
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 March 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.856106
This article is part of the Research Topic Pathogen co-Infections and Plant Diseases View all 5 articles
 Junjie Xu1
Junjie Xu1 Yilin Li2
Yilin Li2 Jinbu Jia1†
Jinbu Jia1† Wenjing Xiong2
Wenjing Xiong2 Chengcheng Zhong1†
Chengcheng Zhong1† Guiyan Huang3†
Guiyan Huang3† Xiuhong Gou1
Xiuhong Gou1 Yuling Meng2
Yuling Meng2 Weixing Shan2*
Weixing Shan2*Oomycetes represent a unique group of plant pathogens that are destructive to a wide range of crops and natural ecosystems. Phytophthora species possess active small RNA (sRNA) silencing pathways, but little is known about the biological roles of sRNAs and associated factors in pathogenicity. Here we show that an AGO gene, PpAGO3, plays a major role in the regulation of effector genes hence the pathogenicity of Phytophthora parasitica. PpAGO3 was unique among five predicted AGO genes in P. parasitica, showing strong mycelium stage-specific expression. Using the CRISPR-Cas9 technology, we generated PpAGO3ΔRGG1-3 mutants that carried a deletion of 1, 2, or 3 copies of the N-terminal RGG motif (QRGGYD) but failed to obtain complete knockout mutants, which suggests its vital role in P. parasitica. These mutants showed increased pathogenicity on both Nicotiana benthamiana and Arabidopsis thaliana plants. Transcriptome and sRNA sequencing of PpAGO3ΔRGG1 and PpAGO3ΔRGG3 showed that these mutants were differentially accumulated with 25–26 nt sRNAs associated with 70 predicted cytoplasmic effector genes compared to the wild-type, of which 13 exhibited inverse correlation between gene expression and 25–26 nt sRNA accumulation. Transient overexpression of the upregulated RXLR effector genes, PPTG_01869 and PPTG_15425 identified in the mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3, strongly enhanced N. benthamiana susceptibility to P. parasitica. Our results suggest that PpAGO3 functions together with 25–26 nt sRNAs to confer dynamic expression regulation of effector genes in P. parasitica, thereby contributing to infection and pathogenicity of the pathogen.
Oomycetes represent a unique group of diploid microorganisms, which resemble but are evolutionarily distant from filamentous fungi (Judelson, 1997). Species in the genus of Phytophthora include many economically important plant pathogens, notably Phytophthora infestans, P. sojae, P. parasitica, and P. ramorum (Tyler, 2007; Fry, 2008; Grünwald et al., 2008).
During infection, Phytophthora secrets a large number of effectors into host plants, including RXLR (Arg–any amino acid-Leu-Arg) effectors and CRN (Crinkling and Necrosis) effectors, to manipulate host physiology and support colonization (Zheng et al., 2014; Fan et al., 2018; Wang and Wang, 2018; Chen et al., 2019). In response, the host plants produce R proteins to recognize the secreted effectors, which are known as avirulence proteins, triggering plant immunity. Nearly all host genotype-specific avirulence factors identified in Phytophthora are RXLR effectors (Armstrong et al., 2005; van Poppel et al., 2008; Bozkurt et al., 2011). At the same time, pathogens have evolved ways to evade the perception of effectors by the host R proteins through sequence variation and deletion of avirulence effectors. Apart from that, polymorphisms in the expression levels of effector genes are common in Phytophthora and contribute to pathogen plasticity in overcoming host genotype-specific resistance (Shan et al., 2004; Dong et al., 2009; Qutob et al., 2009; Wang et al., 2011a, 2020; Cui et al., 2012; Gijzen et al., 2014; Pais et al., 2018). Increasing evidence suggests that 25–26 nt small RNA (sRNA), the dominant size class of sRNAs in Phytophthora, are involved in the regulation of effector gene expression (Vetukuri et al., 2012; Qutob et al., 2013; Jia et al., 2017; Wang et al., 2019). For example, in P. sojae, 25 nt sRNAs are associated with PsAvr3a gene silencing and the silencing status could be “inherited” to the next generation (Qutob et al., 2013). In addition, up to 125 RXLR effector genes are associated with homologous sRNAs, which correlate with the silencing of the corresponding RXLR effector genes at the mycelium stage (Wang et al., 2019). Similarly, 25–26 nt sRNAs are implicated in the silencing of 40% RXLR and 50% CRN effector genes in P. parasitica during vegetative mycelial growth (Jia et al., 2017). However, very little is known about the mechanism and function of sRNA-associated silencing of RXLR effector genes.
The sRNA pathways are conserved in eukaryotes and play important roles in various cellular processes. Depending on the biogenesis pathway and functions, sRNAs are classified into microRNA (miRNA), small interfering RNA (siRNA), and PIWI-interacting RNAs (piRNAs; Girard et al., 2006; Carthew and Sontheimer, 2009; Ye et al., 2016). miRNAs are processed by Dicer or Dicer-like (DCL) proteins from self-folding RNA transcripts and have been widely found in plants and animals (Millar and Waterhouse, 2005), but very few miRNA-like genes exist in fungi and oomycetes (Lee et al., 2010; Fahlgren et al., 2013). piRNAs are only found in animals (Vagin et al., 2006; Brennecke et al., 2007; Czech et al., 2018), whereas siRNAs are present in most eukaryotes, and processed by Dicer or DCL from long double-stranded RNAs (dsRNAs) or hairpin structure (Katiyar-Agarwal et al., 2006; Huang et al., 2019b). To direct gene silencing, sRNAs are loaded to Argonaute (AGO) protein to form an RNA-induced silencing complex (RISC), which then uses the sRNA as guide to direct mRNA cleavage or DNA methylation/histone modification (Kobayashi and Tomari, 2016).
AGO proteins are present in both prokaryotes and eukaryotes (Bohmert et al., 1998; Makarova et al., 2009; Xue et al., 2012; Cervantes et al., 2013; Swarts et al., 2014; Nguyen et al., 2018; Yin et al., 2020), but the number and type of AGO proteins are diverse in different organisms (Tolia and Joshua-Tor, 2007; Hutvagner and Simard, 2008). There are primarily four AGO homologs, including AGO-like, PIWI, Worm-specific AGO (WAGO), and Trypanosoma AGO families (Durand-Dubief et al., 2003; Yigit et al., 2006; Hutvagner and Simard, 2008; Garcia–Silva et al., 2010; Bollmann et al., 2018). Members of the AGO-like family contain primarily the PAZ (PIWI–ARGONAUTE–ZWILLE) and PiWi domain, which contribute to the binding of sRNA 3′ end and the cleavage of complementary mRNA, respectively (Lingel et al., 2003; Song et al., 2004). The AGO-like family is primarily involved in miRNA and siRNA-directed silencing and plays a vital role in transcriptional and translational regulation, heterochromatin assembly, and alternative splicing (Baulcombe, 2004; Buker et al., 2007; Azzam et al., 2012; Wei et al., 2012; Marasovic et al., 2013; Huang and Li, 2014). The TbAGO1 in Trypanosoma brucei is required for RNAi and plays a role in mitosis and chromosome segregation (Durand-Dubief and Bastin, 2003; Garcia-Silva et al., 2010, 2014). Interestingly, besides the well-known PAZ and Piwi domains, TbAGO1 also contains an N-terminal RGG (arginine–glycine–glycine) repeat motif, which is required for RNA silencing (Shi et al., 2004). Loss of the N-terminal RGG domain can strongly block the association of TbAGO1 with polyribosomes and affect mRNA cleavage (Shi et al., 2004, 2009). The RGG domain of TgAGO in Toxoplasma gondii is also functional in the RNA silencing pathway (Musiyenko et al., 2012).
Several AGO proteins have been identified in Phytophthora and are implicated in sRNA-mediated regulation of effector genes. For example, in P. infestans, silencing of PiAgo4 or PiAgo5 caused reduced accumulation of 32 nt sRNAs homologous to PiAvrblb1, while silencing of PiAgo1 resulted in increased accumulation of 32 nt sRNAs to PiAvrblb1. Interestingly, the PiAvrblb1-derived 32 nt sRNAs were not affected in PiDCL1 silencing strain (Vetukuri et al., 2011, 2012). Furthermore, co-immunoprecipitation assays showed that CRN effector genes and pseudo-CRN-derived 18–30 nt sRNA were significantly enriched to PiAGO1 and PiAGO5 proteins (Åsman et al., 2016). However, little is known on the specific roles of AGOs in Phytophthora biology and pathology.
In this study, we investigated the function of PpAGO3 in the model oomycete organism P. parasitica (Meng et al., 2014), where the efficient, complete silencing of many effector genes is associated with accumulation of their homologous 25–26 nt sRNAs during the mycelium stage (Jia et al., 2017). We created mutations in the N-terminal RGG domain repeat region of PpAGO3 by using the CRISPR-Cas9 technology. PpAGO3 mutants (PpAGO3ΔRGG1-3), with the deletion of 1–3 copies of RGG domain (QRGGYD), showed enhanced pathogenicity and examined for sRNA accumulation and gene expression. In addition, two PpAGO3/sRNA-regulated RXLR effector genes were further analyzed for their virulence function. Our results provide compelling evidence that PpAGO3 and 25–26 nt sRNAs function together to regulate effector gene expression.
