
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 March 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.854131
This article is part of the Research TopicAssembly and Functions of Gut Microbiota in Aquatic AnimalsView all 28 articles
 Cong Wei1,2
Cong Wei1,2 Kai Luo1,2
Kai Luo1,2 Mingyang Wang1,2
Mingyang Wang1,2 Yongmei Li1,2
Yongmei Li1,2 Miaojun Pan1,2
Miaojun Pan1,2 Yumeng Xie1,2
Yumeng Xie1,2 Guangcai Qin1,2
Guangcai Qin1,2 Yijun Liu1,2
Yijun Liu1,2 Li Li1,2
Li Li1,2 Qingbing Liu3
Qingbing Liu3 Xiangli Tian1,2*
Xiangli Tian1,2*
In recent years the safety of probiotics has received increasing attention due to the possible transfer and spread of virulence factors (VFs) and antibiotic resistance genes (ARGs) among microorganisms. The safety of a strain of Lactobacillus plantarum named W2 was evaluated in phenotype and genotype in the present study. Its probiotic properties were also evaluated both in vivo and in vitro, including adherence properties, antibacterial properties and beneficial effects on the growth and immunity of Pacific white shrimp, Penaeus vannamei. Hemolysis tests, antibiotic resistance tests and whole genome sequence analysis showed that W2 had no significant virulence effects and did not carry high virulence factors. W2 was found to be sensitive to chloramphenicol, clindamycin, gentamicin, kanamycin and tetracycline, and to be resistant to ampicillin and erythromycin. Most ARGs have no transfer risk and a few have transfer risk but no significant enrichment in human-associated environments. The autoaggregation of W2 was 82.6% and the hydrophobicity was 81.0%. Coaggregation rate with Vibrio parahaemolyticus (24.9%) was significantly higher than Vibrio’s autoaggregation rate (17.8%). This suggested that W2 had adhesion potential to mucosal/intestinal surfaces and was able to attenuate the adherence of V. parahaemolyticus. In addition, several adhesion-related protein genes, including 1 S-layer protein, 1 collagen-binding protein and 9 mucus-binding proteins were identified in the W2 genome. W2 had efficiently antagonistic activity against 7 aquatic pathogenic strains. Antagonistic components analysis indicated that active antibacterial substances might be organic acids. W2 can significantly promote the growth of shrimp when supplemented with 1 × 1010 cfu/kg live cells. Levels of 7 serological immune indicators and expression levels of 12 hepatopancreatic immune-related genes were up-regulated, and the mortality of shrimp exposed to V. parahaemolyticus was significantly reduced. Based on the above, L. plantarum W2 can be applied safely as a potential probiotic to enhance the growth performance, immunity capacity and disease resistance of P. vannamei.
Lactobacillus plantarum is a kind of highly adaptable lactobacillus that is widely found in various fermented products of plants and animals and occurs naturally in the intestinal tract of animals including humans (Xu et al., 2015). Some strains have already been used as probiotics and play a crucial role in human health, livestock rearing and aquaculture. For example, L. plantarum 0612 competes for surface receptors on human intestinal Caco-2 epithelial cells and significantly inhibits the adhesion of Escherichia coli and Listeriosis monocytogenes (Lau and Chye, 2018). L. plantarum CAM6 can be used as an antibiotic alternative for weaned pigs (Betancur et al., 2020). L. plantarum is generally used as an effective probiotic in aquaculture, and has been widely studied and applied in shrimp and fish culture. Several strains of L. plantarum have been shown to enhance resistance to V. alginolyticus (Chiu et al., 2007), V. harveyi (Vieira et al., 2010; Kongnum and Hongpattarakere, 2012), V. parahaemolyticus (Thammasorn et al., 2017) infections in shrimp. L. plantarum FNCC 226 can alleviate parasitism and growth inhibition of Pangasius hypophthalamus by Saprolegnia parasitica (Nurhajati et al., 2012). Several strains have been found to have beneficial effects on the growth, immunity and disease resistance in various fish species, such as Anguilla japonicav (Lee et al., 2017), Oncorhynchus mykiss (Vendrell et al., 2008), Sparus aurata (Picchietti et al., 2007), Labeo rohita (Giri et al., 2013), Oreochromis niloticus (Van Doan et al., 2017), and Paralichthys olivaceus (Beck et al., 2015).
Safety assessment is a vitally important task for the screening of probiotics to exclude adverse effects (FAO/WHO, 2002). Accurate identification, adequate phenotypic characterization and assessment of potential risks are indispensable steps in the safety assessment of probiotics (Miquel et al., 2015; Reid et al., 2019). Generally, Lactobacillus can be used safely in most cases. For example, there are only sporadic cases of Lactobacillus acting as a clinical infection pathogen in humans, with a very small proportion of L. plantarum involved (Haghighat and Crum-Cianflone, 2016; Nayeem et al., 2018; Koyama et al., 2019). Bernardeau et al. (2006) estimated that the risk of Lactobacillus infection was extremely low, with approximately one case per 10 million people over a period of more than a century in France.
Until now, no cases of animal infection have been reported. However, safety of probiotics has received increasing attention in recent years due to the potential transfer and spread of antibiotic resistance genes (ARGs) among microorganisms (Wang et al., 2008; Doron and Snydman, 2015). Previous studies have shown that the extensive use of antibiotics not only enhances antibiotic resistance in bacteria, but also promotes the transfer of ARGs (Santajit and Indrawattana, 2016). A probiotic should have a low risk of transferring ARG to the environment. However, an antibiotic resistance profile of Lactobacillus demonstrated that acquired resistance genes were occasionally present in Lactobacillus species (Campedelli et al., 2019).
L. plantarum is included in the Qualified Presumption of Safety (QPS) list of European Food Safety Authority (EFSA, 2008). When supplementing with live cells, it is recommended to assess the safety of the strain in use by QPS. EFSA guidance specifies four main aspects of bacterial safety assessment: strain identification; strain toxicity and pathogenicity; antibiotic resistance; and antimicrobial substance identification (EFSA, 2018). It is clear in the guidance that virulence factors (VFs) should be excluded in Enterococcus faecalis (IS16, hylEfm, esp) and Bacillus (enterotoxin, cereulide synthase), while those in L. plantarum are not listed. In response to the transfer of ARGs, EFSA delineated the cut-off value of L. plantarum to 7 antibiotics.
In terms of biological and ecological safety, probiotic screening should be carried out through a careful, phenotypically and genotypically based strategy according to the EFSA guidance (EFSA, 2018). In addition, it is essential to consider the beneficial effects of a probiotic candidate in both laboratory and practical applications, and to uncover the related mechanisms. Unfortunately, due to limited research and knowledge, there are still no internationally accepted screening criteria for probiotics in aquaculture which balance safety and beneficial effects. Therefore, it is important to develop standardized evaluation criteria to screen probiotics for application in aquaculture.
A strain of Lactobacillus was isolated from the intestine of Pacific white shrimp, Penaeus vannamei in our laboratory and named W2. In accordance with the latest EFSA guidelines, this study investigates the safety of the W2 strain in genotype and phenotype, including the assessment of virulence and VFs, antibiotic resistance and ARGs, the origin of VFs and ARGs, and the possible transfer potential of acquired genes. The potential probiotic properties of the strain for shrimp farming are also investigated, including the potential for adhesion, antagonistic activity in vitro and any modulatory effects on shrimp growth, immunity and disease resistance when supplemented in feed. The results from this study would be helpful to develop criteria for the screening and evaluation of probiotics in aquaculture.
The strain of L. plantarum named W2 was isolated from the intestinal tract of Pacific white shrimp, P. vannamei, and was obtained from the Microbial Culture Centre, Laboratory of Aquaculture Ecology, Ocean University of China, China. W2 was identified using physiological and biochemical assays and 16S rRNA sequencing. The 16S rRNA was amplified with primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). 16S rRNA sequences were available through Sanger. 19 strains were selected in the NCBI database to construct a phylogenetic tree (MEGAX, Neighbor-Joining).
The microbial adhesion to solvents (MATS) method was employed to determine the hydrophobicity and electron donor-acceptor properties of W2 (Bellon-Fontaine et al., 1996). The autoaggregation and coaggregation assays of bacteria were as described by Kos et al. (2003) with some modifications. Hydrophobicity, autoaggregation, coaggregation were calculated using the following formulas:
W2 was cultured in MRS broth at 37°C for 20 h and pathogens (V. splendens, V. parahaemolyticus, V. vulnificus) were cultured in broth medium at 26°C for 24 h, then, centrifuged at 5,000 g for 10 min and the supernatant discarded. The bacteria were diluted with sterile saline until reaching an absorbance value of around 0.8 (OD 600), recorded as Ax and Ay. The W2 suspension was fully mixed 3:1 with hydrophobic agent (n-dodecane, xylene, chloroform), allowed to settle for 20 min at 25°C and the OD600 of the aqueous phase was measured and recorded as At. Four ml of bacterial suspension was allowed to stand for 5 h at 25°C and the OD600 of the surface suspension measured and recorded as Ap. The W2 suspension was fully mixed 1:1 with the pathogens, allowed to settle for 5 h at 25°C, and the OD600 of the surface suspension measured and recorded as A(x+y). Each treatment consisted of three replicates and was repeated three times.
