
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 04 May 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.864178
This article is part of the Research TopicBiological Rotary NanomotorsView all 21 articles
Most motile bacteria utilize the flagellar type III secretion system (fT3SS) to construct the flagellum, which is a supramolecular motility machine consisting of basal body rings and an axial structure. Each axial protein is translocated via the fT3SS across the cytoplasmic membrane, diffuses down the central channel of the growing flagellar structure and assembles at the distal end. The fT3SS consists of a transmembrane export complex and a cytoplasmic ATPase ring complex with a stoichiometry of 12 FliH, 6 FliI and 1 FliJ. This complex is structurally similar to the cytoplasmic part of the FOF1 ATP synthase. The export complex requires the FliH12-FliI6-FliJ1 ring complex to serve as an active protein transporter. The FliI6 ring has six catalytic sites and hydrolyzes ATP at an interface between FliI subunits. FliJ binds to the center of the FliI6 ring and acts as the central stalk to activate the export complex. The FliH dimer binds to the N-terminal domain of each of the six FliI subunits and anchors the FliI6-FliJ1 ring to the base of the flagellum. In addition, FliI exists as a hetero-trimer with the FliH dimer in the cytoplasm. The rapid association-dissociation cycle of this hetero-trimer with the docking platform of the export complex promotes sequential transfer of export substrates from the cytoplasm to the export gate for high-speed protein transport. In this article, we review our current understanding of multiple roles played by the flagellar cytoplasmic ATPase complex during efficient flagellar assembly.
Pathogenic bacteria use virulence-associated type III secretion systems (vT3SS), also known as the injectisomes, to inject virulence effector proteins directly into eukaryotic host cells as part of their infection process. Motile bacteria employ the flagellar type III secretion system (fT3SS) to construct a supramolecular motility machine, the flagellum, on the cell surface (Wagner and Diepold, 2020). A remarkable feature of both the vT3SS and fT3SS is that the protein export apparatus is capable of translocating export substrates across the cytoplasmic membrane at a rate of tens of thousands of amino acids per second (Iino, 1974; Chen et al., 2017; Renault et al., 2017). The protein export apparatus of the T3SS is composed of a transmembrane export complex powered by the proton motive force (PMF) across the cytoplasmic membrane and a cytoplasmic ATPase ring complex (Figure 1). The transmembrane export complex is composed of five conserved membrane proteins: FlhA, FlhB, FliP, FliQ, and FliR in the fT3SS; SctV, SctU, SctR, SctS, and SctT in the vT3SS. The cytoplasmic ATPase ring complex is composed of three cytoplasmic proteins, FliH, FliI, and FliJ in the fT3SS and SctL, SctN, and SctO in the vT3SS. The ATPase ring complex is structurally similar to the cytoplasmic part of the FOF1 ATP synthase, which is a rotary motor that couples proton (H+) flow through FO with ATP synthesis by F1 (Minamino, 2014; Minamino et al., 2020b).
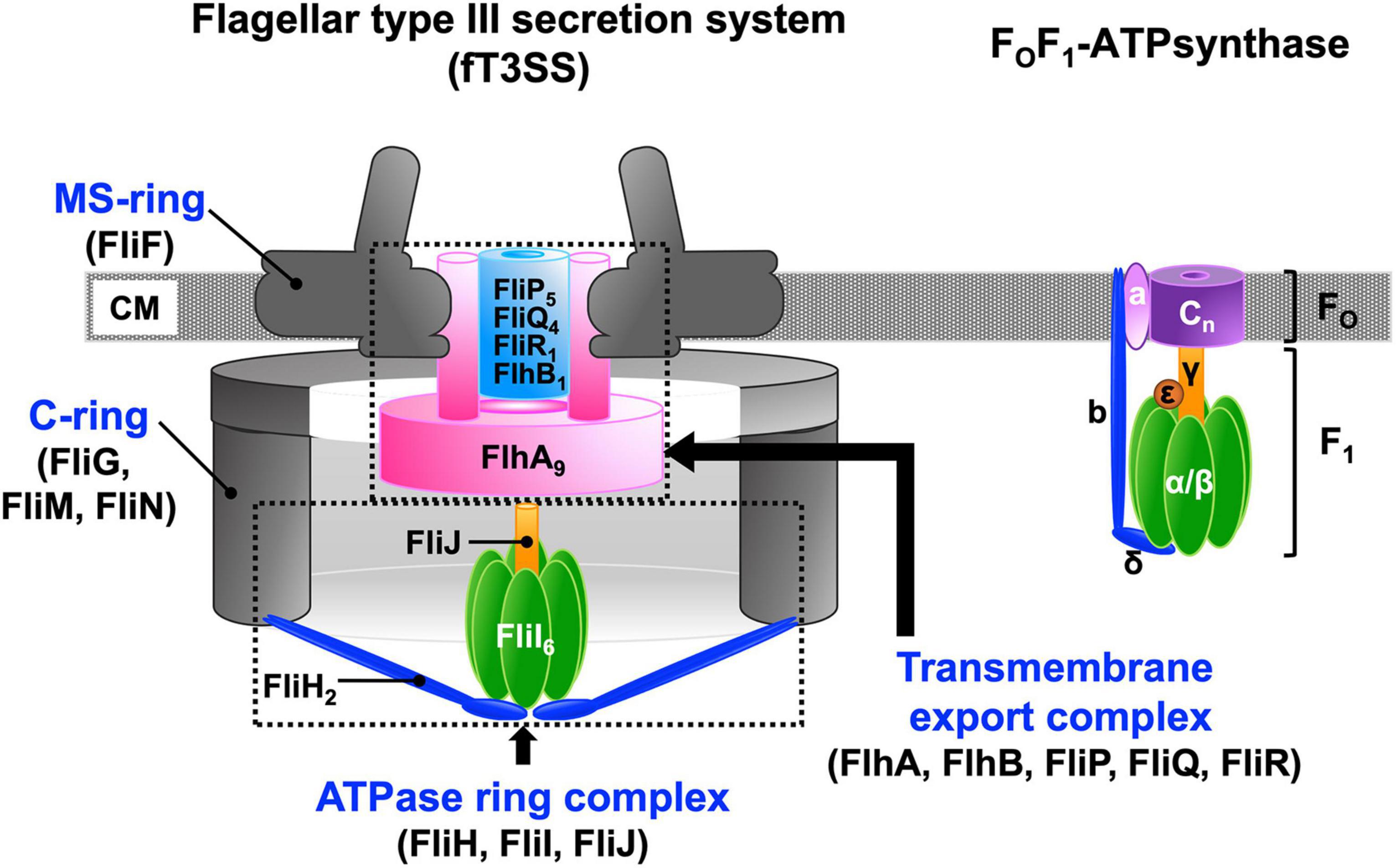
Figure 1. Schematic diagrams of the flagellar type III export apparatus and FOF ATP synthase. The flagellar type III secretion system (fT3SS) is composed of five membrane proteins, FlhA, FlhB, FliP, FliQ, and FliR and three cytoplasmic proteins, FliH, FliI, and FliJ. FlhA, FlhB, FliP, FliQ and FliR assembles into a transmembrane export complex within the MS-ring of the basal body of the flagellum. FliH, FliI, and FliJ form a cytoplasmic ATPase ring. The FliI6-FliJ1 ring complex is structurally similar to the α3β3γ1 ring complex of the FOF1 ATP synthase. The N-terminal and C-terminal domains of FliH structurally are similar in structure to the b and δ subunits, respectively, of the FOF1 ATP synthase. The FliH dimer acts as a peripheral stalk that anchors the FliI6-FliJ ring complex to the base of the flagellum in a similar manner as the b and δ subunits of the FOF1 ATP synthase connect the α3β3γ ring complex to membrane-embedded FO. The stoichiometry of the c-ring varies dramatically from c8 up to at least c15. CM, cytoplasmic membrane.
The flagellum of Salmonella enterica serovar Typhimurium (hereafter referred to as Salmonella) is composed of about 30 different proteins whose copy numbers range from a few to tens of thousands. The Salmonella flagellum is divided into three main structural parts: the basal body, the hook, and the filament (Figure 2). The basal body is located within the cell envelop and serves as a bi-directional rotary motor fueled by the PMF across the cytoplasmic membrane. The hook and filament extend into the cell exterior. The filament functions as a helical propeller to produce the thrust that pushes the cell body forward. The hook between the basal body and filament acts as a universal joint to transmit torque produced by the motor to the filament (Nakamura and Minamino, 2019).
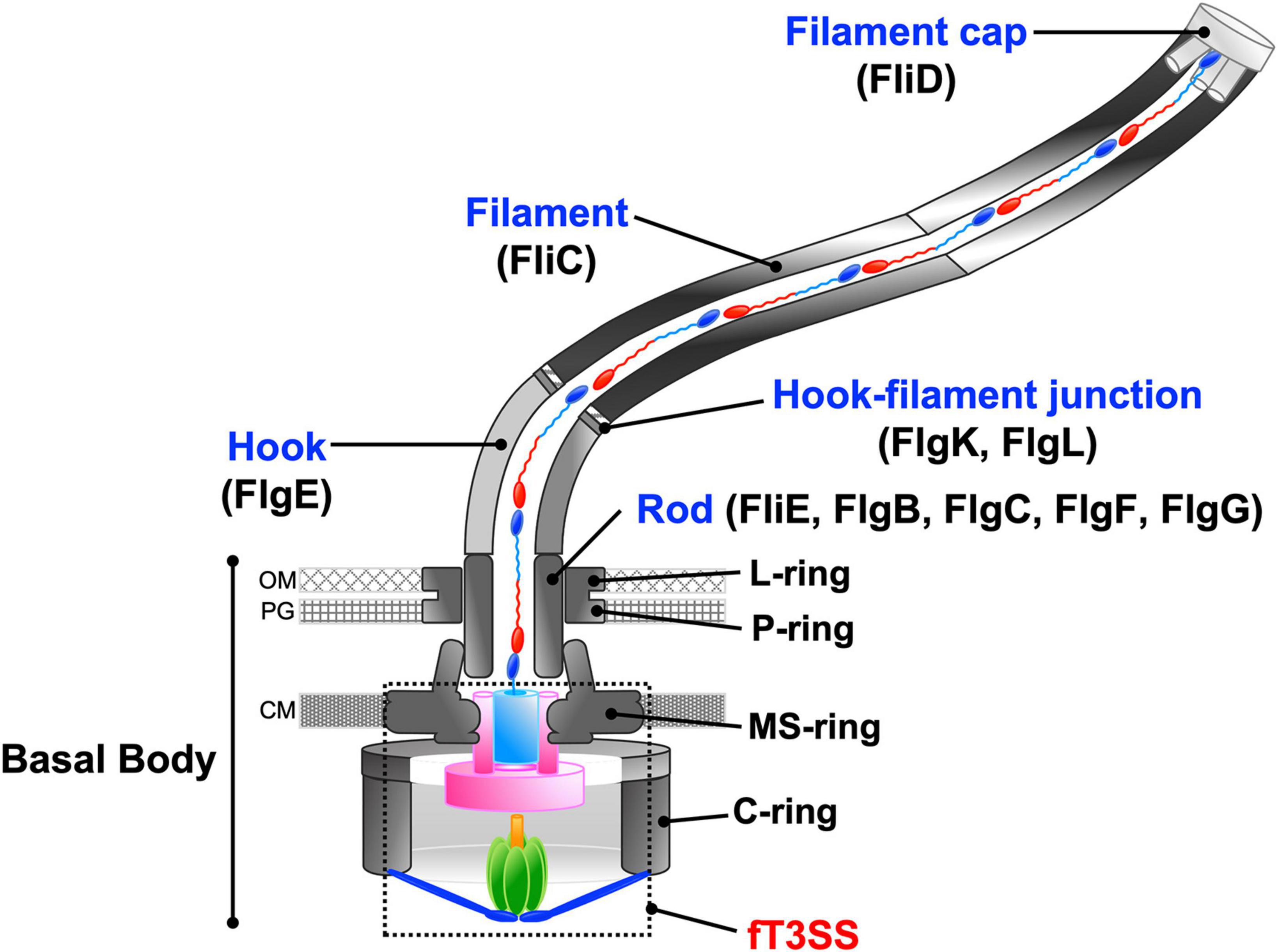
Figure 2. Schematic diagram of the bacterial flagellum. The bacterial flagellum is composed of basal body rings, namely the C-ring, MS-ring, L-ring, and P-ring, and an axial structure consisting of the rod, the hook, the hook-filament junction, the filament, and the filament cap. To construct the axial structure beyond the cytoplasmic membrane, flagellar axial proteins are translocated through the fT3SS, diffuse down a narrow central channel, and assemble at the tip of the growing structure. OM, outer membrane; PG, peptidoglycan layer; CM, cytoplasmic membrane.
