- 1State Key Laboratory of Livestock and Poultry Breeding, Guangdong Key Laboratory of Animal Breeding and Nutrition, Institute of Animal Science, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Institute of Animal Health, Guangdong Academy of Agricultural Sciences, Key Laboratory of Livestock Disease Prevention of Guangdong Province, Scientific Observation and Experiment Station of Veterinary Drugs and Diagnostic Techniques of Guangdong Province, Ministry of Agriculture and Rural Affairs, Guangzhou, China
- 3Sericultural & Agri-Food Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 4Institute of Laboratory Animal Sciences, Chinese Academy of Medical Sciences and Comparative Medicine Center, Peking Union Medical College, Beijing, China
- 5Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Guangdong, China
Host’s innate immunity is the front-line defense against viral infections, but some viruses have evolved multiple strategies for evasion of antiviral innate immunity. The porcine enteric coronaviruses (PECs) consist of porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), transmissible gastroenteritis coronavirus (TGEV), and swine acute diarrhea syndrome-coronavirus (SADS-CoV), which cause lethal diarrhea in neonatal pigs and threaten the swine industry worldwide. PECs interact with host cells to inhibit and evade innate antiviral immune responses like other coronaviruses. Moreover, the immune escape of porcine enteric coronaviruses is the key pathogenic mechanism causing infection. Here, we review the most recent advances in the interactions between viral and host’s factors, focusing on the mechanisms by which viral components antagonize interferon (IFN)-mediated innate antiviral immune responses, trying to shed light on new targets and strategies effective for controlling and eliminating porcine enteric coronaviruses.
Introduction
As the largest positive-sense RNA viruses that exist widely in nature, coronaviruses have genetic diversity and host diversity. Specific coronavirus populations have been found in humans, mice, bats, pigs, chickens, cows, cats, dogs, and many other animals, some of which are zoonotic and pose serious threats to human health and livestock safety. Due to the mild clinical manifestations after infection, coronaviruses have long been ignored by people. However, the outbreak of Severe Acute Respiratory Syndromes (SARS) in 2003 caused a total of 8,000 cases of infection worldwide, including 774 deaths, with a mortality rate of about 10% (Drosten et al., 2003). Even worse, the worldwide spread of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) since 2019 caused hundreds of millions of infections and millions of deaths. Its high infectivity and transmission speed and the lack of specific medicines and vaccines caused a worldwide panic. In the past few decades, with the continuous development of the breeding mode, various coronavirus, including porcine epidemic diarrhea virus (PEDV) and infectious chicken bronchial virus, have been world-widely prevalent and pose great challenges to the health and safety of the breeding industry. Some animals have been proved to carry coronaviruses that can spread from one species to another. A most recent research has revealed a surprising result showing that porcine deltacoronavirus (PDCoV) strains exist in plasma samples of three Haitian children with acute undifferentiated febrile illness (Lednicky et al., 2021). Therefore, systematic analysis of animal coronavirus, especially porcine enteric coronavirus (PEC), appeared to be extremely necessary.
Interferons (IFNs) are key components of the host’s antiviral innate immunity. IFNs are consisted of type I IFNs, type II IFNs, and type III IFNs. Type I IFN is a non-glycosylated protein composed of 165–300 amino acids. Almost all cells can produce type I IFNs when pattern recognition receptors (PRRs) recognize the microbial pathogen-associated molecular patterns (PAMPs). IFN-I binds to type IFN-I receptor (IFNAR) to induce a powerful antiviral defense program involving hundreds of interferon-stimulated genes (ISGs) by activating the JAK–STAT pathway. Furthermore, ISGs are capable of interfering with every step of viral replication (Schoggins and Rice, 2011). Like IFN-I, type III IFNs bind to the type III IFN receptor (IFNLR) and share the same pathway to induce a similar antiviral transcriptional program (Kotenko et al., 2019). It is now well-recognized that the IFN-λ-based antiviral system plays a major role in the antiviral protection of epithelial barriers. Due to the different expression of receptors, IFN-I signaling leads to a more rapid induction and decline of ISG expression. In contrast, IFN-III signaling induces the expression of ISGs in a more sustained way (Lazear et al., 2019). IFNs establish the cellular state of viral resistance and activate the adaptive immune responses to viruses. However, some viruses have evolved quite complicated mechanisms to escape immune recognition and antagonize the effects of IFNs and ISGs. The mechanisms by which different components of these viruses antagonize immune responses are also different. In the present review, the characteristics of PEC biology are elucidated, the mechanisms by which viruses antagonize immune responses are illustrated, and finally, the potential targets and strategies effective for controlling and eliminating porcine enteric coronaviruses are discussed.
Overview of Porcine Enteric Coronaviruses
In 2019, the International Committee on Taxonomy of Viruses divided the Coronaviridae into Letovirinae and Orthocoronavirinae, containing five genuses: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, Deltacoronavirus, and Alphaletovires. To date, there are six known swine coronaviruses, including four alphacoronavirus, one betacoronavirus, and one deltacoronavirus. Transmissible gastroenteritis coronavirus (TGEV), porcine respiratory coronavirus (PRCV), PEDV, and swine acute diarrhea syndrome-coronavirus (SADS-CoV) belong to the alphacoronavirus. Porcine hemagglutinating encephalomyelitis virus (PHEV) belongs to the betacoronavirus, and PDCoV belongs to the deltacoronavirus. Furthermore, the evolutionary genetic analysis suggested that PEDV and SADS-CoV were thought to originate from the bat CoVs and PDCoV from a sparrow CoV (Zhou et al., 2018; Wang et al., 2019), which suggested coronaviruses could spread from species to species. PEDV, PDCoV, and TGEV SADS-CoV can cause gastrointestinal infections and similar characteristics (Table 1). These porcine enteric coronaviruses mainly affect the digestive tract of piglets, and the clinical symptoms include weight loss, lethargy, vomiting, anorexia, watery diarrhea, and even death. The pathological features were necrosis and shedding of intestinal cells and intestinal villi injury (Jung et al., 2014; Pan et al., 2017; Suzuki et al., 2018; Xia et al., 2018). PEDV was first reported in the United Kingdom in the 1970s but was not found in the United States until 2013 (Wood, 1977; Stevenson et al., 2013). In the short time that followed, PEDV spread worldwide, with high morbidity and mortality rates, and caused huge economic losses to the global pig industry (Jung et al., 2015). The incubation period of the virus is generally 5–8 days. In addition, PEDV can infect pigs of all ages, but the severity and mortality of infected pigs are inversely proportional to pigs. The morbidity and mortality of suckling piglets within 7 days were up to 100% (Li et al., 2020). So far, PEDV has only been found to infect pigs and has no impact on public health.
Since 2012, PDCoV has been detected in several countries, including China, the United States, Japan, and Canada (Woo et al., 2012; Wang et al., 2014b; Ajayi et al., 2018). PDCoV can cause diarrhea of piglets in different degrees and PDCoV disease, and the incidence and mortality of PDCOV disease in suckling piglets are about 50%–100%. The ability to spread across species is the most obvious feature of coronavirus. Researchers believed that PDCoV could only infect chickens and calves but not humans for a long time. Furthermore, the virus does not cause serious health problems in these animals (Jung et al., 2017; Liang et al., 2019; Boley et al., 2020). However, a research team has identified porcine deltacoronavirus strains in plasma samples of three Haitian children with acute undifferentiated febrile illness (Lednicky et al., 2021). This discovery makes us aware that these porcine coronaviruses may cause threats to public health.
Transmissible gastroenteritis coronavirus was first reported in the United States in 1946 and then broke out worldwide (Doyle and Hutchings, 1946; Kim et al., 2000). TGEV infection mainly causes infectious gastroenteritis, which leads to vomiting, watery diarrhea, and even death in piglets (Garwes, 1988). TGEV infection is fatal to piglets born less than 1 week, since the mortality rate can reach high up to 100%. Although the mortality rate of infected pigs over 2 weeks old is low, their growth and development are slow, which can cause economic losses to the pig breeding industry (Saif, 1999; Penzes et al., 2001). Significantly, TGEV is a highly contagious disease with a short incubation period (usually 1–3 days) and can quickly affect the entire pig population (Liu and Wang, 2021). Pigs are the only host of TGEV, and no human infection has been reported to date. SADS-CoV was first reported in the southeast of China in 2016, which is the sixth porcine coronavirus identified so far. The SADS-CoV infection leads to acute diarrhea, acute vomiting, and even acute death in piglets, and the mortality rate of virus infection in piglets within 5 days of age is more than 90% (Zhou et al., 2018). Tests on 35 people who had close contact with infected pigs found no evidence of human infection, suggesting the virus may not be capable of transmitting to humans.
The Genome Structure and Function of Porcine Enteric Coronaviruses
The PEDV genome is about 28 kb in length and consists of 3′, 5′ untranslated regions (UTR) and seven open reading frames (ORFs). The ORF sequence is ORF1a, ORF1b, spike protein (S), ORF3, envelope protein (E), membrane protein (M), and nucleoprotein (N; Kocherhans et al., 2001). ORF1 occupies two-thirds of the length of the genome at the end of the 5′ UTR and encodes two proteins (pp1a and pp1ab), which can be hydrolyzed by papain-like protease and serine type 3C-like protease to non-structural proteins (NSP) 1α, NSP1β, and NSP3-16. And then, these NSPs participate in virus replication, transcription, translation, and viral protein processing (Kadoi et al., 2002; Huang et al., 2013). The S protein of PEDV consists of S1 (1-789aa) and S2 (790-1383aa) subunits. The S1 helps PEDV bind to host receptors, and S2 induces membrane fusion and PEDV invasion (Liu et al., 2015). In addition, PEDV S protein also induces neutralizing antibodies in the host body (Song and Park, 2012). PEDV ORF3 sits between the S and E genes and encodes ORF3 protein, which plays an essential role in virulence (Park et al., 2011). The E gene of PEDV is only 231 nt in length and encodes E protein, which is essential for virus assembly and budding (Brian and Baric, 2005). The M gene of PEDV is 681 nt in length and can encode M protein (226aa), which is a transmembrane protein on the viral envelope (Narayanan et al., 2000). It has been reported that M protein is involved in virion assembly, budding, and host innate immune induction (Utiger et al., 1995). The PEDV N protein is a highly conserved protein that consists of 441aa and is involved in the survival of the virus (Wang et al., 2020c).
