- 1College of Life Sciences, Hebei University, Baoding, China
- 2Institute of Life Sciences and Green Development, Hebei University, Baoding, China
- 3College of Veterinary Medicine, Hebei Agricultural University, Baoding, China
Posttranscriptional modifications have been implicated in regulation of nearly all biological aspects of cellular RNAs, from stability, translation, splicing, nuclear export to localization. Chemical modifications also have been revealed for virus derived RNAs several decades before, along with the potential of their regulatory roles in virus infection. Due to the dynamic changes of RNA modifications during virus infection, illustrating the mechanisms of RNA epigenetic regulations remains a challenge. Nevertheless, many studies have indicated that these RNA epigenetic marks may directly regulate virus infection through antiviral innate immune responses. The present review summarizes the impacts of important epigenetic marks on viral RNAs, including N6-methyladenosine (m6A), 5-methylcytidine (m5C), 2ʹ-O-methylation (2ʹ-O-Methyl), and a few uncanonical nucleotides (A-to-I editing, pseudouridine), on antiviral innate immunity and relevant signaling pathways, while highlighting the significance of antiviral innate immune responses during virus infection.
Introduction
Chemical modifications of RNA, also be designated as epitranscriptomic marks of RNA, are considered common features in most natural RNAs. To date, more than 140 posttranscriptional modifications have been discovered to function in the structural diversity and metabolism of RNAs (Zhao et al., 2017). While chemical modifications mainly appear in cellular RNAs such as messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA) as well as other non-coding RNAs, numerous studies have indicated the pivotal roles of RNA epigenetic regulations in virus infection (McIntyre et al., 2018; Netzband and Pager, 2020). The most prevalent modifications in the virus genome include methylation of adenine and cytidine residues, such as N6-methyladenosine (m6A), 5-methylcytidine (m5C), or 7-methylguanosine (m7G), 2ʹ-O-methylation (2ʹ-O-Methyl), as well as uncanonical nucleotides like A-to-I editing and pseudouridine (McIntyre et al., 2018). Although these chemical modifications are generally formed by cellular enzymes, virus-encoded methyltransferases have been implicated in several methylation modifications. Nearly all chemical modifications that are mediated by enzymes undergo dynamic and reversible changes during virus infection, which makes it difficult to define roles of epigenetic modifications in viral RNA metabolism or virus infection. Nevertheless, due to the rapid development of RNA biology, numbers of RNA modifications have been found in genome of various viruses, which are supposed to influence virus infection to some extent (Courtney, 2021; Marchand and Motorin, 2021).
As the primary antiviral strategies, innate immune responses are invariably activated at the early stage of virus infection. Through recognizing the exogenous nucleic acids including virus-derived RNAs or DNAs by Toll-like receptors (TLRs; Creagh and O’Neill, 2006; Beutler, 2009; Lavelle et al., 2010), which belong to pattern-recognition receptors (PRRs), cytoplasmic receptors/adapters like myeloid differentiation factor-88 (MyD-88) or TIR-domain-containing adaptor protein inducing interferon-beta (TRIF) is recruited and in turn activates TNF receptor-associated factors (TRAFs; Creagh and O’Neill, 2006). Activation of TRAFs then gives rise to the activation of IFN response factor 3/7 (IRF3/7) and nuclear factor-κB (NF-κB) signaling pathways that induces type I interferons (IFNs) and proinflammatory cytokines expression (Bonizzi and Karin, 2004; Rius et al., 2008; Dev et al., 2011). Aside from the TLR pathway, another kind of PRRs named as retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) family also has been identified as crucial cytosolic sensors of viral nucleic acids (Schustak et al., 2021). The mitochondrial antiviral-signaling protein (MAVS) is located in mitochondria or endoplasmic reticulum (ER) and considered as the receptor protein of RLR signaling pathway, by which IFN-β is effectively expressed at the early stage of virus infection (Hwang et al., 2013; Tong et al., 2021b). Both IFNs and proinflammatory cytokines have strong antiviral activities. A battery of studies have recently indicated the emerging roles of RNA modifications in regulating antiviral innate immune responses (Thompson et al., 2021). The present review will focus on the impacts of these epigenetic marks, especially on antiviral innate immunity and its relevant signaling pathways, while highlighting the significance of antiviral innate immune responses during virus infection.
Prevalent RNA Modifications in Virus
N6-Methyladenosine
N6-Methyladenosine modification affects nearly all aspects of RNA biology, including stability, translation, splicing, nuclear export, and localization. Methylation modification, adding adenosine to N6 to form m6A, is catalyzed by a large heterogeneous complex of proteins that are named as “writer,” including METTL3, METTL14, or Wilms tumor 1-associated protein and KIAA1429 (Meyer and Jaffrey, 2017; Shi et al., 2019). In contrast, demethylases enzymes like fat mass and obesity-associated protein (FTO) or α-ketoglutarate-dependent dioxygenase AlkB homology 5 (ALKBH5) designated as “eraser” remove the methyl group (Jia et al., 2011; Zheng et al., 2013). The YTH domain family of proteins (YTHDC1, YTHDC2, YTHDF1, YTHDF3, and YTHDF3) and others named as “reader” recognize and bind to the m6A modification site to directly regulate the posttranscriptional functions of modified RNAs (Shi et al., 2019). m6A modifications are typically identified within the DRAmCH motif (D = G/A/U, R = A/G, and H = A/C/U); however, given the fact that only some of the DRACH motifs in eukaryote transcriptome are modified, there might exist some mechanisms for site-selective modification (Dominissini et al., 2012).
5-Methylcytidine
Another kind of RNA base methylation is the C5-methylation of RNA cytosine-m5C. m5C widely exists in cytoplasmic and ribosomal RNA (rRNA), tRNA, mRNA, and some non-coding RNAs (Lewis et al., 2017; Bohnsack et al., 2019). In eukaryotes, m5C is catalyzed by enzymes of the NOL1/NOP2/SUN domain (NSUN) family and DNA methyltransferase family protein (DNMT2), a homolog of DNA methyltransferase (Reid et al., 1999). Recent studies showed m5C was present in numerous virus genomes and might have non-negligible effects on antiviral innate immunity (Winans and Beemon, 2019; Wnuk et al., 2020). For instance, a high level of m5C modifications in HIV-1 genomic RNA (gRNA) promoted the expression of viral genes by regulating splicing and the translation efficiency of viral mRNAs (Courtney et al., 2019). Silencing or inactivation of the major writer NSUN2 of m5C reduced the m5C abundance in HIV-1 transcripts and inhibited virus replication by disrupting the alternative splicing and the followed translation of HIV-1 mRNA (Kong et al., 2020).
2ʹ-O-Methylation and 7-Methylguanosine
Cellular mRNA conventionally has a triphosphate at 5ʹ end (5ʹ-ppp), which is converted to 5ʹ-diphosphate (5ʹ-pp) by RNA triphosphatase (Ramanathan et al., 2016). This conversion resulted in mRNA capping by guanylyltransferase and guanine-N7 methyltransferase (Shatkin, 1976). After adding a terminal guanosine base, the mRNA transcripts possess an m7G joined via a 5ʹ, 5ʹ-triphosphate bridge, designated as cap-0 (Shatkin, 1976). When a cellular 2ʹ-O-methyltransferase, CMTR1, further modifies the mRNA, a methyl group is added at the 2ʹ-O-hydroxyl position of the first nucleotide to form cap-1 RNA structure (Belanger et al., 2010). Meanwhile, a second 2′-O-methyl group can be added at the second nucleotide to form cap-2, catalyzed by another cellular methyltransferase CMTR2 (Werner et al., 2011; Smietanski et al., 2014). mRNA capping is considered one of the key factors in regulating RNA metabolism and function (Topisirovic et al., 2011), including stabilizing the mRNA and serving as a chemical marker to discriminate self from foreign RNA, the latter of which may interfere with the innate immune sensing of viral derived RNA (Hocine et al., 2010; Ramanathan et al., 2016). Some viruses, such as West Neil virus (WNV) or Dengue virus (DENV), encode 2ʹ-O MTases that catalyze 2ʹ-O-methyl adenosines inside the virus genome (Dong et al., 2012; Chang et al., 2016). Interestingly, this internal adonosine 2ʹ-O-methyl activity requires the same K-D-K-E motif as that for 2ʹ-O methylation of the 5ʹcap (Yap et al., 2010; Dong et al., 2012). Given that many viruses possess cap structures in their RNA components, this type of modification is supposed to play a pivotal role in antiviral innate immunity.
Uncanonical Nucleotides
After the pseudouridine (ψ) was firstly identified in plant turnip yellow mosaic virus (TYMV) in 1998 (Becker et al., 1998), the follow-up research continuously indicated the abundant ψ in RNA viruses, especially in positive-sense RNA viruses (McIntyre et al., 2018). Psedouridine occurs through isomerization of uridine-to-5-ribosyl uracil by pseudouridine synthases (PUS; Markham and Smith, 1951). Similar to ψ, the deamination of adenosine to inosine (A-to-I) that depends on the catalyzing of adenosine deaminase acting on RNA (ADAR) family is also considered RNA editing or uncanonical nucleotides (Chen et al., 2000; Bass, 2002; George et al., 2014; Chung et al., 2018; Eisenberg and Levanon, 2018). The difference is that inosine generally acts similarly to guanosine (G), whereas ψ remains the original capacity of uridine to some extent. Although U to ψ conversion does not change the Watson–Crick base-pairing with adenosine, in certain cases, ψ enables base pairing with any other nucleotides (Samuel, 2011; Pfaller et al., 2021). Both ψ and A-to-I editing may significantly convert RNA biology, including changing the coding preference of viral RNA dependent RNA polymerases, mediating alternative splice and even affecting RNA structures (Netzband and Pager, 2020; Pfaller et al., 2021).
