
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 May 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.841920
This article is part of the Research Topic Microbial interactions of Clostridioides difficile View all 7 articles
 Zhengjie Wu1†
Zhengjie Wu1† Qiaomai Xu1†
Qiaomai Xu1† Silan Gu1†
Silan Gu1† Yunbo Chen1
Yunbo Chen1 Longxian Lv1
Longxian Lv1 Beiwen Zheng1
Beiwen Zheng1 Qiangqiang Wang1
Qiangqiang Wang1 Kaicen Wang1
Kaicen Wang1 Shuting Wang1
Shuting Wang1 Jiafeng Xia1
Jiafeng Xia1 Liya Yang1
Liya Yang1 Xiaoyuan Bian1
Xiaoyuan Bian1 Xianwan Jiang1
Xianwan Jiang1 Lisi Zheng1
Lisi Zheng1 Lanjuan Li1,2*
Lanjuan Li1,2*Clostridioides difficile is a common cause of nosocomial infection. Antibiotic-induced dysbiosis in the intestinal microbiota is a core cause of C. difficile infection (CDI). Akkermansia muciniphila plays an active role in maintaining gastrointestinal balance and might offer the protective effects on CDI as probiotics. Here, we investigated the effects and mechanisms of A. muciniphila on CDI. C57BL/6 mice (n = 29) were administered A. muciniphila MucT (3 × 109 CFUs, 0.2 mL) or phosphate-buffered saline (PBS) by oral gavage for 2 weeks. Mice were pretreated with an antibiotic cocktail and subsequently challenged with the C. difficile strain VPI 10463. A. muciniphila treatment prevented weight loss in mice and reduced the histological injury of the colon. And it also alleviated inflammation and improved the barrier function of the intestine. The administration effects of A. muciniphila may be associated with an increase in short-chain fatty acid production and the maintenance of bile acids’ steady-state. Our results provide evidence that administration of A. muciniphila to CDI mice, with an imbalance in the microbial community structure, lead to a decrease in abundance of members of the Enterobacteriaceae and Enterococcaceae. In short, A. muciniphila shows a potential anti-CDI role by modulating gut microbiota and the metabolome.
Clostridioides difficile is a Gram-positive and spore-forming anaerobic bacteria, which can cause diarrhea and severe complications such as pseudomembranous colitis (Gerding et al., 1995; Fletcher et al., 2021). The pathogenesis of C. difficile is mainly derived by two toxins, toxin A and toxin B, which induce intestinal inflammation and tissue damage (Kuehne et al., 2010). The indiscriminate use of antibiotics is currently regarded as the most common reason for gut microbiota imbalance. If this disruption reaches a certain level, the host can develop C. difficile-associated disease (CDAD) (Tang et al., 2016). In general, C. difficile infections (CDI) alter the gut microbiota, decreasing colonization resistance against C. difficile. Antibiotics such as MTZ, vancomycin, and fidaxomicin are still the main therapy for C. difficile (Mushtaq, 2018). In some cases, antibiotics even promote CDIs, which can take over after the symbiotic microflora is damaged by antibiotics (Theriot et al., 2014). These broad-spectrum antibiotics affect the healthy gut microbiota and hinder colonization resistance by reducing the abundance of intestinal microbes that resist C. difficile (van Werkhoven et al., 2021). In turn, the reduced microbiota diversity induces an elevated level of primary bile acids (BAs), which facilitate the germination of C. difficile spores and reduce the concentration of secondary BAs that inhibit the growth of C. difficile (de Aguiar Vallim et al., 2013). In addition to gut microbiota disorders, intestinal inflammation plays a complex role in CDI-mediated diseases (Shen, 2012). Though antibiotics are considered the most effective therapy in clinical practice, the risks of developing antibiotics resistance and further gut microbiota perturbation are important (McDonald et al., 2018b; Rao and Malani, 2020).
Further, the diversity of gut microbes is related to the recurrence of CDI and the severity of the disease. Fecal microbiota transplantation (FMT) can effectively treat recurrent CDI by recovering the gut microbiome (van Nood et al., 2013), but there are concerns regarding its long-term safety. An integrated gut barrier with a healthy microbiota is vital to resist C. difficile pathogenesis. Therefore, supplementing bacteria in a timely manner to establish a health-associated microbiome seems more reasonable. The regulation of the gut microbiome may be critical for preventing and treating CDI. Probiotics improve the balance of microorganisms that populate the gut and reduce CDI in high-risk patients receiving antibiotics (Maziade et al., 2015). Thus, effective alternative non-antibiotic therapies are urgently needed that will be helpful to design strategies to maintain the normal balance of gut microflora. Recently, the focus has shifted to bacteriotherapy which has gradually become an accepted alternative for CDI treatment (Lawley et al., 2012).
Akkermansia muciniphila – a mucophilic anaerobic bacteria, is one such bacterium representing 3–5% of the microbial community in healthy individuals (Belzer and de Vos, 2012; Becken et al., 2021). It has several phenotypes such as oxygen tolerance, adherence to epithelial cells, and bacterial aggregation. It is the first and representative member of the Verrucomicrobia found in the human gut (Derrien et al., 2010), whose metabolic byproducts, such as short-chain fatty acids (SCFAs), benefit other gut bacteria. Additionally, this bacterium is associated with the gut barrier, inflammation, and immune response (Reunanen et al., 2015). A. muciniphila stimulates mucin production (van der Lugt et al., 2019) and is associated with inflammatory bowel disease (Png et al., 2010), metabolic syndrome (Zhang et al., 2009; Santacruz et al., 2010), liver injury (Wu W. et al., 2017), and regulation of gut homeostasis (Derrien et al., 2010). Although A. muciniphila is a mucin-degrading bacterium, it can stimulate mucin production, increase intestinal mucous layer thickness and intestinal barrier integrity (Everard et al., 2013), protect the intestinal tract from pathogens through competitive rejection (van der Lugt et al., 2019). A. muciniphila and its membrane protein Amuc-1100 played an important probiotic role, is considered a very promising probiotic. However, the protective effects of A. muciniphila and its microbial and immunomodulatory properties have not been extensively studied. Therefore, the aim of this study was to explore the protective effects of A. muciniphila on CDI colitis and the potential underlying mechanisms.
Akkermansia muciniphila MucT (= ATCC BAA-835T = CIP 107961T) was grown anaerobically in a BHI medium at 37°C for 48 h (Derrien et al., 2004). The broth was centrifuged at 8,000 g at 4°C for 10 min, washed, and re-suspended in sterile phosphate-buffered saline (PBS). We used a non-hyper virulent strain of C. difficile (strain VPI 10463) known to produce a high-level of toxin (Merrigan et al., 2010). The C. difficile VPI 10463 (ATCC 43255, binary toxin-negative, toxin A-positive, and toxin B-positive) was cultured anaerobically at 37°C for 24 h in anaerobic conditions in Difco cooked meat medium (BD Diagnostic Systems, United States). The culture was centrifuged (3,200 g, 10 min, 4°C). The sediments were washed twice with sterile PBS and re-suspended for further use (Chen et al., 2008).
Female C57BL/6 mice (6–8 weeks old, SLAC Lab, Shanghai, China) were housed under controlled conditions (specific pathogen free). We used a uniform CDI mouse model (Chen et al., 2008). Briefly, mice were grouped randomly: 0.2 mL PBS [CDI group, n = 13; standard control [NC] group, n = 8] and 0.2 mL A. muciniphila suspension [3 × 109 CFUs, A. muciniphila (AKK) group, n = 8] once daily by oral gavage from day-8 to day 5 (Figure 1A). For C. difficile in vivo infection, AKK group and CDI group received antibiotics cocktail (0.4 mg/mL kanamycin, 0.035 mg/mL gentamicin, 850 U/mL colistin, 0.215 mg/mL metronidazole, and 0.045 mg/mL vancomycin) in drinking water for 5 days followed by 2 days of normal water. Mice were intraperitoneally injected with clindamycin (10 mg/kg) 1 day before infection. On day 0, mice were given oral gavage of C. difficile (108 CFUs, 0.2 mL). The diarrhea was graded as 0, normal stool; 1, loose stool; 2, liquid feces or soiled tail (Tam et al., 2018). According to the manufacturers’ instructions, toxins A and B were detected in feces using an enzyme immunoassay (VIDAS C. difficile Toxin A and B; bioMerieux SA, Marcy-l’Étoile, France). The experiment was approved by the Animal Experimental Ethical Inspection of The First Affiliated Hospital, Zhejiang University School of Medicine (Zhejiang, China) (1-7-2021).
