- 1Meat Processing Key Laboratory of Sichuan Province, College of Food and Biological Engineering, Chengdu University, Chengdu, China
- 2College of Science and Technology, Hebei Agricultural University, Cangzhou, China
- 3College of Biological and Chemical Engineering, Guangxi University of Science and Technology, Liuzhou, China
- 4Chongqing Academy of Metrology and Quality Inspection, Chongqing, China
N-hydroxy-pipecolic acid (NHP) is a hydroxylated product of pipecolic acid and an important systemic acquired resistance signal molecule. However, the biosynthesis of NHP does not have a natural metabolic pathway in microorganisms. Here, we designed and constructed a promising artificial pathway in Escherichia coli for the first time to produce NHP from biomass-derived lysine. This biosynthesis route expands the lysine catabolism pathway and employs six enzymes to sequentially convert lysine into NHP. This artificial route involves six functional enzyme coexpression: lysine α-oxidase from Scomber japonicus (RaiP), glucose dehydrogenase from Bacillus subtilis (GDH), Δ1-piperideine-2-carboxylase reductase from Pseudomonas putida (DpkA), lysine permease from E. coli (LysP), flavin-dependent monooxygenase (FMO1), and catalase from E. coli (KatE). Moreover, different FMO1s are used to evaluate the performance of the produce NHP. A titer of 111.06 mg/L of NHP was yielded in shake flasks with minimal medium containing 4 g/L of lysine. By this approach, NHP has so far been produced at final titers reaching 326.42 mg/L by 48 h in a 5-L bioreactor. To the best of our knowledge, this is the first NHP process using E. coli and the first process to directly synthesize NHP by microorganisms. This study lays the foundation for the development and utilization of renewable resources to produce NHP in microorganisms.
Introduction
Plant metabolites play an important role in plant defense, since they can directly harm attacking pathogens, preventing pathogens from entering plant tissues (Chezem et al., 2017). Systemic acquired resistance (SAR) is one of the inducible forms of defense in plants, which produces long-lasting and broad-spectrum immunity against secondary infections at remote locations other than the initial infection site (Fu and Dong, 2013). In dicotyledonous and monocotyledonous plants, enhanced disease resistance is very effective in suppressing infections caused by biological and semibiological vegetative pathogens (Li et al., 2016). Many compounds have been identified as mobile signals of SAR, including salicylic acid (Metraux et al., 1990), methyl salicylate, pipecolic acid (Pip) (Ding et al., 2016), azelaic acid (Vicente et al., 2012), glycerol-3-phosphate (Chanda et al., 2011), nicotinamide adenine dinucleotide, N-hydroxy-pipecolic acid (NHP) (Guerra and Romeis, 2020), nicotinamide adenine dinucleotide phosphate, and dehydroabietinal (Chaturvedi et al., 2012).
Expanding the product range of renewable feedstock is critical to achieving a viable bio-based economy. With the great development of metabolic engineering and synthetic biology, more and more high-value chemicals can be produced from renewable raw materials by natural or artificial pathways in microorganisms (Cheng et al., 2021a). Recently, many important useful chemicals, such as ectoine (Zhang S. et al., 2021), itaconic acid (Elmore et al., 2021), naringenin (Zhang Q. et al., 2021), ε-caprolactone (Xiong et al., 2021), curcuminoids (Rodrigues et al., 2020), cadaverine (Ting et al., 2021), 5-hydroxyvaleric acid (Sohn et al., 2021), tyrian purple (Lee et al., 2021), 1,5-pentanediol (Cen et al., 2021), isobutanol (Yu et al., 2019), and pyruvic acid (Luo et al., 2020) have been obtained in microorganisms. There are many effective strategies that have been developed and used to improve the production of target chemicals, such as enzyme engineering (Ting et al., 2021), cofactor engineering (You et al., 2021), transcription factor engineering (Kang et al., 2021), promoter engineering (Gao et al., 2020), modularity engineering (Cen et al., 2021; Osire et al., 2021), ribosome binding site engineering (Wang et al., 2018), pathway engineering (Wang et al., 2021), fine-tuning gene expression (Wang et al., 2015), biosensor technology (Li et al., 2019), high-through screening (Zeng et al., 2020), and so on.
Recent research have shown that the lysine-derived metabolites Pip and SAR signal molecule NHP are key activators of the Arabidopsis system immunity, and their endogenous biosynthesis are essential for pathogen-induced SAR (Hartmann and Zeier, 2018). Pip is a cyclic amino acid, a common lysine metabolite in plants and animals (Pérez-García et al., 2019). Pip is an important precursor of some drugs, such as bupivacaine, rapamycin (Eggeling and Bott, 2015), and sandramycin (He, 2006). Importantly, Pip was a compatible solute for Escherichia coli, Silicibacter pomeroyi, and Sinorhizobium meliloti (Gouesbet et al., 1994; Gouffi et al., 2000).
Three synthetic pathways for producing Pip from L-lysine have been established (Han et al., 2020). The first synthetic route for Pip production was Δ1-piperidine-2-carboxylic acid (Pip2C)-mediated route (Tani et al., 2015). One-pot process of Pip production was established for expression of lysine α-oxidase (RaiP), Pip2C reductase (DpkA), and glucose dehydrogenase (GDH), which produced 45.1 g/L of Pip (Tani et al., 2015). Cheng et al. (2018) reported that the expression of lysine permease (LysP) can effectively increase the titer of Pip to 46.7 g/L. The second synthetic pathway was the Δ1-piperidine-6-carboxylic acid (P6C)-mediated route (Perez-Garcia et al., 2016). Pip (1.81 g/L) was accumulated in Corynebacterium glutamicum with overexpression of L-lysine 6-dehydrogenase (LysDH) and pyrroline 5-carboxylate reductase (ProC) (Perez-Garcia et al., 2016). Moreover, Perez-Garcia et al. (2017) found that C. glutamicum PIPE4 could utilize glycerol, starch to produce 14.4 g/L of Pip. The third synthetic pathway for Pip synthesis was the lysine cyclodeaminase (LCD)-mediated route (Ying et al., 2019). Han et al. (2020) expressed the LCD and optimized the expression vector and culture conditions, which produced 93.51 g/L of Pip.
