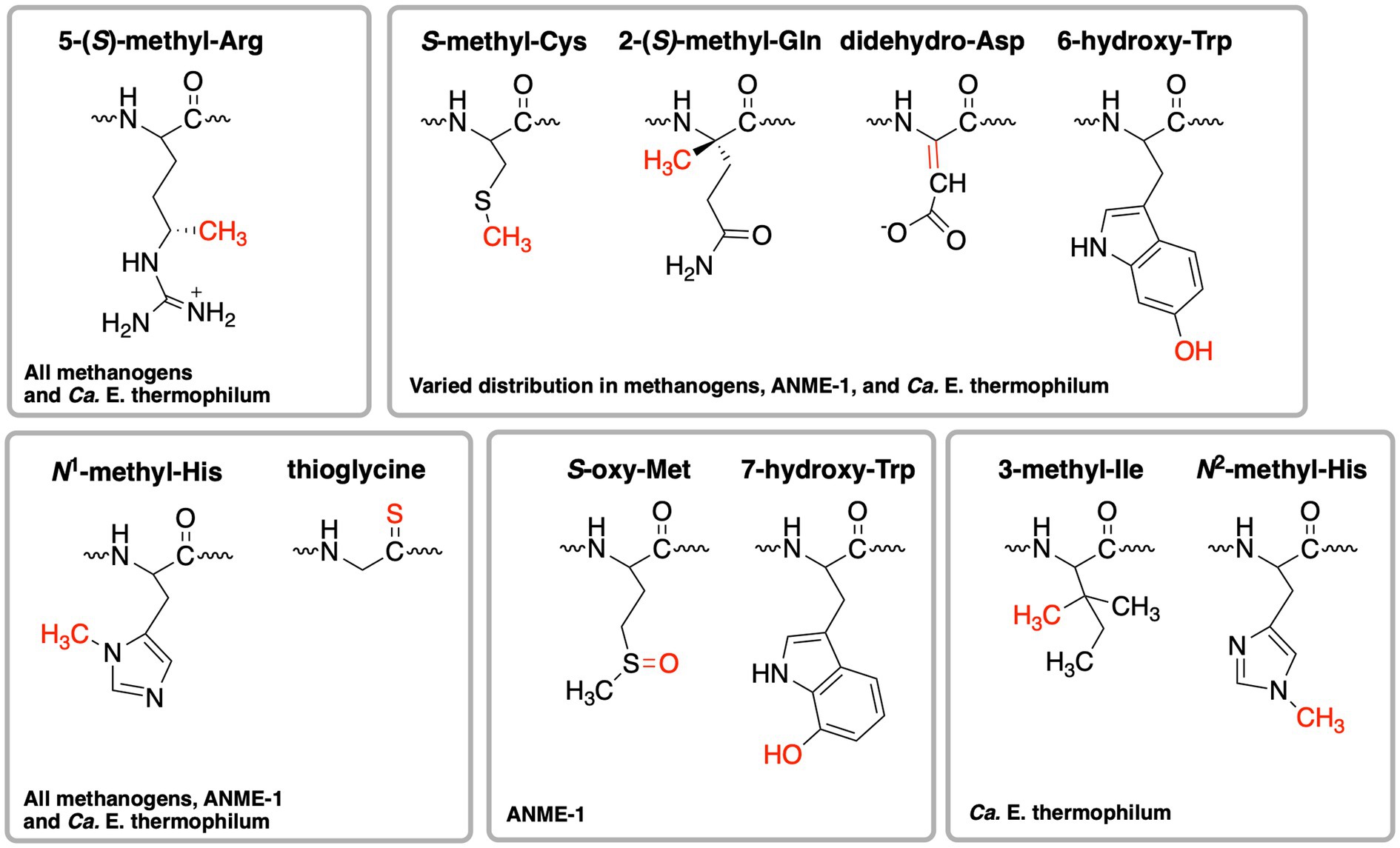
Figure 3. Structures of post-translational modifications identified in the active sites of various MCRs and ECR. The distribution of these PTMs is further summarized in Table 1.
Significant progress has been made toward identifying the enzymes involved in installing MCR PTMs, including identification of the radical S-adenosylmethionine (SAM) enzyme catalyzing the difficult methylation reaction to produce 5-(S)-methylarginine (Deobald et al., 2018; Radle et al., 2019; Lyu et al., 2020) and identification of the enzymes responsible for the thioglycine transformation (Nayak et al., 2017). Both of these PTMs appear to at least be important for the stability of MCR from Methanosarcina acetivorans, especially under thermal stress (Nayak et al., 2017, 2020; Deobald et al., 2018). The methylated arginine seems to have a more significant impact on Methanococcus maripaludis MCR, where a deletion strain lacking this modification showed a highly impaired growth rate and the rate of methanogenesis was only about half the rate of wild type (Lyu et al., 2020). Most recently, the methyltransferase necessary for the synthesis of S-methylcysteine was identified (Nayak et al., 2020). Through the production of a M. acetivorans deletion strain lacking the genes involved in 5-(S)-methylarginine, thioglycine, and S-methylcysteine biosynthesis, the associated MCR variant was produced and its crystal structure was solved, which was surprisingly indistinguishable from the wild-type structure (Nayak et al., 2020). Growth studies with the associated deletion strain suggested that epistatic interactions among MCR PTMs influence the stability and in vivo activity of the enzyme (Nayak et al., 2020). However, in vitro kinetic studies have not yet been carried out on MCR variants lacking one or more PTMs, which will be required to make any conclusions about the specific functions and importance of the respective PTMs.
ANME-1 is the only ANME clade for which an MCR crystal structure has been obtained. By purifying and crystallizing the enzyme directly from a Black Sea mat sample, the crystal structure of the ANME-1 MCR was solved to 2.1 Å resolution (Shima et al., 2012). This ANME MCR possesses the same overall structure as methanogenic MCRs. In particular, the active site channel seems be strictly conserved in both ANME-1 and methanogens, with HS-CoM and HS-CoB holding virtually the same position and conformation (Figure 2C). This result excludes the possibility of ANME MCR using different substrates/products, further supporting that methane oxidation catalyzed by MCR in ANME is the reverse of the methane-generating step in methanogens (Figure 1). The ANME-1 MCR structure does possess a few notable differences. First, the active site contains a modified F430, 172-methylthio F430 (Figures 2C, 4), which was previously structurally characterized by mass spectrometry and NMR (Mayr et al., 2008). This modified F430 appears to be accommodated by the replacement of the bulky 2-(S)-methylglutamine found in methanogens with Val419 in ANME-1. Second, ANME-1 MCR contains five distinct cysteines between F430 and the protein surface, suggesting a potential redox-relay system that could be used to reduce F430 to the active Ni(I) state (Shima et al., 2012). Third, PTM patterns vary between ANME-1 MCR and methanogenic MCR. ANME-1 does not contain the highly conserved arginine methylation seen in methanogens, however, ANME-1 MCR does contain two unique PTMs, 7-hydroxytryptophan and S-oxymethionine. Also present in ANME-1 MCR are N1-methylhistidine and thioglycine (Shima et al., 2012; Table 1 and Figure 3).
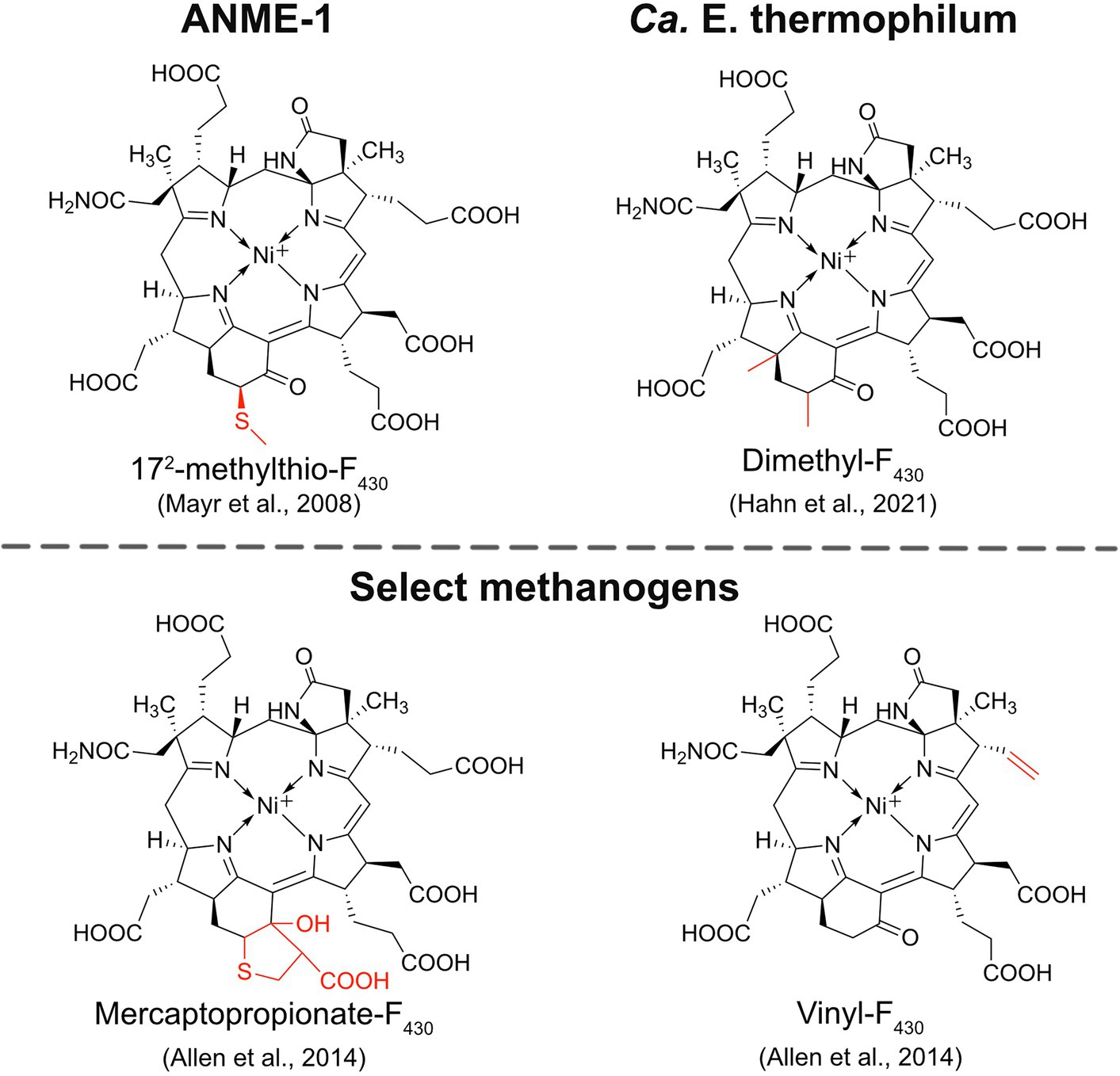
Figure 4. Structures of modified F430s. The structures of the modified F430s in ANME-1 and Ca. E. thermophilum are confirmed based on NMR and/or crystal structures while the modifications in select methanogens are proposed based on mass spectrometry data.
