
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 March 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.857442
This article is part of the Research Topic Metabolic Flexibility of Microbial Methane Oxidation View all 8 articles
 Rob A. Schmitz1,2
Rob A. Schmitz1,2 Sepehr S. Mohammadi1
Sepehr S. Mohammadi1 Timo van Erven1
Timo van Erven1 Tom Berben1
Tom Berben1 Mike S. M. Jetten1
Mike S. M. Jetten1 Arjan Pol1
Arjan Pol1 Huub J. M. Op den Camp1*
Huub J. M. Op den Camp1*
Methanotrophs aerobically oxidize methane to carbon dioxide to make a living and are known to degrade various other short chain carbon compounds as well. Volatile organic sulfur compounds such as methanethiol (CH3SH) are important intermediates in the sulfur cycle. Although volatile organic sulfur compounds co-occur with methane in various environments, little is known about how these compounds affect methanotrophy. The enzyme methanethiol oxidase catalyzing the oxidation of methanethiol has been known for decades, but only recently the mtoX gene encoding this enzyme was identified in a methylotrophic bacterium. The presence of a homologous gene in verrucomicrobial methanotrophs prompted us to examine how methanotrophs cope with methanethiol. Here, we show that the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV consumes methanethiol and produces H2S, which is concurrently oxidized. Consumption of methanethiol is required since methanethiol inhibits methane oxidation. Cells incubated with ∼15 μM methanethiol from the start clearly showed inhibition of growth. After depletion of methanethiol, growth resumed within 1 day. Genes encoding a putative methanethiol oxidase were found in a variety of methanotrophs. Therefore, we hypothesize that methanethiol degradation is a widespread detoxification mechanism in methanotrophs in a range of environments.
The potent greenhouse gas methane (CH4) is one of the key components of the global carbon cycle (Dean et al., 2018). Methane is emitted in large amounts from a variety of natural sources (e.g., wetlands, geothermal environments, and termites) and from industrial and agricultural sources (Etiope, 2009; Brune, 2010; Bridgham et al., 2013; Heede, 2014; Dean et al., 2018). Microorganisms living in natural or man-made environments can respire methane both aerobically (Hanson and Hanson, 1996; Murrell and Jetten, 2009) and anaerobically (Knittel and Boetius, 2009; Ettwig et al., 2010; Haroon et al., 2013). Aerobic methanotrophs are either members of the subphyla Alpha- and Gammaproteobacteria or the phylum Verrucomicrobia (Dunfield et al., 2007; Pol et al., 2007a; Islam et al., 2008; Op den Camp et al., 2009; van Teeseling et al., 2014; Schmitz et al., 2021). These verrucomicrobial methanotrophs are found in acidic geothermal habitats such as fumaroles and mudpots (Dunfield et al., 2007; Pol et al., 2007a; Islam et al., 2008; Erikstad et al., 2019; Awala et al., 2021). They share a low pH optimum (2.0–3.5) and all known isolates are part of the genus Methylacidimicrobium (optimum growth temperature at 30–50°C) or the genus Methylacidiphilum (optimum growth temperature at 50–60°C).
Methylotrophs are organisms that use reduced one-carbon compounds as energy and carbon source (Chistoserdova and Kalyuzhnaya, 2018). Methanotrophs are a special type of methylotrophs that possess a methane monooxygenase to oxidize methane to methanol (CH3OH) (Ross and Rosenzweig, 2017). Interestingly, verrucomicrobial methanotrophs possess the gene homolog mtoX, which was recently revealed to encode a copper-dependent methanethiol oxidase (MTO) in Hyphomicrobium sp. VS (Eyice et al., 2018). The presence of this gene suggests that the one-carbon compound methanethiol (CH3SH) could be a source of energy, carbon and sulfur (Eyice et al., 2018). Methanethiol is a foul-smelling volatile organic sulfur compound (VOSC) primarily degraded by microorganisms and a key intermediate of the global sulfur cycle (Lomans et al., 2002; Schäfer and Eyice, 2019). Moreover, methanethiol is toxic to animals and VOSCs in general are known to impact the environment in various ways, for instance through acid precipitation (Roman et al., 2016; Schäfer and Eyice, 2019; Kiragosyan et al., 2020; Maddry et al., 2020). Still, little is known about the effect of methanethiol on microorganisms (van den Bosch et al., 2009).
In nature, multiple biotic pathways lead to the production of methanethiol (Schäfer and Eyice, 2019). In marine systems, plankton produce the osmolyte dimethylsulfoniopropionate (DMSP), which can be degraded to methanethiol (Kiene, 1996). In both anoxic and oxic environments, the methylation of H2S and the degradation of sulfur-containing amino acids lead to methanethiol production (Lomans et al., 2001; Eyice et al., 2018). Moreover, methanethiol is produced from the degradation of dimethylsulfide (DMS) and dimethylsulfoxide (DMSO) and several methylotrophic methanogens were shown to grow on methanethiol and DMS (Finster et al., 1992; Lomans et al., 1999; Lyimo et al., 2000; Lyimo et al., 2009; Schäfer et al., 2010). The presence of methanethiol in acidic geothermal environments from which verrucomicrobial methanotrophs were isolated from is unresolved. These environments are characterized by emissions of various sulfur compounds such as H2S (Schmitz et al., 2021). Methanethiol can be formed abiotically (Heinen and Lauwers, 1995; Reeves et al., 2014) and in addition, biotic methanethiol production by Archaea in acidic environments has been observed (Baumler et al., 2007).
The gene encoding a putative MTO is found in all known verrucomicrobial methanotrophs (Schmitz et al., 2021). Several Hyphomicrobium strains were shown to degrade methanethiol using MTO to oxidize methanethiol to formaldehyde (CH2O), H2S and hydrogen peroxide (H2O2) (Suylen et al., 1987; Eyice et al., 2018). Interestingly, methylotrophs such as those of the genus Hyphomicrobium are effectively applied in biofilters to remove VOSCs from polluted industrial air (Pol et al., 1994; Pol et al., 2007b). On the contrary, very little is known about the mechanism through which methanotrophs cope with VOSCs such as methanethiol. Here, we show that methanethiol has an inhibitory effect on methanotrophy. Methylacidiphilum fumariolicum SolV grown on methane has a prolonged lag phase in the presence of methanethiol. M. fumariolicum SolV can degrade low concentrations of methanethiol, leading to the production and concurrent oxidation of H2S.
Methylacidiphilum fumariolicum SolV isolated from a hot and acidic mud pool near Naples (Italy) was grown in a continuous bioreactor under methanol limitation. The medium composition and chemostat operation were performed as described before by Picone et al. (2020), without the addition of ethane. Briefly, the cells (OD600∼0.9 or 1.0) grew in a 300 mL chemostat at 55°C and pH 2.2 in medium supplemented with 50 mM methanol at a dilution rate D of 0.013 h–1. To grow the cells continuously, 3.9 mL medium per hour and 10.6 mL gas per minute (10% O2 (v/v) and 5% CO2 (v/v) in argon) were added.
Cell fractions of M. fumariolicum SolV were obtained as described before (Schmitz et al., 2020). Briefly, cells were lysed using a French pressure cell and the crude extract (CE) was obtained after centrifugation at 10,000 × g for 10 min at 4°C. Subsequently, the CE was centrifuged at 137,000 × g for 1 h at 4°C, leading to the separation of the soluble proteins in the supernatant (soluble fraction, SF) and the membrane proteins in the pellet (membrane fraction, MF). The pellet was subsequently homogenized, mixed with buffer containing the detergent n-dodecyl-β-D-maltoside and again centrifuged at 137,000 × g for 1 h at 4°C to obtain the solubilized membrane fraction (SMF) in the supernatant.