The natural occurrence of SAR signal molecule NHP in organisms was first discovered in Arabidopsis (Hartmann and Zeier, 2018). NHP accumulated significantly after Pseudomonas syringae infection in locally inoculated Arabidopsis (Hartmann and Zeier, 2018). The biosynthesis of NHP in Arabidopsis involved AGD2-like defense response protein 1 (ALD1), SAR-deficient 4 (SARD4), and flavin-dependent monooxygenase 1 (FMO1) (Hartmann and Zeier, 2019). External application of NHP can improve the disease resistance of crops and achieve the purpose of disease prevention and control (Holmes et al., 2019). The phylogenetic analysis of the open genome based on the sequence information of Arabidopsis shows that homologs of the core genes of the NHP biosynthesis pathway are widespread throughout the plant kingdom (Holmes et al., 2019). Holmes et al. (2019) found that NHP accumulated in tobacco and tomato when infected with DC3000 bacteria. Therefore, monocots and dicots have the ability to trigger the biosynthesis of NHP in response to the infection of exogenous microorganism.
H2O2 could be exported from the cell, but its rapid production could lead to excessive accumulation of H2O2 in host cells, which would bring harmful effects (Korshunov and Imlay, 2010). Lv et al. (2018) reported that the activity of vanillyl alcohol oxidase PsVAO was significantly decreased over 20 μM of H2O2 by incubating PsVAO in H2O2. Hossain et al. (2016) established a whole-cell biotransformation system to form pyruvic acid in E. coli expressing L-amino acid deaminase. Finally, 14.57 g/L of pyruvic acid was achieved in the biocatalytic system (Hossain et al., 2016). Zhou et al. (2021) introduced a catalase ScCTA1 to scavenge H2O2 to further improve the titer of coniferyl alcohol to 53.90 g/L. Moreover, some catalases from Saccharomyces cerevisiae are introduced into Hansenula polymorpha and Pichia pastoris to scavenge harmful H2O2 (Gellissen et al., 1996; Payne et al., 1997).
In this work, an artificial route for the production of SAR signal molecule NHP with RaiP, DpkA, GDH, LysP, FMO1, and catalase (KatE) overexpression was first successfully established in E. coli, as seen in Figure 1. First, lysine was oxidatively deammoniated, cyclized spontaneously, and reduced to generate Pip by RaiP and DpkA (Cheng et al., 2018). Subsequently, Pip was hydroxylated by FMO1 to form NHP. Engineering NHP biosynthesis into microorganism would be an attractive approach to enhance a plant’s exogenous ability to respond to pathogens.
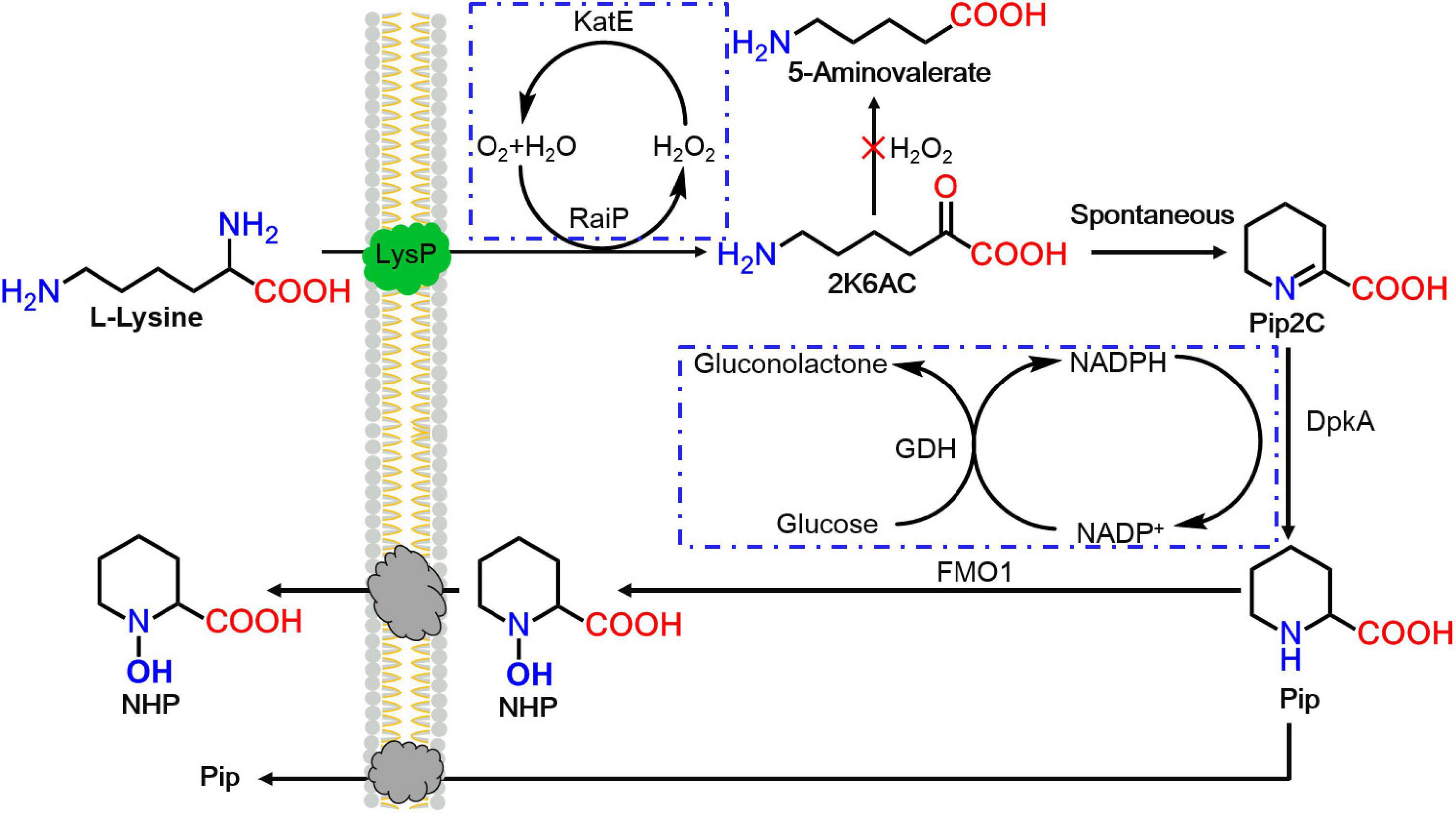
Figure 1. The heterogeneous pathway for N-hydroxy-pipecolic acid (NHP) production from lysine in Escherichia coli. 2K6AC, 2-keto-6-aminocaproate.
