- 1School of Biological Science and Technology, University of Jinan, Jinan, China
- 2Liaoning Academy of Forestry Sciences, Shenyang, China
- 3Faculty of Agricultural Sciences, Luis Vargas Torres de Esmeraldas University of Technology, Esmeraldas, Ecuador
- 4Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
Declining soil quality and microecological imbalances were evaluated in larch plantations in this study. One potential solution to this problem is the cultivation of mixed coniferous and broad-leaved plantations. However, it is unclear whether and how soil microbial community structure and nutrient cycling function would be affected by mixed plantations and soil depths. In this study, we used high-throughput sequencing technology to investigate bacterial 16S and fungal ITS regions for comparisons of soil microbial diversity among plantation types (a Larix gmelinii pure plantation, a Fraxinus mandshurica pure plantation, a Larix–Fraxinus mixed plantation within the Larix row, the Fraxinus row, and between the Larix and Fraxinus rows) and soil depths (0–10, 10–20, and 20–40 cm). These data were used to evaluate variations in microbial communities and nutrient cycling function with the determining environmental factors. Our results indicated that bacteria had a stronger spatial dependence than did fungi, while plantation types significantly affected the fungal community. The relative abundance of Gaiellaceae, as well as bacterial ligninolysis, nitrate ammonification, and nitrite ammonification functions significantly increased with increasing soil depth. Compared with other plantations, the relative abundance of Inocybaceae was significantly higher in the Larix plantation. Distance-based redundancy analysis (db-RDA) showed that Gaiellaceae and Inocybaceae abundances were positively correlated with ammonium nitrogen content, available phosphorus content, and phosphatase activity. Our findings indicate that variations in soil available phosphorus are closely related to the relative abundances of Gaiellaceae at different soil depths and Inocybaceae in different plantation types. Mixed plantations might change the availability of soil phosphorus by controlling the relative abundance of Inocybaceae. We recommend that fungal community changes be considered in the sustainable management of mixed plantations.
Introduction
The long-term management of pure coniferous plantations has resulted in poor nutrient turnover efficiency, low bio-diversity, and a poor soil ecosystem (Liu et al., 2018). It has been reported that cultivating mixed coniferous and broad-leaved plantations could change stand biomass (Wang et al., 2008), soil microbial community catabolic diversity (Jiang et al., 2012), soil carbon storage (He et al., 2013), litter decomposition, and nutrient return rate (Wang et al., 2007, 2019). Despite some studies reporting on soil bacterial and fungal community composition in different plantation types, there were few studies on the differences of microbial community structure and nutrient cycling function among different tree species belt of mixed plantation. And it remains poorly understood how mixed plantation and soil depth influence specific microbial groups relevant to soil nutrient cycling.
As essential participants in the nutrient cycling and energy flow, soil microbial communities have important roles in reestablishing function and biodiversity of ecosystem restoration (Harris, 2009; Jiang et al., 2012). Furthermore, changes in soil microbial community composition and function can sensitively reflect soil environmental quality changes, an important component of soil productivity (Vasconcellos et al., 2013). The soil nutrient content and enzyme activities are reportedly higher in mixed plantations than in pure plantations (Li et al., 2015). Soil microbial abundance, catabolic potentials, and versatility are significantly influenced by forest vegetation (Garau et al., 2019). Previous studies find that compared with a pure coniferous plantation, mixed coniferous and broad-leaved plantations have been shown to significantly increase soil microbial Shannon diversity, OTU richness, and biological functions involved in carbon (C) and nitrogen (N) cycles (i.e., enzyme activities, carbon utilization patterns, and functional gene abundance; Jiang et al., 2012; Pereira et al., 2018). The study showed that the dominant microbial flora and C, N cycling functional groups in different plantation types of soil were diverse. These differences in microbial composition led to soil acidification and a large decline in available soil nutrient (Zhang et al., 2016). It is critical to study the relative contribution of specific microbial groups to nutrient content and enzyme activities in mixed broad-leaved plantations to understanding the potential microbial mechanism involved with soil nutrient cycling and maintain soil fertility of plantation.
Larix is one of the main afforestation tree species in China. Pure larch plantations have some problems, such as soil degradation and acidification (Yang et al., 2010). To improve the ecological stability and soil environment of pure larch plantations, some scholars have investigated combinations of Larix with other broad-leaved tree species. The results have shown that mixed plantations are superior to pure plantations (Liu et al., 1998; Wang et al., 2019). Furthermore, the catabolic diversity and function of soil microbial communities are much better under broadleaf tree species and mixed broadleaf–conifer plant species than under coniferous tree species (Jiang et al., 2012). Fraxinus mandshurica is an important native broad-leaved tree species in the Eastern Liaoning Province, China. To our knowledge, there are few reports regarding mixed plantations of Larix and Fraxinus in this area.
The distributions of soil bacteria and fungi are significantly affected by soil depths (Zhang et al., 2016; Sun et al., 2018). In 0–40-cm soil layer, gram-positive bacteria and actinomycetes reportedly exhibited increased abundance with increasing soil depth (Li et al., 2017). In contrast, Gram-negative bacteria and fungi are most abundant at the soil surface and demonstrate substantially lower abundances in subsurface soil layers (Fierer et al., 2003). Although some studies have demonstrated changes in soil microbial communities among forest types and soil depths, little is known regarding the relationships of specific microbial groups with soil nutrient contents in mixed plantations. Hence, there is a need for a thorough understanding of microbial distributions among plantation types and soil depths, as well as complex linkages among microbial consortia, soil characteristics, and environmental conditions; such information may provide novel insights that support the improvement of Larix plantation management practices.
Here, we selected a pure Larix plantation, a pure F. mandshurica plantation, and a mixed Larix and Fraxinus plantation to compare soil nutrient content, enzyme activity, microbial community composition, and nutrient cycling functional characteristics in five inter-forest zones among three plantation types. This study was performed to explore the structural and nutrient cycling functional characteristics of bacterial and fungal communities among plantation types and soil depths; it was also performed to clarify the relationships among soil microbial communities, enzyme activities, and nutrient contents according to forest type and soil depth. The results of this study will provide a theoretical basis for the scientific evaluation of land productivity in mixed plantations. We hypothesized that responses to plantation types and soil depths would have different effects on bacterial and fungal communities; moreover, we presumed that mixed plantations would influence the abundances of specific microbial groups, leading to alterations in soil nutrient contents.
Materials and Methods
Study Sites and Sample Collection
The research site was located in the Douling forest farm in the south-central part of Xinbin Manchu Autonomous County, Liaoning Province, China (geographical coordinates: 124° 52.37′E and 41° 30.41’N). This site is in the Longgang mountain range extension of the Changbai Mountain system. The mean altitude of the territory is approximately 500 m, the mountain slopes are gentle (between 10° and 25°), and the mountains and hills are low. The study area falls within the north temperate seasonal continental climate with four distinct seasons, a pleasant climate, and abundant rainfall (mean annual rainfall of 750–850 mm). The main tree species are Larix gmelinii, Pinus sylvestris var. mongolica Litv., Pinus thunbergii Parl, Pinus koraiensis Sieb.et Zucc., Quercus mongolica Fisch. ex Ledeb., Juglans mandshurica Maxim., and F. mandshurica Rupr. Shrubs in the plantations mainly include Euonymus alatus (Thunb.) Sieb, Lonicera japonica Thunb., Corylus L., Aralia elata (Miq.) Seem., and Acanthopanax senticosus (Rupr. et Maxim.) Harms.
Three plantation types of pure L. gmelinii, pure F. mandshurica, and mixed L. gmelinii and F. mandshurica were selected in August 2019 as the experimental plots for this study. The selected plots had similar slope aspects, stand age, and stand density. The mixed Larix and Fraxinus plantation was cultivated in mixed row mode (5 × 5). Three 20 m × 30 m standard plots were established for each forest type using the typical plot method, with a total of nine completely independent and randomly distributed sample plots (Table 1). Samples were randomly collected from 10 points at each sample plot. Soil samples were acquired at depths of 0–10, 10–20, and 20–40 cm in each sample plot. Additionally, in the mixed Larix and Fraxinus plantation, soil samples were acquired from within the Larix row, the Fraxinus row, and between the Larix and Fraxinus rows. After removal of visible plant roots and understory litter, the soil material was sieved through a 2-mm sieve. Aliquots for DNA extraction were stored at −20°C and at 4°C for the complete determination of enzyme activities within a 1-month period. The remaining samples were dried at 65°C to constant weight for the determination of the soil physicochemical properties.
Soil Physicochemical Property Analysis
All samples were analyzed for total nitrogen content, nitrate nitrogen content, ammonium nitrogen content, pH, organic matter content, total phosphorus content, available phosphorus content, and moisture content. Analysis of total N content was carried out using an elemental analyzer based on dry combustion (Vario EL III, Germany). Soil organic matter was measured by the external-heat potassium dichromate oxidation method; available nitrogen was measured by the alkaline hydrolysis diffusion method. Total phosphorus was determined by molybdenum antimony resistance colorimetry. Available phosphorus was measured by the NaHCO3 extraction method (Yang and Yang, 2020). The moisture content of the soil was determined by drying to a constant weight at 65°C. pH values were determined in deionized water (1:2.5 for soil sample, m/v; Fioretto et al., 2000).
Soil Enzymes Assays
We examined the activities of five extracellular enzymes involved in soil C, N, and P degradation: β-glucosidase, sucrase, urease, acid phosphatase, and alkaline phosphatase. All soil samples were compared without the matrix, and a soil control was not used in the experiments. Soil enzyme activities were defined as mg of products produced over 24 h per gram of dried sample.
The activity of β-glucosidase was determined using the p-nitrophenol matrix method (Kourtev et al., 2002). Soil sucrase activity was measured by the 3,5-dinitrosalicylic acid method, urease activity was measured by the phenol sodium hypochlorite colorimetric method, acid and alkaline phosphatase activity was estimated using the disodium phenyl phosphate colorimetric method (Ge et al., 2017).
Total DNA Extraction
In accordance with the manufacturer’s instructions, total microbial community genomic DNA was extracted from 500 mg of soil material using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, United States). There were three duplications for each sample. Duplicate DNA extractions were performed for each sample and pooled. DNA extraction results were tested on 1% agarose gels; DNA concentration and purity were determined with a NanoDrop 2000 UV–Vis spectrophotometer (Thermo Scientific, Wilmington, United States).