Batch incubations were performed in 120 mL serum bottles containing 10 mL cells (OD600∼0.9 or 1.0) and air. The bottles were closed with a rubber stopper and incubated at 55°C and 350 rpm with different concentrations of methanethiol, methane, or hydrogen sulfide. Methanethiol and methane were obtained from pure stocks, whereas hydrogen sulfide was prepared by mixing sodium sulfide with hydrochloric acid in a closed bottle to create hydrogen sulfide in the gas phase. To quantify methanethiol and hydrogen sulfide, 100 μl from the headspace of the bottles was injected with a glass Hamilton syringe into a gas chromatograph (7890B GC systems Agilent technologies, Santa Clara, CA, United States) equipped with a Carbopack BHT100 glass column (2 m, ID 2 mm) and a flame photometric detector (FPD) (Pol et al., 2018). Methane was measured as described before (Mohammadi et al., 2019). The areas obtained through GC injections were used to calculate the methanethiol, hydrogen sulfide and methane concentrations using standard curves. It was experimentally determined that the methanethiol concentration in the liquid is about 1.5 times higher than the methanethiol concentration in the gas phase at 55°C. Dry weight of the cells was determined as described by Picone et al. (2020).
To investigate growth of M. fumariolicum SolV on methane in the presence of methanethiol, cells from the methanol-limited continuous culture were diluted to an OD600 of 0.01 in a sterilized 120 mL serum bottle, containing 10 mL medium and a headspace containing air, CO2 (5%), CH4 (7.5%), and with or without 1200 nmol methanethiol. All experiments were performed in triplicates. The optical density of the culture and the consumption of methanethiol and methane were routinely measured.
A representative set of genomes of alpha- and gammaproteobacterial methanotrophs was obtained from GenBank. The MTO sequence of Hyphomicrobium sp. VS (Eyice et al., 2018) was blasted against these proteobacterial genomes and Candidatus Methylomirabilis, using 10e-3 as the e-value threshold. Putative MTO sequences were added to the tree of Eyice et al. (2018), which already contained sequences of the verrucomicrobial methanotrophs Methylacidiphilum fumariolicum SolV and Methylacidiphilum infernorum V4. Sequences were aligned using Muscle 3.8.1551 (Madeira et al., 2019) and the tree was calculated using RAxML 8.2.10 (Stamatakis, 2014) with the rapid bootstrapping method and the PROTGAMMALGF substitution model. SignalP v. 5.0 was used to predict the cellular location of the putative MTO of 77 different methanotrophs (Almagro Armenteros et al., 2019).
To show that strain SolV is able to oxidize methanethiol, cells from the methanol-limited continuous bioreactor (OD600∼0.9) were used for activity tests with different methanethiol concentrations in batch cultures. At a starting liquid concentration of approximately 0.5 μM methanethiol, the substrate was completely consumed (Figure 1A). Cells of M. fumariolicum SolV incubated with a starting liquid concentration of approximately 5 μM methanethiol consumed this compound at a constant rate of 0.58 nmol ⋅ min–1 ⋅mg DW–1 in the initial phase of the incubation (Figure 1B). However, after about half of the initial methanethiol amount was consumed, the consumption rate of methanethiol severely decreased, suggesting inhibition. Addition of oxygen did not enhance methanethiol consumption, excluding that oxygen was limiting (Figure 1B). When cells were incubated with an initial concentration of approximately 2.5 μM, the substrate was fully consumed. Therefore, the cells seem unable to completely consume methanethiol concentrations of approximately 3 μM or higher.
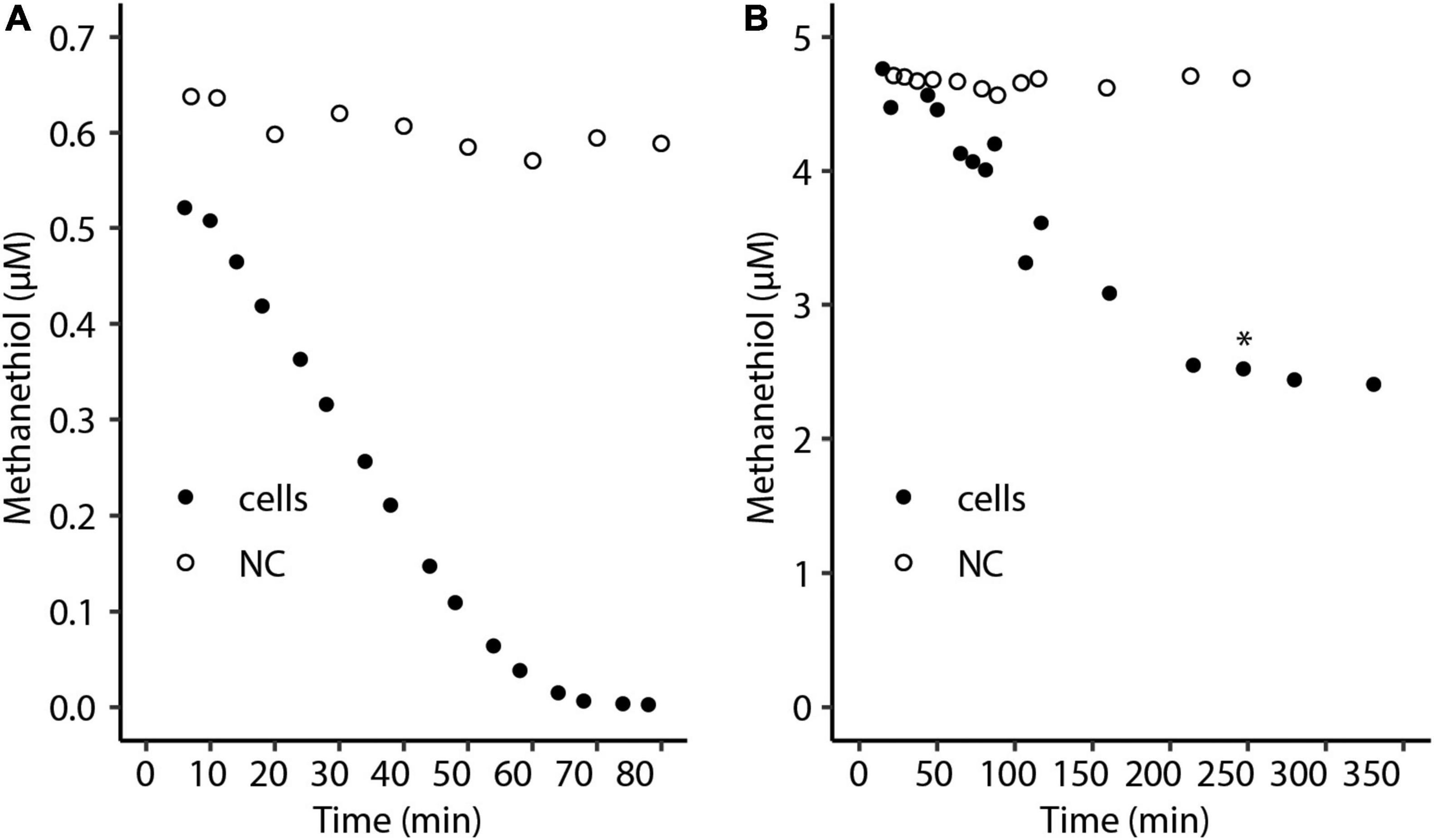
Figure 1. Consumption of a liquid concentration of approximately (A) 0.5 μM and (B) 5 μM methanethiol by Methylacidiphilum fumariolicum SolV cells over time. The asterisk indicates the time point at which additional oxygen was added. NC, negative control with medium only.