Mechanisms of Epigenetic Regulation in RNA Metabolism
Modified nucleotides may stabilize the functional RNA structures by reinforcing the hydrogen bond between Watson–Crick pairs, resulting in augmented thermal stability and reduced dynamics (Serra et al., 2004; Zhou et al., 2016; Frye et al., 2018). Under other circumstances, Watson–Crick base pairs consisting of modified nucleotides may induce an alternative folding representing significant alterations in the RNA secondary or tertiary structures, negatively affecting RNA stability (Durbin et al., 2016; Zhou et al., 2016; Roundtree et al., 2017; Wei and He, 2021). Moreover, the posttranscriptional introduction of modified nucleotides can affect RNA intermolecular interactions with other encountered molecules such as DNA partners, RNA binding proteins, or other RNAs (Zhao et al., 2017; Nachtergaele and He, 2018; Shi et al., 2019). Since all functions are regulated by structure to a certain degree, viral RNAs carrying modified nucleotides (or uncanonical nucleotides) commonly represent functional differences in virus life-cycle, thus mediating virus infection in host cells. Besides, some RNA viruses those complete their life-cycle in cytoplasm influence host cell genomic transcription inside the cell nucleus, for example, ZIKV infection affects some endogenous genes’ trancription which occured inside the cell nucleus (Gokhale et al., 2020). In this case, epigenetic regulations may facilitate the virus to overcome the spatial barrier. To sum up, some of the demonstrated RNA modifications in virus genome as well as the correlative functions are listed in Figure 1. Notably, many viruses use epigenetic modifications as crucial tools to evade antiviral innate immune response.
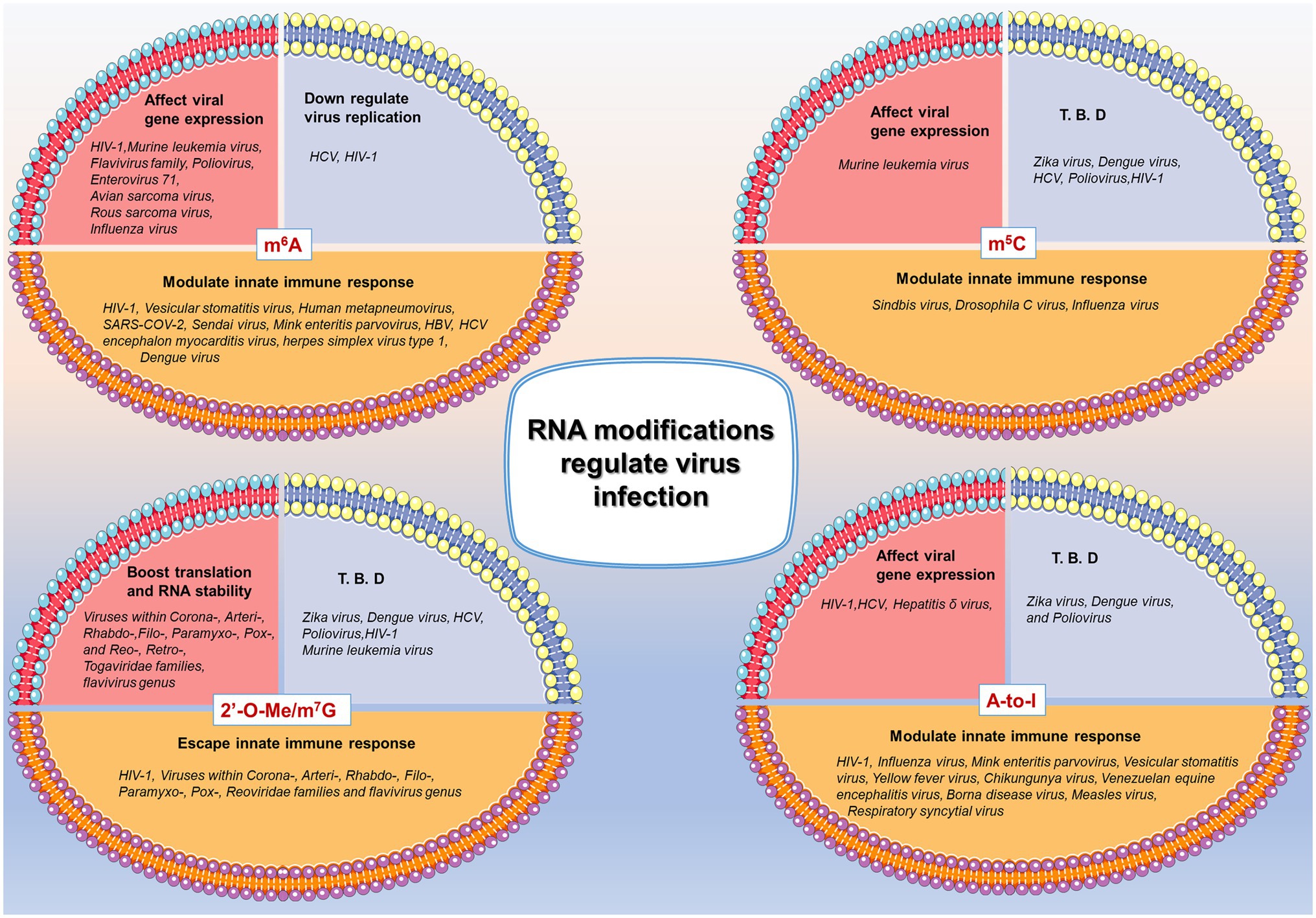
Figure 1. Brief summary of RNA modifications in regulating virus infection. N6-methyladenosine (m6A), 5-methylcytidine (m5C), 2ʹ-O-methylation (2ʹ-O-Me)/7-methylguanosine (m7G), and A-to-I editing (A-to-I) are listed as example of epigenetic regulations in virus infection. Three major effects are summarized in diverse virus infection, among which the modulation of innate immune response is the focus of the present review. T. B. D: To be determined. References are listed in the Supplementary Table S1.
RNA Modifications in Sensing of Foreign Nucleic Acids
Sensing the foreign molecules by the PRRs of the innate immune system serves as the initial step of the innate immune response (Akira et al., 2006; Takeuchi and Akira, 2010). Different PRRs must distinguish the non-self molecules from the self through chemical patterns. To date, several kinds of PRRs, including TLRs, RLRs, cyclic GMP-AMP synthase (cGAS; Schoggins et al., 2014; Aguirre et al., 2017; Ma et al., 2021; Yu et al., 2021), C-type lectin receptors (CLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), and AIM2-like receptors (ALRs), have been utilized by host cells in recognition of viral PAMPs (Akira et al., 2006; Crowl et al., 2017; Babamale and Chen, 2021; Chou et al., 2021; de Oliveira Mann and Hornung, 2021). As the most important component in virus particles, viral-derived RNAs/DNAs are released into the host cell cytoplasm at the early stage of infection. Thus, recognizing the distinction of epitranscriptomic modifications between cellular and pathogen nucleic acids is supposed to regulate antiviral innate immunity at the early stage of virus infection (Figure 2).

Figure 2. Schematic diagram of mechanisms by which RNA modification regulating viral-derived RNA recognition and innate immune responses. N6-methyladenosine (m6A), 5-methylcytidine (m5C), 2ʹ-O-methylation (2ʹ-O-Me), and pseudouridine (ψ) are demonstrated to inhibit melanoma differentiation-associated protein 5 (MDA5) or retinoic acid-inducible gene I (RIG-I) mediated sensing. METTL and YTHDF proteins are involved in these process. Inside the endosomes, 2′-O-Me are identified to block the TLR7-dependent type I interferon (IFN-I) response. Meanwhile, m6A, m5C, and ψ also prevent the Toll-like receptors (TLRs) activation inside the endosomes, although the relevance to virus infection still remains ambiguous. Moreover, m6A may regulate antiviral innate immunity through stress granules or endoplasmic reticulum (ER)-stress pathways, the latter of which has already been illustrated in Flavivirus infection.
