
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 April 2022
Sec. Microbiological Chemistry and Geomicrobiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.859921
 Yonggan Chen1,2
Yonggan Chen1,2 Minjing Zheng1,2
Minjing Zheng1,2 Yue Qiu1
Yue Qiu1 Hong Wang1
Hong Wang1 Haonan Zhang3
Haonan Zhang3 Qiongren Tao1
Qiongren Tao1 Hongwei Luo1
Hongwei Luo1 Zhenhua Zhang1,3*
Zhenhua Zhang1,3*
Coastal lagoon is an important productive ecosystem on the Earth. In this study, we compared microbial community in the sediments between lagoon and surrounding areas, and explored mechanism for the variation of microbial community. As a result, the sediment of surrounding area showed significantly higher organic matter and total nitrogen than that of the lagoon. The linear regression analysis revealed that organic matter and total nitrogen are positively correlated with Xenococcus. Bacterial and fungal PCoA1 showed significantly positive relationships with the relative abundance of Xenococcus, indicating that Xenococcus affects the bacterial and fungal community in the sediments of both the lagoon and surrounding area. ANOSIM analysis demonstrated that there were significant differences in bacterial and fungal community structure in the sediments between the lagoon and surrounding areas. Therefore, organic matter and total nitrogen affect the microbial community structure in the sediments of lagoon and surrounding areas by regulating the abundance of Xenococcus.
Lagoon is a highly productive ecosystem that plays important ecological roles, such as biogeochemical element cycling, coastal biodiversity and blue carbon sequestration (Bhatnagar et al., 2020). The status of lagoon may vary in response to natural events and human activities (Duran et al., 2015). Microbial community shaped biogeochemical cycle, they drive carbon cycling in lagoon sediments, mediate mineral nutrition of sea plants, and alleviate carbon limitations of other sediment organisms. The activity of microbial community is crucial for carbon, nitrogen, and phosphorus cycling (Mohit et al., 2014), as well as determines the fate of nutrition (Bordenave et al., 2004a,b; Pringault et al., 2008; Stauffert et al., 2013; M’rassi et al., 2015).
More insights in the microbial community are critical for understanding the functions of lagoon ecosystem. For instance, Sáenz de Miera et al. (2021) reported that the seasonal disturbance of water level cycles is the major determinant of microbial community structure in sediments of saline lagoons located in the northwest of Spain. Ben Salem et al. (2019) revealed bacterial community assemblages in sediments under high anthropogenic pressure at Bizerte Lagoon hydrological system, Tunisia. Proteobacteria, Bacteroidetes, Chloroflexi, Planctomycetes, and Nitrospirae are the major phyla, which is consistent with previous observations in sediment environments (Behera et al., 2017; Fan et al., 2017). Besides the popularly abundant phyla Proteobacteria, Chloroflexi and Planctomycetes, Spirochaetes were also found in the sediment of an enclosed shallow coastal lagoon, Magdalen Islands, Gulf of St. Lawrence, Canada (Mohit et al., 2015).
As a matter of fact, coastal lagoons usually have strong physicochemical gradient of salinity, nutrient concentration, turbidity, and organic matter content. The diversity of microorganisms in these ecosystems indicates that microbial community can be shaped by a variety of environmental factors, such as salinity, temperature, pH, hydrology, and latitude (Mohit et al., 2014). A previous study has revealed the environmental factors controlling the bacterial abundance in the sediments of a Mediterranean lagoon ecosystem. In this study, the variation of bacterial distribution was related to abiotic (temperature, ammonium, and pH) and trophic parameters (such as chlorophyll a) (Pala et al., 2018). An association analysis between sediment bacterial communities and environmental factors in Intermittently Closed and Open Lakes and Lagoons (ICOLLs) revealed that the bacterial diversity in Manly may be due to eutrophication, and that in Narrabeen may be attributed to the variety of organic matter sources (Filippini et al., 2019). Another study investigated the bacterial diversity and microbial functional response to organic matter composition in deltaic lagoon sediments, finding that at low organic contamination levels, the riverine inputs provide additional bioavailable carbon sources without evident adverse effects (Zoppini et al., 2020).
Coastal lagoon and the surrounding area are important aquatic systems with strong physicochemical gradients, where microorganisms have been well recognized to participate in biogeochemical cycles (Mary et al., 2014). However, the microbial composition, distribution and function in these environments remain largely unknown in the South China Sea. In this study, we investigated the microbial community in Qilianyu lagoon of China by analyzing the microorganisms involved in the regulation of microbial community structure with high-throughput sequencing, and systematically studying the correlation between microorganisms and physicochemical characteristics.
Qilianyu lagoon is located at 16.9703–16.9830N 112.2293–112.2585E. Two areas were selected for analysis, including lagoon zone (IL) and surrounding area (OL). The samples were collected in August 2020. Ten different sites were selected in the lagoon and surrounding areas for the study, respectively. Quadruplicate 1 × 1 m2 plots were constructed in each site. For each plot, five soil cores (5–10 cm in diameter; 1–10 cm in depth) were taken to make a composite sample.
Sediment samples were air-dried and sieved through a 100-mesh sieve for determination of organic matter (OM). pH, total nitrogen (TN), sodium (Na), nitrate (-N) and ammonium (-N) were determined after sieving through a 10-mesh sieve. Available potassium was determined using the method described by Easton and Lovering (1964). Other physicochemical characteristics were tested according to the method described by Chen et al. (2020).
Genomic DNA was extracted from soil samples using the OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, United States). NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and agarose gel electrophoresis were used to measure the quantity and quality of DNA. Bacterial and fungal abundance was determined by real-time quantitative polymerase chain reaction (Chen et al., 2021). The primers 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 519R (5’-ATTACCGCGGCTGCTGG-3’) (Zhang et al., 2017) and the primers ITS5 (5’-GGAAGTAAAAGTCGTAACAAGG-3’) and ITS2 (5’-GCTGCGTTCTTCATCGATGC-3’) (White et al., 1990) were used to evaluate the 16S rRNA and ITS gene copy number, respectively.
Primer 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’) were used to amplify 16S rDNA V3–V4 region (Hong et al., 2020). Primers ITS5 (5’-GGAAGTAAAAGTCGTAACAAGG-3’) and ITS2 (5’-GCTGCGTTCTTCATCGATGC-3’) were used to amplify the ITS1 region (White et al., 1990). PCR amplification and product purification were carried out as described by Chen et al. (2021). Sequencing was performed using the Illumina MiSeq platform at Shanghai Personal Biotechnology Co., Ltd.
QIIME 2 was used to process the sequencing data (Bolyen et al., 2018). DADA2 plugin was then used for sequence quality filtering, denoising, merging and chimera removing (Callahan et al., 2016). Non-singleton amplicon sequence variants (ASVs) were aligned with MAFFT (Katoh et al., 2002) and used to construct phylogenetic trees with FastTree2 (Price et al., 2009). Bacterial and fungal taxonomy was assigned to ASVs using the classify-sklearn naïve Bayes taxonomy classifier in feature-classifier plugin (Bokulich et al., 2018) against the SILVA Release 132 /UNITE Release 8.0 database, respectively (Kõljalg et al., 2013).