Very recently, the crystal structure of the MCR homolog from an anaerobic ethane oxidizing archaeon, Ca. Ethanoperedens thermophilum, was determined (Hahn et al., 2021). This enzyme apparently does not take other alkane substrates outside of ethane (Hahn et al., 2020) and thus has been designated as ethyl-coenzyme M reductase (ECR; Hahn et al., 2021). Although ECR follows the overall structural trend of known MCRs, there are notable differences to consider. First, ECR is 20 kDa larger than canonical MCRs, mainly due to three insertions in the α subunit, one insertion in the β subunit, and one insertion in the γ subunit. These insertions impact surface charges and contribute to the unique architecture of the ethane tunnel, which provides a 33 Å hydrophobic path to the active site (Hahn et al., 2021). The tunnel is flanked by post-translationally modified amino acids, including two unique PTMs—N2-methylhistidine and 3-methylisoleucine (Table 1 and Figure 3). Notably, the ECR structure revealed that the F430 nickel is coordinated by methionine rather than the canonical glutamine, and a modified version of F430 with two methyl groups is present (dimethyl-F430; Figures 2D, 4). Additionally, an active site loop (α367-374) contains a tryptophan residue (αTrp373) instead of the canonical phenylalanine found in other MCRs. This loop shifts the position of F430, which is stabilized by hydrogen bonds with αAsn375 along with a clamping effect from αTyr376 and αPhe441, resulting in a 11.4° tilt on the porphinoid ring. Consequently, the active site volume is increased to adequately accommodate ethane. The authors propose that the F430 methylations likely serve to maintain the structure and reactivity of the cofactor in the expanded active site (Hahn et al., 2021).
An interesting and underexplored area in the MCR field is the potential functions and importance of F430 modifications. F430 is only known to function with MCRs and ACRs, indicating that nature has evolved a specialized coenzyme to catalyze the difficult reactions of methane formation and methane/alkane activation. The first modified F430 to be discovered was 172-methylthio-F430 (Figure 4), which was originally identified and structurally characterized from Black Sea mat samples enriched with ANME-1 (Mayr et al., 2008). This modified F430 is presumed to be the primary physiologically active version of F430 in ANME-1 since it was later characterized in the crystal structure of ANME-1 MCR (Shima et al., 2012; Figure 2C). ANME-2 organisms apparently do not contain 172-methylthio-F430 (Mayr et al., 2008; Kaneko et al., 2014), and the potential F430 modifications present in ANME-3 have not yet been reported to our knowledge. In 2014, additional F430 modifications were identified in select Methanococcales methanogens (Allen et al., 2014). Mercaptopropionate-F430, containing a cyclized mercaptopropionate moiety bound as a thioether, was identified in Methanocaldococcus jannaschii and Methanococcus maripaludis, while vinyl-F430 was observed in M. maripaludis and Methanococcus vannielii (Figure 4). These structures were proposed based on mass spectrometry data, and thus still need to be confirmed by NMR or crystallography. Finally, most recently, the dimethyl-F430 present in ECR was discovered (Hahn et al., 2021; discussed above; Figure 4). One important aspect to note is that the F430 modifications in methanogens are not always present and the modifications generally exist as a minor component compared to the unmodified F430 (Allen et al., 2014). The situation appears to be different in ANME-1, where 172-methylthio-F430 is the predominant form (Kaneko et al., 2014). Additionally, for both ANME-1 and Ca. E. thermophilum, 172-methylthio-F430, and dimethyl-F430 are confirmed to function with the respective MCR/ECR since they were identified in the crystal structures (Shima et al., 2012; Hahn et al., 2021). In contrast, the modified F430s in methanogens have only so far been identified in small molecule cell extracts and have not been observed in any methanogen MCR crystal structures.
Understanding how modified F430s affect MCR will be important for the design of optimized heterologous systems for recombinant MCR production, especially for anaerobic methane/alkane oxidation applications. It will also be necessary to identify the enzymes involved in their biosynthesis. The complete biosynthetic pathway for F430 has been described (Zheng et al., 2016; Moore et al., 2017), but the enzymes required for the installation of F430 modifications are currently unknown.
The three MCR subunits are encoded by mcrA, mcrB, and mcrG, which are usually present in an MCR operon along with two other genes (mcrD and mcrC). The most common MCR operon across all methanogens is mcrBDCGA (Figure 5). McrD and McrC are accessory proteins whose functions have yet to be confirmed. McrC was a component of the large complex of proteins identified as being responsible for the reduction of F430 to its Ni(I) active state; thus McrC likely plays a role in MCR activation (Prakash et al., 2014). McrD may serve as a chaperone protein that binds F430 for subsequent delivery to the MCR active site (Zheng et al., 2016). Early studies demonstrated that McrD interacts with MCR (Sherf and Reeve, 1990) and it also co-purified with a recombinant MCR expressed in M. maripaludis (Lyu et al., 2018b). Finally, McrD was shown to alleviate product inhibition in the final step of F430 biosynthesis in vitro, presumably due to its ability to bind the newly synthesized F430 coenzyme (Zheng et al., 2016).
An interesting aspect of MCR operons is the variability in the presence of accessory proteins, or the presence of additional operons. In some methanogens, including M. marburgensis, there is a second MCR operon that encodes for MCR isozyme II (Rospert et al., 1990). Most work on MCR has been performed on MCR isozyme I, whose operon in M. marburgensis is the typical mcrBDCGA operon. Less studied is MCR II, whose operon lacks mcrC (Figure 5). The two isozymes are highly similar in sequence as well as overall structure and active site architecture (Wagner et al., 2016), but some notable differences have been reported. MCR I and MCR II show significant differences in electrostatic surface potentials, which allows for convenient separation of the two isozymes via anion exchange chromatography (Rospert et al., 1990; Duin et al., 2011). Because MCR II was purified in larger quantities from cells in log phase, and MCR I was purified in larger quantities from cells at the end of growth, it was concluded that MCR II is expressed in non-gas-limiting conditions, while MCR I is expressed during gas-limiting conditions (Rospert et al., 1990; Bonacker et al., 1992). Further studies on both isoenzymes revealed that MCR I and MCR II expression is affected by pH, temperature, and availability of both H2 and CO2 (Bonacker et al., 1992), and in vitro kinetic studies revealed differing catalytic properties (Bonacker et al., 1993).
In addition to M. marburgensis, many other members of Methanobacteriales, and some members of Methanococcales and Methanomicrobiales contain two MCR isozymes. The presence of two MCR isozymes may present a possible evolutionary advantage which could allow methanogens to express different versions of MCR when faced with changes in environmental conditions, such as substrate limitation. The fact that the MCR II gene cluster lacks mcrC would indicate that either McrC is not necessary for the function of MCR II or that the McrC encoded in the MCR I gene cluster can be utilized.
The difference between MCR I and MCR II gene clusters is not the only instance in which the MCR operon displays variability. The MCR operon from ANME-1 isolated from a Black Sea mat lacks both mcrD and mcrC (Meyerdierks et al., 2010; Figure 5). A McrC homolog is present outside of the MCR operon in ANME-1 (BSM_08630, 51% identity to McrC from M. marburgensis), but McrD appears to be completely absent. The ANME-2d organism, Ca. Methanoperedens nitroreducens, contains an MCR operon like that of MCR II in M. marburgensis, where only mcrD is present without mcrC (Haroon et al., 2013; Figure 5). Similar to ANME-1, a likely mcrC exists outside of the MCR operon (ANME2D_00875, 53% identity to McrC from M. marburgensis, 50% identity to McrC from ANME-1 mentioned above). This poses interesting questions regarding the high conservation of the primary MCR I operon mcrBDCGA across different methanogenic species, whereas ANME MCR operons lack one or more of the accessory proteins in the operon. Additionally, ANME-1 organisms seem to completely lack mcrD. A better understanding of the functions and specificity of these and potentially additional yet to be discovered accessory proteins is crucial for the development of optimized recombinant MCR expression systems.
MCR variants known as alkyl-coenzyme M reductases (ACRs) carry out the anaerobic oxidation of various non-methane alkane substrates. The first report of ACR-dependent anaerobic oxidation of an alkane other than methane was published in 2016 when members of the GoM-Arch87 clade, closely related to Methanosarcinales, were proposed to carry out the anaerobic oxidation of butane to CO2 (Laso-Pérez et al., 2016). Similar to ANME-1, these organisms form consortia with HotSeep-1 sulfate-reducing bacteria and couple butane oxidation to sulfate reduction. Two archaeal genomes were assembled from the butane enrichment cultures that resulted in the proposed names for two new organisms, Ca. Syntrophoarchaeum butanivorans and Ca. Syntrophoarchaeum caldarius (Laso-Pérez et al., 2016). Intriguingly, these organisms contain four different mcrBGA genes clusters—three of the four ACR gene sets are arranged in operons in Ca. S. butanivorans, while all four are arranged in operons in Ca. S. caldarius. The butane-dependent formation of butyl-S-CoM was confirmed in these cultures, suggesting that butane oxidation involves the use of the MCR homolog(s). The GoM-Arch87 enrichment cultures were also tested with other hydrocarbons and, interestingly, propane enriched cultures resulted in propane dependent sulfate reduction. As with the butane cultures, propyl-S-CoM was detected, indicating the involvement of an ACR in propane oxidation. Other alkane oxidizers include Ca. Argoarchaeum ethanivorans (Chen et al., 2019) and Ca. E. thermophilum (Hahn et al., 2020), archaeal species that activate ethane using ECR (discussed in MCR structures and post-translational modifications section) to form ethyl-S-CoM, which is subsequently oxidized to CO2. These ethane oxidizers only utilize ethane and cannot metabolize other alkanes.