Materials and Methods
Strains and Plasmids
Bacterial strains and plasmids used in this study are shown in Table 1. E. coli BL21(DE3) was used as the host strain to produce NHP. Strain BL21(DE3) with cadA gene knocked out was conducted in our previous studies (Cheng et al., 2018). The plasmid pTrc99a-raip-dpkA-gdh-lysP was constructed in our previous work (Cheng et al., 2018). The nucleotide sequences of lysine α-oxidase gene raip from Scomber japonicus, Pip2C reductase gene dpkA from Pseudomonas putida, glucose dehydrogenase gene gdh from Bacillus subtilis, and lysine permease gene lysP from E. coli are available in the GenBank with accession numbers MG423617, MG423618, MG425967, and WP_000253273.1, respectively. Codon-optimized flavin-dependent monooxygenase 1 gene fmo1 from Arabidopsis (Arfmo1) (Accession No. NP_173359.3), Brassica rapa (Brfmo1) (Accession No. XP_009149401.1), Glycine max (Gmfmo1) (Accession No. XP_003541317.1), and Zea mays (Zmfmo1) (Accession No. XP_008660479.1) were chemically synthesized and inserted into pZA22 to form plasmid pZA22-Arfmo1, pZA22-Brfmo1, pZA22-Gmfmo1, and pZA22-Zmfmo1 with Acc65I restriction site, respectively. The katE gene was amplified from E. coli MG1655 with SphI and XbaI restriction site, ligated to the pZA22-Afmo1 vector to generate recombinant pZA22-Afmo1-katE. The sequences of all vector constructs were verified by Sanger sequencing (Sangon Biotech Co., Ltd., Shanghai, China). The recombinant strains are listed in Table 1.
Culture Medium and Conditions
Escherichia coli ML03 cells carrying the corresponding plasmids were cultured in 100-ml flasks containing 10 ml of Luria–Bertani (LB) medium (containing 10 g/L of tryptone, 5 g/L of yeast extract, and 10 g/L of NaCl) with appropriate antibiotics for 12 h at 37°C and 220 rpm. Then 20 μl of the culture was transferred into a 100-ml flask containing 20 ml of medium (composed of 15 g/L of glucose, 10 g/L of tryptone, 5 g/L of yeast extract, 0.5 g/L of K2PO4⋅3H2O, 3 g/L of KH2PO4, 2.5 g/L of (NH4)2SO4, 0.5 g/L of MgSO4⋅7H2O, 0.1 g/L of FeCl3, 2.1 g/L of citric acid⋅H2O, 40 mM α-ketoglutarate, and 100 μg/ml of Amp and/or 50 μg/ml of kanamycin) at 37°C and 220 rpm. After the OD600 reached 0.6, 0.5 mM IPTG was added, and culture was continued at 30°C for 48 h. L-lysine, Pip, and NHP were measured by HPLC.
Biotransformation of N-Hydroxy-Pipecolic Acid in a 5-L Bioreactor
The biotransformation of engineering strains for producing NHP were conducted in a 5-L bioreactor. The formula of the medium was 40 g/L of glucose, 7.5 g/L of K2HPO4⋅3H2O, 1.6 g/L of (NH4)2SO4, 1.6 g/L of MgSO4⋅7H2O, 0.00756 g/L of FeSO4⋅7H2O, 2 g/L of citric acid, 0.02 g/L of Na2SO4, 0.0064 g/L of ZnSO4, 40 mM α-ketoglutarate, 0.0006 g/L of Cu2SO4⋅5H2O, 0.004 g/L of CoCl2⋅6H2O, 100 μg/ml of Amp, and 50 μg/ml of kanamycin. For the biotransformation in a 5-L bioreactor, an E. coli colony was seeded in 25 ml of LB medium in a 250-ml flask and incubated at 37°C and 220 rpm for 12 h. A 1-L flask containing 280 ml of LB medium was inoculated with the preculture and incubated at 37°C for 12 h with shaking at 220 rpm. The medium [2.8 L in a 5-L fermenter (Baoxing, Shanghai, China)] was then inoculated with an aliquot (10%) of the seed culture for biotransformation at 30°C. The pH of the medium was adjusted at 6.8 by NH3⋅H2O during the biotransformation process. The biotransformation was carried out at 30°C. After the OD600 reached 18, 0.5 mM IPTG and 40 g/L of lysine were added. The samples were taken at specific time intervals, and the residual lysine, Pip, and NHP were analyzed using HPLC.
Analytical Methods
Lysine, Pip, and NHP were analyzed and quantitated by HPLC (Agilent Technologies 1200 series, Hewlett-Packard). The sample was derived with phenyl isothiocyanate (PITC) for the detection of lysine (Cheng et al., 2020). Pip and NHP contents were analyzed by HPLC with a Chirex® 3126 (D)-penicillamine LC column (4.6 × 250 mm) as described by Cheng et al. (2018). Samples were centrifuged and filtered using a 0.22-μm membrane.
Results and Discussion
Construction of a Heterogeneous Pathway for N-Hydroxy-Pipecolic Acid Production in Escherichia coli
In this study, a functional heterogeneous route for NHP production from lysine was first established in E. coli by a multienzyme expression system. The biosynthesis route is employed in Figure 1. Strain ML07 for the production of NHP precursor Pip was constructed previously (Cheng et al., 2018). The designed heterogeneous route of NHP consists of two steps. The first step was to convert lysine into Pip in strain ML07 with coexpression of RaiP, DpkA, GDH, and LysP. The second step was to convert Pip into NHP mediated by FMO1. Although NHP can be produced in Arabidopsis, tomato (Holmes et al., 2019), and cucumber (Schnake et al., 2020), it has not been reported in microorganisms. Hence, we tried to introduce a plant FMO1 from Arabidopsis (ArFMO1) in combination with Raip, DpkA, GDH, and LysP for NHP production from lysine. First, a plasmid pZA22-Arfmo1 was constructed. Then plasmids pTrc99a-raiP-dpkA-gdh-lysP and pZA22-Arfmo1 were introduced into E. coli ML03 to obtain the engineered strain ML071. As shown in Table 2, we investigated the production of NHP from lysine in strain ML071. When 2 g/L of L-lysine was fed to the recombinant E. coli ML071, 29.36 mg/L of NHP was produced after 12 h (Table 2). A titer of 86.48 mg/L of NHP could be obtained with 4 g/L of lysine addition after 48 h. Our data clearly proved that the heterogeneous pathway of NHP in E. coli is feasible.
