- 1Ministry of Education Key Laboratory of Ecology and Resource Use of the Mongolian Plateau and Inner Mongolia Key Laboratory of Grassland Ecology, School of Ecology and Environment, Inner Mongolia University, Hohhot, China
- 2The High School Affiliated to Minzu University of China, Hohhot, China
- 3Inner Mongolia Key Laboratory of Environmental Pollution Control and Waste Resource Reuse, Inner Mongolia University, Hohhot, China
- 4Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
Root-associated aerobic methanotroph plays an important role in reducing methane emissions from wetlands. In this study, we examined the activity of methane-dependent nitrogen fixation and active nitrogen-fixing bacterial communities on the roots of Typha angustifolia and Scirpus triqueter using a 15N-N2 feeding experiment and a cDNA-based clone library sequence of the nifH gene, respectively. A 15N-N2 feeding experiment showed that the N2 fixation rate of S. triqueter (1.74 μmol h–1 g–1 dry weight) was significantly higther than that of T. angustifolia (0.48 μmol h–1 g–1 dry weight). The presence of CH4 significantly increased the incorporation of 15N-labeled N2 into the roots of both plants, and the rate of CH4-dependent N2 fixation of S. triqueter (5.6 μmol h–1 g–1 dry weight) was fivefold higher than that of T. angustifolia (0.94 μmol h–1 g–1 dry weight). The active root-associated diazotrophic communities differed between the plant species. Diazotrophic Methylosinus of the Methylocystaceae was dominant in S. triqueter, while Rhizobium of the Rhizobiaceae was dominant in T. angustifolia. However, there were no significant differences in the copy numbers of nifH between plant species. These results suggest that N2 fixation was enhanced by the oxidation of CH4 in the roots of macrophytes grown in natural wetlands and that root-associated Methylocystacea, including Methylosinus, contribute to CH4 oxidation-dependent N2 fixation.
Introduction
Methane (CH4) is an important greenhouse gas, and natural wetlands and paddy fields are major sources of CH4 emissions that contribute to the global CH4 budget (Conrad, 2009). The Intergovernmental Panel on Climate Change (IPCC, 2007) reported that natural wetlands emit 100–231 Tg of methane to the atmosphere yearly, which represents 20–39% of the global CH4 emissions (Laanbroek, 2010). In wetlands dominated by emergent plants, more than 70% of the CH4 produced is transported through plants (Van der Nat and Middelburg, 1998; Laanbroek, 2010), and up to 90% is oxidized by aerobic methanotrophs from the root zone (Laanbroek, 2010). Aerobic methanotrophs in wetland ecosystems have been analyzed using culture-dependent and –independent approaches (Calhoun and King, 1998; Yun et al., 2013; Cai et al., 2016; Cui et al., 2020; Liu et al., 2020). Most aerobic methanotrophic strains oxidize CH4 using oxygen (O2), and they can fix N2 to ammonia (NH4+; Auman et al., 2001). Recently, diazotrophic methanotrophs have been reported in N-deficient environments, such as peatlands (Larmola et al., 2014; Reumer et al., 2018), forest soil (Buckley et al., 2008; Mäkipää et al., 2018), and rice paddies (Bao et al., 2014a; Shinoda et al., 2019). For example, Larmola et al. (2014) reported that Sphagnum-associated methanotrophs contribute to N2 fixation in peatland ecosystems. In addition, N2 fixation by Methylosinus (Type II methanotrophs) has been detected in rice roots without fertilization using metaproteomic approaches (Bao et al., 2014a; Minamisawa et al., 2016). Therefore, aerobic methanotrophs are not only important to reducing CH4 emissions, but also play a role in sustaining soil fertility (Shinoda et al., 2019).
As an important component of natural wetlands, emergent plants have a crucial impact on greenhouse gas emissions (budget; Zhang et al., 2018; Chen et al., 2019). Emergent plants, such as Typha angustifolia, Scirpus triqueter, and Phragmites australis, are widely distributed throughout the world and are commonly found in eutrophic wetlands (Duan et al., 2005; Laanbroek, 2010). The roots of these plant species not only play an important role in transporting the CH4 that is produced from anaerobic sediment but also provide a habitat for microorganisms that include methanotrophs (Nat and Middelburg, 1998; Pietrangelo et al., 2018). Studies have shown that root-associated Methylomonas (Type I; Gammaproteobacteria) is dominant in the roots of three emergent plants (P. australis, T. angustifolia and S. triqueter) using in situ hybridization or next-generation sequencing amplicons of pmoA, which encodes the particulate methane monooxgynase, based on their DNA (Fausser et al., 2012; Liu et al., 2020). However, root-associated Methylocystis (Type II methanotrophs, Alphaproteobacteria) is dominant in S. triqueter and T. angustifolia by sequencing pmoA gene amplicons based on cDNA (Cui et al., 2020). Owing to rapid economic development, many wetlands or lakes in the world have become eutrophic because of the excessive application of nitrogen (Bhagowati and Ahamad, 2018). Methanotrophs or diazotrophic bacteria are often affected by environmental factors, such as nitrogen, oxygen and Cu2+ (Hanson and Hanson, 1996; Bodelier and Laanbroek, 2004; Cheng, 2008). Whether methanotroph-mediated N2 fixation could occur in the roots of emergent macrophytes that grow in eutrophic natural wetlands remains unclear.
There have been methodological issues in the estimation of CH4 oxidation-dependent N2 fixation by methanotrophy (Larmola et al., 2014; Ho and Bodelier, 2015). The acetylene (C2H2) reduction assay (ARA) has been used to determine nitrogen fixation by methanotrophs (Takeda et al., 2008; Khadem et al., 2010). However, CH4 monooxygenase, a key enzyme for the oxidation of CH4 in methanotrophs, is strongly inhibited by C2H2, and CH4 oxidation-dependent N2 fixation by methanotrophs has been underestimated (Dalton and Whittenbury, 1976; Takeda, 1988). 15N2-feeding is a powerful approach to estimate the fixation of N2 by microorganisms (Larmola et al., 2014) and has been utilized to estimate the CH4 oxidation-dependent N2 fixation of rice plants (Shinoda et al., 2019) and the sediment of deep seas (Dekas et al., 2009).
Our goal was to demonstrate whether methanotroph-mediated nitrogen fixation occurs in the roots of S. triqueter and T. angustifolia in natural wetlands. This study used (i) 15N-N2 feeding experiments with and without CH4 to analyze the CH4 oxidation-dependent nitrogen fixation of S. triqueter and T. angustifolia, (ii) cDNA-based nifH gene sequencing to analyze the active root-associated nitrogen-fixing bacterial community and abundance of diazotrophic methanotrophs, and (iii) quantitative PCR of nifH to analyze the abundance of gene expression in the roots of the two emergent plants grown in a natural wetland.