The axial structure of the Salmonella flagellum is composed of the rod (FliE, FlgB, FlgC, FlgF, FlgG), the hook (FlgE), the hook-filament junction (FlgK, FlgL), the filament (flagellin, FliC or FljB) and the filament cap (FliD) (Figure 2). The assembly of the axial structure begins with the rod, followed by the hook with the help of the hook cap (FlgD). Upon completion of hook assembly, the hook cap is replaced by FlgK, and then FlgK and FlgL self-assemble into the hook-filament junction structure at the hook tip. FliD forms the filament cap at the tip of the junction structure and promotes the assembly of newly transported flagellin molecules into the long helical filament (Macnab, 2003).
To construct the axial structure beyond the cellular membranes, fourteen different proteins are translocated across the cytoplasmic membrane via the fT3SS, diffuse down the narrow central channel, and assemble at the tip of the growing structure (Figure 2). They can be classified into two export classes: one is the rod-type (FliE, FlgB, FlgC, FlgF, FlgG, FlgJ) and hook-type (FlgD, FlgE, FliK) class needed for assembly of the rod and hook. The other is the filament-type class (FlgK, FlgL, FlgM, FliC, FliD) responsible for filament assembly. The fT3SS secrets a molecular ruler protein, FliK, to measure the length of the hook during hook assembly and switches its substrate specificity from rod/hook-type proteins to filament-type proteins when the hook reaches its mature length of about 55 nm. At that point hook assembly terminates and filament assembly initiates (Minamino, 2018).
The fT3SS and vT3SS utilize the PMF across the cytoplasmic membrane and ATP hydrolysis to drive protein translocation across the cytoplasmic membrane (Minamino and Namba, 2008; Paul et al., 2008; Lee et al., 2014). The Salmonella fT3SS has a backup engine powered by a sodium (Na+) motive force (SMF) across the cytoplasmic membrane to continue flagellar assembly when the cytoplasmic ATPase ring complex does not work properly, as during biofilm development (Minamino et al., 2016b,2021a).
Once the transmembrane export complex of the Salmonella fT3SS is activated by ATP hydrolysis in the cytoplasmic ATPase ring complex, it becomes an active H+/protein antiporter that couples inward-directed H+ flow with outward-directed protein export (Minamino et al., 2011). Furthermore, the cytoplasmic ATPase complex allows the export complex to coordinate flagellar protein export with assembly in Salmonella (Minamino et al., 2016a; Inoue et al., 2018). Thus, the cytoplasmic ATPase ring complex acts as an activator of the H+-driven export engine and also contributes to efficient and robust protein export by the export complex. This review describes our current understanding of the structure and function of the flagellar cytoplasmic ATPase complex in Salmonella.
The transmembrane export complex of the fT3SS is located inside the MS-ring formed by the transmembrane protein FliF (Figure 1; Johnson et al., 2021; Kawamoto et al., 2021; Takekawa et al., 2021; Tan et al., 2021). It consists of nine copies of FlhA, a single copy of FlhB, five copies of FliP, four copies of FliQ, and a single copy of FliR (Abrusci et al., 2013; Kuhlen et al., 2018, 2020; Johnson et al., 2019).
FliP and FliR assemble into the FliP5-FliR1 complex with the help of the FliO scaffolding protein and form the polypeptide channel for the translocation of export substrates across the cytoplasmic membrane (Figure 3, left panel) (Fabiani et al., 2017; Fukumura et al., 2017). Four FliQ subunits bind to the outside of the FliP5-FliR1 complex to form the FliP5-FliQ4-FliR1 complex (Figure 3, middle panel). A flexible loop formed by the highly conserved Met-209, Met-210, and Met-211 residues of FliP (the M-loop) on the cytoplasmic side of the polypeptide channel and a plug loop composed of residues 106–122 of FliR (the R-plug) seem to prevent the leakage of small molecules during high-speed protein translocation (Figure 3, right panel) (Ward et al., 2018; Hüsing et al., 2021). The FliP5-FliQ4-FliR1 complex has a helical arrangement of subunits similar to the rod (Figure 3), so FliE, which is the first export substrate transported by the fT3SS (Minamino and Macnab, 1999; Minamino et al., 2000), can directly assemble at the distal end of the FliP5-FliQ4-FliR1 complex to form the most proximal part of the rod. Interactions between FliE and FliF not only firmly connect the rod with the MS ring but also open the exit gate of the polypeptide channel through conformational changes of FliP and FliR (Hendriksen et al., 2021).
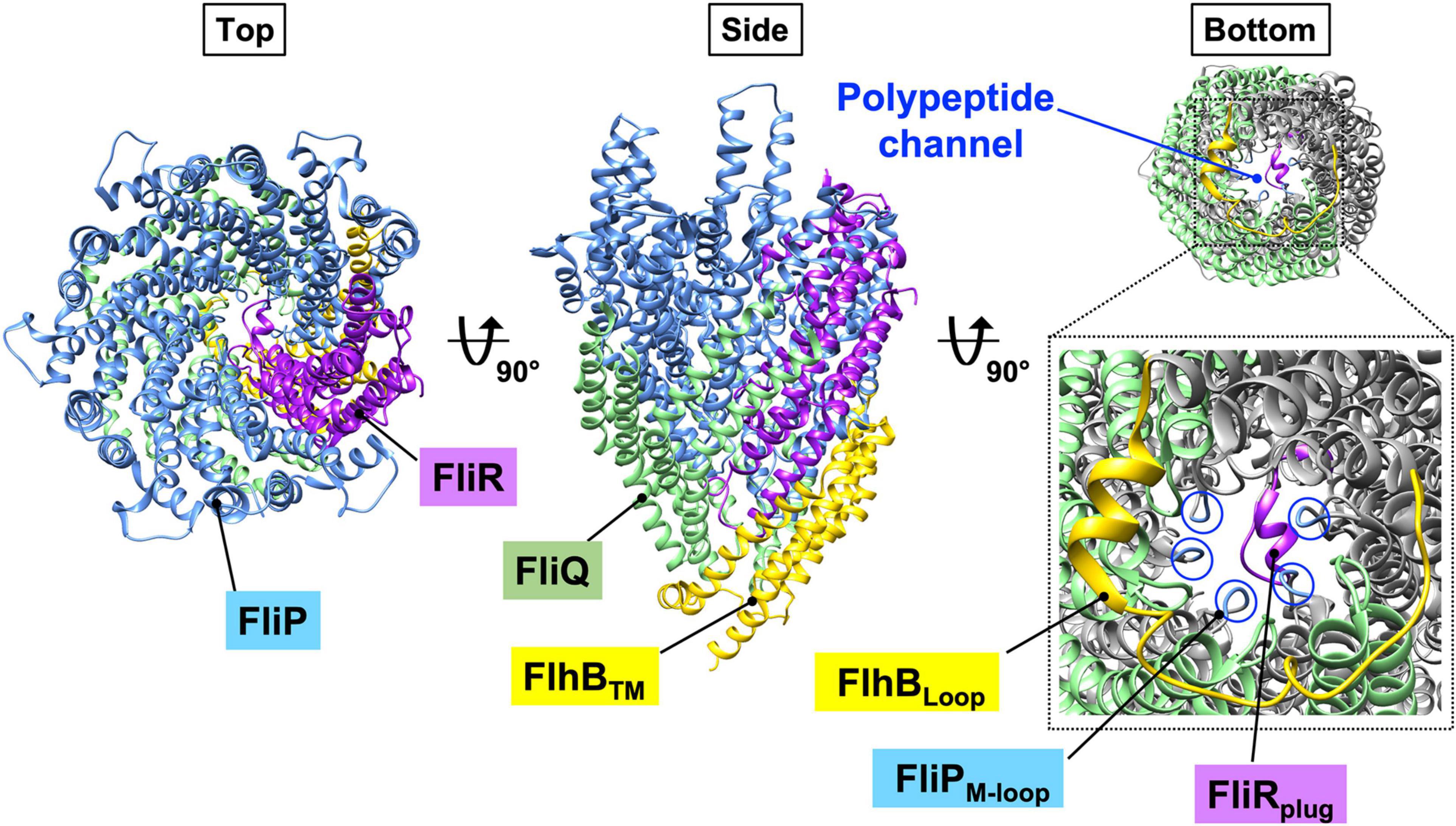
Figure 3. CryoEM structure of the FlhB1-FliP5-FliQ4-FliR1 complex (PDB ID: 6S3L). FliP and FliR assemble into the FliP5-FliR1 complex with the help of the flagellum-specific transmembrane protein, FliO. Four copies of FliQ associates with the outside of the FliP5-FliR1 complex. The central pore of the FliP5-FliR1 complex is thought to be a polypeptide channel. The FliP5-FliQ4-FliR1 complex adopts a right-handed helix similar to that of the flagellar axial structure. The transmembrane domain of FlhB (FlhBTM) associates with the FliP5-FliQ4-FliR1 complex. The highly conserved M-loop formed by Met-209, Met-210, and Met-211 of FliP (FliPM–loop) and the plug loop composed of residues 106–122 of FliR (FliRplug) block leakage of any small molecules during protein translocation. The cytoplasmic loop of FlhB FlhBLoop) interacts with all four FliQ subunits. Because the entrance gate of the FlhB1-FliP5-FliQ4-FliR1 complex is closed, FlhB is proposed to regulate opening of the gate to the polypeptide channel. Cyan, FliP; green, FliQ; magenta, FliR; yellow, FlhBTM.
Salmonella FlhB consists of an N-terminal transmembrane domain (FlhBTM) with four transmembrane helices (TMHs) (residues 1–211) and a large C-terminal cytoplasmic domain (FlhBC) (residues 212–383) (Minamino et al., 1994; Kinoshita et al., 2021). FlhBTM associates with the FliP5-FliQ4-FliR1 complex to form the FliP5-FliQ4-FliR1-FlhB1 complex (Figure 3, middle panel), and the cytoplasmic loop connecting TMH-2 and TMH-3 (FlhBLoop) wraps around the entrance gate of the FliP5-FliQ4-FliR1 complex through interactions of the loop with each FliQ subunit (Figure 3, right panel). It is thus plausible that FlhB may coordinate gate opening for substrate entry into the polypeptide channel. Recent genetic analysis has suggested that the N-terminal cytoplasmic tail of FlhB and FlhBC are involved, along with the cytoplasmic ATPase complex, in the gating function of FlhB (Kinoshita et al., 2021).
