
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 27 June 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.859990
This article is part of the Research Topic"One Health" Approach For Revealing Reservoirs And Transmission Of Antimicrobial Resistance, Volume IIView all 22 articles
 Sani-e-Zehra Zaidi1,2
Sani-e-Zehra Zaidi1,2 Rahat Zaheer1
Rahat Zaheer1 Ruth Barbieri1
Ruth Barbieri1 Shaun R. Cook1
Shaun R. Cook1 Sherry J. Hannon3
Sherry J. Hannon3 Calvin W. Booker3
Calvin W. Booker3 Deirdre Church4,5
Deirdre Church4,5 Gary Van Domselaar6
Gary Van Domselaar6 Athanasios Zovoilis2
Athanasios Zovoilis2 Tim A. McAllister1*
Tim A. McAllister1*Enterococci are commensal bacteria of the gastrointestinal tract of humans, animals, and insects. They are also found in soil, water, and plant ecosystems. The presence of enterococci in human, animal, and environmental settings makes these bacteria ideal candidates to study antimicrobial resistance in the One-Health continuum. This study focused on Enterococcus hirae isolates (n = 4,601) predominantly isolated from beef production systems including bovine feces (n = 4,117, 89.5%), catch-basin water (n = 306, 66.5%), stockpiled bovine manure (n = 24, 0.5%), and natural water sources near feedlots (n = 145, 32%), and a few isolates from urban wastewater (n = 9, 0.2%) denoted as human-associated environmental samples. Antimicrobial susceptibility profiling of a subset (n = 1,319) of E. hirae isolates originating from beef production systems (n = 1,308) showed high resistance to tetracycline (65%) and erythromycin (57%) with 50.4% isolates harboring multi-drug resistance, whereas urban wastewater isolates (n = 9) were resistant to nitrofurantoin (44.5%) and tigecycline (44.5%) followed by linezolid (33.3%). Genes for tetracycline (tetL, M, S/M, and O/32/O) and macrolide resistance erm(B) were frequently found in beef production isolates. Antimicrobial resistance profiles of E. hirae isolates recovered from different environmental settings appeared to reflect the kind of antimicrobial usage in beef and human sectors. Comparative genomic analysis of E. hirae isolates showed an open pan-genome that consisted of 1,427 core genes, 358 soft core genes, 1701 shell genes, and 7,969 cloud genes. Across species comparative genomic analysis conducted on E. hirae, Enterococcus faecalis and Enterococcus faecium genomes revealed that E. hirae had unique genes associated with vitamin production, cellulose, and pectin degradation, traits which may support its adaptation to the bovine digestive tract. E. faecium and E. faecalis more frequently harbored virulence genes associated with biofilm formation, iron transport, and cell adhesion, suggesting niche specificity within these species.
Antimicrobial resistance (AMR) is recognized as one of the major global health challenges of the 21st century. The interconnected microbiomes between humans, animals, and the environment contribute to the emergence, acquisition, and spread of AMR (Hiltunen et al., 2017). A One-health approach provides an in-depth knowledge of the evolution of AMR by focusing on those biological elements that influence the emergence of antimicrobial resistance genes (ARGs) within the microorganism and their dissemination among hosts (human and animals) and the environment (Hernando-Amado et al., 2019). Gram-positive enterococci are core members of the gastrointestinal microbiota of humans and animals and are frequently isolated from soil and water (Byappanahalli et al., 2012; Gilmore et al., 2014). Enterococci often carry ARGs as they compete within complex microbial communities and are exposed to antimicrobials in clinical settings and during livestock production (Murray, 1990; Moreno et al., 2006). Furthermore, depending on the species, enterococci exhibit intrinsic resistance to several antibiotics including cephalosporins, anti-staphylococcal penicillins, aztreonam, aminoglycosides, lincosamides, and streptogramins (Miller et al., 2014). Enterococci are typically commensals, but they can cause nosocomial infections in humans including septicemia, endocarditis, and urinary tract infections (Barnes et al., 2021). There are over 50 species of enterococci with E. faecalis and E. faecium most frequently linked to human infections. Occasionally, other species including E. hirae, E. avium, E. durans, E. gallinarum, E. casseliflavus, and E. raffinosus may also be associated with infections in people (Brayer et al., 2019; Pinkes et al., 2019; Winther et al., 2020). Due to their widespread occurrence and persistence in the environment, enterococci are considered indicators of fecal contamination (Byappanahalli and Fujioka, 2004; Yan et al., 2011) and also serve as key indicator bacteria for AMR surveillance systems in humans and animals (Harwood et al., 2000; Layton et al., 2010).
Studies have indicated that E. faecium and E. faecalis are more prevalent in human-associated environments, whereas E. hirae are prevalent in beef cattle production systems (Zaheer et al., 2020). E. hirae only accounts for 1% of enterococcal infections in humans (Heval Can et al., 2020) and is mainly linked to pyelonephritis (Chan et al., 2012; Pãosinho et al., 2016; Nakamura et al., 2021), endocarditis (Talarmin et al., 2011; Pinkes et al., 2019), and biliary tract infections (Tan et al., 2010; Bourafa et al., 2015). As with E. faecalis and E. faecium, E. hirae infections are typically treated with ampicillin, gentamicin, or vancomycin (Nakamura et al., 2021).
The focus of this study was to investigate the genomic relatedness of E. hirae across the environmental continuum and to identify the genetic nature of AMR in E. hirae. Furthermore, we applied a pan-genome analysis to identify genes that may account for the predominance of E. hirae within beef cattle production systems.
A total of 8,430 Enterococcus strains were isolated in a One-Health surveillance study from different segments of the environmental continuum using samples collected from beef production systems (i.e., feedlot cattle feces, catch-basin water, and manure), natural water sources, urban wastewater, and human clinical samples (Zaheer et al., 2020). Bovine fecal samples came from four feedlots in southern Alberta over two years (March 2014–April 2016). Wastewater samples were collected from catch basins that accumulate runoff from the feedlots. Natural surface water samples came from up-stream and down-stream of the feedlots. Urban wastewater samples came from two wastewater plants located in southern Alberta. Enterococcus spp. recovered from patients with clinical infections were obtained through the Division of Medical Microbiology, Calgary Laboratory Services (now Alberta Precision Laboratories, Alberta Health Services) (Zaheer et al., 2020). This study focuses on Enterococcus hirae, collected as the most prevalent species from beef production system (n = 4,601 isolate) (Zaheer et al., 2020). Figure 1 represents the prevalence of E. hirae isolates in the sampled sources.
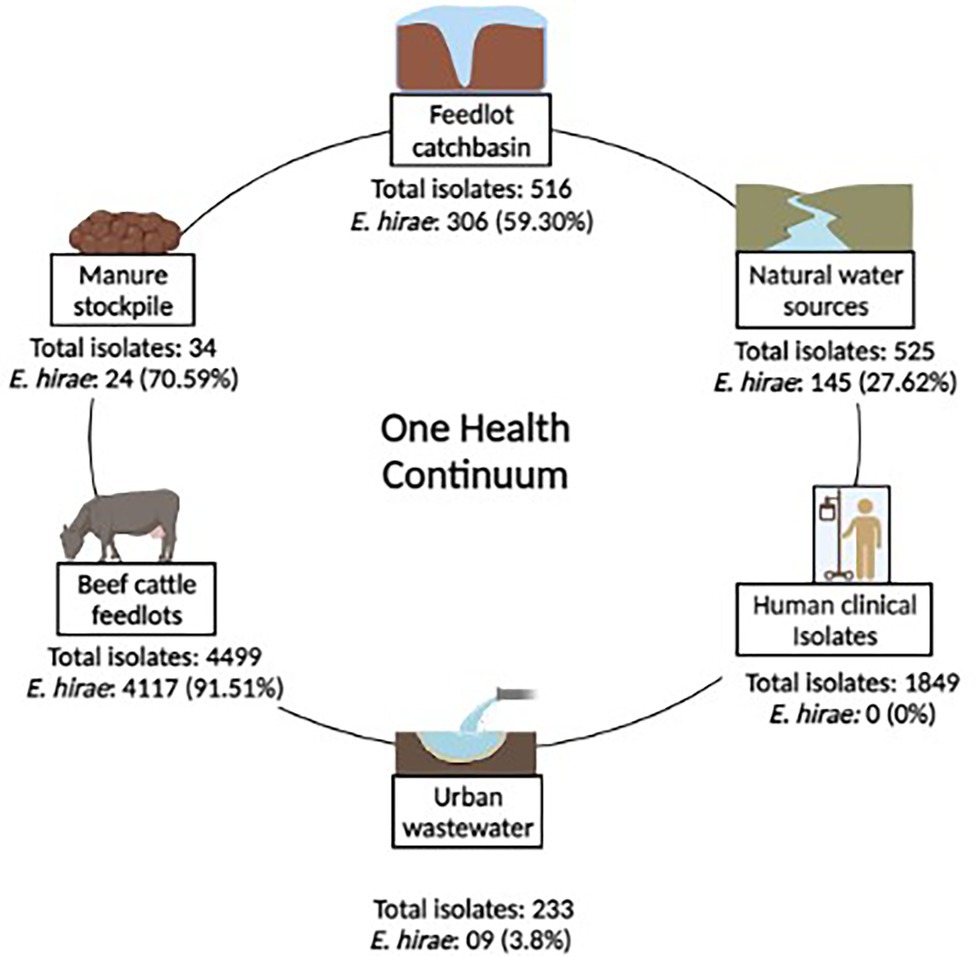
Figure 1. Prevalence of Enterococcus hirae isolates (n = 4,601) identified across a One-health continuum.
Enterococci were recovered in parallel from two different media types including Bile Esculin Azide (BEA) agar without antibiotic and BEA supplemented with 8 μg/ml erythromycin, followed by species identification. E. hirae were identified via multiplex PCR targeting groES-EL and muramidase genes (Zaheer et al., 2020). As E. hirae was absent among clinical Enterococcus isolates (n = 1892; Figure 1), complete genomes (n = 3) of clinical E. hirae were retrieved from NCBI database for comparative genomic analysis (Supplementary Table S1).
Antimicrobial susceptibility testing was performed on a randomly selected subset (n = 1319, 29%) of E. hirae isolates using the disk diffusion method, as per the Clinical and Laboratory Standards Institute (CLSI) documents M02-A12 and M100-S24. A panel of twelve antibiotics was used for testing based on their common usage for treatment of human enterococcal infections that included those drugs of critical importance (levofloxacin, linezolid, quinupristin/dalfopristin, teicoplanin, vancomycin, and tigecycline), high importance (erythromycin, ampicillin, gentamicin, and streptomycin), and medium importance (nitrofurantoin and tetracycline). Staphylococcus aureus ATCC 25923 and E. faecalis ATCC 29212 were used as reference quality controls (Zaheer et al., 2020). The BioMic V3 imaging system (Giles Scientific, Inc., Santa Barbara, CA, USA) was used to read zones of inhibition. Isolates were categorized based on CLSI interpretive criteria, except for tigecycline for which EUCAST interpretive criteria (The European Committee on Antimicrobial Susceptibility Testing, 2014) were used.
