- 1Key Laboratory of Infectious Immune and Antibody Engineering of Guizhou Province, School of Basic Medical Sciences/School of Biology and Engineering, Guizhou Medical University, Guiyang, China
- 2Key Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, Guizhou Medical University, Guiyang, China
- 3Immune Cells and Antibody Engineering Research Center of Guizhou Province, Cellular Immunotherapy Engineering Research Center of Guizhou Province, Guizhou Medical University, Guiyang, China
- 4State Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University, Guiyang, China
- 5Wheat Anti-toxin Breeding Laboratory, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
Fusarium is one of the most important phytopathogenic and mycotoxigenic fungi that caused huge losses worldwide due to the decline of crop yield and quality. To systematically investigate the infections of Fusarium species in ear rot of maize in the Guizhou Province of China and analyze its population structure, 175 samples of rotted maize ears from 76 counties were tested by combining immunoassays and molecular identification. Immunoassay based on single-chain variable fragment (scFv) and alkaline phosphatase (AP) fusion protein was first employed to analyze these samples. Fusarium pathogens were isolated and purified from Fusarium-infected samples. Molecular identification was performed using the partial internal transcribed spacer (ITS) and translation elongation factor 1α (TEF-1α) sequences. Specific primers were used to detect toxigenic chemotypes, and verification was performed by liquid chromatography tandem mass spectrometry (LC–MS/MS). One-hundred and sixty three samples were characterized to be positive, and the infection rate was 93.14%. Sixteen species of Fusarium belonging to six species complexes were detected and Fusarium meridionale belonging to the Fusarium graminearum species complex (FGSC) was the dominant species. Polymerase chain reaction (PCR) identification illustrated that 69 isolates (56.10%) were potential mycotoxin-producing Fusarium pathogens. The key synthetic genes of NIV, NIV + ZEN, DON + ZEN, and FBs were detected in 3, 35, 7, and 24 isolates, respectively. A total of 86.11% of F. meridionale isolates carried both NIV- and ZEN-specific segments, while Fusarium verticillioides isolates mainly represented FBs chemotype. All the isolates carrying DON-producing fragments were FGSC. These results showed that there are different degrees of Fusarium infections in Guizhou Province and their species and toxigenic genotypes display regional distribution patterns. Therefore, scFv-AP fusion-based immunoassays could be conducted to efficiently investigate Fusarium infections and more attention and measures should be taken for mycotoxin contamination in this region.
Introduction
Maize (Zea mays L.) is an important food and feed crop worldwide and one of the main crop in Guizhou Province, China. Numerous pathogenic fungi can infect maize and cause Gibberella ear and stalk rot, resulting in yield and quality reductions and a threat to global food security (Pechanova and Pechan, 2015). Fusarium is one of the most economically destructive and species-rich groups of large-scale pathogenic fungi in the world. These pathogens are present in various natural environments and can infect host crops throughout the growth cycle, causing seed rot, seedling rot, stem rot and panicle rot (Karlsson et al., 2021; Palacios et al., 2021). Fungal crop diseases not only seriously affect the growth and development of plants and product quality but also produce a variety of mycotoxins, such as nivalenol (NIV), zealenone (ZEN), deoxynivalenol (DON), and fumonisins (FBs) (Torres et al., 2019). More seriously, these mycotoxins are detrimental to human and animal health due to their serious acute toxicity, cytotoxicity, immunotoxicity, teratogenicity, mutagenicity and carcinogenicity (Lee and Ryu, 2017; Sun et al., 2017).
The production of food and feed crops in China mainly includes rice, wheat, and maize. Fusarium can invade at multiple stages of plant growth and is widely present in these important food crops (Torres et al., 2019; Palacios et al., 2021). Fusarium pathogens often appear in high-humidity and high-heat areas and Fusarium head blight (FHB) has reached historically high epidemic acreages in the middle and lower reaches of the Yangtze River (Wang et al., 2011; Yang et al., 2021). It is also one of the major diseases in wheat and maize planting areas in the Northeast Plain, North China Plain, and Sichuan Basin (Feng et al., 2011; Wei et al., 2013; Wang et al., 2021). Thus, Fusarium poses a great threat to China’s food production (Gong et al., 2009; Qiu and Shi, 2014; Qiu et al., 2019). Simultaneously, the suitable living environment of different Fusarium species varies and is influenced by ecological factors such as geography and climate. The dominant species of pathogens isolated in different countries, different regions, and different years display significant discrepancies (Zhang et al., 2007). Guizhou is located in the Yunnan-Kweichow Plateau, and the temperature is relatively low. Theoretically, it is not a high-incidence area of Fusarium diseases, and rarely are concerns focused on these regions. In recent years, some investigations have shown that Fusarium can cause plant diseases in this area, such as maize ear rot, Pinellia tuber rot, and tobacco root rot (Shi et al., 2015; Yu and Yao, 2018; Shang et al., 2019). However, the infections, distribution and mycotoxin production of Fusarium pathogens in this area have not been systematically investigated.
Fusarium species have some discrepancies in their housekeeping genes, and molecular identification based on these genes has been widely used for further identification of morphological Fusarium species. DNA sequences, such as translation elongation factor 1-α (TEF-1α), β-tubulin (β-TUB), calmodulin (CAM), mitochondrial small subunit rDNA (mtSSU), 28S rDNA, and internal transcribed spacer (ITS) regions, are widely used for phylogenetic analysis to assess the genetic relationships of Fusarium species (Wang et al., 2014b). O’Donnell et al. (2013, 2015) divided Fusarium into 20 species complexes based on Genealogical Concordance Phylogenetic Species Recognition (GCPSR) analyses of more than 300 pathogens, which provided the basis for the identification and classification of Fusarium species based on DNA sequences. Unfortunately, the pretreatment of molecular identification is labor intensive and time-consuming because the pathogenic fungi in each sample must be subcultured, purified, and polymerase chain reaction (PCR) amplified (Santos et al., 2016; Cambaza et al., 2019; Schiwek et al., 2020). Enzyme-linked immunosorbent assay (ELISA) has distinctive characteristics of simple operation, low price, high sensitivity, good specificity and simple pretreatment of samples (Saccon et al., 2017; Rahman et al., 2019). At present, many fungi-specific antibodies have been exploited to develop immunoassays for rapid detection and monitoring of fungal infections (Wang et al., 2017; He et al., 2018). Therefore, the application of immunoassays for preliminary screening of Fusarium-infected samples is more efficient and convenient for further molecular identification. In our previous study, a single chain variable fragment (scFv) named FvSG7 has been selected from a phage display library and its fusion protein with alkaline phosphatase (FvSG7-AP) has been verified to efficiently detect Fusarium pathogens in cereal grains (Hu et al., 2013). Therefore, this study aimed to first investigate the Fusarium infections in maize and geographic distribution in Guizhou Province of China by using the established rapid immunoassay method. Furthermore, their population structure and toxigenic chemotypes were analyzed by PCR identification and liquid chromatography tandem mass spectrometry (LC–MS/MS) detection. Our results will lay a foundation for effective identification of Fusarium pathogens in the field and further understanding the distribution characteristics of Gibberella ear rot and mycotoxin chemotypes.
Materials and Methods
Experimental Materials
Diseased maize ears with similar symptoms, including kernels covered with white, pink or salmon-colored mold or exhibiting a white streaking (“starburst”) symptom, were collected from counties in Guizhou Province, China, in the maize-harvesting period.
In order to quickly analyze the Fusarium infections in ear rot of maize samples, FvSG7-AP fusion was used for Fusarium detection with one-step ELISA. The recombinant Escherichia coli strain XL1-Blue/pDAP2/S-FvSG7 was obtained by transforming the recombinant plasmid pDAP2/S-FvSG7 containing the FvSG7-AP fusion gene into E. coli XL1-Blue competent cells. The FvSG7 gene (GenBank accession number KC304795) encodes an anti-Fusarium scFv antibody isolated previously (Hu et al., 2013), and the pDAP2/S vector contains a gene encoding AP enzyme (Kerschbaumer et al., 1997).
Expression of FvSG7-AP Fusion Protein
Twenty microliters of recombinant strain XL1-Blue/pDAP2/S-FvSG7 were inoculated into 20 mL of 2 × TY medium (16 g/L tryptone, 10 g/L yeast extract, and 5 g/L NaCl, pH 7.0) supplemented with 100 μg/mL Amp. After incubation overnight at 37°C and 200 r/min, 10 mL of the culture was inoculated into 200 mL of 2 × TY medium supplemented with 100 μg/mL Amp and cultured at 37°C and 200 r/min until the OD600 nm reached 0.5–0.6. A final concentration of 0.1 mmol/L isopropyl-β-D-thiogalactopyranoside (IPTG) was added for 20 h of induction at 16°C and 200 r/min. The FvSG7-AP fusion protein was extracted by ultrasonication, and the enzyme activity of AP was tested using p-nitrophenyl phosphate (pNPP) solution (Wang et al., 2015). The fusion protein was purified by Ni-NTA chromatography, and its concentration was determined by the Brandford method.
Immunoassay Detection of Samples
The collected maize samples were crushed to power and detected by using ELISA method based on the FvSG7-AP fusion protein (Hu et al., 2013). In detail, 0.2 g of each sample was weighed and transferred to 1.5-mL Eppendorf tubes. Then, 1 mL of phosphate-buffered saline (PBS) was added and incubated for 30 min at room temperature with shaking. The homogenates were left standing for 10 min, and then, 100 μL of supernatant was pipetted into the ELISA plate wells. After incubation at 37°C for 2 h, the wells were washed three times with PBS buffer. Next, 200 μL of 2% skimmed milk was added to each well and incubated at 37°C for 2 h. After three washes with PBS, 100 μL of purified FvSG7-AP fusion protein was added to each well. The plates were placed in a 37°C incubator for 1.5 h and washed three times with PBST (PBS containing 0.1% Tween-20) and PBS buffer. Finally, 100 μL of 0.2% pNPP solution was added, and the absorbance was recorded at 405 nm by a microplate reader. Negative controls coated with healthy maize were set up, and each example was repeated in three wells.
