
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 May 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.845601
 Peijun Zhang2†
Peijun Zhang2† Haoxiang Su1†
Haoxiang Su1† Ruoyan Peng3,4,5†
Ruoyan Peng3,4,5† Jasper Fuk-Woo Chan6,7†
Jasper Fuk-Woo Chan6,7† Shijie Bai8
Shijie Bai8 Gaoyu Wang3,4,5
Gaoyu Wang3,4,5 Yi Huang3,4,5
Yi Huang3,4,5 Xiaoyuan Hu3,4,5
Xiaoyuan Hu3,4,5 Jun Luo9
Jun Luo9 Sisi Liu10
Sisi Liu10 Youyou Li3,4,5
Youyou Li3,4,5 Liying Xue3,4,5
Liying Xue3,4,5 Fan Yang1
Fan Yang1 Mingming Zhao11
Mingming Zhao11 Yun Zhang3,4,5
Yun Zhang3,4,5 Chuanning Tang3,4,5
Chuanning Tang3,4,5 Shu Shen12
Shu Shen12 Xiuji Cui3,4,5
Xiuji Cui3,4,5 Lina Niu3,4,5
Lina Niu3,4,5 Gang Lu3,4,5
Gang Lu3,4,5 Kwok-Yung Yuen6,7
Kwok-Yung Yuen6,7 Fei Deng12‡
Fei Deng12‡ Weijia Zhang4*‡
Weijia Zhang4*‡ Feifei Yin3,4,5*‡
Feifei Yin3,4,5*‡ Jiang Du1,3,4*‡
Jiang Du1,3,4*‡
Astroviruses infect human and animals and cause diarrhea, fever, and vomiting. In severe cases, these infections may be fatal in infants and juvenile animals. Previous evidence showed that humans in contact with infected animals can develop serological responses to astroviruses. Mamastrovirus 11 is a species of Mamastrovirus and was first reported in 2018. It was detected in the fecal samples of a California sea lion. The genome sequence of its capsid protein (CP) was submitted to GenBank. However, the genome sequence of its non-structural protein region was not elucidated. In the present study, we characterized the genome sequences of the novel astroviruses AstroV-HMU-1 and AstroV-like-HMU-2. These were obtained from California sea lions (Zalophus californianus) and walruses (Odobenus rosmarus) presenting with loose stools. A phylogenetic analysis revealed that the CP of AstroV-HMU-1 closely clustered with Mamastrovirus 11 while its RNA-dependent RNA polymerase (RdRp) and serine protease (SP) were closely related to the mink astrovirus in the genus Mamastrovirus. The genome of AstroV-HMU-1 provided basic information regarding the NS protein regions of Mamastrovirus 11. Recombination analyses showed that the genomes of Z. californianus AstroV-HMU-1, VA2/human and the mink astrovirus may have recombined long ago. The NS of AstroV-like-HMU-2 segregated from the Astroviridae in the deep root of the phylogenetic tree and exhibited 36% amino acid identity with other mamastroviruses. Thus, AstroV-like-HMU-2 was proposed as a member of a new genus in the unclassified Astroviridae. The present study suggested that that the loose stools of pinnipeds may be the result of occasional infection by this novel astrovirus. This discovery provides a scientific basis for future investigations into other animal-borne infectious diseases.
Astroviruses are leading causes of infectious diarrhea in children, the elderly, immunocompromised individuals, and a wide range of animals. The major clinical symptom is watery diarrhea. The diseases may be fatal in infants and juvenile animals (Boros et al., 2017; Vu et al., 2017). HAstV-1 was first detected in humans in 1975 (Appleton and Higgins, 1975). Since then, the reported incidence of astrovirus infection in humans and animals has increased. Astrovirus infections are ubiquitous and ∼90% of the human population aged >9 years presents with anti-HAstV-1 antibodies (Mitchell et al., 1999). High-throughput next-generation sequencing (NGS) methods have recently disclosed that astroviruses may be associated with aseptic encephalitis, meningitis, and meningoencephalomyelitis in humans and animals (Calistri and Palu, 2015; Cortez et al., 2019; Roach and Langlois, 2021).
Astroviruses are non-enveloped, single-stranded, positive-sense RNA viruses of the Astroviridae. Mamastrovirus includes 19 species while Avastrovirus includes three species. Four Mamastrovirus species are known to infect humans while the other 15 only infect mammals. The prototype strain is Mamastrovirus 1 (MAstV1) which corresponds to human astrovirus genotypes 1–8 (HAstV 1–8). Mamastrovirus 6 contains the human astrovirus clades Melbourne 1–3 (MLB1–3) (Finkbeiner et al., 2008). Mamastrovirus 8 (MAstV8) and Mamastrovirus 9 (MastV9) include the human astrovirus clades Virginia 2 and 4 (VA2, 4) and Virginia 1 and 3 (VA1, 3) (Finkbeiner et al., 2009), respectively. Astrovirus genomes are 6.8–7 kb long and contain three overlapping open reading frames (ORF1a, ORF1b, and ORF2). ORF1a and ORF1b encode a viral protease and a polymerase, respectively. ORF2 is expressed by a subgenomic RNA and encodes a VP90 capsid precursor protein. The 5′-terminus is linked to a viral protein genome-linked (VPg) protein while the 3′-terminus contains a poly (A) tract.
Humans in contact with turkeys may present with serological responses to turkey astrovirus (Meliopoulos et al., 2014). Furthermore, astrovirus strains previously limited to humans have also been detected in non-human primates (Karlsson et al., 2015). Hence, astroviruses may be able to cross species barriers (Johnson et al., 2017). Recent research has focused on invertebrates and vertebrates in ocean ecosystems to study the origins, evolution, and emergence of terrestrial viruses. Astroviruses in marine mammal hosts including California sea lions (Zalophus californianus), Stellar sea lions (Eumetopias jubatus), and common bottlenose dolphins (Tursiops truncatus) were first reported in 2009 (Rivera et al., 2010). Bayesian and maximum likelihood (ML) phylogenetic methods were recently used to demonstrate that the novel flaviviruses detected in crustaceans are more closely related to terrestrial vector-borne than classical insect-specific flaviviruses (Parry and Asgari, 2019). Therefore, the astroviruses in marine mammals may be closely related to those in terrestrial animals according to phylogenetic analysis. The discovery of new marine mammal viruses is significant for viral phylogenetic analysis and taxonomic research. Here, we used NGS and discovered two novel astroviruses in oral and anal swab samples of walruses and California sea lions. It was established that AstroV-like-HMU-2 is phylogenetically distinct from all other known astroviruses, and the nearly complete genome sequence of AstroV-HMU-1 was determined and characterized.
Between January and December 2018, 32 oral, nasal, and anal swabs were collected from six walruses (Odobenus rosmarus) and six California sea lions (Z. californianus) presenting with loose stools and housed at the aquaria of Qingdao Polar Haichang Ocean Park and Dalian Sunasia Tourism Holding Co., Ltd., China. The samples were immersed in maintenance medium in virus-sampling tubes (Yocon, Beijing, China), transported on ice to the laboratory within 24 h, and stored at −80°C.