Whole-genomic sequencing of a subset of E. hirae isolates (n = 286), including isolates originating from bovine feces (n = 168), feedlot catch basin (n = 62), bovine manure stockpiles (n = 8), natural water sources (n = 42), and urban wastewater (n = 7), was performed using next-generation sequencing technology. Briefly, genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Montreal, QC, Canada) with modifications (Zaheer et al., 2020), followed by DNA quality assessment and quantification using a Nanodrop 2000 spectrophotometer and a Qubit Fluorometer with PicoGreen (Thermo Fisher Scientific, Mississauga, ON, Canada). Isolates were sequenced on an Illumina MiSeq platform using the MiSeq Reagent Kit V3 to generate 2 × 300 bp paired-end reads. Raw read FASTQ files were assessed for the quality of sequence data using FastQC (Galaxy Version 0.72 + galaxy1) (Wingett and Andrews, 2018) and de novo assemblies were performed using Shovill (Bankevich et al., 2012). Assembled contigs were then annotated by Prokka to identify all gene-coding sequences (Seemann, 2014).
Assembled genomes were screened for the presence of AMR determinants, virulence genes, and plasmids using ABRicate (https://github.com/tseemann/abricate/) against the NCBI Bacterial Antimicrobial Resistance Reference Gene database (NCBI BioProject ID: PRJNA313047), VirulenceFinder [pmid 15,608,208], and PlasmidFinder databases (Zankari et al., 2012), respectively. Intact prophage were identified using PHASTER tool (Arndt et al., 2016).
A total of 289 genomes including 286 assembled genomes from this study and 3 complete E. hirae genomes of clinical isolates retrieved from NCBI database (strain: 708, accession: NZ_CP055232.1; strain: FDAARGOS_234, accession: NZ_CP023011.2; and strain: 13344, accession: NZ_CP055229.1) were subjected to phylogenomic analysis. A core-genome phylogenomic tree was constructed using the (SNVPhyl v 1.0) pipeline (Petkau et al., 2017). Briefly, all paired-end reads were mapped against the E. hirae reference genome (strain R17; GenBank accession: CP015516.1) to produce read pileups (SMALT v.0.7.5; https://www.sanger.ac.uk/tool/smalt-0/). The read pileups were evaluated for mapping quality (minimum mean mapping quality score of 30), coverage cut offs (15X minimum depth of coverage), and a single-nucleotide variant (SNV) abundance ratio of 0.75 to generate a multiple sequence alignment of SNV containing sites. The final maximum likelihood-based phylogeny was generated by PhyML using unfiltered SNV alignment. Phylogenomic trees and associated metadata were visualized using Interactive Tree Of Life (iTOL) v5 tool (Letunic and Bork, 2021).
Comparative genome analysis was done using the Roary v3.12.0 pipeline with default parameters (Page et al., 2015). Genes identified by Prokka were used to construct pan-genomes. A pan-genome of 289 E. hirae isolates was reconstructed to identify core and accessory genes present in E. hirae. Furthermore, comparative analysis was performed between E. faecium and E. faecalis, which are predominantly associated with humans infections, and E. hirae. For this purpose, a small subset of E. hirae isolates (n = 16) representative of the various sources and phylogenetic clades were randomly selected. Similarly, a subset of E. faecium (n = 26) and E. faecalis (n = 24) isolates were randomly selected on the same bases from our previous study (Zaheer et al., 2020; BioProject PRJNA604849). The phandango interactive viewer tool (Hadfield et al., 2017) was used to interpret pan-genome data obtained from Roary analysis. This tool utilizes two of the Roary output files: one is a gene absence and presence matrix file that creates a heat map based on the number of genes present or absent in each isolate and a Newick-formatted tree file of accessory genomes used to plot a relatedness dendrogram of the accessory genes present in all isolates.
A pan-genome plot was generated using ggplot2 package of R Studio Version 1.4.1103 (R Studio Inc., Boston, MA, USA) based on two Roary output files (the number of conserved genes and the number of total genes). The number of conserved genes represented the size of the core genome. The number of total genes represented both the core and accessory genomes, creating a curve based on the pan-genome completeness. The pan-genome of an organism is considered “closed” if the curve is predicted to plateau or “open” if the curve is predicted to continue to rise. In contrast to a closed genome, the number of new gene families in an open genome increases with the inclusion of new genomes in the analysis.
Discriminatory genomic signatures between E. hirae, E. faecium, and E. faecalis were identified using Neptune v1.2.5 with default parameter (Marinier et al., 2017). The signature discovery process using Neptune identifies sequences that are sufficiently common to a group of target sequences (inclusion group) and sufficiently absent from non-targets (exclusion group) using probabilistic models. Analyses was done using E. hirae genomes as the inclusion group and E. faecium and E. faecalis as independent exclusion groups, respectively. The genomic signature found in ≥90% of isolates in the inclusion group were selected and annotated using Prokka (Seemann, 2014).
Phenotypic susceptibility testing was conducted on 1,319 E. hirae isolates originating from bovine feces, feedlot catch-basin water, stockpiled bovine manure, and natural and urban wastewater sources. (Figure 2). Fifty-one different resistance profiles were identified with the most frequent being resistant to doxycycline and erythromycin (364/1319, 27.6%) followed by resistance to doxycycline alone (242/1319, 18.3%) (Supplementary Table S2). Across all tested isolates, 14.1% (186/1319) were multi-drug resistant (resistant to ≥3 tested antimicrobials). Antimicrobial susceptibility profiles of all tested isolates are presented in Supplementary Figure S2.
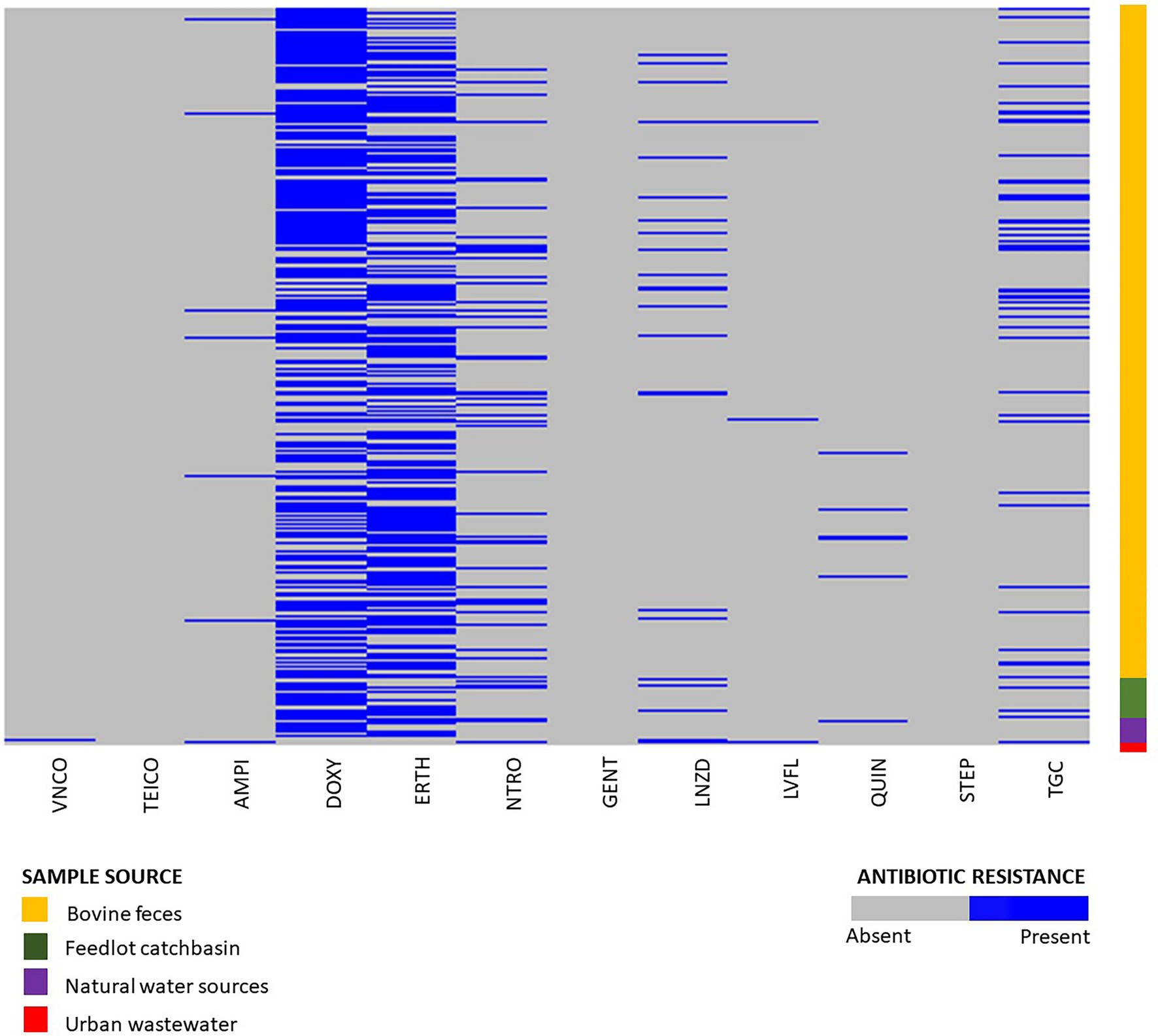
Figure 2. Phenotypic resistance profiles of Enterococcus hirae isolated from beef production system (n = 1,264) including bovine feces and feedlot catch basin, natural water source (n = 45), and urban wastewater sample (n = 9).
Out of 1,319 total isolates tested for antimicrobial susceptibility, 666 isolates were recovered from BEA plates without erythromycin. From these, isolates originating from beef production systems (i.e., bovine feces, catch basin, and stockpiled bovine manure; n = 632) exhibited a high prevalence of resistance to tetracycline (376/632, 59.4%), followed by macrolides (200/632, 31.6%), nitrofurantoin (102/632, 16.1%), tigecycline (76/632, 12.0%), linezolid (40/632, 6.32%), ampicillin (9/632, 1.42%), quinupristin/dalfopristin (8/632, 1.26%), vancomycin (1/632, 0.15%), and teicoplanin (1/632, 0.15%) (Supplementary Figure S1).
The natural water source isolates recovered from BEA plates without antibiotics (n = 28) also showed a high prevalence of tetracycline resistance (22/28, 78.5%), followed by macrolides (8/28, 28.5%), nitrofurantoin (5/28, 17.8%), and tigecycline (1/28, 3.57%). Resistance against quinupristin/dalfopristin, linezolid, ampicillin, and fluoroquinolones was not detected (Supplementary Figure S1).
Of the 233 Enterococcus spp. isolates recovered from urban wastewater (Zaheer et al., 2020), only nine were identified as E. hirae. Six of those were recovered from media without erythromycin. Four of the six isolates exhibited resistance to tigecycline (4/6, 66.6%) followed by nitrofurantoin (3/6, 50%), linezolid (3/7, 48.85%), fluoroquinolones (2/6, 33.3%), vancomycin (1/6, 16.6%), and ampicillin (1/7, 14.2%) (Supplementary Figure S1).