Isolation of Fusarium Pathogens From Maize Kernels
Symptomatic kernels were soaked in 70% alcohol for 30 s and transferred into 2% sodium hypochlorite solution for another 2 min of immersion. After five washes with sterile water, the kernels were dried on sterile filter paper. Then, each seed was cut in half and placed on PDA medium for a 5-day incubation at 28°C in the dark. The mycelia were observed under a microscope, and colonies displaying morphological characteristics of Fusarium were subcultured onto fresh PDA medium (Nelson et al., 1983; Leslie and Summerell, 2006). The putative Fusarium colonies were purified using a single-sporing method.
Genomic DNA Extraction and PCR Amplification
Each isolate was inoculated on fresh PDA medium and cultured at a constant temperature of 28°C for 5 days, and then, the mycelial mass was harvested by scraping. The genomic DNA of Fusarium pathogens was extracted by using a fungal genomic DNA extraction kit (Solarbio, Beijing, China) and stored at −20°C. PCR amplification and sequencing of the ITS and TEF-1α genes were achieved using the primer pairs ITS4/ITS5 (White et al., 1990) and EF1T/EF2T (Mirete et al., 2004), respectively (Table 1). The PCR products were detected by 1% agarose gel electrophoresis and sequenced by Sangon Biological (Shanghai) Co., Ltd.
Phylogenetic Analysis
DNA sequences were aligned and adjusted manually using DNAStar-SeqMan software1. Sequence similarity searches were performed with the BLAST network service based on the FUSARIUM-ID database2 and NCBI GenBank.3 NRRL, MRC or CBS strains were taken as standard controls, and their TEF-1α sequences were downloaded (Supplementary Table 1). Phylogenetic analysis was performed with MEGA-X-10.0.5 software (Kumar et al., 2018), and phylogenetic trees were constructed by using the maximum-likelihood method. Numbers above branches were signed to indicate bootstrap values based on 1,000 replications. The best-fit model of molecular evolution was selected based on the estimation of Bayesian information criterion scores.
Molecular Identification of Toxigenic Chemotypes
Single and/or multiplex PCR were performed to analyze the genes involved in NIV, ZEN, DON, and FBs synthesis using chemotype-specific primer pairs (Table 1). Briefly, PCR amplification was conducted in a final volume of 25 μL containing 1 μL of genomic DNA, 1 μL of each primer (10 μmol/L), 12.5 μL of 2 × Taq PCR starMix kit (GenStar, Beijing, China). A negative control omitting DNA template was used in every set of reactions. The PCR products were detected by 1% agarose gel electrophoresis. Each toxigenic chemotype was identified using two or more primer pairs and repeated three times.
Liquid Chromatography Tandem Mass Spectrometry Detection of Mycotoxin Production
Two Fusarium isolates of each toxigenic chemotype (NIV: LPS-LZ-01, ZY-SY-01; ZEN: QXN-XR-01, GY-GSH-01; DON: QXN-ZF-01, BJ-HZ-02; and FBs: GY-XF-02, QN-LD-03) were selected as representative isolates. Also, Fusarium miscanthi represent isolates of GY-HX-03 and GY-HX-212 and Fusarium concentricum isolate QDN-RJ-01 were cultured. The mycotoxin production was measured by LC–MS/MS and repeated twice. In detail, the Fusarium isolates were inoculated on PDA and cultured at 28°C in the dark for 3–5 days. For NIV and ZEN, 4 mycelium-agar plugs with a 5-mm diameter were placed in a sterilized conical flask containing niblet medium culture (100 g of niblet, 50 mL of deionized water) and statically cultured at 28°C in the dark for 2 weeks. For DON, the mycelia were inoculated in half-strength CM-cellulose-yeast extract (CMC) broth to prepare the spore suspension (Xu et al., 2010), and then, 1 mL of spore fluid was inoculated into the niblet medium culture and statically cultured at 28°C in the dark for 2 weeks. For FBs, the mycelia were inoculated into a sterilized conical flask containing mung bean broth (2 g of mung bean, 200 mL of deionized water) to prepare the spore suspension, and then, 1 mL of spore fluid was inoculated into niblet medium culture for 3 weeks. Grains were dried in a 55°C incubator and finely ground to powder. Five grams of the samples were weighed and supplemented with 10 mL of water and 10 mL of acetonitrile containing 10% formic acid. After treatment with ultrasonication for 15 min, 1 g of citric acid, 1 g of NaCl and 4 g of MgSO4 were added. The mixture was vortexed for a few seconds, followed by centrifugation at 5,000 r/min for 5 min. Then, 5 mL of supernatant was pipetted into a tube, and 0.75 g of MgSO4, 0.25 g of primary secondary amine (PSA) and 0.15 g of octadecyl silane bonded phase (C18) were added, mixed and centrifuged at 5,000 r/min for 5 min. For each sample, 0.5 mL of the supernatant was pipetted, and 0.5 mL of 1% formic acid water was added. Subsequently, the fluid was passed through 0.22-μm nylon filters and quantified using an external standard method at Zhongke Youlong (Hangzhou) Food Safety Standard Technology Co., Ltd.
Chromatographic separation was studied on CNW Athena ultra high performance liquid chromatography (UHPLC) C18 Column (100 mm × 2.1 mm, 1.8 μm). The flow rate was set as 0.25 mL/min; the injection volume was 10.0 μL; and the column temperature was kept constant at 35°C. The mobile phase consisted of water containing 0.1% formic acid (A) and acetonitrile (B). The gradient elution program was performed as follows: 5% B from 0 to 2 min, 5–95% B from 2 to 12 min, 95–99% B from 12 to 12.1 min, 99% B from 12.1 to 14 min, 99–5% B from 14 to 14.1 min, 5% B from 14 to 16 min. The injection volume was 10 μL. MS/MS detection was performed on a triple quadrupled mass spectrometer detector equipped with a jet stream electrospray ionization (ESI) source under multi-reaction monitoring (MRM) conditions. ESI positive (ESI+) and negative (ESI–) subsection acquisition modes were used for the quantification with a capillary voltage of 5.5 kV. The specific MS parameters for mycotoxin analyses in this study are displayed in Supplementary Table 2.
Results
Expression of Fusion Protein and Enzyme-Linked Immunosorbent Assay Detection
FvSG7-AP fusions were induced for expression based on the optimization conditions, and a considerable quantity of soluble protein with high activity was obtained. A total of 175 diseased maize ears with similar symptoms, including kernels covered with white, pink or salmon-colored mold, were collected from 76 counties in Guizhou Province during the maize harvesting period. These kernels were pretreated and detected by rapid immunoassay using FvSG7-AP fusion proteins. As shown in Figure 1, only 5.14% (9/175) of samples displayed no difference compared to controls, 24% (42/175) had mild infections, 33.14% (58/175) had moderate infections, 18.86% (33/175) had serious infections, and 17.14% (30/175) had severe infections. In addition, 1.71% (3/175) of samples showed a small color reaction and were considered suspected of infections. Taken together, the incidence of Fusarium infections on collected maize samples was as high as 93.14%.
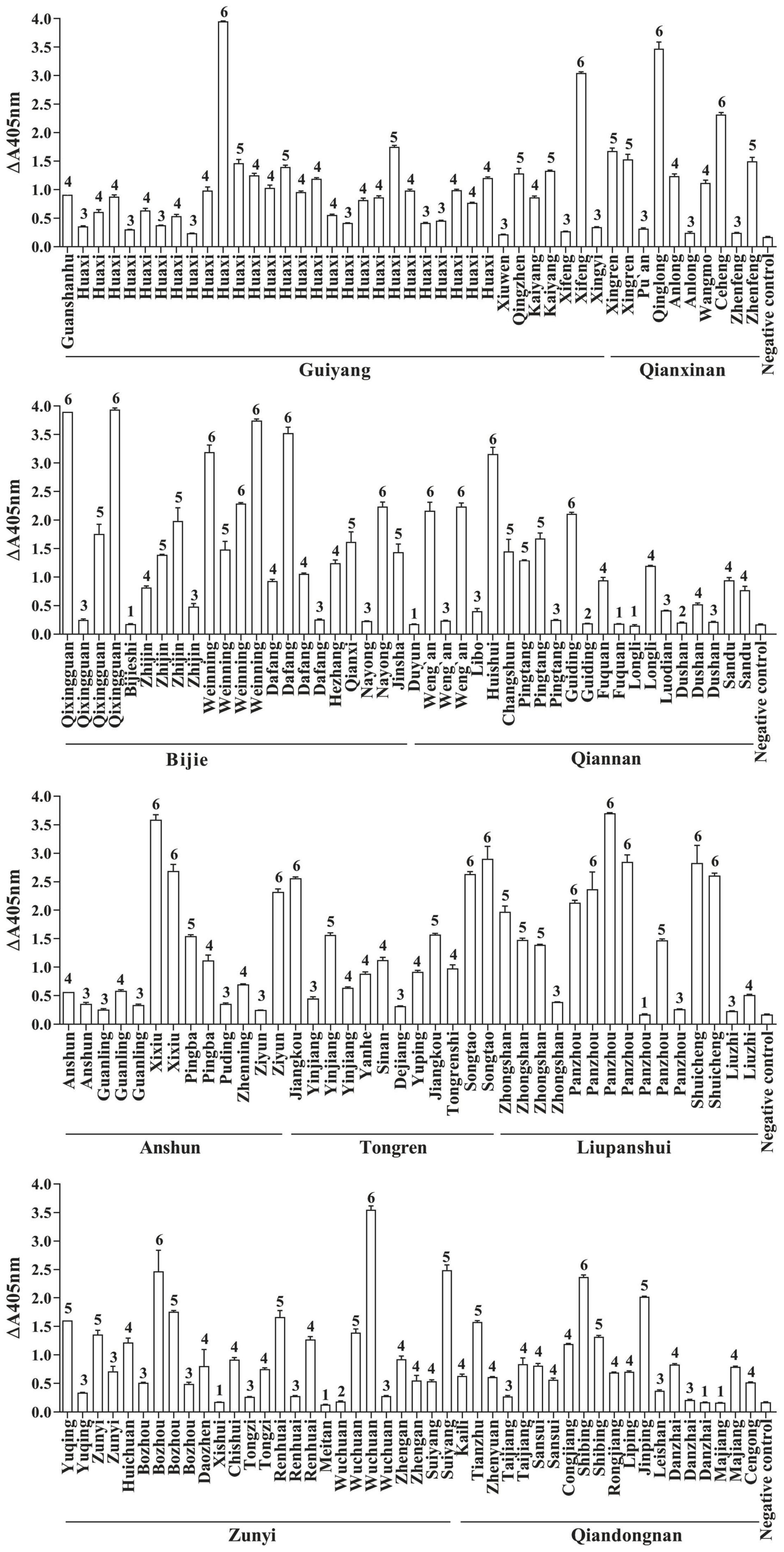
Figure 1. Fusarium infections in maize samples detected by rapid immunoassay. 1: n < 1.5, samples without Fusarium infections; 2: 1.5 ≤ n < 2, suspected infection samples; 3: 2 ≤ n < 10, mild infection samples; 4: 10 ≤ n < 30, moderate infection samples; 5: 30 ≤ n < 50, severe infection samples; 6: n ≥ 50, extreme infection samples; n = (OD405 nm value of sample–OD405 nm value of blank control)/(OD405 nm value of negative control–OD405 nm value of blank control).