All animals were treated according to the guidelines of the Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People’s Republic of China, 1988). The sampling procedures were approved by the Ethics Committee of Hainan Medical University (Approval No. HMUEC20180059).
All 32 samples were combined by species into pool 30 for O. rosmarus and pool 35 for Z. californianus. The samples were passed through 0.45-μm filters. The filtrates were digested with DNase (Applied Biosystems, Santa Clara, CA, United States) and RNase One (Promega, Madison, WI, United States) to remove any unprotected nucleic acids. Total RNA was extracted with a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany, United States) according to the manufacturer’s instructions. The cDNA was generated with Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, United States) as previously described (Wu et al., 2018). The amplified viral nucleic acid libraries were analyzed with an Illumina HiSeq 2500 sequencer (Illumina, San Diego, CA, United States). The sequence data were deposited to the National Center for Biotechnology Information (NCBI; Bethesda, MD, United States) sequence reads archive under accession No. PRJNA650224. The raw sequence reads were filtered and valid sequences were obtained using previously described criteria (Yang et al., 2011).
Sequence similarity-based taxonomic assignments were conducted as previously described (Yang et al., 2011). Briefly, the viral origin of each read was determined via alignments with the NCBI non-redundant nucleotide (NT) and protein (NR) databases using BLASTn and BLASTx (−E: expected value < 10–5; −F: filter query sequence; default = T). The taxonomies of the aligned reads from all lanes with the highest BLAST scores (E-value < 10––5) were parsed and exported with a MEGAN6 MetaGenome Analyzer1 (Tamura et al., 2013).
Molecular clues from the metagenomic analyses were used to classify the sequence reads into virus families or genera with MEGA 6.2 Representative reads for the novel astrovirus were selected for genome sequencing and used in read-based PCR. Reads with accurate genomic locations were used to design specific nested PCR primers and identify partial genomes. PCR was performed with 2× Taq PCR Mastermix (Tiangen Biotech Co., Ltd., Beijing, China). Two microliters first-round PCR product was used as the template for the second PCR round. The thermal cycling conditions for PCR were 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 45 s, and a final elongation step at 72°C for 10 min. Pool samples 30 and 35 were selected for sequencing. The remaining genomic sequences were analyzed by genome walking and 5′- and 3′-rapid amplification of cDNA ends (RACE; Invitrogen, Carlsbad, CA, United States; TaKaRa Bio Inc., Kusatsu, Shiga, Japan). All primer sequences were based on the newly obtained reads and amplified sequences. The primers used are listed in Supplementary Tables 1, 2).
Viral RNA was isolated with a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) using individual samples. The cDNA was generated using random primers and Superscript III Reverse Transcriptase (Invitrogen). The nearly complete genomic sequences of the viruses obtained by end amplification were used to design specific nested primers targeting the non-structural gene for PCR and screen for astroviruses in the oral, nasal, and anal swabs of walruses and California sea lions (Supplementary Table 3). PCR was performed using 2× Taq PCR Mastermix (Tiangen Biotech Co., Ltd., Beijing, China). Two microliters first-round PCR product was used as the template for the second PCR round. The thermal cycling conditions were 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 45 s, and a final elongation step at 72°C for 10 min. The PCR products were analyzed by 1.5% agarose gel electrophoresis and ultraviolet imaging.
The nucleotide sequences of the genomes and the amino acid sequences of the open reading frames (ORFs) were deduced by comparing the sequences against those of other astroviruses. The conserved protein families and domains were predicted with Pfam,3 Blastp,4 and InterProScan 5 (see text footnote 3). Routine sequence alignments were performed with Clustal Omega.5
MEGA6.0 (see text footnote 2), the MUSCLE package,6 and their default parameters were used to align the nucleotide sequences and deduce the amino acid sequences. A phylogenetic tree was constructed by the ML method. The substitution model rtREV and the Freqs (+F) model were run using the model selection function in MEGA6.0 and 1,000 bootstrap replicates. Pairwise amino acid alignment between the novel pinniped astrovirus and the other Astroviridae was performed using the NCBI Basic Local Alignment Search Tool (BLAST).7 The alignments were analyzed with SimPlot v. 3.5.18 using a sliding window of 1,000 nucleotides (nt) in 100-nt steps. Each strain was a query for each run.
We obtained 3.6 Gb of nucleotide data (34,870,258 valid reads, 150 bp long) from two pools according to species. Reads from archaea, bacteria, microbial eukaryotes such as fungi, and those with no significant similarities to the amino acid sequences in the non-redundant (NR) proteins database were removed. Pool 30 of O. rosmarus with 124,219 reads and pool 35 of Z. californianus with 5,026 reads most closely matched viral proteins from the NR database (∼0.37% of the total). A comparison of the virome composition between walrus and California sea lions is shown in Figure 1. We focused mainly on the mammalian viruses within these families. After excluding phages and insect, fungal, plant, and unclassified viruses, the proportion of reads corresponding to astroviruses accounted for 40.11 and 43.64% of the total mammalian viruses in pools 30 and 35, respectively.
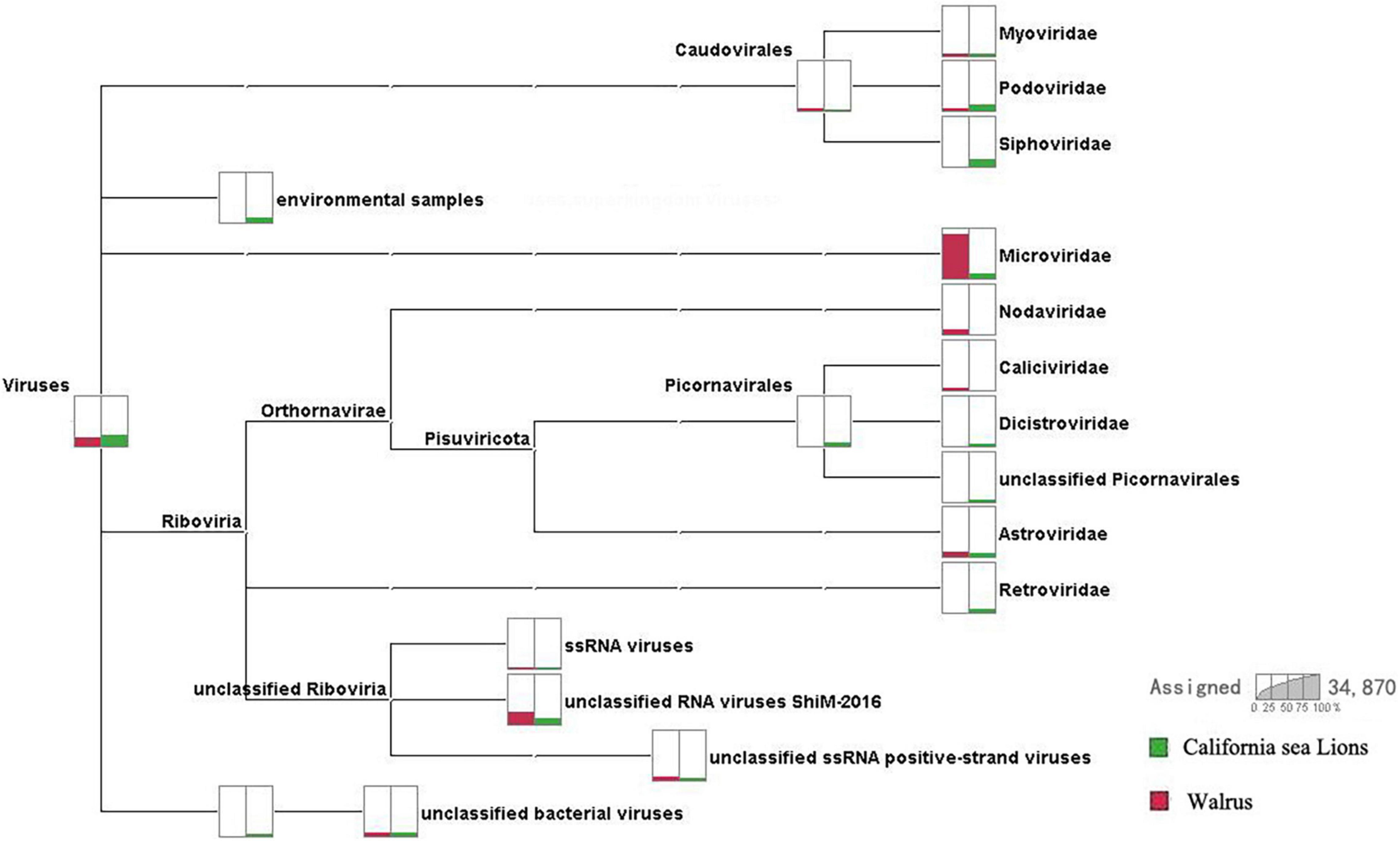
Figure 1. Comparison of virome compositions of walruses and California sea lions. The proportions of viral reads associated with California sea lions and walruses are highlighted in green and red, respectively.