A total of 652 isolates from erythromycin plates were selected for phenotypic antimicrobial testing. Of these isolates, 632 originated from beef production (i.e., bovine feces, catch basin, and stockpiled bovine manure). Tetracycline resistance (437/632, 69%) was the most prevalent resistance in the beef isolates from production systems, followed by macrolides (525/632, 83%), tigecycline (67/632, 10.6%), nitrofurantoin (60/632, 9.5%), linezolid (33/632, 5.2%), quinupristin/dalfoprstin (13/632, 2.0%), ampicillin (9/632, 1.42%), fluoroquinolones (5/632, 0.79%), and gentamicin (1/632, 0.15%) (Supplementary Figure S1).
The isolates recovered from natural water sources (n = 17) showed a high prevalence of resistance to macrolides (15/17, 88.2%), followed by tetracycline (13/17, 76.4%), tigecycline (2/17, 11.76%), quinupristin/dalfoprstin (1/17, 5.88%), and nitrofurantoin (1/17, 5.88%). None of the isolates were resistant to linezolid, ampicillin, or fluoroquinolones (Supplementary Figure S1).
A total of three E. hirae isolates were recovered from urban waste sources on erythromycin plates. Two of those isolates showed macrolide resistance (2/3, 66.6%), followed by tetracycline (1/3, 33.3%), nitrofurantoin (1/3, 33.3%), quinupristin/dalfoprstin (1/3, 33.3%), and streptomycin (1/3, 33.3%). These isolates were sensitive to teicoplanin, ampicillin, vancomycin, gentamicin, tigecycline, fluoroquinolones, and linezolid (Supplementary Figure S2). Overall, 16.8% of isolates (110/652) recovered from erythromycin plates showed intermediate resistance to erythromycin.
Of the E. hirae isolates tested for antimicrobial susceptibility, 286 randomly selected isolates were used for whole-genome sequencing. The size of E. hirae genomes as interpreted from the assembled sequence read data ranged from 2,307,753 bp to 3,200,875 bp, with a GC content of 36.7%. Detailed assembly statistics are provided in Supplementary Table S3.
Assembled genomes (n = 286) were screened for the presence of AMR determinants using the Abricate tool along with the NCBI AMR gene database. Ten different ARGs, including aminoglycosides ARGs aac(6′)-Iid, ant(6)-Ia and aph(3)-III, streptothricin sat4, tetracycline ARGs tet (L, M, O, S/M, (O/32/O)), and macrolide ARG erm(B) were identified across the examined genomes (Figure 3). Overall, nineteen different resistance genotypes were identified with the most frequent being aac(6′)-lid-tet(L)-erm(B) (87/286, 30.41%) followed by aac(6′)-lid-tet(L)-tet(M)-erm(B) (64/286, 22.37%) (Supplementary Table S4). The aminoglycoside resistance gene aac(6′)-lid was identified in all but two of the E. hirae genomes (284/286, 99.30%).
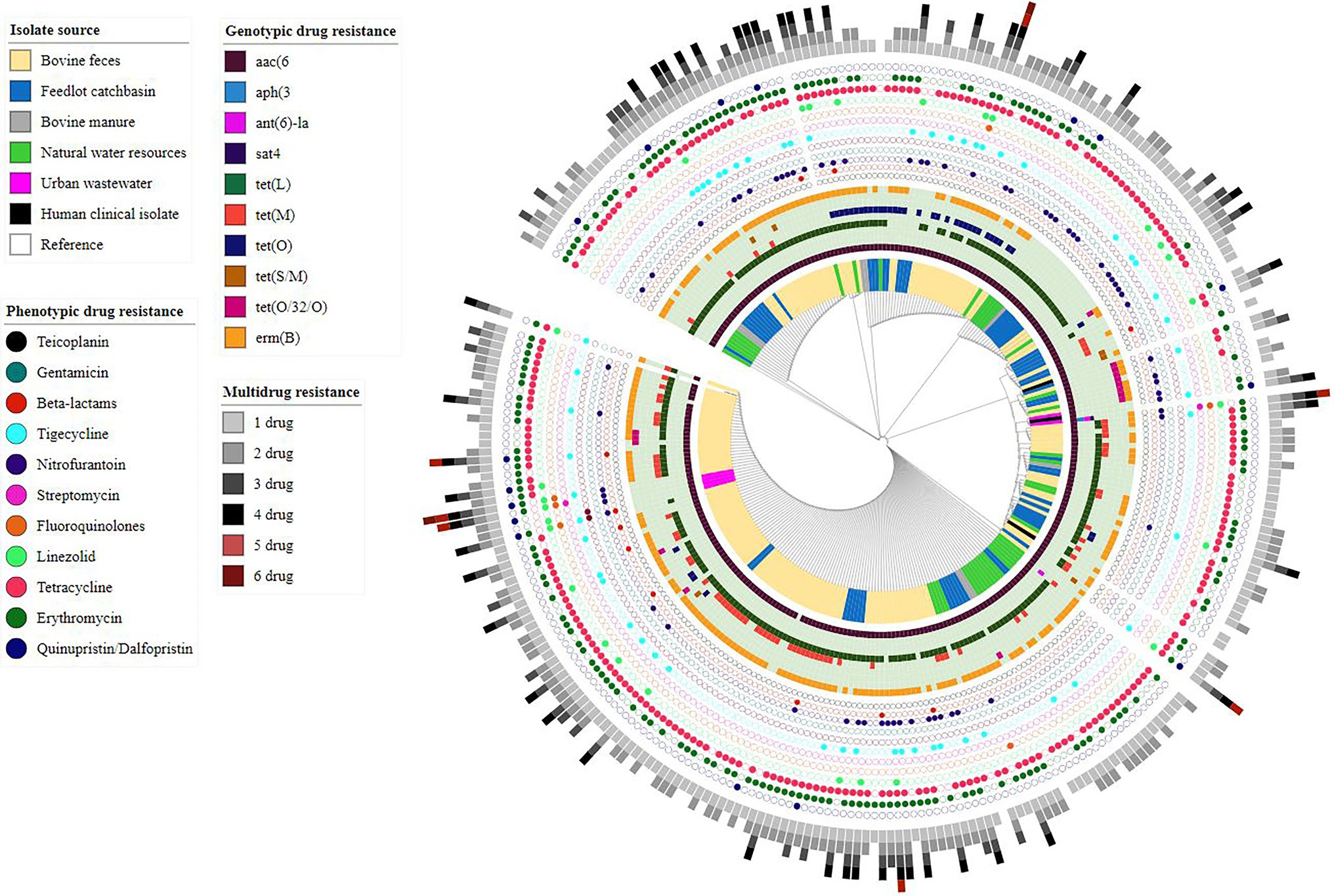
Figure 3. Core-genome phylogenetic tree based on analysis of single-nucleotide polymorphisms (SNPs) of Enterococcus hirae genomes (n = 291) isolated from different environmental settings including beef production systems and human-related isolates. The genomes were compared using E. hirae OG1RF genome (GenBank accession # NZ_CP015516.1/CP015516.1) as a reference.
Of the 286 sequenced isolates, 238 were recovered from beef production systems (i.e., bovine feces, feedlot catch basin, and stockpiled bovine manure). tet(L) (199/238, 83.61%) was the most prevalent ARG identified in these isolates, followed by erm(B) (179/238, 75.21%) and the tetracycline resistance genes, tet (M) (73/238, 30.67%), tetO (36/238, 15.12%), tet(O/32/O) (13/238, 5.46%), and tet(S/M) (03/238, 1.26%).
Similar to beef production system isolates, E. hirae isolates recovered from natural water sources located near feedlots showed a high prevalence of tet(L) (38/41, 92.68%) followed by erm(B) (27/41, 65.85%). Occasionally, tet(M) (4/41, 9.75%), tet(O) (4/41, 9.75%), and tet(O/32/O) (2/41, 4.87%) were also present in these isolates.
Among seven E. hirae isolates recovered from urban wastewater, the streptomycin resistance gene ant(6)-la was present in two isolates (2/7, 28.57%). The kanamycin/neomycin aph(3′)-III and streptothricin sat4 resistance genes were both found in a single urban wastewater isolate (1/7, 14.28%). Tetracycline resistance gene(s) were not found in any of these isolates, whereas erm(B) was only detected in one isolate (1/7, 14.28%).
tet(L) and erm(B) were found together in 63.63% of total isolates (182/286) indicating a strong correlation. Similarly, tet (L) and tet (M) coexisted in 24.12% of isolates (69/286) and in most cases were found on the same contig (60/69, 87%) in assembled genomes.
E. hirae genotypes generally associated with quinolone resistance (i.e., presence of quinolones resistant gene (qnr) or DNA gyrase and DNA topoisomerase IV genes mutations) and linezolid resistance (i.e., mutations in the 23S ribosomal RNA gene or presence of resistance genes including cfr, cfrB, optrA, and poxtA) were not identified.
Within the 286 E. hirae isolates, we identified nine different virulence genes associated with biofilm formation (bopD), capsular polysaccharides biosynthesis (cpsA, cpsB, and cap8E), hyaluronic acid production (hasC), proteolytic activity/chaperones (clpP), fibrinogen adhesions protein (fss3), bile salt hydrolase (bsh), and listeria adhesion protein (lap). All isolates carried cap8E, clpP, cpsA, cpsB, bopD, and lap genes, while hasC, bsh, and fss3 were found in 98.95% (283/286), 89.86% (257/286), and 2.44% (7/286) of total isolates, respectively. These genes were also identified in publicly available clinical E. hirae genomes from humans. Pili protein-encoding gene ebpC was only detected in one of the human clinical isolates retrieved from NCBI. Detailed information is provided in Supplementary Table S5.
Among all E. hirae isolates (n = 286), 16% carried plasmids. Seven different plasmids (rep1, rep2, rep11, rep17, rep18, repUS7, and repUS12) were identified. Among these, rep2 and rep17 were recovered from all sample types except urban wastewater. In contrast, rep1, rep18, and repUS7 were recovered from urban wastewater samples. Two out of fifteen rep17 plasmids carried erm(B), whereas one out of twenty rep2 plasmids carried tet(L). Of six repUS12 plasmids, five carried tet (L) and were recovered from bovine feces.
A total of 30 genomes were randomly selected from all sample sources to identify bacteriophage using PHASTER. All isolates contain at least one prophage ranging from 7 to 48 kb in size. Twenty-four intact prophage sequences were identified, with 95.5% identified as members of the family Siphoviridae. None of the identified prophages harbored ARGs.
Core-genome phylogenomic analysis was conducted on the 286 E. hirae isolated in this study and the three E. hirae genomes retrieved from NCBI. The E. hirae isolates clustered into six different clades, with no obvious segregation by source (Figure 3).
Pan-genome analysis of E. hirae isolates identified 1,427 core genes (99 to 100% of strains), 358 soft core genes (95 to 99% of strains), 1701 shell genes (15 to 95% of strains), and 7,969 cloud genes (0 to 15% of strains) (Figure 4A). The pan-genome of E. hirae is open as the number of accessory genes progressively increased with increasing genomes (Figure 4B). In addition, the gene presence and absence heat map showed that the accessory genome constituted a large part of the pan-genome, indicative of a high level of genomic diversity within this species (Figure 4C). Cross-species comparative analysis of E. hirae, E. faecium, and E. faecalis genomes, highlighted the genomic diversity within Enterococcus spp. as the total core genome shared between three species was small (Figure 5). Furthermore, it also demonstrated the distinct genomic traits of each species as illustrated by the gene absence and presence heat map (Figure 5).