Materials and Methods
Sampling Sites and Plant Materials
T. angustifolia and S. triqueter were located in shallow water (50–70 cm) and deep water (80–100 cm) areas in the Wuliangsuhai wetland (N 40°52′36″, E 108°51′16″; Figure 1) in the Inner Mongolia Autonomous Region, China, respectively, and three to four individual plants of each species were collected on July 15, 2017 (Cui et al., 2020). The physicochemical properties of the sediments are shown in Supplementary Table 1. The roots of plants were washed with sterilized water to ensure that all of the sediment was removed, and the whole plants were divided vertically into two equal parts to collect the roots. Some of the exposed roots were collected with sterilized forceps and transferred into 50 mL Falcon tubes that contained sterile pure water (Bao et al., 2014b). All the tubes with roots were quickly snap-frozen in liquid nitrogen, immediately brought back to the laboratory, and stored at –80°C for molecular analyses.
Figure 1. Sampling site (a) of Scirpus triqueter (b) and Typhus angustifolia (c) in the natural wetland of Wuliangsuhai.
RNA Isolation, Preparation of cDNA Libraries and Sequencing
Total RNA was extracted from approximately 0.5 g of roots using an RNAprep Pure Plant Kit (Tiangen Biotech Co., Beijing, China) according to the manufacturer’s instructions. Since this study primarily focused on the transcriptional level, DNA contamination in the extracted RNA would affect the final experimental results. Therefore, it was imperative that the RNA be examined for possible DNA contamination before reverse transcription was conducted. The extracted total RNA was used as the template, and the 16S rRNA gene primer 27F/1492R (Martin-Laurent et al., 2001) was chosen for PCR amplification. DNA from the roots served as a positive control. PCR products were detected using 1.0% agarose gel electrophoresis and NanoVue™ Plus Spectrophotometry (GE Healthcare, Chicago, IL, United States) to ensure that there was no residual microbial DNA in the total RNA. The extracted RNA was reverse transcribed using a PrimeScript RT Reagent Kit with a gDNA Eraser (TaKaRa, Kyoto, Japan). The kit removes genomic DNA before reverse transcription, and the RT primer mixture (olige dT and random 6 mers) was used. All the samples were stored at –80°C until use.
The nifH gene in cDNA samples from the roots of both plants was amplified with the primers PolF (TGCGAYCCSAARGCBGACTC; Poly et al., 2001) and AQER (GACGATGTAGATYTCCTG; Wartiainen et al., 2008). Amplification and purification were conducted as previously described (Cui et al., 2020). The purified DNA fragments were linked to the pEASY-T1 vector (TransGen, Beijing, China). The products of ligation were transformed into competent cells, mixed with IPTG and X-Gal, and then coated on LB plates that contained kanamycin (100 ng/mL). White colonies were selected after overnight culture. The universal primers M13F and M13R of the vector were used for PCR detection to remove the false positive clones. Approximately 120 nifH positive clones for both plant samples were randomly picked from each sample for sequencing (Sangon Biotec, Beijing, China) using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, United States) to construct the libraries. Sequences with 94% identity (Liu et al., 2021) were classified into the same operational taxonomic units (OTUs) using MOTHUR software (Schloss et al., 2009). The taxonomy of OTUs was identified by comparing the representative OTU sequences to reference nifH sequences using the basic local alignment search tool (BLAST) within GenBank. Each representative of the OTUs was translated to its amino acid sequence using MEGA X software (Kumar et al., 2018). After the alignment of amino acid sequences with the ClustalW program (Thompson et al., 1994), a neighbor-joining phylogenetic tree was constructed using MEGA X software (Kumar et al., 2018). The bootstrap values for each branch were determined using 1,000 iterations.
Quantification of the nifH Gene
The abundances of nifH were quantified using quantitative PCR with a CFX Connect Optical Real-Time Detection System (Bio-Rad Laboratories, Hercules, CA, United States) with the primer set PolF/AQER (Poly et al., 2001; Wartiainen et al., 2008) for the nifH gene based on a cDNA library from the roots of both plants. The reactions were performed in volumes of 20 μL and contained approximately 50 ng extracted cDNA, 10 μL 2 × SYBR Premix Ex Taq (TaKaRa Biotech, Dalian, China), and 500 nM of the primers PolF and AQER (Liu et al., 2021). The PCR conditions included 40 cycles of denaturation at 94°C for 10 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s. Clones of the nifH genes from Methylosinus trichosporium NCIMB 11131 (U31650) were used as the standard references.
15N2 Feeding Experiment for the Macrophyte Roots
Three individual plants of T. angustifolia and S. triqueter were sampled. The root systems were rinsed with water until the sediment had been completely removed. The root samples were placed in a 1 L sealing bag with inflation valves (Supplementary Figure 1), and the gas phase in the assembly was replaced with an argon (Ar)-balanced mixture of 32% (v/v) 15N-N2 (99.9 atom%; Wuhan Newradar Special Gas Co., Ltd., Wuhan, China) and 5% (v/v) O2 with or without 5% (v/v) CH4. The roots were incubated in the bag assembly at 28°C for 48 h in the dark, dried at 80°C for 3–5 days, and then powdered in a blender (Shinoda et al., 2019). Root samples without 15N2 feeding served as the negative control and were dried at 80°C immediately after they were rinsed with water. The 15N concentration and total N content were determined using a Stable Isotope Ratio Mass Spectrometer (MAT253, Thermo Fisher Scientific, Bremen, Germany).
The nitrogen fixation rate was calculated as follows: RW × TN/100 × (15Nc1 – 15Nc2)/15Ng × 100/MW, where RW is the root dry weight (g); TN is the average N content of dried root (%, w/w), and MW is the average molecular weight of N2. 15Nc1 and 15Nc2 represent the respective initial and final 15N concentrations (atom% excess) in the roots, respectively. 15Ng is the 15N concentration (atom% excess) in the N2 gas (Shinoda et al., 2019).
Statistical Analysis
To test for differences in the rate of nitrogen fixation by T. angustifolia or S. triqueter roots under air, 15N2, and 15N2+CH4 conditions, Tukey’s honestly significant difference (HSD) test was performed using R software (ver. 3.3.21) with the “multcomp” package for comparisons of multiple test samples. Significance was defined as p < 0.05 (Shinoda et al., 2019).
Sequence Data Accession Numbers
The sequence data for the nifH gene clones from the roots of S. triqueter and T. angustifolia in this study have been deposited in the NCBI database under accession numbers MZ208474∼MZ208567 and MZ208569∼MZ208680, respectively.
Results
15N2 Feeding of the Root Systems of Plants
To estimate the ability of methanotrophs that inhabit the root systems of macrophytes grown in wetlands to fix nitrogen, the roots of S. triqueter and T. angustifolia were exposed to 15N-labeled N2 gas in the presence and absence of CH4 (Figure 2A and Supplementary Table 2). In the absence of CH4, the 15N atom percentage of S. triqueter roots that had been incubated was significantly higher than that of T. angustifolia. In contrast, the presence of CH4 significantly enhanced the concentration of 15N in the roots of both plants (Figure 2A). In particular, the 15N concentration of S. triqueter roots increased by more than threefold, which was significantly higher than that of T. angustifolia. The CH4-dependent nitrogen fixation of the S. triqueter roots was markedly higher than that of T. angustifolia. The rate of 15N-labeled N2 assimilation was calculated on the basis of the total root N content, dry weight and concentration of 15N (Figure 2B and Supplementary Table 2). The rate of incorporation of 15N-labeled N2 into the S. triqueter roots (5.58 μmol h–1 g–1 dry weight) was significantly higher than that into the T. angustifolia roots (0.94 μmol h–1 g–1 dry weight). This result suggests that the presence of a high concentration of CH4 (5%, v/v) could stimulate nitrogen fixation in the macrophyte roots, and the effect may differ among different plant species.