The TGEV genome is about 28.5 kb in length, consisting of 3′, 5′ UTR, and seven open reading frames. The gene sequence arrangement is 5′-UTR-ORF1a-ORF1b-S-ORF3a-ORF3b-E-M-N-NS7-3′-UTR (Figure 1). TGEV ORF1a and ORF1b encode pp1a and pp1b, respectively, which can be hydrolyzed by papain-like protease and 3C-like protease to NSP1-16 (Van Reeth et al., 2002; Wang et al., 2018). Moreover, ORF3a/b and NS7 encode accessory proteins which are also involved in virus infection and virulence (Park et al., 2008). The TGEV S gene is about 4,344 bp and encodes S protein (1447aa), which has multiple functions, including inducing neutralizing antibodies, influencing host cell affinity, and determining virus activity (Gack et al., 2007). E protein of TGEV is a kind of membrane-associated small structural protein. A study has reported that 64-AYKNF-68 residues are the core sequences for binding E monoclonal antibodies (Yachdav et al., 2014). The M protein of TGEV consists of 263aa, is a kind of glycosylated protein that plays an important role in virus assembly. TGEV M protein is involved in inducing interferons (Sawicki et al., 2005). The N protein of TGEV is a conserved phosphorylated protein that binds to the genome to form an RNA complex. Because of its conserved nature, N is often used as an antigen for PEDV detection.
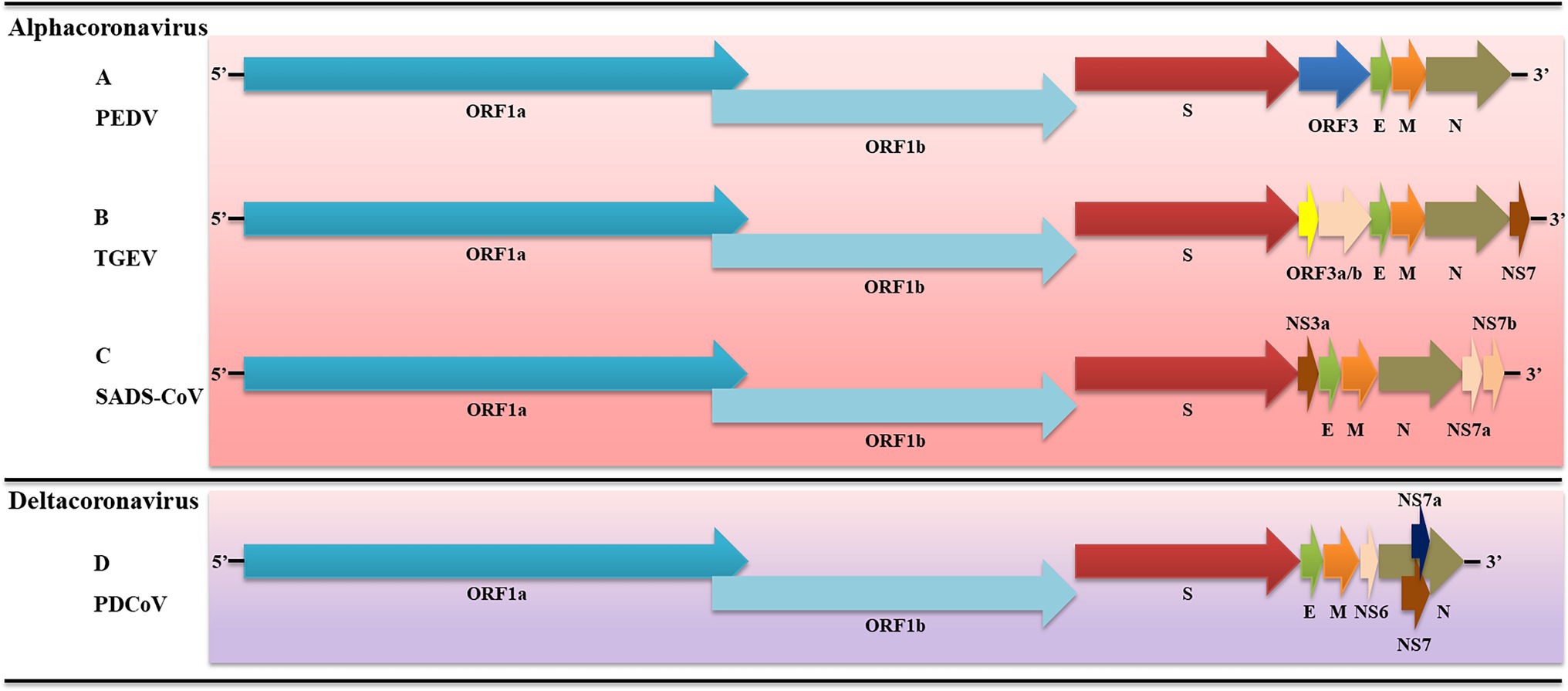
Figure 1. Schematic diagram of porcine enteric coronaviruses (PECs). (A) Genome structure of PDEV; (B) Genome structure of transmissible gastroenteritis coronavirus (TGEV); (C) Genome structure of swine acute diarrhea syndrome-coronavirus (SADS-CoV); (D) Genome structure of porcine deltacoronavirus (PDCoV). S, spike; E, envelope; M, membrane; N, nucleoprotein; and Ns, accessory genes.
The SADS-CoV genome is approximately 27.2 kb in length and consists of 3′, 5′-UTR, and nine open reading frames, which are ORF1a, ORF1b, S, NS3a, E, M, N, NS7a, and NS7b in sequence (Pan et al., 2017; Xu et al., 2019a). SADS-CoV ORF1a encodes pp1a, which can be hydrolyzed to NSP 1–11. Moreover, ORF1b encodes pp1b, which can be hydrolyzed to NSP 12–16. NS3a, NS7a, and NS7b encode accessory proteins that affect viruses’ virulence (Xu et al., 2019a). The membrane protein interacts with the nucleoprotein of SADS-CoV during viral assembly and enhances viral transcription and assembly efficiency. The spike of SADS-CoV (1130aa) determines viral host range and tissue tropism. Notably, SADS-CoV S is highly homologous (95%) to bat α-coronavirus HKU2, which suggests that it may have the potential to spread across species (Yu et al., 2020).
The length of the PDCoV genome is approximately 25.4 kb, which is the smallest known coronavirus. PDCoV genome consists of 3′, 5′ UTR, and eight open reading frames, which are ORF1a, ORF1b, S, E, M, NS6, N, NS7a, and NS7 in order (Woo et al., 2010; Chen et al., 2015; Fang et al., 2017; Figure 1). PDCoV ORF1a and ORF1ab encode 1a polyprotein (3627aa) and 1ab polyprotein (6268aa), which can be hydrolyzed to NSP 2–16. It is widely accepted that PDCoV did not encode non-structural protein 1 (nsp1). These NSPs are associated with virus transcription, replication, and host immune response (Zhang, 2016). In addition, there are two ORFs between the M gene and N gene and within the N gene, which encode NS6, NS7a, and NS7. When PDCoV infects host cells, they are located in the endoplasmic reticulum (ER) and mitochondria, respectively (Fang et al., 2016; Choi and Lee, 2019). The structural analysis of PDCoV S protein showed that it was composed of S1 and S2 subunits. The N-terminal domain of PDCoV S1 recognizes carbohydrates as potential receptors, and the C-terminal domain of PDCoV S1 binds to receptors on the surface of mammalian cells (Shang et al., 2018). PDCoV N protein is located in the cytoplasm and nucleus of the host cell and participates in viral RNA synthesis by interacting with ribosomal subunits or nucleoproteins (Lee and Lee, 2015). In addition, PDCoV N protein is also involved in influencing the immune response of host cells (Likai et al., 2019).
Innate Recognition of Porcine Enteric Coronavirus
Innate immunity is the first line of the host to defense against virus infection. The host cells recognize the invading pathogens through the interactions between PAMPs and host PRRs and induce the production of pro-inflammatory cytokines and interferons to elicit antiviral responses (Akira et al., 2006). When coronaviruses invade cells, PRRs, such as retinoic acid-inducible gene I (RIG-I)-like receptors (RLR) and Toll-like receptors (TLRs), are essential for the innate recognition of viral RNAs and are involved in the restriction of viral replication and dissemination. RLR, a family of cytoplasmic RNA helicases, including RIG-I, melanoma differentiation-associated gene 5 (MDA5), and Laboratory of Genetics and Physiology 2 (LGP2; Liu and Cao, 2016). Activation of RIG-I and MDA5 by double-stranded RNA (dsRNA) from coronaviruses leads to recruitment of the caspase recruitment domain (CARD)-containing adaptor protein mitochondrial antiviral signaling (MAVS) protein to activate TANK-binding kinase 1 (TBK1)/inhibitor-kb kinase ε (IKKε) kinases (Seth et al., 2005). Activated TBK1 and IKKε induce type I and type III IFNs production through phosphorylating interferon regulatory factors (IRFs; Meylan et al., 2006).
Other RNA sensors, TLR3, TLR7, and TLR8, located in the endosomal membrane, also recognize viral nucleotides, among which TLR3 recognizes dsRNA, TLR7 and TLR8 recognize ssRNA. The expression of TLR7 was significantly upregulated in PEDV-infected IPEC-J2 cells (Wang et al., 2020a). Cao et al. (2015) reported that PEDV infection induces nuclear factor-κB (NF-κB) activation through the TLR2, TLR3, and TLR9 pathways in porcine intestinal epithelial cells. PDCoV infection also significantly upregulates the mRNA transcription level of TLR3 and IFN-α in vivo (Xu et al., 2019b). In addition, TLR7 recognizes SARS-CoV, MERS-CoV, and MHV and induces IFN-a production in plasmacytoid dendritic cells (Cervantes-Barragan et al., 2007; Scheuplein et al., 2015). The receptors that sit on the surface of certain cells, especially TLR4, recognize MHV, SARS-CoV, SARS-CoV-2, and respiratory syncytial virus (Kurt-Jones et al., 2000; Khanolkar et al., 2009; Choudhury and Mukherjee, 2020). It has been reported that TLR4 also participates in PEDV infection-related pathogenesis (Huan et al., 2017). Once TLRs recognize the PAMPs of the virus, the factors NF-κB signaling will be activated to stimulate the production of pro-inflammatory cytokines and type I IFNs.
