
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 07 July 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.844997
This article is part of the Research Topic Insights in Microbiotechnology: 2021 View all 9 articles
Bacteria form biofilms on material surfaces within hours. Biofilms are often considered problematic substances in the fields such as biomedical devices and the food industry; however, they are beneficial in other fields such as fermentation, water remediation, and civil engineering. Biofilm properties depend on their genome and the extracellular environment, including pH, shear stress, and matrices topography, stiffness, wettability, and charges during biofilm formation. These surface properties have feedback effects on biofilm formation at different stages. Due to emerging technology such as synthetic biology and genome editing, many studies have focused on functionalizing biofilm for specific applications. Nevertheless, few studies combine these two approaches to produce or modify biofilms. This review summarizes up-to-date materials science and synthetic biology approaches to controlling biofilms. The review proposed a potential research direction in the future that can gain better control of bacteria and biofilms.
Bacteria exist ubiquitously in nature and can adhere to nearly any type of surface, including biotic, abiotic, and natural surfaces. In many cases, it is beneficial for bacteria to transform from a single-cell planktonic lifestyle to a multicellular assembled community mode—biofilm. The biofilm is not only simply clusters of bacteria but also involves essential physiology and phenotypic changes. It is a highly structured multicellular microbial community composed of bacteria cells and an extracellular polymeric substance (EPS) matrix (Verderosa et al., 2019; Arnaouteli et al., 2021). Biofilms can be found on the surface of river rocks, seaside reefs, the roots of plants, deep-sea animal epidermis, water pipelines, food processing equipment, and even medical facilities. The problems caused by biofilms include economic loss and health-related issues since bacteria in a biofilm can be protected from the harsh environment, including the mechanical shear stress of fluid flow, antibiotics, or chemical interference (Schultz et al., 2011; Hall and Mah, 2017; Del Pozo, 2018; Cheng et al., 2019; Abebe, 2020). It is reported that the annual cost for cleaning and coating surfaces of navy ships alone in America is about US$56 million due to damage caused by matured biofilms (Hall and Mah, 2017). In the food-processing field, biofilms form on the processing line is a severe public health concern due to the potential causes of food contamination and foodborne diseases (Abebe, 2020). In addition, infections associated with biofilms developed on medical devices are also a persistent challenge for health care since the antibiotic resistance of biofilm may increase approx. 100- to 1,000-fold compared to their planktonic state (Hall and Mah, 2017; Del Pozo, 2018). The requirements for preventing biofilm formation or removing matured biofilm have been a long-standing focus of research efforts. Techniques used for antimicrobial purposes, including surface modifications and drug developments, have been well documented in some reviews (Rigo et al., 2018; Kyzioł et al., 2020; Valenzuela et al., 2021).
However, biofilms do not always cause negative implications. They can become a powerful tool in industries or agriculture by taking advantage of their “intelligent” characteristics such as systematic growth, self-repairing, and response to environmental signals to switch their growing state (Todhanakasem, 2017; Romero et al., 2018; Zhao et al., 2019; Pandit et al., 2020; Singh and Kaushik, 2021). For example, a biofilm’s ability to biotransformation can be used for wastewater treatment in bioremediation (Zhao et al., 2019; Singh and Kaushik, 2021). It has been demonstrated that biofilm’s high-cell density, immobilization, and stability could be ideal features. Moreover, since biofilms have little sensitivity to medical drugs and toxins and are highly active organized reaction systems, they can act as a biocatalyst in fabricating many bio-based products, such as antibiotics, enzymes, exopolysaccharides, bioenergy, and biorefinery (Todhanakasem, 2017; Romero et al., 2018).
To control biofilm formation, a thorough knowledge of factors influencing the biofilm formation process is crucial for both prevention and application. In addition to its genome, biofilm formation can also be affected by environmental factors, including flow rate, environmental pH, temperature, gravitational force, and substratum surface properties (Linklater et al., 2018; Ren et al., 2018; Achinas et al., 2019; Secchi et al., 2020; Krsmanovic et al., 2021; Nakanishi et al., 2021). In this review, we neither treat biofilms as threats nor beneficial but focus on controlling biofilm formation from synthetic biology and substratum surface properties. Both of these aspects could influence biofilm formation to some extent. We propose that combining these two techniques might better control the biofilm and thus benefit a range of applications (Figure 1). We have structured this review into two parts: the first part will discuss recent updates on the effects of substratum surfaces’ properties on biofilm formation. In contrast, the second part will give representative applications using synthetic biology methods to control biofilm formation.
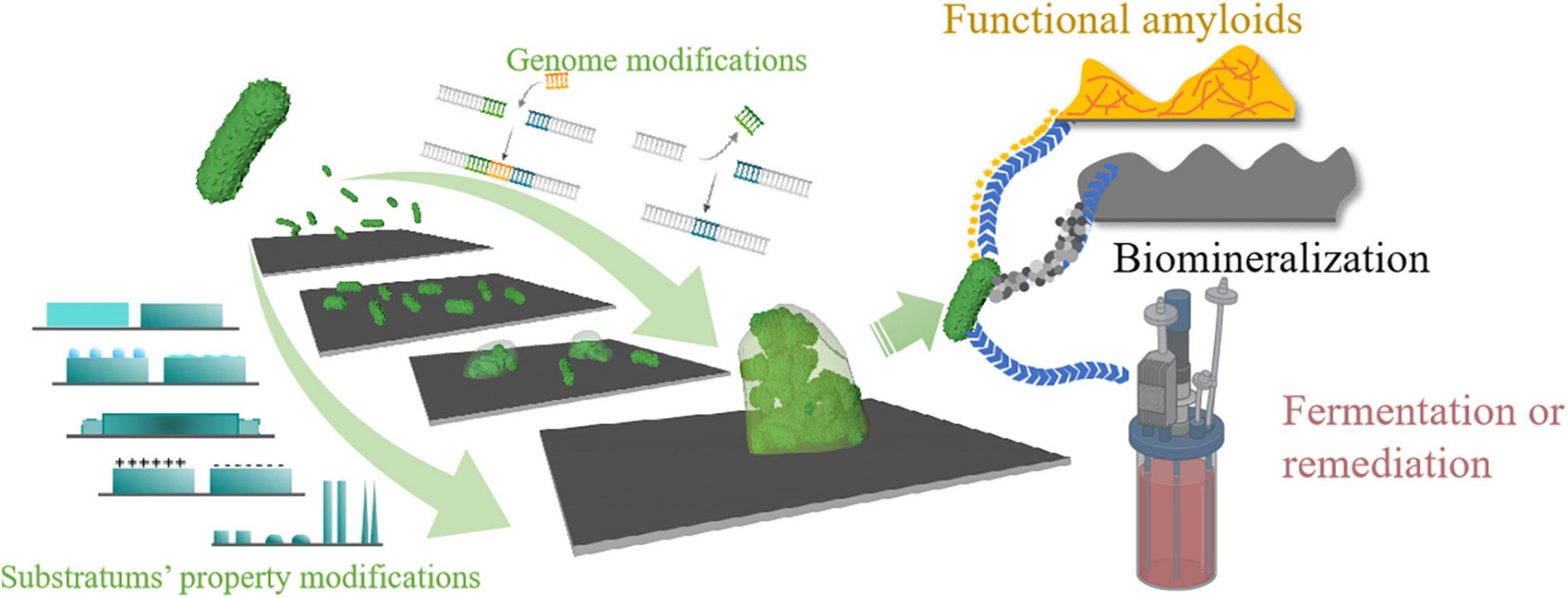
Figure 1. Schematic illustration of the review. Bacteria can form biofilms once attached to a substrate that can be beneficial or problematic depending on specific situations. A thorough understanding of biofilm formation factors and their properties is necessary for better application. Both genomic modifications of the bacteria and substratum’s properties can affect biofilm’s properties. Currently, little research has focused on combining these two methods to control biofilm formation. In this article, we propose that by taking advantage of both synthetic biology and materiobiology, it is possible to gain better control over biofilm formation.
It is widely accepted that the development of biofilms includes the following four stages: (1) initial attachment; (2) microcolony formation; (3) biofilm maturation; and (4) biofilm dispersal (Zhao et al., 2017; Arciola et al., 2018). Bacterial adhesion is the first step for biofilm formation on the surface. Both reversible and irreversible attachment appear at this stage where only the latter can yield those attached bacterial continue to grow into microcolonies. Microcolonies, the basic units in most biofilms, start to form when cells proliferate on surfaces. The biological processes of the bacteria dominate this period and are characterized by the excretion of EPSs and signaling molecules. EPS, consisting of various extracellular polysaccharides, DNA, and proteins, is critical for bacteria to construct and maintain the biofilm structure. Still, the composites and properties of EPS vary significantly by species and environment (Zhao et al., 2017; Arciola et al., 2018). The biofilm then starts to mature with the microcolonies growing and expanding to reach a steady state where nutrient transportation and cellular activities are balanced. The fluid-phase channels contain either nutrients or wastes interspersed within the mature biofilm, creating pillars segregating different microenvironments that translate to various cell activities, making it a complex, highly differentiated cell community (Zhao et al., 2017; Arciola et al., 2018). The last stage of the biofilm development is dispersal, in which cell departs from biofilm and converts to the planktonic mode of growth (Zhao et al., 2017; Arciola et al., 2018). In addition to the bacteria’s genome and environmental factors, the substratum’s surface properties also play an important role during biofilm formation. These surface properties include topography, roughness, stiffness, surface charge, and hydrophobicity. Here, we summarized some representative studies on how substratum properties affect biofilm formation.
Specific patterns and roughness changes can characterize surface topography changes. By specific patterns, we refer to the topography containing certain shapes and the pattern size, usually at micro and sub-micro scale-like pillars, grooves, wire, etc. Roughness changes are irregular shapes at the nanoscale level (Tables 1, 2).
Surface patterns can alter bacterial adhesion behaviors (Table 1). Specific-sized patterns like nanoneedles or pillars can affect bacterial viability (Wu et al., 2018b; Ivanova et al., 2020; Jiang et al., 2020; Xiao et al., 2020). These antimicrobial patterns have been well summarized in other literature, which is not our focus (Linklater et al., 2020; Mahanta et al., 2021). In this discussion, we focus on the bacteria behaviors when adhering to “mild patterns,” i.e., the patterns are not fatal to the majority of attached bacteria (Table 1). Gu et al. studied Escherichia coli’s adhesion behaviors on gratings. They found that the orientation of attached bacteria was more perpendicular to the orientation of gratings when the widths decreased. The attached cell shape was longer and their transcription activity was higher on the narrow gratings (Gu et al., 2016a). Silica and polystyrene particle’s self-assembled surfaces showed that E. coli and Staphylococcus aureus preferred attaching to the valley area instead of growing across the particle (Shi et al., 2021). In addition to attachment locations, patterns also affect the number of bacteria attached. Pattern size and spacing were critical because smaller-sized pillar patterns showed low fouling effects for S. aureus and bactericidal effects for E. coli. In contrast, larger-sized patterns showed low fouling effects for E. coli and patterning toward S. aureus (Heckmann and Schiffman, 2020). By fabricating either protruding or recessing grooves, square and circular features on PDMS surfaces via soft lithography with various parameters, it was demonstrated that patterned surfaces not only reduced Staphylococcus epidermidis and S. aureus attachment but can also alter the bacteria attachment locations. Bacteria actively choose their settling position based on the cell-surface contact points maximization principle (Vadillo-Rodriguez et al., 2018). The authors ruled out the possibility that the low-fouling effects were due to physical constraints or surface hydrophobicity. They proposed that it may be caused due to the nanoscale surface roughness-induced interaction energies (Vadillo-Rodriguez et al., 2018). Other than highly ordered patterns, random surface features, such as filaments and irregular protruding features, can also reduce the attachment of bacteria (Cao et al., 2018; Meier et al., 2018; Peter et al., 2020), but not all patterns have a passive effect on bacterial adhesion. Encinas et al. (2020) showed that there were more bacteria colonies adhered on micropillars than on flat and nanofilament surfaces, and explained that this association is due to the size of micropillars being larger than bacteria, which gives sufficient space for bacteria to anchor and adhere.