QIIME2 and R packages (v3.2.0) were used to analyze the sequence data. The observed species, Chao1, Shannon, Simpson, Faith’s PD, Pielou’s evenness, Good’s coverage, and beta diversity metrics Bray-Curtis dissimilarity were estimated by the diversity plugin. The alpha-diversity was visualized as box plots, and the beta diversity was visualized via principal coordinate analysis (PCoA). Significant differences in microbial structure were evaluated based on ANOSIM (Analysis of similarities) (Clarke, 1993; Warton et al., 2012) performed by QIIME2. Venn diagrams were generated using the R package “VennDiagram” (Zaura et al., 2009). LEfSe (Linear discriminant analysis effect size) was performed to detect differentially abundant taxa across groups using the default parameters (Segata et al., 2011). Linear relationships analysis was performed by the genescloud tools, a free online platform for data analysis1. The environmental factors affecting microbial community was analyzed by Canonical Correlation Analysis (CCA) using the vif.cca, step model, and anova functions within the R package (Oksanen et al., 2016).
The bacterial and fungal sequence data are deposited in China National Microbiology Data Center (NMDC) with accession numbers NMDC10017942 and NMDC10017943, respectively.
The sampling sites were distributed in the lagoon and surrounding areas (Supplementary Figure 1). Physicochemical details of the selected sampling sites are presented in Table 1, which can reflect the ecosystem characteristics of the lagoon. No significant difference was observed in sodium, -N and -N between the samples from the two kinds of sites (P > 0.05). Furthermore, OL sample showed higher organic matter (0.46 ± 0.05%), total nitrogen (0.33 ± 0.05 g/kg), pH (8.12 ± 0.05) and available potassium (174.95 ± 15.76 mg/kg) than IL sample (P < 0.05). A detailed analysis of the data indicated that sediments in the lagoon and surrounding areas were nutrient-deficient and pollution-free.
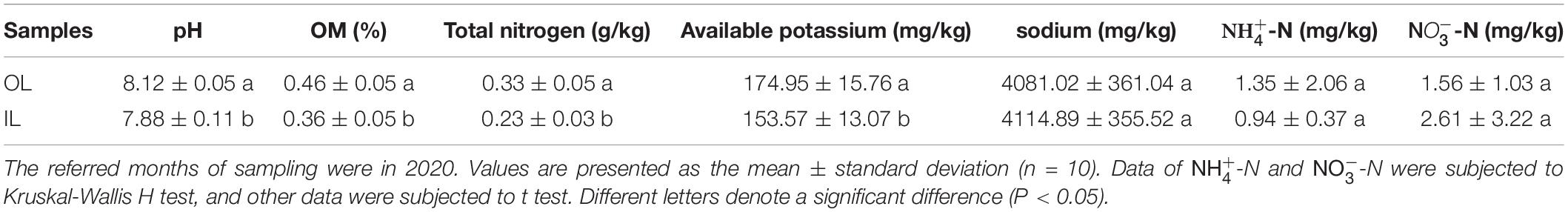
Table 1. Physicochemical parameters measured for the sample sites in Qilianyu lagoon and surrounding areas.
No significant differences were observed in fungal abundance of the two samples, whereas IL sample 3.55 × 109 copies/g was higher than OL sample 1.94 × 109 copies/g in bacterial abundance (P < 0.05). The rarefaction curves were smooth for both the OL and IL samples, indicating that the sequencing depth could well reflect the structural characteristics of the microorganisms collected from two different sites. In addition, Figure 1 shows that the Good’s coverage values of sediments all reached above 0.980, suggesting that the sequencing results can represent the real situation of microorganisms in the sediments of different sampling sites. Shannon, Simpson and Pielou’s evenness index of bacteria (including Faith’s PD) and fungi showed no difference in two kinds of samples. However, the Chao1 and Observed species index indicated that the total number of fungal species in the IL sample was larger than that in the OL sample (P < 0.05) (Figure 1B).
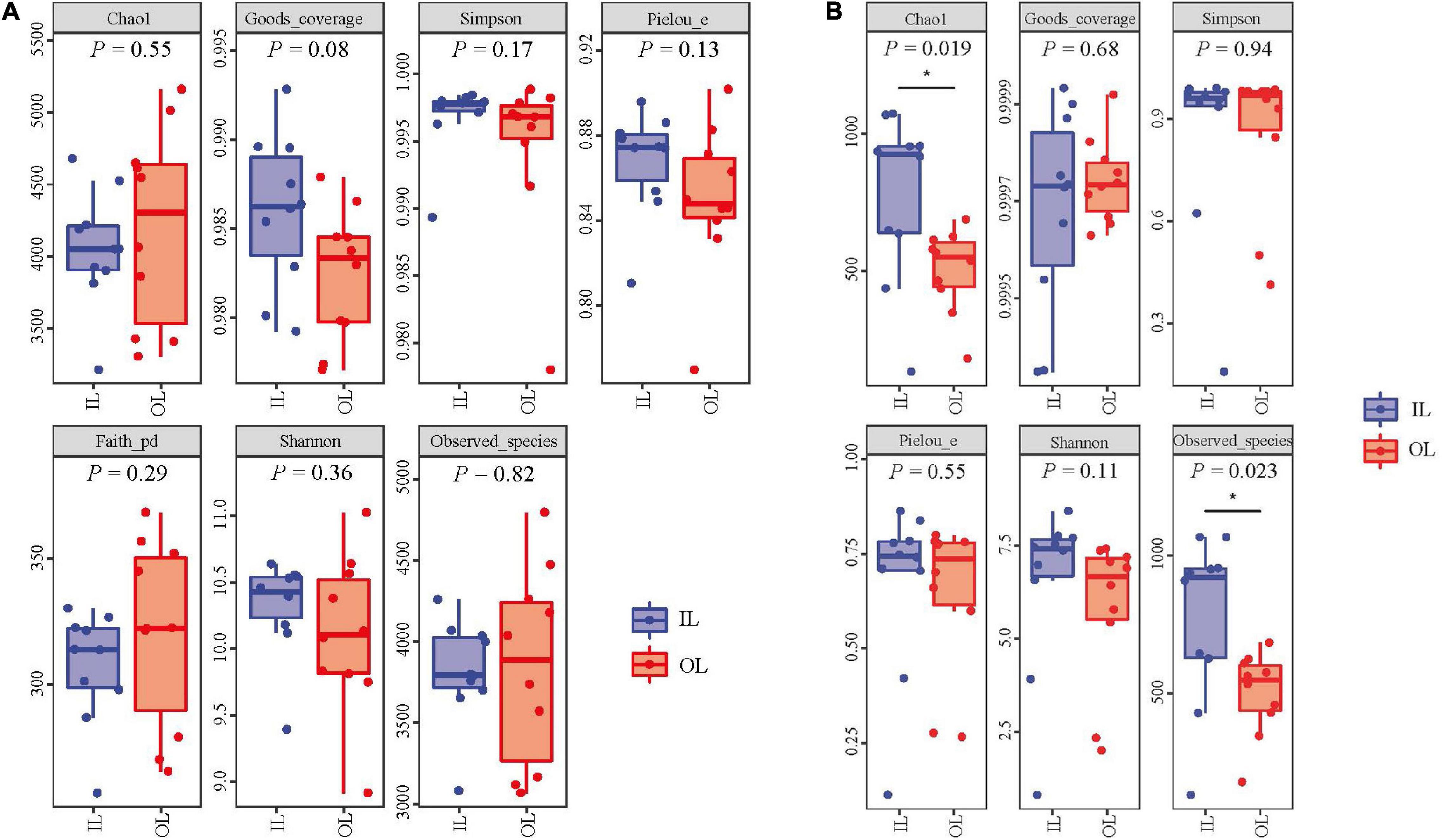
Figure 1. Bacterial (A) and fungal (B) α-diversity indices of the sediments from Qilianyu lagoon and surrounding area.