Salmonella FlhA is divided into two distinct regions: an N-terminal transmembrane region (FlhATM) with eight TMHs (residues 1–327) and a large C-terminal cytoplasmic region (residues 328–692) (Figure 4A; Minamino et al., 1994; Kinoshita et al., 2021). The crystal structure of the C-terminal cytoplasmic region is composed of a compactly folded domain (FlhAC, residues 362–692) and a flexible linker (FlhAL, residues 328–361) connecting FlhAC with FlhATM (Figure 4A; Saijo-Hamano et al., 2010). FlhA assembles into a homo-nonamer through intermolecular interactions between FlhAC subunits, and the interactions of FlhAL with its neighboring FlhAC subunit stabilize the FlhAC-ring structure (Figure 4B; Terahara et al., 2018; Kuhlen et al., 2021). FlhATM associates not only with the FliP5-FliQ4-FliR1 complex but also with the MS-ring (Kihara et al., 2001).
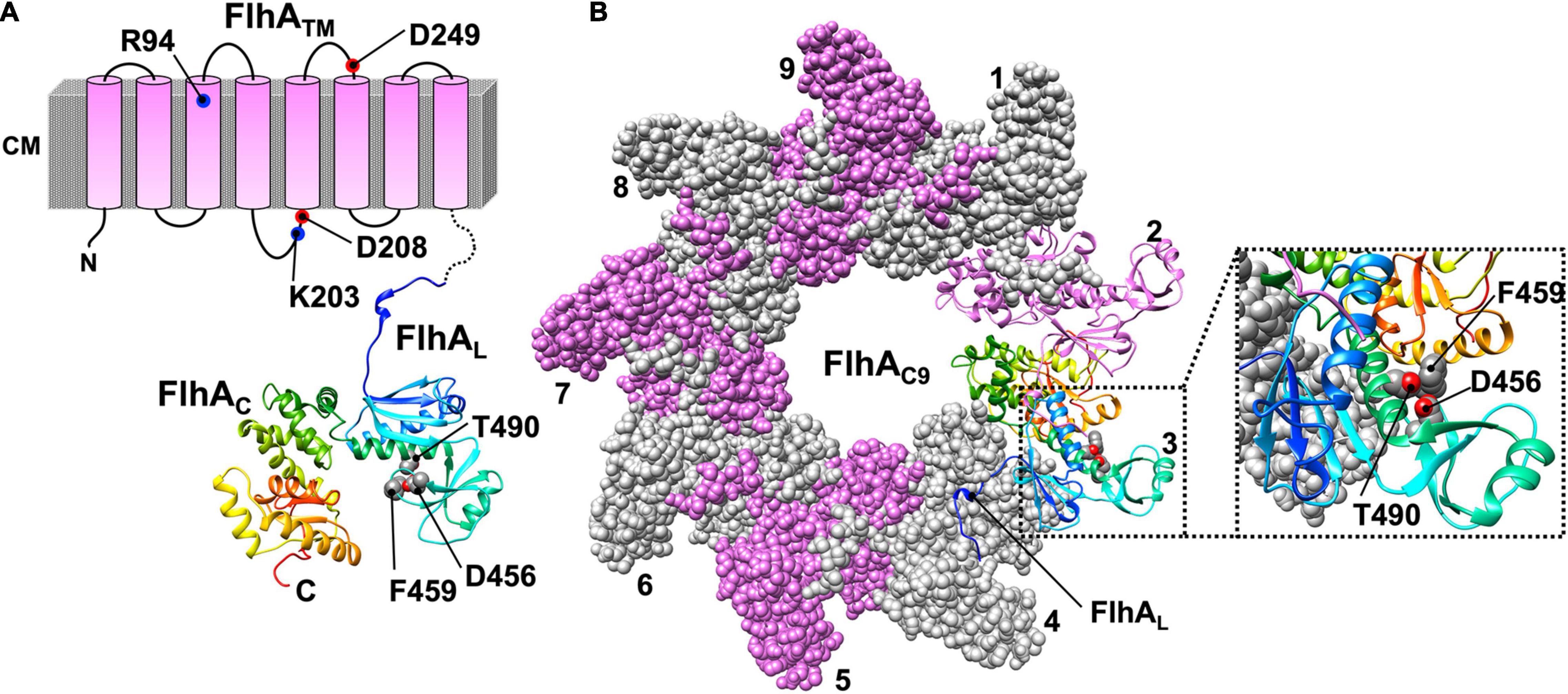
Figure 4. Atomic model of the cytoplasmic domain of FlhA (PDB ID: 3A5I). (A) Topological model of FlhA. FlhA is composed of an N-terminal transmembrane region with eight transmembrane helices (FlhATM) and a large C-terminal cytoplasmic domain (FlhAC). FlhATM acts as a dual-ion channel that can conduct both H+ and Na+. The highly conserved charged residues R94, K203, D208 and D249 are involved in H+-coupled protein export. The highly conserved residues D456, F459 and T490 of FlhAC are critical for substrate recognition. A flexible linker region of FlhA (FlhAL), which connects FlhAC with FlhATM, is involved in the interaction with FliJ. The interaction between FlhAL and FliJ activates the FlhA ion channel. (B) Model of the FlhAC-ring. FlhAC forms a homo-nonameric ring structure. The C-terminal part of FlhAL binds to the neighboring FlhAC subunit to stabilize the open conformation of FlhAC, allowing flagellar export chaperones in complex with their cognate substrates to bind to a chaperone-binding site of FlhAC, which includes the D456, F459, and T490 residues.
If either MS-ring or the FliP5FliQ4FliR1 complex is missing in Salmonella cells, FlhA cannot efficiently form the oligomer at the flagellar base as monitored with FlhA labeled with yellow fluorescent protein (YFP), suggesting that FlhA assembles into the export complex along with other export-gate proteins during MS-ring formation (Morimoto et al., 2014). The highly conserved Arg-94, Lys-203, Asp-208, and Asp-249 residues of FlhATM are critical for H+-coupled protein export (Hara et al., 2011; Erhardt et al., 2017). Over-expression of FlhA in Escherichia coli decreases the intracellular pH. Furthermore, over-expression of FlhA increases intracellular Na+ concentration in the presence of 100 mM NaCl. These observations suggest that FlhA forms a pathway for the transit of both H+ and Na+ across the cytoplasmic membrane. The flhA(D208A) mutation facilitates the H+-channel activity of FlhA, suggesting that Asp-208 of FlhA may coordinate H+ flow though the FlhA channel with protein export. However, this mutation does not affect the Na+-channel activity of FlhA at all, suggesting that the Na+ pathway is different from the H+ pathway (Minamino et al., 2016b).
FlhAC and FlhBC project into the cytoplasmic cavity of the basal body C-ring and form a docking platform for the cytoplasmic ATPase complex, flagellar export chaperones, and export substrates (Minamino and Macnab, 2000c; Minamino et al., 2003, 2010, 2012a; Bange et al., 2010). The FlhAC-FlhBC docking platform determines the order of substrate export to facilitate efficient flagellar assembly and also regulates gate opening of the FlhA ion channel and the FliP5-FliQ4-FliR polypeptide channel (Minamino and Macnab, 2000a; Kinoshita et al., 2013; Inoue et al., 2019; Minamino et al., 2020a,2021b).
A highly conserved hydrophobic dimple including Phe-459, Asp-456, and Thr-490 of FlhA is critical for substrate recognition by the fT3SS during flagellar assembly (Figure 4A; Xing et al., 2018). The C-terminal part of FlhAL binds to its neighboring FlhAC subunit to stabilize the open conformation of FlhAC in the nonameric ring, allowing flagellar export chaperones associated with their cognate substrates to bind to the conserved hydrophobic dimple with a nanomolar affinity (Inoue et al., 2021).
The F1 ATPase is composed of three copies of the α subunit, three copies of the β subunit, a single copy of the γ subunit and a single copy of the ε subunit (Figure 1). The α and β subunits form a hetero-hexameric α3β3 ring, and the γ subunit binds within the central pore of the α3β3 ring (Abrahams et al., 1994). The ε subunit binds to the γ subunit to control the ATP hydrolysis activity of the F1 ATPase in an ATP-dependent manner (Kato-Yamada et al., 2000). The α3β3γ1 subcomplex is the minimum unit that can function as an ATP-driven rotary motor to couple ATP hydrolysis with the rotation of the γ subunit within the α3β3 ring. ATP binds to three catalytic sites in the α3β3 ring, each of which is located at an interface between the α and β subunits. Three catalytic β subunits in the α3β3 ring undergo highly cooperative and sequential conformational changes in their C-terminal domains during ATP hydrolysis. These conformational changes drive the γ subunit to rotate within the α3β3 ring (Watanabe and Noji, 2013). The FliI6-FliJ1 subcomplex of the fT3SS, which looks similar to the α3β3γ1 subcomplex, can act as the ATPase at the base of the flagellum (Figure 1; Ibuki et al., 2011).
FliI is the flagellum-specific ATPase. It has highly conserved Walker A and B motifs (Vogler et al., 1991; Fan and Macnab, 1996). Salmonella FliI consists of three domains: N-terminal (residues 2–97, FliIN), ATPase (residues 109–380, FliICAT) and C-terminal (residues 381–456, FliIC) (Figure 5A; Imada et al., 2007). Residues 98–105, most of which are invisible in the electron density map, form a flexible hinge connecting FliIN and FliICAT, and this flexible hinge loop undergoes conformational changes during ATP binding and hydrolysis (Minamino et al., 2001).
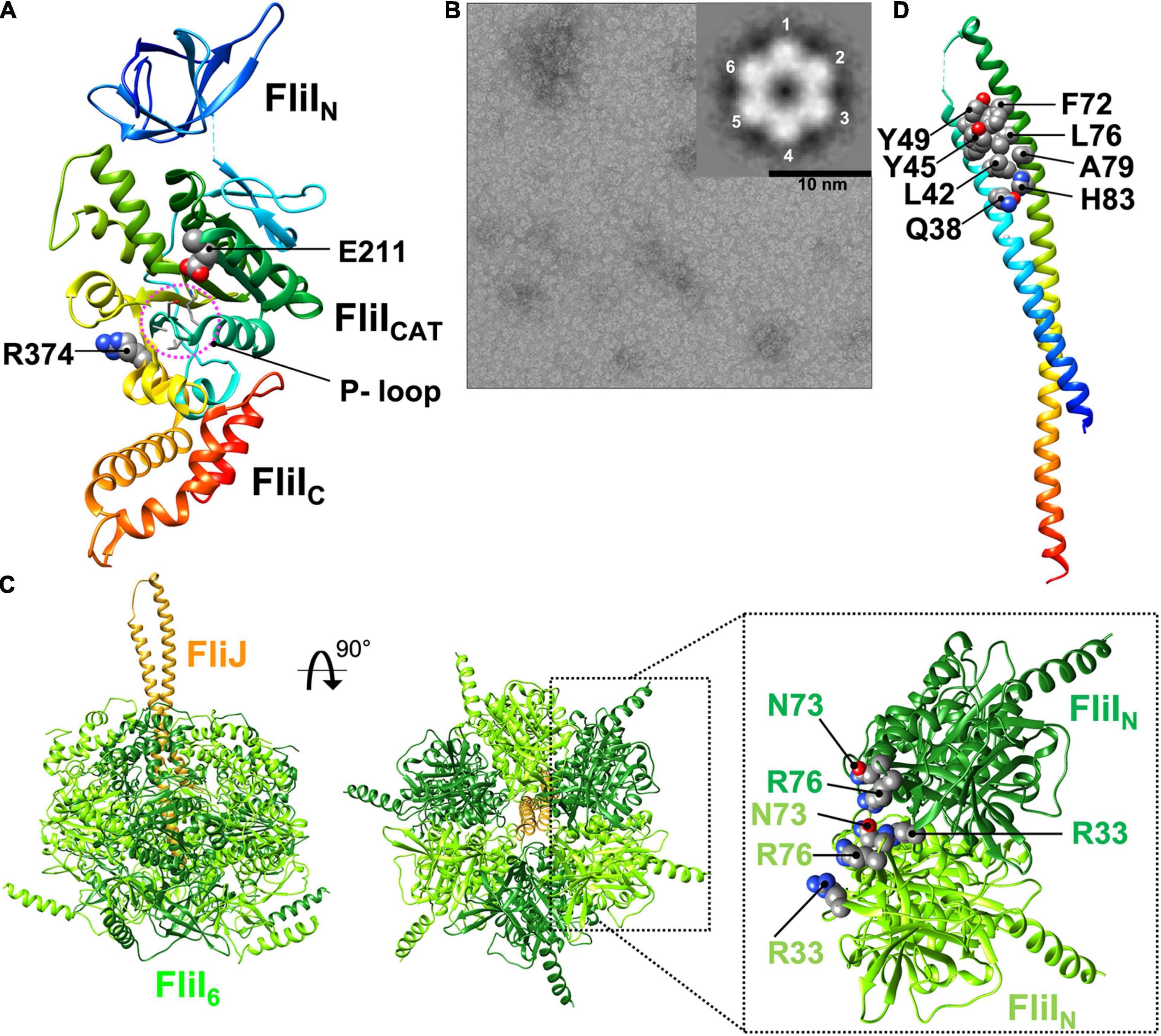
Figure 5. Atomic model of the FliI6-FliJ1 ATPase ring complex. (A) Cα ribbon representation of FliI (PDB ID: 5B0O). FliI consists of an N-terminal (FliIN), an ATPase (FliICAT), and a C-terminal (FliIC) domain. FliIN is involved in formation of the FliI6 ring. FliICAT contains the highly conserved P-loop, the catalytic glutamate (E211), and an arginine finger (R374), which are all involved in ATP hydrolysis. The ATP catalytic cycle induces sequential and cooperative conformational changes of FliIC, which interacts with FliJ. (B) Electron micrograph of negatively stained FliI ring-like structures in complex with Mg2+-ADP-AlF4. The inset shows a 2D class average of the FliI ring structure. (C) Model of the FliI6-FliJ1 ring model. R33, N73, and R76 of FliIN regulate FliI ring formation. FliJ binds to the center of the FliI6 ring. (D) Cα ribbon representation of FliJ (PDB ID: 3AJW). FliJ forms a two stranded coiled-coil structure. The highly conserved Q38, L42, Y45, Y49, F72, L76, A79 and H83 residues of FliJ extends out of the FliI6 ring and are interact with FlhAL.
