
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 March 2022
Sec. Systems Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.848964
This article is part of the Research Topic A Focus on Actinobacteria: Diversity, Distribution, and Secondary Metabolites View all 10 articles
Pseudonocardia species are emerging as important microorganisms of global concern with unique and increasingly significant ecological roles and represent a prominent source of bioactive natural products, but genetic engineering of these organisms for biotechnological applications is greatly hindered due to the limitation of efficient genetic manipulation tools. In this regard, we report here the establishment of an efficient genetic manipulation system for a newly isolated strain, Pseudonocardia alni Shahu, based on plasmid conjugal transfer from Escherichia coli to Pseudonocardia. Conjugants were yielded upon determining the optimal ratio between the donor and recipient cells, and designed genome modifications were efficiently accomplished, including exogenous gene integration based on an integrative plasmid and chromosomal stretch removal by homologous recombination using a suicidal non-replicating vector. Collectively, this work has made the P. alni Shahu accessible for genetic engineering, and provided an important reference for developing genetic manipulation methods in other rare actinomycetes.
Bacteria in the rare genus of Pseudonocardia belong to Actinomycetes that are represented by Gram-positive bacteria with high G + C DNA content (Tiwari and Gupta, 2012). With recent rapid developments in genome sequencing, more and more Pseudonocardia genomic data have been accumulated in the database. Many Pseudonocardia-specific gene clusters are now available, and their functions, concerning, for example, synthesis of many natural bioactive products (Miyamoto et al., 2014; Caffrey et al., 2016; Won et al., 2017; Geddis et al., 2018; Fang et al., 2019; Strecker et al., 2019; Subramani and Sipkema, 2019), biodegradation of large amounts of industrially discharged environmental pollutants (Grostern et al., 2012; Sales et al., 2013; Inoue et al., 2016; Yamamoto et al., 2018; Qi et al., 2019), defense against invading genetic elements (Doron et al., 2018), have been and will continue to be deduced or demonstrated, showing great application potentials in human medicine, animal health, and crop protection, etc. Indeed, current applications of Pseudonocardia have been summarized very recently (Riahi et al., 2021). These beneficial potentials have attracted a great deal of interest in the Pseudonocardia genus. However, the generally poor efficiency or perhaps lack of genetic manipulation tools for Pseudonocardia has largely impeded deeper research and further application of this genus.
Introduction of DNAs into actinomycetes is generally difficult, possibly owing to the strong cell wall of such bacteria. Although strategies including protoplast transformation and electroporation of plasmids have been successfully used to introduce DNAs into some Streptomyces species, they are either of low efficiency or not adaptable for many of the less easy-to-handle non-model actinomycetes. Fortunately, easy and efficient DNA introduction from Echerichia coli to actinomycetes can be achieved via conjugal transfer. This method represents probably the most straightforward strategy for genetic manipulation in actinomycetes.
Conjugal transfer of different types of vectors, including replicative plasmids, integrative plasmids, and suicidal non-replicating vectors, has been applied in various actinomycetes. The replicative plasmids are useful for overproduction of heterologous proteins, but their host range is usually restricted to native strains and permanent selective pressure is required for their stable maintenance in the hosts. Advantageously, the other two types of vectors, integrative plasmids and suicidal non-replicating vectors, can introduce stable genomic alterations in many actinomycetes. Integrative plasmids contain genes encoding attachment/integration (att/int) functions, therein being capable of incorporating DNAs into chromosomes of a range of actinomycete species. Mechanistically, integrases derived from bacteriophages catalyze site-specific recombination between attachment sites, respectively, located on the genomes of phage (attP) and bacteria (attB), forming attL and attR hybrid sites (Combes et al., 2002). Currently, the suicidal non-replicating vectors have been extensively used to generate stable DNA inserts into any neutral genomic region, or deletion of genes of interest, via homologous recombination.
Here we report a rare actinomycete strain, Pseudonocardia alni Shahu, recently isolated from the sediment of a lake in the Shahu Park, Wuhan, China. In order to manipulate P. alni Shahu, we established an efficient conjugal transfer system, and via which, conjugal transfer of the pSET152 plasmid (Flett et al., 1997) and its derivatives from E. coli cells to P. alni spores were successfully achieved. Moreover, upon conjugation, the pSET152 derivatives have mediated the accomplishment of stable genomic alterations, including integration of an exogenous gfp gene into the chromosome and deletion of the pgl genes encoding key elements of a phage growth limitation antiviral defense system (Goldfarb et al., 2015). Collectively, these findings have allowed us to conduct genetic analyses of gene or system functions in P. alni Shahu, which, potentially, may serve as an important resource for both theoretical research and practical application in the future.
Pseudonocardia alni Shahu and derivatives constructed in this work, and E. coli strains, were listed in Supplementary Table S1. Pseudonocardia alni Shahu cells were grown at 30°C in an ISP4 medium (10 g/L soluble starch, 1 g/L K2HPO4, 1 g/L MgSO4·7H2O, 1 g/L NaCl, 2 g/L (NH4)2SO4, 2 g/L CaCO3, 0.001 g/L FeSO4·7H2O, 0.001 g/L MnCl2·4H2O, and 0.001 g/L ZnSO4·7H2O, and pH 7.0–7.4) or on ISP4 agar plates (15 g/L agar in ISP4), while E. coli strains at 37°C in Luria broth or on Luria agar plates. When required, the following antibiotics were used: 50 mg/L of apramycin, 25 mg/L of chloramphenicol, and 50 mg/L of kanamycin.