Retinoic Acid-Inducible Gene I-Like Receptors
Retinoic acid-inducible gene I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) are the three major homologous helicases of RLRs (Wu et al., 2013; Yu et al., 2018; Thoresen et al., 2021). RIG-I and MDA5 displayed similar component structures, including caspase activation domain in N-terminal, recruitment domains (CARDs) for communicating with downstream signals, a DExD/H-box helicase domain with RNA binding and ATP hydrolysis activity, and a C-terminal domain (CTD; Yoneyama et al., 2004; Jiang et al., 2011; Kowalinski et al., 2011; Yu et al., 2018). The pathogen-associated molecular patterns (PAMPs) motifs of RIG-I include exposed 5ʹtriphosphate (5ʹppp) or diphosphate of double-strand RNAs (dsRNA), panhandle structures of viral genomic RNA, and uridine-rich sequences, while MDA5 recognizes long dsRNA such as poly (I:C; Yoneyama et al., 2004; Hornung et al., 2006; Kato et al., 2006; Cui et al., 2008; Liu et al., 2018). RIG-I is expressed at a low level in non-infected cells, usually referred to as a resting state with RNA-binding and helicase domains covered by RDs (Kowalinski et al., 2011; Luo et al., 2011). Following recognition of PAMPs, RIG-I undergoes a conformational change that provides room for closer interaction with more PAMPs RNAs and begins to release the CARDs for MAVS interaction and signaling (Pichlmair et al., 2006; Peisley et al., 2013; Thoresen et al., 2021). When the complex consisting of RIG-I, MAVS, and other cytosolic proteins translocate from the cytoplasm to the associated mitochondrial membrane, RIG-I CARDs interact with the MAVS CARD to catalyze the filament formation of MAVS and then activates TBK1 and IKKε to initiate downstream signaling (Pichlmair et al., 2006; Peisley et al., 2013; Goubau et al., 2014).
m6A and RLRs
As one of the most common RNA modifications, m6A has been widely involved in the innate immune sensing process and thus regulates viral pathogenesis. In some cases, viral RNA-loaded m6A modification dampens the RIG-I mediated RNA sensing and activation of the downstream transcription factors such as IRF3 and IRF7, which depresses the type I interferon (IFN-I) gene expression (Lu et al., 2020; Ge et al., 2021; Xue et al., 2021). One suggested mechanism is that m6A modification might harbor viral PAMPs motifs for RIG-I recognition and innate immune signaling. For example, m6A modifications in HIV RNAs undermined RIG-I sensing and type-I interferon induction in differentiated monocytic cells, while m6A-deficient HIV-1 virions produced from FTO-overexpressing HEK293T cells induced high levels of IFN-I expression in a RIG-I-dependent manner (Chen et al., 2021). Consistently, in several families of negative-sense RNA viruses, such as Pneumoviridae (hMPV), Paramyxoviridae (SeV and MeV), and Rhabdoviridae (VSV), m6A-deficient viral RNAs universally triggered RIG-I-dependent innate immune response much more efficiently compared to the m6A-sufficient viral RNAs, suggesting a crucial role of m6A marker in RIG-I sensing process (Kim et al., 2020; Lu et al., 2020, 2021).
Further investigation demonstrated that these negative effects might be induced by m6A related enzymes, including YTHDFs and METTLs. Instead of encoding innate immune antagonist proteins, m6A modifications in viral RNAs enable the recruitment of the m6A enzymes, which subsequently sequestrates viral ds/ssRNA through their RNA binding ability to prevent RIG-I recognition. Specifically, m6A modification of hepatitis B and C viral RNAs suppressed the activation of RIG-I signaling, whereas single nucleotide mutation of m6A motif of viral RNAs (A8766C) enhanced RIG-I sensing activity (Kim et al., 2020). In this case, YTHDF2 was found to regulate HBV pgRNAs and HCV genomic RNAs to evade RIG-I recognition. Besides YTHDF itself, diverse RNA-binding proteins (RBPs) were identified to interact with YTHDF proteins. The interactions might also regulate RIG-I access to viral RNAs, which mediates the activation of RIG-I signaling pathways through indirect influences (Luo et al., 2012).
Moreover, reducing the m6A “writer” enzyme METTL3 not only downregulates the m6A levels in the 3ʹ end of SARS-COV-2 genome, but also improves the RIG-I binding to enhance the downstream innate immune signaling pathway and inflammatory gene expressions (Li et al., 2021). Similarly, as for vesicular stomatitis virus (VSV) infection, METTL3 decreases viral dsRNA formation, thereby impeding virus-sensing efficacy by RIG-I and dampening antiviral immune signaling (Qiu et al., 2021). However, due to the lack of precise information about RIG-I PAMPs, the universal mechanisms of viral RNA m6A modification inhibiting RIG-I activation remains unclear. A potential clue has been elucidated in human metapneumovirus (hMPV) infection. Due to the indispensable role of conformational change in RIG-I activation, m6A modifications in virus genome might block the binding of viral RNAs to RIG-I, which disabled the conformational change of RIG-I, as well as the subsequent MAVS-TBK1 pathways. In this case, m6A-deficient hMPV virion RNA induced much higher RIG-I expression (Lu et al., 2020).
2ʹ-O-Methylation and RLRs
Besides m6A modifications, RNA 2ʹ-O-methyl is a highly conserved process used by RNA viruses to evade sensing by cytosolic RNA sensor proteins (Daffis et al., 2010; Decroly et al., 2011; McFadden et al., 2017). Early studies indicated that the 2ʹ-O-methyl commonly marks viral RNA as “self,” which prevents RLRs and downstream signaling pathways (Hyde and Diamond, 2015; McFadden et al., 2017; Jaafar and Kieft, 2019). During HIV-1 infection, viral RNAs were methylated to carry internal 2ʹ-O-methylations by the cellular methyltransferase FTSJ3 (Ringeard et al., 2019). When HIV-1 viruses were produced in FRSJ3 knock-out cells, the induction of IFNs was heavily enhanced in an MDA5-dependent manner (Zust et al., 2011; Ringeard et al., 2019). Similarly, 2ʹ-O-methyls on the coronavirus family viral RNAs also perturbed type I interferon production that is dependent on either the MDA5 or RIG-I sensing process (Zust et al., 2011; Devarkar et al., 2016).
Additionally, the virus facilitated the capping of viral RNAs at the 5ʹ terminal to disturb the innate immune sensing process (Bradrick, 2017; De Vlugt et al., 2018). Unlike cellular mRNA transcripts, some viruses, including flaviviruses and coronaviruses, encode enzymes with m7G and 2ʹ-O-methyltransferase (2ʹ-O-MTases) activity to cap their RNA, It has been showed that 2ʹ-O-MTases-deficient virus are highly sensitive to IFN-I (Chen and Guo, 2016; Bradrick, 2017). Although, the precise factors that sense unmethylated RNAs as invading nucleic acid are still unclear, the interferon-induced protein with tetratricopeptide repeats (IFIT) family has been discovered to function in West neil virus, poxvirus, and coronavirus infection (Daffis et al., 2010). Interestingly, instead of encoding 2-O-MTases, the influenza virus applies a “cap-snatching” strategy to ensure the viral RNA 5ʹ end modifications that prevent the viral RNA from being sensed by IFIT proteins (De Vlugt et al., 2018).
Other RNA Modifications and RLRs
Along with m6A modifications and 2ʹ-O-methyl, other RNA chemical modifications also participate in RLRs sensing-dependent innate immune response (Ahmad et al., 2018). For example, RIG-I and MDA5 detection of dsRNA is blocked by adenosine deaminase acting on RNA (ADAR1), which catalyzes RNA A-to-I modification (Mannion et al., 2014; Yang et al., 2014; Ahmad et al., 2018; Tang et al., 2021). Although it is well-demonstrated how ADAR1-mediated A-to-I modifications impeded MDA5 activation in the mouse study (Liddicoat et al., 2015; de Reuver et al., 2021), MDA5-dependent sensing has rarely been found in A-to-I editing-induced innate immune response. In other cases of A-to-I editing in virus infection, suppression of innate immune IFN responses after virus infection is mainly mediated by cytoplasmic dsRNA sensors protein kinase R (PKR) and oligoadenylate synthetase (OAS; Yang et al., 2014; Radetskyy et al., 2018; Lamers et al., 2019). Rather than upstream dsRNA sensors, PKR, and OAS are identified as pivotal antiviral IFN stimulated genes (ISG). Thus, more details about ADAR1-mediated A-to-I modifications in antiviral innate immune response will be further discussed below.
Similar to A-to-I editing, pseudouridine modifications were shown to abolish RIG-I’s filament formation and PAMPs RNA’s binding (Peisley et al., 2013). Given the abundant pseudouridine modifications in RNA viruses, especially the positive-sense RNA viruses (McIntyre et al., 2018), this type of uncanonical nucleotides is suggested to regulate various aspects of the antiviral response.