The structures of FliI and its fT3SS homolog SctN are remarkably similar to the α and β subunits of the F1 ATPase (Zarivach et al., 2007). However, in contrast to the F1 ATPase, FliI and SctN form homo-hexamers in an ATP-dependent manner (Figure 5B, C), and both hexamers themselves can hydrolyze ATP at the interface between FliI/SctN subunits (Claret et al., 2003; Kazetani et al., 2009). Thus, the ATPase ring complex of the T3SS has six catalytic sites. The FliI6 and SctN6 ring structures have been identified at the base of the flagellum and injectisome, respectively, by electron cryotomography and sub-tomogram averaging (Chen et al., 2011; Kawamoto et al., 2013).
Intermolecular interactions between FliIN domains are required for FliI ring formation (Figure 5C; Okabe et al., 2009). The core structure of FliIN can be superimposed onto the N-terminal domains of the α and β subunits of the F1 ATPase within α3β3 hetero-hexamer. In the FliI6-ring model, which was generated by fitting the crystal structure of FliI into the structures of the α and β subunits, FliIN shows steric hindrance at the subunit interfaces, suggesting that a conformational change in FliIN is required for FliI ring formation. Deletion of residues 2–7 of FliIN suppresses FliI hexamerization and decreases the ATPase activity of FliI (Minamino et al., 2006), suggesting that the extreme N-terminal region of FliI regulates FliI oligomerization. Recently, it has been reported that Arg-33, Asn-73, and Arg-76 are also responsible for well-regulated FliI ring formation (Figure 5C; Kinoshita et al., 2021).
Amino acid residues in the F1 ATPase that are known to be involved in ATP hydrolysis are highly conserved in the FliI/SctN family. FliICAT contains the highly conserved P-loop (residues 182–188), the catalytic glutamate (Glu-211), and the arginine finger (Arg-374) (Figure 5A; Walker, 2013). ADP binds to the P-loop of FliI, as it does in the F1 ATPase. The carboxyl group of Glu-190 in the β subunit of the thermophilic Bacillus F1 ATPase, which corresponds to Glu-211 of FliI, polarizes a water molecule for the nucleophilic attack on the γ-phosphate of ATP, and the G190Q substitution results in a complete loss of ATPase activity (Shimabukuro et al., 2003). The fliI(E211Q) mutation completely abolishes ATPase activity but does not affect the binding of ATP to the P-loop, and FliI with the E211Q substitution can form the hexamer ring in the presence of Mg2+-ATP. Thus, Glu-211 of FliICAT acts as the catalytic glutamate.
Arg-373 in the α subunit of the F1 ATPase, which corresponds to Arg-374 of FliI, functions as the arginine finger that protrudes into the nucleotide-binding site of the adjacent β-subunit. The side chain of this arginine residue forms a positively charged binding pocket for the negative charge of the γ-phosphate of ATP (Rees et al., 2012). The fliI(R374A) mutation inhibits FliI ring formation significantly and decreases ATPase activity. This effect indicates that Arg-374 of FliI stabilizes the binding of ATP to the P-loop in a way similar to Arg-373 of the α subunit. These observations suggest that FliI and the F1 ATPase share a similar catalytic pathway for ATP hydrolysis.
The binding of ADP to the P-loop induces a conformational change in FliIC relative to FliICAT, suggesting that the FliI hexamer may undergo conformational changes in its C-terminal domains that are coupled with the catalytic reaction cycle in the same way as in the F1 ATPase. This idea is supported by the asymmetric cryoEM structure of the SctN6-SctO1 ring complex with a non-hydrolyzable ATP analog (Majewski et al., 2019).
FliJ and its vT3SS homolog SctO adopt an antiparallel coiled-coil structure that is similar to the two-stranded α-helical coiled-coil part of the γ subunit of the F1 ATPase (Figure 5D; Ibuki et al., 2011). FliJ binds to the C-terminal region of the first α-helix of FliIC (residues 382–406 of Salmonella FliI), which corresponds to the region of the β subunit that is responsible for interaction with the γ subunit. This interaction facilitates FliI ring formation and increases the ATPase activity of FliI. FliJ penetrates the central cavity of the FliI6 ring like the γ subunit in the F1 ATPase (Figure 5C). These observations have been confirmed by the cryoEM structure of the SctN6-SctO1 ring complex. FliJ has been shown to exert a rotor-like function in both rotary F1 and V1 ATPases (Kishikawa et al., 2013; Baba et al., 2016). Thus, the FliI6-FliJ1 ring complex may function as an ATP-driven rotary motor that couples ATP hydrolysis with the rotation of FliJ within the FliI hexamer.
The b and δ subunits of the FOF1 ATP synthase form the peripheral stalk that connects the α3β3γ1ε1 ring complex to the membrane-embedded FO unit (Figure 1). The extreme N-terminal region of the b subunit binds to FO, whereas the δ subunit interacts with the extreme N-terminal region of the α subunit of F1 (Walker and Dickson, 2006). The N-terminal and C-terminal regions of FliH and its vT3SS homolog SctL are homologous to the b and δ subunits of the ATP synthase (Pallen et al., 2006). This is confirmed by the crystal structure of an N-terminally truncated variant of Salmonella FliH consisting of residues 99–235 in complex with FliI (Figure 6; Imada et al., 2016).
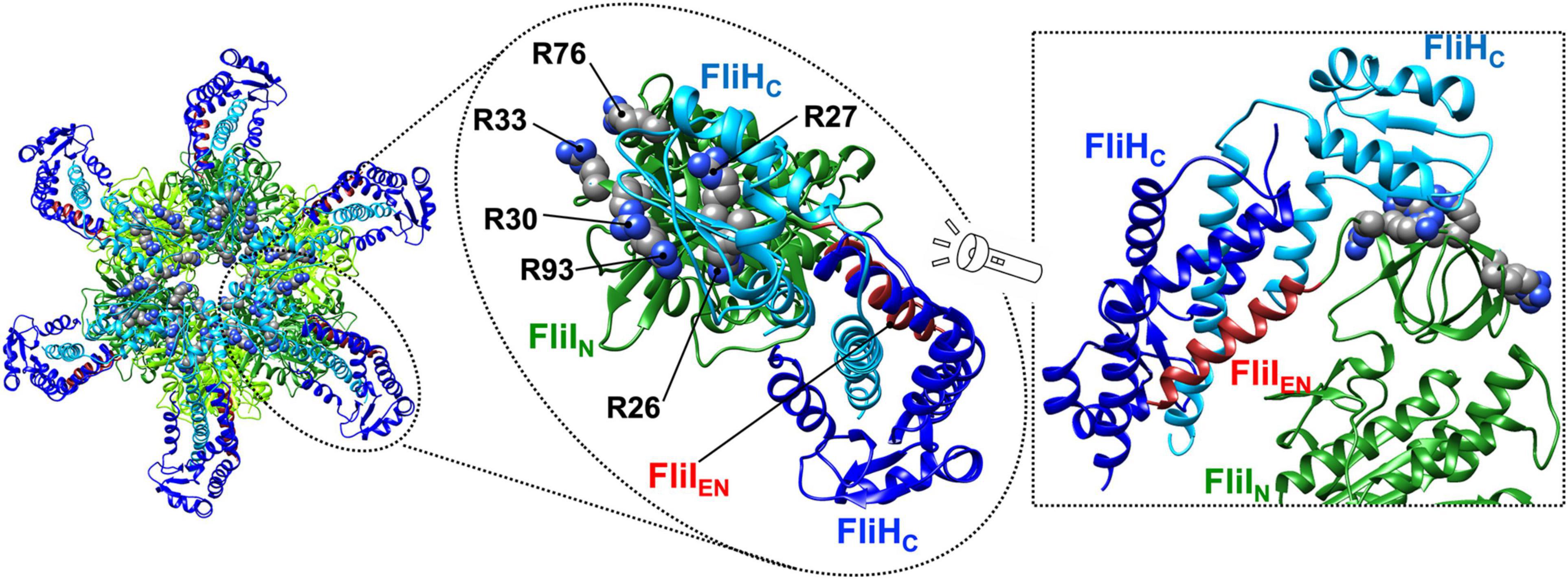
Figure 6. Atomic model of the FliHC12-FliI6 ring complex. Cα ribbon representation of the FliHC2-FliI1 complex (PDB ID: 5B0O) is shown. The C-terminal domain of FliH (residues 141–235, FliHC) forms a dimer via an interaction of residues 101–140, which adopt a coiled-coil structure. The FliHC dimer binds to each N-terminal domain (FliIN) of the FliI6 ring. Interestingly, one FliHC domain (blue) binds to the N-terminal α-helix consisting of residues 2–21 of FliI (brown, FliIEN), and the other (cyan) binds to a positively charged region formed by R26, R27, R30, R33, R76, and R93 of FliI. These two domains adopt conformations that are completely different from each other.
