- 1State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng, China
- 2Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
Insect herbivores can adversely impact terrestrial plants throughout ontogeny and across various ecosystems. Simultaneously, the effects of foliar herbivory may extend belowground, to the soil microbial community. However, the responses in terms of the diversity, assembly, and stability of rhizosphere fungi to aboveground herbivory remain understudied. Here, using high-throughput sequencing, the effects of foliar insect herbivory on rhizosphere fungal microbes were investigated in a common garden experiment that manipulated herbivory intensity and time from herbivore removal. The number of observed fungal species was reduced by a greater herbivory intensity, with some species evidently sensitive to herbivory intensity and time since herbivore removal. Rhizofungal assembly processes were altered by both herbivory intensity and time since herbivore removal. Further, we found evidence that both factors strongly influenced fungal community stability: a high intensity of herbivory coupled with a shorter time since herbivore removal resulted in low stability. These results suggest that foliar herbivory can adversely alter fungal diversity and stability, which would in turn be harmful for plant health. Fortunately, the effect seems to gradually diminish with time elapsed after herbivore removal. Our findings provide a fresh, in-depth view into the roles of rhizofungi in enhancing the adaption ability of plants under environmental stress.
Introduction
All plants inevitably face various biotic and abiotic stresses (Bagchi et al., 2014; Kaisermann et al., 2017; Li et al., 2021). As an important biotic stress, insect herbivores may threaten plants in agricultural settings, forests, and other terrestrial ecosystems (Schoonhoven et al., 1998). During herbivore–plant coevolution, plants gained the ability to enhance their interaction with beneficial soil microbes to overcome this problem (Pineda et al., 2010; Tedersoo et al., 2020) by producing various metabolites (Howe and Jander, 2008; McCormick et al., 2012; Mithoefer and Boland, 2012) that can regulate plant–microbe interactions belowground (Biere and Bennett, 2013; Pangesti et al., 2013; Humphrey and Whiteman, 2020). Accordingly, soil microorganisms can be manipulated to help plants counter attacks and fitness impacts from aboveground insect herbivores (Pineda et al., 2017; Jiang et al., 2020). The rhizosphere is a major source of the plant microbiome and is considered as a “second genome” of plants due to its importance for their health (Lu et al., 2018). In particular, fungi are critical participants in soil biochemical cycles and they have been proven crucial for promoting plant growth and health in various environments (Bahram et al., 2013; van der Heijden et al., 2016; Fisher and Garner, 2020; Yuan et al., 2021b). The rhizosphere fungal community may undergo differential responses in the face of aboveground herbivory due to cascading effects spanning from herbivores to plants to the soil (Cai et al., 2016; Ma et al., 2018a). However, how the rhizofungal diversity and community shift under plant stress caused by insect herbivory remains understudied.
Currently, a key unresolved issue in microbial ecology is our understanding of microbial community assembly processes (Zhou et al., 2008; Stegen et al., 2012; Yang, 2021). To the best of our knowledge, microbial assembly can entail the following five processes: dispersal limitation, drift, homogenous selection, heterogeneous selection, and homogenous dispersal; all these scenarios were carefully described by Shi et al. (2019a). These five scenarios have been extensively studied and revealed in natural (Dini-Andreote et al., 2015; Stegen et al., 2015) and agricultural ecosystems (Shi et al., 2018; Dong et al., 2020; Jiao et al., 2020), with literature now available that reports on the response mechanism of assembly processes to long term fermentation (Fan et al., 2017), climate warming (Howe and Jander, 2008), succession of soil (23), and so forth. Yet that body of work has mainly investigated the bacterial community assembly processes in different habitats. For the fungi, there is a paucity of similar research, likely because of their highly divergent phylogenetic tree (Ning et al., 2020). The high sequence variability of the ITS marker gene can lead to more accurate taxonomic identification than is possible with SSU rRNA, but it makes sequence alignment a lot more unreliable (Xue et al., 2019; Ning et al., 2020). Fortunately, an approach named Phylogenetic-Bin-Based Null Model Analysis (iCAMP) provides an opportunity to explore fungal community assembly by using a “ghost” tree based on ITS fungal sequences (Ning et al., 2020), which is more reliable than a fast tree. It was suggested that iCAMP had higher specificity and accuracy than the entire community-based method and provides an effective tool to quantify microbial assembly processes (Ning et al., 2020). Therefore, it is now possible to survey how rhizofungal community assembly processes vary under herbivory stresses using this newly developed iCAMP approach.
Mounting evidence has revealed that species interactions in biological community networks are crucial for their stability (Neutel et al., 2002; Coux et al., 2016). Likewise, the capacity of soil microbial communities to maintaining stability under disturbances largely depends on the complexity of interactions between species within that community (Steele et al., 2011; Schmitt et al., 2012; Wu et al., 2021). Recently, co-occurrence network approaches have been increasingly applied to investigate in detail the association relationships among microbial individuals (Faust et al., 2012; Ma et al., 2018b; Shi et al., 2020a; Yuan et al., 2021a). In addition, network robustness, calculated as the degree of natural connectivity determined by “attacking” the edges and nodes characterizing the network (removing the nodes and edges in the network randomly; Albert et al., 2000; Peng and Wu, 2016) is frequently being used (Fan et al., 2018; Shi et al., 2019b; Wu et al., 2021); higher robustness of networks (greater resistance of natural connectivity under attack) indicates that the community is more stable, while a sharply declining slope indicates lower network stability. It is conceivable that rhizofungal community networks may be sensitive to aboveground insect herbivory, as a form of biotic disturbance; nevertheless, their property changes and stability in response to insect herbivory remain largely unknown.
To investigate how the rhizofungal community, assembly, and stability each responded to insect herbivory, Chinese tallow (Triadica sebifera L.) and larvae of Spodoptera litura Fabricius (a generalist herbivorous insect that attack Chinese tallow) were selected as a model system (Zhang et al., 2015). Chinese tallow, a perennial tree native to Asia, can produce numerous defensive metabolites, such as tannins and flavonoids, to resist aboveground insect herbivory (Wang et al., 2012; Huang et al., 2014). Recently, studies have reported that root-associated (mycorrhizal) fungi were strongly influenced by the flavonoids in both the roots and root exudates of Chinese tallow (Pei et al., 2020; Tian et al., 2021). These metabolites of Triadica plants in roots can be induced by foliar insect herbivory (Xiao et al., 2019). This indicates that plant metabolites may be a bridge between foliar insect herbivores and root-associated microbes. In addition, the self-recovery and self-maintenance of soil microbes after disturbance are important for soil health (Shade et al., 2012; Lehmann et al., 2020). Based on the knowledge above, here, we hypothesized that foliar insect herbivory and their elapse time would affect overall rhizofungal community composition, and we expected that the rhizofungal association network and its stability would be altered.
Materials and Methods
Plants and Insects
To investigate the response of the rhizofungal community to foliar herbivory, we used Chinese tallow [T. sebifera ‘Small’ (Euphorbiaceae)] and the armyworm S. litura (Lepidoptera: Noctuidae; a leaf-eating caterpillar) as our study system. Chinese tallow seeds were collected in November 2018 from a natural population near Wuhan, China (31°33′N, 114°07′E). Spodoptera litura larvae were obtained from Henan Jiyuan Baiyun Industry Co., Ltd., and reared on an artificial diet (Tu et al., 2010).
Experimental Design
Triadica seeds were sown in an unheated greenhouse at Henan University, Henan, China (34°49′N, 114°18′E), in April 2019. Four weeks later, similar-sized plants (with four or five leaves on them) were transplanted into individual pots (height: 15 cm, upper diameter: 19 cm, lower diameter: 12 cm). These pots were filled with a commercial soil and topsoil (surface litter was removed and then the topsoil was passed through a 1 cm mesh screen) that came from an abandoned field that had been used to grow tallow trees at Henan University (1:1 v:v). To diminish the space effects, we randomly shifted the position of the plants every day. To protect against natural insect herbivores, we enclosed all the prepared tallow plants within a nylon mesh cage (6 m × 4 m × 2 m, 0.1 mm mesh size). Then, 50-day-old healthy and uniform plants were used for the following herbivory treatments.