Geographic Distribution of Fusarium-Infected Samples
The maize samples with or without Fusarium infections were marked on the map of Guizhou Province according to the results of immunoassay detection (Figure 2). The results showed that mild infected samples were detected in 10.53% (8/76) of regions, moderate infected samples were detected in 36.84% (28/76) of regions, severe and extreme infected samples were detected in 48.68% (37/76) of regions, and samples with Fusarium infections were not detected in 3.95% (3/76) of regions. Therefore, maize samples infected by Fusarium were found in 96.05% of counties of Guizhou Province, indicating a very wide distribution of Fusarium infections (Figure 2).
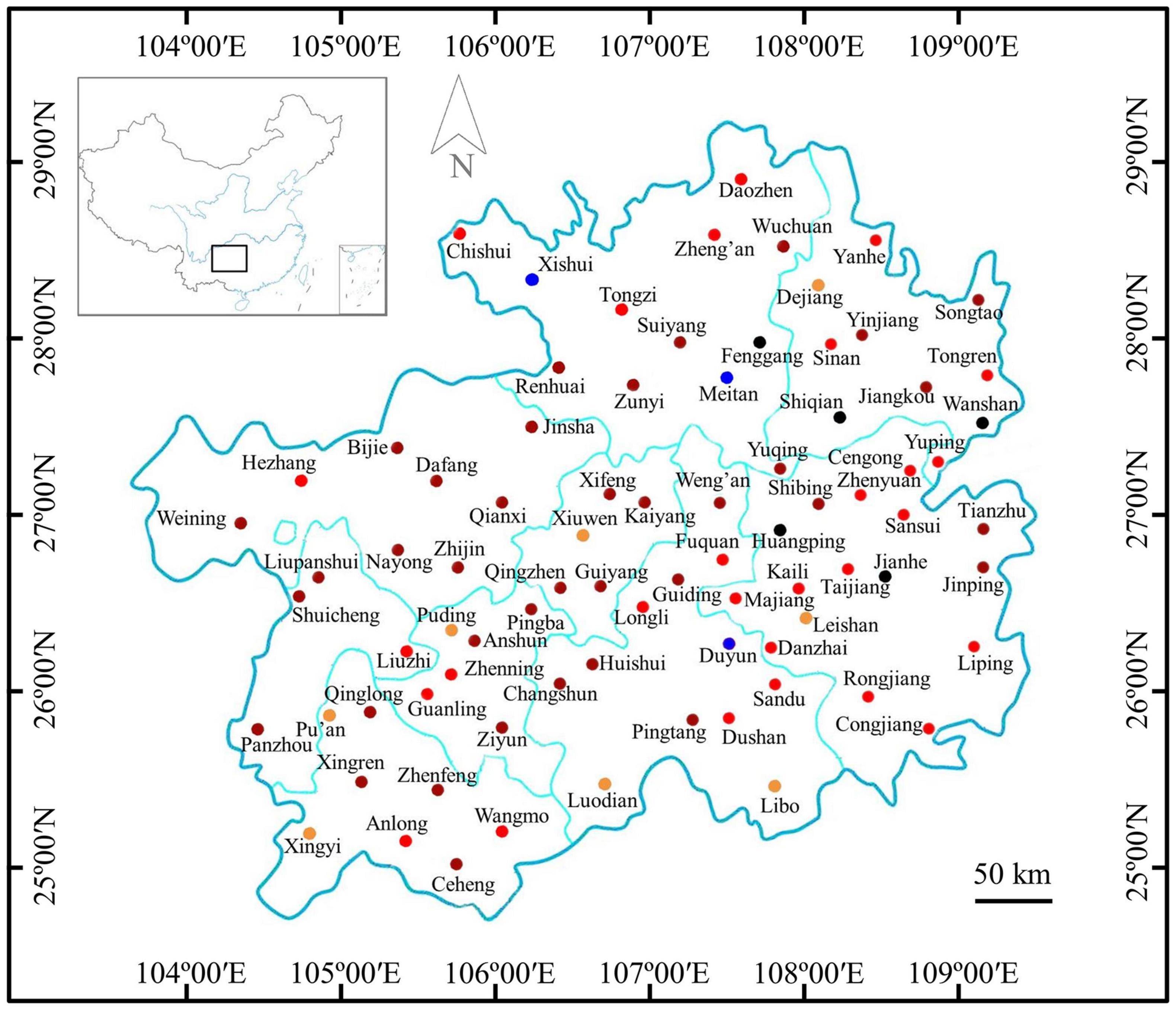
Figure 2. Distribution of Fusarium-infected samples in the Guizhou Province of China.  : no maize sample collected;
: no maize sample collected;  : samples without Fusarium infections in these regions;
: samples without Fusarium infections in these regions;  : samples with suspected infections in these regions;
: samples with suspected infections in these regions;  : samples with mild infections in these regions;
: samples with mild infections in these regions;  samples with moderate infections in these regions; and
samples with moderate infections in these regions; and  : samples with severe and extreme infections in these regions.
: samples with severe and extreme infections in these regions.
Isolation and Molecular Identification of Fusarium Pathogens
The 163 samples that tested positive by immunoassay were biocultured, isolated and purified based on morphological characteristics (Nelson et al., 1983; Leslie and Summerell, 2006), and 139 isolates were tentatively identified as Fusarium species. The ITS and TEF-1α sequences were amplified, and agarose gel electrophoresis was performed to confirm these isolates. As shown in Figure 3, the ITS sequence was approximately 550 bp, while the fragment length of the TEF-1α gene was approximately 650 bp. BLSATn searches of sequence similarity identified 123 Fusarium isolates with an areal distribution rate of 82.19% (60/73).
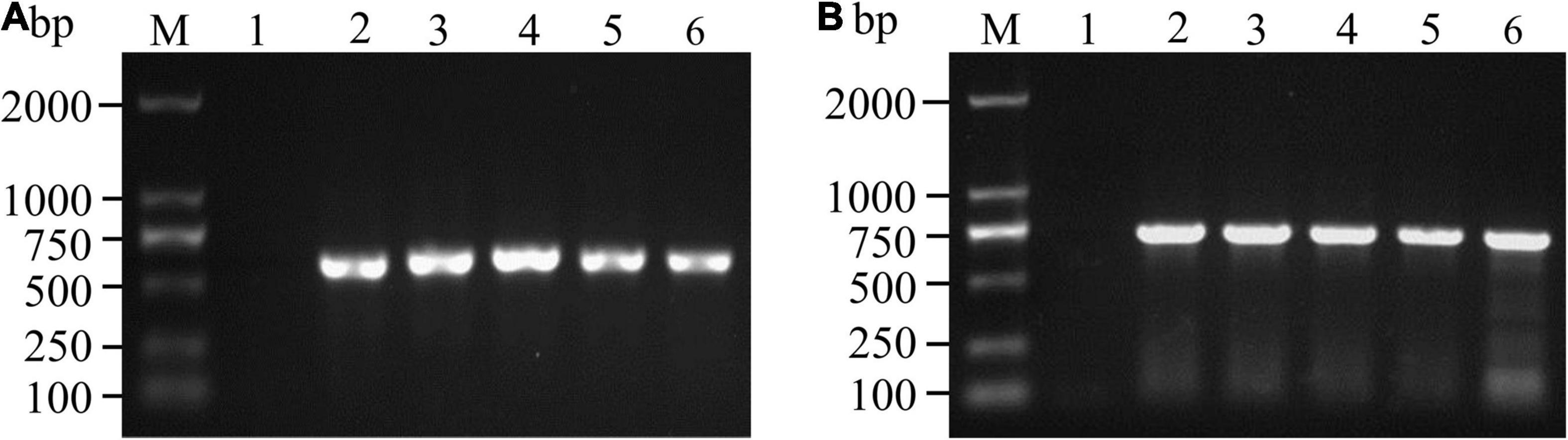
Figure 3. PCR amplification and agarose gel detection of ITS (A) and TEF-1α (B) genes. M: DNA molecular weight standard; Lane 1: negative control; Lanes A2-6: PCR products of the ITS gene of some isolates; Lanes B2-6: PCR products of the TEF-1α gene of some isolates.