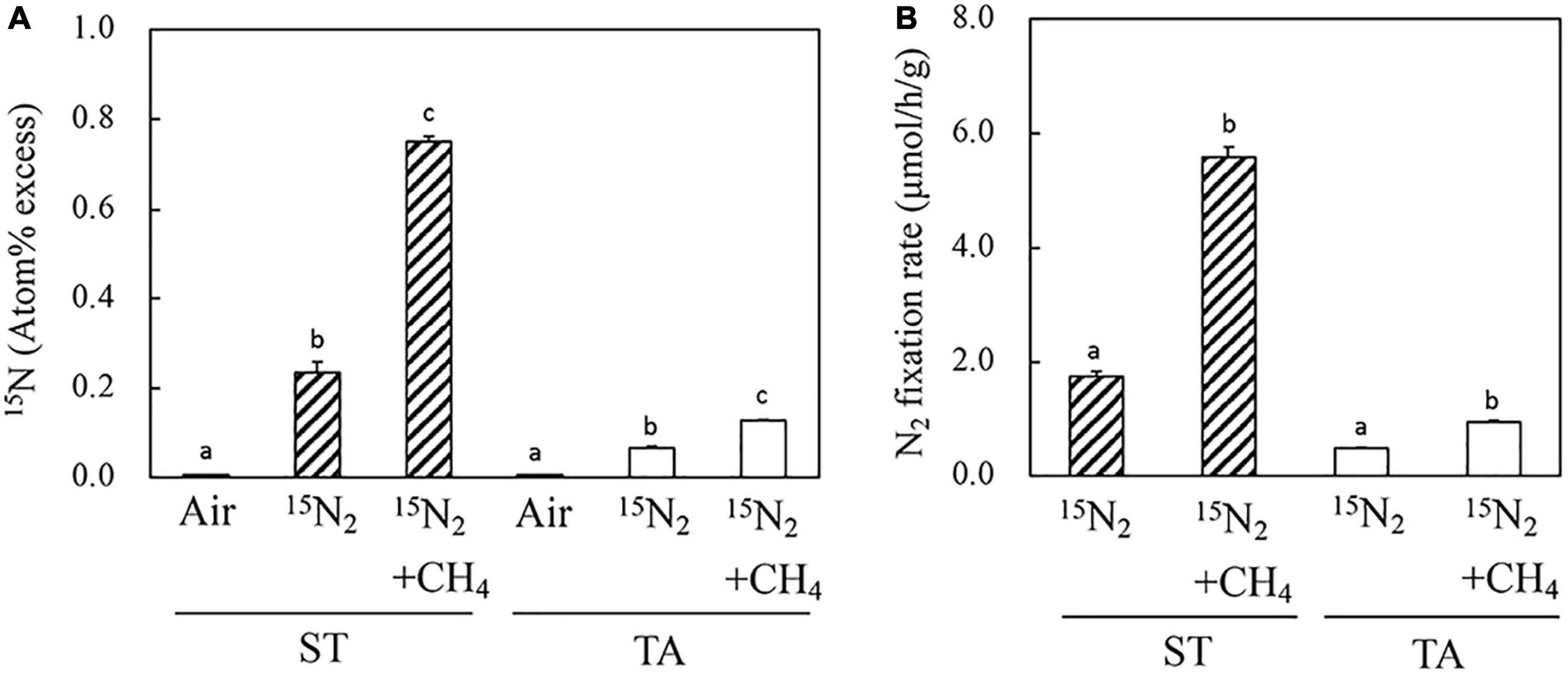
Figure 2. 15N incorporation from 15N2 into roots (A) and the N2 fixation rate (B) of wetland-grown macrophytes in the presence and absence of 5% CH4. Air and 15N2, 15N concentrations before and after root exposure to a gas mixture of 32% (v/v) 15N2 and 5% (v/v) O2 in argon balance, respectively. +CH4, addition of 5% (v/v) CH4 to the gas phase for 48 h. Bars bearing the same letter (a or b) within a panel do not differ significantly according to Tukey’s test for pairwise mean comparison at α = 0.05. ST: Scirpus triqueter; TA: Typha angustifolia.
Diversity of the cDNA Clone Library
A total of 240 nifH positive clones from both types of plant roots were sequenced (Supplementary Table 3). The library coverage was 93.6 and 86.4% in S. triqueter and T. angustifolia, respectively. S. triqueter had the highest number of OTUs and slightly higher diversity of all the indices compared with those of T. angustifolia.
Phylogenetic Diversities of Root-Associated Diazotrophs
The assessment of phylogenetic compositions of diazotrophic communities revealed that Proteobacteria (93.8–98.9%) was dominant in both plant species at the phylum level (Figure 3A). Rhizobiales (52.7–54.3%) of Alphaproteobacteria was dominant, and Desulfobacterales (10.7–11.7%), Rhodocyclales (9.8–10.6%), Gallionellales (6.5–8.3%), and Rhodospirillales (4.3–5.4%) were minor in both libraries at the order level (Supplementary Figure 2A,B). At the family level, the diazotrophic composition clearly differed between both plant species. The relative abundances of Methylocystaceae (35.1%) and Rhizobiaceae (29.5%) indicated that they were dominant in S. triqueter and T. angustifolia, respectively. The relative abundances of Desulfobacteraceae (9.8–11.7%), Rhodocyclaceae (8.0–10.6%), Callionellaceae (6.3–8.5%) and Rhodospirillaceae (4.3–5.4%) were relatively similar in both plants, and the abundances of Methylocystaceae (0.9–35.1%), Rhizobiaceae (6.4–29.5%), Bradyrhizobiaceae (1.8–9.6%), and Phyllobacteriaceae (3.2–13.4%) clearly differed in both plants. In addition, Beijerinckiaceae (5.4%) and Azonexaceae (4.5%) were only detected in T. angustifolia (Supplementary Figure 2C).