According to the PCoA analysis (Figures 2A,B), the first two principal components of bacteria account for 19.2 and 13.0% based on the Bray-curtis distances. For fungi, the first two principal components could account for 16.6 and 13.1%. According to the ANOSIM analysis, there were significant differences in bacterial and fungal structure between the OL and IL samples (P < 0.05). These results indicated that the microorganisms existing in sediments are significantly different in the lagoon and surrounding areas.
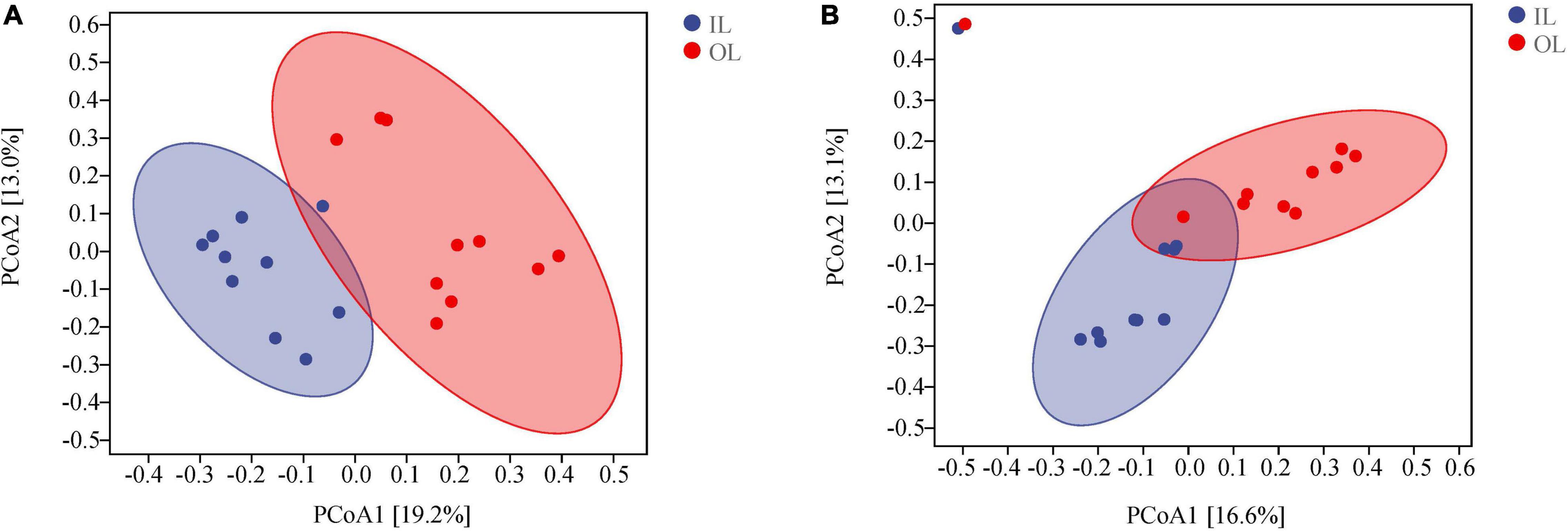
Figure 2. Microbial community structures in the sediments of Qilianyu lagoon and surrounding area. Bray-curtis principal coordinate analysis of bacterial (A) and fungal (B) community structures.
Figures 3A,B show the bacterial community composition at the phylum and genus levels. Proteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria and Acidobacteria were predominant in the sediments of the lagoon and surrounding areas. Compared with that in IL samples (1.8945 ± 1.3869%), the relative abundance of Cyanobacteria in OL samples was significantly increased to 12.0615 ± 9.7312% (P < 0.05). Typically, Xenococcus which belongs to Cyanobacteria was present in both the IL and OL samples, accounting for 0.1675 ± 0.1123% and 1.0191 ± 0.7042% of the bacterial community, respectively (P < 0.05). Besides, Pleurocapsa was also present in both the IL and OL samples, accounting for 0.0228 ± 0.0207% and 1.0605 ± 1.7925% of the bacterial community, respectively (P < 0.05).
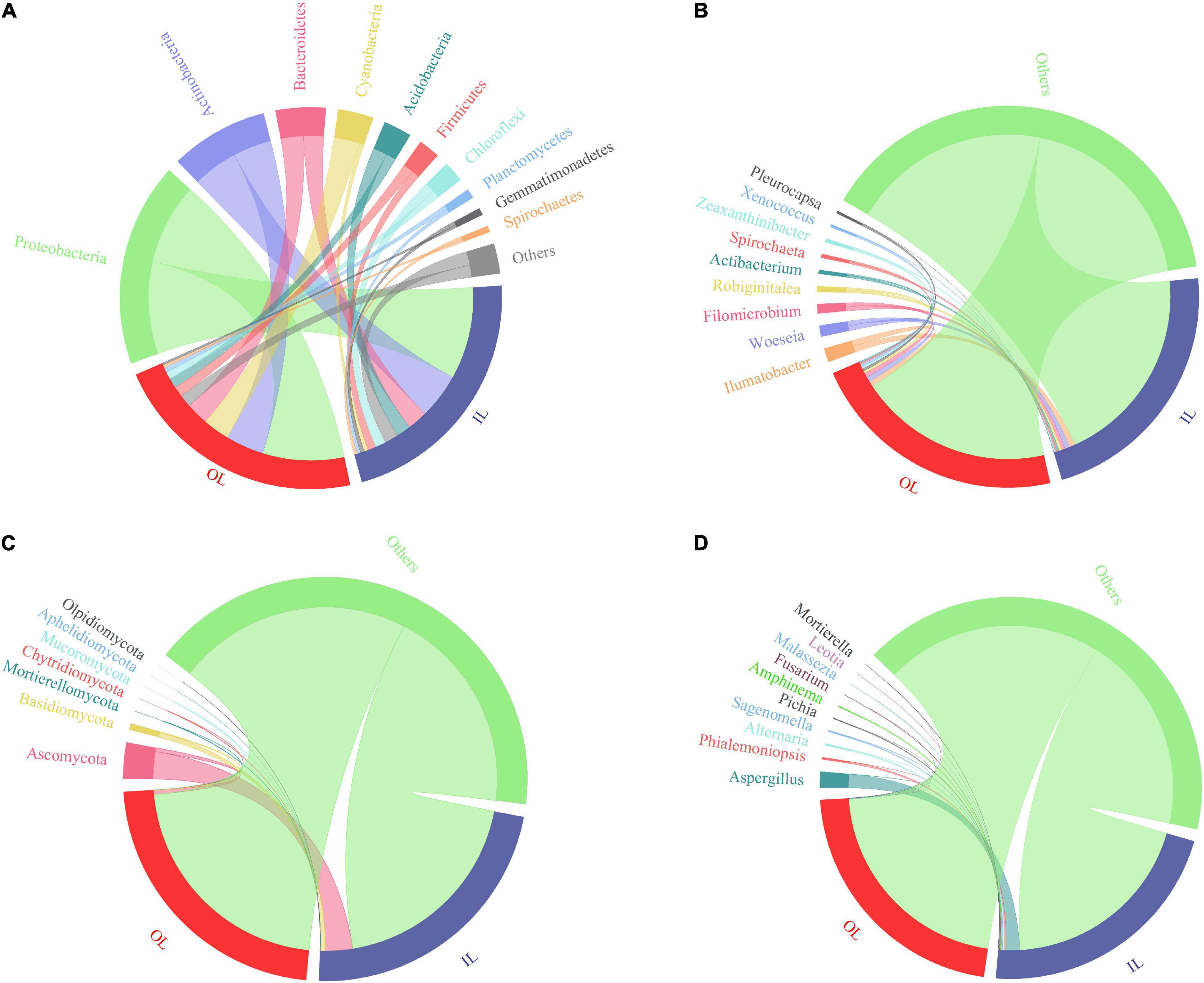
Figure 3. Relative abundance of microbial taxa in the sediments from Qilianyu lagoon and surrounding area. Bacterial phyla (A) and genera (B); and fungal phyla (C) and genera (D).