In addition to the involvement of ACRs in short-chain alkane oxidation, recently published work suggests the use of ACRs to activate long-chain alkanes in oil reservoirs (Zhou et al., 2022). Ca. Methanoliparum couples the degradation of long-chain alkanes to methanogenesis in a process that is independent of syntrophic partners. Metagenomic and transcriptomic data indicated the presence of several different species, which contain and expresses genes encoding putative ACRs as well as MCRs. Additionally, the presence of hexadecyl-S-CoM and other long-chain alkane R-S-CoM derivatives were confirmed by mass spectrometry, thus indicating the involvement of an ACR in alkane activation (Zhou et al., 2022).
Traditionally, all methanogens were thought to belong to the Euryarchaeota phylum, however, this definition was challenged with the discovery of putative methane metabolism in the Bathyarchaeota phylum (Evans et al., 2015). Metagenome data revealed the presence of mcrABG genes and putatively mcrCD, as well as several other genes for methylotrophic methanogenesis. The MCR primary sequence and predicted structure analysis showed putative binding sites for HS-CoM, HS-CoB, and F430, suggesting that the enzyme utilizes the same substrates and coenzyme (Evans et al., 2015). In addition to Bathyarchaeota, five metagenomes from a proposed new archaeal phylum termed Ca. Vestraetearchaeota were shown to contain divergent mcrA sequences (Vanwonterghem et al., 2016). Metabolic reconstructions of the assembled genomes revealed the presence of key genes associated with methylotrophic methanogenesis, including a complete mcrBDCGA operon. It is hypothesized that these organisms would perform H2-dependent methylotrophic methanogenesis. However, it is still unclear if this new phylum is comprised of organisms that perform methanogenesis as a preferential metabolism since the assembled genomes in this study also showed that these organisms likely have the capacity to perform fermentative metabolism (Vanwonterghem et al., 2016).
Archaeoglobi is a class of thermophilic organisms within the Euryarchaeota that were generally believed to be non-methanogenic (Bapteste et al., 2005; Hartzell and Reed, 2006; Boyd et al., 2019). This is because complete MCR-encoding genes and methyl-H4M(S)PT:coenzyme M methyltransferase (MTR) MtrABCDEFGH complex genes had never been identified, even though other characteristic hydrogenotrophic methanogenesis genes as well as archaeal type Wood-Ljungdahl pathway genes have been identified in some Archaeoglobi genomes (Klenk et al., 1997; Bapteste et al., 2005). However, recent metagenome data have demonstrated that some Archaeoglobi contain genes encoding MCR. Ca. Polytropus marinifundus contains two divergent mcrABG operons similar to Bathyarchaeota and Syntrophoarchaeum as well as other potential genes for alkanotrophic metabolism (Boyd et al., 2019). Another new genus of Archaeoglobi, Ca. Methanomixophus, contains MTR complex genes as well as a complete mcrBDCGA operon, along with predicted ligand binding sites for HS-CoM, HS-CoB, and F430 (Liu et al., 2020). Metatranscriptomic experiments showed active hydrogen-dependent methylotrophic methanogenesis as well as heterotrophic fermentation. Additionally, one of the new proposed organisms, Ca. Methanomixophus hydrogenotrophicum, possesses the genes to conserve energy via AOM coupled to syntrophic sulfate reduction, while Ca. Methanomixophus dulitatem contains its own sulfate reduction genes that would allow for a methane oxidizing lifestyle (Liu et al., 2020).
The recent and continuing discoveries of diverse putative MCRs and ACRs highlight the need to develop effective tools to study the catalytic capabilities of these enzymes with likely very different enzymatic properties. Additionally, MCR is a highly attractive, yet challenging, target for potential use in bioengineering applications for biofuel production, either for methane generation or methane/alkane conversion applications (Conrado and Gonzalez, 2014; Haynes and Gonzalez, 2014). Since ANME utilize MCR in the methane oxidation direction, ANME MCRs are especially appealing biocatalysts for potentially converting abundant methane reserves into more usable liquid fuels and other value-added chemicals (Mueller et al., 2015; Lawton and Rosenzweig, 2016a,b). However, as mentioned previously, no ANME MCR has been studied in vitro and thus it is unclear whether ANME MCRs will be better suited for this purpose compared to methanogenic MCRs.
Since AOM consortia grow slowly, with doubling times on the month timescale (Laso-Pérez et al., 2018; Bhattarai et al., 2019), and to low cell densities, obtaining enough cells to purify the native MCR from ANME organisms in sufficient quantities for kinetic and mechanistic studies is not very feasible. Additionally, ANME organisms are not yet genetically tractable and thus are not amenable to bioengineering applications. Thus, the development of heterologous expression systems for ANME MCRs as well as other diverse MCRs from unculturable or difficult-to-culture archaea would be highly advantageous. Traditional hosts, such as Escherichia coli, seem to pose currently unsurmountable challenges to achieve this goal since they do not possess the biochemical machinery for F430 biosynthesis, post-translational modifications, or MCR assembly and activation. Thus, the current likely best option for a heterologous host is a fast-growing methanogen for which genetic manipulation methods exist. The two highly studied model methanogens for which well-established and robust genetic tools exist are Methanosarcina acetivorans and Methanococcus maripaludis. The following paragraphs and Table 2 summarize the major available genetic tools in these organisms as well as recently described genetic tools in Methanocaldococcus jannaschii and Methanothermobacter thermautotrophicus.
Methanosarcina species are cytochrome-containing methanogens that are capable of using the widest variety of methanogenic substrates compared to other genera. Depending on the growth substrate, M. acetivorans exhibits doubling times as low as 8 h at 37°C. Compared to other methanogens with available genetic tools, Methanosarcina are phylogenetically most closely related to ANME (Knittel et al., 2005), thus making Methanosarcina the most logical choice as potential heterologous hosts for ANME MCRs. M. acetivorans C2A is most commonly utilized for genetic experiments, but Methanosarcina barkeri Fusaro is also used successfully and the same basic tools have been developed for both species (Buan et al., 2011). Both organisms are mesophilic and require that they are maintained in high-salt medium to prevent cells from growing in clumps, which can pose issues for genetic experiments. Routine genetic experiments normally involve a parental strain containing a deletion of the hypoxanthine phosphoribosyltransferase hpt gene that is used as a counterselection marker (Pritchett et al., 2004), and often also contain a ΦC31 attP site inserted at the hpt locus, allowing for insertion of plasmids containing the complementary attB sequence into the host chromosome via recombination (Guss et al., 2008). A comprehensive description of the different plasmids and strains of Methanosarcina that have been used for various purposes, including gene deletions and recombinant expression, has been reported (Buan et al., 2011). Transforming Methanosarcina species involves the well-established liposome-mediated transformation, where the transformation efficiency is as high as 20% in M. acetivorans, but is substantially lower in other Methanosarcina species (Metcalf et al., 1997). For inducible expression of recombinant proteins in Methanosarcina, the tetracycline-inducible system can be used, which allows for the expression of proteins at a desired time point during growth (Guss et al., 2008). The cdh operon promoter has also been used successfully to drive acetate-dependent overexpression of a carbonic anhydrase in M. acetivorans (Macauley et al., 2009). More recent work has generated a new series of suicide plasmids that simplify the cloning process and allow for facile expression and purification of tagged proteins in Methanosarcina (Shea et al., 2016). Additionally, an efficient CRISPR/Cas9 system has been developed for M. acetivorans (Nayak and Metcalf, 2017). Notably, this system has been used to add a tandem affinity purification tag to the N-terminus of the native McrG gene in M. acetivorans, thus greatly facilitating MCR purification (Nayak and Metcalf, 2018). Finally, genetic tools have also been utilized for Methanosarcina mazei (Ehlers et al., 2005), including markerless genetic exchange using similar methods developed for M. acetivorans and M. barkeri (Pritchett et al., 2004; Ehlers et al., 2011). A trimethylamine inducible promoter has also been used to successfully heterologously overexpress a fusion-tagged protein in M. mazei (Mondorf et al., 2012).
Methanococcus maripaludis is a hydrogenotrophic mesophilic methanogen with a relatively fast doubling time of 2 h at 37°C (Whitman et al., 1986). The rapid growth of M. maripaludis compared to Methanosarcina is advantageous for genetic experiments and recombinant protein expression. M. maripaludis S2 is the strain for which most reports of genetic manipulation utilize; however, M. maripaludis JJ has also been used successfully for this purpose. Methanococcus maripaludis can be grown on either H2/CO2 or formate, the latter of which provides an easier and safer alternative to dealing with high-pressure gases (Long et al., 2017). Genetic studies often utilize M. maripaludis strains lacking the gene for uracil phosphoribosyltransferase, which confers sensitivity to the base analog 6-azauracil and serves as a marker for negative selection (Costa et al., 2010; Sarmiento et al., 2011). Several shuttle vectors have been reported, and well-established methods exist for markerless mutagenesis and recombinant protein expression (Whitman et al., 1997; Sarmiento et al., 2011). M. maripaludis plasmids utilize puromycin or neomycin resistance for positive selection. Transformation methods for M. maripaludis S2 utilize polyethylene glycol-mediated transformation (Tumbula et al., 1994; Sarmiento et al., 2011). However, natural transformation facilitated by type IV-like pili has been reported for M. maripaludis JJ (Fonseca et al., 2020). Notably, a highly efficient CRISPR/Cas system has recently been developed for M. maripaludis JJ that utilizes a bacterial Cas12a along with the native homology directed repair machinery (Bao and Scheller, 2021). The capacity for natural transformation as well as the available CRISPR/Cas technology for genetic manipulation makes M. maripaludis JJ an especially attractive host for future metabolic engineering applications. Finally, natural transformation via type IV-like pili has also been demonstrated in Methanoculleus thermophilus (Fonseca et al., 2020). Using an established plasmid employed for generating M. maripaludis gene deletions, the authors generated a M. thermophilus deletion strain lacking genes for pili to demonstrate that pili are essential for natural transformation (Fonseca et al., 2020). This is the first report of genetic manipulation in a methanogen from the order Methanomicrobiales.
