- 1State Key Laboratory of Dao-di Herbs, National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 2School of Pharmaceutical Sciences, Peking University, Beijing, China
Gastrodia elata is an entirely heterotrophic plant, the growth of which is completely reliant on Armillaria gallica, an orchid mycorrhizal fungus. To avoid damaging ecosystems, G. elata cultivation is shifting from woodland to farmland. However, whether the microbial community structure remains stable during this conversation is unknown. Here, we cultivated G. elata in woodland or farmland and found that woodland-cultivated G. elata produced a greater yield and larger tuber size. The relative abundance of Rahnella was 22.84- and 122.25-fold higher in woodland- and farmland-cultivated soil samples, respectively, than that in uncultivated soil samples. To investigate how Rahnella impacts the growth of G. elata and establishes symbiosis with Armillaria gallica, three Rahnella spp. strains (HPDA25, SBD3, and SBD11) were isolated from mycorrhizosphere soil samples. It was found that these strains, especially HPDA25, promoted the growth of A. gallica. Ultra-performance liquid chromatography coupled to a triple quadrupole mass spectrometry analysis detected the indole-3-acetic acid with 16.24 ng/ml in HPDA25 fermentation solution. Co-culturing with the strain HPDA25 or exogenous indole-3-acetic acid increased the branching and fresh weight of rhizomorphs and the growth rate and extracellular laccase activity of A. gallica, compared with A. gallica cultured alone. The results of RNA-seq and quantitative real-time polymerase chain reaction analysis showed that co-culturing A. gallica with HPDA25 increased the expression level of the genes including hydrophobin, SUR7/PalI family, and pectin methylesterase, whereas decreased the expression levels of glycolysis-related genes. Furthermore, co-culturing with the strain HPDA25, A. gallica promotes the growth of G. elata and enhances the tuber size of G. elata. These results provide new insights into an orchid mycorrhizal symbiosis and the cultivation of G. elata.
Introduction
Plants establish associations with mutualistic fungi and bacteria to exchange key nutrients and thrive (Wang et al., 2021). Orchidaceae, one of the most diverse and widely distributed plant families, is partially or fully dependent on mycorrhizal fungi during the life cycle (Lu et al., 2019). The popular herb Gastrodia elata, a member of the orchid family, is widely distributed in Asian countries such as South Korea, Japan, and China (Chen et al., 2019). As a completely heterotrophic plant, G. elata spends more than 80% of its life underground as a tuber, associating with the mycorrhizal fungus Armillaria to supply the required nutrition (Yuan et al., 2018). Armillaria is well known as a contributor to carbon cycling via woody tissue breakdown (Alveshere et al., 2021) and is used in G. elata cultivation in China.
Abundant biodiversity in the plant environment leads to a productive ecosystem, improving yield and agricultural sustainability (Reiss and Drinkwater, 2018). For example, core microbial communities increased the drought tolerance of Adenium obesum, Aloe dhufarensis, and Cleome austroarabica (Khan et al., 2020). The isolation and culture of potentially beneficial microorganisms could increase Panax notoginseng yields in a continuous cropping system (Li et al., 2020). G. elata interacts with a diverse spectrum of bacteria during its growth before the arrival of Armillaria (Yuan et al., 2018). We hypothesize that bacteria also play an important role in the establishment of a symbiotic relationship between Armillaria gallica and G. elata. However, the dynamics of bacterial communities during the growth of G. elata under different environments is unknown.
Plant growth-promoting bacteria (PGPB) have been extensively described in the context of sustainable agricultural systems, which directly or indirectly promote plant growth (Hassan, 2017). As important beneficial microorganisms, PGPB play vital roles in improving plant growth and crop production (Ha-Tran et al., 2021). The mycorrhizosphere’s bacterial and fungal communities are extremely varied and complicated (Wagg et al., 2019), influencing plant development and symbiosis (Herrera et al., 2020). Co-inoculation with PGPB and arbuscular mycorrhizal (AM) fungi increased nutrient uptake in plants, leading to high crop yields (Emmanuel and Babalola, 2020), supporting the idea that crop yields may be positively correlated with soil microbial richness (Kieck et al., 2016). However, how mycorrhizosphere bacteria affect the growth of G. elata and A. gallica is poorly understood.
Apart from their roles in the plant–fungus symbiosis, root-associated bacteria also contribute to plant fitness, variety, and coexistence via bi- or tripartite interactions between plant hosts and mycorrhizal fungi (Kaur and Sharma, 2021). Temperature, pH, soil type, vegetation, and landscapes could influence the microbial populations in the soil (Finkel et al., 2017), and the plant diversity also increases the diversity of soil microorganisms (Stefan et al., 2021). Till now, a final conclusion has not yet been reached on the function of the bacterial communities during the development phases of G. elata and A. gallica.
Here, we compared the mycorrhizosphere bacterial communities, agronomic traits, and yield of G. elata cultivated in woodland or farmland. We aimed to understand the dynamics of the mycorrhizosphere bacteria associated with G. elata and A. gallica under different cultivation environments and the function of mycorrhizosphere bacteria in the growth of G. elata and A. gallica. We also tried to isolate growth-promoting mycorrhizosphere bacteria and to investigate the mechanisms of orchid mycorrhizal (OM) microbiome associations.
Materials and Methods
Cultivation, Harvest, and Agronomic Traits Analysis of G. elata
Gastrodia elata was planted and harvested in woodland or farmland landscapes (33°309″N, 108°828″E) in Ningshan (Shaanxi Province, China) in 2019. A total of 16 plant holes, 30 cm (length) × 30 cm (width) × 70 cm (depth), were dug in each landscape. In March, five wood sticks (diameter = 5–12 cm, length = 30 cm) fully infected with A. gallica were placed in parallel at the bottom of each plant hole, in which 0.2 kg/m2 of G. elata tubers were planted. The cultivation and production processes are shown in Figure 1A. In November, mycorrhizosphere soil samples, including the surface soil samples of farmland (FC)- and woodland (WC)-cultivated G. elata mature tuber and A. gallica rhizomorphs, were collected. Uncultivated soil samples from farmland (UNFC) and woodland (UNWC) were collected as control. Meanwhile, the yield of G. elata was evaluated, and agronomic traits such as weight, length, and width in each mature tuber (Figure 1B) were measured. For agronomic characteristics analysis, a total of 121 mature tubers were measured in woodland, and 119 mature tubers were measured in farmland.
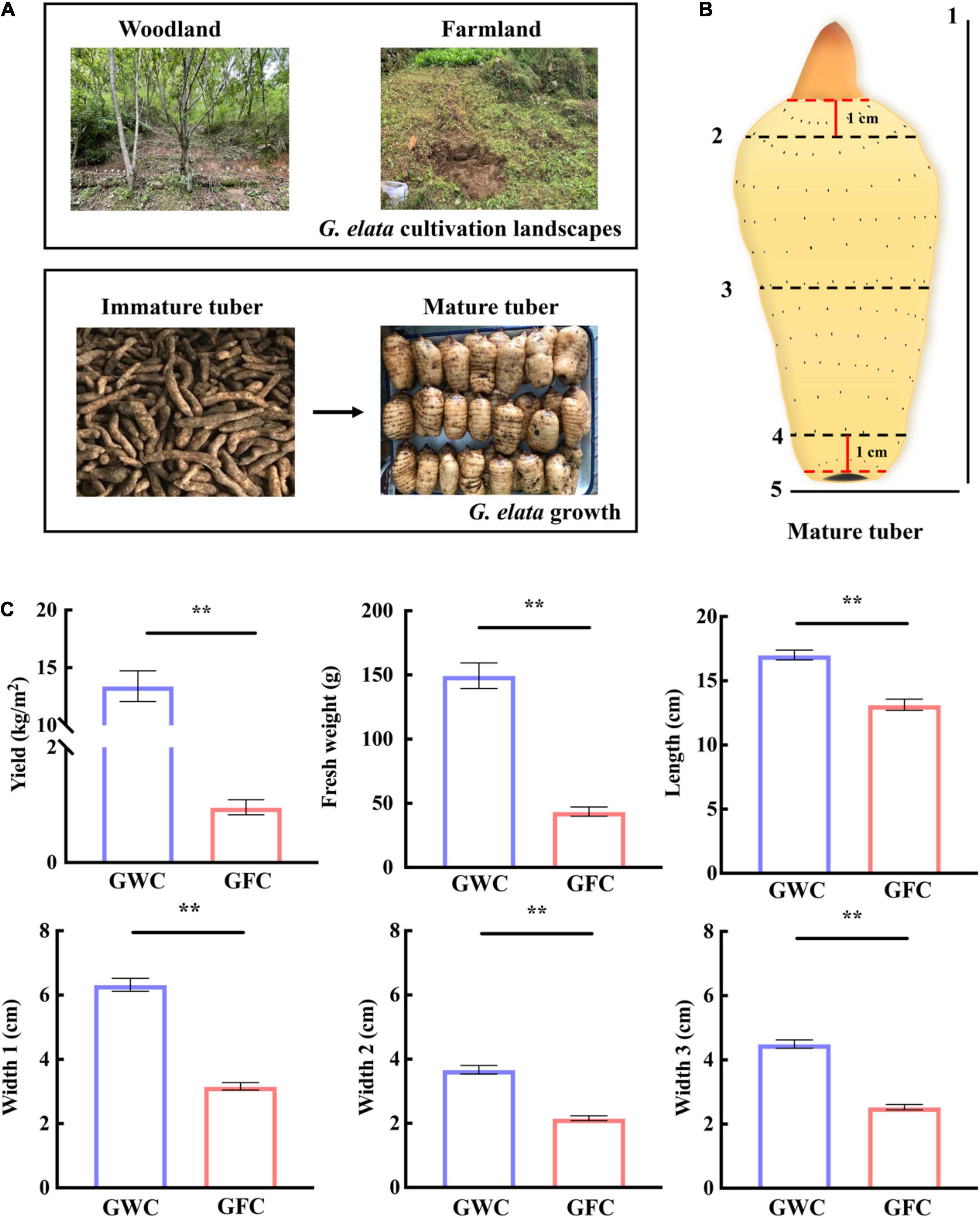
Figure 1. G. elata cultivation, growth, and evaluation of mature tuber agronomic traits. (A) Cultivation of G. elata in woodland or farmland. (B) Agronomic traits of G. elata mature tubers. 1, length; 2, width at 1 cm from top bud (Width 2); 3, half of tuber width (Width 1); 4, width at 1 cm from tuber bottom (Width 3); 5, weight. (C) Yield, tuber weight, and agronomic trait evaluation of G. elata. GWC, G. elata cultivated in woodland; GFC, G. elata cultivated in farmland. Data are shown as means ± SE. **P < 0.01.
