
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 March 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.852434
This article is part of the Research TopicAntimicrobial Resistance in Foodborne Pathogens: ecology, epidemiology, and mechanismsView all 12 articles
 Jianzhong Fan1†
Jianzhong Fan1† Heng Cai2,3,4†
Heng Cai2,3,4† Youhong Fang5†
Youhong Fang5† Jintao He2,3,4
Jintao He2,3,4 Linghong Zhang2,3,4
Linghong Zhang2,3,4 Qingye Xu1
Qingye Xu1 Yunxing Yang1
Yunxing Yang1 Sebastian Leptihn2,6
Sebastian Leptihn2,6 Yunsong Yu2,3,4
Yunsong Yu2,3,4 Dongdong Zhao2,3,4*
Dongdong Zhao2,3,4* Xiaoting Hua2,3,4*
Xiaoting Hua2,3,4*Salmonella enterica is a zoonotic food-borne pathogen threatening public health around the world. As is the case with many other pathogens, the spread of mobilized colistin resistance (mcr) alleles is of grave concern. In this study, totally 689 clinical Salmonella isolates were collected from a local hospital in Hangzhou, Zhejiang Province, China between 2009 and 2018. Resistance genes were screen by PCR. Two mcr-9-positive Salmonella strains S15 and S639 were identified which belong to serotype Typhimurium and Thompson, respectively. We observed that both mcr-9 genes were located on conjugative IncHI2 plasmids which encoded numerous resistance genes, likely facilitating the dissemination of mcr-9 by co-resistance mechanisms. The mcr-9 cassettes encoded on the two plasmids were not identical: downstream of the mcr-9 genes, we found IS1 on one plasmid (pS15), while the other had a WbuC-IS26 (pS639). Despite the presence of mcr-9 cassettes, the strains were not rendered colistin resistant. Yet, it is of epidemiological importance to implement surveillance to be able to observe and possibly control the spread of mcr-9 due to its potential to mediate resistance to the last-resort antibiotic colistin.
Colistin is an effective antibiotic for the treatment of infections caused by multidrug-resistant Gram-negative bacteria as one of the last-resort therapeutic options (Nation and Li, 2009). Since the plasmid-encoding colistin-mediated resistance gene mcr-1 was reported in Escherichia coli of animal origin in China (Liu et al., 2016), plasmid-borne mcr alleles have gained increasing attention and have been extensively researched. Successively, mcr-2 to mcr-10 have been identified, most from animals (Carroll et al., 2019; Lima et al., 2019; Wang et al., 2020). According to the current data, mcr-1 and mcr-9 are the most common colistin resistance cassettes with mcr-9 prevalent in Salmonella enterica (Ling et al., 2020). Salmonella enterica is an important zoonotic pathogen, which can disseminate between animals and people through contaminated food (Lima et al., 2019). Nontyphoidal Salmonella usually causes self-limited enterocolitis with diarrhea. Occasionally an infection with the pathogen can result in more severe diseases including bloodstream infections especially in young children, the elderly, and immunocompromised people (Crump et al., 2015). Thus, the increasing antimicrobial resistance in Salmonella species needs to be monitored (Lozano-Leon et al., 2019).
In a previous study, we focused on the prevalence of the mcr-1 gene in 689 clinical Salmonella isolates in a local hospital and six mcr-1 positive strains were identified (Fan et al., 2020). Five strains belonged to S. Typhimurium and one belonged to S. Indiana. In this work, we have screened the Salmonella isolates for other mcr alleles (mcr-2 to mcr-10). Here, we identified two plasmid-borne mcr-9 in Salmonella Typhimurium and Salmonella Thompson. To our knowledge, this is the first detailed description of mcr-9 plasmid of Salmonella Thompson. In this work, we characterized the composition of the mcr-9 carrying plasmids and the genetic environment surrounding the mcr-9 cassettes, which differed in the two plasmids.
Salmonella clinical isolates were isolated from patients’ specimens such as blood, feces, synovial fluid and pus from abdominal and skin and soft tissue infections in the First People’s Hospital of Hangzhou, Zhejiang Province, China, between 2009 and 2018. Bacterial species were identified by the automated Vitek 2 system (BioMérieux, Marcy-l’Étoile, France) and matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) (Bruker, Bremen, Germany). Salmonella serotyping was identified by slide agglutination with specific antisera (Tianrun Bio-Pharmaceutical Co., Ltd., Ningbo, China) according to the White-Kauffmann-Le Minor scheme (9th edition).
