
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 09 May 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.860992
This article is part of the Research TopicAdvances and Challenges in the Detection and Treatment of Pathogenic Microorganisms in Infectious Disease ControlView all 25 articles
 Pengya Gao1,2†
Pengya Gao1,2† Changde Wu2†
Changde Wu2† Jin Zhang3
Jin Zhang3 Shuping Wang2
Shuping Wang2 Ying Huang1
Ying Huang1 Yinping Dong4
Yinping Dong4 Tingting Liu5
Tingting Liu5 Changyun Ye1
Changyun Ye1 Xuefang Xu1*
Xuefang Xu1* Wenwen Xin5*
Wenwen Xin5*Clostridium botulinum is the causative pathogen of botulism. Laboratory detection of C. botulinum is essential for clinical therapy treatment of botulism due to the difficulty in diagnosis, especially in infant botulism. The extreme toxicity of botulinum neurotoxin (BoNT) requires a sensitive detection method. Due to the detection limit of real-time quantitative PCR (q-PCR), a more sensitive detection method, micro-drop digital PCR (ddPCR) was applied in C. botulinum main serotypes A and B. The following performance criteria were evaluated by ddPCR: analytical sensitivity; repeatability; and diagnostic specificity. The limit of detection (LOD) was 0.84 and 0.88 copies/μl for BoNT A and B genes, respectively, by ddPCR with high specificity, compared to 5.04×102 and 6.91×102 copies/μl by q-PCR. It was increased 10 times compared with q-PCR in spiked stool samples. This improvement in sensitivity was especially important in clinical samples as more positive samples were detected by digital PCR compared with q-PCR. Meanwhile, enrichment time for low bacteria content samples was shortened by four hours both in serotypes A and B C. botulinum by ddPCR compared with q-PCR, which are important for laboratory diagnosis and epidemiology work.
Botulism is a life-threatening disease caused by the action of BoNTs produced by Clostridium botulinum (C. botulinum) (Weigand et al., 2015). The lethal amount of botulinum toxin in mice is 0.5~5 ng/kg, and about 1 ng/kg in humans, which is the strongest natural biological toxin known (Gill, 1982; Arnon et al., 2001). In recent years, many cases of botulism including infant botulism and food-related botulism have been diagnosed in China (Zhang et al., 2020; Lu et al., 2021; Zhu and Fu, 2021). The high mortality of botulism makes rapid diagnosis critical for treatment. Apart from the clinical symptoms and toxin exposure history, positive laboratory results are essential for clinical diagnosis. Laboratory detection of botulinum toxin and C. botulinum is also a growing concern due to the increasing cases of botulism in China (Xin et al., 2019). The only currently admissive standard method for detection and identification of botulinum neurotoxin is the mouse bioassays (MBAs) which cause animal ethics issue and are time-consuming (Ferreira et al., 2004). At present, the detection methods for toxin-producing species mainly include isolation and culture (CfDCaP (CDC), 2016), PCR methods (Cordoba et al., 2001; Akbulut and Grant, 2004; Heffron and Poxton, 2007; Kasai et al., 2007; Dahlsten et al., 2008; Joshy et al., 2008; Fach et al., 2009; Hill et al., 2010; Kirchner et al., 2010; Lindberg et al., 2010; Peck et al., 2010; Satterfield et al., 2010; Anniballi et al., 2013; Fohler et al., 2016; Le Marechal et al., 2018; Masters and Palmer, 2021), sequencing (Gonzalez-Escalona et al., 2018; Gonzalez-Escalona and Sharma, 2020), and matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy (MALDI-TOF MS) based bacterial identification (Kalb et al., 2015; Bano et al., 2017; Xin et al., 2019; Drigo et al., 2020; Tevell Aberg et al., 2021). Most of these are time-consuming, labor-intensive, and not sensitive enough. LOD of q-PCR which is popularly used is normally between 101 and 102 copies in C. botulinum (Hill et al., 2010; Kirchner et al., 2010; Huang et al., 2019). However, clinical samples mostly contain low number of DNA molecules and below the LOD of q-PCR that can lead to false negative results by q-PCR. The highly toxic characterization of BoNT requires a more sensitive laboratory approach. Although ddPCR is widely used in many pathogens, there is no application in BoNT gene detection.
Micro-drop digital PCR is a fundamentally different method to quantifying the number of DNA compared with q-PCR (Gutierrez-Aguirre et al., 2015; Kuypers and Jerome, 2017; Maheshwari et al., 2017; Sun et al., 2018; Wang et al., 2018; Dupas et al., 2019; Capobianco et al., 2020; Cho et al., 2020; Liu et al., 2020; Xie et al., 2020; Yang et al., 2020). In ddPCR, the amplification reaction is compartmentalized into millions of independent partitions. Each partition as an individual reaction mixture contains either a single target molecule or none. The partitioned reactions are then amplified to the endpoint, which displays a positive or negative result. The absolute concentration of the target copies in the initial sample is gained from the number of positive and negative partitions (Kuypers and Jerome, 2017). Apart from the absolute quantification without reliance on a calibration curve, ddPCR has advantages not only in being less affected by sample inhibitors but also in better detection of low-copy-number samples and more precision (Morley, 2014).