Arabidopsis thaliana and Nicotiana benthamiana seeds were sown in a matrix containing soil and vermiculite, and cultured in a phytotron (23°C) with a photoperiod of 14 h light per day for about 4–5 weeks. Phytophthora parasitica strain PpBS042 was isolated from diseased tobacco plant collected from Chongqing, China (Zhang et al., 2020). It was routinely cultured on 5% CA (carrot juice agar) medium with 0.01% CaCO3 and 0.002% β-sitosterol, in darkness for 3–4 days (23°C). To induce the sporangia production, the culture medium with the fresh mycelia were transformed into 5% CA broth with Petri solution for another 5 days as described before (Huang et al., 2019a). For the zoospore release, it was done as described previously (Wang et al., 2011b).
The five AGO protein sequences of P. infestans were BLASTP aligned with the P. parasitica INRA-310 (taxid:761204) database on NCBI, and the five predicted AGO proteins sequences of P. parasitica were BLASTP aligned to P. infestans T30-4 (taxid:403677) database to ensure the consistency of the sequence alignment. The sequence ID of five AGO proteins in P. infestans were PiAGO1 (XP_002906080.1), PiAGO2 (XP_002906081.1), PiAGO3 (XP_002908068.1), PiAGO4 (XP_002908108.1), PiAGO5 (XP_002908109.1). The domain architecture was predicted by using Pfam database and displayed by software IBS (Liu et al., 2015). For gene expression pattern analysis, it was conducted by using the RNA-seq data obtained by Jia (2017), and the RNA-seq data analysis would be described later.
The PpAGO3 mutants were obtained through three main steps: CRISPR-Cas9 vector construction for gene PpAGO3; the plasmid transformation in P. parasitica; transformants selection and identification. Firstly, the CRISPR-Cas9 plasmid was constructed and modified using “all-in-one” vector pYF515 developed for P. sojae (Fang et al., 2017). The sgRNA for PpAGO3 was designed by using EuPaGDT1; then, off-target was analyzed by using FungiDB2; finally, RNA secondary structure was predicted online.3 For PpAGO3, one sgRNA (total score > 0.5) without off-target site and weak RNA secondary structure was chosen for primer design. Through annealing and extension, the synthesized primers were made to dsDNA fragment and were ligated to NheI/BsaI (NEB) digested vector PYF515 by using T4 DNA ligase (Promega). The recombination vector was then transformed into Escherichia coli DH5α cells. The sequence-verified plasmid was extracted and concentrated for transformation.
Secondly, CRISPR-Cas9 plasmid for PpAGO3 was transformed into P. parasitica strain PpBS042. The transformation was conducted by using PEG-CaCl2 mediated method as described (Bottin et al., 1999; Meng et al., 2015). The transformation protoplasts were recovered overnight and then were cultured on the 5% CA medium with 13.6 μg/ml G418, 200 μg/ml ampicillin, 20 μg/ml nystatin, and 20 μg/ml rifampicin. Finally, it was the selection and identification for PpAGO3 mutants. Through 3–7 days selective cultivation (23°C), regenerated mycelium colonies were isolated and transformed to a fresh plate with the same selective medium. Three days later, the G418-resistant transformants were cultivated in 5% CA broth for mycelium collection. Genomic DNA was extracted following the protocol as described previously (Zhang et al., 2020) from each candidate transformants and was examined for target sites by sequence amplification and sequencing.
Phytophthora parasitica was cultivated on 5% CA plates as described (Huang et al., 2019a; Zhang et al., 2020). Detached leaves of the 5-week-old N. benthamiana and 4-week-old A. thaliana Col-0 leaves were used for pathogenicity assays as described previously (Huang et al., 2019a; Zhang et al., 2020). On N. benthamiana leaves, the developed lesions were measured 60 h post-inoculation with P. parasitica mycelial discs, and the expansion of P. parasitica hyphae was visualized by trypan blue staining. For each assay, more than eight leaves were used. On A. thaliana, the disease severity index (DSI) was recorded 48 h after inoculation with P. parasitica mycelial discs, as described previously (Huang et al., 2019a). For each assay, more than 15 leaves were used. Statistical analysis was performed based on Student’s t-test between samples and based on a one-way ANOVA.
For library construction, P. parasitica strain PpBS042 was firstly cultured on 5% CA solid medium with 0.01% CaCO3 and 0.002% β-sitosterol, in darkness for 3–4 days (23°C). Then, the culture medium with the fresh mycelia were transformed into 5% CA broth with Petri solution for another 3 days. The total RNA was extracted from fresh mycelia of both the wild-type and PpAGO3ΔRGG mutants by using the RNA extract kit (Aidlab, RN40). The RNA concentration and quality were examined by using NanoDrop 2000 and gel electrophoresis. Construction and sequencing of small RNA and RNA libraries were performed by Biomarker Technologies (Beijing, China) with Illumina novaseq 6000 platform. For construction of sRNA library, only 18–45 nt size small RNAs were used for sequencing. The wild-type and the two PpAGO3ΔRGG mutants contain three biological replicates, respectively.
According to sRNA raw data, adaptor sequence (AGATCGGAAGAGCACACGTCTG) was first filtered. Then, clean reads were mapped to the P. parasitica genome (INRA-310 version 3.0, Assembly Dev initiative, Broad Institute) with Bowtie with no mismatches (-v 0 -a; Langmead et al., 2009; Jia et al., 2017). Reads mapped to rRNA, tRNA, mitochondrial DNA were also filtered by using bowtie (-v 0). The sRNAs that mapped to the gene locus (gene body plus 500 bp upstream and downstream region) were counted and regarded as the sRNA accumulation. Depending on the strand (sense/antisense) the sRNAs derived and the sRNA size, the detailed analysis was further divided. The sRNA counts were calculated by using self-write perl scripts. For sRNAs differentially expressed between the wild-type and PpAGO3ΔRGG mutants, the mapping counts were analyzed by using R package, DESeq2 (Love et al., 2014). For each gene, the corresponding sRNA expression level was calculated by using RPKM, reads Per Kilobase per Million.
According to the RNA-seq raw data, the adapter sequence and low-quality reads were filtered by using software Trimmomatic (-phred33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36; Bolger et al., 2014). The clean reads were mapped to P. parasitica genome (INRA-310-version 3.0) by using software Hisat2 (-min-intronlen 20 -max-intronlen 3,000; Kim et al., 2015). Software featureCounts was used for mapping counts calculation (Liao et al., 2014). The differential expression analysis was conducted by using DESeq2 (Love et al., 2014). Gene annotation of differentially expressed genes (DEGs) was done by Blast2go (Conesa et al., 2005), and GO enrichment analysis was realized by using software Tbtools (Chen et al., 2020). FeatureCounts output data were used to calculate gene expression level, FPKM. The heatmap for gene/sRNA expression level was performed by using Tbtools (Chen et al., 2020). The expression pattern of PpAGO family genes was obtained by analyzing the RNA-seq data obtained previously (Jia, 2017). The RNA sequencing was conducted by collecting the 6-week-old N. benthamiana leaves infected with P. parasitica strain Pp016 at 3, 6, 12, 24, and 48 h post-inoculation (Jia, 2017). The FPKM values were calculated and extracted by using Cuffquant and Cuffnorm (Trapnell et al., 2012).
RXLR effector genes PPTG_01869 and PPTG_15425 were fused with GFP. The effector gene fragment and GFP were both amplified by using P. parasitica cDNA and DNA polymerase FastPfu (TransGen Biotech). The generated fragments were ligated to the restriction sites (KpnI and XbaI) in the vector pKannibal (Wesley et al., 2001) by using T4 DNA ligase (Promega). The fusion construct was further transferred to the NotI sites in vector pART27 (Gleave, 1992). The resulted constructs PPTG_01869-GFP, PPTG_15425-GFP, and GFP were transferred into Agrobacterium tumefaciens GV3101 cells, respectively, and cultured in Luria Bertani (LB) liquid broth, harvested and suspended in infiltration buffer as described (Huang et al., 2019a). The agroinfiltration was performed at concentration (OD600 0.04) on N. benthamiana leaves by using needleless syringes (Meng et al., 2015). The infiltrated N. benthamiana leaves were detached and gene expression state was examined by using the fluorescence microscope (Olympus-BX-51TRF) 24–36 h post-infiltration, and pathogenicity assays were performed using procedures as described (Huang et al., 2019a). Eight leaves were used for each experiment and the experiments were repeated three times.
Phytophthora genomes encode several AGO homologs that phylogenetically clustered with the AGO-like family (Åsman et al., 2016; Bollmann et al., 2016, 2018). In P. parasitica, five AGO proteins, designated as PpAGO1-5 (Supplementary Figure 1), were shown to have high sequence homologies to PiAGOs of P. infestans, a sister species of P. parasitica. Searching the Pfam database (Finn et al., 2010) revealed that they all contain the typical domains of AGO, including the N-terminal PAZ, Mid (middle), and Piwi domains with one or two linkers (L1 and L2; Supplementary Figure 1). In addition, they all contain the arginine–glycine–glycine (RGG) repeat motif at the N terminus. However, the number of RGG motif is highly variable among five PpAGO proteins, with one for PpAGO1, three for PpAGO2, 24 for PpAGO3, two for PpAGO4, and 13 for PpAGO5 (Supplementary Figure 1).
We examined the expression pattern of five PpAGO genes using RNA-seq data of P. parasitica Pp016 before and after infection of N. benthamiana (Jia, 2017). The results showed that PpAGO3 is highly expressed in mycelia (Supplementary Figure 1) during which the class of the 25–26 nt sRNA is abundant (Jia et al., 2017).