To understand the effects of foliar insect herbivory on soil fungi dynamics, we established treatment groups consisting of three different herbivory intensities (no herbivory, NH; low herbivory, LH; and high herbivory, HH) and three different durations of elapsed time since herbivore removal (2 weeks since herbivore removal, 2 W; 4 weeks since herbivore removal, 4 W; and 6 weeks since herbivore removal, 6 W). The herbivory treatments were implemented as follows: LH: One S. litura larva was placed on one healthy plant and removed after 7 days of feeding (~10% leaf area consumed); HH: Four S. litura larvae were placed on one healthy plant and removed after 7 days of feeding (~60% leaf area consumed). The control group (NH) comprised healthy plants with no herbivores. Each treatment had three replicates. To investigate the effects of time since herbivore removal upon soil fungal communities, we timed our harvests for 2 W, 4 W, and 6 W post-herbivore removal.
Soil Sampling, DNA Extraction, and Amplicon Sequencing
Holding their shoots, the experimental plants were gently shaken to remove any loosely bound soil, and then the soil that remained tightly bound to the surface of the fine roots was collected by manually brushing the soils and designated the rhizosphere soil (Philippot et al., 2013; Fan et al., 2017). In total, 27 rhizosphere soil samples were collected. All of these samples were stored at −20°C, and their DNA was extracted within 2 weeks.
Using a Power Soil DNA kit (MO BIO, Carlsbad, CA, United States), total DNA from each soil sample was extracted and stored at −40°C. To quantify the DNA concentration, a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, United States) was used; the DNA was diluted to approximately 25 ng/μl using distilled water and stored at −20°C until analysis.
The nuclear ribosomal internal transcribed spacer region (ITS rRNA gene) was amplified by polymerase chain reaction (PCR), using the fungal primer set ITS5-F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2-R (5′-GCTGCGTTCTTCATCGATGC-3′; Bokulich and Mills, 2013; Shi et al., 2020b). For the PCR, the samples were initially denatured at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, primer annealing at 50°C for 30 s, and extension at 72°C for 30 s; a final extension step of 5 min at 72°C was added, to ensure complete amplification of the target region. Each sample was amplified in triplicate, and the ensuing PCR products from each sample were pooled and purified. Next, the PCR products from each sample were combined in equimolar ratios in one tube and sequenced using an Illumina NovaSeq 6,000 platform at NovoGene, Beijing, China.1
Rhizosphere Soil Fungal Community Structure Analyses
Using QIIME 2 software (version 2019.7; Bolyen et al., 2019), the raw data sequences were processed and analyzed by following the workflow at https://qiime2.org. Briefly, to obtain the sub-operational taxonomic unit (OTU) table, quality control of the raw sequencing data was performed using the Deblur tool (Amir et al., 2017). To remove low-quality regions from the sequences, each sequence was truncated at 120 bp according to a sequence quality plot. De novo chimera filtering by the VSEARCH tool (Rognes et al., 2016) was used to identify and filter out chimeras. Although the high sequence variability of the ITS marker gene can lead to a more accurate taxonomic identification than is possible using SSU rRNA, it also makes sequence alignment a lot more unreliable. To access reliable phylogenetic information from the ITS region, ghost-tree (Fouquier et al., 2016) was used to create a phylogenetic tree. For this, a pre-built version (ghost_tree_90_qiime_ver7_dynamic_10.10.2017) was used and the command “qiime vsearch cluster-features-closed-reference” was applied, to filter the feature table so as to retain those IDs that matched the ghost tree, and nearly 65% of high quality sequences (1,005,160/1,546,429) were retained. The downstream data analysis was based on the retained sequences. Subsequent taxonomy assignment was done using the Sklearn-based taxonomy classifier with the dynamic Unite database from 10 October 2017.2 To compare fungal diversity metrics between the six treatments, 7,017 high-quality sequences were randomly selected for each sample.
Analyzing the Rhizosphere Soil Fungal Assembly Processes
The assembly of rhizosphere soil fungal communities under different foliar herbivory treatments was estimated using iCAMP, which is based on the turnover of each bin across samples (Ning et al., 2020). The five assembly processes—drift (DR), homogenizing dispersal (HD), dispersal limitation (DL), heterogeneous selection (HeS), and homogeneous selection (HoS)—were quantified in this study. Briefly, running the iCAMP procedure entails three major steps: the first is phylogenetic binning (dividing the observed data into different groups, namely “bins”); the second is the bin-based null model analysis to partition deterministic and stochastic contributions to HoS, HeS, HD, DL, and DR; and the third is to robustly integrate the results of different bins to assess the relative importance of each process (Ning et al., 2020). The minimal size of all bins in this study was 12, with 49 fungal bins obtained. A confidence index (percentage of null values less extreme than the observed value, i.e., a non-parametric one-side confidence level) was applied for the null model significance test.
Analyzing the Rhizosphere Soil Fungal Stability
Following the approach described by Weiss, co-occurrence networks were constructed using the SparCC package (Weiss et al., 2016). To improve the reliability of these networks, the OTU table was first filtered. Here we constructed six networks corresponding to the six treatment groups: NH, LH, HH, 2 W, 4 W, and 6 W. First, any singletons were removed, and then we retained only those OTUs present in more than 1/3 of all samples per treatment. In this way, 228 OTUs in NH, 203 OTUs in LH, 169 OTUs in HH, 187 OTUs in 2 W, 184 OTUs in 4 W, and 188 OTUs in 6 W were ultimately obtained and used to build the networks [following Weiss et al. (2016)].
Robustness testing is a powerful method to measure a network’s stability and has been used to investigate microbial community stability in multiple studies (Fan et al., 2017; Shi et al., 2019a; Wu et al., 2021). To do that in our study, each network natural connectivity was estimated by “attacking” the nodes (May, 1973) or edges (Jordan, 2009) in the SparCC network. In addition, following Banerjee et al. (2018), in our study network hubs, module hubs, and connectors were defined as keystone species (Shi et al., 2020b). To identify these hubs and connectors, the z-score (within-module degree) and c-score (participation coefficient) of each node within each network were calculated according to the method described by Guimera and Amaral (2005). On the basis of threshold values of the within-module degree (z-score) and participation coefficient (c-score) of nodes, those nodes with a z-score > 2.5 and c-score > 0.6 were designated network hubs, while those with a z-score > 2.5 and c-score < 0.6 were defined as module hubs; nodes with a z-score < 2.5 and c-score > 0.6 were deemed connectors; and nodes with a z-score < 2.5 and c-score < 0.6 were denoted as peripherals. The respective roles of these types of node in community networks has been described at length by Shi et al. (Shi et al., 2020b).
Results
Rhizosphere Soil Fungal Community Composition
Using high-throughput sequencing, we obtained between 7,071 and 44,737 high-quality fungal sequences per sample (mean value: 27,921), of which >99% were classified into 1,569 distinct OTUs. Over 70% of the total OTU sequences belonged to the Acomycotas (Figure 1). The number of observed OTUs was not influenced by the stage of herbivore removal, but it did decrease with increasing herbivory intensity (Figure 2). The soil fungal communities were similar under the differing herbivory intensities and the time since herbivore removal (Table 1). These findings suggested that the rhizosphere soil fungal community was not affected by how long the plants were free of herbivore pressure or the herbivore intensity, but the number of observed species was influenced by herbivory intensity.