All Salmonella isolates were screened for mcr-2 to mcr-10 by using PCR with corresponding pairs of primers (Table 1). The amplification products were subsequently sequenced by Sanger sequencing for confirmation.
Antimicrobial susceptibility testing was performed by broth microdilution, including colistin, ampicillin, amoxicillin, piperacillin-tazobactam, cefazolin, cefoxitin, ceftriaxone, cefepime, ceftazidime, aztreonam, ertapenem, imipenem, meropenem, amikacin, gentamicin, kanamycin, ciprofloxacin, levofloxacin, tigecycline, tetracycline, trimethoprim-sulfamethoxazole, and fosfomycin. The minimum inhibitory concentration (MIC) of nitrofurantoin was performed using E-test method. Antimicrobial susceptibility testing of the transconjugants was performed by broth microdilution, including colistin, ampicillin, amoxicillin, piperacillin-tazobactam, amikacin, gentamicin, kanamycin, and tetracycline.
The results of antimicrobial susceptibility testing were interpreted by Clinical and Laboratory Standards Institute guidelines (CLSI) (M100, 30th ed.; CLSI, 2020), except that colistin and tigecycline were used the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints v8.1. The quality control strain was E. coli ATCC 25922.
Genomic DNA of two mcr-9-positive strains S15 and S639 were sequenced by HiSeq (Illumina, San Diego, CA, United States) and MinION sequencer (Oxford Nanopore Technologies, Oxford, United Kingdom). The short read and long read sequence data were hybrid de novo assembled by Unicycler v0.4.8 (Wick et al., 2017). The gene sequences were annotated by Prokka (Seemann, 2014) and NCBI Blast (Camacho et al., 2009). Resistance genes and insertion sequence (IS) were identified by BacAnt (Hua et al., 2021). Multi-locus sequence typing (MLST) was identified by using mlst.1 The gene sequences were compared and visualized by Easyfig 2.2.5 (Sullivan et al., 2011) and BRIG-0.95 (Alikhan et al., 2011).
Conjugation assays were conducted by using rifampicin-resistant Salmonella strain XH1984 and E. coli strain EC600 as the recipient strain and strain S15 as the donor. The Mueller-Hinton agar plates containing rifampicin (100 μg/ml) and ampicillin (4 μg/ml for S15 and XH1984; 32 μg/ml for S15 and EC600) were used for selection. The successful transconjugants of S15 and XH1984 were verified by PCR using two pairs of primers: XH104-F and XH104-R; S15-mcr9.1-F and S15-mcr9.1-R (Table 1). The former pair of primers was used to identify XH1984 and the latter was used to identify pS15. The transconjugants of S15 and EC600 were verified by S15-mcr9.1 primers and MALDI-TOF MS. The conjugation frequency of pS15 was determined.
The assembled mcr-9 carrying S. Thompson and S. Typhimurium genomes were downloaded from NCBI and annotated using prokka 1.13 (Seemann, 2014). The maximum likelihood phylogenetic tree was constructed with IQTree 2.1.2 (Nguyen et al., 2015) from a multiple alignment of the core genomes generated by Roary 3.7.0 (Page et al., 2015). The trees were visualized with ggtree (Yu, 2020) and ggtreeExtra (Xu et al., 2021) in R.
We previously screened 689 clinical Salmonella isolates from hospital patient specimens for the presence of the colistin resistance gene mcr-1 (Fan et al., 2020). This follow-up study, we have screened all isolates for other types of mcr genes, including mcr-2 to mcr-10. While none of the strains contained any mcr-2 to mcr-8 or mcr-10 genes, we found two (0.29%) mcr-9-positive Salmonella spp. strains, S15 and S639. The strain S15 was isolated from the stool of a 53-year-old female patient in 2011, while S639 was a stool sample isolate from a 24-year-old woman obtained in 2018. Both patients came to the outpatient service with symptoms of diarrhea.
The antimicrobial susceptibility results are displayed in Table 2. Two mcr-9-positive strains were both resistant to ampicillin, amoxicillin, and tetracycline. S639 was additionally resistant to cefazolin, cefoxitin, ceftriaxone, ceftazidime, aztreonam, amikacin, kanamycin, ciprofloxacin, levofloxacin, and trimethoprim-sulfamethoxazole. Both strains were sensitive to colistin, cefepime, ertapenem, imipenem, meropenem, gentamicin, tigecycline, and nitrofurantoin.