To investigate the potential biological function of PpAGO, we employed CRISPR-Cas9 gene editing system (Fang et al., 2017; Zhang et al., 2020) to mutate PpAGO3. The N-terminal sequence of PpAGO3 contains 24 copies of the RGG motif. Through two independent transformation experiments, we obtained nine PpAGO3 mutants all with three types of mutations that carry a deletion of 1–3 copies of the RGG domain in the N terminus without frame shifts or complete knockout (Figures 1A,B), suggesting PpAGO3 is vital to P. parasitica.
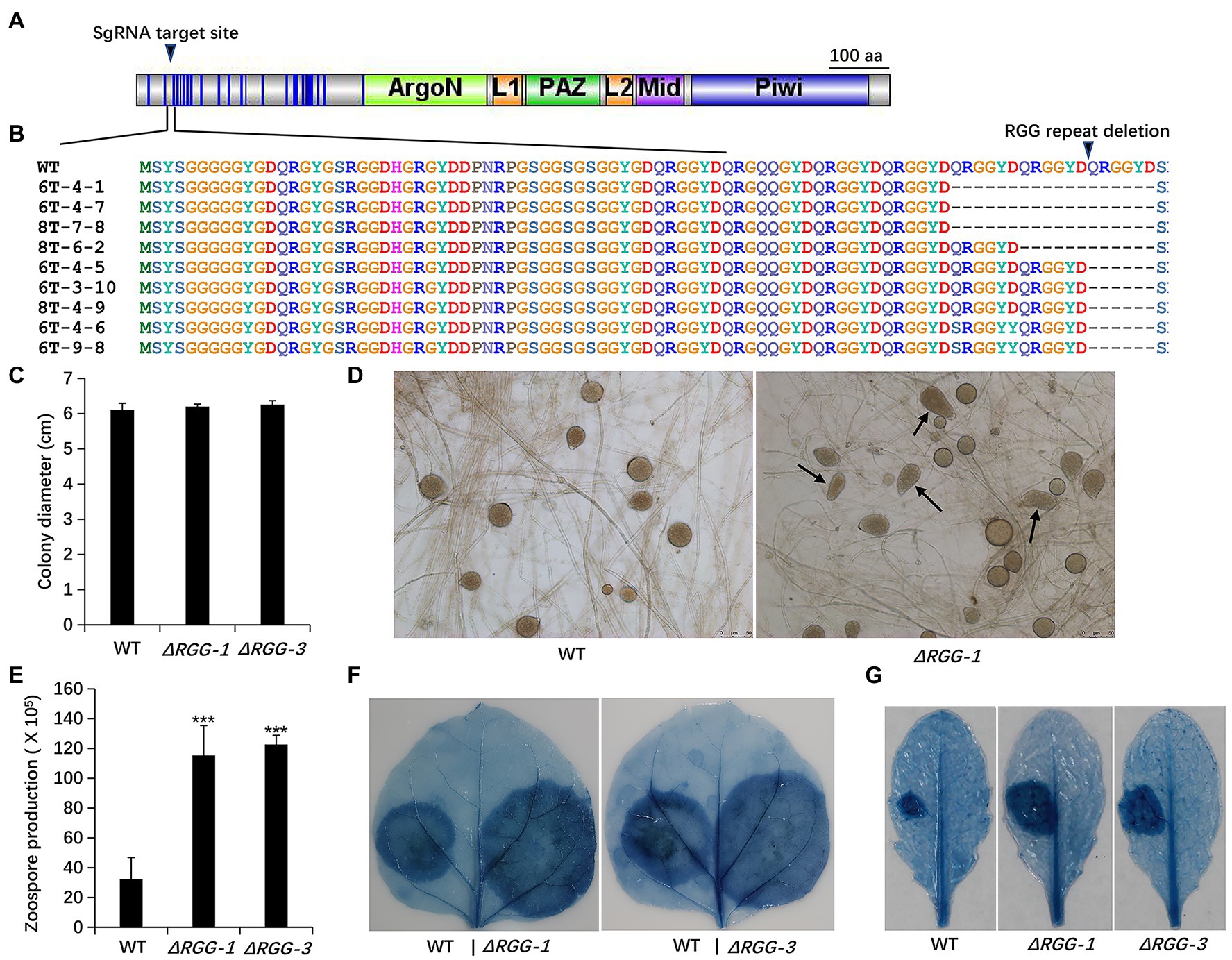
Figure 1. Generation and phenotypic analysis of Phytophthora parasitica mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. (A) The designed sgRNA targeting sites on PpAGO3 protein. (B) Mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 were generated by CRISPR-Cas9 technology. Three types of mutation were obtained: an 18 aa (amino acids) deletion (three QRGGYD repeats) in mutants 6T-4-1, 6T-4-7, and 8T-7-8; a 12 aa deletion (two repeats) for mutant 8T-6-2; and a six aa deletion (one repeat) for mutants 6T-4-5, 6T-3-10, 8T-4-9, 6T-4-6, and 6T-9-8. Besides that, a single amino acid substitution within RGG motif was notable for mutants 6T-4-6 and 6T-9-8. Mutants PpAGO3ΔRGG1 (ΔRGG-1) and PpAGO3ΔRGG3 (ΔRGG-3) showed similar growth rate to the wild-type, as shown by t-test (C), produced abnormal sporangium (D) and more zoospores than the wild-type (Student test: ***p < 0.001) (E), and caused larger lesions than the wild-type on Nicotiana benthamiana (F) and on Arabidopsis thaliana (G). The detached 5-week-old N. benthamiana and 4-week-old A. thaliana leaves were inoculated with mycelial discs and the water-soaked lesions were examined by trypan blue staining at 48 hpi. Similar results were obtained from three independent experiments.
Phenotypic analysis showed that the PpAGO3ΔRGG mutants displayed stronger growth vigor than the wild-type strain, showing more dense and thicker mycelia on 5% CA agar plates, though the colony diameters remained unchanged (Figure 1C). In addition, PpAGO3ΔRGG mutants always produce some abnormal sporangia (Figure 1D) and released many more zoospores than wild-type strain (Figure 1E). More interestingly, PpAGO3ΔRGG mutants were more invasive than the wild-type P. parasitica on N. benthamiana leaves (Figure 1F). Similarly, A. thaliana leaves infected with PpAGO3ΔRGG mutants also showed stronger disease phenotypes (Figure 1G). These results suggest that PpAGO3 plays vital role in the pathogenicity and development of P. parasitica.
Our previous study showed that the 25–26 nt sRNAs are enriched at the vegetative mycelium stage and is associated with high level of silencing of numerous effector genes in P. parasitica, including 40% RXLR (226) and 50% CRN (147) effector genes (Jia et al., 2017). The high level of PpAGO3 expression in vegetative hyphae, together with the enhanced virulence of the PpAGO3ΔRGG mutants, led us to assume that PpAGO3 may interact with 25–26 nt sRNAs to regulate expression of effector genes. Small RNA sequencing was therefore conducted by using RNA isolated from fresh hyphae tissues of the wild-type and mutants PpAGO3ΔRGG1 and PpAGOΔRGG3.
Clean reads of 18–45 nt sRNAs that mapped to P. parasitica genome (P. parasitica INRA-310 version 3.0, Assembly Dev initiative, Broad Institute)4 excluding the rDNA, mtDNA, and tRNA sequences, were obtained for further analysis. The overall sRNA counts and 25–26 nt sRNA percent did not seem to be affected in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 (Supplementary Table 1). Importantly, this analysis detected differential accumulation of the 25–26 nt sRNAs specific to some RXLR and CRN effector genes (Supplementary Table 2) compared to the wild-type strain. Of the 226 RXLR effector genes, 36 (15.9%) contained differentially accumulated 25–26 nt sRNAs in the gene body and 500 nt upstream and downstream flanking sequences in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3, of which 24 genes showed increased and 12 decreased sRNA accumulation (Figure 2A). For the 147 predicted CRN effector genes, 34 (23.13%) displayed differential 25–26 nt sRNA accumulation in the mutants compared to the wild-type strain (Supplementary Table 2), with 17 showing increased and 17 reduced sRNA accumulation (Figure 2A).