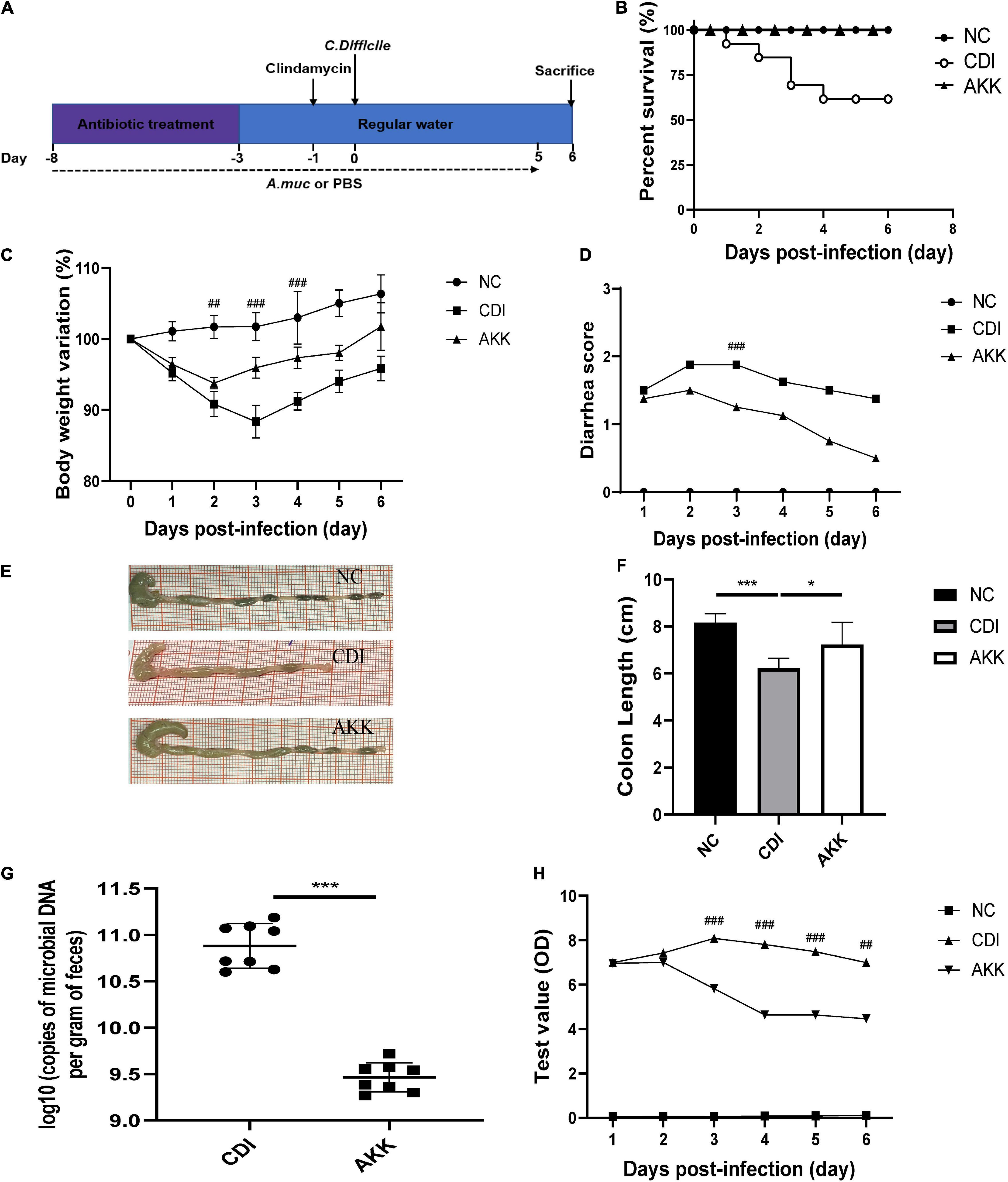
Figure 1. Akkermansia muciniphila administration reduced C. difficile-induced colon damage. (A) Overall schematic experimental plan. (B) Kaplan–Meier survival curve of NC, CDI, and AKK groups. (C) Body weight changes, (D) diarrhea scores during infection. (E) Representative colon pictures and (F) colon length after A. muciniphila administration. (G) C. difficile load in feces was detected on day 6. (H) Fecal toxins A and B were detected on post-infection days and expressed as test values (ODs). *P < 0.05, **P < 0.01, ***P < 0.001, ##P < 0.01, ###P < 0.001 for CDI group vs. AKK group.
Upon euthanasia, distal colon segments were immediately postfixed in 10% formalin. The tissue sample was embedded in paraffin and stained with hematoxylin and eosin. The degree of enteritis, which uses a scoring system as previously described (Chen et al., 2008), consists of epithelial cell damage (score: 0–3), congestion/edema (score: 0–3), and neutrophil infiltration (score: 0–3). Two independent pathology professors double-blindly evaluated these sections. Alcian blue and periodic acid-Schiff (AB-PAS) staining were performed with the instructions (Solarbio). The sections were stained with p-mTOR, beclin1, zonula occludens-1 (ZO-1), occludin, and claudin-1 antibodies as previously described (Chung et al., 2014). Images were managed using the Zeiss LSM T-PMT confocal microscope (Zeiss, Jena, Germany).
The proximal colon samples were collected and immediately transferred to glutaraldehyde (2.5%) at 4°C for 4 h, postfixed with 1% OsO4 and dehydrated in graded alcohol concentrations, and embedded in Spurr resin (SPI-CHEM). The specimen was sectioned using the LEICA EM UC7 ultratome (Leica Microsystems GmbH, Wetzlar, Germany), and sections were stained with uranyl acetate (SPI-CHEM) and alkaline lead citrate. These sections were observed by transmission electron microscopy (TEM) (H-7650; Hitachi, Tokyo, Japan).
The limulus amebocyte lysate (LAL) assay detected and quantitated the endotoxins. Serum LAL levels were measured using the LAL Chromogenic Endpoint Assay (Hycult Biotechnology, Uden, Netherlands). Lipopolysaccharide (LPS)-binding protein (LBP) concentrations were detected using an enzyme-linked immunosorbent assay (ELISA) kit (Abcam, Cambridge, MA, United States). The serum and colon cytokines were measured with the Bio-Plex Pro Mouse Cytokine 23-Plex Assay Kit and Chemokine 31-Plex Assay Kit (Bio-Rad, Hercules, CA, United States). The 23-plex contains granulocyte colony-stimulating factor (G-CSF), monocyte chemoattractant protein-1 (MCP-1), interleukin 17A (IL-17A), IL-1α, IL-6, and tumor necrosis factor-alpha (TNF-α). The 31-plex contains: IL-1β, IL-6, IP-10/C-X-C motif chemokine ligand 10 (CXCL10), MCP-1/CCL2, MDC/CCL22, BCA-1/CXCL13, I-309/C-C motif chemokine ligand 1 (CCL1), MIP-1α/CCL3, MIP-1β/CCL4, RANTES/CCL5, TARC/CCL17, and TNF-α.
Total RNA of colorectal tissue was reverse transcribed into cDNA using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA, United States) and PrimeScript RT Master Kit (Takara Biomedicals, Kusatsu, Japan). The mRNA relative abundance was measured in triplicate using the Premix Ex Taq (Takara Biomedicals) using the VIIA7 Real-time PCR System (Applied Biosystems, Beverly, MA, United States). The relative mRNA expression was determined using the 2–ΔΔCT method and normalized to the internal control β-actin. The primer sequences are listed in Supplementary Table 1.
Fecal samples and extracted DNA through the DNeasy PowerSoil Pro Kit (Qiagen). DNA from fecal samples was used in quantitative PCR. Previously reported primers were used to detect and quantify C. difficile (Pereira et al., 2020) and A. muciniphila (Everard et al., 2013). The standard curve used serial dilutions of isolating DNA to quantify the DNA. The samples were tested in duplicate.
DNA from fecal samples on day 6 was extracted by the DNeasy PowerSoil Pro Kit (Qiagen) (NC = 8, CDI = 8, AKK = 8). The 16S rRNA was amplified using primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) corresponding to the V3–V4 region. Sequencing was performed using the Illumina MiSeq PE300 system and was analyzed on Majorbio Cloud Platform1 as previously described (Gu et al., 2020). Briefly, operational taxonomic units (OTUs) were clustered using UPARSE version 7.1 (Edgar, 2013) with a 0.97 threshold. Representative reads of each OTU were selected, and taxonomic data were then assigned using the RDP classifier and display a 70% confidence threshold. Alpha diversity and beta diversity were calculated using QIIME software. Alpha diversity was estimated with the Chao 1 index and Shannon index. Using the Adonis function, we performed a permutational multivariate analysis of variance (PERMANOVA) based on Bray–Curtis distances. Beta diversity was assessed by the Bray–Curtis distance and presented by principal coordinate analysis (PCoA). The differences between groups in taxonomic composition taxa were analyzed using the linear discriminant analysis (LDA) effect size (LEfSe) analysis, and LDA scores greater >4 were defined as discriminative taxa. The sequencing data has been uploaded to the NCBI Sequence Read Archive (SRA) database (PRJNA 766049).
Bile acids (TCA, taurocholic acid; βMCA, β-muricholic acid; CA, cholic acid; αMCA, α-muricholic acid; UDCA, ursodeoxycholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; ωMCA, ω-muricholic acid; HDCA, hyodeoxycholic acid; muroCA, murocholic acid; βDCA, β-deoxycholic acid; and LCA, lithocholic acid) in colon contents were detected according to previously described protocol (Xie et al., 2015; Yang et al., 2021) and purchased from Steraloids, Inc. (Newport, RI, United States). Briefly, colon contents were mixed with 200 μL acetonitrile/methanol (volume 8:2), which contained internal standards (GCA-d4, TCA-d4, GDCA-d4, CA-d4, DCA-d4, and LCA-d4). After homogenization and centrifugation (13,500 rpm, 20 min, 4°C), 10 μL supernatant was diluted with 90 μL acetonitrile/methanol (80/20) and ultrapure water mixture solution (volume 1:1). Colon contents BAs were detected by ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, United States).
Short-chain fatty acids in feces were detected according to the previous method (Bian et al., 2020). Stool samples were mixed with 500 μl water (hexanoic acid-d3, internal standard, 10 μg/ml). After homogenization, samples were centrifugated at 15,000 rpm and 4°C for 5 min and mixed the supernatant with the equivalent volume mixture of ethyl acetate/sulfuric acid (v/v = 10:1). After centrifugation and derivatization, the supernatant was run and analyzed by GC/MS (Agilent Technologies, Santa Clara, CA, United States).
GraphPad Prism (GraphPad Software, Inc., CA, United States) and SPSS 20.0 (SPSS, Inc., Chicago, IL, United States) was used for statistical analyses. For statistical significance, either one-way ANOVA followed by the Tukey’s test or Kruskal–Wallis test was used. Kaplan–Meier survival curves used the log-rank (Mantel–Cox) test. Spearman’s rank correlation test analyzed correlations between relevant variables. P < 0.05 was considered statistically significant.
As previously reported, body weight loss during CDI-induced infection (Xu et al., 2018) can be a marker of disease progression. Therefore, we used weight loss during the disease course to measure CDI disease severity. The CDI group showed distinct weight loss after infection (D3, CDI group vs. NC group, P < 0.001; Figure 1C), and five mice died by day 4 (Figure 1B; P < 0.05). All mice in the NC group and AKK group survived. Oral supplementation of A. muciniphila reduced body weight loss after infection (Figure 1C), alleviated the diarrhea score (Figure 1D), relieved colon shortening (NC vs. CDI groups, P < 0.001; AKK group vs. NC group, P < 0.05; AKK group vs. CDI group, P < 0.05; Figures 1E,F). Post-infection fecal samples showed a reduction of C. difficile burden and its toxins with AKK treatment (P < 0.001; Figures 1G,H). Therefore, probiotics significantly reduced CDI-induced clinical symptoms. To establish the mechanism of protection induced by A. muciniphila in CDI- induced mice we studied histopathological analysis and immune response to treat such infection.