High-Throughput Sequencing of Soil Bacterial 16S rDNA and Fungal ITS Region
The hypervariable V3–V4 region of the bacterial 16S rRNA gene was amplified with the primer pairs 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5’-CCGTCAATT CMTTTRAGTTT-3′) by an ABI GeneAmp® 9,700 polymerase chain reaction (PCR) thermocycler (ABI, CA, United States). The primers ITS3/ITS4 were amplified with the primer pairs ITS3F (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′). The PCR amplification conditions were as follows: 95°C for 3 min; 27 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s; 72°C for 10 min; and indefinite hold at 4°C upon completion. The PCR condition was the same except that the number of fungal PCR cycles was 35. The PCR mixtures contained 2 μl of 10 × TransSTART FastPfu buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of forward primer (5 μM), 0.8 μl of reverse primer (5 μM), 0.2 μl of TaKaRa rTaq DNA Polymerase, BSA 0.2 μl, 10 ng of template DNA, and sufficient ddH2O to achieve a final volume of 20 μl. PCRs were carried out in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States), in accordance with the manufacturer’s instructions, and then quantified using a Quantus™ Fluorometer (Promega, United States).
Bioinformatics Analyses
The original data were allocated to each sample according to a specific barcode, and then, tags and primer sequences were removed. The FLASH software (V1.2.11) was used to splice paired-end reads (Chen et al., 2020). The Qiime software (V1.9.1) was performed to quality control and filter the spliced sequences (Whelan and Surette, 2017). Chimeric sequences were detected and removed by UCHIME (Edgar et al., 2011). Paired-end reads were obtained by MiSeq-sequencing and then spliced on the basis of their overlapping relationships. Quality control and filtering were carried out on the resulting sequences. After samples had been differentiated, operational taxonomic unit (OTU) clustering analysis and species taxonomy analysis were performed; the OTUs were subjected to multiple diversity index analysis and sequencing depth detection. On the basis of taxonomic information, OTUs were included in statistical analysis of community structure at each classification level. To obtain species classification information regarding each OTU, an RDP classifier Bayesian algorithm was used to analyze representative OTU sequences with a 97% similarity level. The bacterial species classification and comparison database were SILVA (Release138)1, while the fungal were Unite (Release 8.0).2 FAPROTAX database was used to predict the functional potential of bacteria (Sansupa et al., 2021). Functions of fungal communities were classified and analyzed by FUNGuild,3 with the fungi divided into pathotrophs, symbiotrophs, and saprotrophs (Nguyen et al., 2016). Each sample’s bacterial and fungal community compositions were determined at all taxonomic levels. Accordingly, various in-depth statistical and visual analyses were performed to determine the community composition and phylogenetic information of multiple samples; these included multivariate analysis and significant difference tests. All the raw datasets in this study were publicly available in the NCBI Sequence Read Achieve (SRA) database with an accession number SRP355360.
Statistical Analysis
All statistical analyses in this study were performed by SPSS software (SPSS Inc., Chicago, IL). In all analyses, differences were considered statistically significant when p < 0.05 and extremely significant when p < 0.01. When data did not conform to assumptions of normality, the values were transformed into logarithm and/or square root before analysis. ANOVA of split plot was used to test the effects of microbial diversity (Chao, Shannon, and Simpson index) plantation type, soil depth, and their interaction on soil chemical properties and extracellular enzyme activities (Jones and Nachtsheim, 2009). The plantation types were the main plot, and soil depths were the split plot. Kruskal–Wallis H test was performed to analyze the difference of microbial groups in different soil samples. Additional statistical analyses were conducted using R software. Differences in soil microbial community composition and functions, based on changes in plantation types, were evaluated using principal coordinates analysis (PCoA), and analysis of similarities (ANOSIM) was performed in R using the “vegan” package based on Bray–Curtis dissimilarity distances (Calderón et al., 2017; Otsing et al., 2018). The sequences for all the samples were unified by minimum number of sample sequences before PCoA. BIO-ENV package and distance-based redundancy analysis (db-RDA) were implemented by the vegan package in R software to examine relevance among microbial composition, enzyme activity, and soil nutrient content (Wang et al., 2019; Testa et al., 2021).
Results
Soil Physicochemical Properties and Extracellular Enzyme Activities
ANOVA of split plot showed that the contents of total and available soil phosphorus were significantly affected by plantation type and soil depth (p < 0.05; Figure 1). With increasing soil depth, the contents of total and available soil phosphorus gradually decreased (p < 0.05). Among the three plantation types, the available soil phosphorus contents were significantly higher in pure Larix and Fraxinus plantations than in the mixed Larix–Fraxinus plantation (p < 0.05).
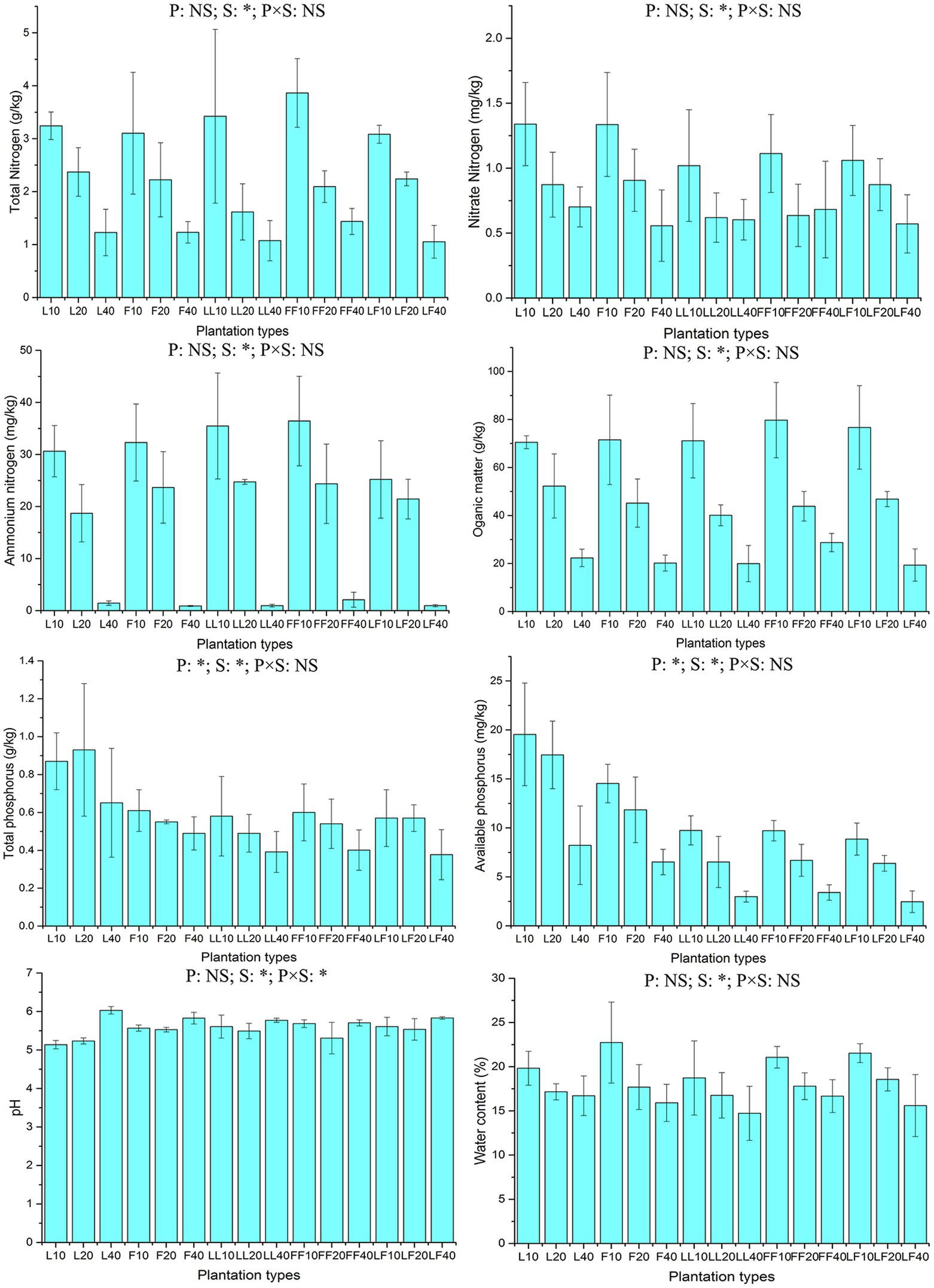
Figure 1. Chemical properties of soil samples. L, Larix plantation; F, Fraxinus plantation; LL, Larix–Larix plantation belt in the mixed plantation; FF, Fraxinus–Fraxinus plantation belt in the mixed plantation; LF, Larix–Fraxinus plantation belt in the mixed plantation; P, plantation types; S, soil depths; NS, not significant. Error bars show standard errors of the mean (n = 3). ANOVA of split plot results are reported. *p < 0.05. Numbers after letters represent soil depths of 0–10, 10–20, 20–40 cm.
Total nitrogen content, nitrate nitrogen content, ammonium nitrogen content, organic matter content, moisture content, and pH in soil were significantly affected by soil depth (p < 0.05). With increasing soil depth, the contents of organic matter, total nitrogen, nitrate nitrogen, ammonium nitrogen, and moisture gradually decreased; in contrast, the pH value gradually increased.
The activities of β-glucosidase, sucrase, and urease were significantly affected by plantation type (p < 0.01; Figure 2). Compared with other soil samples, the β-glucosidase activities were significantly improved in the mixed Larix–Larix plantation soil. The sucrase, alkaline phosphatase, and acid phosphatase activities were significantly affected by soil depth (p < 0.01). The alkaline phosphatase and acid phosphatase activities significantly decreased in all three plantation types at increased soil depths. There were significant differences in sucrase activities among plantation types and soil depths (p < 0.05).