Toll-Like Receptors and Other PRRs
Another well-characterized PRRs, TLRs, are widely distributed invertebrates. TLRs are anchored in the cell membrane as type I transmembrane proteins (Akira et al., 2006). The ectodomain (N-terminal) of TLRs consists of several leucine-rich repeat (LRR), which connect to the C-terminal Toll/interleukin-1 receptor (TIR) domain by transmembrane (TM) domain (Akira et al., 2001; Kawasaki and Kawai, 2014). Studies have shown that most TLRs function as homology dimers (Kawai and Akira, 2011). Two TIR domains became close to forming a competent signaling state that recruits the adapter proteins (O’Neill et al., 2013). Nearly all the activated TLRs can trigger proinflammatory gene expression despite functioning in specific aspects of antiviral immunity (Kawai and Akira, 2011). To date, 10 TLRs have been identified in human cells. Four of them functioned as immune sensors by detecting pathogens-derived nucleotides (Kawasaki and Kawai, 2014). TLR3 recognizes long dsRNA and recruits TRIF as its dedicated adapter protein. Phosphorylated TRIF provides a signaling hub for IRF3 phosphorylation by TBK1, which then activates downstream signaling pathways of TRIF (Matsumoto et al., 2011; Oshiumi et al., 2011; Liu et al., 2015). TLR7 and TLR8 detect RNA debris as short RNA segments, while TLR9 enables sensing short DNA fragments that contain CG dinucleotide motifs (Chan et al., 2015; de Oliveira Mann and Hornung, 2021). TLR7, 8, and 9 can recruit the adapter protein MyD88 to form a complex known as the Myddosome. Myddsosome interact with IκB kinase and TGF-beta-activated kinase 1 (TAK1) complex to initiate NF-κB and MAPK signaling, respectively (Motshwene et al., 2009). Interestingly, the complex can also trigger IRF activation that depends on TASL that is only expressed in specific cells (Heinz et al., 2020). TASL is also capable of IRF phosphorylation, while in this case, IRF5 and IRF7, as well as IRF3, may be activated to drive antiviral gene expression (Wust et al., 2021).
Although several studies indicate the important roles of TLRs that usually sense long dsRNA inside endolysosome or outside the cells in antiviral innate immune response, they have rarely been found to be regulated by RNA modifications, partly because many RNA viruses expose their genomic dsRNA in the cytoplasm (Akira et al., 2001; Alexopoulou et al., 2001; Heil et al., 2004). Some studies have implied that the epigenetic marks of viral RNA interfere with the innate immune signaling pathway by preventing TLRs activation. For instance, the 2ʹ-O-methyl marks on coronavirus RNAs avoid the recognition of TLR7 to evade the activation of the IFN signaling pathway, while this effect may also be achieved through MDA5 sensing signals (Zust et al., 2011). Coronaviruses replicating in MDA5 or TLR7 deficient mice are detected to the same extent as in IFNR-deficient mice. By employing in vitro modified RNA oligos, an early study showed that m6A limited the capacity of RNAs to activate TLR3, TLR7, and TLR8, while m5C and Ψ blocked the activation of TLR7 and TLR8 (Kariko et al., 2005). Recent studies applied CRISPER tools to map the function of m6A and demonstrated that m6A could suppress macrophage activation through TLR mediated signaling (Tong et al., 2021a). However, more evidence of virus RNA modifications regulating TLR mediated pathways in innate immune response remains to be discovered.
RNA Modifications in Regulating IFN Signaling Pathway
Interferon is a group of signal proteins synthesized and released by host cells in response to stress and infections. Interferon exists widely in human and other animal organisms with highly species specifictiy (Crow and Stetson, 2021). According to the types of corresponding receptors, interferon can be divided into three types: IFN-I, type II interferon (IFN-II), and type III interferon (IFN-III; Hervas-Stubbs et al., 2011; Stanifer et al., 2019). After infected with viruses, cells release IFNs to restrict the virus infection and even degrade the virions. Although IFNs do not kill the virus directly, IFNs enable the transcription and production of several enzymes that interfere with the viral genome transcription or translation of viral protein components (Sadler and Williams, 2008).
Meanwhile, IFNs also improve the antiviral ability of the surrounding cells. Therefore, IFNs are commonly considered powerful tools and key components in the first line of innate immune defense against viruses infection.
Interferons function mainly through the interactions between IFN molecules and cell surface receptors. Upon specifical recognition and binding by IFNs, the IFN receptors undergo conformational changes, activating the JAK family proteins and promoting the recruitment and phosphorylation of signal transduction and transcriptional activation (STAT) proteins. The phosphorylated STAT is then dimerized and binds to IRF9 to form an ISGF3 complex, a transcriptional factor after transfer into the nucleus. The ISGF3 regulates the expression of numerous kinds of IFN stimulating genes ISGs, which exert strong antiviral effects (Darnell, 2012; Raftery and Stevenson, 2017). However, many viruses (e.g., SARA-COV-2 or influenza virus) encode structural and non-structural viral proteins that ablate the IFN signaling pathways through interaction with other cellular signaling pathways. This usually results in invalid STAT that fails to form phosphorylated ISGF3 complex, further abolishing the expression of antiviral ISGs (Mazewski et al., 2020; Yin et al., 2020; Jung and Lee, 2021). This process is concluded as an evasion of the innate immune response. Evading of the IFN-dependent innate immune response also relates to persistent infections. For example, direct binding of the Borna disease virus (BoDV) encoded P protein to TBK1 can antagonize the IRF3 activation, which prevents IFNβ induction (Unterstab et al., 2005). It is hypothesized that the ability of BoDV to prevent IRF3-dependent genes transcription might prevent the virus from activating the RLR signaling pathway and give rise to persistent BoDV infections in mammalian and avian hosts (Peng et al., 2007).
Whenever ISGs are successfully expressed, they will perform diverse antiviral effects. More than an important effector in IFN-dependent antiviral immune response, some ISGs can also be upregulated directly and independent of IFNs after virus infection. Although ISGs have different effects, on the whole, they all can resist or control infectious pathogens (Schneider et al., 2014; Fensterl et al., 2015). Previous studies showed that ISGs generally functioned by interacting with different co-factors, mediating antiviral effects by promoting viral RNA degradation, abrogating viral proteins translation, or combining both (Nguyen et al., 2001; Bick et al., 2003; Yang and Li, 2020). Moreover, secreted IFNs and induced ISGs may also activate NF-κB or other related innate immune signaling pathways to improve the release of proinflammatory cytokines and/or induce apoptosis that further restricts virus infection (Peteranderl and Herold, 2017).
m6A in IFN Producing and Effecting
The biological function of m6A is mainly regulated by a methyltransferase (writer), demethylase (eraser), and m6A binding protein (reader; Tong et al., 2018). Many studies have shown that RNA m6A modification plays an important role in innate immune response, while the exact roles of m6A in regulating antiviral IFN signaling displays in opposite aspect (Gokhale et al., 2016; Guo et al., 2020). In some cases, m6A modifications in the virus genome promote the IFN and ISGs induction, whereas, under other circumstances, m6A modification occurs to turn off the antiviral innate immune response. For example, the m6A modifications at specific sites in the HBV transcript restricts the virus replication through IFN α-mediated response. Although HBV is a DNA virus, it replicates through transitional pre-genomic RNA (pgRNA). m6A modification of A1907 in HBV pgRNA is the key regulator of IFN α-mediated pgRNA decay. Further investigation showed that ISG20 selectively degraded the m6A HBV transcripts that are strictly regulated by m6A reader YTHDF2 (Liu et al., 2017; Imam et al., 2020).
Contrary effects were found in encephalon myocarditis virus (EMCV), herpes simplex virus type 1 (HSV-1), and VSV infection. In these cases, YTHDF3 can inhibit the expression of ISGs by promoting the translation of transcriptional inhibitor FOXO3 (Zhang et al., 2019). RAW264.7 cells with YTHDF3 gene deletion have extensive antiviral activity against RNA and DNA virus, and this activity is mediated by the IFNAR1 signal (Zhang et al., 2019). Notably, m6A modification, in this case, regulated the host cell transcripts to inhibit antiviral innate immune response instead of affecting viral RNAs. Indeed, this viral infection-induced host cell m6A epitranscriptome diversity has commonly been found to regulate the antiviral innate immune response. During VSV infection, m6A modifications in MAVS, TRAF3, and TRAF6 are demethylated by ALKBH5 through interacting with the RNA helicase DDX46, which leads these three transcripts to retention in nuclei. Abolished expression of these three transcripts prevents efficient IFN induction (Zheng et al., 2017). Similarly, human cytomegalovirus (hCMV) infection affects host m6A modification machinery, including METTL14 and ALKBH5, reducing the IFNβ production. When knocking down the expression of METTL14, the production of IFNβ and subsequent signaling depending on the JAK/STAT pathway are enhanced, which decreases the production of infectious hCMV virion in infected cells (Rubio et al., 2018).
Interestingly, besides the direct influences of m6A modification on HBV pgRNA, it also has been indicated that m6A modification of tumor suppressor phosphatase and tensin homolog (PTEN) transcript is affected by HBV infection through invaliding PI3K/AKT pathway and inhibiting IRF-3 nuclear export (Kim et al., 2021). Other studies also indicated that DHX58, p65, and IKKγ, which bind to YTHDF2, are mediated by m6A modification, potentially interfering with IFN induction during virus infection (Lichinchi et al., 2016). Besides, YTHDF1, METTL3, and METTL14 have also been found to increase the expression of ISGs like IFITM1 in an m6A binding-dependent manner, which further indicated the m6A methyltransferase complex might promote the antiviral activity of type I IFN (McFadden et al., 2021).
Excluding the direct regulations of host transcripts by m6A modification, interactions between RNA and RBPs may also be affected by m6A modifications that subsequently affect antiviral IFN response (Bidet et al., 2014). For example, it has been found that during DENV-2 infection, three conserved RBPs, G3BP1, G3BP2, and CAPRIN1, are regulatory factors necessary for antiviral IFN response by promoting the efficient translation of PKR and IFITM2 mRNAs (Arguello et al., 2017; Edupuganti et al., 2017).