The antagonistic activity of W2 was evaluated using the agar diffusion method (Oxford cup method) (Kuang et al., 2018). The selected indicator bacteria were 7 common aquatic pathogenic strains, including V. vulnificus S01P2, V. splendidus BSD11, V. harveyi SRTT9 (Wang et al., 2020), V. parahaemolyticus 20130629002S01 (Dong et al., 2017), Streptococcus iniae NUF849 (Sheng et al., 2018), Aeromonas hydrophila AP40301 (Xu et al., 2018), and Shewanella marisflavi AP629 (Li et al., 2010). The culture methods of W2 and pathogens were similar to those described in the adhesion assay. Pathogenic bacteria fermentation broth was centrifuged at 5,000 g for 10 min and the supernatant discarded. It was then diluted with broth medium to a final concentration of 1 × 106 cfu/ml. Different pathogen suspensions were coated onto agar plates and dried for 15 min. An Oxford cup was vertically placed on the plate, and then 200 μl of W2 suspension was dispensed into the cup and allowed to diffuse at 26°C for 24 h. A control experiment was conducted using MRS broth. The inhibition zone diameters were then measured.
For screening for antibacterial activity, the culture broth (CB), cell free supernatant (CFS), and biomass suspension (BS) were used in an antagonistic assay, with V. vulnificus S01P2 and V. parahaemolyticus 20130629002S01 as the indicator bacteria. The CFS was further processed as follows: 1 g/ml Tripsin was added to deproteinated supernatant (CFT). The supernatant was heated to 80°C for 10 min to remove H2O2 (CFH), then the pH was adjusted to 6.0 to exclude the antimicrobial effect of organic acids (CF6.0). Each treatment consisted of three replicates and was repeated three times.
The hemolytic activity was tested using the Oxford cup diffusion test (Kuang et al., 2018). Colombian blood plate (5% sheep blood) was purchased from ELITE-MEDIA. MRS broth was used as the negative control. W2 was incubated in MRS broth for 24 h and the supernatant was obtained by centrifugation. The pH of the supernatant and broth was adjusted to 6.0 to exclude the effects of acidity. The Oxford cups were placed on the blood plate and filled with 200 μl supernatant or broth. The plate was incubated at 28°C for 24 h, then observed for the hemolytic type. Each consisted of three replicates and was repeated three times.
Based on the EFSA (2018) guidelines, this study determined the minimum inhibitory concentration (MIC) of 7 antibiotics (ampicillin, kanamycin, chloramphenicol, clindamycin, erythromycin, gentamicin, and tetracycline) against W2 using the micro-broth dilution method (El Shazely et al., 2020). Escherichia coli ATCC 25,922 was used as the standard strain. Separate solutions of 512 μg/ml of the 7 antibiotics mentioned above were prepared. The concentrations of antibiotics in wells 1–11 of the 96-well plate were 128, 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0.0625 μg/ml, respectively. 100 μl bacterial suspension (OD600: 0.1) was added to each well and incubated for 20 h in 37°C. The results were then observed. Each treatment consisted of three replicates and was repeated three times.
Whole genome sequencing (WGS) was performed on the Illumina Hiseq and PacBio platforms. Sequencing quality control was done with Fastp (Chen et al., 2018), using an average quality score of 20 and a minimum read length threshold of 30 bp. Assembly was performed using Unicycler v0.4.8 (Wick et al., 2017), with parameters that mainly included kmer values ranging from 21 to 41. The softwares used for gene annotation were DIAMOND v0.8.22 (Buchfink et al., 2015) for NR, KOG, Swiss-Prot, and KEGG (e-value = 1e-5); blast2GO (Conesa and Götz, 2008) for GO (e-value = 1e-5); HMMER 3.0 (Eddy, 2009) for Pfam (e-value = 0.01). ARGs and VFs were obtained by comparison in the CARD (The Comprehensive Antibiotic Research Database) and the VFDB (Virulence Factor Database). Mobile genetic elements (MGEs) were analyzed in W2. Genomic island (GEIs) prediction was performed using IslandViewer (Bertelli et al., 2017), pre-phages were predicted using Phage_Finder (Fouts, 2006), CRISPR-Cas was predicted using MinCED (Bland et al., 2007), integrons in DNA sequences were detected using Integron_Finder (Cury et al., 2016), and insertion sequence (IS) elements in the genome were identified using ISEScan (Siguier et al., 2014). The sequencing data were deposited in the Sequence Read Archive (SRA) repository under the accession number PRJNA797524.
To determine the probiotic effects of W2 on aquaculture animals, a 42-day feeding trial of P. vannamei was designed. 225 healthy juvenile shrimps were randomly divided into three treatments. Five replicates were established for each treatment. The three treatments were as follows: (1) shrimp fed basal diet (the control, CON); (2) shrimp fed basal diet supplemented with 1 × 1010cfu/kg live cells (LLP); (3) shrimp fed basal diet supplemented with 15 mg/kg florfenicol for 7 d in a 14-day interval (FLI). Florfenicol treatment was set up as a positive control to compare with the effects of W2 in shrimp. Nutritional compositions of the basal diet are given in Supplementary Table 1. During the experiment, the water was exchanged one third of the aquarium volume (53 × 28 × 34 cm, 50 L) and the shrimp were fed four times per day (08:00, 12:00, 16:00, and 20:00).
Uneaten feed was collected 1 h after feeding, dried at 60°C and weighed. At the end of the feeding trial, the shrimp were starved for 24 h. Then the shrimp were counted, weighed and their tissues were collected. The hemolymph of the shrimp was collected without anticoagulants and stored at 4°C. After 24 h, serum was obtained by centrifugation at 3,000 g for 10 min. Hepatopancreas was collected and immediately frozen in liquid nitrogen. Both serum and hepatopancreas were stored at −80°C waiting for determination of the activities of non-specific immune enzymes activity and expression levels of immune-related genes.
Based on the innate immune properties of crustaceans, the following 8 serological indicators of immune function of shrimp were measured: Alkaline phosphatase (AKP), acid phosphatase (ACP), total nitric oxide synthase (T-NOS), peroxidase (POD), superoxide dismutase (SOD), phenol oxidase (PO) activities, total antioxidant capacity (T-AOC), and lysozyme (LZM) content. They were quantified using commercial detection kits obtained from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The relative expression levels of the following 12 immune genes in the hepatopancreas of the shrimp were determined: Imd, Toll, Relish, TOR, 4E-BP, eIF4E1α, eIF4E2, SOD, LZM, proPO, HSP70, and LGBP. All the above immune-related genes are showed in Supplementary Table 2, with gene description and primers used. Total RNA was extracted using Trizol Reagent (Takara, Japan). The quantity and purity of RNA samples were examined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, United States), followed by 1.5% agarose gel electrophoresis. Total RNA was reverse-transcribed to cDNA using the PrimeScript^TM RT reagent Kit (Takara, Japan). Real-time quantitative RT-qPCR was performed using the QuantStudio^TM 5 real-time PCR system (Applied Biosystems, United States).
After the feeding trial, the remaining shrimp were fed continually for 5 additional days. 24 shrimp were randomly selected from each treatment and divided into 3 replicates for the challenge experiment. Vibrio parahaemolyticus 20130629002S01 were provided by the Microbial Culture Centre, Laboratory of Aquaculture Ecology, Ocean University of China, China. The LC50 of V. parahaemolyticus on 14th day was 5 × 107 cfu/ml determined during the pre-experiment. Shrimp were injected intramuscularly in the third abdominal segment with live V. parahaemolyticus at the concentration of 5 × 107 cfu/ml. Daily management during the challenge test was the same as that during the feeding trial. Cumulative mortality of shrimp was calculated on the 14th day after injection.
The growth performance of shrimp is described by the survival rate (SR, %), specific growth rate (SGR, %/d) and feed efficiency rate (FER). These formulas are defined as follows:
where N0 and Nt represent the number of shrimp at the beginning and end of the culture, respectively; W0 and Wt represent the initial and final weights of the shrimp (g); t refers to the duration of the experiment (d); and F0 refers to the feed intake (dry weight, g).
All non-genomic data in this study were subjected to one-way ANOVA (SPSS 22.0). Data were tested for normality, homogeneity and independence prior to ANOVA. Results are represented as mean ± standard error.
The physiological and biochemical properties of W2 are presented in Supplementary Table 3. W2 is a Gram-positive bacillus, non-budding, non-motile, catalase negative, and produces acid but not acetyl methyl methanol and not hydrogen sulfide. W2 can ferment most monosaccharides and oligosaccharides except xylose. The identification was consistent with the basic characteristics of L. plantarum. A phylogenetic tree constructed based on 16S rRNA sequences confirmed that W2 is a strain of L. plantarum (Figure 1).
The adhesion properties of W2 are shown in Figure 2. The autoaggregation rate of W2 was 82.6% compared to less than 50.0% for the three Vibrio strains. The coaggregation rates of W2 with the three Vibrio strains were 31.2% for V. splendidus, 20.4% for V. vulnificu, and 24.9% for V. parahaemolyticus. This indicated that the interaction between W2 and Vibrio significantly enhanced the aggregation of V. parahaemolyticus (P < 0.05) and significantly reduced the aggregation of V. splendens but had no significant effect on the aggregation of V. vulnificu (P > 0.05). In the hydrophobicity assays, W2 showed the highest hydrophobicity in chloroform (81.0%), which was 66.5 and 61.8% higher than n-dodecane and xylene, respectively. Thus, W2 has a high autoaggregation rate (82.6%), a high hydrophobicity (chloroform: 81.0%), and it is strain-specific in co-coaggregation with different Vibrio strains. It can significantly reduce the adhesion of V. parahaemolyticus.