Escape From Innate Immunity by Porcine Enteric Coronaviruses Structural Proteins
The structural proteins of porcine enteric coronaviruses consist of spike protein, envelope protein, membrane protein, and nucleocapsid protein. These proteins are essential components of viral structure and play important roles in fighting against the host’s immune responses (Figure 2). It is generally believed that the S protein of coronaviruses mainly plays a key role in the invasion of host cells and the induction of neutralizing antibodies (Song and Park, 2012; Li, 2015; Guan et al., 2020). However, a study has found that compared with other structural proteins and NSPs of PEDV, S protein has the strongest ability to induce apoptosis. Similarly, the S protein of TGEV can also strongly induce Vero-E6 cells apoptosis (Chen et al., 2018). Studies have suggested that some viruses actively induce apoptosis to promote the release of virus progeny and spread to neighboring cells for further invasion (Favreau et al., 2012; Lan et al., 2013). Thus, S protein probably helps these porcine enteric coronaviruses evade the host immune response by regulating apoptosis, although the exact mechanism is unclear. Moreover, another recent research has found that PEDV S protein directly interacts with epidermal growth factor receptor (EGFR) and activates EGFR downstream signal transduction, inhibiting IFN and exacerbating viral infection (Yang et al., 2018). Still, more evidence is needed to explore whether porcine enteric coronaviruses S protein is directly involved in viral immune escape or S protein mediates NSPs to realize virus immune escape.
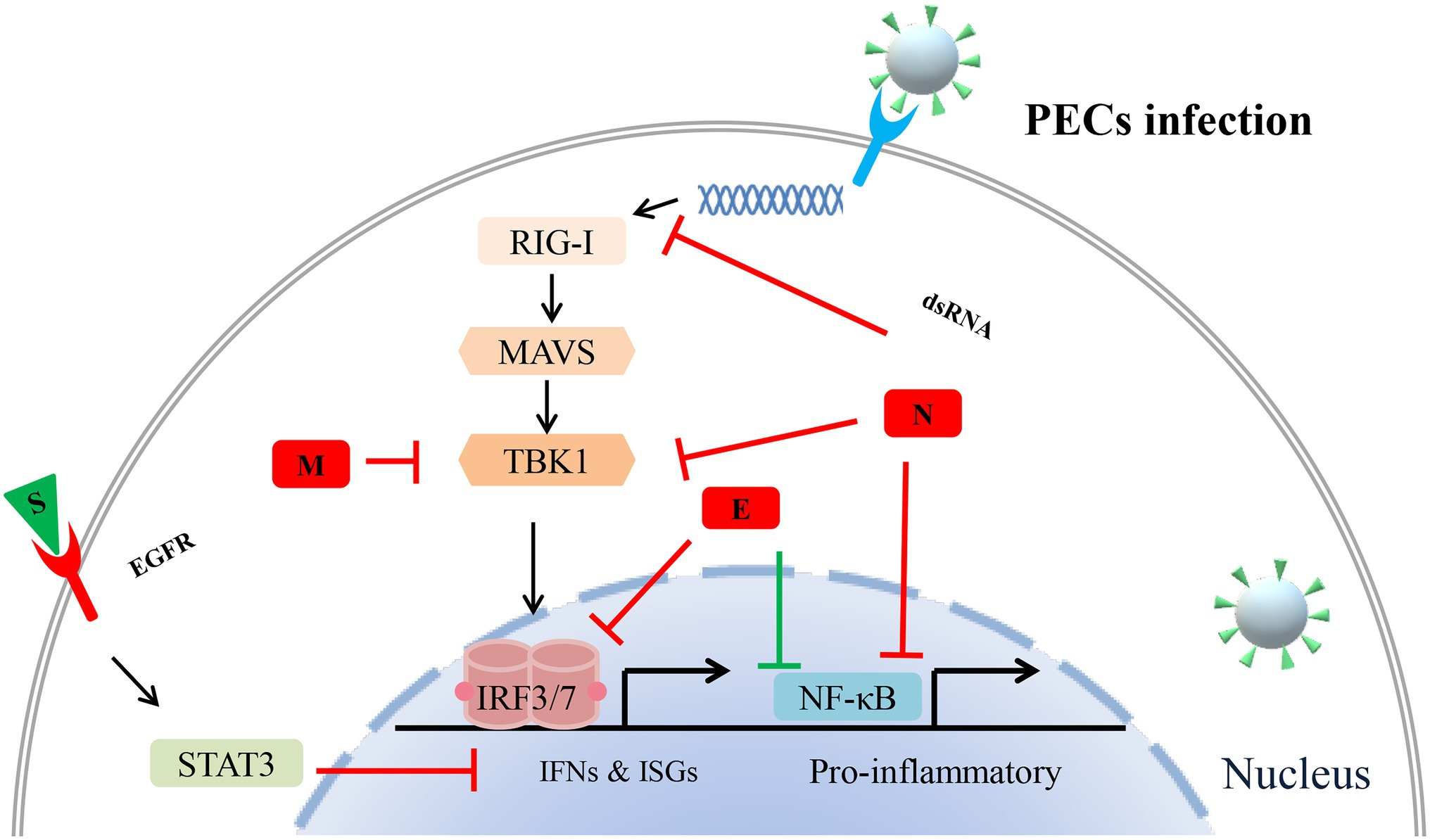
Figure 2. Potential mechanisms of porcine enteric coronavirus (PEC) structural proteins antagonize innate antiviral immune response. Different structural proteins of different porcine enteric coronaviruses used different strategies to antagonize the host’s immune responses. During PEC infection, interferons (IFNs) and pro-inflammatory are activated to fight against invading virus. It is noteworthy that STAT, retinoic acid-inducible gene I (RIG-I), and nuclear factor-κB (NF-κB) signalings are involved in this structural proteins-induced immune evasion.  stands for negative regulation,
stands for negative regulation,  stands for positive regulation.
stands for positive regulation.
The nucleocapsid protein is the most abundant protein of the known porcine enteric coronavirus components. It performs various functions, including viral genome transcription, translation, viral replication, and virus assembly (McBride et al., 2014). The PEDV N protein suppresses NF-κB nuclear translocation and further antagonizes Type II interferon production (Shan et al., 2018). Furthermore, the PEDV N protein targets TBK1 by direct interaction to inhibit IRF3 activation, further antagonizing type I interferon production. PDCoV N protein inhibits the activation of porcine IFN-β promoter by competing with dsRNA for porcine RIG-I binding (Chen et al., 2019). Moreover, the N-terminal region (1-246aa) of PDCoV N protein is the key part of interacting with porcine RIG-I (Likai et al., 2019). In addition, the N protein of PDCoV and SADS-CoV mediates K63-linked ubiquitination of porcine RIG-I, thereby, inhibit the host IFN-β production (Likai et al., 2019; Liu et al., 2021). The expression of the TGEV N gene promotes the accumulation of p53 and p21 and suppresses the expression of cyclin B1, cdc2, and cdk2. Meanwhile, TGEV N protein induces Bax mitochondria translocation and results in the activation of caspase-3, leading to apoptosis (Eleouet et al., 2000; Ding et al., 2014). In contrast, another study found that TGEV N protein, located in mitochondria, may contribute to mitophagy and suppress oxidative stress and apoptosis (Zhu et al., 2016). These different results may be due to their use of different cell lines for infection. These studies also suggested that the N proteins of different porcine enteric coronaviruses used different strategies to antagonize the host’s immune responses.
The member protein of porcine coronaviruses is a kind of transmembrane glycoprotein, which plays an important role in virion assembly, budding, and host immune regulation (Nguyen and Hogue, 1997; Riffault et al., 1997). Though the PEDV M protein affects cell cycle and interleukin 8 expressions, it does not induce ER stress and activation of NF-κB (Xu et al., 2015). Moreover, the PEDV M protein can form a complex with heat shock protein 70, affecting the host’s innate immune response and virus replication (Park et al., 2021). A recent study has identified 218 host cell proteins directly interacting with PDEV M protein. Moreover, these proteins were mainly associated with multiple biological processes such as immune response, apoptosis, and cell cycle (Wang et al., 2020b). In addition, some researches have reported that M proteins of TGEV and PDCoV help virus replication, which may be related to its regulation of IFN expression (Riffault et al., 1997; Gu et al., 2019; Li et al., 2019). So far, there are few studies on M proteins of porcine enteric coronavirus. M protein mediating porcine enteric coronavirus antagonism against host innate immune responses remains to be further studied.
The envelope protein is the smallest structural protein in porcine enteric coronavirus, involved in virus-host interactions. When the viruses invade the host cell, the E proteins are mainly located in the ER and play an essential role in virion assembly and budding (Xu et al., 2013a; Mora-Diaz et al., 2019). Based on its specific location in the cell, a study has found that PEDV E protein-induced ER stress and NF-κB activation upregulate the expression of IL-8 and Bcl-2. On the other hand, PEDV E protein directly interacts with IRF3 to inhibit its nuclear translocation, which further antagonizes interferon-β production (Zheng et al., 2021). The results of protein structure analysis suggest that porcine enteric coronavirus E protein may be involved in inducing humoral and cellular immunity during viral infection (Escors et al., 2001).
