- 1Institute of Biomedical Sciences, Shanxi University, Taiyuan, China
- 2Shanxi Provincial Key Laboratory of Medical Molecular Cell Biology, Shanxi University, Taiyuan, China
- 3Shanxi Provincial Key Laboratory for Prevention and Treatment of Major Infectious Diseases, Taiyuan, China
Porcine hemagglutinating encephalomyelitis virus (PHEV) is a Betacoronavirus characterized by neurological symptoms and a worldwide prevalence. Although PHEV is one of the earliest discovered porcine coronaviruses, it remains poorly studied. The full-length genome of the earliest PHEV strain collected in 1970 in the United States (PHEV/67 N/US/1970) was determined in October 2020. Using this virus as a prototype, we comparatively analyzed all available PHEV full-length sequences during 1970–2015. In phylogenetic trees based on PHEV full-length or spike glycoprotein open reading frame genomic sequences, PHEV/67 N/US/1970 was sorted into a clade different from that of viruses isolated in the United States in 2015. Intriguingly, United States and Belgium viruses isolated in 2015 and 2005, respectively, revealed multiple deletion mutation patterns compared to the strain PHEV/67 N/US/1970, leading to a truncated or a non-functional NS2A coding region. In addition, the genomic similarity analysis showed a hypervariability of the spike glycoprotein coding region, which can affect at least eight potential linear B cell epitopes located in the spike glycoprotein. This report indicates that PHEVs in the United States underwent a significant genetic drift, which might influence PHEV surveillance in other countries.
Introduction
Coronaviruses (CoVs) rank among the largest groups of enveloped positive-sense, single-stranded RNA viruses, which fall under the Coronavirinae subfamily, Coronaviridae family, in Nidovirales order (Payne, 2017). In humans and vertebrates, the clinical picture of CoVs is dominated by the infection of three major systems, including the respiratory, digestive, and central nervous systems (CNS; Chen et al., 2020). CoVs are classified into four genera: Alpha-CoV, Beta-CoV, Gamma-CoV, and Deltacoronavirus (de Groot et al., 2012). Human CoVs are generally attributed to a zoonotic origin before leaping into an intermediate host or directly to humans (Guan et al., 2003). The severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) in 2002/2003 (Ksiazek et al., 2003), the Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 and the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first identified in December 2019 in Wuhan, China (Zhou et al., 2020), confirmed that CoVs are likely originated in animals. The emergence of these three zoonotic severe Betacoronavirus outbreaks in less than two decades indicates the adaptation potentials of CoVs, their genetic plasticity, and the expansion of host range, which prompts us to explore the evolutionary characters of another mammal Betacoronavirus: the porcine hemagglutinating encephalomyelitis virus (PHEV).
Pigs are a natural host of six viruses from the Coronaviridae family (Wang et al., 2019), four of which were identified as Alpha-CoVs, including transmissible gastroenteritis virus (TGEV; Doyle and Hutchings, 1946), porcine epidemic diarrhea virus (PEDV; Wood, 1977), porcine respiratory coronavirus (PRCV; Pensaert et al., 1986), and the newly discovered swine acute diarrhea syndrome coronavirus (SADS-CoV; Gong et al., 2017; Pan et al., 2017; Zhou et al., 2018); one Beta-CoV PHEV (Greig et al., 1962); and the porcine Deltacoronavirus (PDCoV; Woo et al., 2012).
Porcine hemagglutinating encephalomyelitis virus is one of the earliest identified and isolated porcine CoVs and the only known neurotropic virus in pigs (Mora-Diaz et al., 2019). PHEV infection was first reported in the Ontario province of Canada in 1957 (Roe and Alexander, 1958) and the viral pathogen was first isolated from the brains of encephalitis piglets in 1962 (Greig et al., 1962). Since then, PHEV infection has been reported in countries of America, Europe, and Asia, including the United States (Cutlip and Mengeling, 1972), Argentina (Quiroga et al., 2008), Great Britain (Cartwright et al., 1969), Belgium (Sasseville et al., 2001), Japan (Hirano and Ono, 1998), South Korea (Rho et al., 2011), and China (Gao et al., 2011; Dong et al., 2014).
In neonatal pigs, PHEV infection causes vomiting and wasting disease (VWD) or encephalomyelitis (Mengeling et al., 1972; Andries and Pensaert, 1981) with approximately 100% mortality rate and a high prevalence in the United States (Mora-Diaz et al., 2020). The primary replication of PHEV starts in the upper respiratory tract and tonsils, with non-specific clinical signs, including sneezing, coughing, and transient fever of 1–2 days (Andries and Pensaert, 1980; Hirahara et al., 1987). During the incubation, PHEV can penetrate the CNS (Andries and Pensaert, 1980) and invade predominantly neurons with a massive activation of the surrounding microglia and an obvious inflammation of CNS (J. Zhang et al., 2021). During the early stage of PHEV infection, microglial activation leads to a monumental proinflammatory cytokine release (IL-1β, IL-6, TNF-α, and IFN-γ) to limit the viral replication (Zhang et al., 2021). However, an excessive microglial activation at a late stage, with a blood–brain barrier (BBB) destruction and a monocytes/macrophages infiltration of CNS, aggravates PHEV infection (Zhang et al., 2021). Furthermore, PHEV causes lysosomal disorders by decreasing the expression of progranulin protein (Lan et al., 2020), a significant accumulation of autophagosomes (APs) in nerve cells (Li et al., 2021) and other neurological dysfunctions (Mora-Diaz et al., 2021a). Although pigs of any age can be infected, the clinical manifestation depends on the age at PHEV-infection time (Hirano, 2004). In older pigs, PHEV is highly seroprevalent and usually subclinical (Dong et al., 2014; Mora-Diaz et al., 2020). However, a recent report showed the persistent virus shedding in feces and oral fluid with an active humoral immune response (IgM, IgG) in subclinical grower pigs suggesting an active viral replication attenuated by the immune response (Mora-Diaz et al., 2021b).
Porcine hemagglutinating encephalomyelitis virus belongs to Betacoronavirus genus, Embecovirus subgenus (Zhi et al., 2005; Llanes et al., 2020). The RNA genome of PHEV is about ~30 kb in length and shows a short untranslated region (UTR) at both ends (Fehr and Perlman, 2015). The genome is flanked with a 5′MET cap and a 3′ polyA tail (Fehr and Perlman, 2015) and contains at least 11 predicted open reading frames (ORFs; Vijgen et al., 2006; Shi et al., 2018). The first two-thirds of the viral genome is occupied by replicase ORF which encodes replicase polyprotein, a precursor for multiple nonstructural proteins (Nsps; Sola et al., 2015). The one-third 3′ region of the viral genome encodes five major structural proteins (hemagglutinin-esterase protein, HE; spike glycoprotein, S; envelope protein, E; membrane protein, M; and nucleocapsid protein, N) and three non-structural proteins (NS2A, NS4.9, and NS12.7; Weiss and Navas-Martin, 2005; Vijgen et al., 2006; Wang et al., 2019). Among them, N protein is the most abundant structural protein that binds to viral genome RNA. M and E are membrane proteins (Wang et al., 2019). Whereas, HE protein which is not present in other Beta-CoVs, is characterized by hemagglutinating (HA), acetyl-esterase (AE) or receptor-destroying and binding activities (Cornelissen et al., 1997).
The S glycoprotein of coronaviruses is a viral envelope protein with a binding specificity regulated by a receptor-binding domain (RBD; Bosch et al., 2003; Dong et al., 2015). S glycoprotein facilitates the attachment of the virion and its penetration into the host cells by recognizing and interacting with a proper cellular receptor (Schultze et al., 1992). During the entry of viral particles, S glycoprotein is cleaved into two subunits, named S1 and S2 subunits (Walls et al., 2016). S glycoprotein of PHEV is composed of about 1,349 amino acids (aa; Dong et al., 2015) and contains several conserved domains as determined in NCBI conserved domain database (CDD; Marchler-Bauer et al., 2017). The S1 subunit contains N-terminal domain (NTD, aa 16–298) and S1 receptor-binding domain (S1-RBD, aa 311–608), whereas the S2 subunit contains an S2 domain (aa 784–1,276), a transmembrane structure (TM, aa 1,294–1,316), and a cytosolic tail at the C-terminus. The S1-RBD of PHEV has been shown to bind specifically to neural cell adhesion molecules (NCAM), also named CD56 (Dong et al., 2015), which is involved in neuronal infection and injuries (Gao et al., 2010). The S2 subunit controls the membrane fusion between virion and cell during viral entry into the host cells (Bosch et al., 2003; Walls et al., 2016).