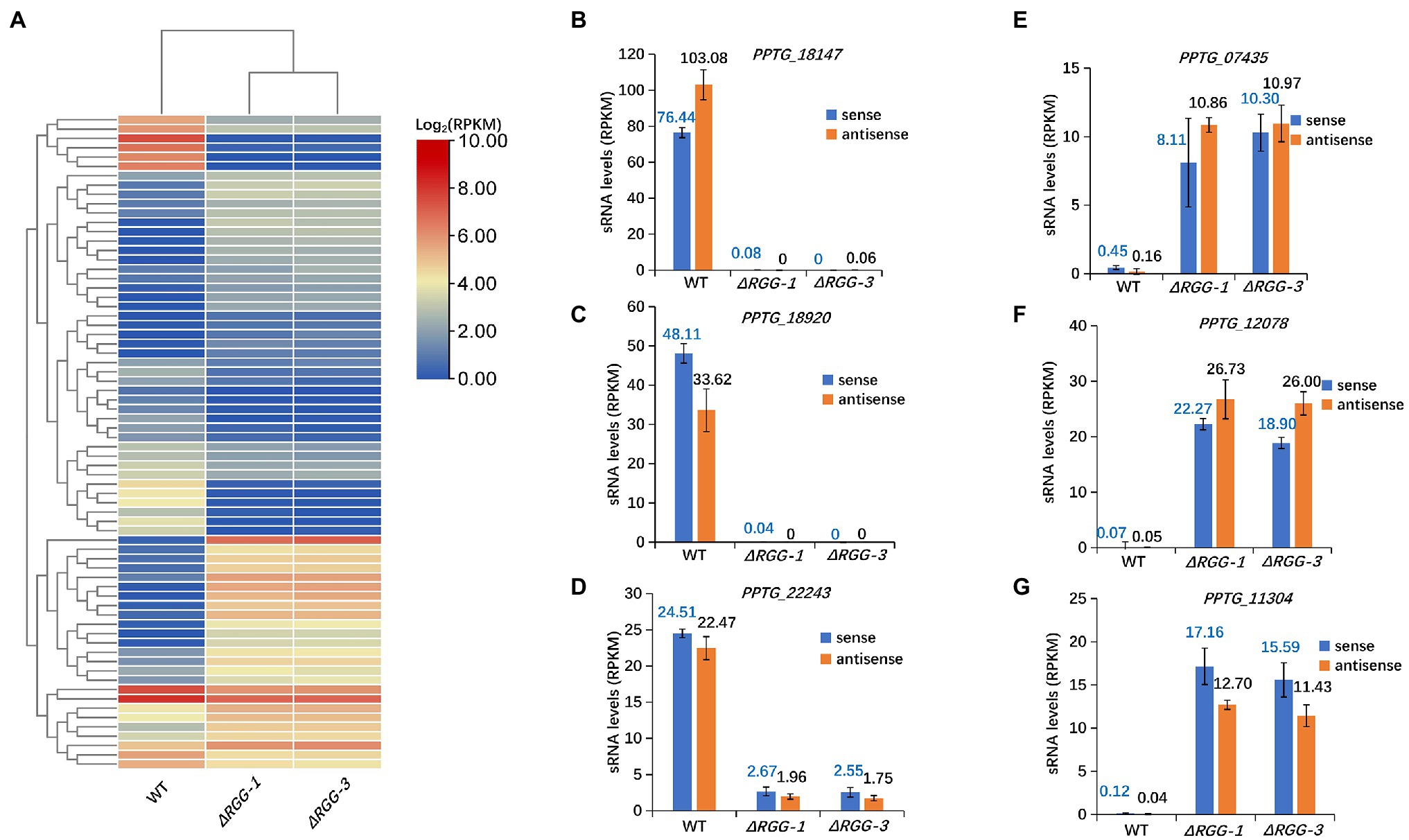
Figure 2. Significant changes in accumulation of 25–26 nt sRNAs homologous to70 cytoplasmic effector genes in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 compared to the wild-type P. parasitica. (A) Heatmap for 25–26 nt sRNA accumulation in wild-type, and mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. The accumulation of 25–26 nt sRNAs was increased for those associated with 24 RXLR and 17 CRN effector genes, and was reduced for those associated with 12 RXLR and 17 CRN effector genes in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. The color represents the transformation value of log2 (RPKM). (B-G) The expression levels of sense and antisense 25–26 nt sRNAs associated with six representative effector genes in wild-type, and mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. Both sense and antisense 25–26 nt sRNAs associated PPTG_18147 (B), PPTG_18920 (C), and PPTG_22243 (D) were highly accumulated in the wild-type but reduced dramatically in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. On the contrary, the sense and antisense 25–26 nt sRNAs associated with PPTG_07435 (E), PPTG_12078 (F), and PPTG_11304 (G) were significantly increased in PpAGO3ΔRGG1 and PpAGO3ΔRGG3 mutants compared to the wild-type.
Our previous studies showed that most of sRNAs in P. parasitica are derived from both sense and antisense strands of the genes typical of double-stranded RNA (dsRNA) processing (Jia et al., 2017). For 49 of the 70 effector genes (36 RXLR and 34 CRN effector genes), 25–26 nt sRNAs of both sense and antisense polarities were differentially expressed in the PpAGO3ΔRGG mutants compared to the wild-type strain. For example, the effector genes PPTG_18147, PPTG_18920, and PPTG_22243, showed a large number of 25–26 nt sense and antisense sRNA in the wild-type strain, but these 25–26 nt sRNAs could not be detected in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 (Figures 2B–D). On the contrary, both the sense and antisense 25–26 nt sRNAs matching the effector genes PPTG_07435, PPTG_12078, and PPTG_11304, showed increased accumulation in the mutants (Figures 2E–G). These results indicated that PpAGO3 is involved in the accumulation of 25–26 nt sRNAs in P. parasitica that have both sense and antisense polarities typical of dsRNA-derived sRNAs.
In addition to effector genes, differential accumulation of 25–26 nt sRNA was also observed for 2,508 non-effector genes. Of these genes, 1,607 showed increased sRNA accumulation and 901 showed reduced accumulation (Supplementary Table 2). The differentially accumulated sRNAs from 1808 genes contained both sense and antisense populations (Figure 3A), which provided more evidence that these differentially expressed sRNAs are generated from dsRNA and associated with PpAGO3 protein.
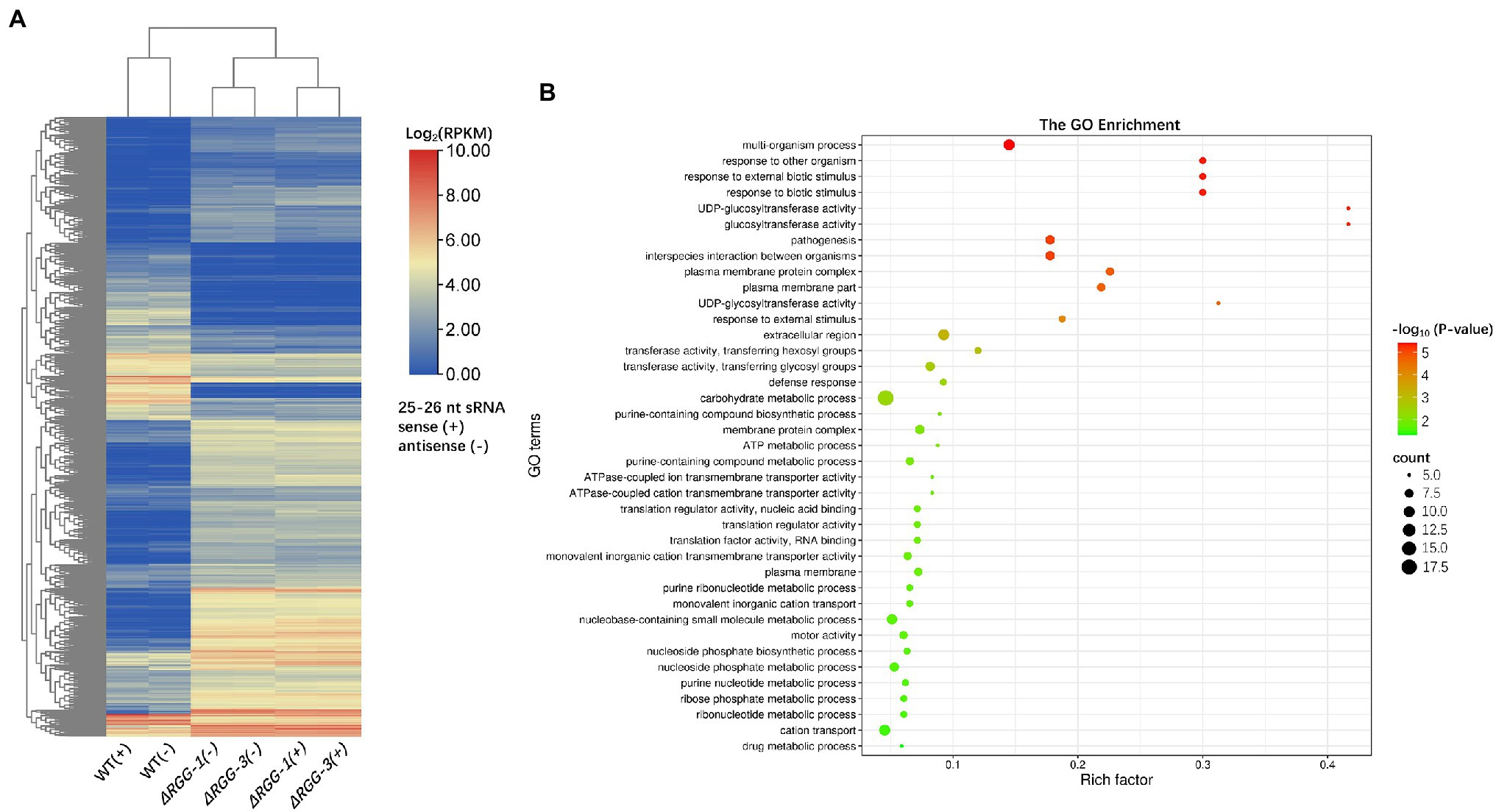
Figure 3. The sRNA distribution of non-effector genes in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. (A) Both sense (+) and antisense (−) strand-derived 25–26 nt sRNAs associated with 1808 non-effector genes were differentially accumulated in mutants compared to the wild-type. RPKM, Reads Per Kilobase per Million mapped reads; the heatmap color represents the transformation value of log2 (RPKM). (B) The GO enrichment analysis for genes with which 25–26 nt sRNAs were downregulated in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. In the bubble plot, x-axis represents the rich factor; the y-axis represents the enriched terms; the bubble size represents the enriched gene count. The larger the bubble is, the more genes enriched; the bubble color represents the enriched significance which depends on the data of –log10 (value of p). GO enrichment was conducted by using software Tbtools and visualized online: http://www.bioinformatics.com.cn/.