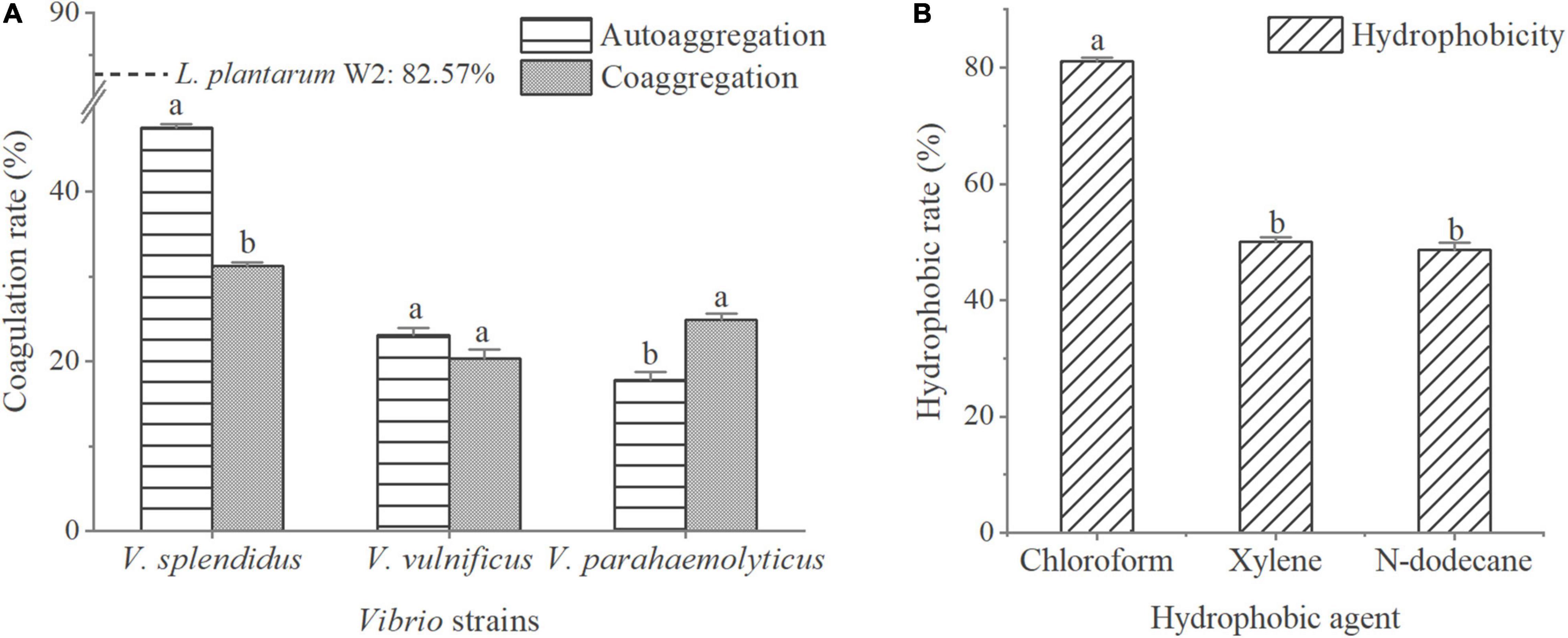
Figure 2. Adhesion properties of L. plantarum W2. (A) Autoaggregation rates of three Vibrio strains (V. splendidus BSD11, V. vulnificus S01P2, V. parahaemolyticus 20130629002S01) and their coaggregation rate with W2. Autoaggregation rate of W2 is indicated by a dotted line. (B) W2 adherence to hydrophobic agent (chloroform, xylene, n-dodecane). P < 0.05.
The extracellular protein/transmembrane protein genes associated with adhesion in W2 are presented in Table 1. 1 S-layer protein, 1 collagen binding protein (CnBP) and 9 mucus binding proteins (Mub) were identified in W2 based on Pfam and NR database annotations.
The antagonistic activity of W2 to seven aquatic pathogenic strains is shown in Figure 3A. The average inhibition zone diameter of W2 against pathogens (V. vulnificus S01P2, V. harveyi SRTT9, V. parahaemolyticus 20130629002S01, Streptococcus iniae NUF849) was > 20 mm. The diameters of the inhibition zones of different fermentation components of W2 against 2 Vibrio strains (V. parahaemolyticus 20130629002S01 and V. vulnificus S01P2) are shown in Figure 3B. The diameter of the inhibition zone of the suspension was 7.8 mm, which is the diameter of the Oxford cup and is considered as indicating no antagonistic activity. The zone diameters of the Fermentation broth and supernatant (V. parahaemolyticus: 26.2, 23.7; V. vulnficus: 27.3, 23.3) were significantly larger than 7.8 mm (P < 0.05), indicating that the antagonistic active substance existed in the supernatant. The diameter of the inhibition zones of the fermentation broth was larger than that of the supernatant, probably due to the continued production of antagonistic substances during the experiment. When the pH was adjusted to 6.0 in the fermentation broth and supernatant, their antagonistic activity disappeared. The inhibition zone diameter of the supernatant did not decrease after the removal of H2O2 and treatment with trypsin.
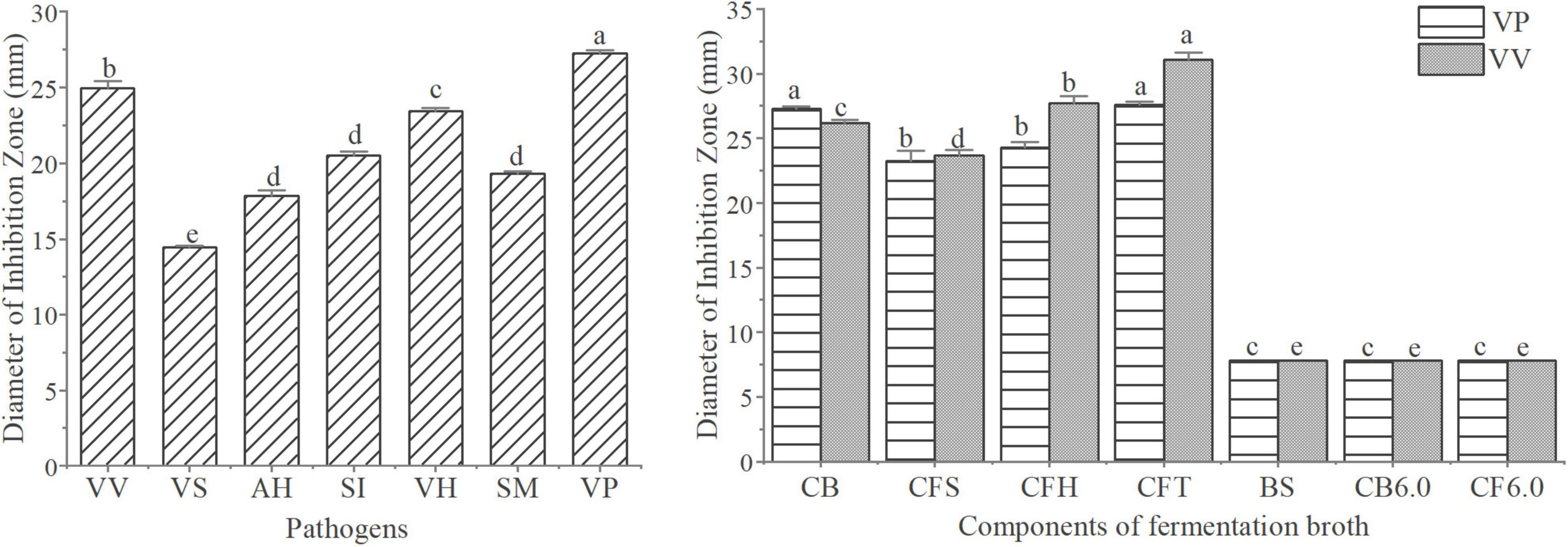
Figure 3. Analysis of the antagonistic activity of W2. (A) Antibacterial activitys of W2 against seven pathogen (V. vulnificus S01P2, V. splendidus BSD11, V. harveyi SRTT9, V. parahaemolyticus 20130629002S01, Streptococcus iniae NUF849, Aeromonas hydrophila AP40301, Shewanella marisflavi AP629). (B) Antibacterial activitys of different fermentation components of W2 against two Vibrios (V. parahaemolyticus 20130629002S01, V. vulnificus S01P2). The groups are as follows: culture broth (CB), cell free supernatant (CFS), cell free supernatant with H2O2 removal (CFH), cell free supernatant treated with tripsin (CFT), biomass suspension (BS), cell free supernatant at pH 6.0 (CB6.0), culture broth at pH 6.0 (CF6.0). P < 0.05.
MICs of seven antibiotics against W2 are presented in Table 2. The cut-off value represents the maximum value of MIC assigned by EFSA, which specifies the maximum resistance level the bacteria group should meet. The resistance of W2 to chloramphenicol (8 μg/ml), clindamycin (4 μg/ml), gentamicin (16 μg/ml) were in accordance with the cut-off values, the resistance to kanamycin (16 μg/ml), tetracycline (2 μg/ml) were below the cut-off values, while the resistance to ampicillin (8 μg/ml) and erythromycin (8 μg/ml) were above the cut-off values. This means that W2 can be inhibited in growth by lower concentrations of kanamycin and tetracycline than the cut-off values, but is resistant to ampicillin and erythromycin at cut-off value concentrations.