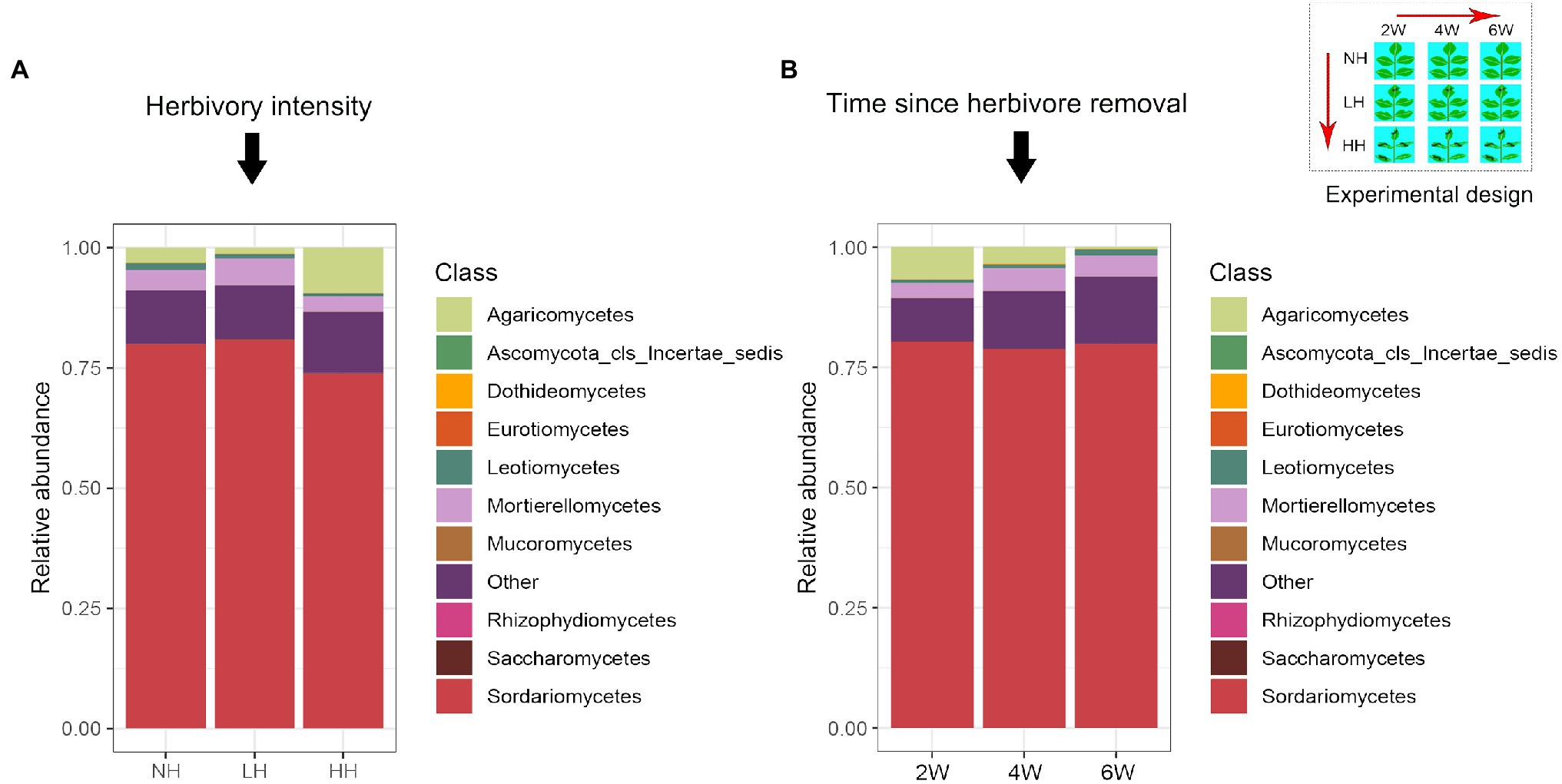
Figure 1. Relative abundance of the dominant fungal classes in rhizosphere soils as affected by different herbivory intensities (A) and time since herbivore removal (B).
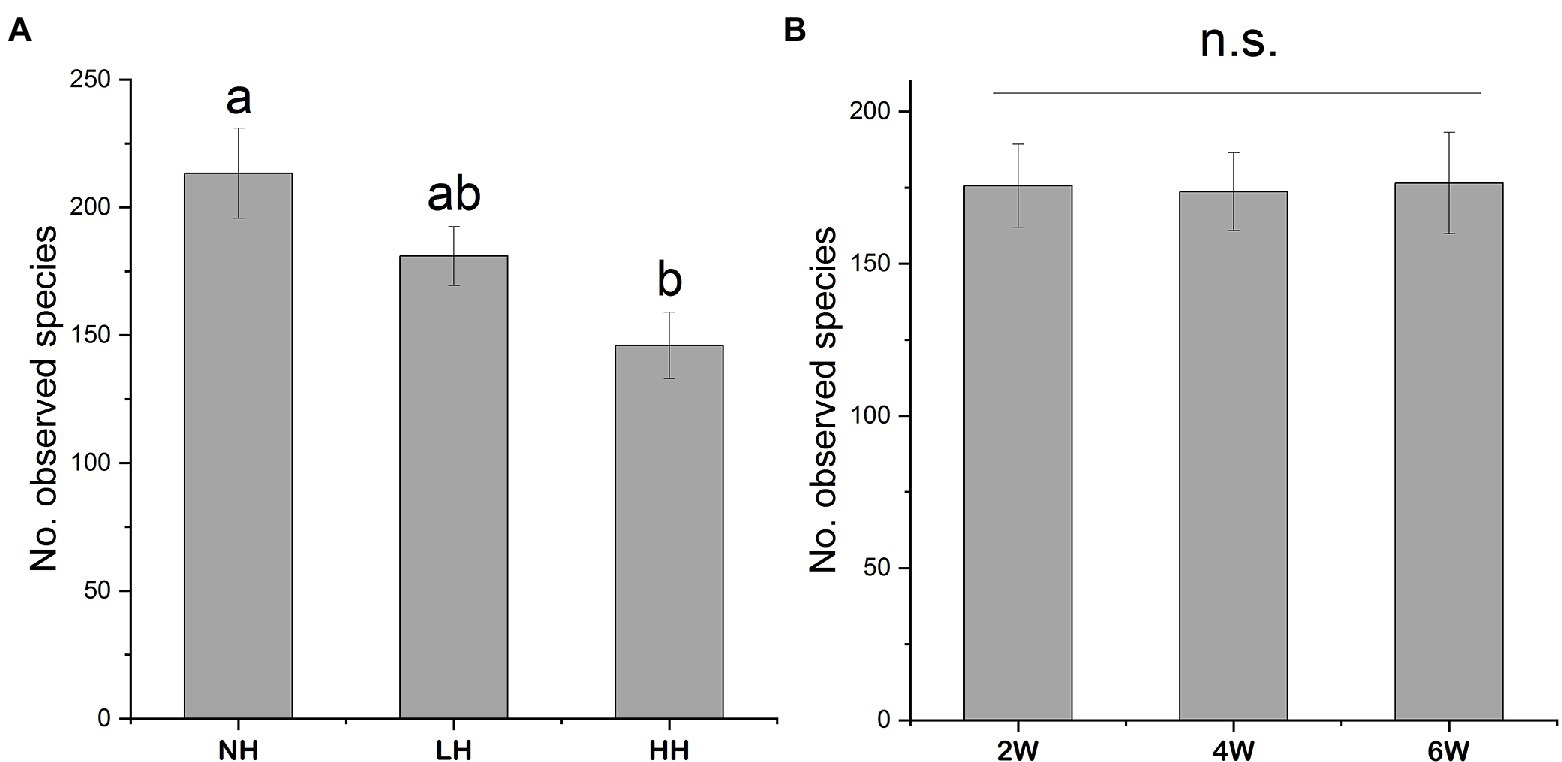
Figure 2. Number of observed rhizosphere fungal operational taxonomic units (OTUs) in rhizosphere soils as affected by different herbivory intensities (A) and time since herbivore removal (B). Different letters indicate statistically significant differences between treatments at p < 0.05. n.s., non-significant.
