
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 April 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.842106
 Pramod Prasad1
Pramod Prasad1 Rajni Kant Thakur1
Rajni Kant Thakur1 Siddanna Savadi2
Siddanna Savadi2 Subhash Chander Bhardwaj1*
Subhash Chander Bhardwaj1* Om Prakash Gangwar1
Om Prakash Gangwar1 Charu Lata1
Charu Lata1 Sneha Adhikari1
Sneha Adhikari1 Subodh Kumar1
Subodh Kumar1Stem rust caused by Puccinia graminis f. sp. tritici (Pgt) is a devastating disease of wheat worldwide since time immemorial. Several wheat stem rust outbreaks have been reported worldwide including India. Approximately 7 mha wheat area in central and peninsular India is highly vulnerable to stem rust epidemics. In this study, a repository of 29 single genotype uredospore pathotypes, representing five geographical regions, was characterized by investigating their virulence phenotype and simple sequence repeat (SSR) genotypes using 37 reproducible polymorphic SSR markers, 32 of which had ≥ 0.50 polymorphic information content (PIC) value. Virulence phenotypes were used to evaluate the virulence frequency (VF) and construct a hypothetical evolutionary hierarchy of these pathotypes. We projected seven lineages to explain the evolutionary pattern of the Pgt population. The VF of these pathotypes ranged between 0% and 100%. The virulence-based neighbor-joining (NJ) cluster analysis grouped Pgt pathotypes into five virulence groups. Likewise, five molecular groups were categorized using molecular genotypes. The molecular grouping was supported by principal coordinate analysis (PCoA), which revealed 25% of the cumulative variance contributed by the first two axes. Analysis of molecular variance (AMOVA) revealed 8 and 92% of the variation among and within the populations, respectively. The Mantel test confirmed a positive but weak correlation (R2 = 0.15) between virulence phenotypes and SSR genotypes. The highest and lowest values of different genetic diversity parameters (Na, Ne, I, He, uHe, and %P) revealed maximum and minimum variability in the Pgt population from Maharashtra and Uttar Pradesh, respectively. The population structure analysis clustered 29 Pgt pathotypes into two subpopulations and an admixture. Our results demonstrated that there was significant genetic diversity among Pgt pathotypes resulting from their long-distance dispersal ability complemented by gene flow. These findings provide insights into the virulence patterns, genetic variations, and possible evolution of Pgt pathotypes, which would support strategic stem rust resistance breeding.
Wheat (Triticum aestivum L.) is one of the most important sources of human nutrition and plays a critical role in global food security. The global wheat production reached 760.7 million tons during 2019, and 31% of it came from the two Asian countries, i.e., China and India, ranked first and second in global wheat production, respectively (FAO, 2021). Wheat production has been continuously threatened by several abiotic and biotic factors including diseases. Stem (black) rust caused by Puccinia graminis f. sp. tritici Eriks. and Henn. (Pgt) is a highly devastating disease of wheat and could result in wheat yield losses up to 100% on susceptible wheat cultivars (Anonymous, 1982; Worku et al., 2016). It is an obligate biotrophic, macrocyclic, and heteroecious fungus. The Pgt population comprises pathotypes with a highly diverse virulence nature or in some cases could be extremely uniform, which can disperse over long distances through wind and evolve new pathotypes that could overcome the stem rust resistance in wheat cultivars (Singh et al., 2015; Prasad et al., 2016).
Stem rust epidemics have occurred in the majority of wheat-growing regions of the world including East Africa, Eastern Europe, Australia, North America, Canada, the United States, and India (Dill-Macky et al., 1990; Eversmeyer and Kramer, 2000; Prasad et al., 2017b,2020). In India, stem rust is known to affect wheat crops in approximately 7 Mha areas covering the Central and Southern parts of the country. Historically, severe stem rust outbreak on Einkorn wheat was reported during 1907, followed by epidemics of 1946–1947 and 1948–1949 in Central and Southern India, respectively (Mehta, 1952; Prasad et al., 2018). With the help of strong disease surveillance, availability of suitable stem rust-resistant wheat cultivars, and high varietal replacement rate, the rusts have been kept under check; and no wheat rust epidemics were reported from any of the wheat-growing areas from India in the post green revolution period. Mehta (1952) reported a total loss of approximately 60 million Indian rupees annually in wheat and barley due to rust diseases in India. Stem rust resistance genes Sr2, Sr5, Sr6, Sr7a, Sr7b, Sr8a, Sr8b, Sr9b, Sr9e, Sr11, Sr12, Sr13, Sr17, Sr21, Sr24, Sr25, Sr30, and Sr31 are very common in Indian wheat cultivars. Among these, Sr2, Sr11, and Sr31 are more frequent in bread wheat, whereas Sr7b, Sr9e, Sr11, and Sr13 have been postulated in many durum cultivars (Jain et al., 2013). The emergence of TTKSK (also known as Ug99), one of the most virulent forms of Pgt, was first identified from Uganda in 1998 (Pretorius et al., 2000) and its variants (15 races), reported in 14 countries including Egypt, Yemen, and Iran (Singh et al., 2015; https://rusttracker.cimmyt.org/?page_id=22) and has rendered 90% of the world wheat varieties susceptible to stem rust (Singh et al., 2015; Worku et al., 2016; Soko et al., 2018). Another stem rust race, namely, Digalu (TKTTF) caused 100% yield loss in many Ethiopian regions during 2013–2014 and 2014–2015 (Olivera et al., 2015). Thus, stem rust has become a massive menace for world wheat production and has threatened global food security (Patpour et al., 2016; Prasad et al., 2016; Olivera et al., 2019).
The stem rust management in wheat primarily relies on the cultivation of disease-resistant varieties in combination with appropriate agronomic practices and the use of fungicides (Bhardwaj et al., 2016; Savadi et al., 2017). However, race-specific resistance is often overcome by the frequent introduction or evolution of new Pgt pathotypes through mutation, somatic recombination, or sexual recombination (Patpour et al., 2016). Therefore, regular monitoring of variations in the Pgt populations is essential for the identification of new resistance sources and anticipatory breeding for rust resistance to effectively manage the newly emerging Pgt isolates. The understanding of the virulence and genetic diversity, distribution, and population structure of the Pgt population allows effective monitoring of the emerging stem rust challenge, which is critical in formulating wheat stem rust management strategies (Abebe et al., 2012).
Virulence-based phenotyping of Pgt races in India started way back in 1932, and since then, 29 pathotypes of Pgt have been identified from the Indian subcontinent based on their pathogenicity on Indian stem rust differentials (Prasad et al., 2018). However, the genetic analysis of these pathotypes using molecular markers is not reported till date. Genetic analysis and genotypic variability of plant-pathogen populations have assisted researchers to reveal the genetic relationship among different pathotypes and track the pathways by which these pathogens evolve over time and space (Olivera et al., 2015; Prasad et al., 2017a). The simple sequence repeat (SSR) or microsatellite markers are most extensively used and established for exploring population dynamics and evolutionary patterns of Puccinia spp. due to the multi-allelic, reproducible, and highly polymorphic nature of these markers (Zhong et al., 2009; Savadi et al., 2020). This study was conducted to explore the virulence-based phenotype and SSR marker-based genotype patterns, population structure, and other diversity parameters in a repository of 29 Pgt pathotypes collected during the past ∼90 years from India and neighboring countries and maintained in vivo at ICAR-Indian Institute of Wheat and Barley Research, Regional Station (ICAR-IIWBR, RS), Shimla, India. Every year 100–200 isolates of Pgt collected from different wheat stem rust-prone areas in the Indian subcontinent are analyzed. Since the alternate host for stem rust is not functional, only 29 pathotypes have been identified in Pgt from this region. These pathotypes have been identified from different locations and years. The information on the distribution of these pathotypes during different years and locations is being published regularly, and the latest was by Prasad et al. (2018).
Twenty-nine P. graminis f. sp. tritici (Pgt) pathotypes, detected from the Indian subcontinent (Figure 1) and maintained at ICAR-IIWBR, RS, Shimla, were selected to study their virulence-based phenotype and SSR marker-based genotype patterns (Table 1). Single pustules, isolated for all the pathotypes to minimize the chances of heterogeneity within individual pathotypes, were multiplied on susceptible wheat variety “Agra local.” The uredospores from all the pathotypes were collected and stored for purity check, virulence phenotype, and SSR genotype analysis.
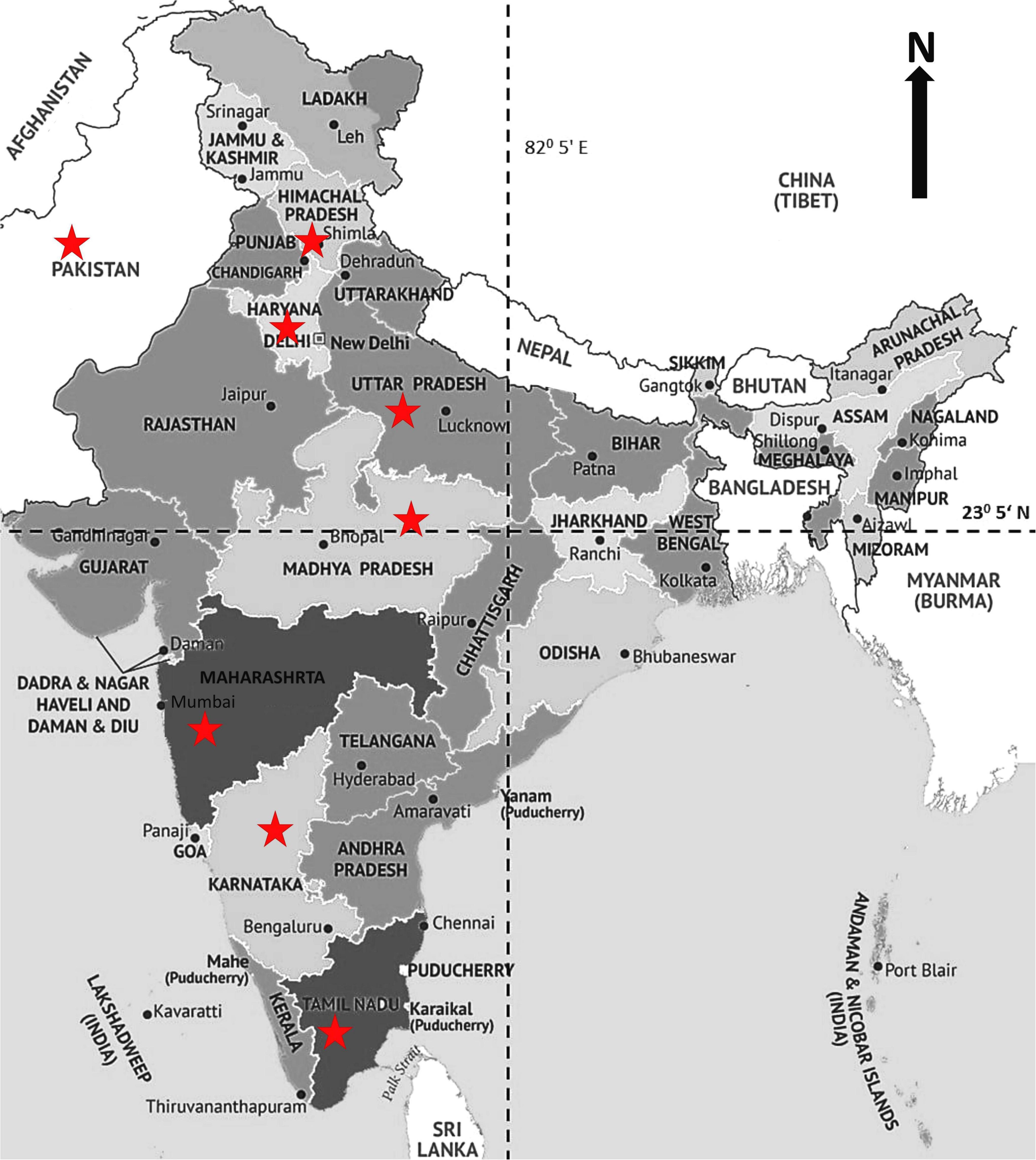
Figure 1. Wheat growing areas of India showing the field locations (indicated by a star), where Puccinia graminis f. sp. tritici pathotypes were initially detected.