By alignment with the CARD database, 155 ARGs were annotated in the W2 WGS. Their detailed distributions in different antibiotic classes are presented in Supplementary Table 4. ARGs in W2 were mainly concentrated in macrolide, tetracycline, fluoroquinolone, phenicol, lincosamide, peptides, penam, streptogramin, and oxazolidinone antibiotic. Based on the MIC results, erythromycin and ampicillin resistance genes were analyzed in this study, and the results are presented in Supplementary Table 5. 50 macrolide resistance genes were identified and they were classified by resistance mechanism into 42 antibiotic effluxs, 7 antibiotic target protection, and 1 antibiotic target alteration. The antibiotic efflux includes 3 efrA, 2 evgA, 25 macB, 1 MexL, 3 mtrA, 5 oleC, 3 Staphylococcus aureus LmrS. Antibiotic target protection includes l mrC, 1 lsaA, 1 lsaC, 2 optrA, 1 tva(A), 1 vmlR. The antibiotic target alteration gene is Erm(K). 11 penam resistance genes were found, grouped into 7 resistance efflux pumps (3 mtrA, mgrA, 2 evgA, golS), 1 antibiotic target protection (mecI) and 3 antibiotic inactivation (Escherichia coli ampH beta-lactamase, y56 beta-lactamase, NmcR). In addition to the CARD, 5 penicillin-binding proteins (PBPs) were annotated using the Swiss-Prot database, 13 β-lactamases using the pfam database, 1 penicillin V acylase using the NR database and 1 penicillin amidase using the GO database. Some of the above genes may have mediated the additional high resistance of W2 to erythromycin and ampicillin. Vancomycin resistance genes are not present in W2.
There were 274 VFs annotated in W2 in the VFDB. Of these, 202 were attribute to specific I/II level, with a few genes in multiple II level functions (Figure 4). Defensive virulence factors, nonspecific virulence factors and regulation of virulence-associated genes accounted for 62.0%, and offensive virulence factors for 38.0%. Toxin accounted for 27.5% (22) of the offensive virulence factors, with the other factors annotated to adhesion, invasion and secretion systems. The 22 toxins included 4 Alpha-hemolysin, 11 Beta-hemolysin/cytolysin, 1 Cytolysin, 1 Colibactin, 1 Hemolysin III, 3 RTX toxins, and 1 TcdA. Their locations on the DNA are presented in Supplementary Table 6. The haemolysis test showed that both the fermentation supernatant of W2 and the MRS broth (negative control) represent γ haemolysis. This suggests that the haemolysin/cytolysin gene of W2 is not expressed, or slightly expressed without haemolytic effect.
In this study, MGEs in W2 (Table 3) were analyzed to assess the potential for horizontal transfer of ARGs and VFs. The W2 genome has only one circular DNA and no plasmids. The following MGEs were identified on the circular DNA: 6 genomic islands (GEIs), comprising a total of 200 genes; 2 prephages (Lactococcus phage P335 and Lactobacillus phage phig1e), comprising a total of 99 genes, overlapping with 2 of the 6 genomic islands; 3 insertion sequences (IS); 10 CRISPR sequences (CRISPRs). In addition, 8 possible integrases and 10 possible transposases (3 of which are insertion sequences) were identified by protein sequence comparison in the Swiss-Prot database. No complete integron was found.
Seven of the 155 ARGs were found located on GEIs. Of these, 4 mediated macrolide resistance (efrA, macB), 3 mediated fluoroquinolone resistance (patB, efrA), 2 mediated rifamycin resistance (efrA), 1 mediated nitroimidazole resistance (msbA), and 1 mediated tetracycline resistance (tetT). 10 of the 274 VFs are located on GEIs/pre-phages, with 2 genes encoding toxins (TcdA, RTX toxin). A total of 11 ARGs/VFs were located on GEIs, due to partial overlap between 7 ARGs and 10 VFs. In addition, 6 VFs and 4 ARGs are located near the integrase/transposase. The location of MGEs such as GEIs, prephages, IS, and the ARGs and VFs carried on them are shown in Figure 5 in circle 5, 4.
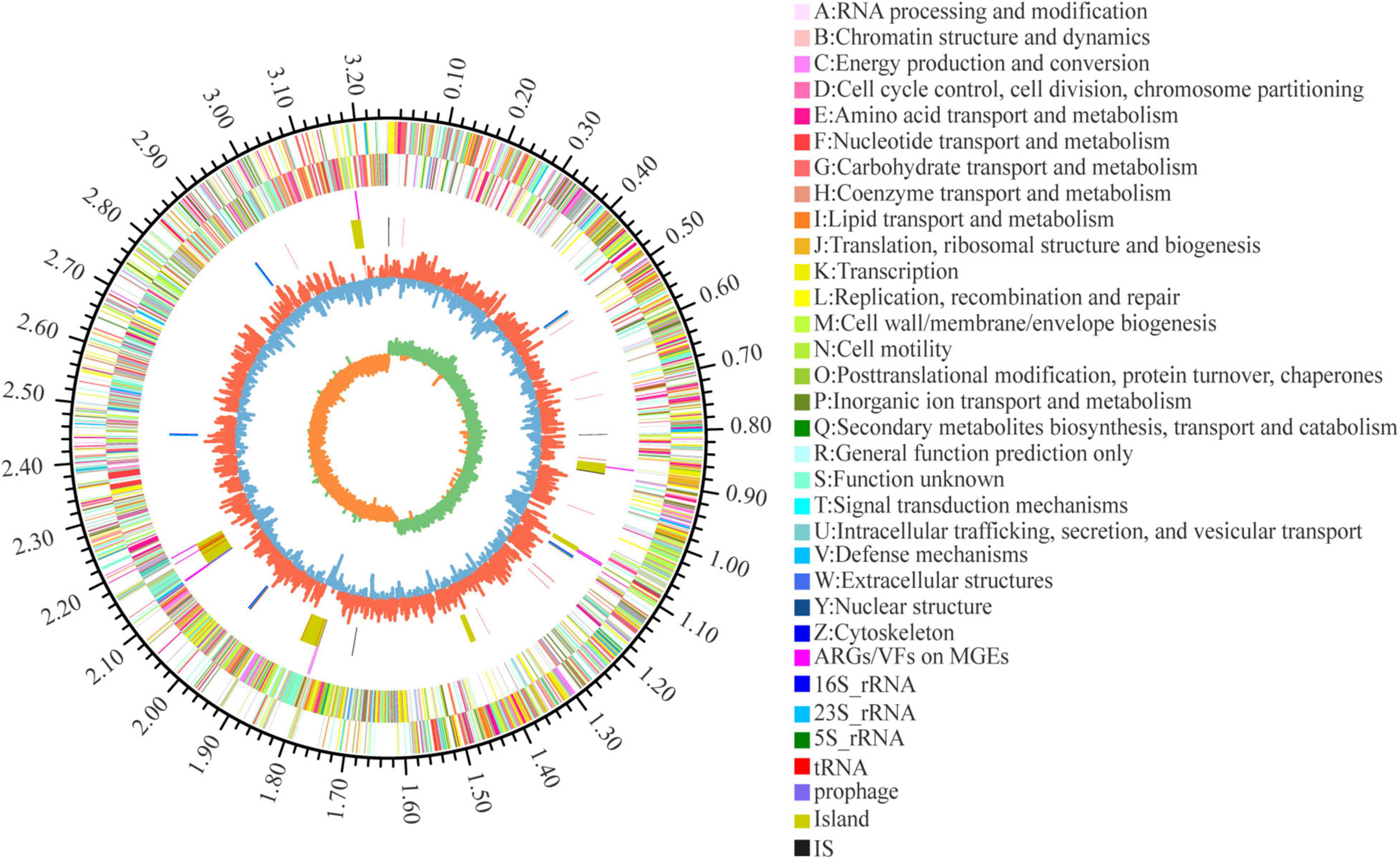
Figure 5. Circular representation of L. plantarum W2 genome. The outer scale is in mega bases (Mb). Circle 1 (from outside to inside), the marker of genome size. Cycles 2 and 3, CDS with positive and negative chain, different colors represent different functional classifications; Circle 4, ARGs and VFs located on genomic islands and prephages; Circle 5, islands and prephages (yellow), insertion sequences (IS, black), rRNA (blue) and tRNA (red); Circle 6, GC content, the higher value makes redder, the lower makes bluer. Circle 7, GC-skew value, skew+ are expressed in green, skew- are expressed in orange.
The growth performance of shrimp in each treatment is presented in Table 4. The data indicated that the specific growth rate (SGR) of shrimp of LLP and FLI were significantly higher than that of the control (P < 0.05). The feed utilization efficiency (FER) was significantly higher in the LLP and FLI than in the control, but there was no significant difference between the two treatments.
