- 1Guangdong Key Laboratory for Veterinary Drug Development and Safety Evaluation, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
- 3Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou, China
IncHI2 plasmids, possessing high flexibility and genetic plasticity, play a vital role in the acquisition and transmission of resistance determinants. Polymorphic mobile genetic elements (MGEs) generated by a chromosomally integrated IncHI2 plasmid in an individual Salmonella isolate have not yet been detected, and the mechanisms of the formation, excision, and dynamic evolution of a multidrug-resistant chromosomally integrated plasmid (MRCP) have remained obscure. Herein, we identified a 260-kb blaCTX–M–55-qnrS1-bearing IncHI2 plasmid within a Salmonella Muenster strain. Plenty of heterogeneous MGEs (new Escherichia coli chromosomally integrated plasmid or circular plasmids with different profiles) were yielded when this MRCP was conjugated into E. coli J53 with a transfer frequency of 10–4–10–5 transconjugants per donor. A bioinformatic analysis indicated that replicative transposition and homologous recombination of IS26 elements were particularly active, and the truncated Tn1721 also played a vital role in the formation of MRCP offspring. More importantly, when released from the chromosome, MRCP could capture and co-transfer adjacent chromosomal segments to form larger plasmid progeny than itself. Stability and growth kinetics assays showed that the biological characteristics of MRCP progeny were differentiated. This study provides an insight into a flexible existence of MRCP. The conversion between vertical and horizontal transmission endowed MRCP with genetic stability as a chromosomal coding structure and transferability as extra-chromosomal elements. This alternation may accelerate the acquisition and persistence of antibiotic resistance of clinical pathogens and enhance their ability to respond to adverse environments, which poses a great challenge to the traditional antibiotic treatment.
Introduction
With the increased usage of antibiotics for the control and treatment of infectious diseases, antibiotic resistance (AR) has emerged rapidly at a global scale and spread faster than previously thought. Mobile genetic elements (MGEs) are crucial transmission factors for antibiotic resistance genes (ARGs). Some MGEs (e.g., insertion sequences and transposons) are capable of transferring ARGs to new locations in the same or different DNA molecules within a single cell, while others are capable of delivering ARGs between host cells (e.g., plasmids, integrative conjugative elements, and prophages). Bacterial genomes exhibiting dynamic plasticity could be shaped by horizontal gene transfer (HGT) and genome rearrangement mediated by MGEs (Brown-Jaque et al., 2015; Delavat et al., 2017; Partridge et al., 2018).
Plasmids are important vehicles for the carriage of other MGEs and usually acquire ARGs associated with these elements (Partridge et al., 2018). IncHI2 plasmids are very flexible and genetically plastic, and the highly active traits in acquiring antibiotic resistance determinants make them the primary vector responsible for ARGs transmission (Garcia et al., 2007; Cain and Hall, 2012; Chen et al., 2016; Guo et al., 2016; Wong et al., 2016; Li et al., 2017b,2020; Fang et al., 2018; Zhao et al., 2018; Wyrsch et al., 2019; Zingali et al., 2020). Cephalosporin resistance genes blaCTX–M have often been found on diverse transmissible plasmids (Canton et al., 2012). In recent years, however, an increasing number of blaCTX–M genes have been identified in chromosomes of Enterobacteriaceae (Fabre et al., 2009; Coelho et al., 2010; Hirai et al., 2013; Hamamoto and Hirai, 2019). The transfer mechanism of ARGs from plasmids to chromosomes leads to the long-term persistence of antibiotic-resistant pathogens and imposes concerns on the potential recurrence of diseases. A thorough understanding of the molecular mechanisms underlying the reshuffling of DNA elements may help to establish new strategies to address the problems of AR.
With the bacterial conjugation discovered in the mid-early twentieth century, high-frequency recombination (Hfr) bacteria possessing the entire F factor integrated into the Escherichia coli chromosome have been deeply studied at that time. Hfr strains can reform the F factor or the F-prime (F’) factor as a free plasmid independent of the chromosome. The difference between F and F’ factor is that the latter harbors a portion of the donor’s chromosome. Researchers showed that a few R factors in E. coli are also capable of transferring chromosomal markers and also form an R’ factor with low frequency (Curtiss, 1969; Arutyunov and Frost, 2013; Villa and Viñas, 2019). The authors suggested that, with the exception of the reported Hfr donors, all other conjugal fertility factors should be referred to as plasmids until a chromosomal integration state can be demonstrated (Curtiss, 1969).
Here, we report the isolation of a multidrug-resistant chromosomally integrated plasmid (MRCP) belonging to the IncHI2 family from a Salmonella strain. According to the definition of Hfr strains, PJM1 can be defined as a SHfrHI2R strain. MRCP possesses the ability of generating heterogeneous forms as well as capturing and co-transferring adjacent chromosomal segments. More importantly, the SHfrHI2R demonstrated the capability to transmit vertically in a host-integrated state or horizontally through excision from the chromosome. This biphasic lifestyle confers MRCP genetic stability and transferability and greatly expands the range of possibilities of AR to be transmitted.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
All bacterial strains used in this study are listed in Table 1. AR phenotypes of these strains and their transconjugants against cephalosporin and ciprofloxacin (Table 1) were determined using the agar dilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2019).
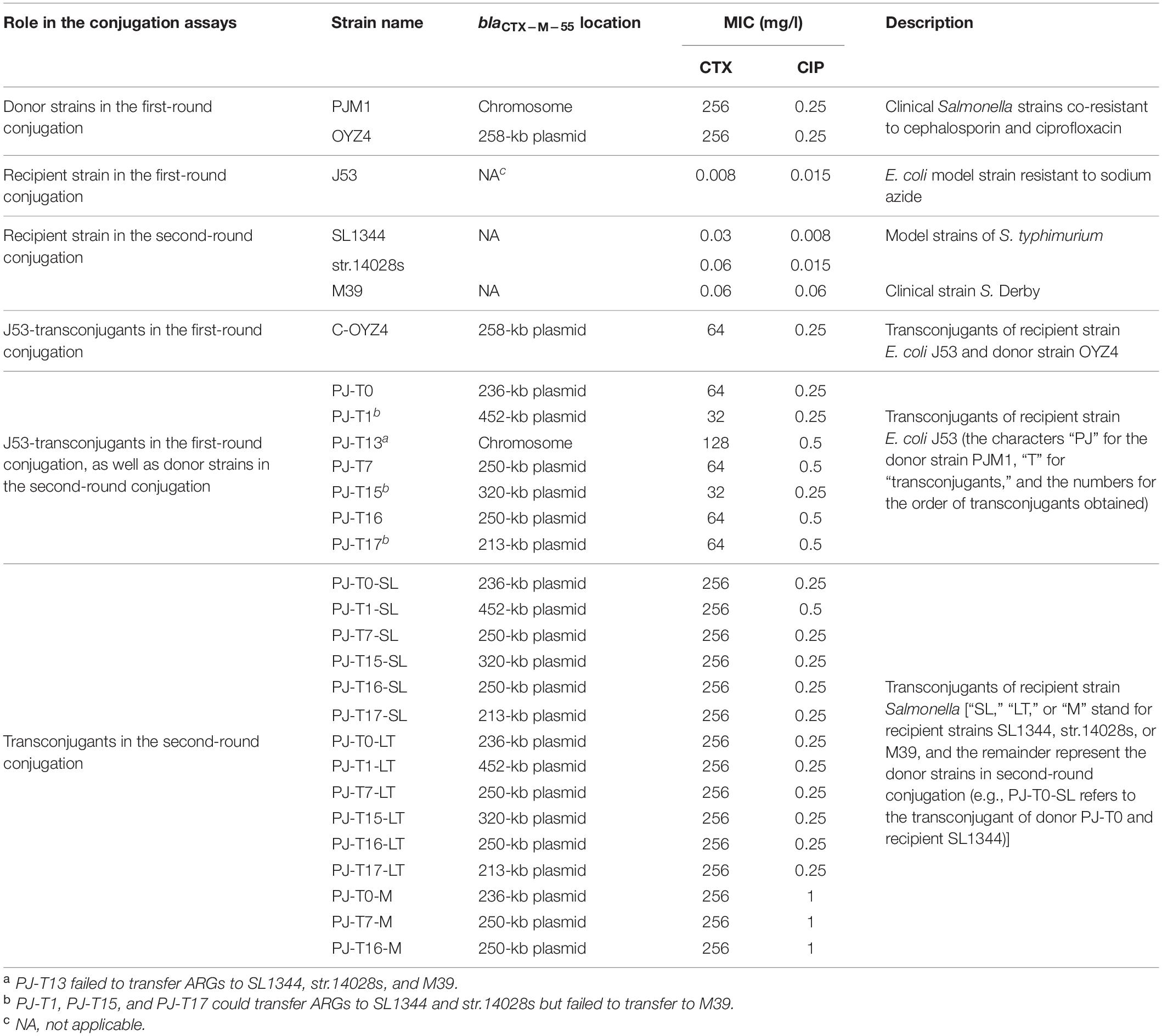
Table 1. Characteristics of blaCTX–M–55-bearing positive strains and their corresponding transconjugants.