Immunohistological staining suggested that A. muciniphila reduced histological injury in the distal colon, such as destroying the histological structure and epithelial barrier (Figure 2A and Supplementary Figure 1). TEM showed that intestinal epithelial cell (IEC) microvilli were ruptured in the CDI group. This disruption across the brush border was reduced in the AKK group (Figure 2A). We speculated that A. muciniphila may play a role by enhancing the mucosal barrier. Immunofluorescence staining and quantitative real-time PCR (qPCR) were used to assess the expression of tight junction proteins (Tjps) and mRNA, respectively. As shown in Supplementary Figure 1A, the AKK group showed stable mucosal integrity of the colon tissue and increased fluorescence intensity of the ZO-1, occludin, and claudin-1 compared to the CDI group. The mRNA expression of Tjps in the colonic tissue of the AKK group was higher than that of the CDI group (occludin, ZO-1/Tjps, claudin-1, P < 0.05 for all; Supplementary Figure 1A). Bacteria and their metabolites, such as LPS, invade the intestines and blood when a leaky gut occurs. LBP is a carrier for LPS and helps its macrophages recognize. ELISA measured serum LBP and LAL to assess the severity of bacterial translocation and inflammation. The results showed that serum LBP and LAL levels in the CDI group were higher than those in the NC and AKK groups (P < 0.05; Figure 2C). In addition, intestinal permeability was evaluated by assessing MUC2 and cannabinoid receptor (CB) levels. CB1 and CB2 had significantly increased expression in the CDI group than the NC group (P < 0.05), meanwhile the expression was downregulated in the AKK group than the CDI group (P < 0.05; Figures 2B,C). We also examined the mucin layer covering the proximal colon by AB-PAS staining in Supplementary Figure 1B. CDI group damaged the mucous layer, while A. muciniphila protected the mucous layer. As noted above, C. difficile caused severe intestinal mucosal injury, while probiotics treatment improved symptoms.
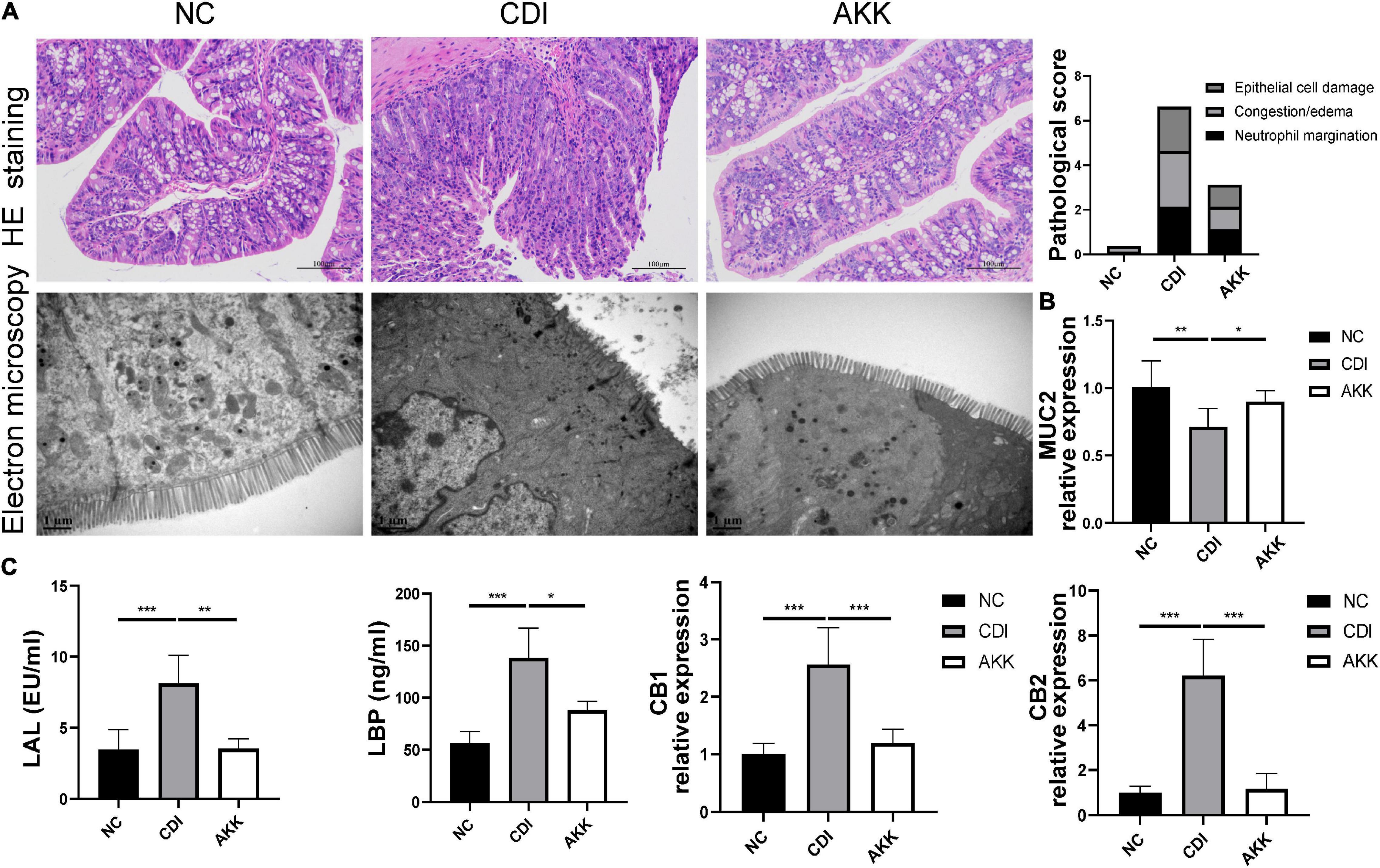
Figure 2. Akkermansia muciniphila reduced colon epithelial injury. (A) Representative images of hematoxylin and eosin staining and ultrastructure under TEM (left panel). Histopathology scores among three groups (right panel). (B) Colon mRNA expression of MUC2. (C) Serum LAL and LBP levels and colon mRNA expression of CB1 and CB2. *P < 0.05, **P < 0.01, ***P < 0.001.
Inflammatory cytokines are related to the pathogenesis mechanism of CDI colitis. We measured serum and colon tissue cytokine levels to explore the protective effects of A. muciniphila on intestinal inflammation. Infected mice were detected with a series of elevated inflammatory cytokines in previous study (Pawlowski et al., 2010), increased levels of serum cytokines G-CSF, MCP-1, IL-17A, IL-1α, IL-6, and TNF-α were observed in CDI colitis mice. While the administration of A. muciniphila significantly reduced the levels of these cytokines (P < 0.05; Figure 3A). In contrast with the NC group, IL-6, TNF-α, IL-1β, CCL1, CCL2, CCL3, CCL4, CCL5, CCL17, CCL22, CXCL10, and CXCL13 were elevated significantly in CDI mice, and are related to intestinal inflammation (P < 0.05; Figure 3B). These cytokine concentrations were downregulated considerably after the administration of A. muciniphila (P < 0.05; Figure 3B). Thus, probiotics reduce inflammation levels and exhibit anti-inflammatory properties.
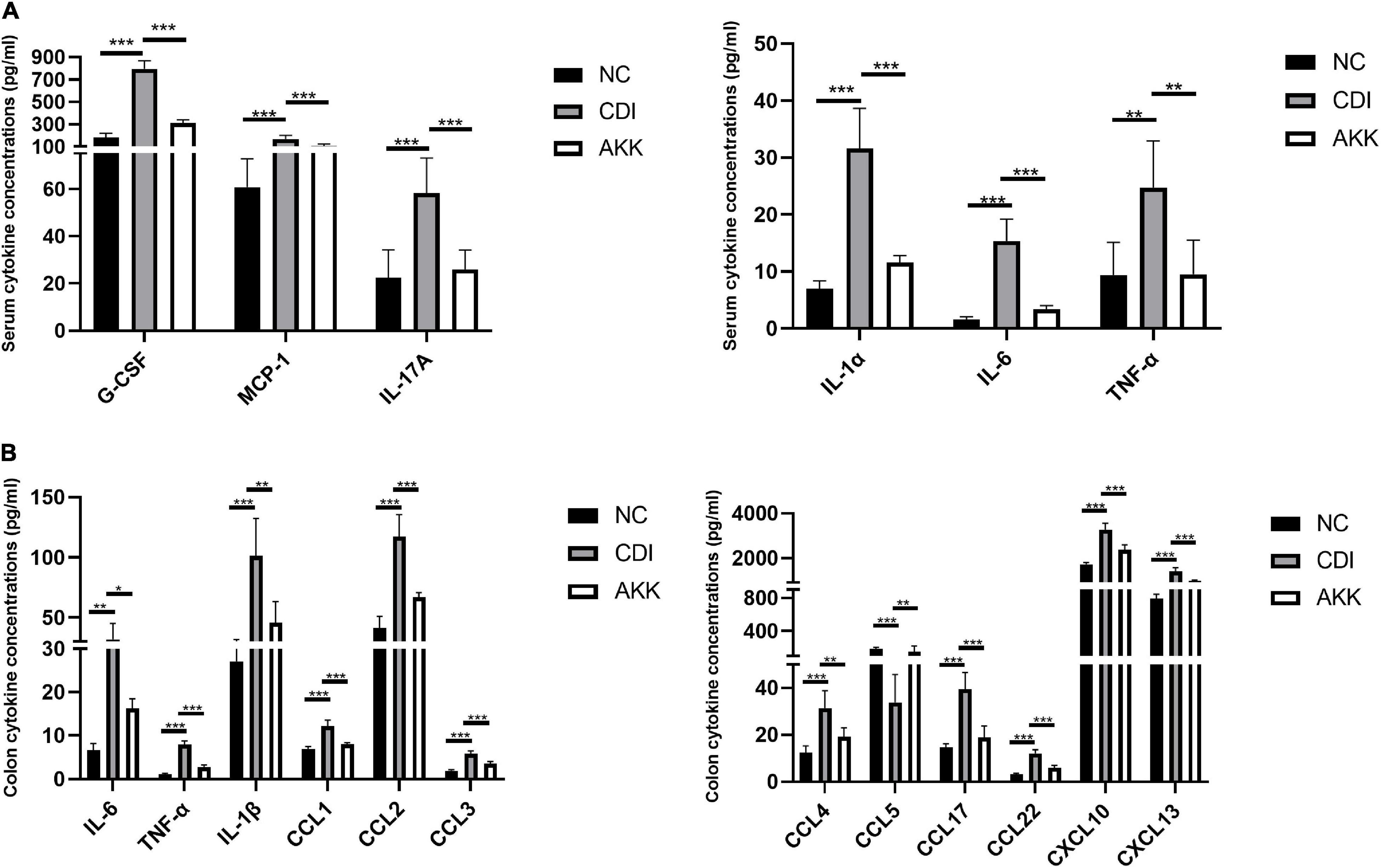
Figure 3. Akkermansia muciniphila ameliorated local and systemic anti-inflammation. (A) Serum concentrations of G-CSF, MCP-1, IL-17A, IL-1α, IL-6, and TNF-α. (B) Colon tissue IL-6, TNF-α, IL-1β, CCL1, CCL2, CCL3, CCL4, CCL5, CCL17, CCL22, CXCL10, and CXCL13. *P < 0.05, **P < 0.01, ***P < 0.001.