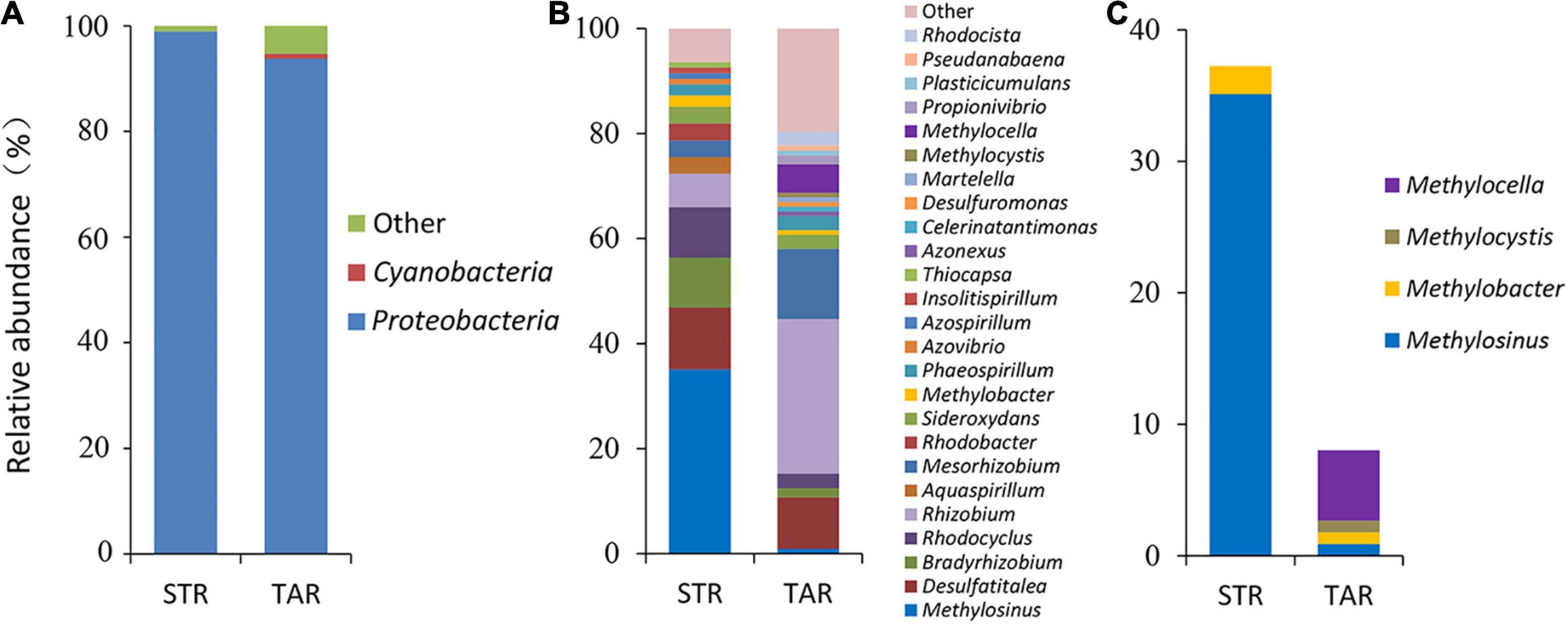
Figure 3. Relative abundance of active root-associated diazotrophic bacterial communities at the phylum (A) and genus (B) level and diazotrophic methanotrophic communities (C) in TAR and STR plants at the genus level based on a cDNA analysis of the nifH gene. TAR, Typha angustifolia roots; STR, Scirpus triqueter roots.
Further analyses of phylogenetic compositions at lower taxonomic levels showed that Methylosinus (35.1%), Desulfatitalea (11.7%), Bradyrhizobium (9.6%), Rhodocyclus (9.6%) and Rhizobium (10.7%) were primarily responsible for the community shifts of Alpha/Gamma/Deltaproteobacteria in S. triqueter (Figure 3B). In contrast, the dominant diazotrophs (> 5%) in T. angustifolia were Rhizobium (29.5%), Mesorhizobium (13.4%), Desulfatitalea (9.8%) and Methylocella (5.4%), while diazotrophic Methylocella (5.4%) affiliated with the methanotrophs also comprised a large proportion (Figures 3B,C). The composition of root-associated diazotrophic methanotrphs clearly differed between the two plants (Figure 3C). Methylosinus of Methylocystaceae was dominant in S. triqueter (35.1%) and minor in T. angustifolia (0.9%). Methylocella (5.4%), Methylobacter (0.9%) and Methylocystis (0.9%) were detected from T. angustifolia, while only Methylobacter (2.1%) was detected from S. triqueter (Figure 3C).
Clustering analysis of the nifH sequences enabled the identification of OTUs responsible for the population shifts of Alpha-Beta and Deltaproteobacteria at the species level (Figure 4). The most abundant OTUs STR18 (S. triqueter) and TAR85 (T. angustifolia) exhibited 99.1% and 99.5% sequence identity to the nifH sequences of Methylosinus sporium and Rhizobium sp. R2-708, respectively (Figure 4). Other clones STR112, STR33, TAR85, and TAR181 were also present at much higher levels in the roots of S. triqueter than those of T. angustifolia and were identified as Rhodocyclus tenuis (97.4%), Bradyrhizobium sp. BRUESC984 (98.2%), and Desulfatitalea sp. Site_C24 (94.7%), respectively. In contrast, clones TAR101, TAR113, and TAR18 were detected more abundantly in the roots of T. angustifolia than those of S. triqueter and were identical to Rhizobium sp. R2-708 (100%), Methylocella tundra (98.2%), Mesorhizobium sp. RITF712 (97.3%), and Desulfatitalea tepidiphila (94.7%), respectively. In addition, a phylogenetic analysis of diazotrophic methanotrophs showed that the representative clones of five OTUs (STR18, TAR101, TAR521, STR54, and TRA38) were affiliated with known genera of methanotrophs, such as Methylosinus, Methylocella, Methylocystis and Methylobacter (Supplementary Figure 3).