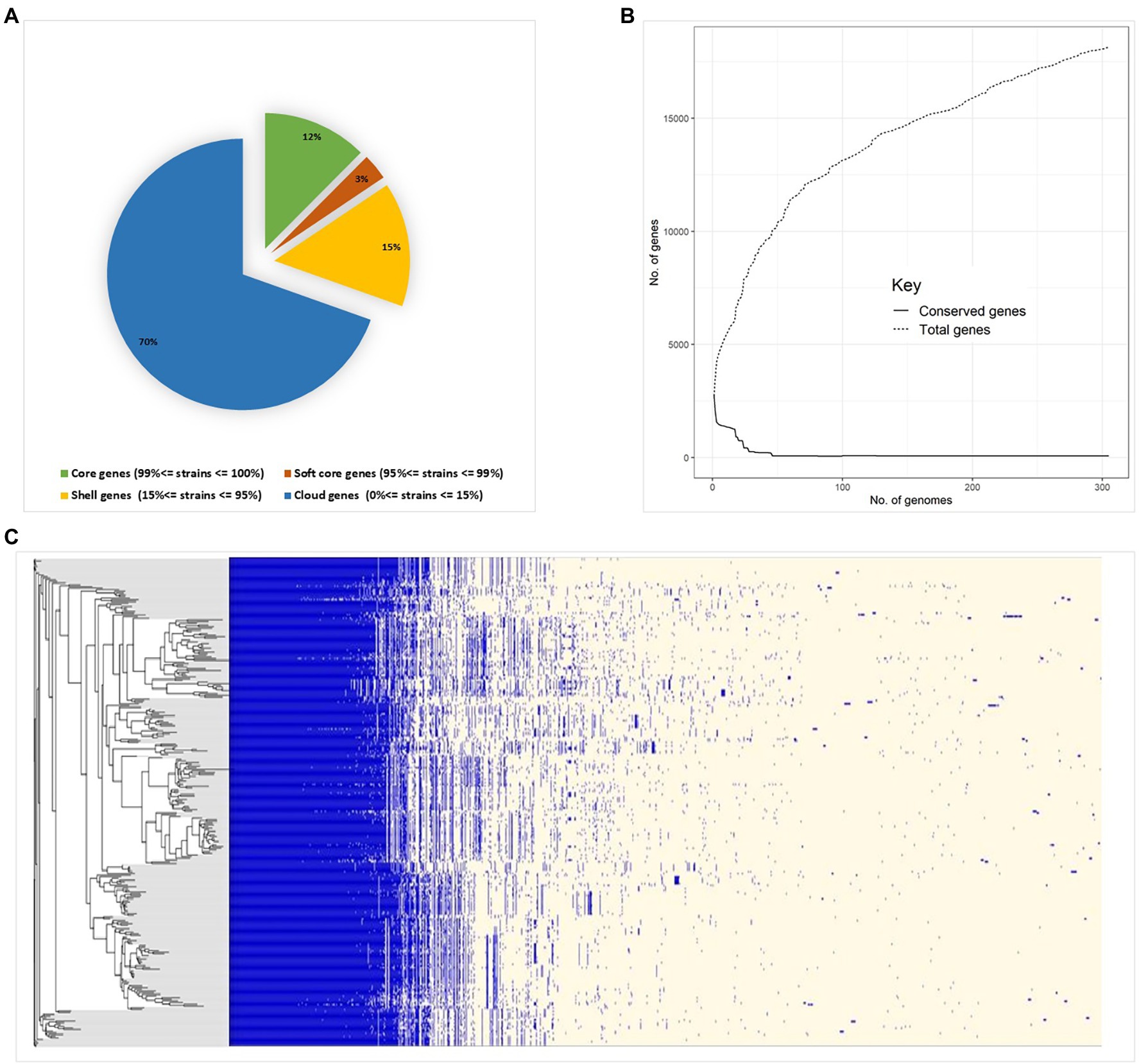
Figure 4. Pan-genome estimation of Enterococcus hirae (A) genomic statistics and pan-genome estimation of 291 isolates. (B) Development of pan- and core genomes, illustrating the open nature of the pan-genome. (C) Heat map representing absence or presence of genes in isolates and the phylogenetic genetic tree generated from accessory genes.
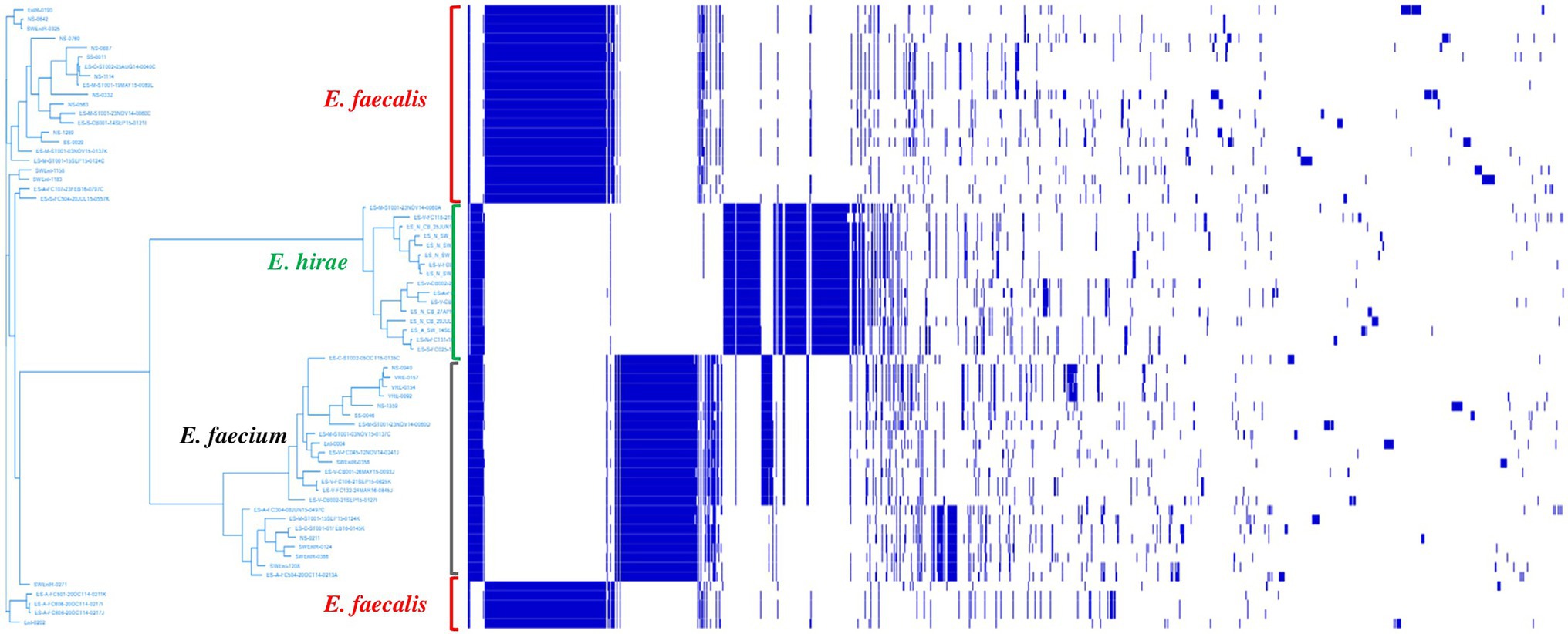
Figure 5. Heat map representing absence or presence of genes and phylogenetic genetic tree generated from accessory genes of Enterococcus hirae (n = 16), Enterococcus faecium (n = 26), and Enterococcus faecalis (n = 24).
A total of 1,069 discriminatory genomic signatures were recognized in E. hirae compared to E. faecalis (n = 808) and E. faecium (n = 261). Most of these genomic signatures encoded for unknown hypothetical proteins (454/1069, 42.46%).
Neptune analysis is capable of identifying inter-species genomic variation, as discriminatory loci were identified in all three species. These loci coded for genes required for the synthesis of aromatic amino-acids including chorismate synthase (aroC), cyclohexadienyl dehydrogenase (tyrC), genes for sugar transport including permease (yteP), transcriptional regulator (mtlR), lichenan permease IIC component (licC), and lipoprotein (lipO).
Signatures found exclusively in E. hirae genomes included genes associated with the phosphotransferase system for galactitol (gatA, B, and C) and fructose (fruA, frwA, and frwD), peptidoglycan synthesis (rodA ftsW, mur E, F, J, and Y), teichoic acid synthesis (tag H, G, and U), coenzyme A biosynthesis (coaD), vitamin B12 synthesis (nrdZ), chitin degradation (chiA), capsule synthesis (epsE, pglF, ywqD, and ywqC), riboflavin synthesis (ribBA, D, E, and H), vitamin B6 synthesis (yvgN), vitamin uptake transporter (queT), gamma-aminobutyric acid (GABA) production (glsA2, gadC, and amt), cardiolipin biosynthesis (clsA), bacitracin export (bceA and B), xenobiotic degradation (nylA), and iron transport (yqgN, feuC, feuB, fepC, and yfiY).
Furthermore, genes that may be associated with antimicrobial resistance were also identified in E. hirae genomes such as those encoding for multi-drug transporters (marA, mepE msrR, and yxlF), doxorubicin resistance (drrA), sulfonamide resistance (bcr), and penicillin-binding protein (pbp). Early secreted antigenic target (ESAT) system genes (eccC, essB, and esxA) were also found in E. hirae genomes.
Compared to E. hirae, 160 and 944 discriminatory genomic signatures were identified in E. faecium and E. faecalis genomes, respectively, with most of these genes associated with various aspects of cellular metabolism, such as molybdopterin biosynthesis (mog, modB, and modB); cadmium, zinc, and cobalt transport (cadA); copper export (copY and copA); phosphotransferase system for glucitol/sorbitol (srlA, srlE, and srlB), sorbose (sorB, sorF, and sorA), mannose (manX and manZ), cellobiose (celA), mannitol (mtlA, mtlF, and mtlD), maltose (malX), ascorbate (ulaC and ulaA), and beta-glucosides (bglF). In addition, genes encoding E. faecalis and E. faecium pathogenesis were also identified, such as biofilm formation genes (brpA, icaA, and lytR); virulence genes, including unsaturated chondroitin disaccharide hydrolase (ugl); anthrax toxin regulator positive (atxA); hemin transport system (hmuU and hmuT); sialic acid TRAP transporter small permease (siaQ); carnitine transport system (opuCB and opuCA); arginine/ornithine system (argR and arcD1); genes encoding the adhesions, including gelatinase (gelE) and collagen (cna); and genes encoding for antimicrobial resistance, including penicillin-binding protein (PbpE, PbpX, and PbpF), multi-drug resistance protein (YkkC, YkkD, and Stp), tetracycline repressor protein (TetR), fluoroquinolones export protein (Rv2688c), and macrolide export protein (MacB) (Supplementary Table S7).