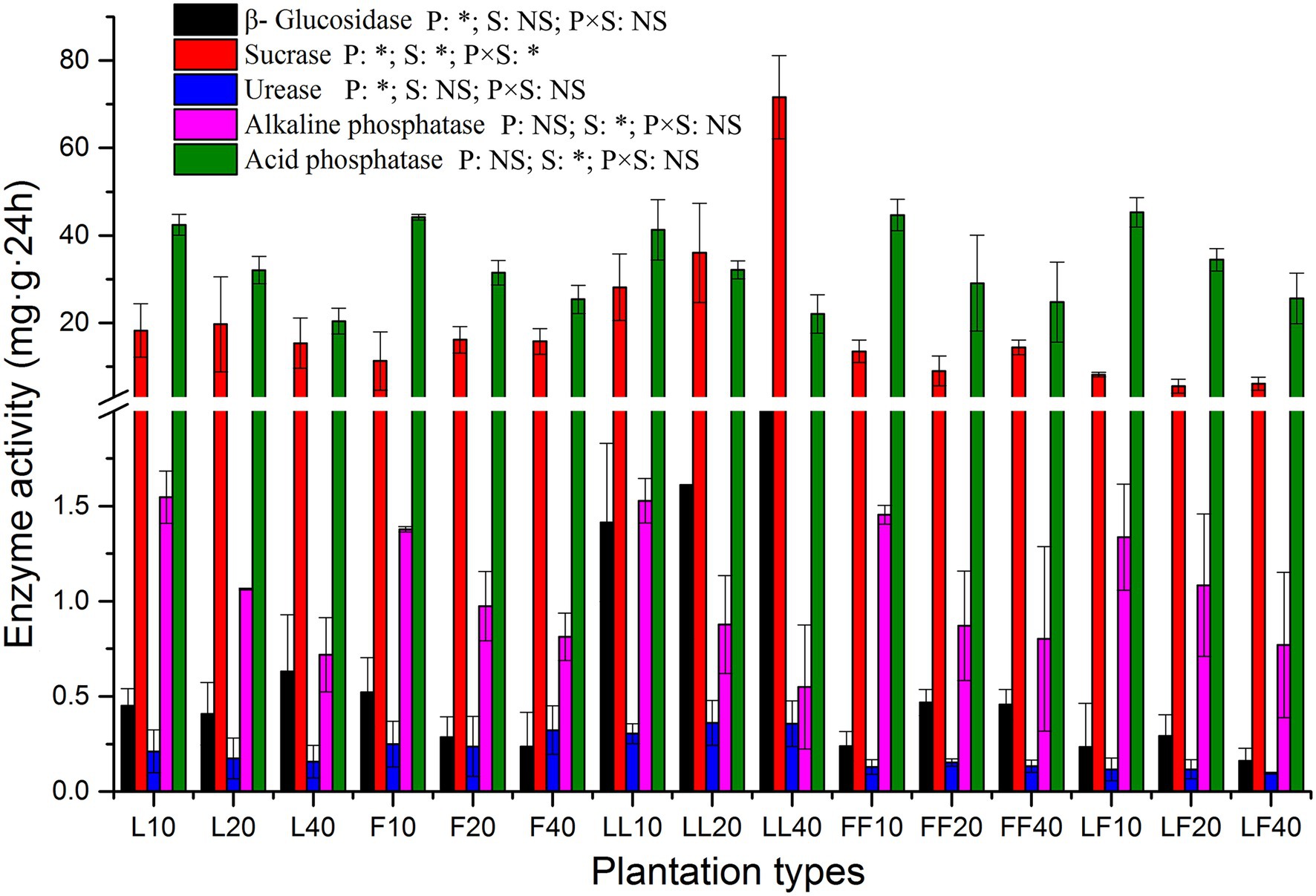
Figure 2. Extracellular enzyme activities in soil samples. Error bars show standard errors of the mean (n = 3). ANOVA of split plot results are reported. Refer to Figure 1 legend for relevant abbreviations.
Soil Bacterial Community Diversity, Composition, and Function
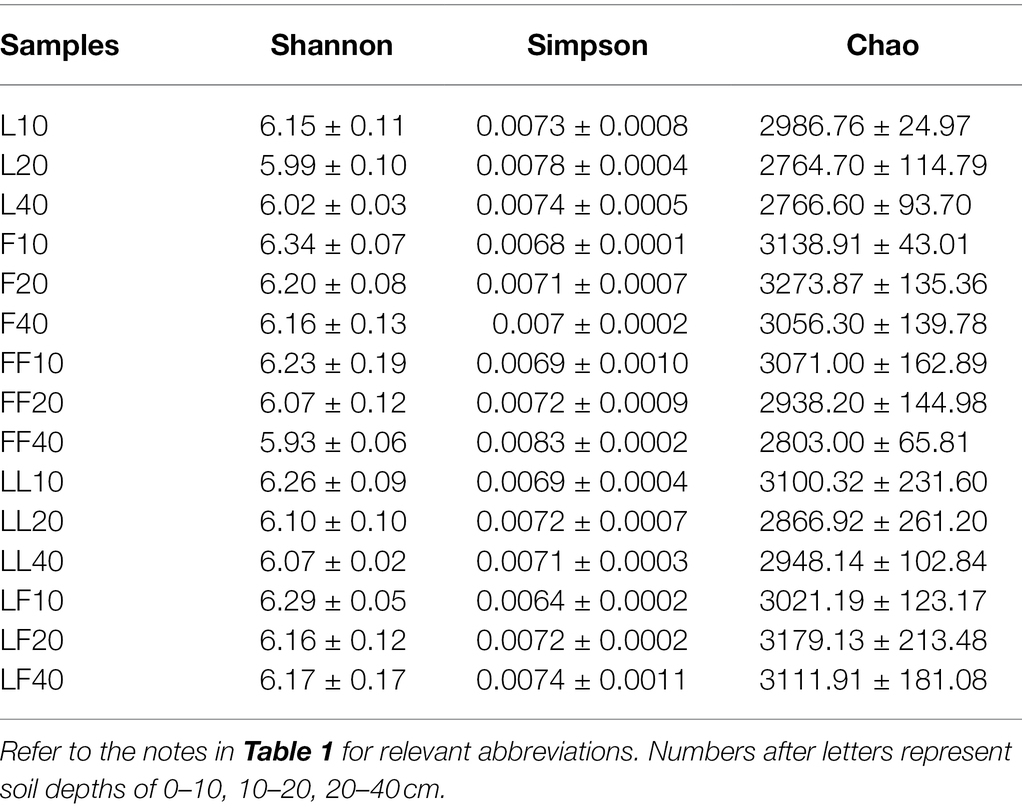
ANOVA of split plot showed that the indices of Shannon were significantly affected by plantation type and soil depth (p < 0.01; Table 2). The index of Simpson was only significantly affected by soil depth (p < 0.05). The index of Chao was only significantly affected by plantation type (p < 0.05). With the increase in soil depth, bacterial community diversity gradually decreased. Compared with Larix plantation, there was higher bacterial community diversity and richness in other plantations.
Principal coordinates analysis showed significant differences in bacterial community composition at family level among soil depth (ANOSIM, R = 0.32, p = 0.001; Figure 3A). There was no significant difference in bacterial community composition among plantation types (ANOSIM, R2 = 0.15, p = 0.059). In total, 12 bacterial families were identified with the relative abundances >3% (Figure 3B). Among them, Gaiellaceae and Acidobacteriales were dominant flora. With increasing soil depths, the relative abundance of Gaiellaceae and Acidobacteriales was gradually increased (Figure 3C). Especially, the relative abundance of Gaiellaceae significantly increased in all soil samples (p < 0.05). Under the same soil depth, there was no significant difference in bacterial community composition among different plantation types.
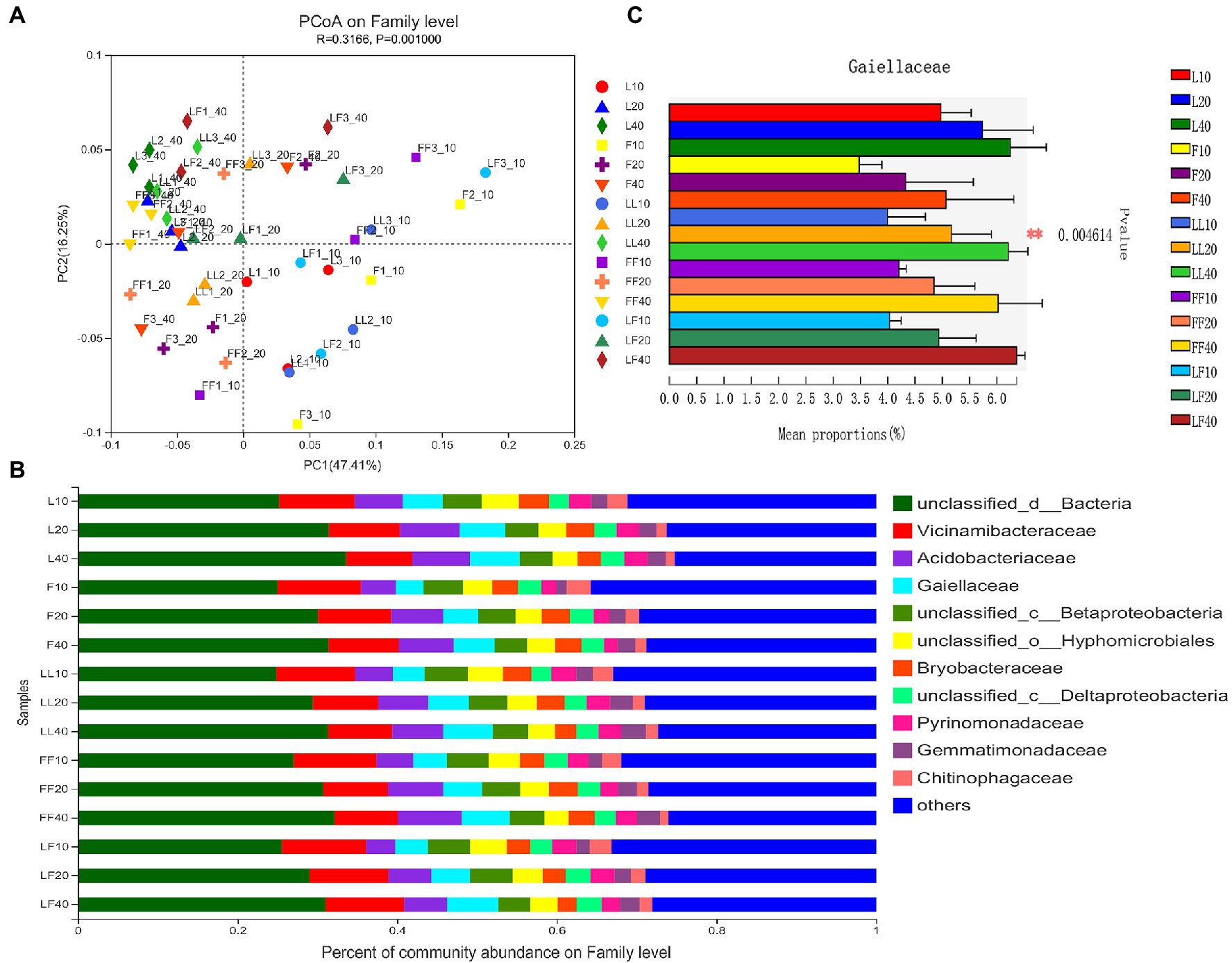
Figure 3. Bacterial community composition in different soil samples. (A) Principal coordinates analysis of bacterial community composition. (B) Bacterial community structure at the level of family. (C) The results of Kruskal–Wallis H test on the difference of Gaiellaceae in different soil samples. Refer to Figure 1 legend for relevant abbreviations.