GO enrichment analysis of these 2,508 non-effector genes showed that the downregulated sRNAs were mainly associated with genes in the Biological Process level category, including multi-organism process, response to external biotic stimulus, response to biotic stimulus and to other organism, pathogenesis, and interspecies interaction between organism terms (Figure 3B). The association of differentially accumulated 25–26 nt sRNAs with biotic response and pathogenesis genes, in addition to effector genes, further suggest the importance of these sRNAs in the regulation of Phytophthora pathogenesis.
The expression variation of effector gene-associated 25–26 nt sRNAs in PpAGO3ΔRGG mutants and increased virulence of the mutants prompted us to examine if expression of effector genes were influenced. RNA sequencing (RNA-seq) was performed on wild-type and mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 using RNA samples extracted from fresh mycelia. Analysis of the RNA-seq data revealed that 24 RXLR effector genes, 10.6% of the 226 predicted RXLR effector genes in P. parasitica, were differentially expressed (|log2 Fold change| > 1 and Padj <0.05; Supplementary Table 3). Of these 24 genes, 12 showed increased mRNA level while the other 12 reduced mRNA level (Supplementary Table 3). For the CRN effector genes, 18.4% (27/147) genes exhibited differential expression with 17 being upregulated and 10 downregulated (Supplementary Table 3). In total, 51 effector genes were differentially expressed in the PpAGO3ΔRGGmutants compared to the wild-type strain.
We then investigated whether the effector genes with differential mRNA level were associated with differentially accumulated 25–26 nt sRNAs. Overlapping the 51 differentially expressed effector genes (Supplementary Table 3) with the 70 effector genes that had differentially accumulated 25–26 nt sRNAs (Figure 2A) identified 14 effector genes that showed significant changes in both 25–26 nt sRNA accumulation and mRNA level. Interestingly, except for PPTG_01844, the remaining 13 effector genes, including 4 RXLR and 9 CRN effector genes, all showed a negative correlation between sRNA accumulation and mRNA expression level (Figure 4A). In particular, 8 genes showed reduced accumulation of 25–26 nt sRNA that correlated with increased gene expression level in PpAGO3ΔRGG mutants compared to the wild-type strain, whereas five genes showed increased sRNA accumulation with reduced gene expression (Figure 4A). For instance, PPTG_22243 and PPTG_09076 both showed a dramatic reduction in 25–26 nt sRNA abundance in PpAGO3ΔRGG mutants (Figure 4B), which correlated with strong upregulation of gene expression at the transcript level (Figure 4C). On the contrary, PPTG_12078 and PPTG_11600 showed an increase in 25–26 nt sRNA level but with reduction on mRNA level (Figures 4D,E). Taken together, these results suggest that PpAGO3 interacts with 25–26 nt sRNA to regulate the expression of the sRNA-associated effector genes.
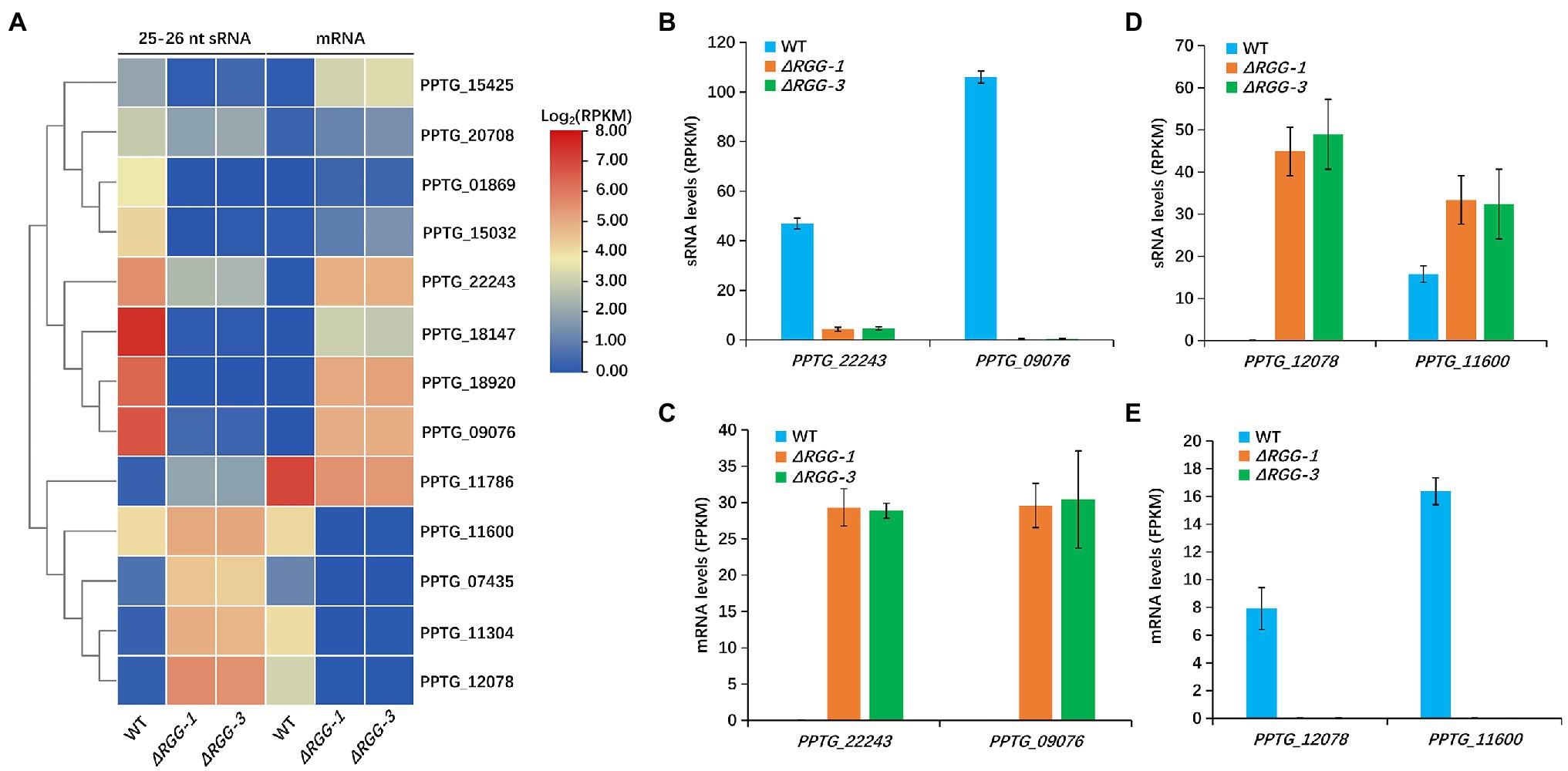
Figure 4. The changed 25–26 nt sRNA accumulation in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 was negatively correlated with expression of the corresponding effector genes. (A) The heatmap for 13 effector genes with which 25–26 nt sRNA and mRNA levels were negatively correlated in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 compared to the wild-type. The color represents the transformation value of log2(RPKM) for sRNA and log2(FPKM) for mRNA. (B–E) The 25–26 nt sRNA and mRNA levels of four representative effector genes, PPTG_22243, PPTG_09076, PPTG_12078, and PPTG_11600. The 25–26 nt sRNA levels were significantly reduced for PPTG_22243 and PPTG_09076 (B) and the mRNA levels were increased obviously (C) in mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3. On the contrary, the 25–26 nt sRNA levels were increased (D) but the mRNA levels were reduced (E) for PPTG_12078 and PPTG_11600.
We next examined if these upregulated effector genes contribute to the enhanced pathogenicity of the mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3, by analyzing the virulence function of the effector genes using A. tumefaciens infiltration-delivered transient expression. We selected two RXLR effector genes, PPTG_01869 and PPTG_15425, which showed strong upregulation of mRNA levels in PpAGO3ΔRGG mutants with dramatic reduction of 25–26 nt sRNA accumulation (Figures 5A–D). Nicotiana benthamiana leaves infiltrated with the PPTG_01869-overexpression construct and inoculated with P. parasitica mycelial disks developed significantly larger lesions at 2 days post-inoculation than leaves infiltrated with the control GFP construct (Figure 5E). Similarly, A. tumefaciens-mediated overexpression of PPTG_15425 strongly enhanced P. parasitica infection (Figure 5F). These results indicated that the PpAGO3-regulated PPTG_01869 and PPTG_15425 effector genes play a positive role in P. parasitica virulence.
