- 1Yunnan Herbal Laboratory, College of Ecology and Environmental Sciences, Yunnan University, Kunming, China
- 2The International Joint Research Center for Sustainable Utilization of Cordyceps Bioresources in China and Southeast Asia, Yunnan University, Kunming, China
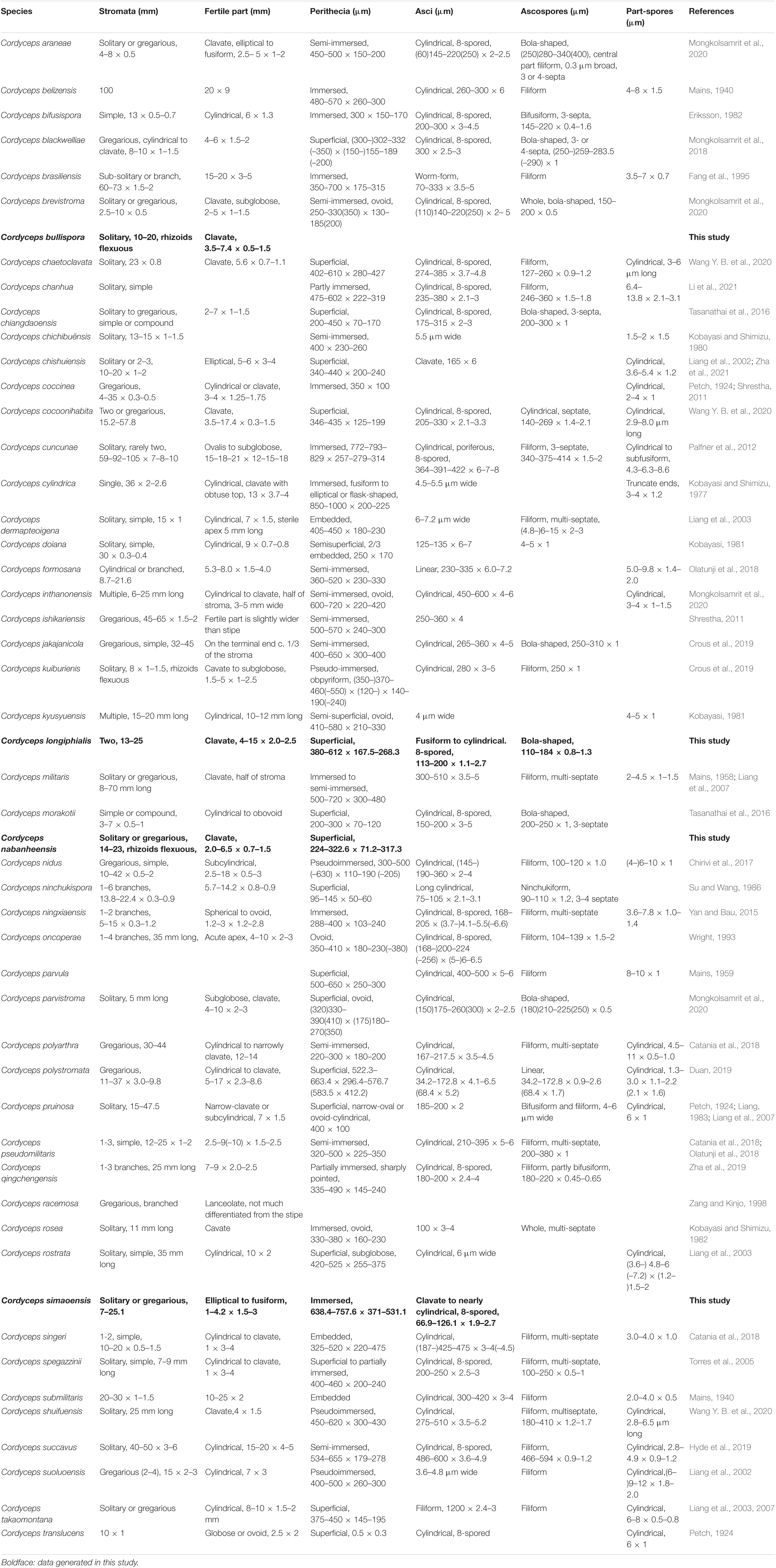
The current study was aimed to introduce five new species of Cordyceps from Yunnan, with morphological descriptions, illustrations, color photographs, phylogenetic placement, associated host, and a comparison with allied taxa. The five new species were morphologically distinct from all other Cordyceps sensu lato species, and it was also suggested that they should differ from other species in the genus Cordyceps based on combined multigene analyses. Employing DNA nucleotide sequences of the nrLSU, nrSSU, tef-1α, rpb1, and rpb2, the five new species were recognized in the clade of Cordyceps by using molecular phylogenetic analyses, including five well-supported subclades: three new species, Cordyceps bullispora, Cordyceps longiphialis, and Cordyceps nabanheensis, were found in the subclade of C. pruinosa, and two new species, Cordyceps pseudotenuipes and Cordyceps simaoensis, were located in the subclade of C. tenuipes. The five novel species shared similar morphologies to other species in the genus Cordyceps, with fleshy and brightly pigmented stromata; perithecia superficial to completely immersed, ordinal in arrangement; and hyaline asci, with thickened cylindrical ascus apex. The morphological characteristics of 66 species in Cordyceps sensu stricto, namely, 5 novel species and 61 known taxa, were also compared.
Introduction
Cordyceps Fr. is a well-known genus of arthropod-pathogenic fungi. It was shown that many species of Cordyceps played a significant role in the cycling of matter in an ecological system, had a high ecological and economic value for biocontrol and bioactive compounds, and served as a model system for research on fungal insect pathology (Zha et al., 2018; Chen W. H. et al., 2019). Complexes of some cordycipitoid fungal species and their natural host, such as C. militaris Fr., C. chanhua Z. Z. Li, F. G. Luan, Hywel-Jones, C. R. Li and S. L. Zhang, and C. kyusyuensis Kawam, have received significant attention in traditional medicine industry due to the detection of bioactive compounds with anti-aging, anti-tumor, antioxidant, anti-inflammatory, and immuno-modulatory effects (Castillo et al., 2018; Zhao et al., 2018; Lou et al., 2019; Li et al., 2021; Zhang et al., 2021). Cordyceps tenuipes (Peck) Kepler, B. Shrestha and Spatafora has been applied in a variety of functional foods in Japan and South Korea, possessing nutritional, immune-regulatory, antitumor, analgesic, antibacterial, and anti-malaria effects (Wang Y. et al., 2020). Cordyceps species reproduce via sexual (ascospores) or asexual (conidia) spores, or both (Mora et al., 2017). The host range of Cordyceps embraces 7 orders of Arthropoda, namely, Araneae, Coleopteran, Dermaptera, Hemiptera, Hymenoptera, Lepidoptera, and Orthoptera, where Coleoptera and Lepidoptera are the two significant orders to host beyond the estimated 200 Cordyceps spp. as recorded (Shrestha et al., 2016; Kepler et al., 2017; Mongkolsamrit et al., 2020; Wang Y. B. et al., 2020).
Fries (1818) was credited with coining Cordyceps as a genus in Pyrenomycetes from a hybrid of the Greek word cordyle and the Latin word caput, meaning a club and a head, respectively. However, this genus was then treated as a tribe level of Sphaeria and described as stroma erect, stipe simple or branching, a sterile stalk supporting the perithecia at the periphery, and projecting openings at the apex (Shrestha et al., 2014). Nearly 20 different genera have been accounted as synonyms of Cordyceps in various sources (Shrestha et al., 2014). Shrestha et al. (2014) provided a comprehensive review regarding the taxonomic history of Cordyceps and concluded that the genus is the oldest valid genus in Cordycipitaceae (Hypocreales, Ascomycota) and is typified by a sexual morph. Owing to the cylindrical shape of the stroma, C. militaris, the type species of Cordyceps, was already described in the 17th- and early 18th-century literature (Shrestha et al., 2014).
Species of Cordycipitaceae produce three types of ascospore, namely disarticulating ascospores (e.g., C. militaris), intact ascospores [e.g., Blackwellomyces cardinalis (G. H. Sung and Spatafora) Spatafora and Luangsa-ard and Blackwellomyces pseudomilitaris Hywel-Jones and Sivichai], and bola-ascospores (e.g., C. bifusispora O. E. Eriksson), with superficial to partially immersed perithecia on fleshy stromata that are pallid to brightly pigmented. Currently, Cordyceps s. l. (Fr) Link (1833) consists of approximately 1,300 known species assigned to three families (Cordycipitaceae, Ophiocordycipitaceae, and partial Clavicipitaceae) in the order Hypocreales (Sung et al., 2007; Zha et al., 2018). Progressive works from researchers worldwide have enriched the account of species diversity within Cordycipitaceae and revealed new genera (Kepler et al., 2017; Mongkolsamrit et al., 2018, 2020; Wang Y. B. et al., 2020). Up to now, the Cordycipitaceae has twenty genera: Akanthomyces Lebert, Amphichorda Fr., Ascopolyporus Möller, Beauveria Vuill, Blackwellomyces Spatafora and Luangsa-ard, Cordyceps s. s., Engyodontium de Hoog, Flavocillium H. Yu, Y. B. Wang, Y. Wang, Q. Fan and Zhu L. Yang, Gamszarea Z. F. Zhang and L. Cai, Gibellula Cavara, Hevansia Luangsa-ard, Hywel-Jones and Spatafora, Hyperdermium J. F. White, R. F. Sullivan, Bills and Hywel-Jones, Lecanicillium W. Gams and Zare, Leptobacillium Zare and W. Gams, Liangia H. Yu, Y. B. Wang, Y. Wang, Z. H. Chen and Zhu L. Yang, Neotorrubiella Tasan., Thanakitp. and Luangsa-ard, Parengyodontium C. C. Tsang, J. F. W. Chan, W. M. Pong, J. H. K. Chen, A. H. Y. Ngan, M. Cheung, C. K. C. Lai, D. N. C. Tsang, S. K. P. Lau and P. C. Y. Woo, Samsoniella (G. Sm.) Mongkols, Noisrip, Thanakitp, Spatafora and Luangsa-ard, Simplicillium W. Gams and Zare, and Torrubiella W. Gams and Zare.
The unispecific genus Phytocordyceps possesses bola-ascospores, and because of its phylogenetic placement, it is also recognized as a member of Cordycipitaceae and thus transferred to Cordyceps s. s. Based on molecular phylogenetic studies or morphological descriptions, 35 names and new combinations were accepted for Cordyceps s. s. (Sung et al., 2007). Concerning the application of Isaria, in an effort to avoid the confusion with Cordyceps, Kepler et al. (2017) proposed the rejection of Isaria and combined 13 species into Cordyceps s. s. To date, together with the species as reported in recent years, there are about 70 species whose phylogenetic positions are known in Cordyceps worldwide (Tasanathai et al., 2016; Chirivi et al., 2017; Catania et al., 2018; Mongkolsamrit et al., 2018, 2020; Olatunji et al., 2018; Crous et al., 2019; Hyde et al., 2019; Li et al., 2020; Wang Y. B. et al., 2020; Hu et al., 2021).