Here, we aim to apply the ddPCR approach in detection of BoNT A and B genes, which are the main toxin serotypes in the clinical botulism. ddPCR assay was compared with q-PCR both in the clinical and spiked contaminated samples first revealing ddPCR assay was more sensitive than q-PCR in both neurotoxin A and B genes. Sensitivity of ddPCR was tested in 59 clinical stool samples which are positive by MBA. In total, 100% detection rate was found in ddPCR. The enrichment time for samples with low colony number of C. botulinum was also shortened.
Bacterial strains and plasmids used in this study are listed in Table 1. C. botulinum, Clostridioides difficile (C. difficile), and Clostridium perfringens (C. perfringens) strains were grown anaerobically at 37 °C in TPGY media (Xin et al., 2019). Escherichia coli (E. coli), Shigella flexneri (S. flexneri), and Shigella sonnei (S. sonnei) strains were grown at 37 °C in lysogeny broth (LB) media. Enterococcus faecium (E. faecium), Enterococcus faecalis (E. faecalis), and Listeria monocytogenes (L. monocytogenes) were grown at 37 °C in brain heart infusion (BHI) broth. For accurate calculating of gene copy numbers, two plasmids containing part of BoNT A and B genes were designated as CTA PMD18-T and CTB PMD18-T with primer pair AF and AR and BF and BR, respectively (Huang et al., 2019).
In total, 51 stool samples from hospitals or local Centers for Disease Control and Prevention confirmed by culture were used in this study. In total, 47 samples contain botulinum toxin B. The other seven are positive with botulinum toxin A. DNA extraction followed by qPCR and ddPCR were routinely immediately carried out after sample reception. Genomic DNA was extracted using QIAamp PowerFecal DNA Kit (catalog no. 51106; Qiagen, Germantown, MD) and stored at−20 °C until use. MBA and enrichment culture were usually performed in 1–2 days when the materials are prepared.
MBA and culture isolation of C. botulinum were both used as reference testing. MBA were carried out as mentioned by Wenwen Xin et al (Xin et al., 2019). Briefly, stool samples were diluted in GelPhos buffer (30 mM sodium phosphate (pH 6.2) and 0.2 % gelatin) in 1:50 and injected into 15–17 g ICR mouse intraperitoneally after centrifugation. Positive samples with classical symptoms were confirmed by antitoxins (Lanzhou Institute of Biological Products co., LTD). Stool samples were also cultured in cooked meat medium and TPGY media as described by Wenwen Xin et al. Identification was confirmed by Gram staining, MBA, and MALDI-TOF MS.
ddPCR is compatible with TaqMan hydrolysis probes as reported (Schaumann et al., 2018). So, same primers and probes were used for q-PCR and ddPCR (Table 1). Primer A–F, A–R, and probe A–P were used for BoNT A genes. Primers B–F, B–R, and probe B–P were used for BoNT B gene. q-PCR was performed as described earlier (Huang et al., 2019). If Ct value ≤ 35 is considered positive. ddPCR was carried out with QX200™ Droplet Generator, QX200™ Droplet Reader, C1000 Touch™Thermal Cycle, PX1™ PCR Plate sealer (Bio-Rad, USA), Microdrop Digital PCR Quantification Kit (Bole Corporation, USA), and ddPCR Super mix (Bio-Rad, USA). The annealing temperature and experimental components of ddPCR for C. botulinum types A and B gene were optimized. For both C. botulinum types A and B gene primers, a series of 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 nmol/L were tested with a probe concentration of 800 nmol/L and 60 °C for annealing temperature. A series of 100, 150, 200, 250, 300, 350, 400, and 450 nmol/L were tested for probes with selected concentration primers and 60°C for annealing temperature. Twelve gradients including 46.0, 47.1, 48.6, 50.4, 52.6, 54.8, 57.3, 59.4, 61.6, 63.4, 64.9, and 66.0 °C were examined with selected concentration of primers and probes.
C. botulinum types A or B strains were inoculated into TPGY medium and incubated for 3 days in anaerobic cabinet at 37 °C. Colony-forming units (CFUs) were calculated on plates. Each 0.25 g of stool suspended in 1 ml gelatin phosphate buffer from healthy infants was added to 6 tubes containing 15 ml TPGY medium. In total, 100 μl C. botulinum types A or B strains of six diluted culture (from 106 to 101) were inoculated to 6 tubes. The blank control was added with 100 μl distilled water. Genomic DNA was extracted as described earlier. ddPCR and q-PCR were both performed in triplicate.
C. botulinum types A and B strains were anaerobically inoculated into TPGY medium at 30 °C for 7 days until 99 % vegetative cells turn into spore-bearing vegetative sporangia. The harvested spores were washed by distilled water. The suspension was heated at 60 °C for 15 min to kill the vegetative cells. A spore suspension containing 106 cfu/ml was obtained. The suspensions were serially diluted 10-fold with sterile saline. The spore numbers were calculated on plates after culture and 10 and 100 CFU/ml were used for inoculation. Since inoculation in TPGY medium, sampling for ddPCR and q-PCR was carried out every 4 h till 72 h.
Each ddPCR experiment should contain at least 10,000 droplets. To optimize the primer and probe concentrations, a series of each content was set as shown in Figure 1. For C. botulinum type A BoNT gene amplification, the optimal concentrations of primer and probe are 900 nmol/L and 250 nmol/L, respectively (Figures 1A,B). The optimized primer and probe concentrations for C. botulinum type B BoNT gene are 800 nmol/L and 450 nmol/L (Figures 1C,D). With the optimized concentrations of primers and probes, reactions at 57.3 °C gave a highest positive droplet proportion (Figures 1E,F).