Research Progress on Accessory Proteins of Porcine Enteric Coronaviruses Antagonizing Antiviral Innate Immune Responses
The porcine coronavirus accessory proteins are unique kinds of protein with special functions. Different numbers of accessory proteins are scattered in different porcine coronavirus genomes. Although they are unnecessary for virus proliferation, they play key roles in regulating innate immunity and viral pathogenicity (Fang et al., 2018; Wu et al., 2020b). PEDV has only one accessory protein, the ORF3. A study showed that ORF3 could suppress IFN-β and IRF3 promoter activities, but a detailed analysis of the certain mechanism is lacking (Zhang et al., 2016). Several studies have reported that PDEV ORF3 can interact with the host’s immune cells. PDEV ORF3 antagonizes the host’s antiviral innate immunity mainly by regulating NF-κB signaling pathway activity. PEDV ORF3 inhibits phosphorylation of IκBα and nuclear factor p65 and interfering p65 nuclear translocation, which in turn reduces the production of pro-inflammatory cytokines such as IL-6 and IL-8 (Wu et al., 2020b). Interestingly, ORF3 directly interacts with the IκB kinase β and upregulates the IκB kinase β-meditated NF-κB promoter activity. However, PEDV ORF3 suppresses the IκB kinase β-meditated IFN-β production (Kaewborisuth et al., 2020). Moreover, PEDV ORF3 induces ER stress via the PERK-eIF2α signaling pathway by upregulating the expression of GRP78, and then inducing autophagy, which benefits viral replication and affects the production of various inflammatory cytokines (Guo et al., 2017; Zou et al., 2019). In addition, conflicting studies have shown that proteins can inhibit or promote apoptosis, which is involved in viral replication and immune escape (Favreau et al., 2012; Si et al., 2020). Cells infected with Ns7-deletion mutant TGEV (TGEV-Δ7) showed an increased cytopathic effect by activation of caspase signaling. Further research found that the C-terminus of accessory protein 7 bound to protein phosphatase 1 catalytic subunit and regulated dephosphorylation of eukaryotic translation initiation factor 2 to counteract the host’s cell defenses (Cruz et al., 2011). Moreover, innate immunity genes such as IL-15, C–C motif chemokine 2/4/5, C–X–C motif chemokine 9/11, tumor necrosis factor, and IFN-β were upregulated during TGEV-Δ7 infection. In vitro and in vivo results suggested that the absence of TGEV accessory protein 7 increased innate immunity responses and acute tissue damage, which proved its antagonistic function from the opposite angle (Cruz et al., 2013). PDCoV accessory protein NS6 cannot prevent RIG-I, MDA5, and their downstream molecules from activating the IFN-β promoter. However, PDCoV NS6 can directly interact with the carboxyl terminus domain of RIG-I and the helicase and carboxyl terminus domains of MDA5 to inhibit dsRNA binding RIG-I/MDA5 and thus antagonize IFN-β production (Fang et al., 2018). PDCoV NS7a can also function as an IFN antagonist. Unlike NS6a, the NS7a inhibits RIG-I, MDA5, and their downstream molecules to activate the IFN-β promoter. Furthermore, NS7a can compete with TRAF3 and IRF3 for binding to IKK, thereby, reducing RLR-mediated IFN-β production. Moreover, the kinase and the scaffold dimerization domains of IKKε are key regions that can directly bond to NS7a (Fang et al., 2020). From those researches, the mechanisms by which different accessory proteins of porcine enteric coronaviruses suppress host antiviral innate immunity are different.
Mechanism of Non-structural Proteins of Porcine Enteric Coronaviruses Antagonizing Innate Immune Response
Non-structural proteins are the earliest expression proteins essential for the virus replication process. They usually act as viruses evade, circumvent, or subvert the host innate immune system roles. During the process of porcine enteric coronavirus infection, NSP1, NSP3, NSP5, NSP15, and NSP16 have been observed to play additional roles in host immune-modulatory functions. Of 16 PEDV NSPs, NSP1, NSP3, NSP7, NSP14, NSP15, and NSP16 were found to inhibit the IFN-β and IRF3 promoter activities (Zhang et al., 2016). In addition, these porcine coronaviruses NSPs are also involved in downregulating the NF-κB activity (Zhang et al., 2017). In 2018, NSP1, NSP3, NSP5, NSP8, NSP14, NSP15, and NSP16 of PEDV were found to suppress type III IFN activities (Zhang et al., 2018). Here, we review the detailed mechanism of these non-structural proteins that antagonize interferon production.
Non-structural protein 1 is only characterized in alphacoronaviruses (α-CoVs) and betacoronaviruses (β-CoVs; Woo et al., 2010). Under the catalysis of the proteasome, the NSP1 of PEDV interrupted the enhanceosome assembly of IRF3 and CREB-binding protein (CBP) by degrading CBP to antagonize IFN-I production (Zhang et al., 2016). The CBP is the key molecular for the activated IRF3 to induce the transcription of IFN-I genes. After IRF3 phosphorylation and dimerization into the nucleus, IRF3 interacts with CBP to form the IRF3-CBP complex. And then, the complex binds to the positive regulatory domain (PRD) I–IV regions of the IFN-β promoter to assemble the enhanceosome with NF-κB and other factors to turn on the transcription of IFN-I genes (Honda and Taniguchi, 2006; Dragan et al., 2007; Panne et al., 2007). Another target gene for NSP1 to inhibit innate immunity is IRF1. IRF1 is the key adaptor protein for type III IFNs production. PEDV NSP1 blocked the nuclear translocation of IRF1 and reduced the number of peroxisomes to suppress IRF1-induced type III IFNs (Zhang et al., 2018). PEDV inhibited both NF-κB and pro-inflammatory cytokines production in porcine epithelial cells. Zhang et al. (2017) found that NSP1 was the most effective NF-κB antagonist among all proteins of PEDV. Moreover, NSP1 suppressed the phosphorylation and degradation of IκBα and blocked the p65 activation (Zhang et al., 2017). It is worth pointing out that the conserved residues of NSP1 were crucial to suppress IRF1-mediated IFN-λ and NF-κB mediated IFN-I and pro-inflammatory cytokines (Zhang et al., 2017, 2018). In addition, Shen et al. (2020) found that seven representative α-CoVs: SADS-CoV, PEDV, HCoV-229E, human NL63 CoV (HCoV-NL63), FIPV, TGEV, and PRCV NSP1s could significantly inhibit the phosphorylation of STAT1-S727 and interfere with the effect of IFN-I. The multiple functions of NSP1 to inhibit innate immune responses through different mechanisms suggest that it is one of the key molecules of porcine coronaviruses to escape innate immunity. Drugs targeting NSP1 conserved sites are likely to prevent and control these viruses.
NSP3 is the largest protein encoded by the porcine coronavirus genome and contains two domains of papain-like protease (PLP1 and PLP2). PLP2 has deubiquitinase (DUB) activity that recognizes and processes K-48 and K-63 linked polyubiquitin chains. Ubiquitin modification is a key mechanism to regulate the activity and stability of the antiviral innate immune. In recent years, several viral DUBs have been found to antagonize IFN-I production by deubiquitination of key host factors, such as the lead protease (Lbpro) of the foot-and-mouth disease virus (FMDV) and the NSP2 of Porcine Reproductive and Respiratory syndrome virus (PRRSV; Sun et al., 2010; Wang et al., 2011). Moreover, the DUB activity is conserved in all members of the arterivirus family. Both arteri- and nairovirus DUBs inhibit RIG-I mediated innate immune signaling (van Kasteren et al., 2012). The PLP2 of HCoV-NL63 and PLPs of SARS-CoV also antagonize IFN induction through disruption of STING dimer and deubiquitination of RIG-I (Chen et al., 2007; Clementz et al., 2010; Sun et al., 2012a). Accordingly, PEDV PLP2 strongly inhibits RIG-I- and STING-activated IFN expression by deubiquitination and co-immunoprecipitating with RIG-I and STING (Xing et al., 2013).
CoVs NSP5 and NSP3 genes encode 3C-like protease (3CLpro) and papain-like proteinase, respectively. These two proteinases can degrade the polyprotein into various non-structural proteins, which further facilitate virus replication. It has been found that many viruses’ 3C protease (3Cpro) antagonizes innate immune signaling pathways dependent on its protease activity. For example, encephalomyocarditis virus (EMCV) 3C protease cleaved TANK and disrupted the TANK-TBK1-IKKε-IRF3 complex, inhibiting IRF3 phosphorylation and IFN-I production (Huang et al., 2017). Coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate IFN-I and apoptotic signaling (Mukherjee et al., 2011). Enterovirus 71 3C protein induces TRIF cleavage to inhibit TLR-mediated antiviral responses (Lei et al., 2011). Similar to the Hepatitis A virus and FMDV 3Cpro, PEDV and PDCoV 3C-like proteases cleave NEMO to impair induction of IFN-β (Wang et al., 2012, 2014a, 2016). The cleave site of NEMO has been identified at Gln231 both in PEDV and PDCoV, suggesting NEMO may be a common target for coronaviruses (Wang et al., 2016; Zhu et al., 2017a). However, it cannot exclude the possibility that other non-active site residues of their NSP5 are also involved. Soon afterward, other target molecules of PDCoV NSP5 inhibit IFN-I signaling was revealed. Like NS5 protein of dengue virus (DENV), Zika virus (ZIKV) and the hepatitis C virus (HCV), PDCoV NSP5 target the JAK–STAT pathway to antagonize IFN-I signaling (Lin et al., 2006; Ashour et al., 2009; Grant et al., 2016). In PDCoV-infected cells, NSP5 cleaved STAT2 at glutamine 685 (Q685) and Q758As to impair ISGs induction (Zhu et al., 2017b). As NSP5 is involved in the cleavage of the viral polyprotein, the inhibitors target its 3C-like protease domain that can suppress porcine enteric coronavirus infection, such as quercetin, GC376 (Zhou et al., 2019; Ye et al., 2020).
NSP15 is identified as a component of the coronavirus replication complex, which has endoribonuclease (EndoU) activity. The role of EndoU was revealed, which showed that EndoU mediates the evasion of viral double-stranded RNA recognition by host sensors in macrophages. In previous studies, SARS-CoV NSP15 was identified as an inhibitor of MAVS-mediated apoptotic responses (Lei et al., 2009). MHV and HCoV-229E NSP15 efficiently prevent simultaneous activation of host cells dsRNA sensors, such as MDA5, OAS, and PKR (Kindler et al., 2017). A study has reported that the EndoU activity of PEDV NSP15 is not required for virus replication. Still, PEDV NSP15 is important for suppressing the type I and type III IFN response in epithelial cells and macrophages. NSP15 facilitates virus replication, shedding, and pathogenesis in vivo (Deng et al., 2019). With the study forward, the mechanism of PEDV NSP15 inhibits the host’s IFN response was found. PEDV NSP15 can directly degrade the mRNA of TBK1 and IRF3 dependent on its EndoU activity to suppress the production of IFN and ISGs, antagonizing the host innate response to facilitate its replication (Wu et al., 2020a). PDCoV NSP15 is also an IFN antagonist. However, PDCoV NSP15 disrupts the phosphorylation and nuclear translocation of the NF-κB p65 subunit but does not antagonize the activation of transcription factor IRF3. Moreover, PDCoV NSP15 inhibits IFN-β production independent of EndoU activity (Liu et al., 2019).