In China, PHEV was first reported in a pig farm in Beijing in 1985, then in Jilin, Liaoning, Shandong, and other provinces (Gao et al., 2011; Chen et al., 2012; Dong et al., 2014). In 2007, two pig farms in Changchun City and Siping City, Jilin Province of China, had outbreaks of PHEV, with mortality rates of 47.6 and 100%, respectively (Gao et al., 2011; Shi et al., 2018). In 2015, the influenza-like respiratory disease occurred in display pigs in Michigan, United States, and PHEV was finally identified in the specimens of clinically sick pigs (Lorbach et al., 2017). Currently, there is no commercially available vaccine against PHEV. The small interfering RNA (siRNA) technique has been suggested to have the potential to partially block the replication of PHEV by targeting the spike glycoprotein and replicase polyprotein genes (Lan et al., 2012) or cell host-specific proteins (Hu et al., 2020). Therefore, surveillance of this virus is the key to preventing the occurrence of an outbreak. In October 2020, the complete genome sequence of the virus PHEV/67 N/US/1970 collected in the United States in 1970 was determined. It represents the earliest full-length genome sequence information so far. In this study, we used this virus as a prototype to compare and analyze the full-length sequences of all other viral isolates and discussed the genome structure, genome similarity, and B cell epitopes to explore the evolutionary relationship and heredity of PHEV diversity, providing critical information for formulating disease control strategies suitable for PHEV.
Materials and Methods
Phylogenetic Tree Construction and Genomic Similarity Analysis
Fourteen full-length genome sequences of PHEVs isolated since 1970 were retrieved from NCBI GenBank. The viruses in this report were identified by their GenBank ID, name, country, and year of collection in a format as [GenBank ID: virus name (country-year of collection)]. The sequence of the virus PHEV/67 N/US/1970 (GenBank ID: MW165134.1) collected in 1970 was included in this analysis as a prototype reference. The phylogenetic trees were constructed using the neighbor-joining method embedded in the MEGA-X software (Kumar et al., 2004, 2018) based on PHEV full-length genomic sequence or the full-length sequence of individual ORFs. 1,000 repeated bootstrap analysis was applied to determine the percentage reliability value of each internal node of the phylogenetic tree. SimPlot ver.3.5.1 (Lole et al., 1999) was used to generate a similarity map of full-length PHEV genome.
Prediction of Linear B Cell Epitopes in S Glycoprotein of PHEV
To analyze the potential linear B cell epitopes in PHEV S glycoprotein, we used the BepiPred-2.0 server, running under IEDB (the immune epitope database, https://www.iedb.org/; Jespersen et al., 2017). The epitope and non-epitope amino acids were determined from the crystal structure using a Random Forest algorithm (Jespersen et al., 2017). Residues with scores higher than the threshold (set to 0.5) were predicted to be part of an epitope.
Three-Dimensional Modeling of PHEV S Glycoprotein Domains
I-TASER (Iterative Threading ASSEmbly Refinement) is a hierarchical method for protein structure modeling and structure-based functional annotation (Roy et al., 2010; Yang et al., 2015). I-TASER first identified structural templates from the RCSB protein data bank (PDB) by multiple threading approach LOMETS, then the construction of the full-length atomic models by iterative template-based fragment assembly simulations. I-TASER was performed to visualize the tertiary structure of three regions of PHEV S glycoprotein, including the NTD-containing region (aa 16–310), S1-RBD-containing region (aa 311–783), and S2 domain (aa 784–1,276).
Results
Phylogenetic Analysis Reveals Two Separate Evolutionary Clades of PHEVs
To determine the genomic characters, variation, and relationship between PHEV strains, we performed a phylogenetic analysis of the entire genomes or individual ORF sequences of PHEV isolates available since 1970 in the NCBI GenBank database using MEGA-X software (Kumar et al., 2004, 2018). We built phylogenetic trees based on 14 variants, isolated from the United States (11), China (2), and Belgium (1). The genomic sequence of PHEV/67 N/US/1970 virus (GenBank ID: MW165134.1) collected in the United States in 1970 was available in the NCBI GenBank database in October 2020, providing the earliest genetic information of PHEV. Therefore, this strain was included as a reference to explore the genetic diversity and evolution of PHEV over time. As indicated in Figure 1A, the phylogenetic tree of PHEV complete genome exhibits two main clades, where PHEV/67 N/US/1970 strain clustered together in one clade with strains isolated from Belgium in 2005 (PHEV/VW572, GenBank ID: DQ011855.1) and China in 2008 (PHEV/JL/2008, GenBank ID: KY994645.1) and 2014 (PHEV/CC14, GenBank ID: MF083115.1). The remaining strains isolated from the United States in 2015 (a total of 10) clustered together into another independent clade, which can be further divided into three lineage groups. To further understand the evolution of PHEV, we proceeded to individual ORF sequence-based phylogenetic analysis. In the phylogenetic tree of S glycoprotein ORF (Figure 1B), PHEV strains were sorted into two different evolutionary clades, similar to that of full-length viral genome (Figure 1A). In replicase polyprotein ORF-based phylogenetic tree, PHEV/67 N/US/1970 is still genetically closer to the strains isolated in China and Belgium (Figure 1G). However, in the phylogenetic trees based on ORFs of HE protein (Figure 1C), membrane protein (Figure 1D), envelop protein (Figure 1E), and nucleocapsid protein (Figure 1F), PHEV strains fell in different lineage groups other than groups indicated in the full-length genome phylogenetic tree (Figure 1A).
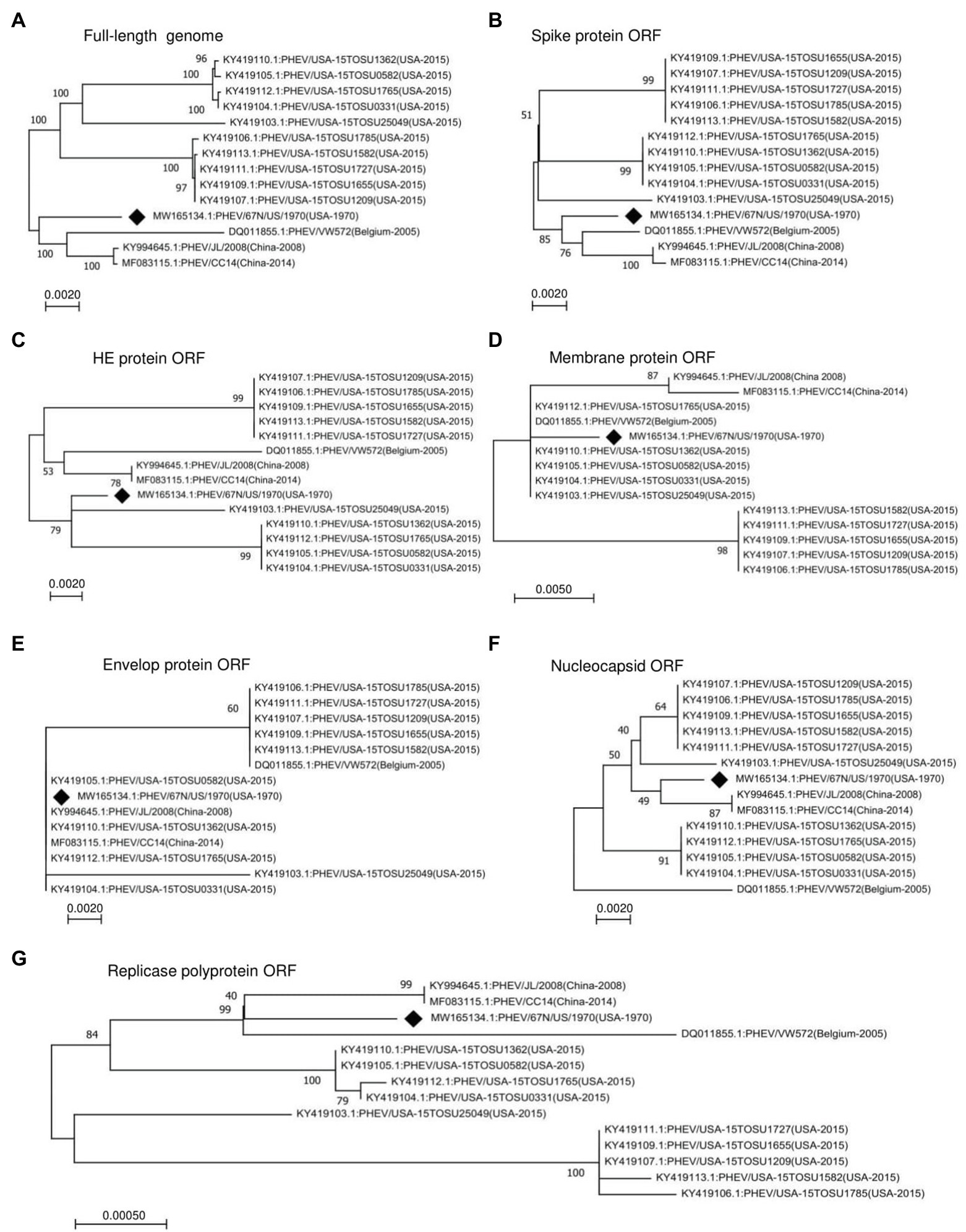
Figure 1. Phylogenetic analysis of 14 PHEV strains isolated between 1970 and 2015. Phylogenetic trees were constructed using the neighbor-joining algorithm in the MEGA-X software and based on (A) Full-length genome, or individual ORFs including (B) spike glycoprotein ORF, (C) HE glycoprotein ORF, (D) membrane protein ORF, (E) envelope protein ORF, (F) nucleocapsid protein ORF, and (G) replicase polyprotein ORF. The numbers on each branch are the bootstrap values (%) in 1000 replicates. The scale bar indicates the amino acid substitution at each position. The strain PHEV/67 N/US/1970 isolated in the United States in 1970 was labeled with a black diamond.