All the conjugation plasmids were constructed based on the pSET152 vector and listed in Supplementary Table S1. The site-specific integration plasmid pInt-green fluorescent protein (GFP) was generated by inserting a GFP-expression cassette into pSET152 at BamH I and Not I sites (Supplementary Figure S1). The GFP-expression DNA fragment was synthesized from GenScript (Nanjing, China). To construct the non-replicating suicidal plasmid, we first amplified three DNA fragments by PCR with each containing either the E. coli ori (primer set P1 + P2), or the aac(3)IV gene encoding the apramycin-resistance marker (P3 + P4), or the elements mediating plasmid transfer, i.e., traJ and oriT of RK2 (P5 + P6), using pSET152 as a template. These fragments were then ligated together using T4 ligase to generate a base vector, pTSF, for plasmid transfer. Subsequently, the upstream (Up, P7 + P8) and downstream (Down, P9 + P10) sequences of the target pgl genes were amplified from the genomic DNA of P. alni Shahu by PCR and inserted into pTSF as homologous recombination arms using EcoR I and Sal I, Hind III, and Pvu II sites, respectively, giving pKO-pgl (Supplementary Figure S1). All oligonucleotides were synthesized from GenScript (Nanjing, China) and listed in Supplementary Table S2. Restriction enzymes and T4 ligase were purchased from New England Biolabs (Beijing) Led (Beijing, China).
Pseudonocardia alni Shahu spores and the methylation-deficient E. coli strain ET12567 containing pUZ8002 were used as the recipient and the donor, respectively, throughout this study. A culture of the donor cells containing a transfer plasmid was grown to an OD600 of 0.4 in the presence of 50 mg/L apramycin, 50 mg/L kanamycin, and 25 mg/L chloramphenicol. The cells were then washed twice with an equal volume of LB to remove the antibiotics and finally resuspended in 0.1 volume of LB. The P. alni Shahu spores were resuspended with 0.5 ml of 2 × YT broth (10 g/L yeast extract, 16 g/L trptone, and 5 g/L NaCl). Recipient spores and donor cells were mixed and spread on ISP4 agar plates. After incubation at 30°C for 12–13 h, the plates were overlaid with 1 ml of distilled water containing 0.75 mg nalidixic acid and 1.25 mg apramycin, and put at 30°C until colonies were observed. Recombinant candidates were screen by colony PCR using primers listed in Supplementary Table S2. The PCR products were analyzed by agarose gel electrophoresis and confirmed by Sanger sequencing (GneScript, Nanjing, China).
The protocol used for FACS analysis was as previously described with slight modifications (Zheng et al., 2019). The P. alni Shahu cells were washed with phosphate buffered saline (PBS) twice and then resuspended into PBS to a concentration of 107 cells/ml. Afterwards, the cells were analyzed via flow cytometry using Beckman CytoFLEX FCM (Beckman Coulter, Inc., United States) with PBS as the sheath fluid. A 488 nm laser was used as an FITC channel to detect the cell fluorescence of GFP.
In order to identify the isolate, based on the complete genome sequence that has been deposited into NCBI with the BioProject accession number PRJNA563468, the 1,540-bp 16S rDNA gene sequence (GenBank accession number MW405797, Supplementary Table S3) was phylogenetically characterized via BLAST (NCBI) search. In the constructed phylogenetic tree, the isolate falls into the cluster including members of P. alni, Pseudonocardia antarctica, and Pseudonocardia carboxydivorans with a reliability of 86% (Figure 1A), suggesting that it belongs to the genus Pseudonocardia. To further detail the species of this isolate, its genome sequence was then subjected to a taxonomic analysis with genomes of Pseudonocardia available in the NCBI Genome database to estimate pairwise Average Nucleotide Identity (ANI) score using the FastANI method (https://github.com/ParBLiSS/FastANI; Jain et al., 2018). The result showed that the genome of the isolate pairing with that of P. alni DSM44104T or that of Pseudonocardia sp. AL041005-10 had ANI values ≥95% (Figure 1B), a criterion determining the same species in an existing genus (Jain et al., 2018). Taken together, the isolate was identified to be P. alni and assigned the strain name Shahu.
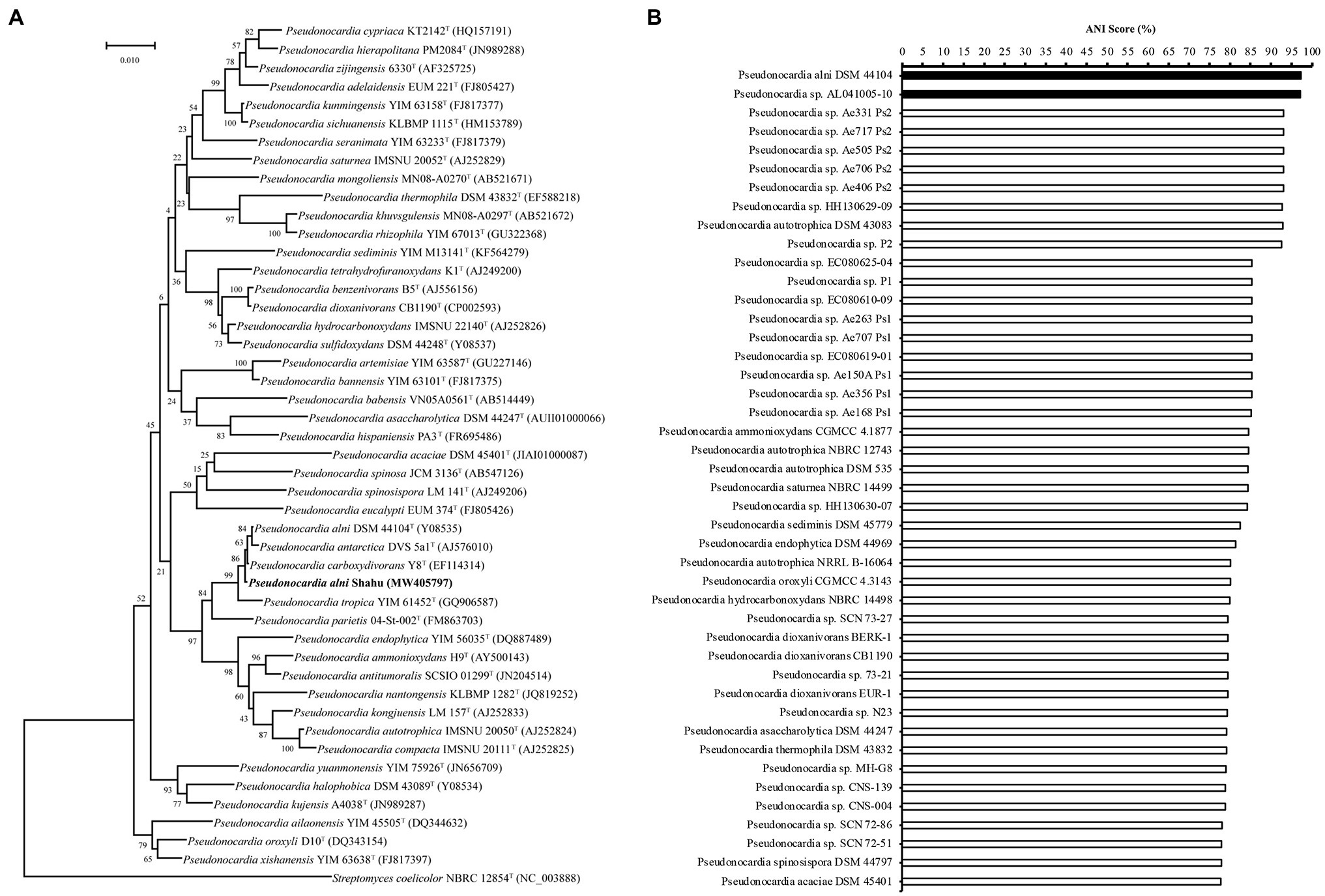
Figure 1. Identification of Pseudonocardia alni Shahu. (A) Neighbor-joining phylogenetic tree based on 16S rDNA sequences showing the phylogenetic relationships of P. anli Shahu and the selected Pseudonocardia species. Streptomyces coelicolor NBRC 12854T represents an outgroup reference. (B) Taxonomic analysis of P. anli Shahu with genomes of Pseudonocardia available in the NCBI Genome database. Pairwise Average Nucleotide Identity (ANI) score was estimated, and ANI score over 95% indicated with filled bars.