NSP16 is one of the RNA modification enzymes involved in forming cap structures in PEDV (Chen et al., 2011). Compared with NSP14, which is another methyltransferase in PEDV, NSP16 is a more efficient regulator in the antagonist of innate immunity. Mechanistically, NSP16 downregulates the activities of RIG-I and MDA5 mediated IFN-β and ISRE dependent on the KDKE tetrad. Moreover, NSP10 enhanced the inhibitory effect of NSP16 on IFN-β (Shi et al., 2019). However, whether NSP16 of PDCoV and SADS-CoV antagonizes interferon production is still unknown.
These studies suggest that the NSPs of porcine enteric coronaviruses antagonize the host’s innate immune responses by regulating IFN signaling pathways (Figure 3). Therefore, further study of the biological functions of NSPs will help us elucidate the pathogenesis of coronaviruses and possibly provide new targets for developing antiviral vaccines and drugs.
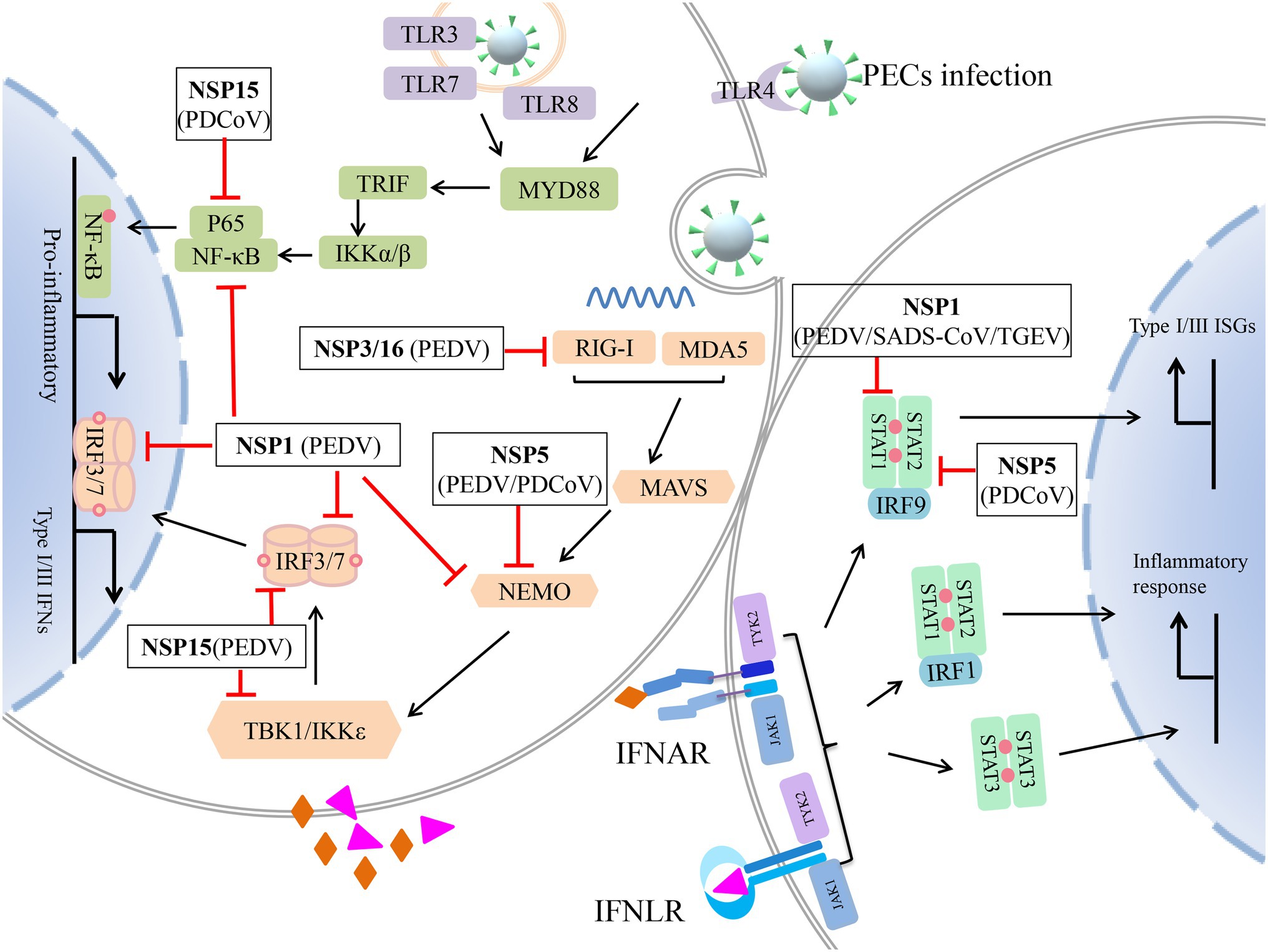
Figure 3. Non-structural protein (NSP) of porcine enteric coronavirus (PEC) antagonizes innate immune response. Retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and Toll-like receptors (TLRs) recognize the invading virus and induce pro-inflammatory cytokines, type I/III interferons (IFNs) by nuclear factor-κB (NF-κB) and (RIG-I)-like receptors (RLRs) signaling pathway, respectively. Extracellular Type I and III IFNs recognized by IFN-I receptor (IFNAR) and type III IFN receptor (IFNLR) to phosphorylate JAK1 and TYK2. And then, STAT1/2 is recruited and phosphorylated to form three STAT complexes. STAT1 and STAT2 form heterodimers and recruit IRF9 or IRF1. These complexes enter the nucleus and induce type I and III ISGs, inflammatory cytokines production. NSPs of PEC antagonize various steps of this antiviral response.  stands for negative regulation;
stands for negative regulation;  stands for type I IFN; and
stands for type I IFN; and  stands for type III IFN.
stands for type III IFN.
Conclusion and Perspectives
The host’s innate immune response protects itself from most pathogenic microorganisms, but some viruses have evolved strategies to antagonize innate immune responses. Coronaviruses are the largest positive-sense RNA viruses that exist widely in nature and are highly genetically variable. This review summarizes how porcine enteric coronaviruses evade the host’s innate immune responses. First, since IFN is the most important regulator of the antiviral innate immunity, these viruses typically inhibit IFN production by various means, including inhibition of RIG-I/TLR signaling and inhibiting dsRNA bind to RIG-I/MDA5 or directly downregulates IFN promoter activity. Second, porcine enteric coronaviruses also attenuate inflammatory response by targeting the NF-κB signaling pathway. In addition, some porcine enteric coronaviruses can regulate apoptosis and evade ISGs to promote virus replication.
Also, some cellular physiological processes, such as autophagy, endoplasmic reticulum stress, programmed cell death, are probably involved in the evasion of the innate immune response of these viruses (Lin et al., 2020; Sun et al., 2021; Wei et al., 2021). Some viruses use autophagy to evade the host immune response and hide in the autophagosome to accumulate RNA and proteins (Sun et al., 2012b; Liu et al., 2016). A recent study has reported that PEDV infection induced autophagy, which promotes virus replication. Moreover, NSP6 and ORF3 of the virus are two of the important inducers of autophagy. Further study showed that PI3K/Akt/mTOR pathway is the key signal of PEDV NSP6-induced autophagy (Lin et al., 2020). During porcine coronavirus infection, some of the viral proteins are located in the endoplasmic reticulum of host cells. E protein, N protein, and ORF3 of PEDV can all induce ER stress via PERK and IRE1 signaling and then upregulate inflammatory factors (Xu et al., 2013a,b; Sun et al., 2021). In addition, PEDV-induced ER stress facilitates autophagy (Zou et al., 2019). Furthermore, TGEV infection in porcine intestinal epithelial cells can induce IL-1β release and pyroptosis, dependent on the expression and assembly of the NOD-like receptor protein 3. The above evidence highlights the importance of investigating virus-host interactions to elucidate viral immune evasion. Some viruses would like to alter the intracellular environment to ensure their survival. Exploring these programs will help us further understand how porcine enteric coronaviruses evade innate immune responses and also provide us with new ideas for developing antiviral vaccines and drugs.
Author Contributions
KZ, SL, SD, JL, JZ, and SW conceived and wrote the manuscript. KZ and SW prepared the figure. All authors approved the final version of this review and agreed to be accountable for the content of the work.