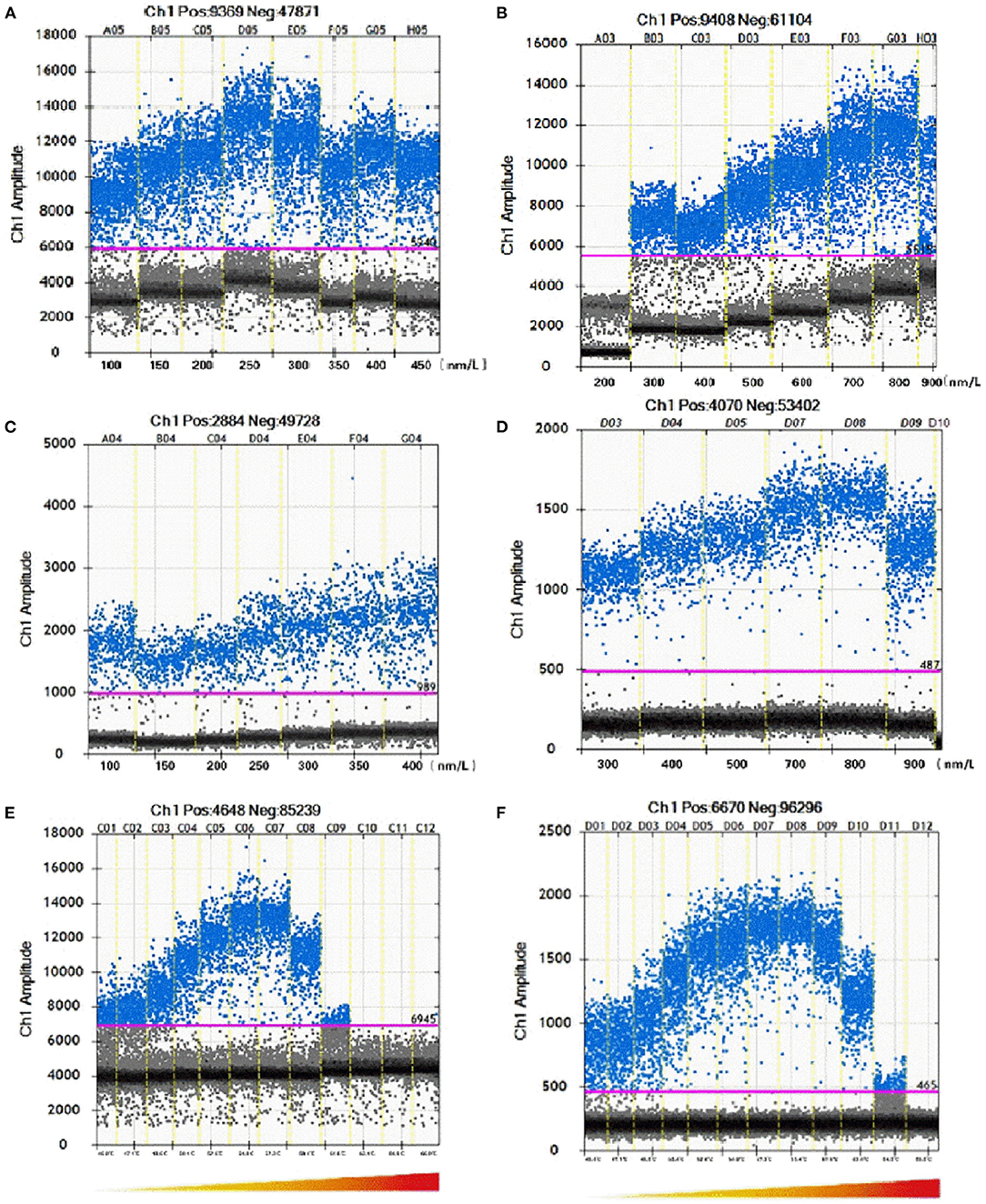
Figure 1. Optimization of ddPCR parameters including concentrations of primers and probes and annealing temperature. The pink line is the threshold. Blue dots represent positive droplets and gray dots represent negative droplets. (A,B) Demonstrated that ddPCR in C. botulinum serotype A with different concentrations of primers and probe. (C,D) Demonstrated that ddPCR in C. botulinum serotype B with different concentrations of primers and probes. (E,F) Demonstrated that ddPCR in C. botulinum serotypes A and B with different annealing temperature.
To test the specificity of ddPCR in C. botulinum types A and B BoNT genes, 2 C. botulinum and 10 other strains were used (Table 1). The target DNA from C. botulinum types A and B strains has been amplified successfully (Figure 2A), while no amplification was detected for the other nine control bacterial strains tested including C. botulinum serotype E (Figure 2A), indicating that primers and probes were specific for C. botulinum types A and B BoNT genes. The sensitivities of q-PCR and ddPCR were compared using constructed plasmid DNA as standard. Serial dilutions of CTA PMD18-T from 8.4 × 105-8.4 × 10−1 and CTB PMD18-T from 8.8 × 105-8.8 × 10−1 with triplicate were tested. The lowest concentration detected by ddPCR was 0.84 and 0.88 copies/μl for toxins A and B, respectively (Figures 2B,C). The LOD of q-PCR in serotypes A and B using same primers and probes were 5.04 × 102 and 6.91 × 102 copies/μl, respectively (Huang et al., 2019).