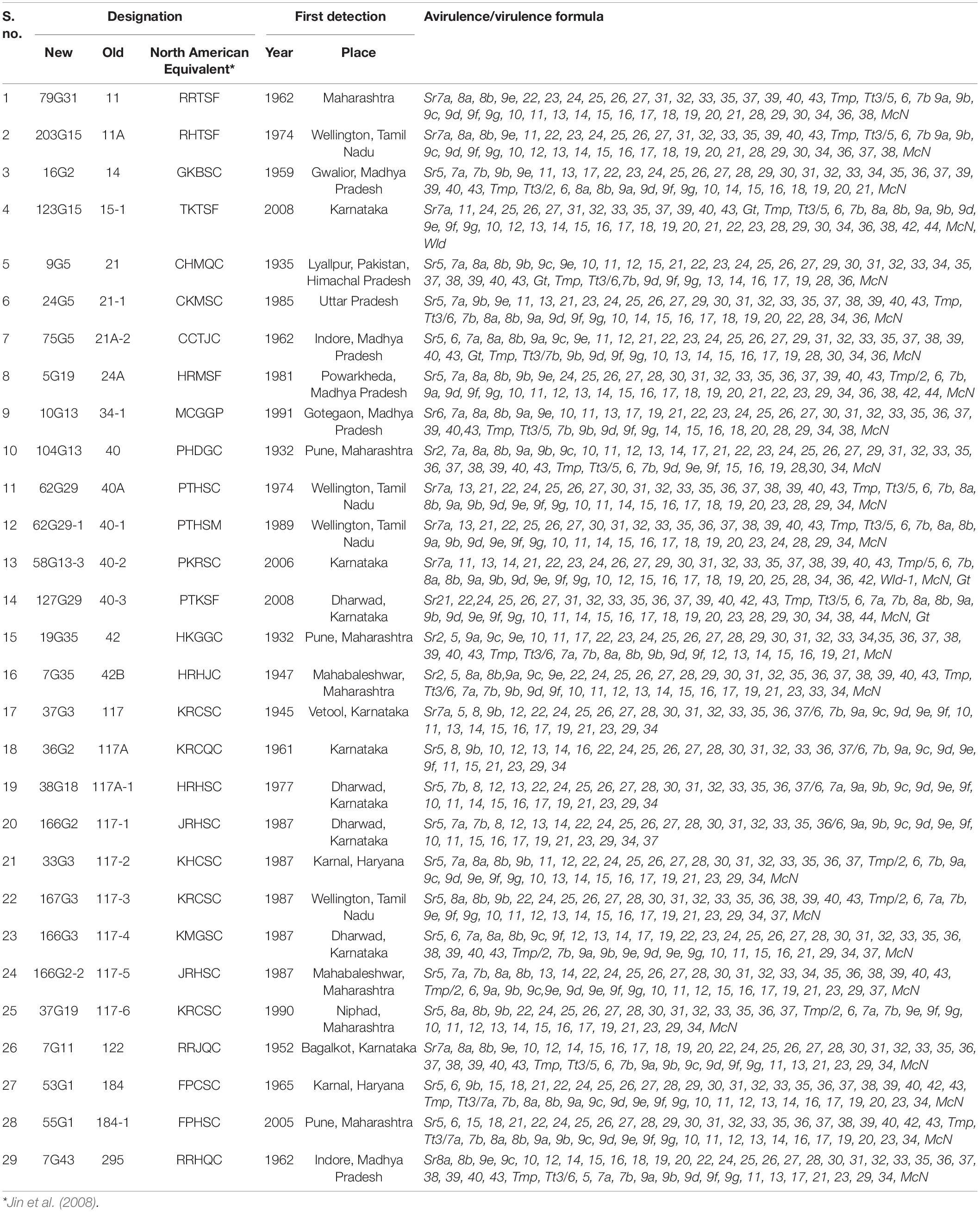
Table 1. Designation, geographic origin, year of identification, and avirulence/virulence formula of 29 pathotypes of Puccinia graminis f. sp. tritici from the Indian subcontinent used to study virulence phenotype and simple sequence repeat (SSR) genotypes.
All the pathotypes were inoculated onto standard Indian stem rust differentials (Supplementary Table 1; Bahadur et al., 1985a; Nayar et al., 1997; Prasad et al., 2018) for screening their purity and virulence phenotypes. Other near isogenic lines and wheat genotypes having known Sr genes were also inoculated with all 29 Pgt pathotypes to derive the avirulence/virulence formula of these pathotypes. The other additional Sr genes that were used to characterize these pathotypes are mentioned against each pathotype (avirulence/virulence formula) in Table 1. The sets of differentials were grown in aluminum bread pans (29 × 12 × 7 cm) containing fresh loam soil and farmyard manure (3:1), autoclaved twice at 60°C for 2 h. Inoculation of each pathotype was performed 10–12 days after sowing, once the primary leaf of seedlings fully emerged. Approximately 50 mg uredospores were suspended in 2 ml Soltrol 170 (Chevron Phillips Chemical Company, United States), a non-phytotoxic isoparaffinic mineral oil, and this suspension was atomized on the differentials using an atomizer in the inoculation chamber. The inoculated seedlings were sprayed with a fine mist of water and then shifted to water-saturated incubation chambers with > 80% relative humidity (RH). After incubation for 48 h, the trays were transferred to the fully air-conditioned greenhouses equipped with separate growth chambers for individual pathotypes and maintained at 25 ± 2°C, 40–60% RH, and 12 h photoperiod. The infection type (IT) of each pathotype on stem rust differentials was scored 15 days after inoculation according to the 0–4 disease rating scale (Stakman et al., 1962), where ITs 0–2 were interpreted as resistant and 3–4 as susceptible. The virulence phenotyping exercise was repeated to confirm the purity of each pathotype. Likewise, the avirulence/virulence formula of each pathotype was assigned based on the ITs of each pathotype on near-isogenic lines or wheat varieties with known stem rust (Sr) resistance genes.
The cetyltrimethylammonium bromide (CTAB) method, as previously described (Kiran et al., 2017), was employed to extract DNA from 50 mg dried uredospores each of 29 Pgt pathotypes. The purity and concentration of the extracted genomic DNA were assessed using the NanoDrop 2000® UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The quality of DNA was also evaluated by running it on 1% agarose gels. One hundred Puccinia spp. specific SSR markers designed previously (Karaoglu et al., 2013; Prasad et al., 2017a,2018; Savadi et al., 2020) were synthesized from Agile Life Science Technologies (New Delhi, India) and screened against selected eleven Pgt pathotypes (11, 14, 15-1, 21, 24A, 34-1, 40A, 42, 117-6, 122, and 184), representing different virulence patterns. Of the 100 markers, 37 (Table 2) with the ability to amplify highly repeatable polymorphic alleles were selected to genotype all 29 pathotypes.
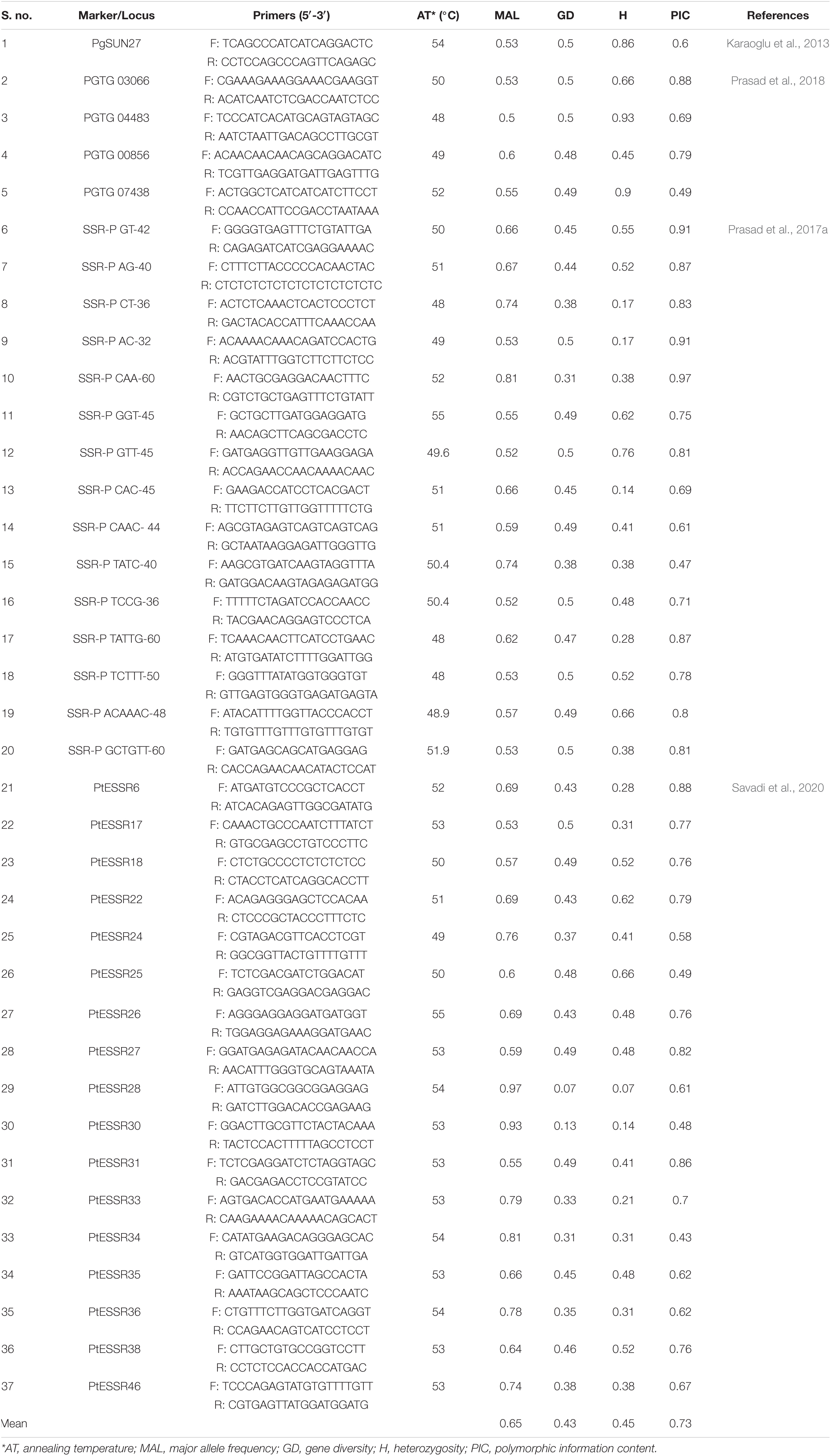
Table 2. List of 37 SSR markers used for population genetic diversity analysis of Puccinia graminis f. sp. tritici and their sequence, annealing temperature, major allele frequency, gene diversity, heterozygosity, and polymorphic information content (PIC) values.