Bacterial functional categories inferred by FAPROTAX were significantly differed among different soil depths (Figure 4). ANOVA of split plot showed that the relative abundances of functional genes for bacterial ligninolysis, nitrate ammonification, and nitrite ammonification were significantly affected by soil depth (p < 0.01). Kruskal–Wallis H test showed that with increasing soil depth, bacterial ligninolysis, nitrate ammonification, and nitrite ammonification functional genes significantly increased (p < 0.05). On the contrary, bacterial denitrification functional genes gradually decreased in different soil samples. There were no significant differences in bacterial metabolism functions in the same soil layer among the three plantation types (p > 0.05).
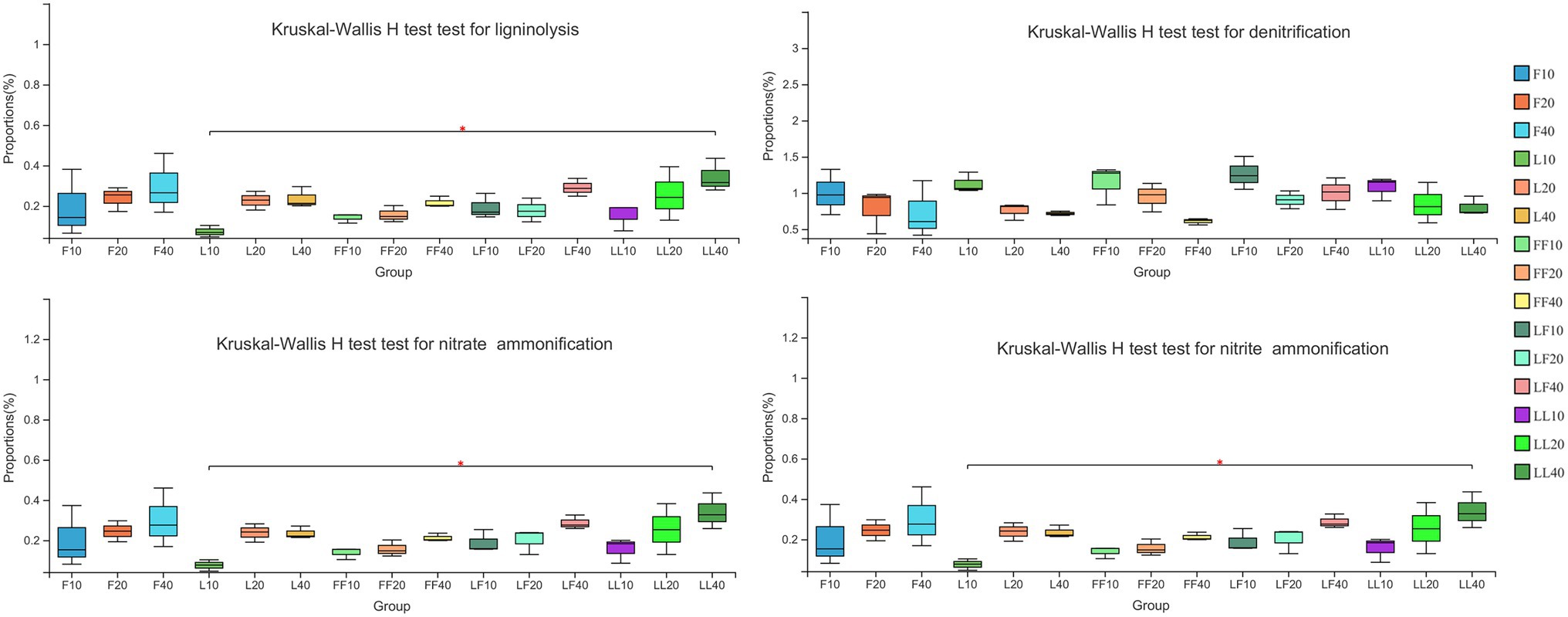
Figure 4. Functional bacterial community categories involved in nutrient cycling function. Error bars show standard errors of the mean (n = 3). Refer to Figure 1 legend for relevant abbreviations.
Soil Fungal Community Diversity, Composition, and Function
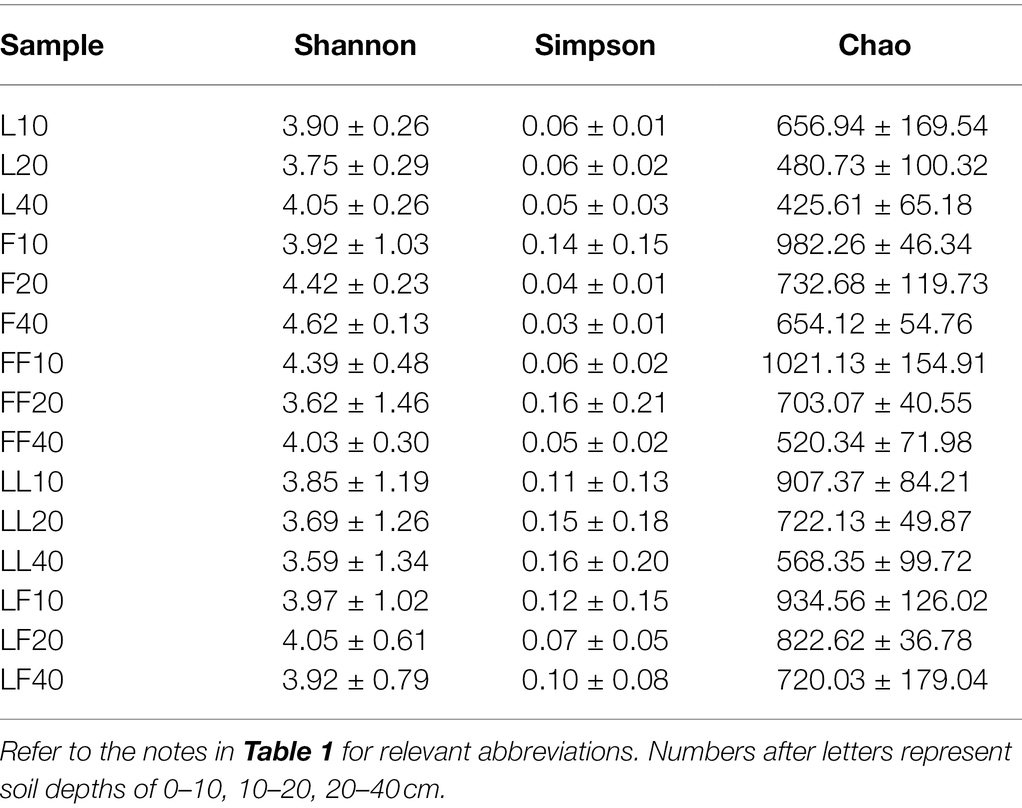
ANOVA of split plot showed that the indices of Chao were significantly affected by plantation type and soil depth (p < 0.05; Table 3). The index of Shannon and Simpson was not significantly affected by plantation type and soil depth (p > 0.05). Compared with Larix plantation, there was higher fungal community richness in other plantations. With the increase in soil depth, fungal community richness gradually decreased in all plantation types.
Principal coordinates analysis showed significant differences in fungal community composition at family level among plantation types (ANOSIM, R = 0.13, p = 0.037; Figure 5A). There were significant differences in fungal community composition among plantation types (ANOSIM, R2 = 0.29, p = 0.001). In total, 20 fungal families were identified with the relative abundances >3% (Figure 5B). Mortierellaceae, Atheliaceae, Hygrophoraceae, and Inocybaceae were dominant flora in all soil samples. Compared with other plantations, the relative abundance of Inocybaceae was significantly higher in the Larix plantation (p < 0.05; Figure 5C). The relative abundances of Atheliaceae were lower in Fraxinus soils than other types of plantation soils. There were no significant differences in fungal community composition with increasing soil depths in the same type of plantation.