Diversity of Bacterial Community
The total genomic DNA was extracted from mycorrhizosphere soil samples using the CTAB method. Each landscape was sampled for a total of 16 soil samples. 16S rRNA genes were amplified using the specific primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Caporaso et al., 2011). All polymerase chain reaction (PCR) reactions were carried out using Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, United States). PCR products were purified with the Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany). Sequencing libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, United States) following the manufacturer’s recommendations. Library quality was assessed using a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). The library was sequenced using the Illumina HiSeq 2500 platform, and 250-bp paired-end reads were generated.
FASTQ files from each sample were processed using Cutadapt version V1.9.1 software1 (Langille et al., 2013) for preliminary quality control of raw data. Briefly, paired-end reads were joined, depleted of barcodes, and trimmed, and chimeras were removed. The clean sequencing reads were clustered into operational taxonomic units (OTUs) with 97% identity using UPARSE version 7.0.1001 software2 (Haas et al., 2011). According to the algorithm principle, sequences with the highest frequencies were selected as the representative sequences of OTUs.
QIIME2 software (Hall and Beiko, 2018) and the SSU rRNA database (Wang et al., 2007) of SILVA 1323 (Edgar, 2013) were used to perform species annotation analysis of the OTU sequences (set threshold 0.8–1) to obtain taxonomic information and statistics of the community composition of each sample at each classification level (kingdom, phylum, class, order, family, genus, and species). MUSCLE version 3.8.31 software (Quast et al., 2013)4 was used to perform rapid multiple sequence alignment to obtain the phylogenetic relationships of all OTU sequences.
Finally, data were normalized to that of the sample with the least amount of data. The subsequent diversity of bacterial community analyses and Spearman correlation analysis between bacterial community and agronomic traits of G. elata tuber were performed in NovoMagic v3.05.
Isolation and Identification of Mycorrhizosphere Bacteria
The soil on the tubers was rinsed off with Milli-Q water, and the tubers were then ultrasonically cleaned three times (120 W, 40 kHz, 5 min). The fresh tubers were subsequently rinsed three times with sterile water, cut into small pieces, and homogenized with 2-ml sterile water. A final 106-fold diluted suspension was prepared using sterile water, and 100 μl of the tissue suspension was plated onto various media, including potato dextrose agar (PDA), Luria-Bertani medium (LB), tryptone soya agar, and nutrient agar. Plates were incubated at 30°C for 2–5 days or until bacterial growth was observed (Luo et al., 2019). All experiments were performed in triplicate. LB medium was used for subsequent purification, growth, and maintenance of bacterial strains. All isolated strains were purified and stored in 50% glycerol at −80°C.
Amplification of the 16S rRNA gene was performed using the primers 27F (5′-CAGAGTTTGATCCTGGCT-3′) and 1492R (5′-AGGAGGTGATCCAGCCGCA-3′) (Ying et al., 2012). The PCR mixture (50 μl) contained 2-μl DNA template, 25-μl 2 × M5 HiPer Taq Mix (Mei5bio, Beijing, China), 19-μl ddH2O, and 2 μl of each primer (10 μM). Amplification of the 16S rRNA genes was performed using the following thermal cycling steps: initial denaturation at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The amplified products were purified and sequenced by BGI Genomics (Beijing, China). The obtained sequences were identified using EzBioCloud6 and the National Center for Biotechnology Information BLAST7. Multiple sequence alignments and genetic distance calculations were performed using the CLUSTAL X program (Larkin et al., 2007), and a neighbor-joining phylogenetic dendrogram was generated using MEGA X software (Kumar et al., 2018). The 16S rRNA gene sequences determined in this study were deposited in GenBank under accession numbers OL700214–OL700216.
Co-cultivation Assay of Mycorrhizosphere Bacterial Strains and A. gallica
Armillaria gallica strain for the co-cultivation assay was previously isolated from G. elata tubers and preserved in the laboratory. A. gallica was incubated at 25°C in the dark for 14 days. The stored bacterial strains were incubated in a solid LB medium at 28°C for 12 h. Fresh rhizomorphs were used for the co-culture experiments. The isolated mycorrhizosphere bacterial strains were cultured in a liquid LB medium at 28°C to an optical density at a wavelength of 600 nm of approximately 0.5 (1.47 × 1010CFU/ml for HPDA25, 1.87 × 1010 CFU/ml for SBD3, and 1.73 × 1010 CFU/ml for SBD11). After that, 100 μl of bacterial liquid was co-cultured with A. gallica on a PDA medium in the dark for 7 days at 25°C. As controls, A. gallica was cultured without bacterial liquid. After separating A. gallica from the medium, the fresh rhizomorph’s weight, growth rate, branching, and extracellular laccase activity of A. gallica were assessed. Six biological replicates were performed for each condition. Separating A. gallica rhizomorphs was used as extracellular laccase activity, indole-3-acetic acid (IAA) content determination, transcriptome sequencing, and quantitative polymerase chain reaction (qPCR).
Extracellular Laccase Activity Analysis
In separating A. gallica from the medium, the medium was weighted (approximately 4 g) and centrifuged at 13,000 rpm for 15 min. The supernatant (1 ml) was then diluted with Milli-Q water to a final volume of 10 ml for extracellular laccase activity determination. The extracellular laccase activity was measured using the 2,2-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid (ABTS) method (Quijada-Morin et al., 2018). The reaction system contained 100 μl of 0.2 mol/L disodium hydrogen phosphate-citric acid buffer (pH = 4.0), 50-μl ABTS, and 50-μl dilute enzyme solution. The absorbance of ABTS and enzyme solution mixture was measured 10 times using a microplate reader (Thermo Fisher Scientific) at 420 nm. One unit of laccase activity was defined as the amount of laccase required to catalyze the oxidation of 1-μmol ABTS per minute in the reaction system. Six biological replicates were performed for each condition.
Indole-3-Acetic Acid Content Determination
HPDA25 was cultured in 20 ml of LB medium at 28°C and 200 rpm until the optical density at a wavelength of 600 nm of approximately 0.5 (1.47 × 1010 CFU/ml). After that, the fermentation solution was centrifuged at 12,000 rpm for 15 min, and the supernatant was removed under nitrogen gas. Subsequently, 70% methanol was added to obtain a final volume of 10 ml, and the mixture was passed through a 0.22-μm filter. Approximately 0.1 g separating A. gallica rhizomorphs was used as IAA content determination.
Indole-3-acetic acid (CAS: 87-51-4) standards were purchased from MedChemExpress (Monmouth Junction, NJ, United States), and hormone levels were determined using ultra-performance liquid chromatography coupled to a triple quadrupole mass spectrometry. The mobile phase consisted of 0.1% formic acid water (B) and acetonitrile (A) (v/v) with the following gradient: 10% A (0–0.3 min), 10–60% A (0.3–3 min), 60–95% A (3–6 min), 10% A (6–6.2 min), and 10% A (6.2–7 min). The equilibration time and the flow rate were set at 7 min and 0.5 ml/min, respectively. The ACQUITY UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm; Waters) was used with a temperature of 40°C, sample plate temperature of 4°C, a sample volume of 1 μl, and analysis time of 7 min. Electrospray ionization was used with 176.000/130.100 (m/z) (ESI+) and 174.000/130.100 (m/z) (ESI−) for IAA (Jiang et al., 2017). The scanning mode captured positive and negative ion data using multi-reaction detection. The pressure of the curtain gas was 30 psi, the ionization voltage was −4,500 V/+5,500 V, the spray gas pressure was 50 psi, the auxiliary heating gas pressure was 50 psi, the acquisition method was +-MRM, and the ion source temperature was 550°C.