WGS, Bioinformatic Analysis, and PCR
Complete genome sequences were annotated using the RAST server (Overbeek et al., 2014). Tools including EasyFig (Sullivan et al., 2011), BRIG (Alikhan et al., 2011), and BWA-MEM (Li et al., 2018) were used for bioinformatic analysis. Primers for PCR are listed in Supplementary Table 1.
Mating Experiments, S1-PFGE, and Southern Hybridization
The transferability of MRCP and its representative progeny were investigated by filter mating. The donor and recipient strains of the first-round conjugation assay were SHfrHI2R strain PJM1 and E. coli J53, respectively. The J53-transconjugants obtained in the first-round conjugation were used as the donor strains of the second-round conjugation and Salmonella strains (SL1344, ATCC 14028, and M39) as recipient strains. Briefly, when grown to log phase in Luria-Bertani (LB) broth, donor and recipient cultures were mixed in a 1:1 ratio; filtered through 0.22-μm pore size membranes (HuanKai Microbial, Guangzhou, China) placed on pre-warmed LB agar plates; and incubated for 16 h at 30, 37, and 42°C, respectively. For E. coli J53, putative J53-transconjugants were selected from MacConkey agar plates supplemented with cefotaxime (4 mg/l) (Sigma-Aldrich, Guangzhou, China) plus sodium azide (100 mg/l) (Sigma-Aldrich, Guangzhou, China), and for Salmonella SL1344, ATCC 14028, and M39, the second-round transconjugants were selected on xylose lysine desoxycholate (XLD) agar (HuanKai Microbial, Guangzhou, China) plates supplemented with cefotaxime (2 μg/ml). Transconjugants were reconfirmed by XbaI-PFGE (XbaI enzyme, Takara Bio Inc., Beijing, China) and PCR screening for blaCTX–M–55 and repHI2. Transfer frequencies were calculated as the number of transconjugants obtained per recipient. S1-PFGE (S1 enzyme, Takara Bio Inc., Beijing, China) followed by southern blot hybridization with digoxigenin-dUTP-labeled probes of blaCTX–M–55 and repHI2 genes were conducted to obtain the localizations and/or profiles of the integrated/free plasmids (Piekarowicz et al., 1968; Yang et al., 2017; Zhang et al., 2019).
Stability and Growth Kinetics Assays
The stability of MRCP in strain PJM1 and its progeny in E. coli J53 were studied by continuous passage for 30 days. Specifically, 5 μl of bacteria culture per strain were diluted in 5 ml of antibiotic-free fresh LB broth (1:1,000) every 24 h and incubated at 30°C and 180 rpm. Three independent experiments were conducted in triplicate under each condition. The serially diluted cultures of PJM1 and its J53-transconjugants grown to 20th and 30th days were plated on XLD and MacConkey agar plates, respectively. Fifty colonies were selected, and the presence of cephalosporin resistance phenotypes and HI2 replicon was confirmed by PCR. Plasmid stability was calculated as the proportion of cells harboring both blaCTX–M–55 and repHI2 in the 50 selected colonies. The growth kinetics of PJM1, OYZ4, E. coli J53, and the J53-transconjugants (PJ-T0, PJ-T7, PJ-T13, PJ-T16, PJ-T17, and C-OYZ4) were studied following the method reported in literature (Lv et al., 2020). The maximum growth rate (μmax), maximum OD600 (ODmax), the area under the growth curve (AUC), and the duration of the lag phase (lag) were calculated to evaluate the growth ability of the J53-transconjugants.
Data Availability
The complete sequences of the plasmids and the chromosomes have been deposited in GenBank under the accession numbers pOYZ4 (MN539018), pPJ-T0-236kb (MN539017), pPJ-T7-250kb (OL598600), pPJ-T16-250kb (OL598601), pPJ-T17-213kb (OL598602), PJ-T1-452kb (OM317741), PJ-T15-320kb (OM317740), PJ-T13-chr (CP087110), and PJM1-chr (CP045038).
Results
The Discovery of Heterogeneous Forms Yielded by a Multidrug-Resistant Chromosomally Integrated Plasmid in PJM1 by Conjugation Assays
A 236-kb IncHI2 plasmid, which carried chromosomally located blaCTX–M–55 and qnrS1 in Salmonella Muenster PJM1, was detected in the corresponding J53-transconjugant C-PJM1 (renamed PJ-T0 in this study) as previously described (Zhang et al., 2019). The unique HI2 replicon located in the PJM1 chromosome was subsequently confirmed by southern hybridization with repHI2 probe (Supplementary Figure 1). Interestingly, when we repeated the conjugation experiment, the size of the plasmid transferred to E. coli J53 was variable, but remained unchanged when re-transferred to Salmonella strains (Table 1). In contrast, another circular IncHI2 plasmid in S. Chester OYZ4 (Zhang et al., 2019) remained constant when transferred to E. coli J53 and other receptor strains by conjugation. Considering the facts above, we suspected that there was a multidrug-resistant plasmid (MRCP) integrated in PJM1 chromosome, resulting in the generation of a SHfrHI2R strain.
To verify the characteristics of this SHfrHI2R strain, 95 and 90 E. coli J53-transconjugants were randomly selected via conjugation at 30 and 37°C, respectively. The cephalosporin and fluoroquinolone co-resistance phenotype, as well as blaCTX–M–55, qnrS1, and repHI2 genes, was detected in all J53-transconjugants. XbaI-PFGE reconfirmed that the transconjugants were indeed E. coli J53. S1-PFGE profiles of the 185 J53-transconjugants showed that the MRCP in PJM1 was transferred to E. coli J53, yielding a wide range of plasmid sizes (187–459-kb) at 30 and 37°C (Supplementary Figure 2).
Three clusters including 14 subtypes were obtained by clustering analysis on the S1-PFGE profiles (Supplementary Figure 3A). Considering the size error margin of S1-PFGE prediction, plasmids showing a similar size (± 20-kb) were regarded as belonging to the same subtype. Cluster B (circular plasmid, sizes < 260-kb) had the highest yields, while cluster A (circular factors, size > 260-kb) were more frequently detected after conjugation at 30°C. Cluster C (the re-formation of plasmid-chromosomal cointegrates in E. coli J53) only seemed to occur at a very low rate at 37°C (Supplementary Figure 3A). Moreover, siiF was detected on pPJ-T1-452kb and pPJ-T15-320kb of cluster A by southern hybridization (Supplementary Figure 3B). Interestingly, the siiABCDEF gene cluster is present in Salmonella sp. but not in E. coli (Morgan et al., 2004; Barlag and Hensel, 2015). These results prompted us to trace why PJM1 has the capability to yield such a wide range of plasmid sizes and how siiF changed from being exclusive to Salmonella to being shared by E. coli and Salmonella.
Genetic Characteristics of Multidrug-Resistant Chromosomally Integrated Plasmid and Its Differentiated Descendants
To investigate the formation mechanism of the MRCP heterogeneous progeny, the accurate genome structures of parental strain PJM1 and its representative progeny (PJ-T13, plasmid integrated into E. coli J53 chromosome; pPJ-T0-230kb, pPJ-T16-250kb, pPJ-T17-213kb, pPJ-T1-452kb, and pPJ-T15-320kb, element as a 230-, 250-, 213-, 452-, and 320-kb circular molecule in J53, respectively) were obtained using deep DNA sequencing. The PJM1 chromosome was 5,000,356-bp in length and composed of 4,725 predicted coding sequences (CDSs) with 51.9% GC content. BLASTN analysis indicated that this chromosome exhibited high homology (94.4% coverage and 99.9% identity) to S. Muenster str. 0315 (NZ_CP019198) isolated from a fecal sample from a dairy farm in the United States. Comparison with the chromosome of str. 0315 showed that the PJM1 chromosome featured an extra DNA fragment, namely, MRCP (Figure 1). MRCP was 261,307-bp with 46.7% GC and 306 CDSs and displayed 99% identity with 78.5 and 88.2% coverage to the IncHI2 plasmids pDGSE139 (KM198330) and pCFSA1096 (CP033347), respectively (Supplementary Figure 4).