In order to determine a proper antibiotic resistance marker for P. alni Shahu, we investigated the resistance profile of this bacterium to common selective antibiotics, including apramycin (50 mg/L), kanamycin (50 mg/L), chloramphenicol (25 mg/L), tetracycline (50 mg/L), streptomycin (50 mg/L), and ampicillin (100 mg/L). About 106 pores were plated onto ISP4 agar containing one of the above antibiotics. At the tested concentrations, P. alni Shahu was sensitive to apramycin, chloramphenicol, and tetracycline, while fully resistant to kanamycin, streptomycin, and ampicillin.
Conjugal transfer represents one of the most straightforward and efficient methods for introducing foreign DNAs into rare Antinomycetes. We initially attempted conjugation between E. coli ET12567 (pUZ8002, pSET152) and P. alni Shahu following a standard conjugation protocol (Rebets et al., 2017), but failed in obtaining any exconjugant on the plate containing apramycin. This suggested that a modified protocol that specifically works for this strain is required to be established.
Previous studies have reported that an appropriate concentration of Mg2+ in the medium, or a suitable heat-shock treatment of the cells, or an optimal ratio of donor-to-recipient could give improved frequency of conjugation (Song et al., 2019; Zhang et al., 2019). We thus individually assessed each of such factors to attain successful conjugation. While, still, no exconjugant could be yielded, neither in the assay examining the various Mg2+ concentrations (0, 5, 10, 15, 30, and 50 mM), nor in that testing the heat-shocks with a temperature range from 37 to 65°C (data not shown). Fortunately, when we attempted changing the ratios of donor cells to recipient spores, we saw that different numbers of exconjugants appeared on the selective plates. The number of recipient cells played a more decisive role than that of the donor cells in the efficiency of conjugal transfer between E. coli and P. alni. When donor and recipient cells were 1 × 108 and 4 × 107, respectively, the highest conjugation efficiency reached 5.53 × 10−6 (Table 1). We therefore took this parameter in our following conjugation experiments.
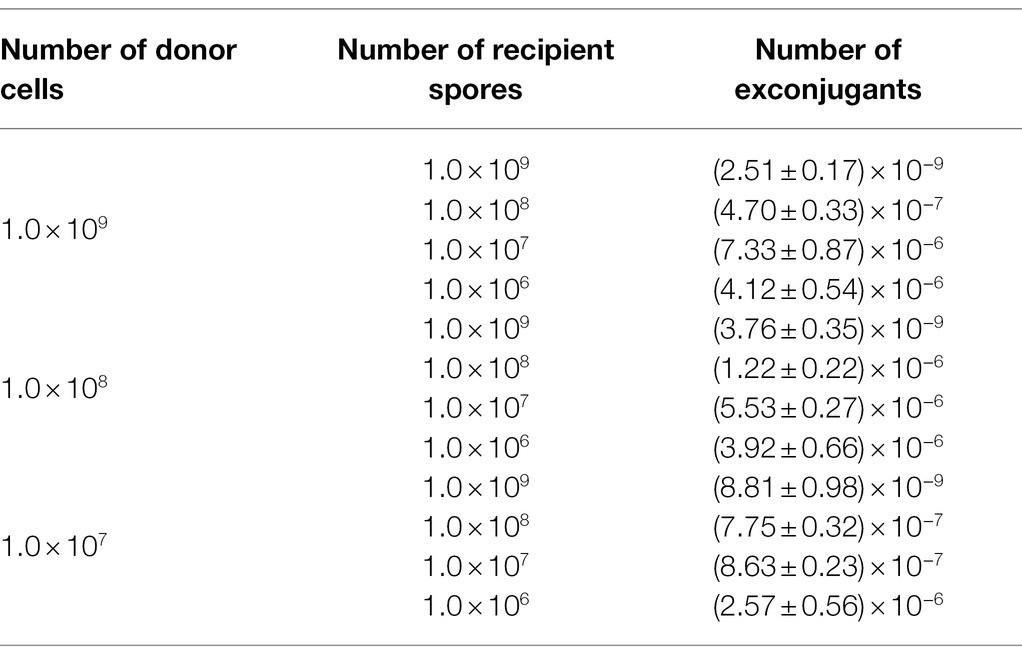
Table 1. Effects of the donor (E. coli cells)-to-recipient (P. alni Shahu spores) ratio on transconjugation efficiency. At ratio, three independent conjugations were analyzed.