Growth of A. gallica in the Medium Containing Indole-3-Acetic Acid
Approximately 16.24 ng/ml IAA was added to the PDA medium, and the A. gallica rhizomorphs were cultured in a 25-ml IAA-containing PDA medium. Also, the control group was cultured in a 25-ml PDA medium supplemented with 70% methanol solution. A. gallica was incubated at 25°C in the dark for 7 days, and the fresh rhizomorph’s weight, growth rate, branching, and extracellular laccase activity of A. gallica were measured. Five biological replicates were performed for each condition.
Transcriptome Sequencing and Quantitative Polymerase Chain Reaction of Co-cultured A. gallica With HPDA25
Fresh A. gallica rhizomorphs were isolated from the medium. RNA was extracted from three biological replicates of cultivated A. gallica rhizomorphs. The RNA integrity was assessed using the Bioanalyzer 2100 system (Agilent Technologies, CA, United States). Sequencing libraries were constructed using the Next Ultra RNA Library Prep Kit for Illumina (New England Biolabs). The 150-bp paired-end reads were then generated by Illumina HiSeq 4000 platform. Quality control of RNA-seq reads was performed using FastQC software (default parameters) (Wang et al., 2015; Ewels et al., 2016). Quality-filtered reads were aligned to the A. gallica genome GWHASIQ00000000 [National Genomics Data Center (NGDC), China]8 using STAR software (Dobin et al., 2013). Read counts were obtained with featureCounts v1.5.0-p3. Differential expression analysis was performed using the DESeq2 R package (1.20.0). The transcript levels with adjusted P-value ≤ 0.05 and a fold change (log2FC) ≥1 were designated as significant differentially expressed genes (DEGs; Fradj et al., 2020). Each DEG was annotated using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes, and the enrichment analysis was performed using the clusterProfiler R package (Wu et al., 2021).
We used quantitative real-time polymerase chain reaction (qRT-PCR) to validate the DEGs that were enriched by GO and Kyoto Encyclopedia of Genes and Genomes annotation. The primers of reference gene (18S) and DEG primers were designed using the Primer3Plus software9 and synthesized by BGI Genomics (Beijing, China) (Supplementary Table 1). A. gallica RNA was reverse transcribed to cDNA using the TransScript II First-Strand cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The cDNA was diluted to 200 μl with sterile water. Tip Green qPCR SuperMix Kit (TransGen Biotech, Beijing, China) was used to conduct RT-PCR on LightCycler 480 II (Roche Applied Science, Mannheim, Germany). Each RT-PCR mixture contained 2-μl cDNA, 1-μl primers, 5-μl Tip Green qPCR SuperMix, and 2-μl nuclease-free water. The qRT-PCR conditions were as follows: initial denaturation at 94°C for 30 s, followed by denaturation at 94°C for 5 s and annealing of primers at 60°C for 20 s, and extension at 72°C for 20 s. The samples were cooled to 60°C and then heated to 94°C by 40 cycles, and the melting curves were generated. Delta delta Ct was used for statistical analysis. Three replicates were performed.
HPDA25 Co-cultured With A. gallica and G. elata
Short-cut woods were sterilized at 121°C for 120 min. Tubers of G. elata were surface sterilized using the following three steps: washing in 75% ethanol for 30 s, in 2% NaOCl for 15 min, followed by rinsing 10 times in sterile distilled water. To verify the effectiveness of the surface sterilization, the distilled water from the final rinse was plated in LB medium and then incubated at 25°C for 5 days. Surface sterilization was considered effective when no microbial growth was observed in the medium. First, HPDA25 fermentation solution (300 μl) and 1% agar were mixed in 100-ml distilled water as the medium. The same volume (300 μl) of LB medium and 1% agar were used as control. Then, the sterilized wood that interacted with A. gallica rhizomorphs for 20 days and surface-sterilized tubers were added to this medium. Finally, another 100-μl HPDA25 fermentation solution was added to the medium surface and cultured at 25°C for 30 days (Khalil et al., 2021). In the control treatment, 100-μl LB medium was also applied to the medium surface. Six biological replicates were performed for each condition.
Statistical Analysis
All statistical analysis was performed using IBM SPSS version 21.0 (IBM Corp., Armonk, NY, United States). Comparisons between control and treatment groups were analyzed by the Student’s T-test, and comparisons for more than two treatments were performed using one-way analysis of variance followed by Duncan’s post hoc test. Data are presented as means ± SEM. The analysis of similarities was performed using NovoMagic v3.0 (see text footnote 5). Graphs were constructed using GraphPad Prism 9 software (GraphPad Software, La Jolla, CA, United States), TBtools (Chen et al., 2020), and Hiplot10, a comprehensive web platform for scientific data visualization.
Data Availability
The 16S sequence datasets are available at the NGDC Genome Sequence Archive BioProject number PRJCA00550611. The transcriptome dataset can be found at the NGDC Genome Sequence Archive BioProject number PRJCA00550812.
Results
Yield and Agronomic Traits of G. elata
Gastrodia elata was first cultivated in woodland and then transferred to farmland for protecting the ecosystem in recent years in China. Here, we tried to compare the effect of two cultivation methods on the growth of G. elata (Figure 1A). For a clear description of the mature tuber, three tuber widths were measured, including the width at half of the tuber (Width 1), the width at 1 cm from the top bud (Width 2), and the width at 1 cm from the tuber bottom (Width 3) (Figure 1B). When cultivated in woodland (GWC), the average fresh weight of one G. elata tuber was 149.51 g, and the yield can reach 13.40 kg/m2, which were 3.43-fold and 13.97-fold higher than those cultivated in farmland (GFC). The lengths and widths (Width 1–3) of woodland-cultivated tubers were also larger than those of farmland-cultivated tubers, and Width 1 of woodland-cultivated tubers were twofold larger than that of farmland-cultivated tubers (Figure 1C). These results showed that G. elata cultivated in woodland produced heavier, longer, and wider tubers than G. elata cultivated in farmland, leading to a higher yield.
Diversity, Isolation, and Identification of Mycorrhizosphere Bacteria
Although the dynamics of fungal communities during G. elata growth have been reported (Chen et al., 2019), the bacterial communities are unknown. To further investigate whether the bacterial communities affect the tuber growth of G. elata, the diversity of mycorrhizosphere bacteria under two cultivation environments was investigated based on 16S rRNA gene sequencing. A total of 5,674 OTUs were used for diversity analysis. The sequenced samples were sufficient to reveal the true diversity because the Good’s coverage was higher than 0.97 in all soil samples (Supplementary Table 2). The result of non-metric multidimensional scaling and analysis of similarities showed a clear separation (stress = 0.098) and the differences of bacterial communities between WC/FC and UNWC/UNFC soil samples (Supplementary Figure 1 and Supplementary Table 3), suggesting G. elata could regulate the composition and abundance of mycorrhizosphere bacterial communities. The results also showed that the bacterial community diversity in uncultivated soil samples (UNWC, UNFC) was significantly higher than that in cultivated soil samples (WC, FC) (Supplementary Table 2).
The relative abundance of the 14 genera, including Pseudomonas, Novosphingobium, Flavobacterium, Rahnella, Variovorax, Bradyrhizobium, Massilia, Collimonas, Duganella, Raoultella, Limnohabitans, Dyella, Rhizobacter, and Mucilaginibacter, were increased in WC soil samples compared with that in UNWC soil samples and were also increased in FC soil samples compared with that in UNFC soil samples (Figures 2A,B and Supplementary Table 4). Furthermore, 11 genera among these 14 genera were associated with the enhancement of tuber growth in G. elata (Supplementary Figure 2). In particular, the relative abundance of genus Rahnella was 22.84-fold higher in WC soil samples than that in UNWC soil samples and 122.25-fold higher in FC soil samples than that in UNFC soil samples (Supplementary Table 4), suggesting that Rahnella could be involved in the growth promotion of G. elata tuber and that the relative abundance of Rahnella is affected by the cultivation environments.
