- Department of Biochemistry, University of Calcutta, Kolkata, India
Macrophomina phaseolina is a global devastating necrotrophic fungal pathogen. It causes charcoal rot disease in more than 500 host plants including major food crops, pulse crops, fiber crops, and oil crops. Despite having the whole-genome sequence of M. phaseolina, understanding the M. phaseolina genome-based plant–pathogen interactions is limited in the absence of direct experimental proof of secretion. Thus, it is essential to understand the host–microbe interaction and the disease pathogenesis, which can ensure global agricultural crop production and security. An in silico–predicted secretome of M. phaseolina is unable to represent the actual secretome. We could identify 117 proteins present in the secretome of M. phaseolina using liquid chromatography–electrospray ionization–tandem mass spectrometry. Data are available via ProteomeXchange with identifier PXD032749. An array of putative virulence factors of M. phaseolina were identified in the present study using solid-state culture. Similar virulence factors have been reported in other plant pathogenic fungi also. Among the secretory fungal proteins with positive economic impacts, lignocellulolytic enzymes are of prime importance. Further, we validated our results by detecting the cell wall–degrading enzymes xylanase, endoglucanase, and amylase in the secretome of M. phaseolina. The present study may provide a better understanding about the necrotrophic fungi M. phaseolina, which modulate the host plant defense barriers using secretory proteins.
Introduction
Macrophomina phaseolina is a global devastating necrotrophic fungal pathogen that causes charcoal rot disease in more than 500 host plants (Wyllie, 1998) including major food crops (Su et al., 2001), pulse crops (Raguchander et al., 1993; Mayek Pérez et al., 2001), fiber crops (De et al., 1992; Aly et al., 2007), and oil crops. Other diseases caused by this pathogen include color rot, root rot, and damping off; stem root and seedling blight in many economically important plants (Kishore Babu et al., 2007). M. phaseolina belongs to the Botryosphaeriaceae family (Crous et al., 2006). The occurrence of sclerotia of M. phaseolina in plant debris allows the fungus to live in soil, even in the absence of a host for 2 or more years, depending on soil conditions. Pathogenicity of M. phaseolina is optimal at 28–35°C, and host water stress is another principal factor favoring development of the disease (Dhingra and Sinclair, 1973). Emergence of M. phaseolina is now a serious concern for the cultivation of soybean under climate change scenario worldwide (Vibha, 2016). In India, this disease became a serious threat due to altered weather condition particularly because of the longer drought spells during crop growth period. Subsequently, the disease started to appear irregularly in some areas, which caused great amount of yield losses (Gupta et al., 2012).
In the last decade, proteomic analysis of secretory proteins of phytopathogenic fungus has been used increasingly to study the plant–pathogen interactions (Gonzalez-Fernandez and Jorrın-Novo, 2012). Proteomic tools such as gel-based or gel-free liquid chromatography–based mass spectrometry (LC-MS/MS) permit identification of key proteins of the secretome during host–microbe interactions. Enzymes secreted by certain fungi hold the key to its pathogenesis. Secretome study of Trichoderma harzianum shows 51.36% presence of glycoside hydrolases (GHs), and the secretome of this filamentous fungus is rich in hydrolytic enzymes (Do Vale et al., 2012). Carbohydrate-degrading enzymes and proteases were found in secretome analysis of Diplodia corticola (Fernandes et al., 2014). However, very little information is available about the secretome of fungi belonging to the Botryosphaeriaceae family due to the unavailability of whole-genome sequence of the fungus. However, this is not the case for M. phaseolina; its genome has already been sequenced. Whole-genome sequence of M. phaseolina shows the presence of 362 putative Cazymes including 219 GHs (Islam et al., 2012). Plant cell wall represents the first barrier to an invading pathogen, and M. phaseolina invades the plant defense by secreting a cocktail of enzymes (Le Marquer et al., 2019). Despite having the whole-genome sequence of M. phaseolina, understanding the M. phaseolina genome-based plant–pathogen interactions is not easy in the absence of direct experimental proof of secretion because an in silico–predicted secretome is unlikely to exactly represent the actual secretome. Thus, proteomic analysis of secretome will become instrumental in designing rational strategies for disease control. Not only that, but it is also essential to understand the host–microbe interaction and the disease pathogenesis, which can ensure global agricultural crop production and security. In our previous study, we observed induction of charcoal rot disease during Corchorus capsularis (jute) JRC 412–M. phaseolina (strain R9) interaction that resulted in elevated nitric oxide, reactive nitrogen species, and S-nitrosothiols production in infected tissues. To mimic the plant–pathogen interaction, M. phaseolina was grown on wheat bran in a solid-state culture, and secretome was collected. Interestingly, nitric oxide was produced in single leaf experiment in the presence of both M. phaseolina and xylanases obtained from fungal secretome. In fact, xylanase is known to play a vital role in presenting pathogen-associated molecular pattern (PAMP) which was also evident in our experimental model system (Sarkar et al., 2014). In the present study also, we used similar solid-state culture where M. phaseolina (strain R9) was grown on wheat bran bed to mimic the plant–pathogen interaction. We identified 117 proteins present in the secretome of M. phaseolina using LC–electrospray ionization–MS/MS (LC-ESI-MS/MS). An array of putative virulence factors of M. phaseolina were identified that have been reported to be involved in other plant pathogenic fungi also. Among the secretory fungal proteins with positive economic impacts, lignocellulolytic enzymes are of prime importance. Further, we validated our results by detecting the cell wall–degrading enzymes xylanase, endoglucanase, and amylase in the secretome of M. phaseolina. The present study may provide a better understanding about the necrotrophic fungi M. phaseolina, which modulate the host plant defense barriers using secretory proteins.
Materials and Methods
Microorganism and Maintenance of the Culture
A virulent isolate of M. phaseolina (strain R9) was collected from Sorbhog, Assam. A pure mycelial culture was generated through single sclerotia of this isolate, maintained in potato dextrose agar (PDA) media at 30°C. PDA slants were inoculated from the stock culture and maintained for several days at 30°C. Wheat bran was selected as the solid state for the experiments. Ten grams of wheat bran was taken in 100-mL conical flask, and it was sterilized. Spores of M. phaseolina were inoculated in wheat bran with the help of Mendel’s Mineral Salt Solution [for 1 L (NH4)2S04 1.4 g, K2HPO4 2 g, CaCl2, 2H2O 0.4 g, MgSO4,7H2O 0.3 g, FeSO4,7H2O 5 mg, MnSO4,7H2O 1.6 mg, ZnSO4,7H2O 1.4 mg, CoCl2,6H2O 2 mg]. The wheat bran–containing conical flasks were then maintained at 30°C and 90% moisture content.
Isolation of Secretory Protein
After specific day of incubation of conical flasks containing M. phaseolina, infected wheat bran was taken for protein isolation. An amount of 30–40 mL of sodium phosphate buffer of pH 7.0 was directly added to the conical flask containing infected wheat bran. The secretome was extracted twice from the infected wheat bran. For the first extraction, the conical flasks were placed in a shaker and rotated at 120 revolutions/min (rpm) for proper shaking at room temperature for 4 h. The buffer containing secretory proteins was extracted from wheat bran by compression using a glass rod from the conical flasks, and the decanted materials were filtered through Whatman filter paper (110 mm). For the second time extraction, an amount of 20–25 mL of 50 mM sodium phosphate buffer of pH 7.0 was again added directly to the wheat bran bed, and the conical flask was kept under shaking condition at 120 rpm for 2 more hours at room temperature. Extraction of the secretory proteins in 50 mM sodium phosphate buffer was done using similar procedure as described before. Concentration of the protein was measured after each extraction using Bradford reagent (Bradford, 1976). Secretory proteins collected after the first and second extractions were pooled, concentrated using 0–80% ammonium sulfate precipitation, and finally dialyzed to remove the salts from the concentrated proteins.
One-Dimensional Gel Electrophoresis
Macrophomina phaseolina secretome was analyzed in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) to observe the protein bands along with the molecular weight markers. Secretome of 5-, 8-, 11-, and 14-day post-inoculated culture was loaded for separation on a 10% SDS-PAGE. Each lane contained 50 μg of protein sample.
Identification of Proteins by LC-ESI-MS/MS
Extracted proteins from M. phaseolina secretome were first lyophilized and then solubilized in 100 mM ammonium bicarbonate. Tetra-fluoroethylene, dithiothreitol, and iodoacetamide were added sequentially to the solution. Then, trypsin was added and kept overnight at 37°C prior to LC-ESI-MS/MS. LC-ESI-MS/MS was done on Waters-Xevo-G2-X’S-QTof machine in ESI-positive mode. BEH C 18 column (Waters) was used in the instrument. The machine used MSE scanning technique where MS and MS/MS took place simultaneously. Sodium formamide was used as primary standard, and leucine enkephalin was used as secondary standard. Ramp collision energy was set at 18–40 V. MS and MS/MS thresholds were set at 150 and 20 counts, respectively; 0.1% formic acid was added to the sample; 10 μL sample was injected finally for detection of the peptides. The run time was set 1 h. In the first 5 min (0–5 min), the column was being equilibrated. In the next 45 min (5–50 min), the peptides were eluted from the column with ACN gradient 0–90%. In the last 10 min (50–60 min), the column was being washed. The peptides detected by LC-MS/MS were matched with M. phaseolina MS6 database downloaded from UniProt in FASTA format. The software used for analysis is Progenesis Qip by Waters.
Enzyme Assay
The activity of xylanase and endoglucanase was measured using dinitrosalicylic acid (DNS) assay method, which measures the total amount of reducing sugars. 3,5-DNS in the presence of reducing sugar forms 3-amino 5-nitrosalicylic acid, and this compound strongly absorbs light at 540 nm (Miller, 1959).
DNS reagent was prepared by dissolving 1 g of 3,5-DNA in 50 mL of water; 30 g of sodium potassium tartrate was added to the solution in small lots, and the solution turns milky yellow in color. Finally, 20 mL of 2(N) NaOH was added, and that turned the solution to transparent orange yellow color.
Xylanase activity was measured by using 1% birchwood xylan as substrate (Yang et al., 1995). Xylanase degrades xylan and forms xylose. The formation of xylose in 10 min at 30°C was measured by the DNS reagent. Xylose standard curve was plotted by taking known concentration of xylose and their corresponding optical density values at 540 nm (OD540). One unit of enzyme activity is defined as the amount of enzyme producing 1 μmol of xylose equivalents per minute under the given conditions.
Endoglucanase activity was determined in accordance with the International Union of Pure and Applied Chemistry recommendations, with a 1% solution of carboxymethyl (CM) cellulose as the substrate. Endoglucanase degrades CM cellulose and forms glucose. The formation of glucose in 10 min at 30°C was measured by the DNS reagent. Standard curve of glucose was obtained by taking known concentrations of glucose followed by addition of DNS reagents and their corresponding OD540 values (Dutta et al., 2008). One unit of enzyme activity is defined as the amount of enzyme producing 1 μmol of glucose equivalents per minute under the given conditions.
Amylase activity was measured by using starch as the substrate. Amylase hydrolyses starch into smaller carbohydrate molecules such as maltose. Iodine reacts with starch to form a starch–iodine complex. The color of the solution turns blue, and it strongly absorbs light at 620 nm (Cochran et al., 2008). Standard curve of starch was plotted by taking known concentrations of starch and their corresponding OD620 values in the presence of iodine. The starting concentration of starch was 250 μg. The degradation of starch occurred when it was incubated with the protein solution for 1 min. Iodine was added to the solution, and OD620 values were taken. The amount of starch degraded can be calculated with the OD620 values and the values from the starch standard curve. The activity of enzyme was calculated as the amount of starch degraded per minute per microgram of protein.
Zymogram Analysis
Zymogram analysis is a great way to visualize the presence of active enzymes in the secretome. Zymogram of xylanase and endoglucanase was performed with a modified method described in Coral et al. (2002). Zymogram of xylanase and endoglucanase was done by casting the resolving gel with 0.1% birchwood xylan and 0.1% CM cellulose. Native-PAGE was done to keep the secretome proteins in active state. Native-PAGE gels (1.5-mm thickness) were prepared with 5% stacking gel and 8% resolving gel for both zymograms. The protein loading samples were prepared without SDS or β-mercaptoethanol to maintain the native state of the protein. Thirty micrograms of protein collected on days 5, 8, 11, and 14 after inoculation was loaded in each of the lanes. The Native-PAGE was run at 4°C. Electrophoresis was done using 30-mA steady current. After the dye front reached the bottom of the gel, it was further electrophoresed for 30 min. The gel was removed from the glass plates and placed in a glass dish. The gel was then stained with 0.1% Congo red and then destained with 1 M NaCl (Dutta et al., 2008). After destaining the gel, white regions can be seen in red color background. The gel pictures were taken on Gel DocTM XR+ BioRad imaging machine.