We obtained a nearly complete genome sequence for the novel California sea lion astrovirus AstroV-HMU-1. We also obtained a complete coding sequence for the non-structural protein of the novel walrus virus AstroV-like-HMU-2 (GenBank accession Nos. MW853971–MW853972). The genome organization of AstroV-HMU-1 was characteristic of a mammalian astrovirus and comparable to those of related mammalian astroviruses (Figure 2). The genome sequence of AstroV-HMU-1 was 5,641 bp long and had 49.6% CG content. The BLAST similarity search revealed reading frame shifts in the translation around 1,642 and 3,205 nt. The capsid protein (CP) consisted of 781 amino acids. The non-structural protein consisted of a viral protease (SP) and a polymerase (Rdrp) which are typical of the astrovirus structure. The partial putative SP was 563 amino acids long and the putative RdRp was 517 amino acids long. The genome identity analysis showed that the CP of AstroV-HMU-1 had 96.71% identity to that of Mamastrovirus 11 (FJ890351) and <63.26% identity with other Mamastroviruses. The partial putative SP of AstroV-HMU-1 shared 58.11% identity with the SP of mink astrovirus (ADR65075) and had <54.20% identity with other Mamastroviruses. The putative RdRp of AstroV-HMU-1 shared 77.50% identity with the RdRp of mink astrovirus (ADR65075) and had <65.03% identity with other Mamastroviruses. However, there were relatively few reads corresponding to the astroviruses of Pool 35. The reads may only be related to the non-structural polyprotein region. We obtained the non-structural polyprotein sequence of AstroV-like-HMU-2 by reads-only PCR, designed specific gene primers, and used the 5′-RACE and 3′-RACE methods to amplify the remaining genome sequence. The amplification results were negative. Hence, we hypothesized that this novel astrovirus may lack poly(A)-tails. We used a Poly(A) Polymerase Kit (TaKaRa Bio Inc., Kusatsu, Shiga, Japan) to add poly(A)-tails to the 3′-hydroxyl end of the viral RNA. We then used the 3′-RACE method to re-amplify the 3′ ends. However, the amplification results remained negative. The non-structural polyprotein of AstroV-like-HMU-2 was 1,247 amino acids long and shared only 36.47% identity with those of bat astrovirus and <36% identity with other mamastroviruses.
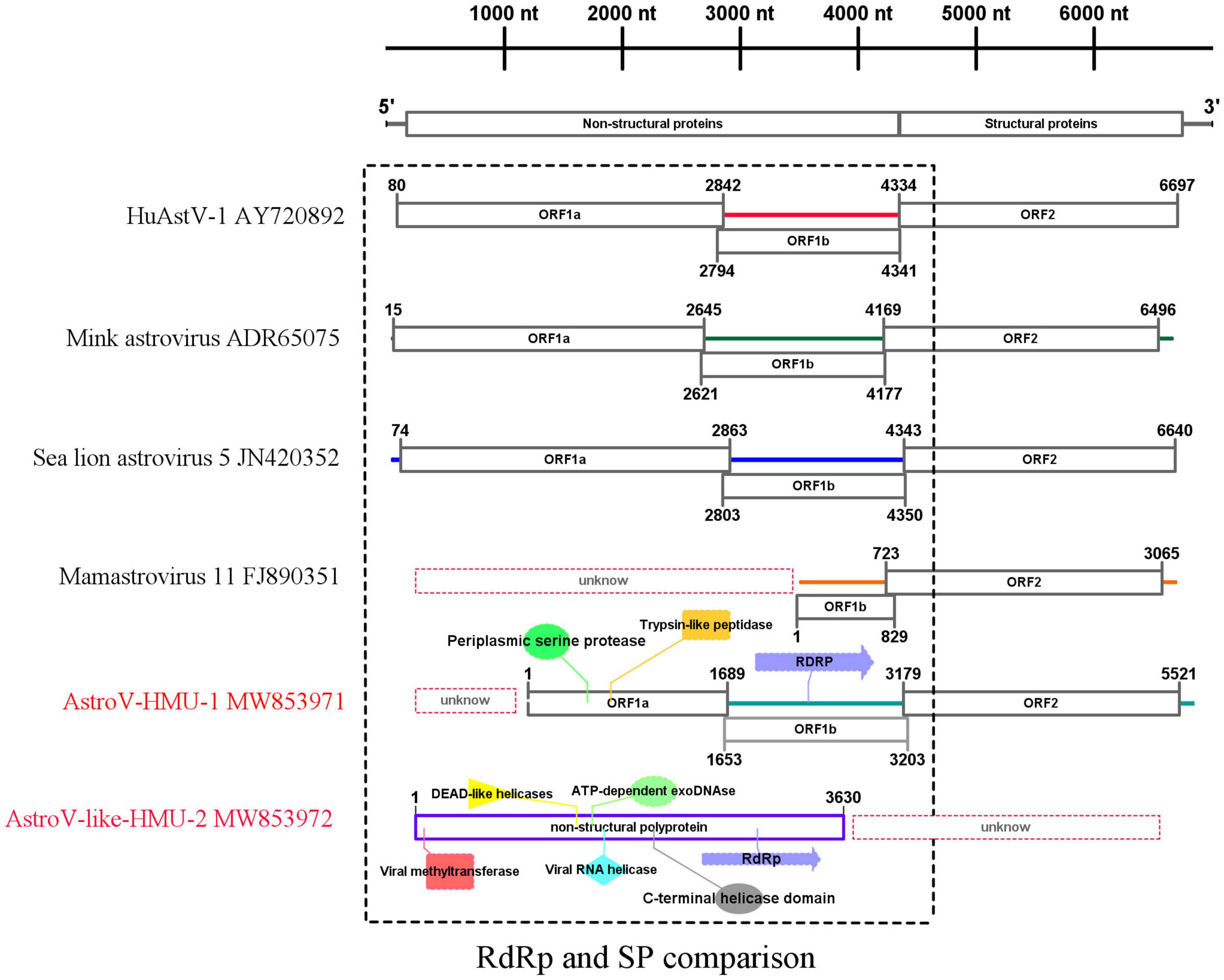
Figure 2. Genome organization and comparison of related astroviruses. Novel astroviruses discovered in the present study are highlighted in red.