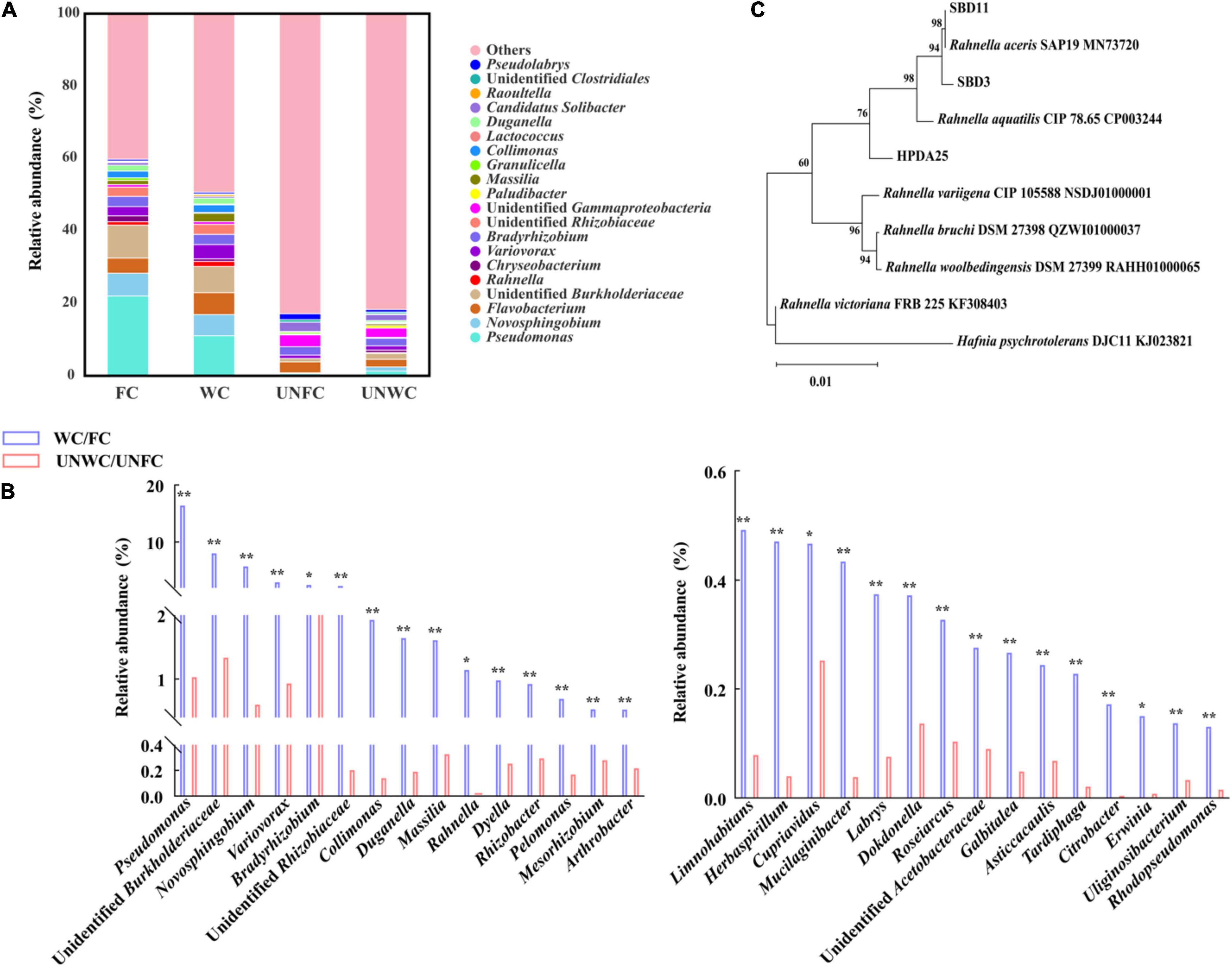
Figure 2. Mycorrhizosphere bacterial diversity. (A) Mycorrhizosphere bacteria whose relative abundance was in top 30 at genus levels. (B) Top 30 genera with a significantly increased relative abundance in WC/FC soil samples compared with that in UNWC/UNFC soil samples. (C) Phylogenetic tree (neighbor-joining) of Rahnella isolates. WC, G. elata cultivated mycorrhizosphere soil from woodland cultivating G. elata; FC, mycorrhizosphere soil from farmland cultivating G. elata; UNWC, mycorrhizosphere soil from woodland uncultivating G. elata; UNFC, G. elata cultivated mycorrhizosphere soil from farmland uncultivating G. elata. *P < 0.05; **P < 0.01.
Rahnella spp. Co-cultured With A. gallica and G. elata
To verify the growth promotion on G. elata tuber of these Rahnella spp., three Rahnella spp. strains (HPDA25, SBD3, and SBD11) were isolated from mycorrhizosphere soils (Figure 2C) and identified using 16S rRNA sequences (Supplementary Table 5). We co-cultured these three Rahnella spp. strains (HPDA25, SBD3, and SBD11) with A. gallica and G. elata, respectively. The results showed that co-cultivation with HPDA25 or SBD3 increased the fresh weight of rhizomorphs (4.62- and 4.03-fold, respectively), growth rate (4.63- and 4.03-fold, respectively), rhizomorph branching (2.74- and 2.48-fold, respectively), and extracellular laccase activity (1.64- and 1.81-fold, respectively). SBD11 increased the fresh weight of rhizomorphs (2.02-fold), growth rate (2.02-fold), and rhizomorph branching (1.95-fold) and decreased extracellular laccase activity (0.93-fold) of A. gallica (Figures 3A,B). Among the three strains of Rahnella spp., HPDA25 was the predominant strain involved in the growth promotion of A. gallica.
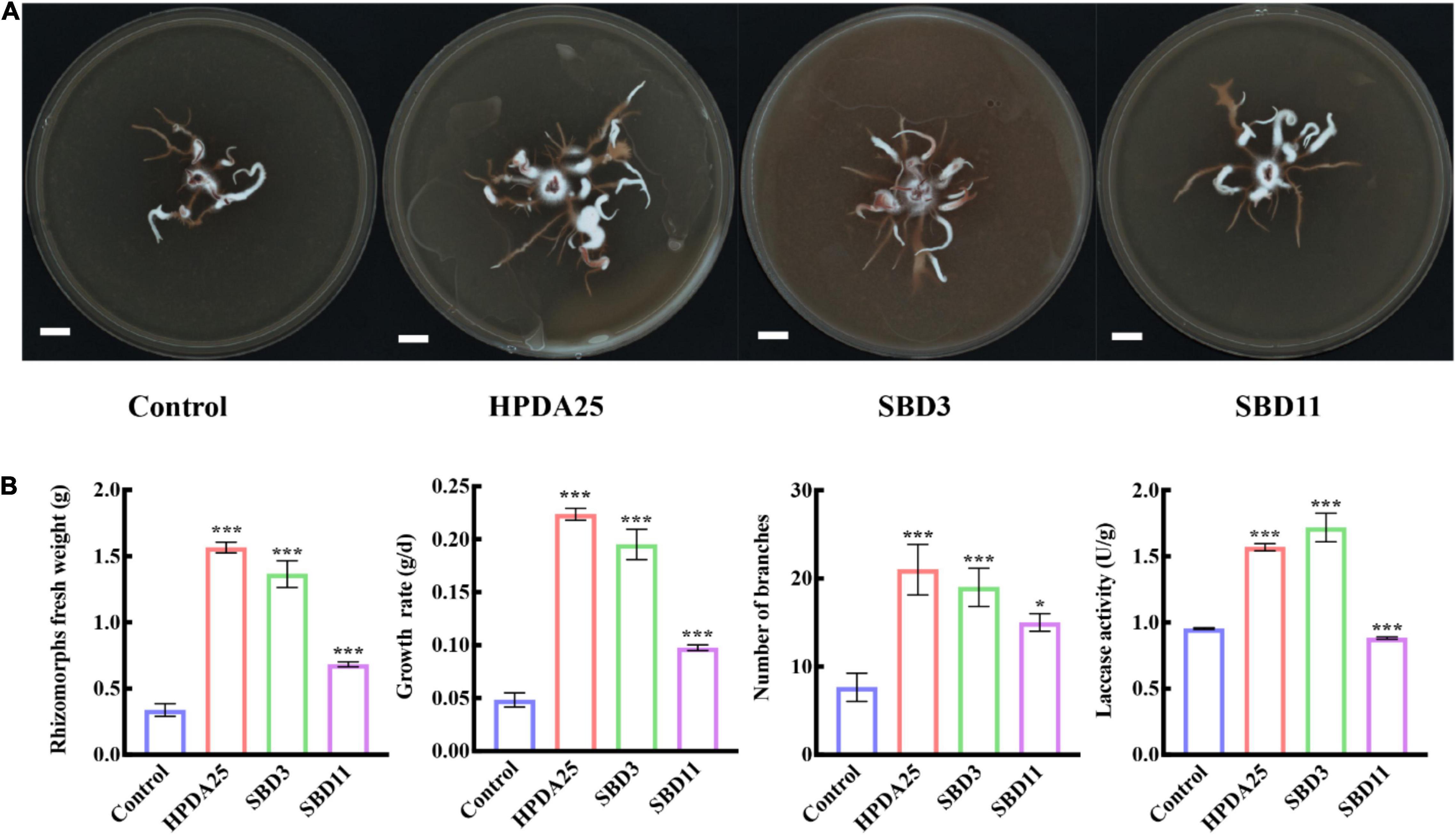
Figure 3. Rahnella spp. co-cultured with A. gallica. (A) A. gallica co-cultured with three Rahnella spp. (B) Rahnella spp. increased rhizomorph’s fresh weight, growth rate, branching, and extracellular laccase activity of A. gallica. Control, A. gallica cultured alone in medium. Data are shown as means ± SE. *P < 0.05; ***P < 0.001.