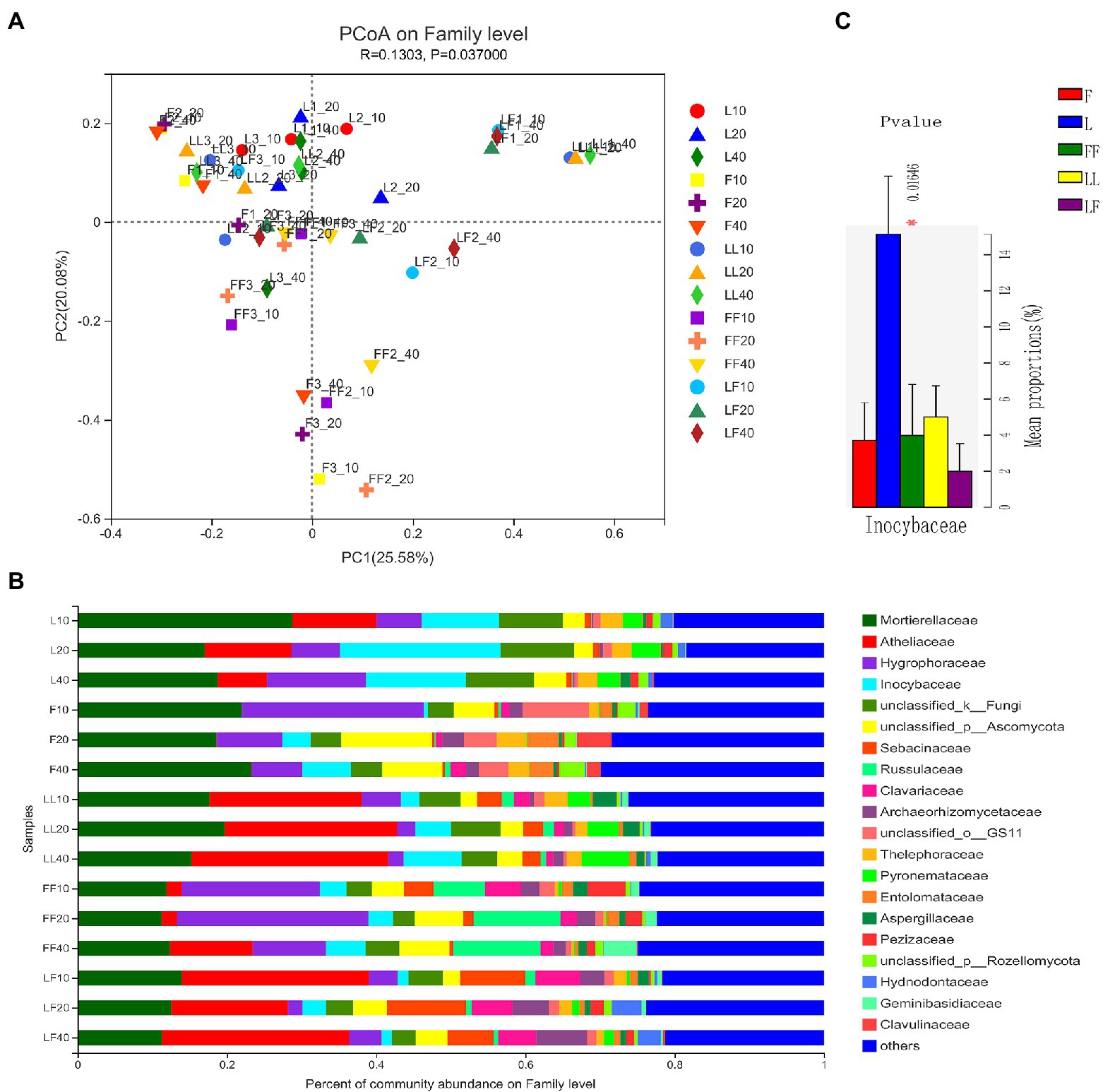
Figure 5. Fungal community composition in different soil samples. (A) Principal coordinates analysis of fungal community composition. (B) Fungal community structure at the level of family. (C) The results of Kruskal–Wallis H test on the difference of Inocybaceae in different soil samples. Refer to Figure 1 legend for relevant abbreviations.
Fungal functions were classified by FUNGuild (Figure 6). The results showed that the mean relative abundance of guild-undefined saprotrophs comprised approximately 48% of all fungi OTUs. The mean relative abundance of Ectomycorrhizae comprised approximately 28% of all fungi OTUs. The relative abundances of saprotrophs were significantly higher in Fraxinus and Fraxinus–Fraxinus plantation soils than in other plantation soils (p < 0.05). In contrast, the relative abundance of Ectomycorrhizae was significantly lower in Fraxinus plantation soil than in other plantation soils (p < 0.05).
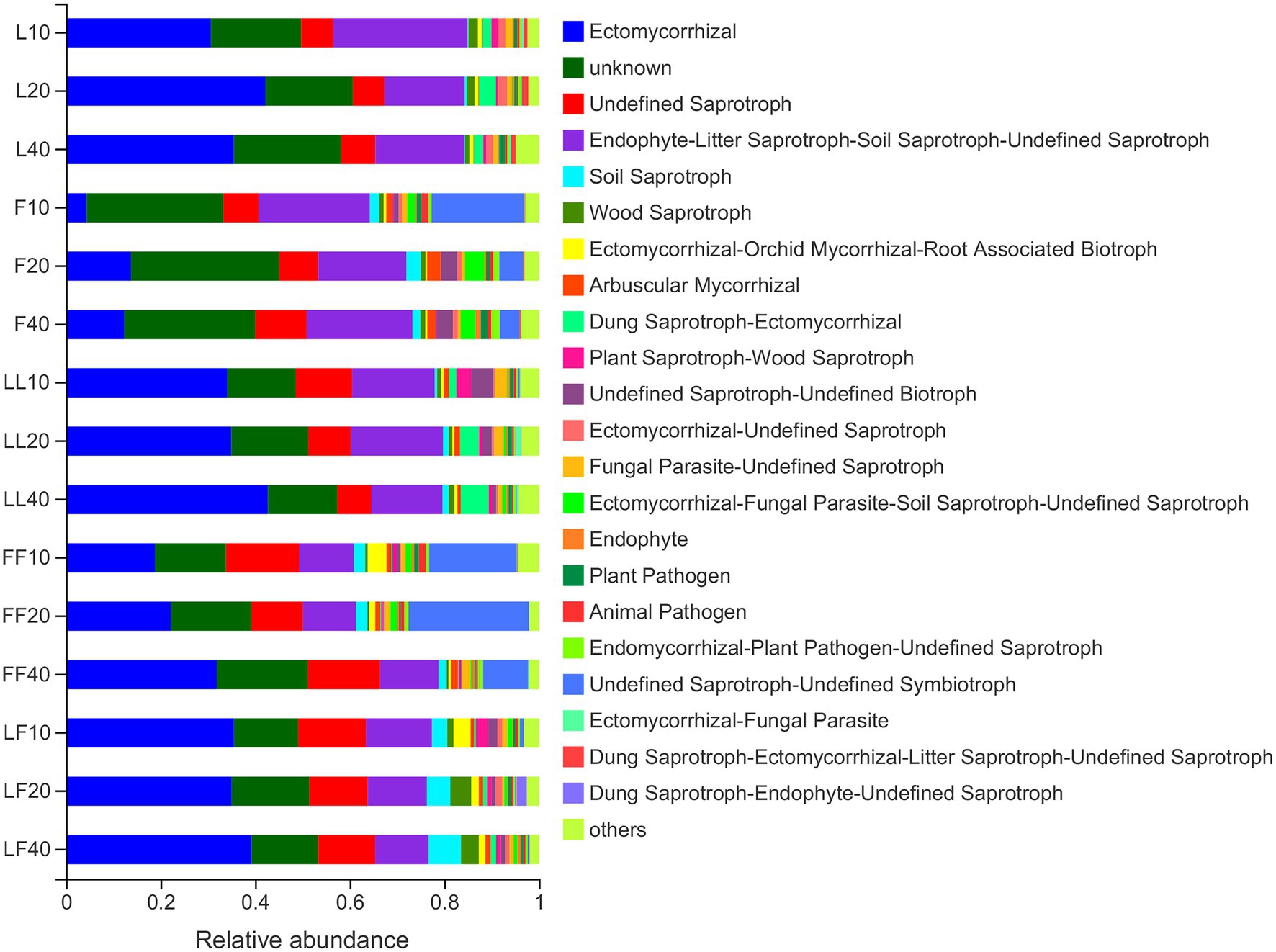
Figure 6. Variations in fungal functional group compositions inferred by FUNGuild. Refer to Figure 1 legend for relevant abbreviations.
Correlation Analysis of Environmental Factors
db-RDA showed that bacterial community compositions at family level were significantly correlated with pH, ammonium nitrogen, and available phosphorus contents (R2 = 0.18, p = 0.017; R2 = 0.18, p = 0.019; R2 = 0.17, p = 0.02; Figure 7A). Among the bacterial families, Gaiellaceae and Acidobacteriales were positively correlated with available phosphorus and ammonium nitrogen contents, while negatively correlated with pH alone. Fungal community compositions were significantly correlated with pH, ammonium nitrogen, total nitrogen, available phosphorus, organic matter, and total phosphorus contents (R2 = 0.44, p = 0.001; R2 = 0.33, p = 0.001; R2 = 0.20, p = 0.01; R2 = 0.40, p = 0.001; R2 = 0.20, p = 0.008; R2 = 0.17, p = 0.017; Figure 7C). Among the fungal families, Inocybaceae was positively correlated with available phosphorus, ammonium nitrogen, and total phosphorus contents and negatively correlated with pH. Atheliaceae was opposite to Inocybaceae.