For the putative non-structural protein, the conserved domains of AstroV-HMU-1 detected by Pfam were trypsin-like peptidase domain (148–268), periplasmic serine protease (123–279), and RNA-dependent RNA polymerase (625–979). The conserved domains of AstroV-like-HMU-2 detected by Pfam were viral methyltransferase (24–222) (involved in mRNA capping), viral RNA helicase (476–726), C-terminal helicase domain (674–726), ATP-dependent exoDNAse (473–739), DEAD-like helicases (456–575), and RdRp (942–1,115). The conserved domains of serine protease and RdRp were typical of astroviruses according to the NS structure but were absent in the NS sequence of AstroV-like-HMU-2. The conserved domains of RdRp were present in both AstroV-HMU-1 and AstroV-like-HMU-2 (Figure 2).
A phylogenetic analysis of AstroV-HMU-1 was conducted using 89, 82, and 65 reference CP, RdRp, and SP sequences, respectively, from the Astroviridae in GenBank (accessed August 1, 2021). Evolutionary trees were constructed for the complete protein sequences of SP, RdRp, and CP (Figures 3, 4). The tree topology showed that the CP of AstroV-HMU-1 clustered with other Z. californianus astroviruses among the Mamastrovirus 11 species in genus Mamastrovirus. The tree topology data showed that the RdRp and SP of AstroV-HMU-1 clustered with mink astroviruses in the same branch and comprised a lineage distinct from other sea lion astroviruses. The recombination analyses (Figure 5) showed that the AstroV-HMU-1, SMS-AstV, and VA2/human viruses underwent substantial recombination events in the NS region.
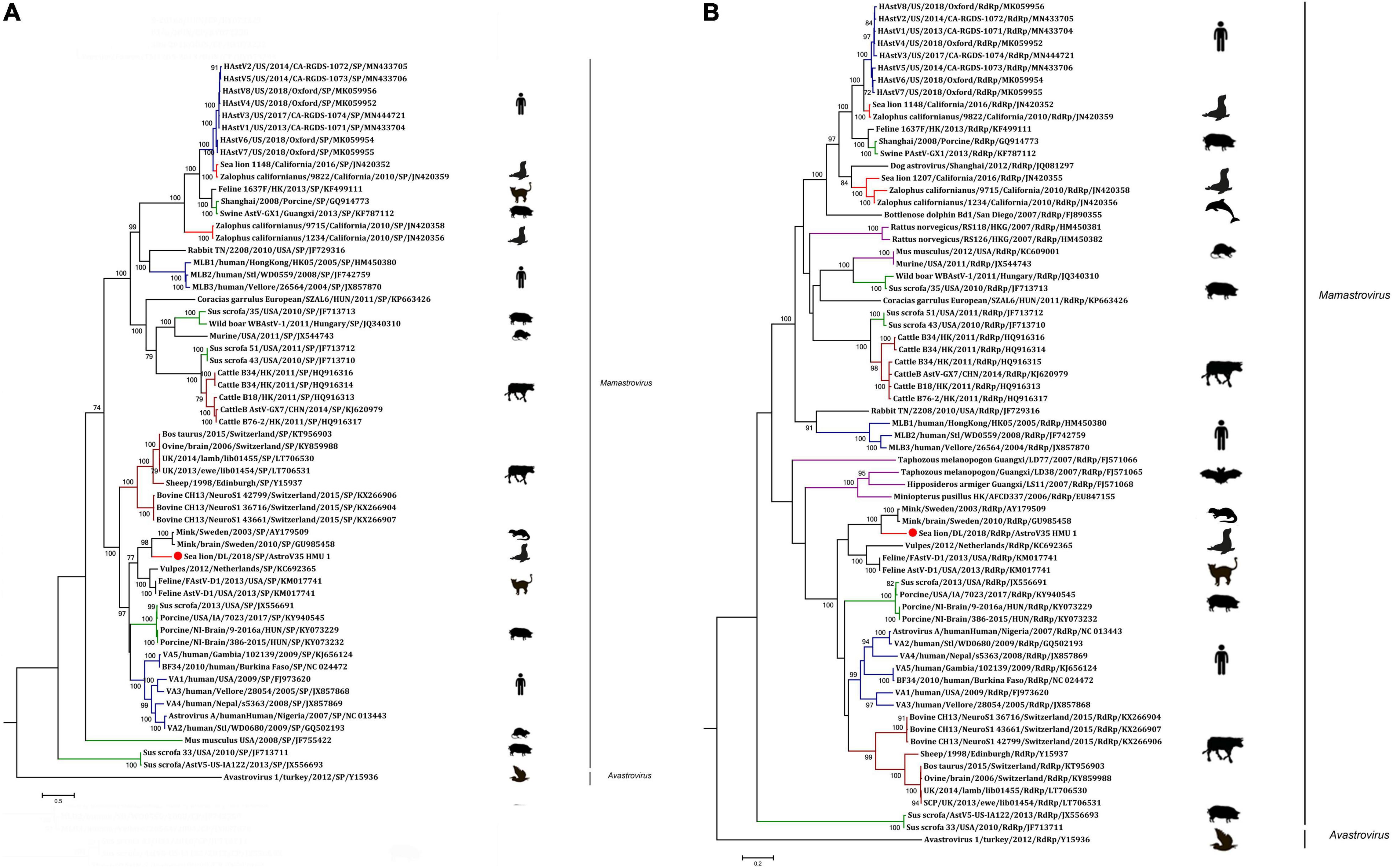
Figure 3. (A) Phylogenetic analyses of SP regions of AstroV-HMU-1 with reference sequences. (B) Phylogenetic analyses of RdRp regions of AstroV-HMU-1 with reference sequences. Maximum likelihood (ML) mtREV with Freqs (+F) model and γ-distributed with invariant sites (G + I) with 1,000 bootstrap replicates performed in MEGA6. Novel astrovirus is indicated by red circles (•). Bootstrap values are shown on branches.