Other RNA Modifications in IFN Producing and Effecting
As one of the most important signaling pathways in innate immune responses, IFN producing and effecting are likely regulated by diversity factors, probably due to numerous protein enzymes evolving in the IFN signaling pathway. For example, NSUN2, the methyltransferase of m5C, has multiple effects on RNA biogenesis, including converting vault ncRNA to vtRNA (Bohnsack et al., 2019; Kong et al., 2020). The vtRNA has been shown to promote Influenza A virus (IAV) replication in A549 cells and mouse lungs through repressing PKR activation and the subsequent effects of interferon (Li et al., 2015; Wnuk et al., 2020). Similarly, DNMT2 has been reported to be required for efficient IFN responses in Drosophila C virus or Sindbis virus infected Drosophila (Durdevic et al., 2013; Bhattacharya et al., 2017).
Other studies also indicate an important role of ADARs, the enzymes mediating A-to-I editing, in modulating innate immune response during virus infection (Pfaller et al., 2021). ADAR1 has a proviral effect on Measles virus (MeV) and VSV infection that depends on PKR activation (Nie et al., 2007; Pfaller et al., 2015), while the suppression of innate immune response by ADAR2 is supposed to rely on STAT1 in the case of Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV; Schoggins et al., 2011; Clavarino et al., 2012). In the case of other viruses, such as BoDV, IAV, and Yellow fever virus (YFV), the mechanisms of ADAR-regulated IFN response remain indistinct (Pfaller et al., 2021). Interestingly, during HIV-1 infection, ADAR1 and ADAR2 may have opposite effects on virus replication, through the forming of DNA:RNA heteroduplex or antiviral innate immune response, respectively (Clerzius et al., 2009; Doria et al., 2009; Pujantell et al., 2017).
RNA Modifications Altered Specific Cellular Transcripts to Regulate Antiviral Responses
Except for the viral RNA modifications, some RNA modification can also directly control the expression of cytokines or specific genes that important for antiviral responses. In Flavivirus infection, the m6A abundance of host cell transcripts CIRBP and RIOK3 are altered through ER stress and RIG-I signaling respectively, which further regulate virus infection through antiviral immune response (Gokhale et al., 2016, 2020). m6A modification also destroyed the binding of stress granules (SGs) proteins to their RNA partners (Arguello et al., 2017; Fu and Zhuang, 2020). These may explain the diverse function of G3BP1, G3BP2, and CAPRIN1 in virus infection. G3BP1 and CAPRIN1 functioned as proviral factors in vaccinia virus (VACV) and respiratory syncytial virus (RSV) infection, while in contrast, G3BP1 and G3BP2 performed antiviral activity against poliovirus (PV) and alphaviruses (Bidet et al., 2014; Eiermann et al., 2020).
5-Methylcytidine has also been described to affect the expresson of host cell genes, which include cell cycle regulator p21 and immunity-related protein IL-17A (Wnuk et al., 2020). There were also studies suggesting a potential role of m5C in regulating other host genes, including those functioning in antiviral response.
How Epigenetic Marks Regulate Virus Infection?
Compared to the heritable evolutions, epitranscriptomic marks on virus genomes that are controlled by various protein factors including endogenous modifying enzymes undergo more dynamic changes. This type of epigentic regulation has been identified to play important roles in virus-host arms race. On one hand, epigenetic modifications of the virus genome prevent the host from recognizing the viral-derived RNAs, thus invaliding the antiviral innate immune response. On the other hand, the host epitranscriptome profiles may vary with virus infection so as to induce expression of uncanonical antiviral genes that restircts virus replication. Notably, the changes to the host transcriptome likely occur in the late stage of virus infection. As a result, the epigenetic machinery tends to facilitate the virus infection at the early stage. However, the dynamic property of RNA modifications on both virus and host transcriptiomes has even complicated epigenetic regulation of the virus-host arms race. Nevertheless, it is worthwhile to harness epigenetic regulations to intervene virus infections and develop antiviral treatments on the future avenue of antiviral research.
Conclusion
Despite the indistinct mechanisms, RNA modifications currently are identified to affect the infection of diverse kinds of viruses, in which the antiviral innate immunity is the most prevalent factor. In the near future, some RNA modifications, including m6A and m5C, may serve as crucial targets for the rational design of improved live attenuated vaccine candidates. Importantly, considering the complex effects of epigenetic modifications in host cell transcriptome, developing these types of antiviral drugs or vaccines still needs additional studies to confirm such assumptions.
Author Contributions
JT, WZ, and GQ: conceptualization and writing, review, and editing. JT and WZ: data curation and writing original draft. YC, QY, and N-NQ: visualization. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Natural Science Foundation of Hebei Province, China (C2021201010), Hundreds Talents Program of Hebei Province, China (E2020050011), and Advanced Talents Incubation Program of the Hebei University (521000981413) to JT, the National Natural Science Foundation of China (31971227), the Science and Technology Project of Hebei Education Department (ZD2021010), and the Natural Science Foundation of Hebei Province, China (C2020201039).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.845625/full#supplementary-material
References
Aguirre, S., Luthra, P., Sanchez-Aparicio, M. T., Maestre, A. M., Patel, J., Lamothe, F., et al. (2017). Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2:17037. doi: 10.1038/nmicrobiol.2017.37
Ahmad, S., Mu, X., Yang, F., Greenwald, E., Park, J. W., Jacob, E., et al. (2018). Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797.e13–810.e13. doi: 10.1016/j.cell.2017.12.016
Akira, S., Takeda, K., and Kaisho, T. (2001). Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680. doi: 10.1038/90609
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-kappaB by toll-like receptor 3. Nature 413, 732–738. doi: 10.1038/35099560
Arguello, A. E., DeLiberto, A. N., and Kleiner, R. E. (2017). RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 139, 17249–17252. doi: 10.1021/jacs.7b09213
Babamale, A. O., and Chen, S. T. (2021). Nod-like receptors: critical intracellular sensors for host protection and cell death in microbial and parasitic infections. Int. J. Mol. Sci. 22:11398. doi: 10.3390/ijms222111398
Bass, B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846. doi: 10.1146/annurev.biochem.71.110601.135501
Becker, H. F., Motorin, Y., Florentz, C., Giegé, R., and Grosjean, H. (1998). Pseudouridine and ribothymidine formation in the tRNA-like domain of turnip yellow mosaic virus RNA. Nucleic Acids Res. 26, 3991–3997. doi: 10.1093/nar/26.17.3991
Belanger, F., Stepinski, J., Darzynkiewicz, E., and Pelletier, J. (2010). Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 285, 33037–33044. doi: 10.1074/jbc.M110.155283
Beutler, B. A. (2009). TLRs and innate immunity. Blood 113, 1399–1407. doi: 10.1182/blood-2008-07-019307
Bhattacharya, T., Newton, I. L. G., and Hardy, R. W. (2017). Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PLoS Pathog. 13:e1006427. doi: 10.1371/journal.ppat.1006427
Bick, M. J., Carroll, J. W., Gao, G., Goff, S. P., Rice, C. M., and MacDonald, M. R. (2003). Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77, 11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003
Bidet, K., Dadlani, D., and Garcia-Blanco, M. A. (2014). G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10:e1004242. doi: 10.1371/journal.ppat.1004242
Bohnsack, K. E., Hobartner, C., and Bohnsack, M. T. (2019). Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes 10:102. doi: 10.3390/genes10020102
Bonizzi, G., and Karin, M. (2004). The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25, 280–288. doi: 10.1016/j.it.2004.03.008
Bradrick, S. S. (2017). Causes and consequences of flavivirus RNA methylation. Front. Microbiol. 8:2374. doi: 10.3389/fmicb.2017.02374
Chan, M. P., Onji, M., Fukui, R., Kawane, K., Shibata, T., Saitoh, S. I., et al. (2015). DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat. Commun. 6:5853. doi: 10.1038/ncomms6853
Chang, D. C., Hoang, L. T., Mohamed Naim, A. N., Dong, H., Schreiber, M. J., Hibberd, M. L., et al. (2016). Evasion of early innate immune response by 2′-O-methylation of dengue genomic RNA. Virology 499, 259–266. doi: 10.1016/j.virol.2016.09.022
Chen, C. X., Cho, D. S., Wang, Q., Lai, F., Carter, K. C., and Nishikura, K. (2000). A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6, 755–767. doi: 10.1017/S1355838200000170
Chen, Y., and Guo, D. (2016). Molecular mechanisms of coronavirus RNA capping and methylation. Virol. Sin. 31, 3–11. doi: 10.1007/s12250-016-3726-4
Chen, S., Kumar, S., Espada, C. E., Tirumuru, N., Cahill, M. P., Hu, L., et al. (2021). N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathog. 17:e1009421. doi: 10.1371/journal.ppat.1009421
Chou, W. C., Rampanelli, E., Li, X., and Ting, J. P.-Y. (2021). Impact of intracellular innate immune receptors on immunometabolism. Cell. Mol. Immunol. doi: 10.1038/s41423-021-00780-y [Epub ahead of print].