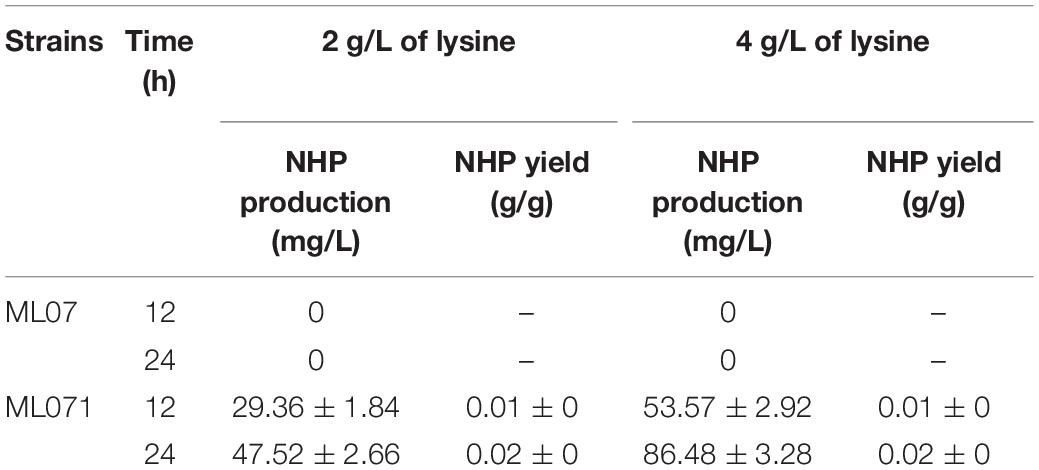
Table 2. Production of N-hydroxy-pipecolic acid (NHP) from lysine by recombinant Escherichia coli ML071.
Comparison of N-Hydroxy-Pipecolic Acid Synthesis by Different Flavin-Dependent Monooxygenases in Escherichia coli Strains
Although the function of AfFMO1 has been primarily verified in this study, the synthesis efficiency of NHP was far from satisfactory. It was presumed that other alternative enzymes from other sources might work better. FMO1s from different sources can catalyze the direct formation of NHP from Pip. To evaluate the performance of the four FMO1s in E. coli, four recombinant strains overexpressing different FMO1s, i.e., ArFMO1 from Arabidopsis, BrFMO1 from B. rapa, GmFMO1 from G. max, and ZmFMO1 from Z. mays, were constructed in this work.
As shown in Figure 2, the strains ML073 (GmFMO1) and ML074 (ZmFMO1) resulted in even less NHP production than ML071 (ArFMO1) (as 55.78 and 39.88 mg/L, respectively). The results improved greatly in ML072 (BrFMO1), with an enhancement to 111.06 mg/L of NHP, which was nearly 28% higher than ML071 (ArFMO1). The results indicated that BrFMO1 is slightly more active than AfFMO1, GmFMO1, and ZmFMO1 toward Pip (Figure 2). As a control, E. coli ML03 carrying the pTrc99a-raiP-dpkA-gdh-lysP vector did not produce any NHP.
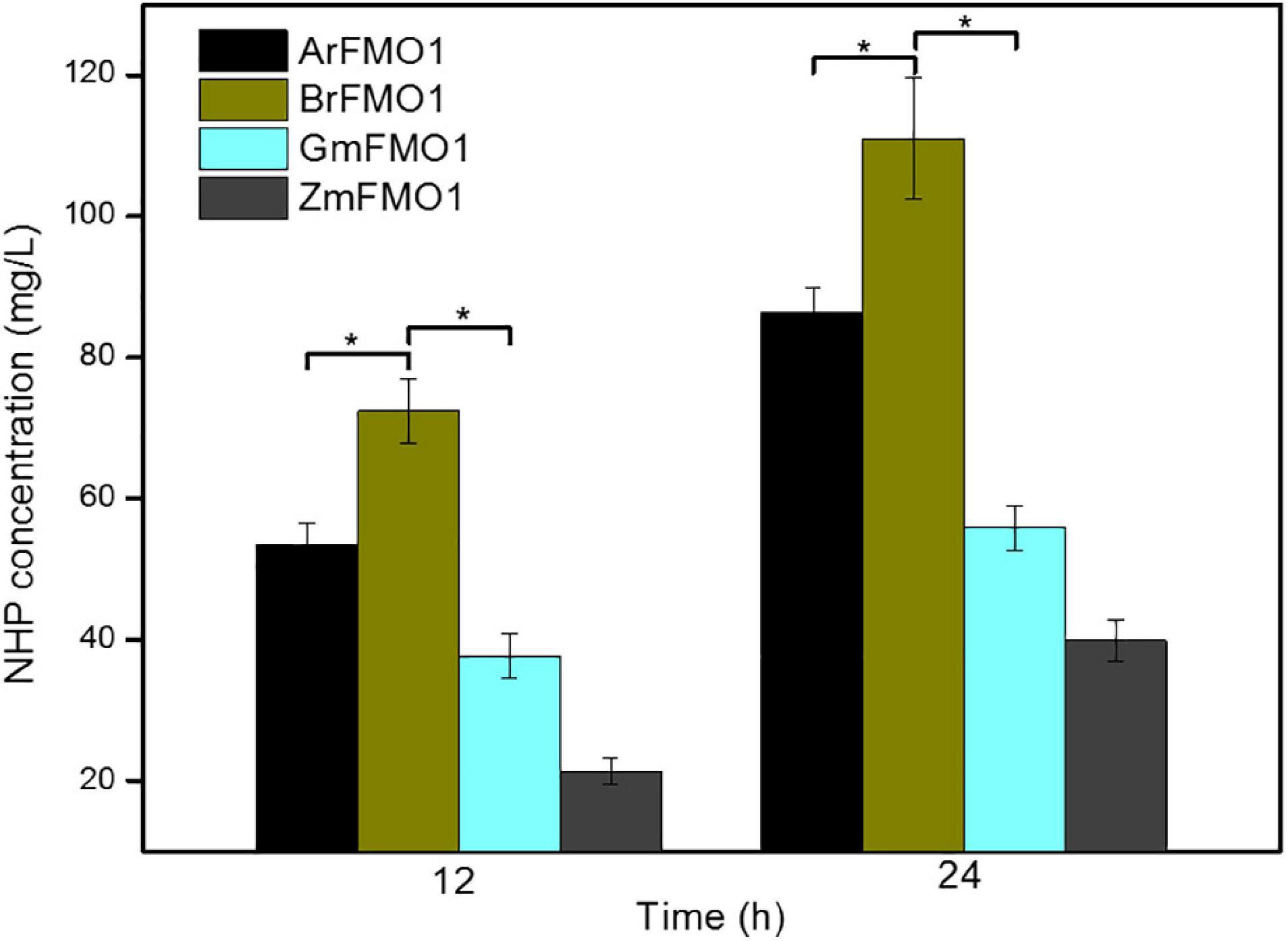
Figure 2. Comparison of NHP synthesis by different FMO1s in E. coli strains. NHP production at 12 and 24 h by strains ML071, ML072, ML073, and ML074, supplemented with 4 g/L of lysine as substrate. Statistics were performed by two-tailed Student’s t-test. *p < 0.05. Each experiment was done at least in triplicate. Error bars indicate standard errors of the means.