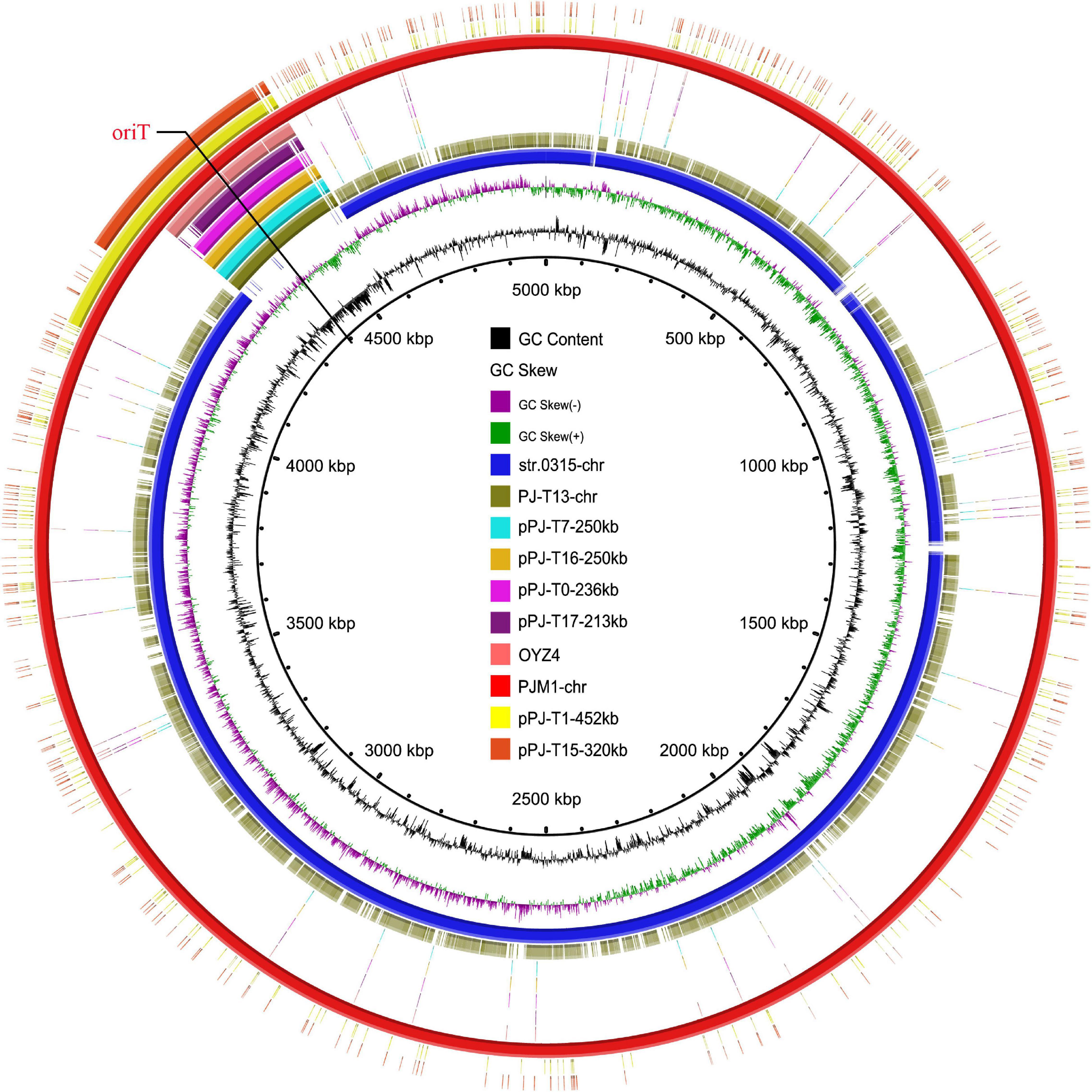
Figure 1. Circular alignment of chromosomes PJM1-chr, str.0315-chr, and PJ-T13-chr and plasmids pPJ-T17-213kb, pPJ-T0-236kb, pPJ-T16-250kb, pPJ-T7-250kb, pOYZ4, pPJ-T1-452kb, and pPJ-T15-320kb. The red circle denotes chromosome PJM1-chr as a reference. Bright blue and olive-green circles represent str.0315-chr (NZ_CP019198) and PJ-T13-chr, respectively; aqua, sallow, violet, deep purple, coral red, yellow, and orange-red circles depict the plasmids pPJ-T7-250kb, pPJ-T16-250kb, pPJ-T0-236kb, pPJ-T17-213kb, pOYZ4, pPJ-T1-452kb, and pPJ-T15-320kb, respectively.
Interestingly, S. Chester OYZ4 obtained from poultry in Guangdong Province, China, at the same time as PJM1 in 2017 carried the circular plasmid pOYZ4, which harbored 327 predicted CDSs distributed among 257,945-bp with a 47% GC content. Importantly, although the two strains belonged to different serotypes and XbaI-PFGE profiles (Zhang et al., 2019), pOYZ4 shared a surprisingly high degree of homology (88.46% coverage; 99.71% identity) with MRCP (Supplementary Figure 4 and Figure 2). Similar to pOYZ4, MRCP also possessed an intact IncHI2-ST2 backbone (190-kb), which contained genes for replication (repHIA and repHI2), partition (parAB and parMR), conjugation (tra, trh, and htd), complete or partial toxin–antitoxin systems (hipA/hipB and higB), an SOS mutagenesis correlative operon (umuCD), and two tellurite resistance operons (terY3Y2XY1W and terZABCDEF) (Figure 3 and Supplementary Figure 4). Interestingly, the backbone was flanked by two variable regions (VRs) containing multi-resistance genes and four genes encoding site-specific recombinase (Figure 2).
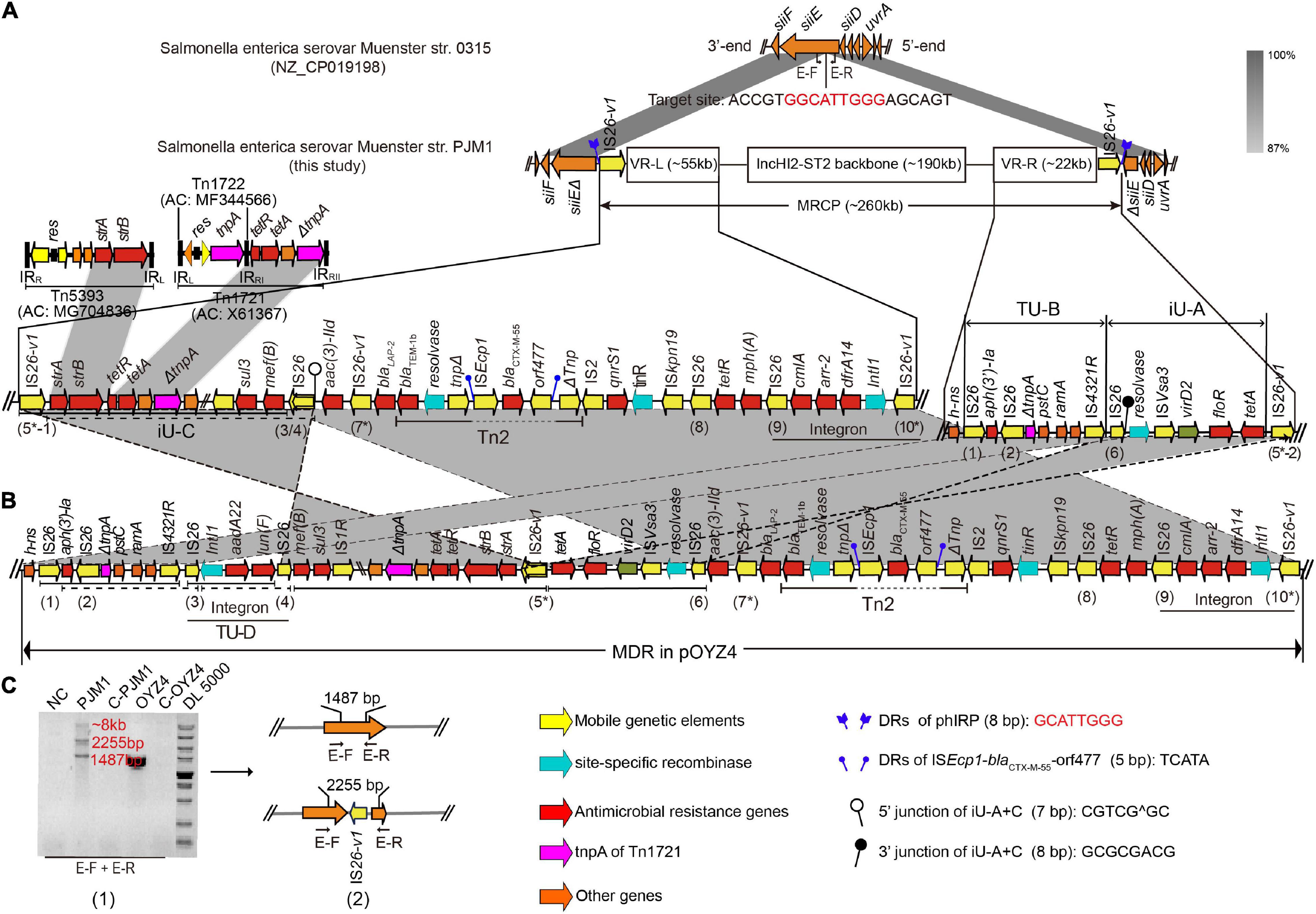
Figure 2. The structure and location of MRCP in PJM1 chromosome. (A) Comparison between the genetic structure of PJM1 chromosome and S. Muenster str. 0315. (B) The left and right variable regions (VR-L and VR-R) of MRCP and the corresponding MDR regions in pOYZ4. The IS26 elements (IS26 and IS26-v1) in pOYZ4 and MRCP are marked with numbers. (C) Gel electrophoretic diagram (1) of PCR amplicons of primer pairs E-F and E-R and the corresponding structure (2) detected. All amplicons were confirmed by sequencing. Primers are listed in Supplementary Table 1.