Methanocaldococcus jannaschii is a hyperthermophilic methanogen (Jones et al., 1983) that was the first archaeal organism to have its genome sequenced (Bult et al., 1996). Methanocaldococcus jannaschii has the fastest doubling time of any methanogen (26 min) and grows optimally at 85°C (Jones et al., 1983). Recently, the first genetic tools for M. jannaschii were reported (Susanti et al., 2019). Methanocaldococcus jannaschii is resistant to antibiotics commonly used with other archaea, such as previously mentioned puromycin or neomycin, as well as the base analogs used for counter selection in other methanogens (Susanti et al., 2019). However, it was found to be sensitive to mevinolin and simvastatin. These compounds are competitive inhibitors of 3-hydroxy-methylglutaryl (HMG)-CoA reductase and thus overexpression of HMG-CoA reductase can be used as a selection marker (Susanti et al., 2019). Based on this, a suicide vector was developed for generating in-frame gene deletions in M. jannaschii, where the gene of interest is replaced with hmgA. Further, a similar strategy was used to place a gene of interest under the control of a strong promoter and to add an affinity tag on the chromosome for subsequent purification of the overexpressed protein (Susanti et al., 2019). M. jannaschii is transformed using heat shock without the need for chemical treatments, such as PEG in M. maripaludis S2 or liposomes in M. acetivorans (Susanti et al., 2019). This system is not yet as advanced as for the methanogens described above since only linearized suicide vectors have been used, which does not allow the investigation of potentially essential genes and is not compatible with most strategies for markerless mutagenesis. Additionally, heterologous protein expression in M. jannaschii has not yet been reported.
Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus ΔH are two highly similar Methanobacteriales methanogens that were used as model hydrogenotrophic methanogens during early biochemical investigation of methanogenesis and MCR (Thauer, 1998). Methanothermobacter species have also been successfully employed in bioreactors for efficient methane production processes (Thema et al., 2019; Pfeifer et al., 2021). Thus, developing genetic manipulation tools for these organisms has been a major area of interest. Very recently, the first reliable system for M. thermautotrophicus ΔH was reported (Fink et al., 2021). Methanothermobacter thermautotrophicus ΔH was found to be amenable to plating on solid medium with good efficiencies, and neomycin was shown to be effective for selection. This allowed for the construction of a shuttle vector containing a full array of cloning sites, selections markers for E. coli and M. thermautotrophicus ΔH, and the β-galactosidase-encoding gene bgaB, which can be used as a reporter (Fink et al., 2021). Transformation of M. thermautotrophicus ΔH is possible via interdomain conjugation with E. coli S17-1. As a proof-of-concept, this system was utilized to heterologously express formate dehydrogenase, thus enabling M. thermautotrophicus ΔH to grow with formate as a substrate (Fink et al., 2021). This genetic system could potentially be very useful for the future heterologous expression of MCRs since established methods for MCR activation in M. marburgensis (see section below) would likely also be effective for MCRs isolated from M. thermautotrophicus ΔH.
An important factor to consider when designing an expression construct for recombinant MCR expression is which promoter to use. Promoters are essential for transcription and translation as, like eukarya, archaea possess more complex machinery for translation compared to bacteria (Lyu and Whitman, 2017). Common promoters used for heterologous protein expression in M. acetivorans include the MCR promoter (PmcrB) and associated tetracycline-inducible forms (Buan et al., 2011), while in M. maripaludis the histone promoter A (PhmvA) is often used (Sarmiento et al., 2011). An inducible nif promoter has also been used for M. maripaludis (Lie and Leigh, 2002; Chaban et al., 2007) and, very recently, an inducible expression system based on phosphate limitation was developed (Akinyemi et al., 2021). Notably, a plasmid containing the Ppst promoter, which becomes activated under phosphate-limiting conditions, was used to express a recombinant M. maripaludis MCR in M. maripaludis, which represented 6% of the total protein content in a cell-free extract (140% increase compared to when using Phmva; Akinyemi et al., 2021). Additionally, the mmpX gene, which encodes the radical SAM methylase responsible for the methyl-arginine modification in McrA was successfully expressed. This is a particularly interesting result, as the expression of this protein using the constitutive promoter Phmva results in very low protein yields since it is apparently toxic to the cells (Akinyemi et al., 2021). Besides using previously employed constitutive or inducible promoters for recombinant MCR expression, another possibility is to utilize the native promoter for the MCR of interest. This has been done previously for the heterologous expression of ANME-1 MCR in M. acetivorans (Soo et al., 2016; discussed more below). If this strategy is chosen, it would be important to consider whether the heterologous host transcription/translation machinery is able to recognize the essential promoter elements present in the foreign promoter.
When designing a plasmid construct for heterologous MCR expression, it is also important to consider operon organization. ANME MCR operons generally lack one or both accessory proteins (e.g., mcrBGA—ANME-1 or mcrBDGA—Ca. M. nitroreducens) in the operon, while methanogens will always contain at least one MCR operon with both accessory proteins within the operon (Figure 5). Although it is still unclear how accessory proteins affect MCR assembly and/or activation, generating constructs for heterologous expression with accessory proteins may be necessary, even in the case where they are not present in the MCR operon of interest, especially since the accessory proteins may be organism-specific. Additionally, considering the post-translational machinery present in the heterologous host of choice will be important, as well as whether those enzymes will effectively recognize and correctly modify the recombinant enzyme. Since not all PTMs are consistent across different MCRs, additional and/or replacement PTM genes may need to be incorporated, which may potentially be toxic to the host.
Assuming successful expression, assembly, and post-translational modification of a recombinant MCR, the next consideration is obtaining the active form of the enzyme. The three oxidation states of F430 are well-described and can be observed via electron paramagnetic resonance (EPR) spectroscopy and UV–Vis spectrophotometry. These include the active MCRred1 in Ni(I) form, the inactive MCRsilent in the Ni(II) form, and the “ready” MCRox1 in the Ni(III) form (Goubeaud et al., 1997; Duin et al., 2011). Many isolation and purification procedures will yield MCR with F430 in an inactive Ni(II) state, thus representing a major limitation for the enzymatic investigation of various MCRs.
The first successful MCR activation procedure involved incubating M. marburgensis cells with 100% H2 prior to harvesting and including 10 mM CH3-S-CoM to stabilize the enzyme during purification (Rospert et al., 1991). This resulted in MCR almost entirely in MCRred1 state, which can be used for subsequent enzymatic assays. Subsequent work has shown that HS-CoM can be used instead of CH3-S-CoM to achieve activation (Duin et al., 2011). Other successful efforts to activate MCR from M. marburgensis involved the treatment of cells with 80% N2/20% CO2 prior to harvesting, which resulted in the MCRox1 form of the enzyme (Goubeaud et al., 1997). Upon incubation of purified MCRox1 with Ti(III) citrate, EPR spectra revealed the conversion from MCRox1 to MCRred1, and specific activity of the enzyme raised from 2 U/mg protein to 100 U/mg protein. The advantage of isolating MCR in the MCRox1 form is that this strategy minimizes oxidation of MCRred1 to the MCRsilent Ni(II) state, which cannot be reduced to the active form in vitro with chemical reductants. Another protocol for activation of MCR from M. marburgensis involved treatment of cells with CO, which was shown to activate MCR at a significantly faster rate than treatment with H2 (Zhou et al., 2013). CO activation resulted in MCRred1 within 1 h of incubation, while H2 treatment required overnight incubation. Although effective, these described MCR activation protocols have primarily only been used successfully for MCR isolated from M. marburgensis.
Toward developing strategies for activating MCR from other organisms, it was reasoned that MCR activation could be achieved by controlling the ligation state of nickel through the addition of different chemical agents (Becker and Ragsdale, 1998). Thus, sodium sulfide was added to Methanosarcina thermophila cells prior to harvesting, which successfully elicited the MCRox1 state (Becker and Ragsdale, 1998). Using 35S-labeled sulfide, the authors demonstrated that the sulfide enters the cell, binds to the nickel site in F430, and remains bound during purification. The amount of MCRox1 was correlated to the amount 35S-labeled sulfide. Thus, based on the available protocols described so far, isolation of MCR as MCRox1 using sodium sulfide treatment will likely be the most widely applicable to different MCRs isolated from various organisms. This is because ligating nickel to control the oxidation state should be independent from the metabolic state of the cell, which would allow for the control of the MCR oxidation state regardless of which organism and/or which methanogenic substrate is used.
In vivo MCR activation remains a poorly understood process. Early work provided initial insights into the cellular components responsible for the reduction of CH3-S-CoM to methane (see (Thauer, 2019) for a comprehensive discussion of these experiments). In 2014, complete activation of MCRox1 and 65% activation of MCRsilent was achieved in the presence of dithiothreitol, ATP, component A2, and component A3a (Prakash et al., 2014). An important discovery for this work was that the heterodisulfide product promotes the inactivation of MCR and thus it is essential to isolate the activation process from the methane formation reaction (Prakash et al., 2014). Further, the authors characterized component A3a as a 700 kDa complex that includes an assortment of redox proteins as well as McrC (Prakash et al., 2014). Any attempts to activate MCR with a smaller version of this complex were unsuccessful. Although much is still unclear about the structural basis of the complex and how it operates to activate MCR, this is a major advancement toward understanding how methanogens are able to supply low potential electrons to reduce F430 to the active Ni(I). Additionally, the presence of McrC in this complex finally linked a functional role to McrC, one of two accessory proteins within methanogenic MCR operons. It will be essential to fully elucidate the activation proteins and cofactors required as well as to obtain information about specificity in order to engineer an effective heterologous host for expression of active MCRs.