One-Pot Biotransformation of N-Hydroxy-Pipecolic Acid Production at the 5-L Scale
However, strain ML072 also produces H2O2 by RaiP, which impairs cells and can further oxidize 2-keto-6-aminocaproate (2K6AC) to 5-aminovalerate (5AVA) as a byproduct (Cheng et al., 2021b). Therefore, we introduced katE encoding a catalase to decompose harmful H2O2 and eliminate its side effects in strain ML08. The biotransformation of NHP in E. coli ML08 was executed in this study, and the results are shown in Figure 3. The data showed that the biomass increased from OD600 5.38 to 65 between 8 and 24 h. Pip (38.42 g/L) was produced, and 286.42 mg/L of NHP was accumulated after 36 h. Further increasing the fermentation time to 48 h could obtain a little higher NHP titer of 326.42 mg/L. At the same time, 36.98 g/L of Pip remained as a byproduct, indicating that the catalytic efficiency of BrFMO1 is not high and needs to be further improved. Under optimal biotransformation conditions, 326.42 mg/L of NHP was obtained in a 5-L bioreactor from 40 g/L of L-lysine in 48 h.
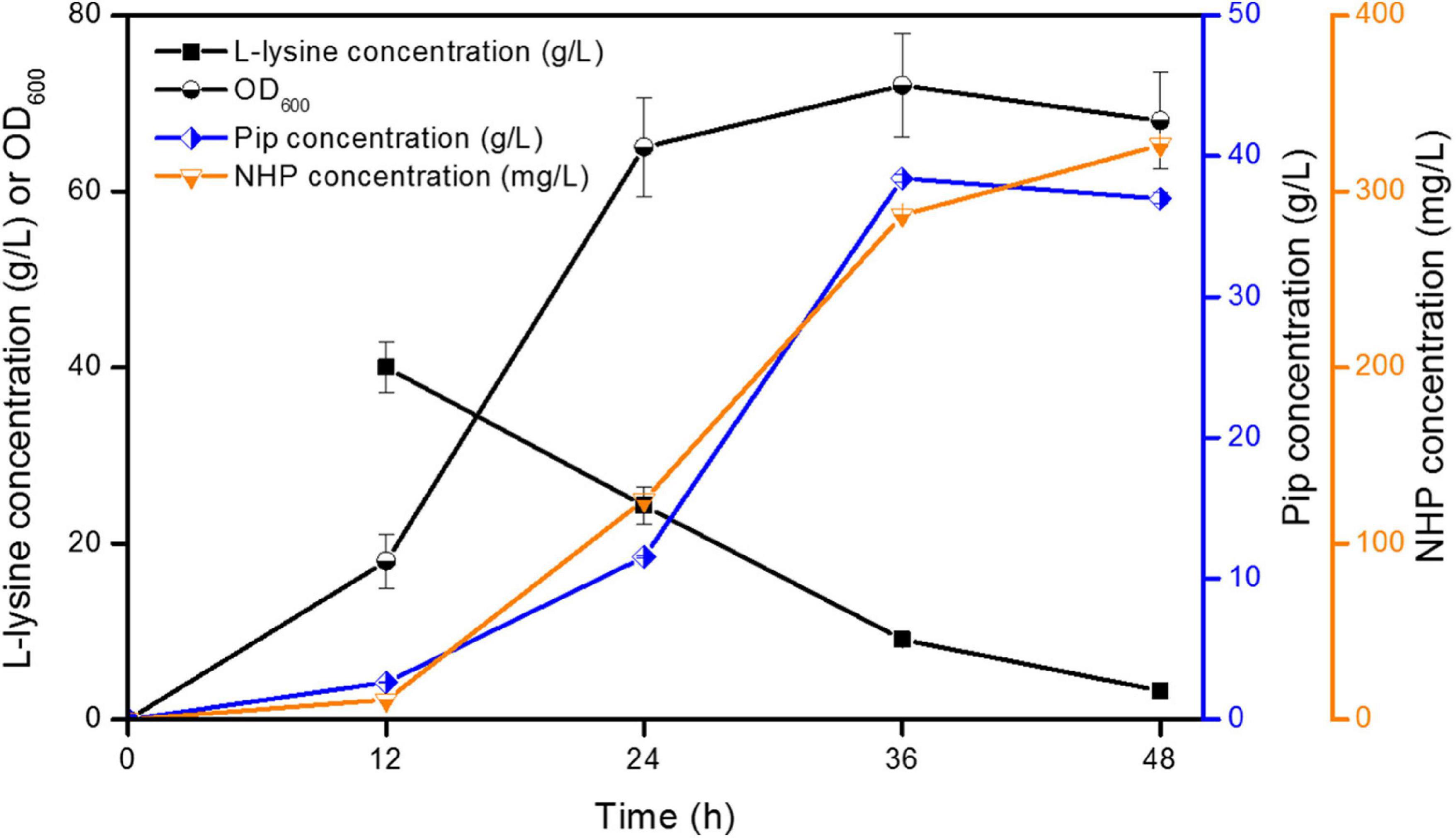
Figure 3. Time profiles of NHP production were investigated in E. coli ML08 in a 5-L fermenter. The experiments were conducted at 40 g/L of L-lysine. Each experiment was done at least in triplicate. Error bars indicate standard errors of the means.