Zymogram of amylase was performed with a modified method described by Andrades and Contreras (2017). Zymogram of amylase was done by casting the resolving gel with 0.4% starch. A native polyacrylamide gel (1.5-mm thickness) was prepared with 5% stacking solution and 8% resolving solution. The protein loading samples were prepared without SDS or β-mercaptoethanol to maintain the native state of the protein. An amount of 10 μg of protein collected on days 5, 8, 11, and 14 after inoculation was loaded in each of the lanes. The gel was prepared and ran on the same day. The Native-PAGE was run at 4°C. Electrophoresis was done using 30-mA steady current. Electrophoresis was stopped after the dye front reached the bottom of the gel. The gel was removed from the glass plates and placed in a glass dish. The gel was stained with iodine solution. Chewed-up region of the gel can be seen as white in a blue/violet color background. The gel picture was taken on Gel DocTM XR+ BioRad imaging machine.
Bioinformatics Tools Used in Analysis of Macrophomina phaseolina Secretome
Signal P is an online tool that predicts the presence of signal peptide in protein sequences. It is a very useful tool for secretome analysis. Proteins found in LC-ESI-MS/MS of M. phaseolina contained a unique UniProt ID. Sequences of specific proteins were collected from UniProt through their unique UniProt ID. Enzyme sequences were searched for secretory signal peptide in SignalP-5.0 server.1 The tool shows the presence or absence of a secretory signal peptide.
Gene Ontology (GO) enrichment analysis was done using another online tool FungiFun2.2 FungiFun2 was used for functional enrichment analysis for fungal genes and proteins. Molecular function and biological process were determined using this tool. A text file was created using UniProt IDs found in LC-ESI-MS/MS of M. phaseolina. M. phaseolina MS6 strain was selected as the reference species, and GO was selected as classification ontology. Significance level was set at 0.05 (=5.00%). Fisher exact test was selected as significance test, and Benjamini–Hochberg procedure was selected as adjustment method.
Statistical Analysis
All experiments were done in biological triplicates, and the results were expressed as mean ± standard error, for number of experiments n = 3.
Results
Isolation and Detection of Macrophomina phaseolina Secretome
It has been reported that M. phaseolina is seed-borne in nature (Kunwar et al., 1986). It is found both on the seed coat and cotyledons (Reuveni et al., 1983), and it can cause charcoal rot by infecting the roots due to the adherence of microsclerotia to the seed coat during germination (De Mooy and Burke, 1990). Temperature near 30°C and dry conditions make this pathogen prevalent in nature during infection process. Plant pathogenic fungi have the largest proportion of secreted proteins. M. phaseolina also infects wheat seeds (Abass et al., 2021). In order to isolate the secretome of M. phaseolina, the fungal spore suspension was inoculated in a wheat bran matrix maintaining optimum temperature and relative humidity. This solid substrate culture actually mimics the natural infection process where M. phaseolina spores germinate and adhere to wheat bran. M. phaseolina secretome was isolated by extracting the wheat bran matrix using phosphate buffer 5, 8, 11, and 14 days after inoculation. Secretome proteins were concentrated using ammonium sulfate precipitation, dialyzed, and run in SDS-PAGE to visualize the secretome profile. Secretome proteins were run three times in SDS-PAGE. Supplementary Figure 1 shows a representative secretome profile of M. phaseolina run in SDS-PAGE where the equal amount of proteins was loaded in each lane collected from 5-, 8-, 11-, and 14-day post-inoculated matrix. The secretome showed differences in their protein components with distinctly different protein bands of varied molecular mass visible when compared between the gel lanes charged with samples collected from different postinoculation time periods. The secretome profile of M. phaseolina of each lane also varied significantly ranging from low-molecular-weight proteins to high-molecular-weight protein when compared with the molecular weight markers (distributed over a molecular weight range of 180–17 kDa). To identify the whole set of secretory proteins of M. phaseolina, the 5-day post-inoculated secretome was selected for LC-ESI-MS/MS analysis because of its low protease secretion in solid substrate culture at that time period. Figure 1 represents the chromatogram of 5-day secretome of M. phaseolina run in Waters-Xevo-G2-X’S-Q-TOF-TOF.
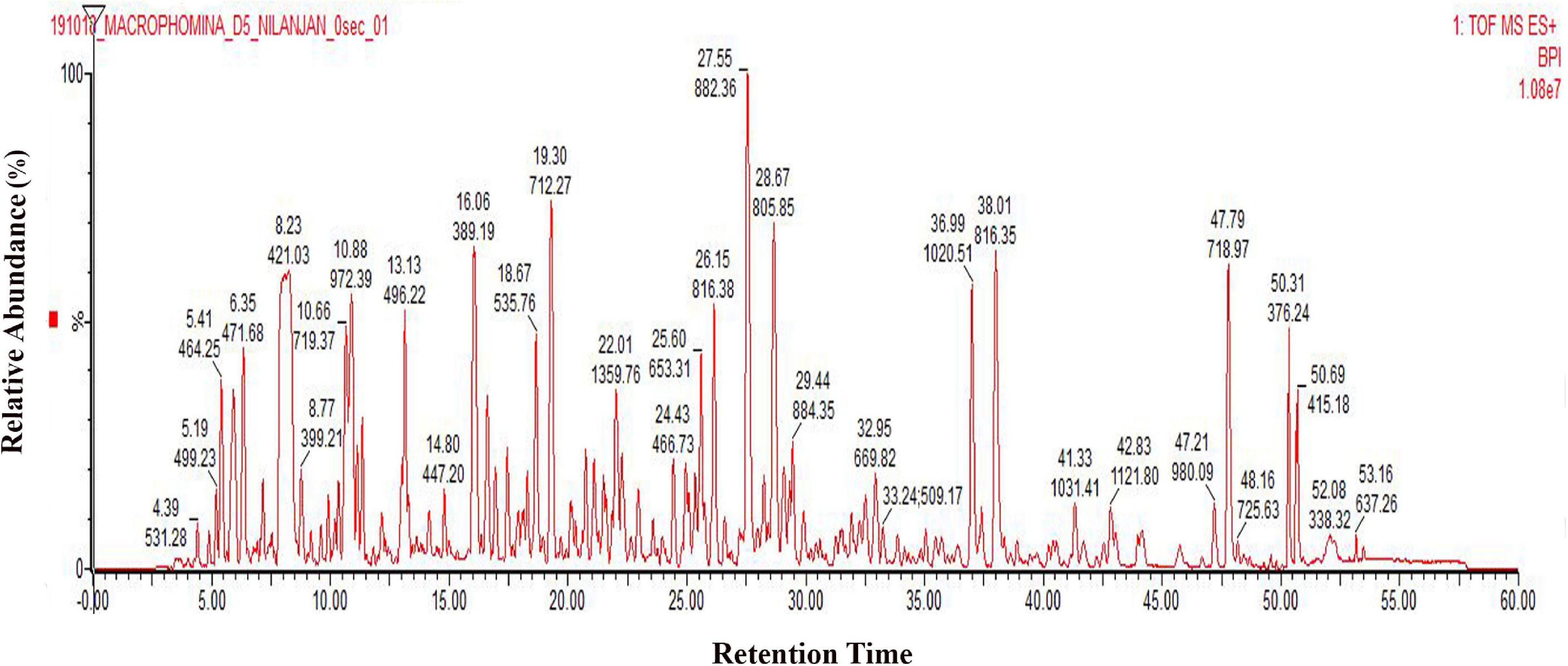
Figure 1. LC-ESI-MS/MS chromatogram of M. phaseolina secretome collected 5 days after inoculation. Chromatogram of secretome was generated using 1 mg/mL protein run in Waters-Xevo-G2-X’S-Q-TOF-TOF. The total MSE scan time was 60 min in which 0–5 min was the equilibration time, and then 5–50 min was the elution of peptides from the column, and 50–60 min was the washing of the column.
Bioinformatics Analysis of Secreted Proteins
Chromatogram profile indicated the presence of a high percentage of peptide abundance, which would become useful for the identification of secretome proteins of M. phaseolina. Whether the chromatogram represents any proteins from wheat bran or not peptide abundance was also searched for Triticum aestivum database. LC-ESI-MS/MS data showed that among 151 identified proteins 117 proteins were matched with M. phaseolina protein database, and the other 34 proteins were matched with T. aestivum protein database (Figure 2A). Supplementary Table 1 represents the list of proteins that matched with T. aestivum. It was reported that the M. phaseolina genome contains 13.07% (1,863) secreted proteins as compared with 7–10% in other plant pathogens (Islam et al., 2012). Among the 117 identified M. phaseolina proteins, 103 proteins were characterized, and 14 proteins were not characterized in the protein database (Figure 2B). Further analysis of the LC-ESI-MS/MS data showed that most of the proteins found in the secretome were enzymes. Among the 103 characterized proteins, 75 proteins were enzymes, and the other 28 proteins were nonenzymatic proteins (Figure 2C). Enzyme classification analysis showed the presence of the following four enzyme classes: (i) isomerases, (ii) hydrolases, (iii) transferases, and (iv) oxidoreductases. Hydrolases are predominantly present (44.66%) in the M. phaseolina secretome (Figure 2D).