Studies have shown that even on the nanoscale, surfaces still can affect bacterial adhesion. Surface roughness was usually impacted by material processing steps such as direct laser interference patterning (DLIP) (Meinshausen et al., 2021), plasma etches (Linklater et al., 2019), and Femtosecond laser (Table 2; Cunha et al., 2016). The roughness of the surface varied in nanoscales, which was much less than the size of bacteria. Wu et al. found that the membrane of bacterial cells on the surface with nanoscale topography would be slightly deformed and elongated. They suggested the nanoscale surface topography could inhibit the bacteria’s adhesion and proliferation (Wu et al., 2018a).
Nevertheless, it is difficult to conclude whether a smooth surface reduces or increases bacterial attachment. Jang et al.’s prepared nanostructured stainless steel 316L by electrochemical etching. They found fewer bacteria were attached to the nanostructured surfaces than the smoother control ones (Jang et al., 2018). Later, in 2020, Bilgili et al. (2020) claimed no statistically significant differences were observed among different bulk-fill composite resins in terms of roughness. However, in 2021, Aouame et al. reported that the bacteria adhesion and biofilm formation mainly depend on surface characteristics. Bacteria are more likely to adhere to and develop biofilm on rough dental surfaces than on smooth stainless-steel surfaces (Aouame et al., 2021). Different materials, strains, and test methods were applied in these studies, making it difficult to compare them.
It is well known that substrates’ stiffness plays a vital role in mammalian cell attachment, migration, differentiation, and tissue homeostasis process (Handorf et al., 2015; Spencer et al., 2017). Fewer studies cover this area regarding their effects on the behavior of bacteria, and the results are still controversial (Figure 2 and Table 3). The discussed substrates here were chemically and mechanically stable.
Lichter et al. were the first to report that substrate stiffness could affect the adhesion of bacteria independent of other physicochemical properties, including roughness, interaction energy, surface charge density, and monovalent ion concentration. By assembling polyelectrolyte multilayers on Ti substrates under different conditions, the stiffness of such hydrated films could be varied between 1 and 100 MPa. Their studies applied the Gram-positive strain S. epidermidis and Gram-negative strain E. coli (wild-type and mreB mutant strains). They concluded that the adhesion of bacteria correlates positively with increasing elastic modulus over the testing range (Linklater et al., 2019). Later, Rachel’s group proved Gram-negative bacterial strain Pseudoalteromonas sp. D41 had similar trends by using agarose hydrogel as a substrate. Besides, they identified 21 proteins differentially regulated when attached to different stiffness via a proteomic approach. Most of these proteins are involved in key metabolic pathways, suggesting the substrate’s stiffness affects both the bacterial adhesion amount and their phenotype. Furthermore, concerning Gram-positive Bacillus sp. 4J6, though the adhesion pattern changed with stiffness, the number of attached bacteria showed no statistical difference (Guegan et al., 2014). Similarly, by using poly-(ethylene glycol) (PEG) hydrogel, Schiffman’s group showed the amount of bacterial attachment (E. coli and S. aureus) was positively correlated with hydrogel stiffness (Kolewe et al., 2015, 2018). Song and Ren (2014) and Song et al. (2017), Moraes (Siddiqui et al., 2019), and Phillips (Wang et al., 2016) research groups drew conflicting findings on how bacteria attached to soft substrates. Phillips’s group used polyacrylamide (PAAm) hydrogel with elastic modulus ranging from 17 to 654 Pa, and found that the adhesion rate and numbers of adherent S. aureus decreased with increased modulus (Wang et al., 2016). The other two groups used PDMS as substrates, and both found a higher number of E. coli attached to soft substrates. In addition to the attached amount, Ren’s group found the length of bacteria attached on the soft surface was longer than stiff ones (Song and Ren, 2014). Moraes’s group studied the effects of shear force and bioactive molecules coating in addition to substrate stiffness. With ECM molecules coating, bacterial adhesion strength generally decreased, but these effects were smaller than those caused by substrate mechanical properties. They demonstrated that bacteria attached evenly on soft and hard surfaces under low shear force, but a higher number of bacteria retained on soft surfaces while exposed to higher shear. They claimed that the sheer force might be one of the reasons causing contradictory results in the literature (Siddiqui et al., 2019).
The studies covering the underlying mechanism of surface stiffness affecting bacteria were even less. By culturing bacteria in different divalent or monovalent ion concentration solutions, the activation of transient receptor potential ion channels was proved not required for S. epidermidis sensing of mechanical stimuli. By comparing mutant strain and wild type together with S. epidermidis, it was shown that the shape of bacteria was also not the reason (Lichter et al., 2008). The behaviors of E. coli RP437 and three of its isogenic mutants of motB, fliC, and fimA genes were compared and confirmed it is the motB gene involved in the response of E. coli to PDMS stiffness during the attachment stage (Song and Ren, 2014).
Other surface properties such as surface charge and hydrophobicity can also affect the bacterial adhesion stage. Highly positively charged surfaces are usually considered bactericidal since they can rupture the cell membrane through electrostatic attractions (Pranantyo et al., 2018; Salama et al., 2020). When surface charge density is below the critical value of bactericidal, it will influence bacteria adhesion behaviors. Layer by layer assembly of cationic/anionic polymers is one of the coating methods for adjusting materials’ surface charges. It was shown that positively charged surfaces attracted bacteria compared to the negative ones due to the original negative potential of the bacterial cell wall (Zhu et al., 2015; Kovačević et al., 2016; Guo et al., 2018). Besides, there was a positive correlation between the adsorption of bacteria and the surface charge (Chen C. et al., 2019). Similar trends were also found by carefully designed polymer chain coating to control surface hydrophobicity and surface potential (Rzhepishevska et al., 2013; Oh et al., 2018). The phenomenon of Pseudomonas aeruginosa oriented vertically on negatively charged surfaces was suggested due to the cell’s intention to minimize the exposure to the repel force between their membrane and the surfaces (Rzhepishevska et al., 2013). However, there are also some controversial conclusions where some studies showed that regardless of the material chosen, the surface charge had no impact on the bacterial attachment (Lorenzetti et al., 2015; Spriano et al., 2017).
Wettability is defined as the ability of a liquid to wet a surface and is usually determined by the contact angle. Though it is generally believed that hydrophobic cells tend to adhere to hydrophobic surfaces while hydrophilic ones to hydrophilic surfaces, the relationship between surface wettability and bacterial adhesion is complicated to discuss on its own because wettability depends on many other surface parameters as well, such as roughness, surface charge, surface chemistry, and so on (Krasowska and Sigler, 2014; Jothi Prakash and Prasanth, 2021).
Conditioning films start to form once substratum surfaces are in contact with aqueous environments where various organic and inorganic compounds are adsorbed onto the surfaces. This conditioning film influenced initial bacterial adhesion by altering substratum surface charge, hydrophobicity, roughness, and chemical composition (Talluri et al., 2020; Bhagwat et al., 2021; Wurzler et al., 2022). Whether this conditioning film promotes or inhibits adhesion highly depends on the status presented on the surfaces (Berne et al., 2018; Wurzler et al., 2022).
It is worth noticing that, when choosing their behaviors, bacteria would seem to comprehensively consider all surface properties. These surface properties might only have one or two factors predominant bacterial behaviors or, sometimes, may all have a contribution (Spriano et al., 2017; Wassmann et al., 2017; Aouame et al., 2021). Besides, the testing conditions, such as culture media used, either flow system or static system, will all affect the bacterial attachment (Senevirathne et al., 2021).
Substrates’ properties can affect bacterial behaviors during biofilm formation. Poly(methyl methacylate) (PMMA) surface with sub-cellular nanopillar topography not only can inhibit P. aeruginosa attachment but also prevent upstream movement and reduce the proliferation of bacteria cells (Figure 3A; Rosenzweig et al., 2019). On a micrometer-scale surface with a crystalline hemispherical pattern, bacteria behave differently depending on the diameter of the raised features. A diameter of 2 μm and larger hemispherical patterns could hinder P. aeruginosa motility, while a 1 μm pattern showed no significant difference compared with a flat surface. In addition, their travel directions also differed by diameters. On 2 μm surfaces, bacteria were more likely to move in a reverse direction. On 4 μm surfaces, they prefer to travel in approximately straight lines in the groove along the crystal axis. In comparison, they were more likely to follow an approximately hexagonal lattice of grooves on 8 μm surfaces since this feature was complex for bacteria to move across the crowns (Figure 3B; Yow-Ren Chang and Ducker, 2018). On a stepped topography, P. aeruginosa drastically reduced the probability of crossing the step compared with crossing a point on a flat surface and significantly reduced the speed perpendicular to the step when they were very close to the step. When step height was similar to the bacterial length, a time penalty to cross the step was found, while no time penalty was resolved to cross tall steps (5–9 μm) (Chang et al., 2019). The critical length-scale for topography affecting P. aeruginosa movements seems to be similar to the dimensions of the bacterium and the length of pili. They proposed that if the topography in a particular direction is not favorable for pili attachment, then that direction of motion may also be disfavored (Chang et al., 2019). Another study also reported this threshold of topographical barriers of ∼1 μm using a different pattern. It was found that a 1 μm depth of micro-fabricated furrows can inhibit the expansion of P. aeruginosa biofilm more effectively than a 0.5 μm depth one (Gloag et al., 2016). For E. coli, a 10 μm tall hexagon-shaped topographic pattern with a side length of 15 μm and inter pattern distance of 2 μm showed the interrupting biofilm formation effects. In contrast to the smooth surfaces, biofilm formation and conjugation were promoted for larger square-shaped patterns with side lengths larger than 20 μm and inter pattern distances no less than 10 μm. Besides, compared to the top and grooves of the pattern, the vertical sidewall was favored for bacterial conjugation (Figure 3C; Gu et al., 2017).