We also determined the affinity constant (Ks) and Vmax using non-inhibitory methanethiol concentrations in a range from 30 to 400 nmol (120-ml serum bottles, 10 ml cell suspension). The control, medium and methanethiol without cells, did not show oxidation of methanethiol. With SolV cells consumption rates of up to 1.7 nmol ⋅ min–1 ⋅ mg DW–1 were measured. From the data a highest rate of methanethiol consumption (Vmax) of about 2.3 nmol ⋅ min–1 ⋅ mg DW–1 and an affinity constant (Ks) of about 0.1 μM was calculated.
We further tested the methanethiol oxidation by feeding it to a methanol limited continuous culture (D = 0.013 h–1). We observed an increase in OD600 from 1.04 to 1.14 (about 10% increase), and the highest consumption rate of methanethiol was measured at 0.21 nmol ⋅ min–1 ⋅ mg DW–1, which is about 1% of the methanol consumption rate. Moreover, similar to our observations in the batch activity experiments, we found that when methanethiol concentrations rose above about 3 nmol per ml gas in the head-space (<4.5 μM in the liquid) of the reactor, a part of the fed methanethiol was detected in the gas outlet of the reactor. Reactor performance became unstable pointing to inhibition.
To investigate the effect of methanethiol on methanotrophy, cells from the methanol-limited continuous culture were transferred to serum bottles with 7.5% methane in the headspace. After 2 h, different amounts of methanethiol were added to create initial liquid concentrations of approximately 1–29 μM methanethiol. A liquid concentration of about 1 μM methanethiol does not affect methane oxidation and is depleted within an hour (Figures 2A,B). Methane oxidation of cells to which higher amounts of methanethiol were added were impeded at least temporarily (Figures 2A,B). When methanethiol was added to create a liquid concentration of approximately 4 μM, methane oxidation by M. fumariolicum SolV continued after more than half the amount of methanethiol was degraded (Figure 2). In the presence of approximately 9 μM methanethiol in the liquid, methane oxidation was inhibited but seems to resume when the concentration methanethiol had dropped below 4 μM. Interestingly, cells that were pre-incubated for 2 h with methane oxidized higher concentrations of methanethiol than cells that were not pre-incubated in batch (Figures 1B, 2B). When incubated with a liquid concentration of about 29 μM methanethiol, methane oxidation did not restore within 4 h (Figure 2A). Since methanethiol consumption continued after 4 h (Figure 2B), methane oxidation may still resume after methanethiol concentrations dropped below 5 μM.
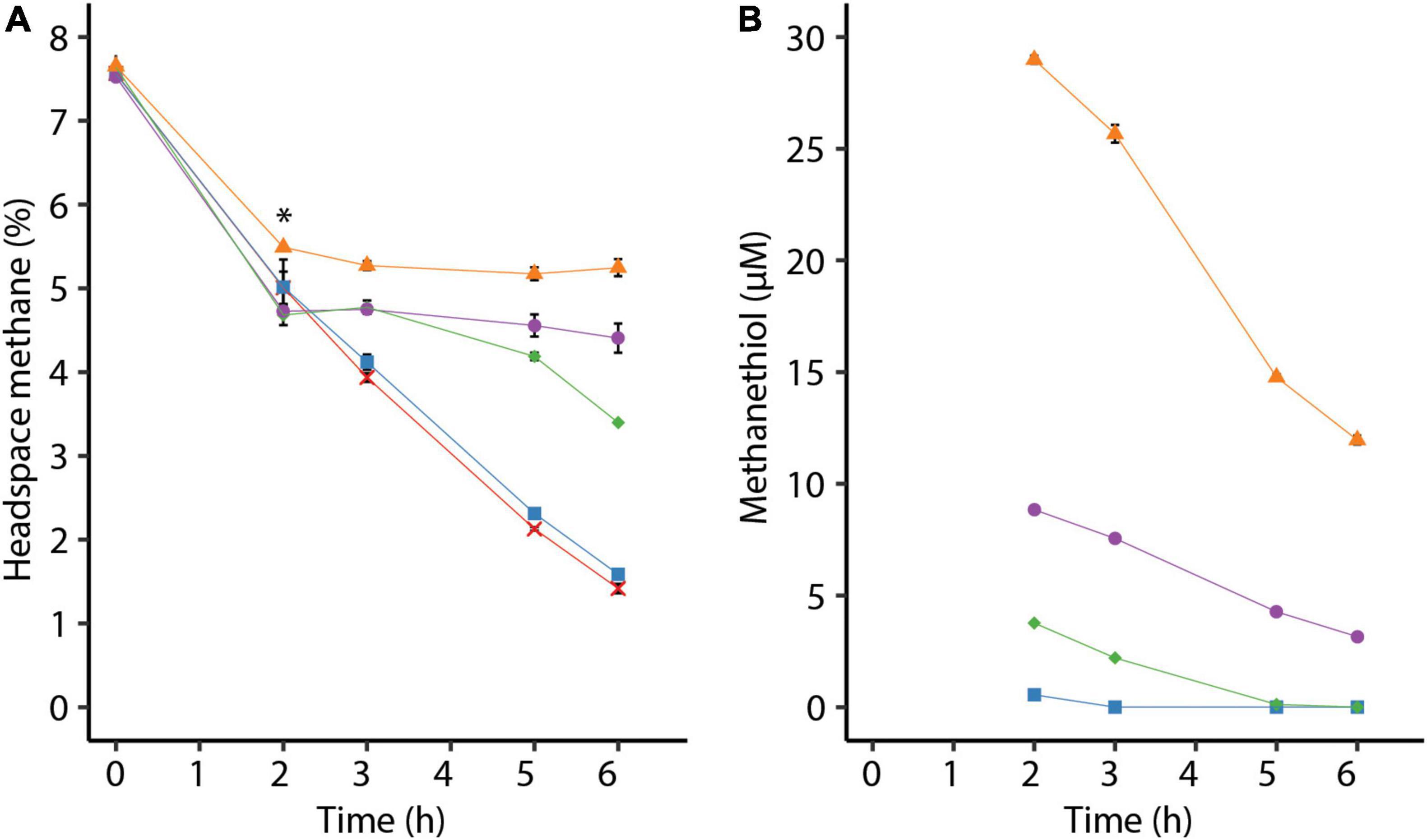
Figure 2. (A) Consumption of methane by Methylacidiphilum fumariolicum SolV cells over time and (B) decrease of methanethiol over time due to microbial and chemical degradation. The asterisk in panel (A) indicates the time point at which different amounts of methanethiol were added, leading to liquid methanethiol concentrations shown in panel (B). To the positive control (red crosses) no methanethiol was added. Error bars indicate standard deviation (n = 3).
To observe the effect of methanethiol on growth of M. fumariolicum SolV on methane, cells from the continuous bioreactor were diluted in growth medium to OD600∼0.01 and grown in batch cultures. Cultures incubated with both methane and methanethiol showed a prolonged lag phase, compared to the control with methane only (Figure 3A). Cells incubated with 1200 nmol methanethiol (∼15 μM in the liquid) from the start clearly showed inhibition of growth with large variations between replicates (Figure 3). After depletion of methanethiol, growth resumed within one day. Cells incubated without methanethiol had a growth rate (μ) of 0.060 h–1 and a doubling time (Td) of 11.5 h, whereas cells incubated with 15 μM methanethiol (depleted after 71 h) had a μ of 0.043 ± 0.003 h–1 (Td of 16.4 ± 1.0 h). It seems that regular growth starts after the high initial concentration of methanethiol is converted. The observed decrease in methanethiol is due a combination of microbial degradation and chemical degradation, as the methanethiol concentration in sterile medium decreases over time as well, leading to the production of mainly dimethyldisulfide (Supplementary Figure 1).