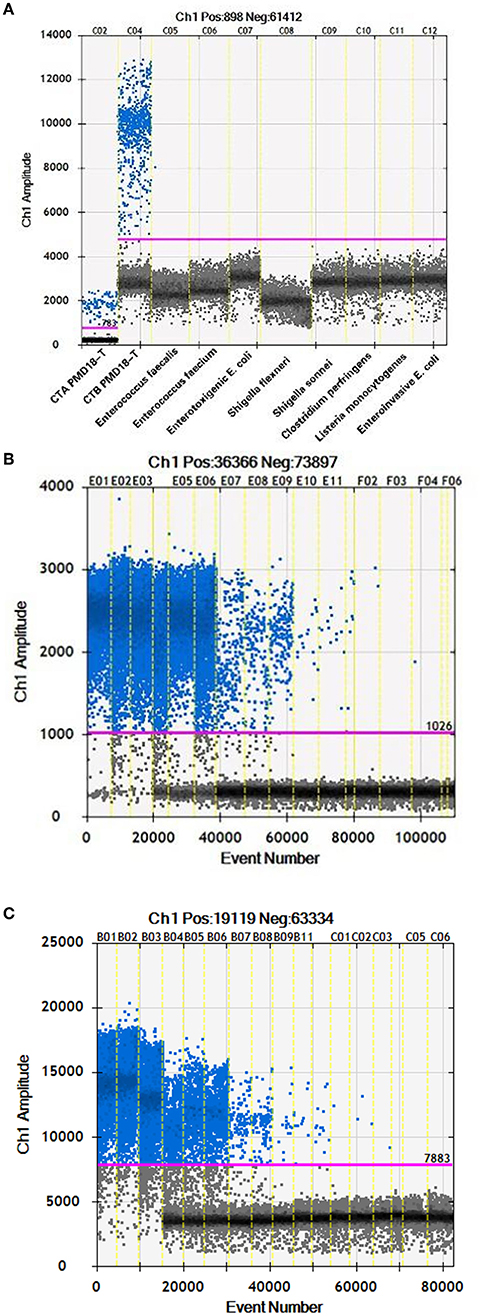
Figure 2. (A) The first two reactions were C. botulinum toxins A and B, respectively. (B) The 18 reactions were serial dilutions of CTA PMD18-T from 8.4×105 to 8.4×10−1 with triplicate. (C) The 18 reactions were serial dilutions of CTB PMD18-T from 8.8 × 105 to 8.8 × 10−1 with triplicate.
Serial dilutions of C. botulinum types A or B strain cultures were added to normal children's stool to mimic the clinical stool samples for sensitivity evaluation. The concentrations of C. botulinum type A strain were 8.1 × 103-8.1 × 100 CFU/100 μl. The diluted concentrations of C. botulinum type A strain were 9.7 × 103-9.7 × 100 CFU/100 μl. The LOD of C. botulinum types A or B in spiked stool is 81 and 97 CFU/μl, respectively, by ddPCR (Tables 2, 3). Consistently, 8.1 × 102 and 9.7 × 102 CFU/μl was the LOD with q-PCR of C. botulinum types A or B, respectively (Tables 2, 3).
All 59 suspected clinical stool samples were tested with MBA, culture isolation, ddPCR, and q-PCR (Supplementary Table S1). In these 59 stool samples, 47 were positive by MBA with 32 isolated strains, 21 are positive by q-PCR and 49 are positive by ddPCR. Among them, four stools were identified toxin A, isolated serotype A strains and confirmed by ddPCR with none was detected by q-PCR. In total, 43 stool samples were detected with toxin B by MBA in which 28 serotype B strains were isolated, 19 were positive by q-PCR and 45 were verified by ddPCR. Interestingly, two stools which are negative by MBA were detected by ddPCR.
The intra-batch reproducibility experiment is to repeat the same sample in the same reaction system for three times. The results showed that the number of positive droplets between the reactions of the same concentration template is similar, and the coefficient of variation of the botulinum toxin type A plasmid is 4.2% and 2.4%, 5.9%; the coefficient of variation of botulinum toxin type B plasmids were 4.5%, 4.9%, and 5.4%, both of which were <6%. It showed that the established ddPCR detection system has good repeatability.
To test the enrichment time of low concentration DNA samples which are below the LOD with q-PCR, two samples containing 10 and 100 spores were cultured in TPGYT medium. In total, 44 and 28 h were the shortest enrichment time for samples containing 10 and 100 spores of C. botulinum type A, respectively, by ddPCR (Tables 4, 5). Consistently, the shortest enrichment time were 48 and 32 h by q-PCR. For C. botulinum type B spores, 48 and 32 h were required for detection by ddPCR (Tables 4, 5) for 10 and 100 spores, respectively. Similarly, 52 and 36 h enrichment time were at least required by q-PCR.
Table 4. The growth of serotypes A and B strains in enrichment culture with 10 spores inoculation detected by ddPCR and q-PCR.
Table 5. The growth of serotypes A and B strains in enrichment culture with 100 spores inoculation detected by ddPCR and q-PCR.