Enterococci are ubiquitous Gram-positive bacteria. They colonize gastrointestinal tracts of most multicellular eukaryotic organisms including humans, animals, and insects and aide in digestion and gut metabolic pathways (De Graef et al., 2003; Farrow and Collins, 1985; Devriese et al., 1990; Andrew and Mitchell, 1997; de Vaux et al., 1998; Muniesa et al., 1999; Fogarty et al., 2003; Law-Brown and Meyers, 2003; Maria da Gloria et al., 2006; Layton et al., 2010; Giraffa, 2014). In addition, they are also found in food, plant, and water ecosystems (Müller et al., 2001; Svec et al., 2001; Klein, 2003; Švec et al., 2006; Byappanahalli et al., 2012). Enterococci are remarkably resilient to broad pH ranges, temperature variation, and osmotic pressure, traits that contribute to their broad distribution in nature (Heim et al., 2002; Caretti and Lubello, 2003; Anderson et al., 2005). Resistant bacterial populations are selected by the exposure of commensal gut microorganisms such as enterococci to antimicrobials that are used for disease treatment and prevention (Francino, 2016). The ubiquitous nature of enterococci may facilitate the dissemination of antimicrobial resistance genes between different environments. For this reason, it is imperative to identify antimicrobial resistance determinants and their role in the spread of antimicrobial resistance (Cameron and McAllister, 2016). Here, we focused on E. hirae isolates recovered from a One-Health surveillance study (Zaheer et al., 2020). The genomic relatedness of E. hirae was examined across various sampling matrices of the continuum and AMR determinants that contribute to antimicrobial resistance were identified. Furthermore, we examined the genomic traits of E. hirae that may facilitate their growth in the cattle gut as compared with other human-associated Enterococcus species.
As described previously, E. hirae is highly prevalent in cattle and thus can be readily isolated from bovine feces, bovine manure, and feedlot catch-basin water samples (Jackson et al., 2011; Zaheer et al., 2013, 2020; Beukers et al., 2015). The number of E. hirae isolates recovered from urban wastewater was low (3%) and most of the Enterococcus spp. from this source were identified as either E. faecalis or E. faecium. Similarly, only E. faecalis or E. faecium was identified among human clinical isolates, confirming that E. hirae is generally not associated with human infections. However, E. hirae have occasionally been isolated from human cases of septicemia (Gilad et al., 1998), endocarditis (Poyart et al., 2002; Talarmin et al., 2011), urinary tract infections (Chan et al., 2012; Bourafa et al., 2015), spondylodiscitis (Canalejo et al., 2008), and acute pancreatitis (Dicpinigaitis et al., 2015). The rarity of this species among clinical enterococci isolates suggests that this species may not be as virulent as E. faecalis and E. faecium. This finding is also evident from our comparative genomic analysis where virulence genes were frequently identified in E. faecalis and E. faecium, but not in E. hirae.
The phenotypic resistance profiles of 1,319 E. hirae isolates showed that antimicrobial use and resistance phenotype were linked within a particular environment. For example, macrolides and tetracyclines are commonly used in beef cattle production systems for disease treatment and prevention including prophylaxis/metaphylaxis (Hurd and Malladi, 2008; Cameron and McAllister, 2016; Vikram et al., 2017). Isolates recovered from bovine feces, feedlot catch basin, stockpiled bovine manure, and natural surface water in the vicinity of the feedlots showed high occurrence of resistance to these antibiotics. Although only nine E. hirae isolates were recovered from urban wastewater, their resistance profiles indicated linkage with drugs commonly used to treat human infections including nitrofurantoin, tigecycline, and linezolid.
Genotypic resistance profiles of E. hirae corroborated to their phenotypic profiles, where tetracycline and macrolide resistance genes were predominantly present in isolates recovered from the beef cattle production system and natural water sources. This is consistent with previous studies where tetracycline and macrolide resistance genotypes were prevalent in beef production systems (Zaheer et al., 2013, 2019; Rovira et al., 2019). Tetracycline resistance was associated with the presence of tetL, M, and O. Two mosaic tetracycline genes tetS/M and tetO/32/O were also identified. tetL confers resistance via an efflux mechanism, while tetM, tetS/M, O, and O/32/O encode for ribosomal protection proteins (Safferling et al., 2003; Kazimierczak et al., 2008; Barile et al., 2012; Crespo et al., 2012; Dönhöfer et al., 2012). These genes are mostly found on transposable elements that are often linked with chloramphenicol and macrolide resistance determinants (Opal and Pop-Vicas, 2015). Macrolide resistance was associated with the presence of erm(B), which confers resistance against macrolide-lincosamide-streptogramin antibiotics (Okitsu et al., 2005). Others have also found erm(B) in E. hirae (Portillo et al., 2000; Chajęcka-Wierzchowska et al., 2016), as well as in E. faecalis and E. faecium isolated from chickens (Kim et al., 2019, 2021), turkies (Tremblay et al., 2011; Demirgül and Tuncer, 2017), pigs (Aarestrup, 2000), fermented food (Sanchez Valenzuela., 2013), and clinical settings (Schmitz et al., 2000; Chen et al., 2010; Wang et al., 2021). The macrolide resistance gene and tetracycline resistance genes in E. hirae appear to be identical to those in E. faecalis and E. faecium (Beukers et al., 2017; Zaheer et al., 2020). Considering that all of these species carry similar AMR determinants, the possibility of horizontal gene transfer across species seems probable (Palmer et al., 2010). Studies have identified the presence of pheromone responsive plasmids in E. faecium and E. faecium that either encode vancomycin resistance or facilitate the transfer of plasmids carrying vancomycin ARGs into recipient cells (Flannagan and Clewell, 2002; Johnson et al., 2021). These plasmids can also transfer between Enterococcus species, as the pMG1 plasmid has been shown to transfer between E. faecium and E. faecalis, and from E. faecium to E. hirae (Costa et al., 1993).
Aminoglycoside gene acc(6′)-lid is known to be intrinsic in E. hirae (Costa et al., 1993) and was detected in all but two genomes, likely as a result of gene coverage and assemblage issues. It is not surprising that vancomycin resistance genes were not identified in E. hirae, as this drug is not approved for veterinary use in North American cattle. Our result is consistent with a previous study where vancomycin resistance genes were not identified in Enterococcus spp. isolated from bovine feces (Beukers et al., 2017). The virulence genes identified in E. hirae were mostly associated with biofilm formation and polysaccharide biosynthesis, as described by others (Hashem et al., 2017). The 10 virulence genes that were identified in E. hirae were similar to those in E. faecalis and E. faecium, but many more (i.e., 49) virulence genes were found in E. faecium and E. faecalis (Zaheer et al., 2020).
Pan-genome analysis is an important comparative analysis tool that allows linkages between genetic changes and specific phenotypes as it describes core- and accessory genomes as well as species-specific genes (Vernikos, 2020). The core genome constituted only 64% of the total genome in E. hirae. Both horizontal and vertical transfer of genes, including those that confer antimicrobial resistance, play a significant role in shaping the pan-genome of a bacterial species (Ding et al., 2018). The pan-genome of E. hirae was considered “open” as there was no sign of saturation and it would be expected to expand with the addition of new genomes as illustrated by the pan-genome curve (Figure 4B). The high presence of cloud genes reflects the heterogeneity of the pan-genome of E. hirae. The existence of E. hirae in diverse environments may increase the chance of gene acquisition, in contrast to other Enterococcus species that may live in more specific environments that require less genomic variation for survival (Costa et al., 2020).
Gram-positive bacteria have sophisticated cell wall structures that ensure bacterial structural integrity and cellular viability and are also a major component of the host defense system (Koch, 2003; Silhavy et al., 2010). For this reason, numerous studies have been conducted to explore components of cell wall synthesis pathways as potential targets for drug therapy. Genes involved in cell wall synthesis were identified as discriminatory genomic signatures between E. faecalis and E. hirae. Identification of these different signatures [peptidoglycan synthesis genes (Mur E, F, and Y), penicillin-binding protein (pbpE and pbpX), teichoic acid synthesis genes (tag H, G, and U), and enterococcal polysaccharide antigen (eps E, D, M, and N)] may identify targets that offer more specific drug development against E. faecalis and E. faecium (Parisien et al., 2008).
Members of gut microflora compete with each other for nutrient availability. Therefore, the ability of one bacterial species to utilize multiple nutrients for energy generation provides an advantage over other species. We found genes involved in the synthesis of cobalamin (vitamin B12), pyridoxine (vitamin B6), riboflavin (vitamin B2), biotin (vitamin B7), and folic acid exclusive to the E. hirae species compared with other enterococci analyzed in this study. With vitamins being undeniably important for both bacteria and the mammalian host, gut bacteria associated with their production directly contribute to the development and welfare of the host and thus may have a specific function within the microbiome of the digestive tract of cattle. In E. hirae genomes, multiple phosphotransferase systems (PTS) for fructose, galactitol, mannose, sorbose, glucose, N-acetyl glucosamine, and cellobiose were identified. The presence of these PTS promotes colonization of these bacterial populations (Jeckelmann and Erni, 2020). In addition, compared to E. faecalis and E. faecium, E. hirae harbored genes that were predicted to contribute to the synthesis of bacterial cellulose. These findings indicate that cellular metabolism genes identified in E. hirae may contribute to fitness within the cattle gut, accounting for its high prevalence in beef cattle.
Analysis of the annotated genomes indicated that E. hirae possessed genes coding for the production of antimicrobial agents like bacilysin, subtilosin, and narbonolide. Bacilysin is a dipeptide antimicrobial with antifungal and antibacterial activity (Özcengiz and Öğülür, 2015). Subtilosin belongs to the lantibiotics class of bacteriocins and has anti-biofilm activity (Babasaki et al., 1985; Algburi et al., 2017). These bacteria are also capable of producing gamma-aminobutyric acid (GABA), an inhibitory neurotransmitter. GABA may increase feed intake in cattle and reduce anxiety and pain (Sarasa et al., 2020; Mamuad and Lee, 2021). These findings suggest that E. hirae may have probiotic properties that could benefit the gastrointestinal environment of cattle (Ben Braïek and Smaoui, 2019). Previously, E. hirae has been employed as a probiotic bacteria in freshwater fish (Adnan et al., 2017). Recent studies have also demonstrated that E. hirae may confer probiotic properties within the intestinal tract of cattle (Arokiyaraj et al., 2014; Daillère et al., 2016).
One of the goals of this study was to identify the niche-specific genes in E. faecalis and E. faecium that may contribute to virulence and infection. Several virulence factors that contribute to the pathogenesis of E. faecalis and E. faecium have been reported (Ali et al., 2017; Zhou et al., 2020). Several virulence genes were unique to E. faecium and/or E. faecalis and were not found in E. hirae. Members of the SlyA/ MarA family of proteins are associated with virulence gene regulation, promote biofilm formation, and act as cell adhesions. The presence of genomic signatures corresponding to these genes may in part account for the higher prevalence of E. faecalis and E. faecium infections than E. hirae infections in humans (Mäkinen et al., 1989; Michaux et al., 2011; Yang et al., 2015). Lipoproteins facilitate intake of nutrients and are often associated with ABC transporters that are linked to pathogenesis. This supports our findings as genes encoding lipoproteins mapped with ABC transport systems for manganese, arabinose, and methionine (Zhang et al., 1998; Hutchings et al., 2009). Furthermore, in E. faecalis and E. faecium, we also identified an arginine-ornithine antiporter which could contribute to cell fitness by facilitating arginine uptake. A study conducted to investigate the role of arginine-ornithine antiporter in Streptococcus suis reported that intercellular survival of this pathogen within epithelial cells was compromised in the absence of the antiporter (Fulde et al., 2014).