Two Highly Variable Regions of PHEV Genome: NS2A ORF and S Glycoprotein ORF
To further analyze the genetic characteristics and connectivity between PHEV strains, we conducted a genomic similarity analysis using SimPlot ver.3.5.1 software (Lole et al., 1999). Six viruses were included as representatives for each lineage group according to the whole genomic sequence-based phylogenetic tree (Figure 1A) and PHEV/USA-15TOSU1209 (GenBank ID: KY419107.1) was used as the query strain. As shown in Figure 2, the complete genome of PHEV/USA-15TOSU1209 (GenBank ID: KY419107.1) showed a great genomic similarity (>96%) with the included strains except for the region containing S glycoprotein and NS2A accessory protein ORFs that have the lowest genomic similarity, indicating their hypervariability. Sequence alignment revealed multiple deletion patterns in NS2A ORF region (Figure 3) of PHEVs isolated in the United States in 2015 and Belgium in 2005. Compared with PHEV/67 N/US/1970 (Figure 3), viruses isolated from China in 2008 (PHEV/JL/2008) and 2014 (PHEV/CC14, Data not shown) have no deletion mutations in the NS2A ORF region. In contrast, NS2A of viruses PHEV/VW572 from Belgium in 2005 and PHEV/USA-15TOSU25049 from the United States in 2015 has small deletions of 39 and 38 amino acids at the C-terminus, respectively. The representative viruses isolated in the United States in 2015 (PHEV/USA-15TOSU1362 and PHEV/USA-15TOSU1209) exhibited multiple deletions all through the NS2A ORF region, resulting in a non-functional expression of NS2A (Figure 3).
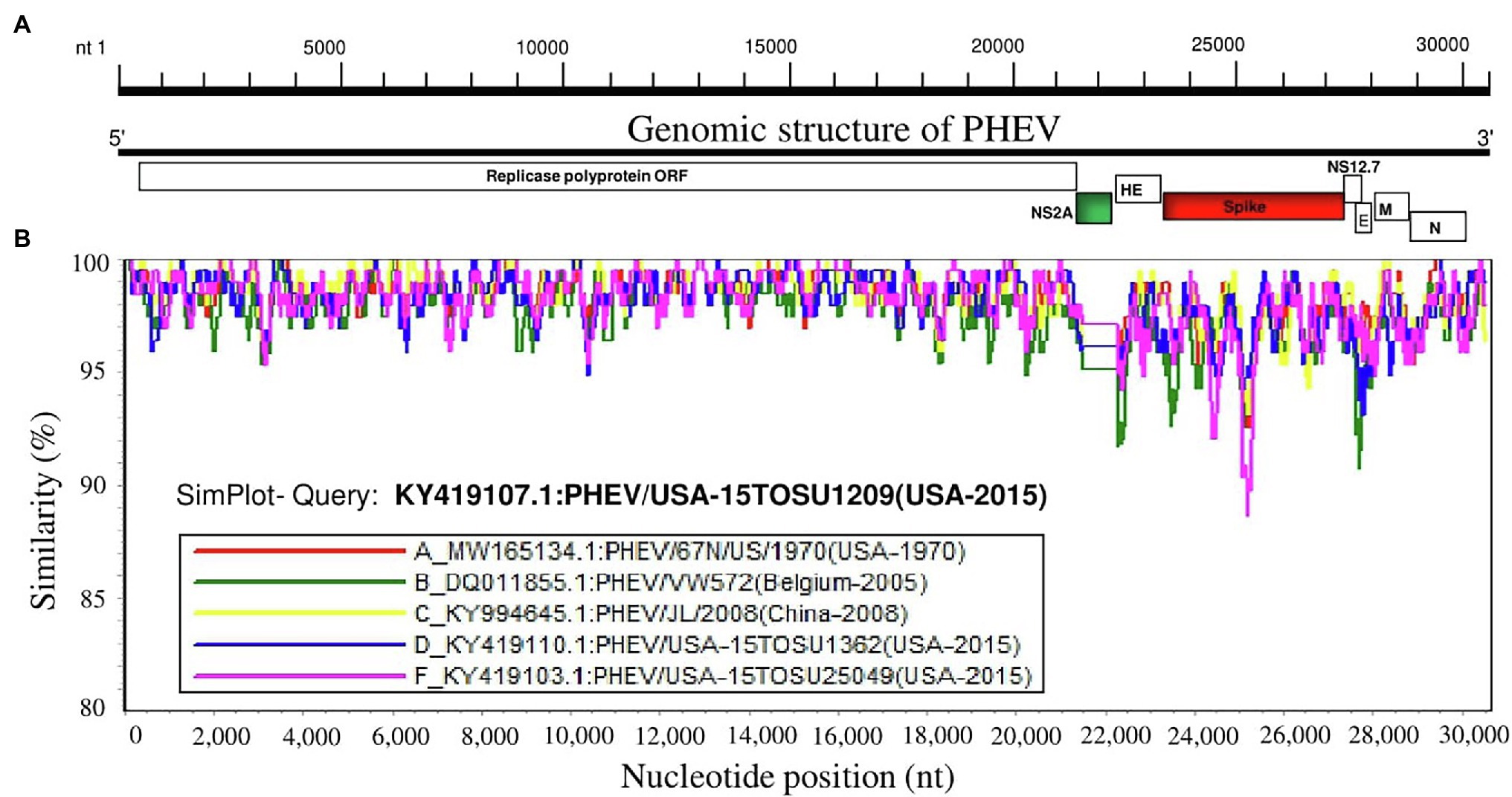
Figure 2. Similarity map of the full-length genome sequence of representative PHEV isolates. (A) Schematic diagram of the complete PHEV genomic structure. From the 5′end to the 3′ end are Replicase polyprotein ORF, accessory protein NS2A ORF, HE protein ORF, spike glycoprotein ORF, accessory protein NS12.7 ORF, envelope protein ORF (E), membrane protein ORF (M) and nucleocapsid protein ORF (N). (B) SimPlot similarity analysis results using SimPlot ver.3.5.1 (Lole et al., 1999). Virus PHEV/USA-15TOSU1209 (GenBank ID: KY419107.1) was used as the query sequence to compare with five other representative strains from the United States, China, and Belgium.