Salmonella FliH consists of 235 amino-acid residues and forms a homo-dimer through residues 101–140, which form a coiled-coil structure (Minamino and Macnab, 2000b; González-Pedrajo et al., 2002). The FliH dimer binds to each FliIN domain of the FliI6 ring (Figure 6, left panel) and also to the FliN protein in the C-ring (González-Pedrajo et al., 2006; McMurry et al., 2006; Paul et al., 2006). The interactions of FliH with FliN and FliIN are required for efficient and robust association of the FliI6-FliJ ring complex with the flagellar basal body (Figure 1; Minamino et al., 2009). The N-terminal domain of FliH (residues 1–140, FliHN) adopts a quite elongated α-helical coiled coil structure similar to that of the b subunit of the ATP synthase, and the extreme N-terminal region of FliH is involved in the interaction with FliN (Hara et al., 2012). Both C-terminal domains (residues 141–235, FliHC) in the FliH dimer are involved in the interaction with FliI (Minamino et al., 2002). These two FliHC domains have completely different conformations; one binds to the extreme N-terminal α-helix of FliI consisting of residues 2–21, and the other binds to a positively charged cluster consisting of Arg-26, Arg-27, Arg-30, Arg-33, Arg-76, and Arg-93 of FliIN (Figure 6, middle and right panels). Because FliI cannot localize to the flagellar base in the absence of FliH, FliH seems to act as a peripheral stalk to firmly anchor the FliI6-FliJ1 ring complex to the C-ring.
The PMF consists of the electric potential difference (Δψ) and the proton concentration difference (ΔpH) across the cytoplasmic membrane. When the cytoplasmic ATPase ring complex works properly for flagellar assembly, the transmembrane export gate complex uses the Δψ component to drive H+-coupled protein export under a variety of environmental conditions (Paul et al., 2008; Minamino et al., 2011, 2021b). However, when the ATPase ring complex becomes non-functional under certain physiological conditions, the export gate complex prefers to use the SMF over a wide range of external pH, indicating that the transmembrane export complex is intrinsically a dual-fuel export engine that can use either H+ or Na+ as the coupling ion (Minamino et al., 2016b,2021a). This in turn suggests that the cytoplasmic ATPase ring complex switches the export gate complex from the dual-fuel engine mode to a highly efficient Δψ-driven one.
FliJ binds to FlhAL with high affinity to activate the H+ channel of FlhATM and to unlock the entrance gate of the polypeptide channel. As a result, the export gate complex becomes an active H+/protein antiporter that couples inward-directed H+ flow through the FlhA ion channel with outward-directed protein translocation across the polypeptide channel (Minamino et al., 2011). An inactive export gate complex can also be activated by an increase in Δψ above a certain threshold through an interaction between FliJ and FlhAL, suggesting that Δψ is required for efficient and stable interaction between FliJ and FlhAL (Minamino et al., 2021b).
A helix-loop-helix formed by Gln-38, Leu-42, Tyr-45, Tyr-49, Phe-72, Leu-76, Ala-79, and His-83 of FliJ, which are highly conserved residues in FliJ homologs, extends out of the FliI6-ring (Figure 5D; Ibuki et al., 2013). This is confirmed by the cryoEM structure of the SctN6-SctO1 ring. Tyr-45, Tyr-49, and Phe-72 of FliJ are also conserved between FliJ and the γ subunit of the F1 ATPase. Among these conserved, surface-exposed residues of FliJ, Phe-72 and Leu-76 are critical for the interaction with FlhAL. Residues in the γ subunit corresponding to Phe-72 and Leu-76 are involved in the interaction with an α-helix of the δ subunit of the bovine mitochondrial F1 ATPase, which is a homolog of the conserved ε subunit in the F1 ATPase family (PDB ID: 1E79) (Gibbons et al., 2000). The flhA(E351A/W354A/D356A) triple mutation significantly reduces the binding affinity of FlhAL for FliJ (Inoue et al., 2021). Because the residues from Val-349 to Val-357 of FlhA form an α-helix, FliJ may bind to this α-helix in FlhAL as is seen in the γ-δ interaction in the bovine mitochondrial F1 ATPase.
ATP hydrolysis by the FliI ATPase and rapid protein translocation by the export complex are both linked to efficient H+ translocation through the FlhA ion channel (Morimoto et al., 2016). Recently, it has been reported that ATP hydrolysis by the FliI ATPase also unlocks the entrance gate of the polypeptide channel formed by FliP, FliQ, and FliR for efficient entry of export substrates into the channel (Kinoshita et al., 2021). Furthermore, the Salmonella ΔfliHIJ flhB(P28T) flhA(T490M) mutant has been isolated as a revertant of the ΔfliHIJ mutant that has increased motility (Minamino et al., 2021a). The protein-export activity of the transmembrane export complex in cells with both the flhA(T490M) and flhB(P28T) mutations is almost at the wild-type level under a variety of experimental conditions even in the absence of the FliH12-FliI6-FliJ1 ring complex (Minamino et al., 2021b). This finding suggests that the export complex normally requires the FliH12-FliI6-FliJ1 complex to serve as a H+-coupled protein transporter. Because FliJ requires FliH and FliI to bind efficiently to FlhAL (Minamino et al., 2011), this observation raises the question of how this ATPase ring complex activates the export complex.
The conserved Glu-211 residue of FliI catalyzes ATP hydrolysis. The E211D substitution decreases FliI ATPase activity by about 100-fold (Minamino et al., 2014). Salmonella wild-type cells produce an average of 4.4 ± 1.6 flagellar filaments per cell. In contrast, more than 90% of Salmonella fliI(E211D) cells have an average of 2.3 ± 1.5 flagellar filaments, and the average length of those filament is only half that of the wild type. Because the fT3SS transports 20,000–30,000 flagellin molecules per flagellum to form a 10–15 μm long helical filament (Minamino and Namba, 2004), the rate of ATP hydrolysis by the FliI ATPase cannot determine the rate of filament assembly, as ATP consumption by the fT3SS during flagellar assembly must be relatively small. Because the export complex still transports flagellar axial proteins even with the infrequent ATP hydrolysis provided by FliI(E211D), ATP hydrolysis appears to be required only for activation of the H+-driven export engine.
The six FliIC domains undergo cooperative and sequential conformational changes triggered by ATP hydrolysis. Deletion of residues 401–410 in the first α-helix of FliIC, which is responsible for the interaction with FliJ, significantly decreases the protein transport activity of the fT3SS although the ATPase activity is still at about 40% of the wild-type level. Because this deletion does not inhibit the interaction between FliI and FliJ, it may affect conformational changes in the FliIC domains that rotate FliJ within the FliI hexamer. Rotation of FliJ may induce conformational changes in the FlhATM domain through an interaction between FliJ and FlhAL, thereby activating the FlhA ion channel and unlocking the entrance gate of the polypeptide channel (Figure 7).
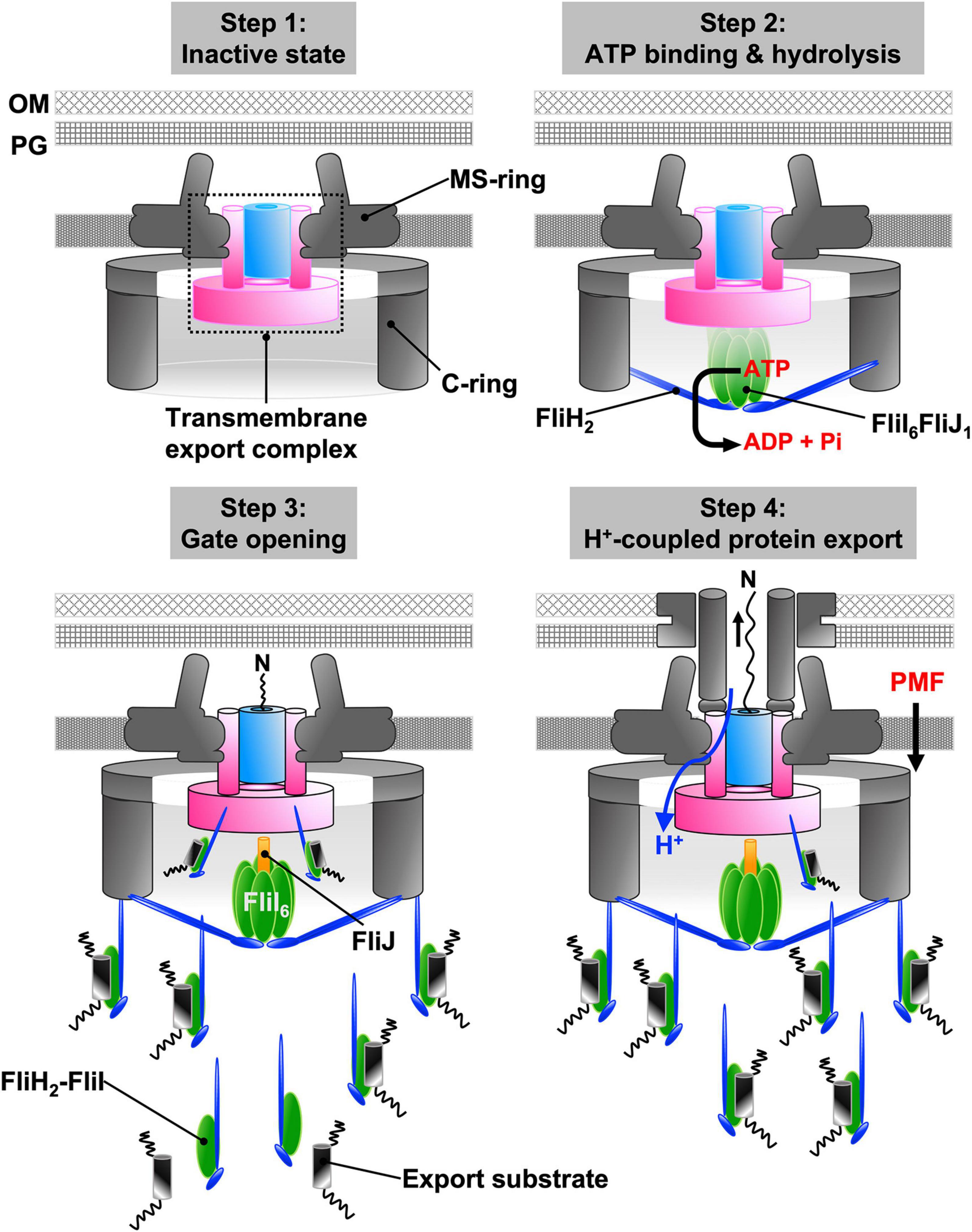
Figure 7. Energy coupling mechanism of the fT3SS. The transmembrane export complex remains inactive until the cytoplasmic ATPase ring complex is formed at the base of the flagellum; the polypeptide and proton channels of the export complex remain closed (step 1). When ATP hydrolysis by FliI induces the rotation of FliJ in the FliI6 ring, interactions between FliJ and FlhAL induces conformational rearrangements of the export complex. As a result, the complex becomes an active protein transporter (step 2). The cytoplasmic FliH2-FliI1 complex acts as a dynamic carrier to deliver export substrates from the cytoplasm to the export complex (step 3). Upon docking of an export substrate to the entrance gate of the polypeptide channel, the gates of both polypeptide and proton channels are opened. The export complex can now act as a H+/protein antiporter that couples H+ flow through the ion channel with protein translocation into the polypeptide channel (step 4).
The elementary step size of γ rotation within the α3β3 ring is 120°, which is composed of 80° and 40° sub-steps driven by ATP binding–ADP release and ATP hydrolysis–Pi release, respectively (Watanabe and Noji, 2013). The cryoEM structure of the SctN6-SctO1 ring complex has suggested a possible rotational mechanism for catalysis. In this model, the SctO stalk rotates in the SctN6 ring through an interaction between each SctNC domain and SctO. Because the SctN6-SctO1 ring complex has six catalytic sites, the elementary step size of SctO rotation within the SctN6 ring is probably 60° (Majewski et al., 2019). The FliI(E211Q) substitution in Salmonella, which completely eliminates ATPase activity but not ATP binding to the P-loop of FliICAT, results in only 17% of cells having one or two flagellar filaments about 25% the length of those of the wild type (Minamino et al., 2014). Thus, ATP binding to the P-loop is sufficient to activate the H+-driven export engine of the fT3SS to some degree. So, the 60° rotation of FliJ may be divided into two sub-steps, and ATP binding may induce the first sub-step, which may be sufficient to activate the H+-driven export engine weakly.