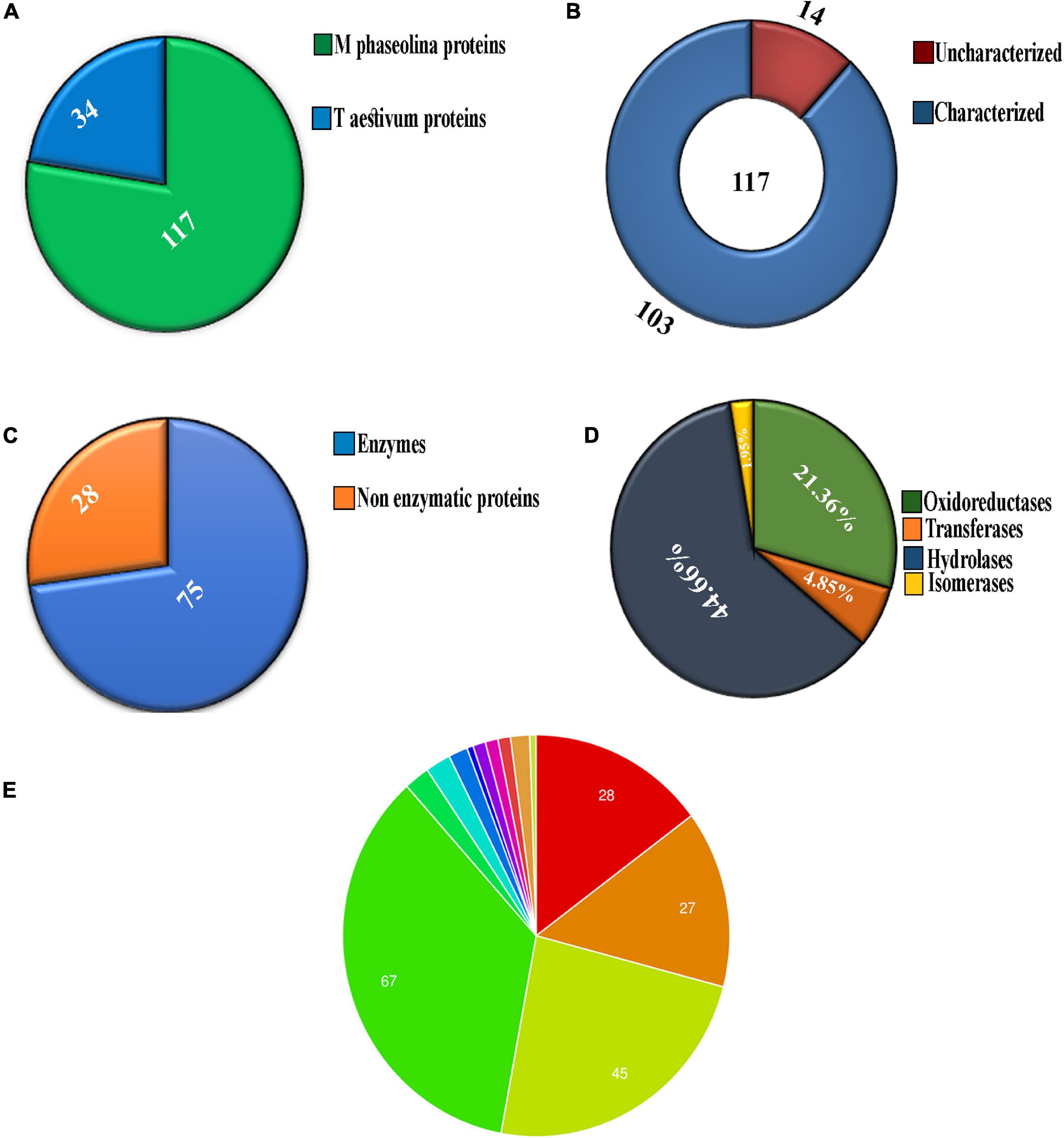
Figure 2. Analysis of secretome obtained from solid-state culture of M. phaseolina grown on wheat bran using LC-ESI-MS/MS. Total 151 proteins were found in LC-ESI-MS/MS analysis. (A) Pie chart represents the number of M. phaseolina and T. aestivum proteins found in LC-ESI-MS/MS. Among 151 proteins, 117 proteins were obtained from M. phaseolina and 34 proteins from T. aestivum. (B) Secretome of M. phaseolina identified by LC-ESI-MS/MS. Among 117 proteins present in the secretome, 14 proteins were found to be uncharacterized and 103 proteins were found to be characterized. (C) Enzymatic and non-enzymatic proteins of M. phaseolina identified by LC-ESI-MS/MS. Among 103 characterized proteins, 28 proteins were nonenzymatic, and the rest 75 proteins were enzymes. (D) Classification of enzymes found in the secretome of M. phaseolina. LC-ESI-MS/MS analysis of the secretome of M. phaseolina identified the following four classes of enzymes: (1) isomerases, (2) hydrolases, (3) transferases, and (4) oxidoreductases. Hydrolases were predominantly present in the aforementioned classification. (E) Molecular function ontology. GO enrichment analysis was performed to get the molecular functions of genes corresponding to the proteins found in the LC-ESI-MS/MS data of M. phaseolina. Most of the genes were found to be involved in different hydrolase activities. Almost 35.5% (67) of the genes corresponding to the proteins are involved in catalytic activity. The involvement of other genes in cellular processes is shown using different color code in the pie chart. All the molecular functions found are as follows:  hydrolase activity, hydrolyzing O-glycosyl compounds,
hydrolase activity, hydrolyzing O-glycosyl compounds,  hydrolase activity, acting on glycosyl bonds,
hydrolase activity, acting on glycosyl bonds,  hydrolase activity,
hydrolase activity,  catalytic activity,
catalytic activity,  endo-1,4-β-xylanase activity,
endo-1,4-β-xylanase activity,  serine-type peptidase activity,
serine-type peptidase activity,  carboxypeptidase activity,
carboxypeptidase activity,  purine ribonucleoside binding,
purine ribonucleoside binding,  glucosidase activity,
glucosidase activity,  polysaccharide binding,
polysaccharide binding,  catalase activity,
catalase activity,  choline dehydrogenase activity,
choline dehydrogenase activity,  purine ribonucleoside triphosphate binding.
purine ribonucleoside triphosphate binding.
Gene ontology annotation analysis classified four main molecular functions, which were as follows: (i) hydrolase activity hydrolyzing O-glycosyl compounds (14.8%), (ii) hydrolase activity acting on glycosyl bond (14.2%), (iii) hydrolase activity (23.8%), and (iv) catalytic activity (35.5%) (Figure 2E). The other molecular functions include endo-1,4-β-xylanase activity, serine-type peptidase activity, carboxypeptidase activity, purine ribonucleoside binding, glucosidase activity, polysaccharide binding, catalase activity, choline dehydrogenase activity, and purine ribonucleoside triphosphate binding.
Identification of Secretome Proteins of Macrophomina phaseolina by LC-ESI-MS/MS Analysis
It was predicted that M. phaseolina contained the lowest number of proteases among ascomycete fungal species (Islam et al., 2012). The list of secretory proteases of M. phaseolina found in the LC-ESI-MS/MS is presented in Table 1. Several types of proteases were present, which include peptidase_S9 domain-containing protein, peptidase S8/S53 subtilisin/kexin/sedolisin, peptidase S28, carboxypeptidase, peptide hydrolase, and peptidase M14 carboxypeptidase A. Identification of secretome proteins of M. phaseolina was based on their conserved domains and function from the genome sequence.
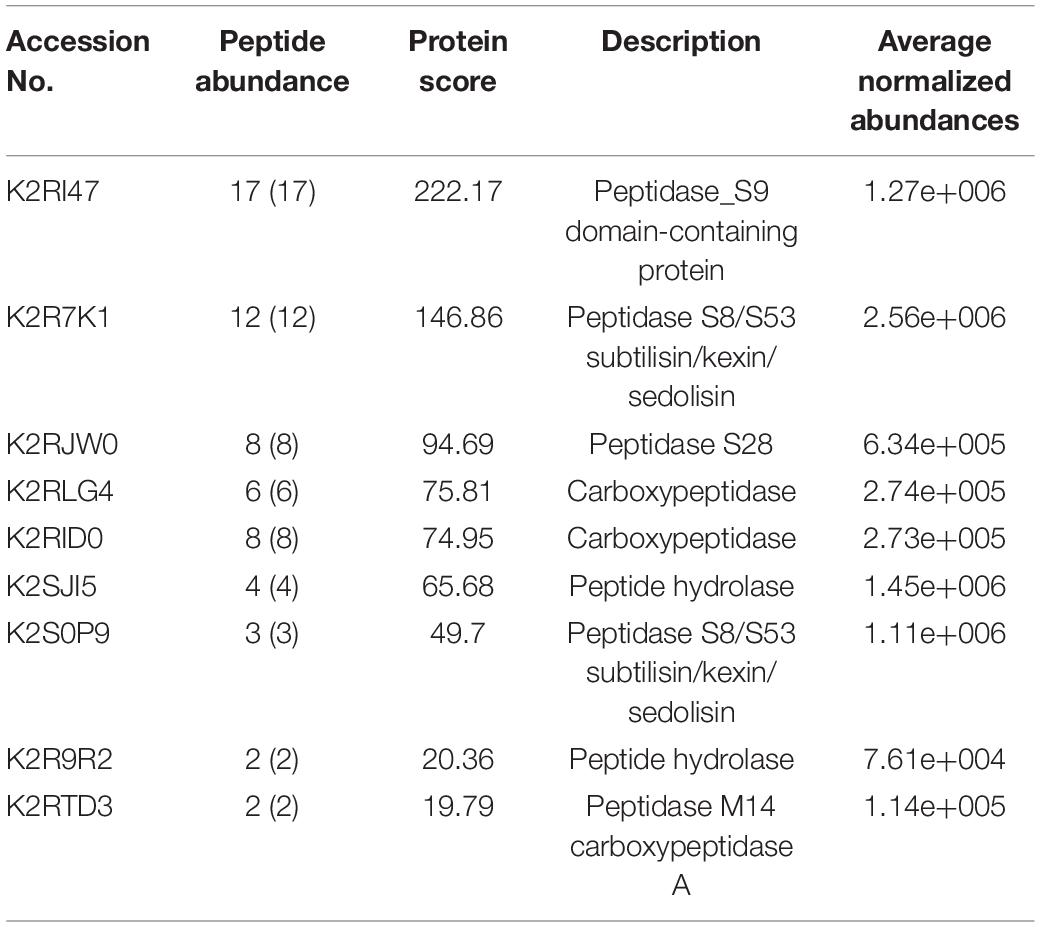
Table 1. List of proteases present in the secretome of M. phaseolina identified using the LC-ESI-MS-MS.
Almost 40% of the enzymes were cell wall–degrading enzymes. Twelve percent of the enzymes were proteases. The rest, 48%, of enzymes do not contribute in degrading host cell wall but have important functions. A significant portion (14.7%) of these enzymes contained a secretory signal sequence. Approximately 33.3% of the enzymes showed no secretory signal sequence analyzed by signal P (Figure 3).
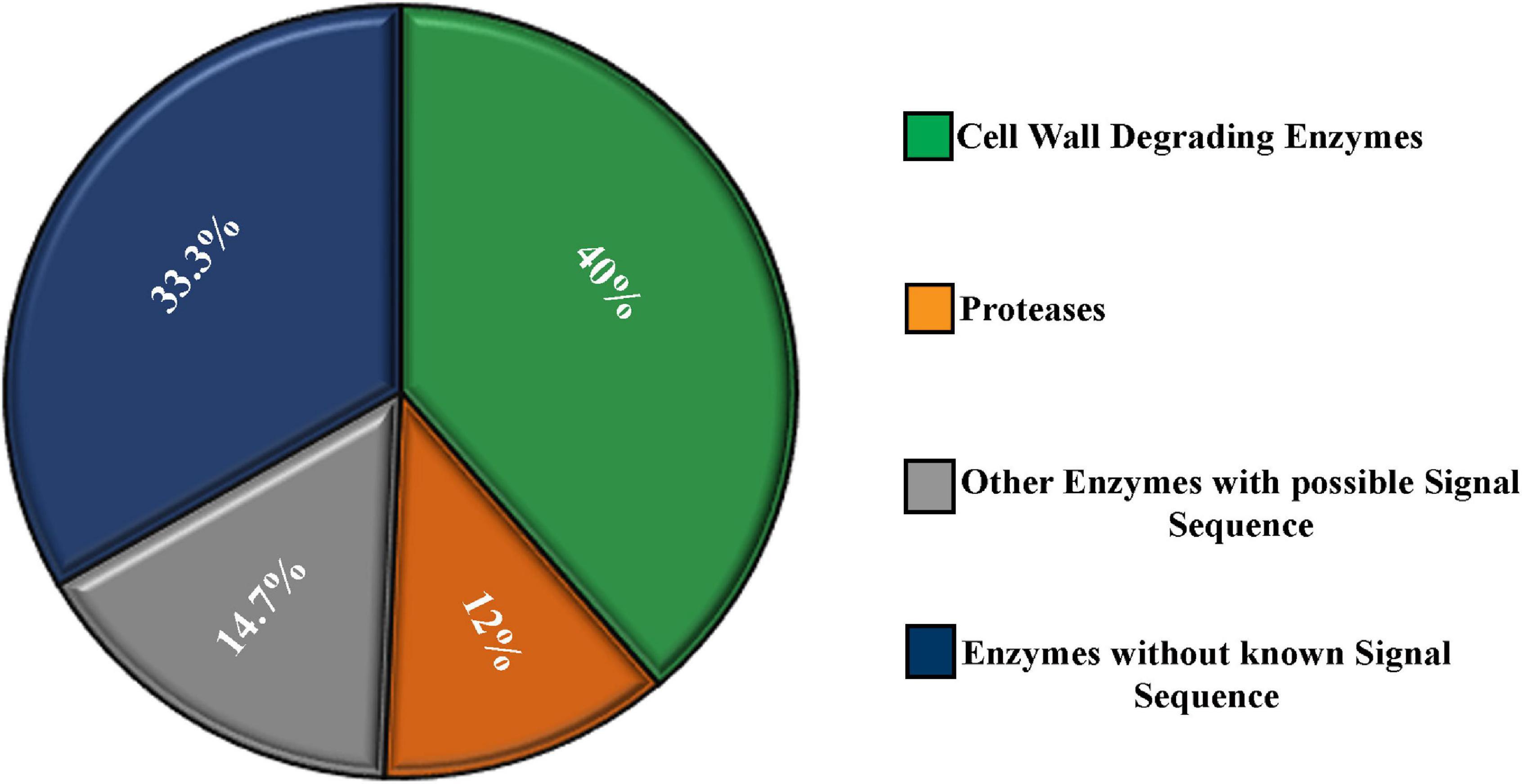
Figure 3. Characterization of the secretome enzymes of M. phaseolina identified by LC-ESI-MS/MS analysis. Most of the proteins found in the secretome were enzymes. Almost 40% of the enzymes were cell wall–degrading enzymes. Twelve percent of the enzymes were proteases. A significant portion (14.7%) of the enzymes contains secretory signal sequence, and the rest 33.3% of the enzymes do not contain secretory signal sequence analyzed by signal P.