The apramycin-resistance ability of the obtained exconjugants is suggestive of the chromosomal integration of the pSET152 plasmid harboring the aac(3)IV apramycin resistance marker gene driven by the ΦC31 integrase (Figure 2A). Therein, integration of exogeneous genes into the P. alni Shahu chromosome could be achievable via the ΦC31 att/int system. To test such capability of the system, an integrative plasmid, pInt-GFP, was constructed by inserting a GFP-expressing cassette to pSET152 (Supplementary Figure S1). Then, conjugal transfer of pInt-GFP from E. coli to P. alni was conducted and its subsequent integration into the P. alni chromosome was examined.
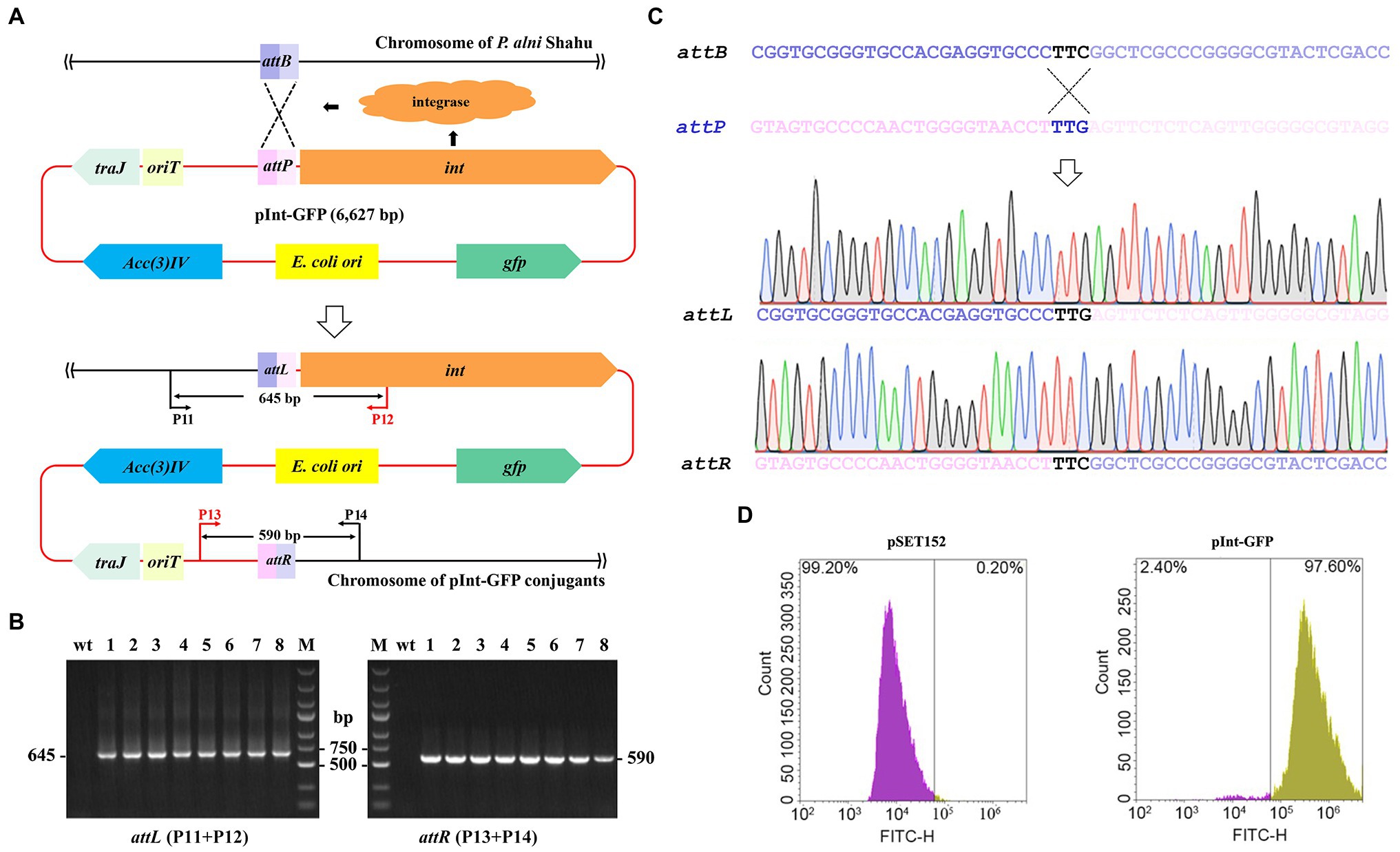
Figure 2. Chromosomal integration of the exogenous gfp gene using the ΦC31 att/int system. (A) Schematic showing integration of the entire pInt-green fluorescent protein (GFP) plasmid into P. anli Shahu chromosome. (B) Colony PCR analyses of eight randomly taken exconjugants, and the wild-type strain (wt) as a reference, using primer sets of P11 + P12 and P13 + P14 as shown in (A). Predicted sizes of PCR products encompassing the created hybrid attL and attR sites after integration are indicated. M, DNA size marker. (C) Representative chromatographs of Sanger sequencing results of the attL and attR sites. Original sequences of attB in P. anli Shahu chromosome and attP in pInt-GFP plasmid, respectively, are shown. (D) Signal of GFP was detected in the pInt-GFP exconjugant whereas not in the pSET152 exconjugant by flow cytometry.
According to the determined optimal ratio, E. coli ET12567 (pUZ8002, pInt-GFP) cells were mixed with P. alni Shahu spores for conjugation. Of the conjugants appeared on the plate with apramycin, eight were randomly picked up and grown up in a liquid ISP4 medium containing 50 mg/L of apramycin. To illustrate the chromosomal integration of pInt-GFP in detail, total DNAs were individually extracted from the selected exconjugants and used as templates for PCR analyses with a primer set of either P11 + P12 or P13 + P14 amplifying DNA fragments encompassing the attL or attR site, respectively (Figure 2A). Since P11 and P14 target the genomic sequences while P12 and P13 are located on the plasmid, PCR products can be amplified only when chromosomal integration of plasmid occurs. Agarose gel electrophoresis of the PCR products suggested successful integration of pInt-GFP into the Shahu genome, as PCR products with the expected sizes were amplified from the chromosomal DNAs of the conjugants but not from that of the wild-type strain (wt; Figure 2B). Subsequent Sanger sequencing of the PCR products confirmed the generation of the hybrid sequences of attL and attR from attB and attP (Figure 2C). In addition, our results detected the fluorescence signal of GFP from all the pInt-GFP exconjugants by flow cytometry, suggesting that the heterologously integrated GFP protein was stably expressed. In contrast, the signal was not detectable in the reference pSET152 exconjugant (Figure 2D). All these combined results suggested that the ΦC31 att/int system could be used for efficient chromosomal incorporation of foreign DNAs in P. alni Shahu.