The FliI monomer interacts with the FliH dimer to form a hetero-trimer in the cytoplasm (Minamino and Macnab, 2000b; Auvray et al., 2002). High-resolution imaging of fluorescently labeled FliI in vivo has revealed that FliH2-FliI1 complexes are associated with the basal body through interactions of FliH with FlhA and FliN. FliI-YFP shows a rapid exchange between the basal body and a freely diffusing cytoplasmic pool. The FliI(K188I) substitution, which inhibits ATP binding to the P-loop in FliICAT, does not affect the exchange rate of FliI-YFP, suggesting that ATP hydrolysis does not drive the association-dissociation cycle (Bai et al., 2014). FliH also suppresses the ATPase activity of the FliH2-FliI1 complex (Minamino and Macnab, 2000b). Deletion of flhA decreases the number of FliI-YFP molecules associated with the basal body but does not affect the exchange rate. The highly conserved Trp-7 and Trp-10 residues of FliHN are directly involved in the interactions of FliH with FliN and FlhATM (Hara et al., 2012; Notti et al., 2015). Because the interaction between FliH and FliN is required for efficient localization of the FliH2-FliI1 complex to the flagellar base, the FliH-FliN interaction must be highly dynamic to achieve rapid and efficient flagellar protein export by the fT3SS. Flagellar chaperones in complex with their cognate substrates both bind to FliIC, suggesting that FliIC is also involved in substrate recognition (Thomas et al., 2004; Imada et al., 2010; Minamino et al., 2012b). Pull-down assays have demonstrated that chaperone-associated export substrates bind to FlhAC and FlhBC even in the absence of FliHI (Evans et al., 2013; Kinoshita et al., 2013; Inoue et al., 2019). However, they require the FliH2-FliI1 complex to efficiently interact with FlhAC and FlhBC in vivo (Minamino et al., 2016b; Inoue et al., 2018; Kinoshita et al., 2021).
In vitro protein transport assays using inverted membrane vesicles have shown that addition of the purified FliH2-FliI1 complex considerably increases the transport of flagellar axial protein into the lumen of the membrane vesicles (Terashima et al., 2018, 2020). Thus, the FliH2-FliI1 complex acts as a dynamic carrier to deliver chaperone-associated export substrates from the cytoplasm to the flagellar base and to facilitate their docking to FlhAC and FlhBC, thereby allowing the activated export complex to unfold and transport export substrates into the central channel of the flagellum.
Salmonella cells lacking the FliH and FliI proteins display a very weak motile phenotype. This defect is considerably alleviated by either an increase in the expression level of export substrates and chaperones or an increase in the PMF (Erhardt et al., 2014). Expression of Vibrio alginolyticus FlhA, which has 73.2% similarity and 52.9% identity in amino acid sequence with Salmonella FlhA, restores motility in the Salmonella ΔflhA mutant but does not increase motility in the Salmonella ΔfliHI flhB(P28T)ΔflhA mutant (Minamino et al., 2016a). Thus, Vibrio FlhA requires FliH and FliI to perform protein export in the Salmonella fT3SS. Deletion of flgM, which encodes the negative regulator of the flagellar regulon, increases the expression levels of FliJ, export substrates and flagellar export chaperones and allows Vibrio FlhA to perform protein transport even in the absence of FliH and FliI. These results suggest that FlhA needs FliH and FliI to buffer protein export against internal perturbations (Minamino et al., 2016a).
The fT3SS utilizes the secreted molecular ruler protein FliK to stop growth of the hook at about 55 nm (Minamino et al., 1999; Erhardt et al., 2011; Kinoshita et al., 2017). The ΔfliHI flhB(P28T) bypass mutant cannot properly control the length of the hook, although it secrets the hook capping protein FlgD and the FliK ruler into the culture media almost at the wild-type level (Inoue et al., 2018). However, secretion level of the hook protein FlgE is about 10-fold lower than the wild-type level. The flhA(F459A) mutation, which targets a residue within FlhAC (Figure 4), significantly increases the secretion of FlgE, so the secreted FliK ruler can measure the length of the hook more precisely. Neither the secretion levels nor control of hook length is affected by the FlhB(P28T) and FlhA(F459A) substitutions when FliH and FliI are present. Because FlgD, FlgE, and FliK bind to FliH and FliI as well as FlhAC and FlhBC (Minamino and Macnab, 2000c), the FliH2-FliI complex may coordinate targeting of FlgD, FlgE, and FliK to the FlhAC-FlhBC docking platform to make control of the hook length more robust.
FlgN, FliS, and FliT act as export chaperones for FlgK/FlgL, FliC, and FliD, respectively (Fraser et al., 1999; Auvray et al., 2001). The chaperone-substrate complexes bind FlhAC with nanomolar affinity (Figure 4; Kinoshita et al., 2013). This strong interaction of the chaperone with FlhAC facilitates protein unfolding and transport by the H+-driven export complex (Furukawa et al., 2016; Minamino et al., 2021c). In wild-type cells, more than 90% of flagellin molecules transported by the fT3SS assemble into the filament. The ΔfliHI flhB(P28T) flhA(F459A) cannot efficiently produce the hook-filament junction and filament cap structures at the hook tip, and hence more than 90% of the flagellin molecules are secreted as monomer into the culture supernatant. Because FlgN and FliT bind to the FliH2-FliI1 complex whereas FliS does not (Thomas et al., 2004; Minamino et al., 2012b; Sajó et al., 2014), the FliH2-FliI1 complex may contribute to hierarchical targeting of the flagellar chaperones to FlhAC, thereby allowing the junction and filament cap structures to be efficiently formed at the hook tip prior to filament formation. Thus, the FliH2-FliI1 complex works with the FlhAC-FlhBC docking platform to ensure the correct order of protein export.
The information thus far summarized allows us to propose a model for the energetics of protein export by the fT3SS (Figure 7). The transmembrane export complex remains inactive until the cytoplasmic ATPase ring complex localizes to the flagellar base through an interaction between FliHN and FlN (Step 1). ATP hydrolysis by the FliI ATPase induces the rotation of FliJ within the FliI6-ring at the FlhAC-FlhBC docking platform. The interactions of FliJ and FlhAL induce conformational changes in the export complex that activate the FlhA ion channel and unlock the entrance gate of the polypeptide channel (Step 2). Then, cytoplasmic FliH2-FliI1 complexes escort export substrates and chaperone-substrate complexes from the cytoplasm to the FlhAC-FlhBC docking platform through the interactions of FliH with FliN and FlhA (Step 3). The binding of the export substrate to the docking platform induces opening of the gate to the polypeptide channel, and the activated export complex acts as an H+/protein antiporter that couples proton flow through the FlhA ion channel with the translocation of export substrates into the polypeptide channel (Step 4). The association-dissociation cycle of the FliH2-FliI complex with the docking platform allows the transport of flagellar axial proteins in a highly controlled manner.
The transmembrane export complex of the fT3SS is a dual-fuel export engine that uses either H+ or Na+ as the coupling ion to drive export of flagellar proteins. Interestingly, when the cytoplasmic ATPase ring complex works properly, the export gate preferentially utilizes the PMF to drive H+-coupled protein export. FlhATM acts as a dual ion channel to conduct both H+ and Na+. Because there is not yet structural information about the FlhATM, it remains unknown how the cytoplasmic ATPase ring complex switches the ion channel mode of FlhATM from an inefficient dual-ion channel to a highly efficient H+ channel.
The export complex couples inward-directed ion flow through FlhA with outward-directed protein translocation through the polypeptide channel. Recently, the cryoEM structure of the FliP5-FliQ4-FliR1 complex associated with the basal body has been obtained with near atomic level (Johnson et al., 2021; Tan et al., 2021). Unfortunately, both FlhA and FlhB are lacking in the structure. To clarify the energy coupling mechanism, high-resolution structures of the entire export complex in different states of substrate export will be required.
The entire structure of the FliI6-FliJ1 ring complex looks similar those of rotary F1 and V1 ATPases. The FliI6-FliJ1 ring complex hydrolyzes ATP at the interfaces between FliI subunits and may induce sequential and cooperative conformational changes in FliIC, which is involved in the interaction with FliJ. These observations lead to the hypothesis that ATP hydrolysis by the FliI ATPase presumably allows FliJ to rotate within the FliI6 ring. This idea is supported by the cryoEM structure of the SctN6-SctO1 ring complex. To demonstrate the rotational catalytic mechanism of the FliI6-FliJ1 ring complex directly will require a biophysical approach.
The fT3SS transports fourteen different flagellar proteins in their copy numbers ranging from a few to tens of thousands in a sequential manner so that the flagellum can be built efficiently. The fT3SS must ensure the correct order of export of flagellar proteins for this to be an efficient process. The FliH2-FliI1 complex is required for efficient and robust flagellar assembly. Although an in vitro protein transport assay using inverted membrane vesicles has been established for the fT3SS, a quantitative measurement of ordered flagellar protein export will be needed to understand how the FliH2-FliI1 complex contributes to hierarchical protein targeting to the export complex.
TM, MK, and KN researched and wrote the review article. All authors contributed to the article and approved the submitted version.
This work was supported in part by JSPS KAKENHI Grant Numbers JP19H03182 (to TM) and JP20K15749 (to MK) and MEXT KAKENHI Grant No. JP20H05532 (to TM). This work has also been supported by Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from AMED under Grant No. JP19am0101117 to KN, by the Cyclic Innovation for Clinical Empowerment (CiCLE) from AMED under Grant No. JP17pc0101020 to KN, and by JEOL YOKOGUSHI Research Alliance Laboratories of Osaka University to KN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge Katsumi Imada for his kind gift of a structural model of the FliHC12FliI6FliJ1 ring complex and Akihiro Kawamoto for his kind gift of an electron micrograph of negatively stained FliI ring structures.