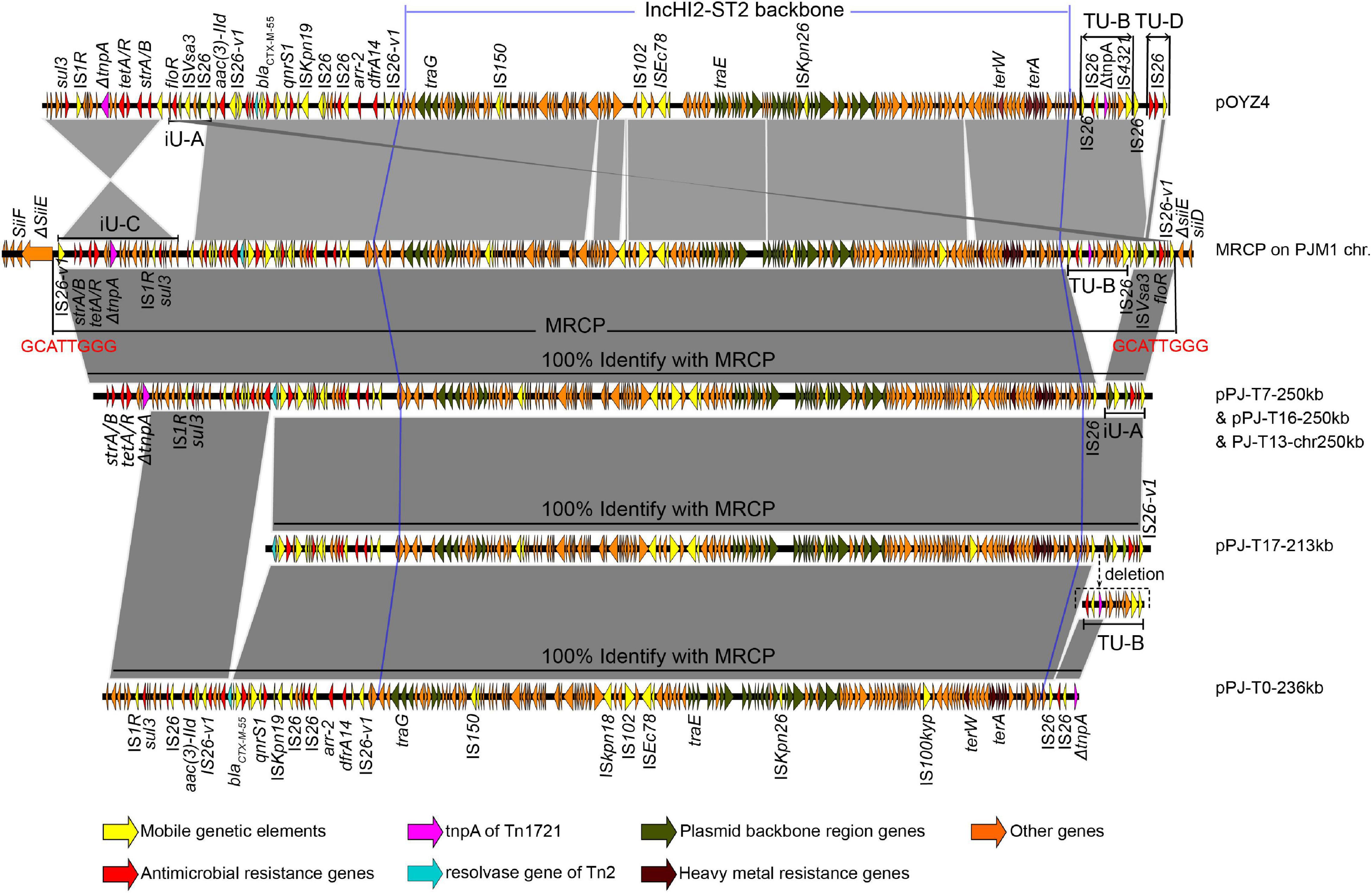
Figure 3. Sequence alignment of the multidrug-resistant chromosomally integrated plasmid (MRCP), its progeny, and pOYZ4. The highlighted GCATTGGG nucleotides represent the duplicated DRs that flanked MRCP. Regions of > 99% identity are marked by gray shading. The positions of the genetic structures iU-A, TU-B, iU-C, and TU-D were marked. iU, inversion unit; TU, translocatable unit.
The left VR (VR-L, 55-kb) clustered numerous mosaic genetic elements. The resistance gene cassettes (cmlA, arr2, and dfrA14) integrated into a class 1 integron were at the end of VR-L adjacent to the plasmid backbone, followed by mph(A), tetR, qnrS1, and a 2,971-bp transposition unit (ISEcp1-blaCTX–M–55-orf477), which truncated a complete composite transposon Tn2. A partial Tn5393 variant was detected at the other end of VR-L, which consisted of its left terminal inverted repeats (IRL5393, 81-bp) and carrying the genes strA/B. Importantly, the sequence adjacent to ΔTn5393 seemed to be the residual variant of Tn1721 after losing its basic transposon Tn1722 (ΔTnp1721, 1,789-bp), tetracycline resistance genes, and two 38-bp right terminal repeat variants (IRRI and IRRII) at both ends (Figure 2). The right VR (VR-R, 22-kb) contained four IS26 elements, a resolvase gene, and many ARGs including aph (3′)-Ia, floR, and tet(A) (Figure 2). Interestingly, a truncated Tn1721 transposase gene (ΔTnp1721, 848-bp) that seemed to be a part of ΔTn1721 in VR-L was also present in VR-R (Figure 2).
The circular plasmid pPJ-T0-230kb in J53-transconjugant PJ-T0 was 236,068-bp with 46% GC and 311 predicted CDSs (Figures 1, 3 and Supplementary Figure 4). Similarly, the pPJ-T16-250kb in PJ-T16 was 247,473-bp with 47% GC and 298 CDSs, and the pPJ-T17-213kb in PJ-T17 was 213,369-bp with 46% GC and 247 CDSs. All the plasmids showed a high homology with the MRCP (100% identity with 95.17, 97.35, and 90.83% coverage, respectively), and they all shared the same IncHI2-ST2 backbone as expected (Figures 1, 3 and Supplementary Figure 4). Interestingly, the fragment PJ-T13-chr250kb inserted into the J53 chromosomal gene bglH was the same as pPJ-T16-250kb (Figures 1, 3 and Supplementary Figure 4). That is, the SHfrHI2R strain could not only form free IncHI2 plasmids but also directly integrate into the chromosome of the recipient, giving place to a new MRCP in E. coli. On the other hand, pPJ-T1-452kb in PJ-T1 was 452,929-bp with 49% GC and 464 CDSs, and pPJ-T15-320kb in PJ-T15 was 331,203-bp with 48% GC and 362 CDSs. Apart from containing the 250-kb MRCP, both pPJ-T1-452kb and pPJ-T15-320kb harbored a segment of PJM1 chromosome (Figure 1), which demonstrated the capability of MRCP to capture and co-transfer adjacent chromosomal segments in PJM1. Therefore, the circular factors (pPJ-T1-452kb and pPJ-T15-320kb) in cluster B corresponded to RHI2’ variants that consisted of MRCP and a portion of a chromosomal fragment.