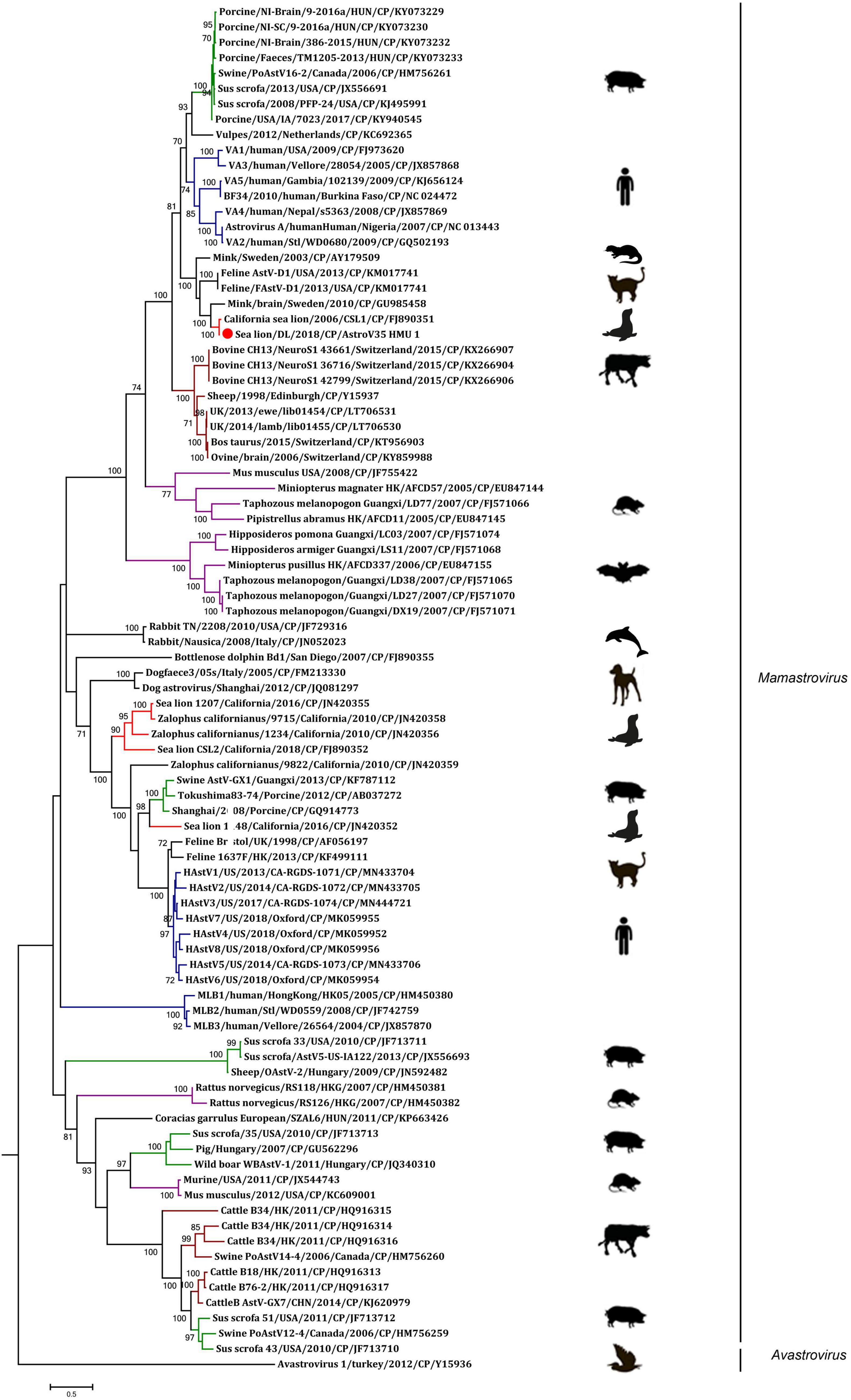
Figure 4. Phylogenetic analyses of CP regions of AstroV-HMU-1 with reference sequences. Maximum likelihood (ML) mtREV with Freqs (+F) model and γ-distributed with invariant sites (G + I) with 1,000 bootstrap replicates performed in MEGA6. Novel astrovirus is indicated by red circles (•). Bootstrap values are shown on branches.
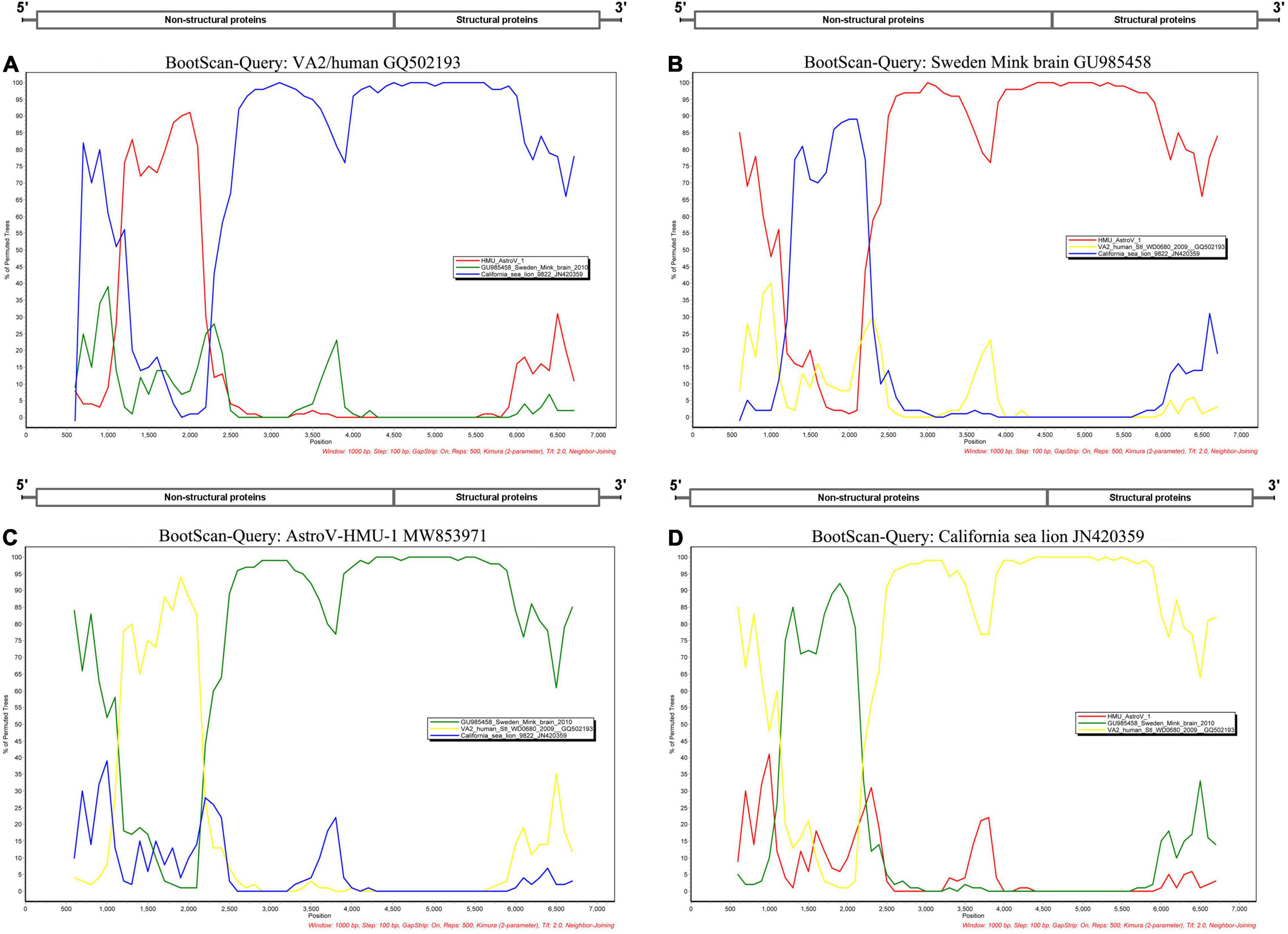
Figure 5. Bootscan analyses based on nearly full-length genomes of AstroV-HMU-1 and Swedish mink brain (GU985458), human VA2/human (GQ502193), and Californian sea lion (JN420359) strains. (A) VA2/human strain used as query sequence in analyses. (B) Swedish mink brain strain used as query sequence in analyses. (C) AstroV-HMU-1 strain used as query sequence in analyses. (D) Californian sea lion strains used as query sequence in analyses. Sliding window of 1,000 nt moving in 100-nucleotide steps.