Due to the mechanism of toxins A/B-induced IEC injury, we investigated the mammalian target of rapamycin (mTOR) signaling pathway to explore intestinal inflammation in CDI mice. The treatment decreased the autophagy-related protein expression (Figure 4A) and mRNA level (Figure 4B). There was reduced expression of phosphorylated mTOR (p-mTOR) and beclin1 in the colon epithelial cells (Figure 4A). The gene expression of autophagy-related genes such as mTOR, beclin1, light chain 3 (LC3)-II, autophagy-related gene 5 (Atg5), Atg7, Atg9a, and Atg12 were more elevated in the CDI group than in the AKK group (P < 0.05; Figure 4B). Similarly, the mRNA expression of immune markers such as Toll-like receptor 4 (TLR4), cluster of differentiation 14 (CD14, a co-receptor of TLR4), and myeloid differentiation 88 (MyD88) were downregulated after A. muciniphila administration (P < 0.001, P < 0.05, and P < 0.05, respectively; Figure 4C).
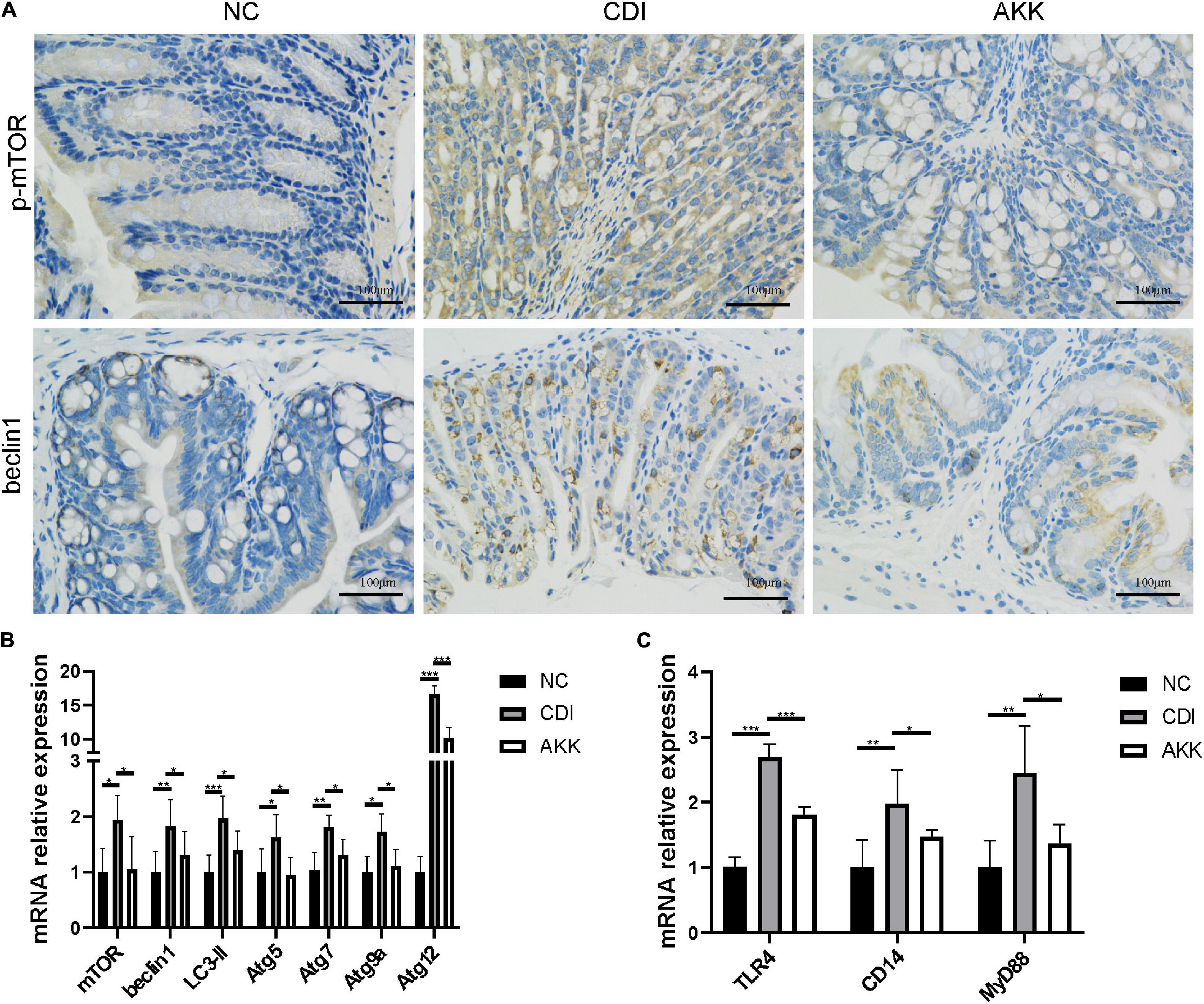
Figure 4. Akkermansia muciniphila relieved CDI-induced inflammation. (A) Representative immunofluorescence staining of p-mTOR and beclin1 in the colon. (B) Relative colon mRNA expression of mTOR, beclin1, LC3-II, Atg5, Atg7, Atg9a, and Atg12 in the colon. (C) Relative mRNA expression of TLR4, CD14, and MyD88 in the colon. *P < 0.05, **P < 0.01, ***P < 0.001.
To study the microbiome composition, we used 16S rRNA gene sequencing and obtained a total of 1,132,433 sequences (47,184 reads per sample). The CDI group showed a decline in microbiome diversity (Shannon index, Figure 5A) and richness (Chao1 index, Figure 5B). Furthermore, the PCoA plot showed significantly different microbiota among the three groups (Figure 5C, PERMANOVA, R2 = 0.8065, p_adjust = 0.001). These results suggested the reduced abundance of A. muciniphila in the CDI group (Figure 5D), with a concomitant increased load of A. muciniphila in the AKK group (Supplementary Figure 2A).
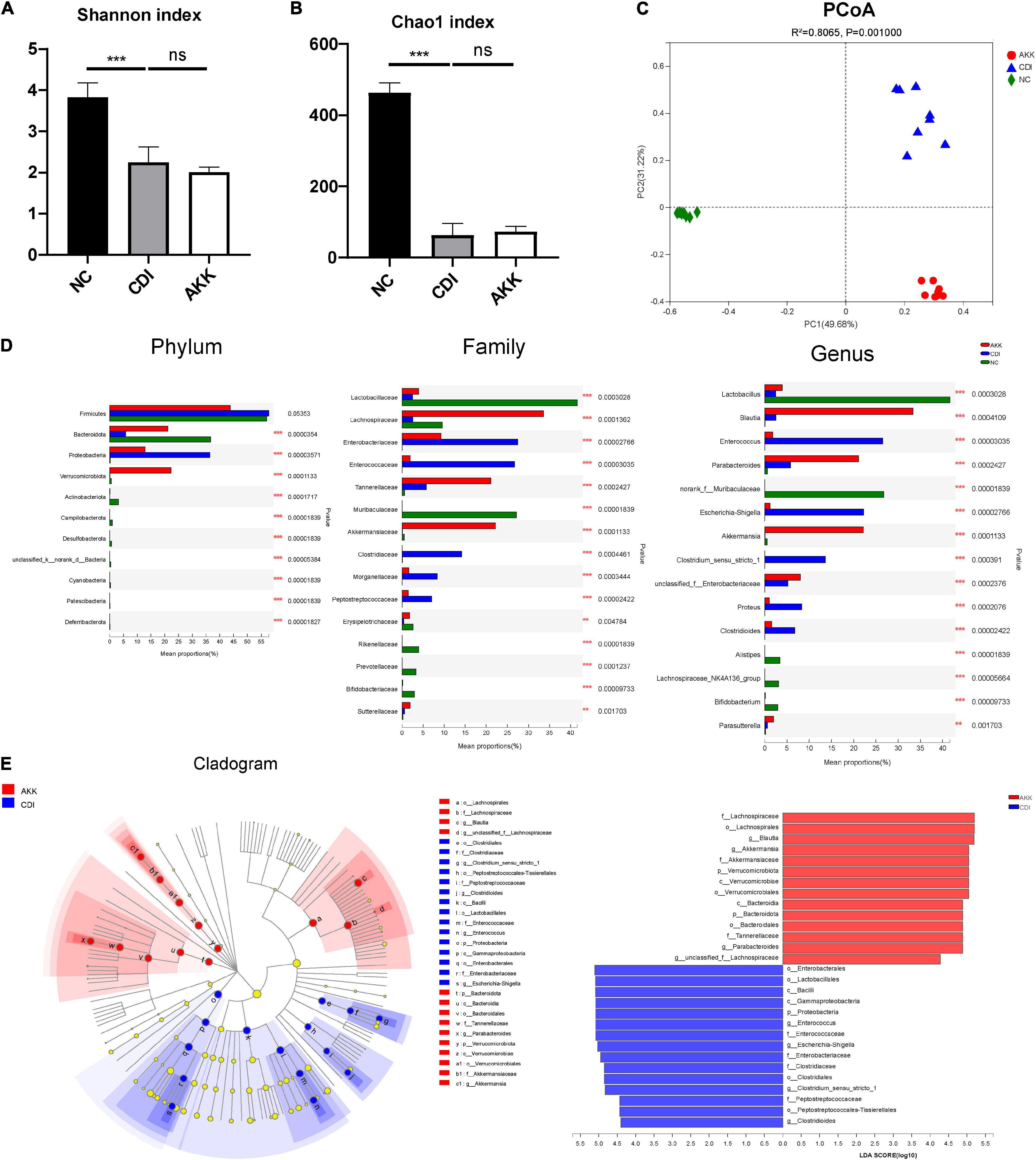
Figure 5. Effects of pretreatment with A. muciniphila on the changes of gut microbial community structure. (A,B) The Shannon index and Chao1 index evaluated alpha diversity. (C) The PCoA plot by Bray–Curtis analysis. (D) Comparison of taxa at the phylum, family, and genus levels among three groups. (E) LEfSe analyses between the CDI group (blue) and the AKK group (red) (LDA score >4). ***P < 0.001.
The relative taxon abundance at the phylum level showed that the CDI group was enriched with Proteobacteria and depleted of Bacteroidota (Figure 5D). While at the family level, the CDI group considerably lacked Lactobacillaceae, Lachnospiraceae, and Akkermansiaceae but was enriched in Enterobacteriaceae, Enterococcaceae, Clostridiaceae, Morganellaceae, and Peptostreptococcaceae. The oral administration of A. muciniphila significantly improved this enrichment and depletion.