The potential of E. hirae as an opportunistic pathogen cannot be ignored, as it is occasionally recovered from both human and animal clinical samples (Nicklas et al., 2010; Dicpinigaitis et al., 2015; Ebeling and Romito, 2019; Pinkes et al., 2019; Bilek et al., 2020). Despite a higher prevalence of virulence genes in E. faecium and E. faecalis, some virulence genes were also identified in E. hirae, like genes associated with the ESX (or Type VII) secretion system, bicyclomycin resistance, capsule biogenesis, quorum sensing system, and an ABC transporter for iron import (Stauff et al., 2008; Rutherford and Bassler, 2012; Borst et al., 2015; Hatosy and Martiny, 2015; Poweleit et al., 2019). In addition, a lipoprotein gene associated with the iron transport system has been identified and is thought to play a role in E. hirae establishing opportunistic infections (Hutchings et al., 2009).
In conclusion, E. hirae has a tremendous ability for survival and adaptation. It has acquired resistance to the most common antimicrobials used in beef production systems. In addition, cellular metabolism genes involved in vitamin biosynthesis, multiple ABC and PTS transport systems, chitin degradation, and cellulose synthesis provide selective advantage and facilitate intestinal colonization of the cattle gut. As E. hirae appears to be uniquely adapted to cattle hosts, this likely limits the extent to which it transfers genes to bacteria that are important in human health. Regardless, the absence of resistance to critical antimicrobials in E. hirae gives credibility to limiting use of these drugs in feedlots and suggests that prudent management of antimicrobials in feedlot settings is an important practice.
The datasets presented in this study can be found in BioProject PRJNA604849 and in online repositories. The names of the repositories and accession number(s) can be found in the article/supplementary material.
The animal study was reviewed and approved by the Lethbridge Research Centre Animal Care and Use Committee and was conducted according to the Canadian Council of Animal Care Guidelines. Sampling procedures were conducted according to the protocol approved by the Animal Care Committee, University of Calgary (Protocol ID: AC14 -0029). Written informed consent was obtained from the owners for the participation of their animals in this study.
RZ and TM designed the study. SH and CB arranged for collection of feedlot samples, metadata, and antimicrobial use data. RB, SC, and RZ isolated and characterized enterococci. RB performed AST. S-e-ZZ, generated figures, analyzed overall data/results, and wrote first draft of the manuscript. GD provided and managed the bioinformatics cluster facility and bioinformatics tools. S-e-ZZ and RZ analyzed sequence data. TM and AZ provided funding and supervision. All authors participated in editing and reviewing the manuscript and approved the final manuscript.
Authors are grateful to the Major Innovation Fund of the Government of Alberta in conjunction with the University of Calgary AMR One Health Consortium, the Beef Cattle Research Council (BCRC) Project FOS 10.13, and Genomics Research and Development Initiative of the Government of Canada for financial support.
CB is part owner and managing partner of Feedlot Health Management Services.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge Noriko Goji for sequencing, and Wendi Smart, Krysty Munns, Reuben Ha, and Taylor Davedow for technical assistance. Cooperation of the feedlot and wastewater treatment plant operators is also gratefully appreciated.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.859990/full#supplementary-material
Aarestrup, F. M. (2000). Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38, 2774–2777. doi: 10.1128/JCM.38.7.2774-2777.2000
Adnan, M., Patel, M., and Hadi, S. (2017). Functional and health promoting inherent attributes of Enterococcus hirae F2 as a novel probiotic isolated from the digestive tract of the freshwater fish Catla catla. PeerJ 5:e3085. doi: 10.7717/peerj.3085
Algburi, A., Zehm, S., Netrebov, V., Bren, A. B., Chistyakov, V., and Chikindas, M. L. (2017). Subtilosin prevents biofilm formation by inhibiting bacterial quorum sensing. Probiotics Antimicrob. Proteins 9, 81–90. doi: 10.1007/s12602-016-9242-x
Ali, L., Goraya, M. U., Arafat, Y., Ajmal, M., Chen, J. L., and Yu, D. (2017). Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int. J. Mol. Sci. 18:960. doi: 10.3390/ijms18050960
Anderson, K. L., Whitlock, J. E., and Harwood, V. J. (2005). Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71, 3041–3048. doi: 10.1128/aem.71.6.3041-3048.2005
Andrew, P. W., and Mitchell, T. J. (1997). The biology of Streptococci and Enterococci. In: Symposium Series-Society for Applied Bacteriology (United Kingdom). Journals Customer Services Blackwell Scientific Publications Ltd, Hoboken.
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Arokiyaraj, S., Islam, V. I. H., Bharanidharan, R., Raveendar, S., Lee, J., Kim, D. H., et al. (2014). Antibacterial, anti-inflammatory and probiotic potential of Enterococcus hirae isolated from the rumen of Bos primigenius. World J. Microbiol. Biotechnol. 30, 2111–2118. doi: 10.1007/s11274-014-1625-0
Babasaki, K., Takao, T., Shimonishi, Y., and Kurahashi, K. (1985). Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 98, 585–603. doi: 10.1093/oxfordjournals.jbchem.a135315
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barile, S., Devirgiliis, C., and Perozzi, G. (2012). Molecular characterization of a novel mosaic tet(S/M) gene encoding tetracycline resistance in foodborne strains of Streptococcus bovis. Microbiology (Reading) 158, 2353–2362. doi: 10.1099/mic.0.058206-0
Barnes, A. M. T., Frank, K. L., and Dunny, G. M. (2021). Enterococcal endocarditis: hiding in plain sight. Front. Cell. Infect. Microbiol. 11:722482. doi: 10.3389/fcimb.2021.722482
Ben Braïek, O., and Smaoui, S. (2019). Enterococci: between emerging pathogens and potential probiotics. Biomed. Res. Int. 5938210–5938213. doi: 10.1155/2019/5938210
Beukers, A. G., Zaheer, R., Cook, S. R., Stanford, K., Chaves, A. V., Ward, M. P., et al. (2015). Effect of in-feed administration and withdrawal of tylosin phosphate on antibiotic resistance in enterococci isolated from feedlot steers. Front. Microbiol. 6:483. doi: 10.3389/fmicb.2015.00483
Beukers, A. G., Zaheer, R., Goji, N., Amoako, K. K., Chaves, A. V., Ward, M. P., et al. (2017). Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC Microbiol. 17:52. doi: 10.1186/s12866-017-0962-1
Bilek, H. C., Deveci, A., Ünal, S., Tanrıverdi Çaycı, Y., and Tanyel, E. (2020). Enterococcus hirae as a cause of bacteremic urinary tract infection: first case report from Turkey. J. Infect. Dev. Ctries. 14, 1780–1482. doi: 10.3855/jidc.12522
Borst, L. B., Suyemoto, M. M., Scholl, E. H., Fuller, F. J., and Barnes, H. J. (2015). Comparative genomic analysis identifies divergent genomic features of pathogenic Enterococcus cecorum including a type IC CRISPR-Cas system, a capsule locus, an epa-like locus, and putative host tissue binding proteins. PLoS One 10:e0121294. doi: 10.1371/journal.pone.0121294
Bourafa, N., Loucif, L., Boutefnouchet, N., and Rolain, J. M. (2015). Enterococcus hirae, an unusual pathogen in humans causing urinary tract infection in a patient with benign prostatic hyperplasia: first case report in Algeria. New Microbes New Infect. 8, 7–9. doi: 10.1016/j.nmni.2015.08.003
Brayer, S., Linn, A., Holt, S., Ellery, K., Mitchell, S., and Williams, J. (2019). Enterococcus hirae bacteremia in an infant: case report and review of the literature. J. Pediatric. Infect. Dis. Soc. 8, 571–573. doi: 10.1093/jpids/piz028
Byappanahalli, M., and Fujioka, R. (2004). Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50, 27–32. doi: 10.2166/wst.2004.0009
Byappanahalli, M. N., Nevers, M. B., Korajkic, A., Staley, Z. R., and Harwood, V. J. (2012). Enterococci in the environment. Microbiol. Mol. Biol. Rev. 76, 685–706. doi: 10.1128/mmbr.00023-12
Cameron, A., and McAllister, T. A. (2016). Antimicrobial usage and resistance in beef production. J. Animal Sci. Biotechnol. 7, 1–22. doi: 10.1186/s40104-016-0127-3
Canalejo, E., Ballesteros, R., Cabezudo, J., García-Arata, M., and Moreno, J. (2008). Bacteraemic spondylodiscitis caused by Enterococcus hirae. Eur. J. Clin. Microbiol. Infect. Dis. 27, 613–615. doi: 10.1007/s10096-008-0476-8
Caretti, C., and Lubello, C. (2003). Wastewater disinfection with PAA and UV combined treatment: a pilot plant study. Water Res. 37, 2365–2371. doi: 10.1016/S0043-1354(03)00025-3
Chajęcka-Wierzchowska, W., Zadernowska, A., and Łaniewska-Trokenheim, Ł. (2016). Diversity of antibiotic resistance genes in Enterococcus strains isolated from ready-to-eat meat products. J. Food Sci. 81, M2799–M2807. doi: 10.1111/1750-3841.13523
Chan, T.-S., Wu, M.-S., Suk, F.-M., Chen, C.-N., Chen, Y.-F., Hou, Y.-H., et al. (2012). Enterococcus hirae-related acute pyelonephritis and cholangitis with bacteremia: an unusual infection in humans. Kaohsiung J. Med. Sci. 28, 111–114. doi: 10.1016/j.kjms.2011.06.027
Chen, L., Ruan, Z., Wu, Q., Zhou, T., and Li, C. (2010). Relationship between ermB gene and transposons Tn1545 and Tn917 in Enterococcus faecalis. Chin. J. Nosocomiol. 20, 763–766.