Phylogenetic analyses of AstroV-like-HMU-2 and AstroV-HMU-1 were conducted using the 16 NS in the positive-sense ssRNA virus related to bastroviruses (Sadeghi et al., 2018). The NS of AstroV-like-HMU-2 was segregated from the Astroviridae in the deep root of the phylogenetic tree (Figure 6) and formed an independent branch with Brazil/sewage, Bastrovirus-like_virus/VietNam/Bat/17819_21, and CAVL/Fresno. AstroV-HMU-1 was closely related to the mink astrovirus. This finding was consistent with results of the phylogenetic analyses of the SP and Rdrp regions.
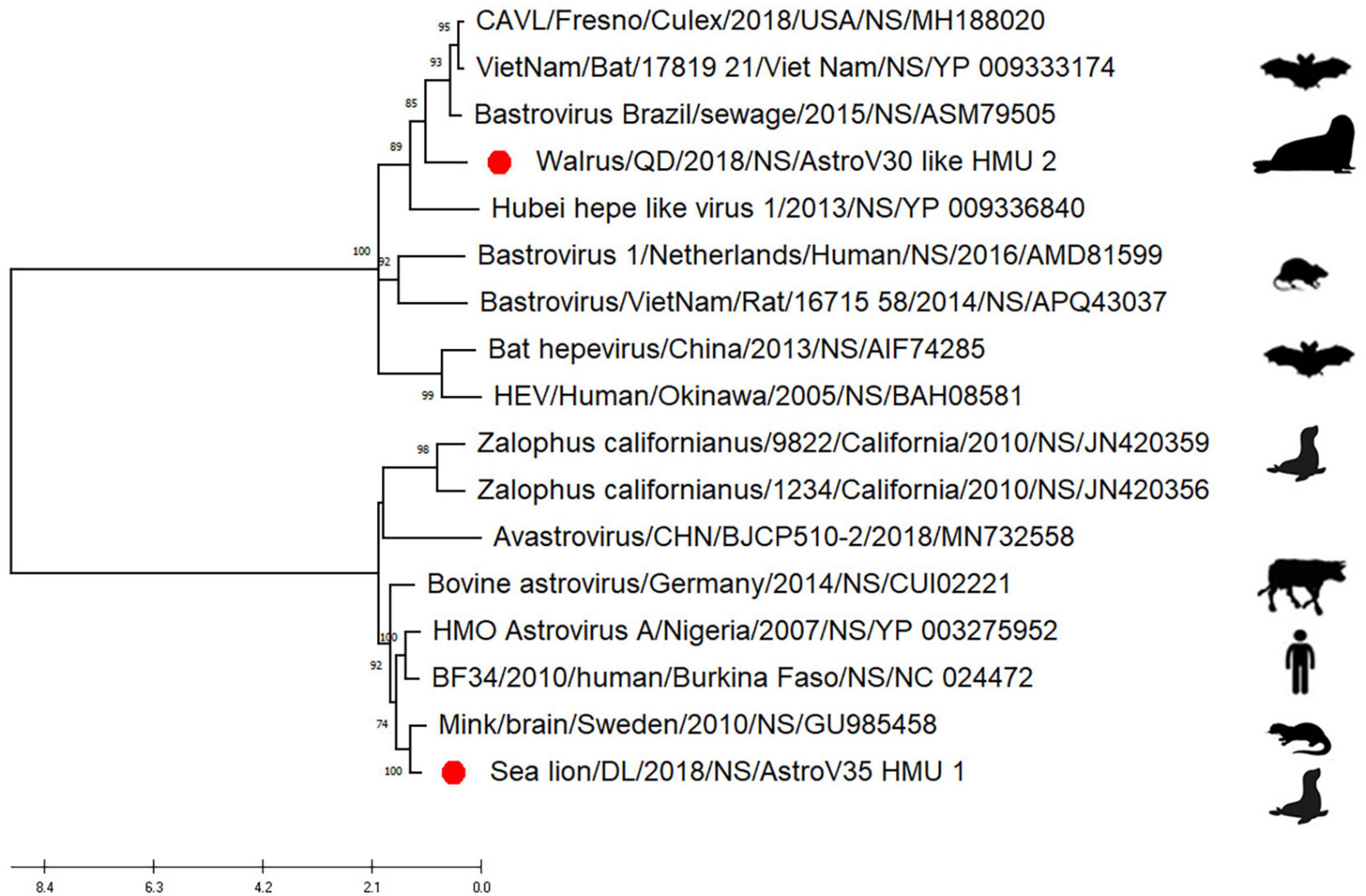
Figure 6. Phylogenetic analyses of NS regions of AstroV-like-HMU-2. Maximum likelihood (ML) mtREV with Freqs (+F) model and γ-distributed with invariant sites (G + I) with 1,000 bootstrap replicates performed in MEGA6. Novel astrovirus is indicated by red circles (•).
Based on the genomic sequences of the novel astroviruses, gene-specific nested primers targeting the non-structural region for PCR were designed to screen for novel astroviruses in the mouths, noses, and anuses of pinnipeds. Two oral and three anal swabs of the California sea lions and one anal swab of the walruses were positive for the novel astrovirus. There was 0% positivity (0/4) for the nasal swabs, 14.3% positivity (2/14) for the oral swabs, and 28.6% (4/14) for the anal swabs (Table 1). The positive samples were identified from 939 bp of amplified products for AstroV-HMU-1 and 845 bp of amplified products for AstroV-like-HMU-2.