Linear discriminant analysis effect size analysis was used to identify taxa, and the most diverse results (LDA score >4) were analyzed. Compared with the NC group, the phylum Bacteroidota, class Bacteroidia, order Bacteroidales, and families Rikenellaceae, Muribaculaceae, and Prevotellaceae in the CDI group were depleted (Supplementary Figure 2B). Meanwhile, the phylum Actinobacteriota and the affiliated class Actinobacteria, order Bifidobacteriales, family Bifidobacteriaceae, and genus Bifidobacterium were also depleted in the CDI group. At the family level, Lachnospiraceae, Lactobacillaceae, and Erysipelotrichaceae in the CDI group were also reduced compared with the NC group. At the same time, the enrichment of families Peptostreptococcaceae, Enterococcaceae, Enterobacteriaceae, and Morganellaceae in the CDI group led to an imbalance of intestinal flora.
Compared with the CDI group, the AKK group showed depletion of opportunistic pathogens in the Enterococcus (belonging to family Enterococcaceae and order Lactobacillales), Escherichia–Shigella (belonging to family Enterobacteriaceae, order Enterobacterales, class Gammaproteobacteria and phylum Proteobacteria), Clostridioides, and Clostridium_sensu_stricto_1 (Figure 5E). In addition, the AKK group was enriched for the genera Blautia (belonging to family Lachnospiraceae and order Lachnospirales), Parabacteroides (belonging to family Tannerellaceae, order Bacteroidales, class Bacteroidia, and phylum Bacteroidota), and Akkermansia compared with the CDI group, showing the role of A. muciniphila in restoring gut microbiota. Next, we studied the biochemical mechanisms associated with CDI’s A. muciniphila mediated protection.
We detected the BA levels in the colon by UPLC-MS/MS. Partial least squares-discriminant analysis (PLS-DA) showed distinct clustering of the BAs among the three groups (Figure 6A), which showed alteration of BA metabolism by CDI. The CDI group showed decreased secondary BAs, DCA, ωMCA, HDCA, muroCA, βDCA, LCA, and UDCA (Figure 6C). Compared to the NC group, the levels of primary BAs were significantly raised in the CDI group. However, the level of secondary BA was restored, and primary BAs such as CDCA and CA were decreased after A. muciniphila administration (Figure 6C). The AKK group had an increased secondary BA to primary BA ratio than the CDI group (Figure 6B).
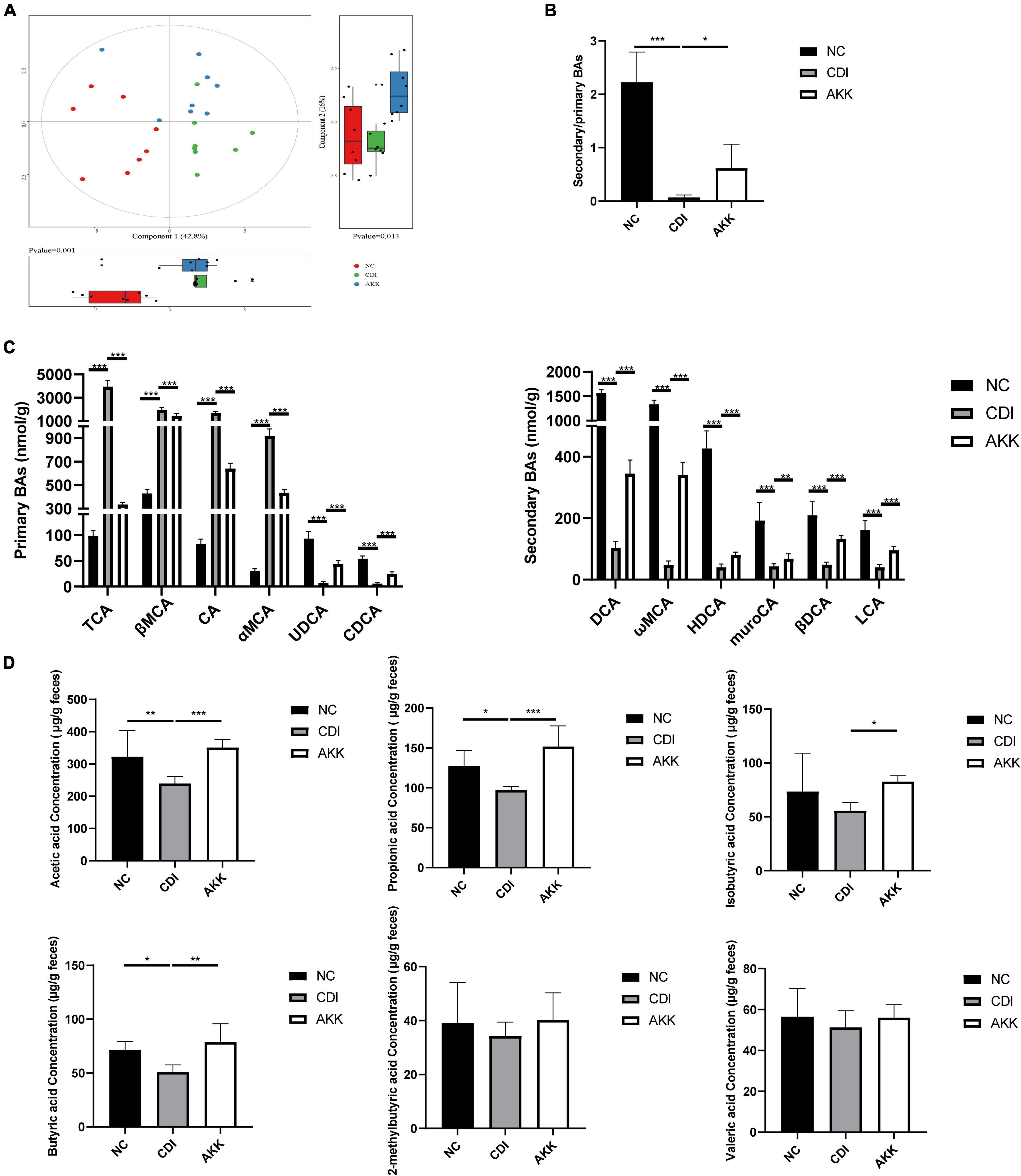
Figure 6. Effects of A. muciniphila on BA and SCFA metabolism. (A) PLS-DA score plot comparing the BA metabolite profile of three groups. (B) The ratio of secondary BAs to primary BAs. (C) Primary BA (left) and secondary BA (right) concentrations. (D) The fecal SCFA concentrations were measured using GC/MS including acetic acid, propionic acid, isobutyric acid, butyric acid,2-methylbutyric acid, and valeric acid. *P < 0.05, **P < 0.01, ***P < 0.001.
As shown in Figure 6D, we determined six major SCFAs in the fecal samples. The AKK group showed a significant increase in SCFA production. The acetic acid, propionic acid, and butyric acid concentrations were significantly decreased in the CDI group (CDI vs. NC: P < 0.05; Figure 6D). Furthermore, A. muciniphila administration increased the concentrations of acetic acid, propionic acid, isobutyric acid, and butyric acid (CDI vs. AKK: P < 0.05; Figure 6D). Thus, probiotics alter metabolic disorders caused by disease.
We performed Spearman’s correlation analysis between discriminative bacteria abundance and metabolic and serum inflammatory indexes to explore the relationship between the changed gut microbiota structures and disease severity. Our results showed that the relative abundance of A. muciniphila-modified bacteria was related to inflammatory indexes and mucosal barrier indicators (Figure 7). Potentially beneficial bacteria such as Bifidobacterium, Bacteroides, and Lactobacillus were negatively correlated with cytokines (e.g., IL-1α, IL-6, and TNF-α) but were positively correlated with SCFAs and secondary BAs. Opportunistic pathogens such as Enterococcus and Escherichia–Shigella were negatively correlated with SCFAs and secondary BAs but positively correlated with inflammatory indexes and primary BAs.
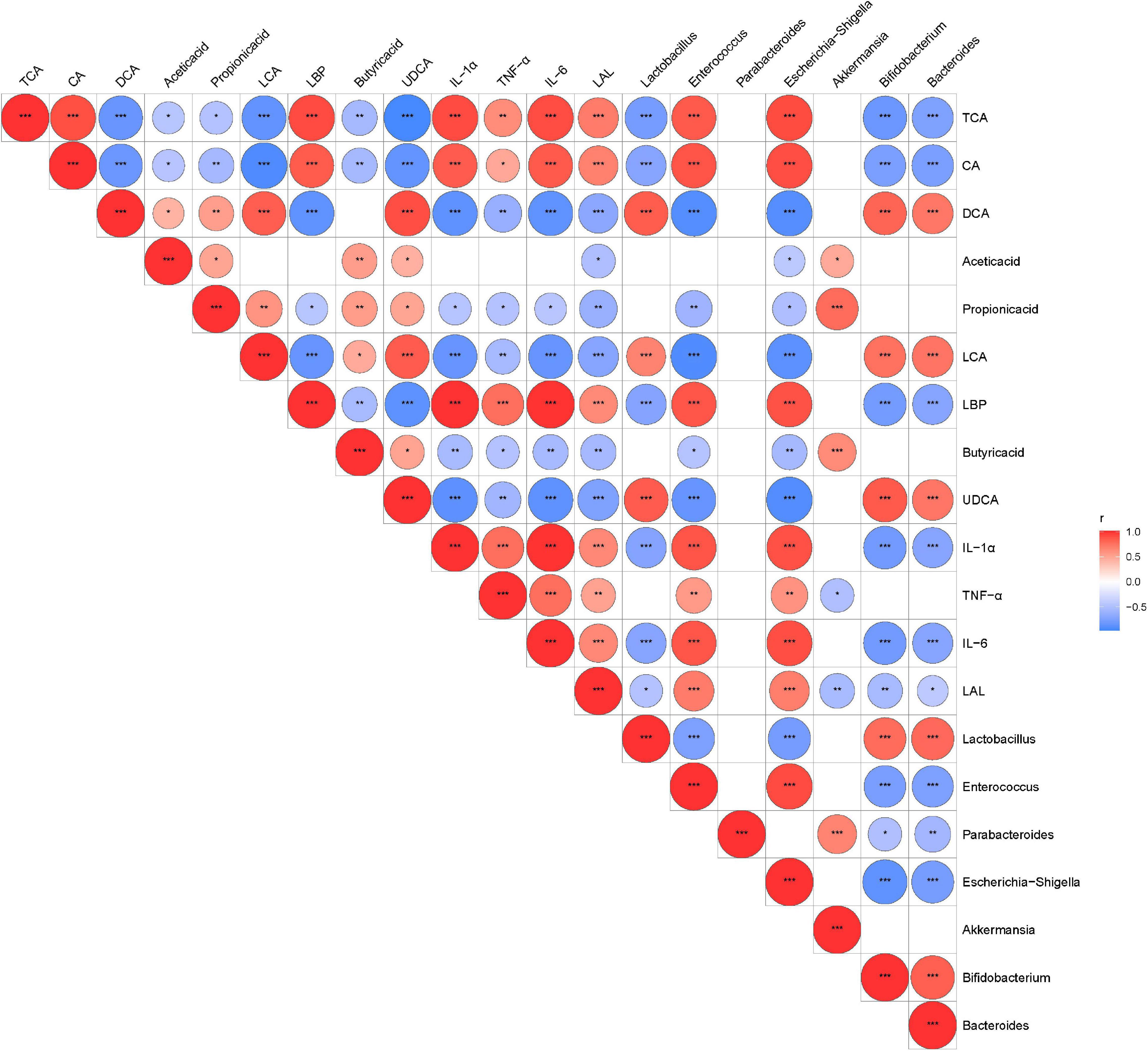
Figure 7. The relationship among the altered intestinal microbiota, metabolites, and inflammatory indexes were evaluated by Spearman’s correlation analysis. Red represents a positive correlation; blue represents a negative correlation. Red and blue represent positive and negative correlation, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
Clostridioides difficile treatment involves using antibiotics against the pathogen, thereby reducing toxin levels and damage to the gut epithelium. Antibiotic treatment does not promote the recovery of the microbiome of CDI patients and may further destroy the local microbiota and cause disease recurrence. This study found that A. muciniphila can reduce CDI mice mortality, significantly ameliorating CD-induced systemic inflammation and the damaged intestinal barrier. Our study further suggests that the potential mechanism might regulate the intestinal microbiota and related microbial metabolism.