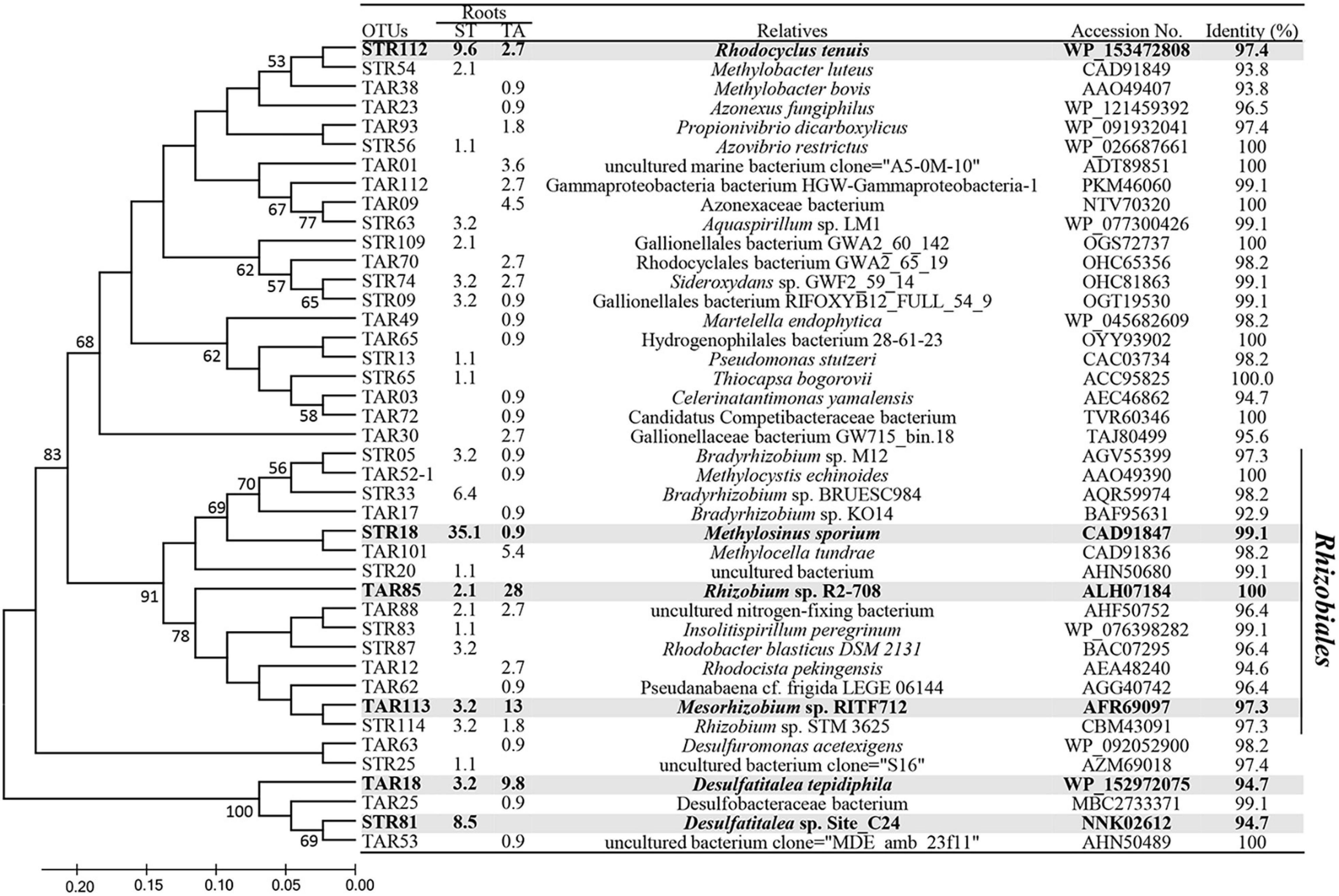
Figure 4. Phylogenetic distribution of representative operational taxonomc units (OTUs) of (≥ 94% amino acid identity) based on translated nifH gene sequences from the roots of Scirpus triqueter and Typha angustifolia. The table shows the relative abundance of OTUs in each library and the results of a BLAST search using the representative sequences. The tree was constructed using the neighbor-joining method, and the bootstrap values (%) for each branch were determined using 1,000 iterations. Bootstrap values (> 50%) are shown to the left of nodes in the tree. The main OTUs (> 5% relative abundance) are shown in gray.
Copy Numbers of the nifH Gene Based on cDNA
To estimate the population levels of active diazotrophs, we conducted real-time quantitative PCR of the root samples of S. triqueter and T. angustifolia from the wetlands based on RNA (cDNA; Figure 5). The copy numbers of nifH in the roots of S. triqueter and T. angustifolia ranged from 105 to 106 and did not differ significantly between these two plants (Figure 4; P > 0.05).
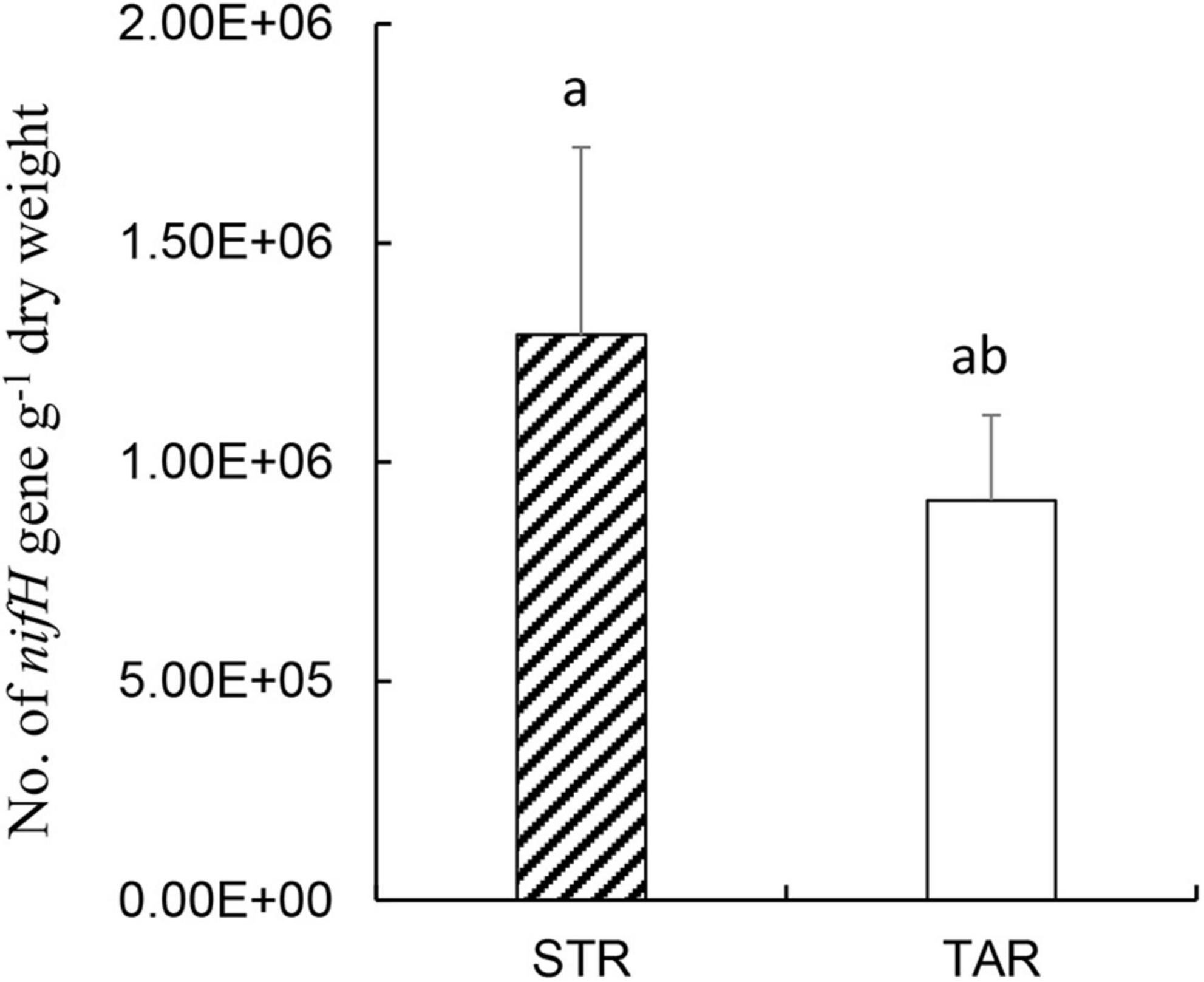
Figure 5. Numbers of nifH gene copies based on cDNA in the root samples from STR and TAR plants during July in the wetlands. Bars represent standard errors (n = 3). Bars with different letters (a, ab) are not significantly different (p < 0.05) according to Tukey’s test for pairwise mean comparison at α = 0.05. STR, Scirpus triqueter roots. TAR, Typha angustifolia roots.
Discussion
In this study, we present direct evidence that shows that CH4-dependent 15N2 fixation occurred in the roots of two different emergent macrophytes (S. triqueter and T. angustifolia) grown in a natural wetland (Figure 2A). CH4-stimulated N2 fixation occurred in an N-sufficient environment, which combined two different processes of research on N2 fixation and methanotrophs in eutrophic wetlands.