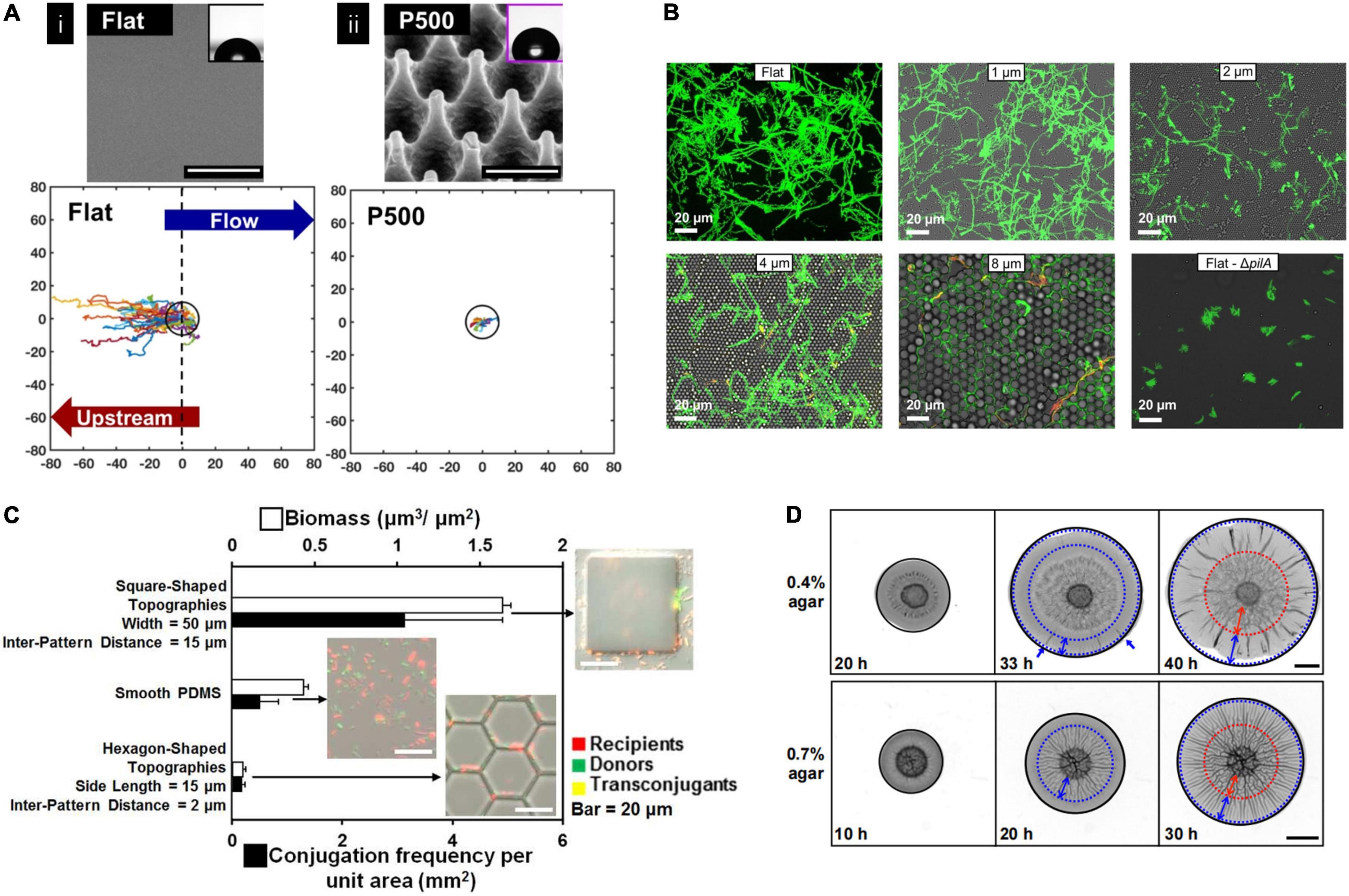
Figure 3. (A) Nanopillared topography inhibited bacteria upstream motility indicated by single-cell trajectories. Single-cell trajectories of Pseudomonas naeruginosa on the flat surfaces originated from 0 and extended to the –80 μm in x-direction indicating upstream motility, while much less motility distance was observed on a nanopillar topography (P500). The black ring is 10 μm radium as a reference (Rosenzweig et al., 2019). Adapted with permission from Rosenzweig et al. (2019). Copyright 2019 American Chemical Society. (B) Hemisphere topography affected bacterial migration. Fluorescence images showing P. aeruginosa movement traces. It showed bacteria appear to explore a smaller fraction of the flat surface compared with hemisphere topography surfaces with 2–8 μm features (Yow-Ren Chang and Ducker, 2018). Adapted with permission from Yow-Ren Chang and Ducker (2018). Copyright 2018 American Chemical Society. (C) Conjugation frequency and biofilm formation of Escherichia coli were influenced by surface topography. The square-shaped pattern showed higher conjugation frequency than smooth and hexagon-shaped surfaces (Gu et al., 2017). Adapted with permission from Gu et al. (2017). Copyright 2017 American Chemical Society. (D) Representative images of Vibrio cholerae biofilm grown on different substrates at different time points. Blue dotted circles mark the boundaries of regions with radical patterns, and red dotted circles mark the boundaries of regions with zigzag patterns. It showed biofilm morphology differed on different substrate’ stiffness (Fei et al., 2020). Adapted with permission from Fei et al. (2020). Copyright 2020 National Academy of Science.
In addition to topography, substrate stiffness also affects the biofilm formation process. It has been found that the average velocity of E. coli movement was higher on rigid PDMS surfaces compared to the soft ones (Song et al., 2017). By establishing an idealized bacteria twitching mathematical model, Sabass’ group analyzed how substrate rigidity influenced bacterial migration and revealed that migration depends on the force-sensitivity of the adhesion bond. The results showed that bacteria migration speed depends non-linearly on substrate rigidity. Bacteria move faster on a rigid surface if their rear adhesion bond is more force-sensitive. However, the deformation of the soft substrate surface may block the pull force produced during migration, which negatively affects migration speed (Simsek et al., 2019).
Furthermore, substrate stiffness can also affect the pattern of growing biofilms. It has been reported that the Vibrio cholerae biofilm spread varied on the surface of different agar concentration hydrogels. The morphology of growing V. cholerae biofilms is fold-shaped, extending from the edge to the center on the soft hydrogel surface of low agar concentration. In contrast, in the high agar concentration hydrogel surface, the folds extend in the opposite direction from the center to out edge. The sliding friction between agar surface and biofilm may contribute to the morphology difference during biofilm expansion (Figure 3D; Fei et al., 2020).
During maturation, bacteria often use quorum sensing to coordinate each other and the community. A cell–cell communication system involves the production, release, accumulation, and detection of autoinducers depending on the cell density, species composition of the microbial community, and the surrounding environment. As the population density of bacteria communities increases, the amount of autoinducer accumulates in the surrounding environment, which further affects the global gene expression patterns and cell-to-cell interactions (Papenfort and Bassler, 2016; Abisado et al., 2018). The key to quorum sensing is that bacteria cells receive, recognize, and respond to these signal molecules. Many studies have demonstrated that using molecular biology techniques to alter the bacterial quorum-sensing system will affect bacterial metabolic activities and biofilm formations. Detailed information has been well-reviewed in other articles, which is not our focus here (Mukherjee and Bassler, 2019; Li and Zhao, 2020; Wang N. et al., 2021).
From a material point of view, autoinducer peptide -I (AIP-I) is an essential inducer of the S. aureus Agr quorum-sensing system and can be detected by a cognate transmembrane bound receptor. This molecule could be immobilized onto a glass substrate via a flexible linker to influence the biofilm formation process (Kim et al., 2017). Similarly, a synthetic quorum sensing inhibitor 5-methylene-1-(prop-2-enoyl)-4-(2-fluorophenyl)-dihydropyrrol-2-one can be covalently incorporated into the surface coating to hindering bacterial communication (Ozcelik et al., 2017).
The substrates’ properties can still have an impact on the matured biofilms. For example, it has been found that micropatterned pillar surfaces could modulate competition dynamics and signaling pathways in the co-culture environment of E. coli and P. aeruginosa. With the increased height of the pillar, the biofilm featured increasing volume fractions of E. coli cells. They explained that this phenomenon resulted from accumulating a signaling molecule indole, which can silence E. coli’s biofilm dispersal and inhibit P. aeruginosa’s formation. Besides, the authors also found that micropatterned pillar surfaces could affect their antibiotic susceptibilities in mono-species culture. Increased susceptibility was found in patterns with the higher pillar (Figure 4A; Bhattacharjee et al., 2017). In addition to topography, the surface charge can influence biofilm structure. For example, P. aeruginosa biofilms form mushroom shapes on negatively charged surfaces while flat on positively charged surfaces. This phenomenon is due to the negatively charged surface inhibiting motility, preventing bacteria from moving over the surface to create a flat biofilm (Rzhepishevska et al., 2013). Besides, substrates’ stiffness also affects biofilm’s susceptibility; biofilms growing on stiff substrates were less susceptible to antibiotics than those on the soft substrates (Song and Ren, 2014).
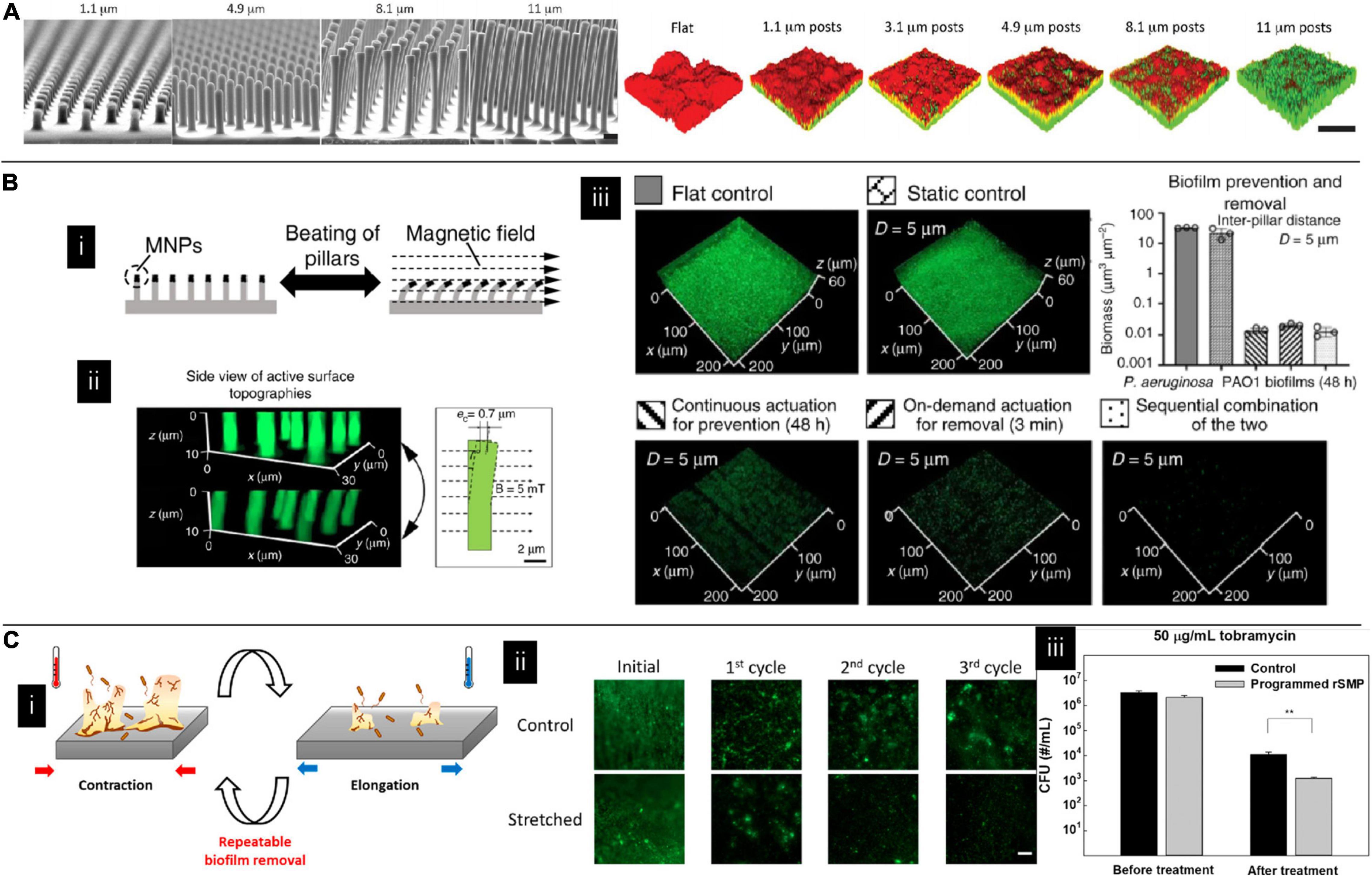
Figure 4. (A) Topography affecting multispecies biofilm morphology and species content where green and red colors indicate Escherichia coli and Pseudomonas aeruginosa, respectively. The fraction of E. coli in the co-cultured film increased with the pillar height (Bhattacharjee et al., 2017). Adapted with permission from Bhattacharjee et al. (2017). Copyright 2017 American Chemical Society. (B) The effect of magnetically driven dynamic pillar pattern on biofilm formation. (i) Schematic image of pillar bending in response to an external magnetic field. (ii) Fluorescent images of the pillar before and after exposure to a magnetic field. (iii) Representative fluorescence images of biofilms on flat controls, static controls, and active surface topographies (Gu et al., 2020). Adapted with permission from Gu et al. (2020). Copyright 2020 Spring Nature. (C) The effect of shape recovery substrate on biofilm formation. (i) Schematic illustration of dynamic substrates. (ii) Biofilm staining on different dynamic substrates. (iii) Detached bacteria were more susceptible to antibiotics on dynamic substrates (Lee et al., 2021).