Some studies show that CDI patients and mouse model have a low abundance of A. muciniphila (Rodriguez et al., 2016; Deng et al., 2018; Pellissery et al., 2021), while an increased number of A. muciniphila was observed in other studies (Sangster et al., 2016; Vakili et al., 2020; Shoaei et al., 2021). This contradictory conclusion associates to various factors. The probiotic/harmful properties of bacteria are strain and dose-specific (Myers et al., 2006; Wu G. et al., 2017). Since the human intestine may be colonized by different Akkermansia-like spp. with specific phenotypes (Becken et al., 2021), recently, 16S rRNA sequences of a set of A. muciniphila strains isolated from humans and mice in China suggested that 97% sequence identity with that of the type strain A. muciniphila MucT isolated from Europe (Guo et al., 2017). In addition to A. muciniphila, only one other Akkermansia-like spp. Akkermansia glycaniphila have been described (Ouwerkerk et al., 2016). Moreover, the average nucleotide identity between the A. glycaniphila and the A. muciniphila MucT genome was found to be 79.7%. The genome of A. muciniphila MucT involves one circular chromosome of 2.66 Mbp, and shared genes (29%) with its closest relatives in the Verrucomicrobia (van Passel et al., 2011). Most current studies on A. muciniphila focus on A. muciniphila MucT. Therefore, we have used a standard A. muciniphila MucT strain which can degrade and mucin and a swell known model for studying the probiotic microbiome in human gastrointestinal tract (Geerlings et al., 2018). Our study demonstrated that the administration of A. muciniphila can improve intestinal inflammation through multiple host regulation, such as microbial-host interaction and SCFAs. At the same time, these conflicting controversies also prompt us to further explore its therapeutic value with better forms and dosages of administration.
Through maintaining the intestinal microbiota and microbiota-derived metabolites, A. muciniphila regulates the immune response in the intestine. The pathophysiology of CDI is closely related to the production of toxins A and B, which are the two main pathogenic enterotoxins and cytotoxins, which mainly target intestinal IECs and cause pseudomembranous colitis (Kuehne et al., 2010). Our results showed that A. muciniphila reduced C. difficile bacterial burden and the production of toxins in feces. The destruction of colonic epithelium and inflammatory activities increases gut bacterial translocation, causing the produce of various cytokines and promoting inflammatory cell migration to the damaged sites. The inflammatory cytokines and genes related to epithelial integrity showed the destruction of the intestinal mucosal barrier in our study. However, in HE staining and immunofluorescence staining, intestine damage was not evident between the CDI and the NC groups. The probable cause is the location of pathological samples was limited, but several parameters, such as inflammatory markers, suggest C. difficile damage to the gut barrier. Our study showed a significant decrease in inflammatory markers associated with the immune response and protection against infection in the AKK group. The CDI group showed elevated serum levels of LBP and LAL. Meanwhile, the serum levels of cytokines G-CSF, MCP-1, IL-17A, IL-1α, IL-6, and TNF-α were considerably increased, with significant elevation in cytokines and chemokines IL-6, TNF-α, IL-1β, CCL1, CCL2, CCL3, CCL4, CCL17, CCL22, CXCL10, and CXCL13 in the colon tissue. The accumulation of LPS (TLR4 agonist) in the blood triggers an inflammatory response through the TLR4 pathway.
Clostridioides difficile Infection-induced intestinal inflammation is associated with the regulation of the gut microbiota and the activation of TLR4/NF-κB signaling (Kim et al., 2006). Our results showed that C. difficile led to a decrease in intestinal cell turnover, reduced number of goblet cells, decreased expression of mucin (MUC2) and Tjps (ZO-1, occludin, and claudin-1), and damage to the integrity of the intestinal mucosa. Meanwhile, activation of the NF-κB pathway usually induces autophagy (Criollo et al., 2012). Toxins induce the necrosis and apoptosis of IECs (Mahida et al., 1996). In this context, our results found that C. difficile significantly upregulated autophagy-related markers, thereby showing increased autophagy. Dysfunctional autophagy may induce chronic intestinal inflammation, leading to severe intestinal damage (Randall-Demllo et al., 2013). A. muciniphila may reduce inflammation by inhibiting the TLR4/MyD88 and mTOR signaling pathways in the intestine and protecting the mucosal immune barrier. We found that the administration of A. muciniphila was associated with reduced pro-inflammatory production in mouse serum and intestinal tissues to alleviate intestinal inflammation. In addition, A. muciniphila improved the morphological architecture of the colon tissue in the CDI group, increased the expression of Tjps and MUC2, and concomitantly decreased the CB1 and CB2 levels in the colonic mucosa. These data show that A. muciniphila may reverse the intestinal mucosal damage caused by CDI by enhancing cell-cell interactions and induce a promoting effect on the gut mucosal barrier (Everard et al., 2013).
Intestinal epithelial cells receive signals from potential pathogens or symbiotic microorganisms and respond to the altered intestine by regulating mucosal barriers and transmitting signaling to IECs (Xu et al., 2021). Bacterial metabolites signal intestinal microbes to immune cells in the intestinal lamina propria, such as SCFAs and BAs. SCFAs, especially acetic acid (produced by multiple bacterial groups like A. muciniphila), propionic acid (produced by some species of Bacteroidetes and Firmicutes), and butyric acid (produced by a small number of Clostridium), are the final products of intestinal bacterial fiber fermentation (Koh et al., 2016; Louis and Flint, 2017). They affect host physiology, including an energy source for colon cells, regulating the intestinal barrier, and affecting inflammatory responses (Correa-Oliveira et al., 2016). SCFAs also inhibit C. difficile toxin production (Yuille et al., 2020), reducing CDI (McDonald et al., 2018a; Fachi et al., 2020) and inhibiting spore growth (Battaglioli et al., 2018; Hryckowian et al., 2018; Neumann-Schaal et al., 2019). Like previous studies (McDonald et al., 2018a), the CDI group showed decreased butyric acid, propionic acid, acetic acid, and A. muciniphila restored SCFA metabolism, possibly by restoring the intestinal flora balance. A. muciniphila acts as an SCFA producer and can enhance barrier immunity (Bian et al., 2019).
The intestinal homeostasis exhibits colonization resistance to C. difficile, and gut microbial-derived secondary BAs exert functions (Thanissery et al., 2017). Some primary BAs, such as taurocholate, promote the spore germination of C. difficile (Sorg and Sonenshein, 2008). DCA, as well as LCA (Buffie et al., 2015), ω-MCA (Francis et al., 2013), and UDCA (Weingarden et al., 2016), inhibit vegetative cell growth and spore outgrowth. Secondary BAs show a positive association with Akkermansia levels (Pierre et al., 2016). Several studies have shown that an antibiotic-induced microbiota imbalance reduces the pool of secondary BAs (Zarrinpar et al., 2018). Similarly, other studies have suggested that the loss of secondary BAs reduces the activation of anti-inflammatory markers such as TGR5 and FXR, which may impair the inflammatory pathways and intestinal barrier function (Sinha et al., 2020). In our studies, primary BAs in CDI mice increased, while secondary BAs such as CA and CDCA decreased. A. muciniphila helps to restore the levels of secondary BAs.
Microbiome regulation has been proposed as a possible mechanism for A. muciniphila to prevent CDI. The complex and stable microbial community in the intestine is a natural barrier against C. difficile. The signs of decreased gut microbial diversity are reduced microbial colonization resistance and increased susceptibility to C. difficile (Chang et al., 2008). The inflammation caused by CDI will, in turn, lead to the loss of microbiota diversity (Theriot et al., 2014). In agreement with the previous report, CDI patients or animal models showed intestinal flora disorders, including decreased relative abundance of Bacteroidota and increased proportion of LPS-producing Proteobacteria in the intestinal flora. The AKK group showed a lower abundance of harmful bacteria Enterococcus and Escherichia–Shigella, which may relate to the regulation of gut microbiota. Escherichia–Shigella (belonging to phylum Proteobacteria) is an adherent-invasive bacteria and may aggravate colon inflammation. Proteobacteria is the primary source of intestinal endotoxin LPS. Its abundance depended on intestinal LPS (Lin et al., 2020). Proteobacteria cause inflammation (Shin et al., 2015) and provide an environment conducive to invading pathogens like C. difficile (Fuentes et al., 2014). Endotoxemia is relevant to the abundance of gut bacteria associated with LPS production. Our results showed these opportunistic pathogens were positively correlated with serum inflammatory indexes, suggesting their harmful role in gut microbiota.
Meanwhile, A. muciniphila significantly improved the abundance of beneficial bacteria such as Blautia, Parabacteroides, and Akkermansia, which alleviated intestinal inflammation in the CDI by ameliorating the intestinal barrier function and microbiota structure. These strains are acetate or butyrate producers, also can inhibit C. difficile in vitro coculture experiments (Ghimire et al., 2020). These strains used mannitol, sorbitol, or succinate as carbon sources, the nutrients used by C. difficile to invade and cause inflammation in the intestine (Theriot et al., 2014; Spiga et al., 2017).