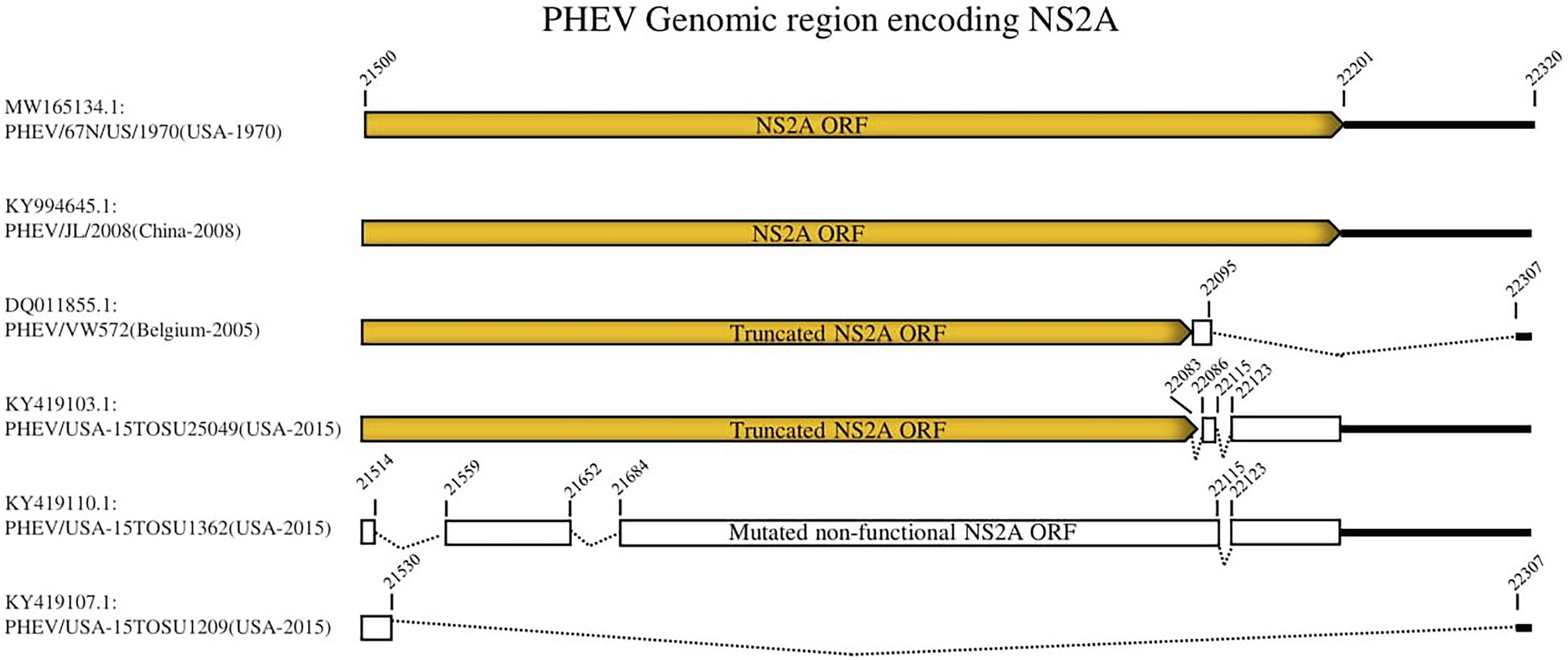
Figure 3. Deletion patterns in NS2A ORF region. The yellow box indicates the expression of full-length or truncated NS2A. The white box indicates the region of non-functional expression. Deleted regions are indicated with dashed lines. The numbers indicate the nucleotide positions relative to the genome of PHEV/67 N/US/1970 strain (GenBank ID: MW165134.1).
Linear B Cell Epitopes in S Glycoprotein of PHEV
S glycoprotein of coronaviruses is the major viral antigen that induces the generation of neutralizing antibodies during host infection, making it a major target protein for vaccine development (Du et al., 2009). Since S glycoprotein ORF of PHEV is found relatively highly variable (Figure 2), we assessed the effects of detected genomic variations on the antigenicity of S glycoprotein. For this aim, the linear B-cell epitopes on S glycoprotein of six representative viruses analyzed in genomic similarity analysis (Figure 2) were predicted using BepiPred-2.0 epitope prediction server (Jespersen et al., 2017). As indicated in Figure 4, a significant difference in the distribution of potential B cell epitopes was seen in S1-RBD (aa 311–608), whereas there were minor differences for the remaining part of S glycoprotein. Viruses PHEV/USA-15TOSU25049 (GenBank ID: KY419103.1) and PHEV/67 N/US/1970 (GenBank ID: MW165134.1) isolated in the same country (United States), were sorted into separate evolutionary clades in either full-length genome-based (Figure 1A) or S glycoprotein ORF-based phylogenetic tree (Figure 1B). However, compared with other viral strains, both viruses were genetically closer to each other in the S glycoprotein ORF-based phylogenetic tree (Figure 1B). Thus, the three-dimensional structures of NTD, S1-RBD, and S2 domains of these two viruses were modeled using I-TASER (Roy et al., 2010; Yang et al., 2015) to visualize the minimal amino acid variations seen in two main evolutionary clades in the potential linear B cell epitopes. For NTD or S2 domains of viruses PHEV/USA-15TOSU25049 and PHEV/67 N/US/1970, only two linear B cell epitopes were affected by amino acid variations (Figures 5 and 6, respectively), whereas four predicted epitopes were affected in S1-RBD (Figure 7). The mutations T571I and R574S eliminated the epitope S1-E4 in PHEV/67 N/US/1970. No amino acid insertions or deletions were seen in the potential linear B cell epitopes. In summary, the S1-RBD may be one of the main factors driving the antigenic drift of S glycoprotein of PHEV.
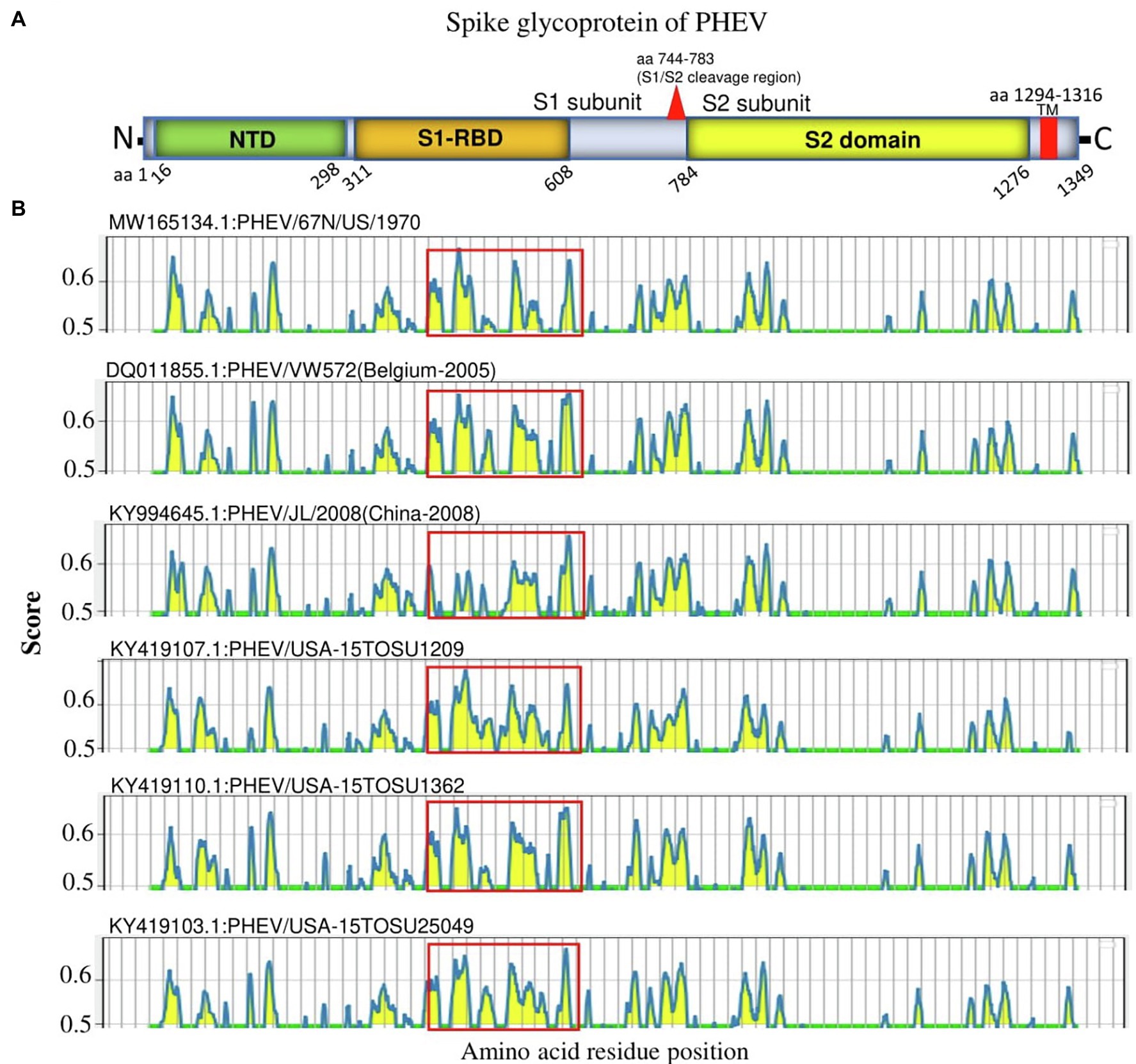
Figure 4. Linear B cell epitope map of the full-length S glycoprotein of PHEV. (A) Diagram of the main features of PHEV S glycoprotein, including NTD (aa 16–298), S1-RBD (aa 311–608), S2 domain (aa 784–1,276), and transmembrane domain (TM; aa 1,294–1,316). Numbers refer to the amino acid positions corresponding to PHEV/67 N/US/1970 S glycoprotein. (B) Mapping of linear B cell epitopes using the BepiPred-2.0 server. The Y-axes represents the residue score, and the X-axes represents the amino acid position, which is also relative to the diagram on the top (A). The predicted epitopes are indicated in yellow.