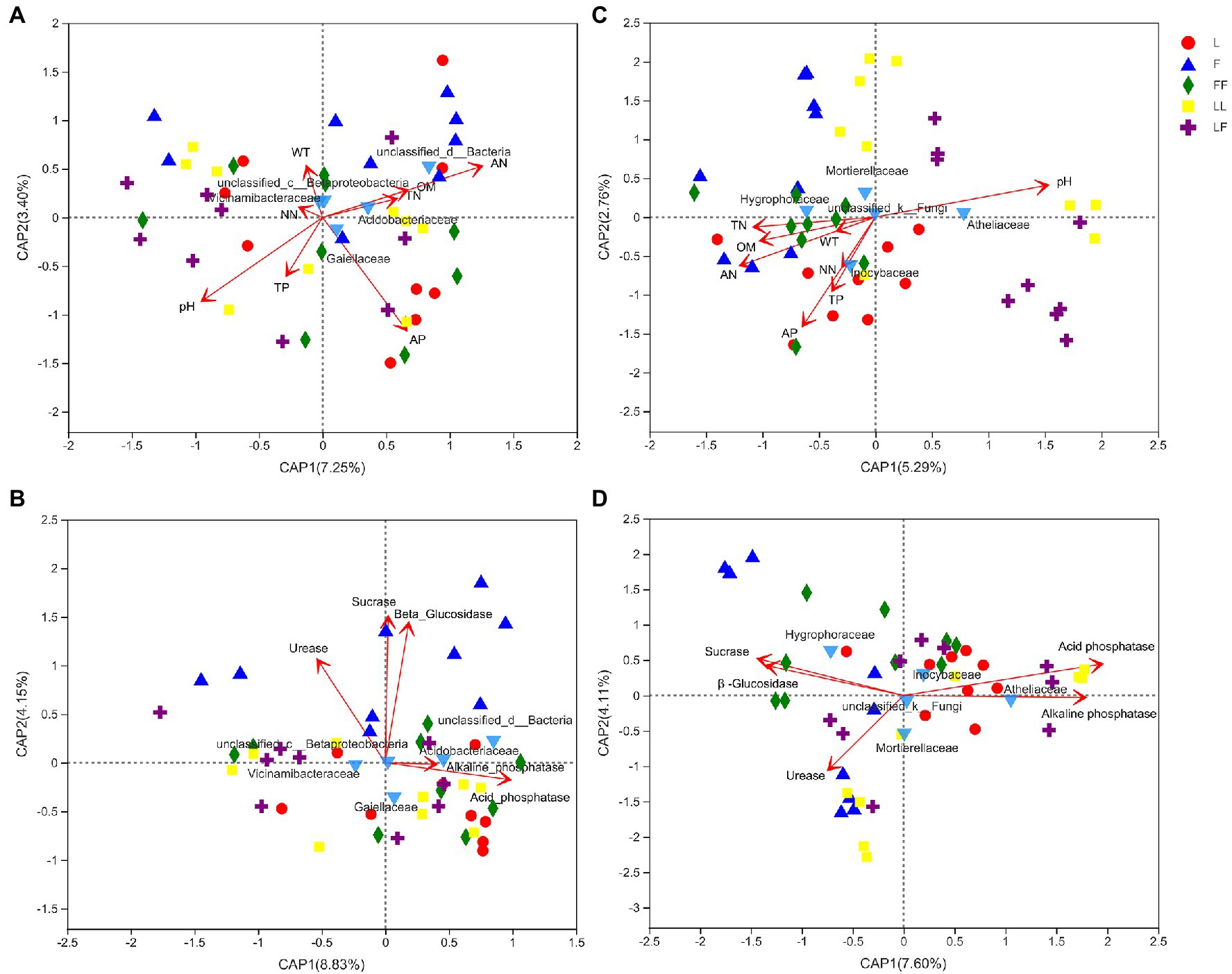
Figure 7. Distance-based redundancy analysis (db-RDA) to show correlation between microbial community and environmental factors in different soil samples. (A) Bacterial families and nutrient; (B) bacterial families and enzymatic activity; (C) fungal families and nutrient; (D) fungal families and enzymatic activity. TN, total nitrogen; AN, ammonium nitrogen; NN, nitrate nitrogen; TP, total phosphorus; AP, available phosphorus; OM, organic matter; WT, water content. Refer to Figure 1 legend for relevant abbreviations.
Bacterial community compositions were significantly correlated with acid phosphatase, β-glucosidase, sucrase, and urease activities (R2 = 0.18, p = 0.013; R2 = 0.58, p = 0.001; R2 = 0.61, p = 0.001; R2 = 0.32, p = 0.001; Figure 7B). Among the bacterial families, Gaiellaceae and Acidobacteriales were positively correlated with acid phosphatase and alkaline phosphatase activities. Fungal community compositions were significantly correlated with β-glucosidase, sucrase, acid phosphatase, alkaline phosphatase, and urease activity (R2 = 0.26, p = 0.007; R2 = 0.30, p = 0.004; R2 = 0.53, p = 0.001; R2 = 0.42, p = 0.001; R2 = 0.19, p = 0.018; Figure 7D). Among the fungal groups, Inocybaceae was positively correlated with acid phosphatase and alkaline phosphatase activities, while negatively correlated with β-glucosidase, sucrase, and urease activities.
Discussion
Effects of Mixed Plantations on Soil Bacterial and Fungal Communities
It was found that vegetation type and soil depth had significant effects on microbial community diversity. In this study, there was lower microbial community diversity and richness in Larix plantation than other plantations. The increase of microbial community biodiversity was conducive to soil health and forest growth (Van Bruggen and Semenov, 2000). Higher microbial diversity can increase the number and resilience of plant beneficial functions, which enable plants to produce beneficial characteristics that are difficult to obtain from any isolated species (Saleem et al., 2019). Therefore, cultivating coniferous and broad-leaved mixed plantation has the potential advantage of improving forest productivity. With the increase in soil depth, bacterial community diversity and fungal richness were gradually decreased. These findings consist with the previous study (Too et al., 2018). The possible reason for the results was that surface soil was rich in nutrients under the action of soil leaching and forest litter decomposition, which could provide niche for the growth of more microbial groups.
Bacterial communities and functions were shaped by soil depth but not plantation type. It is widely accepted that mixed plantations exhibit significant effects on soil microbial community structure (Chen et al., 2013; Pereira et al., 2017), although this effect might depend on soil depth. For example, Zhang et al. (2021) showed that mixed plantations did not significantly alter bacterial community structure in the upper soil. In this study, ANOSIM analysis revealed that there were significant differences in bacterial community composition among different soil depths rather plantation type. One possible reason was that the composition of soil bacterial community was mainly driven by soil nutrients (Dong et al., 2021). In this study, except phosphorus, other nutrient content had no significant differences among plantation types. Compared with other soil environmental factors such as pH, organic C, and inorganic N, phosphorus has a relatively low weight in shaping bacterial community structure (Hu et al., 2014; Wang et al., 2018). With the increasing soil depth, the nutrient content and pH significantly changed. This led to significant changes in bacterial community composition. These results were in agreement with the research of Zhang et al. (2016) and Li et al. (2019); the changes of total carbon, total nitrogen, and pH in different soil depths led to significant differences in bacterial community composition. Huang et al. (2013) showed that soil edaphic characteristics and soil profile might shape bacterial community functions; metabolic profiles revealed the carbon source utilization capacity in the surface layer. Using the BIOLOG system, Griffiths et al. (2003) and Braun et al. (2006) showed that the bacterial capability for utilizing diverse substrates decreased with soil depth. Our study showed that except for Larix plantation, ligninolysis, nitrate ammonification, and nitrite ammonification functional gene abundances significantly decreased with increasing soil depth in other plantations. This suggests that soil depth, rather than plantation type, significantly affected bacterial community structure and functional potential of nutrient cycling.
The abundance and diversity of fungal communities are closely related to tree species composition, and microbial communities associated with mixed species are more active (Khlifa et al., 2017). Notably, plant species, phylogenetic relationships, and plant traits affect the compositions of rhizosphere fungal communities (Sweeney et al., 2020). Our results suggested that soil fungal community composition significantly differed among the three plantation types. The main soil fungal families were Atheliaceae, Hygrophoraceae, and Inocybaceae, which belonged to the phylum of Basidiomycota. Most Basidiomycota were soil saprotrophs, and the relative abundances of soil saprotrophs were significantly higher in Fraxinus and Fraxinus–Fraxinus plantations than in other plantations. One possible explanation is that Fraxinus litter contains more nutrients than Larix litter, thus providing more niches for soil Basidiomycota. This was confirmed by Wu D. et al. (2019a), who showed that saprophytic fungi were more abundant in plantations with high soil nutrients. The mixed Larix and F. mandshurica plantation also had a higher relative abundance of ectomycorrhizal fungi than did a pure F. mandshurica plantation. Ectomycorrhizal fungi commonly occur in conifer roots, and the mixture of broadleaf trees with conifers would likely increase ectomycorrhizal fungal diversity (Ishida et al., 2007). Wu W. et al. (2019b) showed that mixed plantations of F. mandshurica and P. koraiensis contained a highly diverse and abundant population of ectomycorrhizal fungi. In that study, most Inocybaceae were ectomycorrhizal fungi (Matheny et al., 2009), and the relative abundance of Inocybaceae was significantly higher in Larix plantation soil. Our study indicated that mixed Larix and F. mandshurica plantations had significant effects on the fungal community. Thus, there is a need to closely monitor the impacts of mixed plantations on fungal communities in future sustainable management efforts.
Relationships Among Microbial Community Composition, Enzyme Activity, and Nutrient Content
This study found that mixed plantations did not significantly change general soil fertility or organic C and total N contents, compared with monoculture plantations. These results were consistent with the findings of previous studies, in which no significant differences were observed in soil organic carbon, total nitrogen, and available nitrogen contents among the three plantation types (Zhang et al., 2021). However, our study showed that plantation types and soil depths significantly affected the total soil phosphorus and available phosphorus contents. Among the three plantation types, the available soil phosphorus contents were significantly higher in Larix and Fraxinus plantations than in the mixed Larix–Fraxinus plantation. With increasing soil depth, available phosphorus content significantly decreased in all three plantation types. In contrast, some studies found significant increases in available phosphorus contents in the mixed-species treatments (Rachid et al., 2013; Zhang et al., 2021). One explanation is that faster growth leads to decreases in available phosphorus contents in mixed Larix–Fraxinus plantations (Feng et al., 2021).
Changes in soil microbial community structure serve as important indicators of soil quality and soil changes (Sharma et al., 2010). Bacteria and fungi reportedly contribute to nutrient bioavailability in degraded soils (Rashid et al., 2016). Dang et al. (2018) showed that soil organic carbon, total nitrogen, and available phosphorus contents were significantly correlated with soil bacterial communities; soil organic carbon, total nitrogen, total phosphorus, available phosphorus, and nitrate nitrogen contents were significantly correlated with soil fungal communities. The present study showed that bacterial and fungal community compositions were significantly correlated with the available phosphorus content. Our results showed that available phosphorus contents were significantly higher in Larix plantations than in the mixed Larix–Fraxinus plantation. Furthermore, higher relative abundances of Inocybaceae were observed in Larix plantation soils. Some fungi in Inocybaceae family have higher phosphate-solubilizing activity; thus, they can dissolve nearly all types of phosphorus in soil (Carrino-Kyker et al., 2016). This indicated that the higher abundances of Inocybaceae might increase inorganic phosphorus contents in Larix plantation soils.
Spatial and temporal variations in soil microorganisms are affected by differences in dominant tree species; these can ultimately change the availability and dynamics of soil nutrients, as well as shifts in microbial community composition, during adaptation to new environmental conditions. Physicochemical changes along the soil profile may be the main factor that controls the properties of bacterial communities (Ranjard and Richaume, 2001; Fierer et al., 2003). In the soil profile, total phosphorus content, available phosphorus content, pH value, organic matter content, and moisture content were significantly correlated with soil depth; these findings were consistent with the results of previous studies (Blume et al., 2002; Griffiths et al., 2003; Eilers et al., 2012). In the present study, the abundance of Gaiellaceae was positively correlated with available phosphorus content and phosphatase activity. It is reported that the members of family Gaiellaceae had high alkaline and acid phosphatase producing activity (Albuquerque and Costa, 2014; López-Lozano et al., 2020). With increasing soil depth, the relative abundance of Gaiellaceae significantly increased. It may be the distribution and physiological characteristic of Gaillaceae that makes the content of soil available phosphorus decrease with the increase in soil depth.