Polymerase chain reaction (PCR) was performed in 20 μl of the PCR reaction mixture containing 25 ng of template DNA, 200 μM of dNTPs, 1 × PCR buffer (10 mM Tris, pH 9.0, and 50 mM KCl), 1.5 mM MgCl2, 2.5 U Taq polymerase (HiMedia Lab., Mumbai, India), and 10 pmol of both forward and reverse primers. The PCR programs were set at 94°C for 2 min (initial denaturation); followed by 35 cycles of 30 s at 94°C, 30 s at marker specific annealing temperature (Table 2), and 1 min at 72°C; with a final extension at 72°C for 7 min in a thermal cycler (Applied Biosystems Veriti™, CA, United States). The amplified products were resolved on 3% Super MT4 Agarose gel (Life Technologies, New Delhi, India) in 1× Tris-Borate-EDTA (TBE) buffer at 65–70 V for 2–3 h. DNA fragments were visualized under UV light and photographed using the gel documentation system (Bio-Rad Laboratories Inc., Hercules, CA, United States). The number of polymorphic alleles for each SSR locus was scored. The genotyping studies were repeated and confirmed before data analysis.
The virulence frequency (VF) of all 29 pathotypes on each of the entry (gene/cultivar) of differential sets was determined as the percentage of the pathotype virulent for a specific gene or cultivar of the differential set from the total of pathotypes under study. The binary data, generated as “1” and “0” to symbolize the virulence and avirulence, respectively, of each pathotype on wheat differential lines, were used to construct the virulence phenotype-based dendrogram of Pgt pathotypes. Likewise, binary codes as “1” and “0” were generated to represent the presence and absence, respectively, of SSR alleles of different sizes from multilocus allelic data matrix for all pathotype–primer combinations. The SSR alleles of different sizes from multilocus allelic data matrix were arranged according to their molecular weights (in base pair) for calculating major allele frequency (MAF), gene diversity, and heterozygosity of each SSR marker using PowerMarker version 3.25 (Lu et al., 2005). Polymorphic information content (PIC) value, discriminating two loci, was calculated using the following formula proposed by Anderson et al. (1993):
where n and Pij are the numbers of alleles and frequency of the jth allele for marker I, respectively.
For the analysis of genetic diversity parameters using GenAlEx, the Pgt population was divided into the following five subpopulations: (i) North India, (ii) Karnataka, (iii) Madhya Pradesh, (iv) Maharashtra, and (v) Tamil Nadu, which correspond to the place of origin of different pathotypes. The Shannon’s information index (I), expected heterozygosity (He), unbiased expected heterozygosity (uHe), percentage of polymorphic loci (%P), principal coordinate analysis (PCoA), Mantel correlation test, and analysis of molecular variance (AMOVA) were determined using GenAlEx 6.5 (Peakall and Smouse, 2012). GenAlEx was also used to calculate Nei’s genetic distance in a pairwise population matrix. Cluster analysis for both virulence phenotype and molecular genotype data was performed to generate dendrograms based on the neighbor-joining (NJ) cluster analysis using DARwin 6.0.14 (Perrier and Jacquemoud-collet, 2006). The population structure data of Pgt pathotypes were generated using STRUCTURE 2.3.4 (Pritchard et al., 2000) with the admixture model-based clustering method with 50,000 burin-in periods, 500,000 iterations, and two replications.
The virulence phenotyping of 29 Pgt pathotypes detected over the 90 years was performed on Indian stem rust differential sets and other Sr genes to explore their avirulence/virulence formula. Avirulence/virulence formula established for each of the 29 Pgt pathotypes against up to 50 Sr genes is presented in Table 1. Ten Sr genes, namely, Sr26, Sr27, Sr31, Sr32, Sr33, Sr35, Sr39, Sr40, Sr43, SrTmp, and SrTt3 were found effective against all 29 pathotypes (Table 1). The Sr24 was effective against all the pathotypes except 34-1 and 40-1, likewise, Sr25 conferred resistance to all the pathotypes except 40-2. The virulence phenotyping observations were used to calculate the VF of all 29 pathotypes on each of the entry of differential sets. The VF of Pgt pathotypes on stem rust differentials varied from 0.00 {DWR195 (Sr31) and HD2189 (Sr11 and some other unknown gene)} to 100% (for Barley Local and Agra Local) (Figure 2). The VF on Khapli (Sr7a, Sr13, Sr14+), Sr7a +, Sr24, HI1077 (Sr9b, 11+), and Sr25 was less than 10.4%, whereas > 40% VF was recorded on Sr13, Sr9b, Sr11, Sr9e, Marquis (Sr7b+), Einkorn (Sr21+), and Kota (Sr28+). Moderate virulence frequencies was observed on Lok 1 (Sr9b, 11+) (13.8%), Sr37 (17.24%), Sr30 (20.68%), Charter (Sr11+) (24.13%), NI 5439 (Sr11+) (27.58%), Sr8b (34.84%), and Reliance (Sr5+) (37.9%). The dendrogram generated by unweighted NJ cluster analysis using virulence phenotype data of each pathotype on all the differentials divided the Pgt pathotypes into five distinct virulence groups (VGs) (I–V) with 10, 2, 12, 3, and 2 pathotypes, respectively, in each group (Figure 3). Group III was further divided into two subgroups IIIa and IIIb with eight and four pathotypes in each, respectively. All nine pathotypes in race 117 lineage were positioned in group I along with 24A, which to a certain extent have similar virulence pattern as 117-6. Only two pathotypes (184 and 184-1) were clustered in VG II. Pathotypes belonging to race 40 lineage were placed in VG III along with other pathotypes, i.e., race 21 group (21, 21-1, and 21A-2), race 11 group (11 and 11A), 15, and 34-1. Pathotypes 40A, 40-1, and 40-3 with a similar avirulence/virulence formula were clustered in subgroup IIIb (Figure 3).
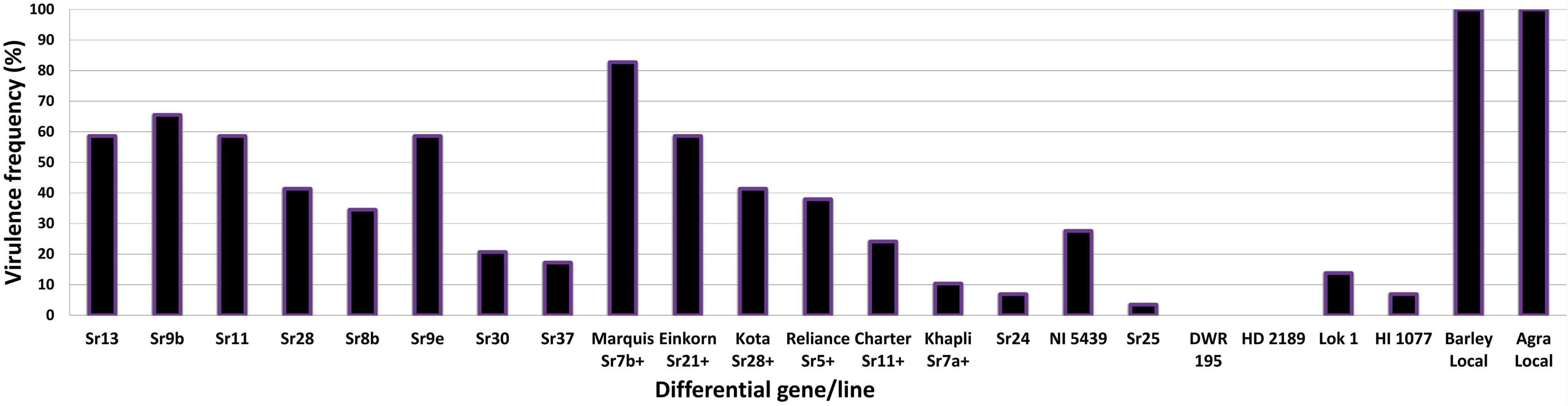
Figure 2. Virulence frequencies (%) of P. graminis f. sp. tritici pathotypes on stem rust differentials.
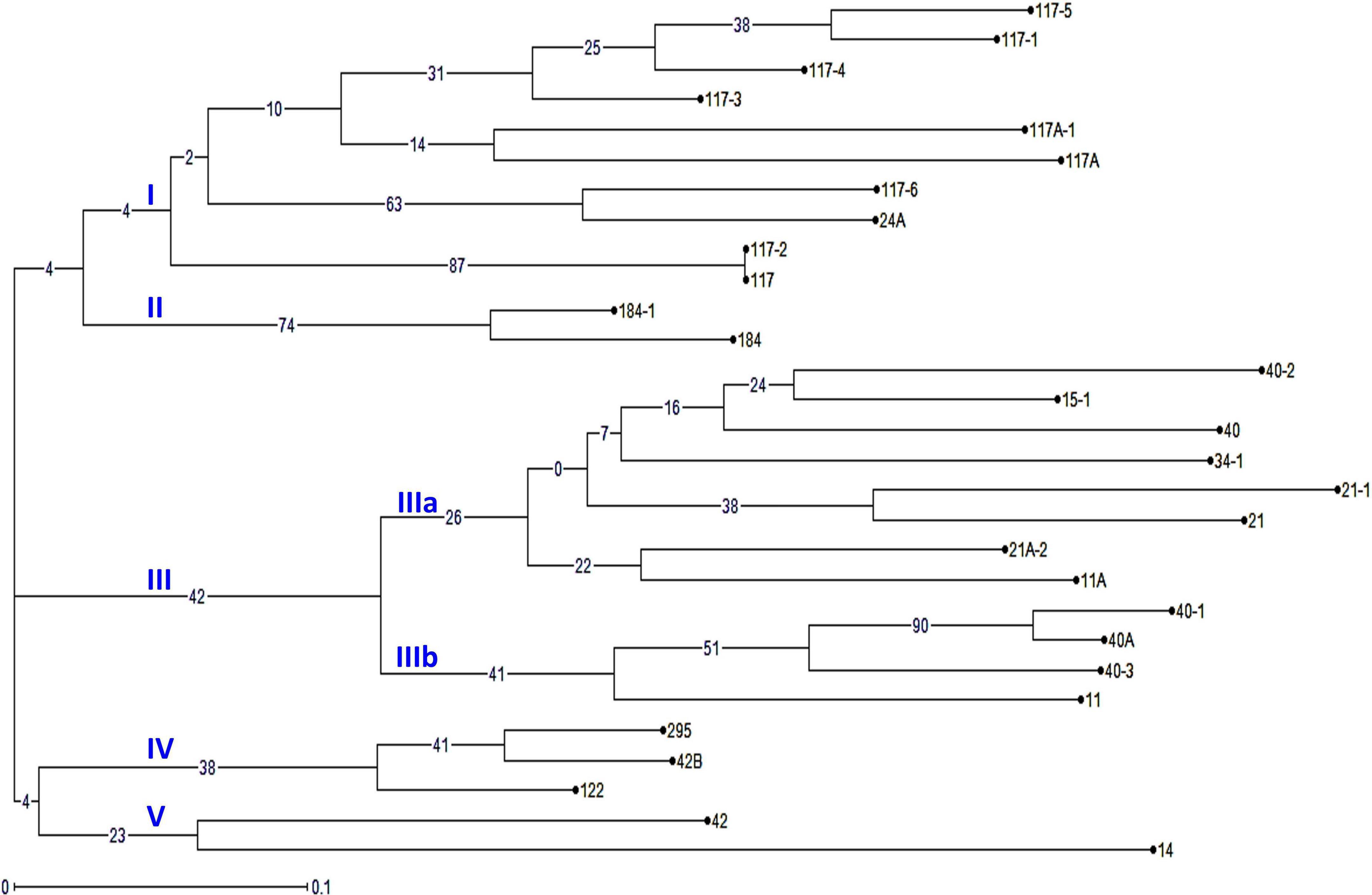
Figure 3. The phylogenetic tree of P. graminis f. sp. tritici pathotypes from the Indian subcontinent based on their avirulence/virulence phenotypes on stem rust differentials. The characters at the base of selected branches indicate the number of the virulence group and subgroups.