The serotype of S15 was Salmonella Typhimurium (O4:Hi), belonging to ST 34. The strain contained a single plasmid only which we called pS15. The plasmid encoded the mcr-9 gene which showed a 100% identity and coverage to the previously reported mcr-9 in Enterobacterales (WP_044704969.1). The genome size of IncHI2 plasmid pS15 was 266,098 bp and the GC content was 46%. This strain contains 12 resistance genes on the plasmid and 20 resistance genes on the chromosome (Table 3).
The serotype of S639 was Salmonella Thompson (O7:Hk:H1,5), belonging to ST 26. Again, this strain contained only a single plasmid which we called pS639. Here, the mcr-9 exhibited 100% identity and coverage to another colistin resistance gene in the Enterobacterales, WP_001572373, mcr-9.1. This sequence was missing a single codon (for tryptophan) right before the STOP codon compared to mcr-9 in S15. The genome size of IncHI2 plasmid pS639 was 308,491 bp and the GC content was 48%. This strain contains 27 resistance genes on the plasmid while only four resistance genes are found on the chromosome (Table 3).
Most of the chromosome-encoded resistance genes in S15 and S639 are related to metal resistance aside from the multidrug efflux RND transporter gene mdsB. The resistance genes in two plasmids differ greatly from each other: for example, pS15 contains genes associated with resistance to tellurium (terW and terZ), mercury (mer gene cluster), β-lactams (blaTEM-1), tetracycline [tet(A)], trimethoprim (dfrA16), streptomycin (aadA2), colistin (mcr-9), and copper (pcoS and pcoE). Plasmid pS639 encodes genes mediating resistance to all substances which are facilitating resistance in pS15, some of which were different alleles like dfrA19 and tet(D). Additionally, pS639 encodes genes associated with resistance to sulfonamide (sul1 and sul2), macrolide [mph(A)], β-lactams (blaOXA-10), aminoglycoside (aacA34), rifamycin (arr-3), streptomycin [aph(6)-ld and aph(3″)-lb], chloramphenicol (catA2), cephalosporin (blaDHA-1), and quinolone (qnrB4). This abundance of additional ARGs might explain why S639 exhibited higher MIC values and a wider resistance to more antibiotics than S15.
The two plasmids we found are similar to other IncHI2 plasmids. Table 4 lists plasmids for comparison, some of which share high query coverage and identity with pS15 and pS639 from different species. The backbone structures of pS15, pEcl10-1 (CP048704), sLN794248 (LN794248), and pC45-VIM4 (LT991958) are closely related (Figure 1). The main differences between them are the resistance gene cluster regions where most insertion elements (ISs) were located. Although pEcl10-1 and sLN794248 were more similar to pS15 in their sequence, they do not contain mcr-9.1 and dfrA16. Plasmid pC45-VIM4 encodes a mcr-9.1 gene but shows more differences in the 90–140 kbp region of pS15 compared to other plasmids. When comparing pS15 to three other plasmids, pS15 contains a gene encoding group II intron reverse transcriptase/maturase (around 210 kb). Two resistance gene clusters are present in pS639, found in two sections, from ~100 to 180 kb and from ~240 to 270 kb, respectively (Figure 2). The first section shares some similarities but also substantial differences with two other plasmid sequences, p48212_MCR (CP059413) and pMOL665_IncHI2 (OU015720). pS639 additionally encodes mph(A), blaOXA-10, aacA34, aar-3, and catA2, genes which are able to facilitate the resistance to macrolide, β-lactam, aminoglycoside, rifamycin, and chloramphenicol. In the other resistance gene cluster which includes mcr-9.1, the main difference is an insertion of the two resistance genes blaDHA-1 and qnrB4 while a gene cluster encoding phage shock protein is also present.