Furthermore, co-culturing HPDA25 with the symbiotic G. elata–A. gallica system (Figure 4A) also increased the fresh weight of A. gallica rhizomorphs (2.15-fold) and G. elata tubers (19.00-fold) (Figures 4B,C). These results demonstrate that HPDA25 could promote the growth of both A. gallica and G. elata.
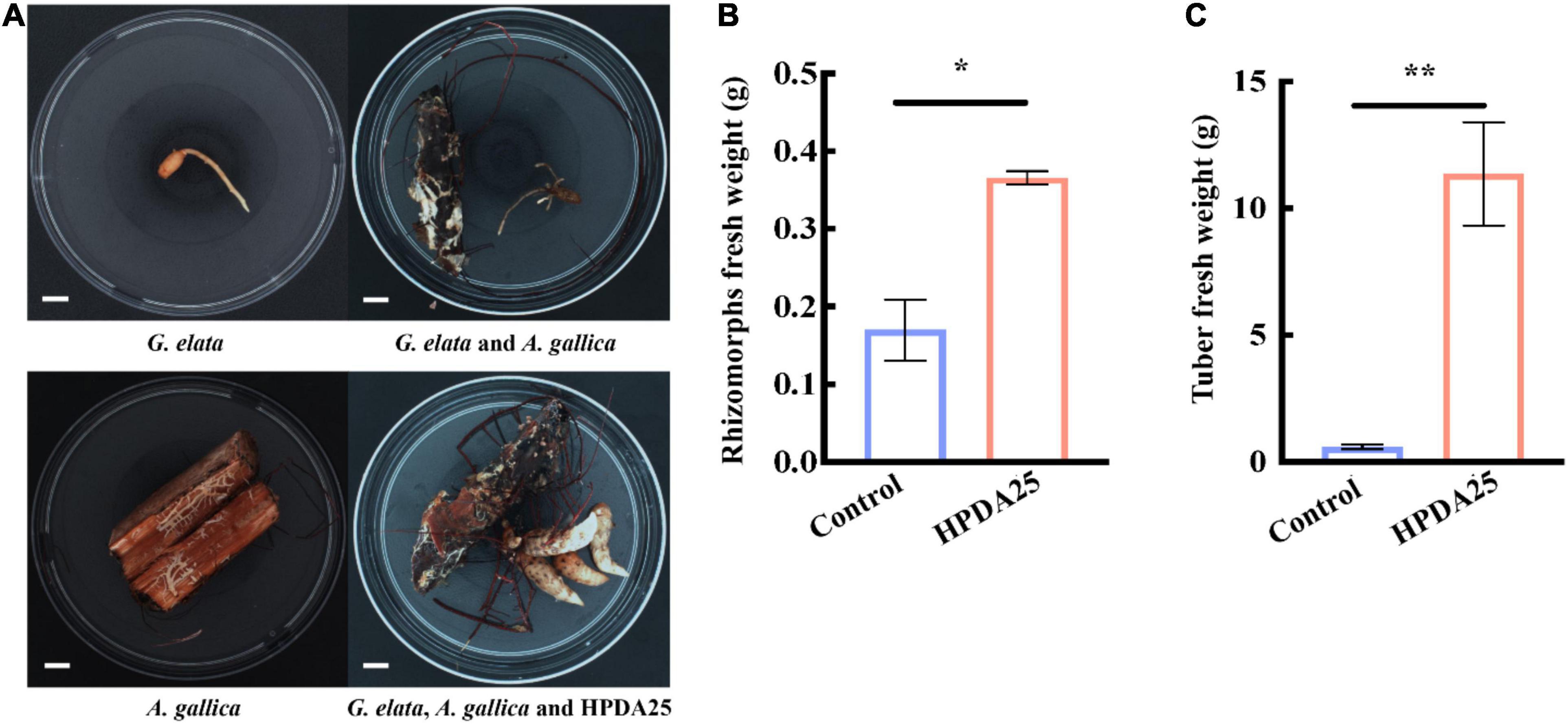
Figure 4. HPDA25 promoted both A. gallica and G. elata growth. (A) HPDA25, A. gallica, and G. elata growth. (B) HPDA25 increased fresh weight of A. gallica rhizomorphs. (C) HPDA25 increased fresh weight of G. elata. Control, G. elata co-cultured with A. gallica. Data are shown as means ± SE. *P < 0.05; **P < 0.01.
HPDA25 Produced Indole-3-Acetic Acid and Promoted the Growth of A. gallica
Auxin treatment stimulates AM fungal growth, mycorrhization formation (Gutjahr, 2014), and arbuscule formation (Guillotin et al., 2017). It has also been reported that Rahnella spp. produced IAA and interacted with AMF extraradical mycelium (Zhang et al., 2019; Yuan et al., 2020). To further investigate the mechanism of HPDA25 promoting the growth of A. gallica and G. elata, IAA content was measured in HPDA25 fermentation solution using ultra-performance liquid chromatography coupled to a triple quadrupole mass spectrometry, and the standard curve data for IAA are shown in Supplementary Table 6. A peak attributed to IAA was detected in the fermentation solution of the HPDA25 (Figure 5A), with 16.24 ng produced in 1 ml of fermentation solution (Figure 5B).
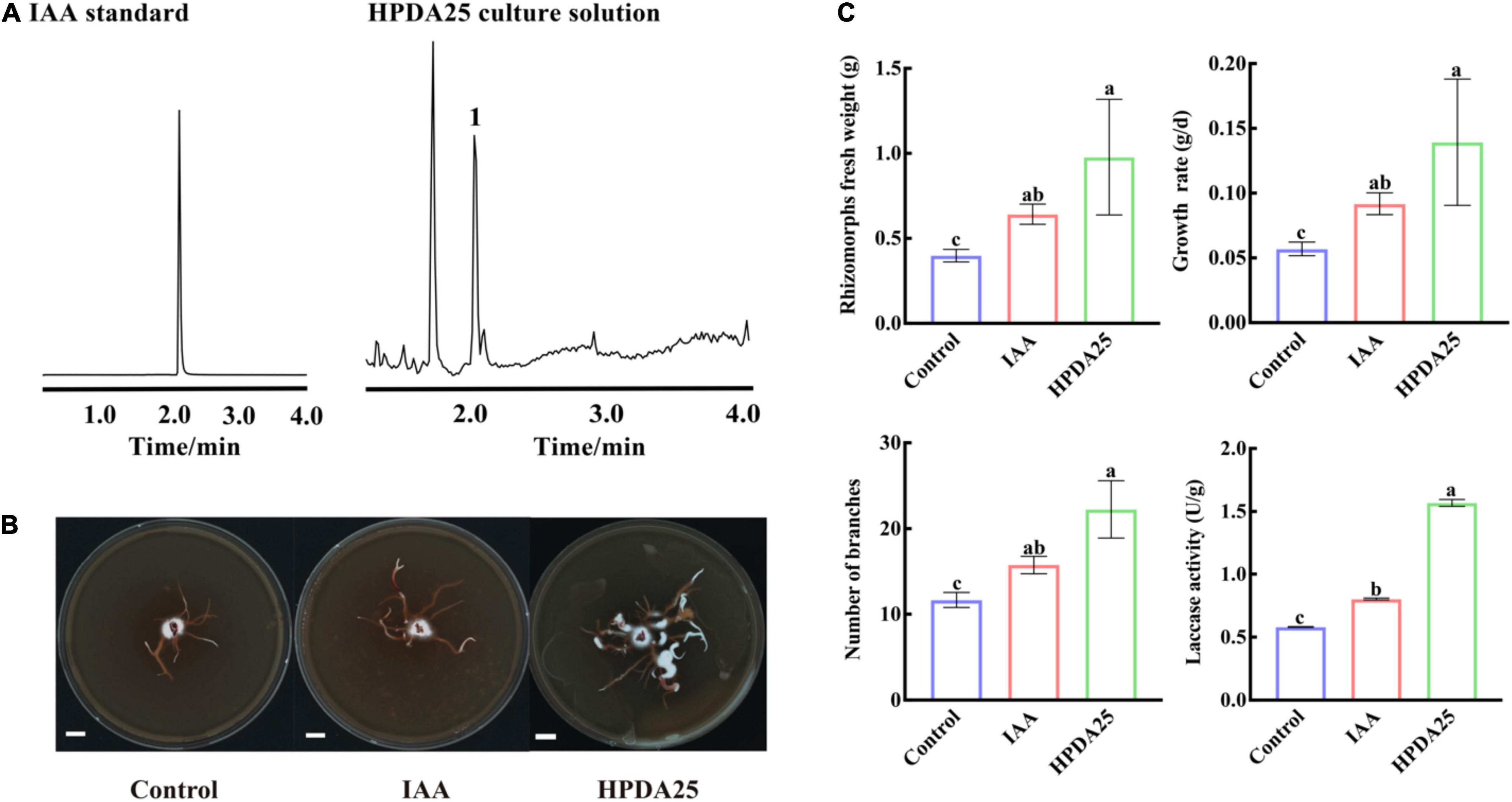
Figure 5. HPDA25 secreted 2.6 indole-3-acetic acid (IAA) and promoted A. gallica growth. (A) IAA was detected by ultra-performance liquid chromatography coupled to a triple quadrupole mass spectrometry. (B) A. gallica cultured in different mediums. (C) IAA increased rhizomorph’s fresh weight, growth rate, branching, and extracellular laccase activity of A. gallica. Control, A. gallica cultured alone in medium. Data are shown as means ± SE. Different letters indicate a significant difference at P < 0.05 level.