Based on the avirulence/virulence assay on the standard differentials possessing different Sr genes, we proposed a possible evolutionary hierarchy among 29 Pgt pathotypes collected from India and neighboring countries over the nine decades (Figure 4). Seven different possible lineages have been projected to explain the evolutionary pattern of these pathotypes over the years: (i) race 11 (2 pathotypes), (ii) race 21 (3 pathotypes), (iii) race 40 (6 pathotypes), (iv) race 42 (2 pathotypes), (v) race 117 (9 pathotypes), (vi) race 122 (2 pathotypes), and (vii) race 184 (2 pathotypes). The remaining three pathotypes, namely, 14, 24A, and 34-1 first discovered in 1959, 1981, and 1991, respectively, may be incursions from elsewhere or emerged independently. No conclusive evidence is available to prove the existence of their variants in the Indian subcontinent.
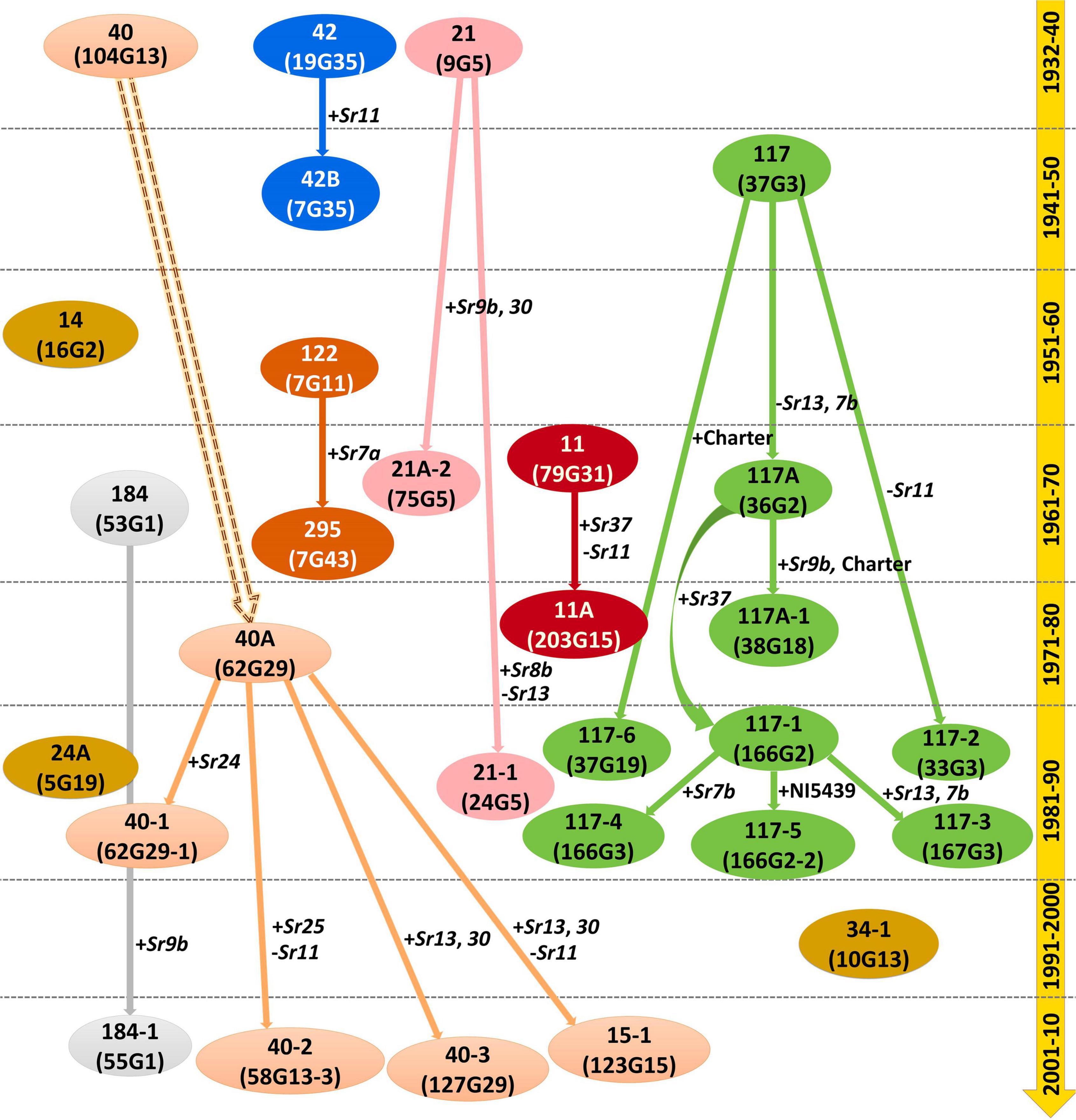
Figure 4. Proposed virulence-based hypothetical evolutionary hierarchy of P. graminis f. sp. tritici pathotypes from the Indian subcontinent based on virulence phenotyping. Solid outlines explain the probability of mutations for gain or loss of virulence for the Sr genes indicated alongside. The parallel dotted lines indicated the timeline of pathotype evolution in decades.
Out of 100 Puccinia spp. SSR markers screened against Pgt pathotypes, 37 were found polymorphic. From 29 Pgt pathotypes used in this study, a total of 175 alleles with 4.7 alleles per marker were amplified. The number of alleles per locus ranged from two to eight. The amplification product sizes of these markers ranged from 0.1 to 1.5 kb (Supplementary Figure 1 and Supplementary Table 2). The MAF of polymorphic markers ranged between 0.50 (PGTG 04483) and 0.97 (PtESSR28), with three other markers (i.e., SSR-P CAA-60, PtESSR30, and PtESSR34) showing ≥ 0.80 MAF (Table 2). Gene diversity ranged from 0.13 (PtESSR30) to 0.50 (Nine markers). The heterozygosity of these markers was in the range of 0.14 (SSR-P CAC-45 and PtESSR30) to 0.97 (PGTG 04483). Three polymorphic markers (PgSUN27, PGTG 04483, and PGTG 07438) had a heterozygosity score ≥0.80. The PIC value ranged from 0.43 (PtESSR34) to 0.97 (SSR-P CAA-60) with thirteen markers showing ≥ 0.80 PIC value (Table 2).
An unrooted dendrogram generated by unweighted NJ cluster analysis of the molecular data generated five distinct molecular groups (MGs I–V) with 12, 4, 8, 1, and 4 pathotypes, respectively, in each group. MGs I and III were further divided into three (i.e., Ia, Ib, and Ic) and two (i.e., IIIa and IIIb) subgroups, respectively (Figure 5). Subgroup Ia had four pathotypes (i.e., 24A, 34-1, 40-2, and 40-3), while Ib had seven pathotypes of which pathotypes 11 and 11A, belonging to race 11 lineage based on their virulence, had identical SSR genotype supported by a bootstrap value of 100%. Likewise, pathotypes 21 and 21A-2, belonging to the race 21 lineage and placed in subgroup Ib, shared 92% similarity in SSR genotype as supported by the bootstrap value. MG II had four pathotypes, namely, 40-1, 42, 184, and 184-1, the latter two of which had 95% similar SSR genotype as indicated by the bootstrap value. The third MG had five pathotypes in subgroup IIIa, all belonging to race 117 lineage, while subgroup IIIb had three pathotypes 117-6, 122, and 295. MG IV and subgroup Ic comprised only a single pathotype, i.e., 117A and 40A, respectively, demonstrating poor genetic similarity of these pathotypes with others. The MG V comprised of four pathotypes, namely 14, 117A-1, 42B, and 117, the latter two had fairly similar SSR genotype with 70% bootstrap value. The scatter plot from the PCoA revealed that the first two axes contributed 25.11% cumulative variance with 13.03% and 12.08% variation explained by the first and second axes, respectively (Figure 6 and Supplementary Table 3). The cumulative variance interpreted by all three axes together was 33.11%. However, this analysis, like virulence-based clustering, could not justify the region-specific grouping of the Pgt pathotypes.
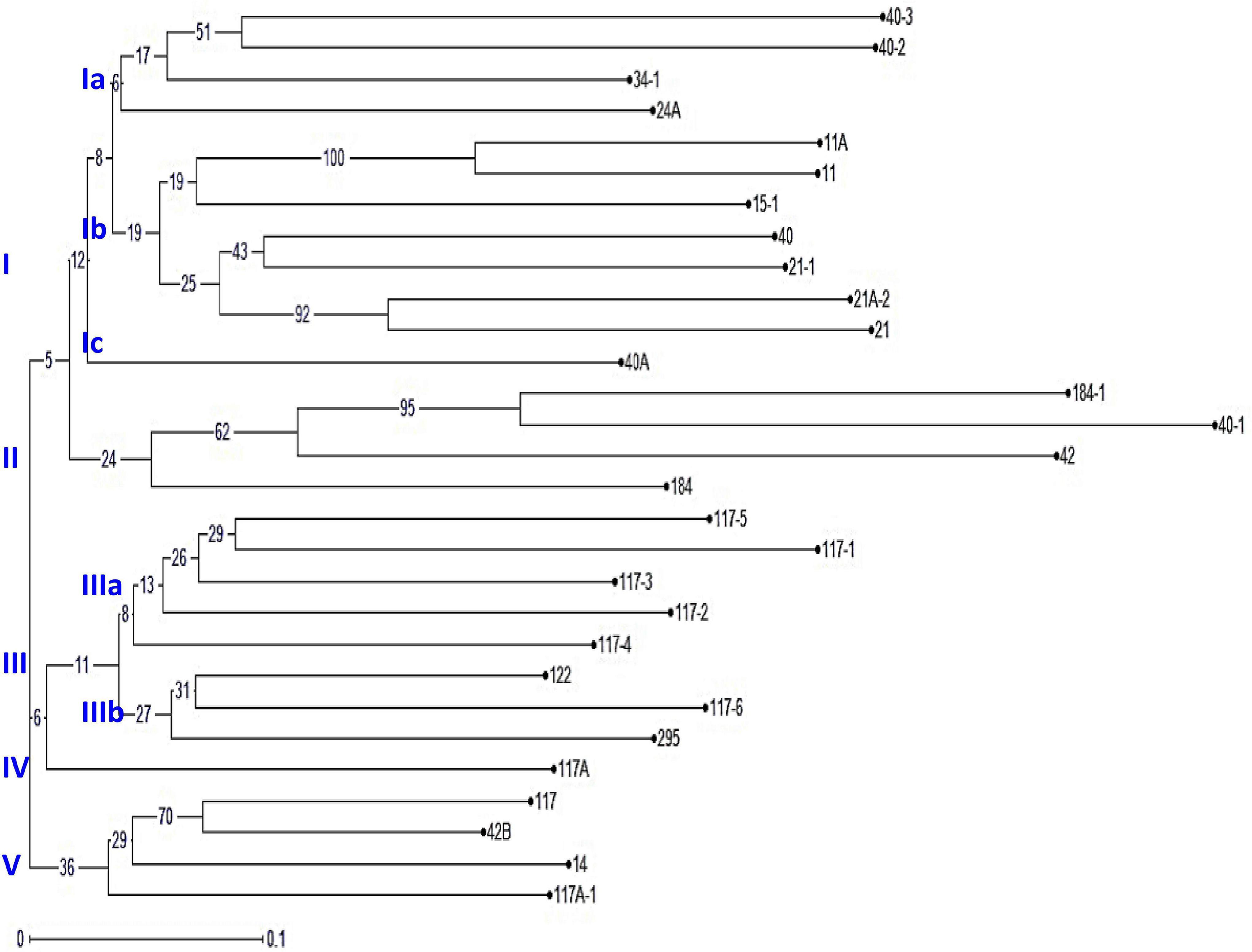
Figure 5. The phylogenetic tree of P. graminis f. sp. tritici pathotypes from the Indian subcontinent constructed using 37 polymorphic simple sequence repeat (SSR) markers. The characters at the base of selected branches indicate the number of the molecular group and subgroups.