In addition to influencing naturally formed biofilms, materials could also assist bacteria in forming biofilms for better performance (Hubenova et al., 2019; Reinhardt et al., 2020). For example, alginate could be functionalized with thiazolyl blue formazan/phenazine methosulfate and showed good biocompatibility of immobilization of bacteria Pseudomonas putida 1046 for constructing artificial biofilms. This artificial biofilm exhibited good electrochemical activities due to the incorporated redox-active cross-linking network in the surrounding polymer matrix around the microbial cells. They were considered good candidates for bioanodes in microbial fuel cells for wastewater treatment (Hubenova et al., 2019). The layer-by-layer technique is another method to manipulate biofilms artificially. Rijavec et al. constructed a protective biofilm coating consisting of two strains on steel surfaces. Gramicidin S producing cells Brevibacillus brevis are encapsulated with artificial polyelectrolytes deposited on steel surfaces and then enclosed within a rubber elastomer layer. On the rubber elastomer layer surfaces, Bacillus pumilus cells were then deposited with the assistance of polyelectrolytes. They demonstrated that when applied, this artificial biofilm could affect the phylogenetic structure of the developing natural biofilm and show a possible way to reduce phylogenetic diversity and exclude undesired bacteria (Rijavec et al., 2019).
Biofilm dispersion is an active process that is usually initiated by native and environmental cues. By leaving bacteria to convert to their planktonic growth mode and leaving behind eroded biofilms, biofilm is believed to be more vulnerable at this stage. Therefore, it has been proposed that promoting biofilm dispersion can be one of the antimicrobial methods. However, more caution needs to be taken by using this method since it has been found the dispersed cells display different phenotypes compared to their planktonic and biofilm counterparts. Dispersing cells without efficient killing might cause a significant problem (Rumbaugh and Sauer, 2020). In addition to natural dispersion, materials can also assist this process. Iron oxide nanoparticles are one of the most studied candidates for assisting in removing established biofilms by either catalyzing free radical generation (Gao et al., 2016; Liu Y. et al., 2018; Hwang et al., 2019) or mechanical force like digging channels or mechanically erasing biofilm structure (Hwang et al., 2019; Quan et al., 2019). In addition to adding external particles, dynamic substrates showed promising results in effectively eliminating matured biofilms (Geilich et al., 2017; Wang W. et al., 2018; Gu et al., 2020). It has been studied on a catheter model. By applying a relatively high strain rate on silicon elastomer-based substrates, biofilms can be readily detached as large pieces once the applied strain reaches a critical value. Interestingly, when the applied strain rate is relatively low, biofilms can still be bound to the substrates even if the substrates are under high strains (Levering et al., 2014). Besides, by incorporating magnetic particles on top of the pillar, micron-sized pillars can be engineered to beat at a programmable frequency and force level when applying an electromagnetic field. These active surfaces effectively removed established biofilms of uropathogenic E. coli, P. aeruginosa, and S. aureus (Figure 4B; Gu et al., 2020). Shape memory polymers are another candidate for fabricating dynamic substrates. Gu et al. (2016b) used shape memory polymer to fabricate recessive hexagonal patterns and found a dynamic change in surface topography could remove the biofilm. However, this is a one-way shape memory polymer, which means that the material can undergo shape change only once. To overcome this drawback, Lee et al. used a reversible shape memory polymer with a shape transition temperature close to body temperature, proving that P. aeruginosa biofilm could be effectively removed. It was also shown that the detached biofilm cell had increased antibiotic susceptibility compared to the static control (Figure 4C; Lee et al., 2021).
The genome of bacteria determines the biofilm formation directly. Synthetic biology is a powerful technique allowing the manipulation of existing biological systems in a controlled manner (El Karoui et al., 2019; McCarty and Ledesma-Amaro, 2019). Bacteria and associated biofilms can be genetically modified to achieve their designed behaviors and functions (McCarty and Ledesma-Amaro, 2019; Mukhi et al., 2022). The following section gives examples of controlling biofilms via synthetic biology techniques for different applications.
The biofilm secretes EPSs containing mainly polysaccharides, eDNA, and proteins. Polysaccharides constitute most biofilm mass composing glucan, fructan homopolymers, glucose/mannose/rhamnose heteropolymers, cellulose, alginate, colonic acid, and N-acetylglucosamine, and are highly heterogeneous across bacterial species (Nguyen, 2017). eDNA primarily relies on cellular lysis, which is an energetically inefficient system for large-scale production (Nguyen, 2017). Functional amyloids are one of the important proteins in the biofilm matrix. They comprise stacks of β-sheets aligned perpendicular to the fibril axis (Cao and Mezzenga, 2019; Gallardo et al., 2020). Unlike amyloid fibers formed upon aberrant misfolding of proteins, which are always associated with several incurable degenerative human diseases, bacterial amyloids benefit the life cycle of the biofilm. They are the key structural components of the biofilm matrix and can mediate toxicity and cell-to-cell interactions within the biofilm. Besides, amyloids are responsible for the spatial structure, morphological differentiation, surface properties, and viral protection of the biofilm (Biesecker et al., 2018; Deshmukh et al., 2018; Erskine et al., 2018; Van Gerven et al., 2018).
Curli are one of the important amyloids that have been used as building blocks for specific applications in nanotechnology due to their high stability and physical robustness (Taglialegna et al., 2016). They are non-covalent heteropolymeric filaments of CsgA and CsgB subunits where CsgA is secreted into the extracellular milieu and self-assembled into nanofibers by seeding onto a membrane-anchored nucleation protein CsgB (Van Gerven et al., 2015; Nguyen, 2017). Therefore, by fusing functional peptides or proteins into the amyloid monomers, a large-scale nanomaterial with programmable functionality can be secreted by cells (Elizabeth et al., 2016; Knowles and Mezzenga, 2016; Li et al., 2018). Nguyen et al. presented a biofilm-integrated nanofiber (BIND) system for precise genetic programming of bacterial matrix by fusing peptide domains onto the amyloid protein CsgA. Three peptides with different functions were fused to CsgA through C-terminal for extracellular self-assembly into functionalized curli nanofibers. They demonstrated the ability to engineer biofilm to template silver nanoparticles, increase adhesion to 304L stainless steel, and covalent immobilize proteins (green fluorescent protein as an example) (Nguyen et al., 2014). Similarly, mussel food protein of Mytilus galloprovincialis can also be fused with CsgA protein to produce adhesives with better properties like strong wet bonding strength, robustness, stability, and intrinsic fluorescence (Zhong et al., 2014). Besides, the BIND platform can also be used to immobilize enzymes for biocatalytic surface application. It has been reported that upon fusing to the SpyCatcher attachment domain, a recombinant α-amylase could be immobilized onto E. coli curli fibers displaying complementary SpyTag capture domains. The obtained enzymes’ immobilized biofilms were shown to be active after exposure to various adverse conditions (Botyanszki et al., 2015). Patterns are always relatively challenging to fabricate and usually require multiple fabrication steps (Shi et al., 2020). Using inducible genetic circuits and cellular communication circuits to regulate E. coli curli amyloid production can render amyloid fibril forming patterns either autonomously or via external control (Chen et al., 2014; Wang X. et al., 2018). It has been demonstrated that amyloid fibrils assembled could co-organize and synthesize inorganic nano-objects like fluorescent quantum dots (QDs) and gold nanowires, nanorods, and nanoparticles (Chen et al., 2014). Wang et al. further pushed these bio-abiotic hybrid materials to a new complexity level by engineering E. coli to harbor a blue-light inducible gene circuit to control the expression of CsgA through programmable light regulation; it is capable of spatiotemporally controlling nano-objects assembly. They demonstrated that discrete nano-objects and complex heterogeneous structures could be assembled hierarchically on different substrates or 3D materials of complex shapes. A minimum patterning resolution of 100 μm was achieved using light-sensing cells (Wang X. et al., 2018). Resettable pressure sensors could be fabricated using amyloid fibrils as the scaffold. With the help of inkjet printing to initiate single colony localization, engineered self-patterned bacteria could grow and subsequently facilitate the assembly of nanoparticles into a 3D dome structure. Their geometry would determine the dome structure’s response to pressure (Cao et al., 2017).
In addition to E. coli cells, Bacillus subtilis is another strain capable of this purpose. B. subtilis’ biofilm amyloid fibers were composites of TasA and TapA as major and minor protein components, respectively. The tapA-sipW-tasA gene operon regulates the production of amyloid fibers. Huang et al. fused the extracellular amyloid-like protein Tas A with various other proteins or protein domains to endow the biofilm with new functionalities. They demonstrated this platform could make programmable materials with various functions, including intrinsic fluorescent, intact enzyme activity, and the capability of templating inorganic nanoparticles. Besides, they were also able to adjust biofilm’s viscoelastic properties to manipulate them into diverse shapes and microstructures independently or using 3D printing techniques (Huang et al., 2019). Similarly, Zhang et al. (2019) engineered amyloid protein with a mussel foot protein and hydrophobin-like protein to control adhesion.
Mineralization is a process in which biology regulates the formation of hierarchical architectural mineralized materials and is vital for nature and human activities. It is a prevalent process in which species capable of forming minerals could be found in six taxonomic kingdoms. Examples of natural mineral products include teeth, coral, protozoan shells, etc. (Yao et al., 2017; Ehrlich et al., 2021). The biomineralization process generally involves an organic matrix to form a framework, and then nucleation sites are constructed, followed by controlling crystal orientation, growth, and termination crystal growth (Sharma et al., 2021). It is generally classified into two classes—biologically induced and biologically controlled mineralization, depending on the degree of biological control exerted. The former refers to the passive interaction of environments that drive the precipitation. The latter means the organism directly controls the precipitation process, producing minerals of a specific size, morphology structure, and orientation. Some literature has already documented the detailed mechanism, which will not be covered here (Qin et al., 2020; Hoffmann et al., 2021). Depending on the strains, bacterium mediates different mineralization, including calcification, silicification, iron, Mn mineralization, and so on (Couasnon et al., 2020; Qin et al., 2020). The application of biomineralization covers many fields such as environmental science for pollutant removal (Long et al., 2021) or water treatment (Arias et al., 2019), civil engineering for artwork conservation or as construction material (Achal et al., 2015; Marvasi et al., 2020), biomedical engineering for cancer therapy, or tissue engineering (Chen Y. et al., 2019; Qin et al., 2020).