To further confirm the effect of IAA on the growth of A. gallica, an equal amount (approximately 16.24 ng/ml) of exogenous IAA was added to the medium to replace HPDA25. The results clearly showed that the supplement of exogenous IAA led to a 1.61-fold increase of the fresh weight of rhizomorphs, 1.61-fold growth rate, 1.35-fold rhizomorph branching, and 1.38-fold extracellular laccase activity of A. gallica (Figure 5C). Both exogenous IAA and co-culturing HPDA25 treatment substantially improve the growth of A. gallica, revealing that IAA produced in HPDA25 may be the major contributor promoting HPDA25-mediated G. elata growth.
Differentially Expressed Genes in HPDA25-Mediated Promotion of A. gallica Growth
To explain the growth-promotion mechanism of HPDA25, we compared gene-expressed levels between A. gallica cultured alone and that co-cultured with HPDA25. A total of 4,658 upregulated DEGs and 2,669 downregulated DEGs of A. gallica co-cultured with HPDA25 were identified (Supplementary Figure 3). The results of GO annotation showed that upregulated DEGs are related to the structural constituent of the cell wall and structural molecule activity in molecular function (Figure 6A and Supplementary Table 7), and downregulated DEGs are related to glycolysis/gluconeogenesis (Supplementary Table 8). Twenty-four DEGs that encode hydrophobin (HFBs), Sur7/PalI family transmembrane protein, pectin methylesterase (PME), enolase (ENO), pyruvate kinase (PYK), aldehyde dehydrogenase (ALDH), or pyruvate decarboxylase (PDC) were selected to perform qRT-PCR (Supplementary Table 9), and the qRT-PCR results were consistent with the results of transcriptome analysis (Supplementary Figure 4).
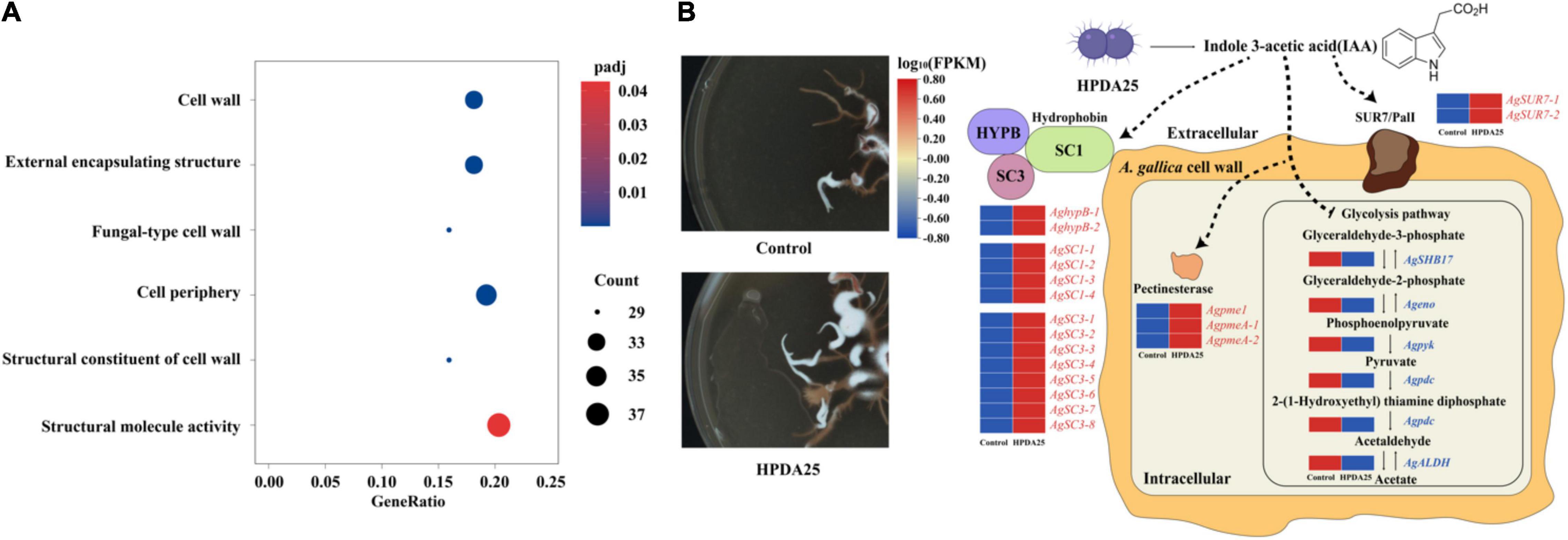
Figure 6. Mechanism of HPDA25 promotes growth of A. gallica. (A) Number of DEGs in A. gallica co-cultured with HPDA25 in GO annotation. (B) Mechanism of HPDA25-mediated promotion of A. gallica growth. Upregulated DEGs were in red words; downregulated DEGs were in blue words.
Discussion
Plants directly face various environmental challenges and thus have to recruit beneficial bacteria to cope with biotic and abiotic stress (Li et al., 2021). Mycorrhizal microbiomes are essential for exploring interactions between mycorrhizal fungi and their surrounding environments (Bahram et al., 2020). In this study, we investigated the mycorrhizosphere bacterial diversity and identified three Rahnella spp. strains (HPDA25, SBD3, and SBD11), which are beneficial for the increase of the yield and tuber size of G. elata (Supplementary Figure 2). Rahnella aquatilis is widely distributed (Yuan et al., 2020), and plants can recruit Rahnella as beneficial bacteria (Li et al., 2014). We also found that cultivating G. elata enhances the relative abundance of Rahnella but reduces bacterial diversity in cultivated soil samples, indicating that G. elata could only recruit beneficial Rahnella for its growth.
Rahnella aquatilis adapts to a variety of environments, which may be related to the ability of resistance (Yuan et al., 2020). To avoid damaging ecosystems, G. elata cultivation is shifting from woodland to farmland, and we also found that the increased relative abundance of Rahnella was higher in soil samples of farmland cultivating G. elata than those of woodland (Supplementary Table 4), suggesting that G. elata could recruit more beneficial Rahnella to accelerate the establishment of a symbiotic relationship between A. gallica and G. elata in an adverse environment.
As the third component of plant–mycorrhizal fungal–bacterial symbiosis, bacteria that are associated with mycorrhizal fungi (hypersphere) also play a critical role in mycorrhizal function (Agnolucci et al., 2020). In recent years, an intimate cooperative relationship between AM fungi mycelium and bacteria has been observed via microscopic observation (Scheublin et al., 2010) and molecular analyses (Wang et al., 2016). These bacterial communities could affect the growth of mycelia and the formation of mycorrhiza (Kim et al., 2013), promote the colonization of AMF (Omirou et al., 2016), and play a role in the material exchange between mycelia and plant host (Emmanuel and Babalola, 2020). However, there are few reports on the function of beneficial bacteria in OMF. G. elata is a completely heterotrophic plant and does not perform photosynthesis, and its growth entirely depends on A. gallica, an OM fungus. The pathogenicity of A. gallica is associated with plant cell wall degrading enzymes and invasive mycelium or rhizomorphs (Sahu et al., 2021). This indicates that A. gallica enter directly into the epidermal cell walls with short invasive mycelium to exchange nutrients from G. elata (Sella et al., 2015). Our results showed that Rahnella HPDA25 isolated from mycorrhizosphere soil samples of G. elata enhanced rhizomorph growth and the extracellular laccase activity of A. gallica, which could help A. gallica to infect wood or G. elata and promote the growth of G. elata.