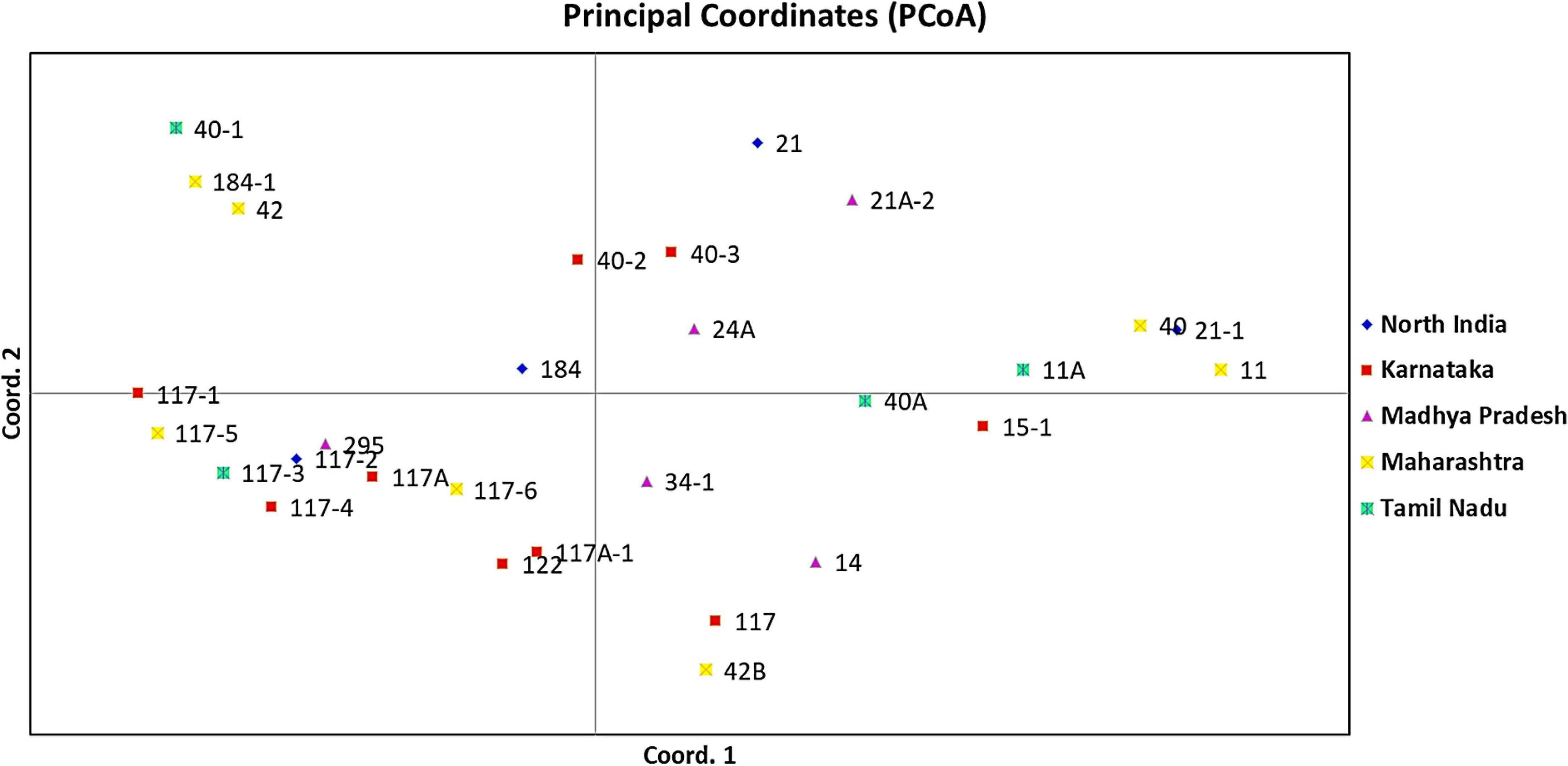
Figure 6. Principal coordinate analysis (PCoA) of P. graminis f. sp. tritici pathotypes from different geographical regions of India using 37 polymorphic SSR markers. The first and second principal axes revealed 13.03% and 12.08% of the total variation, respectively.
The genetic diversity of different Pgt populations was assessed using several parameters including the number of observed (Na) and effective (Ne) alleles, Shannon’s information index (I), expected heterozygosity (He), and percentage of polymorphic loci (%P) (Table 3). Comparing the different regions, Karnataka and Maharashtra had the highest Na (1.64), followed by Tamil Nadu (1.48), Madhya Pradesh, and North India (both 1.46). The Ne ranged between 1.39 (North India) and 1.47 (Maharashtra). The Ne for other regions, i.e., Karnataka, Madhya Pradesh, and Tamil Nadu was 1.46, 1.43, and 1.43, respectively. The highest I-value was observed for Maharashtra (0.41) and the lowest for North India (0.35). Maharashtra and Karnataka had the highest (both 0.27) while North India (0.23) had the lowest He. Equal He (0.25) was observed for Madhya Pradesh and Tamil Nadu. The highest uHe score was 0.29 for both Maharashtra and Tamil Nadu, while lowest for North India (0.26). The %P ranged from 65.71 (North India) to 79.43 (Maharashtra). It was 66.29, 70.86, and 78.29 for Madhya Pradesh, Tamil Nadu, and Karnataka, respectively (Table 3). The Nei’s genetic distance in pairwise population matrix was very low for all the geographical combinations, and it ranged from 0.041 between Tamil Nadu and Maharashtra to 0.087 between Karnataka and North India (Table 4).
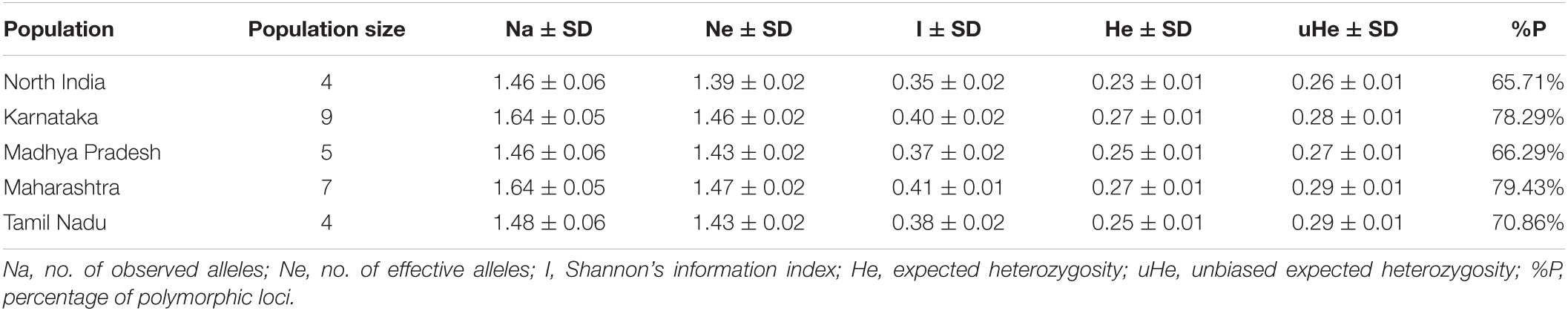
Table 3. Values of different genetic diversity parameters (number of observed and effective alleles, Shannon information index, expected and unbiased expected heterozygosity, and percentage of polymorphic loci) of Puccinia graminis f. sp. tritici groups belonging to five different geographic regions.
The AMOVA carried out by segregating the total variation among and within the populations explained that the maximum genetic variation was within the populations rather than among the Pgt populations belonging to different geographic regions. Genetic variations within the populations were 92%, and the remaining 8% (at P = 0.001) genetic variations were among the Pgt populations belonging to different geographic regions (Supplementary Figure 2 and Supplementary Table 4).
The SSR marker data-based population structure analysis was carried out to understand the genetic association among 29 Pgt pathotypes using STRUCTURE version 2.3.4. The maximum likelihood values and the mode of the distribution of the ΔK index were 2, indicating the existence of two subpopulations. However, seven pathotypes (184, 117, 184-1, 34-1, 14, 40-1, and 42) were designated as admixture individuals and anticipated to have mixed ancestry. The K = 2 divided all the pathotypes into two subpopulations (S1 and S2) consisting of 11 Pgt pathotypes in each that were originated in 5 different geographical regions (Figure 7 and Supplementary Table 5). However, we did not observe any geographical region-specific grouping of Pgt pathotypes in both the subpopulations, signifying that the geographical distances have very less influence on genetic variations in Pgt populations.
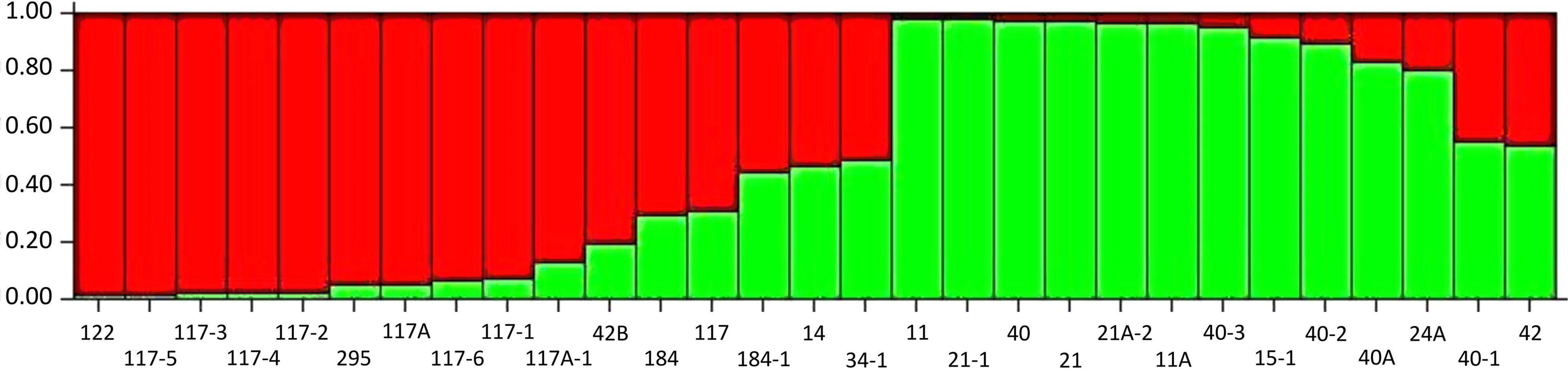
Figure 7. Population grouping of P. graminis f. sp. tritici pathotypes at estimated membership fraction for K = 2 using STRUCTURE. The grouping indicates two different populations in green and red with allele sharing more than 70% and an admixture with allele sharing less than 70%.
Correlation analysis was performed to examine the relationship between virulence phenotypes and SSR genotypes of the Pgt population consisting of 29 individuals using the Mantel correlation test. The analysis revealed that there was a positive but weak (R2 = 0.1546) correlation between these two variables. The linear regression equation of correlation analysis was y = 0.0843x + 1.8141 (Supplementary Figure 3).