Dynamic Evolution and Prevalence of Multidrug-Resistant Chromosomally Integrated Plasmid
To verify the dynamic evolution of MRCP during bacterial division, primers E-F and E-R from the 3’-end and 5’-end of siiE were used to amplify the probable residual regions after the excision of diverse MRCP descendants in the evolutionary PJM1 population (Figure 2A). Three products (1.5, 2.3, and > 5-kb) were obtained in PJM1. DNA sequencing showed that the 1.5-kb product, which was the same as the product in OYZ4, aligned with siiE exactly. The 2.3-kb product consisted of the truncated siiE and a IS26-v1 that interrupted siiE (Figure 2C). Despite many attempts, we still could not obtain the complete sequence of product > 5-kb by Sanger sequencing, which seemed to be a mixture of different sequences with the same adjacent sequence. This phenomenon was consistent with a diversity of J53-transconjugants obtained in the conjugation assays, indicating that the MRCP in the chromosomes of the PJM1 population were in dynamic equilibrium during bacterial division. If MRCP is deleted from the PJM1 chromosome, some sequence traces would be left behind (e.g., if MRCP is deleted in the form of pPJ-T7, it leaves a trace siiEΔ-IS26-v1-ΔsiiE in the PJM1 chromosome; if MRCP is deleted as pPJ-T7, it leaves siiEΔ-IS26-v1-strA/B-tetA/R-ΔTn1721-ISVsa3-floR-ΔsiiE; and in the case of pPJT17, siiEΔ-IS26-v1-strA/B-tetA/R-ΔTn1721-IS1R-sul3-IS26-aac(3)-IId- IS26-v1-Tn2Δ-ΔsiiE would be left, etc.). Due to the heterogeneous forms generated by MRCP, the sequence traces left changed with different deletions. Accordingly, quantitative traces (i.e., decayed MRCP) should theoretically be left in the PJM1 subpopulation. To further verify the dynamic evolution mechanisms of MRCP, the raw data of PJM1 nanopore (ONT) long-reads have also been reanalyzed, but no reads covering the decayed MRCP were found.
The Molecular Mechanisms of Formation and Dynamic Transmission of Multidrug-Resistant Chromosomally Integrated Plasmid
In the two VRs of MRCP, 10 copies of IS26 elements (IS26 or IS26 variant, IS26-v1) were identified (Supplementary Table 2), corresponding to the 10 copies in pOYZ4 (Figures 2B, 4). Almost all IS26s copies were intact, except that copy 8 in MRCP and pOYZ4 lacked the right terminal inverted repeat (IRR), and copy 2 (present in MRCP) featured a single-base deletion (T) at position + 376, which resulted in a frameshift mutation (Figures 2B, 4). None of the IS26s were flanked by target site duplications (TSDs) that are typical for insertion sequences of this family. The 8-bp sequence 5’-CGTCGCGC-3’ (Figure 2B, black lollipop) downstream of IS26 copy 6 and the 7-bp incomplete reverse complement sequence 5’- GC^CGACG-3’ (Figure 2B, white lollipop) upstream of IS26 copy 3/4 of MRCP (“copy 3/4” represents IS26 in MRCP and corresponds with the IS26 copy 3 or IS26 copy 4 in pOYZ4) provided evidence for inverse intramolecular replicative transposition, which reversed the 40-kb segment-designated iU-A + C (Figures 2B, 4A). The IS26-v1 copy 5* (an asterisk was used to distinguish IS26-v1 from IS26) in iU-A + C interacted with the target site (GCATTGGG) in chromosomal siiE, resulting in the integration of the entire plasmid into the PJM1 chromosome to form the MRCP and the splitting of iU-A + C into iU-A and iU-C. Meanwhile, IS26-v1 copies 5*-1 and 5*-2 at each junction between MRCP and the PJM1 chromosome were generated by direct replication of IS26-v1 copy 5* (Figure 4B).
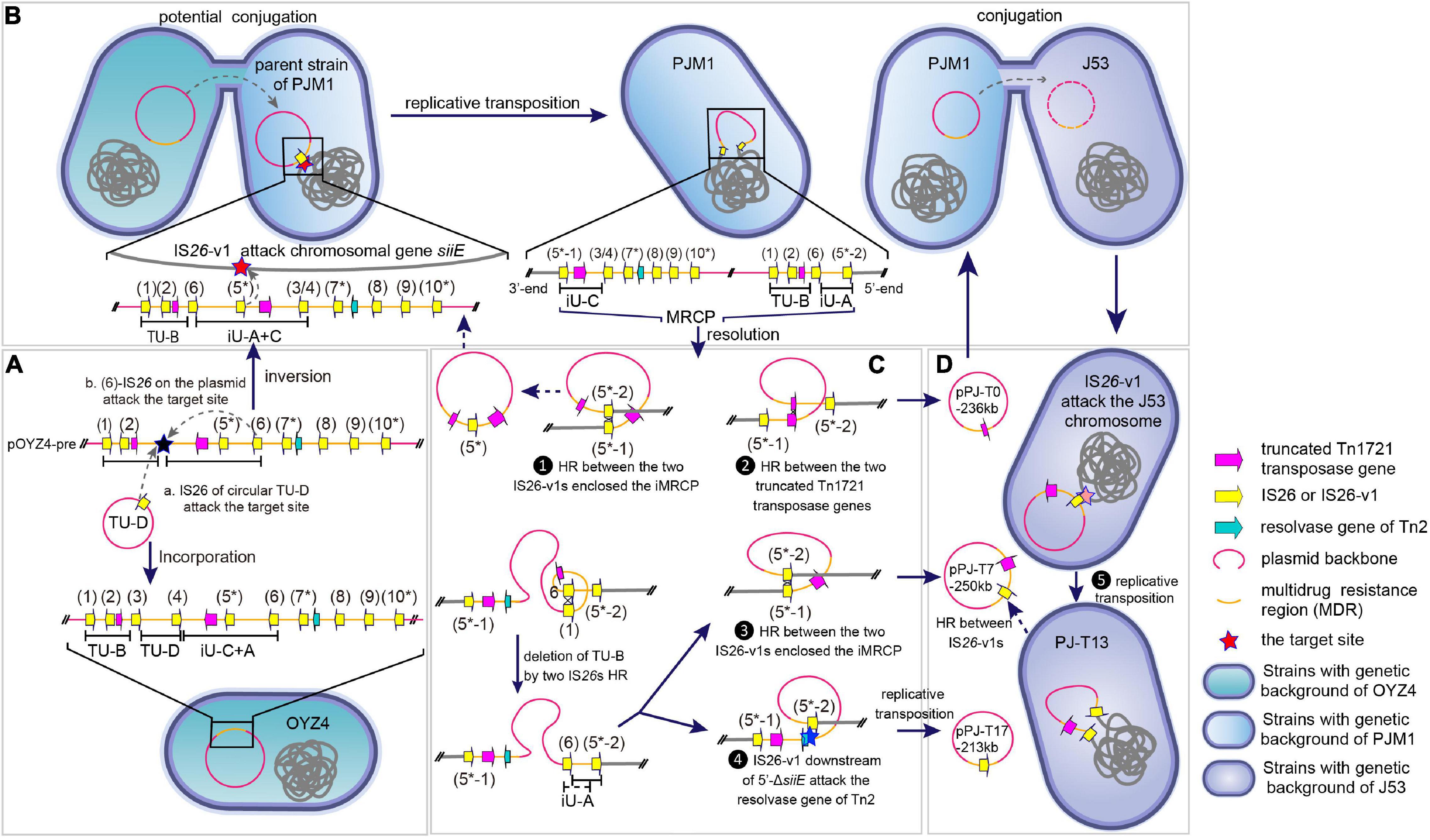
Figure 4. Presumptive formation and multiple transmission form of MRCP. (A) a. TU-D was inserted into the precursor plasmid pOYZ4-pre via conservative transposition, forming pOYZ4. b. The IS26 copy 6 in pOYZ4-pre attacked the target site (black star) in the opposite DNA strand, causing reversal of iU-C + A segment. (B) IS26-v1 in iU-A + C segment attacked the chromosomal siiE of PJM1 during conjugation, forming the cointegrate by intermolecular replicative transposition. The plasmid in PJM1 chromosome was renamed MRCP according to its characteristics. (C) Diverse resolution of MRCP. ❶ The direct homologous recombination between IS26-v1 copies 5*-1 and 5*-2 mediated the generation of a plasmid identical to pPOYZ4-pre except for reversed iU-C + A. ❷ The direct homologous recombination between two truncated Tn1721 led to plasmid pPJ-T0-236kb. ❸ Deletion of TU-B and homologous recombination between IS26-v1 copies 5*-1 and 5*-2 yielded plasmid pPJ-T7-250kb or pPJ-T16-250kb. ❹ Intramolecular replicative transposition resulted in the formation of plasmid pPJ-T17-213kb. After TU-B being deleted by homologous recombination, IS26-v1 copy 5*-2 attacked the target site (blue star) in the resolvase gene of Tn2, forming plasmid pPJ-T17-213kb. (D) ❺ Intermolecular replicative transposition led to PJ-T13-chr250kb. When a plasmid such as pPJ-T7-250kb was transferred into E. coli J53 by conjugation, IS26-v1 copy 5 attacked J53 chromosome, and PJ-T13-chr250kb in PJ-T13 was formed.