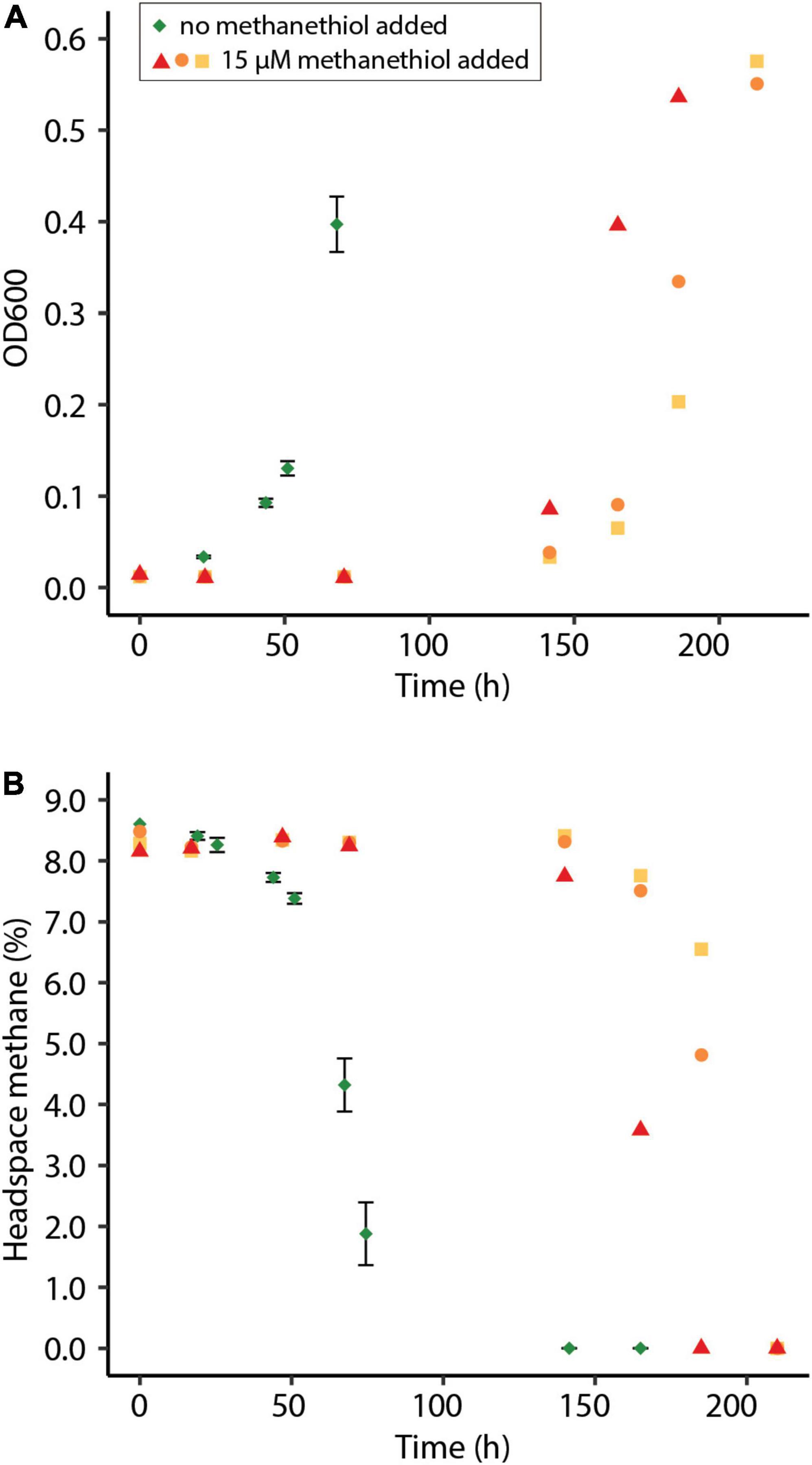
Figure 3. (A) Optical density measured at 600 nm (OD600) of methane-oxidizing Methylacidiphilum fumariolicum SolV cells in serum bottles and (B) the percentage methane in the headspace of the bottles over time. Green diamonds indicate average of positive controls to which no methanethiol was supplemented. Error bars indicate standard deviation (n = 3). Red triangles, orange dots, and yellow squares indicate incubations to which 1200 nmol methanethiol was added at the start of the experiment, to create approximately 15 μM methanethiol in the liquid. Methanethiol was depleted after 71 h. Experiments were performed in triplicate.
The putative methanethiol oxidase encoded by M. fumariolicum SolV is predicted to be a cytoplasmic protein by SignalP 5.0. Nevertheless, proteins could be predicted to be cytoplasmic based on amino acid sequence and still be associated with the membrane (Schmitz et al., 2020). To determine the cellular location of methanethiol consumption, the soluble proteins were separated from the membrane proteins using ultracentrifugation. Clearly, the entire capacity to degrade methanethiol is found in the soluble fraction, suggesting that the putative methanethiol oxidase could be responsible for the observed methanethiol degradation (Figure 4A). Interestingly, during methanethiol consumption by the crude extract the stoichiometry of methanethiol to H2S was never 1:1, caused by simultaneous production and consumption of H2S (Figure 4B). H2S oxidation was enhanced when methanethiol becomes depleted, suggesting substrate competition (Figure 4C). When H2S was added directly instead, a liquid concentration of 3 μM H2S is completely consumed.
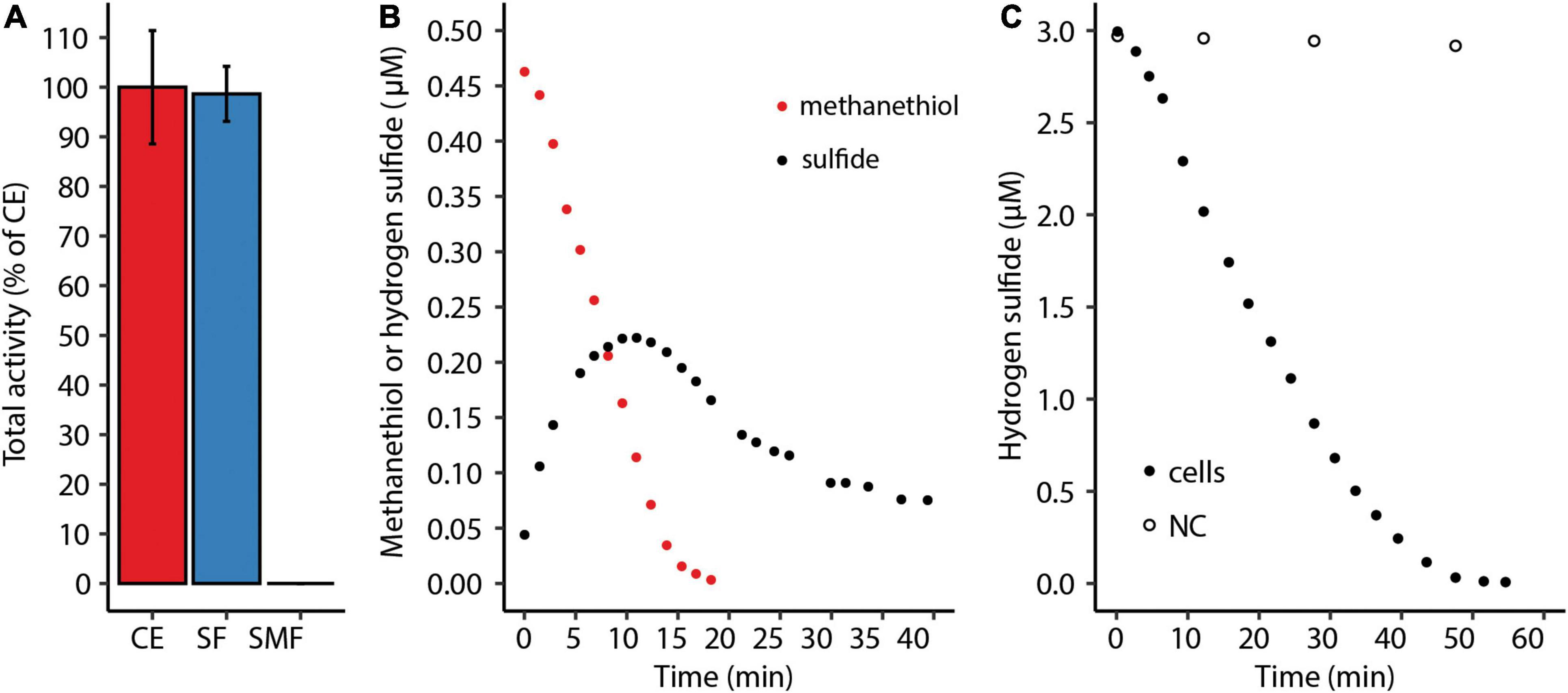
Figure 4. (A) Total methanethiol-degrading activity of the soluble fraction (SF) and the solubilized membrane fraction (SMF) as percentage of the total activity of the crude extract (CE). Error bars indicate standard deviations (n = 2). (B) Methanethiol consumption and H2S production by crude extract of Methylacidiphilum fumariolicum SolV. (C) Consumption of 3 μM H2S by M. fumariolicum SolV cells in a serum bottle over time. NC, negative control with medium only.