Population Structure Analysis of Fusarium Pathogens
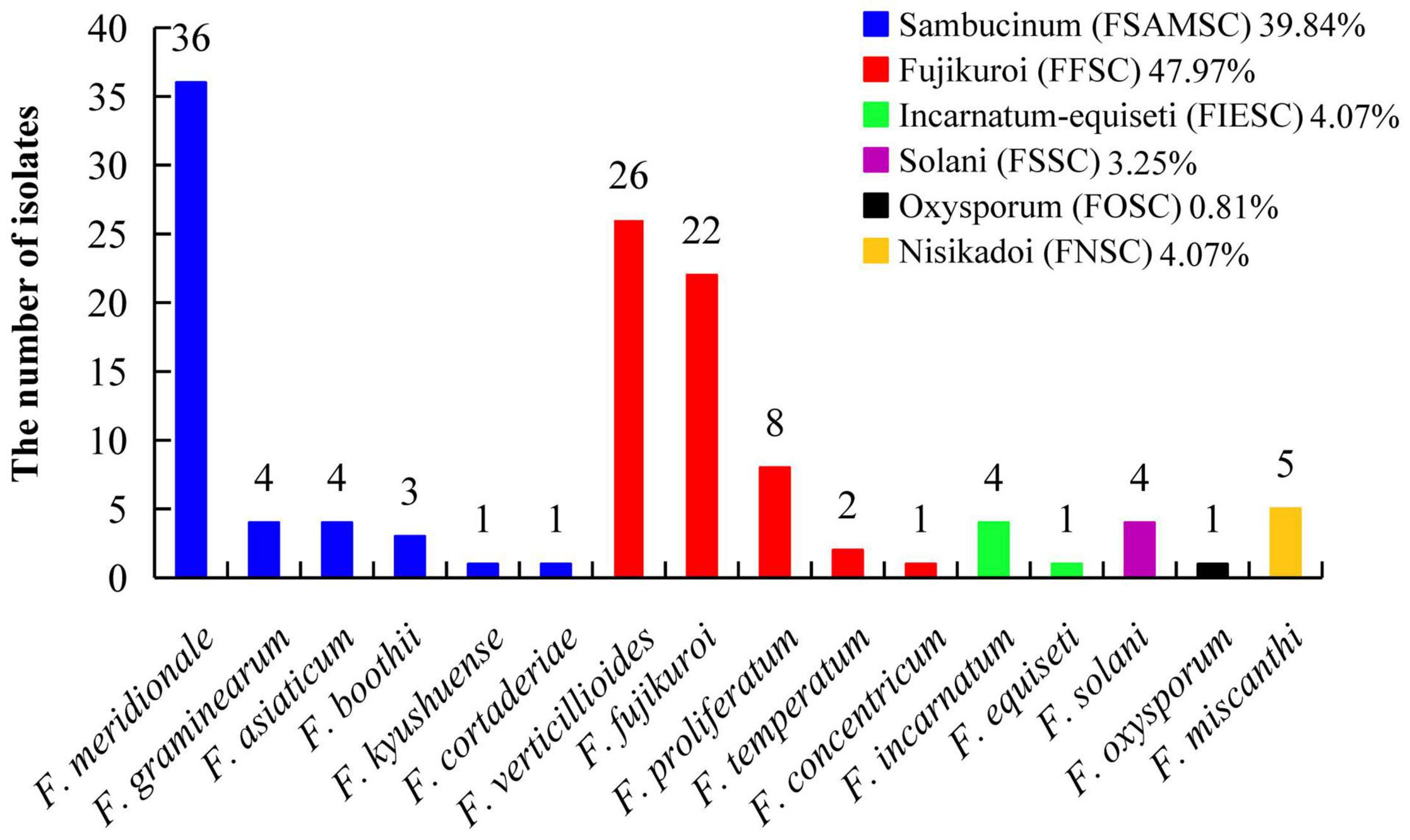
A total of 123 TEF-1α sequences of Fusarium pathogens were successfully sequenced. The nucleotide sequences have been deposited in the GenBank database and the assigned accession numbers were listed in Supplementary Table 3. These sequences were aligned with MEGA-X-10.0.5 software, and then, the phylogenetic tree was constructed by using the maximum-likelihood method (Figure 4). As shown in Figure 5, 16 Fusarium species were characterized, including Fusarium meridionale (29.27%), Fusarium verticillioides (21.14%), Fusarium fujikuroi (17.89%), Fusarium proliferatum (6.50%), Fusarium graminearum (3.25%), F. miscanthi (4.07%), Fusarium solani (3.25%), Fusarium incarnatum (3.25%), Fusarium asiaticum (3.25%), Fusarium temperatum (1.63%), Fusarium boothii (2.44%), F. concentricum (0.81%), Fusarium oxysporum (0.81%), Fusarium kyushuense (0.81%), F. cortaderiae (0.81%), and Fusarium equiseti (0.81%). The 16 Fusarium species belonged to six species complexes, including the Fusarium sambucinum species complex (FSAMSC), F. fujikuroi species complex (FFSC), Fusarium incarnatum-equiseti species complex (FIESC), F. solani species complex (FSSC), F. oxysporum species complex (FOSC) and Fusarium nisikadoi species complex (FNSC) (Figures 4 and 5). At the same time, F. meridionale was identified as the dominant species causing maize ear rot in Guizhou Province, China. Moreover, four species, F. kyushuense, F. concentricum, F. miscanthi, and F. cortaderiae were first isolated from diseased maize samples in this area.
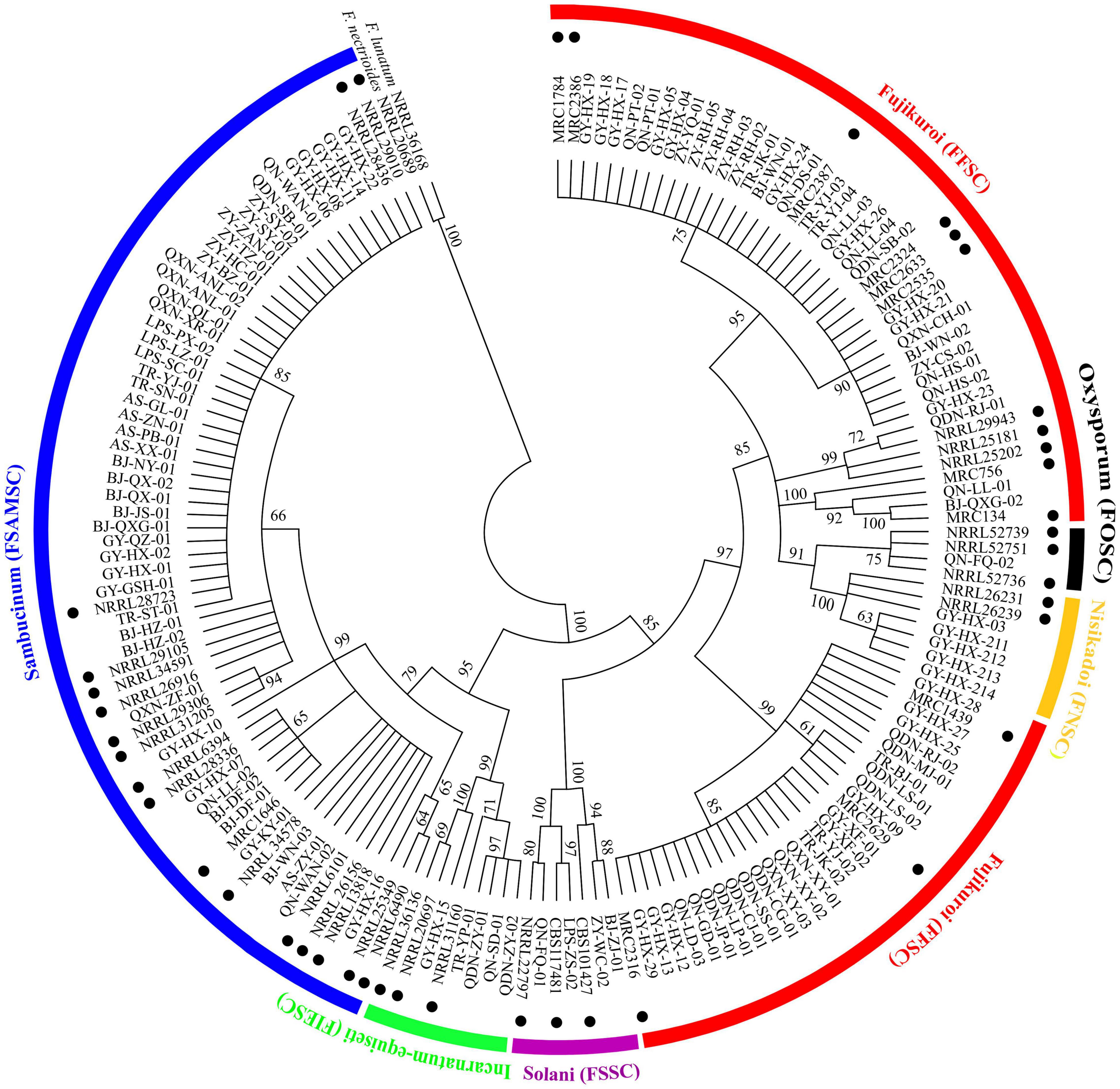
Figure 4. Phylogenetic tree of 123 Fusarium isolates based on the TEF-1α gene. Bootstrap values (≥60%) of a bootstrap test of 1,000 replicates are shown on branching nodes. Reference TEF-1α sequences of the NRRL/MRC/CBS strains were downloaded from the FUSARIUM-ID database and GenBank database. Sequences of Fusarium nectrioides and Fusarium lunatum were used as outgroups.