Plant hosts have a substantial influence on the distribution of microorganisms. The bacterial communities of different plants, including roots, stems and leaves, vary between plant species (Bulgarelli et al., 2013; Pietrangelo et al., 2018). The same type of regulation was found in studies of root methanotrophs; the difference in plant species strongly affected the community structure (Yoshida et al., 2014; Liu et al., 2020) and level of expression of the methanotrophs (Cui et al., 2020). In this study, we compared the structure of diazotrophic communities in the roots of S. triqueter and T. angustifolia. Although the habitats of the two plants were similar, the activity of communities on their roots differed notably. S. triqueter primarily used methanotrophs for nitrogen fixation, while T. angustifolia depended on Rhizobium for nitrogen fixation. This suggests that the species of plant also affected the diazotrophic community.
Type II methanotrophs, particularly Methylosinus, have been proven to fix N2 in rice roots (Bao et al., 2014a; Shinoda et al., 2019). In this study, we found that the abundance of Methylosinus in S. triqueter was much higher than that in T. angustifolia, and the ability of the microorganisms on S. triqueter to fix N2 was also significantly higher according to the 15N2 feeding experiment. Therefore, the abundance of Methylosinus positively correlated with the rate of fixation of N2 on the roots. This was consistent with the research of Bao et al. (2014a) on CH4-dependent 15N2 fixation flora in rice. These results showed that CH4-dependent nitrogen fixation is common in the roots of many emergent plants, and Methylosinus played a significant role in CH4-dependent nitrogen fixation.
Nitrogen fixation, the process by which N2 is converted into ammonia (NH3) via the enzyme nitrogenase, is often inhibited by O2 (Cheng, 2008; Reed et al., 2011). Compared with T. angustifolia, S. triqueter was primarily distributed in shallow water areas in wetlands in this study. Thus, the high concentration of oxygen in the root system favors the habitat of rhizospheric diazotrophic methane-oxidizing bacteria. Moreover, the ability of Methylosinus to fix nitrogen was resistant to high concentrations of O2 (Shinoda et al., 2019). In contrast, T. angustifolia is primarily distributed in deep waters in wetlands (Cui et al., 2020) where the root system has relatively oxic conditions because of the large amount of O2 diffusion (Clevering et al., 1996). This could explain why the root-associated Methylosinus of S. triqueter was found in abundance, and the plant had a greater ability to fix nitrogen compared with T. angustifolia (Figures 2–4). Moreover, other minor methanotrophs, such as Methylocella, Methylocystis and Methylobacter, may also contribute to CH4 oxidation-dependent nitrogen fixation in this study. Type II methanotrophs, such as Methylosinus and Methylocella, are likely to contribute to nitrogen fixation in forest soil and rice plants (Buckley et al., 2008; Bao et al., 2014a; Mäkipää et al., 2018; Shinoda et al., 2019). These results were consistent with those of this study and suggest that aerobic Type II methanotrophs are widely distributed diazotrophs in natural conditions. However, the dominant diazotrophic Methylosinus in this study did not clearly correspond with the root-associated Methylocystis (Type II methanotrophs) that were dominant in S. triqueter and T. angustifolia based on cDNA amplicons of the pmoA gene (Cui et al., 2020). This may be owing to the fact that the amplified sequence of pmoA gene (510 bp) exceeds the sequence range of the MiSeq sequencing platform (< 500 bp), resulting in inaccurate BLAST results of the sequences. Moreover, it is well known that the PCR-based amplification approach also results in bias (Suzuki and Giovannoni, 1996; Sipos et al., 2007). To elucidate this methodological issue, a metatranscriptomic and/or metaproteomic analysis of root-associated microbiome should be conducted to validate the hypothesis that Methylosinus simultaneously contributes to both CH4 oxidation and N2 fixation in natural wetlands.
The concentration of mineral nitrogen (NH4+ or NO3–) is also one of the main factors that affects nitrogen fixation (Reed et al., 2011). Previous studies on nitrogen fixation by methane-oxidizing bacteria have primarily focused on nitrogen-deficient environments (Bao et al., 2014a,b; Larmola et al., 2014; Shinoda et al., 2019). Interestingly, although Wuliangsuhai is a relatively eutrophic wetland (Cui et al., 2020), methane oxidation-dependent nitrogen fixation still occurred in the roots of emergent macrophytes (Figure 1). Nitrogen fixation has recently been found to occur under high nitrogen conditions in forest soils because of the high C/N ratio (Zheng et al., 2020). In wetlands, the CH4 produced from anaerobic sediment as carbon and energy sources aids in nitrogen fixation by root-associated methanotrophs (Bao et al., 2014a). In addition, the concentrations of NH4+ are very low in the root zone of macrophytes, such as S. triqueter, during the growing season (Bodelier et al., 1996). Therefore, the plants might require more nitrogen for growth, and low N availability is conducive to N2 fixation (Reed et al., 2011). This result suggests that the nitrogen fixation of root-associated methanotrophs depends on plant species and can occur in nitrogen-sufficient environments, such as eutrophic wetlands.
In addition to methanotrophs, other diazotrophs, including Rhizobium, Bradyrhizobium and Mesorhizobium, are well known to fix nitrogen and are frequently detected together with methanotrophs from rice (Bao et al., 2014a; Ikeda et al., 2014; Liu et al., 2021) and other plants (Hara et al., 2019; Yoneyama et al., 2019). These nitrogen-fixing bacteria are methylotrophs and can utilize the methanol produced from plants (Hiroyuki et al., 2015; Macey et al., 2020) or from the methane oxidation process by methanotrophs (Hanson and Hanson, 1996). In addition, methylotrophs are dominant in land plant-associated soil (Macey et al., 2020), and over 40% of the diazotrophs are methylotrophs in rice roots (Liu et al., 2021). Methanotrophs typically produce methanol via the first step of methane oxidation process, and diazotrophic methylotrophs may utilize the methanol as carbon and energy sources to fix N2 (Bao et al., 2014a; Liu et al., 2021). Therefore, root-associated methanol-utilizing nitrogen-fixing bacteria cannot be ignored in wetlands. These results suggest that C1-cycling bacteria, including methanotrophs and methylotrophs, in the root zones of aquatic plants are important for the reduction of greenhouse gas methane and increase the benefits for stimulation of plant growth by biofertilizers.