Chung, H., Calis, J. J. A., Wu, X., Sun, T., Yu, Y., Sarbanes, S. L., et al. (2018). Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811.e14–824.e14. doi: 10.1016/j.cell.2017.12.038
Clavarino, G., Claudio, N., Couderc, T., Dalet, A., Judith, D., Camosseto, V., et al. (2012). Induction of GADD34 is necessary for dsRNA-dependent interferon-beta production and participates in the control of chikungunya virus infection. PLoS Pathog. 8:e1002708. doi: 10.1371/journal.ppat.1002708
Clerzius, G., Gelinas, J. F., Daher, A., Bonnet, M., Meurs, E. F., and Gatignol, A. (2009). ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J. Virol. 83, 10119–10128. doi: 10.1128/JVI.02457-08
Courtney, D. G. (2021). Post-transcriptional regulation of viral RNA through epitranscriptional modification. Cells 10:1129. doi: 10.3390/cells10051129
Courtney, D. G., Tsai, K., Bogerd, H. P., Kennedy, E. M., Law, B. A., Emery, A., et al. (2019). Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26, 217.e6–227.e6. doi: 10.1016/j.chom.2019.07.005
Creagh, E. M., and O’Neill, L. A. (2006). TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 27, 352–357. doi: 10.1016/j.it.2006.06.003
Crow, Y. J., and Stetson, D. B. (2021). The type I interferonopathies: 10 years on. Nat. Rev. Immunol. 20, 1–13. doi: 10.1038/s41577-021-00633-9
Crowl, J. T., Gray, E. E., Pestal, K., Volkman, H. E., and Stetson, D. B. (2017). Intracellular nucleic acid detection in autoimmunity. Annu. Rev. Immunol. 35, 313–336. doi: 10.1146/annurev-immunol-051116-052331
Cui, S., Eisenacher, K., Kirchhofer, A., Brzózka, K., Lammens, A., Lammens, K., et al. (2008). The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29, 169–179. doi: 10.1016/j.molcel.2007.10.032
Daffis, S., Szretter, K. J., Schriewer, J., Li, J., Youn, S., Errett, J., et al. (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. doi: 10.1038/nature09489
de Oliveira Mann, C. C., and Hornung, V. (2021). Molecular mechanisms of nonself nucleic acid recognition by the innate immune system. Eur. J. Immunol. 51, 1897–1910. doi: 10.1002/eji.202049116
de Reuver, R., Dierick, E., Wiernicki, B., Staes, K., Seys, L., de Meester, E., et al. (2021). ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep. 36:109500. doi: 10.1016/j.celrep.2021.109500
De Vlugt, C., Sikora, D., and Pelchat, M. (2018). Insight into influenza: a virus cap-snatching. Viruses 10:641. doi: 10.3390/v10110641
Decroly, E., Ferron, F., Lescar, J., and Canard, B. (2011). Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 10, 51–65. doi: 10.1038/nrmicro2675
Dev, A., Iyer, S., Razani, B., and Cheng, G. (2011). NF-kappaB and innate immunity. Curr. Top. Microbiol. Immunol. 349, 115–143. doi: 10.1007/82_2010_102
Devarkar, S. C., Wang, C., Miller, M. T., Ramanathan, A., Jiang, F., Khan, A. G., et al. (2016). Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U. S. A. 113, 596–601. doi: 10.1073/pnas.1515152113
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Dong, H., Chang, D. C., Hua, M. H. C., Lim, S. P., Chionh, Y. H., Hia, F., et al. (2012). 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 8:e1002642. doi: 10.1371/journal.ppat.1002642
Doria, M., Neri, F., Gallo, A., Farace, M. G., and Michienzi, A. (2009). Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 37, 5848–5858. doi: 10.1093/nar/gkp604
Durbin, A. F., Wang, C., Gallo, A., Farace, M. G., and Michienzi, A. (2016). RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7, e00833–e00916. doi: 10.1128/mBio.00833-16
Durdevic, Z., Hanna, K., Gold, B., Pollex, T., Cherry, S., Lyko, F., et al. (2013). Efficient RNA virus control in drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 14, 269–275. doi: 10.1038/embor.2013.3
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi: 10.1038/nsmb.3462
Eiermann, N., Haneke, K., Sun, Z., Stoecklin, G., and Ruggieri, A. (2020). Dance with the devil: stress granules and signaling in antiviral responses. Viruses 12:984. doi: 10.3390/v12090984
Eisenberg, E., and Levanon, E. Y. (2018). A-to-I RNA editing—immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490. doi: 10.1038/s41576-018-0006-1
Fensterl, V., Chattopadhyay, S., and Sen, G. C. (2015). No love lost between viruses and interferons. Annu. Rev. Virol. 2, 549–572. doi: 10.1146/annurev-virology-100114-055249
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA modifications modulate gene expression during development. Science 361, 1346–1349. doi: 10.1126/science.aau1646
Fu, Y., and Zhuang, X. (2020). m(6)A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16, 955–963. doi: 10.1038/s41589-020-0524-y
Ge, Y., Ling, T., Wang, Y., Jia, X., Xie, X., Chen, R., et al. (2021). Degradation of WTAP blocks antiviral responses by reducing the m(6)A levels of IRF3 and IFNAR1 mRNA. EMBO Rep. 22:e52101. doi: 10.15252/embr.202052101
George, C. X., John, L., and Samuel, C. E. (2014). An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1). J. Interf. Cytokine Res. 34, 437–446. doi: 10.1089/jir.2014.0001
Gokhale, N. S., McIntyre, A. B. R., Mattocks, M. D., Holley, C. L., Lazear, H. M., Mason, C. E., et al. (2020). Altered m(6)A modification of specific cellular transcripts affects flaviviridae infection. Mol. Cell 77, 542.e8–555.e8. doi: 10.1016/j.molcel.2019.11.007
Gokhale, N. S., McIntyre, A. B. R., McFadden, M. J., Roder, A. E., Kennedy, E. M., Gandara, J. A., et al. (2016). N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665. doi: 10.1016/j.chom.2016.09.015
Goubau, D., Schlee, M., Deddouche, S., Pruijssers, A. J., Zillinger, T., Goldeck, M., et al. (2014). Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514, 372–375. doi: 10.1038/nature13590
Guo, G., Wang, H., Shi, X., Ye, L., Yan, K., Chen, Z., et al. (2020). Disease activity-associated alteration of mRNA m(5)C methylation in CD4(+) T cells of systemic lupus erythematosus. Front. Cell Dev. Biol. 8:430. doi: 10.3389/fcell.2020.00430
Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F., Kirschning, C., Akira, S., et al. (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529. doi: 10.1126/science.1093620
Heinz, L. X., Lee, J., Kapoor, U., Kartnig, F., Sedlyarov, V., Papakostas, K., et al. (2020). TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7-9. Nature 581, 316–322. doi: 10.1038/s41586-020-2282-0
Hervas-Stubbs, S., Perez-Gracia, J. L., Rouzaut, A., Sanmamed, M. F., le Bon, A., and Melero, I. (2011). Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17, 2619–2627. doi: 10.1158/1078-0432.CCR-10-1114
Hocine, S., Singer, R. H., and Grünwald, D. (2010). RNA processing and export. Cold Spring Harb. Perspect. Biol. 2:a000752. doi: 10.1101/cshperspect.a000752
Hornung, V., Ellegast, J., Kim, S., Brzózka, K., Jung, A., Kato, H., et al. (2006). 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. doi: 10.1126/science.1132505
Hwang, S. Y., Hur, K. Y., Kim, J.-R., Cho, K.-H., Kim, S.-H., and Yoo, J.-Y. (2013). Biphasic RLR-IFN-beta response controls the balance between antiviral immunity and cell damage. J. Immunol. 190, 1192–1200. doi: 10.4049/jimmunol.1202326
Hyde, J. L., and Diamond, M. S. (2015). Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology 479–480, 66–74. doi: 10.1016/j.virol.2015.01.019
Imam, H., Kim, G. W., Mir, S. A., Khan, M., and Siddiqui, A. (2020). Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified hepatitis B virus transcripts. PLoS Pathog. 16:e1008338. doi: 10.1371/journal.ppat.1008338
Jaafar, Z. A., and Kieft, J. S. (2019). Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 17, 110–123. doi: 10.1038/s41579-018-0117-x
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jiang, F., Ramanathan, A., Miller, M. T., Tang, G. Q., Gale, M., Patel, S. S., et al. (2011). Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427. doi: 10.1038/nature10537
Jung, H. E., and Lee, H. K. (2021). Current understanding of the innate control of toll-like receptors in response to SARS-CoV-2 infection. Viruses 13:2132. doi: 10.3390/v13112132
Kariko, K., Buckstein, M., Ni, H., and Weissman, D. (2005). Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. doi: 10.1016/j.immuni.2005.06.008
Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. doi: 10.1038/nature04734
Kawai, T., and Akira, S. (2011). Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34, 637–650. doi: 10.1016/j.immuni.2011.05.006