The first reported example of the heterologous expression of a recombinant MCR in a methanogen was for the ANME-1 MCR from the Black Sea mat (the same ANME-1 MCR for which the crystal structure is solved), which was expressed in M. acetivorans to engineer the methanogen to perform reverse methanogenesis using Fe(III) as an electron acceptor (Soo et al., 2016). The authors found that the strain expressing ANME-1 MCR consumed almost two times more methane compared to M. acetivorans with an empty vector. A further engineered air-adapted strain of M. acetivorans (Jasso-Chávez et al., 2015) containing ANME-1 MCR was used to generate electricity from methane in a microbial fuel cell containing other engineered microbes (McAnulty et al., 2017). Despite the critical importance of these studies toward the goal of activating methane for a range of biotechnology applications, it is still unclear whether the recombinant ANME MCR was necessary to significantly facilitate methane oxidation or, on a more fundamental level, how much of the recombinant MCR was produced in a complete and active form, especially since the methanogen lacks the 172-methylthio-F430 utilized by ANME-1 MCR. Notably, another investigation demonstrated that wild-type M. acetivorans is also capable of growth on methane using Fe(III) as an electron acceptor (Yan et al., 2018), indicating that the native methanogenic MCR is also capable of methane oxidation.
Another significant study described the heterologous expression of the MCR from M. okinawensis in M. maripaludis (Lyu et al., 2018b). The recombinant MCR was cloned into the traditional M. maripaludis protein expression plasmid under the control of PhmvA and with a his-tag on the C-terminus of McrA. This resulted in a highly expressed and uniformly assembled recombinant MCR as determined via SDS-PAGE and MALDI-MS. Additionally, expression of a MCR hybrid construct, consisting of mcrBDCG from M. okinawensis and mcrA from M. maripaludis resulted in a MCR consisting of the exact gene products from the hybrid construct without any components from the chromosomally encoded MCR. This supports the idea of an ordered MCR assembly, where MCR is simultaneously transcribed and translated (Lyu et al., 2018b). The PTMs were shown to be installed correctly for the heterologously produced M. okinawensis MCR and ~ 20% of the recombinant enzyme contained F430. Interestingly, the portion of the recombinant MCR that did not contain F430 was found to be associated with McrD, while the portion that did have F430 largely lacked McrD. This further supports a role for McrD in F430 delivery. Since only a fraction of the recombinant MCR contained F430, this suggests that potentially the F430 biosynthesis machinery cannot keep up with supplying F430 to the additional MCR being produced in the cell. Alternatively, the protein(s) potentially required to interact with McrD and/or MCR for F430 incorporation may not recognize the non-native McrD/MCR. If F430 biosynthesis was the bottleneck, one may expect that the native MCR would also have lower F430 incorporation. The authors found that the native MCR expression was not significantly affected in the presence of the recombinant MCR, but they did not report whether the F430 incorporation into native MCR was impaired. Finally, the purified heterologously produced MCR was found to exhibit low but detectable methane formation activity, where the authors point out that methods to activate M. marburgensis MCR in vitro do not appear to be effective for methanococcal MCR (Lyu et al., 2018b). It is important to note that M okinawensis and M. maripaludis are very closely related organisms, so it is unclear what the threshold of relatedness will be with respect to heterologous expression of MCRs from diverse organisms in model methanogens, such as M. maripaludis and M. acetivorans.
Although the field has seen many significant advancements since the initial discovery of MCR, many questions remain that need to be addressed, especially toward the development of robust heterologous expression systems for diverse MCRs and ACRs. Specifically, the functions of unique PTMs and F430 modifications need to be elucidated, which will require in vitro kinetic studies with mutated MCRs in the presence vs. absence of modified F430s. Further, the activation of MCR remains a poorly understood process, including the roles of specific proteins identified in complex A3a (Prakash et al., 2014) as well as the ATP costs of this process. There is still very little known about the putative accessory proteins necessary for MCR assembly—McrD may serve as an F430 chaperone for delivery to the MCR active site, but no other proteins potentially involved in assembly have been discovered. In terms of MCR kinetics and mechanism, it is important to emphasize that the vast majority of what is known results from studies on a single MCR from M. marburgensis. Since the substrate specificity and catalytic efficiency of even closely related enzymes can vary, it will be important to develop tools to study the enzymatic capabilities of other MCRs and related ACRs.
AG and KA wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
MCR research in the Allen lab is funded by the DOE Office of Science (DE-SC0022338).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Thank you to the reviewers for their critical comments and helpful suggestions that greatly improved the manuscript.
Adam, P. S., Borrel, G., Brochier-Armanet, C., and Gribaldo, S. (2017). The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11, 2407–2425. doi: 10.1038/ismej.2017.122
Akinyemi, T. S., Shao, N., Lyu, Z., Drake, I. J., Liu, Y., and Whitman, W. B. (2021). Tuning gene expression by phosphate in the methanogenic archaeon Methanococcus maripaludis. ACS Synth. Biol. 10, 3028–3039. doi: 10.1021/acssynbio.1c00322
Allen, K. D., Wegener, G., and White, R. H. (2014). Discovery of multiple modified F430 coenzymes in methanogens and anaerobic methanotrophic archaea suggests possible new roles for F430 in nature. Appl. Environ. Microbiol. 80, 6403–6412. doi: 10.1128/AEM.02202-14
Bao, J., and Scheller, S. (2021). Efficient CRISPR/Cas12a-based genome editing toolbox for metabolic engineering in Methanococcus maripaludis. [BioRxiv] doi: 10.1101/2021.12.29.474413
Bapteste, É., Brochier, C., and Boucher, Y. (2005). Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea 1:859728, 353–363. doi: 10.1155/2005/859728
Battistuzzi, F. U., Feijao, A., and Hedges, S. B. (2004). A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4:44. doi: 10.1186/1471-2148-4-44
Becker, D. F., and Ragsdale, S. W. (1998). Activation of methyl-SCoM reductase to high specific activity after treatment of whole cells with sodium sulfide. Biochemistry 37, 2639–2647. doi: 10.1021/bi972145x
Bhattarai, S., Cassarini, C., and Lens, P. N. L. (2019). Physiology and distribution of Archaeal Methanotrophs that couple anaerobic oxidation of methane with sulfate reduction. Microbiol. Mol. Biol. Rev. 83:e00074-18. doi: 10.1128/MMBR.00074-18
Bobik, T. A., Olson, K. D., Noll, K. M., and Wolfe, R. S. (1987). Evidence that the heterodisulfide of coenzyme M and 7-mercaptoheptanoylthreonine phosphate is a product of the methylreductase reaction in Methanobacterium. Biochem. Biophys. Res. Commun. 149, 455–460. doi: 10.1016/0006-291X(87)90389-5
Bonacker, L. G., Baudner, S., Mörschel, E., Bocher, R., and Thauer, R. K. (1993). Properties of the two isoenzymes of methyl-coenzyme M reductase in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 217, 587–595. doi: 10.1111/j.1432-1033.1993.tb18281.x
Bonacker, L. G., Baudner, S., and Thauer, R. K. (1992). Differential expression of the two methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum as determined immunochemically via isoenzyme-specific antisera. Eur. J. Biochem. 206, 87–92. doi: 10.1111/j.1432-1033.1992.tb16904.x
Borrel, G., Adam, P. S., McKay, L. J., Chen, L.-X., Sierra-García, I. N., Sieber, C. M. K., et al. (2019). Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat. Microbiol. 4, 603–613. doi: 10.1038/s41564-019-0363-3