Funding
The authors thank the following funding sources: Independent Research and Development Projects of Maoming Laboratory (2021ZZ003), the Special Fund for Scientific Innovation Strategy-Construction of High-Level Academy of Agriculture Science (R2021PY–QF006, R2019YJ–YB2005, R2019YJ–YB2004, and R2019YJ–YB3002), and the Science and Technology Planning Project of Guangzhou (202103000096).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ajayi, T., Dara, R., Misener, M., Pasma, T., Moser, L., and Poljak, Z. (2018). Herd-level prevalence and incidence of porcine epidemic diarrhoea virus (PEDV) and porcine deltacoronavirus (PDCoV) in swine herds in Ontario, Canada. Transbound. Emerg. Dis. 65, 1197–1207. doi: 10.1111/tbed.12858
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Ashour, J., Laurent-Rolle, M., Shi, P. Y., and Garcia-Sastre, A. (2009). NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83, 5408–5418. doi: 10.1128/JVI.02188-08
Boley, P. A., Alhamo, M. A., Lossie, G., Yadav, K. K., Vasquez-Lee, M., Saif, L. J., et al. (2020). Porcine deltacoronavirus infection and transmission in poultry, United States(1). Emerg. Infect. Dis. 26, 255–265. doi: 10.3201/eid2602.190346
Brian, D. A., and Baric, R. S. (2005). Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287, 1–30. doi: 10.1007/3-540-26765-4_1
Cao, L., Ge, X., Gao, Y., Ren, Y., Ren, X., and Li, G. (2015). Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 96, 1757–1767. doi: 10.1099/vir.0.000133
Cervantes-Barragan, L., Zust, R., Weber, F., Spiegel, M., Lang, K. S., Akira, S., et al. (2007). Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109, 1131–1137. doi: 10.1182/blood-2006-05-023770
Chen, J., Fang, P., Wang, M., Peng, Q., Ren, J., Wang, D., et al. (2019). Porcine deltacoronavirus nucleocapsid protein antagonizes IFN-beta production by impairing dsRNA and PACT binding to RIG-I. Virus Genes 55, 520–531. doi: 10.1007/s11262-019-01673-z
Chen, Y., Su, C., Ke, M., Jin, X., Xu, L., Zhang, Z., et al. (2011). Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2’-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 7:e1002294. doi: 10.1371/journal.ppat.1002294
Chen, Z., Wang, Y., Ratia, K., Mesecar, A. D., Wilkinson, K. D., and Baker, S. C. (2007). Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81, 6007–6018. doi: 10.1128/JVI.02747-06
Chen, Y., Zhang, Z., Li, J., Gao, Y., Zhou, L., Ge, X., et al. (2018). Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol. J. 15:170. doi: 10.1186/s12985-018-1078-4
Chen, F., Zhu, Y., Wu, M., Ku, X., Yao, L., and He, Q. (2015). Full-length genome characterization of chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome Announc. 3, e01284–e01315. doi: 10.1128/genomeA.01284-15
Choi, S., and Lee, C. (2019). Functional characterization and proteomic analysis of porcine deltacoronavirus accessory protein NS7. J. Microbiol. Biotechnol. 29, 1817–1829. doi: 10.4014/jmb.1908.08013
Choudhury, A., and Mukherjee, S. (2020). In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 92, 2105–2113. doi: 10.1002/jmv.25987
Clementz, M. A., Chen, Z., Banach, B. S., Wang, Y., Sun, L., Ratia, K., et al. (2010). Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84, 4619–4629. doi: 10.1128/JVI.02406-09
Cruz, J. L., Becares, M., Sola, I., Oliveros, J. C., Enjuanes, L., and Zuniga, S. (2013). Alphacoronavirus protein 7 modulates host innate immune response. J. Virol. 87, 9754–9767. doi: 10.1128/JVI.01032-13
Cruz, J. L., Sola, I., Becares, M., Alberca, B., Plana, J., Enjuanes, L., et al. (2011). Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 7:e1002090. doi: 10.1371/journal.ppat.1002090
Deng, X., van Geelen, A., Buckley, A. C., O’Brien, A., Pillatzki, A., Lager, K. M., et al. (2019). Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 93, e02000–e02018. doi: 10.1128/JVI.02000-18
Ding, L., Huang, Y., Du, Q., Dong, F., Zhao, X., Zhang, W., et al. (2014). TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem. Biophys. Res. Commun. 445, 497–503. doi: 10.1016/j.bbrc.2014.02.039
Doyle, L. P., and Hutchings, L. M. (1946). A transmissible gastroenteritis in pigs. J. Am. Vet. Med. Assoc. 108, 257–259.
Dragan, A. I., Hargreaves, V. V., Makeyeva, E. N., and Privalov, P. L. (2007). Mechanisms of activation of interferon regulator factor 3: the role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucleic Acids Res. 35, 3525–3534. doi: 10.1093/nar/gkm142
Drosten, C., Gunther, S., Preiser, W., van der Werf, S., Brodt, H. R., Becker, S., et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976. doi: 10.1056/NEJMoa030747
Eleouet, J. F., Slee, E. A., Saurini, F., Castagne, N., Poncet, D., Garrido, C., et al. (2000). The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol. 74, 3975–3983. doi: 10.1128/JVI.74.9.3975-3983.2000
Escors, D., Ortego, J., Laude, H., and Enjuanes, L. (2001). The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75, 1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001
Fang, P., Fang, L., Hong, Y., Liu, X., Dong, N., Ma, P., et al. (2017). Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 98, 173–178. doi: 10.1099/jgv.0.000690
Fang, P., Fang, L., Liu, X., Hong, Y., Wang, Y., Dong, N., et al. (2016). Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology 499, 170–177. doi: 10.1016/j.virol.2016.09.015
Fang, P., Fang, L., Ren, J., Hong, Y., Liu, X., Zhao, Y., et al. (2018). Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 92, e00712–e00718. doi: 10.1128/JVI.00712-18
Fang, P., Fang, L., Xia, S., Ren, J., Zhang, J., Bai, D., et al. (2020). Porcine deltacoronavirus accessory protein NS7a antagonizes IFN-beta production by competing with TRAF3 and IRF3 for binding to IKKepsilon. Front. Cell. Infect. Microbiol. 10:257. doi: 10.3389/fcimb.2020.00257
Favreau, D. J., Meessen-Pinard, M., Desforges, M., and Talbot, P. J. (2012). Human coronavirus-induced neuronal programmed cell death is cyclophilin d dependent and potentially caspase dispensable. J. Virol. 86, 81–93. doi: 10.1128/JVI.06062-11
Gack, M. U., Shin, Y. C., Joo, C. H., Urano, T., Liang, C., Sun, L., et al. (2007). TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920. doi: 10.1038/nature05732
Garwes, D. J. (1988). Transmissible gastroenteritis. Vet. Rec. 122, 462–463. doi: 10.1136/vr.122.19.462
Grant, A., Ponia, S. S., Tripathi, S., Balasubramaniam, V., Miorin, L., Sourisseau, M., et al. (2016). Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19, 882–890. doi: 10.1016/j.chom.2016.05.009
Gu, W. Y., Li, Y., Liu, B. J., Wang, J., Yuan, G. F., Chen, S. J., et al. (2019). Short hairpin RNAs targeting M and N genes reduce replication of porcine deltacoronavirus in ST cells. Virus Genes 55, 795–801. doi: 10.1007/s11262-019-01701-y
Guan, H., Wang, Y., Perculija, V., Saeed, A., Liu, Y., Li, J., et al. (2020). Cryo-electron microscopy structure of the swine acute diarrhea syndrome coronavirus spike glycoprotein provides insights into evolution of unique coronavirus spike proteins. J. Virol. 94, e01301–e01320. doi: 10.1128/JVI.01301-20
Guo, X., Zhang, M., Zhang, X., Tan, X., Guo, H., Zeng, W., et al. (2017). Porcine epidemic diarrhea virus induces autophagy to benefit its replication. Viruses 9:53. doi: 10.3390/v9030053
Honda, K., and Taniguchi, T. (2006). IRFs: master regulators of signalling by toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6, 644–658. doi: 10.1038/nri1900
Huan, C. C., Wang, H. X., Sheng, X. X., Wang, R., Wang, X., and Mao, X. (2017). Glycyrrhizin inhibits porcine epidemic diarrhea virus infection and attenuates the proinflammatory responses by inhibition of high mobility group box-1 protein. Arch. Virol. 162, 1467–1476. doi: 10.1007/s00705-017-3259-7
Huang, Y. W., Dickerman, A. W., Pineyro, P., Li, L., Fang, L., Kiehne, R., et al. (2013). Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4, e00737–e00713. doi: 10.1128/mBio.00737-13
Huang, L., Xiong, T., Yu, H., Zhang, Q., Zhang, K., Li, C., et al. (2017). Encephalomyocarditis virus 3C protease attenuates type I interferon production through disrupting the TANK-TBK1-IKKepsilon-IRF3 complex. Biochem. J. 474, 2051–2065. doi: 10.1042/BCJ20161037
Jung, K., Annamalai, T., Lu, Z., and Saif, L. J. (2015). Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 178, 31–40. doi: 10.1016/j.vetmic.2015.04.022
Jung, K., Hu, H., and Saif, L. J. (2017). Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 162, 2357–2362. doi: 10.1007/s00705-017-3351-z
Jung, K., Wang, Q., Scheuer, K. A., Lu, Z., Zhang, Y., and Saif, L. J. (2014). Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 20, 662–665. doi: 10.3201/eid2004.131685
Kadoi, K., Sugioka, H., Satoh, T., and Kadoi, B. K. (2002). The propagation of a porcine epidemic diarrhea virus in swine cell lines. New Microbiol. 25, 285–290.