Costa, Y., Galimand, M., Leclercq, R., Duval, J., and Courvalin, P. (1993). Characterization of the chromosomal aac(6′)-ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37, 1896–1903. doi: 10.1128/aac.37.9.1896
Costa, S. S., Guimarães, L. C., Silva, A., Soares, S. C., and Baraúna, R. A. (2020). First steps in the analysis of prokaryotic pan-genomes. Bioinf. Biol. Insights 14:1177932220938064. doi: 10.1177/1177932220938064
Crespo, M. D., Olson, J. W., Altermann, E., Siletzky, R. M., and Kathariou, S. (2012). Chromosomal tet(O)-harboring regions in Campylobacter coli isolates from turkeys and swine. Appl. Environ. Microbiol. 78, 8488–8491. doi: 10.1128/aem.02258-12
Daillère, R., Vétizou, M., Waldschmitt, N., Yamazaki, T., Isnard, C., Poirier-Colame, V., et al. (2016). Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 45, 931–943. doi: 10.1016/j.immuni.2016.09.009
De Graef, E. M., Devriese, L. A., Vancanneyt, M., Baele, M., Collins, M. D., Lefebvre, K., et al. (2003). Description of Enterococcus canis sp. nov. from dogs and reclassification of Enterococcus porcinus Teixeira et al. 2001 as a junior synonym of Enterococcus villorum Vancanneyt et al 2001. Int. J. Syst. Evol. Microbiol. 53, 1069–1074. doi: 10.1099/ijs.0.02549-0
de Vaux, A., Laguerre, G., Diviès, C., and Prévost, H. (1998). Enterococcus asini sp. nov. isolated from the caecum of donkeys (Equus asinus). Int. J. Syst. Bacteriol. 48, 383–387. doi: 10.1099/00207713-48-2-383
Demirgül, F., and Tuncer, Y. (2017). Detection of antibiotic resistance and resistance genes in enterococci isolated from Sucuk, a traditional Turkish dry-fermented sausage. Korean J. Food Sci. Anim. Resour. 37, 670–681. doi: 10.5851/kosfa.2017.37.5.670
Devriese, L. A., Ceyssens, K., Rodrigues, U. M., and Collins, M. D. (1990). Enterococcus columbae, a species from pigeon intestines. FEMS Microbiol. Lett. 71, 247–251. doi: 10.1016/0378-1097(90)90228-i
Dicpinigaitis, P. V., De Aguirre, M., and Divito, J. (2015). Enterococcus hirae bacteremia associated with acute pancreatitis and septic shock. Case Rep. Infect. Dis. 2015, 1–3. doi: 10.1155/2015/123852
Ding, W., Baumdicker, F., and Neher, R. A. (2018). panX: pan-genome analysis and exploration. Nucleic Acids Res. 46:e5. doi: 10.1093/nar/gkx977
Dönhöfer, A., Franckenberg, S., Wickles, S., Berninghausen, O., Beckmann, R., and Wilson, D. N. (2012). Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. U. S. A. 109, 16900–16905. doi: 10.1073/pnas.1208037109
Ebeling, C. G., and Romito, B. T. (2019). Aortic valve endocarditis from Enterococcus hirae infection. Proc. (Bayl. Univ. Med. Cent.) 32, 249–250. doi: 10.1080/08998280.2018.1551698
Farrow, J. A., and Collins, M. D. (1985). Enterococcus hirae, a new species that includes amino acid assay strain NCDO 1258 and strains causing growth depression in young chickens. Int. J. Syst. Evol. Microbiol. 35, 73–75.
Flannagan, S. E., and Clewell, D. B. (2002). Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44, 803–817. doi: 10.1046/j.1365-2958.2002.02922.x
Fogarty, L. R., Haack, S. K., Wolcott, M. J., and Whitman, R. L. (2003). Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces. J. Appl. Microbiol. 94, 865–878. doi: 10.1046/j.1365-2672.2003.01910.x
Francino, M. P. (2016). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front. Microbiol. 6:1543. doi: 10.3389/fmicb.2015.01543
Fulde, M., Willenborg, J., Huber, C., Hitzmann, A., Willms, D., Seitz, M., et al. (2014). The arginine-ornithine antiporter ArcD contributes to biological fitness of Streptococcus suis. Front. Cell. Infect. Microbiol. 4:107. doi: 10.3389/fcimb.2014.00107
Gilad, J., Borer, A., Riesenberg, K., Peled, N., Shnaider, A., and Schlaeffer, F. (1998). Enterococcus hirae septicemia in a patient with end-stage renal disease undergoing hemodialysis. Eur. J. Clin. Microbiol. Infect. Dis. 17, 576–577. doi: 10.1007/BF01708623
Gilmore, M. S., Clewell, D. B., Ike, Y., and Shankar, N. (Ed.) (2014): Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary, Boston.
Giraffa, G. (2014). “Enterococcus,” in Encyclopedia of Food Microbiology (2nd Edn.). eds. C. A. Batt and M. L. Tortorello (Oxford: Academic Press).
Hadfield, J., Croucher, N. J., Goater, R. J., Abudahab, K., Aanensen, D. M., and Harris, S. R. (2017). Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34, 292–293. doi: 10.1093/bioinformatics/btx610
Harwood, V. J., Whitlock, J., and Withington, V. (2000). Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66, 3698–3704. doi: 10.1128/AEM.66.9.3698-3704.2000
Hashem, Y. A., Amin, H. M., Essam, T. M., Yassin, A. S., and Aziz, R. K. (2017). Biofilm formation in enterococci: genotype-phenotype correlations and inhibition by vancomycin. Sci. Rep. 7:5733. doi: 10.1038/s41598-017-05901-0
Hatosy, S. M., and Martiny, A. C. (2015). The ocean as a global reservoir of antibiotic resistance genes. Appl. Environ. Microbiol. 81, 7593–7599. doi: 10.1128/AEM.00736-15
Heim, S., Lleo, M., Bonato, B., Guzman, C. A., and Canepari, P. (2002). The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J. Bacteriol. 184, 6739–6745. doi: 10.1128/jb.184.23.6739-6745.2002
Hernando-Amado, S., Coque, T. M., Baquero, F., and Martínez, J. L. (2019). Defining and combating antibiotic resistance from one health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442. doi: 10.1038/s41564-019-0503-9
Heval Can, B., Aydın, D., Sema, Ü., Yeliz Tanrıverdi, Ç., and Esra, T. (2020). Enterococcus hirae as a cause of bacteremic urinary tract infection: first case report from Turkey. J. Infect. Dev. Countries 14, 1780–1482. doi: 10.3855/jidc.12522
Hiltunen, T., Virta, M., and Laine, A.-L. (2017). Antibiotic resistance in the wild: an eco-evolutionary perspective. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 372:20160039. doi: 10.1098/rstb.2016.0039
Hurd, H. S., and Malladi, S. (2008). A stochastic assessment of the public health risks of the use of macrolide antibiotics in food animals. Risk Anal.: Int. J. 28, 695–710. doi: 10.1111/j.1539-6924.2008.01054.x
Hutchings, M. I., Palmer, T., Harrington, D. J., and Sutcliffe, I. C. (2009). Lipoprotein biogenesis in gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 17, 13–21. doi: 10.1016/j.tim.2008.10.001
Jackson, C. R., Lombard, J. E., Dargatz, D. A., and Fedorka-Cray, P. J. (2011). Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett. Appl. Microbiol. 52, 41–48. doi: 10.1111/j.1472-765x.2010.02964.x
Jeckelmann, J.-M., and Erni, B. (2020). Transporters of glucose and other carbohydrates in bacteria. Pflügers Archiv-Eur. J. Physiol. 472, 1129–1153. doi: 10.1007/s00424-020-02379-0
Johnson, C. N., Sheriff, E. K., Duerkop, B. A., Chatterjee, A., and Margolin, W. (2021). Let me upgrade you: impact of mobile genetic elements on Enterococcal adaptation and evolution. J. Bacteriol. 203:e0017721. doi: 10.1128/JB.00177-21
Kazimierczak, K. A., Rincon, M. T., Patterson, A. J., Martin, J. C., Young, P., Flint, H. J., et al. (2008). A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob. Agents Chemother. 52, 4001–4009. doi: 10.1128/aac.00308-08
Kim, Y. B., Seo, K. W., Jeon, H. Y., Lim, S.-K., Sung, H. W., and Lee, Y. J. (2019). Molecular characterization of erythromycin and tetracycline-resistant Enterococcus faecalis isolated from retail chicken meats. Poult. Sci. 98, 977–983. doi: 10.3382/ps/pey477
Kim, Y. B., Yoon, S., Seo, K. W., Shim, J. B., Noh, E. B., and Lee, Y. J. (2021). Detection of linezolid-resistant Enterococcus faecalis and Enterococcus faecium isolates from the layer operation system in Korea. Microb. Drug Resist. 27, 1443–1449. doi: 10.1089/mdr.2020.0028
Klein, G. (2003). Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 88, 123–131. doi: 10.1016/s0168-1605(03)00175-2
Koch, A. L. (2003). Bacterial wall as target for attack: past, present, and future research. Clin. Microbiol. Rev. 16, 673–687. doi: 10.1128/CMR.16.4.673-687.2003
Law-Brown, J., and Meyers, P. R. (2003). Enterococcus phoeniculicola sp. nov., a novel member of the enterococci isolated from the uropygial gland of the red-billed Woodhoopoe, Phoeniculus purpureus. Int. J. Syst. Evol. Microbiol. 53, 683–685. doi: 10.1099/ijs.0.02334-0
Layton, B., Walters, S., Lam, L., and Boehm, A. (2010). Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109, 539–547. doi: 10.1111/j.1365-2672.2010.04675.x
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Mäkinen, P. L., Clewell, D. B., An, F., and Mäkinen, K. K. (1989). Purification and substrate specificity of a strongly hydrophobic extracellular Metalloendopeptidase (“Gelatinase”) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 264, 3325–3334. doi: 10.1016/S0021-9258(18)94069-X
Mamuad, L. L., and Lee, S.-S. (2021). The role of glutamic acid-producing microorganisms in rumen microbial ecosystems. J. Life Sci. 31, 520–526.
Maria da Gloria, S. C., Shewmaker, P. L., Steigerwalt, A. G., Morey, R. E., Sampson, A., Joyce, K., et al. (2006). Enterococcus caccae sp. nov., isolated from human stools. Int. J. Syst. Evol. Microbiol. 56, 1505–1508.
Marinier, E., Zaheer, R., Berry, C., Weedmark, K. A., Domaratzki, M., Mabon, P., et al. (2017). Neptune: a bioinformatics tool for rapid discovery of genomic variation in bacterial populations. Nucleic Acids Res. 45:e159. doi: 10.1093/nar/gkx702
Michaux, C., Sanguinetti, M., Reffuveille, F., Auffray, Y., Posteraro, B., Gilmore, M. S., et al. (2011). SlyA is a transcriptional regulator involved in the virulence of Enterococcus faecalis. Infect. Immun. 79, 2638–2645. doi: 10.1128/IAI.01132-10
Miller, W. R., Munita, J. M., and Arias, C. A. (2014). Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 12, 1221–1236. doi: 10.1586/14787210.2014.956092
Moreno, M. F., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026
Müller, T., Ulrich, A., Ott, E. M., and Müller, M. (2001). Identification of plant-associated enterococci. J. Appl. Microbiol. 91, 268–278. doi: 10.1046/j.1365-2672.2001.01373.x
Muniesa, M., Jofre, J., and Lucena, F. (1999). Occurrence and numbers of bacteriophages and bacterial indicators in faeces of yellow-legged seagull (Larus cachinnans). Lett. Appl. Microbiol. 29, 421–423. doi: 10.1046/j.1365-2672.1999.00666.x
Murray, B. E. (1990). The life and times of the Enterococcus. Clin. Microbiol. Rev. 3, 46–65. doi: 10.1128/cmr.3.1.46
Nakamura, T., Ishikawa, K., Matsuo, T., Kawai, F., Uehara, Y., and Mori, N. (2021). Enterococcus hirae bacteremia associated with acute pyelonephritis in a patient with alcoholic cirrhosis: a case report and literature review. BMC Infect. Dis. 21:999. doi: 10.1186/s12879-021-06707-2
Nicklas, J. L., Moisan, P., Stone, M. R., and Gookin, J. L. (2010). In situ molecular diagnosis and histopathological characterization of enteroadherent Enterococcus hirae infection in pre-weaning-age kittens. J. Clin. Microbiol. 48, 2814–2820. doi: 10.1128/jcm.00916-09
Okitsu, N., Kaieda, S., Yano, H., Nakano, R., Hosaka, Y., Okamoto, R., et al. (2005). Characterization of ermB gene transposition by Tn1545 and Tn917 in macrolide-resistant Streptococcus pneumoniae isolates. J. Clin. Microbiol. 43, 168–173. doi: 10.1128/JCM.43.1.168-173.2005
Opal, S. M., and Pop-Vicas, A. (2015). “18 - molecular mechanisms of antibiotic resistance in bacteria”. in Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, eds. J. E. Bennett, R. Dolin, M. J. Blaser, and W. B. Saunders. (8th Edn.), Elsevier Saunders Philadelphia 235–251.