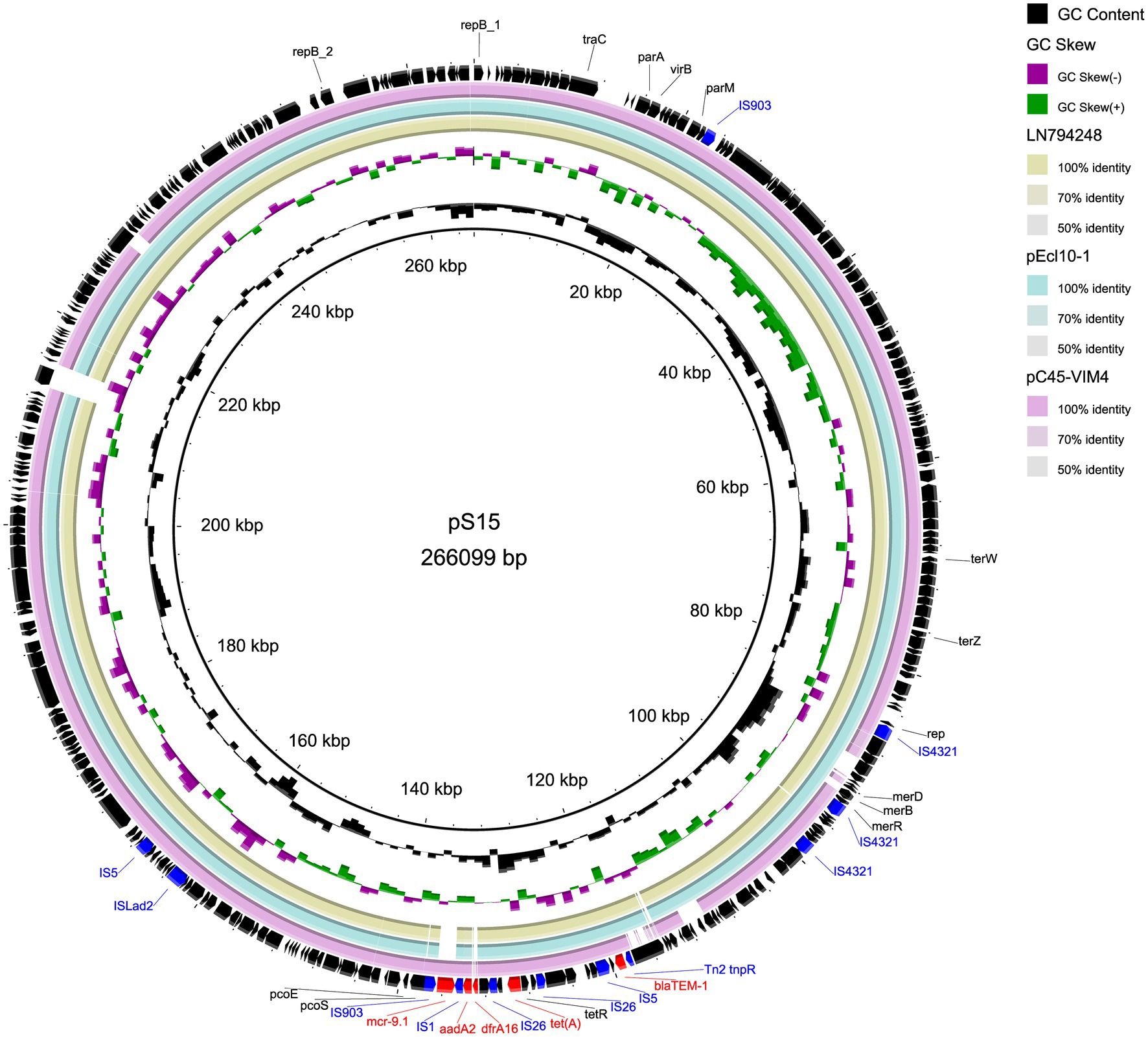
Figure 1. Comparison of circular structures of pS15 and its similar plasmids. Arrows indicate the direction of predicted transcription of each gene. Red arrows indicate antibiotic resistance genes. Blue arrows indicate insertion sequences.