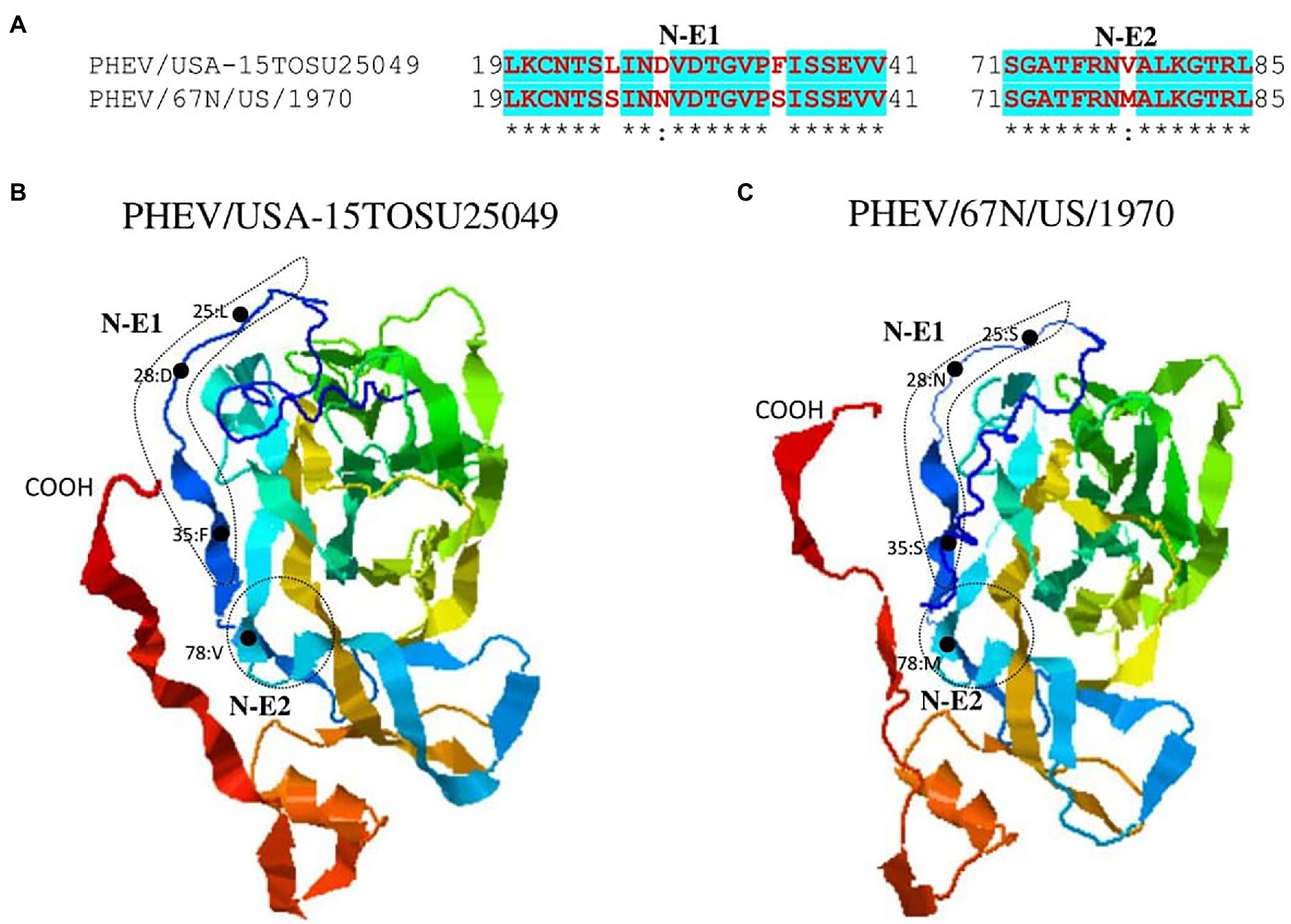
Figure 5. Comparison of linear B cell epitopes affected by amino acid variations in the NTD of S glycoprotein between PHEV/USA-15TOSU25049 and PHEV/67 N/US/1970. (A) Amino acids of the predicted linear B cell epitope are indicated with bold red letters, while numbers refer to their positions relative to the corresponding full-length S glycoprotein. Three-dimensional view of epitopes of PHEV/USA-15TOSU25049 (B) and PHEV/67 N/US/1970 (C). The tertiary structure was modeled using I-TASER. The predicted epitopes are circled, and amino acid variations are marked with black dots.
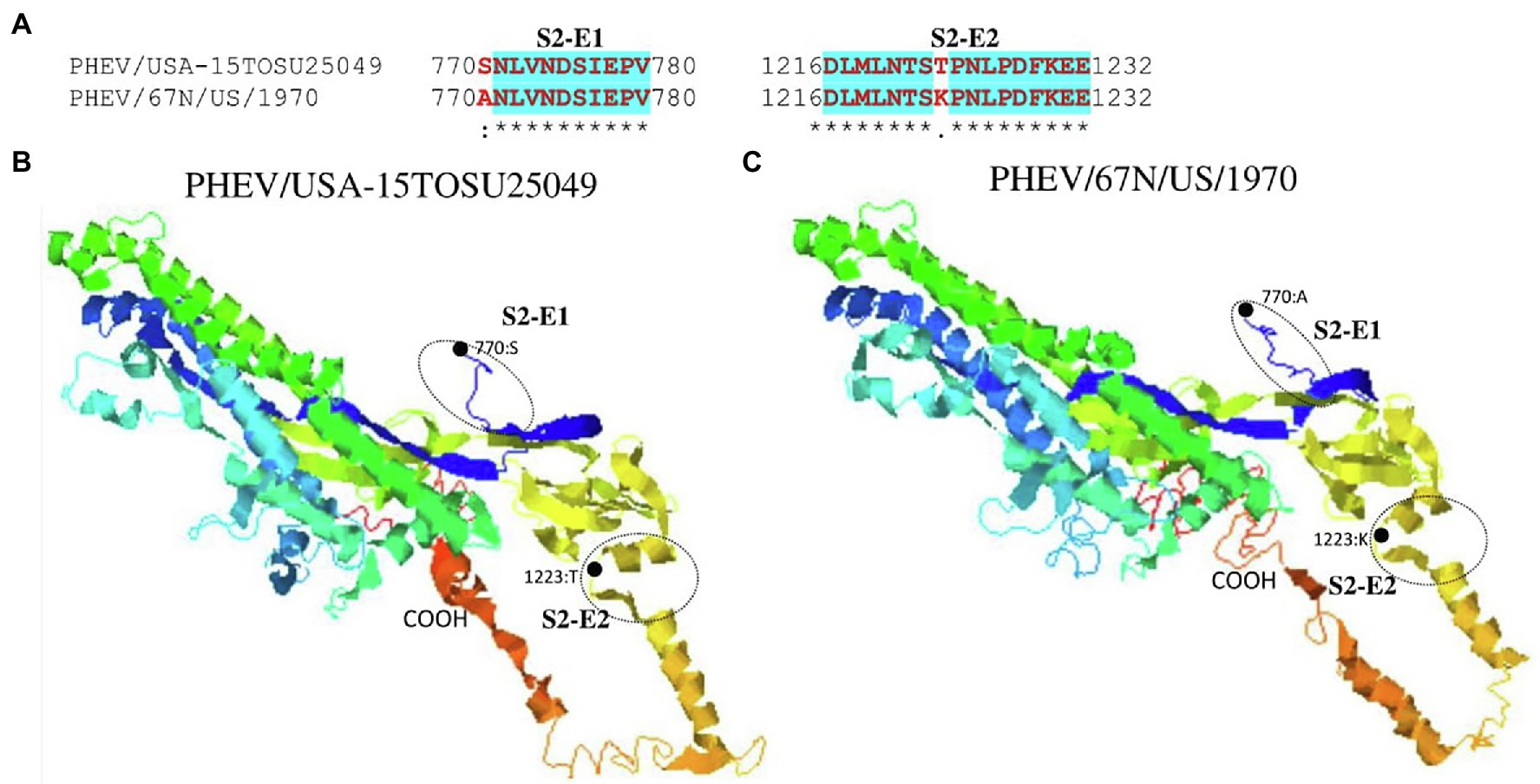
Figure 6. Comparison of linear B cell epitopes affected by amino acid variations in the S2 domain between PHEV/USA-15TOSU25049 and PHEV/67 N/US/1970. (A) Amino acids of the predicted linear B cell epitope are indicated with bold red letters, while numbers refer to their positions relative to the full-length S glycoprotein. Three-dimensional view of epitopes in the S2 domain of PHEV/USA-15TOSU25049 (B) and PHEV/67 N/US/1970 (C). The predicted epitopes are circled, and the amino acid variations are marked with black dots.