After the discovery of the gene that encodes MTO in Hyphomicrobium sp. VS (annotated as selenium-binding protein 56) it became apparent that this gene is found in a wide range of environments (Eyice et al., 2018). Interestingly, specific mutations in the gene encoding MTO in humans cause extra-oral halitosis since methanethiol cannot be degraded (Pol et al., 2018). The putative MTO of M. fumariolicum SolV is 28% identical (43% positives; 1e-29) in amino acid sequence to MTO of Hyphomicrobium sp. VS (KY242492.1) and 36% identical (53% positives; 2e-89) to MTO of Homo sapiens. mtoX is present in all known verrucomicrobial methanotrophs, suggesting the capacity of methanethiol consumption as was shown for M. fumariolicum SolV in this study. Interestingly, we detected putative MTO in a range of methanotrophs. Strains that possess pMMO and/or sMMO in combination with a putative MTO were found in the alphaproteobacterial family Beijerinckiaceae and genera Methylobacterium, Methylocapsa, and Methylocystis, and in the gammaproteobacterial genera Crenothrix, Methylobacter, Methylocaldum, Methylococcus, Methylicorpusculum, Methyloglobus, Methylohalobius, Methylomagnum, Methylomarinum, Methylomicrobium, Methylomonas, Methyloprofundus, Methylosarcina, Methylospira, Methyloterricola, Methylotetracoccus, and Methylotuvimicrobium (Figure 5). In addition, putative MTO was found in the genus Candidatus Methylomirabilis sp., known for anaerobic methane oxidation through an intra-aerobic pathway (Ettwig et al., 2010; Versantvoort et al., 2018). Interestingly, all MTO homologs are predicted be periplasmic proteins, except for those found in verrucomicrobial methanotrophs and Methylospira mobilis, Methylocaldum szegediense, and Methylotuvimicrobium alcaliphilum.
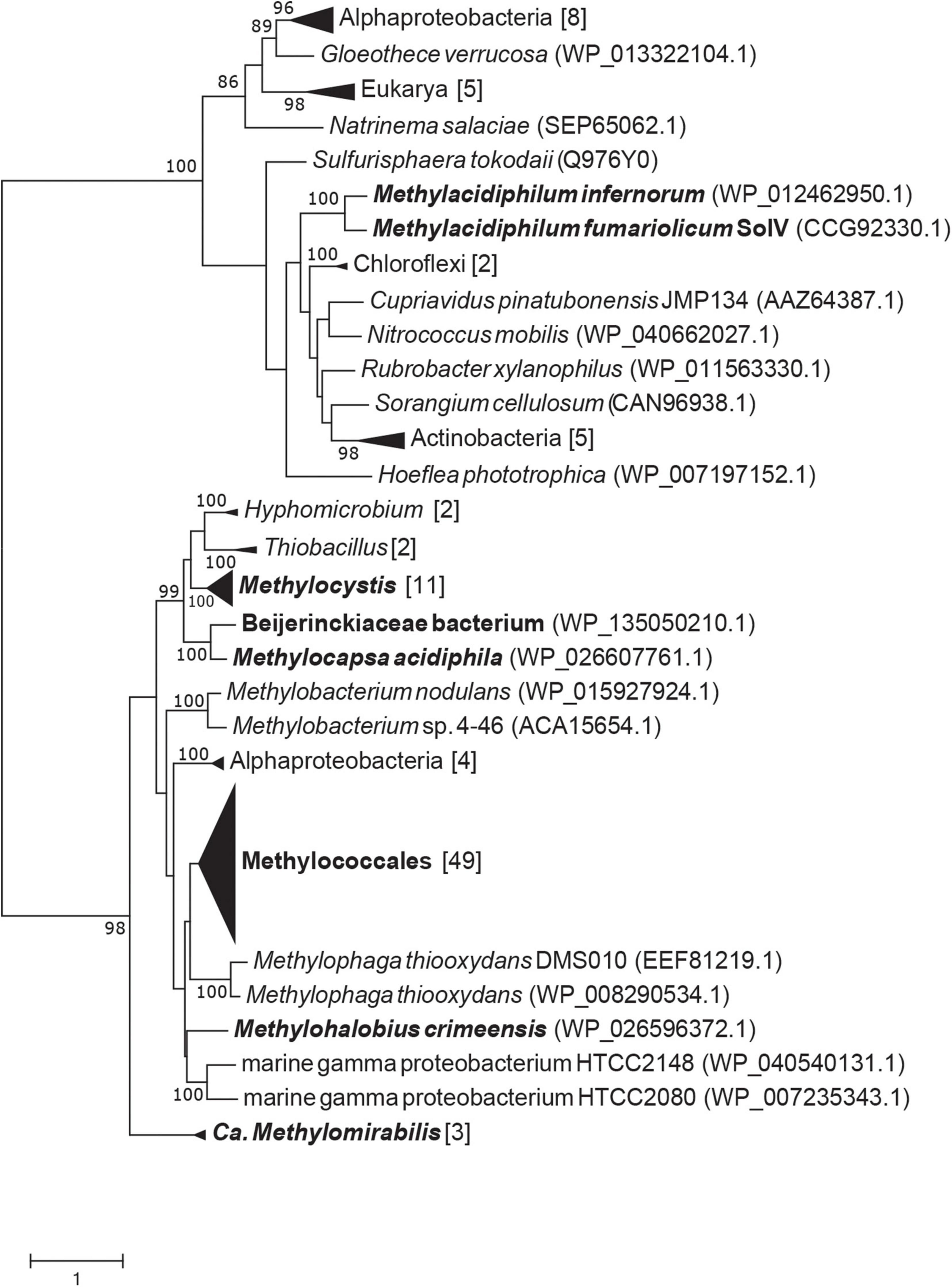
Figure 5. RAxML maximum likelihood tree of putative methanethiol oxidase amino acid sequences from methanotrophs. Bootstrap values above 80 are shown, based on 500 replicates. Black names indicate methanotrophic clades or strains.