Although H2O2 is distributed in almost all organisms, when its concentration exceeds a certain threshold, it could cause adverse effects, such as cytotoxicity and enzyme inhibition (Valderrama, 2010). A high concentration of H2O2 would affect cell growth and limited the production of target products (Niu et al., 2014; Cheng et al., 2021b). Liu et al. (2017) found that catalase KatE was introduced to decompose H2O2, which increased the titer of α-ketoglutaric acid to 77.4 g/L. The concentration of 5-aminovalerate was further improved by expressing catalase KatE in a novel synthetic pathway in our previous study (Cheng et al., 2021b). Therefore, the elimination of H2O2 in situ is conducive to the production of the target product.
Conclusion
In conclusion, we described a biotransformation system utilizing RaiP, DpkA, GDH, LysP, KatE, and BrFMO1 to convert L-lysine to SAR signal molecule NHP in E. coli. This artificial pathway in this work announces a greener and healthier production of SAR signal molecule NHP. An engineered E. coli strain with RaiP, DpkA, GDH, LysP, KatE, and BrFMO1 overexpression can produce 326.42 mg/L of NHP from L-lysine in a 5-L bioreactor. This microbial transformation system utilizes a renewable substrate and has simple culture conditions. As shown in Figure 3, Pip is produced as the main byproduct, which is not an efficient way to produce NHP. So the next goal is to improve the catalytic activity of BrFMO1, then to achieve a more efficient NHP production.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZL and ZW performed the experiments, analyzed the data, and drafted the manuscript. BW, LY, TB, JZ, and HL analyzed the data. ZZ, WW, and JC coordinated the study and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (2210081508), the Key Research and Development Program of Sichuan Province (2020YFN0147), the Open Funding Project of Meat Processing Key Laboratory of Sichuan Province (20-R-06), the Natural Science Foundation of Hebei Province (C2020204013), and the Specialized Research Fund for the Doctoral Program of Hebei Agricultural University (YJ201950).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cen, X., Liu, Y., Chen, B., Liu, D., and Chen, Z. (2021). Metabolic engineering of Escherichia coli for de novo production of 1,5-pentanediol from glucose. ACS Synth. Biol. 10, 192–203. doi: 10.1021/acssynbio.0c00567
Chanda, B., Xia, Y., Mandal, M. K., Yu, K., Sekine, K. T., Gao, Q. M., et al. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427. doi: 10.1038/ng.798
Chaturvedi, R., Venables, B., Petros, R. A., Nalam, V., Li, M., Wang, X., et al. (2012). An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 71, 161–172. doi: 10.1111/j.1365-313X.2012.04981.x
Cheng, J., Huang, Y., Mi, L., Chen, W., Wang, D., and Wang, Q. (2018). An economically and environmentally acceptable synthesis of chiral drug intermediate L-pipecolic acid from biomass-derived lysine via artificially engineered microbes. J. Ind. Microbiol. Biot. 45, 405–415. doi: 10.1007/s10295-018-2044-2
Cheng, J., Luo, Q., Duan, H., Peng, H., Zhang, Y., Hu, J., et al. (2020). Efficient whole-cell catalysis for 5-aminovalerate production from L-lysine by using engineered Escherichia coli with ethanol pretreatment. Sci. Rep. 10:990. doi: 10.1038/s41598-020-57752-x
Cheng, J., Luo, Q., Ren, Y., Luo, Z., Liao, W., Wang, X., et al. (2021a). Panorama of intron dynamics and gene rearrangements in the phylum basidiomycota as revealed by the complete mitochondrial genome of Turbinellus floccosus. Appl. Microbiol. Biotechnol. 105, 2017–2032. doi: 10.1007/s00253-021-11153-w
Cheng, J., Tu, W., Luo, Z., Gou, X., Li, Q., Wang, D., et al. (2021b). A high-efficiency artificial synthetic pathway for 5-aminovalerate production from biobased L-lysine in Escherichia coli. Front. Bioeng Biotechnol. 9:633028. doi: 10.3389/fbioe.2021.633028
Chezem, W. R., Memon, A., Li, F. S., Weng, J. K., and Clay, N. K. (2017). SG2-Type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in arabidopsis. Plant Cell 29, 1907–1926. doi: 10.1105/tpc.16.00954
Ding, P., Rekhter, D., Ding, Y., Feussner, K., Busta, L., Haroth, S., et al. (2016). Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28, 2603–2615. doi: 10.1105/tpc.16.00486
Eggeling, L., and Bott, M. (2015). A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 99, 3387–3394. doi: 10.1007/s00253-015-6508-2
Elmore, J. R., Dexter, G. N., Salvachua, D., Martinez-Baird, J., Hatmaker, E. A., Huenemann, J. D., et al. (2021). Production of itaconic acid from alkali pretreated lignin by dynamic two stage bioconversion. Nat. Commun. 12:2261. doi: 10.1038/s41467-021-22556-8
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gao, S., Xu, X., Zeng, W., Xu, S., Lyv, Y., Feng, Y., et al. (2020). Efficient biosynthesis of (2S)-eriodictyol from (2S)-naringenin in saccharomyces cerevisiae through a combination of promoter adjustment and directed evolution. ACS Synth. Biol. 9, 3288–3297. doi: 10.1021/acssynbio.0c00346
Gellissen, G., Piontek, M., Dahlems, U., Jenzelewski, V., Gavagan, J. E., DiCosimo, R., et al. (1996). Recombinant hansenula polymorpha as a biocatalyst: coexpression of the spinach glycolate oxidase (GO) and the S. cerevisiae catalase T (CTT1) gene. Appl. Microbiol. Biotechnol. 46, 46–54. doi: 10.1007/s002530050781