In summary, this study revealed that CH4 oxidation-dependent nitrogen fixation clearly occurred in the root tissues of emergent plants (S. triqueter and T. angustifolia) in natural wetlands, and it differed between plant species. A cDNA-based nifH gene sequencing analysis suggests that root-associated aerobic methanotrophs, in particular Methylosinus (type II methanotroph), contributed to CH4 oxidation-dependent N2 fixation. In addition, the plant species had a significant effect on the root-associated diazotrophic communities. Following our previous studies on the CH4 oxidation-dependent N2 fixation of rice fields, this study provides evidence that aerobic methanotrophs fix N2 in the roots of emergent plants, and they have a potential role in the reduction of CH4 emissions and enhancement of plant growth in natural wetlands.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZB and JZ designed the study. ZB, JC, MZ, LC, and SZ performed the experiments. JC, YL, WC, LW, ZJ, JZ, and ZB analyzed the data. ZB and JC wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (Nos. 41563009 and 32160028), the Science and Technology Major Project on Lakes of Inner Mongolia (No. ZDZX2018054), and the Natural Science Foundation of Inner Mongolia (No. 2019MS04005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.851424/full#supplementary-material
Footnotes
References
Auman, A. J., Speake, C. C., and Lidstrom, M. E. (2001). nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ. Microbiol. 67, 4009–4016. doi: 10.1128/AEM.67.9.4009-4016.2001
Bao, Z. H., Okubo, T., Kubota, K., Kasahara, Y., Tsurumaru, H., Anda, M., et al. (2014a). Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl. Environ. Microbiol. 80, 5043–5052. doi: 10.1128/AEM.00969-14
Bao, Z., Watanabe, A., Sasaki, K., Okubo, T., Tokida, T., Liu, D., et al. (2014b). A rice gene for microbial symbiosis, OsCCaMK, reduces CH4 flux in a paddy field with low nitrogen input. Appl. Environ. Microbiol. 80, 1995–2003. doi: 10.1128/AEM.03646-13
Bhagowati, B., and Ahamad, K. U. (2018). A review on lake eutrophication dynamics and recent 4 developments in lake modeling. Ecohydrol. Hydrobiol. 19, 155–166. doi: 10.1016/j.ecohyd.2018.03.002
Bodelier, P. L. E., and Laanbroek, H. J. (2004). Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS. Microbiol. Ecol. 47, 265–277. doi: 10.1016/S0168-6496(03)00304-0
Bodelier, P. L. E., Libochant, J. A., Blom, C. W. P. M., and Laanbroek, H. J. (1996). Dynamics of nitrification and denitrification in root-oxygenated sediments and adaptation of ammonia-oxidizing bacteria to low-oxygen or anoxic habitats. Appl. Environ. Microbiol. 62, 4100–4107. doi: 10.1128/aem.62.11.4100-4107.1996
Buckley, D. H., Huangyutitham, V., Hsu, S. F., and Nelson, T. A. (2008). 15N2–DNA–stable isotope probing of diazotrophic methanotrophs in soil. Soil. Biol. Biochem. 40, 1272–1283. doi: 10.1007/978-1-4939-9721-3_2
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver Loren, van Themaat, E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant. Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Cai, Y., Zheng, Y., Bodelier, P. L., Conrad, R., and Jia, Z. (2016). Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat. Commun. 7:11728. doi: 10.1038/ncomms11728
Calhoun, A., and King, G. M. (1998). Characterization of root-associated methanotrophs from three freshwater macrophytes: Pontederia cordata, Sparganium eurycarpum, and Sagittaria latifolia. Appl. Environ. Microbiol. 64, 1099–1105. doi: 10.1128/AEM.64.3.1099-1105.1998
Chen, X., Zhu, H., Yan, B., Shutes, B., Xing, D., Banuelos, G., et al. (2019). Greenhouse gas emissions and wastewater treatment performance by three plant species in subsurface flow constructed wetland mesocosms. Chemosphere 239:124795. doi: 10.1016/j.chemosphere.2019.124795
Cheng, Q. (2008). Perspectives in biological nitrogen fixation research. J. Integr. Plant. Biol. 50, 786–798. doi: 10.1111/j.1744-7909.2008.00700.x
Clevering, O. A., Blom, C. W. P. M., and Van Vierssen, W. (1996). Growth and morphology of Scirpus lacustris and S. maritimus seedlings as affected by water level and light availability. Functl. Ecol. 10, 289–296. doi: 10.2307/2389855
Conrad, R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Env. Microbiol. Rep. 1, 285–292. doi: 10.1111/j.1758-2229.2009.00038.x
Cui, J., Zhao, J., Wang, Z., Cao, W., Zhang, S., Liu, J., et al. (2020). Diversity of active root-associated methanotrophs of three emergent plants in a eutrophic wetland in northern China. AMB Express 10:48. doi: 10.1186/s13568-020-00984-x
Dalton, H., and Whittenbury, R. (1976). The acetylene reduction technique as an assay for nitrogenase activity in the methane oxidizing bacterium Methylococcus capsulatus strain bath. Arch. Microbiol. 109, 147–151. doi: 10.1007/bf00425127
Dekas, A. E., Poretsky, R. S., and Orphan, V. J. (2009). Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326, 422–426. doi: 10.1126/science.1178223
Duan, X. N., Wang, X. K., Mu, Y. J., and Ouyang, Z. Y. (2005). Seasonal and diurnal vari- ations in methane emissions from Wuliangsu Lake in arid regions of China. Atmos. Environ. 39, 4479–4487. doi: 10.1016/j.atmosenv.2005.03.045
Fausser, A. C., Hoppert, M., Walther, P., and Kazda, M. (2012). Roots of the wetland plants Typha latifolia and Phragmites australis are inhabited by methanotrophic bacteria in biofilms. Flora 207, 775–782. doi: 10.1016/j.flora.2012.09.002
Hara, S., Morikawa, T., Wasai, S., Kasahara, Y., Koshiba, T., Yamazaki, K., et al. (2019). Identification of nitrogen-fixing Bradyrhizobium associated with roots of field-grown Sorghum by metagenome and proteome analyses. Front. Microbiol. 10:407. doi: 10.3389/fmicb.2019.00407
Hiroyuki, I., Hiroya, Y., and Yasuyoshi, S. (2015). Interactions of methylotrophs with plants and other heterotrophic bacteria. Microorganisms 3, 137–151. doi: 10.3390/microorganisms3020137
Ho, A., and Bodelier, P. L. E. (2015). Diazotrophic methanotrophs in peatlands: the missing link? Plant. Soil 389, 419–423. doi: 10.1007/s11104-015-2393-9
Ikeda, S., Sasaki, K., Okubo, T., Yamashita, A., Terasawa, K., Bao, Z., et al. (2014). Low nitrogen fertilization adapts rice root microbiome to low nutrient environment by changing biogeochemical functions. Microbes Environ. 29, 50–59. doi: 10.1264/jsme2.me13110
Khadem, A. F., Pol, A., Jetten, M. S. M., and Op den Camp, H. J. M. (2010). Nitrogen fixation by the verrucomicrobial methanotroph ‘Methylacidiphilum fumariolicum’. Sol. Microbiol. 156, 1052–1059. doi: 10.1099/mic.0.036061-0