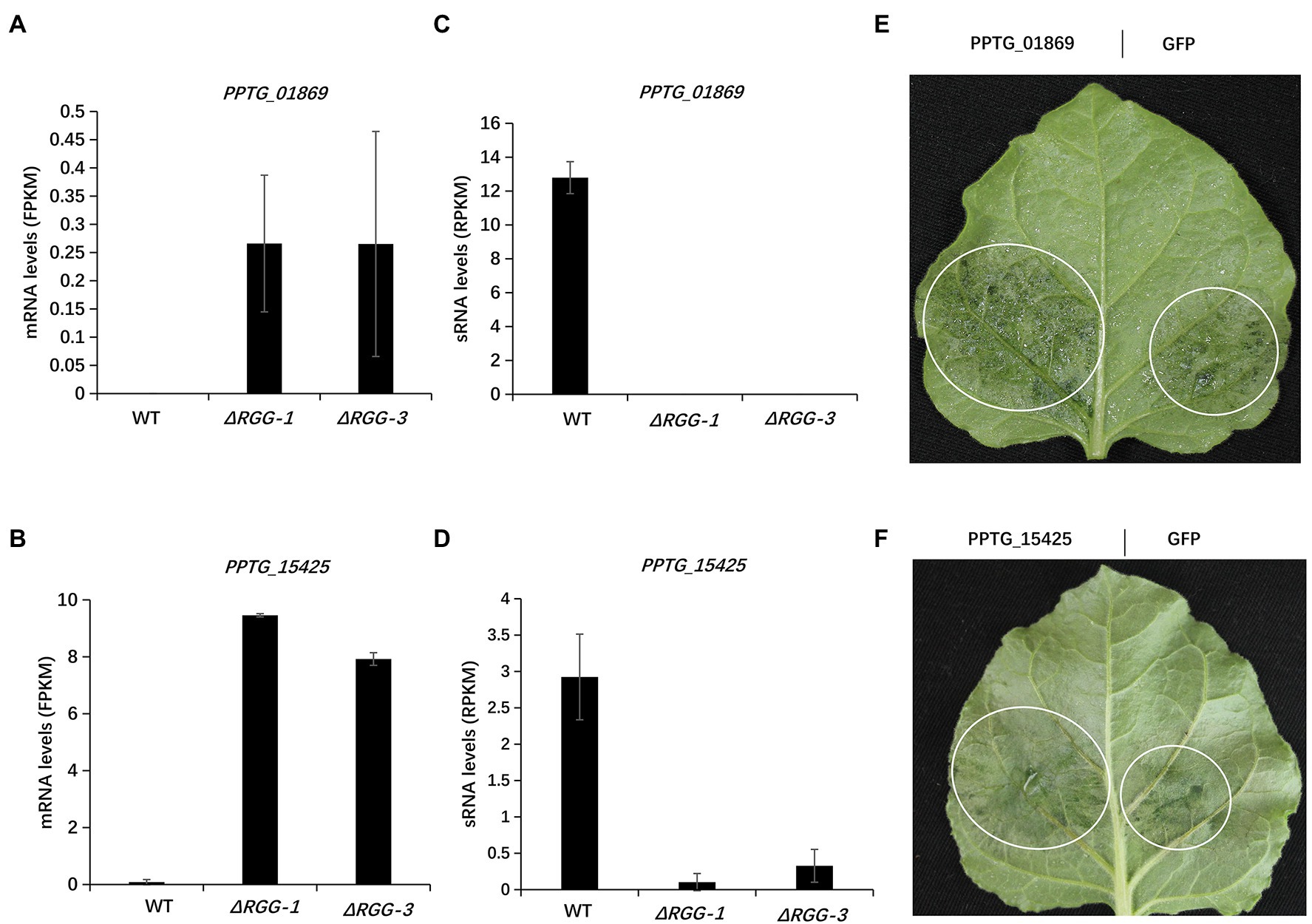
Figure 5. The enhanced pathogenicity of mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 was likely resulted from activation of effector genes. (A,B) The mRNA levels of two upregulated RXLR effector genes, PPTG_01869 and PPTG_15425, in PpAGO3ΔRGG1 and PpAGO3ΔRGG3 mutants. (C,D) The changes of PPTG_01869 and PPTG_15425 mRNA levels were negatively with the corresponding 25–26 nt sRNA levels variation in PpAGO3ΔRGG1 and PpAGO3ΔRGG3 mutants. (E,F) Phytophthora parasitica caused much larger lesion on PPTG_01869 and PPTG_15425 transient expressed N. benthamiana leaves than control GFP, respectively. The white circle marked the water-soaked lesion region. Similar results were obtained from three independent experiments.
A number of reports have suggested that sRNA pathways participate in the silencing of effector genes in Phytophthora species (Vetukuri et al., 2012; Qutob et al., 2013; Jia et al., 2017; Wang et al., 2019). AGO proteins, as the key component of RNAi pathway, were reported to be associated with effector gene-derived sRNA (Vetukuri et al., 2011, 2012; Åsman et al., 2016). These findings suggested the potential function of AGO proteins and sRNAs in the effector genes regulation in Phytophthora but the real functional role of the RNA silencing pathways in Phytophthora pathogenicity has yet to be confirmed.
In this study, we used the model organism P. parasitica to examine the function of AGO in effector gene regulation and pathogenicity. Through sequence analysis, five AGO proteins were identified which all consisted of the conserved domains, PAZ, Mid and Piwi. Functional studies focused on PpAGO3, a unique member with 24 RGG repeats at the beginning of the N terminus and highly expressed in vegetative hyphae.
Evidence for the involvement of PpAGO3 in the pathogenicity of P. parasitica was first suggested by the increased growth vigor of the mutants PpAGO3ΔRGG1-3 deleted with 1–3 copies of the RGG domain, which were generated using the CRISPR-Cas9 system. These mutants displayed increased growth on medium and produced abnormal sporangia and greatly increased number of zoospores. More significantly, the PpAGO3ΔRGG mutants were much more invasive in the tested host plants N. benthamiana and A. thaliana than the wild-type strain.
AGO proteins regulate gene expression through interactions with sRNAs. Consistent with this, our sRNA sequencing analysis of the mutants PpAGO3ΔRGG1 and PpAGO3ΔRGG3 both detected altered accumulation of 25–26 nt sRNAs homologous to 70 effector genes, and 13 of these effector genes showed an inverse correlation between their expression levels and the abundance of the associated 25–26 nt sRNAs. The role of PpAGO3 in regulating effector gene expression is further supported by the opposite developmental stage expression profiles of the PpAGO3 and some of the effector genes. Three such effector genes showed low expression levels at the mycelium stage where the level of PpAGO3 expression was the highest. In contrast, these genes were induced during the infection where the expression of PpAGO3 is reduced.
The functional significance of PpAGO3 regulation of effector genes was shown by the transient overexpression assay of two PpAGO3-regulated effector genes, PPTG_01869 and PPTG_15425. These two genes were strongly upregulated in the PpAGO3ΔRGG mutants, which correlated inversely with reduced abundance of 25–26 nt sRNAs. Overexpression of both of these two effector genes, using agrobacterium infiltration in N. benthmiana leaves, enhanced the infection of P. parasitica. This result suggests that the enhanced virulence of the PpAGO3ΔRGG mutants is related to the activated expression of the sRNA-regulated effector genes. Taken together, our results provided compelling evidence that PpAGO3, in conjunction with 25–26 nt sRNAs, plays a major role in the regulation of effector gene expression in P. parasitica, and this regulation contributes to the virulence of the pathogen in host plants. However, how PpAGO3 interact with 25–26 nt sRNAs and how the effector genes are regulated, remain to be further studied.
One question is why these effector genes need to be regulated by the sRNA silencing pathways. Here we propose three possible scenarios. First, it was considered that some effectors could trigger plant immunity as avirulence factors, and suppression of these effectors would help to evade the perception by host plants. Consistently, some avirulence effector genes are suppressed in P. infestans and P. sojae (Shan et al., 2004; Wang et al., 2019), and the silencing of PiAGO1, PiAGO4, or PiAGO5 in P. infestans is associated with the accumulation of PiAvrblb1 homologous 32 nt sRNA (Vetukuri et al., 2011, 2012). Second, some effectors might be super-virulent to host plants, and silencing of these effector genes is required to establish a balance between successful infection and continuous evolution in the host. The last possible scenario is that, in order to conserve energy, some effectors are suppressed at the mycelium stage but only derepressed when needed during the infection process, like the effector genes PPTG_01869 and PPTG_15425 shown in this study. Further studies are needed to investigate these hypotheses.
Besides the predicted effector genes, PpAGO3 appears to regulate many other genes in P. parasitica. The RNA-seq analysis identified a total of 3,047 genes with differential expression in PpAGO3ΔRGG mutants (Supplementary Table 3), and sRNA-seq data detected 2,578 genes with differential accumulation of 25–26 nt sRNA (Supplementary Table 2). In total, across the P. parasitica genome, 509 non-cytoplasmic effector genes were identified to show negative correlation between 25 and 26 nt sRNA abundance and mRNA level (Supplementary Figure 2A). These genes include three INF-like genes (Supplementary Figures 2B,C), one of which could induce cell death shown by our unpublished work. The function of these non-cytoplasmic effector genes in Phytophthora–host plant interactions is worth further investigation.
Some of the genes downregulated in PpAGO3ΔRGG mutants were associated with increased abundance of 25–26 nt sRNA accumulation, suggesting a role of these sRNAs in the high-level silencing of these genes. It is possible that other AGO members also interact with the 25–26 nt sRNAs, and in the PpAGO3ΔRGG mutants, these AGOs function to repress the expression of these genes. Future studies should examine the interactions of 25–26 nt sRNAs with the different AGO members in P. parasitica.