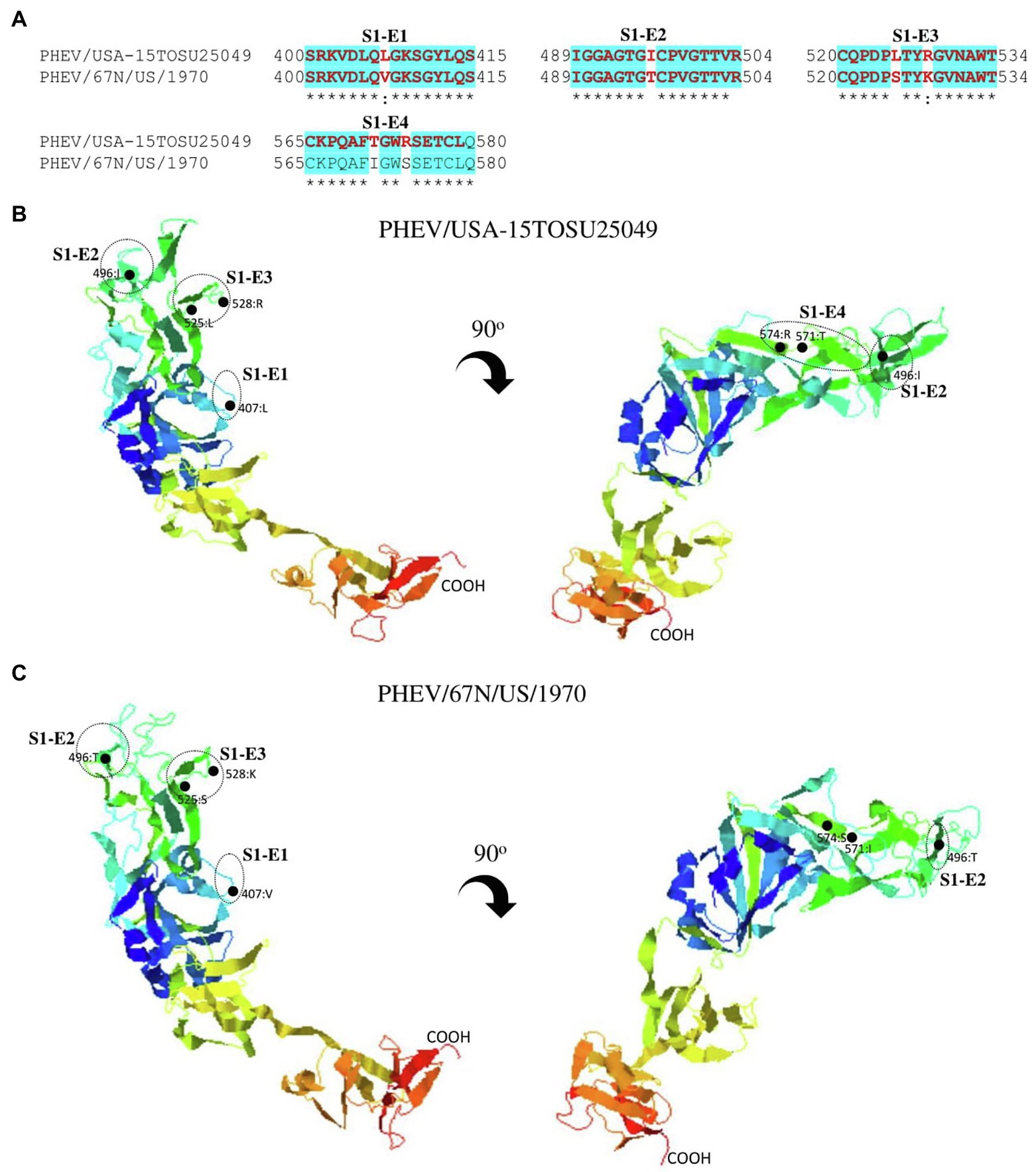
Figure 7. Comparison of linear B cell epitopes affected by amino acid variations in the S1-RBD between PHEV/USA-15TOSU25049 and PHEV/67 N/US/1970. (A) Amino acids of the predicted linear B cell epitope are indicated with bold red letters, while numbers refer to their positions relative to the corresponding full-length S glycoprotein. Three-dimensional view of epitopes in the S1-RBD of PHEV/USA-15TOSU25049 (B) or PHEV/67 N/US/1970 (C). The predicted epitopes are circled, and amino acid variations are marked with black dots.
Discussion
Although PHEV was one of the earliest identified and isolated swine CoVs, it remains after decades poorly documented and less studied with limited availability of genome sequences. In this report, we conducted a phylogenetic analysis based on the complete genome of the earliest isolated PHEV strain in the United States in 1970, available in October 2020, with 13 other isolates from different countries, including 10 from the United States, two from China, and one from Belgium. The phylogenetic tree showed two main clades, where the earliest United States strain (PHEV/67 N/US/1970) clustered together with the two strains from China and one from Belgium, whereas the other 10 United States variants (isolated in 2015) clustered together in another independent clade (Figures 1A,B), demonstrating the existence of two different circulating PHEV groups in the United States. Furthermore, NS2A and S glycoprotein ORFs were highly variable in PHEV genome, altering the expression of a normal NS2A and the composition of linear B cell epitopes in the S glycoprotein, particularly S1-RBD.
Compared with other Beta-CoVs from a supposed common ancestor such as human coronavirus OC43 and bovine coronavirus (Vijgen et al., 2006), NS2A of PHEV is shorter with a missing of ~84 amino acids at the C-terminus (Vijgen et al., 2006; Lau et al., 2012). NS2A belongs to the 2H phosphoesterase superfamily (Mazumder et al., 2002; Snijder et al., 2003), but its function in coronavirus replication is still unknown. In previous reports, mutation deletions in NS2A of PHEVs have been linked to severe respiratory manifestations in pigs (Lorbach et al., 2017; Wang et al., 2019), suggesting that the detected NS2A deletions might be involved in the pathogenicity of PHEV-mediated respiratory syndrome.
Besides NS2A, the S glycoprotein of coronaviruses is a direct target of cellular and humoral immune responses and a vaccine design focus since it represents the main viral surface antigen (Buchholz et al., 2004; Zhi et al., 2005; Tortorici et al., 2019; Dai and Gao, 2021). S glycoprotein mutations help escape the immune response and promote animal-human transmission through adaptation potentials (Zhang et al., 2006). The hypervariability of S glycoprotein is the main cause of changes in tissue tropism (Gallagher and Buchmeier, 2001; Hulswit et al., 2016). The deletion of about 216 or 227 amino acid residues in the N-terminus of S glycoprotein of TGEV has causally changed the enteric tissue tropism to a respiratory tissue tropism (named PRCV; Laude et al., 1993; Vaughn et al., 1995), which may be linked to the distribution of receptors over tissues or their different usage by different CoV strains (Whitworth et al., 2019). PDEV with the large deletion in the N terminus of S glycoprotein (about 194–216 amino acid residues) has shown a partial attenuation, although the tissue tropism was not altered (Hou et al., 2017). During PHEV infection, a crucial interaction between S glycoprotein and NCAM was identified (Gao et al., 2010), with a fragment (aa 291–548) in S1 subunit as a minimum number of amino acids essential to binding NCAM (Dong et al., 2015). Contrary to NS2A ORF region of PHEV, which showed significant deletions (Figure 3), S glycoprotein ORF did not undergo any deletion pattern.
Porcine hemagglutinating encephalomyelitis virus is the unique porcine neurotropic CoV with the potential to invade the central nervous system. Neurologic symptoms in PHEV-infected piglets with respiratory or digestive signs indicate the involvement of neurobiological pathways, which might be related to multiple cellular receptors in tissues of three major above mentioned systems. In the latest report, cell-surface glycans, including sialic acid (SA) and heparan sulfate (HS) were found to act as critical cellular factors involved in the attachment of PHEV (Hu et al., 2022). Altogether, this indicates the complexity of the underlying mechanisms of PHEV S glycoprotein interaction with cellular receptors.
S glycoprotein is a great inducer of neutralizing antibodies and encompasses multiple epitopes, making it an important target and informative source for CoVs vaccine design (Du et al., 2009), including the licensed vaccines against SARS-CoV-1 (Du et al., 2009) and the SARS-CoV-2 vaccines used to fight against the ongoing COVID-19 pandemic (Samrat et al., 2020; Dai and Gao, 2021). Like multiple coronaviruses, the hypervariability and the high dissimilarity of PHEV S glycoprotein ORF detected in this study impacted the potential B cell epitopes. The mutations affected at least eight potential linear B cell epitopes on S glycoprotein, half of which were located in the S1-RBD. This information is of great value for the vaccine design and the surveillance of genetic diversity of PHEV.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: Porcine hemagglutinating encephalomyelitis virus strain PHEV/67 N/US/1970, complete genome GenBank: MW165134.1.