Fungal ASVs were predominantly associated with the phyla of Ascomycota, Basidiomycota, Mortierellomycota, Chytridiomycota and Mucoromycota (Figure 3C). Basidiomycota was more abundant in the IL sample than in the OL sample (P < 0.05), and other phyla were similar in the IL and OL samples. Phialemoniopsis was relatively more abundant (P < 0.05) in the lagoon sediments (0.8639 ± 1.2013%) than in the surrounding sediments (0.0806 ± 0.2467%). Differently, Aspergillus, Alternaria, Sagenomella, Pichia, Amphinema, and Fusarium showed no obvious difference in the two kinds of samples (Figure 3D).
LEfSe was used to further analyze bacterial and fungal genera in the sediments of the lagoon and surrounding areas. The threshold of 3.5 was employed on the logarithmic LDA score to determine the discriminative feature. Cyanobacteria and Bacteroidia were hyper-dominant in the sediments of the surrounding area, whereas Gemmatimonadetes, Acidobacteria, Actinobacteria, Chloroflexi and Proteobacteria were the most dominant phyla (accounting for 75.84% of the total bacterial phyla) in the sediments of the lagoon (Figure 4A). The bacterial genera Xenococcus and Pleurocapsa were more abundant in the sediments of the surrounding area. As for fungi, Basidiomycota and Phialemoniopsis were more abundant in the sediment of lagoon. However, there was no discrepancy in fungal genera in the two kinds of sediments (Figure 4B).

Figure 4. Clade diagram of the LDA scores computed for differentially abundant bacterial (A) and fungal (B) genera in the sediments between Qilianyu lagoon and the surrounding area.
In the linear regression analysis, organic matter and total nitrogen exhibited positive relationships with Xenococcus abundance (Figures 5A,B). However, a bacterial genus Pleurocapsa exhibited no linear relationship with organic matter and total nitrogen (Figures 5C,D). PCoA1 was selected as a microbial indicator in the linear model. As a result, the bacterial and fungal PCoA1 (P < 0.05) showed significant positive relationships with the relative abundance of Xenococcus. These results indicated that organic matter and total nitrogen could significantly regulate the relative abundance of Xenococcus, but have no correlation with Pleurocapsa. Furthermore, Xenococcus affected the bacterial and fungal community structure in the sediments of the lagoon and surrounding areas (Figures 6A,B). To identify the relationship between environmental variables and bacterial community, CCA was applied to interpret the pattern for sediment samples at the genus levels. As shown in Figure 7, 46.41% (P = 0.001 permutation test) and 19.97% (P = 0.005 permutation test) of the cumulative variance of the relationship were explained by the first and second axes of CCA, respectively, which indicated that the environmental variables could mostly explain (66.38%) the distribution of the genus in our study. In addition, organic matter, total nitrogen, and pH significantly affected (P < 0.05) Xenococcus which consistent with the above linear regression analysis. Furthermore, -N and -N contents significantly affected (P < 0.05) Haliangium, Silicimonas and Methyloceanibacter, indicating that above genus may related to the nitrification processes in lagoons sediments. The samples from OL and IL cannot be separated from CCA analysis. However, samples from IL clustered more together than OL, which indicated that the IL bacterial community structure remained relatively stable.