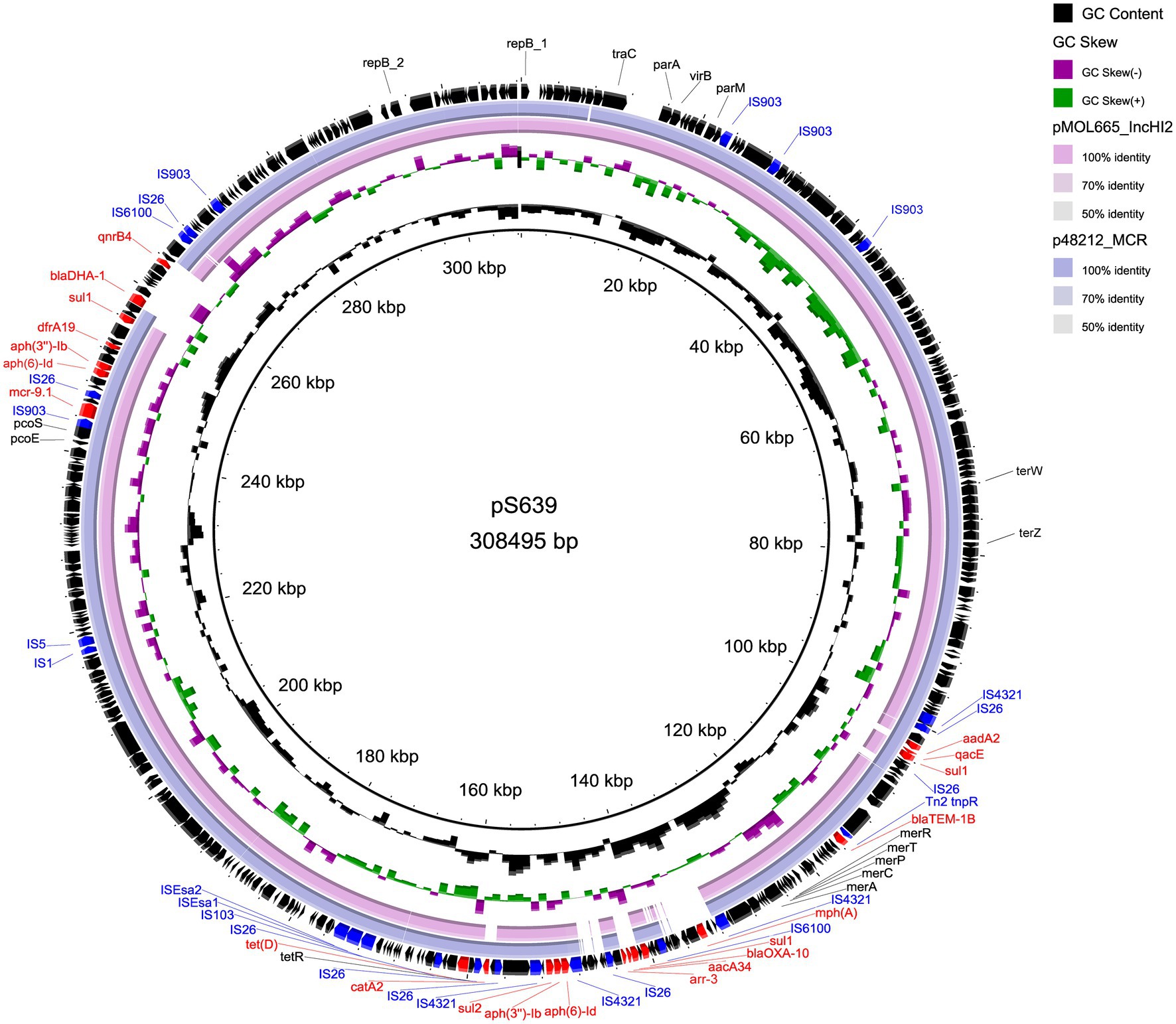
Figure 2. Comparison of circular structures of pS639 and its similar plasmids. Arrows indicate the direction of predicted transcription of each gene. Red arrows indicate antibiotic resistance genes. Blue arrows indicate insertion sequences.
Our genetic analyses regarding the sequences surrounding the mcr-9 genes revealed two types (Figure 3). The mcr-9 surrounding structure of pS639 was pcoE-pcoS-IS903B-mcr-9-WbuC-IS26 similar to p48212_MCR and pMOL665_IncHI2. However, in our case, the genetic context that embedded the mcr-9 gene was pcoE-pcoS-IS903B-mcr-9-IS1 which is present in both, the Salmonella plasmid pS15 and pC45-VIM4, a plasmid found in a bacterium of the Enterobacter cloacae complex. There was an insertion of IS1 and catA in the position of mcr-9 and IS903B.
We also tested the ability of the plasmids to be transferred to other strains and their ability to convey antibiotic resistance. We first tested the plasmid pS15 which was successfully transferred to the rifamycin-resistant Salmonella strain XH1984 and E. coli strain EC600. When determining the MICs for ampicillin, amoxicillin, piperacillin-tazobactam, and tetracycline, we found increased resistance likely conferred by the presence of the plasmid-encoded blaTEM-1 and tet(A) genes (Table 2). However, mcr-9 in pS15 could not confer colistin resistance in neither Salmonella nor E. coli strains. The conjugation efficiency was calculated in pS15 plasmid conjugation assays, which was 1.9 × 10−6 transconjugants per donor when pS15 was transferred to XH1984 and 2.1 × 10−8 transferred to EC600 (Table 5).