In addition, to use bacteria individually for nanoparticle fabrication, mineral generation, or pollutant degradation, there are studies treating biofilm as a type of material. Zeng et al. discovered two Pesudoalteromonas lipolytic variants are capable of forming wrinkled and translucent biofilms. The wrinkled biofilm was due to a nonsense mutation in AToo_o8765, causing overproduction of extracellular polysaccharides, while the translucent ones were due to a point mutation in AToo_17125. Biofilm formed by either of these two strains was proved to have antifouling activities toward larval settlement and metamorphosis of the mussel Mytilus corucus (Zeng et al., 2015). Liu et al. further applied the strain capable of overexpression cellulose for steel protection. This strain can effectively bind Ca2+ and form biomineralization organic-calcite hybrid film on the steel surface to provide protection. The film formed by living bacteria could also provide in situ self-healing activity (Liu T. et al., 2018). In addition to selecting natural strains, Yang et al. genetically engineered E. coli bacteria to express DDDEEK peptide, having a strong ability to absorb mineral ions and induce the formation of a biomineral. This engineered biofilm was shown to be capable of tolerance shear force and stayed on virtually any type of material surface. Interestingly, this biofilm-based coating not only exhibited good mineralization performance and better stability compared to the hydroxyapatite spray sample but also showed no extra immunogenicity and better osteogenicity and osseointegration after 12 weeks of implantation, demonstrating their potential applications in the biomedical field (Yang et al., 2018). Wang et al. genetically engineered E. coli to achieve more control over the mineralization process. CasA and mussel foot protein Mfp were fused to render higher hydroxyapatite mineralization, capacity, and interfacial binding strength. To enable E. coli biofilms to respond to environmental stimuli, such as light, they encode the expression of CsgA-Mfp3S-pep in the Dawn plasmid, which renders the biomass density and spatial pattern controllable. Interestingly, bacteria cells inside the composite remained alive and capable of responding to environmental stimuli after mineralization (Wang Y. et al., 2021).
Bacterial metabolic activities are important when they are applied for fermentation or remediation. Bacteria in their biofilm form have been reported to increase the production of value-added products and secondary metabolites. Biofilm reactors, where biofilms were immobilized onto surfaces, have been widely accepted as a promising technology in wastewater treatment, pollutant removal, and production of alcohol, organic acid, enzymes, syngas, and many other value-added products (Mahdinia et al., 2019; Abdelfattah et al., 2020; Gunes, 2021). Biofilm’s properties such as adhesion strength, biomass, and population are closely related to its efficiency. Synthetic biology techniques can be applied to genetically modify strains to significantly improve their performance. For example, the poor adhesion capacity of stain B. Subtilis 168 was the main issue that limited their application in biofilm reactors. After insertion of the sfp and epsC genes together with the deletion of the sepF gene, the biofilm’s form by this mutant strain showed increased adhesion and surfactin production (Brück et al., 2019). Pseudomonas putida is a widely used strain in bioremediation due to its remarkable ability to degrade pollutants; however, most of the laboratory-adapted P. putida strains suffer from weak biofilm formation abilities. Benedetti et al. genetically programmed their intracellular c-di-GMP levels using an inducible and tightly controlled genetic device for heterologous expression of diguanylate cyclase and phosphodiesterase genes. They then proved this biofilm formed by engineered strain displayed high dehalogenase activity (Benedetti et al., 2016). In addition to monostrain biofilms, microbial consortia behaviors could also be controlled. By manipulating communication networks, regulating gene expressions, and engineering syntrophic interactions, it is possible to control individual species’ population, distribution, and spatial organization in a microbial consortium (McCarty and Ledesma-Amaro, 2019).
Bacteria tend to form biofilm as their preferred life form compared to planktonic ones once attached to a surface. Hence, manipulating the substratum properties and bacteria’s genome can both influence bacterial behavior and biofilm formation. Substrate’s properties have effects on four stages of biofilm formation. For example, surface topography, stiffness, wettability, and zeta potential can affect bacteria adhesion, such as the amount and location of the attached bacteria, the direction of movement, and viability. These factors, in turn, affect bacteria proliferation, biofilm morphology, antibiotic resistance, and biofilm dispersion. On the other hand, synthetic biology techniques are powerful tools capable of genetically modifying bacteria. These techniques directly change bacteria’s functions and metabolic activities and then generate the designed biofilms. Amyloids and biomineralization are two typical biofilms that can be designed and used as materials.
This review discussed the possibilities of controls and uses of biofilms from materiobiology and synthetic biology perspectives. To date, these two perspectives are rarely combined to produce unique biofilms. The main reason is that the two areas claim different standpoints, i.e., materials researchers usually see biofilms as toxins while synthetic biologists see these as biomaterials. In addition, while a vast amount of literature in materials science tries to avoid biofilm formation, the understanding of manipulating biofilms using the materiobiology approach is still insufficient, especially in the mechanism part. Besides, many of these studies only examined the biofilm’s adhesion stage. Consequently, controversial conclusions were obtained, such as the effect of stiffness and roughness on biofilm formation. Thus, further studies on the bacteria-surface interactions and long-term studies are necessary to gain better control properties of biofilm. On the other hand, designing biofilms using the synthetic biology approach is insufficient. We propose that combining these two techniques in the future could give better control over biofilm formation and properties.
P-YW conceived the manuscript. YS and P-YW designed the structure of the review. YS and TC collected the literature. YS, TC, and P-YW wrote the manuscript. YS, PS, and P-YW proofread the manuscript. All authors approved the final version of the manuscript.
The Ministry of Science and Technology of China (2019YFE0113000); the National Natural and Science Foundation of China (31870988); the Chinese Academy of Sciences (172644KYSB20200002 and 172644KYSB20200048); the Department of Science and Technology of Guangdong Province (2021A0505030055); the Science, Technology, and Innovation Commission of Shenzhen (GJHZ20180928115804736 and ZDSYS20190902093409851).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the Shenzhen Institute of Advanced Technology, the Chinese Academy of Sciences, and the Oujiang Laboratory for providing a good research environment.
Abdelfattah, A., Hossain, M. I., and Cheng, L. (2020). High-strength wastewater treatment using microbial biofilm reactor: a critical review. World J. Microbiol. Biotechnol. 36:75. doi: 10.1007/s11274-020-02853-y
Abebe, G. M. (2020). The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020:1705814. doi: 10.1155/2020/1705814
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9:e02331-17. doi: 10.1128/mBio.02331-17
Achal, V., Mukherjee, A., Kumari, D., and Zhang, Q. (2015). Biomineralization for sustainable construction – a review of processes and applications. Earth Sci. Rev. 148, 1–17. doi: 10.1016/j.earscirev.2015.05.008
Achinas, S., Charalampogiannis, N., and Euverink, G. J. (2019). A brief recap of microbial adhesion and biofilms. Appl. Sci. 9:2801. doi: 10.3390/app9142801
Aouame, A. E., Quars, F. E., Bentahar, Z., Zerouali, K., and Sidqui, M. (2021). In Vitro evaluation of bacterial adhesion to dental and stainless-steel surfaces. Open J. Med. Microbiol. 11, 176–197. doi: 10.4236/ojmm.2021.113014
Arciola, C. R., Campoccia, D., and Montanaro, L. (2018). Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409. doi: 10.1038/s41579-018-0019-y
Arias, D., Cisternas, L. A., Miranda, C., and Rivas, M. (2019). Bioprospecting of ureolytic bacteria from laguna salada for biomineralization applications. Front. Bioeng. Biotechnol. 6:209. doi: 10.3389/fbioe.2018.00209
Arnaouteli, S., Bamford, N. C., Stanley-Wall, N. R., and Kovács, Á. T. (2021). Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 19, 600–614. doi: 10.1038/s41579-021-00540-9
Benedetti, I., de Lorenzo, V., and Nikel, P. I. (2016). Genetic programming of catalytic Pseudomonas putida biofilms for boosting biodegradation of haloalkanes. Metab. Eng. 33, 109–118. doi: 10.1016/j.ymben.2015.11.004
Berne, C., Ellison, C. K., Ducret, A., and Brun, Y. V. (2018). Bacterial adhesion at the single-cell level. Nat. Rev. Microbiol. 16, 616–627. doi: 10.1038/s41579-018-0057-5
Bhagwat, G., O’Connor, W., Grainge, I., and Palanisami, T. (2021). Understanding the fundamental basis for biofilm formation on plastic surfaces: role of conditioning films. Front. Microbiol. 12:687118. doi: 10.3389/fmicb.2021.687118
Bhattacharjee, A., Khan, M., Kleiman, M., and Hochbaum, A. I. (2017). Effects of growth surface topography on bacterial signaling in coculture biofilms. ACS Appl. Mater. Interfaces 9, 18531–18539. doi: 10.1021/acsami.7b04223
Biesecker, S. G., Nicastro, L. K., Wilson, R. P., and Tukel, C. (2018). The functional amyloid curli protects Escherichia coli against complement-mediated bactericidal activity. Biomolecules 8:5. doi: 10.3390/biom8010005