Harvesting wild specimens have become a challenge as natural fungi populations are declining and large-scale cultivation has not been achieved. Recently, the demand for cultivable fungal species with medicinal applications has been increasing worldwide. Yunnan Province and the Tibetan plateau are home to a large diversity of entomogenous fungi (Chen Z. H. et al., 2019). C. chaetoclavata H. Yu, Y.B. Wang, Y. Wang, Q. Fan and Zhu L. Yang, C. cocoonihabita H. Yu, Y.B. Wang, Y. Wang, Q. Fan and Zhu L. Yang, C. shuifuensis H. Yu, Y. B. Wang, Y. Wang and Zhu L. Yang, and C. subtenuipes H. Yu, Y. B. Wang, Y. Wang, D. E. Duan and Zhu L. Yang have been described from this area (Wang Y. B. et al., 2020). In this study, based on macro- and micro-morphological characteristics, ecological data together with DNA nucleotide sequence analyses of the nuclear ribosomal large subunit (nrLSU) and small subunit (nrSSU), the genes encoding translation elongation factor 1-α (tef-1α), the largest subunit of RNA polymerase II (rpb1), and the second-largest subunit of RNA polymerase II (rpb2), the fungus’ phylogenetic position was assessed. Furthermore, we have also compared the morphological characteristics of 66 species in Cordyceps s. s., consisting of 5 novel species and 61 known taxa.
Materials and Methods
Sampling
Cordyceps samples were newly collected from Kunming, Weishan, Jinghong, and Pu’er of Yunnan, southwestern China. Voucher specimens were deposited in the YHH (Yunnan Herbal Herbarium) of Yunnan University. The isolated strains were deposited in YFCC (Yunnan Fungal Culture Collection) of Yunnan University.
Fungal materials, including the hosts, were photographed and recorded. Isolation of the fungi was achieved as per Wang Y. B. et al. (2020). The stromata or synnemata were cut into 5-mm-long segments, followed by surface sterilization with 30% H2O2 for 30 s to 1 min, and then rinsed with sterile water five times, dried with sterilized filter paper. Then, a part of the insect body was cut off, and the resulting segments and insect body were inoculated onto potato dextrose agar (PDA: potato 200 g/L, dextrose 20 g/L, and agar 20 g/L) plates containing 0.1 g/L streptomycin and 0.05 g/L tetracycline.
Morphological Studies
Given the field notes, color images of the materials, and complementary literature data, macro-morphological characteristics, such as the host, fungi location, color, and shape of the stromata, and perithecial orientation (superficial, immersed, semi-immersed; ordinal or oblique) were examined under a dissecting microscope (Olympus SZ61), where the insect hosts were recognized with the support from professional entomologists.
The sexual characteristics, such as perithecia, asci, and ascospores, were firstly mounted on glass slides with lactophenol cotton blue solution after removing from the stroma, whereas the asexual characteristics, such as phialides and conidia, were firstly inoculated on glass slides with a thin layer of PDA medium block (5–10 mm in diameter) and overlaid by a cover slip, and cultivated in Petri dishes with a small amount of water under room temperature until further development of phialides and conidia. Then, the micro-morphological descriptions in both size and shape were determined with Olympus CX40 and BX53 microscopes, and FEI Quanta 200 scanning electron microscope. Twenty to thirty individual length and width measurements were taken, with absolute minima and maxima. Mycelia were inoculated on PDA plates and incubated at 25°C for a couple of days, and the colonies were photographed and measured every week.
Molecular Studies
DNA Extraction and PCR Amplification
Total DNA was extracted from the fungal mycelia on PDA plates or herbarium materials using the modified CTAB procedure (Doyle and Doyle, 1987). For DNA amplification, the primer pairs nrSSU-CoF and nrSSU-CoR (Wang Y. B. et al., 2015) and LR5 and LR0R (Vilgalys and Hester, 1990; Rehner and Samuels, 1994) were used for the nrSSU and nrLSU, EF1α-EF and EF1α-ER (Bischoff et al., 2006; Sung et al., 2007) were used to amplify the translation elongation factor 1α (tef-1α), and primers RPB1-5’F and RPB1-5’R, and RPB2-5’F and RPB2-5’R (Bischoff et al., 2006; Sung et al., 2007) were used to amplify the largest and second-largest subunits of RNA polymerase II (rpb1 and rpb2), respectively.
The polymerase chain reaction (PCR) matrix was composed of 2.5 μl of PCR 10 × Buffer (2 mmol/L Mg2+) (Transgen Biotech, Beijing, China), 0.25 μl of Taq DNA polymerase (Transgen Biotech, Beijing, China), 2 μl of dNTP (2.5 mmol/L), 1 μl of DNA template (500 ng/μl), 1 μl of forward primers (10 μmol/L), 1 μl of reverse primers (10 μmol/L), and 17.25 μl of sterile ddH2O. Amplification reactions were performed in a BIO-RAD T100™ thermal cycler (BIO-RAD Laboratories, Hercules, CA, United States). The PCR program was performed as described by Sung et al. (2007) and Wang Y. B. et al. (2020). Products of PCR were purified with the Gel Extraction and PCR Purification Combo Kit Beijing Genomics Institute (Shenzhen, China) and then sequenced on an automatic sequence analyzer (BGI Co., Ltd, Shenzhen, China) using the same primers as those used in amplification.
DNA Sequence Alignments
The samples’ nrSSU, nrLSU, tef-1α, rpb1, and rpb2 nucleotide sequences were compared with those deposited in the GenBank database. To understand the relationship of our sample with those in the GenBank, nrSSU, nrLSU, tef-1α, rpb1, and rpb2 sequences of the representative Cordyceps s. s. species available in GenBank were retrieved and combined with our sequences (Table 1). Five datasets, the nrSSU, nrLSU, tef-1α, rpb1, and rpb2 sequences, were aligned and manually checked on Bioedit v7.0.9 (Hall, 1999). In order to examine phylogenetic conflicts among these datasets, the partition homogeneity (PH) test was performed with 1,000 randomized replicates, using heuristic searches with simple addition of sequences in PAUP* 4.0b10 (Swofford, 2002); the phylogenetic signals in the five gene markers showed no conflict.
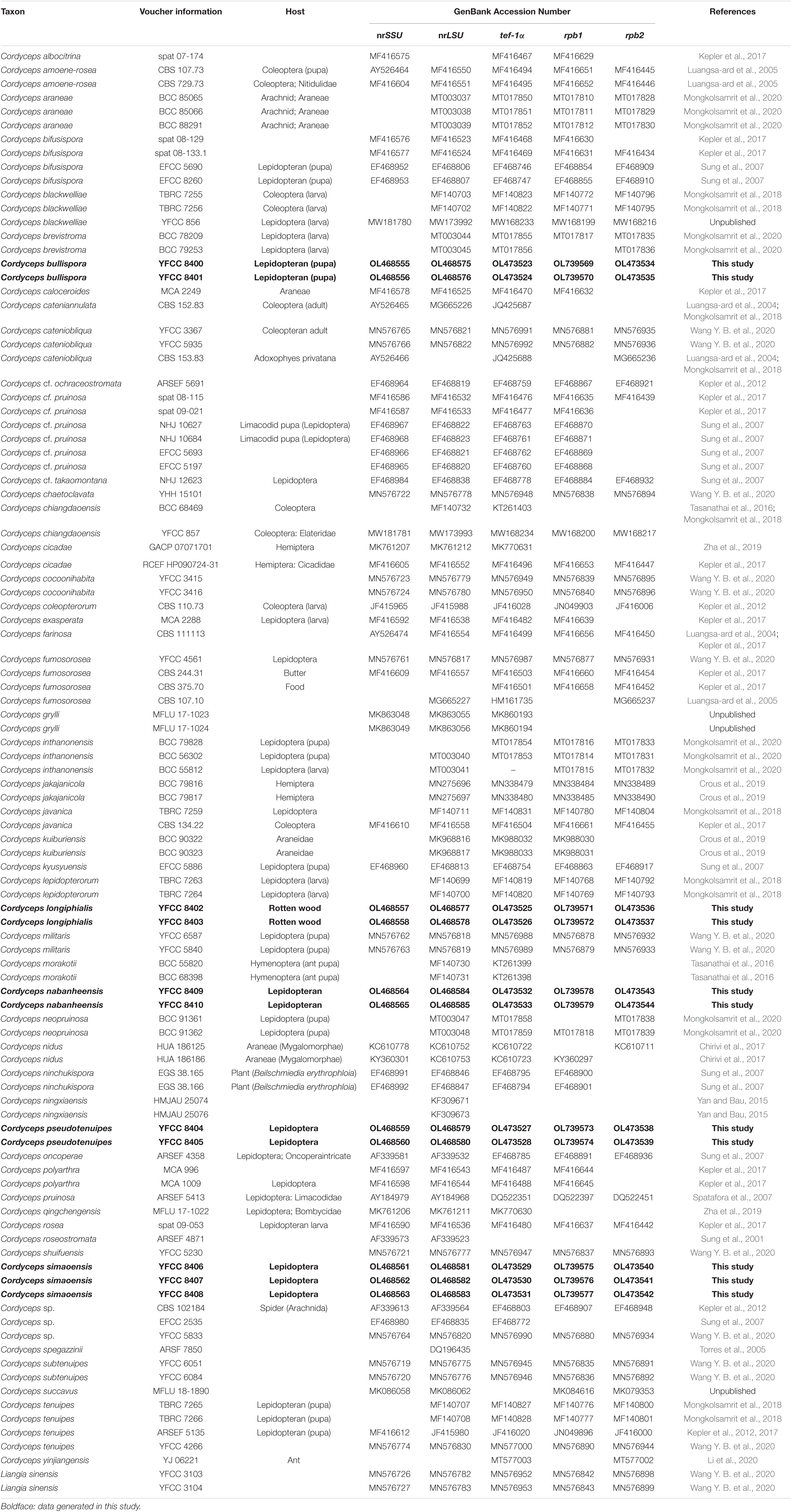
Table 1. Names, voucher information, host, and corresponding GenBank accession numbers of the taxa used in this study.
Phylogenetic Analyses
Phylogenetic trees were visualized with FigTree v1.4.0 (Rambaut, 2006) and edited in Microsoft PowerPoint, then saved as .PDF format and finally converted to .JPG format using Adobe Illustrator CS6 (Adobe Systems Inc., United States). The finalized alignments and trees were submitted in TreeBASE (Submission ID: 29339).
Phylogenetic analyses of the concatenated five-gene datasets were conducted using ML and BI methods. The GTR + I + G were chosen as the best models for nrSSU-nrLSU-tef-1α-rpb1-rpb2, using the Akaike Information Criterion (AIC) implemented in MrModeltest v 2.3 (Nylander, 2004), and then the partitioned analyses were separately conducted. For ML analyses, raxml v 8.2.7 was employed. All parameters were kept as default with an exception that the model was chosen as GTRGAMMAI. The statistic supports were calculated using 1,000 replicates of non-parametric bootstrapping. BI analysis was carried out with MrBayes v 3.2.6 using the selected models for 5 million generations with the value of stopval set to 0.01 via the stoprul command. At the same time, other parameters were kept as default and trees were summarized. Statistic supports were obtained using sumt command complemented in MrBayes by discarding the first 25% generations as burn-ins. The Bayesian trees were sampled every 100 generations. The first 25% trees were discarded as burn-ins, and the remaining trees were employed to create a consensus tree using sumt command.
Results
Sequence Alignment
The combined 101-taxon 5-gene dataset consisted of 4,627 base pairs of sequence data (nrSSU 1060 bp, nrLSU 877 bp, tef-1α 999 bp, rpb1 719 bp, and rpb2 972 bp). A total of 1,039 were parsimony-informative (nrSSU 45 bp, nrLSU 77 bp, tef-1α 370 bp, rpb1 234 bp, and rpb2 313 bp). A total of 101 taxa were complete for all five genes, and the number of taxa for each gene was as follows: nrSSU 71 taxa, nrLSU 97 taxa, tef-1α 95 taxa, rpb1 82 taxa, and rpb2 72 taxa (Table 1).