Our attempt in transferring the plasmid pS639 was unsuccessful. The reason for this is that we did not have a suitable recipient strain available which would allow the use of an antibiotic selection marker, as our strains exhibited resistance to the antibiotics encoded on the plasmid. Also, the possibility that the recipient strains were genetically not suitable to receive this specific plasmid cannot be excluded.
A total of 175 S. Typhimurium strains and 21 S. Thompson strains carrying mcr-9 have been deposited in NCBI till today (February 2022). The phylogenetic trees of two serotypes were displayed in Figures 4, 5, respectively. S. Typhimurium strains were mostly isolated from clinical samples. Australia and the United Kingdom were the countries where the most assembled sequences were uploaded from, which does not necessarily reflect the prevalence of the strains in these countries. S. Typhimurium S15 was most similar to FSIS32003798 isolated from pork in the United States. As for S. Thompson, only three of 21 strains were clinical origin including S639, which had the closest relationship with 813,389 isolated in the United Kingdom. Interestingly, both S15 and S639 were more closely related to strains isolated outside China, despite being isolated within the country.
With multidrug resistance continuously increasing, colistin now belongs to the last-resort antibiotics. Plasmid-bound mcr alleles that mediate resistance to the antimicrobial compound are of great concern in particular if plasmid encoded due to the risk of rapid spread (Smelikova et al., 2021). Several mcr genes have been found, with mcr-9 first identified in Salmonella Typhimurium (Carroll et al., 2019). To date, Salmonella strains have been reported worldwide to carry various mcr alleles, with mcr-1 being the most common and Typhimurium being the most prevalent serotype (Lima et al., 2019; Paveenkittiporn et al., 2021; Portes et al., 2022). As a zoonotic food-borne pathogen, mcr-positive S. enterica strains were mostly isolated from livestock, including pork and poultry, due to the fact that colistin has and continues to be used in animal husbandry (Lima et al., 2019). Therefore, it is important to monitor the spread of mcr alleles in S. enterica.
In this study, we identified two mcr-9-positive Salmonella enterica from a total of 689 clinical Salmonella isolates. The positive rate of mcr-9 (0.29%) might be lower than mcr-1 (0.87%) as we previously reported (Fan et al., 2020) although due to the low numbers (of two and six strains, respectively), statistically sound conclusions are not possible. The two mcr-9-positive strains belong to a different serotype, Typhimurium and Thompson. Salmonella Typhimurium ST34 is most commonly prevalent in causing food-borne infections in China (Wong et al., 2013). While Salmonella Thompson is the main serotype isolated from poultry-based products (Yang et al., 2020; Elbediwi et al., 2021b). In this study, both strains were isolated from stools of patients with diarrhea caused by Salmonella enterica infections, likely to have been exposed to food contaminated with the pathogen. The surprising similarity between the strains we isolated in China and those found outside the country can be explained by the rapid development of international agricultural products trade.
IncHI2 plasmids were the predominant plasmid type carrying mcr-9 (Li et al., 2020). The two plasmids that we characterized in our study, pS15 and pS639, also belonged to the IncHI2 type, both of them having IncHI2 and IncHI2A replicons, which indicates that they are hybrid plasmids. This type of plasmid is conjugative which can result in extensive spread of the mcr-9 gene in recipient hosts (Ai et al., 2021; Elbediwi et al., 2021a; Khodor et al., 2021; Wang et al., 2021). Testing whether the plasmids we discovered can be transmitted to other strains, we found that pS15 was indeed conjugative to both, Salmonella and E. coli. The efficiency of conjugation to Salmonella was about 90 times higher than that to E. coli, indicating pS15 was easier to spread within species. The conjugation experiments using the plasmid pS639 could not be performed due to the fact that we did not have a suitable recipient strain to our disposal as we could not select for an antibiotic encoded on the plasmid. However, pS15 and pS639 were predicted to contain similar conjugative apparatus components by oriTfinder (Li et al., 2018). Thus, it is reasonable to conclude that pS639 is likely to be conjugative since the components of the two plasmids show strong similarities.