Gouesbet, G., Jebbar, M., Talibart, R., Bernard, T., and Blanco, C. (1994). Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology (Reading) 140, 2415–2422. doi: 10.1099/13500872-140-9-2415
Gouffi, K., Bernard, T., and Blanco, C. (2000). Osmoprotection by pipecolic acid in sinorhizobium meliloti: specific effects of D and L isomers. Appl. Environ. Microbiol. 66, 2358–2364. doi: 10.1128/AEM.66.6.2358-2364.2000
Guerra, T., and Romeis, T. (2020). N-hydroxypipecolic acid: a general and conserved activator of systemic plant immunity. J. Exp. Bot. 71, 6193–6196. doi: 10.1093/jxb/eraa345
Han, Y.-H., Choi, T.-R., Park, Y.-L., Park, J. Y., Song, H.-S., Kim, H. J., et al. (2020). Enhancement of pipecolic acid production by the expression of multiple lysine cyclodeaminase in the Escherichia coli whole-cell system. Enzyme Microbial Technol. 140:109643. doi: 10.1016/j.enzmictec.2020.109643
Hartmann, M., and Zeier, J. (2018). l-lysine metabolism to N-hydroxypipecolic acid: an integral immune-activating pathway in plants. Plant J. 96, 5–21. doi: 10.1111/tpj.14037
Hartmann, M., and Zeier, J. (2019). N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr. Opin. Plant Biol. 50, 44–57. doi: 10.1016/j.pbi.2019.02.006
He, M. (2006). Pipecolic acid in microbes: biosynthetic routes and enzymes. J. Ind. Microbiol. Biotechnol. 33, 401–407. doi: 10.1007/s10295-006-0078-3
Holmes, E. C., Chen, Y. C., Sattely, E. S., and Mudgett, M. B. (2019). An engineered pathway for N-hydroxy-pipecolic acid synthesis enhances systemic acquired resistance in tomato. Sci. Signal. 12:eaay3066. doi: 10.1126/scisignal.aay3066
Hossain, G. S., Shin, H.-D., Li, J., Du, G., Chen, J., and Liu, L. (2016). Transporter engineering and enzyme evolution for pyruvate production from d/l-alanine with a whole-cell biocatalyst expressing l-amino acid deaminase from Proteus mirabilis. RSC Adv. 6, 82676–82684.
Kang, Z., Zhang, M., Gao, K., Zhang, W., Meng, W., Liu, Y., et al. (2021). An l-2-hydroxyglutarate biosensor based on specific transcriptional regulator LhgR. Nature Communications 12:3619. doi: 10.1038/s41467-021-23723-7
Korshunov, S., and Imlay, J. A. (2010). Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 75, 1389–1401. doi: 10.1111/j.1365-2958.2010.07059.x
Lee, J., Kim, J., Song, J. E., Song, W. S., Kim, E. J., Kim, Y. G., et al. (2021). Production of tyrian purple indigoid dye from tryptophan in Escherichia coli. Nat. Chem. Biol. 17, 104–112. doi: 10.1038/s41589-020-00684-4
Li, L., Tu, R., Song, G., Cheng, J., Chen, W., Li, L., et al. (2019). Development of a synthetic 3-dehydroshikimate biosensor in Escherichia coli for metabolite monitoring and genetic screening. ACS Synth. Biol. 8, 297–306. doi: 10.1021/acssynbio.8b00317
Li, X., Yang, D. L., Sun, L., Li, Q., Mao, B., and He, Z. (2016). The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-amido synthase expression. Plant Physiol. 172, 546–558. doi: 10.1104/pp.16.00129
Liu, Q., Ma, X., Cheng, H., Xu, N., Liu, J., and Ma, Y. (2017). Co-expression of L-glutamate oxidase and catalase in Escherichia coli to produce alpha-ketoglutaric acid by whole-cell biocatalyst. Biotechnol. Lett. 39, 913–919. doi: 10.1007/s10529-017-2314-5
Luo, Z., Zeng, W., Du, G., Chen, J., and Zhou, J. (2020). Enhancement of pyruvic acid production in candida glabrata by engineering hypoxia-inducible factor 1. Bio. Technol. 295:122248. doi: 10.1016/j.biortech.2019.122248
Lv, Y., Cheng, X., Wu, D., Du, G., Zhou, J., and Chen, J. (2018). Improving bioconversion of eugenol to coniferyl alcohol by in situ eliminating harmful H2O2. Bio. Technol. 267, 578–583. doi: 10.1016/j.biortech.2018.07.104
Metraux, J. P., Signer, H., Ryals, J., Ward, E., Wyss-Benz, M., Gaudin, J., et al. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006. doi: 10.1126/science.250.4983.1004
Niu, P., Dong, X., Wang, Y., and Liu, L. (2014). Enzymatic production of alpha-ketoglutaric acid from l-glutamic acid via l-glutamate oxidase. J. Biotechnol. 179, 56–62.
Osire, T., Yang, T., Xu, M., Zhang, X., Long, M., Ngon, N. K. A., et al. (2021). Integrated gene engineering synergistically improved substrate-product transport, cofactor generation and gene translation for cadaverine biosynthesis in E. coli. Int. J. Biol. Macromol. 169, 8–17. doi: 10.1016/j.ijbiomac.2020.12.017
Payne, M. S., Petrillo, K. L., Gavagan, J. E., diCosimo, R., Wagner, L. W., and Anton, D. L. (1997). Engineering Pichia pastoris for biocatalysis: co-production of two active enzymes. Gene 194, 179–182. doi: 10.1016/s0378-1119(97)00120-0
Pérez-García, F., Brito, L. F., and Wendisch, V. F. (2019). Function of L-pipecolic acid as compatible solute in corynebacterium glutamicum as basis for its production under hyperosmolar conditions. Front. Microbiol. 10:340. doi: 10.3389/fmicb.2019.00340
Perez-Garcia, F., Max Risse, J., Friehs, K., and Wendisch, V. F. (2017). Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 12, 646–657. doi: 10.1002/biot.201600646
Perez-Garcia, F., Peters-Wendisch, P., and Wendisch, V. F. (2016). Engineering Corynebacterium glutamicum for fast production of L-lysine and L-pipecolic acid. Appl. Microbiol. Biotechnol. 100, 8075–8090.