Özcengiz, G., and Öğülür, i. (2015). Biochemistry, genetics and regulation of bacilysin biosynthesis and its significance more than an antibiotic. New Biotechnol. 32, 612–619. doi: 10.1016/j.nbt.2015.01.006
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics (Oxford, England) 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Palmer, K. L., Kos, V. N., and Gilmore, M. S. (2010). Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13, 632–639. doi: 10.1016/j.mib.2010.08.004
Pãosinho, A., Azevedo, T., Alves, J. V., Costa, I. A., Carvalho, G., Peres, S. R., et al. (2016). Acute pyelonephritis with bacteremia caused by Enterococcus hirae: a rare infection in humans. Case reports in infectious diseases 2016:4698462. doi: 10.1155/2016/4698462
Parisien, A., Allain, B., Zhang, J., Mandeville, R., and Lan, C. Q. (2008). Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 104, 1–13. doi: 10.1111/j.1365-2672.2007.03498.x
Petkau, A., Mabon, P., Sieffert, C., Knox, N. C., Cabral, J., Iskander, M., et al. (2017). SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb. Genom. 3:e000116. doi: 10.1099/mgen.0.000116
Pinkes, M. E., White, C., and Wong, C. S. (2019). Native-valve Enterococcus hirae endocarditis: a case report and review of the literature. BMC Infect. Dis. 19:891. doi: 10.1186/s12879-019-4532-z
Portillo, A., Ruiz-Larrea, F., Zarazaga, M., Alonso, A., Martinez, J. L., and Torres, C. (2000). Macrolide Resistance Genes in Enterococcus spp. Antimicrob. Agents Chemother. 44, 967–971. doi: 10.1128/AAC.44.4.967-971.2000
Poweleit, N., Czudnochowski, N., Nakagawa, R., Trinidad, D. D., Murphy, K. C., Sassetti, C. M., et al. (2019). The structure of the endogenous ESX-3 secretion system. eLife 8:e52983. doi: 10.7554/eLife.52983
Poyart, C., Lambert, T., Morand, P., Abassade, P., Quesne, G., Baudouy, Y., et al. (2002). Native valve endocarditis due to Enterococcus hirae. J. Clin. Microbiol. 40, 2689–2690. doi: 10.1128/JCM.40.7.2689-2690.2002
Rovira, P., McAllister, T., Lakin, S. M., Cook, S. R., Doster, E., Noyes, N. R., et al. (2019). Characterization of the microbial resistome in conventional and “raised without antibiotics” beef and dairy production systems. Front. Microbiol. 10:1980. doi: 10.3389/fmicb.2019.01980
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Safferling, M., Griffith, H., Jin, J., Sharp, J., De Jesus, M., Ng, C., et al. (2003). TetL tetracycline efflux protein from Bacillus subtilis is a dimer in the membrane and in detergent solution. Biochemistry 42, 13969–13976. doi: 10.1021/bi035173q
Sanchez Valenzuela, A., Lavilla Lerma, L., Benomar, N., Gálvez, A., Perez Pulido, R., and Abriouel, H. (2013). Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne Pathog. Dis. 10, 143–149. doi: 10.1089/fpd.2012.1279
Sarasa, S. B., Mahendran, R., Muthusamy, G., Thankappan, B., Selta, D. R. F., and Angayarkanni, J. (2020). A brief review on the non-protein amino acid, gamma-amino butyric acid (GABA): its production and role in microbes. Curr. Microbiol. 77, 534–544. doi: 10.1007/s00284-019-01839-w
Schmitz, F.-J., Sadurski, R., Kray, A., Boos, M., Geisel, R., Köhrer, K., et al. (2000). Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45, 891–894. doi: 10.1093/jac/45.6.891
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. doi: 10.1101/cshperspect.a000414
Stauff, D. L., Bagaley, D., Torres, V. J., Joyce, R., Anderson, K. L., Kuechenmeister, L., et al. (2008). Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J. Bacteriol. 190, 3588–3596. doi: 10.1128/jb.01921-07
Svec, P., Devriese, L. A., Sedlácek, I., Baele, M., Vancanneyt, M., Haesebrouck, F., et al. (2001). Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov., isolated from water. Int. J. Syst. Evol. Microbiol. 51, 1567–1574. doi: 10.1099/00207713-51-4-1567
Švec, P., Vancanneyt, M., Sedláček, I., Naser, S. M., Snauwaert, C., Lefebvre, K., et al. (2006). Enterococcus silesiacus sp. nov. and Enterococcus termitis sp. nov. Int. J. Syst. Evol. Microbiol. 56, 577–581. doi: 10.1099/ijs.0.63937-0
Talarmin, J.-P., Pineau, S., Guillouzouic, A., Boutoille, D., Giraudeau, C., Reynaud, A., et al. (2011). Relapse of Enterococcus hirae prosthetic valve endocarditis. J. Clin. Microbiol. 49, 1182–1184. doi: 10.1128/JCM.02049-10
Tan, C.-K., Lai, C.-C., Wang, J.-Y., Lin, S.-H., Liao, C.-H., Huang, Y.-T., et al. (2010). Bacteremia caused by non-faecalis and non-faecium Enterococcus species at a medical center in Taiwan, 2000 to 2008. J. Infect. 61, 34–43. doi: 10.1016/j.jinf.2010.04.007
Tremblay, C.-L., Letellier, A., Quessy, S., Boulianne, M., Daignault, D., and Archambault, M. (2011). Multiple-antibiotic resistance of Enterococcus faecalis and Enterococcus faecium from Cecal contents in broiler chicken and Turkey flocks slaughtered in Canada and plasmid Colocalization of tetO and ermB genes. J. Food Prot. 74, 1639–1648. doi: 10.4315/0362-028x.Jfp-10-451
Vernikos, G. S. (2020) “A review of Pangenome tools and recent studies,” in The Pangenome: Diversity, Dynamics and Evolution of Genomes. eds. H. Tettelin and D. Medini (Springer, Berlin).
Vikram, A., Rovira, P., Agga, G. E., Arthur, T. M., Bosilevac, J. M., Wheeler, T. L., et al. (2017). Impact of “raised without antibiotics” beef cattle production practices on occurrences of antimicrobial resistance. Appl. Environ. Microbiol. 83, e01682–e01617. doi: 10.1128/AEM.01682-17
Wang, Y., Xiong, Y., Wang, Z., Zheng, J., Xu, G., Deng, Q., et al. (2021). Comparison of solithromycin with erythromycin in Enterococcus faecalis and Enterococcus faecium from China: antibacterial activity, clonality, resistance mechanism, and inhibition of biofilm formation. J. Antibiot. 74, 143–151. doi: 10.1038/s41429-020-00374-2
Wingett, S. W., and Andrews, S. (2018). FastQ screen: a tool for multi-genome mapping and quality control. F1000Research 7:7. doi: 10.12688/f1000research.15931.2
Winther, M., Dalager-Pedersen, M., Tarpgaard, I. H., and Nielsen, H. L. (2020). First Danish case of infective endocarditis caused by Enterococcus hirae. BMJ Case Rep. 13:e237950. doi: 10.1136/bcr-2020-237950
Yan, T., Goto, D. K., and Feng, F.: Concentration Dynamics of Fecal Indicators in Hawaiian Coastal and Inland Sand, Soil, and Water during Rainfall Events. Water Environment Research Foundation Alexandria, VA, (2011).
Yang, J., Tan, G., Zhang, T., White, R. H., Lu, J., and Ding, H. (2015). Deletion of the proposed iron chaperones IscA/SufA results in accumulation of a red intermediate cysteine Desulfurase IscS in Escherichia coli. J. Biol. Chem. 290, 14226–14234. doi: 10.1074/jbc.M115.654269
Zaheer, R., Cook, S. R., Barbieri, R., Goji, N., Cameron, A., Petkau, A., et al. (2020). Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a one-health continuum. Sci. Rep. 10:3937. doi: 10.1038/s41598-020-61002-5
Zaheer, R., Cook, S., Klima, C., Stanford, K., Alexander, T., Topp, E., et al. (2013). Effect of subtherapeutic vs. therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front. Microbiol. 4:133. doi: 10.3389/fmicb.2013.00133
Zaheer, R., Lakin, S. M., Polo, R. O., Cook, S. R., Larney, F. J., Morley, P. S., et al. (2019). Comparative diversity of microbiomes and Resistomes in beef feedlots, downstream environments and urban sewage influent. BMC Microbiol. 19, 1–17. doi: 10.1186/s12866-019-1548-x
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, H., Niesel, D. W., Peterson, J. W., and Klimpel, G. R. (1998). Lipoprotein release by bacteria: potential factor in bacterial pathogenesis. Infect. Immun. 66, 5196–5201. doi: 10.1128/iai.66.11.5196-5201.1998
Keywords: cattle production, antimicrobial resistance, enterococci, genomic signatures, pan-genome
Citation: Zaidi S-e-Z, Zaheer R, Barbieri R, Cook SR, Hannon SJ, Booker CW, Church D, Van Domselaar G, Zovoilis A and McAllister TA (2022) Genomic Characterization of Enterococcus hirae From Beef Cattle Feedlots and Associated Environmental Continuum. Front. Microbiol. 13:859990. doi: 10.3389/fmicb.2022.859990
Received: 22 January 2022; Accepted: 08 April 2022;
Published: 27 June 2022.
Edited by:
Ziad Daoud, Central Michigan University, United StatesReviewed by:
Jørgen J Leisner, University of Copenhagen, DenmarkCopyright © 2022 Sherry J. Hannon, Calvin W. Booker, Deirdre Church, Gary Van Domselaar, Athanasios Zovoilis and Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Tim A. McAllister, Sani-e-Zehra Zaidi, Rahat Zaheer, Ruth Barbieri and Shaun R. Cook. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tim A. McAllister, dGltLm1jYWxsaXN0ZXJAYWdyLmdjLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.