We next demonstrated removal of exogeneous genes from the P. alni Shahu chromosome by using a non-replicating vector to facilitate homologous recombination. A DNA stretch containing the pgl genes was chosen as an editing target, since it was reported to be non-essential for cell viability (Goldfarb et al., 2015). The entire sequence of the ΦC31 int gene, together with that of the attP site, was removed from pSET152; while homologous arms of 633 and 733 bp were inserted upstream and downstream, respectively, of the acc(3)IV gene, giving the suicidal plasmid, pKO-pgl, for knockout of the pgl genes (Figure 3A).
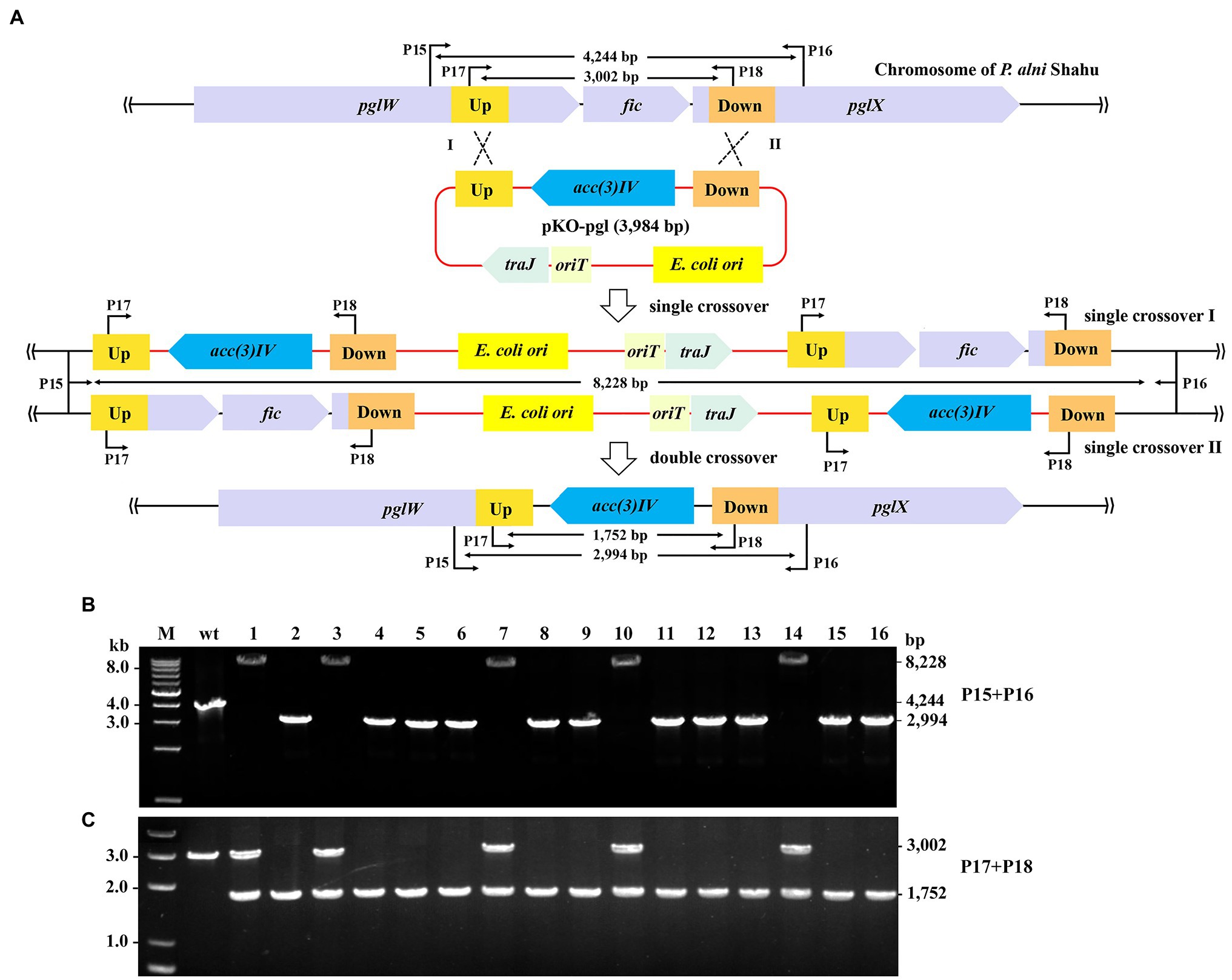
Figure 3. Gene deletion in P. alni Shahu via homologous recombination. (A) Scheme of deletion of the pgl genes using a suicidal non-replicating plasmid, pKO-pgl. Recombination between either the homologous upstream (Up) arms (Single crossover I), or downstream (Down) arms (Single crossover II), would result in integration of the entire plasmid into to the chromosome; while double crossover recombination was expected to occur, eventually leading to replacement of the target genes with the apramycin resistance marker, acc(3)IV. (B,C) PCR screening of pgl::acc(3)IV recombinants using primer sets of P15 + P16 (B) and P17 + 18 (C). Predicted sizes of PCR products in wild-type (wt) and recombinants of single or double crossover recombination are indicated. M, DNA size marker.