Levels of 8 serological immune indicators and expression levels of 12 hepatopancreatic immune-related genes of shrimp are shown in Figures 6, 7. AKP, ACP, TNOS, PO, LZM, POD, and T-AOC were significantly higher in the LLP than in the control. PO was significantly higher in the FLI than in the control, but significantly lower than LLP. SOD was not significantly different in the three groups. The relative expression levels of all 12 immune genes were significantly higher in the LLP group than in the control. Imd, Relish, TOR, proPO, and LGBP were significantly higher in the FLI group than in the control. In summary, levels of serological immune indicators showed the same trend as the relative expression levels of hepatopancreatic immune-related genes. W2 supplementary to the diet significantly improved the immune performance of shrimp (enhanced immune enzyme activity and up-regulation of immune gene expression). Antibiotics florfenicol supplementary to the diet also improved the immune performance of shrimp to some extent, but to a lesser extent than W2.
The cumulative mortality of P. vannamei in the challenge test is shown in Figure 8. The LLP group had the lowest cumulative mortality rate (25.0%) and was not significantly different from the FLI group (29.2%). The control group had a high cumulative mortality rate of 54.2%, which was significantly higher than the LLP and FLI (P > 0.05). Compared to the control, calculations showed a 53.8 and 46.2% reduction in cumulative mortality in the LLP and FLI groups, respectively.
As one of the eco-friendly alternatives to antibiotics, probiotics have been widely considered and their probiotic effect on aquaculture animals have been confirmed by previous studies (e.g., Li et al., 2019a,b; Tran et al., 2019; Yang et al., 2019). L. plantarum was included in the list of microorganisms that can be used in food in many countries and regions such as China, Europe, United States, Australia, and Malaysia (EFSA, 2007; Ministry of Health China, 2010). In this study, the probiotic properties of a L. plantarum W2 strain, isolated from the intestine of healthy Pacific white shrimp, were evaluated including the potential for adhesion, antagonistic activity in vitro and modulatory effects to growth, immunity and disease resistance of P. vannamei.
The adherence of probiotic to epithelial cells is considered to be an important requirement for their colonization and delivery of health effects (Collado et al., 2005; Mohanty et al., 2019). Bacteria develop a cluster through autoaggregation, which facilitates their effective adherence to the intestinal surface (Trunk et al., 2018). Meanwhile, coaggregation between probiotic and pathogen can indicate their intimate interactions (Mohanty et al., 2019). Strain W2 performed high autoaggregation (82.6%) and hydrophobicity (chloroform: 81.0%), which indicated a certain adhesion potential. Notably, its coaggregation with V. parahaemolyticus was significantly greater than the autoaggregation of V. parahaemolyticus, which might mean W2 could make an intimate connection with V. parahaemolyticus, allowing it to release antagonistic substances in a close position (Bujnakova and Kmet, 2002). As a result, W2 might be able to attenuate the adherence and colonization of V. parahaemolyticus to some extent.
Adhesion of Lactobacillus to mucin/epithelial cells is associated with a variety of non-specific and specific ligand-receptor interactions (Boks et al., 2008; Berne et al., 2018). It is generally agreed that the initial mechanism is linked to physicochemical interactions between bacteria and mucin/epithelial cells. Certain features of bacterial surfaces, such as electron-donor properties, surface charges and hydrophobicity, influence the forces of attraction and repulsion between bacteria and surfaces (Humphries et al., 2017; Ren et al., 2018). Xylene and n-dodecane are non-polar solvents and reflect the hydrophobicity of the bacteria. Chloroform is a monopolar and acidic solvent and it served as an indicator of the electron acceptor feature of the bacteria (Bellon-Fontaine et al., 1996). W2 demonstrated a significantly higher hydrophobicity toward chloroform, which meant it was a strong electron donor. This might play an important role in the early stages of adhesion. Meanwhile, special bacterial surface components, such as extracellular proteins/transmembrane proteins, can strengthen the initial contact and play a major role in adhesion (Bath et al., 2005; Tallon et al., 2007; Berne et al., 2013). Previous studies have revealed a variety of proteins associated with Lactobacillus adhesion, including S-layer protein (Mohanty et al., 2019), collagen-binding protein (CnBP) (Roos et al., 1996), mucus-binding protein (Mub) (Roos and Jonsson, 2002), mannose-specific adhesin (Msa) (Pretzer et al., 2005), mucus-binding flagellin (SpaC), and cell wall-anchored protein CwaA (Buntin et al., 2017). In this study, 1 S-layer protein, 1 CnBP and 9 Mubs were annotated in W2 genome. These proteins may help W2 to adhere to mucus/intestinal epithelium and further research into this aspect is needed for validation.
In vitro antagonistic activity against pathogens is one of the more important aspects for the evaluation of beneficial properties of probiotics (Fijan et al., 2018; Doan et al., 2021). In this study, L. plantarum W2 showed strong antagonistic activity against seven common aquatic pathogenic bacteria, with inhibition zone diameters ranging from 13.6 to 27.3 mm. Typical antagonistic substances found in lactic acid bacteria include hydrogen peroxide (Bao et al., 2010), proteins and peptides (Selegard et al., 2019), organic acids (Sanchez-Maldonado et al., 2011; Arena et al., 2016), diacetyl and other compounds (Yan et al., 2019). The effective antibacterial substances of W2 existed in the supernatant and antagonistic activity was pH-dependent. Such pH-dependent antagonistic substances were also found in other strains of L. plantarum. Russo et al. (2017) demonstrated that the antagonistic ability of a L. plantarum strain against 6 filamentous fungi, including Aspergillus niger, derived from organic acids, with lactic acid playing an important role. Gerez et al. (2010) found that the antifungal activity of L. plantarum SL778 was associated with acetic acid, phenyllactic acid, and lactic acid. They also found that higher concentrations of lactic acid production favored the combined antagonistic activity of organic acids. In addition, the accumulation of antagonistic activity occurred when more than one organic acid was involved (Sun et al., 2003). Accordingly, it could be presumed that the antagonistic activity of W2 against the seven pathogens might be derived from organic acids. The exact components of these organic acids still need to be further analyzed in future study.
Until now, the in vitro efficacy and potency of many probiotics against numerous pathogens have been investigated under various conditions (Fijan et al., 2018; Doan et al., 2021). There are, however, many gaps and questions remaining to be clarified, especially the determination of the level of correlation of the in vitro antagonistic activity and the in vivo disease resistance of putative probiotics against pathogenic strains. Strain W2 was found to significantly increase the immune performance of shrimp, as evidenced by a general increase in serum immune enzyme activities and hepatopancreas immune-related gene expression levels. In the challenge test, W2 supplementation significantly enhanced in vivo pathogen resistance of shrimp under V. parahaemolyticus exposure. These results were in accordance with other studies. For example, Zheng et al. (2017) measured the expression levels of non-specific immune genes before and after acute hyposalinity stress in shrimp and showed that dietary supplementation with L. plantarum significantly improved the stress resistance and immune performance of shrimp. Dash et al. (2015) demonstrated that dietary inactivated L. plantarum could enhance non-specific immunity in P. vannamei. Chomwong et al. (2018) found that the use of L. plantarum promoted the immune response of shrimp to VPAHPND infection and improved survival. Based on the above, L. plantarum W2 revealed better performance in vivo efficacy and potency of improving the immune capacity and disease resistance in shrimp.
In recent years, there has been increasing concern about the safety of probiotics due to the potential transfer and spread of ARGs and VFs among microorganisms (Wang et al., 2008; Doron and Snydman, 2015). A systematic safety assessment of L. plantarum W2 was conducted based on the latest EFSA guidelines for feed-added probiotics (EFSA, 2018) and other relevant studies (e.g., FAO/WHO, 2002; EFSA, 2008; WHO, 2014). The evaluation mainly focused on phenotypic and genotypic studies of antibiotic resistance and virulence, and to some extent on the biological and ecological risks of these genes. In this study, 274 VFs were found in the W2 genome although, fortunately, none was any of the genes that need to be excluded according to EFSA guidelines. The 274 VFs mainly perform auxiliary functions such as adhesion, anti-phagocytosis, ion uptake, secretion, and regulation. For pathogenic strains, adhesion and iron uptake are important for their colonization and invasion (Becker and Skaar, 2014; Carpenter and Payne, 2014). However, for probiotics, numerous studies have shown that adhesion to the intestinal surface is essential to its developing probiotic effects. L. plantarum is a kind of bacteria that does not need to absorb iron from its environment (Archibald, 2006). Therefore, some of the VFs might be important for probiotics to exert beneficial effects and should not be considered as malignant virulence factors.
There were 22 toxins in the 274 VFs, including 17 haemolysins/cytolysins, but strain W2 showed no hemolytic activity. These genes may not be expressed in W2, or weakly expressed. Other non-haemolytic toxins, including Colibactin, RTX toxin and TcdA, were difficult to identify as toxic to organisms if they were not highly expressed. Some studies have shown that the expression of some toxins, such as TcdA (Kuehne et al., 2010), can activate host innate immunity to some extent. VFs may be transferred from non-pathogenic bacteria to pathogens through MGEs-mediated Horizontal Gene Transfer (HGT) (von Wintersdorff et al., 2016; Brito, 2021). Gene mapping indicated that 10 VFs of W2 were located on GEIs and 6 were located near integrase/transposases, including no haemolysins/cytolysins genes. These genes may have some risk of HGT but have no predictable virulence to biology (Kuehne et al., 2010; Satchell, 2015).