Phytopathogenic fungi are known to secrete a cocktail of hydrolytic enzymes (including carbohydrate-active enzymes; CAZymes) for degrading the plant cell wall and penetrating into the host tissue (Gibson et al., 2011). M. phaseolina genome encodes 362 putative CAZymes including 219 GHs, 56 glycosyltransferases, 65 carbohydrate esterases, 6 carbohydrate binding modules, and 16 polysaccharide lyases comprising more than 80 distinct families (Islam et al., 2012).
Detection of Cell Wall–Degrading Enzymes Xylanase, Endoglucanase, and Amylase in the Secretome of Macrophomina phaseolina
The GHs family was highly represented in the M. phaseolina secretome. Specific activity of xylanase was detected in the M. phaseolina secretome at 5-, 8-, 11-, and 14-day postinoculation time period (Figure 4A). The highest xylanase-specific activity (2.33 ± 0.000026 mM of xylose formed per minute per milligram of protein) was found at 5-day postinoculation time period. It was gradually decreased at 8-, 11-, and 14-day postinoculation time period. Xylanase activity was further detected in zymogram (Figure 5A). Zymogram profile of xylanase activity present in the secretome of M. phaseolina was corroborated well with the specific activity determination. Endoglucanase-specific activity of M. phaseolina secretome was highest (366.11 ± 16.1 μM of glucose formed per minute per milligram of protein) at 8-day postinoculation time period (Figure 4B). There was no significant change in endoglucanase-specific activity of M. phaseolina secretome in the 11- and 14-day postinoculation time period compared with the 8-day postinoculation time period. Similar trend was observed in zymogram profile of endoglucanase activity present in the secretome of M. phaseolina (Figure 5B). Amylase-specific activity of M. phaseolina secretome was the highest (6 ± 0.15 μg of starch degraded per minute per microgram of protein) at the 8-day postinoculation time period (Figure 4C). There was a significant decrease in the amylase-specific activity of M. phaseolina secretome at the 14-day time period. Zymogram profile of amylase activity also supported the trend observed in specific activity determination (Figure 5C). Nine families of GHs present in the M. phaseolina secretome were found to be involved in the degradation of carbohydrate complexes. Figure 6 represents the profile of GHs found in the secretome proteins of M. phaseolina identified by LC-ESI-MS/MS.
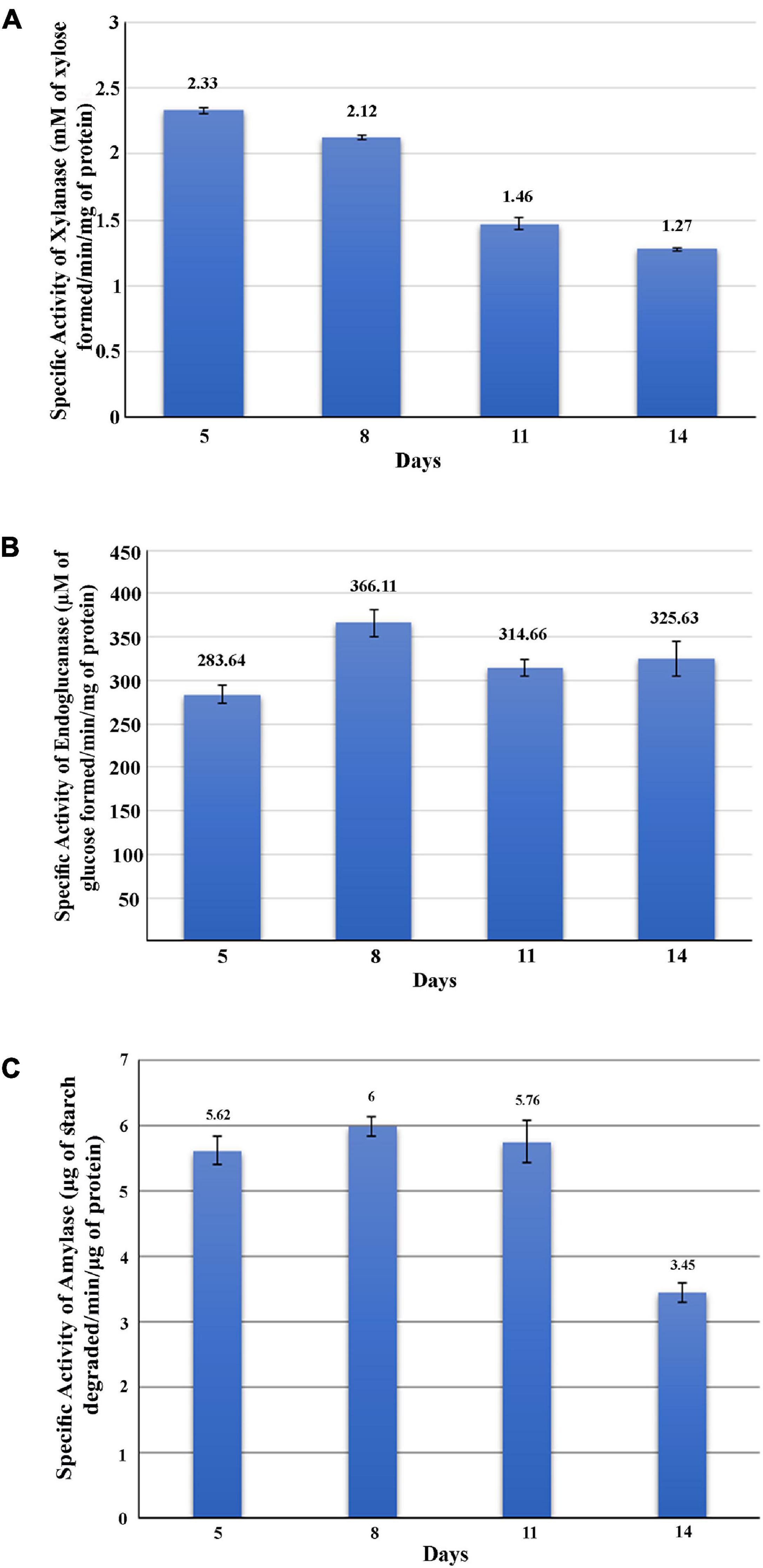
Figure 4. Activity analysis of xylanase, endoglucanase, and amylase enzymes. Secretome of M. phaseolina was collected 5, 8, 11, and 14 days after inoculation in solid-state culture, and xylanase, endoglucanase, and amylase activities were determined using spectrophotometric assay. (A) Determination of xylanase activity. Specific activity of xylanase was expressed as mM of xylose formed per minute per milligram of secretome protein. (B) Determination of endoglucanase activity. Specific activity of endoglucanase was expressed as μM of glucose formed per minute per milligram of secretome protein. (C) Determination of amylase activity. Specific activity of amylase was expressed as micrograms of starch degraded per minute per milligram of secretome protein.
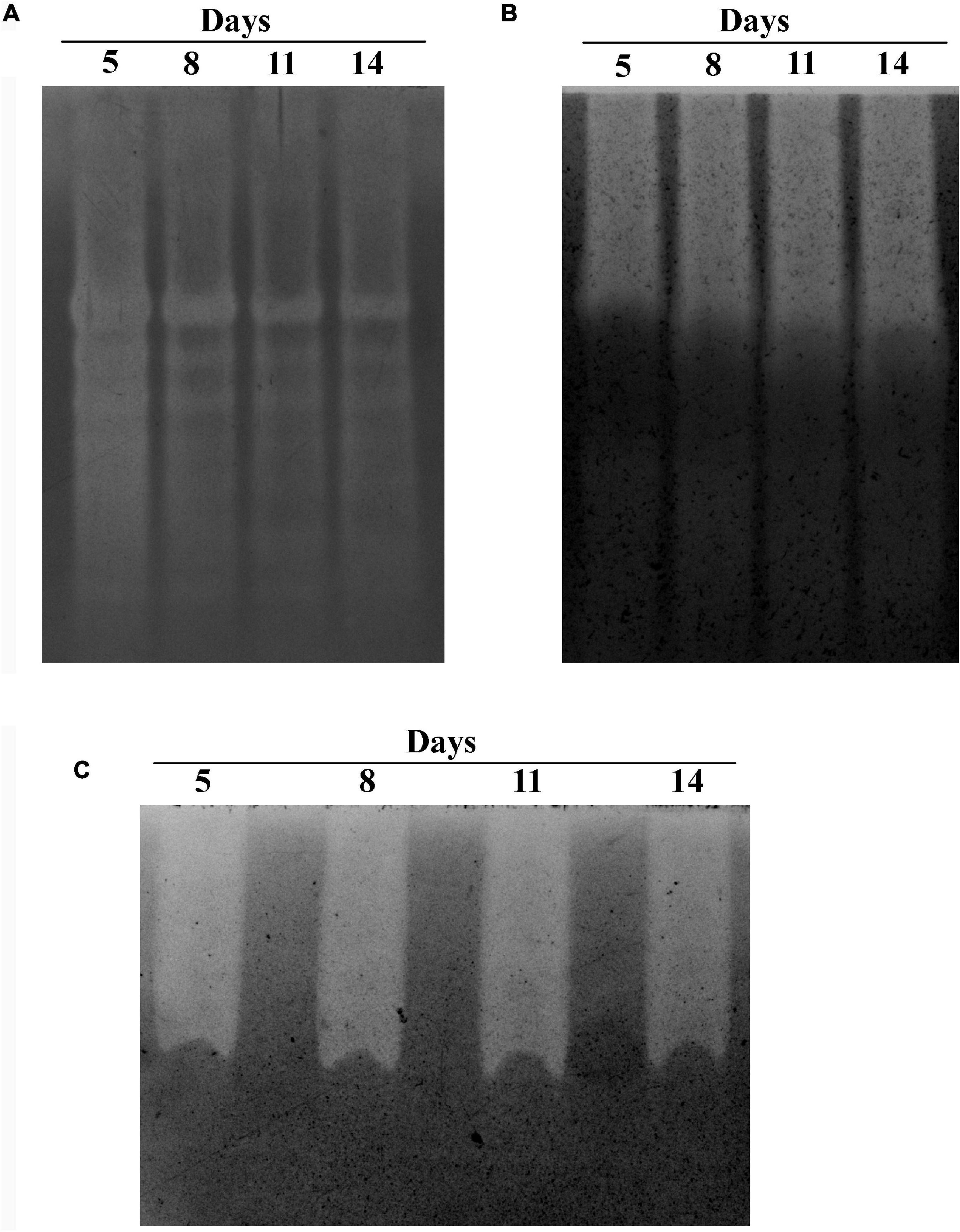
Figure 5. Zymogram analysis of secretory xylanase, endoglucanase, and amylase enzymes. Secretome of M. phaseolina was collected 5, 8, 11, and 14 days after inoculation in solid-state culture, and zymogram was studied for xylanase, endoglucanase, and amylase. (A) Zymogram analysis of xylanase. The resolving gel was casted with 0.1% birchwood xylan. The gel was stained with 0.1% Congo red and destained with 1 M NaCl solution. The decolorized region in the gel picture indicated the presence of xylanase. (B) Zymogram analysis of endoglucanase. The resolving gel was casted with 0.1% CM cellulose. The gel was stained with 0.1% Congo red and destained with 1 M NaCl solution. The decolorized region in the gel picture indicated the presence of endoglucanase. (C) Zymogram analysis of amylase. The resolving gel was casted in with 0.1% starch. The gel is stained with iodine solution. The decolorized region in the gel picture indicated the presence of amylase.