Author Contributions
AB, YG, and LX: conceptualization. AB, YG, PW, YL, and RS: data analysis. AB, YG, and LX: visualization and writing. CW and LX: administration. AB, YG, and LX: manuscript revision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare no conflict of interest concerning the research, authorship, or publication of the article.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all the persons who contributed to the collection and production of PHEV genomic sequences in GenBank.
References
Andries, K., and Pensaert, M. B. (1980). Immunofluorescence studies on the pathogenesis of hemagglutinating encephalomyelitis virus infection in pigs after oronasal inoculation. Am. J. Vet. Res. 41, 1372–1378.
Andries, K., and Pensaert, M. (1981). Vomiting and wasting disease, a coronavirus infection of pigs. Adv. Exp. Med. Biol. 142, 399–408. doi: 10.1007/978-1-4757-0456-3_33
Bosch, B. J., van der Zee, R., de Haan, C. A., and Rottier, P. J. (2003). The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003
Buchholz, U. J., Bukreyev, A., Yang, L., Lamirande, E. W., Murphy, B. R., Subbarao, K., et al. (2004). Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U. S. A. 101, 9804–9809. doi: 10.1073/pnas.0403492101
Cartwright, S. F., Lucas, M., Cavill, J. P., Gush, A. F., and Blandford, T. B. (1969). Vomiting and wasting disease of piglets. Vet. Rec. 84, 175–176. doi: 10.1136/vr.84.7.175
Chen, Y., Liu, Q., and Guo, D. (2020). Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423. doi: 10.1002/jmv.25681
Chen, K., Zhao, K., Song, D., He, W., Gao, W., Zhao, C., et al. (2012). Development and evaluation of an immunochromatographic strip for rapid detection of porcine hemagglutinating encephalomyelitis virus. Virol. J. 9:172. doi: 10.1186/1743-422X-9-172
Cornelissen, L. A., Wierda, C. M., van der Meer, F. J., Herrewegh, A. A., Horzinek, M. C., Egberink, H. F., et al. (1997). Hemagglutinin-esterase, a novel structural protein of torovirus. J. Virol. 71, 5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997
Cutlip, R. C., and Mengeling, W. L. (1972). Lesions induced by hemagglutinating encephalomyelitis virus strain 67N in pigs. Am. J. Vet. Res. 33, 2003–2009.
Dai, L., and Gao, G. F. (2021). Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 21, 73–82. doi: 10.1038/s41577-020-00480-0
de Groot, R., Baker, S., Baric, R., Enjuanes, L., Gorbalenya, A., Holmes, K., et al. (2012). “Virus taxonomy” in Ninth Report of the International Committee on Taxonomy of Viruses: Family Coronaviridae. eds. A. King, M. Adams, E. Cartens, and E. Lefkowitz (San Diego, CA: Elsevier Academic Press), 806–828.
Dong, B., Gao, W., Lu, H., Zhao, K., Ding, N., Liu, W., et al. (2015). A small region of porcine hemagglutinating encephalomyelitis virus spike protein interacts with the neural cell adhesion molecule. Intervirology 58, 130–137. doi: 10.1159/000381060
Dong, B., Lu, H., Zhao, K., Liu, W., Gao, W., Lan, Y., et al. (2014). Identification and genetic characterization of porcine hemagglutinating encephalomyelitis virus from domestic piglets in China. Arch. Virol. 159, 2329–2337. doi: 10.1007/s00705-014-2070-y
Doyle, L. P., and Hutchings, L. M. (1946). A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 108, 257–259.
Du, L., He, Y., Zhou, Y., Liu, S., Zheng, B.-J., and Jiang, S. (2009). The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 7, 226–236. doi: 10.1038/nrmicro2090
Fehr, A. R., and Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23. doi: 10.1007/978-1-4939-2438-7_1
Gallagher, T. M., and Buchmeier, M. J. (2001). Coronavirus spike proteins in viral entry and pathogenesis. Virology 279, 371–374. doi: 10.1006/viro.2000.0757
Gao, W., He, W., Zhao, K., Lu, H., Ren, W., Du, C., et al. (2010). Identification of NCAM that interacts with the PHE-CoV spike protein. Virol. J. 7, 254. doi: 10.1186/1743-422X-7-254
Gao, W., Zhao, K., Zhao, C., Du, C., Ren, W., Song, D., et al. (2011). Vomiting and wasting disease associated with hemagglutinating encephalomyelitis viruses infection in piglets in Jilin. China. Virol J 8, 130. doi: 10.1186/1743-422x-8-130
Gong, L., Li, J., Zhou, Q., Xu, Z., Chen, L., Zhang, Y., et al. (2017). A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 23, 1607–1609. doi: 10.3201/eid2309.170915
Greig, A. S., Mitchell, D., Corner, A. H., Bannister, G. L., Meads, E. B., and Julian, R. J. (1962). A hemagglutinating virus producing encephalomyelitis in baby pigs. Can. J. Comp. Med. Vet. Sci. 26, 49–56.
Guan, Y., Zheng, B. J., He, Y. Q., Liu, X. L., Zhuang, Z. X., Cheung, C. L., et al. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278.
Hirahara, T., Yasuhara, H., Kodama, K., Nakai, M., and Sasaki, N. (1987). Isolation of hemagglutinating encephalomyelitis virus from respiratory tract of pigs in Japan. Nihon Juigaku Zasshi 49, 85–93. doi: 10.1292/jvms1939.49.85
Hirano, N. (2004). Neurotropism of swine Haemagglutinating encephalomyelitis virus (coronavirus) in mice depending upon host age and route of infection. J. Comp. Pathol. 130, 58–65. doi: 10.1016/S0021-9975(03)00083-5
Hirano, N., and Ono, K. (1998). A serological survey of human coronavirus in pigs of the Tohoku District of Japan. Adv. Exp. Med. Biol. 440, 491–494. doi: 10.1007/978-1-4615-5331-1_63
Hou, Y., Lin, C. M., Yokoyama, M., Yount, B. L., Marthaler, D., Douglas, A. L., et al. (2017). Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 91. doi: 10.1128/JVI.00227-17
Hu, S., Li, Z., Lan, Y., Guan, J., Zhao, K., Chu, D., et al. (2020). MiR-10a-5p-mediated Syndecan 1 suppression restricts porcine Hemagglutinating encephalomyelitis virus replication. Front. Microbiol. 11, 105. doi: 10.3389/fmicb.2020.00105
Hu, S., Zhao, K., Lan, Y., Shi, J., Guan, J., Lu, H., et al. (2022). Cell-surface glycans act as attachment factors for porcine hemagglutinating encephalomyelitis virus. Vet. Microbiol. 265:109315. doi: 10.1016/j.vetmic.2021.109315
Hulswit, R. J., de Haan, C. A., and Bosch, B. J. (2016). Coronavirus spike protein and tropism changes. Adv. Virus Res. 96, 29–57. doi: 10.1016/bs.aivir.2016.08.004
Jespersen, M. C., Peters, B., Nielsen, M., and Marcatili, P. (2017). BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 45, W24–W29. doi: 10.1093/nar/gkx346
Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966. doi: 10.1056/NEJMoa030781
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163. doi: 10.1093/bib/5.2.150
Lan, Y., Li, Z., Wang, Z., Wang, X., Wang, G., Zhang, J., et al. (2020). An experimental model of neurodegenerative disease based on porcine Hemagglutinating encephalomyelitis virus-related Lysosomal abnormalities. Mol. Neurobiol. 57, 5299–5306. doi: 10.1007/s12035-020-02105-y
Lan, Y., Lu, H., Zhao, K., He, W., Chen, K., Wang, G., et al. (2012). In vitro inhibition of porcine hemagglutinating encephalomyelitis virus replication with siRNAs targeting the spike glycoprotein and replicase polyprotein genes. Intervirology 55, 53–61. doi: 10.1159/000323523
Lau, S. K., Woo, P. C., Yip, C. C., Fan, R. Y., Huang, Y., Wang, M., et al. (2012). Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J. Virol. 86, 5481–5496. doi: 10.1128/JVI.06927-11
Laude, H., Van Reeth, K., and Pensaert, M. (1993). Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet. Res. 24, 125–150.