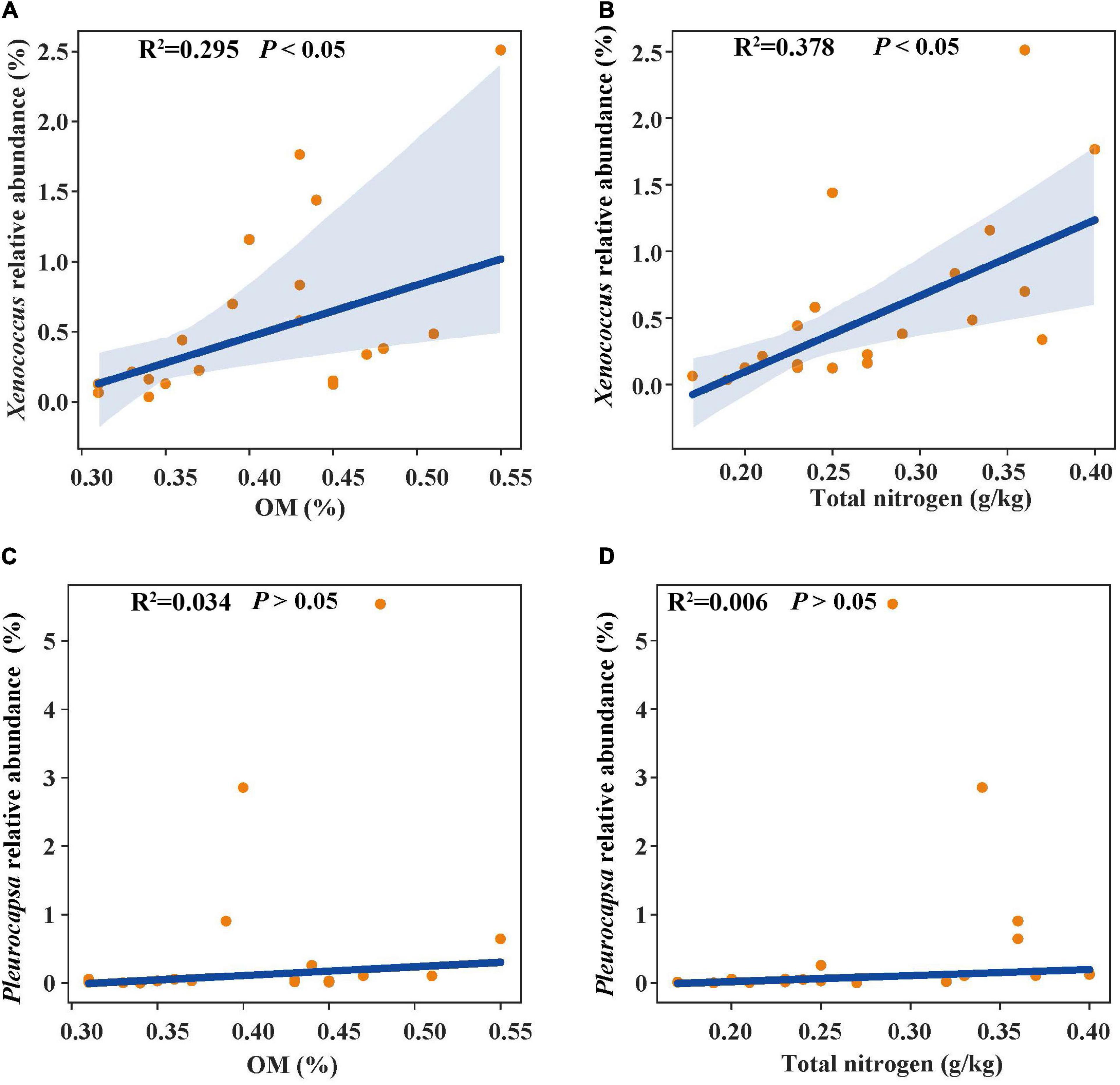
Figure 5. Linear regression relationship between organic matter (A), total nitrogen (B) and Xenococcus relative abundance; organic matter (C), total nitrogen (D) and Pleurocapsa relative abundance.
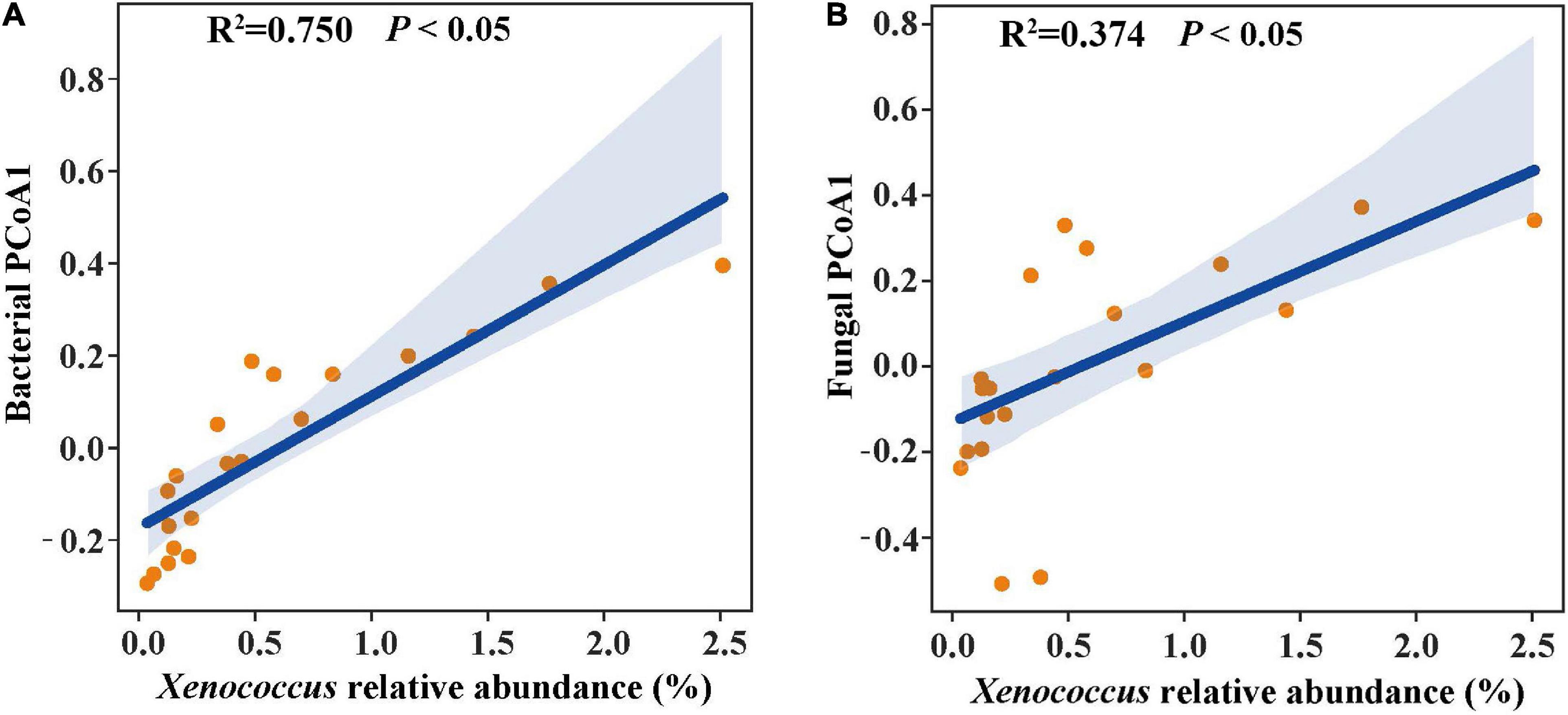
Figure 6. Linear regression relationship of the relative abundance of Xenococcus with bacterial structure (A) and fungal structure (B).