Compared to the low level colistin resistance mediated by mcr-1, most of the mcr-9-carrying strains do not present resistance to colistin (Luo et al., 2017; Wang et al., 2021). We also observed this to be the case with the strains S15 and S639, which we described in this study. However, the inducible expression of mcr-9 could potentially lead to an increasing of colistin MIC after exposure to low concentrations of colistin, mediated by the qseC and qseB genes (Kieffer et al., 2019). This makes mcr-9 a gene that should not be disregarded when addressing antimicrobial resistance. Apart from this, our study identified numerous resistance genes located in the two plasmids in addition to mcr-9, which were responsible for the drug resistance spectrum of two strains. A total of 12 resistance genes are found in pS15 and 27 in pS639. S15 and S639 were resistant to broad spectrum penicillin and tetracycline because they both had plasmid-encoded blaTEM-1 and tet, which was verified by the transconjugants of pS15. In addition, pS639 encoded genes sul, blaOXA-10, blaDHA-1, aacA34, and qnrB4, accounting for the resistance to sulfonamides, cephalosporins, aminoglycosides, and quinolones. Since there are multiple resistance genes encoded on mcr-9 plasmids, it is a matter of concern that co-resistance mechanism could facilitate the spread of mcr-9. The two types of mcr-9 cassettes in our study, pcoE-pcoS-IS903B-mcr-9-WbuC-IS26 and pcoE-pcoS-IS903B-mcr-9-IS1, did not include the qseC-qseB regulatory genes, indicating they might circulate silently. However, there might be other undetermined genes or molecules regulating mcr-9 expression (Kananizadeh et al., 2020). Therefore, it is important to investigate the silent spread of mcr-9 further and to monitor the dissemination of plasmids containing the colistin resistance gene.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The studies obtained ethical approval from the Ethics Committee of Hangzhou First People’s Hospital (2020103-1).
JF, YF, DZ, and XH designed the study. JF, HC, YF, LZ, JH, and YYa performed the experiments. JF, YF, LZ, JH, and HC analyzed the bioinformatics data. JF, YF, and HC wrote the manuscript. QX, DZ, SL, YYu, and XH revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ai, W., Zhou, Y., Wang, B., Zhan, Q., Hu, L., Xu, Y., et al. (2021). Corrigendum: first report of coexistence of bla SFO-1 and bla NDM-1 beta-lactamase genes as well as colistin resistance gene mcr-9 in a transferrable plasmid of a clinical isolate of Enterobacter hormaechei. Front. Microbiol. 12:741628. doi: 10.3389/fmicb.2021.741628
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio 10, e00853–e00919. doi: 10.1128/mBio.00853-19
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing. 30th Edn. Wayne, PA, USA: Clinical and Laboratory Standards Institute.
Crump, J. A., Sjolund-Karlsson, M., Gordon, M. A., and Parry, C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28, 901–937. doi: 10.1128/CMR.00002-15
Elbediwi, M., Pan, H., Zhou, X., Rankin, S. C., Schifferli, D. M., and Yue, M. (2021a). Detection of mcr-9-harbouring ESBL-producing Salmonella newport isolated from an outbreak in a large-animal teaching hospital in the USA. J. Antimicrob. Chemother. 76, 1107–1109. doi: 10.1093/jac/dkaa544
Elbediwi, M., Tang, Y., Shi, D., Ramadan, H., Xu, Y., Xu, S., et al. (2021b). Genomic investigation of antimicrobial-resistant Salmonella enterica isolates from dead chick embryos in China. Front. Microbiol. 12:684400. doi: 10.3389/fmicb.2021.684400
Fan, J., Zhang, L., He, J., Zhao, M., Loh, B., Leptihn, S., et al. (2020). Plasmid dynamics of mcr-1-positive Salmonella spp. in a general hospital in China. Front. Microbiol. 11:604710. doi: 10.3389/fmicb.2020.604710
Hua, X., Liang, Q., Deng, M., He, J., Wang, M., Hong, W., et al. (2021). BacAnt: a combination annotation server for bacterial DNA sequences to identify antibiotic resistance genes, integrons, and transposable elements. Front. Microbiol. 12:649969. doi: 10.3389/fmicb.2021.649969
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible Enterobacter cloacae complex co-harboring blaIMP-1 and mcr-9 in Japan. BMC Infect. Dis. 20:282. doi: 10.1186/s12879-020-05021-7