One limitation of our study is that we only used female mice for CDI model as suggested in previous studies (Chen et al., 2008; Buffie et al., 2015). Though several studies have indicated that male and female mice showed clinical manifestations (Chen et al., 2008), performing equivalent studies in males will be important, and further experimental studies would further refine the issue.
In summary, A. muciniphila treatment ameliorated CDI by modulating intestine barrier function and maintaining the steady state of intestine microbiota, immunological function, and metabolite profiles. Our work puts forth a critical role of A. muciniphila as a sentinel of intestinal homeostasis during CDI. Therefore, A. muciniphila may be a promising therapeutic target for C. difficile-related colitis via modulation of the immune response. Further work should be directed toward understanding the precise molecular mechanisms underlying the protective function of A. muciniphila in microbe-microbe interactions.
Clostridioides difficile is a common cause of hospital infection following antibiotic treatment, inducing a heavy burden on most hospitals. A. muciniphila has showed its active role in maintaining gastrointestinal balance and might prevent and control CDI as probiotics. Our study found that A. muciniphila can reduce CDI mice mortality, significantly ameliorating C. difficile-induced systemic inflammation and the damaged intestinal barrier.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA 766049.
The animal study was reviewed and approved by the Animal Experimental Ethical Inspection of The First Affiliated Hospital, Zhejiang University School of Medicine (Zhejiang, China).
LJL, ZW, QX, and SG designed the experiments. ZW and QX analyzed the data. ZW wrote the manuscript. All authors reviewed the manuscript.
This work was supported by the National Natural Science Foundation of China (82000491, 82073609, 81800457, and 81790631); the National Key R&D Program of China (2018YFC2000500); and Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022045D).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We gratefully acknowledge the support and help with the TEM from the Bio-Ultrastructure Analysis Laboratory of Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.841920/full#supplementary-material
Battaglioli, E. J., Hale, V. L., Chen, J., Jeraldo, P., Ruiz-Mojica, C., Schmidt, B. A., et al. (2018). Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci. Transl. Med. 10:eaam7019. doi: 10.1126/scitranslmed.aam7019
Becken, B., Davey, L., Middleton, D. R., Mueller, K. D., Sharma, A., Holmes, Z. C., et al. (2021). Genotypic and Phenotypic Diversity among Human Isolates of Akkermansia muciniphila. mBio 12, e478–e421. doi: 10.1128/mBio.00478-21
Belzer, C., and de Vos, W. M. (2012). Microbes inside–from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458. doi: 10.1038/ismej.2012.6
Bian, X., Wu, W., Yang, L., Lv, L., Wang, Q., Li, Y., et al. (2019). Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front. Microbiol. 10:2259. doi: 10.3389/fmicb.2019.02259
Bian, X., Yang, L., Wu, W., Lv, L., Jiang, X., Wang, Q., et al. (2020). Pediococcus pentosaceus LI05 alleviates DSS-induced colitis by modulating immunological profiles, the gut microbiota and short-chain fatty acid levels in a mouse model. Microb. Biotechnol. 13, 1228–1244. doi: 10.1111/1751-7915.13583
Buffie, C. G., Bucci, V., Stein, R. R., McKenney, P. T., Ling, L., Gobourne, A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi: 10.1038/nature13828
Chang, J. Y., Antonopoulos, D. A., Kalra, A., Tonelli, A., Khalife, W. T., Schmidt, T. M., et al. (2008). Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438. doi: 10.1086/525047
Chen, X., Katchar, K., Goldsmith, J. D., Nanthakumar, N., Cheknis, A., Gerding, D. N., et al. (2008). A mouse model of Clostridium difficile-associated disease. Gastroenterology 135, 1984–1992. doi: 10.1053/j.gastro.2008.09.002
Chung, C. Y., Alden, S. L., Funderburg, N. T., Fu, P., and Levine, A. D. (2014). Progressive proximal-to-distal reduction in expression of the tight junction complex in colonic epithelium of virally-suppressed HIV+ individuals. PLoS Pathog. 10:e1004198. doi: 10.1371/journal.ppat.1004198
Correa-Oliveira, R., Fachi, J. L., Vieira, A., Sato, F. T., and Vinolo, M. A. (2016). Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 5:e73. doi: 10.1038/cti.2016.17
Criollo, A., Chereau, F., Malik, S. A., Niso-Santano, M., Marino, G., Galluzzi, L., et al. (2012). Autophagy is required for the activation of NF kappa B. Cell Cycle 11, 194–199. doi: 10.4161/cc.11.1.18669
de Aguiar Vallim, T. Q., Tarling, E. J., and Edwards, P. A. (2013). Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669. doi: 10.1016/j.cmet.2013.03.013
Deng, H., Yang, S., Zhang, Y., Qian, K., Zhang, Z., Liu, Y., et al. (2018). Bacteroides fragilis Prevents Clostridium difficile Infection in a Mouse Model by Restoring Gut Barrier and Microbiome Regulation. Front. Microbiol. 9:2976. doi: 10.3389/fmicb.2018.02976
Derrien, M., van Passel, M. W., van de Bovenkamp, J. H., Schipper, R. G., de Vos, W. M., and Dekker, J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1, 254–268. doi: 10.4161/gmic.1.4.12778
Derrien, M., Vaughan, E. E., Plugge, C. M., and de Vos, W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54(Pt 5), 1469–1476. doi: 10.1099/ijs.0.02873-0
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 10, 996–998. doi: 10.1038/nmeth.2604
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Fachi, J. L., Secca, C., Rodrigues, P. B., de Mato, F. C. P., Di Luccia, B., Felipe, J. D., et al. (2020). Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J. Exp. Med. 217:jem.20190489. doi: 10.1084/jem.20190489
Fletcher, J. R., Pike, C. M., Parsons, R. J., Rivera, A. J., Foley, M. H., McLaren, M. R., et al. (2021). Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat. Commun. 12:462. doi: 10.1038/s41467-020-20746-4
Francis, M. B., Allen, C. A., and Sorg, J. A. (2013). Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One 8:e73653. doi: 10.1371/journal.pone.0073653
Fuentes, S., van Nood, E., Tims, S., Heikamp-de Jong, I., ter Braak, C. J., Keller, J. J., et al. (2014). Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. ISME J. 8, 1621–1633. doi: 10.1038/ismej.2014.13
Geerlings, S. Y., Kostopoulos, I., de Vos, W. M., and Belzer, C. (2018). Akkermansia muciniphila in the Human Gastrointestinal Tract: when, Where, and How? Microorganisms 6:75. doi: 10.3390/microorganisms6030075
Gerding, D. N., Johnson, S., Peterson, L. R., Mulligan, M. E., and Silva, J. Jr (1995). Clostridium difficile-associated diarrhea and colitis. Infect. Control Hosp. Epidemiol. 16, 459–477. doi: 10.1086/648363
Ghimire, S., Roy, C., Wongkuna, S., Antony, L., Maji, A., Keena, M. C., et al. (2020). Identification of Clostridioides difficile-Inhibiting Gut Commensals Using Culturomics, Phenotyping, and Combinatorial Community Assembly. Msystems 5, e620–e619. doi: 10.1128/mSystems.00620-19
Gu, S., Chen, Y., Wu, Z., Chen, Y., Gao, H., Lv, L., et al. (2020). Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 71, 2669–2678. doi: 10.1093/cid/ciaa709
Guo, X., Li, S., Zhang, J., Wu, F., Li, X., Wu, D., et al. (2017). Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genom. 18:800. doi: 10.1186/s12864-017-4195-3
Hryckowian, A. J., Van Treuren, W., Smits, S. A., Davis, N. M., Gardner, J. O., Bouley, D. M., et al. (2018). Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat. Microbiol. 3, 662–669. doi: 10.1038/s41564-018-0150-6
Kim, J. M., Lee, J. Y., Yoon, Y. M., Oh, Y. K., Youn, J., and Kim, Y. J. (2006). NF-kappa B activation pathway is essential for the chemokine expression in intestinal epithelial cells stimulated with Clostridium difficile toxin A. Scand. J. Immunol. 63, 453–460. doi: 10.1111/j.1365-3083.2006.001756.x
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Backhed, F. (2016). From Dietary Fiber to Host Physiology: short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Kuehne, S. A., Cartman, S. T., Heap, J. T., Kelly, M. L., Cockayne, A., and Minton, N. P. (2010). The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713. doi: 10.1038/nature09397
Lawley, T. D., Clare, S., Walker, A. W., Stares, M. D., Connor, T. R., Raisen, C., et al. (2012). Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8:e1002995. doi: 10.1371/journal.ppat.1002995
Lin, T. L., Shu, C. C., Chen, Y. M., Lu, J. J., Wu, T. S., Lai, W. F., et al. (2020). Like Cures Like: pharmacological Activity of Anti-Inflammatory Lipopolysaccharides From Gut Microbiome. Front. Pharmacol. 11:554. doi: 10.3389/fphar.2020.00554
Louis, P., and Flint, H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41. doi: 10.1111/1462-2920.13589
Mahida, Y. R., Makh, S., Hyde, S., Gray, T., and Borriello, S. P. (1996). Effect of Clostridium difficile toxin A on human intestinal epithelial cells: induction of interleukin 8 production and apoptosis after cell detachment. Gut 38, 337–347. doi: 10.1136/gut.38.3.337
Maziade, P. J., Pereira, P., and Goldstein, E. J. C. (2015). A Decade of Experience in Primary Prevention of Clostridium difficile Infection at a Community Hospital Using the Probiotic Combination Lactobacillus acidophilus CL1285, Lactobacillus casei LBC80R, and Lactobacillus rhamnosus CLR2 (Bio-K+). Clin. Infect. Dis. 60, S144–S147. doi: 10.1093/cid/civ178
McDonald, L. C., Gerding, D. N., Johnson, S., Bakken, J. S., Carroll, K. C., Coffin, S. E., et al. (2018b). Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 66, e1–e48. doi: 10.1093/cid/cix1085