Puccinia graminis tritici (Pgt), causing wheat stem rust, is highly destructive and one of the most extensively studied plant pathogens. Stem rust is a serious threat to world wheat production and food security. The emergence and dispersal of Pgt races, such as TTKSK (Ug99), TTRTF, TKTTF (Digalu race), and their variants, have posed a serious risk to global wheat production (Pretorius et al., 2000; Olivera et al., 2015; Patpour et al., 2020). The occurrence of such virulence has not been reported from any wheat-growing regions in the Indian subcontinent; however, strategic monitoring to track the change in the virulence pattern of Pgt is in place for early detection of likely new virulence (s), which might be introduced from elsewhere or evolved locally.
This study reports virulence phenotype and SSR genotype-based diversity analysis of Pgt repository (29 pathotypes) from the Indian subcontinent. Virulence phenotype data revealed both inter- and intra-lineage diverse nature of all 29 pathotypes on stem rust differentials and other Sr genes (Table 1). The virulence frequencies of Pgt pathotypes on stem rust differentials ranged between 0% and 100%. It was in the range of 20%–60% for the majority of the differentials. Ten Sr genes (i.e., Sr26, Sr27, Sr31, Sr32, Sr33, Sr39, Sr40, Sr43, SrTmp, and SrTt3) were found effective to the Indian population of Pgt, whereas virulence for Sr24 and Sr25 was found only in two (34-1 and 40-1) and one pathotypes (40-2), respectively. Among the effective all stage resistance (ASR) genes in India, seven (Sr26, Sr27, Sr32, Sr33, Sr39, Sr40, and Sr43) confer resistance to Ug99 and other variants in its lineage and are possibly the most useful ASR genes (Singh et al., 2015). Some of these genes, Sr26 in particular, have performed well against stem rust pathotypes worldwide and, therefore, could be a valuable asset for utilization in breeding programs if used in combinations with other effective genes (Singh et al., 2015; Terefe et al., 2016). Moreover, the availability of promising slow-rusting adult plant resistance (APR) genes such as Sr2, Sr55, Sr56, Sr57, Sr58, and others, and their pyramiding with effective ASR genes in cultivars with superior agronomic traits could further improve the effectiveness and durability of stem rust resistance (Huerta-Espino et al., 2020).
Virulence phenotype-based cluster analysis grouped Pgt pathotypes into five distinct VGs (I–V). All the pathotypes in race 117 lineage were in VG I (Figure 3). The clustering of pathotypes 40A, 40-1, and 40-3 justifies their similar virulence phenotypes on majority of the differentials except for: (i) Sr24 (virulent: 40-1; avirulent: 40A and 40-3), (ii), Sr13, and (iii) Sr30 (virulent: 40-3; avirulent: 40A and 40-1; for both Sr13 and Sr30). The identical virulence phenotypes of 40A and 40-1, except for Sr24 (virulent: 40-1), suggested and have been established that pathotype 40-1 had evolved through a single-step gain of virulence for Sr24 in 40A (Bhardwaj et al., 1990). Other pathotypes in the same race lineage and belonging to one VG include 184 and 181-1 in VG II, 122 and 295 in VG IV, and 21, 21A-2, and 21-1 in subgroup IIIa. Likewise, pathotypes 11 and 11A were clustered in subgroups IIIa and IIIb (VG III), respectively.
The possible evolutionary lineages deduced based on the virulence-avirulence assays of 29 Pgt pathotypes classified them into seven lineages. In race 11 lineage, the first pathotype was 11 (virulence for Sr11 and Sr30), which was detected in 1962 from Maharashtra (India). Subsequently, another pathotype in its lineage with additional virulence on Sr37 was identified in 1974 as 11A (Sinha et al., 1976). Other difference between pathotypes 11 and 11A is that the latter has an avirulence phenotype on Sr11. In race 21 lineage, the first pathotypes to be identified was 21, which was simultaneously detected from Lyallpur, Pakistan, and Himachal Pradesh (India) in 1935. The one-step gain in virulence for Sr9b and Sr30 in pathotype 21 caused the emergence of 21A-2 in 1962 from Madhya Pradesh (India). Likewise, additional virulence for Sr8b and loss of virulence for Sr13 in pathotype 21 resulted in the emergence of 21-1 in 1985 from Uttar Pradesh (India) (Bahadur et al., 1985b). Pathotype 40 in race 40 lineage was detected in 1932 from Pune (India; Mehta, 1940), while a new independent pathotype 40A emerged in the Nilgiri Hills (Wellington, India) in 1974 (Sharma et al., 1975). Additional virulence for Sr24 in pathotype 40A resulted in the emergence of 40-1 (Bhardwaj et al., 1990), while simultaneous gain and loss of virulence for Sr25 and Sr11, respectively, in 40A resulted in the emergence of 40-2 (Jain et al., 2009). Other pathotypes 40-3 and 15-1 in race 40 lineage emerged from Dharwad (Karnataka) in 2008, resulting from the gain of virulence for Sr13 and Sr30 (for both 15-1 and 40-3) and loss of virulence for Sr11 (for 15-1) in 40A (Jain et al., 2013).
Race 117 lineage has been the most variable collection of Pgt pathotypes in the Indian subcontinent. Pathotype 117 in this lineage was first detected from Madhya Pradesh in 1945 (Prasada and Lele, 1952). Since then, eight additional variants of this pathotype have been detected. The loss of virulence for Sr7b and Sr13 in pathotype 117 resulted in the emergence of 117A in 1960 from Karnataka (Patil et al., 1963). Consequently, pathotypes 117-2 and 117-6 evolved as a result of loss of virulence to Sr11 (Bhardwaj et al., 1990) and one step gain of virulence on Charter (Bhardwaj et al., 1994), respectively, in pathotype 117. Additional virulence in pathotype 117A for Sr9b and Charter resulted in the emergence of 117A-1 (Sharma et al., 1979), while the gain in virulence for Sr37 in 117A contributed to the advent of 117-1 (Bhardwaj et al., 1989). Added virulence for Sr7b and NI5439 in pathotype 117-1 resulted in the emergence of 117-4 and 117-5, respectively, whereas pathotype 117-1 advanced to 117-3 after gaining additional virulence for Sr7b and Sr13 (Bhardwaj et al., 1994). The gain in virulence was the principal mode of evolution in the race 117 lineage except for the emergence of 117A (117-Sr13 and Sr7b) and 117-2 (117-Sr11), where the loss of virulence in pathotype 117 resulted in their evolution. The virulence-based hypothetical evolutionary hierarchy proposed in this study is to a great extent in agreement with previously discussed virulence phenotype data-based NJ cluster analysis. The evolutionary behavior of Pgt isolates reveals that the gain of virulence for Sr13 was most frequent. The resistance genes present in the most widely used cultivars always play a critical role in influencing the prevalence of pathogen races in a specific region or location. The Sr13, one of the most widely used Sr genes in durum wheat, is very common in Indian cultivars as revealed by gene postulation using the gene matching technique. Moreover, the areas covering Madhya Pradesh, Karnataka, Maharashtra, and Tamil Nadu states, where most of the new pathotypes were initially detected, have all three types of wheat (i.e., aestivum, durum, and dicoccum) in cultivation. Adequate host diversity among aestivum, durum, and dicoccum wheat cultivars might be facilitating easier survival for newly evolved rust isolates.
The SSR genotype pattern of each of 29 Pgt pathotypes was estimated using 37 Puccinia spp. specific SSR markers developed in previous studies (Karaoglu et al., 2013; Prasad et al., 2017a, 2018; Savadi et al., 2020). Four of these markers (i.e., PGTG 03066, PGTG 04483, PGTG 00856, and PGTG 07438) were gene-based functional SSRs, which are known to be most reliable and reproducible for genetic diversity analysis (Anderson and Lubberstedt, 2003; Zhong et al., 2009). The MAF, gene diversity, heterozygosity, and PIC values indicated that the SSR markers used in this study were highly informative and, therefore, yielded good quality genetic diversity analysis data. Thirty-two SSRs had PIC values more than 0.50, which is considered the most appropriate for conducting genetic diversity analysis (Botstein et al., 1980; Szabo, 2007).
The cluster analysis based on the SSR genotype data grouped 29 Pgt pathotypes into five MGs, of which MGs I and III were further divided into three and two subgroups, respectively (Figure 5). The results of the PCoA explaining 25.11% cumulative variance corroborated the NJ cluster analysis. The Pgt pathotype grouping as explained through NJ cluster analysis and PCoA was entirely independent of their geographical distribution. For instance, the pathotypes from different regions, i.e., 117/117A-1 (Karnataka), 42B (Maharashtra), and 14 (Madhya Pradesh) were grouped in cluster MG-V, which reflected high genetic similarity among them (Figure 5). Similarly, pathotypes 21, 21-1, and 21A-2 shared almost analogous virulence phenotype and SSR genotypes but detected in distal geographical regions, i.e., 21 and 21-1 in North India and 21A-2 in Central India. This relationship of the geographical distribution of Pgt pathotypes with their SSR genotypes could also be endorsed by the findings of AMOVA (Supplementary Figure 2 and Supplementary Table 4). An explanation to justify such distribution of Pgt pathotypes could be the existence of gene flow (the transfer of genetic information from one population to the other) over long distances. Similar findings for cereal rust pathogens are reported from India, South America, North America, and Europe (McCallum et al., 1999; Ordonez and Kolmer, 2007; Prasad et al., 2017a). Conversely, clonal lineages were described for Puccinia spp. evolution from Australia {Pgt (Zhang et al., 2017), P. coronata f. sp. avenae (Brake et al., 2001), and P. graminis f. sp. avenae (Keiper et al., 2006)} and South Africa {Pgt (Visser et al., 2009)}.
The correlation analysis of virulence phenotype and SSR genotype data of all Pgt pathotypes revealed a positive (R2 = 0.1546) correlation between these two variables, which suggests a poor correlation between the virulence phenotypes and SSR genotypes. This is logical because the SSRs used in this study were unrelated to pathogenicity/virulence-specific genes. Similar findings were observed in our earlier studies (Prasad et al., 2017a,2018; Gangwar et al., 2021). Such correlation was also witnessed in the Puccinia striiformis population by Chen et al. (2010), who clarified that SSR genotypes were independent of virulence and that the whole genomic region of the pathogen evolved much faster than its pathogenicity genes. In contrast, a strong correlation between virulence phenotype and SSR genotype data was observed in a population of Puccinia triticina from China (Kolmer, 2015) and Canada (Wang et al., 2010) and of P. striiformis from Europe (Hovmoller et al., 2002) populations. Such a positive strong correlation could be the result of SSR genotypic data acquired by the markers designed from virulence/pathogenicity genes, which are distributed throughout the pathogen genome (Ordonez and Kolmer, 2009; Kolmer, 2015). Moderate correlation (0.43; P = 1.0) between virulence phenotypes and SSR genotypes was reported for the P. triticina population from North America (Ordonez and Kolmer, 2009).