Currently, antimicrobial resistance has been considered to be a global public health threat (WHO, 2014; CDC, 2019). W2 was considered as innately resistant to kanamycin, chloramphenicol, clindamycin, gentamicin, and tetracycline. However, the MICs of erythromycin and ampicillin against W2 were higher than their cut-off values (erythromycin: 1 μg/ml, ampicillin: 2 μg/ml). A new phenotype may signal a newly acquired gene. Gene mapping showed that none of the penicillin ARGs were located on GEIs/pre-phages or in the near vicinity of integrases and transposases. Thus, penicillin resistance of W2 might be innate resistance. Similarly, ampicillin resistance in L. plantarum was also described as innate resistance in other studies. For example, laboratory adaptive evolution experiments showed that chronic exposure to ampicillin up-regulated MIC of L. plantarum strain, but ampicillin ARGs transfer risk was low (Cao et al., 2020). Gene mapping showed that there are 4 macrolide efflux pump genes located on GEIs in W2. These genes might be acquired genes. HGT is a powerful motivator for bacterial evolution. It is not surprising that L. plantarum, a class of metabolically rich and adaptive bacteria, has acquired new genes from the environment. The increased resistance of W2 to erythromycin might derive from these genes in part, for which further evidence is still needed in future research.
Overall, a total of 11 ARGs, in which 7 are located on GEIs and 4 are located near integrases or transposases, were at some risk of gene transfer. However, on the one hand not all ARGs pose a severe threat to public health. Some genes or sequence homology predicted genes that have conferred antibiotic resistance are widespread in bacteria and play biological roles, such as efflux systems (Blair et al., 2015) and intercellular signaling (Davies and Davies, 2010). On the other hand, genes with transfer potential are not always prevalent on a large scale or have detrimental effects to humans. For example, a recent study by Zhang et al. (2021) suggested that the risk assigned to vanA and sul1 resistance families (resistant to vancomycin and sulforaphane, respectively) should be weakened. These two resistant families belong to the 37 ARG families of Rank I listed by WHO (2019). Studies have shown that they are characterized by gene transfer and host pathogenicity but show weak correlation with human activity. Because gene transfer is constrained by the influence of complex regulatory factors, there may be many unknown environmental and host factors (Wagner et al., 2017). Zhang et al. (2021) classified high-risk mobile ARGs into two classes: Rank I, containing 73 gene families that currently have a high risk of conferring new or multiple drug resistance to pathogen; Rank II, containing 19 gene families that may be transferred to pathogens in new forms of resistance in the future. In this study, none of the ARGs in W2 belonged to Rank I and 3 genes belonged to the mdtG family of Rank II, which conferred multi-drug resistance. However, the 3 genes were neither located on GEIs nor in the vicinity of integrases/transposases. Based the above, W2 may have high biological and ecological safety profiles, in terms of virulence and antimicrobial resistance.
Generally, L. plantarum is a common lactic acid bacterium with a long history of safe use and rarely leads to human infection (Bernardeau et al., 2006; Haghighat and Crum-Cianflone, 2016; Le and Yang, 2018; Nayeem et al., 2018; Koyama et al., 2019). As a potential probiotic strain for shrimp, W2 did not carry highly virulent factors by genome analysis, and its non-haemolytic activity was confirmed by haemolysis tests. Meanwhile, according to the omics-based framework for assessing the health risk of antimicrobial resistance genes (Zhang et al., 2021), W2 also has a low risk of facilitating the formation of multidrug resistant pathogens by conferring ARGs to pathogen both in the present and foreseeable future. Hence, based on the above collectively, it could be believed that W2 should have no or low potential risk to public health.
The probiotic properties and safety of L. plantarum W2 were evaluated in this study. Strain W2 represented no significant virulence effect. W2 showed antibiotic resistance to ampicillin and erythromycin to a certain degree, i.e., inherently ampicillin resistant but might acquire resistance to erythromycin. However, it was confirmed that none of the ARGs in W2 belong to the high-risk mobile ARGs. Strain W2 has the ability to adhere to surfaces and can attenuate the adhesion of V. parahaemolyticus. Seven aquatic pathogenic strains were in vitro inhibited efficiently, and organic acids might be the main antagonistic substance produced by W2. Dietary W2 with 1 × 1010 cfu/kg live cells significantly improved the growth performance, enhanced levels of serological immune indicators and the expression levels of hepatopancreatic immune genes of shrimp and reduced the mortality of shrimp exposed to V. parahaemolyticus. Based on these results, L. plantarum W2 can be applied safely as a potential probiotic in shrimp farming to enhance growth performance, immunity capacity and disease resistance of P. vannamei. Additionally, the results from this study would be helpful to develop criteria for the screening and evaluation of probiotics in aquaculture.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA797524.
All animal experiments were conducted in accordance with the guidelines and approval of the respective Animal Research and Ethics Committees of Ocean University of China (Permit Number: 20141201; http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
CW performed the experiments, analyzed the data, and wrote the manuscript with assistance of KL, MW, YML, MP, YX, GQ, YJL, and LL. QL helped to collect and maintain the animals. XT provided the experimental ideas and the design of this study. All authors contributed to the article and approved the submitted version.
This study was funded by the National Key Research and Development Program of China (2020YFD0900201 and 2019YFD0900403), the Joint Fund of Think Tank for Biomanufacturing Industry of Qingdao (QDSWZK202111) and Joint Fund of Ocean University of China—Tarim University.
QL was employed by the Qingdao Ruizi Group Co.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Laboratory of Aquaculture Ecology, Ocean University of China, for the support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.854131/full#supplementary-material
Archibald, F. (2006). Lactobacillus plantarum, an organism not requiring Iron. FEMS Microbiol. Lett. 19, 29–32. doi: 10.1111/j.1574-6968.1983.tb00504.x
Arena, M. P., Silvain, A., Normanno, G., Grieco, F., Drider, D., Spano, G., et al. (2016). Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 7:464. doi: 10.3389/fmicb.2016.00464
Bao, Y., Zhang, Y., Zhang, Y., Liu, Y., Wang, S., Dong, X., et al. (2010). Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 21, 695–701. doi: 10.1016/j.foodcont.2009.10.010
Bath, K., Roos, S., Wall, T., and Jonsson, H. (2005). The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 253, 75–82. doi: 10.1016/j.femsle.2005.09.042
Beck, B. R., Kim, D., Jeon, J., Lee, S.-M., Kim, H. K., Kim, O.-J., et al. (2015). The effects of combined dietary probiotics Lactococcus lactis BFE920 and Lactobacillus plantarum FGL0001 on innate immunity and disease resistance in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 42, 177–183. doi: 10.1016/j.fsi.2014.10.035
Becker, K. W., and Skaar, E. P. (2014). Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 38, 1235–1249. doi: 10.1111/1574-6976.12087
Bellon-Fontaine, M. N., Rault, J., and van Oss, C. J. (1996). Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B Biointerfaces 7, 47–53. doi: 10.1016/0927-7765(96)01272-6
Bernardeau, M., Guguen, M., and Vernoux, J. P. (2006). Beneficial lactobacilli in food and feed: long-term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol. Rev. 30, 487–513. doi: 10.1111/j.1574-6976.2006.00020.x
Berne, C., Ellison, C. K., Ducret, A., and Brun, Y. V. (2018). Bacterial adhesion at the single-cell level. Nat. Rev. Microbiol. 16, 616–627. doi: 10.1038/s41579-018-0057-5
Berne, C., Ma, X., Licata, N. A., Neves, B. R. A., Setayeshgar, S., Brun, Y. V., et al. (2013). Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J. Phys. Chem. B 117, 10492–10503. doi: 10.1021/jp405802e
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Betancur, C., Martinez, Y., Merino-Guzman, R., Hernandez-Velasco, X., Castillo, R., Rodriguez, R., et al. (2020). Evaluation of oral administration of Lactobacillus plantarum cam6 strain as an alternative to antibiotics in weaned pigs. Anim. Basel 10:1218. doi: 10.3390/ani10071218
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. V. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Bland, C., Ramsey, T. L., Sabree, F., Lowe, M., Brown, K., Kyrpides, N. C., et al. (2007). CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209
Boks, N. P., Norde, W., van der Mei, H. C., and Busscher, H. J. (2008). Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiol. Soc. 154, 3122–3133. doi: 10.1099/mic.0.2008/018622-0
Brito, I. L. (2021). Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 19, 442–453. doi: 10.1038/s41579-021-00534-7