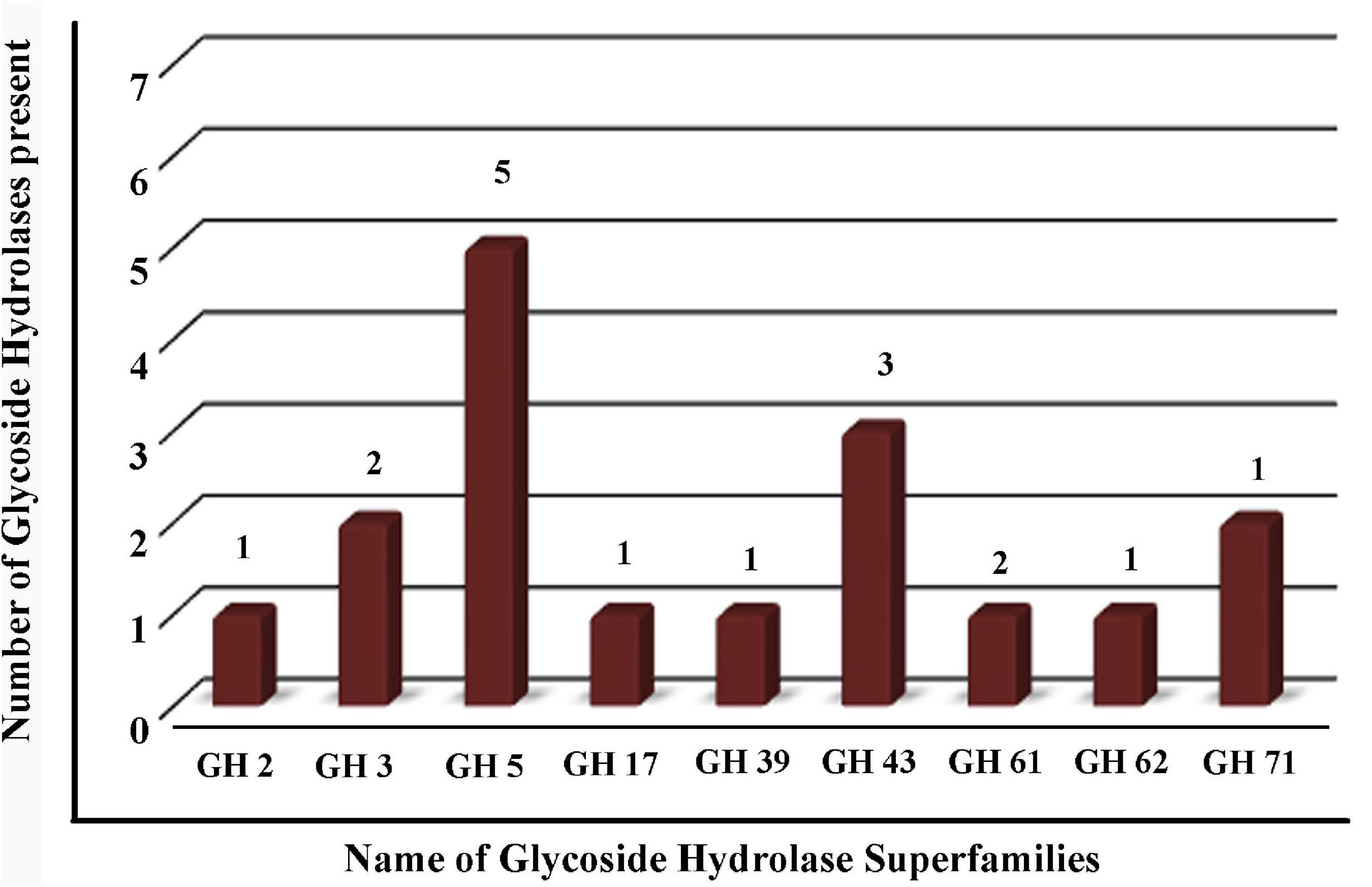
Figure 6. Profile of glycoside hydrolases (GHs) found in the secretome proteins of M. phaseolina identified by LC-ESI-MS/MS. Bar graph shows the distribution of 17 GHs identified by LC-ESI-MS/MS. Nine different types of GHs were found in multiple numbers in the secretome proteins of M. phaseolina.
Identification of Secretome Proteins in the LC-ESI-MS/MS Based on the Presence or Absence of Secretory Signal Sequence
Table 2 represents list of enzymes without having known signal sequence found in the LC-ESI-MS/MS protein identification. Table 3 represents the list of enzymes with signal sequence as identified by signal P found in the LC-ESI-MS/MS data of the secretome proteins of M. phaseolina. Figures 7A,B show the molecular function ontology of the genes corresponding to their proteins presented in Tables 2, 3. Most of the proteins showed catalytic activity, and they belong to the group of virulence factors. The number of genes involved in the molecular function has been shown within the color code. A diverse group of enzymes was found in the secretome starting from metabolic enzyme, redox active enzymes, to genotoxic compound detoxifying enzymes. Proteins that provide protection against reactive oxygen species (ROS)–based plant defenses may be responsible for stronger pathogenicity of M. phaseolina. ROS generation is the earliest response of plant toward the invading pathogen. Production of ROS alters the host cells’ redox status and activates various plant defense responses, including production of pathogenesis related proteins, phytoalexins, and signaling molecules (Huang et al., 2019). This creates an inhospitable situation for the pathogens, and thus pathogens must degrade or neutralize such ROS in order to overcome the host defense responses. A recent study published from our laboratory showed the presence of catalase, catalase peroxidase, and superoxide dismutase in M. phaseolina secretome. In that study, proteins such as superoxide dismutase [Cu-Zn] and catalase–peroxidase were found without having known signal sequence. We also identified a catalase protein with signal sequence as identified by signal P found in the LC–ESI-MS/MS data of the secretome proteins of M. phaseolina (Sinha et al., 2021). These proteins play an essential role in protecting the pathogen against the host-derived ROS and therefore overcoming the host defenses and creating a successful infection. Antioxidant proteins such as thioredoxins and glutathione S-transferase were also identified in the secretome of M. phaseolina. Identified enzymes such as glucose–methanol–choline (GMC) oxidoreductase, aldo/keto reductase (AKR), formate dehydrogenase, alcohol dehydrogenase (ADH) superfamily zinc-containing, malate dehydrogenase, peptidyl-prolyl-cis-trans isomerase, AKR, homogentisate 1 2-dioxygenase, lipase GDSL, inorganic pyrophosphatase, nucleoside diphosphate kinase (NDK), citrate synthase, transaldolase, triosephosphate isomerase, short-chain dehydrogenase/reductase, peptidylprolyl isomerase, dienelactone hydrolase, and epoxide hydrolase are also known virulence factors present in many plant pathogenic fungi.
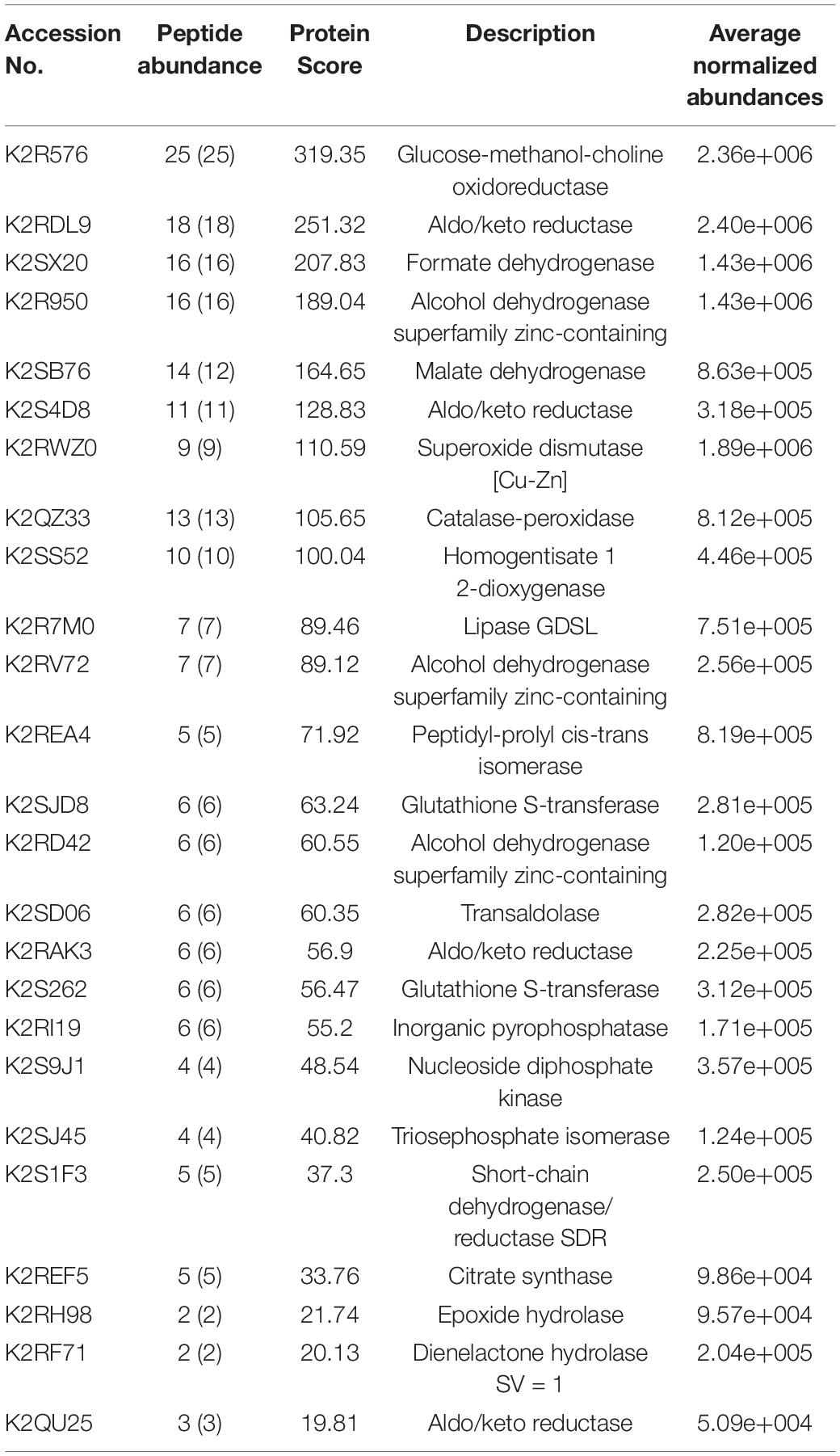
Table 2. List of enzymes without having known signal sequence as identified by Signal P found in the LC-ESI-MS-MS of M. phaseolina secretome.
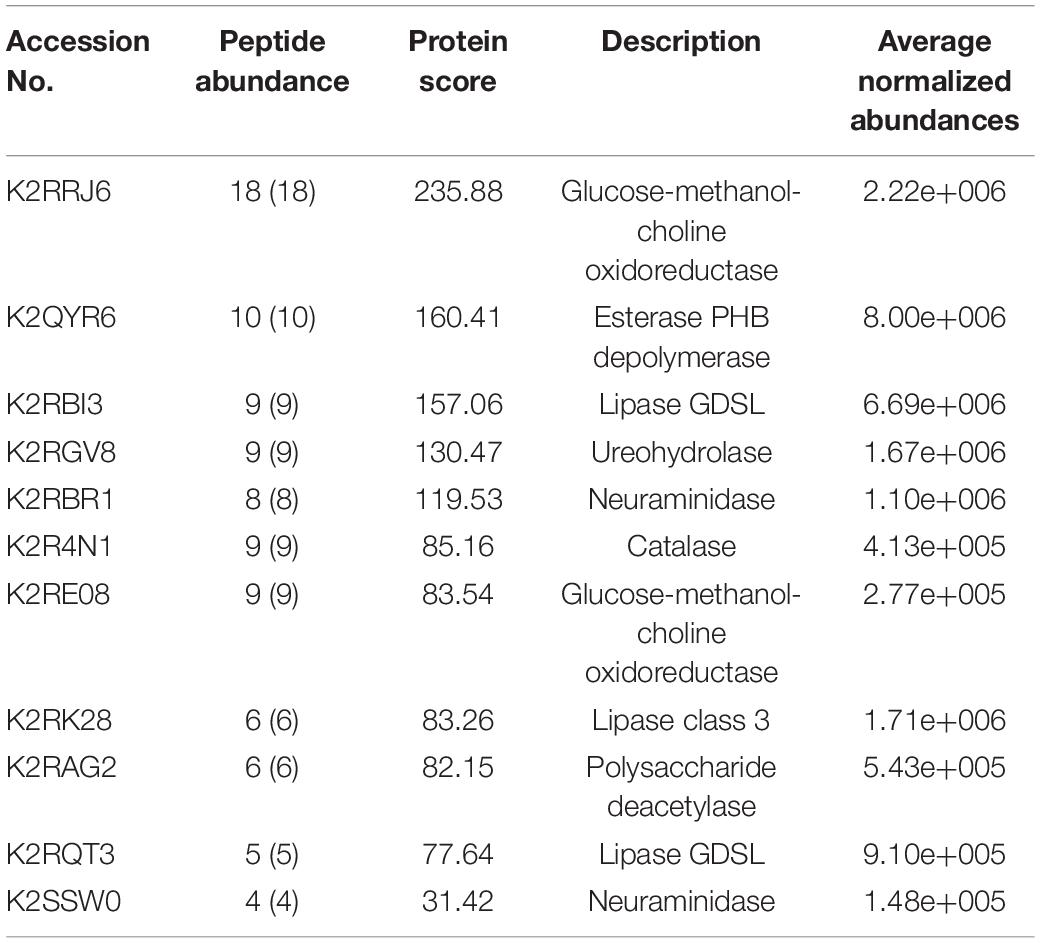
Table 3. List of enzymes with signal sequence as identified by Signal P found in the LC–ESI-MS-MS data of M. phaseolina secretome.