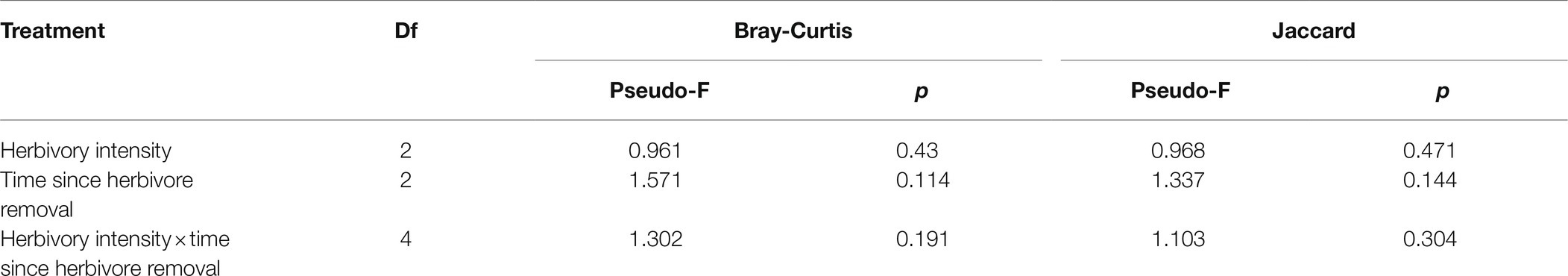
Table 1. PERMANOVA analysis of the dissimilarity of soil fungal communities as affected by different herbivory intensities and time since herbivore removal.
Not only the fungal community as a whole but also the individual species members exhibited variability in sensitivity to time since herbivore removal and herbivore intensity (Table 2). The relative abundance of many species of Agaricomycetes and Sordariomycetes was significantly increased the longer the plants remained free of herbivory. Among the three herbivory intensities, the HH treatment increased the relative abundance of several fungal species and decreased one species in the Agaricales. Furthermore, when compared with LH, the relative abundance of several species in the Ascomycota was decreased by the HH treatment.

Table 2. Significantly altered fungal OTUs between treatments, as affected by a different herbivory intensities and time since herbivore removal.
Rhizosphere Soil Fungal Assembly Processes
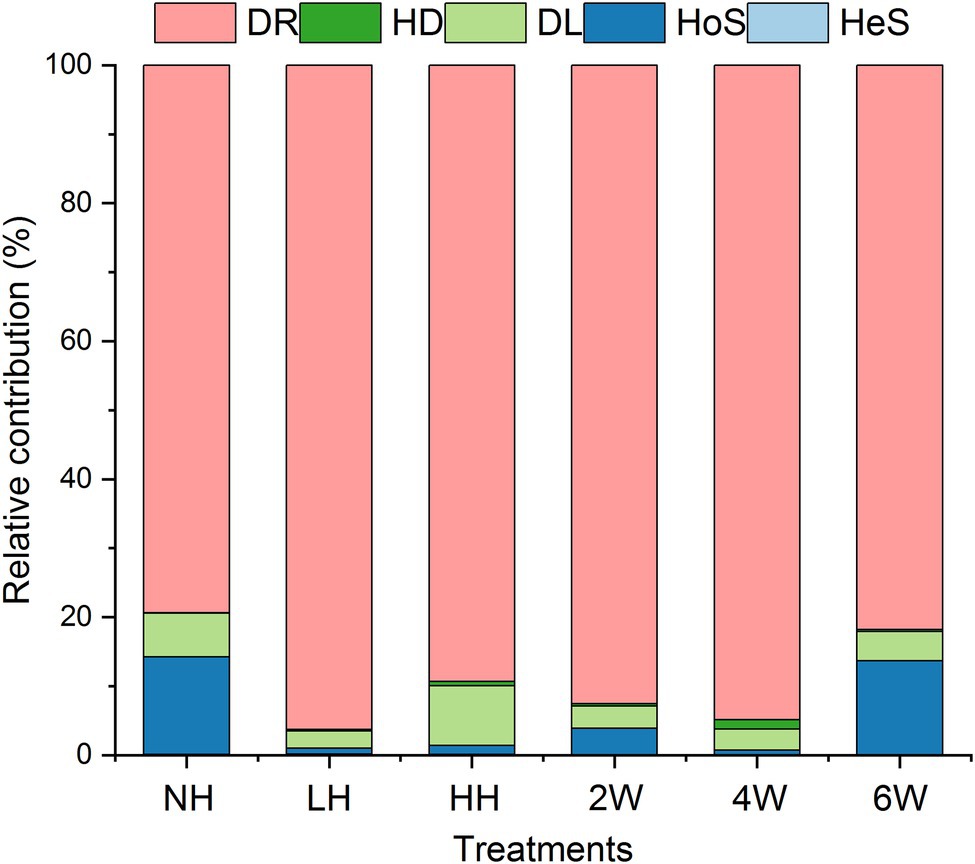
The iCAMP analysis revealed that drift was the most important process among the five responsible for rhizosphere fungal assembly, with an average relative importance of 79.4%–96.3% (Figure 3; Supplementary Table S1). However, foliar herbivory evidently altered the relative importance of the five assembly processes. The LH and HH treatments both increased the relative importance of drift and decreased the ratio of homogeneous selection, and HH also increased the ratio of dispersal limitation. Interestingly, the ratio of homogeneous selection was enhanced under the 6 W treatment; this suggested that how processes contribute to fungal assembly depends on the time elapsed since herbivore removal. In short, both insect herbivory and times since herbivore removal could alter the relevance of the five processes for rhizosphere soil fungal assembly.
Using iCAMP enabled us to better detect and infer the assembly mechanisms, by providing information on the relative importance of different ecological processes in individual lineages (i.e., bins). Here, the observed 1,569 OTUs were divisible into 49 phylogenetic bins (Supplementary Table S2), yet we found that the relative importance of a given assembly process was independent of the relative abundance in bins (Figure 4). Our results also revealed that the relative importance of each bin not only differed within the same treatment but also among the treatments (Figure 5; Supplementary Table S3).
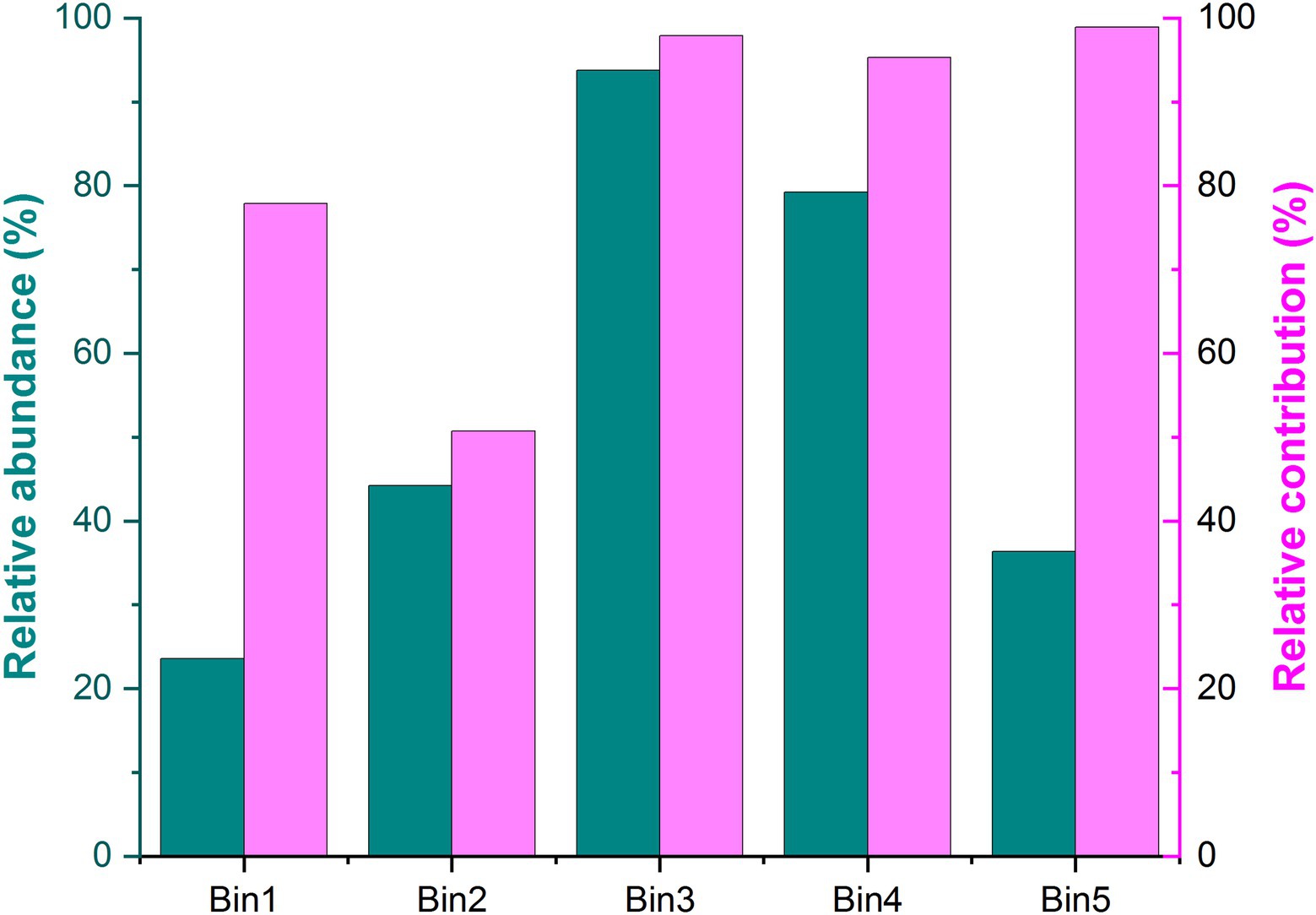
Figure 4. Relative abundance and relative contribution of each bin. Only the five most abundant bins are shown in this figure. Source data can be found in Supplementary Table S3.