Molecular Identification of Toxigenic Chemotypes
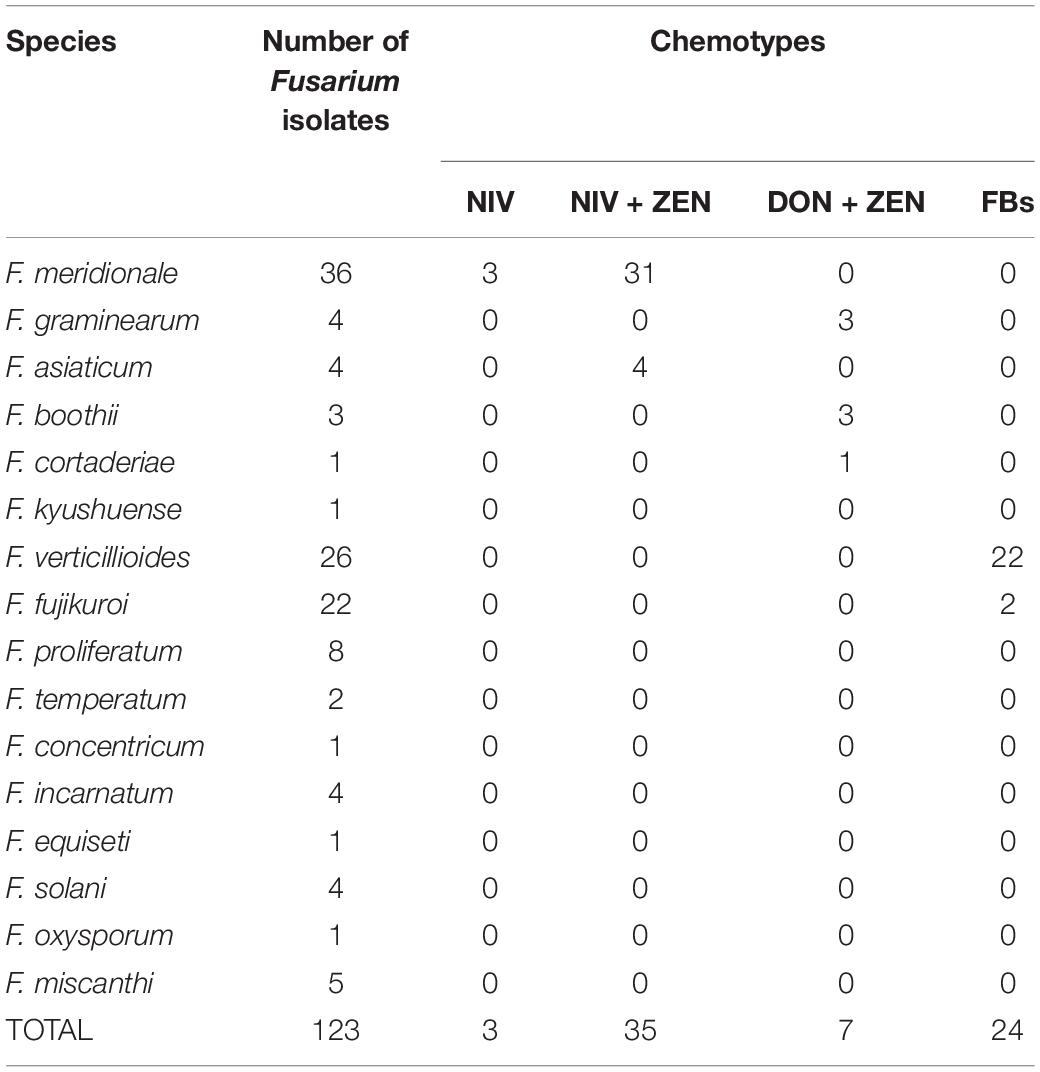
The genes involved in NIV, ZEN, DON, and FBs synthesis were detected by PCR amplification, and DNA fragments with the expected size were amplified from 56.10% (69/123) of isolates (Table 2). Among them, 31 isolates of F. meridionale and four isolates of F. asiaticum had NIV and ZEN genotypes; 34 isolates of F. meridionale had NIV or NIV+ZEN genotype; three isolates of F. graminearum, three isolates of F. boothii and one isolate of F. cortaderiae had DON+ZEN genotypes; 22 isolates of F. verticillioides and two isolates of F. fujikuroi had FBs genotype. The genes involved in DON synthesis were all detected from the F. graminearum species complex (FGSC), such as F. graminearum, F. boothii, and F. cortaderiae. The key genes for the synthesis of DON and ZEN were detected in F. cortaderiae, while the genes related to the synthesis of the four mycotoxins were not detected in F. kyushuense, F. miscanthi and F. concentricum.
The distribution of toxigenic chemotypes of the Fusarium isolates showed that the Fusarium isolates with the potential to produce both NIV and ZEN were largely distributed in western and northern Guizhou Province, whereas the Fusarium isolates with FBs genotype were mainly concentrated in southeastern Guizhou (Figure 6). Eleven representative isolates were cultured, pretreated and then subjected to LC-MS/MS detection for the confirmation of mycotoxin chemtypes. The results of the representative isolates were generally consistent with the PCR results. After inoculated with two F. verticillioides isolates, the contents of FB1, FB2, and FB3 in the cultures reached 270.7281–2269.3430 μg/g, 117.1875–1326.5330 μg/g, and 143.0563–1038.0720 μg/g, respectively. Intriguingly, FB1 and FB2 ranging from 0.0854 to 0.3653 μg/g were measured in the medium that separately inoculated the isolates of F. miscanthi and F. concentricum (Supplementary Table 4).
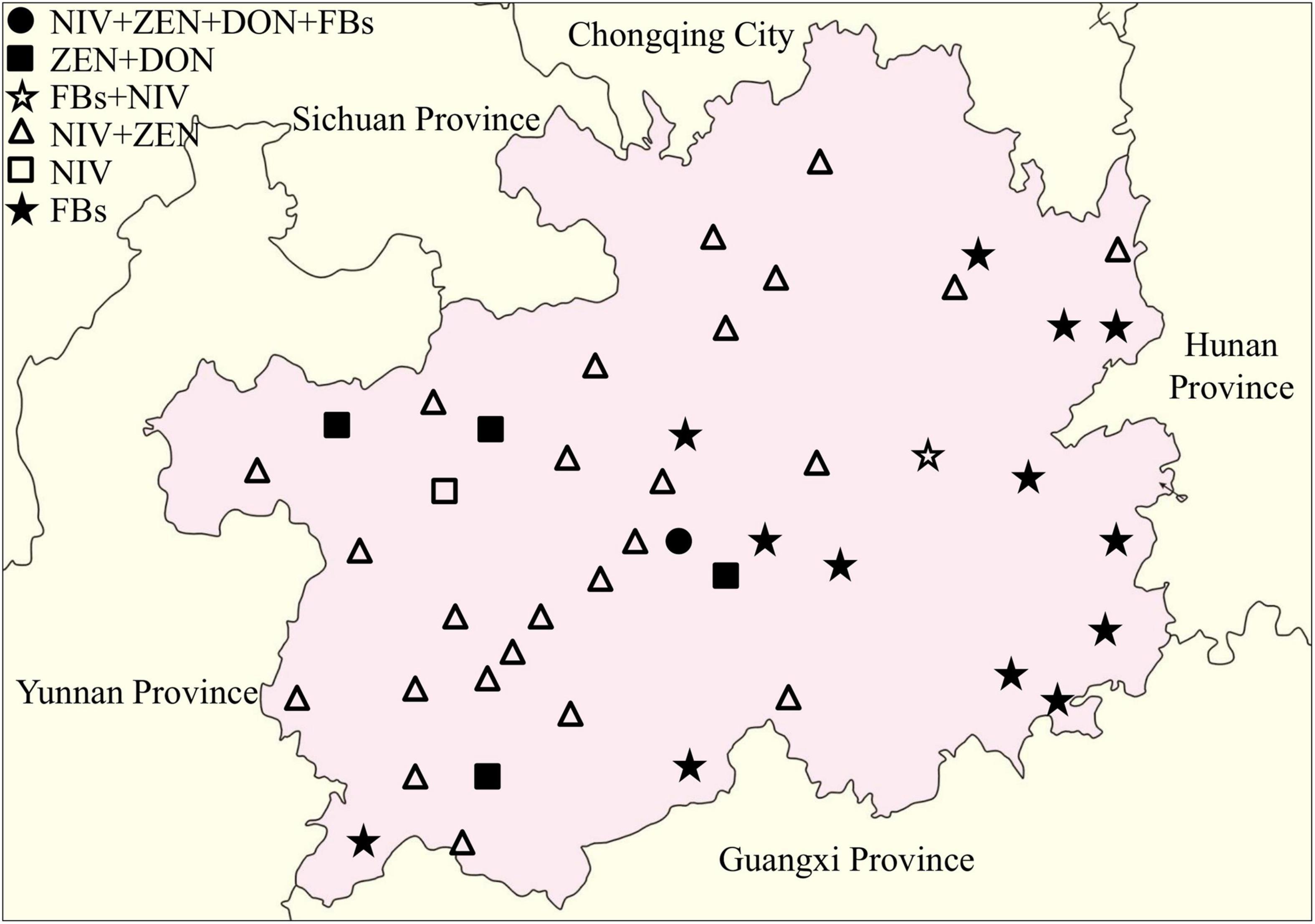
Figure 6. Distribution of toxigenic genotypes of Fusarium isolates in the Guizhou Province of China.
Discussion
Fusarium pathogens show rapid mycelial growth under hot and humid environments, but the optimal temperature for toxin production is 15∼25°C (Samapundo et al., 2005; Rybecky et al., 2018). Thus, Fusarium infection of crops is often asymptomatic with high levels of mycotoxin production (Munkvold and Desjardins, 2007). Guizhou is located on the Yunnan-Kweichow Plateau, which has a plateau humid subtropical monsoon climate with the characteristics of high humidity, low temperature, and diversity. Theoretically, this is not satisfactory for the rapid growth of hyphae, but it may be suitable for the production of mycotoxins. At present, the methods applied to monitor Fusarium are mainly biological characterization and molecular detection. However, these methods require the cultivation of fungi before detection, and the sample pretreatment process is complicated, time-consuming and laborious. Remarkably, immunoassays not only have high specificity and sensitivity but are also adaptable to high-throughput screening and simple sample preparation. The prokaryotic expression of scFv-AP fusion proteins by genetic engineering technology is considered a good choice for rapid immunological detection (Hu et al., 2013). Based on the formerly constructed scFv-AP fusion and optimal expression conditions (Hu et al., 2013; Wang et al., 2015), this study completed large-scale expression of the FvSG7-AP fusion protein for detection of Fusarium infections. A rapid immunoassay was deployed to measure 175 rotted maize ear samples collected from 76 counties in Guizhou Province, China. The results showed that the incidence rate of Fusarium infections in these samples reached 93.14% and the distribution was widespread (Figure 2). Therefore, the status of Fusarium infections in this area needs to be given sufficient attention.