Molecular Phylogeny
In this study, we generated nrSSU, nrLSU, tef-1α, rpb1, and rpb2 sequences by ten living cultures and one wild material, and their accession numbers are shown in Table 1. Sequences of Liangia sinensis YFCC 3103 and YFCC 3104 in the Cordycipitaceae were chosen as outgroups in the phylogenetic analyses.
Five major (I–IV) clades and five new species could be recognized in Cordyceps s. s. (Figure 1); collections from southwestern China were grouped into five separate species (in boldface, see below) (C. bullispora, C. longiphialis, and C. nabanheensis in clade I, and C. pseudotenuipes and C. simaoensis in clade III). Clade I included C. pruinose and 19 other species, with 98% bootstrap support and 1 Bayesian PP support (Figure 1). In clade II, C. militaris and eight other species were grouped together (BS = 70%, PP = 1) (Figure 1). Clade III harbored C. tenuipes and 18 other taxa (BS = 97%, PP = 1) (Figure 1). Clade IV included C. cf. takaomontana NHJ 12623, C. javanica, C. amoenerosea, and C. cateniobliqua (BS = 91%, PP = 1) (Figure 1). Clade V included only two exemplars of the species, C. grylli (Figure 1).
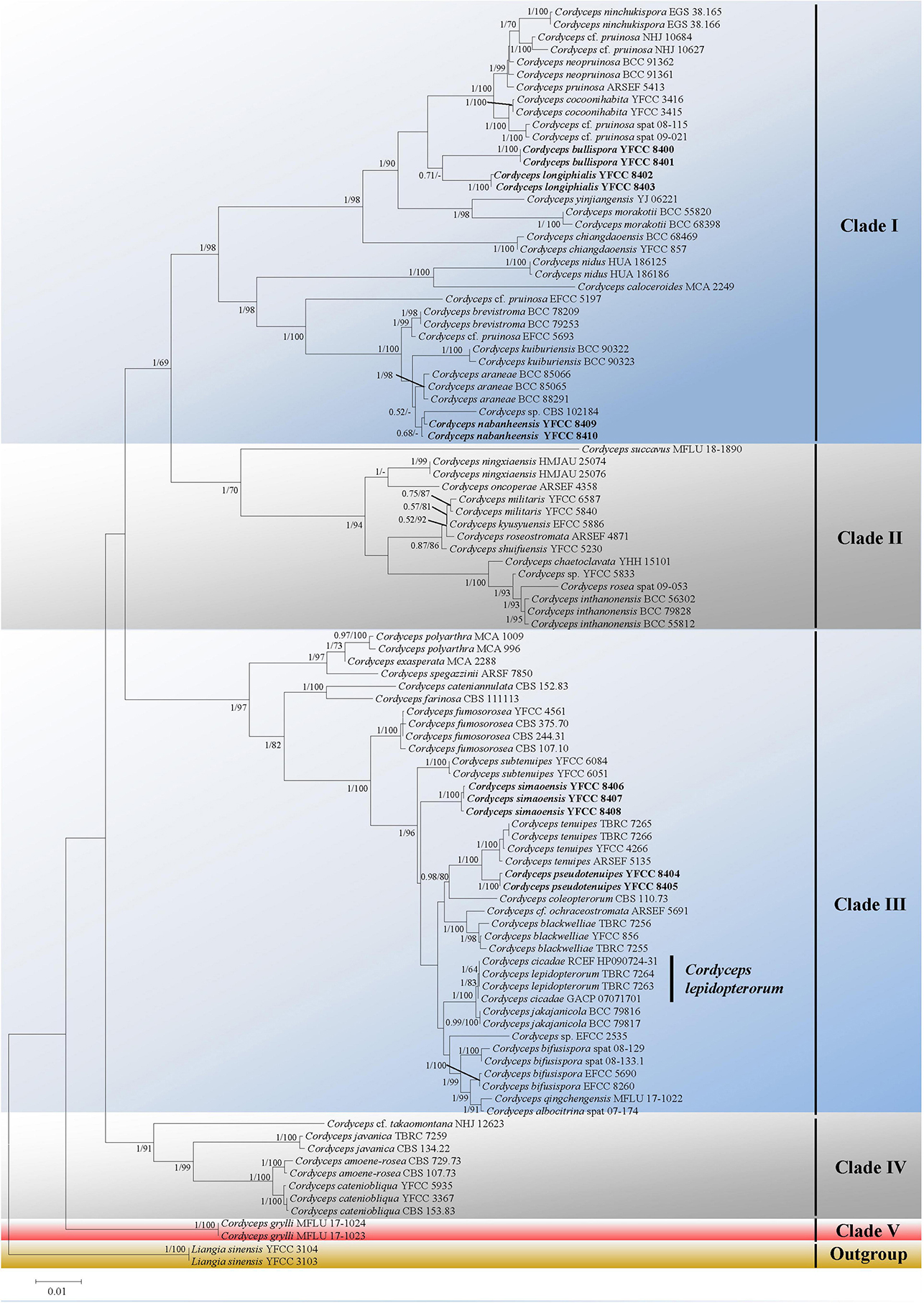
Figure 1. Both ML and BI analyses generate a phylogenetic tree from the concatenated nrSSU, nrLSU, tef-1α, rpb1, and rpb2 datasets. There were no discrepancies between the topology resulting from Bayesian and ML analysis for supported nodes. Bootstrap values (≥60%) derived from ML analyses and posterior probabilities from Bayesian inference (≥0.50) are shown either above or beneath the branches of the nodes. Isolates in bold type are those analyzed in this study.
Taxonomy
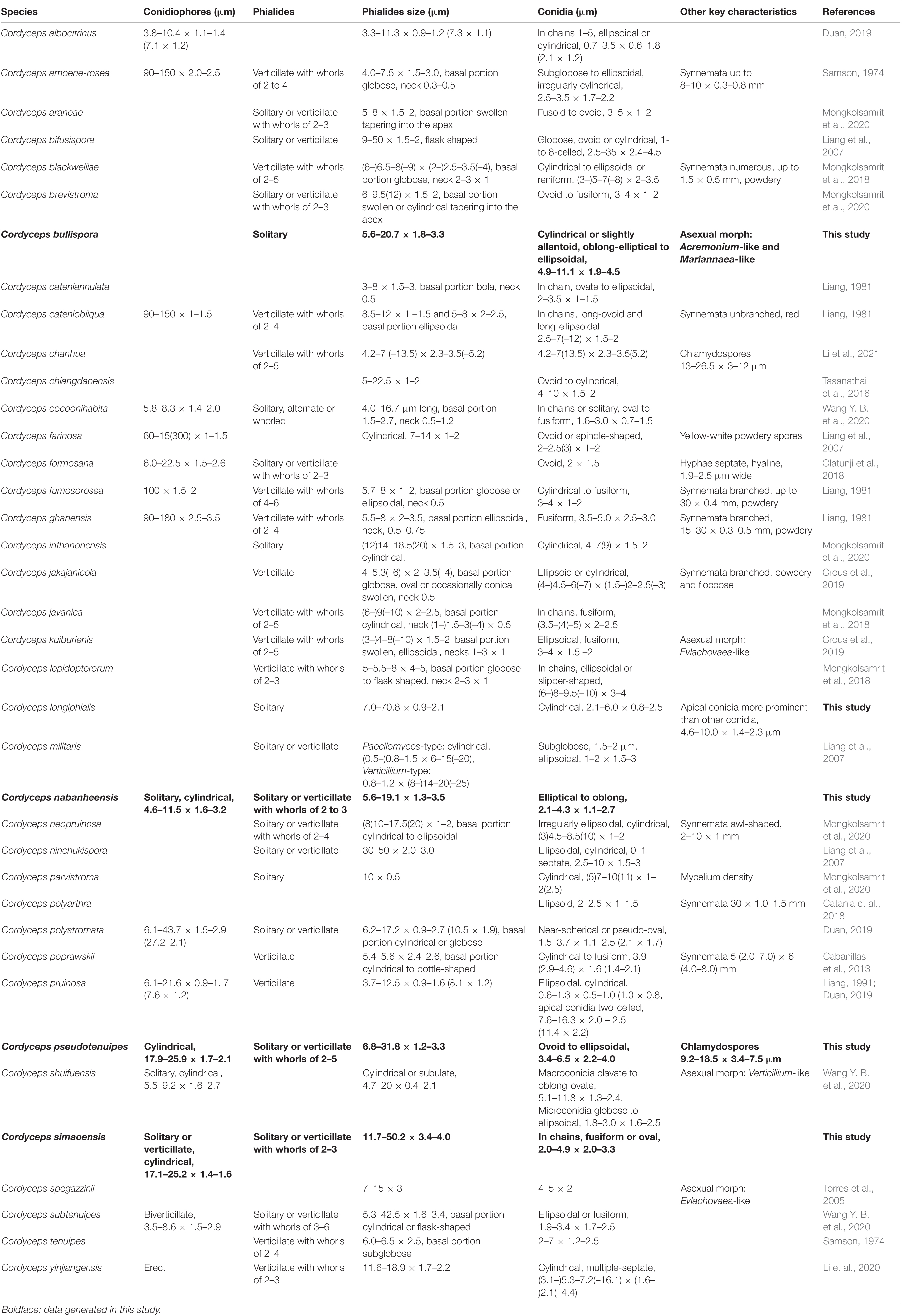
The key morphological characteristics that distinguish current Cordyceps s. s. species are summarized in the literature (Tables 2, 3). Including the five new species, there are 66 species of Cordyceps s. s. involved in the current study, among which we have compared 51 species of the sexual morphs in Cordyceps s. s. in Table 2 and 38 species of the asexual morphs in Cordyceps s. s. in Table 3.