For AGO proteins, a diverse number of RGG motifs was found localized in the N-terminal region of each PpAGO proteins (Supplementary Figure 1). The N-terminal RGG domain was reported to play an important role in AGO-mediated RNA silencing in T. brucei and T. gondii (Shi et al., 2004, 2009; Musiyenko et al., 2012), and is also important for the function of other RGG-containing proteins in pre-mRNA splicing, transcription and mRNA translation processes (Rajyaguru and Parker, 2012; Thandapani et al., 2013; Chong et al., 2018). In this study, our results suggested that the RGG domain is also functionally important in P. parasitica.
In conclusion, the RGG domain-changed PpAGO3ΔRGG mutants have allowed us to generate compelling evidence that PpAGO3 plays an important role in regulating effector gene expression in P. parasitica through interaction with the 25–26 nt class of sRNAs. Future studies should investigate the interactions between PpAGO3 and the 25–26 nt sRNAs and how the 25–26 nt sRNAs are generated and how they direct silencing of the effector genes. Moreover, a detailed functional analysis of the RGG and other domains as well as the full-length protein of PpAGO3 were also necessary, especially for the role in Phytophthora–host plant interactions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WS, YM, and JX designed the experiment. JX, YL, JJ, WX, CZ, GH, XG, and YM performed the experiment. JX, WS, JJ, and CZ analyzed the data. JX and WS wrote the paper with suggestions from all authors. All authors contributed to the article and approved the submitted version.
This study received funding from National Natural Science Foundation of China (31561143007), China Agriculture Research System (CARS-09), and the State Administration of Foreign Experts Affairs (#B18042). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Brett Tyler (Oregon State University, United States) for providing the CRISPR-Cas9 vector and helpful suggestions, Ming-Bo Wang (CSIRO Agriculture and Food, Canberra, Australia) for suggestions and manuscript revision, and Patrick Schäfer (Ulm University, Germany) for useful suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.856106/full#supplementary-material
1. ^http://grna.ctegd.uga.edu/
2. ^https://fungidb.org/fungidb/
3. ^http://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html
Armstrong, M. R., Whisson, S. C., Pritchard, L., Bos, J. I., Venter, E., Avrova, A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102, 7766–7771. doi: 10.1073/pnas.0500113102
Åsman, A. K., Fogelqvist, J., Vetukuri, R. R., and Dixelius, C. (2016). Phytophthora infestans Argonaute 1 binds microRNA and small RNAs from effector genes and transposable elements. New Phytol. 211, 993–1007. doi: 10.1111/nph.13946
Azzam, G., Smibert, P., Lai, E. C., and Liu, J. L. (2012). Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division. Dev. Biol. 365, 384–394. doi: 10.1016/j.ydbio.2012.03.005
Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. doi: 10.1093/emboj/17.1.170
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bollmann, S. R., Fang, Y., Press, C. M., Tyler, B. M., and Grünwald, N. J. (2016). Diverse evolutionary trajectories for small RNA biogenesis genes in the oomycete genus Phytophthora. Front. Plant Sci. 7:284. doi: 10.3389/fpls.2016.00284
Bollmann, S. R., Press, C. M., Tyler, B. M., and Grünwald, N. J. (2018). Expansion and divergence of Argonaute genes in the oomycete genus Phytophthora. Front. Microbiol. 9:2841. doi: 10.3389/fmicb.2018.02841
Bottin, A., Larche, L., Villalba, F., Gaulin, E., Esquerré-Tugayé, M.-T., and Rickauer, M. (1999). Green fluorescent protein (GFP) as gene expression reporter and vital marker for studying development and microbe-plant interaction in the tobacco pathogen Phytophthora parasitica var. nicotianae. FEMS Microbiol. Lett. 176, 51–56. doi: 10.1111/j.1574-6968.1999.tb13641.x
Bozkurt, T. O., Schornack, S., Win, J., Shindo, T., Ilyas, M., Oliva, R., et al. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U. S. A. 108, 20832–20837. doi: 10.1073/pnas.1112708109
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi: 10.1016/j.cell.2007.01.043
Buker, S. M., Iida, T., Bühler, M., Villén, J., Gygi, S. P., Nakayama, J., et al. (2007). Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 14, 200–207. doi: 10.1038/nsmb1211
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Cervantes, M., Vila, A., Nicolás, F. E., Moxon, S., de Haro, J. P., Dalmay, T., et al. (2013). A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus Mucor circinelloides. PLoS One 8:e69283. doi: 10.1371/journal.pone.0069283
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X. R., Zhang, Y., Li, H. Y., Zhang, Z. H., Sheng, G. L., Li, Y. P., et al. (2019). The RXLR effector PcAvh1 is required for full virulence of Phytophthora capsici. Mol. Plant-Microbe Interact. 32, 986–1000. doi: 10.1094/MPMI-09-18-0251-R
Chong, P. A., Vernon, R. M., and Forman-Kay, J. D. (2018). RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665. doi: 10.1016/j.jmb.2018.06.014
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Cui, L., Yin, W., Dong, S., and Wang, Y. (2012). Analysis of polymorphism and transcription of the effector gene Avr1b in Phytophthora sojae isolates from China virulent to Rps1b. Mol. Plant Pathol. 13, 114–122. doi: 10.1111/j.1364-3703.2011.00733.x
Czech, B., Munafò, M., Ciabrelli, F., Eastwood, E. L., Fabry, M. H., Kneuss, E., et al. (2018). piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52, 131–157. doi: 10.1146/annurev-genet-120417-031441
Dong, S., Qutob, D., Tedman-Jones, J., Kuflu, K., Wang, Y., Tyler, B. M., et al. (2009). The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen strains. PLoS One 4:e5556. doi: 10.1371/journal.pone.0005556
Durand-Dubief, M., and Bastin, P. (2003). TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 1:2. doi: 10.1186/1741-7007-1-2
Durand-Dubief, M., Kohl, L., and Bastin, P. (2003). Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 129, 11–21. doi: 10.1016/s0166-6851(03)00071-9
Fahlgren, N., Bollmann, S. R., Kasschau, K. D., Cuperus, J. T., Press, C. M., Sullivan, C. M., et al. (2013). Phytophthora have distinct endogenous small RNA populations that include short interfering and microRNAs. PLoS One 8:e77181. doi: 10.1371/journal.pone.0077181
Fan, G., Yang, Y., Li, T., Lu, W., Du, Y., Qiang, X., et al. (2018). A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum-mediated immunity. Mol. Plant 11, 1067–1083. doi: 10.1016/j.molp.2018.05.009
Fang, Y., Cui, L., Gu, B., Arredondo, F., and Tyler, B. M. (2017). Efficient genome editing in the oomycete Phytophthora sojae using CRISPR/Cas9. Curr. Protoc. Microbiol. 44:21A.1.1-21A.1.26. doi: 10.1002/cpmc.25
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Fry, W. (2008). Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol. 9, 385–402. doi: 10.1111/j.1364-3703.2007.00465.x
Garcia-Silva, M. R., Tosar, J. P., Frugier, M., Pantano, S., Bonilla, B., Esteban, L., et al. (2010). Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene 466, 26–35. doi: 10.1016/j.gene.2010.06.012
Garcia-Silva, M. R., Sanguinetti, J., Cabrera-Cabrera, F., Franzén, O., and Cayota, A. (2014). A particular set of small non-coding RNAs is bound to the distinctive Argonaute protein of Trypanosoma cruzi: insights from RNA-interference deficient organisms. Gene 538, 379–384. doi: 10.1016/j.gene.2014.01.023
Gijzen, M., Ishmael, C., and Shrestha, S. D. (2014). Epigenetic control of effectors in plant pathogens. Front. Plant Sci. 5:638. doi: 10.3389/fpls.2014.00638
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202. doi: 10.1038/nature04917
Gleave, A. P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. doi: 10.1007/BF00028910
Grünwald, N. J., Goss, E. M., and Press, C. M. (2008). Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 9, 729–740. doi: 10.1111/j.1364-3703.2008.00500.x
Huang, V., and Li, L. C. (2014). Demystifying the nuclear function of Argonaute proteins. RNA Biol. 11, 18–24. doi: 10.4161/rna.27604
Huang, G., Liu, Z., Gu, B., Zhao, H., Jia, J., Fan, G., et al. (2019a). An RXLR effector secreted by Phytophthora parasitica is a virulence factor and triggers cell death in various plants. Mol. Plant Pathol. 20, 356–371. doi: 10.1111/mpp.12760
Huang, C. Y., Wang, H., Hu, P., Hamby, R., and Jin, H. (2019b). Small RNAs: big players in plant-microbe interactions. Cell Host Microbe 26, 173–182. doi: 10.1016/j.chom.2019.07.021
Hutvagner, G., and Simard, M. J. (2008). Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32. doi: 10.1038/nrm2321
Jia, J. (2017). Diversity of Small RNAs and Their Potential Roles in the Regulation of Gene Expression in Phytophthora Parasitica. Ph.D. dissertation, Yangling, Shaanxi, China: Northwest A&F University.