After conjugal transfer of pKO-PGL into P. alni Shahu, colonies were observed on the selective plate containing apramycin, indicative of integration of the acc(3)IV gene into the host chromosome. We picked up 16 of the appeared colonies and analyzed their genotypes by PCR and Sanger sequencing. Using the primer set of P15 + P16, DNA products were amplified from total DNAs extracted from the strains that were expected to contain mutant alleles with a predicted size of 2,994 bp. However, the 2,994-bp band was amplified in only 11 of the tested colonies, while, interestingly, a predicted 8,228 bp band was amplified in the remainder (Figure 3B). Reasonably, double crossover has resulted in the replacement of the target region (2,260 bp) with the expression cassette of the acc(3)IV gene (1,010 bp), allowing for amplification of the 2,994-bp fragments (vs. the wild-type 4,244 bp); whereas a single crossover through either of the homologous arms would occur, leading to chromosomal integration of the entire plasmid and hence explaining the detected 8,228-bp DNA products.
To further corroborate the above observations, we designed an additional primer set, P17 + P18 with each, respectively, targeting a sequence located within the homologous arms, for PCR analyses of the same samples. As expected, PCR products of 3,002 and 1,752-bp were yielded in the wild-type cells and the double crossover recombinants, respectively; while both fragments were amplified in the colonies where single crossovers occurred (Figure 3C). These results thus confirmed the homologous recombination events deduced and showcased in Figure 3A.
Although successful examples of genetic engineering of some model actinobacteria have been accumulated in the literature (Deng et al., 2017; Kormanec et al., 2019), it is, in general, still more difficult to generically manipulate many rare actinomycetes. Methods to genetically manipulate the genome of rare actinomycetes, for example Pseudonocardia, might open new avenues toward the demonstration, development, and application of the valuable bacteria. Despite of the importance, until this work, there had been no detailed description of establishing a genetic manipulation system for genome modifications in any Pseudonocardia species.
The major challenges could include the transfer of the vector harboring the foreign DNAs into the host and the incorporation of DNA into the host chromosome. Intergeneric conjugation has been proven to be one of the most efficient approaches for gene transfer into actinomycetes, and has been already successfully applied to Streptomyces species as an example (Netzker et al., 2016; Kormanec et al., 2019). However, it was suggested that conjugation protocols have to be customized for individual species before accomplishing successful conjugation (Marcone et al., 2010), and indeed, our work has confirmed that the standard conjugation protocol (Rebets et al., 2017) is not suitable for P. alni Shahu. We figured out the optimal parameters for conjugation and successfully achieved transfer of the pSET152 plasmid and its derivatives from E. coli into P. alni, therein, making it possible to deliver the powerful CRISPR-Cas machinery and others of interest, into this bacterium by this very approach for high-throughput genome engineering in the future.
We did observe the integration of the transferred pInt-GFP, a pSET152 derivative, into the genome at an attB site. Based on the confirmed attL and attR sequences, we deduced the 51-nt sequence of the attB site (Figure 2C) and traced it back to the genome of P. alni Shahu, seeing that it laid within an OFR coding for a pirin-homolog. This was very similarly to the case of Streptomyces species where attB sites were exclusively located within pirin-encoding ORFs. We found that this attB sequence showed an 89% nt identity to the canonical attB sequence possessed by Streptomyces coelicolor A3(2) from which the antinophage ΦC31 was originated (Lomovskaya et al., 1972). The sequence identity between the attB site of S. coelicolor A3(2) and that of other actinomycetes seemed to play an important role in determining conjugation efficacy (Supplementary Figure S2). For example, plasmid transconjugation into S. ambofaciens harboring an attB sequence identical to that of S. coelicolor A3(2) (100% identity) yielded an efficiency of 1.4 × 10−2, while into S. clavligerus yielded an efficiency of 3 × 10−5 as the attB identity was 96% (Kim et al., 2008). Furthermore, when the plasmid was transconjugated into a non-Streptomyces species Kitasatospora setae with a 78% attB identity, the efficiency was dropped to 2 × 10−7 (Choi et al., 2004). This thus explained the observed efficiency of foreign DNA integration into P. alni Shahu genome (5.5 × 10−6, Table 1) via the ΦC31 int/att system in our work, and might provide a possibility to enhance the conjugation efficiency via engineering the attB sequence of P. alni Shahu to make it as identical as that of S. coelicolor A3(2).
An obvious shortcoming of this genetic manipulation system could be the lack of a counterselection marker. Currently, several suicide genes, e.g., sacB, glkA, pyrF, and rpsL, have been available as counterselection markers in different bacteria. The sacB gene, conferring sucrose sensitivity, was ever used for unmarked gene deletion in actinobacteria species (van der Geize et al., 2001, 2007). However, it was reported that this method was not applicable for S. lividans due to lack of sucrose sensitivity of this actinobacterium (Jager et al., 1992). The glkA, pyrF, and rpsL, and others, did play their roles in counterselection but only in the corresponding null mutants (van Wezel and Bibb, 1996; Hosted and Baltz, 1997; Knipfer et al., 1997). Additionally, some toxic genes, such as mazF (Zhang et al., 2006), can also be used as suicidal markers, which, however, rely on tightly controlled inducible promoters, thus limiting their applicability in strains with poorly developed molecular toolkits. Other methods may also include the application of temperature-sensitive replicons. For instance, the replicon of pIJ101 has been widely applied in some Streptomyces species (Mo et al., 2019). These methods might be also possibly applicable in P. alni Shahu, albeit, of course, necessitating the examination and determination of optimal parameters.
It would be also interesting to introduce the advanced CRISPR-Cas-based technologies into this actinobacterium. One of advantages of the technologies is that no selection marker is required due to the potent killing effect of the CRISPR-Cas systems on the bacterial cells. However, we could not get any transconjugant in the conjugation using a plasmid carrying a Cas9-expression cassette, even without a targeting guide RNA (data not shown). Possibly, the Cas9 nuclease per se is toxic to the host cells. Similar observations have been reported for other bacteria (Zheng et al., 2020; Hao et al., 2022). Interesting, we found that the chromosome of P. alni Shahu encodes a native CRISPR-Cas system. We believe that in the near future this system would be harnessed as a powerful genetic manipulation tool, although thorough demonstration of this system would be essentially needed.