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Bujnakova, D., and Kmet, V. (2002). Aggregation of animal lactobacilli with O157 enterohemorrhagic Escherichia coli. J. Vet. Med. B Infect. Dis. Vet. Public Health 49, 152–154. doi: 10.1046/j.1439-0450.2002.00526.x
Buntin, N., de Vos, W. M., and Hongpattarakere, T. (2017). Variation of mucin adhesion, cell surface characteristics, and molecular mechanisms among Lactobacillus plantarum isolated from different habitats. Appl. Microbiol. Biotechnol. 101, 7663–7674. doi: 10.1007/s00253-017-8482-3
Campedelli, I., Mathur, H., Salvetti, E., Clarke, S., Rea, M. C., Torriani, S., et al. (2019). Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 85:e01738-18. doi: 10.1128/AEM.01738-18
Cao, C., Wang, J., Liu, Y., Kwok, L. Y., Zhang, H., and Zhang, W. (2020). Adaptation of Lactobacillus plantarum to ampicillin involves mechanisms that maintain protein homeostasis. mSystems 5:e00853-19. doi: 10.1128/mSystems.00853-19
Carpenter, C., and Payne, S. M. (2014). Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem. 133, 110–117. doi: 10.1016/j.jinorgbio.2014.01.007
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chiu, C. H., Guu, Y. K., Liu, C. H., Pan, T. M., and Cheng, W. (2007). Immune responses and gene expression in white shrimp, Litopenaeus vannamei, induced by Lactobacillus plantarum. Fish Shellfish Immunol. 23, 364–377. doi: 10.1016/j.fsi.2006.11.010
Chomwong, S., Charoensapsri, W., Amparyup, P., and Tassanakajon, A. (2018). Two host gut-derived lactic acid bacteria activate the proPO system and increase resistance to an AHPND-causing strain of Vibrio parahaemolyticus in the shrimp Litopenaeus vannarnei. Dev. Comp. Immunol. 89, 54–65. doi: 10.1016/j.dci.2018.08.002
Collado, M. C., Gueimonde, M., HernÁNdez, M., Sanz, Y., and Salminen, S. (2005). Adhesion of selected Bifidobacterium strains to human intestinal mucus and the role of adhesion in enteropathogen exclusion. J. Food Prot. 68, 2672–2678. doi: 10.4315/0362-028X-68.12.2672
Conesa, A., and Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. doi: 10.1155/2008/619832
Cury, J., Jove, T., Touchon, M., Neron, B., and Rocha, E. P. C. (2016). Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 44, 4539–4550. doi: 10.1093/nar/gkw319
Dash, G., Raman, R. P., Prasad, K. P., Makesh, M., Pradeep, M. A., and Sen, S. (2015). Evaluation of paraprobiotic applicability of Lactobacillus plantarum in improving the immune response and disease protection in giant freshwater prawn, Macrobrachium rosenbergii (de Man, 1879). Fish Shellfish Immunol. 43, 167–174. doi: 10.1016/j.fsi.2014.12.007
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Doan, H. V., Soltani, M., and Ringø, E. (2021). In vitro antagonistic effect and in vivo protective efficacy of Gram-positive probiotics versus Gram-negative bacterial pathogens in finfish and shellfish. Aquaculture 540:736581. doi: 10.1016/j.aquaculture.2021.736581
Dong, X., Bi, D., Wang, H., Zou, P., Xie, G., Wan, X., et al. (2017). pirABvp-bearing Vibrio parahaemolyticus and Vibrio campbellii pathogens isolated from the same AHPND-affected pond possess highly similar pathogenic plasmids. Front. Microbiol. 8:1859. doi: 10.3389/fmicb.2017.01859
Doron, S., and Snydman, D. R. (2015). Risk and safety of probiotics. Clin. Infect. Dis. 60(Suppl. 2), S129–S134. doi: 10.1093/cid/civ085
Eddy, S. R. (2009). A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211. doi: 10.1142/9781848165632_0019
EFSA (2007). Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA – Opinion of the Scientific Committee. EFSA J. 5:587. doi: 10.2903/j.efsa.2007.587
EFSA (2008). The maintenance of the list of QPS microorganisms intentionally added to food or feed-Scientific opinion of the panel on biological hazards. EFSA J. 6:923. doi: 10.2903/j.efsa.2008.923
EFSA (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:e05206. doi: 10.2903/j.efsa.2018.5206
El Shazely, B., Yu, G., Johnston, P. R., and Rolff, J. (2020). Resistance evolution against antimicrobial peptides in Staphylococcus aureus alters pharmacodynamics beyond the MIC. Front. Microbiol. 11:103. doi: 10.3389/fmicb.2020.00103
Fijan, S., Šulc, D., and Steyer, A. (2018). Study of the in vitro antagonistic activity of various single-strain and multi-strain probiotics against Escherichia coli. Int. J. Environ. Res. Public Health 15:1539. doi: 10.3390/ijerph15071539
Fouts, D. E. (2006). Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 34, 5839–5851. doi: 10.1093/nar/gkl732
Gerez, C. L., Torino, M. I., Obregozo, M. D., and Font de Valdez, G. (2010). A ready-to-use antifungal starter culture improves the shelf life of packaged bread. J. Food Prot. 73, 758–762. doi: 10.4315/0362-028x-73.4.758
Giri, S. S., Sukumaran, V., and Oviya, M. (2013). Potential probiotic Lactobacillus plantarum VSG3 improves the growth, immunity, and disease resistance of tropical freshwater fish, Labeo rohita. Fish Shellfish Immunol. 34, 660–666. doi: 10.1016/j.fsi.2012.12.008
Haghighat, L., and Crum-Cianflone, N. F. (2016). The potential risks of probiotics among HIV-infected persons: bacteraemia due to Lactobacillus acidophilus and review of the literature. Int. J. STD AIDS 27, 1223–1230. doi: 10.1177/0956462415590725
Humphries, J., Xiong, L., Liu, J., Prindle, A., Yuan, F., Arjes, H. A., et al. (2017). Species-independent attraction to biofilms through electrical signaling. Cell 168, 200–209. doi: 10.1016/j.cell.2016.12.014
Kongnum, K., and Hongpattarakere, T. (2012). Effect of Lactobacillus plantarum isolated from digestive tract of wild shrimp on growth and survival of white shrimp (Litopenaeus vannamei) challenged with Vibrio harveyi. Fish Shellfish Immunol. 32, 170–177. doi: 10.1016/j.fsi.2011.11.008
Kos, B., Šušković, J., Vuković, S., Šimpraga, M., Frece, J., and Matošić, S. (2003). Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94, 981–987. doi: 10.1046/j.1365-2672.2003.01915.x
Koyama, S., Fujita, H., Shimosato, T., Kamijo, A., Ishiyama, Y., Yamamoto, E., et al. (2019). Septicemia from Lactobacillus rhamnosus GG, from a probiotic enriched yogurt, in a patient with autologous stem cell transplantation. Probiotics Antimicrob. Proteins 11, 295–298. doi: 10.1007/s12602-018-9399-6
Kuang, C. L., Lv, D., Shen, G. H., Li, S. S., Luo, Q. Y., and Zhang, Z. Q. (2018). Chemical composition and antimicrobial activities of volatile oil extracted from Chrysanthemum morifolium Ramat. J. Food Sci. Technol. 55, 2786–2794. doi: 10.1007/s13197-018-3203-1
Kuehne, S. A., Cartman, S. T., Heap, J. T., Kelly, M. L., Cockayne, A., and Minton, N. P. (2010). The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713. doi: 10.1038/nature09397
Lau, L. Y. J., and Chye, F. Y. (2018). Antagonistic effects of Lactobacillus plantarum 0612 on the adhesion of selected foodborne enteropathogens in various colonic environments. Food Control 91, 237–247. doi: 10.1016/j.foodcont.2018.04.001
Le, B., and Yang, S. H. (2018). Efficacy of Lactobacillus plantarum in prevention of inflammatory bowel disease. Toxicol. Rep. 5, 314–317. doi: 10.1016/j.toxrep.2018.02.007
Lee, S., Katya, K., Park, Y., Won, S., Seong, M., Hamidoghli, A., et al. (2017). Comparative evaluation of dietary probiotics Bacillus subtilis WB60 and Lactobacillus plantarum KCTC3928 on the growth performance, immunological parameters, gut morphology and disease resistance in Japanese eel, Anguilla japonica. Fish Shellfish Immunol. 61, 201–210. doi: 10.1016/j.fsi.2016.12.035
Li, H., Qiao, G., Li, Q., Zhou, W., Won, K. M., Xu, D., et al. (2010). Biological characteristics and pathogenicity of a highly pathogenic Shewanella marisflavi infecting sea cucumber, Apostichopus japonicus. J. Fish Dis. 33, 865–877. doi: 10.1111/j.1365-2761.2010.01189.x
Li, H., Tian, X., Zhao, K., Jiang, W., and Dong, S. (2019a). Effect of Clostridium butyricum in different forms on growth performance, disease resistance, expression of genes involved in immune responses and mTOR signaling pathway of Litopenaeus vannamei. Fish Shellfish Immunol. 87, 13–21. doi: 10.1016/j.fsi.2018.12.069
Li, H., Tian, X., and Dong, S. (2019b). Growth performance, non-specific immunity, intestinal histology and disease resistance of Litopenaeus vannamei fed on a diet supplemented with live cells of Clostridium butyricum. Aquaculture 498, 470–481. doi: 10.1016/j.aquaculture.2018.09.003
Ministry of Health China (2010). List of Strains that can be Used for Food. No. 65. Beijing: Ministry of Health of the People’s Republic of China.
Miquel, S., Beaumont, M., Martín, R., Langella, P., Braesco, V., and Thomas, M. (2015). A proposed framework for an appropriate evaluation scheme for microorganisms as novel foods with a health claim in Europe. Microb. Cell Fact. 14:48. doi: 10.1186/s12934-015-0229-1
Mohanty, D., Panda, S., Kumar, S., and Ray, P. (2019). In vitro evaluation of adherence and anti-infective property of probiotic Lactobacillus plantarum DM 69 against Salmonella enterica. Microb. Pathog. 126, 212–217. doi: 10.1016/j.micpath.2018.11.014
Nayeem, M., Firdous, N., Dang, M.-T., Hafeez, W., and Arsene, C. (2018). When the good becomes the bad: a case of Lactobacillus rhamnosus septicemia unrelated to probiotic use. Am. J. Gastroenterol. 113:S1220.