Cordyceps bullispora H. Yu, Q. Y. Dong and Z. Y. Zhao, sp. nov. (Figure 2)
Mycobank: MB 842328
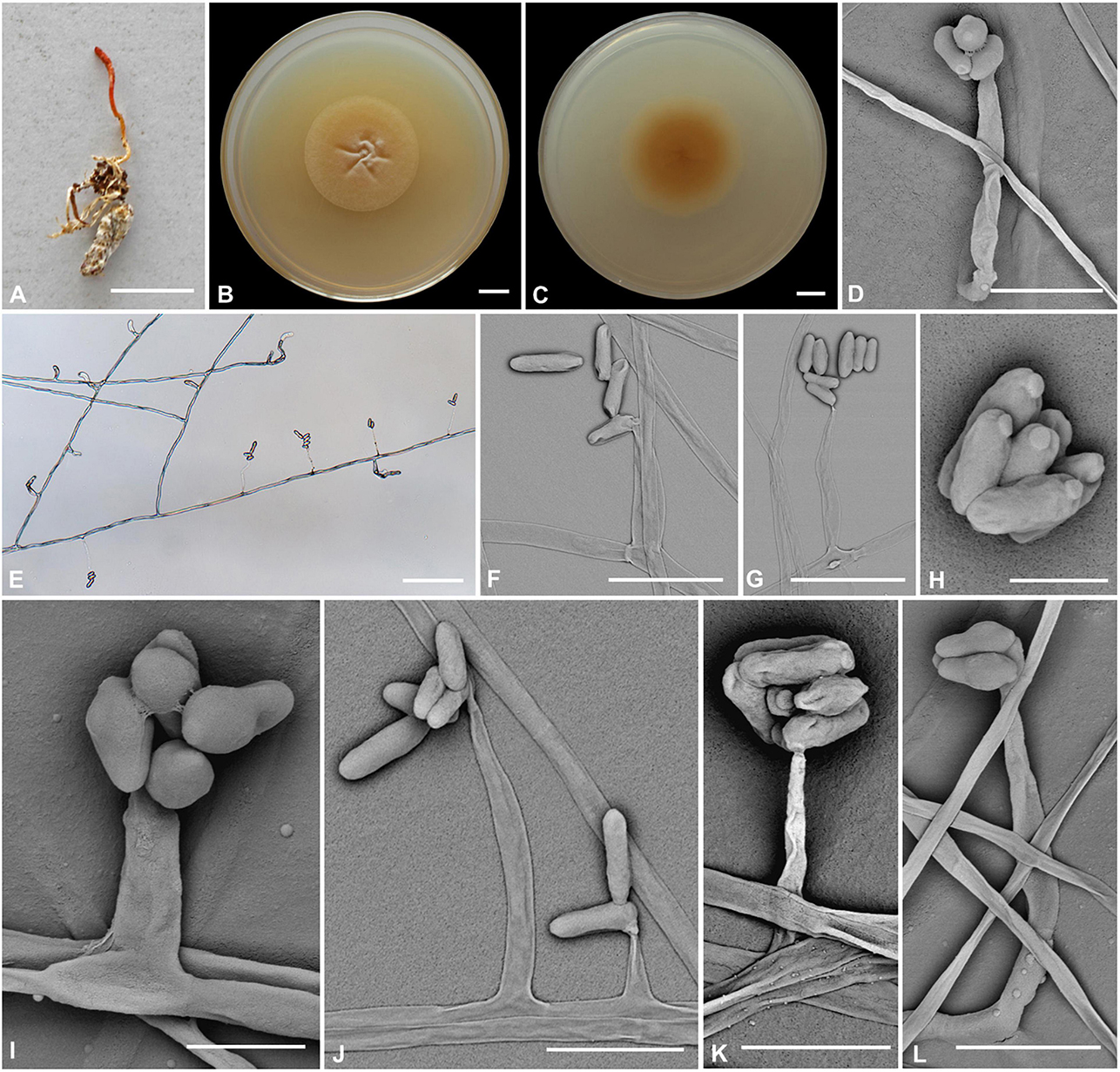
Figure 2. Cordyceps bullispora. (A) Fungus on the pupa of Lepidoptera. (B,C) Colony obverse (B) and reverse (C) on PDA at 21 days. (D–G,I–L) Phialides. (H) Conidia. Scale bars: (A) = 10 mm; (B,C) = 10 mm; (D) = 25 μm; (E) = 50 μm; (F,G) = 15 μm; (H) = 3 μm; (I) = 5 μm; (J) = 15 μm; (K,L) = 10 μm.
Etymology: Referring to button-like structures on the spores.
Type: The Taiji Mountains Nature Reserve, Mizhi Town, Midu County, Yunnan, China. September 20, 2019, H. Yu (YHH 20011, holotype; YFCC 8400, ex-holotype living culture).
Teleomorph: Stromata solitary, 10–20 mm long, unbranched, orange-yellow, cylindrical to enlarging apically. The host is covered by a white mycelial surface. Rhizoids flexuous, arising from the head region of host larva buried in soil, 7–10 mm deep under the ground. Stipes cylindrical, white to reddish-orange, 0.1–1.2 mm wide. Fertile parts clavate, orange to reddish-orange, 3.5–7.4 × 0.5–1.5 mm.
Anamorph: Two types of conidial arrangement. Acremonium-like conidia aggregated in heads at the apex of phialides; Mariannaea-like, conidia in imbricate chains, connected laterally.
Colonies on PDA are moderately fast-growing, attaining a diameter of 31–34 mm in 21 days at 25°C, pulvinate, with high mycelial density, Whitish to orange-yellow, reverse deep yellow. Hyphae smooth-walled, branched, septate, hyaline, 1.1–2.7 μm wide. Cultures readily produced phialides and conidia after 2 weeks on potato dextrose agar at room temperature. Phialides arising from aerial hyphae, solitary, 5.6–20.7 × 1.8–3.3 μm, cylindrical, tapering gradually or abruptly toward the apex. Conidia hyaline, one-celled, cylindrical or slightly allantoid, oblong-elliptical to ellipsoidal, 4.9–11.1 × 1.9–4.5 μm.
Habitat and known distribution: On larva of Lepidoptera buried below ground at elevation 2000 m in northwestern Yunnan, China.
Additional specimens examined: The Taiji Mountains Nature Reserve, Mizhi Town, Midu County, Yunnan, China. On pupae of Lepidoptera, September 20, 2019. YHH 20012, YFCC 8401.
Comments: Cordyceps bullispora was characterized by unbranched stromata, with cylindrical and orange to reddish-orange fertile parts, Rhizoids flexuous, and the host was the lepidopteran larva. For timing reasons, the fertile part of the specimen was not yet mature at the time of collection in the field. The asexual morph of PDA culture produces cylindrical phialides, which are monothetic, oblong-elliptical to ellipsoidal conidia with a button-like shape.
Based on nrLSU, nrSSU, tef-1α, rpb1, and rpb2 multigene analyses, C. cocoonihabita was revealed to have a close relationship with C. pruinosa and C. ninchukispora (Wang Y. B. et al., 2020). Multigene analyses of ITS, nrLSU, rpb1, rpb2, and tef-1α revealed that C. neopruinosa had a close relationship with C. pruinosa and C. ninchukispora (Mongkolsamrit et al., 2020). C. cocoonihabita, C. neopruinosa, C. pruinosa, and C. ninchukispora all had close relationships, where they shared many similar morphological characteristics, such as they were all characterized by orange- to red-colored stromata and superficial perithecia. C. bullispora shared such features, and our phylogenetic analysis indeed indicated that C. bullispora was closely relevant to a previously undescribed taxon C. cf. pruinosa (spat 08-115, spat 09-221) and was separated from C. cocoonihabita, C. neopruinosa. C. pruinosa, and C. ninchukispora in this subclade. However, the perithecia in C. neopruinosa were more prolonged and broader than those reported in Cordyceps pruinosa and C. ninchukispora (330–450 × 150–240 μm vs. 400 × 100 μm vs. 95–145 × 50–60 μm, respectively). C. cocoonihabita had a longer stroma. The micromorphological arrangement of conidia was Isaria-like characteristics and was significantly different from C. pruinosa and C. ninchukispora, which had respective morphs of Mariannaea G. Arnaud and Acremonium Link. C. bullispora had rhizoid stromata, superficial perithecia, wider phialides, and longer conidia 4.9–11.1 × 1.9–4.5 μm. The insect host of C. bullispora and C. neopruinosa all occurred on lepidopteran pupae, and the host of C. ninchukispora was the seed of Beilschmiedia Nees (Liang, 1983, 1991; Su and Wang, 1986; Mongkolsamrit et al., 2020).
Cordyceps yinjiangensis Li et al. (2020) was recently described from Guizhou. Morphologically, it differed from C. bullispora by cylindrical and orange to reddish-orange fertile parts, rhizoids several, flexuous, and the host was the lepidopteran larva. The asexual morph from PDA culture produced cylindrical phialides, which were monothetic and oblong-elliptical to ellipsoidal conidia with a button-like character. Tasanathai et al. (2016) reported an anti-pathogenic species, C. morakotii. C. yinjiangensis had a close relationship with C. morakotii with ant host and conidia formed in an imbricate chain. However, C. yinjiangensis was distinct from C. morakotii, which had longer phialides (16–20 × 2–3 μm) and bigger aseptate conidia (4–12 × 1–2 μm) (Li et al., 2020).
Cordyceps longiphialis H. Yu, Q. Y. Dong and D. X Tang, sp. nov. (Figure 3)
MycoBank: MB 842329
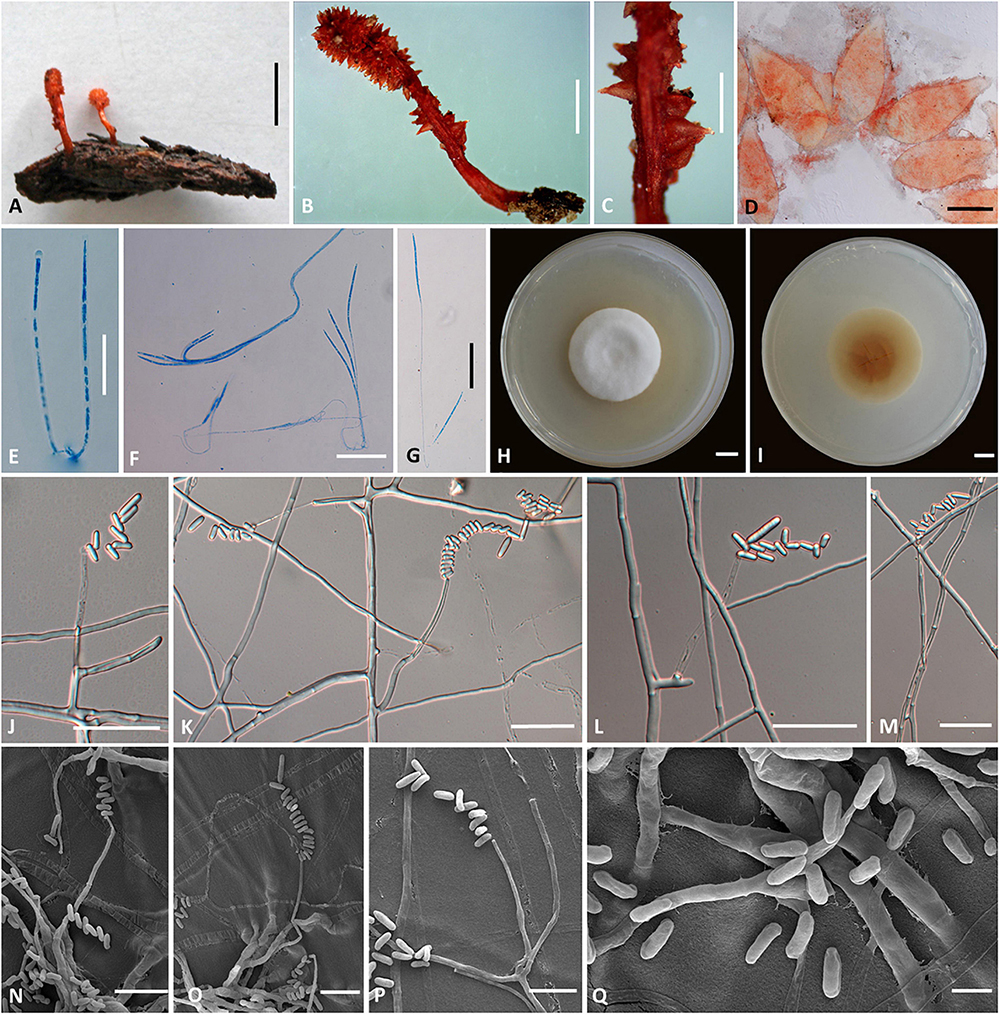
Figure 3. Cordyceps longiphialis. (A,B) Fungus on the host of Lepidoptera. (C) Fertile part. (D) Perithecia. (E) Asci. (F,G) Ascospore. (H,I) Colony obverse (H) and reverse (I) on PDA at 14 days. (J–P) Phialide. (Q) Conidia. Scale bars: (A) = 5 mm; (B) = 2 mm; (C) = 1 mm; (D) = 200 μm; (E–G,J–M) = 20 μm; (H,I) = 10 mm; (N–P) = 10 μm; (Q) = 5 μm.
Etymology: Referring to its longer phialide than close relationship species in this genus.
Type: Xinfang Reservoir, Simao District, Pu’er City, Yunnan, China. September 28, 2020, H. Yu (YHH 20013, holotype; YFCC 8402, ex-holotype living culture).