Khodor, R., Salloum, T., El Jisr, T., El Chaar, M., and Tokajian, S. (2021). Detection and genomic characterization of mcr-9 in Enterobacter hormaechei recovered from a pediatric patient in Lebanon. Infect. Genet. Evol. 94:105014. doi: 10.1016/j.meegid.2021.105014
Kieffer, N., Royer, G., Decousser, J. W., Bourrel, A. S., Palmieri, M., Ortiz De La Rosa, J. M., et al. (2019). mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63, e00965–e01019. doi: 10.1128/AAC.00965-19
Li, Y., Dai, X., Zeng, J., Gao, Y., Zhang, Z., and Zhang, L. (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 10:8113. doi: 10.1038/s41598-020-65106-w
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234. doi: 10.1093/nar/gky352
Lima, T., Domingues, S., and Da Silva, G. J. (2019). Plasmid-mediated colistin resistance in Salmonella enterica: a review. Microorganisms 7:55. doi: 10.3390/microorganisms7020055
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lozano-Leon, A., Garcia-Omil, C., Dalama, J., Rodriguez-Souto, R., Martinez-Urtaza, J., and Gonzalez-Escalona, N. (2019). Detection of colistin resistance mcr-1 gene in Salmonella enterica serovar Rissen isolated from mussels, Spain, 2012- to 2016. Eurosurveillance 24:1900200. doi: 10.2807/1560-7917.ES.2019.24.16.1900200
Luo, Q., Yu, W., Zhou, K., Guo, L., Shen, P., Lu, H., et al. (2017). Molecular epidemiology and colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front. Microbiol. 8:2262. doi: 10.3389/fmicb.2017.02262
Nation, R. L., and Li, J. (2009). Colistin in the 21st century. Curr. Opin. Infect. Dis. 22, 535–543. doi: 10.1097/QCO.0b013e328332e672
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paveenkittiporn, W., Kamjumphol, W., and Kerdsin, A. (2021). Draft genome sequence of invasive Salmonella enterica serovar cannstatt harboring mcr-1.1, isolated from a fatal sepsis case. Microbiol. Resour. Announc. 10, e01270–e01320. doi: 10.1128/MRA.01270-20
Portes, A. B., Rodrigues, G., Leitao, M. P., Ferrari, R., Conte Junior, C. A., and Panzenhagen, P. (2022). Global distribution of plasmid-mediated colistin resistance mcr gene in salmonella: a systematic review. J. Appl. Microbiol. 132, 872–889. doi: 10.1111/jam.15282
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Smelikova, E., Tkadlec, J., and Krutova, M. (2021). How to: screening for mcr-mediated resistance to colistin. Clin. Microbiol. Infect. 28, 43–50. doi: 10.1016/j.cmi.2021.09.009
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, X., Ling, Z., Sun, N., Liu, Y., Huang, J., and Wang, L. (2021). Molecular genetic characteristics of mcr-9-harbouring Salmonella enterica serotype Typhimurium isolated from raw milk. Int. J. Antimicrob. Agents 57:106332. doi: 10.1016/j.ijantimicag.2021.106332
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, M. H., Yan, M., Chan, E. W., Liu, L. Z., Kan, B., and Chen, S. (2013). Expansion of Salmonella enterica serovar Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob. Agents Chemother. 57, 4599–4601. doi: 10.1128/AAC.01174-13
Xu, S., Dai, Z., Guo, P., Fu, X., Liu, S., Zhou, L., et al. (2021). ggtreeExtra: compact visualization of richly annotated phylogenetic data. Mol. Biol. Evol. 38, 4039–4042. doi: 10.1093/molbev/msab166
Yang, X., Huang, J., Zhang, Y., Liu, S., Chen, L., Xiao, C., et al. (2020). Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci. Total Environ. 713:136385. doi: 10.1016/j.scitotenv.2019.136385
Keywords: mcr-9, colistin, IncHI2 plasmid, Salmonella Typhimurium, Salmonella Thompson
Citation: Fan J, Cai H, Fang Y, He J, Zhang L, Xu Q, Yang Y, Leptihn S, Yu Y, Zhao D and Hua X (2022) Molecular Genetic Characteristics of Plasmid-Borne mcr-9 in Salmonella enterica Serotype Typhimurium and Thompson in Zhejiang, China. Front. Microbiol. 13:852434. doi: 10.3389/fmicb.2022.852434
Received: 11 January 2022; Accepted: 28 February 2022;
Published: 17 March 2022.
Edited by:
Guojie Cao, United States Food and Drug Administration, United StatesReviewed by:
Qi Wang, Peking University People’s Hospital, ChinaCopyright © 2022 Fan, Cai, Fang, He, Zhang, Xu, Yang, Leptihn, Yu, Zhao and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongdong Zhao, MzMxMzAwNEB6anUuZWR1LmNu; Xiaoting Hua, eGlhb3RpbmdodWFAemp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.