Figure 7. Molecular function ontology of enzymes analyzed by signal P found in LC-ESI-MS/MS of M. phaseolina secretome. The enzymes other than the cell wall–degrading enzymes and proteases found in LC-ESI-MS/MS were first categorized based on the presence of the secretory signal peptide by signal P online tool. GO enrichment analysis was done using the UniProt IDs of the enzymes. (A) Molecular function of the genes corresponding to the enzymes that contain a signal sequence found in the LC-ESI-MS/MS. Almost 37.5% of the genes corresponding to the enzymes are involved in hydrolase activity. All the molecular functions found are as follows:  catalytic activity,
catalytic activity,  oxidoreductase activity,
oxidoreductase activity,  inorganic diphosphatase activity,
inorganic diphosphatase activity,  nucleoside diphosphate kinase activity,
nucleoside diphosphate kinase activity,  glutathione transferase activity,
glutathione transferase activity,  citrate (Si)-synthase activity,
citrate (Si)-synthase activity,  L-malate dehydrogenase activity,
L-malate dehydrogenase activity,  sedoheptulose-7-phosphate:D-glycreraldehyde-3-phosphate glyceronetransferase activity,
sedoheptulose-7-phosphate:D-glycreraldehyde-3-phosphate glyceronetransferase activity,  phosphatidylininositol-3-phosphate binding,
phosphatidylininositol-3-phosphate binding,  triose-phosphate isomerase activity,
triose-phosphate isomerase activity,  phosphatidylinositol phosphate binding,
phosphatidylinositol phosphate binding,  antioxidant activity,
antioxidant activity,  homogentisate 1,2-dioxygenase activity,
homogentisate 1,2-dioxygenase activity,  isomerase activity,
isomerase activity,  oxidoreductase activity, acting on CH-OH group of donors,
oxidoreductase activity, acting on CH-OH group of donors,  malate dehydrogenase activity,
malate dehydrogenase activity,  oxidoreductase activity, acting on superoxide radicals as acceptor,
oxidoreductase activity, acting on superoxide radicals as acceptor,  superoxide dismutase activity,
superoxide dismutase activity,  transferase activity, transferring aldehyde or ketonic groups,
transferase activity, transferring aldehyde or ketonic groups,  pyrophosphatase activity,
pyrophosphatase activity,  nucleobase-containing compound kinase activity. (B) Molecular function of the genes corresponding to the enzymes that do not contain a signal sequence found in the LC-ESI-MS/MS. Almost 36.9% of the genes corresponding to the enzymes are involved in catalytic activity, and 22.8% of the genes corresponding to the enzymes are involved in oxidoreductase activity. All the molecular functions found are as follows:
nucleobase-containing compound kinase activity. (B) Molecular function of the genes corresponding to the enzymes that do not contain a signal sequence found in the LC-ESI-MS/MS. Almost 36.9% of the genes corresponding to the enzymes are involved in catalytic activity, and 22.8% of the genes corresponding to the enzymes are involved in oxidoreductase activity. All the molecular functions found are as follows:  choline dehydrogenase activity,
choline dehydrogenase activity,  hydrolase activity,
hydrolase activity,  hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds,
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds,  catalase activity,
catalase activity,  hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines,
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines,  flavin adenine dinucleotide binding,
flavin adenine dinucleotide binding,  oxidoreductase activity, acting on CH-OH group of donors.
oxidoreductase activity, acting on CH-OH group of donors.
Plant cell wall–degrading enzymes require the assistance of carbohydrate esterases (Aspeborg et al., 2012) to deacetylate the substituted saccharides (esters or amides) of celluloses (Biely, 2012). Our LC-ESI-MS/MS data showed the presence of polysaccharide deacetylase. Secretome data showed the presence of the GHs, which include choline dehydrogenase activity, hydrolase activity, β-galactosidase activity, catalase activity, hydrolase activity acting on carbon–nitrogen bond but not on peptide bonds, and oxidoreductase activity acting on CH-OH group of donors (Figure 7B).
Discussion
Necrotrophic fungal pathogens have evolved various mechanisms to kill their host quickly to feed themselves and complete their lifecycle. They are generally resistant to a hypersensitive response, indicating that they possess a repertoire of effectors to counteract host-generated oxidative stress and to induce host cell death. Among the necrotrophic fungal pathogens, M. phaseolina is one important soil-borne necrotrophic pathogen that infects more than 500 plant species around the world (Wyllie, 1998). Thus, it is very important to identify the M. phaseolina secretome, which is used as tool box to destroy the host plant. The term fungal secretome now refers to a set of secretory proteins and enzymes with or without signal peptide, proteins from exosomes (Meijer et al., 2014; Vincent et al., 2019). Studies on the whole-genome sequence of M. phaseolina predicted that this phytopathogen has an abundance of secreted oxidases, peroxidases, and hydrolytic enzymes for degrading cell wall polysaccharides and lignocelluloses to penetrate into the host tissue (Islam et al., 2012). For the identification of M. phaseolina secretome, we developed a successful and reproducible bioprocess in this study. By virtue of its secreted enzymes, M. phaseolina are potent decomposers of plant cell walls. Plant cell walls are composed mainly of cellulose, lignin, and hemicellulose. This composite is often referred to as lignocelluloses. As the cell wall imparts a significant barrier to pathogen entrance, necrotrophs, especially M. phaseolina, have evolved mechanisms to overcome this barrier. Our secretome analysis showed that M. phaseolina secretes numerous cell wall–degrading enzymes to disrupt the cell and for absorptive nutrition. Similar observation was found in necrotrophic fungi Botrytis cinerea, which penetrate the host cell wall by releasing the cell wall–degrading enzymes rather than by physical force (Choquer et al., 2007). GHs are the most diverse group of enzymes used by microbes in the degradation of lignocelluloses (Murphy et al., 2011). It is evident from our study that M. phaseolina also relies on producing different family of GH when it was grown in wheat bran matrix. Plant cell wall–degrading enzymes require the assistance of carbohydrate esterases (Aspeborg et al., 2012) to deacetylate the substituted saccharides (esters or amides) of celluloses (Yagminas et al., 1990). Our LC-ESI-MS/MS data showed the presence of polysaccharide deacetylase. β-Glucosidase has been identified as a virulence factor in other microorganisms: it is directly involved in the virulence of the phytopathogenic Pyrenophora tritici-repentis (anamorph: Drechslera tritici-repentis), the necrotrophic fungus responsible for tan spot (Fu et al., 2013). The presence of β-glucosidase in M. phaseolina secretome is in agreement with the previously published reports by Reuveni et al. (2007). Therefore, GHs appeared to be the putative virulence factors for M. phaseolina. A short-chain dehydrogenase/reductase was identified in M. phaseolina secretome grown in wheat bran matrix. It has been reported that a short-chain dehydrogenase/reductase gene is required for infection-related development and pathogenicity in Magnaporthe oryzae, a phytopathogenic fungus (Kwon et al., 2010).
In this study, we identified a list of proteases in solid-state culture of M. phaseolina. During encounter with the host plant, pathogenic fungi produce various cell wall–degrading enzymes and proteases to fragment the plant cell wall polymers and thereby facilitate penetration into the host cells (Doi and Kosugi, 2004; Kubicek et al., 2014; Jashni et al., 2015; Quoc and Chau, 2017). Thus, these M. phaseolina proteases may have certain role in cell wall degradation and as putative virulence factors during plant–pathogen interactions.
We identified neuraminidase in the M. phaseolina secretome collected from wheat bran matrix. Neuraminidase is an exoglycosidase that cleaves glycoconjugates, releasing the terminal sialic acid residues (Warwas et al., 2010). The identification of neuraminidase in M. phaseolina secretome corroborated well with the previous finding where it was shown that M. phaseolina recognizes and infects host plants via a sialic acid–binding lectin (Bhowal et al., 2005, 2006). These enzymes are involved in the virulence of several microorganisms (Jermyn and Boyd, 2002; Zhou et al., 2009; Warwas et al., 2010).
In the present study, GMC oxidoreductase was identified in the M. phaseolina secretome collected from wheat bran matrix. GMC oxidoreductases are flavoenzymes that catalyze the electron transfer between molecules by utilizing diverse substrates and are exclusively found in members of Basidiomycota and Pezizomycotina. In phytopathogenic fungi, these enzymes play important role in various processes, including browning, pigmentation, pathogen defense, virulence mechanisms, and melanin production. The identified GMC oxidoreductase in M. phaseolina secretome may play a dual role. First, they may defend the fungal pathogens from oxidative stress by acting as scavenger of ROS generated by the host plant. Second, they may prevent the activation of host defense mechanisms downstream to ROS production as found in Tilletia indica, which is smut fungus that causes Karnal bunt (KB) disease in wheat (Pandey et al., 2018). AKR is found to play an adaptive role in another necrotrophic pathogen Beauveria bassiana (Wang et al., 2018).
In this study, thioredoxin was identified in M. phaseolina secretome collected from wheat bran matrix. Thioredoxins are small disulfide containing redox proteins. They are protein disulfide oxidoreductases that regulate the cellular redox status and defend the fungal pathogen against ROS accumulation (Li et al., 2016). Thioredoxins are highly conserved among living organisms. In chestnut blight pathogen, corn smut fungus Ustilago maydis is found to secrete a pathogenicity-related thioredoxin (Muller et al., 2008). In Alternaria brassicicola, which are the causative agent of brassica dark leaf spot, thioredoxin gene expression has been detected following exposure to defense metabolites (Kang et al., 2020). That is why thioredoxin has been classified as stress, oxidative burst, and defense gene in causal agent of brassica dark leaf spot.
Triose phosphate isomerase was identified in the secretome of M. phaseolina grown in wheat bran matrix. It catalyzes the reversible conversion of dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate in glycolysis. In pathogenic fungus Paracoccidioides brasiliensis, triose phosphate isomerase has been found to play a crucial role in pathogen adherence to host and involved in host cell invasion (Pereira et al., 2007). It was also found in highly virulent T. indica fungus that causes KB disease in wheat (Pandey et al., 2018). Triose phosphate isomerase may play similar function in M. phaseolina during pathogenesis.
Plant pathogenic fungi secrete several lipases that hydrolyze the carboxyl ester bonds from fatty acid polymers. Fungal lipases are known to be involved in fungal penetration through the plant barriers such as cuticle and waxes (Serrano et al., 2014). The potential role of secreted lipase as fungal virulence factor has been identified first in necrotrophic fungus B. cinerea, a causal agent of gray mold disease of grapes (Reis et al., 2005). In U. maydis, secreted lipase activity has been found to promote the transition of the fungus to pathogenic filamentous state (Klose et al., 2004). We identified the presence of secreted lipase in the M. phaseolina secretome, which was grown in wheat bran matrix. Thus, it seems that secreted lipases may play a crucial role in host–pathogen interactions and are involved in fungal pathogenesis. Esterase PHB depolymerase is involved in lipid degradation. It is involved in the hydrolysis of carboxylic ester. PHB depolymerase was identified in the secretome of M. phaseolina grown in wheat bran matrix. It has also been reported in the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola (Morais do Amaral et al., 2012).