The value of different genetic diversity parameters, namely, Na, Ne, I, He, uHe, and %P was maximum for the Pgt population from Maharashtra and minimum for North India. These results suggest that the Pgt population is highly diverse in Maharashtra followed by Tamil Nadu, while the North Indian population has poor genetic diversity. These findings support our previous results for P. triticina and Pgt populations in the Indian subcontinent, wherein we documented that wheat cultivation round the year and availability of green bridges in higher hills of Tamil Nadu (Nilgiris), which receives high UV intensity sunlight, might be helping these pathogens to mutate and survive without difficulty (Prasad et al., 2017a,2018). All these factors are known to be critical for the evolution and survival of new virulences in nature (Cheng et al., 2014; Singh et al., 2015; Zhao et al., 2020). The new Pgt isolates evolved in these hills migrate to the foothills of Tamil Nadu and Karnataka, and subsequently, to plains of peninsular India (Karnataka, Maharashtra) and Central India (Madhya Pradesh), where these are established on susceptible wheat cultivars (Mehta, 1952). There are speculations that the primary source of inoculum of Pgt is also present in parts of Karnataka other than Nilgiris (Bhardwaj et al., 1989). Furthermore, the cultivation of susceptible bread, durum, and diacoccum wheat species in these areas might be helping an array of Pgt pathotypes with a diverse avirulence/virulence formula to survive and evolve locally. In contrast, cultivation of genetically uniform wheat species (mostly aestivum) in a small stem rust-prone geographical area in North India together with a short favorable period for stem rust development followed by harsh summers prohibits Pgt isolates to evolve and survive until the next crop season, which is reflected by poor genetic diversity within Pgt population in this region (Bhardwaj et al., 2019). The SSR genotype-based population structure analysis using a model-based tool clustered 29 Pgt pathotypes into two major subpopulations (i.e., SP1 and SP2), containing eleven pathotypes in each, and an admixture comprising seven pathotypes (i.e., 184, 117, 184-1, 34-1, 14, 40-1, and 42). Similar differentiation comprising subpopulations and admixture, using population structure analysis, was reported for the Pgt population from South Africa (Terefe et al., 2016; Boshoff et al., 2018) and P. graminis f. sp. avenae population from Australia, Brazil, and Sweden (Gnocato et al., 2018). However, four subpopulations were explained by a combined analysis of Pgt isolates from South Africa and Australia (Visser et al., 2019). Like cluster analysis, PCoA, and AMOVA, the grouping of Pgt pathotypes explained by population structure analysis was not related to their geographical distribution. For example, pathotypes 117-2, 21, and 184 belonged to the North India region while population structure analysis classified them in SP1, SP2, and admixture, respectively. Our results comprehend that the Pgt population in the Indian subcontinent might have evolved from two clonal events, and subsequently, gene flow, long-distance migration, and somatic hybridizations could have contributed to the development of the admixture population, which is not exceptional in Puccinia spp. (Park et al., 1999; Park and Wellings, 2012). The low Nei’s genetic distance between different population combinations indicated that the Pgt population belonging to different geographical regions in the Indian subcontinent are closely related and thus explain poor genetic divergence. Similar findings are reported for two P. striiformis populations from Bajaur and Malakand in Pakistan, while substantial genetic divergence was observed between P. striiformis populations from Nepal, Pakistan, and Bhutan (Khan et al., 2019). Likewise, genetic divergence among P. triticina isolates collected from Aegilops speltoides, durum, and bread wheat; delimited by host specificity was reported from Israel (Liu et al., 2014). Genetic drift, gene flow, accumulated mutation, adaptation, and host specificity are suggested as the driving forces behind genetic divergence among Puccinia spp. (Kohn, 2005; Kolmer et al., 2020).
The repository of 29 Pgt pathotypes collected during the last ∼90 years exhibited significant variability for virulence phenotype and SSR genotypes. The SSR markers used in this study were highly informative in explaining the genetic diversity among Pgt pathotypes. As revealed by different statistical programs and hypothetical evolutionary hierarchy, the gene flow, long-distance dispersal, and single-step mutation for gain and/or rare loss of virulence could have contributed to cryptic genetic variation and evolution in the Pgt population from the Indian subcontinent. The findings of population structure and genetic diversity analysis of Pgt population, supplemented by deployment of effective Sr genes (Sr26, Sr27, Sr31, Sr32, Sr33, Sr35, Sr39, Sr40, Sr43, SrTmp, and SrTt3) in combination with other APR genes, could be utilized for strategic wheat stem rust management.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
PP, SS, and SB conceived the idea and designed the experiments. PP, RT, and SK performed the virulence phenotyping. PP and RT performed SSR genotyping. PP, SS, OG, CL, and SA performed the data analysis and prepared first draft of the manuscript. PP and SB reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We sincerely thank the Director, Indian Council of Agricultural Research (ICAR)-Indian Institute of Wheat and Barley Research (IIWBR), Karnal, India for financial and liberal support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.842106/full#supplementary-material
Abebe, T., Woldeab, G., and Dawit, W. (2012). Distribution and physiologic races of wheat stem rust in Tigray, Ethiopia. J. Plant Pathol. Microbiol. 3, 1–6.
Anderson, J., and Lubberstedt, T. (2003). Functional markers in plants. Trends Plant Sci. 8, 554–560. doi: 10.1016/j.tplants.2003.09.010
Anderson, J. A., Churchil, G. A., Autrique, J. E., Tanksley, S. O., and Sorrels, M. E. (1993). Optimizing parent selection for genetic linkage maps. Genome 36, 181–186. doi: 10.1139/g93-024
Bahadur, P., Nagarajan, S., and Nayar, S. K. (1985a). A proposed system for virulence designation in India to Puccinia graminis f. sp. tritici. Proc. Indian Acad. Sci. 95, 29–33.
Bahadur, P., Nagarajan, S., and Nayar, S. K. (1985b). Identification of new virulence of Puccinia graminis f. sp. tritici in India during 1985. Cereal rusts and Powdery Mildew Bull. 16, 6–7.
Bhardwaj, S. C., Nayar, S. K., Kumar, J., and Prashar, M. (1989). Occurrence of a new virulent form 117-1 in Race 117 of Puccinia graminis f. sp. tritici. Cereal Rusts Powdery Mildews Bull. 17, 1–5.
Bhardwaj, S. C., Nayar, S. K., Prashar, M., Kumar, J., Menon, M. K., and Singh, S. B. (1990). A Pathotype of Puccinia graminis f. sp. tritici on Sr24 in India. Cereal Rusts Powdery Mildews Bull. 18, 35–37.
Bhardwaj, S. C., Nayar, S. K., Prashar, M., Kumar, J., Menon, M. K., and Singh, S. B. (1994). Further Variation in 117 Group of Pathotypes in Puccinia graminis tritici in India. Plant Dis. Res. 9, 91–96.
Bhardwaj, S. C., Prasad, P., Gangwar, O. P., Khan, H., and Kumar, S. (2016). Wheat rust research-then and now. Indian J. Agric. Sci. 86, 1231–1244.
Bhardwaj, S. C., Singh, G. P., Gangwar, O. P., Prasad, P., and Kumar, S. (2019). Status of wheat rust research and progress in rust management-Indian context. Agronomy 9:892. doi: 10.3390/agronomy9120892
Boshoff, W. H. P., Pretorius, Z. A., Terefe, T. G., Bender, C. M., Maree, G. J., Visser, B., et al. (2018). Phenotypic and genotypic description of Puccinia graminis f. sp. tritici race 2SA55 in South Africa. Eur. J. Plant Pathol. 152, 783–789. doi: 10.1007/s10658-018-1527-3
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331.
Brake, V. M., Irwin, J. A. G., and Park, R. F. (2001). Genetic variability in Australian isolates of Puccinia coronate f. sp. avenae assessed with molecular and pathogenicity markers. Aust. Plant Pathol. 30, 259–266.
Chen, X. M., Penman, L., Wan, A. M., and Cheng, P. (2010). Virulence races of Puccinia striiformis f. sp. tritici in 2006 and 2007 and development of wheat stripe rust and distributions, dynamics, and evolutionary relationships of races from 2000 to 2007 in the United States. Can. J. Plant Pathol. 32, 315–333. doi: 10.1080/07060661.2010.499271
Cheng, P., Ma, Z., Wang, X., Wang, C., Li, Y., Wang, S., et al. (2014). Impact of UV-B radiation on aspects of germination and epidemiological components of three major physiological races of Puccinia striiformis f. sp. tritici. Crop Protect. 65, 6–14. doi: 10.1016/j.cropro.2014.07.002
Dill-Macky, R., Rees, R. G., and Platz, G. J. (1990). Stem rust epidemics and their effects on grain yield and quality in Australian barley cultivars. Aust. J. Agric. Res. 41, 1057–1063. doi: 10.1071/ar9901057
Eversmeyer, M. G., and Kramer, C. L. (2000). Epidemiology of wheat leaf and stem rust in the central great plains of the USA. Ann. Rev. Phytopathol. 38, 491–513. doi: 10.1146/annurev.phyto.38.1.491
FAO (2021). Crop Prospects and Food Situation - Quarterly Global Report No. 1. Rome: FAO. doi: 10.4060/cb3672en
Gangwar, O. P., Kumar, S., Bhardwaj, S. C., Prasad, P., Kashyap, P. L., Khan, H., et al. (2021). Virulence and molecular diversity among Puccinia striiformis f. sp. tritici pathotypes identified in India between 2015 and 2019. Crop Protect. 148, 105717. doi: 10.1016/j.cropro.2021.105717
Gnocato, F. S., Dracatos, P. M., Karaoglu, H., Zhang, P., Berlin, A., and Park, R. F. (2018). Development, characterization and application of genomic SSR markers for the oat stem rust pathogen Puccinia graminis f. sp. avenae. Plant Pathol. 67, 457–466.
Hovmoller, M. S., Justesen, A. F., and Brown, J. K. M. (2002). Clonality and long-distance migration of Puccinia striiformis f. sp. tritici in northwest Europe. Plant Pathol. 51, 24–32. doi: 10.1046/j.1365-3059.2002.00652.x
Huerta-Espino, J., Singh, R., Crespo-Herrera, L. A., Villaseñor-Mir, H. E., Rodriguez-Garcia, M. F., Dreisigacker, S., et al. (2020). Adult plant slow rusting genes confer high levels of resistance to rusts in bread wheat cultivars from Mexico. Front. Plant Sci. 11:824. doi: 10.3389/fpls.2020.00824
Jain, S. K., Prashar, M., Bhardwaj, S. C., Singh, S. B., and Sharma, Y. P. (2009). Emergence of virulence to Sr25 of Puccinia graminis f. sp. tritici on wheat in India. Plant Dis. 93:840. doi: 10.1094/PDIS-93-8-0840B
Jain, S. K., Bhardwaj, S. C., Prashar, M., and Singh, S. B. (2013). Physiologic specialization and new virulences of Puccinia graminis f sp tritici causing black rust of wheat (Triticum aestivum) in India during 2005-2009. Indian J. Agr. Sci. 83, 1058–1063.