The results of RNA-seq and qRT-PCR showed that Rahnella HPDA25 promoted the expression level of hydrophobin genes. Among HFBs, hydrophobin SC3 mediated the formation of aerial mycelium (Van Wetter et al., 1996) and aided the maturation of the fungal cell wall (Whiteford and Spanu, 2002). Therefore, Rahnella HPDA25 may promote cell wall maturation and rhizomorph growth of A. gallica by increasing the gene expression level of hydrophobin SC3 protein. Hydrophobin HYPB was first identified in Agaricus bisporus (Van Wetter et al., 1996), which enhanced the adherence of fungal structures to the surface of a host organism, thereby facilitating pathogenesis (Piscitelli et al., 2017). Hydrophobins HYPB may be involved in the symbiosis process, and the expression level of the hydrophobin gene was increased when mycorrhizal symbiosis was established in Tricholoma terreum–Pinus sylvestris symbiosis and Pisolithus tinctorius–Eucalyptus globulus symbiosis (Mankel et al., 2000, 2002). Fungal pectinases are also important during the infection of plants (Sella et al., 2015). Hence, the stimulated expression of hydrophobin HYPB and PME by Rahnella HPDA25 may help establish the symbiosis of A. gallica and G. elata. The downregulated DEGs, including ENO, PYK, ALDH, and PDC, were the key enzymes in the glycolysis pathway (Supplementary Table 8). A recent study reported that accelerated glycolysis might inhibit fungi mycelial growth (Yan et al., 2020). Thus, Rahnella HPDA25 may also promote A. gallica rhizomorph growth by inhibiting the glycolysis of A. gallica. Rahnella HPDA25 has been shown to stimulate mycelial development and fungal pathogenicity, suggesting that it may aid G. elata and A. gallica in improving nutrient absorption and increasing both yield and tuber size in G. elata.
The bacteria may promote the growth of plants and fungi through secreting IAA (Defez et al., 2019). Low concentrations of exogenous IAA modulate arbuscule formation in AM symbiosis (Chen et al., 2021). In our study, 16.24 ng/ml IAA was detected in the fermentation solution of R. aquatilis HPDA25 isolated from mycorrhizosphere soil samples of G. elata. Co-cultured A. gallica with HPDA25 or 16.24 ng/ml exogenous IAA increased the rhizomorph branching and growth rate of A. gallica, and HPDA25 co-cultured with A. gallica significantly increased the IAA content of A. gallica (Supplementary Figure 5). Thus, we speculate that the mechanisms of HPDA25 improving the growth of A. gallica and the nutrient uptake of G. elata are underlying the IAA regulation (Figure 6B).
Conclusion
Orchid mycorrhizal fungi form strict symbiotic relationships with most orchid plants and play critical roles in plant growth and nutrition. In this study, we quantified the diversity of OM bacteria in the symbiosis system of G. elata and Armillaria and found that co-inoculation of both mycorrhizal fungi and PGPB was beneficial to the growth of A. gallica and G. elata. G. elata could regulate the mycorrhizosphere bacterial communities, especially by recruiting Rahnella HPDA25. The IAA-secreted HPDA25 promoted the growth of A. gallica, regulated the nutrient uptake of A. gallica and G. elata, and enhanced the yield and tuber size of G. elata. These results provide new insights into OM symbiosis and the cultivation of G. elata.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YY and LH designed the experiments. TL conducted the experiments. YZ and JZ prepared samples for amplicon sequencing. ZH performed the bioinformatics. TL and PH contributed to data analysis. TL and YY wrote the manuscript. YJ and XL performed the identification of the plant resource. All authors read and approved the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (81891013 and 81891010), Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences (C12021A041), Key Project at Central Government Level for the Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (2060302), Fundamental Research Funds for the Central Public Welfare Research Institutes (ZZ11-116), and Major Science and Technology Projects in Guangxi Province (AA18242040).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ningshan Monyan Pharmaceutical Prepared Slices Co., Ltd., Shaanxi, for the cultivation of G. elata.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.842893/full#supplementary-material
Footnotes
- ^ http://cutadapt.readthedocs.io/en/stable/
- ^ http://www.drive5.com/uparse/
- ^ http://www.arb-silva.de/
- ^ http://www.drive5.com/muscle/
- ^ https://magic.novogene.com
- ^ https://www.ezbiocloud.net/
- ^ https://blast.ncbi.nlm.nih.gov
- ^ https://ngdc.cncb.ac.cn/
- ^ http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi
- ^ https://hiplot.com.cn
- ^ https://ngdc.cncb.ac.cn/gsa/browse/CRA004378
- ^ https://ngdc.cncb.ac.cn/gsa/browse/CRA004382
References
Agnolucci, M., Avio, L., Palla, M., Sbrana, C., Turrini, A., and Giovannetti, M. (2020). Health-promoting properties of plant products: he role of mycorrhizal fungi and associated bacteria. Agronomy 10:1864. doi: 10.3390/agronomy10121864
Alveshere, B. C., McMurtrey, S., Bennett, P., Kim, M. S., Hanna, J. W., Klopfenstein, N. B., et al. (2021). Phylogeography and host range of Armillaria gallica in riparian forests of the northern Great Plains, USA. For. Pathol. 51:e12663. doi: 10.1111/efp.12663
Bahram, M., Netherway, T., Hildebrand, F., Pritsch, K., Drenkhan, R., Loit, K., et al. (2020). Plant nutrient-acquisition strategies drive topsoil microbiome structure and function. New Phytol. 227, 1189–1199. doi: 10.1111/nph.16598
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4622. doi: 10.1073/pnas.1000080107
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Wang, Y. C., Qin, L. Y., He, H. Y., Yu, X. L., Yang, M. Z., et al. (2019). Dynamics of fungal communities during Gastrodia elata growth. BMC Microbiol. 19, 1–11. doi: 10.1186/s12866-019-1501-z
Chen, X., Chen, J., Liao, D., Ye, H., Li, C., Luo, Z., et al. (2021). Auxin-mediated regulation of arbuscular mycorrhizal symbiosis: a role of SlGH3.4 in tomato. Plant Cell Environ. 45, 955–968. doi: 10.1111/pce.14210
Defez, R., Andreozzi, A., Romano, S., Pocsfalvi, G., Fiume, I., Esposito, R., et al. (2019). Bacterial IAA-delivery into medicago root nodules triggers a balanced stimulation of C and N metabolism leading to a biomass increase. Microorganisms 7:403. doi: 10.3390/microorganisms7100403
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Emmanuel, O. C., and Babalola, O. O. (2020). Productivity and quality of horticultural crops through co-inoculation of arbuscular mycorrhizal fungi and plant growth promoting bacteria. Microbiol. Res. 239:126569. doi: 10.1016/j.micres.2020.126569
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I., and Dangl, J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Fradj, N., de Montigny, N., Mérindol, N., Awwad, F., Boumghar, Y., Germain, H., et al. (2020). A first insight into North American plant pathogenic fungi Armillaria sinapina transcriptome. Biology (Basel). 9, 1–20. doi: 10.3390/biology9070153
Guillotin, B., Etemadi, M., Audran, C., Bouzayen, M., Bécard, G., and Combier, J. P. (2017). Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). New Phytol. 213, 1124–1132. doi: 10.1111/nph.14246
Gutjahr, C. (2014). Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 20, 26–34. doi: 10.1016/j.pbi.2014.04.003
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hall, M., and Beiko, R. G. (2018). 16S rRNA gene analysis with QIIME2. Methods Mol. Biol. 1849, 113–129. doi: 10.1007/978-1-4939-8728-3_8
Hassan, S. E. D. (2017). Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. Adv. Res. 8, 687–695. doi: 10.1016/j.jare.2017.09.001
Ha-Tran, D. M., Nguyen, T. T. M., Hung, S.-H., Huang, E., and Huang, C.-C. (2021). Roles of plant growth-promoting Rhizobacteria (PGPR) in stimulating salinity stress defense in plants: a review. Int. J. Mol. Sci. 22:3154. doi: 10.3390/ijms22063154
Herrera, H., Novotná, A., Ortiz, J., Soto, J., and Arriagada, C. (2020). Isolation and identification of plant growth-promoting bacteria from rhizomes of Arachnitis uniflora, a fully mycoheterotrophic plant in southern Chile. Appl. Soil Ecol. 149:103512. doi: 10.1016/j.apsoil.2020.103512
Jiang, Z., Li, J., and Qu, L. J. (2017). “Auxins,” in Hormone Metabolism and Signaling in Plants, eds J. Li, C. Li, and P. Smith (Cambridge, MA: Academic Press), 39–76. doi: 10.1016/B978-0-12-811562-6.00002-5
Kaur, J., and Sharma, J. (2021). Orchid root associated bacteria: linchpins or accessories? Front. Plant Sci. 12:661966. doi: 10.3389/fpls.2021.661966
Khalil, A. M. A., Hassan, S. E. D., Alsharif, S. M., Eid, A. M., Ewais, E. E. D., Azab, E., et al. (2021). Isolation and characterization of fungal endophytes isolated from medicinal plant ephedra pachyclada as plant growth-promoting. Biomolecules 11, 1–18. doi: 10.3390/biom11020140
Khan, A. L., Asaf, S., Abed, R. M. M., Chai, Y. N., Al-Rawahi, A. N., Mohanta, T. K., et al. (2020). Rhizosphere microbiome of arid land medicinal plants and extra cellular enzymes contribute to their abundance. Microorganisms 8:213. doi: 10.3390/microorganisms8020213
Kieck, J. S., Zug, K. L. M., Huamaní Yupanqui, H. A., Gómez Aliaga, R., and Cierjacks, A. (2016). Plant diversity effects on crop yield, pathogen incidence, and secondary metabolism on cacao farms in Peruvian Amazonia. Agric. Ecosyst. Environ. 222, 223–234. doi: 10.1016/j.agee.2016.02.006
Kim, M., Yoon, H., You, Y. H., Kim, Y. E., and Kim, J. G. (2013). Metagenomic analysis of fungal communities inhabiting the fairy ring zone of tricholoma matsutake. J. Microbiol. Biotechnol. 23:1347. doi: 10.4014/jmb1306.06068
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, H., La, S., Zhang, X., Gao, L., and Tian, Y. (2021). Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 15, 2865–2882. doi: 10.1038/s41396-021-00974-2
Li, L., Jiao, Z., Hale, L., Wu, W., and Guo, Y. (2014). Disruption of gene pqqA or pqqB reduces plant growth promotion activity and biocontrol of crown gall disease by Rahnella aquatilis HX2. PLoS One 9:e115010. doi: 10.1371/journal.pone.0115010
Li, M., Chen, Z., Qian, J., Wei, F., Zhang, G., Wang, Y., et al. (2020). Composition and function of rhizosphere microbiome of Panax notoginseng with discrepant yields. Chinese Med. 15:85. doi: 10.1186/s13020-020-00364-4
Lu, H., Liu, Z., and Lan, S. (2019). Genome sequencing reveals the role of MADS-box gene families in the floral morphology evolution of orchids. Hortic. Plant J. 5, 247–254. doi: 10.1016/j.hpj.2019.11.005
Luo, L., Guo, C., Wang, L., Zhang, J., Deng, L., Luo, K., et al. (2019). Negative plant-soil feedback driven by re-assemblage of the Rhizosphere microbiome with the growth of panax notoginseng. Front. Microbiol. 10:1597. doi: 10.3389/fmicb.2019.01597
Mankel, A., Krause, K., and Kothe, E. (2002). Identification of a hydrophobin gene that is developmentally regulated in the Ectomycorrhizal Fungus Tricholoma terreum. Appl Environ. Microbiol. 68, 1408–1413. doi: 10.1128/AEM.68.3.1408-1413.2002
Mankel, A., Krause, K., Genenger, M., Kost, G., and Kothe, E. (2000). A hydrophobin accumulated in the Hartig’ net of ectomycorrhiza formed between Tricholoma terreum and its compatible host tree is missing in an incompatible association. J. Appl. Bot. 74, 95–99.