In this study we have shown that the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV consumes methanethiol, which leads to the production and subsequent consumption of H2S. All known verrucomicrobial methanotrophs encode for a putative cytoplasmic methanethiol oxidase, which could be responsible for the observed methanethiol consumption. Verrucomicrobial methanotrophs may utilize the methanethiol oxidase to detoxify methanethiol, since methanotrophy and growth on methane are inhibited by this organic sulfur compound. The methanethiol consumption rate is about 0.33% of methane consumption rates reported before (Mohammadi et al., 2017).
Methanethiol is an important volatile organic sulfur compound in the global sulfur cycle (Lomans et al., 2002). Recently, methanethiol was shown to be produced through thermogenic processes in seafloor hydrothermal systems (Reeves et al., 2014). Furthermore, it was shown that methanethiol can be formed abiotically from H2S, CO2, and H2 (Heinen and Lauwers, 1995). In view of this, the occurrence of methanethiol production in acidic geothermal environments in which verrucomicrobial methanotrophs thrive is very likely. The observation that M. fumariolicum SolV produces H2S from the degradation of methanethiol is in line with studies of MTO in Hyphomicrobium sp. EG and sp. VS and Thiobacillus thioparus (Suylen et al., 1987; Gould and Kanagawa, 1992; Eyice et al., 2018) and humans (Pol et al., 2018). Also in these organisms produced H2S is simultaneously oxidized. Alternatively, recently microorganisms were found to methylate methanethiol and produce dimethylsulfide aerobically and anaerobically (Lomans et al., 2001; Carrión et al., 2017). However, mddA encoding a methyltransferase catalyzing this reaction is absent in verrucomicrobial methanotrophs. Interestingly, mtoX homologs encoding methanethiol oxidase are found in proteobacterial methanotrophs of various genera. In addition, a study showed that the facultative methanotroph Sphingopyxis sp. MD2, isolated from a landfill in South Korea, is able to degrade methanethiol through an uninvestigated mechanism (Lee et al., 2012). However, molecular evidence of culture purity is lacking and the pmoA gene sequence is 98% identical to that of Methylocystis sp. 39 (AJ459045). In addition, several other methanotrophic strains were shown to be inhibited by methanethiol rather than be stimulated by it (Börjesson, 2001; Lee et al., 2011; Lee et al., 2015). Considering the toxicity of methanethiol, possessing an MTO could be beneficial in environments where methane and methanethiol are known to co-occur, such as aquatic sediments and landfills (Lomans et al., 1997; Kim et al., 2005; Mayr et al., 2020).
Methanethiol clearly inhibits methane oxidation and growth of M. fumariolicum SolV, but the underlying mechanism is unclear. In addition, cells of strain SolV are unable to completely consume methanethiol at concentrations above 3 μM, pointing to inhibition by toxic degradation product as described before (Suylen et al., 1987). Sulfide is a toxic compound that is known to inhibit the respiratory chain and several enzymes by binding to the active site (Bagarinao, 1992; Landry et al., 2021). In microorganisms, methanethiol could have a similar mode of inhibition. Alternatively, if MTO is the enzyme dedicated to the degradation of methanethiol in verrucomicrobial methanotrophs, one or more products of this catalysis, formaldehyde (CH2O), sulfide, and hydrogen peroxide (H2O2), could account for the observed inhibition. The product formaldehyde is a central intermediate in carbon assimilation in the majority of methylotrophs (Suylen et al., 1987; Chistoserdova, 2011). Through the serine cycle and the RuMP cycle in Alpha- and Gammaproteobacteria, formaldehyde is fixed whereas verrucomicrobial methanotrophs oxidize formaldehyde to CO2 through an unresolved pathway. MTO was shown to oxidize H2S, but verrucomicrobial methanotrophs also possess a sulfide:quinone oxidoreductase (SQR) that could be dedicated to this catalysis (Suylen et al., 1987; Schmitz et al., 2021). In addition, exogenous H2S and H2S produced by MTO in the cytoplasm could be used for sulfur assimilation. Finally, catalases detoxify hydrogen peroxide to water and oxygen, but these enzymes are not found in all verrucomicrobial methanotrophs, although enzymes with similar functions could be used, such as peroxidases (Schäfer et al., 2010; Schmitz et al., 2021).
The finding that cell pre-incubated with methane for 2 h can oxidize much higher concentrations of methanethiol is interesting. When methanethiol is supplemented to these energized cells, methane oxidation ceases while methanethiol is degraded. This observation may indicate that also pMMO is involved in the degradation of methanethiol. Partitioning of pMMO in methanethiol degradation cannot be observed through cell fractionation, as this procedure disrupts pMMO activity. pMMO is a monooxygenase and therefore needs reducing equivalents to catalyze a reaction. Accordingly, the cells pre-incubated with methane have synthesized a relatively high concentration of reducing equivalents that could subsequently be used to reduce pMMO for the degradation of methanethiol. Indeed, Börjesson (2001) showed that methane oxidation in landfill soils is inhibited by methanethiol and that methane and methanethiol seem to compete for the same enzymes, which could be pMMO. Whether pMMO is indeed involved in methanethiol degradation in methanotrophs remains to be investigated and could be resolved by using a pMMO-specific inhibitor.
In conclusion, we show that M. fumariolicum SolV is able to consume methanethiol and concurrently produce and consume hydrogen sulfide. H2S is known to be emitted from terrestrial volcanic ecosystems such as mud pools and oxidation of this compound can lead to severe acidification of the environment (Spiro et al., 1992; Lomans et al., 2002). Emissions of methanethiol from the habitat of verrucomicrobial methanotrophs are unknown, but it may be produced both chemically and by microorganisms. Methanethiol has an inhibitory effect on methane oxidation, which is presumably alleviated through MTO in M. fumariolicum SolV. Since putative MTOs are found in a range of methanotrophs, we propose these enzymes to be a widespread mechanism for methanethiol degradation in methanotrophs. Future studies are needed to observe whether methanethiol has an inhibitory effect on methanotrophs in general and whether cells can be adapted to conserve energy from methanethiol.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
RS, SM, TE, AP, and HO designed the project and experiments. RS, SM, TE, and AP performed the experimental work. RS, SM, and AP maintained the chemostat cultures. TB performed the phylogenetic analysis. RS, SM, AP, and HO performed data analysis and data interpretation. RS, SM, and HO wrote the manuscript with feedback from TE, TB, MJ, and AP. HO, AP, and MJ supervised the research. All authors contributed to the article and approved the submitted version.