Given that accurate classification of Fusarium species is the basis for further monitoring and controlling of pathogen infections and mycotoxin contamination, morphological identification and molecular phylogenetic analysis were applied to identify the collected Fusarium isolates. Traditional morphological identification has difficulty accurately distinguishing Fusarium isolates at the species level. Phylogenetic analysis of DNA sequences has been widely used to assess the genetic relationship of Fusarium species (Wang et al., 2014b). The TEF-1α gene provides much better identification among and within lineages than other loci, such as the β-TUB, CAM and ITS regions. In addition, there are no orthologous copies of TEF-1α in this genus, making this locus a better candidate for distinguishing phylogenetic relationships (Amarasinghe et al., 2019). Therefore, classification and identification of Fusarium species based on TEF-1α gene sequences has become the most common method (Mirete et al., 2004; Hafez et al., 2020). Geiser et al. (2004) established a Fusarium database based on the partial sequence of the TEF-1α gene, which allows researchers to easily identify species under Fusarium spp. based on DNA sequencing results. In this study, the subculture and purification of Fusarium pathogens from 163 positive samples were performed according to the results of immunoassays, making the performance more accurate and efficient. According to morphological characteristics (Nelson et al., 1983; Leslie and Summerell, 2006), 139 isolates were initially identified. Further molecular identification by ITS and TEF-1α sequences showed that 123 isolates were characterized as Fusarium spp. at the species level. The Fusarium spp. were not successfully characterized in 40 samples, which might due to the competitive inhibition of other fungi and bacteria in biological culture and inaccurate morphological identification. The nucleotide sequences of the GY-HX-10 isolate had the highest identity (98.3%) with the sequence of F. boothii NRRL 29105 (GenBank accession no. AF212446), and it was separated as a single branch. Further identification using the RPB2 gene confirmed its high identity (99.8%) with the sequence of F. boothii NRRL 26916 (GenBank accession no. GQ915487). In this study, F. kyushuense, F. concentricum, F. miscanthi, and F. cortaderiae were first isolated from rotted maize ears in the Guizhou area. F. kyushuense was first isolated from diseased wheat in Japan (Aoki and O’Donnell, 1998), and first reported to cause tobacco wilt in the Guizhou Province of China (Wang H. C. et al., 2013) and cause maize ear rot in the Anhui, Hubei, and Yunnan provinces of China (Wang et al., 2014a). F. concentricum was first reported to cause pepper fruit rot in China (Wang J. H. et al., 2013) and cause maize ear rot in the Guangxi Zhuang Autonomous Region of China (Du et al., 2020). F. miscanthi was first isolated from the straw of Japanese silver grass, Miscanthus sinensis, in Denmark (Gams et al., 1999) and identified to be infectious agent of maize ear rot (Shang et al., 2021). F. cortaderiae was first isolated from pampas grass (Cortaderia jubata) (O’Donnell et al., 2004) and subsequently separated from maize grain in New Zealand (Monds et al., 2005). F. cortaderiae was first reported to cause head blight on annual ryegrass in Brazil (Machado et al., 2015) and found to be the causal agent of maize stalk rot disease in the Yunnan Province of China (Xi et al., 2021).
According to the classification of Fusarium by O’Donnell et al. (2013, 2015), FSAMSC covers approximately 50 different species, including FGSC. FGSC is considered to be a worldwide population consisting of at least 16 phylogenetic species, some of which have a specific geographical distribution (Chiotta et al., 2016; Hao et al., 2017). In this study, six different phylogenetic FSAMSCs were classified, of which 97.96% belonged to FGSC (F. graminearum, F. meridionale, F. boothii, F. asiaticum, and F. cortaderiae). This is consistent with the results that maize ear rot in northern China was mainly caused by F. verticillioides and that, in the southern area, it was primarily infected by a compound species of FGSC (Qin et al., 2014). This study showed that the dominant pathogen of maize ear rot in Guizhou Province is F. meridionale, followed by F. verticillioides and F. fujikuroi. Zhou et al. (2018) reported that F. verticillioides, F. proliferatum, and F. meridionale were the predominant fungi causing maize ear and kernel rot in Chongqing City, China. Fu et al. (2015) reported that F. graminearum was the dominant species in Yunnan, Guizhou and Shanxi Provinces. However, Xi et al. (2021) reported that F. meridionale was the dominant pathogen of maize stalks in Yunnan Province, China, and F. graminearum was only 0.5%. This difference may be related to the specific geographical environment and annual climate differences.
Nivalenol, ZEN, DON, and FBs are the major mycotoxin contaminants caused by Fusarium species (Zhou et al., 2018). This study applied PCR amplification to identify the chemotypes of isolates based on specific genes involved in NIV, ZEN, DON and FBs synthesis. A total of 69 isolates were identified as potential mycotoxin-producing species, of which 55.07% (38/69) were NIV and 60.87% (42/69) were the ZEN chemotype because the dominant species in the region was F. meridionale. Previous studies have reported that F. meridionale isolated from soybean in Argentina can simultaneously produce DON and NIV toxins (Rybecky et al., 2018), whereas F. meridionale isolated from maize ear rot in Argentina only produces NIV (Sampietro et al., 2011). This study found that both NIV and ZEN synthesis-related genes were amplified from 86.11% (31/36) of F. meridionale, but the Tri5–Tri6 gene for DON synthesis was not detected. The description of mycotoxin chemotypes may be associated with many factors, such as the host and environmental conditions. According to other reports, the optimal conditions for F. meridionale growth were 25°C and high water activity, which is also suitable for toxin production (Rybecky et al., 2018; Belizan et al., 2019). The climatic characteristics of warm, humid and abundant rainfall during the maize-harvesting period (August to October) in Guizhou Province may be one of the reasons for serious Fusarium infection and mycotoxin accumulation. Fumonisin synthesis-related genes of FUM are located in the same gene family. In the biosynthetic pathway of fumonisins, some of the products of FUM genes play a major catalytic role, and the rest play indirect roles (Alexander et al., 2009). The FUM genes were mostly detected in the isolates of F. verticillioides, which is consistent with previous studies (Duan et al., 2016; Zhou et al., 2018). Unusually, the FUM genes were not amplified from the isolates of F. proliferatum and F. temperatum. Although F. proliferatum is one of the main FBs-producing species and verified to be completely synthetic with the fumonisin gene cluster of F. verticillioides (Sun et al., 2019), some studies have shown that the deletion of FUM1 gene can reduce FB1 production by 99% (Duan et al., 2016). At the same time, a PCR analysis of 20 F. proliferatum isolates by using FUM1 gene-specific primers has been conducted and the gene fragments were obtained only from thirteen isolates (Dissanayake et al., 2009). Scauflaire et al. (2011) first discovered F. temperatum in Belgium and identified it as a new species, but only one of the eleven F. temperatum species produced a small amount of FB1 (Scauflaire et al., 2012). Fumero et al. (2020) conducted chemical and genotype identification of 12 F. temperatum isolates from Argentina and showed that neither fumonisin production nor FUM biosynthesis genes were detected. Wang et al. (2014b) reported that 10 isolates of F. temperatum isolated in Zunyi and Guiyang of Guizhou Province can produce FB1 and FB2. The differences in the mycotoxin spectrum may reflect the genetic variation between F. temperatum isolates in different geographic regions.
Although the pretreatment process is complex, LC–MS/MS is widely used for the accurate determination of mycotoxin chemotypes (Duan et al., 2016). In this study, some isolates were randomly selected to culture and measure their mycotoxin production by LC–MS/MS. The results confirmed the molecular identification, suggesting that PCR amplification is a rapid and effective method. Also, our result showed that F. verticillioides has the potential to produce high levels of FBs and is one of the dominant species of maize ear rot pathogens in this area. Therefore, it is necessary to strengthen the monitoring of FBs contamination in food in this area. In addition, a certain amount of FBs was detected in the cultures of F. miscanthi and F. concentricum isolates, from which the FUM gene fragment was not amplified. Certainly, the content of FBs was far lower than that produced by the molecularly identified isolates, which may be correlated with the expression and regulation of toxin synthesis-related genes (Zhou et al., 2018). This study is the first report that F. miscanthi and F. concentricum can produce FBs.
Conclusion
This study combined immunoassays and molecular identification for the first time to systematically investigate maize ear rot caused by Fusarium pathogens as well as their mycotoxin chemotypes in Guizhou Province, China. The results displayed a variety of Fusarium species distributed almost all over the whole area, and F. meridionale was the predominant species. F. kyushuense, F. concentricum, F. miscanthi, and F. cortaderiae were first isolated from maize ear rot in the Guizhou area. Further molecular identification and LC–MS/MS confirmation showed that most of these Fusarium isolates have the potential to produce mycotoxins with typical geographical distribution features. Together, scFv-AP fusion-based immunoassay is proven an effective method to detect fungal infections in crops, and more attention and measures should be taken to ensure human and animal health in Guizhou Province.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession number of 123 genes from 123 Fusarium isolates are listed in Supplementary Table 3.