Kawasaki, T., and Kawai, T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5:461. doi: 10.3389/fimmu.2014.00461
Kim, G. W., Imam, H., Khan, M., Mir, S. A., Kim, S. J., Yoon, S. K., et al. (2021). HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73, 533–547. doi: 10.1002/hep.31313
Kim, G. W., Imam, H., Khan, M., and Siddiqui, A. (2020). N(6)-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 295, 13123–13133. doi: 10.1074/jbc.RA120.014260
Kong, W., Biswas, A., Zhou, D., Fiches, G., Fujinaga, K., Santoso, N., et al. (2020). Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with tat for TAR binding and methylation. PLoS Pathog. 16:e1008430. doi: 10.1371/journal.ppat.1008430
Kowalinski, E., Lunardi, T., McCarthy, A. A., Louber, J., Brunel, J., Grigorov, B., et al. (2011). Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435. doi: 10.1016/j.cell.2011.09.039
Lamers, M. M., van den Hoogen, B. G., and Haagmans, B. L. (2019). ADAR1: “editor-in-chief” of cytoplasmic innate immunity. Front. Immunol. 10:1763. doi: 10.3389/fimmu.2019.01763
Lavelle, E. C., Murphy, C., O’Neill, L. A. J., and Creagh, E. M. (2010). The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 3, 17–28. doi: 10.1038/mi.2009.124
Lewis, C. J., Pan, T., and Kalsotra, A. (2017). RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 18, 202–210. doi: 10.1038/nrm.2016.163
Li, F., Chen, Y., Zhang, Z., Ouyang, J., Wang, Y., Yan, R., et al. (2015). Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res. 43, 10321–10337. doi: 10.1093/nar/gkv1078
Li, N., Hui, H., Bray, B., Gonzalez, G. M., Zeller, M., Anderson, K. G., et al. (2021). METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 35:109091. doi: 10.1016/j.celrep.2021.109091
Lichinchi, G., Gao, S., Saletore, Y., Gonzalez, G. M., Bansal, V., Wang, Y., et al. (2016). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1:16011. doi: 10.1038/nmicrobiol.2016.11
Liddicoat, B. J., Piskol, R., Chalk, A. M., Ramaswami, G., Higuchi, M., Hartner, J. C., et al. (2015). RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120. doi: 10.1126/science.aac7049
Liu, S., Cai, X., Cong, Q., Chen, X., Li, T., Du, F., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630
Liu, G., Lu, Y., Thulasi Raman, S. N., Xu, F., Wu, Q., Li, Z., et al. (2018). Nuclear-resident RIG-I senses viral replication inducing antiviral immunity. Nat. Commun. 9:3199. doi: 10.1038/s41467-018-05745-w
Liu, Y., Nie, H., Mao, R., Mitra, B., Cai, D., Yan, R., et al. (2017). Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 13:e1006296. doi: 10.1371/journal.ppat.1006296
Lu, M., Xue, M., Wang, H.-T., Kairis, E. L., Ahmad, S., Wei, J., et al. (2021). Nonsegmented negative-sense RNA viruses utilize N(6)-methyladenosine (m(6)A) as a common strategy to evade host innate immunity. J. Virol. 95, e01939–e02120. doi: 10.1128/JVI.01939-20
Lu, M., Zhang, Z., Xue, M., Zhao, B. S., Harder, O., Li, A., et al. (2020). N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598. doi: 10.1038/s41564-019-0653-9
Luo, D., Ding, S. C., Vela, A., Kohlway, A., Lindenbach, B. D., and Pyle, A. M. (2011). Structural insights into RNA recognition by RIG-I. Cell 147, 409–422. doi: 10.1016/j.cell.2011.09.023
Luo, D., Kohlway, A., Vela, A., and Pyle, A. M. (2012). Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure 20, 1983–1988. doi: 10.1016/j.str.2012.08.029
Ma, Y., Wang, X., Luo, W., Xiao, J., Song, X., Wang, Y., et al. (2021). Roles of emerging RNA-binding activity of cGAS in innate antiviral response. Front. Immunol. 12:741599. doi: 10.3389/fimmu.2021.741599
Mannion, N. M., Greenwood, S. M., Young, R., Cox, S., Brindle, J., Read, D., et al. (2014). The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494. doi: 10.1016/j.celrep.2014.10.041
Marchand, V., and Motorin, Y. (2021). Detection and functions of viral RNA modifications: perspectives in biology and medicine. Virologie 25, 5–20. doi: 10.1684/vir.2021.0884
Markham, R., and Smith, J. D. (1951). Structure of ribonucleic acid. Nature 168, 406–408. doi: 10.1038/168406a0
Matsumoto, M., Oshiumi, H., and Seya, T. (2011). Antiviral responses induced by the TLR3 pathway. Rev. Med. Virol. 21, 67–77. doi: 10.1002/rmv.680
Mazewski, C., Perez, R. E., Fish, E. N., and Platanias, L. C. (2020). Type I interferon (IFN)-regulated activation of canonical and non-canonical signaling pathways. Front. Immunol. 11:606456. doi: 10.3389/fimmu.2020.606456
McFadden, M. J., Gokhale, N. S., and Horner, S. M. (2017). Protect this house: cytosolic sensing of viruses. Curr. Opin. Virol. 22, 36–43. doi: 10.1016/j.coviro.2016.11.012
McFadden, M. J., McIntyre, A. B. R., Mourelatos, H., Abell, N. S., Gokhale, N. S., Ipas, H., et al. (2021). Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep. 34:108798. doi: 10.1016/j.celrep.2021.108798
McIntyre, W., Netzband, R., Bonenfant, G., Biegel, J. M., Miller, C., Fuchs, G., et al. (2018). Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res. 46, 5776–5791. doi: 10.1093/nar/gky029
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi: 10.1146/annurev-cellbio-100616-060758
Motshwene, P. G., Moncrieffe, M. C., Grossmann, J. G., Kao, C., Ayaluru, M., Sandercock, A. M., et al. (2009). An oligomeric signaling platform formed by the toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 284, 25404–25411. doi: 10.1074/jbc.M109.022392
Nachtergaele, S., and He, C. (2018). Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52, 349–372. doi: 10.1146/annurev-genet-120417-031522
Netzband, R., and Pager, C. T. (2020). Epitranscriptomic marks: emerging modulators of RNA virus gene expression. Wiley Interdiscip. Rev. RNA 11:e1576. doi: 10.1002/wrna.1576
Nguyen, L. H., Espert, L., Mechti, N., and Wilson, D. M. (2001). The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 40, 7174–7179. doi: 10.1021/bi010141t
Nie, Y., Hammond, G. L., and Yang, J. H. (2007). Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81, 917–923. doi: 10.1128/JVI.01527-06
O’Neill, L. A., Golenbock, D., and Bowie, A. G. (2013). The history of toll-like receptors—redefining innate immunity. Nat. Rev. Immunol. 13, 453–460. doi: 10.1038/nri3446
Oshiumi, H., Okamoto, M., Fujii, K., Kawanishi, T., Matsumoto, M., Koike, S., et al. (2011). The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J. Immunol. 187, 5320–5327. doi: 10.4049/jimmunol.1101503
Peisley, A., Wu, B., Yao, H., Walz, T., and Hur, S. (2013). RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell 51, 573–583. doi: 10.1016/j.molcel.2013.07.024
Peng, G., Zhang, F., Zhang, Q., Wu, K., Zhu, F., and Wu, J. (2007). Borna disease virus P protein inhibits nitric oxide synthase gene expression in astrocytes. Virology 366, 446–452. doi: 10.1016/j.virol.2007.04.031
Peteranderl, C., and Herold, S. (2017). The impact of the interferon/TNF-related apoptosis-inducing ligand signaling axis on disease progression in respiratory viral infection and beyond. Front. Immunol. 8:313. doi: 10.3389/fimmu.2017.00313
Pfaller, C. K., George, C. X., and Samuel, C. E. (2021). Adenosine deaminases acting on RNA (ADARs) and viral infections. Annu. Rev. Virol. 8, 239–264. doi: 10.1146/annurev-virology-091919-065320
Pfaller, C. K., Mastorakos, G. M., Matchett, W. E., Ma, X., Samuel, C. E., and Cattaneo, R. (2015). Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J. Virol. 89, 7735–7747. doi: 10.1128/JVI.01017-15
Pichlmair, A., Schulz, O., Tan, C. P., Näslund, T. I., Liljeström, P., Weber, F., et al. (2006). RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314, 997–1001. doi: 10.1126/science.1132998
Pujantell, M., Riveira-Munoz, E., Badia, R., Castellví, M., Garcia-Vidal, E., Sirera, G., et al. (2017). RNA editing by ADAR1 regulates innate and antiviral immune functions in primary macrophages. Sci. Rep. 7:13339. doi: 10.1038/s41598-017-13580-0
Qiu, W., Zhang, Q., Zhang, R., Lu, Y., Wang, X., Tian, H., et al. (2021). N(6)-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 12:1582. doi: 10.1038/s41467-021-21904-y
Radetskyy, R., Daher, A., and Gatignol, A. (2018). ADAR1 and PKR, interferon stimulated genes with clashing effects on HIV-1 replication. Cytokine Growth Factor Rev. 40, 48–58. doi: 10.1016/j.cytogfr.2018.03.007
Raftery, N., and Stevenson, N. J. (2017). Advances in anti-viral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci. 74, 2525–2535. doi: 10.1007/s00018-017-2520-2