McDonald, J. A. K., Mullish, B. H., Pechlivanis, A., Liu, Z., Brignardello, J., Kao, D., et al. (2018a). Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota. Gastroenterology 155, 1495–1507e1415. doi: 10.1053/j.gastro.2018.07.014
Merrigan, M., Venugopal, A., Mallozzi, M., Roxas, B., Viswanathan, V. K., Johnson, S., et al. (2010). Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192, 4904–4911. doi: 10.1128/JB.00445-10
Mushtaq, A. (2018). New clinical recommendations for Clostridium difficile. Lancet Infect. Dis. 18:384. doi: 10.1016/S1473-3099(18)30180-4
Myers, G. S., Rasko, D. A., Cheung, J. K., Ravel, J., Seshadri, R., DeBoy, R. T., et al. (2006). Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genom. Res. 16, 1031–1040. doi: 10.1101/gr.5238106
Neumann-Schaal, M., Jahn, D., and Schmidt-Hohagen, K. (2019). Metabolism the Difficile Way: the Key to the Success of the Pathogen Clostridioides difficile. Front. Microbiol. 10:219. doi: 10.3389/fmicb.2019.00219
Ouwerkerk, J. P., Aalvink, S., Belzer, C., and de Vos, W. M. (2016). Akkermansia glycaniphila sp. nov., an anaerobic mucin-degrading bacterium isolated from reticulated python faeces. Int. J. Syst. Evol. Microbiol. 66, 4614–4620. doi: 10.1099/ijsem.0.001399
Pawlowski, S. W., Calabrese, G., Kolling, G. L., Platts-Mills, J., Freire, R., AlcantaraWarren, C., et al. (2010). Murine model of Clostridium difficile infection with aged gnotobiotic C57BL/6 mice and a BI/NAP1 strain. J. Infect. Dis. 202, 1708–1712. doi: 10.1086/657086
Pellissery, A. J., Vinayamohan, P. G., Kuttappan, D. A., Mishra, N., Fragomeni, B. O., Maas, K., et al. (2021). Protective Effect of Baicalin against Clostridioides difficile Infection in Mice. Antibiotics 10:926. doi: 10.3390/antibiotics10080926
Pereira, F. C., Wasmund, K., Cobankovic, I., Jehmlich, N., Herbold, C. W., Lee, K. S., et al. (2020). Rational design of a microbial consortium of mucosal sugar utilizers reduces Clostridiodes difficile colonization. Nat. Commun. 11:5104. doi: 10.1038/s41467-020-18928-1
Pierre, J. F., Martinez, K. B., Ye, H., Nadimpalli, A., Morton, T. C., Yang, J., et al. (2016). Activation of bile acid signaling improves metabolic phenotypes in high-fat diet-induced obese mice. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G286–G304. doi: 10.1152/ajpgi.00202.2016
Png, C. W., Linden, S. K., Gilshenan, K. S., Zoetendal, E. G., McSweeney, C. S., Sly, L. I., et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428. doi: 10.1038/ajg.2010.281
Randall-Demllo, S., Chieppa, M., and Eri, R. (2013). Intestinal epithelium and autophagy: partners in gut homeostasis. Front. Immunol. 4:301. doi: 10.3389/fimmu.2013.00301
Rao, K. N., and Malani, P. N. (2020). Diagnosis and Treatment of Clostridioides (Clostridium) difficile Infection in Adults in 2020. JAMA 323, 1403–1404. doi: 10.1001/jama.2019.3849
Reunanen, J., Kainulainen, V., Huuskonen, L., Ottman, N., Belzer, C., Huhtinen, H., et al. (2015). Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Environ. Microbiol. 81, 3655–3662. doi: 10.1128/AEM.04050-14
Rodriguez, C., Taminiau, B., Korsak, N., Avesani, V., Van Broeck, J., Brach, P., et al. (2016). Longitudinal survey of Clostridium difficile presence and gut microbiota composition in a Belgian nursing home. BMC Microbiol. 16:229. doi: 10.1186/s12866-016-0848-7
Sangster, W., Hegarty, J. P., Schieffer, K. M., Wright, J. R., Hackman, J., Toole, D. R., et al. (2016). Bacterial and Fungal Microbiota Changes Distinguish C. difficile Infection from Other Forms of Diarrhea: results of a Prospective Inpatient Study. Front. Microbiol. 7:789. doi: 10.3389/fmicb.2016.00789
Santacruz, A., Collado, M. C., Garcia-Valdes, L., Segura, M. T., Martin-Lagos, J. A., Anjos, T., et al. (2010). Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 104, 83–92. doi: 10.1017/S0007114510000176
Shen, A. (2012). Clostridium difficile toxins: mediators of inflammation. J. Innate. Immun. 4, 149–158. doi: 10.1159/000332946
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Shoaei, P., Shojaei, H., Siadat, S. D., Moshiri, A., Vakili, B., Yadegari, S., et al. (2021). Gut microbiota in burned patients with Clostridioides difficile infection. Burns [Epub online ahead of print] doi: 10.1016/j.burns.2021.11.023
Sinha, S. R., Haileselassie, Y., Nguyen, L. P., Tropini, C., Wang, M., Becker, L. S., et al. (2020). Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe 27, 659–670e655. doi: 10.1016/j.chom.2020.01.021
Sorg, J. A., and Sonenshein, A. L. (2008). Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512. doi: 10.1128/Jb.01765-07
Spiga, L., Winter, M. G., Furtado de Carvalho, T., Zhu, W., Hughes, E. R., Gillis, C. C., et al. (2017). An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe 22, 291–301e296. doi: 10.1016/j.chom.2017.07.018
Tam, J., Hamza, T., Ma, B., Chen, K., Beilhartz, G. L., Ravel, J., et al. (2018). Host-targeted niclosamide inhibits C. difficile virulence and prevents disease in mice without disrupting the gut microbiota. Nat. Commun. 9:5233. doi: 10.1038/s41467-018-07705-w
Tang, C., Cui, L., Xu, Y., Xie, L., Sun, P., Liu, C., et al. (2016). The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Sci. Rep. 6:37865. doi: 10.1038/srep37865
Thanissery, R., Winston, J. A., and Theriot, C. M. (2017). Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45, 86–100. doi: 10.1016/j.anaerobe.2017.03.004
Theriot, C. M., Koenigsknecht, M. J., Carlson, P. E. Jr, Hatton, G. E., Nelson, A. M., Li, B., et al. (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114. doi: 10.1038/ncomms4114
Vakili, B., Fateh, A., Asadzadeh Aghdaei, H., Sotoodehnejadnematalahi, F., and Siadat, S. D. (2020). Intestinal Microbiota in Elderly Inpatients with Clostridioides difficile Infection. Infect. Drug Resist. 13, 2723–2731. doi: 10.2147/IDR.S262019
van der Lugt, B., van Beek, A. A., Aalvink, S., Meijer, B., Sovran, B., Vermeij, W. P., et al. (2019). Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1 (-/Delta7) mice. Immun. Ageing 16:6. doi: 10.1186/s12979-019-0145-z
van Nood, E., Vrieze, A., Nieuwdorp, M., Fuentes, S., Zoetendal, E. G., de Vos, W. M., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. doi: 10.1056/NEJMoa1205037
van Passel, M. W., Kant, R., Zoetendal, E. G., Plugge, C. M., Derrien, M., Malfatti, S. A., et al. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. doi: 10.1371/journal.pone.0016876
van Werkhoven, C. H., Ducher, A., Berkell, M., Mysara, M., Lammens, C., Torre-Cisneros, J., et al. (2021). Incidence and predictive biomarkers of Clostridioides difficile infection in hospitalized patients receiving broad-spectrum antibiotics. Nat. Commun. 12:2240. doi: 10.1038/s41467-021-22269-y
Weingarden, A. R., Chen, C., Zhang, N., Graiziger, C. T., Dosa, P. I., Steer, C. J., et al. (2016). Ursodeoxycholic Acid Inhibits Clostridium difficile Spore Germination and Vegetative Growth, and Prevents the Recurrence of Ileal Pouchitis Associated With the Infection. J. Clin. Gastroenterol. 50, 624–630. doi: 10.1097/MCG.0000000000000427
Wu, G., Zhang, C., Wu, H., Wang, R., Shen, J., Wang, L., et al. (2017). Genomic Microdiversity of Bifidobacterium pseudocatenulatum Underlying Differential Strain-Level Responses to Dietary Carbohydrate Intervention. mBio 8:e02348-16. doi: 10.1128/mBio.02348-16
Wu, W., Lv, L., Shi, D., Ye, J., Fang, D., Guo, F., et al. (2017). Protective Effect of Akkermansia muciniphila against Immune-Mediated Liver Injury in a Mouse Model. Front. Microbiol. 8:1804. doi: 10.3389/fmicb.2017.01804
Xie, G., Wang, Y., Wang, X., Zhao, A., Chen, T., Ni, Y., et al. (2015). Profiling of serum bile acids in a healthy Chinese population using UPLC-MS/MS. J. Proteome Res. 14, 850–859. doi: 10.1021/pr500920q
Xu, Q., Gu, S., Chen, Y., Quan, J., Lv, L., Chen, D., et al. (2018). Protective Effect of Pediococcus pentosaceus LI05 Against Clostridium difficile Infection in a Mouse Model. Front. Microbiol. 9:2396. doi: 10.3389/fmicb.2018.02396
Xu, Y., Zhu, J., Feng, B., Lin, F., Zhou, J., Liu, J., et al. (2021). Immunosuppressive effect of mesenchymal stem cells on lung and gut CD8(+) T cells in lipopolysaccharide-induced acute lung injury in mice. Cell Prolif. 54:e13028. doi: 10.1111/cpr.13028
Yang, L., Li, Y., Wang, S., Bian, X., Jiang, X., Wu, J., et al. (2021). Western Diet Aggravated Carbon Tetrachloride-Induced Chronic Liver Injury by Disturbing Gut Microbiota and Bile Acid Metabolism. Mol. Nutr. Food Res. 65:e2000811. doi: 10.1002/mnfr.202000811
Yuille, S., Mackay, W. G., Morrison, D. J., and Tedford, M. C. (2020). Drivers of Clostridioides difficile hypervirulent ribotype 027 spore germination, vegetative cell growth and toxin production in vitro. Clin. Microbiol. Infect. 26, e941–e941. doi: 10.1016/j.cmi.2019.11.004
Zarrinpar, A., Chaix, A., Xu, Z. Z., Chang, M. W., Marotz, C. A., Saghatelian, A., et al. (2018). Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 9:2872. doi: 10.1038/s41467-018-05336-9
Keywords: Clostridioides difficile, probiotic, Akkermansia muciniphila, intestinal microbiota, cytokines
Citation: Wu Z, Xu Q, Gu S, Chen Y, Lv L, Zheng B, Wang Q, Wang K, Wang S, Xia J, Yang L, Bian X, Jiang X, Zheng L and Li L (2022) Akkermansia muciniphila Ameliorates Clostridioides difficile Infection in Mice by Modulating the Intestinal Microbiome and Metabolites. Front. Microbiol. 13:841920. doi: 10.3389/fmicb.2022.841920
Received: 23 December 2021; Accepted: 25 March 2022;
Published: 18 May 2022.
Edited by:
Luis Caetano Martha Antunes, Oswaldo Cruz Institute (FIOCRUZ), BrazilReviewed by:
Daniel Paredes-Sabja, Andres Bello University, ChileCopyright © 2022 Wu, Xu, Gu, Chen, Lv, Zheng, Wang, Wang, Wang, Xia, Yang, Bian, Jiang, Zheng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanjuan Li, bGpsaUB6anUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.