Kaewborisuth, C., Koonpaew, S., Srisutthisamphan, K., Viriyakitkosol, R., Jaru-Ampornpan, P., and Jongkaewwattana, A. (2020). PEDV ORF3 independently regulates IkappaB kinase beta-mediated NF-kappaB and IFN-beta promoter activities. Pathogens 9:376. doi: 10.3390/pathogens9050376
Khanolkar, A., Hartwig, S. M., Haag, B. A., Meyerholz, D. K., Harty, J. T., and Varga, S. M. (2009). Toll-like receptor 4 deficiency increases disease and mortality after mouse hepatitis virus type 1 infection of susceptible C3H mice. J. Virol. 83, 8946–8956. doi: 10.1128/JVI.01857-08
Kim, L., Chang, K. O., Sestak, K., Parwani, A., and Saif, L. J. (2000). Development of a reverse transcription-nested polymerase chain reaction assay for differential diagnosis of transmissible gastroenteritis virus and porcine respiratory coronavirus from feces and nasal swabs of infected pigs. J. Vet. Diagn. Invest. 12, 385–388. doi: 10.1177/104063870001200418
Kindler, E., Gil-Cruz, C., Spanier, J., Li, Y., Wilhelm, J., Rabouw, H. H., et al. (2017). Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 13:e1006195. doi: 10.1371/journal.ppat.1006195
Kocherhans, R., Bridgen, A., Ackermann, M., and Tobler, K. (2001). Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 23, 137–144. doi: 10.1023/A:1011831902219
Kotenko, S. V., Rivera, A., Parker, D., and Durbin, J. E. (2019). Type III IFNs: beyond antiviral protection. Semin. Immunol. 43:101303. doi: 10.1016/j.smim.2019.101303
Kurt-Jones, E. A., Popova, L., Kwinn, L., Haynes, L. M., Jones, L. P., Tripp, R. A., et al. (2000). Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1, 398–401. doi: 10.1038/80833
Lan, Y., Zhao, K., Wang, G., Dong, B., Zhao, J., Tang, B., et al. (2013). Porcine hemagglutinating encephalomyelitis virus induces apoptosis in a porcine kidney cell line via caspase-dependent pathways. Virus Res. 176, 292–297. doi: 10.1016/j.virusres.2013.05.019
Lazear, H. M., Schoggins, J. W., and Diamond, M. S. (2019). Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923. doi: 10.1016/j.immuni.2019.03.025
Lednicky, J. A., Tagliamonte, M. S., White, S. K., Elbadry, M. A., Alam, M. M., Stephenson, C. J., et al. (2021). Independent infections of porcine deltacoronavirus among Haitian children. Nature 600, 133–137. doi: 10.1038/s41586-021-04111-z
Lee, S., and Lee, C. (2015). Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 208, 136–145. doi: 10.1016/j.virusres.2015.06.013
Lei, Y., Moore, C. B., Liesman, R. M., O’Connor, B. P., Bergstralh, D. T., Chen, Z. J., et al. (2009). MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One 4:e5466. doi: 10.1371/journal.pone.0005466
Lei, X., Sun, Z., Liu, X., Jin, Q., He, B., and Wang, J. (2011). Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by toll-like receptor 3. J. Virol. 85, 8811–8818. doi: 10.1128/JVI.00447-11
Li, F. (2015). Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 89, 1954–1964. doi: 10.1128/JVI.02615-14
Li, K., Li, H., Bi, Z., Song, D., Zhang, F., Lei, D., et al. (2019). Significant inhibition of re-emerged and emerging swine enteric coronavirus in vitro using the multiple shRNA expression vector. Antivir. Res. 166, 11–18. doi: 10.1016/j.antiviral.2019.03.010
Li, Z., Ma, Z., Li, Y., Gao, S., and Xiao, S. (2020). Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb. Pathog. 149:104553. doi: 10.1016/j.micpath.2020.104553
Liang, Q., Zhang, H., Li, B., Ding, Q., Wang, Y., Gao, W., et al. (2019). Susceptibility of chickens to porcine deltacoronavirus infection. Viruses 11:573. doi: 10.3390/v11060573
Likai, J., Shasha, L., Wenxian, Z., Jingjiao, M., Jianhe, S., Hengan, W., et al. (2019). Porcine deltacoronavirus nucleocapsid protein suppressed IFN-beta production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitination. Front. Immunol. 10:1024. doi: 10.3389/fimmu.2019.01024
Lin, W., Kim, S. S., Yeung, E., Kamegaya, Y., Blackard, J. T., Kim, K. A., et al. (2006). Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J. Virol. 80, 9226–9235. doi: 10.1128/JVI.00459-06
Lin, H., Li, B., Liu, M., Zhou, H., He, K., and Fan, H. (2020). Nonstructural protein 6 of porcine epidemic diarrhea virus induces autophagy to promote viral replication via the PI3K/Akt/mTOR axis. Vet. Microbiol. 244:108684. doi: 10.1016/j.vetmic.2020.108684
Liu, J., and Cao, X. (2016). Cellular and molecular regulation of innate inflammatory responses. Cell. Mol. Immunol. 13, 711–721. doi: 10.1038/cmi.2016.58
Liu, X., Fang, P., Fang, L., Hong, Y., Zhu, X., Wang, D., et al. (2019). Porcine deltacoronavirus nsp15 antagonizes interferon-beta production independently of its endoribonuclease activity. Mol. Immunol. 114, 100–107. doi: 10.1016/j.molimm.2019.07.003
Liu, Y., Liang, Q. Z., Lu, W., Yang, Y. L., Chen, R., Huang, Y. W., et al. (2021). A comparative analysis of coronavirus nucleocapsid (N) proteins reveals the SADS-CoV N protein antagonizes IFN-beta production by inducing ubiquitination of RIG-I. Front. Immunol. 12:688758. doi: 10.3389/fimmu.2021.688758
Liu, C., Tang, J., Ma, Y., Liang, X., Yang, Y., Peng, G., et al. (2015). Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 89, 6121–6125. doi: 10.1128/JVI.00430-15
Liu, Q., and Wang, H. Y. (2021). Porcine enteric coronaviruses: an updated overview of the pathogenesis, prevalence, and diagnosis. Vet. Res. Commun. 45, 75–86. doi: 10.1007/s11259-021-09808-0
Liu, G., Zhong, M., Guo, C., Komatsu, M., Xu, J., Wang, Y., et al. (2016). Autophagy is involved in regulating influenza A virus RNA and protein synthesis associated with both modulation of Hsp90 induction and mTOR/p70S6K signaling pathway. Int. J. Biochem. Cell Biol. 72, 100–108. doi: 10.1016/j.biocel.2016.01.012
McBride, R., van Zyl, M., and Fielding, B. C. (2014). The coronavirus nucleocapsid is a multifunctional protein. Viruses 6, 2991–3018. doi: 10.3390/v6082991
Meylan, E., Tschopp, J., and Karin, M. (2006). Intracellular pattern recognition receptors in the host response. Nature 442, 39–44. doi: 10.1038/nature04946
Mora-Diaz, J. C., Pineyro, P. E., Houston, E., Zimmerman, J., and Gimenez-Lirola, L. G. (2019). Porcine hemagglutinating encephalomyelitis virus: a review. Front. Vet. Sci. 6:53. doi: 10.3389/fvets.2019.00053
Mukherjee, A., Morosky, S. A., Delorme-Axford, E., Dybdahl-Sissoko, N., Oberste, M. S., Wang, T., et al. (2011). The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 7:e1001311. doi: 10.1371/journal.ppat.1001311
Narayanan, K., Maeda, A., Maeda, J., and Makino, S. (2000). Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74, 8127–8134. doi: 10.1128/JVI.74.17.8127-8134.2000
Nguyen, V. P., and Hogue, B. G. (1997). Protein interactions during coronavirus assembly. J. Virol. 71, 9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997
Pan, Y., Tian, X., Qin, P., Wang, B., Zhao, P., Yang, Y. L., et al. (2017). Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 211, 15–21. doi: 10.1016/j.vetmic.2017.09.020
Panne, D., Maniatis, T., and Harrison, S. C. (2007). An atomic model of the interferon-beta enhanceosome. Cell 129, 1111–1123. doi: 10.1016/j.cell.2007.05.019
Park, J. E., Cruz, D. J., and Shin, H. J. (2011). Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 156, 1749–1756. doi: 10.1007/s00705-011-1044-6
Park, S. J., Moon, H. J., Luo, Y., Kim, H. K., Kim, E. M., Yang, J. S., et al. (2008). Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes 36, 95–104. doi: 10.1007/s11262-007-0164-2
Park, J. Y., Ryu, J., Park, J. E., Hong, E. J., and Shin, H. J. (2021). Heat shock protein 70 could enhance porcine epidemic diarrhoea virus replication by interacting with membrane proteins. Vet. Res. 52:138. doi: 10.1186/s13567-021-01006-9
Penzes, Z., Gonzalez, J. M., Calvo, E., Izeta, A., Smerdou, C., Mendez, A., et al. (2001). Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes 23, 105–118. doi: 10.1023/A:1011147832586
Riffault, S., Carrat, C., Besnardeau, L., La Bonnardiere, C., and Charley, B. (1997). In vivo induction of interferon-alpha in pig by non-infectious coronavirus: tissue localization and in situ phenotypic characterization of interferon-alpha-producing cells. J. Gen. Virol. 78, 2483–2487. doi: 10.1099/0022-1317-78-10-2483
Saif, L. J. (1999). Comparative pathogenesis of enteric viral infections of swine. Adv. Exp. Med. Biol. 473, 47–59. doi: 10.1007/978-1-4615-4143-1_4
Sawicki, S. G., Sawicki, D. L., Younker, D., Meyer, Y., Thiel, V., Stokes, H., et al. (2005). Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. doi: 10.1371/journal.ppat.0010039
Scheuplein, V. A., Seifried, J., Malczyk, A. H., Miller, L., Hocker, L., Vergara-Alert, J., et al. (2015). High secretion of interferons by human plasmacytoid dendritic cells upon recognition of middle east respiratory syndrome coronavirus. J. Virol. 89, 3859–3869. doi: 10.1128/JVI.03607-14
Schoggins, J. W., and Rice, C. M. (2011). Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525. doi: 10.1016/j.coviro.2011.10.008
Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682. doi: 10.1016/j.cell.2005.08.012
Shan, Y., Liu, Z. Q., Li, G. W., Chen, C., Luo, H., Liu, Y. J., et al. (2018). Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-lambda production by blocking the nuclear factor-kappaB nuclear translocation. J. Zhejiang Univ. Sci. B 19, 570–580. doi: 10.1631/jzus.B1700283
Shang, J., Zheng, Y., Yang, Y., Liu, C., Geng, Q., Tai, W., et al. (2018). Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J. Virol. 92, e01556–e01617. doi: 10.1128/JVI.01556-17
Shen, Z., Yang, Y., Yang, S., Zhang, G., Xiao, S., Fu, Z. F., et al. (2020). Structural and biological basis of alphacoronavirus nsp1 associated with host proliferation and immune evasion. Viruses 12:812. doi: 10.3390/v12080812
Shi, P., Su, Y., Li, R., Liang, Z., Dong, S., and Huang, J. (2019). PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 265, 57–66. doi: 10.1016/j.virusres.2019.03.005
Si, F., Hu, X., Wang, C., Chen, B., Wang, R., Dong, S., et al. (2020). Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses 12:214. doi: 10.3390/v12020214