RS, TB, and HO were supported by the European Research Council (ERC Advanced Grant project VOLCANO 669371). SM was supported by the Spinoza grant of MJ (Netherlands Organization for Scientific Research) and the European Research Council (ERC Advanced Grant project VOLCANO 669371). MJ was supported by the European Research Council (ERC Advanced Grant Eco_MoM 339880 and ERC Synergy Grant Marix 854088).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.857442/full#supplementary-material
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Awala, S. I., Gwak, J. H., Kim, Y. M., Kim, S. J., Strazzulli, A., Dunfield, P. F., et al. (2021). Verrucomicrobial methanotrophs grow on diverse C3 compounds and use a homolog of particulate methane monooxygenase to oxidize acetone. ISME J. 15, 3636–3647. doi: 10.1038/s41396-021-01037-2
Bagarinao, T. (1992). Sulfide as an environmental factor and toxicant: tolerance and adaptations in aquatic organisms. Aquat. Toxicol. 24, 21–62. doi: 10.1016/0166-445X(92)90015-F
Baumler, D. J., Hung, K.-F., Jeong, K. C., and Kasper, C. W. (2007). Production of methanethiol and volatile sulfur compounds by the archaeon “Ferroplasma acidarmanus”. Extremophiles 11, 841–851. doi: 10.1007/s00792-007-0108-8
Börjesson, G. (2001). Inhibition of methane oxidation by volatile sulfur compounds (CH3SH and CS2) in landfill cover soils. Waste Manage. Res. 19, 314–319. doi: 10.1177/0734242X0101900
Bridgham, S. D., Cadillo-Quiroz, H., Keller, J. K., and Zhuang, Q. (2013). Methane emissions from wetlands: biogeochemical, microbial, and modelling perspectives from local to global scales. Glob. Chang. Biol. 19, 1325–1346. doi: 10.1111/gcb.12131
Brune, A. (2010). “Methanogens in the digestive tract of termites,” in (Endo)symbiotic Methanogenic Archaea, ed. J. Hackstein (Heidelberg: Springer), 81–100. doi: 10.1007/978-3-319-98836-8_6
Carrión, O., Pratscher, J., Curson, A. R. J., Williams, B. T., Rostant, W. G., Murrell, J. C., et al. (2017). Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 11, 2379–2390. doi: 10.1038/ismej.2017.105
Chistoserdova, L. (2011). Modularity of methylotrophy, revisited. Environ. Microbiol. 13, 2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x
Chistoserdova, L., and Kalyuzhnaya, M. G. (2018). Current trends in methylotrophy. Trends Microbiol. 26, 703–714. doi: 10.1016/j.tim.2018.01.011
Dean, J. F., Middelburg, J. J., Röckmann, T., Aerts, R., Blauw, L. G., Egger, M., et al. (2018). Methane feedbacks to the global climate system in a warmer world. Rev. Geophys. 56, 207–250. doi: 10.1002/2017RG000559
Dunfield, P. F., Yuryev, A., Senin, P., Smirnova, A. V., Stott, M. B., Hou, S., et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–882. doi: 10.1038/nature06411
Erikstad, H.-A., Ceballos, R. M., Smestad, N. B., and Birkeland, N.-K. (2019). Global biogeographic distribution patterns of thermoacidophilic Verrucomicrobia methanotrophs suggest allopatric evolution. Front. Microbiol. 10:1129. doi: 10.3389/fmicb.2019.01129
Etiope, G. (2009). Natural emissions of methane from geological seepage in Europe. Atmos. Environ. 43, 1430–1443. doi: 10.1016/j.atmosenv.2008.03.014
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548. doi: 10.1038/nature08883
Eyice, Ö., Myronova, N., Pol, A., Carrión, O., Todd, J. D., Smith, T. J., et al. (2018). Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 12, 145–160. doi: 10.1038/ismej.2017.148
Finster, K., Tanimoto, Y., and Bak, F. (1992). Fermentation of methanethiol and dimethylsulfide by a newly isolated methanogenic bacterium. Arch. Microbiol. 157, 425–430. doi: 10.1007/BF00249099
Gould, W. D., and Kanagawa, T. (1992). Purification and properties of methyl mercaptan oxidase from Thiobacillus thioparus TK-m. J. Gen. Microbiol. 138, 217–221. doi: 10.1099/00221287-138-1-217
Hanson, R. S., and Hanson, T. E. (1996). Methanotrophic bacteria. Microbiol. Rev. 60, 439–471. doi: 10.1128/mr.60.2.439-471.1996
Haroon, M. F., Hu, S., Shi, Y., Imelfort, M., Keller, J., Hugenholtz, P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570. doi: 10.1038/nature12375
Heede, R. (2014). Tracing anthropogenic carbon dioxide and methane emissions to fossil fuel and cement producers, 1854–2014. Clim. Change 122, 229–241. doi: 10.1007/s10584-013-0986-
Heinen, W., and Lauwers, A. M. (1995). Organic sulfur compounds resulting from the interaction of iron sulfide, hydrogen sulfide and carbon dioxide in an anaerobic aqueous environment. Orig. Life Evol. Biosph. 26, 131–150. doi: 10.1007/BF01809852
Islam, T., Jensen, S., Reigstad, L. J., Larsen, O., and Birkeland, N.-K. (2008). Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A. 105, 300–304. doi: 10.1073/pnas.0704162105
Kiene, R. P. (1996). Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 54, 69–83. doi: 10.1016/0304-4203(96)00006-0
Kim, K.-H., Choi, Y., Jeon, E., and Sunwoo, Y. (2005). Characterization of malodorous sulfur compounds in landfill gas. Atmos. Environ. 39, 1103–1112. doi: 10.1016/j.atmosenv.2004.09.083
Kiragosyan, K., Picard, M., Timmers, P. H. A., Sorokin, D. Y., Klok, J. B. M., Roman, P., et al. (2020). Effect of methanethiol on process performance, selectivity and diversity of sulfur-oxidizing bacteria in a dual bioreactor gas biodesulfurization system. J. Hazard. Mater. 398:123002. doi: 10.1016/j.jhazmat.2020.123002
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Landry, A. P., Ballou, D. P., and Banerjee, R. (2021). Hydrogen sulfide oxidation by sulfide quinone oxidoreductase. ChemBioChem 22, 949–960. doi: 10.1002/cbic.202000661
Lee, E.-H., Moon, K.-E., Kim, T. G., Lee, S.-D., and Cho, K.-S. (2015). Inhibitory effects of sulfur compounds on methane oxidation by a methane-oxidizing consortium. J. Biosci. Bioeng. 120, 670–676. doi: 10.1016/j.jbiosc.2015.04.006
Lee, E.-H., Yi, T., Moon, K.-E., Park, H., Ryu, H. W., and Cho, K.-S. (2011). Characterization of methane oxidation by a methanotroph isolated from a landfill cover soil, South Korea. J. Microbiol. Biotechnol. 21, 753–756. doi: 10.4014/jmb.1102.01055
Lee, J.-H., Kim, T. G., and Cho, K.-S. (2012). Isolation and characterization of a facultative methanotroph degrading malodor-causing volatile sulfur compounds. J. Hazard. Mater. 235–236, 224–229. doi: 10.1016/j.jhazmat.2012.07.047
Lomans, B. P., Leijdekkers, P., Wesselink, J. J., Bakkes, P., Pol, A., van der Drift, C., et al. (2001). Obligate sulfide-dependent degradation of methoxylated aromatic compounds and formation of methanethiol and dimethyl sulfide by a freshwater sediment isolate, Parasporobacterium paucivorans gen. nov., sp. nov. Appl. Environ. Microbiol. 67, 4017–4023. doi: 10.1128/AEM.67.9.4017-4023.2001
Lomans, B. P., Maas, R., Luderer, R., Op den Camp, H. J. M., Pol, A., van der Drift, C., et al. (1999). Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl. Environ. Microbiol. 65, 3641–3650. doi: 10.1128/AEM.65.8.3641-3650.1999