As recently published, when a central fragment is flanked by two identical IS26s, this fragment with one adjacent IS constitutes a translocatable unit (TU), which results in the excision and movement of ARGs between different replicons (from one plasmid to another or to the chromosome, and vice versa) (Harmer et al., 2020). Accordingly, the inversion of iU-A + C also led to the formation of TU-B [IS26-aph (3′)-Ia-IS26-ΔTn1721-IS4321R] in MRCP and in the same orientation of ΔTn1721 within TU-B and iU-C (Figure 4B). A single homologous recombination (HR) event between two ΔTn1721 released pPJ-T0-236kb (Figure 4C). Besides, the HR between two IS26s (copies 1 and 6) mediated the deletion of TU-B also carrying ΔTn1721. Subsequently, two parallel events may have taken place on the chromosome lacking TU-B: (i) The HR between IS26-v1 copies 5*-1 and 5*-2 released pPJ-T7-250kb, resulting in the chromosome modified by the IS26-v1 flanked by the TSD. (ii) IS26-v1 copy 5*-2 downstream of 5′-ΔsiiE interacted with the gene encoding Tn2 resolvase on the same DNA strand giving place to pPJ-T17-213kb (Figure 4C). The IS26-v1 copy 5* on pPJ-T7-250kb, on the other hand, interacted with the bgIH gene on the E. coli J53 chromosome and generated the PJ-T13 cointegrate through intermolecular replicative transposition (Figure 4D). Conversely, the HR between IS26-v1 copies 5*-1 and 5*-2 converted chromosomal fragment PJ-T13-chr250kb into the circular plasmid pPJ-T7-250kb (Figure 4D).
Transferability, Stability, and/or Growth Kinetics of Multidrug-Resistant Chromosomally Integrated Plasmid and Its Progeny
To determine the transferability of the SHfrHI2R strain PJM1, conjugation experiments were performed at 30, 37, and/or 42°C. Plasmid pOYZ4 was used as a control and E. coli J53 as recipient. Compared with the other two culture temperatures, both MRCP and pOYZ4 had the highest transfer frequency at 30°C (Figure 5A). However, the transfer frequency of MRCP (10–4–10–5 transconjugants per donor) was always significantly lower than that of pOYZ4 (10–2–10–3 transconjugants per donor) at selected temperatures (p < 0.0001) (Figure 5A). Interestingly, after 30 days of passage without antibiotics, MRCP and pOYZ4 were stably maintained in parental strain PJM1 and OYZ4, respectively (Figure 5B). Besides, there was no significant difference in the maximum growth rate (μmax, p > 0.05) and AUC (p > 0.05) between PJM1 and OYZ4, but the duration of the lag phase and maximum OD600 of PJM1 was significantly higher than that of OYZ4 (lag, p < 0.01 and ODmax, p < 0.001).
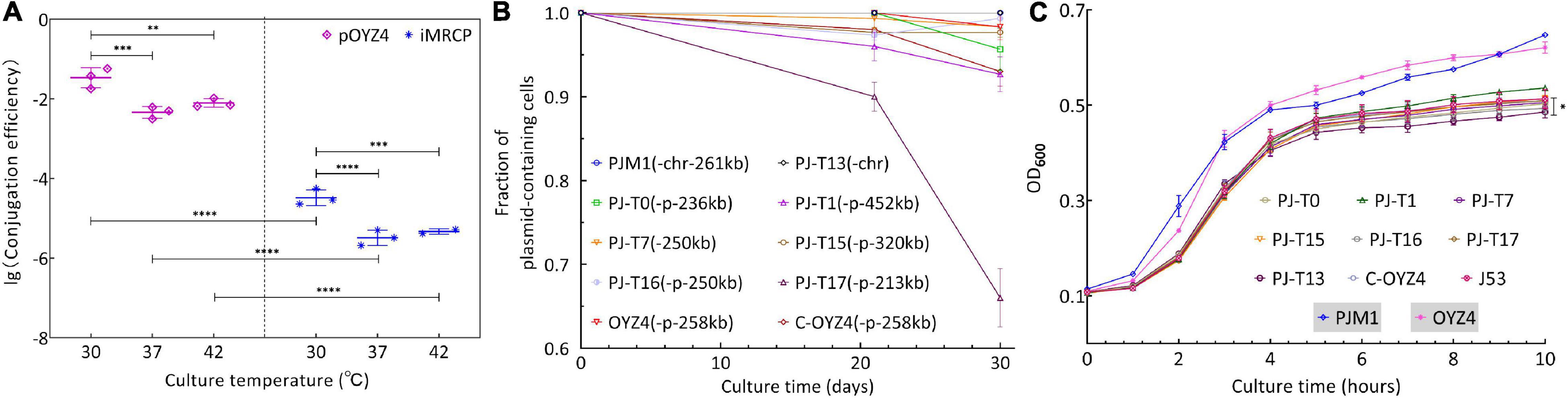
Figure 5. (A) Comparison of transfer efficiencies of MRCP and IncHI2 plasmid pOYZ4 into E. coli J53. Transfer efficiency (mean ± standard deviation) is calculated based on colony counts of the transconjugant and recipient cells in triplicate. (B) Stability of MRCP in PJM1 and its progeny in E. coli J53 for 30 days. (C) Growth curves of representative J53-transconjugants, recipient strain E. coli J53, and donor strain PJM1. Salmonella OYZ4 serves here as a contrast strain for PJM1. Considering the interspecific difference, comparisons were performed between the J53-transconjugant and E. coli J53, and between PJM1 and OYZ4, respectively. All experiments were repeated independently in triplicate. The error bars denote the standard deviations (n = 3). Two-way analysis of variance (ANOVA) with Tukey’s multiple comparison was used to evaluate statistical significance. Significantly different at *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.00001.
After 30 days of passage, pPJ-T1-452kb, pPJ-T15-320kb, pPJ-T7-250kb, pPJ-T16-250kb, pPJ-T0-236kb, and the chromosomal PJ-T13-chr250kb were stably maintained (plasmid-containing populations fraction > 90%) in their host strains (E. coli J53-transconjugants PJ-T1, pPJ-T15, PJ-T7, PJ-T16, and PJ-T0), whereas the stability of plasmid pPJ-T17-213kb was 66% maintained in its host strain PJ-T17 (Figure 5B). As for the growth ability, although there were no significant differences in the maximum growth rate (μmax, p > 0.05) between J53-transconjugants (PJ-T0, PJ-T7, PJ-T13, PJ-T16, PJ-T17, and C-OYZ4) and E. coli J53, there were significant differences between PJ-T13 and E. coli J53 in the duration of the lag phase (lag, p < 0.05), AUC (p < 0.05), and maximum OD600 (ODmax, p < 0.001), implying no significant differences between the other J53-transconjugants and E. coli J53 during 10-h assessment, except for a significant difference between PJ-T13 and E. coli J53 (Figure 5C). To test the transferability of different progenies of MRCP to other host strains, second-round conjugation assays were performed at 37°C with the representative J53-transconjugants as the donor strains and three Salmonella strains (SL1344, str.14028s, and M39) as the recipient strains (Table 1). Plasmids pPJ-T7-250kb, pPJ-T16-250kb, and pPJ-T0-236kb in J53-transconjugants were transferred to the aforementioned Salmonella strains; pPJ-T1-452kb, pPJ-T15-320kb, and pPJ-T17-213kb were transferred to SL1344 and str.14028s, but not M39, while chromosomal MGE PJ-T13-chr250kb could not be transferred to any of the three.
Discussion
Conjugative plasmids are the most important drivers of the evolution of antibiotic-resistant bacteria (Partridge et al., 2018). IncHI2 plasmids are known to be significant vectors of clinically important ARGs encoding resistance to carbapenems (Carattoli, 2013; Zhang et al., 2019), colistin (Li et al., 2017b,2020), and quinolones (Carattoli, 2013; Wong et al., 2016). The high flexibility and genetic plasticity of IncHI2 plasmids can confer their hosts a great capability of acquiring ARGs (Garcia et al., 2007; Guo et al., 2016; Li et al., 2017b,2020; Fang et al., 2018; Zhao et al., 2018; Wyrsch et al., 2019; Zingali et al., 2020) or to form cointegrates with other plasmids (Garcia et al., 2007; Schluter et al., 2014; Li et al., 2017a; Wong et al., 2017; Fang et al., 2018; Wang et al., 2018; Xie et al., 2018; Chen et al., 2019). In this study, we identified a multidrug-resistant chromosomally integrated plasmid that produced many heterogeneous MGEs during the conjugation process, and the strain PJM1 was defined as SHfrHI2R strains according to the definition of Hfr strains.