Borrel, G., Brugère, J. F., Gribaldo, S., Schmitz, R. A., and Moissl-Eichinger, C. (2020). The host-associated archaeome. Nat. Rev. Microbiol. 18, 622–636. doi: 10.1038/s41579-020-0407-y
Boyd, J. A., Jungbluth, S. P., Leu, A. O., Evans, P. N., Woodcroft, B. J., Chadwick, G. L., et al. (2019). Divergent methyl-coenzyme M reductase genes in a deep-subseafloor Archaeoglobi. ISME J. 13, 1269–1279. doi: 10.1038/s41396-018-0343-2
Buan, N., Kulkarni, G., and Metcalf, W. (2011). Genetic methods for Methanosarcina species. Methods Enzymol. 494, 23–42. doi: 10.1016/B978-0-12-385112-3.00002-0
Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., et al. (1996). Complete genome sequence of the Methanogenic Archaeon, Methanococcus jannaschii. Science 273, 1058–1073. doi: 10.1126/science.273.5278.1058
Cedervall, P. E., Dey, M., Li, X., Sarangi, R., Hedman, B., Ragsdale, S. W., et al. (2011). Structural analysis of a Ni-methyl species in methyl-coenzyme M reductase from Methanothermobacter marburgensis. J. Am. Chem. Soc. 133, 5626–5628. doi: 10.1021/ja110492p
Chaban, B., Ng, S. Y., Kanbe, M., Saltzman, I., Nimmo, G., Aizawa, S., et al. (2007). Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol. 66, 596–609. doi: 10.1111/j.1365-2958.2007.05913.x
Chen, S. L., Blomberg, M. R., and Siegbahn, P. E. (2012). How is methane formed and oxidized reversibly when catalyzed by Ni-containing methyl-coenzyme M reductase? Chemistry 18, 6309–6315. doi: 10.1002/chem.201200274
Chen, S. L., Blomberg, M. R., and Siegbahn, P. E. (2014). An investigation of possible competing mechanisms for Ni-containing methyl-coenzyme M reductase. Phys. Chem. Chem. Phys. 16, 14029–14035. doi: 10.1039/c4cp01483a
Chen, H., Gan, Q., and Fan, C. (2020). Methyl-coenzyme M Reductase and its post-translational modifications. Front. Microbiol. 11:578356. doi: 10.3389/fmicb.2020.578356
Chen, S. C., Musat, N., Lechtenfeld, O. J., Paschke, H., Schmidt, M., Said, N., et al. (2019). Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep. Nature 568, 108–111. doi: 10.1038/s41586-019-1063-0
Conrad, R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292. doi: 10.1111/j.1758-2229.2009.00038.x
Conrado, R. J., and Gonzalez, R. (2014). Envisioning the bioconversion of methane to liquid fuels. Science 343, 621–623. doi: 10.1126/science.1246929
Costa, K. C., and Leigh, J. A. (2014). Metabolic versatility in methanogens. Curr. Opin. Biotechnol. 29, 70–75. doi: 10.1016/j.copbio.2014.02.012
Costa, K. C., Wong, P. M., Wang, T., Lie, T. J., Dodsworth, J. A., Swanson, I., et al. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U. S. A. 107, 11050–11055. doi: 10.1073/pnas.1003653107
Deobald, D., Adrian, L., Schöne, C., Rother, M., and Layer, G. (2018). Identification of a unique radical SAM methyltransferase required for the sp(3)-C-methylation of an arginine residue of methyl-coenzyme M reductase. Sci. Rep. 8:7404. doi: 10.1038/s41598-018-25716-x
Dey, M., Li, X., Zhou, Y., and Ragsdale, S. W. (2010). Evidence for organometallic intermediates in bacterial methane formation involving the nickel coenzyme F430. Met. Ions Life Sci. 7, 71–110. doi: 10.1039/BK9781847551771-00071
Dolfing, J., Jiang, B., Henstra, A. M., Stams, A. J., and Plugge, C. M. (2008). Syntrophic growth on formate: a new microbial niche in anoxic environments. Appl. Environ. Microbiol. 74, 6126–6131. doi: 10.1128/AEM.01428-08
Duin, E. C., Prakash, D., and Brungess, C. (2011). Methyl-coenzyme M reductase from Methanothermobacter marburgensis. Methods Enzymol. 494, 159–187. doi: 10.1016/B978-0-12-385112-3.00009-3
Duin, E. C., Wagner, T., Shima, S., Prakash, D., Cronin, B., Yáñez-Ruiz, D. R., et al. (2016). Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. Proc. Natl. Acad. Sci. U. S. A. 113, 6172–6177. doi: 10.1073/pnas.1600298113
Ehlers, C., Jäger, D., and Schmitz, R. A. (2011). Establishing a markerless genetic exchange system for Methanosarcina mazei strain Gö1 for constructing chromosomal mutants of small RNA genes. Archaea 2011:439608. doi: 10.1155/2011/439608
Ehlers, C., Weidenbach, K., Veit, K., Deppenmeier, U., Metcalf, W. W., and Schmitz, R. A. (2005). Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol. Gen. Genomics. 273, 290–298. doi: 10.1007/s00438-005-1128-7
Ellefson, W. L., Whitman, W. B., and Wolfe, R. S. (1982). Nickel-containing factor F430: chromophore of the methylreductase of Methanobacterium. Proc. Natl. Acad. Sci. U. S. A. 79, 3707–3710. doi: 10.1073/pnas.79.12.3707
Ellermann, J., Hedderich, R., Böcher, R., and Thauer, R. K. (1988). The final step in methane formation. Investigations with highly purified methyl-CoM reductase (component C) from Methanobacterium thermoautotrophicum (strain Marburg). Eur. J. Biochem. 172, 669–677. doi: 10.1111/j.1432-1033.1988.tb13941.x
Ellermann, J., Kobelt, A., Pfaltz, A., and Thauer, R. K. (1987). On the role of N-7-mercaptoheptanoyl-O-phospho-L-threonine (component B) in the enzymatic reduction of methyl-coenzyme M to methane. FEBS Lett. 220, 358–362. doi: 10.1016/0014-5793(87)80846-3
Ermler, U., Grabarse, W., Shima, S., Goubeaud, M., and Thauer, R. K. (1997). Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278, 1457–1462.
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., Golding, S. D., et al. (2015). Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438. doi: 10.1126/science.aac7745
Farber, G., Keller, W., Kratky, C., Jaun, B., Pfaltz, A., Spinner, C., et al. (1991). Coenzyme F430 from methanogenic bacteria–complete assignment of configuration based on an X-ray-analysis of 12,13-Diepi-F430 Pentamethyl Ester and on Nmr-spectroscopy. Helv. Chim. Acta 74, 697–716. doi: 10.1002/hlca.19910740404
Fink, C., Beblawy, S., Enkerlin, A. M., Mühling, L., Angenent, L. T., Molitor, B., et al. (2021). A shuttle-vector system allows heterologous gene expression in the thermophilic methanogen Methanothermobacter thermautotrophicus ΔH. mBio 12:e0276621. doi: 10.1128/mBio.02766-21
Fonseca, D. R., Halim, M. F. A., Holten, M. P., and Costa, K. C. (2020). Type IV-like Pili facilitate transformation in naturally competent Archaea. J. Bacteriol. 202:e00355-20. doi: 10.1128/JB.00355-20
Goenrich, M., Duin, E. C., Mahlert, F., and Thauer, R. K. (2005). Temperature dependence of methyl-coenzyme M reductase activity and of the formation of the methyl-coenzyme M reductase red2 state induced by coenzyme B. J. Biol. Inorg. Chem. 10, 333–342. doi: 10.1007/s00775-005-0636-6
Goubeaud, M., Schreiner, G., and Thauer, R. K. (1997). Purified methyl-coenzyme-M reductase is activated when the enzyme-bound coenzyme F430 is reduced to the nickel(I) oxidation state by titanium(III) citrate. Eur. J. Biochem. 243, 110–114. doi: 10.1111/j.1432-1033.1997.00110.x
Grabarse, W., Mahlert, F., Shima, S., Thauer, R. K., and Ermler, U. (2000). Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J. Mol. Biol. 303, 329–344. doi: 10.1006/jmbi.2000.4136.
Gunsalus, R. P., and Wolfe, R. S. (1976). Components and cofactors for enzymatic formation of methane from methyl-coenzyme-M. Fed. Proc. 35:1547
Guss, A. M., Rother, M., Zhang, J. K., Kulkkarni, G., and Metcalf, W. W. (2008). New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2, 193–203. doi: 10.1155/2008/534081
Hahn, C. J., Laso-Pérez, R., Vulcano, F., Vaziourakis, K. M., Stokke, R., Steen, I. H., et al. (2020). “Candidatus Ethanoperedens,” a Thermophilic genus of Archaea mediating the anaerobic oxidation of ethane. mBio 11:e00600-20. doi: 10.1128/mBio.00600-20
Hahn, C. J., Lemaire, O. N., Kahnt, J., Engilberge, S., Wegener, G., and Wagner, T. (2021). Crystal structure of a key enzyme for anaerobic ethane activation. Science 373, 118–121. doi: 10.1126/science.abg1765
Hallam, S. J., Putnam, N., Preston, C. M., Detter, J. C., Rokhsar, D., Richardson, P. M., et al. (2004). Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462. doi: 10.1126/science.1100025
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Hartzell, P., and Reed, D. W. (2006). “The genus Archaeoglobus,” in The Prokaryotes: Archaea bacteria: Firmicutes, Actinomycetes. Vol. 3, eds. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 82–100.