Ramanathan, A., Robb, G. B., and Chan, S.-H. (2016). mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526. doi: 10.1093/nar/gkw551
Reid, R., Greene, P. J., and Santi, D. V. (1999). Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 27, 3138–3145. doi: 10.1093/nar/27.15.3138
Ringeard, M., Marchand, V., Decroly, E., Motorin, Y., and Bennasser, Y. (2019). FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565, 500–504. doi: 10.1038/s41586-018-0841-4
Rius, J., Guma, M., Schachtrup, C., Akassoglou, K., Zinkernagel, A. S., Nizet, V., et al. (2008). NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature 453, 807–811. doi: 10.1038/nature06905
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Rubio, R. M., Depledge, D. P., Bianco, C., Thompson, L., and Mohr, I. (2018). RNA m(6)A modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. 32, 1472–1484. doi: 10.1101/gad.319475.118
Sadler, A. J., and Williams, B. R. (2008). Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568. doi: 10.1038/nri2314
Samuel, C. E. (2011). Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411, 180–193. doi: 10.1016/j.virol.2010.12.004
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Schoggins, J. W., MacDuff, D. A., Imanaka, N., Gainey, M. D., Shrestha, B., Eitson, J. L., et al. (2014). Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695. doi: 10.1038/nature12862
Schoggins, J. W., Wilson, S. J., Panis, M., Murphy, M. Y., Jones, C. T., Bieniasz, P., et al. (2011). A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485. doi: 10.1038/nature09907
Schustak, J., Twarog, M., Wu, X., Wu, H. Y., Huang, Q., and Bao, Y. (2021). Mechanism of nucleic acid sensing in retinal pigment epithelium (RPE): RIG-I mediates type I interferon response in human RPE. J. Immunol. Res. 2021:9975628. doi: 10.1155/2021/9975628
Serra, M. J., Smolter, P. E., and Westhof, E. (2004). Pronounced instability of tandem IU base pairs in RNA. Nucleic Acids Res. 32, 1824–1828. doi: 10.1093/nar/gkh501
Shatkin, A. J. (1976). Capping of eucaryotic mRNAs. Cell 9, 645–653. doi: 10.1016/0092-8674(76)90128-8
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Smietanski, M., Werner, M., Purta, E., Kaminska, K. H., Stepinski, J., Darzynkiewicz, E., et al. (2014). Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 5:3004. doi: 10.1038/ncomms4004
Stanifer, M. L., Pervolaraki, K., and Boulant, S. (2019). Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 20:1445. doi: 10.3390/ijms20061445
Takeuchi, O., and Akira, S. (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. doi: 10.1016/j.cell.2010.01.022
Tang, Q., Rigby, R. E., Young, G. R., Hvidt, A. K., Davis, T., Tan, T. K., et al. (2021). Adenosine-to-inosine editing of endogenous Z-form RNA by the deaminase ADAR1 prevents spontaneous MAVS-dependent type I interferon responses. Immunity 54, 1961.e5–1975.e5. doi: 10.1016/j.immuni.2021.08.011
Thompson, M. G., Sacco, M. T., and Horner, S. M. (2021). How RNA modifications regulate the antiviral response. Immunol. Rev. 304, 169–180. doi: 10.1111/imr.13020
Thoresen, D., Wang, W., Galls, D., Guo, R., Xu, L., and Pyle, A. M. (2021). The molecular mechanism of RIG-I activation and signaling. Immunol. Rev. 304, 154–168. doi: 10.1111/imr.13022
Tong, J., Flavell, R. A., and Li, H.-B. (2018). RNA m(6)A modification and its function in diseases. Front. Med. 12, 481–489. doi: 10.1007/s11684-018-0654-8
Tong, J., Wang, X., Liu, Y., Ren, X., Wang, A., Chen, Z., et al. (2021a). Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci. Adv. 7:eabd4742. doi: 10.1126/sciadv.abd4742
Tong, J. F., Zhou, L., Li, S., Lu, L.-F., Li, Z.-C., Li, Z., et al. (2021b). Two duplicated Ptpn6 homeologs cooperatively and negatively regulate RLR-mediated IFN response in hexaploid gibel carp. Front. Immunol. 12:780667. doi: 10.3389/fimmu.2021.780667
Topisirovic, I., Svitkin, Y. V., Sonenberg, N., and Shatkin, A. J. (2011). Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298. doi: 10.1002/wrna.52
Unterstab, G., Ludwig, S., Anton, A., Planz, O., Dauber, B., Krappmann, D., et al. (2005). Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-{kappa}B activator (TANK)-binding kinase-1. Proc. Natl. Acad. Sci. U. S. A. 102, 13640–13645. doi: 10.1073/pnas.0502883102
Wei, J., and He, C. (2021). Chromatin and transcriptional regulation by reversible RNA methylation. Curr. Opin. Cell Biol. 70, 109–115. doi: 10.1016/j.ceb.2020.11.005
Werner, M., Purta, E., Kaminska, K. H., Cymerman, I. A., Campbell, D. A., Mittra, B., et al. (2011). 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 39, 4756–4768. doi: 10.1093/nar/gkr038
Winans, S., and Beemon, K. (2019). m(5)C goes viral. Cell Host Microbe 26, 154–155. doi: 10.1016/j.chom.2019.07.019
Wnuk, M., Slipek, P., Dziedzic, M., and Lewinska, A. (2020). The roles of host 5-methylcytosine RNA methyltransferases during viral infections. Int. J. Mol. Sci. 21:8176. doi: 10.3390/ijms21218176
Wu, B., Peisley, A., Richards, C., Yao, H., Zeng, X., Lin, C., et al. (2013). Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289. doi: 10.1016/j.cell.2012.11.048
Wust, S., Schad, P., Burkart, S., and Binder, M. (2021). Comparative analysis of six IRF family members in alveolar epithelial cell-intrinsic antiviral responses. Cells 10:2600. doi: 10.3390/cells10102600
Xue, M., Zhang, Y., Wang, H., Kairis, E. L., Lu, M., Ahmad, S., et al. (2021). Viral RNA N6-methyladenosine modification modulates both innate and adaptive immune responses of human respiratory syncytial virus. PLoS Pathog. 17:e1010142. doi: 10.1371/journal.ppat.1010142
Yang, S., Deng, P., Zhu, Z., Zhu, J., Wang, G., Zhang, L., et al. (2014). Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J. Immunol. 193, 3436–3445. doi: 10.4049/jimmunol.1401136
Yang, E., and Li, M. M. H. (2020). All about the RNA: interferon-stimulated genes that interfere with viral RNA processes. Front. Immunol. 11:605024. doi: 10.3389/fimmu.2020.605024
Yap, L. J., Luo, D., Chung, K. Y., Lim, S. P., Bodenreider, C., Noble, C., et al. (2010). Crystal structure of the dengue virus methyltransferase bound to a 5′-capped octameric RNA. PLoS One 5:e12836. doi: 10.1371/journal.pone.0012836
Yin, X., Langer, S., Herbert, K. M., Yoh, S., König, R., and Chanda, S. K. (2020). Sensor sensibility-HIV-1 and the innate immune response. Cells 9:254. doi: 10.3390/cells9010254
Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., et al. (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737. doi: 10.1038/ni1087
Yu, P., Miao, Z., Li, Y., Bansal, R., Peppelenbosch, M. P., and Pan, Q. (2021). cGAS-STING effectively restricts murine norovirus infection but antagonizes the antiviral action of N-terminus of RIG-I in mouse macrophages. Gut Microbes 13:1959839. doi: 10.1080/19490976.2021.1959839
Yu, Q., Qu, K., and Modis, Y. (2018). Cryo-EM structures of MDA5-dsRNA filaments at different stages of ATP hydrolysis. Mol. Cell 72, 999.e6–1012.e6. doi: 10.1016/j.molcel.2018.10.012
Zhang, Y., Wang, X., Zhang, X., Wang, J., Ma, Y., Zhang, L., et al. (2019). RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U. S. A. 116, 976–981. doi: 10.1073/pnas.1812536116
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi: 10.1038/nrm.2016.132
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, Q., Hou, J., Zhou, Y., Li, Z., and Cao, X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103. doi: 10.1038/ni.3830
Zhou, K. I., Parisien, M., Dai, Q., Liu, N., Diatchenko, L., Sachleben, J. R., et al. (2016). N(6)-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J. Mol. Biol. 428, 822–833. doi: 10.1016/j.jmb.2015.08.021
Keywords: RNA modification, viral infection, RIG-I, IFN-I, antiviral innate immunity
Citation: Tong J, Zhang W, Chen Y, Yuan Q, Qin N-N and Qu G (2022) The Emerging Role of RNA Modifications in the Regulation of Antiviral Innate Immunity. Front. Microbiol. 13:845625. doi: 10.3389/fmicb.2022.845625
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Yue Ma, Van Andel Institute, United StatesYan-Dong Tang, Harbin Veterinary Research Institute (CAAS), China
Copyright © 2022 Tong, Zhang, Chen, Yuan, Qin and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Tong, tongjie2019@hbu.edu.cn; Guosheng Qu, guo_sheng_qu@126.com
†These authors have contributed equally to this work
 Jie Tong
Jie Tong