Omirou, M., Fasoula, D. A., and Ioannides, I. M. (2016). Bradyrhizobium inoculation alters indigenous AMF community assemblages and interacts positively with AMF inoculum to improve cowpea performance. Appl. Soil Ecol. 108:18. doi: 10.1016/j.apsoil.2016.09.018
Piscitelli, A., Cicatiello, P., Gravagnuolo, A. M., Sorrentino, I., Pezzella, C., and Giardina, P. (2017). Applications of functional amyloids from fungi: surface modification by class I hydrophobins. Biomolecules 7:45. doi: 10.3390/biom7030045
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quijada-Morin, N., Garcia, F., Lambert, K., Walker, A. S., Tiers, L., Viaud, M., et al. (2018). Strain effect on extracellular laccase activities from Botrytis cinerea. Aust. J. Grape Wine Res. 24, 241–251. doi: 10.1111/ajgw.12322
Reiss, E. R., and Drinkwater, L. E. (2018). Cultivar mixtures: a meta-analysis of the effect of intraspecific diversity on crop yield. Ecol. Appl. 28, 62–77. doi: 10.1002/eap.1629
Sahu, N., Merényi, Z., Bálint, B., Kiss, B., Sipos, G., Owens, R. A., et al. (2021). Hallmarks of basidiomycete soft-and white-rot in wood-decay-omics data of two armillaria species. Microorganisms 9, 1–20. doi: 10.3390/microorganisms9010149
Scheublin, T. R., Sanders, I. R., Keel, C., and Meer, J. (2010). Characterisation of microbial communities colonizing the hyphal surface of arbuscular mycorrhizal fungi. ISME J. 4, 752–763. doi: 10.1038/ismej.2010.5
Sella, L., Castiglioni, C., Paccanaro, M. C., Janni, M., Schäfer, W., D’Ovidio, R., et al. (2015). Involvement of fungal pectin methylesterase activity in the interaction between Fusarium graminearum and wheat. Mol. Plant Microbe Interact. 29, 258–267. doi: 10.1094/mpmi-07-15-0174-r
Stefan, L., Hartmann, M., Engbersen, N., Six, J., and Schöb, C. (2021). Positive effects of crop diversity on productivity driven by changes in soil microbial composition. Front. Microbiol. 12:660749. doi: 10.3389/fmicb.2021.660749
Van Wetter, M. A., Schuren, F. H. J., Schuurs, T. A., and Wessels, J. G. H. (1996). Targeted mutation of the SC3 hydrophobin gene of Schizophyllum commune affects formation of aerial hyphae. FEMS Microbiol. Lett. 140, 265–269. doi: 10.1016/0378-1097(96)00192-9
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. A. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10:4841. doi: 10.1038/s41467-019-12798-y
Wang, F., Shi, N., Jiang, R., Zhang, F., and Feng, G. (2016). In situ stable isotope probing of phosphate-solubilizing bacteria in the hyphosphere. J. Exp. Bot. 67, 1689–1701. doi: 10.1093/jxb/erv561
Wang, L., Wang, S., and Li, W. (2015). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. doi: 10.1093/bioinformatics/bts356
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wang, X., Feng, H., Wang, Y., Wang, M., Xie, X., Chang, H., et al. (2021). Mycorrhizal symbiosis modulates the rhizosphere microbiota to promote rhizobia–legume symbiosis. Mol. Plant 14, 503–516. doi: 10.1016/j.molp.2020.12.002
Whiteford, J. R., and Spanu, P. D. (2002). Hydrophobins and the interactions between fungi and plants. Mol. Plant Pathol. 3, 391–400. doi: 10.1046/j.1364-3703.2002.00129.x
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). ClusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation(China) 2:100141. doi: 10.1016/j.xinn.2021.100141
Yan, Z., Wu, X., Zhao, M., and Zhang, J. (2020). Lactic acid accumulation under heat stress related to accelerated glycolysis and mitochondrial dysfunction inhibits the mycelial growth of Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 104, 6767–6777. doi: 10.1007/s00253-020-10718-5
Ying, Y. X., Ding, W. L., and Li, Y. (2012). Characterization of soil bacterial communities in rhizospheric and nonrhizospheric soil of panax ginseng. Biochem. Genet. 50, 848–859. doi: 10.1007/s10528-012-9525-1
Yuan, L., Li, L., Zheng, F., Shi, Y., Xie, X., Chai, A., et al. (2020). The complete genome sequence of Rahnella aquatilis ZF7 reveals potential beneficial properties and stress tolerance capabilities. Arch. Microbiol. 202, 483–499. doi: 10.1007/s00203-019-01758-1
Yuan, Y., Jin, X., Liu, J., Zhao, X., Zhou, J., Wang, X., et al. (2018). The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 9:1615. doi: 10.1038/s41467-018-03423-5
Zhang, L., Fan, J., Feng, G., and Declerck, S. (2019). The Arbuscular Mycorrhizal fungus Rhizophagus irregularis MUCL 43194 induces the gene expression of citrate synthase in the tricarboxylic acid cycle of the phosphate-solubilizing bacterium Rahnella aquatilis HX2. Mycorrhiza 29, 69–75. doi: 10.1007/s00572-018-0871-7
Keywords: plant growth-promoting bacteria, orchid mycorrhizal symbiosis, cultivation methods, Rahnella, Armillaria
Citation: Liu T, Hua Z, Han P, Zhao Y, Zhou J, Jin Y, Li X, Huang L and Yuan Y (2022) Mycorrhizosphere Bacteria, Rahnella sp. HPDA25, Promotes the Growth of Armillaria gallica and Its Parasitic Host Gastrodia elata. Front. Microbiol. 13:842893. doi: 10.3389/fmicb.2022.842893
Received: 24 December 2021; Accepted: 07 February 2022;
Published: 17 March 2022.
Edited by:
Mohamed Hijri, Université de Montréal, CanadaReviewed by:
Soon-Jae Lee, University of Lausanne, SwitzerlandZineb Rchiad, Mohammed VI Polytechnic University, Morocco
Copyright © 2022 Liu, Hua, Han, Zhao, Zhou, Jin, Li, Huang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Yuan, eV95dWFuMDczMkAxNjMuY29t; Luqi Huang, aHVhbmdsdXFpMDFAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Tianrui Liu
Tianrui Liu Zhongyi Hua
Zhongyi Hua Pengjie Han1,2
Pengjie Han1,2 Xiaolin Li
Xiaolin Li Luqi Huang
Luqi Huang Yuan Yuan
Yuan Yuan