Teleomorph: Stromata (Figures 3A,B) arising from rotten wood, two, unbranched, 13–25 mm long. Stipe cylindrical, 4–10 mm long, 1.5–2 mm in diameter, orange-red to crimson, fleshy, glabrous, smooth. Fertile part single, clavate, covered by a spinous surface, 4–15 mm long, 2.0–2.5 mm in diameter, reddish-orange. Perithecia (Figure 3D) crowded or sparse, crimson-yellowish, superficial, vase-form to oblong, 380–612 × 167.5–268.3 μm. Asci (Figure 3E) 8-spored, fusiform to cylindrical, 113–200 × 1.1–2.7 μm when mature; Ascus caps hemispherical, 1.6–3.6 μm in height, 1.7–3.3 μm in width. Ascospores hyaline, bola-shaped, septate, 110–184 × 0.8–1.3 μm, central region filiform, terminal region narrowly fusiform, do not disarticulate into part-spores.
Anamorph: Conidial arrangement Mariannaea-like. Colonies on PDA are fast-growing, attaining a diameter of 35–37 mm in 14 days at 25°C, white to pale yellow, cottony, with high mycelial density at the centrum, reverse white to pale yellow. Hyphae smooth-walled, septate, hyaline, 0.8–2.5 μm wide. Cultures readily produced phialides and conidia after 2 weeks on potato dextrose agar at room temperature. Phialides usually solitary on hyphae, basal portion cylindrical to clavate, tapering gradually toward the apex; 7.0–70.8 μm long, 0.6–2.1 μm wide at the base, 0.9–2.1 μm at the middle, and 0.6–1.8 μm wide at the apex. Conidia one-celled, smooth-walled, hyaline, cylindrical, 2.1–6.0 × 0.8–2.5 μm, often formed in an imbricate chain, the size of apical conidia significantly more prominent than other conidia in the chain, 4.6–10.0 × 1.4–2.3 μm.
Habitat and known distribution: Buried in rotten logs below ground, in northwestern Yunnan, China.
Additional specimens examined: Xinfang Reservoir, Simao District, Pu’er City, Yunnan, China, isolated from stromata of rotten wood at elevation 1,350 m. September 28, 2020, H. Yu (YFCC 8403, ex-holotype living culture).
Comments: Phylogenetic analyses showed that C. longiphialis was closely related to C. bullispora; however, the independent phylogenetic position and different physiological characteristics could distinguish C. longiphialis from its sister species, C. bullispora (as mentioned above). The distinctive characteristics of Cordyceps longiphialis were the cylindrical stromata with a spinous surface, superficial perithecia, much shorter ascospores (110–184 × 0.8–1.3 μm), and much longer phialides (7.0–70.8 μm long). Ascospores of C. longiphialis were shorter than C. neopruinosa (135–275 × 0.5 μm) and C. pruinose (185–200 × 2 μm), but longer than C. ninchukispora (90–110 × 1.2 μm). The size of the conidia in C. longiphialis was relatively shorter than C. bullispora.
Cordyceps nabanheensis H. Yu and Q. Y. Dong, sp. nov. (Figure 4)
MycoBank: MB 842341
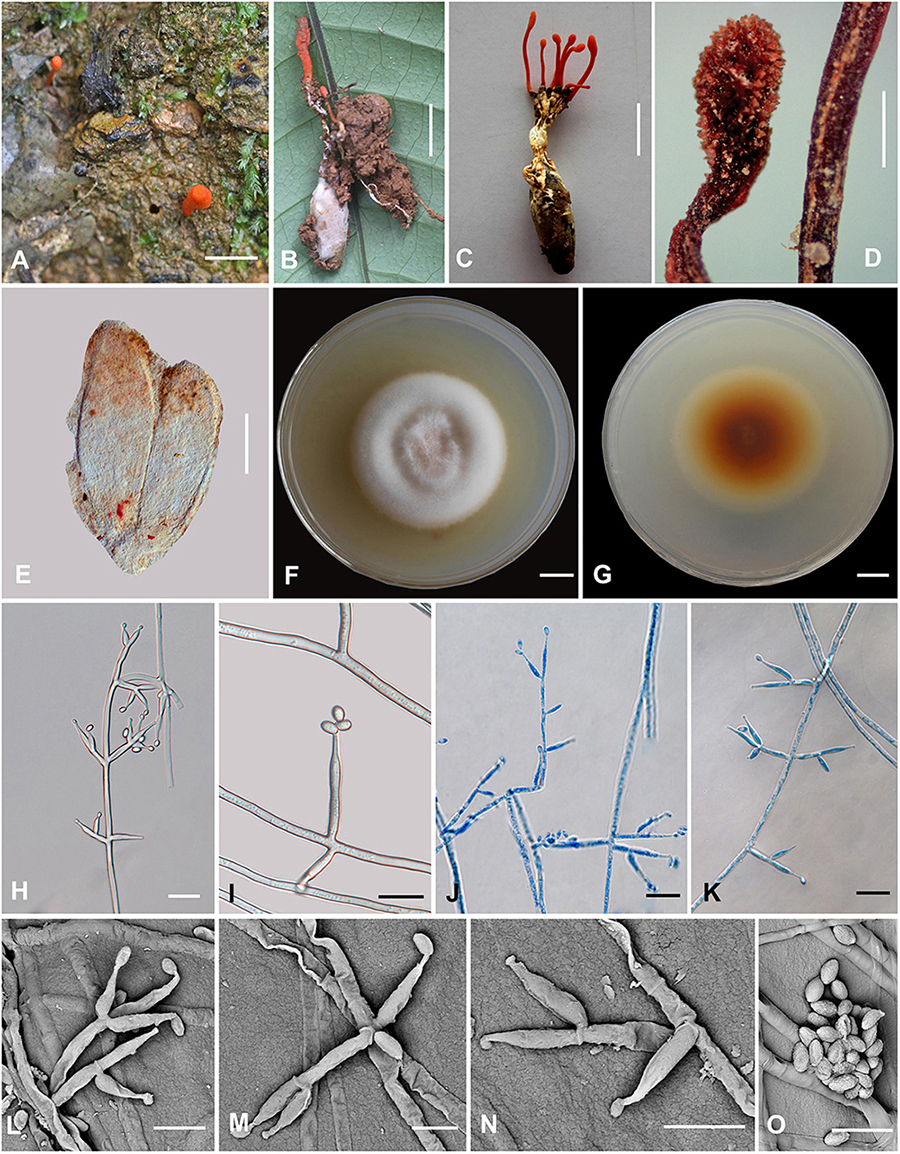
Figure 4. Cordyceps nabanheensis. (A–C) Fungus on the pupa of Lepidoptera. (D) Fertile part. (E) Perithecia. (F,G) Colony obverse (F) and reverse (G) on PDA at 14 days. (I) Solitary phialide on hyphae. (H,J–N) Opposite conidiophore and verticillate phialides. (O) Conidia. Scale bars: (A–C) = 10 mm; (D) = 1 mm; (E) = 100 μm; (F,G) = 10 mm; (H,J,K) = 20 μm; (I) = 10 μm; (L–O) = 5 μm.
Etymology: The location in Nabanhe National Nature Reserve where the species was collected.
Type: Manlv village, Nabanhe National Nature Reserve, Jinghong City, Yunnan, China. August 16, 2020, H. Yu (YHH 20019, holotype; YFCC 8409, ex-holotype living culture).
Teleomorph: Stromata arising from the pupa of Lepidoptera buried in soil, the host covered by a white mycelial surface, solitary or gregarious, cylindrical to enlarging apically, reddish-orange to crimson, 1.4–2.3 cm long. Rhizoids flexuous, arising from the head region of host larva buried in soil, 7–12 mm deep under the ground. Stipes cylindrical, 12.0–20.9 × 0.6–1.3 mm, fertile parts clavate, 2.0–6.5 × 0.7–1.5 mm. Perithecia superficial, oblong-ovate, 224–322.6 × 71.2–317.3 μm. Asci and ascospores were not observed.
Anamorph: Conidial arrangement Evlachovaea-like. Colonies on PDA moderately fast-growing, 48–51 mm diameter in 14 days at 25°C, floccose, with high mycelium density; white to orange pinkish, reverse orange-brown. Hyphae smooth-walled, branched, septate, hyaline, septate, 3.5–9.3 μm wide. Cultures readily produced phialides and conidia after 1 week on potato dextrose agar at room temperature showing a powdery appearance due to profuse conidiation. Conidiophores smooth-walled, solitary, cylindrical, 4.6–11.5 × 1.6–3.2 μm. Phialides arising from aerial hyphae, cylindrical or clavate, solitary or in whorls of two to three, tapering abruptly into a narrow neck, 5.6–19.1 × 1.3–3.5 μm. Conidia one-celled, smooth-walled, hyaline, elliptical to oblong, 2.1–4.3 × 1.1–2.7 μm, often formed in an imbricate chain.
Habitat and known distribution: On larvae of Lepidoptera buried below ground at elevation 600 m in northeastern Yunnan, China.
Additional specimens examined: Manlv village, Nabanhe National Nature Reserve, Jinghong City, Yunnan, China, on larvae of Lepidoptera. August 16, 2020 (YHH 20020, paratype; YFCC 8410 ex-paratype living culture).
Comments: Cordyceps araneae was firstly reported from Khon Kaen Province, northeastern Thailand by Mongkolsamrit et al. (2020); C. araneae was a spider cocoon pathogenetic fungus producing pale orange stromata, perithecia semi-immersed, narrowly ovoid, 450–500 × 150–200 μm with whole bola-shaped ascospores breaking into part-spores 30–65 × 0.5 μm, and developed the Evlachovaea-like anamorph, phialides solitary or in whorls of two to three, 5–8 × 1.5–2 μm, conidia fusoid to ovoid, 3–5 × 1–2 μm (Mongkolsamrit et al., 2020).
Based on the ITS, ribosomal large subunit, rpb1, rpb2, and tef-1α genes, multigene analyses revealed that C. araneae had a close relationship with C. kuiburiensis, C. brevistroma, and C. nidus, and they were all characterized by orange to reddish-orange, cylindrical to enlarging apically stromata and a conidial arrangement Evlachovaea-like (Chirivi et al., 2017; Crous et al., 2019; Mongkolsamrit et al., 2020). Interestingly, C. nabanheensis shared such features, and our phylogenetic analysis indeed indicated that C. nabanheensis had a close relationship to C. araneae and C. brevistroma. However, C. nabanheensis and C. brevistroma differed from C. araneae and C. kuiburiensis regarding their hosts. Both C. nabanheensis and C. brevistroma occur on Lepidoptera larvae, whereas both C. araneae and C. kuiburiensis occurred on spiders. C. brevistroma had bola-shaped whole ascospores, which was the same shape but shorter than C. araneae reported (150–200 μm vs. 250–400 μm). In the natural specimen, C. kuiburiensis developed the anamorph.