Abrahams, J. P., Leslie, A. G., Lutter, R., and Walker, J. E. (1994). Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628. doi: 10.1038/370621a0
Abrusci, P., Vergara-Irigaray, M., Johnson, S., Beeby, M. D., Hendrixson, D. R., Roversi, P., et al. (2013). Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104. doi: 10.1038/nsmb.2452
Auvray, F., Ozin, A. J., Claret, L., and Hughes, C. (2002). Intrinsic membrane targeting of the flagellar export ATPase FliI: interaction with acidic phospholipids and FliH. J. Mol. Biol. 318, 941–950. doi: 10.1016/S0022-2836(02)00172-9
Auvray, F., Thomas, J., Fraser, G. M., and Hughes, C. (2001). Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308, 221–229. doi: 10.1006/jmbi.2001.4597
Baba, M., Iwamoto, K., Iino, R., Ueno, H., Hara, M., Nakanishi, A., et al. (2016). Rotation of artificial rotor axles in rotary molecular motors. Proc. Natl. Acad. Sci. U. S. A. 113, 11214–11219. doi: 10.1073/pnas.1605640113
Bai, F., Morimoto, Y. V., Yoshimura, S. D. J., Hara, N., Kami-Ike, N., Namba, K., et al. (2014). Assembly dynamics and the roles of FliI ATPase of the bacterial flagellar export apparatus. Sci. Rep. 4:6528. doi: 10.1038/srep06528
Bange, G., Kümmerer, N., Engel, C., Bozkurt, G., Wild, K., and Sinning, I. (2010). FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 107, 11295–11300. doi: 10.1073/pnas.1001383107
Chen, M., Zhao, Z., Yang, J., Peng, K., Baker, M. A., Bai, F., et al. (2017). Length-dependent flagellar growth of Vibrio alginolyticus revealed by real time fluorescent imaging. Elife 6:e22140. doi: 10.7554/eLife.22140
Chen, S., Beeby, M., Murphy, G. E., Leadbetter, J. R., Hendrixson, D. R., Briegel, A., et al. (2011). Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981. doi: 10.1038/emboj.2011.186
Claret, L., Calder, S. R., Higgins, M., and Hughes, C. (2003). Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol. Microbiol. 48, 1349–1355. doi: 10.1046/j.1365-2958.2003.03506.x
Erhardt, M., Mertens, M. E., Fabiani, F. D., and Hughes, K. T. (2014). ATPase-independent type-III protein secretion in Salmonella enterica. PLoS Genet. 10:e1004800. doi: 10.1371/journal.pgen.1004800
Erhardt, M., Singer, H. M., Wee, D. H., Keener, J. P., and Hughes, K. T. (2011). An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30, 2948–2961. doi: 10.1038/emboj.2011.185
Erhardt, M., Wheatley, P., Kim, E. A., Hirano, T., Zhang, Y., Sarkar, M. K., et al. (2017). Mechanism of type-III protein secretion: regulation of FlhA conformation by a functionally critical charged-residue cluster. Mol. Microbiol. 104, 234–249. doi: 10.1111/mmi.13623
Evans, L. D., Poulter, S., Terentjev, E. M., Hughes, C., and Fraser, G. M. (2013). A chain mechanism for flagellum growth. Nature 504, 287–290. doi: 10.1038/nature12682
Fabiani, F. D., Renault, T. T., Peters, B., Dietsche, T., Gálvez, E. J. C., Guse, A., et al. (2017). A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol. 15:e2002267. doi: 10.1371/journal.pbio.2002267
Fan, F., and Macnab, R. M. (1996). Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271, 31981–31988. doi: 10.1074/jbc.271.50.31981
Fraser, G. M., Bennett, J. C. Q., and Hughes, C. (1999). Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32, 569–580. doi: 10.1046/j.1365-2958.1999.01372.x
Fukumura, T., Makino, F., Dietsche, T., Kinoshita, M., Kato, T., Wagner, S., et al. (2017). Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 15:e2002281. doi: 10.1371/journal.pbio.2002281
Furukawa, Y., Inoue, Y., Sakaguchi, A., Mori, Y., Fukumura, T., Miyata, T., et al. (2016). Structural stability of flagellin subunit affects the rate of flagellin export in the absence of FliS chaperone. Mol. Microbiol. 102, 405–416. doi: 10.1111/mmi.13469
Gibbons, C., Montgomery, M. G., Leslle, A. G., and Walker, J. E. (2000). The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 7, 1055–1061. doi: 10.1038/80981
González-Pedrajo, B., Fraser, G. M., Minamino, T., and Macnab, R. M. (2002). Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol. Microbiol. 45, 967–982. doi: 10.1046/j.1365-2958.2002.03047.x
González-Pedrajo, B., Minamino, T., Kihara, M., and Namba, K. (2006). Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60, 984–998. doi: 10.1111/j.1365-2958.2006.05149.x
Hara, N., Morimoto, Y. V., Kawamoto, A., Namba, K., and Minamino, T. (2012). Interaction of the extreme N-terminal region of FliH with FlhA is required for efficient bacterial flagellar protein export. J. Bacteriol. 194, 5353–5360. doi: 10.1128/JB.01028-12
Hara, N., Namba, K., and Minamino, T. (2011). Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One 6:e22417. doi: 10.1371/journal.pone.0022417
Hendriksen, J. J., Lee, H. J., Bradshaw, A. J., Namba, K., Chevance, F. F. V., Minamino, T., et al. (2021). Genetic analysis of the Salmonella FliE protein that forms the base of the flagellar axial structure. mBio 12:e0239221. doi: 10.1128/mBio.02392-21
Hüsing, S., Halte, M., van Look, U., Guse, A., Gálvez, E. J. C., Charpentier, E., et al. (2021). Control of membrane barrier during bacterial type-III protein secretion. Nat. Commun. 12:3999. doi: 10.1038/s41467-021-24226-1
Ibuki, T., Imada, K., Minamino, T., Kato, T., Miyata, T., and Namba, K. (2011). Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat. Struct. Mol. Biol. 18, 277–282. doi: 10.1038/nsmb.1977
Ibuki, T., Uchida, Y., Hironaka, Y., Namba, K., Imada, K., and Minamino, T. (2013). Interaction between FliJ and FlhA, components of the bacterial flagellar type III export apparatus. J. Bacteriol. 195, 466–473. doi: 10.1128/JB.01711-12
Iino, T. (1974). Assembly of Salmonella flagellin in vitro and in vivo. J. Supramol. Struct. 2, 372–384. doi: 10.1002/jss.400020226
Imada, K., Minamino, T., Kinoshita, M., Furukawa, Y., and Namba, K. (2010). Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners. Proc. Natl. Acad. Sci. U. S. A. 107, 8812–8817. doi: 10.1073/pnas.1001866107
Imada, K., Minamino, T., Tahara, A., and Namba, K. (2007). Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc. Natl. Acad. Sci. U. S. A. 104, 485–490. doi: 10.1073/pnas.0608090104
Imada, K., Minamino, T., Uchida, Y., Kinoshita, M., and Namba, K. (2016). Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator. Proc. Natl. Acad. Sci. U. S. A. 113, 3633–3638. doi: 10.1073/pnas.1524025113
Inoue, Y., Kinoshita, M., Kida, M., Takekawa, N., Namba, K., Imada, K., et al. (2021). The FlhA linker mediates flagellar protein export switching during flagellar assembly. Commun. Biol. 4:646. doi: 10.1038/s42003-021-02177-z
Inoue, Y., Morimoto, Y. V., Namba, K., and Minamino, T. (2018). Novel insights into the mechanism of well-ordered assembly of bacterial flagellar proteins in Salmonella. Sci. Rep. 8:1787. doi: 10.1038/s41598-018-20209-3
Inoue, Y., Ogawa, Y., Kinoshita, M., Terahara, N., Shimada, M., Kodera, N., et al. (2019). Structural insight into the substrate specificity switching mechanism of the type III protein export apparatus. Structure 27, 965–976. doi: 10.1016/j.str.2019.03.017
Johnson, S., Furlong, E. J., Deme, J. C., Nord, A. L., Caesar, J. J. E., Chevance, F. F. V., et al. (2021). Molecular structure of the intact bacterial flagellar basal body. Nat. Microbiol. 6, 712–721. doi: 10.1038/s41564-021-00895-y
Johnson, S., Kuhlen, L., Deme, J. C., Abrusci, P., and Lea, S. M. (2019). The structure of an injectisome export gate demonstrates conservation of architecture in the core export gate between flagellar and virulence type III secretion systems. mBio 10, e00818–19. doi: 10.1128/mBio.00818-19
Kato-Yamada, Y., Yoshida, M., and Hisabori, T. (2000). Movement of the helical domain of the epsilon subunit is required for the activation of thermophilic F1-ATPase. J. Biol. Chem. 275, 35746–35750. doi: 10.1074/jbc.M006575200
Kawamoto, A., Miyata, T., Makino, F., Kinoshita, M., Minamino, T., Imada, K., et al. (2021). Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries. Nat. Commun. 12:4223. doi: 10.1038/s41467-021-24507-9
Kawamoto, A., Morimoto, Y. V., Miyata, T., Minamino, T., Hughes, K. T., Kato, T., et al. (2013). Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci. Rep. 3:3369. doi: 10.1038/srep03369
Kazetani, K., Minamino, T., Miyata, T., Kato, T., and Namba, K. (2009). ATP-induced FliI hexamerization facilitates bacterial flagellar protein export. Biochem. Biophys. Res. Commun. 388, 323–327. doi: 10.1016/j.bbrc.2009.08.004
Kihara, M., Minamino, T., Yamaguchi, S., and Macnab, R. M. (2001). Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183, 1655–1662. doi: 10.1128/JB.183.5.1655-1662.2001
Kinoshita, M., Aizawa, S.-I., Inoue, Y., Namba, K., and Minamino, T. (2017). The role of intrinsically disordered C-terminal region of FliK in substrate specificity switching of the bacterial flagellar type III export apparatus. Mol. Microbiol. 105, 572–588. doi: 10.1111/mmi.13718
Kinoshita, M., Hara, N., Imada, K., Namba, K., and Minamino, T. (2013). Interactions of bacterial chaperone-substrate complexes with FlhA contribute to coordinating assembly of the flagellar filament. Mol. Microbiol. 90, 1249–1261. doi: 10.1111/mmi.12430
Kinoshita, M., Namba, K., and Minamino, T. (2021). A positive charge region of Salmonella FliI is required for ATPase formation and efficient flagellar protein export. Commun. Biol. 4:464. doi: 10.1038/s42003-021-01980-y
Kishikawa, J., Ibuki, T., Nakamura, S., Nakanishi, A., Minamino, T., Miyata, T., et al. (2013). Common evolutionary origin for the rotor domain of rotary ATPases and flagellar protein export apparatus. PLoS One 8:e64695. doi: 10.1371/journal.pone.0064695
Kuhlen, L., Abrusci, P., Johnson, S., Gault, J., Deme, J., Caesar, J., et al. (2018). Structure of the core of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 25, 583–590. doi: 10.1038/s41594-018-0086-9
Kuhlen, L., Johnson, S., Cao, J., Deme, J. C., and Lea, S. M. (2021). Nonameric structures of the cytoplasmic domain of FlhA and SctV in the context of the full-length protein. PLoS One 16:e0252800. doi: 10.1371/journal.pone.0252800
Kuhlen, L., Johnson, S., Zeitler, A., Bäurle, S., Deme, J. C., Caesar, J. J. E., et al. (2020). The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion. Nat. Commun. 11:1296. doi: 10.1038/s41467-020-15071-9
Lee, P. C., Zmina, S. E., Stopford, C. M., Toska, J., and Rietsch, A. (2014). Control of type III secretion activity and substrate specificity by the cytoplasmic regulator PcrG. Proc. Natl. Acad. Sci. U. S. A. 111, E2027–E2036. doi: 10.1073/pnas.1402658111
Macnab, R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832
Majewski, D. D., Worrall, L. J., Hong, C., Atkinson, C. E., Vuckovic, M., Watanabe, N., et al. (2019). Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry. Nat. Commun. 10:626. doi: 10.1038/s41467-019-08477-7
McMurry, J. L., Murphy, J. W., and González-Pedrajo, B. (2006). The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45, 11790–11798. doi: 10.1021/bi0605890
Minamino, T. (2014). Protein export through the bacterial flagellar type III export pathway. Biochim. Biophys. Acta 1843, 1642–1648. doi: 10.1016/j.bbamcr.2013.09.005
Minamino, T. (2018). Hierarchical protein export mechanism of the bacterial flagellar type III protein export apparatus. FEMS Microbiol. Lett. 365:fny117. doi: 10.1093/femsle/fny117