C. botulinum serotypes A and B are the two main serotype causing botulism in China. As the difficulty in diagnosis, many cases of botulism are misdiagnosed which can be life-threatening (Arnon et al., 2001; CfDCaP (CDC), 2016). So, it is critically important to develop a rapid and sensitive method for the detection of botulinum toxin or BoNT-producing bacteria. Many methods have been developed to detect C. botulinum. MALDI-TOF MS-based bacterial identification is a rapid method providing robust accuracy in C. botulinum identification (Fenicia et al., 2007; Xin et al., 2019). However, MALDI-TOF MS-based bacterial identification requires enrichment and isolation of bacteria, which commonly takes days to complete, and this method cannot identify all BoNT-producing species or discriminate them from the related species (Xin et al., 2019). High-throughput sequencing and single molecule real-time sequencing could provide excellent phylogenetic information for typing and tracing the source of C. botulinum. Nevertheless, they are not used widely in clinical due to the requirements of highly trained staff and expensive instruments. q-PCR of toxin genes can serotype strains of C. botulinum and are widely used in clinical settings due to robustness, low cost, and simplicity. Kirchner et al. reported an LOD of 7–287 genomes by q-PCR in BoNT A–F in 95% possibilities (Kirchner et al., 2010). However, the detection probability in single PCR raised above 10 times in 100% possibilities. Fenicia et al. showed a LOD of 60 copies of C. botulinum type A by SYBR green real-time PCR (Barker et al., 2016). The other researchers demonstrated an LOD between 16–200 copies for BoNT gene by q-PCR (Akbulut and Grant, 2004; Kasai et al., 2007; Fach et al., 2009; Takahashi et al., 2010; Malakar et al., 2013). A minimum of 100 copies BoNT Agene was detected in spiked rice (Sedlak et al., 2014). In the laboratory, stool samples are more likely to produce a higher positive rate than serum. The gene copy number in the clinical stool DNA samples very likely below the LOD of q-PCR. Furthermore, inhibitors in stool samples can affect q-PCR efficiency (Mazaika and Homsy, 2014; Morley, 2014; Wang et al., 2016). ddPCR is an assay that combines state-of-the-art microfluidics technology with TaqMan-based PCR to achieve precise target DNA quantification at high levels of sensitivity and specificity. Because quantification is achieved without the need for standard assays in an easy to interpret, unambiguous digital readout, ddPCR is far simpler, faster, and less error prone than real-time qPCR (Mazaika and Homsy, 2014). In this study, we evaluated ddPCR in detection of C. botulinum toxin A and toxin B genes by comparing with q-PCR. It is seen that ddPCR not only can raise the sensitivity and shorten the enrichment time with high specificity, but also can increase the positive rate of spiked stool samples and clinical stool samples.
With optimized ddPCR, the sensitivity was increased by nearly 100 times with high specificity in constructed plasmids containing C. botulinum toxins A and B genes by comparing with q-PCR. In addition, the LOD was increased in spiked stool samples both in toxins A and B genes by ddPCR. This sensitivity increase remained by comparing with q-PCR and was quite critical in the clinical stool samples as proved. All 47 clinical stool samples were detected by MBA assay and were confirmed by ddPCR. Two suspected stool samples negative with MBA were identified by ddPCR indicating that ddPCR was more sensitive than MBA or none active toxin was absent in the two samples whereas toxin genes were present. Another possibility is the false positive or potential contamination in ddPCR in these two stool samples. However, as blank controls were included in every trial, this possibility is very low. Only 32 strains were isolated from 32 stool samples showing a low-isolation ratio or no live spores were existed in the other samples. Interestingly, only 21 samples were positive by qPCR suggesting a low-positive rate. This might due to the low sensitivity of qPCR and inhibitors factors in stool samples (Mazaika and Homsy, 2014; Morley, 2014; Wang et al., 2016). These results indicated that ddPCR can be used as a potential alternative diagnostic method for MBA.
Enrichment time for low colony number samples were evaluated here. Briefly, the earliest detection time was shortened by 4 h both in 10 and 100 spores samples in C. botulinum serotypes A and B strains with ddPCR. By sampling every 4 h since inoculation, samples containing 10 and 100 C. botulinum serotype A spores can be detected as early as 44 and 28 h, respectively, by ddPCR. This detection time was delayed by 4 h in C. botulinum serotype B spores. This indicated that growth rate differs in C. botulinum serotypes A and B strains with low-colony samples. An important consideration is the use of ddPCR in the isolation of C. botulinum. The good aspect is the sensitivity in the earlier detection from enrichment culture. As most enrichment cultures will be tested by qPCR or ddPCR and only positive cultures will be streaked in the plate. ddPCR shows the potential ability in detecting samples with few cells by its sensitivity. However, lack of DNA standards hinders the application of ddPCR in the clinical and laboratory tests.
In conclusion, here dd-PCR was demonstrated it can be used as a more accurate detection method in the clinical diagnosis by increasing sensitivity in stool samples and culture isolation by shortening enrichment time.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Laboratory Animal Welfare and Ethics Committee in IVDC, China CDC.
PG, CW, JZ, SW, YH, XX, and WX performed the experiments, analyzed the data, and wrote the manuscript. XX and WX conceptualized and designed the study. YD, TL, and CY provided material and samples. All authors reviewed and edited the manuscript and read and approved the final manuscript.