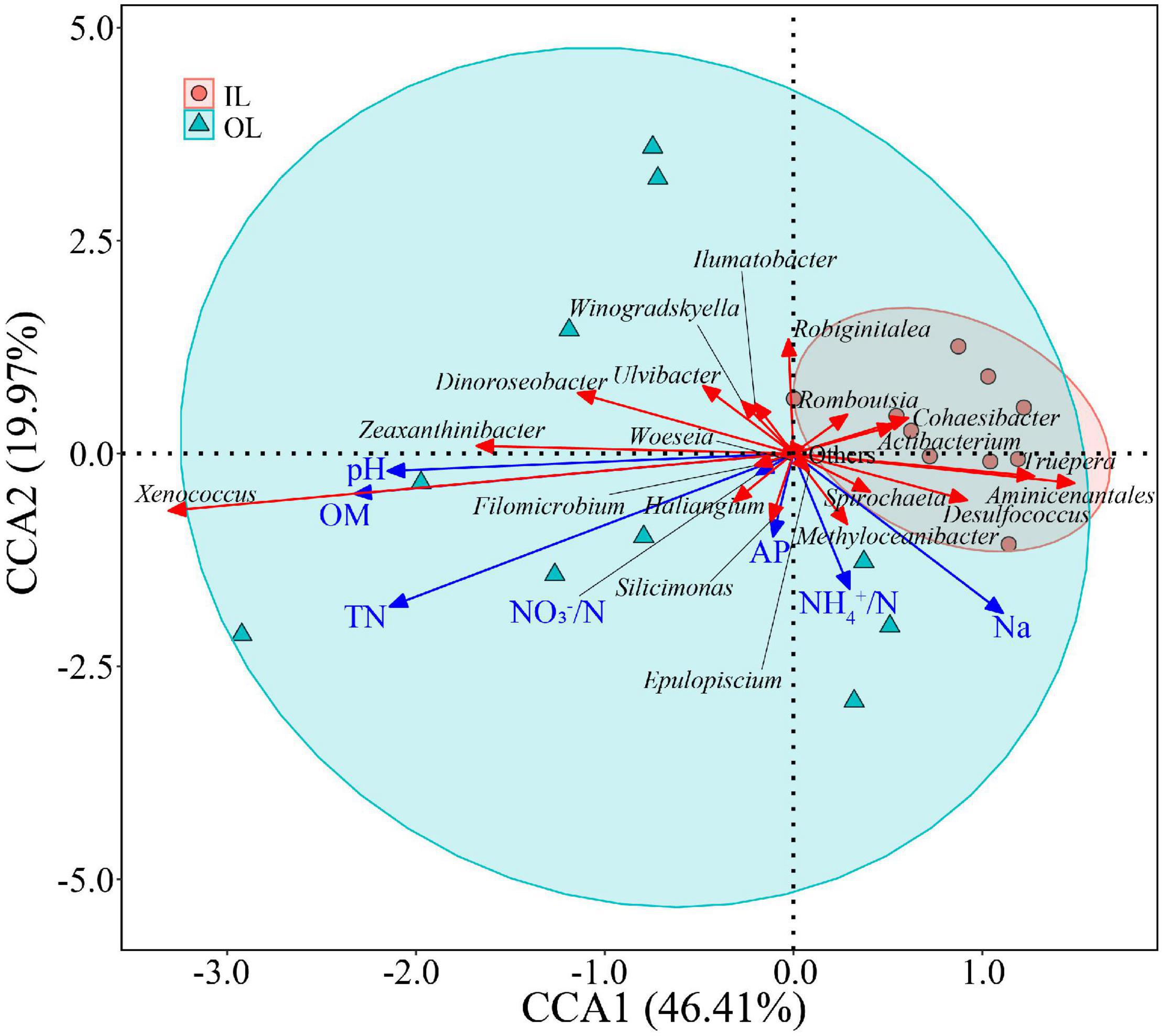
Figure 7. Relationships between sediment microbial communities and environmental factors indicated by Canonical Correlation Analysis (CCA). IL (red circle) and OL (blue triangle) represent sediments from lagoon zone and surrounding area, respectively.
Proteobacteria are frequently reported as the dominant phyla in lagoon sediments, and microbial composition will change according to their geographical location (Behera et al., 2017). Zoppini et al. (2020) found that Proteobacteria and Bacteroidetes were dominant in the deltaic lagoon sediments of Italy, followed by Epsilonbacteraeota, Firmicutes, Chloroflexi, Planctomycetes and Verrucomicrobia. A previous study investigated the benthic sediment bacteria in Celestún Lagoon in the Southern Gulf of Mexico. The results revealed that Proteobacteria, Chloroflexi, Bacteroidetes and Planctomycetes are the dominant taxa, followed by Spirochaetae, Acidobacteria, Actinobacteria, Cyanobacteria and Verrucomicrobia (Cadena et al., 2019). Sáenz de Miera et al. (2021) collected sediments from four lagoons of Spain, including Barillos, Salina Grande, Villarrin and Villalpando. Proteobacteria was identified as the most abundant phylum in all lagoons, except for Grande, where Chloroflexi was more abundant than Proteobacteria despite insignificant differences. The popular marine bacteria Proteobacteria were also found to be dominant in the sediments of Qilianyu lagoon. However, the order of other bacterial phyla in Qilianyu lagoon was different from that in the aforementioned lagoons. For fungi, ITS1 sequences of sediments collected from a coastal lagoon connected as a subestuary to Waquoit Bay (Cape Cod, MA, United States) were identified to be associated with Ascomycete, Chytridiomycota, Sordariomycetes, Pleosporales and Agaricomycetes. Typically, the sediments were dominated by the ascomycete genus Orbilia, which is different from the case of Qilianyu lagoon where Aspergillus is the main genus (Ortega-Arbulú et al., 2019). These results indicate that the divergence of bacterial and fungal composition in the sediments is affected by the geographical location.
Previous studies have identified total organic carbon as one of the most important factors affecting the bacterial composition in different aquatic habitats (Lallias et al., 2015; Highton et al., 2016). The relative abundance of bacterial taxa increased with total organic carbon increasing. These bacteria include Candidatus Giovannonibacteria (GW2011_GWB1_47_6b), Candidate division CPR3 (GW2011_GWF2_35_18), Parcubacteria (group bacterium ADurb. Bin326), Deferribacteraceae, Chlorobiaceae and Bradyamonadaceae (Filippini et al., 2019). A study of Chilika Lagoon located along the east coast of India also revealed the environmental conditions affected the microbial difference. The bacterial taxa include α-Proteobacteria, γ-Proteobacteria, Sphingobacteria, Bacilli, Nitrospira and Acidobacteria_Gp10, which prefer the higher salinity regime in the lagoon. In contrast, β-Proteobacteria, Clostridia and Actinobacteria prefer the freshwater-brackish salinity regime. However, none of the sediment parameters (total organic carbon, total nitrogen, total phosphorus, and pH) measured in the Chilika Lagoon was significantly correlated with the bacterial community composition (Behera et al., 2017). Here, we found that organic matter and total nitrogen exhibited positive relationships with Xenococcus, whereas had no linear relationship with Pleurocapsa. Our results suggest that natural changes in organic and nitrogen enrichment in the sediments can regulate the Xenococcus in Qilianyu lagoon.
Xenococcus classified in the phyla Cyanobacteria, it is widely found in the ocean ecology, but their biogeography and functions in lagoon environments is poorly understood. A case study of the Golden Bay mangrove in China demonstrated that cyanobacterial assemblages were dominated by Synechococcus and Xenococcaceae, and there was a clear shift of dominance from Synechococcus to Xenococcaceae toward the sea (Zhu et al., 2018). Rigonato et al. (2013) revealed that most of cyanobacteria were uncultivated (unclassified) and only a few Xenococcus sequences were detected in the mangrove soils in south-east Brazil. However, Xenococcus species have been detected on the trunks, roots, and pneumatophores of several mangrove trees (including Avicennia marina) with morphology-based methods (Alvarenga et al., 2015). Xenococcus was also found in the intertidal zones of the Portuguese coast, and in cyanobacterial mats retrieved from the La Réunion lagoon (Brito et al., 2012; Echenique-Subiabre et al., 2015). These results indicate the ubiquity of Xenococcus in the marine environment. Xenococcus one of genus of Cyanobacteria, and many research showed that Cyanobacteria play an important role in balance of nutrients, primarily carbon and nitrogen of marine ecosystems (Al-Thukair et al., 2007; Charpy et al., 2012). In this research, we also found that organic matter and total nitrogen could significantly regulate the relative abundance of Xenococcus. Furthermore, Xenococcus was found to affect the bacterial and fungal community structure in the sediments. Therefore, changes in organic matter and total nitrogen may lead to the differences in microbial community structure of the sediments between the lagoon and surrounding areas through the regulation of Xenococcus abundance.