Rodrigues, J. L., Gomes, D., and Rodrigues, L. R. (2020). A Combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front. Bioeng. Biotechnol. 8:59. doi: 10.3389/fbioe.2020.00059
Schnake, A., Hartmann, M., Schreiber, S., Malik, J., Brahmann, L., Yildiz, I., et al. (2020). Inducible biosynthesis and immune function of the systemic acquired resistance inducer N-hydroxypipecolic acid in monocotyledonous and dicotyledonous plants. J. Exp. Bot. 71, 6444–6459. doi: 10.1093/jxb/eraa317
Sohn, Y. J., Kang, M., Baritugo, K.-A., Son, J., Kang, K. H., Ryu, M.-H., et al. (2021). Fermentative high-level production of 5-hydroxyvaleric acid by metabolically engineered Corynebacterium glutamicum. ACS Sust. Chem. Eng. 9, 2523–2533.
Tani, Y., Miyake, R., Yukami, R., Dekishima, Y., China, H., Saito, S., et al. (2015). Functional expression of L-lysine alpha-oxidase from scomber japonicus in Escherichia coli for one-pot synthesis of L-pipecolic acid from DL-lysine. Appl. Microbiol. Biotechnol. 99, 5045–5054. doi: 10.1007/s00253-014-6308-0
Ting, W.-W., Huang, C.-Y., Wu, P.-Y., Huang, S.-F., Lin, H.-Y., Li, S.-F., et al. (2021). Whole-cell biocatalyst for cadaverine production using stable, constitutive and high expression of lysine decarboxylase in recombinant Escherichia coli W3110. Enzyme Microbial Technol. 148:109811. doi: 10.1016/j.enzmictec.2021.109811
Valderrama, B. (2010). “Deactivation of hemeperoxidases by hydrogen peroxide: focus on compound III,” in Biocatalysis Based on Heme Peroxidases, eds E. Torres and M. Ayala 291–314.
Vicente, J., Cascon, T., Vicedo, B., Garcia-Agustin, P., Hamberg, M., and Castresana, C. (2012). Role of 9-lipoxygenase and alpha-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol. Plant 5, 914–928. doi: 10.1093/mp/ssr105
Wang, J., Gao, C., Chen, X., and Liu, L. (2021). Engineering the cad pathway in Escherichia coli to produce glutarate from L-lysine. Appl. Microbiol. Biotechnol. 105, 3587–3599. doi: 10.1007/s00253-021-11275-1
Wang, J., Qin, D., Zhang, B., Li, Q., Li, S., Zhou, X., et al. (2015). Fine-tuning of ecaA and pepc gene expression increases succinic acid production in Escherichia coli. Appl. Microbiol. Biotechnol. 99, 8575–8586. doi: 10.1007/s00253-015-6734-7
Wang, J., Wang, H., Yang, L., Lv, L., Zhang, Z., Ren, B., et al. (2018). A novel riboregulator switch system of gene expression for enhanced microbial production of succinic acid. J. Ind. Microbiol. Biotechnol. 45, 253–269. doi: 10.1007/s10295-018-2019-3
Xiong, J., Chen, H., Liu, R., Yu, H., Zhuo, M., Zhou, T., et al. (2021). Tuning a bi-enzymatic cascade reaction in Escherichia coli to facilitate NADPH regeneration for ε-caprolactone production. Bio. Bioproc. 8:32.
Yan, X., Dong, Q., Zheng, M., and Yang, Z. (2013). Characterization of a family I-liked alkaline-resistant inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus furiosus for rapid determination of sugar nucleotidyltransferase at high temperature. J. Mol. Catal. B Enzymatic 98, 15–20.
Ying, H., Wang, J., Shi, T., Zhao, Y., Ouyang, P., and Chen, K. (2019). Engineering of lysine cyclodeaminase conformational dynamics for relieving substrate and product inhibitions in the biosynthesis of l-pipecolic acid. Catal. Sci. Technol. 9, 398–405.
You, Z.-N., Zhou, K., Han, Y., Yang, B.-Y., Chen, Q., Pan, J., et al. (2021). Design of a self-sufficient hydride-shuttling cascade for concurrent bioproduction of 7,12-dioxolithocholate and l-tert-leucine. Green Chem. 23, 4125–4133.
Yu, H., Wang, N., Huo, W., Zhang, Y., Zhang, W., Yang, Y., et al. (2019). Establishment of BmoR-based biosensor to screen isobutanol overproducer. Microb Cell Fact 18:30. doi: 10.1186/s12934-019-1084-2
Zeng, W. Z., Guo, L. K., Xu, S., Chen, J., and Zhou, J. W. (2020). High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 38, 888–906. doi: 10.1016/j.tibtech.2020.01.001
Zhang, Q., Yu, S., Lyu, Y., Zeng, W., and Zhou, J. (2021). Systematically engineered fatty acid catabolite pathway for the production of (2S)-naringenin in saccharomyces cerevisiae. ACS Synth Biol. 10, 1166–1175. doi: 10.1021/acssynbio.1c00002
Zhang, S., Fang, Y., Zhu, L., Li, H., Wang, Z., Li, Y., et al. (2021). Metabolic engineering of Escherichia coli for efficient ectoine production. Syst. Microbiol. Biomanufact. 1, 444–458.
Keywords: N-hydroxy-pipecolic acid, pipecolic acid, monooxygenase, artificial pathway, hydroxylate
Citation: Luo Z, Wang Z, Wang B, Lu Y, Yan L, Zhao Z, Bai T, Zhang J, Li H, Wang W and Cheng J (2022) An Artificial Pathway for N-Hydroxy-Pipecolic Acid Production From L-Lysine in Escherichia coli. Front. Microbiol. 13:842804. doi: 10.3389/fmicb.2022.842804
Received: 24 December 2021; Accepted: 26 January 2022;
Published: 08 March 2022.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2022 Luo, Wang, Wang, Lu, Yan, Zhao, Bai, Zhang, Li, Wang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiping Zhao, emhhb3poaXBpbmdAY2R1LmVkdS5jbg==; Wei Wang, d2FuZ3dlaTg2MTlAMTYzLmNvbQ==; Jie Cheng, Y2hlbmdqaWVAY2R1LmVkdS5jbg==
†These authors have contributed equally to this work
 Zhou Luo1†
Zhou Luo1† Zhen Wang
Zhen Wang Zhiping Zhao
Zhiping Zhao Ting Bai
Ting Bai Jie Cheng
Jie Cheng