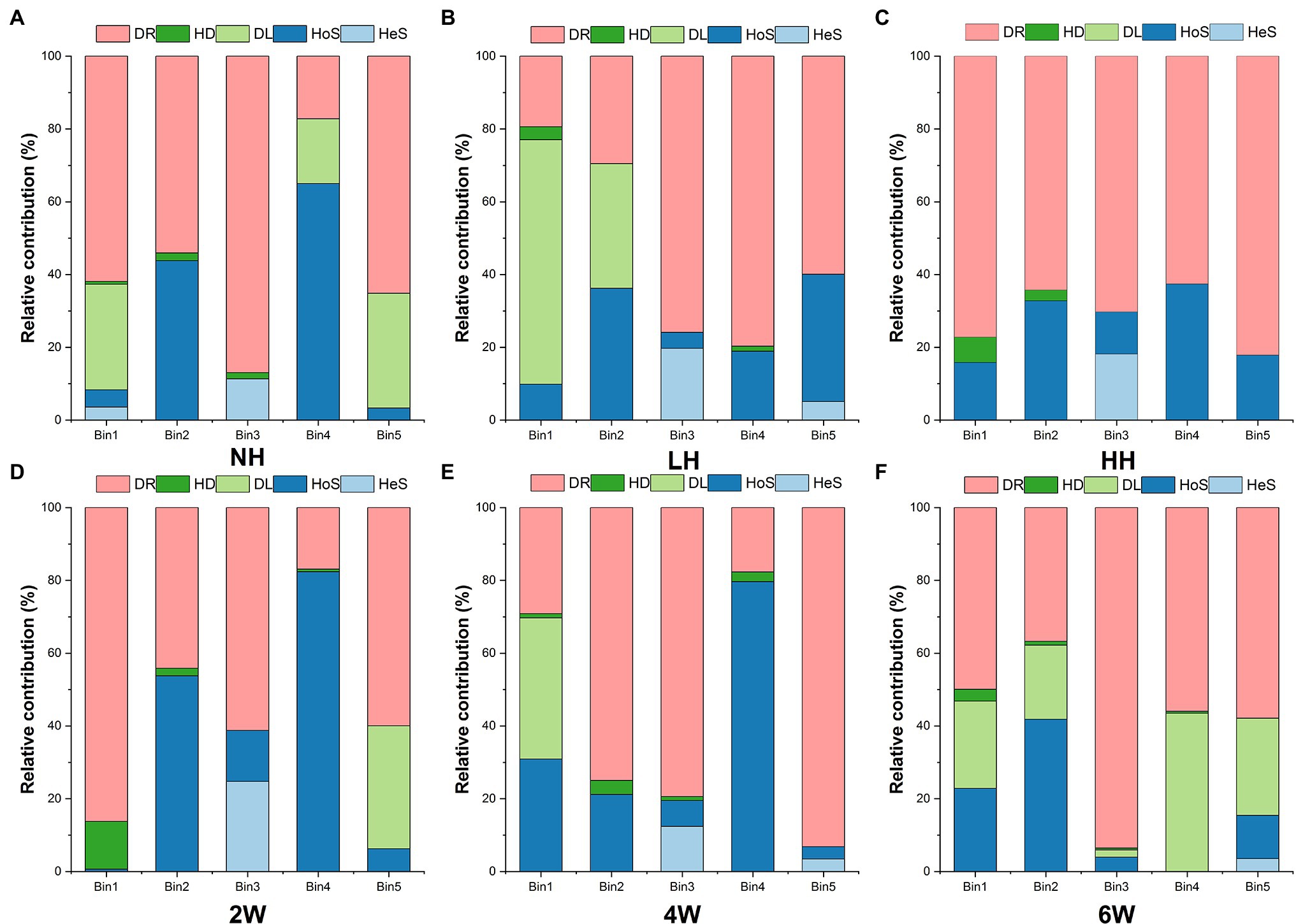
Figure 5. Relative abundance of each bin across the different treatment groups (A-C: different herbivory intensities; D-F: different time since herbivore removal). Only the five most abundant bins are shown in this figure. Source data can be found in Supplementary Table S4.
Rhizosphere Soil Fungal Stability
Robustness analysis is a powerful way to test a network’s stability. Accordingly, robustness of fungal networks to the treatments was also tested here, by altering the amplitude of natural connectivity via the deletion of nodes and edges (Figure 6). Nodes and edges were discarded in declining order of nodes’ betweenness. Doing this, we found that the natural connectivity of fungal networks presented sharp slopes in HH and 2 W treatment samples, indicating they had weak stability (Figure 6).
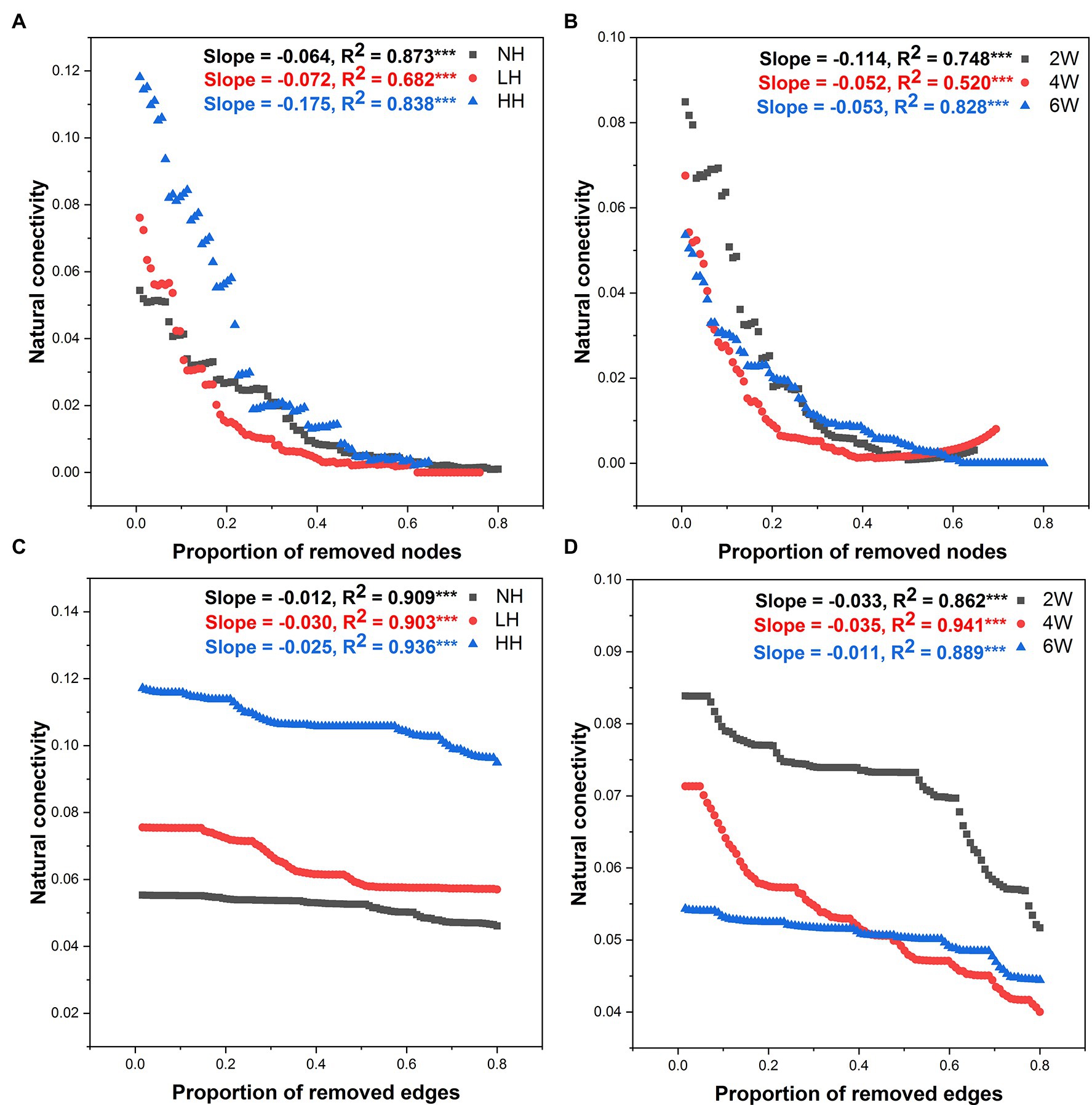
Figure 6. Robustness analysis for the rhizosphere soil fungal community as affected by different lengths of elapsed time since herbivore removal (A,C) and differing intensities of herbivory (B,D). Robustness analysis is depicted as the relationships between microbial natural connectivity and the proportion of excluded nodes and edges, such that larger shifts for the same proportion indicate less robustness or stability within the fungal networks. *** p < 0.001.