Bilgili, D., Dündar, A., Barutçugil, Ç, Tayfun, D., and Özyurt, Ö. K. (2020). Surface properties and bacterial adhesion of bulk-fill composite resins. J. Dent. 95:103317. doi: 10.1016/j.jdent.2020.103317
Botyanszki, Z., Tay, P. K., Nguyen, P. Q., Nussbaumer, M. G., and Joshi, N. S. (2015). Engineered catalytic biofilms: site-specific enzyme immobilization onto E. coli curli nanofibers. Biotechnol. Bioeng. 112, 2016–2024. doi: 10.1002/bit.25638
Brück, H. L., Delvigne, F., Dhulster, P., Jacques, P., and Coutte, F. (2019). Molecular strategies for adapting Bacillus subtilis 168 biosurfactant production to biofilm cultivation mode. Bioresour. Technol. 293:122090. doi: 10.1016/j.biortech.2019.122090
Cao, Y., Feng, Y., Ryser, M. D., Zhu, K., Herschlag, G., Cao, C., et al. (2017). Programmable assembly of pressure sensors using pattern-forming bacteria. Nat. Biotechnol. 35, 1087–1093. doi: 10.1038/nbt.3978
Cao, Y., and Mezzenga, R. (2019). Food protein amyloid fibrils: origin, structure, formation, characterization, applications and health implications. Adv. Colloid Interface Sci. 269, 334–356. doi: 10.1016/j.cis.2019.05.002
Cao, Y. Y., Su, B., Chinnaraj, S., Jana, S., Bowen, L., Charlton, S., et al. (2018). Nanostructured titanium surfaces exhibit recalcitrance towards Staphylococcus epidermidis biofilm formation. Sci. Rep. 8:1071. doi: 10.1038/s41598-018-19484-x
Chang, Y. R., Weeks, E. R., Barton, D., Dobnikar, J., and Ducker, W. A. (2019). Effect of topographical steps on the surface motility of the bacterium Pseudomonas aeruginosa. ACS Biomater. Sci. Eng. 5, 6436–6445. doi: 10.1021/acsbiomaterials.9b00729
Chen, A. Y., Deng, Z., Billings, A. N., Seker, U. O., Lu, M. Y., Citorik, R. J., et al. (2014). Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523. doi: 10.1038/nmat3912
Chen, C., Petterson, T., Illergård, J., Ek, M., and Wågberg, L. (2019). Influence of cellulose charge on bacteria adhesion and viability to PVAm/CNF/PVAm-modified cellulose model surfaces. Biomacromolecules 20, 2075–2083. doi: 10.1021/acs.biomac.9b00297
Chen, Y., Feng, Y., Deveaux, J. G., Masoud, M. A., Chandra, F. S., Chen, H., et al. (2019). Biomineralization forming process and bio-inspired nanomaterials for biomedical application: a review. Minerals 9:68. doi: 10.3390/min9020068
Cheng, Y., Feng, G., and Moraru, C. I. (2019). Micro- and nanotopography sensitive bacterial attachment mechanisms: a review. Front. Microbiol. 10:191. doi: 10.3389/fmicb.2019.00191
Couasnon, T., Alloyeau, D., Ménez, B., Guyot, F., Ghigo, J.-M., and Gélabert, A. (2020). In situ monitoring of exopolymer-dependent Mn mineralization on bacterial surfaces. Sci. Adv. 6:eaaz3125. doi: 10.1126/sciadv.aaz3125
Cunha, A., Elie, A. M., Plawinski, L., Serro, A. P., Rego, A. M. B., Almeida, A., et al. (2016). Femto second laser surface texturing of titanium as a method to reduce the adhesion of Staphylococcus aureus and biofilm formation. Appl. Surf. Sci. 360, 485–493. doi: 10.1016/j.apsusc.2015.10.102
Del Pozo, J. L. (2018). Biofilm-related disease. Expert Rev. Anti Infect. Ther. 16, 51–65. doi: 10.1080/14787210.2018.1417036
Deshmukh, M., Evans, M. L., and Chapman, M. R. (2018). Amyloid by design: intrinsic regulation of microbial amyloid assembly. J. Mol. Biol. 430, 3631–3641. doi: 10.1016/j.jmb.2018.07.007
Ehrlich, H., Bailey, E., Wysokowski, M., and Jesionowski, T. (2021). Forced biomineralization: a review. Biomimetics 6:46. doi: 10.3390/biomimetics6030046
El Karoui, M., Hoyos-Flight, M., and Fletcher, L. (2019). Future trends in synthetic biology—a report. Front. Bioeng. Biotechnol. 7:175. doi: 10.3389/fbioe.2019.00175
Elizabeth, J. L., DeBenedictis, P., and Keten, S. (2016). Adhesion mechanisms of curli subunit CsgA to abiotic surfaces. Sci. Adv. 2:e1600998. doi: 10.1126/sciadv.1600998
Encinas, N., Yang, C. Y., Geyer, F., Kaltbeitzel, A., Baumli, P., Reinholz, J., et al. (2020). Submicrometer-sized roughness suppresses bacteria adhesion. ACS Appl. Mater. Interfaces 12, 21192–21200. doi: 10.1021/acsami.9b22621
Erskine, E., MacPhee, C. E., and Stanley-Wall, N. R. (2018). Functional amyloid and other protein fibers in the biofilm matrix. J. Mol. Biol. 430, 3642–3656. doi: 10.1016/j.jmb.2018.07.026
Fei, C., Mao, S., Yan, J., Alert, R., Stone, H. A., Bassler, B. L., et al. (2020). Nonuniform growth and surface friction determine bacterial biofilm morphology on soft substrates. Proc. Natl. Acad. Sci. U.S.A. 117, 7622–7632. doi: 10.1073/pnas.1919607117
Gallardo, R., Ranson, N. A., and Radford, S. E. (2020). Amyloid structures: much more than just a cross-β fold. Curr. Opin. Struct. Biol. 60, 7–16. doi: 10.1016/j.sbi.2019.09.001
Gao, L., Liu, Y., Kim, D., Li, Y., Hwang, G., Naha, P. C., et al. (2016). Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo. Biomaterials 101, 272–284. doi: 10.1016/j.biomaterials.2016.05.051
Geilich, B. M., Gelfat, I., Sridhar, S., van de Ven, A. L., and Webster, T. J. (2017). Superparamagnetic iron oxide-encapsulating polymersome nanocarriers for biofilm eradication. Biomaterials 119, 78–85. doi: 10.1016/j.biomaterials.2016.12.011
Gloag, E. S., Elbadawi, C., Zachreson, C. J., Aharonovich, I., Toth, M., Charles, I. G., et al. (2016). Micro-patterned surfaces that exploit stigmergy to inhibit biofilm expansion. Front. Microbiol. 7:2157. doi: 10.3389/fmicb.2016.02157
Gu, H., Chen, A., Song, X., Brasch, M. E., Henderson, J. H., and Ren, D. (2016a). How Escherichia coli lands and forms cell clusters on a surface: a new role of surface topography. Sci. Rep. 6:29516. doi: 10.1038/srep29516
Gu, H., Lee, S. W., Buffington, S. L., Henderson, J. H., and Ren, D. (2016b). On-demand removal of bacterial biofilms via shape memory activation. ACS Appl. Mater. Interfaces 8, 21140–21144. doi: 10.1021/acsami.6b06900
Gu, H., Kolewe, K. W., and Ren, D. (2017). Conjugation in Escherichia coli biofilms on poly(dimethylsiloxane) surfaces with microtopographic patterns. Langmuir 33, 3142–3150. doi: 10.1021/acs.langmuir.6b04679
Gu, H., Lee, S. W., Carnicelli, J., Zhang, T., and Ren, D. (2020). Magnetically driven active topography for long-term biofilm control. Nat. Commun. 11:2211. doi: 10.1038/s41467-020-16055-5
Guegan, C., Garderes, J., Le Pennec, G., Gaillard, F., Fay, F., Linossier, I., et al. (2014). Alteration of bacterial adhesion induced by the substrate stiffness. Colloids Surf. B Biointerfaces 114, 193–200. doi: 10.1016/j.colsurfb.2013.10.010
Gunes, B. (2021). A critical review on biofilm-based reactor systems for enhanced syngas fermentation processes. Renew. Sustain. Energy Rev. 143:110950. doi: 10.1016/j.rser.2021.110950
Guo, S., Kwek, M. Y., Toh, Z. Q., Pranantyo, D., Kang, E.-T., Loh, X. J., et al. (2018). Tailoring polyelectrolyte architecture to promote cell growth and inhibit bacterial adhesion. ACS Appl. Mater. Interfaces 10, 7882–7891. doi: 10.1021/acsami.8b00666
Hall, C. W., and Mah, T.-F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Handorf, A. M., Zhou, Y., Halanski, M. A., and Li, W. J. (2015). Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis 11, 1–15. doi: 10.1080/15476278.2015.1019687
Heckmann, T. S., and Schiffman, J. D. (2020). Spatially organized nanopillar arrays dissimilarly affect the antifouling and antibacterial activities of Escherichia coli and Staphylococcus aureus. ACS Appl. Nano Mater. 3, 977–984. doi: 10.1021/acsanm.9b01942
Helbig, R., Gunther, D., Friedrichs, J., Rossler, F., Lasagni, A., and Werner, C. (2016). The impact of structure dimensions on initial bacterial adhesion. Biomater. Sci. 4, 1074–1078. doi: 10.1039/c6bm00078a
Hoffmann, T. D., Reeksting, B. J., and Gebhard, S. (2021). Bacteria-induced mineral precipitation: a mechanistic review. Microbiology 167:001049. doi: 10.1099/mic.0.001049
Huang, J., Liu, S., Zhang, C., Wang, X., Pu, J., Ba, F., et al. (2019). Programmable and printable Bacillus subtilis biofilms as engineered living materials. Nat. Chem. Biol. 15, 34–41. doi: 10.1038/s41589-018-0169-2
Hubenova, Y., Hubenova, E., Burdin, B., Vladikova, D., and Mitov, M. (2019). Development of coupled redox active network in Ca-alginate polymer for immobilization of Pseudomonas putida 1046 on electrode surface. Electrochim. Acta 312, 432–440. doi: 10.1016/j.electacta.2019.05.012
Hwang, G., Paula, A. J., Hunter, E. E., Liu, Y., Babeer, A., Karabucak, B., et al. (2019). Catalytic antimicrobial robots for biofilm eradication. Sci. Robot. 4:eaaw2388. doi: 10.1126/scirobotics.aaw2388
Ivanova, E. P., Linklater, D. P., Werner, M., Baulin, V. A., Xu, X., Vrancken, N., et al. (2020). The multi-faceted mechano-bactericidal mechanism of nanostructured surfaces. Proc. Natl. Acad. Sci. U.S.A. 117, 12598–12605. doi: 10.1073/pnas.1916680117
Jang, Y., Choi, W. T., Johnson, C. T., Garcia, A. J., Singh, P. M., Breedveld, V., et al. (2018). Inhibition of bacterial adhesion on nanotextured stainless steel 316L by electrochemical etching. ACS Biomater. Sci. Eng. 4, 90–97. doi: 10.1021/acsbiomaterials.7b00544
Jiang, R., Hao, L., Song, L., Tian, L., Fan, Y., Zhao, J., et al. (2020). Lotus-leaf-inspired hierarchical structured surface with non-fouling and mechanical bactericidal performances. Chem. Eng. J. 398:125609. doi: 10.1016/j.cej.2020.125609
Jothi Prakash, C. G., and Prasanth, R. (2021). Approaches to design a surface with tunable wettability: a review on surface properties. J. Mater. Sci. 56, 108–135. doi: 10.1007/s10853-020-05116-1
Kim, M. K., Zhao, A., Wang, A., Brown, Z. Z., Muir, T. W., Stone, H. A., et al. (2017). Surface-attached molecules control Staphylococcus aureus quorum sensing and biofilm development. Nat. Microbiol. 2:17080. doi: 10.1038/nmicrobiol.2017.80
Knowles, T. P. J., and Mezzenga, R. (2016). Amyloid fibrils as building blocks for natural and artificial functional materials. Adv. Mater. 28, 6546–6561. doi: 10.1002/adma.201505961
Kolewe, K. W., Peyton, S. R., and Schiffman, J. D. (2015). Fewer bacteria adhere to softer hydrogels. ACS Appl. Mater. Interfaces 7, 19562–19569. doi: 10.1021/acsami.5b04269
Kolewe, K. W., Zhu, J., Mako, N. R., Nonnenmann, S. S., and Schiffman, J. D. (2018). Bacterial adhesion is affected by the thickness and stiffness of poly(ethylene glycol) hydrogels. ACS Appl. Mater. Interfaces 10, 2275–2281. doi: 10.1021/acsami.7b12145