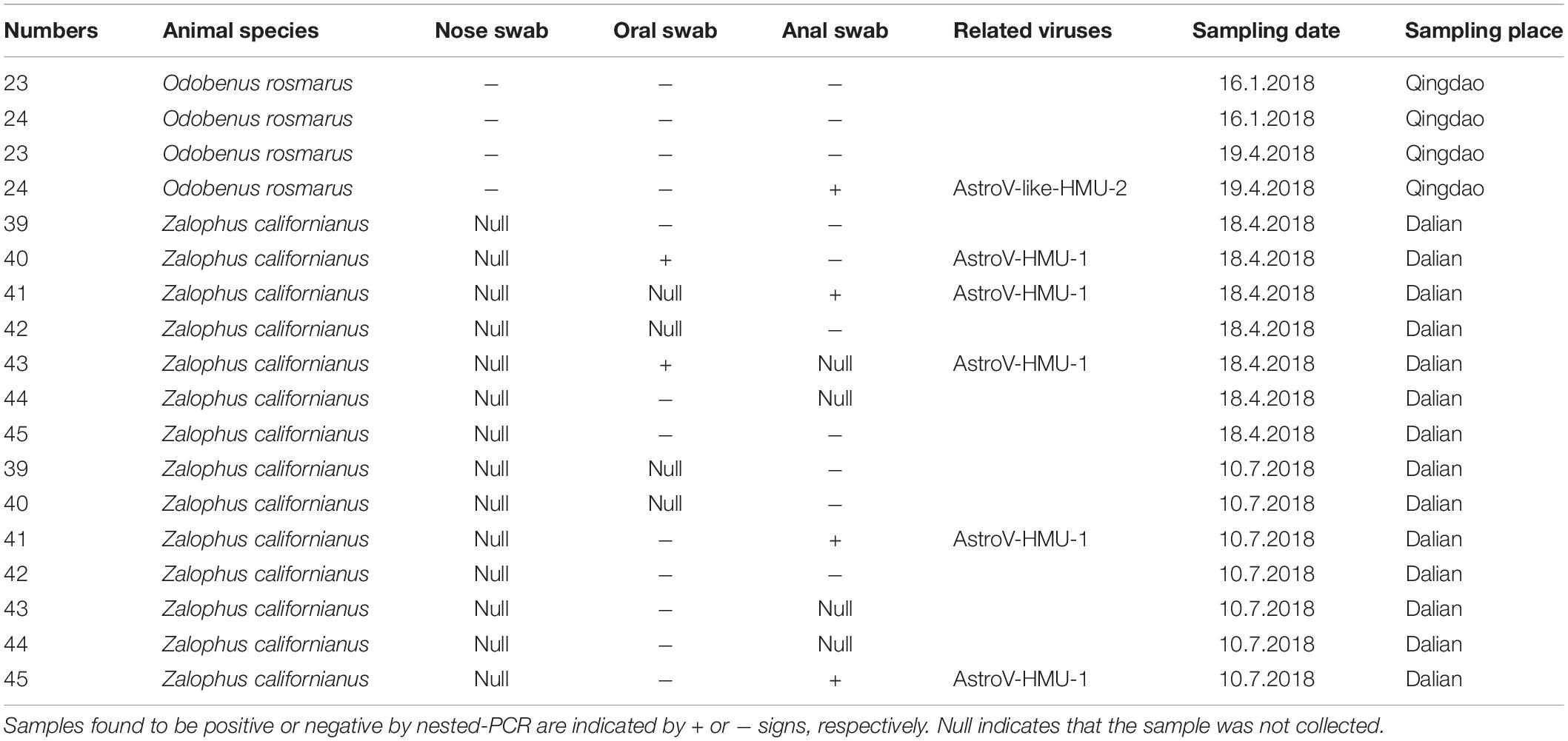
Table 1. Prevalence of AstroV-HMU-1, AstroV-like-HMU-2 in samples of captive walrus, California sea lions from Qingdao and Dalian Ocean Park.
The virome was composed of double-stranded (ds) DNA, dsRNA, retro-transcribing, single-stranded (ss) DNA, and ssRNA viruses and differed between walruses and California sea lions. The virus-associated reads of the California sea lion samples were either assigned to the families Myoviridae, Podoviridae, Siphoviridae, Microviridae, Dicistroviridae, Picornaviridae, Astroviridae, or Retroviridae or were unclassified. The virus-associated reads of the walrus samples were assigned to the families Myoviridae, Podoviridae, Microviridae, Caliciviridae, Dicistroviridae, Astroviridae or unclassified Riboviria viruses. Caliciviridae, Astroviridae, and Picornaviridae include numerous important mammalian viruses. Astrovirus-associated reads were detected in both the walrus and California sea lion samples. The positive rates of astrovirus infection determined for the individual samples reflected the possibility of a high incidence of astrovirus infection in pinnipeds.
The phylogenetic analysis of AstroV-HMU-1 was consistent with the genomic amino acid identity and recombination analyses and showed that the CP was assigned to Mamastrovirus 11 (California sea lion astroviruses) while the RdRp and SP clustered with the mink astroviruses. Hence, the genomes of AstroV-HMU-1, California sea lion 9,822, VA2/human, and mink astrovirus might have recombined long ago and later independently evolved into their natural hosts. Furthermore, Mamastrovirus 11 (FJ890351) is not associated with any genomic information for the NS protein regions in GenBank. Thus, Z. californianus AstroV-HMU-1 was proposed as a new member of the Mamastrovirus 11 species and provided genomic information for the NS protein regions.
The incidence of AstroV-like-HMU-2 was sporadic in samples from captive animals. AstroV-like-HMU-2 was detected in a single anal swab from O. rosmarus. However, the oral and nasal swab samples of this animal were negative. The oral and nasal swabs may not have collected sufficient virus-infected exfoliated host cells. Moreover, AstroV-like-HMU-2 only infected intestinal epithelial cells. The incidence of AstroV-like-HMU-1 was also sporadic. It was detected in only two oral and three anal swabs from Z. californianus. On April 18 and July 10, 2018, both anal swabs for Z. californianus No. 41 were positive. Hence, this individual might have been persistently infected with AstroV-HMU-1. Both oral swabs were also positive for AstroV-HMU-1. The route of astrovirus transmission may be fecal-oral. The pathogen could also be food- or water-borne (Walter and Mitchell, 2000). However, both corresponding anal swabs were negative for AstroV-HMU-1. In general, the positive rate for AstroV-HMU-1 was relatively low in Z. californianus even when positive oral or anal swabs were obtained. There was no direct evidence that AstroV-HMU-1 was associated with diarrhea, but it was nonetheless a possibility as California sea lions normally produce strip-like feces rather than loose stools.
According to the ICTV (Monroe et al., 2005), walruses are hosts of AstroV-like-HMU-2. Furthermore, the non-structural polyprotein of AstroV-like-HMU-2 shared <36.47% identity with those of other astroviruses. The NS of AstroV-like-HMU-2 segregated from the Astroviridae in the deep root of the phylogenetic tree. We propose that AstroV-like-HMU-2 is a novel species among the unclassified astroviruses. However, the astroviruses were classified on the basis of ORF2. Complete genome sequence information is required to determine whether AstroV-like-HMU-2 is, in fact, a new species. Future research should continue to elucidate the genomes of these viruses. Current astrovirus classification criteria do not accommodate the categorization of novel astroviruses. The Astroviridae may also include other genera besides Mamastrovirus and Avastrovirus. To confirm this theory, complete genome sequences of novel astroviruses should be uploaded to GenBank.
The novel pinniped astrovirus discovered and characterized here is a source of new genomic information that expands virological classification and provides a scientific basis for further investigations into animal-borne viral diseases.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA650224.
The animal study was reviewed and approved by the Ethics Committee of the Hainan Medical University.