Segregational instability is the term used to describe the loss of plasmids that occurs during cell division, regardless of whether plasmid carriage is costly or not (Carroll and Wong, 2018). The formation of SHfrHI2R strain may be a strategy employed by bacteria to avoid segregational loss while maintaining mobility of the resistance factors contained therein. Thus, the MRCP can be stably inherited in a host-integrated state and spread horizontally by excision from chromosome. On the other hand, although the available experimental methods cannot eliminate duplicate strains in E. coli J53-transconjugants harboring heterogeneous MGEs and the errors in plasmid size estimates that may arise after S1-PFGE assays, we confirmed the presence of two different sizes of plasmids in one subgroup by deep DNA sequencing (pPT-T0-230kb and pPT-T17-213kb were clustered in the 211–230-kb subgroup), indicating that MRCP progeny size indeed covered a broad range far beyond our expectations. Besides, we speculated that the temperature may not affect the size range of MRCP progeny, but it would affect the distribution of specific plasmids. In other words, the excision, transmission, and re-integration of MRCP (forming clusters A, B, and C) may be mediated by different enzyme systems, and their corresponding activities may be affected by temperature. The re-formation rate (1.6%) of plasmid-chromosomal cointegrates in E. coli J53 is very low and seems to be temperature dependent, consistent with the view of Carroll and Wong (2018) that host chromosome integration is considered an exception because it rarely occurs in the laboratory. More importantly, the capability of MRCP to capture and co-transfer adjacent chromosomal segments was unexpected. Some studies indicated that site-specific recombinase might be responsible for the shuffling of large DNA fragments (Ravatn et al., 1998; Groth and Calos, 2004; Miyazaki and van der Meer, 2013; Lv et al., 2020). However, it seems that the transposase encoded by IS26-v1 in MRCP, rather than the site-specific recombinases, mediated the conjugative mobilization of adjacent chromosomal segments in cis. To the best of our knowledge, IS26-mediated plasmid capture of chromosomal fragments has not been reported in Salmonella, and the translocation mechanisms of PJM1 chromosomal DNA fragments remain to be further investigated.
No plasmid derivatives containing two ΔTn1721 were detected, indicating that the deletion of a ΔTn1721 copy seemed to be an essential prerequisite. HR between two partially identical ΔTn1721 appeared to be the most straightforward way to resolve MRCP from the PJM1 chromosome. Moreover, IS26-v1 HR and IS26-v1-mediated reactions resulted in the formation or polymorphic resolution of the cointegrate. Meanwhile, we found that IS26-mediated inverse intramolecular replicative transposition and TU-B deletion were also involved. We speculate that different IS26 variants tend to participate in different pathways. In this regard, IS26-v1 tends to form and resolve the cointegrate between different replicons, whereas IS26 tends to cause structural variations within a single replicon. An in vitro study supports this view that the IS26 variant with G184N substitution (such as IS26-v1) increased Tnp26 activity and the cointegration frequency (Pong et al., 2019).
In the present study, MRCP was autonomously transferred to other organisms at a much lower frequency than pOYZ4. Moreover, the transfer frequency of MRCP was possibly the final result of multi-type excision, conjugation, and plasmid-chromosomal transposition. Additionally, the significantly lower growth ability of PJ-T13 than that of E. coli J53 indicated that the initial fitness cost of MRCP progeny increased in a cointegrated state, which is in agreement with the statement that any plasmid integrating into and interrupting important host genes can increase fitness costs (Carroll and Wong, 2018). However, whether the plasmid existed independently in OYZ4 or integrated into PJM1 chromosome, the growth ability of bacteria appeared to be unaffected, suggesting that cointegrate will not pose significant fitness cost to PJM1. This seems to be a paradox, but we rationally believe that the cointegrate is probably species specific or requires a longer evolutionary time to adapt to its bacterial host (Wein et al., 2019). Also, as Carroll and Wong (2018) stated, the genetic background of the host might be an important determinant of plasmid fitness. Namely, a single plasmid beneficial in a strain with a certain genetic background could decrease fitness in the other. Furthermore, we consider that the balance of maintaining the fitness of the host and MRCP offspring might govern the transfer types and transfer frequencies (Delavat et al., 2017). On the other hand, despite the growth disadvantages of PJ-T13, its cointegrate could be maintained stably just like MRCP in PJM1. However, a large proportion of the terminal bacterial population (in the stability assays) had lost pPJ-T17-213kb. The divergence of the polymorphic MGEs indicated that the MRCP progeny differed in their ability to survive after being transferred to a new host.
When reanalyzing the raw data of PJM1 nanopore (ONT) long-reads, no reads covering the decayed MRCP were found. We speculate that the inherently low excision frequency of MRCP along with multiple excision types helped in reducing the content of residual reads below a detectable threshold. Meanwhile, we also noticed that the gene siiE truncated by the MRCP on the chromosome is a large open reading frame (16-kb), which encodes a highly repetitive giant protein SiiE (5,559 aa) composed of 53 Ig domains (Barlag and Hensel, 2015). Such a long, repetitive sequence may have also played a part in hindering the detection of the degradation structure of MRCP in chromosome.
There is scarce up-to-date literature regarding Hfr strains, especially concerning multi-resistant Hfr strains. However, the ability of Hfr strains to spread ARGs should not be ignored. In this regard, strain SHfrHI2R was able to generate free IncHI2 plasmids featuring the same size as the chromosomal integration state or generate plasmid featuring smaller sizes. Besides, the presence of RHI2’ factors and of the chromosomal integrated state was observed in the E. coli receptor, albeit in high or low frequencies, respectively. Notably, although MRCP was integrated into the chromosome of recipient E. coli J53, the “pseudo-Hfr” transconjugant was not transferrable to other bacterial hosts. Hence, E. coli strains presenting the chromosomal integrated state (i.e., PJ-T13) should not be regarded as Hfr strain in spite of harboring mating-related genes. Taken together, the data pertaining to the strain SHfrHI2R reported in this study further expands the scope of Hfr strains.
Conclusion
The emergence of polymorphic MGEs in a given strain caused by MRCP, may accelerate the bacterial response to adverse environments. The MRCP possesses conjugative traits but endows the host cell with a biphasic lifestyle, which ensured MRCP maintained a semi-equilibrium state in PJM1, increasing its persistence in the host cell. In this regard, PJM1 could act as plasmid reservoir and transfer ARGs to other strains continuously. The relationship between the mechanisms of lateral and vertical transmission of bacterial resistance may be more complex than previously perceived. Understanding the role and clinical significance of MGEs in the spread of antibiotic resistance is essential to prevent the global spread of AMR.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
M-XC and H-XJ conceived and designed the experiments. M-XC, JNZ, X-MD, JEZ, and YL performed the experiments. M-XC and J-FZ analyzed the data and drafted the manuscript. M-XC, RL, and H-XJ revised the manuscript. All authors approved the final draft and contributed to the article and approved the submitted version.
Funding
We gratefully appreciate the financial support of the project by the National Natural Science Foundation of China (Grant No. 31972734) and the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (Grant No. 2019BT02N054).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.846954/full#supplementary-material
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Arutyunov, D., and Frost, L. S. (2013). F conjugation: back to the beginning. Plasmid 70, 18–32. doi: 10.1016/j.plasmid.2013.03.010
Barlag, B., and Hensel, M. (2015). The giant adhesin SiiE of Salmonella enterica. Molecules 20, 1134–1150. doi: 10.3390/molecules20011134
Brown-Jaque, M., Calero-Caceres, W., and Muniesa, M. (2015). Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7. doi: 10.1016/j.plasmid.2015.01.001
Cain, A. K., and Hall, R. M. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carroll, A. C., and Wong, A. (2018). Plasmid persistence: costs, benefits, and the plasmid paradox. Can. J. Microbiol. 64, 293–304. doi: 10.1139/cjm-2017-0609
Chen, K., Dong, N., Chan, E. W., and Chen, S. (2019). Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microbes Infect. 8, 857–865. doi: 10.1080/22221751.2019.1626197
Chen, W., Fang, T., Zhou, X., Zhang, D., Shi, X., and Shi, C. (2016). IncHI2 plasmids are predominant in antibiotic-resistant Salmonella isolates. Front. Microbiol. 7:1566. doi: 10.3389/fmicb.2016.01566
CLSI (2019). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-nine Informational Supplement M100-S29. Wayne, PA: Clinical and Laboratory Standards Institute.