Author Contributions
ZH, JZ, and ZZ: conceptualization, project administration, and funding acquisition. ZH, JZ, and GS: methodology. GS and SQL: data curation. GS and YW: software. GS, SQL, and HY: formal analysis. GS, SML, JY, HY, YY, and JW: investigation. JY, HY, SML, YY, and JW: validation. ZH, ZZ, JZ, and YW: resources and writing-review and editing. GS, SQL, and HY: writing—original draft preparation. ZH and YW: supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant nos. 21906036 and 12132006), the Guizhou Provincial Natural Science Foundation (grant nos. 2021-5637, ZK2021-029, and 2016-5676), the Excellent Young Talents Plan of Guizhou Medical University (grant no. 2021-101), and the Project B21HJUS02 of Hainan Yazhou Bay Seed Lab.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Research Center for Basic Medical Sciences of Guizhou Medical University for providing large-scale instrument and equipment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.849698/full#supplementary-material
Footnotes
References
Alexander, N. J., Proctor, R. H., and Mccormick, S. P. (2009). Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 28, 198–215. doi: 10.1080/15569540903092142
Amarasinghe, C., Sharanowski, B., and Fernando, W. G. D. (2019). Molecular phylogenetic relationships, trichothecene chemotype diversity and aggressiveness of strains in a global collection of Fusarium graminearum species. Toxins 11, 263–280. doi: 10.3390/toxins11050263
Aoki, T., and O’Donnell, K. (1998). Fusarium kyushuense sp. nov. from Japan. Mycoscience 39, 1–6. doi: 10.1007/BF02461571
Belizan, M. M. E., Gomez, A. L. A., Teran, B. Z. P., Jimenez, C. M., Sánchez, M. M. D. H., Catalán, C. A. N., et al. (2019). Influence of water activity and temperature on growth and production of trichothecenes by Fusarium graminearum sensu stricto and related species in maize grains. Int. J. Food Microbiol. 305, 108242–108247. doi: 10.1016/j.ijfoodmicro.2019.108242
Bluhm, B. H., Flaherty, J. E., Cousin, M. A., and Woloshuk, C. P. (2002). Multiplex polymerase chain reaction assay for the differential detection of trichothecene- and fumonisin-producing species of Fusarium in cornmeal. J. Food Prot. 65, 1955–1961. doi: 10.4315/0362-028x-65.12.1955
Cambaza, E., Koseki, S., and Kawamura, S. (2019). Why RGB imaging should be used to analyze Fusarium graminearum growth and estimate deoxynivalenol contamination. Methods Protoc. 2:25. doi: 10.3390/mps2010025
Chiotta, M. L., Alaniz Zanon, M. S., Palazzini, J. M., Scandiani, M. M., Formento, A. N., Barros, G. G., et al. (2016). Pathogenicity of Fusarium graminearum and F. meridionale on soybean pod blight and trichothecene accumulation. Plant Pathol. 65, 1492–1497. doi: 10.1111/ppa.12532
Dissanayake, M. L., Tanaka, S., and Ito, S. (2009). Fumonisin B1 production by Fusarium proliferatum strains isolated from Allium fistulosum plants and seeds in Japan. Lett. Appl. Microbiol. 48, 598–604. doi: 10.1111/j.1472-765X.2009.02576.x
Du, Q., Duan, C. X., Li, S. C., Tang, Z. L., and Luo, J. Y. (2020). First report of maize ear rot caused by Fusarium concentricum in China. Plant Dis. 104:1539. doi: 10.1094/PDIS-07-19-1515-PDN
Duan, C., Qin, Z., Yang, Z., Li, W., Sun, S., Zhu, Z., et al. (2016). Identification of pathogenic Fusarium spp. causing maize ear rot and potential mycotoxin production in China. Toxins 8, 186–203. doi: 10.3390/toxins8060186
Feng, Y. Z., Lu, X. H., Tao, B., Pang, M. H., Liu, Y. C., and Dong, J. G. (2011). Natural occurrence of fumonisins B1 and B2 in corn from three main production provinces in China. J. Food Prot. 74, 1374–1378. doi: 10.4315/0362-028X.JFP-11-103
Fu, M., Li, R. J., Guo, C. C., Pang, M. H., Liu, Y. C., and Dong, J. G. (2015). Natural incidence of Fusarium species and fumonisins B1 and B2 associated with maize kernels from nine provinces in China in 2012. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 32, 503–511. doi: 10.1080/19440049.2014.976846
Fumero, M. V., Villani, A., Susca, A., Haidukowski, M., Cimmarusti, M. T., Toomajian, C., et al. (2020). Fumonisin and beauvericin chemotypes and genotypes of the sister species Fusarium subglutinans and Fusarium temperatum. Appl. Environ. Microbiol. 86:e00133-20. doi: 10.1128/AEM.00133-20
Gams, W., Klamer, M., and O’Donnell, K. (1999). Fusarium miscanthi sp. nov. from Miscanthus litter. Mycologia 91, 263–268. doi: 10.1080/00275514.1999.12061016
Geiser, D. M., Jiménez-Gasco, M. D. M., Kang, S., Makalowska, I., Veeraraghavan, N., and Ward, T. J. (2004). Fusarium-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110, 473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0
Gong, H. Z., Ji, R., Li, Y. X., Zhang, H. Y., Li, B., Zhao, Y., et al. (2009). Occurrence of fumonisin B1 in corn from the main corn-producing areas of China. Mycopathologia 167, 31–36. doi: 10.1007/s11046-008-9146-8
Hafez, M., Abdelmagid, A., Adam, L. R., and Daayf, F. (2020). Specific detection and identification of Fusarium graminearum sensu stricto using a PCR-RFLP tool and specific primers targeting the translational elongation factor 1α gene. Plant Dis. 104, 1076–1086. doi: 10.1094/PDIS-03-19-0572-RE
Hao, J. J., Xie, S. N., Sun, J., Yang, G. Q., Liu, J. Z., Xu, F., et al. (2017). Analysis of Fusarium graminearum species complex from wheat-maize rotation regions in Henan (China). Plant Dis. 101, 720–725. doi: 10.1094/PDIS-06-16-0912-RE
He, T., Zhu, J., Nie, Y., Hu, R., Wang, T., Li, P., et al. (2018). Nanobody technology for mycotoxin detection: current status and prospects. Toxins 10, 180–199. doi: 10.3390/toxins10050180
Hu, Z. Q., Li, H. P., Zhang, J. B., Huang, T., Liu, J. L., Xue, S., et al. (2013). A phage-displayed chicken single-chain antibody fused to alkaline phosphatase detects Fusarium pathogens and their presence in cereal grains. Anal. Chim. Acta 764, 84–92. doi: 10.1016/j.aca.2012.12.022
Karlsson, I., Persson, P., and Friberg, H. (2021). Fusarium head blight from a microbiome perspective. Front. Microbiol. 12:628373. doi: 10.3389/fmicb.2021.628373
Kerschbaumer, R. J., Hirschl, S., Kaufmann, A., Ibl, M., Koenig, R., and Himmler, G. (1997). Single-chain Fv fusion proteins suitable as coating and detecting reagents in a double antibody sandwich enzyme-linked immunosorbent assay. Anal. Biochem. 249, 219–227. doi: 10.1006/abio.1997.2171
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, H. J., and Ryu, D. (2017). Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: public health perspectives of their co-occurrence. J. Agric. Food Chem. 65, 7034–7051. doi: 10.1021/acs.jafc.6b04847
Leslie, J. F., and Summerell, B. A. (2006). The Fusarium Laboratory Manual. Oxford: Blackwell Publishing, doi: 10.1002/9780470278376
Li, H. P., Wu, A. B., Zhao, C. S., Scholten, O., Löffler, H., and Liao, Y. C. (2005). Development of a generic PCR detection of deoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum. FEMS Microbiol. Lett. 243, 505–511. doi: 10.1016/j.femsle.2005.01.015
Li, J. H., Ji, L. J., Chai, Z. X., Knight, T. E., and Burgess, L. W. (2009). A study on the variance of Fusarium graminearum sampled from central Gansu. Acta Pratacult. Sinica 18, 118–124. doi: 10.11686/cyxb20090118
Machado, F. J., Möller, P. A., Nicolli, C. P., Del Ponte, E. M., and Ward, T. J. (2015). First report of Fusarium graminearum, F. asiaticum, and F. cortaderiae as head blight pathogens of annual ryegrass in Brazil. Plant Dis. 99, 1859–1859. doi: 10.1094/PDIS-04-15-0376-PDN
Mirete, S., Vázquez, C., Mulè, G., Jurado, M., and González-Jaén, M. T. (2004). Differentiation of Fusarium verticillioides from banana fruits by IGS and EF-1α sequence analyses. Eur. J. Plant Pathol. 110, 515–523. doi: 10.1023/B:EJPP.0000032391.38512.f8
Monds, R. D., Cromey, M. G., Lauren, D. R., di Menna, M., and Marshall, J. (2005). Fusarium graminearum, F. cortaderiae and F. pseudograminearum in New Zealand: molecular phylogenetic analysis, mycotoxin chemotypes and co-existence of species. Mycol. Res. 109, 410–420. doi: 10.1017/s0953756204002217
Munkvold, G. P., and Desjardins, A. E. (2007). Fumonisins in maize: can we reduce their occurrence? Plant Dis. 81, 556–565. doi: 10.1094/PDIS.1997.81.6.556
Nelson, P. E., Toussoun, T. A., and Marasas, W. F. O. (1983). Fusarium Species: An Illustrated Manual for Identification. University Park, PA: Pennsylvania State University Press.