Haynes, C. A., and Gonzalez, R. (2014). Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 10, 331–339. doi: 10.1038/nchembio.1509
He, Z., Zhang, Q., Feng, Y., Luo, H., Pan, X., and Gadd, G. M. (2018). Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci. Total Environ. 610-611, 759–768. doi: 10.1016/j.scitotenv.2017.08.140
Hinrichs, K.-U., Hayes, J. M., Sylva, S. P., Brewer, P. G., and DeLong, E. F. (1999). Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805. doi: 10.1038/19751
Holler, T., Widdel, F., Knittel, K., Amann, R., Kellermann, M. Y., Hinrichs, K. U., et al. (2011). Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 5, 1946–1956. doi: 10.1038/ismej.2011.77
Jackson, R. B., Saunois, M., Bousquet, P., Canadell, J. G., Poulter, B., Stavert, A. R., et al. (2020). Increasing anthropogenic methane emissions arise equally from agricultural and fossil fuel sources. Environ. Res. Lett. 15:071002. doi: 10.1088/1748-9326/ab9ed2
Jasso-Chávez, R., Santiago-Martinez, M. G., Lira-Silva, E., Pineda, E., Zepeda-Rodriguez, A., Belmont-Diaz, J., et al. (2015). Air-adapted Methanosarcina acetivorans shows high methane production and develops resistance against oxygen stress. PLoS One 10:e0117331. doi: 10.1371/journal.pone.0117331
Jones, W. J., Leigh, J. A., Mayer, F., Woese, C. R., and Wolfe, R. S. (1983). Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136, 254–261. doi: 10.1007/BF00425213
Kahnt, J., Buchenau, B., Mahlert, F., Krüger, M., Shima, S., and Thauer, R. K. (2007). Post-translational modifications in the active site region of methyl-coenzyme M reductase from methanogenic and methanotrophic archaea. FEBS J. 274, 4913–4921. doi: 10.1111/j.1742-4658.2007.06016.x
Kaneko, M., Takano, Y., Chikaraishi, Y., Ogawa, N. O., Asakawa, S., Watanabe, T., et al. (2014). Quantitative analysis of coenzyme F430 in environmental samples: a new diagnostic tool for methanogenesis and anaerobic methane oxidation. Anal. Chem. 86, 3633–3638. doi: 10.1021/ac500305j
Kaster, A. K., Goenrich, M., Seedorf, H., Liesegang, H., Wollherr, A., Gottschalk, G., et al. (2011). More than 200 genes required for methane formation from H2 and CO2 and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea 2011:973848. doi: 10.1155/2011/973848
Kirschke, S., Bousquet, P., Ciais, P., Saunois, M., Canadell, J. G., Dlugokencky, E. J., et al. (2013). Three decades of global methane sources and sinks. Nat. Geosci. 6, 813–823. doi: 10.1038/ngeo1955
Klenk, H.-P., Clayton, R. A., Tomb, J.-F., White, O., Nelson, K. E., Ketchum, K. A., et al. (1997). The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390, 364–370. doi: 10.1038/37052
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479. doi: 10.1128/AEM.71.1.467-479.2005
Kurth, J. M., Muller, M. C., Welte, C. U., and Wagner, T. (2021b). Structural insights into the methane-generating enzyme from a methoxydotrophic methanogen reveal a restrained gallery of post-translational modifications. Microorganisms. 9:837. doi: 10.3390/microorganisms9040837
Kurth, J. M., Nobu, M. K., Tamaki, H., de Jonge, N., Berger, S., Jetten, M. S. M., et al. (2021a). Methanogenic archaea use a bacteria-like methyltransferase system to demethoxylate aromatic compounds. ISME J. 15, 3549–3565. doi: 10.1038/s41396-021-01025-6
Kurth, J. M., Op den Camp, H. J. M., and Welte, C. U. (2020). Several ways one goal-methanogenesis from unconventional substrates. Appl. Microbiol. Biotechnol. 104, 6839–6854. doi: 10.1007/s00253-020-10724-7
Laso-Pérez, R., Krukenberg, V., Musat, F., and Wegener, G. (2018). Establishing anaerobic hydrocarbon-degrading enrichment cultures of microorganisms under strictly anoxic conditions. Nat. Protoc. 13, 1310–1330. doi: 10.1038/nprot.2018.030
Laso-Pérez, R., Wegener, G., Knittel, K., Widdel, F., Harding, K. J., Krukenberg, V., et al. (2016). Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401. doi: 10.1038/nature20152
Lawton, T. J., and Rosenzweig, A. C. (2016a). Biocatalysts for methane conversion: big progress on breaking a small substrate. Curr. Opin. Chem. Biol. 35, 142–149. doi: 10.1016/j.cbpa.2016.10.001
Lawton, T. J., and Rosenzweig, A. C. (2016b). Methane-oxidizing enzymes: an upstream problem in biological gas-to-liquids conversion. J. Am. Chem. Soc. 138, 9327–9340. doi: 10.1021/jacs.6b04568
Lie, T. J., and Leigh, J. A. (2002). Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source. J. Bacteriol. 184, 5301–5306. doi: 10.1128/JB.184.19.5301-5306.2002
Liu, Y. F., Chen, J., Zaramela, L. S., Wang, L. Y., Mbadinga, S. M., Hou, Z. W., et al. (2020). Genomic and Transcriptomic evidence supports methane metabolism in Archaeoglobi. mSystems 5:e00651-19. doi: 10.1128/mSystems.00651-19
Liu, Y., and Whitman, W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Livingston, D. A., Pfaltz, A., Schreiber, J., Eschenmoser, A., Ankelfuchs, D., Moll, J., et al. (1984). Factor-F430 from Methanogenic bacteria-structure of the protein-free factor. Helv. Chim. Acta 67, 334–351. doi: 10.1002/hlca.19840670141
Long, F., Wang, L. L., Lupa, B., and Whitman, W. B. (2017). A flexible system for cultivation of Methanococcus and other formate-utilizing methanogens. Archaea 2017, 1–12. doi: 10.1155/2017/7046026
Lyu, Z., Chou, C. W., Shi, H., Wang, L., Ghebreab, R., Phillips, D., et al. (2018b). Assembly of methyl coenzyme M Reductase in the Methanogenic Archaeon Methanococcus maripaludis. J. Bacteriol. 200:e00746-17. doi: 10.1128/JB.00746-17
Lyu, Z., Shao, N., Akinyemi, T., and Whitman, W. B. (2018a). Methanogenesis. Curr. Biol. 28, R727–R732. doi: 10.1016/j.cub.2018.05.021
Lyu, Z., Shao, N., Chou, C. W., Shi, H., Patel, R., Duin, E. C., et al. (2020). Posttranslational methylation of arginine in methyl coenzyme M Reductase has a profound impact on both methanogenesis and growth of Methanococcus maripaludis. J. Bacteriol. 202:e00654-19. doi: 10.1128/JB.00654-19
Lyu, Z., and Whitman, W. B. (2017). Evolution of the archaeal and mammalian information processing systems: towards an archaeal model for human disease. Cell. Mol. Life Sci. 74, 183–212. doi: 10.1007/s00018-016-2286-y
Macauley, S. R., Zimmerman, S. A., Apolinario, E. E., Evilia, C., Hou, Y. M., Ferry, J. G., et al. (2009). The archetype gamma-class carbonic anhydrase (cam) contains iron when synthesized in vivo. Biochemistry 48, 817–819. doi: 10.1021/bi802246s
Mayr, S., Latkoczy, C., Krüger, M., Günther, D., Shima, S., Thauer, R. K., et al. (2008). Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J. Am. Chem. Soc. 130, 10758–10767. doi: 10.1021/ja802929z
Mayumi, D., Mochimaru, H., Tamaki, H., Yamamoto, K., Yoshioka, H., Suzuki, Y., et al. (2016). Methane production from coal by a single methanogen. Science 354, 222–225. doi: 10.1126/science.aaf8821
McAnulty, M. J., Poosarla, V. G., Kim, K. Y., Jasso-Chavez, R., Logan, B. E., and Wood, T. K. (2017). Electricity from methane by reversing methanogenesis. Nat. Commun. 8:15419. doi: 10.1038/ncomms15419
McBride, B. C., and Wolfe, R. S. (1971). A new coenzyme of methyl transfer, coenzyme M. Biochemistry 10, 2317–2324. doi: 10.1021/bi00788a022
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
McKay, L. J., Dlakić, M., Fields, M. W., Delmont, T. O., Eren, A. M., Jay, Z. J., et al. (2019). Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat. Microbiol. 4, 614–622. doi: 10.1038/s41564-019-0362-4
Metcalf, W. W., Zhang, J. K., Apolinario, E., Sowers, K. R., and Wolfe, R. S. (1997). A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. U. S. A. 94, 2626–2631.
Meyerdierks, A., Kube, M., Kostadinov, I., Teeling, H., Glöckner, F. O., Reinhardt, R., et al. (2010). Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 12, 422–439. doi: 10.1111/j.1462-2920.2009.02083.x
Moissl-Eichinger, C., Pausan, M., Taffner, J., Berg, G., Bang, C., and Schmitz, R. A. (2018). Archaea are interactive components of complex microbiomes. Trends Microbiol. 26, 70–85. doi: 10.1016/j.tim.2017.07.004
Mondorf, S., Deppenmeier, U., and Welte, C. (2012). A novel inducible protein production system and neomycin resistance as selection marker for Methanosarcina mazei. Archaea 2012:973743. doi: 10.1155/2012/973743
Montzka, S. A., Dlugokencky, E. J., and Butler, J. H. (2011). Non-CO2 greenhouse gases and climate change. Nature 476, 43–50. doi: 10.1038/nature10322
Moore, B. C., and Leigh, J. A. (2005). Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187, 972–979. doi: 10.1128/JB.187.3.972-979.2005
Moore, S. J., Sowa, S. T., Schuchardt, C., Deery, E., Lawrence, A. D., Ramos, J. V., et al. (2017). Elucidation of the biosynthesis of the methane catalyst coenzyme F430. Nature 543, 78–82. doi: 10.1038/nature21427
Mueller, T. J., Grisewood, M. J., Nazem-Bokaee, H., Gopalakrishnan, S., Ferry, J. G., Wood, T. K., et al. (2015). Methane oxidation by anaerobic archaea for conversion to liquid fuels. J. Ind. Microbiol. Biotechnol. 42, 391–401. doi: 10.1007/s10295-014-1548-7
Nauhaus, K., Boetius, A., Kruger, M., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4, 296–305. doi: 10.1046/j.1462-2920.2002.00299.x
Nayak, D. D., Liu, A., Agrawal, N., Rodriguez-Carerro, R., Dong, S. H., Mitchell, D. A., et al. (2020). Functional interactions between posttranslationally modified amino acids of methyl-coenzyme M reductase in Methanosarcina acetivorans. PLoS Biol. 18:e3000507. doi: 10.1371/journal.pbio.3000507
Nayak, D. D., Mahanta, N., Mitchell, D. A., and Metcalf, W. W. (2017). Post-translational thioamidation of methyl-coenzyme M reductase, a key enzyme in methanogenic and methanotrophic Archaea. elife 6. doi: 10.7554/eLife.29218
Nayak, D. D., and Metcalf, W. W. (2017). Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc. Natl. Acad. Sci. U. S. A. 114, 2976–2981. doi: 10.1073/pnas.1618596114
Nayak, D. D., and Metcalf, W. W. (2018). Genetic techniques for studies of methyl-coenzyme M reductase from Methanosarcina acetivorans C2A. Methods Enzymol. 613, 325–347. doi: 10.1016/bs.mie.2018.10.012
Niemann, H., Lösekann, T., de Beer, D., Elvert, M., Nadalig, T., Knittel, K., et al. (2006). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858. doi: 10.1038/nature05227
Orphan, V. J., Hinrichs, K. U., Ussler, W. 3rd, Paull, C. K., Taylor, L. T., Sylva, S. P., et al. (2001). Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D., and DeLong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U. S. A. 99, 7663–7668. doi: 10.1073/pnas.072210299
Patwardhan, A., Sarangi, R., Ginovska, B., Raugei, S., and Ragsdale, S. W. (2021). Nickel-Sulfonate mode of substrate binding for forward and reverse reactions of methyl-SCoM Reductase suggest a radical mechanism involving Long-range electron transfer. J. Am. Chem. Soc. 143, 5481–5496. doi: 10.1021/jacs.1c01086
Pelmenschikov, V., Blomberg, M. R., Siegbahn, P. E., and Crabtree, R. H. (2002). A mechanism from quantum chemical studies for methane formation in methanogenesis. J. Am. Chem. Soc. 124, 4039–4049. doi: 10.1021/ja011664r
Pelmenschikov, V., and Siegbahn, P. E. M. (2003). Catalysis by methyl-coenzyme M reductase: a theoretical study for heterodisulfide product formation. J. Biol. Inorg. Chem. 8, 653–662. doi: 10.1007/s00775-003-0461-8
Pfaltz, A., Jaun, B., Fassler, A., Eschenmoser, A., Jaenchen, R., Gilles, H. H., et al. (1982). Factor-F430 from Methanogenic bacteria–structure of the porphinoid ligand system. Helv. Chim. Acta 65, 828–865. doi: 10.1002/hlca.19820650320
Pfeifer, K., Ergal, İ., Koller, M., Basen, M., Schuster, B., and Simon, K.-M. R. (2021). Archaea biotechnology. Biotechnol. Adv. 47:107668. doi: 10.1016/j.biotechadv.2020.107668
Prakash, D., Wu, Y., Suh, S. J., and Duin, E. C. (2014). Elucidating the process of activation of methyl-coenzyme M reductase. J. Bacteriol. 196, 2491–2498. doi: 10.1128/JB.01658-14
Pritchett, M. A., Zhang, J. K., and Metcalf, W. W. (2004). Development of a Markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for Methanogenic Archaea. Appl. Environ. Microbiol. 70, 1425–1433. doi: 10.1128/AEM.70.3.1425-1433.2004
Radle, M. I., Miller, D. V., Laremore, T. N., and Booker, S. J. (2019). Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical S-adenosylmethionine methylase that unexpectedly requires cobalamin. J. Biol. Chem. 294, 11712–11725. doi: 10.1074/jbc.RA119.007609
Raghoebarsing, A. A., Pol, A., van de Pas-Schoonen, K. T., Smolders, A. J. P., Ettwig, K. F., Rijpstra, W. I. C., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921. doi: 10.1038/nature04617
Ragsdale, S. W., Raugei, S., Ginovska, B., and Wongnate, T. (2017). Biochemistry of methyl-coenzyme M Reductase. Rsc. Metallobio. Ser. 10, 149–169. doi: 10.1039/9781788010580-00149
Rospert, S., Bocher, R., Albracht, S. P., and Thauer, R. K. (1991). Methyl-coenzyme M reductase preparations with high specific activity from H2-preincubated cells of Methanobacterium thermoautotrophicum. FEBS Lett. 291, 371–375.