Soil enzyme activity plays a vital role in the turnover of carbon, nitrogen, and phosphorus; it is closely related to the distribution of soil microbial communities (Midgley and Phillips, 2016; Xu et al., 2017). Our results indicated that plantation type and soil depth had distinct effects on enzyme activities. In general, broad-leaved forests and mixed coniferous broad-leaved forests have higher soil enzyme activities than do coniferous forests alone (Hu et al., 2006; Xing et al., 2009). Similar to the previous results, our study showed significantly increased sucrase and β-glucosidase activities in Larix–Larix plantation soil. Alkaline phosphatase and acid phosphatase activities were significantly decreased with soil depth in the three plantation types, which is consistent with findings by Samuel et al. (2010). Distance-based redundancy analysis revealed that Gaiellaceae and Inocybaceae abundances were positively correlated with available phosphorus content and phosphatase activity. This indicated that soil available phosphorus variations were closely related to the relative abundance of Gaiellaceae at different soil depths and to the relative abundance of Inocybaceae in different plantation types. In general, our study indicated that mixed Larix–Fraxinus plantations resulted in distinct microbial groups, relative to the respective monocultures; these groups had positive effects on soil phosphorus content, potentially increasing the need for phosphorus anthropogenic fertilization.
Conclusion
This study characterized microbial communities and nutrient cycling function in different plantation type and soil depth. Notably, available soil phosphorus was significantly affected by plantation type and soil depth. The mixed Larix–Fraxinus plantation had a significant effect on soil fungal community structure and nutrient cycling function. In contrast, bacterial community characteristics were closely related to soil depth. Variations in soil available phosphorus among plantation types and soil depths were correlated with the relative abundances of Inocybaceae and Gaiellaceae respectively. These findings provided novel insights into the relationships among soil microbial communities, enzyme activities, and nutrient contents according to plantation type and soil depth. There is a need for closer assessment of changes in fungal communities during future efforts to achieve sustainable management of mixed plantations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession numbers can be found at: SRA, NCBI; accession: SRP355360; bioproject: PRJNA797909.
Author Contributions
WW and JF performed conceptualization. WW did methodology, software, writing—original draft preparation, and visualization. WW, JW, and JF done validation. RB performed formal analysis. WW, JW, QW, and JF contributed to investigation. JF contributed to re-sources, funding acquisition, and data curation. JW, QW, and JF were involved in writing—review and editing. SY, PB, ZW, and DC supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by National Key Research Program of China (2017YFD060040103), the Shandong Province Double Hundred Talent Plan (WSG20200001), and Doctoral Fund Project of Jinan University (XBS2103).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Albuquerque, L., and Costa, M. S. (2014). “The family gaiellaceae,” in The Prokaryotes: Actinobacteria. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Heidelberg: Springer), 357–360.
Blume, E., Bischoff, M., Reichert, J. M., Moorman, T., Konopka, A., and Turco, R. F. (2002). Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl. Soil Ecol. 20, 171–181. doi: 10.1016/s0929-1393(02)00025-2
Braun, B., Böckelmann, U., Grohmann, E., and Szewzyk, U. (2006). Polyphasic characterization of the bacterial community in an urban soil profile with in situ and culture-dependent methods. Appl. Soil Ecol. 31, 267–279. doi: 10.1016/j.apsoil.2005.05.003
Calderón, K., Spor, A., Breuil, M. C., Calderón, K., Spor, A., Breuil, M.-C., et al. (2017). Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME J. 11, 272–283. doi: 10.1038/ismej.2016.86
Carrino-Kyker, S. R., Kluber, L. A., Petersen, S. M., Coyle, K. P., Hewins, C. R., DeForest, J. L., et al. (2016). Mycorrhizal fungal communities respond to experimental elevation of soil pH and P availability in temperate hardwood forests. FEMS Microbiol. Ecol. 92:fiw024. doi: 10.1093/femsec/fiw024
Chen, X., Sun, H., Jiang, F., Shen, Y., and Wei, P. (2020). Alteration of the gut microbiota associated with childhood obesity by 16S rRNA gene sequencing. PeerJ 8:e8317. doi: 10.7717/peerj.8317
Chen, F., Zheng, H., Zhang, K., Ouyang, Z., Lan, J., Li, H., et al. (2013). Changes in soil microbial community structure and metabolic activity following conversion from native Pinus massoniana plantations to exotic eucalyptus plantations. Forest Ecol. Manag. 291, 65–72. doi: 10.1016/j.foreco.2012.11.016
Dang, P., Gao, Y., Liu, J., Yu, S., and Zhao, Z. (2018). Effects of thinning intensity on understory vegetation and soil microbial communities of a mature Chinese pine plantation in the loess plateau. Sci. Total Environ. 630, 171–180. doi: 10.1016/j.scitotenv.2018.02.197
Dong, X., Du, X., Sun, Z., and Chen, X. (2021). Effects of residue retention and removal following thinning on soil bacterial community composition and diversity in a Larix olgensis plantation, Northeast China. Forests 12:559. doi: 10.3390/f12050559
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eilers, K. G., Debenport, S., Anderson, S., and Fierer, N. (2012). Digging deeper to find unique microbial communities: the strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 50, 58–65. doi: 10.1016/j.soilbio.2012.03.011
Feng, J., Gao, H. L., Wang, Q. C., Cao, Y., Bu, P. T., Wang, Z. W., et al. (2021). Growth and biomass allocation of mixed larch-ash forest and its pure stand in the eastern mountainous area of Liaoning Province. J. Northeast For. Univ. 49, 22–27. doi: 10.3969/j.issn.1000-5382.2021.07.004
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35, 167–176. doi: 10.1016/s0038-0717(02)00251-1
Fioretto, A., Papa, S., Curcio, E., Sorrentino, G., and Fuggi, A. (2000). Enzyme dynamics on decomposing leaf litter of Cistus incanus and Myrtus communis in a Mediterranean ecosystem. Soil Biol. Biochem. 32, 1847–1855. doi: 10.1016/s0038-0717(00)00158-9
Garau, G., Morillas, L., Roales, J., Castaldi, P., Mangia, N. P., Spano, D., et al. (2019). Effect of monospecific and mixed Mediterranean tree plantations on soil microbial community and biochemical functioning. Appl. Soil Ecol. 140, 78–88. doi: 10.1016/j.apsoil.2019.04.005
Ge, Y., Wang, Q., Wang, L., Liu, W., Liu, X., Huang, Y., et al. (2017). Response of soil enzymes and microbial communities to root extracts of the alien Alternanthera philoxeroides. Arch. Agron. Soil Sci. 64, 708–717. doi: 10.1080/03650340.2017.1373186
Griffiths, R., Whiteleya, A., O’Donnellb, A., and Bailey, M. (2003). Influence of depth and sampling time on bacterial community structure in an upland grassland soil. FEMS Microbiol. Ecol. 43, 35–43. doi: 10.1111/j.1574-6941.2003.tb01043
Harris, J. (2009). Soil microbial communities and restoration ecology: facilitators or followers? Science 325, 573–574. doi: 10.1126/science.1172975
He, Y., Qin, L., Li, Z., Liang, X., Shao, M., and Tan, L. (2013). Carbon storage capacity of monoculture and mixed-species plantations in subtropical China. Forest Ecol. Manag. 295, 193–198. doi: 10.1016/j.foreco.2013.01.020
Hu, Y. L., Wang, S. L., and Zeng, D. H. (2006). Effects of single Chinese fir and mixed leaf litters on soil chemical, microbial properties and soil enzyme activities. Plant and Soil 282, 379–386. doi: 10.1007/s11104-006-0004-5
Hu, Y., Xiang, D., Veresoglou, S. D., Chen, F., Chen, Y., Hao, Z., et al. (2014). Soil organic carbon and soil structure are driving microbial abundance and community composition across the arid and semi-arid grasslands in northern China. Soil Biol. Biochem. 77, 51–57. doi: 10.1016/j.soilbio.2014.06.014
Huang, J., Sheng, X., He, L., Huang, Z., Wang, Q., and Zhang, Z. (2013). Characterization of depth-related changes in bacterial community compositions and functions of a paddy soil profile. FEMS Microbiol. Lett. 347, 33–42. doi: 10.1111/1574-6968.12218
Ishida, T. A., Nara, K., and Hogetsu, T. (2007). Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol. 174, 430–440. doi: 10.1111/j.1469-8137.2007.02016.x
Jiang, Y., Chen, C., Xu, Z., and Liu, Y. (2012). Effects of single and mixed species forest ecosystems on diversity and function of soil microbial community in subtropical China. J. Soil. Sediment. 12, 228–240. doi: 10.1007/s11368-011-0442-4
Jones, B., and Nachtsheim, C. J. (2009). Split-plot designs: what, why, and how. J. Qual. Technol. 41, 340–361. doi: 10.1016/j.jmva.2009.06.012
Khlifa, R., Paquette, A., Messier, C., Reich, P. B., and Munson, A. D. (2017). Do temperate tree species diversity and identity influence soil microbial community function and composition? Ecol. Evol. 7, 7965–7974. doi: 10.1002/ece3.3313
Kourtev, P. S., Ehrenfeld, J. G., and Huang, W. Z. (2002). Enzyme activities during litter decomposition of two exotic and two native plant species in hardwood forests of New Jersey. Soil Biol. Biochem. 34, 1207–1218. doi: 10.1016/s0038-0717(02)00057-3