JD, JF-WC, PZ, and K-YY designed the study. PZ, HS, GW, YH, SB, JL, SL, RP, XH, WZ, FY, XC, LN, YZ, YL, LX, CT, and GL collected the specimens and performed the experiments. JD, FFY, WZ, FD, and MZ analyzed the data. JD and FFY wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the Key Research and Development Plan of Hainan Province (No. ZDYF2020150), the National Natural Science Foundation of China (Nos. 32060015, 819MS058, and 82060378), CAMS Innovation Fund for Medical Sciences (CIFMS) (grant 2021-I2M-1-037), the Major Science and Technology Program of Hainan Province (No. ZDKJ202003), the Hainan Provincial Natural Science Foundation of China (Nos. 820RC628, 2019RC218, and 819MS058), Open Foundation of NHC Key Laboratory of Tropical Disease Control (Nos. 2021NHCTDCKFKT11006), the Scientific Research Foundation of the Higher Education Institutions of Hainan Province, China (No. Hnky2021-40), the Research Project of the Hainan Academician Innovation Platform (No. YSPTZX202004), the Hainan Talent Development Project (No. SRC200003), the National S&T Major Project “China Mega-Project for Infectious Disease” (No. 2018ZX10711001), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (No. 2020363), and the Scientific and Technological Program of Strategic Priority in the Chinese Academy of Sciences (No. XDB42030403), Graduate student innovation grant of Hainan Medical University (Hys2020-385 and Hys2020-33).
JL is employed by Dalian Sunasia Tourism Holding Co., Ltd. SL is employed by Qingdao Polar Haichang Ocean Park.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.845601/full#supplementary-material
Appleton, H., and Higgins, P. G. (1975). Letter: viruses and gastroenteritis in infants. Lancet 1:1297. doi: 10.1016/s0140-6736(75)92581-7
Boros, A., Albert, M., Pankovics, P., Bíró, H., Pesavento, P. A., Phan, T. G., et al. (2017). Outbreaks of neuroinvasive astrovirus associated with encephalomyelitis, weakness, and paralysis among weaned pigs, hungary. Emerg. Infect. Dis. 23, 1982–1993. doi: 10.3201/eid2312.170804
Calistri, A., and Palu, G. (2015). Editorial commentary: unbiased next-generation sequencing and new pathogen discovery: undeniable advantages and still-existing drawbacks. Clin. Infect. Dis. 60, 889–891. doi: 10.1093/cid/ciu913
Cortez, V., Margolis, E., and Schultz-Cherry, S. (2019). Astrovirus and the microbiome. Curr. Opin. Virol. 37, 10–15. doi: 10.1016/j.coviro.2019.05.002
Finkbeiner, S. R., Allred, A. F., Tarr, P. I., Klein, E. J., Kirkwood, C. D., and Wang, D. (2008). Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4:e1000011. doi: 10.1371/journal.ppat.1000011
Finkbeiner, S. R., Li, Y., Ruone, S., Conrardy, C., Gregoricus, N., Toney, D., et al. (2009). Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J. Virol. 83, 10836–10839. doi: 10.1128/JVI.00998-09
Johnson, C., Hargest, V., Cortez, V., Meliopoulos, V. A., and Schultz-Cherry, S. (2017). Astrovirus pathogenesis. Viruses 9:22. doi: 10.3390/v9010022
Karlsson, E. A., Small, C. T., Freiden, P., Feeroz, M. M., Matsen, F. A., San, S., et al. (2015). Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog. 11:e1005225. doi: 10.1371/journal.ppat.1005225
Meliopoulos, V. A., Kayali, G., Burnham, A., Oshansky, C. M., Thomas, P. G., Gray, G. C., et al. (2014). Detection of antibodies against Turkey astrovirus in humans. PLoS One 9:e96934. doi: 10.1371/journal.pone.0096934
Mitchell, D. K., Matson, D. O., Cubitt, W. D., Jackson, L. J., Willcocks, M. M., Pickering, L. K., et al. (1999). Prevalence of antibodies to astrovirus types 1 and 3 in children and adolescents in Norfolk. Virginia. Pediatr. Infect. Dis. J. 18, 249–254. doi: 10.1097/00006454-199903000-00008
Monroe, S. S., Carter, M. J., Hermann, J., Mitchell, D. K., and Sanchez-Fauquier, A. (2005). “Astroviridae,”. in Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, eds C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger and L. A. Ball, Amsterdam (San Diego: Academic Press), 859–864.
Parry, R., and Asgari, S. (2019). Discovery of novel crustacean and cephalopod flaviviruses: insights into the evolution and circulation of flaviviruses between marine invertebrate and vertebrate hosts. J. Virol. 93:e00432-19. doi: 10.1128/JVI.00432-19
Rivera, R., Nollens, H. H., Venn-Watson, S., Gulland, F. M., and Wellehan, J. F. (2010). Characterization of phylogenetically diverse astroviruses of marine mammals. J. Gen. Virol. 91, 166–173. doi: 10.1099/vir.0.015222-0
Roach, S. N., and Langlois, R. A. (2021). Intra- and cross-species transmission of astroviruses. Viruses 13:1127. doi: 10.3390/v13061127
Sadeghi, M., Altan, E., Deng, X., Barker, C. M., Fang, Y., Coffey, L. L., et al. (2018). Virome of > 12 thousand Culex mosquitoes from throughout California. Virology 523, 74–88. doi: 10.1016/j.virol.2018.07.029
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vu, D. L., Bosch, A., Pinto, R. M., and Guix, S. (2017). Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses 9:33. doi: 10.3390/v9020033
Walter, J. E., and Mitchell, D. K. (2000). Role of astroviruses in childhood diarrhea. Curr. Opin. Pediatr. 12, 275–279. doi: 10.1097/00008480-200006000-00018
Wu, Z., Lu, L., Du, J., Yang, L., Ren, X., Liu, B., et al. (2018). Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6:178. doi: 10.1186/s40168-018-0554-9
Keywords: genetic diversity, novel astrovirus, pinnipeds, evolution analysis, recombination
Citation: Zhang P, Su H, Peng R, Chan JF-W, Bai S, Wang G, Huang Y, Hu X, Luo J, Liu S, Li Y, Xue L, Yang F, Zhao M, Zhang Y, Tang C, Shen S, Cui X, Niu L, Lu G, Yuen K-Y, Deng F, Zhang W, Yin F and Du J (2022) Identification of a Novel Astrovirus in Pinnipeds. Front. Microbiol. 13:845601. doi: 10.3389/fmicb.2022.845601
Received: 30 December 2021; Accepted: 03 March 2022;
Published: 04 May 2022.
Edited by:
Jesus L. Romalde, University of Santiago de Compostela, SpainReviewed by:
João Mesquita, Universidade do Porto, PortugalCopyright © 2022 Zhang, Su, Peng, Chan, Bai, Wang, Huang, Hu, Luo, Liu, Li, Xue, Yang, Zhao, Zhang, Tang, Shen, Cui, Niu, Lu, Yuen, Deng, Zhang, Yin and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijia Zhang, d3poYW5nQGlkc3NlLmFjLmNu; Feifei Yin, eWluZmVpZmVpZmZAMTYzLmNvbQ==; Jiang Du, ZHVqaWFuZ2VtYWlsQHFxLmNvbQ==
†These authors have contributed equally to this work
‡These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.