Minamino, T., and Macnab, R. M. (1999). Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181, 1388–1394. doi: 10.1128/JB.181.5.1388-1394.1999
Minamino, T., and Macnab, R. M. (2000c). Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35, 1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x
Minamino, T., and Macnab, R. M. (2000a). Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182, 4906–4914. doi: 10.1128/JB.182.17.4906-4914.2000
Minamino, T., and Macnab, R. M. (2000b). FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37, 1494–1503. doi: 10.1046/j.1365-2958.2000.02106.x
Minamino, T., and Namba, K. (2004). Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7, 5–17. doi: 10.1159/000077865
Minamino, T., and Namba, K. (2008). Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451, 485–488. doi: 10.1038/nature06449
Minamino, T., González-Pedrajo, B., Kihara, M., Namba, K., and Macnab, R. M. (2003). The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185, 3983–3988. doi: 10.1128/JB.185.13.3983-3988.2003
Minamino, T., González-Pedrajo, B., Oosawa, K., Namba, K., and Macnab, R. M. (2002). Structural properties of FliH, an ATPase regulatory component of the Salmonella type III flagellar export apparatus. J. Mol. Biol. 322, 281–290. doi: 10.1016/s0022-2836(02)00754-4
Minamino, T., González-Pedrajo, B., Yamaguchi, K., Aizawa, S.-I., and Macnab, R. M. (1999). FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34, 295–304. doi: 10.1046/j.1365-2958.1999.01597.x
Minamino, T., Iino, T., and Kutsukake, K. (1994). Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 176, 7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994
Minamino, T., Kawamoto, A., Kinoshita, M., and Namba, K. (2020b). Molecular organization and assembly of the export apparatus of flagellar type III secretion systems. Curr. Top. Microbiol. Immunol. 427, 91–107. doi: 10.1007/82_2019_170
Minamino, T., Inoue, Y., Kinoshita, M., and Namba, K. (2020a). FliK-driven conformational rearrangements of FlhA and FlhB are required for export switching of the flagellar protein export apparatus. J. Bacteriol. 202, e00637–19. doi: 10.1128/JB.00637-19
Minamino, T., Kazetani, K., Tahara, A., Suzuki, H., Furukawa, Y., Kihara, M., et al. (2006). Oligomerization of the bacterial flagellar ATPase FliI is controlled by its extreme N-terminal region. J. Mol. Biol. 360, 510–519. doi: 10.1016/j.jmb.2006.05.010
Minamino, T., Kinoshita, M., Hara, N., Takeuchi, S., Hida, A., Koya, S., et al. (2012a). Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 83, 1775–1788. doi: 10.1111/j.1365-2958.2011.07964.x
Minamino, T., Kinoshira, M., Imada, K., and Namba, K. (2012b). Interaction between FliI ATPase and a flagellar chaperone FliT during bacterial flagellar export. Mol. Microbiol. 83, 168–178. doi: 10.1111/j.1365-2958.2011.07924.x
Minamino, T., Morimoto, Y. V., Hara, N., Aldridge, P. D., and Namba, K. (2016b). The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog. 12:e1005495. doi: 10.1371/journal.ppat.1005495
Minamino, T., Kinoshita, M., Inoue, Y., Morimoto, Y. V., Ihara, K., Koya, S., et al. (2016a). FliH and FliI ensure efficient energy coupling of flagellar type III protein export in Salmonella. Microbiologyopen 5, 424–435. doi: 10.1002/mbo3.340
Minamino, T., Morimoto, Y. V., Hara, N., and Namba, K. (2011). An energy transduction mechanism used in bacterial flagellar type III protein export. Nat. Commun. 2:475. doi: 10.1038/ncomms1488
Minamino, T., Morimoto, Y. V., Kinoshita, M., Aldridge, P. D., and Namba, K. (2014). The bacterial flagellar protein export apparatus processively transports flagellar proteins even with extremely infrequent ATP hydrolysis. Sci. Rep. 4:7579. doi: 10.1038/srep07579
Minamino, T., Kinoshita, M., Morimoto, Y. V., and Namba, K. (2021a). The FlgN chaperone activates the Na+-driven engine of the Salmonella flagellar protein export apparatus. Commun. Biol. 4:335. doi: 10.1038/s42003-021-01865-0
Minamino, T., Morimoto, Y. V., Kinoshita, M., and Namba, K. (2021b). Membrane voltage-dependent activation mechanism of the bacterial flagellar protein export apparatus. Proc. Natl. Acad. Sci. U. S. A. 118:e2026587118. doi: 10.1073/pnas.2026587118
Minamino, T., Morimoto, Y. V., Kinoshita, M., and Namba, K. (2021c). Multiple roles of flagellar export chaperones for efficient and robust flagellar filament formation in Salmonella. Front. Microbiol. 12:756044. doi: 10.3389/fmicb.2021.756044
Minamino, T., Shimada, M., Okabe, M., Saijo-Hamano, Y., Imada, K., Kihara, M., et al. (2010). Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagellar type III protein export. J. Bacteriol. 192, 1929–1936. doi: 10.1128/JB.01328-09
Minamino, T., Tame, J. R., Namba, K., and Macnab, R. M. (2001). Proteolytic analysis of the FliH/FliI complex, the ATPase component of the type III flagellar export apparatus of Salmonella. J. Mol. Biol. 312, 1027–1036. doi: 10.1006/jmbi.2001.5000
Minamino, T., Yamaguchi, S., and Macnab, R. M. (2000). Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182, 3029–3036. doi: 10.1128/JB.182.11.3029-3036.2000
Minamino, T., Yoshimura, S. D., Morimoto, Y. V., González-Pedrajo, B., Kami-Ike, N., and Namba, K. (2009). Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol. Microbiol. 74, 1471–1483. doi: 10.1111/j.1365-2958.2009.06946.x
Morimoto, Y. V., Ito, M., Hiraoka, K. D., Che, Y.-S., Bai, F., Kami-Ike, N., et al. (2014). Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol. Microbiol. 91, 1214–1226. doi: 10.1111/mmi.12529
Morimoto, Y. V., Kami-Ike, N., Miyata, T., Kawamoto, A., Kato, T., Namba, K., et al. (2016). High-resolution pH imaging of living bacterial cells to detect local pH differences. mBio 7, e01911–16. doi: 10.1128/mBio.01911-16
Nakamura, S., and Minamino, T. (2019). Flagella-driven motility of bacteria. Biomolecules 9:279. doi: 10.3390/biom9070279
Notti, R. Q., Bhattacharya, S., Lilic, M., and Stebbins, C. E. (2015). A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat. Commun. 6:7125. doi: 10.1038/ncomms8125
Okabe, M., Minamino, T., Imada, K., Namba, K., and Kihara, M. (2009). Role of the N-terminal domain of FliI ATPase in bacterial flagellar protein export. FEBS Lett. 583, 743–748. doi: 10.1016/j.febslet.2009.01.026
Pallen, M. J., Bailey, C. M., and Beatson, S. A. (2006). Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FOF1 and vacuolar ATPases. Protein Sci. 15, 935–941. doi: 10.1110/ps.051958806
Paul, K., Erhardt, M., Hirano, T., Blair, D. F., and Hughes, K. T. (2008). Energy source of flagellar type III secretion. Nature 451, 489–492. doi: 10.1038/nature06497
Paul, K., Harmon, J. G., and Blair, D. F. (2006). Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J. Bacteriol. 188, 5240–5248. doi: 10.1128/JB.00110-06
Rees, D. M., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2012). Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc. Natl. Acad. Sci. U. S. A. 109, 11139–11143. doi: 10.1073/pnas.1207587109
Renault, T. T., Abraham, A. O., Bergmiller, T., Paradis, G., Rainville, S., Charpentier, E., et al. (2017). Bacterial flagella grow through an injection-diffusion mechanism. Elife 6:e23136. doi: 10.7554/eLife.23236
Saijo-Hamano, Y., Imada, K., Minamino, T., Kihara, M., Shimada, M., Kitao, A., et al. (2010). Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol. Microbiol. 76, 260–268. doi: 10.1111/j.1365-2958.2010.07097.x
Sajó, R., Liliom, K., Muskotál, A., Klein, A., Závodszky, P., Vonderviszt, F., et al. (2014). Soluble components of the flagellar export apparatus, FliI, FliJ, and FliH, do not deliver flagellin, the major filament protein, from the cytosol to the export gate. Biochim. Biophys. Acta 1843, 2414–2423. doi: 10.1016/j.bbamcr.2014.07.004
Shimabukuro, K., Yasuda, R., Muneyuki, E., Hara, K. Y., Kinosita, K. Jr., and Yoshida, M. (2003). Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. U. S. A. 100, 14731–14736. doi: 10.1073/pnas.2434983100
Takekawa, N., Kawamoto, A., Sakuma, M., Kato, T., Kojima, S., Kinoshita, M., et al. (2021). Two distinct conformations in 34 FliF subunits generate three different symmetries within the flagellar MS-ring. mBio 12, e03199–20. doi: 10.1128/mBio.03199-20
Tan, J., Zhang, X., Wang, X., Xu, C., Chang, S., Wu, H., et al. (2021). Structural basis of assembly and torque transmission of the bacterial flagellar motor. Cell 184, 2665–2679.e19. doi: 10.1016/j.cell.2021.03.057
Terahara, N., Inoue, Y., Kodera, N., Morimoto, Y. V., Uchihashi, T., Imada, K., et al. (2018). Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export. Sci. Adv. 4:eaao7054. doi: 10.1126/sciadv.aao7054
Terashima, H., Kawamoto, A., Tastumi, C., Namba, K., Minamino, T., and Imada, K. (2018). In vitro reconstitution of functional type III protein export and insights into flagellar assembly. mBio 9, e00988–18. doi: 10.1128/mBio.00988-18
Terashima, H., Tastumi, C., Kawamoto, A., Namba, K., Minamino, T., and Imada, K. (2020). In vitro autonomous construction of the flagellar axial structure in inverted membrane vesicles. Biomolecules 10:126. doi: 10.3390/biom10010126
Thomas, J., Stafford, G. P., and Hughes, C. (2004). Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. U. S. A. 101, 3945–3950. doi: 10.1073/pnas.0307223101
Vogler, A. P., Homma, M., Irikura, V. M., and Macnab, R. M. (1991). Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J. Bacteriol. 173, 3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991
Wagner, S., and Diepold, A. (2020). A unified nomenclature for injectisome-type type III secretion systems. Curr. Top. Microbiol. Immunol. 427, 91–107. doi: 10.1007/82_2020_210
Walker, J. E. (2013). The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 41, 1–16. doi: 10.1042/BST20110773
Walker, J. E., and Dickson, V. K. (2006). The peripheral stalk of the mitochondrial ATP synthase. Biochim. Biophys. Acta 1757, 286–296. doi: 10.1016/j.bbabio.2006.01.001
Ward, E., Renault, T. T., Kim, E. A., Erhardt, M., Hughes, K. T., and Blair, D. F. (2018). Type-III secretion pore formed by flagellar protein FliP. Mol. Microbiol. 107, 94–103. doi: 10.1111/mmi.13870
Watanabe, R., and Noji, H. (2013). Chemomechanical coupling mechanism of F1-ATPase: catalysis and torque generation. FEBS Lett. 587, 1030–1035. doi: 10.1016/j.febslet.2013.01.063
Xing, Q., Shi, K., Portaliou, A., Rossi, P., Economou, A., and Kalodimos, C. G. (2018). Structure of chaperone-substrate complexes docked onto the export gate in a type III secretion system. Nat. Commun. 9:1773. doi: 10.1038/s41467-018-04137-4
Keywords: ATPase, bacterial flagella, F0F1 ATP synthase, flagellar assembly, proton motive force (pmf), protein translocation, type III secretion system (T3SS)
Citation: Minamino T, Kinoshita M and Namba K (2022) Insight Into Distinct Functional Roles of the Flagellar ATPase Complex for Flagellar Assembly in Salmonella. Front. Microbiol. 13:864178. doi: 10.3389/fmicb.2022.864178
Received: 28 January 2022; Accepted: 13 April 2022;
Published: 04 May 2022.
Edited by:
Michael Manson, Texas A&M University, United StatesReviewed by:
Tom Duncan, Upstate Medical University, United StatesCopyright © 2022 Minamino, Kinoshita and Namba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tohru Minamino, dG9ocnVAZmJzLm9zYWthLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.