To identify the keystone species, z-scores and c-scores were calculated for the nodes in the network for each treatment. There were 4, 2, and 1 keystone species in the NH, LH, and HH treatment groups, respectively, and 7, 0, and 2 keystone species in the 2 W, 4 W, and 6 W treatment groups, respectively (Supplementary Table S4). This pattern suggested foliar insect herbivory as well as time since herbivore removal could slightly reduce the number of keystone species present in the soil fungal community of tallow plants.
Discussion
Foliar Herbivory Impact on Rhizofungal Community Composition
In contrast to our hypothesis, herbivory intensity and elapsed time since herbivore removal did not significantly influence the fungal community dissimilarity. This suggests the rhizosphere fungal community may not sensitive to herbivory from leaf-eating insects. However, the fungi are sensitive to partial defoliation is evident by the shift upwards or downwards seen in certain fungal species under the experimentally imposed herbivory conditions. In particular, the number of observed rhizofungal species was found to be significantly lower in the HH than the NH and LH treatments. A previous study showed that aboveground herbivory could affect soil pH (Custer et al., 2020). In addition, myriad studies have reported that the microbial community is strongly related to soil pH (Chu et al., 2010; Fierer, 2017; Xue et al., 2018; Gong et al., 2019). Therefore, the lower number of observed species in HH treatment may be related to altered soil pH, indirectly caused by foliar insect herbivory. We thus suspect that insect herbivory aboveground would enhance root exudation levels, and this could lead to a changed rhizosphere soil pH that influences fungal diversity (Rudrappa et al., 2008; Hoysted et al., 2018).
Foliar Herbivory Impact on Rhizofungal Assembly Process
Although the drift process drove most of the variation in fungal community composition across the samples from all treatments, slight changes in homogenous selection were discernible in some samples. For example, LH and HH treatments both reduced the relative contribution of the HoS process, while the DL process increased under HH. HoS refers to selection occurring under homogeneous abiotic and biotic conditions in both space and time (Dini-Andreote et al., 2015; Ning et al., 2020). So far as we know, deterministic processes (including HoS) which are mainly driven by biotic and abiotic factors are crucial for microbial community structure, therefore, the disturbed HoS process probably result from altered rhizosphere environments by foliar herbivory (Fan et al., 2017; Zhalnina et al., 2018; Shi et al., 2019b). This suggests herbivory has the potential for reducing the rhizosphere selection ability and limiting the community-level dispersal. This might be harmful for plant health, because the growth of plants is strongly related to their recruited microbes, and diminishing their dispersal ability would likely inhibit their recruitment ability (Bakker et al., 2018). Moreover, we found that the relative contribution of HoS increased at 6 W after herbivore removal. Probably, rhizosphere fungi possess self-regulatory ability after herbivory, which might be helpful for plant growth.
The iCAMP approach let us gain insight into the contribution of each fungal group by bin. Our results suggested that the contribution of each phylum or class in each bin was independent of their relative abundance (Figure 4), and the bin groupings contributed differentially to the five assembly scenarios in each treatment. Although other research has focused on the relative contribution of these processes to variation in soil microbial communities, they often neglect the critical role of each phylum or class. But, the role of these species is very important, because it is beneficial to supplement our knowledge of how to manage key microbial assembly processes by mediating the activity of the highly-contributing microbes (Ning et al., 2020).
Foliar Herbivory Impact on Rhizofungal Network Stability
Microbial community stability is crucial to ensure ecosystem functioning (Wu et al., 2021). Our study found that aboveground herbivores were quite capable of disturbing rhizofungal community stability. In particular, a high herbivory level significantly reduced the network stability, and this may portend a certain loss of rhizosphere ecological functioning. Further, the stability of the rhizofungal community was lowest at 2 W post-herbivory, and it tended to be stable at 6 W after herbivore removal. This result implied that the plant rhizosphere might exercise a self-recovery function. Numerous studies indicate microbial community stability can be affected by one or more local environmental stresses. For instance, Wu et al. (2021) found that enhanced warming could significantly reduce the microbial stability in the active layer, and Fan et al. found lower network stability in the microbial network of the rhizosphere than in bulk soil (Fan et al., 2017); however, few studies have considered the possibility of temporal or lag effects of biotic or abiotic disturbances to plants. This is the first study we know of that tested rhizofungal community stability under different herbivory intensities and at differing times after herbivore removal from the host plants. Given that plant roots have ability in filtering rhizosphere microbial communities and root exudation can mediate plant-microbe interactions (Fan et al., 2017; Zhalnina et al., 2018), while the root traits can be altered by aboveground herbivory (Karssemeijer et al., 2020), we suspect that foliar herbivory impacts on rhizofungal community network may mediated by the cascading effects on plants (Friman et al., 2021).
Concluding Remarks
Collectively, our findings reveal that leaf herbivory from an aboveground insect herbivore can affect rhizofungal diversity, with potential weakening of fungal community stability under high herbivory intensity and soon after herbivore removal. The reduced fungal diversity and stability and disrupted assembly processes due to foliar herbivory may impose detrimental effects on plant health in various systems. However, these detrimental effects could be diminished with the extension of herbivore removal, it means the rhizofungal community may possess a self-recovery ability. Our results indicate that aboveground insect herbivores not only influence the rhizosphere microbial community diversity and stability, but might also trigger cascading effects on rhizosphere ecosystem function and cause diminished vitality in this active region belowground. Foliar herbivory should therefore be taken into consideration in future field and greenhouse studies that manipulate soil microbes to improve plant health. Terrestrial ecosystem includes a vast variety of herbivore guilds, plant species and soil types, whether foliar herbivores trigger similar fungi responses in other systems remain unclear, and needs to be further investigated in future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA753601.
Author Contributions
ZX designed the study. YS, ZX, KZ, and TM performed research and analyzed data. YS and ZX wrote the manuscript. ZZ and PL edited the manuscript. JD, YS, and ZX finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFD1900100), the National Natural Science Foundation of China (31770414), and the Natural Science Foundation of Henan (212300410114 and 222300420035).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the native English-speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.846332/full#supplementary-material
Abbreviations
NH, No herbivory; LH, Low herbivory; HH, High herbivory; 2 W, 2 Weeks after herbivore removal; 4 W, 4 Weeks after herbivore removal; 6 W, 6 Weeks after herbivore removal; DR, Drift; HD, Homogenizing dispersal; DL, Dispersal limitation; HeS, Heterogeneous selection; HoS, Homogeneous selection.