Coelho, A., Gonzalez-Lopez, J. J., Miro, E., Alonso-Tarres, C., Mirelis, B., Larrosa, M. N., et al. (2010). Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int. J. Antimicrob. Agents 36, 73–78. doi: 10.1016/j.ijantimicag.2010.03.005
Curtiss, R. III (1969). Bacterial conjugation. Annu. Rev. Microbiol. 23, 69–136. doi: 10.1146/annurev.mi.23.100169.000441
Delavat, F., Miyazaki, R., Carraro, N., Pradervand, N., and van der Meer, J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41, 512–537. doi: 10.1093/femsre/fux008
Fabre, L., Delaune, A., Espie, E., Nygard, K., Pardos de la Gandara, M., Polomack, L., et al. (2009). Chromosomal integration of the extended-spectrum beta-lactamase gene blaCTX-M-15 in Salmonella enterica serotype Concord isolates from internationally adopted children. Antimicrob. Agents Chemother. 53, 1808–1816. doi: 10.1128/AAC.00451-08
Fang, L. X., Li, X. P., Deng, G. H., Li, S. M., Yang, R. S., Wu, Z. W., et al. (2018). High genetic plasticity in multidrug-resistant sequence type 3-IncHI2 plasmids revealed by sequence comparison and phylogenetic analysis. Antimicrob. Agents Chemother. 62:e02068-17. doi: 10.1128/AAC.02068-17
Garcia, A., Navarro, F., Miro, E., Villa, L., Mirelis, B., Coll, P., et al. (2007). Acquisition and diffusion of bla CTX-M-9 gene by R478-IncHI2 derivative plasmids. FEMS Microbiol. Lett. 271, 71–77. doi: 10.1111/j.1574-6968.2007.00695.x
Groth, A. C., and Calos, M. P. (2004). Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678. doi: 10.1016/j.jmb.2003.09.082
Guo, Q., Ding, B., Jove, T., Stoesser, N., Cooper, V. S., Wang, M., et al. (2016). Characterization of a novel IncHI2 plasmid carrying tandem copies of blaCTX-M-2 in a fosA6-harboring Escherichia coli sequence type 410 strain. Antimicrob. Agents Chemother. 60, 6742–6747. doi: 10.1128/AAC.01173-16
Hamamoto, K., and Hirai, I. (2019). Characterisation of chromosomally-located blaCTX-M and its surrounding sequence in CTX-M-type extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Glob. Antimicrob. Resist. 17, 53–57. doi: 10.1016/j.jgar.2018.11.006
Harmer, C. J., Pong, C. H., and Hall, R. M. (2020). Structures bounded by directly-oriented members of the IS26 family are pseudo-compound transposons. Plasmid 111, 102530. doi: 10.1016/j.plasmid.2020.102530
Hirai, I., Fukui, N., Taguchi, M., Yamauchi, K., Nakamura, T., Okano, S., et al. (2013). Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int. J. Antimicrob. Agents 42, 500–506. doi: 10.1016/j.ijantimicag.2013.08.005
Li, R., Xie, M., Dong, N., Lin, D., Yang, X., Wong, M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7, 1–9. doi: 10.1093/gigascience/gix132
Li, R., Xie, M., Lv, J., Wai-Chi Chan, E., and Chen, S. (2017a). Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J. Antimicrob. Chemother. 72, 696–699. doi: 10.1093/jac/dkw509
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017b). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Li, Y., Dai, X., Zeng, J., Gao, Y., Zhang, Z., and Zhang, L. (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 10:8113. doi: 10.1038/s41598-020-65106-w
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio 11:e002930-19. doi: 10.1128/mBio.02930-19
Miyazaki, R., and van der Meer, J. R. (2013). A new large-DNA-fragment delivery system based on integrase activity from an integrative and conjugative element. Appl. Environ. Microbiol. 79, 4440–4447. doi: 10.1128/AEM.00711-13
Morgan, E., Campbell, J. D., Rowe, S. C., Bispham, J., Stevens, M. P., Bowen, A. J., et al. (2004). Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54, 994–1010. doi: 10.1111/j.1365-2958.2004.04323.x
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Piekarowicz, A., Wlodarczyk, M., and Kunicki-Goldfinger, W. (1968). Mechanism of conjugation and recombination in bacteria. I. The role of DNA synthesis in Hfr and F- cells during mating in E. coli K-12. Mol. Gen. Genet. 101, 131–139. doi: 10.1007/BF00336579
Pong, C. H., Harmer, C. J., Ataide, S. F., and Hall, R. M. (2019). An IS26 variant with enhanced activity. FEMS Microbiol. Lett. 366:fnz031. doi: 10.1093/femsle/fnz031
Ravatn, R., Studer, S., Zehnder, A. J., and van der Meer, J. R. (1998). Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. Strain B13. J. Bacteriol. 180, 5505–5514. doi: 10.1128/JB.180.21.5505-5514.1998
Schluter, A., Nordmann, P., Bonnin, R. A., Millemann, Y., Eikmeyer, F. G., Wibberg, D., et al. (2014). IncH-type plasmid harboring bla CTX-M-15, bla DHA-1, and qnrB4 genes recovered from animal isolates. Antimicrob. Agents Chemother. 58, 3768–3773. doi: 10.1128/AAC.02695-14
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Villa, T. G., and Viñas, M. (2019). 2019_Book_HorizontalGeneTransfer_Breaking Borders Between Living Kingdoms. Berlin: Springer.
Wang, J., Huang, X. Y., Xia, Y. B., Guo, Z. W., Ma, Z. B., Yi, M. Y., et al. (2018). Clonal spread of Escherichia coli ST93 carrying mcr-1-Harboring IncN1-IncHI2/ST3 plasmid among companion animals, China. Front. Microbiol. 9:2989. doi: 10.3389/fmicb.2018.02989
Wein, T., Hulter, N. F., Mizrahi, I., and Dagan, T. (2019). Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 10:2595. doi: 10.1038/s41467-019-10600-7
Wong, M. H., Chan, E. W., and Chen, S. (2017). IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J. Antimicrob. Chemother. 72, 2750–2754. doi: 10.1093/jac/dkx238
Wong, M. H., Chan, E. W., Xie, L., Li, R., and Chen, S. (2016). IncHI2 plasmids are the key vectors responsible for oqxAB transmission among Salmonella species. Antimicrob. Agents Chemother. 60, 6911–6915. doi: 10.1128/AAC.01555-16
Wyrsch, E. R., Reid, C. J., DeMaere, M. Z., Liu, M. Y., Chapman, T. A., Roy Chowdhury, P., et al. (2019). Complete sequences of multiple-drug resistant IncHI2 ST3 plasmids in Escherichia coli of porcine origin in Australia. Front. Sustain. Food Syst. 3:18. doi: 10.3389/fsufs.2019.00018
Xie, M., Li, R., Liu, Z., Chan, E. W. C., and Chen, S. (2018). Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. J. Antimicrob. Chemother. 73, 1230–1234. doi: 10.1093/jac/dkx540
Yang, L., Li, W., Jiang, G. Z., Zhang, W. H., Ding, H. Z., Liu, Y. H., et al. (2017). Characterization of a P1-like bacteriophage carrying CTX-M-27 in Salmonella spp. resistant to third generation cephalosporins isolated from pork in China. Sci. Rep. 7:40710. doi: 10.1038/srep40710
Zhang, C. Z., Ding, X. M., Lin, X. L., Sun, R. Y., Lu, Y. W., Cai, R. M., et al. (2019). The emergence of chromosomally located bla CTX-M-55 in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Zhao, H., Chen, W., Xu, X., Zhou, X., and Shi, C. (2018). Transmissible ST3-IncHI2 plasmids are predominant carriers of diverse complex IS26-Class 1 Integron arrangements in multidrug-resistant Salmonella. Front. Microbiol. 9:2492. doi: 10.3389/fmicb.2018.02492
Keywords: multidrug resistance, cointegrate, homologous recombination, Hfr strain, heterogeneous mobile genetic elements
Citation: Chang M-X, Zhang J, Zhang J-F, Ding X-M, Lu Y, Zhang J, Li R and Jiang H-X (2022) Formation, Transmission, and Dynamic Evolution of a Multidrug-Resistant Chromosomally Integrated Plasmid in Salmonella Spp. Front. Microbiol. 13:846954. doi: 10.3389/fmicb.2022.846954
Received: 31 December 2021; Accepted: 17 February 2022;
Published: 06 April 2022.
Edited by:
Andrew Paul Jackson, University of Liverpool, United KingdomReviewed by:
Nicolás Francisco Cordeiro, Universidad de la República, UruguayXavier Bellanger, Université de Lorraine, France
Copyright © 2022 Chang, Zhang, Zhang, Ding, Lu, Zhang, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruichao Li, cmNobDg4QHllYWgubmV0; Hong-Xia Jiang, aHhqaWFuZ0BzY2F1LmVkdS5jbg==
 Man-Xia Chang
Man-Xia Chang Jing Zhang1
Jing Zhang1 Jin-Fei Zhang
Jin-Fei Zhang Ruichao Li
Ruichao Li Hong-Xia Jiang
Hong-Xia Jiang