Rospert, S., Linder, D., Ellermann, J., and Thauer, R. K. (1990). Two genetically distinct methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum strain Marburg and ΔH. Eur. J. Biochem. 194, 871–877. doi: 10.1111/j.1432-1033.1990.tb19481.x
Sarmiento, F., Leigh, J. A., and Whitman, W. B. (2011). Genetic systems for hydrogenotrophic methanogens. Methods Enzymol. 494, 43–73. doi: 10.1016/B978-0-12-385112-3.00003-2
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608. doi: 10.1038/nature09015
Scheller, S., Goenrich, M., Thauer, R. K., and Jaun, B. (2013). Methyl-coenzyme M reductase from methanogenic archaea: isotope effects on the formation and anaerobic oxidation of methane. J. Am. Chem. Soc. 135, 14975–14984. doi: 10.1021/ja406485z
Shea, M. T., Walter, M. E., Duszenko, N., Ducluzeau, A.-L., Aldridge, J., King, S. K., et al. (2016). pNEB193-derived suicide plasmids for gene deletion and protein expression in the methane-producing archaeon, Methanosarcina acetivorans. Plasmid 84-85, 27–35. doi: 10.1016/j.plasmid.2016.02.003
Sherf, B. A., and Reeve, J. N. (1990). Identification of the mcrD gene product and its association with component C of methyl coenzyme M reductase in Methanococcus vannielii. J. Bacteriol. 172, 1828–1833.
Shima, S., Krueger, M., Weinert, T., Demmer, U., Kahnt, J., Thauer, R. K., et al. (2012). Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481, 98–101. doi: 10.1038/nature10663
Shima, S., and Thauer, R. K. (2005). Methyl-coenzyme M reductase and the anaerobic oxidation of methane in methanotrophic Archaea. Curr. Opin. Microbiol. 8, 643–648. doi: 10.1016/j.mib.2005.10.002
Soo, V. W., McAnulty, M. J., Tripathi, A., Zhu, F., Zhang, L., Hatzakis, E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Factories 15:11. doi: 10.1186/s12934-015-0397-z
Stokke, R., Roalkvam, I., Lanzen, A., Haflidason, H., and Steen, I. H. (2012). Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments. Environ. Microbiol. 14, 1333–1346. doi: 10.1111/j.1462-2920.2012.02716.x
Susanti, D., Frazier, M. C., and Mukhopadhyay, B. (2019). A genetic system for Methanocaldococcus jannaschii: An evolutionary deeply rooted Hyperthermophilic Methanarchaeon. Front. Microbiol. 10:1256. doi: 10.3389/fmicb.2019.01256
Thauer, R. K. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson prize lecture. Microbiology 144, 2377–2406. doi: 10.1099/00221287-144-9-2377
Thauer, R. K. (2019). Methyl (alkyl)-coenzyme M Reductases: nickel F430-containing enzymes involved in anaerobic methane formation and in anaerobic oxidation of methane or of short chain alkanes. Biochemistry 58, 5198–5220. doi: 10.1021/acs.biochem.9b00164
Thema, M., Weidlich, T., Hörl, M., Bellack, A., Mörs, F., Hackl, F., et al. (2019). Biological CO2-methanation: an approach to standardization. Energies 12:1670. doi: 10.3390/en12091670
Timmers, P. H., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S., and Stams, A. J. (2017). Reverse Methanogenesis and respiration in Methanotrophic Archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237
Tumbula, D. L., Makula, R. A., and Whitman, W. B. (1994). Transformation of Methanococcus maripaludis and identification of a Psti-Like restriction system. FEMS Microbiol. Lett. 121, 309–314. doi: 10.1111/j.1574-6968.1994.tb07118.x
Vanwonterghem, I., Evans, P. N., Parks, D. H., Jensen, P. D., Woodcroft, B. J., Hugenholtz, P., et al. (2016). Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 1:16170. doi: 10.1038/nmicrobiol.2016.170
Wagner, T., Kahnt, J., Ermler, U., and Shima, S. (2016). Didehydroaspartate modification in methyl-coenzyme M Reductase catalyzing methane formation. Angew. Chem. Int. Ed. Engl. 55, 10630–10633. doi: 10.1002/anie.201603882
Wagner, T., Wegner, C.-E., Kahnt, J., Ermler, U., and Shima, S. (2017). Phylogenetic and structural comparisons of the three types of methyl coenzyme M Reductase from Methanococcales and Methanobacteriales. J. Bacteriol. 199:e00197-17. doi: 10.1128/JB.00197-17
Wang, Y., Wegener, G., Ruff, S. E., and Wang, F. (2021). Methyl/alkyl-coenzyme M reductase-based anaerobic alkane oxidation in archaea. Environ. Microbiol. 23, 530–541. doi: 10.1111/1462-2920.15057
Wegener, G., Krukenberg, V., Riedel, D., Tegetmeyer, H. E., and Boetius, A. (2015). Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590. doi: 10.1038/nature15733
Wegener, G., Krukenberg, V., Ruff, S. E., Kellermann, M. Y., and Knittel, K. (2016). Metabolic capabilities of microorganisms involved in and associated with the anaerobic oxidation of methane. Front. Microbiol. 7:46. doi: 10.3389/fmicb.2016.00046
Whitman, W. B., Shieh, J., Sohn, S., Caras, D. S., and Premachandran, U. (1986). Isolation and characterization of 22 Mesophilic Methanococci. Syst. Appl. Microbiol. 7, 235–240. doi: 10.1016/S0723-2020(86)80012-1
Whitman, W. B., Tumbula, D. L., Yu, J. P., and Kim, W. (1997). Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors 6, 37–46.
Wongnate, T., Sliwa, D., Ginovska, B., Smith, D., Wolf, M. W., Lehnert, N., et al. (2016). The radical mechanism of biological methane synthesis by methyl-coenzyme M reductase. Science 352, 953–958. doi: 10.1126/science.aaf0616
Yan, Z., and Ferry, J. G. (2018). Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front. Microbiol. 9:1322. doi: 10.3389/fmicb.2018.01322
Yan, Z., Joshi, P., Gorski, C. A., and Ferry, J. G. (2018). A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 9:1642. doi: 10.1038/s41467-018-04097-9
Yang, N., Reiher, M., Wang, M., Harmer, J., and Duin, E. C. (2007). Formation of a nickel-methyl species in methyl-coenzyme m reductase, an enzyme catalyzing methane formation. J. Am. Chem. Soc. 129, 11028–11029. doi: 10.1021/ja0734501
Yu, G., Beauchemin, K. A., and Dong, R. (2021). A review of 3-Nitrooxypropanol for enteric methane mitigation from ruminant livestock. Animals 11:11. doi: 10.3390/ani11123540
Zheng, K., Ngo, P. D., Owens, V. L., Yang, X. P., and Mansoorabadi, S. O. (2016). The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science 354, 339–342. doi: 10.1126/science.aag2947
Zhou, Y., Dorchak, A. E., and Ragsdale, S. W. (2013). In vivo activation of methyl-coenzyme M reductase by carbon monoxide. Front. Microbiol. 4:69. doi: 10.3389/fmicb.2013.00069
Keywords: methyl-coenzyme M reductase, MCR, methanogens, anaerobic methanotrophic archaea, ANME
Citation: Gendron A and Allen KD (2022) Overview of Diverse Methyl/Alkyl-Coenzyme M Reductases and Considerations for Their Potential Heterologous Expression. Front. Microbiol. 13:867342. doi: 10.3389/fmicb.2022.867342
Received: 01 February 2022; Accepted: 01 April 2022;
Published: 25 April 2022.
Edited by:
Cornelia Welte, Radboud University Nijmegen, NetherlandsReviewed by:
Tristan Wagner, Max Planck Society, GermanyCopyright © 2022 Gendron and Allen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kylie D. Allen, a2RhbGxlbkB2dC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.