Feruloyl esterases (FAEs, EC 3.1.1.73) are auxiliary enzymes in the degradation of lignocellulosic complexes of the cell wall, through the hydrolysis of the ester bonds that join hydroxycinnamic acids (e.g., ferulic acid) and lignin–carbohydrate complexes. We identified feruloyl esterase in the secretome of M. phaseolina grown in wheat bran matrix. Feruloyl esterase has been identified in the secretome of Alternaria alternata, which is capable of damaging a wide spectrum of plant hosts (Garcia-Calvo et al., 2018). Epoxide hydrolase was also identified in secretome of M. phaseolina grown in wheat bran matrix. The function of epoxide hydrolase (EC 3.3.2.3) is to catalyze the hydrolysis of epoxides or arene oxides to their corresponding diols by the addition of water (Pinot et al., 1997). Little is known about epoxide hydrolases from filamentous fungi. An epoxide hydrolase was found to be induced by Fusarium solani pisi by a cutin extract of plants (Kolattukudy and Brown, 1975). This enzyme is apparently related to the ability of the mycelium to infect some plants (Morisseau et al., 1999). The AKRs are a superfamily of enzymes with diverse functions in the reduction of aldehydes and ketones. AKR was identified in the secretome of M. phaseolina grown in wheat bran matrix. This enzyme has been reported in B. bassiana, a necrotrophic fungus that has evolved the ability to infect and parasitize insect, which is also a saprophyte found in soil, and can form endophytic relationships with various plants (Behie and Bidochka, 2014). M. phaseolina secretome was found to contain formaldehyde dehydrogenase. It has been reported that formaldehyde dehydrogenase was also expressed in Sclerotinia sclerotiorum Fdh1 in fungal pathogenesis (Zhu et al., 2019). ADHs are a group of oxidoreductases that occur in many organisms facilitating the conversion between alcohols and aldehydes with the reduction of NAD+. Alcohol dehydrogenase gene (ADH1) disruption in plant pathogenic fungus Fusarium oxysporum impaired growth under hypoxic conditions, diminished production of ethanol, and affected the fungal disease development in tomato plants (Corrales Escobosa et al., 2011). ADH could be induced by hypoxic condition and influence fungal growth and pathogenesis in Aspergillus fumigatus (Grahl et al., 2011). Interestingly, ADH was also identified in M. phaseolina secretome. ADHs may play a similar role in M. phaseolina during growth and pathogenesis. We identified malate dehydrogenase enzyme in the secretome of M. phaseolina. Malate dehydrogenase enzyme catalyzes the reversible conversion of oxalacetate and malate. In B. cinerea, oxalacetate has been described as a pathogenicity factor (Lyon et al., 2004), as the secretion of oxaloacetic acid may create an acidic environment that facilitates the pathogenic activity of the fungus. Homogentisate 1 2-dioxygenase was also identified in M. phaseolina secretome. Homogentisate 1,2-dioxygenase catalyzes the conversion of homogenistic acid into maleylacetoacetic acid in the tyrosine metabolism pathway. Tyrosine is assimilated via a conserved catabolic pathway that provides the fungus with both nitrogen, from a transamination reaction with α-ketoglutarate to produce glutamate, and carbon via production of fumarate and acetoacetate, which is used to generate acetyl-CoA that can then feed into the TCA cycle. Tyrosine is an important precursor in the formation of two different types of melanin: DOPA melanin via the metabolism of tyrosine to form DOPA and pyomelanin through the oxidation and polymerization of homogentisate during tyrosine catabolism (Fernandez-Canon and Penalva, 1995; Schmaler-Ripcke et al., 2009). Pyomelanin has been reported to protect Penicillium marneffei against oxidative stress. The fundamental biological function of NDK is to catalyze the reversible exchange of the -phosphate between nucleoside triphosphate (NTP) and nucleoside diphosphate (NDP). NDK was identified in M. phaseolina secretome collected from wheat bran matrix. NDK family proteins are ubiquitous enzymes that exchange the -phosphate between NTP and NDP, preserving NTPs in the cell. NDK has been known to play an important role in spore development, sclerotia production, and pathogenicity on seeds In Aspergillus flavus. Similar function of this enzyme can be expected in M. phaseolina. The usefulness of the present experimental setup is to get high-yielding secretory proteins from M. phaseolina when it was grown in wheat bran matrix. Moreover, downstream processing of the secretome requires less purification steps. It has also been reported that M. phaseolina also infects wheat seeds (Abass et al., 2021). Thus, the putative virulence factors that were identified in the present study may help to deduce the infection mechanisms involved in other susceptible host plants. It is very difficult to get the pure fungal secretome from the mixed system of the infected plant tissues. That is why apoplasts have been used in secretome analysis in a few cases. However, solid-state culture for M. phaseolina can provide similar pathological settings when seed cotyledons are infected. Although few wheat proteins have been identified in this secretome, that is small in numbers in the context of whole secretome.
Conclusion
Although during the last five decades, extensive work has been made by the researchers in the areas of etiology, epidemiology, biology, and biocontrol of the ascomycete fungus M. phaseolina, still control of charcoal rot diseases is difficult. Based on the whole-genome sequence, it is possible to examine secretome using analytical and computational tools. For predicted proteins, the accuracy of a signal peptide prediction heavily relies on an accurate gene model, which provides a correct N-terminal end sequence of the encoded protein. However, some proteins will not actually be secreted, even with a correctly predicted signal peptide, for instance, because they are resident endoplasmic reticulum proteins. On the other hand, the presence of intracellular proteins might be explained by exosomes and microvesicles, which actually add complexity to secretome studies. That is why the proteomic analysis of secretome of pathogens may identify the actual process or the involvement of proteins related to pathogenicity or putative virulence factors. ROS seems to be induced following M. phaseolina infection in seed coats of wheat as several proteins involved in oxidative stress defense such as superoxide dismutase, catalase, and thioredoxins were identified in the secretome analysis.
Data Availability Statement
The data presented in the study are deposited in the ProteomeXchange repository, accession number PXD032749.
Author Contributions
NS and SP performed the experiments and analyzed data. NS and SG wrote the manuscript with the contribution of SP. All authors conceived the research and approved the submitted version.
Funding
This research was supported by West Bengal Department of Biotechnology (WB-DBT), India [Grant No. 573 (sanc)/BT/RD-36/2015].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge the support of Bose Institute LC-MS facility and Souvik Roy for his excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.847832/full#supplementary-material
Supplementary Figure 1 | Secretome profile of M. phaseolina grown in solid state culture using wheat bran as substrate. Secretome of M. phaseolina was collected after 5, 8, 11 and 14 days post inoculation in solid state culture, and 50 μg extracted protein was run in SDS-PAGE. Molecular weight markers were also run in the same gel.
Footnotes
References
Abass, M. H., Madhi, Q. H., and Matrood, A. A. A. (2021). Identity and prevalence of wheat damping-off fungal pathogens in different fields of Basrah and Maysan provinces. Bull. Natl. Res. Cent. 45, 51.
Aly, A. A., Abdel-Sattar, M. A., Omar, M. R., and Abd-Elsalam, K. A. (2007). Differential antagonism of Trichoderma sp. against Macrophomina phaseolina. J. Plant Protect. Res. 47, 91–102.
Andrades, A., and Contreras, L. M. (2017). Amylase zymography. Methods Mol. Biol. 1626, 301–308. doi: 10.1007/978-1-4939-7111-4_29
Aspeborg, H., Coutinho, P. M., Wang, Y., Brumer, H. III, and Henrissat, B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. doi: 10.1186/1471-2148-12-186
Behie, S. W., and Bidochka, M. J. (2014). Ubiquity of insect-derived nitrogen transfer to plants by endophytic insect-pathogenic fungi: an additional branch of the soil nitrogen cycle. Appl. Environ. Microbiol. 80, 1553–1560. doi: 10.1128/AEM.03338-13
Bhowal, J., Ghosh, S., Guha, A., and Chatterjee, B. (2006). Infection of jute seedlings by the phytopathogenic fungus Macrophomina phaseolina mediated by endogenous lectin. Res. J. Microbiol. 1, 51–60. doi: 10.3923/jm.2006.51.60
Bhowal, J., Guha, A. K., and Chatterjee, B. P. (2005). Purification and molecular characterization of a sialic acid specific lectin from the phytopathogenic fungus Macrophomina phaseolina. Carbohydr. Res. 340, 1973–1982. doi: 10.1016/j.carres.2005.06.009
Biely, P. (2012). Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 30, 1575–1588. doi: 10.1016/j.biotechadv.2012.04.010
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Choquer, M., Fournier, E., Kunz, C., Levis, C., Pradier, J. M., Simon, A., et al. (2007). Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. doi: 10.1111/j.1574-6968.2007.00930.x
Cochran, B., Lunday, D., and Miskevich, F. (2008). Kinetic analysis of amylase using quantitative Benedict’s and iodine starch reagents. J. Chem. Educ. 85:401. doi: 10.1021/ed085p401
Coral, G., Arikan, B., Naldi, M. N., and Venmez, H. G. (2002). Some properties of crude carboxymethyl cellulase of Aspergillus niger Z10 wild-type strain. Turk. J. Biol. 26, 209–213.
Corrales Escobosa, A. R., Rangel Porras, R. A., Meza Carmen, V., Gonzalez Hernandez, G. A., Torres Guzman, J. C., Wrobel, K., et al. (2011). Fusarium oxysporum Adh1 has dual fermentative and oxidative functions and is involved in fungal virulence in tomato plants. Fungal. Genet. Biol. 48, 886–895. doi: 10.1016/j.fgb.2011.06.004
Crous, P. W., Slippers, B., Wingfield, M. J., Rheeder, J., Marasas, W. F., Philips, A. J., et al. (2006). Phylogenetic lineages in the Botryosphaeriaceae. Stud. Mycol. 55, 235–253. doi: 10.3114/sim.55.1.235
De Mooy, C. J., and Burke, D. W. (1990). External infection mechanism of hypocotyls and cotyledons of cowpea seedling by Macrophomina phaseolina. Plant Dis. 74:720. doi: 10.1094/pd-74-0720d
De, B. K., Chattopadhya, S. B., and Arjunan, G. (1992). Effect of potash on stem rot diseases of jute caused by Macrophomina phaseolina. J. Mycopathol. Res. 30, 51–55.