This study analyzed the physicochemical characteristics and microorganisms in the lagoon and surrounding areas, which is beneficial to further understand the ecological characteristics of Qilianyu lagoon. The results indicate that organic matter and total nitrogen lead to the differences in microbial community structure in sediments between the lagoon and surrounding areas through the regulation of Xenococcus abundance. In the future, further research can be focused on whether fungi are involved in the regulation of the microbial community structure and the bacterial regulation mechanism.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YC: software. YQ, HW, and HL: formal analysis. YC, MZ, YQ, HW, and QT: investigation. YC, MZ, and HZ: writing – original draft preparation. YC and ZZ: writing, review, editing, project administration, and funding acquisition. ZZ: supervision. All authors contributed to the article and approved the submitted version.
This research was supported by the Hainan Province Science and Technology Special Fund (ZDYF2019212).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.859921/full#supplementary-material
Supplementary Figure 1 | Geographic information system map of Qilianyu lagoon, China. IL (red curve) and OL (blue curve) represent sediments from lagoon zone and surrounding area, respectively.
Al-Thukair, A. A., Abed, R. M. M., and Mohamed, L. (2007). Microbial community of cyanobacteria mats in the intertidal zone of oil-polluted coast of Saudi Arabia. Mar. Pollut. Bull. 54, 173–179. doi: 10.1016/j.marpolbul.2006.08.043
Alvarenga, D. O., Rigonato, J., Branco, L. H. Z., and Fiore, M. F. (2015). Cyanobacteria in mangrove ecosystems. Biodivers. Conserv. 24, 799–817. doi: 10.1007/s10531-015-0871-2
Behera, P., Mahapatra, S., Mohapatra, M., Kim, J. Y., Adhya, T. K., Raina, V., et al. (2017). Salinity and macrophyte drive the biogeography of the sedimentary bacterial communities in a brackish water tropical coastal lagoon. Sci. Total Environ. 595, 472–485. doi: 10.1016/j.scitotenv.2017.03.271
Ben Salem, F., Ben Said, O., Cravo-Laureau, C., Mahmoudi, E., Bru, N., Monperrus, M., et al. (2019). Bacterial community assemblages in sediments under high anthropogenic pressure at Ichkeul Lake/Bizerte Lagoon hydrological system, Tunisia. Environ. Pollut. 252, 644–656. doi: 10.1016/j.envpol.2019.05.146
Bhatnagar, S., Cowley, E. S., Kopf, S. H., Castro, S. P., Kearney, S., Dawson, S. C., et al. (2020). Microbial community dynamics and coexistence in a sulfide-driven phototrophic bloom. Environmen. Microbiome. 15, 111–125. doi: 10.1186/s40793-019-0348-0
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al-Ghalith, G. A., et al. (2018). QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ. Preprints 6:e27295v2. doi: 10.7287/peerj.preprints.27295v2
Bordenave, S., Fourçans, A., Blanchard, S., Goñi, M. S., Caumette, P., and Duran, R. (2004a). Structure and functional analyses of bacterial communities changes in microbial mats following petroleum exposure. Ophelia 58, 195–203. doi: 10.1080/00785236.2004.10410227
Bordenave, S., Jézéquel, R., Fourçans, A., Budzinski, H., Merlin, F. X., Fourel, T., et al. (2004b). Degradation of the “Erika” oil. Aquat. Living Resour. 17, 261–267. doi: 10.1051/alr:2004027
Brito, A., Ramos, V., Seabra, R., Santos, A., Santos, C. L., Lopo, M., et al. (2012). Culture-dependent characterization of cyanobacterial diversity in the intertidal zones of the Portuguese coast: a polyphasic study. Syst. Appl. Microbiol. 35, 110–119. doi: 10.1016/j.syapm.2011.07.003
Cadena, S., Aguirre-Macedo, M. L., Cerqueda-García, D., Cervantes, F. J., Herrera-Silveira, J. A., and García-Maldonado, J. Q. (2019). Community structure and distribution of benthic bacteria and archaea in a stratified coastal lagoon in the Southern Gulf of Mexico. Estuar. Coast. Shelf Sci. 230:106433. doi: 10.1016/j.ecss.2019.106433
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583. doi: 10.1038/nmeth.3869
Charpy, L., Casareto, B. E., Langlade, M. J., and Suzuki, Y. (2012). Cyanobacteria in coral reef ecosystems: a review. J. Mar. Biol. 19:259571. doi: 10.1155/2012/259571
Chen, Y. G., Zhang, Z. H., Luo, H. W., Li, Z., Zhang, L. J., and Huang, H. (2020). Distinct characteristics of bacterial community in the soil of Nanshazhou Island, South China Sea. Curr. Microbiol. 77, 1292–1300. doi: 10.1007/s00284-020-01933-4
Chen, Y. G., Zhang, Z. H., Zhang, H. N., Luo, H. W., and Li, Z. (2021). Characteristics of soil bacterial and fungal communities on interval seawater covering Linchang Island, China. Arch. Microbiol. 203, 2453–2461. doi: 10.1007/s00203-021-02268-9
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Duran, R., Cuny, P., Bonin, P., and Cravo-Laureau, C. (2015). Microbial ecology of hydrocarbon-polluted coastal sediments. Environ. Sci. Pollut. Res. Int. 22, 15195–15199. doi: 10.1007/s11356-015-5373-y
Easton, A. J., and Lovering, J. K. (1964). Determination of small quantities of potassium and sodium in stony meteoritic material, rocks and minerals. Anal. Chim. Acta 30, 543–548. doi: 10.1016/s0003-2670(00)88764-4
Echenique-Subiabre, I., Villeneuve, A., Golubic, S., Turquet, J., Humbert, J.-F., and Gugger, M. (2015). Influence of local and global environmental parameters on the composition of cyanobacterial mats in a tropical lagoon. Microb. Ecol. 69, 234–244. doi: 10.1007/s00248-014-0496-0
Fan, L., Barry, K., Hu, G., Meng, S., Song, C., Qiu, L., et al. (2017). Characterizing bacterial communities in tilapia pond surface sediment and their responses to pond differences and temporal variations. World J. Microbiol. Biotechnol. 33:1. doi: 10.1007/s11274-016-2144-y
Filippini, G., Bugnot, A. B., Johnston, E. L., Ruszczyk, J., Potts, J., Scanes, P., et al. (2019). Sediment bacterial communities associated with environmental factors in intermittently closed and open lakes and lagoons (ICOLLs). Sci. Total Environ. 693:133462. doi: 10.1016/j.scitotenv.2019.07.268