This work was supported by the National Key Research and Development Program of China (No. 2018YFC1603800).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JH declared a shared affiliation with several of the authors, YH, CY, and XX, to the handling editor at time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.860992/full#supplementary-material
Akbulut, D., and Grant, K. A. (2004). McLauchlin J: Development and application of Real-Time PCR assays to detect fragments of the Clostridium botulinum types A, B, and E neurotoxin genes for investigation of human foodborne and infant botulism. Foodborne Pathog. Dis. 1, 247–257. doi: 10.1089/fpd.2004.1.247
Anniballi, F., Auricchio, B., Woudstra, C., Fach, P., Fiore, A., Skarin, H., et al. (2013). D: Multiplex real-time PCR for detecting and typing Clostridium botulinum group III organisms and their mosaic variants. Biosecurity and bioterrorism: biodefense strategy, practice, and science 11, S207–214. doi: 10.1089/bsp.2012.0084
Arnon, S. S., Schechter, R., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., et al. (2001). Botulinum toxin as a biological weapon: medical and public health management. JAMA. 285, 1059–1070. doi: 10.1001/jama.285.8.1059
Bano, L., Drigo, I., Tonon, E., Pascoletti, S., Puiatti, C., Anniballi, F., et al. (2017). Identification and characterization of Clostridium botulinum group III field strains by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe. 48, 126–134. doi: 10.1016/j.anaerobe.2017.08.004
Barker, G. C., Malakar, P. K., Plowman, J., and Peck, M. W. (2016). Quantification of nonproteolytic Clostridium botulinum spore loads in food materials. Appl. Environm. Microbiol. 82, 1675–1685. doi: 10.1128/AEM.03630-15
Capobianco, J. A., Clark, M., Cariou, A., Leveau, A., Pierre, S., Fratamico, P., et al. (2020). Detection of shiga toxin-producing Escherichia coli (STEC) in beef products using droplet digital PCR. Int. J. Food Microbiol. 319, 108499. doi: 10.1016/j.ijfoodmicro.2019.108499
CfDCaP (CDC) (2016). Botulism. Available onlne at: http://wwwcdcgov/botulism/
Cho, S. M., Shin, S., Kim, Y., Song, W., Hong, S. G., Jeong, S. H., et al. (2020). A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR. Clin. Microbiol. Infect. e941-−942 e945. doi: 10.1016/j.cmi.2019.11.012
Cordoba, M. G., Aranda, E., Medina, L. M., Jordano, R., and Cordoba, J. J. (2001). Differentiation of Clostridium perfringens and Clostridium botulinum from non-toxigenic clostridia, isolated from prepared and frozen foods by PCR-DAN based methods. Die Nahrung. 45, 125–128. doi: 10.1002/1521-3803(20010401)45:2<125::AID-FOOD125>3.0.CO;2-8
Dahlsten, E., Korkeala, H., Somervuo, P., and Lindstrom, M. (2008). PCR assay for differentiating between Group I (proteolytic) and Group II (nonproteolytic) strains of Clostridium botulinum. Int. J. Food Microbiol. 124, 108–111. doi: 10.1016/j.ijfoodmicro.2008.02.018
Drigo, I., Tonon, E., Pascoletti, S., Anniballi, F., Kalb, S. R., and Bano, L. (2020). Detection of active BoNT/C and D by EndoPep-MS using MALDI biotyper instrument and comparison with the mouse test bioassay. Toxins. 13. doi: 10.3390/toxins13010010
Dupas, E., Legendre, B., Olivier, V., Poliakoff, F., Manceau, C., and Cunty, A. (2019). Comparison of real-time PCR and droplet digital PCR for the detection of Xylella fastidiosa in plants. J. Microbiol. Methods. 162, 86–95. doi: 10.1016/j.mimet.2019.05.010
Fach, P., Micheau, P., Mazuet, C., Perelle, S., and Popoff, M. (2009). Development of real-time PCR tests for detecting botulinum neurotoxins A, B, E, F producing Clostridium botulinum, Clostridium baratii and Clostridium butyricum. J. Appl. Microbiol. 107, 465–473. doi: 10.1111/j.1365-2672.2009.04215.x
Fenicia, L., Anniballi, F., Medici, D.e, Delibato, D., and Aureli, E. (2007). P: SYBR green real-time PCR method to detect Clostridium botulinum type A. Appl. Environm. Microbiol. 73, 2891–2896. doi: 10.1128/AEM.02234-06