Kovačević, D., Pratnekar, R., Godić Torkar, K., Salopek, J., Dražić, G., Abram, A., et al. (2016). Influence of polyelectrolyte multilayer properties on bacterial adhesion capacity. Polymers 8:345. doi: 10.3390/polym8100345
Krasowska, A., and Sigler, K. (2014). How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 4:112. doi: 10.3389/fcimb.2014.00112
Krsmanovic, M., Biswas, D., Ali, H., Kumar, A., Ghosh, R., and Dickerson, A. K. (2021). Hydrodynamics and surface properties influence biofilm proliferation. Adv. Colloid Interface Sci. 288:102336. doi: 10.1016/j.cis.2020.102336
Kyzioł, A., Khan, W., Sebastian, V., and Kyzioł, K. (2020). Tackling microbial infections and increasing resistance involving formulations based on antimicrobial polymers. Chem. Eng. J. 385:123888. doi: 10.1016/j.cej.2019.123888
Lee, S. W., Carnicelli, J., Getya, D., Gitsov, I., Phillips, K. S., and Ren, D. (2021). Biofilm removal by reversible shape recovery of the substrate. ACS Appl. Mater. Interfaces 13, 17174–17182. doi: 10.1021/acsami.0c20697
Levering, V., Wang, Q., Shivapooja, P., Zhao, X., and Lopez, G. P. (2014). Soft robotic concepts in catheter design: an on-demand fouling-release urinary catheter. Adv. Healthc. Mater. 3, 1588–1596. doi: 10.1002/adhm.201400035
Li, C., Qin, R., Liu, R., Miao, S., and Yang, P. (2018). Functional amyloid materials at surfaces/interfaces. Biomater. Sci. 6, 462–472. doi: 10.1039/C7BM01124E
Li, J., and Zhao, X. (2020). Effects of quorum sensing on the biofilm formation and viable but non-culturable state. Food Res. Int. 137:109742. doi: 10.1016/j.foodres.2020.109742
Lichter, J. A., Thompson, M. T., Delgadillo, M., Nishikawa, T., Rubner, M. F., and Van Vliet, K. J. (2008). Substrata mechanical stiffness can regulate adhesion of viable bacteria. Biomacromolecules 9, 1571–1578. doi: 10.1021/bm701430y
Linklater, D. P., Baulin, V. A., Juodkazis, S., Crawford, R. J., Stoodley, P., and Ivanova, E. P. (2020). Mechano-bactericidal actions of nanostructured surfaces. Nat. Rev. Microbiol. 19, 8–22. doi: 10.1038/s41579-020-0414-z
Linklater, D. P., Baulin, V. A., Juodkazis, S., and Ivanova, E. P. (2018). Mechano-bactericidal mechanism of graphene nanomaterials. Interface Focus 8:20170060. doi: 10.1098/rsfs.2017.0060
Linklater, D. P., Juodkazis, S., Crawford, R. J., and Ivanova, E. P. (2019). Mechanical inactivation of Staphylococcus aureus and Pseudomonas aeruginosa by titanium substrata with hierarchical surface structures. Materialia 5:100197. doi: 10.1016/j.mtla.2018.100197
Liu, T., Guo, Z., Zeng, Z., Guo, N., Lei, Y., Liu, T., et al. (2018). Marine bacteria provide lasting anticorrosion activity for steel via biofilm-induced mineralization. ACS Appl. Mater. Interfaces 10, 40317–40327. doi: 10.1021/acsami.8b14991
Liu, Y., Naha, P. C., Hwang, G., Kim, D., Huang, Y., Simon-Soro, A., et al. (2018). Topical ferumoxytol nanoparticles disrupt biofilms and prevent tooth decay in vivo via intrinsic catalytic activity. Nat. Commun. 9:2920. doi: 10.1038/s41467-018-05342-x
Long, M., Long, X., Zheng, C.-W., Luo, Y.-H., Zhou, C., and Rittmann, B. E. (2021). Para-Chlorophenol (4-CP) removal by a palladium-coated biofilm: coupling catalytic dechlorination and microbial mineralization via denitrification. Environ. Sci. Technol. 55, 6309–6319. doi: 10.1021/acs.est.0c08307
Lorenzetti, M., Dogša, I., Stošicki, T., Stopar, D., Kalin, M., Kobe, S., et al. (2015). The influence of surface modification on bacterial adhesion to titanium-based substrates. ACS Appl. Mater. Interfaces 7, 1644–1651. doi: 10.1021/am507148n
Lu, A., Gao, Y., Jin, T., Luo, X., Zeng, Q., and Shang, Z. (2020). Effects of surface roughness and texture on the bacterial adhesion on the bearing surface of bio-ceramic joint implants: an in vitro study. Ceram. Int. 46, 6550–6559. doi: 10.1016/j.ceramint.2019.11.139
Mahanta, U., Khandelwal, M., and Deshpande, A. S. (2021). Antimicrobial surfaces: a review of synthetic approaches, applicability and outlook. J. Mater. Sci. 56, 17915–17941. doi: 10.1007/s10853-021-06404-0
Mahdinia, E., Demirci, A., and Berenjian, A. (2019). Biofilm reactors as a promising method for vitamin K (menaquinone-7) production. Appl. Microbiol. Biotechnol. 103, 5583–5592. doi: 10.1007/s00253-019-09913-w
Marvasi, M., Mastromei, G., and Perito, B. (2020). Bacterial calcium carbonate mineralization in situ strategies for conservation of stone artworks: from cell components to microbial community. Front. Microbiol. 11:1386. doi: 10.3389/fmicb.2020.01386
McCarty, N. S., and Ledesma-Amaro, R. (2019). Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 37, 181–197. doi: 10.1016/j.tibtech.2018.11.002
Meier, M., Dubois, V., and Seeger, S. (2018). Reduced bacterial colonisation on surfaces coated with silicone nanostructures. Appl. Surf. Sci. 459, 505–511. doi: 10.1016/j.apsusc.2018.08.003
Meinshausen, A. K., Herbster, M., Zwahr, C., Soldera, M., Muller, A., Halle, T., et al. (2021). Aspect ratio of nano/microstructures determines Staphylococcus aureus adhesion on PET and titanium surfaces. J. Appl. Microbiol. 131, 1498–1514. doi: 10.1111/jam.15033
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Mukhi, M., Vishwanathan, A. S., and Nojiri, H. (2022). Beneficial biofilms: a minireview of strategies to enhance biofilm formation for biotechnological applications. Appl. Environ. Microbiol. 88:e01994-21. doi: 10.1128/AEM.01994-21
Nakanishi, E. Y., Palacios, J. H., Godbout, S., and Fournel, S. (2021). Interaction between biofilm formation, surface material and cleanability considering different materials used in pig facilities—an overview. Sustainability 13:5836. doi: 10.3390/su13115836
Nguyen, D. H. K., Pham, V. T. H., Truong, V. K., Sbarski, I., Wang, J., Balcytis, A., et al. (2018). Role of topological scale in the differential fouling of Pseudomonas aeruginosa and Staphylococcus aureus bacterial cells on wrinkled gold-coated polystyrene surfaces. Nanoscale 10, 5089–5096. doi: 10.1039/c7nr08178b
Nguyen, P. Q. (2017). Synthetic biology engineering of biofilms as nanomaterials factories. Biochem. Soc. Trans. 45, 585–597. doi: 10.1042/BST20160348
Nguyen, P. Q., Botyanszki, Z., Tay, P. K., and Joshi, N. S. (2014). Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 5:4945. doi: 10.1038/ncomms5945
Oh, J. K., Yegin, Y., Yang, F., Zhang, M., Li, J., Huang, S., et al. (2018). The influence of surface chemistry on the kinetics and thermodynamics of bacterial adhesion. Sci. Rep. 8:17247. doi: 10.1038/s41598-018-35343-1
Ozcelik, B., Ho, K. K. K., Glattauer, V., Willcox, M., Kumar, N., and Thissen, H. (2017). Poly(ethylene glycol)-based coatings combining low-biofouling and quorum-sensing inhibiting properties to reduce bacterial colonization. ACS Biomater. Sci. Eng. 3, 78–87. doi: 10.1021/acsbiomaterials.6b00579
Pandit, A., Adholeya, A., Cahill, D., Brau, L., and Kochar, M. (2020). Microbial biofilms in nature: unlocking their potential for agricultural applications. J. Appl. Microbiol. 129, 199–211. doi: 10.1111/jam.14609
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Peter, A., Lutey, A. H. A., Faas, S., Romoli, L., Onuseit, V., and Graf, T. (2020). Direct laser interference patterning of stainless steel by ultrashort pulses for antibacterial surfaces. Opt. Laser Technol. 123:105954. doi: 10.1016/j.optlastec.2019.105954
Pranantyo, D., Xu, L. Q., Kang, E.-T., and Chan-Park, M. B. (2018). Chitosan-based peptidopolysaccharides as cationic antimicrobial agents and antibacterial coatings. Biomacromolecules 19, 2156–2165. doi: 10.1021/acs.biomac.8b00270
Qin, W., Wang, C. Y., Ma, Y. X., Shen, M. J., Li, J., Jiao, K., et al. (2020). Microbe-mediated extracellular and intracellular mineralization: environmental, industrial, and biotechnological applications. Adv. Mater. 32:e1907833. doi: 10.1002/adma.201907833
Quan, K., Zhang, Z., Chen, H., Ren, X., Ren, Y., Peterson, B. W., et al. (2019). Artificial channels in an infectious biofilm created by magnetic nanoparticles enhanced bacterial killing by antibiotics. Small 15:e1902313. doi: 10.1002/smll.201902313
Reinhardt, T., Moelzner, J., Neu, T. R., and Fink, P. (2020). Biofilm pads—an easy method to manufacture artificial biofilms embedded in an alginate polymer matrix. Limnol. Oceanogr. Methods 18, 1–7.
Ren, Y. J., Wang, C., Chen, Z., Allan, E., van der Mei, H. C., and Busscher, H. J. (2018). Emergent heterogeneous microenvironments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing. FEMS Microbiol. Rev. 42, 259–272. doi: 10.1093/femsre/fuy001
Rigo, S., Cai, C., Gunkel-Grabole, G., Maurizi, L., Zhang, X., Xu, J., et al. (2018). Nanoscience-based strategies to engineer antimicrobial surfaces. Adv. Sci. 5:1700892. doi: 10.1002/advs.201700892
Rijavec, T., Zrimec, J., van Spanning, R., and Lapanje, A. (2019). Natural microbial communities can be manipulated by artificially constructed biofilms. Adv. Sci. 6:1901408. doi: 10.1002/advs.201901408
Romero, C. M., Martorell, P. V., Lopez, A. G., Penalver, C. G. N., Chaves, S., and Mechetti, M. (2018). Architecture and physicochemical characterization of Bacillus biofilm as a potential enzyme immobilization factory. Colloids Surf. B Biointerfaces 162, 246–255. doi: 10.1016/j.colsurfb.2017.11.057
Rosenzweig, R., Perinbam, K., Ly, V. K., Ahrar, S., Siryaporn, A., and Yee, A. F. (2019). Nanopillared surfaces disrupt Pseudomonas aeruginosa mechanoresponsive upstream motility. ACS Appl. Mater. Interfaces 11, 10532–10539. doi: 10.1021/acsami.8b22262
Rzhepishevska, O., Hakobyan, S., Ruhal, R., Gautrot, J., Barbero, D., and Ramstedt, M. (2013). The surface charge of anti-bacterial coatings alters motility and biofilm architecture. Biomater. Sci. 1, 589–602. doi: 10.1039/c3bm00197k
Salama, A., Hasanin, M., and Hesemann, P. (2020). Synthesis and antimicrobial properties of new chitosan derivatives containing guanidinium groups. Carbohydr. Polym. 241:116363. doi: 10.1016/j.carbpol.2020.116363
Schultz, M. P., Bendick, J. A., Holm, E. R., and Hertel, W. M. (2011). Economic impact of biofouling on a naval surface ship. Biofouling 27, 87–98.