Li, Z., Gao, F., Lan, Y., Guan, J., Zhang, J., Lu, H., et al. (2021). Porcine Hemagglutinating encephalomyelitis virus triggers neural autophagy independently of ULK1. J. Virol. 95:e0085121. doi: 10.1128/JVI.00851-21
Llanes, A., Restrepo, C. M., Caballero, Z., Rajeev, S., Kennedy, M. A., and Lleonart, R. (2020). Betacoronavirus genomes: how genomic information has been used to Deal with past outbreaks and the COVID-19 pandemic. Int. J. Mol. Sci. 21:24546. doi: 10.3390/ijms21124546
Lole, K. S., Bollinger, R. C., Paranjape, R. S., Gadkari, D., Kulkarni, S. S., Novak, N. G., et al. (1999). Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73, 152–160. doi: 10.1128/JVI.73.1.152-160.1999
Lorbach, J. N., Wang, L., Nolting, J. M., Benjamin, M. G., Killian, M. L., Zhang, Y., et al. (2017). Porcine Hemagglutinating encephalomyelitis virus and respiratory disease in exhibition swine, Michigan, USA, 2015. Emerg. Infect. Dis. 23, 1168–1171. doi: 10.3201/eid2307.170019
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Mazumder, R., Iyer, L. M., Vasudevan, S., and Aravind, L. (2002). Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 30, 5229–5243. doi: 10.1093/nar/gkf645
Mengeling, W. L., Boothe, A. D., and Ritchie, A. E. (1972). Characteristics of a coronavirus (strain 67N) of pigs. Am. J. Vet. Res. 33, 297–308.
Mora-Diaz, J. C., Magtoto, R., Houston, E., Baum, D., Carrillo-Avila, J. A., Temeeyasen, G., et al. (2020). Detecting and monitoring porcine Hemagglutinating encephalomyelitis virus: an underresearched betacoronavirus. mSphere 5:199. doi: 10.1128/mSphere.00199-20
Mora-Diaz, J. C., Pineyro, P. E., Houston, E., Zimmerman, J., and Gimenez-Lirola, L. G. (2019). Porcine Hemagglutinating encephalomyelitis virus: a review. Front Vet Sci 6, 53. doi: 10.3389/fvets.2019.00053
Mora-Diaz, J. C., Pineyro, P. E., Rauh, R., Nelson, W., Sankoh, Z., Gregg, E., et al. (2021a). Porcine Hemagglutinating encephalomyelitis virus infection In vivo and ex vivo. J. Virol. 95. doi: 10.1128/JVI.02335-20
Mora-Diaz, J. C., Temeeyasen, G., Magtoto, R., Rauh, R., Nelson, W., Carrillo-Avila, J. A., et al. (2021b). Infection and immune response to porcine hemagglutinating encephalomyelitis virus in grower pigs. Vet. Microbiol. 253:108958. doi: 10.1016/j.vetmic.2020.108958
Pan, Y., Tian, X., Qin, P., Wang, B., Zhao, P., Yang, Y. L., et al. (2017). Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 211, 15–21. doi: 10.1016/j.vetmic.2017.09.020
Payne, S. (2017). “Chapter 17 – family coronaviridae” in Viruses. ed. S. Payne (Cambridge, MA: Academic Press), 149–158.
Pensaert, M., Callebaut, P., and Vergote, J. (1986). Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 8, 257–261. doi: 10.1080/01652176.1986.9694050
Quiroga, M. A., Cappuccio, J., Piñeyro, P., Basso, W., Moré, G., Kienast, M., et al. (2008). Hemagglutinating encephalomyelitis coronavirus infection in pigs. Argentina. Emerg Infect Dis 14, 484–486. doi: 10.3201/eid1403.070825
Rho, S., Moon, H. J., Park, S. J., Kim, H. K., Keum, H. O., Han, J. Y., et al. (2011). Detection and genetic analysis of porcine hemagglutinating encephalomyelitis virus in South Korea. Virus Genes 42, 90–96. doi: 10.1007/s11262-010-0551-y
Roe, C. K., and Alexander, T. J. (1958). A disease of nursing pigs previously unreported in Ontario. Can. J. Comp. Med. Vet. Sci. 22, 305–307.
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Samrat, S. K., Tharappel, A. M., Li, Z., and Li, H. (2020). Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus Res. 288:198141. doi: 10.1016/j.virusres.2020.198141
Sasseville, M. J., Gélinas, A., Sawyer, N., Boutin, M., and Dea, S. J. S. U. (2001). Biological and molecular characteristics of an HEV isolate associated with recent acute outbreaks of encephalomyelitis in Quebec pig farms. Adv. Exp. Med. Biol. 494, 57–62. doi: 10.1007/978-1-4615-1325-4_8
Schultze, B., Cavanagh, D., and Herrler, G. (1992). Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology 189, 792–794. doi: 10.1016/0042-6822(92)90608-R
Shi, J., Zhao, K., Lu, H., Li, Z., Lv, X., Lan, Y., et al. (2018). Genomic characterization and pathogenicity of a porcine hemagglutinating encephalomyelitis virus strain isolated in China. Virus Genes 54, 672–683. doi: 10.1007/s11262-018-1591-y
Snijder, E. J., Bredenbeek, P. J., Dobbe, J. C., Thiel, V., Ziebuhr, J., Poon, L. L., et al. (2003). Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004. doi: 10.1016/S0022-2836(03)00865-9
Sola, I., Almazán, F., Zúñiga, S., and Enjuanes, L. (2015). Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2, 265–288. doi: 10.1146/annurev-virology-100114-055218
Tortorici, M. A., Walls, A. C., Lang, Y., Wang, C., Li, Z., Koerhuis, D., et al. (2019). Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 26, 481–489. doi: 10.1038/s41594-019-0233-y
Vaughn, E. M., Halbur, P. G., and Paul, P. S. (1995). Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J. Virol. 69, 3176–3184. doi: 10.1128/jvi.69.5.3176-3184.1995
Vijgen, L., Keyaerts, E., Lemey, P., Maes, P., Van Reeth, K., Nauwynck, H., et al. (2006). Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 80, 7270–7274. doi: 10.1128/JVI.02675-05
Walls, A. C., Tortorici, M. A., Bosch, B.-J., Frenz, B., Rottier, P. J. M., DiMaio, F., et al. (2016). Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531, 114–117. doi: 10.1038/nature16988
Wang, L., Eggett, T. E., Lanka, S., Fredrickson, R. L., Li, G., Zhang, Y., et al. (2019). Development of a triplex real-time RT-PCR assay for detection and differentiation of three US genotypes of porcine hemagglutinating encephalomyelitis virus. J. Virol. Methods 269, 13–17. doi: 10.1016/j.jviromet.2019.04.008
Wang, Q., Vlasova, A. N., Kenney, S. P., and Saif, L. J. (2019). Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 34, 39–49. doi: 10.1016/j.coviro.2018.12.001
Weiss, S. R., and Navas-Martin, S. (2005). Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69, 635–664. doi: 10.1128/MMBR.69.4.635-664.2005
Whitworth, K. M., Rowland, R. R. R., Petrovan, V., Sheahan, M., Cino-Ozuna, A. G., Fang, Y., et al. (2019). Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 28, 21–32. doi: 10.1007/s11248-018-0100-3
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/jvi.06540-11
Wood, E. N. (1977). An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 100, 243–244. doi: 10.1136/vr.100.12.243
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Zhang, J., Li, Z., Lu, H., Shi, J., Gao, R., Ma, Y., et al. (2021). Evidence of microglial immune response following coronavirus PHEV infection of CNS. Front. Immunol. 12:804625. doi: 10.3389/fimmu.2021.804625
Zhang, C. Y., Wei, J. F., and He, S. H. (2006). Adaptive evolution of the spike gene of SARS coronavirus: changes in positively selected sites in different epidemic groups. BMC Microbiol. 6:88. doi: 10.1186/1471-2180-6-88
Zhi, Y., Kobinger, G. P., Jordan, H., Suchma, K., Weiss, S. R., Shen, H., et al. (2005). Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology 335, 34–45. doi: 10.1016/j.virol.2005.01.050
Zhou, P., Fan, H., Lan, T., Yang, X. L., Shi, W. F., Zhang, W., et al. (2018). Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556, 255–258. doi: 10.1038/s41586-018-0010-9
Keywords: porcine hemagglutinating encephalomyelitis virus, coronavirus, spike glycoprotein, receptor-binding domain, genomic mutation, B cell epitope
Citation: Bahoussi AN, Guo Y-Y, Shi R-Z, Wang P-H, Li Y-Q, Wu C-X and Xing L (2022) Genetic Characteristics of Porcine Hemagglutinating Encephalomyelitis Coronavirus: Identification of Naturally Occurring Mutations Between 1970 and 2015. Front. Microbiol. 13:860851. doi: 10.3389/fmicb.2022.860851
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Daxin Peng, Yangzhou University, ChinaHongjie Fan, Nanjing Agricultural University, China
Copyright © 2022 Bahoussi, Guo, Shi, Wang, Li, Wu and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Xing, eGluZ2xpMTA3QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Amina Nawal Bahoussi1†
Amina Nawal Bahoussi1† Li Xing
Li Xing