Ferreira, J. L., Eliasberg, S. J., Edmonds, P., and Harrison, M. A. (2004). Comparison of the mouse bioassay and enzyme-linked immunosorbent assay procedures for the detection of type A botulinal toxin in food. J. Food Protect. 67, 203–206. doi: 10.4315/0362-028X-67.1.203
Fohler, S., Discher, S., Jordan, E., Seyboldt, C., Klein, G., Neubauer, H., et al. (2016). Detection of Clostridium botulinum neurotoxin genes (A-F) in dairy farms from Northern Germany using PCR: a case-control study. Anaerobe. 39, 97–104. doi: 10.1016/j.anaerobe.2016.03.008
Gill, D. M (1982). Bacterial toxins: a table of lethal amounts. Microbiol. Rev. 46, 86–94. doi: 10.1128/mr.46.1.86-94.1982
Gonzalez-Escalona, N., Haendiges, J., Miller, J. D., and Sharma, S. K. (2018). Closed genome sequences of two clostridium botulinum strains obtained by nanopore sequencing. Microbiol. Resour. Announc. 7. doi: 10.1128/MRA.01075-18
Gonzalez-Escalona, N., and Sharma, S. K. (2020). Closing Clostridium botulinum Group I genomes using a combination of short- and long-reads. Front. Microbiol. 11, 239. doi: 10.3389/fmicb.2020.00239
Gutierrez-Aguirre, I., Racki, N., Dreo, T., and Ravnikar, M. (2015). Droplet digital PCR for absolute quantification of pathogens. Methods Mol. Biol. 1302, 331–347. doi: 10.1007/978-1-4939-2620-6_24
Heffron, A., and Poxton, I. R. (2007). A PCR approach to determine the distribution of toxin genes in closely related Clostridium species: Clostridium botulinum type C and D neurotoxins and C2 toxin, and Clostridium novyi alpha toxin. J. Medical Microbiol. 56, 196–201. doi: 10.1099/jmm.0.46802-0
Hill, B. J., Skerry, J. C., Smith, T. J., Arnon, S. S., and Douek, D. C. (2010). Universal and specific quantitative detection of botulinum neurotoxin genes. BMC Microbiol. 10, 267. doi: 10.1186/1471-2180-10-267
Huang, Y., Shi, Y., Ye, C., and Xu, X. (2019). Establishment of real-time PCR assays for rapid detection of Clostridium botulinum type A and B. Disease Surveillance 34, 844–848. doi: 10.3784/j.issn.1003-9961.2019.09.015
Joshy, L., Chaudhry, R., and Chandel, D. S. (2008). Multiplex PCR for the detection of Clostridium botulinum and C. perfringens toxin genes. Indian J. Med. Res. 128, 206–208.
Kalb, S. R., Boyer, A. E., and Barr, J. R. (2015). Mass spectrometric detection of bacterial protein toxins and their enzymatic activity. Toxins. 7, 3497–3511. doi: 10.3390/toxins7093497
Kasai, Y., Kimura, B., Tajima, Y., and Fujii, T. (2007). Quantitative duplex PCR of Clostridium botulinum types A and B neurotoxin genes. Shokuhin eiseigaku zasshi. 48, 19–26. doi: 10.3358/shokueishi.48.19
Kirchner, S., Kramer, K. M., Schulze, M., Pauly, D., Jacob, D., Gessler, F., et al. (2010). Pentaplexed quantitative real-time PCR assay for the simultaneous detection and quantification of botulinum neurotoxin-producing clostridia in food and clinical samples. Appl. Environ. Microbiol. 76, 4387–4395. doi: 10.1128/AEM.02490-09
Kuypers, J., and Jerome, K. R. (2017). Applications of digital PCR for clinical microbiology. J. Clin. Microbiol. 55, 1621–1628. doi: 10.1128/JCM.00211-17
Le Marechal, C., Fourour, S., Ballan, V., Rouxel, S., Souillard, R., and Chemaly, M. (2018). Detection of Clostridium botulinum group III in environmental samples from farms by real-time PCR using four commercial DNA extraction kits. BMC Res. Notes. 11, 441. doi: 10.1186/s13104-018-3549-5
Lindberg, A., Skarin, H., Knutsson, R., Blomqvist, G., and Baverud, V. (2010). Real-time PCR for Clostridium botulinum type C neurotoxin (BoNTC) gene, also covering a chimeric C/D sequence–application on outbreaks of botulism in poultry. Vet. Microbiol. 146, 118–123. doi: 10.1016/j.vetmic.2010.04.030
Liu, X., Feng, J., Zhang, Q., Guo, D., Zhang, L., Suo, T., et al. (2020). Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes & Infect. 9, 1175–1179. doi: 10.1080/22221751.2020.1772679
Lu, K., Wang, Y., Zhang, L., Li, C., Xing, F., Ye, H., et al. (2021). Analysis of 2 cases of foodborne botulism. J. Pract. Shock. 5, 59–60.