Nevertheless, this work has enabled the generation of stable genome modifications in a rare actinomycetes P. alni Shahu with a considerably high efficiency, and thus making this strain amenable to the discovery and design of novel bioactive natural products by genetic engineering.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA563468; https://www.ncbi.nlm.nih.gov/genbank/, MW405797.
WP and YZe designed the research. JL, BW, QY, HS, and YZa performed the experiments. WP, JL, BW, and YZe wrote the manuscript. All authors contributed to data analyses, read, revised, and approved the final manuscript.
This work was supported by the Open Funding Project of the State Key Laboratory of Biocatalysis and Enzyme Engineering (SKLBEE2021019), and the Innovation Base for Introducing Talents of Discipline of Hubei Province (2019BJH021). YZe and WP acknowledge the support from the State Key Laboratory of Biocatalysis and Enzyme Engineering.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.848964/full#supplementary-material
Caffrey, P., De Poire, E., Sheehan, J., and Sweeney, P. (2016). Polyene macrolide biosynthesis in streptomycetes and related bacteria: recent advances from genome sequencing and experimental studies. Appl. Microbiol. Biotechnol. 100, 3893–3908. doi: 10.1007/s00253-016-7474-z
Choi, S. U., Lee, C. K., Hwang, Y. I., Kinoshita, H., and Nihira, T. (2004). Intergeneric conjugal transfer of plasmid DNA from Escherichia coli to Kitasatospora setae, a bafilomycin B1 producer. Arch. Microbiol. 181, 294–298. doi: 10.1007/s00203-004-0654-8
Combes, P., Till, R., Bee, S., and Smith, M. C. (2002). The streptomyces genome contains multiple pseudo-attB sites for the (phi)C31-encoded site-specific recombination system. J. Bacteriol. 184, 5746–5752. doi: 10.1128/JB.184.20.5746-5752.2002
Deng, Y., Zhang, X., and Zhang, X. (2017). Recent advances in genetic modification systems for Actinobacteria. Appl. Microbiol. Biotechnol. 101, 2217–2226. doi: 10.1007/s00253-017-8156-1
Doron, S., Melamed, S., Ofir, G., Leavitt, A., Lopatina, A., Keren, M., et al. (2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. doi: 10.1126/science.aar4120
Fang, Z., Chen, S., Zhu, Y., Li, J., Khan, I., Zhang, Q., et al. (2019). A new uridine derivative and a new indole derivative from the coral-associated actinomycete Pseudonocardia sp. SCSIO 11457. Nat. Prod. Res. 35, 188–194. doi: 10.1080/14786419.2019.1616729
Flett, F., Mersinias, V., and Smith, C. P. (1997). High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155, 223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x
Geddis, S. M., Coroama, T., Forrest, S., Hodgkinson, J. T., Welch, M., and Spring, D. R. (2018). Synthesis and biological evaluation of 1,2-disubsubstituted 4-quinolone analogues of Pseudonocardia sp. natural products. Beilstein J. Org. Chem. 14, 2680–2688. doi: 10.3762/bjoc.14.245
Goldfarb, T., Sberro, H., Weinstock, E., Cohen, O., Doron, S., Charpak-Amikam, Y., et al. (2015). BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183. doi: 10.15252/embj.201489455
Grostern, A., Sales, C. M., Zhuang, W. Q., Erbilgin, O., and Alvarez-Cohen, L. (2012). Glyoxylate metabolism is a key feature of the metabolic degradation of 1,4-dioxane by Pseudonocardia dioxanivorans strain CB1190. Appl. Environ. Microbiol. 78, 3298–3308. doi: 10.1128/AEM.00067-12
Hao, Y., Wang, Q., Li, J., Yang, S., Zheng, Y., and Peng, W. (2022). Double nicking by RNA-directed Cascade-nCas3 for high-efficiency large-scale genome engineering. Open Biol. 12:210241. doi: 10.1098/rsob.210241
Hosted, T. J., and Baltz, R. H. (1997). Use of rpsL for dominance selection and gene replacement in Streptomyces roseosporus. J. Bacteriol. 179, 180–186. doi: 10.1128/jb.179.1.180-186.1997
Inoue, D., Tsunoda, T., Sawada, K., Yamamoto, N., Saito, Y., Sei, K., et al. (2016). 1,4-Dioxane degradation potential of members of the genera Pseudonocardia and Rhodococcus. Biodegradation 27, 277–286. doi: 10.1007/s10532-016-9772-7
Jager, W., Schafer, A., Puhler, A., Labes, G., and Wohlleben, W. (1992). Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J. Bacteriol. 174, 5462–5465. doi: 10.1128/jb.174.16.5462-5465.1992
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kim, M. K., Ha, H. S., and Choi, S. U. (2008). Conjugal transfer using the bacteriophage phiC31 att/int system and properties of the attB site in Streptomyces ambofaciens. Biotechnol. Lett. 30, 695–699. doi: 10.1007/s10529-007-9586-0
Knipfer, N., Seth, A., and Shrader, T. E. (1997). Unmarked gene integration into the chromosome of mycobacterium smegmatis via precise replacement of the pyrF gene. Plasmid 37, 129–140. doi: 10.1006/plas.1997.1286
Kormanec, J., Rezuchova, B., Homerova, D., Csolleiova, D., Sevcikova, B., Novakova, R., et al. (2019). Recent achievements in the generation of stable genome alterations/mutations in species of the genus Streptomyces. Appl. Microbiol. Biotechnol. 103, 5463–5482. doi: 10.1007/s00253-019-09901-0
Lomovskaya, N. D., Mkrtumian, N. M., Gostimskaya, N. L., and Danilenko, V. N. (1972). Characterization of temperate actinophage phi C31 isolated from Streptomyces coelicolor A3(2). J. Virol. 9, 258–262. doi: 10.1128/jvi.9.2.258-262.1972