Lomans, B. P., Smolders, A. J. P., Intven, L. M., Pol, A., Op den Camp, H. J. M., and van der Drift, C. (1997). Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl. Environ. Microbiol. 63, 4741–4747. doi: 10.1128/aem.63.12.4741-4747.1997
Lomans, B. P., van der Drift, C., Pol, A., and Op den Camp, H. J. M. (2002). Microbial cycling of volatile organic sulfur compounds. Cell. Mol. Life Sci. 59, 575–588. doi: 10.1007/s00018-002-84
Lyimo, T. J., Pol, A., Harhangi, H. R., Jetten, M. S. M., and Op den Camp, H. J. M. (2009). Anaerobic oxidation of dimethylsulfide and methanethiol in mangrove sediments is dominated by sulfate-reducing bacteria. FEMS Microbiol. Ecol. 70, 483–492. doi: 10.1111/j.1574-6941.2009.00765.x
Lyimo, T. J., Pol, A., Op den Camp, H. J. M., Harhangi, H. R., and Vogels, G. D. (2000). Methanosarcina semesiae sp. nov., a dimethylsulfide-utilizing methanogen from mangrove sediment. Int. J. Syst. Evol. Microbiol. 50, 171–178. doi: 10.1099/00207713-50-1-171
Maddry, J. K., Paredes, R. M., Rebeles, J., Olson, G., Castaneda, M., Canellis, K., et al. (2020). Efficacy of intravenous hydroxocobalamin for treatment of sodium methanethiolate exposure in a swine model (Sus scrofa) of severe methanethiol toxicity. J. Med. Toxicol. 16, 388–397. doi: 10.1007/s13181-020-00767-7
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mayr, M. J., Zimmermann, M., Dey, J., Brand, A., Wehrli, B., and Bürgmann, H. (2020). Growth and rapid succession of methanotrophs effectively limit methane release during lake overturn. Commun. Biol. 3:108. doi: 10.1038/s42003-020-0838-z
Mohammadi, S., Pol, A., van Alen, T. A., Jetten, M. S. M., and Op den Camp, H. J. M. (2017). Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 11, 945–958. doi: 10.1038/ismej.2016.171
Mohammadi, S. S., Schmitz, R. A., Pol, A., Berben, T., Jetten, M. S. M., and Op den Camp, H. J. M. (2019). The acidophilic methanotroph Methylacidimicrobium tartarophylax 4AC grows as autotroph on H2 under microoxic conditions. Front. Microbiol. 10:2352. doi: 10.3389/fmicb.2019.02352
Murrell, J. C., and Jetten, M. S. M. (2009). The microbial methane cycle. Environ. Microbiol. Rep. 1, 279–284. doi: 10.1111/j.1758-2229.2009.00089.x
Op den Camp, H. J. M., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Picone, N., Mohammadi, S. S., Waajen, A. C., van Alen, T. A., Jetten, M. S. M., Pol, A., et al. (2020). More than a methanotroph: a broader substrate spectrum for Methylacidiphilum fumariolicum SolV. Front. Microbiol. 11:604485. doi: 10.3389/fmicb.2020.604485
Pol, A., Heijmans, K., Harhangi, H. R., Tedesco, D., Jetten, M. S. M., and Op den Camp, H. J. M. (2007a). Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450, 874–878. doi: 10.1038/nature06222
Pol, A., Op den Camp, H. J. M., and van der Drift, C. (2007b). Isolation of a carbon disulfide utilizing Thiomonas sp. and its application in a biotrickling filter. Appl. Microbiol. Biotechnol. 74, 439–446. doi: 10.1007/s00253-006-0663-4
Pol, A., Op den Camp, H. J. M., Mees, S. G. M., Kersten, M. A., and van der Drift, C. (1994). Isolation of a dimethyl sulfide utilizing Hyphomicrobium species and its application in biofiltration of polluted air. Biodegradation 5, 105–112. doi: 10.1007/BF00700635
Pol, A., Renkema, G. H., Tangerman, A., Winkel, E. G., Engelke, U. F., de Brouwer, A. P. M., et al. (2018). Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 50, 120–129. doi: 10.1038/s41588-017-0006-7
Reeves, E. P., McDermott, J. M., and Seewald, J. S. (2014). The origin of methanethiol in midocean ridge hydrothermal fluids. Proc. Natl. Acad. Sci. U.S.A. 111, 5474–5479. doi: 10.1073/pnas.1400643111
Roman, P., Bijmans, M. F. M., and Janssen, A. J. H. (2016). Influence of methanethiol on biological sulphide oxidation in gas treatment system. Environ. Technol. 37, 1693–1703. doi: 10.1080/09593330.2015.1128001
Ross, M. O., and Rosenzweig, A. C. (2017). A tale of two methane monooxygenases. J. Biol. Inorg. Chem. 22, 307–319. doi: 10.1007/s00775-016-1419-y
Schäfer, H., and Eyice, Ö. (2019). Microbial cycling of methanethiol. Curr. Issues Mol. Biol. 33, 173–182. doi: 10.21775/cimb.033.173
Schäfer, H., Myronova, N., and Boden, R. (2010). Microbial degradation of dimethylsulfide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J. Exp. Bot. 61, 315–334. doi: 10.1093/jxb/erp355
Schmitz, R. A., Peeters, S. H., Versantvoort, W., Picone, N., Pol, A., Jetten, M. S. M., et al. (2021). Verrucomicrobial methanotrophs: ecophysiology of metabolically versatile acidophiles. FEMS Microbiol. Rev. 45:fuab007. doi: 10.1093/femsre/fuab007
Schmitz, R. A., Pol, A., Mohammadi, S. S., Hogendoorn, C., van Gelder, A. H., Jetten, M. S. M., et al. (2020). The thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV oxidizes subatmospheric H2 with a high-affinity, membrane-associated [NiFe] hydrogenase. ISME J. 14, 1223–1232. doi: 10.1038/s41396-020-0609-3
Spiro, P. A., Jacob, D. J., and Logan, J. A. (1992). Global inventory of sulfur emissions with 1°x1° resolution. J. Geophys. Res. Atmos. 97, 6023–6036. doi: 10.1029/91JD03139
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Suylen, G. M. H., Large, P. J., van Dijken, J. P., and Kuenen, J. G. (1987). Methyl mercaptan oxidase, a key enzyme in the metabolism of methylated sulfur-compounds by Hyphomicrobium EG. J. Gen. Microbiol. 133, 2989–2997. doi: 10.1099/00221287-133-11-2989
van den Bosch, P. L. F., de Graaff, M., Fortuny-Picornell, M., van Leerdam, R. C., and Janssen, A. J. H. (2009). Inhibition of microbiological sulfide oxidation by methanethiol and dimethyl polysulfides at natronalkaline conditions. Appl. Microbiol. Biotechnol. 83, 579–587. doi: 10.1007/s00253-009-1951-6
van Teeseling, M. C., Pol, A., Harhangi, H. R., van der Zwart, S., Jetten, M. S. M., Op den Camp, H. J. M., et al. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 80, 6782–6791. doi: 10.1128/AEM.01838-14
Keywords: Methylacidiphilum, methanotrophs, methanethiol, hydrogen sulfide, thermoacidophile, sulfur cycle
Citation: Schmitz RA, Mohammadi SS, van Erven T, Berben T, Jetten MSM, Pol A and Op den Camp HJM (2022) Methanethiol Consumption and Hydrogen Sulfide Production by the Thermoacidophilic Methanotroph Methylacidiphilum fumariolicum SolV. Front. Microbiol. 13:857442. doi: 10.3389/fmicb.2022.857442
Received: 18 January 2022; Accepted: 28 February 2022;
Published: 28 March 2022.
Edited by:
Peng Xing, Nanjing Institute of Geography and Limnology (CAS), ChinaReviewed by:
Karen Houghton, GNS Science, New ZealandCopyright © 2022 Schmitz, Mohammadi, van Erven, Berben, Jetten, Pol and Op den Camp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huub J. M. Op den Camp, aC5vcGRlbmNhbXBAc2NpZW5jZS5ydS5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.