Maheshwari, Y., Selvaraj, V., Hajeri, S., and Yokomi, R. (2017). Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PloS ONE. 12, e0184751. doi: 10.1371/journal.pone.0184751
Malakar, P. K., Plowman, J., Aldus, C. F., Xing, Z., Zhao, Y., and Peck, M. W. (2013). Detection limit of Clostridium botulinum spores in dried mushroom samples sourced from China. Int. J. Food Microbiol. 166, 72–76. doi: 10.1016/j.ijfoodmicro.2013.06.010
Masters, A. M., and Palmer, D. G. (2021). Confirmation of botulism diagnosis in Australian bird samples by ELISA and RT rtPCR. J. Vet. Diagnostic Investigat. 10406387211014486. doi: 10.1177/10406387211014486
Mazaika, E., and Homsy, J. (2014). Digital droplet PCR: CNV analysis and other applications. Curr. Protoc. Hum. Genet. 82, 24 21–13. doi: 10.1002/0471142905.hg0724s82
Morley, A. A (2014). Digital PCR: A brief history. Biomol. Detect. Quantif. 1, 1–2. doi: 10.1016/j.bdq.2014.06.001
Peck, M. W., Plowman, J., Aldus, C. F., Wyatt, G. M., Izurieta, W. P., Stringer, S. C., et al. (2010). Development and application of a new method for specific and sensitive enumeration of spores of nonproteolytic Clostridium botulinum types B, E, and F in foods and food materials. Appl. Environ. Microbiol. 76, 6607–6614. doi: 10.1128/AEM.01007-10
Satterfield, B. A., Stewart, A. F., Lew, C. S., Pickett, D. O., Cohen, M. N., Moore, E. A., et al. (2010). A quadruplex real-time PCR assay for rapid detection and differentiation of the Clostridium botulinum toxin genes A, B, E and F. J. Med. Microbiol. 59, 55–64. doi: 10.1099/jmm.0.012567-0
Schaumann, R., Dallacker-Losensky, K., Rosenkranz, C., Genzel, G. H., Stingu, C. S., Schellenberger, W., et al. (2018). Discrimination of Human Pathogen Clostridium Species Especially of the Heterogeneous C. sporogenes and C. botulinum by MALDI-TOF Mass Spectrometry. Curr. Microbiol. 75, 1506–1515. doi: 10.1007/s00284-018-1552-7
Sedlak, R. H., Kuypers, J., and Jerome, K. R. (2014). A multiplexed droplet digital PCR assay performs better than qPCR on inhibition prone samples. Diagn. Microbiol. Infect. Dis. 80, 285–286. doi: 10.1016/j.diagmicrobio.2014.09.004
Sun, L., Talarico, S., Yao, L., He, L., Self, S., You, Y., et al. (2018). Droplet digital PCR-based detection of clarithromycin resistance in helicobacter pylori isolates reveals frequent heteroresistance. J. Clin. Microbiol. 56. doi: 10.1128/JCM.00019-18
Takahashi, H., Takakura, C., and Kimura, B. (2010). A quantitative real-time PCR method for monitoring Clostridium botulinum type A in rice samples. J. Food Protect. 73, 688–694. doi: 10.4315/0362-028X-73.4.688
Tevell Aberg, A., Karlsson, I., and Hedeland, M. (2021). Modification and validation of the Endopep-mass spectrometry method for botulinum neurotoxin detection in liver samples with application to samples collected during animal botulism outbreaks. Anal. Bioanal. Chem. 413, 345–354. doi: 10.1007/s00216-020-03001-z
Wang, D., Yamahara, K. M., Cao, Y., and Boehm, A. B. (2016). Absolute quantification of enterococcal 23S rRNA gene using digital PCR. Environm. Sci. Technol. 50, 3399–3408. doi: 10.1021/acs.est.5b05747
Wang, M., Yang, J., Gai, Z., Huo, S., Zhu, J., Wang, L., et al. (2018). Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. Int. J. Food Microbiol. 266, 251–256. doi: 10.1016/j.ijfoodmicro.2017.12.011
Weigand, M. R., Pena-Gonzalez, A., Shirey, T. B., Broeker, R. G., Ishaq, M. K., Konstantinidis, K. T., et al. (2015). Implications of genome-based discrimination between clostridium botulinum group i and clostridium sporogenes strains for bacterial taxonomy. Appl. Environ. Microbiol. 81, 5420–5429. doi: 10.1128/AEM.01159-15
Xie, S., Wang, Y. u. H, Cheng, Q., and Ding, Y. (2020). T: Two rapid and sensitive methods based on TaqMan qPCR and droplet digital PCR assay for quantitative detection of Bacillus subtilis in rhizosphere. J. Appl. Microbiol. 128, 518–527. doi: 10.1111/jam.14481
Xin, W., Huang, Y., Li, J. i. B, Wu, P., Liu, Y., et al. (2019). S et al: Identification and characterization of Clostridium botulinum strains associated with an infant botulism case in China. Anaerobe. 55, 1–7. doi: 10.1016/j.anaerobe.2018.06.015
Yang, J., Zhang, N., Lv, J., Zhu, P., Pan, X., Hu, J., et al. (2020). Comparing the performance of conventional PCR, RTQ-PCR, and droplet digital PCR assays in detection of Shigella. Mol. Cell. Probes. 51, 101531. doi: 10.1016/j.mcp.2020.101531
Zhang, A., Sun, Q., Huang, Y., Gao, P., Liu, Z., Peng, X., et al. (2020). Laboratory identification of an infant botulism B case. Dis. Surveill. 35, 557–562.
Keywords: Clostridium botulinum, droplet digital PCR, rapid clinical diagnosis, neurotoxin, q-PCR
Citation: Gao P, Wu C, Zhang J, Wang S, Huang Y, Dong Y, Liu T, Ye C, Xu X and Xin W (2022) Evaluation and Optimization of Microdrop Digital PCR for Detection of Serotype A and B Clostridium botulinum. Front. Microbiol. 13:860992. doi: 10.3389/fmicb.2022.860992
Received: 24 January 2022; Accepted: 11 February 2022;
Published: 09 May 2022.
Edited by:
Tianmu Chen, Xiamen University, ChinaReviewed by:
Jun Han, Chinese Center for Disease Control and Prevention, ChinaCopyright © 2022 Gao, Wu, Zhang, Wang, Huang, Dong, Liu, Ye, Xu and Xin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuefang Xu, eHV4dWVmYW5nQGljZGMuY24=; Wenwen Xin, eGlud3dAaG90bWFpbC5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.