Dhingra, O. D., and Sinclair, J. B. (1973). Variation among isolates of Macrophomina phaseolina (Rhizoctonia bataticola) from different regions. J. Phytopathol. 76, 200–204. doi: 10.1111/j.1439-0434.1973.tb02665.x
Do Vale, L. H., Gomez-Mendoza, D. P., Kim, M. S., Pandey, A., Ricart, C. A., Ximenes, F. F. E., et al. (2012). Secretome analysis of the fungus Trichoderma harzianum grown on cellulose. Proteomics 12, 2716–2728. doi: 10.1002/pmic.201200063
Doi, R. H., and Kosugi, A. (2004). Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2, 541–551. doi: 10.1038/nrmicro925
Dutta, T., Sahoo, R., Sengupta, R., Ray, S. S., Bhattacharjee, A., and Ghosh, S. (2008). Novel cellulases from an extremophilic filamentous fungi Penicillium citrinum: production and characterization. J. Ind. Microbiol. Biotechnol. 35, 275–282. doi: 10.1007/s10295-008-0304-2
Fernandes, I., Alves, A., Correia, A., Devreese, B., and Esteves, A. C. (2014). Secretome analysis identifies potential virulence factors of Diplodia corticola, a fungal pathogen involved in cork oak (Quercus suber) decline. Fungal Biol. 118, 516–523. doi: 10.1016/j.funbio.2014.04.006
Fernandez-Canon, J. M., and Penalva, M. A. (1995). Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J. Biol. Chem. 270, 21199–21205. doi: 10.1074/jbc.270.36.21199
Fu, H., Feng, J., Aboukhaddour, R., Cao, T., Hwang, S. F., and Strelkov, S. E. (2013). An exo-1,3-beta-glucanase GLU1 contributes to the virulence of the wheat tan spot pathogen Pyrenophora tritici-repentis. Fungal Biol. 117, 673–681. doi: 10.1016/j.funbio.2013.07.003
Garcia-Calvo, L., Ullan, R. V., Fernandez-Aguado, M., Garcia-Lino, A. M., Balana-Fouce, R., and Barreiro, C. (2018). Secreted protein extract analyses present the plant pathogen Alternaria alternata as a suitable industrial enzyme toolbox. J. Proteomics 177, 48–64. doi: 10.1016/j.jprot.2018.02.012
Gibson, D. M., King, B. C., Hayes, M. L., and Bergstrom, G. C. (2011). Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr. Opin. Microbiol. 14, 264–270. doi: 10.1016/j.mib.2011.04.002
Gonzalez-Fernandez, R., and Jorrın-Novo, J. (2012). Contribution of proteomics to the study of plant pathogenic fungi. J. Proteome Res. 11, 3–16. doi: 10.1021/pr200873p
Grahl, N., Puttikamonkul, S., Macdonald, J. M., Gamcsik, M. P., Ngo, L. Y., Hohl, T. M., et al. (2011). In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 7:e1002145. doi: 10.1371/journal.ppat.1002145
Gupta, G. K., Sharma, S. K., and Ramteke, R. (2012). Biology, epidemiology and management of the pathogenic fungus Macrophomina phaseolina (Tassi) goid with special reference to charcoal rot of soybean (Glycine max (L.) Merrill). J. Phytopathol. 160, 167–180. doi: 10.1111/j.1439-0434.2012.01884.x
Huang, H., Ullah, F., Zhou, D. X., Yi, M., and Zhao, Y. (2019). Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 10:800. doi: 10.3389/fpls.2019.00800
Islam, M. S., Haque, M. S., Islam, M. M., Emdad, E. M., Halim, A., Hossen, Q. M., et al. (2012). Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13:493. doi: 10.1186/1471-2164-13-493
Jashni, M. K., Mehrabi, R., Collemare, J., Mesarich, C. H., and De Wit, P. J. (2015). The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 6:584. doi: 10.3389/fpls.2015.00584
Jermyn, W. S., and Boyd, E. F. (2002). Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology (Reading) 148, 3681–3693. doi: 10.1099/00221287-148-11-3681
Kang, C. H., Park, J. H., Lee, E. S., Paeng, S. K., Chae, H. B., Hong, J. C., et al. (2020). Redox-dependent structural modification of nucleoredoxin triggers defense responses against Alternaria brassicicola in Arabidopsis. Int. J. Mol. Sci. 21:9196. doi: 10.3390/ijms21239196
Kishore Babu, B., Saxena, A. K., Srivastava, A. K., and Arora, D. K. (2007). Identification and detection of Macrophomina phaseolina by using species-specific oligonucleotide primers and probe. Mycologia 99, 797–803. doi: 10.3852/mycologia.99.6.797
Klose, J., De Sa, M. M., and Kronstad, J. W. (2004). Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 52, 823–835. doi: 10.1111/j.1365-2958.2004.04019.x
Kolattukudy, P. E., and Brown, L. (1975). Fate of naturally occurring epoxy acids: a soluble epoxide hydrase, which catalyzes cis hydration, from Fusarium solani pisi. Arch. Biochem. Biophys. 166, 599–607. doi: 10.1016/0003-9861(75)90425-7
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Kunwar, I. R., Sinyh, T., Machado, C. C., and Sinclair, J. B. (1986). Histopathology of soybean seed and seedling infection by Macrophomina phaseolina. Phytopathology 76, 532–535. doi: 10.1094/phyto-76-532
Kwon, M. J., Kim, K. S., and Lee, Y. H. (2010). A Short-chain dehydrogenase/reductase gene is required for infection-related development and pathogenicity in Magnaporthe oryzae. Plant Pathol. J. 26, 8–16. doi: 10.5423/ppj.2010.26.1.008
Le Marquer, M., San Clemente, H., and Roux, C. (2019). Identification of new signalling peptides through a genome-wide survey of 250 fungal secretomes. BMC Genomics 20:64. doi: 10.1186/s12864-018-5414-2
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Lyon, G. D., Goodman, B. A., and Willianson, B. (2004). Botrytis Biology, Pathology, and Control. Dordrecht: Kluwer, 119–142.
Mayek Pérez, N., López-Castañeda, C., López Salinas, E., Cumpián Gutiérrez, J., and Acosta-Gallegos, J. A. (2001). Macrophomina phaseolina resistance in common bean under field conditions in Mexico. Agrociencia 46, 649–661.
Meijer, H. J., Mancuso, F. M., Espadas, G., Seidl, M. F., Chiva, C., Govers, F., et al. (2014). Profiling the secretome and extracellular proteome of the potato late blight pathogen Phytophthora infestans. Mol. Cell. Proteomics 13, 2101–2113. doi: 10.1074/mcp.M113.035873
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426. doi: 10.1021/ac60147a030
Morais do Amaral, A., Antoniw, J., Rudd, J. J., and Hammond-Kosack, K. E. (2012). Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola. PLoS One 7:e49904. doi: 10.1371/journal.pone.0049904
Morisseau, C., Ward, B. L., Gilchrist, D. G., and Hammock, B. D. (1999). Multiple epoxide hydrolases in Alternaria alternata f. sp. lycopersici and their relationship to medium composition and host-specific toxin production. Appl. Environ. Microbiol. 65, 2388–2395. doi: 10.1128/AEM.65.6.2388-2395.1999
Muller, O., Schreier, P. H., and Uhrig, J. F. (2008). Identification and characterization of secreted and pathogenesis-related proteins in Ustilago maydis. Mol. Genet. Genomics 279, 27–39. doi: 10.1007/s00438-007-0291-4
Murphy, C., Powlowski, J., Wu, M., Butler, G., and Tsang, A. (2011). Curation of characterized glycoside hydrolases of fungal origin. Database (Oxf.) 2011:bar020. doi: 10.1093/database/bar020
Pandey, V., Singh, M., Pandey, D., Marla, S., and Kumar, A. (2018). Secretome analysis identifies potential pathogenicity/virulence factors of Tilletia indica, a quarantined fungal pathogen inciting karnal bunt disease in wheat. Proteomics 18:e1700473. doi: 10.1002/pmic.201700473
Pereira, L. A., Bao, S. N., Barbosa, M. S., Da Silva, J. L., Felipe, M. S., De Santana, J. M., et al. (2007). Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 7, 1381–1388. doi: 10.1111/j.1567-1364.2007.00292.x
Pinot, F., Caldas, E. D., Schmidt, C., Gilchrist, D. G., Jones, A. D., Winter, C. K., et al. (1997). Characterization of epoxide hydrolase activity in Alternaria alternata f. sp. lycopersici. Possible involvement in toxin production. Mycopathologia 140, 51–58. doi: 10.1023/a:1006829330490
Quoc, N. B., and Chau, N. N. B. (2017). The role of cell wall degrading enzymes in pathogenesis of Magnaporthe oryzae. Curr. Protein Pept. Sci. 18, 1019–1034. doi: 10.2174/1389203717666160813164955
Raguchander, T., Samiappan, R., and Arjunan, G. J. I. P. (1993). Biocontrol of Macrophomina root rot of mungbean. Ind. Phytopathol. 46, 379–382.
Reis, H., Pfiffi, S., and Hahn, M. (2005). Molecular and functional characterization of a secreted lipase from Botrytis cinerea. Mol. Plant Pathol. 6, 257–267. doi: 10.1111/j.1364-3703.2005.00280.x
Reuveni, M., Sheglov, N., Eshel, D., Prusky, D., and Ben-Arie, R. (2007). Virulence and the production of endo-1,4-β-glucanase by isolates of Alternaria alternata involved in the moldy-core disease of apples. J. Phytopathol. 155, 50–55. doi: 10.1111/j.1439-0434.2006.01201.x
Reuveni, R., Nachmias, A., and Krikun, J. (1983). The role of seed borne inoculum on the development of Macrophomina phaseolina on melon. Plant Dis. 67, 280–281.
Sarkar, T. S., Biswas, P., Ghosh, S. K., and Ghosh, S. (2014). Nitric Oxide production by necrotrophic pathogen Macrophomina phaseolina and the host plant in charcoal rot disease of Jute: complexity of the interplay between necrotroph-host plant interactions. PLoS One 9:e107348. doi: 10.1371/journal.pone.0107348
Schmaler-Ripcke, J., Sugareva, V., Gebhardt, P., Winkler, R., Kniemeyer, O., Heinekamp, T., et al. (2009). Production of pyomelanin, a second type of melanin, via the tyrosine degradation pathway in Aspergillus fumigatus. Appl. Environ. Microbiol. 75, 493–503. doi: 10.1128/AEM.02077-08
Serrano, M., Coluccia, F., Torres, M., L’haridon, F., and Metraux, J. P. (2014). The cuticle and plant defense to pathogens. Front. Plant Sci. 5:274. doi: 10.3389/fpls.2014.00274
Sinha, N., Patra, S. K., Sarkar, T. S., and Ghosh, S. (2021). Secretome analysis identified extracellular superoxide dismutase and catalase of Macrophomina phaseolina. Arch. Microbiol. 204:62. doi: 10.1007/s00203-021-02631-w
Su, G., Suh, S. O., Schneider, R. W., and Russin, J. S. (2001). Host specialization in the charcoal rot fungus, Macrophomina phaseolina. Phytopathology 91, 120–126. doi: 10.1094/PHYTO.2001.91.2.120
Vibha. (2016). “Macrophomina phaseolina: the most destructive soybean fungal pathogen of global concern,” in Current Trends in Plant Disease Diagnostics and Management Practices, eds P. Kumar, V. K. Gupta, A. K. Tiwari, and M. Kamle (Cham: Springer International Publishing), 193–205. doi: 10.1007/978-3-319-27312-9_8
Vincent, D., Rafiqi, M., and Job, D. (2019). The multiple facets of plant-fungal interactions revealed through plant and fungal secretomics. Front. Plant Sci. 10:1626. doi: 10.3389/fpls.2019.01626
Wang, H., He, Z., Luo, L., Zhao, X., Lu, Z., Luo, T., et al. (2018). An aldo-keto reductase, Bbakr1, is involved in stress response and detoxification of heavy metal chromium but not required for virulence in the insect fungal pathogen, Beauveria bassiana. Fungal Genet. Biol. 111, 7–15. doi: 10.1016/j.fgb.2018.01.001
Warwas, M. L., Yeung, J. H., Indurugalla, D., Mooers, A. O., Bennet, A. J., and Moore, M. M. (2010). Cloning and characterization of a sialidase from the filamentous fungus, Aspergillus fumigatus. Glycoconj J. 27, 533–548. doi: 10.1007/s10719-010-9299-9
Wyllie, T. D. (1998). Soybean Diseases of the North Central Region. Charcoal rot of soybean-current status. St. Paul, MN: APS, 106–113.
Yagminas, A. P., Franklin, C. A., Villeneuve, D. C., Gilman, A. P., Little, P. B., and Valli, V. E. (1990). Subchronic oral toxicity of triethyl lead in the male weanling rat. Clinical, biochemical, hematological, and histopathological effects. Fundam. Appl. Toxicol. 15, 580–596. doi: 10.1016/0272-0590(90)90043-j
Yang, V. W., Zhuang, Z., Elegir, G., and Jeffries, T. W. (1995). Alkaline-active xylanase produced by an alkaliphilic Bacillus sp isolated from kraft pulp. J. Ind. Microbiol. 15, 434–444. doi: 10.1007/bf01569971
Zhou, H., Yu, Z., Hu, Y., Tu, J., Zou, W., Peng, Y., et al. (2009). The special neuraminidase stalk-motif responsible for increased virulence and pathogenesis of H5N1 influenza A virus. PLoS One 4:e6277.
Keywords: Macrophomina phaseolina, charcoal rot disease, solid-state culture, secretome analysis, putative virulence factors, LC-MS analysis, cell wall degrading enzymes, proteases
Citation: Sinha N, Patra SK and Ghosh S (2022) Secretome Analysis of Macrophomina phaseolina Identifies an Array of Putative Virulence Factors Responsible for Charcoal Rot Disease in Plants. Front. Microbiol. 13:847832. doi: 10.3389/fmicb.2022.847832
Received: 03 January 2022; Accepted: 14 February 2022;
Published: 05 April 2022.
Edited by:
Mohammad Arif, University of Hawai’i at Mānoa, United StatesReviewed by:
Sowmya Ramachandran, Foreign Disease-Weed Science Research (USDA-ARS), United StatesJose M. Diaz-Minguez, University of Salamanca, Spain
Copyright © 2022 Sinha, Patra and Ghosh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjay Ghosh, c2diaW9jQGNhbHVuaXYuYWMuaW4=; c2diaW9jQGdtYWlsLmNvbQ==
 Nilanjan Sinha
Nilanjan Sinha Sourav Kumar Patra
Sourav Kumar Patra Sanjay Ghosh
Sanjay Ghosh