Cordyceps pseudotenuipes H. Yu, Q. Y. Dong, and Y. Wang, sp. nov. (Figure 5)
Mycobank: MB 842330
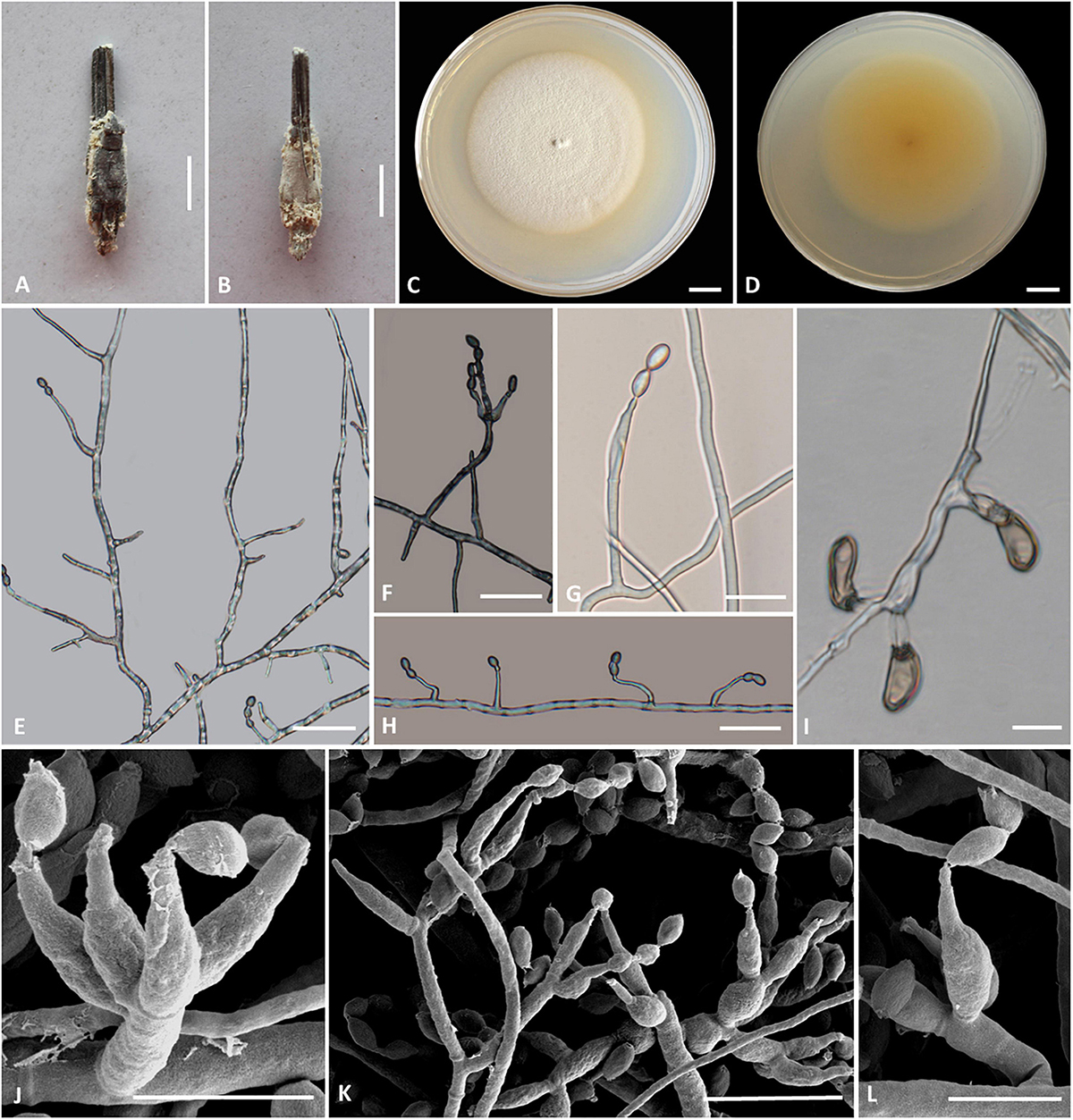
Figure 5. Cordyceps pseudotenuipes. (A,B) Fungus on the host of Coleopteran. (C,D) Colony obverse (C) and reverse (D) on PDA at 14 days. (E,G,H,L) Solitary phialides on hyphae. (F,J,K) Conidiophore and verticillate phialides. (I) Chlamydospore. Scale bars: (C,D) = 5 mm; (E,F,G) = 25 μm, (H,I) = 20 μm; (J,L) = 5 μm; (K) = 10 μm.
Etymology: Referring to morphological resemblance of Cordyceps tenuipes and Cordyceps subtenuipes but phylogenetically distinct, “not” C. tenuipes.
Type: Wild Duck Lake Forest Park, Shuanglong Town, Panlong District, Kunming City, Yunnan, China. September 10, 2019, H. Yu and Y. Wang (YHH 20014, holotype; YFCC 8404, ex-holotype living culture).
Teleomorph: Undetermined.
Anamorph: Conidial arrangement Isaria-like. Synnemata arising from the pupae of Lepidoptera buried in soil, synnemata erect, solitary, flexuous, white, up to 0.5 cm long. Stipes cylindrical, 0.5 mm wide, producing a mass of conidia at the branches of synnemata, powdery.
Colonies on PDA attaining a diameter of 53–55 mm in 14 days at 25°C, white to cream-colored, soft cottony aerial mycelium, reverse pale yellow. Hyphae smooth-walled, branched, septate, hyaline. Synnemata arising from the entire body of larvae were irregularly branched, 0.2–1.0 cm long, 0.1–0.3 mm wide; cylindrical or clavate stipes with powdery white heads. Cultures readily produced phialides and conidia after 2 weeks on potato dextrose agar at room temperature showing a granular appearance due to profuse conidiation. Conidiophores cylindrical, hyaline, smooth-walled, 17.9–25.9 × 1.7–2.1 μm. Phialides from aerial mycelium straight to slightly flexuose, solitary or in whorls of two to five on each branch, cylindrical, usually with a slightly swollen basal part, tapering into the apex, 6.8–31.8 × 1.2–3.3 μm. Conidia hyaline, ovoid to ellipsoidal, smooth, one-celled, 3.4–6.5 × 2.2–4.0 μm. Chlamydospores present, one-celled, solitary, eggplant shape or oval to pyriform, 9.2–18.5 × 3.4–7.5 μm, hyaline becoming brown, thick and smooth-walled.
Habitat and known distribution: On the pupa of Lepidoptera buried in the soil. Kunming City, China.
Additional specimens examined: Wild Duck Lake Forest Park, Shuanglong Town, Panlong District, Kunming City, Yunnan, China. September 10, 2019, H. Yu (YHH 20015, paratype; YFCC 8405, ex-paratype living culture).
Comments: Phylogenetically, C. pseudotenuipes formed a separate subclade from the other species of Cordyceps with high credible support (100%). C. pseudotenuipes was similar to C. tenuipes (Peck) Kepler et al. (2017) based on its conspicuous synnemata and Isaria-like asexual conidiogenous structure forming phialides with a swollen basal portion. It differed from C. tenuipes by its unbranched synnemata, white color, phialides with a globose basal part, and smaller ovoid to ellipsoidal wider conidia measuring 3.4–6.5 × 2.2–4.0 μm. C. tenuipes had multiple synnemata, more giant cylindrical to botuliform conidia with a size of 2.0–7.0 × 1.2–2.5 μm (Samson, 1974). The sexual morph of C. tenuipes was proposed as C. takaomontana Yakush and Kumaz, with yellowish stromata and being often concurrent with its asexual morph (Liang et al., 2007). However, the sexual morph of C. pseudotenuipes was not found in the field.
Cordyceps simaoensis H. Yu, Q. Y. Dong and Z. Q. Wang, sp. nov. (Figure 6)
MycoBank: MB 842331
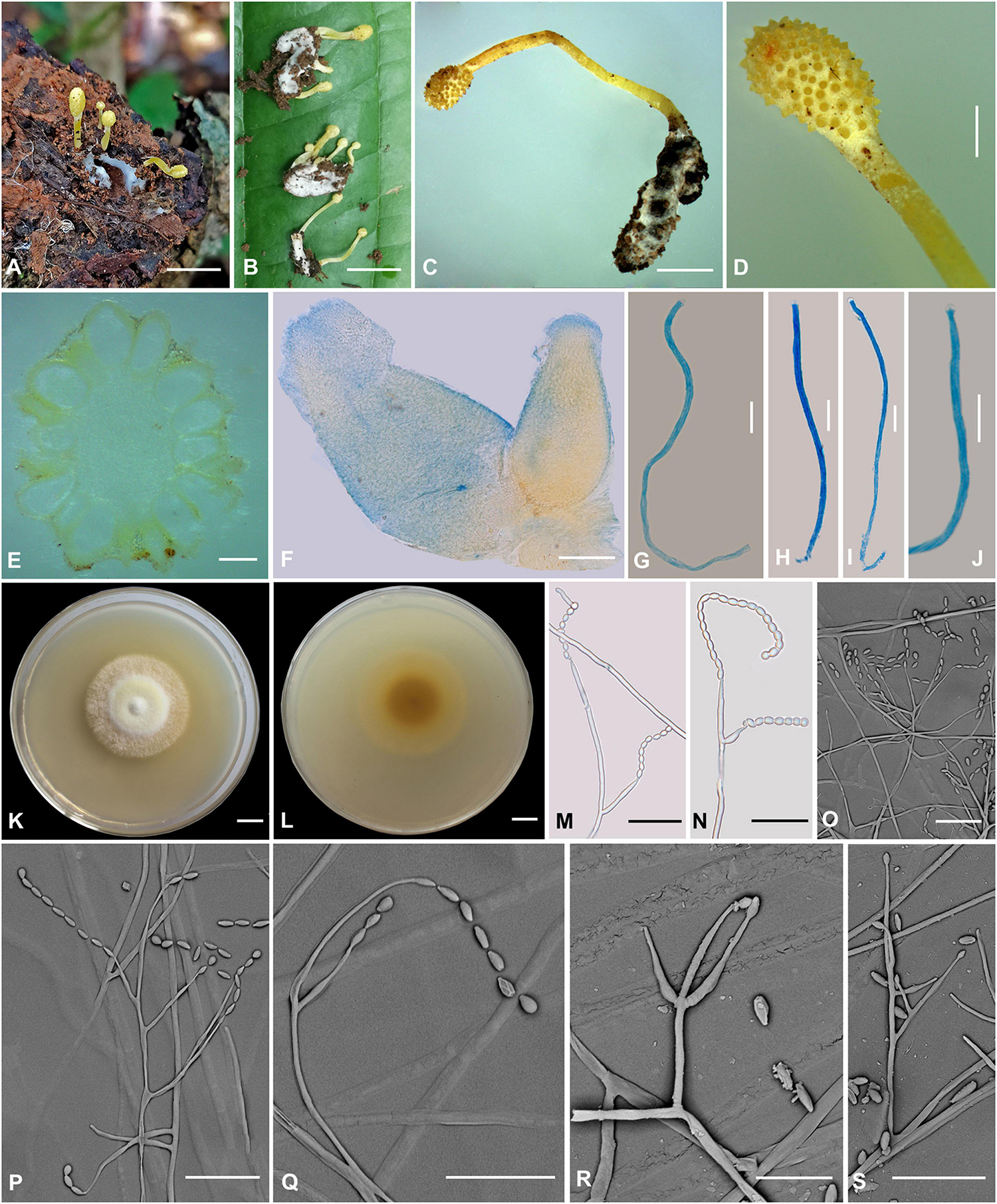
Figure 6. Cordyceps simaoensis. (A–C) Fungus on the host of Lepidoptera. (D) Fertile part. (E,F) Perithecia. (G–J) Asci. (K,L) Colony obverse (K) and reverse (L) on PDA at 21 days. (M–S) Conidiophore and phialide. Scale bars: (A,B) = 10 mm; (C) = 5 mm; (D) = 2 mm; (E) = 400 μm; (F) = 100 μm; (G–J) = 10 μm; (K,L) = 10 mm; (M–S) = 20 μm.
Etymology: The location in Simao District where the species was collected.
Type: Xinfang Reservoir, Simao District, Pu’er City, Yunnan, China. September 28, 2020. H. Yu (YHH 20016, holotype; YFCC 8406, ex-holotype living culture).