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Laanbroek, H. J. (2010). Methane emission from natural wetlands: interplay between emergent macrophytes and soil microbial processes. a mini-review. Ann. Botany 105, 141–153. doi: 10.1093/aob/mcp201
Larmola, T., Leppanen, S. M., Tuittila, E. S., Aarva, M., Merila, P., Fritze, H., et al. (2014). Methanotrophy induces nitrogen fixation during peatland development. Proc. Natl. Acad. Sci. U S A. 111, 734–739. doi: 10.1073/pnas.1314284111
Liu, J., Bao, Z., Cao, W., Han, J., Zhao, J., Kang, Z., et al. (2020). Enrichment of type I methanotrophs with nirS genes of three emergent macrophytes in a eutrophic wetland in China. Microbes. Environ. 35:1. doi: 10.1264/jsme2.ME19098
Liu, J., Han, J., Zhu, C., Cao, W., Luo, Y., Zhang, M., et al. (2021). Elevated atmospheric CO2 and nitrogen fertilization affect the abundance and community structure of rice root-associated nitrogen-fixing bacteria. Front. Microbiol. 12:628108. doi: 10.3389/fmicb.2021.628108
Macey, M. C., Pratscher, J., Crombie, A. T., and Murrell, J. C. (2020). Impact of plants on the diversity and activity of methylotrophs in soil. Microbiome 8:31. doi: 10.1186/s40168-020-00801-4
Mäkipää, R., Leppnen, S. M., Muoz, S. S., Smolander, A., and Fritze, H. (2018). Methanotrophs are core members of the diazotroph community in decaying norway spruce logs. Soil. Biol. Biochem. 120, 230–232. doi: 10.1016/j.soilbio.2018.02.012
Martin-Laurent, F., Philippot, L., Hallet, S., Chaussod, R., Germon, J. C., Soulas, G., et al. (2001). DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67, 2354–2363. doi: 10.1128/AEM.67.5.2354-2359.2001
Minamisawa, K., Imaizumi-Anraku, H., Bao, Z., Shinoda, R., Okubo, T., and Ikeda, S. (2016). Are symbiotic methanotrophs key microbes for N acquisition in paddy rice root? Microbes Environ. 31, 4–10. doi: 10.1264/jsme2.ME15180
Nat, F., and Middelburg, J. J. (1998). Seasonal variation in methane oxidation by the rhizosphere of phragmites australis and scirpus lacustris. Aquat. Bot. 61, 95–110. doi: 10.1016/s0304-3770(98)00072-2
Pietrangelo, L., Bucci, A., Maiuro, L., Bulgarelli, D., and Naclerio, G. (2018). Unraveling the composition of the root-associated bacterial microbiota of Phragmites australis and Typha latifolia. Front. Microbiol. 9:1650. doi: 10.3389/fmicb.2018.01650
Poly, F., Monrozier, L. J., and Bally, R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. doi: 10.1016/s0923-2508(00)01172-4
Reed, S. C., Cleveland, C. C., and Townsend, A. R. (2011). Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annual. Rev. Ecol. Evol. S. 42, 489–512. doi: 10.1146/annurev-ecolsys-102710-145034
Reumer, M., Harnisz, M., Lee, H. J., Reim, A., Grunert, O., Putkinen, A., et al. (2018). Impact of peat mining and restoration on methane turnover potential and methane-cycling microorganisms in a northern bog. Appl. Environ. Microbio. 84:e002218–17. doi: 10.1128/AEM.02218-17
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shinoda, R., Bao, Z. H., and Minamisawa, K. (2019). CH4 oxidation-dependent 15N2 fixation in rice roots in a low-nitrogen paddy field and in Methylosinus sp. strain 3S-1 isolated from the roots. Soil. Biol. Biochem. 132, 40–46. doi: 10.1016/j.soilbio.2019.01.021
Sipos, R., Székely, A. J., Palatinszky, M., Révész, S., Márialigeti, K., and Nikolausz, M. (2007). Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS. Microbiol. Ecol. 60, 341–350. doi: 10.1111/j.1574-6941.2007.00283.x
Suzuki, M. T., and Giovannoni, S. J. (1996). Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62, 625–630. doi: 10.1128/aem.62.2.625-630.1996
Takeda, K. (1988). Characteristics of a nitrogen-fixing methanotroph, Methylocystis T-1. Antonie. Van. Leeuwenhoek 54, 521–534. doi: 10.1007/BF00588388
Takeda, K., Tonouchi, A., Takada, M., Suko, T., Suzuki, S., Kimura, Y., et al. (2008). Characterization of cultivable methanotrophs from paddy soils and rice roots. Soil Sci. Plant Nutr. 54, 876–885. doi: 10.1111/j.1747-0765.2008.00318.x
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weigh matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Van der Nat, F., and Middelburg, J. J. (1998). Effects of two common macrophytes onmethane dynamics in freshwater sediments. Biogeochemistry 43, 79–104. doi: 10.1002/eap.2160
Wartiainen, I., Eriksson, T., Zheng, W. W., and Rasmussen, U. (2008). Variation in the active diazotrophic community in rice paddy—nifH PCR-DGGE analysis of rhizosphere and bulk soil. Appl. Soil Ecol. 39, 65–75. doi: 10.1016/j.apsoil.2007.11.008
Yoneyama, T., Terakado-Tonooka, J., Bao, Z., and Minamisawa, K. (2019). Molecular analyses of the distribution and function of diazotrophic rhizobia and methanotrophs in the tissues and rhizosphere of non-leguminous plants. Plants (Basel) 8:408. doi: 10.3390/plants8100408
Yoshida, N., Iguchi, H., Yurimoto, H., Murakami, A., and Sakai, Y. (2014). Aquatic plant surface as a niche for methanotrophs. Front. Microbiol. 5:30. doi: 10.3389/fmicb.2014.00030
Yun, J., Yu, Z., Ke, L., and Zhang, H. (2013). Diversity, abundance and vertical distribution of methane-oxidizing bacteria (methanotrophs) in the sediments of the xianghai wetland, songnen plain, northeast china. J. Soil. Sediment. 13, 242–252. doi: 10.1007/s11368-012-0610-1
Zhang, K., Luo, H., Zhu, Z., Chen, W., Chen, J., and Mo, Y. (2018). CH4 flux and methanogen community dynamics from five common emergent vegetations in a full-scale constructed wetland. Environ. Sci. Pollut. Res. Int. 25, 26433–26445. doi: 10.1007/s11356-018-2692-9
Keywords: natural wetland, stable isotope analysis, nitrogen fixation, diazotrophic methanotroph, emergent plant
Citation: Cui J, Zhang M, Chen L, Zhang S, Luo Y, Cao W, Zhao J, Wang L, Jia Z and Bao Z (2022) Methanotrophs Contribute to Nitrogen Fixation in Emergent Macrophytes. Front. Microbiol. 13:851424. doi: 10.3389/fmicb.2022.851424
Received: 09 January 2022; Accepted: 14 March 2022;
Published: 11 April 2022.
Edited by:
Anne Bernhard, Connecticut College, United StatesReviewed by:
Anandham Rangasamy, Tamil Nadu Agricultural University, IndiaIrina Kravchenko, Winogradsky Institute of Microbiology (RAS), Russia
Copyright © 2022 Cui, Zhang, Chen, Zhang, Luo, Cao, Zhao, Wang, Jia and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihua Bao, emhpaHVhX2Jhb0BpbXUuZWR1LmNu
 Jing Cui1,2
Jing Cui1,2 Meng Zhang
Meng Zhang Shaohua Zhang
Shaohua Zhang Ying Luo
Ying Luo Weiwei Cao
Weiwei Cao Zhongjun Jia
Zhongjun Jia Zhihua Bao
Zhihua Bao