Secchi, E., Vitale, A., Mino, G. L., Kantsler, V., Eberl, L., Rusconi, R., et al. (2020). The effect of flow on swimming bacteria controls the initial colonization of curved surfaces. Nat. Commun. 11:2851. doi: 10.1038/s41467-020-16620-y
Senevirathne, S. W. M. A. I., Hasan, J., Mathew, A., Woodruff, M., and Yarlagadda, P. K. D. V. (2021). Bactericidal efficiency of micro- and nanostructured surfaces: a critical perspective. RSC Adv. 11, 1883–1900. doi: 10.1039/d0ra08878a
Sharma, V., Srinivasan, A., Nikolajeff, F., and Kumar, S. (2021). Biomineralization process in hard tissues: the interaction complexity within protein and inorganic counterparts. Acta Biomater. 120, 20–37. doi: 10.1016/j.actbio.2020.04.049
Shi, Y., Lin, J., Tao, X., Qu, J., Liao, S., Li, M., et al. (2021). Harnessing colloidal self-assembled patterns (cSAPs) to regulate bacterial and human stem cell response at biointerfaces in vitro and in vivo. ACS Appl. Mater. Interfaces 13, 20982–20994. doi: 10.1021/acsami.1c02591
Shi, Y., Liu, K., Zhang, Z., Tao, X., Chen, H.-Y., Kingshott, P., et al. (2020). Decoration of material surfaces with complex physicochemical signals for biointerface applications. ACS Biomater. Sci. Eng. 6, 1836–1851. doi: 10.1021/acsbiomaterials.9b01806
Siddiqui, S., Chandrasekaran, A., Lin, N., Tufenkji, N., and Moraes, C. (2019). Microfluidic shear assay to distinguish between bacterial adhesion and attachment strength on stiffness-tunable silicone substrates. Langmuir 35, 8840–8849. doi: 10.1021/acs.langmuir.9b00803
Simsek, A. N., Braeutigam, A., Koch, M. D., Shaevitz, J. W., Huang, Y., Gompper, G., et al. (2019). Substrate-rigidity dependent migration of an idealized twitching bacterium. Soft Matter. 15, 6224–6236. doi: 10.1039/c9sm00541b
Singh, A., and Kaushik, A. (2021). Removal of Cd and Ni with enhanced energy generation using biocathode microbial fuel cell: insights from molecular characterization of biofilm communities. J. Clean. Prod. 315:127940.
Song, F., and Ren, D. (2014). Stiffness of cross-linked poly(dimethylsiloxane) affects bacterial adhesion and antibiotic susceptibility of attached cells. Langmuir 30, 10354–10362. doi: 10.1021/la502029f
Song, F., Brasch, M. E., Wang, H., Henderson, J. H., Sauer, K., and Ren, D. (2017). How bacteria respond to material stiffness during attachment: a role of Escherichia coil flagellar motility. ACS Appl. Mater. Interfaces 9, 22176–22184. doi: 10.1021/acsami.7b04757
Spencer, K. C., Sy, J. C., Ramadi, K. B., Graybiel, A. M., Langer, R., and Cima, M. J. (2017). Characterization of mechanically matched hydrogel coatings to improve the biocompatibility of neural implants. Sci. Rep. 7:1952.
Spriano, S., Sarath Chandra, V., Cochis, A., Uberti, F., Rimondini, L., Bertone, E., et al. (2017). How do wettability, zeta potential and hydroxylation degree affect the biological response of biomaterials? Mater. Sci. Eng. C Mater. Biol. Appl. 74, 542–555. doi: 10.1016/j.msec.2016.12.107
Taglialegna, A., Lasa, I., and Valle, J. (2016). Amyloid structures as biofilm matrix scaffolds. J. Bacteriol. 198, 2579–2588.
Talluri, S. N. L., Winter, R. M., and Salem, D. R. (2020). Conditioning film formation and its influence on the initial adhesion and biofilm formation by a cyanobacterium on photobioreactor materials. Biofouling 36, 183–199.
Todhanakasem, T. (2017). Developing microbial biofilm as a robust biocatalyst and its challenges. Biocatal. Biotransformation 35, 86–95.
Truong, V. K., Lapovok, R., Estrin, Y. S., Rundell, S., Wang, J. Y., Fluke, C. J., et al. (2010). The influence of nano-scale surface roughness on bacterial adhesion to ultrafine-grained titanium. Biomaterials 31, 3674–3683. doi: 10.1016/j.biomaterials.2010.01.071
Truong, V. K., Pham, V. T., Medvedev, A., Lapovok, R., Estrin, Y., Lowe, T. C., et al. (2015). Self-organised nanoarchitecture of titanium surfaces influences the attachment of Staphylococcus aureus and Pseudomonas aeruginosa bacteria. Appl. Microbiol. Biotechnol. 99, 6831–6840. doi: 10.1007/s00253-015-6572-7
Vadillo-Rodriguez, V., Guerra-Garcia-Mora, A. I., Perera-Costa, D., Gonzalez-Martin, M. L., and Fernandez-Calderon, M. C. (2018). Bacterial response to spatially organized microtopographic surface patterns with nanometer scale roughness. Colloids Surf. B Biointerfaces 169, 340–347. doi: 10.1016/j.colsurfb.2018.05.038
Valenzuela, L., Faraldos, M., Bahamonde, A., and Rosal, R. (2021). Critical review on the use of photocatalysis and photoelectrocatalysis to create antimicrobial surfaces. Curr. Opin. Chem. Eng. 34:100762.
Van Gerven, N., Klein, R. D., Hultgren, S. J., and Remaut, H. (2015). Bacterial amyloid formation: structural insights into curli biogensis. Trends Microbiol. 23, 693–706. doi: 10.1016/j.tim.2015.07.010
Van Gerven, N., Van der Verren, S. E., Reiter, D. M., and Remaut, H. (2018). The role of functional amyloids in bacterial virulence. J. Mol. Biol. 430, 3657–3684.
Verderosa, A. D., Totsika, M., and Fairfull-Smith, K. E. (2019). Bacterial biofilm eradication agents: a current review. Front. Chem. 7:824. doi: 10.3389/fchem.2019.00824
Wang, N., Gao, J., Liu, Y., Wang, Q., Zhuang, X., and Zhuang, G. (2021). Realizing the role of N-acyl-homoserine lactone-mediated quorum sensing in nitrification and denitrification: a review. Chemosphere 274:129970. doi: 10.1016/j.chemosphere.2021.129970
Wang, W., Timonen, J. V. I., Carlson, A., Drotlef, D. M., Zhang, C. T., Kolle, S., et al. (2018). Multifunctional ferrofluid-infused surfaces with reconfigurable multiscale topography. Nature 559, 77–82. doi: 10.1038/s41586-018-0250-8
Wang, X., Pu, J., An, B., Li, Y., Shang, Y., Ning, Z., et al. (2018). Programming cells for dynamic assembly of inorganic nano-objects with spatiotemporal control. Adv. Mater. 30:e1705968. doi: 10.1002/adma.201705968
Wang, Y., An, B., Xue, B., Pu, J., Zhang, X., Huang, Y., et al. (2021). Living materials fabricated via gradient mineralization of light-inducible biofilms. Nat. Chem. Biol. 17, 351–359. doi: 10.1038/s41589-020-00697-z
Wang, Y., Guan, A., Isayeva, I., Vorvolakos, K., Das, S., Li, Z., et al. (2016). Interactions of Staphylococcus aureus with ultrasoft hydrogel biomaterials. Biomaterials 95, 74–85. doi: 10.1016/j.biomaterials.2016.04.005
Wassmann, T., Kreis, S., Behr, M., and Buergers, R. (2017). The influence of surface texture and wettability on initial bacterial adhesion on titanium and zirconium oxide dental implants. Int. J. Implant Dent. 3:32. doi: 10.1186/s40729-017-0093-3
Wu, S., Zuber, F., Maniura-Weber, K., Brugger, J., and Ren, Q. (2018b). Nanostructured surface topographies have an effect on bactericidal activity. J. Nanobiotechnol. 16:20.
Wu, S., Altenried, S., Zogg, A., Zuber, F., Maniura-Weber, K., and Ren, Q. (2018a). Role of the surface nanoscale roughness of stainless steel on bacterial adhesion and microcolony formation. ACS Omega 3, 6456–6464. doi: 10.1021/acsomega.8b00769
Wurzler, N., Hidde, G., Schenderlein, M., and Ozcan, O. (2022). Effect of organic conditioning layers adsorbed on stainless steel AISI 304 on the attachment and biofilm formation of electroactive bacteria Shewanella putrefaciens CN32. Eng. Rep. 4:e12458.
Xiao, K., Cao, X., Chen, X., Hu, H., and Wu, C. (2020). Bactericidal efficacy of nanopatterned surface tuned by topography. J. Appl. Phys. 128:064701.
Yang, X., Li, Z., Xiao, H., Wang, N., Li, Y., Xu, X., et al. (2018). A Universal and ultrastable mineralization coating bioinspired from biofilms. Adv. Funct. Mater. 28:1802730.
Yao, S., Jin, B., Liu, Z., Shao, C., Zhao, R., Wang, X., et al. (2017). Biomineralization: from material tactics to biological strategy. Adv. Mater. 29:1605903.
Yow-Ren Chang, E. R. W., and Ducker, W. A. (2018). Surface topography hinders bacterial surface motility. ACS Appl. Mater. Interfaces 10, 9225–9234.
Zeng, Z., Guo, X.-P., Li, B., Wang, P., Cai, X., Tian, X., et al. (2015). Characterization of self-generated variants in Pseudoalteromonas lipolytica biofilm with increased antifouling activities. Appl. Microbiol. Biotechnol. 99, 10127–10139. doi: 10.1007/s00253-015-6865-x
Zhang, C., Huang, J., Zhang, J., Liu, S., Cui, M., An, B., et al. (2019). Engineered Bacillus subtilis biofilms as living glues. Mater. Today 28, 40–48.
Zhao, X., Zhao, F., Wang, J., and Zhong, N. (2017). Biofilm formation and control strategies of foodborne pathogens: food safety perspectives. RSC Adv. 7, 36670–36683. doi: 10.1007/s00253-012-4611-1
Zhao, Y., Liu, D., Huang, W., Yang, Y., Ji, M., Nghiem, L. D., et al. (2019). Insights into biofilm carriers for biological wastewater treatment processes: current state-of-the-art, challenges, and opportunities. Bioresour. Technol. 288:121619. doi: 10.1016/j.biortech.2019.121619
Zhong, C., Gurry, T., Cheng, A. A., Downey, J., Deng, Z., Stultz, C. M., et al. (2014). Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 9, 858–866. doi: 10.1038/nnano.2014.199
Keywords: biofilm, bacteria, materiobiology, extracellular matrix, gene editing
Citation: Shi Y, Chen T, Shaw P and Wang P-Y (2022) Manipulating Bacterial Biofilms Using Materiobiology and Synthetic Biology Approaches. Front. Microbiol. 13:844997. doi: 10.3389/fmicb.2022.844997
Received: 29 December 2021; Accepted: 13 June 2022;
Published: 07 July 2022.
Edited by:
Sanket J. Joshi, Sultan Qaboos University, OmanReviewed by:
David Karig, Clemson University, United StatesCopyright © 2022 Shi, Chen, Shaw and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng-Yuan Wang, cHkud2FuZ0BvamxhYi5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.