Jin, Y., Szabo, L. J., Pretorius, Z. A., Singh, R. P., Ward, R., and Fetch, T. (2008). Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 92, 923–926. doi: 10.1094/PDIS-92-6-0923
Karaoglu, H., Lee, C. M. Y., and Park, R. (2013). Simple sequence repeats in Puccinia graminis: abundance, cross-formae speciales and intra-species utility, and development of novel markers. Aust. Plant Pathol. 42, 271–281. doi: 10.1007/s13313-013-0199-x
Keiper, F. J., Haque, M. S., Hayden, M. J., and Park, R. F. (2006). Genetic diversity in Australian populations of Puccinia graminis f. sp. avenae. Phytopathology 96, 96–104. doi: 10.1094/PHYTO-96-0096
Khan, M. R., Rehman, Z. U., Nazir, S. N., Tshewang, S., Baidya, S., Hodson, D., et al. (2019). Genetic divergence and diversity in Himalayan Puccinia striiformis populations from Bhutan, Nepal, and Pakistan. Phytopathology 109, 1793–1800. doi: 10.1094/PHYTO-01-19-0031-R
Kiran, K., Rawal, H. C., Dubey, H., Jaswal, R., Bhardwaj, S. C., Prasad, P., et al. (2017). Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci. Rep. 7:42419. doi: 10.1038/srep42419
Kohn, L. M. (2005). Mechanisms of fungal speciation. Annu. Rev. Phytopathol. 43, 279–308. doi: 10.1146/annurev.phyto.43.040204.135958
Kolmer, J. A. (2015). Collections of Puccinia triticina in different provinces of China are highly related for virulence and molecular genotype. Phytopathology 105, 700–706. doi: 10.1094/PHYTO-11-14-0293-R
Kolmer, J. A., Herman, A., Ordoñez, M. E., German, S., Morgounov, A., Pretorius, Z., et al. (2020). Endemic and panglobal genetic groups, and divergence of host-associated forms in worldwide collections of the wheat leaf rust fungus Puccinia triticina as determined by genotyping by sequencing. Heredity 124, 397–409. doi: 10.1038/s41437-019-0288-x
Liu, M., Rodrigue, N., and Kolmer, J. (2014). Population divergence in the wheat leaf rust fungus Puccinia triticina is correlated with wheat evolution. Heredity 112, 443–453. doi: 10.1038/hdy.2013.123
Lu, H., Redus, M. A., Coburn, J. R., Rutger, J. N., McCouch, S. R., and Tai, T. H. (2005). Population structure and breeding patterns of 145 US rice cultivars based on SSR marker analysis. Crop Sci. 45, 66–76. doi: 10.2135/cropsci2005.0066
McCallum, B. D., Roelfs, A. P., Szabo, L. J., and Groth, J. V. (1999). Comparison of Puccinia graminis f. sp. tritici from South America and Europe. Plant Pathol. 48, 574–581. doi: 10.1046/j.1365-3059.1999.00367.x
Mehta, K. C. (1940). Further studies on Cereal Rusts in India (Vol I). Imperial Council Agricultural Research. New Delhi. Sci. Monogr. 14:224.
Mehta, K. C. (1952). Further Studies on Cereal Rusts of India, Part II. Scientific Monograph, Vol. 18. New Delhi: ICAR.
Nayar, S. K., Prashar, M., and Bhardwaj, S. C. (1997). Manual of Current Techniques in Wheat Rusts. Research Bulletin No.2. Flowerdale: DWR Regional Station, 32.
Olivera, P., Newcomb, M., Szabo, L. J., Rouse, M., Johnson, J., Gale, S., et al. (2015). Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in Southern Ethiopia in 2013-14. Phytopathology 105, 917–928. doi: 10.1094/PHYTO-11-14-0302-FI
Olivera, P. D., Sikharulidze, Z., Dumbadze, R., Szabo, L. J., Newcomb, M., Natsarishvili, K., et al. (2019). Presence of a sexual population of Puccinia graminis f. sp. tritici in Georgia provides a hotspot for genotypic and phenotypic diversity. Phytopathology 109, 2152–2160. doi: 10.1094/PHYTO-06-19-0186-R
Ordonez, M. E., and Kolmer, J. A. (2007). Simple sequence diversity of a worldwide collection of Puccinia triticina from durum wheat. Phytopathology 97, 574–583. doi: 10.1094/phyto-97-5-0574
Ordonez, M. E., and Kolmer, J. A. (2009). Differentiation of molecular genotypes and virulence phenotypes of Puccinia triticina from common wheat in North America. Phytopathology 99, 750–758. doi: 10.1094/PHYTO-99-6-0750
Park, R. F., Burdon, J. J., and Jahoor, A. (1999). Evidence for somatic hybridization in nature in Puccinia recondita f. sp. tritici, the leaf rust pathogen of wheat. Mycol. Res. 103, 715–723. doi: 10.1017/s0953756298007631
Park, R. F., and Wellings, C. R. (2012). Somatic hybridization in the Uredinales. Annu. Rev. Phytopathol. 50, 219–239. doi: 10.1146/annurev-phyto-072910-095405
Patil, B. P., Mutkekar, M. L., and Sulaiman, M. (1963). On the Occurrence of Biotype 117A of Puccinia graminis (Pers.) Eriks. and Henn. in India. Indian Phytopathol. 16, 244–245.
Patpour, M., Hovmoller, M. S., Justesen, A. F., Newcomb, M., Shahin, A., and Szabo, L. J. (2016). Emergence of virulence to SrTmp in the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis. 100:522. doi: 10.1094/pdis-06-15-0668-pdn
Patpour, M., Justesen, A. F., Tecle, A. W., Yazdani, M., Yasaie, M., and Hovmøller, M. S. (2020). First report of race TTRTF of wheat stem rust (Puccinia graminis f. sp. tritici) in Eritrea. Plant Dis. 104:973. doi: 10.1094/pdis-10-19-2133-pdn
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5, Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Perrier, X., and Jacquemoud-collet, J. P. (2006). DARwinsoftware. Available online at: http://darwin.cirad.fr [accessed on Auguest 28, 2021]
Prasad, P., Bhardwaj, S. C., Gangwar, O. P., and Kumar, S. (2020). “Wheat rust research: impact, thrusts, and roadmap to sustained wheat production,” in Improving Cereal Productivity through Climate Smart Practices, eds S. Sindhu Sareen, P. Sharma, C. Singh, P. Jasrotia, G. P. Singh, and A. K. Sarial (Amsterdam: Elsevier), 177–203. doi: 10.1016/b978-0-12-821316-2.00011-x
Prasad, P., Bhardwaj, S. C., Khan, H., Gnangwar, O. P., Savadi, S., and Kumar, S. (2017b). “Stem rust pathotype Ug99: an Indian context,” in Management of Wheat and Barley Diseases, ed. D. P. Singh (New Jersy, NJ: Apple Academic Press), 553–578. doi: 10.1201/9781315207537-21
Prasad, P., Bhardwaj, S. C., Gangwar, O. P., Kumar, S., Khan, H., Kumar, S., et al. (2017a). Population differentiation of wheat leaf rust fungus Puccinia triticina in South Asia. Curr. Sci. 112, 2073–2084. doi: 10.18520/cs/v112/i10/2073-2084
Prasad, P., Bhardwaj, S. C., Khan, H., Gangwar, O. P., Kumar, S., and Singh, S. B. (2016). Ug99: saga, reality and status. Curr. Sci. 110, 1614–1616.
Prasad, P., Bhardwaj, S. C., Savadi, S., Kashyap, P. L., Gangwar, O. P., Khan, H., et al. (2018). Population distribution and differentiation of Puccinia graminis tritici detected in the Indian subcontinent during 2009-2015. Crop Protect. 108, 128–136. doi: 10.1016/j.cropro.2018.02.021
Prasada, R., and Lele, V. C. (1952). New physiologic races in India. Indian Phytopathol. 5, 128–129.
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., and Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 84:203. doi: 10.1094/pdis.2000.84.2.203b
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Savadi, S., Prasad, P., Kashyap, P. L., and Bhardwaj, S. C. (2017). Molecular breeding technologies and strategies for rust resistance in wheat (Triticum aestivum) for sustained food security. Plant Pathol. 67, 771–791. doi: 10.1111/ppa.12802
Savadi, S., Prasad, P., Sharma, K., Rathore, R., Bhardwaj, S., Gangwa, O. P., et al. (2020). Development of novel transcriptome-based SSR markers in Puccinia triticina and their potential application in genetic diversity studies. Trop. Plant Pathol. 45, 499–510. doi: 10.1007/s40858-020-00347-8
Sharma, S. K., Ruikar, S. K., Goel, L. B., and Kulkarni, S. (1979). A New Virulence 117A-1 of stem rust of wheat and its sources of resistance. Indian Phytopathol. 32, 498–499.
Sharma, S. K., Singh, S. D., and Nayar, S. K. (1975). A new virulence of stem rust of wheat attacking chhoti lerma. Curr. Sci. 44:486.
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Sinha, V. C., Upadhyaya, Y. M., and Bahadur, P. (1976). A new virulence in race 11 of stem rust of wheat and some sources of resistance. Indian J. Genet. Plant Br. 36, 345–346.
Soko, T., Bender, C. M., Prins, R., and Pretorius, Z. A. (2018). Yield loss associated with different levels of stem rust resistance in bread wheat. Plant Dis. 102, 2531–2538. doi: 10.1094/PDIS-02-18-0307-RE
Stakman, E. C., Stewart, D. M., and Loegering, W. Q. (1962). Identification of physiologic races of Puccinia graminis var. tritici. U. S. Agric. Res. Serv. 617, 1–53.
Szabo, L. J. (2007). Development of simple sequence repeat markers for the plant pathogenic rust fungus, Puccinia graminis. Mol. Ecol. Notes 7, 92–94. doi: 10.1111/j.1471-8286.2006.01540.x
Terefe, T. G., Visser, B., and Pretorius, Z. A. (2016). Variation in Puccinia graminis f. sp. tritici detected on wheat and triticale in South Africa from 2009 to 2013. Crop Protect. 86, 9–16. doi: 10.1016/j.cropro.2016.04.006
Visser, B., Herselman, L., and Pretorius, Z. A. (2009). Genetic comparison of Ug99 with selected South African races of Puccinia graminis f. sp. tritici. Mol. Plant Pathol. 10, 213–222. doi: 10.1111/j.1364-3703.2008.00525.x
Visser, B., Meyer, M., Park, R. F., Gilligan, C. A., Burgin, L. E., Hort, M. C., et al. (2019). Microsatellite analysis and urediniospore dispersal simulations support the movement of Puccinia graminis f. sp. tritici from Southern Africa to Australia. Phytopathology 109, 133–144. doi: 10.1094/PHYTO-04-18-0110-R
Wang, X., Mulock, B., Guus, B., and McCallum, B. (2010). Development of EST-derived simple sequence repeat markers for wheat leaf rust fungus, Puccinia triticina Eriks. Can. J. Plant Pathol. 32, 98–107. doi: 10.1080/07060661003594133
Worku, D., Zerihun, T., Daniel, K., Habtemariam, Z., Dawit, A., and Wanyera, R. (2016). Development of wheat germplasm for stem rust resistance in eastern Africa. Afric. Crop Sci. J. 24, 25–33. doi: 10.4314/acsj.v24i1.3s
Zhang, J., Zhang, P., Karaoglu, H., and Park, R. F. (2017). Molecular characterization of Australian isolates of Puccinia graminis f. sp. tritici supports long-term clonality but also reveals cryptic genetic variation. Phytopathology 107, 1032–1038. doi: 10.1094/PHYTO-09-16-0334-R
Zhao, Y., Cheng, P., Zhang, Y., and Wang, H. (2020). Proteomic Analysis of UV-B-Induced Virulence-Mutant Strains of Puccinia striiformis f. sp. tritici based on iTRAQ Technology. Front. Microbiol. 11:542961. doi: 10.3389/fmicb.2020.542961
Keywords: wheat, stem rust, virulence phenotype, SSR genotype, genetic diversity, evolution
Citation: Prasad P, Thakur RK, Savadi S, Bhardwaj SC, Gangwar OP, Lata C, Adhikari S and Kumar S (2022) Genetic Diversity and Population Structure Reveal Cryptic Genetic Variation and Long Distance Migration of Puccinia graminis f. sp. tritici in the Indian Subcontinent. Front. Microbiol. 13:842106. doi: 10.3389/fmicb.2022.842106
Received: 23 December 2021; Accepted: 16 March 2022;
Published: 13 April 2022.
Edited by:
Peng Zhang, The University of Sydney, AustraliaReviewed by:
Pablo Daniel Olivera, University of Minnesota Twin Cities, United StatesCopyright © 2022 Prasad, Thakur, Savadi, Bhardwaj, Gangwar, Lata, Adhikari and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subhash Chander Bhardwaj, c2NiZmRsQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.