Jia, J., Lu, W., Zhong, C., Zhou, R., Xu, J., Liu, W., et al. (2017). The 25-26 nt small RNAs in Phytophthora parasitica are associated with efficient silencing of homologous endogenous genes. Front. Microbiol. 8:773. doi: 10.3389/fmicb.2017.00773
Judelson, H. S. (1997). The genetics and biology of Phytophthora infestans: modern approaches to a historical challenge. Fungal Genet. Biol. 22, 65–76. doi: 10.1006/fgbi.1997.1006
Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A. Jr., Zhu, J. K., et al. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U. S. A. 103, 18002–18007. doi: 10.1073/pnas.0608258103
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kobayashi, H., and Tomari, Y. (2016). RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 1859, 71–81. doi: 10.1016/j.bbagrm.2015.08.007
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lee, H. C., Li, L., Gu, W., Xue, Z., Crosthwaite, S. K., Pertsemlidis, A., et al. (2010). Diverse pathways generate microRNA-like RNAs & Dicer-independent small interfering RNAs in fungi. Mol. Cell 38, 803–814. doi: 10.1016/j.molcel.2010.04.005
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lingel, A., Simon, B., Izaurralde, E., and Sattler, M. (2003). Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426, 465–469. doi: 10.1038/nature02123
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Makarova, K. S., Wolf, Y. I., van der Oost, J., and Koonin, E. V. (2009). Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4:29. doi: 10.1186/1745-6150-4-29
Marasovic, M., Zocco, M., and Halic, M. (2013). Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol. Cell 52, 173–183. doi: 10.1016/j.molcel.2013.08.046
Meng, Y., Zhang, Q., Ding, W., and Shan, W. (2014). Phytophthora parasitica: a model oomycete plant pathogen. Mycology 5, 43–51. doi: 10.1080/21501203.2014.917734
Meng, Y., Zhang, Q., Zhang, M., Gu, B., Huang, G., Wang, Q., et al. (2015). The protein disulfide isomerase 1 of Phytophthora parasitica (PpPDI1) is associated with the haustoria-like structures and contributes to plant infection. Front. Plant Sci. 6:632. doi: 10.3389/fpls.2015.00632
Millar, A. A., and Waterhouse, P. M. (2005). Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics 5, 129–135. doi: 10.1007/s10142-005-0145-2
Musiyenko, A., Majumdar, T., Andrews, J., Adams, B., and Barik, S. (2012). PRMT1 methylates the single Argonaute of Toxoplasma gondii and is important for the recruitment of Tudor nuclease for target RNA cleavage by antisense guide RNA. Cell. Microbiol. 14, 882–901. doi: 10.1111/j.1462-5822.2012.01763.x
Nguyen, Q., Iritani, A., Ohkita, S., Vu, B. V., Yokoya, K., Matsubara, A., et al. (2018). A fungal Argonaute interferes with RNA interference. Nucleic Acids Res. 46, 2495–2508. doi: 10.1093/nar/gkx1301
Pais, M., Yoshida, K., Giannakopoulou, A., Pel, M. A., Cano, L. M., Oliva, R. F., et al. (2018). Gene expression polymorphism underpins evasion of host immunity in an asexual lineage of the Irish potato famine pathogen. BMC Evol. Biol. 18:93. doi: 10.1186/s12862-018-1201-6
Qutob, D., Chapman, B. P., and Gijzen, M. (2013). Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat. Commun. 4:1349. doi: 10.1038/ncomms2354
Qutob, D., Tedman-Jones, J., Dong, S., Kuflu, K., Pham, H., Wang, Y., et al. (2009). Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One 4:e5066. doi: 10.1371/journal.pone.0005066
Rajyaguru, P., and Parker, R. (2012). RGG motif proteins: modulators of mRNA functional states. Cell Cycle 11, 2594–2599. doi: 10.4161/cc.20716
Shan, W., Cao, M., Leung, D., and Tyler, B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant-Microbe Interact. 17, 394–403. doi: 10.1094/MPMI.2004.17.4.394
Shi, H., Chamond, N., Djikeng, A., Tschudi, C., and Ullu, E. (2009). RNA interference in Trypanosoma brucei: role of the N-terminal RGG domain and the polyribosome association of argonaute. J. Biol. Chem. 284, 36511–36520. doi: 10.1074/jbc.M109.073072
Shi, H., Ullu, E., and Tschudi, C. (2004). Function of the trypanosome Argonaute 1 protein in RNA interference requires the N-terminal RGG domain and arginine 735 in the Piwi domain. J. Biol. Chem. 279, 49889–49893. doi: 10.1074/jbc.M409280200
Song, J. J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Swarts, D. C., Makarova, K., Wang, Y., Nakanishi, K., Ketting, R. F., Koonin, E. V., et al. (2014). The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753. doi: 10.1038/nsmb.2879
Thandapani, P., O’Connor, T. R., Bailey, T. L., and Richard, S. (2013). Defining the RGG/RG motif. Mol. Cell 50, 613–623. doi: 10.1016/j.molcel.2013.05.021
Tolia, N. H., and Joshua-Tor, L. (2007). Slicer and the argonautes. Nat. Chem. Biol. 3, 36–43. doi: 10.1038/nchembio848
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. doi: 10.1126/science.1129333
van Poppel, P. M., Guo, J., van de Vondervoort, P. J., Jung, M. W., Birch, P. R., Whisson, S. C., et al. (2008). The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol. Plant-Microbe Interact. 21, 1460–1470. doi: 10.1094/MPMI-21-11-1460
Vetukuri, R. R., Åsman, A. K., Tellgren-Roth, C., Jahan, S. N., Reimegård, J., Fogelqvist, J., et al. (2012). Evidence for small RNAs homologous to effector-encoding genes and transposable elements in the oomycete Phytophthora infestans. PLoS One 7:e51399. doi: 10.1371/journal.pone.0051399
Vetukuri, R. R., Avrova, A. O., Grenville-Briggs, L. J., Van West, P., Söderbom, F., Savenkov, E. I., et al. (2011). Evidence for involvement of dicer-like, Argonaute and histone deacetylase proteins in gene silencing in Phytophthora infestans. Mol. Plant Pathol. 12, 772–785. doi: 10.1111/j.1364-3703.2011.00710.x
Wang, L., Chen, H., Li, J., Shu, H., Zhang, X., Wang, Y., et al. (2020). Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res. 48, 1790–1799. doi: 10.1093/nar/gkz1160
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011a). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wang, Q., Li, T., Zhong, C., Luo, S., Xu, K., Gu, B., et al. (2019). Small RNAs generated by bidirectional transcription mediate silencing of RXLR effector genes in the oomycete Phytophthora sojae. Phytopathol. Res. 1:18. doi: 10.1186/s42483-019-0026-6
Wang, Y., Meng, Y., Zhang, M., Tong, X., Wang, Q., Sun, Y., et al. (2011b). Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol. Plant Pathol. 12, 187–201. doi: 10.1111/j.1364-3703.2010.00659.x
Wang, Y., and Wang, Y. (2018). Phytophthora sojae effectors orchestrate warfare with host immunity. Curr. Opin. Microbiol. 46, 7–13. doi: 10.1016/j.mib.2018.01.008
Wei, W., Ba, Z., Gao, M., Wu, Y., Ma, Y., Amiard, S., et al. (2012). A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112. doi: 10.1016/j.cell.2012.03.002
Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M. B., Rouse, D. T., Liu, Q., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. doi: 10.1046/j.1365-313x.2001.01105.x
Xue, Z., Yuan, H., Guo, J., and Liu, Y. (2012). Reconstitution of an Argonaute-dependent small RNA biogenesis pathway reveals a handover mechanism involving the RNA exosome and the exonuclease QIP. Mol. Cell 46, 299–310. doi: 10.1016/j.molcel.2012.03.019
Ye, R., Chen, Z., Lian, B., Rowley, M. J., Xia, N., Chai, J., et al. (2016). A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell 61, 222–235. doi: 10.1016/j.molcel.2015.11.015
Yigit, E., Batista, P. J., Bei, Y., Pang, K. M., Chen, C. C., Tolia, N. H., et al. (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757. doi: 10.1016/j.cell.2006.09.033
Yin, W., Xiao, Y., Niu, M., Meng, W., Li, L., Zhang, X., et al. (2020). ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 32, 2292–2306. doi: 10.1105/tpc.19.00542
Zhang, Q., Li, W., Yang, J., Xu, J., Meng, Y., and Shan, W. (2020). Two Phytophthora parasitica cysteine protease genes, PpCys44 and PpCys45, trigger cell death in various Nicotiana spp. and act as virulence factors. Mol. Plant Pathol. 21, 541–554. doi: 10.1111/mpp.12915
Keywords: Phytophthora parasitica, AGO3, small RNA, RXLR effector, RGG domain
Citation: Xu J, Li Y, Jia J, Xiong W, Zhong C, Huang G, Gou X, Meng Y and Shan W (2022) Mutations in PpAGO3 Lead to Enhanced Virulence of Phytophthora parasitica by Activation of 25–26 nt sRNA-Associated Effector Genes. Front. Microbiol. 13:856106. doi: 10.3389/fmicb.2022.856106
Received: 16 January 2022; Accepted: 03 March 2022;
Published: 24 March 2022.
Edited by:
Xiaofan Zhou, South China Agricultural University, ChinaReviewed by:
Wei Guo, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Xu, Li, Jia, Xiong, Zhong, Huang, Gou, Meng and Shan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weixing Shan, d3hzaGFuQG53YWZ1LmVkdS5jbg==
†Present addresses: Jinbu Jia, Department of Biology, Southern University of Science and Technology, Shenzhen, China
Chengcheng Zhong, CSIRO Agriculture and Food, Canberra, ACT, Australia
Guiyan Huang, College of Life Sciences, Gannan Normal University, Ganzhou, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.