Marcone, G. L., Foulston, L., Binda, E., Marinelli, F., Bibb, M., and Beltrametti, F. (2010). Methods for the genetic manipulation of Nonomuraea sp. ATCC 39727. J. Ind. Microbiol. Biotechnol. 37, 1097–1103. doi: 10.1007/s10295-010-0807-5
Miyamoto, K. T., Komatsu, M., and Ikeda, H. (2014). Discovery of gene cluster for mycosporine-like amino acid biosynthesis from Actinomycetales microorganisms and production of a novel mycosporine-like amino acid by heterologous expression. Appl. Environ. Microbiol. 80, 5028–5036. doi: 10.1128/AEM.00727-14
Mo, J., Wang, S., Zhang, W., Li, C., Deng, Z., Zhang, L., et al. (2019). Efficient editing DNA regions with high sequence identity in actinomycetal genomes by a CRISPR-Cas9 system. Synth. Syst. Biotechnol. 4, 86–91. doi: 10.1016/j.synbio.2019.02.004
Netzker, T., Schroeckh, V., Gregory, M. A., Flak, M., Krespach, M. K. C., Leadlay, P. F., et al. (2016). An efficient method to generate gene deletion mutants of the rapamycin-producing bacterium Streptomyces iranensis HM 35. Appl. Environ. Microbiol. 82, 3481–3492. doi: 10.1128/AEM.00371-16
Qi, M., Huang, H., Zhang, Y., Wang, H., Li, H., and Lu, Z. (2019). Novel tetrahydrofuran (THF) degradation-associated genes and cooperation patterns of a THF-degrading microbial community as revealed by metagenomic. Chemosphere 231, 173–183. doi: 10.1016/j.chemosphere.2019.05.137
Rebets, Y., Kormanec, J., Luzhetskyy, A., Bernaerts, K., and Anne, J. (2017). Cloning and expression of metagenomic DNA in Streptomyces lividans and subsequent fermentation for optimized production. Methods Mol. Biol. 1539, 99–144. doi: 10.1007/978-1-4939-6691-2_8
Riahi, H. S., Heidarieh, P., and Fatahi-Bafghi, M. (2021). Genus Pseudonocardia: what we know about its biological properties, abilities and current application in biotechnology. J. Appl. Microbiol. 132, 890–906. doi: 10.1111/jam.15271
Sales, C. M., Grostern, A., Parales, J. V., Parales, R. E., and Alvarez-Cohen, L. (2013). Oxidation of the cyclic ethers 1,4-dioxane and tetrahydrofuran by a monooxygenase in two Pseudonocardia species. Appl. Environ. Microbiol. 79, 7702–7708. doi: 10.1128/AEM.02418-13
Song, Z. Q., Liao, Z. J., Hu, Y. F., Ma, Z., Bechthold, A., and Yu, X. P. (2019). Development and optimization of an intergeneric conjugation system and analysis of promoter activity in Streptomyces rimosus M527. J Zhejiang Univ Sci B 20, 891–900. doi: 10.1631/jzus.B1900270
Strecker, J., Jones, S., Koopal, B., Schmid-Burgk, J., Zetsche, B., Gao, L., et al. (2019). Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 10:212. doi: 10.1038/s41467-018-08224-4
Subramani, R., and Sipkema, D. (2019). Marine rare actinomycetes: a promising source of structurally diverse and unique novel natural products. Mar. Drugs 17:249. doi: 10.3390/md17050249
Tiwari, K., and Gupta, R. K. (2012). Rare actinomycetes: a potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 32, 108–132. doi: 10.3109/07388551.2011.562482
van der Geize, R., Hessels, G. I., van Gerwen, R., van der Meijden, P., and Dijkhuizen, L. (2001). Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205, 197–202. doi: 10.1111/j.1574-6968.2001.tb10947.x
Van der Geize, R., Yam, K., Heuser, T., Wilbrink, M. H., Hara, H., Anderton, M. C., et al. (2007). A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104, 1947–1952. doi: 10.1073/pnas.0605728104
van Wezel, G. P., and Bibb, M. J. (1996). A novel plasmid vector that uses the glucose kinase gene (glkA) for the positive selection of stable gene disruptants in Streptomyces. Gene 182, 229–230. doi: 10.1016/S0378-1119(96)00563-X
Won, H. J., Kim, H. J., Jang, J. Y., Kang, S. H., Choi, S. S., and Kim, E. S. (2017). Improved recovery and biological activities of an engineered polyene NPP analogue in Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 44, 1293–1299. doi: 10.1007/s10295-017-1954-8
Yamamoto, N., Saito, Y., Inoue, D., Sei, K., and Ike, M. (2018). Characterization of newly isolated Pseudonocardia sp. N23 with high 1,4-dioxane-degrading ability. J. Biosci. Bioeng. 125, 552–558. doi: 10.1016/j.jbiosc.2017.12.005
Zhang, S., Chen, T., Jia, J., Guo, L., Zhang, H., Li, C., et al. (2019). Establishment of a highly efficient conjugation protocol for Streptomyces kanamyceticus ATCC12853. Microbiology 8:e00747. doi: 10.1002/mbo3.747
Zhang, X. Z., Yan, X., Cui, Z. L., Hong, Q., and Li, S. P. (2006). mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 34:e71. doi: 10.1093/nar/gkl358
Zheng, Y., Han, J., Wang, B., Hu, X., Li, R., Shen, W., et al. (2019). Characterization and repurposing of the endogenous type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 47, 11461–11475. doi: 10.1093/nar/gkz940
Keywords: Pseudonocardia, genetic manipulation, conjugal transfer, gene integration, gene deletion
Citation: Li J, Wang B, Yang Q, Si H, Zhao Y, Zheng Y and Peng W (2022) Enabling Efficient Genetic Manipulations in a Rare Actinomycete Pseudonocardia alni Shahu. Front. Microbiol. 13:848964. doi: 10.3389/fmicb.2022.848964
Received: 05 January 2022; Accepted: 01 February 2022;
Published: 03 March 2022.
Edited by:
Yu-Qin Zhang, Institute of Medicinal Biotechnology (CAS), ChinaReviewed by:
Nan Peng, Huazhong Agricultural University, ChinaCopyright © 2022 Li, Wang, Yang, Si, Zhao, Zheng and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanli Zheng, WWFubGkuWmhlbmdAd2hwdS5lZHUuY24=; Wenfang Peng, d2VuZmFuZ0BodWJ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.