Highton, M. P., Roosa, S., Crawshaw, J., Schallenberg, M., and Morales, S. E. (2016). Physical factors correlate to microbial community structure and nitrogen cycling gene abundance in a nitrate fed eutrophic lagoon. Front. Microbiol. 7:1691. doi: 10.3389/fmicb.2016.01691
Hong, Y. H., Huang, Y., Wu, S., Yang, X. Z., Dong, Y. Z., Xu, D. Y., et al. (2020). Effects of imidacloprid on the oxidative stress, detoxification and gut microbiota of Chinese mitten crab, Eriocheir sinensis. Sci. Total Environ 729:138276. doi: 10.1016/j.scitotenv.2020.138276
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast fsourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Lallias, D., Hiddink, J. G., Fonseca, V. G., Gaspar, J. M., Sung, W., Neill, S. P., et al. (2015). Environmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems. ISME J. 9, 1208–1221. doi: 10.1038/ismej.2014.213
Mary, I., Irma, G., Leonor, B., Vanessa, A., and Luis, T. (2014). Microbial activity in surface sediments of chacopata-bocaripo lagoon axis, Sucre State, Venezuela. Mar. Pollut. Bull. 23, 483–490. doi: 10.1016/j.marpolbul.2014.10.028
Mohit, V., Archambault, P., and Lovejoy, C. (2015). Resilience and adjustments of surface sediment bacterial communities in an enclosed shallow coastal lagoon, Magdalen Islands, Gulf of St. Lawrence, Canada. FEMS Microbiol. Ecol. 91:fiv038. doi: 10.1093/femsec/fiv038
Mohit, V., Archambault, P., Toupoint, N., and Lovejoy, C. (2014). Phylogenetic differences in attached and free-living bacterial communities in a temperate coastal lagoon during summer, revealed via high throughput 16S rRNA gene sequencing. Appl. Environ. Microb. 80, 1–14. doi: 10.1128/AEM.02916-13
M’rassi, A. G., Bensalah, F., Gury, J., and Duran, R. (2015). Isolation and characterization of different bacterial strains for bioremediation of n-alkanes and polycyclic aromatic hydrocarbons. Environ. Sci. Pollut. 22, 15332–15346. doi: 10.1007/s11356-015-4343-8
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2016). Vegan: Community Ecology Package. R Package Version 2.3-3. Available online at: http://cran.r-project.org/web/packages/vegan/index.html. (accessed November 28, 2020)
Ortega-Arbulú, A.-S., Pichler, M., Vuillemin, A., and Orsi, W. D. (2019). Effects of organic matter and low oxygen on the mycobenthos in a coastal lagoon. Environ. Microbiol. 21, 374–388. doi: 10.1111/1462-2920.14469
Pala, C., Molari, M., Nizzoli, D., Bartoli, M., Viaroli, P., and Manini, E. (2018). Environmental drivers controlling bacterial and archaeal abundance in the sediments of a Mediterranean lagoon ecosystem. Curr. Microbiol. 75, 1147–1155. doi: 10.1007/s00284-018-1503-3
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Pringault, O., Duran, R., Jacquet, S., and Torréton, J. P. (2008). Temporal variations of microbial activity and diversity in marine tropical sediments (New Caledonia lagoon). Microb. Ecol. 55, 247–258. doi: 10.1007/s00248-007-9272-8
Rigonato, J., Kent, A. D., Alvarenga, D. O., Andreote, F. D., Beirigo, R. M., Vidal-Torrado, P., et al. (2013). Drivers of cyanobacterial diversity and community composition in mangrove soils in south-east Brazil. Environ. Microbiol. 15, 1103–1114. doi: 10.1111/j.1462-2920.2012.02830.x
Sáenz de Miera, L. E., Gutiérrez-González, J. J., Arroyo, P., Falagán, J., and Ansola, G. (2021). Prokaryotic community diversity in the sediments of saline lagoons and its resistance to seasonal disturbances by water level cycles. J. Soils Sediments. 21, 3169–3184. doi: 10.1007/s11368-021-03026-6
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Stauffert, M., Cravo-Laureau, C., Jezequel, R., Barantal, S., Cuny, P., Gilbert, F., et al. (2013). Impact of oil on bacterial community structure in bioturbated sediments. PLoS One. 8:e65347. doi: 10.1371/journal.pone.0065347
Warton, D. I., Wright, S. T., and Wang, Y. (2012). Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol. Evol. 3, 89–101. doi: 10.1111/j.2041-210X.2011.00127.x
White, T. J., Bruns, T., Lee, S. J. W. T., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, Vol. 18, eds M. A. Innis, D. H. Gelfand, J. J. Snisky, and T. J. White (San Diego, CA: Academic Press), 315–322. 8.50042-1 doi: 10.1016/b978-0-12-372180-
Zaura, E., Keijser, B. J. F., Huse, S. M., and Crielaard, W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. doi: 10.1186/1471-2180-9-259
Zhang, X., Zhang, Q., Liang, B., and Li, J. (2017). Changes in the abundance and structure of bacterial communities in the greenhouse tomato cultivation system under long-term fertilization treatments. Appl. Soil Ecol. 121, 82–89. doi: 10.1016/j.apsoil.2017.08.016
Zhu, P., Wang, Y., Shi, T., Zhang, X., Huang, G., and Gong, J. (2018). Intertidal zonation affects diversity and functional potentials of bacteria in surface sediments: a case study of the golden bay mangrove, China. Appl. Soil Ecol. 130, 159–168. doi: 10.1016/j.apsoil.2018.06.003
Zoppini, A., Bongiorni, L., Ademollo, N., Patrolecco, L., Cibic, T., Franzo, A., et al. (2020). Bacterial diversity and microbial functional responses to organic matter composition and persistent organic pollutants in deltaic lagoon sediments. Estuar. Coast. Shelf Sci. 233:106508. doi: 10.1016/j.ecss.2019.106508
Keywords: organic matter, total nitrogen, microbial community, sediments, lagoon, Xenococcus
Citation: Chen Y, Zheng M, Qiu Y, Wang H, Zhang H, Tao Q, Luo H and Zhang Z (2022) Organic Matter and Total Nitrogen Lead to Different Microbial Community Structure in Sediments Between Lagoon and Surrounding Areas by Regulating Xenococcus Abundance. Front. Microbiol. 13:859921. doi: 10.3389/fmicb.2022.859921
Received: 22 January 2022; Accepted: 25 March 2022;
Published: 21 April 2022.
Edited by:
Anhuai Lu, Peking University, ChinaReviewed by:
Yizhi Sheng, Miami University, United StatesCopyright © 2022 Chen, Zheng, Qiu, Wang, Zhang, Tao, Luo and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenhua Zhang, emhhbmcxOTg0OTIyQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.