Song, D., and Park, B. (2012). Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44, 167–175. doi: 10.1007/s11262-012-0713-1
Stevenson, G. W., Hoang, H., Schwartz, K. J., Burrough, E. R., Sun, D., Madson, D., et al. (2013). Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 25, 649–654. doi: 10.1177/1040638713501675
Sun, Z., Chen, Z., Lawson, S. R., and Fang, Y. (2010). The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 84, 7832–7846. doi: 10.1128/JVI.00217-10
Sun, M. X., Huang, L., Wang, R., Yu, Y. L., Li, C., Li, P. P., et al. (2012b). Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 8, 1434–1447. doi: 10.4161/auto.21159
Sun, P., Jin, J., Wang, L., Wang, J., Zhou, H., Zhang, Q., et al. (2021). Porcine epidemic diarrhea virus infections induce autophagy in vero cells via ROS-dependent endoplasmic reticulum stress through PERK and IRE1 pathways. Vet. Microbiol. 253:108959. doi: 10.1016/j.vetmic.2020.108959
Sun, L., Xing, Y., Chen, X., Zheng, Y., Yang, Y., Nichols, D. B., et al. (2012a). Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One 7:e30802. doi: 10.1371/journal.pone.0030802
Suzuki, T., Shibahara, T., Imai, N., Yamamoto, T., and Ohashi, S. (2018). Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 61, 176–182. doi: 10.1016/j.meegid.2018.03.030
Utiger, A., Tobler, K., Bridgen, A., Suter, M., Singh, M., and Ackermann, M. (1995). Identification of proteins specified by porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 380, 287–290. doi: 10.1007/978-1-4615-1899-0_46
van Kasteren, P. B., Beugeling, C., Ninaber, D. K., Frias-Staheli, N., van Boheemen, S., Garcia-Sastre, A., et al. (2012). Arterivirus and nairovirus ovarian tumor domain-containing deubiquitinases target activated RIG-I to control innate immune signaling. J. Virol. 86, 773–785. doi: 10.1128/JVI.06277-11
Van Reeth, K., Van Gucht, S., and Pensaert, M. (2002). In vivo studies on cytokine involvement during acute viral respiratory disease of swine: troublesome but rewarding. Vet. Immunol. Immunopathol. 87, 161–168. doi: 10.1016/S0165-2427(02)00047-8
Wang, L., Byrum, B., and Zhang, Y. (2014b). Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 20, 1227–1230. doi: 10.3201/eid2007.140296
Wang, D., Fang, L., Li, P., Sun, L., Fan, J., Zhang, Q., et al. (2011). The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 85, 3758–3766. doi: 10.1128/JVI.02589-10
Wang, D., Fang, L., Li, K., Zhong, H., Fan, J., Ouyang, C., et al. (2012). Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 86, 9311–9322. doi: 10.1128/JVI.00722-12
Wang, D., Fang, L., Shi, Y., Zhang, H., Gao, L., Peng, G., et al. (2016). Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 90, 2090–2101. doi: 10.1128/JVI.02514-15
Wang, D., Fang, L., Wei, D., Zhang, H., Luo, R., Chen, H., et al. (2014a). Hepatitis A virus 3C protease cleaves NEMO to impair induction of beta interferon. J. Virol. 88, 10252–10258. doi: 10.1128/JVI.00869-14
Wang, L., Qiao, X., Zhang, S., Qin, Y., Guo, T., Hao, Z., et al. (2018). Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-kappaB activation. Virulence 9, 1685–1698. doi: 10.1080/21505594.2018.1536632
Wang, Q., Vlasova, A. N., Kenney, S. P., and Saif, L. J. (2019). Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 34, 39–49. doi: 10.1016/j.coviro.2018.12.001
Wang, F., Wang, S. Q., Wang, H. F., Wu, Z. C., Bao, W. B., and Wu, S. L. (2020a). Effects of porcine epidemic diarrhea virus infection on toll-like receptor expression and cytokine levels in porcine intestinal epithelial cells. Pol. J. Vet. Sci. 23, 119–126. doi: 10.24425/pjvs.2020.132755
Wang, X. W., Wang, M., Zhan, J., Liu, Q. Y., Fang, L. L., Zhao, C. Y., et al. (2020c). Pathogenicity and immunogenicity of a new strain of porcine epidemic diarrhea virus containing a novel deletion in the N gene. Vet. Microbiol. 240:108511. doi: 10.1016/j.vetmic.2019.108511
Wang, R., Yu, R., Chen, B., Si, F., Wang, J., Xie, C., et al. (2020b). Identification of host cell proteins that interact with the M protein of porcine epidemic diarrhea virus. Vet. Microbiol. 246:108729. doi: 10.1016/j.vetmic.2020.108729
Wei, G., Luo, S., Wu, W., Hu, J., and Zhou, R. (2021). Activation of interleukin-1beta release and pyroptosis by transmissible gastroenteritis virus is dependent on the NOD-like receptor protein 3 inflammasome in porcine intestinal epithelial cell line. Viral Immunol. 34, 401–409. doi: 10.1089/vim.2020.0227
Woo, P. C., Huang, Y., Lau, S. K., and Yuen, K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses 2, 1804–1820. doi: 10.3390/v2081803
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/JVI.06540-11
Wood, E. N. (1977). An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 100, 243–244. doi: 10.1136/vr.100.12.243
Wu, Z., Cheng, L., Xu, J., Li, P., Li, X., Zou, D., et al. (2020b). The accessory protein ORF3 of porcine epidemic diarrhea virus inhibits cellular interleukin-6 and interleukin-8 productions by blocking the nuclear factor-kappaB p65 activation. Vet. Microbiol. 251:108892. doi: 10.1016/j.vetmic.2020.108892
Wu, Y., Zhang, H., Shi, Z., Chen, J., Li, M., Shi, H., et al. (2020a). Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA degradation of TBK1 and IRF3. Viruses 12:599. doi: 10.3390/v12060599
Xia, L., Yang, Y., Wang, J., Jing, Y., and Yang, Q. (2018). Impact of TGEV infection on the pig small intestine. Virol. J. 15:102. doi: 10.1186/s12985-018-1012-9
Xing, Y., Chen, J., Tu, J., Zhang, B., Chen, X., Shi, H., et al. (2013). The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 94, 1554–1567. doi: 10.1099/vir.0.051169-0
Xu, Z., Zhang, Y., Gong, L., Huang, L., Lin, Y., Qin, J., et al. (2019a). Isolation and characterization of a highly pathogenic strain of porcine enteric alphacoronavirus causing watery diarrhoea and high mortality in newborn piglets. Transbound. Emerg. Dis. 66, 119–130. doi: 10.1111/tbed.12992
Xu, X. G., Zhang, H. L., Zhang, Q., Dong, J., Huang, Y., and Tong, D. W. (2015). Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virol. 59, 265–275. doi: 10.4149/av_2015_03_265
Xu, X., Zhang, H., Zhang, Q., Dong, J., Liang, Y., Huang, Y., et al. (2013a). Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 10:26. doi: 10.1186/1743-422X-10-26
Xu, X., Zhang, H., Zhang, Q., Huang, Y., Dong, J., Liang, Y., et al. (2013b). Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 164, 212–221. doi: 10.1016/j.vetmic.2013.01.034
Xu, Z., Zhong, H., Huang, S., Zhou, Q., Du, Y., Chen, L., et al. (2019b). Porcine deltacoronavirus induces TLR3, IL-12, IFN-alpha, IFN-beta and PKR mRNA expression in infected peyer’s patches in vivo. Vet. Microbiol. 228, 226–233. doi: 10.1016/j.vetmic.2018.12.012
Yachdav, G., Kloppmann, E., Kajan, L., Hecht, M., Goldberg, T., Hamp, T., et al. (2014). PredictProtein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42, W337–W343. doi: 10.1093/nar/gku366
Yang, L., Xu, J., Guo, L., Guo, T., Zhang, L., Feng, L., et al. (2018). Porcine epidemic diarrhea virus-induced epidermal growth factor receptor activation impairs the antiviral activity of type I interferon. J. Virol. 92, e02095–e02117. doi: 10.1128/JVI.02095-17
Ye, G., Wang, X., Tong, X., Shi, Y., Fu, Z. F., and Peng, G. (2020). Structural basis for inhibiting porcine epidemic diarrhea virus replication with the 3C-like protease inhibitor GC376. Viruses 12:240. doi: 10.3390/v12020240
Yu, J., Qiao, S., Guo, R., and Wang, X. (2020). Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat. Commun. 11:3070. doi: 10.1038/s41467-020-16876-4
Zhang, J. (2016). Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 226, 71–84. doi: 10.1016/j.virusres.2016.05.028
Zhang, Q., Ke, H., Blikslager, A., Fujita, T., and Yoo, D. (2018). Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 92, e01677–e01717. doi: 10.1128/JVI.01677-17
Zhang, Q., Ma, J., and Yoo, D. (2017). Inhibition of NF-kappaB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 510, 111–126. doi: 10.1016/j.virol.2017.07.009
Zhang, Q., Shi, K., and Yoo, D. (2016). Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489, 252–268. doi: 10.1016/j.virol.2015.12.010
Zheng, L., Wang, X., Guo, D., Cao, J., Cheng, L., Li, X., et al. (2021). Porcine epidemic diarrhea virus E protein suppresses RIG-I signaling-mediated interferon-beta production. Vet. Microbiol. 254:108994. doi: 10.1016/j.vetmic.2021.108994
Zhou, P., Fan, H., Lan, T., Yang, X. L., Shi, W. F., Zhang, W., et al. (2018). Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556, 255–258. doi: 10.1038/s41586-018-0010-9
Zhou, J., Fang, L., Yang, Z., Xu, S., Lv, M., Sun, Z., et al. (2019). Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB J. 33, 14575–14587. doi: 10.1096/fj.201901624RR
Zhu, X., Fang, L., Wang, D., Yang, Y., Chen, J., Ye, X., et al. (2017a). Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology 502, 33–38. doi: 10.1016/j.virol.2016.12.005
Zhu, L., Mou, C., Yang, X., Lin, J., and Yang, Q. (2016). Mitophagy in TGEV infection counteracts oxidative stress and apoptosis. Oncotarget 7, 27122–27141. doi: 10.18632/oncotarget.8345
Zhu, X., Wang, D., Zhou, J., Pan, T., Chen, J., Yang, Y., et al. (2017b). Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 91, e00003–e00017. doi: 10.1128/JVI.00003-17
Keywords: porcine enteric coronaviruses, innate immunity response, immune evasion, PEDV, PDCoV, TGEV, SADS-CoV
Citation: Zhang K, Lin S, Li J, Deng S, Zhang J and Wang S (2022) Modulation of Innate Antiviral Immune Response by Porcine Enteric Coronavirus. Front. Microbiol. 13:845137. doi: 10.3389/fmicb.2022.845137
Edited by:
Cao Yong Chang, Sun Yat-sen University, ChinaReviewed by:
Zheng Chen, Jiangxi Agricultural University, ChinaPuxian Fang, Huazhong Agricultural University, China
Copyright © 2022 Zhang, Lin, Li, Deng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sutian Wang, d3N0bHl0QDEyNi5jb20=; Jianfeng Zhang, emhhbmctamlhbmZlbmdAMTM5LmNvbQ==
 Kunli Zhang
Kunli Zhang Sen Lin
Sen Lin Jianhao Li
Jianhao Li Shoulong Deng
Shoulong Deng Jianfeng Zhang
Jianfeng Zhang Sutian Wang
Sutian Wang