Teleomorph: Stroma arising from the host’s head, solitary or gregarious, mace-shaped, unbranched, 7–25.1 mm in height. Stipe cylindrical, 22 mm long, 1.1–1.2 mm in diameter, bright yellow, fleshy, glabrous, smooth, enlarging abruptly at fertile portion. Fertile portion single, elliptical to fusiform, 1–4.2 mm long, 1.5–3 mm in diameter, bright yellow. Perithecia (Figure 6E) crowded, nearly fully immersed, vase-form, oval to oblong, 638.4–757.6 × 371–531.1 μm, ostioles protruding. Asci (Figures 6G–J) 8-spored, narrowly clavate to nearly cylindrical, 66.9–126.1 × 1.9–2.7 μm; cap 1.2–2.3 μm in height, 2.1–3.5 μm in width. Ascospores not observed.
Anamorph: Conidial arrangement Isaria-like. Colonies on PDA are fast-growing, attaining a diameter of 40–43 mm at 25°C in 21 days, white to bright yellow, cottony, with high mycelial density at the centrum, forming concentric rings around the inoculum, reverse pale yellow to deep yellow. Hyphae smooth-walled, branched, septate, hyaline, 2.1–2.9 μm wide. Cultures produced phialides and conidia after 45 days on potato dextrose agar at room temperature. Conidiophores are smooth-walled, cylindrical, solitary, or verticillate, 17.1–25.2 × 1.4–1.6 μm. Phialides solitary or verticillate, in whorls of two to three, usually solitary on hyphae, basal portion cylindrical to narrow lageniform, gradually or abruptly tapering toward the apex; 11.7–50.2 μm long, 1.5–3.1 μm wide at the base, 3.4–4.0 μm at the middle, and 0.9–2.0 μm wide at the apex. Conidia is one-celled, hyaline, smooth-walled, fusiform or oval, 2.0–4.9 × 2.0–3.3 μm, often in chains.
Habitat and known distribution: On the pupa of Lepidoptera buried in soil.
Additional specimens examined: Xinfang Reservoir, Simao District, Pu’er City, Yunnan, China. October 6, 2019, H. Yu (YHH 20017 paratype, YFCC 8407 ex-paratype living culture; YHH 20018 paratype, YFCC 8408 ex-paratype living culture).
Comments: Our phylogenetic results demonstrated that the isolated position of this collection was within the Cordyceps genus. C. simaoensis formed a separate subclade from other species of Cordyceps with moderately high credible support (100%). This species was mainly isolated from soil and produced Isaria-like asexual morphs and fusiform or oval conidia. C. simaoensis was closely related to C. tenuipes (Peck) Kepler et al. and might be identified by the size and presence of conidia and its distinct sexual morph. The conidial size of Cordyceps tenuipes was 2.0–7.0 × 1.2–2.5 μm, the wider conidia of Cordyceps pseudotenuipes was 3.4–6.5 × 2.2–4.0 μm, that of Cordyceps subtenuipes was 1.9–3.4 × 1.5–2.7 μm, and that of Cordyceps simaoensis was 2.0–4.9 × 2.0–3.3 μm. Cultures of C. simaoensis produced phialides and conidia, and it took more days than C. tenuipes. Our field observations and herbaria record also indicated that the C. tenuipes was widely distributed and significantly ecologically diverse in China, with small bright yellow fleshy stromata as teleomorph and Isaria-like anamorph.
Discussion
Considerable changes to the taxonomy of Cordyceps have occurred since the research on entomogenous fungi entered the molecular era. At present, multi-locus phylogenetic analyses have gained importance in delimiting Cordyceps species (Tasanathai et al., 2016; Zha et al., 2019; Li et al., 2020, 2021; Wang Y. B. et al., 2020). In this study, most species of the newly circumscribed genus Cordyceps were analyzed based on phylogenetic inferences of five nuclear molecular markers (nrSSU, nrLSU, tef-1α, rpb1, and rpb2). Both ML and BI analyses produced trees with similar topologies that resolved most Cordyceps lineages in separate terminal branches. Cordyceps s. s. was recognized by five statistically well-supported clades, designated as clade I, clade II, clade III, clade IV, and clade V (Figure 1). There were 20 species in clade I. Morphologically, the 20 species shared relatively complicated host, such as spider, Coleoptera, Lepidoptera, Limacodidae, ant, and even plants. They were also complex and varied in shape (ascospores bola-shaped, filiform, ninchukiform, or bifusiform). Clade II was made up of C. militaris and other closely related species. With the exception of C. kyusyuenis and C. rosea, all other species had filiform ascospores; in addition, unlike C. oncoperae and C. rosea, the rest of the species could easily disarticulate into part-spores. It was found that all the members in clade III had Isaria-like anamorphs and all four species in clade IV were described as conidiophores verticillate with phialides in whorls of 2 to 4 or 5, conidia in chains (Table 3). Clade V consisted solely of Cordyceps grylli, characterized as pathogenic on Gryllidae adults and relatively larger perithecia (up to 650–810 × 270–370 μm) (Teng, 1963; Liang et al., 2007).
Phylogenetic classifications of cordycepitoid fungi showed that most diagnostic characteristics used in current classifications of Cordyceps species (e.g., host, arrangement of perithecia, ascospores fragmentation, conidiogenous structures, conidial shape and size) were not phylogenetically informative (Sung et al., 2007; Kepler et al., 2017; Mongkolsamrit et al., 2018; Wang Y. B. et al., 2020). Cordyceps lepidopterorum Mongkolsamrit, Noisripoom, Thanakitpipattana, Spatafora and Luangsa-ard was firstly reported from Chiang Mai Province Thailand by Mongkolsamrit et al. (2018). Cordyceps chanhua, which had long been mistaken as Isaria (Paecilomyces) cicadae of a Brazilian specimen, was recently reported as a new species in the genus Cordyceps s. s., for the discovery of its teleomorph in Mt. Jinggang, Jiangxi, China and analyses of both morphological and phylogenetical evidence (Li et al., 2021). Our phylogenetic trees suggested that C. chanhua (see Cordyceps cicadae in Figure 1) could not be distinguished from C. lepidopterorum (Figure 1). Regarding the morphology, there were no significant differences in the morphological characteristics of anamorph between the two species except for their host (Table 3). Because C. lepidopterorum was described earlier than C. chanhua, C. lepidopterorum should be recommended as the scientific name for this species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
Q-YD and HY: conceptualization. Q-YD: methodology, writing—original draft preparation, and formal analysis. Q-YD and YW: software and resources.. Z-QW and H-JW: validation. Q-YD, YW, Z-QW, D-XT, Z-YZ, H-JW, and HY: investigation. HY: writing—review and editing and funding acquisition. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31870017 and 32060007) and the Department of Science and Technology of Yunnan Province [No. 2018FY001(-006)].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are very grateful to Yuan-Bing Wang and Nian-Kai Zeng (College of Pharmacy, Hainan Medical University, Haikou, China) for help in naming the new fungal species. Special thanks are due to the reviewers for constructive comments and suggestions for improving our work.
References
Bischoff, J. F., Rehner, S. A., and Humber, R. A. (2006). Metarhizium frigidum sp. nov.: a cryptic species of M. anisopliae and a member of the M. flavoviride Complex. Mycologia 98, 737–745. doi: 10.3852/mycologia.98.5.737
Cabanillas, H. E., De Leon, J. H., Humber, R. A., Murray, K. D., and Jones, W. A. (2013). Isaria poprawskii sp. nov. (Hypocreales: Cordycipitaceae), a new entomopathogenic fungus from Texas affecting sweet potato whitefly. Mycoscience 54, 158–169. doi: 10.1016/j.myc.2012.09.009
Castillo, L. P., Osorio, A., Vargas, N., Sanjuan, T., Grajales, A., and Restrepo, S. (2018). Genetic diversity of the entomopathogenic fungus Cordyceps tenuipes in forests and butterfly gardens in Quindio, Colombia. Fungal Biol. 122, 891–899. doi: 10.1016/j.funbio.2018.05.003
Catania, M., del, V., Sanjuan, T. I., and Robledo, G. L. (2018). South American Cordyceps s. l. (Hypocreales, Ascomycota): first assessment of species diversity in Argentina. Nova Hedwig 106, 261–281. doi: 10.1127/nova_hedwigia/2017/0434
Chen, W. H., Liu, C., Han, Y. F., Liang, J. D., Tian, W. Y., and Liang, Z. Q. (2019). Three novel insect-associated species of Simplicillium (Cordycipitaceae. Hypocreales) from Southwest China. MycoKeys 58, 83–102. doi: 10.3897/mycokeys.58.37176
Chen, Z. H., Wang, Y. B., Dai, Y. D., Chen, K., Xu, L., and He, Q. C. (2019). Species diversity and seasonal fluctuation of entomogenous fungi of Ascomycota in Taibaoshan Forest Park in western Yunnan. Biodivers. Sci. 27, 993–1001. doi: 10.17520/biods.2019135
Chirivi, J., Danies, G., Sierra, R., Schauer, N., Trenkamp, S., Restrepo, S., et al. (2017). Metabolomic profile and nucleoside composition of Cordyceps nidus sp. nov. (Cordycipitaceae): a new source of active compounds. PLoS One 12:e0179428. doi: 10.1371/journal.pone.0179428
Crous, P. W., Wingfield, M. J., Lombard, L., Roets, F., Swart, W. J., Alvarado, P., et al. (2019). Fungal Planet description sheets: 951–1041. Persoonia 43, 223–425. doi: 10.3767/persoonia.2019.43.06
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Duan, D. E. (2019). Taxonomy and Phylogeny of Cordycipitaceae from Vietnam. Master’s thesis. Kunming: Yunnan University.
Fang, D. J., Zheng, X. M., Liao, S. T., Chen, W. D., and Huang, Q. P. (1995). Studies on artificial culture of Cordyceps brasiliensis using the pupa of Bombyx mori. J. South. China Agri. Univ. 16, 103–108.
Fries, E. M. (1818). Observationes mycologicae Praecipue ad Illustrandam Floram Suecicam. Pars secunda (Cancellans issue). Copenhagen: G. Bonnieri.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hu, J. J., Zhao, G. P., Tuo, Y. L., Dai, D., Guo, D. Z., Rao, G., et al. (2021). Morphology and molecular study of three new Cordycipitoid fungi and its related species collected from Jilin Province, northeast China. MycoKeys 83, 161–180. doi: 10.3897/mycokeys.83.72325
Hyde, K. D., Tennakoon, D. S., Jeewon, R., Bhat, D. J., Maharachchikumbura, S. S., Rossi, W., et al. (2019). Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 96, 1–242. doi: 10.1007/s13225-019-00429-2
Kepler, R. M., Luangsa-Ard, J. J., Hywel-Jones, N. L., Quandt, C. A., Sung, G. H., Rehner, S. A., et al. (2017). A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8, 335–353. doi: 10.5598/imafungus.2017.08.02.08
Kepler, R. M., Sung, G. H., Ban, S., Nakagiri, A., Chen, M. J., Huang, B., et al. (2012). New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 104, 182–197. doi: 10.3852/11-070
Kobayasi, Y. (1981). Revision of the genus Cordyceps and its allies 1. Bull. Natn. Sci. Mus. Tokyo Ser. B 7, 1–13. doi: 10.5479/si.00963801.74-2752.1
Kobayasi, Y., and Shimizu, D. (1977). Some species of Cordyceps and its allies on spiders. Kew Bull. 31, 557–566. doi: 10.2307/4119402
Kobayasi, Y., and Shimizu, D. (1980). Cordyceps species from Japan 2. Bull. Natn. Sci. Mus. Tokyo Ser. B (Bot.) 6, 43–63.