Footnotes
References
Albert, R., Jeong, H., and Barabasi, A. L. (2000). Error and attack tolerance of complex networks. Nature 406, 378–382. doi: 10.1038/35019019
Amir, A., McDonald, D., Navas-Molina, J. A., Kopylova, E., Morton, J. T., Zech Xu, Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. msystems 2, e00191–e00116. doi: 10.1128/mSystems.00191-16
Bagchi, R., Gallery, R. E., Gripenberg, S., Gurr, S. J., Narayan, L., Addis, C. E., et al. (2014). Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature 506, 85–88. doi: 10.1038/nature12911
Bahram, M., Koljalg, U., Courty, P. E., Diedhiou, A. G., Kjoller, R., Polme, S., et al. (2013). The distance decay of similarity in communities of ectomycorrhizal fungi in different ecosystems and scales. J. Ecol. 101, 1335–1344. doi: 10.1111/1365-2745.12120
Bakker, P., Pieterse, C. M. J., de Jonge, R., and Berendsen, R. L. (2018). The soil-borne legacy. Cell 172, 1178–1180. doi: 10.1016/j.cell.2018.02.024
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Biere, A., and Bennett, A. E. (2013). Three-way interactions between plants, microbes and insects. Funct. Ecol. 27, 567–573. doi: 10.1111/1365-2435.12100
Bokulich, N. A., and Mills, D. A. (2013). Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79, 2519–2526. doi: 10.1128/AEM.03870-12
Bolyen, E., Rideout, J. R., and Dillon, M. R. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 1091–1091. doi: 10.1038/s41587-019-0252-6
Cai, Y., Chang, S. X., Ma, B., and Bork, E. W. (2016). Watering increased DOC concentration but decreased N2O emission from a mixed grassland soil under different defoliation regimes. Biol. Fertil. Soils 52, 987–996. doi: 10.1007/s00374-016-1135-3
Chu, H., Fierer, N., Lauber, C. L., Caporaso, J. G., Knight, R., and Grogan, P. (2010). Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 12, 2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x
Coux, C., Rader, R., Bartomeus, I., and Tylianakis, J. M. (2016). Linking species functional roles to their network roles. Ecol. Lett. 19, 762–770. doi: 10.1111/ele.12612
Custer, G. F., van Diepen, L. T. A., and Stump, W. L. (2020). Structural and functional dynamics of soil microbes following spruce beetle infestation. Appl. Environ. Microbiol. 86, e01984–e01919. doi: 10.1128/AEM.01984-19
Dini-Andreote, F., Stegen, J. C., van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U. S. A. 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Dong, M., Zhao, M., Shen, Z., Deng, X., Ou, Y., Tao, C., et al. (2020). Biofertilizer application triggered microbial assembly in microaggregates associated with tomato bacterial wilt suppression. Biol. Fertil. Soils 56, 551–563. doi: 10.1007/s00374-020-01459-8
Fan, K., Cardona, C., Li, Y., Shi, Y., Xiang, X., Shen, C., et al. (2017). Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol. Biochem. 113, 275–284. doi: 10.1016/j.soilbio.2017.06.020
Fan, K. K., Weisenhorn, P., Gilbert, J. A., and Chu, H. Y. (2018). Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 125, 251–260. doi: 10.1016/j.soilbio.2018.07.022
Faust, K., Sathirapongsasuti, J. F., Izard, J., Segata, N., Gevers, D., Raes, J., et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. doi: 10.1371/journal.pcbi.1002606
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fisher, M. C., and Garner, T. W. J. (2020). Chytrid fungi and global amphibian declines. Nat. Rev. Microbiol. 18, 332–343. doi: 10.1038/s41579-020-0335-x
Fouquier, J., Rideout, J. R., Bolyen, E., Chase, J., Shiffer, A., McDonald, D., et al. (2016). Ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses. Microbiome 4:11. doi: 10.1186/s40168-016-0153-6
Friman, J., Karssemeijer, P. N., Haller, J., de Kreek, K., van Loon, J. J. A., and Dicke, M. (2021). Shoot and root insect herbivory change the plant rhizosphere microbiome and affects cabbage-insect interactions through plant-soil feedback. New Phytol. 232, 2475–2490. doi: 10.1111/nph.17746
Gong, X., Wang, S., Wang, Z., Jiang, Y., Hu, Z., Zheng, Y., et al. (2019). Earthworms modify soil bacterial and fungal communities through enhancing aggregation and buffering pH. Geoderma 347, 59–69. doi: 10.1016/j.geoderma.2019.03.043
Guimera, R., and Amaral, L. A. N. (2005). Functional cartography of complex metabolic networks. Nature 433, 895–900. doi: 10.1038/nature03288
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Hoysted, G. A., Bell, C. A., Lilley, C. J., and Urwin, P. E. (2018). Aphid colonization affects potato root exudate composition and the hatching of a soil borne pathogen. Front. Plant Sci. 9:1278. doi: 10.3389/fpls.2018.01278
Huang, W., Siemann, E., Xiao, L., Yang, X. F., and Ding, J. Q. (2014). Species-specific defence responses facilitate conspecifics and inhibit heterospecifics in above-belowground herbivore interactions. Nat. Commun. 5:4851. doi: 10.1038/ncomms5851
Humphrey, P. T., and Whiteman, N. K. (2020). Insect herbivory reshapes a native leaf microbiome. Nat. Ecol. Evol. 4, 221–229. doi: 10.1038/s41559-019-1085-x
Jiang, L., Bonkowski, M., Luo, L., Kardol, P., Zhang, Y., Chen, X., et al. (2020). Combined addition of chemical and organic amendments enhances plant resistance to aboveground herbivores through increasing microbial abundance and diversity. Biol. Fertil. Soils 56, 1007–1022. doi: 10.1007/s00374-020-01473-w
Jiao, S., Yang, Y. F., Xu, Y. Q., Zhang, J., and Lu, Y. H. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Jordan, F. (2009). Keystone species and food webs. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 364, 1733–1741. doi: 10.1098/rstb.2008.0335
Kaisermann, A., de Vries, F. T., Griffiths, R. I., and Bardgett, R. D. (2017). Legacy effects of drought on plant-soil feedbacks and plant-plant interactions. New Phytol. 215, 1413–1424. doi: 10.1111/nph.14661
Karssemeijer, P. N., Reichelt, M., Gershenzon, J., van Loon, J., and Dicke, M. (2020). Foliar herbivory by caterpillars and aphids differentially affects phytohormonal signalling in roots and plant defence to a root herbivore. Plant Cell Environ. 43, 775–786. doi: 10.1111/pce.13707
Lehmann, J., Bossio, D. A., Kogel-Knabner, I., and Rillig, M. C. (2020). The concept and future prospects of soil health. Nat. Rev. Earth Environ. 1, 544–553. doi: 10.1038/s43017-020-0080-8
Li, H., La, S. K., Zhang, X., Gao, L. H., and Tian, Y. Q. (2021). Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 15, 2865–2882. doi: 10.1038/s41396-41021-00974-41392
Lu, T., Ke, M., Lavoie, M., Jin, Y., Fan, X., Zhang, Z., et al. (2018). Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome 6:231. doi: 10.1186/s40168-018-0615-0
Ma, B., Cai, Y., Bork, E. W., and Chang, S. X. (2018a). Defoliation intensity and elevated precipitation effects on microbiome and interactome depend on site type in northern mixed-grass prairie. Soil Biol. Biochem. 122, 163–172. doi: 10.1016/j.soilbio.2018.04.015
Ma, B., Lv, X., Cai, Y., Chang, S. X., and Dyck, M. F. (2018b). Liming does not counteract the influence of long-term fertilization on soil bacterial community structure and its co-occurrence pattern. Soil Biol. Biochem. 123, 45–53. doi: 10.1016/j.soilbio.2018.05.003
May, R.M. (1973). Stability and Complexity in Model Ecosystems. Princeton University Press: Princeton, New Jersey.