O’Donnell, K., Rooney, A. P., Proctor, R. H., Brown, D. W., McCormick, S. P., Ward, T. J., et al. (2013). Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 52, 20–31. doi: 10.1016/j.fgb.2012.12.004
O’Donnell, K., Ward, T. J., Geiser, D. M., Corby Kistler, H., and Aoki, T. (2004). Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 41, 600–623. doi: 10.1016/j.fgb.2004.03.003
O’Donnell, K., Ward, T. J., Robert, V. A. R. G., Crous, P. W., Geiser, D. M., and Kang, S. (2015). DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43, 583–595. doi: 10.1007/s12600-015-0484-z
Palacios, S. A., Del Canto, A., Erazo, J., and Torres, A. M. (2021). Fusarium cerealis causing Fusarium head blight of durum wheat and its associated mycotoxins. Int. J. Food Microbiol. 346:109161. doi: 10.1016/j.ijfoodmicro.2021.109161
Pechanova, O., and Pechan, T. (2015). Maize-pathogen interactions: an ongoing combat from a proteomics perspective. Int. J. Mol. Sci. 16, 28429–28448. doi: 10.3390/ijms161226106
Qin, Z., Xu, R., Kai, J., Wu, X., Yang, Z., and Wang, X. (2014). Identification of Fusarium species and F. graminearum species complex causing maize ear rot in China. Acta Phytophys. Sin. 41, 589–596. doi: 10.13802/j.cnki.zwbhxb.2014.05.032
Qiu, J., and Shi, J. (2014). Genetic relationships, carbendazim sensitivity and mycotoxin production of the Fusarium graminearum populations from maize, wheat and rice in eastern China. Toxins 6, 2291–2309. doi: 10.3390/toxins6082291
Qiu, J., Xu, J., and Shi, J. (2019). Fusarium toxins in Chinese wheat since the 1980s. Toxins 11:248. doi: 10.3390/toxins11050248
Rahman, H. U., Yue, X., Yu, Q., Xie, H., Zhang, W., Zhang, Q., et al. (2019). Specific antigen-based and emerging detection technologies of mycotoxins. J. Sci. Food Agric. 99, 4869–4877. doi: 10.1002/jsfa.9686
Ramana, M. V., Balakrishna, K., Murali, K. C. S., and Batra, H. V. (2011). Multiplex PCR-based strategy to detect contamination with mycotoxigenic Fusarium species in rice and fingermillet collected from southern India. J. Sci. Food Agric. 91, 1666–1673. doi: 10.1002/jsfa.4365
Rybecky, A. I., Chulze, S. N., and Chiotta, M. L. (2018). Effect of water activity and temperature on growth and trichothecene production by Fusarium meridionale. Int. J. Food Microbiol. 285, 69–73. doi: 10.1016/j.ijfoodmicro.2018.07.028
Saccon, F. A. M., Parcey, D., Paliwal, J., and Sherif, S. (2017). Assessment of Fusarium and deoxynivalenol using optical methods. Food Bioproc. Technol. 10, 34–50. doi: 10.1007/s11947-016-1788-9
Samapundo, S., Devliehgere, F., De, M. B., and Debevere, J. (2005). Effect of water activity and temperature on growth and the relationship between fumonisin production and the radial growth of Fusarium verticillioides and Fusarium proliferatum on corn. J. Food Prot. 68, 1054–1059. doi: 10.4315/0362-028x-68.5.1054
Sampietro, D. A., Díaz, C. G., Gonzalez, V., Vattuone, M. A., Ploper, L. D., Catalan, C. A., et al. (2011). Species diversity and toxigenic potential of Fusarium graminearum complex isolates from maize fields in northwest Argentina. Int. J. Food Microbiol. 145, 359–364. doi: 10.1016/j.ijfoodmicro.2010.12.021
Santos, C., Ventura, J. A., and Lima, N. (2016). New insights for diagnosis of pineapple fusariosis by MALDI-TOF MS technique. Curr. Microbiol. 73, 206–213. doi: 10.1007/s00284-016-1041-9
Scauflaire, J., Gourgue, M., Callebaut, A., and Munaut, F. (2012). Fusarium temperatum, a mycotoxin-producing pathogen of maize. Eur. J. Plant Pathol. 133, 911–922. doi: 10.1007/s10658-012-9958-8
Scauflaire, J., Gourgue, M., and Munaut, F. (2011). Fusarium temperatum sp. nov, from maize, an emergent species closely related to Fusarium subglutinans. Mycologia 103, 586–597. doi: 10.3852/10-135
Schiwek, S., Beule, L., Vinas, M., Pfordt, A., von Tiedemann, A., and Karlovsky, P. (2020). High-Resolution Melting (HRM) curve assay for the identification of eight Fusarium species causing ear rot in maize. Pathogens 9:270. doi: 10.3390/pathogens9040270
Shang, G. F., Yu, H., Yang, J., Zeng, Z., and Hu, Z. Q. (2021). First report of Fusarium miscanthi causing ear rot on maize in China. Plant Dis. 105, 1565–1565. doi: 10.1094/PDIS-10-20-2182-PDN
Shang, G. F., Yu, H., Zhao, X., Xia, X., Liu, J. L., Peng, X. Y., et al. (2019). Investigation of Fusarium contamination in maize kernels from Guizhou province. Microbiol. China 46, 752–759. doi: 10.13344/j.microbiol.china.180229
Shi, J. L., Li, Y. Q., Hu, K. M., Ren, J. G., and Liu, H. M. (2015). Isolation and identification of pathogen of tuber rot of Pinellia ternata in Guizhou. Microbiol. China 42, 289–299. doi: 10.13344/j.microbiol.china.140391
Sim, J. H., Tian, F., Jung, S. Y., Auh, J. H., and Chun, H. S. (2018). Multiplex polymerase chain reaction assays for the detection of the zearalenone chemotype of Fusarium species in white and brown rice. Int. J. Food Microbiol. 269, 120–127. doi: 10.1016/j.ijfoodmicro.2018.02.003
Sun, L., Chen, X., Gao, J., Zhao, Y., Liu, L., Hou, Y., et al. (2019). Effects of disruption of five FUM genes on fumonisin biosynthesis and pathogenicity in Fusarium proliferatum. Toxins 11:327. doi: 10.3390/toxins11060327
Sun, X. D., Su, P., and Shan, H. (2017). Mycotoxin contamination of rice in China. J. Food Sci. 82, 573–584. doi: 10.1111/1750-3841.13631
Torres, A. M., Palacios, S. A., Yerkovich, N., Palazzini, J. M., Battilani, P., Leslie, J. F., et al. (2019). Fusarium head blight and mycotoxins in wheat: prevention and control strategies across the food chain. World Mycotoxin J. 12, 333–355. doi: 10.3920/WMJ2019.2438
Wang, H. C., Wang, M. S., Xia, H. Q., Yang, S. J., and Shi, J. X. (2013). First report of Fusarium wilt of tobacco caused by Fusarium kyushuense in China. Plant Dis. 97:424. doi: 10.1094/PDIS-09-12-0835-PDN
Wang, J. H., Feng, Z. H., Han, Z., Song, S. Q., Lin, S. H., and Wu, A. B. (2013). First report of pepper fruit rot caused by Fusarium concentricum in China. Plant Dis. 97:1657. doi: 10.1094/PDIS-03-13-0325-PDN
Wang, J. H., Li, H. P., Zhang, J. B., Wang, B. T., and Liao, Y. C. (2014a). First report of Fusarium maize ear rot caused by Fusarium kyushuense in China. Plant Dis. 98:279. doi: 10.1094/PDIS-05-13-0558-PDN
Wang, J. H., Zhang, J. B., Li, H. P., Gong, A. D., Xue, S., Agboola, R. S., et al. (2014b). Molecular identification, mycotoxin production and comparative pathogenicity of Fusarium temperatum isolated from maize in China. J. Phytopathol. 162, 147–157. doi: 10.1111/jph.12164
Wang, J. H., Ndoye, M., Zhang, J. B., Li, H. P., and Liao, Y. C. (2011). Population structure and genetic diversity of the Fusarium graminearum species complex. Toxins 3, 1020–1037. doi: 10.3390/toxins3081020
Wang, T., Li, P., Zhang, Q., Zhang, W., Zhang, Z., Wang, T., et al. (2017). Determination of Aspergillus pathogens in agricultural products by a specific nanobody-polyclonal antibody sandwich ELISA. Sci. Rep. 7, 4348–4359. doi: 10.1038/s41598-017-04195-6
Wang, W., Wang, B., Sun, X., Qi, X., and Gong, G. (2021). Symptoms and pathogens diversity of corn Fusarium sheath rot in Sichuan province, China. Sci. Rep. 11:2835. doi: 10.1038/s41598-021-82463-2
Wang, Y., Zhou, J., Liu, L. N., Zhong, N. F., Wang, Y. L., and Hu, Z. Q. (2015). Activity analysis of single-chain Fv antibody-alkaline phosphatase fusion protein constructed in different ways. Genom. Appl. Biol. 34, 1798–1803.
Wei, T. S., Zhu, W. F., Pang, M. H., Liu, Y. C., and Dong, J. G. (2013). Natural occurrence of fumonisins B1 and B2 in corn in four provinces of China. Food Addit.Contam. Part B 6, 270–274. doi: 10.1080/19393210.2013.819816
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct seqencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Xi, K., Shan, L., Yang, Y., Zhang, G., Zhang, J., and Guo, W. (2021). Species diversity and chemotypes of Fusarium species associated with maize stalk rot in Yunnan province of southwest China. Front. Microbiol. 12:652062. doi: 10.3389/fmicb.2021.652062
Xu, Y. B., Li, H. P., Zhang, J. B., Song, B., Chen, F. F., Duan, X. J., et al. (2010). Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genet. Biol. 47, 205–215. doi: 10.1016/j.fgb.2009.11.003
Yang, P., Yi, S. Y., Nian, J. N., Yuan, Q. S., He, W. J., Zhang, J. B., et al. (2021). Application of double-strand RNAs targeting chitin synthase, glucan synthase, and protein kinase reduces Fusarium graminearum spreading in wheat. Front. Microbiol. 12:660976. doi: 10.3389/fmicb.2021.660976
Yu, Q. T., and Yao, T. S. (2018). Research progress on Fusarium root rot of tobacco. J. Anhui Agric. Sci. 46, 34–36. doi: 10.13989/j.cnki.0517-6611.2018.17.010
Zhang, J. B., Li, H. P., Dang, F. J., Qu, B., Xu, Y. B., Zhao, C. S., et al. (2007). Determination of the trichothecene mycotoxin chemotypes and associated geographical distribution and phylogenetic species of the Fusarium graminearum clade from China. Mycol. Res. 111, 967–975. doi: 10.1016/j.mycres.2007.06.008
Zhang, R., Ma, Q., and Wang, X. (2009). Detection of zearalenone produced by Fusarium graminearum using PCR assay. Plant Protect. 35, 94–98. doi: 10.3969/j.issn.0529-1542.2009.06.021
Keywords: Fusarium infections, enzyme-linked immunosorbent assay (ELISA), molecular identification, population structure, mycotoxin chemotype
Citation: Shang G, Li S, Yu H, Yang J, Li S, Yu Y, Wang J, Wang Y, Zeng Z, Zhang J and Hu Z (2022) An Efficient Strategy Combining Immunoassays and Molecular Identification for the Investigation of Fusarium Infections in Ear Rot of Maize in Guizhou Province, China. Front. Microbiol. 13:849698. doi: 10.3389/fmicb.2022.849698
Received: 06 January 2022; Accepted: 31 January 2022;
Published: 14 March 2022.
Edited by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Zahoor Ul Hassan, Qatar University, QatarNarayan Chandra Paul, Chonnam National University, South Korea
Copyright © 2022 Shang, Li, Yu, Yang, Li, Yu, Wang, Wang, Zeng, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zuquan Hu, aHV6dXF1YW5AZ21jLmVkdS5jbg==; Jingbo Zhang, amluZ2JvemhhbmdAbWFpbC5oemF1LmVkdS5jbg==; Zhu Zeng, emVuZ3podUBnbWMuZWR1LmNu
†These authors have contributed equally to this work
 Guofu Shang
Guofu Shang Shuqin Li
Shuqin Li Huan Yu
Huan Yu Jie Yang
Jie Yang Shimei Li
Shimei Li Yanqin Yu
Yanqin Yu Jianman Wang
Jianman Wang Yun Wang
Yun Wang Zhu Zeng
Zhu Zeng Jingbo Zhang
Jingbo Zhang Zuquan Hu
Zuquan Hu