Kobayasi, Y., and Shimizu, D. (1982). Cordyceps species from Japan 5. Bull. Natn. Sci. Mus. Tokyo Ser. B (Bot.) 8, 111–123.
Li, Y. P., Chen, W. H., Han, Y. F., Liang, J. D., and Liang, Z. Q. (2020). Cordyceps yinjiangensis, a new ant-pathogenic fungus. Phytotaxa 453, 284–292. doi: 10.11646/phytotaxa.453.3.10
Li, Z. Z., Luan, F. G., Hywel-Jones Nigel, L., Zhang, S. L., Chen, M. J., Huang, B., et al. (2021). Biodiversity of cordycipitoid fungi associated with Isaria cicadae Miquel II: Teleomorph discovery and nomenclature of Chanhua, an important medicinal fungus in China. Mycosystema 40, 95–107.
Liang, Z. Q. (1981). Entomogenous fungi from the pests of tea-plants. Acta Phytopathol. Sin. 11, 9–16.
Liang, Z. Q. (1983). A record and description on Cordyceps pruinosa Petch and its conidial state. J. Guizhou Agric. Coll. 2, 72–80.
Liang, Z. Q. (1991). Verification and identification of the anomorph of Cordyceps pruinosa Petch. Acta Mycol. Sin. 10, 104–107.
Liang, Z. Q., Liu, A. Y., Huang, J. Z., and Jiao, Y. C. (2002). Some Cordyceps species and their allies from Suoluo Nature Preserve in Guizhou. Mycosystema 21, 9–14.
Liang, Z. Q., Liu, A. Y., Liu, M. H., and Kang, J. C. (2003). The genus Cordyceps and its allies from the Kuankuoshui Reserve in Guizhou III. Fungal Divers. 14, 95–101.
Liang, Z. Q., Liu, A. Y., and Liu, Z. Y. (2007). Cordyceps. In: Flora Fungorum Sinicorum. Beijing: Science Press.
Lou, H. W., Lin, J. F., Guo, L. Q., Wang, X. W., Tian, S. Q., Liu, C. X., et al. (2019). Advances in research on Cordyceps militaris degeneration. Appl. Microbiol. Biotechnol. 103, 7835–7841. doi: 10.1007/s00253-019-10074-z
Luangsa-ard, J. J., Hywel-Jones, N. L., Manoch, L., and Samson, R. A. (2005). On the relationships of Paecilomyces sect. Isarioidea species. Mycol. Res. 109, 581–589. doi: 10.1017/s0953756205002741
Luangsa-ard, J. J., Hywel-Jones, N. L., and Samson, R. A. (2004). The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 96, 773–780. doi: 10.1080/15572536.2005.11832925
Mains, E. B. (1940). Cordyceps Species from British Honduras. Mycologia 32, 16–22. doi: 10.1080/00275514.1940.12017390
Mains, E. B. (1958). North American entomogenous species of Cordyceps. Mycologia 50, 169–222. doi: 10.1080/00275514.1958.12024722
Mongkolsamrit, S., Noisripoom, W., Tasanathai, K., Khonsanit, A., Thanakitpipattana, D., Himaman, W., et al. (2020). Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol. Prog. 19, 957–983. doi: 10.1007/s11557-020-01615-2
Mongkolsamrit, S., Noisripoom, W., Thanakitpipattana, D., Wutikhun, T., Spatafora, J. W., and Luangsa-ard, J. J. (2018). Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia 110, 230–257. doi: 10.1080/00275514.2018.1446651
Mora, M. A. E., Castilho, A. M. C., and Fraga, M. E. (2017). Classification and infection mechanism of entomopathogenic fungi. Arq. Inst. Biol 84, 1–10. doi: 10.1590/1808-1657000552015
Nylander, J. A. A. (2004). MrModeltest v2. 3. Program Distributed by the Author. Uppsala: Uppsala University.
Olatunji, O. J., Tang, J., Tola, A., Auberon, F., Oluwaniyi, O., and Ouyang, Z. (2018). The genus Cordyceps: an extensive review of its traditional uses, phytochemistry and pharmacology. Fitoterapia 129, 293–316. doi: 10.1016/j.fitote.2018.05.010
Palfner, G., Valenzuela-Muñoz, V., Gallardo-Escarate, C., Parra, L. E., Becerra, J., and Silva, M. (2012). Cordyceps cuncunae (Ascomycota, Hypocreales), a new pleoanamorphic species from temperate rainforest in southern Chile. Mycol. Prog. 11, 733–739. doi: 10.1007/s11557-011-0784-8
Petch, T. (1924). Studies in entomogenous fungi, IV, some Ceylon Cordyceps. Trans. Brit. Myc. Socy. 10, 28–45. doi: 10.1016/S0007-1536(24)80005-0
Rambaut, A. (2006). FigTree. Tree Figure Drawing Tool Version 1. 3. 1. Edinburgh: University of Edinburgh.
Rehner, S. A., and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/S0953-7562(09)80409-7
Samson, R. A. (1974). Paecilomyces and some allied Hyphomycetes. Stud. Mycol. 6, 1–119. doi: 10.1186/1471-2180-12-3
Shrestha, B. (2011). Diversity of Cordyceps fungi in Nepal. Nepal J. Sci. Tech. 12, 103–110. doi: 10.3126/njst.v12i0.6487
Shrestha, B., Tanaka, E., Han, J. G., Oh, J. S., Han, S. K., Lee, K. H., et al. (2014). A brief chronicle of the genus Cordyceps Fr., the oldest valid genus in Cordycipitaceae (Hypocreales, Ascomycota). Mycobiology 42, 93–99. doi: 10.5941/myco.2014.42.2.93
Shrestha, B., Tanaka, E., Hyun, M. W., Han, J. G., Kim, C. S., Jo, J. W., et al. (2016). Coleopteran and lepidopteran hosts of the entomopathogenic genus Cordyceps sensu lato. J. Mycol. 2016, 1–14. doi: 10.1155/2016/7648219
Spatafora, J. W., Sung, G. H., Hywel-Jones, N. L., and White, J. F. (2007). Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 16, 1701–1711. x doi: 10.1111/j.1365-294X.2007.03225
Su, C. H., and Wang, H. H. (1986). Phytocordyceps, a new genus of the Clavicipitaceae. Mycotaxon 26, 337–344.
Sung, G. H., Hywel-Jones, N. L., Sung, J. M., Luangsa-ard, J. J., Shrestha, B., and Spatafora, J. W. (2007). Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 57, 5–59. doi: 10.3114/sim.2007.7.01
Sung, G. H., Spatafora, J. W., Zare, R., and Gams, W. (2001). A revision of Verticillium sect. Prostrata. II. Phylogenetic analyses of SSU and LSU nuclear rDNA sequences from anamorphs and teleomorphs of the Clavicipitaceae. Nova Hedwig 72, 311–328. doi: 10.1127/nova.hedwigia/72/2001/311
Swofford, D. L. (2002). PAUP: Phylogenetic Analysis Using Parsimony, Version 4. 0b10. Sunderland, MA: Sinauer Associates.
Tasanathai, K., Thanakitpipattana, D., Noisripoom, W., Khonsanit, A., Kumsao, J., and Luangsa-ard, J. J. (2016). Two new Cordyceps species from a community forest in Thailand. Mycol. Prog. 15:28. doi: 10.1007/s11557-016-1170-3
Torres, M. S., White, J., and Bischoff, J. F. (2005). Cordyceps spegazzinii sp. nov., a new species of the C. militaris group. Mycotaxon 94, 253–263.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, Y., Liu, Y. F., Fan, Q., Wang, Y. B., and Yu, H. (2020). Cordycipitoid fungi powders promote mycelial growth and bioactive-etabolite production in liquid cultures of the stout camphor medicinal mushroom Taiwanofungus camphoratus (Agaricomycetes). Int. J. Med. Mushrooms 22, 615–626. doi: 10.1615/IntJMedMushrooms.2020035440
Wang, Y. B., Wang, Y., Fan, Q., Duan, D. E., Zhang, G. D., Dai, R. Q., et al. (2020). Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 103, 1–46. doi: 10.1007/s13225-020-00457-3
Wang, Y. B., Yu, H., Dai, Y. D., Wu, C. K., Zeng, W. B., Yuan, F., et al. (2015). Polycephalomyces agaricus, a new hyperparasite of Ophiocordyceps sp. infecting melolonthid larvae in southwestern China. Mycol. Prog. 14:70. doi: 10.1007/s11557-015-1090-7
Wright, P. J. (1993). Cordyceps oncoperae sp. nov. (Ascomycota) Infecting oncopera spp. (Lepidoptera: Hepialidae). J. Invertebr. Pathol. 61, 211–213. doi: 10.1006/jipa.1993.1038
Yan, J. Q., and Bau, T. (2015). Cordyceps ningxiaensis sp. nov., a new species from dipteran pupae in Ningxia Hui Autonomous Region of China. Nova hedwig 100, 251–258. doi: 10.1127/nova_hedwigia/2014/0222
Zang, M., and Kinjo, N. (1998). Notes on the alpine Cordyceps of China and nearby nations. Mycotaxon 66, 215–229.
Zha, L. S., Huang, S. K., Xiao, Y. P., Boonmee, S., Eungwanichayapant, P. D., McKenzie, E. H., et al. (2018). An evaluation of common Cordyceps (Ascomycetes) species found in Chinese markets. Int. J. Med. Mushrooms 20, 1149–1162. doi: 10.1615/IntJMedMushrooms.2018027330
Zha, L. S., Kryukov, V. Y., Ding, J. H., Jeewon, R., and Chomnunti, P. (2021). Novel taxa and species diversity of Cordyceps sensu lato (Hypocreales, Ascomycota) developing on wireworms (Elateroidea and Tenebrionoidea, Coleoptera). MycoKeys 78, 79–117. doi: 10.3897/mycokeys.78.61836
Zha, L. S., Wen, T. C., Huang, S. K., Boonmee, S., and Eungwanichayapant, P. D. (2019). Taxonomy and biology of Cordyceps qingchengensis sp. nov. and its allies. Phytotaxa 416, 14–24. doi: 10.11646/phytotaxa.416.1.2
Zhang, J. Y., Jian, T. T., Zhang, Y., Zhang, G. Y., and Ling, J. Y. (2021). Dynamic content changes of cordycepin and adenosine and transcriptome in Cordyceps kyushuensis Kob at different fermentation stages. Bioprocess Biosyst. Eng. 44, 1793–1803. doi: 10.1007/s00449-021-02561-3
Keywords: entomopathogenic fungi, multilocus phylogeny, new taxon, species diversity, taxonomy
Citation: Dong Q-Y, Wang Y, Wang Z-Q, Tang D-X, Zhao Z-Y, Wu H-J and Yu H (2022) Morphology and Phylogeny Reveal Five Novel Species in the Genus Cordyceps (Cordycipitaceae, Hypocreales) From Yunnan, China. Front. Microbiol. 13:846909. doi: 10.3389/fmicb.2022.846909
Received: 31 December 2021; Accepted: 16 February 2022;
Published: 13 April 2022.
Edited by:
Rajesh Jeewon, University of Mauritius, MauritiusReviewed by:
Sanjay K. Singh, Agharkar Research Institute, IndiaSamantha Chandranath Karunarathna, Qujing Normal University, China
Tingchi Wen, Guizhou University, China
Copyright © 2022 Dong, Wang, Wang, Tang, Zhao, Wu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Yu, aG9uZ3l1QHludS5lZHUuY24=
 Quan-Ying Dong1,2
Quan-Ying Dong1,2 Yao Wang
Yao Wang Hong Yu
Hong Yu