Nurhajati, J., Atira, Aryantha, I. N. P., and Indah, K. D. G. (2012). The curative action of Lactobacillus plantarum FNCC 226 to Saprolegnia parasitica A3 on catfish (Pangasius hypophthalamus Sauvage). Int. Food Res. J. 19, 1723–1727.
Picchietti, S., Mazzini, M., Taddei, A. R., Renna, R., Fausto, A. M., Mulero, V., et al. (2007). Effects of administration of probiotic strains on GALT of larval gilthead seabream: immunohistochemical and ultrastructural studies. Fish Shellfish Immunol. 22, 57–67. doi: 10.1016/j.fsi.2006.03.009
Pretzer, G., Snel, J., Molenaar, D., Wiersma, A., Bron, P. A., Lambert, J., et al. (2005). Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187, 6128–6136. doi: 10.1128/jb.187.17.6128-6136.2005
Reid, G., Gadir, A. A., and Dhir, R. (2019). Probiotics: reiterating what they are and what they are not. Front. Microbiol. 10:424. doi: 10.3389/fmicb.2019.00424
Ren, Y., Wang, C., Chen, Z., Allan, E., van der Mei, H. C., and Busscher, H. J. (2018). Emergent heterogeneous microenvironments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing. FEMS Microbiol. Rev. 42, 259–272. doi: 10.1093/femsre/fuy001
Roos, S., Aleljung, P., Robert, N., Lee, B., Wadstrom, T., Lindberg, M., et al. (1996). A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol. Lett. 144, 33–38. doi: 10.1111/j.1574-6968.1996.tb08505.x
Roos, S., and Jonsson, H. (2002). A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148, 433–442. doi: 10.1099/00221287-148-2-433
Russo, P., Arena, M. P., Fiocco, D., Capozzi, V., Drider, D., and Spano, G. (2017). Lactobacillus plantarum with broad antifungal activity: a promising approach to increase safety and shelf-life of cereal-based products. Int. J. Food Microbiol. 247, 48–54. doi: 10.1016/j.ijfoodmicro.2016.04.027
Sanchez-Maldonado, A. F., Schieber, A., and Gaenzle, M. G. (2011). Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J. Appl. Microbiol. 111, 1176–1184. doi: 10.1111/j.1365-2672.2011.05141.x
Santajit, S., and Indrawattana, N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016:2475067. doi: 10.1155/2016/2475067
Satchell, K. J. F. (2015). Multifunctional-autoprocessing repeats-in-toxin (MARTX) toxins of Vibrios. Microbiol. Spectr. 3:3. doi: 10.1128/microbiolspec.VE-0002-2014
Selegard, R., Musa, A., Nystroem, P., Aili, D., Bengtsson, T., and Khalaf, H. (2019). Plantaricins markedly enhance the effects of traditional antibiotics against Staphylococcus epidermidis. Future Microbiol. 14, 195–206. doi: 10.2217/fmb-2018-0285
Sheng, X., Gao, J., Liu, H., Tang, X., Xing, J., and Zhan, W. (2018). Recombinant phosphoglucomutase and CAMP factor as potential subunit vaccine antigens induced high protection against Streptococcus iniae infection in flounder (Paralichthys olivaceus). J. Appl. Microbiol. 125, 997–1007. doi: 10.1111/jam.13948
Siguier, P., Gourbeyre, E., and Chandler, M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891. doi: 10.1111/1574-6976.12067
Sun, C. Q., O’Connor, C. J., and Roberton, A. M. (2003). Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Med. Microbiol. 36, 9–17. doi: 10.1016/s0928-8244(03)00008-7
Tallon, R., Arias, S., Bressollier, P., and Urdaci, M. C. (2007). Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J. Appl. Microbiol. 102, 442–451. doi: 10.1111/j.1365-2672.2006.03086.x
Thammasorn, T., Jitrakorn, S., Charoonnart, P., Sirimanakul, S., Rattanarojpong, T., Chaturongakul, S., et al. (2017). Probiotic bacteria (Lactobacillus plantarum) expressing specific double-stranded RNA and its potential for controlling shrimp viral and bacterial diseases. Aquac. Int. 25, 1679–1692. doi: 10.1007/s10499-017-0144-z
Tran, T. T., Bott, N. J., Lam, N. D., Nguyen, N. T., and Chu, H. H. (2019). The role of pseudomonas in heterotrophic nitrification: a case study on shrimp ponds (Litopenaeus vannamei) in Soc Trang province. Microorganisms 7:155. doi: 10.3390/microorganisms7060155
Trunk, T., Khalil, H. S., and Leo, J. C. (2018). Bacterial autoaggregation. AIMS Microbiol. 4, 140–164. doi: 10.3934/microbiol.2018.1.140
Van Doan, H., Hoseinifar, S. H., Dawood, M. A. O., Chitmanat, C., and Tayyamath, K. (2017). Effects of Cordyceps militaris spent mushroom substrate and Lactobacillus plantarum on mucosal, serum immunology and growth performance of Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 70, 87–94. doi: 10.1016/j.fsi.2017.09.002
Vendrell, D., Balcazar, J. L., de Blas, I., Ruiz-Zarzuela, I., Girones, O., and Muzquiz, J. L. (2008). Protection of rainbow trout (Oncorhynchus mykiss) from lactococcosis by probiotic bacteria. Comp. Immunol. Microbiol. Infect. Dis. 31, 337–345. doi: 10.1016/j.cimid.2007.04.002
Vieira, F. N., Buglione, C. C., Mourino, J. P. L., Jatoba, A., Martins, M. L., Schleder, D. D., et al. (2010). Effect of probiotic supplemented diet on marine shrimp survival after challenge with Vibrio harveyi. Arq. Bras. Med. Vet. Zootec. 62, 631–638. doi: 10.1590/s0102-09352010000300019
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Wagner, A., Whitaker, R. J., Krause, D. J., Heilers, J.-H., van Wolferen, M., van der Does, C., et al. (2017). Mechanisms of gene flow in archaea. Nat. Rev. Microbiol. 15, 492–501. doi: 10.1038/nrmicro.2017.41
Wang, D. D., Li, J. H., Zhao, K., Jiang, W. W., Li, H. D., Wang, W. J., et al. (2020). Mechanism of the potential therapeutic candidate Bacillus subtilis BSXE-1601 against shrimp pathogenic Vibios and multifunctional metabolites biosynthetic capability of the strain a s predicted by genome analysis. Front. Microbiol. 11:581802. doi: 10.3389/fmicb.2020.581802
Wang, Y. B., Li, J. R., and Lin, J. (2008). Probiotics in aquaculture: challenges and outlook. Aquaculture 281, 1–4. doi: 10.1016/j.aquaculture.2008.06.002
WHO (2014). Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization.
WHO (2019). Antibacterial Agents in Clinical Development an Analysis of the Antibacterial Clinical Development Pipeline. Geneva: World Health Organization.
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xu, H. Y., Liu, W. J., Zhang, W. Y., Yu, J., Song, Y. Q., Menhe, B., et al. (2015). Use of multilocus sequence typing to infer genetic diversity and population structure of Lactobacillus plantarum isolates from different sources. BMC Microbiol. 15:241. doi: 10.1186/s12866-015-0584-4
Xu, Y., Yao, H., Guo, Q., Li, H., Ye, S., Li, R., et al. (2018). Isolation, virulence and ERIC-PCR genotyping of Aeromonas hydrophila in farmed turbot Scophthalmus maximus (L.). J. Dalian Ocean Univ. 33, 430–434. doi: 10.16535/j.cnki.dlhyxb.2018.04.003
Yan, X., Gu, S., Cui, X., Shi, Y., Wen, S., Chen, H., et al. (2019). Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 127, 12–20. doi: 10.1016/j.micpath.2018.11.039
Yang, G., Tian, X., and Dong, S. (2019). Bacillus cereus and rhubarb regulate the intestinal microbiota of sea cucumber (Apostichopus japonicus Selenka): species-species interaction, network, and stability. Aquaculture 512:734284. doi: 10.1016/j.aquaculture.2019.734284
Zhang, A. N., Gaston, J. M., Dai, C. L., Zhao, S., Poyet, M., Groussin, M., et al. (2021). An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 12:4765. doi: 10.1038/s41467-021-25096-3
Zheng, X., Duan, Y., Dong, H., and Zhang, J. (2017). Effects of dietary Lactobacillus plantarum in different treatments on growth performance and immune gene expression of white shrimp Litopenaeus vannamei under normal condition and stress of acute low salinity. Fish Shellfish Immunol. 62, 195–201. doi: 10.1016/j.fsi.2017.01.015
Keywords: probiotic safety, virulence, antibiotic resistance, genomic analysis, probiotics, Lactobacillus plantarum, Penaeus vannamei
Citation: Wei C, Luo K, Wang M, Li Y, Pan M, Xie Y, Qin G, Liu Y, Li L, Liu Q and Tian X (2022) Evaluation of Potential Probiotic Properties of a Strain of Lactobacillus plantarum for Shrimp Farming: From Beneficial Functions to Safety Assessment. Front. Microbiol. 13:854131. doi: 10.3389/fmicb.2022.854131
Received: 13 January 2022; Accepted: 28 February 2022;
Published: 24 March 2022.
Edited by:
Qingyun Yan, Sun Yat-sen University, ChinaCopyright © 2022 Wei, Luo, Wang, Li, Pan, Xie, Qin, Liu, Li, Liu and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangli Tian, eGlhbmdsaXRpYW5Ab3VjLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.