Li, X., Sun, J., Wang, H., Li, X., Wang, J., and Zhang, H. (2017). Changes in the soil microbial phospholipid fatty acid profile with depth in three soil types of paddy fields in China. Geoderma 290, 69–74. doi: 10.1016/j.geoderma.2016.11.006
Li, X., Wang, H., Li, X., Li, X., and Zhang, H. (2019). Shifts in bacterial community composition increase with depth in three soil types from paddy fields in China. Pedobiologia 77:150589. doi: 10.1016/j.pedobi.2019.150589
Li, J., Zhou, X., Yan, J., Li, H., and He, J. (2015). Effects of regenerating vegetation on soil enzyme activity and microbial structure in reclaimed soils on a surface coal mine site. Appl. Soil Ecol. 87, 56–62. doi: 10.1016/j.apsoil.2014.11.010
Liu, C. L. C., Kuchma, O., and Krutovsky, K. V. (2018). Mixed-species versus monocultures in plantation forestry: development, benefits, ecosystem services and perspectives for the future. Glob. Ecol. Conserv. 15:e00419. doi: 10.1016/j.gecco.2018.e00419
Liu, S., Li, X., and Niu, L. (1998). The degradation of soil fertility in pure Larix plantations in the northeastern part of China. Ecol. Eng. 10, 75–86. doi: 10.1016/S0925-8574(97)10024-6
López-Lozano, N. E., Echeverría Molinar, A., Ortiz Durán, E. A., Hernández Rosales, M., and Souza, V. (2020). Bacterial diversity and interaction networks of agave lechuguilla rhizosphere differ significantly from bulk soil in the oligotrophic basin of cuatro cienegas. Front. Plant Sci. 11:1028. doi: 10.3389/fpls.2020.01028
Matheny, P. B., Aime, M. C., Bougher, N. L., Buyck, B., Desjardin, D. E., Horak, E., et al. (2009). Out of the Palaeotropics? Historical biogeography and diversification of the cosmopolitan ectomycorrhizal mushroom family Inocybaceae. J. Biogeogr. 36, 577–592. doi: 10.1111/j.1365-2699.2008.02055.x
Midgley, M. G., and Phillips, R. P. (2016). Resource stoichiometry and the biogeochemical consequences of nitrogen deposition in a mixed deciduous forest. Ecology 97, 3369–3378. doi: 10.1002/ecy.1595
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Otsing, E., Barantal, S., Anslan, S., Koricheva, J., and Tedersoo, L. (2018). Litter species richness and composition effects on fungal richness and community structure in decomposing foliar and root litter. Soil Biol. Biochem. 125, 328–339. doi: 10.1016/j.soilbio.2018.08.006
Pereira, A. P.d. A., Andrade, P. A. M.d., Bini, D., Durrer, A., Robin, A., Bouillet, J. P., et al. (2017). Shifts in the bacterial community composition along deep soil profiles in monospecific and mixed stands of Eucalyptus grandis and Acacia mangium. PLoS One 12:e0180371. doi: 10.1371/journal.pone.0180371
Pereira, A. P. A., Durrer, A., Gumiere, T., Nçalvesa, J. L. M., Robin, A., Bouillet, J. P., et al. (2018). Mixed eucalyptus plantations induce changes in microbial communities and increase biological functions in the soil and litter layers. Forest Ecol. Manag. 433, 332–342. doi: 10.1016/j.foreco.2018.11.018
Rachid, C. T. C. C., Balieiro, F. C., Peixoto, R. S., Pinheiro, Y. A. S., Piccolo, M. C., Chaer, G. M., et al. (2013). Mixed plantations can promote microbial integration and soil nitrate increases with changes in the N cycling genes. Soil Biol. Biochem. 66, 146–153. doi: 10.1016/j.soilbio.2013.07.005
Ranjard, L., and Richaume, A. (2001). Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152, 707–716. doi: 10.1016/s0923-2508(01)01251-7
Rashid, M. I., Mujawar, L. H., Shahzad, T., Almeelbi, T., Ismail, I. M. I., and Oves, M. (2016). Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 183, 26–41. doi: 10.1016/j.micres.2015.11.007
Saleem, M., Hu, J., and Jousset, A. (2019). More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annu. Rev. Ecol. Evol. S. 50, 145–168. doi: 10.1146/annurev-ecolsys-110617-062605
Samuel, A. D., Domua, C., Andor, M., Vucan, A., and Domua, C. (2010). The estimation of phosphatase activity in soil. Res. J. Agric. Sci. 42, 311–314.
Sansupa, C., Wahdan, S. F. M., Hossen, S., Disayathanoowat, T., Wubet, T., and Purahong, W. (2021). Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl. Sci. 11:688. doi: 10.3390/app11020688
Sharma, S. K., Ramesh, A., Sharma, M. P., Joshi, O. P., Govaerts, B., Steenwerth, K. L., et al. (2010). “Microbial community structure and diversity as indicators for evaluating soil quality,” in Biodiversity, Biofuels, Agroforestry and Conservation Agriculture. eds. E. Lichtfouse (Springer, Dordrecht), 317–358.
Sun, R., Li, W., Dong, W., Tian, Y., Hu, C., and Liu, B. (2018). Tillage changes vertical distribution of soil bacterial and fungal communities. Front. Microbiol. 9:699. doi: 10.3389/fmicb.2018.00699
Sweeney, C. J., de Vries, F. T., van Dongen, B. E., and Bardgett, R. D. (2020). Root traits explain rhizosphere fungal community composition among temperate grassland plant species. New Phytol. 229, 1492–1507. doi: 10.1111/nph.16976
Testa, G., Piñones, A., and Castro, L. R. (2021). Physical and biogeochemical regionalization of the Southern Ocean and the CCAMLR zone 48.1. Front. Mar. Sci. 8:592378. doi: 10.3389/fmars.2021.592378
Too, C. C., Keller, A., Sickel, W., Lee, S. M., and Yule, C. M. (2018). Microbial community structure in a malaysian tropical peat swamp forest: the influence of tree species and depth. Front. Microbiol. 9:2859. doi: 10.3389/fmicb.2018.02859
Van Bruggen, A. H. C., and Semenov, A. M. (2000). In search of biological indicators for soil health and disease suppression. Appl. Soil Ecol. 15, 13–24. doi: 10.1016/s0929-1393(00)00068-8
Vasconcellos, R. L. F., Bonfim, J. A., Andreote, F. D., Mendes, L. W., and Cardoso, E. J. B. N. (2013). Microbiological indicators of soil quality in a riparian forest recovery gradient. Ecol. Eng. 53, 313–320. doi: 10.1016/j.ecoleng.2012.12.067
Wang, W., Chen, D., Sun, X., Zhang, Q., Koide, R. T., Insam, H., et al. (2019). Impacts of mixed litter on the structure and functional pathway of microbial community in litter decomposition. Appl. Soil Ecol. 144, 72–82. doi: 10.1016/j.apsoil.2019.07.006
Wang, X., Fang, J., and Zhu, B. (2008). Forest biomass and root–shoot allocation in Northeast China. Forest Ecol. Manag. 255, 4007–4020. doi: 10.1016/j.foreco.2008.03.055
Wang, Q., Wang, S., Fan, B., and Yu, X. (2007). Litter production, leaf litter decomposition and nutrient return in Cunninghamia lanceolata plantations in South China: effect of planting conifers with broadleaved species. Plant and Soil 297, 201–211. doi: 10.1007/s11104-007-9333-2
Wang, Q., Wang, C., Yu, W., Turak, A., Chen, D., Huang, Y., et al. (2018). Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese fir plantations. Front. Microbiol. 9:1543. doi: 10.3389/fmicb.2018.01543
Whelan, F. J., and Surette, M. G. (2017). A comprehensive evaluation of the sl1p pipeline for 16S rRNA gene sequencing analysis. Microbiome 5:100. doi: 10.1186/s40168-017-0314-2
Wu, D., Zhang, M., Peng, M., Sui, X., Li, W., and Sun, G. (2019a). Variations in soil functional fungal community structure associated with pure and mixed plantations in typical temperate forests of China. Front. Microbiol. 10:1636. doi: 10.3389/fmicb.2019.01636
Wu, W., Zhou, X., Wen, Y., Zhu, H., You, Y., Qin, Z., et al. (2019b). Coniferous-broadleaf mixture increases soil microbial biomass and functions accompanied by improved stand biomass and litter production in subtropical China. Forests 10:879. doi: 10.3390/f10100879
Xing, S., Chen, C., Zhou, B., Zhang, H., Nang, Z., and Xu, Z. (2009). Soil soluble organic nitrogen and active microbial characteristics under adjacent coniferous and broadleaf plantation forests. J. Soil. Sediment. 10, 748–757. doi: 10.1007/s11368-009-0159-9
Xu, Z., Yu, G., Zhang, X., He, N., Wang, Q., Wang, S., et al. (2017). Soil enzyme activity and stoichiometry in forest ecosystems along the north-south transect in eastern China (NSTEC). Soil Biol. Biochem. 104, 152–163. doi: 10.1016/j.soilbio.2016.10.020
Yang, L., and Yang, K. (2020). Biological function of Klebsiella variicola and its effect on the rhizosphere soil of maize seedlings. Peer J 8:e9894. doi: 10.7717/peerj.9894
Yang, K., Zhu, J., Zhang, M., Yan, Q. L., and Sun, O. J. (2010). Soil microbial biomass carbon and nitrogen in forest ecosystems of Northeast China: a comparison between natural secondary forest and Larix plantation. J. Plant Ecol. 3, 175–182. doi: 10.1093/jpe/rtq022
Zhang, W., Liu, W., He, S., Chen, Q., Han, J., and Zhang, Q. (2021). Mixed plantations of Metasequoia glyptostroboides and Bischofia polycarpa change soil fungal and archaeal communities and enhance soil phosphorus availability in Shanghai, China. Ecol. Evol. 11, 7239–7249. doi: 10.1002/ece3.7532
Keywords: mixed plantation, nutrient cycling function, soil microbial community, soil depth, enzyme activity
Citation: Wang W, Wang J, Wang Q, Bermudez RS, Yu S, Bu P, Wang Z, Chen D and Feng J (2022) Effects of Plantation Type and Soil Depth on Microbial Community Structure and Nutrient Cycling Function. Front. Microbiol. 13:846468. doi: 10.3389/fmicb.2022.846468
Edited by:
Yongchun Li, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Xin Sui, Heilongjiang University, ChinaTouqeer Abbas, University of Minnesota Twin Cities, United States
Copyright © 2022 Wang, Wang, Wang, Bermudez, Yu, Bu, Wang, Chen and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Feng, bG5sa3lmakAxNjMuY29t
 Wenbo Wang
Wenbo Wang Jianjun Wang2
Jianjun Wang2 Ramon Santos Bermudez
Ramon Santos Bermudez