McCormick, A. C., Unsicker, S. B., and Gershenzon, J. (2012). The specificity of herbivore-induced plant volatiles in attracting herbivore enemies. Trends Plant Sci. 17, 303–310. doi: 10.1016/j.tplants.2012.03.012
Mithoefer, A., and Boland, W. (2012). Plant defense against herbivores: chemical aspects. Annu. Rev. Plant Biol. 63, 431–450. doi: 10.1146/annurev-arplant-042110-103854
Neutel, A. M., Heesterbeek, J. A. P., and de Ruiter, P. C. (2002). Stability in real food webs: weak links in long loops. Science 296, 1120–1123. doi: 10.1126/science.1068326
Ning, D., Yuan, M., Wu, L., Zhang, Y., Guo, X., Zhou, X., et al. (2020). A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11:4717. doi: 10.1038/s41467-020-18560-z
Pangesti, N., Pineda, A., Pieterse, C. M. J., Dicke, M., and van Loon, J. J. A. (2013). Two-way plant-mediated interactions between root-associated microbes and insects: from ecology to mechanisms. Front. Plant Sci. 4:414. doi: 10.3389/fpls.2013.00414
Pei, Y. C., Siemann, E., Tian, B. L., and Ding, J. Q. (2020). Root flavonoids are related to enhanced AMF colonization of an invasive tree. AoB Plants 12:Plaa002. doi: 10.1093/aobpla/plaa002
Peng, G. S., and Wu, J. (2016). Optimal network topology for structural robustness based on natural connectivity. Phys. A: Stat. Mech. Appl. 443, 212–220. doi: 10.1016/j.physa.2015.09.023
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pineda, A., Kaplan, I., and Bezemer, T. M. (2017). Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 22, 770–778. doi: 10.1016/j.tplants.2017.07.002
Pineda, A., Zheng, S.-J., van Loon, J. J. A., Pieterse, C. M. J., and Dicke, M. (2010). Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci. 15, 507–514. doi: 10.1016/j.tplants.2010.05.007
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rudrappa, T., Czymmek, K. J., Paré, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Schmitt, S., Tsai, P., Bell, J., Fromont, J., Ilan, M., Lindquist, N., et al. (2012). Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 6, 564–576. doi: 10.1038/ismej.2011.116
Schoonhoven, L. M., Jermey, T., and van Loon, J. J. A. (1998). Insect-Plant Biology: From Physiology to Evolution. London: Chapman & Hall.
Shade, A., Peter, H., Allison, S. D., Baho, D. L., Berga, M., Burgmann, H., et al. (2012). Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:417. doi: 10.3389/fmicb.2012.00417
Shi, Y., Dang, K. K., Dong, Y. H., Feng, M. M., Wang, B. R., Li, J. G., et al. (2019a). Soil fungal community assembly processes under long-term fertilization. Eur. J. Soil Sci. 71, 716–726. doi: 10.1111/ejss.12902
Shi, Y., Delgado-Baquerizo, M., Li, Y., Yang, Y., Zhu, Y.-G., Peñuelas, J., et al. (2020a). Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ. Int. 142:105869. doi: 10.1016/j.envint.2020.105869
Shi, Y., Fan, K., Li, Y., Yang, T., He, J.-S., and Chu, H. (2019b). Archaea enhance the robustness of microbial co-occurrence networks in Tibetan plateau soils. Soil Sci. Soc. Am. J. 83, 1093–1099. doi: 10.2136/sssaj2018.11.0426
Shi, Y., Li, Y. T., Xiang, X. J., Sun, R. B., Yang, T., He, D., et al. (2018). Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China plain. Microbiome 6:27. doi: 10.1186/s40168-018-0409-4
Steele, J. A., Countway, P. D., Xia, L., Vigil, P. D., Beman, J. M., Kim, D. Y., et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425. doi: 10.1038/ismej.2011.24
Stegen, J. C., Lin, X. J., Fredrickson, J. K., and Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6:370. doi: 10.3389/fmicb.2015.00370
Stegen, J. C., Lin, X. J., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Tedersoo, L., Bahram, M., and Zobel, M. (2020). How mycorrhizal associations drive plant population and community biology. Science 367:eaba1223. doi: 10.1126/science.aba1223
Tian, B. L., Pei, Y. C., Huang, W., Ding, J. Q., and Siemann, E. (2021). Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J. 15, 1919–1930. doi: 10.1038/s41396-021-00894-1
Tu, Y. G., Wu, K. M., Xue, F. S., and Lu, Y. H. (2010). Laboratory evaluation of flight activity of the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Insect Sci. 17, 53–59. doi: 10.1111/j.1744-7917.2009.01281.x
van der Heijden, M. G., de Bruin, S., Luckerhoff, L., van Logtestijn, R. S., and Schlaeppi, K. (2016). A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399. doi: 10.1038/ismej.2015.120
Wang, Y., Siemann, E., Wheeler, G. S., Zhu, L., Gu, X., and Ding, J. Q. (2012). Genetic variation in anti-herbivore chemical defences in an invasive plant. J. Ecol. 100, 894–904. doi: 10.1111/j.1365-2745.2012.01980.x
Weiss, S., Van Treuren, W., Lozupone, C., Faust, K., Friedman, J., Deng, Y., et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681. doi: 10.1038/ismej.2015.235
Wu, M. H., Chen, S. Y., Chen, J. W., Xue, K., Chen, S. L., Wang, X. M., et al. (2021). Reduced microbial stability in the active layer is associated with carbon loss under alpine permafrost degradation. Proc. Natl. Acad. Sci. U. S. A. 118:e2025321118. doi: 10.1073/pnas.2025321118
Xiao, L., Carrillo, J., Siemann, E., and Ding, J. Q. (2019). Herbivore-specific induction of indirect and direct defensive responses in leaves and roots. AoB Plants 11:plz003. doi: 10.1093/aobpla/plz003
Xue, C., Hao, Y., Pu, X., Penton, C. R., Wang, Q., Zhao, M., et al. (2019). Effect of LSU and ITS genetic markers and reference databases on analyses of fungal communities. Biol. Fertil. Soils 55, 79–88. doi: 10.1007/s00374-018-1331-4
Xue, C., Penton, C. R., Zhu, C., Chen, H., Duan, Y., Peng, C., et al. (2018). Alterations in soil fungal community composition and network assemblage structure by different long-term fertilization regimes are correlated to the soil ionome. Biol. Fertil. Soils 54, 95–106. doi: 10.1007/s00374-017-1241-x
Yang, Y. (2021). Emerging patterns of microbial functional traits. Trends Microbiol. 29, 874–882. doi: 10.1016/j.tim.2021.1004.1004
Yuan, M. M., Guo, X., Wu, L. W., Zhang, Y., Xiao, N. J., Ning, D. L., et al. (2021a). Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 11, 343–348. doi: 10.1038/s41558-021-00989-9
Yuan, X., Hong, S., Xiong, W., Raza, W., Shen, Z., Wang, B., et al. (2021b). Development of fungal-mediated soil suppressiveness against Fusarium wilt disease via plant residue manipulation. Microbiome 9:200. doi: 10.1186/s40168-021-01133-7
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, J. L., Wang, Y., and Ding, J. Q. (2015). List of pest insects on Triadica sebifera. Forest Pest and Disease 34, 25–35. doi: 10.3969/j.issn.1671-0886.2015.05.006
Keywords: foliar chewing herbivore, rhizosphere fungal community, Triadica sebifera, diversity, assembly processes, stability
Citation: Shi Y, Zhang K, Ma T, Zhang Z, Li P, Xing Z and Ding J (2022) Foliar Herbivory Reduces Rhizosphere Fungal Diversity and Destabilizes the Co-occurrence Network. Front. Microbiol. 13:846332. doi: 10.3389/fmicb.2022.846332
Edited by:
Yurong Liu, Huazhong Agricultural University, ChinaReviewed by:
Shi Huang, University of California, San Diego, United StatesCongcong Shen, Research Center for Eco-environmental Sciences (CAS), China
Copyright © 2022 Shi, Zhang, Ma, Zhang, Li, Xing and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenlong Xing, enhpbmdAdmlwLmhlbnUuZWR1LmNu
 Yu Shi
Yu Shi Kaoping Zhang2
Kaoping Zhang2 Zhenlong Xing
Zhenlong Xing Jianqing Ding
Jianqing Ding